- 1Ulm University, Core Facility Small Animal Imaging, Ulm, Germany
- 2Department of Cardiology, The Second Hospital of Shandong University, Jinan, China
- 3Department of Internal Medicine II, Ulm University Medical Center, Ulm, Germany
Cardiovascular magnetic resonance (CMR) imaging has become an accurate and versatile imaging modality to visualize the cardiovascular system in normal or abnormal conditions. In preclinical research, small rodent animal models of human cardiovascular diseases are frequently used to investigate the basic underlying mechanism of normal and abnormal cardiac function and for monitoring the disease progression under therapy. Technical improvements have enabled the transfer of CMR to small animal research, and as such made this non-invasive technique available to provide insights into cardiac morphology, function, perfusion, and pathophysiology in small animal cardiac disease models. This article reviews the basic technical approaches to in vivo small animal magnetic resonance imaging and its variants for the most promising applications.
Introduction
Cardiovascular disease is considered the leading cause of death in the developed world, with high morbidity and mortality [1, 2]. In preclinical research, small rodent animal models of human cardiovascular disease are frequently used to investigate the basic underlying mechanism of normal and abnormal cardiac functions and for monitoring the disease progression under therapy [3]. Over the past decades, the use of small animal models has provided improved understanding of cardiac diseases [4, 5].
Translational research accounts for a large proportion of animal studies performed each year. Since the implementation of the 3 Rs principles (replacement, refinement, reduction) into the European Directive 2010/63/EU, medical imaging is highly recommended in translational research, because it can visualize the progression of the disease longitudinally often in a quantitative and non-invasive way. It thus has the potential to significantly reduce the number of experimental animals. Due to the high spatial and temporal resolutions, a versatile image contrast, and access to metabolic information, magnetic resonance imaging (MRI), and spectroscopy (MRS) have been proven to be promising diagnostic tools to monitor disease progression and response to treatment in small animal models. Its versatility, accuracy and high reproducibility has made cardiovascular magnetic resonance (CMR) imaging the non-invasive reference modality in preclinical research. MRI offers exceptional accuracy in the investigation of cardiac anatomy, perfusion, wall motion, and contractility, and its excellent soft-tissue contrast enables advanced myocardial tissue characterization [6–9].
However, the small size of the mouse heart [5–6 mm left ventricle (LV) diameter, ~0.2 g of heart weight] [10], high heart (about 250–600 beats per minute [bpm]), respiratory (about 60–160 cycles per minute [cpm]) rates, and fast systemic blood circulation times (4–5 heartbeats) impose substantial challenges for functional assessment by MRI. Over the past decades, major improvements in MRI methodology and instrumentation have been achieved, including rapidly switching high-performance gradient systems, ultra-high magnetic fields (>7 T), and non-Cartesian k-space encoding strategies such as spiral [11, 12] and radial trajectories [13, 14]. In combination with advanced mathematical concepts for image reconstruction [15–17] rapid data acquisition techniques, providing a high temporal resolution while preserving adequate spatial resolution and sufficient volume coverage, have been realized.
The main objective of this manuscript is to summarize the most important technical aspects of in vivo small animal CMR imaging and the required variants for specific applications. For the physiological, biological and pharmacological applications and perspective, please refer to comprehensive recent reviews [18–22]. With the included brief description of hardware demands and animal handling strategies, the reader will get familiar with the basic principle and challenges of small animal CMR imaging.
Technical Considerations and Methodologies
Hardware
Small animal MRI systems usually possess high field strength ranging from 4.7 up to 21 Tesla and strong gradient systems with bore diameters typically ranging between 10 and 40 cm. Higher field strengths provide an increased signal-to-noise ratio (SNR), thus enabling a higher spatial resolution. Disadvantages arise from changes in relaxation parameters, a higher chemical shift, a higher magnetic field inhomogeneity, susceptibility to artifacts, and increased specific absorption rate (SAR) [23], which often limits the translation of the results to clinical settings. Even though the lower SNR can partly be compensated with dedicated coils at 1.5T and 3T scanner [24, 25], and high-resolution mouse embryonic cardiac images were acquired at 17.6T [26], due to the high costs of ultra-high field instruments, field strengths between 7 and 11.7 T are most commonly used in small animal CMR imaging [7, 27, 28].
Further SNR improvement is achieved with dedicated radio frequency (RF) coils [29, 30]. Especially phased-array coils, additionally enabling parallel imaging [31, 32], have become standard in small animal CMR imaging. Further improvements have been reported by Wagenhaus et al. [33] and Dieringer et al. [34], who showed a 3–5-fold gain in SNR when comparing a 4-element room-temperature heart coil with a cryogenic coil.
As a further major progress, gradient systems that, depending on their inner diameters, provide amplitudes of up to 1,000 mT/m with slew rates of up to 9,000 T/m/s, have been introduced. In combination with efficient water-cooling high-duty cycles have been achieved, thus enabling rapid imaging at a high spatial resolution and a large field-of-view (FOV).
Animal Handling
Motion is one of the main challenges for in vivo imaging. In contrast to human studies, suitable anesthesia is generally required in small animal imaging to prevent animal motion. During scanning, the physiological status of the animal needs to be carefully monitored (e.g., ECG, respiration sensor, rectal temperature probe) and heating blankets are required to avoid temperature loss of the animal during anesthesia. General anesthesia is achieved using injectable or inhalational agents, or a combination of the two methods, to achieve the loss of consciousness, analgesia, suppression of reflex activity, and muscle relaxation [35]. An ideal anesthetic agent for animal CMR imaging should be easy to administer, and provide adequate and reproducible immobilization, preferably without changes in cardiac function and heart rate. However, since such agents are not available, the impact of the agent on cardiovascular physiology, especially depression of cardiac function and heart rate, respiratory function and induced hypothermia [36], has to be carefully considered.
Compared with inhalant agents, injectable anesthetics have several advantages. Their administration is more convenient to perform, and complex and expensive equipment such as precision vaporizers and specific breathing systems are not required. Due to its large safety threshold and compatibility with other drugs, ketamine is one of the most widely used anesthetics in animal research [37]. However, because of muscle rigidity, it is usually combined with xylazine, resulting in cardiac and respiratory depression. Berry et al. [38] reported deep sedation by subcutaneous injection of morphine and midazolam causing significantly less depression of heart rate and ejection fraction than imaging during general anesthesia with isoflurane in mice with normal cardiac function. Pachon et al. [39] suggested that ketamine was the least effective on the LV function and the heart rate, followed by Avertin, isoflurane (see below), and ketamine-xylazine combination. Other agents such as pentobarbitone, fentanyl/fluanisone, and urethane were shown to result in profound cardiovascular depression or prolonged recovery time and are not recommended in cardiac imaging. A general major drawback of injectable agents results from the limited anesthesia duration and adjustment possibilities during scanning.
Even though more complicated, in most cases inhalation anesthesia by halothanes is suggested. Here, the preferred approach is isoflurane inhalation [40]. Major advantages comprise short induction and fast recovery times, convenient adjustment during scanning, rather low hemodynamic depression, and flexible maintenance. In order to achieve high reproducible experiments, isoflurane (5% for induction, 1–1.5% for maintenance) in medical air (0.1 L/min) [7] is normally used.
Cine Imaging
For the quantification of cardiac function by CMR, rapid data acquisition techniques are required. Where for clinical applications mainly steady-state-free-procession (SSFP) sequences are applied for improving myocardium-blood contrast. In small animal CMR, fast low-angle shot (FLASH) gradient-echo [41] techniques are applied to avoid banding artifacts caused by off-resonances. Because of high heart and respiratory rates, real-time imaging of cardiac function is normally not possible and CMR imaging generally requires synchronization of the data acquisition to the cardiac and respiratory motion. In current practice, prospective triggering and retrospective/self-gated gating are used.
Prospective Trigger
For the prospective triggering, the ECG signals are simultaneously recorded for synchronization [42, 43]. ECG signals are normally drawn from two electrodes attached to one front and rear paw. After detecting the R wave, data acquisition is performed either at a specific phase of the heart (single-phase imaging) or continuously over multiple consecutive phases of the cardiac cycle (multi-phase or cine imaging, Figure 1A). Simultaneously, the respiration phase is measured with a balloon pressure sensor, and respiratory gating is additionally applied to avoid respiratory motion artifacts.
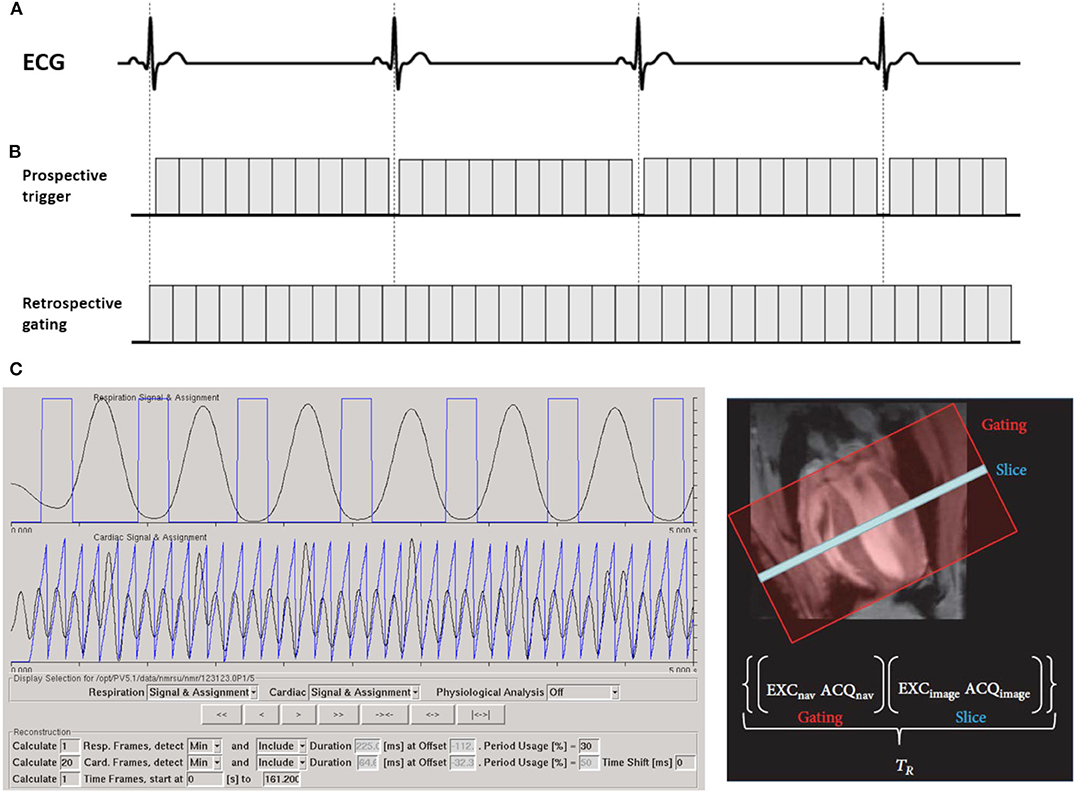
Figure 1. Strategies for the synchronization of cardiac cine MRI with the cardiac cycle. (A) In the prospective triggering scheme, the acquisition is synchronized with the ECG signal. After detecting the R wave, a suitable trigger delay is set due to application, then cardiac acquisitions can be performed at a specific phase of the heartbeat or to acquire multiple consecutive cardiac phases of the cardiac cycle. (B) In the retrospective gating scheme, data are acquired continuously while simultaneously recording the ECG and respiratory signals. (C) As an alternative, an additional navigator signal is acquired prior to each echo. Primary signals are calculated from navigator scans. The navigator signals are automatically analyzed and weighted according to their contribution to respiration and cardiac signals. The weighted physiological signals are retrieved (with demerging) and/or frequency filtered (with Fourier Filter) to create respiration and cardiac signal, which are used to assign each echo to its correct position in the cardiac and respiratory cycles. Retrospective gating signal is acquired with ParaVision 5.1 (IntraGate©, Bruker Biospin, Ettlingen, Germany). The partial figure is reproduced with permission from Zuo et al. [7].
However, radiofrequency pulses and gradient switching induce interferences in the ECG signal and may cause false triggers, which are more prominent at ultra-high magnetic fields. Further, the magneto-hydrodynamic effect is more prominent at higher field-strengths, often limiting the detectability of the R-peak. To avoid severe susceptibility artifacts, non-metallic materials like carbon wires have been introduced almost 30 years ago [44]. Recently, Choquet et al. [45] tested carbon wire electrodes in an 11.7 T magnetic field. Even though R-peaks were usually detectable, a clear distortion in the ECG signal still persisted. Other types of trigger devices such as optical fiber-based gating were developed, and excellent ECG signal quality has been reported [46]. Meanwhile, different filter techniques were developed to reduce ECG distortion during MRI examinations [47–49]. In principle, these filter techniques allow electrical ECG recordings for scan synchronization even at high field strengths. However, a principal limitation of prospective triggering is the required gap prior to the R-peak to ensure its proper detection. This causes loss of the end-diastolic phase or, in case only every second heartbeat is used for triggering, doubling of the scan times.
Retrospective Gating
In the clinical setting retrospective gating refers to simultaneous recording of the ECG signal with the data acquisition and subsequent reordering of the data into different cardiac phases according to the recorded ECG (Figure 1B). In small animal research, the term is related to self-gating techniques. Here, data synchronization is not based on physiological signals and no ECG recording is required. Instead, an additional navigator signal is acquired prior to each data acquisition (echo). After the scan, this navigator signal is analyzed and used to assign each echo to its correct position in the cardiac and respiratory cycles (Figure 1C).
Two of the most widely used retrospective gating sequences are self-gated FLASH (IgFLASH) and ultra-short echo time (IgUTE). The advantage of IgUTE compared to its Cartesian counterpart IgFLASH is the shorter achievable echo time (TE), which helps to reduce flow artifacts. Hoerr et al. [50] reported an IgUTE sequence with a minimal TE of 314 μs, clearly showing superior image quality especially for the delineation of small morphological structures like valves or papillary muscles. When compared with igFLASH, a substantial reduction of flow artifacts but maintained functional parameters were reported for IgUTE by Motaal et al. [13].
Compared with prospective triggering, retrospective approaches yielded similar global cardiac function data [43, 51]. As the data are acquired continuously, a flexible number of reconstructed cardiac phases and different temporal resolution can be achieved. However, the data acquired during certain periods of motion are not used for reconstruction. Thence, data oversampling is required to compensate for excluded data [52]. More importantly, the retrospective gating allows for steady-state acquisition, which is essential for mapping myocardial relaxation times [53, 54] or for quantifying image contrast as required in molecular CMR imaging with contrast agents [53, 55].
Real-Time Imaging
The self-gating techniques enable high-quality cardiac MRI with high reproducibilities [7]. However, the acquisition times in the minute range for a single slice still limit their applications in time critical settings such as pharmacological stress or first-pass perfusion imaging. Recently, based on technical progress in hardware and software, real-time concepts have been introduced and applied in CMR imaging. Real-time imaging refers to the rapid and continuous data acquisition followed by image reconstruction and visualization. To reduce acquisition times, dedicated real-time methods, including parallel imaging [56–58], k-t acceleration methods [59–61], and compressed sensing (CS) [15, 62, 63], have been suggested and initially evaluated [64] for the rapid and continuous acquisition of image series. Real-time methods utilize undersampling techniques, thus reducing the amount of acquired data for a single frame of an image series. Iterative reconstruction algorithms ensure image fidelity, e.g., by adding spatial and temporal regularization. Recent progress in real-time MRI results in high-quality images with high SNR, adequate spatial resolution and unsurpassed temporal resolution [65–68].
Dai et al. [69] first reported real-time cine MRI in mice with a single-shot echo-planar sequence with the Karhunen-Loeve transform (KLT) filter. Radial trajectories have shown favorable properties for real-time imaging by their intrinsic low motion artifact level. Further, due to the continuous recording of all spatial frequencies with every single spoke, undersampling results in almost incoherent artifacts, often showing no noticeable effect on the reconstructed images [70]. Winkelmann et al. [71] showed that a uniform profile distribution is guaranteed with a constant golden angle (111.246°) increment. The concept of Golden Angle (GA) angular spacing enabled data acquisition with optimal k-space coverage almost independent on the number of projections (Fibonacci sequence) and ensured incoherent undersampling artifacts [72]. Wech et al. [64] investigated the application of radial generalized autocalibrating partially parallel acquisitions (GRAPPA) with large golden angle (111.25°) real-time imaging in mice. However, only mid-ventricular slices were reported, and fully left-ventricular functions were not assessed. Its extension to the tiny golden angle (tyGA) [72] enabled the translation of the GA principle to higher field strength and provided even larger flexibility in the selection of the number of projections used for reconstruction of a single frame (generalized Fibonacci sequence). Our group investigates the feasibility of tyGA radial sparse MRI for real-time imaging of cardiac function in healthy mice [73] and a nexilin induced heart failure model [74]. Real-time cardiac tyGA radial sparse sense (tyGRASP) MRI in mice appears feasible with sufficient image quality for the quantification of global functional parameters. It enables a flexible number of projections for image reconstruction and thus offers the possibility for cardiac-phase dependent adjustment of the temporal resolution.
Accelerated Methods for CMR Imaging
MRI is an essential medical imaging tool with inherently slow data acquisition (Figure 2A), which imposes limitations to spatial and temporal resolution and volumetric coverage for dynamic cardiac imaging. The reduction of acquisition times can be achieved by incomplete sampling of k-space data. However, related aliasing artifacts often cause degrading of the diagnostic quality. During recent years, various techniques have been developed and applied to increased undersampling, while maintaining diagnostic image quality by advanced reconstruction techniques.
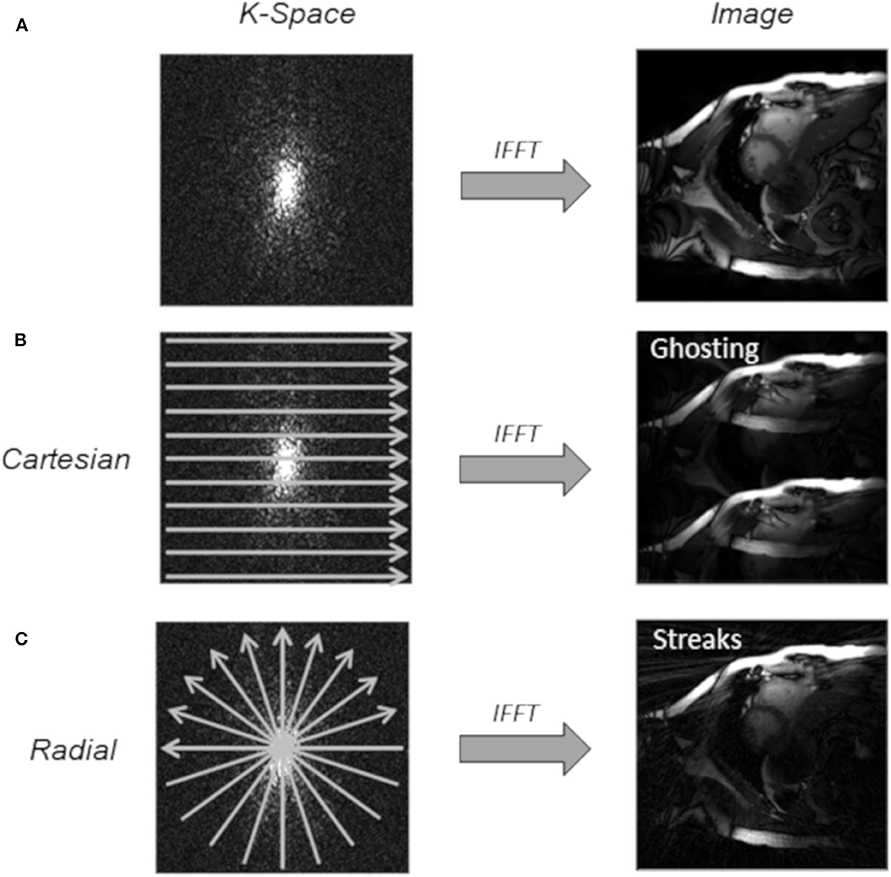
Figure 2. Undersampling techniques used for cardiac imaging. (A) Fully sampled k-space. (B) Regular Cartesian undersampling (2-fold), which results in discrete aliasing artifacts in the reconstructed image with an inverse fast Fourier transform (IFFT). (C) Uniform radial undersampling (2-fold) leads to low-intensity streaking artifacts.
