- 1School of Biological and Behavioural Sciences, Queen Mary University of London, London, United Kingdom
- 2School of Science, University of Greenwich, Chatham, United Kingdom
The abundance and subcellular location of specific mRNA molecules can give rich information on bacterial cell biology and gene expression at the single-cell level. We have been using RNA Fluorescent in situ Hybridization (RNA-FISH) to probe for specific mRNA species in both unicellular and filamentous cyanobacteria. We have shown that the technique can be used to reveal the locations of membrane protein production and can also reveal heterogeneity in gene expression at the single-cell level, including patterns of gene expression within the filaments of heterocyst-forming cyanobacteria as they differentiate in diazotrophic conditions. However, the background fluorescence from pigments in cyanobacteria can cause problems, as can the resistance of heterocysts to permeabilization. Here we discuss the potential and the pitfalls of RNA-FISH as applied to cyanobacteria. We compare the information that can be obtained from RNA-FISH with that available from other techniques for probing gene expression.
1 Introduction to RNA-FISH
RNA Fluorescent in situ Hybridization (RNA-FISH) is a technique that allows the visualization of the abundance and location of RNA molecules in their native context in the cell. Synthetic single-stranded and fluorescently-tagged DNA molecules are used as probes that will hybridize with single-stranded RNA molecules that have complementary sequences, allowing the presence of the RNA to be detected in a fluorescence microscope. RNA-FISH differs from DNA-FISH in that the sample preparation does not include a step to denature double-stranded nucleic acids. Therefore, genomic DNA sequences generally do not hybridize with the fluorescent probe DNA, which will only find single-stranded RNA molecules. In order to allow the DNA probes into the cell, the cells must first be fixed with formaldehyde or a similar crosslinking agent, and then permeabilized with ethanol solution. So, RNA-FISH is not a live-cell technique but can be used to give “snapshots” of an mRNA species within the cell under particular conditions. Formaldehyde crosslinking is not instantaneous, but it is rapid, effectively preserving interactions with lifetimes longer than about 5 s (Schmiedeberg et al., 2009).
We have employed a variant known as single-molecule (sm) RNA-FISH (Skinner et al., 2013), in which an RNA species is detected by its hybridization to a set of short single-stranded DNA probes, each 20 bases long and designed to bind at sequential positions along the RNA, with binding sites separated by short intervals of RNA sequence (Orjalo et al., 2011). We designed probe sets using the proprietary Stellaris™ RNA–FISH Probe Designer program (https://www.biosearchtech.com/stellaris-designer) with a minimum separation of 2 bases between the probe binding sites and a GC content of around 50% (Mahbub et al., 2020). Each probe is linked to a fluorophore at its 3′ end, and the high specificity and sensitivity of the technique comes from the large number of fluorescent DNA molecules that can associate with each RNA molecule (Orjalo et al., 2011). A set of 40–48 probes is recommended, which in practice means that the RNA molecule to be detected should be at least 1,000 bases long. In principle the technique gives sufficient sensitivity to detect single mRNA molecules, and it is possible to count the number of mRNA molecules in each cell using a statistical approach that estimates the minimum step size in the fluorescent signals from many cells (Skinner et al., 2013). Here, we discuss what we have learned so far from applying RNA-FISH to cyanobacteria, including information on the sub-cellular location of RNA species and heterogeneity in gene expression at the cellular level. We discuss some specific issues with the use of RNA-FISH in cyanobacteria and we compare the information that can be obtained from RNA-FISH with the information provided by other approaches to probing gene expression.
2 Practicalities and problems with applying RNA-FISH to cyanobacteria
We found that the RNA-FISH labeling protocol developed by Skinner et al. for Escherichia coli (Skinner et al., 2013) works well in cyanobacteria with only minor modifications. To get consistent labeling, we found it beneficial to increase the exposure of the formaldehyde-fixed cells to 70% ethanol for permeabilization. For the unicellular cyanobacteria Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942 we incubated the cells with 70% ethanol for 1 h at room temperature followed by 2 h at 4°C (Mahbub et al., 2020). For the filamentous Anabaena sp. PCC 7120 we further increased the exposure to 70% ethanol (1 h at room temperature, 3–5 h at 4°C and then a further 1 h at room temperature (Wang et al., 2024).
