- 1Centre for Life Sciences, Mahindra University, Hyderabad, India
- 2Interdisciplinary Centre for Nanosensors and Nanomedicine, Mahindra University, Hyderabad, India
ATP-binding cassette transporters represent a superfamily of dynamic membrane-based proteins with diverse yet common functions such as use of ATP hydrolysis to efflux substrates across cellular membranes. Three major transporters—P-glycoprotein (P-gp or ABCB1), multidrug resistance protein 1 (MRP1 or ABCC1), and breast cancer resistance protein (BCRP or ABCG2) are notoriously involved in therapy resistance in cancer patients. Despite exhaustive individual characterizations of each of these transporters, there is a lack of understanding in terms of the functional role of mutations in substrate binding and efflux, leading to drug resistance. We analyzed clinical variations reported in endometrial cancers for these transporters. For ABCB1, the majority of key mutations were present in the membrane-facing region, followed by the drug transport channel and ATP-binding regions. Similarly, for ABCG2, the majority of key mutations were located in the membrane-facing region, followed by the ATP-binding region and drug transport channel, thus highlighting the importance of membrane-mediated drug recruitment and efflux in ABCB1 and ABCG2. On the other hand, for ABCC1, the majority of key mutations were present in the inactive nucleotide-binding domain, followed by the drug transport channel and membrane-facing regions, highlighting the importance of the inactive nucleotide-binding domain in facilitating indirect drug efflux in ABCC1. The identified key mutations in endometrial cancer and mapped common mutations present across different types of cancers in ABCB1, ABCC1, and ABCG2 will facilitate the design and discovery of inhibitors targeting unexplored structural regions of these transporters and re-engineering of these transporters to tackle chemoresistance.
1 Introduction
Despite tremendous advancements in the treatment of clinical cancer via chemotherapy, radiotherapy, immunotherapy, etc., the development of therapy resistance remains a major challenge and concern for clinicians and researchers alike. The most common mechanisms of resistance include drug efflux, DNA repair, alteration of drug targets, inactivation or sequestration of drug candidates, apoptosis blockage, and genetic/epigenetic alterations across a wide spectrum of anticancer agents (Sajid et al., 2023). Resistance toward anticancer drugs can be classified as either intrinsic or extrinsic, implying that the factors facilitating resistance in cells were present before or after drugs were administered. Of these resistance mechanisms, the ability of cancer cells to identify and selectively efflux a wide spectrum of mechanistically and structurally unrelated drugs is conferred by ATP-binding cassette (ABC) transporters (Juan-Carlos et al., 2021), which can occur due to either intrinsic or extrinsic factors of cancer cells.
ABC transporters are a superfamily of integral membrane proteins with a tissue-specific expression and are responsible for the transfer of many biological substrates across the cell membrane (Robey et al., 2018). In addition to niche biological functions such as export of metabolic end-products and cholesterol in the liver (Wlcek and Stieger, 2014), ABC transporters are involved in efflux of a wide range of chemotherapeutic agents. Consequently, this has serious clinical implications on therapeutic success in patients undergoing chemotherapy.
ABC transporters mainly consist of four domains—two nucleotide-binding domains (NBDs) and two transmembrane domains (TMDs). It can act as both an exporter and importer or may not participate in any exchange at all depending on its location in the body. Usually, ATP or nucleotide binding to NBD causes conformational changes in such a manner that the substrate molecule is captured in its cavity and released on the other side. (Rees et al., 2009; Vasiliou et al., 2009; Tarling et al., 2013; Alam et al., 2019; Di et al., 2022). Of the 48 classified ABC transporters, the three most widely studied and characterized efflux glycoproteins, namely, P-glycoprotein (also known as Pgp or ABCB1), multidrug resistance protein 1 (also known as MRP1 or ABCC1), and breast cancer resistance protein (BCRP) (also known as BCRP or ABCG2), have significant roles in causing multidrug resistance in cancer. Although each of the transporters has been extensively studied and characterized individually, there are gaps in the understanding and comparative analysis of clinically significant mutations at the structural level and its possible implications in chemoresistance.
The NBD regions of ABC transporters are the motor domains where ATP and nucleotide binding occurs. It has already been studied that the ATP-binding regions of these ABC transporters are highly conserved. They consist of several conserved motifs that are involved in ATP binding, ATP hydrolysis, and in facilitating important interfaces in active ABC transporters (Walker et al., 1982; Jones and George, 2004; Hollenstein et al., 2007).
These motifs include the “Walker A motif” also known as p-loop (represented by G-X-X-G-X-G-K-S/T, where X represents any amino acid and common among nucleotide-binding proteins), the core fold of which is characterized by centrally placed parallel beta-sheets flanked by helices, and this motif is responsible for the binding of nucleotides and is essential for ATP hydrolysis; the “LSGGQ motif,” also known to be the “C-loop” or “signature motif” for ABC transporters, interacts with the nucleotide in the ATP-bound state and is essential in ATP hydrolysis as well along with the p-loop; the “Walker-B motif” (represented by h-h-h-h-D, where h signifies hydrophobic residues) offers a conserved glutamate residue that is responsible for the nucleophilic attack on ATP using a water molecule; the “A-loop” consisting of stacked aromatic residues is hypothesized to form a p–p interaction with the bound ATP’s adenine ring; the “Q-loop” is hypothesized to detect the g-phosphate moiety and hence has a role in the contact interface with the transmembrane domain; the “D-loop” is thought to be involved in an interaction between the two NBD regions; and the “switch motif” containing a histidine is said to be involved in a catalytic reaction that takes place in the NBD (Oldham et al., 2008; Zolnerciks et al., 2011; Wilkens, 2015).
Since the NBDs are arranged in a head-to-toe manner, the two nucleotide molecules bound in the NBD region tend to interact with Walker A and Walker B motifs from one NBD and the LSGGQ motif from the other NBD. This arrangement leads to the formation of two ATP-binding sites between Walker A of one NBD and the LSGGQ motif of the other, proving cooperativity in ATP binding and its catalytic activity. In the bound state, two ATP molecules are sandwiched at these interfaces (Walker et al., 1982; Hollenstein et al., 2007).
ABC transporters play a significant role in reproduction. They modulate steroidogenesis, fertilization, fetus implantation, and play a crucial role as a barrier during pregnancy to protect the fetus against the onslaught of harmful xenobiotics, drugs, and environmental toxins (Bloise et al., 2015). Endometrial tissue is known to express all three major ABC transporters—ABCB1, ABCC1, and ABCG2, due to its direct involvement during peri-implantation leading up to placental/fetal development. Hence, in case of endometrial uterine cancer, intrinsic overexpression of all three ABC transporters interferes throughout single-/combination chemotherapy (Arain et al., 2021; Takagi et al., 2023). The response rate in chemotherapy-naïve endometrial carcinoma patients to single-agent chemotherapy (either of doxorubicin, paclitaxel, cisplatin, or carboplatin) is not good and is reportedly below 40% (Moxley and McMeekin, 2010). Furthermore, in case of relapse, there is limited, if any, established second-line chemotherapy/agent to administer to women with an MDR endometrial cancer profile.
In this study, we have focused on endometrial cancer due to the abundance of mutation data, which were relatively higher than those of other cancer types, as shown in Figure 1, Figure 2, and Figure 3 for ABCG2, ABCB1, and ABCC1, respectively, taken from cBioPortal for cancer genomics (Gao et al., 2013). The data are part of TCGA PanCancer data for uterine corpus endometrial carcinoma. Out of a total of 529 cases, the ABCB1, ABCC1, and ABCG2 transporters were found to be mutated in 10.96%, 9.45%, and 6.62% of the cases, respectively (Berger et al., 2018; Hoadley et al., 2018).
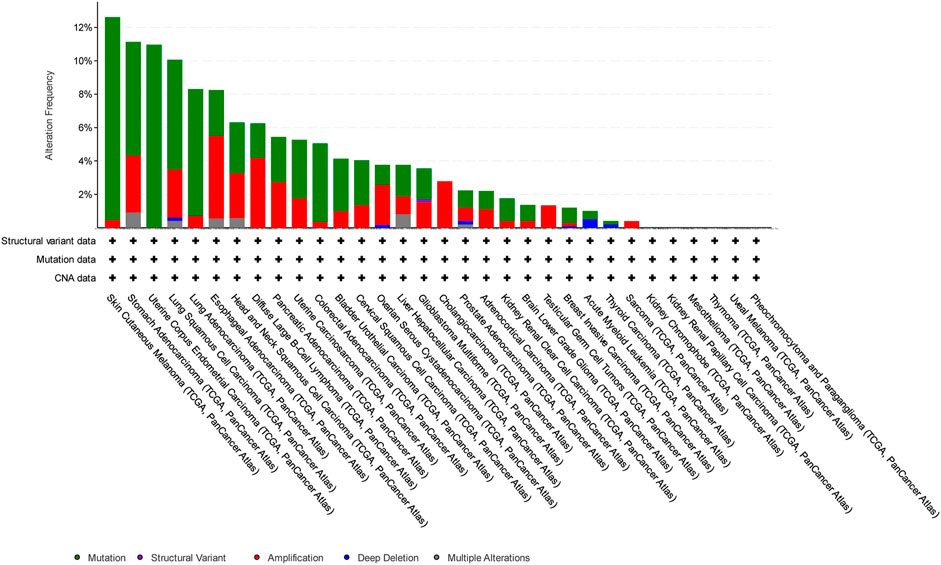
Figure 1. Overview of the frequency of clinical variations observed in ABCB1 in different cancers. The uterine corpus endometrial carcinoma was among the top two cancers with respect to mutation frequency for ABCB1.
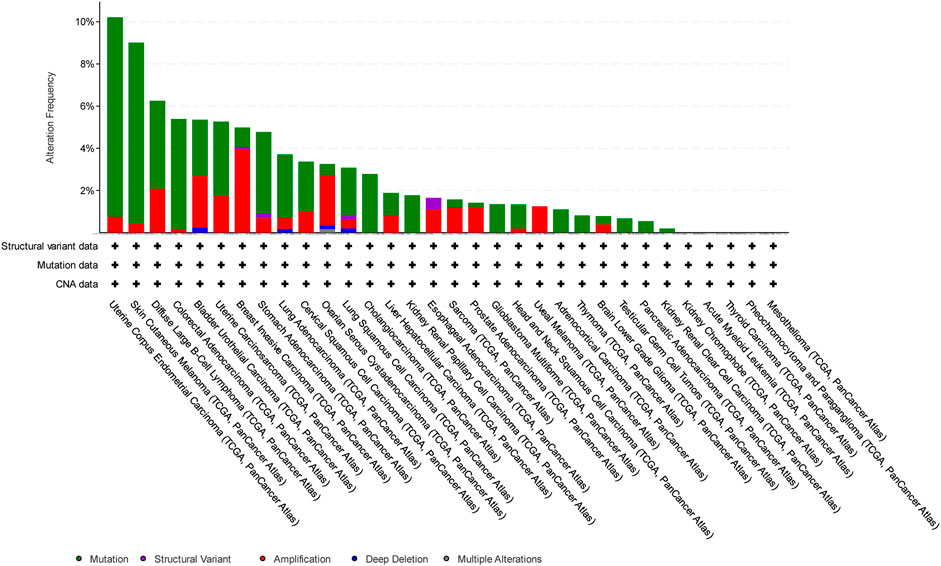
Figure 2. Overview of the frequency of clinical variations observed in ABCC1 in different cancers. The uterine corpus endometrial carcinoma shows the highest mutation frequency for ABCC1.
