
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Phys., 08 September 2021
Sec. Medical Physics and Imaging
Volume 9 - 2021 | https://doi.org/10.3389/fphy.2021.684184
This article is part of the Research TopicArtificial Intelligence and MRI: Boosting Clinical DiagnosisView all 28 articles
 Hassan Haji-Valizadeh†‡
Hassan Haji-Valizadeh†‡ Rui Guo‡
Rui Guo‡ Selcuk Kucukseymen
Selcuk Kucukseymen Yankama Tuyen
Yankama Tuyen Jennifer Rodriguez
Jennifer Rodriguez Amanda Paskavitz
Amanda Paskavitz Patrick Pierce
Patrick Pierce Beth Goddu
Beth Goddu Long H. Ngo
Long H. Ngo Reza Nezafat*
Reza Nezafat*Propose: The purpose of this study was to compare the performance of deep learning networks trained with complex-valued and magnitude images in suppressing the aliasing artifact for highly accelerated real-time cine MRI.
Methods: Two 3D U-net models (Complex-Valued-Net and Magnitude-Net) were implemented to suppress aliasing artifacts in real-time cine images. ECG-segmented cine images (n = 503) generated from both complex k-space data and magnitude-only DICOM were used to synthetize radial real-time cine MRI. Complex-Valued-Net and Magnitude-Net were trained with fully sampled and synthetized radial real-time cine pairs generated from highly undersampled (12-fold) complex k-space and DICOM images, respectively. Real-time cine was prospectively acquired in 29 patients with 12-fold accelerated free-breathing tiny golden-angle radial sequence and reconstructed with both Complex-Valued-Net and Magnitude-Net. Cardiac function, left-ventricular (LV) structure, and subjective image quality [1(non-diagnostic)-5(excellent)] were calculated from Complex-Valued-Net– and Magnitude-Net–reconstructed real-time cine datasets and compared to those of ECG-segmented cine (reference).
Results: Free-breathing real-time cine reconstructed by both networks had high correlation (all R2 > 0.7) and good agreement (all p > 0.05) with standard clinical ECG-segmented cine with respect to LV function and structural parameters. Real-time cine reconstructed by Complex-Valued-Net had superior image quality compared to images from Magnitude-Net in terms of myocardial edge sharpness (Complex-Valued-Net = 3.5 ± 0.5; Magnitude-Net = 2.6 ± 0.5), temporal fidelity (Complex-Valued-Net = 3.1 ± 0.4; Magnitude-Net = 2.1 ± 0.4), and artifact suppression (Complex-Valued-Net = 3.1 ± 0.5; Magnitude-Net = 2.0 ± 0.0), which were all inferior to those of ECG-segmented cine (4.1 ± 1.4, 3.9 ± 1.0, and 4.0 ± 1.1).
Conclusion: Compared to Magnitude-Net, Complex-Valued-Net produced improved subjective image quality for reconstructed real-time cine images and did not show any difference in quantitative measures of LV function and structure.
Cardiovascular MR (CMR) is the clinical gold-standard imaging modality for evaluation of cardiac function and structure. Breath-hold ECG-segmented cine imaging using balanced steady-state free-procession readout (bSSFP) allows for accurate and reproducible measurement of left-ventricular (LV) and right-ventricular (RV) function and volume [1–3]. In this technique, k-space is divided into different segments collected over consecutive cardiac cycles within a single breath-hold scan. However, ECG-segmented cine acquisition has limited spatial and temporal resolution, is sensitive to changes in heart rate, and requires repeated breath-holds [4–6]. Alternatively, free-breathing real-time cine has been proposed and pursued using rapid real-time imaging or multiple averaging with or without motion correction [7–12]. Using free-breathing real-time cine is advantageous because it does not require multiple breath-holds and is insensitive to heart rate variations. However, real-time cine has lower temporal and spatial resolution than ECG-segmented cine [10, 11]. Therefore, there is a need to further accelerate data collection for real-time cine MRI.
Over the past three decades, there has been considerable progress in the development of accelerated real-time cine imaging including parallel imaging and compressed sensing [13–18]. Parallel imaging is almost always used in cine imaging for both real-time and ECG-segmented acquisition with robust and highly reliable image quality [13]. However, the acceleration rate of parallel imaging cannot be more than three without compromising image quality [19–21]. Compressed sensing has recently been integrated into applications by vendors enabling higher acceleration rates than parallel imaging; however, reconstruction time is long, and acceleration rates beyond four can result in degradation of image quality [17]. Alternative techniques that exploit spatial–temporal correlation and sparsity of cine data have also been explored [22–26]; however, these approaches can suffer from temporal data filtering, often removing information that is crucial to cardiac cine evaluation. Therefore, despite considerable interest from the image reconstruction community, these techniques are rarely clinically used.
Deep learning–based reconstruction has been recently proposed to enable rapid reconstruction of accelerated cine MRI. Hauptmann et al. [27] showed that a 3D U-net was capable of reconstructing accelerated (acceleration rate = 13) real-time cine MRI. Schlemper et al. [28] showed that a trained cascade network was able to rapidly reconstruct accelerated (acceleration rate = 11) cine MRI. Kustner et al. [29] showed that (3 + 1)-dimensional complex-valued spatio-temporal convolutions and multi-coil data processing (CINENet) could reconstruct accelerated (9 ≤ acceleration rate ≤15) 3D ECG-segmented cine. El-Rewaidy et al. [30] reconstructed accelerated radial cine MRI (acceleration rate = 14) using a complex-valued network (MD-CNN) designed to process MR data in both k-space and image space. Daming et al. [31] used a complex U-net with a combined mean-squared error and perceptual loss (PCNN) to reconstruct real-time cine MRI (acceleration rate = 15).
While promising, popular deep learning–based reconstructions methods [27–32] for cine MRI rely on supervised learning and, as such, require training with large and diverse patient datasets. However, prospectively acquiring large patient datasets within a clinical setting can be difficult due to long scanning times, respiratory/cardiac motion, or contrast washout. To overcome these limitations, Hauptmann et al. proposed training a deep learning network using synthetic data generated from DICOMs (Digital Imaging and Communications in Medicine) [27]. The use of DICOM imaging is advantageous because it is readily available in large numbers at centers with cardiac MR expertise. While promising, DICOM usage during training is theoretically non-optimal given that DICOM images are magnitude images, which lack phase and multi-coil information; furthermore, vendors often apply different filtering techniques to improve image quality in the DICOM creation process. The effect of using DICOM images for training on the performance of a deep learning model has not yet been rigorously studied.
