
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol. , 15 July 2024
Sec. Pharmacology of Infectious Diseases
Volume 15 - 2024 | https://doi.org/10.3389/fphar.2024.1410453
This article is part of the Research Topic Emerging and Reemerging Neglected Tropical Diseases: Epidemiology, Transmission, Mitigation, and Vaccines and Chemotherapy Advancements View all 9 articles
The liver fluke O. viverrini (Opisthorchis viverrini), a neglected tropical disease (NTD), endemic to the Great Mekong Subregion (GMS), mainly afflicts the northeastern region of Thailand. It is a leading cause of cholangiocarcinoma (CCA) in humans. Presently, the treatment modalities for opisthorchiasis incorporate the use of the antihelminthic drug praziquantel, the rapid occurrence of reinfection, and the looming threat of drug resistance highlight the urgent need for vaccine development. Recent advances in “omics” technologies have proven to be a powerful tool for such studies. Utilizing candidate proteins identified through proteomics and refined via immunoproteomics, reverse vaccinology (RV) offers promising prospects for designing vaccines targeting essential antibody responses to eliminate parasite. Machine learning-based computational tools can predict epitopes of candidate protein/antigens exhibiting high binding affinities for B cells, MHC classes I and II, indicating strong potential for triggering both humoral and cell-mediated immune responses. Subsequently, these vaccine designs can undergo population-specific testing and docking/dynamics studies to assess efficacy and synergistic immunogenicity. Hence, refining proteomics data through immunoinformatics and employing computational tools to generate antigen-specific targets for trials offers a targeted and efficient approach to vaccine development that applies to all domains of parasite infections. In this review, we delve into the strategic antigen selection process using omics modalities for the O. viverrini parasite and propose an innovative framework for vaccine design. We harness omics technologies to revolutionize vaccine development, promising accelerated discoveries and streamlined preclinical and clinical evaluations.
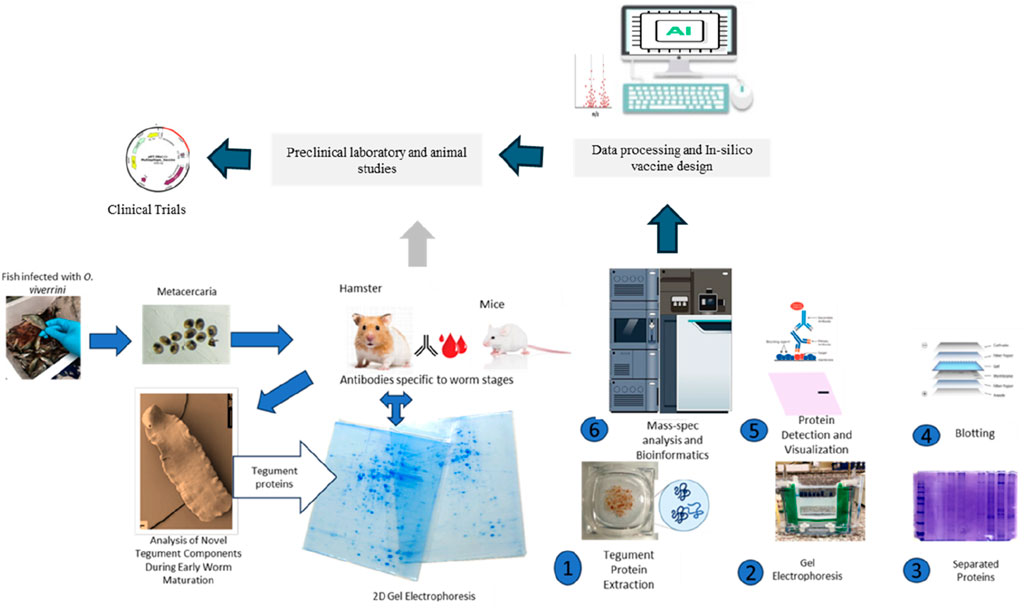
GRAPHICAL ABSTRACT | Sincere gratitude to Prof. Jennifer Keiser for generously granting permission for the use of tegument picture in this graphical abstract.
Opisthorchiasis caused by O. viverrini (Opisthorchis viverrini; Ov) infection is a significant public health issue in many Southeast Asian countries, including Thailand, Lao PDR, Vietnam, and Cambodia, infecting more than 12 million people (Kaewpitoon et al., 2008; Sripa et al., 2021). The liver fluke infection is caused by eating raw or uncooked fish products, which is common in Thailand’s northeastern and northern regions, particularly in rural areas. Infection progression can result in cholangitis, obstructive jaundice, hepatomegaly, cholecystitis, cholelithiasis, and cholangiocarcinoma (CCA), among other hepatic disorders (Sripa et al., 2007). Biliary damage caused by O. viverrini examined in the hamster model showed that the parasite infection can solely cause CCA in the long term (Songserm et al., 2009). Hence, O. viverrini and C. sinensis liver fluke parasites have been classified as “Group 1” biological carcinogens (World Health Organization, 1994).
Upon ingesting raw infected fish, O. viverrini metacercariae excyst in the duodenum. The newly excysted juveniles (NEJs) start to migrate towards the ampulla of Vater, relying on chemotaxis and glycogen reserves for the movement (Tielens et al., 1982). Once within the bile duct, they mature into self-fertilizing adult flukes in the bile ducts, thereby acquiring the reproductive capabilities necessary to release eggs (Harinasuta and Harinasuta, 1984; Orido, 1990). The pathogenic mechanisms of O. viverrini-induced CCA (Ov-CCA) involve parasite-feeding-triggered inflammation, host-helminth interactions at the tegument interface, and immunopathology driven by secreted ES (Excretory Secretory products) products and EVs (Extracellular Vesicles) effector repertoire (Chaiyadet et al., 2019; Glaser, Gaudio et al., 2009; Sripa, 2003; Sripa et al., 2012).
Understanding parasite biology and host interactions better is imperative to identify new drug targets or potential vaccine candidates. The analysis of genomic (Laha et al., 2007; Young et al., 2014), transcriptomic (Jex et al., 2012), and proteomic (Mulvenna et al., 2010) data of O. viverrini has provided us with a deeper understanding of Ov-tegument and secreted proteins. These proteins may function both protectively and as immunogens in host-specific interactions. Despite substantial research efforts and omics advancement, no vaccine conferring efficacious protection against trematode infections has achieved the level of effectiveness required for commercial deployment (McManus, 2020). Furthermore, the lack of interest from pharmaceutical industries and attention from health agencies and policies is related to the fact that no vaccine against opisthorchiasis is available for clinical use. This can also be attributed partially to the abundance of proteomics data and findings, yet relatively less focus on filtering and analysing these datas for wet lab and translational investigations. Hence, incorporating immunoproteomics is essential in this context, as it facilitates a filter in the rigorous validation of potential markers, thereby bridging the gap between discovery and clinical verification (Pedersen et al., 2005; Ejazi et al., 2018). Moreover, previous investigations exploring antibody responses in the context of parasitic infections have demonstrated the value and feasibility of such immunoproteomics-based studies (Jedlina-Panasiuk, 2002; Pleasance et al., 2011; Zhang et al., 2015; Driguez et al., 2016).
Antigens identified through immunoproteomics analysis spanning from the tegument to ES molecules and EVs of O. viverrini can be computationally evaluated. Furthermore, assessing vaccine solubility, antigenicity, immunogenicity, and allergenicity predictions informs the selection of epitopes optimally suited to activating robust host defences (Kapuganti et al., 2022). For the last few years, advanced Artificial intelligence (AI) and Machine Learning (ML) algorithms to predict protein structures and modelling have permitted structural prediction with about 90% accuracy in silico (Stevens and He, 2022). Moreover, molecular docking simulations yield insights into projected binding dynamics between prioritised epitopes and immune receptors (Sahoo et al., 2022; Che et al., 2023). Access to these two web-based servers alone holds immense potential to significantly streamline academic research efforts and lead to substantial cost savings. We believe it is now imperative to incorporate immunoinformatics in the prioritisation process before in vitro and in vivo studies, thereby propelling the progression toward clinical trials more efficiently (Figure 1). Hence, this review highlights the initial phases in vaccine design, which involves the integration of immunoproteomics utilising omics data and computational bioinformatics tools for the systematic studies on helminthic parasites. This integration has the potential to identify numerous protein marker candidates suitable for clinical investigations and to advance the rational design of multi-epitope vaccines against opisthorchiasis.
