- 1Division of Colorectal and Anal Surgery, Department of Gastrointestinal Surgery, Guangxi Medical University Cancer Hospital, Nanning, China
- 2Department of Experimental Research, Guangxi Medical University Cancer Hospital, Nanning, China
- 3Guangxi Clinical Research Center for Colorectal Cancer, Nanning, China
Recent studies have identified the reprogramming of lipid metabolism as a critical hallmark of malignancy. Enhanced cholesterol uptake and increased cholesterol biosynthesis significantly contribute to the rapid growth of tumors, with cholesterol also playing essential roles in cellular signaling pathways. Targeting cholesterol metabolism has emerged as a promising therapeutic strategy in oncology. The sterol regulatory element-binding protein-2 (SREBP2) serves as a primary transcriptional regulator of genes involved in cholesterol biosynthesis and is crucial for maintaining cholesterol homeostasis. Numerous studies have reported the upregulation of SREBP2 across various cancers, facilitating tumor progression. This review aims to provide a comprehensive overview of the structure, biological functions, and regulatory mechanisms of SREBP2. Furthermore, we summarize that SREBP2 plays a crucial role in various cancers and tumor microenvironment primarily by regulating cholesterol, as well as through several non-cholesterol pathways. We also particularly emphasize therapeutic agents targeting SREBP2 that are currently under investigation. This review seeks to enhance our understanding of SREBP2’s involvement in cancer and provide theoretical references for cancer therapies that target SREBP2.
1 Introduction
Lipids comprise a diverse array of molecules that serve as essential components of biological membranes and are widely distributed across cellular organelles (Pomorski et al., 2001; Efeyan et al., 2015). Cholesterol is a lipid that primarily regulates the rigidity, fluidity, and permeability of the lipid bilayer in cell membranes, and also plays a critical role in signal transduction, promoting diverse cellular functions (Goldstein et al., 2006; Menendez and Lupu, 2007). Dysregulation of cholesterol metabolism can initiate or exacerbate the progression of numerous diseases (Cohen et al., 2011; Schwartz et al., 2013; Goldstein and Brown, 2015). Recent reports indicate a significant upregulation of cholesterol biosynthesis in human cancers, as elevated synthesis and uptake of cholesterol are necessary to meet the demands of membrane biogenesis and support ongoing cellular replication (Gruenbacher and Thurnher, 2015; Bathaie et al., 2017). In the tumor microenvironment, cancer cells exploit cholesterol metabolism to facilitate rapid migration, invasion, and metastasis (Bian et al., 2021).
Lipid metabolism is transcriptionally regulated by sterol regulatory element-binding proteins (SREBPs) (Horton et al., 2002). First isolated from the nuclei of HeLa cells in 1993, SREBPs belong to the basic helix-loop-helix–leucine zipper (bHLH-Zip) family of transcription factors (Briggs et al., 1993; Wang et al., 1993; Brown and Goldstein, 1997). The SREBPs family comprises three subtypes, SREBP1a and SREBP1c, SREBP2(Brown and Goldstein, 1997). SREBP2 predominantly activates the transcription of key genes in the mevalonate (MVA) pathway, thereby regulating cholesterol synthesis (Hua et al., 1993; Horton et al., 2002; Horton et al., 2003). SREBP2-mediated cholesterol metabolism plays a crucial role in various cancers, including lung cancer, colorectal cancer, and breast cancer, among others. In this review, we first introduce the structure, biological processes, and recent advances in the regulation of SREBP2. Subsequently, we summarize the role of SREBP2 in different types of cancer and tumor microenvironment. Given the emergence of SREBP2 as a significant target for cancer therapy, we focus on and discuss several important SREBP2-targeting drugs. Through these discussions, this article aims to provide new insights into potential cancer therapies.
2 SREBP2 structure
The human SREBP2 gene is located on chromosome 22q13 and encodes a protein consisting of 1,141 amino acids. It was first cloned and characterized by Xianxin Hua et al., in 1993, who isolated cDNA from cultured HeLa cells (Hua et al., 1993; Miserez et al., 1997). The SREBP2 protein comprises three main segments: (a) an NH2-terminal domain of approximately 480 amino acids, which contains an acidic region responsible for transcriptional activation, along with a basic helix-loop-helix-leucine zipper (bHLH-Zip) motif that specifically binds to DNA sequences; (b) a middle hydrophobic region of approximately 80 amino acids, which includes two hydrophobic transmembrane segments; and (c) a COOH-terminal domain of approximately 590 amino acids (Brown and Goldstein, 1997) (Figure 1).
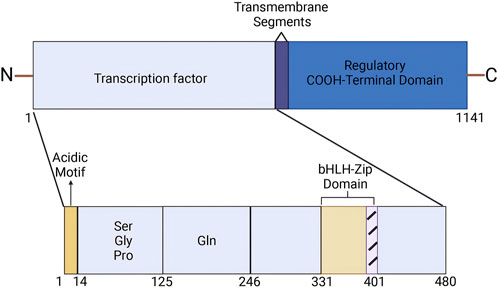
Figure 1. Structure of SREBP2. (Created in BioRender. Chen, R. (2025) https://BioRender.com/r73a111). The SREBP2 protein comprises three main segments: an NH2-terminal domain of approximately 480 amino acids, a middle hydrophobic region of approximately 80 amino acids, and a COOH-terminal domain of approximately 590 amino acids. The NH2-terminal domain contains an acidic region that is responsible for transcriptional activation, as well as a basic helix-loop-helix-leucine zipper (bHLH-Zip) motif that specifically binds to DNA sequences.
The hydrolyzed activated NH2-terminal of SREBP2 binds to sterol regulatory element (SRE) sequences in the promoters of target genes in the nucleus, thereby upregulating their transcription (Goldstein et al., 2006). The middle hydrophobic region features two hydrophobic membrane-spanning sequences and a hydrophilic lumenal loop that separates these segments (Luo et al., 2020). This hydrophilic luminal loop extends into the endoplasmic reticulum (ER) lumen (Weber et al., 2004; Luo et al., 2020). The COOH-terminal domain of SREBP2, referred to as the regulatory domain, interacts with the WD-repeat domain of SREBP-cleavage activating protein (SCAP) (Gong et al., 2016; Brown et al., 2018).
3 Regulation of SREBP2
The SREBP2 gene is transcribed and translated into SREBP2 precursor (Pre -SREBP2), which is then anchored in the endoplasmic reticulum. To become active, Pre -SREBP2 must exit the ER and undergo cleavage in the Golgi apparatus. This cleavage releases the N-terminal domain (n-SREBP2), which then translocates to the nucleus to activate the transcription of downstream target genes. This process is regulated by various mechanisms, as discussed below.
3.1 Transcription regulation of the SREBP2
Notably, because there is a 10-base pair SRE upstream of the transcription start site of SREBP2, it is also regulated by n-SREBP2(Sato et al., 1996). In addition, the region upstream contains binding sites for the transcription factors SP1 and NF-Y (Sato et al., 1996). Both of them cooperate with n-SREBP2 to upregulate the transcription of SREBP2. The key negative regulators of SREBP2 gene expression include SIRT6 and forkhead box O (FOXO3) (Tao et al., 2013). FOXO3 binds to a conserved insulin response element (IRE) in the SREBP2 gene and recruits SIRT6(Tao et al., 2013). Subsequently, SIRT6 deacetylates histone H3 at lysine residues 9 and 56 on the SREBP2 gene promoter (Tao et al., 2013). This modification promotes a repressive chromatin state, thereby inhibiting the expression of SREBP2 and its target genes (Figure 2).
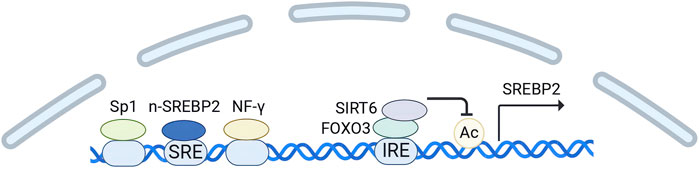
Figure 2. Transcription regulation of SREBP2. (Created in BioRender. Chen, R. (2024) https://BioRender.com/h24n110). The transcription start site of SREBP2 contains binding sites for the transcription factors SP1 and NF-Y, along with a 10-base pair SRE. SP1 and NF-Y cooperate with n-SREBP2 to upregulate the transcription of SREBP2. FOXO3 acts as a negative regulator by binding to a conserved insulin response element (IRE) in the SREBP2 gene and recruiting SIRT6. Subsequently, SIRT6 deacetylates histone H3 in the SREBP2 gene promoter, thereby suppressing SREBP2 expression.
3.2 Post-transcriptional regulation of the SREBP2
SREBP2 is post-transcriptionally regulated by microRNAs (miRNAs), which are small endogenous non-coding RNAs that exert their effects by binding to target mRNAs (Bartel, 2004). Studies demonstrated that miR-185 significantly reduced the levels of both full-length and mature SREBP2 proteins in liver cancer and prostate cancer cells by binding to the 3′untranslated region (3′UTR) of SREBP2 mRNA (Li et al., 2013; Yang et al., 2014; Chen et al., 2021). In vivo experiments also revealed that the miR-185 sequences resulted in decreased SREBP2 levels (Yang et al., 2014). However, inhibiting miR-185 did not lead to increased mRNA levels of SREBP2 or its downstream targets in vivo and vitro, suggesting that basal miR-185 may not significantly repress SREBP2 in these contexts (Chen et al., 2021). Additionally, several oncological studies have demonstrated that miR-185-5p, miR-195, miR-130b, miR-328-3p, and miR-98 directly target SREBP2 gene, leading to the inhibition of its expression (Geng et al., 2018; Yu et al., 2019; Huang et al., 2022; Tan et al., 2022; Tang et al., 2023). Overexpression of miR-27a and miR-10a was also found to decrease SREBP2 levels, although the mechanisms underlying these effects remain to be elucidated (Shirasaki et al., 2013; Horii et al., 2019) (Figure 3).
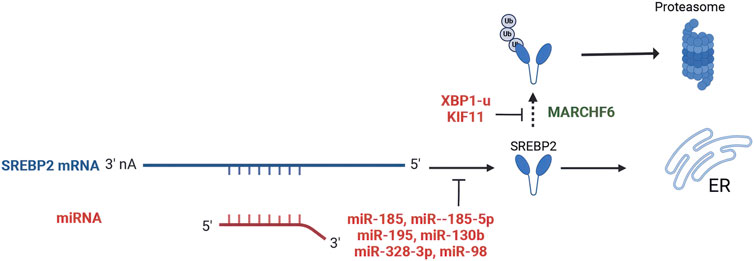
Figure 3. Post-transcriptional regulation and post-translational modifications of SREBP2. (Created in BioRender. Chen, R. (2024) https://BioRender.com/q25n513). miR-185, miR-185-5p, miR-195, miR-130b, miR-328-3p, and miR-98 directly bind to the 3′-untranslated region (3′UTR) of SREBP2 mRNA, leading to the inhibition of its expression. MARCHF6 serves as a primary ubiquitin ligase that promotes SREBP2 degradation. XBP1-u and KIF11 inhibit the ubiquitination and proteasomal degradation of SREBP2.
3.3 Post-translational modification of the SREBP2
Ubiquitination of the SREBP2 protein is a key post-translational modification. MARCHF6 serves as a primary ubiquitin ligase SREBP2 degradation (Dickson et al., 2023). However, XBP1-u inhibits the ubiquitination and proteasomal degradation of SREBP2, then stabilizing the protein in HCC(Wei et al., 2022). In pancreatic cancer, kinesin family member 11 (KIF11) interacts with SREBP2, increasing its protein levels by attenuating ubiquitination-mediated degradation (Gu et al., 2022) (Figure 3). The degradation of SREBP2 is regulated by its binding to SCAP. Early studies demonstrated that SREBP2 precursor levels are markedly reduced in the absence of SCAP (Rawson et al., 1999; Shao and Espenshade, 2014). Recent investigations have identified two distinct motifs within the carboxyl-terminal domain (CTD) of SREBP2: one functioning as a protective signal and the other as a degradation signal. When SREBP2 dissociates from SCAP, the degradation signal triggers its proteasomal degradation in the endoplasmic reticulum. Conversely, the protective signal motif enables SREBP2 to bind SCAP, thereby masking the degradation signal and stabilizing the protein (Kober et al., 2020).
