
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol., 09 October 2024
Sec. Pharmacology of Anti-Cancer Drugs
Volume 15 - 2024 | https://doi.org/10.3389/fphar.2024.1466424
 Haiyan Zhang1,2†
Haiyan Zhang1,2† Shanshan Pei3†
Shanshan Pei3† Jiaxuan Li1†
Jiaxuan Li1† Jiajie Zhu1
Jiajie Zhu1 Hongyu Li1
Hongyu Li1 Guangshang Wu1
Guangshang Wu1 Ruiqi Weng1
Ruiqi Weng1 Ruyi Chen1
Ruyi Chen1 Zhongbiao Fang1
Zhongbiao Fang1 Jingbo Sun3*
Jingbo Sun3* Keda Chen1*
Keda Chen1*One of the most prevalent pathological types of Primary Liver Cancer (PLC) is the Hepatocellular Carcinoma (HCC) poses a global health issue. The high recurrence and metastasis rate of HCC, coupled with a low 5-year survival rate, result in a bleak prognosis. Exosomes, small extracellular vesicles released by various cells, contain diverse non-coding RNA molecules, including circular RNAs (circRNAs), which play a significant role in intercellular communication and can impact HCC progression. Studies have revealed the potential clinical applications of exosomal circRNAs as biomarkers and therapeutic targets for HCC. These circRNAs can be transferred via exosomes to nearby non-cancerous cells, thereby regulating HCC progression and influencing malignant phenotypes, such as cell proliferation, invasion, metastasis, and drug resistance. This review provides a comprehensive overview of the identified exosomal circRNAs, highlighting their potential as non-invasive biomarkers for HCC, and suggesting new perspectives for HCC diagnosis and treatment. The circRNA from exosomal organelles promotes metastasis and immune scape because of their unique chirality which is different from the Biomolecular Homochirality.
Primary liver cancer (PLC), which is one of the most prevalent gastrointestinal tumors globally, ranks fifth in terms of incidence worldwide and is among the top three causes of cancer-related deaths (Sung et al., 2021). In 2020, There were over 900,000 new cases and about 830,000 deaths globally (Sung et al., 2021), with hepatocellular carcinoma (HCC) accounting for approximately 75%–90% of PLC cases (Center and Jemal, 2011). The major risk factors for HCC include chronic infection with hepatitis viruses, such as hepatitis B virus (HBV) or hepatitis C virus (HCV), obesity, aflatoxin exposure, and excessive alcohol consumption (EATSLEORTC, 2004; Heilig et al., 2004; Marcon et al., 2018). Alpha-fetoprotein (AFP) is the most commonly used serum biomarker for HCC diagnosis in clinical practice; however, it has poor sensitivity and specificity for early diagnosis (Omar et al., 2023). Clinical studies have found that approximately 40% of patients with HCC are negative for AFP (Daniele et al., 2004), and even lower positive rates are observed in postoperative patients with metastatic HCC (Huang et al., 2020). AFP combined with ultrasound can be used for early HCC screening; however, ultrasound diagnosis relies heavily on the operator’s subjective judgment and might result in a low detection rate for early HCC (Hartke et al., 2017). In addition, des-gamma-carboxy prothrombin (DCP) and Lens culinaris agglutinin-reactive fraction of alpha-fetoprotein (AFP-L3) are also used to diagnose early HCC, especially in patients with negative imaging results; their sensitivity is low (Sterling et al., 2009). For the early HCC, the means of diagnosis recommended by all clinical guidelines remains unsatisfactory (Wang and Wei, 2020).
The progression of HCC correlates negatively with prognosis. Radical treatment options, including surgical resection, radiofrequency ablation (RFA), and liver transplantation, are suitable for only approximately one-third of early-stage patients, with a 5-year survival rate exceeding 50% (2012) The majority of advanced-stage patients undergo non-curative treatments, such as transarterial chemoembolization (TACE), chemotherapy, immunotherapy, and targeted therapy (Anwanwan et al., 2020). However, these treatments are associated with high rates of recurrence and metastasis (Allaire et al., 2020), leading to a 5-year survival rate of only about 18% (Jiang et al., 2019). Moreover, increased resistance to therapeutic drugs, such as immune checkpoint inhibitors, sorafenib, and levatinib, further decreases the efficacy of treatment for patients with advanced-stage HCC (Anwanwan et al., 2020). Multi-drug combination therapy and special drug delivery technology, such as nanotechnology, aim to reduce drug resistance and improve curative effects (Xia et al., 2017); however, the incidence of HCC is still increasing (Sung et al., 2021). Further research is needed to find better ways to diagnose and treat HCC.
With the advances in high-throughput chips and second-generation sequencing technologies, an increasing number of non-coding RNAs have been discovered. Circular RNAs (circRNAs) are non-coding RNA molecules that form a closed loop via covalent binding, which can regulate gene expression at the transcriptional and post-transcriptional levels, acting as miRNA sponges, encoding peptides or proteins, and forming stable RNA-protein complexes to regulate downstream biological processes (Meng et al., 2017; Kos et al., 1986). CircRNAs are stable, conserved, abundant, and tissue or stage-specific, allowing them to play a crucial role in various diseases, especially in tumors (Memczak et al., 2013; Meng et al., 2017; Chen and Shan, 2021). Studies found that circRNAs loaded in exosomes can promote metastasis between tumor cells and non-tumor cells, enhance or inhibit tumor progression, and can be detected using liquid biopsies (Pang et al., 2020). Exosomal circSHKBP1 promotes gastric cancer progression by regulating the miR-582-3p/Hu antigen R (HUR)/vascular endothelial growth factor (VEGF) pathway) (Xie et al., 2020). Exosomal ciRS-122 induced an increase of chemotherapy resistance in colorectal cancer, and exosomal has-circ-0001380 affects the occurrence and development of HCC (Wu et al., 2022). In recent years, research around exosomal circRNAs has gradually increased, and these molecules have emerged as attractive candidates for the early diagnosis and treatment of HCC. In this review, we summarize the exosomal circRNAs that might be developed to improve the prognosis of HCC.
Exosomes are small extracellular vesicles (EVs) enclosed by a lipid bilayer and measuring between 40 and 120 nm in diameter (S et al., 2013). They were initially discovered in sheep reticulocytes in 1983 and can be released by various cells under both normal and pathological conditions (Harding et al., 1983).[1] Exosomes contain DNA, proteins, lipids, circRNAs, and other functional molecules, and can remain stable in bodily fluids such as blood, urine, cerebrospinal fluid, amniotic fluid, and breast milk (Xu et al., 2016). The process of exosome formation begins with the formation of vesicles via endocytosis from the cell membrane, followed by the inward budding of vesicles to create multivesicular bodies (MVBs). Eventually, the MVBs fuse with the cytoplasmic membrane, thereby releasing exosomes into the extracellular fluid (Kowal et al., 2014). Exosomes can transfer certain functional molecules, such as proteins and circRNAs, between different cells to mediate cell-to-cell communication, which regulates protein synthesis, cell growth and differentiation, antiviral activity, and numerous other physiological and pathological activities (Wang et al., 2019b). Exosomes mediate cell communication through four main mechanisms: (1) The binding of exosomal membrane proteins to target cells, which activates intracellular signaling pathways; (2) the delivery of functional proteins or infectious particles to recipient cells; (3) the transfer of receptors between cells; and (4) the transfer of genetic information through mRNA, miRNA, circRNA, or transcription factors between cells (as shown in Figure 1). (Masyuk et al., 2013).