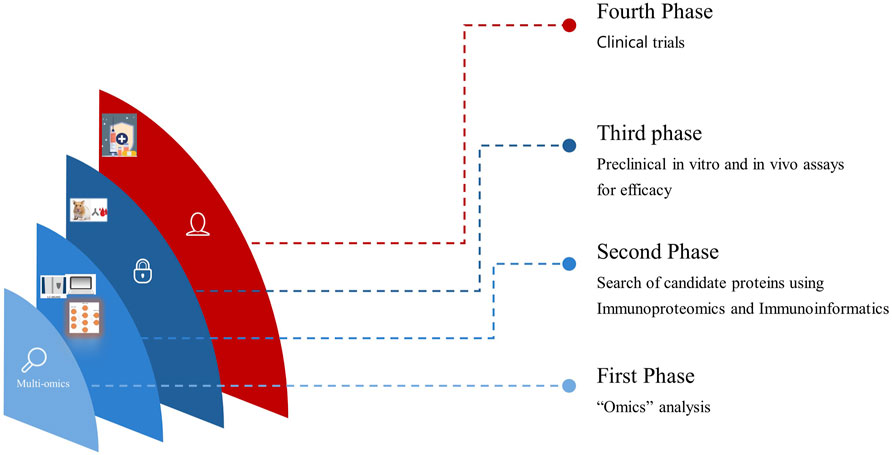
Figure 1. Translational progression by integrating Omics, Immunoproteomics, and in vitro and in vivo studies seamlessly accelerates vaccine development from discovery to clinical application.
The multi-omics journey for O. viverrini started over 15 years ago by generating and analysing so-called expressed sequence tags (EST) that profiled the first description of gene discovery for the liver fluke O. viverrini (Laha et al., 2007). Since 2010, the integration of multi-omics strategies and methodologies, including liquid chromatography-mass spectrometry (LC-MS), next-generation sequencing, and various -omics based technologies, has significantly enhanced our understanding of trematode biology-spanning across the field of genomics, transcriptomics, proteomics, metabolomics, and glycomics (Table 1) (Mulvenna et al., 2010; Jex et al., 2012; Talabnin et al., 2013; Young et al., 2014; Prasopdee et al., 2019; Kokova et al., 2020). Therefore, research efforts focused on the genomes, transcriptomes, and proteomes of O. viverrini isolates at a global level hold great potential to significantly advance our understanding of the molecular pathogenesis of O. viverrini associated CCA. Such investigations are expected to lay the groundwork for identifying critical targets for designing new interventions.
Before exploring the O. viverrini genome and transcriptome, molecular-based studies identified, characterised, and proposed potential drug targets and pathways for exploration. The groundbreaking work of Young et al. (2014) in elucidating the draft genome of O. viverrini provided an advanced understanding of the genetic makeup of the liver fluke parasite (Young et al., 2014). This study provided the first global understanding of the biology of O. viverrini during its establishment and adaptation to life within the bile duct, helping researchers know how the parasite persists in this unique ecological niche.
Genomic findings provided insights into the developmental stages of O. viverrini, revealing that juvenile and immature forms exhibit increased molecular activity and cellular growth. These processes are crucial for their maturation into reproductively competent adults. The expression of distinct genes helps the O. viverrini parasite facilitate tissue differentiation, development, and immune evasion, which are functions that become less active in adulthood. The genomic study utilised existing O. viverrini transcriptomic data at both the assembly and annotation stages. It continued to map RNA-seq to infer relative expression levels of predicted genes and explore developmental expression changes (Jex et al., 2012). This mapping of transcripts to genes allowed researchers to annotate the predicted functional pathways of these genes based on sequence homology to characterised genes in other species, thereby tentatively linking genes to relevant biological processes such as metabolism, growth, and reproduction, even in the absence of complete ortholog characterisation in O. viverrini.
Hence, comparing transcript expression profiles across developmental stages of O. viverrini may help identify genes involved in immune evasion and tissue-specific functions, offering potential intervention targets to block the establishment during early parasite infections. Moreover, the transcriptome analysis of adult and juvenile O. viverrini worms revealed that immature worms exhibited upregulation in the transcription of peptides associated with energy metabolism. This included oxidative phosphorylation, fatty acid metabolism, and amino acid degradation. In contrast, adult parasites showed increased levels of transcripts involved in nucleotide metabolic processes and processing, oocyte meiosis, and egg-specific antigens. The elevated metabolic activity-related genes observed in the juvenile and immature stages may indicate an anabolic state aimed at acquiring sufficient nutrients from the host to support rapid growth and development. Growth hormones likely play a crucial role in driving tissue proliferation, elongation, and differentiation in juvenile and immature parasites. However, the expression of growth hormones may be downregulated once the adult form is attained. In contrast, mature adults may shift towards a relatively catabolic state focused on survival and reproduction (Jex et al., 2012; Young et al., 2014). Thus, genomic and transcriptomic data availability can help us compare gene expression across developmental stages, reveal the stage-specific biological processes, indicate potential therapeutic vulnerabilities, and offer a more holistic view of the molecular adaptions that enable the parasite’s complex life cycle.
In 2009, Boonmee et al. employed two-dimensional gel electrophoresis to analyse the protein secretion of O. viverrini at various maturation stages (Boonmee et al., 2003). These distinctions were identified in the 4-week adult fluke compared to the first-week juveniles, indicating an age-dependent augmentation in protein secretion during the liver fluke maturation. This study provides insights into the protein expression patterns of O. viverrini during development, which can significantly contribute to our understanding of the pathogenesis of opisthorchiasis.
In 2010, Mulvenna et al. conducted the first detailed proteomic analysis of O. viverrini, using sequential solubilisation of isolated teguments, and a subset of these was localised to the surface membrane of the tegument by labelling O. viverrini flukes with biotin and confirming surface localisation with fluorescence microscopy (Mulvenna et al., 2010). This fractionation approach identified five differentially expressed proteins between juvenile and immature stages, of which two were uncharacterised proteins, Experimental autoimmune prostatitis antigen 2 (EAP-2) involved in calcium homeostasis, the cysteine protease legumain, and two granulins that function as growth factors. The study unveiled two granulins, OvGRN-1 and OvGRN-2, as growth factors expressed in both juvenile and adult parasite stages. Alongside identifying various other peptidases and potential markers and targets, their involvement in crucial processes is implied, with certain protein expressions persisting throughout immature and adult life stages (Table 2). This underscores the potential of omics, advanced informatics technologies, and the increasing accessibility of annotated helminth genomes to substantially expedite progress in computational and practical inquiries within NTDs-based studies.
These omics fields are in their early stages of research, and ample exploration is yet to be undertaken in this domain. O. viverrini helminth can express distinctive and highly immunogenic glycans; these glycan antigens share structural features with those present in the intermediate and vertebrate hosts, thus resembling host-like glycans (van Die and Cummings, 2009). The immunogenicity of glycans present on the surface proteins of parasites, especially on the O. viverrini tegument, is a critical factor in the parasite’s ability to establish infection and modulate the host immune response (Talabnin et al., 2013). Thus, identifying tegument glycoproteins and their glycan structures in developmental stages can help discover targets that elicit a protective immune response during the early stages of infection (Garcia-Campos et al., 2016).
Metabolomics is another crucial facet of omics research. Studying changes in the parasite’s metabolome during different life stages and in response to the host environment can provide clues about metabolic pathways vital to the parasite’s survival. In the case of Opisthorchis felinues, a plasma metabolomics study of the temporal response to opisthorchiasis in an animal model revealed an altered metabolic response in the host, characterised by amino acid depletion and alterations in lipoprotein and cholesterol levels (Kokova et al., 2020). These alterations likely represent a host defence mechanism against the infection, offering perspectives/understanding that could guide therapeutic approaches.
