
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol., 04 January 2024
Sec. Experimental Pharmacology and Drug Discovery
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1329244
This article is part of the Research TopicOld Drugs: Confronting Recent Advancements and ChallengesView all 11 articles
 Nayeralsadat Fatemi1
Nayeralsadat Fatemi1 Mina Karimpour2
Mina Karimpour2 Hoda Bahrami2
Hoda Bahrami2 Mohammad Reza Zali3
Mohammad Reza Zali3 Vahid Chaleshi1
Vahid Chaleshi1 Andrea Riccio4,5*
Andrea Riccio4,5* Ehsan Nazemalhosseini-Mojarad3
Ehsan Nazemalhosseini-Mojarad3 Mehdi Totonchi1,4,6*
Mehdi Totonchi1,4,6*Gastrointestinal (GI) cancers comprise a significant number of cancer cases worldwide and contribute to a high percentage of cancer-related deaths. To improve survival rates of GI cancer patients, it is important to find and implement more effective therapeutic strategies with better prognoses and fewer side effects. The development of new drugs can be a lengthy and expensive process, often involving clinical trials that may fail in the early stages. One strategy to address these challenges is drug repurposing (DR). Drug repurposing is a developmental strategy that involves using existing drugs approved for other diseases and leveraging their safety and pharmacological data to explore their potential use in treating different diseases. In this paper, we outline the existing therapeutic strategies and challenges associated with GI cancers and explore DR as a promising alternative approach. We have presented an extensive review of different DR methodologies, research efforts and examples of repurposed drugs within various GI cancer types, such as colorectal, pancreatic and liver cancers. Our aim is to provide a comprehensive overview of employing the DR approach in GI cancers to inform future research endeavors and clinical trials in this field.
Gastrointestinal (GI) cancers, a group of malignancies occurring within the digestive system including colorectal cancer (CRC), esophageal cancer (EC), gastric cancer (GC), liver cancer (LC), and pancreatic cancer (PC) comprised a substantial proportion of global cancer cases. They were responsible for approximately 27.7% of cancer cases (5.5 million cases out of 18.1 million worldwide) and 35.8% of global cancer-related deaths in 2020 (Sung et al., 2021). GI cancers have different global prevalence and mortality rates. CRC is the most frequently occurring type, ranking third in prevalence and the second in cancer-related deaths. GC, LC and EC rank as the fifth, sixth and eighth most commonly diagnosed cancers, with mortality rates ranking fourth, third and sixth, respectively. PC which has a poor prognosis with low curability rates compared to the other GI cancers, ranks 12th in frequency and the seventh in cancer death (Ferlay et al., 2021).
GI cancers exhibit diverse incidence patterns across regions, with PC and CRC being more prevalent in Europe and Northern America, and GC, LC, and EC in Asia. Interestingly, the countries with high human development indicators (HDIs) report a higher incidence of PC cases (Li et al., 2021a; Huang et al., 2021). In addition, the incidence of GI cancers is influenced by several factors such as lifestyle habits and dietary choices as well as age, with elderly males having a higher risk of incidence and mortality compared to females. This emphasizes the complex interplay of environmental and biological factors in the development of GI cancers (Sung et al., 2021).
The existing standard treatment options for GI cancers comprise surgery, chemotherapy, radiation therapy, targeted therapy, and immunotherapy (Nakayama et al., 2013; Anwanwan et al., 2020; Biller and Schrag, 2021; Joshi and Badgwell, 2021). Although combination regimens offer higher response rates and improved survival rates than single-agent therapy, it is crucial to consider the toxicity profile of these regimens closely (Chakraborty and Rahman, 2012; Miller et al., 2016; Jin and Mills, 2020). Reducing GI cancer mortality involves identifying and implementing more efficient therapy strategies that lead to superior prognosis and/or fewer side effects. Although the discovery and development of novel drugs are crucial for halting and reversing disease effects, they demand substantial funding, broad experimentation, and subsequent investigations into efficacy, pharmacokinetics, and toxicity. Moreover, only a small percentage of these drugs, approximately 5%, undergo clinical trials which may be approved for clinical use after successful results from the three phases of clinical trials (Gupta et al., 2013). Thus, developing new drugs is a costly and time-consuming procedure that requires extensive resources and expertise.
Drug repurposing (DR), also known as drug repositioning, is an alternative and promising strategy for cancer treatment. It involves exploring the potential therapeutic applications of already approved drugs or withdrawn/outdated agents in the clinic (Tables 1–4; Supplementary Table S2; Figure 1). DR offers a substantial asset in drug development, uses the prior knowledge of the pharmacodynamics, pharmacokinetics and toxicity of already approved drugs which have undergone extensive animal and human studies (Gupta et al., 2013; Bertolini et al., 2015). Accordingly, repurposed compounds can be authorized for use in cancer treatment and other therapeutic applications rapidly, relying on the established safety profiles and efficacy data. Moreover, the regulatory approval process for repurposed drugs is often faster and less expensive, making DR an attractive alternative strategy for drug development (Chong and Sullivan, 2007). In this review, we aim to provide a critical overview of the current therapies for GI cancers, as well as examples of repositioned components, and the most promising candidate for drugs repurposing in GI cancers.
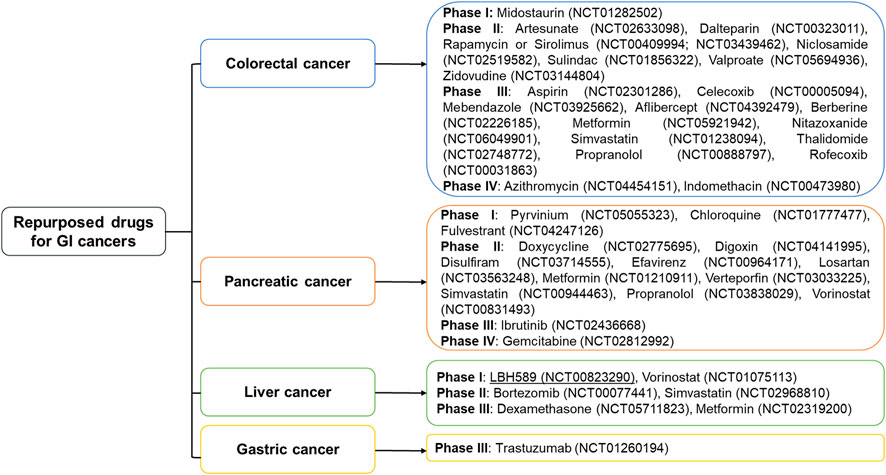
FIGURE 1. Repurposed drugs currently being evaluated against gastrointestinal cancers in various clinical phases. Metformin, simvastatin, and propranolol are entering phases 2 and 3 to treat more than 2 gastrointestinal cancers. Two drugs indomethacin and azithromycin for colorectal cancer, and gemcitabine for pancreatic cancer have reached phase IV.
The treatment approaches for GI cancer patients are diverse and contingent upon factors such as the patient’s performance status, medical comorbidities, cancer type, stage, potential side effects and overall health (Joshi and Badgwell, 2021; Catalano et al., 2022; Ducreux et al., 2023). Treatment strategies encompass adjuvant chemotherapy, adjuvant chemoradiotherapy, preoperative chemoradiotherapy, endoscopic/colonoscopy resection, surgical resection, and perioperative chemotherapy. A comprehensive therapy regimen involves a combination of suitable treatment options to effectively address GI cancers (Sugarbaker, 2005).
Targeted therapy serves as a viable treatment option within various therapeutic regimens for GI cancers. Notably, for GC, HER2-targeted therapy (trastuzumab) and anti-angiogenesis therapy (ramucirumab) are major targeted therapies employed (Joshi and Badgwell, 2021). Ramucirumab is also considered as a targeted therapy alternative for patients with EC who have not responded well to initial treatment approaches (Fuchs et al., 2014). Erlotinib, an epidermal growth factor receptor (EGFR) blocking agent, has obtained Food and Drug Administration (FDA) approval for advanced PC patients (Moore et al., 2007). In the treatment of CRC, the commonly employed anti-angiogenesis agent, bevacizumab, in combination with chemotherapy can prolong the survival rate of advanced CRC patients (Xiong et al., 2021). Unresectable or metastatic hepatocellular carcinoma (HCC) patients commonly undergo a combination of anti-angiogenesis targeted therapy and immunotherapy as the predominant treatment strategies for HCC, while systemic chemotherapy proves ineffective in such cases (Greten et al., 2008).
Immunotherapy, aimed at restoring immune system function, represents another line of therapy for patients with advanced GI cancer. An alternative treatment for metastatic CRC, HER2-positive EC, and advanced GC patients who exhibit Programmed Cell Death Ligand 1 (PD-L1) or microsatellite instability-high (MSI-H) biomarkers and are resistant to chemotherapy is pembrolizumab, an anti-PD-1 antibody, which functions by targeting the PD-1 receptor on tumor cells and preventing their evasion from the immune system. In addition, dostarlimab is another alternative option for the treatment of PCs with MSI-H or deficient mismatch repair (dMMR). Nivolumab either alone or in combination with ipilimumab is employed to treat adults with MSI-H or dMMR who have metastatic and drug-resistant CRC or EC (Zhang X. et al., 2022; Teixeira Farinha et al., 2022). It is worth noting that dostarlimab and nivolumab act upon the PD-1 receptor, whereas ipilimumab specifically targets CTLA-4. For additional information on this subject, refer to the Supplementary Material, Supplementary Table S1.
Despite the range of therapy options available for GI cancers, the high mortality rate highlights the limited efficiency of current treatments for these malignancies (Sung et al., 2021). Several challenges impede the improvement of existing therapeutic strategies, including chemotherapy resistance, tumor heterogeneity, late diagnosis, and limited efficiency of certain treatments, all contributing to treatment failure in GI cancer patients (Au et al., 2017; Parikh et al., 2019; Raziq et al., 2020). The development of drug resistance involves a complex multi-step process influenced by a variety of contributing factors. Numerous studies have shown that intensified DNA repair, apoptosis or autophagy disorders, epithelial-mesenchymal transition, inactivation of drug-metabolizing enzymes, and changes in expression or activity of membrane transporters are potential factors that promote chemotherapy resistance (Zheng, 2017). Furthermore, In the context of GI cancers, the unique characteristics of certain gastrointestinal tumors pose significant challenges to their effective treatment. For instance, the pancreas is anatomically situated in a hard-to-reach location, leading to difficulties in early diagnosis and the absence of effective screening techniques presents a significant challenge in detecting PC during its initial stages (McGuigan et al., 2018). Despite endeavors to implement personalized treatments, PC has not exhibited convincing outcomes comparable to those seen in other cancer types (Chantrill et al., 2015). The efficiency of radiotherapy in EC is limited due to the prevention of TAZ (Transcriptional Activator with PDZ-Binding Motif) ubiquitination and degradation (He et al., 2021). Furthermore, epigenetics and non-coding RNAs play a critical role in EC-related multidrug resistance and affect the effectiveness of therapies. In fact, EC’s susceptibility to recurrence, metastasis, and drug resistance development after first-line treatment, highlights the urgent requirement for optimizing the medicine regimen (Liu et al., 2021; Wei et al., 2021).
DR, is a strategic approach aimed at investigating alternative therapeutic applications of existing approved medications, beyond their originally intended uses. This method offers significant advantages in improving treatment outcomes, primarily by circumventing several essential stages of drug development. This results in reduced expenditure, shorter clinical trial durations, and mitigated risks associated with clinical trial failures due to adverse reactions (Ashburn and Thor, 2004; Chong and Sullivan, 2007; Zamami et al., 2017).
