
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol., 25 July 2024
Sec. Neuropharmacology
Volume 15 - 2024 | https://doi.org/10.3389/fphar.2024.1428242
This article is part of the Research TopicTargeting Mitochondrial Dysfunction for the Discovery and Development of Novel CNS Therapies in Rare and Neurodegenerative DiseasesView all 8 articles
 Kassandra A. Zachos1,2
Kassandra A. Zachos1,2 Jann Aldrin Gamboa1
Jann Aldrin Gamboa1 Aleena S. Dewji1
Aleena S. Dewji1 Jocelyn Lee2
Jocelyn Lee2 Sonya Brijbassi2
Sonya Brijbassi2 Ana C. Andreazza1,3,2*
Ana C. Andreazza1,3,2*The various roles of the mitochondria and the microbiome in health and disease have been thoroughly investigated, though they are often examined independently and in the context of chronic disease. However, the mitochondria and microbiome are closely connected, namely, through their evolution, maternal inheritance patterns, overlapping role in many diseases and their importance in the maintenance of human health. The concept known as the “mitochondria-microbiome crosstalk” is the ongoing bidirectional crosstalk between these two entities and warrants further exploration and consideration, especially in the context of primary mitochondrial disease, where mitochondrial dysfunction can be detrimental for clinical manifestation of disease, and the role and composition of the microbiome is rarely investigated. A potential mechanism underlying this crosstalk is the role of metabolites from both the mitochondria and the microbiome. During digestion, gut microbes modulate compounds found in food, which can produce metabolites with various bioactive effects. Similarly, mitochondrial metabolites are produced from substrates that undergo biochemical processes during cellular respiration. This review aims to provide an overview of current literature examining the mitochondria-microbiome crosstalk, the role of commonly studied metabolites serve in signaling and mediating these biochemical pathways, and the impact diet has on both the mitochondria and the microbiome. As a final point, this review highlights the up-to-date implications of the mitochondria–microbiome crosstalk in mitochondrial disease and its potential as a therapeutic tool or target.
It is widely understood that mitochondria are essential to normal body function and human health. Their role in the production and maintenance of energy is noteworthy, yet this organelle is imperative in many other mechanisms. Mitochondria are dynamic entities that act as signalling and integration hubs mediating biochemical pathways throughout the entire body (Whitaker et al., 2016). Due to their complexity, dysfunctional mitochondria are capable of provoking and contributing to profound cellular changes that can cause complications at the biochemical level (Aguilar-López et al., 2020). Causative factors of mitochondrial dysfunction encompass a wide array of possibilities: 1) damaged mitochondrial membranes, 2) the number of functional mitochondria, 3) an impairment in provision of substrates required for energy production or 4) genetic mutations that modulate the complexes along the electron transport chain. Since mitochondrial function is influenced by both the nuclear and mitochondrial genome, the precise mechanism is highly dependent on the individual, their environment, and their genetic makeup (Garagnani et al., 2014; Nicolson et al., 2018). Mitochondrial dysfunction may have systemic effects, however its damage is most prominent in organs with a high energy demand, such as the brain, heart, and muscles. Thus, it is plausible to assert that mitochondrial health plays a central role in brain health, aging, longevity, and immune response (Nicolson et al., 2018; Chen and Vitetta, 2019).
Growing evidence supports the pivotal role of the microbiome in maintaining human health and facilitating the development of chronic conditions, such as neurovegetative diseases (Forsythe et al., 2010; Moos et al., 2018). As exemplified in the Human Microbiome Project, the diversity of microbes within and amongst even the healthiest of individuals is remarkable (Human Microbiome Project Consortium, 2012). The link between the microbiome and human disease often stems from its diversity or lack thereof (Anhê et al., 2020). Many factors, including, but not limited to, our diet, environment, lifestyle, medications, and genetics, contribute to the development and maturation of microbes colonizing our gut. Evidence suggests that specific genes within our nuclear genome, particularly those associated with metabolism and immune function, contribute to the determination of bacterial taxa in the gut (Foster and McVey Neufeld, 2013; Garagnani et al., 2014). Improved understanding of the microbiome has led scientists to consider humans as metaorganisms, leading to scientific advancements in new study areas, such as “the gut-brain axis,” and “nutritional mental health.” Communication between the microbiome and the nuclear genome has been established and many mechanisms in the areas above are still being explored (Kaplan et al., 2015; González Olmo et al., 2021).
It is theorized that if both the mitochondrial genome and the microbiome interact with the nuclear genome, they are also likely to interact with each other, giving rise to the phenomenon known as mitochondria-microbiome interplay. (Figure 1) (Garagnani et al., 2014). From an evolutionary perspective, the mitochondrion is similar to microbes and is a known descendant of alpha-proteobacteria (Bajpat et al., 2018). Furthermore, both mitochondria and the microbiome are inherited maternally, mitochondria through the transmission of the mitochondrial genome, and the microbiome through prenatal colonization (Moos et al., 2018). This literature review aims to elucidate the intricate relationship between mitochondria and the microbiome by exploring their interplay, emphasizing the significance of their respective metabolites, and investigating the implications of diet, with a particular focus on mitochondrial disease.
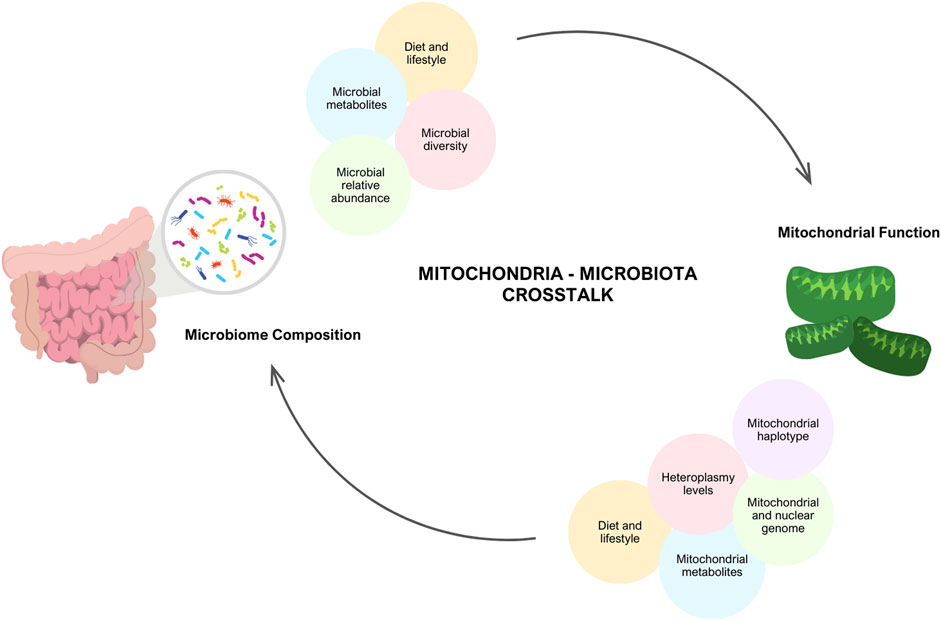
Figure 1. Interplay between Mitochondria and Gut Microbiome. There is evidence that there is communication between the mitochondria and the microbiome. Mitochondrial function is dependent on an individual’s genetics (mitochondrial and nuclear DNA mutations, heterpolasmy levels and the haplotype) and environmental factors (diet, lifestyle and stress). In turn, these factors have the potential to alter microbial diversity, relative abundance and microbial metabolites produced. Similarly, the microbiome composition and lifestyle factors (diet, exercise, medications) modulate mitochondrial function. Created with BioRender.com.
Mitochondrial disease is an umbrella term for diseases and disorders in which mitochondrial dysfunction occurs; however, these conditions can be further classified into primary and secondary mitochondria diseases (Niyazov et al., 2016; Craven et al., 2017). Mitochondrial diseases that occur due to genetic mutations in the mitochondrial genome or nuclear genome, and directly code for mitochondrial function, are considered primary mitochondrial diseases (Schapira, 2006; Alston et al., 2017). Primary mitochondrial diseases are thought to affect one in 5,000 individuals, although the incidence is expected to be higher due to challenges in proper diagnoses and limited reported data (Horan et al., 2012). They differ from other monogenic disorders in that they lack a consistent genotype-phenotype correlation (Alston et al., 2017). This inconsistency can be attributed to a combination of many factors: the inheritance pattern of the reported genetic mutation, the interactions between the mitochondrial and nuclear genome, an individual’s haplogroup (specific patterns of inherited polymorphisms that are commonly used to track ancestry), and an individual’s tissue-specific heteroplasmy (Niyazov et al., 2016; Alston et al., 2017). Heteroplasmy is especially pivotal in primary mitochondrial diseases, as there is a variable threshold at which the cell can tolerate defective mitochondrial DNA. It is when the mutation load exceeds this threshold, we see metabolic dysfunction and clinical symptoms associated with a disease state (Niyazov et al., 2016). Secondary mitochondrial diseases are more loosely defined and even harder to diagnose as they can arise from environmental triggers and mutations that do not directly encode mitochondrial function. In many cases, secondary mitochondrial diseases occur when mitochondrial dysfunction is a downstream consequence of the initiating disease processes (Niyazov et al., 2016; Murphy and Hartley, 2018).
