- Naturwissenschaftliche Fakultät, Institut für Botanik, Pflanzenphysiologie, Gottfried-Wilhelm-Leibniz-Universität Hannover, Hannover, Germany
Light from Sun has tremendously shaped the evolution of plants and represents one of their key triggers for proper morphogenesis and development. Energy from Sun light is converted by photosynthesis into chemical energy that ultimately drives all energy consuming processes in plants. Besides, Sun light provides information about environmental conditions or constraints and signals important parameters such as day length, time point of season, direction and intensity of illumination or spatial competition with neighbouring plants. Plants possess a sophisticated array of photoreceptors that perceive this information (photoperception) and initiate signalling pathways that control appropriate responses at developmental or physiological level. While the primary processes of photoperception are largely understood, many aspects of the subsequent signalling networks are still elusive and especially the interaction with other signalling networks is far from understood. Light represents also a highly versatile tool for scientists to study morphogenesis and development of plants by a steadily increasing number of remote sensing technologies that allow to observe plants in real time and high resolution (photodetection). Further, scientists now can even use the knowledge about photobiology and photoreceptors to construct synthetic tools that can be genetically introduced into plants to monitor internal processes (so-called biosensors). Recent technological developments in optogenetics even allow to generate tools that actively regulate gene expression or metabolism by selective illumination (photocontrol). In this perspective article we highlight progress in our understanding of light signalling and a number of selected technological improvements in photocontrol with a special focus on the areas of phytochrome signalling and plant optogenetics.
Introduction
Light is one of the most fundamental abiotic factors controlling and influencing growth, development and survival of plants. It is, however, of dual significance as it serves as reporter of environmental conditions that are detected by specific photoreceptors and, at the same time, as energy source for photosynthesis. Thus, it triggers multiple developmental responses and provides also the required energy for the underlying cellular activities. Further, both photo-controlled development and photosynthesis are affected in parallel by other external factors, notably temperature, which may change, modify or even block light-triggered cellular activities. This asks for integrated research approaches that respect the manifold spatio-temporal interactions and interceptions of various external factors with the light-controlled processes in plants. While the molecular structure and physiological involvement of photoreceptors is largely understood, many aspects of their regulation and their effective role in molecular signalling networks are still under investigation. Here we highlight some recent breakthroughs of our knowledge in these fields of research and provide a number of perspectives on promising technological improvements in the area of plant photobiology.
Novel factors in phytochrome signalling networks
Plants sense environmental light information via an array of photoreceptors that detect specific wavelengths in the UV/blue and red light range which largely overlap with the photosynthetic active radiation (400–700 nm). The group of UV/blue light receptors comprises UVR8, Cry1-3, Phot1 and 2 and members of the ZTL family which are involved in a vast range of physiological responses including UV stress response, light intensity reactions, chloroplast movements, stomata opening control, circadian rhythmicity and photoperiodic control of flowering and development (Ponnu and Hoecker, 2022). Many of these photoreceptors have orthologues in bacteria, fungi and animals indicating a common evolutionary origin. The group of red light photoreceptors comprises the family of phytochromes, a plant specific photoreceptor family with cyanobacterial ancestry (Lamparter, 2004). They control essential developmental steps such as seed germination, photomorphogenic programs of seedling development or shade avoidance reactions in dense plant stands with high competition for incident light (Cheng et al., 2021).
Upon illumination, phytochromes (Phys) convert from their inactive Pr-to their active Pfr-form, enter the nucleus and form so-called photobodies (PBs) via interaction with other proteins (Figure 1). Phy target proteins, such as Phytochrome interaction factors (PIFs) 1 and 3, are then degraded releasing the PIF-dependent repression of photomorphogenesis (PM) genes. Simultaneously, Phys disrupt the CONSTITUTIVE PHOTOMORPHOGENIC1/SUPPRESSOR OF PHYA-105 (COP1/SPA1) mediated degradation of PM-promoting factors, such as ELONGATED HYPOCOTYL 5 (HY5) and HY5-HOMOLOG (HYH) (Lu et al., 2015; Sheerin et al., 2015). PBs can be directly observed by microscopy in Arabidopsis lines stably transformed with green fluorescence protein (GFP) tagged PhyB (PBG) (Yamaguchi et al., 1999). They exhibit dynamic changes in size and number during PM and likely form microdomains where regulator proteins condense and jointly participate in the initiation and regulation of PM. The physical nature and protein composition of PBs remained elusive for long time, but recent reports shed more light on their structure. PBs appear as discrete, but not membrane-delineated bodies within the nucleus and are formed due to liquid-liquid phase separation (LLPS) of PhyB (Chen et al., 2022). Isolation of PBG-particles using fluorescence-activated particle sorting (FAPS) combined with mass spectrometry identified proteins associated with PB formation or marked for PB-mediated degradation (Kim et al., 2023). This included direct PhyB interaction partners as well as proteins indirectly recruited to PBs. However, several PhyB interaction partners known to influence PB-formation (see below) where not identified. Either i) these proteins were too low abundant under the experimental conditions, or ii) the size-selection of FAPS was too stringent to deal with the size dynamics of PBs, or iii) PB protein composition varies in response to internal or external stimuli (light conditions/shading/daytime/daylength/temperature/hormones/tissue) (Geilen and Bohmer, 2015; Legris et al., 2016; Hahm et al., 2020) or iv) they are involved in PB-formation without being a structural component of it. Future PB-isolation from plants grown under different conditions, thus, is needed to solve these questions.