The major issue in cyanobacteria as compared to E. coli, for example, is the high level of background fluorescence in cyanobacterial cells, which complicates the detection and quantitation of the FISH signal. FISH probes can be supplied with a range of fluorescent dyes, and it is obviously advisable to choose dyes whose fluorescence emission is as far removed as possible from the peak fluorescence of the photosynthetic pigments. We have been able to measure FISH signals in cyanobacteria using probes labeled with fluorescein (emission maximum about 515 nm) and carboxytetramethylrhodamine (TAMRA: emission maximum about 578 nm), with TAMRA giving more stable and consistent signals (Mahbub and Mullineaux, 2023). Fluorescein and TAMRA can be readily distinguished in a confocal microscope, so that it is possible to probe for two RNA species simultaneously (Mahbub and Mullineaux, 2023). Although both fluorophores fluoresce at shorter peak wavelengths than the photosynthetic pigments, background fluorescence remains an issue. The background comes mainly from the thylakoid membrane region, and probably comes from a mixture of photosynthetic pigments and other thylakoid-associated components such as cytochromes (Mahbub et al., 2020). Much of the chlorophyll in the cell is washed out during the permeabilization step of sample preparation, which involves incubation in 70% ethanol (Mahbub et al., 2020). However, the phycobilins, and likely other fluorescent thylakoid components, remain in place in the fixed and permeabilized cells, with fluorescence that extends right across the green window where we look for FISH signals. An advantage of this is that the thylakoid membranes can still be visualized very clearly in the treated cells, which allows determination of the positions of mRNA molecules in relation to the thylakoids (Mahbub et al., 2020; Mahbub and Mullineaux, 2023; Wang et al., 2024). However, the drawback is that quantitation of the FISH signal becomes harder. The problem can be reduced, but not eliminated, by careful choice of emission detection windows in the microscope: this is more easily achieved in laser scanning confocal microscopes where the emission window is set by a monochromator. In some circumstances, the background signal in the FISH detection window can be reliably computed as a predictable fraction of the longer-wavelength fluorescence from photosynthetic pigments, which means that “background” images can be calculated and subtracted from the FISH images to give a cleaner FISH signal (Mahbub et al., 2020). However, this method breaks down in some circumstances where the cells are undergoing changes in their pigment content, for example, in developing heterocysts (Wang et al., 2024). In any case, the background fluorescence means that we can only be confident about FISH signals from abundant RNA species, such as photosynthetic mRNAs (Mahbub et al., 2020; Mahbub and Mullineaux, 2023; Wang et al., 2024) and ribosomal RNAs (Hemm et al., 2024). So far, the background fluorescence has prevented us from applying the elegant statistical approach for counting mRNA molecules (Skinner et al., 2013). FISH signals from abundant RNA species can give a good indication of relative RNA levels in different cells and under different conditions, but we have not been able to quantify absolute numbers of RNA molecules per cell in cyanobacteria (Mahbub et al., 2020; Mahbub and Mullineaux, 2023).
Another FISH variant, Catalyzed Reporter Deposition or CARD-FISH (Kubota, 2013), offers a potential route to enhancing the brightness of FISH signals. It has been employed in environmental samples to identify cells of particular species (Ramm et al., 2012), however, the technique has rarely been employed for RNA detection at the sub-cellular level. CARD-FISH relies on the deposition of fluorophore-linked tyramide species catalyzed by horseradish peroxidase conjugated to the FISH probe (Kubota, 2013). The indirect detection method is liable to result in a loss of quantitative information and spatial resolution, and it necessitates a harsher permeabilization treatment to allow entry of the horseradish peroxidase (Kubota, 2013).
In most circumstances in cyanobacteria, we have not found permeabilization of cells to allow entry of the FISH probes to be an issue. We have successfully probed several species of cyanobacteria under multiple growth conditions, obtaining very consistent and reliable signals. Penetration of the probes through the thylakoid system into the central cytoplasm does not seem to be a problem, since some RNA species specifically light up in the central cytoplasm or at the proximal thylakoid membrane surface (Mahbub et al., 2020; Mahbub and Mullineaux, 2023; Hemm et al., 2024; Wang et al., 2024). However, we have found that penetration of FISH probes into the cell abruptly ceases in developing Anabaena heterocysts 20–24 h after nitrogen stepdown, although we can again detect FISH signals in some older heterocysts 48 h after nitrogen stepdown (Wang et al., 2024). The problem presumably arises from the thick laminated heterocyst surface layers, and we were not able to resolve it with longer exposure to 70% ethanol. Although we have not found the same issue in any other circumstances, it might potentially arise in other cases where the cell surrounds itself with particularly impenetrable surface layers. The difficulty of FISH labeling in heterocysts could potentially be addressed by using mutants deficient in heterocyst glycolipid production, however, such mutations can strongly affect heterocyst function (Garg and Maldener, 2021).