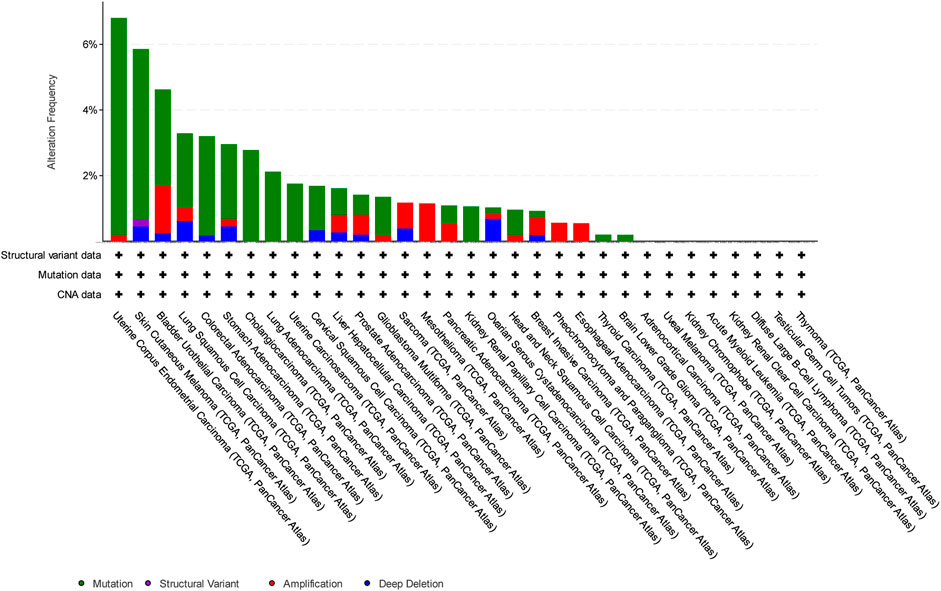
Figure 3. Overview of the frequency of clinical variations observed in ABCG2 in different cancers. The uterine corpus endometrial carcinoma shows the highest mutation frequency for ABCG2.
Mitoxantrone is a topoisomerase type IIα inhibitor, extensively used for the treatment of acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), and advanced prostate cancer, besides other conditions. The mechanism of action in cancer cells involves disruption of DNA synthesis and prevention of re-ligation of broken DNA strands, thereby impairing DNA repair function. For this work, we have taken mitoxantrone as the model substrate as it is known to confer resistance and lead to upregulation of all the three ABC transporters of interest: ABCG2, ABCB1, and ABCC1 (Nieth and Lage, 2005). Stefan et al. curated a list of functional tracers based on extensive literature search of experimental data. Of the various molecules, only Calcein AM, Rhodamine-123, and mitoxantrone were used to evaluate potential inhibitors against all three transporters—ABCB1, ABCC1, and ABCG2. Out of these compounds, since only mitoxantrone had both functions—as a tracer and chemotherapeutic agent, therefore we decided to perform molecular docking with mitoxantrone for all the three transporters (Stefan et al., 2022). Mutagenesis studies in the drug-binding pockets across ABC-transporter variants have demonstrated the presence of multiple overlapping sites where drug or inhibitors can bind, referred to as polyspecificity (Sajid et al., 2023). This leads to multidrug resistance, and hence, it is imperative to evaluate the common binding points, which can aid in better designing of therapeutics (small-molecule inhibitors/peptides, etc.) which can evade efflux by ABC transporters.
2 Materials and methods
2.1 Data collection and pre-processing
Mutation data for this study were collected from cBioPortal (Cerami et al., 2012) developed at Memorial Sloan Kettering Cancer Center (MSKCC) which hosts data from The Cancer Genome Atlas (TCGA) (Weinstein et al., 2013), National Institute of Health (NIH). TCGA catalogs the genetic mutations responsible for different cancers using genome sequencing. TCGA was supervised by the National Cancer Institute’s Center for Cancer Genomics and the National Human Genome Research Institute. Out of all the listed mutations in ABCB1, ABCC1, and ABCG2 for endometrial cancer, only mutations supported by at least two mutation functional impact prediction methods (Mutation Assessor, SIFT and PolyPhen-2) for their deleterious or damaging impact were selected (Ng and Henikoff, 2001; Adzhubei et al., 2010; Reva et al., 2011) (Supplementary Material).
2.2 Molecular docking and visualization
The three-dimensional structures for the ABCB1, ABCC1, and ABCG2 transporters were taken from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank. (Gao et al., 2013; Kowal et al., 2021; de Bruijn et al., 2023). PDB files for docking were taken from the RCSB with ID 7NFD(31) for ABCG2, 7A69 (Nosol et al., 2020) for ABCB1, and 6UY0 (Wang et al., 2020) for ABCC1. The mitoxantrone structure was taken from ChEMBL (Gaulton et al., 2012; Zdrazil et al., 2024). The list of residues that are known to interact with mitoxantrone or other drug molecules for ABCG2, ABCB1, and ABCC1 was used during molecular docking. Molecular docking was performed between ABC transporters and mitoxantrone individually using LeDock (Liu and Xu, 2019), and the complex structures were visualized for interactions in the 2D mode using LigPlus (Laskowski and Swindells, 2011). Three-dimensional visualization was performed using PyMOL (PyMOL, 2015; Yuan et al., 2017).
2.3 Classification of key residues
All the mutations that were considered for the analysis based on their possible functional impact in ABCB1, ABCC1, and ABCG2 were further classified based on their location in the three-dimensional structure as follows: in the drug binding/transport channel, at the entry or exit, the membrane-facing regions, and the nucleotide- or ATP-binding regions. We have also taken inputs from the annotations provided at the cBioPortal for this purpose (Gao et al., 2013).
3 Results
Mutation data collected from the cBioPortal contained a total of 891 mutations across three ABC transporter sub-families, 480 in ABCB1, 294 in ABCC1, and 117 in ABCG2. Out of these, mutations observed in uterine corpus endometrial carcinoma were taken into account for further analysis, which narrowed down to a total of 248 mutations: 118 in ABCB1, 82 in ABCC1, and 48 in ABCG2.
3.1 Key mutations involved in drug binding
After review of the literature, a list of residues that have been shown to interact with the drugs or substrate molecules was curated for all the three ABC transporters. We found a total of 22 residues in ABCB1, 27 residues in ABCC1, and 28 residues in ABCG2, as shown in the first column of Table 1, Table 2, and Table 3, respectively. These residues were then compared with the TCGA mutation data for any potential matches, wherein we observed six matches in ABCB1 (W232L, A987T, X235_splice, G872V, A230T, and S993A), six matches in ABCC1 (M1092V, G1319R, A438V, V1325M, C1208Y, and V776M), and three matches in ABCG2 (L405M, E446N, and L595I), as shown in the second column of Tables 1–3, respectively. These matches were significant for determining the location of our grid box for molecular docking.
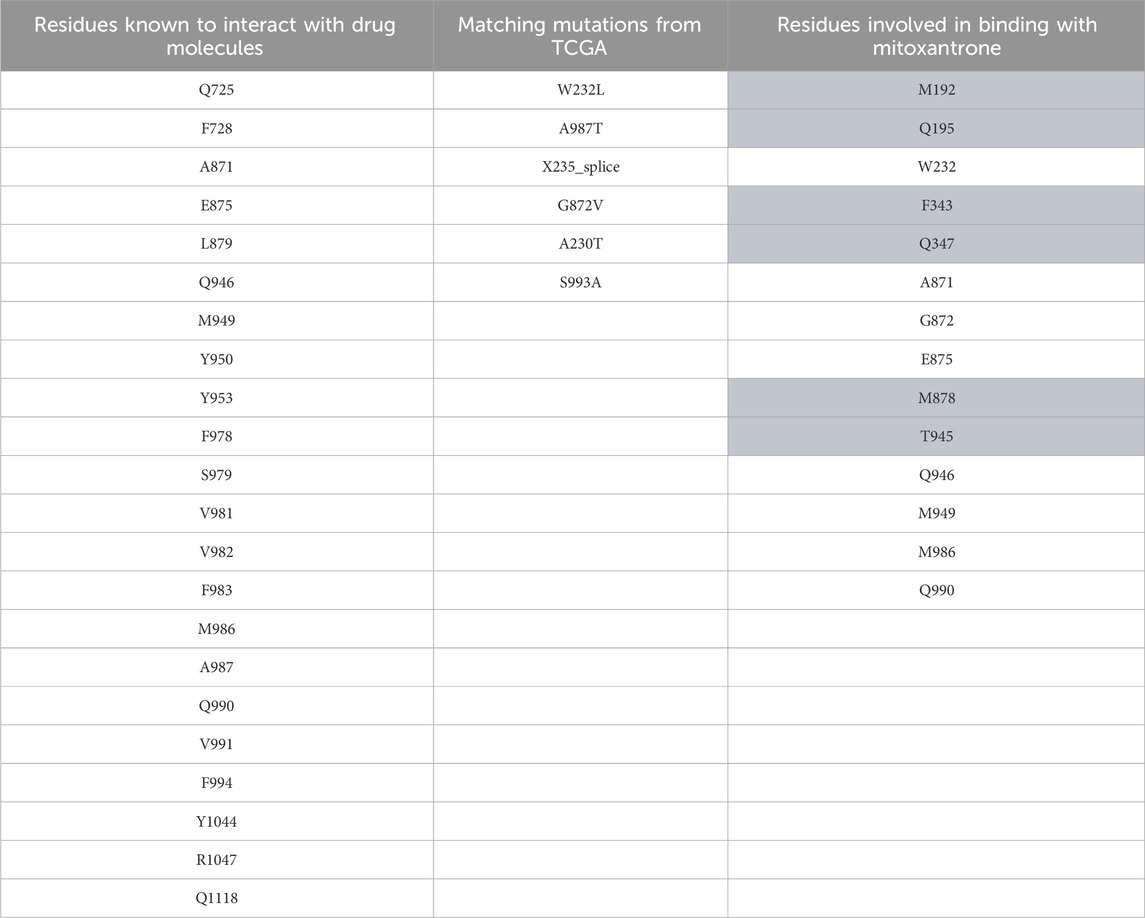
Table 1. ABCB1 mutation data. The first column represents residues known to interact with drug molecules from the literature survey; the second column represents the matches between known residues and TCGA dataset/mutations; and the third column represents the residues found to be interacting after docking of mitoxantrone to ABCB1 using LeDock. The highlighted residues are the novel residues observed to be interacting with mitoxantrone.