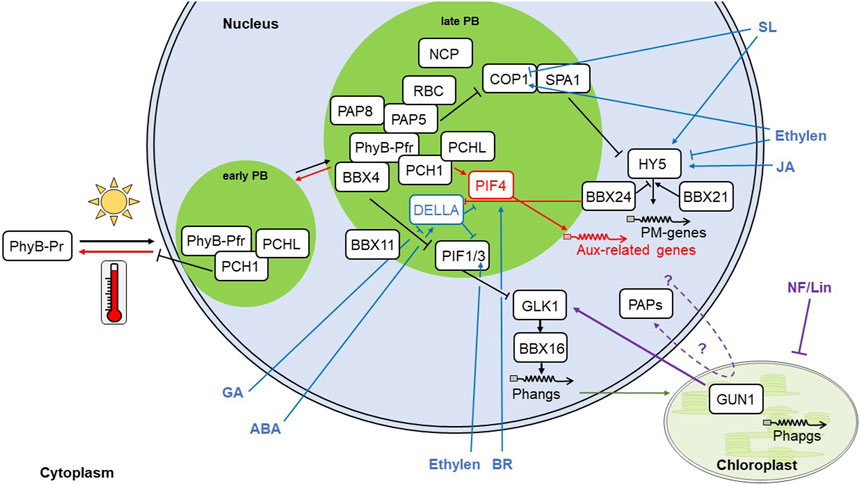
FIGURE 1. Regulators and signalling pathways in PM. Light triggers PhyB-Pr-photoconversion into its active form Pfr. Pfr translocates to the nucleus and forms early nuclear PBs. PB-formation is supported by PCH1 and PCHL, which prevents thermal backconversion of Pfr into Pr. Prolonged illumination initiates late PB formation, which is dependent on PAP5 and its interaction partners PAP8, RBC and NCP as well as the B-Box proteins BBX 4 and, likely, BBX11. Within PBs, PIFs are hampered in binding to their target genes and become degraded, thus, releasing their repression of PM genes, such as GLK1, which mediate gene expression of Phangs through BBX16. Simultaneously, COP1/SPA1 become inactivated, leading to the stabilization of HY5 protein and subsequent expression PM genes. HY5 expression is positively regulated by BBX21 and negatively by BBX24. The concerted expression of PIF and HY5 target genes leads to repression of hypocotyl elongation and activates cotyledon opening. Further, nuclear and plastid encoded photosynthesis associated genes (Phangs, green arrow; and Phapgs) are expressed and induce the build-up of chloroplasts. The PM pathway (black arrows) can be altered through other pathways that converge at distinct points. High ambient temperatures induce thermomorphogenesis (red symbols) by scaling up thermal conversion of Pfr, thus stabilizing PIF4, which in turn induces expression of auxin-related genes leading to cell elongation. This pathway largely coincides with the shade avoidance response. Retrograde signalling (violett arrow) is mediated through GUN1, which, together with GLK1 act on BBX16 and Phang expression. Phytohormones are not only regulated by the PM pathway, but regulate also PM (dark blue arrows). GA (gibberellic acid) reduces and ABA (abscisic acid) induces DELLAs, which in turn repress PIFs. Under high ambient temperatures, however, BBX24 binds to DELLAs and derepresses PIF4 action. Through BRZ1, BR promotes PIF4 action. SL (strigolactones) retain COP1 in the cytoplasm and promote HY5 expression. Ethylene promotes nuclear COP1 localization, stabilizes PIFs and leads to HY5 degradation. JA (jasmonic acid) through the MYC2 transcription factor promotes HY5 expression. Protein symbols that are in contact represent experimentally shown interaction. Lines with arrow heads represent promotion, lines with block represent repression. NF, Norflurazon; Lin, Lincomycin; Aux, auxin; BR, brassinosteroids. For other abbreviations see text.
During PM-initiation, early PB formation relies on the presence of PHOTOPERIODIC CONTROL OF HYPOCOTYL1 (PCH1) and likely its homologue PCH1-LIKE (PCHL) (Huang et al., 2019). PCH1/PCHL promote PM through the stabilization of PBs by inhibiting the thermal reversion of Pfr to Pr (Enderle et al., 2017; Huang et al., 2019). Further, the formation of late PBs depends on the presence of a number of novel regulators including PLASTID-ENCODED RNA POLYMERASE (PEP) ASSOCIATED PROTEIN 5 (PAP5)/Hemera and PAP8/pTAC6 (Chen et al., 2010; Liebers et al., 2020). Inactivation of these regulators blocks the transition from early to late PBs indicating that the structural dynamics of PBs are required for PM. PAPs are 12 nuclear-encoded plastid proteins that are crucial for the formation of the PEP-complex and the proper build-up of chloroplasts (Pfannschmidt et al., 2015). However, six PAPs, including PAP5 and PAP8, contain a nuclear localization signal that permits them to enter the nucleus and to participate in PB-mediated PM control providing nucleo-plastidic regulation of chloroplast build-up (Pfannschmidt et al., 2015; Chambon et al., 2022). Interestingly, both nuclear and plastid versions of PAP5/Hemera and PAP8/pTAC6 display similar sizes suggesting a plastid passage for removal of the chloroplast transit peptide in both protein versions (Chen et al., 2010; Liebers et al., 2020). Similar observations were reported also for other proteins (Grabowski et al., 2008; Isemer et al., 2012; Yang et al., 2019; Yoo et al., 2019). Whether or not this hypothesis holds true requires further investigations and a number of potential export mechanisms from plastids have been discussed (Krause et al., 2012).
Two further dually localized proteins, NUCLEAR CONTROL OF PEP ACTIVITY and REGULATOR OF CHLOROPLAST BIOGENESIS (NCP and RCB), were identified in a genetic screen (Yang et al., 2019; Yoo et al., 2019). Both are connected to PB-controlled PM since RCB was shown to interact with PAP5/Hemera, and ncp as well as rcb mutants lost the ability to form late PBs. Furthermore, null alleles of these mutants display phenocopies of PAP-inactivation mutants. Interestingly, inactivation of all four PIFs (pifq) releases nuclear repression of hypocotyl elongation of pap5/hemera, rcb and ncp (Qiu et al., 2015; Yang et al., 2019; Yoo et al., 2019). The build-up of the PEP-complex, however, remains impaired in pap and ncp mutants and the plants stay albino (Qiu et al., 2015; Yang et al., 2019; Liebers et al., 2020). Rcb-mutants, in contrary, are able to form a PEP-complex and start to green (Yoo et al., 2019). Thus, PAPs and NCP affect PM both, through nuclear and plastid signaling events. RCB on the contrary seems to modulate PM mainly from the nucleus.
Another group of recently identified proteins involved in PM are B-box-type zinc finger domain proteins (BBX). Arabidopsis contains 32 BBX, contributing to various pathways, such as seed germination, flowering and circadian rhythmicity, and are involved in light signaling. BBX4 and 11 are able to interact with PhyB and PIFs (Heng et al., 2019; Song et al., 2021) pointing to a basal function in early PM. Interestingly, BBX4 is recruited to PBs through its interaction with PhyB, enhancing PhyB-promoted PIF3 degradation (Heng et al., 2019). In addition, BBX4 and 11 directly interact with PIFs preventing their binding to target genes. Similar interference with transcription factors was found further downstream in PM-signaling. The positive regulator of PM, BBX21, binds to the HY5 promoter and promotes HY5 expression, while the negative regulator of PM, BBX24 interacts with HY5 in order to prevent the HY5 association to its target gene chalcone synthase (Job et al., 2018; Xu et al., 2018).
