- 1Institute of Biomedical Engineering, Chinese Academy of Medical Sciences and Peking Union Medical College, Tianjin, China
- 2Department of Gastroenterology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 3School of Life Sciences, Tiangong University, Tianjin, China
- 4Liang Xiang Hospital Affiliated to Capital Medical University, Beijing, China
- 5Department of Gastroenterology, Ningcheng Central Hospital, Chifeng, China
Magnification endoscopy with narrow-band imaging (ME-NBI) technology is widely used in the early diagnosis of precancerous lesions and gastric cancer, which is critical to reducing the incidence of gastric cancer and improving the survival rate of gastric cancer patients. The diagnosis based on ME-NBI image is mostly in manual way in clinics. In order to improve its objectivity and efficiency, here we proposed a lightweight attention mechanism deep learning model to automatically classify ME-NBI images for artificial intelligence (AI) diagnosis of early gastric cancer, low-grade intraepithelial neoplasia, and non-neoplasm. We collected 4,098 images from 990 patients for model training and validation and evaluated the performance of our model by comparisons with that of other models, such as the benchmark model. An additional 587 images from 296 patients were collected as an independent test set to further evaluate our method’s performance. The validation set showed that the overall accuracy, recall, precision, F1 score, and the area under the curve of our method were higher than those of other methods. The independent test set showed that our method achieved state-of-the-art classification for low-grade intraepithelial neoplasia (accuracy = 93.9%, sensitivity = 92.6%). Our method displayed the advantages of lightweight and high effectiveness in classifying effectiveness, which is the potential for AI diagnosis of early gastric cancer, low-grade intraepithelial neoplasia, and non-neoplasm.
1 Introduction
Gastric cancer (GC) is the third leading cause of cancer death worldwide, with one million new cases and 783,000 additional deaths reported globally in 2021 [1]. GC has a series of evolution processes, gradually developing from chronic gastritis (CGT), intestinal metaplasia (IM), and low-grade intraepithelial neoplasia (LGIN) [2]. With the progression of lesions, the risk of cancer significantly increases. For example, patients with IM and LGIN are about 10 and 25 times more likely to develop GC than ordinary people[2, 3]. The 5-year overall survival rate of patients with pathological early gastric cancer (EGC) is higher than 90% [4], while in Asian countries such as South Korea and Japan, the survival rate for patients with advanced gastric cancer is less than 20% [5, 6]. Therefore, timely detection and accurate classification of precancerous lesions and EGC are crucial for providing proper treatment, reducing the incidence of gastric cancer, and improving the survival rate of EGC patients.
Endoscopy is the primary tool for examining and diagnosing various gastric injuries [7]. Standard endoscopes include white-light imaging (WLI) endoscopy, narrow-band imaging (NBI) endoscopy, and magnification endoscopy with narrow-band imaging (ME-NBI). Compared with WLI and NBI, ME-NBI can observe the morphology of gastric mucosal surface microstructures and microvessels in detail, which has been widely used in diagnosing clinical gastric lesions, especially EGC [8, 9]. However, there are many problems in clinical gastroscopy. First, the diagnosis of endoscopic images is often subjective and depends on the professional knowledge and experience of endoscopists; when endoscopists are inexperienced, misdiagnosis or missed diagnosis will be caused[10, 11]. Secondly, endoscopy will produce many images, so the artificial classification of endoscopic images is a labor-intensive process; when the number of patients is large, the lack of endoscopists and fatigue will further aggravate the missed diagnosis or misdiagnosis of patients[12]. Computer-aided diagnosis (CAD) provides an objective and automatic classification method; it can help doctors make more effective decisions in a shorter diagnosis cycle. Studies have shown that CAD can improve the efficiency and accuracy of diagnosing gastrointestinal lesions [13–15].
Early CAD systems were usually based on manual feature extraction algorithms[16–19]. Kanesaka et al.[14] designed 8 gray level co-occurrence matrix (GLCM) features and developed a CAD system based on these manual features and the coefficient of variation of feature vectors for GC detection. Van D S F et al. [16] developed a CAD system for early esophageal cancer detection in high-definition endoscopic images by calculating local color and texture features based on the original and Gabor-filtered images. However, these methods rely on human-designed algorithms for feature extraction and fail to realize automatic identification of gastric lesions. Moreover, the manual features are insufficient to fit the diversified features of actual lesions[20] and cannot be generalized to practical diagnostic applications.
Deep learning (DL) can automatically capture the subtle features in images and has better accuracy and flexibility than manual feature extraction methods. It has made significant progress in the application of computer vision[21–24] and has been widely used in the field of medical images to solve automatic classification [25–28], segmentation [29–31], localization [32–34], and other tasks. Several scholars have recently demonstrated DL’s applicability in automatic endoscopic image analysis[35–38]. Horiuchi et al.[39] proposed a classification method based on GoogleNet, which could automatically identify EGC and gastritis in ME-NBI images with an accuracy of 85.3%. Yan T et al. [40] developed a diagnostic system for IM detection based on the EfficientNet B4 network, and the accuracy for patients reached 88.8%. In terms of detecting LGIN, Cho et al.[41] developed a classification system for gastric lesions based on 5017 WLI images and the DL model, and the accuracy for LGIN was 78.5%, lower than that of endoscopists. Lui et al.[42] developed a classification system based on 3000 NBI images that could automatically classify LGIN, high-grade intraepithelial neoplasia (HGIN), and GC and achieved better classification performance than primary endoscopists. Liu et al.[43] automatically classified gastritis, LGIN, and EGC based on the transfer learning method and ME-NBI images and achieved an average accuracy of 96%.
However, there are still some problems in the automatic classification of gastric lesions based on deep learning. First, existing studies use transfer learning methods, but there is no study on using an end-to-end training model to classify images in small sample datasets. The transfer learning method can solve the problem that training on small sample datasets is challenging to fit, but this method assumes that the source domain datasets and the target domain datasets are correlated, which may reduce the accuracy when the images of the two datasets are quite different [44]. In addition, the traditional DL model has relatively high structural complexity and large volume; even when combined with transfer learning, it still has problems of slow convergence or overfitting on small sample datasets. Second, no research group has classified non-neoplasm (gastritis, IM), LGIN, and EGC based on ME-NBI images and deep learning. Third, the classification performance of precancerous lesions in existing studies needs to be improved.
The attention mechanism can improve the classification performance of deep learning models [45], which has been confirmed in medical image analysis tasks [46, 47]. Inspired by F. Wang et al. [48], this study proposed a deep learning model of attention mechanism. Based on this model, an automatic classification framework for non-neoplasm, low-grade intraepithelial neoplasia, and early gastric cancer based on ME-NBI images was developed. The main contributions of this paper are as follows:
1) A deep learning model with the attention mechanism and cost-sensitive learning was proposed, which can strengthen the discrimination ability of subtle feature differences of gastric lesions and solve the problem of class imbalance in the dataset. This is the first time an attention mechanism model has been introduced into the endoscopic analysis of gastric lesions.
2) The lightweight model was implemented, which allows the model to be trained from scratch on the small sample gastroscopy dataset and does not require pre-trained weights, and significantly reduces the training and deployment time of the model.
3) An automatic classification method was developed based on the proposed model, and the classification of non-neoplasm (including gastritis and IM), LGIN, and EGC based on ME-NBI images were performed for the first time, and state-of-the-art performances were obtained in the classification of LGIN. This method can be used as an additional diagnostic tool in diagnosing gastric lesions under clinical endoscopy.
2 Materials and methods
2.1 Materials
This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Peking Union Medical College Hospital, Beijing, China. This was a retrospective study, and the data were analyzed anonymously, so informed consent from patients was not required. Endoscopic images were captured using the GIF-H260 Z endoscope with an EVIS LUCERA CV-290 endoscopic video imaging system (Olympus OpticalCorp, Tokyo, Japan), and saved as graphic files of type JPEG (Joint Photographic Experts Group) with two resolutions: 1920 × 1080 pixels and 1440 × 1080 pixels.
