- 1Institute for Regenerative Medicine, Shanghai East Hospital, School of Life Sciences and Technology, Tongji University, Shanghai, China
- 2Department of Anesthesiology, The First Affiliated Hospital of Naval Medical University, Shanghai, China
- 3Tongji Alpha Natural Medicine Research Institute, Tongji University, Shanghai, China
Alzheimer’s disease (AD) is a complex neurodegenerative disease that affects elderly individuals, characterized by senile plaques formed by extracellular amyloid beta (Aβ). Autophagy dysfunction is a manifestation of protein homeostasis imbalance in patients with AD, but its relationship with Aβ remains unclear. Here, we showed that in Aβ transgenic Caenorhabditis elegans, Aβ activated the TOR pathway and reduced the nuclear entry of HLH-30, leading to autophagy dysfunction characterized by autophagosome accumulation. Then, utilizing RNA-seq, we investigated the regulatory mechanisms by which HLH-30 modulates autophagy in C. elegans. We found that HLH-30 elevated the transcript levels of v-ATPase and cathepsin, thus enhancing lysosomal activity. This led to an increase in autophagic flux, facilitating more pronounced degradation of Aβ. Moreover, HLH-30 reduced the level of ROS induction by Aβ and enhanced the antioxidant stress capacity of the worms through the gsto-1 gene. Additionally, we identified two HLH-30/TFEB activators, saikosaponin B2 and hypericin, that improved autophagic flux, thereby enhancing protein homeostasis in C. elegans. Overall, our findings suggested that HLH-30/TFEB plays a key role in modulating autophagy and can be considered a promising drug target for AD treatments.
Introduction
Alzheimer’s disease (AD) is characterized by the accumulation of amyloid beta (Aβ) peptides and hyperphosphorylated tau (p-Tau) protein in senile plaques and neurofibrillary tangles, respectively, which cause neurotoxicity and neuron loss, leading to memory and learning impairments (Silva et al., 2019). Therefore, the removal of misfolded proteins plays a significant role in maintaining protein homeostasis in the treatment of AD (Lukiw, 2022). The autophagy–lysosome pathway (ALP) is a cellular pathway in which cells utilize lysosomes to degrade long-lived proteins and organelles. The inability of the ALP to fully clear misfolded proteins has been recognized as important in the pathophysiology of AD (Cheng et al., 2023). Correspondingly, autophagy dysfunction is prevalent in AD patients (Ghavami et al., 2014); however, the mechanisms underlying this dysfunction are poorly understood.
The transcription factor EB (TFEB, known as HLH-30 in Caenorhabditis elegans), a member of the MiT/TFE family of transcription factors, is a key regulator of ALP (Martini-Stoica et al., 2016). It is translocated into the nucleus to coordinate the expression of autophagy and lysosomal target genes (Napolitano and Ballabio, 2016). The subcellular localization and activity of TFEB are regulated by mammalian target of rapamycin (mTOR)-mediated phosphorylation, which occurs at the lysosome surface (Zhu et al., 2019a). Phosphorylated TFEB is retained in the cytoplasm, whereas dephosphorylated TFEB translocates to the nucleus to induce the transcription of target genes (Puertollano et al., 2018). In AD patients, the level of TFEB in the nucleus is significantly reduced in cells in the hippocampus (Wang et al., 2016), which is the main brain region where autophagy dysfunction occurs (Nixon and Yang, 2011). Therefore, inhibition of TFEB nuclear translocation may be closely related to autophagy dysfunction. In our previous work, we reported impaired autophagic flux and abnormally elevated LET-363 (a mammalian ortholog of TOR) in Aβ transgenic Caenorhabditis elegans (C. elegans) (Lin et al., 2022). It has been reported that LET-363 can affect the nuclear localization of HLH-30 (Nakamura et al., 2016). However, the correlation between the aberrant upregulation of LET-363 expression and impaired autophagic function in Aβ transgenic worms has not yet been tested.
Accumulating studies have suggested that targeting TFEB is promising for the treatment of AD (Gu et al., 2022). However, there are very few effective and specific small molecule TFEB activators at present. The main known activators of TFEB are mostly mTOR inhibitors (Song et al., 2016). mTOR is well-known as a major regulator of cell growth and metabolism and is involved in a wide range of biological functions. Therefore, identifying TFEB-specific activators would be an important step in developing better therapeutics.
In this study, we determined that Aβ could activate TOR activity, inhibit the nuclear localization of HLH-30, and block the fusion of autophagosomes with lysosomes, thus leading to autophagy dysfunction in C. elegans. In addition, RNA sequencing to investigate the molecular mechanisms by which HLH-30 regulates autophagy revealed a close association between lysosomal function and autophagy dysfunction. Finally, we searched for HLH-30/TFEB-specific activators by molecular docking and found two active compounds, saikosaponin B2 and hypericin, that could specifically promote the nuclear entry of HLH-30 without inhibiting TOR activity, thereby enhancing autophagy and facilitating the degradation of Aβ.
Materials and methods
Strains and maintenance conditions
All strains used in this study are detailed in Supplementary Table S1. All strains were maintained and cultured under standard conditions at 20°C using E. coli OP50 as a food source, except CL4176, which was maintained at 16°C according to standard procedures.
Transgenic strain construction
The plasmid pMH878 (lgg-1p::mcherry::gfp::lgg-1) was donated by Malene Hansen (Chang et al., 2017). PHX3392, PHX3636 and PHX6883 were generated by SunyBiotech at our request. We knocked out the first four exons of the hlh-30 gene to construct PHX6883. Other strains were obtained by hybridization.
Gene expression analysis by quantitative PCR (qPCR)
Total RNA was extracted using TRIzol A+ and reverse transcribed into cDNA. The expressed genes were amplified in triplicate and quantified by PCR using SYBR Green PCR Mix (B21702, Bimake, China) with a Roche LightCycler system. The data were analyzed using the 2−ΔΔCT method (Lin et al., 2022). The primer list has been added to Supplementary Table S2.
Paralysis assay
Aβ transgenic CL4176 worms were maintained on NGM at 16°C and synchronized. L1 animals were treated with drugs or RNAi-engineered bacteria (or left untreated) for 36 h and then transferred to 23°C for transgene induction. This temperature shift stimulated Aβ expression, causing Aβ aggregation and leading to paralysis. Scoring began 27 h after the temperature shift, and the worms were considered paralyzed if they failed to move their bodies when touched and produced a “halo” of cleared bacterial lawn because they moved only their heads while feeding. We used PT50 (the time interval from the onset of paralysis at which 50% of the worms were paralyzed) as an index (Wu et al., 2006). For example, the PT50 of 4.2 h for the control was obtained by subtracting the time at which paralysis was first observed (29 h) from the time when 50% of the worms were paralyzed (33.2 h). The assay was performed at least three times. Each individual group contained more than 30 worms. Statistical analysis was conducted with GraphPad Prism 6.0 software, and p values were calculated using the log-rank test.
LysoTracker red staining
The acidophilic dye LysoTracker Red (C1046; Beyotime Biotechnology, Shanghai, China) was used at a final concentration of 15 μM. Synchronized strains maintained at 20°C were treated with LysoTracker from the L1 to the young adult stage and then transferred to 25°C for transgene induction. After 24 h, the worms were washed twice with fresh M9 (without LysoTracker) and imaged using a Revolve microscope (Imanikia et al., 2019).
RNA interference (RNAi)
RNAi experiments were performed according to a previously reported protocol. In brief, RNA was delivered to the worms by feeding, gravid adults were bleached, and the eggs were placed on NGM dishes supplemented with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). An HT-115(DE3) bacterial colony containing L4440 or the target gene plasmid was inoculated in LB broth supplemented with 100 μg/mL ampicillin and 12.5 μg/mL tetracycline and grown for 8 h in a 37°C shaker. The bacteria were plated on an NGM dish containing IPTG 1 h prior to the addition of the worms. The synchronized strain was grown on RNAi NGM plates from the egg stage to the adult stage for two generations. Synchronized L1 worms were treated with or without drugs for paralysis assays as described above (Zhang et al., 2016).
Measurement of reactive oxygen species (ROS)
Endogenous ROS levels were measured using 2′,7′-dichlorofluorescein diacetate (H2DCF–DA), which reacts with endogenous ROS to generate a fluorescent product (Yang et al., 2018). Worms were treated as in the paralysis assay, and ROS were measured at the point of paralysis. The cells were then incubated with 50 μM H2DCF–DA for 30 min at 37°C, and the fluorescence intensity was measured at excitation and emission wavelengths of 485 and 535 nm, respectively. The assay was performed in triplicate.
Oxidative stress resistance assay
Synchronized worms were cultured to the young adult stage. Then, the worms were transferred to a new 96-well plate containing 1 mM H2O2. The worms were monitored every 12 h and scored as dead when they no longer moved through the liquid medium (Chen et al., 2018).