Repurposing FDA-approved medications is an efficient and inexpensive method to address oncology needs, including cancer treatment and reducing problems from existing anticancer treatments or radiation therapy (Ciociola et al., 2014). The drug-repurposing strategy has various advantages, including a shorter development period (usually 3–5 years), reduced costs (under $10 million), and higher success rates than original drug research (Morgan et al., 2011). Furthermore, DR can offer numerous benefits in overcoming therapeutic challenges by targeting various components both inside and outside of cancer cells (Würth et al., 2016). The concomitant use of multiple drugs that can target different tumor subtypes simultaneously can be a remarkable plan to overcome tumor heterogeneity (Li and Jones, 2012). Moreover, DR reveals the anticancer potential of non-oncology drugs with fewer side effects than traditional chemotherapy, making it a valuable option for cancer treatment (Crawford, 2014; Ferioli et al., 2018). Repositioned drugs can be used in combination with regular chemotherapy, and some demonstrate selectivity in causing cytotoxicity to cancer cells while sparing non-cancerous cells (Foglietta et al., 2021).
Drug repositioning involves a multi-step process. Initially, drug selection can start with in silico methods like molecular docking, pathway matching, and genome-wide association studies to create a ranked list of compounds. The next step is secondary analysis, which includes experimental techniques to refine and prioritize these compounds. Tertiary analysis aims to validate these compounds using cell cultures and animal models. Finally, the chosen drugs are advanced to clinical trials, and successful ones are repurposed for new uses (Shameer et al., 2015; Kulkarni et al., 2023). The key stages of DR are elucidated in (Figure 2).
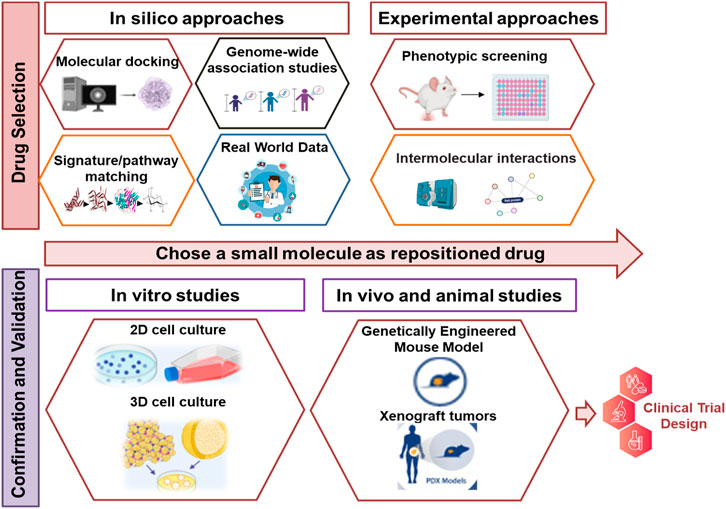
FIGURE 2. The key stages of drug repurposing. The process of DR begins with the use of computational or experimental techniques to select candidate drugs. Subsequently, these selected drugs are subjected to additional validation through potential evidence and additional experimental methods. Compounds that successfully pass through this rigorous evaluation then advance to clinical trials to obtain the FDA approval. Upon approval, these drugs are introduced to the market and their labeling is updated to reflect their new applications.
To evaluate the repurposing of an existing drug as a potentially effective anti-cancer agent, a mechanistic assessment of the drug’s action in preclinical models is crucial and requires systematic approaches (El-Hachem et al., 2017; Nagaraj et al., 2018). These approaches can be classified into two categories, computational and experimental, which use available data and biochemical experiments, respectively, to explore the potential of repurposing existing drugs as novel cancer treatments (Luo et al., 2017; Fernández-Torras et al., 2019). Indeed, successful DR is contingent upon the integrated and synergistic use of both approaches (Mottini et al., 2021; Palve et al., 2021).
Data-driven computational approaches involve the systematic analysis of data from different sources such as gene expression, chemical structure, genotype or proteomic data, or electronic health records (EHRs), which lead to the establishment of hypotheses on DR (Hurle et al., 2013). Carla Mottini et al. (2021) thoroughly reviewed computer-aided DR strategies for cancer therapy, offering examples of this approach in cancer studies within their article (Mottini et al., 2021). Future research should improve advanced computational tools, like machine learning and artificial intelligence, to improve therapeutic efficacy and safety prediction (Prasad and Kumar, 2021). Furthermore, computational methods can be used to address current anticancer medications or radiation therapy issues in addition to cancer treatment (Dalwadi et al., 2023). The most commonly used computational approaches are discussed below.
Molecular docking, a structure-based computational strategy, predicts complementarity of binding sites between the ligand (drug) and the receptor (target) (Hurle et al., 2013; Knapp, 2018; Mottini et al., 2021; Palve et al., 2021). If prior knowledge is available about a receptor target involved in cancer, multiple drugs could be investigated against that specific target, or drug libraries can be screened for a collection of target receptors to identify new interactions suitable for repurposing (Honarparvar et al., 2014; Nero et al., 2014). Dakshanamurthy and others performed molecular fit computations on a list of 3,671 FDA-approved drugs against 2,335 crystal structures of human proteins. They experimentally validated that mebendazole, an anti-parasitic agent, shows significant potential to bind to the vascular endothelial growth factor receptor 2 (VEGFR2) and effectively block angiogenesis (Dakshanamurthy et al., 2012). Furthermore, it has been predicted that levosimendan, a heart failure drug, could serve as a potential inhibitor of several kinases including RIO Kinase 1 (RIOK1), through ligand-binding site comparison and protein-ligand docking. Levosimendan shows anti-cancer activity against various cancers by directly inhibiting RIOK1 and RNA processing enzymes (Lim et al., 2019). Similarly, Arulanandam CD et al. (2021) suggested itraconazole as a better inhibitor of platelet-derived growth factor receptor alpha (PDGFRA) compared to other antifungal drugs for treating gastrointestinal stromal tumors (GISTs) (Arulanandam et al., 2021).
Genome-Wide Association Studies (GWAS) aim to identify variants associated with common genetic disorders, thereby providing new insights into the biology of the disease. The novel associations between genes and cancer through GWAS and Phenome-Wide Association Studies (PheWAS) enable the identification of new targets for existing drugs leading to the repositioning of drugs (Zhang et al., 2015; Sud et al., 2017; Kim et al., 2018). Grover et al. (2015) employed a bioinformatics strategy to match gene targets for coronary artery disease (CAD) with drug information collected from three different drug–target databases (DrugBank, Therapeutic Target Database, and PharmGKB) to identify potential therapeutics for repositioning toward treatment of CAD (Grover et al., 2015). Another valuable resource for medication indications, called Medication Indication Resource (MEDI), can validate the plausibility of inferring novel drug indications with clinical potential (Wei et al., 2013; Bejan et al., 2015). Furthermore, a genome-wide positioning systems network algorithm uncovered the antitumor activity of ouabain, an approved drug for cardiac arrhythmia and heart failure, in lung adenocarcinoma cells (Cheng et al., 2019).
GWAS results may pose challenges when applied to DR due to several reasons. Firstly, GWAS signals in gene-rich loci, where linkage disequilibrium commonly occur, can complicate the identification of the associated genes and their specific variants. Secondly, the direction of the gene variant’s effect may not be immediately apparent, necessitating functional studies to determine whether they act as activators or suppressors for disease control (Sanseau et al., 2012). Moreover, GWAS results may not provide detailed pathogenic information on genetic diseases (Wang and Zhang, 2013). It is important to acknowledge that understanding of the human genome is continually evolving, and new genes may continue to be discovered (Willyard, 2018). Therefore, careful consideration and further research are necessary when leveraging GWAS data for DR endeavors.
The signature matching method represents an innovative and promising approach within the realm of DR, as it involves comparing the distinct characteristics of a drug with those of other drugs, disease or clinical phenotype (Hieronymus et al., 2006; Keiser et al., 2009). The drug signatures can be derived from three different sources of data including omics data, chemical structures and adverse event profiles (Pushpakom et al., 2019). As a specific example, a study employed an optimal approach including two novel benchmarking standards, namely, area under the curve (AUC)-based standard and Kolmogorov-Smirnov (KS) statistic-based standard for signature-based DR and reported homoharringtonine (HHT) as a potential agent in the treatment and prevention of LC (Yang et al., 2022). Furthermore, various omics studies (transcriptomic, proteomic, or metabolomics) in the field of cancer not only provide large-scale data for supporting DR through applying advanced bioinformatics, but also expand our knowledge of hallmarks of cancer at the molecular level (Fernandez-Banet et al., 2016; Chen B. et al., 2020). Chemical structures serve as another type of signature matching used in DR; where the comparison of the chemical features of each drug with others helps identify possible chemical similarities that could signify shared biological activity (Pushpakom et al., 2019). A recent research employed this method along with other conventional DR methods to create a multilayer network algorithm. This algorithm was used to rank drugs that possibly can be repurposed for various types of cancer, including GI cancers and to explore new therapeutic possibilities for existing drugs (Cheng et al., 2021).
The unique adverse event profile of a drug can serve as a proxy for its related phenotypic effects, and drugs with similar side effects may act on the same target protein or pathway (Dudley et al., 2011; Cheng et al., 2021). In addition, if a drug’s phenotypic response resembles that of a disease, it may indicate that the drug and disease share pathways and physiological mechanisms (Pushpakom et al., 2019). Pathway and network-based approaches have been broadly used to identify drugs or drug targets with the potential for repurposing (Smith et al., 2012). Moreover, despite some of the targets identified by GWAS or other analytical networks appearing potent for drug targets, many of these genes may not be ideal in practical drug targeting applications. In such circumstances, a pathway-based approach can provide insights into genes upstream or downstream of the GWAS-associated target and could be considered as potential repurposing opportunities (Greene and Voight, 2016). In a recent study, the network-wide association study (NetWAS) technique has been employed, combining GWAS-identified genetic variant information with tissue-specific functional networks to identify disease-relevant genes more accurately than GWAS alone. Using NetWAS on the concept of hypertension and incorporating drug–target data from the DrugBank, Greene et al. (2015) observed an expansion in the number of the top target genes for the anti-hypertensive drugs compared to the GWAS (Greene et al., 2015). Additionally, pathway analysis of gene expression data obtained from a wide range of studies of human viral respiratory diseases identified 67 signaling pathways which may play important roles in respiratory viral infections (Smith et al., 2012). In summary, DR candidates can be identified through construction of drug or disease networks using gene expression patterns, protein interactions, disease pathology or GWAS data along with the signature matching studies to complement the network analysis approach (Iorio et al., 2010, 2013).
Real World Data (RWD) includes information from electronic health records (EHRs) of patients, characterized by large and complex datasets (Booth et al., 2019; Eichler et al., 2019). EHRs store patient and population health information in digital format, providing a wealth of data on patient outcomes (Miriovsky et al., 2012; Luhn et al., 2019). While diagnostic and pathophysiological data including experimental and drug prescribing data are structured, a significant portion of EHR data, such as clinical descriptions of patient symptoms and signs as well as imaging data remains unstructured (Khozin et al., 2017; Booth et al., 2019). Both structured and unstructured data from patients can serve as valuable sources to identify consistent signals for drug repurposing (Hurle et al., 2013; Low et al., 2017).
The retrospective clinical trial analysis is a commonly used computational approach based on RWD, with data commonly extracted from EHRs (Gray et al., 2019; Zheng et al., 2020). Sildenafil is an interesting example of retrospective clinical analysis which led to the repurposing of a candidate molecule (Ashburn and Thor, 2004). A classical example of repurposing a noncancer drug for cancer therapy through retrospective clinical analysis is metformin, which has shown to reduce cancer mortality in a dose-dependent manner (Sadeghi et al., 2012; Chaiteerakij et al., 2016). Furthermore, Xu et al. (2015) conducted a clinical cohort study using EHRs from Vanderbilt University Medical Center and Mayo Clinic, confirming the favorable effects of metformin in cancer survival including breast, colorectal, lung and prostate cancers in independent populations (Xu et al., 2015). Other successful examples of repurposing opportunities through retrospective clinical and/or pharmacological analyses include raloxifene in breast cancer, aspirin in CRC, propranolol in osteoporosis, and valproate in acute myeloid leukemia and glioblastoma (Cavalla and Singal, 2012; Happold et al., 2016; Lübbert et al., 2020).
