- 1Department of Experimental and Clinical Biomedical Sciences “Mario Serio”, Section of Experimental Pathology and Oncology, University of Florence, Florence, Italy
- 2Department of Chemistry “Ugo Schiff”, University of Florence, Florence, Italy
- 3Department of Health Sciences, Section of Clinical Pharmacology and Oncology, University of Florence, Florence, Italy
Ovarian cancer is the deadliest gynaecologic malignancies worldwide. Platinum based chemotherapy is the mainstay treatment for ovarian cancer; however, frequent recurrence and chemoresistance onset in patients with advanced diseases remain a therapeutic challenge. Although mechanisms underlying the development of chemoresistance are still ambiguous, the B-cell lymphoma-2 (Bcl-2) family is closely associated with chemoresistance in ovarian cancer. We previously disclosed that Zeta-Crystallin (CryZ) is a post-transcriptional regulator of Bcl-2 gene expression, by binding to Bcl-2 mRNA and increasing its half-life. Here, we investigated the role of CryZ as a novel therapeutic target in A2780 ovarian carcinoma cells by modulating the protein activity with acetylsalicylic acid (ASA) to restore chemosensitivity. Molecular docking and fragment-mapping based approach revealed potential interaction of ASA within CryZ protein. Inhibition of CryZ binding activity to Bcl-2 and Bcl-xl mRNA targets by ASA was demonstrated in A375 cells. Cytotoxicity assays were conducted in A2780S and A2780R ovarian cancer cells to evaluate if CryZ binding activity inhibition and CryZ silencing were able to reverse cisplatin resistance. ASA-treatment determined a downregulation of Bcl-2 and Bcl-xl mRNA levels in A2780S and A2780R cells. ASA-treatment or CryZ silencing were able to increase and restore the chemosensitivity in both sensitive and resistant A2780 ovarian cancer cells, respectively. In this research article we demonstrated that the pharmacological or genetic inhibition of CryZ restores the sensitivity to cisplatin in a model of sensitive or resistant ovarian cancer cells. These findings suggest a new gene-targeted chemotherapeutic approach to restore the cytotoxicity in drug-resistant ovarian cancers and increase the sensitivity in non-resistant cells.
Background
Ovarian cancer is the deadliest gynaecological tumour. In agreement with the National Comprehensive Cancer Network (NCCN) Guidelines for Ovarian Cancer and with European Society for Medical Oncology (ESMO) and European Society of Gynaecological Oncology (ESGO), the standard of care treatment of ovarian cancer is a full-stage surgery and platinum-based systemic chemotherapy (Morgan et al., 2016; Colombo et al., 2019). However, inherent or acquired resistance is the main cause of treatment failure. Even today, the mechanisms responsible for resistance in ovarian cancer treatment are poorly understood (Cornelison et al., 2017). Regarding acquired resistance, the deregulation of anti-apoptotic genes Bcl-2 and Bcl-xL has been well established (Dai et al., 2023; Karademir and Özgür, 2023). Expression of both genes is regulated by post-transcriptional mechanisms, including the interactions between stabilizing or destabilizing RNA-binding proteins (RBP) and their mRNAs (Cui and Placzek, 2018). In the last few years, several aspects of the post-transcriptional control of Bcl-2 have been characterized, and it has been demonstrated the existence of a complex regulative network controlled by the interaction of an adenylate-uridylate-rich element (ARE) in the 3′UTR region of Bcl-2 mRNA, with different ARE-binding proteins (AUBPs) (Lapucci et al., 2002; Donnini et al., 2004). Among the Bcl–2 AUBPs we identified Zeta-Crystallin (CryZ), demonstrating its role in increasing Bcl-2 mRNA stability in human leukaemia cell lines (Lapucci et al., 2010; Lulli et al., 2020). CryZ exhibits enzymatic activity, particularly acting as NADPH dependent quinone reductase. In the effort to identify pharmacological inhibitors of CryZ, the group of Bazzi MD found that aspirin-like analgesics are potent inhibitors of its enzymatic activity (Bazzi, 2002; Bazzi et al., 2002). Paracetamol (acetaminophen), ibuprofen (2-(4-isobutyl phenyl)-propionic acid), salicylic acid and aspirin (ASA) inhibit the enzyme to varying degrees; the latter being the most potent CryZ inhibitors. The inhibition of CryZ enzymatic activity by ASA occurs through the interaction with a specific binding site, closed to that of NADPH (Trabert et al., 2014; Huang et al., 2016), therefore it appears to be a non-competitive inhibitor with respect to NADPH, interacting with both the free form of CryZ as well as the CryZ/NADPH complex.
The most relevant data on the antitumor role of ASA and other non-steroidal anti-inflammatory drugs (NSAIDs) derive from epidemiologic studies, in a large panel of tumour types (Elwood et al., 2024). Furthermore, a synergistic effect of classical chemotherapeutic agents and ASA on cancer growth and apoptosis has been observed in a human myeloma xenograft model (Mortensen et al., 1998). Also, a reduction in incidence of ovarian cancer has been observed in patients who take ASA regularly (Trabert et al., 2014). In preclinical ovarian cancer models, ASA treatment determined a significant reduction in Bcl-2 protein level with concomitant increase in caspase 3, revealing its antitumor and antiangiogenic effects (Huang et al., 2016). Bcl-2 and Bcl-xl play major roles in the pathobiology and chemoresistance of ovarian cancer, and their inhibitions were useful for treatment (Suh et al., 2023).