3.4 Regulation of SREBP2 protein egress from the ER
The SREBP2 precursor binds to sterol regulatory element-binding protein-cleavage activating protein (SCAP) via its C-terminal domain, anchoring it within the ER. The transfer of the SREBP2-SCAP complex is regulated by cholesterol levels. SCAP undergoes conformational changes in response to fluctuations in ER cholesterol levels (Brown et al., 2018). Recent cryoelectron microscopy (cryo-EM) structures suggest that Scap’s two ER luminal loops (loop 1 and loop 7), which flank an intramembrane sterol-sensing domain (SSD), intertwine tightly to form a stable domain (Kober et al., 2021; Yan et al., 2021a; Yan et al., 2021b). The domain may interact with the membrane to sense cholesterol (Kober et al., 2021). When cholesterol levels exceed 5% of the molar content of membrane lipids in the endoplasmic reticulum (ER), the interaction between SCAP’s loop 1 and loop 7 dissociates, allowing SCAP to bind to cholesterol, inducing SCAP to bind INSIG. (Radhakrishnan et al., 2008). This interaction promotes the binding of SCAP to INSIG, an ER-retention membrane protein, which inhibits the association of the SCAP-SREBP2 complex with COPII vesicles (Sun et al., 2005). Consequently, this effectively prevents the exit of the SCAP-SREBP2 complex from the ER. Additionally, Cholesterol not only promotes the binding of SCAP to INSIG proteins but also stabilizes INSIG, preventing its degradation via the ubiquitin-proteasome pathway (Gong et al., 2006; Lee et al., 2006). Conversely, when cholesterol levels decrease, the first and seventh loops of SCAP interconnect, enabling the SCAP-SREBP2 complex to bind to COPII vesicles, which facilitates its translocation to the Golgi apparatus (Sun et al., 2005; Brown et al., 2018) (Figure 4). Moreover, the transport of the SREBP2 is also regulated by other molecules. Multimeric endoplasmic reticulum (ER) proteins known as ERLINs, large tumor suppressor kinase 2 (LATS2), along with two ubiquitin ligases, TRC8 and RNF145, negatively regulate the egress of SREBP2 from the ER. ERLINs tightly interact with the SCAP-SREBP2-INSIG complex, while TRC8 directly binds to SREBP2 and SCAP (Irisawa et al., 2009; Huber et al., 2013). RNF145 ubiquitylates SCAP within a critical loop essential for COPII binding (Zhang L. et al., 2017). These events impair COPII binding and hinder the transport of SREBP2. LATS2 directly interacts with SREBP2 to retain it in the ER. However, MiR-96 reduced the levels of the SREBP2 anchor protein INSIG2 (Aylon et al., 2016). Phosphoenolpyruvate carboxykinase 1 (PCK1) can phosphorylate the Ser207 of INSIG1 and the Ser151 of INSIG2, thereby disrupting the interaction between INSIG proteins and SCAP. These result in an increase in transport of SREBP2(Xu et al., 2020). However, another study found that SREBP2 processing unperturbed by phosphorylated INSIG2, which exhibits exclusive inhibitory specificity toward SREBP1(Tian et al., 2024).
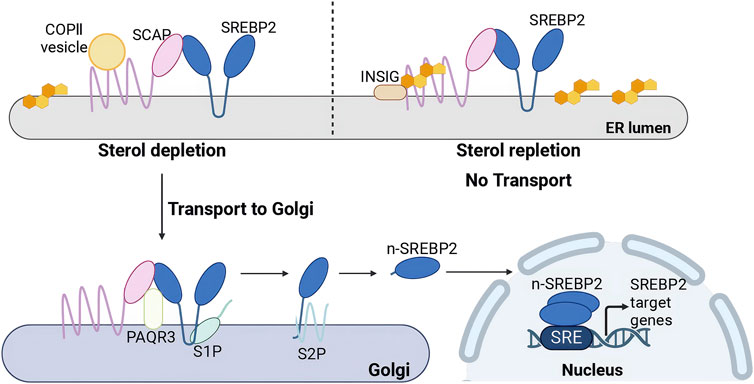
Figure 4. Activation regulation of SREBP2. (Created in BioRender. Chen, R. (2025) https://BioRender.com/xugbutl). Under cholesterol depletion: The SCAP-SREBP2 complex binds to COPII vesicles, facilitating its translocation to the Golgi apparatus. PAQR3 interacts with the SCAP-SREBP2 complex in the Golgi, retaining it within this organelle. Sequential proteolytic cleavage by Site-1 protease (S1P) and Site-2 protease (S2P) generates nuclear SREBP2 (n-SREBP2), which translocates to the nucleus as a homodimer to bind sterol regulatory elements (SREs) and activate transcription of target genes. Under cholesterol repletion: SCAP binds to INSIG, forming an ER-anchored INSIG/SCAP/SREBP complex that inhibits the transport of SREBP2 to the Golgi apparatus.
3.5 SREBP2 proteolytic activation in the Golgi
The SCAP–SREBP2 complex binds to COPII and is transported from the endoplasmic reticulum (ER) to the Golgi apparatus. Within the Golgi, the PAQR3 protein interacts with the SCAP–SREBP2 complex, retaining it in this organelle. Under cholesterol-depleting conditions, the transcription of the PAQR3 gene is upregulated, facilitating the retention process (Xu et al., 2015). SREBP2 undergoes proteolytic activation in a two-step process. The first step involves the cleavage of the luminal loop of SREBP2 by site 1 protease (S1P), producing a cleaved form of SREBP2 that is approximately half the size of the original protein; this cleavage is crucial for the subsequent activation step (Sakai et al., 1998). SPRING acts as an activating cofactor for S1P to involve in the proteolytic cleavage of SREBP2. Multiple studies have established that SPRING (previously C12ORF29) deficiency impairs SREBP2 processing (Bayraktar et al., 2020; Loregger et al., 2020). Current works demonstrate that SPRING facilitates S1P maturation to regulate SREBP2 activation (Bayraktar et al., 2020; Xiao et al., 2021; Hendrix et al., 2024a). Crucially, SPRING specifically enables a specific pool of S1P to execute SREBPs proteolytic cleavage, rather than being universally involved in all S1P-dependent pathways (Hendrix et al., 2024b). Additionally, SPRING has been reported to regulate the transport of SCAP (Aregger et al., 2020; Loregger et al., 2020). Following cleavage of SREBP2 by S1P, the N-terminal region of the SREBP2 protein undergoes a second proteolytic processing by site 2 protease (S2P) (Rawson et al., 1997). These cleavage events release a soluble N-terminal transcription factor known as n-SREBP2, which translocates to the nucleus as a homodimer (Brown et al., 2018). In the nucleus, n-SREBP2 binds to SRE sequences, thereby activating the transcription of target genes involved in cholesterol metabolism (Goldstein et al., 2006) (Figure 4).
3.6 Regulation of n-SREBP2 protein
The level and transcriptional activity of n-SREBP2 protein are regulated by various pathways. The mechanistic target of rapamycin complex 1 (mTORC1) upregulates n-SREBP2 protein level through two primary mechanisms. mTORC1 increases n-SREBP2 proteins by decreasing the nuclear entry of lipin 1, a phosphatidic acid phosphatase that downregulates the protein levels of n-SREBP2 in the nucleus (Peterson et al., 2011). mTORC1 can also activate SREBP2 by inhibiting cholesterol trafficking from lysosomes to the ER, which increases n-SREBP2 proteins level (Eid et al., 2017). Conversely, carbohydrate response element-binding protein (ChREBP) enhances the ubiquitination and proteasomal degradation of n-SREBP2, although the underlying mechanism remains unclear (Zhang D. et al., 2017). Phosphorylation of n-SREBP2 by serine/threonine protein kinase GSK3 targets it for proteasomal degradation via the SCF-FBW7 ubiquitin ligase complex, resulting in a reduction in n-SREBP2 levels (Sundqvist et al., 2005). However, miR-182 binds to the 3′UTR of FBW7, leading to a decrease in FBXW7 mRNA levels and a reduction in the degradation of n-SREBP2 protein (Jeon et al., 2013). Notably, the deubiquitinating enzyme USP28 has been shown to stabilize n-SREBP2 by reversing ubiquitination (Maier et al., 2023).
Importantly, the n-SREBP2 undergoes various post-translational modifications, including acetylation, phosphorylation, and sumoylation, which modulates the transcriptional activity of n-SREBP2. The histone acetyltransferase p300 and its related protein CBP can acetylate n-SREBP2 and increased its transcriptional activity, whereas deacetylation of Sirtuin1 (SIRT1) can deacetylate n-SREBP2 and counteract the effects of p300 and CBP(Giandomenico et al., 2003; Walker et al., 2010). SREBP2 has also been identified as a phosphorylation target of extracellular signal-regulated kinase (Erk) at Ser-455 and Ser-432, with Erk1/2-dependent phosphorylation upregulating SREBP2’s N-terminal transactivation (Bennett and Osborne, 2000; Kotzka et al., 2004). Conversely, AMP-activated protein kinase (AMPK) interacts with and directly phosphorylates n-SREBP2, inhibiting its nuclear translocation and transcriptional activity (Meng et al., 2024). SUMO1-mediated sumoylation at the sumoylation site of SREBP2 (Lys 464) also inhibits the transcriptional activity of n-SREBP2(Hirano et al., 2003) (Figure 5).
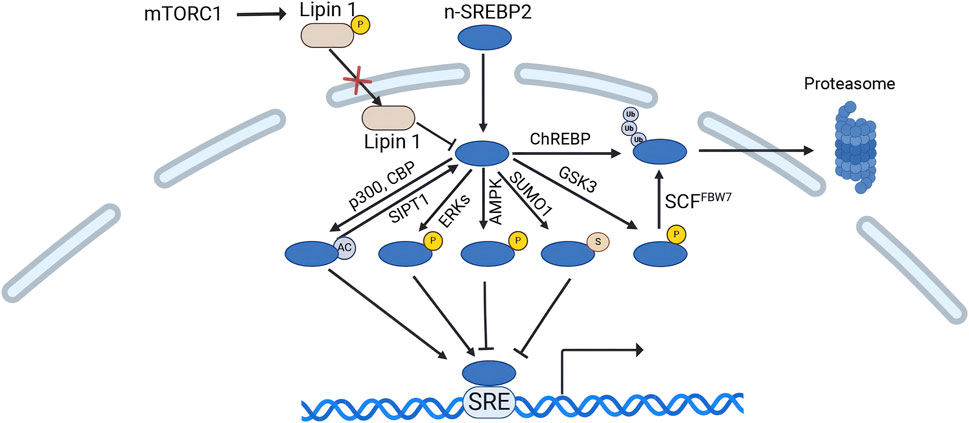
Figure 5. Regulation of n-SREBP2 protein. (Created in BioRender. Chen, R. (2024) https://BioRender.com/z88u544). mTORC1 phosphorylates lipin 1, reducing its nuclear entry. Lipin 1 decreases n-SREBP2 levels. The histone acetyltransferase p300 and its related protein CBP acetylate n-SREBP2 to enhance its transcriptional activity, while Sirtuin1 (SIRT1) deacetylates n-SREBP2. Additionally, phosphorylation of n-SREBP2 by ERK proteins also increases its transcriptional activity. Conversely, AMP-activated protein kinase (AMPK) phosphorylates n-SREBP2, inhibiting its nuclear translocation and transcriptional activity. SUMO1-mediated SUMOylation of n-SREBP2 also suppresses its transcriptional activity. Serine/threonine protein kinase GSK3 phosphorylates n-SREBP2, mediating its proteasomal degradation via the SCF-FBW7 ubiquitin ligase complex, thereby reducing n-SREBP2 levels. Carbohydrate response element-binding protein (ChREBP) also promotes n-SREBP2 ubiquitination and proteasomal degradation.
4 SREBP2 in cancer
Cholesterol homeostasis is crucial for both cellular and systemic functions. In cancer cells, cholesterol levels are significantly elevated to support rapid proliferation, resulting in increased uptake and storage of cholesterol within malignant tumors. As a key transcription factor regulating the expression of genes involved in cholesterol synthesis and uptake, SREBP2 connects oncogenic signaling with alterations in cholesterol metabolism, thereby playing a vital role in cancer development.
4.1 Liver cancer
There is a mounting body of evidence suggesting that SREBP2-mediated cholesterol biosynthesis plays a pivotal role in hepatocellular carcinoma (HCC) tumorigenesis (Calvisi et al., 2011; Che et al., 2020; Chen W. et al., 2022; Saito et al., 2023; Wang et al., 2023). In HCC, SREBP2 upregulates TAZ expression and increases TAZ interaction with TEAD2 by mediating cholesterol metabolism, thereby promotes HCC through enhanced DNA damage and associated proliferation (Saito et al., 2023). Furthermore, in the context of gut microbiota dysregulation, the increased levels of SREBP2 and heightened expression of cholesterol synthesis-related genes are associated with the reduction of tryptophan metabolites and weakened AhR activation, contributing to the initiation of liver cancer (Chen W. et al., 2022). Conversely, impaired SREBP2 maturation and suppressed cholesterol biosynthesis inhibit HCC cell proliferation, which is associated with a favorable prognosis in HCC patients (Feng et al., 2017; Liang et al., 2019; Xiang et al., 2022; He et al., 2024). In addition, SREBP2 plays a significant role in promoting the metastasis of liver cancer by enhancing the expression of genes involved in cellular migration and invasion. Nuclear translocation of SREBP2 is promoted by inhibition of LATS, significantly reducing E-cadherin expression while upregulating N-cadherin, Snail, and Vimentin, thereby facilitating HCC cell migration and invasion (Zhang et al., 2023). Beyond its role in promoting the progression of HCC, SREBP2 also significantly contributes to drug resistance in this malignancy. Cleavage of SREBP2 from the ER by Caspase-3 (CASP3) activates cholesterol biosynthesis, subsequently activating the sonic hedgehog signaling pathway, which renders cancer stem cell populations in HCC resistant to sorafenib and lenvatinib treatment (Mok et al., 2022). The study highlighting the need for therapeutic strategies that can effectively target SREBP2 to overcome resistance and improve patient responses to treatment. In conclusion, these studies suggest that SREBP2 participates in hepatocellular carcinogenesis and progression and targeting SREBP2 may represent a promising therapeutic strategy for liver cancer.
4.2 Pancreatic cancer
In pancreatic cancer, a growing body of research indicates that the activation of SREBP2 leads to alterations in cholesterol metabolism, thereby enhancing cancer cell resistance to apoptosis and promoting tumorigenesis and progression. KRAS mutations are prevalent in promoting pancreatic ductal adenocarcinoma (PDAC) (Duan et al., 2024). A study employing scRNA-seq revealed that the cholesterol synthesis pathway, including SREBP2, is specifically upregulated in KRAS mutant pancreatic organoids. These findings support a model in which oncogenic KRAS mutations activate SREBP2, leading to the reprogramming of cholesterol metabolism (Duan et al., 2024). The resultant cholesterol accumulation provides energy for pancreatic cancer growth and enhances resistance to apoptosis. Reducing the expression levels of SREBP2 can reverse the reprogramming of cholesterol metabolism induced by oncogenic KRAS (Zhang D. et al., 2021). Furthermore, SREBP2 interacts with transcription factor CP2 (TFCP2) in pancreatic cancer cells, allowing them to overcome KRAS mutation-induced senescence (Zhang D. et al., 2021). This interaction results in enhanced growth and metastasis of pancreatic cancer cells (Zhang D. et al., 2021). SREBP2 also forms a complex with β-catenin on the promoters of MVA pathway genes following the disruption of ciliogenesis, activating the transcription of cholesterol metabolism-related genes and further promoting PDAC development (Deng et al., 2018). Together, these findings underscore the significant role of SREBP2 in the tumorigenesis and progression of pancreatic cancer.