Phenotypic screening, a direct method of DR, involves identifying compounds based on their effects in model systems without prior knowledge of candidate drug targets (Moffat et al., 2014, 2017; Kim et al., 2019). This approach typically uses a wide range of cell-based in vitro assays in a 96- or 384-well format (Sala et al., 2010). Iljin and others (2009) have performed high-throughput cell-based screening of a library including 4,910 drug-like small molecules against prostate cancer and non-malignant prostate epithelial cell lines using proliferation as the primary phenotypic criteria. They found an anticancer effect of disulfiram, a medication used to treat alcohol abuse, which was subsequently validated through genome-wide-gene expression studies (Iljin et al., 2009). Moreover, whole animal screening assays can be employed for DR which offer insights into efficient anticancer drugs, as well as pharmacokinetic and organ-toxicity potential results (Yadav et al., 2016; Baxendale et al., 2017). Cousin et al. (2014) evaluated 39 FDA-approved medications using a zebrafish model for tobacco dependence treatment and identified compounds like apomorphine and topiramate that modulate the behavioral effects of nicotine and ethanol in this model (Cousin et al., 2014). Similarly, over 26,000 small molecules were evaluated for their efficacy against leukemia using a genetically engineered T-cell reporting zebrafish model and discovered the remarkable activity of lenaldekar, against various hematologic neoplasms (Clements and Traver, 2012; Ridges et al., 2012).
Proteomic techniques including affinity chromatography and mass spectrometry reveal protein-protein interactions based on the intermolecular force. These techniques have been used to identify binding partners for various drugs, thus facilitating DR due to an experimentally based pharmacological analysis (Brehmer et al., 2005; Shantikumar et al., 2015). In this approach, drug treatments are administered to cells or animals, followed by a deep proteome analysis to identify protein changes (Savitski et al., 2018). An early successful example of this technique involves the validation of more than 20 cellular targets for the epidermal growth factor receptor kinase inhibitor gefitinib using mass spectrometry (Brehmer et al., 2005).
The Cellular Thermo Stability Assay (CETSA) has been introduced as a method based on altered protein thermal stabilization/destabilization in response to ligand binding which can predict drug targets when combined with thermal proteome profiling (Martinez Molina et al., 2013; Li J. et al., 2020; Mateus et al., 2020; Perrin et al., 2020). Additionally, chemical genetic approaches such as kinase drug discovery also rely on intermolecular forces and can provide insights into the relationship between binding and drug efficacy (Sloane et al., 2010; Cong et al., 2012; Wong et al., 2016). These findings can be interpreted quickly into new clinical applications or to improve drug resistance outcomes which are near-inevitable phenotypic responses to protein kinase inhibitors for the cancer treatment (Carter et al., 2005; Bago et al., 2016). Karaman et al. (2008) used a competition binding assay in vitro to evaluate 38 protein kinase inhibitors against a panel of 317 pathologically significant human protein kinases. Their analysis identified 3,175 binding interactions, revealing that some drugs such as sorafenib and dasatinib showed higher affinity to other kinase targets than their known target (Karaman et al., 2008). The development of chemical-genetic approaches, particularly non-kinase targets of small molecules planned to inhibit kinases is becoming increasingly recognized (Munoz, 2017) resulting in new repurposing opportunities for cancer treatment, such as the use of anthelmintic drug niclosamide to treat Zika virus infection (Xu et al., 2016) and the potential to treat drug-resistant pathogens (Sun et al., 2016).
In order to understand the mechanism of action of repurposed drugs for cancer treatment, it is often essential to evaluate and validate their effects within a comprehensive system that considers safety, dosage and toxicity before advancing to clinical trials. This evaluation can be accomplished using various models, which are divided into two main categories: in vivo and in vitro models. These models should be reproducible, cost-effective, and quickly constructed (Würth et al., 2016).
In in vitro tumor models, cancer cell lines are the predominant choice, followed by primary cells, although these models may also incorporate immune cells, stem cells, and stromal cells alongside cancer cells (Martinez-Pacheco and O’Driscoll, 2021). When using these cell types for detecting repositioning activity, in vitro assays offer several advantages including the ability to examine multiple substances with distinct mechanisms of action across a wide concentration-effect range, depending on the throughput of the experiment. Furthermore, these assays provide direct knowledge about potential new disease settings and allow testing of different drugs with novel mechanisms of action (Wilkinson and Pritchard, 2015).
In vitro screening approaches have been used for medication and DR in the various GI cancer types such as CRC, PC, GC and LC. A deep understanding of cancer progression and treatment has prompted the development of accurate in vitro tumor models that better represent the physiological features of the tumor microenvironment. Consequently, recent in vitro tumor models have become increasingly complex, extending beyond monitoring primary cell behaviors such as proliferation, invasion and cytotoxicity. These advanced in vitro models recapitulate important metastasis steps including angiogenesis, extracellular matrix remodeling and tumor cell metabolism reprogramming and dormancy (Wu and Swartz, 2014).
Cell culture plates and Transwell-based models are widely employed to assess viability, apoptosis, intra/extravasation and matrix remodeling of cancer cells. Two-dimensional (2D) models have been extensively used for drug screening and drug repurposing (Hulkower and Herber, 2011; Yu et al., 2020; Tomi-Andrino et al., 2022). In recent years, three-dimensional (3D) cell culture methods have become popular as they replicate in vivo microenvironmental providing data with greater predictive value for clinical outcomes. These authentic 3D cell culture models using human cells can overcome the limitations of mice models, which in addition to their high cost and ethical implications, are not always capable of accurately mimicking human illnesses or capturing medication side effects such as liver damage (Sivaraman et al., 2005). Moreover, by simulating the cell culture environment, 3D cell cultures can encourage specific cell activity, allowing drug discovery to target cell behavior with greater precision, such as enhancing cellular motility, promoting epithelial cell proliferation and differentiation, inducing cell dormancy, supporting of stem cell-like characteristics or mimicking desired microenvironment, like metastatic niches (Valastyan and Weinberg, 2011). According to their importance and applications in cancer research or DR studies, 3D cell culture models can be categorized into two major groups: Spheroids and Organoids.
Spheroids are cell aggressions that grow in suspension or are embedded in a three-dimensional matrix using three-dimensional culture methods. Cancer cell spheroids represent avascular tumor nodules, also known as micro-metastases and are widely used for drug screening despite being more expensive and time-consuming than 2D cell cultures. Spheroids also recapitulate interaction between cells and matrix in the tumor microenvironment and their size-dependent structure includes a necrotic central nucleus, resembling tumors with poor angiogenesis. Moreover, tumor spheroids offer valuable insights into how tumors respond to candidate drugs and combination therapies which reduces the need for animal testing and providing a more realistic representation of the tumor microenvironment (El Harane et al., 2023).
Organoids are novel ex vivo tumor models composed of self-organized three-dimensional multicellular tissue cultures derived from stem cells, primary tissue specimens or cancer cell lines. These models are capable of mimicking the in vivo organ (Simian and Bissell, 2017). Patient-derived cancer organoids provide a promising opportunity to predict drug efficiency and treatment response in GI cancers. Furthermore, incorporating 3D cell culture technology with primary patient-derived cancer cells, molecular characterization data, or the establishment of GI organoid banks representing molecular tumor subtypes could lead to preclinical assessment of personalized drug targets to improve treatment outcomes by reducing side effects in cancer therapy (Perrone and Zilbauer, 2021).
The first CRC organoid biobank was established in 2015 for drug screening, enabling the study of gene-drug interactions to recognize potential treatment response biomarkers and understand the molecular basis of drug response (van de Wetering et al., 2015). Notably, numerous studies have reported a significant overlap between esophageal adenocarcinoma organoids and tumor response to standard chemotherapy (Donohoe and Reynolds, 2017; Li et al., 2018) and a similar approach yielded comparable outcomes using GC organoids (Verduin et al., 2021). These results highlight the benefits and advantages of 3D ex vivo models in DR and repositioned drug efficiency assessments.
Animal models offer a powerful tool to simulate physiological conditions and complicated interactions, as well as the responses of reactions of different cell types and tissues to chemicals (Ruggeri et al., 2014). During the drug development process, in vivo testing plays a crucial role as it allows the study of interactions between the drug and both target and non-target cells.
Based on the premise of evolutionarily conserved pathogenetic mechanisms, animal models such as zebrafish, mice, fruit flies, worms and yeast have been used to give a comprehensive understanding of the biological mechanisms underlying the effect of drug administration. Xenograft models and genetically engineered mouse cancer models were among the in vivo models used in drug repurposing (Freires et al., 2017).
The human tumor xenograft is a widely used in vivo model where human cancer cells are transferred into immunocompromised (nude) mice through either ectopic or orthotopic implantation. Although the cell line-derived xenograft (CDX) model is considered as the gold-standard model for cancer research and investigation of anti-tumor therapies, patient-derived xenograft (PDX) tumors can also be used for this purpose (Tentler et al., 2012).
Xenograft tumors are highly used as animal models in GI cancer and DR research (Onaciu et al., 2020). In situ GI cancer can be induced by locally injecting of cell lines or implantation of tumor cells, while the metastatic models are established by injection of the tumor cells through the tail vein or into the specific organ (Morton and Houghton, 2007; Tentler et al., 2012). For decades, athymic nude mice and mouse xenograft models using human tumor cell lines have been used to study tumor progression factors. In the DR approaches, the use of xenograft tumors can be very helpful in assessing the accuracy of in vitro study results and the effectiveness of drugs on cancer cells in safe doses under in vivo situations. Doxycycline, a tetracycline-class antibiotic commonly used to treat bacterial and parasitic infections, has demonstrated to reduce tumor growth by ∼80% in pancreatic tumor xenografts (Son et al., 2009; Liu H. et al., 2020).
The Genetically Engineered Mouse Model (GEMM) is another animal model for studying human cancer and conducting preclinical study of repurposed drugs to target special gene-derived GI tumors. Genetic technologies have been recently applied by an increasing number of studies to introduce oncogenes into mouse embryonic or somatic cells through tissue-specific promoters targeting the GI tract and inducing GI cancers (Kersten et al., 2017). Transgenic, gene knock-in, and gene knock-out techniques are used to modify the genetic sequence of these mice in order to transfer, mutate, delete or overexpress one or more genes associated with transformation or malignancy. For instance, transgenic mice overexpressing KRAS mutant genes can mimic pancreatic tumorigenesis. Indeed, while general single-gene modified models may not fully represent the entire process of GI tumorigenesis, it has been discovered that physiological levels of KRAS G12D induce ductal lesions that serve as putative precursors to invasive PC (Westphalen and Olive, 2012). Additional genetic modifications, such as P53 mutation, can promote tumorigenesis and metastasis (Walrath et al., 2010). Another gene that can be engineered to produce GI cancer GEMM is carbonic anhydrase, present in the basolateral membranes of gastrointestinal epithelial cells and Its overexpression has been reported in many carcinomas including GC. Mice with null mutations of the Car9 gene develop gastric hyperplasia in glandular epithelium after 1 month (Gut et al., 2002). Additionally, ApcMin/Pten−/− mice are developed to study CRC (Stastna et al., 2019).