Wherever we have been able to verify RNA-FISH labeling in cyanobacteria with positive and negative controls, the technique has proved specific and reliable. Negative controls have included the use of null mutants for non-essential genes, including psbA in Synechocystis (Mahbub et al., 2020) and cox2 in Anabaena (Wang et al., 2024). Note that such negative controls are only reliable where the open reading frame has been completely removed: for example, we observed an RNA-FISH signal for Anabaena cox3 in a mutant where the gene had been interrupted rather than deleted, presumably due to production of a residual mRNA species with sufficient stability for detection (Wang et al., 2024). Positive controls have included the expected upregulation of psbA expression upon high light exposure in Synechocystis and Synechococcus (Mahbub et al., 2020) and the specific induction of cox2 and cox3 after nitrogen stepdown in Anabaena pro-heterocysts (Wang et al., 2024).
3 RNA-FISH in comparison to other techniques for probing mRNA and gene expression
3.1 RNA-FISH in comparison to transcriptomics
RNA-Seq and related approaches give rich information on changing patterns of gene expression [for examples in cyanobacteria, see (Voigt et al., 2014; Saha et al., 2016)]. Obviously, a full transcriptome gives a much more complete picture of gene expression than an RNA-FISH experiment probing only one, or at most two, mRNA species. However, RNA-FISH experiments provide complementary information that can fill some of the gaps in transcriptomics.
Firstly, RNA-FISH gives information on cell-to-cell variability in transcript levels. Such variations can arise stochastically, but nevertheless have strong biological implications, as has been shown for E. coli, for example (Engl et al., 2020). Cell-to-cell differences in gene expression can also arise in a more deterministic way as part of a developmental program in filamentous bacteria, for which heterocyst differentiation in the Nostocales is a classic example (Flores et al., 2019). It should be noted that many “unicellular” cyanobacteria are often found in colonies, flocs, biofilms or other assemblages (Rossi and De Philippis, 2015; Xiao et al., 2018; Conradi et al., 2019) and distinct patterns of gene expression according to position in the assemblage are almost inevitable. Emerging methods for single-cell bacterial transcriptomics (Homberger et al., 2022) potentially offer another way to probe heterogeneity in gene expression, but, to our knowledge, these methods have not yet been applied to cyanobacteria.
A second way in which RNA-FISH complements transcriptomics is by providing direct visualization of the (relative) level of an RNA species per cell. Transcriptomic data (and also data obtained from RT-PCR and Northern blots) are problematic to relate to expression at the cellular level – generally expression is judged to have changed if the level of a transcript changes relative to the total RNA pool or relative to the level of a “housekeeping” transcript that is presumed to be constant. However, in reality, both the cellular level of any individual RNA species and the total cellular RNA pool might well be variable depending on conditions: for example in cyanobacteria the level of transcriptional activity and the total transcript pool can decrease drastically during the night (Takano et al., 2015; Welkie et al., 2019). For the selected transcripts that are probed, RNA-FISH gives a direct readout of the transcript level per cell: it could be employed to normalize transcriptomic data to give a more accurate picture of which transcripts are actually increasing or decreasing per cell. For example, Figure 1A clearly demonstrates the increase in average cellular content of psbA transcripts following high light exposure, although the increase is not seen in every cell.
