- 1Department of Pharmacy Practice, College of Pharmacy, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia
- 2Department of Pediatric Behavior and Development and Adolescent Medicine, King Fahad Medical City, Riyadh, Saudi Arabia
- 3Pharmaceutical Analysis Section, King Abdullah International Medical Research Center (KAIMRC), King Abdulaziz Medical City, Ministry of National Guard - Health Affairs, Riyadh, Saudi Arabia
- 4Molecular Pathology Laboratory, Pathology and Clinical Laboratory Medicine Administration, King Fahad Medical City, Riyadh, Saudi Arabia
- 5Health Sciences Research Center, King Abdullah Bin Abdulaziz University Hospital, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia
- 6Pharmaceutical Care Department, Ministry of National Guard-Health Affairs, Jeddah, Saudi Arabia
- 7Independent researcher, Malaga, Spain
Background: Autism spectrum disorders (ASDs) encompass a broad range of phenotypes characterized by diverse neurological alterations. Genomic studies have revealed considerable overlap between the molecular mechanisms implicated in the etiology of ASD and genes involved in the pharmacokinetic (PK) and pharmacodynamic (PD) pathways of antipsychotic drugs employed in ASD management. Given the conflicting data originating from candidate PK or PD gene association studies in diverse ethnogeographic ASD populations, dosage individualization based on “actionable” pharmacogenetic (PGx) markers has limited application in clinical practice. Additionally, off-label use of different antipsychotics is an ongoing practice, which is justified given the shortage of approved cures, despite the lack of satisfactory evidence for its safety according to precision medicine. This exploratory study aimed to identify PGx markers predictive of risperidone (RIS) exposure in autistic Saudi children.
Methods: This prospective cohort study enrolled 89 Saudi children with ASD treated with RIS-based antipsychotic therapy. Plasma levels of RIS and 9-OH-RIS were measured using a liquid chromatography–tandem mass spectrometry system. To enable focused exploratory testing, genotyping was performed with the Axiom PharmacoFocus Array, which included a collection of probe sets targeting PK/PD genes. A total of 720 PGx markers were included in the association analysis.
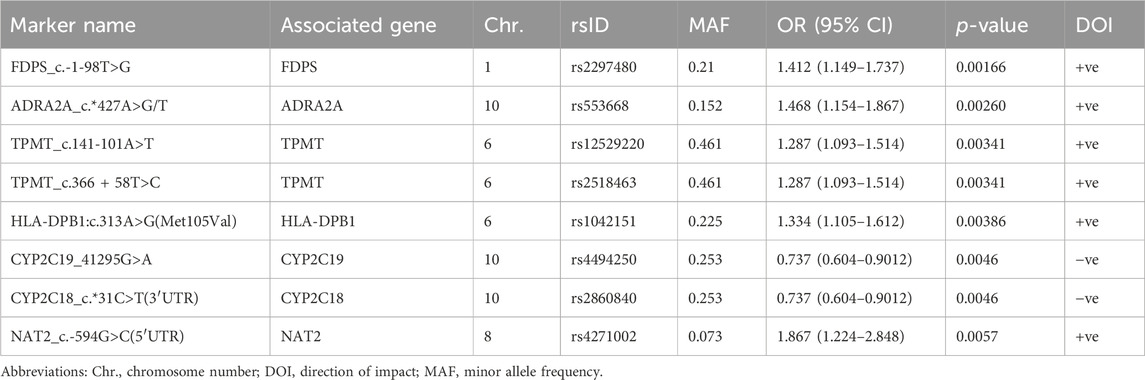
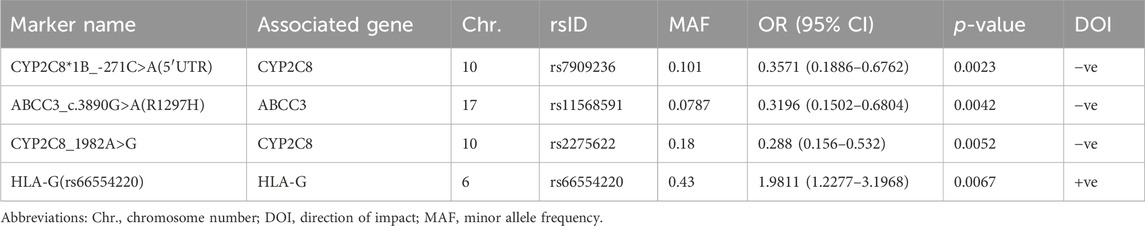
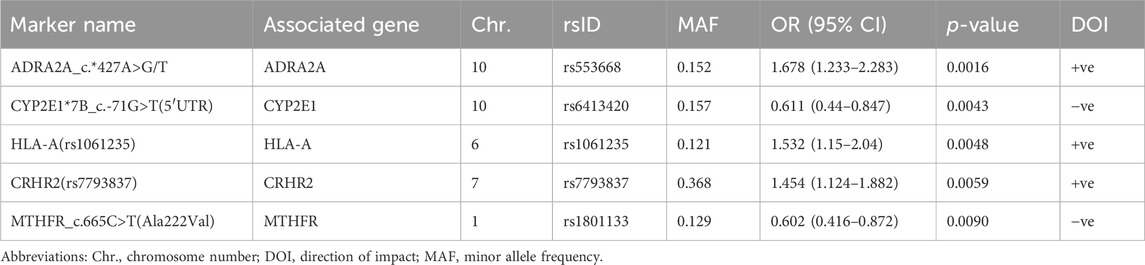
Results: A total of 27 PGx variants were found to have a prominent impact on various RIS PK parameters; most were not located within the genes involved in the classical RIS PK pathway. Specifically, 8 markers in 7 genes were identified as the PGx markers with the strongest impact on RIS levels (p < 0.01). Four PGx variants in 3 genes were strongly associated with 9-OH-RIS levels, while 5 markers in 5 different genes explained the interindividual variability in the total active moiety. Notably, 6 CYP2D6 variants exhibited strong linkage disequilibrium; however, they significantly influenced only the metabolic ratio and had no considerable effects on the individual estimates of RIS, 9-OH-RIS, or the total active moiety. After correction for multiple testing, rs78998153 in UGT2B17 (which is highly expressed in the brain) remained the most significant PGx marker positively adjusting the metabolic ratio. For the first time, certain human leukocyte antigen (HLA) markers were found to enhance various RIS exposure parameters, which reinforces the gut–brain axis theory of ASD etiology and its suggested inflammatory impacts on drug bioavailability through modulation of the brain, gastrointestinal tract and/or hepatic expression of metabolizing enzymes and transporters.
Conclusion: Our hypothesis-generating approach identified a broad spectrum of PGx markers that interactively influence RIS exposure in ASD children, which indicated the need for further validation in population PK modeling studies to define polygenic scores for antipsychotic efficacy and safety, which could facilitate personalized therapeutic decision-making in this complex neurodevelopmental condition.
1 Introduction
Autism spectrum disorder (ASD) is a complex neurodevelopmental disorder characterized by early onset in youth. However, its exact etiology, involving genetic and nongenetic (i.e., environmental) factors acting either alone or in combination, is still not clear. With the evolution in genomic technology and bioinformatics analysis techniques, several genetic mutations at both the gene and chromosome levels have been identified to be associated with different ASD phenotypes (Genovese and Butler, 2023). Recent reviews of ASD genomics using genome-wide association studies (GWASs) revealed considerable overlap between the molecular mechanisms implicated in the etiology of ASD and certain common genes involved in drug absorption, distribution, metabolism, and excretion (ADME) (Khanzada et al., 2017; Sundararajan et al., 2018; Fang et al., 2023). For example, 11 genes (SLC6A3, UGT, GSK3B, HTR2A, MAOA, NOS1AP, PDE4B, TPH2, CACNA1C, CHRNA7 and DRD2) that influence serotonin and dopamine homeostasis and signal transduction pathways affecting mood, behavior and physical activity in ASD are also known to be associated with the pharmacokinetic (PK) and pharmacodynamic (PD) pathways of drugs employed in ASD management (Butler et al., 2016; Sundararajan et al., 2018; Sheng et al., 2021).
Therefore, it may be possible to observe variations in the PK and PD parameters of drugs in specific ASD patients when compared to other disease populations or normal volunteers (Genovese and Butler, 2020).
As a spectrum disorder, individuals with ASD usually exhibit variable degrees of behavioral and psychiatric manifestations, reflecting heterogeneity in the underlying etiology, which results in the segregation into different ASD phenotypes. This fact highlights the need for the implementation of precision medicine to enable individualization of psychopharmacological drug therapy regimens based on ASD subtype manifestations (Genovese and Butler, 2020). According to recent updates, the medications commonly used to address the comorbidities associated with ASD are atypical antipsychotics, which are frequently employed on a chronic basis according to standard dosing guidelines, which might not fit all etiological subtypes (Aishworiya et al., 2022; Ooi et al., 2023). Additionally, off-label antipsychotic use is still an ongoing clinical practice, despite the lack of evidence for its safety and tolerability (Højlund et al., 2021; Wang et al., 2021; Carthy et al., 2023). This practice was thought to be justified given the shortage of approved clinical cures and was even found to be motivated by advancements in diagnostic and clinician recognition of disparity in ASD and cooccurring mental health issues (Gupta and Gupta, 2023).
Risperidone (RIS) is a U.S. Food and Drug Administration (FDA)-approved atypical antipsychotic medication to target irritability often associated with autistic children (Lamy and Erickson, 2018). However, interindividual variability in RIS effectiveness and safety profiles has been reported in adults and children, even patients with similar diagnoses of psychiatric disorders, including ASD (Lamy and Erickson, 2018; Taurines et al., 2022; Liang et al., 2023). Moreover, RIS PK (exposure) parameters demonstrated wide interindividual variability within children with ASD (Dodsworth et al., 2018; Maruf et al., 2021).
RIS is mainly metabolized in the liver by the CYP450 isoenzyme CYP2D6; however, CYP3A4 and CYP3A5 have also been reported to be partially involved in the 9-hydroxylation of RIS (Fang et al., 1999). 9-OH-RIS is a pharmacologically active metabolite that is approximately equipotent to the parent drug; therefore, both concentrations are collectively referred to as the total active moiety. 9-OH-RIS was later approved by itself as the antipsychotic paliperidone (Clarke et al., 2013). RIS and 9-OH-RIS efflux from cells are affected by certain transporter proteins, such as adenosine triphosphate-binding cassette subfamily B member 1 (ABCB1) (Yasui-Furukori et al., 2004; Saiz-Rodríguez et al., 2018).
Since the transformation of RIS to 9-OH-RIS is mainly mediated by CYP2D6, the ratio of the two molecules (RIS/9-OH-RIS ratio) in blood was classically suggested to be proportional to the CYP2D6 metabolic phenotype (Huang et al., 1993; Cho and Lee, 2006). Therefore, it is assumed that normal healthy subjects with a poor metabolizer (PM) status will have a higher metabolic ratio (less metabolic conversion of RIS) than extensive metabolizers (EMs; usually designated as normal metabolizers [NMs]) and ultrarapid metabolizers (UMs), as both conditions will result in a greater quantity of 9-OH-RIS (Huang et al., 1993; Cho and Lee, 2006; Novalbos et al., 2010). However, a large-scale study involving psychiatric patients of various ages revealed that the positive predictive value of an RIS/9-OH-RIS ratio >1 to predict CYP2D6 PMs or <1 to predict CYP2D6 UMs (95% CI) was 35% (26%–46%) and 9% (5%–14%), respectively (Mannheimer et al., 2016). Another pharmacogenetic clinical trial in healthy subjects demonstrated that CYP2D6 predicted only 65% of RIS metabolism variability and highlighted the demand for exploring pharmacogenetic predictors considering the complexity of its PK and PD pathway relationships (Gassó et al., 2014).
Collectively, these results indicated the presence of a potential research scope for examining other genetic markers of non-CYP2D6 variants (such as single-nucleotide polymorphisms (SNPs) involved in genes encoding transporters) (Yoo et al., 2011), which may affect RIS ADME and more comprehensively predict the extent of RIS and OH-RIS exposure (plasma levels) in healthy (Yoo et al., 2011) or unhealthy subjects, such as children with ASD (Troost et al., 2007; Sherwin et al., 2012; Roke et al., 2013; Youngster et al., 2014; Medhasi et al., 2016; Vanwong et al., 2016, 2017; Nuntamool et al., 2017; Rafaniello et al., 2017; Hongkaew et al., 2021; Kloosterboer et al., 2021).
Within the context of ASD, a systematic review of the current state of knowledge regarding CYP2D6 genetic variation and its impact on RIS PK and the propensity for adverse drug reactions in children and adolescents has suggested that CYP2D6 metabolic status was not consistently the sole genetic factor explaining the variabilities within these age groups (Dodsworth et al., 2018; Maruf et al., 2021). Despite the observed trend for a positive association between higher CYP2D6 activity and lower RIS concentration and RIS/9-OH-RIS ratios in some of the included studies (Troost et al., 2007; Youngster et al., 2014; Vanwong et al., 2016, 2017; Nuntamool et al., 2017), there was a consistent nonsignificant difference in the total active moiety concentrations among the different CYP2D6 phenotypes (PMs, EMs, and UMs) in all studies (Troost et al., 2007; Youngster et al., 2014; Vanwong et al., 2016; Nuntamool et al., 2017; Rafaniello et al., 2017; Vanwong et al., 2017). Additionally, these studies were conducted in ASD children of either Caucasian (Troost et al., 2007; Sherwin et al., 2012; Roke et al., 2013; Youngster et al., 2014; Rafaniello et al., 2017) or South Asian (Vanwong et al., 2016; Nuntamool et al., 2017; Vanwong et al., 2017) backgrounds, which could limit extrapolation of the results to other ethnogeographic groups (for example, Saudi Arabians) (McLellan et al., 1997; Al-Dosari et al., 2013) that may carry rare variants that are relatively less frequent in Europeans or South Asians.
Additionally, the few previous studies (Nuntamool et al., 2017; Rafaniello et al., 2017) that attempted to examine a limited number of other typical candidate pharmacogenetic (PGx) markers in RIS ADME (ABCB1, ABCG2, CYP3A4, DRD2, DRD3, and HTR2A) did not have a large enough sample size to make robust conclusions about a panel of genes or their variants or SNPs that should be included for individualized RIS therapy in ASD patients. Given the currently available conflicting data originating from candidate gene association study methods, “actionable” PGx markers related to antipsychotic dosing and selection, in general and with respect to different psychiatric conditions, have a limited application in routine clinical practice, even in developing countries, due to imperfect guidelines for interpretation and implementation (Eap, 2016; Eum et al., 2022).