GEMM animals can be used to study the impact of a specific gene on tumorigenesis, or to investigate whether a de novo/repositioned drug acts through predicted pathways or genes to control the disease (Richmond and Su, 2008). Kobayashi et al. (2017) used mogp-TAg transgenic mice for DR to target the mevalonate pathway as a key tumorigenesis pathway in ovarian cancer (Kobayashi et al., 2017). Furthermore, the effect of metformin cancer initiation and progression suppression was studied using transgenic KPC mice (Chen K. et al., 2017). This animal model was also employed to evaluate the efficiency of repurposed histone deacetylase (HDAC) and mammalian target of rapamycin (mTOR) inhibitors on PC treatment (Biermann et al., 2022). Transgenic animals can be used to evaluate safe, first-in-human (FIH) doses during preclinical studies in drug development. For example, human-CYP3A4-expressing transgenic (Cyp3aXAV) mouse serve as practical model to evaluate the safe dosage and efficiency of CYP3A4-metabolized small-molecule drugs (Damoiseaux et al., 2022). The application of GEMM animal models can prove advantageous in specialized DR studies and allows researchers to focus on specific molecular pathways.
Chemical agents can be used to induce GI cancers in mice, potentially leading to mutations in relevant human cancer genes. The most well-established chemically induced GC model is produced by the administration of N-Methyl-N-nitrosourea (MNU) (Tomita et al., 2011; Ji et al., 2020; Rabben et al., 2021b). MNU is an N-nitroso compound mostly generated by anaerobic gut bacteria in the presence of nitrates and nitrites (Sobko et al., 2005; Zhuang et al., 2017). Furthermore, 1,2-dimethylhydrazine (DMH) and its metabolite, azoxymethane (AOM), are the most commonly used chemical compounds and carcinogens to induce CRC in mice. It has also shown that intraperitoneal injection of azaserine in rats induces metastatic pancreatic acinar cell carcinoma, although 10% of animals develop tumors in other organs (Rosenberg et al., 2009). Another method to produce a chemically induced PC model is through topical application of benzopyrene which induces adenocarcinoma (Kong et al., 2020). This group of animal models is widely employed in DR studies for GI cancers. Kannen et al. (2012) used C57BL/6 mice exposed to the carcinogen N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) as a colonic carcinogen mouse model to evaluate the antiproliferative effect of fluoxetine, an antidepressant medicine, on colon cancer (Kannen et al., 2012). In a similar study, the inhibitory effects of Liuwei Dihuang Pill (LDP) were investigated on MNU-induced gastric tumorigenesis in diabetic mice (Zhuang et al., 2017).
Researchers in the field of Gl cancer treatment are exploring innovative therapeutic strategies through drug repurposing, with an extensive focus on leveraging existing medications for the management of CRC. Among the studies of significance, Antona et al. (2022) have recently showed that spiperone, an approved drug for schizophrenia treatment, triggers apoptosis in CRC cells by activating phospholipase C, disrupting intracellular calcium balance, inducing irreversible endoplasmic reticulum stress, causing lipid metabolism changes, and damaging the Golgi apparatus (Antona et al., 2022). Furthermore, ten small molecules/drugs were identified via bioinformatic techniques for treating CRC, specifically targeting the upregulated Tissue Inhibitor of Matrix Metalloproteinases-1 (TIMP1) gene; these included established agents like formaldehyde and paclitaxel, as well as promising new drug candidates (Leng et al., 2022). Another study discovered that ebselen effectively inhibits Autophagy related protease 4B (ATG4B) through oxidative modification. This was based on the FDA-approved drug library, using Fluorescence Resonance Energy Transfer (FRET)-based high-throughput screening and gel-based analysis. The study showcased the potential of ebselen as an anti-CRC agent by influencing autophagy and tumor suppression (Xie et al., 2022). Mao et al. (2023) developed an efficient method for identifying repurposed drugs for CRC using organoid-based screening and computational drug prediction. Out of 335 tested drugs, 34 showed anti-CRC effects, with distinct transcriptome patterns including differentiation induction, growth inhibition, metabolism inhibition, immune response promotion, and cell cycle inhibition. Validation in patient-derived organoid-based xenograft (PDOX) systems demonstrated the anticancer effectiveness of drugs like fedratinib, trametinib, and bortezomib. (Mao et al., 2023). It has been revealed that the antifungal agent oxiconazole induces anti-tumor effects in CRC cells by inhibiting autophagy through downregulation of peroxiredoxin-2 (PRDX2), leading to growth suppression, and suggests its potential therapeutic use in combination with oxaliplatin for CRC treatment (Shi J. et al., 2022). Moreover, Dhakal et al. (2022) investigated the cytotoxic effects of the anti-anginal drug perhexiline and its enantiomers on CRC cells and demonstrated their ability to induce apoptosis and reduce cell viability in both monolayers and spheroids, as well as patient-derived organoids (Dhakal et al., 2022). Liñares-Blanco et al. (2020) proposed abemaciclib, an inhibitor of the CDK4/6 protein, as a promising option for the treatment of colon cancer (Liñares-Blanco et al., 2020). Furthermore, mebendazole, an antihelminthic medication used to treat gut worm infections, exhibited a cytotoxic effect on the RKO and HCT-116 colon cancer cell lines. Mebendazole was evaluated against a panel of kinases to determine the mechanism of its cytotoxic effect, which indicated significant inhibitory action against Abl and BRAF proteins. Additionally, in a case study of a patient with resistant metastatic colon cancer, twice-daily therapy with the normal antihelminthic dose of mebendazole led to a substantial reduction in metastasis (Nygren et al., 2013). A comprehensive investigation introduced several promising repurposing drugs (crizotinib, arsenic trioxide, vorinostat, dasatinib, estramustine, and tamibarotene) for CRC by prioritizing candidate genes obtained from the GWAS data (Zhang et al., 2015). Zhao P et al. (2022) highlighted the integration of metabolomics and transcriptomics as a powerful approach to gain insights into the antitumor mechanism of tadalafil, a phosphodiesterase type 5 (PDE5) inhibitor, in patients with CRC (Zhao et al., 2022). The anti-cancer effect of niclosamide was confirmed in nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice implanted with human CRC xenografts from patients with metastatic CRC and remarkable results were obtained with a non-lethal dose of 200 mg/kg (Osada et al., 2011). Tioconazole, originally used to treat vaginal yeast infections, was reported to enhance chemotherapy efficacy in colorectal tumor xenografts (Liu et al., 2018). Table 1 provides a comprehensive summary of various repurposed drugs in CRC.
In the field of PC treatment research, recent developments in DR have yielded promising strategies and novel candidates for therapeutic intervention. Pham et al. (2022) have recently introduced a deep learning framework that employs various genome-wide chemical-induced gene expression datasets to predict gene rankings in expression profiles induced by de novo chemicals, based on their chemical structures. They used this model for DR to identify potential treatments for PC from all existing drugs in DrugBank, and proposed candidates including dipyridamole, AZD-8055, linagliptin, and preladenant, which were subsequently validated in vitro (Pham et al., 2022). A recent study established a biobank of over 30 genetically distinct human pancreatic ductal adenocarcinoma (PDAC) organoid lines, demonstrating their correlation with the molecular and phenotypic heterogeneity observed in primary PDAC tissue and in vivo drug responses. Using a fully automated screening platform, this study conducted a DR analysis covering 1,172 FDA-approved drugs. Among the in vivo validated hits were several drugs currently approved for non-cancer indications, including emetine and ouabain. These drugs were found to specifically target PDAC organoids by disrupting their response to hypoxia. Notably, a dose of 0.56 mg/kg/d of ouabain significantly reduced PDAC xenograft growth in mice (Hirt et al., 2022). Mugiyanto et al. (2022) used genomic data from the cBio Cancer Genomics Portal to identify PC-associated and drug target genes. Through functional annotations, they prioritized 318 PC risk genes, of which 216 were druggable according to DrugBank. The Connectivity Map (CMap) Touchstone analysis revealed 13 potential PC drugs, including midostaurin and fulvestrant, which target PRKA and ESR1 respectively, as promising candidates for PC treatment (Mugiyanto et al., 2022). Another study investigated whether aspirin (ASA) and oseltamivir phosphate (OP) treatment could enhance PC cell sensitivity to gemcitabine-induced cytotoxicity and hinder chemoresistance development. The combination of ASA and OP with gemcitabine significantly disrupted PC cell viability, clonogenicity, ECM protein expression, migration, and induced apoptosis in MiaPaCa-2 and PANC-1 cells (Qorri et al., 2022). Several studies have recently identified Meflin, a glycosylphosphatidylinositol-anchored membrane molecule, as a functional maker of cancer-restraining CAFs (rCAFs) in PDAC (Kobayashi et al., 2019, 2021; Mizutani et al., 2019; Takahashi et al., 2021). Lida et al. conducted a screening of nuclear receptor ligands and identified Am580, a synthetic retinoid and RARα-selective agonist, as a compound that upregulates Meflin expression in both human pancreatic stellate cells (PSCs) and mouse mesenchymal stem cells (MSCs). Furthermore, Increasing Meflin enhances tumor sensitivity to chemotherapy in a PDAC xenograft model (Iida et al., 2022). The phase I and II clinical trials are conducting to investigate the efficacy of a combination of AM80 and gemcitabine and nab-paclitaxel in patients with advanced PC (Mizutani et al., 2022). Molecular docking technique confirmed that ZINC000001612996, ZINC000052955754, ZINC000003978005, and ZINC000006716957 could potentially act as small molecule drugs and co-ligands for TRPC3 and TRPC7, both of which are part of the Transient Receptor Potential Channels (TRPs)-related gene signature in PDAC (Shi W. et al., 2022). Chen et al. (2020) reported dose-dependent negative effects of albendazole, an anthelmintic drug, on proliferation, migration and viability of the human PC cell lines SW1990 and PANC-1. The cytotoxicity of albendazole was further confirmed in vivo using a nude mouse xenograft model, showing a significant reduction in tumor growth (Chen H. et al., 2020). Table 2 presents details on various repurposed drugs for PC.
Recently, several innovative transcriptomics-based DR methods were employed to uncover novel therapeutic candidates and combinations for the treatment of hepatocellular carcinoma (HCC or LC). Regan-Fendt K et al. (2020) showed that fostamatinib and dasatinib could be effective for sorafenib-resistant HCC (Regan-Fendt et al., 2020). Additionally, an mRNA expression profile-based DR method revealed that TOP2A has consistently unfavorable association with HCC patient survival and successfully repositioned withaferin-a (WFA) and mitoxantrone (MTX) through molecular docking studies as potential inhibitors of HepG2 cell proliferation for HCC treatment (Yuan et al., 2022). Tan et al. (2023) have screened a compound library containing 419 FDA-approved drugs and discovered that desloratadine, an antiallergic drug, inhibits proliferation in HCC cell lines as well as CDX, patient-derived organoid (PDO) and PDX. The study also identified N-myristoyl transferase 1 (NMT1) as its target, linking high NMT1 and VILIP3 expression to advanced HCC stages and poor survival (Tan et al., 2023). Furthermore, the combined effects of sorafenib, raloxifene, and loratadine on LC cells were assessed, finding that these two- or three-drug combinations significantly reduced metabolic activity, increased apoptosis, and decreased colony formation compared to single-drug treatments, suggesting the potential of the triple combination as a promising approach for LC treatment (Villarruel-Melquiades et al., 2023). We have summarized several repurposed drugs for HCC in Table 3.
