
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 23 August 2024
Sec. Pharmacogenetics and Pharmacogenomics
Volume 15 - 2024 | https://doi.org/10.3389/fphar.2024.1431923
This article is part of the Research Topic Use of Bioinformatics in Pharmacogenetics to Optimize Drug Efficacy View all 8 articles
 Ruixue Yuan1
Ruixue Yuan1 Yaojing Li2
Yaojing Li2 Xiangyi Li3
Xiangyi Li3 Yingmei Fu1
Yingmei Fu1 Ailing Ning1
Ailing Ning1 Dongxiang Wang1
Dongxiang Wang1 Ran Zhang1
Ran Zhang1 Shunying Yu1*
Shunying Yu1* Qingqing Xu1*
Qingqing Xu1*Background: Risperidone is one of the most reliable and effective antipsychotics for schizophrenia treatment. However, the mechanism of action of risperidone is not yet fully understood. Traf2 and Nck-interacting protein kinase (TNIK), a schizophrenia susceptibility gene, is associated with risperidone treatment response. Our previous in vitro experiments confirmed that downregulated TNIK affected the effect of risperidone on downstream targets. However, the effect of downregulated TNIK on risperidone-induced molecular expression remains to be further explored.
Methods: Transcriptome analysis was performed on U251 cells subjected to risperidone, TNIK siRNA, and no treatment, respectively. Compared to the no-treatment group, two groups of DEGs were screened out and then intersected with the schizophrenia-related genes to screen the cross-talk genes. Those DEGs were analyzed using GO and KEGG. STRING and Cytoscape were used to construct a protein-protein interaction (PPI) network for the cross-talk gene.
Results: The results showed that the parathyroid hormone synthesis, secretion, and action were significantly enriched after risperidone treatment. Downregulated TNIK could have an impact on the collagen-containing extracellular matrix, signaling receptor activator activity, and PI3K-Akt signaling pathway. Interestingly, bone mineralization function and calcium signaling pathway were enriched in the cross-talk genes. Additionally, FGFR2, FGF1, and FGFR might be the potential targets for TNIK affecting the effects of risperidone.
Conclusion: The study indicated that risperidone primarily influences functions and/or pathways associated with bone metabolism, potentially contributing to the adverse effect of osteoporosis. Our study may offer a novel perspective on investigating the mechanisms underlying the adverse effects of risperidone.
Schizophrenia (SCZ) is a chronic psychiatric illness characterized by psychotic symptoms including hallucinations, cognitive impairment, and delusions, leading to a social and occupational decline in many case (Hung et al., 2021; Jauhar et al., 2022). Patients usually need long-term medication to relieve the symptoms. Currently, antipsychotic drugs are the primary treatment for patients with SCZ via blocking dopaminergic and serotonergic receptors, but there is high inter-individual variability in terms of therapeutic benefits and side effects (Heide et al., 2016; Morozova et al., 2022). Approximately 20%–30% of SCZ patients are resistant to antipsychotic treatment and less than 40% achieve remission (Lisoway et al., 2021).
Risperidone (RPD), one of the widely prescribed antipsychotics for schizophrenia treatment, is a selective monoaminergic antagonist drug. It can be used to treat both positive and negative symptoms of SCZ, as well as alleviate the emotional symptoms associated with SCZ (Arani et al., 2023). However, there are a variety of side effects associated with RPD treatment, including weight gain, extrapyramidal symptoms, prolactin elevation, and osteoporosis (Zheng et al., 2019; Arani et al., 2023). For osteoporosis, many studies have suggested that long-term treatment with RPD increases the risk of osteoporosis by elevating serum prolactin levels (De Hert et al., 2016; Zheng et al., 2019; Chen Y. et al., 2021). Specifically, a recent study about 250 inpatients with schizophrenia receiving risperidone monotherapy has shown that patients have a high rate of osteoporosis (Chen Y. et al., 2021). However, Clapham et al. argued that RPD does not appear to be linked to an increased risk of osteoporosis-related fractures based on a population-based study (Clapham et al., 2020). Therefore, there is inconsistency among the available studies regarding the causal relationship between RPD and osteoporosis. The complex of therapeutic and adverse effects of RPD may derive from the variety of receptor targets. RPD treatment has multiple receptor actions, including blockade of serotonin 2A, 2C and 7 receptors and dopamine D2 receptor, α1 and α2-adrenergic receptors antagonism, and inverse agonist actions at histamine H1 receptors (Amodeo et al., 2014; Zheng et al., 2019). The blocking effect on dopamine D2 receptor can elevate prolactin secretion (Peuskens et al., 2014), which may be linked to elevated risk of osteoporosis. However, the effects of RPD are also influenced by other genetic factors. For example, Heide J et al. have reported that genotypes associated with increased Kv11.1-3.1 expression play a positive role in the therapeutic action of RPD (Heide et al., 2016). Two single-nucleotide polymorphisms (SNPs) rs155333 and rs6465938 of RELN were found to be associated with RPD response in the Chinese Han population (Xu et al., 2020). The study also revealed that apart from the previously identified genes CYP2D6, ABCB1, DRD3, and HTR2C, several new candidate genes, including TNIK, RELN, NOTCH4, and SLC6A2, were also identified to be associated with antipsychotic treatment response, including for RPD (Xu et al., 2016).
Since it is well established that genetic factors can influence the efficacy of RPD and may also partially explain the variation of treatment between patients (Yu et al., 2018), a better understanding of the genetic risk factors involved in medication response may offer potential guidance for clinicians in choosing a personalized and optimized medication based on their genetic profiles. Traf2 and Nck-interacting protein kinase (TNIK) may be a promising target to influence the efficacy of antipsychotics, including RPD. A genome-wide association study (GWAS) about Chinese Han inpatients with SCZ revealed five novel loci has shown that significant genome-wide association with general antipsychotic treatment response, one of which was rs6444970 in TNIK (p = 4.85 × 10−8) (Yu et al., 2018). Based on our preliminary study, we identified an association between several genes, including TNIK, and four antipsychotic drugs: risperidone, clozapine, quetiapine, and chlorpromazine (Xu et al., 2016). Subsequently, we specifically confirmed that variants rs2088885 and rs7627954 in TNIK were significantly associated with risperidone treatment response in a cohort of 148 unrelated inpatients who received risperidone monotherapy (Xu et al., 2022). Moreover, our in vitro cell experiments suggested that downregulated TNIK indeed affected the effects of RPD on downstream targets (Yuan et al., 2021). However, there is limited research investigating the mechanism by which the downregulation of TNIK impacts the effect of RPD.
TNIK encodes a serine/threonine kinase of the germinal center kinase (GCK) family and contains a scaffold domain that is important in the regulation of cell proliferation and postsynaptic signaling (Coba et al., 2012; Nie et al., 2021). It is found to be highly expressed in the human nervous system, and in vivo animal study has shown that Tnik plays a critical role in synapse and nucleus function within the nervous system and is a crucial regulator for cognitive function (Coba et al., 2012). In patients with SCZ, TNIK mRNA has been confirmed to be upregulated in the dorsolateral prefrontal cortex (Potkin et al., 2010). TNIK is also found as a crucial synaptic partner for the known psychiatric risk-associated protein DISC1 to regulate synapse composition and activity via DISC1–TNIK interaction (Wang et al., 2011). In addition, the methylome-wide association study of first-onset SCZ patients in Chinese Han population integrated with GWAS has identified TNIK as a susceptibility gene of SCZ (Nie et al., 2021). Hence, these reports support a role for TNIK as a risk factor for SCZ, as well as a potential target for RPD.