Compressed Sensing
The mathematical foundation of compressed sensing (CS) was first introduced by Donoho [75] and Candès et al. [76] and translated to MRI by Lustig et al. [15]. CS aims to reconstruct signals and images from a reduced number of k-space samples not following the Nyquist criterion. As the MRI data are redundant and naturally compressible by sparse coding in some appropriate transform domain, CS has the potential to significantly reduce scan time.
A successful application of CS has three basic requirements [16]. First, the desired images have a sparse representation in a known transform domain, and they can be characterized by only a small number of non-zero coefficients. Especially for dynamic CMR imaging, the quasi-periodicity of the heartbeat causes a sparse temporal Fourier transform, and data are as such highly compressible. This enables higher acceleration for dynamic imaging than static imaging [77]. Second, the aliasing artifacts should be incoherent, i.e., that they must manifest as noise-like patterns. Regular undersampling patterns lead to fold over artifacts, known as coherent aliasing (Figure 2B). In the Cartesian sampling, some k-space lines can be pseudo-randomly omitted, resulting in more incoherent artifacts. Alternatively, due to its highly incoherent undersampling property, many non-Cartesian trajectories, such as radial trajectories [64, 73] (Figure 2C), are widely used in CS applications. Especially radial trajectories with golden or tiny golden angle angular increments have proven to be beneficial undersampling properties since almost any arbitrary number of projections provides uniform k-space coverage [78]. Motaal et al. [13] reported UTE Cine images in the rat model, and successfully reconstructed from up to 5-fold undersampled kt-space data utilizing a CS algorithm. Wech et al. [64] and Li et al. [73] investigated CS with golden angle radial real-time imaging in the mouse model. Third, a non-linear reconstruction algorithm should be applied to enforce sparsity constrains and data consistency, to suppress incoherent aliasing artifacts [64, 72].
Several studies have shown the feasibility and accuracy of CS to accelerate CMR imaging [79–81]. Further acceleration was reported by combining CS and parallel imaging [64, 82].
Parallel Imaging
Parallel imaging is a robust method to accelerate the acquisition of MRI data using a reduced number of k-space data simultaneously acquired with multiple receive coils. However, simply reducing the number of measured k-space data results in aliasing artifacts, mainly represented as well-known ghosting artifacts in Cartesian sampling and streaks in radial sampling [83]. The sensitivity encoding (SENSE) and GRAPPA are two principal methods in parallel imaging to correct for aliasing artifacts. The separate images reconstructed from the signals acquired with the different receive coil elements will have different relative intensities of the aliased component of the image. SENSE makes use of this property to separate the aliased components from the true structures in the image [84]. GRAPPA uses autocalibration signals and neighboring points in k-space to perform reconstruction of the missing portion in the k-space [85]. For the proper application of SENSE and GRAPPA, the signal at each point in the FOV has to be simultaneously recorded by several independent receive coil elements and careful alignment of the coil-array with the FOV is required. Theoretically, the maximum acceleration is only limited by the number of independent coils, but SNR limitations normally restrict the possible acceleration [83]. Wagenhaus et al. [33] investigated cardiac functional imaging of mice using a cryogenic quadrature RF coil with parallel imaging of an acceleration factor of 2. Ratering et al. [86] reported accelerated CMR of the mouse heart using self-gated parallel imaging strategies with a high acceleration factor up to 3.
k-t Acceleration
Cardiac dynamic images are sparse in an appropriate transform domain and exhibit correlations in k-space and time, enabling the recovery of missing data in undersampled data acquisitions. Based on this hypothesis, the k-t broad-use linear acquisition speed-up technique (k-t BLAST) and k-t sensitivity encoding (k-t SENSE) were developed to improve the performance of dynamic cardiac imaging. As a further generalization of the original method, k-t principal component analysis (k-t PCA) constrains the reconstruction using a standard data compression technique, thus enabling higher acceleration [87]. Signal correlations are learned from a small set of training data with low spatial but high temporal resolution, with subsequent reconstruction using temporal correlations [88].
Marshall et al. [89] investigated the k-t BLAST with an acceleration factor of three in healthy and myocardial infarction mice. Compared with the gradient echo cine sequence, the k-t BLAST scanning showed no significant differences. Makowski et al. [90] reported first-pass perfusion imaging with 10-fold undersampling in mice on a clinical 3 Tesla MR scanner using the k-t PCA technique.
Preclinical CMR Imaging Application
Cardiac Function
The heart can be considered as a central circulatory pump, generating the driving force to pump the blood through the vascular system. The visualization and quantification of the cardiac function are crucial, as many diseases have an impact on the performance of the heart.
Global ventricular performance parameters such as end diastolic/systolic volume (EDV/ESV), stroke volume (SV), ejection fraction (EF), and left ventricle mass (LVM) are widely used to evaluate cardiac function, which means the capability of the ventricles to eject blood into the great vessels. To derive accurate and reproducible volumetric quantification from a stack of parallel slices entirely covering the whole ventricle, the Simpson's rule is applied (Figure 3). The CMR assessment of the biventricular function is normally acquired with ECG-triggered or self-gated bright-blood contrast gradient-echo techniques [7, 43, 50]. Recently, the concept of real-time imaging was translated to preclinical research [64, 73, 91].
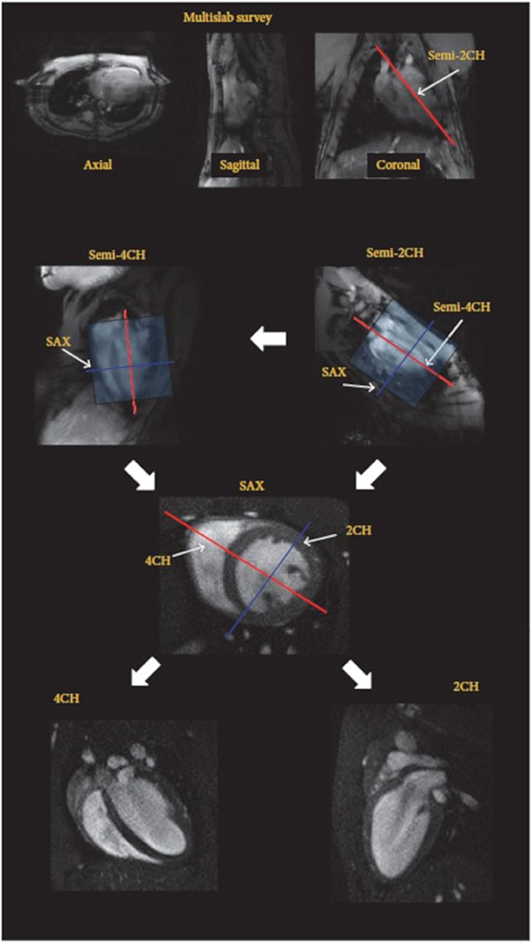
Figure 3. Imaging protocol for highly reproducible imaging of cardiac function and anatomy (2/4CH: 2/4-chamber, SAX: short axes orientations). After the acquisition of a fast survey in axial, sagittal, and coronal orientation a long axis (LAX) scans are acquired in semi-2 chamber (2CH) and semi-4 chamber (4CH) orientation, which are used for accurate planning of the short axis stack (SAX). The final 2CH and 4CH orientations are planned on the SAX stack. Reproduced with permission from Zuo et al. [7].
The assessment of systolic function in different mouse models has been investigated and the impact on, e.g., the EF, has been validated [92–94]. In contrast, the assessment of diastolic function in rodents is challenging. Normally the quantification is based on the filling rates at early left ventricular relaxation and late atrial contraction derived from time-volume curves of the left ventricle. Thus, a high temporal resolution acquisition (>60 frames per cardiac cycle) is highly recommended for reliable assessment. Coolen et al. [52] investigated diastolic dysfunction in diabetic mice with frame rates up to 80 frames per cardiac cycle. Later, a temporal resolution of 1 ms was reported by Roberts et al. [95] and the diastolic function was comparable to ultrasound analysis in normal mice.
Pharmacological Stress Imaging
Assessment of the left ventricular function under pharmacological stress is widely used in cardiovascular research to detect myocardial viability, ischemia or cardiac reserve and to determine the risk of subsequent cardiovascular event. However, due to the fast half-life and rapid metabolism of special drugs, a rapid quantification technique is highly required to visualize acute changes of cardiac morphology and function during pharmacological stress in the preclinical research.
Vasodilator and dobutamine are the main pharmacological agents used in stress CMR imaging [96]. By activating the adenosine receptors, the vasodilator agents can trigger coronary vasodilation and directly increase coronary flow [97]. Dobutamine is a synthetic catecholamine that primarily stimulates β1-adrenergic receptor and mildly stimulates α1, β2-adrenergic receptor, and augments myocardial contractility [98]. Wiesmann et al. [99] firstly used dobutamine stress MRI to reveal the loss of inotropic and lusitropic response in transgenic heart failure mice with myocardial infarction and diastolic dysfunction as an early sign of cardiac dysfunction. Since then, more and more studies using the dobutamine stress MRI to investigate cardiac function in the mouse model have been reported [93, 100, 101]. Different dobutamine doses and ways of administration were reported (4 to 40 μg/min/kg intravenous infusion [i.v.] or 1.5 μg/g body weight, intra-peritoneal bolus injection [i.p.]). To comply with the rapid imaging demands, real-time techniques combined with tyGA radial sparse MRI have been reported to characterize the acute changes of murine cardiac function from baseline to physiologically stress conditions in vivo without ECG and respiration synchronization [102].
Myocardial Strain
Myocardial strain is defined as the percentage change of myocardial length compared with the initial state in a certain direction by an internal or external force, and it is a well-validated parameter for evaluating myocardial performance. Several approaches are currently available for myocardial strain assessments.
Tagging
With the introduction of myocardial tagging in the late 1980s [103, 104], quantification of myocardial strain by CMR imaging became possible. Later, it was introduced by de Crespigny et al. [105] into rodent research. Tagging MRI applies a series of short RF saturation pulses to spatially modulate the longitudinal magnetization prior to the conventional image acquisition, which generates regional tags with stripes or grids on the myocardium (Figure 4). The measurement of myocardial strain is derived from the tag deformation over the cardiac cycle.
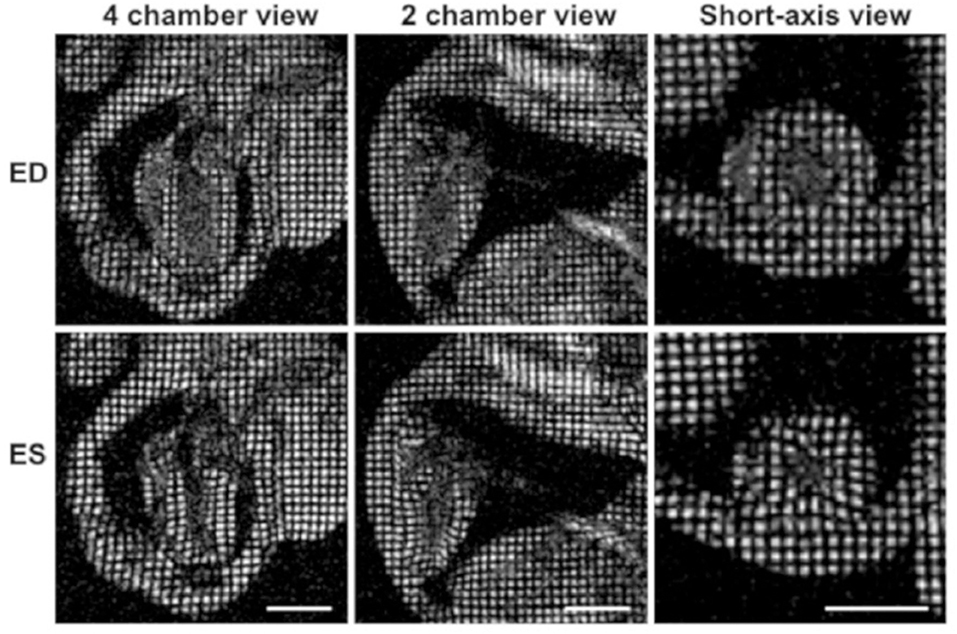
Figure 4. Myocardial tagging in the mouse. Four-chamber, two-chamber, and short-axis views of a mouse heart at the end-diastole are shown in the top row, and the end-systolic frames are depicted in the lower row. The movement of the tagged myocardial tissue can be tracked to calculate myocardial strain. Reproduced with permission from Price [106].
Initially, spatial modulation of the magnetization (SPAMM) was used to generate a one- or two-dimensional grid of saturated spins [104]. However, due to longitudinal relaxation, tagging contrast is weakened in later cardiac phases. A complementary SPAMM technique (CSPAMM) was introduced by Fischer et al. [107] to improve the grid contrast by using the difference image of two acquisitions with inverted tagging grid phase.
After the tagged image acquisition, quantitative analysis of the strain is achieved by a variety of fast and accurate analysis methods, such as detecting and tracking the tag lines in the images [108], assessing temporal and spatial changes in image intensities by optical flow [109] and harmonic phase (HARP) analysis [110].
Even though tagging MRI is the most validated CMR technique to assess myocardial strain, the requirement for extra image acquisition sequences, influence of tags fading (less relevant for high field-strength and heart rates), and time-consuming post-processing limits its application.
Tissue Phase Mapping
Velocity encoded tissue phase mapping (TPM) is a valuable technique to evaluate myocardial strain, strain rate and displacement with high spatial and temporal resolutions [111–113]. TPM is based on bipolar gradients to encode myocardial velocity and enables quantitative assessments of the myocardial velocity in three directions over the whole cardiac cycle. TPM is susceptible to eddy-currents and phase distortion induced by concomitant gradient fields [114, 115], and several approaches have been developed to minimize these errors [115–117]. However, TPM-encoded acquisitions are sensitive to flow- and motion-related artifacts and require long acquisition times. Several feasible methods have been developed to accelerate image acquisition and compensate for motion artifacts. Espe et al. [112] introduced in-plane rotation of the FOV in rats to reduce directional-dependent artifacts by acquiring each slice twice. Recently, McGinley et al. [118] investigated accelerated TPM imaging by applying compressed sensing in the myocardial infarction rat model.
Displacement Encoding With Stimulated Echoes
Displacement encoding with stimulated echoes (DENSE) was introduced for high-resolution myocardial displacement mapping via stimulated echoes with a bipolar gradient by Aletras et al. [119]. It has the distinct advantage of encoding tissue displacements into the pixel phase, thus encoding motion over long periods while maintaining high spatial resolution. However, due to the stimulated echoes, DENSE has the disadvantage of a relatively low SNR. Kim et al. [120] increased SNR of 15–34% by extracting a pair of subsampled DENSE images with uncorrelated noise from the CSPAMM image, and combined them during image reconstruction. After initially being implemented as single-frame imaging, DENSE was subsequently extended to cine imaging. In diet-induced obesity mice, high reproducibility was reported in the quantification of the LV function, including strain, torsion, and measures of synchrony [121]. Zhong et al. [122] investigated a 3D cine DENSE sequence with a spiral k-space trajectory in mice. Vandsburger et al. [123] combined DENSE with pharmacological stimulation to investigate the mechanics of endothelial nitric oxide synthase and neuronal nitric oxide synthase in modulating contractions and calcium influx in mice. DENSE has also been shown to be benefited from acceleration techniques, such as parallel imaging and compressed sensing [124, 125]. However, the feasibility of acceleration techniques in preclinical research needs further investigation.
Feature Tracking
Recently, CMR feature tracking (CMR-FT) was introduced for deriving global and regional myocardial strain [126]. It is mainly based on a block-matching approach. After manual defining endocardial and epicardial borders at end-diastole, in CMR-FT the borders are automatically tracked over the cardiac cycle by correlation of similar regions in the subsequent images (Figure 5). As CMR-FT is a promising novel method for the quantification of myocardial strain from routinely acquired cine CMR images without excessive post-processing times, it has the potential for a fast assessment of myocardial mechanics. Its clinical potential has recently been described [128–130], and excellent inter- and intra-observer agreement and high inter-study reproducibility were reported. However, to the best of our knowledge, only one preliminary study with limited temporal resolution of 15 phases per cardiac cycle has been reported in preclinical research [131]. There, feature tracking showed high reproducibility in left ventricle global circumferential and longitudinal strain in healthy mice, whereas reproducibility of radial strain was limited.
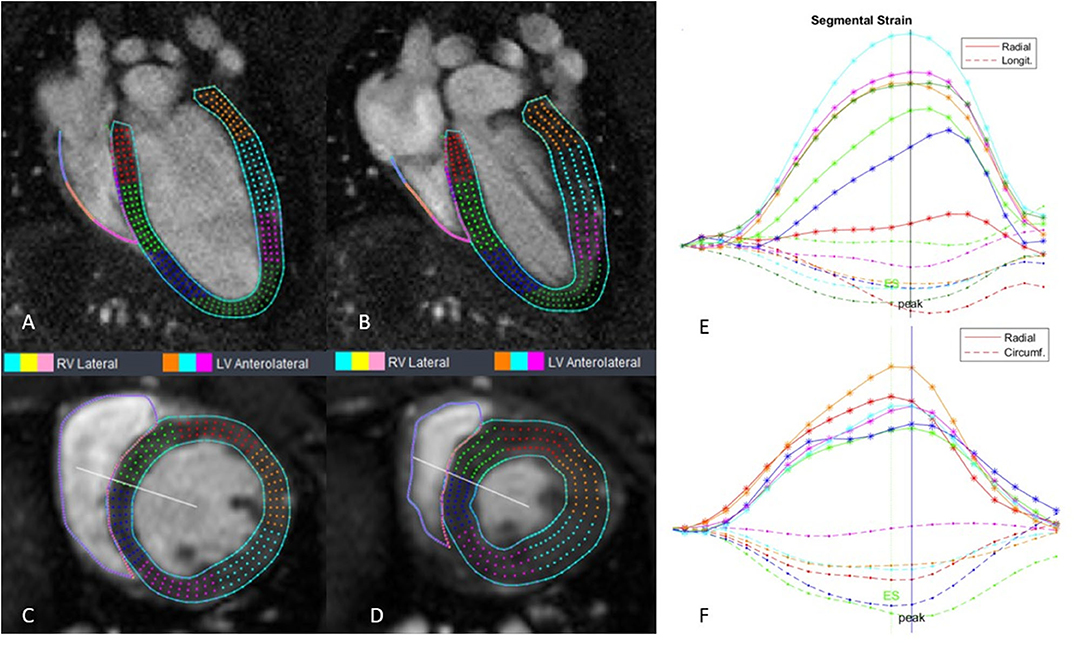
Figure 5. Cine CMR images with CMR-FT myocardial strain curves in mouse. (A) 4-chamber at end-diastole; (B) 4-chamber at end-systole; (C) short axis at the end-diastole; (D) short axis at the end-systole; (E) segmental strain for 4-chamber; (F) segmental strain for short axis. Stain analysis was done using Segment v3.0 R7732 [127].
