- 1Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Southwest Medical University, Luzhou, China
- 2Department of Pathology, The Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan, China
- 3Department of Urology, The Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan, China
Prostate cancer (PCa) is the second leading cause of cancer-related death among men in western countries. Evidence has indicated the significant role of the androgen receptor (AR) as the main driving factor in controlling the development of PCa, making androgen receptor inhibition (ARI) therapy a pivotal management approach. In addition, AR independent signaling pathways also contribute to PCa progression. One such signaling pathway that has garnered our attention is N6-Methyladenosine (m6A) signaling, which refers to a chemical modification on RNA with crucial roles in RNA metabolism and disease progression, including PCa. It is important to comprehensively summarize the role of each individual m6A regulator in PCa development and understand its interaction with AR signaling. This review aims to provide a thorough summary of the involvement of m6A regulators in PCa development, shedding light on their upstream and downstream signaling pathways. This summary sets the stage for a comprehensive review that would benefit the scientific community and clinical practice by enhancing our understanding of the biology of m6A regulators in the context of PCa.
1 Introduction
Prostate cancer (PCa), a malignancy originating from epithelial cells in the peripheral zone of prostate (Zhou et al., 2023), remains the second commonly diagnosed adenocarcinoma and the leading cause of cancer related deaths among men worldwide. The World Cancer Research Fund International survey estimated that 1,467,854 new cases of PCa were reported globally in 2022, resulting in approximately 397,430 deaths (Bray et al., 2024). Epidemiological studies have established that age (Godtman et al., 2022; Choi et al., 2018), race (Akaza et al., 2011; Jeong et al., 2016) and genetic factors (Bratt, 2002; Rebbeck, 2017; Thalgott et al., 2018) as the significant risk factors for PCa. PCa progresses through four stages, as determined by digital rectal examination (DRE) (Mottet et al., 2017), serum prostate specific antigen (PSA) level (Mottet et al., 2017) and pathological examination of biopsy samples (Kwon et al., 2020). Generally, low-grade and early localized PCa patients (PSA ≤10, Gleason score ≤6, or clinical stage T1-2a) are often managed by either radiotherapy or surgery. However, approximately 8% of PCa patients are viewed as advanced disease at their first diagnosis (Siegel et al., 2022). The cancer cells may spread from the prostate to other parts of the body, particularly the bones (Peng et al., 2017) and lymph nodes (Cai et al., 2011). In advanced stage, it may lead to urinary difficulty, hematuria, or pelvic pain. Targeting the androgen receptor (AR) signaling axis with androgen deprivation therapy (ADT) has been a primary treatment approach, showing favorable outcome (Dai et al., 2017; Davies et al., 2021; Guan et al., 2022; Jeon et al., 2023). Unfortunately, ADT is not curative and most patients will relapse within 2 years despite the low castrated level of serum testosterone. These patients are then considered to acquire castration-resistant PCa (CRPC), a highly lethal disease that accounts for the main mortality (Shigeta et al., 2019; Cai et al., 2023; Cheng et al., 2022; Wang et al., 2023a). Increasing evidence suggest that the reactivation of AR signaling plays a critical role in CRPC development, leading to the clinical approval of the second-generation AR antagonists such as enzalutamide (Enz) for managing this disease (de Bono et al., 2011; Agarwal et al., 2023; Powles et al., 2022; Wenzel et al., 2022). Despite the initial responses to this therapy, patients will eventually become Enz resistance owing to various mechanisms (Bennett et al., 2024; Liu et al., 2019; Zhang et al., 2020; Zheng et al., 2022). Additionally, approximately 30% of patients exhibit primary resistance to Enz treatment. These clinical findings collectively indicate limitations in the application of Enz.
Although AR is the main driving force for PCa progression, other signaling pathways, such as m6A signaling, are also involved in the regulation of PCa carcinogenesis and therapy resistance (He et al., 2022; Han et al., 2023). This review aims to comprehensively summarize the current understanding of the roles of RNA m6A regulators in PCa development and offer insights for further scientific research and clinical strategies.
2 Epitranscriptome and RNA m6A modification
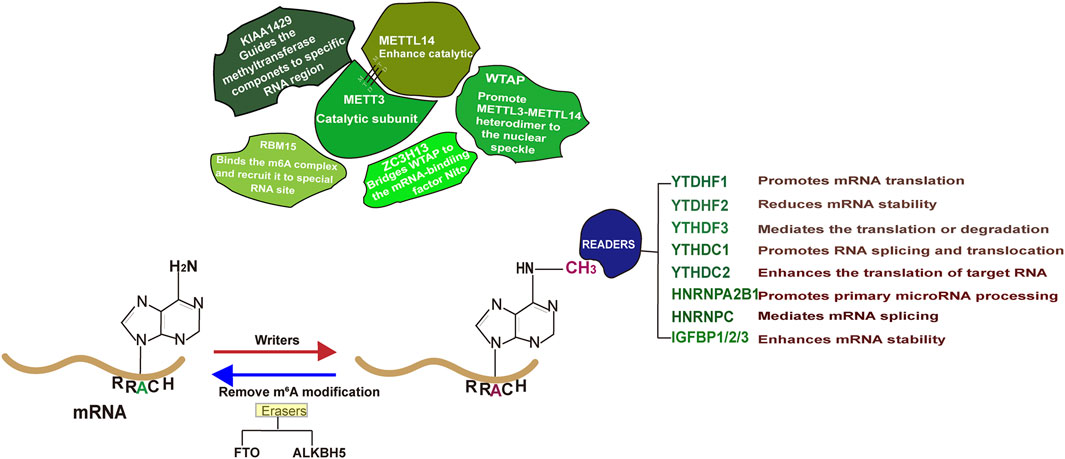
Epitranscriptome, a biochemical modification on RNA, has received significant attention from scientists due to its critical roles in determining RNA metabolism as well as disease progression (Murakami and Jaffrey, 2022; Wang Y. et al., 2014; He et al., 2018; Bokar et al., 1997; Clancy et al., 2002; Sommer et al., 1978; Zhong et al., 2008). It is estimated that over 170 types of biochemical modifications occur in RNAs, with m6A as the major form (Wiener and Schwartz, 2021). Early identified in 1970s, m6A, the methyl-nitrogen at the position six of adenylate (Figure 1), has been reported to be functional (Wei et al., 1976; Desrosiers et al., 1974). The enzyme responsible for catalyzing RNA m6A modification, known as “Writer,” includes methyltransferase-like protein 3 (METTL3), METTL16, METTL5 and zinc finger CCHC type containing 4 (ZCCHC4) (Jiang et al., 2021). Among them, METTL3 methyltransferase complex, consisting of METTL3, METT14, WTAP (Wilms tumor 1 associated protein), Zinc finger CCCH-type containing 13 (ZC3H13), RNA-binding motif protein 15 (RBM15) and VIRMA (Vir Like M6A Methyltransferase Associated), is mainly responsible for the RNA m6A modification on the consensus sequence DRACH (D = A/G/U, R = A/G, H = A/C/U) (Linder et al., 2015; Zaccara et al., 2019; Huang et al., 2022; Raj et al., 2022; Wei et al., 2022; Ma et al., 2019). It is noting that the m6A modification is a reversible process and the methyl group can be removed by demetyltransferase (Eraser) such as obesity-associated protein (FTO) and Human AlkB homolog H5 (ALKBH5) (He et al., 2019). Once an RNA molecule is m6A modified, it becomes prone to recognition by a variety of proteins (Readers) and undergoes distinct fate (Zaccara et al., 2019). In general, m6A modification on mRNA enables to influence its splicing, stability or translation. Recent advances in this area suggest that m6A regulators play vital roles in various human cancers, including PCa (Zhu W. et al., 2023). Studies have demonstrated that m6A level in PCa is disease stage dependent and m6A regulators are causally related to PCa growth, metastasis and targeted therapy resistance (Lothion-Roy et al., 2022). Therefore, there is a need to comprehensively summarize the molecular basis of m6A regulator mediated PCa carcinogenesis, which will definitely provide valuable insights for future scientific investigations and clinical applications.
2.1 METTL3/METTL14 m6A writer in PCa
METTL3/METTL14 methyltransferase complex is primarily responsible for the m6A modification of RNAs (Wang P. et al., 2016; Wang X. et al., 2016; Śledź and Jinek, 2016; Choe et al., 2018; Geula et al., 2015; Lin et al., 2016). Several studies have demonstrated that the expression levels of METTL3 and METTL14 are elevated in PCa as compared to normal tissues, acting as tumor promoting driver (Xu and Ge, 2022). Additionally, castration resistance perpetuates the increased expression levels of these two proteins (Wu et al., 2021). Supported by in vitro and in vivo evidence, METT3 complex promotes PCa growth and metastasis via catalyzing m6A modification of various mRNAs and non-coding RNAs (ncRNAs).
2.1.1 The targets and biological functions of METTL3 in PCa
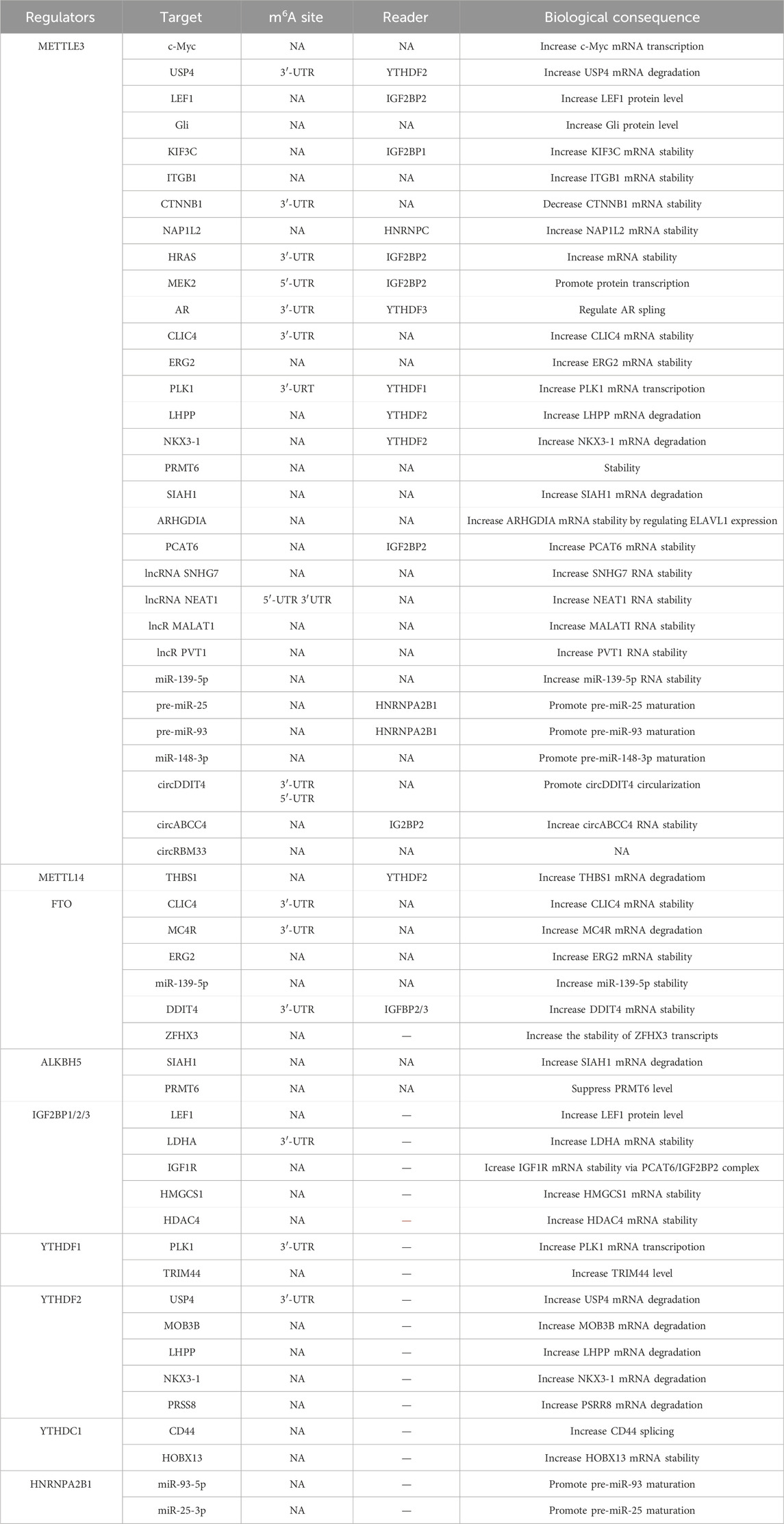
Advance in this field has led to the identification of a wide range of m6A targets. To date, mRNAs including c-Myc (Liu et al., 2022), USP4 (Chen et al., 2021a), LEF1 (Ma et al., 2020), DDIT4 (Zhao Y. et al., 2024), PRSS8 (Zhao X. et al., 2024), ZFHX3 (Hu et al., 2024) and others have been viewed as m6A targets in PCa (Table 1). In addition, ncRNAs, a class of RNAs without protein coding potential but proven to be physiologically and pathologically functional in a variety of disease models, are also potential targets of METTL3 complex in PCa. Specifically, lncRNAs (NEAT1 (Wen et al., 2020), MALAT1 (Mao et al., 2022), SNHG7 (Liu et al., 2022), PVT1 (Chen B. et al., 2023)), miRNAs (miR-139-5p (Azhati et al., 2023), pre-miR-25 (Qi et al., 2023), pre-miR-93 (Qi et al., 2023), miR-148-3p (Li G. et al., 2023)) and circRNAs (circDDIT4 (Kong et al., 2023), circABCC4 (Huang C. et al., 2023), circRBM33 (Zhong et al., 2023) and hsa_circ_0003258 (Yu et al., 2022)) have been reported as the substrates of METTL3. The m6A modification site, the RNA fate, the specific reader and the biological consequence of each individual RNA molecule are summarized and listed in Table 1. The literature illustrate a high expression level of METTL3 in PCa, implying it may contribute to PCa development. Indeed, by catalyzing m6A modifications of RNAs, METTL3 promotes PCa survival, metastasis and therapy resistance. For example, ubiquitin-specific protease 4 (USP4) was identified by Chen et al. as one target of METTL3 by the m6A-RIP (RNA immunoprecipitation) qPCR. Upon being m6A modified at the A2696, USP4 mRNA is recognized by YTH N (6)-Methyladenosine RNA Binding Protein 2 (YTHDF2) and undergoes degradation, subsequently leading to the protein degradation of ELAV like RNA-binding protein 1 (ELAV1). As a consequence, METTL3 mediated ELAV1 degradation increases ARHGDIA expression and promotes PCa growth and metastasis. Thus, targeting METTL3 by shRNAs powerfully attenuated PCa development in vitro and in vivo (Chen et al., 2021a).
