- Department of Animal Science and Technology, Chung-Ang University, Anseong-si, Republic of Korea
The use of Bisphenol A (BPA) and its analogs in industries, as well as the products made from them, is becoming a significant concern for human health. Scientific studies have revealed that BPA functions as an endocrine disruptor. While some analogs of BPA (bisphenols) have been used for a longer time, it was later discovered that they are toxic, similar to BPA. Their widespread use ensures their presence in the environment, and thus, everyone is exposed to them. Scientific research has shown that BPA interacts with estrogen-related receptor gamma (ERRγ), affecting its normal function. ERRγ is involved in biological processes including energy metabolism and mitochondrial function. Therefore, continuous exposure to bisphenols increases the risk of various diseases. In our previous study, we observed that some analogs of BPA had a higher binding affinity to ERRγ compared to BPA itself and analyzed the amino acid residues involved in this interaction. We hypothesized that by antagonizing the interaction between bisphenols and ERRγ, we could neutralize their toxic effects. Taking into account the health benefits of millets and their toxin removal properties, virtual screening of millet-derived compounds was conducted along with prediction of their ADMET profiles. Top five candidates were prioritized for Density Functional Theory (DFT) calculations and further analyses. Long-term molecular dynamics simulation (1 µs) were utilized to evaluate their binding, stability, and antagonizing abilities. Furthermore, reevaluation of their binding energy was conducted using the MM-PBSA method. This study reports millet-derived compounds, namely, Tricin 7-rutinoside, Tricin 7-glucoside, Glucotricin, Kaempferol, and Setarin. These compounds are predicted to be potent competitive inhibitors that can antagonize the interactions between bisphenols and ERRγ. These compounds could potentially assist in the development of future therapeutics. They may also be considered for use as food supplements, although further investigations, including wet-lab experiments and clinical studies, are needed.
1 Introduction
Bisphenols, especially bisphenol A (BPA), are well-known hazardous chemicals, and their role as endocrine disruptors is well established. Additionally, several analogs of BPA are present in the environment, and these have been found to be harmful to human health. Data indicate that the annual production of BPA amounted to approximately 4.85 million tons (Eladak et al., 2015). Beginning in the 1960s, BPA found utility in the production of epoxy resins and polycarbonate plastics. These materials are utilized in the development of common items used daily, such as containers for food and drinks, toys, tooth sealants, paints, medical tools, thermal receipts, and CDs/DVDs (Huo et al., 2015; Sirasanagandla et al., 2022). Consequently, people are frequently exposed to BPA and its analogs through several routes, including oral, transdermal, and respiratory. These chemicals have been detected in urine, milk, blood, serum, plasma, hair, saliva, sweat, placenta, and amniotic fluid in different studies (Akhbarizadeh et al., 2020; Rahman et al., 2021; Tschersich et al., 2021). They are accountable for a wide range of pathological conditions in both humans and animals, including reproductive abnormalities, developmental disorders, neurobehavioral disorders, metabolic diseases, and cancers (Siracusa et al., 2018; Ma et al., 2019; Mustieles et al., 2020). Despite the adverse effects associated with BPA, its utilization in consumer products complicates efforts to reduce its prevalence and alleviate its damaging consequences. As of now, no alternative to BPA that is both safer and more cost-effective has been identified (Eladak et al., 2015; Moon, 2019).
Since the 1930s, researchers have noted that BPA can mimic the hormone estradiol. Therefore, many studies have investigated how BPA can disrupt hormone systems (Yuan et al., 2023). Estradiol is the most potent estrogen hormone and plays a crucial role in various essential physiological processes (Yasar et al., 2017). These functions include the development of reproductive organs, as well as the regulation of cardiovascular, musculoskeletal, immune, and central nervous system balance (Nelson and Bulun, 2001; Gruber et al., 2002; Yasar et al., 2017). The cellular effects of estradiol are mediated by transcription factors and the estrogen receptor (Yasar et al., 2017). Estrogen-related receptor gamma (ERRγ) is classified as an orphan nuclear receptor and functions as a constant transcription activator, playing a crucial role in regulating both mitochondrial energy generation and overall energy metabolism (Matsushima et al., 2007; Matsushima et al., 2022). The estrogenic activity of BPA is well documented, and it has been found that BPA interacts with ERRγ more strongly than the natural hormone estradiol (Babu et al., 2012). This interaction causes several dysfunctions in the normal activity of ERRγ, leading to various diseases (Delfosse et al., 2014; Tohmé et al., 2014; Gao et al., 2015). Therefore, there is a pressing need for a concerted effort to eliminate the toxic effects of bisphenols in order to maintain good health.
A previous study conducted in our lab analyzed the interaction between BPA and its analogs (bisphenols; n = 22) with ERRγ, as well as the key amino acid residues involved in these interactions (Pathak and Kim, 2024). Antagonizing these interactions can help mitigate the toxicity of bisphenols. Millets are recognized as nutri-cereal crops, commonly referred to as smart food (Lydia Pramitha et al., 2022), and they play a role in detoxification by eliminating harmful toxins and free radicals while neutralizing enzymes within organs. This contributes to the prevention of various health concerns, including cancer, diabetes, and cardiovascular diseases (Peterson et al., 2010; Rodriguez et al., 2020; Saini et al., 2021; Babele et al., 2022). Since millets are abundant sources of phytochemicals with significant nutraceutical potential, there is an urgent need for comprehensive and collaborative research dedicated to exploring the phytochemicals present in these cereals. Currently, the majority of research efforts have primarily focused on whole grains and extracts derived from millets (Kumar et al., 2021b; Shah et al., 2021). To raise awareness about the advantages of millets, the United Nations has designated 2023 as the International Year of Millets (https://www.fao.org/millets-2023/en; accessed on 7/9/2023).
Based on our previous study on the interactions of BPA and its analogs with ERRγ, as well as the importance of millets in human health, we hypothesized that phytochemicals from millets can be utilized to counteract the interactions between bisphenols and ERRγ. In this study, we conducted virtual screening of 59 millet phytochemicals targeting the interactions between BPA and ERRγ, to predict their antagonistic activity. Furthermore, we have selected the top ten millet compounds with strong binding affinity and subjected them to ADMET prediction. The top five were then prioritized for Density Functional Theory (DFT) calculations, molecular dynamics simulations, Gibbs free energy landscape analysis, and binding energy calculations using the MM-PBSA method. Through these analyses, we deciphered the mechanisms by which each of these compounds antagonize the interactions, identified the amino acid residues of ERRγ that contribute to the binding, and estimated their binding free energies. These findings will be valuable in optimizing these millet-derived compounds for further investigation and the development of therapeutics aimed at mitigating the toxicity of bisphenols (Figure 1).
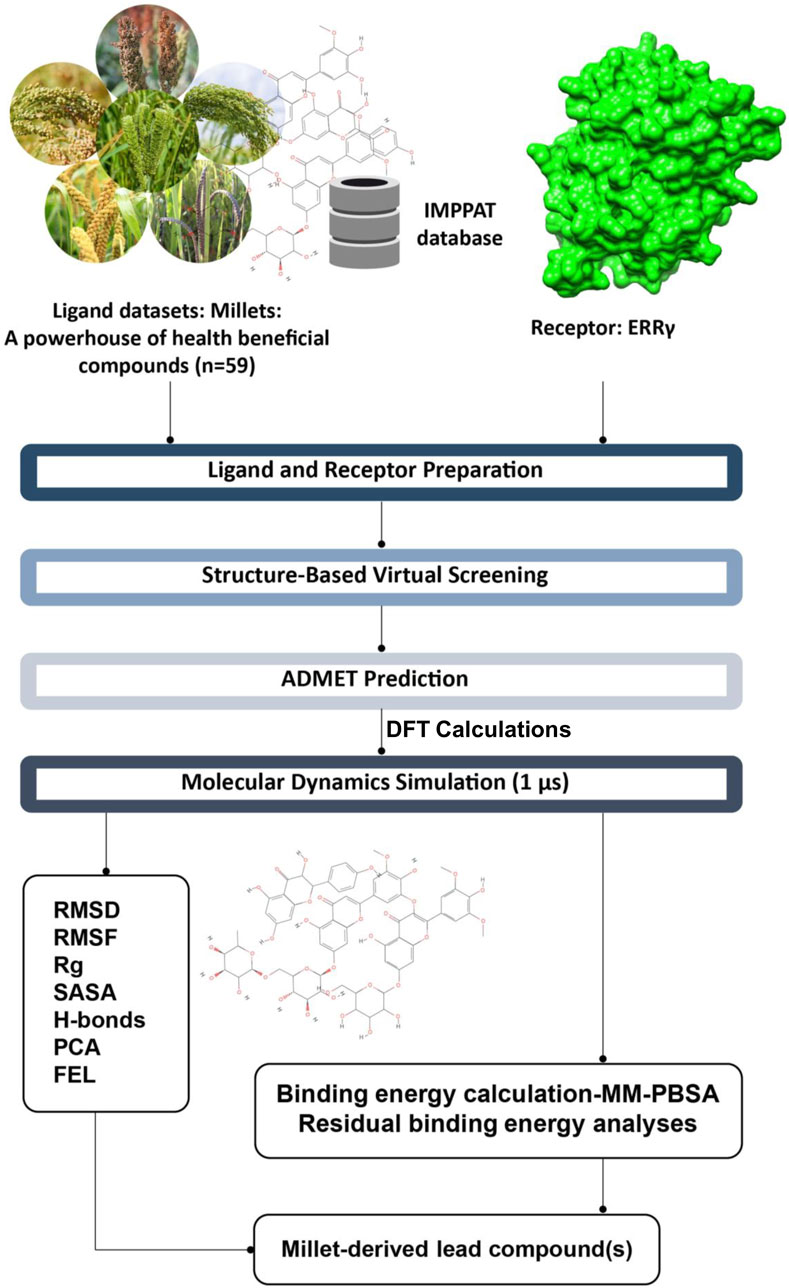
Figure 1. Summary of the study and the computational approaches used to identify millet-derived compounds that counteract the interactions between bisphenols and ERRγ.
2 Materials and methods
2.1 Retrieval of millet-derived compounds and the structure of estrogen-related receptor gamma
Based on information about millet-derived compounds documented in scientific literature, the 3D structures of 59 millet-derived compounds were obtained from the Indian Medicinal Plants, Phytochemistry, and Therapeutics 2.0 (IMPPAT 2.0) database (https://cb.imsc.res.in/imppat/) in protein data bank (PDB) file format (Mohanraj et al., 2018; Pathak et al., 2018; Kumar et al., 2021b; Shah et al., 2021; Kumar and Kotwal, 2023; Priya et al., 2023; Vivek-Ananth et al., 2023). OpenBabel (https://openbabel.org/wiki/Main_Page) was employed to convert the compounds into Protein Data Bank (PDB), partial charge (Q), and atom type (T) (PDBQT) files. The structure of ERRγ (PDB ID: 2E2R) was retrieved from the PDB database (https://www.rcsb.org). The protein structure was visualized and prepared with UCSF Chimera by removing all non-standard residues and co-crystallized ligand (Matsushima et al., 2022). A PDBQT file of ERRγ was then generated using AutoDock tools-1.5.6 by adding charges and polar hydrogens (https://autodock.scripps.edu/). This was followed by generating the grid box size that incorporated the amino acid residues of ERRγ interacting with bisphenols for virtual screening (Trott and Olson, 2010).