In the present research study, we demonstrated that ASA inhibits the binding of CryZ to the mRNAs of the anti-apoptotic Bcl-2 and Bcl-xL genes. In addition, we revealed that modulation of CryZ activity using ASA or its downregulation by specific siRNA increases sensitivity in both sensitive and drug-resistant human ovarian cancer cells in response to cisplatin treatment.
Materials and methods
Cell culture
The human A2780S and A2780R ovarian cancer cell lines and the A375 melanoma cell line were purchased from ATCC (ATCC Manassas, United States). A2780S and A2780R cells or A375 cells were maintained in RPMI-1640 or DMEM medium, respectively, supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 U/mL) (Euroclone, Milan, Italy) at 37 C in a humidified incubator with 5% CO2. Cells were periodically screened for mycoplasma contamination using PCR.
RNA-Amply-Seq analysis
Total RNA was isolated from A2780S and A2780R cells by means of NucleoSpin Total RNA kit according to the manufacturer’s protocol (Macherey-Nagel, PA, United States). RNA was quantified by Qubit™ 3.0 Fluorometer (Invitrogen, MA, United States). After quantification, the quality of RNA was evaluated using the Agilent RNA 6000 Nano LabChip® kit with the Agilent 2100 Bioanalyzer (Agilent Technologies, CA, United States). RNA-Sequencing (RNA-Seq) analysis was performed by Novogene (Cambridge, United Kingdom) on an Illumina NovaSeq X Plus system (Illumina, San Diego, CA, United States). Briefly, for each cell line, an experimental triplicate was analyzed. mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. After fragmentation, the first strand cDNA was synthesized using random hexamer primers, followed by the second strand cDNA synthesis using either dUTP for directional library or dTTP for non-directional library. Raw data (raw reads) of fastq format were firstly processed through fastp software. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N and low-quality reads from raw data. Paired-end clean reads were aligned to the reference genome (hg38) using Hisat2 v2.0.5. feature Counts v1.5.0-p3 was used to count the reads numbers mapped to each gene and then Fragments Per Kilobase Million (FPKM) of each gene was calculated based on the length of the gene and reads count mapped to this gene.
Western blot assay
For Western blot analysis, cells were lysed in cold radioimmunoprecipitation assay (RIPA) lysis buffer (1% NP-40, 150 mM NaCl, 5 mM EDTA, 0.25% NaDOC, 50 mM Tris-HCl pH 7.5, 0.1% SDS) supplemented with protease and phosphatase inhibitors (Merck Life Science, Milan, Italy). Total proteins extracted from the cells were quantified with Bradford reagent (Merck Life Science, Milan, Italy). Aliquots of total protein (30 μg) were electrophoresed on 8%–12% (v/v) SDS-PAGE gel (Thermo Fischer Scientific, Milan, Italy), transferred onto nitrocellulose membranes (Bio-Rad, CA, United States) and probed overnight at 4 °C with primary antibodies against CryZ (2A9DD1, Abcam, United States), Bcl-2 (clone 100, Thermo Fisher Scientific), Bcl-xl (SC-8392, Santa Cruz Biotechnologies) and beta-actin (β-actin Thermo Fisher Scientific). Bound primary antibodies were detected using goat anti-mouse or goat anti-rabbit IgG Alexa Fluor 680 or 800 secondary antibodies (LI-COR® Bioscience, Lincoln, NE, United States). Membranes were visualized at the Odyssey Infrared Imaging System (LI-COR® Bioscience, Lincoln, NE, USAEuroclone, Milan, Italy) and densitometric analysis was performed by Odyssey Image studio software (LI-COR® Bioscience, Lincoln, NE, United States Euroclone, Milan, Italy) Quantity One software (Bio-Rad, CA, United States).
Identification of potential ligand binding sites and hot sposts
A fragment-mapping based approach (FTMAP) was used to locate potential small molecule binding hot spots on each of the Rho crystallographic structures. FTMAP has been shown as a powerful tool for identifying druggable hot spots in proteins (Brenke et al., 2009; Kozakov et al., 2015), as it correlates pocket druggability with propensity to bind clusters of small organic compound fragments.
We used the FTMap method of Vajda et al. to highlight protein surface regions that have the potential to bind the highest number of small molecular probes. The available crystal structure of CryZ (PDB: 1yb5) was subjected to fragment mapping and the resulting hot-spot residues (those that comprise prominent fragment binding sites) were analyzed. Then, complimentary to FTMap studies, we applied FTSite to find likely ligand-binding sites. FTSite uses the hot spots detected by FTMap to identify and rank binding sites, on the basis that the binding site of a protein generally consists of a strong main hot spot plus some other adjacent hot spots (Ngan et al., 2012).