In this study, we sought to investigate differences in performance between two deep learning–based models trained to suppress artifacts in 12-fold accelerated real-time cine. Paired complex-valued k-space data and DICOM images of ECG-segmented cine (n = 503) were used to synthetize highly undersampled radial real-time cine data. Both artifact suppression models were made using 3D U-net architectures. One model was trained with synthetic radial real-time cine images generated from complex k-space data (Complex-Valued-Net), while the other model was trained with synthetic radial real-time cine images generated from DICOM images (Magnitude-Net). The performance of the two models was evaluated against prospectively collected free-breathing real-time cine CMR with radial acquisition.
Figure 1 summarizes our study which was designed to compare the performance of deep learning–based networks trained to suppress aliasing artifacts in highly accelerated real-time cine using complex-valued images (derived from k-space data) and magnitude-only images (derived from DICOM images). We prepared a dataset containing both complex k-space data and corresponding magnitude images (i.e., DICOM) scanned by breath-holding ECG-segmented cine using a Cartesian trajectory to synthesize radial real-time cine data (Figure 1A) [27]. Two 3D U-net models [33], Complex-Valued-Net and Magnitude-Net, were developed to remove aliasing artifacts in complex-valued and magnitude images of highly accelerated radial real-time cine, respectively. Complex-Valued-Net and Magnitude-Net were trained using synthetized radial real-time cine with aliasing artifacts generated from complex-valued k-space and magnitude-only images, respectively. “Artifact-free” images used to produce synthetized radial cine were used as the ground truth (Figure 1B). Finally, the performance of both networks was compared using prospectively acquired free-breathing highly accelerated (12x) radial real-time cine in 29 patients. Quantitative functional and structural parameters of the LV and qualitative visual assessments of the LV were compared against reference values derived from ECG-segmented cine images (Figure 1C).
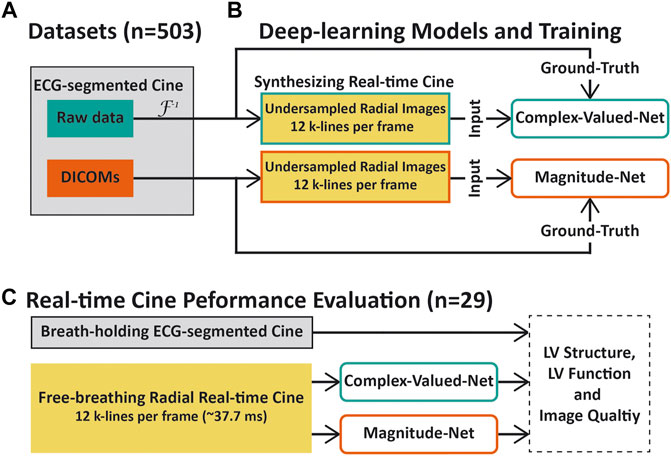
FIGURE 1. Overview of this study. (A) Cine images of 503 patients with both raw k-space data and DICOMs were collected. These images were scanned using a breath-holding cine sequence with a Cartesian trajectory. (B) Raw k-space data and DICOMs of ECG-segmented cine were used to synthesize highly accelerated radial real-time cine datasets for training Complex-Valued-Net and Magnitude-Net, respectively. (C) Performance comparison between the two neural networks. Real-time radial cine and corresponding ECG-segmented cine images were collected from 29 patients. The left-ventricular function, structural parameters, and subjective image scores were used to compare the performance of both deep learning models with respect to aliasing artifact suppression. For quantitative and qualitative evaluation, Magnitude-Net reconstruction, Complex-Valued-Net reconstruction, and ECG-segmented cine were compared to one another in pairs.
We retrospectively collected short-axis (SAX) cine data from 503 patients (286 males, 55.4 ± 15.8 years) who underwent clinical scans at BIDMC from October 2018 to May 2020. Imaging was performed on a 3T MR scanner (MAGNETOM Vida Siemens Healthineers, Erlangen, Germany) using a breath-hold ECG-segmented sequence with the following parameters: bSSFP readout, FOV = 355 × 370 mm2, in-plane resolution = 1.7 × 1.4 mm2, slice thickness = 8 mm, TE/TR = 1.41/3.12 ms, flip angle = 42°, GRAPPA acceleration rate = 2–3, ∼18 cardiac phases at a temporal resolution of ∼55.3 ms, receiver bandwidth = 1,502 Hz/pixel, Cartesian sampling pattern, and slices per volume = 11 ± 1 (from 9 to 17). Cine’s paired raw k-space data and DICOM images were used in this study. This study protocol was approved by the institutional review board, and written consent was waived. Patient information was handled in compliance with the Health Insurance Portability and Accountability Act.
Supplementary Figure S1 shows the data preparation workflow for producing synthetic accelerated radial real-time cine datasets from ECG-segmented cine data acquired using the Cartesian trajectory. The complex-valued multi-coli k-space data with an acceleration rate of 2–3 were first reconstructed by GRAPPA [21] offline. Offline GRAPPA reconstruction was implemented with the code made available by Dr. Chiew (https://users.fmrib.ox.ac.uk/∼mchiew/Teaching.html).
Then, GRAPPA-reconstructed images and the original DICOM images exported from the scanner were interpolated to achieve 2 × 2 mm2 in-plane resolution with a temporal resolution of 37.7 ms. We chose these interpolated spatial and temporal resolutions to match the temporal and spatial resolutions used during prospective real-time cine scanning (see below). These GRAPPA-reconstructed or DICOM images were also used as the ground truth in training of two neural networks, respectively. Subsequently, backward non-uniform fast Fourier transform (NUFFT) [34] was applied to GRAPPA-reconstructed and DICOM images to produce complex-valued radial k-space. Twelve lines per frame, which were distributed over the whole k-space with a tiny golden-angle rotation (32.049°) [35, 36], were chosen to simulate highly accelerated radial k-space of real-time cine.