HLH-30 translocation assay
The transgenic strain was used to observe the intracellular localization of HLH-30. Age-synchronized L1 worms were treated with or without 100 μM rapamycin for 3 days, mounted on 2% agarose pads and visualized under a fluorescence microscope (Revolve FL, Echo Laboratories, United States) for GFP localization. The nuclear accumulation of HLH-30::GFP was quantified by ImageJ (Visvikis et al., 2014). We use 4% paraformaldehyde as anesthetic. The assay was performed three times independently.
Western blotting
The worms were synchronized, and eggs were allowed to hatch and develop to the L4 stage on NGM plates at 20°C. Then, the temperature was increased to 25°C for 24 h. The worms were then collected from the plates with M9 buffer and washed twice to remove bacteria. Nuclear protein extraction was performed using a nuclear extract kit (Active Motif). The samples were heated at 100°C in sample loading buffer for 10 min and centrifuged at 10,000 × g for 10 min (Sangha et al., 2015). The collected supernatant was boiled with loading buffer at 100°C for 5 min before being loaded into the gel, which also included a 10–180 kDa protein molecular weight marker (PR1910, Solarbio, China) (Zhu et al., 2019b). The samples were run at 40 V for 40 min on a stacking gel and at 80 V for 120 min on a separating gel. The gel was then transferred to a polyvinylidene fluoride membrane using 20% methanol transfer buffer at 100 V for 1 h. The membranes were then blocked in Tris-buffered saline with Tween 20 + 5% skim milk for 1 h. The level of phosphorylated RSKS-1 protein was detected with an anti-phospho-p70 S6 kinase antibody (dilution 1:2,000; 9205, Cell Signaling Technology), with an anti-β-actin antibody (dilution 1:2,000; 60008, Proteintech) as a control. The RSKS-1 protein level was detected with an anti-p70 S6 kinase antibody (1:2,000 dilution; 9202, Cell Signaling Technology). The Aβ protein level was detected with the 6E10 monoclonal antibody (dilution 1:500; 803014, BioLegend). The protein level of mCherry-GFP-LGG-1 was detected with an anti-GFP antibody (dilution 1:500 Beyotime Technology). Histone H3 (1:2,000 dilution; ab1791 dilution, Abcam) served as an internal reference protein for the nuclear protein experiments. The membrane was incubated with the primary antibody overnight at 4°C and then with the secondary antibody for 2 h at room temperature. Images were captured using a Chemiscope 3400 mini Western blot imaging system (Amersham Imager 600, GE, United States). The mean densities of the Aβ bands were analyzed using ImageJ software.
Hybridization experiment
Five to eight worms in the L3 stage were selected and heat shock cultured for 3.5 h at 33°C, after which they were transferred to 20°C for further culture. A number of males could be found among the offspring. The male worms were hybridized with hermaphroditic worms, and homozygous worms were identified by agarose electrophoresis screening (Fay, 2006).
Molecular docking
Core targets (large molecular receptors) and the corresponding active compounds (small molecule ligands) were selected for molecular docking analysis. The 3D structures of the core targets were downloaded from the PDB database (https://www.rcsb.org/). The 2D structures of the corresponding active compounds were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The raw files of the core targets and the corresponding active components were processed with the AutoDock (version 4.2) tool (Morris et al., 2009) and converted to PDBQT format for molecular docking. Binding activity was expressed as binding energy, with lower bonding energy corresponding to more stable docking (Morris et al., 2008). In general, a docking energy < −5 kcal/mol was considered to indicate good docking.
RNA sequencing (RNA-seq)
The worms were synchronized (Fabian and Johnson, 1994), and their eggs were allowed to hatch and develop to the young adult stage on NGM plates at 20°C. Then, the temperature was increased to 25°C to induce Aβ expression for 24 h. Gene expression was assessed by Novo Gene Corporation (Beijing, China). Sequencing libraries were generated using the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, United States). To preferentially select cDNA fragments 250∼300 bp in length, the library fragments were purified with the AMPure XP system (Beckman Coulter, Beverly, United States). Then, 3 µL of USER Enzyme (NEB, United States) was incubated with size-selected adaptor-ligated cDNA at 37°C for 15 min and then 5 min at 95°C. Then, PCR was performed with Phusion High-Fidelity DNA polymerase, universal PCR primers and Index (X) Primer. Finally, the PCR products were purified (AMPure XP system), and library quality was assessed on an Agilent Bioanalyzer 2100 system. Clustering of the index-coded samples was performed on a cBot Cluster Generation System using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the libraries were sequenced on an Illumina NovaSeq platform to generate 150 bp paired-end reads. Up- or downregulated genes were identified by filtering RNA-seq data with the following criteria: twofold change in expression level and a false discovery rate analog of p < 0.05. KEGG pathway enrichment analysis was performed using “clusterProfiler” with the criterion of p < 0.05.
Statistical analysis
All quantified data are presented as the mean ± SD. The data were analyzed by Student’s t test to determine statistical significance with GraphPad Prism 6.0 software (GraphPad, La Jolla, United States). For multiple comparisons, the results were analyzed by one-way analysis of variance (ANOVA). p < 0.05 was considered to indicate statistical significance.
Results
Aβ expression affects TOR-mediated HLH-30 nuclear localization
In previous studies, we found that both the transcription and protein levels of let-363 were significantly increased in Aβ transgenic C. elegans (Lin et al., 2022). Thus, we hypothesized that Aβ expression might affect TOR activity in C. elegans. To test this hypothesis, we evaluated the transcript levels of lmtr-2 and ragc-1, which regulate TOR activation (Charar et al., 2021; Blackwell et al., 2019), and found that they were obviously upregulated (Figure 1A), suggesting that Aβ expression induced TOR activation. To further explore the regulatory role of Aβ expression in TOR activation, we examined TOR activity by measuring the phosphorylation level of its substrate RSKS-1 (an ortholog of S6K). The results showed that RSKS-1 phosphorylation in the Aβ strain GMC101 was significantly greater than that in the control strain CL2122, while the total protein level of RSKS-1 remained unchanged (Figures 1B, C). These results suggested that Aβ expression could activate the TOR pathway. TOR activation prevents the nuclear translocation of HLH-30 (Blackwell et al., 2019). The strain JIN1821 is often used to observe the nuclear localization of HLH-30 (Visvikis et al., 2014). We crossed JIN1821 with CL4176 (a transgenic strain expressing Aβ) to observe the nuclear localization of HLH-30 in the presence of Aβ. Compared with that in the control group, the nuclear entry of HLH-30::GFP was obviously reduced in the Aβ expressing group (Figures 1D, E). Moreover, the nuclear localization of HLH-30::GFP was significantly restored after the worms were treated with the mTOR inhibitor rapamycin at 100 μM (Figures 1D, E). We also observed a significant decrease in the nuclear level of the HLH-30 protein in the Aβ group compared to that in the control group (Figures 1F, G). The above results suggested that Aβ affected the entry of HLH-30 into the nucleus by activating TOR and that this may be one of important factor causing the abnormal autophagy in Aβ transgenic worms.