Perfusion
Assessment of myocardial perfusion is considered to be a key parameter in the characterization of cardiac pathology, especially in ischemic heart disease or microvascular dysfunction. In normal conditions, a myocardial oxygen supply is balanced to the continuously changing myocardial oxygen demand. The imbalance of supply and consumption may result in myocardial ischemia. Currently, myocardial perfusion in rodents is typically assessed using arterial spin labeling (ASL) or first-pass perfusion imaging.
Arterial Spin Labeling
ASL is a valuable CMR technique utilizing arterial blood water protons as an endogenous diffusible tracer to non-invasively quantify regional myocardial blood flow without contrast agents [132]. RF pulses are applied to label arterial blood which then acts as endogenous tracer. ASL requires the subtraction of two images. One image is acquired after the labeled blood flow into the target tissue and the other one is acquired without labeling. The difference between both images can be utilized to calculate the tissue's blood perfusion. The signal difference depends on labeling decays with the time-constant equal to the blood T1 relaxation time. It can be made directly proportional to myocardial blood flow in units of ml-blood per g-tissue per minute. However, the contrast differences created by magnetic labeling of blood are inherently limited, which results in relatively low SNR. To compensate for low SNR, high field strength is beneficial.
ASL was firstly demonstrated in the rodent brain by Detre et al. [133]. Later, it was applied to the perfused excised rat heart by Williams et al. [134]. However, ASL suffers from relatively long acquisition times and it is sensitive to variations in heart and respiratory rates [135].
The flow-sensitive alternating inversion recovery (FAIR) combined with the Look-Locker readout scheme is the most widely used technique in ASL (Figure 6). Belle et al. [136] employed an LLFAIR-FLASH sequence to quantify myocardial blood flow (MBF) in rats. Due to blood flow, non-excited spins enter the detection slice, which leads to an increase in the relaxation rate. The relative difference in the apparent T1 relaxation times corresponding to selective and non-selective inversion is related to perfusion via a two-compartment tissue model. Kober et al. [137] improved MBF quantification using a respiration and ECG gated LLFAIR single-gradient echo technique in a mouse model. A nearly 10-fold increase in spatial resolution was achieved with respect to previous study in rats [138]. However, this led to a significantly increased acquisition time of about 25 min. Vandsburger et al. [139] investigated a cardio-respiratory gated ASL sequence using a fuzzy C-means algorithm to better cope with respiratory motion and heart rate variations in a myocardial infarction mice model. Abeykoon et al. [140] introduced an ASL method based on the signal intensity of flow sensitized CMR to shorten scan time to 2–4 min. In order to benefit from sensitivity advantages of continuous ASL, Troalen et al. [141] proposed cine-ASL, which is based on an ECG-gated steady-pulsed labeling approach combined with simultaneous readout over the cardia cycle. The cine-ASL led to shorter acquisition time than the LLFAIR technique while preserving spatial resolution and robustness with respect to cardiac motion.
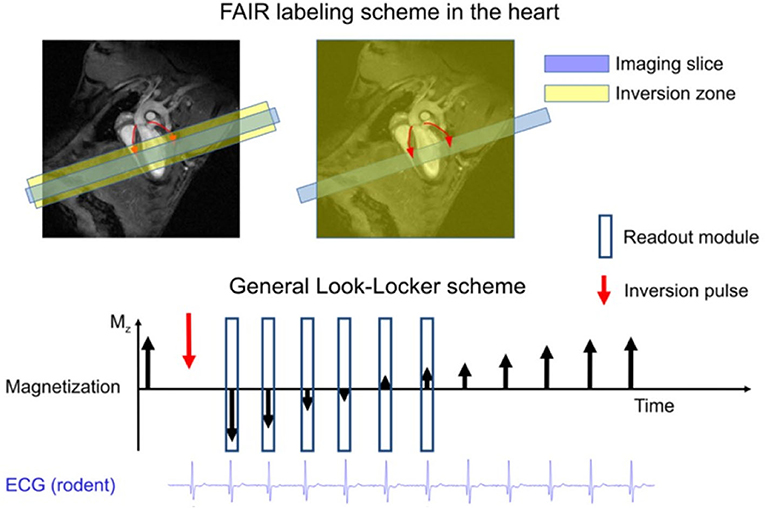
Figure 6. The FAIR labeling scheme (upper part) consists of two inversion recovery measurements. In the first measurement the labeled zone is only around the imaging slice, and in the second measurement the inversion is global. FAIR is the most commonly used ASL technique in the heart. The timing of the inversion and that of the readout modules should be ECG-gated and occur in the same cardiac phase. The Look-Locker inversion-recovery readout (lower part) has been used mainly in the rodent heart, where the rapid heart rate permits dense sampling of the magnetization recovery curve. Reproduced with permission from Kober et al. [132].
First-Pass Perfusion
The basic principle of first-pass perfusion imaging involves intravenous injection of a bolus of a suitable contrast agent (CA), with subsequent monitoring of the passage of the CA through the heart. However, the high heart rate (400–600 bpm) and fast systemic blood circulation time limit its application in rodents. Recently, the major progress in data acquisition acceleration has rendered small animal first-pass perfusion imaging feasible.
Makowski et al. [90] firstly proposed first-pass perfusion imaging in mice using a k-t SENSE technique with an acceleration factor of 10 on a clinical 3T scanner. Later, Coolen et al. [142] introduced a segmented ECG-triggered acquisition combined with parallel imaging acceleration to capture the first pass of contrast agent in healthy and myocardial infarction mice. A temporal resolution of one image per three heartbeats was reached. The same group further investigated the quantification of regional perfusion values using a dual-bolus approach in combination with a Fermi-constrained deconvolution model [143]. Later they applied this dual-bolus approach to test the feasibility of first-pass perfusion in pressure overload induced hypertrophy and heart failure mouse models after transverse aortic constriction [144]. The hypertrophic mice revealed reduced myocardial perfusion proportional to LV volume and mass, and a related decrease in LV ejection fraction. Naresh et al. [28] used k-t undersampled dual-contrast first-pass MRI with motion-compensated compressed sensing reconstruction to study myocardial blood flow in a high-fat diet mouse model. They also investigated the repeatability and variability of first-pass perfusion imaging and ASL. They concluded that first-pass MRI shows better repeatability and variability in low MBF conditions such as myocardial infarction. Due to better image quality and lower user variability, ASL is more suitable at high MBF. Recently, the tyGRASP sequence with block-wise cardiac synchronization was reported for first-pass perfusion imaging in nexilin knock-out mice [74]. During each cardiac cycle, a single block of projections with acquisition duration of t = 31.5ms was acquired. For suppression of the background signal and coping with arrhythmic cycles, a non-selective saturation pulse was additionally applied prior to each acquisition block with a saturation recovery time TSAT of 60 ms. The trigger delay was chosen so that the acquisition was performed during end-diastole to further minimize the motion artifacts (Figure 7). A temporal resolution of one image per cardiac cycle was achieved. Thus, an improved temporal fidelity of the inflow and washout curves was shown.
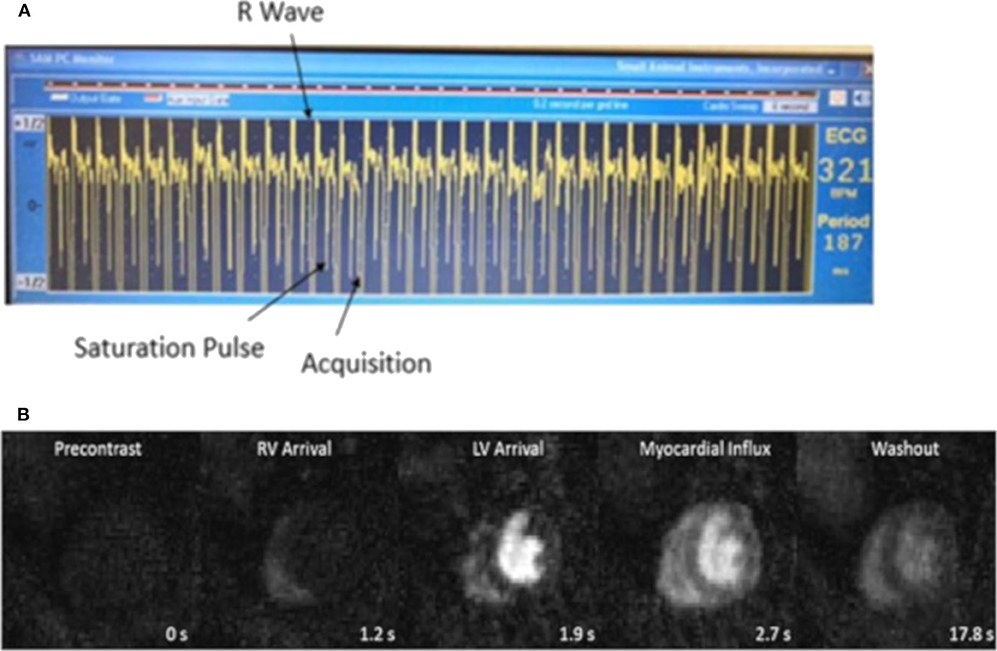
Figure 7. (A) ECG signal during scan. According to the heart rate, a trigger delay was set after the detection of the R wave, then the blood and myocardial signal were approximately nulled by a non-selective saturation pulse. The acquisition was performed during end-diastole. (B) Time series of myocardial first-pass perfusion images of a nexilin wild-type mouse. From left to right: precontrast, RV arrival, LV arrival, myocardial influx, and washout.
Tissue Characterization
Because of its ability to provide superior soft tissues contrast in exquisite detail, MRI has exceeded other imaging modalities in its multi-parametric capabilities for a comprehensive myocardial tissue characterization. It can provide superior and well-validated biomarkers of important pathophysiological processes in cardiac diseases based on intrinsic relaxation properties T1, T2, and T2* with or without contrast agent. Tissue contrast is tailored by adjusting acquisition parameters such as flip angle, TE, repetition time (TR), and inversion delay (TI). Mapping techniques can further provide direct pixel-by-pixel quantitative myocardial tissue characterization to depict small variations of relaxation properties and to highlight tissue pathology.
T1 Mapping
The T1 relaxation time is the longitudinal relaxation time, describing the return of the magnetization to thermodynamic equilibrium after excitation. The native T1 value is a tissue specific constant which changes in some pathologic conditions, such as diffuse myocardial fibrosis, hemorrhage, edema, inflammation, and acute infarction [145]. Thus, the quantitation of T1 value can be used to distinguish different soft tissues and pathologies. The general principle for T1 mapping is to acquire multiple images at different inversion times in order to assess the T1 recovery curve.
Several T1 mapping methods have been proposed in preclinical research. One of the most widely used methods for measuring myocardial T1 is a single slice inversion recovery Look-Locker technique [146–148]. With this technique, image datasets are acquired repeatedly after an inversion pulse to create multiple images along the recovery curve. However, this method has several drawbacks. The long TR results in relatively long total acquisition times. Poor ECG signals and variations in the heart rate cause inversion times to differ for subsequent acquisition thus causing k-space inconsistencies. In-plane and through-plane motion between excitations reduce accuracy [54]. An ECG-triggered saturation-recovery Look-Locker (SRLL) method has been proposed by Li et al. [149] to acquire a single-slice T1 map in about 3 min. By combining the SRLL method with a modified model-based compressed sensing method, a 50% reduction in imaging time was achieved for the in vivo T1 mapping of the mouse heart [150]. Later, Jiang et al. [151] further extended the method to multi-slice T1 mapping (MRLL) allowing for more coverage without increasing scan time. However, the SRLL sequence yields relatively low SNR and a lower accuracy compared with the inversion recovery. The single shot 2D modified Look-Locker (MOLLI) technique has shown high reproducibility and high SNR in humans [152]. However, it is challenging in mice due to the high heart rates and requirements on spatial resolution. Recently, Nezafat et al. [153] proposed a multi-shot 2D modified Look-Locker sequence for high-resolution T1 mapping in mice at a 3T MRI clinical scanner. A further method for myocardial T1 mapping in mice has been proposed by Coolen et al. [54] using a 3D intra-gate FLASH sequence in combination with a variable flip angle DESPOT1 (driven equilibrium single-pulse observation of T1) analysis. With this protocol, 3D T1 maps of the heart ventricles could be obtained in 20 min with a sufficient spatial resolution (Figure 8). Castets et al. [154] accelerated 3D T1 mapping measurements using spiral encoding with a higher spatial resolution (208 × 208 × 315 μm3) in a 10–12 min acquisition time.
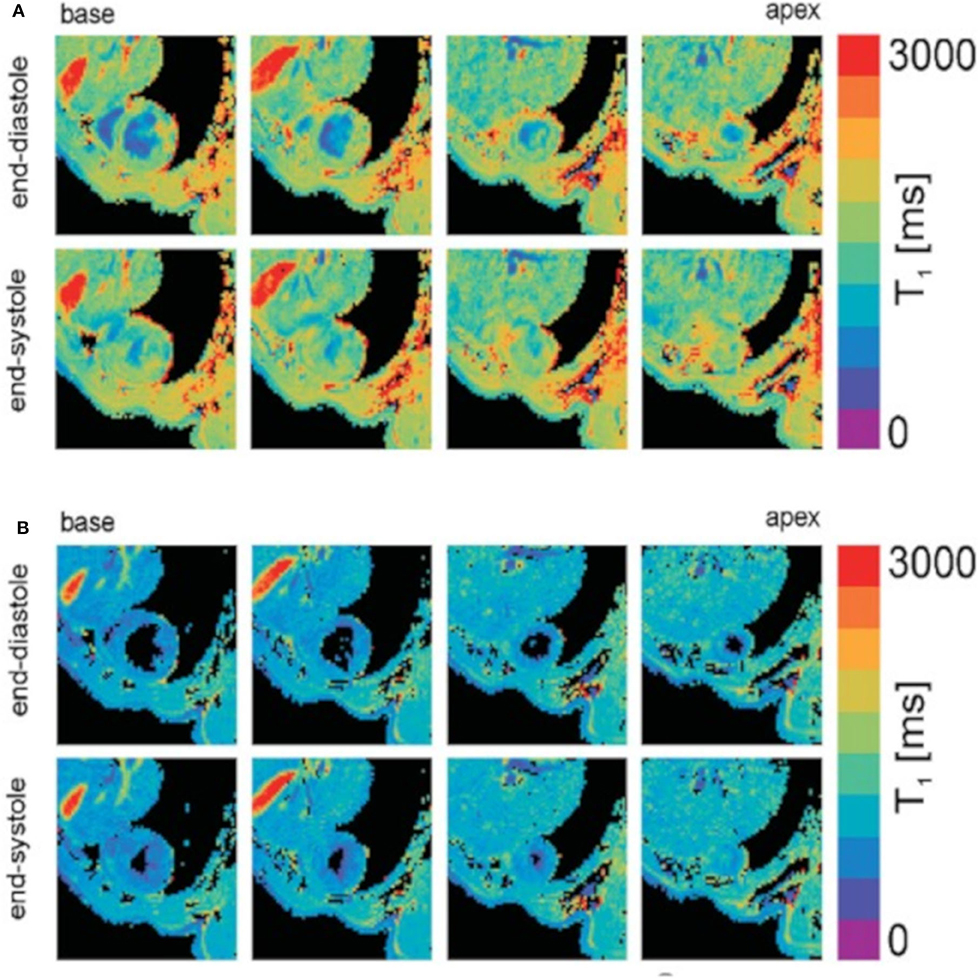
Figure 8. T1 mapping of the mouse heart resulting from DESPOT1 analysis of both black-blood (A) and bright-blood (B) data. T1 maps are shown for four different slices covering the base to apex and for both the end-diastolic and end-systolic cardiac phases. Reproduced with permission from Coolen et al. [54].
A promising application for T1 mapping is the quantification of myocardial extracellular volume (ECV) fraction. ECV contains the interstitial and intravascular spaces. Many disease processes affecting the myocardium can be understood through ECV changes, such as diffuse or interstitial myocardial fibrosis. ECV can be measured by combining native and post-contrast T1 mapping of blood and myocardium. Neilan et al. [155] applied and validated a Look-Locker FLASH sequence to quantify ECV in juvenile and aged mice.
T2 and T2* Mapping
The T2 relaxation time, also referred to as spin-spin or transverse relaxation time, is describing the tissue-specific decay of the transversal magnetization after excitation. Myocardial edema is the main pathology responsible for changes in the T2 value, and it was first demonstrated in a canine model of acute myocardial infarction (AMI) by Higgins et al. [156].
Two variants of T2 mapping techniques are usually used in CMR imaging: dark blood turbo spin-echo (TSE), and bright blood T2 preparation. Schneider et al. [147] mapped T2 in mice by acquiring the spin echo data with echo times ranging from 3.7 to 24 ms. Later, Bun et al. [157] performed T2 measurements with a similar sequence to quantify myocardial fibrosis in diabetic mice at 11.75 T. However, the TSE-based T2 mapping suffered from ghosting artifacts caused by blood flow and signal loss due to through-plane motion [158]. The T2 preparation-based method is less prone to TSE-associated artifacts [159]. It mainly contains a T2 preparation module and a rapid imaging sequence such as FLASH. The T2-preparation module contains non-selective 90 and 180° pulses to create spin-spin relaxation followed by a −90° restore pulse. After the restore pulse, the longitudinal magnetization depends on the tissue T2 value. Beyers et al. [55] proposed a T2 preparation module containing a Carr-Purcell-Meiboom-Grill and Malcolm Levitt (CPMG-MLEV) weighted series of composite 180° pulses, followed by a multi-slice gradient echo readout. In this way, the resulting T2 values are less sensitive to effects of B0 and B1 inhomogeneities. Coolen et al. [160] extended this preparation module with fast steady-state-free-precession (SSFP, FISP) readout. However, the total acquisition time for one T2 measurement with three slices was about 30 min, which limited its application in fast dynamic contrast agent enhanced MRI studies. Chen et al. [161] performed rapid T2 mapping of the mouse heart using the CPMG sequence and compressed sensing reconstruction, which allowed T2 quantification at a temporal resolution of 1 min per slice (Figure 9).
