- 1Clinical School of Medicine, Jiangxi University of Chinese Medicine, Nanchang, Jiangxi, China
- 2Department of Andrology, Affiliated Reproductive Hospital of Jiangxi University of Chinese Medicine, Nanchang, Jiangxi, China
Background: Prostate cancer (PC) is the most frequently diagnosed cancer in men and continues to be a major cause of cancer-related mortality worldwide. In recent years, non-coding RNAs (ncRNAs) have emerged as a significant focus in molecular biology research, playing a pivotal role in the development and progression of PC. This study employed bibliometric analysis to explore the global outputs, research hotspots, and future trends in ncRNA-related PC research over the past 20 years.
Methods: Publications on PC-related ncRNAs from 2004 to 2023 were retrieved from Web of Science Core Collection. The co-operation network of countries, institutions, and authors on this topic was analyzed using CiteSpace (version 6.2. R6). In addition, co-occurrence analysis of keywords and co-citation analysis of references were performed using CiteSpace, and emergent detection was also performed.
Results: A total of 2,951 articles on PC-related ncRNAs were finally included in this study for analysis. China contributed the largest number of publications, while the United States was the most influential country in this field, with collaborative ties to 26 other countries. Fudan University was identified as the most active institution in this field. Rajvir Dahiya was the most prolific and influential author. Within the co-citation network, clusters labeled “EVs,” “circRNA,” and “ceRNA” represented current research directions. The cluster labeled “gene” dominated the co-occurrence keywords. “circRNA” showed the highest burst strength among keywords, with “circRNA,” “EVs” and “exosome” maintaining sustained burst strength, suggesting these are the emerging research frontiers in this field.
Conclusion: Investigating ncRNAs as pivotal research subjects in PC is essential for addressing the public health impact of the disease and advancing innovative diagnostic and targeted therapeutic strategies. This study provides a comprehensive bibliometric analysis of research related to PC-associated ncRNAs, delivering a scientific perspective and identifying potential research directions for scholars in this field.
1 Introduction
Prostate cancer (PC) is one of the most common malignancies threatening men’s health (Litwin and Tan, 2017). According to GLOBOCAN cancer statistics, PC had an estimated 1.5 million new cases and 397,000 deaths globally in 2022, making it the second most common cancer and the fifth leading cause of cancer-related mortality among men (Bray et al., 2024). As an age-related malignancy of the male genitourinary system, PC has a higher incidence in elderly populations and continues to rise with the increasing aging of the population (Siegel et al., 2023). Family history, race, and genetic syndromes are established risk factors for PC, while smoking and obesity have a suspected role in modulating PC-specific mortality (Gandaglia et al., 2021). In its early stages, PC is asymptomatic, with early detection primarily achieved through blood tests for prostate-specific antigen (PSA) and confirmed via tissue biopsy (Van Poppel et al., 2022). Although radiation therapy and radical prostatectomy are effective treatments for localized PC, they may increase the risk of treatment-related complications, including incontinence and erectile dysfunction from surgery, and gastrointestinal and erectile dysfunction from radiation therapy (Pinsky and Parnes, 2023). Androgen deprivation therapy (ADT) is the standard treatment for recurrent or metastatic PC patients, encompassing both surgical and pharmacological castration, with the latter being more commonly employed (Feldman and Feldman, 2001). The goal of ADT is to lower serum testosterone levels in PC patients and maintain this suppression to induce cancer cell death and achieve transient clinical remission (Schröder et al., 2012). Nevertheless, the vast majority of PC patients subjected to ADT will ultimately develop castration-resistant PC (CRPC), which is a leading cause of mortality among these patients (Cai et al., 2023).
For decades, the functional focus of PC research has primarily been on protein-coding genes. However, these genes constitute only about 2%–3% of the human genome, while the diverse and ubiquitous non-coding RNAs (ncRNAs) originate from the remaining nucleotides (Rinn and Guttman, 2014). With the advent of high-throughput sequencing technologies, most of the non-coding genome has been characterized as functional transcripts, playing crucial roles in various biological processes and pathological states (Slack and Chinnaiyan, 2019). ncRNAs are localized in both the nucleus and cytoplasm and can also be found within extracellular vesicles (EVs), such as exosomes, or other microvesicles in bodily fluids. Based on their length, ncRNAs can be broadly categorized into two main families: small ncRNAs, which are shorter than 200 nucleotides and include microRNAs, transfer RNA-derived small RNAs, and Piwi-interacting RNAs, and long ncRNAs (lncRNAs), which exceed 200 nucleotides and encompass pseudogenes, chimeric RNAs, and circular RNAs (circRNAs) (Brosnan and Voinnet, 2009; Shi et al., 2021). As competing endogenous RNAs (ceRNAs), ncRNAs interact with protein-coding messenger RNAs (mRNAs) in complex ways, offering new insights into the gene regulatory networks of humans. Notably, recent studies have demonstrated that certain ncRNAs, initially considered incapable of protein-coding, contribute critically to disease development and progression through the production of derived peptides or proteins (Wang et al., 2019; Yi et al., 2024). ncRNA therapies have progressed relatively slowly on a global scale, with most ncRNA-related drugs still in the early phases of human clinical trials. For example, a 13-mer locked nucleic acid (LNA) inhibitor of miR-221, known as LNA-i-miR-221, demonstrated promising anti-tumor activity and safety in a Phase I clinical study (Tassone et al., 2023). The miR-122 inhibitor miravirsen resulted in a dose-dependent reduction of hepatitis C virus RNA levels in a Phase II clinical trial (Janssen et al., 2013). Similarly, CDR132L, a miR-132 inhibitor, has demonstrated effectiveness in improving cardiac function in patients with heart failure (Täubel et al., 2021). Additionally, remlarsen (miR-29b mimic) was found to inhibit collagen expression and the development of fibroproliferation in incisional skin wounds among healthy volunteers (Gallant-Behm et al., 2019). Recent studies have extensively reported abnormal expression of ncRNAs as a significant feature of PC, identifying them as key players in the onset and progression of the disease (Ramalho-Carvalho et al., 2016; Mugoni et al., 2022). Research into the biology of ncRNAs not only reveals their potential roles in PC pathology but also provides a theoretical foundation for specific diagnostic, therapeutic, and preventive strategies targeting ncRNAs in the human genome.
Bibliometrics offers an objective approach for objectively reflecting the knowledge structure and emerging trends in a particular field through quantitative analysis of published scientific literature (Nicolaisen, 2010). CiteSpace, as a tool for visualizing scientific knowledge, aids researchers in examining contributions from authors, countries, and institutions, identifying rapidly evolving topics, and forecasting future research directions (Chen, 2004; Synnestvedt et al., 2005). For instance, Zhong et al. (2022) utilized bibliometric analysis to evaluate global scientific output on immunotherapy for PC from 1999 to 2021, summarizing future research trends. Similarly, Chen et al. (2022) investigated the research status, hotspots, and trends in bone metastases of PC through bibliometric analysis. Despite the rapid advancement of research on ncRNAs in recent years, the impact of these studies on the PC field has not yet been fully assessed. Therefore, this study aimed to employ bibliometric analysis and knowledge domain mapping to evaluate the publications on ncRNAs in PC published over the past 20 years in the Web of Science Core Collection (WoSCC), with the goal of identifying the knowledge domain and emerging trends in this field.
2 Methods
2.1 Data collection
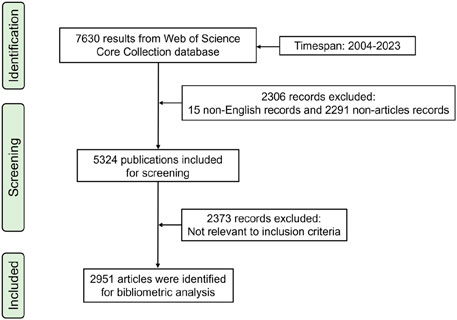
The Science Citation Index-Expanded (SCI-Expanded) of WoSCC was searched for publications related to ncRNAs in PC. The retrieval strategy used in this study was “TS = [(“miRNA*” OR “microRNA*” OR “miR-*” OR “lncRNA*” OR “long noncoding RNA*” OR “long non-coding RNA*” OR “circRNA*” OR “circular RNA*” OR “circ_*” OR “non-coding RNA*” OR “noncoding RNA*”) AND (“prostate cancer” OR “prostate neoplasm”)].” The search time span was set to between 1 January 2004 and 31 December 2023. In the first stage of screening, the type of publication was restricted to Article and the language was limited to English. In the second stage, irrelevant publications were excluded based on the title, keyword, abstract, and full text. The full records and cited references of eligible articles were exported from SCI-Expanded in plain text format. The flow chart of research steps of this study is shown in Figure 1.
2.2 Bibliometric analysis
All valid data were imported into Origin (version 2021) or CiteSpace (version 6.2. R6) for analysis and visualization. Origin was used to conduct statistical analysis on the annual number of publications and their citations, and to visualize the collaborations between different countries. CiteSpace was performed for co-operation analysis of countries, institutions, and authors. In addition, we used CiteSpace to draw a dual-map overlay of journals to investigate the evolutionary relationship between knowledge topics in directly citing journals and cited journals. In keyword analysis, we merged keywords with the same meaning to get a better perspective. The co-occurrence analysis of keywords and the co-citation analysis of references were performed by CiteSpace. Moreover, burst detection in CiteSpace was used to detect sudden surges in references and keywords at a certain time.
3 Results
3.1 Publication outputs and trends
A total of 2,951 articles on ncRNAs in PC were screened from 7,630 records retrieved from WoSCC. According to the number of citations, the total citations of the top 10 most cited papers accounted for 43.87% of all papers (Supplementary Table S1). The most cited article was titled “c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism,” which had 1,641 citations in Nature. The article published in Cell titled “The Landscape of Circular RNA in Cancer” had the highest annual citation frequency. The distribution of publications and their citations is presented in Figure 2. From 2004 to 2023, the cumulative number of global publications showed a consistent upward trend. Specifically, the evolution of PC-related ncRNA research consisted of threee stages. The field was in its early stages from 2004 to 2008, when fewer than 20 papers were published each year. The second stage was from 2009 to 2021, with a rapid increase in the number of annual publications. As the third stage from 2022 to 2023, the number of annual publications was in a steady growth, and the cumulative number of publications in 2023 was 113.5 times that of 2008. The cumulative number of citations for all articles in 2023 was 21,846, with an average of 7 citations per article. These results indicate that ncRNA research is an active field in PC and has received extensive attention from scholars.
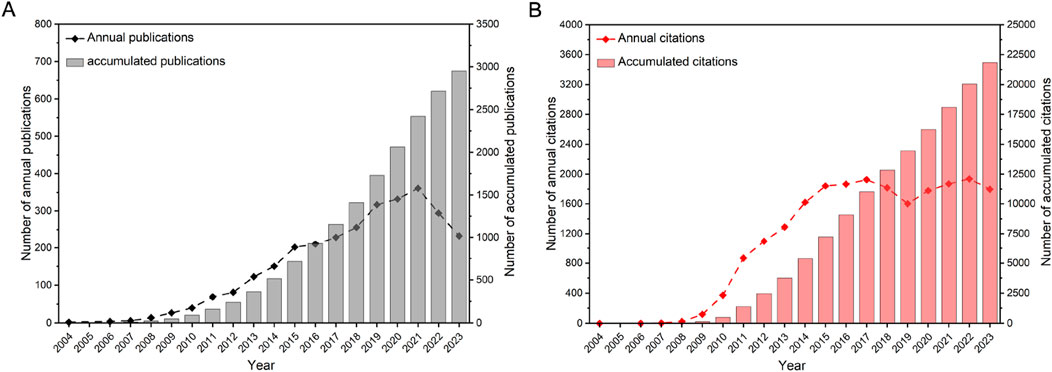
Figure 2. The distribution of publications (A) and their citations (B) from 2004 to 2023. The data were extracted using CiteSpace, and the figure was created with Origin.