Zhang et al. (2022) identified 6-Thioguanine (6-TG) as a potential therapeutic agent for GC by inducing ferroptosis through inactivation of system xc−, inhibition of glutathione (GSH) production, downregulation of GPX4, and elevation of lipid reactive oxygen species (ROS) levels in MGC-803 and AGS cell lines, with in vivo data supporting its anti-tumor activity (Zhang et al., 2022a). Furthermore, it has been shown that HC-056456, a CatSper channel blocker, as a novel ferroptosis-inducing compound inhibits GC cell growth by reducing GSH through the p53/SLC7A11 pathway, leading to increased Fe2+ and lipid peroxides in vitro and in vivo (Zhang et al., 2022b). In a comprehensive bioinformatic analysis of highly differentially expressed genes in GC, Hossain et al. (2023) found that CDH2, COL4A1, and COL5A are associated with the survival of GC patients. They also identified docetaxel, lanreotide, venetoclax, temsirolimus, and nilotinib as the top six candidate drugs, targeting aforementioned proteins, for the treatment of GC patients (Hossain et al., 2023). Nitazoxanide also yielded favorable outcomes, as it exhibited activity across GC cell lines tested (Ribeiro et al., 2023). In addition, Rabben et al. (2021a) computationally predicted the repositioning of ivermectin for the treatment of GC based on gene expression profiles of both human and mouse models of GC. They further validated their in silico prediction used human GC cell lines MKN74 and KATO-III in vitro. Transgenic insulin–gastrin (INS-GAS) mice were employed for experimental validation of ivermectin in GC treatment (Rabben et al., 2021a). Furthermore, in vivo and in vitro anti-tumor and growth suppression effects of ivermectin were demonstrated on GC, showing that ivermectin suppressed MKN1 cells growth through yes-associated protein 1 (YAP1) downregulation (Nambara et al., 2017). To identify drugs capable of inhibiting the DNA-binding activity of the helicobacter pylori transcription factor HP104, a combination of computational and in vitro methods led to the discovery of three promising drugs, including temoporfin, trientine, and tetraethylenepentamine, for potential antibacterial applications (Antoniciello et al., 2022). Table 4 provides a summary of available repurposed drugs in GC.
Surveillance for individuals at risk of GI cancers is essential for early diagnosis and prognosis improvement, and in choosing long-term chemoprotective drugs, approved molecules with well-known long-term effects are preferred. Aspirin (acetylsalicylic acid), introduced at the end of the 19th century, has been proposed for various diseases, such as cardiovascular diseases, strokes (Brighton et al., 2012; Dimitriadis et al., 2022; Gdovinova et al., 2022) and the chronic treatment of Fabry Disease (Monticelli et al., 2022). Specifically, aspirin has been shown to prevent CRC (Baron et al., 2003; Benamouzig et al., 2012; Guirguis-Blake et al., 2022) and PC (Streicher et al., 2014). Other non-steroid anti-inflammatory molecules like celecoxib (Arber et al., 2006; Bertagnolli et al., 2006) and sulindac (Long et al., 2020) have also been suggested for CRC.
Recent studies have highlighted the promising potential of repurposing non-oncology drugs for future cancer therapy. Examples include anticoagulant agents (warfarin (Rebelo et al., 2021) and dalteparin (Agnelli et al., 2022)), anti-fungi (itraconazole (Shen et al., 2021)), antidiabetic drugs (metformin (Cunha Júnior et al., 2021) and linagliptin (Li Y. et al., 2020)), antiparasitic (ivermectin (Nambara et al., 2017)), anthelminthic (parbendazole (Son et al., 2020)), antibiotics (nitroxoline (Mitrović and Kos, 2019), doxycycline (Ghasemi and Ghasemi, 2022), azithromycin (Qiao et al., 2018) and tigecycline (ElHefnawi et al., 2022)).
Brefeldin A, originally used as a macrolide antibiotic, has shown significant induction of autophagy in CRC cells both in vitro and in vivo (Bei et al., 2022). It functions by provoking endoplasmic reticulum stress (ER-stress) and upregulating Bip which decreases Akt phosphorylation through increased Bip/Akt interaction leading to autophagy induction in CRC cells (Zhou L. et al., 2019). In addition, antifungal drug ketoconazole has been reported to induce PINK1/Parkin-mediated mitophagy and accelerate apoptosis in HCC cells via COX-2 downregulation (Chen Y. et al., 2019).
Genistein, originally prescribed for reducing symptoms of menopause, osteoporosis, and obesity, has shown promising effects in cancer therapy. Genistein has been reported to inhibit proliferation, induce apoptosis and cell cycle arrest in G2 by inhibiting the Wnt/β-catenin signaling pathway in CRC cells (Oliveira et al., 2022). It also promotes apoptosis in HT29 CRC cells by modulating the caspase-3 and p38 MAPK signaling pathways (Shafiee et al., 2016). Furthermore, genistein inhibits glycolysis and induces mitochondrial apoptosis through downregulating of HIF-1α which leads to GLUT1 and HK2 inactivation and apoptosis induction in drug-resistant HCC cells (Li J. et al., 2017). Genistein modulates telomerase activity and reduces tumorigenesis by hTERT downregulation as well as modulation of Gli1 gene expression to weaken cancer stem-like properties in GC cells (Jian-Hui et al., 2016). Genistein, which shows promise as an anticancer drug candidate, is currently in phase II of clinical trials (Cao et al., 2022; Chu et al., 2023).
Metformin is another successful repositioned drug for GI cancers which is currently in phase II of the clinical trial. Metformin reduces cell survival and tumorigenesis by lowering serum insulin levels and downregulation of IGF-1 (Sarfstein et al., 2013). Additionally, it induces G1-arrest via AMPK activation and cyclin D1 downregulation (Wang Y. et al., 2018), inhibits proliferation through mTOR signaling pathway regulation regardless of AMPK dependency (Demaré et al., 2021). It has been shown that metformin inhibits the progression of GC through the inhabitation of HIF1α/PKM2 signaling (Chen G. et al., 2015). Furthermore, it inactivates RAS/ERK and AKT/mTOR signaling pathways and reduces proliferation in KRAS-derived tumors. Metformin has been reported to selectively inhibit KRAS-driven metastatic CRC by silencing MATE1 (Xie et al., 2020).
Several Studies have demonstrated that the combination of repurposed drugs with cytotoxic drugs, radiotherapy or even the combination of multiple repositioned drugs can exhibit synergistic antitumor effects. A recent meta-analysis by Heer et al. (2022) revealed that aspirin when co-administered with sulindac and difluoromethylornithine (DFMO), an inhibitor of ornithine decarboxylase used to treat facial hirsutism, showed significantly more effective results in protecting against CRC adenomas (Heer et al., 2022). The combination of bortezomib and chloroquine has been shown to suppress proliferation and induce apoptosis in human liver tumors, whether orthotopically or subcutaneously xenografted in mice (Hui et al., 2012). In clinical research, combining nelfinavir with a short course of hypofractionated radiotherapy (SCHRT) showed increased sensitivity of CRC tumors to radiotherapy (Meyn et al., 2016). In addition, FOLFIRINOX, a chemotherapy regimen comprising leucovorin calcium (folinic acid), fluorouracil, irinotecan hydrochloride and oxaliplatin, is used for the treatment of advanced PC. In a phase II clinical trial, FOLFIRINOX is combined with losartan as neoadjuvant therapy, followed by chemoradiotherapy, for locally advanced PC (Murphy et al., 2019). Another successful example of DR is a combination of metformin, digoxin and somatostatin, which has shown significant suppression of PC cell proliferation in clinically relevant animal models. Currently, this combination is being evaluated in a clinical trial (Liu S.-H. et al., 2020). Tables 1–4 provide comprehensive information on various drugs that have been successfully repurposed to treat GI cancers and approved for clinical use by the FDA. Furthermore, we have prepared a Supplementary Table S2, which presents a list of repurposed unapproved or withdrawn drugs/natural components targeting GI-related cancers.
Drug repurposing for cancer therapy is widely used to discover new indications for existing compounds; However, only a few repurposed drugs have been formally subjected to the clinical treatment guidelines. Despite advantages such as anticancer pharmacokinetic parameters, acceptable safety and tolerability in humans, there is still a risk of late-phase clinical trial failure due to competition with new drug development. The legal and regulatory barriers such as patents issues and prescription charges must also be addressed (Vickers, 2017; Breckenridge and Jacob, 2019; Pushpakom et al., 2019).
The financial factors significantly influence the clinical development and approval of repurposing drugs, as private sector organizations prioritize higher returns due to intellectual property rights (Vickers, 2017). Most registered clinical trials listed in the Repurposing Drugs in Oncology (ReDo) project are sponsored by Universities or Hospitals (67%), research institutes or non-profit organizations (28%), and only a small percentage by pharmaceutical companies (Pantziarka et al., 2018). Patent considerations for off-patent drugs can pose significant barriers to DR, requiring credible and statistically strong evidence for a new indication and legislative efforts to address this issue (Pushpakom et al., 2019).
Physicians prescribe drugs based on scientific evidence from clinical trials, and both generic and repurposed drugs should be used when suitable. Nevertheless, the pharmaceutical industry can exert substantial investments on drug promotion, physician marketing and consumer advertising. A clear example is thalidomide, originally used as a sedative or antiemetic, which has been repurposed for multiple myeloma. Despite phase III clinical trials showing no survival advantage for the combination of melphalan-prednisone-lenalidomide over melphalan-prednisone-thalidomide (Stewart et al., 2015; Zweegman et al., 2016) lenalidomide became the standard treatment approach, even with a higher estimated cost than thalidomide. Thus, underfunding for clinical trials in oncology and prescribing rejection bias remain significant challenges for cancer drug repurposing.
Another challenge lies in the genetic diversity of individuals and the complex nature of disease (Pritchard et al., 2017). While there are many common pathways involved in cancer development, differences in pathways and genes exist among subgroups and individuals. This genetic diversity results in varying side effects and treatment responses to routine drugs and therapies.
Moreover, current GI cancer therapies may be ineffective for specific patients or cancer types due to inadequate drug targeting or inefficient drug interactions (Apicella et al., 2017). In rectal cancer, personalized DR based on gene expression signatures and reverse drug-induced gene expression profiles has shown promising results. For instance, Carvalho et al. (2021) identified potential topoisomerase II inhibitors like doxorubicin, teniposide, idarubicin, mitoxantrone and epirubicin for CRC therapy, leading to a significant reversal of rectal cancer gene expression signatures (Carvalho et al., 2021). Given the significant differences in drug efficacy among individuals due to gene profiles and tumor heterogeneity, it becomes crucial to focus on DR based on tumor/subject molecular profiles to reduce inefficiencies in cancer treatment (Li and Jones, 2012).
While drug repurposing offers various benefits compared to the conventional de novo approach, it may not always lead to success due to lack of efficacy or toxicity issues. Bevacizumab, initially developed to treat CRC, was determined to be a strong candidate to treat other kinds of cancer such as colon, rectal, brain, lung, and kidney through drug repositioning. However, it failed in phase III trials despite positive results (Kim and Oh, 2018). These failed drug candidates in clinical trials still represent an affluent resource for repositioning, as they are well studied pharmacokinetically and clinically. Personalized genomics studies focusing on patient and disease heterogeneities may reveal that many of these failures were tested in inappropriate subject groups, making them practical options for future personalized medicine approaches, particularly for subjects with limited treatment options.
Drug repurposing is increasingly considered by both academia and the pharmaceutical industry as a cost and time-saving alternative to de novo drug development. Repurposing non-oncology drugs in cancer therapy provides a promising therapeutic opportunity, especially for patients with rare cancers, advanced diseases, or chemo-resistant tumors. In the present review, we have explored the potential of DR approaches, with a particular focus on their application in GI cancers. Repurposed drugs can target known pathways and key molecular targets in cancer biology due to their established functional mechanisms. DR has received attention owing to its potential to enhance treatment effectiveness and ability to overwhelm resistance to standard chemotherapy as well as improve therapy outcomes in tumors with limited response to conventional treatments. Additionally, when repurposed drugs are used in combination with routine oncology drugs, they offer a unique opportunity to target multiple pathways and molecular targets in cancer cells, going beyond the scope of traditional chemotherapy drugs and modulating diverse cancer-relevant pathways. However, it should be noted that the interaction of repurposed drugs with standard cancer drugs may pose challenges during the clinical trials.
