- 1Department of Surgical Oncology and General Surgery, First Hospital of China Medical University, Shenyang, Liaoning, China
- 2Department of Thoracic Surgery, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 3Department of Thoracic Surgery, First Hospital of China Medical University, Shenyang, Liaoning, China
- 4Department of Breast Surgery, Liaoning Cancer Hospital and Institute, Cancer Hospital of Dalian University of Technology, Cancer Hospital of China Medical University, Shenyang, Liaoning, China
Esophageal squamous cell carcinoma (ESCC) is a malignancy with high incidence in China. Due to the lack of effective molecular targets, the prognosis of ESCC patients is poor. It is urgent to explore the pathogenesis of ESCC to identify promising therapeutic targets. Metabolic reprogramming is an emerging hallmark of ESCC, providing a novel perspective for revealing the biological features of ESCC. In the hypoxic and nutrient-limited tumor microenvironment, ESCC cells have to reprogram their metabolic phenotypes to fulfill the demands of bioenergetics, biosynthesis and redox homostasis of ESCC cells. In this review, we summarized the metabolic reprogramming of ESCC cells that involves glucose metabolism, lipid metabolism, and amino acid metabolism and explore how reprogrammed metabolism provokes novel opportunities for biomarkers and potential therapeutic targets of ESCC.
Introduction
Esophageal cancer is a common malignancy of the digestive tract. The new cases of esophageal cancer have exceeded 6,00,000 annually, ranking the seventh among all cancers. The death cases of esophageal cancer are about 5,40,000 annually, ranking the sixth among all cancer types (Sung et al., 2021). Esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) are the two main histological subtypes of esophageal cancer (Rogers et al., 2022). As a high-risk country of esophageal cancer, more than 90% of the cases diagnosed in China are ESCC (Liu CC et al., 2019). With the improvement of early diagnosis and comprehensive therapeutic strategies for ESCC, the therapeutic effect of esophageal cancer has been improved, but the 5-year overall survival rate of ESCC is only 15%∼20% (Huang and Yu, 2018; Zhu et al., 2023).
Metabolic reprogramming is an emerging hallmark of tumors (Kroemer and Pouyssegur, 2008). To adapt to hypoxia and nutrient deprivation, proliferating tumor cells need to reprogram metabolic pathways to fulfill the increasing demands of energetic production, cellular building components and redox balance of tumor cells (Pavlova and Thompson, 2016). A series of genetic alteration and cellular signaling are involved in the regulation of diverse aspects of cellular metabolism (DeBerardinis and Chandel, 2016). Studies have identified profound metabolic reprogramming in aggressive ESCC that involves glycolysis, tricarboxylic acid (TCA) cycle, lipid metabolism, and glutamine addiction, etc. Spatially resolved metabolomics in 256 ESCC patients revealed that multiple altered metabolic pathways, including proline biosynthesis, glutamine metabolism, uridine metabolism, histidine metabolism, fatty acid biosynthesis and polyamine biosynthesis participate in the pathogenesis of ESCC. Notably, metabolic enzymes pyrroline-5-carboxylate reductase 2 and uridine phosphorylase 1 are therefore identified as potential therapeutic targets. Huang et al. (2024) integrated the single cell RNA-seq with metabolomics of ESCC tissues and plasma samples, indicating that nicotinate and nicotinamide metabolism pathway was dysregulated in ESCC patients with lymphatic metastasis with significant 1-methylnicotinamide upregulation. In this review, we primarily discussed the advantages of metabolic reprogramming for ESCC cells and explored how reprogrammed metabolism provokes novel opportunities for biomarkers and potential therapeutic targets of ESCC.
Reprogrammed metabolic pathways in ESCC
Glucose metabolism and ESCC
Altered glucose metabolism is commonly observed in ESCC cells (Figure 1). Like other tumors, glycolysis is the preferred energetic pathway in ESCC cells (Liu et al., 2021; Su et al., 2022). Depending on glycolysis, ESCC cells convert glucose to lactate to rapidly produce ATP and promote tumor progression (Wang et al., 2021; Li et al., 2023). The glycolytic pathway plays an essential role in ESCC metabolism, as shown by the deregulation of multiple metabolic enzymes. As the first rate-limiting enzyme in glycolysis, hexokinases (HKs) catalyze the phosphorylation of glucose to glucose-6-phosphate. Among the four isoforms, HK2 exerts greater effect in promoting aerobic glycolysis than other isoforms (Ciscato F et al., 2021). Liu K et al. (2019) found that HK2 is essential for maintaining esophageal cancer stem cell (CSC) phenotypes. Esophageal CSCs exhibit higher glycolysis, which are regulated via the small heat shock protein 27 (Hsp27)-AKT-HK2 axis. Hsp27 could increase the expression of HK2 through activating AKT-mTOR in esophageal CSCs. Quercetin, the Hsp27 inhibitor, could augment the anti-ESCC effects of adjuvant chemotherapy. Phosphofructokinases (PFKs) catalyze the phosphorylation of fructose-6-phosphate to fructose-1, 6-bisphosphate, and has three PFK isozymes including liver type (PFKL), platelet type (PFKP) and muscle type (PFKM) (Feng et al., 2020). PFKL overexpression is associated with reduced survival of ESCC patients, and its inhibition suppresses ESCC growth (Zheng et al., 2022). Antipsychotic drug penfluridol directly targets PFKL to impair glycolytic pathway and only exhibits its anti-tumor effect in PFKL-proficient tumors (Zheng et al., 2022). Under hypoxia, GHRH-R splice variant 1 (SV1) is elevated to upregulate PFKM to promote glycolytic pathway and enhance development of ESCC (Gu et al., 2022). MIA-602 (a SV1 inhibitor) could block the oncogenic role of PFKM in ESCC. 6-phosphofructo-2-kinase/fructose-2, 6-biphosphatase 3 (PFKFB3) mediates the synthesis and degradation of fructose 2, 6-bisphosphate. PFKFB3 could increase glycolytic flux in ESCC cells, as well as ESCC cell cycle progression and growth. lncRNA Actin Gamma 1 Pseudogene (AGPG) has been identified to bind to and stabilize PFKFB3 by preventing APC/C-mediated ubiquitination of PFKFB3 and stabilizing PFKFB3 protein (Liu et al., 2020). Pyruvate kinases (PKs), the last rate-limiting enzymes in glycolysis, catalyze the conversion of phosphoenolpyruvate to pyruvate. There are four isoforms of PKs, including PKL (found in liver), PKR (red blood cell), PKM1 and PKM2 (found in muscle) (Israelsen and Vander Heiden, 2015). The critical role of glycolysis in ESCC development is further supported by the correlation between increased PKM2 expression and poor prognosis. PKM2 has been found to be upregulated in ESCC patients (Ma et al., 2019). circCYP24A1 interacts with PKM2 to enhance C-C chemokine ligand 5 (CCL5) production and development of ESCC (Gu et al., 2022). Methyltransferase-like 3 induces the m6A modification of APC by recruiting YTH domain family (YTHDF) for APC mRNA degradation, which further upregulates β-catenin and its downstream targets PKM2 to enhance aerobic glycolysis and promote ESCC progression. PKM2 has also been found to be a target of photodynamic therapy (PDT) (Li B et al., 2021). PDT inhibits PKM2 and activates caspase-3/8 to release Gasdermin E-N (N-GSDME) and induce pyroptosis in ESCC cells. Lysyl-oxidase like-2 (LOXL2) and its catalytically inactive L2Δ13 splice variant interact physically with aldolase A, glyceraldehyde-3-phosphate dehydrogenase and enolase to boost glycolysis of ESCC cells, therefore promoting ESCC progression (Jiao et al., 2022). Pyruvate dehydrogenase kinase 1 (PDK1) is a Ser/Thr kinase that inactivates mitochondrial pyruvate dehydrogenase, leading to a metabolic reprogramming from oxidative phosphorylation (OXPHOS) to glycolytic pathway. PDK1 is significantly overexpressed in ESCC tissues and cell lines compared with the normal tissues or cells (Ma et al., 2023). PDK1 could suppress cell proliferation of ESCC by blocking glycolytic pathway (Ma et al., 2023). Hypoxia-inducible factor 1α (HIF-1α) is the key transcriptional regulator of glycolysis, mediating cellular adaptation to hypoxia. In ESCC, long intergenic noncoding RNA for kinase activation (LINK-A) disrupts the interaction between MCM3 and HIF-1α, reducing MCM3-mediated HIF-1α transcriptional inhibition to enhance glycolytic phenotype and chemoresistance. m6A demethylase fat mass and obesity-associated protein (FTO) could stabilize LINK-A, making the FTO/LINK-A/MCM3/HIF-1α axis as a potential target for anti-ESCC strategy (Nan et al., 2023). LncRNA MALR upregulation leads to ILF3 liquid-liquid phase separation and activates HIF1α to promote ESCC development (Liu et al., 2023). Hypoxia-induced lncRNA G077640 upregulation promotes ESCC progression by stabilizing HIF1α to upregulate glucose transporter 4 (GLUT4), HK2 and PDK1 (Huang et al., 2023). Targeting glycolytic rate-limiting enzymes, glucose transporters and other metabolic enzymes are considered targets for screening potential anti-tumor drugs. However, based on in vitro or in vivo experiments, most anti-ESCC drugs that target glucose metabolism are still in the development stage.
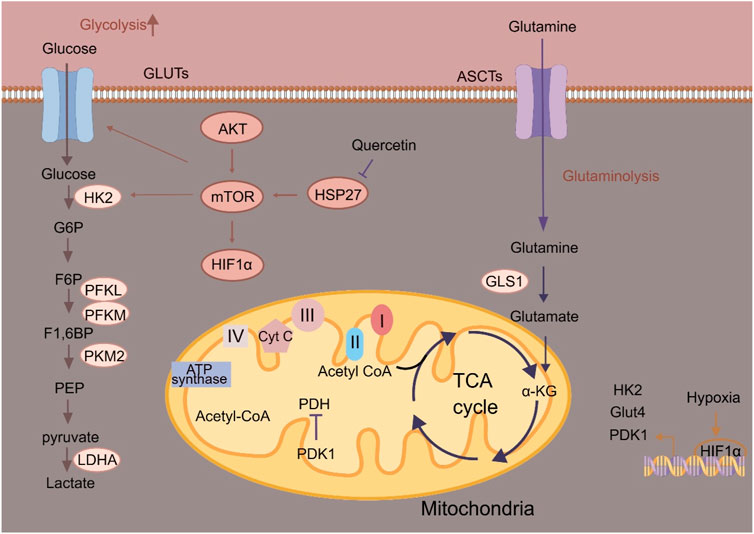
Figure 1. Schematic illustration of glucose metabolism in ESCC cells. α-KG, α-ketoglutarate; ASCT, Alanine serine cysteine transporter; Cyt C, Cytochrome c; F-1, 6-BP, Fructose-1, 6-bisphosphatase; G6P, Glucose-6-phosphate; Glut, Glucose transporter; GLS1, Glutaminase 1; HK2, Hexokinase 2; HIF-1α, Hypoxia-inducible factor 1α; HSP27, Heat shock protein 27; LDHA, Lactate dehydrogenase A; PDK1, 3-phosphoinositide-dependent kinase 1; PFK, Phosphofructokinase; PKM2, Pyruvate kinase M2.