Overall, RPD, an atypical antipsychotic agent, has limited research on drug metabolic pathways and genetic risk factors influencing drug effect. TNIK, as a susceptibility gene of SCZ, has been reported to associate with RPD treatment response, but the potential mechanisms underlying downregulated TNIK affects RPD’s effect have been not or rarely investigated. Therefore, the purposes of this study were to discover the transcriptome profile of RPD and TNIK knockdown respectively, and then further explore the potential mechanism of TNIK influencing the molecular expression of RPD. To address this issue, we intervened pharmacologically with the appropriate concentration of RPD and downregulated the expression of TNIK by siRNA in human glioma U251 cells, respectively. Finally, we extracted the RNA of different groups for ribonucleic acid sequencing (RNA-seq) to identify the differentially expressed genes (DEGs) and gain more insight into the potential mechanisms.
U251 (human glioma) cell line was purchased from the Cell Bank of Shanghai Institute of the Chinese Academy of Sciences and cultured in a humidified 5% CO2 incubator at 37°C in Dulbecco’s Modified Eagle’s Medium (DMEM, Thermo Scientific) supplemented with 10% fetal bovine serum (FBS, Thermo Scientific) and 1% penicillin/streptomycin (GIBCO). For investigating the effect of downregulated TNIK, the cells were cultured in 6-well plates and transiently transfected with hTNIK small interfering RNAs (siRNAs, Shanghai dnabio company) (knockdown group) using Lipofectamine 3,000 kit (Invitrogen) at 70% confluency. To identify the role of RPD, we determined the half inhibitory concentration (IC50) of RPD using MTT cell proliferation assay kit (Thermo Scientific) as described previously (Yuan et al., 2021), and the result showed the IC50 of RPD was 111.3 μM. Then, RPD treatment with 111.3 μM was performed on U251 cells (RPD group) for 24 h. The control group was constructed without treatment. All assays were performed in triplicate in at least three independent experiments.
The TNIK siRNA sequences were: 5′-GCAAGAAAGAGACUACUUATT-3′ (sense) and 5′-UAAGUAGUCUCUUUCUUGCTT-3′ (anti-sense). To assess the knockdown efficiency of TNIK siRNA, we quantified the expression level of TNIK mRNA using qRT-PCR, with GAPDH serving as the endogenous reference gene. The primers used were as follows: TNIK, forward 5′- GGTAGAAGAACGGTCAAGGCTCAAC-3′ and reverse 5′- GGCTGAACTCCACTAATGCTGAAGG-3′; GAPDH, forward 5′- CAGCCTCAAGATCATCAGCAATG-3′ and reverse 5′- CATGAGTCCTTCCACGATACC-3′. Total RNA was extracted using RNA extraction kit (DP419, TIANGEN). Next, RNA was reverse transcribed into cDNA with PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa) according to the instructions. SYBR® Green Realtime PCR Master Mix (TOYOBO) was applied for PCR amplification with Roche LightCycler 480. Reactions were carried out with the following amplification conditions: 95°C for 30 s, followed by 40 cycles of reaction at 95°C for 5 s, 55°C for 10 s, and 72°C for 15 s. Last, a melting curve was performed after the amplification cycle to confirm the specificity of the amplification reaction. The relative mRNA expression levels of genes were calculated using the 2−ΔΔCT method. PCR reactions were performed in triplicate.
After transfection or drug treatment, total cell RNA from the three groups (control, knockdown, and RPD) was extracted respectively using RNA extraction kit (DP419, TIANGEN) according to the manufacturer’s instructions. 1% agarose gels were used to monitor the degradation and contamination of RNA samples. The purity of RNA was examined by NanoPhotometer® spectrophotometer (IMPLEN, CA, United States) and the integrity was accurately determined using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, United States).
The RNA-seq library was constructed in line with the manufacturer’s protocol of NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, United States) and index codes were added to attribute sequences to each sample. After the purification of total RNA with poly-T oligo-attached magnetic beads, mRNA was obtained and subsequently fragmented into small pieces using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (×5). First-strand complementary DNA (cDNA) was transcribed from the fragmented mRNA using random hexamer primer and M-MuLV Reverse Transcriptase. Second-strand cDNA was synthesized with dNTPs in DNA polymerase I system after the degradation of the RNA strand with RNase H. Purified double-stranded cDNA then went through end reparation and 3′ ends adenylation, followed by ligation of NEBNext adaptor with hairpin loop structure. DNA fragments around 250–300 bp were screened for PCR amplification with AMPure XP beads. PCR products were purified using AMPure XP beads to create the final complementary DNA library, and library quality was assessed on the Agilent Bioanalyzer 2100 system. Lastly, after clustering the index-coded samples on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia), paired-end sequencing of the sequencing library was performed in Illumina Novaseq platform and 150 bp paired-end reads were generated.
Before data analysis, raw reads should be pre-processed to remove reads containing adapter, reads containing poly-N, and low-quality to obtain clean reads with high quality for subsequent analyses. Reference genome and gene model annotation files were downloaded from the genome website, and Hisat2 v2.0.5 was used to build the index of the reference genome and align paired-end clean reads to the reference genome. The expression level of genes in each sample was estimated using Fragments Per Kilobase Per Million Mapped Fragments (FPKM) values. Differential expression analysis was performed in edgeR package, which is based on the statistical test of the dispersion of negative binomial distributions of genes by fitting a generalized linear model with likelihood ratios to identify DEGs. Bonferroni correction was used for multiple tests and p.adj < 0.05 was considered significant. Furthermore, we also calculated the fold change by comparing two different groups, and |log2 (fold change)| > 1 was set as an additional condition to screen for DEGs. In general, the screening criterion for DEGs was to satisfy both p.adj < 0.05 and |log2 (fold change)| > 1. The ggplot2 package was used to draw the volcano plot to display the distribution of DEGs.
To better investigate the underlying biological mechanism and pathways, all DEGs were subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses via clusterProfiler packages in R platform. For GO analysis, it was classified into three subgroups, including biological process (BP), cellular component (CC), and molecular function (MF). p.adj < 0.05 was set as the threshold.
Firstly, we obtained the SCZ-related gene from the following sites: GeneCards (https://www.genecards.org/), OMIM (https://www.omim.org/) and DisGeNet (https://www.disgenet.org/). We subsequently made the Venn diagram in R platform by intersecting the SCZ-associated genes with the DEGs of RPD vs. control (R vs. C) and knockdown vs. control (K vs. C). The cross-talk genes of these three groups were then used to perform the GO and KEGG enrichment analysis. Subsequently, the STRING database (https://cn.string-db.org/) was used to construct the PPI network of cross-talk genes. After that, the PPI network was revisualized using Cytoscape 3.10.1 software. Firstly, we hid the unconnected genes and then calculated the degree score with the network analysis function. Finally, the PPI network was re-adjusted based on the degree score. The larger the circle and the darker the color, the higher the degree score.