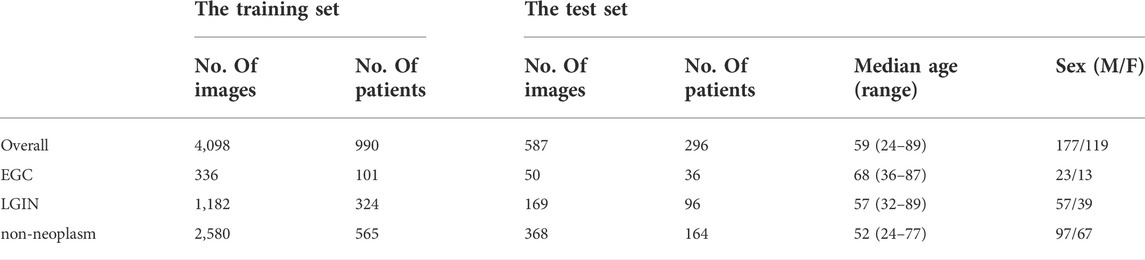
All ME-NBI images were collected retrospectively from patients admitted to the Department of Gastroenterology, Peking Union Medical College Hospital, from February 2014 to February 2020. Images of poor quality due to under-inflation, defocus, mucus, blur, and lack of pathological diagnosis were excluded from the study. Finally, 4098 ME-NBI images from 990 patients were collected, including 336 EGC images from 101 patients, 1182 LGIN images from 324 patients, and 2580 non-neoplasm images from 565 patients.
All patients were confirmed by biopsy pathology, with pathological diagnosis as the gold standard. The final pathological results were determined by endoscopic submucosal dissection or surgical resection for suspected cancerous lesions during endoscopy. Two Peking Union Medical College Hospital pathologists made the pathological diagnosis based on tissue sections. Patients with EGC, LGIN, and non-neoplasm confirmed by histology were eligible for this study. Three endoscopists from the Department of Gastroenterology, Peking Union Medical College Hospital, evaluated and classified the gastroscopic images: first, two endoscopists (with more than 7 years of gastroscopy experience) carefully reviewed all ME-NBI images according to the pathology report, those images that did not match the anatomical location in the pathology report were discarded, and the final retained images were captured at almost the exact location as the biopsy or surgical location where the doctor suspected the abnormality, and have corresponding tissue samples; later, according to pathological diagnosis, two endoscopists divided the remaining images into EGC, LGIN, and non-neoplasm (including gastritis and IM); when two endoscopists have different opinions on image classification, the third endoscopist (with more than 10 years of gastroscopy experience) will review, verify, and determine the image category.
In this study, a 5-fold cross-validation was performed on the training set. The training set was divided into five groups using a patient-based random sampling method. The images of a single patient with a type of lesion were only assigned to one group. When a patient had different lesions, the images of each type of lesion might appear in different groups, and the number of patients with the same type of lesion in different groups was the same. Then, five iterations of training and validation were performed, with one different group for validation in each iteration and the remaining four groups for training. The number of images acquired in this study is minor. In order to improve the robustness of the system, the data augmentation strategy was implemented for the training group. We augmented the training group by rotation (±15°), flip (vertical and horizontal), and other transformations that did not affect the image features, while the validation group was not augmented.
Another new dataset was collected and used as the independent test set. All ME-NBI images with pathologically confirmed were collected from consecutive patients who underwent gastroscopy at Peking Union Medical College Hospital from March 2020 to December 2020. With the same exclusion criteria as above, a total of 587 ME-NBI images from 296 patients were finally collected. The overall median age of the test set was 56, with a range of 24–89, and the sex ratio between males and females was 177/119. Table 1 shows the image category composition of the datasets used in this study and the population characteristics of the patients in the test set.
2.2 Methods
2.2.1 Data preprocessing and cost-sensitive learning
The original gastroscopy image contains a black border and text information that does not contribute to disease identification and may contain patient information. Therefore, in the preprocessing process, the black border of the original image is firstly removed by cropping, then the text information is removed to protect the patient’s privacy. After that, the image size was uniformly adjusted to 224 × 224 pixels.
There is a severe class imbalance problem in our dataset; that is, the number of samples in the minority class is significantly less than that in the majority class, which will make the model obtain more prior information from the majority class samples, resulting in the classification results biased toward the majority class, and thus affecting the classification accuracy. Cost-sensitive learning uses a unique penalty term or weight value for each class to characterize the importance of different classes. Usually, it uses a smaller weight value for the majority class and a larger weight value for the minority class to adjust the loss value of each class to minimize the cost of misclassification and alleviate the bias of class imbalance. In order to solve the problem of class imbalance in our dataset, this study introduced cost-sensitive learning into the model and redesigned the loss function. We first set each category’s penalty or weight value; the method for obtaining the weight value is shown in Equation 1:
In the above equation, j represents the category,
In the above equation, N represents the number of batch samples,i represents the sample,
In Equation 3,
2.2.2 Lesion classification framework
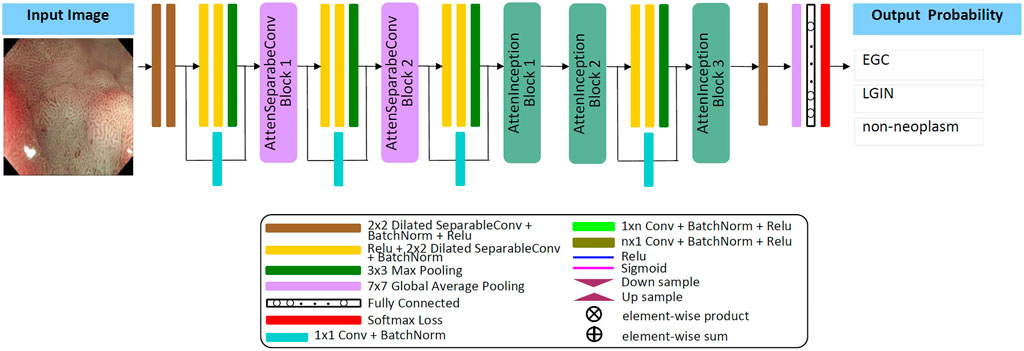
The Attention Module [48] adopts the (bottom-up, top-down) encoding and decoding structure, which can refine the attention to the image in feedforward learning, highlight the subtle feature differences between different lesions, enhance feature selection, and promote gradient update in feedback learning. The separable convolution layer [49] maps cross-channel correlation and spatial correlation separately, improving the model’s performance without increasing the number of model parameters and computational complexity. The Inception module [50] can effectively reduce parameters, extend network depth through factorizing convolution, and combine multi-layer feature fusion technology to achieve better performance with lower computational cost. Dilated convolution [51, 52] expands the convolution range by inserting spaces between the elements of the small-size convolution kernel; it can obtain the same “receptive field” and feature learning ability as the large-size convolution kernel under the condition of occupying a few parameters. Inspired by these techniques, this study designs a new lightweight convolution neural network model and develops a framework for automatically classifying EGC, LGIN, and non-neoplasm gastroscopy images based on this model. The details are as follows:
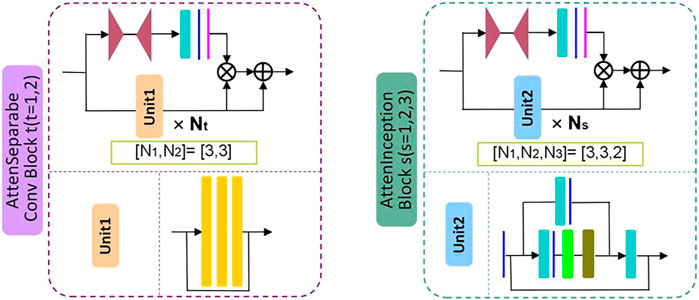
Firstly, by referring to the structural characteristics of the attention module, this study designs two types of attention modules, the AttenSeparableConv block and the AttenInception block. The two attention modules have the same branching structure. AttenSeparableConv block uses stacked separable convolution layers as convolution units of trunk branches, while AttenInception block uses stacked Inception units as convolution units of trunk branches. Secondly, the unit composed of two separable convolution layers and the Maxpooling layer is used as the dimension reduction unit to reduce the dimension of the feature map, and the trade-off between the bottleneck of the feature map and pooling can be effectively balanced. In order to reduce the number of model parameters and computational complexity, we replace the convolution kernels of all separable convolution layers in the model with the dilated convolution kernels of size 2 × 2 and dilated rate 2 and name the new convolution layer as Dilated SeparableConv layer. The final model structure is shown in Figure 1. The model contains four dimension reduction units, two AttenSeparableConv blocks, and three AttenInception blocks. The dimension reduction units are located at the starting position and the low, middle, and high-level feature extraction positions to reduce the dimension of the feature map timely. Affected by structural differences, at the same position of the model, the number of parameters occupied by the separable convolution layer far exceeds that occupied by the factorization convolutional layer. For example, the number of parameters occupied by AttenInception block 1 is 1.28 million. If an AttenSeparableConv block 3, in which the structure is the same as AttenSeparableConv block1-2, is set at this location, the number of parameters occupied by this block is 2.38 million. And the parameters difference between the two blocks increases with the depth of the position. Therefore, we only use AttenSeparableConv blocks in the shallow layer of the model and use AttenInception blocks in the deeper layer of the model to extend the network depth, to effectively balance the number of parameters and performance. We set the AttenSeparableConv block after the first and second dimension reduction units for low-level feature extraction. The AttenInception block is set after the third and fourth dimension reduction units for middle and high-level feature extraction. Another three separate Dilated SeparableConv layers are used for head and tail feature learning, respectively. The above components make up the feature extractor. The global average pooling layer, the fully connected layer, and the softmax loss function compose the classifier. After the ME-NBI image is input to the network, the feature extractor identifies the feature, and the classifier outputs the probabilities that the image belongs to three gastric lesions.