2.2 Structure-based virtual screening
Structure-based virtual screening is used to screen a database of small molecules against a target to identify low-energy binding modes of a ligand within the active site of a receptor (Pathak et al., 2023). This computational technique is widely employed in the identification of lead compounds in drug discovery (Singh and Pathak, 2021). In this study, we performed virtual screening of 59 millet-derived compounds, targeting BPA-ERRγ interactions using AutoDock vina (Trott and Olson, 2010). The protein-ligand complexes were generated using UCSF Chimera for visualization and analysis (Pettersen et al., 2004). Furthermore, protein-ligand interaction diagrams in both 2D and 3D were plotted using Discovery Studio Visualizer to determine the key amino acid residues contributing to the binding through various types of interactions (https://discover.3ds.com/discovery-studio-visualizer-download).
2.3 ADMET prediction and PAINS analysis
Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties and Pan Assay Interference Structures (PAINS) analyses of the top ten millet-derived compounds were performed using the pkCSM (https://biosig.lab.uq.edu.au/pkcsm/prediction) and SwissADME (http://www.swissadme.ch/index.php) tools. The SMILES of the selected compounds were obtained from the IMPPAT database and used as the input file for ADMET and PAINS analysis. The following were assessed: molecular properties (molecular weight, logP, rotatable bonds, hydrogen bond donor, hydrogen bond acceptor), absorption (water solubility and intestinal absorption), distribution (blood-brain barrier [BBB] permeability, central nervous system [CNS] permeability), metabolism (CYP2D6 substrate, CYP3A4 substrate, and CYP1A2 inhibitor), excretion (total clearance), toxicity (AMES toxicity and oral rat acute toxicity), and PAINS alert (Pires et al., 2015; Daina et al., 2017).
2.4 Density function theory (DFT) calculation
The top 5 millet-derived compounds were prioritized based on their predicted effectiveness through binding energy with ERRγ for downstream analysis. Density Functional Theory (DFT) calculations were used to evaluate the chemical reactivity of these millet compounds. Specifically, these calculations helped identify key molecular orbitals, such as the Highest Occupied Molecular Orbital (HOMO) and the Lowest Unoccupied Molecular Orbital (LUMO). This provided insights into how these millet compounds interact with ERRγ. The DFT calculations were carried out using the Jaguar-v12.2 module in Schrödinger maestro, with default settings (Schrödinger Release 2023–4; Schrödinger, LLC, New York, NY) (Bochevarov et al., 2013).
2.5 Molecular dynamics (MD) simulation
MD simulations were carried out using the GROMACS package, version 2018.1, with GPU acceleration (Abraham et al., 2015; Pall et al., 2020). The top five millet-derived compound-ERRγ complexes and apo-ERRγ were selected for molecular dynamics (MD) simulations. The system setup followed established protocols from previous studies (Pathak et al., 2022; Pathak et al., 2023). Ligand topology was generated using PRODRG, while protein topology was generated using pdb2gmx (Oostenbrink et al., 2004; Schuttelkopf and Van Aalten, 2004). Solvation was achieved using the simple point charge water model. To create topologies for the protein-ligand complexes, the generated protein and ligand topologies were merged. A cubic simulation box was created, and electroneutrality was ensured by adding ions. Additionally, the steepest descent minimization algorithm was employed to minimize the energy of both the protein and protein-ligand complexes. Subsequently, the system underwent equilibration through NVT and NPT simulations to maintain volume, temperature, and pressure. Finally, all systems were simulated for 1 µs. The structural stability of apo-ERRγ and millet-derived compound-ERRγ complexes was assessed through root-mean-square deviation (RMSD). The RMSD can be calculated as follows.
In this equation, n represents the total count of atoms, ri(t) denotes the current position of an atom at a given time t, and riref signifies the initial positions of the atom. Essentially, t represents the various time points being considered.The flexibility was evaluated using the root-mean-square fluctuation (RMSF).The RMSF can be calculated as follows.
Here,T represents the total duration of the analysis, ri(t) denotes the current positions of an atom at a given time t, and riref signifies the initial positions of the atom. The structural compactness was analyzed using the radius of gyration (Rg).The Rg value can be calculated as follows.
In the equation, mi denotes the atomic mass, Ri signifies various atom coordinates relative to the center of mass, and Rg(z) indicates the radius of gyration around an axis (Sen Gupta et al., 2020). Furthermore, we performed protein folding and stability analysis using solvent-accessible surface area (SASA), followed by analysis of protein–ligand interactions through hydrogen bonding, and essential dynamics using principal component analysis (PCA). The GROMACS utilities ‘gmx rms’, ‘gmx rmsf’, ‘gmx gyrate’, ‘gmx sasa’, ‘gmx hbond’, ‘gmx covar’, and ‘gmx anaeig’ facilitated this analysis (https://manual.gromacs.org/documentation/2019/reference-manual/analysis.html). The 2D plotting tool Grace (https://plasma-gate.weizmann.ac.il/Grace/) was employed for graphical representation and data analysis.
2.6 Free energy landscape (FEL) and binding energy calculation
The analysis of the free energy landscape (FEL) was employed to evaluate the lowest energy states of ERRγ and ERRγ-millet-derived compound complexes (Pathak et al., 2023). The FEL calculations were performed using the ‘gmx sham’ utility in GROMACS. Additionally, the binding free energy of the top five millet-derived compounds complexed with ERRγ was determined using the g_mmpbsa tool, utilizing data from high-throughput molecular dynamics simulations (Kumari et al., 2014).The binding energy (Gbinding) of a protein–ligand complex can be expressed using the following equation.
In the equation, Gcomplex represents the overall binding free energy of the complex, Gprotein corresponds to the receptor in its unbound state, and Gligand represents the ligand in its unbound form. The calculation of the energy contribution of amino acid residue ‘x' involved in an interaction was performed as follows.
In this equation, n represents the total residues count, while Aibound and Aifree represent the bound and free energy of the ith atom within each x residue, respectively.
3 Results
3.1 Predicting millet-derived compounds that inhibit the interaction between bisphenols and ERRγ
Structure-based virtual screening can be employed to identify the most favorable interactions between a receptor and ligand by utilizing a chemical compound database. This method is valuable for evaluating which ligand would effectively interact with a target to form a complex. The ligands are then ranked based on their binding free energy with the target. In this study, we employed 59 compounds derived from millet to conduct virtual screening against the interaction of Bisphenols with ERRγ, in search of potential hits. We assessed the binding free energy of each compound to identify these potential hits for further assessment. Typically, a protein-ligand complex with a low binding energy indicates a strong binding affinity. Therefore, we selected the top 10 screened compounds with the lowest binding energies, ranging from −8.5 to −7.7 kcal/mol, as potential lead compounds. The names of the millet-derived compounds, their IMPPAT and PubChem IDs, binding energy, type of interaction, and interacting amino acid residues are listed in Table 1. The binding free energies of all selected compounds with ERRγ are listed in Supplementary Table S1.
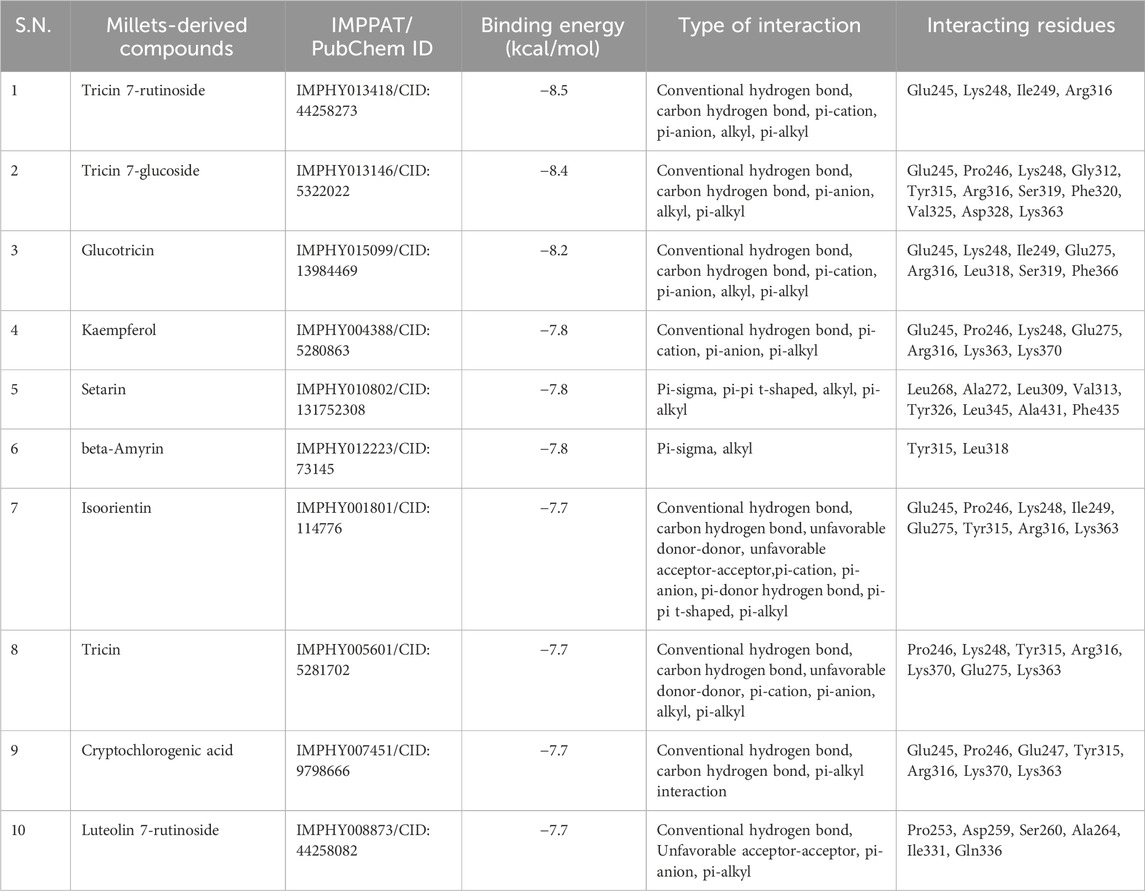
Table 1. List of the top 10 screened millet-derived compounds, with their IMPPAT/PubChem ID, binding free energies, types of bonding interactions, and interacting amino acid residues of ERRγ.
