- 1College of Medicine, University of Sharjah, Sharjah, United Arab Emirates
- 2Sharjah Institute for Medical Research, University of Sharjah, Sharjah, United Arab Emirates
The CYP2C19 gene is frequently included in different pharmacogenomic panels tested in clinical practice, due to its involvement in the metabolism of a myriad of frequently prescribed medications. Accordingly, CYP2C19 genotyping can promote precise therapeutic decisions and avoid the occurrence of significant drug-drug-gene interactions in the clinical setting. A comprehensive examination of the role of the CYP2C19 gene in real-world medical settings is presented in this review. This review summarizes the most recent information on how genetic variants in CYP2C19 affect drug metabolism and therapeutic outcomes. It goes into the wide range of CYP2C19 phenotypes, with different degrees of metabolizing activity, and their implications for customized medication response through a review of the literature. The review also analyzes the clinical significance of CYP2C19 in several medical specialties, including cardiology, psychiatry, and gastro-enterology clinics, and illuminates how it affects pharmacological efficacy, safety, and adverse effects. Finally, CYP2C19-supported clinical decision-making is outlined, highlighting the possibility of improving therapeutic outcomes and achieving more affordable treatment options, a step towards optimizing healthcare provision through precision medicine.
1 Introduction
With the advancements in pharmacogenetics, CYP2C19 has emerged as a gene for personalized drug prescriptions that serve many medical specialties, by considering the effect of genetic variants on the expected drug response (Naujokaitis et al., 2021). Therapeutic guidelines from the Clinical Pharmacogenetics Implementation Consortium (CPIC) are commonly used to recommend an appropriate treatment regimen based on the genotype test, specifically for patients in need of antiplatelet medication (Lee et al., 2022). Understanding the relevance of CYP2C19 is essential for realizing the full promise of personalized medicine to improve medication use and transform contemporary healthcare (Zanger and Schwab, 2013). The insight gained from the diverse genetic alterations in CYP2C19 and their consequences on enzyme action has ushered in a significant transformation in the approach to drug prescription and distribution. Healthcare professionals can maximize treatment efficacy while lowering the risk of adverse medication responses by implementing pharmacogenomic testing (Lee, 2013). This article will explore the practical applications of CYP2C19 genotyping, particularly in handling cardiovascular (such as Clopidogrel), psychiatric (such as sertraline, fluoxetine, citalopram, escitalopram, and others), and gastrointestinal (such as Pantoprazole, Lansoprazole and Omeprazole (Esomeprazole)) conditions and drugs. The review aims to assess the role and impact of CYP2C19 status in current clinical settings. Furthermore, the challenges and benefits of integrating CYP2C19 genotyping into healthcare strategies are discussed, considering elements such as cost-efficiency and prospective advances in personalized medicine. By meticulously analyzing both established literature and recent studies, this review seeks to contribute to the growing compendium of knowledge regarding the assimilation of CYP2C19 genotyping into routine clinical practice.
2 Methodology of searching
Q.S. and Am.A. conducted an extensive search of PubMed and MEDLINE databases, covering literature from 2010 until October 2023. They independently screened titles, abstracts, and full texts to assess the eligibility of articles. The search methodology adhered to the Preferred Reporting Items for Systematic Reviews and Review Articles. The number of results was 44,613. However, after excluding review articles, animal studies and case reports, as well as duplications of data, they reached to the number of references used in this review article. The search strategy included the following relevant terms: “pharmacogenomics AND CYP2C19”, “clinical practice AND CYP2C19”, “ethnic variation AND CYP2C19”, “cardiology AND CYP2C19”, “psychiatry AND CYP2C19, “gastroenterology AND CYP2C19”. The inclusion criteria were English language, clinical trials, observational studies, pharmacokinetics studies, and epidemiological studies.
3 CYP2C19 gene, and the role of CYP2C19 as a drug-metabolizing enzyme
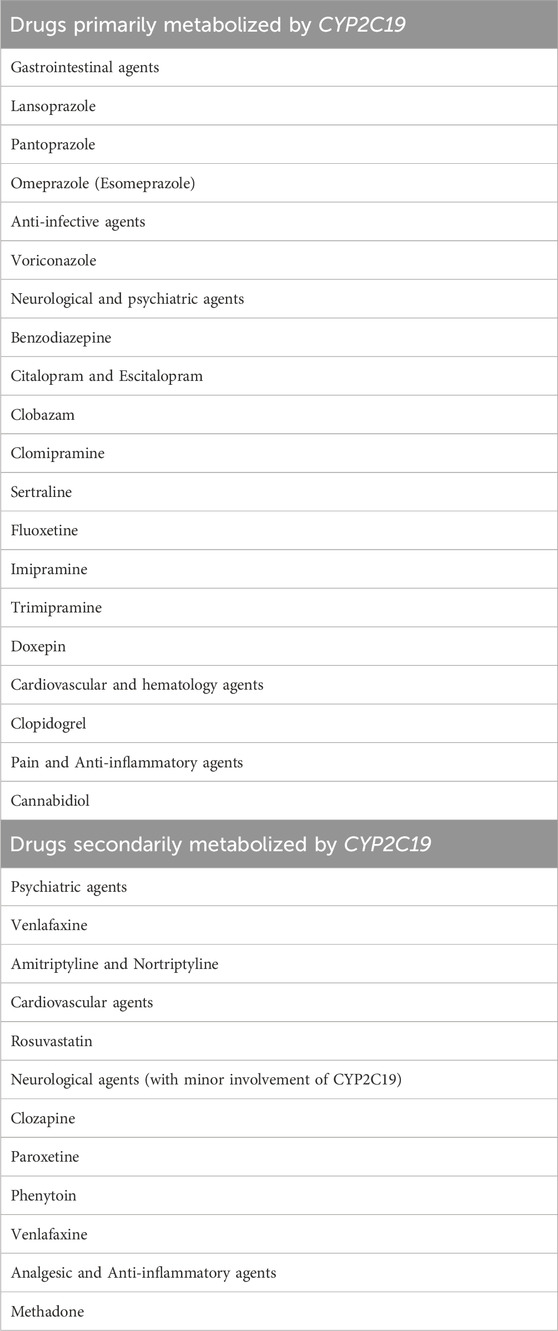
The CYP450 superfamily is a large and diverse group of enzymes whose main function is to metabolize many drugs. CYP2C19 is a member of the CYP2C subfamily of cytochromes which are involved in the metabolism of a range of clinically important compounds, such as anticoagulants, proton pump inhibitors (PPIs), benzodiazepines, anticonvulsants, and tricyclic antidepressants (Saeed and Mayet, 2013). The drugs that are primarily metabolized by the CYP2C19 enzyme are listed in Table 1. The CYP2C19 gene is located on chromosome 10q23.33 and to date, 39 alleles and 2000 SNPs have been identified (Shao et al., 2020). Among these variants, CYP2C19*2 and CYP2C19*3 are the most frequent and have received the greatest attention, as they identify poor metabolizers (Ionova et al., 2020) Understanding CYP2C19 as a metabolizing enzyme will enable healthcare professionals to make decisions regarding drug selection and dosing based on each individual’s genetic makeup to reach a safe and effective treatment (Pierre-François et al., 2022). Intriguingly, the CYP2C19 gene is highly polymorphic, leading to changes in enzymatic activity, therapeutic responses, and/or adverse drug reactions. Within the CPIC guidelines, the system used to translate genotype to phenotype depends on the star (*) allele nomenclature (Botton et al., 2021). In particular, an individual is categorized as a normal [previously described as extensive metabolizer (EM)], intermediate metabolizer (IM), poor metabolizer (PM), rapid metabolizer (RM), or ultra-rapid metabolizer (UM) based on CYP2C19 metabolizing activity which is determined by their genetic profile. The most prevalent phenotype is the normal metabolizer with CYP2C19*1 genotype, in which individuals would be predicted to have full CYP2C19 functioning enzyme, allowing them to metabolize medicines efficiently. Poor metabolizers (PM) with CYP2C19*2 or CYP2C19*3 genotypes have restricted or absent CYP2C19 enzymatic activity, resulting in a delay in drug metabolism and probable drug toxicity, which can lead to unpleasant adverse effects. Intermediate metabolizers (IM) carry one loss-of-function allele such as the genotype *1/*2, allowing for intermediate drug metabolism. Ultra-rapid metabolizers (UM) with the CYP2C19*17 genotype, on the other hand, have overactive variations of the CYP2C19 gene, resulting in faster medication metabolism and clearance. Patients with ultra-rapid metabolizer phenotype may require higher doses of drugs to reach the intended therapeutic response (Rollinson et al., 2020).