Docking calculations
Automated docking studies were carried out using the Lamarckian Genetic Algorithm (LGA) as a search engine implemented in the Autodock 4.0.1 program (Morris et al., 1998). The AutoDockTools 1.4.5 (ADT) graphical interface (Gillet et al., 2005) was used to prepare CryZ and ligand PDBQT files. Coordinates of ASA were generated using Spartan (version 5.147), and then energy-minimized through the AM1 semi-empirical method to calculate the equilibrium geometry. The coordinates of CryZ were retrieved from the Protein Data Bank (PDB code: 1yb5), and protein-ligand complex was unmerged for achieving the free protein structure. Water molecules were removed. Hydrogen atoms were added to the enzyme and the ligand, Gasteiger charges were added and non-polar hydrogens were merged. Three-dimensional energy scoring grid of 0.375 Å resolution and 126 Å × 126 Å × 126 Å dimension was computed to include the whole protein. The center of the grid was set on the center of the protein. A total of 50 runs with a maximum of 2500000 energy evaluations were carried out for ASA, using the default parameters for LGA. Low energy ligand-protein complexes were subjected to AMMP energy minimization using VegaZZ (Pedretti et al., 2004), then Cluster analysis was performed on docked results using a root-mean-square (rms) tolerance of 1.5 Å. The analysis of the binding mode of the docked conformations was carried out using the Autodock plugin within PyMol software v0.99 (Alexander et al., 2011).
Silencing of CryZ
The A2780S and A2780R were transfected with 100 nM specific siRNA (IDT, Leuven, Belgium) against human CryZ by means of jetPRIME® (Polyplus, Illkirch, France) according to the instruction manual. Scramble siRNA was used as control (si-CTR).
In vitro transcription
Renilla luciferase RNA was in vitro transcribed to be used as internal RT-qPCR control. A PCR product containing the ORF of Renilla luciferase was obtained from pGL4.71 plasmid, inserting the T7 RNA polymerase consensus by means of PCR reaction. This product was used as a template in the in vitro transcription reaction using MEGAscript T7 Transcription kit (Ambion, United States). Obtained RNA was purified using the RNAeasy kit (Qiagen, United States) e quantified by Qubit™ 3.0 Fluorometer.
RNA immunoprecipitation assay
RNA protein immunoprecipitation (RIP) was performed based on Tenenbaum et al., 2002. In brief, A375 cells were treated with ASA 1 mM the concentration previously demonstrated to inhibit CryZ enzymatic activity in cell free biochemical assays (Bazzi et al., 2002) for 24 h, then cells were lysed in non-denaturing lysis buffer (20 mM Tris-HCl pH 8.0, 137 mM NaCl, 1% Nonidet P-40, 2 mM EDTA) in presence of protease inhibitors cocktail (Merck, United States) and 100 U/mL RNasein ribonuclease inhibitor (Promega, United States). 1 mg of protein extracts were incubated with 15 μL of Dynabeads protein G (ThermoFisher, United States), which were pre-washed with wash buffer (100 mM Tris-HCl pH 7.4, 2 mM EDTA, 150 mM NaCl, 1% Triton X100, 0.2 mM sodium orthovanadate), and with 1 ug of mouse anti-CryZ antibody (2A9DD1, Abcam) or mouse IgG (Santa Cruz, United States) for 18 h at 4°C. Supernatants were then collected, as internal protein control samples, and beads were washed four times using wash buffer. After washing, 30% of beads were used to elute protein for subsequent Western Blotting analysis of immunoprecipitation, and the remaining 70% of beads were subjected to RNA extraction. To this aim, beads were incubated for 15 min at 55 °C in 100 μL of NT2 buffer (50 mM Tris-HCl pH 7.4, 1 mM MgCl2, 150 mM NaCl, 0.05% Nonidet P-40) supplemented with 0.5 mg/mL of proteinase K and 0.1% SDS. 1 ng of in vitro transcribed R. luciferase RNA (as internal control) was added, and total RNA was extracted using RNAqueous total RNA Isolation kit (ThermoFisher, United States). Equal volumes of RNA were retrotranscribed using iScript kit (Biorad). Quantification of Bcl-2, Bcl-xl, beta-2 microglobulin and R. luciferase levels was performed through real-time PCR with specific primers (listed in Supplementary Table S1) carried out using SsoAdvancedTM universal SYBR® Green Supermix (Bio-Rad, United States) in a CFX96 Real Time Detection System instrument (Bio-Rad, United States).
Quantitative real-time PCR
Total RNA was isolated from A2780S and A2780R cells by means of NucleoSpin Total RNA kit according to the manufacturer’s protocol (Macherey-Nagel). RNA was quantified by Qubit™ 3.0 Fluorometer (Invitrogen). 500 ng of total RNA were retrotranscribed using iScript kit (Biorad). Quantification of Bcl-2, Bcl-xl and beta-2 microglobulin was performed through real-time PCR with specific primers (listed in Supplementary Table S1) carried out using SsoAdvancedTM universal SYBR® Green Supermix (Bio-Rad) in a CFX96 Real Time Detection System instrument (Bio-Rad). mRNA was quantified with the ΔΔCt method. mRNA levels were normalized to beta-2 microglobulin as an endogenous control.
