
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol. , 27 June 2024
Sec. Pharmacology of Anti-Cancer Drugs
Volume 15 - 2024 | https://doi.org/10.3389/fphar.2024.1367981
This article is part of the Research Topic Metabolic reprogramming in cancer View all 13 articles
Cancer stem cells (CSCs) represent a small subset of heterogeneous cells within tumors that possess the ability to self-renew and initiate tumorigenesis. They serve as potential drivers for tumor initiation, metastasis, recurrence, and drug resistance. Recent research has demonstrated that the stemness preservation of CSCs is heavily reliant on their unique lipid metabolism alterations, enabling them to maintain their own environmental homeostasis through various mechanisms. The primary objectives involve augmenting intracellular fatty acid (FA) content to bolster energy supply, promoting β-oxidation of FA to optimize energy utilization, and elevating the mevalonate (MVA) pathway for efficient cholesterol synthesis. Additionally, lipid droplets (LDs) can serve as alternative energy sources in the presence of glycolysis blockade in CSCs, thereby safeguarding FA from peroxidation. Furthermore, the interplay between autophagy and lipid metabolism facilitates rapid adaptation of CSCs to the harsh microenvironment induced by chemotherapy. In this review, we comprehensively review recent studies pertaining to lipid metabolism in CSCs and provide a concise overview of the indispensable role played by LDs, FA, cholesterol metabolism, and autophagy in maintaining the stemness of CSCs.
The global burden of cancer in 2019 was estimated at 23.6 million new cases (95% UI, 22.2–24.9 million) and 10 million deaths (95% UI, 9.36–10.6 million), making it the leading cause of mortality worldwide (Global Burden of Disease Cancer et al., 2022). Despite advancements in cancer prevention, it remains a formidable challenge for medical professionals due to the scarcity of cures and the prevalence of metastasis or recurrence among patients. The term “stem cells” refers to a unique population of cells that possess the remarkable abilities of self-renewal and differentiation, enabling them to give rise to diverse cell lineages (Al-Hajj and Clarke, 2004). The majority of cancers are composed of heterogeneous cell populations with varying capacities to induce tumor growth. Only the less differentiated and distinct cell populations within the tumor exhibit a high ability to self-renew and initiate tumorigenesis, which are commonly known as cancer stem cells (CSCs), tumor-propagating cells, or tumor initiating cells (TICs), possessing pluripotency and the capacity to repopulate tumors (Reya et al., 2001) (Figure 1). The CSCs represent a limited population of tumor cells exhibiting stem cell characteristics. CSCs serve as the primordial cells responsible for tumorigenesis, recurrence, and metastasis, with numerous CSCs or CSC-like cells having been identified and isolated from diverse tumors (Prasetyanti and Medema, 2017).The identification of CSCs in leukemia was initially achieved through the experimental technique of xenotransplantation (Bonnet and Dick, 1997). The presence of distinct populations of CSCs has been demonstrated in the majority of blood cancers and solid tumors, including breast, brain, colon cancers, and melanoma (Al-Hajj et al., 2003; Singh et al., 2004; Galli et al., 2004; OBrien et al., 2007; Schatton et al., 2008). The presence of CSCs renders them resistant to cytotoxic treatment and significantly contributes to the tumor’s resistance against radio/chemotherapy (Shiozawa et al., 2013). CSCs also initiate metastasis and drive cancer relapse by their capacity for self-renewal and proliferation, enabling them to expand into the bulk of the tumor (Reya et al., 2001). The pressing demand for the advancement of innovative therapies that efficiently eliminate CSCs remains paramount.
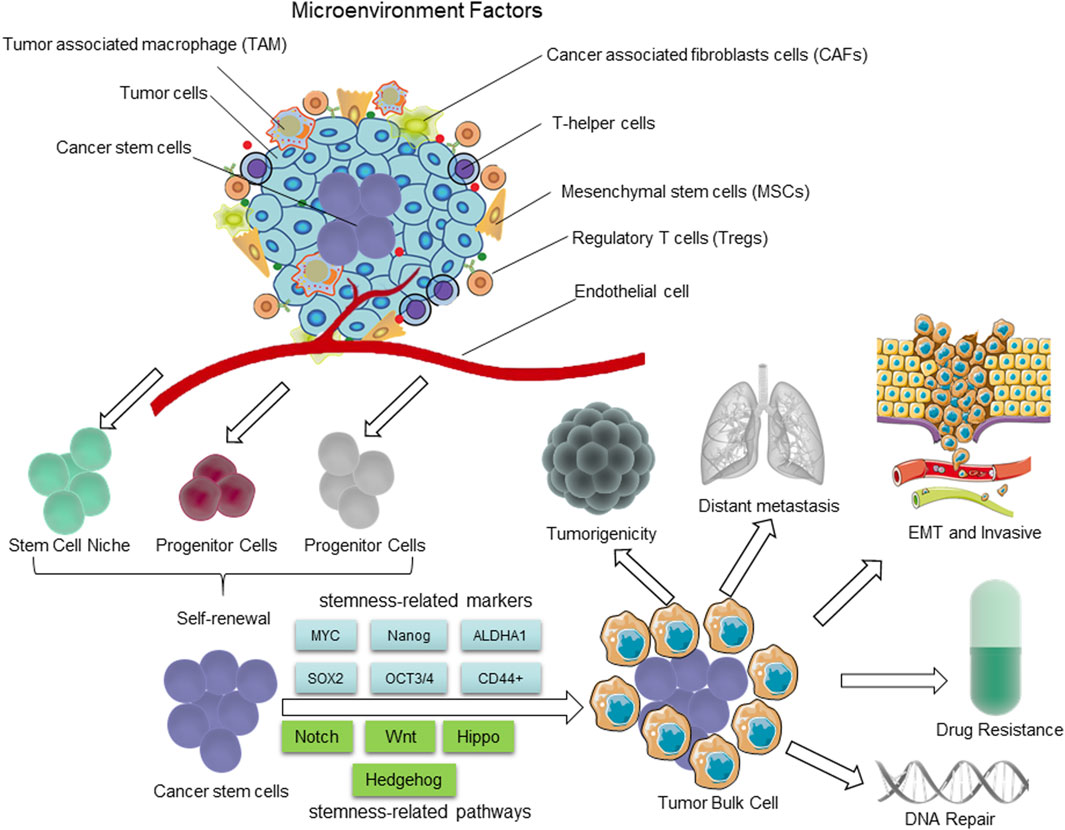
Figure 1. The distinguishing characteristics of CSCs. The presence of CSCs is closely linked to the tumor microenvironment (TME). The self-renewal capability of CSCs is significantly enhanced to certain degrees, comparable to or even surpassing that of normal pluripotent and multipotent stem cells; however, there is a partial loss in their differentiation capacity. During the process of epithelial-mesenchymal transition (EMT), cancer cells acquire characteristics resembling those of stem cells, enabling them to migrate and invade foreign tissues. The hierarchical plasticity subpopulations of CSCs are the primary drivers of tumorigenesis, EMT, tumor progression, metastasis, and drug resistance. The regulation of these CSC characters is mediated by multiple signaling pathways, including Notch, Wnt, Hippo cascades, Hh, etc.
The metabolic reprogramming has emerged as a pivotal hallmark of cancer in recent years (Hanahan and Weinberg, 2011). It has been demonstrated that the initiation and progression of cancer are frequently accompanied by profound metabolic alterations (Cao and Yan, 2020; Martinez-Reyes and Chandel, 2021). Notably, the biological characteristics of TICs and CSCs differ from those of non-CSCs (Dando et al., 2015). The metabolic regulation of ATP synthesis and bioconstruct formation in CSCs differs from that of non-stem cancer cells, but resembles that of stem cells derived from normal tissue (Sancho et al., 2016). The role of metabolism in the biology of CSCs has emerged as a prominent area of research over the past decade, with particular emphasis on lipid metabolism. Various studies have demonstrated the crucial role of lipid metabolism in preserving the stemness of CSCs and fulfilling their energy demands, ultimately contributing to cancer progression (Martinez-Outschoorn et al., 2017). For example, the overexpression of acetyl coenzyme a synthase (ACSL1 and ACSL4) and sterol coenzyme a desaturase (SCD) induces epithelial-mesenchymal transition (EMT) in colorectal cancer, thereby enhancing the migratory and invasive capabilities of tumor cells (Sanchez-Martinez et al., 2015). The overexpression of CD36 enhances the uptake of fatty acids and activates Wnt-dependent EMT in hepatocellular carcinoma (HCC) (Nath et al., 2015). Notably, CSCs may originate from normal stem cells or tissue progenitor cells due to stochastic genetic mutations and epigenetic alterations, with tumor progression associated with genome-wide epigenetic regulation influencing CSC maintenance and survival via diverse pathways (Peiris-Pages et al., 2016). The occurrence of abnormal epigenetic modifications can induce the conversion of normal stem cells into CSCs, for example, the processes of DNA methylation and histone modification play crucial roles in guiding the differentiation of stem cells into specific cell and tissue types (Toh et al., 2017). The differentiation of CSCs can be inhibited through the suppression of gene expression via H3K27me3 modification and/or DNA methylation, in a manner analogous to that observed in ESCs (Easwaran et al., 2012). The crucial role of DNA methylation in maintaining the properties of CSCs in leukemia, lung, and colon has been extensively reported (Broske et al., 2009; Brunetti et al., 2017; Maiuri et al., 2019; Liang et al., 2021). Moreover, DNA methylation plays a pivotal role in facilitating this transformation process through the involvement of DNA methyltransferases (Wongtrakoongate, 2015). Notably, these CSC can maintain their stemness through their specific epigenetic alteration by regulating lipid metabolism, for example, CSCs can sustain their stemness through their super-enhancers by promoting polyunsaturated fatty acid (FA) (PUFA) synthesis (Gimple et al., 2019).
Besides, the epigenetic mechanisms also play a crucial role in regulating several key pathways of CSCs, including the Wnt/β-catenin, Hedgehog (Hh), and Notch signaling pathways. These pathways play a pivotal role in the development and maintenance of normal tissues, as well as in the self-renewal and differentiation of hematopoietic stem cells (Hoffmeyer et al., 2012; Myant et al., 2013) (Figure 1). Additionally, they also regulate the proliferation and maintain the stemness of progenitor cells and CSCs in a variety of tissues through modulating lipid metabolic process (Beachy et al., 2004; Yang et al., 2020; Wang et al., 2022). Furthermore, these pathways through which stem cells can be derived via genetic mutations and epigenetic alterations have a significant potential to be exploited for the maintenance of unrestricted proliferation, invasion, and drug resistance (Reya et al., 2001; Wang et al., 2022). This review presents a comprehensive overview of the metabolism of lipid droplets (LDs), FA, and cholesterol, as well as the impact of autophagy on maintaining stemness in CSCs. Furthermore, we investigate the characteristics and mechanisms of lipid metabolism in CSCs and their role in conferring resistance to radiotherapy.
Over the past decade, numerous studies have been conducted to assess the expression profiles of cancer cells exhibiting stem cell properties in various solid tumors, leading to the identification of a plethora of biomarkers, pathways, and therapeutic targets against CSCs (Medema, 2013; Wang et al., 2014). The principal characteristics of CSCs encompass cell surface adhesion molecules, cytoprotective enzymes, transcription factors, and drug efflux pumps (Medema, 2013). However, the markers of CSCs in one organ or tissue differ from those in other organs or tissues, with only a few shared markers between them. Table 1 provides an overview of the most prevalent molecules that can serve as CSC biomarkers. Recently, the involvement of CSCs is pivotal in driving cancer progression, facilitating metastasis, promoting recurrence, and conferring resistance to cytotoxic therapies. The primary targets of classical radiotherapy and chemotherapy are predominantly fast-proliferating cells (Gerlinger et al., 2012). Unlike normal stem cells, cancer stem cells are believed to be responsible for tumor growth, recurrence, and drug resistance. One of the key characteristics of stem cells is their quiescent or dormant state, indicating infrequent division and prolonged periods in a dormant state. This attribute renders them less susceptible to conventional cancer therapies that primarily target rapidly dividing cells. However, recent studies have indicated that certain CSCs expressing leucine-rich repeat G protein-coupled receptor 5 (LGR5+) may not exhibit complete quiescence (Shiokawa et al., 2020). LGR5+ CSCs have been identified in various types of cancers, including colorectal, liver, and pancreatic cancers. It is believed that these cells undergo regular cell division cycles, indicating their cyclic nature. These findings hold significant implications as they challenge the previous notion of cancer stem cells being quiescent and suggest that targeting these cells may be more effective than conventional therapies aimed at rapidly dividing cells (Higa and Nakayama, 2024). Furthermore, it underscores the importance of comprehending the heterogeneity within the CSC population since different subpopulations may exhibit varying degrees of quiescence and sensitivity to treatment. In conclusion, while the concept of CSCs existing in a dormant state has long been fundamental in cancer research, recent evidence suggests that at least some CSCs, such as LGR5+ CSCs, are not entirely dormant. This discovery carries crucial implications for developing novel therapeutic strategies to target these cells and enhance cancer prognosis.
It is widely recognized that one of the primary challenges in cancer treatment lies in the development of drug resistance. Cellular plasticity has been identified as a pivotal factor contributing to the emergence of drug resistance (Boumahdi and de Sauvage, 2020). Cancer cell plasticity refers to a state wherein non-transformed differentiated cells exhibit adaptive plastic behavior under oncogenic stress (Figure 2). This phenomenon promotes diversity and heterogeneity among cancer cells within tumors, serving as a bypass mechanism to evade therapeutic agents. In the context of Darwinian selection, a vast number of non-CSCs conceals a small population of CSCs. According to the coexisting model theory, dynamic transcriptional fluctuations in individual cells give rise to therapeutic resistance markers, leading to resistance against treatment. The Lamarckian induction concept suggests that epigenetic modifications occurring in a small subset of cancer cells result in alterations of their drug-refractory phenotype and subsequently enhance therapeutic resistance. These changes in phenotype are commonly referred to as cellular plasticity (Vergara et al., 2019; Paul et al., 2022). This phenomenon promotes diversity and heterogeneity of CSCs within the tumor as a bypass mechanism to evade therapeutic agents.
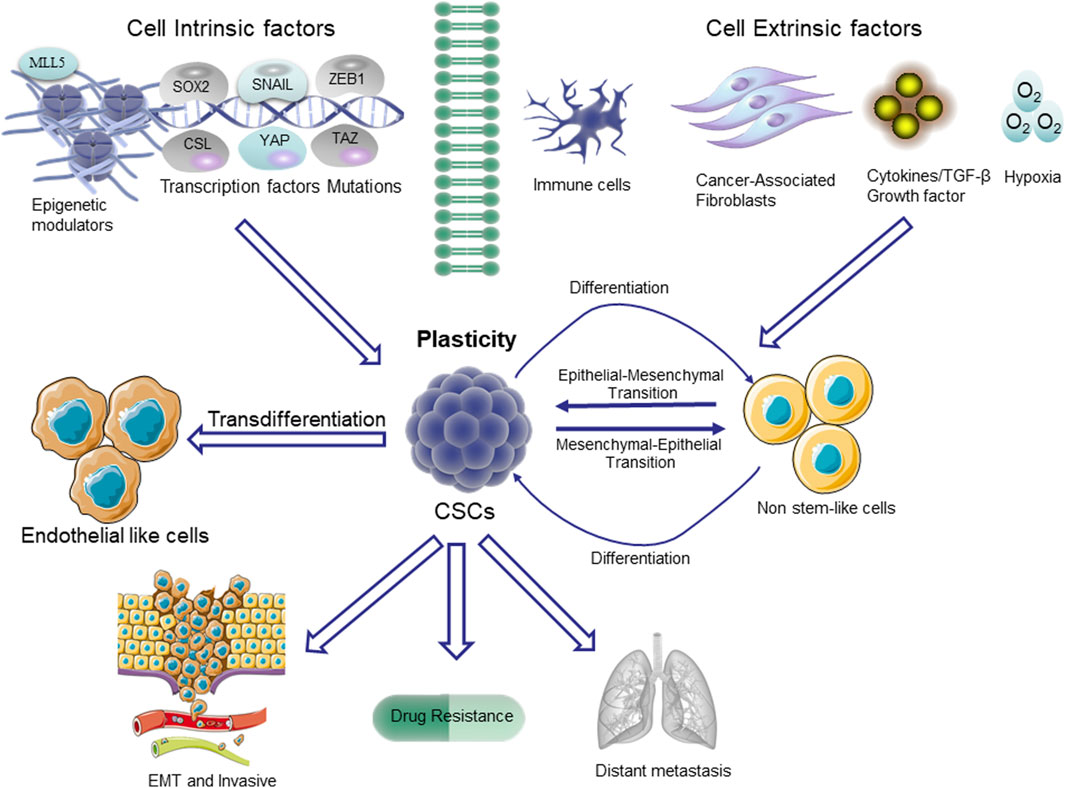
Figure 2. Plasticity in CSCs. The non-stem cells depicted in the figure typically exhibit restricted tumorigenic capacity and maintain their differentiated state. The upregulation of the stem cell pathway in these cells enables them to undergo a transition to a higher level of cellular state, referred to as plasticity. The pathways that trigger stem cell differentiation are mediated through transcription factors involved in epithelial-mesenchymal transition (EMT), such as SNAIL and ZEB1, as well as stem cell-specific transcription factors like SOX2. Additionally, the Hippo/YES-associated protein (YAP)/tafazzin (TAZ) signaling pathway, NOTCH/CSL pathway, and the epigenetic regulator MLL5 also play crucial roles in this process.
In addition, CSCs express high levels of ATP-binding cassette transporters (ABC transporters), which contribute to the efflux of chemotherapeutic agents, leading to multidrug resistance (Dean et al., 2005; Robey et al., 2018). In order to develop novel and efficacious therapeutic strategies targeting the stem cell-like subpopulations of tumor cells, it is imperative to gain comprehensive insights into the characteristics of CSCs and elucidate the underlying mechanisms responsible for their acquired resistance and stem cell-like properties, which are closely associated with CSCs plasticity, senescence, and quiescence. The concept of plasticity refers to the phenomenon that stem cells can generate tumor cells through asymmetric division (Vlashi and Pajonk, 2015). However, this hierarchy is not unidirectional; tumor cells have the ability to undergo de-differentiation and acquire stem-like properties (Figure 2). The occurrence of plasticity can be attributed to genetic and epigenetic alterations. For example, the non-CSC tumor cells in the basal-like subtype of breast cancer have the ability to undergo a transition to a CSC-like state through ZEB1 (Chaffer et al., 2013). In glioblastoma (GBM), cancer cells expressing CSC markers do not represent a functionally distinct clonal entity but rather exhibit a phenotypic plasticity that can be induced by microenvironmental cues (Dirkse et al., 2019). Recent findings from single-cell RNA sequencing analyses of human GBM tumors have revealed intratumoral heterogeneity, indicating the simultaneous presence of three cancer grades and cellular plasticity within the tumors (Patel et al., 2014; Neftel et al., 2019; Couturier et al., 2020; Lopes and Vinga, 2020). The study conducted by Neftel reveals that plasticity encompasses not only the process of de-cellularization but also the dynamic transition between distinct cell states, including intratumoral heterogeneity. This is exemplified by the simultaneous presence of 4 cell states within a single GBM, namely, neural progenitor cells, oligodendrocytes, astrocytes, and mesenchymal-like cells, which can interconvert among each other (Neftel et al., 2019). The rapid changes in cellular state can significantly impede therapies aimed at targeting specific tumor cell states. Cellular senescence is a stress response triggered by various molecular damages, resulting in cell cycle arrest, and characterized by diverse phenotypic alterations, including the secretion of bioactive molecules. Senescent cells progressively accumulate during the aging process and are observed in cancerous and fibrotic lesions (Bousset and Gil, 2022). The process of cellular senescence exerts a well-established tumor-suppressive effect by constraining the tumorigenic potential of cancer cells and enhancing the efficacy of cytotoxic therapies (Braig et al., 2005; Michaloglou et al., 2005; Dorr et al., 2013). However, the co-regulation of senescence and stemness functions through overlapping signaling pathways such as p16, p21, and p53 suggests that senescence may trigger genetic reprogramming and activate stemness, thereby contributing to CSC-mediated tumor progression, metastasis, and therapy resistance (Zon, 2008; Milanovic et al., 2018). A recent study has also provided insights into dysfunctional aging from a molecular perspective: in response to genotoxic damage or oncogenic stresses, tumor cells activate extensive chromatin remodeling that involves the addition of repressive methylation marks. The aforementioned marks exert a stable repression on S-phase promoter genes, while concurrently enhancing the secretion of pro-tumorigenic factors and activating stem cell transcription factors such as WNT/lymphocyte enhancer factor 1 (LEF1) (Lecot et al., 2016; Milanovic et al., 2018). The elucidation of the pathways involved in this reprogramming mechanism during cellular senescence will facilitate the overcoming of challenges posed by current therapeutic approaches through targeted CSCs.