METTL3 has also been implicated in the regulation of glycolysis in PCa by adding methyl groups to lncRNA SNHG7, thereby enhancing its stability. Consequently, SNHG7 interacts with SRSF1 to promote the expression of c-Myc, a transcription factor related to glycolysis by regulating the expression of various genes (Liu et al., 2022). Furthermore, Li et al. observed an increased level of METTL3 in enzalutamide resistant PCa cells, implying it may be a causal factor determining enzalutamide resistance. Indeed, METTL3 could activate MAPK signaling via catalyzing the m6A modifications of HRAS and MEK2 mRNAs to bypass AR inhibition therapy (Li Y. et al., 2023). Based on this, we can envision a potential combined therapy involving enzalutamide and a specific METTL3 inhibitor for the treatment of CRPC patients.
To summarize, METTL3 plays a tumor promoting role in PCa progression and targeted therapy resistance at least by catalyzing some oncogenes (c-Myc) (Liu et al., 2022) and core component of multiple signaling pathways including WNT signaling (CTNNB1) (Zhang S. et al., 2023), Hedgehog signaling (Gli) (Cai et al., 2019), MAPK signaling (HRAS, MEK2) (Li Y. et al., 2023). Whether METTL3 has an impact on other signaling pathways that influence PCa remains to be further explored through the continuous identification of its targets.
2.1.2 The role of METTL14 in PCa
As a critical component of METTL3 complex (Liu et al., 2014), METTL14 is also clinically correlated to PCa prognosis. Functionally, METT14 increases PCa proliferation in vitro and in vivo, largely through its regulation of thrombospondin 1 (THBS1) mRNA based on the analysis of RNA-seq and MeRIP (Methylated RNA Immunoprecipitation)-seq. Mechanistically, the m6A mark of THBS1 mRNA in the presence of METTL14 is recognized by YTHDF2, predisposing THBS1 mRNA to degrade (Wang Y. et al., 2022). However, in our opinion, the observed phenotype caused by METTL14 knockdown may be METTL3 complex dependent since the main role of METTL14 is to enhance METTL3 activity. It is anticipated that METTL14 deficiency severely impairs the enzymatic activity of METTL3 complex, leading to abnormal m6A modifications and impeding PCa growth. Nevertheless, it is plausible that METTL14 may have a METTL3 complex independent role in PCa, and this hypothesis can be tested by proposing experiments in METTL13-KO cells.
2.1.3 Other m6A writers in PCa
METTL16, another methyltransferase responsible for the m6A modifications of snRNAs and some lncRNAs (Pendleton et al., 2017; Shima et al., 2017; Warda et al., 2017), has not been investigated in PCa yet. It is noteworthy that the splicing events in PCa, especially CRPC, are highly active, leading to the generation of splicing products such as androgen receptor variant 7 (ARv7). Given the facts that 1) METTL16 is a m6A writer of MALAT1 (Ruszkowska et al., 2018); 2) MALAT1 mediated ARv7 signaling contribute to enzalutamide resistance (Wang et al., 2017), it would be interesting enough to explore the potential connections of METTL16 with anti-androgen resistance. Besides, whether METTL5 and ZCCHC4, the enzymes adding methyl group to ribosome RNAs (rRNAs), play contributing roles in PCa development is worthy of future investigations (van Tran et al., 2019).
2.2 M6A eraser
As mentioned above, it is important to note that m6A modification is a reversible process. FTO and ALKBH5 are the two well-known demethylases responsible for the removal of m6A in RNA molecule.
2.2.1 FTO in PCa
FTO was initially viewed as a demethylase of methylated DNAs (Gerken et al., 2007). However, subsequent studies have unraveled its preference for selecting RNAs, especially snRNAs (small nuclear RNAs), as substrates. Specifically, FTO recognizes m6Am (N6,2′-O-dimethyladenosine) in snRNAs and removes the methyl base (Wei et al., 2018; Mauer et al., 2017; Mauer et al., 2019) (Figure 2). Nevertheless, upcoming evidence suggests that FTO also holds a weak activity towards m6A, indicating its abnormal expression may impair the mRNAs metabolism (Li Y. et al., 2022).
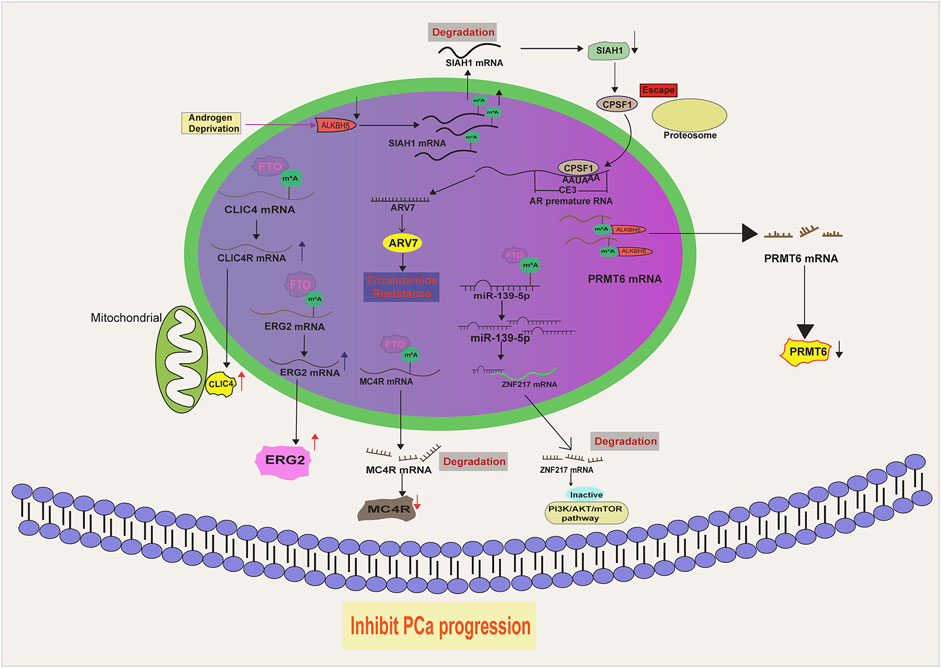
FTO is expressed at a lower level in PCa as compared to normal prostate tissues (Zhu et al., 2021). Moreover, PCa patients with low FTO expression often experience advanced disease and poor survival, suggesting that it acts as a tumor suppressor during PCa development (Wang Z. et al., 2022). Indeed, FTO depletion remarkably facilitates PCa malignancy in vitro and in vivo by increasing the total m6A level. Mechanistically, the loss of FTO increases the m6A levels of chloride intracellular channel 4(CLIC4) and ERG2, which are two tumor suppressors in PCa, accelerating their degradation (Zou et al., 2022). Moreover, melanocortin 4 receptor (MC4R), identified as another substrate of FTO in PCa, exhibits a high expression level owing to its abundant m6A mark resulting from FTO loss (Li and Cao, 2022). A recent literature has also demonstrated that FTO enables to decrease Zinc Finger Protein (ZNF217) expression by stabilizing miR-139-5p level via an m6A dependent manner. Consequently, FTO mediated ZNF217 reduction inactivates PI3K/AKT/mTOR signaling, impeding PCa progression. Collectively, these results suggest that FTO exerts a tumor-suppressing role in PCa progression via altering the m6A level of a specific RNA population (Figure 2). Intriguingly, the biological function of FTO is cancer context dependent. For instance, in renal cell carcinoma (Zhang et al., 2022), bladder cancer (Tao et al., 2021), breast cancer (Xu et al., 2020) and leukemia (Li et al., 2017), FTO functions as a tumor promoting factor. We postulate that the targets of FTO in different cancer models vary and determine the its functional identity. Therefore, it will be necessary to devote more efforts to identify the substrates of FTO in order to fully understand its biology in PCa.
2.2.2 ALKBH5 in PCa
ALKBH5, a member of the ALKB Family, specifically catalyzes the removal of the m6A modification on small nuclear RNAs (Figure 2). In contrast to FTO, ALKBH5 does not exhibit activity towards m6Am (Mauer et al., 2017; Mauer et al., 2019; Koh et al., 2019). Despite appearing to be an oncogene in cancer development due to its reported induction by hypoxia (Dong et al., 2021; Thalhammer et al., 2011), ALKBH5 actually functions to attenuate PCa growth. A study by Li et al. revealed that ALKBH5 has a marginal expression in PCa tissues and its overexpression apparently suppresses PCa cell growth and cell invasion via reducing the expression level of protein arginine methyltransferase 6 (PRMT6) via an m6A dependent manner (Li X. et al., 2023) (Figure 2). Similarly, Xia et al. (2022) observed a reduction of ALKBH5 in PCa cells with androgen deprivation. Consequently, SIAH1 mRNA is degraded due to the elevated m6A level resulted from the reduction of ALKBH5. Being a target of SIAH1, cleavage and polyadenylation specificity factor 1 (CPSF1) evades the proteosomal degradation and binds to the enriched AAUAAA sequence in the CE3 (cryptic exon 3) region of the AR premature mRNA, thereby facilitating its splicing to ARv7, a potent AR variant playing a critical role in castration resistance (Figure 2). These evidence suggest that ALKBH5 is a tumor suppressor in PCa. Again, the identification of ALKBH5 targets should be pursued if we want to fully understand its PCa associated biology. Potentially, an ALKBH5 agonist, if available in the future, may offer clinical benefits for PCa patients.
2.3 M6A readers
2.3.1 YTHDF family proteins
The YTHDF family consists of YTHDF1, YTHDF2 and YTHDF3 (Patil et al., 2018). Although sharing similar identity at the amino acid sequence, they have distinct biological effects on their targets (Patil et al., 2018; Chen L. et al., 2023). An early study has demonstrated that YTHDF1 binds to the m6A modified 3′-UTR of mRNAs, enhancing their translation (Wang et al., 2015; Wang X. et al., 2014). In contrast, YTHDF2 binds to its targets, leading to their instability and degradation (Li et al., 2020). While YTHDF3 has the capacity to influence both translation and stability of its bound targets (Shi et al., 2017).