3.2 Structural visualization and interaction analysis
Structural visualization and interaction analysis play a significant role in investigating the interaction of amino acid residues in the target and types of bonding interactions with the ligand (Figure 2). The interaction analysis of the top ten millet-derived compounds was conducted. Tricin 7-rutinoside is one of the promising candidates that interacted with the ERRγ amino acid residues Ile249 and Arg316 through conventional hydrogen bonds. Ile249 also formed a carbon hydrogen bond, and Arg316 interacted with a pi-cation interaction; Lys248 interacted with alkyl and pi-alkyl interaction; Glu245 interacted with a pi-anion interaction. The binding energy of the Tricin 7-rutinoside with the bisphenols–ERRγ interaction was predicted to be −8.5 kcal/mol. Tricin 7-glucoside interacted with Glu245, Pro246, Arg316, Ser319, and Lys363 through conventional hydrogen bonds. Glu245 and Arg316 also interacted with pi-anion and alkyl/pi-alkyl interactions, respectively; Gly312 and Asp328 formed carbon hydrogen bonds; Lys248 formed an alkyl and pi-alkyl interaction; Tyr315, Phe320, and Val325 interacted with alkyl interactions. The binding energy of Tricin 7-glucoside was predicted to be −8.4 kcal/mol. Glucotricin interacted with Glu275, Ile249, Leu318, and Ser319 through conventional hydrogen bonds; Ser319 formed two conventional bonds, and Ile249 formed one conventional and one carbon hydrogen bond. The other mentioned amino acid residues formed single conventional bonds. Lys248 interacted with alkyl, pi-alkyl, and pi-cation interactions; Glu245 and Arg316 interacted with pi-cation and pi-anion bonding; Phe366 interacted through alkyl interaction. The binding energy of Glucotricin was predicted to be −8.2 kcal/mol. Kaempferol interacted with Arg316 through conventional hydrogen bonds, pi-anion, and pi-alkyl interaction. Pro246 and Lys248 interacted with pi-alkyl interaction; Glu245 interacted with pi-anion bonding; Glu275 and Lys363 interacted with pi-anion bonding; Lys370 interacted with pi-cation and pi-anion bonding. The binding energy of Kaempferol was predicted to be −7.8 kcal/mol. Setarin interacted with Leu345 and Ala431 through alkyl interaction; Leu268, Ala272, and Val313 interacted with pi-alkyl interaction; Tyr326 interacted with pi-pi t-shaped bonding; Leu309 interacted with pi-sigma and pi-alkyl interaction; Phe435 interacted with pi-pi t-shaped and pi-sigma bonding. The binding energy of Setarin was predicted to be −7.8 kcal/mol (Figure 3). The binding energy, type of interaction, and interacting amino acid residues of the remaining selected compounds, namelybeta-Amyrin, Isoorientin, Tricin, Cryptochlorogenic acid, and Luteolin 7-rutinoside, are provided in Table 1. Additionally, the binding interactions of bisphenol A (BPA) and the millet-derived compounds Tricin 7-rutinoside and Setarin with ERRγ are shown in Supplementary Figure S1.
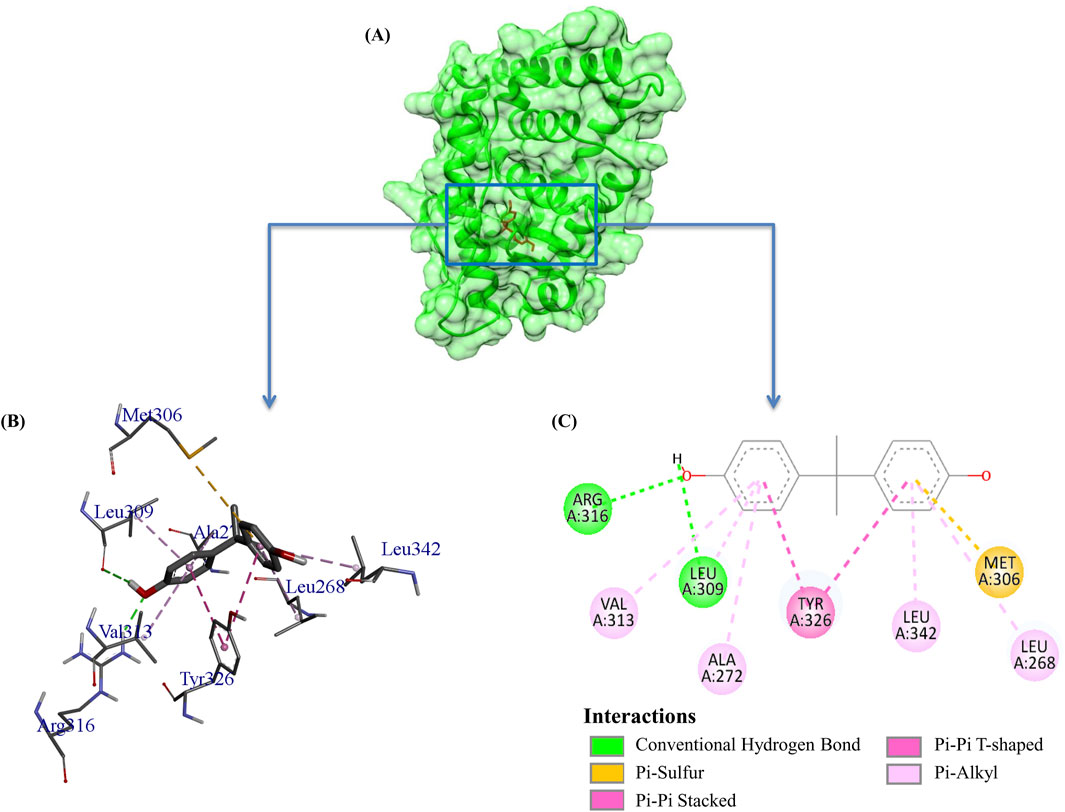
Figure 2. Visualization of the interaction between Bisphenol A (BPA) and ERRγ, highlighting key amino acid residues involved in the interactions. The complex was obtained by redocking the co-crystallized ligand BPA with its receptor ERRγ from our previous study. (A) Surface view; (B) 3D view; and (C) 2D view illustrate different types of bonding interactions.
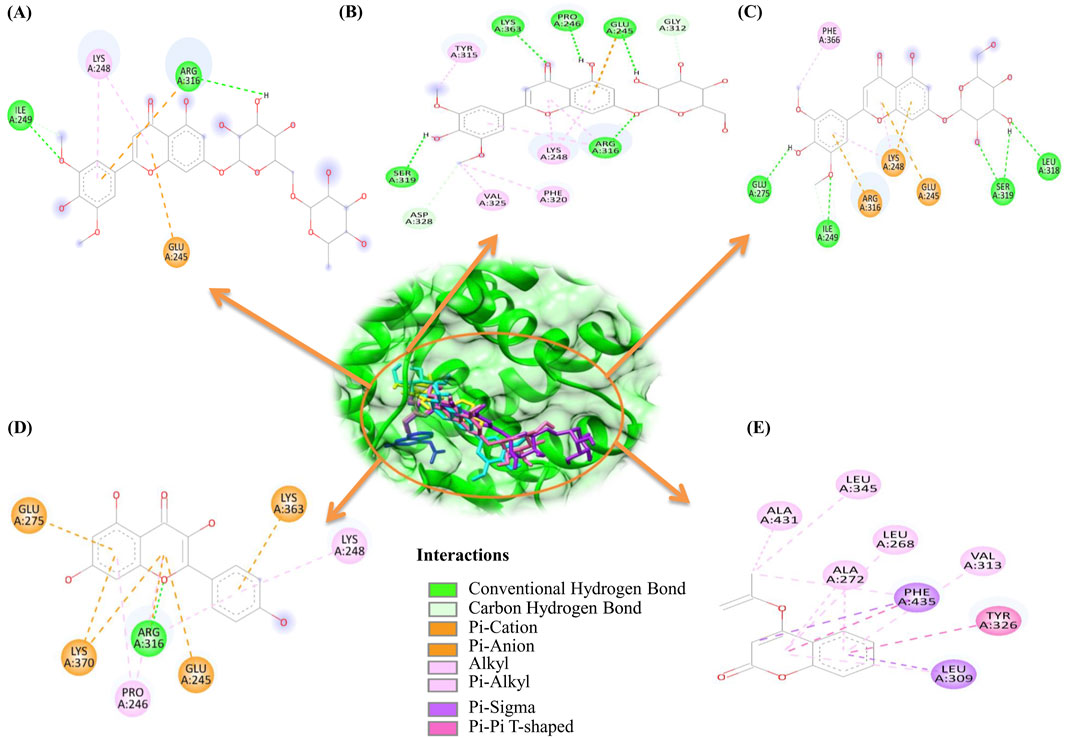
Figure 3. 2D representations of the binding interactions of the top five millet-derived compounds at the site where bisphenols bind to ERRγ. The obtained positions of these top-interacting compounds are shown within the ERRγ binding pocket. These figures illustrate key amino acid residues that contribute to the protein–ligand interactions for antagonizing bisphenol binding. (A) ERRγ–Tricin 7-rutinoside. (B) ERRγ–Tricin 7-glucoside. (C) ERRγ–Glucotricin. (D) ERRγ–Kaempferol. (E) ERRγ–Setarin.