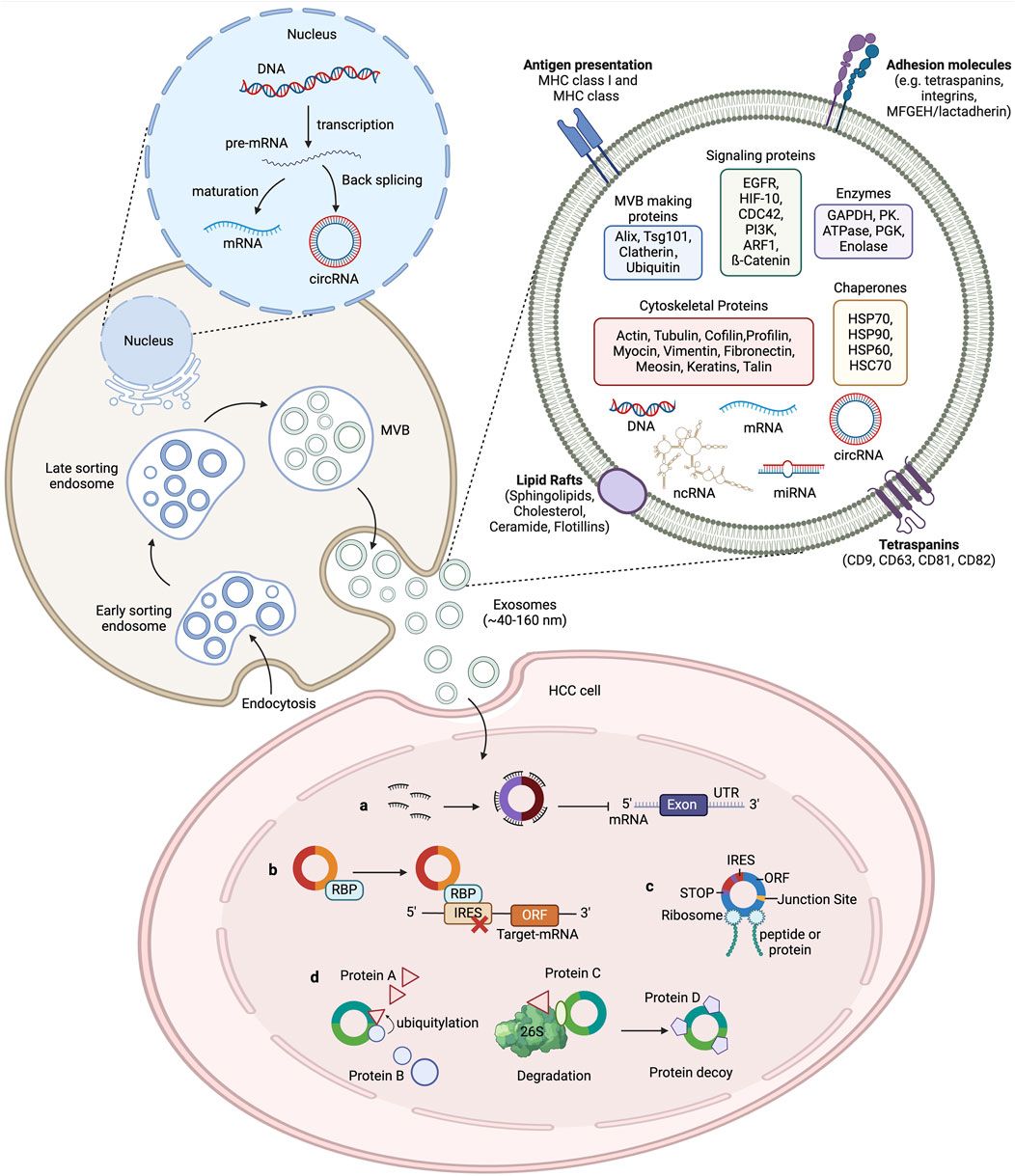
Figure 1. The biogenesis of exosomes and the function of circRNAs in HCC. The cytoplasmic membrane internalizes extracellular components through endocytosis, forming early endosomes. During endosome maturation, they are classified as intraluminal vesicles (ILV) within the endosome. The endosomal membrane further folds to form multivesicular bodies (MVB) that contain ILVs. Selectively or passively, various cellular contents such as RNA (including mRNA, circRNA, and other ncRNAs), DNA, and lipids are integrated into the vesicles. Subsequently, MVBs can either fuse with lysosomes for degradation or merge with the plasma membrane to release the ILVs, now called exosomes, into the extracellular fluid, where they perform their physiological functions. Exosomal circRNAs play an important role in tumour development. CircRNAs regulate the transcription of HCC target genes by acting as a microRNA sponges to impair microRNA function and protect target mRNAs from degradation. CircRNAs also bind to RNA-binding proteins (RBPs) to regulate the expression of relevant genes. CircRNAs interact with proteins to affect their structure and activity. Some circRNAs contain an internal ribosome entry site (IRES) element, in which the AUG site serves as a template for coding peptides or proteins.
Exosomes are involved in tumor initiation, immune escape, drug resistance, and drug delivery, making them highly valuable in the diagnosis, prognosis, and treatment of tumors (Wu et al., 2022; Kalluri and LeBleu, 2020). In addition, because of their small size and ability to evade elimination by other cells, exosomes can serve as carriers for drugs and functional RNAs (2004) Nanotechnology-based delivery of exosomes as carriers for drugs and functional RNAs to tumor cells has shown great potential for precise tumor treatment without causing adverse reactions, thereby offering promising clinical applications (Ha et al., 2016).
In the human genome sequence, approximately 98% of genes do not encode proteins (2004) Non-coding RNAs can be classified into linear and circular forms based on their structural characteristics (Yu and Shan, 2016). Linear non-coding RNAs can be further categorized into small non-coding RNAs and long non-coding RNAs (lncRNAs). MicroRNAs, siRNAs, and small nucleolar RNAs (snoRNAs) are examples of small non-coding RNAs, with microRNAs (miRNAs) being the most extensively studied (Hombach and Kretz, 2016). The presence of circular RNAs (circRNAs) was initially discovered in RNA viruses, such as retroviruses and hepatitis viruses, in 1976 (Sanger et al., 1976). It was later observed that circRNAs are also produced in eukaryotes during transcription. Initially regarded as “byproducts” of transcription because of their lack of protein-coding ability, circRNAs were not extensively investigated by researchers (Cocquerelle et al., 1993). CircRNAs are typically formed through the reverse splicing of precursor mRNAs, resulting in a single-stranded closed-loop structure held together by covalent bonds (Chen and Yang, 2015). Based on their origin, circRNAs are classified into four categories: exonic circRNAs, intronic circRNAs, circRNAs formed by introns and exons, and intergenic circRNAs (Wang et al., 2016). Compared with linear RNAs, circular RNAs possess greater resistance to degradation by nucleases and RNA enzymes because of the absence of free 5′ cap structures and 3′ poly(A) tails (Yu and Shan, 2016). CircRNAs containing miRNA binding sites (MREs) can directly regulate gene expression by binding to miRNAs and indirectly influence the expression levels of downstream target genes by interacting with RNA-binding proteins and affecting mRNA stability (Feng et al., 2019). Moreover, CircRNAs with RNA protein binding sites can interact with proteins, and serve as protein decoys, scaffolds and recruiters to interact with one or more proteins, protein conformations are influenced to alter protein-protein interactions, but the mechanisms need to be further studied (Zhou et al., 2020). Differential expression of circRNAs has been observed in various malignant tumor cells and tissues and has shown significant correlations with tumor staging and prognosis, implying their potential involvement in tumor initiation and progression (Wang et al., 2018).
The mechanisms of circRNA action vary depending on their subcellular location. CircRNAs derived from exonic regions are predominantly found in the cytoplasm and primarily function as miRNA sponges, thereby modulating the expression of downstream target genes. An example is has-circ-0000519, which is enriched in the cytoplasm and promotes HCC angiogenesis by influencing the miR-1296/E2F transcription factor 7 (E2F7) axis (Liu et al., 2023b). Conversely, circRNAs localized in the nucleus typically originate from introns or consist of both exonic and intronic sequences. They regulate gene expression at the transcriptional or post-transcriptional level (Memczak et al., 2013). Exosomal circ-ZNF652 is upregulated in the serum and cells of patients with HCC, promoting HCC cell proliferation, invasion, and metastasis through targeting the miR-29a-3p/guanylyl cyclase domain containing 1 (GUCD1) axis (Li et al., 2020). Numerous studies have demonstrated the oncogenic or tumor suppressive roles of circRNAs in HCC development, with some circRNAs being closely associated with HCC prognosis. In malignant tumors like HCC, circRNAs often act as competitive endogenous RNAs (ceRNAs) by interacting with miRNAs in a circRNA/miRNA/mRNA axis to regulate downstream gene expression (Ng et al., 2018). Additionally, circRNAs can interact with transcription factors, mediating their role in epithelial-mesenchymal transition (EMT) and promoting HCC progression (Xu et al., 2020b).
In 2015, circRNAs were first discovered in exosomes, and subsequent research revealed that circulating exogenous circRNAs could differentiate non-patients from patients based on their expression levels (Li et al., 2015). Further investigations confirmed the stable existence and wide enrichment of circRNAs in exosomes, particularly those derived from tumor cells, thus exosomal circRNAs have been identified as potential biomarkers for tumor diagnosis and prognosis (Li et al., 2015). In malignant tumors, most exosomal circRNAs function as miRNA sponges, exerting oncogenic or tumor suppressive effects. For instance, exosomal circ-PDE8A, derived from pancreatic ductal adenocarcinoma, regulates metastasis-associated in colon cancer 1 (MACC1) expression by binding to miR-338, stimulating cancer cell invasive growth via the MACC/MET/extracellular regulated kinase (ERK) or protein kinase B (AKT) pathway, and the expression levels of exosomal circRNAs closely correlate with patient prognosis (Li et al., 2018). Exosomal circLPAR1 inhibits bromodomain containing 4 (BRD4) expression through its interaction with Methyltransferase 3, N6adenosine-methyltransferase complex catalytic subunit (METTL3)-eukaryotic translation initiation factor 3 subunit H (eIF3h), playing a crucial role in the diagnosis and treatment of colorectal cancer (Zheng et al., 2022).
The tumor microenvironment (TME) is formed through the interaction of various cells, including both tumor and non-tumor cells (Hanahan and Coussens, 2012). It consists of extracellular matrix components (ECM), stromal cells (such as cancer-associated fibroblasts, mesenchymal stem cells, pericytes, and endothelial cells), different immune cells (including T and B lymphocytes, natural killer (NK) cells, regulatory T cells, and tumor-associated macrophages) (Xiao and Yu, 2021). The TME plays a crucial role in tumor progression, and understanding the TME and its intricate interactions opens up new possibilities for anti-cancer treatments, making significant contributions to improving treatment efficacy and reducing mortality rates (Xiao and Yu, 2021). Recent studies have highlighted the role of circRNAs in regulating the TME (Zhang et al., 2020b). CircRNAs are abundant and stable in exosomes derived from HCC cells, which can be secreted into the TME and delivered to different target cells through systemic circulation, affecting malignant characteristics such as tumor proliferation, metastasis, invasion, angiogenesis, immune response, and drug resistance (Li et al., 2015). Accumulating studies have shown that exosomes can remodel the TME to induce an inhibitory immune microenvironment to promote the progression of HCC (Li et al., 2023). Thus, exosomal circRNAs play an essential role in regulating communication between cancer cells and their surrounding cells in the TME, either promoting or inhibiting the invasion and metastasis of HCC. In addition, exosomal circRNAs have emerged as a promising therapeutic strategy for malignant tumors, including HCC, utilizing the exosomal RNA delivery system; the use of RNA-based targeted therapy has shown potential as a novel treatment modality for cancer (Zhang et al., 2016). Thus, circRNAs are expected to serve as effective biomarkers for the diagnosis, treatment, and prognosis of HCC, particularly those found in exosomes.
