- 1Department of Pharmacy, Xinhua Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
- 2School of Traditional Chinese Medicine, Jilin Agriculture Science and Technology University, Jilin, China
Objectives: To explore the risk factors and clinical outcomes of carbapenem-resistant Klebsiella pneumoniae (CRKP) infection and establish nomograms to predict the probability of CRKP infection and mortality in adult patients.
Methods: Patients infected with KP from August 2019 to April 2021 in a tertiary hospital in Shanghai were enrolled. Risk factors associated with CRKP and 30-day mortality were identified using multivariate logistic regression analysis and Cox regression analysis.
Results: Overall, 467 patients with KP infection were enrolled, wherein 210 (45.0%) patients were infected with CRKP and 257 (55.0%) patients with carbapenem-susceptible K. pneumoniae (CSKP). Five factors, namely Charlson’s Comorbidity Index (CCI) ≥ 3, the use of central venous catheterization, prior hospitalization during the 3 months before infection, and previous exposure to carbapenems and broad-spectrum β-lactams, were found to be independently associated with CRKP infection. Based on these parameters, the nomogram showed a better performance as indicated by C-index of 0.94 (95% confidence interval [CI]: 0.92–0.96) and well-fitted calibration curves. CRKP was independently associated with 30-day mortality. Multivariate Cox regression analysis revealed that age ≥65 years, higher CCI scores, higher Sequential Organ Failure Assessment scores, the presence of respiratory failure, albumin levels ≤30 g/L, and non-appropriate treatments in 3 days, were associated with 30-day mortality.
Conclusion: The predictive nomogram established in this study can facilitate the clinicians to make better clinical decisions when treating patients with KP infection.
Introduction
Klebsiella pneumoniae (KP) is emerging as one of the most common Gram-negative pathogens causing a significant amount of mortality and morbidity in hospitals, with a wide variety of infections (Bengoechea and Sa Pessoa, 2019; Rodríguez-Medina et al., 2019). In clinical practice, carbapenems, broad-spectrum β-lactams, fluoroquinolones and aminoglycosides are commonly used antibiotics to treat severe KP infections. However, with the increasing prevalence of carbapenem-resistant K. pneumoniae (CRKP), treatment options have become severely limited, leading to the use of alternative agents like tigecycline, polymyxin B, and ceftazidime-avibactam. During the last 10-year period in China, the cases of CRKP have increased from 14.1% in 2013 to 26.0% in 2023 (http://www.chinets.com/Data/GermYear). Due to high mortality rates and the limited antibiotic therapy options, CRKP infections have emerged as an important problem to public health (Durante-Mangoni et al., 2019; Tilahun et al., 2021). Recent studies demonstrated the association of CRKP with prolonged hospital stays, increased medical costs, and higher mortality for patients (Del Puente et al., 2019).
Since CRKP infection can lead to fatal clinical outcomes, early and accurate evaluation is essential in managing its treatment. Several studies so far have evaluated the risk factors contributing to the acquisition of CRKP and mortality (Li et al., 2020; Xiao et al., 2020; Wu et al., 2022) by simply analyzing the associated factors; however, further predictive models are warranted to identify high-risk patients for clinical applications. Moreover, metagenomic next-generation sequencing (mNGS), a powerful method for pathogen detection, is increasingly being used. This method showed higher sensitivity and specificity, which could markedly improve the rate of early diagnosis of KP infection (Forbes et al., 2017; Leguia et al., 2020). However, mNGS cannot determine the antibiotic resistance of causative pathogens accurately relative to that by the conventional culture methods (Liu et al., 2022), which emphasizes the need for personalized prediction models.
Recently, the nomogram has been widely used to visualize the predictive model (Park, 2018). In this retrospective study, we conducted an extensive analysis of the clinical data pertaining to adult patients afflicted with KP infection. Utilizing this data, we successfully formulated a practical and operable nomogram scoring system. This system is designed to assist clinicians in accurately predicting the likelihood of CRKP infection, as well as the associated prognosis, thereby contributing to more informed and targeted therapeutic interventions.
Materials and methods
Study design
The present retrospective study was carried out in a 2600-bed tertiary-care hospital, conducted from August 2019 to October 2021. Data from August 2019 to March 2021 were used for modeling, while data from April 2021 to October 2021 were used for validation. Patients who were 18 years of age and older, with proven KP infection (defined as the presence of a positive culture, associated with clinical signs of systemic inflammatory response syndrome) were enrolled. Individuals exhibiting KP colonization (characterized by positive KP cultures, yet devoid of any clinical or laboratory evidence of infection) or those engaged in an end-of-life care pathway were excluded from the study.
Definitions
The relevant demographic and clinical data on the day of KP detection were extracted and reviewed from patients’ charts and electronic medical records and documented in the data collection form.
Comorbid conditions were quantified using the Charlson’s Comorbidity Index (CCI), a widely used tool that predicts mortality risk by assigning weighted scores to comorbidities, with higher scores indicating worse outcomes (Charlson et al., 1994). The use of immunosuppressant was defined as receipt of corticosteroids at a dosage equivalent to or higher than 10 mg of prednisolone daily for more than 2 weeks, or antineoplastic chemotherapy within 4 weeks prior to the onset of KP infection. Respiratory failure was defined as a partial pressure of arterial oxygen/fraction of inspired oxygen PaO2/FiO2 ratio <200 mmHg, and/or the need for mechanical ventilation (either non-invasive positive pressure ventilation or invasive mechanical ventilation). Antimicrobial drug exposure was defined as the use of antibiotics for >72 h within the 30-day prior to KP infection. Appropriate therapy in 3 days was defined as treatment that was initiated before or within 72 h, with at least one antimicrobial agent that has been proven in vitro activity against the KP strains.
Antimicrobial susceptibility testing
KP isolates were identified using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF), and antimicrobial minimum inhibitory concentrations (MICs) were determined using the Vitek 2 system (bioMerieux, France) and the AST-GN card, as described previously (Chen et al., 2021). Carbapenem resistance was defined as resistant to any carbapenem antimicrobial (a MIC of ≥2 μg/mL for ertapenem or a MIC of ≥4 μg/mL for imipenem/meropenem) in accordance with the CLSI criteria (CALS Institute, 2021).