4 CYP2C19 testing
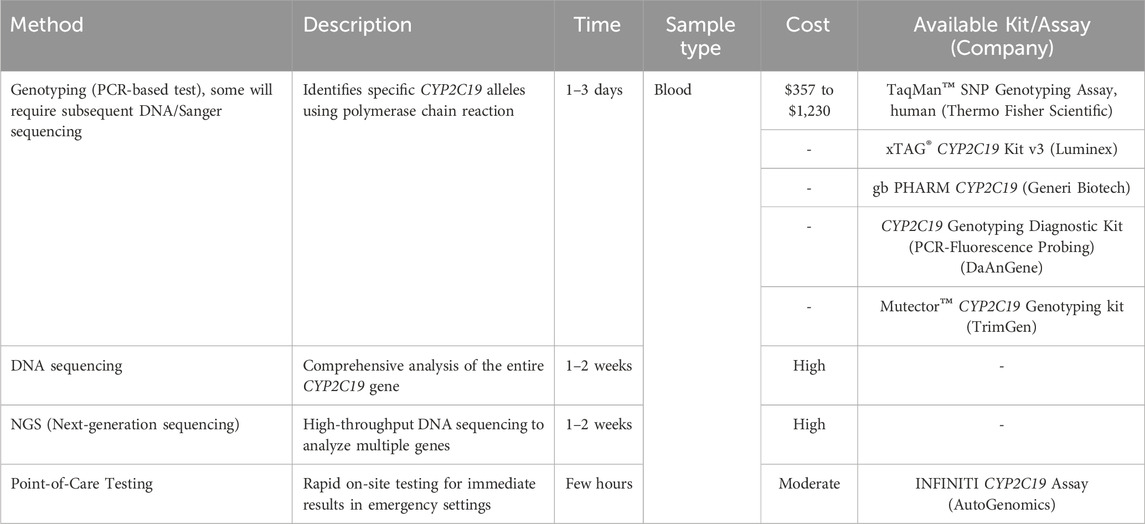
Pharmacogenomics (PGx) testing can involve a single gene or a panel of multiple genes. Evaluations of commercial PGx testing panels have revealed that gene composition as well as variant composition varies from test to test and frequently contains genes for which evidence is lacking to recommend prescription in various clinics, besides the fact that the CYP2C19 gene is so large that, whole gene cannot be routinely performed. As a result, the number of genes on a testing panel is insufficient as a criterion for test selection. Even though the same genes appear on a testing panel, the number of sequence variants, or alleles, tested within those genes might vary significantly between tests. To achieve a high level of analytical validity (i.e., the capacity of a test to identify whether a certain genetic variation is present or missing), PGx testing should preferably be done in laboratories that have been certified and accredited under national regulations. According to the Clinical Laboratory Improvement Amendments (CLIA) and the College of American Pathologists (CAP), a validation process must be carried out to evaluate the accuracy, precision, reference interval, sensitivity, and specificity of a test (Black et al., 2020) When a person undergoes CYP2C19 genetic testing, the analysis aims to identify and characterize specific genetic variations or alleles within the CYP2C19 gene. The CYP2C19 gene can have different forms, known as alleles, due to genetic variations among individuals. CYP2C19 testing provides specific alleles. Different methods and approaches for CYP2C19 testing are shown in Table 2.
5 CYP2C19 genotype-guided therapy (individualized prescription)
Genotype-guided treatment, also known as tailored prescription, is an advanced strategy in personalized medicine that optimizes drug selection and administration based on a patient’s exact genetic composition. By evaluating genetic variations of the CYP2C19 gene, healthcare practitioners can determine an individual’s drug metabolism phenotype and classify them as normal metabolizer (NM), poor metabolizer (PM), intermediate metabolizer (IM), rapid metabolizer (RM) or ultra-rapid metabolizer (UM) (El Rouby et al., 2018)Clinicians may adjust medicine prescriptions based on a patient’s specific metabolic profile, ensuring that patients receive the most effective treatment, safely. Genotype-guided treatment is very useful when prescribing drugs with proven CYP2C19 involvement, such as clopidogrel for cardiovascular illnesses or PPIs for digestive disorders. CYP2C19 genotype-guided drug prescribing has been proven in several trials to increase pharmacological effectiveness and safety. For example, CYP2C19 PMs have been observed to have a greater risk of cardiovascular events in patients receiving antiplatelet treatment with clopidogrel following stent installation. Switching PMs to alternative antiplatelet medications, such as ticagrelor or prasugrel, based on their genotype, has been associated with better clinical outcomes (Castrichini et al., 2023). A summary of studies on CYP2C19 Genotype-Guided therapy in cardiovascular practice is shown in Table 3. CYP2C19 genotype-guided medication has the potential to increase treatment efficacy, minimize adverse drug responses, and improve overall patient outcomes by incorporating genetic information into the treatment decision-making process, ushering in a new age of personalized medicine. Other aspects, such as drug interactions, co-existing illnesses, and lifestyle issues, must also be considered to achieve thorough and holistic patient treatment.
6 CYP2C19 in clinical practice
6.1 CYP2C19 in cardiology
i. Significant recommendations in clinical practice according to the gene variant:
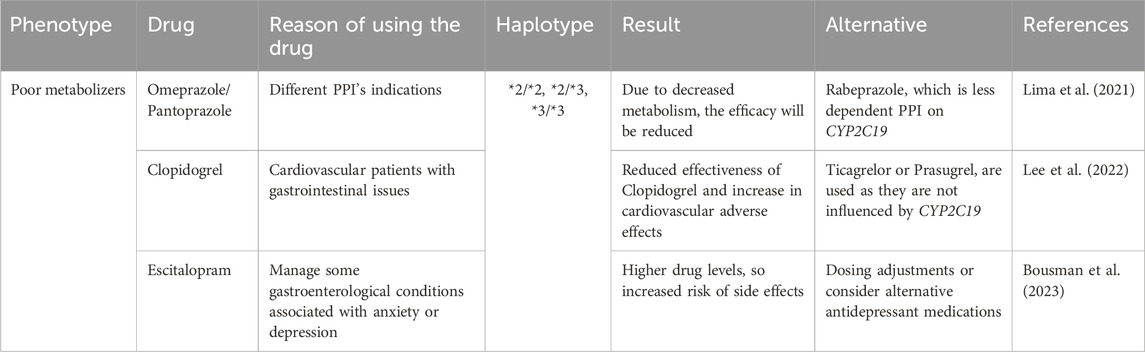
CYP2C19 gene variants play an important role in cardiology, primarily considering the metabolism of commonly used medications. These gene variants can significantly influence how patients metabolize drugs, especially, clopidogrel, a widely used antiplatelet drug (Pereira et al., 2019). Poor metabolizers may exhibit reduced conversion of clopidogrel to its active form, thus compromising its effectiveness in preventing cardiovascular events. In consideration of this, cardiology practice recommendations emphasize the importance of genotyping CYP2C19 variants to identify patients at higher risk and personalize their medications accordingly (Turner and Pirmohamed, 2014). On the other hand, concomitant use of clopidogrel and PPIs may reduce the activation and it will affect the efficacy of clopidogrel, hence the need for clinicians to consider PPIs with minimal CYP2C19 dependent (Kenngott et al., 2010). Table 4 summarizes CYP2C19 gene variation based on the following recommendations that are relevant in the cardiology clinic.
ii. Recommended panels:
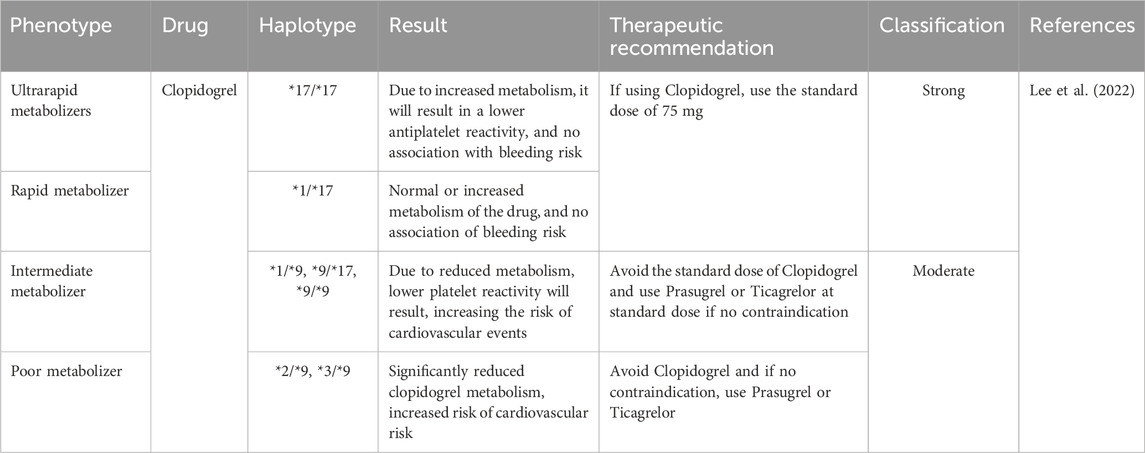
TABLE 4. Dosing recommendations for clopidogrel based on CYP2C19 phenotype. Data with strong evidence was retrieved from CPIC guidelines.