The ability of CSCs to transition into a dormant or quiescent state, specifically entering the reversible G0 phase of the cell cycle and remaining dormant, is another distinguishing characteristic (Chen W. et al., 2016). This state transition is typically induced by the microenvironment, such as hypoxia, nutrient deprivation, or oxidative stress, or by selective pressure from chemotherapeutic agents. The fact that most conventional cancer treatments primarily target actively dividing cells allows quiescent CSCs to evade the effects of such therapies and tend to transition back into a proliferative state when favorable conditions arise (Sosa et al., 2014; De Angelis et al., 2019). The resistance of slow-circulating CSCs to temozolomide treatment in GBM was determined through strain tracing. Interestingly, the restoration of sensitivity to temozolomide was observed upon ablation of this specific population (Chen et al., 2012). The future plays a crucial role in the successful treatment of cancer through the identification of CSCs derived from quiescent or dormant cell populations and their associated maintenance pathways.
Due to the heterogeneity of tumor cells, cancer cells heavily rely on glucose and aerobic oxidation for energy supply, distinguishing their energy metabolism from that of normal cells (Liberti and Locasale, 2016). The reprogramming of metabolism is a hallmark feature exhibited by cancer cells (Hanahan and Weinberg, 2011). The metabolic characteristics of cancer cells differ significantly from those of normal cells. Given the hypoxic, highly oxidized, acidic, and nutrient-deprived TME resulting from rapid tumor cell proliferation and inadequate blood vessel formation, cancer cells must effectively adapt their cellular bioenergetics to survive in this unfavorable milieu (Viale et al., 2014; Lue et al., 2017). This adaptive process is commonly referred to as metabolic reprogramming. Metabolic reprogramming becomes indispensable for sustaining cancer cell proliferation and survival when oncogenic signaling is obstructed. The majority of human cancers exhibit aerobic glycolysis even in the presence of abundant oxygen, a phenomenon commonly referred to as the Warburg effect (Danhier et al., 2017; Strickland and Stoll, 2017). The Warburg effect characterizes the metabolic shift from oxidative phosphorylation (OXPHOS) to glycolysis in the pentose phosphate pathway, accompanied by the substitution of lactate accumulation for sustained ATP production in the TME (Koren and Fuchs, 2016). The metabolic rewiring not only fulfills the energy demands for sustained proliferation, but also generates a substantial amount of substrate for cellular compartments. It has been shown that poorly differentiated cancers exhibit higher glucose uptake than differentiated cancers, suggesting that the high glycolytic flux in tumor tissues arises mainly from impaired differentiation of CSCs (Riester et al., 2018). The activation of mitochondrial metabolism, in contrast, results in the loss of pluripotent potential and triggers the differentiation of P19 embryonal carcinoma stem cells (Vega-Naredo et al., 2014). The emerging evidence suggests that the Warburg effect of glycolytic metabolism is implicated in the processes of stemness and EMT (Aguilar et al., 2016). Further findings also provided compelling evidence that the regulation of stem cell metabolism plays a pivotal role in governing the control of stem cell fate. For instance, the compound R406 functions as a Syk inhibitor in immune thrombocytopenia by inducing a metabolic shift from glycolysis to OXPHOS in glioma stem cells (GSCs). This metabolic alteration subsequently leads to an excessive production of reactive oxygen species (ROS), ultimately triggering apoptosis in GBM cells (Sun et al., 2019). Peng et al. demonstrated that the overexpression of pyruvate dehydrogenase kinase 1 (PDK1) in breast CSCs (BCSCs) leads to the inhibition of aerobic glycolysis in mitochondria. The depletion of PDK1 resulted in a significant reduction in ALDH1-positive BCSCs, thereby impairing their ability to form spheroids (Peng et al., 2018). During the process of EMT in basal-like breast cancer, SNAIL-mediated methylation of the fructose-1,6-bisphosphatase promoter enhances the characteristics of CSCs and tumorigenicity by increasing glucose uptake and macromolecule biosynthesis. Additionally, it inhibits oxygen consumption through the inhibition of mitochondrial complex I activity (Dong et al., 2013). The presence of mutations in the internal tandem duplication (ITD) of the Fms-like tyrosine kinase 3 gene (FLT3/ITD) in acute myeloid leukemia (AML) is considered an unfavorable genetic alteration associated with a poor prognosis (Burchert, 2021). H-Q Ju et al. reported that FLT3/ITD induces a significant increase in aerobic glycolysis through akt-mediated upregulation of mitochondrial hexokinase 2, leading to heightened reliance on glycolysis and enhanced sensitivity of leukemic cells to pharmacological inhibition of glycolytic activity. The preferential inhibition of glycolysis results in severe depletion of ATP and extensive cell death in FLT3/ITD leukemia cells (Ju et al., 2017).
Notably, although the utilization of glycolysis for survival has been reported in some studies, others have suggested that CSCs may also rely on OXPHOS for their survival (Ciavardelli et al., 2014; Pasto et al., 2014). The transfer of electrons and H+ from the donor molecules, reduced nicotinamide adenine dinucleotide or reduced flavin adenine dinucleotide, to the acceptor molecule O2 is accompanied by the liberation of energy for ATP synthesis, involving a series of protein complexes located in the inner mitochondrial membrane. LAGADINOU et al. discovered that AML cells enriched with low levels of ROS from leukemia stem cells (LSCs) exhibited elevated expression of B-cell lymphocytoma-2 (Bcl-2), an antiapoptotic protein involved in mitochondrial regulation. Furthermore, Bcl-2 inhibition hampers ATP production in leukemia stem cells (LSCs) by hindering oxidative phosphorylation. Unlike AML cells and normal CD34+ cells, LSCs lack efficient backup glycolysis, making this metabolic vulnerability a promising target for selective elimination in clinical applications (Lagadinou et al., 2013). Additionally, through isotope tracing combined with metabolomics, researchers have demonstrated that LSCs exhibit enhanced efficiency in converting stearic acid and glucose into intermediates of the tricarboxylic acid cycle compared to other chronic myeloid leukemia cells, indicating a high dependence on OXPHOS (Kuntz et al., 2017). Similarly, the crucial significance of OXPHOS in solid tumors persists for CSCs. Ciavardelli et al., 2014 demonstrated that the proliferation of CD44+/CD24-breast CSCs could be reduced by inhibiting their glycolysis, indicating a glycolytic nature of this specific population. The proto-oncogene-encoded transcription factor MYC and the anti-apoptotic protein MCL1 synergistically enhance OXPHOS in CSCs, thereby promoting chemoresistance maintenance in triple-negative breast cancer (TNBC) (Lee et al., 2017). The results of clinical studies also indicate that well-differentiated tumors exhibit decreased uptake levels of 18F-fluorodeoxyglucose, whereas poorly differentiated tumors demonstrate elevated uptake levels of 18F-fluorodeoxyglucose (Riester et al., 2018). Additionally, the levels of oxidative metabolism and ATP in GSCs are higher compared to differentiated tumor cells and inhibiting OXPHOS, but not glycolysis, significantly impairs the tumorigenic potential and survival ability of GSCs in xenograft models (Minami et al., 2021). Notably, the nutrient-deprived CSCs in GBMs preferentially utilize the pentose phosphate shunt, thereby facilitating the self-renewal, proliferation, and survival of CSCs (Kathagen et al., 2013). Furthermore, the regulation of OXPHOS in GSCs is mediated by endogenous insulin-like growth factor 2 mRNA binding protein 2 (IMP2), which plays a crucial role in the transportation of mRNAs encoding respiratory chain-associated components to the mitochondria and facilitates OXPHOS maintenance through its involvement in the assembly of respiratory chain complexes (Janiszewska et al., 2012). In another study, based on the energy metabolism characteristics of pancreatic CSCs, a shift in the carbon source was employed by replacing glucose with galactose to induce enhanced OXPHOS activity in pancreatic cancer cells under ex vivo conditions. Consequently, this approach led to the enrichment of pancreatic CSCs, which exhibit upregulated expression of CSC surface antigens, heightened tumorigenicity, and immune evasion properties (Valle et al., 2020). Importantly, nasopharyngeal carcinoma, ovarian cancer, osteosarcoma, GBM, and colon cancer heavily rely on mitochondrial OXPHOS for energy generation (Zhou et al., 2011; Liao et al., 2014; Palorini et al., 2014; Shen et al., 2015; Song et al., 2015). These findings suggest that targeting aerobic glycolysis or OXPHOS could be a potential strategy for eradicating CSCs.
The alteration of cellular metabolism, particularly in lipid metabolism, has recently been acknowledged as a distinctive characteristic of various cancer cells (Yi et al., 2018). The lipid category encompasses various types of lipids, such as phospholipids, cholesterol and cholesterol esters, while fats primarily refer to triglycerides (TG). Lipids play a crucial role in numerous cellular functions, including membrane formation, signaling pathways, and other biological activities, while TG serves as a significant source of cellular energy (Lingwood and Simons, 2010). A growing body of evidence suggests that cancer cells undergo alterations in various aspects of cell membrane formation, lipid synthesis and degradation, as well as lipid-driven cellular signaling. A hallmark of cancer metabolism is the upregulation of de novo lipogenesis (Kinlaw et al., 2016), as illustrated in (Figure 3). The energy requirements of cancer cells are primarily met through de novo lipogenesis, as dietary lipids are limited in availability, unlike most non-malignant cells. The metabolic intermediates of glycolysis may be redirected towards enhanced lipid biosynthesis by CSCs, thereby promoting self-renewal growth (Corominas-Faja et al., 2014). The expression of various lipid synthases is elevated in cancer cells, including sterol regulatory element binding proteins (SREBPs), ATP-citrate lyase (ACLY), acetyl-CoA carboxylase (ACC), fatty acid synthase (FASN), and stearoyl-CoA desaturase 1 (SCD1) (Currie et al., 2013; Sun et al., 2016; Geng et al., 2016; Rohrig and Schulze, 2016; Shimano and Sato, 2017). The citric acid (TCA) cycle also plays a crucial role in lipid metabolism by providing acetyl groups for FA synthesis, thereby contributing to the maintenance of malignant biological behavior in cancer cells (Williams and ONeill, 2018). Here, we will concentrate the roles of these master regulatory elements in CSC progression.
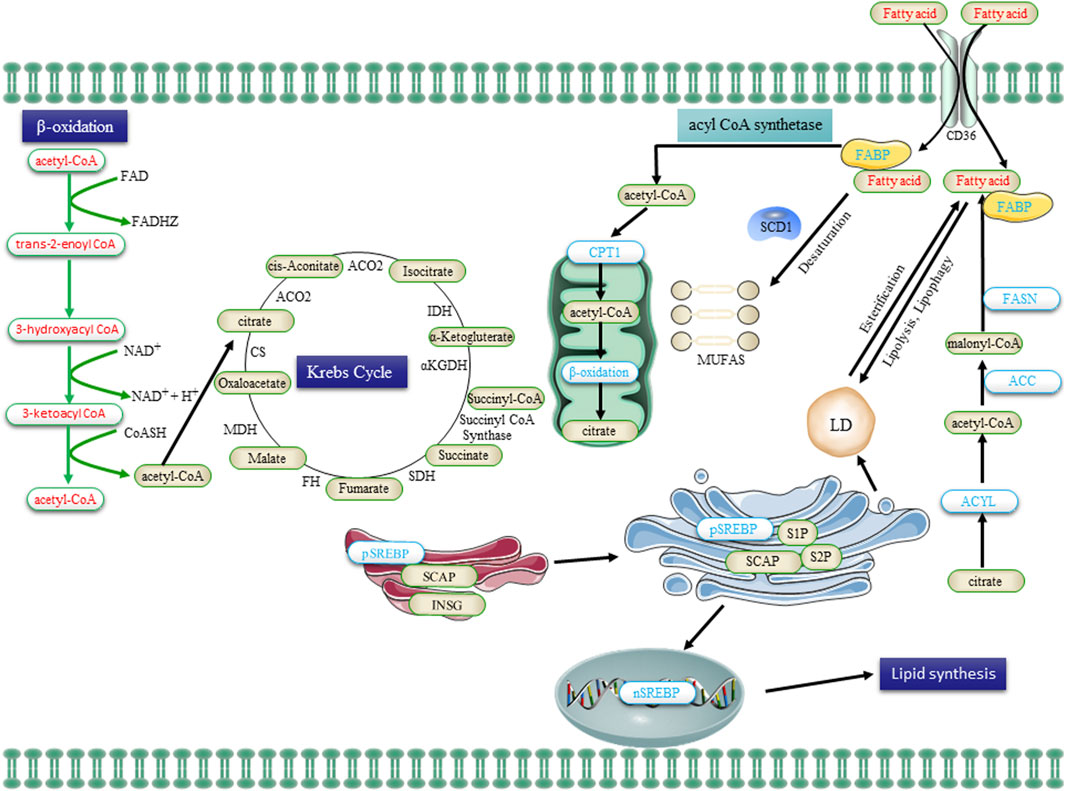
Figure 3. The lipid metabolism in CSCs, encompassing uptake, lipogenesis, and utilization, undergoes alterations. Extracellular FA is transported into cells via the CD36 receptor and subsequently undergone β-oxidation in mitochondria to generate acetyl-CoA. Acetyl-CoA is then converted to citrate by the enzyme citrate synthase, which enters the Krebs cycle for complete oxidation. The de novo synthesis of FA initiates with acetyl-CoA and proceeds through the sequential addition of two-carbon units, a process tightly regulated by the key enzyme FASN. Transcriptional control exerted by SREBPs governs the majority of enzymes involved in FA synthesis. Saturated FA undergoes desaturation to form monounsaturated FA (MUFA) catalyzed by SCD1. Additionally, FA is esterified to glycerol and subsequently stored as triglycerides in LDs. FA, obtained from the uptake of exogenous lipids or through de novo lipogenesis, is released to provide cellular fuel via FAO in the mitochondria, with CPTI catalyzing the rate-limiting step. Abbreviations: FA, fatty acid; FABP, FA binding protein; FASN, FA synthase; ACC, acetyl-CoA carboxylase; SREBP, sterol regulatory element binding protein; SCAP, SREBP cleavage-activating protein; INSG, insulin-induced gene protein; SCD1, stearoyl-CoA desaturase 1; MUFAs, mono-unsaturated FA; CPT1, carnitine palmitoyltransferase 1; and LDs, lipid droplets.
The activation of ACC leads to the catalysis of the conversion from acetyl coenzyme A (AcCoA) to malonyl-coenzyme A, which plays a crucial role in ab initio FA synthesis, particularly when cells require additional FA for energy demands or other biosynthetic processes. CSCs typically exhibit a heightened capacity for FA synthesis owing to their substantial energy demands required to sustain their highly active metabolic state (Liu et al., 2022). Additionally, ACC exhibits high expression levels in induced pluripotent stem cells (iPSCs) and suppression of ACC expression significantly diminishes the reprogramming efficiency of iPSCs (Vazquez-Martin et al., 2013). As normal stem cells always share the similar reprogramming procedure with that of CSCs (Zheng et al., 2019), this pronounced upregulation of ACC and FASN in iPSC underscores the significance of adipogenesis in stem cells and paves the way for potential therapeutic applications for CSCs. Thus, targeting ACC has been shown to suppress CSC progression, such as the ACC inhibitors in vitro reinstated the tumor cells to a histological epithelial phenotype (Petrova et al., 2017), and the inhibition of ACC in pancreatic cancer cells effectively suppresses both in vivo and in vitro pancreatic tumor growth by attenuating the ligand palmitoylation of Wnt and Hh, thereby inhibiting the signaling pathways of Wnt and Hh (Petrova et al., 2017), both of which are also implicated in the regulation of ACC in CSCs, as evident by that β-catenin knockdown leads to an upregulation of ACC expression (Vergara et al., 2017). Additionally, the inhibition of ACC activation effectively restored intracellular lipid levels, attenuated EMT, and suppressed the characterization of CSCs (Bort et al., 2020). Currently, given its pivotal role in lipid metabolism within CSCs, ACC represents a promising target for potential therapeutic interventions against numerous tumors (Moncur et al., 1998; Fang et al., 2014; Svensson et al., 2016).
Located predominantly in the cytoplasm, ACLY facilitates the enzymatic conversion of citric acid into AcCoA and oxaloacetate, thereby playing a crucial role in FA and cholesterol synthesis within CSCs (Feng et al., 2020). AcCoA serves as a crucial substrate not only for FA and cholesterol synthesis, but also plays an essential role in protein acetylation reactions, consequently emerging as a pivotal enzyme in lipid synthesis and connecting catabolic pathways with biosynthesis (Metallo et al., 2011). The upregulation and activation of ACLY have been extensively documented in various malignancies, as evidenced by the association with an increased malignant phenotype and a poor prognosis (Hatzivassiliou et al., 2005; Qian et al., 2015). Additionally, ACLY can facilitate tumor stemness through the downstream effectors, such as overexpression of ACLY enhances the expression of Snail, an EMT master regulator, thereby promoting EMT and stemness (Hanai et al., 2013). Conversely, inhibition of ACLY diminishes the invasiveness of breast cancer cells, while targeting ACLY reduces the proliferative potential and cisplatin resistance of ovarian cancer cells (Lucenay et al., 2016; Wei et al., 2021). Notably, Migita T et al. discovered a positive correlation between phosphorylation of ACLY in the PI3K/AKT pathway and the stage, grade of tumor differentiation, as well as poor prognosis in non-small cell lung cancer. This particular form of phosphorylation is believed to significantly enhance the role of ACLY in CSCs (Migita et al., 2008).
The transmembrane glycolipid protein CD36 is extensively expressed in various cell types, encompassing adipocytes, myocytes, endothelial cells, macrophages, and hepatocytes (Yang P. et al., 2022). CD36 protein exhibits numerous biological functions, encompassing lipid metabolism, inflammatory response, apoptosis, cell migration, and tumorigenesis (Zhao L. et al., 2018; Wang and Li, 2019). Elevated CD36 expression in tumor cells plays a pivotal role in lipid metabolism by facilitating the uptake and utilization of saturated fatty acids, such as palmitic acid, thereby contributing to both lipid synthesis and catabolism processes and augmenting proliferation, invasion, and metastatic potential (Li et al., 2022; Yuan et al., 2022). Additionally, CD36 is implicated in the crucial FA metabolic pathway known as β-oxidation (Zeng et al., 2022). Consistently, the blockade of β-oxidation through the targeting of CD36 with neutralizing antibodies has the potential to completely eradicate metastasis in melanoma and breast cancer (Pascual et al., 2017). In GSCs, CD36 facilitates the uptake of oxidized phospholipids, thereby promoting the proliferation of GSCs (Hale et al., 2014). Specifically, CD36 recognizes and binds to oxidized phospholipids, internalizing them into the intracellular compartment, thereby facilitating the proliferation and self-renewal of GSCs, this is attributed to the inhibitory effect exerted by elevated CD36 levels on the activation of apoptotic signaling pathways, thereby safeguarding GSCs against death-inducing stimuli. Additionally, with respect to non-solid tumor hematopoietic stem cells, CD36-enriched LSCs derived from gonadal adipose tissue exhibit enhanced survival advantages and resistance to treatment (Ye et al., 2016).