The repositioning of drugs to treat GI cancers presents an attractive option given the increasing number of new cases, annual deaths, and the challenges in treating certain tumors. This review outlines various DR approaches that can be used to improve the efficiency of existing GI therapies. However, further clinical studies are needed to determine their potential for clinical adoption.
NF: Conceptualization, Visualization, Writing–original draft. MK: Conceptualization, Visualization, Writing–original draft. HB: Conceptualization, Visualization, Writing–original draft. MZ: Conceptualization, Supervision, Writing–review and editing. VC: Conceptualization, Visualization, Writing–original draft. AR: Conceptualization, Supervision, Writing–original draft. EN-M: Conceptualization, Writing–original draft, Visualization. MT: Conceptualization, Supervision, Writing–review and editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Royan Institute in Tehran and AIRC-Associazione Italiana Ricerca sul Cancro IG 2020 ID 24405 (AR).
The authors would like to thank all individuals whose fruitful research has contributed in any way for the elucidation of drugs, their repositioning, and their activity in GI cancers.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2023.1329244/full#supplementary-material
Acedo, P., Fernandes, A., and Zawacka-Pankau, J. (2019). Activation of TAp73 and inhibition of TrxR by Verteporfin for improved cancer therapy in TP53 mutant pancreatic tumors. Future Sci. OA 5, FSO366. doi:10.4155/fsoa-2018-0082
Agnelli, G., Muñoz, A., Franco, L., Mahé, I., Brenner, B., Connors, J. M., et al. (2022). Apixaban and dalteparin for the treatment of venous thromboembolism in patients with different sites of cancer. Thromb. Haemost. 122, 796–807. doi:10.1055/s-0041-1735194
Ahmed, M., Jinks, N., Babaei-Jadidi, R., Kashfi, H., Castellanos-Uribe, M., May, S. T., et al. (2019). Repurposing antibacterial AM404 as a potential anticancer drug for targeting colorectal cancer stem-like cells. Cancers 12, 106. doi:10.3390/cancers12010106
Alburquerque-González, B., Bernabé-García, Á., Bernabé-García, M., Ruiz-Sanz, J., López-Calderón, F. F., Gonnelli, L., et al. (2021). The FDA-approved antiviral raltegravir inhibits fascin1-dependent invasion of colorectal tumor cells in vitro and in vivo. Cancers 13, 861. doi:10.3390/cancers13040861
Al-Wadei, H. A., Al-Wadei, M. H., and Schuller, H. M. (2009). Prevention of pancreatic cancer by the beta-blocker propranolol. Anticancer Drugs 20, 477–482. doi:10.1097/CAD.0b013e32832bd1e3
Anselmino, L. E., Baglioni, M. V., Reynoso, G., Rozados, V. R., Scharovsky, O. G., Rico, M. J., et al. (2023). Potential effect of chloroquine and propranolol combination to treat colorectal and triple-negative breast cancers. Sci. Rep. 13, 7923–7927. doi:10.1038/s41598-023-34793-6
Antona, A., Varalda, M., Roy, K., Favero, F., Mazzucco, E., Zuccalà, M., et al. (2022). Dissecting the mechanism of action of spiperone-A candidate for drug repurposing for colorectal cancer. Cancers 14, 776. doi:10.3390/cancers14030776
Antoniciello, F., Roncarati, D., Zannoni, A., Chiti, E., Scarlato, V., and Chiappori, F. (2022). Targeting the essential transcription factor HP1043 of Helicobacter pylori: a drug repositioning study. Front. Mol. Biosci. 9, 887564. doi:10.3389/fmolb.2022.887564
Anwanwan, D., Singh, S. K., Singh, S., Saikam, V., and Singh, R. (2020). Challenges in liver cancer and possible treatment approaches. Biochim. Biophys. Acta Rev. Cancer 1873, 188314. doi:10.1016/j.bbcan.2019.188314
Apicella, M., Corso, S., and Giordano, S. (2017). Targeted therapies for gastric cancer: failures and hopes from clinical trials. Oncotarget 8, 57654–57669. doi:10.18632/oncotarget.14825
Arber, N., Eagle, C. J., Spicak, J., Rácz, I., Dite, P., Hajer, J., et al. (2006). Celecoxib for the prevention of colorectal adenomatous polyps. N. Engl. J. Med. 355, 885–895. doi:10.1056/NEJMoa061652
Arulanandam, C. D., Prathiviraj, R., and Kaveriyappan, G. R. (2021). Repurposing of an antifungal drug against gastrointestinal stromal tumors. bioRxiv. doi:10.1101/2021.01.15.426618
Ashburn, T. T., and Thor, K. B. (2004). Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 3, 673–683. doi:10.1038/nrd1468
Attia, Y. M., Tawfiq, R. A., Ali, A. A., and Elmazar, M. M. (2017). The FXR agonist, obeticholic acid, suppresses HCC proliferation & metastasis: role of IL-6/STAT3 signalling pathway. Sci. Rep. 7, 12502. doi:10.1038/s41598-017-12629-4
Au, T. H., Wang, K., Stenehjem, D., and Garrido-Laguna, I. (2017). Personalized and precision medicine: integrating genomics into treatment decisions in gastrointestinal malignancies. J. Gastrointest. Oncol. 8, 387–404. doi:10.21037/jgo.2017.01.04
Ayoub, B. M., Attia, Y. M., and Ahmed, M. S. (2018). Structural re-positioning, in silico molecular modelling, oxidative degradation, and biological screening of linagliptin as adenosine 3 receptor (ADORA3) modulators targeting hepatocellular carcinoma. J. Enzyme Inhib. Med. Chem. 33, 858–866. doi:10.1080/14756366.2018.1462801
Bago, R., Sommer, E., Castel, P., Crafter, C., Bailey, F. P., Shpiro, N., et al. (2016). The hVps34-SGK3 pathway alleviates sustained PI3K/Akt inhibition by stimulating mTORC1 and tumour growth. EMBO J. 35, 2263. doi:10.15252/embj.201670010
Bahmad, H. F., Demus, T., Moubarak, M. M., Daher, D., Alvarez Moreno, J. C., Polit, F., et al. (2022). Overcoming drug resistance in advanced prostate cancer by drug repurposing. Med. Sci. (Basel) 10, 15. doi:10.3390/medsci10010015
Baron, J. A., Cole, B. F., Sandler, R. S., Haile, R. W., Ahnen, D., Bresalier, R., et al. (2003). A randomized trial of aspirin to prevent colorectal adenomas. N. Engl. J. Med. 348, 891–899. doi:10.1056/NEJMoa021735
Batchu, R. B., Gruzdyn, O. V., Bryant, C. S., Qazi, A. M., Kumar, S., Chamala, S., et al. (2014). Ritonavir-mediated induction of apoptosis in pancreatic cancer occurs via the RB/E2F-1 and AKT pathways. Pharmaceuticals 7, 46–57. doi:10.3390/ph7010046
Baxendale, S., van Eeden, F., and Wilkinson, R. (2017). The power of zebrafish in personalised medicine. Adv. Exp. Med. Biol. 1007, 179–197. doi:10.1007/978-3-319-60733-7_10
Bei, S., Xu, Q., Li, F., Wu, C., Sun, Q., and Feng, L. (2022). Brefeldin A: a newly identified cell death inducer selectively targets radio-resistant colorectal cancer cells by direct interacting with caspase-3. J. King Saud. Univ. Sci. 34, 101728. doi:10.1016/j.jksus.2021.101728
Bejan, C. A., Wei, W.-Q., and Denny, J. C. (2015). Assessing the role of a medication-indication resource in the treatment relation extraction from clinical text. J. Am. Med. Inf. Assoc. 22, e162–e176. doi:10.1136/amiajnl-2014-002954
Benamouzig, R., Uzzan, B., Deyra, J., Martin, A., Girard, B., Little, J., et al. (2012). Prevention by daily soluble aspirin of colorectal adenoma recurrence: 4-year results of the APACC randomised trial. Gut 61, 255–261. doi:10.1136/gutjnl-2011-300113
Bertagnolli, M. M., Eagle, C. J., Zauber, A. G., Redston, M., Solomon, S. D., Kim, K., et al. (2006). Celecoxib for the prevention of sporadic colorectal adenomas. N. Engl. J. Med. 355, 873–884. doi:10.1056/NEJMoa061355
Bertolini, F., Sukhatme, V. P., and Bouche, G. (2015). Drug repurposing in oncology--patient and health systems opportunities. Nat. Rev. Clin. Oncol. 12, 732–742. doi:10.1038/nrclinonc.2015.169
Biermann, M., Quintero, C., Ferguson, P., Rajbhandari, N., Park, D. E., Patel, H., et al. (2022). Repurposing HDAC and mTOR inhibitors for pancreatic cancer. J. Clin. Oncol. 40, e16234, doi:10.1200/jco.2022.40.16_suppl.e16234
Biller, L. H., and Schrag, D. (2021). Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA 325, 669–685. doi:10.1001/jama.2021.0106
Booth, C. M., Karim, S., and Mackillop, W. J. (2019). Real-world data: towards achieving the achievable in cancer care. Nat. Rev. Clin. Oncol. 16, 312–325. doi:10.1038/s41571-019-0167-7
Breckenridge, A., and Jacob, R. (2019). Overcoming the legal and regulatory barriers to drug repurposing. Nat. Rev. Drug Discov. 18, 1–2. doi:10.1038/nrd.2018.92
Brehmer, D., Greff, Z., Godl, K., Blencke, S., Kurtenbach, A., Weber, M., et al. (2005). Cellular targets of gefitinib. Cancer Res. 65, 379–382. doi:10.1158/0008-5472.379.65.2
Brighton, T. A., Eikelboom, J. W., Mann, K., Mister, R., Gallus, A., Ockelford, P., et al. (2012). Low-dose aspirin for preventing recurrent venous thromboembolism. N. Engl. J. Med. 367, 1979–1987. doi:10.1056/NEJMoa1210384
Cao, X., Ren, K., Song, Z., Li, D., Quan, M., Zheng, Y., et al. (2022). [Corrigendum] 7-Difluoromethoxyl-5,4’-di-n-octyl genistein inhibits the stem-like characteristics of gastric cancer stem-like cells and reverses the phenotype of epithelial-mesenchymal transition in gastric cancer cells. Oncol. Rep. 48, 176. doi:10.3892/or.2022.8391
Carter, T. A., Wodicka, L. M., Shah, N. P., Velasco, A. M., Fabian, M. A., Treiber, D. K., et al. (2005). Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc. Natl. Acad. Sci. U. S. A. 102, 11011–11016. doi:10.1073/pnas.0504952102
Carvalho, R. F., do Canto, L. M., Cury, S. S., Frøstrup Hansen, T., Jensen, L. H., and Rogatto, S. R. (2021). Drug repositioning based on the reversal of gene expression signatures identifies TOP2A as a therapeutic target for rectal cancer. Cancers 13, 5492. doi:10.3390/cancers13215492
Catalano, M., Aprile, G., Conca, R., Petrioli, R., Ramello, M., and Roviello, G. (2022). The impact of age, performance status and comorbidities on nab-paclitaxel plus gemcitabine effectiveness in patients with metastatic pancreatic cancer. Sci. Rep. 12, 8244. doi:10.1038/s41598-022-12214-4
Cavalla, D., and Singal, C. (2012). Retrospective clinical analysis for drug rescue: for new indications or stratified patient groups. Drug Discov. Today 17, 104–109. doi:10.1016/j.drudis.2011.09.019
Chaiteerakij, R., Petersen, G. M., Bamlet, W. R., Chaffee, K. G., Zhen, D. B., Burch, P. A., et al. (2016). Metformin use and survival of patients with pancreatic cancer: a cautionary lesson. J. Clin. Oncol. 34, 1898–1904. doi:10.1200/JCO.2015.63.3511
Chakraborty, S., and Rahman, T. (2012). The difficulties in cancer treatment. Ecancermedicalscience 6, ed16. doi:10.3332/ecancer.2012.ed16
Chantrill, L. A., Nagrial, A. M., Watson, C., Johns, A. L., Martyn-Smith, M., Simpson, S., et al. (2015). Precision medicine for advanced pancreas cancer: the individualized molecular pancreatic cancer therapy (IMPaCT) trial. Clin. Cancer Res. 21, 2029–2037. doi:10.1158/1078-0432.CCR-15-0426
Chao, M.-W., Chen, T.-H., Huang, H.-L., Chang, Y.-W., HuangFu, W.-C., Lee, Y.-C., et al. (2017). Lanatoside C, a cardiac glycoside, acts through protein kinase Cδ to cause apoptosis of human hepatocellular carcinoma cells. Sci. Rep. 7, 46134. doi:10.1038/srep46134
Chen, B., Garmire, L., Calvisi, D. F., Chua, M.-S., Kelley, R. K., and Chen, X. (2020a). Publisher Correction: harnessing big 'omics' data and AI for drug discovery in hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 17, 238–251. doi:10.1038/s41575-020-0288-6
Chen, B., Wei, W., Ma, L., Yang, B., Gill, R. M., Chua, M.-S., et al. (2017a). Computational discovery of niclosamide ethanolamine, a repurposed drug candidate that reduces growth of hepatocellular carcinoma cells in vitro and in mice by inhibiting cell division cycle 37 signaling. Gastroenterology 152, 2022–2036. doi:10.1053/j.gastro.2017.02.039
Chen, C.-T., Chen, Y.-C., Yamaguchi, H., and Hung, M.-C. (2015a). Carglumic acid promotes apoptosis and suppresses cancer cell proliferation in vitro and in vivo. Am. J. Cancer Res. 5, 3560–3569.