For both Complex-Valued-Net and Magnitude-Net, simulated highly accelerated radial k-space data were transformed into image space using forward NUFFT. Specifically, for complex-valued multi-coil k-space, the above procedures were performed on a coil-by-coil basis. Finally, a coil-combined image was generated using sensitivity-encoding coil combination [37]. An auto-calibrated sensitivity profile for each coil was produced as previously described [38]. Note that a GPU-based implementation of NUFFT (https://cai2r.net/resources/gpunufft-an-open-source-gpu-library-for-3d-gridding-with-direct-matlab-interface/) was used for synthetic MRI generation.
Supplementary Figure S2 presents an in-depth description of the 3D residual U-net architecture used for Complex-Valued-Net and Magnitude-Net. The U-net architecture of both networks comprised five million kernels and two max-pooling layers/up-convolutional layers. Each convolutional processing layer consisted of 3 × 3 × 3 kernels, batch normalization, and rectified linear activation function (ReLU) [33].
The input/output of each network consisted of paired artifact-free ground truth images and their corresponding undersampled, artifact-contaminated images (size: M × N × T = 144 × 144 × 20). Specifically, for Complex-Valued-Net, we concatenated real and imaginary components of complex-valued input/output pairs to enable real-valued deep learning model processing of complex-valued data (size: 2M × N × T = 288 × 144 × 20) [39]. For Magnitude-Net, a ReLU operator was positioned at the final layer to force the output to be non-negative [27]. L2 loss function was used to train both networks.
Both networks were implemented using PyTorch (Facebook, Menlo Park, California) and trained on a DGX-1 workstation (NVIDIA Santa Clara, California, United States) equipped with 88 Intel Xeon central processing units (2.20 GHz), eight NVIDIA Tesla V100 graphics processing units (GPUs), and 504 GB RAM. Each GPU has 32 GB memory and 5120 Tensor cores. Each network was trained with 2,900 iterations using an ADAM optimizer and with a 15% drop-out rate. Each iteration randomly chose cine images of 16 LV slices from different patients (batch size). For synthetized real-time cine with ≥20 frames, the starting frame was randomly selected to achieve 20 consecutive frames. For <20 timeframes, the dynamic series was circularly padded to 20. Both input and output images were normalized by the 95th percentile magnitude pixel intensity within the central region (i.e., 48 × 48) across 20 frames. The initial learning rate was 0.001, which decreased by 5% after every 100 iterations. The cost function and optimizer were selected to match parameters proposed by Hauptmann et al. [27] for neural network training using DICOM-derived simulated real-time cine.
Twenty-nine patients (16 males, 58 ± 16 years) were prospectively recruited. Free-breathing radial real-time cine research sequences in addition to clinically indicated CMR sequences were collected from each patient. Written informed consent was obtained from each patient prior to CMR imaging. Clinical indications and characteristics of these patients are listed in Supplementary Table S1. Breath-hold ECG-segmented cine was performed using the same imaging parameters as those detailed in Training Datasets. Free-breathing radial real-time cine was collected with the following parameters: bSSFP readout, FOV = 288 × 288 mm2, resolution = 2 × 2 mm2, slice thickness = 8 mm, TE/TR = 1.3/3.2 ms, flip angle = 43°, receiver bandwidth = 1,085 Hz/pixel, radial lines per phase = 12, and temporal resolution = 37.7 ms. The rotating angle of the radial line was 32.049° [36]. Both sequences imaged a stack of 14 SAX slices covering the entire LV. Breath-holding ECG-segmented cine was reconstructed by the scanner. For free-breathing real-time cine, NUFFT first transformed radial k-space data into complex-valued and magnitude images. Subsequently, two neural networks were used to remove aliasing artifacts.
We used both quantitative imaging parameters and qualitative assessments of image quality to compare the performance of both deep learning reconstructions. ECG-segmented cine images collected using the standard clinical protocol were used as a reference. For each patient in our independent validation dataset, one reader (HH), trained by a clinical reader (SK) with 5 years of experience, calculated the following cardiac function and structural parameters: LV ejection fraction (LVEF), LV end-diastolic volume (LVEDV), LV end-systolic volume (LVESV), LV stroke volume (LVSV), and LV mass (LVMass). All quantifications were performed using CVI42 (v5.9.3, Cardiovascular Imaging, Calgary, Canada). Linear regression and Bland–Altman analysis were performed to evaluate correlation and agreement between real-time cine and ECG-segmented cine. A paired Student’s t-test was conducted to compare the difference between two approaches in measures of LV function and structural parameters. p < 0.05 was considered statistically significant. Three pairwise group comparisons were assessed using the t-test with Bonferroni correction, with p less than 0.0167 considered significant.
Subjective image quality was graded by one reader (SK) with 5 years of CMR experience. Cine images of all patients obtained from the three methods were randomized and de-identified. For each method, whole LV cine images from each subject were scored with respect to conspicuity of endocardial borders (1: non-diagnostic, 2: poor, 3: adequate, 4: good, 5: excellent), temporal fidelity of wall motion (1: non-diagnostic, 2: poor, 3: adequate, 4: good, 5: excellent), and artifact level on the myocardium (1: non-diagnostic, 2: severe, 3: moderate, 4: mild, 5: minimal). Supplementary Figure S3 shows representative graded images. The z-test was used to compare image quality between every two methods, and a p-value < 0.05 was considered significant. SAS version 9.4 (SAS Institute, Cary, NC, United States) was utilized for all above analyses. Note that we elected not to quantitatively and qualitatively analyze real-time cine reconstructed with gridding because gridding alone did not produce diagnostic image quality.
Figures 2A,B show images obtained from the basal, mid, and apical cavities of one subject at end-systole and -diastole by ECG-segmented cine and free-breathing real-time cine via gridding, Complex-Valued-Net, and Magnitude-Net reconstruction. Supplementary Videos S1–S4 show the corresponding movies for dynamic display. We also show representative end-systolic images for three patients in Supplementary Figure S4. In both Figure 2 and Supplementary Figure S4, free-breathing real-time cine reconstructed by Magnitude-Net had more artifacts in the myocardial wall and greater blurring than ECG-segmented cine and real-time cine by Complex-Valued-Net.