Recently, it has been demonstrated that lipids and lipoproteins, both acquired through exogenous (or dietary) uptake and synthesized endogenously, exert a profound influence on the maintenance of CSCs’ stemness during tumorigenesis. FASN, a key enzyme in de novo lipid synthesis, has consistently been identified as a facilitator in various types of CSCs (Ali et al., 2018; Rabionet et al., 2021; Castagnoli et al., 2023). Interestingly, overexpression of FASN in patient-derived GSCs was significantly diminished during serum-induced differentiation, indicating that augmented de novo adipogenesis contributes to the maintenance of the undifferentiated state of GSCs. After treatment with 20 μM cerulenin, a pharmacological inhibitor of FASN, the proliferation and migration of GSCs were significantly suppressed, and de novo lipogenesis was reduced. Additionally, the expression levels of nestin, Sox2, and FABP7, which are markers of GCSs, are decreased while the expression level of glial fibrillary acidic protein increased (Yasumoto et al., 2016). However, to date, limited research has been conducted regarding the involvement of FABPs in CSCs. The FABPs, functioning as lipid chaperones, are believed to bind and transport FA across various cellular compartments and organelles including plasma membranes, mitochondria, LDs, endoplasmic reticulum, and nuclei. A notable exception is FABP7, a widely recognized neural stem cell marker that exhibits high enrichment in GSCs and demonstrates significant downregulation in differentiated GSCs (Morihiro et al., 2013). Furthermore, Antonella De Rosa et al. showed that FABP7 downregulation in GSCs is associated with decreased proliferation and migration ability (De Rosa et al., 2012). Through proteomic and metabolomic analyses, Brandi et al. demonstrated that pancreatic CSCs exhibit elevated glycolysis levels and increased de novo adipogenic activity, while displaying reduced mitochondrial OXPHOS levels compared to a significant number of parental cancer cells. The authors discovered that FASN exhibited higher expression levels in Panc1 CSCs and displayed increased sensitivity to cerulenin inhibition compared to parental non-stem cell cancer cells (Brandi et al., 2017). Additionally, the expression level of FASN is regulated by β-catenin and exhibits a positive correlation with the expression levels of stem cell markers (SOX2, CD133, and Nestin) in GSCs (Yasumoto et al., 2016; El-Sahli et al., 2019). Importantly, FASN inhibitors have been shown to decrease the expression of stemness markers in GSCs (Yasumoto et al., 2016). For instance, resveratrol has the ability to induce apoptosis in CSCs by inhibiting adipogenesis through the downregulation of FASN expression (Pandey et al., 2011). Notably, CSCs often show a positive correlation between the levels of FASN and ACC expression under specific conditions, and FASN is more vulnerable to attacks in CSCs compared to regular cancer cells. Ongoing molecular and cell-based preclinical studies have focused on the development and characterization of various FASN blockers. Despite these efforts, translating these promising findings into clinical applications remains a challenging endeavor (Menendez and Lupu, 2017; Schcolnik-Cabrera et al., 2018). Therefore, numerous efforts should be directed towards several key areas. Firstly, there is a need for a comprehensive investigation into the interplay between FASN and ACC expression in CSCs to enhance our understanding of their correlation in CSCs. Additionally, a deeper exploration of the mechanisms underlying the increased vulnerability of FASN in CSCs compared to normal cancer cells is essential for the development of more effective therapeutic strategies. Optimization of existing FASN blockers is crucial, ensuring they exhibit high selectivity and efficacy in inhibiting FASN within CSCs. Moreover, emphasis should be placed on translating molecular and cell-based preclinical research findings into clinical applications by designing improved drug delivery systems and refining precision treatment protocols. Finally, addressing challenges associated with clinical applications, such as formulating suitable treatment regimens, ensuring drug safety and efficacy, and accommodating individual patient variations, is vital for the successful implementation of these research outcomes in the clinical setting. Through these concerted efforts, a better understanding of FASN’s role in CSCs can be achieved, leading to improved treatment strategies and ultimately facilitating the translation of these research findings into clinical applications.
Recently, multiple studies have demonstrated that an elevation in FA within cancer cells may also heighten their reliance on desaturase activity (Peck and Schulze, 2016). The promotion of CSCs through the regulation of unsaturated FA has been demonstrated in several studies, highlighting the significance of signaling pathways in this process, for example, NF-κB, a pivotal regulator of tumors and CSCs, directly governs the expression and activation of lipid desaturases (Li et al., 2017). Moreover, inhibition of adipogenesis through desaturase inactivation effectively disrupts the AKT/ERK-mediated NF-κB signaling pathway (Fritz et al., 2010; Li et al., 2017). Similarly, the levels of SCD-dependent MUFAs directly regulate CSCs through the Wnt/β-catenin pathway, which is a pivotal signaling pathway in stem cells and CSCs (Zheng et al., 2016; Lai et al., 2017). The two isoforms of SCDs in humans are SCD1 and SCD5, with SCD1 being the predominant enzyme responsible for desaturation in all tissues, while SCD5 is primarily expressed in the pancreas and brain (Wang et al., 2005). The expression of SCD-1, is significantly upregulated and contributes to the progression of cancer. It catalyzes the conversion of saturated FA into ∆9-MUFA (AM et al., 2017). Noto et al. demonstrated that the gene encoding SCD1 exhibited the highest level of upregulation in lung tumor spheroidal cells with adherent cultures, and further revealed that SCD1 inhibitors selectively eradicated cells possessing stem-like properties (Noto et al., 2013). Additionally, their subsequent investigation demonstrated that SCD1 governs the regulation of stem cells in lung cancer by stabilizing and localizing transcriptional co-activators of the Hippo pathway effector yes-related proteins and PDZ-binding motifs (Noto et al., 2017a). The Hippo pathway, which is regulated by YAP and TAZ, has been shown to facilitate the renewal and differentiation of both embryonic and somatic stem cells (Cao et al., 2020). Meanwhile, another one of their studies confirmed that the expression of SCD1 was associated with a poor prognosis in lung adenocarcinoma, and inhibiting the activity of SCD1 reversed resistance to cisplatin in lung CSCs (Pisanu et al., 2017). The significance of MUFAs is further underscored by the heightened levels of SCD1 expression in lung, ovarian, breast, and GSCs, which aligns with the presence of MUFAs in CSCs (Li L. et al., 2013; Colacino et al., 2016; Lobello et al., 2016; Noto et al., 2017b). Furthermore, SCD1 also governs the Wnt signaling pathway in CSCs (El-Sahli et al., 2019) and has been observed to play a crucial role in the maintenance of stem cells in various other cancers, including melanoma, hepatocellular carcinoma (HCC), and colon cancer (Pisanu et al., 2018; Choi et al., 2019; Ma et al., 2019). However, it must be noted that certain cancer cells utilize an alternative pathway for FA desaturation and sapienate biosynthesis, bypassing the established SCD-dependent pathway and diminishing the relevance of SCD, thereby questioning its suitability as a therapeutic target (Vriens et al., 2019).
The synthesis and activation of FA in cancer cells can be accomplished through various mechanisms, including transcriptional induction. The transcriptional regulation of SREBPs governs the majority of enzymes involved in FA synthesis. The SREBPs are a group of transmembrane transcription factors that activate the expression of genes encoding enzymes essential for cholesterol synthesis and the production of UFA. Human cells contain three isoforms of SREBP, namely, SREBPla, SREBPic, and SREBP2. Among these isoforms, both SREBPla and SREBP1c are derived from individual genes through distinct transcription start sites (Shimano and Sato, 2017; DeBose-Boyd and Ye, 2018). The SREBP1 protein is a member of the SREBP family of transcription factors and serves as a key transcriptional regulator in adipogenesis, controlling the synthesis of FA and cholesterol (Li L. et al., 2013). SREBP1 is essential for maintaining lipogenesis in lipid and hypoxic conditions, and it directly regulates several key lipogenic enzymes, including ACLY, ACC1, and FASN (Pandey et al., 2013; Shimano and Sato, 2017) (Figure 4). Overexpression of SREBP1 has been observed in various human cancers, promoting the growth of a wide range of tumors and playing an essential role in maintaining the stemness of CSCs (Pandey et al., 2013). For instance, PR Pandey et al. demonstrated that the ectopic expression of SREBP1 in MCF10A cells significantly augmented stem cell adipogenesis and facilitated cellular proliferation and mammosphere formation (Pandey et al., 2013). The expression of oncogenic PI3K (H1047R) or K-Ras (G12V) in mammary epithelial cells induces de novo synthesis of adipose tissue, which necessitates the activation of sterol-regulatory element binding proteins (SREBP1 and SREBP2) within the PI3K/AKT/mTOR signaling pathway (Ricoult et al., 2016). In addition to promoting lipogenesis, SREBP1 also stimulates the expression of SCD1, thereby facilitating the production of MUFA (Lewis et al., 2015) (Figure 4). The silencing of SREBP1 results in the inhibition of proliferation and the induction of apoptosis in pancreatic cancer cells, thereby further suppressing lipid metabolism and impeding tumor growth in vivo (Sun et al., 2015). Furthermore, the growth of glioblastoma spheroids was significantly inhibited by blocking SREBP1 (Lewis et al., 2015). Mechanistically, during mitosis, SREBP1 protein hinders the interaction between the ubiquitin ligase Fbw7 and SREBP1, thereby suppressing the phosphorylation-mediated degradation of SREBP1 by Cdk1 and Plk1 (Bengoechea-Alonso and Ericsson, 2006; Bengoechea-Alonso and Ericsson, 2009; Bengoechea-Alonso and Ericsson, 2016). Furthermore, the PI3-kinase/Akt/rapamycin target (mTOR) C1 signaling pathway additionally promotes the nuclear accumulation of mature SREBP1 (Porstmann et al., 2008). Activation of EGFR signaling triggers the translocation of pyruvate kinase M2 (PKM2) into the nucleus, thereby inducing the Warburg effect (Christofk et al., 2008; Yang et al., 2011). Notably, nuclear PKM2 physically engages with SREBP1, contributing to enhanced lipid biosynthesis by stabilizing SREBP-1 proteins (Figure 4) (Zhao X. et al., 2018). These findings provide further evidence for the interplay between glycolysis and FA metabolism.
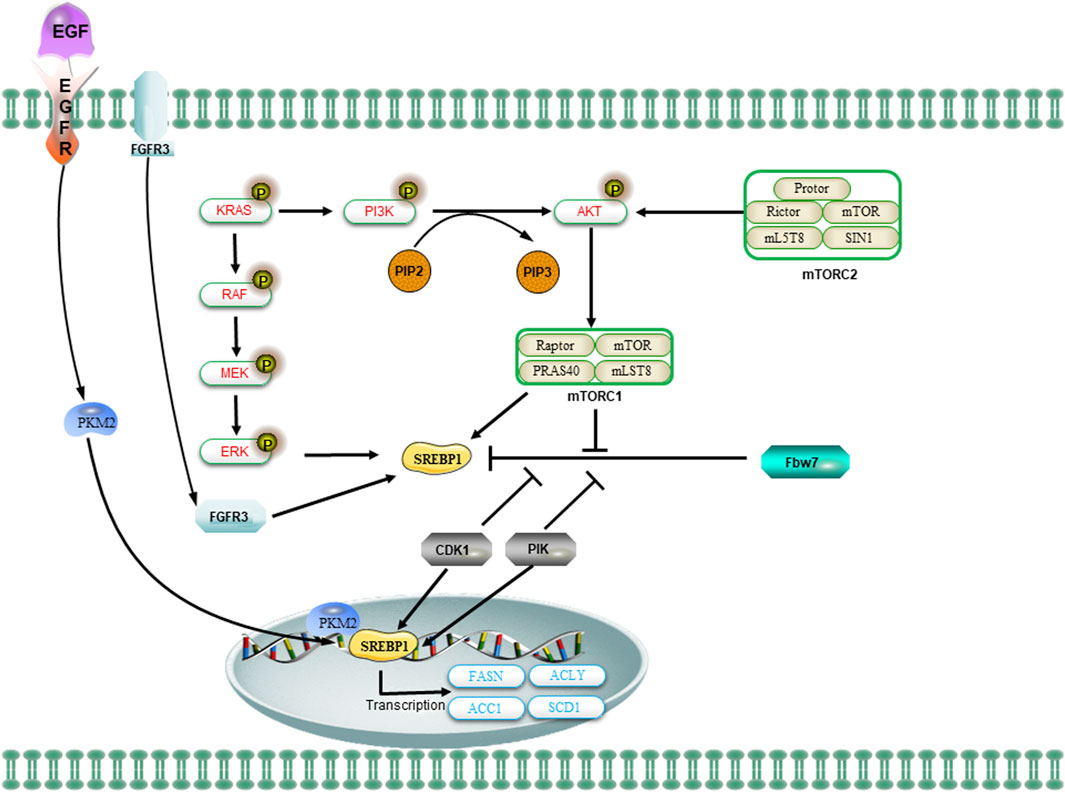
Figure 4. The regulation of SREBP1 and lipid metabolism in CSCs is mediated by oncogenic signaling. The oncogenic PI3K/Akt/mTORC1 and KRAS signaling pathways activate SREBP1 to facilitate de novo lipid synthesis and promote cellular growth. The mTOR signaling regulates the levels of SREBP1 through transcriptional or translational mechanisms. The activation of the PI3K/AKT/mTOR signaling pathway or FGFR3 results in the stabilization of SREBP1 proteins and facilitates their translocation to the nucleus. Additionally, the nuclear SREBP1 protein interacts with mitotic kinases Cdk1 and Plk1. The sequential phosphorylation of SREBP1 by Cdk1 and Plk1 impedes the interaction between the ubiquitin ligase Fbw7 and SREBP1, thereby attenuating the degradation of SREBP1. Upon activation of EGFR signaling, the nuclear form of PKM2 interacts with SREBP to facilitate the activation of SREBP target gene expression and promote lipid biosynthesis.
Recently, the significance of lipid metabolism in cancer cells has been consistently emphasized, leading to a series of notable advancements that offer valuable reference indices and guidance for cancer therapy (Luo X. et al., 2017). The energy metabolism of CSCs is mainly carried out in mitochondria, and they are able to efficiently utilize nutrients, such as FA, to produce ATP to provide the energy they need in the harsh microenvironment. The high level of UFA in CSCs has been demonstrated, and it has been shown that inhibiting the activities of SCD1 and acetaldehyde dehydrogenase 1A1 (ALDH1A1), or reducing the level of NF-κB in CSCs, can significantly decrease UFA content, diminish the stemness of CSCs, and impede tumor formation (Sun and Yang, 2019). The level of unsaturated fatty acid (UFA) in ovarian CSCs is significantly elevated compared to non-CSCs, and key lipases involved in FA synthesis, including ACLY, ACC, and FASN, exhibited elevated levels (Li et al., 2017). These lipases were regulated by the lipid-generating transcription factor SREBP1c, which has gradually emerged as a reliable marker of stemness in CSCs (Visweswaran et al., 2020). Additionally, the activity of lipase activity was modulated by other protein kinases, such as the reduction in AMP kinase (AMPK) phosphorylation, and content in CSCs not only elevated lipase activity but also increased malonyl coenzyme A levels, a precursor for fatty acid synthesis, resulting in enhanced fatty acid synthesis and mitochondrial β-oxidation (Bort et al., 2020). Furthermore, increased FA synthesis facilitates the uptake and utilization of lipids by CSCs (Peck and Schulze, 2016). Conversely, inhibition of the NF-κB-regulated lipid desaturase signaling pathway can effectively eradicate CSCs and impede their tumorigenic potential (Li et al., 2017). Additionally, the dynamic equilibrium of FA is pivotal for lipid metabolism in CSCs, and maintaining a stable metabolic state in CSCs contributes to chemoresistance and the acquisition of stem cell-like properties (Wang et al., 2018).
The connection between glycolipid metabolism in CSCs lies in the fact that AcCoA, generated through the oxidation of pyruvate—an intermediate product of glycolipid metabolism—can be utilized for FA synthesis, thereby facilitating the self-renewal of CSCs and contributing to the maintenance of their stemness (Pizer et al., 2000). Studies have demonstrated that maintaining a well-balanced ratio between FA and glycerophospholipids can effectively impede the progression of HCC (Lin et al., 2017). The elevated glucose metabolism and β-oxidation in CSCs of HCC and leukemia ensure the provision of alternative energy sources under extreme conditions, thereby sustaining their stemness (Chen C. L. et al., 2016). The intermediates generated through glycolysis can also serve as substrates for the synthesis of FA, thereby facilitating the self-renewal of CSCs (Corominas-Faja et al., 2014). Thus, the maintenance of pluripotency, self-renewal, proliferation, and formation of CSCs relies on the delicate balance of FA homeostasis or the state of catabolism/anabolism.
The sources of cholesterol can be categorized as either exogenous or endogenous, with exogenous cholesterol originating from dietary intake and endogenous cholesterol being synthesized within the body. The maintenance of cholesterol homeostasis is dependent on two primary mechanisms (Hafiane et al., 2019). On one hand, cholesterol levels can be elevated through the re-synthesis of AcCoA provided by glycolysis, glutamine metabolism, the TCA cycle, or exogenous uptake of low-density lipoprotein (LDL) receptors. Additionally, peripheral cholesterol can return to the liver in the form of LDL via the cholesterol reverse transporter, as well (Munir et al., 2019). On the contrary, cholesterol levels can be downregulated through inhibition of the MVA pathway or activation of liver X receptors (LXRs). The MVA pathway can be attenuated by protein hydrolysis or nuclear importation of SREBP2, while LXRs can be stimulated by the conversion of cholesterol to oxysterols (Ahmad et al., 2019). The activation of the LXRs/PPAR pathway subsequently induces the transcription of the E3 ubiquitin ligase IDOL, which in turn facilitates the ubiquitination of LDLR and enhances the expression of cholesterol efflux pumps ABCA1 and ABCG1 (Figure 5) (Spite, 2014). The MVA pathway plays a crucial role in the biosynthesis of steroid hormones, cholesterol, and nonsteroidal isoprenoids. The pathway primarily maintains homeostasis in the microenvironment of CSCs through protein geranylgeranylation. Additionally, 3-hydroxy-3-methylglutaryl monoacyl-coenzyme A (HMG-CoA) serves as the rate-limiting enzyme in the MAV pathway and represents a molecular target for statin drugs. The HMG-CoA reductase, which is the rate-limiting enzyme in the MAV pathway, serves as a molecular target for statin-type cholesterol-lowering drugs. The administration of statin drugs disrupts the MAV pathway, thereby inhibiting geranylgeranylation of proteins and disrupting the homeostasis of the microenvironment in CSCs, ultimately leading to their eradication (Mullen et al., 2016). For example, an overexpression of HMG-CoA is revealed in basal-like tumors, and the inhibition of the MAV pathway through simvastatin demonstrated a significant reduction in the number of CSCs within the tumors (Mancini et al., 2018). The combination of valproic acid and simvastatin can concurrently modulate the MAV pathway and AMPK phosphorylation level, thereby inhibiting the YAP oncogene. This enhances the sensitivity of denervation-tolerant prostate cancer cells to doxorubicin while reducing drug resistance caused by CSCs (Iannelli et al., 2020). Furthermore, the treatment of colorectal cancer cell lines with metformin, an AMPK activator, an HMG-CoA reductase inhibitor, or a mammalian target of rapamycin (mTOR) inhibitor significantly decreased the population of CSCs; however, the number of CSCs rebounded after treatment with mevalonic acid, indicating that mevalonic acid attenuates the inhibitory effect of these treatments on CSCs (Sharon et al., 2015). Moreover, bile acids and oxysterols serve as two chemical by-products of the MVA pathway. They function as ligands for a variety of nuclear receptors (including FXR, VDR, LXR, and PXR) as well as G-protein coupled receptors (Joyce and Gahan, 2016; Hafiane et al., 2019). Ting Fu et al. discovered that in colorectal cancer, a high-fat diet and dysregulated WNT signaling pathway led to alterations in bile acid profiles, activation of FXR, and the initiation of malignant transformation in Lgr5+ subpopulation CSCs (Fu et al., 2019). Similarly, the MAV pathway is found to enhance the proliferation of pancreatic CSCs, while the administration of atorvastatin effectively inhibits this proliferative effect (Brandi et al., 2017). Notably, in the MAV pathway, the inhibition of geranylgeranyltransferase can effectively reduce CSCs by suppressing protein isoprenylation. However, it has been observed that inhibiting cholesterol synthesis with squalene synthase inhibitors does not lead to a decrease in the number of CSCs (Brandi et al., 2017). The formation of breast tumorspheres occurs during the culture of breast CSCs, giving rise to both breast cancer cells and breast CSCs (Gupta et al., 2009). The inhibition of cholesterol synthesis effectively reduces the formation of breast tumorspheres, indicating that targeting the cholesterol synthesis pathway is a promising therapeutic strategy for suppressing the development of breast cancer by specifically targeting CSCs (Ehmsen et al., 2019). Some researchers have also demonstrated that synthetic progestins, such as medroxyprogesterone acetate (MPA), widely utilized in clinical practice, expedite the formation of breast tumorspheres while simultaneously increasing the activity of ALDH1A1 in CSCs and augmenting the tumorigenicity of CSCs (Goyette et al., 2017). Furthermore, it has been found that cholesterol synthesis inhibitors can mitigate this undesirable induction of breast tumorspheres by MPA (Liang et al., 2017).