As the qRT-PCR result indicated, transfection of TNIK siRNA in U251 cells significantly reduced the mRNA levels of TNIK (Supplementary Figure S1).
This study aimed to investigate the molecular mechanism of RPD and downregulated TNIK. Additionally, we also explored the potential functions and/or pathways involved in the downregulation of TNIK that may impact the effects of RPD. High-throughput sequencing was conducted, followed by a DEG analysis to identify changes in gene expression. Compared with the control group, the volcano plot showed that a total 2,126 DEGs with p.adj < 0.05 and |log2 (fold change)| > 1 were obtained in RPD group, of which 1,157 were upregulated while 969 downregulated (Figure 1A). In the meanwhile, data analysis of control and knockdown groups obtained 316 significantly upregulated genes and 339 significantly downregulated genes (Figure 1B). The results indicated that RPD treatment induced greater changes in gene expression than TNIK knockdown in U251 cells.
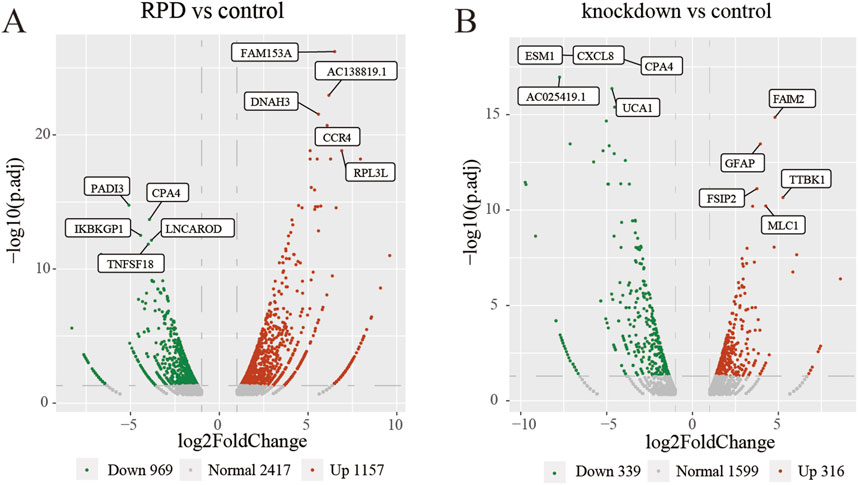
Figure 1. Volcano plots of DEGs. The top 5 upregulated and downregulated genes with the smallest p-adj value were marked on the plot. (A) Volcano plot of all DEGs between RPD group and control group. (B) Volcano plot of all DEGs between TNIK knockdown group and control group. p.adj < 0.05 was considered significant.
To explore the biological functions and pathways of these DEGs, we performed GO and KEGG enrichment analyses. Firstly, we observed the comparison between RPD and control groups. According to the GO enrichment result, 32 BP terms, 1 CC term, and 3 MF terms were significantly enriched in DEGs. The most significantly enriched terms include nucleoside diphosphate metabolic process and modified amino acid binding (Figure 2A). In addition, a total of 697 genes were mapped to the signaling pathways. Only two KEGG pathways were significantly enriched, including galactose metabolism and parathyroid hormone synthesis, secretion, and action (Figure 2B). Then, for the comparison of knockdown and control groups, 289 BP terms, 19 CC terms, and 26 MF terms were identified to be significantly enriched in GO analysis, and the top 10 processes among these three term were displayed in Figure 3A. Among the BP terms, most genes were enriched in cell chemotaxis, cell-substrate adhesion, and leukocyte migration. Among the CC terms, the most enriched terms were mainly related to collagen-containing extracellular matrix, synaptic membrane, and endoplasmic reticulum lumen. Among the MF terms, most genes were categorized as receptor-ligand activity, signaling receptor activator activity, glycosaminoglycan binding, and sulfur compound binding. Further to this, 21 KEGG pathways were enriched in total, mainly including focal adhesion, PI3K-Akt signaling pathway, and Ras signaling pathway (Figure 3B).
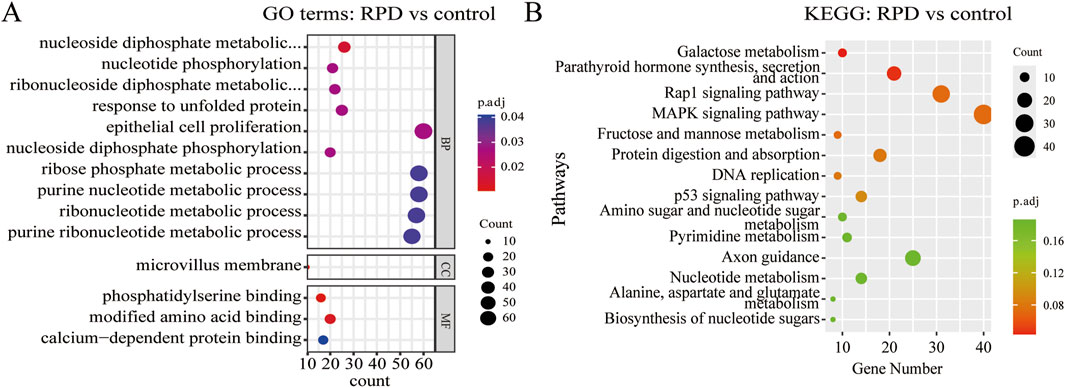
Figure 2. GO and KEGG enrichment analysis of DEGs between RPD group and control group. (A) The bubble plot of GO analysis showed the top 10 processes in BP, the only 1 process in CC, and the only three processes in MF. (B) The bubble plot of KEGG analysis showed only the first two pathways were significantly enriched by DEGs. p.adj < 0.05 was considered significant.
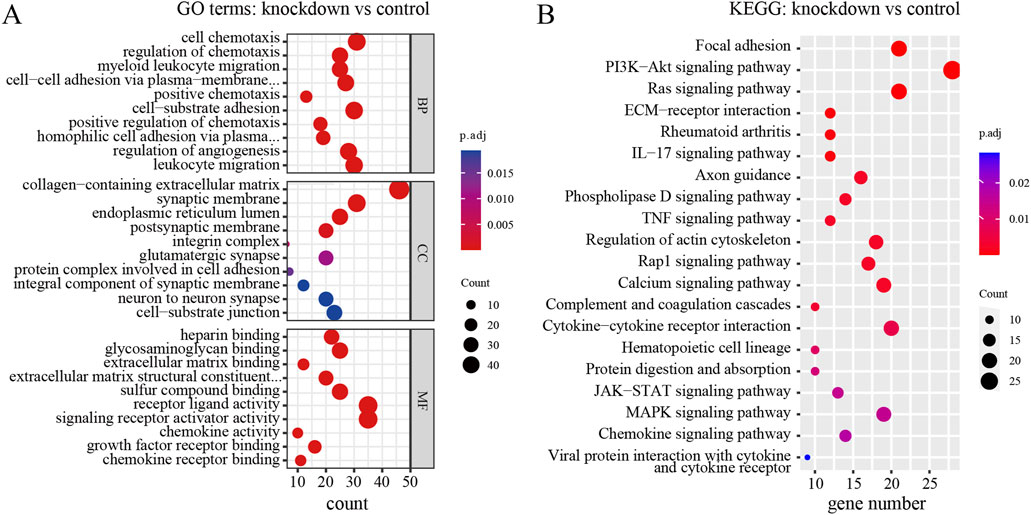
Figure 3. GO and KEGG enrichment analysis of DEGs between TNIK knockdown group and control group. (A) The result of GO analysis showed the top 10 processes enriched by DEGs in BPs, CCs, and MFs. (B) The result of KEGG analysis showed the top 20 pathways enriched by DEGs. p.adj < 0.05 was considered significant.