The schematic diagrams of the AttenSeparableConv block and AttenInception block are shown in Figure 2. The trunk of the AttenSeparableConv block is composed of two stacked Unit1 units, each of which contains three Dilated SeparableConv layers. We added a RELU activation function and a batch normalization layer before and after each Dilated SeparableConv layer. Relevant studies have shown that a short-time connection can improve the classification performance of attention-oriented structures [48], so we added a short-time connection to Unit1 to improve the classification performance of AttenSeparableConv blocks. The trunk of the AttenInception block is composed of stacked Unit2 units, and different AttenInception blocks contain different amounts of Unit2 units in the trunk. The trunk of AttenInception block1-2 at the middle-level feature extraction position contains three Unit2 units, and the trunk of AttenInception block3 at the high-level feature extraction position contains two Unit2 units. Unit2 adopts the Inception block. In the original literature, the Inception block has convolution kernels of various sizes (1×n, n×1, n = 3,5,7) [50]. In this study, a relatively large convolution kernel (n = 5 or 7) is used in the Unit2 of AttenInception block1-2, while a small convolution kernel (n = 3) is used in the Unit2 of AttenInception block3. The attention branches of the two types of attention modules both use the Maxpooling layer as the bottom-up structure and the linear interpolation layer as the top-down structure to retain the characteristics of the original attention branch structure, and the Sigmoid function is retained for the mixed attention constraint. Unlike the original attention branch structure, this study only retained one convolution layer and added the RELU function to this convolution layer to enhance the learnable feature change space. The weight of the trunk feature map is realized by multiplying and then adding the output of the attention branch and the output of the trunk feature map.
2.2.3 Training details
Adam was used as the network optimizer for all CNN models with a learning rate of 0.0001, training epochs of 150, and a batch size of 8. All experiments were performed on an AMD Ryzen 7-1700X eight-core processor CPU and a GeForce GTX 1080 Ti GPU(graphics processing units).
2.2.4 Evaluation metrics
Accuracy (ACC), recall (RE), precision (PRE), F1 score (F1), and the area under the curve (AUC) were used to evaluate the classification performance of the model, and each metric was calculated for an independent category. ACC and F1 evaluation comprehensive classification ability, RE represents the susceptibility to disease, PRE representative disease recognition accuracy, and receiver operating characteristic (ROC) curve can measure classifier robustness. The AUC value is automatically calculated according to the ROC curve, which can intuitively reflect the comprehensive classification ability of the model, and its range is between 0 and 1. The larger the value, the better the performance of the classification model. The calculation of each metric is shown in equations (4)–(7):
TP, TN, FP, and FN stand for True Positive, True Negative, False Positive, and False Negative, respectively. To evaluate the overall performance of the model, the overall accuracy (OA), recall (OR), precision (OP), F1 Score (OF1), and AUC (O-AUC) were obtained by adding and calculating the average metric of each category. For the 5-fold cross-validation experiments, the average results of multiple cross-validation experiments were evaluated using the evaluation metrics with 95% confidence intervals (CI).
3 Results
We first evaluated the model’s performance on the validation set. In order to demonstrate the effectiveness of the proposed method for three types of gastric lesions classification, we performed ablation studies, cost-sensitive learning tests, and comparison tests with other advanced methods on the validation set. We used the 5-fold cross-validation method; the whole training set was divided into five groups and performed five experiments. In each experiment, a different group was used as the validation set, and the remaining four groups were used for training. We evaluated the model performance on the validation set. The final result was the average of five experiments. OA, OR, OP, OF1, and O-AUC were used as evaluation metrics to evaluate the classification performance. The number of parameters of each model was counted to indicate the computational complexity of different models. In addition, the classification performance of the proposed method was further evaluated on an independent test set.
3.1 Ablation studies
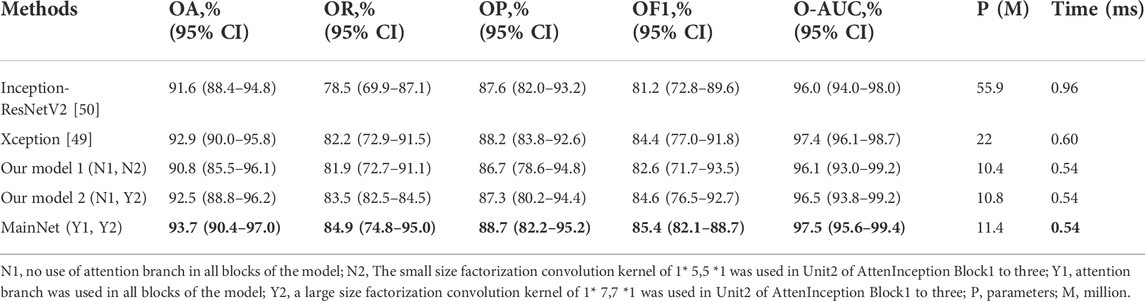
We first performed ablation studies to demonstrate the proposed attention mechanism model’s effectiveness. Xception [49]and Inception-ResNetV2 [50] with ImageNet pre-trained weights were used as benchmark models and compared with our three models: Our model 1 and Our model 2, which were obtained by removing the attention branch, and MainNet (the model in Figure 1), which was obtained by adding attention branch based on Our model 2. The difference between Our model 1 and Our model 2 is the factorization convolution kernel in Unit2 of AttenInception block1 to 2 (Figure 1). In Our model 1, the convolution kernel size was 1 × 5,5 × 1, while in Our model 2, the convolution kernel size was 1 × 7,7 × 1. The classification performance of our three models (Our model 1, Our model 2, and MainNet) was compared with that of the benchmark model to prove the performance of the designed models, and the proposed models can be compared with each other to show the role of attention mechanism further. In order to ensure the fairness of the comparison, the training conditions of all models were the same (learning rate = 1E-4, batch size = 8, epochs = 150), and the input image size was consistent with the original network, that was, the input image size of Inception-ResNetV2 and Xception model was 299 × 299, while the input image size of our three models was 224 × 224.