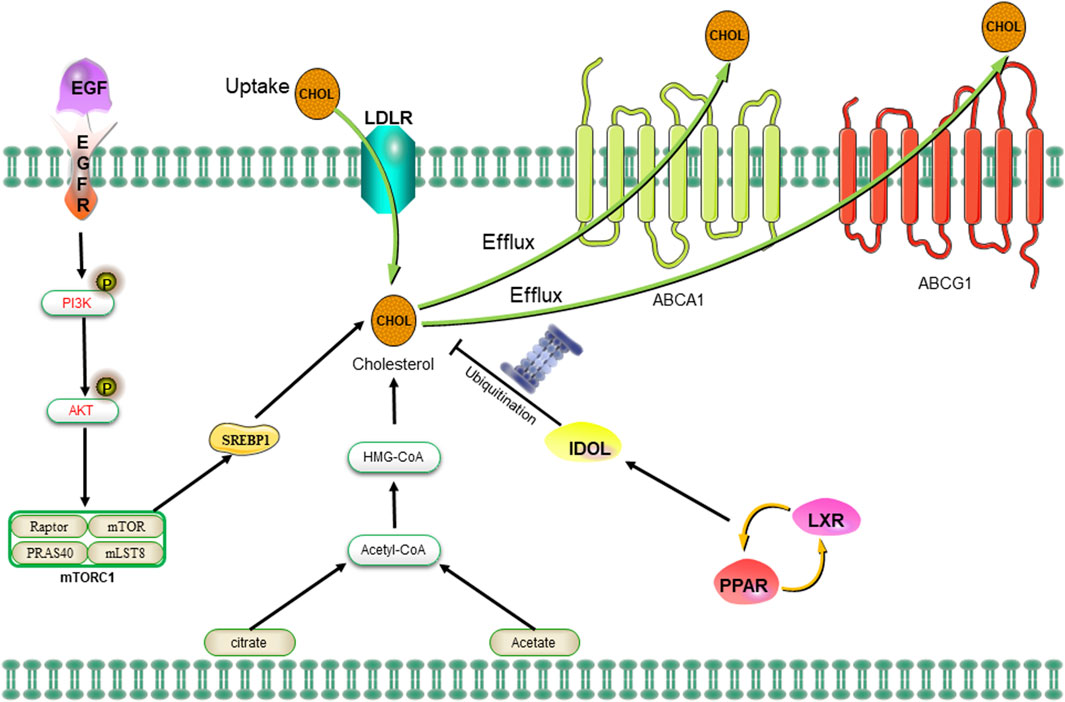
Figure 5. The uptake of cholesterol is facilitated by LDLR via the epidermal growth factor receptor-dependent pathway. The MVA pathway is responsible for cholesterol synthesis, while LDLR plays a crucial role in both the negative regulation of cholesterol uptake and the control of cholesterol efflux. Abbreviation: EGFR, epidermal growth factor receptor; LDLR, low density lipoprotein receptors; ABCA1, ATP-binding cassette transporter 1; PI3K/Akt, phosphatidylinositol 3-kinase/protein kinase B; SREBP-1, sterol regulatory element binding proteins; MVA, mevalonate; IDOL, inducible degrader of the low-density lipoprotein receptor; PPAR/LXR, lipid-activated transcription factors/LXRs.
Autophagy is a natural and highly conserved cellular degradation process that involves the lysosome-mediated breakdown of unwanted or dysfunctional intracellular components, including molecules and organelles (Mahapatra et al., 2021). The process of autophagy is crucial for maintaining cellular homeostasis, and any disruption to this mechanism may potentially facilitate the development of tumorigenesis. The process of lipophagy involves the fusion of LDs phagosomes with lysosomes to form autophagolysosomes, followed by the breakdown of LDs to generate free FA (Zhang et al., 2022). These FAs are then transported to the mitochondria for β-oxidation, resulting in energy production (Singh et al., 2009). The process of autophagy serves as a metabolic adaptation employed by tumor cells to surmount nutrient deprivation (Sinha et al., 2017). Additionally, autophagy facilitates the mobilization of nutrients and confers survival advantages to cancer cells, particularly under conditions of cellular stress such as hypoxia, chemotherapy, and radiotherapy (Lin et al., 2023). It plays a crucial role in maintaining cellular homeostasis and enables cells to withstand disturbances within the TME (Camuzard et al., 2020). The process of autophagy is crucial for maintaining lipid homeostasis, and the inhibition of autophagy leads to a decrease in the rate of β-oxidation in LDs, thereby reducing energy utilization (Li et al., 2018). Furthermore, autophagy facilitates the survival of CSCs by providing energy, enabling them to swiftly adapt to the challenging microenvironment post-chemotherapy, such as CSCs exhibit a higher autophagic rate compared to normal stem cells, and inhibition of autophagy compromises the stemness and tumorigenicity of CSCs (Visweswaran et al., 2020). The induction of autophagy is governed by the UNC-51-like autophagy-activated kinase 1 (ULK1) complex, and both mTOR and AMPK have the ability to modulate the expression level of ULK1. The activation of AMPK and the inhibition of mTOR can effectively suppress cholesterol synthesis and downregulate ULK1, thereby inducing apoptosis in CSCs (Bu et al., 2020). Moreover, the basal level of autophagy/mitophagy is higher in BCSCs compared to normal tissue-specific stem cells and autophagy induces the upregulation of CD44 and vimentin, both of which are recognized as stem cell markers (Cufi et al., 2011). GSCs also exhibit high expression levels of the autophagy regulators SQSTM1 and DRAM1, which are positively correlated with the expression of mesenchymal factors such as c-MET (Galavotti et al., 2013). The involvement of CD133, another stem cell marker, in the regulation of autophagy in GSC has been observed, as evidenced by the fact that the cytoplasmic localization of CD133 is relatively enhanced under conditions of glucose deprivation, while it remains membrane-bound in the presence of normal glucose levels within the cells (Sun H. et al., 2016). Autophagy involves several specific membrane structures, including phagosomes, autophagolysosomes, and autolysosomes. These structures are capable of engulfing isolated cytoplasmic constituents and subsequently degrading intracellular substances through hydrolytic enzymes (Klionsky et al., 2014). The lipid metabolism alterations in CSCs facilitate the regeneration of these membrane structures, and reciprocally, the regenerated membrane structures exert an influence on the CSCs, thereby establishing a positive feedback loop (Snaebjornsson et al., 2020). Moreover, the maintenance of pluripotency in breast hematopoietic stem cells under various pathophysiological conditions is also attributed to the crucial role played by autophagic homeostasis (Han et al., 2018). Interestingly, in addition to providing energy to CSCs, autophagy also facilitates the lipid peroxidation of UFA, which is abundantly expressed on the cellular membrane, leading to programmed cell death - ferroptosis (Xie et al., 2020). Ours and other studies have indicated that facilitating ferroptosis can specifically kill CSCs (Mai et al., 2017; Polewski et al., 2017; Ni et al., 2021; Yang Y. et al., 2022). However, it must be admitted that the potential benefits and drawbacks of autophagy in promoting the survival of CSCs necessitate further investigation: On the one hand, autophagy may facilitate CSC survival during periods of nutrient deprivation by recycling cellular components, providing an alternative energy source for their maintenance; On the other hand, excessive or dysregulated autophagy could lead to the degradation of essential cellular components in CSCs, compromising their functionality and survival.
The maintenance of stemness, survival, proliferation, and invasion in CSCs involves a variety of lipid metabolic pathways, including the Notch, Hippo, Hh, and Wnt signaling pathways (Wang et al., 2022). The pathways through which stem cells can be derived via genetic mutations and epigenetic alterations have a significant potential to be exploited for the maintenance of unrestricted proliferation, invasion, and drug resistance (Reya et al., 2001; Wang et al., 2022).
The Wnt signaling cascade comprises three primary pathways: the canonical Wnt pathway (which results in β-catenin accumulation, activation of transcription-activation complexes, and involvement in tumorigenesis), the non-canonical planar cell polarity pathway, and the non-canonical Wnt-calcium pathway (Takebe et al., 2015). In the typical Wnt pathway, Wnt ligands bind to the frizzled family of transmembrane receptors, thereby activating disheveled. This activation then collaborates with the T-cell factor/LEF family to induce stabilization and accumulation of nuclear β-catenin transcriptional activity. The Wnt signaling pathway exhibits a high degree of evolutionary conservation during the development of embryonic proliferative tissues, such as the hematopoietic system, skin, and intestines. This conservation is evident in terms of somatotaxis stereotypy, cell fate specification, cell proliferation, and migration (Teo et al., 2006). During tumorigenesis, the Wnt signaling facilitates tumor migration and invasion by upregulating genes involved in cell adhesion, such as Eph/ephrin, E-cadherin, and MMPs (Lang et al., 2019). Thus, the Wnt signaling pathway plays a pivotal role in the regulation of CSCs (Patel et al., 2019; Fendler et al., 2020). The Wnt signaling pathway, for instance, is also implicated in lipid synthesis within CSCs. Specifically, the canonical Wnt/β-catenin pathway governs subordinate lipogenesis and MUFA production (Bagchi et al., 2020). Additionally, the Wnt/β-catenin signaling significantly regulates the process of de novo adipogenesis in breast cancer cells, characterized by a substantial upregulation of ACC, FASN, and SREBP1-c expression (Vergara et al., 2017). Another study suggests that SCD potentially links Wnt signaling and lipid metabolism as in mouse liver hematopoietic stem cells, the Wnt/β-catenin pathway regulates SCD expression, and SCD-derived monounsaturated fatty acids create a positive feedback loop, reinforcing Wnt signaling through enhanced Lrp5/6 mRNA stability and expression (Lai et al., 2017). Furthermore, MUFAs play a crucial role in the synthesis and release of Wnt ligands (Rios-Esteves and Resh, 2013). The metabolism of FA in YAP/TAZ signaling, specifically the function of SCD1, is reliant on the activity of the β-catenin pathway in CSCs (Noto et al., 2017a). The dysregulation of the Wnt signaling pathway and a high-fat diet in colorectal cancer lead to alterations in bile acid distribution, activation of FXR, and subsequent malignant transformation of the Lgr5+ subpopulation of CSCs. This process promotes the progression from adenomas to adenocarcinomas (Fu et al., 2019).
The Notch signaling pathway comprises of Notch receptors, DSL proteins (Notch ligands), CSLs (CBF-1, Hairless Suppressor Factor, Lag), DNA-binding proteins, other effectors, and Notch-regulated molecules. The Notch signaling pathway is a highly conserved signal transduction pathway that plays a crucial role in various biological processes, including tumor metastasis and immune evasion (Takebe et al., 2015; Bocci et al., 2019). The activation of the Notch pathway in CSCs has been demonstrated by numerous studies to promote cell survival, self-renewal, and metastasis while inhibiting apoptosis. The aberrant activation of Notch signaling (Notch1 and Notch4) facilitates the self-renewal and metastasis processes in breast cancer and HCC stem cells (Stylianou et al., 2006; Harrison et al., 2010). In lipid metabolism, the Notch signaling pathway regulates the expression of peroxisome proliferator-activated receptor α and lipid oxidation genes to maintain lipid homeostasis and redox homeostasis in lipid metabolism (Song et al., 2016). The selective elimination of colon CSCs through inhibition of Notch signaling is achieved by targeting scd1-dependent lipid desaturation (Yu et al., 2021). Furthermore, in Drosophila, the Notch signaling pathway is responsive to environmental sterol levels and its expression is regulated by dietary cholesterol, ultimately leading to the differentiation of enterocytes from a stem-like state (Obniski et al., 2018). The activation of the Notch signaling is also regulated by dietary cholesterol, thereby inducing the differentiation of enterocytes from a stem-like state. Additionally, the Notch pathway plays a crucial role in angiogenesis, EMT, and the proliferation of CSCs within cancer cells. It has been observed that a low-sterol diet can potentially restrict the growth of enteroendocrine tumors by attenuating the Notch response (Obniski et al., 2018). Kalucka et al., 2018 discovered that Notch1 regulates FAO to maintain intermediate lipid homeostasis and redox homeostasis in CSCs. Importantly, the Notch signaling in humans is influenced by the lipid composition of the cellular membrane (Sorrentino et al., 2014).
The evolutionarily conserved Hippo signaling pathway is initiated by Hippo kinase, a serine/threonine kinase. In normal conditions, Hippo kinase is inactive due to phosphorylation. However, external stimuli activate Hippo kinase, leading to the phosphorylation and activation of downstream molecules MST1/2. Activated MST1/2, in turn, phosphorylate LATS1/2, exerting inhibitory effects on the activities of YAP and transcriptional coactivator with PDZ-binding motif (Mo et al., 2014). The activation of YAP or TAZ in CSCs has been confirmed to sustain the self-renewal and tumor-initiating capacity (Bhat et al., 2011), promote cellular pluripotency (Cordenonsi et al., 2011) and drug resistance (Huo et al., 2013), and exhibits a strong correlation with the process of EMT (Kulkarni et al., 2018; Sundqvist et al., 2018). Emerging evidence suggests a close and significant association between the Hippo-YAP/TAZ signaling and the regulation of lipid metabolism in cancer stemness (Shu et al., 2019). For example, SCD1, a major regulator of MUFA, contributes to the maintenance of cancer stemness by modulating the expression and nuclear localization of YAP/TAZ (Noto et al., 2017a). GGPP, an intermediate in the control of the MAV pathway, stabilizes YAP/TAZ (Sorrentino et al., 2014). As previously mentioned, SCD1 plays a crucial role in regulating the stemness of lung CSCs by stabilizing YAP/TAZ and facilitating their nuclear localization (Noto et al., 2017a). Notably, a positive feedback loops involving LATS2 and p53 has been identified to inhibit cholesterol synthesis (Aylon et al., 2016). Additionally, LATS2 binds to endoplasmic reticulum tethered precursors (P-SREBP) of SREBP1 and SREBP2, thereby suppressing the transcription of SREBP mRNAs and subsequently inhibiting cellular SREBP activity (Aylon and Oren, 2016). Recent studies have demonstrated that the pro-carcinogenic properties of YAP/TAZ are contingent upon cholesterol biosynthesis activity and MVA-dependent nuclear localization and activity of YAP/TAZ (Sorrentino et al., 2014). The lipid synthesis mediated by YAP/TAZ may serve as a crucial factor influencing metabolic alterations in CSCs (Koo and Guan, 2018).
The Hh signaling network is intricate and encompasses extracellular Hh ligands, the transmembrane protein receptor PTCH, the transmembrane protein SMO, intermediate signal transduction molecules, and the downstream effector molecule GLI (Yang et al., 2020). The membrane protein SMO exerts a positive regulatory function, while the transmembrane protein PTCH plays a negative regulatory role. The GLI protein acts as an effector, and in vertebrates, it exists in three isoforms: Gli1, Gli2, and Gli3. These effector proteins exhibit distinct functionalities: Gli1 exerts a robust transcriptional activation effect, whereas Gli3 functions as a transcriptional inhibitor. On the other hand, Gli2 displays dual functionality by both activating and inhibiting transcription but primarily serves as a transcriptional activator (Sasaki et al., 1999; Li S. H. et al., 2013). The Hh signaling pathway also plays a pivotal role in the regulation of CSCs. It governs the proliferation of postnatal mammary stem cells and orchestrates the intricate ductal architecture formation in the adult mammary gland (Miyazaki et al., 2016). The existing evidence suggests that lipid metabolism also plays a regulatory role in the Hh signaling and its ligand properties, highlighting the crucial involvement of lipids as key regulators in Hh signaling (Blassberg and Jacob, 2017). The covalent modification of SMO by cholesterol is regulated by the Hh signaling pathway and plays a crucial role in mediating Hh signaling and cellular biological functions (Radhakrishnan et al., 2020). PTCH1 inhibits the cholesterol modification of SMO, while SHH overexpression enhances it (Hu and Song, 2019). Furthermore, SMO exerts direct or indirect inhibition on FA and cholesterol synthesis by activating adenosine monophosphate kinase through a non-classical pathway (Blassberg and Jacob, 2017). Notably, recent clinical trials have utilized SMO and GLI inhibitors to target the Hh signaling pathway (Long et al., 2016; Zhang et al., 2016). Thus, the Hh signaling pathway, intricately linked with lipid metabolism and CSCs, demonstrates the regulatory role of lipids in signaling cascades, offering potential therapeutic targets for cancer intervention.
The targeted eradication of CSCs can be accomplished by intervening in various aspects of their lipid metabolism, including lipogenesis, lipid uptake, lipid desaturation, and FA oxidation (Yi et al., 2018). Due to the exorbitant cost and inherent risks associated with the discovery and development of novel therapeutic agents, there has been a growing interest in drug repositioning strategies for hard-to-treat diseases. This approach offers an opportunity to establish effective targeting strategies aimed at eradicating CSCs. The targeted elimination of CSCs can be achieved by disrupting various aspects of lipid synthesis, including FA synthesis, lipid desaturation, and cholesterol synthesis (Table 2).
The FASN gene is the primary focus among adipogenic genes and the antitumor activity of several FASN inhibitors has been demonstrated in preclinical cancer models. The expression of invasiveness, sphere formation, and stemness markers is effectively reduced by both FASN inhibitors and RNA silencing, leading to the eradication of various CSCs (Table 2) (Yasumoto et al., 2016; Brandi et al., 2017). The development of novel FASN inhibitors is underway, and preliminary clinical trial data on TVB-2640 demonstrate its potential to mitigate tumor response in patients with non-small cell lung cancer and breast cancer when used in combination with paclitaxel (Jones and Infante, 2015). Similarly, the formation of mammospheres was inhibited by Soraphen A, an inhibitor of ACC (Corominas-Faja et al., 2014). Mechanistically, Soraphen A inhibits the self-renewal and growth of CSC-like cells by blocking FA synthesis and abolishes the promotion of CSC proliferation mediated by human epidermal growth factor receptor 2 (HER2) (Corominas-Faja et al., 2014). The inhibition of ACC not only suppressed tumor growth, metastasis, and recurrence in non-small cell lung and breast cancers, but also underscored the significance and potential of ACC in suppressing CSCs and combating cancer (Schug et al., 2015; Svensson et al., 2016).
Additionally, the selective elimination of CSCs can be achieved by targeting SCD1, an enzyme responsible for the conversion of fully saturated FA into MUFAs. The recent demonstration of SCD in a genetic mouse model is particularly noteworthy, as it is essential for the emergence of tumor-initiating cells (Lai et al., 2017). The significant reliance on UFA renders SCD a promising target for the eradication of CSCs (Peck and Schulze, 2016). The SCD1 inhibitors, such as CAY10566 and A939572, have been documented to effectively suppress cancer stemness, inhibit tumorigenesis, and overcome chemoresistance in cancer cells (Tracz-Gaszewska and Dobrzyn, 2019). Notably, MF-438 and PluriSIn #1, as inhibitors of SCD1, exhibited selective eradication of colonic CSCs while showing no efficacy against cancer cells (Yu et al., 2021). Furthermore, the inhibition of SCD1 expression enhances the sensitivity of CSCs to cisplatin and reduces the occurrence of drug resistance (Pisanu et al., 2017). The SSI-4 compound is a novel inhibitor of SCD1 that effectively overcomes sorafenib resistance in hepatic CSCs. The antitumor activity of SSI-4 against hepatic CSCs was also observed in animal models, with no significant adverse effects reported (Ma et al., 2017). Importantly, the combination of SSI-4 and sorafenib demonstrated the most significant inhibition of tumorigenesis in the sorafenib-resistant PDTX model. The combination of SCD1 inhibitors and chemotherapy may potentially offer a more efficacious therapeutic approach. The effective SCD1 inhibitors utilized in the preclinical phase are summarized in Table 1.
The reliance of CSCs on FAO justifies the targeting of these cells with FAO inhibitors. The compound Etomoxir functions as a highly specific inhibitor of mitochondrial CPT1A (Holubarsch et al., 2007) (as shown in Table 1). The inhibition of FAO by etomoxir in leukemic CSCs leads to the suppression of cell proliferation and enhances the susceptibility of human leukemic cells to apoptosis (Samudio et al., 2010; Estan et al., 2014). ST1326 is a CPT1A inhibitor that suppresses FAO and exhibits cytotoxic activity against leukemia cell lines, while sparing normal CD34+ myeloid cells (Ricciardi et al., 2015) (Table 1). ST1326 also exhibits preventive effects against MYC-driven lymphomagenesis in an Eμ-myc transgenic mouse model (Pacilli et al., 2013).