3.2 Analysis of countries/regions and institutions
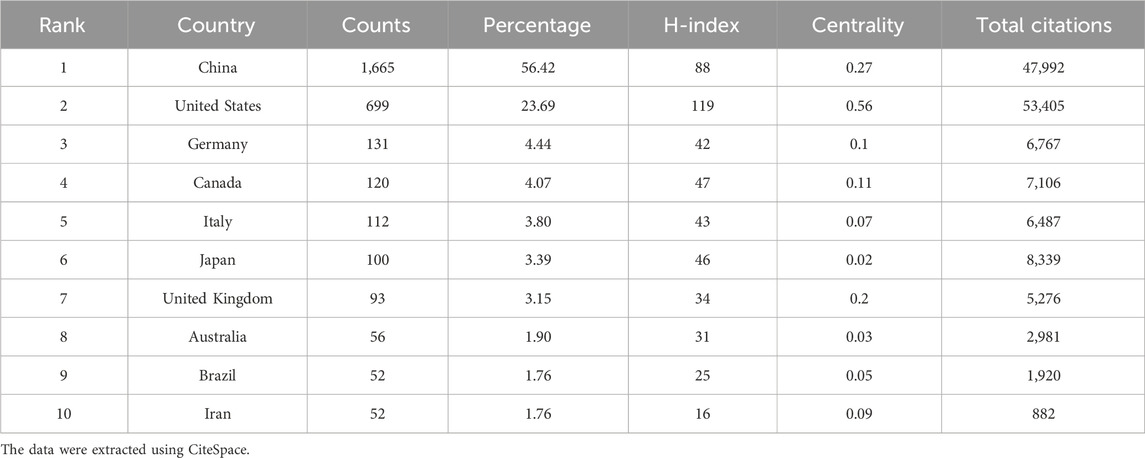
Analyzing the research output of countries/regions and institutions provides insights into the global distribution and trends within a specific field, enabling the identification of influential countries/regions and institutions. A total of 78 countries/regions were involved in the publications of ncRNAs in PC. Among them, China (1,665), the United States (699), Germany (131), Canada (120), and Italy (112) were the top five productive countries/regions in terms of the number of publications (Table 1). The H-index is a hybrid quantitative metric used to measure academic impact. The United States (119) has the highest H-index, indicating its leading academic influence in PC-related ncRNA research, followed by China (88), Canada (47), Japan (46), and Italy (43). Centrality helps identify key roles in information flow and knowledge dissemination within a network. The United (0.56) States exhibited the highest centrality, followed by China (0.27), the United Kingdom (0.2), Canada (0.11), and Germany (0.1). As depicted in Figure 3A, the United States collaborated with 26 other countries, notably including China, Canada, and the United Kingdom, which underscores its significant role as a central hub in international collaborations.
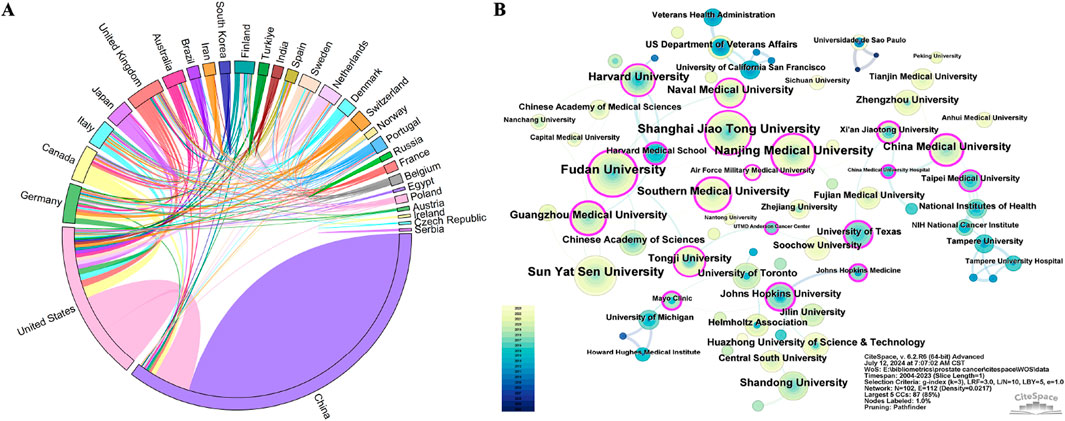
Figure 3. Collaboration network of countries/regions and institutions of ncRNA research in PC from 2004 to 2023. (A) The cooperation string diagram between top 30 countries/regions. (B) The network visualization map of different institutions. Each node represents an institution, and the size of the node corresponds to the frequency of publications produced by that institution. Each link between two nodes indicates a collaborative relationship between the respective institutions. Nodes with purple rings denote higher centrality, signifying their prominent role in the collaboration network. The data were extracted using CiteSpace. Panel (A) was created using Origin, while Panel (B) was generated with CiteSpace.
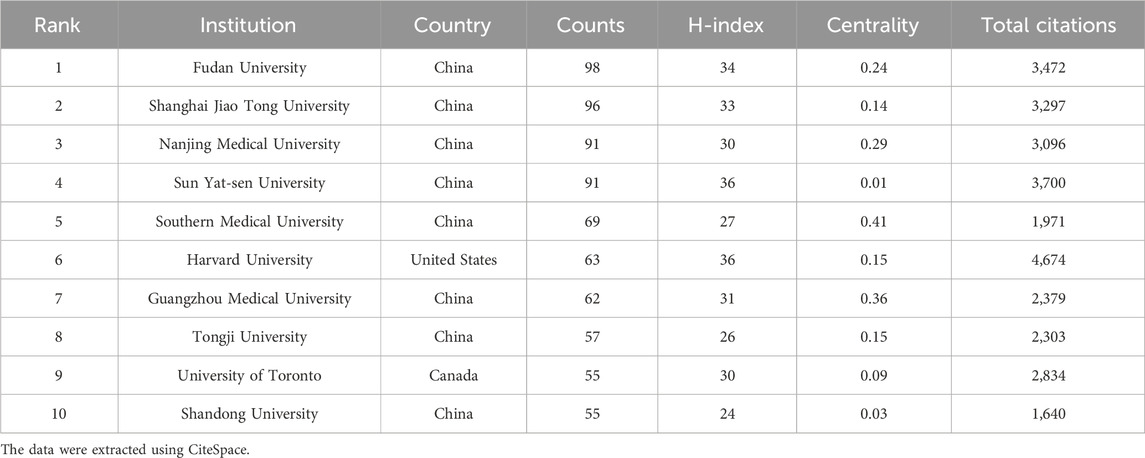
Based on the number of publications, Table 2 presents the top 10 institutions of ncRNA research in PC, with 9 of them located in China. Fudan University (98) had the highest number of publications, followed by Shanghai Jiao Tong University (96), Nanjing Medical University (91), and Sun Yat-sen University (91). Furthermore, the H-index of Sun Yat-sen University (36) and Harvard University (36) was the highest, followed by Fudan University (34) and Shanghai Jiao Tong University (33). Visualized with CiteSpace, we mapped an institutional collaboration network consisting of 102 nodes and 112 linkages (Figure 3B). Southern Medical University (0.41) had the highest centrality, followed by Guangzhou Medical University (0.36) and Nanjing Medical University (0.29). As the institution with the most publications, Fudan University primarily had close collaborations with Naval Medical University, Tongji University, and Chinese Academy of Sciences.
3.3 Analysis of authors
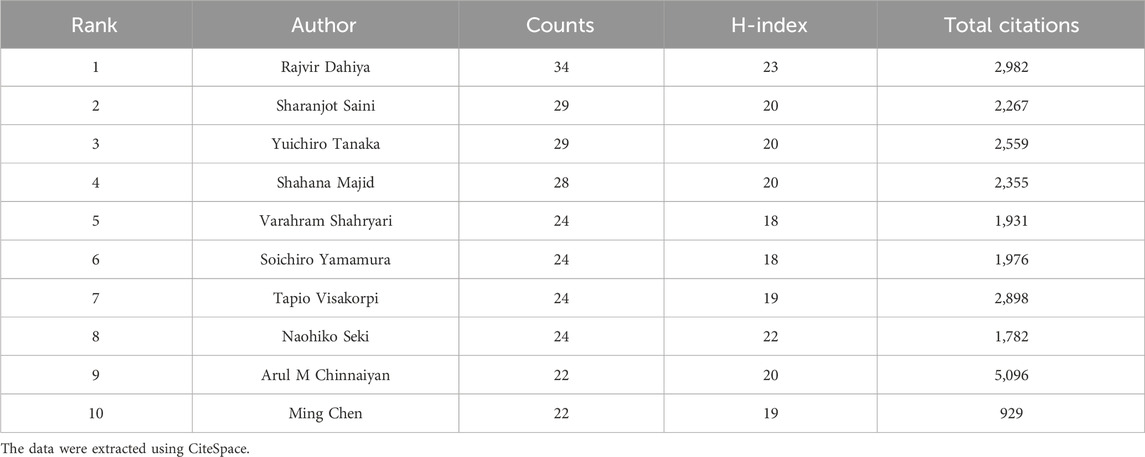
Author analysis facilitates the identification of pivotal contributors within a field and offers insights to guide academic exchanges and project collaborations. The top 10 active authors are listed in Table 3. Rajvir Dahiya contributed the most research with 34 publications, followed by Sharanjot Saini (29) and Yuichiro Tanaka (29). According to authors’ H-index, Rajvir Dahiya (23) ranked first, followed by Naohiko Seki (22), Sharanjot Saini (20), Yuichiro Tanaka (20), Shahana Majid (20), and Arul M Chinnaiyan (20). Rajvir Dahiya made significant contributions, with his research emphasizing the regulatory roles of miRNAs in the proliferation and metastasis of PC (Noonan et al., 2009; Saini et al., 2011). Co-cited authors are those cited together in one or more publications, used to identify key researchers in a particular academic field. Supplementary Table S2 shows that Rebecca L Siegel (978) had the most co-citations, followed by David P Bartel (451) and Ahmedin Jemal (394). These scholars have established the groundwork for research on ncRNAs in PC.
3.4 Analysis of journals
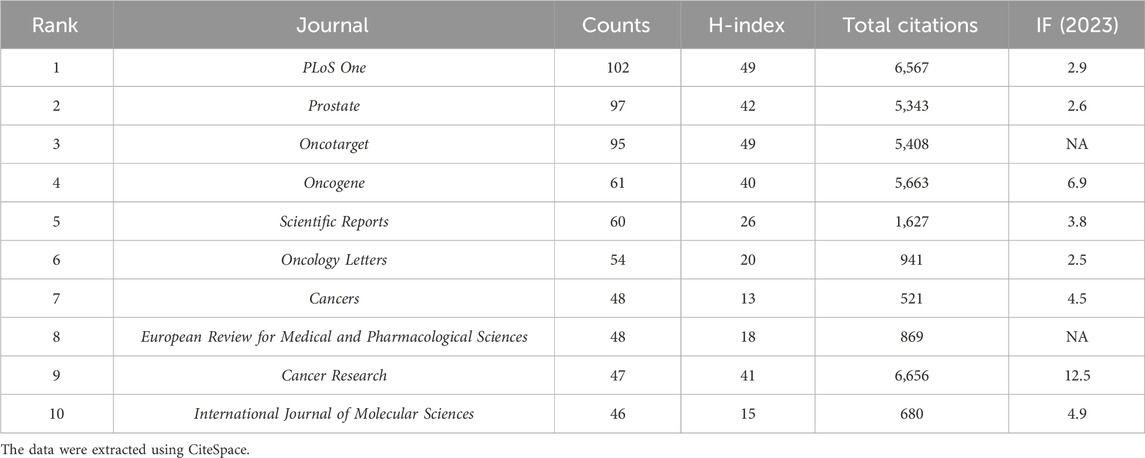
Journal analysis provides researchers with prioritized options for submitting their work and accessing articles within a specific field. A total of 556 academic journals included articles on PC-related ncRNAs. The top 10 journals included 658 papers in the field, accounting for 22.30% of all publications (Table 4). PLoS One (102) was the most published journal, followed by Prostate (97), Oncotarget (95), Oncogene (61), and Scientific Reports (60). Cancer Research (6,656), PLoS One (6,567), and Oncogene (5,663) were the top three most frequently cited journals.