The precise functional significance of PBs, however, remains unclear, since mutants lacking PCH1, PAP5, PAP8, NCP or RCB are still able to repress hypocotyl growth, de-hook, and open cotyledons upon illumination (Chen et al., 2010; Huang et al., 2016; Yang et al., 2019; Yoo et al., 2019; Liebers et al., 2020). It is feasible that PBs represent a protein interaction platform that may facilitate rapid reactions and a lack maybe only decelerates PM. Further, similar PB dynamics can be involved in apparently conflicting physiological reactions. Light and high ambient temperatures decrease the number of PBs, while their size increases. However, light triggers also formation of Pfr, which represses elongation growth, while higher ambient temperatures promote conversion to Pr and, thus, elongation (Van Buskirk et al., 2012; Hahm et al., 2020). Shade avoidance on the other hand involves hypocotyl elongation, similar to thermomorphogenesis (TM), however, in stark contrast to TM with a multitude of small PBs, comparable to the mutants of hmr-5, pap8, rcb and ncp (Chen et al., 2010; Trupkin et al., 2014; Yang et al., 2019; Yoo et al., 2019; Hahm et al., 2020; Liebers et al., 2020). In addition, PB dynamics seem also to depend on tissue-identity (Hahm et al., 2020). It, thus, remains to be investigated whether PBs of same size but found in different conditions are comprised of identical components.
Integration of light control with other signalling networks
Light signaling is intimately linked to phytohormone signaling at multiple levels. For instance, Phys mediate the breaking of seed dormancy through PHYTOCHROME INTERACTING FACTOR 3-LIKE 5 (PIL5) that directly controls transcription regulators of gibberellic acid (GA) and abscisic acid (ABA) (Oh et al., 2009; Seo et al., 2009). Phototropism relies on Phototropin-induced changes in auxin transport and might be enhanced by PhyA (Han et al., 2008; Liscum et al., 2014) and COP1 modulates root growth through changes in shoot-to-root auxin transport by repressing PIN1 transcription, as well as intercellular distribution of PIN 1 and 2 (Sassi et al., 2012). Interestingly, several important components of the light signaling network do not only combine light and hormone signaling, but are also responsive to both stimuli. PIF3 and PIF4 are post-translationally regulated by GA through DELLAs (de Lucas et al., 2008; Feng et al., 2008; Li et al., 2016) and by ABA through modulation of GA and DELLAs (Lorrai et al., 2018). At the transcriptional level, PIF4 was shown to be regulated by brassinosteroids via the transcription factor BRASSINAZOLE RESISTANT 1 (BZR1) (Bernardo-Garcia et al., 2014; Ibanez et al., 2018). Strigolactones retain COP1 in the cytoplasm and promote HY5 expression via MORE AXILLARY GROWTH 2 (MAX2) (Tsuchiya et al., 2010; Jia et al., 2014) and ethylene stabilizes PIFs and nuclear localization of COP1 in the light and induces HY5 degradation (Zhong et al., 2012; Yu et al., 2013). Jasmonic acid regulates HY5 transcription via the transcription factor MYC2 (Ortigosa et al., 2020). In roots, auxin negatively regulates greening due to the repression of HY5, while cytokinin promotes greening through the transcription factors GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM INVOLVED -like (GNL) and GOLDEN-like 2 (GLK2) (Kobayashi et al., 2017). Further, HY5 is also involved in germination (Chen et al., 2008; Yadukrishnan et al., 2020), entrainment of the circadian clock (Andronis et al., 2008; Hajdu et al., 2018), root development (Zhang et al., 2019; Duan et al., 2021) and flowering (Bhagat et al., 2021; Chu et al., 2022).
Also BBX have multiple connections to phytohormones. For instance, BBX24 mediates shade avoidance by binding to DELLA-proteins, which are negative regulators of GA signaling, and thus, prevent PIF4 repression. Furthermore, GA-treatment can even rescue the short hypocotyl-phenotype of bbx24 under low R:FR (Crocco et al., 2015). In rice, BBX8 and 30 expression is not only regulated in response to light-dark-cycles, but also to GA, auxin and cytokinin (Huang et al., 2012). BBX21 antagonizes ABA-induced stomata closure, since bbx21 mutants are hypersensitive to ABA and show reduced stomata opening (Xu et al., 2014).
BBX connect light signaling also to retrograde signaling from plastids. BBX16 was reported to be regulated by GUN1 and GLK1 and to promote the expression of nuclear photosynthesis genes (Veciana et al., 2022). Interestingly, this protein was also found to be downregulated, whenever chloroplast biogenesis is blocked, either by chemical treatment or genetic inactivation (Grubler et al., 2021). Additionally, BBX16 also integrates shading signals. Thus, many PM regulators are active at key events of the light signaling cascade and seem to integrate several external and internal signals, such as light, shading, phytohormones and biogenic retrograde signals in order to fine-tune the growth response of the plant in accordance with the prevailing conditions.
Another important signal, that is integrated into the light signaling network are changes in temperatures. Thermomorphogenesis (TM) is an architectural reprogramming initiated upon high ambient temperatures that induces morphological changes, such as increased hypocotyl and petiole elongation, hyponastic growth as well as early flowering, in order to circumvent negative heat effects through cooling (Quint et al., 2023). High ambient temperatures can be sensed by PhyB due to increased kinetics of their thermal back conversion from the active Pfr to the inactive Pr form. As a consequence, repression on PIF4 is released and emanates in the PIF4-mediated elongation of hypocotyls and petioles due to the induction of auxin biosynthesis and signalling, therefore, coupling TM and hormone signalling. PIF4 regulation, however, is influenced by many factors both on the transcriptional and the (post)-translational level and many signalling pathways converge. Thus, PIF4 might represent another integrator for biotic signals and fine-tune their responses.
As an important consequence, the different signaling networks should not be viewed separately. In laboratory conditions often only one parameter (the one of researcher´s interest) is changed, while the others are kept constant by active technological regulation. In nature, however, major environmental parameters often change in conjunction with others, a connection that often is not reflected in experimental set-ups generating potentially conflicting or misleading results. For instance a prolonged increase in ambient temperature in nature is often accompanied by low water availability. This could highly affect the physiological output, because TM is accompanied by stomata opening to provide cooling (Kostaki et al., 2020). Open stomata, however, would favor higher transpiration rates, which is counterintuitive under drought. Similarly, increased temperatures have strong negative effects on light-induced repression of hypocotyl elongation (Gray et al., 1998). Also circadian gating (Salter et al., 2003) and day length impacts positively hypocotyl elongation. The pap5/hmr mutant exhibited strong repression of TM-induced hypocotyl elongation in continuous red light, which was partially abolished in short and long day conditions (Qiu et al., 2019).