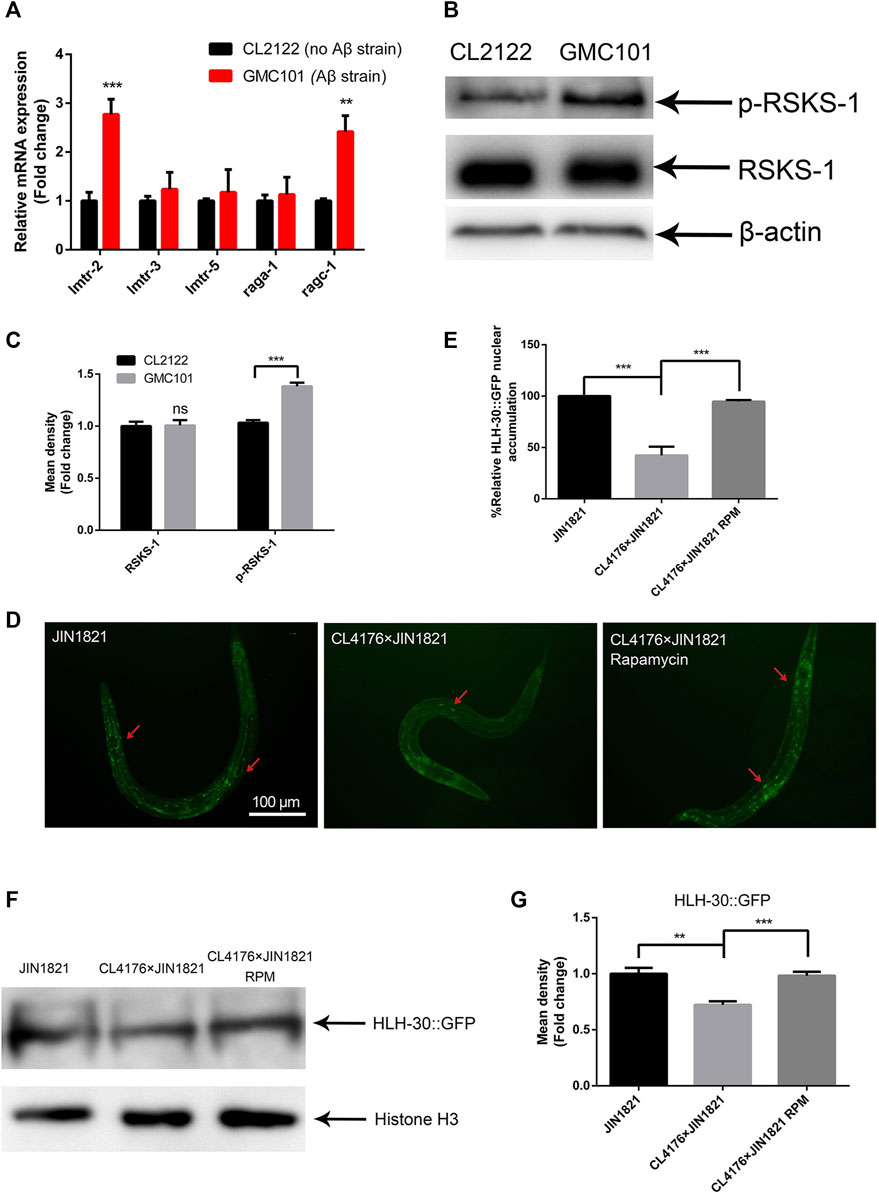
Figure 1. Aβ affects the nuclear localization of HLH-30 by activating mTOR. (A) mRNA expression of genes related to mTOR (strains GM101 and CL2122). (B) Representative Western blot showing phospho-RSKS-1 and total RSKS-1 in the CL2122 and GMC101 strains, three biological replicates. (C) Quantified Western blot gel intensities, as determined by ImageJ software. (D) GFP fluorescence images of HLH-30 entering the nucleus (n = 10 for each group, repeated three times). The dose of rapamycin was 100 μM. (E) Fluorescence signal ratio quantification. RPM, rapamycin. The HLH-30::GFP accumulation in JIN1821 was defined as 100% for quantitative comparisons. (F) Representative Western blot showing the content of HLH-30::GFP in the nucleus. Each experiment was repeated three times. (G) Quantified Western blot gel intensities, as determined by ImageJ software. **p < 0.01; ***p < 0.001. Detailed genotypic descriptions of strains CL2122, GMC101, and JIN1821 are listed in Supplementary Material.
The stability of autophagic flux in Aβ transgenic worms depends on HLH-30
Previous studies have shown that Aβ causes the accumulation of autophagosomes in C. elegans (Florez-McClure et al., 2007); however, the cause of this phenomenon is unknown. The transcription factor HLH-30 plays a crucial role in the autophagy‒lysosome pathway (Napolitano and Ballabio, 2016). To explore the relationship between HLH-30 and autophagy dysfunction in Aβ transgenic worms, we performed hlh-30 RNAi on the PHX3392 (Aβ) and PHX3636 (no Aβ) strains, which express mCherry::GFP::LGG-1 (genotypes are shown in Supplementary Table S1). Thus, the autophagosomes in these strains are labeled by both mCherry and GFP (Chang et al., 2017). The number of autophagosomes in the PHX3636 (no Aβ) group was significantly greater than that in the control group after hlh-30 RNAi treatment (Figures 2A, B). In addition, we found that the protein level of mCherry::GFP::LGG-1-Ⅱ was significantly increased (Figures 2C, D). This suggests that hlh-30 RNAi may result in autophagy dysfunction in worms. To further investigate the mechanism by which HLH-30 regulates autophagy, we generated hlh-30-overexpressing (hlh-30 ox) strains and hlh-30 (syb6883) knockout mutants on the basis of the Aβ strains. The use of GFP labeling in the construction of transgenic nematodes is necessary for the microscopic observation of mCherry::GFP::LGG-1. Therefore, we detected the expression level of mCherry::GFP::LGG-1 to evaluate autophagosome accumulation in worms. The results showed that overexpression of hlh-30 alleviated the accumulation of autophagosomes induced by Aβ, while hlh-30 knockout did not increase the accumulation of autophagosomes in Aβ worms (Figures 2E, F). Then, we compared the paralysis rates of hlh-30-overexpressing and hlh-30 knockout worms. Overexpression of hlh-30 obviously decreased the paralysis rate, while hlh-30 knockout accelerated the paralysis rate (Figure 2G). Finally, we found that the protein level of Aβ was significantly decreased in hlh-30-overexpressing worms (Figures 2H, I). These results indicated that the overexpression of hlh-30 could lead to the degradation of Aβ by enhancing autophagy.
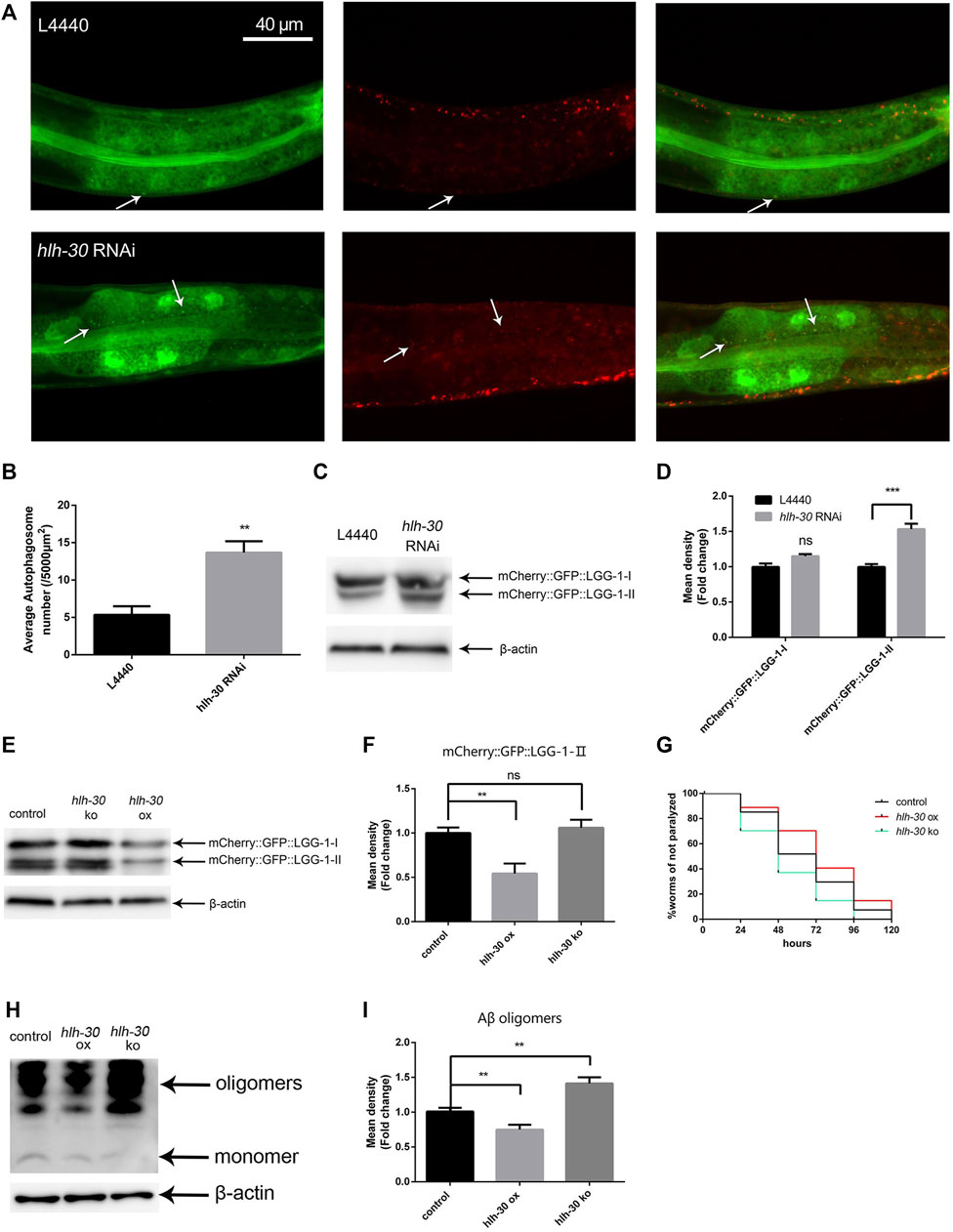
Figure 2. The stability of autophagic flux in Aβ transgenic worms depends on HLH-30. (A) Puncta formation in the PHX3636 strain as shown by fluorescence microscopy (strain PHX3636). (B) Scoring of punctae (n = 10 for each group, repeated three times). (C) Immunoblot analysis to evaluate the effects of hlh-30 RNAi on mCherry::GFP::LGG-1 expression (strain PHX3636). (D) Quantified Western blot gel intensities, as determined by ImageJ software. (E) Effects of hlh-30 overexpression or knockout on mCherry::GFP::LGG-1, as measured by immunoblot analysis (strains HBW003 and HBW004). “ox”, overexpression; “ko”, knockout. Each experiment was repeated three times. (F) Quantified Western blot gel intensities, as determined by ImageJ software. (G) Paralysis assay of hlh-30-overexpressing or hlh-30 knockout worms (n > 30 for each group, repeated three times). (H) Immunoblot analysis to evaluate the effects of hlh-30 overexpression or knockout on Aβ species. Each experiment was repeated three times. (I) Quantified Western blot gel intensities, as determined by ImageJ software. “ns”, not significant; **p < 0.01; ***p < 0.001. Detailed genotypic descriptions of strains PHX3636, HBW003, and HBW004 are listed in Supplementary Material.