When considering whether to include CYP2C19 genotyping with other genes in the cardiology clinic, it is vital to concentrate on genes that can alter how drugs are metabolized, especially for medications that are often prescribed. Together with CYP2C19, several genes may be taken into consideration for panels, such as CYP2C9, another essential gene in drug metabolism that is particularly important for drugs like warfarin and non-vitamin K oral anticoagulants (NOACs). VKORC1 is another crucial gene, as it influences the dose needed to achieve the desired anticoagulation effect of warfarin. Combining CYP2C19, CYP2C9, and VKORC1 genotyping helps researchers to have a full picture of how patients metabolize these drugs (De Lara et al., 2022). The ADRB1 gene, which encodes for the beta-1 adrenergic receptor, is one of the genes that have an impact on the response of beta blockers which are commonly used in the treatment of heart failure and hypertension. Considering patients’ ADRB1 genotype will provide clinicians with guidance for the selection and dose of these medications (Howaidi and Lababidi, 2022). The ADRA2A gene which encodes for alpha-2A receptor, has a vital role in the regulation of the sympathetic nervous system. Variation in the ADRA2A gene can also influence the patient’s response to beta-blocker drugs. The precision of cardiovascular therapy could be improved by including these genes in a curated panel as it would enable a more individualized approach that considers a person’s genetic variables, lowering the chance of adverse reactions and ensuring that patients receive the best possible care.
iii. Adverse effects:
In a study on 168 patients with coronary heart disease who received clopidogrel/dual antiplatelet therapy (DAPT) after percutaneous coronary intervention (PCI), the incidence of cardiovascular adverse events was recorded by the high-on treatment-platelet reactivity (HPR) at 1-year follow-up visits. HPR was measured using thrombo-elastography which is a test used to assess the efficiency of blood coagulation. Moreover, PCR was done at the beginning of the study to determine CYP2C19 and ABCB1 3435−ΔΔCT gene polymorphisms. The study concluded that the non-functional CYP2C19*3 variant was associated with a higher incidence of HPR which was correlated with a higher incidence of cardiovascular adverse events. On the other hand, the non-functional allele CYP2C19*2 and ABCB1 3435−ΔΔCT were not significantly associated with HPR or cardiovascular events. This suggests that CYP2C19 and ABCB1 3435−ΔΔCT genotyping before initiation of clopidogrel therapy can be a significant predictive factor for treatment failure and the development of adverse effects (Mega et al., 2010). DAPT with aspirin and a P2Y12 inhibitor (clopidogrel, prasugrel, or ticagrelor) is known to be the standard care therapy for patients following PCI. The CYP2C19 enzyme is responsible for the activation of the prodrug “clopidogrel” into its active metabolite to carry out its antiplatelet activity. Patients who carry the non-functioning allele of CYP2C19 and receive clopidogrel as anti-platelet therapy are at a higher risk of treatment failure and development of major adverse cardiovascular and cerebrovascular events. Prasugrel and ticagrelor have not been linked with CYP2C19 activation like clopidogrel, which makes them better options for patients with non-functioning CYP2C19 alleles. Therefore, CYP2C19 genotype-guided anti-platelet therapy is believed to be beneficial for the prevention of major adverse cardiovascular and cerebrovascular events, which is further reinforced by multiple data emerging from cardiovascular and neurology clinical studies (Sanderson et al., 2005). Multi-factorial drug-gene interaction is the umbrella term used to describe the cumulative effects of both drug-drug interactions and drug-gene interactions. This phenomenon can be applied to patients who inherited the CYP2C19 loss-of-function allele and are receiving clopidogrel with concomitant PPI administration. In a systematic review and meta-analysis, five studies were included, comprising 8,802 patients of coronary heart disease or stroke. 3,767 were prescribed clopidogrel alone, 1,931 were concomitantly taking clopidogrel and PPIs, 2,146 were carrying CYP2C19 loss-of-function alleles and 958 were taking both clopidogrel and PPIs while also carrying CYP2C19 loss-of-function alleles. Patients with coronary heart disease or stroke who are receiving clopidogrel and concomitant proton pump inhibitor (PPI) therapy while inheriting loss-of-function alleles (CYP2C19*2 or CYP2C19*3) had a 63% higher risk of developing major cardiovascular adverse events (Biswas et al., 2021). In another systematic review and meta-analysis, 12.2% carried the CYP2C9*2 variant and 7.9% carried the CYP2C9*3 variant. Previous reports showed a 17% reduction in the original warfarin dose for CYP2C9*2 carriers and 37% for CYP2C9*3 carriers with a relative bleeding risk of 1.91 and 1.77, respectively. The study concluded that patients who carry CYP2C9*2 and CYP2C9*3 have a lower mean daily warfarin dose and a higher bleeding risk, which speculates that genotype-guided warfarin therapy could markedly alter the management of patients being started on warfarin (Barbarino et al., 2018). However, it showed that there is a marked difference in the enzymatic activity between CYP2C9*2 and CYP2C9*3 carriers. Poor metabolizer patients on Clopidogrel carrying CYP2C19 *3/*9 genotype may experience diminished antiplatelet effects, potentially increasing the risk of cardiovascular events, thus, they may use alternative drugs such as Prasugrel or Ticagrelor. The diplotype is more commonly seen in African American/Afro-Caribbean and Sub-Saharan African populations as the frequency is 0.01% according to CPIC, while in Latino population its frequency is 0.0001%. Such meta-analyses signify the critical role of CYP2C19 as a key pharmacogene in cardiology practice.
iv. Drug-drug interactions:
Co-occurrence of drug-drug interaction (DDI) with drug-gene interaction (DGI) might alter drug biotransformation pathways and produce drug-drug-gene-interaction (DDGI). PPIs are commonly used in the treatment of gastric disorders. Omeprazole is among the PPIs which poses the highest propensity to interact with other drugs compared to other PPIs like pantoprazole, rabeprazole, and lansoprazole. This is explained by its high affinity for CYP2C19 and moderate affinity for CYP3A4. Studies published since 2006 have shown clinically significant interaction between the antiplatelet medication, clopidogrel, and omeprazole which is mediated by CYP2C19 (Barbarino et al., 2018). Patients receiving DAPT often receive PPI therapy to reduce the risk of bleeding. Clopidogrel, the antiplatelet agent most commonly used in DAPT poses a challenge when concomitant PPI therapy is given due to their conflicting pharmacokinetic interaction via CYP2C19. Even though concomitant use of PPI and DAPT has been shown to decrease active metabolites of clopidogrel and ex vivo-measured platelet inhibition, there is still a conflict about whether this interaction has a significant effect on clinical outcomes (Saven et al., 2022).
