
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 14 June 2024
Sec. Ethnopharmacology
Volume 15 - 2024 | https://doi.org/10.3389/fphar.2024.1371890
 Luyi Shen1†
Luyi Shen1† Min Zhang1†
Min Zhang1† Yanmei Qiu2
Yanmei Qiu2 Lin Yang3
Lin Yang3 Yiwen Lu1
Yiwen Lu1 Hua Li4
Hua Li4 Leilei Zhang1
Leilei Zhang1 Fan Tang1
Fan Tang1 Feijuan Wang1
Feijuan Wang1 Cheng Zhu1
Cheng Zhu1 Hexigeduleng Bao1,4*
Hexigeduleng Bao1,4* Yanfei Ding1*
Yanfei Ding1*Introduction: Rhubarb is a frequently used and beneficial traditional Chinese medicine. Wild resources of these plants are constantly being depleted, meaning that rhubarb products have been subjected to an unparalleled level of adulteration. Consequentially, reliable technology is urgently required to verify the authenticity of rhubarb raw materials and commercial botanical drugs.
Methods: In this study, the barcode-DNA high-resolution melting (Bar-HRM) method was applied to characterize 63 rhubarb samples (five Polygonaceae species: Rheum tanguticum, Rh. palmatum, Rh. officinale, Rumex japonicus and Ru. sp.) and distinguish the rhubarb contents of 24 traditional Chinese patent medicine (TCPM) samples. Three markers, namely ITS2, rbcL and psbA-trnH, were tested to assess the candidate DNA barcodes for their effectiveness in distinguishing rhubarb from its adulterants. A segment from ITS2 was selected as the most suitable mini-barcode to identify the botanical drug rhubarb in TCPMs. Then, rhubarbs and TCPM samples were subjected to HRM analysis based on the ITS2barcode.
Results: Among the tested barcoding loci, ITS2 displayed abundant sites of variation and was effective in identifying Polygonaceae species and their botanical origins. HRM analysis based on the ITS2 mini-barcode region successfully distinguished the authenticity of five Polygonaceae species and eight batches of TCPMs. Of the 18 TCPM samples, 66.7 % (12 samples) were identified as containing Rh. tanguticum or Rh. officinale. However, 33.3 % were shown to consist of adulterants.
Conclusions: These results demonstrated that DNA barcoding combined with HRM is a specific, suitable and powerful approach for identifying rhubarb species and TCPMs, which is crucial to guaranteeing the security of medicinal plants being traded internationally.
Rheum (Polygonaceae), commonly known as rhubarb, or Radix et Rhizoma Rhei, grows widely in Asia’s high mountainous areas under temperate and subtropical climates (Zhou et al., 2020). The Shennong Herbal Classic, written in 270 BC, contains the first recorded uses of rhubarb as a traditional Chinese medicine (TCM) (Zhou et al., 2018; Xiang et al., 2020). Modern pharmacological research suggests that rhubarb exhibits antitumor, anti-inflammatory, anti-diabetic and neuroprotective effects (Deitersen et al., 2019; Dong et al., 2020; Zhao et al., 2021). Although the genus contains about 60 species, according to the Chinese Pharmacopoeia (2020), rhubarb is defined as the dried rhizomes and roots of Rheum tanguticum Maxim.ex Balf., Rheum palmatum L. and Rheum officinale Baill. (State Pharmacopoeia Committee, 2020). Conversely, the underground parts of Rheum rhabarbarum L., Rheum hotaoense C.Y.Cheng & T.C.Kao, Rheum austral D.Don, Rheum undulatum L. and Rumex japonicus Houtt. are used as folk medicines under the name Tu-dahuang in China (Yang et al., 2004). Due to the confusion that stems from these different definitions, the rhubarb raw materials available in China are of varying quality. Since the majority of botanical drugs in the consumer market are composed of plant fragments, slices, or even powders, it has frequently been observed that rhubarb products are purposely or unintentionally blended with species that are closely related. Hence, there is a growing need for trustworthy techniques that provide the ability to verify the authenticity of the raw botanical materials and commercial botanical drugs.
Identification of rhubarb is based primarily on morphological traits, microscopic identification, and chemical profiling, which are often affected by plant growth and environmental conditions, causing difficulty in distinguishing rhubarb in processed botanical drugs (Yang et al., 2004; Zhou et al., 2018). As DNA is present in all tissues and unaffected by external or physiological conditions, molecular biology techniques have proven to be effective in the clear identification of various plant species (Sucher and Maria, 2008). DNA barcoding, which entails the sequencing of a specific and standard DNA segment, was originally proposed by Hebert et al. (2003) and is effective when used for species identification. Because of its high diversity and mutation rate, ITS2 has been shown in recent research to be a promising candidate DNA barcode for the identification and classification of many different plant taxa (Zhao et al., 2015; Zhu et al., 2022). The use of ITS2 region is 73.68% effective in identifying species and has been shown to perform best in the characterization of Bupleurum L. members. Moreover, psbA-trnH can be used as a supplementary site to improve the success rate in Bupleurum L. identification (Chao et al., 2014). The chloroplast matK region was shown to be effective for use in the identification of rhubarb species (Yang et al., 2004; Zhang et al., 2014). Chen et al. (2022) found that 14 sites containing nucleotide alterations in matK and 8 in rpl32, which were found to be valuable in the identification of four Rheum species (Rh. tanguticum, Rh. palmatum, Rh. officinale and Rh. undulatum L. Since DNA barcoding is based on sequencing results, it is an accurate, albeit costly, method.