The TCA cycle is indispensable for OXPHOS to fulfill the needs of bioenergetics and biosynthesis (Nolfi-Donegan et al., 2020). The regulation of mitochondrial biogenesis is essential for sustaining redox homeostasis to boost anti-immunity and inhibit tumor progression (Zong et al., 2016). Mitochondrial transcription factor A (TFAM) has been found to be downregulated in ESCC, and TFAM downregulation is correlated with poor survival of ESCC patients. TFAM is essential for replication and stability of mitochondrial genome, and TFAM inhibition induces mitochondrial DNA (mtDNA) release into the cytosol and activates the cGAS-STING signaling to induce autophagy and ESCC proliferation. Further, STING abrogation or mtDNA degradation by DNase I impairs TFAM-depleted ESCC cell proliferation (Li et al., 2022). CircPUM1 is positively associated with the expression of HIF1α under hypoxia in ESCC cells. CircPUM1 regulates the mitochondrial complex III assembly through interacting with mitochondrial ubiquinol-cytochrome c reductase core protein (UQCRC), which ultimately promotes OXPHOS for ATP production to facilitate proliferation and inhibit pyroptosis of ESCC cells (Gong et al., 2022). In ESCC, oncogenic STAT3 signaling pathway participates in regulating the activity of electron transport chain (ETC). STAT3β could inhibit the phosphorylation of STAT3α at S727 in mitochondria via ERK1/2 to impair the activity of ETC, therefore activating GSDME for pyroptosis and sensitizing ESCC cells to cisplatin (Zheng et al., 2021). Targeting OXPHOS in ESCC cells as an anti-ESCC strategy deserves further investigations.
The pentose phosphate pathway is a branch from glycolysis and serves as a major source of ribonucleotides and NADPH. In ESCC, downregulation of polo like kinase 1 (PLK1) could block the pentose phosphate pathway to impair the production of NADPH, thereby enhancing ferroptosis and promoting the sensitivity of ESCC cells to chemotherapy (Zhao et al., 2023).
Lipid metabolism and ESCC
Altered lipid metabolism is a prominent metabolic reprogramming in ESCC (Figure 2). Enhanced lipid biosynthesis or uptake is required for ESCC progression (Cheng et al., 2018). Fatty acid synthesis is a process that begins with the carboxylation of acetyl-CoA to malonyl-CoA by acetyl-CoA carboxylase. Malonyl-CoA is further committed to fatty acid synthesis by producing mainly 16-C palmitate via fatty acid synthase (FASN). FASN has been found to promote the progression of ESCC. circHIPK3 acts as a ceRNA by sponging miR-637 to upregulate FASN expression and fatty acid biosynthesis of ESCC cells, indicating circHIPK3/miR-637/FASN axis as a promising therapeutic target for anti-ESCC strategy (Cao et al., 2024). De novo lipogenesis is transcriptionally regulated by sterol regulatory element binding protein (SREBP) (Eberlé et al., 2004). Overexpression of SREBP1 is correlated with poor prognosis in ESCC patients, and supports ESCC progression by enhancing fatty acid biosynthesis. In ESCC, pre-mRNA processing factor 19 (PRP19) enhances the stability of SREBP1 mRNA in an N6-methyladenosine-dependent manner to mediate SREBP-dependent fatty acid synthesis and ESCC progression. SREBP could also cooperate with TP63/Kruppel like factor 5 (KLF5) to regulate the biosynthesis of fatty acids (Zhang et al., 2023).
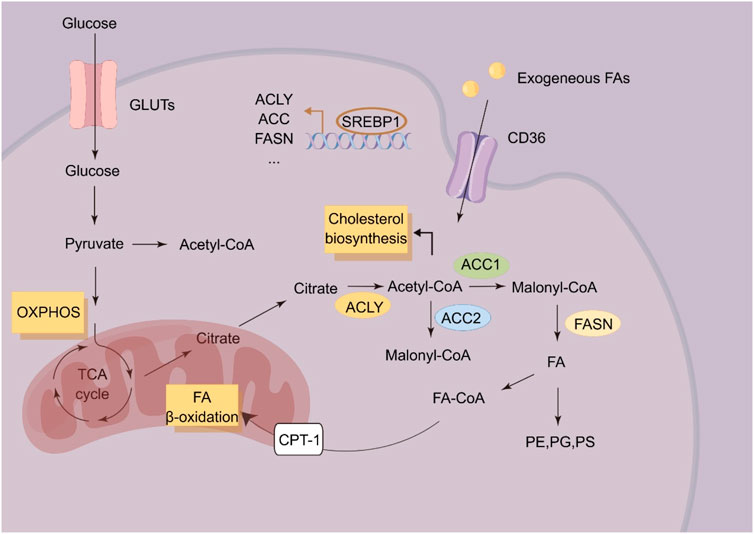
Figure 2. Schematic illustration of lipid metabolism in ESCC cells. ACC, acetyl-CoA; ACLY, ATP citrate lyase carboxylase; CPT-1, Carnitine palmitoyl transferase 1; FA, Fatty acid; FASN, Fatty acid synthase; PE, Phosphatidylethanolamine; PS, Phosphatidylserine; PG, Phosphatidylglycerol; SREBP1, Sterol response element binding protein-1.
Fatty acid oxidation is the process involving shortening of fatty acids and producing in acetyl-CoA, NADH and FADH2 (Ma et al., 2018). Carnitine O-palmitoyl transferase 1 (CPT1A), the key rate-limiting enzyme in fatty acid oxidation, is upregulated in ESCC, which is correlated with poor survival of ESCC patients (Tian et al., 2022). In ESCC, CPT1A could inhibit cellular apoptosis by providing GSH and NADPH to sustain redox homeostasis. Genetic or pharmacologic inhibition of CPT1A reduces the NADPH supply to impair anchorage-independent growth of ESCC cells in vitro and in vivo (Tian et al., 2022).