Mitochondrial dysfunction has been reported to influence microbiome composition and thus, it is function. Yardeni et al. (2019) detailed that the gut microbiome compositions of C57BL/6J mouse models, each characterized by distinct mitochondrial mutations, exhibited correlations with their respective genotypes (Yardeni et al., 2019). Using a cross-fostering approach, the microbial profile of pups were shown to closely resemble that of the foster mother, reflecting environmental influence. However, as the pups matured, the microbial profile reverted toward the distinctive microbial signature associated with their mitochondrial genotype. A separate study found that mitochondrial dysfunction promotes gut dysbiosis in the context of IBD. Specifically, these authors found that transplantation of fecal matter from mice deficient in the mitochondrial protein methylation-controlled J (MCJ) protein to germ free mice resulted in the transferring of the increased enteric inflammation that is observed in the MCJ deficient mice (Peña-Cearra et al., 2023). Other experiments supported that increased mitochondrial reactive oxygen species (ROS) production was associated with reduced microbial diversity, which then improved when ROS production decreased, as highlighted in Table 1. The connection between the gut microbiome, the mitochondria and ROS production has been further explored in a separate review (please refer to (Ballard and Towarnicki, 2020) for details).
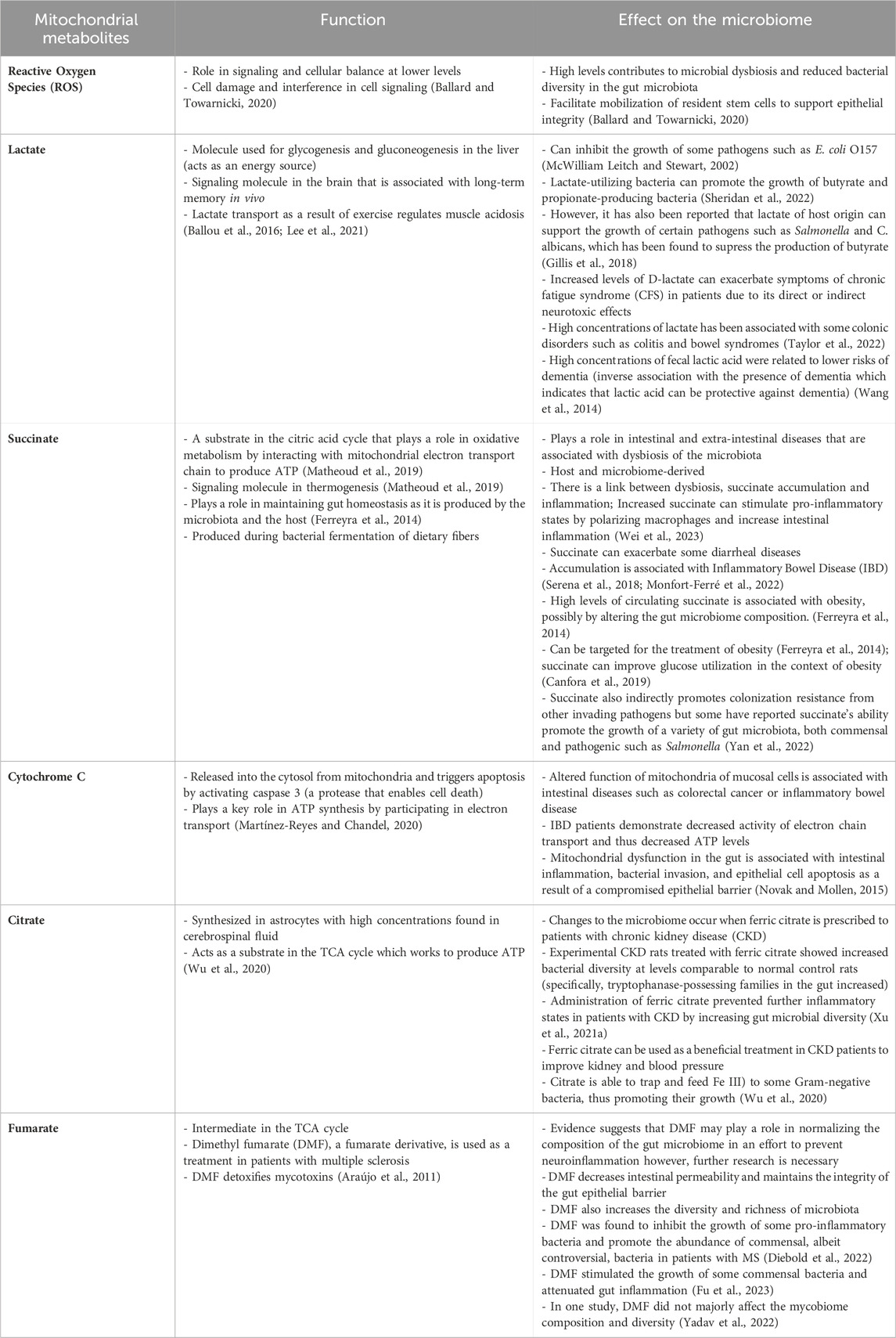
Table 1. Critical mitochondrial metabolites with various signaling functions and their potential implication on the microbiota composition and function.
Other mitochondrial metabolites include those produced in the tricarboxylic acid cycle (TCA), which play a role in thermogenesis, tumorigenesis and immune modulation (Martínez-Reyes and Chandel, 2020). Some examples of these metabolites include succinate, fumarate, citrate, succinate, cytochrome c and lactate (Table 1). Some of these metabolites, like succinate and lactate have been more thoroughly examined in the literature, while for the others, data is more limited. Despite succinate’s critical role in cell signaling, an accumulation, often caused by the inactivation of succinate dehydrogenase (SDH), has been associated with a pro-inflammatory state (Martínez-Reyes and Chandel, 2020). For instance, individuals with Inflammatory Bowel Disease (IBD) were observed to have increased concentrations of succinate-producing bacteria (Connors et al., 2018) which correlated with increased inflammatory abrasions (Macias-Ceja et al., 2019). Strikingly, some IBD patients were found to have a diminished abundance of succinate-feeding bacteria (Morgan et al., 2012). Another research group found comparable results in the context of obesity, showing increased succinate levels (Serena et al., 2018). Succinate-induced stimulation of HIF-1α, a critical curator of gut health (Kumar et al., 2020; Pral et al., 2021), is one of the key regulators of the inflammatory state (Connors et al., 2018). In addition, increased succinate produced by Bacteroides thetaiotaomicron has also been found to be a catalyst of pathogenic bacterial colonization (Curtis et al., 2014; Ferreyra et al., 2014). Together, these results suggest that high inflammatory phenotypes induced by succinate, which can be evident in various mitochondrial diseases, contribute to microbial dysbiosis.
Lactate is a unique metabolite, that was considered a by-product of metabolism for many years. It is now better understood that lactate is a critical component in metabolic homeostasis, acts to signal and mediate biochemical pathways and is generated under in several different conditions, including the gut microbiome composition (Brooks, 2020). There are several lactate-producing bacteria, that typically ferment various monosaccharides and this lactate is commonly utilized by short chain fatty acids as an energy source. There are several well-established reviews and reports on the role of lactate in the gut microbiome and thus, will not be further outlined here (Wang et al., 2020; Louis et al., 2022; McGuinness et al., 2022). However, is critical to highlight lactate in this review as lactate is very tightly linked to mitochondrial function. Typically, elevated lactate is linked to a shift from aerobic respiration, due to mitochondrial dysfunction or damage, and this lactate signals that the organism is utilizing other methods, such as anaerobic respiration, to compensate for the depletion in energy production (Brooks, 2020). Elevated lactate is also a biomarker assessed in the diagnostic process for individuals with primary mitochondrial diseases and thus is worth highlighting and conducting further studies of lactate-producing bacteria in this population (Schapira, 2006).
Fumarate, a metabolite of the TCA cycle, has been highlighted for its role in immune modulation and cancer (Araújo et al., 2011; Diebold et al., 2022). However, in the context of the gut microbiome, it has been examined the most in disease such as psoriasis, multiple sclerosis, covid, and even some cardiovascular diseases, and mostly in the form of dimethyl fumarate (DMF) (Diebold et al., 2022). DMF has some controversial findings as in some cases it has been highlighted to attenuate growth of some commensal bacteria, ultimately contributing to gut inflammation. Alternatively, in other findings it inhibits the growth of pro-inflammatory bacteria (Yadav et al., 2022; Fu et al., 2023). Cytochrome c has limited findings in the context of microbiome studies, but it is known for its role in activating caspases, encouraging cell death and releasing ROS(Martínez-Reyes and Chandel, 2020). It has been linked to altered mitochondrial function in inflammatory bowel diseases, which are known to have microbial dysbiosis, but no further links have been well established (Novak and Mollen, 2015). Lastly, citrate, a key mediator of the TCA cycle and a rate-limiting metabolite known for its role in lipid synthesis (Martínez-Reyes and Chandel, 2020). In the context of the mitochondria and the microbiome, citrate has been examined in chronic kidney diseases, where supplementation of ferric citrate has improved inflammatory states and led to increased diversity of the gut microbiome. This is potentially due to its ability to chelate iron (Fe III), which feeds Gram-negative bacteria and increase richness (Wu et al., 2020).
In a separate study using mice strains with an identical nuclear genetic background, C57BL/6J (B6.B6), and varying mitochondrial genetic backgrounds (B6. NZB, B6NOD and B6. AKR) demonstrated that improved oxidative phosphorylation resulted in increased ATP levels and was protective against colitis (Bär et al., 2013). A separate study took the opposite approach and utilized knockout mice lacking the OCTN2 carnitine transporter, which was reported to cause spontaneous colitis and inflammation. This carnitine transporter is a critical transporter of long-chain fatty acids into the mitochondria to be metabolized via beta-oxidation (Shekhawat et al., 2007). Results from Ma et al. (2014), indicate that there are specific mitochondrial haplogroups associated with microbiome compositions polymorphisms in the ND5 and cytochrome b mitochondrial genes, as well as the D-loop region, exhibited associations with stool taxonomies such as Roseburia, Eubacterium and Deltaproteobacteria, respectively.