DNA barcoding and a non-sequencing-based methodology like high-resolution melting (HRM) analysis combine to provide an analytical method that is more dependable, practical, and cost-effective (Chen et al., 2021). Barcode-DNA HRM (Bar-HRM) analysis is noted for its use in the detection of botanical drug adulterants. The identification of species contained in crude drug samples of Senna alexandrina Mill. using HRM based on the ITS1 region was attempted initially by Mishra et al. (2018). Furthermore, Xiong et al. (2016) assessed Bar-HRM’s effectiveness at differentiating between Hyoscyamus niger L. and its adulterants. Their findings demonstrated that the method could be used to identify adulterants effectively and determine the amount of H. niger DNA extract in an admixture.
Rhubarb is a valuable botanical drug and health supplement that is traded throughout the world. However, the contents of commercial rhubarb products, such as traditional Chinese patent medicines (TCPMs), are difficult to identify following the multiple processing steps that take place when creating the formulations, causing difficulties for customs inspectors and trade supervisors. It is crucial to authenticate rhubarb quickly and accurately to guarantee drug safety in the global botanical drugs market. There are currently no reports of Rheum species identification using the HRM technique. In our study, we aimed to create suitable rhubarb DNA barcodes for creating Bar-HRM markers for the quick identification of rhubarb and its adulterants, as well as the contents of rhubarb-based TCPMs.
Three official Rheum species and their adulterants were represented by 63 fresh root, leaf and stem samples gathered from various parts of China, including Gansu, Qinghai, Sichuan, Zhejiang, and Beijing (Table 1). All samples were authenticated by Prof. Dequan Zhang from the College of Life Science and Technology, Southwest University of Science and Technology. All samples were stored at −20°C for subsequent use. In addition, eight batches of TCPMs, comprising 24 samples containing rhubarb, were obtained as tablets, honeyed pills, water pills, concentrated pills, and granules from drug stores (Table 2). The samples of TCPM were ground and stored in a desiccator for further use.
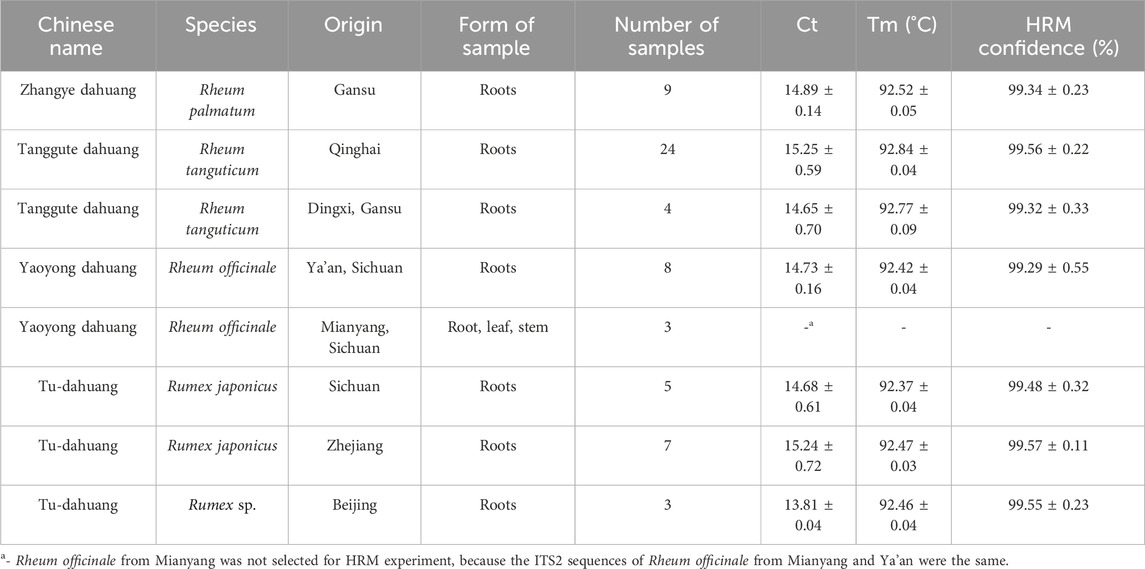
Table 1. Information of rhubarb species used in our study, and the results of real-time PCR and HRM analysis using a ITS2 mini-barcode.