The study of exosomal circRNAs can help predict and assess the early progression of HCC without the need for invasive clinical procedures, which is a significant advantage for clinical application. Recent research has shown that exosomal hsa-circ-0028861 and hsa-circ-0070396 have excellent diagnostic value in HBV-related HCC. They can effectively differentiate patients with HCC from those with chronic hepatitis B and cirrhosis, which is of great significance in improving the diagnosis rate and prognosis of early-stage HCC (Wang et al., 2021b; Lyu et al., 2021). The combination of serum AFP and exosomal circRNA might further improve the early diagnosis rate of HCC and have critical clinical significance in the diagnosis of HBV-derived HCC.
The exosomal circRNAs derived from HCC cells and adjacent non-cancerous cells might have different mechanisms of action (Shi et al., 2020). Hsa-circ-0051443, which is highly expressed in normal cells, can be transferred to HCC cells via exosomes. It can competitively bind to miR-331-3p leading to decreased expression of BCL2 antagonist/killer 1(BAK1), thus inhibiting HCC cell growth and promoting apoptosis. Moreover, the level of hsa-circ-0051443 could differentiate between patients with HCC and non-HCC individuals, with an area under the curve (AUC) of 0.8089 (Chen et al., 2020). However, further clinical validation is required for accurate diagnosis.
Macrophages play a well-established role in HCC, and understanding their mechanism is essential for HCC treatment (Tian et al., 2019). Recent studies have demonstrated that exosomes derived from macrophages with high expression of recombination signal-binding protein Jk (RBPJ) can effectively inhibit the progression of HCC, and further investigation has revealed that exosomes carry hsa-circ-0004658, which is abundantly expressed in macrophages overexpressing RBPJ. This circRNA sponges miR-499b-5p, leading to an upregulation of junctional adhesion molecule 3 (JAM3). As a result, HCC cell proliferation is inhibited, and apoptosis is promoted (Zhang et al., 2022a). However, the results still need to be validated in a clinical cohort. In addition, it should be noted that not all exosomal circRNAs derived from non-cancerous cells have inhibitory effects on HCC. CircWDR25 facilitated HCC cell proliferation and invasion by regulating the circWDR25/miR-4474-3p/arachidonate 15-lipoxygenase (ALOX15) and EMT axes, in which it enhances the expression of cytotoxic T-lymphocyte associated protein 4 (CTLA-4) in HSCs and programmed cell death 1 ligand 1 (PD-L1) in HCC cells, thereby influencing the immune response in the TME and ultimately affecting HCC prognosis and recurrence (Liu et al., 2022).
Understanding the mechanisms of cell cycle regulation is crucial in studying the transformation of normal cells into malignant tumors, because abnormal cell proliferation plays a vital role in this process (Fernández et al., 2002). Researchers have discovered that prolonged exposure to arsenic trioxide increases the exosome levels of circRNA-100284 and facilitates its transfer to normal LO-2 cells, in which the circRNA interacts with miRNA-217, leading to accelerated cell cycle progression and promotion of hepatocellular malignancy (Dai et al., 2018). Similarly, circ-002136 is upregulated in HCC tissues and cells and can be transferred via cancer cell-derived exosomes. It promotes HCC cell proliferation, migration, and invasion via the miR-19a-3p/Rab GTPase YPT1 homolog (RAB1A) pathway, is associated with poor prognosis, and correlates positively with disease staging (Yuan et al., 2022). However, it's worth noting that the study lacked clinical validation.
Exosomal has-circ-0061395 is also significantly upregulated in the serum of patients with HCC, and silencing its expression led to cell cycle arrest and promotion of apoptosis. It interacts with the 3′untranslated region of miR-877-5p, resulting in increased expression of phosphatidylinositol 3-kinase regulatory subunit gamma (PIK3R3), thereby promoting HCC cell proliferation, migration, invasion, and malignant behavior (Yu et al., 2021). Circ-0072088 is widely enriched in exosomes derived from HCC cells and can be secreted into surrounding normal cells through blood circulation, promoting cell transfer. It has a high diagnostic value in patients with HCC (AUC: 0.899), and high levels of circ-0072088 are associated with poor prognosis. Knocking out circ-0072088 abolished the sponging of miR-375 and downregulated matrix metalloproteinase 16 (MMP-16) expression, thus inhibiting HCC cell invasion and metastasis (Lin et al., 2021). CircANTXR1 is highly abundant in exosomes isolated from the serum of patients with HCC and plays a crucial role in promoting HCC cell proliferation, migration, invasion, and tumorigenicity. It directly interacts with miR-532-5p, which in turn regulates the expression of X-ray repair cross-complementing 5 (XRCC5) to facilitate HCC development and worsen prognosis. Inhibiting circANTXR1 could potentially serve as a novel therapeutic approach for HCC and as an early diagnostic marker (Huang et al., 2021). Matrix metalloproteinase 2 (MMP2) is closely associated with metastasis and malignant tumor dissemination (Zhu et al., 2017a). A recent study found that exosomal circMMP2 (has-circ-0039411) can be transmitted to normal liver cell line L02, acting as a ceRNA to enhance the expression of MMP2 by sequestering miR-136-5p, thereby promoting the metastasis of HCC cells (Liu et al., 2020). Conversely, suppressing the expression of circMMP2 reversed the aggressive behavior of L02 cells, and its increased expression is associated with a lower overall survival rate in patients. Exosomal circ-0006602 is specifically expressed in the plasma of patients with HCC and enhances the expression of proteins associated with tumor proliferation in HCC cell lines; its diagnostic performance surpasses that of AFP, with an AUC of 0.907 compared with 0.694 (Guo et al., 2021). Combining circ-0006602 and AFP greatly improved the early detection rate of HCC (AUC: 0.942).
A previous study reported that cytoplasmic circRNA Cdr1as functions as an oncogene by targeting the miR-7/epidermal growth factor receptor (EGFR) axis to promote HCC development (Yang et al., 2017). Further investigations revealed that Cdr1as is highly abundant in exosomes derived from HCC cells, acting as a ceRNA to promote HCC cell proliferation and migration by sequestering miR-1270 and upregulating AFP expression. In vitro experiments also demonstrated that exosomes derived from HCC cells overexpressing Cdr1as can directly transfer to adjacent normal cells, enhancing their proliferation and invasion abilities (Su et al., 2019). Other animal experiments showed that exosomal circular RNA tubulin tyrosine ligase-like family member 5 (circTTLL5) promotes mouse hepatocellular carcinoma cell proliferation and metastasis through the miR-136-5p/KIAA1522 axis, thereby suppressing tumor growth. This suggests that blocking exosome-mediated circTTLL5 transfer might be a therapeutic target for HCC (Liu et al., 2023a). CircTMEM45A (has-circ-0066659) acts as a sponge for miR-665, regulating the expression of downstream insulin-like growth factor 2 (IGF2) to promote cell migration in vitro and the occurrence of HCC in vivo, and its expression correlates with tumor size, TNM staging, and vascular invasion. Patients with low circTMEM45A expression have longer survival (Zhang et al., 2020c). Another study has revealed that circPTGR1 is abundant in exosomes released by highly metastatic HCC cells, and the presence of high levels of circPTGR1 enhances EMT processes in low or non-metastatic cells through its interaction with miR-449a, thereby disrupting TME homeostasis, and is closely related to the clinical staging of HCC (Wang et al., 2019a). Has-circ-0004001, has-circ-0004123, and has-circ-0075792 were found to be upregulated in HCC serum exosomes according to bioinformatic analysis, and they demonstrated high sensitivity and specificity in distinguishing between healthy controls and patients with HCC. When combined, these three circRNAs achieved an AUC of 0.885 for HCC diagnosis, indicating their potential as biomarkers (Sun et al., 2020). Furthermore, exosomal circAKT3 is associated with a high risk of HCC recurrence and death. Patients with high circAKT3 expression have lower overall survival and disease-free survival rates, suggesting a poor prognosis (Luo et al., 2020).
EMT plays a crucial role in HCC metastasis, and exosomal circRNAs have been found to promote HCC progression by influencing EMT processes (Liu et al., 2022). Overexpression of HUR, an RNA-binding protein involved in maintaining mRNA stability, has been associated with poor prognosis in HCC (Song et al., 2020). Has-circ-0074854 is highly expressed in both HCC cells and tissues, and downregulation of its expression affects the stability of the HUR protein. This attenuates zinc finger E-box binding homeobox 1 (ZEB1) signal transduction in HCC cells, which suppresses malignant behaviors, including cell proliferation, migration, and EMT, and can induce M2 polarization in macrophages, thereby promoting HCC progression (Wang et al., 2021a). There are experiments have revealed that has-circ-0004277 competes to bind with HUR, leading to reduced expression of zona occludens-1 (ZO-1) and upregulation of the EMT-related transcription factor ZEB1 to promote HCC progression (Zhu et al., 2020). Notably, exosomal has-circ-0004277 can be used to diagnose HCC, with a diagnostic area AUC reaching 0.816 (Zhu et al., 2020).