Chen, G., Feng, W., Zhang, S., Bian, K., Yang, Y., Fang, C., et al. (2015b). Metformin inhibits gastric cancer via the inhibition of HIF1α/PKM2 signaling. Am. J. Cancer Res. 5, 1423–1434.
Chen, H., Weng, Z., and Xu, C. (2020b). Albendazole suppresses cell proliferation and migration and induces apoptosis in human pancreatic cancer cells. Anticancer Drugs 31, 431–439. doi:10.1097/CAD.0000000000000914
Chen, H.-N., Chen, Y., Zhou, Z.-G., Wei, Y., and Huang, C. (2019a). A novel role for ketoconazole in hepatocellular carcinoma treatment: linking PTGS2 to mitophagy machinery. Autophagy 15, 733–734. doi:10.1080/15548627.2019.1569934
Chen, K., Cheng, L., Qian, W., Jiang, Z., Sun, L., Zhao, Y., et al. (2018). Itraconazole inhibits invasion and migration of pancreatic cancer cells by suppressing TGF-β/SMAD2/3 signaling. Oncol. Rep. 39, 1573–1582. doi:10.3892/or.2018.6281
Chen, K., Qian, W., Jiang, Z., Cheng, L., Li, J., Sun, L., et al. (2017b). Metformin suppresses cancer initiation and progression in genetic mouse models of pancreatic cancer. Mol. Cancer 16, 131. doi:10.1186/s12943-017-0701-0
Chen, V. C.-H., Lin, C.-F., Hsieh, Y.-H., Liang, H.-Y., Huang, K.-Y., Chiu, W.-C., et al. (2017c). Hepatocellular carcinoma and antidepressants: a nationwide population-based study. Oncotarget 8, 30464–30470. doi:10.18632/oncotarget.12826
Chen, W., Chen, F., Gong, M., Ye, L., Weng, D., Jin, Z., et al. (2023). Fenofibrate suppresses the progression of hepatoma by downregulating osteopontin through inhibiting the PI3K/AKT/Twist pathway. Naunyn. Schmiedeb. Arch. Pharmacol., 1–11. doi:10.1007/s00210-023-02604-4
Chen, Y., Chen, H.-N., Wang, K., Zhang, L., Huang, Z., Liu, J., et al. (2019b). Ketoconazole exacerbates mitophagy to induce apoptosis by downregulating cyclooxygenase-2 in hepatocellular carcinoma. J. Hepatol. 70, 66–77. doi:10.1016/j.jhep.2018.09.022
Cheng, F., Lu, W., Liu, C., Fang, J., Hou, Y., Handy, D. E., et al. (2019). A genome-wide positioning systems network algorithm for in silico drug repurposing. Nat. Commun. 10, 3476. doi:10.1038/s41467-019-10744-6
Cheng, X., Zhao, W., Zhu, M., Wang, B., Wang, X., Yang, X., et al. (2021). Drug repurposing for cancer treatment through global propagation with a greedy algorithm in a multilayer network. Cancer Biol. Med. 19, 74–89. doi:10.20892/j.issn.2095-3941.2020.0218
Chong, C. R., and Sullivan, D. J. (2007). New uses for old drugs. Nature 448, 645–646. doi:10.1038/448645a
Chu, Y.-D., Chen, C.-W., Lai, M.-W., Lim, S.-N., and Lin, W.-R. (2023). Bioenergetic alteration in gastrointestinal cancers: the good, the bad and the ugly. World J. Gastroenterol. 29, 4499–4527. doi:10.3748/wjg.v29.i29.4499
Ciociola, A. A., Cohen, L. B., and Kulkarni, P. (2014). How drugs are developed and approved by the FDA: current process and future directions. Am. J. Gastroenterol. 109, 620–623. doi:10.1038/ajg.2013.407
Clements, W. K., and Traver, D. (2012). Fish pharming: zebrafish antileukemia screening. Blood 119, 5614–5615. doi:10.1182/blood-2012-04-425249
Cong, F., Cheung, A. K., and Huang, S.-M. A. (2012). Chemical genetics-based target identification in drug discovery. Annu. Rev. Pharmacol. Toxicol. 52, 57–78. doi:10.1146/annurev-pharmtox-010611-134639
Cousin, M. A., Ebbert, J. O., Wiinamaki, A. R., Urban, M. D., Argue, D. P., Ekker, S. C., et al. (2014). Larval zebrafish model for FDA-approved drug repositioning for tobacco dependence treatment. PLoS One 9, e90467. doi:10.1371/journal.pone.0090467
Crawford, S. (2014). Anti-inflammatory/antioxidant use in long-term maintenance cancer therapy: a new therapeutic approach to disease progression and recurrence. Ther. Adv. Med. Oncol. 6, 52–68. doi:10.1177/1758834014521111
Cunha Júnior, A. D., Bragagnoli, A. C., Costa, F. O., and Carvalheira, J. B. C. (2021). Repurposing metformin for the treatment of gastrointestinal cancer. World J. Gastroenterol. 27, 1883–1904. doi:10.3748/wjg.v27.i17.1883
Dakshanamurthy, S., Issa, N. T., Assefnia, S., Seshasayee, A., Peters, O. J., Madhavan, S., et al. (2012). Predicting new indications for approved drugs using a proteochemometric method. J. Med. Chem. 55, 6832–6848. doi:10.1021/jm300576q
Dalwadi, S. M., Hunt, A., Bonnen, M. D., and Ghebre, Y. T. (2023). Computational approaches for drug repurposing in oncology: untapped opportunity for high value innovation. Front. Oncol. 13, 1198284. doi:10.3389/fonc.2023.1198284
Damoiseaux, D., Li, W., Martínez-Chávez, A., Beijnen, J. H., Schinkel, A. H., Huitema, A. D. R., et al. (2022). Predictiveness of the human-CYP3A4-transgenic mouse model (Cyp3aXAV) for human drug exposure of CYP3A4-metabolized drugs. Pharmaceuticals 15, 860. doi:10.3390/ph15070860
Dehnavi, S., Kiani, A., Sadeghi, M., Biregani, A. F., Banach, M., Atkin, S. L., et al. (2021). Targeting AMPK by statins: a potential therapeutic approach. Drugs 81, 923–933. doi:10.1007/s40265-021-01510-4
Demaré, S., Kothari, A., Calcutt, N. A., and Fernyhough, P. (2021). Metformin as a potential therapeutic for neurological disease: mobilizing AMPK to repair the nervous system. Expert Rev. Neurother. 21, 45–63. doi:10.1080/14737175.2021.1847645
Dhakal, B., Li, C. M. Y., Li, R., Yeo, K., Wright, J. A., Gieniec, K. A., et al. (2022). The antianginal drug perhexiline displays cytotoxicity against colorectal cancer cells in vitro: a potential for drug repurposing. Cancers 14, 1043. doi:10.3390/cancers14041043
Díaz-Carballo, D., Acikelli, A. H., Klein, J., Jastrow, H., Dammann, P., Wyganowski, T., et al. (2015). Therapeutic potential of antiviral drugs targeting chemorefractory colorectal adenocarcinoma cells overexpressing endogenous retroviral elements. J. Exp. Clin. Cancer Res. 34, 81. doi:10.1186/s13046-015-0199-5
Dimitriadis, K., Lazarou, E., Tsioufis, P., Soulaidopoulos, S., and Tsioufis, K. (2022). Aspirin for primary prevention of cardiovascular diseases: “WALTZ” with the evidence. Curr. Cardiol. Rep. 24, 1139–1147. doi:10.1007/s11886-022-01740-2
Diop-Frimpong, B., Chauhan, V. P., Krane, S., Boucher, Y., and Jain, R. K. (2011). Losartan inhibits collagen I synthesis and improves the distribution and efficacy of nanotherapeutics in tumors. Proc. Natl. Acad. Sci. U. S. A. 108, 2909–2914. doi:10.1073/pnas.1018892108
Donohoe, C. L., and Reynolds, J. V. (2017). Neoadjuvant treatment of locally advanced esophageal and junctional cancer: the evidence-base, current key questions and clinical trials. J. Thorac. Dis. 9, S697–S704. doi:10.21037/jtd.2017.03.159
Dou, C., Mo, H., Chen, T., Liu, J., Zeng, Y., Li, S., et al. (2021). ZMYND8 promotes the growth and metastasis of hepatocellular carcinoma by promoting HK2-mediated glycolysis. Pathol. Res. Pract. 219, 153345. doi:10.1016/j.prp.2021.153345
Du, D., Liu, C., Qin, M., Zhang, X., Xi, T., Yuan, S., et al. (2022). Metabolic dysregulation and emerging therapeutical targets for hepatocellular carcinoma. Acta Pharm. Sin. B 12, 558–580. doi:10.1016/j.apsb.2021.09.019
Ducreux, M., Abou-Alfa, G. K., Bekaii-Saab, T., Berlin, J., Cervantes, A., de Baere, T., et al. (2023). The management of hepatocellular carcinoma. Current expert opinion and recommendations derived from the 24th ESMO/World Congress on Gastrointestinal Cancer, Barcelona. ESMO Open 8, 101567. doi:10.1016/j.esmoop.2023.101567
Dudley, J. T., Deshpande, T., and Butte, A. J. (2011). Exploiting drug-disease relationships for computational drug repositioning. Brief. Bioinform. 12, 303–311. doi:10.1093/bib/bbr013
Eichler, H.-G., Bloechl-Daum, B., Broich, K., Kyrle, P. A., Oderkirk, J., Rasi, G., et al. (2019). Data rich, information poor: can we use electronic health records to create a learning healthcare system for pharmaceuticals? Clin. Pharmacol. Ther. 105, 912–922. doi:10.1002/cpt.1226
El-Hachem, N., Gendoo, D. M. A., Ghoraie, L. S., Safikhani, Z., Smirnov, P., Chung, C., et al. (2017). Integrative cancer pharmacogenomics to infer large-scale drug taxonomy. Cancer Res. 77, 3057–3069. doi:10.1158/0008-5472.CAN-17-0096
El Harane, S., Zidi, B., El Harane, N., Krause, K.-H., Matthes, T., and Preynat-Seauve, O. (2023). Cancer spheroids and organoids as novel tools for research and therapy: state of the art and challenges to guide precision medicine. Cells 12. doi:10.3390/cells12071001
ElHefnawi, M., Jo, E., Tolba, M. M., Fares, M., Yang, J., Shahbaaz, M., et al. (2022). Drug repurposing through virtual screening and in vitro validation identifies tigecycline as a novel putative HCV polymerase inhibitor. Virology 570, 9–17. doi:10.1016/j.virol.2022.02.006
Emamzaden, F. N., Word, B., Hammons, G., and Lyn-Cook, B. D. (2022). Abstract 1631: anti-cancer effects of vorinostat on 3D cultured pancreatic cancer cells. Cancer Res. 82, 1631. doi:10.1158/1538-7445.am2022-1631