The activation of cholesterol synthesis has been previously suggested to be linked with the invasive and metastatic capabilities of CSCs. The inhibition of SREBP activation by 25-HC or adiponectin hampers adipogenesis, including FA and cholesterol synthesis, and disrupts the expression of genes associated with CSCs (Sun H. W. et al., 2016). Furthermore, Dattilo R not only significantly inhibits lipid anabolism in CSCs but also exhibits a cytotoxic effect on TNBC -CSCs by suppressing cholesterol synthesis in TNBC (Dattilo et al., 2020). The inhibition of cholesterol biosynthesis by Simvastatin significantly suppresses the formation and growth of mammospheres (Gu et al., 2019). Additionally, statins exert their effects on CSCs by inhibiting signaling pathways associated with protein farnesylation and protein geranylation in the MVA pathway (Ginestier et al., 2012; Iannelli et al., 2020). Similarly, metformin suppresses colorectal CSCs by inhibiting protein prenylation of the MVA pathway (Seo et al., 2020). Taken together, these emerging drugs targeting lipid metabolism in CSCs offer promising avenues for selective elimination of CSCs, focusing on disrupting various aspects such as lipogenesis, lipid uptake, desaturation, and fatty acid oxidation, and presenting potential strategies to suppress CSC proliferation and enhance therapeutic outcomes.
In conclusion, the intricate interplay between lipid metabolism dynamics and CSCs has emerged as a pivotal determinant in the initiation, progression, and therapeutic response of various cancers. The preservation of CSC stemness relies heavily on unique alterations in lipid metabolism, encompassing strategies to increase intracellular fatty acid content, promote β-oxidation for optimized energy utilization, and upregulate the MAV pathway for efficient cholesterol synthesis. Additionally, the versatile role of LDs as alternative energy sources in the context of glycolysis blockade underscores their significance in safeguarding FAs from peroxidation. The symbiotic relationship between autophagy and lipid metabolism not only facilitates the rapid adaptation of CSCs to the harsh microenvironment induced by chemotherapy but also presents a complex network of potential therapeutic targets. As we delve into the molecular intricacies of lipid metabolism in CSCs, a myriad of possibilities emerges for innovative cancer therapies. Identifying and targeting key components within this dynamic metabolic landscape holds promise for disrupting CSC functions, curbing tumor initiation, and overcoming drug resistance.
In recent years, the rapid advancement of high-throughput technologies has significantly enhanced our comprehension of the role of metabolic reprogramming in oncogenesis, which is now widely acknowledged as a fundamental characteristic of cancer. CSCs play a pivotal role in the initiation, progression, distant dissemination, and acquisition of drug resistance in tumors. Metabolic alterations have been shown to serve as the primary mechanism through which cancer cells and CSCs evade adverse environmental influences. In lipid metabolism, such as augmented FA uptake, neoadipogenesis, LDL formation, FAO, and lipid desaturation, play a significant role in the generation and sustained maintenance of CSCs. Moreover, lipid synthesis plays a crucial role in the activation of several pivotal oncogenic signaling pathways, including the Notch, Wnt/β-catenin, Hippo, and HH signaling. The targeting of crucial lipid synthesis molecules holds great potential in the elimination of CSCs. In addition, the heterogeneity and metabolic plasticity of CSCs pose a dilemma, despite the promising opportunity to target cellular metabolism for their elimination (Wang et al., 2013; Thomas and Majeti, 2016; Krstic et al., 2017). The metabolic profiles of CSCs and tumor cells can be modulated in response to nutrient availability. For instance, when cetuximab counteracted the Warburg effect, HNSCC cells exhibited elevated levels of ACC, thereby reprogramming cancer metabolism from glycolysis to adipogenesis in order to support energy demands and facilitate proliferation (Luo J. et al., 2017). The enhanced uptake of FA, neoadipogenesis, LDL formation, FAO, and lipid desaturation has been extensively documented in CSCs across various cancer types. The majority of the targeted compounds, however, have not demonstrated therapeutic efficacy in preclinical cancer models, with only a limited number of inhibitors progressing to clinical trials. The metabolic flexibility of CSCs poses a challenge in effectively eliminating these cells through a single metabolic pathway, necessitating the exploration of synergistic targeting of multiple metabolic pathways in future studies.
In conclusion, the intricate interplay between lipid metabolism dynamics and CSCs has emerged as a critical determinant in various cancers’ initiation, progression, and therapeutic response. The preservation of CSC stemness heavily relies on unique alterations in lipid metabolism, including strategies to increase intracellular fatty acid content, promote β-oxidation for optimized energy utilization, and upregulate the mevalonate pathway for efficient cholesterol synthesis. Additionally, the versatile role of LDs as alternative energy sources in the context of glycolysis blockade underscores their significance in safeguarding fatty acids from peroxidation. The symbiotic relationship between autophagy and lipid metabolism not only facilitates the rapid adaptation of CSCs to the harsh microenvironment induced by chemotherapy but also presents a complex network of potential therapeutic targets. As we delve into the molecular intricacies of lipid metabolism in CSCs, a myriad of possibilities emerges for innovative cancer therapies. Identifying and targeting key components within this dynamic metabolic landscape holds promise for disrupting CSC functions, curbing tumor initiation, and overcoming drug resistance.
All in all, this comprehensive review underscores the potential of lipid metabolism as a therapeutic nexus, offering valuable insights into the vulnerabilities of CSCs. As we move forward, unraveling the complexities of lipid-mediated stemness maintenance opens avenues for the development of targeted interventions, marking a significant stride in the pursuit of effective cancer treatments.
JD: Data curation, Funding acquisition, Methodology, Resources, Software, Visualization, Writing–review and editing. HQ: Conceptualization, Formal Analysis, Investigation, Project administration, Supervision, Validation, Writing–original draft.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aguilar, E., Marin de Mas, I., Zodda, E., Marin, S., Morrish, F., Selivanov, V., et al. (2016). Metabolic reprogramming and dependencies associated with epithelial cancer stem cells independent of the epithelial-mesenchymal transition program. Stem Cells 34, 1163–1176. doi:10.1002/stem.2286
Ahmad, F., Sun, Q., Patel, D., and Stommel, J. M. (2019). Cholesterol metabolism: a potential therapeutic target in glioblastoma. Cancers (Basel) 11, 146. doi:10.3390/cancers11020146
Al-Hajj, M., and Clarke, M. F. (2004). Self-renewal and solid tumor stem cells. Oncogene 23, 7274–7282. doi:10.1038/sj.onc.1207947
Al-Hajj, M., Wicha, M. S., Benito-Hernandez, A., Morrison, S. J., and Clarke, M. F. (2003). Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 100, 3983–3988. doi:10.1073/pnas.0530291100
Ali, A., Levantini, E., Teo, J. T., Goggi, J., Clohessy, J. G., Wu, C. S., et al. (2018). Fatty acid synthase mediates EGFR palmitoylation in EGFR mutated non-small cell lung cancer. EMBO Mol. Med. 10, e8313. doi:10.15252/emmm.201708313
Am, A. L., Syed, D. N., and Ntambi, J. M. (2017). Insights into stearoyl-CoA desaturase-1 regulation of systemic metabolism. Trends Endocrinol. Metab. 28, 831–842. doi:10.1016/j.tem.2017.10.003
Aylon, Y., Gershoni, A., Rotkopf, R., Biton, I. E., Porat, Z., Koh, A. P., et al. (2016). The LATS2 tumor suppressor inhibits SREBP and suppresses hepatic cholesterol accumulation. Genes. Dev. 30, 786–797. doi:10.1101/gad.274167.115
Aylon, Y., and Oren, M. (2016). The Hippo pathway, p53 and cholesterol. Cell. Cycle 15, 2248–2255. doi:10.1080/15384101.2016.1207840
Bagchi, D. P., Nishii, A., Li, Z., DelProposto, J. B., Corsa, C. A., Mori, H., et al. (2020). Wnt/β-catenin signaling regulates adipose tissue lipogenesis and adipocyte-specific loss is rigorously defended by neighboring stromal-vascular cells. Mol. Metab. 42, 101078. doi:10.1016/j.molmet.2020.101078
Beachy, P. A., Karhadkar, S. S., and Berman, D. M. (2004). Tissue repair and stem cell renewal in carcinogenesis. Nature 432, 324–331. doi:10.1038/nature03100
Belkaid, A., Duguay, S. R., Ouellette, R. J., and Surette, M. E. (2015). 17β-estradiol induces stearoyl-CoA desaturase-1 expression in estrogen receptor-positive breast cancer cells. BMC Cancer 15, 440. doi:10.1186/s12885-015-1452-1
Bengoechea-Alonso, M. T., and Ericsson, J. (2006). Cdk1/cyclin B-mediated phosphorylation stabilizes SREBP1 during mitosis. Cell. Cycle 5, 1708–1718. doi:10.4161/cc.5.15.3131
Bengoechea-Alonso, M. T., and Ericsson, J. (2009). A phosphorylation cascade controls the degradation of active SREBP1. J. Biol. Chem. 284, 5885–5895. doi:10.1074/jbc.M807906200
Bengoechea-Alonso, M. T., and Ericsson, J. (2016). The phosphorylation-dependent regulation of nuclear SREBP1 during mitosis links lipid metabolism and cell growth. Cell. Cycle 15, 2753–2765. doi:10.1080/15384101.2016.1220456
Bhat, K. P., Salazar, K. L., Balasubramaniyan, V., Wani, K., Heathcock, L., Hollingsworth, F., et al. (2011). The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes. Dev. 25, 2594–2609. doi:10.1101/gad.176800.111
Blassberg, R., and Jacob, J. (2017). Lipid metabolism fattens up hedgehog signaling. BMC Biol. 15, 95. doi:10.1186/s12915-017-0442-y
Bocci, F., Gearhart-Serna, L., Boareto, M., Ribeiro, M., Ben-Jacob, E., Devi, G. R., et al. (2019). Toward understanding cancer stem cell heterogeneity in the tumor microenvironment. Proc. Natl. Acad. Sci. U. S. A. 116, 148–157. doi:10.1073/pnas.1815345116
Bonnet, D., and Dick, J. E. (1997). Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3, 730–737. doi:10.1038/nm0797-730
Bort, A., Sanchez, B. G., de Miguel, I., Mateos-Gomez, P. A., and Diaz-Laviada, I. (2020). Dysregulated lipid metabolism in hepatocellular carcinoma cancer stem cells. Mol. Biol. Rep. 47, 2635–2647. doi:10.1007/s11033-020-05352-3
Boumahdi, S., and de Sauvage, F. J. (2020). The great escape: tumour cell plasticity in resistance to targeted therapy. Nat. Rev. Drug Discov. 19, 39–56. doi:10.1038/s41573-019-0044-1
Bousset, L., and Gil, J. (2022). Targeting senescence as an anticancer therapy. Mol. Oncol. 16, 3855–3880. doi:10.1002/1878-0261.13312
Braig, M., Lee, S., Loddenkemper, C., Rudolph, C., Peters, A. H., Schlegelberger, B., et al. (2005). Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436, 660–665. doi:10.1038/nature03841
Brandi, J., Dando, I., Pozza, E. D., Biondani, G., Jenkins, R., Elliott, V., et al. (2017). Proteomic analysis of pancreatic cancer stem cells: functional role of fatty acid synthesis and mevalonate pathways. J. Proteomics 150, 310–322. doi:10.1016/j.jprot.2016.10.002
Broske, A. M., Vockentanz, L., Kharazi, S., Huska, M. R., Mancini, E., Scheller, M., et al. (2009). DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat. Genet. 41, 1207–1215. doi:10.1038/ng.463
Brunetti, L., Gundry, M. C., and Goodell, M. A. (2017). DNMT3A in leukemia. Cold Spring Harb. Perspect. Med. 7, a030320. doi:10.1101/cshperspect.a030320
Bu, H., Liu, D., Zhang, G., Chen, L., and Song, Z. (2020). AMPK/mTOR/ULK1 axis-mediated pathway participates in apoptosis and autophagy induction by oridonin in colon cancer DLD-1 cells. Onco Targets Ther. 13, 8533–8545. doi:10.2147/OTT.S262022
Burchert, A. (2021). Maintenance therapy for FLT3-ITD-mutated acute myeloid leukemia. Haematologica 106, 664–670. doi:10.3324/haematol.2019.240747
Camarda, R., Zhou, A. Y., Kohnz, R. A., Balakrishnan, S., Mahieu, C., Anderton, B., et al. (2016). Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat. Med. 22, 427–432. doi:10.1038/nm.4055
Camuzard, O., Santucci-Darmanin, S., Carle, G. F., and Pierrefite-Carle, V. (2020). Autophagy in the crosstalk between tumor and microenvironment. Cancer Lett. 490, 143–153. doi:10.1016/j.canlet.2020.06.015
Cao, J., and Yan, Q. (2020). Cancer epigenetics, tumor immunity, and immunotherapy. Trends Cancer 6, 580–592. doi:10.1016/j.trecan.2020.02.003
Cao, X., Wang, C., Liu, J., and Zhao, B. (2020). Regulation and functions of the Hippo pathway in stemness and differentiation. Acta Biochim. Biophys. Sin. (Shanghai) 52, 736–748. doi:10.1093/abbs/gmaa048
Castagnoli, L., Corso, S., Franceschini, A., Raimondi, A., Bellomo, S. E., Dugo, M., et al. (2023). Fatty acid synthase as a new therapeutic target for HER2-positive gastric cancer. Cell. Oncol. (Dordr) 46, 661–676. doi:10.1007/s13402-023-00769-x
Chaffer, C. L., Marjanovic, N. D., Lee, T., Bell, G., Kleer, C. G., Reinhardt, F., et al. (2013). Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 154, 61–74. doi:10.1016/j.cell.2013.06.005
Chen, C. L., Uthaya Kumar, D. B., Punj, V., Xu, J., Sher, L., Tahara, S. M., et al. (2016b). NANOG metabolically reprograms tumor-initiating stem-like cells through tumorigenic changes in oxidative phosphorylation and fatty acid metabolism. Cell. Metab. 23, 206–219. doi:10.1016/j.cmet.2015.12.004
Chen, J., Li, Y., Yu, T. S., McKay, R. M., Burns, D. K., Kernie, S. G., et al. (2012). A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 488, 522–526. doi:10.1038/nature11287
Chen, L., Ren, J., Yang, L., Li, Y., Fu, J., Li, Y., et al. (2016c). Stearoyl-CoA desaturase-1 mediated cell apoptosis in colorectal cancer by promoting ceramide synthesis. Sci. Rep. 6, 19665. doi:10.1038/srep19665
Chen, W., Dong, J., Haiech, J., Kilhoffer, M. C., and Zeniou, M. (2016a). Cancer stem cell quiescence and plasticity as major challenges in cancer therapy. Stem Cells Int. 2016, 1740936. doi:10.1155/2016/1740936
Choi, S., Yoo, Y. J., Kim, H., Lee, H., Chung, H., Nam, M. H., et al. (2019). Clinical and biochemical relevance of monounsaturated fatty acid metabolism targeting strategy for cancer stem cell elimination in colon cancer. Biochem. Biophys. Res. Commun. 519, 100–105. doi:10.1016/j.bbrc.2019.08.137
Christofk, H. R., Vander Heiden, M. G., Harris, M. H., Ramanathan, A., Gerszten, R. E., Wei, R., et al. (2008). The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233. doi:10.1038/nature06734
Ciavardelli, D., Rossi, C., Barcaroli, D., Volpe, S., Consalvo, A., Zucchelli, M., et al. (2014). Breast cancer stem cells rely on fermentative glycolysis and are sensitive to 2-deoxyglucose treatment. Cell. Death Dis. 5, e1336. doi:10.1038/cddis.2014.285
Colacino, J. A., McDermott, S. P., Sartor, M. A., Wicha, M. S., and Rozek, L. S. (2016). Transcriptomic profiling of curcumin-treated human breast stem cells identifies a role for stearoyl-coa desaturase in breast cancer prevention. Breast Cancer Res. Treat. 158, 29–41. doi:10.1007/s10549-016-3854-4
Cordenonsi, M., Zanconato, F., Azzolin, L., Forcato, M., Rosato, A., Frasson, C., et al. (2011). The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 147, 759–772. doi:10.1016/j.cell.2011.09.048
Corominas-Faja, B., Cuyas, E., Gumuzio, J., Bosch-Barrera, J., Leis, O., Martin, A. G., et al. (2014). Chemical inhibition of acetyl-CoA carboxylase suppresses self-renewal growth of cancer stem cells. Oncotarget 5, 8306–8316. doi:10.18632/oncotarget.2059
Couturier, C. P., Ayyadhury, S., Le, P. U., Nadaf, J., Monlong, J., Riva, G., et al. (2020). Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat. Commun. 11, 3406. doi:10.1038/s41467-020-17186-5