Inducing neovascularization is a crucial step in the invasion and metastasis of HCC, and exosomes released by cancer cells play a significant role in stimulating the formation of new blood vessels through signaling between HCC cells and endothelial cells (Morse et al., 2019). In the stimulation of tumor angiogenesis, exosomes can directly interact with vascular endothelial growth factor (VEGF)/VEGF receptors or act as carriers to transport circRNAs that target downstream molecules, indirectly contributing to the generation of new blood vessels (Zhang et al., 2022b). Huang et al. discovered that circRNA-100338 is highly abundant in exosomes of HCC cells with high metastatic potential, and the mechanism is that it can promote the proliferation of human umbilical vein endothelial cells (HUVECs) and regulates angiogenesis by interacting with neuro-oncological ventral antigen 2 (NOVA2), an RNA-binding protein associated with vascular development and lumen formation. Thus, the level of circRNA-100338 in postoperative HCC serum can predict the occurrence of postoperative metastasis (Huang et al., 2020). Another study reported that exosomal circCMTM3 directly binds to miR-3619-5p and targets the downstream SRY-box transcription factor 9 (SOX9) to promote the proliferation, migration, invasion, and angiogenesis of HUVECs, enhancing their survival capacity and inducing tumor growth. The level of exosomal circCMTM3 also correlates positively with tumor stage and lymph node metastasis (Hu et al., 2021). Besides, exosomal circPAK1 has also been found to promote angiogenesis and metastasis in HCC cells (Hao et al., 2022).
Cytokine Storm is involved in the occurrence and development of many diseases and may be one of the important causes of poor prognosis (Fajgenbaum and June 2020). Studies have found that circRNAs may be involved in the development of cytokine storms, for example, has-circ-0004812 acts on the JAK/STAT, STAT3 signaling pathway through hsa-mir-1287-5p/IL6R, RIG-I axis, and causes a in COVID-19 inflammatory responses (Mohammadisoleimani et al., 2022). Similarly, circRNAs may also contribute to HCC by promoting inflammatory cytokine storm processes. Tumor cells can modify their surface antigens and change the TME of the surrounding tissue, enabling them to evade recognition and attack by immune cells, which ultimately leads to tumor immune escape (Villalba et al., 2013). Programmed cell death receptor 1 (PD-1) is an inhibitory receptor expressed on the surface of active lymphocytes such as T cells, B cells, and NK cells (Greten and Sangro, 2017). Overexpression of PD-1 can lead to dysfunction of these cells and facilitate tumor immune evasion (Gao et al., 2009). In recent years, immune checkpoint blockade therapy, particularly anti-PD-1 therapy targeting the PD-1/PD-L1 axis and immune cell exhaustion, has been extensively used to treat various malignancies, including HCC. It has also been approved as a second-line treatment for advanced HCC (El-Khoueiry et al., 2017). However, less than 20% of patients with advanced HCC respond effectively to anti-PD-1 treatment because of innate or acquired resistance (Zhu et al., 2018). The specific mechanisms underlying resistance to anti-PD-1 immune therapy in HCC are still unclear. Nevertheless, some studies have demonstrated the involvement of exosomal circRNAs in regulating cancer immune evasion, promoting exhaustion of NK cells and CD8+ effector T cells, expanding regulatory T cells (Tregs), driving cancer progression, and inducing resistance to anti-PD-1 therapy (Chen et al., 2019).
NK cells play an important role in the antitumor immune response, and T cell immunoglobulin and mucin domain-containing protein 3 (TIM-3) is expressed on the surface of NK cells as an inhibitory molecule. Increased expression of TIM-3 is associated with NK cell dysfunction and death (Zheng et al., 2019). Exosomal circUHRF1 (has-circ-0048677) derived from HCC cells can be delivered to NK cells in the body fluid circulation where it induces NK cell dysfunction and exhaustion by upregulating the expression of downstream target gene TIM3 through adsorption of miR-449c-5p. This promotes cancer immune escape and drives resistance to anti-PD-1 immune therapy in patients with HCC (Zhang et al., 2020a). Therefore, analyzing the expression levels of circUHRF1 can identify patients who are resistant to PD-1 therapy and improve the clinical efficacy in patients with HCC.
CD8+ effector T cells also play a key role in the immune system (Greten and Sangro, 2017). Recent studies have shown that circCCAR1 is significantly enriched in exosomes derived from HCC and it promotes immune evasion of cancer cells and resistance to anti-PD-1 immune therapy by acting on the miR-127-5p/WT1 associated protein (WTAP) axis, increasing the stability of PD-1 protein, and causing dysfunction of CD8+ T cells (Hu et al., 2023). Additionally, circRNAs in exosomes derived from HCC cells, specifically circGSE1, act on the miR-324-5p/transforming growth factor beta receptor 1 (TGFBR1)/SMAD family member 3 (Smad3) axis to induce expansion of regulatory Tregs and facilitate immune evasion of HCC cells (Huang et al., 2022).
Macrophages can also influence the liver immune response in HCC (Tian et al., 2019). Exosomal circRNAs mediate intercellular communication between macrophages and HCC cells, thereby mediating HCC progression (Han et al., 2019). Depending on the stimuli, macrophages can polarize into two phenotypes: classical (M1) with anti-tumor properties or alternative (M2), with pro-tumor properties. During HCC progression, tumor cells often induce M2 polarization of macrophages to promote tumor growth (Komohara et al., 2016). Research has identified the overexpression of circtMEM181 in HCC tissues, particularly in patients with a poor PD-1 response. The mechanism involves HCC cells delivering circtMEM181 to macrophages via exosomes, where it binds to miR-488-3p and upregulates CD39 expression. This cooperative action between macrophages and HCC cells activates the ATP-adenosine pathway in the HCC TME, which interferes with the proliferation of CD8+ T cells, inducing CD8+ T cell exhaustion, and promoting resistance to PD-1 antibodies (Lu et al., 2021). As a result, the responsiveness to PD-1 therapy is limited. These findings highlight the potential of exosomal circRNAs in HCC immunotherapy and their potential as therapeutic targets. Furthermore, the ratio of Tregs to CD8+ T cells can serve as a predictive marker for the anti-tumor response to PD-1/PD-L1 inhibitors (Huang et al., 2022).
Malignant tumors rely on aerobic glycolysis to generate the substantial amount of energy required for the rapid proliferation of tumor cells; thus, glycolysis plays a significant role in the accelerated progression of malignant tumors (Lunt and Vander Heiden, 2011). Exosomal circRNAs have been found to influence the glycolytic process in HCC. The level of CircFBLIM1 is significantly elevated in the serum exosomes of patients with HCC and HCC cells. It can be transferred to HCC cells through exosomes and acts on the miR-338/low-density lipoprotein receptor-related protein 6 (LRP6) axis, inducing the glycolytic process in HCC (Lai et al., 2023). Another study discovered that in HCC, exosomal circ-ZNF652 can adsorb miR-29a-3p, leading to increased expression of the downstream target gene GUCD1. This, in turn, promotes cancer cell glycolysis and affects malignant phenotypes like proliferation, migration, and invasion. Silencing the expression of circ-ZNF652 could inhibit HCC progression (Li et al., 2020).
Drug resistance is a significant factor contributing to the poor prognosis of HCC, and the dysregulation of extracellular vesicle circRNAs has been identified to play a role in tumor drug resistance (Li et al., 2019). Sorafenib, a Food and Drug Administration (FDA)-approved first-line treatment for advanced HCC, has shown reduced efficacy in recent years because of the emergence of drug resistance, thus limiting its clinical utility (Wen et al., 2022; Zhu et al., 2017b). A study has revealed that circRNA-SORE interacts with Y-box binding protein 1 (YBX1) in the cytoplasm, inhibiting the translocation of YBX1 to the cell nucleus, preventing pre-mRNA processing factor 19 (PRP19)-mediated ubiquitination and nuclear degradation of YBX1, and consequently affecting the expression of downstream target genes controlled by YBX1 (including AKT, RAF1 (encoding Raf-1 proto-oncogene), ERK, MYC (encoding c-Myc), and TGFB1 (encoding transforming growth factor beta 1)). Silencing the expression of circRNA-SORE was demonstrated to enhance the efficacy of sorafenib (Xu et al., 2020a). Lenvatinib, a tyrosine kinase inhibitor, has been approved by the FDA as a targeted therapy drug for advanced HCC alongside sorafenib; however, drug resistance remains a significant obstacle to its clinical use (Personeni et al., 2019). In studies investigating lenvatinib resistance mechanisms, it was discovered that the levels of circPAK1 were significantly increased in extracellular vesicles derived from lenvatinib-resistant HCC cells. The primary mechanism involves circPAK1 competitively binding to the cytoskeletal protein 14-3-3ζ, which promotes the nuclear translocation and inactivation of yes associated protein (YAP) and the Hippo signaling pathway (Hao et al., 2022). The Hippo signaling pathway is an inhibitory pathway primarily responsible for regulating cell size and tumor volume, with YAP serving as a downstream effector molecule. YAP, considered an oncoprotein, is regulated by phosphorylation and its subcellular localization. Phosphorylated YAP interacts with 14-3-3ζ, exits the cell nucleus, and undergoes ubiquitination and degradation mediated by beta-transducin repeat containing E3 ubiquitin protein ligase (β-Trcp). However, when the Hippo signaling pathway is inhibited, YAP remains non-phosphorylated, enters the cell nucleus through the nuclear membrane, interacts with transcription factors, and initiates the transcription of downstream target genes (Zhao et al., 2007). Furthermore, HCC-derived circPAK1 can transfer from extracellular vesicles to lenvatinib-sensitive HCC cells, thereby reducing their drug sensitivity (Hao et al., 2022).