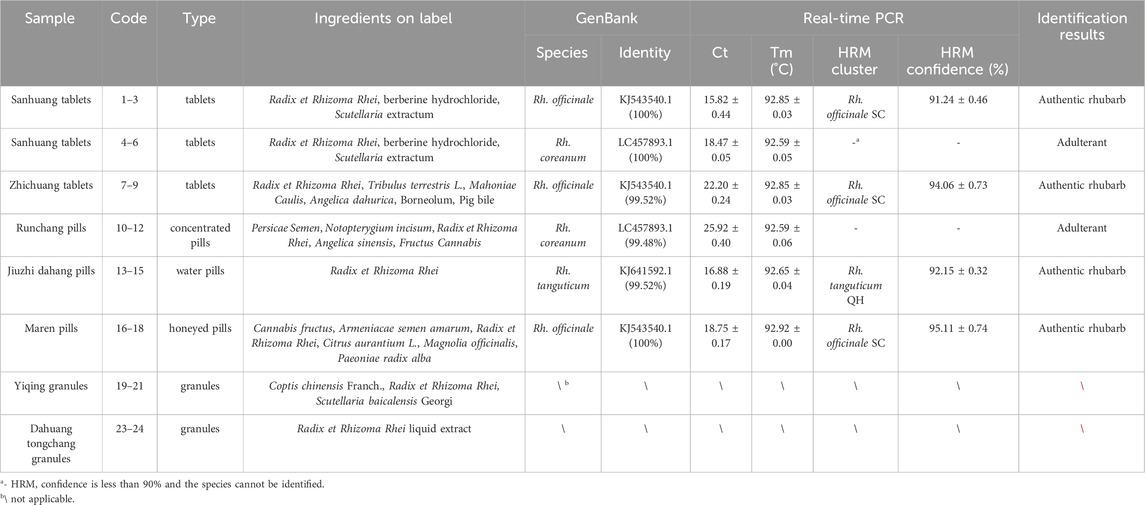
Table 2. Information of the traditional Chinese patent medicines containing rhubarb, and the results of real-time PCR and HRM analysis targeting ITS2 mini-barcode.
Fresh rhubarb plant samples were chopped and thoroughly ground in a mortar and pestle, and then 100 mg was taken for DNA extraction using the method of Cui et al. (2006). 2 g of powdered rhubarb TCPMs was taken for DNA extraction by the method of Cheng et al. (2015). The purity and concentration of the DNA was measured using a standard spectrophotometric method at 260 and 280 nm UV using a Nano-100 spectrophotometer (Aosheng Instrument Company, Hangzhou, China). All DNA samples were stored at −20 °C for subsequent analysis after being diluted to 50 ng/μL.
Three candidate barcodes, ITS2, psbA-trnH and rbcL, were chosen for PCR amplification utilizing universal primers (Supplementary Table S1). A total PCR reaction volume of 25 μL consisted of 50 ng of DNA, 1 μL of each primer (10 μmol/L), and 12.5 μL of 2X SanTaq PCR Mix (Shanghai sangon Biotech Co., Shanghai, China). The reactions were performed with a Thermal Cycler Block 5020 (Thermo Fisher scientific, United States). A 1% agarose gel stained with GoldView II was used to separate the amplified products for 25 min at 120 V and visualized under Tanon-5200 Multi (Tanon, Shanghai, China).
Beijing Tsingke Biotech Co. sequenced the PCR products. Chromas software was used to remove low quality sequences. BLAST tools were used to compare the sequences in GenBank and determine whether they showed high similarity. Then, the obtained DNA sequences, ITS2, rbcL and psbA-trnH, were compared for sequence lengths, variant sites and guanine-cytosine (GC) contents differences using MEGA 11.0 software. Intraspecific variation and interspecific differences were also calculated using the Kimura 2-parameter (K2P) model. In addition, we downloaded a total of 11 sequences of six Rheum species from GenBank. The GenBank accession numbers of the downloaded sequences have been listed in the Supplementary Table S2. The K2P model was utilized in MEGA 11.0 to construct a phylogenetic tree by neighbor-joining (NJ) with bootstrap testing of 1,000 replicates.
The TCPMs’ DNA was severely damaged, making it challenging to extract complete ITS2 sequences. To conduct the HRM experiments, mini-barcode primers (ITS2-39F-293R) were designed in conserved regions based on ITS2 sequence using Primer Premier 5 software (Figure 2). The real-time PCR reaction mixtures contained 10 μL of 2× EvaGreen HRM premix (TIANGEN Biotech), 0.4 μL of each primer (10 μmol/L), and 50 ng of DNA. DNA amplification was performed in a Rotor-Gene Q (QIAGEN, Germany) with real-time PCR conditions: 95 °C 10 min, followed by 45 cycles of 95 °C 5 s and 59 °C 10 s. At the conclusion of every cycle, the fluorescence signals were recorded. The temperature was set to increase from 80°C to 97°C at 0.1°C every 2 s. Each sample had three replicates. The melting curves were analyzed by Rotor-Gene Q Series software, and the normalized and differential melting curves were obtained by setting the confidence interval to 90% (Xiong et al., 2016).
High sequencing efficiency, good PCR amplification, notable inter-species divergences, and low intra-species variance are the ideal characteristics of DNA barcodes (Chao et al., 2014). In our study, the A260/A280 ratios of all DNAs were between 1.78 and 2.04 to satisfy the requirements of subsequent experiments (Supplementary Table S1). Three DNA barcodes, ITS2, psbA-trnH and rbcL, were amplified from 63 rhubarb individuals, encompassing five Polygonaceae species. The amplified samples were all obtained successfully according to the PCR results (Supplementary Figure S1). The sequencing efficiencies of ITS2, rbcL, and psbA-trnH were 83.87%, 90.32%, and 66.12%, respectively. The sequence lengths in alignment were 233-384bp for ITS2, 824 bp for rbcL, and 311–434 bp for psbA-trnH (Supplementary Table S3). After calculations, the GC contents of the psbA-trnH and rbcL sequences were found to range from 32.26% to 32.80% and 41.38%–41.99%, respectively. Additionally, there were 27 (3.26%) and 96 (21.92%) variant sites in the psbA-trnH and rbcL sequences, respectively. The ITS2 sequence had 146 variation sites (37.34%) with a range of GC contents 57.26% to 68.67%. Yao et al. (2010) and Liu et al. (2023) showed that the distribution of GC contents on the plant phylogeny was heterogeneous. In our study, ITS2 contained the highest percentage of variable sites, indicating that this sequence was the most suitable for distinguishing rhubarb species.