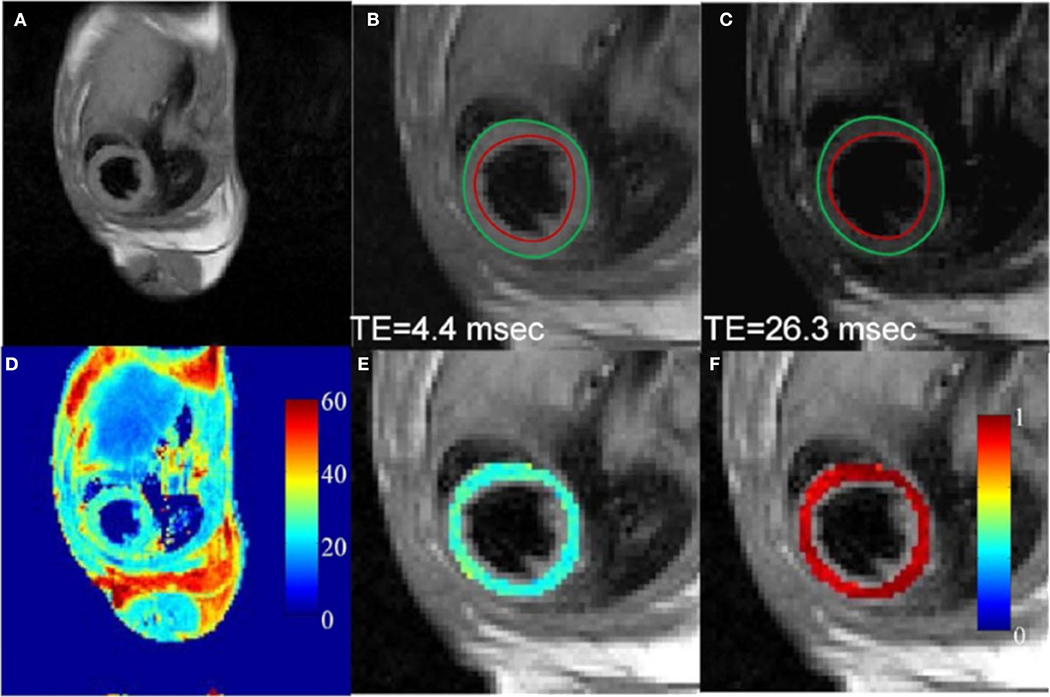
Figure 9. In vivo T2 mapping of a mouse heart using a fully-sampled dataset. (A) Full FOV image of the first echo (TE: 4.4 msec). (B,C) Enlarged images of the heart from the first and sixth echo, respectively. Red and green circles represent endocardial and epicardial contours drawn from the first echo image, respectively. (D) The corresponding T2 map for the whole image. (E) Enlarged T2 map for the mouse heart. (F) Map of squared correlation coefficient for the exponential T2 fitting. Reproduced with permission from Chen et al. [161].
T2* relaxation refers to decay of transverse magnetization caused by a combination of spin-spin relaxation and magnetic field inhomogeneity. Gradient-echo MRI with T2*-based contrast can be used to depict different lesions in diseased mouse heart. T2* cardiac MRI has been utilized to evaluate myocardial iron overload and collagen deposits. T2* is reduced in iron- and collagen-laden tissues due to increased magnetic field inhomogeneity. Jackson et al. [162] measured T2* relaxation using two multi-echo gradient echo sequences with 15 echo times in the range of 0.9–14.9 ms at 1 ms intervals in a mouse model of β-thalassemia. Over 10-fold decreased myocardial T2* (0.7 ± 0.2 ms) was observed in the iron-loaded thalassemia group, which was consistent with histological results. van Nierop et al. [163] reported reduced T2* value in myocardial infarction (MI) and transverse aortic constriction (TAC) mice using an ECG-triggered T2*-weighted 3D center-out radial sequence with TE ranging from 21 μs to 4 ms. T2*-shortening contrast agents, such as iron oxide nanoparticles (IONPs), have been used to exploit T2* contrast in mouse cardiovascular system [164, 165]. Sosnovik et al. [164] reported an iron oxide–based particle targeted at apoptotic cells in an ischemia/reperfusion injury mice model. Significantly decreased signal intensity was observed in T2*-weighted FLASH images. Zhou et al. [166] used the Sweep Imaging with Fourier Transformation (SWIFT) technique to visualize iron oxide particle labeled stem cells in the rat heart. Due to its zero echo-time properties, SWIFT images showed reduced blooming artifacts compared to gradient-echo images, and enhancement of off-resonance signals relative to the background. However, a major remaining challenge for IONPs application is to distinguish regions of signal void due to IONPs from those due to low signal tissues or susceptibility artifacts. Even though several bright-iron methods such as off-resonance techniques [167], gradient-compensation techniques [168], and post-processing methods to identify IONP-introduced magnetic field inhomogeneities [169] have been introduced, IONP-induced off-resonances can hardly be distinguished from other sources. This yields a rather low specificity of the IONP approach. Similar to SWIFT, ultrashort echo time (UTE) techniques have been used for providing endogenous T2* contrast [163, 170].
Contrast Enhanced MRI
Contrast enhanced MRI (CE-MRI) is a widely used imaging technique to investigate microvascular structure and function by injecting a contrast agent. MR contrast agents work by modifying the tissue relaxation properties and thus directly affect the image contrast. This effect is known as relaxivity and enables better visualization of tissues in which the agent accumulates. The contrast agents can be classified as T1-weighted (i.e., gadolinium chelates, manganese chelates) or T2*-weighted (i.e., iron oxide particles). Here, we mainly discuss T1-weighted CE-MRI.
Late Gadolinium Enhancement MRI
Late gadolinium enhancement (LGE) CMR is a non-invasive reference standard for investigating myocardial viability. In AMI, cellular necrosis, lysis, and edema are the main pathological changes. In chronic infarcted tissues, fibrous scar tissues with increased extracellular space form. Compared with healthy tissues, the Gd-agent was slowly washed in and washed out in pathological tissues with expanded extracellular space. Analysis of the CA dynamics allows to distinguish between healthy and high-risk tissues and myocardial scars [171].
Kim et al. [172] initially applied LGE to study myocardial viability in a rabbit model. A clear difference in contrast kinetics between normal tissue, infarct rim, and infarct core regions could be observed. Yang et al. [173] validated the LGE technique for accurate assessment of infarct size in MI mice. After intravenously injecting a 0.3–0.6 mmol/kg bolus of Gd-DTPA, CA-enhanced images of the entire heart were acquired in 15 to 30 min. Great agreements were achieved for infarct size between LGE images and 2,3,5-triphenyl tetrazolium chloride (TTC) staining. The results were in excellent concordance with many other studies [174–176], which clearly demonstrated LGE cardiac imaging to be a reference standard to follow up myocardial viability in vivo.
Two main methods for LGE imaging are the inversion recovery (IR) fast gradient echo sequence and the T1-weighted cine FLASH sequence. Due to the ability to null the signal of remote myocardium, IR shows better contrast between the infarcted and non-infarcted myocardium and is widely used in humans. In preclinical research, Price et al. [177] applied LGE in small animals using a multi-slice IR gradient-echo sequence in combination with a Look-Locker sequence for assessing the optimal inversion point to null the signal from healthy myocardium (300–450 ms for rats, around 330 ms for mice, Figure 10). Thomas et al. [174] and Protti et al. [178] compared IR LGE and T1-weighted cine FLASH LGE imaging at 4.7 and 7 T. Both protocols produced reliable results for the assessment of infarction size. Cine FLASH was found to be a more robust, faster and less user dependent method for visualizing infarct size and recommended as the more promising technique in small rodents.
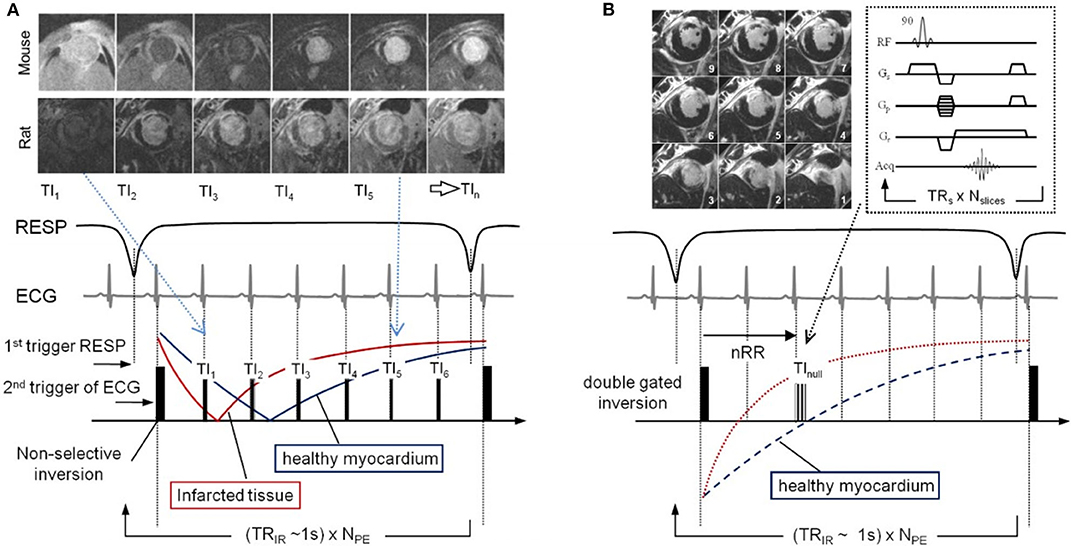
Figure 10. Pulse sequence diagrams for LGE imaging. (A) Look-Locker pulse sequence and gating strategy used to acquire multiple inversion time (TI). Images from TI1–TI6 time points are shown for both mouse and rat, where the indices are related to the number of R-wave triggers given the double gated inversion pulse. (B) The LGE IR sequence diagram follows the same gating and acquisition scheme but acquires a 90° flip angle multislice GRE stack at the nearest TI value to null healthy myocardium. TRs is the time between slice acquisition pulses, TRIR is the time between inversion pulses, NPE is the number of phase-encoding steps, and Nslices is the number of slices. Reproduced with permission from Price et al. [177].
Manganese-Enhanced MRI (MEMRI)
Unlike gadolinium, which only allows extracellular space imaging for assessment of myocardial viability, manganese provides T1-weighted intracellular contrast through calcium handling. During myocardial contraction, Ca2+ enters cardiomyocytes primarily by conduction through voltage-gated L-type calcium channel that causes increase in cytosolic Ca2+ concentration, which then binds to troponin C and activates myocardial contraction (excitation-contraction coupling) [179]. After systolic contraction, Ca2+ is actively transported into the sarcoendoplasmic reticulum (SR) by Ca-ATPase and excreted from the cell through the sarcolemmal Na+-Ca2+ exchanger [179]. Alterations in calcium handling may impair the Ca2+ cycling between the extracellular space, cytosol, and SR, resulting in systolic or diastolic dysfunction. Manganese is a T1 shortening compound taken up through voltage-gated calcium channels into cells with active calcium handling. It has the ability to measure Ca2+ channel activity and myocardial viability, and has been validated in various clinical [180–182] and preclinical [183–185] studies. However, its application in clinical routine was limited by its potential toxicity causing myocardial depression. Manganese formulations using chelation or linkage to a calcium compound [186] were developed. Spath et al. [184] investigated manganese-based contrast media in healthy and infarcted rat models (Figure 11). The investigated manganese agents resulted in calcium channel-dependent myocardial T1 shortening. A good agreement between infarct size on MEMRI T1 mapping and Masson's trichrome (MTC) staining (bias 1.4, 95% CI −14.8 to 17.1, p > 0.05) was reported. In contrast, standard gadolinium delayed enhancement MRI (DEMRI) with the inversion recovery technique overestimated the infarct size (bias 11.4, 95% CI −9.1 to 31.8, p = 0.0002), as did DEMRI T1 mapping (bias 8.2, 95% CI −10.7 to 27.2, p = 0.008). Recently, Toma et al. [183] proposed a dual contrast MEMRI and DEMRI technique to evaluate the physiologically unstable peri-infarct region and to track the therapeutic effects of telmisartan on the injured myocardium longitudinally with attenuation of the peri-infarct region.
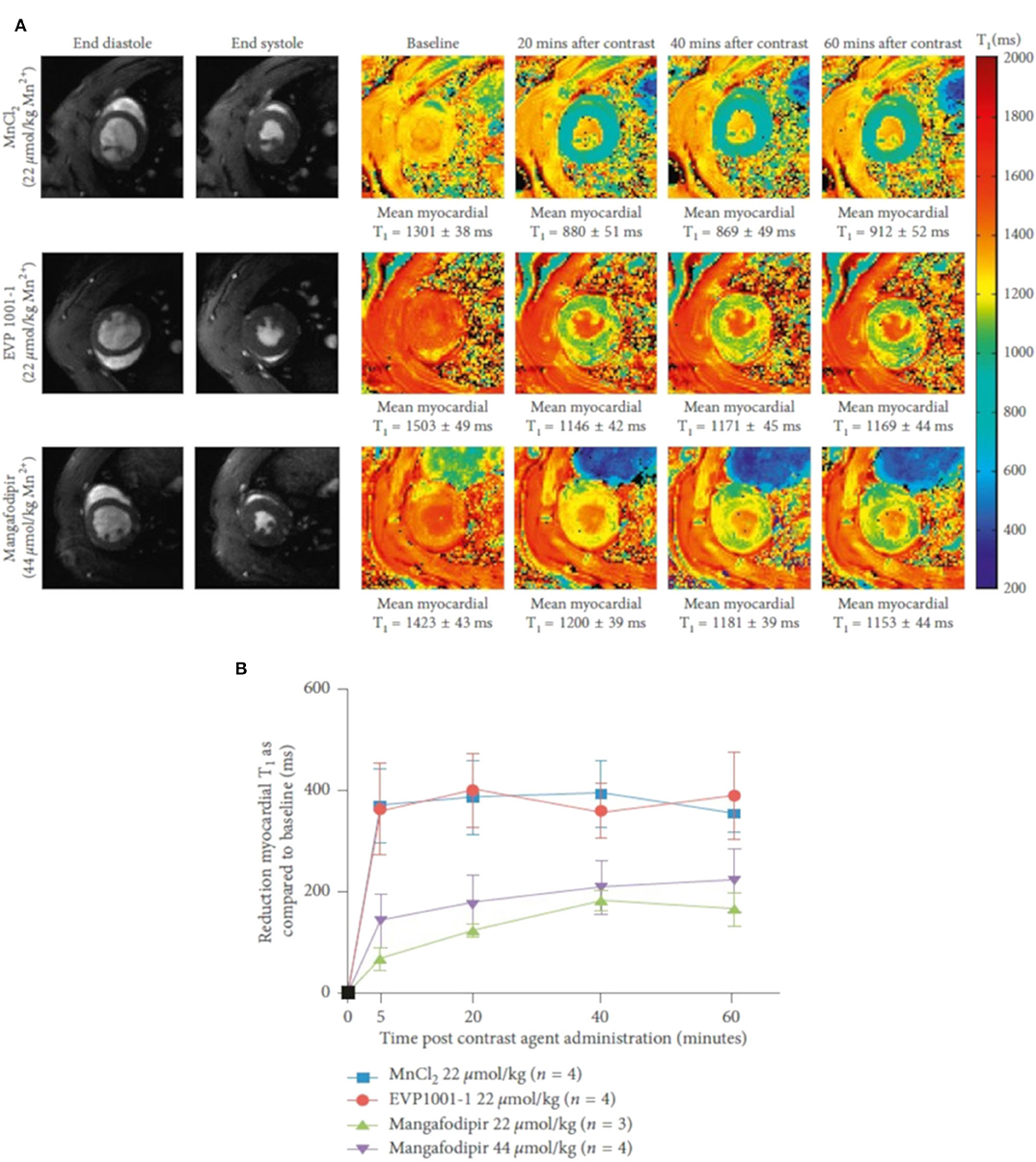
Figure 11. T1 shortening with different manganese contrast media over time. (A) Normalized T1 maps acquired subsequent to infusion of MnCl2, EVP1001-1, and mangafodipir at 20 min intervals up to 60 min. A clear decreased myocardial T1 value could be observed in all groups. (B) Reduction in mean left ventricular T1 values over 60 min with MnCl2, EVP1001-1 and different concentrations of mangafodipir. Two-way ANOVA confirmed a dependence of mean myocardial T1 shortening between each of the contrast agents (P < 0.0001). Reproduced with permission from Spath et al. [184].
Diffusion Tensor Imaging
The myocardial fiber anatomy underlies the mechanical and electrical properties of the heart [187]. Within the normal LV, the myofibers follow left-handed helices in the epicardium, and transit smoothly through a circumferential orientation at the mesocardium to right-handed helices in the endocardium. A feasible and accurate technique to characterize and identify fiber structure changes can contribute to elucidating the complex cardiac structure-function relationships. MR diffusion tensor imaging (DTI) has been validated as a valuable tool to obtain non-invasive measures of myocardial microstructure in both clinical and preclinical studies [187–189]. Several quantitative parameters derived from DTI, such as mean diffusivity (MD) and fractional anisotropy (FA), helix angle (HA) and second eigenvector angulation (E2A), can be used to describe myocardial microstructural organization.
After first developed in 1990s [190], DTI has been widely used in brain imaging to study the spatial organization of white matter fiber tracts [191, 192]. The preclinical application of cardiac DTI is more challenging, due to cardiac and respiratory motion, the diffusion signal attenuation, and a short T2 relaxation time causing low SNR condition. Further, high-resolution DTI of the mouse heart is more complicated due to a low degree of diffusion anisotropy in the tissue [188]. With the developments of pulse sequence design and gradient systems, cardiac DTI has become feasible for in vitro and in vivo applications.
In the beginning, cardiac DTI was mainly performed on isolated hearts. Scollan et al. [193] introduced two-dimensional stimulated echo acquisition mode (STEAM) in perfused rabbit hearts using standard and fast spin-echo pulse sequence with half-sine diffusion gradients in six non-collinear directions. The inclination angle derived from both protocols showed high agreement with histological results. Jiang et al. [188] firstly extended a similar protocol to acquire 3D diffusion-weighted images of 12 directions in a fixed mouse heart. High spatial resolution (isotropic 100 μm) images were acquired in 9.1 h and allowed the quantification of the myocardial structure at more than 170,000 locations throughout the mouse heart, which is about 10 times better than histological studies. The FA value of 0.27 ± 0.06 and the mean diffusion tensor eigenvalues of 0.75 ± 0.13, 0.60 ± 0.13, and 0.51 ± 0.13 x 10−3 mm2/s are in good agreement with other studies [193, 194]. Huang et al. [195] firstly investigated in vivo DTI-tractography of the mouse heart to follow myocardial microstructural changes in ischemia/reperfusion mouse models. It showed increased MD and FA in acute ischemia (24 h after injury), but decreased values during myocardial healing (2–3 weeks after reperfusion) (Figure 12). Later, the same group applied 3D DTI-tractography in vivo to study ischemic myocardium and assessed the cell therapy effect [196]. With the velocity-compensated Stejskal-Tanner diffusion sequence, they were able to obtain high-resolution 3D reconstructions of the myofibrillar tracts in the mouse heart. The derived parameters clearly showed that DTI could non-invasively reveal the microstructural features of the myocardium.