RNA interference of hlh-30 reduces the transcript level of syx-17 and leads to autophagosome accumulation
In mammals, TFEB can regulate hundreds of ALP genes at the transcriptional level, so we hypothesized that HLH-30 may also regulate the transcription levels of ALP genes in worms. The expression levels of rab-7 and syx-17, which are related to autophagosome–lysosome fusion (Zhao et al., 2021), were found to be significantly decreased by hlh-30 RNAi (Figure 3A). It was reported that syx-17 RNAi could block autophagosome–lysosome fusion (Gkikas et al., 2023). Therefore, we investigated the association between the reduced expression of syx-17 and autophagy dysfunction caused by the decreased expression of HLH-30. We found that the number of autophagosomes in PHX3636 cells (without Aβ) increased significantly in response to syx-17 RNAi (Figures 3B, C). However, autophagosome accumulation after syx-17 and hlh-30 dual gene RNAi was not significantly greater than that after syx-17 RNAi in PHX3636. Similarly, the accumulation of autophagosomes in PHX3392 (Aβ) cells was not further elevated by syx-17 RNAi (Figures 3B, C). These results indicated that the decreased expression of HLH-30 reduced the expression of syx-17, resulting in the abnormal fusion of autophagosomes and lysosomes in Aβ nematodes and subsequent autophagy dysfunction.
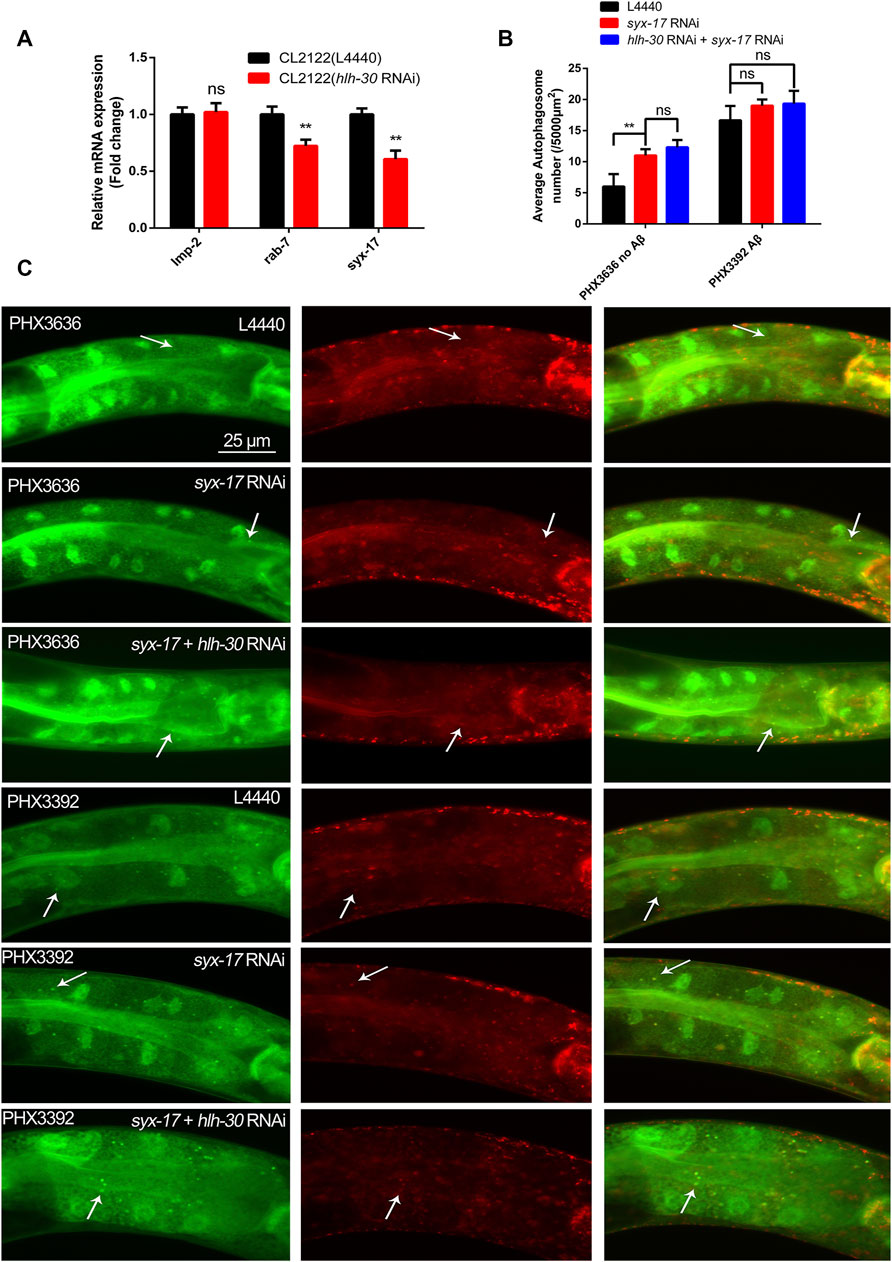
Figure 3. Downregulation of syx-17 at the transcriptional level leads to the accumulation of autophagosomes in worms. (A) Effects of hlh-30 RNAi on the mRNA expression of genes related to autophagosome‒lysosome fusion. (B) Effects of RNAi of syx-17 or hlh-30 on puncta formation in the PHX3636 (no Aβ) or PHX3392 (Aβ) strains, as shown by fluorescence microscopy. (C) Scoring of punctae (n = 10 for each group, repeated three times). “ns”, not significant; **p < 0.01. Detailed genotypic descriptions of strains PHX3636 and PHX3392 are listed in Supplementary Material.
HLH-30 maintains the stability of autophagic flux by enhancing lysosomal activity
To further explore how hlh-30 regulates autophagy to stabilize protein homeostasis in worms, we conducted RNA-seq analysis on strains in which hlh-30 was overexpressed or knocked out (Figure 4A). Through KEGG enrichment, we found that both the overexpression and knockout of hlh-30 affected the transcript levels of lysosome-related genes (Figures 4B, C), which was verified by qPCR (Supplementary Figure S1). The results showed that the v-ATPase and cathepsin B genes were significantly upregulated in the hlh-30 overexpression group (Supplementary Figure S1). To further evaluate the effect of hlh-30 overexpression on lysosomal function, we utilized LysoTracker Red to analyze lysosome acidification. The results showed that hlh-30 overexpression obviously enhanced lysosomal activity (Figures 4D, E).
