
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Pharmacol., 27 June 2024
Sec. Pharmacology of Anti-Cancer Drugs
Volume 15 - 2024 | https://doi.org/10.3389/fphar.2024.1428755
This article is part of the Research TopicDrug Discovery in Cancer Research: Success Stories and Open ChallengesView all 26 articles
 Tedi Rustandi1*†
Tedi Rustandi1*† Abdul Mahmud Yumassik1
Abdul Mahmud Yumassik1 Fitrah Shafran Ilahi1
Fitrah Shafran Ilahi1 Riza Alfian1†
Riza Alfian1† Erna Prihandiwati1
Erna Prihandiwati1 Yugo Susanto1
Yugo Susanto1 Yudi Hardi Susilo1
Yudi Hardi Susilo1 Maria Ulfah1
Maria Ulfah1 Faizatun Faizatun2
Faizatun Faizatun2The prevalence of cancer in 2022, according to World Health Organization (WHO) data, is 20 million new cases and 9.7 deaths. The comparison of death rates based on gender is that 1 in 9 men and 2 in 12 women die from cancer (WHO, 2024). New cancer cases in the United States (US) in 2024 will be 2,001,140, with 611,720 resulting in death (Siegel et al., 2024).
Cancer is one of the leading causes of death worldwide, with the rate of adoption of new drugs likely to be slower in clinical practice than expected. New drug development takes a long time, with an average of 13 years at a cost of ∼USD 2–3 billion (Zhang et al., 2020). This condition has global health and financial burdens (Roth et al., 2018). Discovery and development of new drugs to overcome this need to be done.
The drug repurposing method is a promising approach that will accelerate the research and development cycle. This approach is more effective in terms of cost and time than drug research and development using the de novo drug discovery approach (Tran and Prasad, 2020). Ibrexafungerp, approved by the FDA in 2021 as an antifungal derived from natural-product-based small compounds, has excellent potential to be developed using repurposing techniques to become a drug with other functions (Xu et al., 2022). The success of repurposing techniques in the development of anticancer drugs that have been approved by the FDA, such as a combination of aspirin, the antibiotic doxycycline, mifepristone, and the amino acid lysine, is used to prevent cancer metastasis (Wan et al., 2015).
The method used in this opinion article is a literature review. The literature review process uses Pubmed, Scopus, and Springer databases with criteria for articles published from 2015–2024. The article search method uses the query “repurposing therapy” AND/OR “ibrexafungerp” AND/OR “vulvovaginal candidiasis” AND/OR “cancer” AND/OR “computational screening” AND/OR “glucan synthase inhibitor” AND/OR “triterpenoid” AND/OR “ROS” AND/OR “siRNA” AND/OR “cancer mechanism” AND/OR “Tools” AND/OR “Computational” AND/OR “Artificial Intelligence” AND/OR “In Silico” AND/OR “Deep Learning” AND/OR “Machine Learning” AND/OR “bioinformatics.”
Ibrexafungerp has antifungal activity by inhibiting (1,3)-β-D-glucan synthase (Apgar et al., 2021). This mechanism gives Ibrexafungerp a good toxicity profile in host cells. The pharmacokinetic profile of Ibrexafungerp is well-classified, with the ability to penetrate tissues and organs, such as the liver, lungs, and skin. This pharmacokinetic profile is influenced by the structure of Ibrexafungerp, which has a core phenanthropyran carboxylic acid ring system at position 15 and 2-amino-2,3,3-trimethyl-butyl ether at position 14, both of which are derivatives of the naturally occurring hemiacetal triterpene glycoside enfumafungin. The pharmacokinetic profile in animals shows that Ibrexafungerp has a 30%–50% bioavailability when administered orally and has poor penetration into the central nervous system. In vitro studies show hydroxylation metabolism by the CYP3A4 isoenzyme with primary excretion via bile. The steady-state volume of distribution (Vss) profile in humans averages 600 L with high binding to protein, mainly albumin (Apgar et al., 2021; Angulo et al., 2022).
The potential of Ibrexafungerp as a cancer therapeutics is based on the use of antifungals, which have been used as anticancer agents. Antifungals with anticancer activity include itraconazole, rapamycin, griseofulvin, clotrimazole, ciclopirox, and nannocystin A (Li et al., 2022; Mohi-ud-din et al., 2023). The mechanisms of antifungal drugs that act as anticancers include the function of increasing autophagy, reducing angiogenesis, increasing tumor regression, and reducing metastasis (Mohi-ud-din et al., 2023).