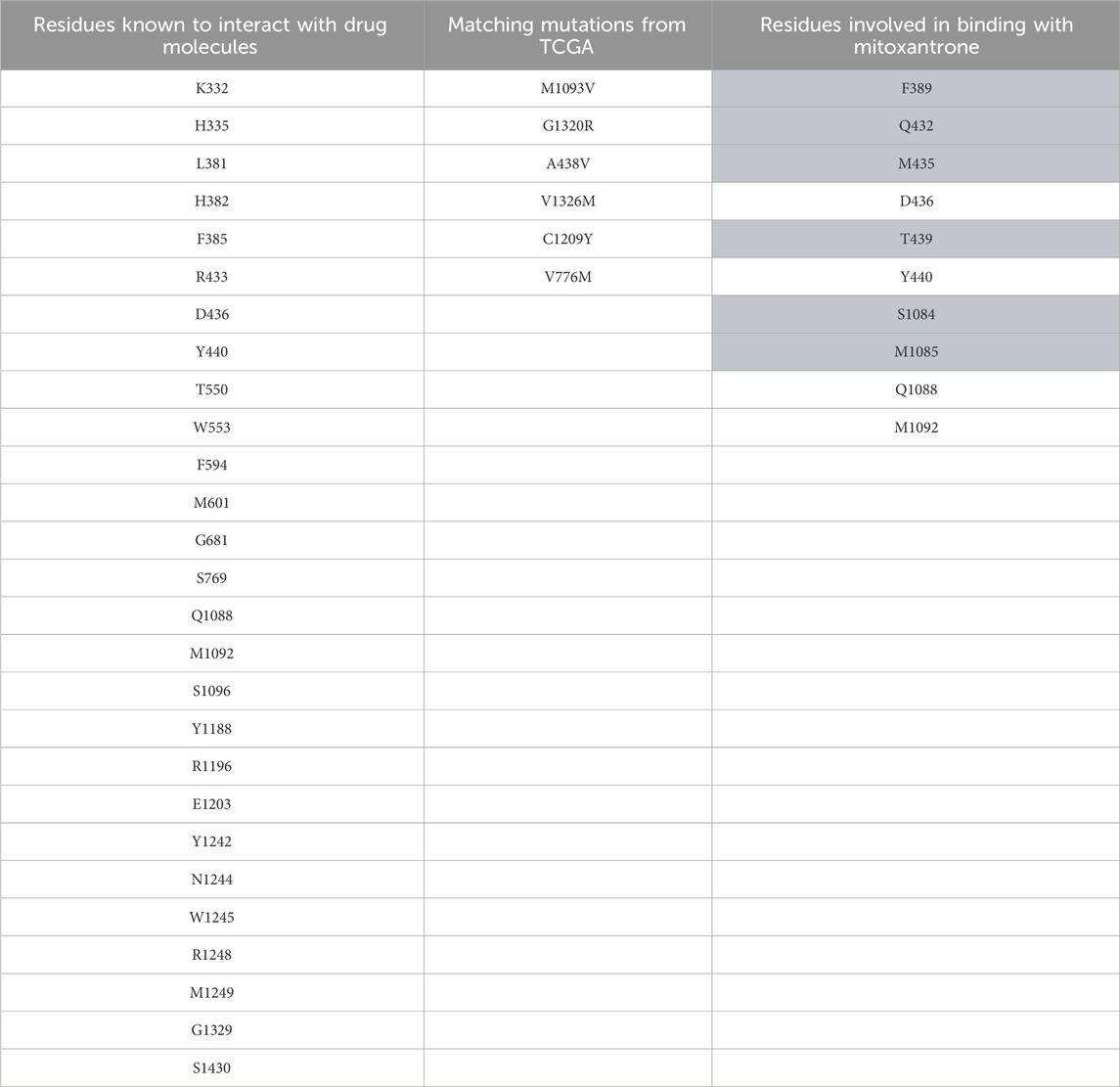
Table 2. ABCC1 mutation data. The first column represents residues known to interact with drug molecules from the literature survey; the second column represents the matches between known residues and TCGA dataset; and the third column represents the residues found to be interacting after docking of mitoxantrone to ABCC1 using LeDock. The highlighted residues are the novel residues observed to be interacting with mitoxantrone.
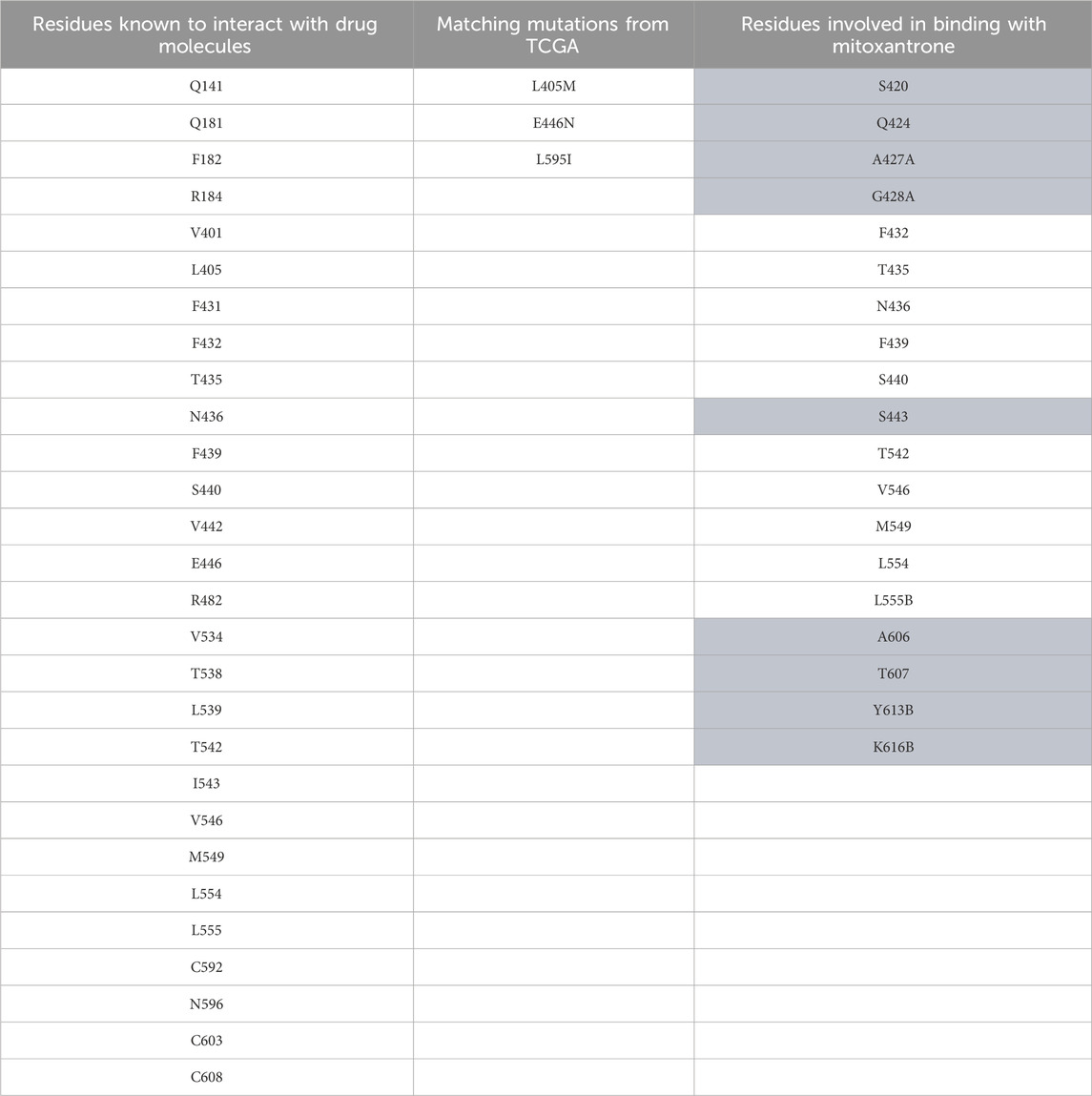
Table 3. ABCG2 mutation data. The first column represents residues known to interact with drug molecules from the literature survey; the second column represents the matches between known residues and TCGA dataset; and the third column represents the residues found to be interacting after docking of mitoxantrone to ABCG2 using LeDock. The highlighted residues are the novel residues observed to be interacting with mitoxantrone.
The PDB structure taken for ABCG2, 7NFD had already mitoxantrone bound to it, making it a reference for our results. The comparison with complex structures generated by LeDock (Liu and Xu, 2019) has shown similar amino acid residues taking part in the molecular interaction. This result allowed us to use the same docking methodology to build all the complex structures.
For structural level interaction studies between ABC transporters and substrate/drug molecules, molecular docking was performed for the selected PDB structures of ABCB1, ABCC1, ABCG2, and mitoxantrone using LeDock (Liu and Xu, 2019). After molecular docking, the complexes were then analyzed for visualizing molecular interactions between mitoxantrone and amino acid residues using a LigPlot (Laskowski and Swindells, 2011)-based two-dimensional interaction diagram (Figure 4; Figure 5; Figure 6).
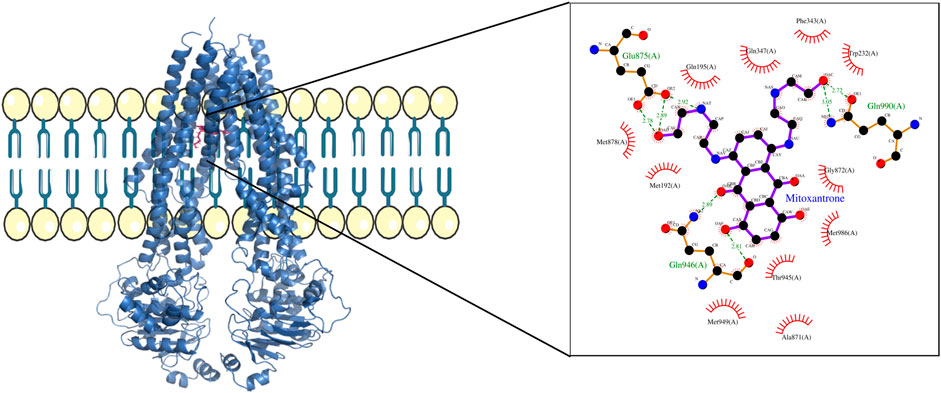
Figure 4. ABCB1–mitoxantrone complex structure highlighting the residues in ABCB1 interacting with mitoxantrone. The molecular interaction representation on the right shows the non-covalent interactions between the key amino acid residues in ABCB1 and mitoxantrone.