Therefore, integrating various “omics” techniques, including genomics, transcriptomics, proteomics, glycomics, and metabolomics, can lead to a more profound understanding of the dynamic interaction between O. viverrini and its host. In-depth Omics analyses literature discussed above indicate juvenile parasites’ role in upregulating energy metabolism to support anabolic processes during development, synthesising complex molecules from simpler precursors. Conversely, adult parasites transition to lower metabolic activity and growth hormone expression, prioritising survival and reproduction over continued somatic growth (Figure 2). Hence, immunoproteomic approaches, integrating host serology, capture real-time dynamics in the interplay between fluke antigens and the immune response across infection stages, providing a system-level understanding of liver fluke parasite-regulated immune modulation and complementing proteomics in action.
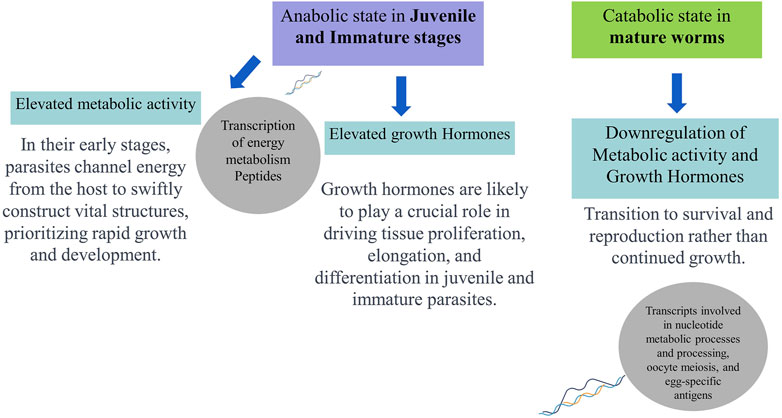
Figure 2. Schematic representation of hypothesised developmental (stage-specific) metabolic profiles and functions of Opisthorchis viverrini based on omics analyses.
As -omics techniques enable the broad discovery of temporally-resolved molecular alterations underlying host-parasite interactions, disease progression, and therapeutic targets during different stages of parasite infection (Hasin et al., 2017; Mitreva, 2022). Considerable progress in parasitology has been made through generating and interrogating omics data (Gardinassi et al., 2018; Liu et al., 2021; Athieniti and Spyrou, 2023). The term “immunoproteomics” is becoming a mature field with potential impact on developing immunoproteomics-based assays and vaccines (Fulton and Walker, 2013). Immunoproteomics explicitly profiles the host immune recognition of protein antigens, aiding the isolation of immunoreactive proteins and leveraging such data to predict and prioritise optimal vaccine candidates efficiently (Dennehy and McClean, 2012).
Immunocapture of antigenic proteins followed by mass spectrometry has proven an effective strategy for learning vaccine targets from trematode parasites (Bennett and Robinson, 2021). The assays have played a crucial role in identifying antigenic proteins from parasitic organisms, including parasitic trematodes (Reamtong, 2013). They leverage an expanding array of technologies to identify and quantify antigenic peptides and proteins that induce immunity (Hayes et al., 2018).
The immunoproteomics techniques include a wide range of gel-based, array-based, mass spectrometry, DNA-based, and in silico methodologies to gain unprecedented insights into disease aetiology and progression and facilitate the identification of novel vaccine candidates and biomarker discovery (Table 3). By comprehensively profiling the host immune response at the protein (antigen) level, immunoproteomics enhances our understanding of pathogenic mechanisms and enables accelerated selection of optimal antigens for rational vaccine design against complex infectious diseases. Therefore, integrating immunoproteomics strategies with omics-related discoveries provides a powerful approach for unravelling the functions of upregulated proteins during the infection and a successful vaccine design.
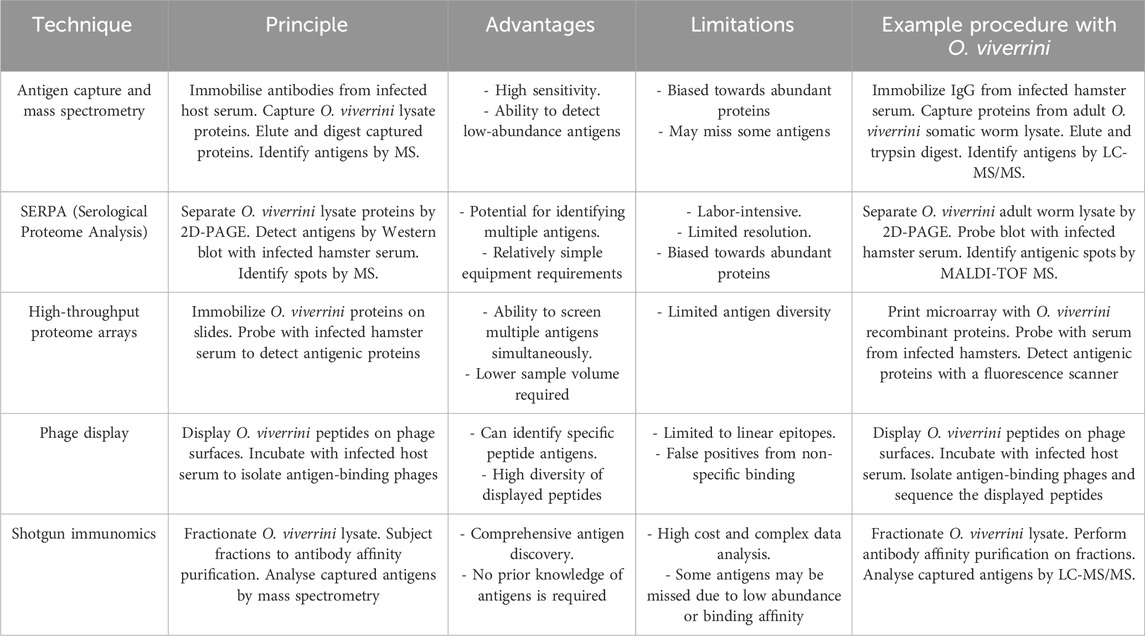
Table 3. This table compares the principles, advantages, and limitations and provides an immunoproteomics workflow technique to identify antigens from the liver fluke Opisthorchis viverrini.
Immunoproteomics, while crucial for vaccine development, also plays a pivotal role in biomarker discovery and diagnostics by identifying antigens that elicit immune responses (Costa et al., 2023; Germanó et al., 2022; Shi et al., 2018; Zawistowska-Deniziak et al., 2021). The tegument of O. viverrini, a multinuclear syncytium, presents an enticing prospect for vaccine development due to its direct contact with the host bile duct and immune system. Vaccination can effectively stimulate the production of both mucosal and systemic antibodies. Moreover, the anatomical arrangement of O. viverrini allows for an immediate response of the tegument muscles to external stimuli such as mechanical pressure, ion gradients, nutrient gradients, and damage by deviating immune response that favours parasite survival in the host or avoiding damage through the release of ES products and EVs from the tegument (Chaiyadet et al., 2017; Sripa et al., 2012; Thompson and Geary, 2003).
The tegument analysis of the array of proteins in O. viverrini is crucial for unravelling the intricate host-parasite relationship, enhancing opisthorchiasis diagnosis, and advancing the development of marker based studies, therapeutic target based studies to manage O. viverrini infection effectively (Liau et al., 2023; Kafle and Suttiprapa, 2024). Studies using serum from O. viverrini-infected animals and humans have shown seroreactivity with antigens of the tegument, eggs, reproductive organs, and secretory products (Wilkins et al., 1999; Kafle and Suttiprapa, 2024). These proteins have the potential to be considered vaccine candidates and are more likely to come into contact with the host and may play roles in the parasite’s survival (Young et al., 2014; Molehin et al., 2022). The scanning electron microscopy of the tegument membrane demonstrated stage-specific morphological alterations on the surface of the O. viverrini parasite (Apinhasmit et la., 1993). Moreover, several studies have shown differences in tegument proteins based on the gender of worms (Pérez-Sánchez et al., 2008; Zhang et al., 2013; Winkelmann et al., 2022). Thus providing robust evidence for the pivotal role of tegument proteins in parasite reproduction, signal transduction, nutrition, pathogenesis, and host immune modulation (Dalton et al., 2004; Loukas et al., 2007). These interactions are postulated to occur through the parasites’ outer tegument layer and the release of soluble metabolites (Suttiprapa et al., 2018), and this intricate crosstalk fosters tumorigenesis through complex intercellular communication (Elinav et al., 2013).