In sum, several parallel factors should be considered in future research including the question whether some signals are more prominent than others. For instance, Pfr converts to Pr under high ambient temperatures. PCH1, however prevents thermal conversion of Pfr (Huang et al., 2019). But how both signals interact and which signal dominates remains to be investigated. It also needs to be considered that also the ecotype of the test organism could have an effect on the results. Indeed, thermal responses were shown to vary between Arabidopsis traits (Sanchez-Bermejo et al., 2015). It is, thus, likely that also crop plants vary in their temperature responses depending on their (ancestral) origin and line of breeding.
Light as technological tool to understand plant development and physiology
Light as carrier of information is used by plants to sense the state of critical environmental conditions and dynamic changes in it (photoperception). Light, however, also provides a tool for researchers to obtain information about endogenous processes in plants (photodetection) and to even gain control over gene expression, metabolism and development (photocontrol). In recent years, there was lot of technological progress in the areas of photodetection and photocontrol that allow to tackle scientific question by fully novel approaches.
Photonic detection technologies based on fluorescence or bioluminescence became of fundamental importance in life sciences as they allow to analyze biological processes in lifetime without destructive impact on the object of study. A well-established method in plants represents the measurement of chlorophyll a (Chl) fluorescence (Maxwell and Johnson, 2000). It exploits the red autofluorescence of illuminated Chl to obtain in vivo information about the functional (and in some aspects structural) state of the photosynthetic apparatus (Kramer et al., 2004) and are also used for the physiological characterization of plants including their responses to environmental fluctuations or stressors (Kalaji et al., 2014; Kalaji et al., 2017). The invention of the pulse-amplitude modulation (PAM) technology even allowed the determination of such information in presence of ambient light (Schreiber, 1983; Bilger et al., 1995), a general problem for the usage of photonic-based research approaches (see also below). Recent development of other remote sensing technologies such as hyperspectral imaging, nuclear magnetic resonance imaging, mass spectrometry imaging and infra-red spectroscopy imaging even further expand the possibilities to investigate endogenous processes of plants providing a whole set of novel approaches for plant phenotyping (Saric et al., 2022; Borisjuk et al., 2023).
Such external photodetection of biological processes is complemented by intracellular in vivo monitoring of spatio-temporal variations using genetically encoded biosensors that typically are based on fluorescent proteins or enzymes catalyzing bioluminescent reactions. Such biosensors can be used to detect for instance the redox state of antioxidant metabolite pools, reactive oxygen species (ROS), Ca2+, and K+ ion concentrations, pH and many more parameters, but also the time-dependent localisation of proteins or hormone effects. This field has seen tremendous progress in the last decade and provides an extensive toolbox to study biological processes in high resolution and in all four dimensions (Isoda et al., 2021; Schwarzlander and Zurbriggen, 2021; Molinari et al., 2023).
Our understanding of functional components and mechanistic connections in light signalling at the molecular level has reached a niveau that allows to get beyond fundamental research of photoperception and to use this knowledge for the construction of optogenetic tools. While this field of science has tremendous success in various areas of the life sciences such as neuroscience or animal research, optogenetics in plants is lagging behind mostly because of interference with plant internal light-dependent processes (Christie and Zurbriggen, 2021; Konrad et al., 2023). Major obstacles in the past were the principle requirement of plants for light as energy source in photosynthesis (requiring growth under light/dark cycles) as well as the sophisticated internal light signalling networks that show high degrees of redundancy providing high compensation capabilities of genetic manipulations. More importantly, the lack of retinal in plants hampered the technological development of rhodopsin-based optical switches, e.g., the implementation of channelrhodopsins that were used with great success in neurosciences (Miller, 2006). Optogenetic tools, however, provide a number of advantages over chemical inducible systems and there is a strong interest in the establishment of such tools for plant research (Omelina et al., 2022).
Recent approaches combining the progress in LED-based illumination technologies with smart molecular solutions of synthetic biology provided several important improvements in plant optogenetics. The availability of cost-efficient LED arrays equipped with combinations of narrow band-width LEDs allowed the establishment of highly flexible and defined illumination solutions in plant growth technologies that are superior to earlier standard light sources with coloured filters. Using these in combination with genetic constructs comprising specific light responsive cis-elements and corresponding trans-acting proteins or protein domains (either as activator or repressor) generate molecular switches that do nicely work in plants. The research field benefits a lot from the progress in synthetic biology and the principles in engineering synthetic orthogonal gene circuits with sensor, processor and actuator modules in which the components of the circuits show only minimal interaction with endogenous signalling pathways or networks (Vazquez-Vilar et al., 2023).
The reversible cis/trans isomerisation of phytochromes upon red/far-red illumination shifts provides a natural optoswitch that was used to engineer a first optogenetic tool for control of gene transcription in yeast (Shimizu-Sato et al., 2002). In plants the first optogenetic tool has been constructed as a split transcription factor module from the N-terminus of PHYB fused to the viral transactivator VP16 and the N-terminus of PIF6 fused to an E. coli repressor protein. The system restores a functional transcription factor upon red light illumination and is switched off by far-red illumination allowing to control gene expression upon corresponding illumination switches (Muller et al., 2014). It bears, however, the risk that the endogenous phytochrome system is also affected by the light regime. This can be overcome by using optogenetic switches that work in the green wavelengths range where plants do exhibit a light detection gap of their photoreceptor systems. The transcription factor CarH from Thermus thermophilus is naturally activated by green light. It was used to engineer a switch in Arabidopsis protoplasts that terminates transcription from a synthetic binding site/promoter construct upon green light illumination (Chatelle et al., 2018). Disadvantage of this system is that CarH requires adenosylcobalamin as chromophore. Since this vitamin B12 derivative is not produced in plants, it must be exogenously added preventing effective use in whole plants unless the corresponding biosynthetic pathway is engineered in them. An alternative might be the orange carotenoid protein family from cyanobacteria that perform conformational changes separating N- and C-terminal domains upon illumination with blue-green light. This behaviour was exploited to engineer a heterodimerization switch in which the domain separation allowed chloroplast transcription initiation by the plastid Sigma factor 2 in Arabidopsis protoplasts. However, also this system requires a chromophore not produced in plants (i.e., keto-carotenoids) and, hence, the engineering of ketocarotenoid production (Piccinini et al., 2022).