Statistical analysis
Statistical analyses were performed with R version 4.1.0 (R Foundation for Statistical Computing, Vienna, Austria). More than 10% of the missing variables were excluded from the analyses. The remaining missing values were interpolated via multiple interpolations (MICE package). Continuous variables were presented as mean ± standard deviation or medians (interquartile range [IQR]), as appropriate, and categorical variables were reported as numbers and percentages. All categorical variables were compared by using the Fisher’s exact test or Chi-square test, and continuous variables were compared using the Student’s t-test or the Mann-Whitney U test, if appropriate. All statistical tests were performed two-tailed, and a P-value <0.05 was considered to indicate statistical significance.
Univariate and multivariate analyses were performed to identify independent risk factors, and the LASSO regression was applied to reduce the data dimensions (glmnet package). Odds ratio (OR) or hazard ratio (HR) with a 95% confidence interval (CI) was employed for measuring the strength of the association. The nomogram for predicting the probability of CRKP infection and prognosis were established based on the logistic regression and Cox regression analyses (rms package), respectively. The predictive performance was evaluated in terms of its discriminative ability, calibration, and clinical usefulness (Steyerberg and Vergouwe, 2014). The bootstrap method was used for repeated sampling 1,000 times to internally evaluate the consistency of model prediction, and the discriminative ability and predictive accuracy of the model were assessed using the area under (AUC) the receiver operating characteristic curve (ROC) and Harrel’s concordance index (C-index), using the “timeROC” and “pROC” R packages. A decision curve analysis (DCA) was employed to evaluate the clinical usefulness and net benefit of the intervention, utilizing the ggDCA package. An online version of the nomogram was established and deployed with reference to the shinyapps online website using “DynNom” and “rsconnect” R packages.
Propensity score matching (PSM) analysis was applied to balance the baseline characteristics between the CRKP and CSKP groups in the ratio of 1:2 (MatchIt package), in accordance with the differences in sex, age, baseline diseases or comorbidities, CCI, clinical status, invasive procedure, and the type of infections. The 30-day survival rate was generated using Kaplan–Meier (K-M) curve and the differences obtained were compared using the log-rank test.