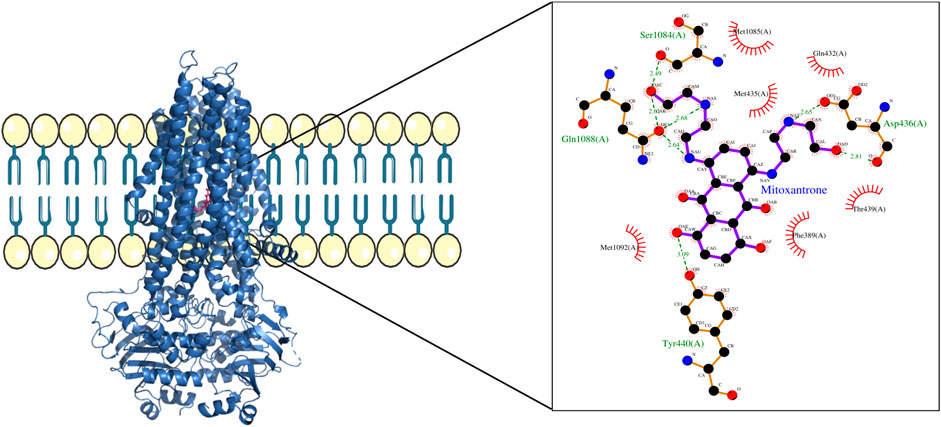
Figure 5. ABCC1–mitoxantrone complex structure highlighting the residues in ABCC1 interacting with mitoxantrone. The molecular interaction representation on the right shows the non-covalent interactions between the key amino acid residues in ABCC1 and mitoxantrone.
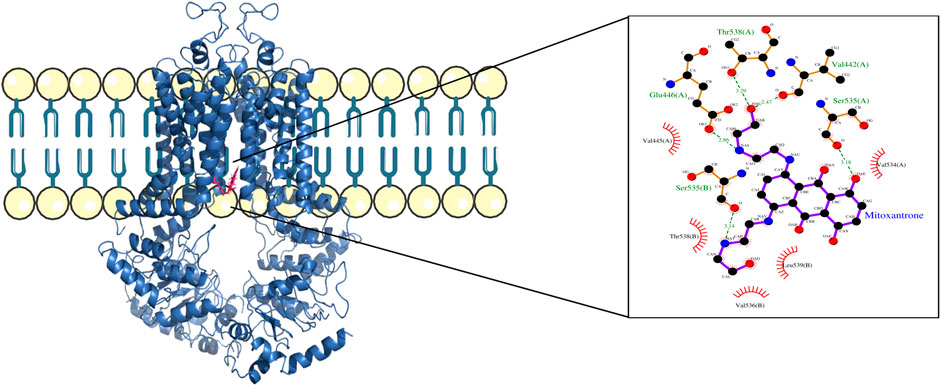
Figure 6. ABCG2–mitoxantrone complex structure highlighting the residues in ABCG2 interacting with mitoxantrone. The molecular interaction representation on the right shows the non-covalent interactions between the key amino acid residues in ABCG2 and mitoxantrone.
In Figure 4, the structure of the complex between ABCB1 and mitoxantrone can be seen. Novel residues found to be interacting with mitoxantrone were M192, Q195, F343, Q347, M878, and T945. All the interacting residues are also listed in the third column of Table 1, and the interactions can be seen in a two-dimensional representation on the right side in Figure 4. Similarly, in Figure 5, the structure of the complex between ABCC1 and mitoxantrone can be seen. Novel residues found to be interacting with mitoxantrone were F389, Q432, M435, T439, S1084, and M1085. All the interacting residues are also listed in the third column of Table 2, and the interactions can be seen in the two-dimensional representation on the right side in Figure 5. Figure 6 shows the structure of the complex formed between ABCG2 and mitoxantrone. The two-dimensional visualization of interactions can be seen on the right side in Figure 6. Out of all the interactions observed, many amino acid residues were similar to the residues already reported to interact with the ABC transporters. However, few novel residues S420, Q424, A427, G428, S443, A606, T607, Y613, and K616 were also shown to interact with mitoxantrone, and these residues along with all the interacting residues that were observed are also listed in the third column of Table 3.
The description of mutations in ABCB1, ABCC1, and ABCG2 that are also present in other different cancers (such as cancers of the brain, lung, colon, prostate, and kidney) in addition to endometrial cancer is provided in Table 4.
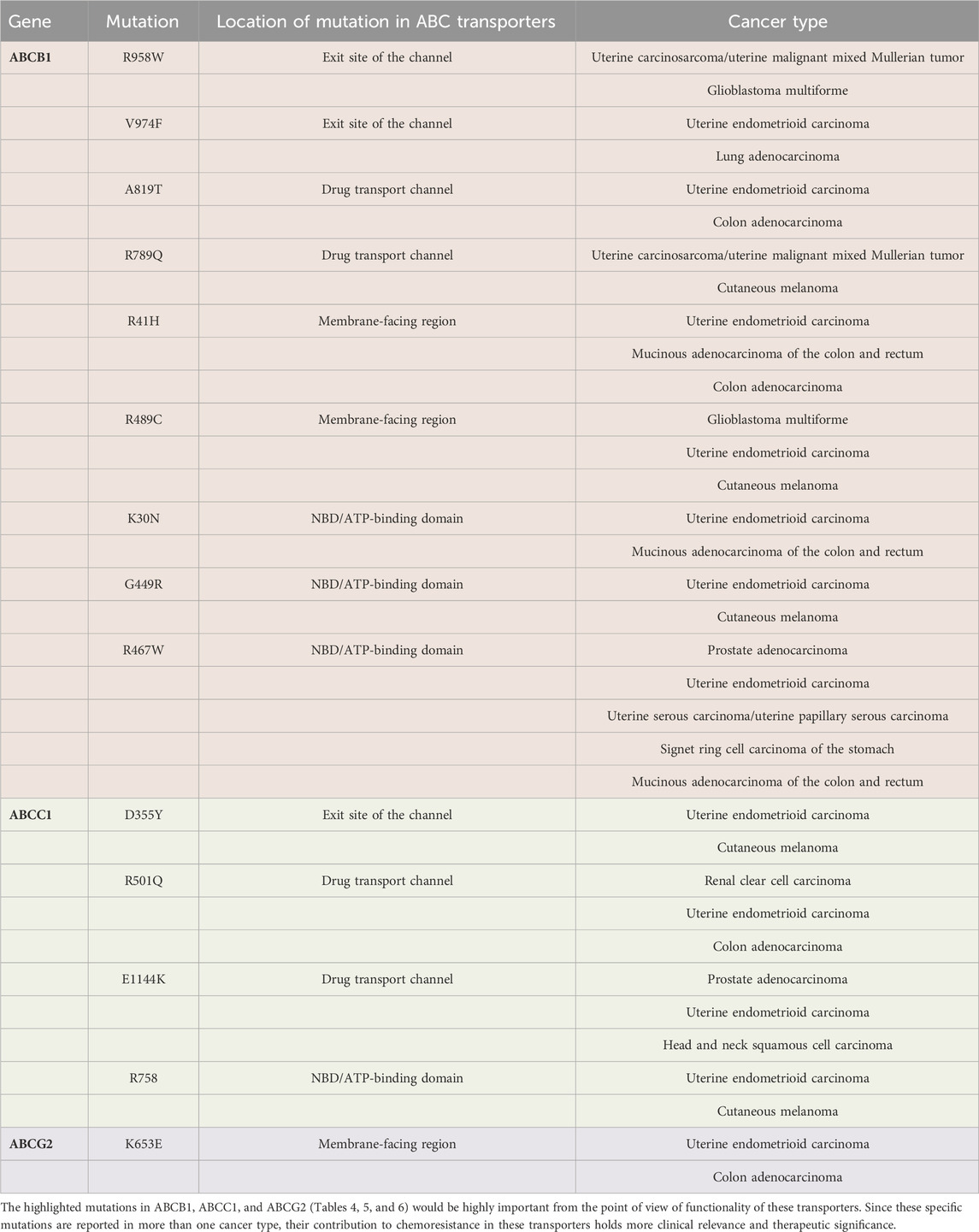
Table 4. Mutations in ABCB1 (nine mutations) are highlighted in pink, ABCC1 (four mutations) are highlighted in green, and ABCG2 (one mutation) is highlighted in purple. These functionally important mutations were present in other different cancers in addition to endometrial cancer. The cancer types for each mutation are listed on the right most column of the table.
3.2 Functional impact of mutations present in ABCB1
We found that three residues D77, V974, and R958 found on the exit site of the channel were shown to be mutated into hydrophobic residues. All these mutations will lead to the increase of drug efflux because it is known that ABCB1 majorly transports hydrophobic and amphipathic compounds, and all these mutations will enhance ABCB1’s capability to transport such compounds that can possibly lead to chemoresistance (Figure 7; Table 5).
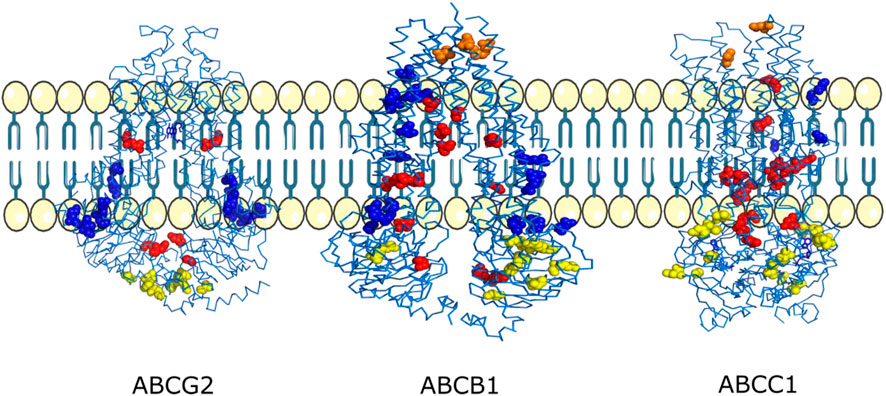
Figure 7. Mutations mapped onto 3D structures of the ABCG2, ABCB1, and ABCC1 transporters. Mutations observed near the exit of the drug transport channel are shown in orange, mutations observed inside the drug transport channel are shown in red, mutations observed on the membrane-facing regions are shown in blue, and mutations observed at the ATP/nucleotide-binding regions are shown in yellow.