Based on background information and consistent with the evolution of advanced technology, various pharmacogenomic approaches (targeted, focused or exploratory) can often be employed in human subjects to characterize the genetic determinants that may play roles in various aspects of drug activity and to support ongoing efforts to identify biomarkers predictive of drug exposure and/or safety in specific subgroups of diseases or certain age categories (Burczynski, 2009). Any of the three strategies can be pursued depending on the scale or number of genes in other ADME pathways that need to be examined in parallel (Table 1 in Burczynski, 2009). In our study, as dozens to thousands of genes are suspected to be involved in the drug metabolism or transport of RIS, both exploratory (nonhypothesis) and focused (guided) approaches were justified for identifying genetic alterations in all ADME genes (known PGx markers). Research with a multiplex genotyping approach is relatively innovative and is assumed to provide evidence for better optimum guidance of RIS dosing in physiologically and genetically modified settings, such as children with ASD, to avoid the risk of adverse drug reactions and/or suboptimal responses, as reported in several previous investigations (Correia et al., 2009; Nuntamool et al., 2017; Oshikoya et al., 2019; Shilbayeh S. A. R. et al., 2023).
Given the continuous growth in the knowledge base of DNA polymorphisms associated with ASD risk and ADME of antipsychotics, the current exploratory pharmacogenetic study was conducted with the aim of investigating potential PGx markers involved in RIS exposure (PK) in Saudi children with ASD and achieving a better understanding and clearer insights into the underlying mechanisms of disease-drug-gene interactions in this setting.
2 Materials and methods
2.1 Study design and participants
This study was a prospective cohort study conducted from November 2020 to February 2021 at three autism centers in Riyadh, Saudi Arabia. All the methodological details, including screening, inclusion of candidate children, and data collection, were described previously in full detail (Shilbayeh S. A. R. et al., 2023).
Blood samples were collected for genotyping and RIS plasma level measurement as described previously (Shilbayeh S. A. R. et al., 2023).
2.2 Assay of plasma drug levels
RIS and 9-OH-RIS were extracted and measured in serum samples using a liquid chromatography tandem mass spectrometry system (LC/MS/MS, Waters, USA) according to a previously developed and validated method (Aravagiri and Marder, 2000). The lower limit of quantification of RIS and 9-OH-RIS was 1 ng/mL, and the lower limit of detection for serum RIS was 0.08 ng/mL and for serum 9-OH-RIS was 0.26 ng/mL.
2.3 Pharmacogenetic analysis
2.3.1 DNA extraction
Genomic DNA was extracted using the QIAsymphonySP automated extraction system and a QIAsymphony® DSP DNA Midi Kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). A Nanodrop spectrophotometer (Thermo Fisher Scientific, Santa Clara, CA, USA) was used to determine the concentration and purity of the extracted DNA.
2.3.2 Axiom PharmacoFocus Array
The Axiom PharmacoFocus Array (Catalog identifier: 952396; Thermo Fisher Scientific) offers comprehensive coverage of more than 2,000 markers (SNPs and insertions and deletions) in 150 genes across diverse populations (Tilleman et al., 2019) and functional variants that influence the ADME of commonly prescribed medications that are curated by the Pharmacogenomics Knowledge Base (PharmGKB) with clinical annotation levels of evidence 1A–2B (Whirl-Carrillo et al., 2021). These variants are commonly termed actionable PGx markers for testing in clinical practice (Whirl-Carrillo et al., 2021). Specifically, the Axiom PharmacoFocus Array facilitates genotyping in regions of high homology of key pharmacogenes (CYP2A6, CYP2D6, GSTM1, GSTT1, UGT2B17, and SULT1A1), which are usually difficult to obtain by complex multistep traditional methods (Tilleman et al., 2019).
According to the manufacturer’s instructions, genomic DNA was amplified by multiplex polymerase chain reaction (PCR) using a QIAGEN Multiplex PCR Kit (Qiagen). These amplified products were then fragmented, pooled, resuspended, and hybridized to the PharmacoFocus Array platform (Thermo Fisher Scientific, Santa Clara, CA, USA). The array was scanned on the automated Applied Biosystems™ GeneTitan™ Multi-Channel (MC) Instrument (Affymetrix Inc., Santa Clara, CA, USA). The genotyping call rates and quality parameters of all available markers and samples were generated using Applied Biosystems Axiom™ Analysis Suite software (version 5.2, Thermo Fisher Scientific, Santa Clara, CA, USA).
To achieve the highest genotyping performance, samples that did not satisfy the Dish QC (DQC) parameters were excluded. Furthermore, individuals with a <90% genotyping call rate were excluded from the analyses. Moreover, all markers with any of the following criteria were not considered for the bioinformatic analyses: genotyping call rate <95%, minor allele frequency (MAF) < 0.05, Hardy–Weinberg equilibrium (HWE) p < 0.001, and located on the X chromosome.
2.4 Statistical and bioinformatic analyses
The identity by descent (IBD) test was performed using PLINK v1.9 (Purcell et al., 2007) to exclude samples with hidden relatedness. To calculate principal components (PCs), we used a total of 8319 probeset markers examined via the PharmacoFocus Array. Subsequently, variants were filtered according to the standard specified criteria: biallelic, passed aligner’s QC, MAF >0.05, HWE p > 0.001, and no evidence of linkage disequilibrium (LD) (r2 < 0.2). The remaining 4000 variants were used to perform PC analysis with PLINK v2.0 (Chang et al., 2015). Since 84% of the variation was explained by the first two PCs, they were used to remove ancestry and hidden relatedness biases in the association analysis (Supplementary Figure S1).
Association analysis was carried out with PLINK v2.0 (Chang et al., 2015). The 720 selected PGx markers were fitted into a generalized linear model with log2-transformed response values and adjusted by covariates including age, sex, PC1, PC2, and RIS medication history (duration, daily dose). The obtained p values were adjusted for multiple testing.
LD analysis was performed with PLINK v1.9 with an r2 threshold of 0.5 and a window of 1,000 Kb. LD figures were generated for markers with significant associations (p-value <0.05) using HaploView v20.0.1 (Barrett et al., 2004), where blocks were defined by solid spine (SS).
3 Results
3.1 Study population
Of the 110 samples from pediatric patients with ASD who underwent clinical and psychological evaluations, 7 individual samples were excluded from genotyping for not meeting the DQC criteria, and 8 samples were excluded due to a call rate of <90% in the genotyping results. Furthermore, 6 patients who did not provide plasma samples for drug concentration determination were excluded. The average QC call rate for the passing samples was 99.2%.
Supplementary Table S1 displays the demographics and clinical criteria of the 89 patients in our study population. Eighty-three (93.3%) patients received RIS monotherapy. The majority of patients were male (N = 67, 75.3%), with a mean age of 9 (standard deviation (SD) = 4.1) years. The median RIS dose was 0.75 (interquartile range (IQR): 0.5–1.5) mg/day, with a median treatment duration of 21.5 (IQR: 3.23–57.9) months. Thirty-two (36%) patients also received concomitant psychotropic medications, primarily psychostimulants and melatonin. The median concentration of RIS was 0.56 ng/mL (IQR: 0.3–2.4) and that of 9-OH-RIS was 7.02 ng/mL (IQR: 2.4–13.4), while the active moiety concentration was 8.18 ng/mL (IQR: 2.8–16.4). The RIS/9-OH-RIS concentration ratio was 0.14 (IQR: 0.07–0.23).
3.2 Selection of PharmacoFocus PGx markers
A total of 8319 probeset markers were obtained via the PharmacoFocus Array, of which 2218 markers were identified as PGx markers. In the process of filtering 2218 PGx markers with the QC parameters, 100 (4.5%) markers were removed due to a global genotyping call rate of less than 95%. Of the remaining 2118 markers, 1,378 (62.13%) markers were excluded from further analysis because their MAF was less than 5%. Furthermore, 8 (0.4%) markers on the X chromosome and 10 (0.45%) markers with HWE p values <0.001 were omitted.
As a result, 722 (32.6%) of 2218 PGx markers were included in the association analysis. The average call rate of the 722 selected markers was 99.7%. The PCA plot did not show any clear clusters, indicating the absence of strong subpopulation stratification (Supplementary Figure S1).
3.3 Association of PGx variants with RIS PK parameters in the ASD cohort
A total of 27 PGx variants in 20 genes were demonstrated by PLINK software to have a significant association (p-value <0.01) with various RIS PK parameters measured in plasma, including RIS, its metabolite (9-OH-RIS), total active moiety, and RIS/9-OH-RIS metabolic ratio (results are displayed in Tables 1–4, respectively). The PGx markers are arranged in the tables according to their strength of association with the response variable. The direction of impact (DOI) of each individual PGx marker was positive or negative, indicating either increasing or decreasing an RIS PK measure, and is presented in its specific table.
Additional PGx markers that revealed potential associations with the RIS PK parameters with a minimum of p < 0.05 are presented in Supplementary Tables S2–S5. Nongenetic confounding variables, including age, sex, self-identified ethnicity, RIS dosage, treatment duration, and concomitant medications, were not significant in any of the models of RIS PK parameters.
First, out of all 722 included variants, only 8 markers in 7 genes were identified as top PGx markers for the RIS plasma level with p < 0.01 (Table 1; Figure 1A). Specifically, two SNPs (1 in CYP2C19 (rs4494250) and 1 in CYP2C18 (rs2860840)) were negatively correlated with the plasma level of RIS. However, the other top identified SNPs in five genes (FDPS, ADRA2A, TPMT, HLA-DPB1, and NAT2) were positively associated, indicating a greater effect on the RIS concentration estimates (p < 0.01). An additional 10 novel markers (i.e., not within the known RIS metabolic pathway) (Whirl-Carrillo et al., 2021) were identified to be associated at the level of p < 0.05, as shown in Supplementary Table S2. However, only one SNP (CYP2D6*4_1847G>A, splice site variant) in the CYP2D6 gene, known to be primarily involved in the established RIS metabolic pathway (Whirl-Carrillo et al., 2021), was identified as significant at the level of p < 0.05 (p = 0.04). Although it had a low prevalence of 5% among the study sample, it was found to increase RIS levels by 1.5-fold (Supplementary Table S2).
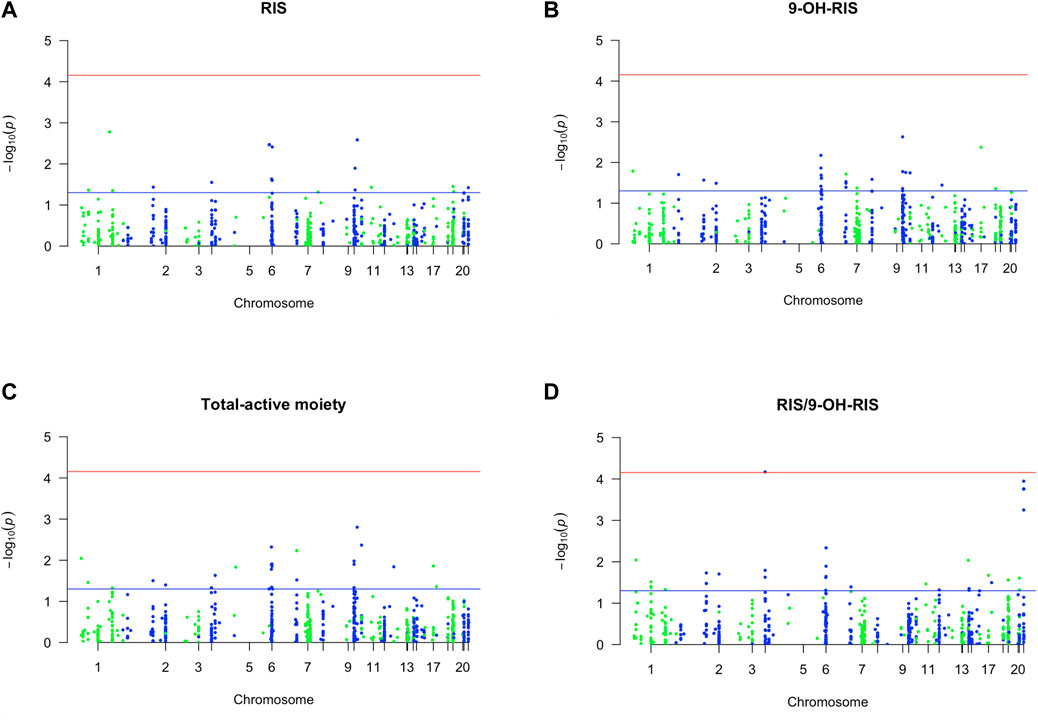
FIGURE 1. Manhattan plot of associations of RIS exposure parameters with 720 PGx PharmacoFocus markers. The horizontal x-axis represents the chromosomal position; the vertical y-axis represents–log10 P from the linear regression. The red horizontal line represents the significance level of p = 7.0 × 10−5 after Bonferroni correction. The horizontal blue line represents the significance level p = 0.05. (A) PGx variants of RIS exposure. (B) PGx variants of 9-OH-RIS. (C) PGx variants of the total active moiety. (D) PGx variants of metabolic ratio (RIS/9-OH-RIS).
Second, 4 PGx variants in 3 genes were found to be strongly associated with the level of the RIS metabolite (9-OH-RIS) at p < 0.01 (Table 2; Figure 1B). Interestingly, 2 of these SNPs are located in the CYP2C8 gene, which encodes a novel CYP450 enzyme [CYP2C8] that was not previously known to influence the metabolism of RIS. The other 2 variants were identified in two different genes (ABCC3 and HLA-G). However, at the level of p < 0.05, a total of 22 supplementary PGx markers in 17 genes were shown to have either a positive or negative effect on the metabolite concentration in plasma (Supplementary Table S3). Notably, among these secondary markers, 2 SNPs in the CYP2C9 gene positively impacted the concentration of 9-OH-RIS by a 1.7-fold increase, reflecting the potential of the CYP2C9 enzyme to play an important role in the conversion of RIS to this metabolite.
Third, 5 markers in 5 different genes were shown to be significant in determining the total active moiety at the level of p < 0.01 (Table 3; Figure 1C). Only one PD marker (ADRA2A_c.*427A>G/T) was found to simultaneously enhance the exposure of RIS (Table 1) and the total active moiety at the level of p < 0.01 (Table 3). In the current study analyses, none of the metabolic PGx markers that affected RIS (Table 1) or 9-OH-RIS levels (Table 2) were simultaneously observed to have a significant impact on the total active moiety at the level of p < 0.01. This result indicated the possibility of their involvement in the conversion of RIS and 9-OH-RIS to other inactive metabolites.
Fourth, 10 SNPs in 5 genes were revealed to have a highly significant impact on the RIS/9-OH-RIS metabolic ratio (Table 4; Figure 1D). Notably, 6 SNPs (rs28360521, rs2267447, rs28588594, rs1080989, rs1065852, and rs3892097) in the CYP2D6 gene (preliminarily identified as the main metabolic enzyme guiding the conversion of RIS to 9-OH-RIS) were observed to significantly influence the metabolic ratio with an average OR = 1.7 (Table 4); no considerable effects related to these markers were found on the individual estimates of RIS, 9-OH-RIS, or the total active moiety plasma concentrations. Several other supplementary PGx variants were also shown to have an influence on the metabolic ratio but at a lower threshold (p < 0.05), as depicted in Supplementary Table S5.