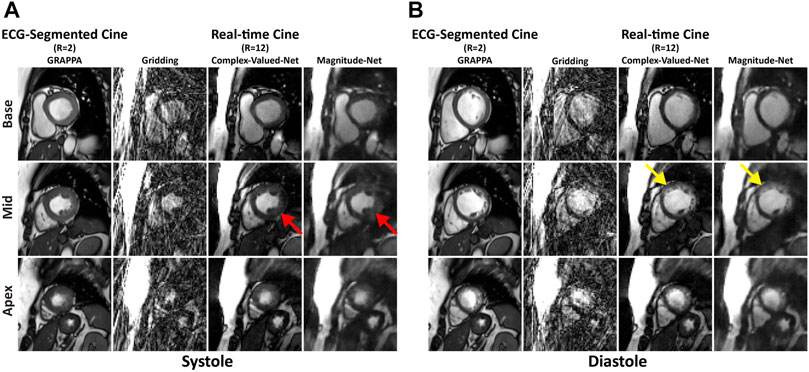
FIGURE 2. Images at end-systolic (A) and end-diastolic (B) phases for three short-axis slices (base, mid, apex) in one patient. Magnitude-Net exhibits more image artifact (red arrow) and greater blurring (yellow arrow) at the myocardial wall than Complex-Valued-Net. Gridding reconstruction produces non-diagnostic image quality.
Supplementary Table S2 summarizes LV structure and cardiac function values from ECG-segmented cine and free-breathing real-time cine in 29 patients. The mean difference and 95% CI between every two methods are listed in Supplementary Table S3. According to Bland–Altman analysis (Figures 3A–C), mean differences between ECG-segmented cine and real-time cine by Complex-Valued-Net reconstruction were −0.9 ± 6.5% (p = 0.48) for LVEF, 0.9 ± 13.6 ml (p = 0.73) for LVEDV, and 2.2 ± 12.5 ml (p = 0.34) for LVESV. Correspondingly, mean differences between real-time cine by Magnitude-Net and ECG-segmented cine images were −2.3 ± 5.1% (p = 0.02), −0.5 ± 15.5 ml (p = 0.85), and 3.7 ± 9.8 ml (p = 0.05) for LVEF, LVEDV, and LVESV, respectively (Figures 3D–F). Supplementary Figure S5 compares real-time cine and ECG-segmented cine according to LVSV and LVMass using Bland–Altman analysis. For real-time cine images reconstructed by Complex-Valued-Net, the mean difference was −1.4 ± 16.3 ml (p = 0.65) for LVSV and 2.2 ± 15.2 g (p = 0.43) for LVMass. For Magnitude-Net real-time cine, the mean difference was −4.2 ± 15.4 ml (p = 0.15) and 1.6 ± 18.0 g (p = 0.64) for LVSV and LVMass, respectively. Free-breathing real-time cine reconstructed by both Complex-Valued-Net and Magnitude-Net had high correlation with ECG-gated segmented cine on quantification of LV function and structure (all R2 ≥ 0.74 and all slope ≥ 0.88) (Figures 3G–I and Supplementary Figures S5C, F). The difference between real-time cine images reconstructed by Complex-Valued-Net and Magnitude-Net in quantification of LV function and structure was 1.4 ± 5.1% (p = 0.15) for LVEF, 1.4 ± 8.1 ml (p = 0.36) for LVEDV, −1.4 ± 8.7 ml (p = 0.39) for LVESV, 2.8 ± 12.1 ml (p = 0.22) for LVSV, and 0.7 ± 10.8 g (p = 0.74) for LVMass.
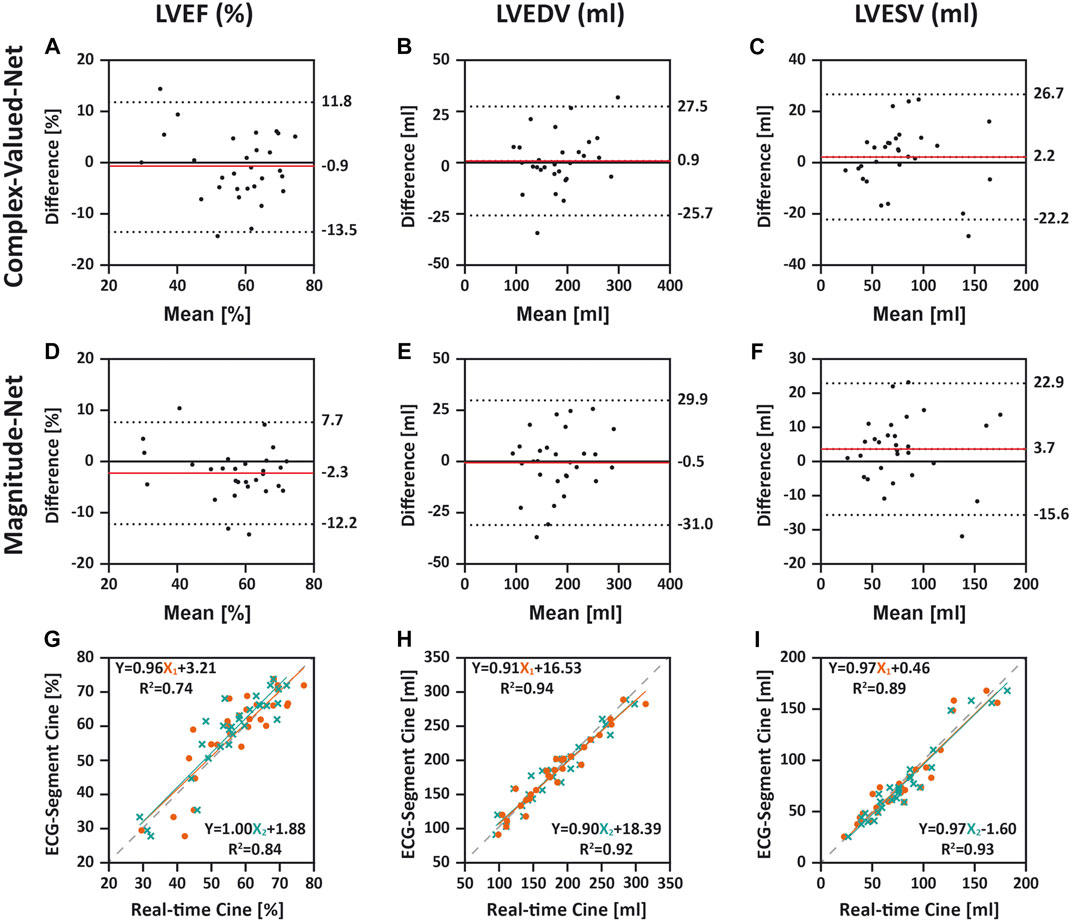
FIGURE 3. Comparison between ECG-segmented cine and real-time cine for quantifying left-ventricular ejection fraction (LVEF), left-ventricular end-diastolic volume (LVEDV), and left-ventricular end-systolic volume (LVESV) using Bland–Altman analysis (A–F) and linear regression (G–I). In Bland–Altman, dotted lines indicate upper and lower 95% limits of agreement and the red line represents the mean difference. The difference was calculated as real-time cine (Complex-Valued-Net and Magnitude-Net) minus ECG-segmented cine. In linear regression, X1 and X2 indicate real-time cine reconstructed by Complex-Valued-Net and Magnitude-Net, respectively. The dashed line shows a reference line with a slope of 1. All three quantifications from real-time cine using both Complex-Valued-Net and Magnitude-Net had good agreement and high correlation with quantifications by ECG-segmented cine (all p > 0.0167).