Moreover, the surface proteins are often considered promising biomarkers due to their heightened specificity for pathogens, particularly in host-parasite interactions. This specificity reduces the likelihood of cross-reactivity with host proteins, making them valuable targets for diagnostic and therapeutic purposes (Retra et al., 2015; Zhang et al., 2015; de Jong and Kocer, 2023).
The survival and pathogenesis of O. viverrini depend on the successful evasion of the host immune response by the juvenile and immature fluke stages, which mature into hermaphrodite adults, eventually causing opisthorchiasis. It has been observed that O. viverrini metacercaria possesses several antigens similar to those found in adult worms and their metabolic products (Wongratanacheewin et al., 1988).
Comparing protein profiles of O. viverrini metacercariae, NEJs to developing immature and mature worms helps reveal stage-specific protein expression. This analysis using LC-MS and bioinformatics may offer insights into the roles of these proteins in the parasite’s development, survival, and reproduction. Several studies have demonstrated the early detection of antibodies in serum as early as 15 days post-infection, with notable inflammatory cell infiltration and the onset of hepatic pathology observed from the third day following infection (Bhamarapravati et al., 1978; Chawengkirttikul and Sirisinha, 1988). This suggests that NEJs and immature O. viverrini flukes interact and activate the host immune system during the initial invasion and establishment phases. Furthermore, the suggestion that O. viverrini induces immune-suppressive responses early on may facilitate parasite persistence, thereby contributing to developing hepatobiliary diseases associated with chronic O. viverrini infection (Sripa et al., 2018). Such investigations identify host-upregulated proteins and signalling pathways detrimental to the parasite, which is crucial for uncovering potential survival strategies in susceptible hosts.
With the plethora of knowledge in the Omics domain, immunoinformatics, combining immunology and bioinformatics, offers innovative solutions in drug design and identifying vaccine candidates (María et al., 2017; Tomar, 2020). Variations in proteins released in the form of EVs (Trelis et al., 2022) and ES proteins (Wang et al., 2013; Yeshi et al., 2022), and tegument surface proteins may help O. viverrini maintain stability, survival, and immune evasion strategies. This can be explored in the systematic vaccine design (Figure 3). Hence, whole proteome analysis in trematode parasitism is beneficial for identifying crucial parasite proteins associated with virulence, exploring essential protein interactions, and uncovering distinctive immuno-modulatory mechanisms contributing to prolonged infection (van der Ree and Mutapi, 2015). Moreover, longitudinal profiling of antibody responses through immunoproteomic tools reveals antigenic proteins expressed in specific parasite developmental stages, from NEJ to immature and adult developmental phases.
The strategy for developing a vaccine against cancer-inducing O. viverrini parasites involves using epitopes from antigens to elicit an immune response. However, the primary challenge in creating a cancer vaccine lies in understanding the immunology of infection and identifying inflammation-inducing antigens that can potentially help parasites in vital functions. These challenges can be addressed by blending omics and immunoproteomics results with immunoinformatics (Figure 4).
The in silico screening and experimental validation process streamlines identifying and evaluating potential antigens from a liver fluke parasite, offering a faster and more efficient alternative to traditional methods. Candidate antigens can be evaluated in silico for population coverage (Bui et al., 2006), antigenicity (Doytchinova and Flower, 2007), physicochemical properties (Wilkins et al., 1999), and ability to induce B- and T-cell epitopes (Bui et al., 2006; Khan et al., 2022). Particularly intriguing are fluke-specific antigens predicted to stimulate both humoral and cellular immune responses in infected hosts, reflecting scientific inclination and necessity. Structural modelling and prediction of immunogenic epitopes using servers like AlphaFold2 (Jumper et al., 2021; Yang, et al., 2023) allows rigorous in vitro testing through molecular docking and dynamics to generate hypotheses regarding immune receptors, proteins and designed vaccine interaction (Soleymani et al., 2022). Recombinant protein expression techniques such as cloning predicted multi-epitope subsequences into prokaryotic vectors like pET28b (+) provide a robust means of antigen production and evaluation in immunogenicity studies. (Martinelli, 2022). Various computational and bioinformatics tools and techniques are available in immunoinformatics, particularly in the context of vaccine development (Table 4), which can be utilized for parasitic and infectious diseases.
The synergistic application of experimental vaccine design using omics and computational immunoinformatics represents a promising approach to develop safe, efficacious multi-antigen helminth vaccines rationally. This integrated framework may help overcome decades of challenges by accelerating the selection of optimized antigens that induce robust, broad, and long-lasting immunity against diverse helminth strains. Such vaccines targeting multiple parasite stages and species could significantly reduce the health burden of soil-transmitted helminthiases and other NTDs that predominantly impact low-resource populations worldwide.
Vaccination has played a pivotal role in mitigating the spread of numerous pathogens and is a cornerstone in disease eradication efforts (Rauch et al., 2018). However, despite its proven efficacy, a notable absence of licensed anti-helminth vaccines for human use remains. Furthermore, helminths, being eukaryotic organisms, present many challenges for vaccination strategies due to their distinct biological characteristics. Hence, immunoinformatics and reverse vaccinology offer transformative avenues in vaccine development. These methodologies leverage computational tools and genomic insights to streamline the identification of potential vaccine candidates from the pool of identified proteins.
Researchers use helminth genomes and proteomes to identify antigenic proteins that initiate immune responses, aiding in the development of customized vaccines. Immunoinformatics methods then prioritize vaccine candidates based on their capacity to elicit protective immune responses, reducing the risk of vaccine failure during advanced clinical trials. Immunoinformatic analysis has been conducted across diverse parasite species. For the focus of our review, we present a curated selection of studies employing the vaccine development pipeline specifically within the domain of helminth parasites, as illustrated in (Table 5). This implies ongoing progress in helminth vaccine research through continual discovery and prioritization of novel candidates.
The lack of attention, prioritization and funding of helminth infections within health organisations and research spheres can be attributed to the absence of opisthorchiasis vaccines currently available for clinical use. Recent strides in transcriptomics, genomics, and proteomics have shed light on the potential engagement of genes and proteins during the early stages of O. viverrini, influencing aspects such as survival, motility, immune evasion, and pathogenicity. However, to obtain a clear view of antigen targets, it is crucial to integrate data across various “omics” modalities (Vahabi and Michailidis, 2022), and immunoinformatics represents a sensitive high-throughput platform for profiling immunogenic antigens from the parasite. Insilico analysis with high predictive outcomes can limit the use of animal models, leading to more sustainable research (Pappalardo et al., 2019). In the future, there is hope that in silico trials can replace human trials (Ebert et al., 2021). Furthermore, in silico analyses in the O. viverrini vaccine design process can significantly reduce costs and expedition timelines. Computational screening of antigen candidates can yield substantial savings by minimising experimental wet-lab validation and animal trials, thereby mitigating financial barriers for this under-resourced yet high-impact disease (Saldanha et al., 2023). This approach expedites the entry of numerous vaccine candidates into clinical trials, often ahead of schedule (Shawan et al., 2023). However, the vaccine design against carcinogenic parasites like O. viverrini must carefully account for the inflammation associated with its infection and the potential presence of undiagnosed tumours in endemic populations. This consideration is vital due to the intricate interplay between tumours and immune cells, whereby various pathways influence each other, ultimately steering toward distinct outcomes (Gonzalez et al., 2018; Bedognetti et al., 2019). Multistage vaccines have been proposed as an effective strategy in complicated pathogens, as this may show a synergistic effect and would be particularly desirable for biliary tissue-dwelling pathogenic flukes (Maizels, 2021). Furthermore, the choice of proper and safe synthetic immunomodulatory adjuvants should be carefully selected/designed (Zhao et al., 2023) and follow-up in vivo studies. Overall, combining immunoproteomics and immunoinformatics offers a compelling strategy to accelerate the rational design and development of vaccines, holding significant potential for preventing and controlling infectious diseases in forthcoming years.