A suitably broad DNA barcoding gap is necessary for accurate DNA barcoding (Kvist, 2016). The genetic divergence between five Polygonaceae species determined using the K2P model was shown in Supplementary Table S4. The genetic distances revealed a glaring barcoding gap, and the maximum intraspecific genetic distance seen using ITS2 was less than the minimum interspecific genetic distance. As for rbcL and psbA-trnH, the minimum interspecific genetic distances were lower than the maximum intraspecific genetic distances; thus, the intraspecific and interspecific genetic distances overlapped. Therefore, the rbcL and psbA-trnH segments were not ideal for use in identifying rhubarb species. We constructed NJ trees of three kinds of sequences using Angelica sinensis (Oliv.) Diels as outgroup (Figure 1). Of the three NJ trees, the majority of branches were independent at the species level in the ITS2 topology. Every rhubarb species was placed in a distinct category that did not overlap with any other species. It is worth noting that the ITS2 sequence distinguished Rh. tanguticum and Rumex from different origins, whereas the rbcL and psbA-trnH segments could not accurately distinguish all species, and the species classification in the phylogenetic trees was chaotic. Based on an analysis of genetic distances using the NJ tree, it was concluded that ITS2 was the most viable barcode for the identification of the Polygonaceae species.
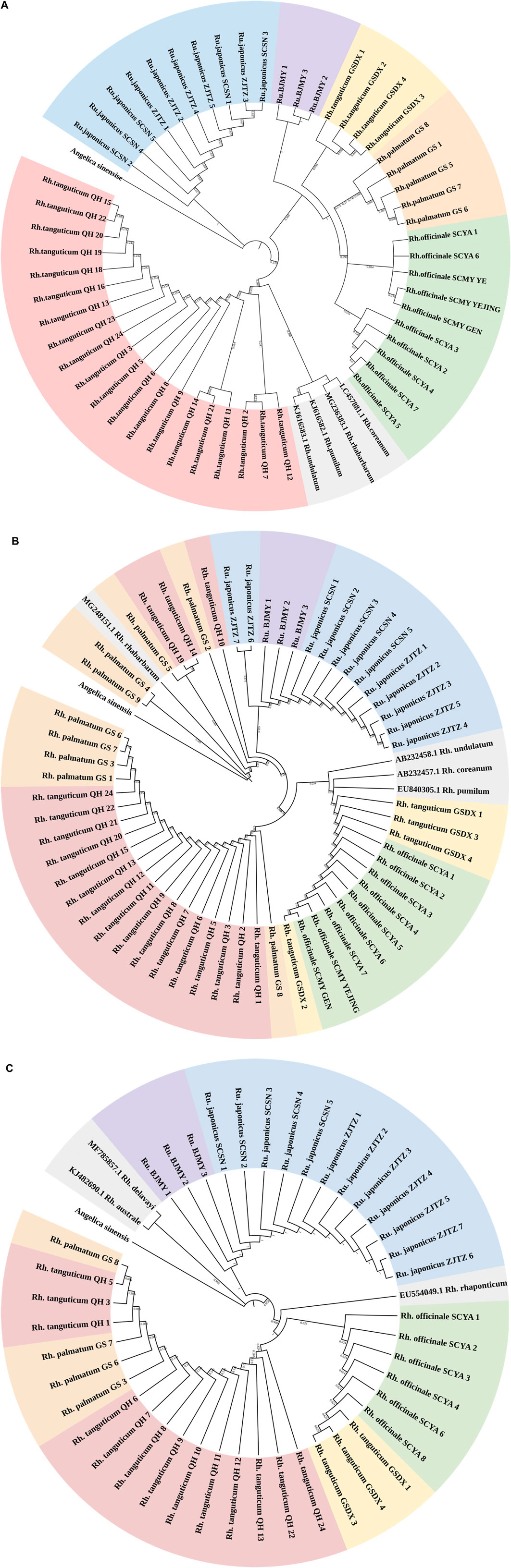
Figure 1. (Continued). NJ tree of Polygonaceae species based on ITS2 (A), rbcL (B) and psbA-trnH (C) sequences. Red, yellow, orange, green, blue, purple, and gray regions represent Rheum tanguticum from Qinghai, Rh. tanguticum from Gansu, Rh. palmatum from Gansu, Rh. officinale from Sichuan, Rumex japonicus from Zhejiang and Sichuan, and Rumex sp. from Beijing, and sequences downloaded from GenBank. Angelica sinensis as an outgroup.
We collected eight batches of TCPMs from drug stores in various geographic areas of China. The DNA of the TCPMs was severely damaged, making it challenging to extract the whole ITS2 sequence. Thus, a variable region of 247 bp was chosen from the ITS2 sequence to produce a mini-barcode. Primers (ITS2-39F-293R) were designed for this region (39–293 bp) of the alignment sequence (Figure 2). ITS2 mini-barcode sequences were amplified successfully in six batches of TCPMs, although not in the remaining batch, Dahuang tongchang (TC) and Yiqing (YQ) granule samples (Supplementary Figure S2). Table 2 showed the all comparisons. The BLAST results showed that tablet samples 1–3, 7–9 and pill samples 16–18 were composed of Rh. officinale, and pill samples 13–15 were composed of Rh. tanguticum. However, tablet samples 4-6 and pill samples 10–12 were composed of Rheum coreanum Nakai, although they were labeled as containing official rhubarb species. These findings showed that the ITS2 mini-barcode could be used to identify TCPMs.