Recently, an optogenetic switch in plants could be constructed that is blind to ambient white light allowing to express genes on demand by illumination with a discrete wavelength (Ochoa-Fernandez et al., 2020). To this end two photoswitches controlling a synthetic promoter construct were combined comprising a blue-light off switch and a red-light on switch. The system allows vegetative and regenerative growth of plants in light chambers and greenhouses since the blue wavelengths in white light prevent any expression. Only under monochromatic red light the system (dubbed plant-useable light switch element (PULSE)) will be activated providing a versatile tool to control signalling and metabolic networks in whole plants and has been also successfully combined with CRISPR-Cas based tools (Ochoa-Fernandez et al., 2020). The PULSE system, therefore, provides a highly promising approach for future fundamental research and biotechnological applications under dark/white light cycles.
One major field in optogenetics is the control of ion fluxes over membranes using channelrhodopsins. Depolarization/hyperpolarization is also in plant cells of high relevance, for instance in the control of stomata aperture and plant transpiration. However, plants do not produce retinal leading to degradation of opsins (Ullrich et al., 2013). Recently, this problem could be overcome by introduction of a plastid-targeted marine bacterium dioxygenase that converts plastidial β-carotene (the precursor) into retinal. The stable chromophor availability allowed the expression of the light-sensitive anion channel GtACR1 and subsequently green-light stimulated membrane potential changes in pollen tubes and guard cells resulting in corresponding physiological reactions (pollen tube bending or stomata closure, respectively) (Zhou et al., 2021).
In sum, major obstacles for wide application of optogenetics in plants has been removed now. One highly interesting future application will be the combination of optoswitches with specific biosensors providing the possibility to study physiological effects of light-induced cellular manipulations in real time, both at cellular and tissue level. Such research may pave the avenue for future applications in the field to control decisive developmental transitions or improved acclimatory responses to environmental constraints in crop plants.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
ML: Conceptualization, Writing–original draft. TP: Conceptualization, Funding acquisition, Writing–original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. DFG PF323-7 and DFG PF323-9.
Acknowledgments
Due to space constraints we could not cover all aspects of photobiology in plants and, therefore, concentrated on selected aspects. We apologize to all colleagues whose work was not cited because of that. Research in our team is funded by the Deutsche Forschungsgemeinschaft (PF323-7).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Andronis, C., Barak, S., Knowles, S. M., Sugano, S., and Tobin, E. M. (2008). The clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis. Mol. Plant 1 (1), 58–67. doi:10.1093/mp/ssm005
Bernardo-Garcia, S., de Lucas, M., Martinez, C., Espinosa-Ruiz, A., Daviere, J. M., and Prat, S. (2014). BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 28 (15), 1681–1694. doi:10.1101/gad.243675.114
Bhagat, P. K., Verma, D., Sharma, D., and Sinha, A. K. (2021). HY5 and ABI5 transcription factors physically interact to fine tune light and ABA signaling in Arabidopsis. Plant Mol. Biol. 107 (1-2), 117–127. doi:10.1007/s11103-021-01187-z
Bilger, W., Schreiber, U., and Bock, M. (1995). Determination of the quantum efficiency of photosystem II and of non-photochemical quenching of chlorophyll fluorescence in the field. Oecologia 102 (4), 425–432. doi:10.1007/BF00341354
Borisjuk, L., Horn, P., Chapman, K., Jakob, P. M., Gundel, A., and Rolletschek, H. (2023). Seeing plants as never before. New Phytol. 238 (5), 1775–1794. doi:10.1111/nph.18871
Chambon, L., Gillet, F. X., Chieb, M., Cobessi, D., Pfannschmidt, T., and Blanvillain, R. (2022). PAP8/pTAC6 is part of a nuclear protein complex and displays RNA recognition motifs of viral origin. Int. J. Mol. Sci. 23 (6), 3059. doi:10.3390/ijms23063059
Chatelle, C., Ochoa-Fernandez, R., Engesser, R., Schneider, N., Beyer, H. M., Jones, A. R., et al. (2018). A green-light-responsive system for the control of transgene expression in mammalian and plant cells. ACS Synth. Biol. 7 (5), 1349–1358. doi:10.1021/acssynbio.7b00450
Chen, D., Lyu, M., Kou, X., Li, J., Yang, Z., Gao, L., et al. (2022). Integration of light and temperature sensing by liquid-liquid phase separation of phytochrome B. Mol. Cell 82 (16), 3015–3029.e6. doi:10.1016/j.molcel.2022.05.026
Chen, H., Zhang, J., Neff, M. M., Hong, S. W., Zhang, H., Deng, X. W., et al. (2008). Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc. Natl. Acad. Sci. U. S. A. 105 (11), 4495–4500. doi:10.1073/pnas.0710778105
Chen, M., Galvao, R. M., Li, M., Burger, B., Bugea, J., Bolado, J., et al. (2010). Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141 (7), 1230–1240. doi:10.1016/j.cell.2010.05.007
Cheng, M. C., Kathare, P. K., Paik, I., and Huq, E. (2021). Phytochrome signaling networks. Annu. Rev. Plant Biol. 72, 217–244. doi:10.1146/annurev-arplant-080620-024221
Christie, J. M., and Zurbriggen, M. D. (2021). Optogenetics in plants. New Phytol. 229 (6), 3108–3115. doi:10.1111/nph.17008
Chu, L., Yang, C., Zhuang, F., Gao, Y., and Luo, M. (2022). The HDA9-HY5 module epigenetically regulates flowering time in Arabidopsis thaliana. J. Cell Physiol. 237 (7), 2961–2968. doi:10.1002/jcp.30761