Results
Characteristics of patients with KP
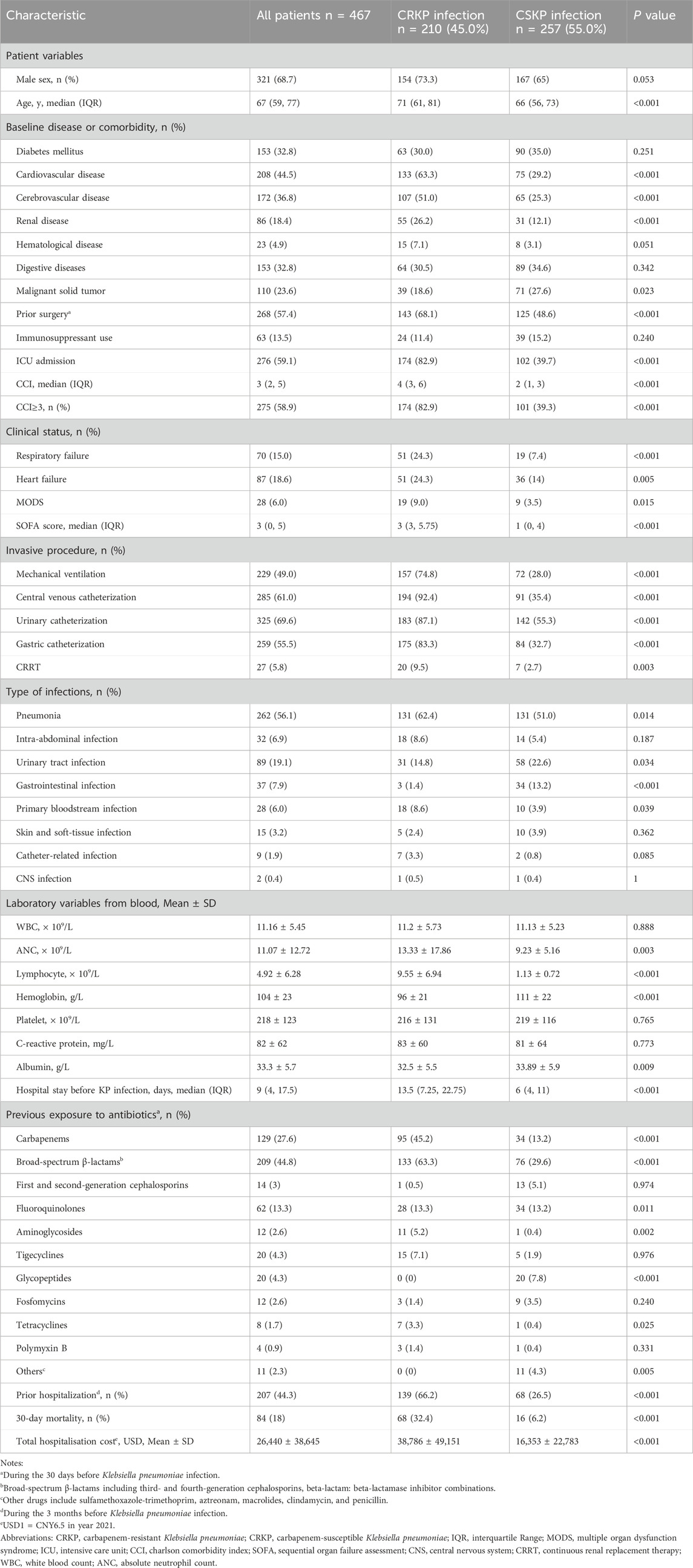
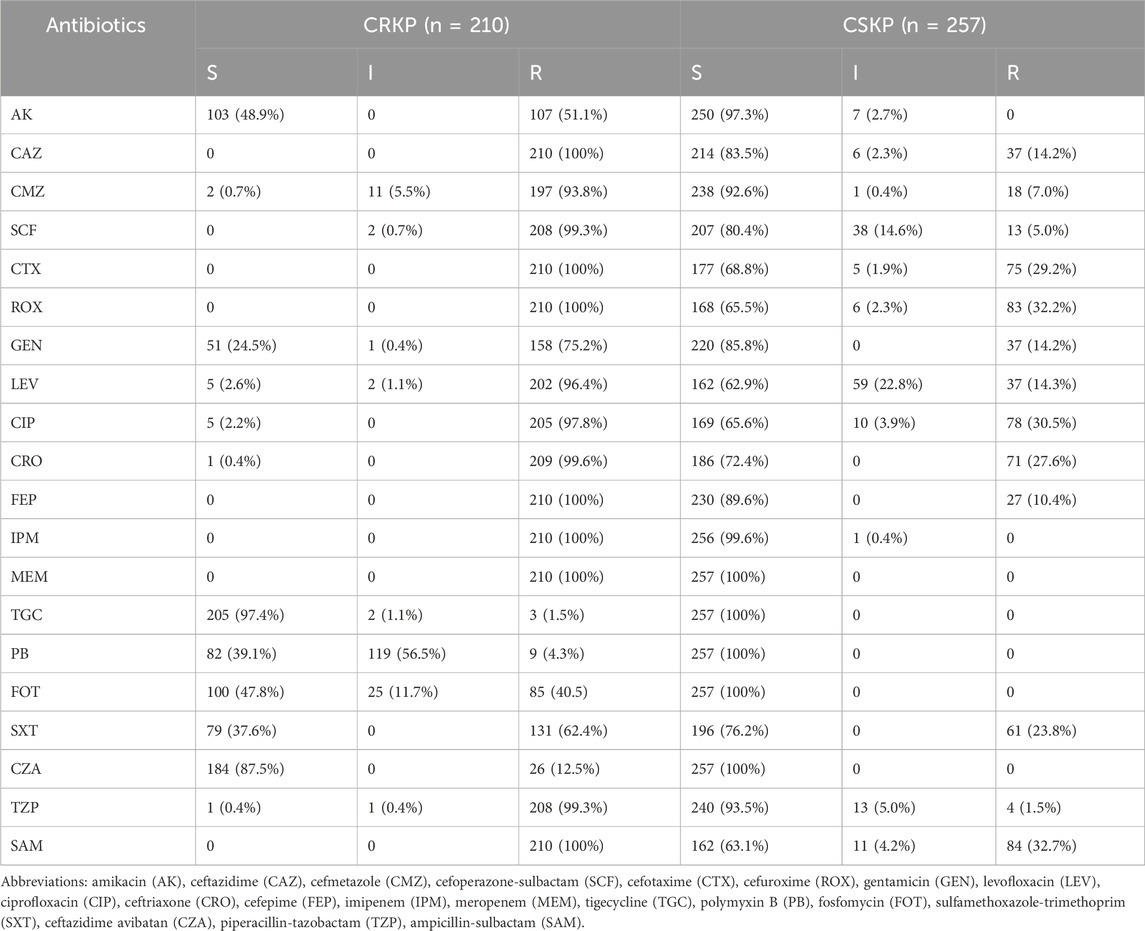
A total of 467 patients with KP infection were enrolled in this study, wherein 210 patients were infected with carbapenem-resistant KP (CRKP) while 257 were with carbapenem-susceptible KP (CSKP), and no significant changes were observed in the proportions of CRKP from 2019 to 2021. The demographic details, clinical characteristics, and laboratory findings were provided in Table 1. The median age was 67 (IQR, 59–77) years and 68.7% of the patients were of the male gender. The detailed antimicrobial susceptibility results are presented in Table 2. 86 patients were included in the validation group, with relevant data available in Supplementary Table S1.
Risk factors associated with CRKP infection and prediction model
The CRKP group had higher values for age, CCI, and Sequential Organ Failure Assessment (SOFA) score, whereas lower values for hemoglobin, albumin, and platelet counts. CRKP was more likely to be observed in patients with ICU admission, longer hospital stays, surgery within the last 30 days, and prior hospitalization during the 3-month before KP infection. With reference to the treatment measurements, the patients in the CRKP group showed greater use of mechanical ventilation, central venous catheterization, urinary catheterization, gastric catheterization, and continuous renal replacement therapy. Moreover, the patients in the CRKP group recorded a higher frequency of previous antibiotic exposure, including those of carbapenems, broad-spectrum β-lactams, fluoroquinolones, aminoglycosides, glycopeptides, and tetracyclines.
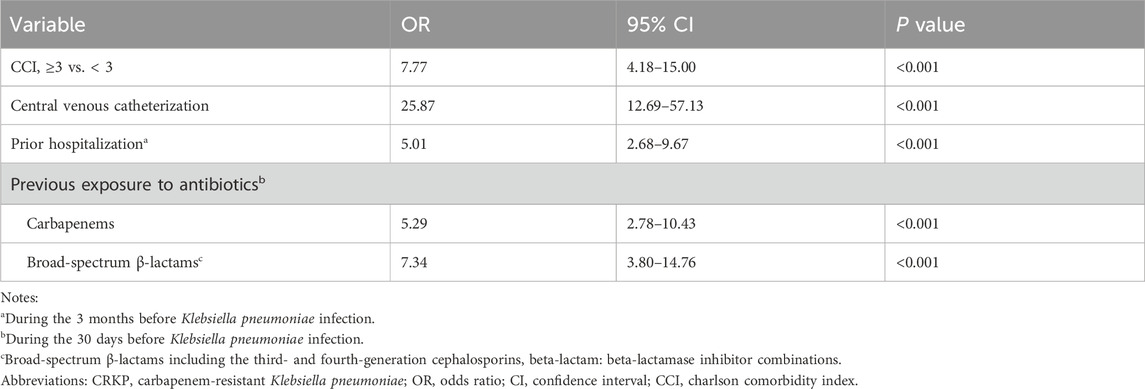
To eliminate the collinearity of the variables and avoid over-fitting, the LASSO regression was employed to reduce the data dimensionality. When the number of variables was reduced to 6, the model conferred good performance, and it was deemed easy to implement. Multivariate logistic regression analysis identified the followings as independent risk factors for CRKP infection: CCI ≥3 (OR, 7.77; 95% CI: 4.18–15.00; P < 0.001), the use of central venous catheterization (OR, 25.87; 95% CI: 12.69–57.13; P < 0.001), prior hospitalization during the last 3 months before infection (OR, 5.01; 95% CI: 2.68–9.67; P < 0.001), and previous exposure to carbapenems (OR, 5.29; 95% CI: 2.78–10.43; P < 0.001) and broad-spectrum β-lactams (OR, 7.34; 95% CI: 3.80–14.76; P < 0.001) (Table 3).