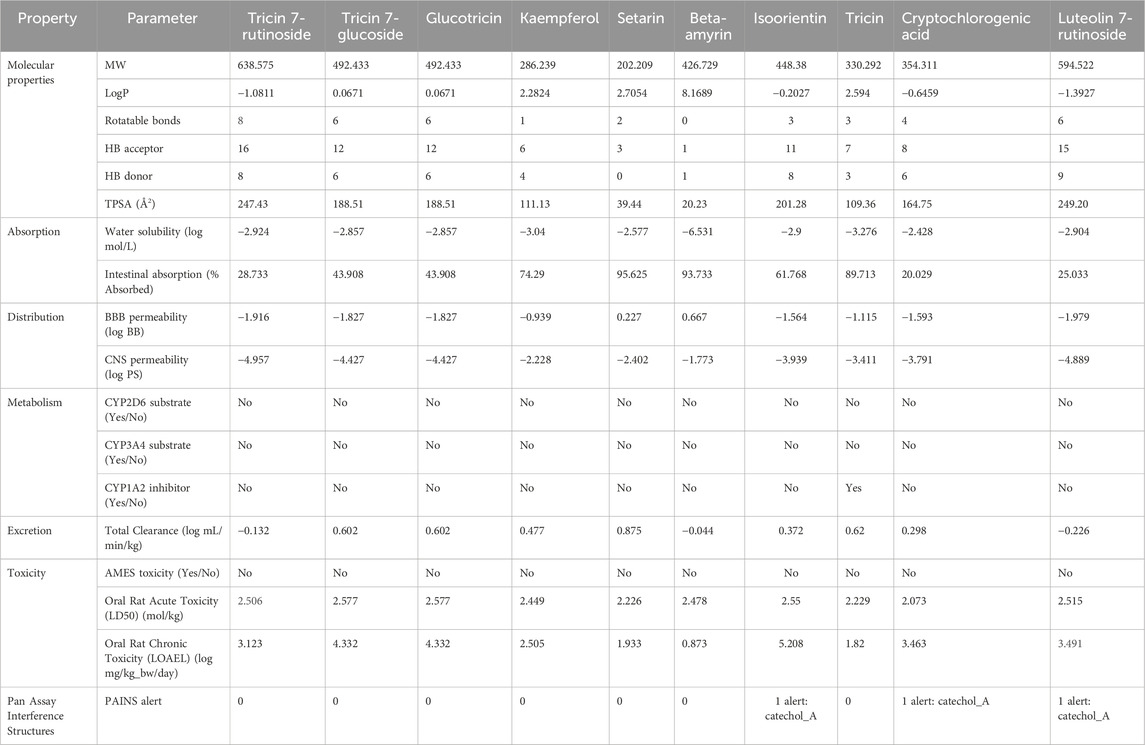
3.3 Assessment of physicochemical properties and toxicity prediction
The assessment of physicochemical properties and prediction of toxicity help in predicting drug-like compounds, significantly reducing the chances of failures in later stages. In this study, pkCSM and SwissADME were used to predict the Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties. The predicted molecular weight of the top screened millet-derived compounds ranged from 202.209 to 638.575 Da. Other parameters were as follows: logP, −1.3927 to 8.1689; rotatable bonds, 0 to 8; hydrogen bond acceptor, 1 to 16; hydrogen bond donor, 0 to 9; topological polar surface area (TPSA), 20.23 to 249.2 Å2. Absorption parameters: water solubility, −6.531 to −2.428; intestinal absorption 20.029 to 95.625. Distribution parameters: BBB permeability, −1.979 to 0.667; CNS permeability, −4.957 to −1.773. Metabolism parameters: CYP2D6 substrate, “No” for all compounds; CYP3A4 substrate, “No” for all compounds; and CYP1A2 inhibitor, “Yes” for Tricin and “No” for all other compounds. The total clearance was predicted in the range of −0.226 to 0.875 for excretion evaluation. In terms of toxicity, none of the compounds were predicted to have AMES toxicity. The values of Oral Rat Acute Toxicity and Oral Rat Chronic Toxicity ranged from 2.073 to 2.577 and from 0.873 to 5.208, respectively. In addition, none of the compounds were found to have Pan Assay Interference Structures (PAINS) except for Isoorientin, Cryptochlorogenic acid, and Luteolin 7-rutinoside, which were predicted to have a PAINS alert for catechol_A. The computed values for each compound are provided in Table 2.
3.4 Exploring molecular properties using DFT calculations
The frontier molecular orbitals, specifically the HOMO and LUMO, of the top five millet-derived compounds selected based on their binding free energy with ERRγ were analyzed using DFT calculations. The analysis was performed using the Jaguar module in Schrödinger Maestro. This method provides insight into the energy levels of the occupied and unoccupied orbitals. Supplementary Figure S2 illustrates the frontier molecular orbitals of the selected compounds. A larger energy gap typically indicates higher chemical stability and lower reactivity, while a smaller energy gap suggests reduced stability and increased reactivity due to easier electron transitions. According to Table 3 the calculated energy gaps range from 3.80 to 4.78 eV. Among the compounds analyzed, Tricin 7-rutinoside, Tricin 7-glucoside, Glucotricin, and Setarin exhibit the largest energy gaps, indicating higher stability. Kaempferol has an energy gap of 3.80 eV, which is lower than the other selected compounds. This suggests that the chemical reactivity of these millet-derived compounds is comparable, allowing favorable interactions with ERRγ. The detailed results are presented in Table 3.
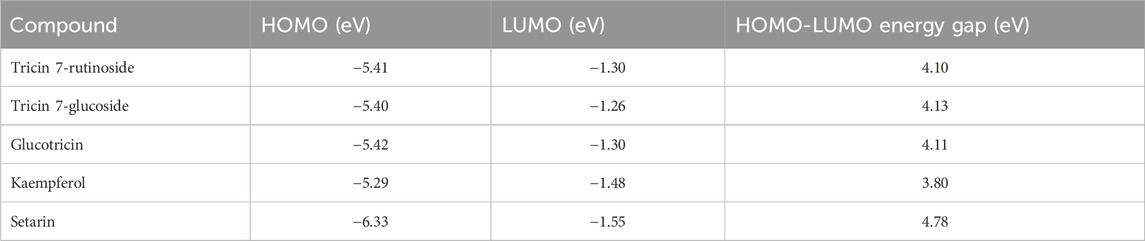
Table 3. Comparison of energy levels for the Highest Occupied Molecular Orbital (HOMO), Lowest Unoccupied Molecular Orbital (LUMO), and the energy gap of compounds from millets.
3.5 Assessing the structural dynamics of ERRγ using MD simulation in both unbound and bound states
In recent years, the significance of MD simulation in drug discovery has greatly increased. These simulations provide in-depth information on the atomic-level behavior of proteins and other biomolecular systems. To evaluate the structural dynamics of ERRγ in both unbound and bound states, MD simulations were carried out for 1 µs. Various parameters were utilized to provide a comprehensive overview of the results, including measures such as root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), solvent accessible surface area (SASA), hydrogen bonding interactions (H-bond), essential dynamics, i.e., principal component analysis (PCA), and free energy landscape (FEL) analysis.
3.5.1 Conformational stability
We assessed the conformational stability of ERRγ through RMSD analysis of a molecular dynamics simulation trajectory. A lower RMSD value indicates a higher degree of conformational stability. We analyzed the RMSD over a duration of 1 µs to gain insights into conformational changes during that time period. The ERRγ protein and all selected docked complexes showed lower RMSD values in the analysis of backbone C-alpha atoms, with the RMSD values of the docked complexes being close to the average RMSD of ERRγ. The average RMSD of ERRγ was calculated as 0.30 nm. However, the RMSD values of the ERRγ-Tricin 7-rutinoside, ERRγ-Tricin 7-glucoside, ERRγ-Glucotricin, ERRγ-Kaempferol, and ERRγ-Setarin complexes were 0.33, 0.30, 0.36, 0.44, and 0.30 nm, respectively. During the simulation, it was observed that all systems reached a stable state (Figure 4A).
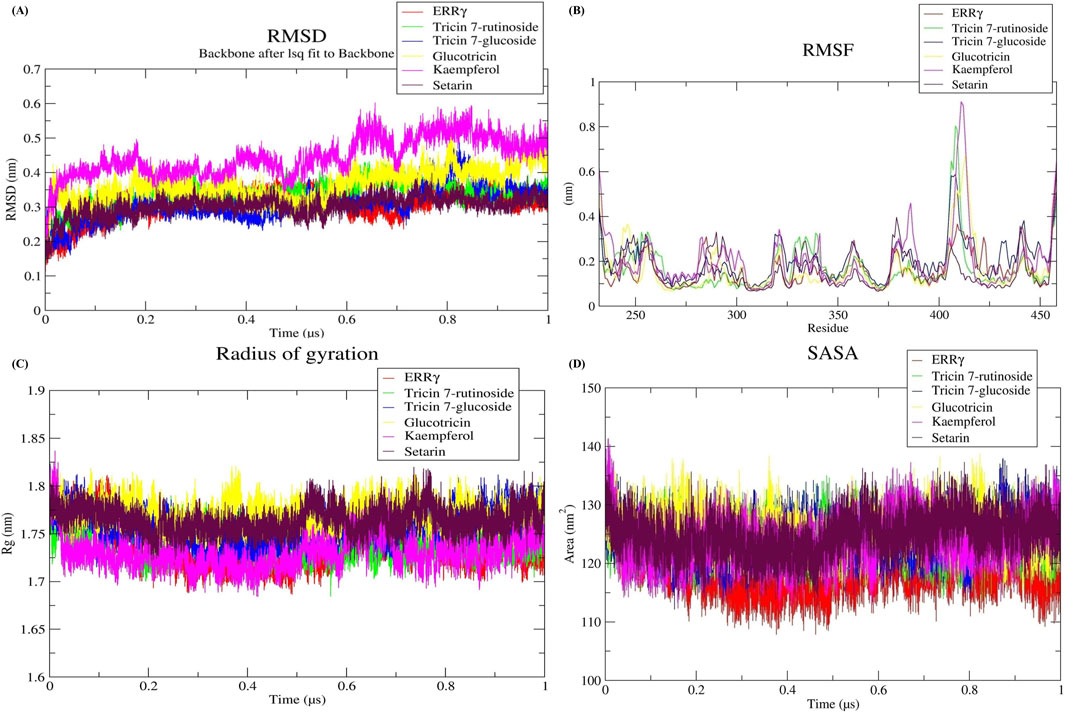
Figure 4. (A) Stability analysis using root mean square deviation (RMSD). (B) Flexibility analysis using root mean square fluctuation (RMSF). (C) Compactness analysis using radius of gyration (Rg). (D) Solvent accessible surface area analysis using SASA values for ERRγ–millet-derived compound complexes during a 1 µs simulation.
3.5.2 Flexibility and residual mobility
We assessed the flexibility and residual mobility of ERRγ to understand the behavior of protein after ligand binding. For this purpose, we conducted RMSF analyses. The average RMSF value for ERRγ was determined to be 0.16 nm. However, the RMSF values for the ERRγ-Tricin 7-rutinoside, ERRγ-Tricin 7-glucoside, ERRγ-Glucotricin, ERRγ-Kaempferol, and ERRγ-Setarin complexes were found to be 0.17, 0.20, 0.16, 0.20, and 0.15 nm, respectively (Figure 4B).
3.5.3 Compactness
To understand the variation in stability and folding of the protein structure over time, the Rg value can be used as an assessment tool. These values provide information about the compactness of the protein structure. We calculated the Rg values for ERRγ and its complexes. The average Rg value for ERRγ was determined to be 1.73 nm. However, the Rg values for the ERRγ-Tricin 7-rutinoside, ERRγ-Tricin 7-glucoside, ERRγ-Glucotricin, ERRγ-Kaempferol, and ERRγ-Setarin complexes were found to be 1.74, 1.75, 1.77, 1.73, and 1.76 nm, respectively (Figure 4C).
3.5.4 Solvent accessible surface area (SASA)
The SASA values were calculated during the entire simulation period to assess the effect of ligand on the solvent accessible surface area. The average SASA value for ERRγ was determined to be 118.59 nm2. However, the SASA values for the ERRγ-Tricin 7-rutinoside, ERRγ-Tricin 7-glucoside, ERRγ-Glucotricin, ERRγ-Kaempferol, and ERRγ-Setarin complexes were found to be 123.89, 124.41, 125.49, 123.83, and 124.79 nm2, respectively (Figure 4D).