Crocco, C. D., Locascio, A., Escudero, C. M., Alabadi, D., Blazquez, M. A., and Botto, J. F. (2015). The transcriptional regulator BBX24 impairs DELLA activity to promote shade avoidance in Arabidopsis thaliana. Nat. Commun. 6, 6202. doi:10.1038/ncomms7202
de Lucas, M., Daviere, J. M., Rodriguez-Falcon, M., Pontin, M., Iglesias-Pedraz, J. M., Lorrain, S., et al. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451 (7177), 480–484. doi:10.1038/nature06520
Duan, X., Xu, S., Xie, Y., Li, L., Qi, W., Parizot, B., et al. (2021). Periodic root branching is influenced by light through an HY1-HY5-auxin pathway. Curr. Biol. 31 (17), 3834–3847.e5. doi:10.1016/j.cub.2021.06.055
Enderle, B., Sheerin, D. J., Paik, I., Kathare, P. K., Schwenk, P., Klose, C., et al. (2017). PCH1 and PCHL promote photomorphogenesis in plants by controlling phytochrome B dark reversion. Nat. Commun. 8 (1), 2221. doi:10.1038/s41467-017-02311-8
Feng, S., Martinez, C., Gusmaroli, G., Wang, Y., Zhou, J., Wang, F., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451 (7177), 475–479. doi:10.1038/nature06448
Geilen, K., and Bohmer, M. (2015). Dynamic subnuclear relocalisation of WRKY40 in response to Abscisic acid in Arabidopsis thaliana. Sci. Rep. 5, 13369. doi:10.1038/srep13369
Grabowski, E., Miao, Y., Mulisch, M., and Krupinska, K. (2008). Single-stranded DNA-binding protein Whirly1 in barley leaves is located in plastids and the nucleus of the same cell. Plant Physiol. 147 (4), 1800–1804. doi:10.1104/pp.108.122796
Gray, W. M., Ostin, A., Sandberg, G., Romano, C. P., and Estelle, M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 95 (12), 7197–7202. doi:10.1073/pnas.95.12.7197
Grubler, B., Cozzi, C., and Pfannschmidt, T. (2021). A core module of nuclear genes regulated by biogenic retrograde signals from plastids. Plants (Basel) 10 (2), 296. doi:10.3390/plants10020296
Hahm, J., Kim, K., Qiu, Y., and Chen, M. (2020). Increasing ambient temperature progressively disassembles Arabidopsis phytochrome B from individual photobodies with distinct thermostabilities. Nat. Commun. 11 (1), 1660. doi:10.1038/s41467-020-15526-z
Hajdu, A., Dobos, O., Domijan, M., Balint, B., Nagy, I., Nagy, F., et al. (2018). ELONGATED HYPOCOTYL 5 mediates blue light signalling to the Arabidopsis circadian clock. Plant J. 96 (6), 1242–1254. doi:10.1111/tpj.14106
Han, I. S., Tseng, T. S., Eisinger, W., and Briggs, W. R. (2008). Phytochrome A regulates the intracellular distribution of phototropin 1-green fluorescent protein in Arabidopsis thaliana. Plant Cell 20 (10), 2835–2847. doi:10.1105/tpc.108.059915
Heng, Y., Jiang, Y., Zhao, X., Zhou, H., Wang, X., Deng, X. W., et al. (2019). BBX4, a phyB-interacting and modulated regulator, directly interacts with PIF3 to fine tune red light-mediated photomorphogenesis. Proc. Natl. Acad. Sci. U. S. A. 116 (51), 26049–26056. doi:10.1073/pnas.1915149116
Huang, H., McLoughlin, K. E., Sorkin, M. L., Burgie, E. S., Bindbeutel, R. K., Vierstra, R. D., et al. (2019). PCH1 regulates light, temperature, and circadian signaling as a structural component of phytochrome B-photobodies in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 116 (17), 8603–8608. doi:10.1073/pnas.1818217116
Huang, H., Yoo, C. Y., Bindbeutel, R., Goldsworthy, J., Tielking, A., Alvarez, S., et al. (2016). PCH1 integrates circadian and light-signaling pathways to control photoperiod-responsive growth in Arabidopsis. Elife 5, e13292. doi:10.7554/eLife.13292
Huang, J., Zhao, X., Weng, X., Wang, L., and Xie, W. (2012). The rice B-box zinc finger gene family: genomic identification, characterization, expression profiling and diurnal analysis. PLoS One 7 (10), e48242. doi:10.1371/journal.pone.0048242
Ibanez, C., Delker, C., Martinez, C., Burstenbinder, K., Janitza, P., Lippmann, R., et al. (2018). Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via BZR1. Curr. Biol. 28 (2), 303–310. doi:10.1016/j.cub.2017.11.077
Isemer, R., Mulisch, M., Schafer, A., Kirchner, S., Koop, H. U., and Krupinska, K. (2012). Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett. 586 (1), 85–88. doi:10.1016/j.febslet.2011.11.029
Isoda, R., Yoshinari, A., Ishikawa, Y., Sadoine, M., Simon, R., Frommer, W. B., et al. (2021). Sensors for the quantification, localization and analysis of the dynamics of plant hormones. Plant J. 105 (2), 542–557. doi:10.1111/tpj.15096
Jia, K. P., Luo, Q., He, S. B., Lu, X. D., and Yang, H. Q. (2014). Strigolactone-regulated hypocotyl elongation is dependent on cryptochrome and phytochrome signaling pathways in Arabidopsis. Mol. Plant 7 (3), 528–540. doi:10.1093/mp/sst093
Job, N., Yadukrishnan, P., Bursch, K., Datta, S., and Johansson, H. (2018). Two B-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol. 176 (4), 2963–2976. doi:10.1104/pp.17.00856
Kalaji, H. M., Schansker, G., Brestic, M., Bussotti, F., Calatayud, A., Ferroni, L., et al. (2017). Frequently asked questions about chlorophyll fluorescence, the sequel. Photosynth Res. 132 (1), 13–66. doi:10.1007/s11120-016-0318-y
Kalaji, H. M., Schansker, G., Ladle, R. J., Goltsev, V., Bosa, K., Allakhverdiev, S. I., et al. (2014). Frequently asked questions about in vivo chlorophyll fluorescence: practical issues. Photosynth Res. 122 (2), 121–158. doi:10.1007/s11120-014-0024-6
Kim, C., Kwon, Y., Jeong, J., Kang, M., Lee, G. S., Moon, J. H., et al. (2023). Phytochrome B photobodies are comprised of phytochrome B and its primary and secondary interacting proteins. Nat. Commun. 14 (1), 1708. doi:10.1038/s41467-023-37421-z
Kobayashi, K., Ohnishi, A., Sasaki, D., Fujii, S., Iwase, A., Sugimoto, K., et al. (2017). Shoot removal induces chloroplast development in roots via cytokinin signaling. Plant Physiol. 173 (4), 2340–2355. doi:10.1104/pp.16.01368