4.3 Colorectal cancer
An increasing body of evidence reveals that SREBP2-mediated cholesterol biosynthesis is involved in colorectal carcinogenesis, progression and change in energy homeostasis. Clinical studies have demonstrated that SREBP2 levels are elevated in early-stage colorectal cancer but diminished in late-stage disease (Sharma et al., 2019). SREBP2-mediated cholesterol metabolism participates in oncogenic pathways that drive and promote colorectal cancer (CRC). Dysregulated activity of Yes-associated protein (YAP) and mechanistic target of rapamycin complex 1 (mTORC1) is associated with tumorigenesis and progression (Pan et al., 2021). SREBP2 interacts with ZMYND8, which is upregulated by YAP, leading to increased cholesterol levels that subsequently activate mTORC1 and drive the colorectal carcinogenesis. This interaction highlights the SREBP2-mediated YAP/ZMYND8/mTORC1 pathway, which endows CRC cells with specific metabolic vulnerabilities (Pan et al., 2021). Similarly, in aggressive mesenchymal CRC, enhanced stabilization of SCAP and activation of SREBP2 also creates a cholesterol metabolic addiction and a therapeutic vulnerability (Muta et al., 2023). It makes statin-based inhibitors strongly suppress tumor growth (Muta et al., 2023). This heightened expression of SREBP2 plays a crucial role in the metabolic adaptations of CRC, highlighting the significance of cholesterol metabolism in this malignancy (Wen et al., 2018; Jin et al., 2023). In addition, SREBP2 also promotes the colorectal cancer metastasis. SREBP2 and SREBP2-dependent cholesterol biosynthesis are activated by c-Met/PI3K/AKT/mTOR axis in CRC, which allows CRC cells to undergo dynamic metabolic adjustments, facilitating adaptation to metastatic conditions (Zhang K. L. et al., 2021). This activation is required for the CRC cells metastasis, colonization and growth (Zhang K. L. et al., 2021). Collectively, these findings clarify that SREBP2 play a vital role in colorectal cancer development. Strategies targeting SREBP2-mediated cholesterol metabolism beneficial for the development of effective therapeutic for CRC.
4.4 Lung cancer
In lung cancer, downregulation of SREBP2 significantly inhibits the proliferation, migration, and invasion of cancer cells (Zhu et al., 2024). In addition, SREBP2 also promotes the occurrence of complication associated with lung cancer treatment. Lung cancer-related pleural effusion (LCPF), a common complication of lung cancer treatment, stimulates expression of SREBP2, which can induce pleural angiogenesis in patients and exacerbate advanced on-small cell lung cancer (NSCLC) with pleural effusion (Tsai et al., 2022). At present, drug resistance brings great challenges to the advanced treatment and prognosis of cancer patients. Activating the cleavage of SREBP2 increased the synthesis of cholesterol, leading to NSCLC cells more resistant to cisplatin (Akman et al., 2024). Another study found that SREBP2 and the cholesterol metabolism was highly activated in NSCLC cells resistance to osimertinib (Cao et al., 2024). Inhibiting SREBP2 is helpful for reversing NSCLC cells osimertinib acquired resistance (Cao et al., 2024). In summary, these results illustrate that SREBP2 participates in lung cancer progression, treatment complications, and drug resistance, and SREBP2 is a potential target for lung cancer treatment.
4.5 Breast cancer
In breast cancer, SREBP2 expression is upregulated, and its elevated levels contribute to tumor progression (Ricoult et al., 2016; Huang et al., 2017; Cai et al., 2019; Chen Y. Y. et al., 2022; Hunt et al., 2023). The transcription of SREBP2 is enhanced through phosphorylation of STAT3 (Tyr705) in triple-negative breast cancer (TNBC) (Chen Y. Y. et al., 2022). Additionally, RORγ mediates the chromatin recruitment and activation of SREBP2 in TNBC cells (Cai et al., 2019). Phosphorylated β-catenin also stimulates SREBP2 expression, promoting circulating tumor cell colony formation and tumor recurrence (Hunt et al., 2023). In contrast, inhibition of SREBP2 protein levels suppresses breast cancer cell proliferation. The cholesterol transporter ABCA9, which accumulates cholesterol in the endoplasmic reticulum (ER), reduces SREBP2 expression, thereby impairing breast cancer cell proliferation (Hwang et al., 2023). Furthermore, elevated SREBP2 expression is induced by CREB signaling, which subsequently upregulates NFATc1 expression required for mature osteoclast formation, contributing critically to breast cancer invasion and bone metastasis (Jie et al., 2019). Another study demonstrated that increased SREBP2 levels, mediated by the mTORC1 pathway, promote metastasis and a more malignant phenotype in breast cancer (Ning et al., 2023). Collectively, these findings indicate that the upregulation of SREBP2 drives tumorigenesis and progression in breast cancer.
4.6 Ovarian cancer
An expanding array of studies demonstrates that SREBP2 expression is increased and that cholesterol synthesis is facilitated in ovarian cancer (OC), which plays a pivotal role in tumorigenesis (Zhao et al., 2020). Decreasing SREBP2 levels can prevent statin-induced sterol feedback, thereby enhancing statin toxicity and efficacy in ovarian cancer cells (Casella et al., 2014). In addition to promoting tumor progression by upregulating cholesterol metabolism, SREBP2 can also promote the proliferation, migration, and epithelial-to-mesenchymal transition (EMT) of OC cells by directly activating the PRSS8/SCNN1A axis (Cai et al., 2021). Moreover, SREBP2 is implicated in mechanisms of drug resistance in OC. One study found that SREBP2 and its downstream genes were upregulated in cisplatin-resistant cells, indicating its involvement in mediating resistance to cisplatin (Zheng et al., 2018). Notably, blocking the SREBP2 pathway has been shown to increase cisplatin sensitivity in OC (Zheng et al., 2018). As such, these findings suggest that targeting SREBP2 and its related pathways may offer a promising strategy for developing effective therapeutic interventions for OC.
4.7 Endometrial cancer
Research generally indicates that inhibition of SREBP2 expression contributes to the suppression of endometrial cancer progression (Gao et al., 2018; Wang et al., 2021). For instance, the downregulation of SREBP2 by BF175, resulting in reduced cholesterol levels in endometrial cancer, enhance anti-tumor effect (Wang et al., 2021). Similarly, fatostatin inhibits the development of endometrial carcinoma by downregulating SREBP2 and interfering with SREBP2-mediated cholesterol metabolic pathways, further demonstrating anti-tumor effect of downregulated SREBP2 (Gao et al., 2018). Contrastingly, another study reported low expression levels of SREBP2 and AMPK in endometrial cancer tissues, noting that SREBP2 is a target gene inhibited by AMPK (Efsun Antmen et al., 2021). This discrepancy may be attributed to variations in the samples studied and the context of SREBP2 regulation in different tumor microenvironments. Overall, these findings highlight the complex role of SREBP2 in endometrial cancer, suggesting that its regulation may have therapeutic implications.
4.8 Prostate cancer
Androgens play a critical role in maintaining the survival and proliferation of prostate cancer (PCa) by binding to and activating the androgen receptor (AR) (Fujita and Nonomura, 2019). Consequently, lipid synthesis and uptake are vital energy resources that support tumor progression in PCa. A study comparing prostate cancer tissue with benign prostate tissue through single-cell sequencing revealed heightened activity in cholesterol metabolism and underscored the essential role of SREBP2 in prostate cancer progression (Wei et al., 2024). In castration-resistant prostate cancer (CRPC), the SREBP2 is activated by PTEN/p53 deficiency, thereby upregulating cholesterol metabolism and facilitating tumor cell survival and growth (Shangguan et al., 2022). Another study also found that increasing SREBP2 transcription contributes to the malignant characteristics of prostate adenocarcinoma (Lin et al., 2022). Conversely, downregulated of SREBP2 in prostate cancer cells leads to suppressed cancer progression (Li et al., 2013; Li et al., 2014; Longo et al., 2019). Additionally, studies have shown that SREBP2 and its downstream effector genes are upregulated in prostate cancer following androgen ablation, triggering the formation of androgen-independent (AI) tumors (Ettinger et al., 2004). Targeting SREBP2 effectively inhibited tumor growth and metastasis (Wei et al., 2024). Collectively, these findings suggest that inhibiting SREBP2 represents a promising therapeutic strategy for prostate cancer.
4.9 Bladder cancer
In bladder cancer, a study indicates that SREBP2 interacts with CBP and NFYC-37, activating the transcription of genes involved in the mevalonate pathway, thereby promoting cholesterol biosynthesis and tumor growth (Liu et al., 2023). In studies utilizing mRNA/miRNA microarrays and protein analysis in T24 bladder cancer cells, archazolid B was found to activate SREBP2, resulting in severely deregulated cholesterol homeostasis and contributing to archazolid B-induced resistance (Hamm et al., 2014). Additionally, SREBP2 plays an important in metastasis of bladder cancer. Decreasing SREBP2 expression by the Farnesoid X Receptor (FXR) has been shown to suppress lung metastasis in bladder cancer (Lai et al., 2022). Combining with cholesterol suppression treatment can further inhibits the migratory, invasive, and angiogenic properties of human urothelial carcinoma, including bladder cancer (Lai et al., 2022). These results suggest that targeting SREPB2 seems to have potential as a strategy for bladder cancer treatment.
4.10 Glioblastoma
In glioblastoma (GBM), SREBP2 is highly expressed and plays a crucial role in upregulating cholesterol biosynthesis in glioblastoma stem-like cells (GSCs), promoting tumor proliferation, self-renewal, and overall tumor growth (Gu et al., 2023). Retention of cholesterol impairs its intracellular delivery in GSCs, subsequently triggering the SREBP2 transcriptional program to meet cholesterol demands (Maghe et al., 2024). Conversely, reduced SREBP2 expression can exacerbate autophagy defects and increase cell death in GSCs, consequently suppressing tumor progression (Maghe et al., 2024). Additionally, oxygen and nutrient limitations in the tumor microenvironment are closely associated with cancer progression (Lewis et al., 2015). In glioblastoma multiforme, inhibiting SREBP2 function has been shown to block lipid biosynthesis in hypoxic cancer cells, impairing their survival under hypoxic conditions (Lewis et al., 2015). These results illustrate that SREBP2-mediated cholesterol metabolism participates in glioblastoma progression. Furthermore, decreasing the expression of the SREBP2 gene in glioblastoma can suppress mesenchymal transformation in non-mesenchymal gliomas, thereby inhibiting tumor metastasis (Ferrarese et al., 2023). Furthermore, lipid synthesis pathways mediated by SREBP2 are implicated in the resistance mechanism to temozolomide (TMZ), a common chemotherapy for GBM (Choo et al., 2023). Inhibiting SREBP2 enhances sensitivity to TMZ therapy, indicating its potential as a therapeutic target (Choo et al., 2023).
4.11 Melanoma
In melanoma, the SREBP2 pathway is activated by different ways, leading to high levels of cholesterol biosynthesis, which in turn sustains the rapid proliferation of melanoma cells both in vivo and in vitro (Yamauchi et al., 2011; Dinavahi et al., 2022). Additionally, in addition to promoting tumor progression by upregulating cholesterol metabolism, SREBP2 can also reduce reactive oxygen species (ROS) and lipid peroxidation, which confers resistance to inducers of ferroptosis in melanoma cells, thereby facilitating tumor progression (Hong et al., 2021).
4.12 Other cancers
In gastric cancer, expression of SREBP2 and cholesterol synthesis are promoted by sterol O-acyltransferase 1 (SOAT1), which facilitates cancer cells lymph node metastasis (Zhu et al., 2021). Another study shows that gastric cancer stem cells enhance the expression of SREBP2 to augment cholesterol metabolism, thereby altering tumor cell membranes and increasing their resistance to perforin released by natural killer (NK) cells (Zhu and Wang, 2024). In esophageal squamous cell carcinoma (ESCC), SREBP2 interacts with c-Myc, synergistically inducing the expression of HMGCR, which promotes ESCC migration, invasiveness, viability, and anchorage-independent growth (Zhong et al., 2019). Additionally, lysophosphatidylcholine acyltransferase 1 (LPCAT1) regulates the nuclear translocation of SREBP2, thereby promoting the proliferation of ESCC cells (Tao et al., 2021). In clear cell renal cell carcinoma (ccRCC), SREBP2 interacts with MED15 that acts as a SREBP2 coactivator to promote cholesterol biosynthesis enzyme expression, resulting in enhancing malignant tumor behavior phenotypes (Hua et al., 2024). PI3K/AKT/mTOR/SREBP2 pathway is upregulated by VHL mutations and the subsequent stabilization of HIFα, contributing to the accumulation of intracellular cholesteryl esters and facilitating ccRCC development (Zhang et al., 2024). In osteosarcoma, research indicates that SREBP2 phosphorylation at T610 by PKM2 enhances its stability and promoting tumorigenesis (Pu et al., 2022). In t (4; 11) leukemia, SREBP2 is significantly overexpressed and correlates with a poorer prognosis (Erkner et al., 2024). In summary, SREBP2 is significantly involved in the occurrence and development of various tumors (Table 1).