The microbiome of a mouse strain (C57BL/6J-mtFVB/NJ) carrying a mitochondrial mutation in the ATP-synthase 8 gene, was characterized and compared to other mice strains (Hirose et al., 2017). These mice had reduced beta diversity and altered microbiomes compared to the other models, specifically showing an increased relative abundance in Bacteroidales, Deferribacteraceae, Desulfovibrionaceae, and Helicobacteraceae. C57BL/6J-mtFVB/NJ mice underwent metabolic characterization, revealing that mitochondrial OXPHOS impairment was compensated by an upregulation in glycolysis. This suggests that mitochondrial impairments play a role in modulating the composition and function of the microbiome, as demonstrated by Hirose et al. (2017). Existing literature regarding the mitochondria’s role warrants further exploration. Studies may consider examining mitochondrial metabolites altered in primary mitochondrial diseases and assessing modulations on the microbiome function.
To our knowledge, one intervention study has been conducted in a population of adults with mitochondrial disease. This study was designed to address gastrointestinal dysmotility, an uncomfortable symptom in many mitochondrial diseases (Houghton et al., 2022). This study was a single-arm pilot trial where 24 patients completed a 12-week low-residue fiber diet. Overall, this diet was well tolerated, and differences were highlighted in improvements in stool consistency, reduced use of laxatives and improved overall GI discomfort. The gut microbiome was evaluated in these participants and in 10 non-disease controls, and while there were observed differences between the patients with mitochondrial disease and controls, there were no significant changes in the patients’ microbiomes over the course of the diet. Finally, there were no observed difference in participants transit time and stool frequency (Houghton et al., 2022).
The body of literature examining the implications of the microbiome on mitochondrial function is growing rapidly, with supporting evidence that mitochondrial modifications are often mediated by bioactive metabolites produced by the microbes within the gut (Bajpat et al., 2018). For instance, short-chain fatty acids (SCFAs) and secondary bile acids have been reported to regulate redox balance, energy production, and activate AMP-activated protein kinases—a proposed connection linking SCFAs to mitochondrial biogenesis (Donohoe et al., 2011; Kim, 2018). SCFAs and mitochondrial health may also be linked through the modulation of synthesis pathways for immune messengers, such as TLR-4, IL-6, TNF-α (Luu and Visekruna, 2019; Dou et al., 2022). In addition, a specific SCFA called butyrate has been demonstrated to rescue the respiration of colonocytes in germ-free mice models as they undergo fatty acid oxidation (FAO) to produce acetyl coenzyme A (acetyl-CoA) and facilitating ATP production (Table 2) (Franco-Obregon and Gilbert, 2017; Mollica et al., 2017).
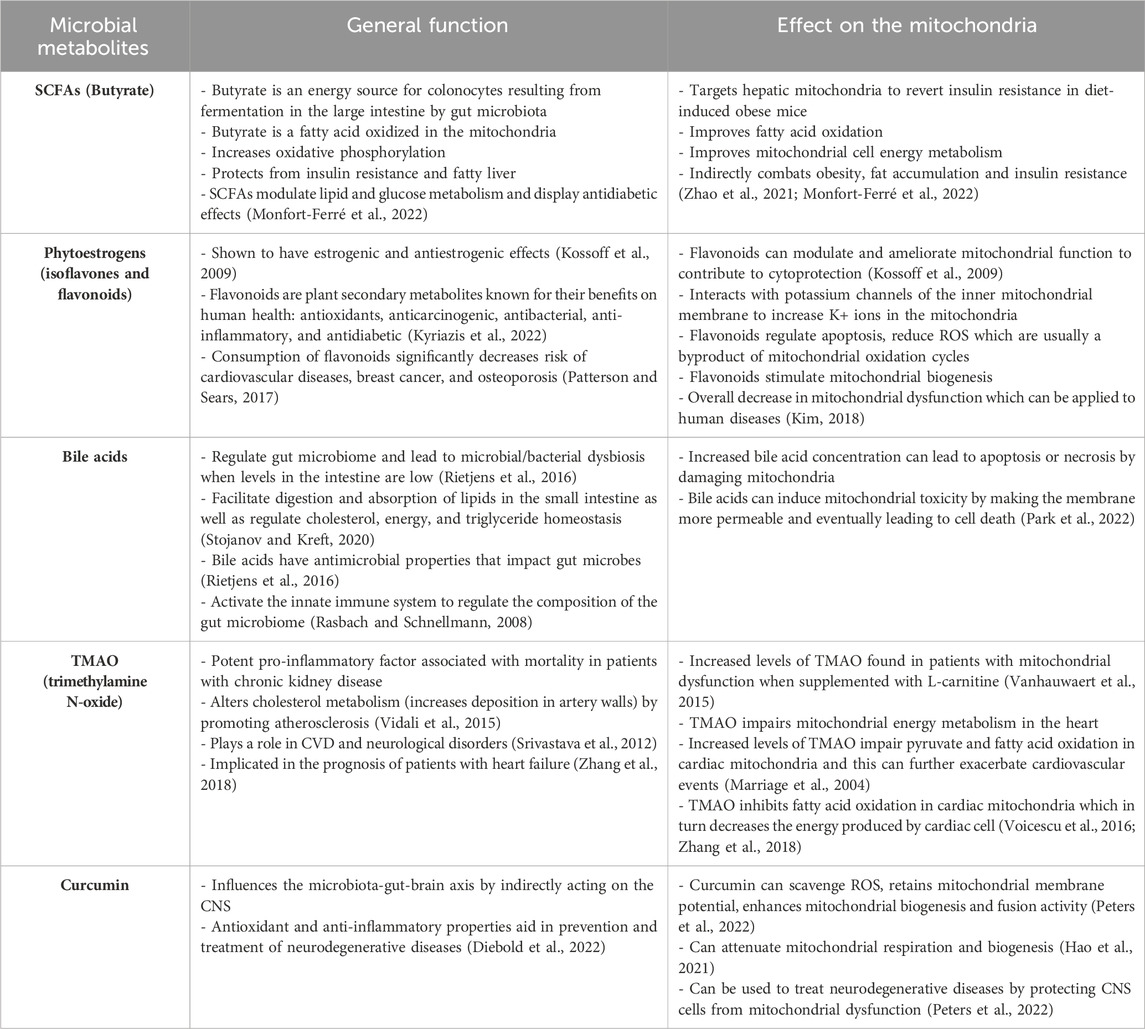
Table 2. Metabolites produced by the gut microbiota that have reported modulations on mitochondrial function.
Similar results were obtained (Hu et al., 2020) but using the combined effects of butyrate and acetate in the context of Islet cell dysfunction. Zhao et al. (2020) also investigated butyrate’s (sodium butyrate) effects on hepatic mitochondria in hopes of finding ways to restore glycemic control in individuals with diabetes. Their team discovered that sodium butyrate was able to enhance mitochondrial biogenesis and bioenergetics by increasing beta-oxidation, counteracting oxidative stress, and modulating the gene expression of several genes implicated in mitochondrial function (Zhao et al., 2020). A study developed a lymphoblastoid cell model of autism spectrum disorder, with a subset of these cells that have a mitochondrial dysfunction, as mitochondrial dysfunction has been highlighted in autism. Butyrate was then introduced at differing concentrations (0.1, 0.5, and 1 mM) for 24 h or 48 h. Interestingly, these authors found that butyrate enhances mitochondrial function as evidenced by increased ATP production in the cell lines that exhibits mitochondrial dysfunction but not in the cell lines that demonstrates normal functioning of the mitochondria. In addition, 1 mM of butyrate also lead to increased expression of genes involved in mitochondrial fission (PINK1, DRP1, and FIS1) (Rose et al., 2018).
Butyrate and urolithin A, a bioactive metabolite of ellagitannins, enhance skeletal muscle respiratory capacity and improve microbiome diversity (Franco-Obregon and Gilbert, 2017; Zhang et al., 2019). Notably, comparable results were also observed across other disease states (Bachem et al., 2019; Tang et al., 2020; Zhao et al., 2020; Hao et al., 2021; Li et al., 2023). The relative concentration of SCFAs in various disease states has also been studied based on the relative abundance of SCFA-producing bacteria. A particular research group found that some patients with Multiple Sclerosis have a diminished abundance of SCFAs-producing gut bacteria, such as Faecalibacterium prausnitzii and Roseburia intestinalis (Melbye et al., 2018). Researchers have observed this trend in other conditions as well, including IBD (Joossens et al., 2011; Kumari et al., 2013) and cerebral hypoperfusion (Su et al., 2023). However, the relative abundance of SCFA and SCFA-producing bacteria in patients with primary mitochondrial disorders is yet to be investigated.