Konrad, K. R., Gao, S., Zurbriggen, M. D., and Nagel, G. (2023). Optogenetic methods in plant biology. Annu. Rev. Plant Biol. 74, 313–339. doi:10.1146/annurev-arplant-071122-094840
Kostaki, K. I., Coupel-Ledru, A., Bonnell, V. C., Gustavsson, M., Sun, P., McLaughlin, F. J., et al. (2020). Guard cells integrate light and temperature signals to control stomatal aperture. Plant Physiol. 182 (3), 1404–1419. doi:10.1104/pp.19.01528
Kramer, D. M., Johnson, G., Kiirats, O., and Edwards, G. E. (2004). New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth Res. 79, 209. doi:10.1023/B:PRES.0000015391.99477.0d
Krause, K., Oetke, S., and Krupinska, K. (2012). Dual targeting and retrograde translocation: regulators of plant nuclear gene expression can be sequestered by plastids. Int. J. Mol. Sci. 13 (9), 11085–11101. doi:10.3390/ijms130911085
Lamparter, T. (2004). Evolution of cyanobacterial and plant phytochromes. FEBS Lett. 573 (1-3), 1–5. doi:10.1016/j.febslet.2004.07.050
Legris, M., Klose, C., Burgie, E. S., Rojas, C. C., Neme, M., Hiltbrunner, A., et al. (2016). Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354 (6314), 897–900. doi:10.1126/science.aaf5656
Li, K., Yu, R., Fan, L. M., Wei, N., Chen, H., and Deng, X. W. (2016). DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 7, 11868. doi:10.1038/ncomms11868
Liebers, M., Gillet, F. X., Israel, A., Pounot, K., Chambon, L., Chieb, M., et al. (2020). Nucleo-plastidic PAP8/pTAC6 couples chloroplast formation with photomorphogenesis. EMBO J. 39 (22), e104941. doi:10.15252/embj.2020104941
Liscum, E., Askinosie, S. K., Leuchtman, D. L., Morrow, J., Willenburg, K. T., and Coats, D. R. (2014). Phototropism: growing towards an understanding of plant movement. Plant Cell 26 (1), 38–55. doi:10.1105/tpc.113.119727
Lorrai, R., Boccaccini, A., Ruta, V., Possenti, M., Costantino, P., and Vittorioso, P. (2018). Abscisic acid inhibits hypocotyl elongation acting on gibberellins, DELLA proteins and auxin. AoB Plants 10 (5), ply061. doi:10.1093/aobpla/ply061
Lu, X. D., Zhou, C. M., Xu, P. B., Luo, Q., Lian, H. L., and Yang, H. Q. (2015). Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 8 (3), 467–478. doi:10.1016/j.molp.2014.11.025
Maxwell, K., and Johnson, G. N. (2000). Chlorophyll fluorescence--a practical guide. J. Exp. Bot. 51 (345), 659–668. doi:10.1093/jxb/51.345.659
Miller, G. (2006). Optogenetics. Shining new light on neural circuits. Science 314 (5806), 1674–1676. doi:10.1126/science.314.5806.1674
Molinari, P. E., Krapp, A. R., Zurbriggen, M. D., and Carrillo, N. (2023). Lighting the light reactions of photosynthesis by means of redox-responsive genetically encoded biosensors for photosynthetic intermediates. Photochem Photobiol. Sci. 22 (8), 2005–2018. doi:10.1007/s43630-023-00425-1
Muller, K., Siegel, D., Rodriguez Jahnke, F., Gerrer, K., Wend, S., Decker, E. L., et al. (2014). A red light-controlled synthetic gene expression switch for plant systems. Mol. Biosyst. 10 (7), 1679–1688. doi:10.1039/c3mb70579j
Ochoa-Fernandez, R., Abel, N. B., Wieland, F. G., Schlegel, J., Koch, L. A., Miller, J. B., et al. (2020). Optogenetic control of gene expression in plants in the presence of ambient white light. Nat. Methods 17 (7), 717–725. doi:10.1038/s41592-020-0868-y
Oh, E., Kang, H., Yamaguchi, S., Park, J., Lee, D., Kamiya, Y., et al. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21 (2), 403–419. doi:10.1105/tpc.108.064691
Omelina, E. S., Yushkova, A. A., Motorina, D. M., Volegov, G. A., Kozhevnikova, E. N., and Pindyurin, A. V. (2022). Optogenetic and chemical induction systems for regulation of transgene expression in plants: use in basic and applied research. Int. J. Mol. Sci. 23 (3), 1737. doi:10.3390/ijms23031737
Ortigosa, A., Fonseca, S., Franco-Zorrilla, J. M., Fernandez-Calvo, P., Zander, M., Lewsey, M. G., et al. (2020). The JA-pathway MYC transcription factors regulate photomorphogenic responses by targeting HY5 gene expression. Plant J. 102 (1), 138–152. doi:10.1111/tpj.14618
Pfannschmidt, T., Blanvillain, R., Merendino, L., Courtois, F., Chevalier, F., Liebers, M., et al. (2015). Plastid RNA polymerases: orchestration of enzymes with different evolutionary origins controls chloroplast biogenesis during the plant life cycle. J. Exp. Bot. 66, 6957–6973. doi:10.1093/jxb/erv415
Piccinini, L., Iacopino, S., Cazzaniga, S., Ballottari, M., Giuntoli, B., and Licausi, F. (2022). A synthetic switch based on orange carotenoid protein to control blue-green light responses in chloroplasts. Plant Physiol. 189 (2), 1153–1168. doi:10.1093/plphys/kiac122
Ponnu, J., and Hoecker, U. (2022). Signaling mechanisms by Arabidopsis cryptochromes. Front. Plant Sci. 13, 844714. doi:10.3389/fpls.2022.844714
Qiu, Y., Li, M., Kim, R. J., Moore, C. M., and Chen, M. (2019). Daytime temperature is sensed by phytochrome B in Arabidopsis through a transcriptional activator HEMERA. Nat. Commun. 10 (1), 140. doi:10.1038/s41467-018-08059-z
Qiu, Y., Li, M., Pasoreck, E. K., Long, L., Shi, Y., Galvao, R. M., et al. (2015). HEMERA couples the proteolysis and transcriptional activity of PHYTOCHROME INTERACTING FACTORs in Arabidopsis photomorphogenesis. Plant Cell 27 (5), 1409–1427. doi:10.1105/tpc.114.136093
Quint, M., Delker, C., Balasubramanian, S., Balcerowicz, M., Casal, J. J., Castroverde, C. D. M., et al. (2023). 25 Years of thermomorphogenesis research: milestones and perspectives. Trends Plant Sci. 28 (10), 1098–1100. doi:10.1016/j.tplants.2023.07.001
Salter, M. G., Franklin, K. A., and Whitelam, G. C. (2003). Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426 (6967), 680–683. doi:10.1038/nature02174