3.5.5 Hydrogen bond-based interaction study
Hydrogen bonds (H-bonds) play a crucial role in stabilizing the interactions between a protein and a ligand. To thoroughly evaluate these interactions, we performed a comprehensive analysis of hydrogen bonding during the simulation. The ERRγ-Tricin 7-rutinoside complex formed 0–4 H-bonds. However, the ERRγ-Tricin 7-glucoside, ERRγ-Glucotricin, ERRγ-Kaempferol, and ERRγ-Setarin complexes formed 0–5, 0–4, 0–5, and 0–2 H-bonds, respectively. This observation suggests that there is consistent stability in the interactions between these millet compounds and the interaction site of bisphenols-ERRγ (Supplementary Figure S3).
3.5.6 Essential dynamics
We conducted a principal component analysis (PCA) to understand the essential dynamics and significant structural changes that occur when a ligand binds to the protein. In general, the initial eigenvectors play a crucial role in governing the overall motion of the protein. Therefore, we utilized the first 50 eigenvectors to study alterations in structural dynamics. To gain a more precise understanding of the movements induced by ligand binding, we computed the proportions of correlated motions from the first ten eigenvectors. The first ten eigenvectors accounted for 76.93% of the motions for ERRγ. However, the ERRγ-Tricin 7-rutinoside, ERRγ-Tricin 7-glucoside, ERRγ-Glucotricin, ERRγ-Kaempferol, and ERRγ-Setarin complexes showed 80.94%, 79.26%, 79.03%, 83.76%, and 77.30% correlated motions, respectively. Here, we observed that the ERRγ-Setarin, ERRγ-Glucotricin, and ERRγ-Tricin 7-glucoside complexes showed lower values compared to ERRγ-Tricin 7-rutinoside (Supplementary Figure S4A). Additionally, the 2D plot of these complexes was also analyzed. The initial eigenvectors of the protein reveal its core dynamics. Consequently, we examined the first two of these eigenvectors in a phase space, which depicted a stable cluster for all the selected complexes (Supplementary Figure S4B).
3.5.7 Gibbs free energy landscape (FEL)
We utilized the initial two principal components to calculate the Gibbs free energy landscape (FEL). The FEL for each system is depicted in Figure 5. In the plot, the deep blue color represents the conformation with the lowest energy, whereas the red color indicates the conformation with the highest energy. The deep blue valleys represent transition states with lower energy. As observed, ERRγ has energy minima enriched with blue color and a wide space. The ERRγ-Tricin 7-rutinosidecomplex showed the deepest minima with two lowest energy funnels, indicating that it is energetically more stable than ERRγ and undergoes less frequent changes in conformational states. Similarly, the ERRγ-Tricin 7-glucoside complex had the deepest minima with three funnels. However, the ERRγ-Glucotricin and ERRγ-Kaempferol complexes spanned a large area of conformational space but still had the deepest minima. The ERRγ-Setarin complex showed two energy funnels connected to each other, covering a larger blue area. Overall, the FEL analysis indicated that all the systems exhibit low energy conformations after ligand binding and form stable complexes (Figure 5).
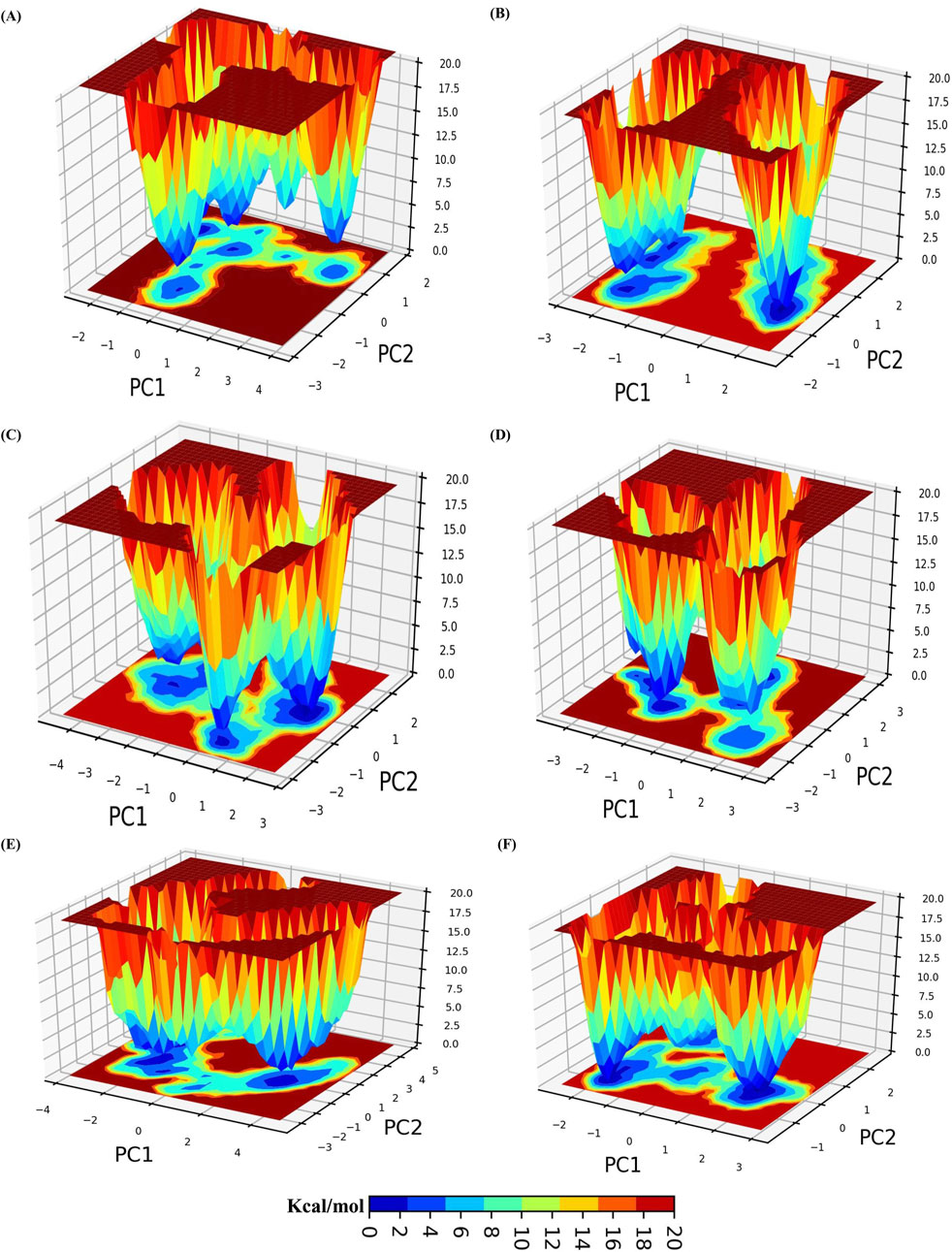
Figure 5. A color-coded representation of the Gibbs free energy landscape (FEL) for ERRγ and ERRγ–millet-derived compounds complexes, generated using the principal components PC1 and PC2. The color scale indicates the Gibbs free energies (kcal/mol) for different conformational states. Blue represents the lowest energy states, while red represents the highest energy states. (A) ERRγ. (B) ERRγ-Tricin 7-rutinoside. (C) ERRγ-Tricin 7-glucoside. (D) ERRγ-Glucotricin. (E) ERRγ-Kaempferol. (F) ERRγ-Setarin.
3.5.8 Binding free energy calculation
We employed the MM-PBSA method to assess the binding free energy of all simulated complexes, thereby confirming their affinities towards bisphenols-ERRγ interactions. The binding free energies were calculated based on the final 10 ns of the MD simulation trajectories. The binding free energy for ERRγ-Tricin 7-rutinoside, ERRγ-Tricin 7-glucoside, ERRγ-Glucotricin, ERRγ-Kaempferol, and ERRγ-Setarin complexes were found to be −134.843, −173.650, −203.530, −113.351, and −124.159 kJ mol-1, respectively. The values for van der Waals, electrostatic, polar solvation, SASA, and binding free energies are provided in Table 4.
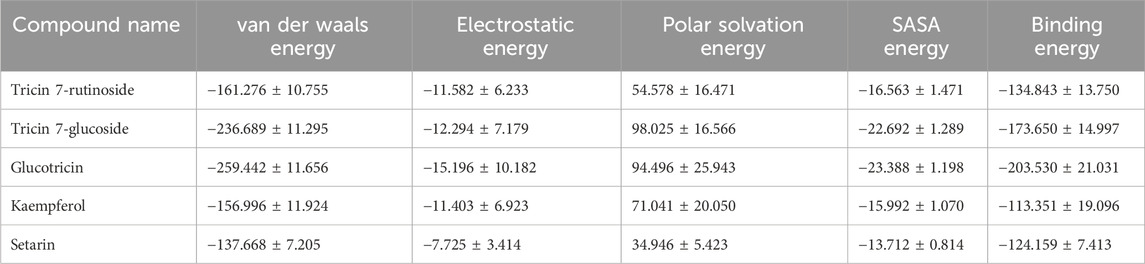
Table 4. Binding energies (kJ/mol) of the top five compounds obtained from millets with ERRγ as determined through molecular dynamics simulations trajectories calculated using the MM-PBSA method (van der Waals and electrostatic forces, polar solvation, SASA, and binding free energy).
To identify the amino acid residues that are crucial for ligand binding, we conducted residual binding energy analyses on the simulated complexes. All of the selected compounds exhibited significant involvement in interactions with amino acid residues of ERRγ, which are associated with interactions with bisphenols, suggesting that these compounds have the potential to act as antagonists for these interactions. Amino acid residues within positions 232 to 245, 260 to 290, and 310 to 375 made significant contributions to these interactions (Figure 6).
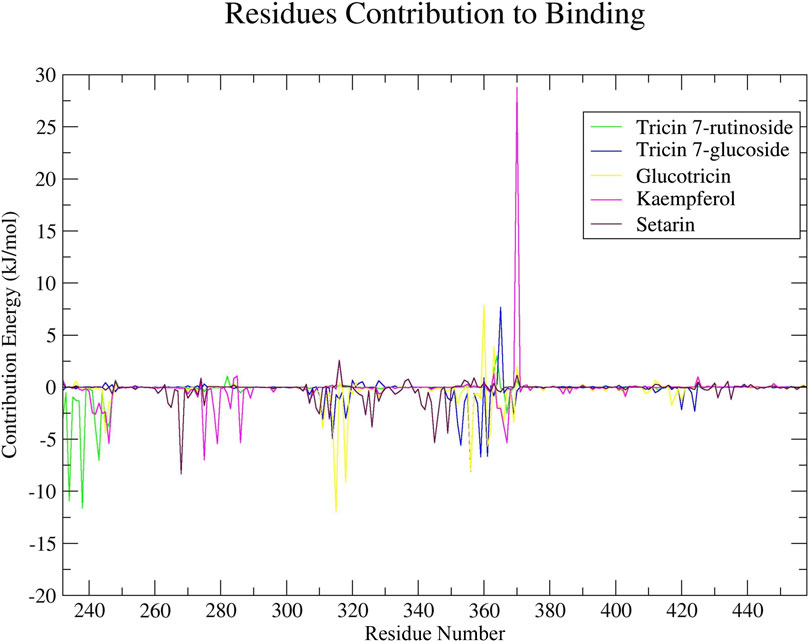
Figure 6. The decomposition of residual binding free energy provides valuable insights into the specific contributions of individual amino acid residues of ERRγ to the binding of millet-derived compounds.