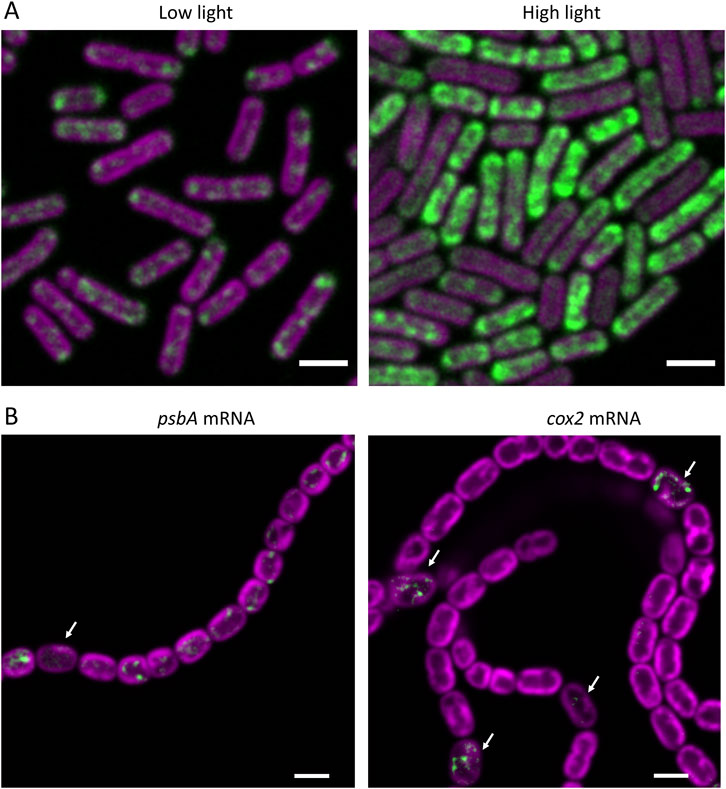
Figure 1. RNA-FISH as a probe of cellular heterogeneity in cyanobacteria. FISH signals are shown in green and fluorescence from the photosynthetic pigments in magenta. (A) psbA mRNA probed in cells of Synechococcus elongatus PCC 7942 grown in low light, or after 1 h exposure to high light. Scalebars 2 µm. Adapted from (Mahbub et al., 2020). (B) psbA and cox2 mRNAs probed in filaments of Anabaena (Nostoc) sp PCC 7120 8 h after transfer to nitrate-free medium to induce heterocyst differentiation. Arrows indicate specific cells (most likely pro-heterocysts) where psbA expression has been repressed or cox2 expression has been induced. Scalebars 3 µm. Adapted from (Wang et al., 2024).
Finally, of course, RNA-FISH gives information on the sub-cellular location of RNA species, as will be discussed below in Section 4.
3.2 RNA-FISH in comparison to live-cell mRNA visualization
An alternative method to visualize mRNA molecules within the cell involves the creation of a mutant in which the RNA species of interest carries a tandem repeat of the binding site for the phage MS2 coat protein. The cells are further engineered to express fluorescent protein-tagged MS2, which can then bind to the mRNA in vivo, generating fluorescent spots that can be observed in the microscope (Golding and Cox, 2004). The great benefit of this technique is that, unlike RNA-FISH, it can be used to track changing mRNA abundance and location with time in live cells (Golding and Cox, 2004). We are not aware of any reports of the use of technique in cyanobacteria, but it has been employed in E. coli to study the factors that lead to location of some mRNA species at the membrane (Nevo-Dinur et al., 2011; Sarmah et al., 2023). Drawbacks in comparison to RNA-FISH are that it requires the construction of a mutant line for every mRNA to be studied, it does not give such a direct measure of mRNA abundance, and the addition of the MS2 binding site array to the mRNA (and the binding of multiple fluorescent proteins) could potentially create artefacts in the lifecycle, motion, interactions and location of the mRNA species. In cyanobacteria, specific regions of some mRNA molecules at the 3′ end of the coding sequence seem to be particularly important for protein interaction, stability and sub-cellular location (Hemm et al., 2024), suggesting a strong potential for artefacts that could arise from adding tags to the mRNA sequence.
3.3 RNA-FISH in comparison to the use of reporter genes
Reporter genes have been extensively used to probe gene expression at the single-cell level in filamentous cyanobacteria (Videau et al., 2014), and gene expression as a function of time in studies of the circadian clock (Kondo et al., 1993). The principle is that the promoter region of a gene of interest is fused to a gene encoding a protein that generates a signal that is readily detectable in live cells. Reporters used have included GFP and its derivatives (Rajagopalan and Callahan, 2010), EcFbFP, a flavin mononucleotide-dependent fluorophore (Videau et al., 2014), and luciferase (Kondo et al., 1993). The great benefit of reporter genes is that they enable continuous monitoring of gene expression in live cells, which is obviously not available through RNA-FISH. However, unlike RNA-FISH, the reporters give no useful information at the sub-cellular level, and they do not give a direct readout of mRNA level. They give a good indicator of promoter activity, but do not give reliable information on factors such as the turnover and stability of the native mRNA, which may also be significant in controlling gene expression (Sakamoto and Bryant, 1997). The maturation of the reporter protein can result in a significant time-lag in observation of the onset of the gene expression [10–30 min for GFP (Drepper et al., 2010)] although luciferase appears significantly faster (Mazo-Vargas et al., 2014). The lifetime of the reporter protein must always be a complicating factor when observing the repression of gene expression. By contrast, mRNA-FISH can give a direct readout of mRNA levels in individual cells with a time resolution that is effectively limited by the timescale of formaldehyde fixation, which is around 5s (Schmiedeberg et al., 2009).