The current period marks an exciting phase for research in this field, with significant advancements seen in recent years, particularly in the structure-based design of new antigens, antibodies, and binders. Traditional vaccine development faces significant challenges, including high costs, hesitancy, stringent safety requirements, and societal demands for perfect efficacy. Additionally, issues like immunosenescence, limited adjuvants, and concurrent health problems in the developing world complicate efforts. Therefore, the global availability of in silico technologies in vaccine design and access to tools utilising supercomputers via internet-enabled interfaces maximises their utilization across diverse research communities internationally. In NTDs based research, merging AI and ML with biological, medical, and biochemistry-based approaches holds immense promise for driving breakthroughs. This convergence is poised to accelerate the discovery of novel biologics, offering new avenues for combating such neglected diseases effectively (Rodrigues and Ersching, 2014; Winkler, 2021). This initiative aims to end the neglect and contribute to achieving the Sustainable Development Goals (World Health Organization, 2020).
While converging experimental and computational vaccine design holds promise for rational helminth vaccine development, certain limitations must also be acknowledged. These in silico based tools require high-quality input data to train algorithms and predict outcomes accurately. Further experimental validation is advised to refine helminth vaccine formulations before proceeding to clinical testing. Moreover, ensuring vaccine effectiveness and safety profiles suitable for use in endemic areas where NTDs are prevalent and infrastructure is limited poses difficulties. In summary, while integrating immunoproteomics and immunoinformatics holds promise for optimising vaccine design, transitioning from theoretical constructs to practical, field-ready vaccines for helminth diseases remains a formidable long-term challenge. Overcoming both natural and technical barriers necessitates sustained dedication from all stakeholders involved.
AK: Data curation, Methodology, Conceptualization, Formal analysis, Validation, Investigation, Visualization, Writing–original draft, Writing–review and editing. SO: Data curation, Supervision, Formal analysis, Project administration, Validation, Funding acquisition, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The study was funded by the Doctoral Research Fund to the corresponding author, SCO. AK was supported by a Postgraduate Study Support Grant of the Faculty of Medicine, Khon Kaen University for his studies.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aarthy, M., Pandiyan, G. N., Paramasivan, R., Kumar, A., and Gupta, B. (2024). Identification and prioritisation of potential vaccine candidates using subtractive proteomics and designing of a multi-epitope vaccine against Wuchereria bancrofti. Sci. Rep. 14 (1), 1970. doi:10.1038/s41598-024-52457-x
Abraham, D., Graham-Brown, J., Carter, D., Gray, S. A., Hess, J. A., Makepeace, B. L., et al. (2021). Development of a recombinant vaccine against human onchocerciasis. Expert Rev. Vaccines 20 (11), 1459–1470. doi:10.1080/14760584.2021.1977125
Akıl, M., Aykur, M., Karakavuk, M., Can, H., and Döşkaya, M. (2022). Construction of a multiepitope vaccine candidate against Fasciola hepatica: an in silico design using various immunogenic excretory/secretory antigens. Expert Rev. Vaccines 21 (7), 993–1006. doi:10.1080/14760584.2022.1996233
Apinhasmit, W., Sobhon, P., Saitongdee, P., and Upatham, E. S. (1993). Opisthorchis viverrini: changes of the tegumental surface in newly excysted juvenile, first-week and adult flukes. Int. J. Parasitol. 23 (7), 829–839. doi:10.1016/0020-7519(93)90046-2
Athieniti, E., and Spyrou, G. M. (2023). A guide to multi-omics data collection and integration for translational medicine. Comput. Struct. Biotechnol. J. 21, 134–149. doi:10.1016/j.csbj.2022.11.050
Bedognetti, D., Ceccarelli, M., Galluzzi, L., Lu, R., Palucka, K., Samayoa, J., et al. (2019). Toward a comprehensive view of cancer immune responsiveness: a synopsis from the SITC workshop. J. Immunother. Cancer 7 (1), 131. doi:10.1186/s40425-019-0602-4
Bennett, A. P. S., and Robinson, M. W. (2021). Trematode proteomics: recent advances and future directions. Pathogens 10 (3), 348. doi:10.3390/pathogens10030348
Bhamarapravati, N., Thammavit, W., and Vajrasthira, S. (1978). Liver changes in hamsters infected with a liver fluke of man, Opisthorchis viverrini. Am. J. Trop. Med. Hyg. 27 (4), 787–794. doi:10.4269/ajtmh.1978.27.787
Boonmee, S., Imtawil, K., Wongkham, C., and Wongkham, S. (2003). Comparative proteomic analysis of juvenile and adult liver fluke, Opisthorchis viverrini. Acta Trop. 88 (3), 233–238. doi:10.1016/j.actatropica.2003.09.006
Bui, H.-H., Sidney, J., Dinh, K., Southwood, S., Newman, M. J., and Sette, A. (2006). Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinforma. 7 (1), 153. doi:10.1186/1471-2105-7-153
Chaiyadet, S., Krueajampa, W., Hipkaeo, W., Plosan, Y., Piratae, S., Sotillo, J., et al. (2017). Suppression of mRNAs encoding CD63 family tetraspanins from the carcinogenic liver fluke Opisthorchis viverrini results in distinct tegument phenotypes. Sci. Rep. 7 (1), 14342. doi:10.1038/s41598-017-13527-5
Chaiyadet, S., Sotillo, J., Krueajampa, W., Thongsen, S., Brindley, P. J., Sripa, B., et al. (2019). Vaccination of hamsters with Opisthorchis viverrini extracellular vesicles and vesicle-derived recombinant tetraspanins induces antibodies that block vesicle uptake by cholangiocytes and reduce parasite burden after challenge infection. PLoS Neglected Trop. Dis. 13 (5), e0007450. doi:10.1371/journal.pntd.0007450
Chawengkirttikul, R., and Sirisinha, S. (1988). Antibodies in serum and bile of hamsters experimentally infected with Opisthorchis viverrini. Int. J. Parasitol. 18 (6), 721–727. doi:10.1016/0020-7519(88)90111-7
Che, X., Liu, Q., and Zhang, L. (2023). An accurate and universal protein-small molecule batch docking solution using Autodock Vina. Results Eng. 19, 101335. doi:10.1016/j.rineng.2023.101335
Costa, S. S., Santos, L. M. O., Freire, L. C., Tedeschi, A. L. F., Ribeiro, N. R., Queiroz, M. H. R., et al. (2023). Immunoproteomics approach for the discovery of antigens applied to the diagnosis of canine visceral leishmaniasis. Acta Trop. 241, 106865. doi:10.1016/j.actatropica.2023.106865
Dalton, J. P., Skelly, P., and Halton, D. W. (2004). Role of the tegument and gut in nutrient uptake by parasitic platyhelminths. Can. J. Zoology 82 (2), 211–232. doi:10.1139/z03-213
de Jong, E., and Kocer, A. (2023). Current methods for identifying plasma membrane proteins as cancer biomarkers. Membranes 13 (4), 409. doi:10.3390/membranes13040409
Dennehy, R., and McClean, S. (2012). Immunoproteomics: the key to discovery of new vaccine antigens against bacterial respiratory infections. Curr. Protein Pept. Sci. 13 (8), 807–815. doi:10.2174/138920312804871184
Doytchinova, I. A., and Flower, D. R. (2007). VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinforma. 8 (1), 4. doi:10.1186/1471-2105-8-4
Driguez, P., McWilliam, H. E., Gaze, S., Piedrafita, D., Pearson, M. S., Nakajima, R., et al. (2016). Specific humoral response of hosts with variable schistosomiasis susceptibility. Immunol. Cell Biol. 94 (1), 52–65. doi:10.1038/icb.2015.61
Ebert, M. A., Gebski, V., and Baldock, C. (2021). In the future simulations will replace clinical trials. Phys. Eng. Sci. Med. 44 (4), 997–1001. doi:10.1007/s13246-021-01079-y