The top 20 active co-cited journals are presented in Supplementary Table S3. Among them, Cancer Research (5,615) had the most co-citations, followed by Oncogene (3,325), PLoS One (3,190), Cell (3,034), and Proceedings of the National Academy of Sciences (2,720). In addition, to investigate the evolutionary relationship between knowledge topics in directly citing journals and cited journals, we used CiteSpace to draw a dual-map overlay of journals, in which the topics of directly citing journals are distributed on the left and the topics of cited journals are represented on the right. The analysis presented in Supplementary Figure S1 revealed a prominent citation pathway, indicating that research published in Molecular/Biology/Genetics journals was predominantly referenced by studies from Molecular/Biology/Immunology journals.
3.5 Co-citation analysis of references
To track the knowledge structure and research trends in the field of PC-related ncRNAs, we conducted a co-citation analysis of the references from 2,951 articles with CiteSpace. Supplementary Table S4 shows the top 10 frequently co-cited references. Among them, “(Siegel et al., 2017)” was the most co-cited reference, with 406 citations. Subsequently, the clustering analysis of CiteSpace was performed to construct a map of co-cited references, which consisted of 842 filtered nodes, 1917 connections, and 10 clusters (Figure 4). The modularity Q score was 0.7094 (>0.3), and the mean silhouette S was 0.867 (>0.7), demonstrating both a significant clustering structure and a highly homogeneous clustering network. Supplementary Figure S2 displays the timeline view of co-cited references, highlighting the evolution of each cluster over time. The largest cluster was “EVs” (cluster #0), followed by “PC” (cluster #1) and “circRNA” (cluster #2). According to the results of cluster analysis, “microarray” (cluster #7), “PC” (cluster #1), and “HRPC” (cluster #8) represented the early knowledge base in the field of PC-related ncRNAs in the past 20 years, while “EVs” (cluster #0), “circRNA” (cluster #2), and “ceRNA” (cluster #4) reflected the current research direction.
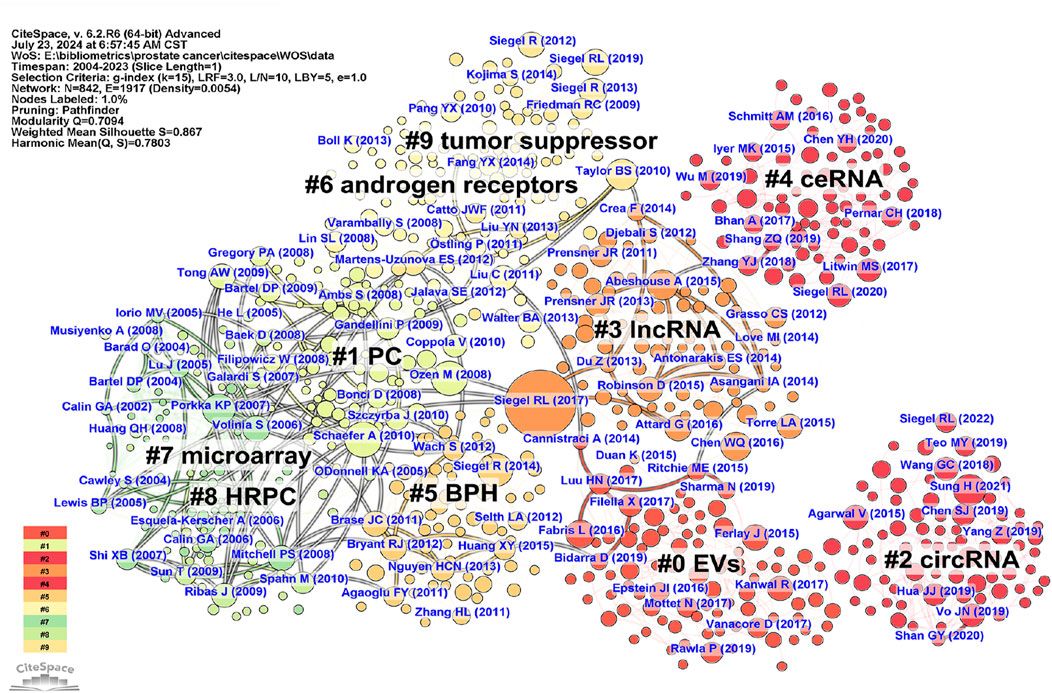
Figure 4. The network map of co-cited references related to ncRNA research in PC from 2004 to 2023. Each node represents a filtered co-cited reference, with the size of the node indicating its co-citation frequency. Each link between two nodes indicates a co-citation relationship between the respective references. Both the data and figures were generated using CiteSpace.
Furthermore, through the burst detection function of CiteSpace, we explored the top 15 co-cited references with strong citation bursts (Table 5). The paper that first exhibited strong citation burst over the past 20 years was “A microRNA expression signature of human solid tumors defines cancer gene targets,” authored by Stefano Volinia and published in 2006. The paper titled “microRNA expression in human prostate cancer” published in 2008 received the strongest citation burst (46.12), followed by “Kati P Porkka, 2007” (42.48), and “Stefan Ambs, 2008” (38.65). These papers constitute significant events in the current knowledge base of PC-related ncRNA research.
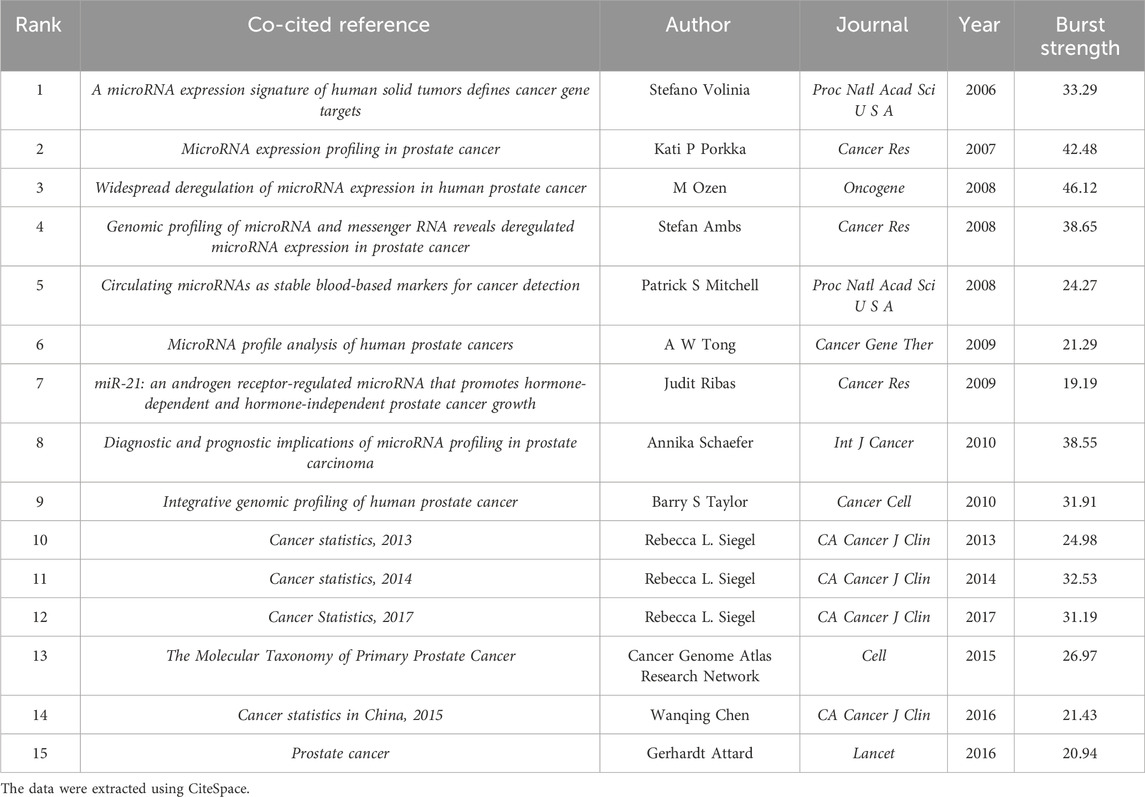
Table 5. Top 15 co-cited references with strong citation bursts related to ncRNA research in PC from 2004 to 2023.
3.6 Analysis of keywords
Identifying the high-frequency keywords in research on PC-associated ncRNAs facilitates the rapid recognition of the hot topics of interest within the academic community. Supplementary Table S5 lists the top 50 most frequent keywords, with the 15 highest-ranking ones being “PC” (1938), “expression” (1,258), “miRNA” (809), “proliferation” (688), “metastasis” (632), “progression” (521), “growth” (492), “cancer cell” (490), “invasion” (447), “gene” (360), “biomarker” (352), “lncRNA” (329), “androgen receptors” (284), “cancer” (268), and “apoptosis” (265). Among these high-frequency keywords, those pertaining to ncRNAs include “miRNA” (809), “lncRNA” (329), and “circRNA” (82). The co-occurrence analysis of keywords helps elucidate the relationships between different knowledge points in PC-associated ncRNA research, thereby contributing to the construction of a knowledge map for the field. To obtain a better network visualization of the keyword co-occurrence, we merged similar keywords from 2,951 articles, resulting in a co-occurrence map with 129 filtered nodes, 137 connections, and 11 clusters (Figure 5A). The modularity Q score was 0.8081, and the mean silhouette S reached 0.9215, indicating the highly convincing keyword network. In addition, we presented a visualization of the evolution of keywords in different clusters over time in the form of a timeline view (Supplementary Figure S3). “gene” (cluster #0) was the largest cluster, encompassing keywords such as “miRNA,” “mRNA,” “polymorphism,” “amplification,” “receptor.” and “activation.” “progression” (cluster #1) was the second largest cluster, including keywords such as “invasion,” “growth,” “signature,” “circRNA,” and “cancer.” Followed by “lncRNA” (cluster #2), including keywords such as “mechanism,” “ceRNA,” “mesenchymal transition,” “chromatin,” and “sequence.”