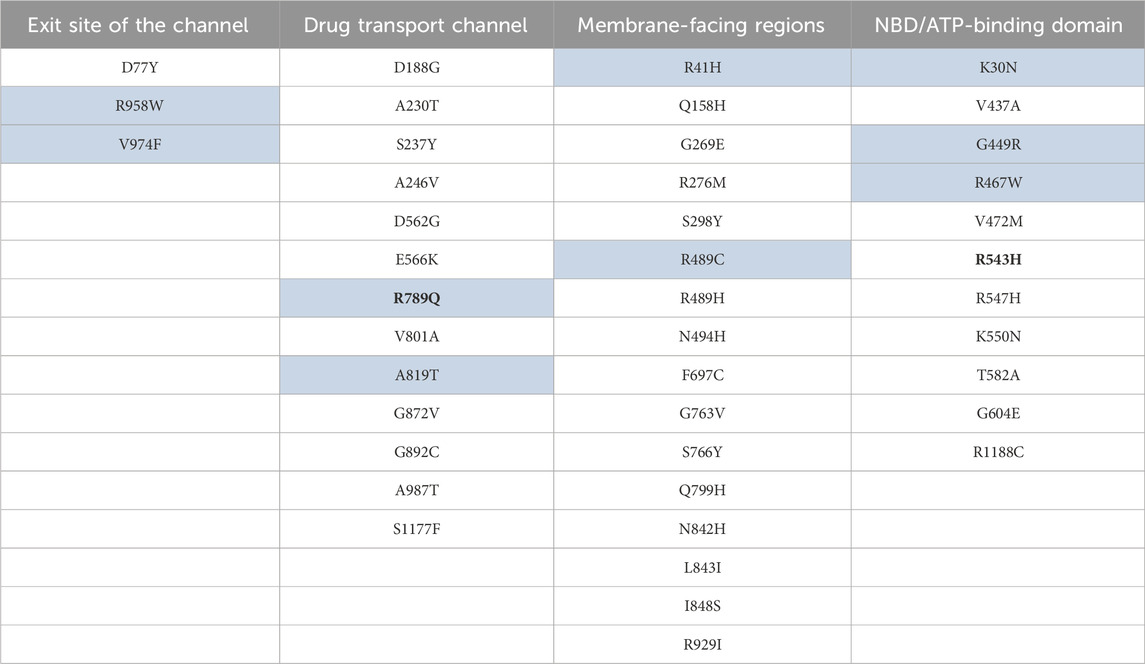
Table 5. Key mutations mapped to the important structural regions of the ABCB1. The highlighted mutations are also present in other cancer(s) in addition to endometrial uterine carcinoma. Mutations shown in bold were reported to be important in drug response as per ClinVar (Landrum et al., 2020).
Mutations A987T, G872V, A230T, S237Y, and D188G are located close to the binding site of the drug molecule inside the channel, and mutations A819T, A246V, and R789Q indicate that it will lead to an increase in the amphiphilic nature of the channel that can lead to binding of diverse drug compounds and seems important in explaining the polyspecificity in ABCB1 important for chemoresistance. The R789Q mutation was also shown to be important in the drug response as per ClinVar (Landrum et al., 2020). Mutations S1177F, V801A, and G892C are present close to the entry and intramembrane region, with the closest being S1177F. S1177F and V801A can lead to an increase in hydrophobicity and entry volume of the channel, which is important in entry of the drug from the intracellular environment, making it more efficient, which in turn helps in chemoresistance (Figure 7; Table 5).
Mutations E566K and D562G are very closely located on the entry from the intracellular side. Common drugs like paclitaxel and cyclosporine A (known to have an effect in ABCB1) contains hydroxyl and keto group and aspartic acid residues, respectively, which are negatively charged; therefore, due to mutations from glutamate to lysine, they can effectively be impacted via charged mutated and hydrogen bonding interactions that can lead to better drug efflux. The other mutation D562G can be important in increasing the volume of the intracellular entry side, important for accessing the drug-binding channel (Figure 7; Table 5) (Sissung et al., 2006; Chang et al., 2009; García et al., 2013; Zhang et al., 2013; Gao et al., 2014; Damont et al., 2016; Vallo et al., 2017; van Eerden et al., 2021).
According to the hydrophobic match/mismatch theory, transmembrane proteins avoid unfavorable exposure of the hydrophobic surface present on the transmembrane region of the protein to a hydrophilic environment present on the lipid bilayer surfaces. Biological membranes tend to adapt to such mismatch by expanding and shrinking. Lipid bilayers get adjusted elastically to the hydrophobic length of the transmembrane helices where a decreased hydrophobic transmembrane length would lead to shrinkage of the lipid bilayer along the transmembrane region, reducing the curvature of the overall membrane, whereas increased hydrophobic transmembrane length would lead to elongation of the bilipid layer along the transmembrane region, increasing the curvature of the overall membrane. We discussed the possible effects of this phenomenon with respect to mutations in the ABCB1, ABCC1, and ABCG2 transporters (Weiss et al., 2003; Janmey and Kinnunen, 2006; Soubias et al., 2008; de Jesus and Allen, 2013; Engberg et al., 2022).
With mutations I848S, L843I, and N842H, here threonine to serine will change from non-polar to polar residues, leucine to isoleucine will change from large hydrophobic residues to smaller hydrophobic residues, and asparagine to histidine will change from polar to positively charged amino acids. All these three mutations can lead to a favorable interaction with the non-polar membrane component and the polar component of the cholesterol molecule. For example, asparagine to histidine mutation will introduce an imidazole side chain which can interact favorably with the polar part cholesterol and non-polar part of the phospholipid membrane. Overall, these mutations will help in increasing the fluidity of the cell membrane.
Other mutations like S766Y, G763V, and S298Y would lead to an increase in the non-polar interaction between these residues and the non-polar part of the membrane. However, the hydroxyl group present in tyrosine can also interact favorably with cholesterol molecules. These mutations can lead to a probable increase in the rigidity of the membrane. With mutations R276M and G269E, arginine to methionine will change from charged to non-polar residues, which will lead to favorable interactions for non-polar membrane components. With mutation G269E, glycine to glutamic acid will lead to favorable interactions with cholesterol molecules present on the membrane, important for maintaining the fluidity and stability of the membrane. With mutation Q799H, glutamine to histidine will favor interactions with cholesterol molecules, leading to an increase in fluidity and stability of the membrane.
Q158H, R489H/C, and N494H mutations from polar to positively charged residues like Q158H and N494H can lead to favorable interactions with cholesterol and other polar molecules present on the membrane, important for maintaining the membrane fluidity and stability. On the other hand, R489H and R41H mutations will lead to a favorable interaction with cholesterol and polar molecules, and also an imidazole ring can interact favorably with the non-polar components of cholesterol and phospholipid membrane, signifying a dual role in histidine. Mutation F697C which is facing outwards toward the membrane may favorably interact with cholesterol and polar molecules required for maintaining the fluidity of the cell membrane and mutation R929I with hydrophobic nature would have favorable interactions with the non-polar component of the membrane and cholesterol.
The mutations L843I, S766Y, G763V, S298Y, R489H/C, F697C, and R929I present in the transmembrane region of ABCB1 would result in a change in hydrophobic interactions, which can modulate the hydrophobic mismatch. This would lead to a change in membrane curvature and/or fluidity affecting the ABCB1 conformational dynamics important for the drug transport and efflux.
In the ATP-binding site 1, we observed mutations V437A and R467W, both of which will lead to an enhanced ATP binding or release through the Walker motif A and B. Since V437A will lead to decrease in the side chain volume, it will allow more conformational freedom for the adjacent residues T435 of the Walker motif A. The R467W mutation will lead to an increase in the volume, and due to its hydrophobic nature, it would lead to reorientation of the R441 residue located opposite to that, which can restrict the conformational freedom of the Walker A motif important in ATP binding and hydrolysis (Loo et al., 2002; Orelle et al., 2003; Rai et al., 2006; Szöllősi et al., 2020).
V472M and G604E mutation will lead to an increase in the side chain volume, leading to the restriction of movement of the Walker A motif, thus favoring the ATP bound conformation which may have importance in chemoresistance. G449R mutation to the positively charged residue can form favorable interactions with adjacent negatively charged glutamic acid residues (E448), which will in turn lead to stabilization of an adjacent long loop which is directly connected to the Walker A motif. Mutations R543H and R547H which are located adjacent to each other upon mutation to histidine will lead to overall greater conformational freedom, allowing an optimal interaction between Walker A motif and Q loop for better ATP binding and hydrolysis. The R543H mutation was also shown to be important in the drug response as per ClinVar (Landrum et al., 2020). Mutations K550N and T582A will lead to a decrease in the overall volume of the side chain of amino acid residues, allowing more conformational freedom for the interaction between Walker A motif and ATP for optimal binding and hydrolysis.
Mutation R1188C located on the ATP-binding domain 2 is present in the vicinity of Q loop and Walker B motif; therefore, the mutation from R to C will allow more conformational freedom for better interaction and thus better binding with ATP molecules. The G604E mutation, which is located close to the cystine C431 residue of the Walker A motif, may lead to formation of polar interactions with C431 and would restrict the movement of the Walker A motif that may stabilize the ATP bound form of the ATP-binding domain. Mutations in NBD regions can be targeted to inhibit or reduce the ATP hydrolysis which is important for drug transport and hence chemoresistance.
3.3 Functional impact of mutations present in ABCC1
D355Y mutation present on the exit site of this transporter may facilitate efficient efflux of diverse drug compounds since the side chain of tyrosine containing the hydroxyl group can act as a hydrogen bond donor or acceptor. Furthermore, the aromatic side chain of tyrosine can facilitate a hydrophobic interaction with diverse drug compounds. V584A mutation which is present on the opposite site of the D335 mutation will lead to increase in exit cavity volume, leading to efficient drug efflux (Figure 7; Table 6).