Despite holding great promise, it is important to consider that high doses of butyrate may not be ideal, as demonstrated by Xu et al. (2021b). This study found that sodium butyrate decreased body temperature of C57BL/6 mice models by altering mitochondrial metabolism in the brain, including the TCA cycle and glycolysis, and resulted in transient mitochondrial swelling. Upon closer examination, they also discovered proton leakage via the opening of the mitochondrial permeability transition pore, which ultimately caused disintegration of the mitochondrial potential (Xu et al., 2021b). Interestingly, they found that the intervention did not alter the gene expression of their models. Rather, they suggested that the above changes occurred due to alterations in post-translational modification or mitochondrial enzymes (Xu et al., 2021b). Additionally, Peters et al. (2022) found that in vitro butyrate administration did not affect mitochondrial bioenergetics and instead, aggravated mitochondrial ROS production and dysfunction and in the context of sepsis. Clinical benefits of intravenous butyrate administration in vivo were not observed (Peters et al., 2022).
In contrast, other phenolic compounds, such as fisetin and quercetin, are converted by the microbiota to alkyl catechols and reported to induce apoptosis in senescent cells, which has been shown to increase the lifespan of mice (Shiels et al., 2019). Other studies have shown that these compounds also counteract oxidative stress and improve mitochondrial health (Karuppagounder et al., 2013; Alexandre et al., 2020; Dai et al., 2022). Curcumin, a well-known polyphenol with antioxidant and anti-inflammatory properties (Perrone et al., 2019), also becomes more biologically active after microbiome metabolism (Table 2) (Di Meo et al., 2019), suggesting that a diverse microbiome plays an important role in achieving maximum effects from such compounds. Importantly, studies demonstrated that curcumin is able to attenuate mitochondrial respiration and biogenesis, possibly via PGC1 pathway, or PPARγ and TFAM activation, among others (Eckert et al., 2013; Hamidie et al., 2015).
Phytoestrogens, which resemble human estrogens in their chemical structure and biological activity, have also been reported to affect mitochondrial function (Zaheer and Akhtar, 2017; Klinge, 2020). The isoflavones commonly found in soy products, enistin and daidzin, are phytoestrogens that are metabolized by bacterial β-glucosidases into bioactive compounds (genistein and daidzein, respectively) and have inconsistent ramifications on mitochondrial function (Kalaiselvan et al., 2010). More specifically, there is evidence of genistein increasing ATP concentrations, restoring complex activity, and maintaining membrane integrity (Rasbach and Schnellmann, 2008; de Oliveira, 2016). These results may be attributed to the compound’s ability to increase the tricarboxylic acid (TCA) cycle turnover and restore redox balance in the mitochondria (Rasbach and Schnellmann, 2008). Other research groups have also investigated the potential applications of genistein in various disease states and found overall positive results (Farruggio et al., 2019; Li et al., 2022). Notably, Li et al. (2022) found that genistein increases Mfn2 gene expression, a regulatory protein of various pathways (e.g., Ras/MAPK, PERK) that promotes mitochondrial health (Li et al., 2022). While genistein’s role in modulating mitochondrial function is more commonly investigated, there is also evidence of daidzein increasing expression of representative OXPHOS genes, such as COX1, CYTB and ATP5 (Rietjens et al., 2016; Cady et al., 2020). Yoshino et al. (2015) determined that daidzein was able to regulate mitochondrial biogenesis through a sirtuin-1 (SIRT-1)-associated pathway whereby it directly activated the mitochondrial transcription factor A (TFAM) promotor (Yoshino et al., 2015; Kicinska and Jarmuszkiewicz, 2020). Isoflavones’ role as an antioxidant is reported to act through different mechanisms. In some studies, daidzein and genistein function as ROS scavengers themselves, however in others, they mediate an increase in the production of other antioxidants such as glutathione (Lee et al., 2006; Rasbach and Schnellmann, 2008; Qian et al., 2012; Kładna et al., 2016; Voicescu et al., 2016).
Negative implications of isoflavones include the inhibition of mitochondrial function through its interaction with complex III, although this has only been examined at the cellular level, as opposed to the whole-body level (Salvi et al., 2002). Genistein was also shown to induce mitochondrial swelling, a loss of the mitochondrial membrane potential, and trigger the release of accumulated calcium (Salvi et al., 2002). Moreover, this compound triggered mitochondrial dysfunction through the upregulation cytochrome C and ROS production (Franco-Obregon and Gilbert, 2017; Lee et al., 2019; Chan et al., 2022). These effects are desirable when targeting malignant cells as they could potentially lead to apoptosis. However, these effects remain harmful to healthy cells and consequently, individuals.
Hydrogen sulfide (H2S) is a by-product of amino acid digestion and negatively impacts mitochondrial function by inhibiting cytochrome oxidase, a complex in the electron transport chain (ETC.) (Leschelle et al., 2005; Franco-Obregon and Gilbert, 2017). However, this is only reported to occur at high concentrations, since low levels of H2S can be reduced to form sulfide and used as a substrate in metabolism (Goubern et al., 2007; Saint-Georges-Chaumet and Edeas, 2015). Bile acids also have varying effects on mitochondrial function and at high levels, can contribute to apoptosis and necrosis through the nucleotide oligomerization domain (NOD)-like receptor protein-3 (NLRP3) inflammasome complex and membrane permeability (Table 2) (Palmeira and Rolo, 2004; Donohoe et al., 2011). Finally, trimethyl N-oxide (TMAO) is a potent pro-inflammatory metabolite typically associated with the consumption of red meat and has demonstrated the ability to impair mitochondrial function through pyruvate and fatty acid oxidation (Table 2) (Velasquez et al., 2016; Vallance et al., 2018; Zhang et al., 2021). In addition to impacting the mitochondria directly, various metabolites produced by pathogenic bacteria have been shown to have significant effects. One notable study by Matheoud et al. (2019) revealed that lipopolysaccharides (LPS) from Gram-negative bacteria can be taken up by antigen-presenting cells, triggering an autoimmune reaction against crucial neurons in the brain. The researchers utilized Pink1−/− mice for their experiments, building on (Matheoud et al., 2016) previous findings that the protein Parkin prevents mitochondrial antigen presentation. The study demonstrated that CD8+ T cells, responsible for the autoimmune response, specifically target mitochondrial antigens expressed by dopaminergic neurons, leading to the induction of parkinsonism (Matheoud et al., 2019). Subsequent experiments confirmed this association, as the administration of L-DOPA reversed the condition (Matheoud et al., 2019). Interestingly, another research group discovered that sodium butyrate administration can alleviate LPS-induced pathologies by modulating T-cell activation, as reported by Dou et al. (2022). Moreover, a separate study (Li et al., 2022)found that butyric acid, a derivative of sodium butyrate, may promote mitochondrial function, potentially by influencing macrophage polarization.
It is clear there is an abundance of literature on microbial metabolites and mitochondrial function. A previous review has summarized the clinical applications of various flavonoids in the context of several diseases where mitochondriopathy is implicated (Koklesova et al., 2021). However, it is limited in its application towards primary mitochondrial diseases, in which the mitochondrial function is greatly impaired and may result in altered effects of such compounds. To our knowledge, these metabolites have not been investigated in interventional studies to improve mitochondrial functions in individuals with mitochondrial disorders. One research group has found elevation in plasma TMAO in patients with mitochondrial disorders treated with oral l-carnitine (Vallance et al., 2018). However, carnitine’s and TMAO’s efficacy in treating mitochondrial dysfunction has not been systematically evaluated. Interestingly, a separate research group investigated blood levels of TMA and TMAO, as well as mitochondrial DNA copy number in persons with healthy and unhealthy western diet (Bordoni et al., 2022). These authors found that there are no significant differences in the levels of TMAO between these two groups, or mitochondrial DNA copy number. Moreover, they found no significant correlation between TMAO and mitochondrial DNA copy number. TMAO along with other enteric microbiome-derived metabolites’ potential in ameliorating mitochondrial dysfunction has not been deeply explored clinically, which should be the focus of future intervention trials.
All literature highlighted and discussed in this review highlights some of the research that has been conducted examining the relationship between mitochondria and the gut microbiome, but it is clear that further research is required before we can utilize this relationship as a therapeutic intervention in primary mitochondrial diseases. In fact, conducting research to assess and improve our understanding on the interplay between the mitochondria and the microbiota is not anticipated to be an easy feat. These two entities are highly complex and challenging to understand and utilize as therapeutic interventions when assessed individually. The mitochondrion has a double-membraned structure, and various heteroplasmy levels in different tissues, which poses challenges in therapeutic design and effectiveness (Tyynismaa and Suomalainen, 2009). The microbiota is highly variable, and the gut microbiota is only one of several sources in the body where bacteria resides (Flint, 2020). Challenges that can be anticipated in future research studies include designing studies that will help elicit causality. Participant enrolment and controlling interventions are only two of several anticipated challenges. Animal studies are a useful tool when assessing causality, however generalizing any findings should be done cautiously (Saccone et al., 1991; Tyynismaa and Suomalainen, 2009). Lastly, off-target effects and systemic consequences are potential therapeutic adverse effects that need to be considered, especially since the mitochondria are found throughout the entire body and both the mitochondria and microbiota have effects on other systems, through the gut-brain axis, immune system, and metabolism, to name a few (Flint, 2020).
Alternatively, improving our understanding on the mitochondria-microbiome crosstalk has the potential for profound effects on personalized therapeutics, and overall efficacy of individualized treatments we design. Firstly, utilizing mitochondrial heteroplasmy, haplogroups, and mitochondrial DNA in the assessment of gut microbiota profiles, may demonstrate trends that we can utilize in our understanding on the development, function and metabolites produced from the microbiome. The pursuit of microbiome studies at the single-cell level, using tools such as microfluidics, fluorescence-activated cell sorting, and laser capture microdissection, can provide detailed insight on the interactions between the mitochondria and specific gut microbes. The utilization of Artificial Intelligence (AI) can also help sort and understand connections with all of these trends we have highlighted in the field up until this point. Lastly, in the context of mitochondrial disease, therapeutics for these individuals are highly limited. The introduction of fecal microbial transplants has the potential to improve the gastrointestinal discomfort, mood related consequences and immune function of these individuals. Understanding the gut microbiota can potentially highlight trends in specific mitochondrial diseases and improve patients stratification in these complex diseases.