4 RNA-FISH as a probe of cellular heterogeneity in cyanobacteria
Our RNA-FISH experiments to date have provided indications of two kinds of cellular heterogeneity: stochastic heterogeneity in gene expression in unicellular cyanobacteria, and systematic heterogeneity as part of a developmental programme in filamentous cyanobacteria. Stochastic heterogeneity is revealed, for example, in RNA-FISH visualization of psbA mRNA encoding the Photosystem 2 D1 protein in the unicellular cyanobacteria Synechocystis sp PCC 6803 and Synechococcus sp PCC 7942. Levels of this mRNA species are rather variable from cell to cell, and differences become extremely pronounced in high light-treated cells (Figure 1A) and in the presence of inhibitors or translation. Both treatments boost mRNA content in some cells and reduce it in others (Mahbub et al., 2020). It is not clear whether the heterogeneity results from differences in transcription, or differences in mRNA stability, or both, but the observations highlight very clear heterogeneity in cellular mRNA content that would not be apparent in transcriptomic or qRT-PCR studies, for example,. The physiological significance of such heterogeneity in mRNA levels remains to be determined: there is no obvious correlation with cell pigmentation or the stage of the cell cycle, and cells were taken from well-shaken planktonic cultures in which all cells should have experienced the same environment (Mahbub et al., 2020). Underlying causes could include stochastic “bursty” transcription as observed in E. coli (Engl et al., 2020) or there might be a connection with circadian control of gene expression, as the circadian clock will be randomized in cells grown in continuous light. Expression of psbA is under strong circadian control (Kondo et al., 1993).
Developmental heterogeneity is illustrated by our RNA-FISH observations of mRNA species in filaments of Anabaena (Nostoc) sp PCC 7120 undergoing heterocyst differentiation following transfer to growth medium lacking combined nitrogen (Wang et al., 2024) (Figure 1B). Here, the RNA-FISH experiments reveal the onset of expression of the cox2 and cox3 genes specifically in developing heterocysts about 8 h after nitrogen stepdown and shutdown of psbA expression in developing heterocysts at a similar time. However, expression of psaA is maintained in developing heterocysts (Wang et al., 2024), underlining the importance of Photosystem I activity in heterocysts (Magnuson and Cardona, 2016). All these results are broadly in line with deductions from other approaches (Flores et al., 2019), but they illustrate the potential of RNA-FISH to probe developmental regulation in filamentous cyanobacteria. The technique could potentially be applied to other developmental processes, such as the generation of akinetes and hormogonia (Meeks et al., 2002). As compared to the use of reporter genes, for example, RNA-FISH does not require the production of mutants and it gives a more direct indicator of RNA levels and better time-resolution, as discussed in Section 3.3 above. However, note that only longer transcripts can be probed reliably, and access of the probes to the cell interior might be lost under some conditions, as in heterocysts at 20–48 h after nitrogen stepdown (see Section 2 above).
5 RNA-FISH as a probe of sub-cellular heterogeneity in cyanobacteria
A crucial feature of RNA-FISH is that it can give information (at optical resolution) on the sub-cellular location of mRNA species. The location of mRNAs can reveal important aspects of sub-cellular organization, including the location of sites of protein production. For membrane-integral thylakoid membrane proteins in cyanobacteria, RNA-FISH studies revealed that protein translation is confined to specific regions of the thylakoid membrane system, corresponding to the proximal thylakoid surfaces adjacent to the central cytoplasm and the nucleoid (Mahbub et al., 2020; Mahbub and Mullineaux, 2023; Wang et al., 2024) (Figure 2A). These specific thylakoid surfaces are also highly enriched in ribosomes, as demonstrated by RNA-FISH (Hemm et al., 2024), GFP-tagging (Mahbub and Mullineaux, 2023) and cryo-electron tomography (Rast et al., 2019). However, protein translation appears surprisingly delocalized on the proximal thylakoid surface, with translation of different mRNA species occurring in different patches of the membrane surface, even when the mRNAs encode different subunits of the same complex (Mahbub and Mullineaux, 2023) (Figure 2B). Thus, RNA-FISH gives a picture of the proximal thylakoid surfaces as specific translation and assembly regions, that are distinct from the mature photosynthetically functional membranes elsewhere in the thylakoid system (Eckardt et al., 2024). More broadly, RNA-FISH experiments suggest the translation of plasma membrane proteins at the plasma membrane (Mahbub and Mullineaux, 2023) and translation of Rubisco large subunits in situ at the carboxysomes (Mahbub et al., 2020). Therefore, FISH studies of RNA location can nicely complement studies of cell organization by cryo-electron tomography and fluorescent protein tagging to reveal how the complex architecture of cyanobacterial cells is established and maintained.