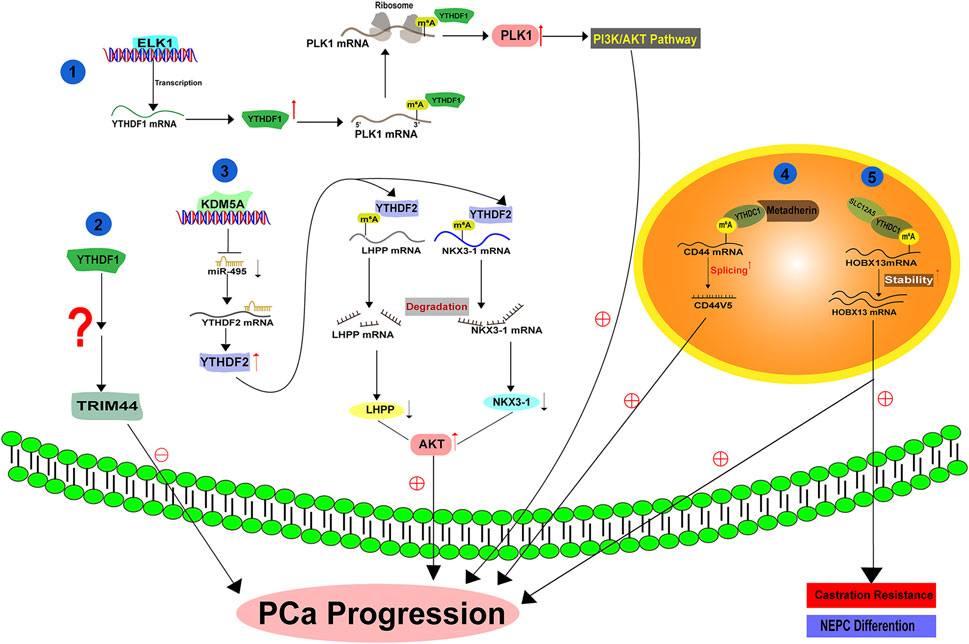
Li et al. (2021) demonstrated that YTHDF1 exhibits high expressionin in PCa and its level is correlated with disease prognosis. Knockdown of YTHDF1 significantly represses PCa survival, migration and invasion by regulating tripartite motif containing 44 (TRIM44) (Figure 3). Agreeably, another literature also suggested that YTHDF1, which is transcriptionally controlled by ELK1, facilitates PCa development in vitro and in vivo by activating polo-like kinase1 (PLK1) mediated PI3K-AKT signaling. Mechanistically, YTHDF1 binds to the m6A modified 3′-UTR of PLK1 mRNA and enhances its translation (Li P. et al., 2022) (Figure 3). YTHDF2 is also increased in PCa and its high expression indicates a poor overall survival. YTHDF2 exerts its oncogenic effect at least by mediating the instability and degradation of Phospholysine Phosphohistidine Inorganic Pyrophosphate Phosphatase (LHPP) and Homeobox Protein NK-3 Homolog A (NKX3–1) mRNAs, leading to the activation of AKT signaling and PCa progression (Li et al., 2020) (Figure 3). Therefore, upregulation of YTHDF2 via Lysine Demethylase 5A (KDM5A) mediated miR-495 reduction could drive PCa progression in vitro and in vivo (Du et al., 2020) (Figure 3). As another YTHDF family protein, YTHDF3 has not been functionally characterized in PCa. A recent literature has illustrated that YTHDF3 can bind the m6A modified AR mRNA and increase its translation in PCa cells (Somasekharan et al., 2022) (Figure 3). Given the significance of AR in PCa, it is tempting to hypothesize that YTHDF3 may act as an oncogenic protein to facilitate PCa growth, although this hypothesis requires experimental supports.
2.3.2 YTHDC1 and YTHDC2 in PCa
Primarily localized in the nucleus (Hartmann et al., 1999), YTHDC1 has been reported to regulate the splicing and nuclear export of the targets with m6A modification (Widagdo et al., 2022; Roundtree et al., 2017). The splicing activity of YTHDC1 is attributed to its association with serine and arginine-rich splicing factor 3 (SRSF3), an important splicing factor that regulates exon inclusion (Xiao et al., 2016). A recent study by Cheng et al. (2021) reported that YTHDC1 undergoes phase separation to control gene expression via various means, suggesting its diverse biological functions. In PCa, YTHDC1 interacts with the oncogene protein MTDH (Metadherin), facilitating the generation of splicing product CD44v5 and promoting PCa malignancy (Luxton et al., 2019) (Figure 3). In addition, YTHDC1 can also complex with SLC12A5 (a neuron-specific potassium-chloride co-transporter) and enhance its oncogenic function. As a result, YTHDC1-SLC12A5 complex promotes PCa progression, castration resistance and neuroendocrine differentiation by recognizing and stabilizing m6A modified Homeobox B13 (HOXB13) mRNA (Yuan et al., 2023) (Figure 3). Considering the highly active splicing process during the progression of PCa to an advanced stage, we surmise that YTHDC1 may hold a fundamental role in the development of PCa by regulating the amount of various splicing products in an m6A dependent manner.
Although YTHDC2 is not ubiquitously expressed and its high abundance is observed in testes (Bailey et al., 2017; Hsu et al., 2017; Jain et al., 2018), it does not exclude the possible causal involvement of YTHDC2 into the development of other diseases. Notably, a high expression of YTHDC2 is observed in PCa as compared to BPH (Benign prostatic hyperplasia) and normal prostate tissues. Experimental results have shown that YTHDC2 induction substantially promotes PCa cell growth and invasion (Song et al., 2023). Nevertheless, the underlying mechanism by which YTHDC2 drives PCa progression has not been investigated, and the exploration of the downstream targets of YTHDC2 in PCa remains an open area. Since the early claim suggested that YTHDC2 exhibits a very weak affinity towards m6A motif (Wojtas et al., 2017), it is reasonable to speculate that YTHDC2 may have non-m6A targets.
2.3.3 IGF2BP family proteins
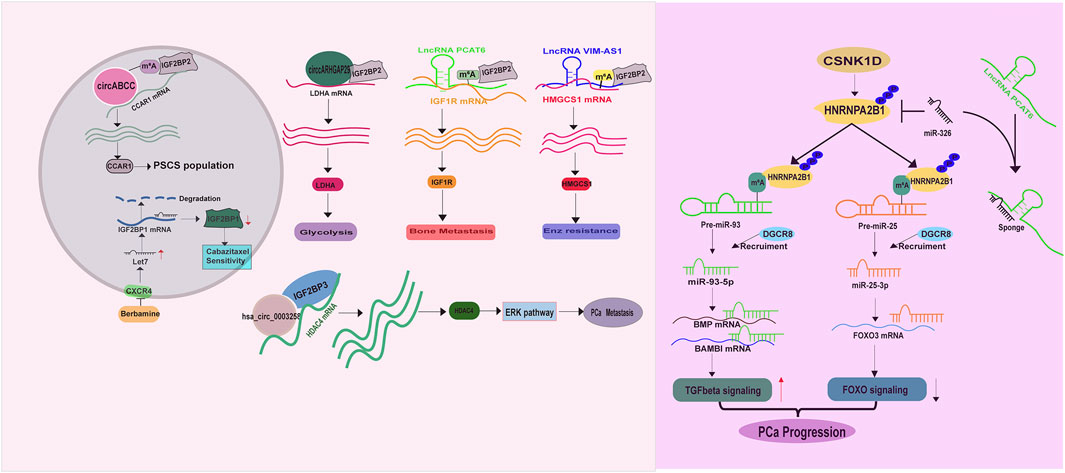
IGF2BP proteins enable to recognize m6A targets or non m6A targets and to increase their stabilities (Jiang et al., 2021; Huang et al., 2018; Lan et al., 2021), thus having a great impact on PCa development. A literature has demonstrated an increase of IGF2BP1 expression in prostate cancer stem cells (PCSCs), contributing to cabazitaxel resistance. Thus targeting CXCR4 (C-X-C chemokine receptor type 4)/let-7 mediated IGF2BP1 induction in PCSCs by Berbamine could restore PCa response to cabazitaxel treatment (Wang et al., 2024). Similarly, IGF2BP2 has been reported to recognize m6A labeled circABCC (circular ATP Binding Cassette Subfamily C Member), a prerequisite for stabilizing Cell Division Cycle And Apoptosis Regulator 1 (CCAR1) mRNA, expanding PCSCs population (Huang C. et al., 2023) (Figure 4). Besides, IGF2BP2 exerts a tumor promoting role in altering PCa metabolism, bone metastasis and targeted therapy resistance. A study from Jiang et al. unraveled that IGF2BP2 increases its binding affinity to Lactate dehydrogenase A (LDHA) mRNA in the presence of circARHGAP29 (circular Rho GTPase Activating Protein 29), thereby enhancing glycolytic metabolism (Jiang et al., 2022) (Figure 4). Another study demonstrated that IGF2BP2 is recruited by m6A modified lncRNA PCAT6 (Prostate Cancer Associated Transcript 6) to interact with IGF1R (Insulin-Like Growth Factor I Receptor) mRNA, resulting in its stabilization and the promotion of PCa bone metastasis (Lang et al., 2021) (Figure 4). Moreover, IGF2BP2 can confer enzalutamide resistance via binding to and stabilizing 3-Hydroxy-3-Methylglutaryl-CoA Synthase 1 (HMGCS1) mRNA in the presence of lncRNA VIM-AS1 (VIM Antisense RNA 1) (Shi et al., 2023) (Figure 4). IGF2BP3 also serves as an oncogene in PCa, as a study illustrated its ability to combine with hsa_circ_0003258 to directly enhance the stability of histone deacetylase 4 (HDAC4) mRNA, consequently activating ERK signaling pathway to drive PCa metastasis (Yu et al., 2022) (Figure 4).
Together, these evidences suggest that IGF2BP proteins support PCa survival and hasten its malignancy via stabilizing a wide range of mRNAs. Furthermore, it is evident that the impact of IGF2BP proteins on mRNA stabilization is m6A and non-m6A dependent, suggesting that classifying and identifying the targets of IGF2BP proteins based on the m6A status may aid in comprehending their biologies in PCa.
2.3.4 HnRNP family proteins
Accumulating evidence have demonstrated that the heterogeneous nuclear ribonucleoproteins (HnRNP) such as HnRNPC, HnRNPG and HNRNPA2B1 are direct or indirect readers of m6A labeled RNAs, especially miRNAs (Wang et al., 2020; Liu et al., 2015; Spitale et al., 2015; Liu et al., 2017; Wu et al., 2018). In PCa, elevated HnRNPC expression is closely correlated with tumor stage, tumor grade and the overall survival (Wang et al., 2021). Functionally, HnRNPC promotes PCa proliferation and metastasis (Cheng et al., 2023). Moreover, a high level of HNRNPA2B1 is also examined in PCa. HNRNPA2B1 binds to the m6A marks in several miRNA precursors (miR-93-5p (Qi et al., 2023; Sun et al., 2023), miR-25-3p (Qi et al., 2023)) and facilitates their processing and maturation via recruiting DGCR8 (DiGeorge syndrome critical region gene 8) (Sun et al., 2023), driving PCa development (Figure 4). For this point of view, molecules enabling to regulate HNRNPA2B1 expression is supposed to have a considerable impact on PCa survival and metastasis. As expected, casein kinase 1 delta (CSNK1D) phosphorylates and stabilizes HNRNPA2B1 protein, exacerbating PCa malignancy (Qi et al., 2023) (Figure 4). The lncRNA PCAT6 also has capacity to increase HNRNPA2B1 expression via acting as sponge of miR-326 to facilitate PCa neuroendocrine differentiation (Liu B. et al., 2021) (Figure 4). However, the role of another m6A reader, HnRNPG, has not been investigated in PCa.