Cisplatin (DDP) is commonly used as a first-line chemotherapy drug for HCC; however, resistance to DDP has a significant impact on the prognosis of patients with HCC (Zhou et al., 2019). Previous studies have demonstrated that CircZFR promotes the progression of HCC by influencing the miR-511/AKT1 axis (Yang et al., 2019). Further investigation into its resistance mechanism revealed that CircZFR is highly expressed in HCC-associated fibroblasts and is abundant in their extracellular vesicles. Through these extracellular vesicles, CircZFR can be transferred to HCC cells, where it inhibits the signal transducer and activator of transcription 3 (STAT3)/nuclear factor kappa B (NF-kβ) signaling pathway, ultimately promoting HCC progression and chemotherapy resistance (Zhou et al., 2022). Transarterial chemoembolization (TACE) is a treatment option for patients with advanced HCCs (Elderkin et al., 2023). In a recent study, researchers examined the variation in the circRNA contents of exosomes during TACE treatment and found that the abundance of exosomal circ-G004213 increased after TACE treatment compared to before treatment. This circRNA acts as a sponge for miR-513b-5p and enhances the expression of pre-mRNA processing factor 39 (PRPF39), thus improving the sensitivity of HepG2 cells to cisplatin and improved survival in vivo. Besides, the circ-G00421 expression level correlated positively with post-TACE prognosis, it may be used as an indicator for predicting the efficacy of TACE (Qin et al., 2021). This research provides a new potential combination treatment strategy to overcome chemotherapy resistance in HCC.
It is predicted that by 2040, there will be 1.3 million deaths from HCC, an increase of over 50% compared with that in 2020 (Rumgay et al., 2022). In recent years, an increasing number of exosomal circRNAs have been found to exhibit significantly changed expression profiles in HCC. The abnormal expression of circRNAs plays an important role in the occurrence and development of HCC. They transfer and transmit biological information between normal cells and HCC cells through exosomes, affecting malignant phenotypes such as HCC cell proliferation, invasion and metastasis, immune escape, cellular metabolism, and drug resistance (as shown in Figure 2; Table 1). Pathological diagnosis remains the gold standard for HCC; however, its early clinical application is limited by invasive procedures. Exosomal circRNAs, as novel biomarkers, exist in various body fluids, with high abundance, stability, and sensitivity, and can be sampled in a relatively non-invasive manner. They can serve as biomarkers for the early diagnosis and prognostication of HCC, as well as representing new therapeutic targets to improve the prognosis of patients with HCC. Based on all the facts about the circRNA in pathophysiology of cell proliferation suggest that some of the biological functions govern by genes via noncoding circRNA in immunocompromised beings. Moreover, current research on exosomes is still in its nascent phase, and there are numerous hurdles to overcome before their clinical application can guide practical treatments, including the efficient isolation and extraction of exosomes. Nevertheless, we maintain that exosomal circRNAs will undoubtedly play a pivotal role in the future diagnosis and treatment of HCC.
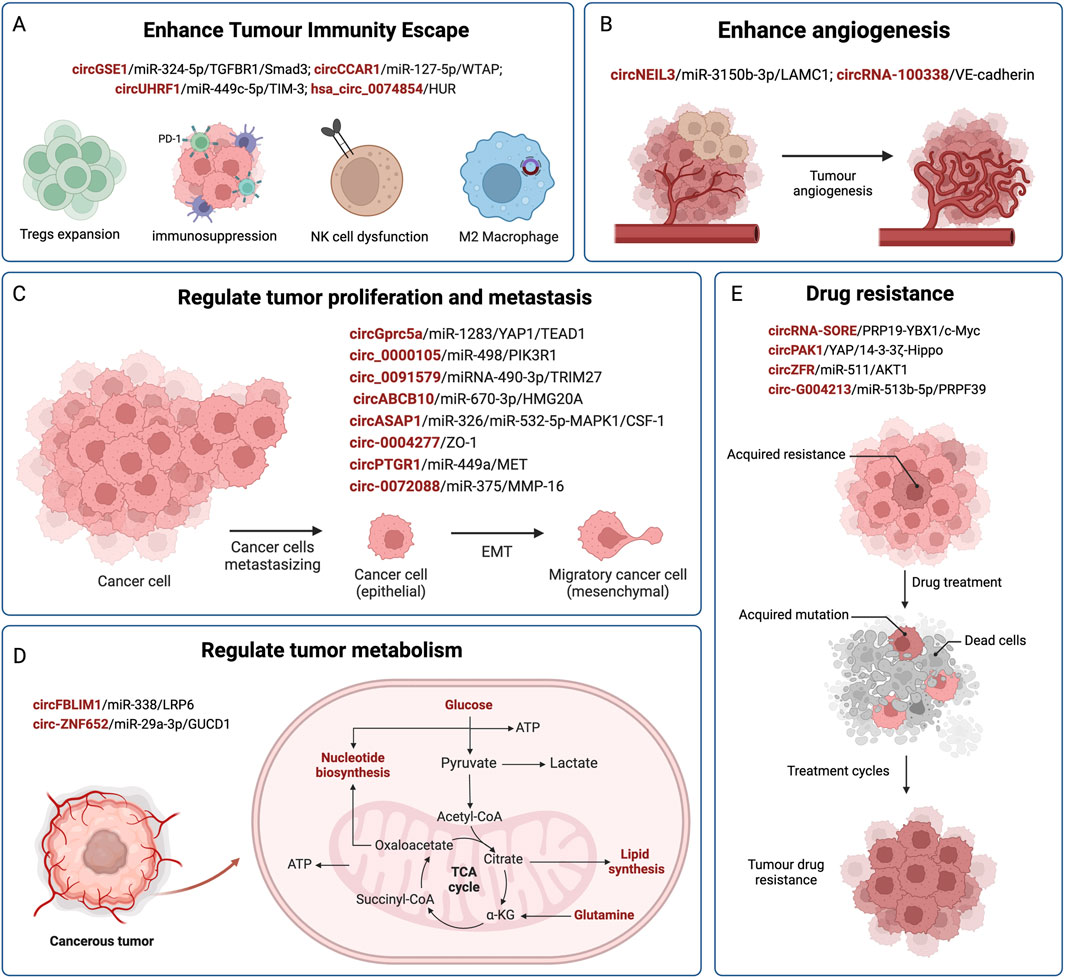
Figure 2. The regulatory role of exosomal circRNAs in HCC. CircRNAs are delivered by exosomes from donor cells to recipient cells, and affect the progression of HCC by competitively binding miRNAs and acting on downstream target genes. Exosomal circRNAs are involved in mediating the malignant phenotype of HCC, such as immune evasion (A), angiogenesis (B), HCC cell proliferation and metastasis (C), metabolism (D), and drug resistance (E).
One potential strategy to enhance the survival rate of patients with (HCC) is effectively delivering drugs into tumor cells. Exosomes, owing to their unique characteristics, can serve as cargo carriers for various functional molecules, including RNA, proteins, lipids, and circRNAs. Exploiting this feature, researchers have explored the use of nanotechnology to harness exosomes as vehicles to deliver drugs and functional RNAs, enabling targeted and precise tumor therapy (Ha et al., 2016). In recent years, RNA-based targeted therapy has emerged as a promising approach in cancer treatment, and circRNAs are anticipated to be valuable biomarkers for diagnosing, treating, and prognosticating liver cancer, particularly when they are encapsulated within exosomes. We believe that the combined utilization of circRNAs within exosomes and traditional biomarkers could significantly enhance their clinical utility in liver cancer, providing precise and personalized treatments for patients with HCC, ultimately leading to improved prognosis.
HZ: Writing–original draft, Writing–review and editing. SP: Writing–original draft, Writing–review and editing. JL: Writing–original draft, Writing–review and editing. JZ: Writing–original draft, Writing–review and editing. HL: Data curation, Writing–original draft. GW: Data curation, Writing–original draft. RW: Data curation, Writing–original draft. RC: Data curation, Writing–original draft. ZF: Data curation, Writing–review and editing. JS: Writing–review and editing. KC: Conceptualization, Funding acquisition, Project administration, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the opening foundation of the State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, grant no (SKLID2020KF042) and the Major horizontal projects of Zhejiang Shuren University (2021D1034).