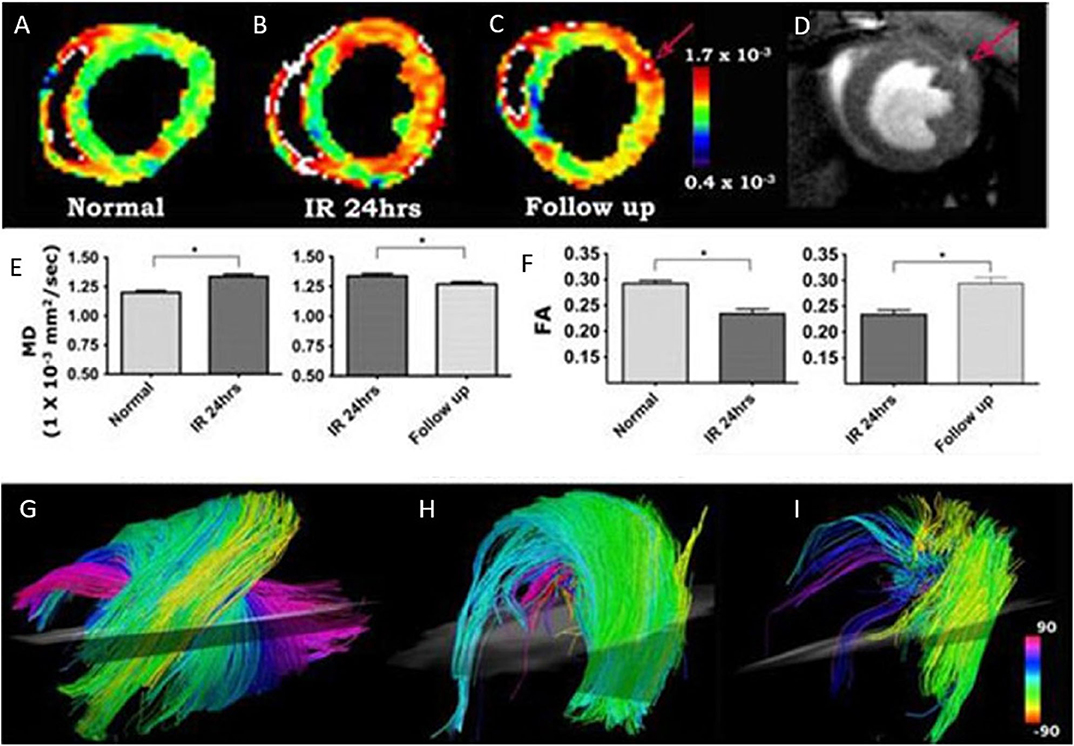
Figure 12. (A–I) In vivo DTI-tractography of mouse heart at baseline, 24 h after injury and 2–3 weeks after ischemia-reperfusion. The mean diffusivity (MD) maps clearly showed that MD in the injured anterolateral wall increased acutely (24 h after injury) and subsequently returned toward baseline as the myocardium healed (A–C,E). Fractional anisotropy (FA) decreased acutely and returned to baseline 2–3 weeks after injury (F). (C,D) The left coronary artery, when present in the image, can be used for internal standardization and control. MD in the vessel and its branches (red arrow, C,D) will be significantly higher than MD in normal or healing myocardium. Myofibers passing through a region of interest placed in the anterolateral wall are shown in (G–I). (G) Normal mouse heart. The characteristic arrangement of myofibers into an array of crossing helices is clearly visible. (H) Mouse exposed to ischemia-reperfusion with severe hypokinesis in the anterolateral wall. However, MD has remained <1.3 × 10× mm2/s and fiber architecture remains reasonably organized. (I) Mouse exposed to ischemia-reperfusion, also presenting with severe hypokinesis of the anterolateral wall. MD in this mouse, however, is <1.3 × 10−3 mm2/s and fiber architecture is severely perturbed. Reproduced with permission from Huang et al. [195].
Magnetic Resonance Spectroscopy and Multi-Nuclei Imaging
CMR spectroscopy (CMRS) is a non-invasive technique to investigate the myocardial metabolism. It can use the signal from different endogenous nuclei, including 31phosphorus (31P), 1Hydrogen (1H), 23Sodium (23Na), and 13Carbon (13C), to quantify myocardial metabolism in vivo [197]. As the resonance frequency of nuclei is dependent on its molecular environment, different metabolites exhibit slightly different frequencies (chemical shift), thus leading to MR frequency spectra according to the chemical composition of the investigated voxel. An analysis of the frequency response (the MR spectrum) allows quantification of different metabolites [198].
31P-MRS is most widely used to quantify high-energy phosphates in the heart, including adenosine triphosphate (ATP) and phosphocreatine (PCr). The mutual transformation of ATP and PCr are necessary to meet the energy consumption required to maintain normal cardiac function. The impairments in energy metabolism reveal pathological processes in cardiac disease. Various preclinical studies showed reduced concentrations of ATP, PCr, and PCr/ATP ratios in cardiomyopathy or heart failure [199–201]. CMRS needs to localize signal to a certain voxel and to exclude signal from nearby structures (e.g., liver, chest skeletal muscle). Localization methods for cardiac MRS mainly contain two approaches, single-voxel and chemical shift imaging (CSI). Both contain advantages and limitations. Single-voxel localization is usually performed with image selected in vivo spectroscopy (ISIS), which consists of three slice-selective 180° pulses for localization and a single nonselective 90° pulse for signal detection [202]. Bakermans et al. [203] assessed myocardial energy status in vivo using single-voxel ISIS-localized 31P-MRS. Accurate localization and decreased PCr/γ-ATP ratios were observed in TAC mice (Figure 13). The 31P-CSI can be performed in 1D (column of voxel), 2D (plane of voxel), or 3D (block of voxels) mode [200, 204, 205]. CSI employs phase-encoding gradients in different directions and resolves spectra from different locations across the myocardium.
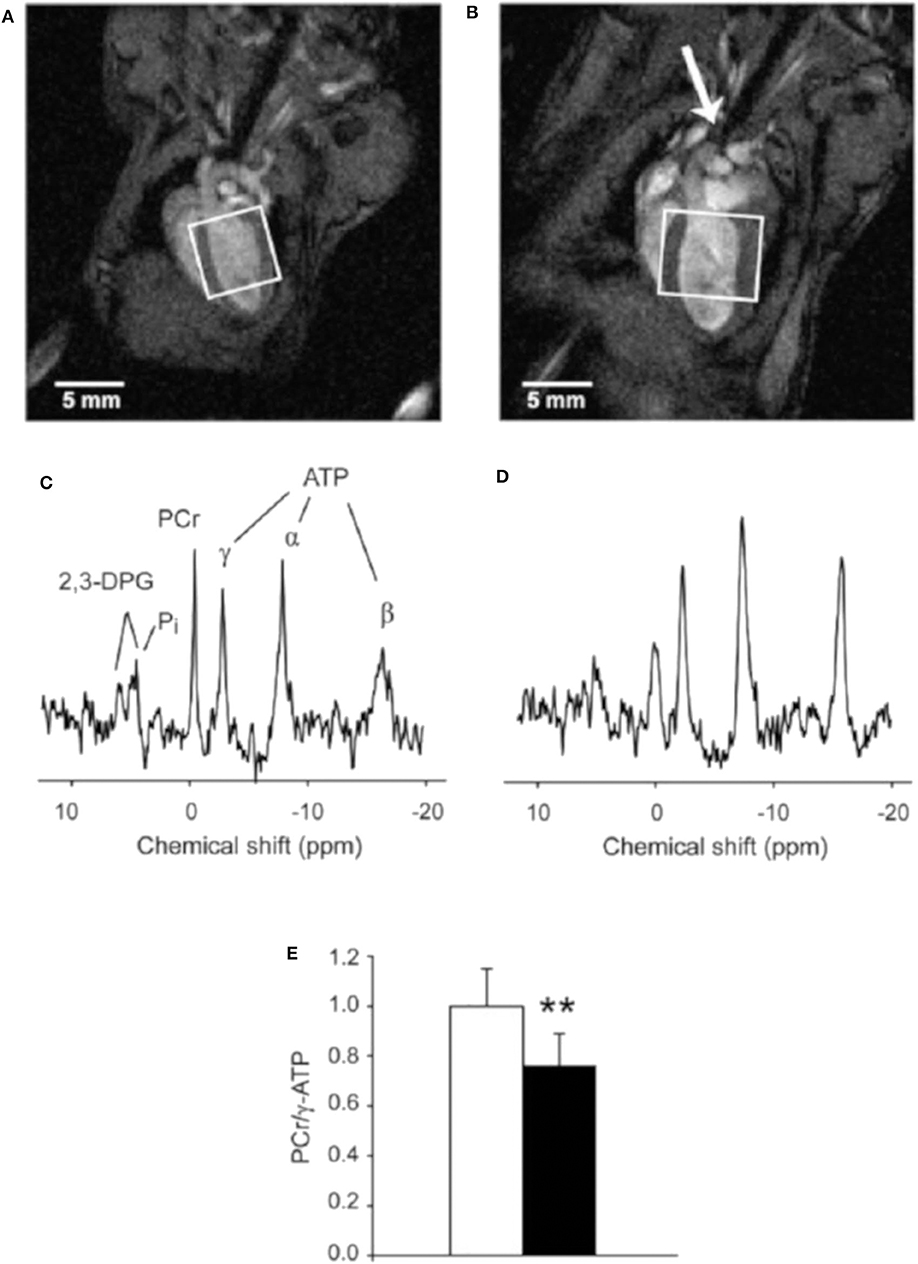
Figure 13. End-diastolic left ventricular MR images obtained from a control mouse (A) and a mouse with a transverse aortic constriction (TAC) (B). The constriction is indicated by the arrow. Dilated hypertrophic cardiomyopathy is evidenced by increased LV wall thickness and LV cavity volume in the TAC mouse. Rectangles indicate the voxels selected for localized 31P-MRS with 3D ISIS. (C,D) display 31P MR spectra acquired in vivo with 3D ISIS in a healthy mouse heart and a TAC heart, respectively. Myocardial PCr/γ-ATP was lower in TAC mice (n = 8) compared to healthy controls (n = 9) (E). Data are expressed as mean ± SD. **P < 0.01. α-, β-, γ-ATP, α-, β-, and γ-phosphate groups in ATP; 2,3-DPG, 2,3-diphosphoglycerate; PCr, phosphocreatine; Pi, inorganic phosphate. Reproduced with permission from Bakermans et al. [203].
The 1H represents the MR-active nuclei with the highest natural abundance and sensitivity in living tissues, and ensures cardiac 1H-MRS to be a useful target for quantifying myocardial metabolites, such as triglycerides, lactate, carnitine, myoglobin, and creatine (Cr) levels [206]. Since creatine plays a key role in the creatine kinase system, its measurement can provide additional information about myocardial energy transportation and storage. The high lipid concentration is associated with atherosclerosis and type 2 diabetes mellitus, which results in impaired cardiac function. Due to the technical restriction, it took decades for translating to in vivo imaging after first implemented 1H-CMRS in a perfused rat heart in Ugurbil et al. [207]. Schneider et al. [208] reported cardiac 1H-MRS in a guanidinoacetate N-methyltransferase (GAMT) deficient mouse model in vivo using a single-voxel point resolved spectroscopy sequence (PRESS). Various cardiac metabolites were detected in voxels of 2 μl and a clear decreased myocardial creatine level was observed. PRESS is the dominant method for 1H-MRS, containing three slice-selective RF pulses (90–180–180°) applied concurrently with three mutually orthogonal field gradients [209]. Another technique for 1H-MRS is STEAM. It is similar to PRESS, but uses three 90° RF pulses. However, 1H-MRS remains technically challenging due to the superabundance of 1H in the water molecule, and it will result in baseline distortion and affect the quantification.
23Na is an important component in the myocardial signal transduction pathway. The 23Na signal is increased in myocardial ischemia. Thus, 23Na-MRS can measure intracellular and extracellular Na+ changes to investigate myocardial viability without external contrast agents [210, 211]. Combination with acceleration methods such as CS has been shown [212].
As many metabolites contain carbon, 13C-MRS is particularly suited to study myocardial metabolism. However, traditional 13C-MRS is limited by low inherent sensitivity. In the last 10 years, in vivo cardiac 13C-MRS has become feasible with the development of dynamic nuclear hyperpolarization (DNP) technique. DNP is based on transferring electron to nuclear spins thus achieving substantially higher polarization through DNP. It requires the presence of unpaired electrons homogeneously distributed within the sample, which can be achieved by mixing the sample with radicals. To achieve almost 100% polarization of the electrons, DNP works at low temperature and high magnetic fields. Electron-nuclear spin transitions are induced through microwave irradiation. In the solid state, the nuclear polarizations of 13C can be increased to over 40% [213]. Subsequently, the solid sample is dissolved rapidly to yield a solution of molecules with hyperpolarized nuclear spins [214]. The sensitivity of 13C-MRS can be increased over 10,000-fold and the metabolism of infused hyperpolarized compounds be visualized in vivo [210]. 1-13C-pyruvate has been the most widely used DNP hyperpolarized molecules. Pyruvate is the primary metabolites of carbohydrates. After being transported into mitochondria, it is irreversibly converted to acetyl-CoA by the pyruvate dehydrogenase enzyme complex (PDH), then participated into the tricarboxylic acid (TCA) cycle [215]. By labeling pyruvate in a unique carbon position and investigating its metabolism by MRS, insights into myocardial metabolism can be derived. Dodd et al. [216] investigated in vivo metabolism of [1-13C]pyruvate in the mouse heart and revealed metabolic differences between different mouse strains. Atherton et al. [217] used [2-13C]pyruvate to evaluate Krebs cycle metabolism in hyperthyroidism rats and increased anaplerosis was observed in the hyperthyroid heart. With infused hyperpolarized pyruvate, myocardial metabolic processes have been quantified in different rodent models, including diabetes [218], ischemia [219], cardiac inflammation [220], etc.
Conclusions
MRI is a tremendously versatile and flexible imaging modality and provides high-resolution images in any arbitrary plane without the risk of ionizing radiation. Especially its accuracy and high reproducibility have made CMR the non-invasive reference modality for deriving myocardial structure, global and regional function, perfusion, tissue characterization and myocardial metabolism in small animal research.
Even though well-established, cardiac imaging in small animals is still challenging especially due to high cardiac and respiratory rates and the small anatomical structures, often demanding dedicated hardware components for providing sufficient image quality in acceptable imaging times. Especially dedicated multielement receiver coils either placed under the chest of the animal or wrapped around its thorax ensure sufficient SNR and reduction of the acquisition time by parallel imaging approaches.
According to the nature of MRI, reliable and highly reproducible information on the cardiac condition of the animal can be obtained. However, the impact of the required general anesthesia and rather long acquisition times on the investigated parameters have to be carefully considered during data interpretation. Inhalation anesthesia is highly recommended, since it provides greater safety, lesser cardiovascular depression, and rapid recovery, and convenient adjustments and maintenance during scanning. The body temperature control of the animal seems mandatory to avoid the deterioration of the derived functional parameters by cardiovascular depression due to cooling out of the animal.
With the availability of reliably working self-gating techniques, assessment of global cardiac function has been proven to be an excellent tool for reliable quantification of systolic dysfunction. However, due to the high heart rates, the assessment of diastolic dysfunction is still challenging and demands long acquisition times to provide sufficient temporal resolution. As in human imaging, regional cardiac function has been addressed by several approaches, and the use of conventional cine images in combination with feature tracking show the potential to enable efficient regional wall motion analysis.
One of the strengths of CMR is its capability for tissue characterization. Identification of scarred tissue by means of delayed contrast enhancement imaging is straight forward using (black-blood) self-gating techniques. Although mapping of relaxation parameters by different means has been reported, it is technically more challenging and well-trained operators are required especially for ensuring accurate and reproducible numbers. More advanced techniques such as (first-pass) perfusion, diffusion-weighted imaging or DTI for deriving myocardial fiber orientation must still be seen as experimental.
Further important information on tissue composition and metabolism can be retrieved from MR spectroscopy and multinuclei imaging. Due to its high natural abundance and sensitivity, 1H spectroscopy has been efficiently applied to quantifying triglycerides, lactate, carnitine, myoglobin, and creatine (Cr) levels. However, to make full use of the technique, different nuclei like phosphorus (31P), Sodium (23Na), and Carbon (13C) need to be investigated to quantify myocardial metabolism in vivo. 31P-MRS can be used to quantify high-energy phosphates in the heart, 23Na is an important component in the myocardial signal transduction pathway, and 13C-MRS is particularly suited to study myocardial metabolism. In general, the multinuclei techniques can be extended to imaging thus providing the chemical composition of each voxel. Due to the low sensitivity and the need to spectrally separate the signal from the different metabolites, multinuclei MRS and MRI are mainly performed at ultra-high field strengths. Even though available in research for decades, due to the long scan times and the expertise required to performing multi-nuclei MRS and MRI in the heart, this technique must still be seen as experimental with unfortunately only little utilization. With the recent introduction of hyperpolarized 13C imaging, limitations from the intrinsic low sensitivity may be solved and rapid access to 13C-labeled metabolites may be enabled at the expense of high related costs.
CMR imaging and spectroscopy is still one of the most promising noninvasive tools for the staging and longitudinal monitoring of cardiovascular diseases in small animals. Due to its versatility, it can be applied to a wide range of translational applications and provide quantitative data from global functional assessment to metabolic characterization of the underlying myocardial substrate. Due to its complexity, the application is currently often limited to global functional assessment and scar imaging. More challenging techniques such as assessing metabolism or the structural organization of the myocardial fibers are still limited by long scan times and may need further technical developments before finally becoming routine in small animal applications.
Author Contributions
HL and VR drafted the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the Ulm University Center for Translational Imaging MoMAN for its support.
References
1. Wang H, Naghavi M, Allen C, Barber RM, Bhutta ZA, Carter A, et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. (2016) 388:1459–544. doi: 10.1016/S0140-6736(16)31012-1
2. Joseph P, Leong D, McKee M, Anand SS, Schwalm J-D, Teo K, et al. Reducing the global burden of cardiovascular disease, part 1: the epidemiology and risk factors. Circ Res. (2017) 121:677–94. doi: 10.1161/CIRCRESAHA.117.308903
3. Camacho P, Fan H, Liu Z, He JQ. Small mammalian animal models of heart disease. Am J Cardiovasc Dis. (2016) 6:70–80.
4. Wang K, Long B, Liu F, Wang J-X, Liu C-Y, Zhao B, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. (2016) 37:2602–11. doi: 10.1093/eurheartj/ehv713
5. Aherrahrou Z, Schlossarek S, Stoelting S, Klinger M, Geertz B, Weinberger F, et al. Knock-out of nexilin in mice leads to dilated cardiomyopathy and endomyocardial fibroelastosis. Basic Res Cardiol. (2016) 111:6. doi: 10.1007/s00395-015-0522-5
6. Vanhoutte L, Gallez B, Feron O, Balligand J-L, Esfahani H, d'Hoore W, et al. Variability of mouse left ventricular function assessment by 11.7 Tesla MRI. J Cardiovasc Trans Res. (2015) 8:362–71. doi: 10.1007/s12265-015-9638-0
7. Zuo Z, Subgang A, Abaei A, Rottbauer W, Stiller D, Ma G, et al. Assessment of longitudinal reproducibility of mice LV function parameters at 11.7 T derived from self-gated CINE MRI. Biomed Res Int. (2017) 2017:8392952. doi: 10.1155/2017/8392952
8. Lottonen-Raikaslehto L, Rissanen R, Gurzeler E, Merentie M, Huusko J, Schneider JE, et al. Left ventricular remodeling leads to heart failure in mice with cardiac-specific overexpression of VEGF-B 167 : echocardiography and magnetic resonance imaging study. Physiol Rep. (2017) 5:e13096. doi: 10.14814/phy2.13096
9. Weisell J, Ruotsalainen A, Laakso H, Ylä-Herttuala E, Näpänkangas J, Levonen A, et al. Characterizing valve dynamics in mice by high-resolution cine-MRI. NMR Biomed. (2019) 32:e4108. doi: 10.1002/nbm.4108
10. Wessels A, Sedmera D. Developmental anatomy of the heart: a tale of mice and man. Physiol Genomics. (2003) 15:165–76. doi: 10.1152/physiolgenomics.00033.2003
11. Janiczek RL, Blackman BR, Roy RJ, Meyer CH, Acton ST, Epstein FH. Three-dimensional phase contrast angiography of the mouse aortic arch using spiral MRI. Magn Reson Med. (2011) 66:1382–90. doi: 10.1002/mrm.22937
12. Chen X, Salerno M, Epstein FH, Meyer CH. Accelerated multi-TI spiral MRI using compressed sensing with temporal constraints. In: 19th Annual Meeting of IS. Montreal, QC (2011). p. 4369.