This review underscores the intricate and dynamic relationship between mitochondrial dysfunction and the gut microbiome, highlighting the potential role of mitochondria-microbiome crosstalk in the pathophysiology of mitochondrial diseases. The bidirectional interactions between mitochondrial metabolites and microbial byproducts not only influence systemic metabolic pathways but also hold significant implications for disease manifestation and progression. Given the emerging evidence, targeting this crosstalk presents a promising therapeutic avenue. Future research should aim to deepen our understanding of these interactions through more detailed mechanistic studies, which could pave the way for novel dietary and microbial-based interventions. Such strategies may offer significant benefits, improving clinical outcomes for individuals suffering from mitochondrial disorders. Embracing a holistic view of these interactions, integrating genomic, proteomic, and metabolomic data, will be essential for developing personalized medicine approaches that can effectively address the complexities of mitochondrial diseases.
KZ: Writing–review and editing, Writing–original draft, Methodology, Data curation, Conceptualization. JG: Writing–review and editing, Writing–original draft. AD: Writing–review and editing, Writing–original draft. JL: Writing–review and editing. SB: Writing–review and editing. AA: Writing–review and editing, Visualization, Resources, Project administration, Methodology, Funding acquisition, Conceptualization.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aguilar-López, B. A., Moreno-Altamirano, M. M. B., Dockrell, H. M., Duchen, M. R., and Sánchez-Garcia, F. J. (2020). Mitochondria: an integrative hub coordinating circadian rhythms, metabolism, the microbiome, and immunity. Front. Cell Dev. Biol. 8 (51), 51. doi:10.3389/fcell.2020.00051
Alexandre, J. V. L., Viana, Y. I. P., David, C. E. B., Cunha, P. L. O., Albuquerque, A. C., Varela, A. L. N., et al. (2020). Quercetin treatment increases H2O2 removal by restoration of endogenous antioxidant activity and blocks isoproterenol-induced cardiac hypertrophy. Berlin, Germany: Naunyn-Schmiedeberg’s Archives.
Alston, C. L., Rocha, M. C., Lax, N. Z., Turnbull, D. M., and Taylor, R. W. (2017). The genetics and pathology of mitochondrial disease. J. Pathol. 241, 236–250. doi:10.1002/path.4809
Anhê, F. F., Jensen, B. A. H., Varin, T. B., Servant, F., Van Blerk, S., Richard, D., et al. (2020). Type 2 diabetes influences bacterial tissue compartmentalisation in human obesity. Nat. Metab. 2, 233–242. doi:10.1038/s42255-020-0178-9
Araújo, W. L., Nunes-Nesi, A., and Fernie, A. R. (2011). Fumarate: multiple functions of a simple metabolite. Phytochemistry 72 (9), 838–843. doi:10.1016/j.phytochem.2011.02.028
Bachem, A., Makhlouf, C., Binger, K. J., de Souza, D. P., Tull, D., Hochheiser, K., et al. (2019). Microbiota-Derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity 51 (2), 285–297. doi:10.1016/j.immuni.2019.06.002
Bajpat, P., Darra, A., and Agrawal, A. (2018). Microbe-Mitochondrion crosstalk and health: an emerging paradigm. Mitochondrion 39, 20–25. doi:10.1016/j.mito.2017.08.008
Ballard, J. W. O., and Towarnicki, S. G. (2020). Mitochondria, the gut microbiome and ROS. Cell Signal 75, 109737. doi:10.1016/j.cellsig.2020.109737
Ballou, E. R., Mol Avelar, G., Childers, D. S., Mackie, J., Bain, J. M., Wagener, J., et al. (2016). Lactate signalling regulates fungal β-glucan masking and immune evasion. Nat. Microbiol. 2 (2), 16238. doi:10.1038/nmicrobiol.2016.238
Bär, F., Bochmann, W., Widok, A., Von Medem, K., Pagel, R., Hirose, M., et al. (2013). Mitochondrial gene polymorphisms that protect mice from colitis. Gastroenterol 145, 1055–1063. doi:10.1053/j.gastro.2013.07.015
Bordoni, L., Malinowska, A., Petracci, I., Szwengiel, A., Gabbianelli, R., and Chmurzynska, A. (2022). Diet, trimethylamine metabolism, and mitochondrial DNA: an observational study. Mol. Nutr. Food Res. 66 (13), e2200003. doi:10.1002/mnfr.202200003
Brooks, G. A. (2020). Lactate as a fulcrum of metabolism. Redox Biol., 35, 101454. doi:10.1016/j.redox.2020.101454
Cady, N., Peterson, S. R., Freedman, S. N., and Mangalam, A. K. (2020). Beyond metabolism: the complex interplay between dietary phytoestrogens, gut bacteria and cells of nervous and immune systems. Front. Neurol. 11 (150), 150. doi:10.3389/fneur.2020.00150
Canfora, E. E., Meex, R. C. R., Venema, K., and Blaak, E. E. (2019). Gut microbial metabolites in obesity, NAFLD and T2DM. Nat. Rev. Endocrin 15 (5), 261–273. doi:10.1038/s41574-019-0156-z
Chan, L., Pang, Y., Wang, Y., Zhu, D., Taledaohan, A., Jia, Y., et al. (2022). Genistein-induced mitochondrial dysfunction and FOXO3a/PUMA expression in non-small lung cancer cells. Pharm. Biol. 60 (10), 1876–1883. doi:10.1080/13880209.2022.2123933
Chen, J., and Vitetta, L. (2019). Mitochondria could be a potential key mediator linking the intestinal microbiota to depression. J. Cell Biochem. 121, 17–24. doi:10.1002/jcb.29311
Connors, J., Dawe, N., and Van Limbergen, J. (2018). The role of succinate in the regulation of intestinal inflammation. Nutrients 11 (25), 25. doi:10.3390/nu11010025
Craven, L., Alston, C. L., Taylor, R. W., and Turnbull, D. M. (2017). Recent advances in mitochondrial disease. Annu. Rev. Genom. Hum. Gen. 18, 257–275. doi:10.1146/annurev-genom-091416-035426
Curtis, M. M., Hu, Z., Klimko, C., Narayanan, S., Deberardinis, R., and Sperandio, V. (2014). The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 16 (6), 759–769. doi:10.1016/j.chom.2014.11.005
Dai, X., Kuang, Q., Sun, Y., Xu, M., Zhu, L., Ge, C., et al. (2022). Fisetin represses oxidative stress and mitochondrial dysfunction in NAFLD through suppressing GRP78-mediated endoplasmic reticulum (ER) stress. J. Funct. Foods 90, 104954. doi:10.1016/j.jff.2022.104954
de Oliveira, M. R. (2016). Evidence for genistein as a mitochondriotropic molecule. Mitochondrion 29, 35–44. doi:10.1016/j.mito.2016.05.005
Diebold, M., Meola, M., Purushothaman, S., Siewert, L. K., Pössnecker, E., Roloff, T., et al. (2022). Gut microbiota composition as a candidate risk factor for dimethyl fumarate-induced lymphopenia in multiple sclerosis. Gut Microbes 14 (1), 2147055. doi:10.1080/19490976.2022.2147055
Di Meo, F., Margarucci, S., Galderisi, U., Crispi, S., and Peluso, G. (2019). Curcumin, gut microbiota and neuroprotection. Nutrients 11, 2426. doi:10.3390/nu11102426
Donohoe, D. R., Garge, N., Zhang, X., Sun, W., O’Connell, T. M., Bunger, M. K., et al. (2011). The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Met. 13, 517–526. doi:10.1016/j.cmet.2011.02.018
Dou, X., Ma, Z., Yan, D., Gao, N., Li, Z., Li, Y., et al. (2022). Sodium butyrate alleviates intestinal injury and microbial flora disturbance induced by lipopolysaccharides in rats. Food and Funct. 13 (3), 1360–1369. doi:10.1039/d1fo03183j
Eckert, G. P., Schiborr, C., Hagl, S., Abdel-Kader, R. M., Müller, W. E., Rimbach, G., et al. (2013). Curcumin prevents mitochondrial dysfunction in the brain of the senescence-accelerated mouse-prone. Neurochem. Int. 62 (5), 595–602. doi:10.1016/j.neuint.2013.02.014
Farruggio, S., Raina, G., Cocomazzi, G., Librasi, C., Mary, D., Gentilli, S., et al. (2019). Genistein improves viability, proliferation and mitochondrial function of cardiomyoblasts cultured in physiologic and peroxidative conditions. Int. J. Mol. Med. 44, 2298–2310. doi:10.3892/ijmm.2019.4365