5 SREBP2 in tumor microenvironment
As previously discussed, SREBP2 signaling pathway is frequently activated in cancer cells, leading to increased cholesterol biosynthesis and uptake that promotes proliferation. However, the tumor microenvironment (TME) is a critical component of cancer, and current understanding of tumorigenesis and progression is increasingly shifting from a tumor cell-centric view to one that embraces the complexity of the tumor ecosystem (de Visser and Joyce, 2023). Consequently, recent many researches have focused on the regulation of SREBP2 within the TME. CD8+ T cells, which are immune cells capable of recognizing and eliminating cancer cells, are exposed to oxidized sterols secreted by cancer cells into the tumor microenvironment (TME) (Yan et al., 2023). These oxidized sterols significantly suppress SREBP2 activity in CD8+ T cells, resulting in cholesterol depletion and autophagy-mediated T cell apoptosis or dysfunction (Yan et al., 2023). Another study employing fatostatin to inhibit SREBP2 found that reduced cholesterol levels in the TME led to a decreased proportion of regulatory T (Treg) cells and alleviated CD8+ T cell exhaustion, thereby enhancing antitumor activity (Zhu et al., 2024). However, recent studies have shown that the upregulation of SREBP2 also supports CD8+ T cell function, not through cholesterol metabolism, but by mediating the synthesis of non-steroidal products such as coenzyme Q (CoQ) (Reina-Campos et al., 2023). This process promotes their differentiation into tissue-resident memory CD8+ T (TRM) cells, thereby enhancing antitumor immunity (Reina-Campos et al., 2023). Natural killer T (NKT) cells can mediate immune responses in cancer. Obesity-induced activation of SREBP2 and altered cholesterol metabolism in liver lead to hypercholesterolemia, resulting in excessive cholesterol uptake by NKT cells (Tang et al., 2022). This leads to the accumulation of lipid peroxides in NKT cells, thereby impairing their antitumor immunosurveillance capabilities (Tang et al., 2022). Furthermore, in melanoma, it has been observed that cancer-derived lactate activates SREBP2 in conventional dendritic cells (DCs), driving their transformation into regulatory DCs (mregDCs) (Plebanek et al., 2024). These mature mregDCs suppress DC antigen cross-presentation, thereby promoting melanoma progression (Plebanek et al., 2024).
6 Targeting SREBP2 for cancer therapy
As outlined above, abnormal expression and activation of SREBP2 play a significant role in the initiation and progression of various cancers. Consequently, targeting SREBP2 maturation or transcriptional activity has emerged as a high priority in cancer therapy. Recent studies have identified various SREBP2 inhibitors, and findings regarding several of these agents for cancer treatment are summarized below (Table 2).
6.1 Fatostatin
Fatostatin, a non-sterol diarylthiazole derivative, has been investigated for the treatment of multiple tumors. Originally developed from a chemical library in 2003 and referred to as 125B11, fatostatin has demonstrated promising antitumor effects (Choi et al., 2003). It inhibited IGF1-induced growth in prostate cancer DU-145 cells with an half-maximal inhibitory concentrations (IC50) of 0.1 µM (Choi et al., 2003). Mechanistically, fatostatin blocks the ER-Golgi translocation of SREBP2 by directly binding to SCAP at a site distinct from the sterol-binding domain, thereby inhibiting the cleavage and activation of SREBP2 (Kamisuki et al., 2009). In prostate cancer, fatostatin reduces the secretion of extracellular vesicles (EVs) from hypoxia-stimulated cancer cells by inhibiting cholesterol biosynthesis, which decreases tumor EMT, invasiveness, and stemness (Schlaepfer et al., 2015). A study utilizing human prostate cancer cell lines demonstrated that fatostatin inhibits cell proliferation, invasion, and migration while inducing caspase-mediated apoptosis, with IC50 (72 h of treatment) of 9.1 µM and 10.4 µM in C4-2B and LNCaP cells, respectively (Li et al., 2014). In vivo, fatostatin exhibited antitumor efficacy in a subcutaneous C4-2B xenograft mouse model when administered at a dose of 15 mg/kg for 42 days (Li et al., 2014). Another study also indicated that treatment with fatostatin (15 mg/kg) for 60 days in the Pmlpc−/− mouse prostate tumor metastatic model effectively suppressed the SREBP pathway and increased antimetastatic activity (Chen et al., 2018). In androgen receptor-negative prostate cancer cells harboring mutant p53, fatostatin also inhibited cell growth and induced apoptosis (Li et al., 2015). Similar to its effects in prostate cancer, fatostatin has been shown to inhibit growth and proliferation while inducing caspase-mediated apoptosis in endometrial carcinoma, with IC50 (after 72 h of treatment) of 17.96 µM and 4.53 µM in Ishikawa and HEC-1A cells, respectively (Gao et al., 2018; Yao et al., 2020). In breast cancer, fatostatin prevents SREBP2 activation, attenuating osteoclastogenesis and breast cancer-induced osteolysis in vivo, thereby providing therapeutic benefits for patients with osteolytic bone lesions (Jie et al., 2019). In HCC, fatostatin markedly suppressed the EMT process (Zhang et al., 2023). Studies focusing on the tumor microenvironment have revealed that fatostatin decreases SREBP2 activation and intracellular cholesterol accumulation, leading to a reduction in regulatory T (Treg) cells—immunosuppressive cells—while alleviating CD8+ T cell exhaustion in the TME (Zhu et al., 2024). These findings confirm fatostatin’s promise in tumor immunotherapy by inhibiting SREBP2 activation. In acute lymphoblastic leukemia (ALL), fatostatin disrupts cholesterol metabolism, effectively counteracting drug tolerance by inducing cell death and repressing stemness (Malyukova et al., 2024). In glioblastoma multiforme, fatostatin has shown potential in overcoming temozolomide resistance (Choo et al., 2023). Despite its potential therapeutic value, fatostatin has been shown to inhibit the growth of control cells due to its toxic effects (Li et al., 2014; Erkner et al., 2024). Consequently, clinical trials involving fatostatin have not advanced, likely due to safety concerns associated with the compound. The in vivo activity and pharmacokinetics of fatostatin are both lower than expected (Peng et al., 2023). In addition, a study has found that fatostatin has additional pathway targets rather than specifically blocking SCAP’s ER-to-Golgi transport (Shao et al., 2016). Therefore, continued efforts should be made to develop specific SCAP inhibitors.
6.2 Betulin
Betulin is a pentacyclic triterpene abundantly found in birch bark (Alakurtti et al., 2006). Unlike fatostatin, which directly binds to SCAP to inhibit the translocation of SREBP2 to the Golgi, the tumor-suppressive effects of betulin primarily rely on enhancing the interaction between SCAP and INSIG (Tang et al., 2011, 2021). This interaction obstructs SCAP from binding to COPII vesicles, preventing the SCAP-SREBP2 complex from exiting the endoplasmic reticulum (ER) and thereby inhibiting SREBP2 activation (Tang et al., 2011, 2021). A study on prostate cancer demonstrated that betulin significantly downregulated SREBP2 and its target genes, exhibiting inhibitory effects on the downstream activity of the androgen receptor (AR), which suppressed prostate cancer progression (Wei et al., 2024). The IC50 values of betulin for LNCaP and PC3 cell lines at 24 h were 4.921 μg/mL and 2.936 μg/mL, respectively. At 48 h, the IC50 values for LNCaP and PC3 cells were 7.347 μg/mL and 3.035 μg/mL, respectively (Wei et al., 2024). In a study focusing on obesity-induced breast cancer, betulin was shown to inhibit SREBP2 processing in MCF-7 cells (McClellan et al., 2022). This reduction led to decreased survival and proliferation of breast cancer cells (McClellan et al., 2022). In HCC, treatment with betulin effectively alleviated inflammatory responses and HCC development in vivo by targeting the SREBP2 pathway (Li et al., 2017). In terms of tumor resistance, betulin enhanced gefitinib sensitivity in lung cancer both in vitro and in vivo (Li et al., 2016). Treatment with betulin and gefitinib significantly reduced tumor growth by inhibiting SREBP2 and targeting tumor metabolism (Li et al., 2016). Studies in rat and dog models indicated that betulin exhibited very low toxicity, with no severe adverse effects observed in vitro (Jäger et al., 2008). However, Betulin has poor water solubility; therefore, preclinical studies on more soluble forms of betulin are required (Adepoju et al., 2023).
6.3 Xanthohumol
Xanthohumol (XN) is a prenylated flavonoid derived from natural food constituents, recognized for its antioxidant and anti-inflammatory properties, which contribute to its antitumor effects (Stevens et al., 2000; Lupinacci et al., 2009). Shingo Miyata et al. demonstrated that XN acts as a novel inactivator of SREBP2. Mechanistically, Sec23/24 is a heterodimer recruited to the ER membrane, driving the formation of COPII vesicles (Espenshade et al., 2002). The binding of XN to Sec23/24 disrupts the incorporation of the SCAP/SREBP2 complex into COPII, preventing its ER-to-Golgi translocation and thus inhibiting SREBP2 activation (Espenshade et al., 2002; Miyata et al., 2015). In vitro, treatment with 10 µM XN significantly decreased the production of mature SREBP2 in human hepatoma Huh-7 cells (Miyata et al., 2015). In vivo, mature SREBP-2 levels were markedly downregulated in apolipoprotein E-deficient (ApoE−/−) mice fed a Western-type diet and treated with XN (300 mg/kg body weight/day) for 8 weeks (Doddapattar et al., 2013). However, current data are insufficient to systematically evaluate the anticancer activity of XN, highlighting the need for further in vivo studies to enhance understanding of its clinical efficacy (Harish et al., 2021). Additionally, XN also suffers from low oral bioavailability (Legette et al., 2014).
6.4 PF-429242
PF-429242 is an aminopyrrolidineamide that inhibits endogenous SREBP2 processing and decreases SREBP2 activation through pharmacological inhibition of S1P activity (Hawkins et al., 2008). PF-429242 inhibited GSK343-induced SREBP2 activation, contributing to its anticancer activity in HCC(Yang et al., 2019). Furthermore, research has demonstrated that PF-429242 suppresses the expression of SREBP2 target genes and cholesterol synthesis in hepatoma HepG2 cells, exhibiting an IC50 of 0.5 µM. In a vivo research indicated that treatment with PF-429242 at a dose of 30 mg/kg for 24 h, administered every 6 h, effectively reduced the expression of hepatic SREBP2 target genes and lowered cholesterol levels in mice (Hawkins et al., 2008). In a series of in vitro experiments, PF-429242 was identified as an anti-glioblastoma agent by suppressing SREBP2 activation, inhibiting cell growth and inducing apoptotic cell death (Caruana et al., 2017). The IC50 of PF-429242 in T98G cells with full serum media was 15 ± 1.3 mM for 72 h, which was higher than the IC50 of 0.32 ± 0.09 mM in lipoprotein-deficient serum, indicating that extracellular cholesterol reduces the potency of PF-429242 (Caruana et al., 2017). They also assessed the IC50 of PF-429242 in two other human glioblastoma cell lines, U87-MG and A172, which were found to be 15.2 ± 3.0 mM and 27.6 ± 5.3 mM, respectively (Caruana et al., 2017). However, PF-429242 presents certain medicinal limitations, including a high clearance rate and poor oral bioavailability (Hawkins et al., 2008).
6.5 BF175
Xiaoping Zhao et al. designed and synthesized several boron-containing bioactive stilbene derivatives, among which BF175 was identified as an inhibitor of SREBP2 expression and transcriptional activity (Zhao et al., 2014). In a high-fat diet mouse model, treatment with BF175 (0.3 mg/g body weight/week) for 1 week significantly decreased the mRNA levels of SREBP2 and cholesterol biosynthesis in vivo (Zhao et al., 2014). Unlike the direct inhibition of SREBP1a by blocking the MED15-KIX and SREBP-1a-TAD interaction, the mechanism by which BF175 suppresses SREBP2 expression and transcriptional activity remains unclear and requires further investigation (Zhao et al., 2014). In endometrial cancer, treatment of AN3CA cells with varying concentrations of BF175 for 24 h resulted in a significant, dose-dependent reduction in SREBP2 expression, demonstrating antitumor activity (Wang et al., 2021). Additionally, BF175 exhibited low toxicity in tissue culture, suggesting it may be an ideal candidate as an SREBP2 inhibitor for cancer treatment (Zhao et al., 2014).
7 Conclusion and future directions
This review has provided an overview of the structure and regulation of SREBP2, highlighting its functions and mechanisms in various cancers. As a central transcription factor in cholesterol metabolism, SREBP2 plays a crucial role in malignancy by linking oncogenic signaling pathways with alterations in lipid metabolism. Additionally, SREBP2 also exerts tumorigenic effects through several non-cholesterol-related pathways. Numerous signaling molecules regulate the transcription, activation, and stability of SREBP2, driving tumorigenesis and progression. Additionally, while only a limited number of studies have explored this aspect, SREBP2 also influence inflammation, immunity and tumor microenvironment—processes that are intricately connected to tumor biology. Future studies should focus on elucidating the relationship between SREBP2-mediated immunity and tumorigenesis, as well as cancer progression. A more comprehensive understanding of these interactions may offer valuable insights into the role of SREBP2 in cancer and uncover novel therapeutic strategies targeting antitumor immunity.