3 The upstream signaling pathways regulating m6A regulators
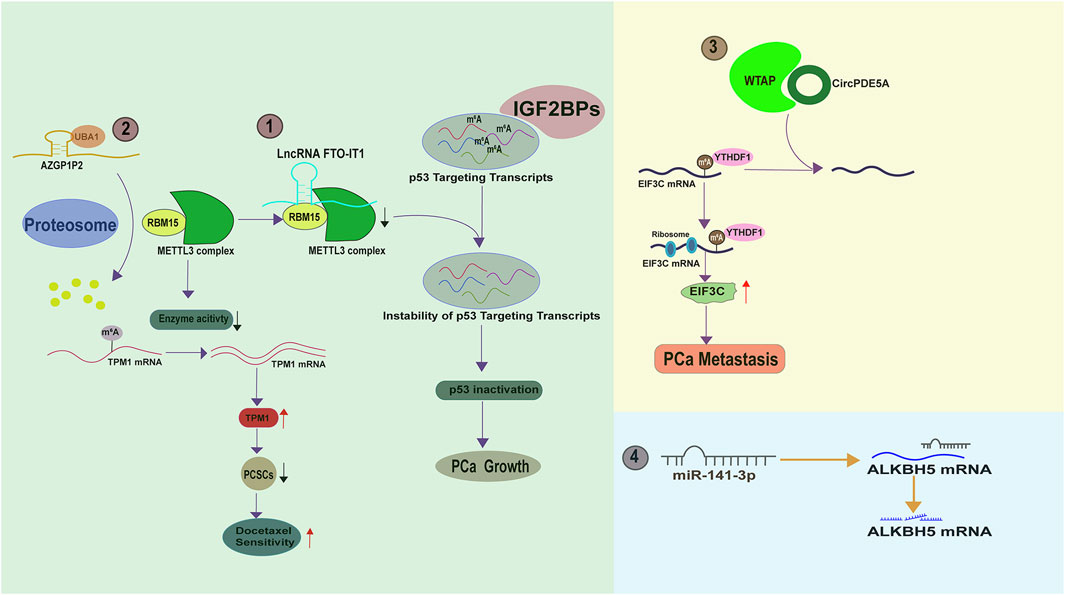
A literature suggest that the total m6A levels are gradually increased as PCa progresses from the localized mass to the metastatic disease (Wan et al., 2022), indicating the existence of a molecular network upstream of m6A regulators in PCa. Understanding this network may provide insight into novel strategies to improve the efficacy of current therapies. A fascinating study by Zhang et al. demonstrated that FTO-IT1 (FTO intronic transcript 1), a lncRNA transcribed from the intron 8 of FTO gene focus, downregulates the transcript levels of several p53 targeting genes such as FAS (Fas Cell Surface Death Receptor), TP53INP1 (Tumor protein p53-inducible nuclear protein 1), SESN2 (Sestrin2), and MDM2 (Mouse double minute 2 homolog), thereby recapitulating p53 inactivation. Results from RNA pull down and subsequent mass spectrum analysis illustrated that FTO-IT1 directly interacts with RBM15 but not other m6A regulator to inhibit the methyltransferase activity of METTL3 complex. As a sequence specific RNA binding protein, RBM15 fails to bind p53 targeting transcripts for m6A modification in the presence of FTO-IT1, leading to their failure to be recognized by IGF2BP proteins. Thus, FTO-IT1 knock-out specifically boosts the m6A levels of p53 targeting transcripts by releasing RBM15 mediated m6A “writer” activity and caused PCa cell growth arrest (Zhang J. et al., 2023) (Figure 5). Another study by Wang et al. also documented that RBM15 can be regulated by AZGP1P2, a pseudogene of AZGP2. According to the data, AZGP1P2 binds and recruits UBA1 (Ubiquitin Like Modifier Activating Enzyme 1) as a E1 conjugating enzyme for RBM15 degradation. As a result, the m6A of RBM15 recognized TPM1 mRNA (tropomyosin 1) at its coding region is erased and TPM1 mRNA is stabilized. TPM1 induction by AZGP1P2 functions as a tumor suppressor to sensitize PCa cells to docetaxel therapy via eradicating the population of prostate cancer stem cells (PCSCs) (Wang et al., 2023b) (Figure 5).
WTAP, a known m6A regulator, is reportedly regulated by circPDE5A, a circular form of exon 2 and exon 3 of PDE5A (Phosphodiesterase 5A). CircPDE5A binds WTAP and disrupts its mediated m6A modification of eukaryotic translation initiation factor 3c (EIF3C) (Figure 5). Therefore, circPDE5A inactivation in CRPC leads to an m6A increase of EIF3C mRNA, which is subsequently recognized by YTHDF1 and has an enhanced translation efficiency, eventually promoting PCa metastasis (Ding et al., 2022). Moreover, it has been documented that the m6A “eraser” ALKBH5 is a direct target of miR-141-3p (Li X. et al., 2023) (Figure 5). In the future, we can anticipate the identification of more upstream molecules that affect m6A regulators. Armed with this knowledge, we can effectively silence m6A signaling by targeting these upstream molecules.
4 The cross-talk between RNA m6A modification and AR signaling
Androgen receptor (AR), a member of steroid hormone receptors, has been acknowledged as the key driving factor determining PCa development for decades (Tang et al., 2021). Structurally consisting of N-terminal, DNA binding domain, Hinge region and Ligand binding domain, AR responds to dihydrotestosterone (DHT) and translocates into nucleus as dimer to regulate the transcription of numerous genes (Tan et al., 2015). Owing to the significant role of AR in PCa development, for a long time, AR signaling inhibition has been the main strategy for PCa management.
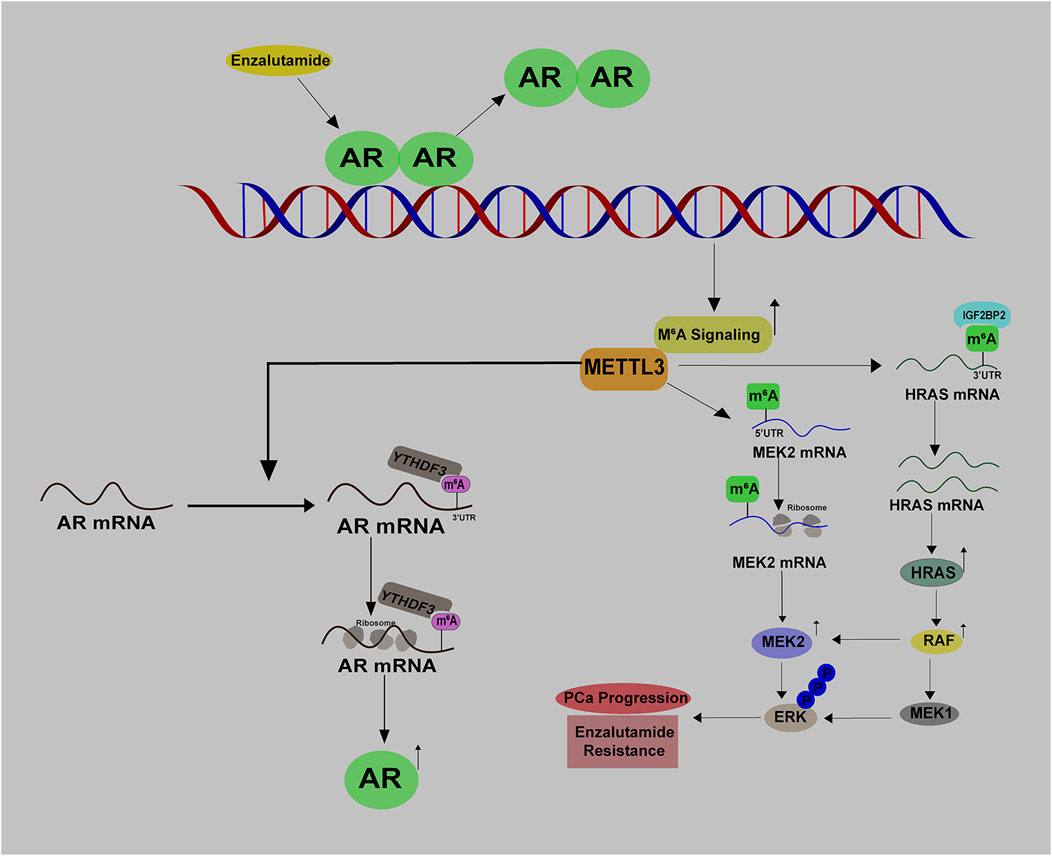
Androgen deprivation therapy (ADT) has been utilized as the golden mean to treat PCa for many decades, with promising clinical outcomes. Li et al. have uncovered a direct link between RNA m6A modification and androgen receptor (AR) signaling. Their research showed that ADT with enzalutamide treatment leads to an increase in METTL3 expression and the total m6A levels, suggesting METTL3 mediated m6A modification may contribute to the acquired Enz resistance (Figure 6). By performing MeRIP-seq and RNA-seq, the authors identified that METTL3 directly mediates m6A modifications of HRAS and MEK2 mRNAs. Mechanistically, HRAS mRNA with m6A at its 3′-UTR is much more stable, and MEK2 mRNA with m6A at 5′-UTR has a higher translation potential as compared to the corresponding non-modified controls. As a result, MAPK signaling is activated and bypasses AR signaling inhibition to promote PCa growth (Li Y. et al., 2023) (Figure 6). Therefore, activation of m6A signaling serves as a self-protective mechanism in response to AR inhibition, providing a non-AR survival source for PCa growth. Given the fact that enzalutamide is an anti-androgen drug specifically preventing the transcription activity of AR, it will be intriguing to explore whether AR enables to control the expression of m6A regulators at the chromatin level, thus affecting the m6A signaling.
Reciprocally, m6A signaling also has a great impact on AR signaling. Evidence from Haigh et al. suggested that METTL3 inhibition by siRNAs could substantially impair androgen regulated transcriptome in PCa (Haigh et al., 2022). Additionally, in early 2022, Somasekharan et al. (2022) discovered that AR mRNA is a direct target of METTL3 and its translation is potentiated with the m6A modification at 8953A (Figure 6). By connecting these two findings, we speculate that METTL3 may affect androgen regulated transcriptome via directly methylating AR mRNA. Given that CRPC expresses more abundant AR protein than primary PCa, it is hypothesized that METTL3-mediated AR mRNA translation at least partially accounts for this phenomenon (Wu et al., 2021). Therefore, targeting METTL3 may alleviate the reactivation of AR signaling and aid in overcoming CRPC progression. In summary, there exists a reciprocal regulation between AR signaling and m6A signaling in PCa.
5 Clinical implications of RNA m6A modification in PCa and future perspectives
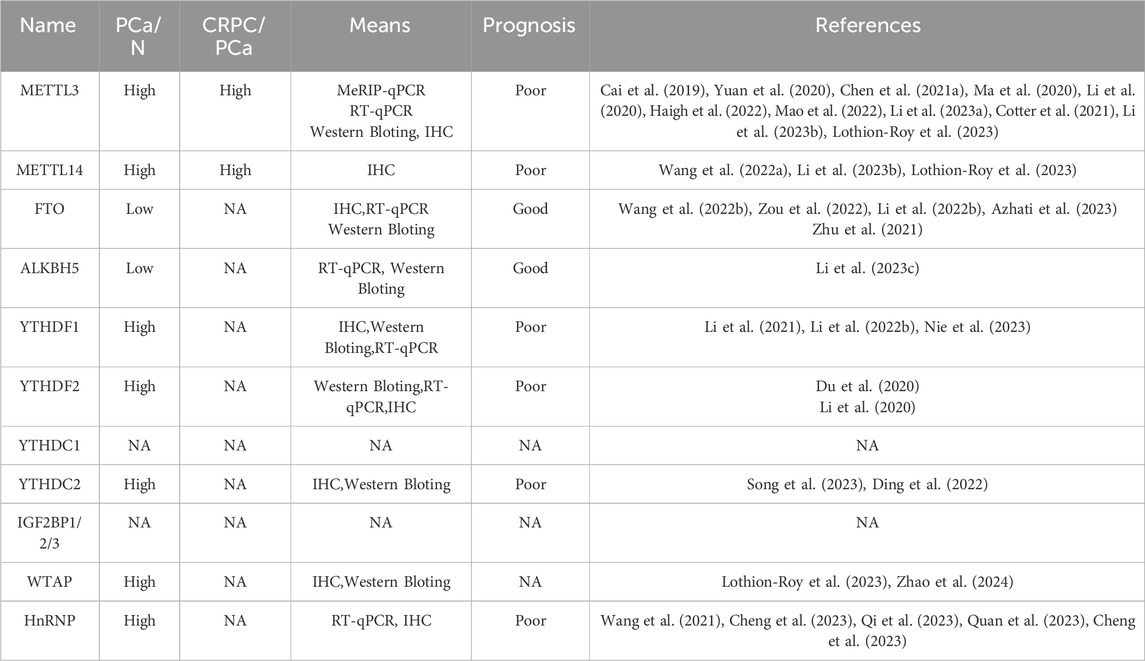
The clinical significance of RNA m6A modification should be acknowledged owing to its close relationship with PCa initiation, progression and therapy resistance. METTL3 and METTL14, the main components of m6A “writer,” elevate their expression when prostate epithelial cells become malignant (Xu and Ge, 2022) (Table 2). A continual rise in METTL3 and METTL14 expression is observed in CRPC disease (Wu et al., 2021). Conversely, the expression levels of m6A “eraser,” FTO and ALKBH5, display an opposite trend (Fang et al., 2022). In line with this, Lu et al. found that m6A modification levels are elevated in metastatic PCa as compared to the primary control, as evidenced by MeRIP-seq and RNA-seq on 4 metastatic PCa, 4 primary PCa tumors and 4 benign prostate hyperplasia (BPH). Importantly, they also reported that PCa patients with high m6A-modified mRNA (MMM) score experience shorter biochemical recurrence free survival and have a poor response to androgen signaling inhibition therapy as compared to the patients with a low MMM score, suggesting m6A modification status is a poor prognostic factor for predicting disease development and therapy resistance. However, their findings also exhibited that the primary PCa harbors a paucity of m6A modified mRNAs as compared to the BPH, implying hypo m6A modification of mRNAs contributes to PCa initiation In this context, a discrepancy is found between the expression pattern of m6A regulators and the m6A levels when the comparison was made between BPH and primary PCa (Lu et al., 2023). We hypothesize that the activities of m6A regulators are inhibited by some proteins so that a hypo m6A levels are observed in PCa.