3.4 Linkage disequilibrium analysis
LD analysis of the genetic markers that were associated with the 4 RIS PK parameters at the level of p < 0.05 is shown in Figure 2. Accordingly, on chromosome 1 (Figure 2A), DPYD rs2152878 and rs4492658 were observed to be in strong LD, while FMO3 rs1736557 and FMO1 rs12954 were noted to be likely in LD. On chromosome 2 (Figure 2B), ABCB11 rs495714, rs473351, and rs497692 were found to be in strong LD. On chromosome 6 (Figure 2C), TPMT rs2518463 and rs12529220 were observed to be likely in LD; HLA-A rs1061235 and intergenic HCG4 (rs1633021) and HLA-G (rs66554220) were noted to be likely in LD; HLA-DQA1-AS rs3129900, rs3129934, and rs9268542 and HLA-DQA1 rs9272346 were in strong LD; SLC22A1 rs1867351 and rs683369 were likely in LD; and SLC22A1 rs683369, rs628031, and rs35854239 were in strong LD. On chromosome 10 (Figure 2D), CYP2C9 rs4918758 and rs1505 and CYP2C8 rs2275622 and rs7909236 were in strong LD and likely in LD with CYP2C19 rs4917623. On chromosome 22 (Figure 2E), 6 CYP2D6 SNPs (rs2267447, rs3892097, rs1065852, rs1080989, rs28588594, and rs28360521) were found to be in strong LD.
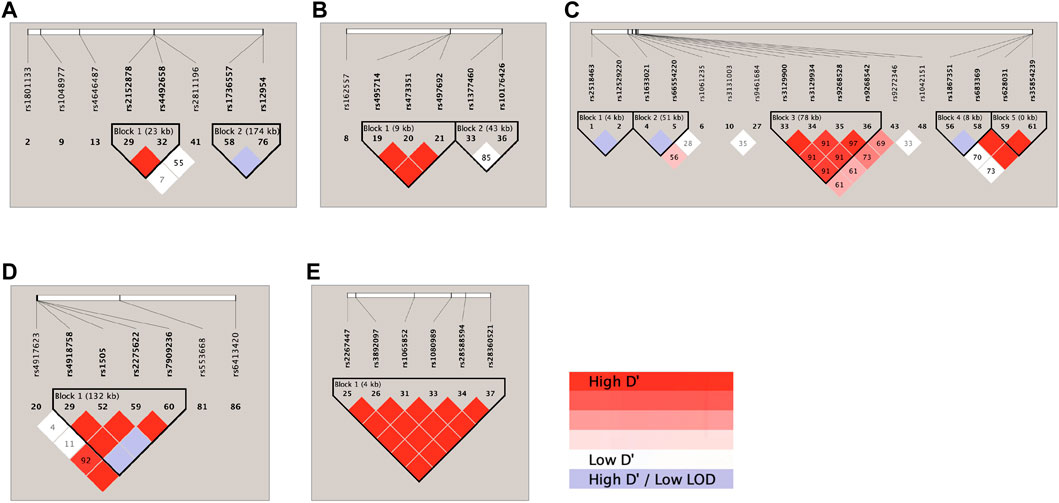
FIGURE 2. Haploview linkage disequilibrium map of SNPs associated with responses: (A) chromosome 1; (B) chromosome 2; (C) chromosome 6; (D) chromosome 10; and (E) chromosome 22. Pairwise linkage disequilibrium (D′) values are given in blocks for each SNP combination. Red indicates high D′ values; white indicates low D′; and violet indicates high D′ with a low logarithm of odds (LOD).
4 Discussion
In this pharmacogenetic study, use of the Axiom PharmacoFocus Array platform revealed various novel PGx markers highly associated with RIS exposure in Saudi children with ASD. The key finding of this exploratory focused study is that most of the PGx markers that showed a prominent impact on various RIS PK parameters (27 out of 722 PGx variants examined) were not located within the genes involved in the classical RIS PK pathway, as previously defined by in vivo studies (Whirl-Carrillo et al., 2021).
4.1 PGx markers encoding phase I metabolic enzymes
4.1.1 CYP2C8 and CYP2C9
CYP2C8 is a phase I metabolizing enzyme that has been recently described by PharmGKB as a very important pseudogene (VIP) (Whirl-Carrillo et al., 2021). Interest in CYP2C8 emerged for several reasons, such as its central role in the biotransformation of structurally dissimilar compounds and endogenous molecules, attributed to its ability to bind divergent substrates without extensive conformational changes (Lai et al., 2009); its wide expression in body tissues other than hepatocytes (Sjostedt et al., 2020; Karlsson et al., 2021); more updates in identification of its substrates and inhibitors; and advanced knowledge in characterization of its SNPs and star alleles (Gaedigk et al., 2022). Additionally, the CYP2C8 gene is positioned on chromosome 10q24 in the CYP2C gene cluster (centromere-CYP2C18-CYP2C19-CYP2C9-CYP2C8-telomere), and given the proximity of CYP2C8 and CYP2C9, LD was previously reported between these genes (Yasar et al., 2002). Interestingly, the present exploratory study revealed strong LD between 2 CYP2C8 (rs7909236 and rs2275622) and 2 CYP2C9 (rs4918758 and rs1505) SNPs, all of which were shown to have significant individual associations with 9-OH-RIS plasma levels with decreasing (CYP2C8 pair) and increasing impacts (CYP2C9 pair). Only the CYP2C8 pair was associated with a decreased total active moiety, possibly indicating increased metabolism of 9-OH-RIS. This assumption may be supported by evidence from previous studies indicating that rs7909236 (-271C>A SNP designated as CYP2C8*1B) is associated with normal enzyme function compared with wild-type (Bahadur et al., 2002; Yasar et al., 2002; Rodríguez-Antona et al., 2007). While this CYP2C8 SNP was reported to be absent in Africans, its prevalence in our population (10%) was similar to that in Asians but lower than that in Caucasians (23%) (Bahadur et al., 2002). The other CYP2C8 SNP (rs2275622) (18% in our population) is a variant that was less commonly reported in clinical studies with contradictory functional effects (associated with higher or lower enzymatic activity) depending on the substrate (Kirchheiner et al., 2008; Grau et al., 2009).
Collectively, the observed negative impact of CYP2C8 on 9-OH-RIS and the total active moiety support the assumption that this enzyme simultaneously acts on RIS and 9-OH-RIS, with a greater reduction in the RIS level (possibly highlighting the influence of CYP2C8 on a second RIS metabolic pathway to an alternative metabolite).
However, our observed increase in 9-OH-RIS plasma levels associated with the 2 CYP2C9 variants (both designated as a CYP2C9*1 allele with normal function) (Gaedigk et al., 2017) could be linked to increased RIS metabolism to 9-OH-RIS mediated by the CYP2C9 enzyme, presumably as a minor non-CYP2D6 pathway. In contrast, previous studies with smaller sample sizes have failed to reveal any CYP2C9 impacts on RIS PK parameters (Llerena et al., 2004; Cabaleiro et al., 2014).
Additional studies are needed to explain the relative contributions of CYP2C8 and CYP2C9 to RIS metabolism and the power of simultaneously existing CYP2C8/CYP2C9 haplotypes to predict RIS efficacy and safety before implementation in clinical practice.
4.1.2 CYP2C19 and CYP2C18
Two SNPs from two genes encoding different CYP450 enzymes (1 in CYP2C19 (rs4494250) and 1 in CYP2C18 (rs2860840)) were observed to equally contribute to reduced concentrations of RIS among carriers compared to noncarriers. Unfortunately, the functional status of those two SNPs was not defined in the known resources of CYP2C19 or CYP2C18 Allele Definition Tables (Whirl-Carrillo et al., 2021; Gaedigk et al., 2022). However, both genes are within the CYP2C subfamily, which is responsible for the metabolism of various drugs, including warfarin, escitalopram and omeprazole; however, the role of CYP2C18 in drug metabolism in general remains unclear (Goldstein and de Morais, 1994). Interestingly, the two genes are closely located on the same chromosome, and previous evidence in Japanese subjects suggested complete linkages between the mutated alleles of CYP2C18 and CYP2C19 (Kubota et al., 1998). Furthermore, our observation of lower RIS concentrations among autistic children carrying rs2860840 (CYP2C18_c.*31C>T(3′UTR)) reinforces a recent novel finding by Bråten et al. (2021), indicating that increased CYP2C19-dependent escitalopram metabolism leads to decreased concentrations in CYP2C19 NMs (*1/*1), similar to levels obtained in patients classified as CYP2C19 UMs (*17/*17) or RMs (*1/*17). The latter findings were mainly attributed to the liver enhancer effect induced by only one SNP, CYP2C18 (rs2860840), which was simultaneously carried by the CYP2C19 NMs. This novel SNP finding was later examined in Native American cohorts, and due to its high frequency and clinical implications for the treatment of >20 drugs with official annotations for CYP2C19 polymorphisms, it was recommended to add this SNP to the PGx practice panel to reveal the mismatching between CYP2C19-predicted and exposure-substantiated CYP2C19 metabolic phenotypes (Fernandes et al., 2023).
Our study is the first to report on the significant individual and combined impact of two SNPs in CYP2C19 and CYP2C18 on reducing RIS plasma levels, which could lead to infectiveness and require a higher dosage to achieve the RIS target therapeutic range. In contrast, Cabaleiro et al. (2014) reported an increased chance of RIS-induced neurologic manifestations among CYP2C19 NM healthy individuals compared to CYP2C19 PMs. However, in the same study, no significant impact of CYP2C19 polymorphisms was found on RIS or 9-OH-RIS PK parameters. Whether this finding could be attributed to the absence of both SNPs identified in our study as enhancing the metabolism of RIS or the smaller sample size in the Cabaleiro et al. (2014) study remains to be examined in future large-scale studies.
4.1.3 CYP2D6
In the classical RIS metabolic pathway (Whirl-Carrillo et al., 2021), CYP2D6 is the primary enzyme involved in RIS metabolism to 9-OH-RIS. The current work reinforced this fact by revealing that 6 mutated CYP2D6 PGx variants have a significant positive impact on the RIS/9-OH-RIS metabolic ratio, which indirectly reflects a substantial increase in the plasma bioavailability of RIS in comparison to its major metabolite. Notably, all these SNPs were in complete LD in our autistic children. However, none of these variants were shown to have an influence on the individual estimates of RIS, RIS metabolite, or total active moiety, which may reflect the presence of other, undiscovered non-CYP2D6 variants (for example, as discussed earlier, CYP2C8/9 haplotypes) that could further modulate global exposure to and the efficacy and safety of RIS therapy in ASD. This finding is consistent with a similar observation in an exploratory study in Thai children with ASD, which revealed that three CYP2D6 variants existing in strong LD significantly influenced the metabolic ratio but not the discrete measurements of RIS, 9-OH-RIS, or total active moiety in plasma (Medhasi et al., 2016).
4.1.4 CYP2E1
CYP2E1 is a member of the CYP450 family that metabolizes relatively few prescription drugs but is better known for the metabolism of toxins and procarcinogens (Hayashi et al., 1991). Several CYP2E1 SNPs in the promoter and intronic regions have been identified; however, luciferase promoter studies have shown that polymorphisms in the *7 haplotype, in particular the rs6413420 variant, increase CYP2E1 transcription (Huang et al., 2012). Interestingly, the current study revealed a significant impact of this SNP (15.7% prevalence) in reducing the total active moiety (at the level of p < 0.01) and 9-OH-RIS (p = 0.018) up to 0.6- and 0.44-fold, respectively, compared to noncarriers. Consistent with this finding, two previous studies highlighted the role of CYP2E1 SNPs in the etiology of schizophrenia and RIS treatment outcomes in the Chinese population (Huo et al., 2012; Shi et al., 2017). Of note, 5 CYP2E1 SNPs (including *5, rs3813867 and rs2031920) were associated with increased total active moiety, suggesting lower enzyme activity (Huo et al., 2012). However, rs2515641 in CYP2E1 was found to be significantly related to nonresponse to RIS treatment for schizophrenia in a Chinese cohort (p = 0.007) (Shi et al., 2017). Additionally, molecular genetic studies found that DNA methylation levels of CYP2E1 in the placenta were associated with children later diagnosed with ASD (Zhu et al., 2019; Bahado-Singh et al., 2022). Consistent with this finding, some molecular expression studies in inflammatory-mediated gastrointestinal diseases reported increased levels of CYP2E1 (122%) (Effinger et al., 2019), which strengthens the evidence of the role of the gut–brain axis in ASD etiology (Lombardi et al., 2023; Morton et al., 2023) and its correlation to our observation of reduced total active moiety and 9-OH-RIS levels in the study.
4.1.5 CYP1A1
The CYP1A1 gene encodes the CYP1A1 enzyme, which is a member of the CYP1A subfamily and is responsible for the metabolism of diverse substrate molecules, such as sex hormones, caffeine, therapeutic drugs, environmental pollutants, toxins, and carcinogens. This diversity of CYP1A1 functions implies its involvement in numerous biological pathways (Kukal et al., 2023). Individual divergences in CYP1A1 expression and activity not only are attributed to genetic polymorphisms in CYP1A genes but can also be up- or downregulated through the interaction of environmental and endogenous physiological factors (Kukal et al., 2023). According to our association analysis, only one CYP1A1 SNP (rs2606345), which was highly prevalent in our ASD population (30.9%), had a significant positive impact on the metabolic ratio. In PGx studies of other medications, this variant produced controversial findings regarding its influence on enzyme activity (Whirl-Carrillo et al., 2021). Despite the SNP’s position in the intronic region of the CYP1A1 gene, some PK studies reported decreased function (Allegra et al., 2016; Talwar et al., 2016), while others reported a gain of function (Grover et al., 2010; Cusato et al., 2014; Allegra et al., 2017). Therefore, it is quite challenging to interpret from current data the actual contribution of this CYP1A1 polymorphism to the elevated metabolic ratio (increased RIS or decreased 9-OH-RIS levels in plasma), particularly because it is not directly involved in the primary RIS metabolic pathway. Moreover, this enzyme function was reported to be modified by a wide range of downstream modifications (genetic or epigenetic) and environmental factors that function together to alter the expression of the underlying genetic variant, leading to the ultimate biological response (Ye et al., 2019; Xu et al., 2023). Notably, another CYP1A1 SNP (rs1048943) was highly prevalent in Thai children with ASD (30.3%) (Sukasem et al., 2016), indicating that further molecular studies are needed to define the mechanisms connecting the CYP1A1 polymorphism to ASD and to modulate RIS PK in this disease population.