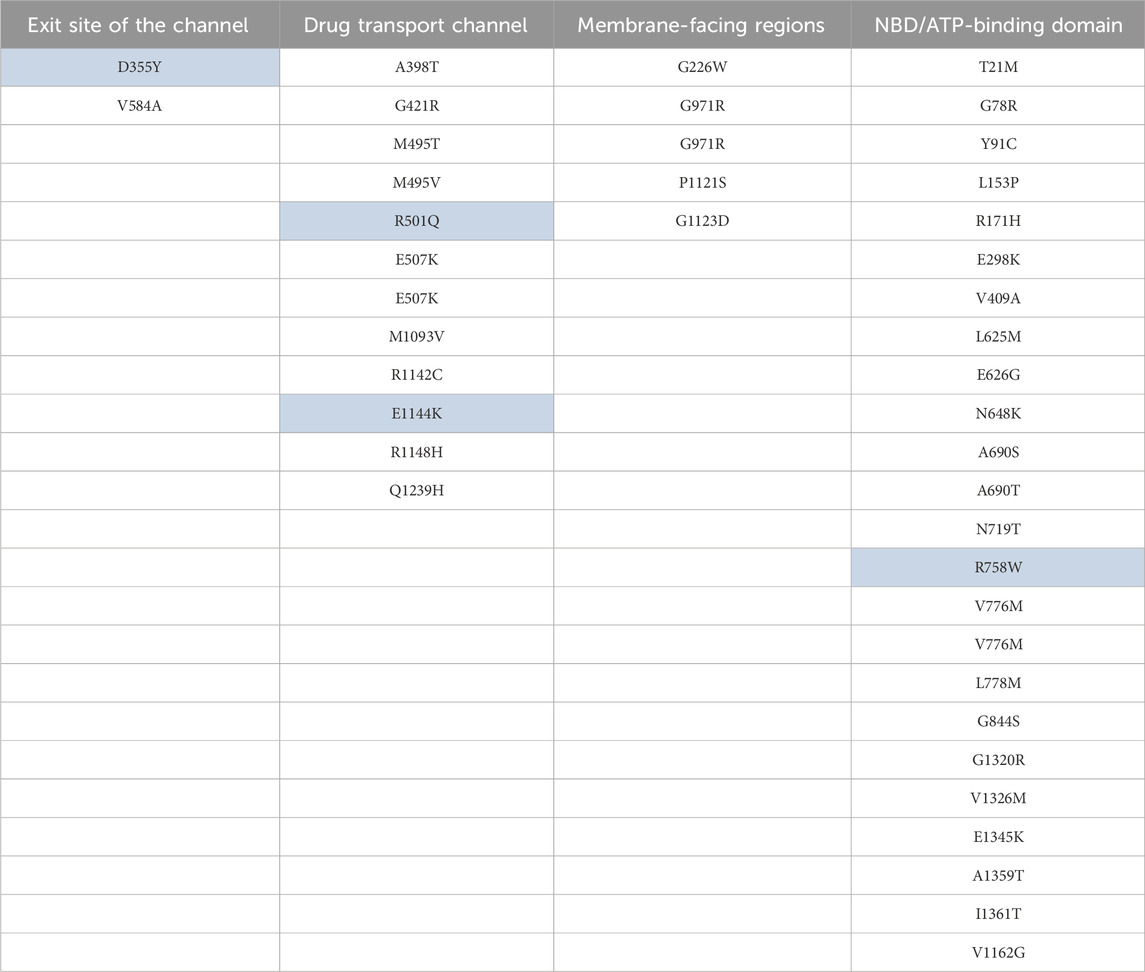
Table 6. Key mutations mapped to the important structural regions of the ABCC1. The highlighted mutations are also present in other cancer(s) in addition to endometrial uterine carcinoma.
Mutation Q1239H located close to the binding site will allow both the charge-mediated and non-polar interaction with diverse drug compounds. Since this mutation is located just above the Y1242 residue of the ABCC1 binding pocket, it can also stabilize the binding pocket through pi–pi stacking between the histidine and tyrosine residues. Thus overall, this mutation may enhance the efflux of diverse drug compounds. Mutation M1093V takes place in the H-pocket which comprises W1245, Y1242, M1093, T550, and W553. Upon mutation to valine, there will be a reduction in the site chain volume; therefore, the H-pocket can accommodate binding of both smaller and larger drug molecules mediated by a hydrophobic interaction (Figure 7; Table 6).
Mutations R1142C present on the channel lining will lead to an increase in the volume of the channel and will also facilitate the transport of diverse amphipathic drug compounds. E1144K mutation, which is located in the vicinity of R1146, K1139, and R1140, will lead to repulsion due to like charges and, thus, may lead to an increase in channel volume and flexibility.
The R1148H mutation is again located on the channel lining. Histidine’s imidazole ring would lead to better non-polar interactions in addition to charge-mediated interactions with diverse amphipathic drug molecules. It would also lead to the reduction in the side chain volume, resulting in a probable increase in channel volume, favoring efficient drug efflux. In residue M495, it is found to be mutated in two different cancer samples to threonine and valine. In both cases, it will lead to a decrease in the volume of the side chain, leading to easy efflux of diverse amphipathic drug molecules. R501Q mutation will lead to a decrease in the side chain volume without losing the capability of forming a polar interaction with drug molecules for efficient efflux. G421R mutation located adjacent to E422 might lead to favorable charge-mediated interaction, leading to the stabilizing of the helix which forms the efflux channel (Figure 7; Table 6).
The A398T mutation is surrounded by residues V253, V256, and L257. So this mutation will change the hydrophobic nature of the side chain to polar and, therefore, may form unfavorable interactions with the surrounding residues while pushing itself toward the drug efflux channel, thus facilitating the interaction with amphipathic drug molecules. E507K mutation is located in close proximity to K503 and is involved in the formation of polar contact. Thus, this mutation leading to a lysine residue in this position pushes this residue toward the entry of the channel, which may be important in regulating the drug axis and efflux.
In the NBD2 region of ABCC1, mutation V1326M will lead to increase in the hydrophobic side chain volume, leading to better packing with the Walker A motif and surrounding hydrophobic residues.
Residues V776 and L778 are sandwiched between LSGGQ and Walker motif B in the NBD1 region. Mutations V776M/M and L778M, where the residues are directly interacting with the LSGGQ motif, upon mutation to methionine, will lead to a better interaction with the LSGGQ motif and the other surrounding residues. G844S, A690S, and A690T are located close to Walker motif A in NBD1. All these mutations will lead to a hydrophobic to polar side chain that will lead to a favorable polar interaction with the charged and polar residue of Walker motif A.
The G971R mutation will lead to formation of polar interactions with cholesterol and other polar molecules present in the phospholipid membrane required for maintaining membrane fluidity. G226W mutation located in the vicinity of two other tryptophan residues W223 and W242 will lead to the formation of strong hydrophobic clusters, thus stabilizing the L0 region (205–268) which is known to be essential for folding of ABCC1 (Weinstein et al., 2013). P1121S and G1123D both are located facing the membrane. These mutations will lead to a better interaction with cholesterol and the polar component of the phospholipid membrane, thus providing membrane fluidity required for efficient drug efflux.
We observed a higher number of key mutations in the NBD1 region as compared to the NBD2 region of ABCC1. It is reported that in ABCC1, only the NBD2 region is catalytically active (Sajid et al., 2023). Therefore, a higher number of mutations in NBD1 might be important in retaining the activity of NBD2, which is important for drug transport, while increasing the binding affinity of the NBD1 region through mutations.
3.4 Functional impact of mutations present in ABCG2
R482K, D217Y, and G80R mutations are located close to the entry site from the intracellular side of ABCG2. R482K will lead to an increase in channel volume without affecting the formation of a charge-mediated interaction with diverse drug compounds, while mutation of D217Y to tyrosine can facilitate the efflux of diverse amphipathic drug molecules through side-chain flipping mediated by favorable interactions. G80R mutation located to arginine provides a stable interaction with the charged substrate and drug molecule (Figure 7; Table 7).
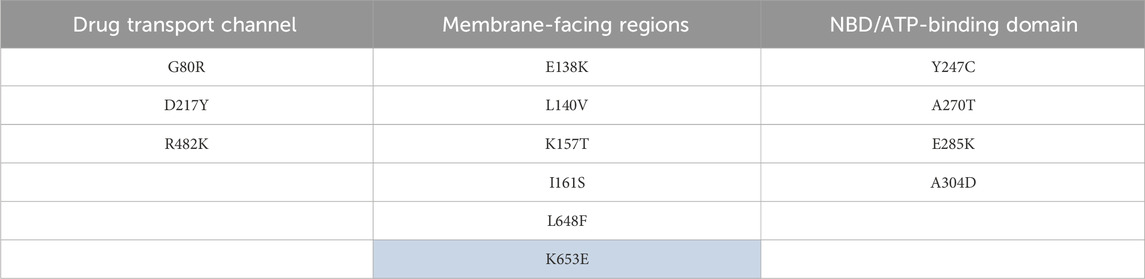
Table 7. Key mutations mapped to the important structural regions of the ABCG2. The highlighted mutations are also present in other cancer(s) in addition to endometrial uterine carcinoma.
The E138K mutation would probably make contact with the hydrophilic head of the membrane which is often negatively charged; therefore, this mutation will facilitate favorable charge-mediated interactions. L140V mutation will allow more flexible hydrophobic interactions with the nearby residues such as I161, thereby helping modulate the flexibility of this region. K157T mutation will lead to formation of a better interaction with nearby residues such as T150, N154, and M152, therefore playing a crucial role for enhancing the stability of this whole region. Mutation I161S will lead to the formation of favorable polar interactions with E138 and K157 (Figure 7; Table 7).
Mutation L648F facing toward the membrane region and located in the vicinity of F650 will lead to stabilization of the helix through hydrophobic interactions and can also interact favorably with the non-polar region of the phospholipid membrane. Mutation K653E facing toward the membrane region may interact favorably with the phospholipid membrane and also the polar molecule present in the phospholipid membrane and therefore may be crucial for membrane stability.
In ABCG2, mutations L140V, K157T, and L648F changing to hydrophobic residues may lead to hydrophobic matching by increasing and elongating the bilipid layer across the transmembrane region, contributing in its flexibility and hence important in drug transport and efflux.
The Y247C, A270T, E285K, and A304D mutations are located on the entry region on the intracellular side. Mutations Y247C and A304D, wherein A304D mutation also becomes larger due to an increase in the side chain volume, will allow flexible polar interactions with diverse drug substrates. The A270T mutation will replace a smaller residue with a larger residue therefore facilitating interaction with amphipathic drug molecules. E285K mutation will lead favorable interactions with nearby negatively charged residues such as D296, D292, and D301.
4 Discussion
We analyzed a total of 248 mutations present in ABCG2, ABCB1, and ABCC1 transporters. The position and physicochemical characteristics of the mutations were investigated to understand their possible impact on the function of transporters and its surroundings. Mutations present inside the binding pocket or channel are likely to affect the binding of substrate molecules. A drug molecule that can competitively bind in the pocket of these transporters with high affinity could block the passage for its natural substrates. This in turn will suppress the efflux of drug molecules from the cell, which is usually how cancer cells showcase multidrug resistance. It was observed that the frequency of mutations is also high in the membrane-facing regions. We believe this is because these mutations are involved in modulating the flexibility of the transmembrane and, in turn, affecting its surroundings, which in turn can affect the lipid and lipid raft composition of the cell membrane in the process. Change in the membrane composition affects the fluidity and surface curvature, which in turn is important for drug transport and efflux.