According to this information, total m6A levels may serve as a diagnostic biomarker to predict disease status of PCa, and the elimination of m6A levels by METTL3/METTL14 inhibitor or others holds promise as a therapeutic strategy to prevent PCa progression. In 2021, Yankova et al identified a small molecule STM2457 as a potent METTL3 inhibitor to suppress acute myeloid leukaemia (AML), opening a new avenue of METTL3 targeted therapy. Additionally, Storm Therapeutics Company has screened another METTL3 inhibitor STC-15, which displays anti-tumor activity across different AML models and is currently being investigated in a clinical trail (NCT05584111) (Yankova et al., 2021). Although currently not available, it would be promising to test the efficacy of METTL3 inhibitors in PCa models and PCa patients. It is noting that some small molecules including curcumin (Chen et al., 2021b), quercetin (Zhu J. et al., 2023), epigallocatechin gallate (EGCG) (Wu et al., 2005) and simvastatin (Chen et al., 2020), have been reported to influence m6A signaling. However, in our opinion, they are not specific for interrupting m6A levels and their contributions to cancer prevention may not be solely due to the m6A alteration. Therefore, the continuous screening of METTL3-specific and potent inhibitors remains a priority for scientists and pharmacologists.
6 Conclusion
PCa is a male carcinoma and its mortality is continuously rising. Despite of the initial response, ADT treatment will lead to the emergence of recurrent tumor, suggesting other signaling pathways actively respond in order to bypass AR inhibition. As a type of epitranscriptomal modifications, m6A is now received much attention and it is indeed implicated into a variety of biological processes including tumorigenesis. Particularly in PCa, abnormal expression levels of m6A regulators are frequently observed by many researchers. The experimental evidence suggest that m6A writers, m6A erasers and m6A readers all contribute to PCa survival and malignancy. Additional evidence also suggest that the total m6A levels and METTL3 are closely related to enzalutamide resistance. These findings provide a strong rationale to propose a therapy using m6A inhibitor, alone or with anti-androgen, to treat CRPC patients.
Although numerous RNAs has been identified to be m6A modified, the blueprint of m6A signaling remains incomplete. In the clinical setting, a comprehensive understanding of m6A targets and their related signaling pathways can guide the discovery of novel targeted therapies to overcome PCa development. To this end, scientists should exert significant efforts to identify functional m6A targets during PCa evolution.
Although FTO inhibitors such as bisisantrene (Su et al., 2020), brequinar (Su et al., 2020), and Dac51 (Wu et al., 2023; Yang and Al-Hendy, 2023; Huang Y. et al., 2023; Liu Y. et al., 2021) have shown potency against several solid tumors, including renal carcinoma, bladder cancer, they may not be the ideal choice for the treatment of PCa model as researchers have confirmed the tumor suppressing role of FTO in PCa models. Alternatively, researchers should consider screening specific inhibitors of m6A readers, as they are positively implicated in PCa development. YTH family proteins, IGF2BP proteins, and other m6A readers have been proven to be oncogenic factors driving PCa progression. From our perspective, m6A reader inhibitors may be more specific than METTL3 inhibitors in suppressing a small population of RNA. While METTL3 has a variety of targets, each m6A reader has its uniquely recognized RNA population. Indeed, IGF2BP1 inhibitors (AVJ16 (Singh et al., 2024), BTYNB (Müller et al., 2020; Mahapatra et al., 2017; Jamal et al., 2023; Hagemann et al., 2023; Xiao et al., 2023; Wang JJ. et al., 2023), and 7773 (Singh et al., 2024)), IGF2BP2 inhibitor CWI1-2 (Weng et al., 2022), and YTHDF proteins inhibitor ebselen (Micaelli et al., 2022) all show promising anti-cancer activity in preclinical models. However, the identification of m6A reader inhibitors is still in the preliminary stage and requires intensive dedication.
Author contributions
YC: Writing–original draft, Writing–review and editing. MJ: Writing–original draft, Writing–review and editing. CD: Writing–original draft, Writing–review and editing. ZY: Writing–original draft. BC: Conceptualization, Funding acquisition, Writing–original draft, Writing–review and editing. RW: Conceptualization, Funding acquisition, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was kindly supported by the Sichuan Science and Technology Program (2022YFS0636) and the Science and Technology Program of Luzhou (2020LZXNYDJ43).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agarwal, N., Azad, A. A., Carles, J., Fay, A. P., Matsubara, N., Heinrich, D., et al. (2023). Talazoparib plus enzalutamide in men with first-line metastatic castration-resistant prostate cancer (TALAPRO-2): a randomised, placebo-controlled, phase 3 trial. Lancet London, Engl. 402, 291–303. doi:10.1016/S0140-6736(23)01055-3
Akaza, H., Kanetake, H., Tsukamoto, T., Miyanaga, N., Sakai, H., Masumori, N., et al. (2011). Efficacy and safety of dutasteride on prostate cancer risk reduction in Asian men: the results from the REDUCE study. Jpn. J. Clin. Oncol. 41, 417–423. doi:10.1093/jjco/hyq221
Azhati, B., Reheman, A., Dilixiati, D., and Rexiati, M. (2023). FTO-stabilized miR-139-5p targets ZNF217 to suppress prostate cancer cell malignancies by inactivating the PI3K/Akt/mTOR signal pathway. Archives Biochem. biophysics 741, 109604. doi:10.1016/j.abb.2023.109604
Bailey, A. S., Batista, P. J., Gold, R. S., Chen, Y. G., de Rooij, D. G., Chang, H. Y., et al. (2017). The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. eLife 6, e26116. doi:10.7554/eLife.26116
Bennett, L., Jaiswal, P. K., Harkless, R. V., Long, T. M., Gao, N., Vandenburg, B., et al. (2024). Glucocorticoid receptor (GR) activation is associated with increased cAMP/PKA signaling in castration-resistant prostate cancer. Mol. cancer Ther. 23, 552–563. doi:10.1158/1535-7163.MCT-22-0479
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G., and Rottman, F. M. (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA (New York, NY) 3, 1233–1247.
Bratt, O. (2002). Hereditary prostate cancer: clinical aspects. J. urology 168, 906–913. doi:10.1097/01.ju.0000024402.67529.ca
Bray, F., Laversanne, M., Sung, H., Ferlay, J., Siegel, R. L., Soerjomataram, I., et al. (2024). Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA a cancer J. Clin. 74, 229–263. doi:10.3322/caac.21834
Cai, J., Yang, F., Zhan, H., Situ, J., Li, W., Mao, Y., et al. (2019). RNA m(6)A methyltransferase METTL3 promotes the growth of prostate cancer by regulating Hedgehog pathway. OncoTargets Ther. 12, 9143–9152. doi:10.2147/OTT.S226796
Cai, M., Song, X. L., Li, X. A., Chen, M., Guo, J., Yang, D. H., et al. (2023). Current therapy and drug resistance in metastatic castration-resistant prostate cancer. Drug Resist. Updat. Rev. Comment. Antimicrob. anticancer Chemother. 68, 100962. doi:10.1016/j.drup.2023.100962
Cai, T., Nesi, G., Tinacci, G., Giubilei, G., Gavazzi, A., Mondaini, N., et al. (2011). Clinical importance of lymph node density in predicting outcome of prostate cancer patients. J. Surg. Res. 167, 267–272. doi:10.1016/j.jss.2009.05.004
Chen, B., Liu, C., Long, H., Bai, G., Zhu, Y., and Xu, H. (2023a). N(6)-methyladenosine-induced long non-coding RNA PVT1 regulates the miR-27b-3p/BLM axis to promote prostate cancer progression. Int. J. Oncol. 62, 16. doi:10.3892/ijo.2022.5464
Chen, L., Gao, Y., Xu, S., Yuan, J., Wang, M., Li, T., et al. (2023b). N6-methyladenosine reader YTHDF family in biological processes: structures, roles, and mechanisms. Front. Immunol. 14, 1162607. doi:10.3389/fimmu.2023.1162607
Chen, W. W., Qi, J. W., Hang, Y., Wu, J. X., Zhou, X. X., Chen, J. Z., et al. (2020). Simvastatin is beneficial to lung cancer progression by inducing METTL3-induced m6A modification on EZH2 mRNA. Eur. Rev. Med. Pharmacol. Sci. 24, 4263–4270. doi:10.26355/eurrev_202004_21006
Chen, Y., Pan, C., Wang, X., Xu, D., Ma, Y., Hu, J., et al. (2021a). Silencing of METTL3 effectively hinders invasion and metastasis of prostate cancer cells. Theranostics 11, 7640–7657. doi:10.7150/thno.61178
Chen, Y., Wu, R., Chen, W., Liu, Y., Liao, X., Zeng, B., et al. (2021b). Curcumin prevents obesity by targeting TRAF4-induced ubiquitylation in m(6) A-dependent manner. EMBO Rep. 22, e52146. doi:10.15252/embr.202052146
Cheng, Q., Butler, W., Zhou, Y., Zhang, H., Tang, L., Perkinson, K., et al. (2022). Pre-existing castration-resistant prostate cancer-like cells in primary prostate cancer promote resistance to hormonal therapy. Eur. Urol. 81, 446–455. doi:10.1016/j.eururo.2021.12.039
Cheng, Y., Li, L., Wei, X., Xu, F., Huang, X., Qi, F., et al. (2023). HNRNPC suppresses tumor immune microenvironment by activating Treg cells promoting the progression of prostate cancer. Cancer Sci. 114, 1830–1845. doi:10.1111/cas.15745
Cheng, Y., Xie, W., Pickering, B. F., Chu, K. L., Savino, A. M., Yang, X., et al. (2021). N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer cell 39, 958–972.e8. doi:10.1016/j.ccell.2021.04.017
Choe, J., Lin, S., Zhang, W., Liu, Q., Wang, L., Ramirez-Moya, J., et al. (2018). mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 561, 556–560. doi:10.1038/s41586-018-0538-8
Choi, J. B., Kim, J. H., Hong, S. H., Han, K. D., and Ha, U. S. (2018). Difference in prostate cancer incidence around sixty years: effects of age and metabolic diseases. Cancer Med. 7, 2736–2743. doi:10.1002/cam4.1462
Clancy, M. J., Shambaugh, M. E., Timpte, C. S., and Bokar, J. A. (2002). Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic acids Res. 30, 4509–4518. doi:10.1093/nar/gkf573
Cotter, K. A., Gallon, J., Uebersax, N., Rubin, P., Meyer, K. D., Piscuoglio, S., et al. (2021). Mapping of m(6)A and its regulatory targets in prostate cancer reveals a METTL3-low induction of therapy resistance. Mol. cancer Res.: MCR 19, 1398–1411.