Cholesterol is an important component of cell and organelle membranes and a precursor of steroid hormones and bile acids (Huang et al., 2020). The rapid proliferation of tumor cells requires enhanced biosynthesis of cell and organelle membranes, and aberrant cholesterol metabolism therefore plays a pivotal role in tumor initiation and progression (Xu et al., 2020). Metastatic ESCC cells display elevated cholesterol accumulation via upregulating the expression of anoctamin 1 (ANO1). ANO1 results in cholesterol accumulation by blocking LXR signaling and inhibiting cholesterol hydroxylation by downregulating hydroxylase CYP27A1 (Singh and Mehla, 2023). Lysophosphatidylcholine acyltransferase 1 (LPCAT1) has been found to be upregulated in ESCC tissues, and LPCAT1 could rewire cholesterol metabolism of ESCC cells. LPCAT1 could upregulate the expression of SQLE to promoting the entry of SP1/SREBP2 into the nucleus via activating PI3K. LPCAT1 also promotes the entry of SREBP1 into the nucleus by EGFR-mediated INSIG-1 downregulation, leading to enhance cholesterol biosynthesis (Tao et al., 2021).
Phospholipid is composed of phosphoglyceride and sphingomyelin. Phosphoglyceride includes phosphatidylcholine, phosphatidylserine, phosphatidylglycerol, diphosphatidylglycerol, phosphatidylethanolamine, and phosphatidylinositol. In ESCC, DNA hypermethylation of otubain 2 (OTUB2) has been found to induce tumor initiation and chemoresistance by regulating biosynthesis of phosphatidylserine. Mechanistically, OTUB2 mediates the deubiquitination and phosphorylation of STAT1 and further promotes calmodulin-like protein 3 (CALML3) upregulation. Subsequently, CALML3 regulates mitochondrial calcium signaling to promote OXPHOS and biosynthesis of phosphatidylserine. More importantly, orally-administered phosphatidylserine impairs tumor initiation of OTUB2-low ESCC cells in mouse models, making phosphatidylserine administration as a potential anti-ESCC strategy (Chang et al., 2022). Fatty acid 2-hydroxylase (FA2H), catalyzing the hydroxylation of fatty acids, has been found to be significantly enriched in a subpopulation of ESCC cells with high metastatic potential, and that FA2H inhibition significantly blocks metastatic potential of ESCC cells. Notably, Ceramide (d18:0/24:0) and Ceramide (d18:0/24:1) are increased in FA2H-knockdown ESCC cells. Upon administration, Cer (d18:0/24:0) and Cer (d18:0/24:1) significantly blocks tumor metastasis in mouse models (Zhou et al., 2022).
Currently, multiple studies have revealed that lipid metabolism in ESCC tissues is significantly altered compared with normal esophageal tissues, which may lead to the occurrence and development of ESCC. Therefore, targeting lipid metabolism as a potential therapeutic approach will provide new opportunities for the treatment of ESCC. However, there are still challenges to overcome in techniques of lipid detection. The accuracy of mass spectrometry-based lipidomics is required to be improved for more accurate qualitative and quantitative analysis. The combined analysis of multi-omics could reveal the biological feature of the occurrence and development of ESCC comprehensively. The combination of lipidomics with genomics, transcriptomics and proteomics may provide more insights for the treatment ESCC.
Amino acid metabolism and ESCC
Cancer cells exhibit elevated demand for amino acids to support their fast proliferation (Sivanand and Vander Heiden, 2020). It has been found that tumor cells exhibit greater dependence on glutamine (Cluntun et al., 2017). Glutamine participates in cellular bioenergetics through α-ketoglutarate in TCA cycle and serves as a major source of nitrogen for nucleotide biosynthesis. In ESCC, genetic alterations including Fbxo4 loss and hyperactivation of cyclin D1-CDK4/6 kinases have been frequently observed, leading to glutamine addiction (Qie et al., 2019). The Fbxo4-cyclin D1 axis could regulate glutamine consumption and mitochondrial dysfunction by suppressing Rb activity and activating mTORC1 to promote ESCC development. Glutaminase 1 (GLS1) is a rate-limiting glutaminolysis enzyme that transforms glutamine into glutamate to support proliferation of ESCC cells. Combined treatment of CB-839 (GLS1 inhibitor) and metformin could synergistically impair proliferation of ESCC cells, providing promising therapeutic strategy for anti-ESCC treatment. Glutamine metabolism is also regulated by RNA binding motif protein 4 (RBM4)-LKB1 axis to enhance ESCC cell survival. RBM4 binds with LKB1 to impair the LKB1/STRAD/MO25 complex and promote TRIM26-mediated LKB1 ubiquitination and degradation to overcome cell senescence (Chen et al., 2023). NEDD4 like E3 ubiquitin protein ligase (NEDD4L) blocks glutamine metabolism in ESCC by ubiquitination of c-Myc to downregulate GLS1 and SLC1A5, which suppresses tumor progression (Cheng et al., 2022). Collectively, targeting glutamine metabolism in cancer therapy brings new opportunities for anti-ESCC strategy.
Arginine functions as the precursor for synthesis of protein, nitric oxide, polyamines, agmatine, creatine, and urea (Chen et al., 2021). Of the enzymes catalyzing rate-controlling steps in arginine metabolism, argininosuccinate synthetase 1 (ASS1) and argininosuccinate lyase (ASL) levels were increased in ESCC tissues, but reduced in metastatic ESCC tissues (Sun et al., 2024). Blocking ASS1 or ASL could impair ESCC growth at the primary site and promote distant metastasis (Sun et al., 2024). The gene encoding 26S proteasome non-ATPase regulatory subunit 2 (PSMD2) activates the mTOR pathway by upregulating ASS1 to inhibit autophagy and promote ESCC progression (Liu et al., 2023). Under hypoxia, receptor tyrosine kinase IGF1R is upregulated in ESCC to elevate the transcription of ASS1 through c-MYC to reprogram arginine metabolism (Fang et al., 2023).