6.2 CYP2C19 in psychiatry
i. Significant recommendations in clinical practice according to the gene variant:
The CYP2C19 enzyme plays a crucial role in the metabolism of many antidepressants, including selective serotonin reuptake inhibitors (SSRIs) such as sertraline, fluoxetine, citalopram, escitalopram, and others. In addition, multiple conventional tricyclic antidepressants such as imipramine, amitriptyline, trimipramine and clomipramine, are known CYP2C19 substrates (Alchakee et al., 2022). Genetic variations of CYP2C19 significantly impact the efficacy and safety of antidepressant medications, thus clinically influencing depression management. Table 5 summarizes dosing recommendations of antidepressants classified as level 1A evidence. Level 1A evidence indicates a specific gene-variant prescribing advice is provided in current clinical guidelines or FDA-approved drug label annotations (Hicks et al., 2015). Based on the CYP2C19 genotype, the CPIC published gene-based therapy recommendations for the SSRIs citalopram and escitalopram. For CYP2C19 ultrarapid and poor metabolizers, it is recommended to use an alternative antidepressant that is not primarily metabolized by CYP2C19 or to adjust the dose according to metabolizer status. Furthermore, people with a CYP2C19 *17/*17 genotype have significantly lower citalopram or escitalopram plasma concentrations at steady state when compared to normal metabolizers, thus it is recommended to titer citalopram to a higher target dose (compared to normal metabolizers) or to initiate an alternative SSRI, such as fluoxetine, fluvoxamine, and paroxetine, which are strongly metabolized by CYP2D6 only (Wong et al., 2023). The choice of an alternative antidepressant medication should be individualized based on the patient’s specific needs and medical history.
ii. Recommended panels:

TABLE 5. Dosing recommendations for antidepressants based on CYP2C19 phenotype. Data with strong evidence were retrieved from PharmGKB and CPIC guidelines.
The cytochrome P450 isoenzymes, mainly CYP2D6, CYP2C9, and CYP2C19 are responsible for the metabolism of the majority of psychotropic medications, including antipsychotics, antidepressants, and mood stabilizers. The highly polymorphic CYP2C19 enzyme plays a crucial role in the metabolism of many antidepressants, including SSRIs such as, sertraline, fluoxetine and (es)citalopram. Psychiatric gene sequencing panels vary depending on the specific focus of a clinic, patient population, psychiatric disorders, or medications of interest (Thiele et al., 2022). CYP2C19 demethylates several tricyclic antidepressants including, clomipramine, amitriptyline, trimipramine and imipramine to pharmacologically active metabolites. These compounds and their metabolites along with nortriptyline and desipramine, are hydroxylated by CYP2C19 enzyme to fewer active metabolites (Hicks et al., 2013). Therefore, combining CYP2D6 and CYP2C19 genomic variants in a single panel can provide a more comprehensive understanding of an individual’s drug metabolism profile that affects drug efficacy and safety (Matthaei et al., 2021). Interestingly, in psychiatry, ADRB1 polymorphisms, along with other genes, increase the risk of developing Alzheimer’s disease and sleep disturbances caused by altered cell responsiveness to adrenergic stimulation (Bullido et al., 2004). Genetic variations of the catechol-O-methyltransferase (COMT) gene, which encodes for an enzyme involved in the metabolism of dopamine and norepinephrine, have been associated with altered response to antipsychotic medications (Nikolac Perkovic et al., 2020). Notably, some studies in cardiology have explored a relationship between COMT variants and hypertension; however, data on this is scarce, and further investigations are required (Xu et al., 2017). In general, combining CYP2C19 with other actionable pharmacogenes in a genotyping test can provide valuable insights into personalized medicine.
iii. Adverse effects:
The discontinuation of antidepressant treatment is a common behavior in people with depression, mainly due to adverse drug reactions. Nearly 50% of undesirable drug reactions can be attributed to the differences in drug metabolism between individuals (Solomon et al., 2019; Kee et al., 2023). A clinical study was conducted to explore the association of CYP2C19 actionable variants translated into phenotypes with suicidal behavior in patients with depression who were using citalopram. The rate of suicide was 2-fold higher in individuals classified as CYP2C19 poor metabolizers compared to those classified as CYP2C19 normal metabolizers (Aldrich et al., 2019; Joas et al., 2023). The association of CYP2C19 metabolism status and side effects including hyperactivity, weight gain, gastrointestinal symptoms and insomnia was also investigated in pediatric patients prescribed escitalopram for anxiety or depressive disorders. The CYP2C19 poor metabolizers experienced more unwanted effects compared to faster metabolizers. In particular, CYP2C19 PMs had more rapid weight gain and hyperactivity (Ramsey, 2018). A recent Australian study consisting of 9,500 participants revealed that escitalopram is more tolerable by rapid CYP2C19 metabolizers while sertraline is more tolerable by poor CYP2C19 metabolizers, compared to normal metabolizers (Campos et al., 2022). On the other hand, a Swedish genetic study has revealed that the incidence of treatment-emergent mania was increased in patients with slower CYP2C19 metabolism status who were using amitriptyline or sertraline to treat bipolar depression with a hazard ratio (1.3, 1.46), respectively (Rahikainen et al., 2019). The Pre-emptive Pharmacogenomic Testing for Preventing Adverse Drug Reactions (PREPARE) study is the first large-scale and randomized clinical trial conducted in Europe to investigate the impact of applying pharmacogenomic test on the incidence of adverse drug reactions. The PREPARE study covered 39 different medications to treat multiple diseases. Notably, preemptively tested participants with actionable variants experienced a remarkable 30% reduction in the incidence rate of clinically relevant adverse drug reactions associated with drug-genotype interactions (Swen et al., 2023). Therefore, determining the patient’s CYP2C19 metabolizing status based on his genetic profile might enhance the safety of using antidepressant medications (Joas et al., 2023).
iv. Drug-drug interactions:
As previously mentioned, significantly altered rates of metabolism may occur due to DDGI. Escitalopram is mainly metabolized by the CYP2C19 and CYP3A4 enzymes and to a lesser extent by the CYP2D6 enzyme. Blood concentration of escitalopram is significantly influenced by the concomitant administration of CYP2C19, CYP3A4, and CYP2D6 modulator drugs (Rochat et al., 1997). The Combination of CYP3A5 and CYP2C19 genetic polymorphisms mediates several DDIs and DGIs. For instance, the co-presence of CYP3A4 EM and CYP2C19 IM/PM increases the risk of (es)citalopram toxicity and hence the urge for dose reduction or drug switching (Bahar et al., 2020). Unfortunately, the recent dosage recommendation for escitalopram is based on DGIs and DDIs separately and a knowledge gap remains regarding In a recent clinical case report, a patient complained of inadequate depression control despite several attempts with multiple antidepressants, including escitalopram, venlafaxine, and bupropion. The patient was phenotypically a CYP2C19 IM and a CYP2D6 PM (due to phenoconversion), and genetic variants in CYP2D6 and CYP2C19 increased their venlafaxine plasma concentration. In addition, the metabolism of other concomitant medications was impacted by the strong CYP2C19 inhibitor, bupropion, which contributed to the treatment failure. Cannabidiol (CBD) and PPIs are clinically known as CYP2C19 inhibitors and hence cause CYP2C19 phenoconversion (Von Moltke et al., 2001; Bousman et al., 2023). In particular, the concomitant use of CYP2C19 inhibitors and psychiatric medications may commonly lead to phenotype conversion from nonpoor metabolizer phenotype to poor metabolizer phenotype (Klieber et al., 2015). In a recently published case report, a patient with intermediate CYP2C19 phenotype who was on sertraline for 20 years developed cognitive dysfunction and hyponatremia due to an increase in sertraline plasma concertation after addition of CBD to their treatment regimen (Nanan et al., 2022). In another clinical case report, a patient complained of inadequate depression control despite several attempts with multiple antidepressants, including escitalopram, venlafaxine, and bupropion. The patient was phenotypically a CYP2C19 IM and a CYP2D6 PM, and genetic variants in CYP2D6 and CYP2C19 increased his venlafaxine plasma concentration. In addition, the metabolism of other concomitant medications was impacted by the strong CYP2C19 inhibitor, bupropion, which contributed to the treatment failure (Nanan et al., 2022).