Sanchez-Bermejo, E., Zhu, W., Tasset, C., Eimer, H., Sureshkumar, S., Singh, R., et al. (2015). Genetic architecture of natural variation in thermal responses of Arabidopsis. Plant Physiol. 169 (1), 647–659. doi:10.1104/pp.15.00942
Saric, R., Nguyen, V. D., Burge, T., Berkowitz, O., Trtilek, M., Whelan, J., et al. (2022). Applications of hyperspectral imaging in plant phenotyping. Trends Plant Sci. 27 (3), 301–315. doi:10.1016/j.tplants.2021.12.003
Sassi, M., Lu, Y., Zhang, Y., Wang, J., Dhonukshe, P., Blilou, I., et al. (2012). COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1- and PIN2-dependent auxin transport in Arabidopsis. Development 139 (18), 3402–3412. doi:10.1242/dev.078212
Schreiber, U. (1983). Chlorophyll fluorescence yield changes as a tool in plant physiology I. The measuring system. Photosynth Res. 4 (4), 361–373. doi:10.1007/BF00054144
Schwarzlander, M., and Zurbriggen, M. D. (2021). Sensors and controllers-for and from plants. Plant Physiol. 187 (2), 473–476. doi:10.1093/plphys/kiab364
Seo, M., Nambara, E., Choi, G., and Yamaguchi, S. (2009). Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 69 (4), 463–472. doi:10.1007/s11103-008-9429-y
Sheerin, D. J., Menon, C., zur Oven-Krockhaus, S., Enderle, B., Zhu, L., Johnen, P., et al. (2015). Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27 (1), 189–201. doi:10.1105/tpc.114.134775
Shimizu-Sato, S., Huq, E., Tepperman, J. M., and Quail, P. H. (2002). A light-switchable gene promoter system. Nat. Biotechnol. 20 (10), 1041–1044. doi:10.1038/nbt734
Song, Z., Heng, Y., Bian, Y., Xiao, Y., Liu, J., Zhao, X., et al. (2021). BBX11 promotes red light-mediated photomorphogenic development by modulating phyB-PIF4 signaling. aBIOTECH 2 (2), 117–130. doi:10.1007/s42994-021-00037-2
Trupkin, S. A., Legris, M., Buchovsky, A. S., Tolava Rivero, M. B., and Casal, J. J. (2014). Phytochrome B nuclear bodies respond to the low red to far-red ratio and to the reduced irradiance of canopy shade in Arabidopsis. Plant Physiol. 165 (4), 1698–1708. doi:10.1104/pp.114.242438
Tsuchiya, Y., Vidaurre, D., Toh, S., Hanada, A., Nambara, E., Kamiya, Y., et al. (2010). A small-molecule screen identifies new functions for the plant hormone strigolactone. Nat. Chem. Biol. 6 (10), 741–749. doi:10.1038/nchembio.435
Ullrich, S., Gueta, R., and Nagel, G. (2013). Degradation of channelopsin-2 in the absence of retinal and degradation resistance in certain mutants. Biol. Chem. 394 (2), 271–280. doi:10.1515/hsz-2012-0256
Van Buskirk, E. K., Decker, P. V., and Chen, M. (2012). Photobodies in light signaling. Plant Physiol. 158 (1), 52–60. doi:10.1104/pp.111.186411
Vazquez-Vilar, M., Selma, S., and Orzaez, D. (2023). The design of synthetic gene circuits in plants: new components, old challenges. J. Exp. Bot. 74 (13), 3791–3805. doi:10.1093/jxb/erad167
Veciana, N., Martin, G., Leivar, P., and Monte, E. (2022). BBX16 mediates the repression of seedling photomorphogenesis downstream of the GUN1/GLK1 module during retrograde signalling. New Phytol. 234 (1), 93–106. doi:10.1111/nph.17975
Xu, D., Jiang, Y., Li, J., Holm, M., and Deng, X. W. (2018). The B-box domain protein BBX21 promotes photomorphogenesis. Plant Physiol. 176 (3), 2365–2375. doi:10.1104/pp.17.01305
Xu, D., Li, J., Gangappa, S. N., Hettiarachchi, C., Lin, F., Andersson, M. X., et al. (2014). Convergence of Light and ABA signaling on the ABI5 promoter. PLoS Genet. 10 (2), e1004197. doi:10.1371/journal.pgen.1004197
Yadukrishnan, P., Rahul, P. V., and Datta, S. (2020). HY5 suppresses, rather than promotes, abscisic acid-mediated inhibition of postgermination seedling development. Plant Physiol. 184 (2), 574–578. doi:10.1104/pp.20.00783
Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S. A., and Nagatani, A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145 (3), 437–445. doi:10.1083/jcb.145.3.437
Yang, E. J., Yoo, C. Y., Liu, J., Wang, H., Cao, J., Li, F. W., et al. (2019). NCP activates chloroplast transcription by controlling phytochrome-dependent dual nuclear and plastidial switches. Nat. Commun. 10 (1), 2630. doi:10.1038/s41467-019-10517-1
Yoo, C. Y., Pasoreck, E. K., Wang, H., Cao, J., Blaha, G. M., Weigel, D., et al. (2019). Phytochrome activates the plastid-encoded RNA polymerase for chloroplast biogenesis via nucleus-to-plastid signaling. Nat. Commun. 10 (1), 2629. doi:10.1038/s41467-019-10518-0
Yu, Y., Wang, J., Zhang, Z., Quan, R., Zhang, H., Deng, X. W., et al. (2013). Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light. PLoS Genet. 9 (12), e1004025. doi:10.1371/journal.pgen.1004025
Zhang, Y., Wang, C., Xu, H., Shi, X., Zhen, W., Hu, Z., et al. (2019). HY5 contributes to light-regulated root system architecture under a root-covered culture system. Front. Plant Sci. 10, 1490. doi:10.3389/fpls.2019.01490
Zhong, S., Shi, H., Xue, C., Wang, L., Xi, Y., Li, J., et al. (2012). A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr. Biol. 22 (16), 1530–1535. doi:10.1016/j.cub.2012.06.039
Keywords: light signalling, photomorphogenesis, molecular networks, photocontrol, optogenetics
Citation: Liebers M and Pfannschmidt T (2024) New horizons in light control of plant photomorphogenesis and development. Front. Photobiol. 1:1346705. doi: 10.3389/fphbi.2023.1346705
Received: 29 November 2023; Accepted: 15 December 2023;
Published: 08 January 2024.
Edited by:
Miroslava Rakocevic, State University of Northern Rio de Janeiro, BrazilReviewed by:
Ute Hoecker, University of Cologne, GermanyCopyright © 2024 Liebers and Pfannschmidt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Pfannschmidt, VC5QZmFubnNjaG1pZHRAYm90YW5pay51bmktaGFubm92ZXIuZGU=
 Monique Liebers
Monique Liebers Thomas Pfannschmidt
Thomas Pfannschmidt