Although nutritional epidemiological study has shown that high methionine intake is not associated with ESCC risk, Jin et al. (2024) found that methionine cycling has been hyperactivated in ESCC tissues and correlated with poor survival outcome. ESCC cells prefers to utilize exogenous methionine to produce S-adenosine methionine (SAM), leading to enhanced ESCC cell proliferation. Mechanistically, methionine enhances METTL3-mediated RNA m6A methylation through SAM and upregulates NR4A2 expression. Celecoxib has been identified as a potent NR4A2 inhibitor with promising anti-ESCC potential.
More comprehensive analysis of amino acid metabolic reprogramming and its related metabolic pathways in ESCC cells should be conducted to assist developing more effective anti-ESCC strategies. However, there are still questions waiting for answer. For example, whether metabolic reprogramming in amino acid cooperates with other related pathways to sustain tumor development? How other metabolic pathways compensate to promote ESCC cell proliferation once amino acid metabolism is dysregulated in ESCC cells? What is the therapeutic effect of the usage of specific amino acid-limited diet to treat ESCC?
Clinical implications of metabolism-targeted therapy in ESCC
Metabolomics cooperates with genomics, transcriptomics and proteomics to form “systems biology,” which plays an important role in cancer research. Metabolomics mainly includes targeted metabolomics and non-targeted metabolomics. Untargeted metabolomics works by detecting all metabolites in the samples to obtain quantitative information to decipher the differences in metabolites between groups. Targeted metabolomics analyzes a limited number of metabolites associated with biological process (Schrimpe-Rutledge et al., 2016). Based on the tissue metabolic profiles consistently identified by nuclear magnetic resonance and targeted mass spectrometry techniques, Zhao et al. (2024) developed nuclear magnetic resonance -based serum and urine metabolic profiles and optimized to reliably reflect the metabolic profiles of ESCC. Due to the metabolic adaptability during tumor progression and treatment, it is important to track and monitor metabolic adaptability of ESCC cells during tumor progression and treatment. Therefore, it is necessary to dynamically and accurately observe metabolic alterations and change treatment strategies accordingly. Optimization of existing detection methods and searching for alternative non-invasive detection methods are key research directions (Xiao et al., 2023).
One strategy for discovering anti-ESCC therapy is to screen FDA-approved drugs. Recent study reported that antitussive agent cloperastine inhibits the proliferation of ESCC in vivo and in vitro by impairing mitochondrial OXPHOS (Li L et al., 2021; Li LY et al., 2021). Liu Y et al. (2023) found that sulconazole can effectively block the growth of ESCC cells by inducing mitochondrial oxidative stress and inhibiting glycolysis. The current reported metabolism-targeted therapy has been summarized in Table 1. Metabolic enzymes are attractive therapeutic targets in anti-tumor therapy, but new drugs targeting metabolism have been limited due to toxicity to normal tissues. And more studies have recognized that some metabolic enzymes drive tumor progression through non-catalytic mechanisms (Pan et al., 2021). Preclinical studies have identified several metabolic molecules that inhibit tumor progression as potential therapeutic targets, and some are already in clinical trials (Xiao et al., 2023). However, at present, metabolic therapy for ESCC patients is still stuck in pre-clinical research, and it is urgent to further explore new targets and optimal treatment strategies. The goal of identifying and tracking metabolic targets is to enable precision therapy. Considering the heterogeneity of metabolism in tumors, individualized metabolism-targeted therapy will be the future development direction.
Collectively, exploring the anti-ESCC strategy in the context of tumor metabolism has unveiled novel and promising opportunities, as well as more comprehensive understandings of metabolic reprogramming of ESCC cells. In the future, more knowledge of the metabolic reprogramming of ESCC cells are required to refine rational anti-ESCC strategies that target tumor metabolism.
Author contributions
ZW: Writing–original draft, Writing–review and editing. XS: Writing–original draft, Writing–review and editing. ZL: Writing–original draft, Writing–review and editing. HY: Writing–original draft, Writing–review and editing. WL: Writing–original draft, Writing–review and editing. YX: Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (82072733) and Technology Program Joint Fund of Liaoning Province (2023-BSBA-207).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Cao, S. Q., Xue, S. T., Li, W. J., Hu, G. S., Wu, Z. G., Zheng, J. C., et al. (2024). CircHIPK3 regulates fatty acid metabolism through miR-637/FASN axis to promote esophageal squamous cell carcinoma. Cell Death Discov. 10 (1), 110. doi:10.1038/s41420-024-01881-z