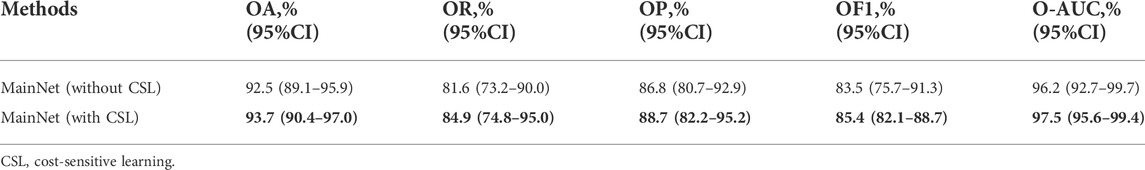
Table 2 summarizes the overall classification performance of five methods for three kinds of gastric lesions. The values in bold in the Table 2 represent the optimal values. Compared with other methods, MainNet obtained the highest OA, OR, OP, OF1, and O-AUC, which were 93.7%, 84.9%, 88.7%, 85.4%, and 97.5%, respectively. MainNet had 11.4M parameters, nearly half the number of parameters for the Xception and less than one-fifth of the number for Inception-ResNetV2. These results showed that our attention mechanism model achieves lightweight while maintaining high classification performance. In addition, Our model 1 and Our mode 2 achieved comparable classification performance with Inception-ResNetV2, proving the superiority of our backbone model. In terms of speed, the prediction time of our three models was 0.54 ms for each image, which was less than the prediction time of the benchmark model, which verified that the model trained from scratch proposed in this study was more conducive to the classification of endoscopic images of gastric lesions than the transfer learning model pre-trained on the natural image dataset. In addition, the overall classification performance of MainNet was better than that of Our model 1 and Our model 2; Our model 2’s classification performance was better than that of Our model 1. These comparison results showed that using the attention mechanism can effectively improve the model’s classification performance, and using a large convolution kernel in the middle-level feature extractor helps improve the model’s classification accuracy.
3.2 Cost-sensitive learning test
In order to prove the effectiveness of cost-sensitive learning in solving the problem of class imbalance in dataset, the performance of MainNet with and without cost-sensitive learning is compared. The results are shown in Table 3. As can be seen from Table 3, the classification performance of MainNet with cost-sensitive learning was significantly better than that of MainNet without cost-sensitive learning, and the difference in OR was particularly prominent, reaching 3.3%. This indicated that combining cost-sensitive learning with our attention mechanism model can effectively improve the model’s sensitivity to minority samples in the imbalanced dataset and the overall classification performance.
3.3 Comparison with other advanced methods
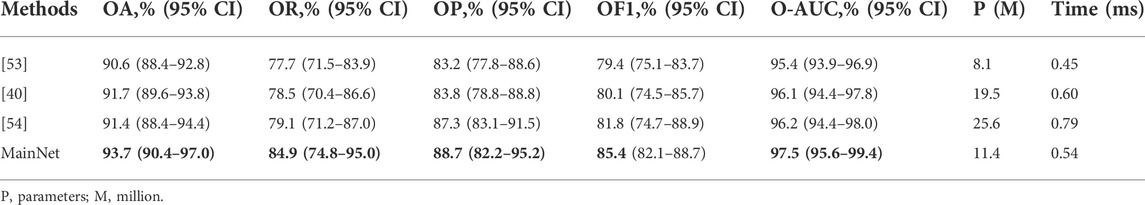
To further verify the proposed method’s effectiveness, we compared our method’s classification performance with other advanced methods on our dataset. We selected some characteristic classification methods for gastric lesions, which either have lighter models[53] or better classification performance [40, 54]. These advanced methods include: the classification method of intestinal metaplasia proposed by Yan et al. [40], the classification method of chronic atrophic gastritis proposed by Zhang et al. [53], and the classification method of atrophic gastritis and intestinal metaplasia proposed by Zheng et al.[54]. The models in these advanced methods are pre-trained in ImageNet. While for our model, we trained it from scratch on the gastric lesion dataset. Other training conditions of all methods are the same to ensure the fairness of comparison. The statistical results of the evaluation metrics are shown in Table 4. As can be seen from Table 4, the OA, OR, OP, OF1, and O-AUC obtained based on MainNet are significantly higher than those obtained by other methods. In terms of the number of parameters, our model’s parameters are higher than that of Zhang et al.’s method (11.4M vs 8.1M), which is much lower than that of Yan T et al.’s 19.5M and Zheng et al.’s 25.6M. In terms of computational efficiency, the average test time of our method is 0.54 ms per image, while the average test time of Zhang et al., Yan T et al., and Zheng et al. are 0.45 ms, 0.60 ms, and 0.79 ms per image, respectively. Zhang et al.’s method show lower parameters and higher computational efficiency. However, compared with their method, the proposed method achieves 3.1%, 7.2%, and 5.5% performance increments in OA, OR, and OP, respectively. The results show that compared with other advanced methods, our method can achieve a better balance among classification performance, number of parameters and computational efficiency.
Supplementary Table S1 summarizes the performance of several methods in per-category classification. It can be seen that in the identification of EGC, the ACC, RE, F1, and AUC obtained based on MainNet were higher than those obtained by other advanced methods, and only the PRE was lower than that obtained by Zheng et al. [54]. In identifying LGIN, our method obtained the highest ACC, PRE, F1, and AUC, and only the RE was lower than that of Zheng et al. [54]. In identifying non-neoplasm, the proposed method performed better than all other methods and achieved the highest values of ACC, RE, PRE, F1, and AUC.
3.4 Model evaluation on the independent test set
We further evaluated MainNet’s classification performance on the independent test set. A total of 587 ME-NBI images from 296 patients were included in the test set, including 50 EGC images from 36 patients, 169 LGIN images from 96 patients, and 368 non-neoplasm images from 164 patients. The classification performance of the MainNet on the test set was compared with the results of other studies, and our best-performing model was compared with human experts.
3.4.1 Comparison with other research results
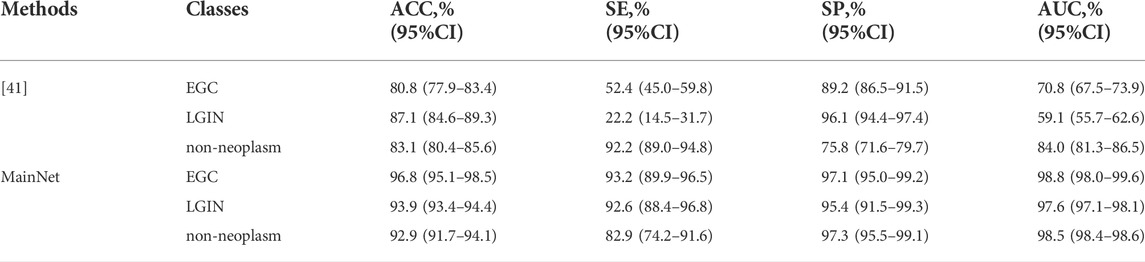
First, MainNet was trained on the training set using the 5-fold cross-validation method, and then the average results on the independent test set were counted and compared with the average results of Cho et al. [41] which was obtained in their independent test based on 5-fold cross-validation. We adopted the same evaluation metric as the literature[41]. The results are shown in Table 5. It can be observed that the accuracy (ACC), sensitivity (SE), specificity (SP) and AUC of the proposed method for each category were higher than those of Cho, et al., and only the SP of LGIN (95.4% vs. 96.1%) and SE of non-neoplasm (82.9% vs. 92.2) were lower than those of Cho, et al. The comparison results showed that the proposed method improved classification performance in classifying EGC, LGIN, and non-neoplasm gastric lesions images. Based on deep learning, Liu et al.[43] divided gastric ME-NBI images into EGC, LGIN, and chronic gastritis (CGT). Their recall (which means the same as our SE) for EGC and LGIN were 99% and 92%, respectively. Compared with the SE of our method for EGC and LGIN(93.2%, 92.6%, respectively), it can be seen that the classification performance of our method for LGIN was better than that of Liu et al., but the SE for EGC was lower than that of Liu et al.