Given that SREBP2 is involved in the progression of various cancers, it holds promise as both a prognostic biomarker and a potential therapeutic target, making the development of SREBP2 inhibitors a key component of future cancer treatment strategies. While several SREBP2 inhibitors have shown anticancer effects in preclinical models, they have not yet entered clinical trials. The development of these inhibitors faces several challenges, including issues related to poor solubility, insufficient potency, and rapid metabolism, which hinder their progression into clinical use. Overcoming these hurdles will require the establishment of efficient high-throughput screening systems for SREBP2 inhibitors and the rational design of compounds with improved pharmacological properties. In addition, current inhibitors lack specificity for SREBP2. In the SREBP family, SREBP1 and SREBP2 have the same activation pathways, which leads to inhibitors targeting both proteins simultaneously. Furthermore, these agents may exert effects on other oncogenic pathways. Fatostatin, Betulin, and Xanthohumol have been shown to suppress cancer progression not only via the SCAP-SREBP pathway but also through inhibition of mTOR, MAPK, and Notch signaling (Kunnimalaiyaan et al., 2015; Han et al., 2019; Cai et al., 2023). These multi-target inhibitors still require more research regarding their efficacy and safety. SREBP2-interacting proteins or therapeutic antibodies may offer superior target selectivity, though these approaches remain unexplored to date. Recent studies identified UT-59 as a SCAP-specific inhibitor that could open the door to developing therapeutic leads for cancer; however, its therapeutic efficacy and mechanistic details require further experimental validation (Xu et al., 2024). Given the critical role of the basic helix-loop-helix (bHLH) domain in SREBP2 function, designing inhibitors targeting this region represents a promising therapeutic development strategy. In conclusion, SREBP2 is a key regulator of cholesterol metabolism and plays an integral role in cancer progression. Targeting SREBP2 directly may present a promising anticancer strategy. Further investigation into its regulatory mechanisms in tumors, coupled with the development of more effective inhibitors, holds the potential to offer new, impactful therapeutic opportunities for cancer treatment.
Author contributions
RC: Writing – original draft, Writing – review and editing. TC: Visualization, Writing – review and editing. XL: Visualization, Writing – review and editing. JY: Writing – review and editing. ML: Writing – review and editing. SW: Writing – review and editing. MZ: Writing – review and editing. JC: Writing – review and editing. BY: Writing – review and editing. HZ: Supervision, Writing – review and editing. ZL: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work is supported by the Youth Project of Guangxi Natural Science Foundation (2023GXNSFBA026004), Baigui youth top talent training program, National Natural Science Foundation of China (82170421 and 82360464), Young leading talents training program of Guangxi Medical University, and Outstanding Young Talent Training Program of Guangxi Medical University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adepoju, F. O., Duru, K. C., Li, E., Kovaleva, E. G., and Tsurkan, M. V. (2023). Pharmacological potential of betulin as a multitarget compound. Biomolecules 13 (7), 1105. doi:10.3390/biom13071105
Akman, M., Monteleone, C., Doronzo, G., Godel, M., Napoli, F., Merlini, A., et al. (2024). TFEB controls sensitivity to chemotherapy and immuno-killing in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 43 (1), 219. doi:10.1186/s13046-024-03142-4
Alakurtti, S., Mäkelä, T., Koskimies, S., and Yli-Kauhaluoma, J. (2006). Pharmacological properties of the ubiquitous natural product betulin. Eur. J. Pharm. Sci. 29 (1), 1–13. doi:10.1016/j.ejps.2006.04.006
Aregger, M., Lawson, K. A., Billmann, M., Costanzo, M., Tong, A. H. Y., Chan, K., et al. (2020). Systematic mapping of genetic interactions for de novo fatty acid synthesis identifies C12orf49 as a regulator of lipid metabolism. Nat. Metab. 2 (6), 499–513. doi:10.1038/s42255-020-0211-z
Aylon, Y., Gershoni, A., Rotkopf, R., Biton, I. E., Porat, Z., Koh, A. P., et al. (2016). The LATS2 tumor suppressor inhibits SREBP and suppresses hepatic cholesterol accumulation. Genes Dev. 30 (7), 786–797. doi:10.1101/gad.274167.115
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 (2), 281–297. doi:10.1016/s0092-8674(04)00045-5
Bathaie, S. Z., Ashrafi, M., Azizian, M., and Tamanoi, F. (2017). Mevalonate pathway and human cancers. Curr. Mol. Pharmacol. 10 (2), 77–85. doi:10.2174/1874467209666160112123205
Bayraktar, E. C., La, K., Karpman, K., Unlu, G., Ozerdem, C., Ritter, D. J., et al. (2020). Metabolic coessentiality mapping identifies C12orf49 as a regulator of SREBP processing and cholesterol metabolism. Nat. Metab. 2 (6), 487–498. doi:10.1038/s42255-020-0206-9
Bennett, M. K., and Osborne, T. F. (2000). Nutrient regulation of gene expression by the sterol regulatory element binding proteins: increased recruitment of gene-specific coregulatory factors and selective hyperacetylation of histone H3 in vivo. Proc. Natl. Acad. Sci. U. S. A. 97 (12), 6340–6344. doi:10.1073/pnas.97.12.6340
Bian, X., Liu, R., Meng, Y., Xing, D., Xu, D., and Lu, Z. (2021). Lipid metabolism and cancer. J. Exp. Med. 218 (1), e20201606. doi:10.1084/jem.20201606
Briggs, M. R., Yokoyama, C., Wang, X., Brown, M. S., and Goldstein, J. L. (1993). Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. I. Identification of the protein and delineation of its target nucleotide sequence. J. Biol. Chem. 268 (19), 14490–14496. doi:10.1016/s0021-9258(19)85265-1
Brown, M. S., and Goldstein, J. L. (1997). The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89 (3), 331–340. doi:10.1016/s0092-8674(00)80213-5
Brown, M. S., Radhakrishnan, A., and Goldstein, J. L. (2018). Retrospective on cholesterol homeostasis: the central role of Scap. Annu. Rev. Biochem. 87, 783–807. doi:10.1146/annurev-biochem-062917-011852
Cai, C., Zhang, Y., and Peng, X. (2021). Knocking down Sterol regulatory element binding protein 2 (SREBF2) inhibits the Serine Protease 8 (PRSS8)/sodium channel epithelial 1alpha subunit (SCNN1A) axis to reduce the cell proliferation, migration and epithelial-mesenchymal transformation of ovarian cancer. Bioengineered 12 (2), 9390–9400. doi:10.1080/21655979.2021.1978615
Cai, D., Wang, J., Gao, B., Li, J., Wu, F., Zou, J. X., et al. (2019). RORγ is a targetable master regulator of cholesterol biosynthesis in a cancer subtype. Nat. Commun. 10 (1), 4621. doi:10.1038/s41467-019-12529-3
Cai, J., Ye, Z., Hu, Y., Ye, L., Gao, L., Wang, Y., et al. (2023). Fatostatin induces ferroptosis through inhibition of the AKT/mTORC1/GPX4 signaling pathway in glioblastoma. Cell Death Dis. 14 (3), 211. doi:10.1038/s41419-023-05738-8
Calvisi, D. F., Wang, C., Ho, C., Ladu, S., Lee, S. A., Mattu, S., et al. (2011). Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology 140 (3), 1071–1083. doi:10.1053/j.gastro.2010.12.006
Cao, L., Qin, Z., Yu, T., Bai, X., Jiang, S., Wang, D., et al. (2024). Tanshinone IIA acts as a regulator of lipogenesis to overcome osimertinib acquired resistance in lung cancer. Biochem. Pharmacol. 224, 116207. doi:10.1016/j.bcp.2024.116207
Caruana, B. T., Skoric, A., Brown, A. J., and Lutze-Mann, L. H. (2017). Site-1 protease, a novel metabolic target for glioblastoma. Biochem. Biophys. Res. Commun. 490 (3), 760–766. doi:10.1016/j.bbrc.2017.06.114
Casella, C., Miller, D. H., Lynch, K., and Brodsky, A. S. (2014). Oxysterols synergize with statins by inhibiting SREBP-2 in ovarian cancer cells. Gynecol. Oncol. 135 (2), 333–341. doi:10.1016/j.ygyno.2014.08.015
Che, L., Chi, W., Qiao, Y., Zhang, J., Song, X., Liu, Y., et al. (2020). Cholesterol biosynthesis supports the growth of hepatocarcinoma lesions depleted of fatty acid synthase in mice and humans. Gut 69 (1), 177–186. doi:10.1136/gutjnl-2018-317581
Chen, C., Matye, D., Wang, Y., and Li, T. (2021). Liver-specific microRNA-185 knockout promotes cholesterol dysregulation in mice. Liver Res. 5 (4), 232–238. doi:10.1016/j.livres.2020.09.001
Chen, M., Zhang, J., Sampieri, K., Clohessy, J. G., Mendez, L., Gonzalez-Billalabeitia, E., et al. (2018). An aberrant SREBP-dependent lipogenic program promotes metastatic prostate cancer. Nat. Genet. 50 (2), 206–218. doi:10.1038/s41588-017-0027-2
Chen, W., Wen, L., Bao, Y., Tang, Z., Zhao, J., Zhang, X., et al. (2022a). Gut flora disequilibrium promotes the initiation of liver cancer by modulating tryptophan metabolism and up-regulating SREBP2. Proc. Natl. Acad. Sci. U. S. A. 119 (52), e2203894119. doi:10.1073/pnas.2203894119
Chen, Y. Y., Ge, J. Y., Zhu, S. Y., Shao, Z. M., and Yu, K. D. (2022b). Copy number amplification of ENSA promotes the progression of triple-negative breast cancer via cholesterol biosynthesis. Nat. Commun. 13 (1), 791. doi:10.1038/s41467-022-28452-z
Choi, Y., Kawazoe, Y., Murakami, K., Misawa, H., and Uesugi, M. (2003). Identification of bioactive molecules by adipogenesis profiling of organic compounds. J. Biol. Chem. 278 (9), 7320–7324. doi:10.1074/jbc.M210283200
Choo, M., Mai, V. H., Kim, H. S., Kim, D. H., Ku, J. L., Lee, S. K., et al. (2023). Involvement of cell shape and lipid metabolism in glioblastoma resistance to temozolomide. Acta Pharmacol. Sin. 44 (3), 670–679. doi:10.1038/s41401-022-00984-6
Cohen, J. C., Horton, J. D., and Hobbs, H. H. (2011). Human fatty liver disease: old questions and new insights. Science 332 (6037), 1519–1523. doi:10.1126/science.1204265
Deng, Y. Z., Cai, Z., Shi, S., Jiang, H., Shang, Y. R., Ma, N., et al. (2018). Cilia loss sensitizes cells to transformation by activating the mevalonate pathway. J. Exp. Med. 215 (1), 177–195. doi:10.1084/jem.20170399
de Visser, K. E., and Joyce, J. A. (2023). The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell 41 (3), 374–403. doi:10.1016/j.ccell.2023.02.016
Dickson, A. S., Pauzaite, T., Arnaiz, E., Ortmann, B. M., West, J. A., Volkmar, N., et al. (2023). A HIF independent oxygen-sensitive pathway for controlling cholesterol synthesis. Nat. Commun. 14 (1), 4816. doi:10.1038/s41467-023-40541-1
Dinavahi, S. S., Chen, Y. C., Gowda, R., Dhanyamraju, P. K., Punnath, K., Desai, D., et al. (2022). Targeting protein translation in melanoma by inhibiting EEF-2 kinase regulates cholesterol metabolism though SREBP2 to inhibit tumour development. Int. J. Mol. Sci. 23 (7), 3481. doi:10.3390/ijms23073481
Doddapattar, P., Radović, B., Patankar, J. V., Obrowsky, S., Jandl, K., Nusshold, C., et al. (2013). Xanthohumol ameliorates atherosclerotic plaque formation, hypercholesterolemia, and hepatic steatosis in ApoE-deficient mice. Mol. Nutr. Food Res. 57 (10), 1718–1728. doi:10.1002/mnfr.201200794
Duan, X., Zhang, T., Feng, L., de Silva, N., Greenspun, B., Wang, X., et al. (2024). A pancreatic cancer organoid platform identifies an inhibitor specific to mutant KRAS. Cell Stem Cell 31 (1), 71–88.e8. doi:10.1016/j.stem.2023.11.011
Efeyan, A., Comb, W. C., and Sabatini, D. M. (2015). Nutrient-sensing mechanisms and pathways. Nature 517 (7534), 302–310. doi:10.1038/nature14190
Efsun Antmen, S., Canacankatan, N., Gürses, İ., Aytan, H., and Erden Ertürk, S. (2021). Relevance of lipogenesis and AMPK/Akt/mTOR signaling pathway in endometrial cancer. Eur. Rev. Med. Pharmacol. Sci. 25 (2), 687–695. doi:10.26355/eurrev_202101_24630