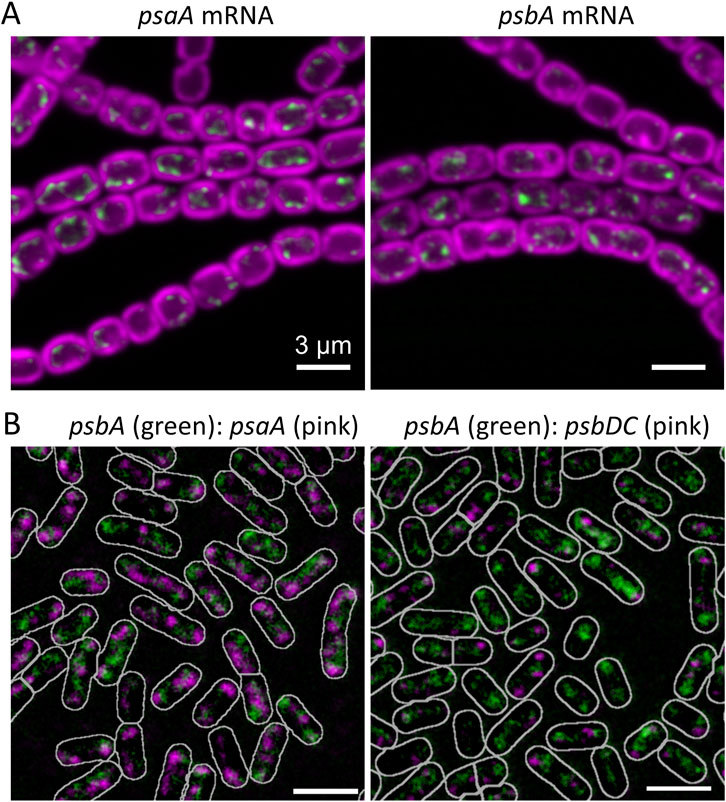
Figure 2. RNA-FISH as a probe of sub-cellular heterogeneity in cyanobacteria. (A) Core photosynthetic mRNAs probed in Anabaena (Nostoc) sp PCC 7120. FISH signals are shown in green and photosynthetic pigments in magenta. The mRNA species are concentrated in patches at the innermost thylakoid surface, marking likely translation/insertion zones for the photosynthetic complexes. Scalebars 3 µm. Adapted from (Wang et al., 2024). (B) Simultaneous FISH probing of pairs of photosynthetic mRNA species in Synechococcus elongatus PCC7942, suggesting that different mRNA species are translated at different places on the innermost thylakoid surface. Scalebars 2 µm and cell outlines shown in white. Adapted from (Mahbub and Mullineaux, 2023).
6 Conclusion
RNA-FISH studies can complement existing techniques for probing gene expression and developmental regulation in cyanobacteria. At the sub-cellular level, they can provide new insights into protein targeting and the biogenesis of protein complexes.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
CM: Writing–original draft, Writing–review and editing. KW: Writing–review and editing. MM: Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Relevant research by the authors was supported by Biotechnology and Biological Sciences Research Council (UKRI BBSRC) research grants BB/W001012/1 and BB/W019698/1 and by a scholarship (#201904910467) to KW from the China Scholarship Council.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Conradi, F. D., Zhou, R.-Q., Oeser, S., Schuergers, N., Wilde, A., and Mullineaux, C. W. (2019). Factors controlling floc formation and structure in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 201, 003444–e419. doi:10.1128/JB.00344-19
Drepper, T., Huber, R., Heck, A., Circolone, F., Hillmer, A.-K., Buchs, J., et al. (2010). Flavin mononucleotide-based fluorescent reporter proteins outperform green fluorescent protein-like proteins as quantitative in vivo real-time reporters. Appl. Environ. Microbiol. 76, 5990–5994. doi:10.1128/AEM.00701-10
Eckardt, N. A., Allahverdiyeva, Y., Alvarez, C. E., Büchel, C., Burlacot, A., Cardona, T., et al. (2024). Lighting the way: compelling open questions in photosynthesis research. Plant Cell 36, 3914–3943. doi:10.1093/plcell/koae203
Engl, C., Jovanovic, G., Brackston, R. D., Kotta-Loizou, I., and Buck, M. (2020). The route to transcription initiation determines the mode of transcriptional bursting in E. coli. Nat. Commun. 11, 2422. doi:10.1038/s41467-020-16367-6
Flores, E., Picossi, S., Valladares, A., and Herrero, A. (2019). Transcriptional regulation of development in heterocyst-forming cyanobacteria. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 1862, 673–684. doi:10.1016/j.bbagrm.2018.04.006
Garg, R., and Maldener, I. (2021). The dual role of the glycolipid envelope in different cell types of the multicellular cyanobacterium Anabaena variabilis ATCC 29413. Front. Microbiol. 12, 645028. doi:10.3389/fmicb.2021.645028
Golding, I., and Cox, E. C. (2004). RNA dynamics in live Escherichia coli cells. Proc. Natl. Acad. Sci. 101, 11310–11315. doi:10.1073/pnas.0404443101
Hemm, L., Lichtenberg, E., Tholen, S., Reimann, V., Kakazu, K., Machida, S., et al. (2024). The RRM domain-containing protein Rbp3 interacts with ribosomes and the 3’ends of mRNAs encoding photosynthesis proteins. bioRxiv, 2007–2024.