Ibrexafungerp has a mechanism as a non-competitive glucan synthase inhibitor and the exact mechanism as echinocandins as an antifungal (Jallow and Govender, 2021; Shi et al., 2023; Kumar et al., 2024). Ibrexafungerp’s activity includes a broad-spectrum anti-candida fungicide against species resistant to azole drugs. Capable Candida species associated with ibrexafungerp activity include auris, dubliniensis, glabrata, guilliermondii, keyfr, krusei, lusitaniae, parapsilosis, and tropicalis (Phillips et al., 2023). Activity as a broad-spectrum antifungal, such as Candida species, indicates that ibrexafungerp may have anticancer activity. The anticancer activity of broad-spectrum antifungals such as the triazole group, namely, itraconazole, is related to the mechanism of molecular smoothened (SMO) D477G mutations, sterol carrier protein 2 (SCP2), voltage-dependent anion channel 1 (VDAC1), and Niemann-Pick Type C 1 (NPC1) (Weng et al., 2023).
The mechanism of ibrexafungerp has the same action as micafungin, which is one of the echinocandin classes of antifungal agents. The mechanisms of action of Ibrexafungerp and micafungin as antifungals may have mechanisms similar to anticancer. The predicted mechanism of ibrexafungerp is to inhibit the neddylation process by stabilizing ubiquitin-conjugating enzyme 2 M (UBE2M). This enzyme is essential in molecular mechanisms such as DNA damage, apoptosis, and cell proliferation (Mamun et al., 2023a). The prediction of the Ibrexafungerp mechanism can be seen in Figure 1A (Mamun et al., 2023b; Mamun et al., 2023a; Yu et al., 2020b; Zheng et al., 2021; Zhou et al., 2023).
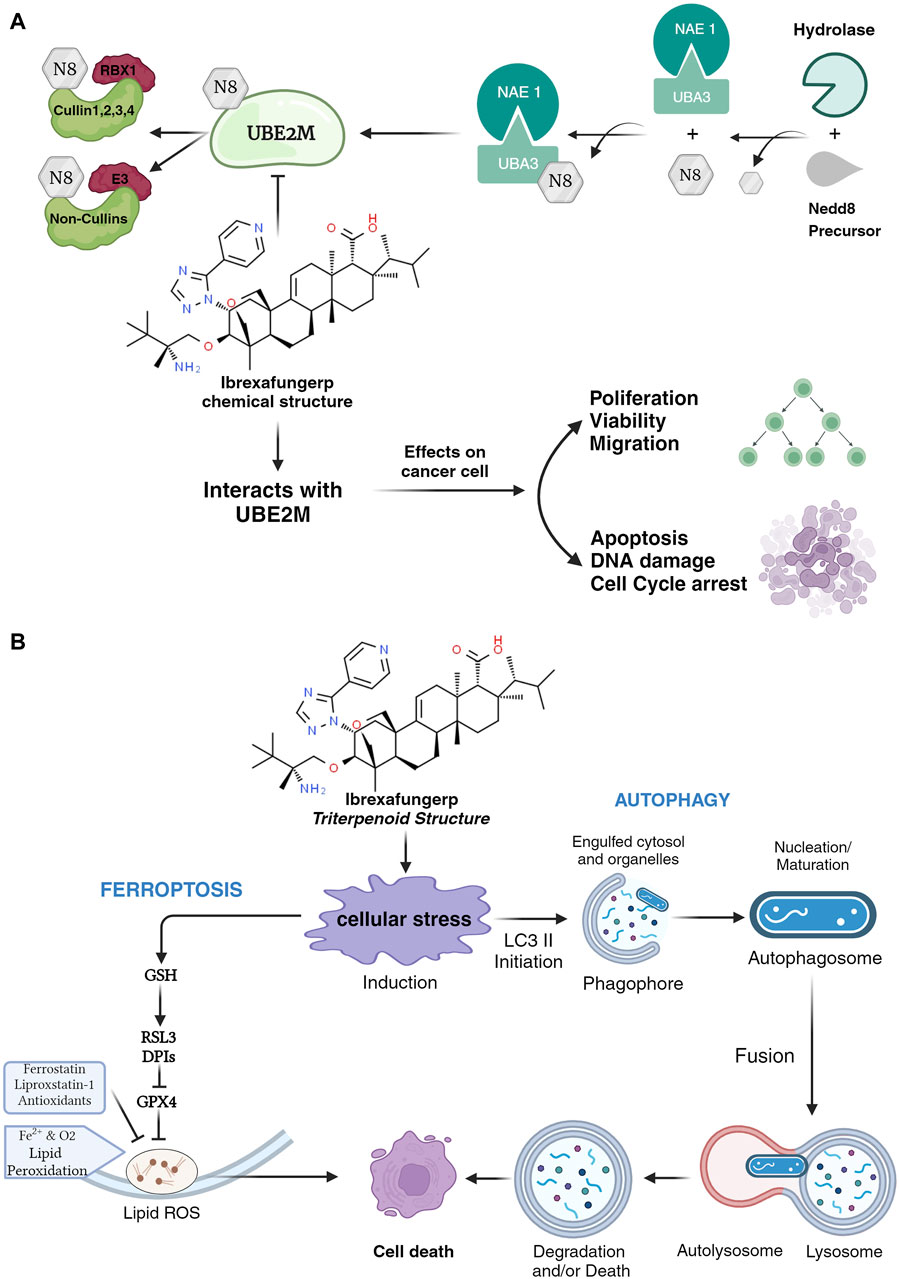
Figure 1. (A) Mechanism of Ibrexafungerp in inhibiting the neddylation process and (B) Mechanism of Ibrexafungerp as a ROS modulator.