4 Discussion
Bisphenol A (BPA) and its analogs are commonly used chemicals in the production of polycarbonate plastics and epoxy resins. These industrial compounds have the ability to mimic estrogen, thereby disrupting the endocrine system (Babu et al., 2012; Nayak et al., 2022; Kodila et al., 2023). BPA is so prevalent that it can be found everywhere. One of the primary ways it enters the human body is through the consumption of water, food, and drinks that have been contaminated with BPA (Agarwal et al., 2016; Begum et al., 2020). Physical contact with thermal printed paper can result in the transfer BPA to human skin, which is detrimental to human health (Bernier and Vandenberg, 2017; Almeida et al., 2018).
In experiments with rodents, exposure to BPA during pregnancy and while nursing made the animals more anxious and prone to depression-like behaviors (Xu et al., 2012). Continuous exposure to BPA may result in the buildup of this hazardous chemical in tissues (Andra et al., 2016; Nayak et al., 2022). Previous studies have demonstrated that ingesting BPA through food can impair the colon’s normal permeability (Feng et al., 2019). Dietary BPA exposure also affects the morphological and biological barriers of the intestine, as well as its chemical environment (Feng et al., 2019). BPA is metabolized by the liver (Nachman et al., 2014). After absorption, certain enzymes, including uridine 5′diphospho-glucuronyl transferase (UGT) and sulfotransferases, convert unbound BPA into BPA-glucuronide and BPA-sulfate (Nachman et al., 2014). After detoxification, the body excretes BPA through urine, with a reported half-life of 2 h in blood (Neri et al., 2015). BPA can also be removed from the body through various routes, including sweat, feces, and bile (Genuis et al., 2012; Nayak et al., 2022). Due to increasing concerns about the safety of BPA, the industry has responded by adopting new, less studied BPA analogues that have similar polymer-forming properties (Kodila et al., 2023). Recent computational studies conducted in our lab have demonstrated that some BPA analogs have a higher binding affinity and toxic effect compared to BPA (Pathak and Kim, 2024). Despite avoiding them, humans can come into contact with BPA and its analog-based products due to their widespread availability (Wang et al., 2019; Sendra et al., 2023). Industry workers are exposed to it daily, and continuous exposure to this chemical is not safe (Konieczna et al., 2015; Vom Saal and Vandenberg, 2021). Therefore, multiple studies have recommended employing antioxidant treatment to reduce the toxic effects of BPA (Nayak et al., 2022). In addition, natural products are also suggested for the development of future therapeutics against BPA exposure (Sirasanagandla et al., 2022).
The interaction of bisphenols with ERRγ has been well documented (Liu et al., 2019; Ohore and Zhang, 2019). ERRγ is involved in the regulation of energy metabolism and mitochondrial function. Additionally, it directly or indirectly regulates numerous target genes (Misra et al., 2017). Mice without ERRγ do not survive their first week due to mitochondrial dysfunction. ERRγ is also vital for heart, muscle, and brain function (Alaynick et al., 2010; Misra et al., 2017). Furthermore, ERRγ is linked to various health issues, such as type 2 diabetes and alcoholic liver disease. In these conditions, inhibiting ERRγ through pharmacological means restores normal glucose levels and improves insulin sensitivity in mice (Kim et al., 2011; Kim et al., 2012; Kim et al., 2013; Misra et al., 2017). Therefore, the interactions of bisphenols with ERRγ interfere with its normal activity, leading to several dysfunctions (Babu et al., 2012; Nayak et al., 2022).
Previous studies on millets led to their recognition as smart food and highlighted the importance of their phytochemicals in human health, as well as their preventive role in pathologies such as cardiovascular diseases, diabetes, osteoporosis, cancer, and neurodegenerative disorders (Anitha et al., 2021; Kumar et al., 2021a; Lydia Pramitha et al., 2022; Kumar and Kotwal, 2023). Furthermore, millets aid in detoxification by removing toxins and free radicals while neutralizing enzymes within organs, thus preventing several health issues (Babele et al., 2022). The present study utilized millet-derived compounds to antagonize bisphenols-ERRγ interactions to mitigate the toxicity of BPA and its analogs.
Structure-based virtual screening is a well-recognized computational approach used to screen a database of small molecules against a molecular target of interest to investigate candidate compounds (Singh and Pathak, 2020; Pant et al., 2022). In the present study, virtual screening of 59 millet-derived phytochemicals was conducted, targeting amino acid residues of ERRγ that are involved in its interactions with bisphenols. During visualization of the protein-ligand complex, it was observed that higher molecular weight millet compounds, such as Tricin 7-rutinoside, adopted different poses, while lower molecular weight compounds, such as Setarin, showed similar poses to BPA. These compounds interacted with some of the residues where BPA binds to ERRγ, potentially antagonizing BPA’s binding to ERRγ. Our aim was to identify suitable millet-derived candidate compounds that can antagonize these interactions. To validate the findings, additional analyses were performed, including ADMET prediction, DFT calculations, molecular dynamics simulation, Gibbs free energy landscape analysis, and binding energy calculation (Pathak and Kim, 2024).
After conducting a rigorous assessment of compounds derived from millets, which were evaluated for their ability to antagonize bisphenols-ERRγ interactions, the ten most promising compounds were selected for detailed evaluation using a range of computational techniques (Maia et al., 2020). Subsequently, we examined the ADMET profile of these top ten compounds obtained from millets (Pires et al., 2015; Daina et al., 2017). It is worth highlighting that while a few of these leading compounds did not follow Lipinski’s Rule of 5, they still have significant potential as natural agents capable of antagonizing bisphenols-ERRγ interactions (Lipinski et al., 2001). We must acknowledge that numerous drug molecules currently in use have been approved despite not conforming to Lipinski’s Rule of 5 (Hartung et al., 2023). The DFT calculation analysis suggested that the chemical reactivity of the top five selected millet-derived compounds is comparable, allowing for favorable interactions with ERRγ (Bochevarov et al., 2013). Therefore, these identified molecules can be considered for further evaluation to determine their practical applicability.
Furthermore, we used an advanced computational method known as molecular dynamics simulation to evaluate the top five docked complexes: ERRγ-Tricin 7-rutinoside, ERRγ-Tricin 7-glucoside, ERRγ-Glucotricin, ERRγ-Kaempferol, and ERRγ-Setarin. This approach allowedus examine the interactions between these millet-derived compounds and ERRγ amino acids, where bisphenols are involved (Pant et al., 2022). By analyzing the RMSD plot, we found that all systems reached a stable state, indicating significant interactions with ERRγ. We also used other metrics such as RMSF, Rg, SASA, H-bonds, PCA, and FEL to investigate the stability of these complexes. The results showed that the millet-derived compounds formed robust and stable bonds with ERRγ, which is important for counteracting bisphenols-ERRγ interactions. This is a promising finding for potential medical applications (Priya et al., 2023).
Additionally, we investigated the binding ability of millet-derived compounds to ERRγ. We used a method called MM-PBSA to determine the strength of binding (Kuhn et al., 2005; Kumari et al., 2014). This approach provides insights into the potency of these compounds in their interactions with ERRγ, which is crucial for lead identification (Kuhn et al., 2005). The binding energy of Tricin 7-rutinoside, Tricin 7-glucoside, Glucotricin, Kaempferol, and Setarin were calculated. A negative binding energy indicates a strong binding between a ligand and receptor, signifying a favorable interaction. On the other hand, a high binding energy suggests a weaker interaction (Wan et al., 2020). Our analysis using MM-PBSA and residual binding energy demonstrated the stability of the complexes formed between millet-derived compounds and ERRγ. This finding highlights Glucotricin as a promising lead compound. However, other compounds such as Tricin 7-rutinoside, Tricin 7-glucoside, Kaempferol, and Setarin also show potential for further development and for antagonizing the interaction between bisphenols and ERRγ, which is important for future therapeutic advancements (Sirasanagandla et al., 2022; Priya et al., 2023).
Computer-assisted drug discovery can accelerate the process of identifying potential compounds by facilitating the analysis of plant-derived compounds against specific drug targets (Pathak et al., 2018; Pathak et al., 2020). This method reduces the time and financial resources required for experiments and enhances research outcomes (Pathak et al., 2020; Singh and Pathak, 2020). It is worth nothing that many of the medications available today are derived from plants or natural sources (Singh et al., 2022). Scientific research has already demonstrated the ability of natural products to mitigate the toxicity of bisphenols (Sirasanagandla et al., 2022). Bisphenols are well-known toxic compounds, but millets have been safely consumed by humans for centuries. While whole millets offer health benefits due to the synergistic effects of their phytochemicals, individual millet-derived compounds may behave differently. However, these compounds could be used in combination if found effective. Therefore, the findings of this study can be applied to the development of therapeutics that counteract the harmful effects of bisphenols by antagonizing their interactions with ERRγ.