3.4.2 Comparison with endoscopists
Our best model’s classification results were further compared with those of two endoscopists with more than 8 years of endoscopy experience on the independent test set. The best model refers to a MainNet model that achieves the best classification performance on the test set by 5-fold cross-validation. Two endoscopists classified the images without knowing the image category labels. Overall accuracy (OA), sensitivity (OS), specificity (O-SP), and Kappa is used to measure the agreement among raters, which can quantify the degree of agreement between the classifier and the gold standard.
Table 6 summarizes the overall classification performance of our best model and two endoscopists on EGC, LGIN, and non-neoplasm. It can be seen that the Kappa, OA, OS, and O-SP of the best model were 0.859, 95.0%, 93.2%, and 96.1%, respectively, which were higher than those of Endoscopist 1 and Endoscopist 2 (the optimal values were bolded). Figure 3 compares accuracy, sensitivity, and specificity for each category between our best model and the two endoscopists. It can be seen that the best model’s accuracy for EGC and LGIN was better than that of the two endoscopists, while the accuracy for non-neoplasm was slightly lower than that of Endoscopist 1 and the same as that of Endoscopist 2. Besides, our best model showed higher sensitivity to EGC and LGIN, but slightly lower sensitivity to non-neoplasm.
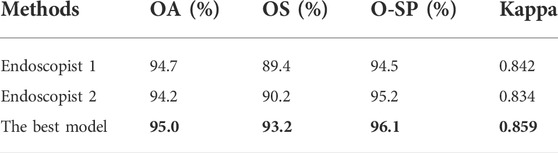
TABLE 6. Statistical comparison of two endoscopists and the best model’s performance in three-category classification.
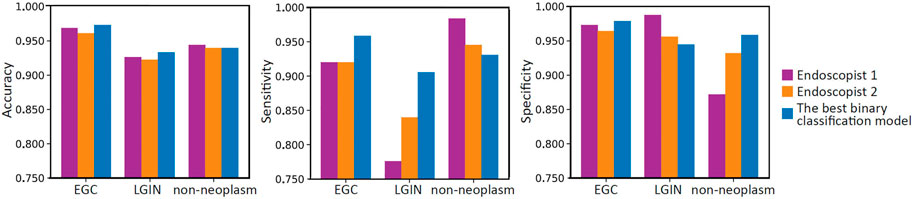
FIGURE 3. Bar charts compare the per-category accuracy, sensitivity, and specificity between two endoscopists and the best model.
We calculated the correlation between the predicted and actual labels for each image by the best model and the two endoscopists and reflected the results in the confusion matrix shown in Figure 4. As can be seen from Figure 4, in the identification of EGC images, the best model had the lowest number of misclassified images; two out of 50 images were misclassified as non-neoplasm, while there were four misclassified images for Endoscopist 1 and Endoscopist 2. In the identification of LGIN, the number of misclassified images of the best model was 16, much lower than the 38 images of Endoscopist 1 and 27 images of Endoscopist 2. In identifying non-neoplasm, the number of images correctly identified by the best model was 342, which was lower than that of Endoscopist 1 and Endoscopist 2 (363 and 348, respectively).
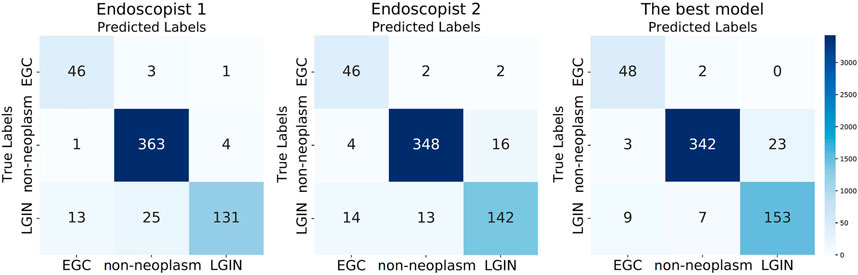
FIGURE 4. Three confusion matrixes for two endoscopists and the best model’s predictions, respectively.
3.4.3 Model binary classification
To further evaluate the binary classification performance of MainNet on gastric lesions, three groups of tests were performed, including classification tests for cancer or non-cancer (EGC vs Others), LGIN or non-LGIN(LGIN vs Others), non-neoplasm or neoplasm (non-neoplasm vs Others). We plotted the ROC curve of the best binary classification model in each group of tests and obtained the AUC, as shown in Figure 5. In the discrimination of cancer or non-cancer, the AUC reached 0.981; In the discrimination of LGIN or non-LGIN, the AUC reached 0.984; In the discrimination of non-neoplasm or neoplasm, the AUC reached 0.988. The binary classification results show that the performance of MainNet was well decomposed.
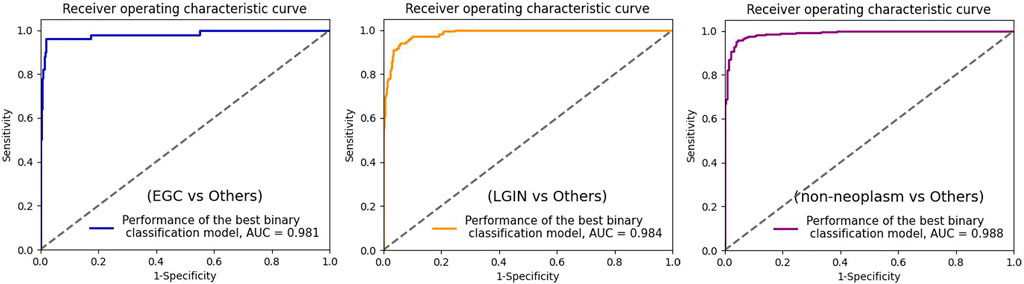
FIGURE 5. ROC curve for the three best binary classification models. ROC, receiver operating characteristic; AUC, area under the curve; EGC, early gastric cancer; LGIN, low-grade intraepithelial neoplasia.
4 Discussion and conclusion
Although ME-NBI can provide imaging of gastric mucosal surface microstructure and microvascular morphology and is widely used in clinical diagnosis of early gastric cancer and precancerous lesions, qualitative assessment of ME-NBI images requires much training, and even experienced endoscopists may misdiagnose or miss a diagnosis. In addition, in the case of limited medical resources, it is unrealistic to manually examine a large number of ME-NBI images, which may cause missed diagnosis or misdiagnosis. Therefore, it is significant to realize the accurate and automatic diagnosis of gastric lesions in clinical practice. In this study, we developed an automated diagnosis system based on a deep learning model with the attention mechanism, which could automatically classify ME-NBI images in a small sample dataset into EGC, LGIN, and non-neoplasm. The classification performance of the proposed method is verified on the validation set and independent test set, respectively.