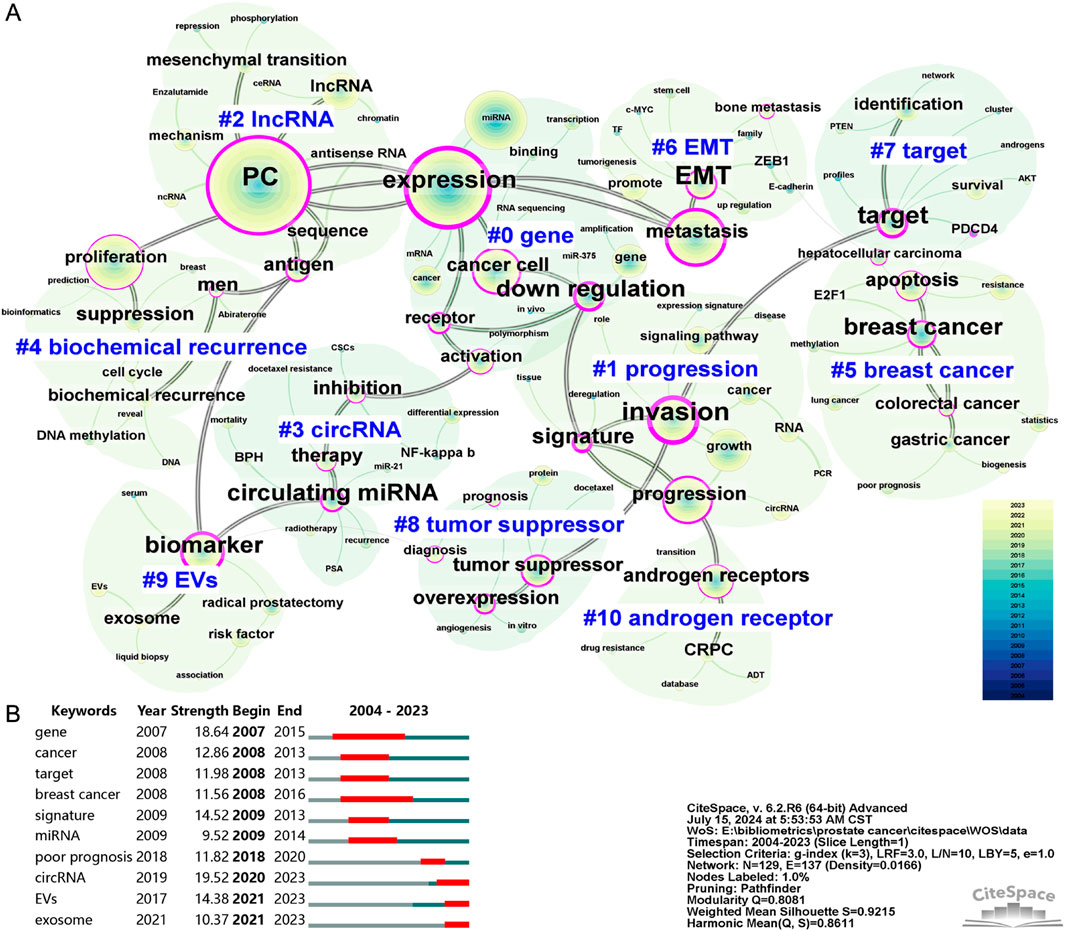
Figure 5. The co-occurrence map (A) and burst detection (B) of keywords related to ncRNA research in PC from 2004 to 2023. Each node represents a filtered co-occurring keyword, with the size of the node indicating its co-occurrence frequency. Each link between two nodes indicates a co-occurrence relationship between the respective keywords. Nodes with purple rings indicate higher centrality, highlighting their importance within the network. Both the data and figures were generated using CiteSpace.
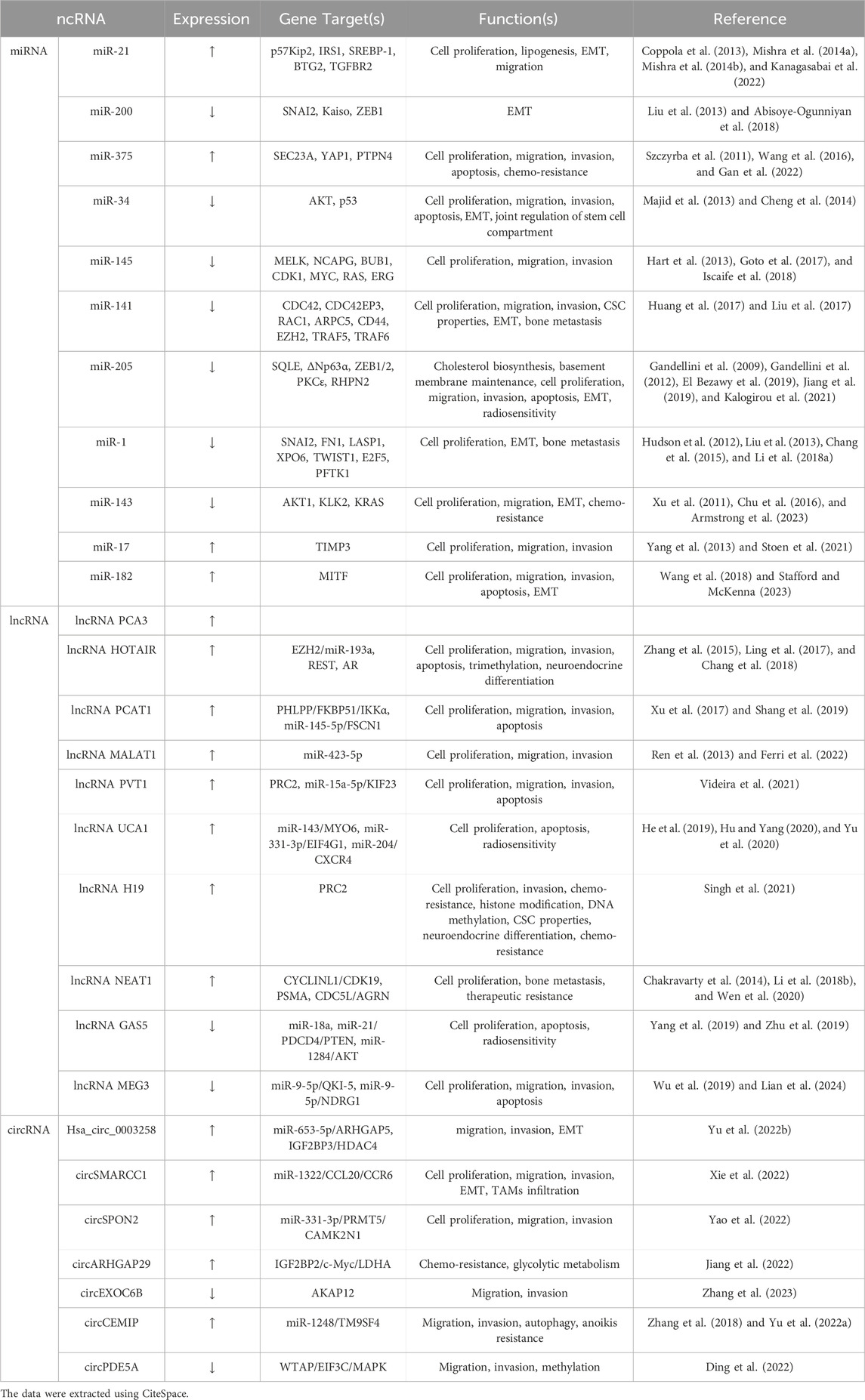
Burst detection of keywords is employed to identify new or rapidly increasing keywords, which may represent cutting-edge topics or novel areas of research. By using CiteSpace, we obtained the top 10 keywords with the strongest bursts (Figure 5B). The top three keywords with the strongest burst were “circRNA” (19.52), “gene” (18.64), and “signature” (14.52). Notably, the keywords “circRNA,” “EVs,” and “exosome” continued to show significant burst activity in 2023, indicating that these topics are key directions for future research. In addition, we extracted all miRNAs, lncRNAs, and circRNAs from the keywords, and merged and standardized similar miRNAs. Table 6 lists the frequently occurring miRNAs, lncRNAs, and circRNAs, all of which have been confirmed to play significant roles in the development of PC.
4 Discussion
4.1 General information
With the rapid development of information technology and increased medical academic papers, bibliometrics has been applied in various medical fields and plays an irreplaceable role in quantitative analysis. In this study, we retrieved publications indexed in WoSCC over the past 20 years and excluded records unrelated to ncRNA research in PC. Ultimately, 2,951 published articles were included in our bibliometric analysis. Between 2009 and 2023, there was a marked increase in both the total number of publications and citations concerning ncRNA research in PC, underscoring its emergence as a rapidly expanding area of interest. China had the highest number of publications and the second-highest number of citations, which shows its significant contribution in this field. As the most influential country, the United States collaborated with 26 countries/regions and maintained its closest partnership with China. In terms of institutional distribution, Fudan University, which had the highest number of publications, primarily collaborated with the Naval Medical University, Tongji University, and Chinese Academy of Sciences. The research focus of these institutions mainly revolves around understanding the mechanisms by which miRNAs (Li et al., 2014; Xiang et al., 2015), lncRNAs (Yao et al., 2020), and circRNAs (Kong et al., 2021) contribute to the progression of PC. According to the author analysis, Rajvir Dahiya was the most active and influential author in this field.
Additionally, we identified the top 10 most cited articles in ncRNA research related to PC. The most cited paper was published in Nature by Gao et al. (2009), which was groundbreaking in linking changes in miR-23a/b expression and glutamine metabolism to PC progression and proposed that c-Myc plays a crucial role in regulating crucial metabolic pathways in PC. The paper with the highest annual citation frequency was published by Vo et al. (2019) in 2019 in Cell, which highlighted the potential of circRNAs as diagnostic biomarkers and therapeutic targets for PC.
4.2 Knowledge base
We further explored the theoretical foundations of PC-related ncRNA research through co-citation analysis of the references. Through CiteSpace clustering analysis, all co-cited references were categorized into a network map containing 10 clusters with diverse labels, where “EVs” (cluster #0), “circRNA” (cluster #2), and “ceRNA” (cluster #4) reflected the current research trends. The top 10 frequently co-cited references included four epidemiological studies and six foundational studies. These epidemiological findings consistently highlighted PC as a leading cause of cancer-related deaths in men and a significant public health concern in China (Siegel et al., 2014; Chen et al., 2016; Siegel et al., 2017; Sung et al., 2021). Among the six foundational studies, four investigated microRNA profiles in PC (Porkka et al., 2007; Ambs et al., 2008; Ozen et al., 2008; Schaefer et al., 2010), whereas the other two focused on exploring the molecular and genomic characteristics of the disease (Taylor et al., 2010; Network, 2015). Moreover, we identified the top 15 popular references in PC-related ncRNA research based on citation bursts, which include most of the highly co-cited references mentioned above. It is worth noting that the study by Mitchell et al. (2008) underscored the potential of circulating miRNAs in serum or plasma as stable blood-based biomarkers for cancer classification and prognostication, specifically demonstrating that miR-141 effectively distinguished PC patients from healthy controls. This research provided the first strong evidence supporting the diagnostic utility of circulating miRNAs in PC. Ribas et al. (2009) were the first to discover that high expression of miR-21 promoted the growth of PC and conferred castration resistance, while inhibition of miR-21 reduced androgen-driven proliferation of PC cells. This study provided crucial molecular insights into targeting miRNAs for the treatment of PC. In summary, co-citation analysis of the references enables us to better understand the knowledge structure of PC-related ncRNAs and to identify the core references within this field.
4.3 Research hotspots
To explore the research hotspots in PC-related ncRNAs, we analyzed the keywords from 2,951 articles. High-frequency keywords with co-occurrence rates exceeding 500 included “PC,” “expression,” “miRNA,” “proliferation,” “metastasis,” and “progression,” indicating that research primarily focused on the molecular mechanisms of miRNAs in PC proliferation and metastasis. Commonly mentioned miRNAs included miR-21, miR-200, miR-375, miR-34, miR-145, miR-141, miR-205, miR-1, miR-143, miR-17, and miR-182. Aside from “miRNA,” “lncRNA” and “circRNA” were also high-frequency keywords associated with ncRNAs, highlighting their significant relevance in the field of PC. We also used CiteSpace for keyword burst detection, which revealed that recent emergent keywords include “circRNA,” “EVs,” and “exosome,” all of which represent cutting-edge areas in ncRNA research related to PC. Here, we provide a brief discussion on the potential of circRNAs and EVs in PC.