Existing inhibitory drugs such as tariquidar known to bind and inhibit P-glycoprotein (ABCB1) were studied for its inhibitory activity and also ABCB1 MDR activity (Ahmed et al., 2022). Discovery of inhibitor drugs for these transporters to tackle MDR has been a challenge. The phenomenon of MDR has been a major challenge for pharmaceutical industry. ABC transporters are often involved in drug efflux in diverse cell types; posing significant challenge in targeting various cancers. The binding sites of ABCB1, ABCC1 and ABCG2 are highly flexible and known to bind and transport diverse compounds including anti-cancer drugs. While this property helps these transporters to perform various important physiological activities, its overexpression has become one of the major cause for MDR in different cancer types. Targeting binding sites in these transporters is generally considered an attractive strategy to overcome drug resistance. However, the mutational analysis presented in this study, also suggests the importance of alternative strategies for addressing the chemoresistance by targeting the transmembrane and ATP binding domains for drug discovery.
Targeting of these specific residues to design an appropriate drug that can target both the binding region in the cavity and the transmembrane region have the potential to overcome MDRs. Designed inhibitors hypothetically will block the pathway, which will restrict the passage for the drug to be thrown out and will also prevent conformational and biochemical changes due to transmembrane mutations.
Research has shown that certain mutations can help reverse the direction of drug transport in these ABC transporters, which also have great potential in preventing MDR in cancer cells (Sajid et al., 2020). A group of 14 conserved residues (seven in both TMHs 6 and 12) were replaced with alanine and generated a mutant that lost the ability to pump most of the substrates tested out of cancer cells, but this reversed the direction of the pump, and the substrate was rather imported. It was able to import four substrates, including Rhodamine-123 and the taxol derivative flutax-1 (Kerb et al., 2001; Uitto, 2005; Seeger and van Veen, 2009; Kueppers et al., 2013; Kadioglu et al., 2020; Sajid et al., 2020).
5 Conclusions and future aspects
We performed a comprehensive analysis of mutations present in ABCB1, ABCC1, and ABCG2 transporters for endometrial cancer. The key mutations have been identified in these transporters and their possible functional impact on the drug binding, transport, and efflux has been discussed at the molecular level. The molecular level explanation of the impact of key mutations identified in ABCB1, ABCC1, and ABCG2 would be important in understanding the mechanisms of chemoresistance, design, and discovery of ABC inhibitors and therapeutic molecules. The key mutations identified in ABCB1, ABCC1, and ABCG2 can also be explored to reengineer these transporters from drug efflux transporters to drug importers to tackle chemoresistance.
Comparison of these mutations with ABC transporters expressed in normal endometrial tissue could be a potential area of exploration, which is currently unavailable on open access platforms. Additionally, there are no curated datasets for mutations present in all three ABC transporters to compare with cancerous and relapse-tissue samples from same patient pre- and post-chemotherapy. Due to the absence of mutational data from normal endometrial tissue (non-cancerous) for the ABC transporters, it is currently out of scope of this work to compare the mutational status between cancerous and normal endometrial tissues. Detailed analysis on these key aspects is important for understanding the intrinsic and acquired resistance and subsequent designing of inhibitors and therapeutic molecules.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
Ethics statement
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
Author contributions
AG: data curation, formal analysis, investigation, methodology, and writing–original draft. Manu SS: conceptualization, investigation, project administration, supervision, and writing–original draft. BS: conceptualization, formal analysis, investigation, project administration, supervision, and writing–original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors acknowledge the computational lab infrastructure support and financial research assistance to AG from Mahindra University, Hyderabad, India.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1380371/full#supplementary-material
References
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A., Bork, P., et al. (2010). A method and server for predicting damaging missense mutations. Nat. Methods 7 (4), 248–249. doi:10.1038/nmeth0410-248
Ahmed, J., Abdul Hamid, A. A., Abd Halim, K. B., and Che, H. A. T. (2022). P-glycoprotein: new insights into structure, physiological function, regulation and alterations in disease. Heliyon 8 (6), e09777. doi:10.1016/j.heliyon.2022.e09777
Alam, A., Kowal, J., Broude, E., Roninson, I., and Locher, K. P. (2019). Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science 363 (6428), 753–756. doi:10.1126/science.aav7102
Arain, A., Muhsen, I. N., Abudayyeh, A., and Abdelrahim, M. (2021). “Chapter 11 - chemoresistance in uterine cancer: mechanisms of resistance and current therapies,” in Overcoming drug resistance in gynecologic cancers. Editors R. Basha,, and S. Ahmad (Academic Press), 267–281.
Berger, A. C., Korkut, A., Kanchi, R. S., Hegde, A. M., Lenoir, W., Liu, W., et al. (2018). A comprehensive pan-cancer molecular study of gynecologic and breast cancers. Cancer Cell 33 (4), 690–705.e9. doi:10.1016/j.ccell.2018.03.014
Bloise, E., Ortiga-Carvalho, T. M., Reis, F. M., Lye, S. J., Gibb, W., and Matthews, S. G. (2015). ATP-binding cassette transporters in reproduction: a new frontier. Hum. Reprod. Update 5, dmv049. doi:10.1093/humupd/dmv049
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2 (5), 401–404. doi:10.1158/2159-8290.CD-12-0095
Chang, H., Rha, S. Y., Jeung, H. C., Im, C. K., Ahn, J. B., Kwon, W. S., et al. (2009). Association of the ABCB1 gene polymorphisms 2677G>T/A and 3435C>T with clinical outcomes of paclitaxel monotherapy in metastatic breast cancer patients. Ann. Oncol. 20 (2), 272–277. doi:10.1093/annonc/mdn624
Damont, A., Goutal, S., Auvity, S., Valette, H., Kuhnast, B., Saba, W., et al. (2016). Imaging the impact of cyclosporin A and dipyridamole on P-glycoprotein (ABCB1) function at the blood-brain barrier: a [11C]-N-desmethyl-loperamide PET study in nonhuman primates. Eur. J. Pharm. Sci. 91, 98–104. doi:10.1016/j.ejps.2016.06.005
de Bruijn, I., Kundra, R., Mastrogiacomo, B., Tran, T. N., Sikina, L., Mazor, T., et al. (2023). Analysis and visualization of longitudinal genomic and clinical data from the AACR Project GENIE biopharma collaborative in cBioPortal. Cancer Res. 83 (23), 3861–3867. doi:10.1158/0008-5472.CAN-23-0816
de Jesus, A. J., and Allen, T. W. (2013). The determinants of hydrophobic mismatch response for transmembrane helices. Biochim. Biophys. Acta BBA - Biomembr. 1828 (2), 851–863. doi:10.1016/j.bbamem.2012.09.012
Di, M. S., Di Sarno, V., Rossi, M., Vestuto, V., Konno, T., Novi, S., et al. (2022). In silico identification and in vitro evaluation of new ABCG2 transporter inhibitors as potential anticancer agents. Int. J. Mol. Sci. 24 (1), 725. doi:10.3390/ijms24010725
Engberg, O., Ulbricht, D., Döbel, V., Siebert, V., Frie, C., Penk, A., et al. (2022). Rhomboid-catalyzed intramembrane proteolysis requires hydrophobic matching with the surrounding lipid bilayer. Sci. Adv. 8 (38), eabq8303. doi:10.1126/sciadv.abq8303
Gao, B., Russell, A., Beesley, J., Chen, X. Q., Healey, S., Henderson, M., et al. (2014). Paclitaxel sensitivity in relation to ABCB1 expression, efflux and single nucleotide polymorphisms in ovarian cancer. Sci. Rep. 4, 4669. doi:10.1038/srep04669
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6 (269), pl1. doi:10.1126/scisignal.2004088
García, M., Macías, R. M., Cubero, J. J., Benítez, J., Caravaca, F., and Gervasini, G. (2013). ABCB1 polymorphisms are associated with cyclosporine-induced nephrotoxicity and gingival hyperplasia in renal transplant recipients. Eur. J. Clin. Pharmacol. 69 (3), 385–393. doi:10.1007/s00228-012-1355-x
Gaulton, A., Bellis, L. J., Bento, A. P., Chambers, J., Davies, M., Hersey, A., et al. (2012). ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40, D1100–D1107. doi:10.1093/nar/gkr777
Hoadley, K. A., Yau, C., Hinoue, T., Wolf, D. M., Lazar, A. J., Drill, E., et al. (2018). Cell-of-Origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173 (2), 291–304.e6. doi:10.1016/j.cell.2018.03.022
Hollenstein, K., Dawson, R. J. P., and Locher, K. P. (2007). Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 17, 412–418. doi:10.1016/j.sbi.2007.07.003
Janmey, P., and Kinnunen, P. (2006). Biophysical properties of lipids and dynamic membranes. Trends Cell Biol. 16, 538–546. doi:10.1016/j.tcb.2006.08.009
Jones, P. M., and George, A. M. (2004). The ABC transporter structure and mechanism: perspectives on recent research. Cell Mol. Life Sci. CMLS 61 (6), 682–699. doi:10.1007/s00018-003-3336-9
Juan-Carlos, P. D. M., Perla-Lidia, P. P., Stephanie-Talia, M. M., Mónica-Griselda, A. M., and Luz-María, T. E. (2021). ABC transporter superfamily. An updated overview, relevance in cancer multidrug resistance and perspectives with personalized medicine. Mol. Biol. Rep. 48 (2), 1883–1901. doi:10.1007/s11033-021-06155-w