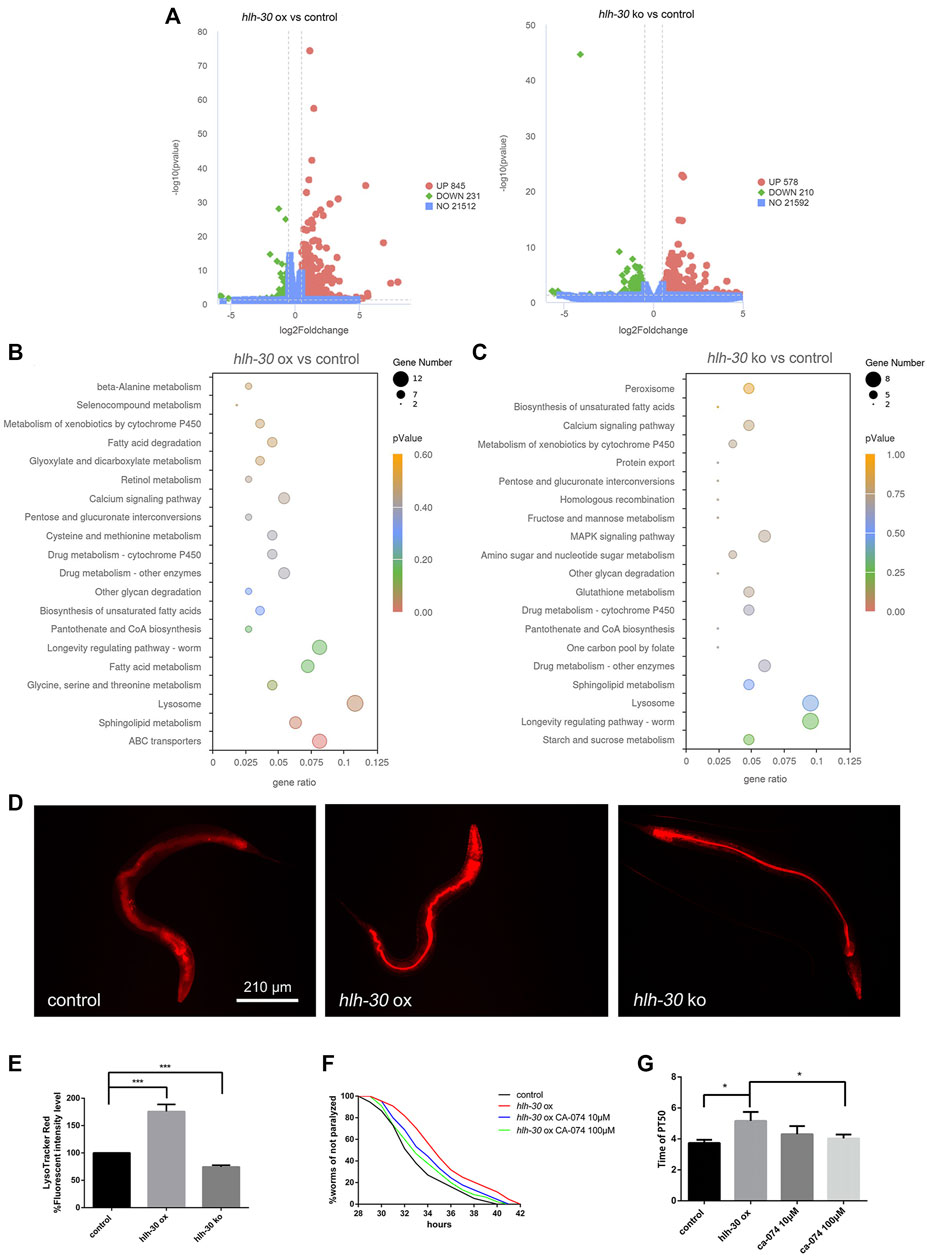
Figure 4. HLH-30 maintains the stability of autophagic flux by enhancing lysosomal activity. (A) RNA-seq differential gene volcano map. “ox”, overexpression; “ko”, knockout. (B) KEGG pathway enrichment of genes differentially expressed in the hlh-30-overexpressing strain. The x- and y-axes represent the GeneRatio and enriched KEGG pathways, respectively. The color represents the significance of the enrichment, and the bubble size represents the gene count. (C) KEGG pathway enrichment of genes differentially expressed in the hlh-30 knockout strain. (D) Representative fluorescence images of strains stained with LysoTracker Red after Aβ induction for 24 h (n = 10 for each group, repeated three times). The worms used in this study were overexpressed and knockout strains based on GMC101. (E) Quantified fluorescence intensities (n = 10 for each group, repeated three times). (F) Overexpression of hlh-30 delayed CL4176 paralysis in a cathepsin B-dependent manner (n > 30 for each group, repeated three times). The worms used in this study were overexpressed strains based on CL4176. (G) Statistical analysis of the half paralysis time of worms. *p < 0.05; ***p < 0.001.
Given the observed upregulation of cathepsin B transcription and enhancement of lysosomal activity, we hypothesized that HLH-30 promotes Aβ degradation by enhancing lysosomal activity and hydrolase expression, thereby improving protein homeostasis in worms. To further confirm the above results, we evaluated the impact of CA-074, a cathepsin-specific inhibitor (Bai et al., 2021), on the paralysis rate in hlh-30 Aβ-overexpressing worms. The results showed that the ability of hlh-30 overexpression to prolong paralysis was offset by the administration of CA-074 (Figures 4F, G). The above results suggested that lysosomes may be involved in the maintenance of protein homeostasis through the overexpression of hlh-30.
HLH-30 maintains protein homeostasis by clearing ROS through gsto-1
In previous studies, we found that Aβ can cause severe oxidative stress, manifested by a large increase in reactive oxygen species (ROS) (Lin et al., 2022). Through an ROS assay, we found that ROS levels were significantly decreased by hlh-30 overexpression and increased in hlh-30(syb6883) mutants (Figure 5A). Furthermore, the survival time of hlh-30-overexpressing worms in an oxidative stress environment induced by hydrogen peroxide was increased by 10.3% compared with that in the control group (Figures 5B, C). It has been reported that the levels of glutathione within brain neurons are reduced in AD patients, which leads to increased sensitivity of the neurons to oxidative damage (Allen et al., 2012). Our RNA-seq analysis indicated that the transcription level of gsto-1, a glutathione transferase that is primarily involved in the metabolism of glutathione within cells, was significantly downregulated in hlh-30 mutants. Paralysis experiments revealed that the paralysis delaying and RO reducing effects of hlh-30 overexpression were reversed by gsto-1 RNAi (Figures 5D, E). The augmented antioxidant capacity following hlh-30 overexpression was also closely associated with gsto-1 (Figure 5F). These findings indicate that gsto-1 could potentially contribute to the regulation of oxidative stress and the maintenance of protein homeostasis in worms by hlh-30.
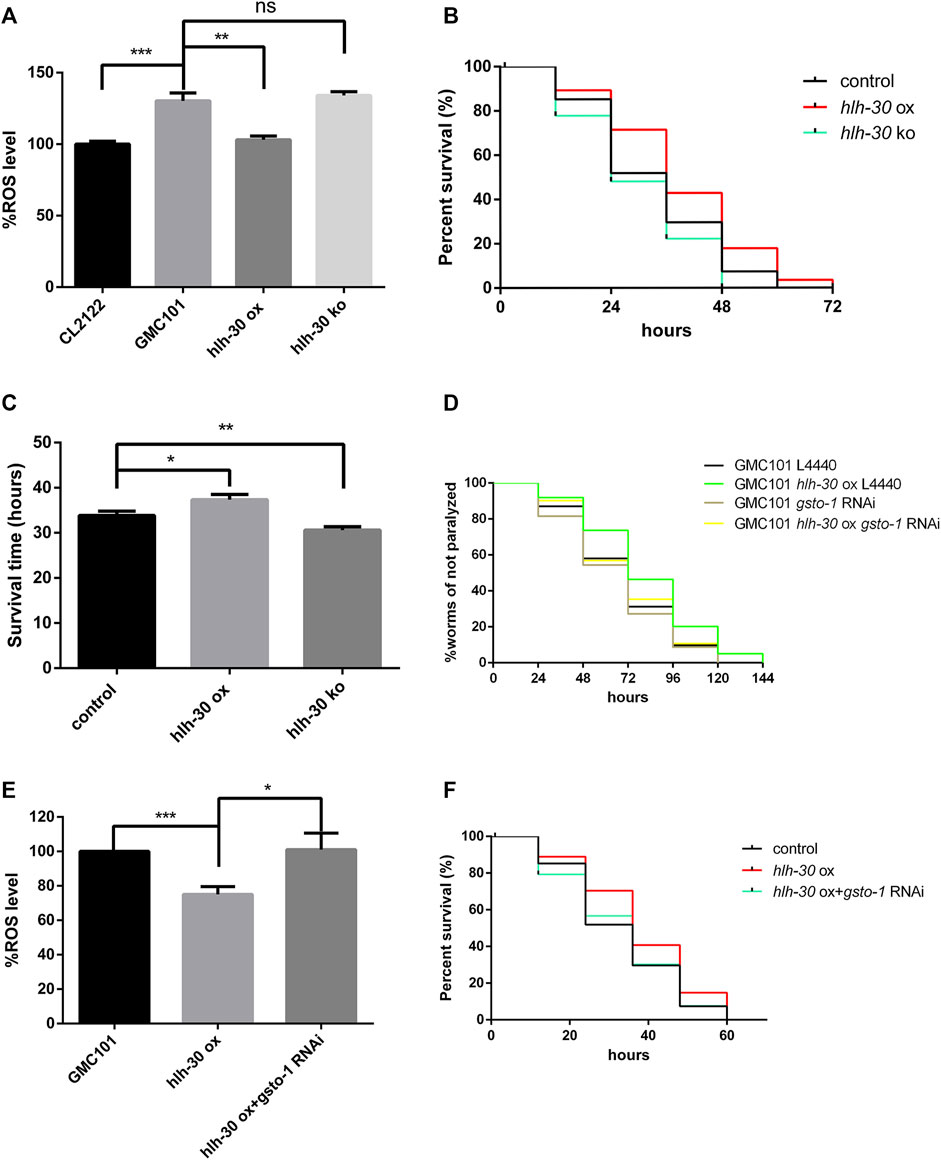
Figure 5. HLH-30 maintains protein homeostasis by clearing ROS through gsto-1. (A) Levels of ROS in transgenic worms (n > 30 for each group, repeated three times). “ox”, overexpression; “ko”, knockout. The worms used in this study were overexpressed strains based on GMC101. (B) Evaluation of the antioxidant capacity of transgenic worms (n > 30 for each group, repeated three times). The worms used in this study were overexpressed strains based on CL4176. (C) Survival time histogram under oxidative stress induced by hydrogen peroxide. (D) Paralysis assay of RNAi worms (n > 30 for each group, repeated three times). (E) Levels of ROS in transgenic worms (n > 30 for each group, repeated three times). (F) Evaluation of the antioxidant capacity of the transgenic worms (n > 30 for each group, repeated three times). “ns”, not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
Saikosaponin B2 and hypericin ameliorate the proteostasis imbalance caused by Aβ by promoting HLH-30 translocation to the nucleus and reducing autophagosome accumulation
TFEB/HLH-30 regulators that do not inhibit the mTOR pathway are preferred as they may have less harmful effects on cells (Song et al., 2016). The curcumin analog C1, a TFEB-specific activator, has been found to significantly reduce Aβ and tau aggregation and improve synaptic and memory functions in different AD animal models (Song et al., 2020). We used C1 as a ligand for molecular docking to identify the active site of TFEB (Figure 6A). Then, based on the predicted active site, we screened thousands of small molecule compounds derived from natural products. Subsequently, we evaluated the impact of the top ten small molecular compounds (Supplementary Table S3), as determined by docking scores, on the paralysis rate of the Aβ strain CL4176 (Supplementary Figure S2).