4.3.1 circRNAs
Early detection of PC remains crucial in reducing cancer-related mortality in men. The identification of potential cancer biomarkers significantly enhances the diagnosis and ongoing monitoring of PC, thereby improving patient outcomes. PSA liquid biopsy is the most common method for PC detection. However, its limited specificity in distinguishing between benign prostatic hyperplasia (BPH) and PC poses significant challenges for early detection and diagnosis (Stenman et al., 1999). circRNA was first discovered in RNA viruses in 1976 and was initially considered as splicing noise based on its biogenesis (Sanger et al., 1976). Until recent years, the rapid development of RNA sequencing technology has led to the identification of numerous circRNAs in eukaryotes, offering promising insights for the discovery of novel PC-related biomarkers based on liquid biopsies (Wang et al., 2014; Maass et al., 2017). As a newly discovered, abundant, and conserved class of ncRNAs, circRNAs originate from precursor mRNA and are characterized by their covalently closed loop structure formed through backsplicing (Jeck and Sharpless, 2014). With a unique structure lacking free ends, circRNAs are less exposed to exoribonuclease degradation (Jeck and Sharpless, 2014). This stability enables their reliable presence in plasma, urine, and saliva, reinforcing their potential as ideal circulating materials for liquid biopsies. Several investigations have emphasized the potential of circRNAs as biomarkers for PC. By utilizing exome capture RNA sequencing, Vo et al. (2019) established the MiOncoCirc database composed of circRNAs detected in tumor tissues, and identified 1,092 circRNAs in urine samples from PC patients. Chen et al. (2019) performed ultra-deep non-poly-A RNA sequencing on tumor samples from 144 patients with localized PC, revealing 76,311 distinct circRNAs. Among these, 171 circRNAs were identified as being essential for the proliferation of PC cells (Chen et al., 2019). These transcriptional profiles contribute to advancing the application of circRNAs in the diagnostic medicine of PC.
Furthermore, since circRNAs were first characterized as transcriptional regulators that control the miRNA-mRNA axis, an increasing body of research have preliminarily confirmed their critical roles in the epigenetic regulation associated with PC (Hansen et al., 2013; Zhou et al., 2021). Yu et al. (2022b) first observed the overexpression of hsa_circ_0003258 in human PC tissues, which was associated with the aggressive progression of the disease. Subsequent investigation revealed that hsa_circ_0003258 enhances the expression of Rho GTPase activating protein 5 by sponging miR-653-5p and forming a complex with insulin-like growth factor 2 mRNA binding protein 3 to stabilize HDAC4 mRNA. This interaction activates the ERK signaling pathway, accelerating the epithelial-mesenchymal transition (EMT) and ultimately promoting PC metastasis (Yu et al., 2022b). Xie et al. (2022) found that circSMARCC1 was significantly upregulated in the plasma and tissues of PC patients, promoting tumor proliferation and metastasis. Mechanistically, circSMARCC1 sponges miR-1322 to regulate the expression of CC-chemokine ligand 20 (CCL20), which activates PC cell proliferation and EMT. Additionally, circSMARCC1 induces the infiltration of tumor-associated macrophages and M2 polarization through the CCL20-CCR6 axis, thereby facilitating the progression of PC. Yu et al. (2022a) reported that the upregulation of circCEMIP in PC tissues promoted the invasion and metastasis of PC cells. CircCEMIP functions as a ceRNA for miR-1248, reducing the inhibitory effects of miR-1248 on its downstream target, transmembrane 9 superfamily member 4, which induces mTOR phosphorylation-mediated anoikis resistance (Yu et al., 2022a). In addition to serving as miRNA sponges, circRNAs interact with RNA-binding proteins and even participate in protein-coding processes, thus modulating the pathological processes of PC. Feng et al. (2019) identified that circ0005276, upregulated in PC tissues, promoted the proliferation, migration, and EMT of PC cells by interacting with the RNA-binding protein FUS to activate the transcription of X-linked inhibitor of apoptosis protein. Wang et al. (2024) discovered that the protein-coding circRNA circCCDC7 was significantly downregulated in PC patients and indicated that it upregulates FLRT3 by encoding the secretory protein circCCDC7-180aa, thereby inhibiting PC cell activity and suggesting its potential role as a tumor suppressor in PC. These studies highlight the critical involvement of circRNAs in the proliferation, metastasis, and treatment resistance associated with PC. Further exploration of circRNAs will enhance our understanding of the pathogenic mechanisms underlying PC and provide therapeutic strategies for future clinical applications.
4.3.2 EVs
EVs are small vesicles naturally released by all cell types into the extracellular space, initially believed to be membranous structures derived from cells for the purpose of clearing metabolic waste (van Niel et al., 2018). Subsequently, extensive research has uncovered that EVs, particularly exosomes, participate in various intercellular communication processes and have emerged as a prominent area of interest in biomedicine and bioengineering (van Niel et al., 2018; Kalluri and LeBleu, 2020). As nanoparticles encapsulated by a lipid bilayer membrane, EVs function as optimal carriers for safely transporting ncRNAs, playing a pivotal role in PC progression and metastasis by transferring these biomolecules to target cells (Mugoni et al., 2022). Liquid biopsies based on ncRNAs in EVs provide a non-invasive alternative to tissue biopsies, offering an abundant source of biological material and enabling the identification of potential tumor markers that can inform the staging and risk prognosis of PC (Hamed et al., 2024; Hu et al., 2024). The miRNA family represents one of the most frequently identified classes of ncRNAs linked to EVs. In a study comparing blood samples from 102 PC patients with those of 50 healthy controls, Souza et al. (2017) found that miR-200b levels in circulating EVs were associated with PSA levels exceeding 10 ng/mL and bone metastasis, whereas miR-200c expression was associated with Gleason score. Similarly, miR-424 (Albino et al., 2021) and miR-1246 (Bhagirath et al., 2018) in circulating EVs from PC patients were correlated with the metastatic spread of tumor cells. Some researchers have attached importance to the value of EVs in predicting the efficacy of drug treatment and radiotherapy in PC patients. For example, Guo et al. (2020) identified miR-423-3p as a potential biomarker for early prediction of castration resistance by analyzing plasma exosomal miRNAs in PC patients and those with CRPC after ADT. Wang et al. (2016) showed that elevated levels of miR-375 were involved in the chemo-resistance to docetaxel in metastatic CRPC patients and were significantly associated with their overall survival. Additionally, Yu et al. (2018) found that 57 miRNAs in serum exosomes exhibited significant changes following carbon ion radiotherapy in localized PC patients, with miR-654-3p and miR-379-5p emerging as potential predictors of therapeutic response to this treatment.
It is evident that PC-derived EVs function as crucial carriers of ncRNAs, facilitating the progression of PC through intercellular communication, including tumor proliferation, metastasis, angiogenesis, immune evasion, and drug resistance (Hu et al., 2024). Recent studies have identified several miRNAs, lncRNAs, and circRNAs within PC-derived EVs as potential inhibitory targets in PC therapy, offering new insights into precision medicine for this disease (Aghdam et al., 2019; Xu et al., 2019; Zhou et al., 2021). Additionally, compared to traditional delivery vehicles, EVs demonstrate superior biocompatibility and delivery efficiency, and their intrinsic advantages of naturally homing to tumor cells offer prospects for PC therapeutic strategies targeting EVs (Hu et al., 2024). Inhibiting the secretion of PC-derived EVs could be a critical step in reducing communication within the tumor microenvironment. Urabe et al. (2020) conducted a high-throughput miRNA-based screening using the ExoScreen assay and identified miR-26a as a regulator of EV secretion in PC cells. The associated mechanism involved the modulation of the SHC4, PFDN4, and CHORDC1 genes. Moreover, limited data confirmed that certain normal cells exhibit antitumor activity against PC. For instance, adipose-derived stromal cells release EVs containing miR-145, which suppress the proliferation of PC cells and induce apoptosis (Takahara et al., 2016). Exosomal miR-205 and miR-99b-5p derived from human bone marrow mesenchymal stem cells inhibit the proliferation, invasion, and migration of PC cells (Jiang et al., 2019). A deeper understanding of the roles of ncRNAs in EVs secreted by different cell types in PC paves the way for novel therapeutic strategies based on engineered EVs.
4.4 Limitations
In this study, we conducted the first bibliometric analysis using data from the WoSCC to objectively assess the research trends and current landscape in the field of PC-related ncRNAs. While our approach offers valuable insights, it is important to recognize certain limitations. First, although WoSCC is widely considered the premier database for bibliometric studies, there remains the possibility that some relevant articles were not captured, potentially introducing minor biases in our findings. Moreover, our analysis was based on data from publications spanning 2004 to 2023, limiting its ability to reflect the most recent advances in the field. Third, the nature of bibliometric analysis tends to prioritize highly cited mainstream research, potentially overlooking less cited but innovative studies. Nevertheless, we believe these limitations do not substantially diminish the relevance or contributions of the publications included in our analysis to the field.