A total of 14,239 genes related to schizophrenia were collected from GeneCards, OMIM, and DisGeNET, deduplicated and combined to form a target gene dataset. Then, the SCZ-related genes were intersected with the above two sets of DEGs, and 82 cross-talk genes were screened out (Figure 4A). Subsequently, GO and KEGG analyses were performed. The GO analysis revealed that these 82 cross-talk genes were mainly enriched in bone mineralization, biomineral tissue development, and biomineralization (Figure 4B). According to KEGG pathway enrichment analysis, only 4 pathways were significantly enriched including Rap1 signaling pathway, Regulation of actin cytoskeleton, Calcium signaling pathway, and JAK-STAT signaling pathway (Figure 4C). To analyze the interaction among the proteins corresponding to the 82 genes, the STRING database was used to construct a PPI network. The software Cytoscape was then used to visualize the PPI network. The PPI network consisted of 43 nodes and 114 edges, 31 nodes and 96 edges were remained after hiding the unconnected genes. The genes with higher degree scores were labeled in darker colors and larger circles (Figure 4D). The hub genes were screened by the degree score, of which the top 10 genes included CSF2, FGFR2, FGF1, FGFR3, HBEGF, CDH5, MMP3, ANGPTL4, F2R, and PTPN6. After reviewing relevant literature, we selected fibroblast growth factors (FGFs)-related genes (including FGFR2, FGF1 and FGFR3) as the key genes that might be the potential targets for TNIK affecting the effect of RPD.
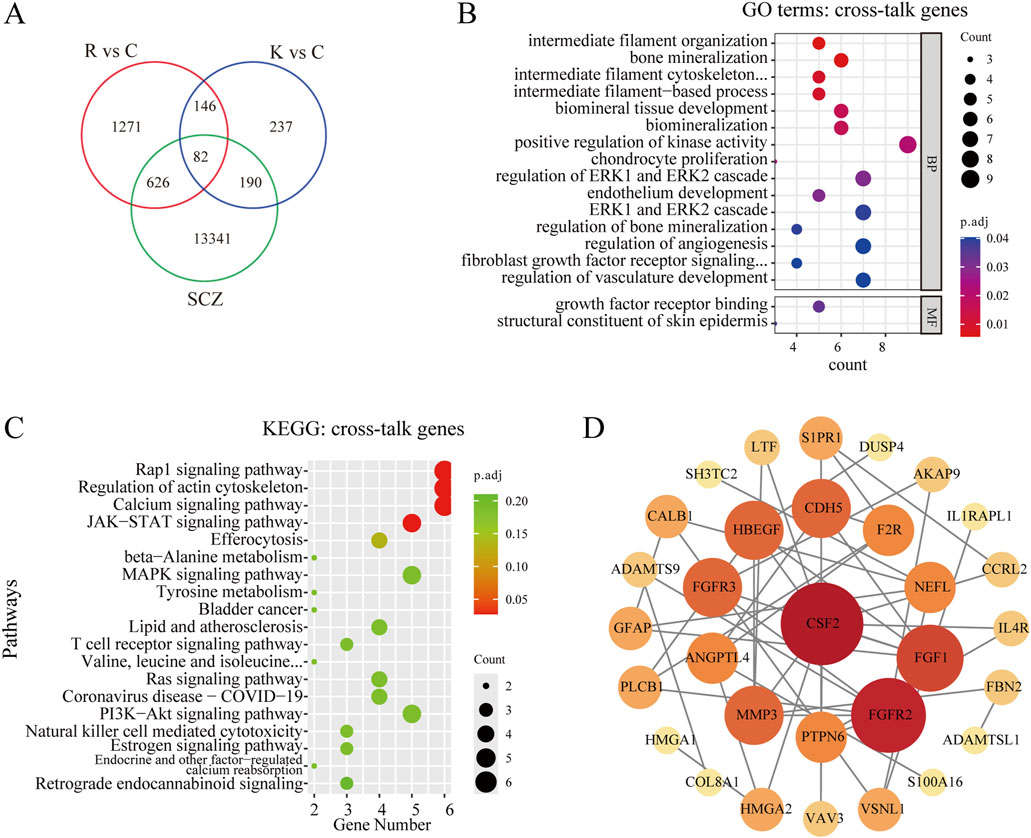
Figure 4. Screening and analyzing the cross-talk genes, and constructing the PPI network. (A) Venn diagram of cross-talk genes from schizophrenia-related genes and two groups of DEGs, and 82 cross-talk genes were screened out. R vs C: RPD vs Control. K vs C: Knockdown vs Control. SCZ: schizophrenia. (B) GO analysis of the cross-talk genes. The bubble plot showed the top 10 processes in BP, and the only 2 processes in MF. There was no process enriched in CC. (C) KEGG enrichment analysis of the cross-talk genes. The bubble plot showed only the top four pathways were significantly enriched. (D) The STRING database was used to construct the PPI network of 82 cross-talk genes and the Cytoscape software was used to visualize the PPI network and screen the hub genes. The degree score was calculated for every gene, and the larger the circle and the darker the color meant the higher the degree score. p.adj < 0.05 was considered significant.
In this study, we identified a total of 2,126 DEGs in RPD group and 655 DEGs in TNIK knockdown group after transcriptomic analysis. After intersecting the SCZ-associated genes group with the above two DEGs groups, we screened out a total of 82 genes. For the first time, we investigated the molecular mechanisms of gene expression and pathways in human glioma cells following the effects of RPD. Through KEGG enrichment analysis, we found a significant enrichment of the pathway for galactose metabolism and parathyroid hormone synthesis, secretion, and action. On the other hand, TNIK, a gene of interest, was first explored to identify the potential mechanisms following its knockdown, and the significant enrichments of the collagen-containing extracellular matrix, signaling receptor activator activity, and PI3K-Akt signaling and Ras signaling pathways were found. More interesting, the study showed that both RPD treatment and TNIK knockdown had a significant impact on bone mineralization and calcium signaling pathway. Furthermore, the PPI network analysis indicated that TNIK might affect the adverse effect of RPD through FGF-related genes, such as FGFR2, FGF1, and FGFR3.