Eid, W., Dauner, K., Courtney, K. C., Gagnon, A., Parks, R. J., Sorisky, A., et al. (2017). mTORC1 activates SREBP-2 by suppressing cholesterol trafficking to lysosomes in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 114 (30), 7999–8004. doi:10.1073/pnas.1705304114
Erkner, E., Hentrich, T., Schairer, R., Fitzel, R., Secker-Grob, K. A., Jeong, J., et al. (2024). The RORɣ/SREBP2 pathway is a master regulator of cholesterol metabolism and serves as potential therapeutic target in t(4;11) leukemia. Oncogene 43 (4), 281–293. doi:10.1038/s41388-023-02903-3
Espenshade, P. J., Li, W. P., and Yabe, D. (2002). Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. Proc. Natl. Acad. Sci. U. S. A. 99 (18), 11694–11699. doi:10.1073/pnas.182412799
Ettinger, S. L., Sobel, R., Whitmore, T. G., Akbari, M., Bradley, D. R., Gleave, M. E., et al. (2004). Dysregulation of sterol response element-binding proteins and downstream effectors in prostate cancer during progression to androgen independence. Cancer Res. 64 (6), 2212–2221. doi:10.1158/0008-5472.can-2148-2
Feng, S., Han, M., Zhou, L., Wang, Q., Li, Z., Li, Y., et al. (2017). NS5ABP37 inhibits liver cancer by impeding lipogenesis and cholesterogenesis. Cancer Sci. 108 (1), 12–22. doi:10.1111/cas.13117
Ferrarese, R., Izzo, A., Andrieux, G., Lagies, S., Bartmuss, J. P., Masilamani, A. P., et al. (2023). ZBTB18 inhibits SREBP-dependent lipid synthesis by halting CTBPs and LSD1 activity in glioblastoma. Life Sci. Alliance 6 (1), e202201400. doi:10.26508/lsa.202201400
Fujita, K., and Nonomura, N. (2019). Role of androgen receptor in prostate cancer: a review. World J. Mens. Health 37 (3), 288–295. doi:10.5534/wjmh.180040
Gao, S., Shi, Z., Li, X., Li, W., Wang, Y., Liu, Z., et al. (2018). Fatostatin suppresses growth and enhances apoptosis by blocking SREBP-regulated metabolic pathways in endometrial carcinoma. Oncol. Rep. 39 (4), 1919–1929. doi:10.3892/or.2018.6265
Geng, C., Dong, T., Jin, W., Yu, B., Yin, F., Peng, F., et al. (2018). MicroRNA-98 regulates hepatic cholesterol metabolism via targeting sterol regulatory element-binding protein 2. Biochem. Biophys. Res. Commun. 504 (2), 422–426. doi:10.1016/j.bbrc.2018.08.205
Giandomenico, V., Simonsson, M., Grönroos, E., and Ericsson, J. (2003). Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol. Cell Biol. 23 (7), 2587–2599. doi:10.1128/mcb.23.7.2587-2599.2003
Goldstein, J. L., and Brown, M. S. (2015). A century of cholesterol and coronaries: from plaques to genes to statins. Cell 161 (1), 161–172. doi:10.1016/j.cell.2015.01.036
Goldstein, J. L., DeBose-Boyd, R. A., and Brown, M. S. (2006). Protein sensors for membrane sterols. Cell 124 (1), 35–46. doi:10.1016/j.cell.2005.12.022
Gong, X., Qian, H., Shao, W., Li, J., Wu, J., Liu, J. J., et al. (2016). Complex structure of the fission yeast SREBP-SCAP binding domains reveals an oligomeric organization. Cell Res. 26 (11), 1197–1211. doi:10.1038/cr.2016.123
Gong, Y., Lee, J. N., Lee, P. C., Goldstein, J. L., Brown, M. S., and Ye, J. (2006). Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell Metab. 3 (1), 15–24. doi:10.1016/j.cmet.2005.11.014
Gruenbacher, G., and Thurnher, M. (2015). Mevalonate metabolism in cancer. Cancer Lett. 356 (2 Pt A), 192–196. doi:10.1016/j.canlet.2014.01.013
Gu, D., Zhou, F., You, H., Gao, J., Kang, T., Dixit, D., et al. (2023). Sterol regulatory element-binding protein 2 maintains glioblastoma stem cells by keeping the balance between cholesterol biosynthesis and uptake. Neuro Oncol. 25 (9), 1578–1591. doi:10.1093/neuonc/noad060
Gu, X., Zhu, Q., Tian, G., Song, W., Wang, T., Wang, A., et al. (2022). KIF11 manipulates SREBP2-dependent mevalonate cross talk to promote tumor progression in pancreatic ductal adenocarcinoma. Cancer Med. 11 (17), 3282–3295. doi:10.1002/cam4.4683
Hamm, R., Chen, Y. R., Seo, E. J., Zeino, M., Wu, C. F., Müller, R., et al. (2014). Induction of cholesterol biosynthesis by archazolid B in T24 bladder cancer cells. Biochem. Pharmacol. 91 (1), 18–30. doi:10.1016/j.bcp.2014.06.018
Han, Y. H., Mun, J. G., Jeon, H. D., Kee, J. Y., and Hong, S. H. (2019). Betulin inhibits lung metastasis by inducing cell cycle arrest, autophagy, and apoptosis of metastatic colorectal cancer cells. Nutrients 12 (1), 66. doi:10.3390/nu12010066
Harish, V., Haque, E., Śmiech, M., Taniguchi, H., Jamieson, S., Tewari, D., et al. (2021). Xanthohumol for human malignancies: chemistry, pharmacokinetics and molecular targets. Int. J. Mol. Sci. 22 (9), 4478. doi:10.3390/ijms22094478
Hawkins, J. L., Robbins, M. D., Warren, L. C., Xia, D., Petras, S. F., Valentine, J. J., et al. (2008). Pharmacologic inhibition of site 1 protease activity inhibits sterol regulatory element-binding protein processing and reduces lipogenic enzyme gene expression and lipid synthesis in cultured cells and experimental animals. J. Pharmacol. Exp. Ther. 326 (3), 801–808. doi:10.1124/jpet.108.139626
He, W., Wang, M., Zhang, X., Wang, Y., Zhao, D., Li, W., et al. (2024). Estrogen induces LCAT to maintain cholesterol homeostasis and suppress hepatocellular carcinoma development. Cancer Res. 84 (15), 2417–2431. doi:10.1158/0008-5472.Can-23-3966
Hendrix, S., Dartigue, V., Hall, H., Bawaria, S., Kingma, J., Bajaj, B., et al. (2024a). SPRING licenses S1P-mediated cleavage of SREBP2 by displacing an inhibitory pro-domain. Nat. Commun. 15 (1), 5732. doi:10.1038/s41467-024-50068-8
Hendrix, S., Tan, J. M. E., Ndoj, K., Kingma, J., Valiloo, M., Zijlstra, L. F., et al. (2024b). SPRING is a dedicated licensing factor for SREBP-specific activation by S1P. Mol. Cell Biol. 44 (4), 123–137. doi:10.1080/10985549.2024.2348711
Hirano, Y., Murata, S., Tanaka, K., Shimizu, M., and Sato, R. (2003). Sterol regulatory element-binding proteins are negatively regulated through SUMO-1 modification independent of the ubiquitin/26 S proteasome pathway. J. Biol. Chem. 278 (19), 16809–16819. doi:10.1074/jbc.M212448200
Hong, X., Roh, W., Sullivan, R. J., Wong, K. H. K., Wittner, B. S., Guo, H., et al. (2021). The lipogenic regulator SREBP2 induces transferrin in circulating melanoma cells and suppresses ferroptosis. Cancer Discov. 11 (3), 678–695. doi:10.1158/2159-8290.Cd-19-1500
Horii, R., Honda, M., Shirasaki, T., Shimakami, T., Shimizu, R., Yamanaka, S., et al. (2019). MicroRNA-10a impairs liver metabolism in hepatitis C virus-related cirrhosis through deregulation of the circadian clock gene brain and muscle aryl hydrocarbon receptor nuclear translocator-like 1. Hepatol. Commun. 3 (12), 1687–1703. doi:10.1002/hep4.1431
Horton, J. D., Goldstein, J. L., and Brown, M. S. (2002). SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest 109 (9), 1125–1131. doi:10.1172/jci15593
Horton, J. D., Shah, N. A., Warrington, J. A., Anderson, N. N., Park, S. W., Brown, M. S., et al. (2003). Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. U. S. A. 100 (21), 12027–12032. doi:10.1073/pnas.1534923100
Hua, X., Ge, S., Zhang, L., Jiang, Q., Chen, J., Xiao, H., et al. (2024). MED15 is upregulated by HIF-2α and promotes proliferation and metastasis in clear cell renal cell carcinoma via activation of SREBP-dependent fatty acid synthesis. Cell Death Discov. 10 (1), 188. doi:10.1038/s41420-024-01944-1
Hua, X., Yokoyama, C., Wu, J., Briggs, M. R., Brown, M. S., Goldstein, J. L., et al. (1993). SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc. Natl. Acad. Sci. U. S. A. 90 (24), 11603–11607. doi:10.1073/pnas.90.24.11603
Huang, L., Luo, J., Song, N., Gao, W., Zhu, L., and Yao, W. (2022). CRISPR/Cas9-Mediated knockout of miR-130b affects mono- and polyunsaturated fatty acid content via PPARG-pgc1α Axis in goat mammary epithelial cells. Int. J. Mol. Sci. 23 (7), 3640. doi:10.3390/ijms23073640
Huang, Y., Jin, Q., Su, M., Ji, F., Wang, N., Zhong, C., et al. (2017). Leptin promotes the migration and invasion of breast cancer cells by upregulating ACAT2. Cell Oncol. (Dordr) 40 (6), 537–547. doi:10.1007/s13402-017-0342-8
Huber, M. D., Vesely, P. W., Datta, K., and Gerace, L. (2013). Erlins restrict SREBP activation in the ER and regulate cellular cholesterol homeostasis. J. Cell Biol. 203 (3), 427–436. doi:10.1083/jcb.201305076
Hunt, B. G., Davis, J. C., Fox, L. H., Vicente-Muñoz, S., Lester, C., Wells, S. I., et al. (2023). RON-augmented cholesterol biosynthesis in breast cancer metastatic progression and recurrence. Oncogene 42 (21), 1716–1727. doi:10.1038/s41388-023-02688-5
Hwang, H. J., Lee, K. H., and Cho, J. Y. (2023). ABCA9, an ER cholesterol transporter, inhibits breast cancer cell proliferation via SREBP-2 signaling. Cancer Sci. 114 (4), 1451–1463. doi:10.1111/cas.15710
Irisawa, M., Inoue, J., Ozawa, N., Mori, K., and Sato, R. (2009). The sterol-sensing endoplasmic reticulum (ER) membrane protein TRC8 hampers ER to Golgi transport of sterol regulatory element-binding protein-2 (SREBP-2)/SREBP cleavage-activated protein and reduces SREBP-2 cleavage. J. Biol. Chem. 284 (42), 28995–29004. doi:10.1074/jbc.M109.041376
Jäger, S., Laszczyk, M. N., and Scheffler, A. (2008). A preliminary pharmacokinetic study of betulin, the main pentacyclic triterpene from extract of outer bark of birch (Betulae alba cortex). Molecules 13 (12), 3224–3235. doi:10.3390/molecules13123224
Jeon, T. I., Esquejo, R. M., Roqueta-Rivera, M., Phelan, P. E., Moon, Y. A., Govindarajan, S. S., et al. (2013). An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell Metab. 18 (1), 51–61. doi:10.1016/j.cmet.2013.06.010
Jie, Z., Xie, Z., Xu, W., Zhao, X., Jin, G., Sun, X., et al. (2019). SREBP-2 aggravates breast cancer associated osteolysis by promoting osteoclastogenesis and breast cancer metastasis. Biochim. Biophys. Acta Mol. Basis Dis. 1865 (1), 115–125. doi:10.1016/j.bbadis.2018.10.026
Jin, H., Zhu, M., Zhang, D., Liu, X., Guo, Y., Xia, L., et al. (2023). B7H3 increases ferroptosis resistance by inhibiting cholesterol metabolism in colorectal cancer. Cancer Sci. 114 (11), 4225–4236. doi:10.1111/cas.15944
Kamisuki, S., Mao, Q., Abu-Elheiga, L., Gu, Z., Kugimiya, A., Kwon, Y., et al. (2009). A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chem. Biol. 16 (8), 882–892. doi:10.1016/j.chembiol.2009.07.007
Kober, D. L., Radhakrishnan, A., Goldstein, J. L., Brown, M. S., Clark, L. D., Bai, X. C., et al. (2021). Scap structures highlight key role for rotation of intertwined luminal loops in cholesterol sensing. Cell 184 (14), 3689–3701.e22. doi:10.1016/j.cell.2021.05.019
Kober, D. L., Xu, S., Li, S., Bajaj, B., Liang, G., Rosenbaum, D. M., et al. (2020). Identification of a degradation signal at the carboxy terminus of SREBP2: a new role for this domain in cholesterol homeostasis. Proc. Natl. Acad. Sci. U. S. A. 117 (45), 28080–28091. doi:10.1073/pnas.2018578117
Kotzka, J., Lehr, S., Roth, G., Avci, H., Knebel, B., and Muller-Wieland, D. (2004). Insulin-activated Erk-mitogen-activated protein kinases phosphorylate sterol regulatory element-binding Protein-2 at serine residues 432 and 455 in vivo. J. Biol. Chem. 279 (21), 22404–22411. doi:10.1074/jbc.M401198200
Kunnimalaiyaan, S., Sokolowski, K. M., Balamurugan, M., Gamblin, T. C., and Kunnimalaiyaan, M. (2015). Xanthohumol inhibits Notch signaling and induces apoptosis in hepatocellular carcinoma. PLoS One 10 (5), e0127464. doi:10.1371/journal.pone.0127464
Lai, C. R., Tsai, Y. L., Tsai, W. C., Chen, T. M., Chang, H. H., Changchien, C. Y., et al. (2022). Farnesoid X receptor overexpression decreases the migration, invasion and angiogenesis of human bladder cancers via AMPK activation and cholesterol biosynthesis inhibition. Cancers (Basel) 14 (18), 4398. doi:10.3390/cancers14184398
Lee, J. N., Song, B., DeBose-Boyd, R. A., and Ye, J. (2006). Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase gp78. J. Biol. Chem. 281 (51), 39308–39315. doi:10.1074/jbc.M608999200