Prediction of the mechanism of ibrexafungerp as a UBE2M inhibitor can inhibit the neddylation pathway which can reduce tumor-promoting factors and increase levels of tumor suppressors thereby improving the occurrence of tumors and prognosis (Zheng et al., 2021). Anticancers that target UBE2M in the neddylation process play a role in posttranslational modification mechanisms and target protein activity. The neddylation process begins with NEDD8 which is activated by E1 NEDD8-activating enzyme (NAE-consists of NAE1 and UBA3). This activation process results in the formation of the thioester-linked E1-NEDD8 complex which is then transferred to the NEDD8-conjugating enzyme (E2)/UBE2M (Yu et al., 2020b; Zheng et al., 2021). Ibrexafungerp inhibits the NEDD8 mechanism in UBE2M so that it cannot proceed to the next stage, namely, transferring NEDD8 from charged E2 to lysine residues in its target (Zhou et al., 2023).
Ibrexafungerp has a structure that belongs to the triterpenoid class (Angulo et al., 2022; Kumar et al., 2024). The triterpenoid group has the potential to be a cancer chemotherapy agent with a mechanism as a reactive oxygen species (ROS) modulator that can regulate cell survival and function. The impact of ROS on cancer cells is the mechanism of autophagy and ferroptosis (Endale et al., 2023; Jiang et al., 2021; Lee et al., 2023; Ling et al., 2022; Zeng et al., 2023). Autophagy works by causing cellular lipid accumulation and, ultimately, cell death. Another mechanism is inducing ferroptosis, which can cause increased chemosensitivity to chemotherapy drugs that are used to treat cancer cells. The mechanism of ibrexafungerp as a ROS modulator can be seen in Figure 1B (Ling et al., 2022).
The development of ibrexafungerp as a cancer therapeutic can be done through 2 methods: experimental screening and computational (virtual) screening (Oliveira et al., 2023; Prada Gori et al., 2023; Weth et al., 2024). Experimental screening involves in vivo and in vitro research with drug-based phenotypic screens and target-based high throughput assays. Computational (virtual) screening methods include signature matching (-omics data), artificial intelligence (machine learning and deep learning), GWAS disease/target associations, and chemical similarity and molecular docking (Weth et al., 2024). A virtual screening server that can be used in computational approaches in the development of drug repurposing research, namely, DrugRep. The use of DrugRep in drug repurposing research uses receptor-based and ligand-based screening systems (Gan et al., 2023). Several tools can be used to develop anticancer from Ibrexafungerp, some of which can be seen in Table 1.
Ibrexafungerp is predicted to have two anticancer mechanisms. The anticancer mechanism is obtained by inhibiting the neddylation stage by stabilizing UBE2M, and Ibrexafungerp acts as a ROS modulator, which acts through cell death mechanisms with autophagy and ferroptosis.