Homberger, C., Barquist, L., and Vogel, J. (2022). Ushering in a new era of single-cell transcriptomics in bacteria. Microlife 3, uqac020. doi:10.1093/femsml/uqac020
Kondo, T., Strayer, C. A., Kulkarni, R. D., Taylor, W., Ishiura, M., Golden, S. S., et al. (1993). Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. 90, 5672–5676. doi:10.1073/pnas.90.12.5672
Kubota, K. (2013). CARD-FISH for environmental microorganisms: technical advancement and future applications. Microbes Environ. 28, 3–12. doi:10.1264/jsme2.me12107
Magnuson, A., and Cardona, T. (2016). Thylakoid membrane function in heterocysts. Biochim. Biophys. Acta - Bioenerg. 1857, 309–319. doi:10.1016/j.bbabio.2015.10.016
Mahbub, M., Hemm, L., Yang, Y., Kaur, R., Carmen, H., Engl, C., et al. (2020). mRNA localization, reaction centre biogenesis and thylakoid membrane targeting in cyanobacteria. Nat. Plants 6, 1179–1191. doi:10.1038/s41477-020-00764-2
Mahbub, M., and Mullineaux, C. W. (2023). Locations of membrane protein production in a cyanobacterium. J. Bacteriol. 205, e0020923–23. doi:10.1128/jb.00209-23
Mazo-Vargas, A., Park, H., Aydin, M., and Buchler, N. E. (2014). Measuring fast gene dynamics in single cells with time-lapse luminescence microscopy. Mol. Biol. Cell 25, 3699–3708. doi:10.1091/mbc.E14-07-1187
Meeks, J. C., Campbell, E. L., Summers, M. L., and Wong, F. C. (2002). Cellular differentiation in the cyanobacterium Nostoc punctiforme. Arch. Microbiol. 178, 395–403. doi:10.1007/s00203-002-0476-5
Nevo-Dinur, K., Nussbaum-Shochat, A., Ben-Yehuda, S., and Amster-Choder, O. (2011). Translation-independent localization of mRNA in E. coli. Science 331, 1081–1084. doi:10.1126/science.1195691
Orjalo, A., Johansson, H. E., and Ruth, J. L. (2011). StellarisTM fluorescence in situ hybridization (FISH) probes: a powerful tool for mRNA detection. Nat. Methods 8, i–ii. doi:10.1038/nmeth.f.349
Rajagopalan, R., and Callahan, S. M. (2010). Temporal and spatial regulation of the four transcription start sites of hetR from Anabaena sp. strain PCC 7120. J. Bacteriol. 192, 1088–1096. doi:10.1128/jb.01297-09
Ramm, J., Lupu, A., Hadas, O., Ballot, A., Rücker, J., Wiedner, C., et al. (2012). A CARD-FISH protocol for the identification and enumeration of cyanobacterial akinetes in lake sediments. FEMS Microbiol. Ecol. 82, 23–36. doi:10.1111/j.1574-6941.2012.01401.x
Rast, A., Schaffer, M., Albert, S., Wan, W., Pfeffer, S., Beck, F., et al. (2019). Biogenic regions of cyanobacterial thylakoids form contact sites with the plasma membrane. Nat. Plants 5, 436–446. doi:10.1038/s41477-019-0399-7
Rossi, F., and De Philippis, R. (2015). Role of cyanobacterial exopolysaccharides in phototrophic biofilms and in complex microbial mats. Life 5, 1218–1238. doi:10.3390/life5021218
Saha, R., Liu, D., Hoynes-O’Connor, A., Liberton, M., Yu, J., Bhattacharyya-Pakrasi, M., et al. (2016). Diurnal regulation of cellular processes in the cyanobacterium Synechocystis sp. strain PCC 6803: insights from transcriptomic, fluxomic, and physiological analyses. MBio 7, e00464–e01128. doi:10.1128/mBio.00464-16