In this paper, we first conduct ablation studies on the validation set to verify the role of the attention module. The attention module in MainNet adopted the structure of the attention branch combined with the trunk. The encoding and decoding structure of the attention branch can refine the attention to the image and use the branch’s output to weight the trunk’s output to enhance feature selection. Then the attention module was embedded into the model in a stacked manner to refine attention to subtle lesion features. We used efficient convolution layers such as the separable and factorization convolutional layers as the basis and introduced a dilated convolution kernel to reduce the parameters further. By adopting this attention-guided feature learning mode, combined with the parameter reduction strategy, MainNet achieves better classification performance than the backbone and the benchmark models when the parameters are only 11.4M. Our proposed three models are relatively lightweight (10.4M, 10.8M, and 11.4M parameters, respectively) but achieve similar or even better classification performance than Inception-ResNetV2. This phenomenon is related to model complexity. Inception-ResNetV2 demonstrates excellent classification performance on large natural image datasets by increasing network complexity and parameters; however, in practical applications, these heavyweight characteristics limit the implementation of Inception-ResNetV2 on small sample datasets (such as most medical image datasets). On the contrary, Our models are relatively simple and effectively balance computational complexity and task requirements, ultimately achieving classification performance comparable to Inception-ResNetV2.
Our dataset has a severe class imbalance problem, and the deep learning model will tend to over-classify the majority category in the imbalanced dataset. When the appropriate solution is not taken, the accuracy of the majority category may be high, while the overall accuracy is low. As seen in Table 3, the classification performance of MainNet using cost-sensitive learning was higher than that of MainNet without cost-sensitive learning. This indicates that the introduction of cost-sensitive learning can effectively solve the problem of class imbalance in data sets and improve the classification accuracy of gastric lesions.
In the comparison experiment with other advanced methods, the overall classification performance of the MainNet was better than other methods, OA was improved by at least 2.0%, OR was improved by at least 5.8%, and most of the evaluation metrics of per-category classification were also significantly improved. It is worth noting that MainNet outperforms other advanced methods even without using cost-sensitive learning. This indicates that the proposed method could effectively improve the classification performance. It is especially noted that for these advanced methods, we adopted the same training mode as in the original literature; we fine-tuned these pre-trained models on the gastric lesion dataset. While for our model, we trained it from scratch on the gastric lesion dataset. Experimental results show that our end-to-end training model outperforms these pre-trained models on the gastric lesions dataset.
The classification performance of MainNet on an independent test set was further compared with the results of related studies. As seen in Table 5, compared with Cho et al. [41], our method’s ACC for EGC, LGIN, and non-neoplasm increased by 16.8%, 6.8%, and 9.8%, respectively. We believe that the model’s superiority and the practical solution to the class imbalance problem are one of the reasons that make our ACC better than those of Cho et al. Still, the most important reason is the difference in image modality. ME-NBI images were used in this study, while Cho et al. conducted their analysis based on WLI images. However, gastric lesions usually show changes in mucosal surface microstructure. It is difficult for conventional WLI to capture subtle disease features, especially for LGIN. Liu et al. [43] performed EGC, LGIN, and chronic gastritis (CGT) classification based on deep learning and ME-NBI images. Liu et al. obtained 92% and 99% sensitivity for LGIN and EGC, respectively, while the sensitivity of our method for LGIN and EGC was 92.6% and 93.2%, respectively. In general, the sensitivity of our method for LGIN exceeds that of Liu et al. and achieves state-of-the-art classification performance. However, the sensitivity of our method to EGC is lower than that of Liu et al., which may be related to the small sample size of EGC in our dataset. In this study, there are only 369 EGC images in the training set and 50 EGC images in the test set. In such conditions, the feature variation space that the model in training can learn is limited, and the generalization effect and accuracy of the test set are affected. This problem can be solved by including more EGC samples.
Besides, the classification performance of the best model was compared with that of human experts in the independent test set. The results in Table 6 showed that the overall classification performance of the proposed method was better than that of the two endoscopists. In addition, it can be observed from Figure 3 that the diagnostic accuracy of EGC, non-neoplasm, and LGIN decreased successively in both the proposed method and the two endoscopists. For endoscopists, EGC is a severe gastric disease with significant imaging features associated with significant mortality, so they will emphasize identifying such lesions. For our method, although the number of EGC samples in our dataset is small, the attention mechanism model can strengthen the learning of subtle feature differences, and cost-sensitive learning can solve the classification bias that tends to the majority classes. Those make our method achieve better classification performance than endoscopists. On the contrary, LGIN is a lesion characterized by cellular atypia of mucosal structure, different degrees of atypia and different feature patterns also make it difficult to identify accurately, so the accuracy of LGIN by both us and endoscopists is relatively low. However, benefiting from the attention-guided feature learning model, the accuracy of our method for LGIN exceeds that of endoscopists.
Although our method performs better than other methods in gastric ME-NBI image lesion classification, this study still has some limitations. First of all, the data of this study were collected in the same hospital, and the endoscopic equipment in different centers and the characteristics of different populations may have an impact on the method’s performance; in the subsequent study, we will collect endoscopic images from different centers and analyze them. Secondly, there are few EGC and non-neoplasm samples in the dataset of this study, which may affect the accuracy; more samples will be included in subsequent studies to improve the system’s accuracy further. In addition, we only analyzed gastric lesions in this study; after more cases were included, esophagitis and early esophageal cancer will be included in the system to increase the clinical application value of the system. Besides, grading the severity of the lesions is crucial to the prognosis and formulation of treatment plans; so far, only the classification of lesions has been completed in this study, and the severity of lesions will be further graded in subsequent studies. Finally, the proposed method is a supervised learning method, which still requires doctors to carry out a large amount of data annotation work in the early stage, causing a specific workload for doctors; in subsequent studies, methods based on self-supervised deep learning models can be used to solve this problem.
In this study, we designed an attention-guided deep learning model and introduced cost-sensitive learning into the model. Based on this model, we developed an automatic classification method for gastric lesions, which achieved good diagnostic performance on a limited number of ME-NBI images and outperformed other advanced methods. In addition, we achieved the most advanced classification performance for LGIN and non-neoplasm. Through an in-depth literature review, we found that this was the first time to automatically classify non-neoplasm, LGIN, and EGC based on the deep learning model and gastric ME-NBI images. And the first time to introduce the attention mechanism model into the automatic classification of gastric lesions. The number of parameters in our model was only 11.4 million, which allowed the model to be trained end-to-end on small sample medical datasets and can shorten the prediction time per image. The prediction time of each image of our method was only 0.54 ms, which met the demand for real-time diagnosis. In conclusion, our approach can provide objective and accurate guidance information for endoscopists in real time and has an excellent clinical application prospect.
Data availability statement
The datasets presented in this article are not readily available due to patient confidentiality. Requests to access the datasets should be directed to the corresponding authors.
Author contributions
TL and AY designed the study. LW, YY, and JL performed the study and carried out the implementation. WT, KH, TX, and ZF analyzed the data. LW and TL wrote the manuscript. TL and AY reviewed the manuscript and were responsible for the master plan.