Figure 4 shows the mean/standard deviation and distribution of image quality scores across all patients. Supplementary Table S4 lists the percentages as two grades (1–3 and 4–5) of image quality scores across all patients by each method. The corresponding differences in the percentage of two grade groups (1–3 and 4–5) among three methods are listed in Table 1. The table shows that 79% of ECG-segment cine images had good or excellent scores (>3) for myocardial edge (4.1 ± 1.4) and temporal fidelity (3.9 ± 1.0). In contrast, 50% of real-time cine images reconstructed by both Complex-Valued-Net and Magnitude-Net scored less than or equal to 3 (myocardial edge: 3.5 ± 0.5 vs 2.6 ± 0.5; temporal fidelity: 3.1 ± 0.4 vs 2.1 ± 0.4), suggesting poor image quality. ECG-segment cine had less artifact (4.0 ± 1.1) than real-time cine (Complex-Valued-Net: 3.1 ± 0.5; Magnitude-Net: 2.0 ± 0.0). All z-tests were found to be significant (p < 0.05).

FIGURE 4. Distribution and average image quality scores across all cine images of 29 patients by three methods: ECG-segmented cine, Complex-Valued-Net real-time cine, and Magnitude-Net real-time cine. The P-values of z-tests between every two methods regarding each criterion are labeled. Real-time cine by Complex-Valued-Net reconstruction yielded superior subjective scores for all three criteria compared to those by Magnitude-Net. ECG-segmented breath-hold cine had the highest score across all three criteria.
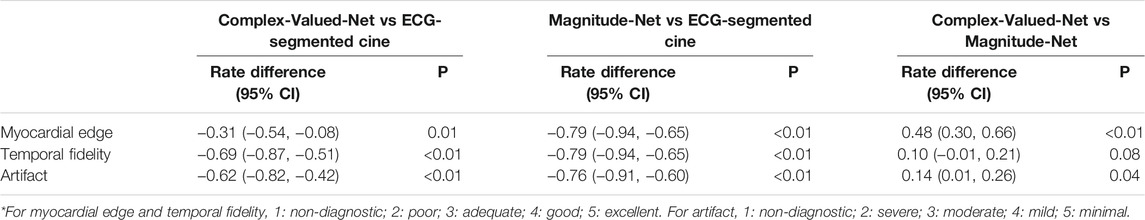
TABLE 1. Differences in percentage of two grades (1–3 and 4–5) of image quality scores between three methods.
This study compares the performance of deep learning approaches for reconstruction of highly accelerated real-time cine using synthetized training data generated from complex-valued multi-coil k-space data (Complex-Valued-Net) and real-valued DICOMs (Magnitude-Net). Our subjective assessment of image quality demonstrates that Complex-Valued-Net yields better image quality than Magnitude-Net. However, the clinically relevant parameters of LV function and structure extracted from real-time cine reconstructed by both Complex-Valued-Net and Magnitude-Net were highly correlated and had excellent agreement with those of clinical breath-holding ECG-segmented cine.
There is a growing body of literature in deep learning, beyond CMR, in which magnitude images are used for training a variety of deep learning techniques [27, 40–42]. However, there is also concern regarding the impact that discarded phase information may have on the clinical interpretation of reconstructed images [27, 29, 43–45]. Our study demonstrates that availability of complex k-space data improves overall image quality; however, these improvements in image quality do not necessary impact clinical interpretation and quantification. This observation is not unique, and it is often debated whether “prettier” images necessarily lead to better diagnostic information. While the resulting data do not show clinically meaningful differences in LV function and structural parameters, an improvement in overall image quality may still be clinically relevant. For example, we often rely on wall motion abnormality to assess the presence of ischemia, which can be visually assessed by reviewing cine images [46]. One can envision that improved image quality may still be clinically relevant and provide additional confidence in image assessments. Further studies in patients with different imaging indications are warranted.
In cine imaging, voxel-values are not meaningful; however, in quantitative CMR imaging (e.g., T1/T2 mapping, quantitative perfusion, or phase-contrast), voxel-values represent a tissue-specific meaning [47]. While qualitative imaging such as cine imaging is more forgiving in terms of artifact and inaccuracy during image reconstruction, quantitative CMR imaging is very sensitive to image artifacts. In addition, complex k-space data carry crucial information in quantitative imaging and cannot simply be discarded. Therefore, complex k-space data will still be needed for quantitative CMR image reconstruction with deep learning, despite our findings showing that magnitude-only images may be sufficient in real-time cine imaging. Further studies are needed to rigorously study other imaging sequences.
For this study, our goal was not necessarily to study or develop a new architecture but was instead motivated by Hauptmann et al. and their important contribution of using readily available DICOMs for network training [27]. Raw complex k-space data will still be needed for deep learning models that integrate complex k-space data for image reconstruction. However, limited availability of complex k-space data will remain a major challenge for training such networks on different applications, diseases, scanner vendors, field strengths, and number of coils. On the contrary, if one can train the model using only DICOM images, there are vast amounts of available data for different organs, sequences, diseases, and vendors that could greatly impact the adoption of deep learning artifact reconstruction techniques.