Figure 2. Alignment of ITS2 sequences among Rh. palmatum, Rh. tanguticum, Rh. officinale, Ru. japonicus, and Rumex sp.
In this study, HRM assays were conducted using the primers ITS2-39F-293R, based on ITS2 sequences. Table 1 showed the detailed identification results. A melting curve is deemed to be from a different species if the similarity between the curves each species is less than 90%, according to the 90% genotype confidence percentage (GCP) threshold (Xiong et al., 2016). For the melting curve analysis, Rh. palmatum was used as the reference cluster. Obvious disparities were shown in the difference curves of the rhubarb species (Figure 3A, B). The rhubarb samples were sorted into seven clusters that were simple to identify. Of note, Rh. tanguticum from two origins (Qinghai and Gansu) fell into different clusters. These results implied that HRM using the ITS2 mini-barcode could be used to differentiate rhubarb from its adulterants and identify the origin of each sample.
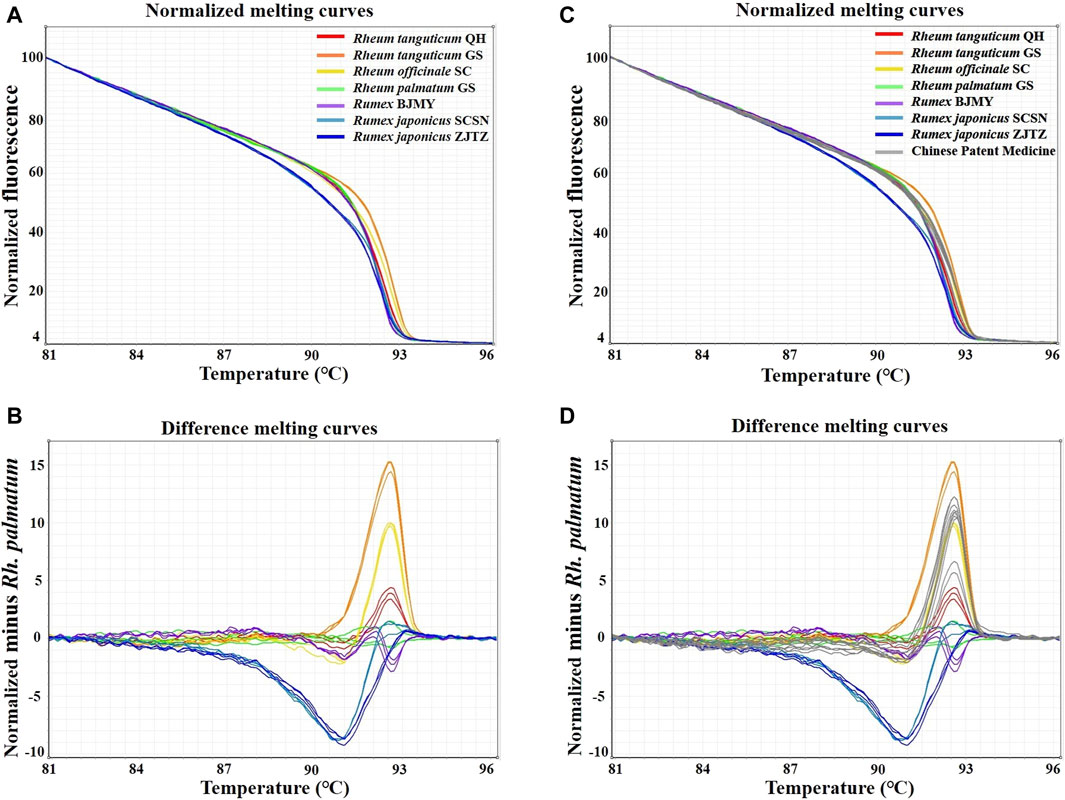
Figure 3. Real-time PCR normalized melting curves and difference melting curves from the HRM analysis of rhubarb species and TCPMs. Rh. palmatum was used as the reference genotype in normalized melting curves (A, B) and difference melting curves (C, D) of all samples. TCPMs include Sanhuang tablets, Zhichuang tablets, Jiuzhi dahang pills, Maren pills and Runchang pills.
Furthermore, the primers ITS2-39F-293R were utilized to carry on HRM analysis on eight batches of TCPMs. All TCPMs can have their resemblances to reference genotypes automatically computed by setting the GCP to 90% and allocating the melting curve of every authenticated rhubarb to the reference genotype. The normalized melting curves and difference melting curves were displayed in Figure 3C, D. The specifics of HRM outcomes were shown in Table 2. Apart from the TC and YQ granule samples, 18 TCPM samples underwent effective amplification, with the Ct values ranging from 15.49 to 26.36. In terms of classification of samples into clusters, the tablet samples 1–3, 7–9 and pill samples 16–18 were classified into the Rh. officinale SC cluster, while the pill samples 13–15 were grouped into the Rh. tanguticum QH cluster. However, tablet samples 4-6 and pill samples 10–12 were not identified as being comprised of official rhubarb species. Based on these findings, 67.7% of the 18 TCPM samples were identified as being composed of official rhubarb, whereas 33.3% of the samples were adulterants or did not contain rhubarb.