Funding
This study was supported by the National Natural Science Foundation of China (No.81971660), Tianjin Outstanding Youth Fund Project (No. 20JCJQIC00230), Capital’s Funds for Health Improvement and Research (2022-4-4019), National High-level Hospital Clinician Research Funding (APL221003101004060, 2022-PUMCH-A-017), Chinese Academy of Medical Science health innovation project (2021-I2M-042, 2021-I2M-058), Program of Chinese Institute for Brain Research in Beijing(2020-NKX-XM-14), Sichuan Science and Technology Program (No. 2021YFH0004), and Basic Research Program for Beijing-Tianjin-Hebei Coordination (19JCZDJC65500(Z)).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphy.2022.1033422/full#supplementary-material
References
1. Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. (2018) 68(6):394-424. doi:10.3322/caac.21492
2. Correa P., Piazuelo M. B. The gastric precancerous cascade. Journal of digestive diseases (2012) 13:2–9. doi:10.1111/j.1751-2980.2011.00550.x
3. Li D., Bautista M. C., Jiang S. F., Daryani P., Brackett M., Armstrong M. A., Hung Y., Postlethwaite D., Ladabaum U., Risks and Predictors of gastric Adenocarcinoma in patients with gastric intestinal metaplasia and Dysplasia: A population-based study. Official journal of the american college of gastroenterology (2016) 111(8):1104–13. doi:10.1038/ajg.2016.188
4. Suzuki H., Oda I., Abe S., Sekiguchi M., Mori G., Nonaka S., Yoshinaga S., Saito Y., High rate of 5-year survival among patients with early gastric cancer undergoing curative endoscopic submucosal dissection. Gastric Cancer (2016) 19(1):198–205. doi:10.1007/s10120-015-0469-0
5. Katai H., Ishikawa T., Akazawa K., Isobe Y., Miyashiro I., Oda I., Tsujitani S., Ono H., Tanabe S., Fukagawa T., Five-year survival analysis of surgically resected gastric cancer cases in Japan: A retrospective analysis of more than 100, 000 patients from the nationwide registry of the Japanese gastric cancer association (2001-2007). Gastric Cancer (2018) 21(1):144–154. doi:10.1007/s10120-017-0716-7
6. Chun H. J., Keum B., Kim J. H., Seol S. Y., Current status of endoscopic submucosal dissection for the management of early gastric cancer: A Korean perspective. World journal of gastroenterology (2011) 17(21):2592–2596. doi:10.3748/wjg.v17.i21.2592
7. Take I., Shi Q., Zhong Y. S. Progress with each passing day: Role of endoscopy in early gastric cancer. Translational Gastrointest Cancer (2015) 4(6):423–428. doi:10.3978/j.issn.2224-4778.2015.09.04
8. Buxbaum J. L., Hormozdi D., Dinis-Ribeiro M., Lane C., Dias-Silva D., Sahakian A., Jayaram P., Pimentel-Nunes P., Shue D., Pepper M., Narrow-band imaging versus white light versus mapping biopsy for gastric intestinal metaplasia: A prospective blinded trial. Gastrointestinal endoscopy (2017) 86(5):857–865. doi:10.1016/j.gie.2017.03.1528
9. Sumiyama K. Past and current trends in endoscopic diagnosis for early stage gastric cancer in Japan. Gastric Cancer (2017) 20(Suppl. 1):20–27. doi:10.1007/s10120-016-0659-4
10. Decharatanachart P., Chaiteerakij R., Tiyarattanachai T., Treeprasertsuk S. Application of artificial intelligence in chronic liver diseases: A systematic review and meta-analysis. BMC Gastroenterol (2021) 21(1):10. doi:10.1186/s12876-020-01585-5
11. Menon S., Trudgill N. How commonly is upper gastrointestinal cancer missed at endoscopy? A metaanalysis. Endoscopy international open (2014) 2(2):E46–50. doi:10.1055/s-0034-1365524
12. Gao Y., Zhang Z. D., Li S., Guo Y. T., Wu Q. Y., Liu S., Yang S., Ding L., Zhao B., Li S., Deep neural network-assisted computed tomography diagnosis of metastatic lymph nodes from gastric cancer. Chinese Medical Journal (2019) 132(23):2804–2811. doi:10.1097/CM9.0000000000000532
13. Ali H., Yasmin M., Sharif M., Rehmani M. H., Computer assisted gastric abnormalities detection using hybrid texture descriptors for chromoendoscopy images. Comput Methods Programs Biomed (2018) 157:39–47. doi:10.1016/j.cmpb.2018.01.013
14. Kanesaka T., Lee T. C., Uedo N., Lin K. P., Chen H. Z., Lee J. Y., Wang H., Chen H. Computer-aided diagnosis for identifying and delineating early gastric cancers in magnifying narrow-band imaging. Gastrointest Endosc (2018) 87(5):1339–1344. doi:10.1016/j.gie.2017.11.029
15. Wang S., Zhu Y., Yu L., Chen H., Lin H., Wan X., Fan X., Heng P. Recalibrated multi-instance deep learning for whole slide gastric image classification. Medical image analysis (2019) 58:101549. doi:10.1016/j.media.2019.101549
16. Van D. S., Zinger S., Schoon E. J., With D. P. Supportive automatic annotation of early esophageal cancer using local Gabor and color features. Neurocomputing (2014) 144:92–106. doi:10.1016/j.neucom.2014.02.066
17. Liu D. Y, Gan T., Rao N. N., Xing Y. W., Zheng J., Li S., Luo C., Zhou Z., Wan Y. Identification of lesion images from gastrointestinal endoscope based on feature extraction of combinational methods with and without learning process. Medical image analysis (2016) 32:281–294. doi:10.1016/j.media.2016.04.007
18. Zhou R., Yang C., Meng M. Q. H., Xu G., Hu C., Li B. Capsule endoscopy images classification by random forests and ferns. In Proceedings of the 4th IEEE International Conference on information Science and technology, Shenzhen, China: IEEE (2014). p. 414–417. doi:10.1109/ICIST.2014.6920505
19. Riaz F., Silva F. B., Ribeiro M. D., Coimbra M. T. Invariant Gabor texture descriptors for classification of gastroenterology images. IEEE Transactions Biomedical Engineering (2012) 59(10):2893–2904. doi:10.1109/TBME.2012.2212440
20. Ali H., Sharif M., Yasmin M., Rehmani M. H., Riaz F. A survey of feature extraction and fusion of deep learning for detection of abnormalities in video endoscopy of gastrointestinal-tract. Artificial Intelligence Review (2020) 53:2635–2707. doi:10.1007/s10462-019-09743-2
21. Gando G., Yamada T., Sato H., Oyama S., Kurihara M. Fine-tuning deep convolutional neural networks for distinguishing illustrations from photographs. Expert Systems with Applications (2016) 66:295–301. doi:10.1016/j.eswa.2016.08.057
22. Zhao Z., Jiao L., Zhao J., Gu J., Zhao J. Discriminant deep belief network for high-resolution SAR image classification. Pattern Recognition (2017) 61:686–701. doi:10.1016/j.patcog.2016.05.028
23. Bochkovskiy A, Wang CY, Liao H. YOLOV4: Optimal speed and accuracy of Object detection (2020). 10934, https://arxiv.org/abs/2004.10934.
24. Lin T. Y., Dollár P., Girshick R., He K., Hariharan B., Belongie S. Feature pyramid networks for object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition. Hawaii: IEEE (2017). p. 2117–25. doi:10.48550/arXiv.1612.03144
25. Shankar K., Zhang Y., Liu Y., Wu L., Chen C. H. Hyperparameter tuning deep learning for diabetic retinopathy fundus image classification. IEEE Access (2020) 8:118164–73. doi:10.1109/ACCESS.2020.3005152
26. Maghdid H. S., Asaad A. T., Ghafoor K. Z., Sadiq A. S., Khan M. K. Diagnosing COVID-19 pneumonia from x-ray and CT images using deep learning and transfer learning algorithms. Multimodal Image Exploitation and Learning, 2021:11734. doi:10.1117/12.2588672
27. Bulten W., Pinckaers H., van Boven H., Vink R., de Bel T., van Ginneken B., van der Laak J., Hulsbergen-van de Kaa C., Litjens G. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: A diagnostic study. Lancet Oncology (2020) 21(2):233–41. doi:10.1016/S1470-2045(19)30739-9
28. Sajjad M., Khan S., Muhammad K., Wu W., Ullah A., Baik S. W. Multi-grade brain tumor classification using deep CNN with extensive data augmentation. Journal of Computer Science (2019) 30:174–182. doi:10.1016/j.jocs.2018.12.003
29. Huo Y., Xu Z., Xiong Y., Aboud K., Parvathaneni P., Bao S., 3D whole brain segmentation using spatially localized atlas network tiles. Neuroimage (2019) 194:105–119. doi:10.1016/j.neuroimage.2019.03.041
30. Xie W., Jacobs C., Charbonnier J. P., van Ginneken B. Relational modeling for Robust and efficient Pulmonary Lobe segmentation in CT Scans. IEEE Transaction Medical Imaging (2020) 39(8):2664–75. doi:10.1109/TMI.2020.2995108
31. Zilly J., Buhmann J. M., Mahapatra D. Glaucoma detection using entropy sampling and ensemble learning for automatic optic cup and disc segmentation. Computerized Medical Imaging Graphics (2017) 55:28–41. doi:10.1016/j.compmedimag.2016.07.012
32. Sun W, Tseng TB, Zhang J, Qian W. Enhancing deep convolutional neural network scheme for breast cancer diagnosis with unlabeled data. Computerized Medical Imaging Graphics (2017) 57:4–9. doi:10.1016/j.compmedimag.2016.07.004
33. Huang R, Xie W, Vp-Nets ANJ. VP-Nets : Efficient automatic localization of key brain structures in 3D fetal neurosonography. Medical Image Analysis (2018) 47:127–39. doi:10.1016/j.media.2018.04.004
34. Alison Q., Kascenas A., Henry J., Wyeth D., Shepherd M., Beverid E., Clunie L., Sansom C. Attaining human-level performance with atlas location autocontext for anatomical landmark detection in 3D CT data. In: Proceedings of the European conference on computer vision (ECCV) Workshops (2019). Available at: https://link.springer.com/conference/eccv.