Legette, L., Karnpracha, C., Reed, R. L., Choi, J., Bobe, G., Christensen, J. M., et al. (2014). Human pharmacokinetics of xanthohumol, an antihyperglycemic flavonoid from hops. Mol. Nutr. Food Res. 58 (2), 248–255. doi:10.1002/mnfr.201300333
Lewis, C. A., Brault, C., Peck, B., Bensaad, K., Griffiths, B., Mitter, R., et al. (2015). SREBP maintains lipid biosynthesis and viability of cancer cells under lipid- and oxygen-deprived conditions and defines a gene signature associated with poor survival in glioblastoma multiforme. Oncogene 34 (40), 5128–5140. doi:10.1038/onc.2014.439
Li, J., Yan, H., Zhao, L., Jia, W., Yang, H., Liu, L., et al. (2016). Inhibition of SREBP increases gefitinib sensitivity in non-small cell lung cancer cells. Oncotarget 7 (32), 52392–52403. doi:10.18632/oncotarget.10721
Li, N., Zhou, Z. S., Shen, Y., Xu, J., Miao, H. H., Xiong, Y., et al. (2017). Inhibition of the sterol regulatory element-binding protein pathway suppresses hepatocellular carcinoma by repressing inflammation in mice. Hepatology 65 (6), 1936–1947. doi:10.1002/hep.29018
Li, X., Chen, Y. T., Hu, P., and Huang, W. C. (2014). Fatostatin displays high antitumor activity in prostate cancer by blocking SREBP-regulated metabolic pathways and androgen receptor signaling. Mol. Cancer Ther. 13 (4), 855–866. doi:10.1158/1535-7163.Mct-13-0797
Li, X., Chen, Y. T., Josson, S., Mukhopadhyay, N. K., Kim, J., Freeman, M. R., et al. (2013). MicroRNA-185 and 342 inhibit tumorigenicity and induce apoptosis through blockade of the SREBP metabolic pathway in prostate cancer cells. PLoS One 8 (8), e70987. doi:10.1371/journal.pone.0070987
Li, X., Wu, J. B., Chung, L. W., and Huang, W. C. (2015). Anti-cancer efficacy of SREBP inhibitor, alone or in combination with docetaxel, in prostate cancer harboring p53 mutations. Oncotarget 6 (38), 41018–41032. doi:10.18632/oncotarget.5879
Liang, B., Chen, R., Song, S., Wang, H., Sun, G., Yang, H., et al. (2019). ASPP2 inhibits tumor growth by repressing the mevalonate pathway in hepatocellular carcinoma. Cell Death Dis. 10 (11), 830. doi:10.1038/s41419-019-2054-7
Lin, G., Huang, T., Zhang, X., and Wang, G. (2022). Deubiquitinase USP35 stabilizes BRPF1 to activate mevalonate (MVA) metabolism during prostate tumorigenesis. Cell Death Discov. 8 (1), 453. doi:10.1038/s41420-022-01231-x
Liu, Z., Zheng, X., Chen, J., Zheng, L., Ma, Z., Chen, L., et al. (2023). NFYC-37 promotes tumor growth by activating the mevalonate pathway in bladder cancer. Cell Rep. 42 (8), 112963. doi:10.1016/j.celrep.2023.112963
Longo, J., Mullen, P. J., Yu, R., van Leeuwen, J. E., Masoomian, M., Woon, D. T. S., et al. (2019). An actionable sterol-regulated feedback loop modulates statin sensitivity in prostate cancer. Mol. Metab. 25, 119–130. doi:10.1016/j.molmet.2019.04.003
Loregger, A., Raaben, M., Nieuwenhuis, J., Tan, J. M. E., Jae, L. T., van den Hengel, L. G., et al. (2020). Haploid genetic screens identify SPRING/C12ORF49 as a determinant of SREBP signaling and cholesterol metabolism. Nat. Commun. 11 (1), 1128. doi:10.1038/s41467-020-14811-1
Luo, J., Yang, H., and Song, B. L. (2020). Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 21 (4), 225–245. doi:10.1038/s41580-019-0190-7
Lupinacci, E., Meijerink, J., Vincken, J. P., Gabriele, B., Gruppen, H., and Witkamp, R. F. (2009). Xanthohumol from hop (Humulus lupulus L.) is an efficient inhibitor of monocyte chemoattractant protein-1 and tumor necrosis factor-alpha release in LPS-stimulated RAW 264.7 mouse macrophages and U937 human monocytes. J. Agric. Food Chem. 57 (16), 7274–7281. doi:10.1021/jf901244k
Maghe, C., Trillet, K., André-Grégoire, G., Kerhervé, M., Merlet, L., Jacobs, K. A., et al. (2024). The paracaspase MALT1 controls cholesterol homeostasis in glioblastoma stem-like cells through lysosome proteome shaping. Cell Rep. 43 (1), 113631. doi:10.1016/j.celrep.2023.113631
Maier, C. R., Hartmann, O., Prieto-Garcia, C., Al-Shami, K. M., Schlicker, L., Vogel, F. C. E., et al. (2023). USP28 controls SREBP2 and the mevalonate pathway to drive tumour growth in squamous cancer. Cell Death Differ. 30 (7), 1710–1725. doi:10.1038/s41418-023-01173-6
Malyukova, A., Lahnalampi, M., Falqués-Costa, T., Pölönen, P., Sipola, M., Mehtonen, J., et al. (2024). Sequential drug treatment targeting cell cycle and cell fate regulatory programs blocks non-genetic cancer evolution in acute lymphoblastic leukemia. Genome Biol. 25 (1), 143. doi:10.1186/s13059-024-03260-4
McClellan, B., Pham, T., Harlow, B., Lee, G., Quach, D., Jolly, C., et al. (2022). Modulation of breast cancer cell FASN expression by obesity-related systemic factors. Breast Cancer (Auckl) 16, 11782234221111374. doi:10.1177/11782234221111374
Menendez, J. A., and Lupu, R. (2007). Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 7 (10), 763–777. doi:10.1038/nrc2222
Meng, C., Zhou, L., Huang, L., Gu, Q., Du, X., Wang, C., et al. (2024). Chlorogenic acid regulates the expression of NPC1L1 and HMGCR through PXR and SREBP2 signaling pathways and their interactions with HSP90 to maintain cholesterol homeostasis. Phytomedicine 123, 155271. doi:10.1016/j.phymed.2023.155271
Miserez, A. R., Cao, G., Probst, L. C., and Hobbs, H. H. (1997). Structure of the human gene encoding sterol regulatory element binding protein 2 (SREBF2). Genomics 40 (1), 31–40. doi:10.1006/geno.1996.4525
Miyata, S., Inoue, J., Shimizu, M., and Sato, R. (2015). Xanthohumol improves diet-induced obesity and fatty liver by suppressing sterol regulatory element-binding protein (SREBP) activation. J. Biol. Chem. 290 (33), 20565–20579. doi:10.1074/jbc.M115.656975
Mok, E. H. K., Leung, C. O. N., Zhou, L., Lei, M. M. L., Leung, H. W., Tong, M., et al. (2022). Caspase-3-Induced activation of SREBP2 drives drug resistance via promotion of cholesterol biosynthesis in hepatocellular carcinoma. Cancer Res. 82 (17), 3102–3115. doi:10.1158/0008-5472.Can-21-2934
Muta, Y., Linares, J. F., Martinez-Ordoñez, A., Duran, A., Cid-Diaz, T., Kinoshita, H., et al. (2023). Enhanced SREBP2-driven cholesterol biosynthesis by PKCλ/ι deficiency in intestinal epithelial cells promotes aggressive serrated tumorigenesis. Nat. Commun. 14 (1), 8075. doi:10.1038/s41467-023-43690-5
Ning, S., Liu, C., Wang, K., Cai, Y., Ning, Z., Li, M., et al. (2023). NUCB2/Nesfatin-1 drives breast cancer metastasis through the up-regulation of cholesterol synthesis via the mTORC1 pathway. J. Transl. Med. 21 (1), 362. doi:10.1186/s12967-023-04236-x
Pan, Q., Zhong, S., Wang, H., Wang, X., Li, N., Li, Y., et al. (2021). The ZMYND8-regulated mevalonate pathway endows YAP-high intestinal cancer with metabolic vulnerability. Mol. Cell 81 (13), 2736–2751.e8. doi:10.1016/j.molcel.2021.04.009
Peng, Z., Chen, L., Wang, M., Yue, X., Wei, H., Xu, F., et al. (2023). SREBP inhibitors: an updated patent review for 2008-present. Expert Opin. Ther. Pat. 33 (10), 669–680. doi:10.1080/13543776.2023.2291393
Peterson, T. R., Sengupta, S. S., Harris, T. E., Carmack, A. E., Kang, S. A., Balderas, E., et al. (2011). mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146 (3), 408–420. doi:10.1016/j.cell.2011.06.034
Plebanek, M. P., Xue, Y., Nguyen, Y. V., DeVito, N. C., Wang, X., Holtzhausen, A., et al. (2024). A lactate-SREBP2 signaling axis drives tolerogenic dendritic cell maturation and promotes cancer progression. Sci. Immunol. 9 (95), eadi4191. doi:10.1126/sciimmunol.adi4191
Pomorski, T., Hrafnsdóttir, S., Devaux, P. F., and van Meer, G. (2001). Lipid distribution and transport across cellular membranes. Semin. Cell Dev. Biol. 12 (2), 139–148. doi:10.1006/scdb.2000.0231
Pu, F., Liu, J., Jing, D., Chen, F., Huang, X., Shi, D., et al. (2022). LncCCAT1 interaction protein PKM2 upregulates SREBP2 phosphorylation to promote osteosarcoma tumorigenesis by enhancing the Warburg effect and lipogenesis. Int. J. Oncol. 60 (4), 44. doi:10.3892/ijo.2022.5334
Radhakrishnan, A., Goldstein, J. L., McDonald, J. G., and Brown, M. S. (2008). Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 8 (6), 512–521. doi:10.1016/j.cmet.2008.10.008
Rawson, R. B., DeBose-Boyd, R., Goldstein, J. L., and Brown, M. S. (1999). Failure to cleave sterol regulatory element-binding proteins (SREBPs) causes cholesterol auxotrophy in Chinese hamster ovary cells with genetic absence of SREBP cleavage-activating protein. J. Biol. Chem. 274 (40), 28549–28556. doi:10.1074/jbc.274.40.28549
Rawson, R. B., Zelenski, N. G., Nijhawan, D., Ye, J., Sakai, J., Hasan, M. T., et al. (1997). Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell 1 (1), 47–57. doi:10.1016/s1097-2765(00)80006-4
Reina-Campos, M., Heeg, M., Kennewick, K., Mathews, I. T., Galletti, G., Luna, V., et al. (2023). Metabolic programs of T cell tissue residency empower tumour immunity. Nature 621 (7977), 179–187. doi:10.1038/s41586-023-06483-w
Ricoult, S. J., Yecies, J. L., Ben-Sahra, I., and Manning, B. D. (2016). Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through mTORC1 and SREBP. Oncogene 35 (10), 1250–1260. doi:10.1038/onc.2015.179
Saito, Y., Yin, D., Kubota, N., Wang, X., Filliol, A., Remotti, H., et al. (2023). A therapeutically targetable TAZ-TEAD2 pathway drives the growth of hepatocellular carcinoma via ANLN and KIF23. Gastroenterology 164 (7), 1279–1292. doi:10.1053/j.gastro.2023.02.043
Sakai, J., Rawson, R. B., Espenshade, P. J., Cheng, D., Seegmiller, A. C., Goldstein, J. L., et al. (1998). Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol. Cell 2 (4), 505–514. doi:10.1016/s1097-2765(00)80150-1
Sato, R., Inoue, J., Kawabe, Y., Kodama, T., Takano, T., and Maeda, M. (1996). Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J. Biol. Chem. 271 (43), 26461–26464. doi:10.1074/jbc.271.43.26461
Schlaepfer, I. R., Nambiar, D. K., Ramteke, A., Kumar, R., Dhar, D., Agarwal, C., et al. (2015). Hypoxia induces triglycerides accumulation in prostate cancer cells and extracellular vesicles supporting growth and invasiveness following reoxygenation. Oncotarget 6 (26), 22836–22856. doi:10.18632/oncotarget.4479
Schwartz, M. W., Seeley, R. J., Tschöp, M. H., Woods, S. C., Morton, G. J., Myers, M. G., et al. (2013). Cooperation between brain and islet in glucose homeostasis and diabetes. Nature 503 (7474), 59–66. doi:10.1038/nature12709
Shangguan, X., Ma, Z., Yu, M., Ding, J., Xue, W., and Qi, J. (2022). Squalene epoxidase metabolic dependency is a targetable vulnerability in castration-resistant prostate cancer. Cancer Res. 82 (17), 3032–3044. doi:10.1158/0008-5472.Can-21-3822
Shao, W., and Espenshade, P. J. (2014). Sterol regulatory element-binding protein (SREBP) cleavage regulates Golgi-to-endoplasmic reticulum recycling of SREBP cleavage-activating protein (SCAP). J. Biol. Chem. 289 (11), 7547–7557. doi:10.1074/jbc.M113.545699
Shao, W., Machamer, C. E., and Espenshade, P. J. (2016). Fatostatin blocks ER exit of SCAP but inhibits cell growth in a SCAP-independent manner. J. Lipid Res. 57 (8), 1564–1573. doi:10.1194/jlr.M069583
Sharma, B., Gupta, V., Dahiya, D., Kumar, H., Vaiphei, K., and Agnihotri, N. (2019). Clinical relevance of cholesterol homeostasis genes in colorectal cancer. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1864 (10), 1314–1327. doi:10.1016/j.bbalip.2019.06.008
Shirasaki, T., Honda, M., Shimakami, T., Horii, R., Yamashita, T., Sakai, Y., et al. (2013). MicroRNA-27a regulates lipid metabolism and inhibits hepatitis C virus replication in human hepatoma cells. J. Virol. 87 (9), 5270–5286. doi:10.1128/jvi.03022-12