Ferreyra, J. A., Wu, K. J., Hryckowian, A. J., Bouley, D. M., Weimer, B. C., and Sonnenburg, J. L. (2014). Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 16 (6), 770–777. doi:10.1016/j.chom.2014.11.003
Flint, H. J. (2020). Variability and stability of the human gut microbiome. In Why gut microbes matter (Cham: Springer), 63–79. doi:10.1007/978-3-030-43246-1_6
Forsythe, P., Sudo, N., Dinan, T., Taylor, V. H., and Bienenstock, J. (2010). Mood and gut feelings. Brain Behav. Immun. 24, 9–16. doi:10.1016/j.bbi.2009.05.058
Foster, J. A., and McVey Neufeld, K. A. (2013). Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 36, 305–312. doi:10.1016/j.tins.2013.01.005
Franco-Obregon, A., and Gilbert, J. A. (2017). The microbiome-mitochondrion connection: common ancestries, common mechanisms, common goals. mSystems 2, 000188-17. doi:10.1128/mSystems.00018-17
Fu, L., Cai, W., Liu, H., Hooft, J. M., Øverland, M., Han, D., et al. (2023). Effects of 4-octyl itaconate and dimethyl fumarate on growth performance, intestinal microbiota, intestinal and hepatopancreas health of juvenile gibel carp (Carassius gibelio). Aquaculture 569, 739376. doi:10.1016/j.aquaculture.2023.739376
Garagnani, P., Pirazzini, C., Giuliani, C., Candela, M., Brigidi, P., Sevini, F., et al. (2014). The three genetics (nuclear DNA, mitochondrial DNA, and gut microbiome) of longevity in humans considered as metaorganisms. Biomed. Res. Int. 2014, 560340. doi:10.1155/2014/560340
Gillis, C. C., Hughes, E. R., Spiga, L., Winter, M. G., Zhu, W., Furtado de Carvalho, T., et al. (2018). Dysbiosis-associated change in host metabolism generates lactate to support Salmonella growth. Cell Host Microbe 23 (4), 54–64. doi:10.1016/j.chom.2017.11.006
González Olmo, B. M., Butler, M. J., and Barrientos, R. M. (2021). Evolution of the human diet and its impact on gut microbiota, immune responses, and brain health. Nutrients 13 (196), 196. doi:10.3390/nu13010196
Goubern, M., Andriamihaja, M., Nubel, T., Blachier, F., and Bouillard, F. (2007). Sulfide, the first inorganic substrate for human cells. FASEB J. 21, 1699–1706. doi:10.1096/fj.06-7407com
Hamidie, R. D. R., Yamada, T., Ishizawa, R., Saito, Y., and Masuda, K. (2015). Curcumin treatment enhances the effect of exercise on mitochondrial biogenesis in skeletal muscle by increasing cAMP levels. Metabolism 64 (10), 1334–1347. doi:10.1016/j.metabol.2015.07.010
Hao, F., Tian, M., Zhang, X., Jin, X., Jiang, Y., Sun, X., et al. (2021). Butyrate enhances CPT1A activity to promote fatty acid oxidation and iTreg differentiation. Proc. Natl. Acad. Sci. 118 (22), e2014681118. doi:10.1073/pnas.2014681118
Hirose, M., Künstner, A., Schilf, P., Sünderhauf, A., Rupp, J., Jöhren, O., et al. (2017). Mitochondrial gene polymorphism is associated with gut microbial communities in mice. Sci. Rep. 7, 15293. doi:10.1038/s41598-017-15377-7
Horan, M. P., Pichaud, N., and Ballard, J. W. O. (2012). Review: quantifying mitochondrial dysfunction in complex diseases of aging. J. Gerontol. A Biol. Sci. Med. Sci. 67, 1022–1035. doi:10.1093/gerona/glr263
Houghton, D., Ng, Y. S., Jackson, M. A., Stefanetti, R., Hynd, P., Aogáin, M. M., et al. (2022). Phase II feasibility study of the efficacy, tolerability, and impact on the gut microbiome of a low-residue (fiber) diet in adult patients with mitochondrial disease. Gastro Hep Adv. 1, 666–677. doi:10.1016/j.gastha.2022.03.007
Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi:10.1038/nature11234
Hu, S., Kuwabara, R., de Haan, B. J., Smink, A. M., and de Vos, P. (2020). Acetate and butyrate improve β-cell metabolism and mitochondrial respiration under oxidative stress. Int. J. Mol. Sci. 21 (4), 1542. doi:10.3390/ijms21041542
Joossens, M., Huys, G., Cnockaert, M., De Preter, V., Verbeke, K., Rutgeerts, P., et al. (2011). Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 60 (5), 631–637. doi:10.1136/gut.2010.223263
Kalaiselvan, V., Kalaivani, M., Vijayakumar, A., Sureshkumar, K., and Venkateskumar, K. (2010). Current knowledge and future direction of research on soy isoflavones as a therapeutic agents. Pharmacogn. Rev. 4, 111–117. doi:10.4103/0973-7847.70900
Kaplan, B. J., Rucklidge, J. J., Romijn, A., and McLeod, K. (2015). The emerging field of nutritional mental health: inflammation, the microbiome, oxidative stress, and mitochondrial function. Clin. Psychol. Sci. 3, 964–980. doi:10.1177/2167702614555413
Karuppagounder, S. S., Madathil, S. K., Pandey, M., Haobam, R., Rajamma, U., and Mohanakumar, K. P. (2013). Quercetin up-regulates mitochondrial complex-I activity to protect against programmed cell death in rotenone model of Parkinson’s disease in rats. Neuroscience 236, 136–148. doi:10.1016/j.neuroscience.2013.01.032
Kicinska, A., and Jarmuszkiewicz, W. (2020). Flavanoids and mitochondria: activation of cytoprotective pathways? Molecules 25 (3060), 3060. doi:10.3390/molecules25133060
Kim, C. H. (2018). Immune regulation by microbiome metabolites. Immunol 154, 220–229. doi:10.1111/imm.12930
Kładna, A., Berczyński, P., Kruk, I., Piechowska, T., and Aboul-Enein, H. Y. (2016). Studies on the antioxidant properties of some phytoestrogens. Luminescence 31, 1201–1206. doi:10.1002/bio.3091
Klinge, C. M. (2020). Estrogenic control of mitochondrial function. Redox Biol. 31, 101435. doi:10.1016/j.redox.2020.101435
Koklesova, L., Liskova, A., Samec, M., Zhai, K., AL-Ishaq, R. K., Bugos, O., et al. (2021). Protective effects of flavonoids against mitochondriopathies and associated pathologies: focus on the predictive approach and personalized prevention. Int. J. Mol. Sci. 22 (16), 8649. doi:10.3390/ijms22168649
Kossoff, E. H., Zupec-Kania, B., and Rho, J. M. (2009). Ketogenic diets: an update for child neurologists. J. Child Neurology 24 (8), 979–988. doi:10.1177/0883073809337162
Kumar, T., Pandey, R., and Chauhan, N. S. (2020). Hypoxia inducible factor-1α: the curator of gut homeostasis. Front. Cell. Infect. Microbiol. 10, 227. doi:10.3389/fcimb.2020.00227
Kumari, R., Ahuja, V., and Paul, J. (2013). Fluctuations in butyrate-producing bacteria in ulcerative colitis patients of North India. World J. Gastroenterology 19 (22), 3404–3414. doi:10.3748/wjg.v19.i22.3404
Kyriazis, I., Vassi, E., Alvanou, M., Angelakis, C., Skaperda, Z., Tekos, F., et al. (2022). The impact of diet upon mitochondrial physiology (Review). Int. J. Mol. Med. 50, 135. doi:10.3892/ijmm.2022.5191
Lee, S. R., Kwon, S. W., Lee, Y. H., Kaya, P., Kim, J. M., Ahn, C., et al. (2019). Dietary intake of genistein suppresses hepatocellular carcinoma through AMPK-mediated apoptosis and anti-inflammation. BMC Cancer 19 (1), 6. doi:10.1186/s12885-018-5222-8
Lee, Y. M., Choi, J. S., Kim, M. H., Jung, M. H., Lee, Y. S., and Song, J. (2006). Effects of dietary genistein on hepatic lipid metabolism and mitochondrial function in mice fed high-fat diets. Nutrients 22, 956–964. doi:10.1016/j.nut.2005.12.014
Lee, Y.-S., Kim, T.-Y., Kim, Y., Kim, S., Lee, S.-H., Seo, S.-U., et al. (2021). Microbiota-derived lactate promotes hematopoiesis and erythropoiesis by inducing stem cell factor production from leptin receptor+ niche cells. Exp. Mol. Med. 53 (9), 1319–1331. doi:10.1038/s12276-021-00667-y
Leschelle, X., Goubern, M., Andriamihaja, M., Blottière, H. M., Couplan, E., Gonzalez-Barroso, M., et al. (2005). Adaptative metabolic response of human colonic epithelial cells to the adverse effects of the luminal compound sulfide. Biochem. Biophys. Acta 1725, 201–212. doi:10.1016/j.bbagen.2005.06.002
Li, Y., Ou, S., Liu, Q., Gan, L., Zhang, L., Wang, Y., et al. (2022). Genistein improves mitochondrial function and inflammatory in rats with diabetic nephropathy via inhibiting MAPK/NF-κB pathway. Acta Cirúrgica Bras. 37 (6), e370601. doi:10.1590/acb370601