Chang, W., Luo, Q., Wu, X., Nan, Y., Zhao, P., Zhang, L., et al. (2022). OTUB2 exerts tumor-suppressive roles via STAT1-mediated CALML3 activation and increased phosphatidylserine synthesis. Cell Rep. 41 (4), 111561. doi:10.1016/j.celrep.2022.111561
Chen, C. L., Hsu, S. C., Ann, D. K., Yen, Y., and Kung, H. J. (2021). Arginine signaling and cancer metabolism. Cancers (Basel) 13 (14), 3541. doi:10.3390/cancers13143541
Chen, L., Zhang, W., Chen, D., Yang, Q., Sun, S., Dai, Z., et al. (2023). RBM4 dictates ESCC cell fate switch from cellular senescence to glutamine-addiction survival through inhibiting LKB1-AMPK-axis. Signal Transduct. Target Ther. 8 (1), 159. doi:10.1038/s41392-023-01367-x
Cheng, C., Geng, F., Cheng, X., and Guo, D. (2018). Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. (Lond). 38 (1), 27. doi:10.1186/s40880-018-0301-4
Cheng, W., Li, G., Ye, Z., Hu, J., Gao, L., Jia, X., et al. (2022). NEDD4L inhibits cell viability, cell cycle progression, and glutamine metabolism in esophageal squamous cell carcinoma via ubiquitination of c-Myc. Acta Biochim. Biophys. Sin. (Shanghai) 54 (5), 716–724. doi:10.3724/abbs.2022048
Ciscato, F., Ferrone, L., Masgras, I., Laquatra, C., and Rasola, A. (2021). Hexokinase 2 in cancer: a prima donna playing multiple characters. Int. J. Mol. Sci. 22 (9), 4716. doi:10.3390/ijms22094716
Cluntun, A. A., Lukey, M. J., Cerione, R. A., and Locasale, J. W. (2017). Glutamine metabolism in cancer: understanding the heterogeneity. Trends Cancer 3 (3), 169–180. doi:10.1016/j.trecan.2017.01.005
DeBerardinis, R. J., and Chandel, N. S. (2016). Fundamentals of cancer metabolism. Sci. Adv. 2 (5), e1600200. eCollection 2016 May. doi:10.1126/sciadv.1600200
Eberlé, D., Hegarty, B., Bossard, P., Ferré, P., and Foufelle, F. (2004). SREBP transcription factors: master regulators of lipid homeostasis. Biochimie 86 (11), 839–848. doi:10.1016/j.biochi.2004.09.018
Fang, K., Sun, M., Leng, Z., Chu, Y., Zhao, Z., Li, Z., et al. (2023). Targeting IGF1R signaling enhances the sensitivity of cisplatin by inhibiting proline and arginine metabolism in oesophageal squamous cell carcinoma under hypoxia. J. Exp. Clin. Cancer Res. 42 (1), 73. doi:10.1186/s13046-023-02623-2
Feng, J., Li, J., Wu, L., Yu, Q., Ji, J., Wu, J., et al. (2020). Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 39 (1), 126. doi:10.1186/s13046-020-01629-4
Gong, W., Xu, J., Wang, Y., Min, Q., Chen, X., Zhang, W., et al. (2022). Nuclear genome-derived circular RNA circPUM1 localizes in mitochondria and regulates oxidative phosphorylation in esophageal squamous cell carcinoma. Signal Transduct. Target Ther. 7 (1), 40. doi:10.1038/s41392-021-00865-0
Gu, L., Sang, Y., Nan, X., Zheng, Y., Liu, F., Meng, L., et al. (2022). circCYP24A1 facilitates esophageal squamous cell carcinoma progression through binding PKM2 to regulate NF-κB-induced CCL5 secretion. Mol. Cancer 21 (1), 217. doi:10.1186/s12943-022-01686-7
Huang, B., Song, B. L., and Xu, C. (2020). Cholesterol metabolism in cancer: mechanisms and therapeutic opportunities. Nat. Metab. 2 (2), 132–141. Epub 2020 Feb 10. doi:10.1038/s42255-020-0174-0
Huang, F. L., and Yu, S. J. (2018). Esophageal cancer: risk factors, genetic association, and treatment. Asian J. Surg. 41 (3), 210–215. doi:10.1016/j.asjsur.2016.10.005
Huang, Q., Chen, H., Yin, D., Wang, J., Wang, S., Yang, F., et al. (2024). Multi-omics analysis reveals NNMT as a master metabolic regulator of metastasis in esophageal squamous cell carcinoma. NPJ Precis. Oncol. 8 (1), 24. doi:10.1038/s41698-024-00509-w
Huang, X., Liu, C., Li, H., Dai, T., Luo, G., Zhang, C., et al. (2023). Hypoxia-responsive lncRNA G077640 promotes ESCC tumorigenesis via the H2AX-HIF1α-glycolysis axis. Carcinogenesis 44 (5), 383–393. doi:10.1093/carcin/bgad036
Israelsen, W. J., and Vander Heiden, M. G. (2015). Pyruvate kinase: function, regulation and role in cancer. Semin. Cell Dev. Biol. 43, 43–51. Epub 2015 Aug 13. doi:10.1016/j.semcdb.2015.08.004
Jiao, J. W., Zhan, X. H., Wang, J. J., He, L. X., Guo, Z. C., Xu, X. E., et al. (2022). LOXL2-dependent deacetylation of aldolase A induces metabolic reprogramming and tumor progression. Redox Biol. 57, 102496. Epub 2022 Oct 3. doi:10.1016/j.redox.2022.102496
Jin, X., Liu, L., Liu, D., Wu, J., Wang, C., Wang, S., et al. (2024). Unveiling the methionine cycle: a key metabolic signature and NR4A2 as a methionine-responsive oncogene in esophageal squamous cell carcinoma. Cell Death Differ. 31 (5), 558–573. Epub 2024 Apr 3. doi:10.1038/s41418-024-01285-7
Kroemer, G., and Pouyssegur, J. (2008). Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell 13 (6), 472–482. doi:10.1016/j.ccr.2008.05.005