4.1.6 CDA
The CDA gene encodes the cytidine deaminase enzyme, which catalyzes the irreversible hydrolytic deamination of cytidine and deoxycytidine to uridine and deoxyuridine, respectively. It is one of numerous deaminases responsible for preserving the cellular pyrimidine pool. It is known that certain drugs can be rapidly metabolized by the CDA enzyme, which can affect their bioavailability and efficacy (Lavelle et al., 2012). In the context of chemotherapy, CDA plays a significant role (Abbaspour et al., 2023), since it metabolizes several chemotherapeutic drugs, including gemcitabine (Pellicer et al., 2017). Gemcitabine interferes with DNA synthesis and replication either by inhibiting enzymes involved in the synthesis of nucleic acid precursors or by misincorporation of nucleic acids into DNA or RNA macromolecules (Ciccolini et al., 2016). Studies have shown that the activity of the CDA enzyme can be a predictive biomarker in gemcitabine-treated cancer patients. Patients with lower CDA activity had significantly longer survival compared to patients with higher CDA activity (Soo et al., 2009). The current data suggested that one CDA variant (CDA_c.435C>T(T145 = ), rs1048977) was a strong predictor for a higher RIS/9-OH-RIS metabolic ratio. However, to date, there is no specific information in the medical literature about the interaction between CDA and RIS (Whirl-Carrillo et al., 2021). In the context of psychiatry and ASD, there is ongoing research into the role of various enzymes and their potential impact on these conditions. Some studies have demonstrated that CDA gene expression in the brain is associated with certain psychiatric disorders by creating DNA mutations via deamination of cytosine bases, which results in uracil (Gavin et al., 2012; Guidotti and Grayson, 2014). The same studies highlighted antipsychotics, including RIS, as potential targets in altering DNA methylation profiles in the brain. One study found that salivary immunoglobulin A (IgA) levels were significantly decreased in patients with ASD, and this correlated with bacteria-induced downregulation of the polymeric immunoglobulin receptor (Pigr) in salivary glands (Gong et al., 2021). However, this study did not specifically mention the CDA enzyme. Another study discussed the role of activation-induced CDA (AID) in the adaptive immune system and its potential implications in various diseases (Rios et al., 2020). However, it did not specifically link AID to autism or psychiatric disorders. While these studies provide valuable insights, more research is needed to fully understand the complex interactions among CDA, AID, and ASD etiology. Subsequently, it is important to investigate how genetic polymorphisms affecting CDA could act as markers for RIS clinical outcome (i.e., toxicity, efficacy) in real clinical practice.
4.2 PGx markers encoding phase II metabolic enzymes
4.2.1 FDPS
Another novel variant that was found to be associated with increased RIS levels was rs2297480 (by 1.4-fold, p = 0.0012) in the FDPS gene, which encodes farnesyl diphosphate synthase (FDPS), an essential enzyme in the mevalonate pathway (cholesterol biosynthesis) (Göbel et al., 2020). The FDPS gene is implicated in various diseases, including neuropsychiatric disorders such as ASDs (Segatto et al., 2019; Lin et al., 2023). Additionally, as an RNA-binding protein, FDPS is also involved in transcriptional and post-transcriptional regulation of many enzymes (Wang L. et al., 2023). A recent in silico analysis revealed that FDPS is an overlapping gene that is involved in CNS disorders and is simultaneously associated with the encoding of enzymes in the lipid and cholesterol metabolic pathways (Ang and Moon, 2022). Of note, antipsychotic drugs were observed to result in upregulated expression of the genes involved in cholesterol biosynthesis (Le Hellard et al., 2008), which was suggested as a potential causal pathway for their role in the pathogenesis of neuropsychiatric disorders (Zhou et al., 2021) as well as their subsequent induced adverse metabolic effects (Le Hellard et al., 2008). However, according to the present association study, how FDPS polymorphisms and expression modulate RIS exposure and response in ASD patients and vice versa remain unknown. However, our results highlight the need for further investigation of the pathways underlying this gene-disease-drug interaction.
4.2.2 TPMT
The TPMT gene encodes the thiopurine S-methyltransferase enzyme, which plays a crucial role in the metabolism of thiopurine drugs (Lee et al., 1995). Moreover, it is dependent on the S-adenosylmethionine (SAM) methyl donor substrate in the methionine pathway (Weinshilboum, 2006). The TPMT enzyme is involved with other conjugation enzymes in phase II detoxification, where liver cells add a substance (such as cysteine, glycine, methyl or a sulfur molecule) to a toxic chemical or drug to make it less harmful and easier for the body to excrete (Zhang, 2011). It has also been implicated in the metabolism of other aromatic and heterocyclic sulfhydryl compounds (Woodson and Weinshilboum, 1983).
Several pharmacogenetic studies have demonstrated that certain polymorphism-induced mutations in the TPMT gene result in completely undetectable TPMT enzyme activity, leading to life-threatening adverse events associated with even normal doses of anticancer drugs, such as azathioprine, cyclosporine, and daunorubicin (Wang et al., 2010). However, TPMT is not involved in the direct metabolic pathway (Phase I) of RIS (Whirl-Carrillo et al., 2021). In our study, two TPMT intronic SNPs (rs12529220 and rs2518463; both known to express the normal functional TPMT allele *1), with a prevalence of 46.1% among Saudi autistic children, were likely to be in LD at chromosome 6. Both were associated with a substantial increase in RIS levels (1.3-fold). Consistent with this finding, a previous RIS PK study in normal volunteers demonstrated significantly higher 9-OH-RIS plasma levels in *1/*1 genotype participants in comparison to mutant genotype carriers (*1/*2,*1/*3C,*1/*3A) (Cabaleiro et al., 2014). Another earlier study reported an association of decreased TPMT activity (mutant genotypes) with olanzapine-induced fatigue and dizziness in healthy volunteers with no significant impact on any of its PK parameters (Cabaleiro et al., 2013). However, no other data are available on their association with the PK of RIS or other antipsychotics (Whirl-Carrillo et al., 2021); therefore, it is challenging to provide a satisfactory interpretation. Further studies are needed to explore the impact of TPMT polymorphisms on chronic RIS therapy in ASD.
4.2.3 NAT2
The NAT2 gene encodes N-acetyltransferase 2 (arylamine N-acetyltransferase), which is a typical xenobiotic metabolizing enzyme (Ackenheil and Weber, 2004) responsible for acetylation as a phase II conjugation reaction. In previous studies, NAT2 was identified to play a role in the metabolism of benzodiazepines (Camargo et al., 2023) and also hypothetically plays a role in the metabolism of some antipsychotics (Ackenheil and Weber, 2004). To date, 88 SNPs have been identified within the NAT2 gene that can affect NAT2 function by resulting in reduced enzyme stability or altered affinity for a substrate. NAT2 genotypes can be divided into three subgroups: “slow acetylator” (two slow alleles), “intermediate acetylator” (1 slow and 1 rapid allele), and “rapid acetylator” (2 rapid alleles, occasionally referred to as “fast”). Out of 38 NAT2 SNPs examined in our exploratory study via the PharmacoFocus array, NAT2_c.-594G>C (5′UTR) (rs4271002) had a significant impact, increasing the RIS plasma level by 1.867-fold compared to noncarriers. Therefore, it is expected that carriers of this mutant allele may require dose modification to avoid RIS-induced adverse effects. Importantly, this is a novel NAT2 variant that is not a part of any named alleles and has been shown in one study to be significantly associated with the risk of aspirin-intolerant asthma (Kim et al., 2010). According to PharmGKB, the functional consequence of this SNP is currently unknown, but it may affect transcription, and its exact role in the RIS metabolic pathway remains unclear (McDonagh et al., 2014). However, our result is consistent with the impact of NAT2 polymorphisms in the study by Cabaleiro et al. (2014) reporting an increased incidence of RIS-induced headache among NAT2 IM and PM healthy individuals in comparison to NAT2 NMs.
4.2.4 UGT2B17
The UGT2B17 gene encodes uridine diphosphate glycosyltransferase 2 family, member B17, which is part of the family of UDP-glucuronosyltransferases (UGTs) (Beaulieu et al., 1996). As part of the phase II liver detoxification system, these genes are responsible for maintaining steady-state levels of a variety of substrates, including steroid hormones, by catalyzing the transfer of glucuronic acid moieties to these molecules and rendering them hydrophilic (Burchell et al., 1995). UGT2B17 is expressed not only in the small intestine and liver but also in steroid target tissues such as the breast, uterus, and prostate, where the extent of glucuronidation can be substantial (Karlsson et al., 2021), indicating its potential role in hormonally induced diseases (Wilson et al., 2004). However, molecular studies reported that UGT2B17 isoforms had a 4.4-fold higher abundance in the intestine than in the liver (Zhang et al., 2018). This fact suggested the potential of UGT2B17 to have a greater first-pass effect on its substrates when orally administered than subsequent liver metabolism, particularly in high UGT2B17-expressing individuals (Zhang et al., 2018). Additionally, in proteome studies, the UGT2B17 isoform, unlike other UGTs, was expressed in different brain regions, particularly in the cerebellum (Karlsson et al., 2021). According to the current study data, UGT2B17 rs78998153, which had a prevalence of 26% among our ASD children, exhibited a very significant effect on RIS exposure. Indeed, after Bonferroni correction for multiple testing, this novel variant is the only PGx marker that still demonstrated a positive impact on the RIS/9-OH-RIS metabolic ratio, indicating a substantial increase in the RIS plasma circulating levels in comparison to its major metabolite. Consistently, an exploratory-based study of RIS PK in Thai children with ASD demonstrated that UGT2B4 c.∗448A>G (rs1131878), an isoform that is more highly expressed in liver than brain tissues (Karlsson et al., 2021), was highly associated with the metabolic ratio (Medhasi et al., 2016). Additional evidence for the impact of UGTs on RIS PK can be drawn from a study in Thai children with ASD, where three SNPs indicating UGT1A1 mutation (an isoform that is highly expressed in brain tissues (Sjostedt et al., 2020)) have shown a significant association with the RIS-induced prolactin response (Hongkaew et al., 2018). Collectively, these results highlight the need for further molecular studies to explore the correlation between various UGT genotypes and their induced modifications in UGT enzyme expression in the brain tissues of patients with ASD. This research topic is anticipated to enable a better understanding of altered RIS disposition in the brain and its precise dose individualization requirements under this central nervous system condition (Sheng et al., 2021).
4.2.5 MTHFR
The MTHFR gene encodes the enzyme methylenetetrahydrofolate reductase (MTHER). MTHER is involved in a chemical reaction involving forms of the vitamin folate. Specifically, this enzyme converts 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate. This reaction is part of the multistep process that converts the amino acid homocysteine to another amino acid, methionine. The body uses methionine to make proteins and other important compounds, such as neurotransmitters (dopamine and serotonin) (Jalgaonkar et al., 2022; Majhi et al., 2023). Individuals homozygous for the SNP rs1801133 (MTHFR_c.665C>T(Ala222Val)) have lower MTHFR activity than CC or CT (heterozygous) individuals and therefore are predisposed to hyperhomocysteinemia associated with lower plasma folate levels (Majhi et al., 2023). A meta-analysis conducted on the association between MTHFR SNPs and ASD susceptibilities indicated that MTHFR rs1801133 was associated with ASD in the five genetic models (Li et al., 2020).
Consistent with this, a growing body of evidence suggests that the severity of autistic symptoms as a whole may be associated with increased levels of homocysteine associated with aggravation of dopamine deficiency (Carpita et al., 2023; Dangmann, 2023; Majhi et al., 2023). According to our data, rs1801133 of MTHFR, despite being less prevalent in our sample (12.9%) than in another cohort of Saudi children with ASD (36%) (Arab and Elhawary, 2019), was associated with a significant decrease in 9-OH-RIS and total active moiety levels, yet no evident impact was observed on RIS plasma exposure. The decreased concentration of the active moiety and a more pronounced effect on 9-OH-RIS could be explained by several factors. 9-OH-RIS undergoes minor hepatic metabolism and is primarily excreted unchanged by the kidney (79.6%). One of the known metabolic pathways for 9-OH-RIS is mediated by oxidative N-dealkylation, forming the acid metabolite M1 (Citrome, 2007; Vermeir et al., 2008). Emerging evidence indicates that high levels of homocysteine may enhance several metabolic pathways, such as oxidation (oxidative stress), nitrosylation, acylation, and hypomethylation (Perna et al., 2003a; 2003b). According to drug metabolism theories, these mechanisms (except hypomethylation) are believed to produce more polar metabolites that cannot diffuse across membranes and may, therefore, be actively transported (Li et al., 2019). Therefore, enhanced 9-OH-RIS excretion linked to hyperhomocysteinemia (induced by the MTHER mutation) via oxidation is assumed to be a superior postulated mechanism. In addition, homocysteine is a sulfur-containing amino acid (Lentz, 2005), which could serve as a co-factor in the conjugation of a drug metabolite by sulphation (Chen and Tang, 2022), leading to increased facilitated excretion in urine (Pan et al., 2020). However, these hypotheses remain uncertain. Clearly, more studies are necessary to elucidate the role of homocysteine in enhancing 9-OH-RIS excretion and its clinical consequences in ASD patients.
4.3 PGx markers encoding transporters
4.3.1 ABCC3
The ABCC3 gene encodes a protein that is a member of the superfamily of ATP-binding cassette (ABC) transporters. These ABC proteins transport various molecules across extra- and intracellular membranes. The specific function of this transporter has not yet been determined; however, it was reported to mediate biliary and intestinal excretion of organic anions (Banach et al., 2022). The functional activity of some ABCC3 variants has been fully examined (Singh et al., 2020). The current study revealed for the first time that a specific variant of the ABCC3 gene (ABCC3_c.3890G>A(R1297H)) had a prominent negative impact on 9-OH-RIS plasma levels, indicating decreased excretion (efflux) from target cells (hepatocytes or brain) to the bile or peripheral blood. The negative impact of this variant on RIS clinical efficacy and safety in ASD carriers warrants further investigation.
4.3.2 ABCB11
The ABCB11 gene encodes the ATP-binding cassette subfamily B member 11 protein, which is another member of the superfamily of ABC transporters. This membrane-associated protein is also named bile salt export pump (BSEP) or sister of P-glycoprotein (sPgp) (Strautnieks et al., 1998). Consistent with a previous exploratory study in Thai children with ASD (Medhasi et al., 2016), the current work detected the same 4 ABCB11 variants (ABCB11 rs495714, rs496550, rs473351 and rs497692), which displayed a significant reducing effect on the RIS/9-OH-RIS metabolic ratio, although at a lower rank of importance (Supplementary Table S4). Notably, our results in this population (Figure 2B) are compatible with the previous LD finding (Medhasi et al., 2016) that 3 ABCB11 SNPs were strongly linked. This observation hypothetically indicates a more predominant influence of these mutations on reducing RIS efflux than 9-OH-RIS efflux, probably from various tissues’ cells into bile or blood. In contrast, previous candidate gene studies in Caucasian children with ASD (Correia et al., 2009; Rafaniello et al., 2017) and adults with schizophrenia (Xing et al., 2006; Kuzman et al., 2008) have linked reduced RIS efflux with other ABC transporter subtype mutations (ABCG2_c.421C>A in ABCG2; c.3435C>T, c.1199G>A, c.1236C>T and c.2677G>T in ABCB1). These inconsistent findings emphasized the importance of genome-wide exploratory studies to reveal disease-specific PGx markers of certain drugs, with priority according to clinical significance and their ethnogeographic frequencies.