The abovementioned independent factors were employed to establish a nomogram to predict the probability of CRKP infection (Figure 1). The C-index and Brier score of the model indicated its good accuracy (C-index 0.94, 95% CI 0.92–0.96; Brier score 0.093). The calibration plots and ROC curves were utilized to validate the predictive accuracy of the model (Figures 2A, B). The nomogram showed a superior net benefit for predicting CRKP in the DCA (Figure 2C). Furthermore, 86 patients were used for external validation of the model. The C-index for the validation set was 0.915 (95% CI 0.857–0.973), and the Brier score was 0.131. The calibration plots indicated that the external validation model was well-calibrated (Supplementary Figure S1). Finally, we developed a dynamic nomogram which is accessible for free on https://jihuichen.shinyapps.io/dynnomapp_for_probability/ (Supplementary Figure S2).

Figure 1. Nomogram for predicting the probability of carbapenem-resistant Klebsiella pneumoniae in adult patients. Points are summed for each risk factor. Broad-spectrum β-lactams included third- and fourth-generation cephalosporins, beta-lactam: beta-lactamase inhibitor combinations.
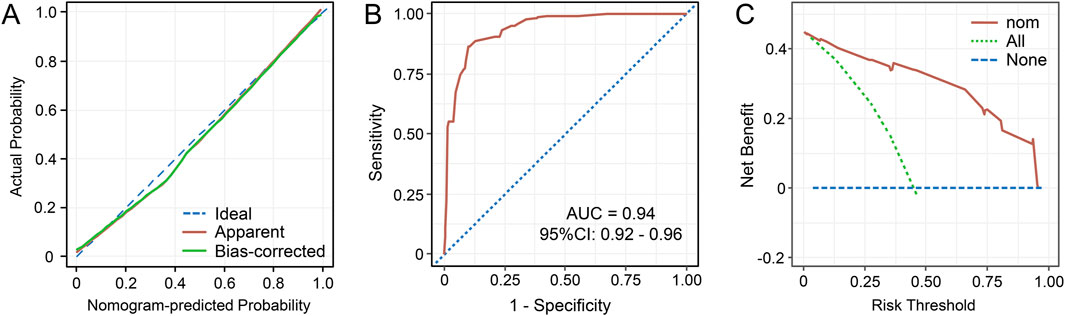
Figure 2. Calibration curves for the nomogram to predict the probability of carbapenem-resistant Klebsiella pneumoniae in adult patients (A). The receiver operating characteristics (ROC) curves (B) and decision curve analysis (DCA) (C) of the prediction model. Abbreviations: AUC, the area under the curve; CI, confidence interval; nom, nomogram.
CRKP infection was independently associated with mortality
PSM analysis was conducted between the CRKP and CSKP groups to estimate the impact of CRKP infection on the 30-day survival rate. After PSM, the 2 groups indicated no significant difference (P > 0.05) in terms of all baseline characteristics and were comparable (Supplementary Table S2). However, the K-M survival analysis indicated significant differences between the CRKP and CSKP groups in terms of the 30-day survival rate (P = 0.002) (Figure 3).
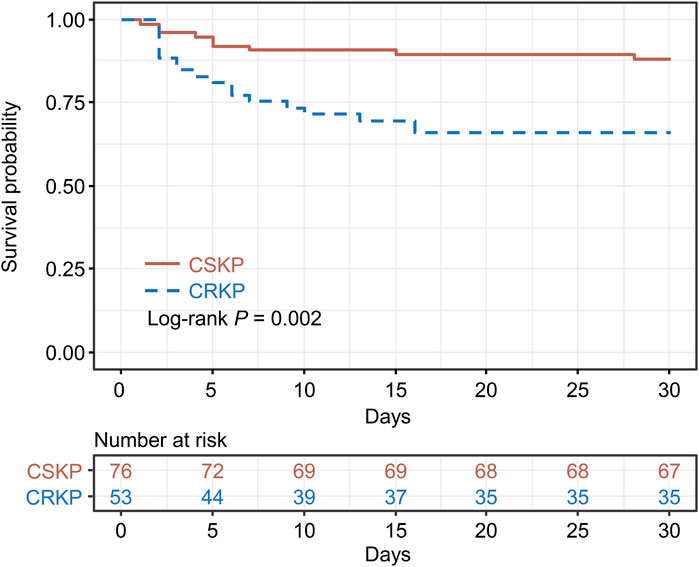
Figure 3. Kaplan–Meier’s survival estimation of the 30-day survival probability of carbapenem-resistant Klebsiella pneumoniae (CRKP) and carbapenem-susceptible Klebsiella pneumoniae (CSKP) patients. Propensity score matching was conducted between the CRKP and CSKP groups, and the CRKP and CSKP groups were matched on propensity scores.
The mortality risk prediction model and nomogram
The demographic details, clinical characteristics, and laboratory findings were compared between the 30-day survivors and non-survivors (Supplementary Table S3). The 30-day mortality rates were found to be 18% (84/467) in the entire study population, 32.4% (68/210) among CRKP patients, and 6.2% (16/257) among CSKP patients.