This study has several limitations. Our training data were not collected using prospectively acquired datasets using radial k-space filling, but instead training data were synthesized in a similar manner as proposed by Hauptmann et al. [27]. We used ECG-gated cine images with Cartesian sampling to extract reference values for different LV functional and structural parameters for comparison with real-time radial imaging [27]. There may be differences between the two approaches due to the k-space sampling scheme. Additionally, ECG-segmented data were collected with breath-holding, while real-time data were collected during free-breathing. The evaluation of deep learning reconstruction methodologies was limited to image quality assessment and quantification of left-ventricular functional and structural parameters (i.e., EF, LVEDV, LVESV, LVSV, and LVMass). We chose these metrics because of their clinical importance. That said, further studies are warranted to evaluate the capacity of the presented methods (Magnitude-Net and Complex-Valued-Net) for diagnosis of cardiovascular diseases. Real-time cine reconstructed with gridding was not quantitatively or qualitatively analyzed because gridding alone produced non-diagnostic image quality. Subjective image assessment was performed by a single reader, and there may be differences in image perception by different reviewers. Both Magnitude-Net and Complex-Valued-Net suffer from reduced temporal fidelity compared to ECG-gated segmented cine. Such a loss of temporal fidelity can be especially problematic during systolic phases and may be a source of error during qualitative and quantitative evaluation. All patients in our testing cohort were in sinus rhythm. Only a single neural network architecture (i.e., 3D U-net) was used to compare the performance of magnitude vs complex-valued synthetic training data. We chose this network architecture because, to the best of our knowledge, it is the only architecture shown to be capable of reconstructing radial real-time cine MRI acquired with bSSFP readout [27, 31]. Other state-of-the-art approaches such as cascade networks [28, 29] have yet to be investigated for radial real-time cine reconstruction. Future collaborations are warranted to first extend other state-of-the-art methods to radial real-time cine reconstruction and then compare the performance of different synthetic training data (i.e., magnitude vs. complex-valued) using these methods. ECG-segmented cine images used for training were gathered from one cardiac MR center. As such, trained networks could contain bias which can prevent generalization. Although we used a relatively large number of patients for training, our testing cohort with real-time radial imaging was relatively small, and images were acquired at a single clinical center. Future studies with more patients and imaging from different centers are required to evaluate proposed deep learning methodologies for real-time cine reconstruction.
Despite improved subjective image quality in real-time cine images reconstructed using a deep learning model trained with complex k-space data when compared to magnitude-only data, there were no differences with respect to quantitative measures of LV function and structural parameters.
The original contributions presented in the study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding author.
This study was approved by the BIDMC Institutional Review Board (IRB) and was Health Insurance Portability and Accountability Act (HIPPA)-compliant. This study was performed under two IRB approved protocols, including one allowing use of retrospective data collected as part of a clinical exam for machine learning research; informed consent was waived for use of previously collected data. In addition, we prospectively recruited subjects for this study, and written-informed consent was obtained from all prospective participants.
HH-V and RN contributed to study design and validation of deep learning models. HH-V contributed to training of deep learning models. RG, SK, YT, and LN performed data analysis, and RG and RN prepared the manuscript. JR, AP, PP, and BG contributed to data collection. RN contributed to data interpretation. All authors critically revised the paper and read and approved the final manuscript.
RN was funded by the NIH (5R01HL127015-02, 1R01HL129157-01A1, 5R01HL129185, and 1R01HL154744) and the American Heart Association (15EIA22710040). HH-V was supported by the NIH (5T32HL007374-41).
HH-V is currently an employee of Canon Medical Research USA Inc. The work presented in this study was performed during his post-doctoral training fellowship at Beth Israel Deaconess Medical Center.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphy.2021.684184/full#supplementary-material
1. Carr JC, Simonetti O, Bundy J, Li D, Pereles S, Finn JP. Cine MR Angiography of the Heart with Segmented True Fast Imaging with Steady-State Precession. Radiology (2001) 219(3):828–34. doi:10.1148/radiology.219.3.r01jn44828
2. Hundley WG, Bluemke DA, Finn JP, Flamm SD, Fogel MA, Friedrich MG, et al. ACCF/ACR/AHA/NASCI/SCMR 2010 Expert Consensus Document on Cardiovascular Magnetic Resonance. J Am Coll Cardiol (2010) 55(23):2614–62. doi:10.1016/j.jacc.2009.11.011
3. Leiner T, Bogaert J, Friedrich MG, Mohiaddin R, Muthurangu V, Myerson S, et al. SCMR Position Paper (2020) on Clinical Indications for Cardiovascular Magnetic Resonance. J Cardiovasc Magn Reson (2020) 22(1):76. doi:10.1186/s12968-020-00682-4
4. Atkinson DJ, Edelman RR. Cineangiography of the Heart in a Single Breath Hold with a Segmented TurboFLASH Sequence. Radiology (1991) 178(2):357–60. doi:10.1148/radiology.178.2.1987592
5. James CC, Orlando S, Jeff B, Debiao L, Scott P, Finn JP. Cine MR Angiography of the Heart with Segmented True Fast Imaging with Steady-State Precession1. Radiology (2001) 219(3):828–34. doi:10.1148/radiology.219.3.r01jn44828
6. Finn JP, Nael K, Deshpande V, Ratib O, Laub G. Cardiac MR Imaging: State of the Technology. Radiology (2006) 241(2):338–54. doi:10.1148/radiol.2412041866