Authors thank all the researchers, doctors, nurses, medical technicians, front-line workers, and public health officials for their hard work during this pandemic.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Allaire, M., Goumard, C., Lim, C., LE Cleach, A., Wagner, M., and Scatton, O. (2020). New frontiers in liver resection for hepatocellular carcinoma. JHEP Rep. 2, 100134. doi:10.1016/j.jhepr.2020.100134
Anwanwan, D., Singh, S. K., Singh, S., Saikam, V., and Singh, R. (2020). Challenges in liver cancer and possible treatment approaches. Biochim. Biophys. Acta Rev. Cancer 1873, 188314. doi:10.1016/j.bbcan.2019.188314
Center, M. M., and Jemal, A. (2011). International trends in liver cancer incidence rates. Cancer Epidemiol. Biomarkers Prev. 20, 2362–2368. doi:10.1158/1055-9965.EPI-11-0643
Chen, L. L., and Yang, L. (2015). Regulation of circRNA biogenesis. RNA Biol. 12, 381–388. doi:10.1080/15476286.2015.1020271
Chen, L., and Shan, G. (2021). CircRNA in cancer: fundamental mechanism and clinical potential. Cancer Lett. 505, 49–57. doi:10.1016/j.canlet.2021.02.004
Chen, W., Quan, Y., Fan, S., Wang, H., Liang, J., Huang, L., et al. (2020). Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 475, 119–128. doi:10.1016/j.canlet.2020.01.022
Chen, X., Yang, T., Wang, W., XI, W., Zhang, T., Li, Q., et al. (2019). Circular RNAs in immune responses and immune diseases. Theranostics 9, 588–607. doi:10.7150/thno.29678
Cocquerelle, C., Mascrez, B., Hétuin, D., and Bailleul, B. (1993). Mis-splicing yields circular RNA molecules. Faseb J. 7, 155–160. doi:10.1096/fasebj.7.1.7678559
Dai, X., Chen, C., Yang, Q., Xue, J., Chen, X., Sun, B., et al. (2018). Exosomal circRNA_100284 from arsenite-transformed cells, via microRNA-217 regulation of EZH2, is involved in the malignant transformation of human hepatic cells by accelerating the cell cycle and promoting cell proliferation. Cell Death Dis. 9, 454. doi:10.1038/s41419-018-0485-1
Daniele, B., Bencivenga, A., Megna, A. S., and Tinessa, V. (2004). Alpha-fetoprotein and ultrasonography screening for hepatocellular carcinoma. Gastroenterology 127, S108–S112. doi:10.1053/j.gastro.2004.09.023
Elderkin, J., AL Hallak, N., Azmi, A. S., Aoun, H., Critchfield, J., Tobon, M., et al. (2023). Hepatocellular carcinoma: surveillance, diagnosis, evaluation and management. Cancers (Basel) 15, 5118. doi:10.3390/cancers15215118
EL-Khoueiry, A. B., Sangro, B., Yau, T., Crocenzi, T. S., Kudo, M., Hsu, C., et al. (2017). Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 389, 2492–2502. doi:10.1016/S0140-6736(17)31046-2
European Association For The Study Of The LiverEuropean Organisation For Research And Treatment Of Cancer (2004). EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J. Hepatol. 56, 908–943. doi:10.1016/j.jhep.2011.12.001
Fajgenbaum, D. C., and June, C. H. (2020). Cytokine storm. N. Engl. J. Med. 383, 2255–2273. doi:10.1056/NEJMra2026131
Feng, C., Li, Y., Lin, Y., Cao, X., Li, D., Zhang, H., et al. (2019). CircRNA-associated ceRNA network reveals ErbB and Hippo signaling pathways in hypopharyngeal cancer. Int. J. Mol. Med. 43, 127–142. doi:10.3892/ijmm.2018.3942
Fernández, P. L., Hernández, L., Farré, X., Campo, E., and Cardesa, A. (2002). Alterations of cell cycle-regulatory genes in prostate cancer. Pathobiology 70, 1–10. doi:10.1159/000065998
Gao, Q., Wang, X. Y., Qiu, S. J., Yamato, I., Sho, M., Nakajima, Y., et al. (2009). Overexpression of PD-L1 significantly associates with tumor aggressiveness and postoperative recurrence in human hepatocellular carcinoma. Clin. Cancer Res. 15, 971–979. doi:10.1158/1078-0432.CCR-08-1608
Greten, T. F., and Sangro, B. (2017). Targets for immunotherapy of liver cancer. J. Hepatol. 68, 157–166. doi:10.1016/j.jhep.2017.09.007
Guo, S., Hu, C., Zhai, X., and Sun, D. (2021). Circular RNA 0006602 in plasma exosomes: a new potential diagnostic biomarker for hepatocellular carcinoma. Am. J. Transl. Res. 13, 6001–6015.
Ha, D., Yang, N., and Nadithe, V. (2016). Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm. Sin. B 6, 287–296. doi:10.1016/j.apsb.2016.02.001
Hanahan, D., and Coussens, L. M. (2012). Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322. doi:10.1016/j.ccr.2012.02.022
Han, Q., Zhao, H., Jiang, Y., Yin, C., and Zhang, J. (2019). HCC-derived exosomes: critical player and target for cancer immune escape. Cells 8, 558. doi:10.3390/cells8060558
Hao, X., Zhang, Y., Shi, X., Liu, H., Zheng, Z., Han, G., et al. (2022). CircPAK1 promotes the progression of hepatocellular carcinoma via modulation of YAP nucleus localization by interacting with 14-3-3ζ. J. Exp. Clin. Cancer Res. 41, 281. doi:10.1186/s13046-022-02494-z
Harding, C., Heuser, J., and Stahl, P. (1983). Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 97, 329–339. doi:10.1083/jcb.97.2.329
Hartke, J., Johnson, M., and Ghabril, M. (2017). The diagnosis and treatment of hepatocellular carcinoma. Semin. Diagn Pathol. 34, 153–159. doi:10.1053/j.semdp.2016.12.011
Heilig, R., Eckenberg, R., Petitv, J., Fonknechten, N., Da Silva, C., and Cattolico, L. (2004). Finishing the euchromatic sequence of the human genome. Nature 431, 931–945. doi:10.1038/nature03001
Hombach, S., and Kretz, M. (2016). Non-coding RNAs: classification, biology and functioning. Adv. Exp. Med. Biol. 937, 3–17. doi:10.1007/978-3-319-42059-2_1
Huang, X. Y., Huang, Z. L., Huang, J., Xu, B., Huang, X. Y., Xu, Y. H., et al. (2020). Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 39, 20. doi:10.1186/s13046-020-1529-9
Huang, C., Yu, W., Wang, Q., Huang, T., and Ding, Y. (2021). CircANTXR1 contributes to the malignant progression of hepatocellular carcinoma by promoting proliferation and metastasis. J. Hepatocell. Carcinoma 8, 1339–1353. doi:10.2147/JHC.S317256
Huang, M., Huang, X., and Huang, N. (2022). Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory T cells. Cancer Sci. 113, 1968–1983. doi:10.1111/cas.15365
Hu, K., Li, N. F., Li, J. R., Chen, Z. G., Wang, J. H., and Sheng, L. Q. (2021). Exosome circCMTM3 promotes angiogenesis and tumorigenesis of hepatocellular carcinoma through miR-3619-5p/SOX9. Hepatol. Res. 51, 1139–1152. doi:10.1111/hepr.13692
Hu, Z., Chen, G., Zhao, Y., Gao, H., Li, L., Yin, Y., et al. (2023). Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol. Cancer 22, 55. doi:10.1186/s12943-023-01759-1
Jiang, Y., Han, Q. J., and Zhang, J. (2019). Hepatocellular carcinoma: mechanisms of progression and immunotherapy. World J. Gastroenterol. 25, 3151–3167. doi:10.3748/wjg.v25.i25.3151
Kalluri, R., and Lebleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Science 367, eaau6977. doi:10.1126/science.aau6977
Komohara, Y., Fujiwara, Y., Ohnishi, K., and Takeya, M. (2016). Tumor-associated macrophages: potential therapeutic targets for anti-cancer therapy. Adv. Drug Deliv. Rev. 99, 180–185. doi:10.1016/j.addr.2015.11.009
Kos, A., Dijkema, R., Arnberg, A. C., Van der Meide, P. H., and Schellekens, H. (1986). The hepatitis delta (delta) virus possesses a circular RNA. Nature 323, 558–560. doi:10.1038/323558a0
Kowal, J., Tkach, M., and Théry, C. (2014). Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 29, 116–125. doi:10.1016/j.ceb.2014.05.004
Lai, Z., Wei, T., Li, Q., Wang, X., Zhang, Y., and Zhang, S. (2023). Exosomal circFBLIM1 promotes hepatocellular carcinoma progression and glycolysis by regulating the miR-338/LRP6 Axis. Cancer Biother Radiopharm. 38, 674–683. doi:10.1089/cbr.2020.3564