Cytotoxicity and drug resistance reversal assays
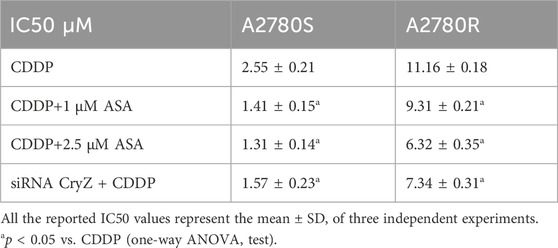
The growth inhibitory effects of ASA and CryZ-siRNA in combination with cisplatin (CDDP) were evaluated in A2780S and A2780R after 72 h treatment by means of the sulforhodamine B (SRB) assay. Briefly, exponentially growing cells were seeded in 96-well plates in complete RPMI 1640 at a plating density of 4 × 103 cells/well. After 24 h, cells were exposed to ASA at 1 and 2.5 µM (concentration doses comparable to those in vivo (Dovizio et al., 2012), and previously shown not affecting A2780 cell viability (Huang et al., 2016)) with an increase ranging of concentrations from 1 nM to 100 μM of CDDP or CryZ-siRNA 100 nM plus CDDP at the same concentration described above. Each concentration was tested in triplicate. In all cases, after 48-h of drugs or siRNA-drug exposure, cells were fixed with 10% trichloroacetic acid and stained with 0.4% SRB in 1% acetic acid. The SRB fixed to the cells was dissolved in 10 mM Tris-HCl and the absorbance was read at 540 nm on Victor X5 (Perkin Elmer). IC50 values were calculated using GraphPad Prism v7 software (GraphPad Prism La Jolla, CA, United States). The IC50 drug concentration resulting in a 50% reduction in the net protein content (as measured by SRB staining) in drug treated cells as compared to untreated control cells was determined. All the reported IC50 values represent the mean ± SD of three independent experiments. One-way ANOVA test was performed to analyze data.
Statistical analysis
t-Test and one-way ANOVA test were used for statistical comparisons of samples. p values <0.05 were considered statistically significant. All data were analyzed using GraphPad Prism v7 software (GraphPad Prism La Jolla, CA, United States).
Results
Bcl-2, Bcl-xl and CryZ expression levels are higher in resistant-compared to sensitive-A2780 cells
We analysed the expression levels of Bcl-2, Bcl-xl and CryZ genes in the A2780 sensitive and A2780 resistant cells (A2780S and A2780R, respectively) at both RNA and protein levels. As shown in Figure 1A, analysis of transcriptomic data reveals that the A2780R cells show higher expression levels of the three genes compared to the sensitive cells. In agreement with mRNA expression levels, Western Blotting analysis revealed that while in sensitive cells the Bcl-2 protein is apparently absent, its levels result dramatically increased in resistant cells (Figure 1B). In addition, Bcl-xl and CryZ protein levels are higher in A2780R compared to A2780S cells (Figure 1B).
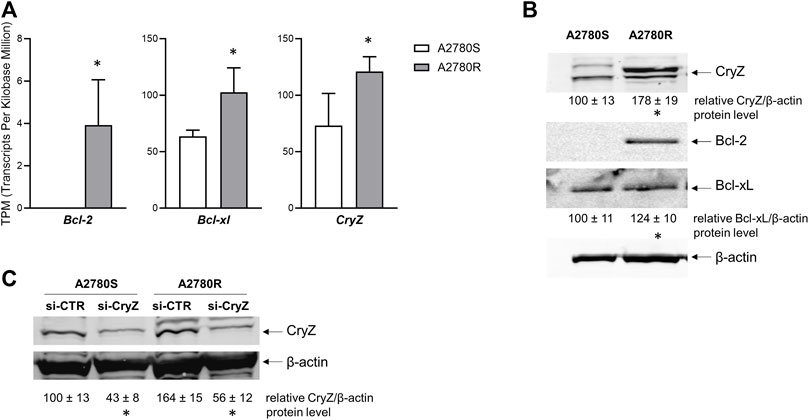
Figure 1. (A) Transcriptomic analysis of Bcl-2, Bcl-xl and CryZ genes in A2780S and A2780R; data represent mean ± SD of at three independent experiments (*p < 0.05 vs. A2780S). (B) Western Blotting analysis of Bcl-2, Bcl-xl and CryZ, including quantitative analysis, in A2780S and A2780R; the image is representative of three different experiments. β-actin was used as housekeeping control (*p < 0.05 vs. A2780S). (C) Western Blotting analysis of CryZ, including quantitative analysis, in A2780S and A2780R transfected with si-CTR or si-CryZ; the image is representative of three different experiments. β-actin was used as housekeeping control (*p < 0.05 vs. si-CTR).
Identification of interaction site of ASA within CryZ
To elucidate a possible interaction site of ASA within CryZ, two computational methods using molecular docking and fragment-mapping based approach were considered, taking advantage of the available crystal structure of the complex formed by CryZ with NADPH (PDB code: 1yb5).
The docking algorithm Autodock 4.0 was used to evaluate the binding energies of minimum-energy conformations of ASA ligand within the whole protein using the largest 3D energy scoring grid available. Docking calculations resulted in a main cluster of 21 out of 50 conformations including the lowest energy one (Supplementary Table S2). All the conformations of the main cluster, including the best docked pose were found within the NADPH binding site, experiencing with the carboxylic group a key salt bridge with Lys324 and hydrogen bonding interactions with Asn48, Met325 and Gly160. The carbonyl oxygen atoms were found establishing a hydrogen bond with NH Val161. As regarding to additional hydrophobic interactions, the best-binding pose of ASA was found in the proximity of the side-chains of Pro49, Gly159, Ile315, Ala321 and Met325 (Figure 2A).