7. Goebel J, Nensa F, Bomas B, Schemuth HP, Maderwald S, Gratz M, et al. Real-Time SPARSE-SENSE Cardiac Cine MR Imaging: Optimization of Image Reconstruction and Sequence Validation. Eur Radiol (2016) 26(12):4482–9. doi:10.1007/s00330-016-4301-y
8. Setser RM, Fischer SE, Lorenz CH. Quantification of Left Ventricular Function with Magnetic Resonance Images Acquired in Real Time. J Magn Reson Imaging (2000) 12(3):430–8. doi:10.1002/1522-2586(200009)12:3<430::aid-jmri8>3.0.co;2-v
9. Plein S, Smith WHT, Ridgway JP, Kassner A, Beacock DJ, Bloomer TN, et al. Qualitative and Quantitative Analysis of Regional Left Ventricular wall Dynamics Using Real-Time Magnetic Resonance Imaging: Comparison with Conventional Breath-Hold Gradient echo Acquisition in Volunteers and Patients. J Magn Reson Imaging (2001) 14(1):23–30. doi:10.1002/jmri.1146
10. Kellman P, Chefd'hotel C, Lorenz CH, Mancini C, Arai AE, McVeigh ER. Fully Automatic, Retrospective Enhancement of Real-Time Acquired Cardiac Cine MR Images Using Image-Based Navigators and Respiratory Motion-Corrected Averaging. Magn Reson Med (2008) 59(4):771–8. doi:10.1002/mrm.21509
11. Kellman P, Chefd'hotel C, Lorenz CH, Mancini C, Arai AE, McVeigh ER. High Spatial and Temporal Resolution Cardiac Cine MRI from Retrospective Reconstruction of Data Acquired in Real Time Using Motion Correction and Resorting. Magn Reson Med (2009) 62(6):1557–64. doi:10.1002/mrm.22153
12. Kaji S, Yang PC, Kerr AB, Tang WHW, Meyer CH, Macovski A, et al. Rapid Evaluation of Left Ventricular Volume and Mass without Breath-Holding Using Real-Time Interactive Cardiac Magnetic Resonance Imaging System. J Am Coll Cardiol (2001) 38(2):527–33. doi:10.1016/s0735-1097(01)01399-7
13. Feng L, Srichai MB, Lim RP, Harrison A, King W, Adluru G, et al. Highly Accelerated Real-Time Cardiac Cine MRI Usingk-tSPARSE-SENSE. Magn Reson Med (2013) 70(1):64–74. doi:10.1002/mrm.24440
14. Tsao J, Boesiger P, Pruessmann KP. K-T BLAST Andk-T SENSE: Dynamic MRI with High Frame Rate Exploiting Spatiotemporal Correlations. Magn Reson Med (2003) 50(5):1031–42. doi:10.1002/mrm.10611
15. Tsao J, Kozerke S, Boesiger P, Pruessmann KP. Optimizing Spatiotemporal Sampling fork-t BLAST Andk-T SENSE: Application to High-Resolution Real-Time Cardiac Steady-State Free Precession. Magn Reson Med (2005) 53(6):1372–82. doi:10.1002/mrm.20483
16. Jung H, Park J, Yoo J, Ye JC. Radial K-T FOCUSS for High-Resolution Cardiac Cine MRI. Magn Reson Med (2010) 63(1):68–78. doi:10.1002/mrm.22172
17. Otazo R, Kim D, Axel L, Sodickson DK. Combination of Compressed Sensing and Parallel Imaging for Highly Accelerated First-Pass Cardiac Perfusion MRI. Magn Reson Med (2010) 64(3):767–76. doi:10.1002/mrm.22463
18. Ding Y, Chung Y-C, Jekic M, Simonetti OP. A New Approach to Autocalibrated Dynamic Parallel Imaging Based on the Karhunen-Loeve Transform: KL-TSENSE and KL-TGRAPPA. Magn Reson Med (2011) 65(6):1786–92. doi:10.1002/mrm.22766
19. Deshmane A, Gulani V, Griswold MA, Seiberlich N. Parallel MR Imaging. J Magn Reson Imaging (2012) 36(1):55–72. doi:10.1002/jmri.23639
20. Blaimer M, Breuer F, Mueller M, Heidemann RM, Griswold MA, Jakob PM. SMASH, SENSE, PILS, GRAPPA: How to Choose the Optimal Method. Top Magn Reson Imaging (2004) 15(4):223–36. doi:10.1097/01.rmr.0000136558.09801.dd
21. Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, et al. Generalized Autocalibrating Partially Parallel Acquisitions (GRAPPA). Magn Reson Med (2002) 47(6):1202–10. doi:10.1002/mrm.10171
22. Otazo R, Candès E, Sodickson DK. Low-Rank Plus Sparse Matrix Decomposition for Accelerated Dynamic MRI with Separation of Background and Dynamic Components. Magn Reson Med (2015) 73(3):1125–36. doi:10.1002/mrm.25240
23. Lingala SG, Hu Y, DiBella E, Jacob M. Accelerated Dynamic MRI Exploiting Sparsity and Low-Rank Structure: K-T SLR. IEEE Trans Med Imaging (2011) 30(5):1042–54. doi:10.1109/tmi.2010.2100850
24. Zhao B, Haldar JP, Christodoulou AG, Liang ZP. Image Reconstruction from Highly Undersampled (K,t)-Space Data with Joint Partial Separability and Sparsity Constraints. IEEE Trans Med Imaging (2012) 31(9):1809–20. doi:10.1109/TMI.2012.2203921
25. Haji-Valizadeh H, Rahsepar AA, Collins JD, Bassett E, Isakova T, Block T, et al. Validation of Highly Accelerated Real-Time Cardiac Cine MRI with Radial K-Space Sampling and Compressed Sensing in Patients at 1.5T and 3T. Magn Reson Med (2018) 79(5):2745–51. doi:10.1002/mrm.26918
26. Uecker M, Zhang S, Voit D, Karaus A, Merboldt K-D, Frahm J. Real-Time MRI at a Resolution of 20 Ms. NMR Biomed (2010) 23(8):986–94. doi:10.1002/nbm.1585
27. Hauptmann A, Arridge S, Lucka F, Muthurangu V, Steeden JA. Real‐Time Cardiovascular MR with Spatio‐Temporal Artifact Suppression Using Deep Learning-Proof of Concept in Congenital Heart Disease. Magn Reson Med (2019) 81(2):1143–56. doi:10.1002/mrm.27480
28. Schlemper J, Caballero J, Hajnal JV, Price AN, Rueckert D. A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction. IEEE Trans Med Imaging (2018) 37(2):491–503. doi:10.1109/tmi.2017.2760978
29. Küstner T, Fuin N, Hammernik K, Bustin A, Qi H, Hajhosseiny R, et al. CINENet: Deep Learning-Based 3D Cardiac CINE MRI Reconstruction with Multi-Coil Complex-Valued 4D Spatio-Temporal Convolutions. Sci Rep (2020) 10(1):13710. doi:10.1038/s41598-020-70551-8
30. El-Rewaidy H, Fahmy AS, Pashakhanloo F, Cai X, Kucukseymen S, Csecs I, et al. Multi-Domain Convolutional Neural Network (MD-CNN) for Radial Reconstruction of Dynamic Cardiac MRI. Magn Reson Med (2021) 85(3):1195–208. doi:10.1002/mrm.28485
31. Shen D, Ghosh S, Haji-Valizadeh H, Pathrose A, Schiffers F, Lee DC, et al. Rapid Reconstruction of Highly Undersampled, Non-Cartesian Real-Time Cine K-Space Data Using a Perceptual Complex Neural Network (PCNN). NMR Biomed (2021) 34(1):e4405. doi:10.1002/nbm.4405
32. Qin C, Schlemper J, Caballero J, Price AN, Hajnal JV, Rueckert D. Convolutional Recurrent Neural Networks for Dynamic MR Image Reconstruction. IEEE Trans Med Imaging (2019) 38(1):280–90. doi:10.1109/tmi.2018.2863670
33. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: N Navab, J Hornegger, WM Wells, and AF Frangi, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015: 18th international conference; October 5-9, 2015; Munich, Germany. Cham: Springer International Publishing (2015).