5 Conclusion
The widespread use of bisphenols in various industries has raised serious concerns for human health due to their well-documented effects as endocrine disruptors. Research has shown that these compounds can interfere with the normal activity of ERRγ, a vital factor in energy metabolism and other biological functions, increasing the risk of various diseases. Our prior study revealed that some BPA analogs exhibit a stronger binding to ERRγ compared to BPA, which motivated us to investigate potential approaches to neutralize their toxic effects by inhibiting their interactions with ERRγ. Based on the health benefits and detoxification properties of millets, we screened millet-derived compounds, selected the top ten, and predicted their ADMET. Furthermore, prioritize the top five compounds to evaluate their reactivity through DFT analysis. Subsequently, molecular dynamics simulations were conducted on docked complexes of the top five millet compounds with ERRγ to assess their binding, stability, and antagonistic nature over time. Additionally, we refined our selections by evaluating their binding energies using the MM-PBSA method. These compounds exhibited potential therapeutic properties that might mitigate the health risks associated with bisphenols. However, it is crucial to emphasize the need for further research including wet-lab experiments and clinical studies to validate their efficacy and safety. These compounds, which might be utilized either as single molecules or in combination, could potentially play a vital role in protecting human health.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
RKP: Conceptualization, Methodology, Software, Formal Analysis, Visualization, Investigation, Writing–original draft, Writing–review and editing. J-MK: Conceptualization, Supervision. Resources, Project administration, Funding acquisition, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2018R1A6A1A03025159). High-performance computing and other necessary facilities provided by the Chung-Ang University are gratefully acknowledged.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1435254/full#supplementary-material
References
Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., et al. (2015). GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25. doi:10.1016/j.softx.2015.06.001
Agarwal, S., Yadav, A., Tiwari, S. K., Seth, B., Chauhan, L. K., Khare, P., et al. (2016). Dynamin-related protein 1 inhibition mitigates bisphenol A-mediated alterations in mitochondrial dynamics and neural stem cell proliferation and differentiation. J. Biol. Chem. 291, 15923–15939. doi:10.1074/jbc.M115.709493
Akhbarizadeh, R., Dobaradaran, S., Schmidt, T. C., Nabipour, I., and Spitz, J. (2020). Worldwide bottled water occurrence of emerging contaminants: a review of the recent scientific literature. J. Hazard Mater 392, 122271. doi:10.1016/j.jhazmat.2020.122271
Alaynick, W. A., Way, J. M., Wilson, S. A., Benson, W. G., Pei, L., Downes, M., et al. (2010). ERRgamma regulates cardiac, gastric, and renal potassium homeostasis. Mol. Endocrinol. 24, 299–309. doi:10.1210/me.2009-0114
Almeida, S., Raposo, A., Almeida-Gonzalez, M., and Carrascosa, C. (2018). Bisphenol A: food exposure and impact on human health. Compr. Rev. Food Sci. Food Saf. 17, 1503–1517. doi:10.1111/1541-4337.12388
Andra, S. S., Austin, C., Yang, J., Patel, D., and Arora, M. (2016). Recent advances in simultaneous analysis of bisphenol A and its conjugates in human matrices: exposure biomarker perspectives. Sci. Total Environ. 572, 770–781. doi:10.1016/j.scitotenv.2016.07.062
Anitha, S., Botha, R., Kane-Potaka, J., Givens, D. I., Rajendran, A., Tsusaka, T. W., et al. (2021). Can millet consumption help manage hyperlipidemia and obesity? a systematic review and meta-analysis. Front. Nutr. 8, 700778. doi:10.3389/fnut.2021.700778
Babele, P. K., Kudapa, H., Singh, Y., Varshney, R. K., and Kumar, A. (2022). Mainstreaming orphan millets for advancing climate smart agriculture to secure nutrition and health. Front. Plant Sci. 13, 902536. doi:10.3389/fpls.2022.902536
Babu, S., Vellore, N. A., Kasibotla, A. V., Dwayne, H. J., Stubblefield, M. A., and Uppu, R. M. (2012). Molecular docking of bisphenol A and its nitrated and chlorinated metabolites onto human estrogen-related receptor-gamma. Biochem. Biophys. Res. Commun. 426, 215–220. doi:10.1016/j.bbrc.2012.08.065
Begum, M., Paul, P., Das, D., and Ghosh, S. (2020). Endocrine-disrupting plasticizer Bisphenol A (BPA) exposure causes change in behavioral attributes in Drosophila melanogaster. Toxicol. Environ. Health Sci. 12, 237–246. doi:10.1007/s13530-020-00052-8
Bernier, M. R., and Vandenberg, L. N. (2017). Handling of thermal paper: implications for dermal exposure to bisphenol A and its alternatives. PLoS One 12, e0178449. doi:10.1371/journal.pone.0178449
Bochevarov, A. D., Harder, E., Hughes, T. F., Greenwood, J. R., Braden, D. A., Philipp, D. M., et al. (2013). Jaguar: a high-performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem. 113, 2110–2142. doi:10.1002/qua.24481
Daina, A., Michielin, O., and Zoete, V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 42717. doi:10.1038/srep42717
Delfosse, V., Grimaldi, M., Le Maire, A., Bourguet, W., and Balaguer, P. (2014). Nuclear receptor profiling of bisphenol-A and its halogenated analogues. Endocr. Disrupters 94, 229–251. doi:10.1016/B978-0-12-800095-3.00009-2
Eladak, S., Grisin, T., Moison, D., Guerquin, M.-J., N'tumba-Byn, T., Pozzi-Gaudin, S., et al. (2015). A new chapter in the bisphenol A story: bisphenol S and bisphenol F are not safe alternatives to this compound. Fertil. Steril. 103, 11–21. doi:10.1016/j.fertnstert.2014.11.005
Feng, L., Chen, S. J., Zhang, L. J., Qu, W., and Chen, Z. G. (2019). Bisphenol A increases intestinal permeability through disrupting intestinal barrier function in mice. Environ. Pollut. 254, 112960. doi:10.1016/j.envpol.2019.112960
Gao, H., Yang, B. J., Li, N., Feng, L. M., Shi, X. Y., Zhao, W. H., et al. (2015). Bisphenol A and hormone-associated cancers: current progress and perspectives. Med. Baltim. 94, e211. doi:10.1097/MD.0000000000000211
Genuis, S. J., Beesoon, S., Birkholz, D., and Lobo, R. A. (2012). Human excretion of bisphenol A: blood, urine, and sweat (BUS) study. J. Environ. Public Health 2012, 185731. doi:10.1155/2012/185731
Gruber, C. J., Tschugguel, W., Schneeberger, C., and Huber, J. C. (2002). Production and actions of estrogens. N. Engl. J. Med. 346, 340–352. doi:10.1056/NEJMra000471
Hartung, I. V., Huck, B. R., and Crespo, A. (2023). Rules were made to be broken. Nat. Rev. Chem. 7, 3–4. doi:10.1038/s41570-022-00451-0
Huo, X., Chen, D., He, Y., Zhu, W., Zhou, W., and Zhang, J. (2015). Bisphenol-A and female infertility: a possible role of gene-environment interactions. Int. J. Environ. Res. Public Health 12, 11101–11116. doi:10.3390/ijerph120911101
Kim, D. K., Gang, G. T., Ryu, D., Koh, M., Kim, Y. N., Kim, S. S., et al. (2013). Inverse agonist of nuclear receptor ERRγ mediates antidiabetic effect through inhibition of hepatic gluconeogenesis. Diabetes 62, 3093–3102. doi:10.2337/db12-0946
Kim, D. K., Kim, J. R., Koh, M., Kim, Y. D., Lee, J. M., Chanda, D., et al. (2011). Estrogen-related receptor γ (ERRγ) is a novel transcriptional regulator of phosphatidic acid phosphatase, LIPIN1, and inhibits hepatic insulin signaling. J. Biol. Chem. 286, 38035–38042. doi:10.1074/jbc.M111.250613
Kim, D. K., Ryu, D., Koh, M., Lee, M. W., Lim, D., Kim, M. J., et al. (2012). Orphan nuclear receptor estrogen-related receptor γ (ERRγ) is key regulator of hepatic gluconeogenesis. J. Biol. Chem. 287, 21628–21639. doi:10.1074/jbc.M111.315168
Kodila, A., Franko, N., and Sollner Dolenc, M. (2023). A review on immunomodulatory effects of BPA analogues. Arch. Toxicol. 97, 1831–1846. doi:10.1007/s00204-023-03519-y
Konieczna, A., Rutkowska, A., and Rachon, D. (2015). Health risk of exposure to Bisphenol A (BPA). Rocz. Panstw Zakl. Hig. 66, 5–11.
Kuhn, B., Gerber, P., Schulz-Gasch, T., and Stahl, M. (2005). Validation and use of the MM-PBSA approach for drug discovery. J. Med. Chem. 48, 4040–4048. doi:10.1021/jm049081q
Kumar, A., Rani, M., Mani, S., Shah, P. L. V., Singh, D. B., Kudapa, H., et al. (2021a). Nutritional significance and antioxidant-mediated antiaging effects of finger millet: molecular insights and prospects. Front. Sustain. Food Syst. 5. doi:10.3389/fsufs.2021.684318
Kumar, A., Tripathi, M. K., Joshi, D., and Kumar, V. (2021b). Millets and millet technology. Springer.
Kumar, S., and Kotwal, N. (2023). Millets (Shrianna) and lifestyle diseases: a healing touch. Med. J. Armed Forces India 79, 249–252. doi:10.1016/j.mjafi.2023.04.001
Kumari, R., Kumar, R., Open Source Drug Discovery, C., and Lynn, A. (2014). g_mmpbsa--a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model 54, 1951–1962. doi:10.1021/ci500020m
Lipinski, C. A., Lombardo, F., Dominy, B. W., and Feeney, P. J. (2001). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26. doi:10.1016/s0169-409x(00)00129-0
Liu, X. H., Sakai, H., Nishigori, M., Suyama, K., Nawaji, T., Ikeda, S., et al. (2019). Receptor-binding affinities of bisphenol A and its next-generation analogs for human nuclear receptors. Toxicol. Appl. Pharmacol. 377, 114610. doi:10.1016/j.taap.2019.114610
Lydia Pramitha, J., Ganesan, J., Francis, N., Rajasekharan, R., and Thinakaran, J. (2022). Revitalization of small millets for nutritional and food security by advanced genetics and genomics approaches. Front. Genet. 13, 1007552. doi:10.3389/fgene.2022.1007552
Ma, Y., Liu, H., Wu, J., Yuan, L., Wang, Y., Du, X., et al. (2019). The adverse health effects of bisphenol A and related toxicity mechanisms. Environ. Res. 176, 108575. doi:10.1016/j.envres.2019.108575
Maia, E. H. B., Assis, L. C., De Oliveira, T. A., Da Silva, A. M., and Taranto, A. G. (2020). Structure-based virtual screening: from classical to artificial intelligence. Front. Chem. 8, 343. doi:10.3389/fchem.2020.00343
Matsushima, A., Kakuta, Y., Teramoto, T., Koshiba, T., Liu, X., Okada, H., et al. (2007). Structural evidence for endocrine disruptor bisphenol A binding to human nuclear receptor ERR gamma. J. Biochem. 142, 517–524. doi:10.1093/jb/mvm158
Matsushima, A., Teramoto, T., and Kakuta, Y. (2022). Crystal structure of endocrine-disrupting chemical bisphenol A and estrogen-related receptor γ. J. Biochem. 171, 23–25. doi:10.1093/jb/mvab145
Misra, J., Kim, D. K., and Choi, H. S. (2017). ERRγ: a junior orphan with a senior role in metabolism. Trends Endocrinol. Metabolism 28, 261–272. doi:10.1016/j.tem.2016.12.005
Mohanraj, K., Karthikeyan, B. S., Vivek-Ananth, R. P., Chand, R. P. B., Aparna, S. R., Mangalapandi, P., et al. (2018). IMPPAT: a curated database of Indian medicinal plants, Phytochemistry and therapeutics. Sci. Rep. 8, 4329. doi:10.1038/s41598-018-22631-z
Moon, M. K. (2019). Concern about the safety of bisphenol A substitutes. Diabetes & Metabolism J. 43, 46–48. doi:10.4093/dmj.2019.0027
Mustieles, V., D'cruz, S. C., Couderq, S., Rodriguez-Carrillo, A., Fini, J. B., Hofer, T., et al. (2020). Bisphenol A and its analogues: a comprehensive review to identify and prioritize effect biomarkers for human biomonitoring. Environ. Int. 144, 105811. doi:10.1016/j.envint.2020.105811
Nachman, R. M., Hartle, J. C., Lees, P. S., and Groopman, J. D. (2014). Early life metabolism of bisphenol A: a systematic review of the literature. Curr. Environ. Health Rep. 1, 90–100. doi:10.1007/s40572-013-0003-7
Nayak, D., Adiga, D., Khan, N. G., Rai, P. S., Dsouza, H. S., Chakrabarty, S., et al. (2022). Impact of bisphenol A on structure and function of mitochondria: a critical review. Rev. Environ. Contam. Toxicol. 260, 10. doi:10.1007/s44169-022-00011-z
Nelson, L. R., and Bulun, S. E. (2001). Estrogen production and action. J. Am. Acad. Dermatol 45, S116–S124. doi:10.1067/mjd.2001.117432
Neri, M., Virzi, G. M., Brocca, A., Garzotto, F., Kim, J. C., Ramponi, F., et al. (2015). In vitro cytotoxicity of bisphenol A in monocytes cell line. Blood Purif. 40, 180–186. doi:10.1159/000437039
Ohore, O. E., and Zhang, S. H. (2019). Endocrine disrupting effects of bisphenol A exposure and recent advances on its removal by water treatment systems. A review. Sci. Afr. 5, e00135. doi:10.1016/j.sciaf.2019.e00135
Oostenbrink, C., Villa, A., Mark, A. E., and Van Gunsteren, W. F. (2004). A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 25, 1656–1676. doi:10.1002/jcc.20090
Pall, S., Zhmurov, A., Bauer, P., Abraham, M., Lundborg, M., Gray, A., et al. (2020). Heterogeneous parallelization and acceleration of molecular dynamics simulations in GROMACS. J. Chem. Phys. 153, 134110. doi:10.1063/5.0018516
Pant, S., Verma, S., Pathak, R. K., and Singh, D. B. (2022). “Structure-based drug designing,” in Bioinformatics (Elsevier), 219–231.