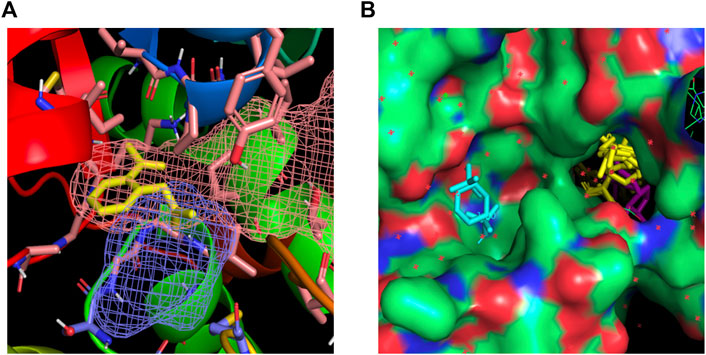
Figure 2. (A) Best-binding pose of ASA found in the proximity of the side-chains of Lys324, Asn48, Met325, Gly160 for polar contacts and of Pro49, Gly159, Ile315, Ala321 and Met325 for hydrophobic interactions. The putative binding site found by FTMap and FTSite evaluations are highlighted in orange and blue mesh and in agreement with docking results. (B) Fragments populating the hotspots identified within the NADPH (magenta) binding region are highlighted in yellow; fragments in cyan are found within an additional hotspot.
FTMap analysis resulted in the identification of six potential hotspots, five of which were found within two regions in the NADPH binding site, specifically in the phosphodiester and nicotinamide sites (Figure 2B). Docking calculation of ASA within the hotspot not found in the NADPH binding region was carried out, too, resulting in ten-fold lower estimated inhibition constant, as a consequence of modest interaction of ASA within this hotspot (best docked pose was found possessing an estimated free energy of binding of −4.1 kcal/mol, results not shown).
The hotspots identified by FTMap were successively computed by FTSite to identify and rank binding sites. The results confirmed the FTMap evaluation and the docking calculation, identifying a binding site within the NADPH binding region. Figure 2A shows the identified binding site highlighted in mesh, along with the key interacting residues that correspond to those found interacting with ASA in the docking calculation.
Such complementary approaches demonstrated ASA interacting within the nicotinamide site of the NADPH binding region of CryZ, that was found by FTMap and FTSite as a strong main hot spot.
ASA impairs the binding of CryZ to Bcl-2 and Bcl-xl mRNAs
As shown in Figures 3A,B, RNA-immunoprecipitation experiments carried out on the A375 cells using the anti-CryZ antibody or unrelated IgG as control demonstrate that treatment with 1 mM ASA for 24 h drastically decreased the amount of Bcl-2 and Bcl-xL mRNAs bound by CryZ, but not b2-microglobulin used as negative control. This data demonstrates that the presence of ASA impairs the binding of CryZ to these mRNAs, probably by interfering with the binding domain of CryZ to the AU-rich regions. In addition, a 72 h treatment of A2780S and A2780R cells with ASA at different concentrations (from 0,2 to 2 mM) determined a significant dose-dependent reduction of Bcl-2 and Bcl-xl mRNA levels (Figure 3C). Based on the evidence that CryZ is a stabilizing post-transcriptional regulator, this result suggests an increase in destabilization of the two mRNAs following ASA treatment, which could affect drug sensitivity.
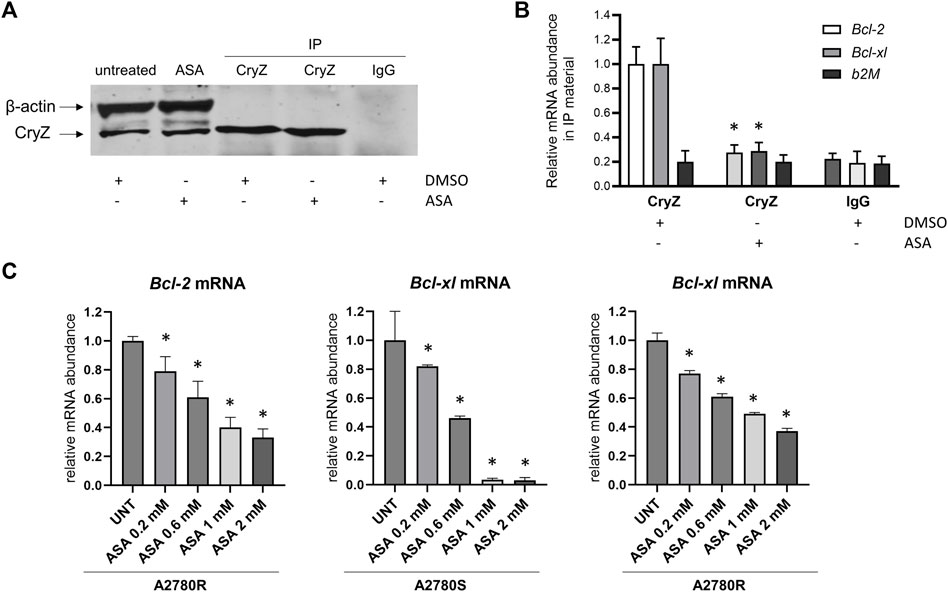
Figure 3. (A) Immunoprecipitation assay of CryZ in extracts of A375 cells treated with ASA or with the DMSO alone; β-actin was used as control. (B) Evaluation of Bcl-2, Bcl-xl and b2M mRNA expression in immunoprecipitated CryZ-RNA complexes by RNA-immunoprecipitation assay. Statistically significant differences from the control detected by one-way ANOVA are shown (*: p < 0.05). (C) Evaluation of Bcl-2 and Bcl-xl mRNA expression in A2780S and A2780R cells following ASA treatment (ranging from 0,2 to 2 mM) for 72 h beta-2 microglobulin has been used as housekeeping gene. Statistically significant differences from the control (UNT) detected by one-way ANOVA are shown (*: p < 0.05).