Risperidone is one of the widely prescribed antipsychotics; however, long-term use is usually accompanied by various side effects, such as weight gain, extrapyramidal symptoms, prolactin elevation, and osteoporosis. The complex mechanism of RPD is not yet fully understood and requires further research. Therefore, we have studied at the cellular level which pathways can be altered after RPD treatment through transcriptomic analysis. As result showed, KEGG pathway analysis only emphasized two pathways: galactose metabolism and parathyroid hormone synthesis, secretion, and action pathway. Regarding galactose metabolism, Yu et al. suggested an association between upregulation of galactose metabolism and postmenopausal osteoporosis by metabolome analysis (Yu et al., 2019). In addition, galactose was used to induce aging and osteoporosis in the rat model, which exhibited marked bone loss (Yu et al., 2019; Li et al., 2020). Regarding parathyroid hormone synthesis, secretion, and action pathway, 20 genes enriched in this pathway, and 18 genes were significantly upregulated, indicating that this pathway might be highly activated during RPD application. Parathyroid hormone (PTH) is a peptide hormone made up of 84 amino acids that is synthesized and secreted by the parathyroid glands. It is essential for the metabolism of calcium and phosphorus through direct effects on bone and kidney and indirect effects on the human intestine (Chen T. et al., 2021). PTH has been shown to result in catabolic effects on the skeleton via continuous stimulation, while intermittent stimulation can lead to osteoanabolic effects (Silva and Bilezikian, 2015). Therefore, PTH has the opposite effects on bone resorption and bone formation in different situations. Overall, our previous description pointed out that long-term use of RPD might promote the risk of osteoporosis due to elevated prolactin levels (Graham et al., 2011; Bishop et al., 2012). However, the present study provided a new perspective that RPD might result in osteoporosis by affecting galactose metabolism and parathyroid hormone synthesis, secretion, and action pathway.
In recent studies, TNIK has been implicated in multiple cellular signaling processes. Among them, the most reported is that TNIK is the downstream signal protein and nuclear coactivator of the Wnt/β-catenin signaling pathway to fully activate Wnt signaling and promote cell growth and invasion (Ma et al., 2022). Larhammar et al. demonstrated that TNIK regulates stress-induced c-Jun-N-terminal kinase (JNK) signaling in neurons, which affects nervous system development, axon regeneration, and neuronal degeneration (Larhammar et al., 2017). In addition, research has also proven a critical role for TNIK in canonical NF-kB signaling (Shkoda et al., 2012). In the present study, GO enrichment showed that DEGs produced by TNIK knockdown were mainly enriched in cellular components about collagen-containing extracellular matrix, cell-substrate adhesion, receptor-ligand activity, and signaling receptor activator activity. TNIK has been implicated in postsynaptic signaling and regulation of cell proliferation (Coba et al., 2012). Therefore, GO analysis results suggested that downregulated TNIK might affect the interaction between extracellular proteins secreted into the extracellular matrix and receptors on the cell membrane, thereby regulating the binding of receptors and ligands, as well as the signal transduction for cell-to-cell communication. KEGG enrichment analysis highlighted PI3K-Akt signaling pathway. Research has found that the PI3K-Akt signaling pathway is mediated by several targets for SCZ and antipsychotics, such as dopamine, serotonin, glutamate, neuregulin 1, dysbindin, and DISC-1 (Liu et al., 2016). For example, the dysfunction of dopamine D1 and/or D5 receptor signaling in SCZ is linked to the activation of PI3K/AKT signaling, which subsequently leads to the inactivation of GSK3 (Matsuda et al., 2019). In addition, study of animal models has suggested that the miR-25-3p-mediated TWIST1/PI3K/Akt/GSK3β signaling pathway is altered in the brain of schizophrenia model rat, and this alteration can be restored by RPD, indicating that this pathway might be a potential biomarker for SCZ and a possible target for antipsychotic treatment (Pan et al., 2023). Furthermore, TNIK has been shown to play a role in the PI3K-Akt signaling pathway by regulating Akt activation in gastric cancer growth (Yu et al., 2014). These results suggested that downregulated TNIK might affect the efficacy of RPD through the PI3K-Akt signaling pathway.
Finally, to further identify the proteins and/or pathways that downregulated TNIK and RPD treatment co-influence in SCZ, we made the intersection among the SCZ-associated genes and the above two sets of DEGs, and a total of 82 cross-talk genes were obtained. According to GO analysis on these 82 genes, it was found that these genes were primarily associated with bone mineralization, biomineral tissue development, and biomineralization, indicating that RPD medication and TNIK knockdown might have a combined effect on biomineralization processes in schizophrenia. Biomineralization is a natural process through which inorganic calcium (Ca) and phosphate (Pi) precipitate to form biological hard tissues such as bones, teeth, and other hard tissues (Bhadada and Rao, 2021). In bone mineralization, calcium phosphate crystals are deposited onto collagen fibrils, providing the bone with hardness and flexibility, respectively (Hasegawa et al., 2022). The accretion of adequate mineral content is essential for normal bone mineralization (Couce and Saenz de Pipaon, 2021). The pathological deficiency in mineralization results in osteoporosis due to the reduction in bone density (Vimalraj, 2020). Moreover, the chronic use of antipsychotics, particularly for RPD, has been associated with impaired bone mineralization, partially mediated by hyperprolactinemia (Calarge et al., 2018). Therefore we hypothesized that RPD may cause abnormal bone mineralization by elevating the level of prolactin, eventually leading to osteoporosis. Regarding TNIK, it is known to be an essential activator of the Wnt signaling pathway. The Wnt signaling pathway has been reported to promote bone formation by enhancing osteoblastic proliferation and differentiation, whereas aberrant activation of this pathway leads to altered osteocyte mineralization (Zhou et al., 2019; Palmer et al., 2021). Thus, the effect of downregulated TNIK on bone mineralization may be achieved by influencing the activation of the Wnt signaling pathway. Through KEGG pathway analysis, calcium signaling pathway was one of the significantly enriched pathways. Calcium signaling pathway is a crucial component of mechanisms that regulate neuronal excitability, information processing, and cognition, and dysregulation of calcium signaling has been implicated in the development of schizophrenia. Study suggested that the DISC1 gene may contribute to schizophrenia by interacting with calcium signaling pathways in the brain (Zhang et al., 2023), and variations in calcium channel signaling genes were also identified to play a role in the pathophysiology of schizophrenia through an extensive GWAS analysis (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013). Additionally, the extracellular level of ionic calcium may be one of the critical determinants for bone mineralization based on the chemical structure of bone minerals (Murshed, 2018). These results collectively suggested that both RPD and downregulated TNIK may affect the biological process of bone mineralization.