4.4 PGx markers encoding PD receptors
4.4.1 ADRA2A
One of the receptors that RIS blocks is the alpha 2A adrenoceptor (α2A-AR), which is an adrenergic receptor that responds to adrenaline and noradrenaline (Shahid et al., 2009). α2A-AR is encoded by the ADRA2A gene and is mainly found in the brain, where it regulates various functions, such as mood, cognition, attention, and sleep (Nyrönen et al., 2001). The role of α2A-AR in the therapeutic effects and side effects of RIS is not fully understood, but some studies have suggested that blocking this receptor may have both positive and negative consequences (Marcus et al., 2009). Additionally, other factors, such as genetic variations, drug interactions, and individual differences, may influence the response to RIS and the α2A-AR antagonism effect (Uys et al., 2017). The current association analysis revealed that ADRA2A SNP rs553668 carriers exhibit significantly increased RIS and total active moiety plasma levels (by 1.468- and 1.678-fold, respectively) in comparison to noncarriers. This finding could be interpreted in light of a previous study involving pheochromocytoma patients reporting that ADRA2A SNPs rs553668/rs521674 were associated with higher dosage requirements of α-adrenergic receptor blockers to control blood pressure (Berends et al., 2022).
Both our results and previous findings suggested that a certain degree of mutation in the α2A-AR receptor (decreased expression and/or density) mediated by these ADRA2A SNPs could lead to decreased drug-receptor occupancy and affinity, which could explain the subsequent higher PK exposure of any drugs targeted to antagonize it, such as that shown in our ASD patients exhibiting higher RIS plasma levels adjusted by the RIS dose. However, this assumption regarding the correlation between receptor affinity and drug plasma level remains speculative, and the functional consequences of the ADRA2A SNP on its receptor need to be examined. To our knowledge, to date, no other antipsychotic PGx studies have addressed the potential clinical consequences of polymorphisms of any of the genes encoding adrenergic receptors (including the ADRA2A gene) on efficacy and safety (Bousman et al., 2023). Therefore, it is important to investigate the current observation of significantly higher RIS plasma levels associated with ADRA2A polymorphisms in terms of clinical impacts in a larger cohort.
4.4.2 CRHR2
Corticotropin-releasing hormone receptor 2 (CRHR2) is a protein that is encoded by the CRHR2 gene, which is highly expressed in the choroid plexus (part of the blood–brain barrier) of the human brain and to a lesser extent in the plasma membranes of hormone-sensitive cells, including those in the gastrointestinal tract and kidney (Pal et al., 2010). CRH is a hormone secreted from the hypothalamus in response to stress, which needs to efficiently bind with the CRHR2 receptor to stimulate its effects (Grammatopoulos et al., 1999). CRF is a key hormone that is involved in the control of various body systems via its mediatory stimulation of the hypothalamic–pituitary–adrenal (HPA) axis. On the other hand, hypothalamic CRF, via its action on the HPA axis, may be partially involved in the reinforcing effects of metabolic enzymes in phases I and II (Mormede et al., 2011; Wójcikowski and Daniel, 2011; Bromek and Daniel, 2021). In addition, recently, increased activation of the HPA axis was suggested to play an important role in ASD-like social behaviors (Jacobson, 2014; Rusch et al., 2023). As CRHR2 is one of the receptors for the hormones involved in the HPA axis, decreased CRHR2 expression levels in the hypothalamus were recently suggested to increase the risk of ASD (Wang X. et al., 2023). Interestingly, the current data revealed that an intronic CRHR2 variant (rs7793837), which mostly causes mutations in the CRHR2 protein, was significantly associated with a positive impact on the total active moiety level in the plasma of children with ASD. These findings suggest that CRH could play a complex role in drug metabolism and possibly in the clinical response to RIS therapy, and further research could clarify its significance as a PGx marker within the context of ASD. A possible explanation for this increased level of RIS and 9-OH-RIS could be attributed to decreased binding of CRH to the mutated CRHR2 variant 3 (rs7793837), leading to its decreased functional impact on various downstream signaling pathways mediating the metabolism and excretion of both molecules. However, this hypothesis needs to be proven in further studies to elucidate its clinical impact on RIS therapy outcomes.
4.5 PGx markers encoding immunity proteins
Growing evidence in recent decades has highlighted the role of alterations in immune function, including heightened inflammation, anti-brain protein antibodies, and changes in T-cell and natural killer (NK) cell function in individuals diagnosed with ASD (Hsiao, 2013; Morton et al., 2023). Given that ASD may be induced by immunological or inflammatory pathological processes within the brain, GWASs have identified various associated human leukocyte antigen (HLA) risk alleles, including those related to HLA-DPB1, that obtained the most significant probabilities (Nudel et al., 2019; Morton et al., 2023). The current study revealed significant associations between HLA-DPB1 (rs1042151), HLA-G (rs66554220), and HLA-A (rs1061235) and increased plasma levels of RIS, 9-OH-RIS, and total active moiety, respectively. A possible explanation for these influences could be attributed to the recently reported potential interplay between gut inflammatory processes mediated by these HLA markers and ASD incidence in children (the gut–brain axis) (Lombardi et al., 2023; Morton et al., 2023), thus leading to the postulation of pathophysiological changes in gastrointestinal permeability with subsequent alterations in RIS absorption and other possible inflammatory process-related consequences on the downregulation of the hepatic expression of its metabolizing enzymes or transporters (Effinger et al., 2019).
The latter hypothesis could be of interest for further examination in molecular expression studies involving ASD patients to confirm the impact of HLA-mediated inflammatory status on any medication’s global PK, particularly those that are indicated for chronic use, such as RIS.
4.6 Strengths and limitations
Our study had several strengths and limitations. The first major strength of the current study compared with earlier candidate gene studies is the employment of an exploratory focused pharmacogenetic approach with a comprehensive array platform, which enables genotyping of most known PGx markers in genes related to the exposure and clinical consequences of most drugs, as curated by PharmGKB. Second, this is the first study to describe RIS PK in a cohort of Arabic children. To date, only a few RIS PK studies have been conducted, and these have focused mostly on children of European backgrounds (Maruf et al., 2021). Third, in addition to characterization of the exposure of RIS in Arabic children based on classical pathways, the modern methodology enabled identification of genetic variants of known association with ASD etiology that could specifically modulate RIS exposure. Fourth, our hypothesis-generating approach in this study revealed several novel PD SNPs that have a significant influence on RIS exposure in ASD children rather than PK SNPs alone, which indicated the need for further population PK modeling studies in this specific population (with more extensive blood sampling at several time points) to re-estimate the other RIS PK parameters such as steady-state volume of distribution and absorption and elimination rates. Modeling studies incorporating the PD, PK, and disease variants as predictors of interindividual variabilities in RIS plasma concentrations are speculated to enable more precise dosing and individualized therapy for children with ASD. Fifth, we employed LD analysis to reveal significant haplotype approximate loci that are not obtained by single genetic variation genotyping alone (Eberle et al., 2006).
However, there are also limitations that should be acknowledged. As detailed earlier, the study revealed significant associations for several PGx variants describing novel pathways (for example, immunity markers), but most of these associations did not remain significant after correction for multiple testing. This could be due to a lack of statistical power, in addition to the large number of markers tested in this study. However, our strict limitation to p values of less than 0.01 (together with 95% CI interval) in the top tier findings of this cohort has added confidence in adjusting for type I error to avoid false-positive findings. In addition, concerns were raised regarding the misuse and overly conservative practice of correction for multiple testing in exploratory studies due its potential to produce false-negative conclusions for significant markers that are certainly important (type II error) (Johnson et al., 2010). Alternatively, to ideally ensure richness in datasets’ information to answer the exploratory research question (finding of innovative unanticipated associations), correction for multiple testing is statistically ill-advised, particularly if the modeling was adjusted by other justifiable techniques (such as PCs and nongenetic variables) (Rothman, 1990; García-Pérez, 2023). Second, since our study population has rarely been explored, our novel findings should be considered hypothesis generating and require validation in diverse ancestry cohorts. Third, in this study, RIS clinical outcomes reflecting effectiveness and safety were not examined; therefore, their relation to the novel variants identified in this study require further validation in future studies to confirm their utility in clinical practice with chronically treated patients with ASD. Finally, markers that failed the DQC parameters were removed from the present analysis. Some of these SNPs are related to the novel genes discovered in this study. Therefore, further association studies including these SNPs could strengthen the evidence of their related genes’ impact on RIS PK.
5 Conclusion
In conclusion, the study provided strong evidence of an interplay of PK (metabolic enzymes), transporters, PD (receptors), and other novel groups of genetic variants (immune markers) in determining the RIS exposure level in ASD patients. The study also demonstrates the importance of an exploratory approach via the Axiom array technique, which has contributed to more precisely revealing and simulating the complex system of the pathophysiology of RIS disposition in children with ASD, in comparison to earlier candidate gene approach studies, where relevant genes were probably not fully addressed. Additionally, there could be physiologically relevant signaling pathways for some of our novel PGx markers that have not yet been revealed, and polymorphisms in genes influencing the signal transduction of these variants could also be of interest to reveal the complicated mechanisms underlying autistic phenotypes. Therefore, future studies in a larger cohort of diverse ancestry groups could confirm our current findings and improve the knowledge base on how these PGx variants could modify the efficacy of RIS or other antipsychotics and the risk of developing side effects in a broader range of ASD phenotypes, characterized by diverse neurological alterations, which could facilitate personalized therapeutic decision-making in this complex neurodevelopmental condition. In addition, the present findings could open a state-of-the-art track for mechanistic research into genetic informers of variability in antipsychotic exposure-mediated responses, which may indicate a novel approach for drug development.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by the Institutional Review Board (IRB) committees of both PNU (IRB Log Number: 20-0321) and KFMC (IRB Log Number: 20-758E). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
SS: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing–original draft, Writing–review and editing. IA: Conceptualization, Data curation, Investigation, Validation, Writing–review and editing. EG: Methodology, Validation, Writing–review and editing. HA: Methodology, Writing–review and editing. KA: Methodology, Writing–review and editing. FA: Visualization, Writing–review and editing. AF: Data curation, Formal Analysis, Methodology, Validation, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Health Sciences Research Center, King Abdullah bin Abdulaziz University Hospital, and Princess Nourah bint Abdulrahman University through the Research Funding Program (grant no. G18-00018).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1356763/full#supplementary-material
Abbreviations
ADME, absorption, distribution, metabolism, and excretion; ASD, autism spectrum disorder; DOI, direction of impact; GWAS, genome-wide association study; HWE, Hardy–Weinberg equilibrium; IQR, interquartile range; LD, linkage disequilibrium; PD, pharmacodynamics; PGx, pharmacogenetic; PharmGKB, Pharmacogenomics Knowledge Base; PK, pharmacokinetics; QC, quality control; MAF, minor allele frequency; UM, ultrarapid metabolizer; NM, extensive or normal metabolizer; IM, intermediate metabolizer; PM, poor metabolizer; RIS, risperidone; 9-OH-RIS, 9-hydroxyrisperidone; SNP, single-nucleotide polymorphism.