5 Conclusion
This study presented a comprehensive bibliometric analysis of 2,951 articles on PC-related ncRNAs over the past 20 years. The global publication output in this field had seen rapid growth since 2011, indicating its emergence as a focal point of scholarly attention. China led in publication output, while the United States was the most influential nation, engaging in collaborating with 26 other countries. Rajvir Dahiya stood out as the most prolific and influential author. The research landscape of PC- related ncRNAs was predominantly focused on elucidating the molecular mechanisms by which miRNAs drive PC proliferation and metastasis. Additionally, circRNAs and EVs are emerging as pivotal areas of future exploration, offering promising avenues for advancing the precise diagnosis and targeted treatment of PC.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
Y-LZ: Conceptualization, Data curation, Writing–original draft, Writing–review and editing. W-LY: Conceptualization, Writing–review and editing, Writing–original draft. S-HC: Conceptualization, Funding acquisition, Writing–review and editing, Supervision. PW: Data curation, Writing–original draft. J-WF: Data curation, Methodology, Writing–original draft. J-QZ: Data curation, Methodology, Software, Writing–original draft. J-YZ: Data curation, Methodology, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Jiangxi Provincial Key Laboratory of Traditional Chinese Medicine Andrology (KP202102007).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1483186/full#supplementary-material
References
Abisoye-Ogunniyan, A., Lin, H., Ghebremedhin, A., Salam, A. B., Karanam, B., Theodore, S., et al. (2018). Transcriptional repressor Kaiso promotes epithelial to mesenchymal transition and metastasis in prostate cancer through direct regulation of miR-200c. Cancer Lett. 431, 1–10. doi:10.1016/j.canlet.2018.04.044
Aghdam, S. G., Ebrazeh, M., Hemmatzadeh, M., Seyfizadeh, N., Shabgah, A. G., Azizi, G., et al. (2019). The role of microRNAs in prostate cancer migration, invasion, and metastasis. J. Cell Physiol. 234 (7), 9927–9942. doi:10.1002/jcp.27948
Albino, D., Falcione, M., Uboldi, V., Temilola, D. O., Sandrini, G., Merulla, J., et al. (2021). Circulating extracellular vesicles release oncogenic miR-424 in experimental models and patients with aggressive prostate cancer. Commun. Biol. 4 (1), 119. doi:10.1038/s42003-020-01642-5
Ambs, S., Prueitt, R. L., Yi, M., Hudson, R. S., Howe, T. M., Petrocca, F., et al. (2008). Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 68 (15), 6162–6170. doi:10.1158/0008-5472.Can-08-0144
Armstrong, L., Willoughby, C. E., and McKenna, D. J. (2023). Targeting of AKT1 by miR-143-3p suppresses epithelial-to-mesenchymal transition in prostate cancer. Cells 12 (18), 2207. doi:10.3390/cells12182207
Bhagirath, D., Yang, T. L., Bucay, N., Sekhon, K., Majid, S., Shahryari, V., et al. (2018). microRNA-1246 is an exosomal biomarker for aggressive prostate cancer. Cancer Res. 78 (7), 1833–1844. doi:10.1158/0008-5472.Can-17-2069
Bray, F., Laversanne, M., Sung, H., Ferlay, J., Siegel, R. L., Soerjomataram, I., et al. (2024). Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74 (3), 229–263. doi:10.3322/caac.21834
Brosnan, C. A., and Voinnet, O. (2009). The long and the short of noncoding RNAs. Curr. Opin. Cell Biol. 21 (3), 416–425. doi:10.1016/j.ceb.2009.04.001
Cai, M., Song, X. L., Li, X. A., Chen, M., Guo, J., Yang, D. H., et al. (2023). Current therapy and drug resistance in metastatic castration-resistant prostate cancer. Drug Resist Updat 68, 100962. doi:10.1016/j.drup.2023.100962
Chakravarty, D., Sboner, A., Nair, S. S., Giannopoulou, E., Li, R., Hennig, S., et al. (2014). The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 5, 5383. doi:10.1038/ncomms6383
Chang, Y. S., Chen, W. Y., Yin, J. J., Sheppard-Tillman, H., Huang, J., and Liu, Y. N. (2015). EGF receptor promotes prostate cancer bone metastasis by downregulating miR-1 and activating TWIST1. Cancer Res. 75 (15), 3077–3086. doi:10.1158/0008-5472.Can-14-3380
Chang, Y. T., Lin, T. P., Tang, J. T., Campbell, M., Luo, Y. L., Lu, S. Y., et al. (2018). HOTAIR is a REST-regulated lncRNA that promotes neuroendocrine differentiation in castration resistant prostate cancer. Cancer Lett. 433, 43–52. doi:10.1016/j.canlet.2018.06.029
Chen, C. (2004). Searching for intellectual turning points: progressive knowledge domain visualization. Proc. Natl. Acad. Sci. U. S. A. 101 (Suppl. 1), 5303–5310. doi:10.1073/pnas.0307513100
Chen, S., Huang, V., Xu, X., Livingstone, J., Soares, F., Jeon, J., et al. (2019). Widespread and functional RNA circularization in localized prostate cancer. Cell 176 (4), 831–843. doi:10.1016/j.cell.2019.01.025
Chen, W., Zheng, R., Baade, P. D., Zhang, S., Zeng, H., Bray, F., et al. (2016). Cancer statistics in China, 2015. CA Cancer J. Clin. 66 (2), 115–132. doi:10.3322/caac.21338
Chen, Y., Tang, C., Shen, Z., Peng, S., Wu, W., Lei, Z., et al. (2022). Bibliometric analysis of the global research development of bone metastases in prostate cancer: a 22-year study. Front. Oncol. 12, 947445. doi:10.3389/fonc.2022.947445
Cheng, C. Y., Hwang, C. I., Corney, D. C., Flesken-Nikitin, A., Jiang, L., Öner, G. M., et al. (2014). miR-34 cooperates with p53 in suppression of prostate cancer by joint regulation of stem cell compartment. Cell Rep. 6 (6), 1000–1007. doi:10.1016/j.celrep.2014.02.023
Chu, H., Zhong, D., Tang, J., Li, J., Xue, Y., Tong, N., et al. (2016). A functional variant in miR-143 promoter contributes to prostate cancer risk. Arch. Toxicol. 90 (2), 403–414. doi:10.1007/s00204-014-1396-2
Coppola, V., Musumeci, M., Patrizii, M., Cannistraci, A., Addario, A., Maugeri-Saccà, M., et al. (2013). BTG2 loss and miR-21 upregulation contribute to prostate cell transformation by inducing luminal markers expression and epithelial-mesenchymal transition. Oncogene 32 (14), 1843–1853. doi:10.1038/onc.2012.194
Ding, L., Wang, R., Zheng, Q., Shen, D., Wang, H., Lu, Z., et al. (2022). circPDE5A regulates prostate cancer metastasis via controlling WTAP-dependent N6-methyladenisine methylation of EIF3C mRNA. J. Exp. Clin. Cancer Res. 41 (1), 187. doi:10.1186/s13046-022-02391-5
El Bezawy, R., Tinelli, S., Tortoreto, M., Doldi, V., Zuco, V., Folini, M., et al. (2019). miR-205 enhances radiation sensitivity of prostate cancer cells by impairing DNA damage repair through PKCε and ZEB1 inhibition. J. Exp. Clin. Cancer Res. 38 (1), 51. doi:10.1186/s13046-019-1060-z
Feldman, B. J., and Feldman, D. (2001). The development of androgen-independent prostate cancer. Nat. Rev. Cancer 1 (1), 34–45. doi:10.1038/35094009
Feng, Y., Yang, Y., Zhao, X., Fan, Y., Zhou, L., Rong, J., et al. (2019). Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell Death Dis. 10 (11), 792. doi:10.1038/s41419-019-2028-9
Ferri, C., Di Biase, A., Bocchetti, M., Zappavigna, S., Wagner, S., Le Vu, P., et al. (2022). MiR-423-5p prevents MALAT1-mediated proliferation and metastasis in prostate cancer. J. Exp. Clin. Cancer Res. 41 (1), 20. doi:10.1186/s13046-021-02233-w
Gallant-Behm, C. L., Piper, J., Lynch, J. M., Seto, A. G., Hong, S. J., Mustoe, T. A., et al. (2019). A MicroRNA-29 mimic (remlarsen) represses extracellular matrix expression and fibroplasia in the skin. J. Invest Dermatol 139 (5), 1073–1081. doi:10.1016/j.jid.2018.11.007
Gan, J., Liu, S., Zhang, Y., He, L., Bai, L., Liao, R., et al. (2022). MicroRNA-375 is a therapeutic target for castration-resistant prostate cancer through the PTPN4/STAT3 axis. Exp. Mol. Med. 54 (8), 1290–1305. doi:10.1038/s12276-022-00837-6
Gandaglia, G., Leni, R., Bray, F., Fleshner, N., Freedland, S. J., Kibel, A., et al. (2021). Epidemiology and prevention of prostate cancer. Eur. Urol. Oncol. 4 (6), 877–892. doi:10.1016/j.euo.2021.09.006
Gandellini, P., Folini, M., Longoni, N., Pennati, M., Binda, M., Colecchia, M., et al. (2009). miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 69 (6), 2287–2295. doi:10.1158/0008-5472.Can-08-2894
Gandellini, P., Profumo, V., Casamichele, A., Fenderico, N., Borrelli, S., Petrovich, G., et al. (2012). miR-205 regulates basement membrane deposition in human prostate: implications for cancer development. Cell Death Differ. 19 (11), 1750–1760. doi:10.1038/cdd.2012.56
Gao, P., Tchernyshyov, I., Chang, T. C., Lee, Y. S., Kita, K., Ochi, T., et al. (2009). c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458 (7239), 762–765. doi:10.1038/nature07823
Goto, Y., Kurozumi, A., Arai, T., Nohata, N., Kojima, S., Okato, A., et al. (2017). Impact of novel miR-145-3p regulatory networks on survival in patients with castration-resistant prostate cancer. Br. J. Cancer 117 (3), 409–420. doi:10.1038/bjc.2017.191
Guo, T., Wang, Y., Jia, J., Mao, X., Stankiewicz, E., Scandura, G., et al. (2020). The identification of plasma exosomal miR-423-3p as a potential predictive biomarker for prostate cancer castration-resistance development by plasma exosomal miRNA sequencing. Front. Cell Dev. Biol. 8, 602493. doi:10.3389/fcell.2020.602493
Hamed, M. A., Wasinger, V., Wang, Q., Graham, P., Malouf, D., Bucci, J., et al. (2024). Prostate cancer-derived extracellular vesicles metabolic biomarkers: emerging roles for diagnosis and prognosis. J. Control Release 371, 126–145. doi:10.1016/j.jconrel.2024.05.029
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495 (7441), 384–388. doi:10.1038/nature11993
Hart, M., Wach, S., Nolte, E., Szczyrba, J., Menon, R., Taubert, H., et al. (2013). The proto-oncogene ERG is a target of microRNA miR-145 in prostate cancer. Febs J. 280 (9), 2105–2116. doi:10.1111/febs.12236
He, C., Lu, X., Yang, F., Qin, L., Guo, Z., Sun, Y., et al. (2019). LncRNA UCA1 acts as a sponge of miR-204 to up-regulate CXCR4 expression and promote prostate cancer progression. Biosci. Rep. 39 (5). doi:10.1042/bsr20181465
Hu, C., Chen, Q., Wu, T., Du, X., Dong, Y., Peng, Z., et al. (2024). The role of extracellular vesicles in the treatment of prostate cancer. Small 20, e2311071. doi:10.1002/smll.202311071
Hu, M., and Yang, J. (2020). Down-regulation of lncRNA UCA1 enhances radiosensitivity in prostate cancer by suppressing EIF4G1 expression via sponging miR-331-3p. Cancer Cell Int. 20, 449. doi:10.1186/s12935-020-01538-8
Huang, S., Wa, Q., Pan, J., Peng, X., Ren, D., Huang, Y., et al. (2017). Downregulation of miR-141-3p promotes bone metastasis via activating NF-κB signaling in prostate cancer. J. Exp. Clin. Cancer Res. 36 (1), 173. doi:10.1186/s13046-017-0645-7