As the TCM industry is developing quickly, it is becoming increasingly crucial to have rigorous means to guarantee the efficacy and security of these medicines. However, the components of TCPMs are diverse and the processing methods are complex. Moreover, there are problems with substitution, adulteration, and counterfeiting of these medicines. In China, rhubarb is one of the most widely utilized botanical drugs. In all, the National Medical Products Administration has recorded over nine hundred proprietary TCMs that contain rhubarb (Li et al., 2021). Thus, it is critical to provide a quick, accurate, and dependable way to guarantee TCPM quality.
DNA barcoding has grown in popularity over recent years as a method for the taxonomic identification the plant-based components of botanical drugs, and the technique is crucial to the authenticity of TCMs. Chen et al. (2022) and Zhang et al. (2014) both found that three official rhubarbs and their adulterants could be distinguished by the nucleotide differences in the matK sequence. However, in this study, we found that the sequencing results from the matK sequence in our samples were poor. Among the remaining three candidate barcodes, psbA-trnH and rbcL sequences contained few variation sites and had poor ability to distinguish rhubarb species. A number of other studies have revealed that, because the rbcL and psbA-trnH regions frequently lack variations in related species, they are useful for solving phylogenetic puzzles at higher taxonomic levels but inadequate for species classification (Dong et al., 2012; Chao et al., 2014; Zhu et al., 2022). ITS2 contains a large number of variable sites and a clear barcoding gap to enable official rhubarb to be distinguished from its adulterants. According to our findings, the ITS2 sequence is the ideal choice for rhubarb species identification and shows great potential for origin identification.
In recent years, there has been a big increase in the successful application of the novel and sophisticated technique known as HRM analysis for the identification of TCMs (Li et al., 2018). According to Osathanunkul et al. (2018), Bar-HRM was used to identify adulterants in Ophiocordyceps sinensis products available on the market and distinguish between O. sinensis and O. militaris species. Importantly, previous studies have focused only on the original rhubarb species used upstream in the TCM production chain, whereas there have been few reports on the identification of rhubarb in TCPMs downstream of the production chain (Zhou et al., 2017; Asanuma et al., 2019). At present, DNA barcoding and DNA metabarcoding have been used to identify rhubarb TCPMs (Tian et al., 2014; Xin et al., 2022), but HRM has not been used to identify rhubarb TCPMs. The DNA of the TCPMs used in this study was found to be highly degraded. A mini-barcode, using a sequence (100–250 bp) that was substantially shorter than the original barcode length, was developed to increase the PCR amplification success rate in the DNA-degraded samples (Yang et al., 2022).
In this study, the mini-barcode designed according to ITS2 sequences successfully amplified the target fragments from 18 TCPM samples and identified the specific species of rhubarb contained in them. It was notable that 33.3% of the TCPMs contained an adulterant (Rh. coreanum), although they were labeled as containing official rhubarb. These findings indicated that ITS2 mini-barcode marker was effective in distinguishing TCPMs containing rhubarb and ensure accuracy in the labeling of the contents. This technique could, therefore, potentially help to prevent consumer fraud and the mislabeling of rhubarb goods. However, HRM also has some limitations. Compared with other new technologies such as DNA metabarcoding, HRM is limited by the deviation generated by PCR amplification process, which is easy to affect the changes of the melting curve. Based on the specificity of primers, only single species sequence information of mixed samples can be analyzed (Zou et al., 2023; Cui and Hou, 2024). In the future, HRM could be combined with other methods such as DNA metabarcoding, microfluidic enrichment barcoding and gold nanoparticles to identify TCPMs.
In this work, we proposed DNA barcoding combined with HRM as a highly specific method for distinguishing various rhubarb species (Rh. tanguticum, Rh. palmatum, Rh. officinale and Rumex) and the components of 18 TCPM samples. ITS2 was the most appropriate DNA barcode for identifying rhubarb species and tracking the geographic origins of the species. Furthermore, Bar-HRM analysis showed that 33.3% of the 18 TCPM samples were composed of adulterants. To our knowledge, this is the first study using Bar-HRM technology for the simple, fast, and accurate discrimination of rhubarb species and TCPM contents, ensuring the safety of TCPMs and promoting development of the rhubarb industry.
The 52 ITS2 sequences and 41 psbA-trnH sequences can be found in the NCBI database; accession numbers—PP860965-PP861016 and PP858630-PP858670, respectively. For further queries, please contact the corresponding authors.