13. Motaal AG, Noorman N, de Graaf WL, Hoerr V, Florack LMJ, Nicolay K, et al. Functional imaging of murine hearts using accelerated self-gated UTE cine MRI. Int J Cardiovasc Imaging. (2015) 31:83–94. doi: 10.1007/s10554-014-0531-8
14. Hiba B, Richard N, Thibault H, Janier M. Cardiac and respiratory self-gated cine MRI in the mouse: comparison between radial and rectilinear techniques at 7T. Magn Reson Med. (2007) 58:745–53. doi: 10.1002/mrm.21355
15. Lustig M, Donoho D, Pauly JM. Sparse MRI: the application of compressed sensing for rapid MR imaging. Magn Reson Med. (2007) 58:1182–95. doi: 10.1002/mrm.21391
16. Lustig M, Donoho DL, Santos JM, Pauly JM. Compressed sensing MRI. IEEE Signal Process Mag. (2008) 25:72–82. doi: 10.1109/MSP.2007.914728
17. Mistretta CA, Wieben O, Velikina J, Block W, Perry J, Wu Y, et al. Highly constrained backprojection for time-resolved MRI. Magn Reson Med. (2006) 55:30–40. doi: 10.1002/mrm.20772
18. Lindsey ML, Kassiri Z, Virag JAI, de Castro Brás LE, Scherrer-Crosbie M. Guidelines for measuring cardiac physiology in mice. Am J Physiol Heart Circ Physiol. (2018) 314:H733–52. doi: 10.1152/ajpheart.00339.2017
19. Curley D, Lavin Plaza B, Shah AM, Botnar RM. Molecular imaging of cardiac remodelling after myocardial infarction. Basic Res Cardiol. (2018) 113:10. doi: 10.1007/s00395-018-0668-z
20. Gyöngyösi M, Winkler J, Ramos I, Do Q, Firat H, McDonald K, et al. Myocardial fibrosis: biomedical research from bench to bedside. Eur J Heart Fail. (2017) 19:177–91. doi: 10.1002/ejhf.696
21. Constantinides C. Study of the murine cardiac mechanical function using magnetic resonance imaging: the current status, challenges, and future perspectives. In: Andrade AO, Alves Pereira A, Naves ELM, Soares AB, editors. Practical Applications in Biomedical Engineering. Rijek: InTech Publications (2012). p. 343–86. doi: 10.5772/51364
22. Epstein FH. MR in mouse models of cardiac disease. NMR Biomed. (2007) 20:238–55. doi: 10.1002/nbm.1152
23. Vanhoutte L, Gerber BL, Gallez B, Po C, Magat J, Jean-Luc B, et al. High field magnetic resonance imaging of rodents in cardiovascular research. Basic Res Cardiol. (2016) 111:46. doi: 10.1007/s00395-016-0565-2
24. Gilson WD, Kraitchman DL. Cardiac magnetic resonance imaging in small rodents using clinical 1.5T and 3.0T scanners. Methods. (2007) 43:35–45. doi: 10.1016/j.ymeth.2007.03.012
25. Bunck AC, Engelen MA, Schnackenburg B, Furkert J, Bremer C, Heindel W, et al. Feasibility of functional cardiac MR imaging in mice using a clinical 3 Tesla whole body scanner. Invest Radiol. (2009) 44:749–56. doi: 10.1097/RLI.0b013e3181b2c135
26. Gabbay-Benziv R, Reece EA, Wang F, Bar-Shir A, Harman C, Turan OM, et al. A step-wise approach for analysis of the mouse embryonic heart using 17.6 Tesla MRI. Magn Reson Imaging. (2017) 35:46–53. doi: 10.1016/j.mri.2016.08.008
27. Felder J, Celik AA, Choi CH, Schwan S, Shah NJ. 9.4 T small animal MRI using clinical components for direct translational studies. J Transl Med. (2017) 15:264. doi: 10.1186/s12967-017-1373-7
28. Naresh NK, Chen X, Roy RJ, Antkowiak PF, Annex BH, Epstein FH. Accelerated dual-contrast first-pass perfusion MRI of the mouse heart: development and application to diet-induced obese mice: dual-contrast first-pass MRI in Mice. Magn Reson Med. (2015) 73:1237–45. doi: 10.1002/mrm.25238
29. Constantinides C, Angeli S, Gkagkarellis S, Cofer G. Intercomparison of performance of RF coil geometries for high field mouse cardiac MRI. Concepts Magn Reson. (2011) 38A:236–52. doi: 10.1002/cmr.a.20225
30. Doty FD, Entzminger G, Kulkarni J, Pamarthy K, Staab JP. Radio frequency coil technology for small-animal MRI. NMR Biomed. (2007) 20:304–25. doi: 10.1002/nbm.1149
31. Gareis D, Wichmann T, Lanz T, Melkus G, Horn M, Jakob PM. Mouse MRI using phased-array coils. NMR Biomed. (2007) 20:326–34. doi: 10.1002/nbm.1156
32. Schneider JE, Lanz T, Barnes H, Stork L-A, Bohl S, Lygate CA, et al. Accelerated cardiac magnetic resonance imaging in the mouse using an eight-channel array at 9.4 Tesla. Magn Reson Med. (2011) 65:60–70. doi: 10.1002/mrm.22605
33. Wagenhaus B, Pohlmann A, Dieringer MA, Els A, Waiczies H, Waiczies S, et al. Functional and morphological cardiac magnetic resonance imaging of mice using a cryogenic quadrature radiofrequency coil. PLoS ONE. (2012) 7:e42383. doi: 10.1371/journal.pone.0042383
34. Dieringer B, Pohlmann A, Dieringer M, Fuchs K, Els A, Waiczies H, et al. Feasibility and benefit of using a cryogenic radiofrequency coil for functional cardiac magnetic resonance imaging of mice at 9.4 T. J Cardiovasc Magn Reson. (2013) 15:W39. doi: 10.1186/1532-429X-15-S1-W39
35. Flecknell PA. Laboratory Animal Anaesthesia. 4th ed. Newcastle upon Tyne: Academic Press (2009).
36. Gargiulo S, Greco A, Gramanzini M, Esposito S, Affuso A, Brunetti A, et al. Mice anesthesia, analgesia, and care, part I: anesthetic considerations in preclinical research. ILAR J. (2012) 53:E55–69. doi: 10.1093/ilar.53.1.55
37. Hildebrandt IJ, Su H, Weber WA. Anesthesia and other considerations for in vivo imaging of small animals. ILAR J. (2008) 49:17–26. doi: 10.1093/ilar.49.1.17
38. Berry CJ, Thedens DR, Light-McGroary K, Miller JD, Kutschke W, Zimmerman KA, et al. Effects of deep sedation or general anesthesia on cardiac function in mice undergoing cardiovascular magnetic resonance. J Cardiovasc Magn Reson. (2009) 11:16. doi: 10.1186/1532-429X-11-16
39. Pachon RE, Scharf BA, Vatner DE, Vatner SF. Best anesthetics for assessing left ventricular systolic function by echocardiography in mice. Am J Physiol Heart Circ Physiol. (2015) 308:H1525–9. doi: 10.1152/ajpheart.00890.2014
40. Constantinides C, Mean R, Janssen BJ. Effects of isoflurane anesthesia on the cardiovascular function of the C57BL/6 mouse. ILAR J. (2011) 52:e21–31.
41. Haase A, Frahm J, Matthaei D, Hänicke W, Merboldt KD. FLASH imaging: rapid NMR imaging using low flip-angle pulses. J Magn Reson. (2011) 213:533–41. doi: 10.1016/j.jmr.2011.09.021
42. Rose SE, Wilson SJ, Zelaya FO, Crozier S, Doddrell DM. High resolution high field rodent cardiac imaging with flow enhancement suppression. Magn Reson Imaging. (1994) 12:1183–90. doi: 10.1016/0730-725X(94)90084-5
43. Heijman E, de Graaf W, Niessen P, Nauerth A, van Eys G, de Graaf L, et al. Comparison between prospective and retrospective triggering for mouse cardiac MRI. NMR Biomed. (2007) 20:439–47. doi: 10.1002/nbm.1110
44. van Genderingen HR, Sprenger M, de Ridder JW, van Rossum AC. Carbon-fiber electrodes and leads for electrocardiography during MR imaging. Radiology. (1989) 171:872. doi: 10.1148/radiology.171.3.2717770
45. Choquet P, Goetz C, Aubertin G, Hubele F. Carbon tube electrodes for electrocardiographygated cardiac multimodality imaging in mice. J Am Assoc Lab Anim Sci. (2011) 50:61–4.
46. Sablong R, Rengle A, Ramgolam A, Saint-Jalmes H, Beuf O. An optical fiber-based gating device for prospective mouse cardiac MRI. IEEE Trans Biomed Eng. (2014) 61:162–70. doi: 10.1109/TBME.2013.2278712
47. Schmidt M, Krug JW, Rosenheimer MN, Rose G. Filtering of ECG signals distorted by magnetic field gradients during MRI using non-linear filters and higher-order statistics. Biomed Tech. (2018) 63:395–406. doi: 10.1515/bmt-2016-0232
48. Schmidt M, Krug JW, Rose G. Reducing of gradient induced artifacts on the ECG signal during MRI examinations using Wilcoxon filter. Curr Dir Biomed Eng. (2016) 2:175–8. doi: 10.1515/cdbme-2016-0040
49. Park H, Park Y, Cho S, Jang B, Lee K. New cardiac MRI gating method using event-synchronous adaptive digital filter. Ann Biomed Eng. (2009) 37:2170–87. doi: 10.1007/s10439-009-9764-4
50. Hoerr V, Nagelmann N, Nauerth A, Kuhlmann MT, Stypmann J, Faber C. Cardiac-respiratory self-gated cine ultra-short echo time (UTE) cardiovascular magnetic resonance for assessment of functional cardiac parameters at high magnetic fields. J Cardiovasc Magn Reson. (2013) 15:59. doi: 10.1186/1532-429X-15-59
51. Bovens SM, te Boekhorst BCM, Ouden K den, van de Kolk KWA, Nauerth A, Nederhoff MGJ, et al. Evaluation of infarcted murine heart function: comparison of prospectively triggered with self-gated MRI. NMR Biomed. (2011) 24:307–15. doi: 10.1002/nbm.1593
52. Coolen BF, Abdurrachim D, Motaal AG, Nicolay K, Prompers JJ, Strijkers GJ. High frame rate retrospectively triggered cine MRI for assessment of murine diastolic function. Magn Reson Med. (2013) 69:648–56. doi: 10.1002/mrm.24287
53. Haberkorn SM, Jacoby C, Ding Z, Keul P, Bönner F, Polzin A, et al. Cardiovascular magnetic resonance relaxometry predicts regional functional outcome after experimental myocardial infarction. Circ Cardiovasc Imaging. (2017) 10:e006025. doi: 10.1161/CIRCIMAGING.116.006025
54. Coolen BF, Geelen T, Paulis LEM, Nauerth A, Nicolay K, Strijkers GJ. Three-dimensional T1 mapping of the mouse heart using variable flip angle steady-state MR imaging. NMR Biomed. (2011) 24:154–62. doi: 10.1002/nbm.1566
55. Beyers RJ, Smith RS, Xu Y, Piras BA, Salerno M, Berr SS, et al. T2-weighted MRI of post-infarct myocardial edema in mice. Magn Reson Med. (2012) 67:201–9. doi: 10.1002/mrm.22975
56. Breuer FA, Kellman P, Griswold MA, Jakob PM. Dynamic autocalibrated parallel imaging using temporal GRAPPA (TGRAPPA). Magn Reson Med. (2005) 53:981–5. doi: 10.1002/mrm.20430
57. Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med. (2002) 47:1202–10. doi: 10.1002/mrm.10171
58. Seiberlich N, Ehses P, Duerk J, Gilkeson R, Griswold M. Improved radial GRAPPA calibration for real-time free-breathing cardiac imaging. Magn Reson Med. (2011) 65:492–505. doi: 10.1002/mrm.22618
59. Feng L, Srichai MB, Lim RP, Harrison A, King W, Adluru G, et al. Highly accelerated real-time cardiac cine MRI using K - T SPARSE-SENSE. Magn Reson Med. (2013) 70:64–74. doi: 10.1002/mrm.24440
60. Bollache E, Barker AJ, Dolan RS, Carr JC, van Ooij P, Ahmadian R, et al. k-t accelerated aortic 4D flow MRI in under two minutes: feasibility and impact of resolution, k-space sampling patterns, and respiratory navigator gating on hemodynamic measurements. Magn Reson Med. (2018) 79:195–207. doi: 10.1002/mrm.26661
61. Schnell S, Markl M, Entezari P, Mahadewia RJ, Semaan E, Stankovic Z, et al. K-T GRAPPA accelerated four-dimensional flow MRI in the aorta: effect on scan time, image quality, and quantification of flow and wall shear stress. Magn Reson Med. (2014) 72:522–33. doi: 10.1002/mrm.24925
62. Kido T, Kido T, Nakamura M, Watanabe K, Schmidt M, Forman C, et al. Compressed sensing real-time cine cardiovascular magnetic resonance: accurate assessment of left ventricular function in a single-breath-hold. J Cardiovasc Magn Reson. (2016) 18:50. doi: 10.1186/s12968-016-0271-0
63. Otazo R, Kim D, Axel L, Sodickson DK. Combination of compressed sensing and parallel imaging for highly accelerated first-pass cardiac perfusion MRI. Magn Reson Med. (2010) 64:767–76. doi: 10.1002/mrm.22463
64. Wech T, Seiberlich N, Schindele A, Grau V, Diffley L, Gyngell ML, et al. Development of real-time magnetic resonance imaging of mouse hearts at 9.4 tesla – simulations and first application. IEEE Trans Med Imaging. (2016) 35:912–20. doi: 10.1109/TMI.2015.2501832
65. Frahm J, Voit D, Uecker M. Real-time magnetic resonance imaging: radial gradient-echo sequences with nonlinear inverse reconstruction. Invest Radiol. (2019) 54:757–66. doi: 10.1097/RLI.0000000000000584
66. Uecker M, Zhang S, Voit D, Karaus A, Merboldt K-D, Frahm J. Real-time MRI at a resolution of 20 ms. NMR Biomed. (2010) 23:986–94. doi: 10.1002/nbm.1585
67. Xue H, Kellman P, LaRocca G, Arai AE, Hansen MS. High spatial and temporal resolution retrospective cine cardiovascular magnetic resonance from shortened free breathing real-time acquisitions. J Cardiovasc Magn Reson. (2013) 15:102. doi: 10.1186/1532-429X-15-102
68. Zhang S, Uecker M, Voit D, Merboldt K-D, Frahm J. Real-time cardiovascular magnetic resonance at high temporal resolution: radial FLASH with nonlinear inverse reconstruction. J Cardiovasc Magn Reson. (2010) 12:39. doi: 10.1186/1532-429X-12-39
69. Dai G, Ding Y, Huang S, simonetti OP, Sosnovik DE. Realtime cine MRI in mice with a single-shot EPI sequence and the karhunen–loeve transform. In: Proceedings of the 18th Annual Meeting of IS. Stockholm (2010). p. 1437.
70. Uecker M, Zhang S, Voit D, Merboldt KD, Frahm J. Real-time MRI: recent advances using radial FLASH. Imaging Med. (2012) 4:461–76. doi: 10.2217/iim.12.32
71. Winkelmann S, Schaeffter T, Koehler T, Eggers H, Doessel O. An optimal radial profile order based on the golden ratio for time-resolved MRI. IEEE Trans Med Imaging. (2007) 26:68–76. doi: 10.1109/TMI.2006.885337
72. Wundrak S, Paul J, Ulrici J, Hell E, Geibel M-A, Bernhardt P, et al. Golden ratio sparse MRI using tiny golden angles: golden ratio sparse MRI using tiny Golden Angles. Magn Reson Med. (2016) 75:2372–8. doi: 10.1002/mrm.25831
73. Li H, Abaei A, Lu Q, Rasche V. Tiny golden angle real-time cardiac MRI in the mouse model. In: 27th Annual Meeting of IS. Montreal, QC (2019). p. 155.
74. Li H, Metze P, Abaei A, Rottbauer W, Just S, Lu Q, et al. Feasibility of real-time cardiac MRI in mice using tiny golden angle radial sparse. NMR Biomed. (2020) e4300. doi: 10.1002/nbm.4300. [Epub ahead of print].
75. Donoho DL. Compressed sensing. IEEE Trans Inform Theory. (2006) 52:1289–306. doi: 10.1109/TIT.2006.871582
76. Candes EJ, Romberg J, Tao T. Robust uncertainty principles: exact signal reconstruction from highly incomplete frequency information. IEEE Trans Inform Theory. (2006) 52:489–509. doi: 10.1109/TIT.2005.862083
77. Feng L, Benkert T, Block KT, Sodickson DK, Otazo R, Chandarana H. Compressed sensing for body MRI: compressed sensing for body MRI. J Magn Reson Imaging. (2017) 45:966–87. doi: 10.1002/jmri.25547
78. Feng L, Grimm R, Block KT, Chandarana H, Kim S, Xu J, et al. Golden-angle radial sparse parallel MRI: combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric MRI. Magn Reson Med. (2014) 72:707–17. doi: 10.1002/mrm.24980
79. Montesinos P, Abascal JFPJ, Cussó L, Vaquero JJ, Desco M. Application of the compressed sensing technique to self-gated cardiac cine sequences in small animals: compressed sensing for self-gated cardiac cine sequences. Magn Reson Med. (2014) 72:369–80. doi: 10.1002/mrm.24936
80. Motaal AG, Coolen BF, Abdurrachim D, Castro RM, Prompers JJ, Florack LMJ, et al. Accelerated high-frame-rate mouse heart cine-MRI using compressed sensing reconstruction. NMR Biomed. (2013) 26:451–7. doi: 10.1002/nbm.2883
81. Wech T, Lygate CA, Neubauer S, Köstler H, Schneider JE. Highly accelerated cardiac functional MRI in rodent hearts using compressed sensing and parallel imaging at 9.4T. J Cardiovasc Magn Reson. (2012) 14:P65. doi: 10.1186/1532-429X-14-S1-P65
82. Buonincontri G, Methner C, Krieg T, Carpenter TA, Sawiak SJ. Functional assessment of the mouse heart by MRI with a 1-min acquisition. NMR Biomed. (2014) 27:733–7. doi: 10.1002/nbm.3116
83. Axel L, Otazo R. Accelerated MRI for the assessment of cardiac function. BJR. (2016) 89:20150655. doi: 10.1259/bjr.20150655
84. Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. (1999) 42:952–62.