After analyzing the PPI network of 82 cross-talk genes and reviewing relevant literature, we screened three FGF-related genes (FGFR2, FGF1, and FGFR3) from the top 10 hub genes, and these three genes might be the potential targets through which TNIK might impact the adverse effect of RPD. In human, FGF family consists of 22 FGFs and 5 fibroblast growth factor receptors (FGFRs). The FGFs and their receptors have been reported to play an essential role in neurodevelopment and psychopathology, including SCZ (Terwisscha van Scheltinga et al., 2013; Talaei et al., 2020). FGF1, widely expressed in the human central nervous system, is located in the regions that have been replicated for linkage with SCZ (Terwisscha van Scheltinga et al., 2010). FGFR2, which is involved in presynaptic organization during early development, as well as neuroprotection and repair during adulthood, has been identified as a potential susceptibility gene for SCZ (O’Donovan et al., 2009). Moreover, a postmortem brain study for patients with SCZ revealed a decrease in the expression of FGFR2 in the hippocampus and cingulate cortex (Terwisscha van Scheltinga et al., 2010). Interestingly, the result of this study showed that the expression of FGFR2 was significantly upregulated with RPD treatment. This indicated that FGFR2 might also be a target for RPD to exert its effect during SCZ treatment. Regarding FGFR3, there is little evidence to discuss the association between FGFR3 and SCZ. According to previous studies, perhaps of most plausible relevance to SCZ is that FGFR3 is expressed in glial cells of the diencephalon and myelencephalon during development, with expression patterns widening during adulthood (Terwisscha van Scheltinga et al., 2010). Furthermore, studies have substantiated the strong interaction between FGFs and the dopamine system, which is strongly implicated in SCZ pathophysiology and is the main target for RPD medication (Grace, 2016; Talaei et al., 2020). For example, in a model of Parkinson’s disease, it has been proven that FGFs can protect against the loss of dopaminergic neurons and promote neurotrophic activity on dopamine neurons both in vitro and in vivo (Liu et al., 2021). Specifically, numerous studies have found that FGF2 is involved in the development and maintenance of the brain’s dopamine system (Even-Chen et al., 2017; Grinchii et al., 2023). On the other hand, we found that FGF, in addition to PTH, is widely expressed in the human bone matrix and plays a role in regulating the formation and development of osteoporosis (Gao et al., 2023). Early studies in animal models found that system administration of FGF1 can prevent bone loss and promote new bone formation in ovariectomized rats (Dunstan et al., 1999). FGFR2, as a cell membrane surface receptor, plays a critical role in bone development and homeostasis. Via stabilizing FGFR2, Otubain1 can positively regulate osteogenic differentiation and mineralization in bone homeostasis (Zhu et al., 2023). Overexpression of this gene in bone marrow mesenchymal stem cells has also been found to enhance osteogenesis and promote regeneration of critical cranial bone defects (Zhou et al., 2023). FGFR3 is a negative regulator of endochondral bone development. In an ovariectomized mouse model, activated FGFR3 can suppress bone regeneration and bone mineralization (Kawashima et al., 2023). Through targeting FGFR3, MiR-99b-5p can exhibit inhibitory effects on osteoblast proliferation and differentiation, thereby affecting bone formation in osteoporosis progression (Ding et al., 2021). As with FGFR2, FGFR3 was also significantly upregulated after RPD treatment. Therefore, RPD medication may also increase the incidence of osteoporosis by affecting the expression levels of FGF-related genes. In summary, the above three genes might be potential targets through which TNIK knockdown might affect the molecular expression of RPD.
However, there are several limitations in this study. Firstly, we conducted this study based on the association studies of TNIK affecting the antipsychotic effect of RPD. Therefore, the neuronal cell lines could be more suitable than the U251 cell line for this study to disentangle the mechanism involved in the antipsychotic effect of RPD. Secondly, the genetic variants of TNIK have been reported to be associated with the treatment response of several antipsychotic drugs, but only RPD was involved in this study. In addition, the use of IC50 as the dose for RPD treatment lacks sufficient empirical support, and determining the optimal concentration of RPD treatment will require further exploration in U251 cells. Last, in this study, our initial goal was to investigate the pathways or targets through which TNIK downregulation might influence the efficacy of RPD. Ultimately, we identified potential targets indicating that TNIK downregulation affects the adverse effect of RPD on osteoporosis. Overall, to determine the exact mechanism of TNIK affecting the efficacy of antipsychotics, further researches are required to explore the biological functions of the potential targets using more scientifically rigorous experimental protocols.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
RY: Data curation, Investigation, Visualization, Writing–original draft. YaL: Investigation, Writing–review and editing. XL: Data curation, Writing–review and editing. YF: Project administration, Writing–review and editing. AN: Project administration, Writing–review and editing. DW: Resources, Writing–review and editing. RZ: Resources, Writing–review and editing. SY: Funding acquisition, Writing–review and editing. QX: Conceptualization, Data curation, Writing–original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported financially by the National Natural Science Foundation of China (No.82401760, No.81971257), the General Program of Shanghai Mental Health Center “Academic project” (2021-YJ04), the Fundamental Research Funds for the Central Universities (YG2024QNB31), the General Program of Shanghai Mental Health Center “Academic project” (2021-YJ07), the Key Program of Shanghai Mental Health Center “Academic project” (2023zd02), and the Key Program of the Clinical Research Center of Shanghai Mental Health Center (CRC2021ZD02).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1431923/full#supplementary-material
Amodeo, D. A., Jones, J. H., Sweeney, J. A., and Ragozzino, M. E. (2014). Risperidone and the 5- HT 2A receptor antagonist M 100907 improve probabilistic reversal learning in BTBR T + tf/J mice. Autism Res. 7, 555–567. doi:10.1002/aur.1395
Arani, Z. M., Heidariyeh, N., Ghavipanjeh, G., Lotfinia, M., and Banafshe, H. R. (2023). Effect of risperidone on morphine-induced conditioned place preference and dopamine receptor D2 gene expression in male rat hippocampus. Brain Behav. 13, e2975. doi:10.1002/brb3.2975