Li, B., Yu, Y., Jiang, Y., Zhao, L., Li, A., Li, M., et al. (2021). Cloperastine inhibits esophageal squamous cell carcinoma proliferation in vivo and in vitro by suppressing mitochondrial oxidative phosphorylation. Cell Death Discov. 7 (1), 166. doi:10.1038/s41420-021-00509-w
Li, L., Jiang, D., Zhang, Q., Liu, H., Xu, F., Guo, C., et al. (2023). Integrative proteogenomic characterization of early esophageal cancer. Nat. Commun. 14 (1), 1666. doi:10.1038/s41467-023-37440-w
Li, L., Song, D., Qi, L., Jiang, M., Wu, Y., Gan, J., et al. (2021). Photodynamic therapy induces human esophageal carcinoma cell pyroptosis by targeting the PKM2/caspase-8/caspase-3/GSDME axis. Cancer Lett. 520, 143–159. Epub 2021 Jul 10. doi:10.1016/j.canlet.2021.07.014
Li, L. Y., Yang, Q., Jiang, Y. Y., Yang, W., Jiang, Y., Li, X., et al. (2021). Interplay and cooperation between SREBF1 and master transcription factors regulate lipid metabolism and tumor-promoting pathways in squamous cancer. Nat. Commun. 12 (1), 4362. doi:10.1038/s41467-021-24656-x
Li, Y., Yang, Q., Chen, H., Yang, X., Han, J., Yao, X., et al. (2022). TFAM downregulation promotes autophagy and ESCC survival through mtDNA stress-mediated STING pathway. Oncogene 41, 3735–3746. doi:10.1038/s41388-022-02365-z
Liu, C. C., Chou, K. T., Hsu, J. W., Lin, J. H., Hsu, T. W., Yen, D. H., et al. (2019). High metabolic rate and stem cell characteristics of esophageal cancer stem-like cells depend on the Hsp27-AKT-HK2 pathway. Int. J. Cancer 145 (8), 2144–2156. Epub 2019 Apr 10. doi:10.1002/ijc.32301
Liu, H., Zhang, Q., Song, Y., Hao, Y., Cui, Y., Zhang, X., et al. (2021). Long non-coding RNA SLC2A1-AS1 induced by GLI3 promotes aerobic glycolysis and progression in esophageal squamous cell carcinoma by sponging miR-378a-3p to enhance Glut1 expression. J. Exp. Clin. Cancer Res. 40 (1), 287. doi:10.1186/s13046-021-02081-8
Liu, J., Liu, Z. X., Wu, Q. N., Lu, Y. X., Wong, C. W., Miao, L., et al. (2020). Long noncoding RNA AGPG regulates PFKFB3-mediated tumor glycolytic reprogramming. Nat. Commun. 11 (1), 1507. doi:10.1038/s41467-020-15112-3
Liu, K., Zhao, T., Wang, J., Chen, Y., Zhang, R., Lan, X., et al. (2019). Etiology, cancer stem cells and potential diagnostic biomarkers for esophageal cancer. Cancer Lett. 458, 21–28. doi:10.1016/j.canlet.2019.05.018
Liu, L. X., Heng, J. H., Deng, D. X., Zhao, H., Zheng, Z. Y., Liao, L. D., et al. (2023). Sulconazole induces PANoptosis by triggering oxidative stress and inhibiting glycolysis to increase radiosensitivity in esophageal cancer. Mol. Cell Proteomics 22 (6), 100551. Epub 2023 Apr 17. doi:10.1016/j.mcpro.2023.100551
Liu, Y., Wu, M., Xu, S., Niu, X., Liu, W., Miao, C., et al. (2023). PSMD2 contributes to the progression of esophageal squamous cell carcinoma by repressing autophagy. Cell Biosci. 13 (1), 67. doi:10.1186/s13578-023-01016-4
Ma, R., Liu, Q., Zheng, S., Liu, T., Tan, D., and Lu, X. (2019). PKM2-regulated STAT3 promotes esophageal squamous cell carcinoma progression via TGF-β1-induced EMT. J. Cell Biochem. 120 (7), 11539–11550. Epub 2019 Feb 12. doi:10.1002/jcb.28434
Ma, Y., Temkin, S. M., Hawkridge, A. M., Guo, C., Wang, W., Wang, X. Y., et al. (2018). Fatty acid oxidation: an emerging facet of metabolic transformation in cancer. Cancer Lett. 435, 92–100. Epub 2018 Aug 10. doi:10.1016/j.canlet.2018.08.006
Ma, Y., Zhang, X., Han, X., Hao, Z., Ji, R., Li, J., et al. (2023). PDK1 regulates the progression of esophageal squamous cell carcinoma through metabolic reprogramming. Mol. Carcinog. 62 (6), 866–881. Epub 2023 Mar 29. doi:10.1002/mc.23531
Nan, Y., Liu, S., Luo, Q., Wu, X., Zhao, P., Chang, W., et al. (2023). m6A demethylase FTO stabilizes LINK-A to exert oncogenic roles via MCM3-mediated cell-cycle progression and HIF-1α activation. Cell Rep. 42 (10), 113273. Epub 2023 Oct 19. doi:10.1016/j.celrep.2023.113273
Nolfi-Donegan, D., Braganza, A., and Shiva, S. (2020). Mitochondrial electron transport chain: oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol. 37, 101674. Epub 2020 Aug 6. doi:10.1016/j.redox.2020.101674
Pan, C., Li, B., and Simon, M. C. (2021). Moonlighting functions of metabolic enzymes and metabolites in cancer. Mol. Cell 81 (18), 3760–3774. doi:10.1016/j.molcel.2021.08.031
Pavlova, N. N., and Thompson, C. B. (2016). The emerging hallmarks of cancer metabolism. Cell Metab. 23 (1), 27–47. doi:10.1016/j.cmet.2015.12.006
Qie, S., Yoshida, A., Parnham, S., Oleinik, N., Beeson, G. C., Beeson, C. C., et al. (2019). Targeting glutamine-addiction and overcoming CDK4/6 inhibitor resistance in human esophageal squamous cell carcinoma. Nat. Commun. 10 (1), 1296. doi:10.1038/s41467-019-09179-w
Rogers, J. E., Sewastjanow-Silva, M., Waters, R. E., and Ajani, J. A. (2022). Esophageal cancer: emerging therapeutics. Expert Opin. Ther. Targets 26 (2), 107–117. doi:10.1080/14728222.2022.2036718