Li, Z., Zhou, E., Liu, C., Wicks, H., Yildiz, S., Razack, F., et al. (2023). Dietary butyrate ameliorates metabolic health associated with selective proliferation of gut Lachnospiraceae bacterium. JCI Insight 8 (4), e166655. doi:10.1172/jci.insight.166655
Louis, P., Duncan, S., Sheridan, P., Walker, A., and Flint, H. (2022). Microbial lactate utilisation and the stability of the gut microbiome. Gut Microbiome 3 (e3), e3–e16. doi:10.1017/gmb.2022.3
Luu, M., and Visekruna, A. (2019). Short-chain fatty acids: bacterial messengers modulating the immunometabolism of T cells. Eur. J. Immunol. 4 (9), 842–848. doi:10.1002/eji.201848009
Ma, J., Coarfa, C., Qin, X., Bonnen, P. E., Milosavljevic, A., Versalovic, J., et al. (2014). mtDNA haplogroup and single nucleotide polymorphisms structure human microbiome communities. BMC Genomics 15 (257), 257. doi:10.1186/1471-2164-15-257
Macias-Ceja, D. C., Ortiz-Masiá, D., Salvador, P., Gisbert-Ferrándiz, L., Hernández, C., Hausmann, M., et al. (2019). Succinate receptor mediates intestinal inflammation and fibrosis. Mucosal Immunol. 12 (1), 178–187. doi:10.1038/s41385-018-0087-3
Marriage, B. J., Clandinin, M. T., Macdonald, I. M., and Glerum, D. M. (2004). Cofactor treatment improves ATP synthetic capacity in patients with oxidative phosphorylation disorders. Mol. Genet. Metabolism 81 (4), 263–272. doi:10.1016/j.ymgme.2003.12.008
Martínez-Reyes, I., and Chandel, N. S. (2020). Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 11 (102), 102. doi:10.1038/s41467-019-13668-3
Matheoud, D., Cannon, T., Voisin, A., Penttinen, A.-M., Ramet, L., Fahmy, A. M., et al. (2019). Intestinal infection triggers Parkinson’s disease-like symptoms in Pink1−/− mice. Nature 571 (7766), 565–569. doi:10.1038/s41586-019-1405-y
Matheoud, D., Sugiura, A., Bellemare-Pelletier, A., Laplante, A., Rondeau, C., Chemali, M., et al. (2016). Parkinson’s disease-related proteins PINK1 and parkin repress mitochondrial antigen presentation. Cell 166 (2), 314–327. doi:10.1016/j.cell.2016.05.039
McGuinness, A., Davis, A., Dawson, S., Loughman, A., Collier, F., O’Hely, M., et al. (2022). A systematic review of gut microbiota composition in observational studies of major depressive disorder, bipolar disorder and schizophrenia. Mol. Psychiatry 27, 1920–1935. doi:10.1038/s41380-022-01456-3
McWilliam Leitch, E. C., and Stewart, C. S. (2002). Escherichia coli O157 and non-O157 isolates are more susceptible to l -lactate than to d -lactate. Appl. Environ. Microbiol. 68 (9), 4676–4678. doi:10.1128/aem.68.9.4676-4678.2002
Melbye, P., Olsson, A., Hansen, T. H., Søndergaard, H. B., and Bang Oturai, A. (2018). Short-chain fatty acids and gut microbiota in multiple sclerosis. Acta Neurol. Scand. 139 (3), 208–219. doi:10.1111/ane.13045
Mollica, P. M., Raso, G. M., Cavaliere, G., Trinchese, G., De Filippo, C., Aceto, S., et al. (2017). Butyrate regulates liver mitochondrial function, efficiency, and dynamics in insulin-resistant obese mice. Diabetes 66 (1045-1418), 1405–1418. doi:10.2337/db16-0924
Monfort-Ferré, D., Caro, A., Menacho, M., Martí, M., Espina, B., Boronat-Toscano, A., et al. (2022). The gut microbiota metabolite succinate promotes adipose tissue browning in crohn’s disease. J. Crohn’s Colitis 16 (10), 1571–1583. doi:10.1093/ecco-jcc/jjac069
Moos, W. H., Faller, D. V., Harpp, D. N., Kanara, I., Pernokas, J., Powers, W. R., et al. (2018). Microbiota and neurological disorders: a gut feeling. Biores Open Access 5, 137–145. doi:10.1089/biores.2016.0010
Morgan, X. C., Tickle, T. L., Sokol, H., Gevers, D., Devaney, K. L., Ward, D. V., et al. (2012). Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13 (9), R79. doi:10.1186/gb-2012-13-9-r79
Murphy, M. P., and Hartley, R. C. (2018). Mitochondria as a therapeutic target for common pathologies. Nat. Rev. Drug Discov. 17, 865–886. doi:10.1038/nrd.2018.174
Nicolson, G. L., Ferreira, G., Settineri, R., Ellithorpe, R. R., Breeding, P., and Ash, M. E. (2018) “Mitochondrial dysfunction and chronic disease: treatment with membrane lipid replacement and other natural supplements,” in Mitochondrial biology and experimental therapeutics, 499–522.
Niyazov, D. M., Kahler, S. G., and Frye, R. (2016). Primary mitochondrial disease and secondary mitochondrial dysfunction: importance of distinction for diagnosis and treatment. Mol. Syndromol. 7, 122–137. doi:10.1159/000446586
Novak, E., and Mollen, K. (2015). Mitochondrial dysfunction in inflammatory bowel disease. Front. Cell Dev. Biol. 3 (62), 62. doi:10.3389/fcell.2015.00062
Palmeira, C. M., and Rolo, A. P. (2004). Mitochondrially-mediated toxicity of bile acids. Toxicol 203, 1–15. doi:10.1016/j.tox.2004.06.001
Park, C., Noh, J. S., Jung, Y., Leem, S.-H., Hyun, J. W., Chang, Y.-C., et al. (2022). Fisetin attenuated oxidative stress-induced cellular damage in ARPE-19 human retinal pigment epithelial cells through nrf2-mediated activation of heme oxygenase-1. Front. Pharmacol. 13, 927898. doi:10.3389/fphar.2022.927898
Patterson, R. E., and Sears, D. D. (2017). Metabolic effects of intermittent fasting. Ann. Rev. Nutr. 37, 371–393. doi:10.1146/annurev-nutr-071816-064634
Peña-Cearra, A., Song, D., Castelo, J., Palacios, A., Lavín, J., Azkargorta, M., et al. (2023). Mitochondrial dysfunction promotes microbial composition that negatively impacts on ulcerative colitis development and progression. npj Biofilms Microbiomes 9 (74), 74. doi:10.1038/s41522-023-00443-y
Perrone, L., Squillaro, T., Napolitano, F., Terracciano, C., Sampaolo, S., and Melone, M. A. B. (2019). The autophagy signaling pathway: a potential multifunctional therapeutic target of curcumin in neurological and neuromuscular diseases. Nutrients 11 (8), 1881. doi:10.3390/nu11081881
Peters, V. B. M., Arulkumaran, N., Melis, M. J., Gaupp, C., Roger, T., Shankar-Hari, M., et al. (2022). Butyrate supplementation exacerbates myocardial and immune cell mitochondrial dysfunction in a rat model of faecal peritonitis. Life Sci. 12, 2034. doi:10.3390/life12122034
Pral, L. P., Fachi, J. L., Corrêa, R. O., Colonna, M., and Vinolo, M. A. R. (2021). Hypoxia and HIF-1 as key regulators of gut microbiota and host interactions. Trends Immunol. 42 (7), 604–621. doi:10.1016/j.it.2021.05.004
Qian, Y., Guan, T., Huang, M., Cao, L., Li, Y., Cheng, H., et al. (2012). Neuroprotection by the soy isoflavone, genistein, via inhibition of mitochondria-dependent apoptosis pathways and reactive oxygen induced-NF-κB activation in a cerebral ischemia mouse model. Neurochem. Int. 60, 759–767. doi:10.1016/j.neuint.2012.03.011
Rasbach, K. A., and Schnellmann, R. G. (2008). Isoflavones promote mitochondrial biogenesis. J. Pharmcol Exp. Ther. 325, 536–543. doi:10.1124/jpet.107.134882
Rietjens, I., Louisse, J., and Beekmann, K. (2016). The Potential health effects of dietary phytoestrogens. Br. J. Pharmacol. 174, 1263–1280. doi:10.1111/bph.13622
Rose, S., Bennuri, S., Davis, J., Wynne, R., Slattery, J., Tippett, M., et al. (2018). Butyrate enhances mitochondrial function during oxidative stress in cell lines from boys with autism. Transl. Psych. 8 (42), 42. doi:10.1038/s41398-017-0089-z
Saccone, C., Pesole, G., and Sbis, E. (1991). The main regulatory region of mammalian mitochondrial DNA: structure-function model and evolutionary pattern. J. Mol. Evol. 33, 83–91. doi:10.1007/BF02100199
Saint-Georges-Chaumet, Y., and Edeas, M. (2015). Microbiota-mitochondria inner-talk: consequence for microbiota-host interaction. FEMS Path Dis. 76.