6.3 CYP2C19 in gastroenterology
i. Significant recommendations in clinical practice according to the gene variant:
Based on the previously described CYP2C19 gene variation, recommendations relevant to Gastroenterology are summarized in Table 6. Individual patient variables, pharmacological interactions, and the exact clinical circumstance should all be considered when deciding on the best treatment approach. PPI dosing recommendations based on CYP2C19 phenotype reflect a prototypical integration of pharmacogenomics into gastroenterology as shown in Table 7. CYP2C19 is a key enzyme in PPI metabolism, CYP2C19 genetic variations have important implications for the therapeutic response and safety of PPI regimens. Substantial data compiled by impactful sources such as PharmGKB (Pharmacogenomics Knowledgebase) and the stringent clinical directions provided by the CPIC guidelines contribute to the cogency of these suggestions.
ii. Recommended panels:
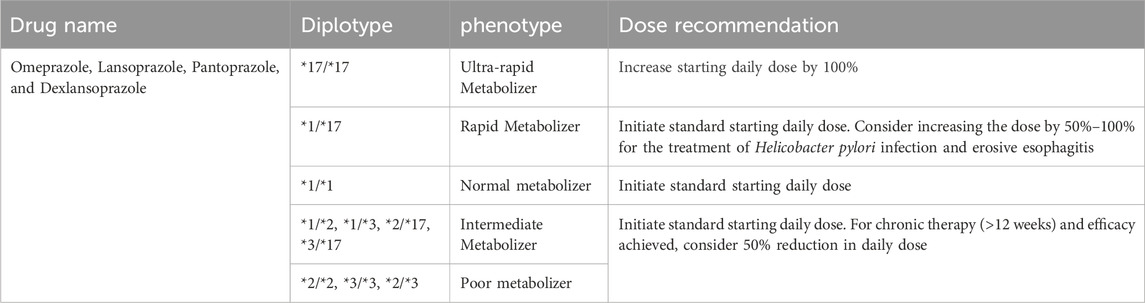
TABLE 7. Dosing recommendations for Proton Pump Inhibitors based on CYP2C19 phenotype. Data with strong evidence were retrieved from PharmGKB and CPIC guidelines.
Genetic testing panels at a gastrointestinal clinic can offer useful information about patients' genetic tendencies and probable drug reactions. Genetic testing can be used to personalize therapy regimens, forecast illness risk, and spot potential negative effects. Although several genetic testing panels could be taken into consideration, advice on the use of particular panels may vary depending on the clinic’s specialty and resources. The Inflammatory Bowel Disease (IBD) Panel, which includes NOD2, IL23R, and ATG16L1 genes along with CYP2C19, which are associated with IBD severity and susceptibility can aid in defining genetic risk and tailoring treatment for patients suffering from Crohn’s disease and ulcerative colitis (Slavin et al., 2019). The Liver Disease and Drug Metabolism Panel which includes CYP2C19, UGT1A1, HFE, and other genes, covers genetic variables impacting liver disorders and drug metabolism, which are relevant in gastrointestinal (Liu et al., 2022) and could be very relevant to clinical practice due to the liver’s crucial function in digestion and drug processing. A collaborative approach could provide an understanding of the intricate genetic framework that underpins gastrointestinal wellbeing, enabling healthcare professionals to define patient-focused alignment of diagnostic and therapeutic strategies.
iii. Adverse effects:
Adverse effects pose a significant concern in the Gastroenterology clinic, and understanding both genophenotypic factors and pharmacogenomics can provide insights into the origins of adverse drug reactions (ADRs). Genophenotypic factors encompass genetic and phenotypic variations that significantly affect an individual’s susceptibility to ADRs. Meanwhile, pharmacogenomics investigates how genetic variants affect medication metabolism, effectiveness, and safety (Chevalier et al., 2023). Adverse consequences of CYP2C19 genetic variants can emerge as impaired drug metabolism and reactions to drugs routinely used in gastrointestinal diseases. The most prevalent side effect related to CYP2C19 genetic variants is an altered response to PPIs in poor metabolizers carrying CYP2C19 loss-of-function variants which leads to reduced efficacy of some PPIs like omeprazole and lansoprazole), due to impaired conversion to their active forms. This would result in diminished acid suppression, which will potentially impact symptom relief of gastrointestinal disorders such as peptic ulcer disease, and gastroesophageal reflux disease (GERD) (Paré et al., 2010). For patients with gastrointestinal and cardiac conditions, CYP2C19 poor metabolizers may have a reduced ability to convert clopidogrel to its active form, thus resulting in decreased antiplatelet activity, and leading to increased adverse cardiovascular events (Chen et al., 2012). Patients with known CYP2C19 genetic variants should be actively examined by gastroenterologists for the risk of these unfavorable outcomes. To minimize these side effects and improve treatment results, genetic testing can disclose important information about a patient’s metabolic profile, which can then be used to guide medication selection and dosing modifications.
7 Pharmacoeconomic (health burden) and CYP2C19
Pharmacoeconomic studies assessing the effect of CYP2C19 genotype-guided treatment have garnered attention in recent years, the purpose of which is to analyze the economic consequences of using pharmacogenetic tests to guide pharmacological therapy selections based on CYP2C19 genetic variations. Pharmacoeconomic studies evaluate the economic effect of various treatment regimens by considering both direct hospital expenses and larger social costs associated with illness management (Sorich et al., 2013). In a meta-analysis of pharmacoeconomic research on CYP2C19 genotype-guided antiplatelet medication in patients with acute coronary syndrome, researchers discovered that genotyping individuals and tailoring antiplatelet medication based on CYP2C19 variations resulted in significant reductions in severe adverse cardiovascular events and total healthcare expenditures (Fu et al., 2019). Lee et al. assessed the cost-effectiveness of CYP2C19 genotype-guided antiplatelet treatment in patients undergoing PCI. The study found that genotype-guided medicine was a more cost-effective choice than the standard treatment, particularly in those at high risk for adverse cardiovascular events. The study highlighted the significance of incorporating pharmacogenetic testing into standard clinical practice to improve treatment results and resource use (Lee et al., 2011). Another research on the cost-effectiveness of CYP2C19 genotype-guided antiplatelet treatment in Korean patients having PCI for acute coronary syndrome found that adding genotyping into clinical decision-making was a cost-effective strategy that resulted in improved clinical outcomes and decreased healthcare costs when compared to standard therapy. The authors emphasized the potential for significant economic gains from genotype-guided treatment (Al-Rubaish et al., 2020). Overall, data suggests that CYP2C19 genotype-guided therapy has the potential to improve patient outcomes and reduce healthcare costs in a range of clinical contexts, notably antiplatelet treatment for cardiovascular cases. Healthcare professionals can personalize medication therapies to optimize therapeutic advantages while avoiding adverse drug responses and treatment inefficiencies by identifying patients with distinct CYP2C19 variations. Despite the positive data, broad implementation of pharmacogenetic testing in ordinary clinical practice remains a challenge, and further research is needed to overcome adoption obstacles and enable fair access to genotype-guided medication. As the field of pharmaco-economics gains more attention, more research and real-world data will be needed to drive policy and procedure that is grounded in evidence in personalized medicine. In another research study, the authors propose that PGx-guided clopidogrel therapy is an affordable choice for ACS patients receiving care in Spain (Koufaki et al., 2023).