Hudson, R. S., Yi, M., Esposito, D., Watkins, S. K., Hurwitz, A. A., Yfantis, H. G., et al. (2012). MicroRNA-1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic Acids Res. 40 (8), 3689–3703. doi:10.1093/nar/gkr1222
Iscaife, A., Reis, S. T., Morais, D. R., Viana, N. I., da Silva, I. A., Pimenta, R., et al. (2018). Treating metastatic prostate cancer with microRNA-145. Apoptosis 23 (7-8), 388–395. doi:10.1007/s10495-018-1461-z
Janssen, H. L., Reesink, H. W., Lawitz, E. J., Zeuzem, S., Rodriguez-Torres, M., Patel, K., et al. (2013). Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368 (18), 1685–1694. doi:10.1056/NEJMoa1209026
Jeck, W. R., and Sharpless, N. E. (2014). Detecting and characterizing circular RNAs. Nat. Biotechnol. 32 (5), 453–461. doi:10.1038/nbt.2890
Jiang, S., Mo, C., Guo, S., Zhuang, J., Huang, B., and Mao, X. (2019). Human bone marrow mesenchymal stem cells-derived microRNA-205-containing exosomes impede the progression of prostate cancer through suppression of RHPN2. J. Exp. Clin. Cancer Res. 38 (1), 495. doi:10.1186/s13046-019-1488-1
Jiang, X., Guo, S., Wang, S., Zhang, Y., Chen, H., Wang, Y., et al. (2022). EIF4A3-Induced circARHGAP29 promotes aerobic glycolysis in docetaxel-resistant prostate cancer through IGF2BP2/c-Myc/LDHA signaling. Cancer Res. 82 (5), 831–845. doi:10.1158/0008-5472.Can-21-2988
Kalluri, R., and LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Science 367 (6478), eaau6977. doi:10.1126/science.aau6977
Kalogirou, C., Linxweiler, J., Schmucker, P., Snaebjornsson, M. T., Schmitz, W., Wach, S., et al. (2021). MiR-205-driven downregulation of cholesterol biosynthesis through SQLE-inhibition identifies therapeutic vulnerability in aggressive prostate cancer. Nat. Commun. 12 (1), 5066. doi:10.1038/s41467-021-25325-9
Kanagasabai, T., Li, G., Shen, T. H., Gladoun, N., Castillo-Martin, M., Celada, S. I., et al. (2022). MicroRNA-21 deficiency suppresses prostate cancer progression through downregulation of the IRS1-SREBP-1 signaling pathway. Cancer Lett. 525, 46–54. doi:10.1016/j.canlet.2021.09.041
Kong, Z., Lu, Y., Wan, X., Luo, J., Li, D., Huang, Y., et al. (2021). Comprehensive characterization of androgen-responsive circRNAs in prostate cancer. Life (Basel) 11 (10), 1096. doi:10.3390/life11101096
Li, S. M., Wu, H. L., Yu, X., Tang, K., Wang, S. G., Ye, Z. Q., et al. (2018a). The putative tumour suppressor miR-1-3p modulates prostate cancer cell aggressiveness by repressing E2F5 and PFTK1. J. Exp. Clin. Cancer Res. 37 (1), 219. doi:10.1186/s13046-018-0895-z
Li, X., Wan, X., Chen, H., Yang, S., Liu, Y., Mo, W., et al. (2014). Identification of miR-133b and RB1CC1 as independent predictors for biochemical recurrence and potential therapeutic targets for prostate cancer. Clin. Cancer Res. 20 (9), 2312–2325. doi:10.1158/1078-0432.Ccr-13-1588
Li, X., Wang, X., Song, W., Xu, H., Huang, R., Wang, Y., et al. (2018b). Oncogenic properties of NEAT1 in prostate cancer cells depend on the cdc5l-AGRN transcriptional regulation circuit. Cancer Res. 78 (15), 4138–4149. doi:10.1158/0008-5472.Can-18-0688
Lian, Z., Tian, P., Ma, S., Chang, T., Liu, R., Feng, Q., et al. (2024). Long noncoding RNA MEG3 regulates cell proliferation and apoptosis by disrupting microRNA-9-5p-mediated inhibition of NDRG1 in prostate cancer. Aging (Albany NY) 16 (2), 1938–1951. doi:10.18632/aging.205472
Ling, Z., Wang, X., Tao, T., Zhang, L., Guan, H., You, Z., et al. (2017). Involvement of aberrantly activated HOTAIR/EZH2/miR-193a feedback loop in progression of prostate cancer. J. Exp. Clin. Cancer Res. 36 (1), 159. doi:10.1186/s13046-017-0629-7
Litwin, M. S., and Tan, H. J. (2017). The diagnosis and treatment of prostate cancer: a review. Jama 317 (24), 2532–2542. doi:10.1001/jama.2017.7248
Liu, C., Liu, R., Zhang, D., Deng, Q., Liu, B., Chao, H. P., et al. (2017). MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat. Commun. 8, 14270. doi:10.1038/ncomms14270
Liu, Y. N., Yin, J. J., Abou-Kheir, W., Hynes, P. G., Casey, O. M., Fang, L., et al. (2013). MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene 32 (3), 296–306. doi:10.1038/onc.2012.58
Maass, P. G., Glažar, P., Memczak, S., Dittmar, G., Hollfinger, I., Schreyer, L., et al. (2017). A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. Berl. 95 (11), 1179–1189. doi:10.1007/s00109-017-1582-9
Majid, S., Dar, A. A., Saini, S., Shahryari, V., Arora, S., Zaman, M. S., et al. (2013). miRNA-34b inhibits prostate cancer through demethylation, active chromatin modifications, and AKT pathways. Clin. Cancer Res. 19 (1), 73–84. doi:10.1158/1078-0432.Ccr-12-2952
Mishra, S., Deng, J. J., Gowda, P. S., Rao, M. K., Lin, C. L., Chen, C. L., et al. (2014a). Androgen receptor and microRNA-21 axis downregulates transforming growth factor beta receptor II (TGFBR2) expression in prostate cancer. Oncogene 33 (31), 4097–4106. doi:10.1038/onc.2013.374
Mishra, S., Lin, C. L., Huang, T. H., Bouamar, H., and Sun, L. Z. (2014b). MicroRNA-21 inhibits p57Kip2 expression in prostate cancer. Mol. Cancer 13, 212. doi:10.1186/1476-4598-13-212
Mitchell, P. S., Parkin, R. K., Kroh, E. M., Fritz, B. R., Wyman, S. K., Pogosova-Agadjanyan, E. L., et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U. S. A. 105 (30), 10513–10518. doi:10.1073/pnas.0804549105
Mugoni, V., Ciani, Y., Nardella, C., and Demichelis, F. (2022). Circulating RNAs in prostate cancer patients. Cancer Lett. 524, 57–69. doi:10.1016/j.canlet.2021.10.011
Network, C. G. A. R. (2015). The molecular taxonomy of primary prostate cancer. Cell 163 (4), 1011–1025. doi:10.1016/j.cell.2015.10.025
Nicolaisen, J. (2010). Bibliometrics and citation analysis: from the science citation index to cybermetrics. J. Am. Soc. Inf. Sci. 61 (1), 205–207. doi:10.1002/asi.21181
Noonan, E. J., Place, R. F., Pookot, D., Basak, S., Whitson, J. M., Hirata, H., et al. (2009). miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene 28 (14), 1714–1724. doi:10.1038/onc.2009.19
Ozen, M., Creighton, C. J., Ozdemir, M., and Ittmann, M. (2008). Widespread deregulation of microRNA expression in human prostate cancer. Oncogene 27 (12), 1788–1793. doi:10.1038/sj.onc.1210809
Pinsky, P. F., and Parnes, H. (2023). Screening for prostate cancer. N. Engl. J. Med. 388 (15), 1405–1414. doi:10.1056/NEJMcp2209151
Porkka, K. P., Pfeiffer, M. J., Waltering, K. K., Vessella, R. L., Tammela, T. L., and Visakorpi, T. (2007). MicroRNA expression profiling in prostate cancer. Cancer Res. 67 (13), 6130–6135. doi:10.1158/0008-5472.Can-07-0533
Ramalho-Carvalho, J., Fromm, B., Henrique, R., and Jerónimo, C. (2016). Deciphering the function of non-coding RNAs in prostate cancer. Cancer Metastasis Rev. 35 (2), 235–262. doi:10.1007/s10555-016-9628-y
Ren, S., Liu, Y., Xu, W., Sun, Y., Lu, J., Wang, F., et al. (2013). Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J. Urol. 190 (6), 2278–2287. doi:10.1016/j.juro.2013.07.001
Ribas, J., Ni, X., Haffner, M., Wentzel, E. A., Salmasi, A. H., Chowdhury, W. H., et al. (2009). miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 69 (18), 7165–7169. doi:10.1158/0008-5472.Can-09-1448
Rinn, J., and Guttman, M. (2014). RNA Function. RNA and dynamic nuclear organization. Science 345 (6202), 1240–1241. doi:10.1126/science.1252966
Saini, S., Majid, S., Yamamura, S., Tabatabai, L., Suh, S. O., Shahryari, V., et al. (2011). Regulatory role of mir-203 in prostate cancer progression and metastasis. Clin. Cancer Res. 17 (16), 5287–5298. doi:10.1158/1078-0432.Ccr-10-2619
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U. S. A. 73 (11), 3852–3856. doi:10.1073/pnas.73.11.3852
Schaefer, A., Jung, M., Mollenkopf, H. J., Wagner, I., Stephan, C., Jentzmik, F., et al. (2010). Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int. J. Cancer 126 (5), 1166–1176. doi:10.1002/ijc.24827
Schröder, F., Crawford, E. D., Axcrona, K., Payne, H., and Keane, T. E. (2012). Androgen deprivation therapy: past, present and future. BJU Int. 109 (Suppl. 6), 1–12. doi:10.1111/j.1464-410X.2012.11215.x
Shang, Z., Yu, J., Sun, L., Tian, J., Zhu, S., Zhang, B., et al. (2019). LncRNA PCAT1 activates AKT and NF-κB signaling in castration-resistant prostate cancer by regulating the PHLPP/FKBP51/IKKα complex. Nucleic Acids Res. 47 (8), 4211–4225. doi:10.1093/nar/gkz108
Shi, X., Singh, S., Lin, E., and Li, H. (2021). Chimeric RNAs in cancer. Adv. Clin. Chem. 100, 1–35. doi:10.1016/bs.acc.2020.04.001
Siegel, R., Ma, J., Zou, Z., and Jemal, A. (2014). Cancer statistics, 2014. CA Cancer J. Clin. 64 (1), 9–29. doi:10.3322/caac.21208
Siegel, R. L., Miller, K. D., and Jemal, A. (2017). Cancer statistics, 2017. CA Cancer J. Clin. 67 (1), 7–30. doi:10.3322/caac.21387
Siegel, R. L., Miller, K. D., Wagle, N. S., and Jemal, A. (2023). Cancer statistics, 2023. CA Cancer J. Clin. 73 (1), 17–48. doi:10.3322/caac.21763
Singh, N., Ramnarine, V. R., Song, J. H., Pandey, R., Padi, S. K. R., Nouri, M., et al. (2021). The long noncoding RNA H19 regulates tumor plasticity in neuroendocrine prostate cancer. Nat. Commun. 12 (1), 7349. doi:10.1038/s41467-021-26901-9
Slack, F. J., and Chinnaiyan, A. M. (2019). The role of non-coding RNAs in oncology. Cell 179 (5), 1033–1055. doi:10.1016/j.cell.2019.10.017
Souza, M. F., Kuasne, H., Barros-Filho, M. C., Cilião, H. L., Marchi, F. A., Fuganti, P. E., et al. (2017). Circulating mRNAs and miRNAs as candidate markers for the diagnosis and prognosis of prostate cancer. PLoS One 12 (9), e0184094. doi:10.1371/journal.pone.0184094
Stafford, M. Y. C., and McKenna, D. J. (2023). MiR-182 is upregulated in prostate cancer and contributes to tumor progression by targeting MITF. Int. J. Mol. Sci. 24 (3), 1824. doi:10.3390/ijms24031824
Stenman, U. H., Leinonen, J., Zhang, W. M., and Finne, P. (1999). Prostate-specific antigen. Semin. Cancer Biol. 9 (2), 83–93. doi:10.1006/scbi.1998.0086
Stoen, M. J., Andersen, S., Rakaee, M., Pedersen, M. I., Ingebriktsen, L. M., Bremnes, R. M., et al. (2021). High expression of miR-17-5p in tumor epithelium is a predictor for poor prognosis for prostate cancer patients. Sci. Rep. 11 (1), 13864. doi:10.1038/s41598-021-93208-6
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Synnestvedt, M. B., Chen, C., and Holmes, J. H. (2005). CiteSpace II: visualization and knowledge discovery in bibliographic databases. AMIA Annu. Symp. Proc. 2005, 724–728.