Kadioglu, O., Saeed, M. E. M., Munder, M., Spuller, A., Greten, H. J., and Efferth, T. (2020). Effect of ABC transporter expression and mutational status on survival rates of cancer patients. Biomed. Pharmacother. 131, 110718. doi:10.1016/j.biopha.2020.110718
Kerb, R., Hoffmeyer, S., and Brinkmann, U. (2001). ABC drug transporters: hereditary polymorphisms and pharmacological impact in MDR1, MRP1 and MRP2. Pharmacogenomics 2 (1), 51–64. doi:10.1517/14622416.2.1.51
Kowal, J., Ni, D., Jackson, S. M., Manolaridis, I., Stahlberg, H., and Locher, K. P. (2021). Structural basis of drug recognition by the multidrug transporter ABCG2. J. Mol. Biol. 433 (13), 166980. doi:10.1016/j.jmb.2021.166980
Kueppers, P., Gupta, R. P., Stindt, J., Smits, S. H. J., and Schmitt, L. (2013). Functional impact of a single mutation within the transmembrane domain of the multidrug ABC transporter Pdr5. Biochemistry 52 (13), 2184–2195. doi:10.1021/bi3015778
Landrum, M. J., Chitipiralla, S., Brown, G. R., Chen, C., Gu, B., Hart, J., et al. (2020). ClinVar: improvements to accessing data. Nucleic Acids Res. 48 (D1), D835–D844. doi:10.1093/nar/gkz972
Laskowski, R. A., and Swindells, M. B. (2011). LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model 51 (10), 2778–2786. doi:10.1021/ci200227u
Liu, N., and Xu, Z. (2019). Using LeDock as a docking tool for computational drug design. IOP Conf. Ser. Earth Environ. Sci. 218 (1), 012143. doi:10.1088/1755-1315/218/1/012143
Loo, T. W., Bartlett, M. C., and Clarke, D. M. (2002). The “LSGGQ” motif in each nucleotide-binding domain of human P-glycoprotein is adjacent to the opposing walker A sequence. J. Biol. Chem. 277 (44), 41303–41306. doi:10.1074/jbc.C200484200
Moxley, K. M., and McMeekin, D. S. (2010). Endometrial carcinoma: a review of chemotherapy, drug resistance, and the search for new agents. Oncol. 15, 1026–1033. doi:10.1634/theoncologist.2010-0087
Ng, P. C., and Henikoff, S. (2001). Predicting deleterious amino acid substitutions. Genome Res. 11, 863–874. doi:10.1101/gr.176601
Nieth, C., and Lage, H. (2005). Induction of the ABC-transporters Mdr1/P-gp (Abcb1), mrpl (Abcc1), and bcrp (Abcg2) during establishment of multidrug resistance following exposure to mitoxantrone. J. Chemother. 17 (2), 215–223. doi:10.1179/joc.2005.17.2.215
Nosol, K., Romane, K., Irobalieva, R. N., Alam, A., Kowal, J., Fujita, N., et al. (2020). Cryo-EM structures reveal distinct mechanisms of inhibition of the human multidrug transporter ABCB1. Proc. Natl. Acad. Sci. 117 (42), 26245–26253. doi:10.1073/pnas.2010264117
Oldham, M. L., Davidson, A. L., and Chen, J. (2008). Structural insights into ABC transporter mechanism. Curr. Opin. Struct. Biol. 18, 726–733. doi:10.1016/j.sbi.2008.09.007
Orelle, C., Dalmas, O., Gros, P., Di Pietro, A., and Jault, J. M. (2003). The conserved glutamate residue adjacent to the walker-B motif is the catalytic base for ATP hydrolysis in the ATP-binding cassette transporter BmrA. J. Biol. Chem. 278 (47), 47002–47008. doi:10.1074/jbc.M308268200
Rai, V., Gaur, M., Shukla, S., Shukla, S., Ambudkar, S. V., Komath, S. S., et al. (2006). Conserved Asp327 of Walker B motif in the N-terminal Nucleotide Binding Domain (NBD-1) of Cdr1p of Candida albicans has acquired a new role in ATP hydrolysis. Biochemistry 45 (49), 14726–14739. doi:10.1021/bi061535t
Rees, D. C., Johnson, E., and Lewinson, O. (2009). ABC transporters: the power to change. Nat. Rev. Mol. Cell Biol. 10 (3), 218–227. doi:10.1038/nrm2646
Reva, B., Antipin, Y., and Sander, C. (2011). Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39 (17), e118. doi:10.1093/nar/gkr407
Robey, R. W., Pluchino, K. M., Hall, M. D., Fojo, A. T., Bates, S. E., and Gottesman, M. M. (2018). Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 18 (7), 452–464. doi:10.1038/s41568-018-0005-8
Sajid, A., Lusvarghi, S., Murakami, M., Chufan, E. E., Abel, B., Gottesman, M. M., et al. (2020). Reversing the direction of drug transport mediated by the human multidrug transporter P-glycoprotein. Proc. Natl. Acad. Sci. 117 (47), 29609–29617. doi:10.1073/pnas.2016270117
Sajid, A., Rahman, H., and Ambudkar, S. V. (2023). Advances in the structure, mechanism and targeting of chemoresistance-linked ABC transporters. Nat. Rev. Cancer 23 (11), 762–779. doi:10.1038/s41568-023-00612-3
Seeger, M. A., and van Veen, H. W. (2009). Molecular basis of multidrug transport by ABC transporters. Biochim. Biophys. Acta BBA - Proteins Proteomics. 1794 (5), 725–737. doi:10.1016/j.bbapap.2008.12.004
Sissung, T. M., Mross, K., Steinberg, S. M., Behringer, D., Figg, W. D., Sparreboom, A., et al. (2006). Association of ABCB1 genotypes with paclitaxel-mediated peripheral neuropathy and neutropenia. Eur. J. Cancer 42 (17), 2893–2896. doi:10.1016/j.ejca.2006.06.017
Soubias, O., Niu, S. L., Mitchell, D. C., and Gawrisch, K. (2008). Lipid−Rhodopsin hydrophobic mismatch alters rhodopsin helical content. J. Am. Chem. Soc. 130 (37), 12465–12471. doi:10.1021/ja803599x
Stefan, S. M., Jansson, P. J., Pahnke, J., and Namasivayam, V. (2022). A curated binary pattern multitarget dataset of focused ATP-binding cassette transporter inhibitors. Sci. Data 9 (1), 446. doi:10.1038/s41597-022-01506-z
Szöllősi, D., Chiba, P., Szakacs, G., and Stockner, T. (2020). Conversion of chemical to mechanical energy by the nucleotide binding domains of ABCB1. Sci. Rep. 10 (1), 2589. doi:10.1038/s41598-020-59403-7
Takagi, S., Onishi, T., Takashima, T., Shibahara, K., and Mori, M. (2023). Acquired AKT-inhibitor resistance is mediated by ATP-binding cassette transporters in endometrial carcinoma. Anticancer Res. 43 (6), 2501–2507. doi:10.21873/anticanres.16417
Tarling, E. J., Vallim, TQDA, and Edwards, P. A. (2013). Role of ABC transporters in lipid transport and human disease. Trends Endocrinol. Metab. 24 (7), 342–350. doi:10.1016/j.tem.2013.01.006
Uitto, J. (2005). The gene family of ABC transporters-novel mutations, new phenotypes. Trends Mol. Med. 11 (8), 341–343. doi:10.1016/j.molmed.2005.06.004
Vallo, S., Köpp, R., Michaelis, M., Rothweiler, F., Bartsch, G., Brandt, M. P., et al. (2017). Resistance to nanoparticle albumin-bound paclitaxel is mediated by ABCB1 in urothelial cancer cells. Oncol. Lett. 13 (6), 4085–4092. doi:10.3892/ol.2017.5986
van Eerden, R. A. G., van Doorn, L., de Man, F. M., Heersche, N., Doukas, M., van den Bosch, T. P. P., et al. (2021). Tissue type differences in ABCB1 expression and paclitaxel tissue pharmacokinetics in patients with esophageal cancer. Front. Pharmacol. 12, 759146. doi:10.3389/fphar.2021.759146
Vasiliou, V., Vasiliou, K., and Nebert, D. W. (2009). Human ATP-binding cassette (ABC) transporter family. Hum. Genomics. 3 (3), 281–290. doi:10.1186/1479-7364-3-3-281
Walker, J. E., Saraste, M., Runswick, M. J., and Gay, N. J. (1982). Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1 (8), 945–951. doi:10.1002/j.1460-2075.1982.tb01276.x
Wang, L., Johnson, Z. L., Wasserman, M. R., Levring, J., Chen, J., and Liu, S. (2020). Characterization of the kinetic cycle of an ABC transporter by single-molecule and cryo-EM analyses. eLife 9, e56451. doi:10.7554/eLife.56451
Weinstein, J. N., Collisson, E. A., Mills, G. B., Shaw, K. M., Ozenberger, B. A., Ellrott, K., et al. (2013). The cancer genome Atlas pan-cancer analysis Project. Nat. Genet. 45 (10), 1113–1120. doi:10.1038/ng.2764
Weiss, T. M., Wel, P. C. A., Killian, J. A., Koeppe, R. E., and Huang, H. W. (2003). Hydrophobic mismatch between helices and lipid bilayers. Biophys. J. 84 (1), 379–385. doi:10.1016/S0006-3495(03)74858-9
Wilkens, S. (2015). Structure and mechanism of ABC transporters. F1000Prime Rep. 7, 14. doi:10.12703/P7-14
Wlcek, K., and Stieger, B. (2014). ATP-binding cassette transporters in liver. BioFactors Oxf Engl. 40 (2), 188–198. doi:10.1002/biof.1136
Yuan, S., Chan, H. S., and Hu, Z. (2017). Using PyMOL as a platform for computational drug design. WIREs Comput. Mol. Sci. 7, e1298. doi:10.1002/wcms.1298
Zdrazil, B., Felix, E., Hunter, F., Manners, E. J., Blackshaw, J., Corbett, S., et al. (2024). The ChEMBL Database in 2023: a drug discovery platform spanning multiple bioactivity data types and time periods. Nucleic Acids Res. 52 (D1), D1180–D1192. doi:10.1093/nar/gkad1004
Zhang, Y., li, Li J., Fu, Q., Wang, X. D., Shan, L. L., Wang, C. X., et al. (2013). Associations of ABCB1, NFKB1, CYP3A, and NR1I2 polymorphisms with cyclosporine trough concentrations in Chinese renal transplant recipients. Acta Pharmacol. Sin. 34 (4), 555–560. doi:10.1038/aps.2012.200
Keywords: cancer, ATP-binding cassette transporters, chemoresistance, mutation, mitoxantrone, drug resistance, efflux transporters
Citation: Gupta A, Singh MS and Singh B (2024) Deciphering the functional role of clinical mutations in ABCB1, ABCC1, and ABCG2 ABC transporters in endometrial cancer. Front. Pharmacol. 15:1380371. doi: 10.3389/fphar.2024.1380371
Received: 01 February 2024; Accepted: 28 March 2024;
Published: 02 May 2024.
Edited by:
Muhammad Rafehi, University Medical Center Göttingen, GermanyReviewed by:
Zufeng Guo, Chongqing Medical University, ChinaKatja Stefan, University of Lübeck, Germany
Copyright © 2024 Gupta, Singh and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manu Smriti Singh, bWFudS5zaW5naEBtYWhpbmRyYXVuaXZlcnNpdHkuZWR1Lmlu; Bipin Singh, YmlwaW4uc2luZ2hAbWFoaW5kcmF1bml2ZXJzaXR5LmVkdS5pbg==
 Aayushi Gupta
Aayushi Gupta Manu Smriti Singh
Manu Smriti Singh Bipin Singh
Bipin Singh