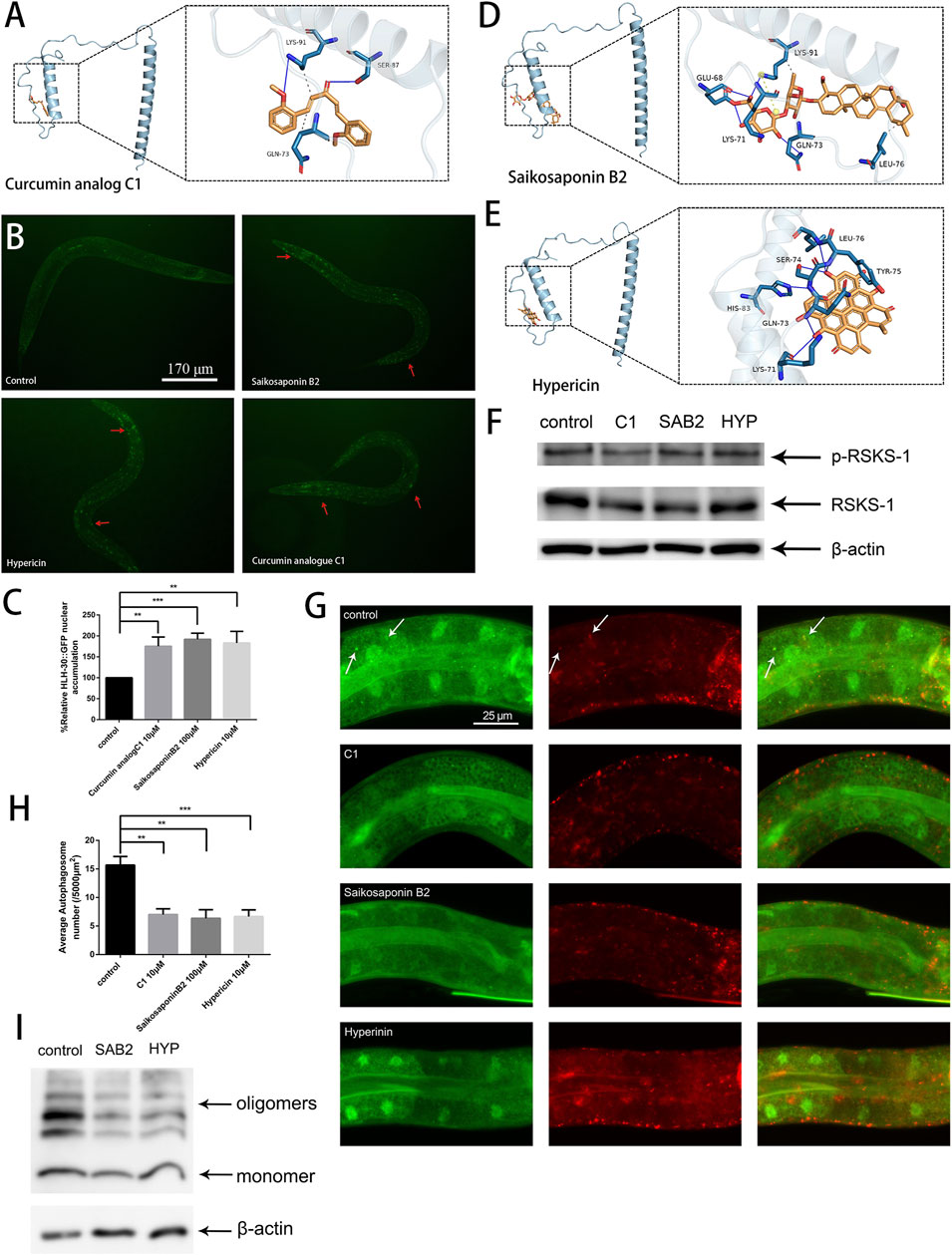
Figure 6. Saikosaponin B2 and hypericin improve protein homeostasis by reducing autophagosome accumulation and alleviating oxidative stress. (A) Possible binding sites of the curcumin analog C1. (B) GFP fluorescence images of HLH-30 in the nucleus (n = 10 for each group, repeated three times, strain HBW001). (C) Fluorescence signal ratio quantification. The HLH-30::GFP accumulation in the control group was defined as 100% for quantitative comparisons. (D) Possible sites of action of saikosaponin B2. (E) Possible sites of action of hypericin. (F) Immunoblot analysis to evaluate the effects of C1 (C1), saikosaponin B2 (SAB2), and hypericin (HYP) on the phosphorylation of RSKS-1 and total RSKS-1. (G) Fluorescence microscopy analysis to evaluate the effects of the C1, saikosaponin B2, and hypericin on puncta formation in the PHX3392 strain. (H) Scoring of punctae (n = 10 for each group, repeated three times). (I) Immunoblot analysis to evaluate the effects of saikosaponin B2 (SAB2) and hypericin (HYP) on Aβ species (strain GMC101). **p < 0.01; ***p < 0.001. Detailed genotypic descriptions of strains HBW001, PHX3636 and GMC101 are listed in Supplementary Material.
By combining the results of the HLH-30::GFP nuclear localization and paralysis assays, we identified two active compounds, saikosaponin B2 and hypericin (Figures 6B, C). Molecular docking revealed that the binding sites of saikosaponin B2, hypericin, and the curcumin analog C1 on TFEB were relatively similar (Figures 6A, D, E). By detecting the phosphorylation levels of TOR and its downstream target protein RSKS-1, we confirmed that neither saikosaponin B2 nor hypericin could inhibit TOR activity (Figure 6F). Further, both saikosaponin B2 and hypericin could reduce autophagosome accumulation, maintain the stability of autophagic flux (Figures 6G, H) and promote Aβ degradation in worms (Figure 6I). Our results suggested that targeting TFEB/HLH-30 presents a promising alternative for discovering new treatments for AD.
Discussion
Previous studies have documented autophagy dysfunction in the brains of Alzheimer’s disease (AD) patients, characterized by the accumulation of autophagosomal vesicles (Yu et al., 2005). However, the underlying causes of this phenomenon remain elusive. Abnormal phosphorylation of the tau protein prevents the fusion of autophagosomes with lysosomes (Hamano et al., 2021). Protein misfolding, immune abnormalities, and loss of lysosomal activity can also lead to impaired autophagy (Cozzi and Ferrari, 2022). In this work, we used transgenic C. elegans to explore the association between Aβ and autophagy dysfunction. We found that the expression of Aβ in C. elegans activates the mTOR pathway, which inhibits the entry of the transcription factor HLH-30 into the nucleus (Figures 1B–D) and ultimately leads to the accumulation of autophagosomes (Figures 2A–C). These observations provide insight into the mechanisms underlying autophagy dysfunction in the brains of Alzheimer’s disease (AD) patients.
Autophagy is a double-edged sword in neurodegenerative diseases (Zhou and Soukas, 2019). Under normal conditions, autophagy is important for the degradation of misfolded proteins (Chua et al., 2022). However, when autophagy is abnormal, its induction alone increases the metabolic burden on brain cells (Zhang et al., 2021). As a transcription factor that can simultaneously induce autophagy and increase lysosomal activity, TFEB/HLH-30 is a worthy target for study. Here, we discovered that hlh-30 is the key gene involved in regulating protein homeostasis through our observations of an hlh-30-overexpressing strain and an hlh-30 (syb6883) mutant strain (Figures 2E, H). Through RNA-seq, we found that hlh-30 overexpression can significantly increase the level of cathepsin B transcripts and promote lysosomal activity. Cathepsin B has been reported to degrade Aβ (Hook et al., 2020). We identified the critical role of cathepsin B in the regulation of protein homeostasis mediated by HLH-30 through the use of specific cathepsin B inhibitors (Figures 4F, G). The lysosome, a key organelle that degrades misfolded proteins, plays a necessary role in maintaining protein homeostasis. With increasing age, lysosome function gradually weakens, resulting in waste materials and aging organelles in the cell not being effectively removed, which in turn accelerates the aging process of the cell (Colacurcio and Nixon, 2016). In this study, we found that overexpressing hlh-30 could enhance lysosomal acidification and the expression of cathepsins, which contribute positively to protein homeostasis. It is necessary to consider age-related lysosomal dysfunction when developing treatment strategies for TFEB-related Alzheimer’s disease.