Variables with P < 0.05 in univariate Cox regression analyses are shown in Table 4. The LASSO regression was applied, and six variables were selected into the final model. Multivariate Cox regression analysis revealed age ≥65 years, higher CCI, presence of respiratory failure, higher SOFA score, albumin ≤30 g/L, and appropriate treatments in 3 days were associated with fatal outcomes. A nomogram was developed to predict the survival probability in adult patients with KP infection based on the associated factors identified in the multivariate Cox regression analysis (Figure 4) (dynamic nomogram: https://jihuichen.shinyapps.io/dynnomapp_for_survival/, Supplementary Figure S3). The established nomogram demonstrated strong accuracy and reliability, as indicated by a C-index of 0.86 and a Brier score of 0.109. The ROC curves, calibration plots, and DCA for 7-, 15-, and 30-day further validated the clinical usefulness and consistency of the model (Figure 5). The validation set yielded a C-index of 0.91 and a Brier score of 0.074. The ROC curves, along with the calibration plots, demonstrated good concordance and reliability (Supplementary Figure S4).
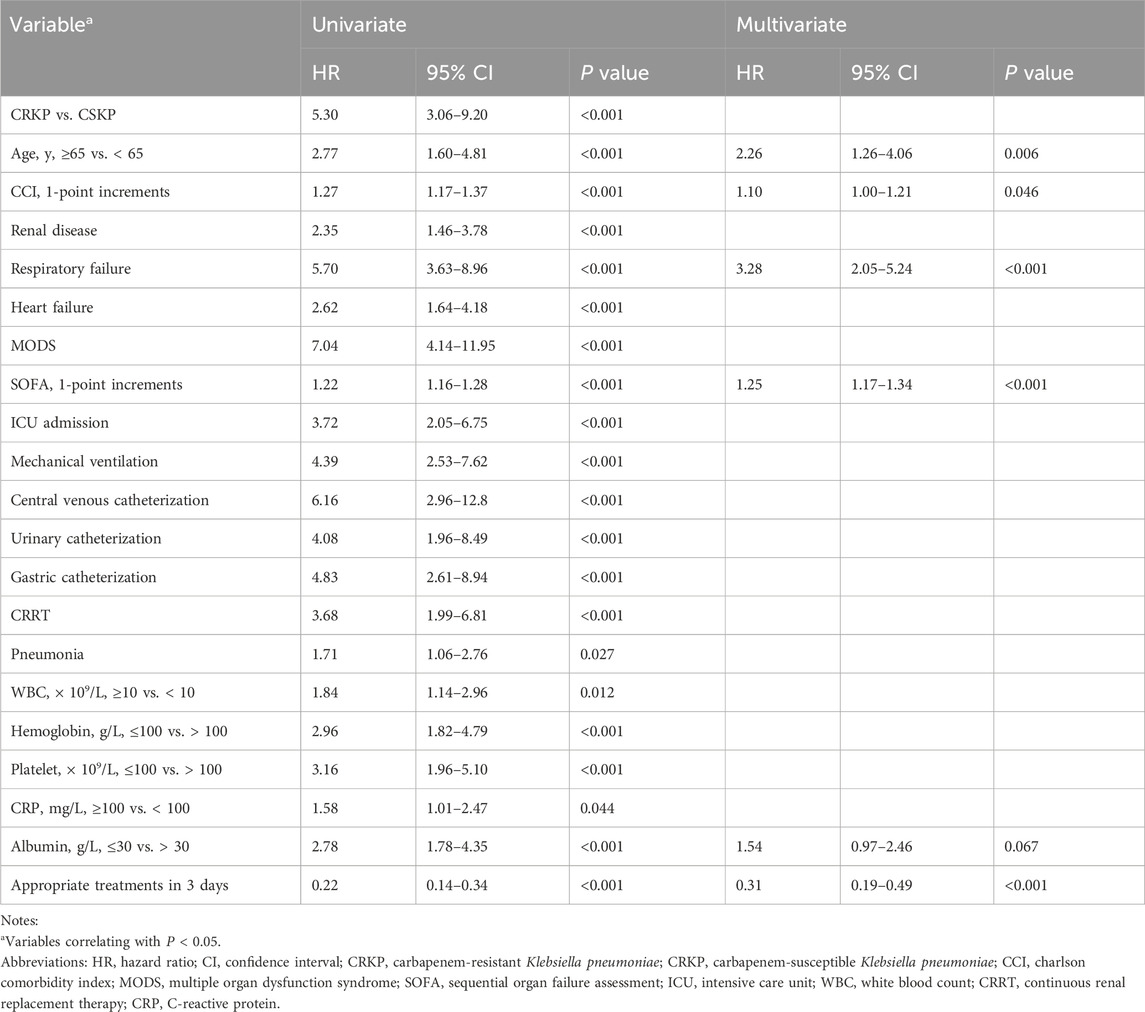
Table 4. Results of univariate and multivariate Cox regression analyses for risk factors associated with 30-day mortality.
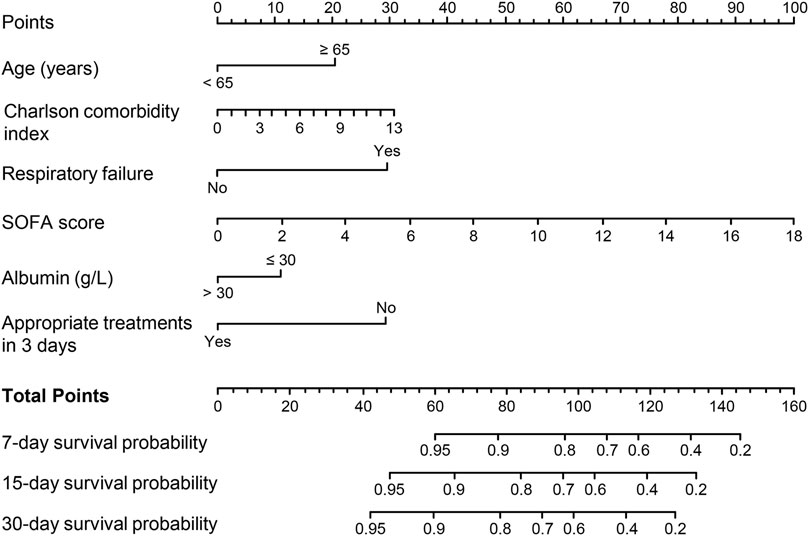
Figure 4. Nomogram for predicting 7-, 15-, and 30-day mortality in adult patients with Klebsiella pneumoniae infection. Abbreviations: SOFA, Sequential Organ Failure Assessment.