Dai, C., Heemers, H., and Sharifi, N. (2017). Androgen signaling in prostate cancer. Cold Spring Harb. Perspect. Med. 7, a030452. doi:10.1101/cshperspect.a030452
Davies, A., Nouruzi, S., Ganguli, D., Namekawa, T., Thaper, D., Linder, S., et al. (2021). An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat. cell Biol. 23, 1023–1034. doi:10.1038/s41556-021-00743-5
de Bono, J. S., Logothetis, C. J., Molina, A., Fizazi, K., North, S., Chu, L., et al. (2011). Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 364, 1995–2005. doi:10.1056/NEJMoa1014618
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 71, 3971–3975. doi:10.1073/pnas.71.10.3971
Ding, L., Wang, R., Zheng, Q., Shen, D., Wang, H., Lu, Z., et al. (2022). circPDE5A regulates prostate cancer metastasis via controlling WTAP-dependent N6-methyladenisine methylation of EIF3C mRNA. J. Exp. and Clin. cancer Res. CR 41, 187. doi:10.1186/s13046-022-02391-5
Dong, F., Qin, X., Wang, B., Li, Q., Hu, J., Cheng, X., et al. (2021). ALKBH5 facilitates hypoxia-induced paraspeckle assembly and IL8 secretion to generate an immunosuppressive tumor microenvironment. Cancer Res. 81, 5876–5888. doi:10.1158/0008-5472.CAN-21-1456
Du, C., Lv, C., Feng, Y., and Yu, S. (2020). Activation of the KDM5A/miRNA-495/YTHDF2/m6A-MOB3B axis facilitates prostate cancer progression. J. Exp. and Clin. cancer Res. CR 39, 223. doi:10.1186/s13046-020-01735-3
Fang, Z., Mei, W., Qu, C., Lu, J., Shang, L., Cao, F., et al. (2022). Role of m6A writers, erasers and readers in cancer. Exp. Hematol. and Oncol. 11, 45. doi:10.1186/s40164-022-00298-7
Gerken, T., Girard, C. A., Tung, Y. C., Webby, C. J., Saudek, V., Hewitson, K. S., et al. (2007). The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Sci. (New York, NY) 318, 1469–1472. doi:10.1126/science.1151710
Geula, S., Moshitch-Moshkovitz, S., Dominissini, D., Mansour, A. A., Kol, N., Salmon-Divon, M., et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Sci. (New York, NY) 347, 1002–1006. doi:10.1126/science.1261417
Godtman, R. A., Kollberg, K. S., Pihl, C. G., Månsson, M., and Hugosson, J. (2022). The association between age, prostate cancer risk, and higher Gleason score in a long-term screening Program: results from the göteborg-1 prostate cancer screening trial. Eur. Urol. 82, 311–317. doi:10.1016/j.eururo.2022.01.018
Guan, X., Polesso, F., Wang, C., Sehrawat, A., Hawkins, R. M., Murray, S. E., et al. (2022). Androgen receptor activity in T cells limits checkpoint blockade efficacy. Nature 606, 791–796. doi:10.1038/s41586-022-04522-6
Hagemann, S., Misiak, D., Bell, J. L., Fuchs, T., Lederer, M. I., Bley, N., et al. (2023). IGF2BP1 induces neuroblastoma via a druggable feedforward loop with MYCN promoting 17q oncogene expression. Mol. cancer 22, 88. doi:10.1186/s12943-023-01792-0
Haigh, D. B., Woodcock, C. L., Lothion-Roy, J., Harris, A. E., Metzler, V. M., Persson, J. L., et al. (2022). The METTL3 RNA methyltransferase regulates transcriptional networks in prostate cancer. Cancers 14, 5148. doi:10.3390/cancers14205148
Han, Z., Yi, X., Li, J., Zhang, T., Liao, D., You, J., et al. (2023). RNA m(6)A modification in prostate cancer: a new weapon for its diagnosis and therapy. Biochimica biophysica acta Rev. cancer 1878, 188961. doi:10.1016/j.bbcan.2023.188961
Hartmann, A. M., Nayler, O., Schwaiger, F. W., Obermeier, A., and Stamm, S. (1999). The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59(fyn). Mol. Biol. cell 10, 3909–3926. doi:10.1091/mbc.10.11.3909
He, L., Li, H., Wu, A., Peng, Y., Shu, G., and Yin, G. (2019). Functions of N6-methyladenosine and its role in cancer. Mol. cancer 18, 176. doi:10.1186/s12943-019-1109-9
He, L., Li, J., Wang, X., Ying, Y., Xie, H., Yan, H., et al. (2018). The dual role of N6-methyladenosine modification of RNAs is involved in human cancers. J. Cell. Mol. Med. 22, 4630–4639. doi:10.1111/jcmm.13804
He, Y., Xu, W., Xiao, Y. T., Huang, H., Gu, D., and Ren, S. (2022). Targeting signaling pathways in prostate cancer: mechanisms and clinical trials. Signal Transduct. Target. Ther. 7, 198. doi:10.1038/s41392-022-01042-7
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127. doi:10.1038/cr.2017.99
Hu, Q., Yin, J., Zhao, S., Wang, Y., Shi, R., Yan, K., et al. (2024). ZFHX3 acts as a tumor suppressor in prostate cancer by targeting FTO-mediated m(6)A demethylation. Cell death Discov. 10, 284. doi:10.1038/s41420-024-02060-w
Huang, C., Xu, R., Zhu, X., and Jiang, H. (2023a). m6A-modified circABCC4 promotes stemness and metastasis of prostate cancer by recruiting IGF2BP2 to increase stability of CCAR1. Cancer gene Ther. 30, 1426–1440. doi:10.1038/s41417-023-00650-x
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. cell Biol. 20, 285–295. doi:10.1038/s41556-018-0045-z
Huang, Q., Mo, J., Liao, Z., Chen, X., and Zhang, B. (2022). The RNA m(6)A writer WTAP in diseases: structure, roles, and mechanisms. Cell death and Dis. 13, 852. doi:10.1038/s41419-022-05268-9
Huang, Y., Xia, W., Dong, Z., and Yang, C. G. (2023b). Chemical inhibitors targeting the oncogenic m(6)A modifying proteins. Accounts Chem. Res. 56, 3010–3022. doi:10.1021/acs.accounts.3c00451
Jain, D., Puno, M. R., Meydan, C., Lailler, N., Mason, C. E., Lima, C. D., et al. (2018). Ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. eLife 7, e30919. doi:10.7554/eLife.30919
Jamal, A., Hassan Dalhat, M., Jahan, S., Choudhry, H., and Imran Khan, M. (2023). BTYNB, an inhibitor of RNA binding protein IGF2BP1 reduces proliferation and induces differentiation of leukemic cancer cells. Saudi J. Biol. Sci. 30, 103569. doi:10.1016/j.sjbs.2023.103569
Jeon, H. Y., Pornour, M., Ryu, H., Khadka, S., Xu, R., Jang, J., et al. (2023). SMAD3 promotes expression and activity of the androgen receptor in prostate cancer. Nucleic acids Res. 51, 2655–2670. doi:10.1093/nar/gkad043
Jeong, I. G., Dajani, D., Verghese, M., Hwang, J., Cho, Y. M., Hong, J. H., et al. (2016). Differences in the aggressiveness of prostate cancer among Korean, Caucasian, and African American men: a retrospective cohort study of radical prostatectomy. Urol. Oncol. 34 (3), e9–e14. doi:10.1016/j.urolonc.2015.08.004
Jiang, X., Guo, S., Wang, S., Zhang, Y., Chen, H., Wang, Y., et al. (2022). EIF4A3-Induced circARHGAP29 promotes aerobic glycolysis in docetaxel-resistant prostate cancer through IGF2BP2/c-Myc/LDHA signaling. Cancer Res. 82, 831–845. doi:10.1158/0008-5472.CAN-21-2988
Jiang, X., Liu, B., Nie, Z., Duan, L., Xiong, Q., Jin, Z., et al. (2021). The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 6, 74. doi:10.1038/s41392-020-00450-x
Koh, C. W. Q., Goh, Y. T., and Goh, W. S. S. (2019). Atlas of quantitative single-base-resolution N(6)-methyl-adenine methylomes. Nat. Commun. 10, 5636. doi:10.1038/s41467-019-13561-z
Kong, Z., Lu, Y., Yang, Y., Chang, K., Lin, Y., Huang, Y., et al. (2023). m6A-Mediated biogenesis of circDDIT4 inhibits prostate cancer progression by sequestrating ELAVL1/HuR. Mol. cancer Res. MCR 21, 1342–1355. doi:10.1158/1541-7786.MCR-22-0271
Kwon, D. H., Borno, H. T., Cheng, H. H., Zhou, A. Y., and Small, E. J. (2020). Ethnic disparities among men with prostate cancer undergoing germline testing. Urol. Oncol. 38, 80.e81–80. doi:10.1016/j.urolonc.2019.09.010
Lan, Q., Liu, P. Y., Bell, J. L., Wang, J. Y., Hüttelmaier, S., Zhang, X. D., et al. (2021). The emerging roles of RNA m(6)A methylation and demethylation as critical regulators of tumorigenesis, drug sensitivity, and resistance. Cancer Res. 81, 3431–3440. doi:10.1158/0008-5472.CAN-20-4107
Lang, C., Yin, C., Lin, K., Li, Y., Yang, Q., Wu, Z., et al. (2021). m(6) A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clin. Transl. Med. 11, e426. doi:10.1002/ctm2.426
Li, G., Liu, J., Wang, Y., Liu, H., Fu, J., Zhao, Y., et al. (2023a). METTL3-mediated m6A modification of pri-miR-148a-3p affects prostate cancer progression by regulating TXNIP. Environ. Toxicol. 38, 2377–2390. doi:10.1002/tox.23874
Li, J., Xie, H., Ying, Y., Chen, H., Yan, H., He, L., et al. (2020). YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol. cancer 19, 152. doi:10.1186/s12943-020-01267-6
Li, P., Shi, Y., Gao, D., Xu, H., Zou, Y., Wang, Z., et al. (2022b). ELK1-mediated YTHDF1 drives prostate cancer progression by facilitating the translation of Polo-like kinase 1 in an m6A dependent manner. Int. J. Biol. Sci. 18, 6145–6162. doi:10.7150/ijbs.75063
Li, S., and Cao, L. (2022). Demethyltransferase FTO alpha-ketoglutarate dependent dioxygenase (FTO) regulates the proliferation, migration, invasion and tumor growth of prostate cancer by modulating the expression of melanocortin 4 receptor (MC4R). Bioengineered 13, 5598–5612. doi:10.1080/21655979.2021.2001936
Li, W., Chen, G., Feng, Z., Zhu, B., Zhou, L., Zhang, Y., et al. (2021). YTHDF1 promotes the proliferation, migration, and invasion of prostate cancer cells by regulating TRIM44. Genes and genomics. 43, 1413–1421. doi:10.1007/s13258-021-01175-z
Li, X., Liu, B., Wang, S., Li, J., and Ge, X. (2023c). MiR-141-3p promotes malignant progression in prostate cancer through AlkB homolog 5-mediated m(6)A modification of protein arginine methyltransferase 6. Chin. J. physiology 66, 43–51. doi:10.4103/cjop.CJOP-D-22-00071
Li, Y., Su, R., Deng, X., Chen, Y., and Chen, J. (2022a). FTO in cancer: functions, molecular mechanisms, and therapeutic implications. Trends cancer 8, 598–614. doi:10.1016/j.trecan.2022.02.010
Li, Y., Zhu, S., Chen, Y., Ma, Q., Kan, D., Yu, W., et al. (2023b). Post-transcriptional modification of m(6)A methylase METTL3 regulates ERK-induced androgen-deprived treatment resistance prostate cancer. Cell death and Dis. 14, 289. doi:10.1038/s41419-023-05773-5
Li, Z., Weng, H., Su, R., Weng, X., Zuo, Z., Li, C., et al. (2017). FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer cell 31, 127–141. doi:10.1016/j.ccell.2016.11.017
Lin, S., Choe, J., Du, P., Triboulet, R., and Gregory, R. I. (2016). The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. cell 62, 335–345. doi:10.1016/j.molcel.2016.03.021
Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., and Jaffrey, S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. methods 12, 767–772. doi:10.1038/nmeth.3453
Liu, B., Jiang, H. Y., Yuan, T., Luo, J., Zhou, W. D., Jiang, Q. Q., et al. (2021a). Enzalutamide-Induced upregulation of PCAT6 promotes prostate cancer neuroendocrine differentiation by regulating miR-326/HNRNPA2B1 Axis. Front. Oncol. 11, 650054. doi:10.3389/fonc.2021.650054
Liu, B., Li, L., Yang, G., Geng, C., Luo, Y., Wu, W., et al. (2019). PARP inhibition suppresses GR-MYCN-CDK5-RB1-E2F1 signaling and neuroendocrine differentiation in castration-resistant prostate cancer. Clin. cancer Res. official J. Am. Assoc. Cancer Res. 25, 6839–6851. doi:10.1158/1078-0432.CCR-19-0317
Liu, J., Yuan, J. F., and Wang, Y. Z. (2022). METTL3-stabilized lncRNA SNHG7 accelerates glycolysis in prostate cancer via SRSF1/c-Myc axis. Exp. cell Res. 416, 113149. doi:10.1016/j.yexcr.2022.113149
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi:10.1038/nchembio.1432
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564. doi:10.1038/nature14234
Liu, N., Zhou, K. I., Parisien, M., Dai, Q., Diatchenko, L., and Pan, T. (2017). N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic acids Res. 45, 6051–6063. doi:10.1093/nar/gkx141
Liu, Y., Liang, G., Xu, H., Dong, W., Dong, Z., Qiu, Z., et al. (2021b). Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance. Cell metab. 33, 1221–1233.e11. doi:10.1016/j.cmet.2021.04.001
Lothion-Roy, J., Haigh, D. B., Harris, A. E., Metzler, V. M., Alsaleem, M., Toss, M. S., et al. (2022). Clinical and molecular significance of the RNA m(6)A methyltransferase complex in prostate cancer. Front. Genet. 13, 1096071. doi:10.3389/fgene.2022.1096071
Lu, J., Chen, J., Lin, Z., Liu, Q., Zhong, C., Cai, Z., et al. (2023). A prognostic signature consisting of N6-methyladenosine modified mRNAs demonstrates clinical potential in prediction of biochemical recurrence and guidance on precision therapy in prostate cancer. Transl. Oncol. 33, 101670. doi:10.1016/j.tranon.2023.101670
Luxton, H. J., Simpson, B. S., Mills, I. G., Brindle, N. R., Ahmed, Z., Stavrinides, V., et al. (2019). The oncogene metadherin interacts with the known splicing proteins YTHDC1, Sam68 and T-STAR and plays a novel role in alternative mRNA splicing. Cancers. 11, 1233. doi:10.3390/cancers11091233
Ma, H., Wang, X., Cai, J., Dai, Q., Natchiar, S. K., Lv, R., et al. (2019). N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 15, 88–94. doi:10.1038/s41589-018-0184-3
Ma, X. X., Cao, Z. G., and Zhao, S. L. (2020). m6A methyltransferase METTL3 promotes the progression of prostate cancer via m6A-modified LEF1. Eur. Rev. Med. Pharmacol. Sci. 24, 3565–3571. doi:10.26355/eurrev_202004_20817
Mahapatra, L., Andruska, N., Mao, C., Le, J., and Shapiro, D. J. (2017). A novel IMP1 inhibitor, BTYNB, targets c-myc and inhibits melanoma and ovarian cancer cell proliferation. Transl. Oncol. 10, 818–827. doi:10.1016/j.tranon.2017.07.008
Mao, Y., Li, W., Weng, Y., Hua, B., Gu, X., Lu, C., et al. (2022). METTL3-Mediated m(6)A modification of lncRNA MALAT1 facilitates prostate cancer growth by activation of PI3K/AKT signaling. Cell Transplant. 31, 9636897221122997. doi:10.1177/09636897221122997
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m(6)A(m) in the 5' cap controls mRNA stability. Nature 541, 371–375. doi:10.1038/nature21022
Mauer, J., Sindelar, M., Despic, V., Guez, T., Hawley, B. R., Vasseur, J. J., et al. (2019). FTO controls reversible m(6)Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 15, 340–347. doi:10.1038/s41589-019-0231-8
Micaelli, M., Dalle Vedove, A., Cerofolini, L., Vigna, J., Sighel, D., Zaccara, S., et al. (2022). Small-molecule ebselen binds to YTHDF proteins interfering with the recognition of N (6)-methyladenosine-modified RNAs. ACS Pharmacol. and Transl. Sci. 5, 872–891. doi:10.1021/acsptsci.2c00008
Mottet, N., Bellmunt, J., Bolla, M., Briers, E., Cumberbatch, M. G., De Santis, M., et al. (2017). EAU-ESTRO-SIOG guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent. Eur. Urol. 71, 618–629. doi:10.1016/j.eururo.2016.08.003
Müller, S., Bley, N., Busch, B., Glaß, M., Lederer, M., Misiak, C., et al. (2020). The oncofetal RNA-binding protein IGF2BP1 is a druggable, post-transcriptional super-enhancer of E2F-driven gene expression in cancer. Nucleic acids Res. 48, 8576–8590. doi:10.1093/nar/gkaa653
Murakami, S., and Jaffrey, S. R. (2022). Hidden codes in mRNA: control of gene expression by m(6)A. Mol. cell 82, 2236–2251. doi:10.1016/j.molcel.2022.05.029
Nie, Q., Wu, X., Huang, Y., Guo, T., Kuang, J., and Du, C. (2023). RNA N6-methyladenosine-modified-binding protein YTHDF1 promotes prostate cancer progression by regulating androgen function-related gene TRIM68. Eur. J. Med. Res.