LS: Data curation, Formal Analysis, Methodology, Writing–original draft. MZ: Formal Analysis, Investigation, Writing–review and editing. YQ: Data curation, Investigation, Writing–review and editing. LY: Data curation, Resources, Writing–review and editing. YL: Investigation, Writing–review and editing. HL: Formal Analysis, Resources, Writing–review and editing. LZ: Formal Analysis, Resources, Writing–review and editing. FT: Formal Analysis, Resources, Writing–review and editing. FW: Data curation, Investigation, Writing–review and editing. CZ: Funding acquisition, Supervision, Writing–review and editing. HB: Conceptualization, Funding acquisition, Project administration, Supervision, Writing–review and editing. YD: Conceptualization, Funding acquisition, Project administration, Supervision, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported through funding from Science and Technology Program of Zhejiang Province (2022C04002), the Natural Science Foundation of Zhejiang Province (LZ22C130003), the National Natural Science Foundation of China (32272051), the Fundamental Research Funds for the Provincial Universities of Zhejiang (2022YW79), and the fellowship of China Postdoctoral Science Foundation (2022M721644).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1371890/full#supplementary-material
Asanuma, M., Zhu, S., Okura, N., Cai, S. Q., Yoshimatsu, K., and Komatsu, K. (2019). Genetic polymorphism of Japanese cultivated Rheum species in the internal transcribed spacer region of nuclear ribosomal DNA. J. Nat. Med. 73 (3), 541–554. doi:10.1007/s11418-019-01298-4
Chao, Z., Zeng, W. P., Liao, J., Liu, L., Liang, Z. B., and Li, X. L. (2014). DNA barcoding Chinese medicinal Bupleurum. Phytomedicine 21 (13), 1767–1773. doi:10.1016/j.phymed.2014.09.001
Chen, C. T., Ding, Y. F., Wang, Y., Jiang, Q., Wang, F. J., Lu, C. Z., et al. (2021). High-resolution melting analysis of COI sequences distinguishes Pufferfish species (Takifugu Spp.) in China. J. Agric. Food Chem. 69 (2), 794–804. doi:10.1021/acs.jafc.0c06584
Chen, D. W., Zhang, H., Chang, L. B., Jia, L. Y., and Sun, K. (2022). A molecular identification of medicinal Rheum species cultivated germplasm from the northwest of China using DNA barcoding. Genet. Resour. Crop Evol. 69, 997–1008. doi:10.1007/s10722-021-01276-4
Cheng, C. S., Tan, T. Q., Long, Z., Liu, Z. Z., Wu, W. R., Wang, Y. P., et al. (2015). Optimization of DNA extraction for Chinese patent medicine and its application on molecular identification of ginseng preparations by MAS-PCR. Chin. Tradit. Herb. Drugs. 46 (17), 2549–2555. doi:10.7501/j.issn.0253-2670.2015.17.008
Cui, G. H., Tang, X. J., and Huang, L. Q. (2006). Study on the extraction method of DNA from Chinese medicinal materials containing starch and polysaccharides. Chin. J. Chin. Mat. Med. (16), 1365–1367.
Cui, J. L., and Hou, L. (2024). Research and application of DNA barcoding technology in traditional Chinese medicine. Chin. Med. J. Res. Prac. 38 (1), 89–94. doi:10.13728/j.1673-6427.2024.01.016
Deitersen, J., El-Kashef, D., Proksch, P., and Stork, B. (2019). Anthraquinones and autophagy-three rings to rule them all? Bioorg Med. Chem. 27 (20), 115042. doi:10.1016/j.bmc.2019.115042
Dong, W., Liu, J., Yu, J., Wang, L., and Zhou, S. (2012). Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One 7, e35071. doi:10.1371/journal.pone.0035071
Dong, X. X., Zeng, Y. W., Liu, Y., You, L. T., Yin, X. B., Fu, J., et al. (2020). Aloe-emodin: a review of its pharmacology, toxicity, and pharmacokinetics. Phytother. Res. 34 (2), 270–281. doi:10.1002/ptr.6532
Hebert, P. D. N., Alina, C., Shelley, L. B., and Jeremy, R. W. (2003). Biological identifications through DNA barcodes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 270 (1512), 313–321. doi:10.1098/rspb.2002.2218
Kvist, S. (2016). Does a global DNA barcoding gap exist in Annelida? Mitochondrial DNA Part a 27 (3), 2241–2252. doi:10.3109/19401736.2014.984166
Li, J. J., Xiong, C., He, X., Lu, Z., Zhang, X., Chen, X., et al. (2018). Using SSR-HRM to identify closely related species in herbal medicine products: a case study on Licorice. Front. Pharmacol. 9, 407. doi:10.3389/fphar.2018.00407
Li, R., Xin, T., Song, L., Yan, H., Liao, H., Zhou, J., et al. (2021). Research progress in original species identification in industry chain of Rhei Radix et Rhizoma. Zhongguo Zhong Yao Za Zhi 46, 1060–1066. doi:10.19540/j.cnki.cjcmm.20201207.601
Liu, Y., Liang, N., Xian, Q., and Zhang, W. (2023). GC heterogeneity reveals sequence-structures evolution of angiosperm ITS2. BMC Plant Biol. 23 (1), 608. doi:10.1186/s12870-023-04634-9
Mishra, P., Ashutosh, K. S., and Velusamy, S. (2018). Candidate DNA barcode tags combined with High resolution melting (Bar-HRM) curve analysis for authentication of Senna Alexandrina Mill. with validation in crude drugs. Front. Plant Sci. 9 (3), 283. doi:10.3389/fpls.2018.00283
Osathanunkul, M., Osathanunkul, K., Wongwanakul, S., Osathanunkul, R., and Madesis, P. (2018). Multiuse of Bar-HRM for Ophiocordyceps sinensis identification and authentication. Sci. Rep. 8 (1), 12770. doi:10.1038/s41598-018-31164-4
State Pharmacopoeia Committee (2020) Pharmacopoeia of the people’s Republic of China (2020 edition, Part I). Beijing: China Medical Science Press, 24–25.