Bhadada, S. K., and Rao, S. D. (2021). Role of phosphate in biomineralization. Calcif. Tissue Int. 108, 32–40. doi:10.1007/s00223-020-00729-9
Bishop, J. R., Rubin, L. H., Reilly, J. L., Pavuluri, M. N., and Sweeney, J. A. (2012). Risperidone-associated prolactin elevation and markers of bone turnover during acute treatment. Ther. Adv. Psychopharmacol. 2, 95–102. doi:10.1177/2045125312442080
Calarge, C. A., Mills, J. A., Ziegler, E. E., and Schlechte, J. A. (2018). Calcium and vitamin D supplementation in boys with risperidone-induced hyperprolactinemia: a randomized, placebo-controlled pilot study. J. Child. Adolesc. Psychopharmacol. 28, 145–150. doi:10.1089/cap.2017.0104
Chen, T., Wang, Y., Hao, Z., Hu, Y., and Li, J. (2021a). Parathyroid hormone and its related peptides in bone metabolism. Biochem. Pharmacol. 192, 114669. doi:10.1016/j.bcp.2021.114669
Chen, Y., Zhang, Y., Fan, K., Xu, W., Teng, C., Wang, S., et al. (2021b). Association between gonadal hormones and osteoporosis in schizophrenia patients undergoing risperidone monotherapy: a cross-sectional study. PeerJ 9, e11332. doi:10.7717/peerj.11332
Clapham, E., Bodén, R., Reutfors, J., Svensson, T., Ramcharran, D., Qiu, H., et al. (2020). Exposure to risperidone versus other antipsychotics and risk of osteoporosis-related fractures: a population-based study. Acta Psychiatr. Scand. 141, 74–83. doi:10.1111/acps.13101
Coba, M. P., Komiyama, N. H., Nithianantharajah, J., Kopanitsa, M. V., Indersmitten, T., Skene, N. G., et al. (2012). TNiK is required for postsynaptic and nuclear signaling pathways and cognitive function. J. Neurosci. 32, 13987–13999. doi:10.1523/JNEUROSCI.2433-12.2012
Couce, M. L., and Saenz de Pipaon, M. (2021). Bone mineralization and calcium phosphorus metabolism. Nutrients 13, 3692. doi:10.3390/nu13113692
Cross-Disorder Group of the Psychiatric Genomics Consortium (2013). Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379. doi:10.1016/S0140-6736(12)62129-1
De Hert, M., Detraux, J., and Stubbs, B. (2016). Relationship between antipsychotic medication, serum prolactin levels and osteoporosis/osteoporotic fractures in patients with schizophrenia: a critical literature review. Expert Opin. Drug Saf. 15, 809–823. doi:10.1517/14740338.2016.1167873
Ding, M., Liu, B., Chen, X., Ouyang, Z., Peng, D., and Zhou, Y. (2021). MiR-99b-5p suppressed proliferation of human osteoblasts by targeting FGFR3 in osteoporosis. Hum. Cell 34, 1398–1409. doi:10.1007/s13577-021-00567-3
Dunstan, C. R., Boyce, R., Boyce, B. F., Garrett, I. R., Izbicka, E., Burgess, W. H., et al. (1999). Systemic administration of acidic fibroblast growth factor (FGF-1) prevents bone loss and increases new bone formation in ovariectomized rats. J. Bone Min. Res. 14, 953–959. doi:10.1359/jbmr.1999.14.6.953
Even-Chen, O., Sadot-Sogrin, Y., Shaham, O., and Barak, S. (2017). Fibroblast growth factor 2 in the dorsomedial striatum is a novel positive regulator of alcohol consumption. J. Neurosci. 37, 8742–8754. doi:10.1523/JNEUROSCI.0890-17.2017
Gao, Y., Chen, N., Fu, Z., and Zhang, Q. (2023). Progress of Wnt signaling pathway in osteoporosis. Biomolecules 13, 483. doi:10.3390/biom13030483
Grace, A. A. (2016). Dysregulation of the dopamine system in the pathophysiology of schizophrenia and depression. Nat. Rev. Neurosci. 17, 524–532. doi:10.1038/nrn.2016.57
Graham, S. M., Howgate, D., Anderson, W., Howes, C., Heliotis, M., Mantalaris, A., et al. (2011). Risk of osteoporosis and fracture incidence in patients on antipsychotic medication. Expert Opin. Drug Saf. 10, 575–602. doi:10.1517/14740338.2011.560112
Grinchii, D., Levin-Greenwald, M., Lezmy, N., Gordon, T., Paliokha, R., Khoury, T., et al. (2023). FGF2 activity regulates operant alcohol self-administration and mesolimbic dopamine transmission. Drug Alcohol Depend. 248, 109920. doi:10.1016/j.drugalcdep.2023.109920
Hasegawa, T., Hongo, H., Yamamoto, T., Abe, M., Yoshino, H., Haraguchi-Kitakamae, M., et al. (2022). Matrix vesicle-mediated mineralization and osteocytic regulation of bone mineralization. Int. J. Mol. Sci. 23, 9941. doi:10.3390/ijms23179941
Heide, J., Zhang, F., Bigos, K. L., Mann, S. A., Carr, V. J., Shannon Weickert, C., et al. (2016). Differential response to risperidone in schizophrenia patients by KCNH2 genotype and drug metabolizer status. Am. J. Psychiatry 173, 53–59. doi:10.1176/appi.ajp.2015.14050653
Hung, C.-C., Lin, C.-H., and Lane, H.-Y. (2021). Cystine/glutamate antiporter in schizophrenia: from molecular mechanism to novel biomarker and treatment. Int. J. Mol. Sci. 22, 9718. doi:10.3390/ijms22189718
Jauhar, S., Johnstone, M., and McKenna, P. J. (2022). Schizophrenia. Lancet 399, 473–486. doi:10.1016/S0140-6736(21)01730-X
Kawashima, I., Matsushita, M., Mishima, K., Kamiya, Y., Osawa, Y., Ohkawara, B., et al. (2023). Activated FGFR3 suppresses bone regeneration and bone mineralization in an ovariectomized mouse model. BMC Musculoskelet. Disord. 24, 200. doi:10.1186/s12891-023-06318-9
Larhammar, M., Huntwork-Rodriguez, S., Rudhard, Y., Sengupta-Ghosh, A., and Lewcock, J. W. (2017). The Ste20 family kinases MAP4K4, MINK1, and TNIK converge to regulate stress-induced JNK signaling in neurons. J. Neurosci. 37, 11074–11084. doi:10.1523/JNEUROSCI.0905-17.2017
Li, N., Dong, X., Ma, F., Tian, X., Bu, R., Li, C., et al. (2020). Pharmacokinetics study of 16 active ingredients from Tabson-2 decoction in normal and d -galactose induced osteoporosis rats by liquid chromatography–tandem mass spectrometry. J. Sep. Sci. 43, 3702–3713. doi:10.1002/jssc.202000355
Lisoway, A. J., Chen, C. C., Zai, C. C., Tiwari, A. K., and Kennedy, J. L. (2021). Toward personalized medicine in schizophrenia: genetics and epigenetics of antipsychotic treatment. Schizophr. Res. 232, 112–124. doi:10.1016/j.schres.2021.05.010
Liu, L., Luo, Y., Zhang, G., Jin, C., Zhou, Z., Cheng, Z., et al. (2016). The mRNA expression of DRD2, PI3KCB, and AKT1 in the blood of acute schizophrenia patients. Psychiatry Res. 243, 397–402. doi:10.1016/j.psychres.2016.07.010
Liu, Y., Deng, J., Liu, Y., Li, W., and Nie, X. (2021). FGF, mechanism of action, role in Parkinson’s disease, and therapeutics. Front. Pharmacol. 12, 675725. doi:10.3389/fphar.2021.675725
Ma, L., Li, R., Yao, Z., Wang, B., Liu, Y., Liu, C., et al. (2022). Computational study on new natural compound inhibitors of Traf2 and Nck-interacting kinase (TNIK). Aging 14, 8394–8410. doi:10.18632/aging.204349
Matsuda, S., Ikeda, Y., Murakami, M., Nakagawa, Y., Tsuji, A., and Kitagishi, Y. (2019). Roles of PI3K/AKT/GSK3 pathway involved in psychiatric illnesses. Diseases 7, 22. doi:10.3390/diseases7010022
Morozova, M. A., Lezheiko, T. V., Lepilkina, T. A., Burminskiy, D. S., Potanin, S. S., Beniashvili, A. G., et al. (2022). Treatment response and GWAS risk allele rs2514218 (C) of the dopamine D2 receptor gene in inpatients with schizophrenia. Neuropsychobiology 81, 149–155. doi:10.1159/000519155