34. Matej S, Fessler JA, Kazantsev IG. Iterative Tomographic Image Reconstruction Using Fourier-Based Forward and Back-Projectors. IEEE Trans Med Imaging (2004) 23(4):401–12. doi:10.1109/tmi.2004.824233
35. Wundrak S, Paul J, Ulrici J, Hell E, Rasche V. A Small Surrogate for the Golden Angle in Time-Resolved Radial MRI Based on Generalized Fibonacci Sequences. IEEE Trans Med Imaging (2015) 34(6):1262–9. doi:10.1109/tmi.2014.2382572
36. Wundrak S, Paul J, Ulrici J, Hell E, Geibel M-A, Bernhardt P, et al. Golden Ratio Sparse MRI Using Tiny golden Angles. Magn Reson Med (2016) 75(6):2372–8. doi:10.1002/mrm.25831
37. Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: Sensitivity Encoding for Fast MRI. Magn Reson Med (1999) 42(5):952–62. doi:10.1002/(sici)1522-2594(199911)42:5<952::aid-mrm16>3.0.co;2-s
38. Walsh DO, Gmitro AF, Marcellin MW. Adaptive Reconstruction of Phased Array MR Imagery. Magn Reson Med (2000) 43(5):682–90. doi:10.1002/(sici)1522-2594(200005)43:5<682::aid-mrm10>3.0.co;2-g
39. Eo T, Jun Y, Kim T, Jang J, Lee HJ, Hwang D. KIKI ‐Net: Cross‐Domain Convolutional Neural Networks for Reconstructing Undersampled Magnetic Resonance Images. Magn Reson Med (2018) 80(5):2188–201. doi:10.1002/mrm.27201
40. Terpstra ML, Maspero M, d’Agata F, Stemkens B, Intven MPW, Lagendijk JJW, et al. Deep Learning-Based Image Reconstruction and Motion Estimation from Undersampled Radial K-Space for Real-Time MRI-Guided Radiotherapy. Phys Med Biol (2020) 65(15):155015. doi:10.1088/1361-6560/ab9358
41. Fahmy AS, Neisius U, Chan RH, Rowin EJ, Manning WJ, Maron MS, et al. Three-Dimensional Deep Convolutional Neural Networks for Automated Myocardial Scar Quantification in Hypertrophic Cardiomyopathy: A Multicenter Multivendor Study. Radiology (2019) 294(1):52–60. doi:10.1148/radiol.2019190737
42. Fahmy AS, El-Rewaidy H, Nezafat M, Nakamori S, Nezafat R. Automated Analysis of Cardiovascular Magnetic Resonance Myocardial Native T1 Mapping Images Using Fully Convolutional Neural Networks. J Cardiovasc Magn Reson (2019) 21(1):7. doi:10.1186/s12968-018-0516-1
43. El-Rewaidy H, Neisius U, Mancio J, Kucukseymen S, Rodriguez J, Paskavitz A, et al. Deep Complex Convolutional Network for Fast Reconstruction of 3D Late Gadolinium Enhancement Cardiac MRI. NMR Biomed (2020) 33(7):e4312. doi:10.1002/nbm.4312
44. Han Y, Sunwoo L, Ye JC. ${k}$ -Space Deep Learning for Accelerated MRI. IEEE Trans Med Imaging (2020) 39(2):377–86. doi:10.1109/tmi.2019.2927101
45. Lee D, Yoo J, Tak S, Ye JC. Deep Residual Learning for Accelerated MRI Using Magnitude and Phase Networks. IEEE Trans Biomed Eng (2018) 65(9):1985–95. doi:10.1109/tbme.2018.2821699
46. Hundley WG, Hamilton CA, Thomas MS, Herrington DM, Salido TB, Kitzman DW, et al. Utility of Fast Cine Magnetic Resonance Imaging and Display for the Detection of Myocardial Ischemia in Patients Not Well Suited for Second Harmonic Stress Echocardiography. Circulation (1999) 100(16):1697–702. doi:10.1161/01.cir.100.16.1697
47. Messroghli DR, Moon JC, Ferreira VM, Grosse-Wortmann L, He T, Kellman P, et al. Clinical Recommendations for Cardiovascular Magnetic Resonance Mapping of T1, T2, T2* and Extracellular Volume: A Consensus Statement by the Society for Cardiovascular Magnetic Resonance (SCMR) Endorsed by the European Association for Cardiovascular Imaging (EACVI). J Cardiovasc Magn Reson (2017) 19(1):75. doi:10.1186/s12968-017-0389-8
Keywords: real-time cine, artifact suppression, deep learning, complex, magnitude
Citation: Haji-Valizadeh H, Guo R, Kucukseymen S, Tuyen Y, Rodriguez J, Paskavitz A, Pierce P, Goddu B, Ngo LH and Nezafat R (2021) Comparison of Complex k-Space Data and Magnitude-Only for Training of Deep Learning–Based Artifact Suppression for Real-Time Cine MRI. Front. Phys. 9:684184. doi: 10.3389/fphy.2021.684184
Received: 22 March 2021; Accepted: 09 August 2021;
Published: 08 September 2021.
Edited by:
Oliver Diaz, University of Barcelona, SpainReviewed by:
Zahra Raisi-Estabragh, Queen Mary University of London, United KingdomCopyright © 2021 Haji-Valizadeh, Guo, Kucukseymen, Tuyen, Rodriguez, Paskavitz, Pierce, Goddu, Ngo and Nezafat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Reza Nezafat, cm5lemFmYXRAYmlkbWMuaGFydmFyZC5lZHU=
†Present Address: Hassan Haji-Valizadeh, Canon Medical Research USA Inc., Mayfield Village, OH, United States
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.