Cufi, S., Vazquez-Martin, A., Oliveras-Ferraros, C., Martin-Castillo, B., Vellon, L., and Menendez, J. A. (2011). Autophagy positively regulates the CD44(+) CD24(-/low) breast cancer stem-like phenotype. Cell. Cycle 10, 3871–3885. doi:10.4161/cc.10.22.17976
Currie, E., Schulze, A., Zechner, R., Walther, T. C., and Farese, R. V. (2013). Cellular fatty acid metabolism and cancer. Cell. Metab. 18, 153–161. doi:10.1016/j.cmet.2013.05.017
Dando, I., Dalla Pozza, E., Biondani, G., Cordani, M., Palmieri, M., and Donadelli, M. (2015). The metabolic landscape of cancer stem cells. IUBMB Life 67, 687–693. doi:10.1002/iub.1426
Danhier, P., Banski, P., Payen, V. L., Grasso, D., Ippolito, L., Sonveaux, P., et al. (2017). Cancer metabolism in space and time: beyond the Warburg effect. Biochim. Biophys. Acta Bioenerg. 1858, 556–572. doi:10.1016/j.bbabio.2017.02.001
Dattilo, R., Mottini, C., Camera, E., Lamolinara, A., Auslander, N., Doglioni, G., et al. (2020). Pyrvinium pamoate induces death of triple-negative breast cancer stem-like cells and reduces metastases through effects on lipid anabolism. Cancer Res. 80, 4087–4102. doi:10.1158/0008-5472.CAN-19-1184
Dean, M., Fojo, T., and Bates, S. (2005). Tumour stem cells and drug resistance. Nat. Rev. Cancer 5, 275–284. doi:10.1038/nrc1590
De Angelis, M. L., Francescangeli, F., La Torre, F., and Zeuner, A. (2019). Stem cell plasticity and dormancy in the development of cancer therapy resistance. Front. Oncol. 9, 626. doi:10.3389/fonc.2019.00626
DeBose-Boyd, R. A., and Ye, J. (2018). SREBPs in lipid metabolism, insulin signaling, and beyond. Trends Biochem. Sci. 43, 358–368. doi:10.1016/j.tibs.2018.01.005
De Rosa, A., Pellegatta, S., Rossi, M., Tunici, P., Magnoni, L., Speranza, M. C., et al. (2012). A radial glia gene marker, fatty acid binding protein 7 (FABP7), is involved in proliferation and invasion of glioblastoma cells. PLoS One 7, e52113. doi:10.1371/journal.pone.0052113
Dirkse, A., Golebiewska, A., Buder, T., Nazarov, P. V., Muller, A., Poovathingal, S., et al. (2019). Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat. Commun. 10, 1787. doi:10.1038/s41467-019-09853-z
Dong, C., Yuan, T., Wu, Y., Wang, Y., Fan, T. W., Miriyala, S., et al. (2013). Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 23, 316–331. doi:10.1016/j.ccr.2013.01.022
Dorr, J. R., Yu, Y., Milanovic, M., Beuster, G., Zasada, C., Dabritz, J. H., et al. (2013). Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature 501, 421–425. doi:10.1038/nature12437
Easwaran, H., Johnstone, S. E., Van Neste, L., Ohm, J., Mosbruger, T., Wang, Q., et al. (2012). A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res. 22, 837–849. doi:10.1101/gr.131169.111
Ehmsen, S., Pedersen, M. H., Wang, G., Terp, M. G., Arslanagic, A., Hood, B. L., et al. (2019). Increased cholesterol biosynthesis is a key characteristic of breast cancer stem cells influencing patient outcome. Cell. Rep. 27, 3927–3938. doi:10.1016/j.celrep.2019.05.104
El-Sahli, S., Xie, Y., Wang, L., and Liu, S. (2019). Wnt signaling in cancer metabolism and immunity. Cancers (Basel) 11, 904. doi:10.3390/cancers11070904
Erhart, F., Blauensteiner, B., Zirkovits, G., Printz, D., Soukup, K., Klingenbrunner, S., et al. (2019). Gliomasphere marker combinatorics: multidimensional flow cytometry detects CD44+/CD133+/ITGA6+/CD36+ signature. J. Cell. Mol. Med. 23, 281–292. doi:10.1111/jcmm.13927
Estan, M. C., Calvino, E., Calvo, S., Guillen-Guio, B., Boyano-Adanez Mdel, C., de Blas, E., et al. (2014). Apoptotic efficacy of etomoxir in human acute myeloid leukemia cells. Cooperation with arsenic trioxide and glycolytic inhibitors, and regulation by oxidative stress and protein kinase activities. PLoS One 9, e115250. doi:10.1371/journal.pone.0115250
Fang, W., Cui, H., Yu, D., Chen, Y., Wang, J., and Yu, G. (2014). Increased expression of phospho-acetyl-CoA carboxylase protein is an independent prognostic factor for human gastric cancer without lymph node metastasis. Med. Oncol. 31, 15. doi:10.1007/s12032-014-0015-7
Fendler, A., Bauer, D., Busch, J., Jung, K., Wulf-Goldenberg, A., Kunz, S., et al. (2020). Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Nat. Commun. 11, 929. doi:10.1038/s41467-020-14700-7
Feng, X., Zhang, L., Xu, S., and Shen, A. Z. (2020). ATP-citrate lyase (ACLY) in lipid metabolism and atherosclerosis: an updated review. Prog. Lipid Res. 77, 101006. doi:10.1016/j.plipres.2019.101006
Fritz, V., Benfodda, Z., Rodier, G., Henriquet, C., Iborra, F., Avances, C., et al. (2010). Abrogation of de novo lipogenesis by stearoyl-CoA desaturase 1 inhibition interferes with oncogenic signaling and blocks prostate cancer progression in mice. Mol. Cancer Ther. 9, 1740–1754. doi:10.1158/1535-7163.MCT-09-1064
Fu, T., Coulter, S., Yoshihara, E., Oh, T. G., Fang, S., Cayabyab, F., et al. (2019). FXR regulates intestinal cancer stem cell proliferation. Cell. 176, 1098–1112. doi:10.1016/j.cell.2019.01.036
Galavotti, S., Bartesaghi, S., Faccenda, D., Shaked-Rabi, M., Sanzone, S., McEvoy, A., et al. (2013). The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells. Oncogene 32, 699–712. doi:10.1038/onc.2012.111
Galli, R., Binda, E., Orfanelli, U., Cipelletti, B., Gritti, A., De Vitis, S., et al. (2004). Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 64, 7011–7021. doi:10.1158/0008-5472.CAN-04-1364
Geng, F., Cheng, X., Wu, X., Yoo, J. Y., Cheng, C., Guo, J. Y., et al. (2016). Inhibition of SOAT1 suppresses glioblastoma growth via blocking SREBP-1-mediated lipogenesis. Clin. Cancer Res. 22, 5337–5348. doi:10.1158/1078-0432.CCR-15-2973
Gerlinger, M., Rowan, A. J., Horswell, S., Math, M., Larkin, J., Endesfelder, D., et al. (2012). Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892. doi:10.1056/NEJMoa1113205
Gimple, R. C., Kidwell, R. L., Kim, L. J. Y., Sun, T., Gromovsky, A. D., Wu, Q., et al. (2019). Glioma stem cell-specific superenhancer promotes polyunsaturated fatty-acid synthesis to support EGFR signaling. Cancer Discov. 9, 1248–1267. doi:10.1158/2159-8290.cd-19-0061
Ginestier, C., Monville, F., Wicinski, J., Cabaud, O., Cervera, N., Josselin, E., et al. (2012). Mevalonate metabolism regulates Basal breast cancer stem cells and is a potential therapeutic target. Stem Cells 30, 1327–1337. doi:10.1002/stem.1122
Global Burden of Disease Cancer, C., Kocarnik, J. M., Compton, K., Dean, F. E., Fu, W., Gaw, B. L., et al. (2022). Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019: a systematic analysis for the global burden of Disease study 2019. JAMA Oncol. 8, 420–444. doi:10.1001/jamaoncol.2021.6987
Goyette, S., Liang, Y., Mafuvadze, B., Cook, M. T., Munir, M., and Hyder, S. M. (2017). Natural and synthetic progestins enrich cancer stem cell-like cells in hormone-responsive human breast cancer cell populations in vitro. Breast Cancer (Dove Med. Press) 9, 347–357. doi:10.2147/BCTT.S135371
Gu, Q., Yang, X., Lv, J., Zhang, J., Xia, B., Kim, J. D., et al. (2019). AIBP-mediated cholesterol efflux instructs hematopoietic stem and progenitor cell fate. Science 363, 1085–1088. doi:10.1126/science.aav1749
Gupta, P. B., Onder, T. T., Jiang, G., Tao, K., Kuperwasser, C., Weinberg, R. A., et al. (2009). Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 138, 645–659. doi:10.1016/j.cell.2009.06.034
Hafiane, A., Gasbarrino, K., and Daskalopoulou, S. S. (2019). The role of adiponectin in cholesterol efflux and HDL biogenesis and metabolism. Metabolism 100, 153953. doi:10.1016/j.metabol.2019.153953
Hale, J. S., Otvos, B., Sinyuk, M., Alvarado, A. G., Hitomi, M., Stoltz, K., et al. (2014). Cancer stem cell-specific scavenger receptor CD36 drives glioblastoma progression. Stem Cells 32, 1746–1758. doi:10.1002/stem.1716
Han, Y., Fan, S., Qin, T., Yang, J., Sun, Y., Lu, Y., et al. (2018). Role of autophagy in breast cancer and breast cancer stem cells (Review). Int. J. Oncol. 52, 1057–1070. doi:10.3892/ijo.2018.4270
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell. 144, 646–674. doi:10.1016/j.cell.2011.02.013
Hanai, J. I., Doro, N., Seth, P., and Sukhatme, V. P. (2013). ATP citrate lyase knockdown impacts cancer stem cells in vitro. Cell. Death Dis. 4, e696. doi:10.1038/cddis.2013.215
Harrison, H., Farnie, G., Howell, S. J., Rock, R. E., Stylianou, S., Brennan, K. R., et al. (2010). Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 70, 709–718. doi:10.1158/0008-5472.CAN-09-1681
Hatzivassiliou, G., Zhao, F., Bauer, D. E., Andreadis, C., Shaw, A. N., Dhanak, D., et al. (2005). ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 8, 311–321. doi:10.1016/j.ccr.2005.09.008
Herpel, E., Jensen, K., Muley, T., Warth, A., Schnabel, P. A., Meister, M., et al. (2011). The cancer stem cell antigens CD133, BCRP1/ABCG2 and CD117/c-KIT are not associated with prognosis in resected early-stage non-small cell lung cancer. Anticancer Res. 31, 4491–4500.
Herreros-Pomares, A., de-Maya-Girones, J. D., Calabuig-Farinas, S., Lucas, R., Martinez, A., Pardo-Sanchez, J. M., et al. (2019). Lung tumorspheres reveal cancer stem cell-like properties and a score with prognostic impact in resected non-small-cell lung cancer. Cell. Death Dis. 10, 660. doi:10.1038/s41419-019-1898-1
Higa, T., and Nakayama, K. I. (2024). Cell cycle heterogeneity and plasticity of colorectal cancer stem cells. Cancer Sci. 115, 1370–1377. doi:10.1111/cas.16117
Hoffmeyer, K., Raggioli, A., Rudloff, S., Anton, R., Hierholzer, A., Del Valle, I., et al. (2012). Wnt/β-catenin signaling regulates telomerase in stem cells and cancer cells. Science 336, 1549–1554. doi:10.1126/science.1218370
Holubarsch, C. J., Rohrbach, M., Karrasch, M., Boehm, E., Polonski, L., Ponikowski, P., et al. (2007). A double-blind randomized multicentre clinical trial to evaluate the efficacy and safety of two doses of etomoxir in comparison with placebo in patients with moderate congestive heart failure: the ERGO (etomoxir for the recovery of glucose oxidation) study. Clin. Sci. (Lond) 113, 205–212. doi:10.1042/CS20060307
Hu, A., and Song, B. L. (2019). The interplay of Patched, Smoothened and cholesterol in Hedgehog signaling. Curr. Opin. Cell. Biol. 61, 31–38. doi:10.1016/j.ceb.2019.06.008
Huo, X., Zhang, Q., Liu, A. M., Tang, C., Gong, Y., Bian, J., et al. (2013). Overexpression of Yes-associated protein confers doxorubicin resistance in hepatocellullar carcinoma. Oncol. Rep. 29, 840–846. doi:10.3892/or.2012.2176
Iannelli, F., Roca, M. S., Lombardi, R., Ciardiello, C., Grumetti, L., De Rienzo, S., et al. (2020). Synergistic antitumor interaction of valproic acid and simvastatin sensitizes prostate cancer to docetaxel by targeting CSCs compartment via YAP inhibition. J. Exp. Clin. Cancer Res. 39, 213. doi:10.1186/s13046-020-01723-7
Janiszewska, M., Suva, M. L., Riggi, N., Houtkooper, R. H., Auwerx, J., Clement-Schatlo, V., et al. (2012). Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes. Dev. 26, 1926–1944. doi:10.1101/gad.188292.112
Jia, Q., Zhang, X., Deng, T., and Gao, J. (2013). Positive correlation of Oct4 and ABCG2 to chemotherapeutic resistance in CD90(+)CD133(+) liver cancer stem cells. Cell. Reprogr. 15, 143–150. doi:10.1089/cell.2012.0048
Jiao, J., Hindoyan, A., Wang, S., Tran, L. M., Goldstein, A. S., Lawson, D., et al. (2012). Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS One 7, e42564. doi:10.1371/journal.pone.0042564
Jones, S. F., and Infante, J. R. (2015). Molecular pathways: fatty acid synthase. Clin. cancer Res. official J. Am. Assoc. Cancer Res. 21, 5434–5438. doi:10.1158/1078-0432.ccr-15-0126
Joyce, S. A., and Gahan, C. G. (2016). Bile acid modifications at the microbe-host interface: potential for nutraceutical and pharmaceutical interventions in host health. Annu. Rev. Food Sci. Technol. 7, 313–333. doi:10.1146/annurev-food-041715-033159
Ju, H. Q., Zhan, G., Huang, A., Sun, Y., Wen, S., Yang, J., et al. (2017). ITD mutation in FLT3 tyrosine kinase promotes Warburg effect and renders therapeutic sensitivity to glycolytic inhibition. Leukemia 31, 2143–2150. doi:10.1038/leu.2017.45
Kalucka, J., Bierhansl, L., Conchinha, N. V., Missiaen, R., Elia, I., Bruning, U., et al. (2018). Quiescent endothelial cells upregulate fatty acid beta-oxidation for vasculoprotection via redox homeostasis. Cell. Metab. 28, 881–894 e813. doi:10.1016/j.cmet.2018.07.016
Kathagen, A., Schulte, A., Balcke, G., Phillips, H. S., Martens, T., Matschke, J., et al. (2013). Hypoxia and oxygenation induce a metabolic switch between pentose phosphate pathway and glycolysis in glioma stem-like cells. Acta Neuropathol. 126, 763–780. doi:10.1007/s00401-013-1173-y
Kinlaw, W. B., Baures, P. W., Lupien, L. E., Davis, W. L., and Kuemmerle, N. B. (2016). Fatty acids and breast cancer: make them on site or have them delivered. J. Cell. Physiol. 231, 2128–2141. doi:10.1002/jcp.25332
Klionsky, D. J., Eskelinen, E. L., and Deretic, V. (2014). Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes. wait, I'm confused. Autophagy 10, 549–551. doi:10.4161/auto.28448
Koo, J. H., and Guan, K. L. (2018). Interplay between YAP/TAZ and metabolism. Cell. Metab. 28, 196–206. doi:10.1016/j.cmet.2018.07.010
Koren, E., and Fuchs, Y. (2016). The bad seed: cancer stem cells in tumor development and resistance. Drug Resist Updat 28, 1–12. doi:10.1016/j.drup.2016.06.006
Krstic, J., Trivanovic, D., Jaukovic, A., Santibanez, J. F., and Bugarski, D. (2017). Metabolic plasticity of stem cells and macrophages in cancer. Front. Immunol. 8, 939. doi:10.3389/fimmu.2017.00939
Kulkarni, M., Tan, T. Z., Syed Sulaiman, N. B., Lamar, J. M., Bansal, P., Cui, J., et al. (2018). RUNX1 and RUNX3 protect against YAP-mediated EMT, stem-ness and shorter survival outcomes in breast cancer. Oncotarget 9, 14175–14192. doi:10.18632/oncotarget.24419
Kuntz, E. M., Baquero, P., Michie, A. M., Dunn, K., Tardito, S., Holyoake, T. L., et al. (2017). Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat. Med. 23, 1234–1240. doi:10.1038/nm.4399
Lagadinou, E. D., Sach, A., Callahan, K., Rossi, R. M., Neering, S. J., Minhajuddin, M., et al. (2013). BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell. Stem Cell. 12, 329–341. doi:10.1016/j.stem.2012.12.013
Lai, K. K. Y., Kweon, S. M., Chi, F., Hwang, E., Kabe, Y., Higashiyama, R., et al. (2017). Stearoyl-CoA desaturase promotes liver fibrosis and tumor development in mice via a Wnt positive-signaling loop by stabilization of low-density lipoprotein-receptor-related proteins 5 and 6. Gastroenterology 152, 1477–1491. doi:10.1053/j.gastro.2017.01.021
Lang, C. M. R., Chan, C. K., Veltri, A., and Lien, W. H. (2019). Wnt signaling pathways in keratinocyte carcinomas. Cancers (Basel) 11, 1216. doi:10.3390/cancers11091216
Lecot, P., Alimirah, F., Desprez, P. Y., Campisi, J., and Wiley, C. (2016). Context-dependent effects of cellular senescence in cancer development. Br. J. Cancer 114, 1180–1184. doi:10.1038/bjc.2016.115
Lee, K. M., Giltnane, J. M., Balko, J. M., Schwarz, L. J., Guerrero-Zotano, A. L., Hutchinson, K. E., et al. (2017). MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell. metab. 26, 633–647. doi:10.1016/j.cmet.2017.09.009
Lewis, C. A., Brault, C., Peck, B., Bensaad, K., Griffiths, B., Mitter, R., et al. (2015). SREBP maintains lipid biosynthesis and viability of cancer cells under lipid- and oxygen-deprived conditions and defines a gene signature associated with poor survival in glioblastoma multiforme. Oncogene 34, 5128–5140. doi:10.1038/onc.2014.439
Li, J., Condello, S., Thomes-Pepin, J., Ma, X., Xia, Y., Hurley, T. D., et al. (2017). Lipid desaturation is a metabolic marker and therapeutic target of ovarian cancer stem cells. Cell. Stem Cell. 20, 303–314. doi:10.1016/j.stem.2016.11.004
Li, L., Wang, C., Calvisi, D. F., Evert, M., Pilo, M. G., Jiang, L., et al. (2013a). SCD1 Expression is dispensable for hepatocarcinogenesis induced by AKT and Ras oncogenes in mice. PLoS One 8, e75104. doi:10.1371/journal.pone.0075104