8 Ethnic variation
In the framework of CYP2C19 genetics and its impact on drug metabolism, ethnic diversity is crucial to explore. Normal metabolizers (NMs) may be more prevalent in some cultures, whilst poor metabolizers (PMs) may be more prevalent in others. Within certain ethnic groups, these variances may affect pharmaceutical reactions and efficacy. It is essential to understand ethnic diversity in CYP2C19 genotypes to tailor pharmacological regimens and improve treatment results while taking into consideration genetic propensity and sensitivity to adverse drug responses (Nguyen et al., 2022). In a study conducted to evaluate the disparity between individuals with different racial backgrounds, when it comes to CYP2C19 genotype-guided P2Y12 antiplatelet therapy, patients from 9 sites that performed genotyping for CYP2C19 following percutaneous coronary intervention was recruited. A total of 3,342 participants were included, out of which 2,448 (73%) were European people and 659 (20%) were African people. The main aim was to compare the rate of prescribing P2Y12 inhibitors between European and African people races following CYP2C19 genotyping to guide antiplatelet therapy selection after PCI. Patients who carried the non-functioning CYP2C19 allele were prescribed alternative P2Y12 inhibitors (Prasugrel and Ticagrelor) instead of clopidogrel since clopidogrel’s effectiveness would be decreased. Choosing between clopidogrel and alternative therapy based on the genotype was the primary outcome. African people had a significantly higher prevalence of carrying the non-functioning allele compared to European people. There was no statistically significant association between race (European and African people with non-functioning alleles) and the prescription of alternative antiplatelet therapy at discharge following PCI and 12 months after the last follow-up visit. According to this study, there is an absence of racial disparity in genotype-guided antiplatelet prescribing among patients receiving CYP2C19 testing (Cavallari et al., 2023). The clinical outcomes of the coadministration of clopidogrel and omeprazole have not been adequately studied in the Asian population. It is believed that concomitant administration of omeprazole decreases the efficacy of clopidogrel due to its inhibition of the CYP450 CYP2C19 variant, which is responsible for the activation of clopidogrel. According to several studies, this interaction has not shown an increase in mortality or incidence of myocardial infarction in Caucasians. Data are scarce regarding this combination of drugs in the Asian population, which is believed to have a high prevalence of the non-functioning allele of CYP2C19. In a retrospective study that utilized the medical records and prescriptions of more than12,000 Asian patients receiving clopidogrel. The study findings revealed that coadministration of clopidogrel and omeprazole had a significant positive association with the incidence of MI, but the association with mortality, cerebrovascular accidents, and coronary interventions deemed to be statistically insignificant. Additionally, there was ethnic variability, with an increased incidence of MI in the Malay and Chinese populations compared to the Indian population (Muthiah et al., 2021). East-Asian populations commonly exhibit a higher prevalence of the CYP2C19*17 allele, a genetic variant associated with increased enzymatic activity, leading to ultra-rapid drug metabolism. However, this genetic trait has implications for the use of proton pump inhibitors (PPIs). The increased enzymatic activity associated with the CYP2C1917 allele may result in faster metabolism of PPIs, potentially leading to reduced drug efficacy (Zhang, 2021). In Oceanian populations, there is a notable increase in the allele frequency of CYP2C19*2 and CYP2C19*3 genetic variants, contributing to a higher prevalence of individuals classified as poor metabolizers of clopidogrel. However, the drug’s activation is heavily dependent on the enzymatic activity of CYP2C19. The *2 and *3 alleles are associated with reduced function of the CYP2C19 enzyme, leading to impaired conversion of clopidogrel into its active form (Helsby, 2016).
9 Conclusion
This review article emphasizes the importance of CYP2C19 in clinical practice across a range of various disciplines. CYP2C19 genotypes should be considered when prescribing drugs in Cardiology and Gastroenterology clinics, such as antiplatelet medicines and PPIs, respectively. The function of CYP2C19 in Psychiatry and its role in individualized medicine is also significant, especially with the long-term use of psychiatry medications. As we are in the era of precision medicine, the integration of CYP2C19 genotyping into clinical decision-making is a crucial first step toward tailoring medications to specific genetic profiles of our patients, ultimately increasing the bar for patient care.
Author contributions
QS: Data curation, Investigation, Visualization, Writing–original draft. AmA: Writing–original draft. KI: Writing–original draft. AAd: Writing–original draft. AS: Writing–original draft, Writing–review and editing. MS-A: Conceptualization, Funding acquisition, Project administration, Supervision, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. MS-A is funded by a collaborative grant provided by the University of Sharjah (Project No #2001090279).
Acknowledgments
The authors would like to acknowledge the support of the University of Sharjah.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alchakee, A., Ahmed, M., Eldohaji, L., Alhaj, H., and Saber-Ayad, M. (2022). Pharmacogenomics in psychiatry practice: the value and the challenges. Int. J. Mol. Sci. 23, 13485. doi:10.3390/ijms232113485
Aldrich, S. L., Poweleit, E. A., Prows, C. A., Martin, L. J., Strawn, J. R., and Ramsey, L. B. (2019). Influence of CYP2C19 metabolizer status on escitalopram/citalopram tolerability and response in youth with anxiety and depressive disorders. Front. Pharmacol. 10 (FEB), 99. doi:10.3389/fphar.2019.00099
Al-Rubaish, A. M., Al-Muhanna, F. A., Alshehri, A. M., Al-Mansori, M. A., Alali, R. A., Khalil, R. M., et al. (2020). Bedside testing of CYP2C19 gene for treatment of patients with PCI with antiplatelet therapy. BMC Cardiovasc Disord. 20 (1), 268. doi:10.1186/s12872-020-01558-2
Bahar, M. A., Lanting, P., Bos, J. H. J., Sijmons, R. H., Hak, E., and Wilffert, B. (2020). Impact of drug-gene-interaction, drug-drug-interaction, and drug-drug-gene-interaction on (Es)citalopram therapy: the pharmlines initiative. J. Pers. Med. 10 (4), 256. doi:10.3390/jpm10040256
Barbarino, J. M., Whirl-Carrillo, M., Altman, R. B., and Klein, T. E. (2018). PharmGKB: a worldwide resource for pharmacogenomic information. Wiley Interdiscip. Rev. Syst. Biol. Med. 10, e1417. doi:10.1002/wsbm.1417
Beitelshees, A. L., Thomas, C. D., Empey, P. E., Stouffer, G. A., Angiolillo, D. J., Franchi, F., et al. (2022). CYP2C19 genotype-guided antiplatelet therapy after percutaneous coronary intervention in diverse clinical settings. J. Am. Heart Assoc. 11 (4), e024159
Biswas, M., Rahaman, S., Biswas, T. K., and Ibrahim, B. (2021). Risk of major adverse cardiovascular events for concomitant use of clopidogrel and proton pump inhibitors in patients inheriting CYP2C19 loss-of-function alleles: meta-analysis. Int. J. Clin. Pharm. 43 (5), 1360–1369. doi:10.1007/s11096-021-01261-y
Black, R. M., Williams, A. K., Ratner, L., Crona, D. J., Wiltshire, T., Weck, K. E., et al. (2020). Projected impact of pharmacogenomic testing on medications beyond antiplatelet therapy in percutaneous coronary intervention patients. Pharmacogenomics 21 (7), 431–441. doi:10.2217/pgs-2019-0185
Botton, M. R., Whirl-Carrillo, M., Del Tredici, A. L., Sangkuhl, K., Cavallari, L. H., Agúndez, J. A. G., et al. (2021). PharmVar GeneFocus: CYP2C19. Clin. Pharmacol. Ther. 109, 352–366. doi:10.1002/cpt.1973
Bousman, C. A., Stevenson, J. M., Ramsey, L. B., Sangkuhl, K., Hicks, J. K., Strawn, J. R., et al. (2023). Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2D6, CYP2C19, CYP2B6, SLC6A4, and HTR2A genotypes and serotonin reuptake inhibitor antidepressants. Clin. Pharmacol. Ther. 114 (1), 51–68. doi:10.1002/cpt.2903
Bullido, M. J., Ramos, M. C., Ruiz-Gómez, A., Tutor, A. S., Sastre, I., Frank, A., et al. (2004). Polymorphism in genes involved in adrenergic signaling associated with Alzheimer’s. Neurobiol. Aging 25 (7), 853–859. doi:10.1016/j.neurobiolaging.2003.10.006