Furthermore, we discovered that the regulation of protein homeostasis by hlh-30 is dependent on the involvement of gsto-1 (Figures 5D, F). The gene gsto-1 encodes glutathione transferase, which is involved in the synthesis and transport of glutathione within cells (Burmeister et al., 2008). Glutathione transferase is also an important factor in the crosstalk between apoptosis and autophagy (Mazari et al., 2023). However, there is insufficient research on the relationship between hlh-30 and gsto-1. Moreover, the interactions among apoptosis, autophagy dysfunction and severe oxidative stress contribute greatly to the difficulty of studying the disease mechanisms of AD (Twarowski and Herbet, 2023); therefore, finding suitable models and research methods is the key to advancing the study of these factors.
The known activators of TFEB/HLH-30 are mainly inhibitors of mTOR, such as rapamycin. As a major regulator of cell growth and metabolism, mTOR is involved in a wide range of biological functions (Saxton and Sabatini, 2017). Therefore, identifying TFEB-specific activators is a promising strategy, as it may offer reduced cytotoxicity to cells (Song et al., 2021). We found that two natural products, saikosaponin B2 and hypericin, have potential as TFEB activators. The binding sites of saikosaponin B2, hypericin, and the curcumin analog C1 are located at the N-terminus of TFEB (Figures 6A, D, E), suggesting that the development of activators targeting TFEB should focus on the N-terminal protein structure of TFEB. Furthermore, both saikosaponin B2 and hypericin improved autophagy dysfunction and promoted the degradation of Aβ (Figures 6G, I) without inhibiting TOR activity (Figure 6F). In summary, our findings support a close association between inhibited nuclear entry of HLH-30/TFEB and autophagy dysfunction in AD and indicate that identifying TFEB-specific activators is a promising strategy for treating AD.
Conclusion
We report that Aβ-induced autophagy dysfunction in C. elegans may be associated with the inhibition of nuclear translocation of the transcription factor HLH-30. This could lead to a reduction in the expression of the membrane fusion gene syx-17, which is a key molecular event in Aβ-induced autophagy dysfunction. Overexpression of the hlh-30 gene appears to enhance autophagy activity and may decrease autophagosome accumulation, potentially promoting Aβ degradation. In addition, two potential HLH-30/TFEB activators, saikosaponin B2 and hypericin, were found by molecular docking and demonstrated to promote the nuclear entry of HLH-30, thereby enhancing autophagy and facilitating the degradation of Aβ.
Data availability statement
The raw data supporting the findings of the study are publicly available and can be accessed through the NCBI Gene Expression Omnibus (GEO) database with the accession number GSE272146: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE272146.
Author contributions
HL: Investigation, Validation, Formal Analysis, Writing–original draft. CZ: Investigation, Data curation, Formal Analysis, Writing–original draft. YG: Data curation, Writing–original draft. YZ: Formal Analysis, Writing–original draft. BM: Data curation, Writing–original draft. JJ: Formal Analysis, Writing–original draft. XL: Data curation, Writing–original draft. NY: Software, Writing–original draft. KZ: Software, Writing–review and editing. YL: Validation, Writing–review and editing. XC: Conceptualization, Writing–review and editing. HW: Project administration, Funding acquisition, Supervision, Validation, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (Grant no. 31670347 and 32370420) and Natural Science Foundation of Shanghai (23ZR1467200).
Acknowledgments
We thank for Springer Nature Author Services for editing the English text of a draft of this manuscript. We are grateful to Jian Fei (Tongji University, China) for their technical assistance. We thanked Ben YZ (Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences) for the strain JIN1821 and Malene Hansen (Sanford Burnham Prebys Medical Discovery Institute, United States) for the plasmid pMH878.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1433030/full#supplementary-material
Abbreviations
AD, Alzheimer’s disease; ALP, Autophagy lysosome pathway; Aβ, Amyloid beta; C1, Curcumin analog C1; C. elegans, Caenorhabditis elegans; GFP, Green fluorescent protein; mCherry, Cherry fluorescent protein; mTOR, Mammalian target of rapamycin; RNAi, RNA interference; ROS, Reactive oxygen species; S6K, Substrate S6 kinase; TFEB, Transcription factor EB; TOR, Target of rapamycin; v-ATPase, Vacuolar ATPase.
References
Allen, M., Zou, F., Chai, H. S., Younkin, C. S., Miles, R., Nair, A. A., et al. (2012). Glutathione S-transferase omega genes in Alzheimer and Parkinson disease risk, age-at-diagnosis and brain gene expression: an association study with mechanistic implications. Mol. Neurodegener. 7, 13. doi:10.1186/1750-1326-7-13
Bai, G., Gao, Y., Liu, S., Shui, S., and Liu, G. (2021). pH-dependent rearrangement determines the iron-activation and antitumor activity of artemisinins. Free Radic. Biol. Med. 163, 234–242. doi:10.1016/j.freeradbiomed.2020.12.024
Blackwell, T. K., Sewell, A. K., Wu, Z., and Han, M. (2019). TOR signaling in Caenorhabditis elegans development, metabolism, and aging. Aging. Genet. 213, 329–360. doi:10.1534/genetics.119.302504
Burmeister, C., Luersen, K., Heinick, A., Hussein, A., Domagalski, M., Walter, R. D., et al. (2008). Oxidative stress in Caenorhabditis elegans: protective effects of the Omega class glutathione transferase (GSTO-1). FASEB J. 22, 343–354. doi:10.1096/fj.06-7426com
Chang, J. T., Kumsta, C., Hellman, A. B., Adams, L. M., and Hansen, M. (2017). Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. Elife 6, e18459. doi:10.7554/eLife.18459
Charar, C., Metsuyanim-Cohen, S., and Bar, D. Z. (2021). Lamin regulates the dietary restriction response via the mTOR pathway in Caenorhabditis elegans. J. Cell Sci. 134, jcs258428. doi:10.1242/jcs.258428
Chen, W., Lin, H. R., Wei, C. M., Luo, X. H., Sun, M. L., Yang, Z. Z., et al. (2018). Echinacoside, a phenylethanoid glycoside from Cistanche deserticola, extends lifespan of Caenorhabditis elegans and protects from Aβ-induced toxicity. Biogerontology 19, 47–65. doi:10.1007/s10522-017-9738-0
Cheng, L., Chen, Y., Guo, D., Zhong, Y., Li, W., Lin, Y., et al. (2023). mTOR-dependent TFEB activation and TFEB overexpression enhance autophagy-lysosome pathway and ameliorate Alzheimer's disease-like pathology in diabetic encephalopathy. Cell Commun. Signal 21, 91. doi:10.1186/s12964-023-01097-1
Chua, J. P., De Calbiac, H., Kabashi, E., and Barmada, S. J. (2022). Autophagy and ALS: mechanistic insights and therapeutic implications. Autophagy 18, 254–282. doi:10.1080/15548627.2021.1926656
Colacurcio, D. J., and Nixon, R. A. (2016). Disorders of lysosomal acidification-The emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res. Rev. 32, 75–88. doi:10.1016/j.arr.2016.05.004
Cozzi, M., and Ferrari, V. (2022). Autophagy dysfunction in ALS: from transport to protein degradation. J. Mol. Neurosci. 72, 1456–1481. doi:10.1007/s12031-022-02029-3
Fabian, T. J., and Johnson, T. E. (1994). Production of age-synchronous mass-cultures of caenorhabditis-elegans. Journals Gerontology 49, B145–B156. doi:10.1093/geronj/49.4.B145
Fay, D. (2006). Genetic mapping and manipulation: chapter 7--Making compound mutants. WormBook 14, 1–5. doi:10.1895/wormbook.1.96.2
Florez-McClure, M. L., Hohsfield, L. A., Fonte, G., Bealor, M. T., and Link, C. D. (2007). Decreased insulin-receptor signaling promotes the autophagic degradation of beta-amyloid peptide in C. elegans. Autophagy 3, 569–580. doi:10.4161/auto.4776
Ghavami, S., Shojaei, S., Yeganeh, B., Ande, S. R., Jangamreddy, J. R., Mehrpour, M., et al. (2014). Autophagy and apoptosis dysfunction in neurodegenerative disorders. Prog. Neurobiol. 112, 24–49. doi:10.1016/j.pneurobio.2013.10.004