85. Deshmane A, Gulani V, Griswold MA, Seiberlich N. Parallel MR imaging. J Magn Reson Imaging. (2012) 36:55–72. doi: 10.1002/jmri.23639
86. Ratering D, Baltes C, Dörries C, Rudin M. Accelerated cardiovascular magnetic resonance of the mouse heart using self-gated parallel imaging strategies does not compromise accuracy of structural and functional measures. J Cardiovasc Magn Reson. (2010) 12:43. doi: 10.1186/1532-429X-12-43
87. Pedersen H, Kozerke S, Ringgaard S, Nehrke K, Kim WY. K-T PCA: temporally constrained K-T BLAST reconstruction using principal component analysis. Magn Reson Med. (2009) 62:706–16. doi: 10.1002/mrm.22052
88. Tsao J, Boesiger P, Pruessmann KP. K-T BLAST and K-T SENSE: dynamic MRI with high frame rate exploiting spatiotemporal correlations. Magn Reson Med. (2003) 50:1031–42. doi: 10.1002/mrm.10611
89. Marshall I, Jansen MA, Tao Y, Merrifield GD, Gray GA. Application of kt-BLAST acceleration to reduce cardiac MR imaging time in healthy and infarcted mice. Magn Reson Mater Phys. (2014) 27:201–10. doi: 10.1007/s10334-013-0392-5
90. Makowski M, Jansen C, Webb I, Chiribiri A, Nagel E, Botnar R, et al. First-pass contrast-enhanced myocardial perfusion MRI in mice on a 3-T clinical MR scanner. Magn Reson Med. (2010) 64:1592–8. doi: 10.1002/mrm.22470
91. Bassett EC, Kholmovski EG, Wilson BD, DiBella EVR, Dosdall DJ, Ranjan R, et al. Evaluation of highly accelerated real-time cardiac cine MRI in tachycardia. NMR Biomed. (2014) 27:175–82. doi: 10.1002/nbm.3049
92. Nahrendorf M, Hiller KH, Hu K, Ertl G, Haase A, Bauer WR. Cardiac magnetic resonance imaging in small animal models of human heart failure. Med Image Anal. (2003) 7:369–75. doi: 10.1016/S1361-8415(03)00011-2
93. Stuckey DJ, Carr CA, Camelliti P, Tyler DJ, Davies KE, Clarke K. In vivo MRI characterization of progressive cardiac dysfunction in the mdx mouse model of muscular dystrophy. PLoS ONE. (2012) 7:e28569. doi: 10.1371/journal.pone.0028569
94. Heijman E, Strijkers GJ, Habets J, Janssen B, Nicolay K. Magnetic resonance imaging of regional cardiac function in the mouse. MAGMA. (2004) 17:170–8. doi: 10.1007/s10334-004-0082-4
95. Roberts TA, Price AN, Jackson LH, Taylor V, David AL, Lythgoe MF, et al. Direct comparison of high-temporal-resolution CINE MRI with doppler ultrasound for assessment of diastolic dysfunction in mice. NMR Biomed. (2017) 30:e3763. doi: 10.1002/nbm.3763
96. Le TT, Huang W, Bryant JA, Cook SA, Chin CWL. Stress cardiovascular magnetic resonance imaging: current and future perspectives. Expert Rev Cardiovasc Ther. (2017) 15:181–9. doi: 10.1080/14779072.2017.1296356
97. Saab R, Hage FG. Vasodilator stress agents for myocardial perfusion imaging. J Nucl Cardiol. (2017) 24:434–8. doi: 10.1007/s12350-016-0408-4
98. Charoenpanichkit C, Hundley WG. The 20 year evolution of dobutamine stress cardiovascular magnetic resonance. J Cardiovasc Magn Reson. (2010) 12:59. doi: 10.1186/1532-429X-12-59
99. Wiesmann F, Ruff J, Engelhardt S, Hein L, Dienesch C, Leupold A, et al. Dobutamine-stress magnetic resonance microimaging in mice: acute changes of cardiac geometry and function in normal and failing murine hearts. Circ Res. (2001) 88:563–9. doi: 10.1161/01.RES.88.6.563
100. Tyrankiewicz U, Skorka T, Jablonska M, Petkow-Dimitrow P, Chlopicki S. Characterization of the cardiac response to a low and high dose of dobutamine in the mouse model of dilated cardiomyopathy by MRI in vivo. J Magn Reson Imaging. (2013) 37:669–77. doi: 10.1002/jmri.23854
101. Palmer OR, Chiu CB, Cao A, Scheven UM, Diaz JA, Greve JM. In vivo characterization of the murine venous system before and during dobutamine stimulation: implications for preclinical models of venous disease. Ann Anat. (2017) 214:43–52. doi: 10.1016/j.aanat.2017.08.004
102. Hao Li, Metze P, Abaei A, Rottbauer W, Just S, Lu Q, Rasche V. Real-time cardiac MRI in the Mice model. In: 23rd Annual Meeting of SCMR. Orlando, FL (2020).
103. Zerhouni EA, Parish DM, Rogers WJ, Yang A, Shapiro EP. Human heart: tagging with MR imaging–a method for noninvasive assessment of myocardial motion. Radiology. (1988) 169:59–63. doi: 10.1148/radiology.169.1.3420283
104. Axel L, Dougherty L. Heart wall motion: improved method of spatial modulation of magnetization for MR imaging. Radiology. (1989) 172:349–50. doi: 10.1148/radiology.172.2.2748813
105. de Crespigny AJS, Carpenter TA, Hall LD. Cardiac tagging in the rat using a DANTE sequence. Magn Reson Med. (1991) 21:151–6. doi: 10.1002/mrm.1910210119
106. Price AN. Cardiovascular magnetic resonance imaging in experimental models. TOCMJ. (2010) 4:278–92. doi: 10.2174/1874192401004010278
107. Fischer SE, McKinnon GC, Maier SE, Boesiger P. Improved myocardial tagging contrast. Magn Reson Med. (1993) 30:191–200. doi: 10.1002/mrm.1910300207
108. Axel L, Gonçalves RC, Bloomgarden D. Regional heart wall motion: two-dimensional analysis and functional imaging with MR imaging. Radiology. (1992) 183:745–50. doi: 10.1148/radiology.183.3.1584931
109. Prince JL, Gupta SN, Osman NF. Bandpass optical flow for tagged MRI. Med Phys. (2000) 27:108–18. doi: 10.1118/1.598862
110. Liu W, Chen J, Ji S, Allen JS, Bayly PV, Yu X. HARP MRI tagging for direct quantification of lagrangian strain in rat hearts after myocardial infarction. J Biomech Eng. (2004) 126:523–8. doi: 10.1115/1.1785811
111. Delfino JG, Fornwalt BK, Eisner RL, Leon AR, Oshinski JN. Determination of transmural, endocardial, and epicardial radial strain and strain rate from phase contrast MR velocity data. J Magn Reson Imaging. (2008) 27:522–8. doi: 10.1002/jmri.21211
112. Espe EK, Aronsen JM, Skårdal K, Schneider JE, Zhang L, Sjaastad I. Novel insight into the detailed myocardial motion and deformation of the rodent heart using high-resolution phase contrast cardiovascular magnetic resonance. J Cardiovasc Magn Reson. (2013) 15:82. doi: 10.1186/1532-429X-15-82
113. Zhu Y, Drangova M, Pelc NJ. Estimation of deformation gradient and strain from cine-PC velocity data. IEEE Trans Med Imaging. (1997) 16:840–51. doi: 10.1109/42.650880
114. Bernstein MA, Zhou XJ, Polzin JA, King KF, Ganin A, Pelc NJ, et al. Concomitant gradient terms in phase contrast MR: analysis and correction. Magn Reson Med. (1998) 39:300–8. doi: 10.1002/mrm.1910390218
115. Walker PG, Cranney GB, Scheidegger MB, Waseleski G, Pohost GM, Yoganathan AP. Semiautomated method for noise reduction and background phase error correction in MR phase velocity data. J Magn Reson Imaging. (1993) 3:521–30. doi: 10.1002/jmri.1880030315
116. Espe EKS, Aronsen JM, Skrbic B, Skulberg VM, Schneider JE, Sejersted OM, et al. Improved MR phase-contrast velocimetry using a novel nine-point balanced motion-encoding scheme with increased robustness to eddy current effects. Magn Reson Med. (2013) 69:48–61. doi: 10.1002/mrm.24226
117. Rolf MP, Hofman MB, Gatehouse PD, Markenroth-Bloch K, Heymans MW, Ebbers T, et al. Sequence optimization to reduce velocity offsets in cardiovascular magnetic resonance volume flow quantification - a multi-vendor study. J Cardiovasc Magn Reson. (2011) 13:18. doi: 10.1186/1532-429X-13-18
118. McGinley G, Bendiksen BA, Zhang L, Aronsen JM, Nordén ES, Sjaastad I, et al. Accelerated magnetic resonance imaging tissue phase mapping of the rat myocardium using compressed sensing with iterative soft-thresholding. PLoS ONE. (2019) 14:e0218874. doi: 10.1371/journal.pone.0218874
119. Aletras AH, Ding S, Balaban RS, Wen H. DENSE: displacement encoding with stimulated echoes in cardiac functional MRI. J Magn Reson. (1999) 137:247–52. doi: 10.1006/jmre.1998.1676
120. Kim D, Epstein FH, Gilson WD, Axel L. Increasing the signal-to-noise ratio in DENSE MRI by combining displacement-encoded echoes. Magn Reson Med. (2004) 52:188–92. doi: 10.1002/mrm.20109
121. Haggerty CM, Kramer SP, Binkley CM, Powell DK, Mattingly AC, Charnigo R, et al. Reproducibility of cine displacement encoding with stimulated echoes (DENSE) cardiovascular magnetic resonance for measuring left ventricular strains, torsion, and synchrony in mice. J Cardiovasc Magn Reson. (2013) 15:71. doi: 10.1186/1532-429X-15-71
122. Zhong X, Gibberman LB, Spottiswoode BS, Gilliam AD, Meyer CH, French BA, et al. Comprehensive cardiovascular magnetic resonance of myocardial mechanics in mice using three-dimensional cine DENSE. J Cardiovasc Magn Reson. (2011) 13:83. doi: 10.1186/1532-429X-13-83
123. Vandsburger MH, French BA, Kramer CM, Zhong X, Epstein FH. Displacement-encoded and manganese-enhanced cardiac MRI reveal that nNOS, not eNOS, plays a dominant role in modulating contraction and calcium influx in the mammalian heart. Am J Physiol Heart Circ Physiol. (2012) 302:H412–9. doi: 10.1152/ajpheart.00705.2011
124. Aletras AH, Ingkanisorn WP, Mancini C, Arai AE. DENSE with SENSE. J Magn Reson. (2005) 176:99–106. doi: 10.1016/j.jmr.2005.05.010
125. Chen X, Yang Y, Cai X, Auger DA, Meyer CH, Salerno M, et al. Accelerated two-dimensional cine DENSE cardiovascular magnetic resonance using compressed sensing and parallel imaging. J Cardiovasc Magn Reson. (2016) 18:38. doi: 10.1186/s12968-016-0253-2
126. Maret E, Todt T, Brudin L, Nylander E, Swahn E, Ohlsson JL, et al. Functional measurements based on feature tracking of cine magnetic resonance images identify left ventricular segments with myocardial scar. Cardiovasc Ultrasound. (2009) 7:53. doi: 10.1186/1476-7120-7-53
127. Morais P, Marchi A, Bogaert JA, Dresselaers T, Heyde B, D'hooge J, et al. Cardiovascular magnetic resonance myocardial feature tracking using a non-rigid, elastic image registration algorithm: assessment of variability in a real-life clinical setting. J Cardiovasc Magn Reson. (2017) 19:24. doi: 10.1186/s12968-017-0333-y
128. Schuster A, Morton G, Hussain ST, Jogiya R, Kutty S, Asrress KN, et al. The intra-observer reproducibility of cardiovascular magnetic resonance myocardial feature tracking strain assessment is independent of field strength. Eur J Radiol. (2013) 82:296–301. doi: 10.1016/j.ejrad.2012.11.012
129. Morton G, Schuster A, Jogiya R, Kutty S, Beerbaum P, Nagel E. Inter-study reproducibility of cardiovascular magnetic resonance myocardial feature tracking. J Cardiovasc Magn Reson. (2012) 14:43. doi: 10.1186/1532-429X-14-43
130. Kempny A, Fernández-Jiménez R, Orwat S, Schuler P, Bunck AC, Maintz D, et al. Quantification of biventricular myocardial function using cardiac magnetic resonance feature tracking, endocardial border delineation and echocardiographic speckle tracking in patients with repaired tetralogy of fallot and healthy controls. J Cardiovasc Magn Reson. (2012) 14:32. doi: 10.1186/1532-429X-14-32
131. Lapinskas T, Grune J, Zamani SM, Jeuthe S, Messroghli D, Gebker R, et al. Cardiovascular magnetic resonance feature tracking in small animals – a preliminary study on reproducibility and sample size calculation. BMC Med Imaging. (2017) 17:51. doi: 10.1186/s12880-017-0223-7
132. Kober F, Jao T, Troalen T, Nayak KS. Myocardial arterial spin labeling. J Cardiovasc Magn Reson. (2016) 18:22. doi: 10.1186/s12968-016-0235-4
133. Detre JA, Leigh JS, Williams DS, Koretsky AP. Perfusion imaging. Magn Reson Med. (1992) 23:37–45. doi: 10.1002/mrm.1910230106
134. Williams DS, Grandis DJ, Zhang W, Koretsky AP. Magnetic resonance imaging of perfusion in the isolated rat heart using spin inversion of arterial water. Magn Reson Med. (1993) 30:361–5. doi: 10.1002/mrm.1910300314
135. Kwiatkowski G, Kozerke S. Extended quantitative dynamic contrast-enhanced cardiac perfusion imaging in mice using accelerated data acquisition and spatially distributed, two-compartment exchange modeling. NMR Biomed. (2019) 32:e4123. doi: 10.1002/nbm.4123
136. Belle V, Kahler E, Waller C, Rommel E, Voll S, Hiller K, et al. In vivo quantitative mapping of cardiac perfusion in rats using a noninvasive MR spin-labeling method. J Magn Reson Imaging. (1998) 8:1240–5. doi: 10.1002/jmri.1880080610
137. Kober F, Iltis I, Cozzone PJ, Bernard M. Myocardial blood flow mapping in mice using high-resolution spin labeling magnetic resonance imaging: influence of ketamine/xylazine and isoflurane anesthesia. Magn Reson Med. (2005) 53:601–6. doi: 10.1002/mrm.20373
138. Kober F, Iltis I, Izquierdo M, Desrois M, Ibarrola D, Cozzone PJ, et al. High-resolution myocardial perfusion mapping in small animals in vivo by spin-labeling gradient-echo imaging. Magn Reson Med. (2004) 51:62–7. doi: 10.1002/mrm.10676
139. Vandsburger MH, Janiczek RL, Xu Y, French BA, Meyer CH, Kramer CM, et al. Improved arterial spin labeling after myocardial infarction in mice using cardiac and respiratory gated look-locker imaging with fuzzy C-means clustering. Magn Reson Med. (2010) 63:648–57. doi: 10.1002/mrm.22280
140. Abeykoon S, Sargent M, Wansapura JP. Quantitative myocardial perfusion in mice based on the signal intensity of flow sensitized CMR. J Cardiovasc Magn Reson. (2012) 14:73. doi: 10.1186/1532-429X-14-73
141. Troalen T, Capron T, Cozzone PJ, Bernard M, Kober F. Cine-ASL: a steady-pulsed arterial spin labeling method for myocardial perfusion mapping in mice. Part I. Experimental study. Magn Reson Med. (2013) 70:1389–98. doi: 10.1002/mrm.24565
142. Coolen BF, Moonen RPM, Paulis LEM, Geelen T, Nicolay K, Strijkers GJ. Mouse myocardial first-pass perfusion MR imaging. Magn Reson Med. (2010) 64:1658–63. doi: 10.1002/mrm.22588
143. van Nierop BJ, Coolen BF, Dijk WJR, Hendriks AD, de Graaf L, Nicolay K, et al. Quantitative first-pass perfusion MRI of the mouse myocardium: MRI. Magn Reson Med. (2013) 69:1735–44. doi: 10.1002/mrm.24424
144. van Nierop BJ, Coolen BF, Bax NA, Dijk WJR, van Deel ED, Duncker DJ, et al. Myocardial perfusion MRI shows impaired perfusion of the mouse hypertrophic left ventricle. Int J Cardiovasc Imaging. (2014) 30:619–28. doi: 10.1007/s10554-014-0369-0
145. Bulluck H, Maestrini V, Rosmini S, Abdel-Gadir A, Treibel TA, Castelletti S, et al. Myocardial T1 mapping. Circ J. (2015) 79:487–94. doi: 10.1253/circj.CJ-15-0054
146. Waghorn B, Edwards T, Yang Y, Chuang K-H, Yanasak N, Hu TCC. Monitoring dynamic alterations in calcium homeostasis by T 1 -weighted and T 1 -mapping cardiac manganese-enhanced MRI in a murine myocardial infarction model. NMR Biomed. (2008) 21:1102–11. doi: 10.1002/nbm.1287
147. Schneider JE, Cassidy PJ, Lygate C, Tyler DJ, Wiesmann F, Grieve SM, et al. Fast, high-resolution in vivo cine magnetic resonance imaging in normal and failing mouse hearts on a vertical 11.7 T system. J Magn Reson Imaging. (2003) 18:691–701. doi: 10.1002/jmri.10411
148. Coelho-Filho OR, Shah RV, Mitchell R, Neilan TG, Moreno H, Simonson B, et al. Quantification of cardiomyocyte hypertrophy by cardiac magnetic resonance: implications for early cardiac remodeling. Circulation. (2013) 128:1225–33. doi: 10.1161/CIRCULATIONAHA.112.000438
149. Li W, Griswold M, Yu X. Rapid T1 mapping of mouse myocardium with saturation recovery look-locker method. Magn Reson Med. (2010) 64:1296–303. doi: 10.1002/mrm.22544
150. Li W, Griswold M, Yu X. Fast cardiac T 1 mapping in mice using a model-based compressed sensing method. Magn Reson Med. (2012) 68:1127–34. doi: 10.1002/mrm.23323
151. Jiang K, Li W, Li W, Jiao S, Castel L, Van Wagoner DR, et al. Rapid multislice T 1 mapping of mouse myocardium: application to quantification of manganese uptake in α-Dystrobrevin knockout mice: myocardial Mn 2+ uptake in α-Dystrobrevin knockout mice by rapid T 1 mapping. Magn Reson Med. (2015) 74:1370–9. doi: 10.1002/mrm.25533
152. Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified look-Locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn Reson Med. (2004) 52:141–6. doi: 10.1002/mrm.20110
153. Nezafat M, Ramos IT, Henningsson M, Protti A, Basha T, Botnar RM. Improved segmented modified look-Locker inversion recovery T1 mapping sequence in mice. PLoS ONE. (2017) 12:e0187621. doi: 10.1371/journal.pone.0187621
154. Castets CR, Ribot EJ, Lefrançois W, Trotier AJ, Thiaudière E, Franconi J-M, et al. Fast and robust 3D T 1 mapping using spiral encoding and steady RF excitation at 7 T: application to cardiac manganese enhanced MRI (MEMRI) in mice: spiral encoding mouse heart 3D T 1 mapping. NMR Biomed. (2015) 28:881–9. doi: 10.1002/nbm.3327