Campos, A. I., Byrne, E. M., Mitchell, B. L., Wray, N. R., Lind, P. A., Licinio, J., et al. (2022). Impact of CYP2C19 metaboliser status on SSRI response: a retrospective study of 9500 participants of the Australian Genetics of Depression Study. Pharmacogenomics J. 22 (2), 130–135. doi:10.1038/s41397-022-00267-7
Castrichini, M., Luzum, J. A., and Pereira, N. (2023). Pharmacogenetics of antiplatelet therapy. Annu. Rev. Pharmacol. Toxicol. 63, 211–229. doi:10.1146/annurev-pharmtox-051921-092701
Cavallari, L. H., Limdi, N. A., Beitelshees, A. L., Lee, J. C., Duarte, J. D., Franchi, F., et al. (2023). Evaluation of potential racial disparities in CYP2C19-guided P2Y12 inhibitor prescribing after percutaneous coronary intervention. Clin. Pharmacol. Ther. 113 (3), 615–623. doi:10.1002/cpt.2776
Chen, M., Wei, J. F., Xu, Y. N., Liu, X. J., and Huang, D. J. (2012). A meta-analysis of impact of proton pump inhibitors on antiplatelet effect of clopidogrel. Cardiovasc. Ther. 30, e227–e233. doi:10.1111/j.1755-5922.2011.00289.x
Chevalier, R., Attard, T., Van Driest, S. L., and Shakhnovich, V. (2023). A fresh look at proton pump inhibitor (PPI)-associated adverse events through a CYP2C19 pharmacogenetic lens. Expert Opin. Drug Metabolism Toxicol. 19, 53–56. doi:10.1080/17425255.2023.2190883
De Lara, D. V., De Melo, D. O., Araújo Silva, L. C., Gonçalves, T. S., and Santos, PCJL (2022). Pharmacogenetics of clopidogrel and warfarin in the treatment of cardiovascular diseases: an overview of reviews. Pharmacogenomics 23, 443–452. doi:10.2217/pgs-2021-0158
El Rouby, N., Lima, J. J., and Johnson, J. A. (2018). Proton pump inhibitors: from CYP2C19 pharmacogenetics to precision medicine. Expert Opin. Drug Metabolism Toxicol. 14, 447–460. doi:10.1080/17425255.2018.1461835
Fu, Y., Zhang, X. Y., Qin, S. B., Nie, X. Y., Shi, L. W., Shao, H., et al. (2019). Cost-effectiveness of CYP2C19 LOF-guided antiplatelet therapy in Chinese patients with acute coronary syndrome. Pharmacogenomics 21 (1), 33–42. doi:10.2217/pgs-2019-0050
Helsby, N. A. (2016). CYP2C19 and CYP2D6 genotypes in Pacificpeoples. Br. J. Clin. Pharmacol. 82, 1303–1307. doi:10.1111/bcp.13045
Hicks, J. K., Bishop, J. R., Sangkuhl, K., Muller, D. J., Ji, Y., Leckband, S. G., et al. (2015). Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 98 (2), 127–134. doi:10.1002/cpt.147
Hicks, J. K., Swen, J. J., Thorn, C. F., Sangkuhl, K., Kharasch, E. D., Ellingrod, V. L., et al. (2013). Clinical pharmacogenetics implementation consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin. Pharmacol. Ther. 93 (5), 402–408. doi:10.1038/clpt.2013.2
Howaidi, J., and Lababidi, H. (2022). Pharmacogenomics of adrenergic receptors from bench to bedside: potential clinical implications in critical care. Saudi Crit. Care J. 6 (1), 1. doi:10.4103/sccj.sccj_19_21
Ionova, Y., Ashenhurst, J., Zhan, J., Nhan, H., Kosinski, C., Tamraz, B., et al. (2020). CYP2C19 allele frequencies in over 2.2 million direct-to-consumer genetics research participants and the potential implication for prescriptions in a large health system. Clin. Transl. Sci. 13, 1298–1306. doi:10.1111/cts.12830
Joas, E., Jonsson, L., Viktorin, A., Smedler, E., Pålsson, E., Goodwin, G. M., et al. (2023). Effect of CYP2C19 polymorphisms on antidepressant prescription patterns and treatment emergent mania in bipolar disorder. Pharmacogenomics J. 23 (1), 28–35. doi:10.1038/s41397-022-00294-4
Kee, P. S., Maggo, S. D. S., Kennedy, M. A., and Chin, P. K. L. (2023). The pharmacogenetics of CYP2D6 and CYP2C19 in a case series of antidepressant responses. Front. Pharmacol. 14, 14. doi:10.3389/fphar.2023.1080117
Kenngott, S., Olze, R., Kollmer, M., Bottheim, H., Laner, A., Holinski-Feder, E., et al. (2010). Clopidogrel and proton pump inhibitor (PPI) interaction: separate intake and a non-omeprazole PPI the solution? Eur. J. Med. Res. 15 (5), 220–224. doi:10.1186/2047-783x-15-5-220
Klieber, M., Oberacher, H., Hofstaetter, S., Beer, B., Neururer, M., Amann, A., et al. (2015). CYP2C19 phenoconversion by routinely prescribed proton pump inhibitors omeprazole and esomeprazole: clinical implications for personalized medicine. Pharmacol. Exp. Ther. 354, 426–430. doi:10.1124/jpet.115.225680
Koufaki, M. I., Fragoulakis, V., Díaz-Villamarín, X., Karamperis, K., Vozikis, A., Swen, J. J., et al. (2023). Economic evaluation of pharmacogenomic-guided antiplatelet treatment in Spanish patients suffering from acute coronary syndrome participating in the U-PGx PREPARE study. Pharmacol. Ther. 17 (1).
Lee, C. R., Luzum, J. A., Gammal, R. S., Sabatine, M. S., Stein, C. M., Kisor, D. F., Limdi, N. A., et al. (2022). Clinical pharmacogenetics implementation consortium guideline for CYP2C19 genotype and clopidogrel therapy: 2022 update. Clin. Pharmacol. Ther. 112, 959–967. doi:10.1002/cpt.2526
Lee, J. B., Lee, K. A., and Lee, K. Y. (2011). Cytochrome P450 2C19 polymorphism is associated with reduced clopidogrel response in cerebrovascular disease. Yonsei Med. J. 52 (5), 734–738. doi:10.3349/ymj.2011.52.5.734
Lee, S. J. (2013). Clinical application of CYP2C19 pharmacogenetics toward more personalized medicine. Front. Genet. 3, 318. doi:10.3389/fgene.2012.00318
Liu, D., Yu, Q., Ning, Q., Liu, Z., and Song, J. (2022). The relationship between UGT1A1 gene and various diseases and prevention strategies. Drug Metab. Rev. 54, 1–21. doi:10.1080/03602532.2021.2001493
Lima, J. J., Thomas, C. D., Barbarino, J., Desta, Z., Van Driest, S. L., El Rouby, N., et al. (2021). Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2C19 and proton pump inhibitor dosing. Clin. Pharmacol. Ther. 109 (6), 1417–1423.
Matthaei, J., Brockmöller, J., Steimer, W., Pischa, K., Leucht, S., Kullmann, M., et al. (2021). Effects of genetic polymorphism in CYP2D6, CYP2C19, and the organic cation transporter OCT1 on amitriptyline pharmacokinetics in healthy volunteers and depressive disorder patients. Front. Pharmacol. 12, 12. doi:10.3389/fphar.2021.688950
Mega, J. L., Close, S. L., Wiviott, S. D., Shen, L., Walker, J. R., Simon, T., et al. (2010). Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: a pharmacogenetic analysis. Lancet 376 (9749), 1312–1319. doi:10.1016/S0140-6736(10)61273-1
Muthiah, M. D., Zheng, H., Chew, N. W. S., Xiao, J., Lim, L. G., Tan, H. C., et al. (2021). Outcomes of a multi-ethnic Asian population on combined treatment with clopidogrel and omeprazole in 12,440 patients. J. Thromb. Thrombolysis 52 (3), 925–933. doi:10.1007/s11239-021-02472-w
Nanan, J., Crosby, S., and Schuh, M. J. (2022). Hyponatremic cognitive dysfunction resulting from drug-drug-gene interaction between sertraline and cannabidiol in an intermediate CYP2C19 metabolizer patient. Innov. Pharm. 13 (3), 2. doi:10.24926/iip.v13i3.4890
Naujokaitis, D., Asmoniene, V., and Kadusevicius, E. (2021). Cytochrome P450 2C19 enzyme, Cytochrome P450 2C9 enzyme, and Cytochrome P450 2D6 enzyme allelic variants and its possible effect on drug metabolism: a retrospective study. Med. (United States) 100 (11), e24545. doi:10.1097/MD.0000000000024545
Nguyen, A. B., Cavallari, L. H., Rossi, J. S., Stouffer, G. A., and Lee, C. R. (2022). Evaluation of race and ethnicity disparities in outcome studies of CYP2C19 genotype-guided antiplatelet therapy. Front. Cardiovasc. Med. 9, 991646. doi:10.3389/fcvm.2022.991646
Nikolac Perkovic, M., Sagud, M., Zivkovic, M., Uzun, S., Nedic Erjavec, G., Kozumplik, O., et al. (2020). Catechol-O-methyltransferase rs4680 and rs4818 haplotype association with treatment response to olanzapine in patients with schizophrenia. Sci. Rep. 10 (1), 10049. doi:10.1038/s41598-020-67351-5
Paré, G., Mehta, S. R., Yusuf, S., Anand, S. S., Connolly, S. J., Hirsh, J., et al. (2010). Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N. Engl. J. Med. 363 (18), 1704–1714. doi:10.1056/NEJMoa1008410
Pereira, N. L., Rihal, C. S., So, D. Y. F., Rosenberg, Y., Lennon, R. J., Mathew, V., et al. (2019). Clopidogrel pharmacogenetics. Circ. Cardiovasc Interv. 12 (4), e007811. doi:10.1161/CIRCINTERVENTIONS.119.007811
Pereira, N. L., Farkouh, M. E., So, D., Lennon, R., Geller, N., Mathew, V., et al. (2021). Effect of genotype-guided oral P2Y12 inhibitor selection vs conventional clopidogrel therapy on ischemic outcomes after percutaneous coronary intervention: the TAILOR-PCI randomized clinical trial. Jama 324 (8), 761–771.