Pathak, R. K., Gupta, A., Shukla, R., and Baunthiyal, M. (2018). Identification of new drug-like compounds from millets as Xanthine oxidoreductase inhibitors for treatment of Hyperuricemia: a molecular docking and simulation study. Comput. Biol. Chem. 76, 32–41. doi:10.1016/j.compbiolchem.2018.05.015
Pathak, R. K., Kim, D. Y., Lim, B., and Kim, J. M. (2022). Investigating multi-target antiviral compounds by screening of phytochemicals from neem (Azadirachta indica) against prrsv: a vetinformatics approach. Front. Vet. Sci. 9, 854528. doi:10.3389/fvets.2022.854528
Pathak, R. K., and Kim, J. M. (2024). Structural insight into the mechanisms and interacting features of endocrine disruptor Bisphenol A and its analogs with human estrogen-related receptor gamma. Environ. Pollut. 345, 123549. doi:10.1016/j.envpol.2024.123549
Pathak, R. K., Kim, W. I., and Kim, J. M. (2023). Targeting the PEDV 3CL protease for identification of small molecule inhibitors: an insight from virtual screening, ADMET prediction, molecular dynamics, free energy landscape, and binding energy calculations. J. Biol. Eng. 17, 29. doi:10.1186/s13036-023-00342-y
Pathak, R. K., Singh, D. B., Sagar, M., Baunthiyal, M., and Kumar, A. (2020). Computational approaches in drug discovery and design. Computer-aided drug Des., 1–21. doi:10.1007/978-981-15-6815-2_1
Peterson, J., Dwyer, J., Adlercreutz, H., Scalbert, A., Jacques, P., and Mccullough, M. L. (2010). Dietary lignans: physiology and potential for cardiovascular disease risk reduction. Nutr. Rev. 68, 571–603. doi:10.1111/j.1753-4887.2010.00319.x
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi:10.1002/jcc.20084
Pires, D. E., Blundell, T. L., and Ascher, D. B. (2015). pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 58, 4066–4072. doi:10.1021/acs.jmedchem.5b00104
Priya, , Verma, R. K., Lakhawat, S., Yadav, V. K., Gacem, A., Abbas, M., et al. (2023). Millets: sustainable treasure house of bioactive components. Int. J. Food Prop. 26, 1822–1840. doi:10.1080/10942912.2023.2236317
Rahman, M. S., Adegoke, E. O., and Pang, M. G. (2021). Drivers of owning more BPA. J. Hazard Mater 417, 126076. doi:10.1016/j.jhazmat.2021.126076
Rodriguez, J. P., Rahman, H., Thushar, S., and Singh, R. K. (2020). Healthy and resilient cereals and pseudo-cereals for marginal agriculture: molecular advances for improving nutrient bioavailability. Front. Genet. 11, 49. doi:10.3389/fgene.2020.00049
Saini, S., Saxena, S., Samtiya, M., Puniya, M., and Dhewa, T. (2021). Potential of underutilized millets as Nutri-cereal: an overview. J. Food Sci. Technology-Mysore 58, 4465–4477. doi:10.1007/s13197-021-04985-x
Schuttelkopf, A. W., and Van Aalten, D. M. (2004). PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D. Biol. Crystallogr. 60, 1355–1363. doi:10.1107/S0907444904011679
Sendra, M., Stampar, M., Fras, K., Novoa, B., Figueras, A., and Zegura, B. (2023). Adverse (geno)toxic effects of bisphenol A and its analogues in hepatic 3D cell model. Environ. Int. 171, 107721. doi:10.1016/j.envint.2022.107721
Sen Gupta, P. S., Bhat, H. R., Biswal, S., and Rana, M. K. (2020). Computer-aided discovery of bis-indole derivatives as multi-target drugs against cancer and bacterial infections: DFT, docking, virtual screening, and molecular dynamics studies. J. Mol. Liq. 320, 114375. doi:10.1016/j.molliq.2020.114375
Shah, P., Kumar, A., Kumar, V., and Tripathi, M. K. (2021). Millets, phytochemicals, and their health attributes. Millets Millet Technol., 191–218. doi:10.1007/978-981-16-0676-2_9
Singh, D. B., and Pathak, R. K. (2020). “Computational approaches in drug designing and their applications,” in Experimental protocols in biotechnology, 95–117.
Singh, D. B., Pathak, R. K., and Rai, D. (2022). From traditional herbal medicine to rational drug discovery: strategies, challenges, and future perspectives. Revista Brasileira De Farmacognosia-Brazilian J. Pharmacogn. 32, 147–159. doi:10.1007/s43450-022-00235-z
Siracusa, J. S., Yin, L., Measel, E., Liang, S., and Yu, X. (2018). Effects of bisphenol A and its analogs on reproductive health: a mini review. Reprod. Toxicol. 79, 96–123. doi:10.1016/j.reprotox.2018.06.005
Sirasanagandla, S. R., Al-Huseini, I., Sakr, H., Moqadass, M., Das, S., Juliana, N., et al. (2022). Natural products in mitigation of bisphenol A toxicity: future therapeutic use. Molecules 27, 5384. doi:10.3390/molecules27175384
Tohmé, M., Prud'homme, S. M., Boulahtouf, A., Samarut, E., Brunet, F., Bernard, L., et al. (2014). Estrogen-related receptor γ is an in vivo receptor of bisphenol A. FASEB J. 28, 3124–3133. doi:10.1096/fj.13-240465
Trott, O., and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. doi:10.1002/jcc.21334
Tschersich, C., Murawski, A., Schwedler, G., Rucic, E., Moos, R. K., Kasper-Sonnenberg, M., et al. (2021). Bisphenol A and six other environmental phenols in urine of children and adolescents in Germany - human biomonitoring results of the German Environmental Survey 2014-2017 (GerES V). Sci. Total Environ. 763, 144615. doi:10.1016/j.scitotenv.2020.144615
Vivek-Ananth, R. P., Mohanraj, K., Sahoo, A. K., and Samal, A. (2023). IMPPAT 2.0: an enhanced and expanded phytochemical atlas of Indian medicinal plants. ACS Omega 8, 8827–8845. doi:10.1021/acsomega.3c00156
Vom Saal, F. S., and Vandenberg, L. N. (2021). Update on the health effects of bisphenol A: overwhelming evidence of harm. Endocrinology 162, bqaa171. doi:10.1210/endocr/bqaa171
Wan, S., Bhati, A. P., Zasada, S. J., and Coveney, P. V. (2020). Rapid, accurate, precise and reproducible ligand-protein binding free energy prediction. Interface Focus 10, 20200007. doi:10.1098/rsfs.2020.0007
Wang, L., Zhang, Y., Liu, Y., Gong, X., Zhang, T., and Sun, H. (2019). Widespread occurrence of bisphenol A in daily clothes and its high exposure risk in humans. Environ. Sci. Technol. 53, 7095–7102. doi:10.1021/acs.est.9b02090
Xu, X., Hong, X., Xie, L., Li, T., Yang, Y., Zhang, Q., et al. (2012). Gestational and lactational exposure to bisphenol-A affects anxiety- and depression-like behaviors in mice. Horm. Behav. 62, 480–490. doi:10.1016/j.yhbeh.2012.08.005
Yasar, P., Ayaz, G., User, S. D., Gupur, G., and Muyan, M. (2017). Molecular mechanism of estrogen-estrogen receptor signaling. Reprod. Med. Biol. 16, 4–20. doi:10.1002/rmb2.12006
Keywords: BPA, ERRγ, millets, virtual screening, admet, molecular dynamics simulation
Citation: Pathak RK and Kim J-M (2024) A computational assay for identifying millet-derived compounds that antagonize the interaction between bisphenols and estrogen-related receptor gamma. Front. Pharmacol. 15:1435254. doi: 10.3389/fphar.2024.1435254
Received: 20 May 2024; Accepted: 21 October 2024;
Published: 31 October 2024.
Edited by:
Dmitry Nilov, Lomonosov Moscow State University, RussiaReviewed by:
Poonam Dhankhar, Cornell University, United StatesRajesh K. Das, University of North Bengal, India
Copyright © 2024 Pathak and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun-Mo Kim, anVubW9raW1AY2F1LmFjLmty
 Rajesh Kumar Pathak
Rajesh Kumar Pathak Jun-Mo Kim
Jun-Mo Kim