Sakamoto, T., and Bryant, D. A. (1997). Temperature-regulated mRNA accumulation and stabilization for fatty acid desaturase genes in the cyanobacterium Synechococcus sp. strain PCC 7002. Mol. Microbiol. 23, 1281–1292. doi:10.1046/j.1365-2958.1997.3071676.x
Sarmah, P., Shang, W., Origi, A., Licheva, M., Kraft, C., Ulbrich, M., et al. (2023). mRNA targeting eliminates the need for the signal recognition particle during membrane protein insertion in bacteria. Cell Rep. 42, 112140. doi:10.1016/j.celrep.2023.112140
Schmiedeberg, L., Skene, P., Deaton, A., and Bird, A. (2009). A temporal threshold for formaldehyde crosslinking and fixation. PLoS One 4, e4636. doi:10.1371/journal.pone.0004636
Skinner, S. O., Sepúlveda, L. A., Xu, H., and Golding, I. (2013). Measuring mRNA copy number in individual Escherichia coli cells using single-molecule fluorescent in situ hybridization. Nat. Protoc. 8, 1100–1113. doi:10.1038/nprot.2013.066
Takano, S., Tomita, J., Sonoike, K., and Iwasaki, H. (2015). The initiation of nocturnal dormancy in Synechococcus as an active process. BMC Biol. 13, 36–11. doi:10.1186/s12915-015-0144-2
Videau, P., Oshiro, R. T., Cozy, L. M., and Callahan, S. M. (2014). Transcriptional dynamics of developmental genes assessed with an FMN-dependent fluorophore in mature heterocysts of Anabaena sp. strain PCC 7120. Microbiology 160, 1874–1881. doi:10.1099/mic.0.078352-0
Voigt, K., Sharma, C. M., Mitschke, J., Lambrecht, S. J., Voß, B., Hess, W. R., et al. (2014). Comparative transcriptomics of two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity. ISME J. 8, 2056–2068. doi:10.1038/ismej.2014.57
Wang, K., Mahbub, M., Mastroianni, G., Valladares, A., and Mullineaux, C. W. (2024). mRNA localization and thylakoid protein biogenesis in the filamentous heterocyst-forming cyanobacterium Anabaena sp. PCC 7120. J. Bacteriol. 0, e0032824. doi:10.1128/jb.00328-24
Welkie, D. G., Rubin, B. E., Diamond, S., Hood, R. D., Savage, D. F., and Golden, S. S. (2019). A hard day’s night: cyanobacteria in diel cycles. Trends Microbiol. 27, 231–242. doi:10.1016/j.tim.2018.11.002
Keywords: cyanobacteria, fluorescent in situ hybridization, gene expression, heterocyst, mRNA, thylakoid membrane
Citation: Mullineaux CW, Wang K and Mahbub M (2024) RNA-FISH as a probe for heterogeneity at the cellular and subcellular levels in cyanobacteria. Front. Photobiol. 2:1511953. doi: 10.3389/fphbi.2024.1511953
Received: 15 October 2024; Accepted: 09 December 2024;
Published: 23 December 2024.
Edited by:
Radek Kana, Academy of Sciences of the Czech Republic (ASCR), CzechiaReviewed by:
Iskander M. Ibrahim, Towson University, United StatesKasia Piwosz, National Marine Fisheries Research Institute, Poland
Meri J. Eichner, Academy of Sciences of the Czech Republic (ASCR), Czechia
Copyright © 2024 Mullineaux, Wang and Mahbub. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Conrad W. Mullineaux, Yy5tdWxsaW5lYXV4QHFtdWwuYWMudWs=
 Conrad W. Mullineaux
Conrad W. Mullineaux Kexin Wang
Kexin Wang Moontaha Mahbub
Moontaha Mahbub