Li, L., Li, W., Chen, N., Zhao, H., Xu, G., Zhao, Y., et al. (2019). FLI1 exonic circular RNAs as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin. Cancer Res. 25, 1302–1317. doi:10.1158/1078-0432.CCR-18-1447
Lin, Y., Zheng, Z. H., Wang, J. X., Zhao, Z., and Peng, T. Y. (2021). Tumor cell-derived exosomal circ-0072088 suppresses migration and invasion of hepatic carcinoma cells through regulating MMP-16. Front. Cell Dev. Biol. 9, 726323. doi:10.3389/fcell.2021.726323
Li, T., Jiao, J., Ke, H., Ouyang, W., Wang, L., Pan, J., et al. (2023). Role of exosomes in the development of the immune microenvironment in hepatocellular carcinoma. Front. Immunol. 14, 1200201. doi:10.3389/fimmu.2023.1200201
Liu, C., Ren, C., Guo, L., Yang, C., and Yu, Q. (2023a). Exosome-mediated circTTLL5 transfer promotes hepatocellular carcinoma malignant progression through miR-136-5p/KIAA1522 axis. Pathol. Res. Pract. 241, 154276. doi:10.1016/j.prp.2022.154276
Liu, D., Kang, H., Gao, M., Jin, L., Zhang, F., Chen, D., et al. (2020). Exosome-transmitted circ_MMP2 promotes hepatocellular carcinoma metastasis by upregulating MMP2. Mol. Oncol. 14, 1365–1380. doi:10.1002/1878-0261.12637
Liu, L., Liao, R., Wu, Z., Du, C., You, Y., Que, K., et al. (2022). Hepatic stellate cell exosome-derived circWDR25 promotes the progression of hepatocellular carcinoma via the miRNA-4474-3P-ALOX-15 and EMT axes. Biosci. Trends 16, 267–281. doi:10.5582/bst.2022.01281
Liu, Y., Tang, H., Zhang, Y., Wang, Q., Li, S., Wang, Z., et al. (2023b). Circular RNA hsa_circ_0000519 contributes to angiogenesis and tumor progression in hepatocellular carcinoma through the miR-1296/E2F7 axis. Hum. Cell 36, 738–751. doi:10.1007/s13577-022-00854-7
Li, Y., Zang, H., Zhang, X., and Huang, G. (2020). Exosomal circ-znf652 promotes cell proliferation, migration, invasion and glycolysis in hepatocellular carcinoma via miR-29a-3p/GUCD1 Axis. Cancer Manag. Res. 12, 7739–7751. doi:10.2147/CMAR.S259424
Li, Y., Zheng, Q., Bao, C., Li, S., Guo, W., Zhao, J., et al. (2015). Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25, 981–984. doi:10.1038/cr.2015.82
Li, Z., Yanfang, W., Li, J., Jiang, P., Peng, T., Chen, K., et al. (2018). Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 432, 237–250. doi:10.1016/j.canlet.2018.04.035
Lu, J. C., Zhang, P. F., Huang, X. Y., Guo, X. J., Gao, C., Zeng, H. Y., et al. (2021). Amplification of spatially isolated adenosine pathway by tumor-macrophage interaction induces anti-PD1 resistance in hepatocellular carcinoma. J. Hematol. Oncol. 14, 200. doi:10.1186/s13045-021-01207-x
Lunt, S. Y., and Vander Heiden, M. G. (2011). Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 27, 441–464. doi:10.1146/annurev-cellbio-092910-154237
Luo, Y., Liu, F., and Gui, R. (2020). High expression of circulating exosomal circAKT3 is associated with higher recurrence in HCC patients undergoing surgical treatment. Surg. Oncol. 33, 276–281. doi:10.1016/j.suronc.2020.04.021
Lyu, L., Yang, W., Yao, J., Wang, H., Zhu, J., Jin, A., et al. (2021). The diagnostic value of plasma exosomal hsa_circ_0070396 for hepatocellular carcinoma. Biomark. Med. 15, 359–371. doi:10.2217/bmm-2020-0476
Marcon, P. D. S., Tovo, C. V., Kliemann, D. A., Fisch, P., and De Mattos, A. A. (2018). Incidence of hepatocellular carcinoma in patients with chronic liver disease due to hepatitis B or C and coinfected with the human immunodeficiency virus: a retrospective cohort study. World J. Gastroenterol. 24, 613–622. doi:10.3748/wjg.v24.i5.613
Masyuk, A. I., Masyuk, T. V., and Larusso, N. F. (2013). Exosomes in the pathogenesis, diagnostics and therapeutics of liver diseases. J. Hepatol. 59, 621–625. doi:10.1016/j.jhep.2013.03.028
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi:10.1038/nature11928
Meng, S., Zhou, H., Feng, Z., Xu, Z., Tang, Y., Li, P., et al. (2017). CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer 16, 94. doi:10.1186/s12943-017-0663-2
Mohammadisoleimani, E., Firoozi, Z., Naghizadeh, M. M., Ghanbari Asad, A., Pezeshki, B., Gholampour, Y., et al. (2022). Upregulation of hsa_circ_0004812 promotes COVID-19 cytokine storm via hsa-miR-1287-5p/IL6R, RIG-I axis. J. Clin. Lab. Anal. 36, e24666. doi:10.1002/jcla.24666
Morse, M. A., Sun, W., Kim, R., He, A. R., Abada, P. B., Mynderse, M., et al. (2019). The role of angiogenesis in hepatocellular carcinoma. Clin. Cancer Res. 25, 912–920. doi:10.1158/1078-0432.CCR-18-1254
Ng, W. L., Mohd Mohidin, T. B., and Shukla, K. (2018). Functional role of circular RNAs in cancer development and progression. RNA Biol. 15, 995–1005. doi:10.1080/15476286.2018.1486659
Omar, M. A., Omran, M. M., Farid, K., Tabll, A. A., Shahein, Y. E., Emran, T. M., et al. (2023). Biomarkers for hepatocellular carcinoma: from origin to clinical diagnosis. Biomedicines 11, 1852. doi:10.3390/biomedicines11071852
Pang, B., Zhu, Y., Ni, J., Thompson, J., Malouf, D., Bucci, J., et al. (2020). Extracellular vesicles: the next generation of biomarkers for liquid biopsy-based prostate cancer diagnosis. Theranostics 10, 2309–2326. doi:10.7150/thno.39486
Personeni, N., Pressiani, T., and Rimassa, L. (2019). Lenvatinib for the treatment of unresectable hepatocellular carcinoma: evidence to date. J. Hepatocell. Carcinoma 6, 31–39. doi:10.2147/JHC.S168953
Qin, L., Zhan, Z., Wei, C., Li, X., Zhang, T., and Li, J. (2021). Hsa-circRNA-G004213 promotes cisplatin sensitivity by regulating miR-513b-5p/PRPF39 in liver cancer. Mol. Med. Rep. 23, 421. doi:10.3892/mmr.2021.12060
Rumgay, H., Arnold, M., Ferlay, J., Lesi, O., Cabasag, C. J., Vignat, J., et al. (2022). Global burden of primary liver cancer in 2020 and predictions to 2040. J. Hepatol. 77, 1598–1606. doi:10.1016/j.jhep.2022.08.021
S, E. L. A., Mäger, I., Breakefield, X. O., and Wood, M. J. (2013). Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 12, 347–357. doi:10.1038/nrd3978
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U. S. A. 73, 3852–3856. doi:10.1073/pnas.73.11.3852
Shi, X., Wang, B., Feng, X., Xu, Y., Lu, K., and Sun, M. (2020). circRNAs and exosomes: a mysterious frontier for human cancer. Mol. Ther. Nucleic Acids 19, 384–392. doi:10.1016/j.omtn.2019.11.023
Song, X., Shi, X., Li, W., Zhang, F., and Cai, Z. (2020). The RNA-binding protein HuR in digestive system tumors. Biomed. Res. Int. 2020, 9656051. doi:10.1155/2020/9656051
Sterling, R. K., Jeffers, L., Gordon, F., Venook, A. P., Reddy, K. R., Satomura, S., et al. (2009). Utility of Lens culinaris agglutinin-reactive fraction of alpha-fetoprotein and des-gamma-carboxy prothrombin, alone or in combination, as biomarkers for hepatocellular carcinoma. Clin. Gastroenterol. Hepatol. 7, 104–113. doi:10.1016/j.cgh.2008.08.041
Sun, X. H., Wang, Y. T., Li, G. F., Zhang, N., and Fan, L. (2020). Serum-derived three-circRNA signature as a diagnostic biomarker for hepatocellular carcinoma. Cancer Cell Int. 20, 226. doi:10.1186/s12935-020-01302-y
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Su, Y., Lv, X., Yin, W., Zhou, L., Hu, Y., Zhou, A., et al. (2019). CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY) 11, 8182–8203. doi:10.18632/aging.102312
Tian, Z., Hou, X., Liu, W., Han, Z., and Wei, L. (2019). Macrophages and hepatocellular carcinoma. Cell Biosci. 9, 79. doi:10.1186/s13578-019-0342-7
Villalba, M., Rathore, M. G., Lopez-Royuela, N., Krzywinska, E., Garaude, J., and Allende-Vega, N. (2013). From tumor cell metabolism to tumor immune escape. Int. J. Biochem. Cell Biol. 45, 106–113. doi:10.1016/j.biocel.2012.04.024
Wang, F., Nazarali, A. J., and Ji, S. (2016). Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. Cancer Res. 6, 1167–1176.