Ebrahimi, M., Seyyedtabaei, S. J, Ranjbar, M. M., Tahvildar-biderouni, F., and Mamaghani, A. J. (2020). Designing and modeling of multi-epitope proteins for diagnosis of Toxocara canis infection. Int. J. Pept. Res. Ther. 26, 1371–1380. doi:10.1007/s10989-019-09940-1
Ejazi, S. A., Bhattacharyya, A., Choudhury, S. T., Ghosh, S., Sabur, A., Pandey, K., et al. (2018). Immunoproteomic identification and characterization of leishmania membrane proteins as non-invasive diagnostic candidates for clinical visceral leishmaniasis. Sci. Rep. 8 (1), 12110. doi:10.1038/s41598-018-30546-y
Elinav, E., Nowarski, R., Thaiss, C. A., Hu, B., Jin, C., and Flavell, R. A. (2013). Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer 13 (11), 759–771. doi:10.1038/nrc3611
Evangelista, F. M. D., van Vliet, A. H. M., Lawton, S. P., and Betson, M. (2022). A reverse vaccinology approach identifies putative vaccination targets in the zoonotic nematode Ascaris. Front. Vet. Sci. 9, 1014198. doi:10.3389/fvets.2022.1014198
Garcia-Campos, A., Ravidà, A., Nguyen, D. L., Cwiklinski, K., Dalton, J. P., Hokke, C. H., et al. (2016). Tegument glycoproteins and cathepsins of newly excysted juvenile fasciola hepatica carry mannosidic and paucimannosidic N-glycans. PLoS Negl. Trop. Dis. 10 (5), e0004688. doi:10.1371/journal.pntd.0004688
Gardinassi, L. G., Arévalo-Herrera, M., Herrera, S., Cordy, R. J., Tran, V., Smith, M. R., et al. (2018). Integrative metabolomics and transcriptomics signatures of clinical tolerance to Plasmodium vivax reveal activation of innate cell immunity and T cell signaling. Redox Biol. 17, 158–170. doi:10.1016/j.redox.2018.04.011
Germanó, M. J., Mackern-Oberti, J. P., Vitório, J. G., Duarte, M. C., Pimenta, D. C., Sanchez, M. V., et al. (2022). Identification of immunodominant antigens from a first-generation vaccine against cutaneous leishmaniasis. Front. Immunol. 13, 825007. doi:10.3389/fimmu.2022.825007
Glaser, S. S., Gaudio, E., Miller, T., Alvaro, D., and Alpini, G. (2009). Cholangiocyte proliferation and liver fibrosis. Expert Rev. Mol. Med. 11, e7. doi:10.1017/s1462399409000994
Gonzalez, H., Hagerling, C., and Werb, Z. (2018). Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 32 (19-20), 1267–1284. doi:10.1101/gad.314617.118
Harinasuta, C., and Harinasuta, T. (1984). Opisthorchis viverrini: life cycle, intermediate hosts, transmission to man and geographical distribution in Thailand. Arzneimittelforschung 34 (9b), 1164–1167.
Hasin, Y., Seldin, M., and Lusis, A. (2017). Multi-omics approaches to disease. Genome Biol. 18 (1), 83. doi:10.1186/s13059-017-1215-1
Hayes, S. A., Clarke, S., Pavlakis, N., and Howell, V. M. (2018). The role of proteomics in the age of immunotherapies. Mamm. Genome 29 (11), 757–769. doi:10.1007/s00335-018-9763-6
Jedlina-Panasiuk, L. (2002). The immune response of various species to Fasciola hepatica infection. Wiad. Parazytol. 48 (1), 11–21.
Jex, A. R., Young, N. D., Sripa, J., Hall, R. S., Scheerlinck, J. P., Laha, T., et al. (2012). Molecular changes in Opisthorchis viverrini (Southeast Asian liver fluke) during the transition from the juvenile to the adult stage. PLoS Negl. Trop. Dis. 6 (11), e1916. doi:10.1371/journal.pntd.0001916
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. doi:10.1038/s41586-021-03819-2
Kaewpitoon, N., Kaewpitoon, S. J., and Pengsaa, P. (2008). Opisthorchiasis in Thailand: review and current status. World J. Gastroenterology 14 (15), 2297–2302. doi:10.3748/wjg.14.2297
Kafle, A., and Suttiprapa, S. (2024). Current state of knowledge on blood and tissue-based biomarkers for Opisthorchis viverrini-induced cholangiocarcinoma: a review of prognostic, predictive, and diagnostic markers. Asian Pac J. Cancer Prev. 25 (1), 25–41. doi:10.31557/apjcp.2024.25.1.25
Kalita, J., Padhi, A. K., and Tripathi, T. (2020). Designing a vaccine for fascioliasis using immunogenic 24 kDa mu-class glutathione s-transferase. Infect. Genet. Evol. 83, 104352. doi:10.1016/j.meegid.2020.104352
Kapuganti, S. K., Bhardwaj, A., Nayak, N., Bhardwaj, T., Kumar, P., and Giri, R. (2022). “Vaccine Omics: role of bioinformatics in vaccinology,” in System vaccinology (Elsevier), 33–54.
Kaur, R., Arora, N., Jamakhani, M. A., Malik, S., Kumar, P., Anjum, F., et al. (2020). Development of multi-epitope chimeric vaccine against Taenia solium by exploring its proteome: an in silico approach. Expert Rev. Vaccines 19 (1), 105–114. doi:10.1080/14760584.2019.1711057
Khan, M. S., Khan, I. M., Ahmad, S. U., Rahman, I., Khan, M. Z., Khan, M. S. Z., et al. (2022). Immunoinformatics design of B and T-cell epitope-based SARS-CoV-2 peptide vaccination. Front. Immunol. 13, 1001430. doi:10.3389/fimmu.2022.1001430
Kokova, D., Verhoeven, A., Perina, E. A., Ivanov, V. V., Knyazeva, E. M., Saltykova, I. V., et al. (2020). Plasma metabolomics of the time resolved response to Opisthorchis felineus infection in an animal model (golden hamster, Mesocricetus auratus). PLoS Negl. Trop. Dis. 14 (1), e0008015. doi:10.1371/journal.pntd.0008015
Laha, T., Pinlaor, P., Mulvenna, J., Sripa, B., Sripa, M., Smout, M. J., et al. (2007). Gene discovery for the carcinogenic human liver fluke, Opisthorchis viverrini. BMC Genomics 8 (1), 189. doi:10.1186/1471-2164-8-189
Liau, M. Y. Q., Toh, E. Q., and Shelat, V. G. (2023). Opisthorchis viverrini-current understanding of the neglected hepatobiliary parasite. Hepatobiliary Parasite 12 (6), 795. doi:10.3390/pathogens12060795
Liu, X., Feng, Y., Bai, X., Wang, X., Qin, R., Tang, B., et al. (2021). Comparative multi-omics analyses reveal differential expression of key genes relevant for parasitism between non-encapsulated and encapsulated Trichinella. Commun. Biol. 4 (1), 134. doi:10.1038/s42003-021-01650-z
Loukas, A., Tran, M., and Pearson, M. S. (2007). Schistosome membrane proteins as vaccines. Int. J. Parasitol. 37 (3-4), 257–263. doi:10.1016/j.ijpara.2006.12.001
Madanagopal, P., Muthusamy, S., Pradhan, S. N., and Prince, P. R. (2023). Construction and validation of a multi-epitope in silico vaccine model for lymphatic filariasis by targeting Brugia malayi: a reverse vaccinology approach. Bull. Natl. Res. Centre 47 (1), 47. doi:10.1186/s42269-023-01013-0
Maizels, R. M. (2021). Identifying novel candidates and configurations for human helminth vaccines. Expert Rev. Vaccines 20 (11), 1389–1393. doi:10.1080/14760584.2021.1999810
María, R., Arturo, C., Alicia, J. A., Paulina, M., and Gerardo, A. O. (2017). The impact of bioinformatics on vaccine design and development. Vaccines 2, 3–6. doi:10.5772/intechopen.69273
Martinelli, D. D. (2022) In silico vaccine design: a tutorial in immunoinformatics. Healthcare Analytics 2, 100044. doi:10.1016/j.health.2022.100044
McManus, D. P. (2020). Recent progress in the development of liver fluke and blood fluke vaccines. Vaccines (Basel) 8 (3), 553. doi:10.3390/vaccines8030553
Mitreva, M. (2022). Parasite OMICS, the grand challenges ahead. Front. Parasitol. 1, 995302. doi:10.3389/fpara.2022.995302
Molehin, A. J., McManus, D. P, and You, H. (2022). Vaccines for human schistosomiasis: recent progress, new developments and future prospects. Int. J. Mol. Sci. 23 (4), 2255–263. doi:10.3390/ijms23042255
Mulvenna, J., Moertel, L., Jones, M. K., Nawaratna, S., Lovas, E. M., Gobert, G. N., et al. (2010). Exposed proteins of the Schistosoma japonicum tegument. Int. J. Parasitol. 40 (5), 543–554. doi:10.1016/j.ijpara.2009.10.002
Nourmohammadi, H., Javanmardi, E., Shams, M., Shamsinia, S., Nosrati, M. C., Yousefi, A., et al. (2020). Multi-epitope vaccine against cystic echinococcosis using immunodominant epitopes from EgA31 and EgG1Y162 antigens. Inf. Med. Unlocked 21, 100464. doi:10.1016/j.imu.2020.100464
Orido, Y. (1990). Development of the excretory bladder of the lung fluke Paragonimus ohirai (Trematoda: troglotrematidae). J. Parasitol. 76, 205–211.