ASA restores the chemosensitivity to CDDP chemotherapeutics in A2780R cells
Table 1 shows how treatment with ASA (1 and 2.5 µM) together with CDDP at different doses for 72 h in A2780S and A2780R cells significantly decreases the apoptotic threshold, increasing sensitivity to CDDP. To highlight the fact that CryZ may be at the basis of the reversion of the resistant phenotype, we performed cytotoxicity experiments following CryZ silencing (Figure 1C), treating A2780S and A2780R cells with CDDP at different doses for 48 h. As reported in Table 1, CryZ silencing in the A2780R and A2780S cells determined an induction of drug sensitivity, highlighting that CryZ has a major role in drug resistance. Furthermore, these experiments highlight that the inhibition of CryZ binding on the Bcl-2 and Bcl-xl mRNAs by ASA has a fundamental relevance in increasing sensitivity in the A2780S cell line and in reversing resistance in the A2780R cell line.
Discussion
The onset of resistance to chemotherapy drugs is the main determinant of the failure of anticancer chemotherapy in tumours initially sensitive to the drugs (Holohan et al., 2013). Human cell lines resistant to anticancer drugs represent an in vitro useful model for studying the mechanisms of action of drugs and the development of tumour resistance. Here, we show that ASA inhibits the binding of CryZ to the mRNAs of Bcl-2 and Bcl-xl, two important anti apoptotic genes and that it is able to revert the drug resistance in ovarian cancer cells. We previously demonstrated that CryZ is a stabilizing RBP of Bcl-2 (Lapucci et al., 2010), while this is the first demonstration that CryZ is a new RBP of Bcl-xl. The mRNAs of both antiapoptotic genes are physiologically able to bind AUBPs by AU-rich sequence element in the 3′-UTR of their mRNAs, thus modulating the recruitment of the degradation machinery constituted by the exosome complex. Therefore, we demonstrated that CryZ could act as Bcl-2 and Bcl-xl AUBP in A2780 ovarian cancer cells. It was previously demonstrated that ASA is a non-competitive inhibitor of CryZ quinone oxidoreductases activity through the interaction with a binding site closed to that of NADPH (Trabert et al., 2014; Huang et al., 2016). Furthermore, Porte et al. demonstrated that NADPH competitively prevents binding of CryZ to synthetic AU-rich RNA sequences, suggesting that the NADPH binding site is involved in CryZ binding to RNA (Porté et al., 2009). Here, by molecular docking and fragment-mapping based approach, we disclose that ASA could interact with CryZ, particularly identifying a binding site within the NADPH binding region. Moreover, we demonstrated that treatment with ASA determines a significant decrease of Bcl-2 and Bcl-xl mRNA expression levels in A2780 cells. In addition, ASA remarkably reduces the ability of CryZ to bind to the target mRNAs, thus it could decrease their stability and ultimately lead to a reversal of the resistant phenotype. This is the first demonstration in ovarian cancer cell lines that chemoresistance can be reverted by the modulation of the post-transcriptional mRNA regulatory pathway.
ASA is the pivotal non-steroidal anti-inflammatory drug (NSAID). To date, NSAIDs are one of the best explored candidate as repurposed drugs for cancer prevention and treatment. In addition to ASA, several commercial NSAIDs, such as ibuprofen, diclofenac, celecoxib and indomethacin, have been qualified as potential antitumor drugs, and some of them are in clinical trials for cancer treatment (Sousa et al., 2023). Furthermore, conjugations of different NSAIDs ligands with platinum-based drugs (such as cisplatin- and oxaliplatin based platinum(IV) complexes) has been recently used to overcome platinum-resistance of ovarian cancer cells (Barth et al., 2023).
We are conscious of the limits of this brief communication in that ASA has a number of other effects that can affect drug sensitivity. However, the fact that CryZ silencing mimics the ASA effect at a similar extent suggests that inhibition of CryZ mRNA binding by means of ASA may play a major role. Further studies are needed to finely address mechanistic issues. Considering that the increase of apoptosis resistance is a major player in chemoresistance of ovarian cancer, the impact of molecular characterization of this phenomenon and the identification of putative molecular druggable-targets is of pivotal importance for clinicians. Accordingly, our results could lay the basis for the design of innovative therapies based on CryZ pharmacological inhibition in combination with approved chemotherapies, with the benefit of increasing overall drug effectiveness and reducing side effects.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation. RNA-seq data are deposited at NCBI GEO data repository (GEO dataset GSE270030).