155. Neilan TG, Coelho-Filho OR, Shah RV, Abbasi SA, Heydari B, Watanabe E, et al. Myocardial extracellular volume fraction from T1 measurements in healthy volunteers and Mice. JACC Cardiovasc Imaging. (2013) 6:672–83. doi: 10.1016/j.jcmg.2012.09.020
156. Higgins CB, Herfkens R, Lipton JM, Sievers R, Sheldon P, Kaufman L, et al. Nuclear magnetic resonance imaging of acute myocardial infarction in dogs: alterations in magnetic relaxation times. Am J Cardiol. (1983) 52:184–8. doi: 10.1016/0002-9149(83)90093-0
157. Bun SS, Kober F, Jacquier A, Espinosa L, Kalifa J, Bonzi MF, et al. Value of in vivo T2 measurement for myocardial fibrosis assessment in diabetic mice at 11.75 T. Invest Radiol. (2012) 47:319–23. doi: 10.1097/RLI.0b013e318243e062
158. Kim PK, Hong YJ, Im DJ, Suh YJ, Park CH, Kim JY, et al. Myocardial T1 and T2 mapping: techniques and clinical applications. Korean J Radiol. (2017) 18:113. doi: 10.3348/kjr.2017.18.1.113
159. Giri S, Chung YC, Merchant A, Mihai G, Rajagopalan S, Raman SV, et al. T2 quantification for improved detection of myocardial edema. J Cardiovasc Magn Reson. (2009) 11:56. doi: 10.1186/1532-429X-11-56
160. Coolen BF, Simonis FFJ, Geelen T, Moonen RPM, Arslan F, Paulis LEM, et al. Quantitative T 2 mapping of the mouse heart by segmented MLEV phase-cycled T 2 preparation: T 2 mapping of the mouse heart. Magn Reson Med. (2014) 72:409–17. doi: 10.1002/mrm.24952
161. Chen Y, Li W, Jiang K, Wang CY, Yu X. Rapid T 2 mapping of mouse heart using the carr-purcell-meiboom-gill sequence and compressed sensing reconstruction: rapid T 2 mapping of mouse heart. J Magn Reson Imaging. (2016) 44:375–82. doi: 10.1002/jmri.25175
162. Jackson LH, Vlachodimitropoulou E, Shangaris P, Roberts TA, Ryan TM, Campbell-Washburn AE, et al. Non-invasive MRI biomarkers for the early assessment of iron overload in a humanized mouse model of β-thalassemia. Sci Rep. (2017) 7:43439. doi: 10.1038/srep43439
163. van Nierop BJ, Bax NAM, Nelissen JL, Arslan F, Motaal AG, de Graaf L, et al. Assessment of myocardial fibrosis in mice using a T2*-weighted 3D radial magnetic resonance imaging sequence. PLoS ONE. (2015) 10:e0129899. doi: 10.1371/journal.pone.0129899
164. Sosnovik DE, Schellenberger EA, Nahrendorf M, Novikov MS, Matsui T, Dai G, et al. Magnetic resonance imaging of cardiomyocyte apoptosis with a novel magneto-optical nanoparticle. Magn Reson Med. (2005) 54:718–24. doi: 10.1002/mrm.20617
165. Sosnovik DE, Nahrendorf M, Deliolanis N, Novikov M, Aikawa E, Josephson L, et al. Fluorescence tomography and magnetic resonance imaging of myocardial macrophage infiltration in infarcted myocardium in vivo. Circulation. (2007) 115:1384–91. doi: 10.1161/CIRCULATIONAHA.106.663351
166. Zhou R, Idiyatullin D, Moeller S, Corum C, Zhang H, Qiao H, et al. SWIFT detection of SPIO-labeled stem cells grafted in the myocardium. Magn Reson Med. (2010) 63:1154–61. doi: 10.1002/mrm.22378
167. Cunningham CH, Arai T, Yang PC, McConnell MV, Pauly JM, Conolly SM. Positive contrast magnetic resonance imaging of cells labeled with magnetic nanoparticles. Magn Reson Med. (2005) 53:999–1005. doi: 10.1002/mrm.20477
168. Mani V, Briley-Saebo KC, Itskovich VV, Samber DD, Fayad ZA. Gradient echo acquisition for superparamagnetic particles with positive contrast (GRASP): sequence characterization in membrane and glass superparamagnetic iron oxide phantoms at 1.5T and 3T. Magn Reson Med. (2006) 55:126–35. doi: 10.1002/mrm.20739
169. Dahnke H, Liu W, Herzka D, Frank JA, Schaeffter T. Susceptibility gradient mapping (SGM): a new postprocessing method for positive contrast generation applied to superparamagnetic iron oxide particle (SPIO)-labeled cells. Magn Reson Med. (2008) 60:595–603. doi: 10.1002/mrm.21478
170. de Jong S, Zwanenburg JJ, Visser F, der Nagel R van, van Rijen HV, Vos MA, et al. Direct detection of myocardial fibrosis by MRI. J Mol Cell Cardiol. (2011) 51:974–9. doi: 10.1016/j.yjmcc.2011.08.024
171. Doltra A, Amundsen B, Gebker R, Fleck E, Kelle S. Emerging concepts for myocardial late gadolinium enhancement MRI. CCR. (2013) 9:185–90. doi: 10.2174/1573403X113099990030
172. Kim RJ, Chen E-L, Lima JAC, Judd RM. Myocardial Gd-DTPA kinetics determine MRI contrast enhancement and reflect the extent and severity of myocardial injury after acute reperfused infarction. Circulation. (1996) 94:3318–26. doi: 10.1161/01.CIR.94.12.3318
173. Yang Z, Berr SS, Gilson WD, Toufektsian M-C, French BA. Simultaneous evaluation of infarct size and cardiac function in intact mice by contrast-enhanced cardiac magnetic resonance imaging reveals contractile dysfunction in noninfarcted regions early after myocardial infarction. Circulation. (2004) 109:1161–7. doi: 10.1161/01.CIR.0000118495.88442.32
174. Thomas D, Dumont C, Pickup S, Misselwitz B, Zhou R, Horowitz J, et al. T1-weighted cine FLASH is superior to IR imaging of post-infarction myocardial viability at 4.7T. J Cardiovasc Magn Reson. (2006) 8:345–52. doi: 10.1080/10976640500451986
175. Ojha N, Roy S, Radtke J, Simonetti O, Gnyawali S, Zweier JL, et al. Characterization of the structural and functional changes in the myocardium following focal ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. (2008) 294:H2435–43. doi: 10.1152/ajpheart.01190.2007
176. Buonincontri G, Methner C, Krieg T, Carpenter TA, Sawiak SJ. A fast protocol for infarct quantification in mice: fast LGE protocol in Mice. J Magn Reson Imaging. (2013) 38:468–73. doi: 10.1002/jmri.24001
177. Price AN, Cheung KK, Lim SY, Yellon DM, Hausenloy DJ, Lythgoe MF. Rapid assessment of myocardial infarct size in rodents using multi-slice inversion recovery late gadolinium enhancement CMR at 9.4T. J Cardiovasc Magn Reson. (2011) 13:44. doi: 10.1186/1532-429X-13-44
178. Protti A, Sirker A, Shah AM, Botnar R. Late gadolinium enhancement of acute myocardial infarction in mice at 7T: cine-FLASH versus inversion recovery. J Magn Reson Imaging. (2010) 32:878–86. doi: 10.1002/jmri.22325
179. Luo M, Anderson ME. Mechanisms of altered Ca 2+ handling in heart failure. Circ Res. (2013) 113:690–708. doi: 10.1161/CIRCRESAHA.113.301651
180. Fernandes JL, Storey P, da Silva JA, de Figueiredo GS, Kalaf JM, Coelho OR. Preliminary assessment of cardiac short term safety and efficacy of manganese chloride for cardiovascular magnetic resonance in humans. J Cardiovasc Magn Reson. (2011) 13:6. doi: 10.1186/1532-429X-13-6
181. Skjold A, Kristoffersen A, Vangberg TR, Haraldseth O, Jynge P, Larsson HB. An apparent unidirectional influx constant for manganese as a measure of myocardial calcium channel activity. J Magn Reson Imaging. (2006) 24:1047–55. doi: 10.1002/jmri.20736
182. Skjold A, Vangberg TR, Kristoffersen A, Haraldseth O, Jynge P, Larsson HBW. Relaxation enhancing properties of MnDPDP in human myocardium. J Magn Reson Imaging. (2004) 20:948–52. doi: 10.1002/jmri.20200
183. Toma I, Kim PJ, Dash R, McConnell MV, Nishimura D, Harnish P, et al. Telmisartan in the diabetic murine model of acute myocardial infarction: dual contrast manganese-enhanced and delayed enhancement MRI evaluation of the peri-infarct region. Cardiovasc Diabetol. (2016) 15:24. doi: 10.1186/s12933-016-0348-y
184. Spath NB, Lilburn DML, Gray GA, Le Page LM, Papanastasiou G, Lennen RJ, et al. Manganese-enhanced T 1 mapping in the myocardium of normal and infarcted hearts. Contrast Media Mol Imaging. (2018) 25:9641527. doi: 10.1155/2018/9641527
185. Hu TC-C, Bao W, Lenhard SC, Schaeffer TR, Yue T, Willette RN, et al. Simultaneous assessment of left-ventricular infarction size, function and tissue viability in a murine model of myocardial infarction by cardiac manganese-enhanced magnetic resonance imaging (MEMRI). NMR Biomed. (2004) 17:620–6. doi: 10.1002/nbm.933
186. Spath NB, Thompson G, Baker AH, Dweck MR, Newby DE, Semple SIK. Manganese-enhanced MRI of the myocardium. Heart. (2019) 105:1695–700. doi: 10.1136/heartjnl-2019-315227
187. Hsu EW, Muzikant AL, Matulevicius SA, Penland RC, Henriquez CS. Magnetic resonance myocardial fiber-orientation mapping with direct histological correlation. Am J Physiol. (1998) 274:H1627–34. doi: 10.1152/ajpheart.1998.274.5.H1627
188. Jiang Y, Pandya K, Smithies O, Hsu EW. Three-dimensional diffusion tensor microscopy of fixed mouse hearts. Magn Reson Med. (2004) 52:453–60. doi: 10.1002/mrm.20191
189. McGill LA, Ferreira PF, Scott AD, Nielles-Vallespin S, Giannakidis A, Kilner PJ, et al. Relationship between cardiac diffusion tensor imaging parameters and anthropometrics in healthy volunteers. J Cardiovasc Magn Reson. (2015) 18:2. doi: 10.1186/s12968-015-0215-0
190. Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys J. (1994) 66:259–67. doi: 10.1016/S0006-3495(94)80775-1
191. Saggu R, Schumacher T, Gerich F, Rakers C, Tai K, Delekate A, et al. Astroglial NF-kB contributes to white matter damage and cognitive impairment in a mouse model of vascular dementia. Acta Neuropathol Commun. (2016) 4:76. doi: 10.1186/s40478-016-0350-3
192. Argyridis I, Li W, Johnson GA, Liu C. Quantitative magnetic susceptibility of the developing mouse brain reveals microstructural changes in the white matter. NeuroImage. (2014) 88:134–42. doi: 10.1016/j.neuroimage.2013.11.026
193. Scollan DF, Holmes A, Winslow R, Forder J. Histological validation of myocardial microstructure obtained from diffusion tensor magnetic resonance imaging. Am J Physiol. (1998) 275:H2308–18. doi: 10.1152/ajpheart.1998.275.6.H2308
194. Chen J, Song S-K, Liu W, McLean M, Allen JS, Tan J, et al. Remodeling of cardiac fiber structure after infarction in rats quantified with diffusion tensor MRI. Am J Physiol Heart Circ Physiol. (2003) 285:H946–54. doi: 10.1152/ajpheart.00889.2002
195. Huang S, Mekkaoui C, Chen HH, Wang R, Ngoy S, Liao R, et al. Serial diffusion tensor MRI and tractography of the mouse heart in-vivo: impact of ischemia on myocardial microstructure. J Cardiovasc Magn Reson. (2011) 13:O28. doi: 10.1186/1532-429X-13-S1-O28
196. Sosnovik DE, Mekkaoui C, Huang S, Chen HH, Dai G, Stoeck CT, et al. Microstructural impact of ischemia and bone marrow–derived cell therapy revealed with diffusion tensor magnetic resonance imaging tractography of the heart in vivo. Circulation. (2014) 129:1731–41. doi: 10.1161/CIRCULATIONAHA.113.005841
197. Akki A, Gupta A, Weiss RG. Magnetic resonance imaging and spectroscopy of the murine cardiovascular system. Am J Physiol Heart Circ Physiol. (2013) 304:H633–48. doi: 10.1152/ajpheart.00771.2011
198. Tognarelli JM, Dawood M, Shariff MIF, Grover VPB, Crossey MME, Cox IJ, et al. Magnetic resonance spectroscopy: principles and techniques: lessons for clinicians. J Clin Exp Hepatol. (2015) 5:320–8. doi: 10.1016/j.jceh.2015.10.006
199. Abdurrachim D, Nabben M, Hoerr V, Kuhlmann MT, Bovenkamp P, Ciapaite J, et al. Diabetic db/db mice do not develop heart failure upon pressure overload: a longitudinal in vivo PET, MRI, and MRS study on cardiac metabolic, structural, and functional adaptations. Cardiovasc Res. (2017) 113:1148–60. doi: 10.1093/cvr/cvx100
200. Chacko VP, Aresta F, Chacko SM, Weiss RG. MRI/MRS assessment of in vivo murine cardiac metabolism, morphology, and function at physiological heart rates. Am J Physiol Heart Circ Physiol. (2000) 279:H2218–24. doi: 10.1152/ajpheart.2000.279.5.H2218
201. Gupta A, Chacko VP, Schär M, Akki A, Weiss RG. Impaired ATP kinetics in failing in vivo mouse heart. Circ Cardiovasc Imaging. (2011) 4:42–50. doi: 10.1161/CIRCIMAGING.110.959320
202. Bakermans AJ, Abdurrachim D, Moonen RPM, Motaal AG, Prompers JJ, Strijkers GJ, et al. Small animal cardiovascular MR imaging and spectroscopy. Prog Nucl Magn Reson Spectrosc. (2015) 88–89:1–47. doi: 10.1016/j.pnmrs.2015.03.001
203. Bakermans AJ, Abdurrachim D, van Nierop BJ, Koeman A, van der Kroon I, Baartscheer A, et al. In vivo mouse myocardial (31) P MRS using three-dimensional image-selected in vivo spectroscopy (3D ISIS): technical considerations and biochemical validations. NMR Biomed. (2015) 28:1218–27. doi: 10.1002/nbm.3371
204. Flögel U, Jacoby C, Gödecke A, Schrader J. In vivo 2D mapping of impaired murine cardiac energetics in NO-induced heart failure. Magn Reson Med. (2007) 57:50–8. doi: 10.1002/mrm.21101
205. Rodgers CT, Clarke WT, Snyder C, Vaughan JT, Neubauer S, Robson MD. Human cardiac 31 P magnetic resonance spectroscopy at 7 tesla: human cardiac 31 P MR spectroscopy at 7T. Magn Reson Med. (2014) 72:304–15. doi: 10.1002/mrm.24922
206. Faller KME, Lygate CA, Neubauer S, Schneider JE. 1H-MR spectroscopy for analysis of cardiac lipid and creatine metabolism. Heart Fail Rev. (2013). 18:657–68. doi: 10.1007/s10741-012-9341-z
207. Ugurbil K, Petein M, Maidan R, Michurski S, Cohn JN, From AH. High resolution proton NMR studies of perfused rat hearts. FEBS Lett. (1984) 167:73–8. doi: 10.1016/0014-5793(84)80835-2
208. Schneider JE, Tyler DJ, ten Hove M, Sang AE, Cassidy PJ, Fischer A, et al. In vivo cardiac1H-MRS in the mouse. Magn Reson Med. (2004) 52:1029–35. doi: 10.1002/mrm.20257
209. Klose U. Measurement sequences for single voxel proton MR spectroscopy. Eur J Radiol. (2008) 67:194–201. doi: 10.1016/j.ejrad.2008.03.023
210. Anzawa R, Bernard M, Tamareille S, Baetz D, Confort-Gouny S, Gascard JP, et al. Intracellular sodium increase and susceptibility to ischaemia in hearts from type 2 diabetic db/db mice. Diabetologia. (2006) 49:598–606. doi: 10.1007/s00125-005-0091-5
211. Imahashi K, London RE, Steenbergen C, Murphy E. Male/female differences in intracellular Na+regulation during ischemia/reperfusion in mouse heart. J Mol Cell Cardiol. (2004) 37:747–53. doi: 10.1016/j.yjmcc.2004.06.010
212. Maguire ML, Geethanath S, Lygate CA, Kodibagkar VD, Schneider JE. Compressed sensing to accelerate magnetic resonance spectroscopic imaging: evaluation and application to 23Na-imaging of mouse hearts. J Cardiovasc Magn Reson. (2015) 17:45. doi: 10.1186/s12968-015-0149-6
213. Hurd RE, Yen YF, Chen A, Ardenkjaer-Larsen JH. Hyperpolarized 13 C metabolic imaging using dissolution dynamic nuclear polarization. J Magn Reson Imaging. (2012) 36:1314–28. doi: 10.1002/jmri.23753
214. Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, et al. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc Natl Acad Sci USA. (2003) 100:10158–63. doi: 10.1073/pnas.1733835100
215. Kolwicz SC, Purohit S, Tian R. Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes. Circ Res. (2013) 113:603–16. doi: 10.1161/CIRCRESAHA.113.302095
216. Dodd MS, Ball V, Bray R, Ashrafian H, Watkins H, Clarke K, et al. In vivo mouse cardiac hyperpolarized magnetic resonance spectroscopy. J Cardiovasc Magn Reson. (2013) 15:19. doi: 10.1186/1532-429X-15-19
217. Atherton HJ, Dodd MS, Heather LC, Schroeder MA, Griffin JL, Radda GK, et al. Role of pyruvate dehydrogenase inhibition in the development of hypertrophy in the hyperthyroid rat heart: a combined magnetic resonance imaging and hyperpolarized magnetic resonance spectroscopy study. Circulation. (2011) 123:2552–61. doi: 10.1161/CIRCULATIONAHA.110.011387
218. Le Page LM, Ball DR, Ball V, Dodd MS, Miller JJ, Heather LC, et al. Simultaneous in vivo assessment of cardiac and hepatic metabolism in the diabetic rat using hyperpolarized MRS: In vivo assessment of both cardiac and hepatic diabetic metabolism. NMR Biomed. (2016) 29:1759–67. doi: 10.1002/nbm.3656
219. Miller JJ, Lau AZ, Nielsen PM, McMullen-Klein G, Lewis AJ, Jespersen NR, et al. Hyperpolarized [1,4-13C2]fumarate enables magnetic resonance-based imaging of myocardial necrosis. JACC Cardiovasc Imaging. (2018) 11:1594–606. doi: 10.1016/j.jcmg.2017.09.020
Keywords: cardiac MRI, cardiac MRS, imaging techniques, small animals, cardiovascular diseases, cardiac function
Citation: Li H, Abaei A, Metze P, Just S, Lu Q and Rasche V (2020) Technical Aspects of in vivo Small Animal CMR Imaging. Front. Phys. 8:183. doi: 10.3389/fphy.2020.00183
Received: 13 December 2019; Accepted: 27 April 2020;
Published: 10 June 2020.
Edited by:
Andreas Hess, University of Erlangen Nuremberg, GermanyReviewed by:
Yu-chung Norman Cheng, Wayne State University, United StatesAlbrecht Ingo Schmid, Medical University of Vienna, Austria
Copyright © 2020 Li, Abaei, Metze, Just, Lu and Rasche. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Volker Rasche, dm9sa2VyLnJhc2NoZUB1bmktdWxtLmRl
 Hao Li
Hao Li Alireza Abaei
Alireza Abaei Patrick Metze
Patrick Metze Steffen Just
Steffen Just Qinghua Lu2
Qinghua Lu2 Volker Rasche
Volker Rasche