TR: Writing–original draft, Writing–review and editing, Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization. AY: Writing–review and editing, Conceptualization, Visualization. FI: Writing–review and editing, Data curation, Methodology, Software. RA: Formal Analysis, Validation, Writing–review and editing. EP: Writing–review and editing. YgS: Supervision, Writing–review and editing. YdS: Writing–review and editing, Formal Analysis. MU: Formal Analysis, Writing–review and editing. Faizatun: Writing–review and editing, Supervision.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The figure was made using BioRender (https://app.biorender.com).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Angulo, D. A., Alexander, B., Rautemaa-Richardson, R., Alastruey-Izquierdo, A., Hoenigl, M., Ibrahim, A. S., et al. (2022). Ibrexafungerp, a novel triterpenoid antifungal in development for the treatment of mold infections. J. Fungi 8 (11), 1121. doi:10.3390/jof8111121
Apgar, J. M., Wilkening, R. R., Parker, D. L., Meng, D., Wildonger, K. J., Sperbeck, D., et al. (2021). Ibrexafungerp: an orally active β-1,3-glucan synthesis inhibitor. Bioorg. Med. Chem. Lett. 32, 127661. doi:10.1016/j.bmcl.2020.127661
Bui, D. C., Song, B., Kim, K., and Kwak, J. T. (2024). DAX-Net: a dual-branch dual-task adaptive cross-weight feature fusion network for robust multi-class cancer classification in pathology images. Comput. Methods Programs Biomed. 248, 108112. doi:10.1016/j.cmpb.2024.108112
Chen, M. J. M., Li, J., Wang, Y., Akbani, R., Lu, Y., Mills, G. B., et al. (2019). TCPA v3.0: an integrative platform to explore the pan-cancer analysis of functional proteomic data. Mol. Cell. Proteomics 18 (8), S15–S25. doi:10.1074/mcp.RA118.001260
Devaraj, V., and Bose, B. (2020). DEBay: a computational tool for deconvolution of quantitative PCR data for estimation of cell type-specific gene expression in a mixed population. Heliyon 6 (7), e04489. doi:10.1016/j.heliyon.2020.e04489
Endale, H. T., Tesfaye, W., and Mengstie, T. A. (2023). ROS induced lipid peroxidation and their role in ferroptosis. Front. Cell Dev. Biol. 11, 1226044. doi:10.3389/fcell.2023.1226044
Firoozbakht, F., Rezaeian, I., Rueda, L., and Ngom, A. (2022). Computationally repurposing drugs for breast cancer subtypes using a network-based approach. BMC Bioinforma. 23 (1), 143. doi:10.1186/s12859-022-04662-6
Gabriel, A. A. G., Mathian, E., Mangiante, L., Voegele, C., Cahais, V., Ghantous, A., et al. (2020). A molecular map of lung neuroendocrine neoplasms. GigaScience 9 (11), giaa112. doi:10.1093/gigascience/giaa112
Gan, J. H., Liu, J. X., Liu, Y., Chen, S. W., Dai, W. T., Xiao, Z. X., et al. (2023). DrugRep: an automatic virtual screening server for drug repurposing. Acta Pharmacol. Sin. 44 (4), 888–896. doi:10.1038/s41401-022-00996-2
Jallow, S., and Govender, N. P. (2021). Ibrexafungerp: a first-in-class oral triterpenoid glucan synthase inhibitor. J. Fungi 7 (3), 163. doi:10.3390/jof7030163
Jiang, M., Hu, R., Yu, R., Tang, Y., and Li, J. (2021). A narrative review of mechanisms of ferroptosis in cancer: new challenges and opportunities. Ann. Transl. Med. 9 (20), 1599. doi:10.21037/atm-21-4863
Kumar, V., Huang, J., Dong, Y., and Hao, G. F. (2024). Targeting Fks1 proteins for novel antifungal drug discovery. Trends Pharmacol. Sci. 45 (4), 366–384. doi:10.1016/j.tips.2024.02.007
Lawarde, A., Sharif Rahmani, E., Nath, A., Lavogina, D., Jaal, J., Salumets, A., et al. (2024). ExplORRNet: an interactive web tool to explore stage-wise miRNA expression profiles and their interactions with mRNA and lncRNA in human breast and gynecological cancers. Non-Coding RNA Res. 9 (1), 125–140. doi:10.1016/j.ncrna.2023.10.006
Lee, S., Hwang, N., Seok, B. G., Lee, S., Lee, S. J., and Chung, S. W. (2023). Autophagy mediates an amplification loop during ferroptosis. Cell Death Dis. 14 (7), 464. doi:10.1038/s41419-023-05978-8
Li, C. L., Fang, Z. X., Wu, Z., Hou, Y. Y., Wu, H. T., and Liu, J. (2022). Repurposed itraconazole for use in the treatment of malignancies as a promising therapeutic strategy. Biomed. Pharmacother. 154, 113616. doi:10.1016/j.biopha.2022.113616
Ling, T., Boyd, L., and Rivas, F. (2022). Triterpenoids as reactive oxygen species modulators of cell fate. Chem. Res. Toxicol. 35 (4), 569–584. doi:10.1021/acs.chemrestox.1c00428