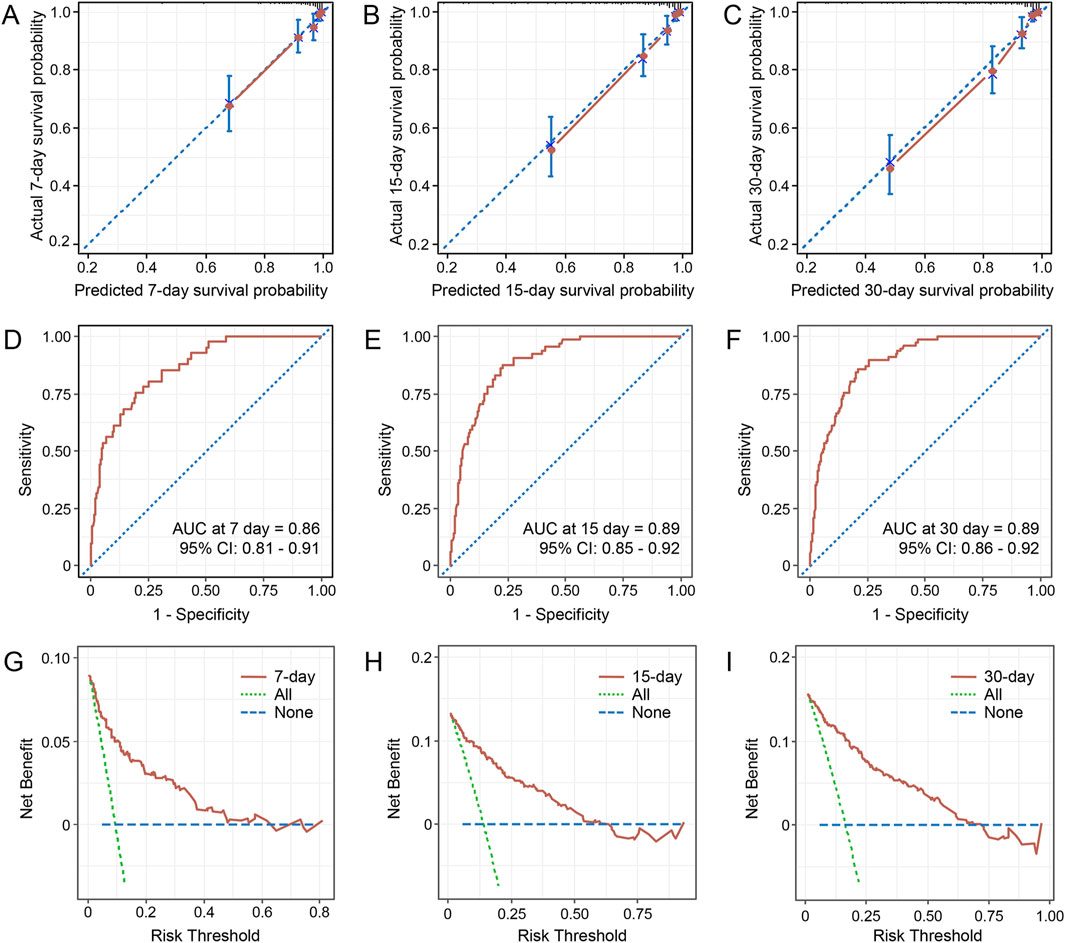
Figure 5. Calibration curves for the nomogram predicting 7-, 15-, and 30-day survival probability (A–C). The time-dependent receiver operating characteristics (ROC) curves for predicting 7-, 15-, and 30-day survival (D–F). Decision curve analysis for the 7-, 15-, and 30-day survival nomogram (G–I). Abbreviations: AUC, the area under the curve; CI, confidence interval.
Discussion
The present study aimed to identify the risk factors and build nomograms to predict the risk of CRKP infection and mortality based on multivariable logistic and Cox regression analyses in adult patients. The nomogram presents a straightforward and pragmatic clinical system, allowing for the identification of high-risk patients who may benefit from intensive medical intervention. The nomogram used to predict CRKP infection was composed of two variables CCI (a tool to measure comorbidity burden) and prior hospitalization (Xiao et al., 2020) (a well-known risk factor for CRKP infection). Similar to the previous reports (Li et al., 2020; Xiao et al., 2020; Dai et al., 2021; Zuo et al., 2021; Wu et al., 2022), prior exposure to carbapenems and broad-spectrum β-lactams were included in the nomogram and acted as independent risk factors. The use of a central venous catheter was also identified as a risk factor in a previous study, (Li et al., 2020), and particularly, it showed the longest line in the nomogram, implying the greatest impact on the CRKP acquisition risk in our study. ICU admission, SOFA score, mechanical ventilation, urinary catheterization, and gastric catheterization were also associated with CRKP acquisition in the univariate analysis and the previous studies (Liu et al., 2018; Li et al., 2020; Xiao et al., 2020). Because of obvious correlations observed between central venous catheterization and ICU admission (r = 0.70), SOFA score (r = 0.49), mechanical ventilation (r = 0.66), urinary catheterization (r = 0.56), and gastric catheterization (r = 0.66), only central venous catheterization with the highest predictive value was included in the final nomogram. These five variables, included in the final nomogram, were obtained through routine examinations, which offered a prominent advantage. Thus, the nomogram could facilitate the identification of patients at high risk of CRKP infection, thereby informing and guiding decision-making in their clinical management. For high-risk patients, antibiotics such as tigecycline, polymyxin, and ceftazidime-avibactam, may be considered.