Murshed, M. (2018). Mechanism of bone mineralization. Cold Spring Harb. Perspect. Med. 8, a031229. doi:10.1101/cshperspect.a031229
Nie, F., Zhang, M., Shang, S., Zhang, Q., Zhang, R., Chen, P., et al. (2021). Methylome-wide association study of first-episode schizophrenia reveals a hypermethylated CpG site in the promoter region of the TNIK susceptibility gene. Prog. Neuropsychopharmacol. Biol. Psychiatry 106, 110081. doi:10.1016/j.pnpbp.2020.110081
O’Donovan, M. C., Norton, N., Williams, H., Peirce, T., Moskvina, V., Nikolov, I., et al. (2009). Analysis of 10 independent samples provides evidence for association between schizophrenia and a SNP flanking fibroblast growth factor receptor 2. Mol. Psychiatry 14, 30–36. doi:10.1038/mp.2008.108
Palmer, N. D., Lu, L., Register, T. C., Lenchik, L., Carr, J. J., Hicks, P. J., et al. (2021). Genome-wide association study of vitamin D concentrations and bone mineral density in the African American-Diabetes Heart Study. PLoS ONE 16, e0251423. doi:10.1371/journal.pone.0251423
Pan, B., Zhu, X., Han, B., Weng, J., Wang, Y., and Liu, Y. (2023). The SIK1/CRTC2/CREB1 and TWIST1/PI3K/Akt/GSK3β signaling pathways mediated by microRNA-25-3p are altered in the schizophrenic rat brain. Front. Cell. Neurosci. 17, 1087335. doi:10.3389/fncel.2023.1087335
Peuskens, J., Pani, L., Detraux, J., and De Hert, M. (2014). The effects of novel and newly approved antipsychotics on serum prolactin levels: a comprehensive review. CNS Drugs 28, 421–453. doi:10.1007/s40263-014-0157-3
Potkin, S. G., Macciardi, F., Guffanti, G., Wang, Q., Turner, J. A., Lakatos, A., et al. (2010). Identifying gene regulatory networks in schizophrenia. NeuroImage 53, 839–847. doi:10.1016/j.neuroimage.2010.06.036
Shkoda, A., Town, J. A., Griese, J., Romio, M., Sarioglu, H., Knöfel, T., et al. (2012). The germinal center kinase TNIK is required for canonical NF-κB and JNK signaling in B-cells by the EBV oncoprotein LMP1 and the CD40 receptor. PLoS Biol. 10, e1001376. doi:10.1371/journal.pbio.1001376
Silva, B. C., and Bilezikian, J. P. (2015). Parathyroid hormone: anabolic and catabolic actions on the skeleton. Curr. Opin. Pharmacol. 22, 41–50. doi:10.1016/j.coph.2015.03.005
Talaei, A., Farkhondeh, T., and Forouzanfar, F. (2020). Fibroblast growth factor: promising target for schizophrenia. Curr. Drug Targets 21, 1344–1353. doi:10.2174/1389450121666200628114843
Terwisscha van Scheltinga, A. F., Bakker, S. C., and Kahn, R. S. (2010). Fibroblast growth factors in schizophrenia. Schizophr. Bull. 36, 1157–1166. doi:10.1093/schbul/sbp033
Terwisscha van Scheltinga, A. F., Bakker, S. C., Kahn, R. S., and Kas, M. J. H. (2013). Fibroblast growth factors in neurodevelopment and psychopathology. Neurosci. Rev. J. Bringing Neurobiol. Neurol. Psychiatry 19, 479–494. doi:10.1177/1073858412472399
Vimalraj, S. (2020). Alkaline phosphatase: structure, expression and its function in bone mineralization. Gene 754, 144855. doi:10.1016/j.gene.2020.144855
Wang, Q., Charych, E. I., Pulito, V. L., Lee, J. B., Graziane, N. M., Crozier, R. A., et al. (2011). The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Mol. Psychiatry 16, 1006–1023. doi:10.1038/mp.2010.87
Xu, Q., Li, M., Qin, S., Li, Y., Ning, A., Fu, Y., et al. (2020). Two novel loci of RELN associated with antipsychotics response in Chinese han population. Front. Pharmacol. 11, 7. doi:10.3389/fphar.2020.00007
Xu, Q., Li, Y., Li, M., Qin, S., Ning, A., Yuan, R., et al. (2022). The influence of TNIK gene polymorphisms on risperidone response in a Chinese Han population. Pharmacogenomics 23, 575–583. doi:10.2217/pgs-2022-0052
Xu, Q., Wu, X., Li, M., Huang, H., Minica, C., Yi, Z., et al. (2016). Association studies of genomic variants with treatment response to risperidone, clozapine, quetiapine and chlorpromazine in the Chinese Han population. Pharmacogenomics J. 16, 357–365. doi:10.1038/tpj.2015.61
Yu, D.-H., Zhang, X., Wang, H., Zhang, L., Chen, H., Hu, M., et al. (2014). The essential role of TNIK gene amplification in gastric cancer growth. Oncogenesis 3, e93. doi:10.1038/oncsis.2014.9
Yu, H., Yan, H., Wang, L., Li, J., Tan, L., Deng, W., et al. (2018). Five novel loci associated with antipsychotic treatment response in patients with schizophrenia: a genome-wide association study. Lancet Psychiatry 5, 327–338. doi:10.1016/S2215-0366(18)30049-X
Yu, L., Qi, H., An, G., Bao, J., Ma, B., Zhu, J., et al. (2019). Association between metabolic profiles in urine and bone mineral density of pre- and postmenopausal Chinese women. Menopause 26, 94–102. doi:10.1097/GME.0000000000001158
Yuan, R., Li, Y., Fu, Y., Ning, A., Wang, D., Zhang, R., et al. (2021). TNIK influence the effects of antipsychotics on Wnt/β-catenin signaling pathway. Psychopharmacol. (Berl.) 238, 3283–3292. doi:10.1007/s00213-021-05943-3
Zhang, Z., Lin, H., Feng, Z., Xie, H., Liu, P., Shu, Y., et al. (2023). Impaired calcium channel function and pronounced hippocampal atrophy in a schizophrenia patient with cognitive impairment carrying Presenilin-2 Ser130Leu mutation: a case report and literature review. Schizophr. Res. 258, 78–80. doi:10.1016/j.schres.2023.07.024
Zheng, L., Yang, L., Zhao, X., Long, N., Li, P., and Wang, Y. (2019). Effect of risperidone on proliferation and apoptosis of MC3T3-E1 cells. Braz. J. Med. Biol. Res. 52, e8098. doi:10.1590/1414-431X20188098
Zhou, Y., Lin, J., Shao, J., Zuo, Q., Wang, S., Wolff, A., et al. (2019). Aberrant activation of Wnt signaling pathway altered osteocyte mineralization. Bone 127, 324–333. doi:10.1016/j.bone.2019.06.027
Zhou, Y., Zhu, P., Shen, S., Wang, Y., Li, B., Guo, B., et al. (2023). Overexpression of fibroblast growth factor receptor 2 in bone marrow mesenchymal stem cells enhances osteogenesis and promotes critical cranial bone defect regeneration. Front. Cell Dev. Biol. 11, 1208239. doi:10.3389/fcell.2023.1208239
Keywords: risperidone, Traf2 and Nck-interacting protein kinase (TNIK), transcriptome analysis, protein-protein interaction, bone mineralization
Citation: Yuan R, Li Y, Li X, Fu Y, Ning A, Wang D, Zhang R, Yu S and Xu Q (2024) Transcriptome analysis to explore the mechanism of downregulated TNIK influencing the effect of risperidone. Front. Pharmacol. 15:1431923. doi: 10.3389/fphar.2024.1431923
Received: 13 May 2024; Accepted: 12 August 2024;
Published: 23 August 2024.
Edited by:
Mounir Tilaoui, Waterford Institute of Technology, IrelandReviewed by:
Rebeca Diez-Alarcia, University of the Basque Country, SpainCopyright © 2024 Yuan, Li, Li, Fu, Ning, Wang, Zhang, Yu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingqing Xu, aHVhbGl4cXFAMTYzLmNvbQ==; Shunying Yu, eXVzaHVueUB5YWhvby5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.