Mamun, M., Liu, Y., Geng, Y. P., Zheng, Y. C., Gao, Y., Sun, J. G., et al. (2023). Discovery of neddylation E2s inhibitors with therapeutic activity. Oncogenesis 12 (1), 45. doi:10.1038/s41389-023-00490-2
Mamun, M. A. A., Liu, S., Zhao, L., Zhao, L., Li, Z.-R., Shen, D., et al. (2023). Micafungin: a promising inhibitor of UBE2M in cancer cell growth suppression. Eur. J. Med. Chem. 260, 115732. doi:10.1016/j.ejmech.2023.115732
Mohi-ud-din, R., Chawla, A., Sharma, P., Mir, P. A., Potoo, F. H., Reiner, Ž., et al. (2023). Repurposing approved non-oncology drugs for cancer therapy: a comprehensive review of mechanisms, efficacy, and clinical prospects. Eur. J. Med. Res. 28 (1), 345. doi:10.1186/s40001-023-01275-4
Oliveira, T., Silva, M., Maia, E., Silva, A., and Taranto, A. (2023). Virtual screening algorithms in drug discovery: a review focused on machine and deep learning methods. Drugs Drug Candidates 2 (2), 311–334. doi:10.3390/ddc2020017
Phillips, N. A., Rocktashel, M., and Merjanian, L. (2023). Ibrexafungerp for the treatment of vulvovaginal Candidiasis: design, development and place in therapy. Drug Des. Dev. Ther. 17, 363–367. doi:10.2147/DDDT.S339349
Prada Gori, D. N., Ruatta, S., Fló, M., Alberca, L. N., Bellera, C. L., Park, S., et al. (2023). Drug repurposing screening validated by experimental assays identifies two clinical drugs targeting SARS-CoV-2 main protease. Front. Drug Discov. 2. doi:10.3389/fddsv.2022.1082065
Roth, G. A., Abate, D., Abate, K. H., Abay, S. M., Abbafati, C., Abbasi, N., et al. (2018). Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392 (10159), 1736–1788. doi:10.1016/S0140-6736(18)32203-7
Salnikov, M., Gameiro, S. F., Zeng, P. Y. F., Barrett, J. W., Nichols, A. C., and Mymryk, J. S. (2022). The HPV Induced Cancer Resource (THInCR): a suite of tools for investigating HPV-Dependent human carcinogenesis. MSphere 7 (4), e0031722. doi:10.1128/msphere.00317-22
Shi, Z., Zhang, J., Tian, L., Xin, L., Liang, C., Ren, X., et al. (2023). A comprehensive overview of the antibiotics approved in the last two decades: retrospects and prospects. Molecules 28 (4), 1762. doi:10.3390/molecules28041762
Siegel, R. L., Giaquinto, A. N., and Jemal, A. (2024). Cancer statistics, 2024. CA A Cancer J. Clin. 74 (1), 12–49. doi:10.3322/caac.21820
Tomczak, K., Czerwińska, P., and Wiznerowicz, M. (2015). The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Wspolczesna Onkol. 1A, A68–A77. doi:10.5114/wo.2014.47136
Tran, A. A., and Prasad, V. (2020). Drug repurposing for cancer treatments: a well-intentioned, but misguided strategy. Lancet Oncol. 21 (9), 1134–1136. doi:10.1016/S1470-2045(20)30424-1
Wan, L., Dong, H., Xu, H., Ma, J., Zhu, Y., Lu, Y., et al. (2015). Aspirin, lysine, mifepristone and doxycycline combined can effectively and safely prevent and treat cancer metastasis: prevent seeds from gemmating on soil. Oncotarget 6 (34), 35157–35172. doi:10.18632/oncotarget.6038
Wang, J., Chen, N., Guo, J., Xu, X., Liu, L., and Yi, Z. (2021). SurvNet: a novel deep neural network for lung cancer survival analysis with missing values. Front. Oncol. 10, 588990. doi:10.3389/fonc.2020.588990
Wang, L. J., Ning, M., Nayak, T., Kasper, M. J., Monga, S. P., Huang, Y., et al. (2024). shinyDeepDR: a user-friendly R Shiny app for predicting anti-cancer drug response using deep learning. Patterns 5 (2), 100894. doi:10.1016/j.patter.2023.100894
Wang, Z., Cao, S., Morris, J. S., Ahn, J., Liu, R., Tyekucheva, S., et al. (2018). Transcriptome deconvolution of heterogeneous tumor samples with immune infiltration. IScience 9, 451–460. doi:10.1016/j.isci.2018.10.028
Weng, N., Zhang, Z., Tan, Y., Zhang, X., Wei, X., and Zhu, Q. (2023). Repurposing antifungal drugs for cancer therapy. J. Adv. Res. 48, 259–273. doi:10.1016/j.jare.2022.08.018
Weth, F. R., Hoggarth, G. B., Weth, A. F., Paterson, E., White, M. P. J., Tan, S. T., et al. (2024). Unlocking hidden potential: advancements, approaches, and obstacles in repurposing drugs for cancer therapy. Br. J. Cancer 130 (5), 703–715. doi:10.1038/s41416-023-02502-9
WHO (2024) Global cancer burden growing, amidst mounting need for services. Lyon, France, Geneva, Switzerland: World Health Organization. Available at: https://www.who.int/news/item/01-02-2024-global-cancer-burden-growing--amidst-mounting-need-for-services.