Pierre-François, M. J. D., Gagné, V., Brukner, I., and Krajinovic, M. (2022). Pharmacogenetic expression of CYP2C19 in a pediatric population. J. Personalized Med. 12, 1383. doi:10.3390/jpm12091383
Rahikainen, A. L., Vauhkonen, P., Pett, H., Ju, P., Haukka, J., Ojanperä, I., et al. (2019). Completed suicides of citalopram users—the role of CYP genotypes and adverse drug interactions. Int. J. Leg. Med. 133 (2), 353–363. doi:10.1007/s00414-018-1927-0
Ramsey, L. B. (2018). 21.3 Cyp2C19 influence on escitalopram efficacy and tolerability in youth with anxiety and depression. J. Am. Acad. Child. Adolesc. Psychiatry 57 (10), S301. doi:10.1016/j.jaac.2018.07.736
Rochat, B., Amey, M., Gillet, M., Meyer, U. A., and Baumann, P. (1997). Identification of three cytochrome P450 isozymes involved in N-demethylation of citalopram enantiomers in human liver microsomes. Pharmacogenetics 7 (1), 1–10. doi:10.1097/00008571-199702000-00001
Rollinson, V., Turner, R., and Pirmohamed, M. (2020). Pharmacogenomics for primary care: an overview. Genes 11, 1337. doi:10.3390/genes11111337
Saeed, L. H., and Mayet, A. Y. (2013). Genotype-phenotype analysis of CYP2C19 in healthy saudi individuals and its potential clinical implication in drug therapy. Int. J. Med. Sci. 10 (11), 1497–1502. doi:10.7150/ijms.6795
Sanderson, S., Emery, J., and Higgins, J. (2005). CYP2C9 gene variants, drug dose, and bleeding risk in warfarin-treated patients: a HuGEnetTM systematic review and meta-analysis. Genet. Med. 7, 97–104. doi:10.1097/01.gim.0000153664.65759.cf
Saven, H., Zhong, L., and McFarlane, I. M. (2022). Co-Prescription of dual-antiplatelet therapy and proton pump inhibitors: current guidelines. Cureus 14, e21885. doi:10.7759/cureus.21885
Shao, Z., Kyriakopoulou, L. G., Ito, S., et al. Chapter 14 - pharmacogenomics, Handbook of analytical separations. Vol. 7. 2020. 321–353 p.
Slavin, T. P., Weitzel, J. N., Neuhausen, S. L., Schrader, K. A., Oliveira, C., and Karam, R. (2019). Genetics of gastric cancer: what do we know about the genetic risks? Transl. Gastroenterology Hepatology 4, 55. doi:10.21037/tgh.2019.07.02
Solomon, H. V., Cates, K. W., and Li, K. J. (2019). Does obtaining CYP2D6 and CYP2C19 pharmacogenetic testing predict antidepressant response or adverse drug reactions? Psychiatry Res. 271, 604–613. doi:10.1016/j.psychres.2018.12.053
Sorich, M. J., Polasek, T. M., and Wiese, M. D. (2013). Challenges and limitations in the interpretation of systematic reviews: making sense of clopidogrel and CYP2C19 pharmacogenetics. Clin. Pharmacol. Ther. 94, 376–382. doi:10.1038/clpt.2013.100
Swen, J. J., van der Wouden, C. H., Manson, L. E., Abdullah-Koolmees, H., Blagec, K., Blagus, T., et al. (2023). A 12-gene pharmacogenetic panel to prevent adverse drug reactions: an open-label, multicentre, controlled, cluster-randomised crossover implementation study. Lancet, 401, 10374. doi:10.1016/S0140-6736(22)01841-4
Thiele, L. S., Ishtiak-Ahmed, K., Thirstrup, J. P., Agerbo, E., Lunenburg, CATC, Müller, D. J., et al. (2022). Clinical impact of functional CYP2C19 and CYP2D6 gene variants on treatment with antidepressants in young people with depression: a Danish cohort study. Pharmaceuticals 15 (7), 870. doi:10.3390/ph15070870
Turner, R. M., and Pirmohamed, M. (2014). Cardiovascular pharmacogenomics: expectations and practical benefits. Clin. Pharmacol. Ther. 95 (3), 281–293. doi:10.1038/clpt.2013.234
Von Moltke, L. L., Greenblatt, D. J., Giancarlo, G. M., Granda, B. W., Harmatz, J. S., and Shader, R. I. (2001). Escitalopram (S-citalopram) and its metabolites in vitro: cytochromes mediating biotransformation, inhibitory effects, and comparison to R-citalopram. Drug Metabolism Dispos. 29 (8), 1102–1109.
Williams, A. K., Klein, M. D., Martin, J., Weck, K. E., Rossi, J. S., Stouffer, G. A., et al. (2021). CYP2C19 genotype-guided antiplatelet therapy and 30-day outcomes after percutaneous coronary intervention. Circ. Genom. Precis. Med. 12 (2), e002441.
Wong, W. L. E., Fabbri, C., Laplace, B., Li, D., van Westrhenen, R., Lewis, C. M., et al. (2023). The effects of CYP2C19 genotype on proxies of SSRI antidepressant response in the UK biobank. Pharmaceuticals 16 (9), 1277. doi:10.3390/ph16091277
Xu, J., Boström, A. E., Saeed, M., Dubey, R. K., Waeber, G., Vollenweider, P., et al. (2017). A genetic variant in the catechol-O-methyl transferase (COMT) gene is related to age-dependent differences in the therapeutic effect of calcium-channel blockers. Med. (United States) 96 (30), e7029. doi:10.1097/MD.0000000000007029
Yao, H., Qin, K., Liu, Y., Yang, Y., Zhu, J., Chen, A., et al. (2022). CYP2C19 genotype and platelet aggregation test-guided dual antiplatelet therapy after off-pump coronary artery bypass grafting: A retrospective cohort study. Front. Cardiovasc. Med. 9, 1023004.
Zanger, U. M., and Schwab, M. (2013). Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 138, 103–141. doi:10.1016/j.pharmthera.2012.12.007
Zhang, G. (2021). “Frequencies of clinically important CYP2C19 and CYP2D6 alleles across east asian populations,” in Proceedings of the 2021 10th International Conference on Bioinformatics and Biomedical Science.
Zhang, Y., Shi, X. J., Peng, W. X., Han, J. L., Lin, B. D., Zhang, R., et al. (2021). Impact of implementing CYP2C19 genotype-guided antiplatelet therapy on P2Y12 inhibitor selection and clinical outcomes in acute coronary syndrome patients after percutaneous coronary intervention: a real-world study in China. Front. Pharmacol. 11, 582929.
Keywords: CYP2C19, pharmacogenetics, precision medicine, gentotype, pharmacoeconomic, CPIC, ethnic variation, gene polymorphism
Citation: Shubbar Q, Alchakee A, Issa KW, Adi AJ, Shorbagi AI and Saber-Ayad M (2024) From genes to drugs: CYP2C19 and pharmacogenetics in clinical practice. Front. Pharmacol. 15:1326776. doi: 10.3389/fphar.2024.1326776
Received: 23 October 2023; Accepted: 25 January 2024;
Published: 14 February 2024.
Edited by:
Francesca Coperchini, University of Pavia, ItalyReviewed by:
Otito Frances Iwuchukwu, Fairleigh Dickinson University, United StatesGeorge P. Patrinos, University of Patras, Greece
Copyright © 2024 Shubbar, Alchakee, Issa, Adi, Shorbagi and Saber-Ayad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maha Saber-Ayad, bXNhYmVyQHNoYXJqYWguYWMuYWU=
 Qamar Shubbar
Qamar Shubbar Aminah Alchakee
Aminah Alchakee Khaled Walid Issa1
Khaled Walid Issa1 Maha Saber-Ayad
Maha Saber-Ayad