References
Abbaspour, A., Dehghani, M., Setayesh, M., Tavakkoli, M., Rostamipour, H. A., Ghorbani, M., et al. (2023). Cytidine deaminase enzyme activity is a predictive biomarker in gemcitabine-treated cancer patients. Cancer Chemother. Pharmacol. 92, 475–483. doi:10.1007/s00280-023-04579-8
Ackenheil, M., and Weber, K. (2004). Differing response to antipsychotic therapy in schizophrenia: pharmacogenomic aspects. Dialogues Clin. Neurosci. 6, 71–77. doi:10.31887/dcns.2004.6.1/mackenheil
Aishworiya, R., Valica, T., Hagerman, R., and Restrepo, B. (2022). An update on psychopharmacological treatment of autism spectrum disorder. Neurotherapeutics 19, 248–262. doi:10.1007/s13311-022-01183-1
Al-Dosari, M. S., Al-Jenoobi, F. I., Alkharfy, K. M., Alghamdi, A. M., Bagulb, K. M., Parvez, M. K., et al. (2013). High prevalence of CYP2D6*41 (G2988A) allele in Saudi Arabians. Environ. Toxicol. Pharmacol. 36, 1063–1067. doi:10.1016/j.etap.2013.09.008
Allegra, S., Cusato, J., De Francia, S., Massano, D., Piga, A., and D’Avolio, A. (2016). Deferasirox AUC efficacy cutoff and role of pharmacogenetics. Eur. J. Clin. Pharmacol. 72, 1155–1157. doi:10.1007/s00228-016-2070-9
Allegra, S., De Francia, S., Cusato, J., Arduino, A., Massano, D., Longo, F., et al. (2017). Deferasirox pharmacogenetic influence on pharmacokinetic, efficacy and toxicity in a cohort of pediatric patients. Pharmacogenomics 18, 539–554. doi:10.2217/pgs-2016-0176
Ang, M. J., and Moon, C. (2022). SREBP and central nervous system disorders: genetic overlaps revealed by in silico analysis. J. Integr. Neurosci. 21, 95. doi:10.31083/j.jin2103095
Arab, A. H., and Elhawary, N. A. (2019). Methylenetetrahydrofolate reductase gene variants confer potential vulnerability to autism spectrum disorder in a Saudi Community. Neuropsychiatr. Dis. Treat. 15, 3569–3581. doi:10.2147/ndt.s230348
Aravagiri, M., and Marder, S. R. (2000). Simultaneous determination of risperidone and 9-hydroxyrisperidone in plasma by liquid chromatography/electrospray tandem mass spectrometry. J. Mass Spectrom. 35, 718–724. doi:10.1002/1096-9888(200006)35:6<718::aid-jms999>3.0.co;2-o
Bahado-Singh, R. O., Vishweswaraiah, S., Aydas, B., and Radhakrishna, U. (2022). Artificial intelligence and placental DNA methylation: newborn prediction and molecular mechanisms of autism in preterm children. J. Matern. Fetal Neonatal Med. 35, 8150–8159. doi:10.1080/14767058.2021.1963704
Bahadur, N., Leathart, J. B. S., Mutch, E., Steimel-Crespi, D., Dunn, S. A., Gilissen, R., et al. (2002). CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6α-hydroxylase activity in human liver microsomes. Biochem. Pharmacol. 64, 1579–1589. doi:10.1016/s0006-2952(02)01354-0
Banach, B., Modrzejewski, A., Juzyszyn, Z., Kurzawski, M., Sroczynski, T., and Pawlik, A. (2022). Association study of SLCO1B3 and ABCC3 genetic variants in gallstone disease. Genes 13, 512. doi:10.3390/genes13030512
Barrett, J. C., Fry, B., Maller, J., and Daly, M. J. (2004). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265. doi:10.1093/bioinformatics/bth457
Beaulieu, M., Lávesque, E., Hum, D. W., and Bálanger, A. (1996). Isolation and characterization of a novel cDNA encoding a human UDP-glucuronosyltransferase active on C19 steroids. J. Biol. Chem. 271, 22855–22862. doi:10.1074/jbc.271.37.22855
Berends, A. M. A., Bolhuis, M. S., Nolte, I. M., Buitenwerf, E., Links, T. P., Timmers, H. J. L. M., et al. (2022). Influence of receptor polymorphisms on the response to α-adrenergic receptor blockers in pheochromocytoma patients. Biomedicines 10, 896. doi:10.3390/biomedicines10040896
Bousman, C. A., Maruf, A. A., Marques, D. F., Brown, L. C., and Müller, D. J. (2023). The emergence, implementation, and future growth of pharmacogenomics in psychiatry: a narrative review. Psychol. Med., 1–11. doi:10.1017/s0033291723002817
Bråten, L. S., Haslemo, T., Jukic, M. M., Ivanov, M., Ingelman-Sundberg, M., Molden, E., et al. (2021). A novel CYP2C-haplotype associated with ultrarapid metabolism of escitalopram. Clin. Pharmacol. Ther. 110, 786–793. doi:10.1002/cpt.2233
Bromek, E., and Daniel, W. A. (2021). The regulation of liver cytochrome P450 expression and activity by the brain serotonergic system in different experimental models. Expert Opin. Drug Metab. Toxicol. 17, 413–424. doi:10.1080/17425255.2021.1872543
Burchell, B., Brierley, C. H., and Rance, D. (1995). Specificity of human UDP-Glucuronosyltransferases and xenobiotic glucuronidation. Life Sci. 57, 1819–1831. doi:10.1016/0024-3205(95)02073-r
Burczynski, M. E. (2009). Pharmacogenomic approaches in clinical studies to identify biomarkers of safety and efficacy. Toxicol. Lett. 186, 18–21. doi:10.1016/j.toxlet.2008.10.019
Butler, M. G., McGuire, A. B., Masoud, H., and Manzardo, A. M. (2016). Currently recognized genes for schizophrenia: high-resolution chromosome ideogram representation. Am. J. Med. Genet. B Neuropsychiatr. Genet. 171, 181–202. doi:10.1002/ajmg.b.32391
Cabaleiro, T., López-Rodríguez, R., Ochoa, D., Román, M., Novalbos, J., and Abad-Santos, F. (2013). Polymorphisms influencing olanzapine metabolism and adverse effects in healthy subjects. Hum. Psychopharmacol. Clin. Exp. 28, 205–214. doi:10.1002/hup.2308
Cabaleiro, T., Ochoa, D., López-Rodríguez, R., Román, M., Novalbos, J., Ayuso, C., et al. (2014). Effect of polymorphisms on the pharmacokinetics, pharmacodynamics, and safety of risperidone in healthy volunteers. Hum. Psychopharmacol. Clin. Exp. 29, 459–469. doi:10.1002/hup.2420
Camargo, B. M. D. O., Ziliani, M. J. D. G. F., Luiza, A., Alvarenga, B. C., Nagao, R. S., Michelin, L. F. G., et al. (2023). Benzodiazepine dependence and genetic factors: a literature review. Rev. Med. (São Paulo. 102 (1), e–203848.
Carpita, B., Massoni, L., Battaglini, S., Palego, L., Cremone, I. M., Massimetti, G., et al. (2023). IL-6, homocysteine, and autism spectrum phenotypes: an investigation among adults with autism spectrum disorder and their first-degree relatives. CNS Spectr. 28, 620–628. doi:10.1017/s1092852923000019
Carthy, E., Ross, C., and Murphy, D. (2023). Psychotropic medication prescribing in people with autism spectrum disorders with and without psychiatric comorbidity. BJPsych Adv. 29, 131–140. doi:10.1192/bja.2021.32
Chang, C. C., Chow, C. C., Tellier, L. C. A. M., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, 7. doi:10.1186/s13742-015-0047-8
Chen, S. M., and Tang, X. Q. (2022). Homocysteinylation and sulfhydration in diseases. Curr. Neuropharmacol. 20, 1726–1735. doi:10.2174/1570159x20666211223125448
Cho, H. Y., and Lee, Y. B. (2006). Pharmacokinetics and bioequivalence evaluation of risperidone in healthy male subjects with different CYP2D6 genotypes. Arch. Pharmacal Res. 29, 525–533. doi:10.1007/bf02969428
Ciccolini, J., Serdjebi, C., Peters, G. J., and Giovannetti, E. (2016). Pharmacokinetics and pharmacogenetics of Gemcitabine as a mainstay in adult and pediatric oncology: an EORTC-PAMM perspective. Cancer Chemother. Pharmacol. 78, 1–12. doi:10.1007/s00280-016-3003-0
Citrome, L. (2007). Paliperidone: quo vadis? Int. J. Clin. Pract. 61, 653–662. doi:10.1111/j.1742-1241.2007.01321.x
Clarke, W. P., Chavera, T. A., Silva, M., Sullivan, L. C., and Berg, K. A. (2013). Signalling profile differences: paliperidone versus risperidone: paliperidone versus risperidone. Br. J. Pharmacol. 170, 532–545. doi:10.1111/bph.12295
Correia, C. T., Almeida, J. P., Santos, P. E., Sequeira, A. F., Marques, C. E., Miguel, T. S., et al. (2009). Pharmacogenetics of risperidone therapy in autism: association analysis of eight candidate genes with drug efficacy and adverse drug reactions. Pharmacogenomics J. 10, 418–430. doi:10.1038/tpj.2009.63
Cusato, J., Allegra, S., Massano, D., De Francia, S., Piga, A., and D'Avolio, A. (2014). Influence of single-nucleotide polymorphisms on deferasirox Ctrough levels and effectiveness. Pharmacogenomics J. 15, 263–271. doi:10.1038/tpj.2014.65
Dangmann, R. (2023). A hypothesis to explain the potential influence of hormones on the severity of autism spectrum conditions in women. Med. Hypotheses 178, 111136. doi:10.1016/j.mehy.2023.111136
Dodsworth, T., Kim, D. D., Procyshyn, R. M., Ross, C. J., Honer, W. G., and Barr, A. M. (2018). A systematic review of the effects of CYP2D6 phenotypes on risperidone treatment in children and adolescents. Child. Adolesc. Psychiatry Ment. Health 12, 37. doi:10.1186/s13034-018-0243-2
Eap, C. B. (2016). Personalized prescribing: a new medical model for clinical implementation of psychotropic drugs. Dialogues Clin. Neurosci. 18, 313–322. doi:10.31887/dcns.2016.18.3/ceap
Eberle, M. A., Rieder, M. J., Kruglyak, L., and Nickerson, D. A. (2006). Allele frequency matching between SNPs reveals an excess of linkage disequilibrium in genic regions of the human genome. PLoS Genet. 2, e142. doi:10.1371/journal.pgen.0020142
Effinger, A., O'Driscoll, C. M., McAllister, M., and Fotaki, N. (2019). Impact of gastrointestinal disease states on oral drug absorption – implications for formulation design – a PEARRL review. J. Pharm. Pharmacol. 71, 674–698. doi:10.1111/jphp.12928
Eum, S., Lee, A. M., and Bishop, J. R. (2022). Pharmacogenetic tests for antipsychotic medications: clinical implications and considerations. Dialogues Clin. Neurosci. 18, 323–337. doi:10.31887/dcns.2016.18.3/jbishop
Fang, J., Bourin, M., and Baker, G. B. (1999). Metabolism of risperidone to 9-hydroxyrisperidone by human cytochromes P450 2D6 and 3A4. Naunyn-Schmiedeb. Arch. Pharmacol. 359, 147–151. doi:10.1007/pl00005334
Fang, Y., Cui, Y., Yin, Z., Hou, M., Guo, P., Wang, H., et al. (2023). Comprehensive systematic review and meta-analysis of the association between common genetic variants and autism spectrum disorder. Gene 887, 147723. doi:10.1016/j.gene.2023.147723
Fernandes, V. C., Pretti, M. A. M., Tsuneto, L. T., Petzl-Erler, M. L., and Suarez-Kurtz, G. (2023). Distribution of a novel CYP2C haplotype in Native American populations. Front. Genet. 14, 1114742. doi:10.3389/fgene.2023.1114742
Gaedigk, A., Boone, E. C., Scherer, S. E., Lee, S.-b., Numanagić, I., Sahinalp, C., et al. (2022). CYP2C8, CYP2C9, and CYP2C19 characterization using next-generation sequencing and haplotype analysis. J. Mol. Diagn. 24, 337–350. doi:10.1016/j.jmoldx.2021.12.011
Gaedigk, A., Ingelman-Sundberg, M., Miller, N. A., Leeder, J. S., Whirl-Carrillo, M., and Klein, T. E. (2017). The pharmacogene variation (PharmVar) consortium: incorporation of the human cytochrome P450 (CYP) allele nomenclature database. Clin. Pharmacol. Ther. 103, 399–401. doi:10.1002/cpt.910
García-Pérez, M. A. (2023). Use and misuse of corrections for multiple testing. Methods Psychol. 8, 100120. doi:10.1016/j.metip.2023.100120
Gassó, P., Mas, S., Papagianni, K., Ferrando, E., de Bobadilla, R. F., Arnaiz, J. A., et al. (2014). Effect of CYP2D6 on risperidone pharmacokinetics and extrapyramidal symptoms in healthy volunteers: results from a pharmacogenetic clinical trial. Pharmacogenomics 15, 17–28. doi:10.2217/pgs.13.204
Gavin, D. P., Sharma, R. P., Chase, K. A., Matrisciano, F., Dong, E., and Guidotti, A. (2012). Growth arrest and DNA-damage-inducible, beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology 37, 531–542. doi:10.1038/npp.2011.221
Genovese, A., and Butler, M. G. (2020). Clinical assessment, genetics, and treatment approaches in autism spectrum disorder (ASD). Int. J. Mol. Sci. 21, 4726. doi:10.3390/ijms21134726
Genovese, A., and Butler, M. G. (2023). The autism spectrum: behavioral, psychiatric and genetic associations. Genes 14, 677. doi:10.3390/genes14030677
Göbel, A., Rauner, M., Hofbauer, L. C., and Rachner, T. D. (2020). Cholesterol and beyond - the role of the mevalonate pathway in cancer biology. Biochim. Biophys. Acta (BBA) Rev. Cancer 1873, 188351. doi:10.1016/j.bbcan.2020.188351
Goldstein, J. A., and de Morais, S. M. F. (1994). Biochemistry and molecular biology of the human CYP2C subfamily. Pharmacogenetics 4, 285–300. doi:10.1097/00008571-199412000-00001
Gong, W., Qiao, Y., Li, B., Zheng, X., Xu, R., Wang, M., et al. (2021). The alteration of salivary immunoglobulin A in autism spectrum disorders. Front. Psychiatry 12, 669193. doi:10.3389/fpsyt.2021.669193
Grammatopoulos, D. K., Dai, Y., Randeva, H. S., Levine, M. A., Karteris, E., Easton, A. J., et al. (1999). A novel spliced variant of the type 1 corticotropin-releasing hormone receptor with a deletion in the seventh transmembrane domain present in the human pregnant term myometrium and fetal membranes. Mol. Endocrinol. 13, 2189–2202. doi:10.1210/mend.13.12.0391