Li, S. H., Fu, J., Watkins, D. N., Srivastava, R. K., and Shankar, S. (2013b). Sulforaphane regulates self-renewal of pancreatic cancer stem cells through the modulation of Sonic hedgehog-GLI pathway. Mol. Cell. Biochem. 373, 217–227. doi:10.1007/s11010-012-1493-6
Li, Y., Chao, X., Yang, L., Lu, Q., Li, T., Ding, W. X., et al. (2018). Impaired fasting-induced adaptive lipid droplet biogenesis in liver-specific atg5-deficient mouse liver is mediated by persistent nuclear factor-like 2 activation. Am. J. Pathol. 188, 1833–1846. doi:10.1016/j.ajpath.2018.04.015
Li, Y., Huang, X., Yang, G., Xu, K., Yin, Y., Brecchia, G., et al. (2022). CD36 favours fat sensing and transport to govern lipid metabolism. Prog. Lipid Res. 88, 101193. doi:10.1016/j.plipres.2022.101193
Liang, R., Li, X., Li, W., Zhu, X., and Li, C. (2021). DNA methylation in lung cancer patients: opening a "window of life" under precision medicine. Biomed. Pharmacother. 144, 112202. doi:10.1016/j.biopha.2021.112202
Liang, Y., Goyette, S., and Hyder, S. M. (2017). Cholesterol biosynthesis inhibitor RO 48-8071 reduces progesterone receptor expression and inhibits progestin-dependent stem cell-like cell growth in hormone-dependent human breast cancer cells. Breast Cancer (Dove Med. Press) 9, 487–494. doi:10.2147/BCTT.S140265
Liao, J., Qian, F., Tchabo, N., Mhawech-Fauceglia, P., Beck, A., Qian, Z., et al. (2014). Ovarian cancer spheroid cells with stem cell-like properties contribute to tumor generation, metastasis and chemotherapy resistance through hypoxia-resistant metabolism. PloS one 9, e84941. doi:10.1371/journal.pone.0084941
Liberti, M. V., and Locasale, J. W. (2016). The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. 41, 211–218. doi:10.1016/j.tibs.2015.12.001
Lin, L., Ding, Y., Wang, Y., Wang, Z., Yin, X., Yan, G., et al. (2017). Functional lipidomics: palmitic acid impairs hepatocellular carcinoma development by modulating membrane fluidity and glucose metabolism. Hepatology 66, 432–448. doi:10.1002/hep.29033
Lin, Y., Yu, B., Fang, P., and Wang, J. (2023). Inhibiting autophagy before it starts. Autophagy 20, 923–924. doi:10.1080/15548627.2023.2197364
Lingwood, D., and Simons, K. (2010). Lipid rafts as a membrane-organizing principle. Science 327, 46–50. doi:10.1126/science.1174621
Liu, H., Zhang, Z., Song, L., Gao, J., and Liu, Y. (2022). Lipid metabolism of cancer stem cells. Oncol. Lett. 23, 119. doi:10.3892/ol.2022.13239
Lobello, N., Biamonte, F., Pisanu, M. E., Faniello, M. C., Jakopin, Z., Chiarella, E., et al. (2016). Ferritin heavy chain is a negative regulator of ovarian cancer stem cell expansion and epithelial to mesenchymal transition. Oncotarget 7, 62019–62033. doi:10.18632/oncotarget.11495
Long, B., Wang, L. X., Zheng, F. M., Lai, S. P., Xu, D. R., Hu, Y., et al. (2016). Targeting GLI1 suppresses cell growth and enhances chemosensitivity in CD34+ enriched acute myeloid leukemia progenitor cells. Cell. Physiol. Biochem. 38, 1288–1302. doi:10.1159/000443075
Lopes, M. B., and Vinga, S. (2020). Tracking intratumoral heterogeneity in glioblastoma via regularized classification of single-cell RNA-Seq data. BMC Bioinforma. 21, 59. doi:10.1186/s12859-020-3390-4
Lucenay, K. S., Doostan, I., Karakas, C., Bui, T., Ding, Z., Mills, G. B., et al. (2016). Cyclin E associates with the lipogenic enzyme ATP-citrate lyase to enable malignant growth of breast cancer cells. Cancer Res. 76, 2406–2418. doi:10.1158/0008-5472.CAN-15-1646
Lue, H. W., Podolak, J., Kolahi, K., Cheng, L., Rao, S., Garg, D., et al. (2017). Metabolic reprogramming ensures cancer cell survival despite oncogenic signaling blockade. Genes. Dev. 31, 2067–2084. doi:10.1101/gad.305292.117
Luo, J., Hong, Y., Lu, Y., Qiu, S., Chaganty, B. K., Zhang, L., et al. (2017b). Acetyl-CoA carboxylase rewires cancer metabolism to allow cancer cells to survive inhibition of the Warburg effect by cetuximab. Cancer Lett. 384, 39–49. doi:10.1016/j.canlet.2016.09.020
Luo, X., Cheng, C., Tan, Z., Li, N., Tang, M., Yang, L., et al. (2017a). Emerging roles of lipid metabolism in cancer metastasis. Mol. Cancer 16, 76. doi:10.1186/s12943-017-0646-3
Ma, M. K. F., Lau, E. Y. T., Leung, D. H. W., Lo, J., Ho, N. P. Y., Cheng, L. K. W., et al. (2017). Stearoyl-CoA desaturase regulates sorafenib resistance via modulation of ER stress-induced differentiation. J. Hepatol. 67, 979–990. doi:10.1016/j.jhep.2017.06.015
Ma, X. L., Sun, Y. F., Wang, B. L., Shen, M. N., Zhou, Y., Chen, J. W., et al. (2019). Sphere-forming culture enriches liver cancer stem cells and reveals Stearoyl-CoA desaturase 1 as a potential therapeutic target. BMC Cancer 19, 760. doi:10.1186/s12885-019-5963-z
Mahapatra, K. K., Mishra, S. R., Behera, B. P., Patil, S., Gewirtz, D. A., and Bhutia, S. K. (2021). The lysosome as an imperative regulator of autophagy and cell death. Cell. Mol. Life Sci. 78, 7435–7449. doi:10.1007/s00018-021-03988-3
Mai, T. T., Hamai, A., Hienzsch, A., Caneque, T., Muller, S., Wicinski, J., et al. (2017). Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat. Chem. 9, 1025–1033. doi:10.1038/nchem.2778
Maiuri, A. R., Savant, S. S., Podicheti, R., Rusch, D. B., and O'Hagan, H. M. (2019). DNA methyltransferase inhibition reduces inflammation-induced colon tumorigenesis. Epigenetics 14, 1209–1223. doi:10.1080/15592294.2019.1634986
Mancini, R., Noto, A., Pisanu, M. E., De Vitis, C., Maugeri-Sacca, M., and Ciliberto, G. (2018). Metabolic features of cancer stem cells: the emerging role of lipid metabolism. Oncogene 37, 2367–2378. doi:10.1038/s41388-018-0141-3
Margaritescu, C., Pirici, D., Cherciu, I., Barbalan, A., Cartana, T., and Saftoiu, A. (2014). CD133/CD166/Ki-67 triple immunouorescence assessment for putative cancer stem cells in colon carcinoma*. J. Gastrointestin Liver Dis. 23, 161–170. doi:10.15403/jgld.2014.1121.232.cm1
Martinez-Outschoorn, U. E., Peiris-Pages, M., Pestell, R. G., Sotgia, F., and Lisanti, M. P. (2017). Cancer metabolism: a therapeutic perspective. Nat. Rev. Clin. Oncol. 14, 11–31. doi:10.1038/nrclinonc.2016.60
Martinez-Reyes, I., and Chandel, N. S. (2021). Cancer metabolism: looking forward. Nat. Rev. Cancer 21, 669–680. doi:10.1038/s41568-021-00378-6
Medema, J. P. (2013). Cancer stem cells: the challenges ahead. Nat. Cell. Biol. 15, 338–344. doi:10.1038/ncb2717
Menendez, J. A., and Lupu, R. (2017). Fatty acid synthase (FASN) as a therapeutic target in breast cancer. Expert Opin. Ther. Targets 21, 1001–1016. doi:10.1080/14728222.2017.1381087
Metallo, C. M., Gameiro, P. A., Bell, E. L., Mattaini, K. R., Yang, J., Hiller, K., et al. (2011). Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384. doi:10.1038/nature10602
Michaloglou, C., Vredeveld, L. C., Soengas, M. S., Denoyelle, C., Kuilman, T., van der Horst, C. M., et al. (2005). BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436, 720–724. doi:10.1038/nature03890
Migita, T., Narita, T., Nomura, K., Miyagi, E., Inazuka, F., Matsuura, M., et al. (2008). ATP citrate lyase: activation and therapeutic implications in non-small cell lung cancer. Cancer Res. 68, 8547–8554. doi:10.1158/0008-5472.CAN-08-1235
Milanovic, M., Fan, D. N. Y., Belenki, D., Dabritz, J. H. M., Zhao, Z., Yu, Y., et al. (2018). Senescence-associated reprogramming promotes cancer stemness. Nature 553, 96–100. doi:10.1038/nature25167
Minami, N., Tanaka, K., Sasayama, T., Kohmura, E., Saya, H., and Sampetrean, O. (2021). Lactate reprograms energy and lipid metabolism in glucose-deprived oxidative glioma stem cells. Metabolites 11, 325. doi:10.3390/metabo11050325
Miyazaki, Y., Matsubara, S., Ding, Q., Tsukasa, K., Yoshimitsu, M., Kosai, K., et al. (2016). Efficient elimination of pancreatic cancer stem cells by hedgehog/GLI inhibitor GANT61 in combination with mTOR inhibition. Mol. Cancer 15, 49. doi:10.1186/s12943-016-0534-2
Mo, J. S., Park, H. W., and Guan, K. L. (2014). The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 15, 642–656. doi:10.15252/embr.201438638
Moncur, J. T., Park, J. P., Memoli, V. A., Mohandas, T. K., and Kinlaw, W. B. (1998). The "Spot 14" gene resides on the telomeric end of the 11q13 amplicon and is expressed in lipogenic breast cancers: implications for control of tumor metabolism. Proc. Natl. Acad. Sci. U. S. A. 95, 6989–6994. doi:10.1073/pnas.95.12.6989
Morihiro, Y., Yasumoto, Y., Vaidyan, L. K., Sadahiro, H., Uchida, T., Inamura, A., et al. (2013). Fatty acid binding protein 7 as a marker of glioma stem cells. Pathol. Int. 63, 546–553. doi:10.1111/pin.12109
Mullen, P. J., Yu, R., Longo, J., Archer, M. C., and Penn, L. Z. (2016). The interplay between cell signalling and the mevalonate pathway in cancer. Nat. Rev. Cancer 16, 718–731. doi:10.1038/nrc.2016.76
Munir, R., Lisec, J., Swinnen, J. V., and Zaidi, N. (2019). Lipid metabolism in cancer cells under metabolic stress. Br. J. Cancer 120, 1090–1098. doi:10.1038/s41416-019-0451-4
Myant, K. B., Cammareri, P., McGhee, E. J., Ridgway, R. A., Huels, D. J., Cordero, J. B., et al. (2013). ROS production and NF-κB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell. Stem Cell. 12, 761–773. doi:10.1016/j.stem.2013.04.006
Nath, A., Li, I., Roberts, L. R., and Chan, C. (2015). Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma. Sci. Rep. 5, 14752. doi:10.1038/srep14752
Neftel, C., Laffy, J., Filbin, M. G., Hara, T., Shore, M. E., Rahme, G. J., et al. (2019). An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 178, 835–849. doi:10.1016/j.cell.2019.06.024
Ni, H., Qin, H., Sun, C., Liu, Y., Ruan, G., Guo, Q., et al. (2021). MiR-375 reduces the stemness of gastric cancer cells through triggering ferroptosis. Stem Cell. Res. Ther. 12, 325. doi:10.1186/s13287-021-02394-7
Nishizawa, S., Sumi, H., Satoh, Y., Yamamoto, Y., Kitazawa, S., Honda, K., et al. (2017). In vitro and in vivo antitumor activities of T-3764518, a novel and orally available small molecule stearoyl-CoA desaturase 1 inhibitor. Eur. J. Pharmacol. 807, 21–31. doi:10.1016/j.ejphar.2017.03.064
Noto, A., De Vitis, C., Pisanu, M. E., Roscilli, G., Ricci, G., Catizone, A., et al. (2017a). Stearoyl-CoA-desaturase 1 regulates lung cancer stemness via stabilization and nuclear localization of YAP/TAZ. Oncogene 36, 4573–4584. doi:10.1038/onc.2017.75
Noto, A., De Vitis, C., Pisanu, M. E., Roscilli, G., Ricci, G., Catizone, A., et al. (2017b). Stearoyl-CoA-desaturase 1 regulates lung cancer stemness via stabilization and nuclear localization of YAP/TAZ. Oncogene 36, 4671–4672. doi:10.1038/onc.2017.212
Noto, A., Raffa, S., De Vitis, C., Roscilli, G., Malpicci, D., Coluccia, P., et al. (2013). Stearoyl-CoA desaturase-1 is a key factor for lung cancer-initiating cells. Cell. Death Dis. 4, e947. doi:10.1038/cddis.2013.444
Novak, D., Huser, L., Elton, J. J., Umansky, V., Altevogt, P., and Utikal, J. (2020). SOX2 in development and cancer biology. Semin. Cancer Biol. 67, 74–82. doi:10.1016/j.semcancer.2019.08.007
Obniski, R., Sieber, M., and Spradling, A. C. (2018). Dietary lipids modulate notch signaling and influence adult intestinal development and metabolism in Drosophila. Dev. Cell. 47, 98–111. doi:10.1016/j.devcel.2018.08.013
OBrien, C. A., Pollett, A., Gallinger, S., and Dick, J. E. (2007). A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445, 106–110. doi:10.1038/nature05372
Pacilli, A., Calienni, M., Margarucci, S., D'Apolito, M., Petillo, O., Rocchi, L., et al. (2013). Carnitine-acyltransferase system inhibition, cancer cell death, and prevention of myc-induced lymphomagenesis. J. Natl. Cancer Inst. 105, 489–498. doi:10.1093/jnci/djt030
Palorini, R., Votta, G., Balestrieri, C., Monestiroli, A., Olivieri, S., Vento, R., et al. (2014). Energy metabolism characterization of a novel cancer stem cell-like line 3AB-OS. J. Cell. Biochem. 115, 368–379. doi:10.1002/jcb.24671
Pandey, P. R., Okuda, H., Watabe, M., Pai, S. K., Liu, W., Kobayashi, A., et al. (2011). Resveratrol suppresses growth of cancer stem-like cells by inhibiting fatty acid synthase. Breast cancer Res. Treat. 130, 387–398. doi:10.1007/s10549-010-1300-6
Pandey, P. R., Xing, F., Sharma, S., Watabe, M., Pai, S. K., Iiizumi-Gairani, M., et al. (2013). Elevated lipogenesis in epithelial stem-like cell confers survival advantage in ductal carcinoma in situ of breast cancer. Oncogene 32, 5111–5122. doi:10.1038/onc.2012.519
Pascual, G., Avgustinova, A., Mejetta, S., Martin, M., Castellanos, A., Attolini, C. S., et al. (2017). Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 541, 41–45. doi:10.1038/nature20791
Pasto, A., Bellio, C., Pilotto, G., Ciminale, V., Silic-Benussi, M., Guzzo, G., et al. (2014). Cancer stem cells from epithelial ovarian cancer patients privilege oxidative phosphorylation, and resist glucose deprivation. Oncotarget 5, 4305–4319. doi:10.18632/oncotarget.2010
Patel, A. P., Tirosh, I., Trombetta, J. J., Shalek, A. K., Gillespie, S. M., Wakimoto, H., et al. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401. doi:10.1126/science.1254257
Patel, S., Alam, A., Pant, R., and Chattopadhyay, S. (2019). Wnt signaling and its significance within the tumor microenvironment: novel therapeutic insights. Front. Immunol. 10, 2872. doi:10.3389/fimmu.2019.02872
Paul, R., Dorsey, J. F., and Fan, Y. (2022). Cell plasticity, senescence, and quiescence in cancer stem cells: biological and therapeutic implications. Pharmacol. Ther. 231, 107985. doi:10.1016/j.pharmthera.2021.107985
Peck, B., and Schulze, A. (2016). Lipid desaturation - the next step in targeting lipogenesis in cancer? FEBS J. 283, 2767–2778. doi:10.1111/febs.13681
Peiris-Pages, M., Martinez-Outschoorn, U. E., Pestell, R. G., Sotgia, F., and Lisanti, M. P. (2016). Cancer stem cell metabolism. Breast cancer Res. BCR 18, 55. doi:10.1186/s13058-016-0712-6
Peng, F., Wang, J. H., Fan, W. J., Meng, Y. T., Li, M. M., Li, T. T., et al. (2018). Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene 37, 1062–1074. doi:10.1038/onc.2017.368
Petrova, E., Scholz, A., Paul, J., Sturz, A., Haike, K., Siegel, F., et al. (2017). Acetyl-CoA carboxylase inhibitors attenuate WNT and Hedgehog signaling and suppress pancreatic tumor growth. Oncotarget 8, 48660–48670. doi:10.18632/oncotarget.12650
Pisanu, M. E., Maugeri-Sacca, M., Fattore, L., Bruschini, S., De Vitis, C., Tabbi, E., et al. (2018). Inhibition of Stearoyl-CoA desaturase 1 reverts BRAF and MEK inhibition-induced selection of cancer stem cells in BRAF-mutated melanoma. J. Exp. Clin. Cancer Res. 37, 318. doi:10.1186/s13046-018-0989-7
Pisanu, M. E., Noto, A., De Vitis, C., Morrone, S., Scognamiglio, G., Botti, G., et al. (2017). Blockade of Stearoyl-CoA-desaturase 1 activity reverts resistance to cisplatin in lung cancer stem cells. Cancer Lett. 406, 93–104. doi:10.1016/j.canlet.2017.07.027
Pizer, E. S., Thupari, J., Han, W. F., Pinn, M. L., Chrest, F. J., Frehywot, G. L., et al. (2000). Malonyl-coenzyme-A is a potential mediator of cytotoxicity induced by fatty-acid synthase inhibition in human breast cancer cells and xenografts. Cancer Res. 60, 213–218.
Polewski, M. D., Reveron-Thornton, R. F., Cherryholmes, G. A., Marinov, G. K., and Aboody, K. S. (2017). SLC7A11 overexpression in glioblastoma is associated with increased cancer stem cell-like properties. Stem Cells Dev. 26, 1236–1246. doi:10.1089/scd.2017.0123
Porstmann, T., Santos, C. R., Griffiths, B., Cully, M., Wu, M., Leevers, S., et al. (2008). SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell. Metab. 8, 224–236. doi:10.1016/j.cmet.2008.07.007
Potze, L., di Franco, S., Kessler, J. H., Stassi, G., and Medema, J. P. (2016). Betulinic acid kills colon cancer stem cells. Curr. Stem Cell. Res. Ther. 11, 427–433. doi:10.2174/1574888x11666151203223512
Prasetyanti, P. R., and Medema, J. P. (2017). Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 16, 41. doi:10.1186/s12943-017-0600-4
Qian, X., Hu, J., Zhao, J., and Chen, H. (2015). ATP citrate lyase expression is associated with advanced stage and prognosis in gastric adenocarcinoma. Int. J. Clin. Exp. Med. 8, 7855–7860.