Stevens, J. F., Taylor, A. W., Nickerson, G. B., Ivancic, M., Henning, J., Haunold, A., et al. (2000). Prenylflavonoid variation in Humulus lupulus: distribution and taxonomic significance of xanthogalenol and 4'-O-methylxanthohumol. Phytochemistry 53 (7), 759–775. doi:10.1016/s0031-9422(00)00005-4
Sun, L. P., Li, L., Goldstein, J. L., and Brown, M. S. (2005). Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J. Biol. Chem. 280 (28), 26483–26490. doi:10.1074/jbc.M504041200
Sundqvist, A., Bengoechea-Alonso, M. T., Ye, X., Lukiyanchuk, V., Jin, J., Harper, J. W., et al. (2005). Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7). Cell Metab. 1 (6), 379–391. doi:10.1016/j.cmet.2005.04.010
Tan, W., Wang, G., Liu, G., You, D., Wei, M., Jin, X., et al. (2022). The elevation of miR-185-5p alleviates high-fat diet-induced atherosclerosis and lipid accumulation in vivo and in vitro via SREBP2 activation. Aging (Albany NY) 14 (4), 1729–1742. doi:10.18632/aging.203896
Tang, J. J., Li, J. G., Qi, W., Qiu, W. W., Li, P. S., Li, B. L., et al. (2011). Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metab. 13 (1), 44–56. doi:10.1016/j.cmet.2010.12.004
Tang, J. J., Li, J. G., Qi, W., Qiu, W. W., Li, P. S., Li, B. L., et al. (2021). Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metab. 33 (1), 222. doi:10.1016/j.cmet.2020.11.013
Tang, W., Zhou, J., Yang, W., Feng, Y., Wu, H., Mok, M. T. S., et al. (2022). Aberrant cholesterol metabolic signaling impairs antitumor immunosurveillance through natural killer T cell dysfunction in obese liver. Cell Mol. Immunol. 19 (7), 834–847. doi:10.1038/s41423-022-00872-3
Tang, X., Qin, Q., Xu, W., and Zhang, X. (2023). Long non-coding RNA TUG1 attenuates insulin resistance in mice with gestational diabetes mellitus via regulation of the MicroRNA-328-3p/SREBP-2/ERK Axis. Diabetes Metab. J. 47 (2), 267–286. doi:10.4093/dmj.2021.0216
Tao, M., Luo, J., Gu, T., Yu, X., Song, Z., Jun, Y., et al. (2021). LPCAT1 reprogramming cholesterol metabolism promotes the progression of esophageal squamous cell carcinoma. Cell Death Dis. 12 (9), 845. doi:10.1038/s41419-021-04132-6
Tao, R., Xiong, X., DePinho, R. A., Deng, C. X., and Dong, X. C. (2013). Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J. Lipid Res. 54 (10), 2745–2753. doi:10.1194/jlr.M039339
Tian, J., Goldstein, J. L., Li, S., Schumacher, M. M., and Brown, M. S. (2024). Phosphorylation of Insig-2 mediates inhibition of fatty acid synthesis by polyunsaturated fatty acids. Proc. Natl. Acad. Sci. U. S. A. 121 (34), e2409262121. doi:10.1073/pnas.2409262121
Tsai, C. L., Changchien, C. Y., Chen, Y., Lai, C. R., Chen, T. M., Chang, H. H., et al. (2022). Survival benefit of statin with anti-angiogenesis efficacy in lung cancer-associated pleural fluid through FXR modulation. Cancers (Basel) 14 (11), 2765. doi:10.3390/cancers14112765
Walker, A. K., Yang, F., Jiang, K., Ji, J. Y., Watts, J. L., Purushotham, A., et al. (2010). Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 24 (13), 1403–1417. doi:10.1101/gad.1901210
Wang, Q., Hu, R., Li, W., Tai, Y., Gu, W., Das, B. C., et al. (2021). BF175 inhibits endometrial carcinoma through SREBP-regulated metabolic pathways in vitro. Mol. Cell Endocrinol. 523, 111135. doi:10.1016/j.mce.2020.111135
Wang, S., Zhou, Y., Yu, R., Ling, J., Li, B., Yang, C., et al. (2023). Loss of hepatic FTCD promotes lipid accumulation and hepatocarcinogenesis by upregulating PPARγ and SREBP2. JHEP Rep. 5 (10), 100843. doi:10.1016/j.jhepr.2023.100843
Wang, X., Briggs, M. R., Hua, X., Yokoyama, C., Goldstein, J. L., and Brown, M. S. (1993). Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. II. Purification and characterization. J. Biol. Chem. 268 (19), 14497–14504. doi:10.1016/s0021-9258(19)85266-3
Weber, L. W., Boll, M., and Stampfl, A. (2004). Maintaining cholesterol homeostasis: sterol regulatory element-binding proteins. World J. Gastroenterol. 10 (21), 3081–3087. doi:10.3748/wjg.v10.i21.3081
Wei, G., Zhu, H., Zhou, Y., Pan, Y., Yi, B., and Bai, Y. (2024). Single-cell sequencing revealed metabolic reprogramming and its transcription factor regulatory network in prostate cancer. Transl. Oncol. 44, 101925. doi:10.1016/j.tranon.2024.101925
Wei, M., Nurjanah, U., Herkilini, A., Huang, C., Li, Y., Miyagishi, M., et al. (2022). Unspliced XBP1 contributes to cholesterol biosynthesis and tumorigenesis by stabilizing SREBP2 in hepatocellular carcinoma. Cell Mol. Life Sci. 79 (9), 472. doi:10.1007/s00018-022-04504-x
Wen, Y. A., Xiong, X., Zaytseva, Y. Y., Napier, D. L., Vallee, E., Li, A. T., et al. (2018). Downregulation of SREBP inhibits tumor growth and initiation by altering cellular metabolism in colon cancer. Cell Death Dis. 9 (3), 265. doi:10.1038/s41419-018-0330-6
Xiang, X., Ohshiro, K., Zaidi, S., Yang, X., Bhowmick, K., Vegesna, A. K., et al. (2022). Impaired reciprocal regulation between SIRT6 and TGF-β signaling in fatty liver. Faseb J. 36 (6), e22335. doi:10.1096/fj.202101518R
Xiao, J., Xiong, Y., Yang, L. T., Wang, J. Q., Zhou, Z. M., Dong, L. W., et al. (2021). POST1/C12ORF49 regulates the SREBP pathway by promoting site-1 protease maturation. Protein Cell 12 (4), 279–296. doi:10.1007/s13238-020-00753-3
Xu, D., Wang, Z., Xia, Y., Shao, F., Xia, W., Wei, Y., et al. (2020). The gluconeogenic enzyme PCK1 phosphorylates INSIG1/2 for lipogenesis. Nature 580 (7804), 530–535. doi:10.1038/s41586-020-2183-2
Xu, D., Wang, Z., Zhang, Y., Jiang, W., Pan, Y., Song, B. L., et al. (2015). PAQR3 modulates cholesterol homeostasis by anchoring Scap/SREBP complex to the Golgi apparatus. Nat. Commun. 6, 8100. doi:10.1038/ncomms9100
Xu, S., Smothers, J. C., Rye, D., Endapally, S., Chen, H., Li, S., et al. (2024). A cholesterol-binding bacterial toxin provides a strategy for identifying a specific Scap inhibitor that blocks lipid synthesis in animal cells. Proc. Natl. Acad. Sci. U. S. A. 121 (7), e2318024121. doi:10.1073/pnas.2318024121
Yamauchi, Y., Furukawa, K., Hamamura, K., and Furukawa, K. (2011). Positive feedback loop between PI3K-Akt-mTORC1 signaling and the lipogenic pathway boosts Akt signaling: induction of the lipogenic pathway by a melanoma antigen. Cancer Res. 71 (14), 4989–4997. doi:10.1158/0008-5472.Can-10-4108
Yan, C., Zheng, L., Jiang, S., Yang, H., Guo, J., Jiang, L. Y., et al. (2023). Exhaustion-associated cholesterol deficiency dampens the cytotoxic arm of antitumor immunity. Cancer Cell 41 (7), 1276–1293.e11. doi:10.1016/j.ccell.2023.04.016
Yan, R., Cao, P., Song, W., Li, Y., Wang, T., Qian, H., et al. (2021a). Structural basis for sterol sensing by Scap and Insig. Cell Rep. 35 (13), 109299. doi:10.1016/j.celrep.2021.109299
Yan, R., Cao, P., Song, W., Qian, H., Du, X., Coates, H. W., et al. (2021b). A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols. Science 371 (6533), eabb2224. doi:10.1126/science.abb2224
Yang, M., Liu, W., Pellicane, C., Sahyoun, C., Joseph, B. K., Gallo-Ebert, C., et al. (2014). Identification of miR-185 as a regulator of de novo cholesterol biosynthesis and low density lipoprotein uptake. J. Lipid Res. 55 (2), 226–238. doi:10.1194/jlr.M041335
Yang, P. M., Hong, Y. H., Hsu, K. C., and Liu, T. P. (2019). p38α/S1P/SREBP2 activation by the SAM-competitive EZH2 inhibitor GSK343 limits its anticancer activity but creates a druggable vulnerability in hepatocellular carcinoma. Am. J. Cancer Res. 9 (10), 2120–2139.
Yao, L., Chen, S., and Li, W. (2020). Fatostatin inhibits the development of endometrial carcinoma in endometrial carcinoma cells and a xenograft model by targeting lipid metabolism. Arch. Biochem. Biophys. 684, 108327. doi:10.1016/j.abb.2020.108327
Yu, Y., Dong, J. T., He, B., Zou, Y. F., Li, X. S., Xi, C. H., et al. (2019). LncRNA SNHG16 induces the SREBP2 to promote lipogenesis and enhance the progression of pancreatic cancer. Future Oncol. 15 (33), 3831–3844. doi:10.2217/fon-2019-0321
Zhang, D., Lu, P., Zhu, K., Wu, H., and Dai, Y. (2021a). TFCP2 overcomes senescence by cooperating with SREBP2 to activate cholesterol synthesis in pancreatic cancer. Front. Oncol. 11, 724437. doi:10.3389/fonc.2021.724437
Zhang, D., Tong, X., VanDommelen, K., Gupta, N., Stamper, K., Brady, G. F., et al. (2017a). Lipogenic transcription factor ChREBP mediates fructose-induced metabolic adaptations to prevent hepatotoxicity. J. Clin. Invest 127 (7), 2855–2867. doi:10.1172/jci89934
Zhang, F., Gao, J., Liu, X., Sun, Y., Liu, L., Hu, B., et al. (2023). LATS-regulated nuclear-cytoplasmic translocation of SREBP2 inhibits hepatocellular carcinoma cell migration and invasion via epithelial-mesenchymal transition. Mol. Carcinog. 62 (7), 963–974. doi:10.1002/mc.23538
Zhang, K. L., Zhu, W. W., Wang, S. H., Gao, C., Pan, J. J., Du, Z. G., et al. (2021b). Organ-specific cholesterol metabolic aberration fuels liver metastasis of colorectal cancer. Theranostics 11 (13), 6560–6572. doi:10.7150/thno.55609
Zhang, L., Rajbhandari, P., Priest, C., Sandhu, J., Wu, X., Temel, R., et al. (2017b). Inhibition of cholesterol biosynthesis through RNF145-dependent ubiquitination of SCAP. Elife 6, e28766. doi:10.7554/eLife.28766
Zhang, S., Fang, T., He, Y., Feng, W., Yu, Z., Zheng, Y., et al. (2024). VHL mutation drives human clear cell renal cell carcinoma progression through PI3K/AKT-dependent cholesteryl ester accumulation. EBioMedicine 103, 105070. doi:10.1016/j.ebiom.2024.105070
Zhao, J., Zhang, X., Gao, T., Wang, S., Hou, Y., Yuan, P., et al. (2020). SIK2 enhances synthesis of fatty acid and cholesterol in ovarian cancer cells and tumor growth through PI3K/Akt signaling pathway. Cell Death Dis. 11 (1), 25. doi:10.1038/s41419-019-2221-x
Zhao, X., Xiaoli, , Zong, H., Abdulla, A., Yang, E. S., Wang, Q., et al. (2014). Inhibition of SREBP transcriptional activity by a boron-containing compound improves lipid homeostasis in diet-induced obesity. Diabetes 63 (7), 2464–2473. doi:10.2337/db13-0835
Zheng, L., Li, L., Lu, Y., Jiang, F., and Yang, X. A. (2018). SREBP2 contributes to cisplatin resistance in ovarian cancer cells. Exp. Biol. Med. (Maywood) 243 (7), 655–662. doi:10.1177/1535370218760283
Zhong, C., Fan, L., Li, Z., Yao, F., and Zhao, H. (2019). SREBP2 is upregulated in esophageal squamous cell carcinoma and co-operates with c-Myc to regulate HMGCR expression. Mol. Med. Rep. 20 (4), 3003–3010. doi:10.3892/mmr.2019.10577
Zhu, L., Shi, Y., Feng, Z., Yuan, D., Guo, S., Wang, Y., et al. (2024). Fatostatin promotes anti-tumor immunity by reducing SREBP2 mediated cholesterol metabolism in tumor-infiltrating T lymphocytes. Eur. J. Pharmacol. 971, 176519. doi:10.1016/j.ejphar.2024.176519
Zhu, L., and Wang, H. (2024). Cholesterol-regulated cellular stiffness may enhance evasion of NK cell-mediated cytotoxicity in gastric cancer stem cells. FEBS Open Bio 14 (5), 855–866. doi:10.1002/2211-5463.13793
Keywords: SREBP2, cholesterol metabolism, cancer, tumor microenvironment, cancer therapy
Citation: Chen R, Chen T, Li X, Yu J, Lin M, Wen S, Zhang M, Chen J, Yi B, Zhong H and Li Z (2025) SREBP2 as a central player in cancer progression: potential for targeted therapeutics. Front. Pharmacol. 16:1535691. doi: 10.3389/fphar.2025.1535691
Received: 27 November 2024; Accepted: 31 March 2025;
Published: 16 April 2025.
Edited by:
Michihisa Umetani, University of Houston, United StatesReviewed by:
Wei Guo, University of North Carolina at Greensboro, United StatesDaniel Kober, University of Texas Southwestern Medical Center, United States
Copyright © 2025 Chen, Chen, Li, Yu, Lin, Wen, Zhang, Chen, Yi, Zhong and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huage Zhong, MjEyMzA0MDNAcXEuY29t; Zhao Li, emhhb2xpMTk4OTA0MTFAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Ruiqi Chen
Ruiqi Chen Tianyu Chen1†
Tianyu Chen1† Siqi Wen
Siqi Wen Huage Zhong
Huage Zhong