Grau, J. J., Caballero, M., Campayo, M., Jansa, S., Vargas, M., Alós, L., et al. (2009). Gene single nucleotide polymorphism accumulation improves survival in advanced head and neck cancer patients treated with weekly paclitaxel. Laryngoscope 119, 1484–1490. doi:10.1002/lary.20254
Grover, S., Talwar, P., Gourie-Devi, M., Gupta, M., Bala, K., Sharma, S., et al. (2010). Genetic polymorphisms in sex hormone metabolizing genes and drug response in women with epilepsy. Pharmacogenomics 11, 1525–1534. doi:10.2217/pgs.10.120
Guidotti, A., and Grayson, D. R. (2014). DNA methylation and demethylation as targets for antipsychotic therapy. Dialogues Clin. Neurosci. 16, 419–429. doi:10.31887/dcns.2014.16.3/aguidotti
Gupta, N., and Gupta, M. (2023). Off-label psychopharmacological interventions for autism spectrum disorders: strategic pathways for clinicians. CNS Spectr., 1–16. doi:10.1017/s1092852923002389
Hayashi, S. I., Watanabe, J., and Kawajiri, K. (1991). Genetic polymorphisms in the 5′-flanking region change transcriptional regulation of the human cytochrome P450IIE1 gene1. J. Biochem. 110, 559–565. doi:10.1093/oxfordjournals.jbchem.a123619
Højlund, M., Andersen, J. H., Andersen, K., Correll, C. U., and Hallas, J. (2021). Use of antipsychotics in Denmark 1997–2018: a nation-wide drug utilisation study with focus on off-label use and associated diagnoses. Epidemiol. Psychiatr. Sci. 30, e28. doi:10.1017/s2045796021000159
Hongkaew, Y., Gaedigk, A., Wilffert, B., Ngamsamut, N., Kittitharaphan, W., Limsila, P., et al. (2021). Relationship between CYP2D6 genotype, activity score and phenotype in a pediatric Thai population treated with risperidone. Sci. Rep. 11, 4158. doi:10.1038/s41598-021-83570-w
Hongkaew, Y., Medhasi, S., Pasomsub, E., Ngamsamut, N., Puangpetch, A., Vanwong, N., et al. (2018). UGT1A1 polymorphisms associated with prolactin response in risperidone-treated children and adolescents with autism spectrum disorder. Pharmacogenomics J. 18, 740–748. doi:10.1038/s41397-018-0031-7
Hsiao, E. Y. (2013). Immune dysregulation in autism spectrum disorder. Int. Rev. Neurobiol. 113, 269–302. doi:10.1016/B978-0-12-418700-9.00009-5
Huang, M. L., Peer, A. V., Woestenborghs, R., De Coster, R., Heykants, J., Jansen, A. A. I., et al. (1993). Pharmacokinetics of the novel antipsychotic agent risperidone and the prolactin response in healthy subjects. Clin. Pharmacol. Ther. 54, 257–268. doi:10.1038/clpt.1993.146
Huang, X., Chen, L., Song, W., Chen, L., Niu, J., Han, X., et al. (2012). Systematic functional characterization of cytochrome P450 2E1 promoter variants in the Chinese Han population. PLoS One 7, e40883. doi:10.1371/journal.pone.0040883
Huo, R., Tang, K., Wei, Z., Shen, L., Xiong, Y., Wu, X., et al. (2012). Genetic polymorphisms in CYP2E1: association with schizophrenia susceptibility and risperidone response in the Chinese han population. PLoS One 7, e34809. doi:10.1371/journal.pone.0034809
Jacobson, L. (2014). Hypothalamic-pituitary-adrenocortical axis: neuropsychiatric aspects. Compr. Physiol. 4, 715–738. doi:10.1002/cphy.c130036
Jalgaonkar, S., Gajbhiye, S., Sayyed, M., Tripathi, R., Khatri, N., Parmar, U., et al. (2022). S-adenosyl methionine improves motor co-ordination with reduced oxidative stress, dopaminergic neuronal loss, and DNA methylation in the brain striatum of 6-hydroxydopamine-induced neurodegeneration in rats. Anat. Rec. 306, 820–830. doi:10.1002/ar.24948
Johnson, R. C., Nelson, G. W., Troyer, J. L., Lautenberger, J. A., Kessing, B. D., Winkler, C. A., et al. (2010). Accounting for multiple comparisons in a genome-wide association study (GWAS). BMC Genom 11, 724. doi:10.1186/1471-2164-11-724
Karlsson, M., Zhang, C., Mear, L., Zhong, W., Digre, A., Katona, B., et al. (2021). A single-cell type transcriptomics map of human tissues. Sci. Adv. 7, eabh2169. doi:10.1126/sciadv.abh2169
Khanzada, N., Butler, M., and Manzardo, A. (2017). GeneAnalytics pathway analysis and genetic overlap among autism spectrum disorder, bipolar disorder and schizophrenia. Int. J. Mol. Sci. 18, 527. doi:10.3390/ijms18030527
Kim, J. M., Park, B. L., Park, S. M., Lee, S. H., Kim, M. O., Jung, S., et al. (2010). Association analysis of N-acetyl transferase-2 polymorphisms with aspirin intolerance among asthmatics. Pharmacogenomics 11, 951–958. doi:10.2217/pgs.10.65
Kirchheiner, J., Meineke, I., Fuhr, U., Rodríguez-Antona, C., Lebedeva, E., and Brockmöller, J. (2008). Impact of genetic polymorphisms in CYP2C8 and rosiglitazone intake on the urinary excretion of dihydroxyeicosatrienoic acids. Pharmacogenomics 9, 277–288. doi:10.2217/14622416.9.3.277
Kloosterboer, S. M., de Winter, B. C. M., Reichart, C. G., Kouijzer, M. E. J., de Kroon, M. M. J., van Daalen, E., et al. (2021). Risperidone plasma concentrations are associated with side effects and effectiveness in children and adolescents with autism spectrum disorder. Br. J. Clin. Pharmacol. 87, 1069–1081. doi:10.1111/bcp.14465
Kubota, T., Hibi, N., and Chiba, K. (1998). Linkage of mutant alleles of CYP2C18 and CYP2C19 in a Japanese population. Biochem. Pharmacol. 55, 2039–2042. doi:10.1016/s0006-2952(98)00022-7
Kukal, S., Thakran, S., Kanojia, N., Yadav, S., Mishra, M. K., Guin, D., et al. (2023). Genic-intergenic polymorphisms of CYP1A genes and their clinical impact. Gene 857, 147171. doi:10.1016/j.gene.2023.147171
Kuzman, M. R., Medved, V., Bozina, N., Hotujac, L., Sain, I., and Bilusic, H. (2008). The influence of 5-HT2C and MDR1 genetic polymorphisms on antipsychotic-induced weight gain in female schizophrenic patients. Psychiatry Res. 160, 308–315. doi:10.1016/j.psychres.2007.06.006
Lai, X. S., Yang, L. P., Li, X. T., Liu, J. P., Zhou, Z. W., and Zhou, S. F. (2009). Human CYP2C8: structure, substrate specificity, inhibitor selectivity, inducers and polymorphisms. Curr. Drug Metab. 10, 1009–1047. doi:10.2174/138920009790711832
Lamy, M., and Erickson, C. A. (2018). Pharmacological management of behavioral disturbances in children and adolescents with autism spectrum disorders. Curr. Probl. Pediatr. Adolesc. Health Care 48, 250–264. doi:10.1016/j.cppeds.2018.08.015
Lavelle, D., Vaitkus, K., Ling, Y., Ruiz, M. A., Mahfouz, R., Ng, K. P., et al. (2012). Effects of tetrahydrouridine on pharmacokinetics and pharmacodynamics of oral decitabine. Blood 119, 1240–1247. doi:10.1182/blood-2011-08-371690
Lee, D., Szumlanski, C., Houtman, J., Honchel, R., Rojas, K., Overhauser, J., et al. (1995). Thiopurine methyltransferase pharmacogenetics. Cloning of human liver cDNA and a processed pseudogene on human chromosome 18q21.1. Drug Metab. Dispos. 23, 398–405.
Le Hellard, S., Theisen, F. M., Haberhausen, M., Raeder, M. B., Fernø, J., Gebhardt, S., et al. (2008). Association between the insulin-induced gene 2 (INSIG2) and weight gain in a German sample of antipsychotic-treated schizophrenic patients: perturbation of SREBP-controlled lipogenesis in drug-related metabolic adverse effects? Mol. Psychiatry 14, 308–317. doi:10.1038/sj.mp.4002133
Lentz, S. R. (2005). Mechanisms of homocysteine-induced atherothrombosis. J. Thromb. Haemost. 3, 1646–1654. doi:10.1111/j.1538-7836.2005.01364.x
Li, Y., Meng, Q., Yang, M., Liu, D., Hou, X., Tang, L., et al. (2019). Current trends in drug metabolism and pharmacokinetics. Acta Pharm. Sin. B 9, 1113–1144. doi:10.1016/j.apsb.2019.10.001
Li, Y., Qiu, S., Shi, J., Guo, Y., Li, Z., Cheng, Y., et al. (2020). Association between MTHFR C677T/A1298C and susceptibility to autism spectrum disorders: a meta-analysis. BMC Pediatr. 20, 449. doi:10.1186/s12887-020-02330-3
Liang, J., Ringeling, L. T., Hermans, R. A., Bayraktar, I., Bosch, T. M., Egberts, K. M., et al. (2023). Clinical pharmacokinetics of antipsychotics in pediatric populations: a scoping review focusing on dosing regimen. Expert Opin. Drug Metab. Toxicol. 19, 501–509. doi:10.1080/17425255.2023.2252340
Lin, J., de Rezende, V. L., da Costa, M. d.A., de Oliveira, J., and Gonçalves, C. L. (2023). Cholesterol metabolism pathway in autism spectrum disorder: from animal models to clinical observations. Pharmacol. Biochem. Behav. 223, 173522. doi:10.1016/j.pbb.2023.173522
Llerena, A., Berecz, R., Dorado, P., and de la Rubia, A. (2004). QTc interval, CYP2D6 and CYP2C9 genotypes and risperidone plasma concentrations. J. Psychopharmacol. 18, 189–193. doi:10.1177/0269881104042618
Lombardi, L., Le Clerc, S., Wu, C. L., Bouassida, J., Boukouaci, W., Sugusabesan, S., et al. (2023). A human leukocyte antigen imputation study uncovers possible genetic interplay between gut inflammatory processes and autism spectrum disorders. Transl. Psychiatry 13, 244. doi:10.1038/s41398-023-02550-y
Majhi, S., Kumar, S., and Singh, L. (2023). A review on autism spectrum disorder: pathogenesis, biomarkers, pharmacological and non-pharmacological interventions. CNS Neurol. Disord. Drug Targets 22, 659–677. doi:10.2174/1871527321666220428134802
Mannheimer, B., Haslemo, T., Lindh, J. D., Eliasson, E., and Molden, E. (2016). Risperidone and venlafaxine metabolic ratios strongly predict a CYP2D6 poor metabolizing genotype. Ther. Drug Monit. 38, 127–134. doi:10.1097/ftd.0000000000000251
Marcus, M. M., Wiker, C., Frånberg, O., Konradsson-Geuken, Å., Langlois, X., Jardemark, K., et al. (2009). Adjunctive α2-adrenoceptor blockade enhances the antipsychotic-like effect of risperidone and facilitates cortical dopaminergic and glutamatergic, NMDA receptor-mediated transmission. Int. J. Neuropsychopharmacol. 13, 891–903. doi:10.1017/s1461145709990794
Maruf, A. A., Stein, K., Arnold, P. D., Aitchison, K. J., Müller, D. J., and Bousman, C. (2021). CYP2D6 and antipsychotic treatment outcomes in children and youth: a systematic review. J. Child. Adolesc. Psychopharmacol. 31, 33–45. doi:10.1089/cap.2020.0093
McDonagh, E. M., Boukouvala, S., Aklillu, E., Hein, D. W., Altman, R. B., and Klein, T. E. (2014). PharmGKB summary: very important pharmacogene information for N-acetyltransferase 2. Pharmacogenetics Genom 24, 409–425. doi:10.1097/fpc.0000000000000062
McLellan, R. A., Oscarson, M., Seidegard, J., Evans, D. A. P., and Ingelman-Sundberg, M. (1997). Frequent occurrence of CYP2D6 gene duplication in Saudi Arabians. Pharmacogenetics 7, 187–191. doi:10.1097/00008571-199706000-00003
Medhasi, S., Pinthong, D., Pasomsub, E., Vanwong, N., Ngamsamut, N., Puangpetch, A., et al. (2016). Pharmacogenomic study reveals new variants of drug metabolizing enzyme and transporter genes associated with steady-state plasma concentrations of risperidone and 9-hydroxyrisperidone in Thai autism spectrum disorder patients. Front. Pharmacol. 7, 475. doi:10.3389/fphar.2016.00475
Mormede, P., Foury, A., Barat, P., Corcuff, J. B., Terenina, E., Marissal-Arvy, N., et al. (2011). Molecular genetics of hypothalamic–pituitary–adrenal axis activity and function. Ann. N. Y. Acad. Sci. 1220, 127–136. doi:10.1111/j.1749-6632.2010.05902.x
Morton, J. T., Jin, D. M., Mills, R. H., Shao, Y., Rahman, G., McDonald, D., et al. (2023). Multi-level analysis of the gut–brain axis shows autism spectrum disorder-associated molecular and microbial profiles. Nat. Neurosci. 26, 1208–1217. doi:10.1038/s41593-023-01361-0
Novalbos, J., López-Rodríguez, R., Román, M., Gallego-Sandín, S., Ochoa, D., and Abad-Santos, F. (2010). Effects of CYP2D6 genotype on the pharmacokinetics, pharmacodynamics, and safety of risperidone in healthy volunteers. J. Clin. Psychopharmacol. 30, 504–511. doi:10.1097/jcp.0b013e3181ee84c7
Nudel, R., Benros, M. E., Krebs, M. D., Allesøe, R. L., Lemvigh, C. K., Bybjerg-Grauholm, J., et al. (2019). Immunity and mental illness: findings from a Danish population-based immunogenetic study of seven psychiatric and neurodevelopmental disorders. Eur. J. Hum. Genet. 27, 1445–1455. doi:10.1038/s41431-019-0402-9
Nuntamool, N., Ngamsamut, N., Vanwong, N., Puangpetch, A., Chamnanphon, M., Hongkaew, Y., et al. (2017). Pharmacogenomics and efficacy of risperidone long-term treatment in Thai autistic children and adolescents. Basic Clin. Pharmacol. Toxicol. 121, 316–324. doi:10.1111/bcpt.12803
Nyrönen, T., Pihlavisto, M., Peltonen, J. M., Hoffrén, A. M., Varis, M., Salminen, T., et al. (2001). Molecular mechanism for agonist-promoted α2A-adrenoceptor activation by norepinephrine and epinephrine. Mol. Pharmacol. 59, 1343–1354. doi:10.1124/mol.59.5.1343
Ooi, A., Banno, B., McFee, K., Elbe, D., and Friedlander, R. (2023). Evaluating and managing irritability and aggression in children and adolescents with autism spectrum disorder: an algorithm. B. C. Med. J. 65, 291–301.