Rabionet, M., Polonio-Alcala, E., Relat, J., Yeste, M., Sims-Mourtada, J., Kloxin, A. M., et al. (2021). Fatty acid synthase as a feasible biomarker for triple negative breast cancer stem cell subpopulation cultured on electrospun scaffolds. Mater Today Bio 12, 100155. doi:10.1016/j.mtbio.2021.100155
Radhakrishnan, A., Rohatgi, R., and Siebold, C. (2020). Cholesterol access in cellular membranes controls Hedgehog signaling. Nat. Chem. Biol. 16, 1303–1313. doi:10.1038/s41589-020-00678-2
Reya, T., Morrison, S. J., Clarke, M. F., and Weissman, I. L. (2001). Stem cells, cancer, and cancer stem cells. Nature 414, 105–111. doi:10.1038/35102167
Ricciardi, M. R., Mirabilii, S., Allegretti, M., Licchetta, R., Calarco, A., Torrisi, M. R., et al. (2015). Targeting the leukemia cell metabolism by the CPT1a inhibition: functional preclinical effects in leukemias. Blood 126, 1925–1929. doi:10.1182/blood-2014-12-617498
Ricoult, S. J., Yecies, J. L., Ben-Sahra, I., and Manning, B. D. (2016). Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through mTORC1 and SREBP. Oncogene 35, 1250–1260. doi:10.1038/onc.2015.179
Riester, M., Xu, Q., Moreira, A., Zheng, J., Michor, F., and Downey, R. J. (2018). The Warburg effect: persistence of stem-cell metabolism in cancers as a failure of differentiation. Ann. Oncol. 29, 264–270. doi:10.1093/annonc/mdx645
Rios-Esteves, J., and Resh, M. D. (2013). Stearoyl CoA desaturase is required to produce active, lipid-modified Wnt proteins. Cell. Rep. 4, 1072–1081. doi:10.1016/j.celrep.2013.08.027
Robey, R. W., Pluchino, K. M., Hall, M. D., Fojo, A. T., Bates, S. E., and Gottesman, M. M. (2018). Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 18, 452–464. doi:10.1038/s41568-018-0005-8
Rohrig, F., and Schulze, A. (2016). The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 16, 732–749. doi:10.1038/nrc.2016.89
Samudio, I., Harmancey, R., Fiegl, M., Kantarjian, H., Konopleva, M., Korchin, B., et al. (2010). Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J. Clin. Investig. 120, 142–156. doi:10.1172/JCI38942
Sanchez-Martinez, R., Cruz-Gil, S., Gomez de Cedron, M., Alvarez-Fernandez, M., Vargas, T., Molina, S., et al. (2015). A link between lipid metabolism and epithelial-mesenchymal transition provides a target for colon cancer therapy. Oncotarget 6, 38719–38736. doi:10.18632/oncotarget.5340
Sancho, P., Barneda, D., and Heeschen, C. (2016). Hallmarks of cancer stem cell metabolism. Br. J. Cancer 114, 1305–1312. doi:10.1038/bjc.2016.152
Sasaki, H., Nishizaki, Y., Hui, C., Nakafuku, M., and Kondoh, H. (1999). Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 126, 3915–3924. doi:10.1242/dev.126.17.3915
Sayd, S., Thirant, C., El-Habr, E. A., Lipecka, J., Dubois, L. G., Bogeas, A., et al. (2014). Sirtuin-2 activity is required for glioma stem cell proliferation arrest but not necrosis induced by resveratrol. Stem Cell. Rev. Rep. 10, 103–113. doi:10.1007/s12015-013-9465-0
Schatton, T., Murphy, G. F., Frank, N. Y., Yamaura, K., Waaga-Gasser, A. M., Gasser, M., et al. (2008). Identification of cells initiating human melanomas. Nature 451, 345–349. doi:10.1038/nature06489
Schcolnik-Cabrera, A., Chavez-Blanco, A., Dominguez-Gomez, G., Taja-Chayeb, L., Morales-Barcenas, R., Trejo-Becerril, C., et al. (2018). Orlistat as a FASN inhibitor and multitargeted agent for cancer therapy. Expert Opin. Investig. Drugs 27, 475–489. doi:10.1080/13543784.2018.1471132
Schug, Z. T., Peck, B., Jones, D. T., Zhang, Q., Grosskurth, S., Alam, I. S., et al. (2015). Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 27, 57–71. doi:10.1016/j.ccell.2014.12.002
Seo, Y., Kim, J., Park, S. J., Park, J. J., Cheon, J. H., Kim, W. H., et al. (2020). Metformin suppresses cancer stem cells through AMPK activation and inhibition of protein prenylation of the mevalonate pathway in colorectal cancer. Cancers (Basel) 12, 2554. doi:10.3390/cancers12092554
Sharon, C., Baranwal, S., Patel, N. J., Rodriguez-Agudo, D., Pandak, W. M., Majumdar, A. P., et al. (2015). Inhibition of insulin-like growth factor receptor/AKT/mammalian target of rapamycin axis targets colorectal cancer stem cells by attenuating mevalonate-isoprenoid pathway in vitro and in vivo. Oncotarget 6, 15332–15347. doi:10.18632/oncotarget.3684
Shen, Y. A., Wang, C. Y., Hsieh, Y. T., Chen, Y. J., and Wei, Y. H. (2015). Metabolic reprogramming orchestrates cancer stem cell properties in nasopharyngeal carcinoma. Cell. Cycle 14, 86–98. doi:10.4161/15384101.2014.974419
Shimano, H., and Sato, R. (2017). SREBP-regulated lipid metabolism: convergent physiology - divergent pathophysiology. Nat. Rev. Endocrinol. 13, 710–730. doi:10.1038/nrendo.2017.91
Shiokawa, D., Sakai, H., Ohata, H., Miyazaki, T., Kanda, Y., Sekine, S., et al. (2020). Slow-cycling cancer stem cells regulate progression and chemoresistance in colon cancer. Cancer Res. 80, 4451–4464. doi:10.1158/0008-5472.CAN-20-0378
Shiozawa, Y., Nie, B., Pienta, K. J., Morgan, T. M., and Taichman, R. S. (2013). Cancer stem cells and their role in metastasis. Pharmacol. Ther. 138, 285–293. doi:10.1016/j.pharmthera.2013.01.014
Shu, Z., Gao, Y., Zhang, G., Zhou, Y., Cao, J., Wan, D., et al. (2019). A functional interaction between Hippo-YAP signalling and SREBPs mediates hepatic steatosis in diabetic mice. J. Cell. Mol. Med. 23, 3616–3628. doi:10.1111/jcmm.14262
Singh, R., Kaushik, S., Wang, Y., Xiang, Y., Novak, I., Komatsu, M., et al. (2009). Autophagy regulates lipid metabolism. Nature 458, 1131–1135. doi:10.1038/nature07976
Singh, S. K., Hawkins, C., Clarke, I. D., Squire, J. A., Bayani, J., Hide, T., et al. (2004). Identification of human brain tumour initiating cells. Nature 432, 396–401. doi:10.1038/nature03128
Sinha, R. A., Singh, B. K., and Yen, P. M. (2017). Reciprocal crosstalk between autophagic and endocrine signaling in metabolic homeostasis. Endocr. Rev. 38, 69–102. doi:10.1210/er.2016-1103
Snaebjornsson, M. T., Janaki-Raman, S., and Schulze, A. (2020). Greasing the wheels of the cancer machine: the role of lipid metabolism in cancer. Cell. Metab. 31, 62–76. doi:10.1016/j.cmet.2019.11.010
Song, I. S., Jeong, Y. J., and Han, J. (2015). Mitochondrial metabolism in cancer stem cells: a therapeutic target for colon cancer. BMB Rep. 48, 539–540. doi:10.5483/bmbrep.2015.48.10.179
Song, N. J., Yun, U. J., Yang, S., Wu, C., Seo, C. R., Gwon, A. R., et al. (2016). Notch1 deficiency decreases hepatic lipid accumulation by induction of fatty acid oxidation. Sci. Rep. 6, 19377. doi:10.1038/srep19377
Sorrentino, G., Ruggeri, N., Specchia, V., Cordenonsi, M., Mano, M., Dupont, S., et al. (2014). Metabolic control of YAP and TAZ by the mevalonate pathway. Nat. Cell. Biol. 16, 357–366. doi:10.1038/ncb2936
Sosa, M. S., Bragado, P., and Aguirre-Ghiso, J. A. (2014). Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat. Rev. Cancer 14, 611–622. doi:10.1038/nrc3793
Spite, M. (2014). Resolving lipids: lipoxins regulate reverse cholesterol transport. Cell. Metab. 20, 935–937. doi:10.1016/j.cmet.2014.11.012
Strickland, M., and Stoll, E. A. (2017). Metabolic reprogramming in glioma. Front. Cell. Dev. Biol. 5, 43. doi:10.3389/fcell.2017.00043
Stylianou, S., Clarke, R. B., and Brennan, K. (2006). Aberrant activation of notch signaling in human breast cancer. Cancer Res. 66, 1517–1525. doi:10.1158/0008-5472.CAN-05-3054
Su, J., Wu, S., Tang, W., Qian, H., Zhou, H., and Guo, T. (2016). Reduced SLC27A2 induces cisplatin resistance in lung cancer stem cells by negatively regulating Bmi1-ABCG2 signaling. Mol. Carcinog. 55, 1822–1832. doi:10.1002/mc.22430
Subramaniam, D., Kaushik, G., Dandawate, P., and Anant, S. (2018). Targeting cancer stem cells for chemoprevention of pancreatic cancer. Curr. Med. Chem. 25, 2585–2594. doi:10.2174/0929867324666170127095832
Sun, H., Zhang, M., Cheng, K., Li, P., Han, S., Li, R., et al. (2016b). Resistance of glioma cells to nutrient-deprived microenvironment can be enhanced by CD133-mediated autophagy. Oncotarget 7, 76238–76249. doi:10.18632/oncotarget.12803
Sun, H. W., Yu, X. J., Wu, W. C., Chen, J., Shi, M., Zheng, L., et al. (2016a). GLUT1 and ASCT2 as predictors for prognosis of hepatocellular carcinoma. PLoS One 11, e0168907. doi:10.1371/journal.pone.0168907
Sun, M., and Yang, Z. (2019). Metabolomic studies of live single cancer stem cells using mass spectrometry. Anal. Chem. 91, 2384–2391. doi:10.1021/acs.analchem.8b05166
Sun, S., Xue, D., Chen, Z., Ou-Yang, Y., Zhang, J., Mai, J., et al. (2019). R406 elicits anti-Warburg effect via Syk-dependent and -independent mechanisms to trigger apoptosis in glioma stem cells. Cell. Death Dis. 10, 358. doi:10.1038/s41419-019-1587-0
Sun, Y., He, W., Luo, M., Zhou, Y., Chang, G., Ren, W., et al. (2015). SREBP1 regulates tumorigenesis and prognosis of pancreatic cancer through targeting lipid metabolism. Tumour Biol. 36, 4133–4141. doi:10.1007/s13277-015-3047-5
Sundqvist, A., Morikawa, M., Ren, J., Vasilaki, E., Kawasaki, N., Kobayashi, M., et al. (2018). JUNB governs a feed-forward network of TGFβ signaling that aggravates breast cancer invasion. Nucleic Acids Res. 46, 1180–1195. doi:10.1093/nar/gkx1190
Svensson, R. U., Parker, S. J., Eichner, L. J., Kolar, M. J., Wallace, M., Brun, S. N., et al. (2016). Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat. Med. 22, 1108–1119. doi:10.1038/nm.4181
Tabe, Y., Saitoh, K., Yang, H., Sekihara, K., Yamatani, K., Ruvolo, V., et al. (2018). Inhibition of FAO in AML co-cultured with BM adipocytes: mechanisms of survival and chemosensitization to cytarabine. Sci. Rep. 8, 16837. doi:10.1038/s41598-018-35198-6
Takebe, N., Miele, L., Harris, P. J., Jeong, W., Bando, H., Kahn, M., et al. (2015). Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat. Rev. Clin. Oncol. 12, 445–464. doi:10.1038/nrclinonc.2015.61
Teo, R., Mohrlen, F., Plickert, G., Muller, W. A., and Frank, U. (2006). An evolutionary conserved role of Wnt signaling in stem cell fate decision. Dev. Biol. 289, 91–99. doi:10.1016/j.ydbio.2005.10.009
Thomas, D., and Majeti, R. (2016). Burning fat fuels leukemic stem cell heterogeneity. Cell. Stem Cell. 19, 1–2. doi:10.1016/j.stem.2016.06.014
Todaro, M., Gaggianesi, M., Catalano, V., Benfante, A., Iovino, F., Biffoni, M., et al. (2014). CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell. Stem Cell. 14, 342–356. doi:10.1016/j.stem.2014.01.009
Toh, T. B., Lim, J. J., and Chow, E. K. (2017). Epigenetics in cancer stem cells. Mol. Cancer 16, 29. doi:10.1186/s12943-017-0596-9
Tracz-Gaszewska, Z., and Dobrzyn, P. (2019). Stearoyl-CoA desaturase 1 as a therapeutic target for the treatment of cancer. Cancers (Basel) 11, 948. doi:10.3390/cancers11070948
Valle, S., Alcala, S., Martin-Hijano, L., Cabezas-Sainz, P., Navarro, D., Munoz, E. R., et al. (2020). Exploiting oxidative phosphorylation to promote the stem and immunoevasive properties of pancreatic cancer stem cells. Nat. Commun. 11, 5265. doi:10.1038/s41467-020-18954-z
Vasefifar, P., Motafakkerazad, R., Maleki, L. A., Najafi, S., Ghrobaninezhad, F., Najafzadeh, B., et al. (2022). Nanog, as a key cancer stem cell marker in tumor progression. Gene 827, 146448. doi:10.1016/j.gene.2022.146448
Vazquez-Martin, A., Corominas-Faja, B., Cufi, S., Vellon, L., Oliveras-Ferraros, C., Menendez, O. J., et al. (2013). The mitochondrial H(+)-ATP synthase and the lipogenic switch: new core components of metabolic reprogramming in induced pluripotent stem (iPS) cells. Cell. Cycle 12, 207–218. doi:10.4161/cc.23352
Vega-Naredo, I., Loureiro, R., Mesquita, K. A., Barbosa, I. A., Tavares, L. C., Branco, A. F., et al. (2014). Mitochondrial metabolism directs stemness and differentiation in P19 embryonal carcinoma stem cells. Cell. Death Differ. 21, 1560–1574. doi:10.1038/cdd.2014.66
Vergara, D., Simeone, P., Damato, M., Maffia, M., Lanuti, P., and Trerotola, M. (2019). The cancer microbiota: EMT and inflammation as shared molecular mechanisms associated with plasticity and progression. J. Oncol. 2019, 1253727. doi:10.1155/2019/1253727
Vergara, D., Stanca, E., Guerra, F., Priore, P., Gaballo, A., Franck, J., et al. (2017). β-Catenin knockdown affects mitochondrial biogenesis and lipid metabolism in breast cancer cells. Front. Physiol. 8, 544. doi:10.3389/fphys.2017.00544
Viale, A., Pettazzoni, P., Lyssiotis, C. A., Ying, H., Sanchez, N., Marchesini, M., et al. (2014). Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628–632. doi:10.1038/nature13611
Visweswaran, M., Arfuso, F., Warrier, S., and Dharmarajan, A. (2020). Aberrant lipid metabolism as an emerging therapeutic strategy to target cancer stem cells. Stem cells 38, 6–14. doi:10.1002/stem.3101
Vlashi, E., and Pajonk, F. (2015). Cancer stem cells, cancer cell plasticity and radiation therapy. Semin. Cancer Biol. 31, 28–35. doi:10.1016/j.semcancer.2014.07.001
von Roemeling, C. A., Marlow, L. A., Wei, J. J., Cooper, S. J., Caulfield, T. R., Wu, K., et al. (2013). Stearoyl-CoA desaturase 1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Clin. Cancer Res. 19, 2368–2380. doi:10.1158/1078-0432.CCR-12-3249
Vriens, K., Christen, S., Parik, S., Broekaert, D., Yoshinaga, K., Talebi, A., et al. (2019). Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature 566, 403–406. doi:10.1038/s41586-019-0904-1
Wang, D., Huang, J., Wang, X., Yu, Y., Zhang, H., Chen, Y., et al. (2013). The eradication of breast cancer cells and stem cells by 8-hydroxyquinoline-loaded hyaluronan modified mesoporous silica nanoparticle-supported lipid bilayers containing docetaxel. Biomaterials 34, 7662–7673. doi:10.1016/j.biomaterials.2013.06.042
Wang, J., and Li, Y. (2019). CD36 tango in cancer: signaling pathways and functions. Theranostics 9, 4893–4908. doi:10.7150/thno.36037
Wang, J., Li, Z. H., White, J., and Zhang, L. B. (2014). Lung cancer stem cells and implications for future therapeutics. Cell. Biochem. Biophys. 69, 389–398. doi:10.1007/s12013-014-9844-4
Wang, J., Yu, L., Schmidt, R. E., Su, C., Huang, X., Gould, K., et al. (2005). Characterization of HSCD5, a novel human stearoyl-CoA desaturase unique to primates. Biochem. Biophys. Res. Commun. 332, 735–742. doi:10.1016/j.bbrc.2005.05.013
Wang, S. Y., Hu, Q. C., Wu, T., Xia, J., Tao, X. A., and Cheng, B. (2022). Abnormal lipid synthesis as a therapeutic target for cancer stem cells. World J. Stem Cells 14, 146–162. doi:10.4252/wjsc.v14.i2.146
Wang, T., Fahrmann, J. F., Lee, H., Li, Y. J., Tripathi, S. C., Yue, C., et al. (2018). JAK/STAT3-Regulated fatty acid beta-oxidation is critical for breast cancer stem cell self-renewal and chemoresistance. Cell. metab. 27, 136–150 e135. doi:10.1016/j.cmet.2017.11.001
Wei, X., Shi, J., Lin, Q., Ma, X., Pang, Y., Mao, H., et al. (2021). Targeting ACLY attenuates tumor growth and acquired cisplatin resistance in ovarian cancer by inhibiting the PI3K-AKT pathway and activating the AMPK-ROS pathway. Front. Oncol. 11, 642229. doi:10.3389/fonc.2021.642229
Wei, Y., Li, Y., Chen, Y., Liu, P., Huang, S., Zhang, Y., et al. (2022). ALDH1: a potential therapeutic target for cancer stem cells in solid tumors. Front. Oncol. 12, 1026278. doi:10.3389/fonc.2022.1026278
Wen, Y. A., Xiong, X., Zaytseva, Y. Y., Napier, D. L., Vallee, E., Li, A. T., et al. (2018). Downregulation of SREBP inhibits tumor growth and initiation by altering cellular metabolism in colon cancer. Cell. Death Dis. 9, 265. doi:10.1038/s41419-018-0330-6
Williams, N. C., and ONeill, L. A. J. (2018). A role for the Krebs cycle intermediate citrate in metabolic reprogramming in innate immunity and inflammation. Front. Immunol. 9, 141. doi:10.3389/fimmu.2018.00141
Wongtrakoongate, P. (2015). Epigenetic therapy of cancer stem and progenitor cells by targeting DNA methylation machineries. World J. Stem Cells 7, 137–148. doi:10.4252/wjsc.v7.i1.137
Xie, Y., Li, J., Kang, R., and Tang, D. (2020). Interplay between lipid metabolism and autophagy. Front. Cell. Dev. Biol. 8, 431. doi:10.3389/fcell.2020.00431
Yan, M., Yang, X., Wang, L., Clark, D., Zuo, H., Ye, D., et al. (2013a). Plasma membrane proteomics of tumor spheres identify CD166 as a novel marker for cancer stem-like cells in head and neck squamous cell carcinoma. Mol. Cell. Proteomics 12, 3271–3284. doi:10.1074/mcp.M112.025460
Yan, X., Luo, H., Zhou, X., Zhu, B., Wang, Y., and Bian, X. (2013b). Identification of CD90 as a marker for lung cancer stem cells in A549 and H446 cell lines. Oncol. Rep. 30, 2733–2740. doi:10.3892/or.2013.2784
Yan, Y., Zuo, X., and Wei, D. (2015). Concise review: emerging role of CD44 in cancer stem cells: a promising biomarker and therapeutic target. Stem Cells Transl. Med. 4, 1033–1043. doi:10.5966/sctm.2015-0048
Yang, L., Shi, P., Zhao, G., Xu, J., Peng, W., Zhang, J., et al. (2020). Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target Ther. 5, 8. doi:10.1038/s41392-020-0110-5
Yang, P., Qin, H., Li, Y., Xiao, A., Zheng, E., Zeng, H., et al. (2022a). CD36-mediated metabolic crosstalk between tumor cells and macrophages affects liver metastasis. Nat. Commun. 13, 5782. doi:10.1038/s41467-022-33349-y
Yang, W., Xia, Y., Ji, H., Zheng, Y., Liang, J., Huang, W., et al. (2011). Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature 480, 118–122. doi:10.1038/nature10598
Yang, Y., Lu, Y., Zhang, C., Guo, Q., Zhang, W., Wang, T., et al. (2022b). Phenazine derivatives attenuate the stemness of breast cancer cells through triggering ferroptosis. Cell. Mol. Life Sci. 79, 360. doi:10.1007/s00018-022-04384-1
Yasumoto, Y., Miyazaki, H., Vaidyan, L. K., Kagawa, Y., Ebrahimi, M., Yamamoto, Y., et al. (2016). Inhibition of fatty acid synthase decreases expression of stemness markers in glioma stem cells. PLoS One 11, e0147717. doi:10.1371/journal.pone.0147717
Ye, H., Adane, B., Khan, N., Sullivan, T., Minhajuddin, M., Gasparetto, M., et al. (2016). Leukemic stem cells evade chemotherapy by metabolic adaptation to an adipose tissue niche. Cell. Stem Cell. 19, 23–37. doi:10.1016/j.stem.2016.06.001
Yi, M., Li, J., Chen, S., Cai, J., Ban, Y., Peng, Q., et al. (2018). Emerging role of lipid metabolism alterations in Cancer stem cells. J. Exp. Clin. cancer Res. CR 37, 118. doi:10.1186/s13046-018-0784-5
Yu, Y., Kim, H., Choi, S., Yu, J., Lee, J. Y., Lee, H., et al. (2021). Targeting a lipid desaturation enzyme, SCD1, selectively eliminates colon cancer stem cells through the suppression of Wnt and NOTCH signaling. Cells 10, 106. doi:10.3390/cells10010106
Yuan, S. F., Hung, A. C., Hsu, C. W., Lan, T. H., Su, C. W., Chi, T. C., et al. (2022). CD44 mediates oral squamous cell carcinoma-promoting activity of MRE11 via AKT signaling. J. Pers. Med. 12, 841. doi:10.3390/jpm12050841
Zeng, S., Wu, F., Chen, M., Li, Y., You, M., Zhang, Y., et al. (2022). Inhibition of fatty acid translocase (FAT/CD36) palmitoylation enhances hepatic fatty acid β-oxidation by increasing its localization to mitochondria and interaction with long-chain acyl-CoA synthetase 1. Antioxid. Redox Signal 36, 1081–1100. doi:10.1089/ars.2021.0157
Zhang, S., Peng, X., Yang, S., Li, X., Huang, M., Wei, S., et al. (2022). The regulation, function, and role of lipophagy, a form of selective autophagy, in metabolic disorders. Cell. Death Dis. 13, 132. doi:10.1038/s41419-022-04593-3
Zhang, Y., Jin, Z., Zhou, H., Ou, X., Xu, Y., Li, H., et al. (2016). Suppression of prostate cancer progression by cancer cell stemness inhibitor napabucasin. Cancer Med. 5, 1251–1258. doi:10.1002/cam4.675
Zhao, L., Varghese, Z., Moorhead, J. F., Chen, Y., and Ruan, X. Z. (2018a). CD36 and lipid metabolism in the evolution of atherosclerosis. Br. Med. Bull. 126, 101–112. doi:10.1093/bmb/ldy006
Zhao, X., Zhao, L., Yang, H., Li, J., Min, X., Yang, F., et al. (2018b). Pyruvate kinase M2 interacts with nuclear sterol regulatory element-binding protein 1a and thereby activates lipogenesis and cell proliferation in hepatocellular carcinoma. J. Biol. Chem. 293, 6623–6634. doi:10.1074/jbc.RA117.000100
Zheng, B., Jarugumilli, G. K., Chen, B., and Wu, X. (2016). Chemical probes to directly profile palmitoleoylation of proteins. Chembiochem 17, 2022–2027. doi:10.1002/cbic.201600403
Zheng, L., Guo, Q., Xiang, C., Liu, S., Jiang, Y., Gao, L., et al. (2019). Transcriptional factor six2 promotes the competitive endogenous RNA network between CYP4Z1 and pseudogene CYP4Z2P responsible for maintaining the stemness of breast cancer cells. J. Hematol. Oncol. 12, 23. doi:10.1186/s13045-019-0697-6
Zhou, Y., Zhou, Y., Shingu, T., Feng, L., Chen, Z., Ogasawara, M., et al. (2011). Metabolic alterations in highly tumorigenic glioblastoma cells: preference for hypoxia and high dependency on glycolysis. J. Biol. Chem. 286, 32843–32853. doi:10.1074/jbc.M111.260935
Keywords: cancer stem cells, lipid metabolism, fatty acid, lipid droplets, autophagy
Citation: Du J and Qin H (2024) Lipid metabolism dynamics in cancer stem cells: potential targets for cancers. Front. Pharmacol. 15:1367981. doi: 10.3389/fphar.2024.1367981
Received: 09 January 2024; Accepted: 10 June 2024;
Published: 27 June 2024.
Edited by:
Elisa Giovannetti, VU Medical Center, NetherlandsReviewed by:
Vasudevarao Penugurti, Duke University, United StatesCopyright © 2024 Du and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Du, amluZGFsYWk2NkAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.