Patil, D. P., Pickering, B. F., and Jaffrey, S. R. (2018). Reading m(6)A in the transcriptome: m(6)a-binding proteins. Trends cell Biol. 28, 113–127. doi:10.1016/j.tcb.2017.10.001
Pendleton, K. E., Chen, B., Liu, K., Hunter, O. V., Xie, Y., Tu, B. P., et al. (2017). The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169, 824–835. doi:10.1016/j.cell.2017.05.003
Peng, S., Yi, Z., and Liu, M. (2017). Ailanthone: a new potential drug for castration-resistant prostate cancer. Chin. J. cancer 36, 25. doi:10.1186/s40880-017-0194-7
Powles, T., Yuen, K. C., Gillessen, S., Kadel, E. E., Rathkopf, D., Matsubara, N., et al. (2022). Atezolizumab with enzalutamide versus enzalutamide alone in metastatic castration-resistant prostate cancer: a randomized phase 3 trial. Nat. Med. 28, 144–153. doi:10.1038/s41591-021-01600-6
Qi, F., Shen, W., Wei, X., Cheng, Y., Xu, F., Zheng, Y., et al. (2023). CSNK1D-mediated phosphorylation of HNRNPA2B1 induces miR-25-3p/miR-93-5p maturation to promote prostate cancer cell proliferation and migration through m(6)A-dependent manner. Cell. Mol. life Sci. CMLS 80, 156. doi:10.1007/s00018-023-04798-5
Quan, Y., Ping, H., Wang, M., and Zhang, X. (2023). RNA-sequencing analysis indicates that N-cadherin promotes prostate cancer progression by the epigenetic modification of key genes. DNA cell biol. 42, 563–577.
Raj, N., Wang, M., Seoane, J. A., Zhao, R. L., Kaiser, A. M., Moonie, N. A., et al. (2022). The Mettl3 epitranscriptomic writer amplifies p53 stress responses. Mol. cell 82, 2370–2384.e10. doi:10.1016/j.molcel.2022.04.010
Rebbeck, T. R. (2017). Prostate cancer genetics: variation by race, ethnicity, and geography. Seminars Radiat. Oncol. 27, 3–10. doi:10.1016/j.semradonc.2016.08.002
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife 6, e31311. doi:10.7554/eLife.31311
Ruszkowska, A., Ruszkowski, M., Dauter, Z., and Brown, J. A. (2018). Structural insights into the RNA methyltransferase domain of METTL16. Sci. Rep. 8, 5311. doi:10.1038/s41598-018-23608-8
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27, 315–328. doi:10.1038/cr.2017.15
Shi, S. J., Han, D. H., Zhang, J. L., Li, Y., Yang, A. G., and Zhang, R. (2023). VIM-AS1 promotes proliferation and drives enzalutamide resistance in prostate cancer via IGF2BP2-mediated HMGCS1 mRNA stabilization. Int. J. Oncol. 62, 34. doi:10.3892/ijo.2023.5482
Shigeta, K., Kosaka, T., Hongo, H., Yanai, Y., Matsumoto, K., Morita, S., et al. (2019). Castration-resistant prostate cancer patients who had poor response on first androgen deprivation therapy would obtain certain clinical benefit from early docetaxel administration. Int. J. Clin. Oncol. 24, 546–553. doi:10.1007/s10147-018-01388-5
Shima, H., Matsumoto, M., Ishigami, Y., Ebina, M., Muto, A., Sato, Y., et al. (2017). S-adenosylmethionine synthesis is regulated by selective N(6)-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 21, 3354–3363. doi:10.1016/j.celrep.2017.11.092
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics. CA a cancer J. Clin. 72, 7–30. doi:10.3322/caac.21332
Singh, A., Singh, V., Wallis, N., Abis, G., Oberman, F., Wood, T., et al. (2024). Development of a specific and potent IGF2BP1 inhibitor: a promising therapeutic agent for IGF2BP1-expressing cancers. Eur. J. Med. Chem. 263, 115940. doi:10.1016/j.ejmech.2023.115940
Śledź, P., and Jinek, M. (2016). Structural insights into the molecular mechanism of the m(6)A writer complex. eLife. 5, e18434. doi:10.7554/eLife.18434
Somasekharan, S. P., Saxena, N., Zhang, F., Beraldi, E., Huang, J. N., Gentle, C., et al. (2022). Regulation of AR mRNA translation in response to acute AR pathway inhibition. Nucleic acids Res. 50, 1069–1091. doi:10.1093/nar/gkab1247
Sommer, S., Lavi, U., and Darnell, J. E. (1978). The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J. Mol. Biol. 124, 487–499. doi:10.1016/0022-2836(78)90183-3
Song, J., You, G., Yin, X., Zhu, G., Wang, W., Yu, Y., et al. (2023). Overexpression of YTHDC2 contributes to the progression of prostate cancer and predicts poor outcomes in patients with prostate cancer. J. Biochem. Mol. Toxicol. 37, e23308. doi:10.1002/jbt.23308
Spitale, R. C., Flynn, R. A., Zhang, Q. C., Crisalli, P., Lee, B., Jung, J. W., et al. (2015). Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519, 486–490. doi:10.1038/nature14263
Su, R., Dong, L., Li, Y., Gao, M., Han, L., Wunderlich, M., et al. (2020). Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer cell 38, 79–96. doi:10.1016/j.ccell.2020.04.017
Sun, M., Shen, Y., Jia, G., Deng, Z., Shi, F., Jing, Y., et al. (2023). Activation of the HNRNPA2B1/miR-93-5p/FRMD6 axis facilitates prostate cancer progression in an m6A-dependent manner. J. Cancer 14, 1242–1256. doi:10.7150/jca.83863
Tan, M. H., Li, J., Xu, H. E., Melcher, K., and Yong, E. L. (2015). Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol. Sin. 36, 3–23. doi:10.1038/aps.2014.18
Tang, Q., Cheng, B., Dai, R., and Wang, R. (2021). The role of androgen receptor in cross talk between stromal cells and prostate cancer epithelial cells. Front. cell Dev. Biol. 9, 729498. doi:10.3389/fcell.2021.729498
Tao, L., Mu, X., Chen, H., Jin, D., Zhang, R., Zhao, Y., et al. (2021). FTO modifies the m6A level of MALAT and promotes bladder cancer progression. Clin. Transl. Med. 11, e310. doi:10.1002/ctm2.310
Thalgott, M., Kron, M., Brath, J. M., Ankerst, D. P., Thompson, I. M., Gschwend, J. E., et al. (2018). Men with family history of prostate cancer have a higher risk of disease recurrence after radical prostatectomy. World J. urology 36, 177–185. doi:10.1007/s00345-017-2122-5
Thalhammer, A., Bencokova, Z., Poole, R., Loenarz, C., Adam, J., O'Flaherty, L., et al. (2011). Human AlkB homologue 5 is a nuclear 2-oxoglutarate dependent oxygenase and a direct target of hypoxia-inducible factor 1α (HIF-1α). PloS one 6, e16210. doi:10.1371/journal.pone.0016210
van Tran, N., Ernst, F. G. M., Hawley, B. R., Zorbas, C., Ulryck, N., Hackert, P., et al. (2019). The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic acids Res. 47, 7719–7733. doi:10.1093/nar/gkz619
Wan, H., Feng, Y., Wu, J., Zhu, L., and Mi, Y. (2022). Functions and mechanisms of N6-methyladenosine in prostate cancer (Review). Mol. Med. Rep. 26, 280. doi:10.3892/mmr.2022.12796
Wang, H., Li, N., Liu, Q., Guo, J., Pan, Q., Cheng, B., et al. (2023a). Antiandrogen treatment induces stromal cell reprogramming to promote castration resistance in prostate cancer. Cancer cell 41, 1345–1362.e9. doi:10.1016/j.ccell.2023.05.016
Wang, H., Liu, J., Zhu, X., Yang, B., He, Z., and Yao, X. (2023b). AZGP1P2/UBA1/RBM15 cascade mediates the fate determinations of prostate cancer stem cells and promotes therapeutic effect of docetaxel in castration-resistant prostate cancer via TPM1 m6A modification. Res. Wash. DC 6, 0252. doi:10.34133/research.0252
Wang, J. J., Chen, D. X., Zhang, Y., Xu, X., Cai, Y., Wei, W. Q., et al. (2023c). Elevated expression of the RNA-binding protein IGF2BP1 enhances the mRNA stability of INHBA to promote the invasion and migration of esophageal squamous cancer cells. Exp. Hematol. and Oncol. 12, 75. doi:10.1186/s40164-023-00429-8
Wang, L., Lyu, C., Stadlbauer, B., Buchner, A., Nößner, E., and Pohla, H. (2024). Berbamine targets cancer stem cells and reverses cabazitaxel resistance via inhibiting IGF2BP1 and p-STAT3 in prostate cancer. Prostate 84, 131–147. doi:10.1002/pros.24632
Wang, P., Doxtader, K. A., and Nam, Y. (2016a). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. cell 63, 306–317. doi:10.1016/j.molcel.2016.05.041
Wang, R., Sun, Y., Li, L., Niu, Y., Lin, W., Lin, C., et al. (2017). Preclinical study using Malat1 small interfering RNA or androgen receptor splicing variant 7 degradation enhancer ASC-j9® to suppress enzalutamide-resistant prostate cancer progression. Eur. Urol. 72, 835–844. doi:10.1016/j.eururo.2017.04.005
Wang, S., Xu, G., Chao, F., Zhang, C., Han, D., and Chen, G. (2021). HNRNPC promotes proliferation, metastasis and predicts prognosis in prostate cancer. Cancer Manag. Res. 13, 7263–7276. doi:10.2147/CMAR.S330713
Wang, T., Kong, S., Tao, M., and Ju, S. (2020). The potential role of RNA N6-methyladenosine in Cancer progression. Mol. cancer 19, 88. doi:10.1186/s12943-020-01204-7
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016b). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578. doi:10.1038/nature18298
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014b). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi:10.1038/nature12730
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399. doi:10.1016/j.cell.2015.05.014
Wang, Y., Chen, J., Gao, W. Q., and Yang, R. (2022a). METTL14 promotes prostate tumorigenesis by inhibiting THBS1 via an m6A-YTHDF2-dependent mechanism. Cell death Discov. 8, 143. doi:10.1038/s41420-022-00939-0
Wang, Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., and Zhao, J. C. (2014a). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. cell Biol. 16, 191–198. doi:10.1038/ncb2902
Wang, Z., Sun, H., Zhu, H., Gu, D., Chen, X., Pan, Y., et al. (2022b). Demethylase FTO inhibits the development of prostate cancer by upregulating EGR2 expression in an m6A manner. Turkish J. Biol. = Turk biyoloji dergisi 46, 426–438. doi:10.55730/1300-0152.2629