In our study, the 30-day mortality of 32.4% for CRKP was much higher than that of the CSKP (6.4%). We performed PSM including variables such as sex, age, baseline disease or comorbidity, CCI, clinical status, invasive procedure, and type of infections to minimize the difference between the two groups and ensure a more accurate assessment of the effects of CRKP. After excluding confounding factors, we found that the 30-day mortality rate was significantly higher in the CRKP group (log-rank P = 0.002). Also, in the original dataset, differences in 30-day mortality between the two groups remained significant (adjust-HR, 2.36; 95% CI: 1.09–5.14; P = 0.030) after adjusting for the confounding factors in Cox multivariate analysis. The abovementioned results indicated that CRKP infection was directly related to poor prognosis, similar to that reported previously (Kohler et al., 2017; Xu et al., 2017; Yuan et al., 2020).
The risk factors for mortality in patients with KP infection in our analysis were found to be similar to those reported in past publications (Xu et al., 2017; Xu et al., 2018; Agyeman et al., 2020). We accordingly developed a nomogram to predict the prognosis of patients with KP infection and observed that the factors age, CCI, respiratory failure, SOFA score, and albumin were associated with 30-day mortality. Consistent with the previous studies (Xiao et al., 2020; Chen et al., 2021), we found that an appropriate antimicrobial regimen in 3 days (HR, 0.31; 95% CI, 0.19–0.49; P < 0.001) significantly improved the prognosis and that the inclusion of the variable significantly improved the predictive power of the model. With the widespread use of mNGS, more KP infections can be diagnosed early, albeit it cannot be confirmed whether the strains are resistant to carbapenems. Therefore, to increase the applicability of the nomogram, we did not include the variable of CRKP, although including CRKP slightly increased the predictive power (C-index, 0.865; 95% CI: 0.832–0.898; P = 0.022 by ANOVA analysis). For patients at high-risk, intensive care management and an aggressive antimicrobial regimen should be considered (Qureshi et al., 2012; Chen et al., 2021). In our previous study (Chen et al., 2021), we identified that antibiotics such as ceftazidime-avibactam, fosfomycin, and amikacin have shown promising effectiveness against CRKP infections. However, while tigecycline improved early treatment outcomes, it was associated with a trend toward increased mortality, necessitating cautious use. Combination therapy, particularly in high-risk patients, has demonstrated significant benefits in reducing mortality, underscoring the importance of prompt and appropriate antibiotic regimens to improve prognosis and clinical outcomes.
The present study has several limitations. First, this is a single-center, retrospective, observational study, and hence involves a certain degree of selection bias. Second, the study did not include a molecular analysis to confirm the presence of genes encoding carbapenemase production. Finally, the established nomograms were not validated based on external data, and dynamic nomograms were plotted for more extensive validation.
Conclusion
We developed nomograms to predict the probability of CRKP infection and mortality in adult patients. CCI ≥3, the use of central venous catheterization, hospitalization during the 3 months before infection, and previous exposure to carbapenems and broad-spectrum β-lactams were found to be independently associated with CRKP infection via logistic analysis. Multivariate Cox regression analysis identified several factors associated with 30-day mortality, including age ≥65 years, higher CCI, the presence of respiratory failure, higher SOFA score, albumin level ≤30 g/L, and appropriate treatments within the first 3 days. The established nomograms indicated good consistency and discrimination that can facilitate clinicians to make better clinical decisions when treating patients with KP infection. Nevertheless, prospective studies of a larger number of patients are needed to validate the predictive value of the models.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Ethics Committee of Xin Hua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
HY (1st author): Funding acquisition, Methodology, Project administration, Software, Writing–original draft. YY: Investigation, Methodology, Project administration, Writing–original draft. HY (3rd author): Investigation, Methodology, Project administration, Writing–original draft. SB: Resources, Writing–review and editing. LL: Formal Analysis, Writing–review and editing. FW: Project administration, Writing–review and editing. JZ: Funding acquisition, Resources, Writing–review and editing. JC: Conceptualization, Funding acquisition, Methodology, Supervision, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by National Natural Science Foundation of China (No. 82003742); the Shanghai “Rising Stars of Medical Talent” Youth Development Program -Youth Medical Talents - Clinical Pharmacist Program (SHWSRS2020_087); Shanghai Municipal Clinical Key Specialty Project (shslczdzk06503).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1439116/full#supplementary-material
References
Agyeman, A. A., Bergen, P. J., Rao, G. G., Nation, R. L., and Landersdorfer, C. B. (2020). Mortality, clinical and microbiological response following antibiotic therapy among patients with carbapenem-resistant Klebsiella pneumoniae infections (a meta-analysis dataset). Data Brief. 28, 104907. doi:10.1016/j.dib.2019.104907
Bengoechea, J. A., and Sa Pessoa, J. (2019). Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol. Rev. 43, 123–144. doi:10.1093/femsre/fuy043
CALS Institute (2021). Performance standards for antimicrobial susceptibility testing. 31st edn. Wayne, PA, USA: Clinical and Laboratory Standard Institute.
Charlson, M., Szatrowski, T. P., Peterson, J., and Gold, J. (1994). Validation of a combined comorbidity index. J. Clin. Epidemiol. 47, 1245–1251. doi:10.1016/0895-4356(94)90129-5
Chen, J., Yang, Y., Yao, H., Bu, S., Li, L., Wang, F., et al. (2021). Prediction of prognosis in adult patients with carbapenem-resistant Klebsiella pneumoniae infection. Front. Cell. Infect. Microbiol. 11, 818308. doi:10.3389/fcimb.2021.818308
Dai, G., Xu, Y., Kong, H., Xie, W., and Wang, H. (2021). Risk factors for carbapenem-resistant Klebsiella pneumoniae infection and associated clinical outcomes. Am. J. Transl. Res. 13, 7276–7281.