Oshikoya, K. A., Neely, K. M., Carroll, R. J., Aka, I. T., Maxwell-Horn, A. C., Roden, D. M., et al. (2019). CYP2D6 genotype and adverse events to risperidone in children and adolescents. Pediatr. Res. 85, 602–606. doi:10.1038/s41390-019-0305-z
Pal, K., Swaminathan, K., Xu, H. E., and Pioszak, A. A. (2010). Structural basis for hormone recognition by the human CRFR2α G protein-coupled receptor. J. Biol. Chem. 285, 40351–40361. doi:10.1074/jbc.m110.186072
Pan, L. L., Yang, Y., Hui, M., Wang, S., Li, C. Y., Zhang, H., et al. (2020). Sulfation predominates the pharmacokinetics, metabolism, and excretion of forsythin in humans: major enzymes and transporters identified. Acta Pharmacol. Sin. 42, 311–322. doi:10.1038/s41401-020-0481-8
Pellicer, M., García-González, X., García, M. I., Robles, L., Grávalos, C., García-Alfonso, P., et al. (2017). Identification of new SNPs associated with severe toxicity to capecitabine. Pharmacol. Res. 120, 133–137. doi:10.1016/j.phrs.2017.03.021
Perna, A. F., Ingrosso, D., and De Santo, N. G. (2003a). Homocysteine and oxidative stress. Amino Acids 25, 409–417. doi:10.1007/s00726-003-0026-8
Perna, A. F., Ingrosso, D., Lombardi, C., Acanfora, F., Satta, E., Cesare, C. M., et al. (2003b). Possible mechanisms of homocysteine toxicity. Kidney Int. 63, S137–S140. doi:10.1046/j.1523-1755.63.s84.33.x
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi:10.1086/519795
Rafaniello, C., Sessa, M., Bernardi, F. F., Pozzi, M., Cheli, S., Cattaneo, D., et al. (2017). The predictive value of ABCB1, ABCG2, CYP3A4/5 and CYP2D6 polymorphisms for risperidone and aripiprazole plasma concentrations and the occurrence of adverse drug reactions. Pharmacogenomics J. 18, 422–430. doi:10.1038/tpj.2017.38
Rios, L. A. D. S., Cloete, B., and Mowla, S. (2020). Activation-induced cytidine deaminase: in sickness and in health. J. Cancer Res. Clin. Oncol. 146, 2721–2730. doi:10.1007/s00432-020-03348-x
Rodríguez-Antona, C., Niemi, M., Backman, J. T., Kajosaari, L. I., Neuvonen, P. J., Robledo, M., et al. (2007). Characterization of novel CYP2C8 haplotypes and their contribution to paclitaxel and repaglinide metabolism. Pharmacogenomics J. 8, 268–277. doi:10.1038/sj.tpj.6500482
Roke, Y., van Harten, P. N., Franke, B., Galesloot, T. E., Boot, A. M., and Buitelaar, J. K. (2013). The effect of the Taq1A variant in the dopamine D2 receptor gene and common CYP2D6 alleles on prolactin levels in risperidone-treated boys. Pharmacogenetics Genom 23, 487–493. doi:10.1097/fpc.0b013e3283647c33
Rothman, K. J. (1990). No adjustments are needed for multiple comparisons. Epidemiology 1, 43–46. doi:10.1097/00001648-199001000-00010
Rusch, J. A., Layden, B. T., and Dugas, L. R. (2023). Signalling cognition: the gut microbiota and hypothalamic-pituitary-adrenal axis. Front. Endocrinol. 14, 1130689. doi:10.3389/fendo.2023.1130689
Saiz-Rodríguez, M., Belmonte, C., Román, M., Ochoa, D., Jiang-Zheng, C., Koller, D., et al. (2018). Effect of ABCB1 C3435T polymorphism on pharmacokinetics of antipsychotics and antidepressants. Basic Clin. Pharmacol. Toxicol. 123, 474–485. doi:10.1111/bcpt.13031
Segatto, M., Tonini, C., Pfrieger, F. W., Trezza, V., and Pallottini, V. (2019). Loss of mevalonate/cholesterol homeostasis in the brain: a focus on autism spectrum disorder and rett syndrome. Int. J. Mol. Sci. 20, 3317. doi:10.3390/ijms20133317
Shahid, M., Walker, G. B., Zorn, S. H., and Wong, E. (2009). Asenapine: a novel psychopharmacologic agent with a unique human receptor signature. J. Psychopharmacol. 23, 65–73. doi:10.1177/0269881107082944
Sheng, Y., Yang, H., Wu, T., Zhu, L., Liu, L., and Liu, X. (2021). Alterations of cytochrome P450s and UDP-glucuronosyltransferases in brain under diseases and their clinical significances. Front. Pharmacol. 12, 650027. doi:10.3389/fphar.2021.650027
Sherwin, C. M. T., Saldaña, S. N., Bies, R. R., Aman, M. G., and Vinks, A. A. (2012). Population pharmacokinetic modeling of risperidone and 9-hydroxyrisperidone to estimate CYP2D6 subpopulations in children and adolescents. Ther. Drug Monit. 34, 535–544. doi:10.1097/ftd.0b013e318261c240
Shi, Y., Li, M., Song, C., Xu, Q., Huo, R., Shen, L., et al. (2017). Combined study of genetic and epigenetic biomarker risperidone treatment efficacy in Chinese Han schizophrenia patients. Transl. Psychiatry 7, e1170. doi:10.1038/tp.2017.143
Shilbayeh, S. A. R., Adeen, I. S., Alhazmi, A. S., Aldilaijan, K. E., and Aloyouni, S. Y. (2023a). Risperidone pharmacogenetics: the impact of star alleles' predicted phenotypes on global safety in autistic children. Int. J. Pharmacol. 19, 485–504. doi:10.3923/ijp.2023.485.50
Shilbayeh, S. A. R., Adeen, I. S., Alhazmi, A. S., Ibrahim, S. F., Al Enazi, F. A. R., Ghanem, E. H., et al. (2023b). The frequency of CYP2D6 and CYP3A4/5 genotypes and the impact of their allele translation and phenoconversion-predicted enzyme activity on risperidone pharmacokinetics in Saudi children with autism. Biochem. Genet., 1–26. doi:10.1007/s10528-023-10580-w
Singh, H., Lata, S., Choudhari, R., and Dhole, T. N. (2020). Prevalence of ABCC3-1767G/A polymorphism among patients with antiretroviral-associated hepatotoxicity. Mol. Genet. Genom. Med. 8, e1124. doi:10.1002/mgg3.1124
Sjostedt, E., Zhong, W., Fagerberg, L., Karlsson, M., Mitsios, N., Adori, C., et al. (2020). An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 367, eaay5947. doi:10.1126/science.aay5947
Soo, R. A., Wang, L. Z., Ng, S. S., Chong, P. Y., Yong, W. P., Lee, S. C., et al. (2009). Distribution of gemcitabine pathway genotypes in ethnic Asians and their association with outcome in non-small cell lung cancer patients. Lung Cancer 63, 121–127. doi:10.1016/j.lungcan.2008.04.010
Strautnieks, S. S., Bull, L. N., Knisely, A. S., Kocoshis, S. A., Dahl, N., Arnell, H., et al. (1998). A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat. Genet. 20, 233–238. doi:10.1038/3034
Sukasem, C., Medhasi, S., Pasomsub, E., Vanwong, N., Ngamsamut, N., Puangpetch, A., et al. (2016). Clinically relevant genetic variants of drug-metabolizing enzyme and transporter genes detected in Thai children and adolescents with autism spectrum disorder. Neuropsychiatr. Dis. Treat. 12, 843–851. doi:10.2147/ndt.s101580
Sundararajan, T., Manzardo, A. M., and Butler, M. G. (2018). Functional analysis of schizophrenia genes using GeneAnalytics program and integrated databases. Gene 641, 25–34. doi:10.1016/j.gene.2017.10.035
Talwar, P., Kanojia, N., Mahendru, S., Baghel, R., Grover, S., Arora, G., et al. (2016). Genetic contribution of CYP1A1 variant on treatment outcome in epilepsy patients: a functional and interethnic perspective. Pharmacogenomics J. 17, 242–251. doi:10.1038/tpj.2016.1
Taurines, R., Fekete, S., Preuss-Wiedenhoff, A., Warnke, A., Wewetzer, C., Plener, P., et al. (2022). Therapeutic drug monitoring in children and adolescents with schizophrenia and other psychotic disorders using risperidone. J. Neural Transm. 129, 689–701. doi:10.1007/s00702-022-02485-6
Tilleman, L., Weymaere, J., Heindryckx, B., Deforce, D., and Nieuwerburgh, F. V. (2019). Contemporary pharmacogenetic assays in view of the PharmGKB database. Pharmacogenomics 20, 261–272. doi:10.2217/pgs-2018-0167
Troost, P. W., Lahuis, B. E., Hermans, M. H., Buitelaar, J. K., van Engeland, H., Scahill, L., et al. (2007). Prolactin release in children treated with risperidone: impact and role of CYP2D6 metabolism. J. Clin. Psychopharmacol. 27, 52–57. doi:10.1097/jcp.0b013e31802e68d5
Uys, M. M., Shahid, M., and Harvey, B. H. (2017). Therapeutic potential of selectively targeting the α2C-adrenoceptor in cognition, depression, and schizophrenia—new developments and future perspective. Front. Psychiatry 8, 144. doi:10.3389/fpsyt.2017.00144
Vanwong, N., Ngamsamut, N., Hongkaew, Y., Nuntamool, N., Puangpetch, A., Chamnanphon, M., et al. (2016). Detection of CYP2D6 polymorphism using Luminex xTAG technology in autism spectrum disorder: CYP2D6 activity score and its association with risperidone levels. Drug Metab. Pharmacokinet. 31, 156–162. doi:10.1016/j.dmpk.2016.01.005
Vanwong, N., Ngamsamut, N., Medhasi, S., Puangpetch, A., Chamnanphon, M., Tan-Kam, T., et al. (2017). Impact of CYP2D6 polymorphism on steady-state plasma levels of risperidone and 9-hydroxyrisperidone in Thai children and adolescents with autism spectrum disorder. J. Child. Adolesc. Psychopharmacol. 27, 185–191. doi:10.1089/cap.2014.0171
Vermeir, M., Naessens, I., Remmerie, B., Mannens, G., Hendrickx, J., Sterkens, P., et al. (2008). Absorption, metabolism, and excretion of paliperidone, a new monoaminergic antagonist, in humans. Drug Metab. Dispos. 36, 769–779. doi:10.1124/dmd.107.018275
Wang, J., Jiang, F., Yang, Y., Zhang, Y., Liu, Z., Qin, X., et al. (2021). Off-label use of antipsychotic medications in psychiatric inpatients in China: a national real-world survey. BMC Psychiatry 21, 375. doi:10.1186/s12888-021-03374-0
Wang, L., Chen, Z., Chen, D., Kan, B., He, Y., and Cai, H. (2023a). Farnesyl diphosphate synthase promotes cell proliferation by regulating gene expression and alternative splicing profiles in HeLa cells. Oncol. Lett. 25, 145. doi:10.3892/ol.2023.13731
Wang, L., Pelleymounter, L., Weinshilboum, R., Johnson, J. A., Hebert, J. M., Altman, R. B., et al. (2010). Very important pharmacogene summary: thiopurine S-methyltransferase. Pharmacogenetics Genom 20, 401–405. doi:10.1097/fpc.0b013e3283352860
Wang, X., Sun, Z., Yang, T., Lin, F., Ye, S., Yan, J., et al. (2023b). Sodium butyrate facilitates CRHR2 expression to alleviate HPA axis hyperactivity in autism-like rats induced by prenatal lipopolysaccharides through histone deacetylase inhibition. mSystems 8, e0091523. doi:10.1128/msystems.00415-23
Weinshilboum, R. M. (2006). Pharmacogenomics: catechol O-methyltransferase to thiopurine S-methyltransferase. Cell. Mol. Neurobiol. 26, 537–559. doi:10.1007/s10571-006-9095-z
Whirl-Carrillo, M., Huddart, R., Gong, L., Sangkuhl, K., Thorn, C. F., Whaley, R., et al. (2021). An evidence-based framework for evaluating pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 110, 563–572. doi:10.1002/cpt.2350
Wilson, W., de Villena, F. P. M., Lyn-Cook, B. D., Chatterjee, P. K., Bell, T. A., Detwiler, D. A., et al. (2004). Characterization of a common deletion polymorphism of the UGT2B17 gene linked to UGT2B15. Genomics 84, 707–714. doi:10.1016/j.ygeno.2004.06.011
Wójcikowski, J., and Daniel, W. A. (2011). The role of the nervous system in the regulation of liver cytochrome p450. Curr. Drug Metab. 12, 124–138. doi:10.2174/138920011795016908
Woodson, L. C., and Weinshilboum, R. M. (1983). Human kidney thiopurine methyltransferase purification and biochemical properties. Biochem. Pharmacol. 32, 819–826. doi:10.1016/0006-2952(83)90582-8
Xing, Q., Gao, R., Li, H., Feng, G., Xu, M., Duan, S., et al. (2006). Polymorphisms of the ABCB1 gene are associated with the therapeutic response to risperidone in Chinese schizophrenia patients. Pharmacogenomics 7, 987–993. doi:10.2217/14622416.7.7.987
Xu, Y., Wang, Y., Xiao, H., and Li, Y. (2023). Hypoxia caused by unilateral nasal obstruction decreases mandibular density in rats through inhibition of Cyp1a1 expression. J. Oral Pathol. Med. 52, 786–794. doi:10.1111/jop.13468
Yasar, U., Lundgren, S., Eliasson, E., Bennet, A., Wiman, B., de Faire, U., et al. (2002). Linkage between the CYP2C8 and CYP2C9 genetic polymorphisms. Biochem. Biophys. Res. Commun. 299, 25–28. doi:10.1016/s0006-291x(02)02592-5
Yasui-Furukori, N., Mihara, K., Takahata, T., Suzuki, A., Nakagami, T., De Vries, R., et al. (2004). Effects of various factors on steady-state plasma concentrations of risperidone and 9-hydroxyrisperidone: lack of impact of MDR-1 genotypes. Int. Clin. Psychopharmacol. 19, 178–179. doi:10.1097/00004850-200405000-00022
Ye, W., Chen, R., Chen, X., Huang, B., Lin, R., Xie, X., et al. (2019). AhR regulates the expression of human cytochrome P450 1A1 (CYP1A1) by recruiting Sp1. FEBS J. 286, 4215–4231. doi:10.1111/febs.14956
Yoo, H. D., Lee, S. N., Kang, H. A., Cho, H. Y., Lee, I. K., and Lee, Y. B. (2011). Influence of ABCB1 genetic polymorphisms on the pharmacokinetics of risperidone in healthy subjects with CYP2D6* 10/* 10. Br. J. Pharmacol. 164, 433–443. doi:10.1111/j.1476-5381.2011.01385.x
Youngster, I., Zachor, D. A., Gabis, L. V., Bar-Chaim, A., Benveniste-Levkovitz, P., Britzi, M., et al. (2014). CYP2D6 genotyping in paediatric patients with autism treated with risperidone: a preliminary cohort study. Dev. Med. Child. Neurol. 56, 990–994. doi:10.1111/dmcn.12470
Zhang, H. Y., Basit, A., Busch, D., Bhatt, D. K., Drozdzik, M., Ostrowski, M., et al. (2018). Investigation of relative contribution of intestinal and hepatic UGT2B17 on testosterone first-pass metabolism. FASEB J. 32, 564–517. doi:10.1096/fasebj.2018.32.1_supplement.564.17
Zhang, Y. (2011). “Phase II enzymes,” in Encyclopedia of cancer. Editor M. Schwab (Berlin, Heidelberg: Springer), 2853–2855.
Zhou, X., Shin, S., He, C., Zhang, Q., Rasband, M. N., Ren, J., et al. (2021). Qki regulates myelinogenesis through Srebp2-dependent cholesterol biosynthesis. Elife 10, e60467. doi:10.7554/eLife.60467
Keywords: exploratory, pharmacogenetic testing, autism, risperidone pharmacokinetics, array genotyping
Citation: Shilbayeh SAR, Adeen IS, Ghanem EH, Aljurayb H, Aldilaijan KE, AlDosari F and Fadda A (2024) Exploratory focused pharmacogenetic testing reveals novel markers associated with risperidone pharmacokinetics in Saudi children with autism. Front. Pharmacol. 15:1356763. doi: 10.3389/fphar.2024.1356763
Received: 16 December 2023; Accepted: 24 January 2024;
Published: 05 February 2024.
Edited by:
Fatih M. Uckun, Ares Pharmaceuticals, LLC, United StatesCopyright © 2024 Shilbayeh, Adeen, Ghanem, Aljurayb, Aldilaijan, AlDosari and Fadda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sireen Abdul Rahim Shilbayeh, c3NhYmR1bHJhaGltQHBudS5lZHUuc2E=
 Sireen Abdul Rahim Shilbayeh
Sireen Abdul Rahim Shilbayeh Iman Sharaf Adeen2
Iman Sharaf Adeen2