Schrimpe-Rutledge, A. C., Codreanu, S. G., Sherrod, S. D., and McLean, J. A. (2016). Untargeted metabolomics strategies-challenges and emerging directions. J. Am. Soc. Mass Spectrom. 27 (12), 1897–1905. Epub 2016 Sep 13. doi:10.1007/s13361-016-1469-y
Singh, P. K., and Mehla, K. (2023). LXR signaling-mediated cholesterol metabolism reprogramming regulates cancer cell metastasis. Cancer Res. 83 (11), 1759–1761. doi:10.1158/0008-5472.CAN-23-0624
Sivanand, S., and Vander Heiden, M. G. (2020). Emerging roles for branched-chain amino acid metabolism in cancer. Cancer Cell 37 (2), 147–156. doi:10.1016/j.ccell.2019.12.011
Su, X., Xue, C., Xie, C., Si, X., Xu, J., Huang, W., et al. (2022). lncRNA-LET regulates glycolysis and glutamine decomposition of esophageal squamous cell carcinoma through miR-93-5p/miR-106b-5p/SOCS4. Front. Oncol. 12, 897751. eCollection 2022. doi:10.3389/fonc.2022.897751
Sun, W., Kou, H., Fang, Y., Xu, F., Xu, Z., Wang, X., et al. (2024). FOXO3a-regulated arginine metabolic plasticity adaptively promotes esophageal cancer proliferation and metastasis. Oncogene 43 (3), 216–223. Epub 2023 Dec 4. doi:10.1038/s41388-023-02906-0
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Tao, M., Luo, J., Gu, T., Yu, X., Song, Z., Jun, Y., et al. (2021). LPCAT1 reprogramming cholesterol metabolism promotes the progression of esophageal squamous cell carcinoma. Cell Death Dis. 12 (9), 845. doi:10.1038/s41419-021-04132-6
Tian, T., Lu, Y., Lin, J., Chen, M., Qiu, H., Zhu, W., et al. (2022). CPT1A promotes anoikis resistance in esophageal squamous cell carcinoma via redox homeostasis. Redox Biol. 58, 102544. Epub 2022 Nov 15. doi:10.1016/j.redox.2022.102544
Wang, W., Shao, F., Yang, X., Wang, J., Zhu, R., Yang, Y., et al. (2021). METTL3 promotes tumour development by decreasing APC expression mediated by APC mRNA N6-methyladenosine-dependent YTHDF binding. Nat. Commun. 12 (1), 3803. doi:10.1038/s41467-021-23501-5
Xiao, Y., Yu, T. J., Xu, Y., Ding, R., Wang, Y. P., Jiang, Y. Z., et al. (2023). Emerging therapies in cancer metabolism. Cell Metab. 35 (8), 1283–1303. doi:10.1016/j.cmet.2023.07.006
Xu, H., Zhou, S., Tang, Q., Xia, H., and Bi, F. (2020). Cholesterol metabolism: new functions and therapeutic approaches in cancer. Biochim. Biophys. Acta Rev. Cancer 1874 (1), 188394. doi:10.1016/j.bbcan.2020.188394
Zhang, G. C., Yu, X. N., Guo, H. Y., Sun, J. L., Liu, Z. Y., Zhu, J. M., et al. (2023). PRP19 enhances esophageal squamous cell carcinoma progression by reprogramming SREBF1-dependent fatty acid metabolism. Cancer Res. 83 (4), 521–537. doi:10.1158/0008-5472.CAN-22-2156
Zhao, M., Lu, T., Bi, G., Hu, Z., Liang, J., Bian, Y., et al. (2023). PLK1 regulating chemoradiotherapy sensitivity of esophageal squamous cell carcinoma through pentose phosphate pathway/ferroptosis. Biomed. Pharmacother. 168, 115711. Epub 2023 Oct 24. doi:10.1016/j.biopha.2023.115711
Zhao, Y., Ma, C., Cai, R., Xin, L., Li, Y., Ke, L., et al. (2024). NMR and MS reveal characteristic metabolome atlas and optimize esophageal squamous cell carcinoma early detection. Nat. Commun. 15 (1), 2463. doi:10.1038/s41467-024-46837-0
Zheng, C., Yu, X., Liang, Y., Zhu, Y., He, Y., Liao, L., et al. (2022). Targeting PFKL with penfluridol inhibits glycolysis and suppresses esophageal cancer tumorigenesis in an AMPK/FOXO3a/BIM-dependent manner. Acta Pharm. Sin. B 12 (3), 1271–1287. Epub 2021 Sep 11. doi:10.1016/j.apsb.2021.09.007
Zheng, Z. Y., Yang, P. L., Li, R. Y., Liu, L. X., Xu, X. E., Liao, L. D., et al. (2021). STAT3β disrupted mitochondrial electron transport chain enhances chemosensitivity by inducing pyroptosis in esophageal squamous cell carcinoma. Cancer Lett. 522, 171–183. Epub 2021 Sep 25. doi:10.1016/j.canlet.2021.09.035
Zhou, X., Huang, F., Ma, G., Wei, W., Wu, N., and Liu, Z. (2022). Dysregulated ceramides metabolism by fatty acid 2-hydroxylase exposes a metabolic vulnerability to target cancer metastasis. Signal Transduct. Target Ther. 7 (1), 370. doi:10.1038/s41392-022-01199-1
Zhu, H., Ma, X., Ye, T., Wang, H., Wang, Z., Liu, Q., et al. (2023). Esophageal cancer in China: practice and research in the new era. Int. J. Cancer 152 (9), 1741–1751. Epub 2022 Oct 5. doi:10.1002/ijc.34301
Keywords: metabolic reprogramming, glucose, fatty acid, amino acid, esophageal cancer
Citation: Wang Z, Sun X, Li Z, Yu H, Li W and Xu Y (2024) Metabolic reprogramming in esophageal squamous cell carcinoma. Front. Pharmacol. 15:1423629. doi: 10.3389/fphar.2024.1423629
Received: 26 April 2024; Accepted: 06 June 2024;
Published: 26 June 2024.
Edited by:
Elisa Giovannetti, VU Medical Center, NetherlandsReviewed by:
Qiang Wang, First Affiliated Hospital of Anhui Medical University, ChinaBaharan Fekry, University of Texas Health Science Center at Houston, United States
Copyright © 2024 Wang, Sun, Li, Yu, Li and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenya Li, c2FpbnQ1Mjg4QGhvdG1haWwuY29t; Yan Xu, eWFueHVAY211LmVkdS5jbg==
 Ziyi Wang
Ziyi Wang Xiangyu Sun
Xiangyu Sun Zehui Li1
Zehui Li1 Wenya Li
Wenya Li