Del Puente, F., Giacobbe, D. R., Salsano, A., Maraolo, A. E., Ong, D. S. Y., Yusuf, E., et al. (2019). Epidemiology and outcome of Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae (KPC-KP) infections in cardiac surgery patients: a brief narrative review. J. Chemother. (Florence, Italy) 31, 359–366. doi:10.1080/1120009X.2019.1685794
Durante-Mangoni, E., Andini, R., and Zampino, R. (2019). Management of carbapenem-resistant Enterobacteriaceae infections. Clin. Microbiol. Infect. official Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 25, 943–950. doi:10.1016/j.cmi.2019.04.013
Forbes, J. D., Knox, N. C., Ronholm, J., Pagotto, F., and Reimer, A. (2017). Metagenomics: the next culture-independent game changer. Front. Microbiol. 8, 1069. doi:10.3389/fmicb.2017.01069
Kohler, P. P., Volling, C., Green, K., Uleryk, E. M., Shah, P. S., and Mcgeer, A. (2017). Carbapenem resistance, initial antibiotic therapy, and mortality inKlebsiella pneumoniaeBacteremia: a systematic review and meta-analysis. Infect. Control and Hosp. Epidemiol. 38, 1319–1328. doi:10.1017/ice.2017.197
Leguia, M., Vila-Sanjurjo, A., Chain, P. S. G., Berry, I. M., Jarman, R. G., and Pollett, S. (2020). Precision medicine and precision public health in the era of pathogen next-generation sequencing. J. Infect. Dis. 221, S289–S291. doi:10.1093/infdis/jiz424
Li, J., Li, Y., Song, N., and Chen, Y. (2020). Risk factors for carbapenem-resistant Klebsiella pneumoniae infection: a meta-analysis. J. Glob. Antimicrob. Resist. 21, 306–313. doi:10.1016/j.jgar.2019.09.006
Liu, H., Zhang, Y., Yang, J., Liu, Y., and Chen, J. (2022). Application of mNGS in the etiological analysis of lower respiratory tract infections and the prediction of drug resistance. Microbiol. Spectr. 10, e0250221. doi:10.1128/spectrum.02502-21
Liu, P., Li, X., Luo, M., Xu, X., Su, K., Chen, S., et al. (2018). Risk factors for carbapenem-ResistantKlebsiella pneumoniaeInfection: a meta-analysis. Microb. Drug Resist. 24, 190–198. doi:10.1089/mdr.2017.0061
Park, S. Y. (2018). Nomogram: an analogue tool to deliver digital knowledge. J. Thorac. Cardiovasc. Surg. 155, 1793. doi:10.1016/j.jtcvs.2017.12.107
Qureshi, Z. A., Paterson, D. L., Potoski, B. A., Kilayko, M. C., Sandovsky, G., Sordillo, E., et al. (2012). Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob. Agents Chemother. 56, 2108–2113. doi:10.1128/AAC.06268-11
Rodríguez-Medina, N., Barrios-Camacho, H., Duran-Bedolla, J., and Garza-Ramos, U. (2019). Klebsiella variicola: an emerging pathogen in humans. Emerg. microbes and Infect. 8, 973–988. doi:10.1080/22221751.2019.1634981
Steyerberg, E. W., and Vergouwe, Y. (2014). Towards better clinical prediction models: seven steps for development and an ABCD for validation. Eur. heart J. 35, 1925–1931. doi:10.1093/eurheartj/ehu207
Tilahun, M., Kassa, Y., Gedefie, A., and Ashagire, M. (2021). Emerging carbapenem-resistant enterobacteriaceae infection, its epidemiology and novel treatment options: a review. Infect. Drug Resist 14, 4363–4374. doi:10.2147/IDR.S337611
Wu, C., Zheng, L., and Yao, J. (2022). Analysis of risk factors and mortality of patients with carbapenem-resistant Klebsiella pneumoniae infection. Infect. Drug Resist. 15, 2383–2391. doi:10.2147/IDR.S362723
Xiao, T., Zhu, Y., Zhang, S., Wang, Y., Shen, P., Zhou, Y., et al. (2020). A retrospective analysis of risk factors and outcomes of carbapenem-resistant Klebsiella pneumoniae bacteremia in nontransplant patients. J. Infect. Dis. 221, S174–S183. doi:10.1093/infdis/jiz559
Xu, L., Sun, X., and Ma, X. (2017). Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann. Clin. Microbiol. Antimicrob. 16, 18. doi:10.1186/s12941-017-0191-3
Xu, M., Fu, Y., Kong, H., Chen, X., Chen, Y., Li, L., et al. (2018). Bloodstream infections caused by Klebsiella pneumoniae: prevalence of blaKPC, virulence factors and their impacts on clinical outcome. BMC Infect. Dis. 18, 358. doi:10.1186/s12879-018-3263-x
Yuan, Y., Wang, J., Yao, Z., Ma, B., Li, Y., Yan, W., et al. (2020). Risk factors for carbapenem-resistant Klebsiella pneumoniae bloodstream infections and outcomes. Infect. Drug Resist. 13, 207–215. doi:10.2147/IDR.S223243
Zuo, Y., Zhao, D., Song, G., Li, J., Xu, Y., and Wang, Z. (2021). Risk factors, molecular epidemiology, and outcomes of carbapenem-resistant Klebsiella pneumoniae infection for hospital-acquired pneumonia: a matched case-control study in eastern China during 2015–2017. Microb. Drug Resist. 27, 204–211. doi:10.1089/mdr.2020.0162
Keywords: carbapenem-resistant, Klebsiella pneumoniae, infection, nomogram, prognosis
Citation: Yao H, Yang Y, Yao H, Bu S, Li L, Wang F, Zhang J and Chen J (2024) Development of prediction models for carbapenem-resistant Klebsiella pneumoniae acquisition and prognosis in adult patients. Front. Pharmacol. 15:1439116. doi: 10.3389/fphar.2024.1439116
Received: 27 May 2024; Accepted: 28 October 2024;
Published: 05 November 2024.
Edited by:
Karunakaran Kalesh, Teesside University, United KingdomReviewed by:
Yunbao Pan, Zhongnan Hospital of Wuhan University, ChinaAnutthaman Parthasarathy, University of Bradford, United Kingdom
Copyright © 2024 Yao, Yang, Yao, Bu, Li, Wang, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jihui Chen, Y2hlbmppaHVpQHhpbmh1YW1lZC5jb20uY24=
†These authors have contributed equally to this work
 Huijuan Yao
Huijuan Yao Yu Yang1†
Yu Yang1† Lixia Li
Lixia Li Jihui Chen
Jihui Chen