Author contributions
ML: Writing–original draft, Writing–review and editing, Methodology. AT: Writing–review and editing, Data curation, Supervision, formal analysis. GR: Writing–original draft, Writing–review and editing, Conceptualization, Validation. MC: Writing–original draft, Writing–review and editing, Data curation. LP: Writing–review and editing, Methodology, Formal analysis. AP: Data curation, Writing–original draft, Writing–review and editing. AM: Writing–original draft, Writing–review and editing, Methodology. CN: Writing–original draft, Writing–review and editing, Formal Analysis. IL: Writing–original draft, Writing–review and editing, Methodology. NS: Writing–original draft, Writing–review and editing. AL: Writing–original draft, Writing–review and editing, Data curation, Investigation, Supervision, Validation.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Open access funding provided by University of Florence within the CRUI-CARE Agreement.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1377028/full#supplementary-material
References
Alexander, N., Woetzel, N., and Meiler, J. (2011). bcl::Cluster: a method for clustering biological molecules coupled with visualization in the Pymol Molecular Graphics System. IEEE Int. Conf. Comput. Adv. Bio Med. Sci. 2011, 13–18. doi:10.1109/ICCABS.2011.5729867
Barth, M. C., Häfner, N., Runnebaum, I. B., and Weigand, W. (2023). Synthesis, characterization and biological investigation of the platinum(IV) tolfenamato prodrug-resolving cisplatin-resistance in ovarian carcinoma cell lines. Int. J. Mol. Sci. 24 (6), 5718. doi:10.3390/ijms24065718
Bazzi, M. D. (2002). Zeta-crystallin displays strong selectivity for salicylic acid over aspirin. Biochem. Biophys. Res. Commun. 293 (1), 440–445. doi:10.1016/S0006-291X(02)00248-6
Bazzi, M. D., Rabbani, N., and Duhaiman, A. S. (2002). Inhibition of camel lens zeta-crystallin by aspirin and aspirin-like analgesics. Int. J. Biochem. Cell Biol. 34 (1), 70–77. doi:10.1016/s1357-2725(01)00099-1
Brenke, R., Kozakov, D., Chuang, G. Y., Beglov, D., Hall, D., Landon, M. R., et al. (2009). Fragment-based identification of druggable ’hot spots' of proteins using Fourier domain correlation techniques. Bioinformatics 25 (5), 621–627. doi:10.1093/bioinformatics/btp036
Colombo, N., Sessa, C., du Bois, A., Ledermann, J., McCluggage, W. G., McNeish, I., et al. (2019). ESMO-ESGO consensus conference recommendations on ovarian cancer: pathology and molecular biology, early and advanced stages, borderline tumours and recurrent disease. Ann. Oncol. 30 (5), 672–705. doi:10.1093/annonc/mdz062
Cornelison, R., Llaneza, D. C., and Landen, C. N. (2017). Emerging therapeutics to overcome chemoresistance in epithelial ovarian cancer: a mini-review. Int. J. Mol. Sci. 18 (10), 2171. doi:10.3390/ijms18102171
Cui, J., and Placzek, W. J. (2018). Post-transcriptional regulation of anti-apoptotic BCL2 family members. Int. J. Mol. Sci. 19 (1), 308. doi:10.3390/ijms19010308
Dai, C., Dai, S. Y., Gao, Y., Yan, T., Zhou, Q. Y., Liu, S. J., et al. (2023). Circ_0078607 increases platinum drug sensitivity via miR-196b-5p/GAS7 axis in ovarian cancer. Epigenetics 18 (1), 2175565. doi:10.1080/15592294.2023.2175565
Donnini, M., Lapucci, A., Papucci, L., Witort, E., Jacquier, A., Brewer, G., et al. (2004). Identification of TINO: a new evolutionarily conserved BCL-2 AU-rich element RNA-binding protein. J. Biol. Chem. 279 (19), 20154–20166. doi:10.1074/jbc.M314071200
Dovizio, M., Tacconelli, S., Sostres, C., Ricciotti, E., and Patrignani, P. (2012). Mechanistic and pharmacological issues of aspirin as an anticancer agent. Pharm. (Basel) 5 (12), 1346–1371. doi:10.3390/ph5121346
Elwood, P., Morgan, G., Watkins, J., Protty, M., Mason, M., Adams, R., et al. (2024). Aspirin and cancer treatment: systematic reviews and meta-analyses of evidence: for and against. Br. J. Cancer 130 (1), 3–8. doi:10.1038/s41416-023-02506-5
Gillet, A., Sanner, M., Stoffler, D., and Olson, A. (2005). Tangible interfaces for structural molecular biology. Structure 13 (3), 483–491. doi:10.1016/j.str.2005.01.009
Holohan, C., Van Schaeybroeck, S., Longley, D. B., and Johnston, P. G. (2013). Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer 13 (10), 714–726. doi:10.1038/nrc3599
Huang, Y., Lichtenberger, L. M., Taylor, M., Bottsford-Miller, J. N., Haemmerle, M., Wagner, M. J., et al. (2016). Antitumor and antiangiogenic effects of aspirin-PC in ovarian cancer. Mol. Cancer Ther. 15 (12), 2894–2904. doi:10.1158/1535-7163.MCT-16-0074
Karademir, D., and Özgür, A. (2023). Small molecule heat shock protein 27 inhibitor J2 decreases ovarian cancer cell proliferation via induction of apoptotic pathways. Med. Oncol. 40 (9), 250. doi:10.1007/s12032-023-02126-2
Kozakov, D., Grove, L. E., Hall, D. R., Bohnuud, T., Mottarella, S. E., Luo, L., et al. (2015). The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat. Protoc. 10 (5), 733–755. doi:10.1038/nprot.2015.043