Pandya, N., and Kumar, A. (2023). Immunoinformatics analysis for design of multi-epitope subunit vaccine by using heat shock proteins against Schistosoma mansoni. J. Biomol. Struct. Dyn. 41 (5), 1859–1878. doi:10.1080/07391102.2021.2025430
Pappalardo, F., Russo, G., Tshinanu, F. M., and Viceconti, M. (2019). In silico clinical trials: concepts and early adoptions. Brief. Bioinform 20 (5), 1699–1708. doi:10.1093/bib/bby043
Pedersen, S. K., Sloane, A. J., Prasad, S. S., Sebastian, L. T., Lindner, R. A., Hsu, M., et al. (2005). An immunoproteomic approach for identification of clinical biomarkers for monitoring disease: application to cystic fibrosis. Mol. Cell. Proteomics 4 (8), 1052–1060. doi:10.1074/mcp.M400175-MCP200
Pérez-Sánchez, R., Valero, M. L., Ramajo-Hernández, A., Siles-Lucas, M., Ramajo-Martín, V., and Oleaga, A. (2008). A proteomic approach to the identification of tegumental proteins of male and female Schistosoma bovis worms. Mol. Biochem. Parasitol. 161 (2), 112–123. doi:10.1016/j.molbiopara.2008.06.011
Pleasance, J., Raadsma, H. W., Estuningsih, S. E., Widjajanti, S., Meeusen, E., and Piedrafita, D. (2011). Innate and adaptive resistance of Indonesian Thin Tail sheep to liver fluke: a comparative analysis of Fasciola gigantica and Fasciola hepatica infection. Vet. Parasitol. 178 (3-4), 264–272. doi:10.1016/j.vetpar.2011.01.037
Pourseif, M. M., Yousefpour, M., Aminianfar, M., Moghaddam, G., and Nematollahi, A. (2019). A multi-method and structure-based in silico vaccine designing against Echinococcus granulosus through investigating enolase protein. Bioimpacts 9 (3), 131–144. doi:10.15171/bi.2019.18
Prasopdee, S., Thitapakorn, V., Sathavornmanee, T., and Tesana, S. (2019). A comprehensive review of omics and host-parasite interplays studies, towards control of Opisthorchis viverrini infection for prevention of cholangiocarcinoma. Acta Trop. 196, 76–82. doi:10.1016/j.actatropica.2019.05.011
Rauch, S., Jasny, E., Schmidt, K. E., and Petsch, B. J. F. (2018). New vaccine technologies to combat outbreak situations. Front. Immunol. 9, 1963. doi:10.3389/fimmu.2018.01963
Rehman, A., Ahmad, S., Shahid, F., Albutti, A., Alwashmi, A. S. S., Aljasir, M. A., et al. (2021). Integrated core proteomics, subtractive proteomics, and immunoinformatics investigation to unveil a potential multi-epitope vaccine against schistosomiasis. Vaccines (Basel). 9 (6), 658. doi:10.3390/vaccines9060658
Retra, K., deWalick, S., Schmitz, M., Yazdanbakhsh, M., Tielens, A. G., Brouwers, J. F., et al. (2015). The tegumental surface membranes of Schistosoma mansoni are enriched in parasite-specific phospholipid species. Int. J. Parasitol. 45 (9-10), 629–636. doi:10.1016/j.ijpara.2015.03.011
Rodrigues, M. M., and Ersching, J. (2014). Neglected tropical diseases, bioinformatics, and vaccines. J. Infect. Dis. 211 (2), 175–177. doi:10.1093/infdis/jiu420
Sahoo, R., Pattanaik, S., Pattnaik, G., Mallick, S., and Mohapatra, R. (2022). Review on the use of molecular docking as the first line tool in drug discovery and development. Indian J. Pharm. Sci. 84 (5). doi:10.36468/pharmaceutical-sciences.1031
Saldanha, L., Langel, Ü., and Vale, N. (2023). In silico studies to support vaccine development. Pharmaceutics 15 (2), 654. doi:10.3390/pharmaceutics15020654
Sanches, R. C. O., Tiwari, S., Ferreira, L. C. G., Oliveira, F. M., Lopes, M. D., Passos, M. J. F., et al. (2021). Immunoinformatics design of multi-epitope peptide-based vaccine against schistosoma mansoni using transmembrane proteins as a target. Front. Immunol. 12, 621706. doi:10.3389/fimmu.2021.621706
Shams, M., Javanmardi, E., Nosrati, M. C., Ghasemi, E., Shamsinia, S., Yousefi, A., et al. (2021). Bioinformatics features and immunogenic epitopes of Echinococcus granulosus Myophilin as a promising target for vaccination against cystic echinococcosis. Infect. Genet. Evol. 89, 104714. doi:10.1016/j.meegid.2021.104714
Shawan, M. M. A. K., Sharma, A. R., Halder, S. K., Arian, T. A., Shuvo, M. N., Sarker, S. R., et al. (2023). Advances in computational and bioinformatics tools and databases for designing and developing a multi-epitope-based peptide vaccine. Int. J. Peptide Res. Ther. 29 (4), 60. doi:10.1007/s10989-023-10535-0
Shi, Y., Yue, T., Zhang, Y., Wei, J., and Yuan, Y. (2018). Surface immunoproteomics reveals potential biomarkers in alicyclobacillus acidoterrestris. Front. Microbiol. 9, 3032. doi:10.3389/fmicb.2018.03032
Soleymani, S., Tavassoli, A., and Housaindokht, M. R. (2022). An overview of progress from empirical to rational design in modern vaccine development, with an emphasis on computational tools and immunoinformatics approaches. Comput. Biol. Med. 140, 105057. doi:10.1016/j.compbiomed.2021.105057
Songserm, N., Prasongwattana, J., Sithithaworn, P., Sripa, B., and Pipitkool, V. (2009). Cholangiocarcinoma in experimental hamsters with long-standing Opisthorchis viverrini infection. Asian Pac. J. Cancer Prev. 10 (2), 299–302.