Szczyrba, J., Nolte, E., Wach, S., Kremmer, E., Stöhr, R., Hartmann, A., et al. (2011). Downregulation of Sec23A protein by miRNA-375 in prostate carcinoma. Mol. Cancer Res. 9 (6), 791–800. doi:10.1158/1541-7786.Mcr-10-0573
Takahara, K., Ii, M., Inamoto, T., Nakagawa, T., Ibuki, N., Yoshikawa, Y., et al. (2016). microRNA-145 mediates the inhibitory effect of adipose tissue-derived stromal cells on prostate cancer. Stem Cells Dev. 25 (17), 1290–1298. doi:10.1089/scd.2016.0093
Tassone, P., Di Martino, M. T., Arbitrio, M., Fiorillo, L., Staropoli, N., Ciliberto, D., et al. (2023). Safety and activity of the first-in-class locked nucleic acid (LNA) miR-221 selective inhibitor in refractory advanced cancer patients: a first-in-human, phase 1, open-label, dose-escalation study. J. Hematol. Oncol. 16 (1), 68. doi:10.1186/s13045-023-01468-8
Täubel, J., Hauke, W., Rump, S., Viereck, J., Batkai, S., Poetzsch, J., et al. (2021). Novel antisense therapy targeting microRNA-132 in patients with heart failure: results of a first-in-human Phase 1b randomized, double-blind, placebo-controlled study. Eur. Heart J. 42 (2), 178–188. doi:10.1093/eurheartj/ehaa898
Taylor, B. S., Schultz, N., Hieronymus, H., Gopalan, A., Xiao, Y., Carver, B. S., et al. (2010). Integrative genomic profiling of human prostate cancer. Cancer Cell 18 (1), 11–22. doi:10.1016/j.ccr.2010.05.026
Urabe, F., Kosaka, N., Sawa, Y., Yamamoto, Y., Ito, K., Yamamoto, T., et al. (2020). miR-26a regulates extracellular vesicle secretion from prostate cancer cells via targeting SHC4, PFDN4, and CHORDC1. Sci. Adv. 6 (18), eaay3051. doi:10.1126/sciadv.aay3051
van Niel, G., D'Angelo, G., and Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19 (4), 213–228. doi:10.1038/nrm.2017.125
Van Poppel, H., Albreht, T., Basu, P., Hogenhout, R., Collen, S., and Roobol, M. (2022). Serum PSA-based early detection of prostate cancer in Europe and globally: past, present and future. Nat. Rev. Urol. 19 (9), 562–572. doi:10.1038/s41585-022-00638-6
Videira, A., Beckedorff, F. C., daSilva, L. F., and Verjovski-Almeida, S. (2021). PVT1 signals an androgen-dependent transcriptional repression program in prostate cancer cells and a set of the repressed genes predicts high-risk tumors. Cell Commun. Signal 19 (1), 5. doi:10.1186/s12964-020-00691-x
Vo, J. N., Cieslik, M., Zhang, Y., Shukla, S., Xiao, L., Zhang, Y., et al. (2019). The landscape of circular RNA in cancer. Cell 176 (4), 869–881. doi:10.1016/j.cell.2018.12.021
Wang, D., Lu, G., Shao, Y., and Xu, D. (2018). MiR-182 promotes prostate cancer progression through activating Wnt/β-catenin signal pathway. Biomed. Pharmacother. 99, 334–339. doi:10.1016/j.biopha.2018.01.082
Wang, J., Zhu, S., Meng, N., He, Y., Lu, R., and Yan, G. R. (2019). ncRNA-Encoded peptides or proteins and cancer. Mol. Ther. 27 (10), 1718–1725. doi:10.1016/j.ymthe.2019.09.001
Wang, P. L., Bao, Y., Yee, M. C., Barrett, S. P., Hogan, G. J., Olsen, M. N., et al. (2014). Circular RNA is expressed across the eukaryotic tree of life. PLoS One 9 (6), e90859. doi:10.1371/journal.pone.0090859
Wang, Q., Cheng, B., Singh, S., Tao, Y., Xie, Z., Qin, F., et al. (2024). A protein-encoding CCDC7 circular RNA inhibits the progression of prostate cancer by up-regulating FLRT3. NPJ Precis. Oncol. 8 (1), 11. doi:10.1038/s41698-024-00503-2
Wang, Y., Lieberman, R., Pan, J., Zhang, Q., Du, M., Zhang, P., et al. (2016). miR-375 induces docetaxel resistance in prostate cancer by targeting SEC23A and YAP1. Mol. Cancer 15 (1), 70. doi:10.1186/s12943-016-0556-9
Wen, S., Wei, Y., Zen, C., Xiong, W., Niu, Y., and Zhao, Y. (2020). Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol. Cancer 19 (1), 171. doi:10.1186/s12943-020-01293-4
Wu, M., Huang, Y., Chen, T., Wang, W., Yang, S., Ye, Z., et al. (2019). LncRNA MEG3 inhibits the progression of prostate cancer by modulating miR-9-5p/QKI-5 axis. J. Cell Mol. Med. 23 (1), 29–38. doi:10.1111/jcmm.13658
Xiang, J., Bian, C., Wang, H., Huang, S., and Wu, D. (2015). MiR-203 down-regulates Rap1A and suppresses cell proliferation, adhesion and invasion in prostate cancer. J. Exp. Clin. Cancer Res. 34 (1), 8. doi:10.1186/s13046-015-0125-x
Xie, T., Fu, D. J., Li, Z. M., Lv, D. J., Song, X. L., Yu, Y. Z., et al. (2022). CircSMARCC1 facilitates tumor progression by disrupting the crosstalk between prostate cancer cells and tumor-associated macrophages via miR-1322/CCL20/CCR6 signaling. Mol. Cancer 21 (1), 173. doi:10.1186/s12943-022-01630-9
Xu, B., Niu, X., Zhang, X., Tao, J., Wu, D., Wang, Z., et al. (2011). miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol. Cell Biochem. 350 (1-2), 207–213. doi:10.1007/s11010-010-0700-6
Xu, W., Chang, J., Du, X., and Hou, J. (2017). Long non-coding RNA PCAT-1 contributes to tumorigenesis by regulating FSCN1 via miR-145-5p in prostate cancer. Biomed. Pharmacother. 95, 1112–1118. doi:10.1016/j.biopha.2017.09.019
Xu, Y. H., Deng, J. L., Wang, G., and Zhu, Y. S. (2019). Long non-coding RNAs in prostate cancer: functional roles and clinical implications. Cancer Lett. 464, 37–55. doi:10.1016/j.canlet.2019.08.010
Yang, J., Hao, T., Sun, J., Wei, P., and Zhang, H. (2019). Long noncoding RNA GAS5 modulates α-Solanine-induced radiosensitivity by negatively regulating miR-18a in human prostate cancer cells. Biomed. Pharmacother. 112, 108656. doi:10.1016/j.biopha.2019.108656
Yang, X., Du, W. W., Li, H., Liu, F., Khorshidi, A., Rutnam, Z. J., et al. (2013). Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res. 41 (21), 9688–9704. doi:10.1093/nar/gkt680
Yao, B., Zhu, S., Wei, X., Chen, M. K., Feng, Y., Li, Z., et al. (2022). The circSPON2/miR-331-3p axis regulates PRMT5, an epigenetic regulator of CAMK2N1 transcription and prostate cancer progression. Mol. Cancer 21 (1), 119. doi:10.1186/s12943-022-01598-6
Yao, M., Shi, X., Li, Y., Xiao, Y., Butler, W., Huang, Y., et al. (2020). LINC00675 activates androgen receptor axis signaling pathway to promote castration-resistant prostate cancer progression. Cell Death Dis. 11 (8), 638. doi:10.1038/s41419-020-02856-5
Yi, Q., Feng, J., Lan, W., Shi, H., Sun, W., and Sun, W. (2024). CircRNA and lncRNA-encoded peptide in diseases, an update review. Mol. Cancer 23 (1), 214. doi:10.1186/s12943-024-02131-7
Yu, Q., Li, P., Weng, M., Wu, S., Zhang, Y., Chen, X., et al. (2018). Nano-vesicles are a potential tool to monitor therapeutic efficacy of carbon ion radiotherapy in prostate cancer. J. Biomed. Nanotechnol. 14 (1), 168–178. doi:10.1166/jbn.2018.2503
Yu, Y., Gao, F., He, Q., Li, G., and Ding, G. (2020). lncRNA UCA1 functions as a ceRNA to promote prostate cancer progression via sponging miR143. Mol. Ther. Nucleic Acids 19, 751–758. doi:10.1016/j.omtn.2019.11.021
Yu, Y., Song, Y., Cheng, L., Chen, L., Liu, B., Lu, D., et al. (2022a). CircCEMIP promotes anoikis-resistance by enhancing protective autophagy in prostate cancer cells. J. Exp. Clin. Cancer Res. 41 (1), 188. doi:10.1186/s13046-022-02381-7
Yu, Y. Z., Lv, D. J., Wang, C., Song, X. L., Xie, T., Wang, T., et al. (2022b). Hsa_circ_0003258 promotes prostate cancer metastasis by complexing with IGF2BP3 and sponging miR-653-5p. Mol. Cancer 21 (1), 12. doi:10.1186/s12943-021-01480-x
Zhang, A., Zhao, J. C., Kim, J., Fong, K. W., Yang, Y. A., Chakravarti, D., et al. (2015). LncRNA HOTAIR enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate cancer. Cell Rep. 13 (1), 209–221. doi:10.1016/j.celrep.2015.08.069
Zhang, C., Wang, S., Chao, F., Jia, G., Ye, X., Han, D., et al. (2023). The short inverted repeats-induced circEXOC6B inhibits prostate cancer metastasis by enhancing the binding of RBMS1 and HuR. Mol. Ther. 31 (6), 1705–1721. doi:10.1016/j.ymthe.2022.08.006
Zhang, P., Song, Y., Sun, Y., Li, X., Chen, L., Yang, L., et al. (2018). AMPK/GSK3β/β-catenin cascade-triggered overexpression of CEMIP promotes migration and invasion in anoikis-resistant prostate cancer cells by enhancing metabolic reprogramming. Faseb J. 32 (7), 3924–3935. doi:10.1096/fj.201701078R
Zhong, W., Shen, Z., Wu, Y., Mao, X., Kong, J., and Wu, W. (2022). Knowledge mapping and current trends of immunotherapy for prostate cancer: a bibliometric study. Front. Immunol. 13, 1014981. doi:10.3389/fimmu.2022.1014981
Zhou, H., Zheng, X. D., Lin, C. M., Min, J., Hu, S., Hu, Y., et al. (2021). Advancement and properties of circular RNAs in prostate cancer: an emerging and compelling frontier for discovering. Int. J. Biol. Sci. 17 (2), 651–669. doi:10.7150/ijbs.52266
Keywords: non-coding RNA, prostate cancer, bibliometric analysis, extracellular vesicle, circular RNA
Citation: Zhou Y-L, Yao W-L, Chen S-H, Wang P, Fu J-W, Zhao J-Q and Zhang J-Y (2025) Global research landscape and emerging trends of non-coding RNAs in prostate cancer: a bibliometric analysis. Front. Pharmacol. 15:1483186. doi: 10.3389/fphar.2024.1483186
Received: 19 August 2024; Accepted: 13 December 2024;
Published: 07 January 2025.
Edited by:
Olivier Cuvillier, UPR8241 Laboratoire de Chimie de Coordination (LCC), FranceReviewed by:
Qiong Wang, Southern Medical University, ChinaTian-Bao Huang, Yangzhou University, China
Copyright © 2025 Zhou, Yao, Chen, Wang, Fu, Zhao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng-Hui Chen, Y2hlbnNoZW5odWkxOTY1QDE2My5jb20=
†These authors have contributed equally to this work
 Yu-Liang Zhou
Yu-Liang Zhou Wen-Liang Yao1,2†
Wen-Liang Yao1,2†