Lapucci, A., Donnini, M., Papucci, L., Witort, E., Tempestini, A., Bevilacqua, A., et al. (2002). AUF1 Is a bcl-2 A + U-rich element-binding protein involved in bcl-2 mRNA destabilization during apoptosis. J. Biol. Chem. 277 (18), 16139–16146. doi:10.1074/jbc.M201377200
Lapucci, A., Lulli, M., Amedei, A., Papucci, L., Witort, E., Di Gesualdo, F., et al. (2010). zeta-Crystallin is a bcl-2 mRNA binding protein involved in bcl-2 overexpression in T-cell acute lymphocytic leukemia. FASEB J. 24 (6), 1852–1865. doi:10.1096/fj.09-140459
Lulli, M., Nencioni, D., Papucci, L., and Schiavone, N. (2020). Zeta-crystallin: a moonlighting player in cancer. Cell Mol. Life Sci. 77 (6), 965–976. doi:10.1007/s00018-019-03301-3
Mini, E., Lapucci, A., Perrone, G., D’Aurizio, R., Napoli, C., Brugia, M., et al. (2019). RNA sequencing reveals PNN and KCNQ1OT1 as predictive biomarkers of clinical outcome in stage III colorectal cancer patients treated with adjuvant chemotherapy. Int. J. Cancer 145 (9), 2580–2593. doi:10.1002/ijc.32326
Morgan, R. J., Armstrong, D. K., Alvarez, R. D., Bakkum-Gamez, J. N., Behbakht, K., Chen, L. M., et al. (2016). Ovarian cancer, version 1.2016, NCCN clinical practice Guidelines in Oncology. J. Natl. Compr. Canc Netw. 14 (9), 1134–1163. doi:10.6004/jnccn.2016.0122
Morris, G. M., Goodsell, D. S., Halliday, R. S., Huey, R., Hart, W. E., Belew, R. K., et al. (1998). Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19, 1639–1662. doi:10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B
Mortensen, B. T., Jensen, P. O., Helledie, N., Iversen, P. O., Ralfkiaer, E., Larsen, J. K., et al. (1998). Changing bone marrow micro-environment during development of acute myeloid leukaemia in rats. Br. J. Haematol. 102 (2), 458–464. doi:10.1046/j.1365-2141.1998.00801.x
Ngan, C. H., Hall, D. R., Zerbe, B. S., Grove, L. E., Kozakov, D., and Vajda, S. (2012). FTSite: high accuracy detection of ligand binding sites on unbound protein structures. Bioinformatics 28, 286–287. doi:10.1093/bioinformatics/btr651
Pedretti, A., Villa, L., and Vistoli, G. (2004). VEGA--an open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J. Comput. Aided Mol. Des. 18 (3), 167–173. doi:10.1023/b:jcam.0000035186.90683.f2
Porté, S., Crosas, E., Yakovtseva, E., Biosca, J. A., Farrés, J., Fernández, M. R., et al. (2009). MDR quinone oxidoreductases: the human and yeast zeta-crystallins. Chemico-biological Interact. 178 (1-3), 288–294. doi:10.1016/j.cbi.2008.10.018
Sousa, S. M., Xavier, C. P. R., Vasconcelos, M. H., and Palmeira, A. (2023). Repurposing some of the well-known non-steroid anti-inflammatory drugs (NSAIDs) for cancer treatment. Curr. Top. Med. Chem. 23 (13), 1171–1195. doi:10.2174/1568026623666230130150029
Suh, D. H., Park, W. H., Kim, M., Kim, K., No, J. H., and Kim, Y. B. (2023). HOXB9 overexpression confers chemoresistance to ovarian cancer cells by inducing ERCC-1, MRP-2, and XIAP. Int. J. Mol. Sci. 24 (2), 1249. doi:10.3390/ijms24021249
Tenenbaum, S. A., Lager, P. J., Carson, C. C., and Keene, J. D. (2002). Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods 26 (2), 191–198. doi:10.1016/S1046-2023(02)00022-1
Trabert, B., Ness, R. B., Lo-Ciganic, W. H., Murphy, M. A., Goode, E. L., Poole, E. M., et al. (2014). Aspirin, nonaspirin nonsteroidal anti-inflammatory drug, and acetaminophen use and risk of invasive epithelial ovarian cancer: a pooled analysis in the Ovarian Cancer Association Consortium. J. Natl. Cancer Inst. 106 (2), djt431. doi:10.1093/jnci/djt431
Keywords: ovarian cancer, chemoresistance, pharmacology, acetylsalicylic acid, RNA binding proteins
Citation: Lulli M, Trabocchi A, Roviello G, Catalano M, Papucci L, Parenti A, Molli A, Napoli C, Landini I, Schiavone N and Lapucci A (2024) Targeting z-Crystallin by aspirin restores the sensitivity to cisplatin in resistant A2780 ovarian cancer cells. Front. Pharmacol. 15:1377028. doi: 10.3389/fphar.2024.1377028
Received: 26 January 2024; Accepted: 16 April 2024;
Published: 03 July 2024.
Edited by:
Qingbin Cui, University of Toledo College of Medicine and Life Sciences, United StatesReviewed by:
Ruo Wang, Shanghai Jiao Tong University, ChinaRasha Cosman, St Vincent’s Hospital Sydney, Australia
Yanning Hao, United States Food and Drug Administration, United States
Copyright © 2024 Lulli, Trabocchi, Roviello, Catalano, Papucci, Parenti, Molli, Napoli, Landini, Schiavone and Lapucci. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Lapucci, andrea.lapucci@unifi.it; Nicola Schiavone, nicola.schiavone@unifi.it
 Matteo Lulli
Matteo Lulli Andrea Trabocchi2
Andrea Trabocchi2 Giandomenico Roviello
Giandomenico Roviello Martina Catalano
Martina Catalano Astrid Parenti
Astrid Parenti Nicola Schiavone
Nicola Schiavone Andrea Lapucci
Andrea Lapucci