Sripa, B. (2003). Pathobiology of opisthorchiasis: an update. Acta Trop. 88 (3), 209–220. doi:10.1016/j.actatropica.2003.08.002
Sripa, B., Brindley, P. J., Mulvenna, J., Laha, T., Smout, M. J., Mairiang, E., et al. (2012). The tumorigenic liver fluke Opisthorchis viverrini--multiple pathways to cancer. Trends Parasitol. 28 (10), 395–407. doi:10.1016/j.pt.2012.07.006
Sripa, B., Jumnainsong, A., Tangkawattana, S., and Haswell, M. R. (2018). Immune response to Opisthorchis viverrini infection and its role in pathology. Adv. Parasitol. 102, 73–95. doi:10.1016/bs.apar.2018.08.003
Sripa, B., Kaewkes, S., Sithithaworn, P., Mairiang, E., Laha, T., Smout, M., et al. (2007). Liver fluke induces cholangiocarcinoma. PLoS Med. 4 (7), e201. doi:10.1371/journal.pmed.0040201
Sripa, B., Suwannatrai, A. T., Sayasone, S., Do, D. T., Khieu, V., and Yang, Y. (2021). Current status of human liver fluke infections in the Greater Mekong Subregion. Acta Trop. 224, 106133. doi:10.1016/j.actatropica.2021.106133
Stevens, A. O., and He, Y. (2022). Benchmarking the accuracy of AlphaFold 2 in loop structure prediction. Biomolecules 12 (7), 985. doi:10.3390/biom12070985
Suttiprapa, S., Sotillo, J., Smout, M., Suyapoh, W., Chaiyadet, S., Tripathi, T., et al. (2018). Opisthorchis viverrini proteome and host-parasite interactions. in Adv. Parasitol., advances in parasitology. Editors B. Sripa, and P. J. Brindley (London: Academic Press), 45–72). doi:10.1016/bs.apar.2018.06.002
Talabnin, K., Aoki, K., Saichua, P., Wongkham, S., Kaewkes, S., Boons, G. J., et al. (2013). Stage-specific expression and antigenicity of glycoprotein glycans isolated from the human liver fluke, Opisthorchis viverrini. Int. J. Parasitol. 43 (1), 37–50. doi:10.1016/j.ijpara.2012.10.013
Thompson, D. P., and Geary, T. G. (2003). “Chapter 13 - helminth surfaces: structural, molecular and functional properties,” in Molecular medical parasitology. Editors J. J. Marr, T. W. Nilsen, and R. W. Komuniecki (London: Academic Press), 297–338.
Tielens, A. G. M., Van Den Heuvel, J. M., and Van Den Bergh, S. G. (1982). Changes in energy metabolism of the juvenile Fasciola hepatica during its development in the liver parenchyma. Mol. Biochem. Parasitol. 6 (5), 277–286. doi:10.1016/0166-6851(82)90060-3
Trelis, M., Sánchez-López, C. M., Sánchez-Palencia, L. F., Ramírez-Toledo, V., Marcilla, A., and Bernal, D. (2022). Proteomic analysis of extracellular vesicles from fasciola hepatica hatching eggs and juveniles in culture. Front. Cell Infect. Microbiol. 12, 903602. doi:10.3389/fcimb.2022.903602
Vahabi, N., and Michailidis, G. (2022). Unsupervised multi-omics data integration methods: a comprehensive review. Front. Genet. 13, 854752. doi:10.3389/fgene.2022.854752
van der Ree, A. M., and Mutapi, F. (2015). The helminth parasite proteome at the host-parasite interface - informing diagnosis and control. Exp. Parasitol. 157, 48–58. doi:10.1016/j.exppara.2015.06.007
van Die, I., and Cummings, R. D. (2009). Glycan gimmickry by parasitic helminths: a strategy for modulating the host immune response? Glycobiology 20 (1), 2–12. doi:10.1093/glycob/cwp140
Wang, T., Van Steendam, K., Dhaenens, M., Vlaminck, J., Deforce, D., Jex, A. R., et al. (2013). Proteomic analysis of the excretory-secretory products from larval stages of Ascaris suum reveals high abundance of glycosyl hydrolases. PLoS Negl. Trop. Dis. 7 (10), e2467. doi:10.1371/journal.pntd.0002467
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi:10.1385/1-59259-584-7:531
Winkelmann, F., Gesell Salazar, M., Hentschker, C., Michalik, S., Macháček, T., Scharf, C., et al. (2022). Comparative proteome analysis of the tegument of male and female adult Schistosoma mansoni. Sci. Rep. 12 (1), 7569. doi:10.1038/s41598-022-11645-3
Winkler, D. A. (2021). Use of artificial intelligence and machine learning for discovery of drugs for neglected tropical diseases. Front. Chem. 9, 614073. doi:10.3389/fchem.2021.614073
Wongratanacheewin, S., Chawengkirttikul, R., Bunnag, D., and Sirisinha, S. (1988). Analysis of Opisthorchis viverrini antigens by immunoprecipitation and polyacrylamide gel electrophoresis. Parasitology 96 (1), 119–128. doi:10.1017/S0031182000081701
World Health Organization (1994). Schistosomes, Liver Flukes and Helicobacter pylori IARC monographs on the evaluation of carcinogenic risks to humans volume 61. IARC, 1–241.
World Health Organization (2020). Ending the neglect to attain the Sustainable Development Goals: a road map for neglected tropical diseases 2021–2030. World Health Organization.
Yang, Z., Zeng, X., Zhao, Y., and Chen, R. (2023). AlphaFold2 and its applications in the fields of biology and medicine. Signal Transduct. Target. Ther. 8 (1), 115. doi:10.1038/s41392-023-01381-z
Yeshi, K., Ruscher, R., Loukas, A., and Wangchuk, P. (2022). Immunomodulatory and biological properties of helminth-derived small molecules: potential applications in diagnostics and therapeutics. Front. Parasitol. 1, 984152. doi:10.3389/fpara.2022.984152
Young, N. D., Nagarajan, N., Lin, S. J., Korhonen, P. K., Jex, A. R., Hall, R. S., et al. (2014). The Opisthorchis viverrini genome provides insights into life in the bile duct. Nat. Commun. 5, 4378. doi:10.1038/ncomms5378
Zawistowska-Deniziak, A., Powązka, K., Pękacz, M., Basałaj, K., Klockiewicz, M., Wiśniewski, M., et al. (2021). Immunoproteomic analysis of dirofilaria repens microfilariae and adult parasite stages. Pathogens 10 (2), 174. doi:10.3390/pathogens10020174
Zhang, M., Fu, Z., Li, C., Han, Y., Cao, X., Han, H., et al. (2015). Screening diagnostic candidates for schistosomiasis from tegument proteins of adult Schistosoma japonicum using an immunoproteomic approach. PLoS Negl. Trop. Dis. 9 (2), e0003454. doi:10.1371/journal.pntd.0003454
Zhang, M., Hong, Y., Han, Y., Han, H., Peng, J., Qiu, C., et al. (2013). Proteomic analysis of tegument-exposed proteins of female and male Schistosoma japonicum worms. J. Proteome Res. 12 (11), 5260–5270. doi:10.1021/pr400476a
Keywords: opisthorchiasis, liver fluke, host-parasite, proteomics, immunoproteomics, immunoinformatics, vaccinology
Citation: Kafle A and Ojha SC (2024) Advancing vaccine development against Opisthorchis viverrini: A synergistic integration of omics technologies and advanced computational tools. Front. Pharmacol. 15:1410453. doi: 10.3389/fphar.2024.1410453
Received: 01 April 2024; Accepted: 10 June 2024;
Published: 15 July 2024.
Edited by:
Chandra Sekhar Sirka, All India Institute of Medical Sciences Bhubaneswar, IndiaReviewed by:
Arijit Bhattacharya, Adamas University, IndiaCopyright © 2024 Kafle and Ojha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suvash Chandra Ojha, c3V2YXNoX29qaGFAc3dtdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.