Xu, Z., Eichler, B., Klausner, E. A., Duffy-Matzner, J., and Zheng, W. (2022). Lead/Drug discovery from natural resources. Molecules 27 (23), 8280. doi:10.3390/molecules27238280
Yu, C., Qi, X., Lin, Y., Li, Y., and Shen, B. (2020). iODA: an integrated tool for analysis of cancer pathway consistency from heterogeneous multi-omics data. J. Biomed. Inf. 112, 103605. doi:10.1016/j.jbi.2020.103605
Yu, Q., Jiang, Y., and Sun, Y. (2020). Anticancer drug discovery by targeting cullin neddylation. Acta Pharm. Sin. B 10 (5), 746–765. doi:10.1016/j.apsb.2019.09.005
Zeng, X. Y., Qiu, X. Z., Liang, S. M., Huang, J. A., Liu, S. Q., and Wu, J. N. (2023). Interaction mechanisms between autophagy and ferroptosis: potential role in colorectal cancer. World J. Gastrointest. Oncol. 15 (7), 1135–1148. doi:10.4251/wjgo.v15.i7.1135
Zhang, Z., Zhou, L., Xie, N., Nice, E. C., Zhang, T., Cui, Y., et al. (2020). Overcoming cancer therapeutic bottleneck by drug repurposing. Signal Transduct. Target. Ther. 5 (1), 113. doi:10.1038/s41392-020-00213-8
Zheng, Y. C., Guo, Y. J., Wang, B., Wang, C., Mamun, M. A. A., Gao, Y., et al. (2021). Targeting neddylation E2s: a novel therapeutic strategy in cancer. J. Hematol. Oncol. 14 (1), 57. doi:10.1186/s13045-021-01070-w
Zhou, L., Lin, X., Zhu, J., Zhang, L., Chen, S., Yang, H., et al. (2023). NEDD8-conjugating enzyme E2s: critical targets for cancer therapy. Cell Death Discov. 9 (1), 23. doi:10.1038/s41420-023-01337-w
Keywords: antifungal, anticancer, computational screening, glucan synthase inhibitor, ROS, triterpenoid, UBE2M, autophagy
Citation: Rustandi T, Yumassik AM, Ilahi FS, Alfian R, Prihandiwati E, Susanto Y, Susilo YH, Ulfah M and Faizatun F (2024) Repurposing therapy of ibrexafungerp vulvovaginal candidiasis drugs as cancer therapeutics. Front. Pharmacol. 15:1428755. doi: 10.3389/fphar.2024.1428755
Received: 07 May 2024; Accepted: 31 May 2024;
Published: 27 June 2024.
Edited by:
Wagdy Mohamed Eldehna, Kafrelsheikh University, EgyptReviewed by:
Haytham O. Tawfik, Tanta University, EgyptCopyright © 2024 Rustandi, Yumassik, Ilahi, Alfian, Prihandiwati, Susanto, Susilo, Ulfah and Faizatun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tedi Rustandi, dGVkaXJ1c3RhbmRpMjZAZ21haWwuY29t
†ORCID: Tedi Rustandi, orcid.org/0000-0002-5322-4476; Riza Alfian, orcid.org/0000-0001-9697-7999
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.