Salvi, M., Brunati, A. M., Clari, G., and Toninello, A. (2002). Interaction of genistein with the mitochondrial electron transport chain results in opening of the membrane transition pore. Biochem. Biophys. Acta Bioenerg. 1556, 187–196. doi:10.1016/s0005-2728(02)00361-4
Schapira, A. H. V. (2006). Mitochondrial disease. Lancet 368, 70–82. doi:10.1016/S0140-6736(06)68970-8
Serena, C., Ceperuelo-Mallafré, V., Keiran, N., Queipo-Ortuño, M. I., Bernal, R., Gomez-Huelgas, R., et al. (2018). Elevated circulating levels of succinate in human obesity are linked to specific gut microbiota. ISME J. 12 (7), 1642–1657. doi:10.1038/s41396-018-0068-2
Shekhawat, P. S., Srinivas, S. R., Matern, D., Bennet, M. J., Boriack, R., George, V., et al. (2007). Spontaneous development of intestinal and colonic atrophy and inflammation in the carnitine-deficient jvs (OCTN2-/-) mice. Molec Gen. Metab. 92, 315–324. doi:10.1016/j.ymgme.2007.08.002
Sheridan, P. O., Louis, P., Tsompanidou, E., Shaw, S., Harmsen, H. J., Duncan, S. H., et al. (2022). Distribution, organization and expression of genes concerned with anaerobic lactate utilization in human intestinal bacteria. Microb. Genomics 8 (1), 000739. doi:10.1099/mgen.0.000739
Shiels, P. G., Buchanan, S., Selman, C., and Stenvinkel, P. (2019). Allostatic load and ageing: linking the microbiome and nutrition with age-related health. Biochem. Soc. Trans. 47, 1165–1172. doi:10.1042/BST20190110
Srivastava, S., Kashiwaya, Y., King, M. T., Baxa, U., Tam, J., Niu, G., et al. (2012). Mitochondrial biogenesis and increased uncoupling protein 1 in brown adipose tissue of mice fed a ketone ester diet. FASEB J. 26 (6), 2351–2362. doi:10.1096/fj.11-200410
Stojanov, S., and Kreft, S. (2020). Gut microbiota and the metabolism of phytoestrogens. Rev. Bras. Farm. 30, 145–154. doi:10.1007/s43450-020-00049-x
Su, S., Chen, M., Wu, Y., Lin, Q., Wang, D., Sun, J., et al. (2023). Fecal microbiota transplantation and short-chain fatty acids protected against cognitive dysfunction in a rat model of chronic cerebral hypoperfusion. CNS Neurosci. Ther. 29, 98–114. doi:10.1111/cns.14089
Tang, Q. Y., Alexander, M., Newman, J. C., Tian, Y., Cai, J., Upadhyay, V., et al. (2020). Ketogenic diets alter the gut microbiome resulting in decreased intestinal Th17 cells. Cell 181, 1263–1275. doi:10.1016/j.cell.2020.04.027
Taylor, S. J., Winter, M. G., Gillis, C. C., da Silva, L. A., Dobbins, A. L., Muramatsu, M. K., et al. (2022). Colonocyte-derived lactate promotes E. coli fitness in the context of inflammation-associated gut microbiota dysbiosis. Microbiome 10 (1), 200. doi:10.1186/s40168-022-01389-7
Tyynismaa, H., and Suomalainen, A. (2009). Mouse models of mitochondrial DNA defects and their relevance for human disease. EMBO Rep. 10 (2), 137–143. doi:10.1038/embor.2008.242
Vallance, H. D., Koochin, A., Branov, J., Rosen-Heath, A., Bosdet, T., Wang, Z., et al. (2018). Marked Elevation in plasma trimethylamine-N-oxide (TMAO) in patients with mitochondrial disorders treated with oral L-carnitine. Molec Gen. Metab. Rep. 15, 130–133. doi:10.1016/j.ymgmr.2018.04.005
Vanhauwaert, E., Matthys, C., Verdonck, L., and De Preter, V. (2015). Low-Residue and low-fiber diets in gastrointestinal disease management. Adv. Nutr. 6 (6), 820–827. doi:10.3945/an.115.009688
Velasquez, M. T., Ramezani, A., Manal, A., and Raj, D. S. (2016). Trimethylamine N-oxide: the good, the bad and the unknown. Toxins 8, 326. doi:10.3390/toxins8110326
Vidali, S., Aminzadeh, S., Lambert, B., Rutherford, T., Sperl, W., Kofler, B., et al. (2015). Mitochondria: the ketogenic diet – a metabolism – based therapy. Int. J. Biochem. Cell Biol. 63, 55–59. doi:10.1016/j.biocel.2015.01.022
Voicescu, M., Hellwig, P., and Meghea, A. (2016). Antioxidant activity of phytoestrogen type isoflavones in biomimetic environments. New J. Chem. 40, 606–612. doi:10.1039/c5nj01568e
Wang, S., Rubio, L., Duncan, S., Donachie, G., Holtrop, G., Lo, G., et al. (2020). Pivotal roles for pH, lactate, and lactate-utilizing bacteria in the stability of a human colonic microbial ecosystem. ASM Journals, mSystems 5 (5), 006455–20. doi:10.1128/mSystems.00645-20
Wang, W., Chen, L., Zhou, R., Wang, X., Song, L., Huang, S., et al. (2014). Increased proportions of bifidobacterium and the lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J. Clin. Microbiol. 52 (2), 398–406. doi:10.1128/JCM.01500-13
Wei, Y., Ma, X., Zhao, J., Wang, X., and Gao, C. (2023). Succinate metabolism and its regulation of host-microbe interactions. Gut Microbes 51 (1). doi:10.1080/19490976.2023.2190300
Whitaker, R. M., Corum, D., Beeson, C. C., and Schnellmann, R. G. (2016). Mitochondrial biogenesis as a pharmacological target: a new approach to acute and chronic diseases. Annu. Rev. Pharmacol. Toxicol. 56, 229–249. doi:10.1146/annurev-pharmtox-010715-103155
Wu, P.-H., Liu, P.-Y., Chiu, Y.-W., Hung, W.-C., Lin, Y.-T., Lin, T.-Y., et al. (2020). Comparative gut microbiome differences between ferric citrate and calcium carbonate phosphate binders in patients with end-stage kidney disease. Microorganisms 8 (12), 2040. doi:10.3390/microorganisms8122040
Xu, S., Tao, H., Cao, W., Cao, L., Lin, Y., Zhao, S.-M., et al. (2021a). Ketogenic diets inhibit mitochondrial biogenesis and induce cardiac fibrosis. Signal Transduct. Target. Ther. 6 (1), 54. doi:10.1038/s41392-020-00411-4
Xu, Y., Peng, S., Cao, X., Qian, S., Shen, S., Luo, J., et al. (2021b). High doses of butyrate induce a reversible body temperature drop through transient proton leak in mitochondria of brain neurons. Life Sci. 278, 119614. doi:10.1016/j.lfs.2021.119614
Yadav, M., Ali, S., Shrode, R. L., Shahi, S. K., Jensen, S. N., Hoang, J., et al. (2022). Multiple sclerosis patients have an altered gut mycobiome and increased fungal to bacterial richness. PLOS ONE 17 (4), e0264556. doi:10.1371/journal.pone.0264556
Yan, X.-L., Liu, X.-C., Zhang, Y.-N., Du, T.-T., Ai, Q., Gao, X., et al. (2022). Succinate aggravates intestinal injury in mice with necrotizing enterocolitis. Front. Cell. Infect. Microbiol. 12, 1064462. doi:10.3389/fcimb.2022.1064462
Yardeni, T., Tanes, C. E., Bittinger, K., Mattei, L. M., Schaefer, P. M., Singh, L. N., et al. (2019). Host mitochondria influence gut microbiome diversity: a role for ROS. Sci. Signal 12, eaaw3159. doi:10.1126/scisignal.aaw3159
Yoshino, M., Naka, A., Sakamoto, Y., Shibasaki, A., Toh, M., Tsukamoto, S., et al. (2015). Dietary isoflavone daidzein promotes Tfam expression that increases mitochondrial biogenesis in C2C12 muscle cells. J. Nutr. Biochem. 25, 1193–1199. doi:10.1016/j.jnutbio.2015.05.010
Zaheer, K., and Akhtar, M. H. (2017). An updated review of dietary isoflavones: nutrition, processing, bioavailability and impacts on human health. Crit. Rev. Food Sci. Nutr. 57, 1280–1293. doi:10.1080/10408398.2014.989958
Zhang, S., Al-Maghout, T., Cao, H., Pelzl, L., Salker, M. S., Veldhoen, M., et al. (2019). Gut bacterial metabolite urolithin A (ua) mitigates Ca2+ entry in T cells by regulating miR-10a-5p. Front. Imm 10, 1737. doi:10.3389/fimmu.2019.01737
Zhang, Y., Wang, Y., Ke, B., and Du, J. (2021). TMAO: how gut microbiota contributes to heart failure. Transl. Res. 228, 109–125. doi:10.1016/j.trsl.2020.08.007
Zhang, Y., Zhou, S., Zhou, Y., Yu, L., Zhang, L., and Wang, Y. (2018). Altered gut microbiome composition in children with refractory epilepsy after ketogenic diet. Epilepsy Res. 145, 163–168. doi:10.1016/j.eplepsyres.2018.06.015
Zhao, T., Gu, J., Zhang, H., Wang, Z., Zhang, W., Zhao, Y., et al. (2020). Sodium butyrate-modulated mitochondrial function in high-insulin induced hepg2 cell dysfunction. Oxidative Med. Cell. Longev. 2020, 1904609. doi:10.1155/2020/1904609
Keywords: mitochondria, mitochondrial disease, microbiome, mitochondria-microbiome crosstalk, diet
Citation: Zachos KA, Gamboa JA, Dewji AS, Lee J, Brijbassi S and Andreazza AC (2024) The interplay between mitochondria, the gut microbiome and metabolites and their therapeutic potential in primary mitochondrial disease. Front. Pharmacol. 15:1428242. doi: 10.3389/fphar.2024.1428242
Received: 06 May 2024; Accepted: 10 July 2024;
Published: 25 July 2024.
Edited by:
Marcos Roberto De Oliveira, Federal University of Rio Grande do Sul, BrazilReviewed by:
Marvin Edeas, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceCopyright © 2024 Zachos, Gamboa, Dewji, Lee, Brijbassi and Andreazza. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana C. Andreazza, YW5hLmFuZHJlYXp6YUB1dG9yb250by5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.