35. de Groof A. J., Struyvenberg M. R., van der Putten J., van der Sommen F., Fockens K. N, Curvers W. L., . Deep-learning system Detects neoplasia in patients with Barrett's Esophagus with higher accuracy than endoscopists in a Multistep training and validation study with benchmarking. Gastroenterology (2020) 158(4):915–29.e4. doi:10.1053/j.gastro.2019.11.030
36. Ohmori M., Ishihara R., Aoyama K., Nakagawa K., Iwagami H., Matsuura N., Endoscopic detection and differentiation of esophageal lesions using a deep neural network. Gastrointest Endosc (2020) 91(2):301–9.e1. doi:10.1016/j.gie.2019.09.034
37. Pannala R., Krishnan K., Melson J., Parsi M. A., Schulman A. R., Sullivan S., Artificial intelligence in gastrointestinal endoscopy. VideoGIE (2020) 5(12):598–613. doi:10.1016/j.vgie.2020.08.013
38. Wang C., Li Y., Yao J., Chen B., Song J., Yang X. Localizing and identifying intestinal metaplasia based on deep learning in Oesophagoscope. In Proceedings of the 8th International Symposium on Next Generation Electronics (ISNE). Zhengzhou, China: IEEE (2019). 1–4. doi:10.1109/ISNE.2019.8896546
39. Horiuchi Y., Aoyama K., Tokai Y., Hirasawa T., Yoshimizu S., Ishiyama A., Convolutional neural network for Differentiating gastric cancer from gastritis using magnified endoscopy with narrow band imaging. Digestive Diseases and Sciences (2020) 65(5):1355–63. doi:10.1007/s10620-019-05862-6
40. Yan T, Wong P. K., Choi I. C., Vong C. M., Yu H. H. Intelligent diagnosis of gastric intestinal metaplasia based on convolutional neural network and limited number of endoscopic images. Computuers in Biology and Medicine (2020) 126:104026 doi:10.1016/j.compbiomed.2020.104026
41. Cho BJ, Bang CS, Park SW, Yang YJ, Seo SI, Lim H. Automated classification of gastric neoplasms in endoscopic images using a convolutional neural network. Endoscopy (2019) 51(12):1121–9. doi:10.1055/a-0981-6133
42. Lui T. K. L., Wong K. K. Y., Mak L. L. Y., To E. W. P., Tsui V. W. M., Deng Z., Guo J., Ni L., Cheung M. K. S., Leung W. K., Feedback from artificial intelligence improved the learning of junior endoscopists on histology prediction of gastric lesions. Endosc Int Open (2020) 8(2):E139–46. doi:10.1055/a-1036-6114
43. Liu X., Wang C., Bai J., Liao G. Fine-tuning pre-trained convolutional neural networks for gastric precancerous disease classification on magnification narrow-band imaging images. Neurocomputing (2020) 392:253–67. doi:10.1016/j.neucom.2018.10.100
44. Weiss K., Khoshgoftaar T. M., Wang D. A survey of transfer learning. Journal of Big Data (2016) 3(1):9–40. doi:10.1186/s40537-016-0043-6
45. Fu J., Zheng H., Mei T. Look Closer to See better: Recurrent attention convolutional neural network for fine-grained image recognition. In: Proceedings -30th IEEE Conference.On computer vision and pattern recognition. Hawaii: IEEE (2017). p. 4476–84. doi:10.1109/CVPR.2017.476
46. Guan H. Y., Zhong Z., Zheng Z., Zheng L., Yang Y. Diagnose like A Radiologist: Attention guided convolutional neural network for Thorax disease classification. arxiv: 1801.09927(2018).
47. Du W., Rao N., Dong C., Wang Y., Hu D., Zhu L., Zeng B., Gan T. Automatic classification of esophageal disease in gastroscopic images using an efficient channel attention deep dense convolutional neural network. Biomedical Optic Express (2021) 12(6):3066–81. doi:10.1364/BOE.420935
48. Wang F., Jiang M., Qian C., Yang S., Li C., Zhang H., Wang X., Tang X. Residual attention network for image classification. In: Proceedings -30th IEEE Conference.On computer vision and pattern recognition. Hawaii: IEEE (2017). p. 6450–8. doi:10.1109/CVPR.2017.683
49. Chollet F. Xception: Deep learning with depthwise separable convolutions (2016). arxiv:1610.02357v2.
50. Szegedy C, Ioffe S, Vanhoucke V. Inception-v4, inception-resnet and the impact of residual connections on learning. arxiv:1602.07261. (2016).
51. Yu F, Koltun V. Multi-scale context aggregation by dilated convolutions. arxiv: 1511.07122, (2015).
52. Zhang Y, Kang B, Hooi B, Yan S., Feng J., Deep long-tailed learning: A survey (2021). Available at: https://arxiv.org/abs/2110.04596.
53. Zhang Y., Li F., Yuan F., Zhang K., Huo L., Dong Z., . Diagnosing chronic atrophic gastritis by gastroscopy using artificial intelligence. Digestive and Liver Disease (2020) 52(5):566–72. doi:10.1016/j.dld.2019.12.146
Keywords: gastric cancer and precancerous lesions, deep learning, magnification endoscopy with narrow-band imaging, artificial Intelligence, automatic classification
Citation: Wang L, Yang Y, Li J, Tian W, He K, Xu T, Fang Z, Yang A and Li T (2022) Automatic classification of gastric lesions in gastroscopic images using a lightweight deep learning model with attention mechanism and cost-sensitive learning. Front. Phys. 10:1033422. doi: 10.3389/fphy.2022.1033422
Received: 31 August 2022; Accepted: 03 October 2022;
Published: 19 October 2022.
Edited by:
Xunbin Wei, Peking University, ChinaReviewed by:
Jinchao Feng, Beijing University of Technology, ChinaXiaohua Qian, Shanghai Jiao Tong University, China
Copyright © 2022 Wang, Yang, Li, Tian, He, Xu, Fang, Yang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ting Li, bGl0aW5nQGJtZS5jYW1zLmNu; Aiming Yang, eWFuZ2FpbWluZ0BtZWRtYWlsLmNvbS5jbg==
†These authors share first authorship
 Lingxiao Wang
Lingxiao Wang Yingyun Yang
Yingyun Yang Jingyang Li3
Jingyang Li3 Kun He
Kun He Tianming Xu
Tianming Xu Ting Li
Ting Li