Gkikas, I., Daskalaki, I., Kounakis, K., Tavernarakis, N., and Lionaki, E. (2023). MitoSNARE assembly and disassembly factors regulate basal autophagy and aging in C. elegans. Int. J. Mol. Sci. 24, 4230. doi:10.3390/ijms24044230
Gu, Z., Cao, H., Zuo, C., Huang, Y., Miao, J., Song, Y., et al. (2022). TFEB in Alzheimer's disease: from molecular mechanisms to therapeutic implications. Neurobiol. Dis. 173, 105855. doi:10.1016/j.nbd.2022.105855
Hamano, T., Enomoto, S., Shirafuji, N., Ikawa, M., Yamamura, O., Yen, S. H., et al. (2021). Autophagy and tau protein. Int. J. Mol. Sci. 22, 7475. doi:10.3390/ijms22147475
Hook, V., Yoon, M., Mosier, C., Ito, G., Podvin, S., Head, B. P., et al. (2020). Cathepsin B in neurodegeneration of Alzheimer's disease, traumatic brain injury, and related brain disorders. Biochim. Biophys. Acta Proteins Proteom 1868, 140428. doi:10.1016/j.bbapap.2020.140428
Imanikia, S., Ozbey, N. P., Krueger, C., Casanueva, M. O., and Taylor, R. C. (2019). Neuronal XBP-1 activates intestinal lysosomes to improve proteostasis in C. elegans. Curr. Biol. 29, 2322–2338. doi:10.1016/j.cub.2019.06.031
Lin, H., Gao, Y., Zhang, C., Ma, B., Wu, M., Cui, X., et al. (2022). Autophagy regulation influences β-amyloid toxicity in transgenic Caenorhabditis elegans. Front. Aging Neurosci. 14, 885145. doi:10.3389/fnagi.2022.885145
Lukiw, W. J. (2022). Recent advances in our molecular and mechanistic understanding of misfolded cellular proteins in Alzheimer's disease (AD) and prion disease (PrD). Biomolecules 12, 166. doi:10.3390/biom12020166
Martini-Stoica, H., Xu, Y., Ballabio, A., and Zheng, H. (2016). The autophagy-lysosomal pathway in neurodegeneration: a TFEB perspective. Trends Neurosci. 39, 221–234. doi:10.1016/j.tins.2016.02.002
Mazari, A. M. A., Zhang, L., Ye, Z. W., Zhang, J., Tew, K. D., and Townsend, D. M. (2023). The multifaceted role of glutathione S-transferases in Health and disease. Biomolecules 13, 688. doi:10.3390/biom13040688
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., et al. (2009). AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. doi:10.1002/jcc.21256
Morris, G. M., Huey, R., and Olson, A. J. (2008). Using AutoDock for ligand-receptor docking. Curr. Protoc. Bioinforma. 8, Unit 8.14–14. Chapter. doi:10.1002/0471250953.bi0814s24
Nakamura, S., Karalay, O., Jager, P. S., Horikawa, M., Klein, C., Nakamura, K., et al. (2016). Mondo complexes regulate TFEB via TOR inhibition to promote longevity in response to gonadal signals. Nat. Commun. 7, 10944. doi:10.1038/ncomms10944
Napolitano, G., and Ballabio, A. (2016). TFEB at a glance. J. Cell Sci. 129, 2475–2481. doi:10.1242/jcs.146365
Nixon, R. A., and Yang, D. S. (2011). Autophagy failure in Alzheimer's disease--locating the primary defect. Neurobiol. Dis. 43, 38–45. doi:10.1016/j.nbd.2011.01.021
Puertollano, R., Ferguson, S. M., Brugarolas, J., and Ballabio, A. (2018). The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 37, e98804. doi:10.15252/embj.201798804
Sangha, J. S., Wally, O., Banskota, A. H., Stefanova, R., Hafting, J. T., Critchley, A. T., et al. (2015). A cultivated form of a red seaweed (chondrus crispus), suppresses β-amyloid-induced paralysis in Caenorhabditis elegans. Mar. Drugs 13, 6407–6424. doi:10.3390/md13106407
Saxton, R. A., and Sabatini, D. M. (2017). mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976. doi:10.1016/j.cell.2017.02.004
Silva, M. V. F., Loures, C. M. G., Alves, L. C. V., de Souza, L. C., Borges, K. B. G., and Carvalho, M. D. G. (2019). Alzheimer's disease: risk factors and potentially protective measures. J. Biomed. Sci. 26, 33. doi:10.1186/s12929-019-0524-y
Song, J. X., Liu, J., Jiang, Y., Wang, Z. Y., and Li, M. (2021). Transcription factor EB: an emerging drug target for neurodegenerative disorders. Drug Discov. Today 26, 164–172. doi:10.1016/j.drudis.2020.10.013
Song, J. X., Malampati, S., Zeng, Y., Durairajan, S. S. K., Yang, C. B., Tong, B. C., et al. (2020). A small molecule transcription factor EB activator ameliorates beta-amyloid precursor protein and Tau pathology in Alzheimer's disease models. Aging Cell 19, e13069. doi:10.1111/acel.13069
Song, J. X., Sun, Y. R., Peluso, I., Zeng, Y., Yu, X., Lu, J. H., et al. (2016). A novel curcumin analog binds to and activates TFEB in vitro and in vivo independent of mTOR inhibition. Autophagy 12, 1372–1389. doi:10.1080/15548627.2016.1179404
Twarowski, B., and Herbet, M. (2023). Inflammatory processes in Alzheimer's disease-pathomechanism, diagnosis and treatment: a review. Int. J. Mol. Sci. 24, 6518. doi:10.3390/ijms24076518
Visvikis, O., Ihuegbu, N., Labed, S. A., Luhachack, L. G., Alves, A. F., Wollenberg, A. C., et al. (2014). Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity 40, 896–909. doi:10.1016/j.immuni.2014.05.002
Wang, H., Wang, R., Xu, S., and Lakshmana, M. K. (2016). Transcription factor EB is selectively reduced in the nuclear fractions of Alzheimer's and amyotrophic lateral sclerosis brains. Neurosci. J. 2016, 4732837. doi:10.1155/2016/4732837
Wu, Y., Wu, Z., Butko, P., Christen, Y., Lambert, M. P., Klein, W. L., et al. (2006). Amyloid-beta-induced pathological behaviors are suppressed by Ginkgo biloba extract EGb 761 and ginkgolides in transgenic Caenorhabditis elegans. J. Neurosci. 26, 13102–13113. doi:10.1523/JNEUROSCI.3448-06.2006
Yang, Z. Z., Yu, Y. T., Lin, H. R., Liao, D. C., Cui, X. H., and Wang, H. B. (2018). Lonicera japonica extends lifespan and healthspan in Caenorhabditis elegans. Free Radic. Biol. Med. 129, 310–322. doi:10.1016/j.freeradbiomed.2018.09.035
Yu, W. H., Cuervo, A. M., Kumar, A., Peterhoff, C. M., Schmidt, S. D., Lee, J. H., et al. (2005). Macroautophagy--a novel Beta-amyloid peptide-generating pathway activated in Alzheimer's disease. J. Cell Biol. 171, 87–98. doi:10.1083/jcb.200505082
Zhang, X. G., Wang, X., Zhou, T. T., Wu, X. F., Peng, Y., Zhang, W. Q., et al. (2016). Scorpion venom heat-resistant peptide protects transgenic Caenorhabditis elegans from β-amyloid toxicity. Front. Pharmacol. 7, 227. doi:10.3389/fphar.2016.00227
Zhang, Z., Yang, X., Song, Y. Q., and Tu, J. (2021). Autophagy in Alzheimer's disease pathogenesis: therapeutic potential and future perspectives. Ageing Res. Rev. 72, 101464. doi:10.1016/j.arr.2021.101464
Zhao, Y. G., Codogno, P., and Zhang, H. (2021). Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat. Rev. Mol. Cell Biol. 22, 733–750. doi:10.1038/s41580-021-00392-4
Zhou, B., and Soukas, A. A. (2019). Suppressing the dark side of autophagy. Autophagy 15, 1852–1853. doi:10.1080/15548627.2019.1644077
Zhu, Z., Yang, C., Iyaswamy, A., Krishnamoorthi, S., Sreenivasmurthy, S. G., Liu, J., et al. (2019a). Balancing mTOR signaling and autophagy in the treatment of Parkinson's disease. Int. J. Mol. Sci. 20, 728. doi:10.3390/ijms20030728
Keywords: HLH-30, amyloid beta, autophagy, HLH-30/TFEB activators, Caenorhabditis elegans
Citation: Lin H, Zhang C, Gao Y, Zhou Y, Ma B, Jiang J, Long X, Yimamu N, Zhong K, Li Y, Cui X and Wang H (2024) HLH-30/TFEB modulates autophagy to improve proteostasis in Aβ transgenic Caenorhabditis elegans. Front. Pharmacol. 15:1433030. doi: 10.3389/fphar.2024.1433030
Received: 15 May 2024; Accepted: 16 August 2024;
Published: 30 August 2024.
Edited by:
Danilo Bilches Medinas, University of São Paulo, BrazilReviewed by:
Evandro A De-Souza, University of São Paulo, BrazilYina Dong, Children’s Hospital of Philadelphia, United States
Copyright © 2024 Lin, Zhang, Gao, Zhou, Ma, Jiang, Long, Yimamu, Zhong, Li, Cui and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongbing Wang, aGJ3YW5nQHRvbmdqaS5lZHUuY24=; Xianghuan Cui, Y3VpeGhAdG9uZ2ppLmVkdS5jbg==
 Hongru Lin
Hongru Lin Chen Zhang1
Chen Zhang1 Yi Zhou
Yi Zhou Hongbing Wang
Hongbing Wang