Sucher, N., and Maria, C. (2008). Genome-based approaches to the authentication of medicinal plants. Planta Medica 74 (6), 603–623. doi:10.1055/s-2008-1074517
Tian, X. X., Liu, J., Dou, Y. J., Zhao, Y. N., Lv, C. G., Zhang, C. H., et al. (2014). The origin identification of Chinese traditional medicine Dahuang Zhechong pill based on metabarcoding method. Tianjin J. Traditional Chin. Med. 31 (4), 234–237. doi:10.11656/j.issn.1672-1519.2014.04.14
Xiang, H., Zuo, J., Guo, F., and Dong, D. (2020). What we already know about rhubarb: a comprehensive review. Chin. Med. 15, 88. doi:10.1186/s13020-020-00370-6
Xin, T., Li, R., Lou, Q., Lin, Y., Liao, H., Sun, W., et al. (2022). Application of DNA barcoding to the entire traditional Chinese medicine industrial chain: A case study of Rhei Radix et Rhizoma. Phytomedicine 105, 154375. doi:10.1016/j.phymed.2022.154375
Xiong, C., Hu, Z. G., Tu, Y., Liu, H. G., Wang, P., Zhao, M. M., et al. (2016). ITS2 barcoding DNA region combined with High resolution melting (HRM) analysis of Hyoscyami Semen, the mature seed of Hyoscyamus Niger. Chin. J. Nat. Med. 14 (12), 898–903. doi:10.1016/S1875-5364(17)30014-6
Yang, D. Y., Hirotoshi, F., Cai, S. Q., and Katsuko, K. (2004). Molecular analysis of Rheum species used as Rhei Rhizoma based on the chloroplast MatK gene sequence and its application for identification. Biol. Pharm. Bull. 27 (3), 375–383. doi:10.1248/bpb.27.375
Yang, X., Yu, X., Zhang, X., Guo, H., Xing, Z., Xu, L., et al. (2022). Development of Mini-barcode based on chloroplast genome and its application in metabarcoding molecular identification of Chinese medicinal material Radix Paeoniae Rubra (Chishao). Front. Plant Sci. 13, 819822. doi:10.3389/fpls.2022.819822
Yao, H., Song, J., Liu, C., Luo, K., Han, J., Li, Y., et al. (2010). Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One 5 (10), e13102. doi:10.1371/journal.pone.0013102
Zhang, X., Liu, Y., Gu, X., Guo, Z., Li, L., Song, X., et al. (2014). Genetic diversity and population structure of Rheum tanguticum (Dahuang) in China. Chin. Med. 9 (1), 26. doi:10.1186/1749-8546-9-26
Zhao, D., Feng, S. X., Zhang, H. J., Zhang, N., Liu, X. F., Wan, Y., et al. (2021). Pharmacokinetics, tissue distribution and excretion of five rhubarb anthraquinones in rats after oral administration of effective fraction of anthraquinones from Rheum Officinale. Xenobiotica 51 (8), 916–925. doi:10.1080/00498254.2021.1940353
Zhao, S., Chen, X., Song, J., Pang, X., and Chen, S. (2015). Internal transcribed spacer 2 barcode: a good tool for identifying Acanthopanacis cortex. Front. Plant Sci. 6, 840. doi:10.3389/fpls.2015.00840
Zhou, T., Zhu, H. H., Wang, J., Xu, Y. X., Xu, F. S., and Wang, X. M. (2020). Complete chloroplast genome sequence determination of Rheum species and comparative chloroplast genomics for the members of Rumiceae. Plant Cell Rep. 39 (6), 811–824. doi:10.1007/s00299-020-02532-0
Zhou, Y., Du, X. L., Zheng, X., Huang, M., Li, Y., and Wang, X. M. (2017). ITS2 barcode for identifying the officinal rhubarb source plants from its adulterants. Biochem. Syst. Ecol. 70, 177–185. doi:10.1016/j.bse.2016.12.004
Zhou, Y. X., Nie, J., Xiao, L., Hu, Z. G., and Wang, B. (2018). Comparative chloroplast genome analysis of Rhubarb botanical origins and the development of specific identification markers. Molecules 23 (11), 2811. doi:10.3390/molecules23112811
Zhu, S., Liu, Q., Qiu, S., Dai, J., and Gao, X. (2022). DNA barcoding: an efficient technology to authenticate plant species of traditional Chinese medicine and recent advances. Chin. Med. 17 (1), 112. doi:10.1186/s13020-022-00655-y
Keywords: rhubarb, DNA barcoding, ITS2, HRM, traditional Chinese patent medicine
Citation: Shen L, Zhang M, Qiu Y, Yang L, Lu Y, Li H, Zhang L, Tang F, Wang F, Zhu C, Bao H and Ding Y (2024) DNA barcoding combined with high-resolution melting analysis to discriminate rhubarb species and its traditional Chinese patent medicines. Front. Pharmacol. 15:1371890. doi: 10.3389/fphar.2024.1371890
Received: 17 January 2024; Accepted: 06 May 2024;
Published: 14 June 2024.
Edited by:
Dâmaris Silveira, University of Brasilia, BrazilReviewed by:
Mengyue Guo, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaCopyright © 2024 Shen, Zhang, Qiu, Yang, Lu, Li, Zhang, Tang, Wang, Zhu, Bao and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hexigeduleng Bao, aHhnQGNqbHUuZWR1LmNu; Yanfei Ding, ZGluZ3lhbmZlaUBjamx1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.