Warda, A. S., Kretschmer, J., Hackert, P., Lenz, C., Urlaub, H., Höbartner, C., et al. (2017). Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18, 2004–2014. doi:10.15252/embr.201744940
Wei, C. M., Gershowitz, A., and Moss, B. (1976). 5'-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry 15, 397–401. doi:10.1021/bi00647a024
Wei, J., Liu, F., Lu, Z., Fei, Q., Ai, Y., He, P. C., et al. (2018). Differential m(6)A, m(6)A(m), and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. cell 71, 973–985. doi:10.1016/j.molcel.2018.08.011
Wei, X., Huo, Y., Pi, J., Gao, Y., Rao, S., He, M., et al. (2022). METTL3 preferentially enhances non-m(6)A translation of epigenetic factors and promotes tumourigenesis. Nat. cell Biol. 24, 1278–1290. doi:10.1038/s41556-022-00968-y
Wen, S., Wei, Y., Zen, C., Xiong, W., Niu, Y., and Zhao, Y. (2020). Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol. cancer 19, 171. doi:10.1186/s12943-020-01293-4
Weng, H., Huang, F., Yu, Z., Chen, Z., Prince, E., Kang, Y., et al. (2022). The m(6)A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer cell 40, 1566–1582.e10. doi:10.1016/j.ccell.2022.10.004
Wenzel, M., Nocera, L., Collà Ruvolo, C., Würnschimmel, C., Tian, Z., Shariat, S. F., et al. (2022). Overall survival and adverse events after treatment with darolutamide vs. apalutamide vs. enzalutamide for high-risk non-metastatic castration-resistant prostate cancer: a systematic review and network meta-analysis. Prostate cancer prostatic Dis. 25, 139–148. doi:10.1038/s41391-021-00395-4
Widagdo, J., Anggono, V., and Wong, J. J. (2022). The multifaceted effects of YTHDC1-mediated nuclear m(6)A recognition. Trends Genet. TIG 38, 325–332. doi:10.1016/j.tig.2021.11.005
Wiener, D., and Schwartz, S. (2021). The epitranscriptome beyond m(6)A. Nat. Rev. Genet. 22, 119–131. doi:10.1038/s41576-020-00295-8
Wojtas, M. N., Pandey, R. R., Mendel, M., Homolka, D., Sachidanandam, R., and Pillai, R. S. (2017). Regulation of m(6)A transcripts by the 3'→5' RNA helicase YTHDC2 is essential for a successful meiotic Program in the mammalian germline. Mol. cell 68, 374–387. doi:10.1016/j.molcel.2017.09.021
Wu, B., Su, S., Patil, D. P., Liu, H., Gan, J., Jaffrey, S. R., et al. (2018). Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 9, 420. doi:10.1038/s41467-017-02770-z
Wu, Q., Xie, X., Huang, Y., Meng, S., Li, Y., Wang, H., et al. (2021). N6-methyladenosine RNA methylation regulators contribute to the progression of prostate cancer. J. Cancer 12, 682–692. doi:10.7150/jca.46379
Wu, R., Yao, Y., Jiang, Q., Cai, M., Liu, Q., Wang, Y., et al. (2005)2018). Epigallocatechin gallate targets FTO and inhibits adipogenesis in an mRNA m(6)A-YTHDF2-dependent manner. Int. J. Obes. 42, 1378–1388. doi:10.1038/s41366-018-0082-5
Wu, T., Shao, Y., Li, X., Wu, T., Yu, L., Liang, J., et al. (2023). NR3C1/Glucocorticoid receptor activation promotes pancreatic β-cell autophagy overload in response to glucolipotoxicity. Autophagy 19, 2538–2557. doi:10.1080/15548627.2023.2200625
Xia, L., Han, Q., Duan, X., Zhu, Y., Pan, J., Dong, B., et al. (2022). m(6)A-induced repression of SIAH1 facilitates alternative splicing of androgen receptor variant 7 by regulating CPSF1. Mol. Ther. Nucleic acids 28, 219–230. doi:10.1016/j.omtn.2022.03.008
Xiao, P., Meng, Q., Liu, Q., Lang, Q., Yin, Z., Li, G., et al. (2023). IGF2BP1-mediated N6-methyladenosine modification promotes intrahepatic cholangiocarcinoma progression. Cancer Lett. 557, 216075. doi:10.1016/j.canlet.2023.216075
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. cell 61, 507–519. doi:10.1016/j.molcel.2016.01.012
Xu, P., and Ge, R. (2022). Roles and drug development of METTL3 (methyltransferase-like 3) in anti-tumor therapy. Eur. J. Med. Chem. 230, 114118. doi:10.1016/j.ejmech.2022.114118
Xu, Y., Ye, S., Zhang, N., Zheng, S., Liu, H., Zhou, K., et al. (2020). The FTO/miR-181b-3p/ARL5B signaling pathway regulates cell migration and invasion in breast cancer. Cancer Commun. Lond. Engl. 40, 484–500. doi:10.1002/cac2.12075
Yang, Q., and Al-Hendy, A. (2023). The functional role and regulatory mechanism of FTO m(6)A RNA demethylase in human uterine leiomyosarcoma. Int. J. Mol. Sci. 24, 7957. doi:10.3390/ijms24097957
Yankova, E., Blackaby, W., Albertella, M., Rak, J., De Braekeleer, E., Tsagkogeorga, G., et al. (2021). Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593, 597–601. doi:10.1038/s41586-021-03536-w
Yu, Y. Z., Lv, D. J., Wang, C., Song, X. L., Xie, T., Wang, T., et al. (2022). Hsa_circ_0003258 promotes prostate cancer metastasis by complexing with IGF2BP3 and sponging miR-653-5p. Mol. cancer 21, 12. doi:10.1186/s12943-021-01480-x
Yuan, S., He, S. H., Li, L. Y., Xi, S., Weng, H., Zhang, J. H., et al. (2023). A potassium-chloride co-transporter promotes tumor progression and castration resistance of prostate cancer through m(6)A reader YTHDC1. Cell death and Dis. 14, 7. doi:10.1038/s41419-022-05544-8
Yuan, Y., Du, Y., Wang, L., and Liu, X. (2020). The M6A methyltransferase METTL3 promotes the development and progression of prostate carcinoma via mediating MYC methylation. J. Cancer 11, 3588–3595.
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. cell Biol. 20, 608–624. doi:10.1038/s41580-019-0168-5
Zhang, D., Wornow, S., Peehl, D. M., Rankin, E. B., and Brooks, J. D. (2022). The controversial role and therapeutic development of the m6A demethylase FTO in renal cell carcinoma. Transl. Oncol. 25, 101518. doi:10.1016/j.tranon.2022.101518
Zhang, J., Wei, J., Sun, R., Sheng, H., Yin, K., Pan, Y., et al. (2023b). A lncRNA from the FTO locus acts as a suppressor of the m(6)A writer complex and p53 tumor suppression signaling. Mol. cell 83, 2692–2708.e7. doi:10.1016/j.molcel.2023.06.024
Zhang, S., Lv, C., Niu, Y., Li, C., Li, X., Shang, Y., et al. (2023a). RBM3 suppresses stemness remodeling of prostate cancer in bone microenvironment by modulating N6-methyladenosine on CTNNB1 mRNA. Cell death and Dis. 14, 91. doi:10.1038/s41419-023-05627-0
Zhang, Z., Zhou, C., Li, X., Barnes, S. D., Deng, S., Hoover, E., et al. (2020). Loss of CHD1 promotes heterogeneous mechanisms of resistance to AR-targeted therapy via chromatin dysregulation. Cancer cell 37, 584–598. doi:10.1016/j.ccell.2020.03.001
Zhao, P., Han, P., Ma, Y., Tian, P., and Li, J. (2024c). Circ_0082878 contributes to prostate cancer progression via the miR-455-3p/WTAP axis. Environ. Toxicol. 39, 979–990.
Zhao, X., Lv, S., Li, N., Zou, Q., Sun, L., and Song, T. (2024b). YTHDF2 protein stabilization by the deubiquitinase OTUB1 promotes prostate cancer cell proliferation via PRSS8 mRNA degradation. J. Biol. Chem. 300, 107152. doi:10.1016/j.jbc.2024.107152
Zhao, Y., Hu, X., Yu, H., Sun, H., Zhang, L., and Shao, C. (2024a). The FTO mediated N6-methyladenosine modification of DDIT4 regulation with tumorigenesis and metastasis in prostate cancer. Res. Wash. DC 7, 0313. doi:10.34133/research.0313
Zheng, Z., Li, J., Liu, Y., Shi, Z., Xuan, Z., Yang, K., et al. (2022). The crucial role of AR-V7 in enzalutamide-resistance of castration-resistant prostate cancer. Cancers 14, 4877. doi:10.3390/cancers14194877
Zhong, C., Long, Z., Yang, T., Wang, S., Zhong, W., Hu, F., et al. (2023). M6A-modified circRBM33 promotes prostate cancer progression via PDHA1-mediated mitochondrial respiration regulation and presents a potential target for ARSI therapy. Int. J. Biol. Sci. 19, 1543–1563. doi:10.7150/ijbs.77133
Zhong, S., Li, H., Bodi, Z., Button, J., Vespa, L., Herzog, M., et al. (2008). MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant cell 20, 1278–1288. doi:10.1105/tpc.108.058883
Zhou, Y., Li, T., Jia, M., Dai, R., and Wang, R. (2023). The molecular biology of prostate cancer stem cells: from the past to the future. Int. J. Mol. Sci. 24, 7482. doi:10.3390/ijms24087482
Zhu, J., Cheng, X., Naumovski, N., Hu, L., and Wang, K. (2023b). Epigenetic regulation by quercetin: a comprehensive review focused on its biological mechanisms. Crit. Rev. food Sci. Nutr., 1–20. doi:10.1080/10408398.2023.2278760
Zhu, K., Li, Y., and Xu, Y. (2021). The FTO m(6)A demethylase inhibits the invasion and migration of prostate cancer cells by regulating total m(6)A levels. Life Sci. 271, 119180. doi:10.1016/j.lfs.2021.119180
Zhu, W., Zhao, R., Guan, X., and Wang, X. (2023a). The emerging roles and mechanism of N6-methyladenosine (m(6)A) modifications in urologic tumours progression. Front. Pharmacol. 14, 1192495. doi:10.3389/fphar.2023.1192495
Zou, L., Chen, W., Zhou, X., Yang, T., Luo, J., Long, Z., et al. (2022). N6-methyladenosine demethylase FTO suppressed prostate cancer progression by maintaining CLIC4 mRNA stability. Cell death Discov. 8, 184. doi:10.1038/s41420-022-01003-7
Glossary
Keywords: PCa, N6-Methyladenosine, androgen deprivation therapy (ADT), androgen recepter, signaling pathway
Citation: Cao Y, Jia M, Duan C, Yang Z, Cheng B and Wang R (2024) The m6A regulators in prostate cancer: molecular basis and clinical perspective. Front. Pharmacol. 15:1448872. doi: 10.3389/fphar.2024.1448872
Received: 14 June 2024; Accepted: 14 August 2024;
Published: 29 August 2024.
Edited by:
Lei Yin, Shanghai Jiaotong University School of Medicine, ChinaCopyright © 2024 Cao, Jia, Duan, Yang, Cheng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ronghao Wang, Z21hY2tAMTYzLmNvbQ==; Bo Cheng, Y2IxOTQ5QHN3bXUuZWR1LmNu
 Yu Cao
Yu Cao Man Jia1
Man Jia1 Zhihui Yang
Zhihui Yang Ronghao Wang
Ronghao Wang