Wang, G., Liu, W., Zou, Y., Wang, G., Deng, Y., Luo, J., et al. (2019a). Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine 40, 432–445. doi:10.1016/j.ebiom.2018.12.062
Wang, M., Yang, Y., Xu, J., Bai, W., Ren, X., and Wu, H. (2018). CircRNAs as biomarkers of cancer: a meta-analysis. BMC Cancer 18, 303. doi:10.1186/s12885-018-4213-0
Wang, W., and Wei, C. (2020). Advances in the early diagnosis of hepatocellular carcinoma. Genes Dis. 7, 308–319. doi:10.1016/j.gendis.2020.01.014
Wang, Y., Gao, R., Li, J., Tang, S., Li, S., Tong, Q., et al. (2021a). Downregulation of hsa_circ_0074854 suppresses the migration and invasion in hepatocellular carcinoma via interacting with HuR and via suppressing exosomes-mediated macrophage M2 polarization. Int. J. Nanomedicine 16, 2803–2818. doi:10.2147/IJN.S284560
Wang, Y., Liu, J., Ma, J., Sun, T., Zhou, Q., Wang, W., et al. (2019b). Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol. Cancer 18, 116. doi:10.1186/s12943-019-1041-z
Wang, Y., Pei, L., Yue, Z., Jia, M., Wang, H., and Cao, L. L. (2021b). The potential of serum exosomal hsa_circ_0028861 as the novel diagnostic biomarker of HBV-derived hepatocellular cancer. Front. Genet. 12, 703205. doi:10.3389/fgene.2021.703205
Wen, N., Cai, Y., Li, F., Ye, H., Tang, W., Song, P., et al. (2022). The clinical management of hepatocellular carcinoma worldwide: a concise review and comparison of current guidelines: 2022 update. Biosci. Trends 16, 20–30. doi:10.5582/bst.2022.01061
Wu, Z. H., Li, C., Zhang, Y. J., and Lin, R. (2022). Bioinformatics study revealed significance of exosome transcriptome in hepatocellular carcinoma diagnosis. Front. Cell Dev. Biol. 10, 813701. doi:10.3389/fcell.2022.813701
Xiao, Y., and Yu, D. (2021). Tumor microenvironment as a therapeutic target in cancer. Pharmacol. Ther. 221, 107753. doi:10.1016/j.pharmthera.2020.107753
Xia, Q., Li, L., and Zhao, L. (2017). Silica nanoparticle-based dual-responsive nanoprodrug system for liver cancer therapy. Exp. Ther. Med. 14, 2071–2077. doi:10.3892/etm.2017.4768
Xie, M., Yu, T., Jing, X., Ma, L., Fan, Y., Yang, F., et al. (2020). Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol. Cancer 19, 112. doi:10.1186/s12943-020-01208-3
Xu, J., Ji, L., Liang, Y., Wan, Z., Zheng, W., Song, X., et al. (2020a). CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct. Target Ther. 5, 298. doi:10.1038/s41392-020-00375-5
Xu, R., Greening, D. W., Zhu, H. J., Takahashi, N., and Simpson, R. J. (2016). Extracellular vesicle isolation and characterization: toward clinical application. J. Clin. Invest 126, 1152–1162. doi:10.1172/JCI81129
Xu, W., Van Knegsel, A., Saccenti, E., Van Hoeij, R., Kemp, B., and Vervoort, J. (2020b). Metabolomics of milk reflects a negative energy balance in cows. J. Proteome Res. 19, 2942–2949. doi:10.1021/acs.jproteome.9b00706
Yang, X., Liu, L., Zou, H., Zheng, Y. W., and Wang, K. P. (2019). circZFR promotes cell proliferation and migration by regulating miR-511/AKT1 axis in hepatocellular carcinoma. Dig. Liver Dis. 51, 1446–1455. doi:10.1016/j.dld.2019.04.012
Yang, X., Xiong, Q., Wu, Y., Li, S., and Ge, F. (2017). Quantitative proteomics reveals the regulatory networks of circular RNA CDR1as in hepatocellular carcinoma cells. J. Proteome Res. 16, 3891–3902. doi:10.1021/acs.jproteome.7b00519
Yuan, P., Song, J., Wang, F., and Chen, B. (2022). Exosome-transmitted circ_002136 promotes hepatocellular carcinoma progression by miR-19a-3p/RAB1A pathway. BMC Cancer 22, 1284. doi:10.1186/s12885-022-10367-z
Yu, B., and Shan, G. (2016). Functions of long noncoding RNAs in the nucleus. Nucleus 7, 155–166. doi:10.1080/19491034.2016.1179408
Yu, Y., Bian, L., Liu, R., Wang, Y., and Xiao, X. (2021). Circular RNA hsa_circ_0061395 accelerates hepatocellular carcinoma progression via regulation of the miR-877-5p/PIK3R3 axis. Cancer Cell Int. 21, 10. doi:10.1186/s12935-020-01695-w
Zhang, P. F., Gao, C., Huang, X. Y., Lu, J. C., Guo, X. J., Shi, G. M., et al. (2020a). Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 19, 110. doi:10.1186/s12943-020-01222-5
Zhang, X. P., Pei, J. P., Zhang, C. D., Yusupu, M., Han, M. H., and Dai, D. Q. (2022b). Exosomal circRNAs: a key factor of tumor angiogenesis and therapeutic intervention. Biomed. Pharmacother. 156, 113921. doi:10.1016/j.biopha.2022.113921
Zhang, H., Bai, M., Deng, T., Liu, R., Wang, X., Qu, Y., et al. (2016). Cell-derived microvesicles mediate the delivery of miR-29a/c to suppress angiogenesis in gastric carcinoma. Cancer Lett. 375, 331–339. doi:10.1016/j.canlet.2016.03.026
Zhang, L., Zhang, J., Li, P., Li, T., Zhou, Z., and Wu, H. (2022a). Exosomal hsa_circ_0004658 derived from RBPJ overexpressed-macrophages inhibits hepatocellular carcinoma progression via miR-499b-5p/JAM3. Cell Death Dis. 13, 32. doi:10.1038/s41419-021-04345-9
Zhang, Q., Wang, W., Zhou, Q., Chen, C., Yuan, W., Liu, J., et al. (2020b). Roles of circRNAs in the tumour microenvironment. Mol. Cancer 19, 14. doi:10.1186/s12943-019-1125-9
Zhang, T., Jing, B., Bai, Y., Zhang, Y., and Yu, H. (2020c). Circular RNA circTMEM45A acts as the sponge of MicroRNA-665 to promote hepatocellular carcinoma progression. Mol. Ther. Nucleic Acids 22, 285–297. doi:10.1016/j.omtn.2020.08.011
Zhao, B., Wei, X., Li, W., Udan, R. S., Yang, Q., Kim, J., et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761. doi:10.1101/gad.1602907
Zheng, R., Zhang, K., Tan, S., Gao, F., Zhang, Y., Xu, W., et al. (2022). Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Mol. Cancer 21, 49. doi:10.1186/s12943-021-01471-y
Zheng, Y., Li, Y., Lian, J., Yang, H., Li, F., Zhao, S., et al. (2019). TNF-α-induced Tim-3 expression marks the dysfunction of infiltrating natural killer cells in human esophageal cancer. J. Transl. Med. 17, 165. doi:10.1186/s12967-019-1917-0
Zhou, W. Y., Cai, Z. R., Liu, J., Wang, D. S., Ju, H. Q., and Xu, R. H. (2020). Circular RNA: metabolism, functions and interactions with proteins. Mol. Cancer 19, 172. doi:10.1186/s12943-020-01286-3
Zhou, Z. J., Xin, H. Y., Li, J., Hu, Z. Q., Luo, C. B., and Zhou, S. L. (2019). Intratumoral plasmacytoid dendritic cells as a poor prognostic factor for hepatocellular carcinoma following curative resection. Cancer Immunol. Immunother. 68, 1223–1233. doi:10.1007/s00262-019-02355-3
Zhou, Y., Tang, W., Zhuo, H., Zhu, D., Rong, D., Sun, J., et al. (2022). Cancer-associated fibroblast exosomes promote chemoresistance to cisplatin in hepatocellular carcinoma through circZFR targeting signal transducers and activators of transcription (STAT3)/nuclear factor -kappa B (NF-κB) pathway. Bioengineered 13, 4786–4797. doi:10.1080/21655979.2022.2032972
Zhu, A. X., Finn, R. S., Edeline, J., Cattan, S., Ogasawara, S., Palmer, D., et al. (2018). Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 19, 940–952. doi:10.1016/S1470-2045(18)30351-6
Zhu, K. P., Ma, X. L., and Zhang, C. L. (2017a). LncRNA ODRUL contributes to osteosarcoma progression through the miR-3182/MMP2 Axis. Mol. Ther. 25, 2383–2393. doi:10.1016/j.ymthe.2017.06.027
Zhu, Y. J., Zheng, B., Wang, H. Y., and Chen, L. (2017b). New knowledge of the mechanisms of sorafenib resistance in liver cancer. Acta Pharmacol. Sin. 38, 614–622. doi:10.1038/aps.2017.5
Keywords: hepatocellular carcinoma, exosomes, circRNA, biomarker, cancer therapy
Citation: Zhang H, Pei S, Li J, Zhu J, Li H, Wu G, Weng R, Chen R, Fang Z, Sun J and Chen K (2024) Insights about exosomal circular RNAs as novel biomarkers and therapeutic targets for hepatocellular carcinoma. Front. Pharmacol. 15:1466424. doi: 10.3389/fphar.2024.1466424
Received: 18 July 2024; Accepted: 30 September 2024;
Published: 09 October 2024.
Edited by:
Zhaofeng Liang, Jiangsu University, ChinaReviewed by:
Li Zhang, Brown University, United StatesCopyright © 2024 Zhang, Pei, Li, Zhu, Li, Wu, Weng, Chen, Fang, Sun and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingbo Sun, c3VuamluZ2JvQGJlaWh1YS5lZHUuY24=; Keda Chen, Y2hlbmtkQHpqc3J1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.