Falcone, A., Lencioni, M., Brunetti, I., Pfanner, E., Allegrini, G., Antonuzzo, A., et al. (1997). Maximum tolerable doses of intravenous zidovudine in combination with 5-fluorouracil and leucovorin in metastatic colorectal cancer patients. Clinical evidence of significant antitumor activity and enhancement of zidovudine-induced DNA single strand breaks in peripheral nuclear blood cells. Ann. Oncol. 8, 539–545. doi:10.1023/a:1008249803523
Favoulet, P., Cercueil, J. P., Faure, P., Osmak, L., Isambert, N., Beltramo, J. L., et al. (2001). Increased cytotoxicity and stability of Lipiodol-pirarubicin emulsion compared to classical doxorubicin-Lipiodol: potential advantage for chemoembolization of unresectable hepatocellular carcinoma. Anticancer Drugs 12, 801–806. doi:10.1097/00001813-200111000-00003
Ferioli, M., Zauli, G., Martelli, A. M., Vitale, M., McCubrey, J. A., Ultimo, S., et al. (2018). Impact of physical exercise in cancer survivors during and after antineoplastic treatments. Oncotarget 9, 14005–14034. doi:10.18632/oncotarget.24456
Ferlay, J., Colombet, M., Soerjomataram, I., Parkin, D. M., Piñeros, M., Znaor, A., et al. (2021). Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 144, 1941–1953. doi:10.1002/ijc.31937
Fernandez-Banet, J., Esposito, A., Coffin, S., Horvath, I. B., Estrella, H., Schefzick, S., et al. (2016). OASIS: web-based platform for exploring cancer multi-omics data. Nat. Methods 13, 9–10. doi:10.1038/nmeth.3692
Fernández-Torras, A., Duran-Frigola, M., and Aloy, P. (2019). Encircling the regions of the pharmacogenomic landscape that determine drug response. Genome Med. 11, 17. doi:10.1186/s13073-019-0626-x
Florio, R., Veschi, S., di Giacomo, V., Pagotto, S., Carradori, S., Verginelli, F., et al. (2019). The benzimidazole-based anthelmintic parbendazole: a repurposed drug candidate that synergizes with gemcitabine in pancreatic cancer. Cancers 11, 2042. doi:10.3390/cancers11122042
Foglietta, F., Pinnelli, V., Giuntini, F., Barbero, N., Panzanelli, P., Durando, G., et al. (2021). Sonodynamic treatment induces selective killing of cancer cells in an in vitro co-culture model. Cancers 13, 3852. doi:10.3390/cancers13153852
Freires, I. A., Sardi, J. de C. O., de Castro, R. D., and Rosalen, P. L. (2017). Alternative animal and non-animal models for drug discovery and development: bonus or burden? Pharm. Res. 34, 681–686. doi:10.1007/s11095-016-2069-z
Frouws, M. A., Rademaker, E., Bastiaannet, E., van Herk-Sukel, M. P. P., Lemmens, V. E., Van de Velde, C. J. H., et al. (2017). The difference in association between aspirin use and other thrombocyte aggregation inhibitors and survival in patients with colorectal cancer. Eur. J. Cancer 77, 24–30. doi:10.1016/j.ejca.2017.02.025
Fuchs, C. S., Tomasek, J., Yong, C. J., Dumitru, F., Passalacqua, R., Goswami, C., et al. (2014). Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): an international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet 383, 31–39. doi:10.1016/S0140-6736(13)61719-5
Gao, F., Wu, J., Niu, S., Sun, T., Li, F., Bai, Y., et al. (2019). Biodegradable, pH-sensitive hollow mesoporous organosilica nanoparticle (HMON) with controlled release of pirfenidone and ultrasound-target-microbubble-destruction (UTMD) for pancreatic cancer treatment. Theranostics 9, 6002–6018. doi:10.7150/thno.36135
Gao, X., Liu, X., Shan, W., Liu, Q., Wang, C., Zheng, J., et al. (2018). Anti-malarial atovaquone exhibits anti-tumor effects by inducing DNA damage in hepatocellular carcinoma. Am. J. Cancer Res. 8, 1697–1711.
Gdovinova, Z., Kremer, C., Lorenzano, S., Dawson, J., Lal, A., and Caso, V. (2022). Aspirin for primary stroke prevention; evidence for a differential effect in men and women. Front. Neurol. 13, 856239. doi:10.3389/fneur.2022.856239
Ghasemi, K., and Ghasemi, K. (2022). A Brief look at antitumor effects of doxycycline in the treatment of colorectal cancer and combination therapies. Eur. J. Pharmacol. 916, 174593. doi:10.1016/j.ejphar.2021.174593
Gong, R.-H., Yang, D.-J., Kwan, H.-Y., Lyu, A.-P., Chen, G.-Q., and Bian, Z.-X. (2022). Cell death mechanisms induced by synergistic effects of halofuginone and artemisinin in colorectal cancer cells. Int. J. Med. Sci. 19, 175–185. doi:10.7150/ijms.66737
Gordon, S. W., McGuire, W. P., Shafer, D. A., Sterling, R. K., Lee, H. M., Matherly, S. C., et al. (2019). Phase I study of sorafenib and vorinostat in advanced hepatocellular carcinoma. Am. J. Clin. Oncol. 42, 649–654. doi:10.1097/COC.0000000000000567
Gou, H., Liu, S., Liu, L., Luo, M., Qin, S., He, K., et al. (2022). Obeticholic acid and 5β-cholanic acid 3 exhibit anti-tumor effects on liver cancer through CXCL16/CXCR6 pathway. Front. Immunol. 13, 1095915. doi:10.3389/fimmu.2022.1095915
Gray, E., Marti, J., Wyatt, J. C., Brewster, D. H., and Hall, P. S.SATURNE advisory group (2019). Chemotherapy effectiveness in trial-underrepresented groups with early breast cancer: a retrospective cohort study. PLoS Med. 16, e1003006. doi:10.1371/journal.pmed.1003006
Greene, C. S., Krishnan, A., Wong, A. K., Ricciotti, E., Zelaya, R. A., Himmelstein, D. S., et al. (2015). Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 47, 569–576. doi:10.1038/ng.3259
Greene, C. S., and Voight, B. F. (2016). Pathway and network-based strategies to translate genetic discoveries into effective therapies. Hum. Mol. Genet. 25, R94–R98. doi:10.1093/hmg/ddw160
Greten, T. F., Manns, M. P., and Korangy, F. (2008). Immunotherapy of HCC. Rev. Recent Clin. Trials 3, 31–39. doi:10.2174/157488708783330549
Grösch, S., Tegeder, I., Niederberger, E., Bräutigam, L., and Geisslinger, G. (2001). COX-2 independent induction of cell cycle arrest and apoptosis in colon cancer cells by the selective COX-2 inhibitor celecoxib. FASEB J. 15, 2742–2744. doi:10.1096/fj.01-0299fje
Grover, M. P., Ballouz, S., Mohanasundaram, K. A., George, R. A., Goscinski, A., Crowley, T. M., et al. (2015). Novel therapeutics for coronary artery disease from genome-wide association study data. BMC Med. Genomics 8 (2), S1. doi:10.1186/1755-8794-8-S2-S1
Guirguis-Blake, J. M., Evans, C. V., Perdue, L. A., Bean, S. I., and Senger, C. A. (2022). Aspirin use to prevent cardiovascular disease and colorectal cancer: updated evidence report and systematic review for the US preventive services task force. JAMA 327, 1585–1597. doi:10.1001/jama.2022.3337
Gulhati, P., Cai, Q., Li, J., Liu, J., Rychahou, P. G., Qiu, S., et al. (2009). Targeted inhibition of mammalian target of rapamycin signaling inhibits tumorigenesis of colorectal cancer. Clin. Cancer Res. 15, 7207–7216. doi:10.1158/1078-0432.CCR-09-1249
Gunderson, A. J., Kaneda, M. M., Tsujikawa, T., Nguyen, A. V., Affara, N. I., Ruffell, B., et al. (2016). Bruton tyrosine kinase-dependent immune cell cross-talk drives pancreas cancer. Cancer Discov. 6, 270–285. doi:10.1158/2159-8290.CD-15-0827
Guo, L., Chen, X.-J., Hu, Y.-H., Yu, Z.-J., Wang, D., and Liu, J.-Z. (2013). Curcumin inhibits proliferation and induces apoptosis of human colorectal cancer cells by activating the mitochondria apoptotic pathway. Phytother. Res. 27, 422–430. doi:10.1002/ptr.4731
Gupta, S. C., Sung, B., Prasad, S., Webb, L. J., and Aggarwal, B. B. (2013). Cancer drug discovery by repurposing: teaching new tricks to old dogs. Trends Pharmacol. Sci. 34, 508–517. doi:10.1016/j.tips.2013.06.005
Gut, M. O., Parkkila, S., Vernerová, Z., Rohde, E., Závada, J., Höcker, M., et al. (2002). Gastric hyperplasia in mice with targeted disruption of the carbonic anhydrase gene Car9. Gastroenterology 123, 1889–1903. doi:10.1053/gast.2002.37052
Hanada, Y., Pereira, S. P., Pogue, B., Maytin, E. V., Hasan, T., Linn, B., et al. (2021). EUS-guided verteporfin photodynamic therapy for pancreatic cancer. Gastrointest. Endosc. 94, 179–186. doi:10.1016/j.gie.2021.02.027
Happold, C., Gorlia, T., Chinot, O., Gilbert, M. R., Nabors, L. B., Wick, W., et al. (2016). Does valproic acid or levetiracetam improve survival in glioblastoma? A pooled analysis of prospective clinical trials in newly diagnosed glioblastoma. J. Clin. Oncol. 34, 731–739. doi:10.1200/JCO.2015.63.6563
He, S., Xu, J., Liu, X., and Zhen, Y. (2021). Advances and challenges in the treatment of esophageal cancer. Acta Pharm. Sin. B 11, 3379–3392. doi:10.1016/j.apsb.2021.03.008
Hecht, M., Harrer, T., Büttner, M., Schwegler, M., Erber, S., Fietkau, R., et al. (2013). Cytotoxic effect of efavirenz is selective against cancer cells and associated with the cannabinoid system. AIDS 27, 2031–2040. doi:10.1097/QAD.0b013e3283625444
Hecht, M., Harrer, T., Körber, V., Sarpong, E. O., Moser, F., Fiebig, N., et al. (2018). Cytotoxic effect of Efavirenz in BxPC-3 pancreatic cancer cells is based on oxidative stress and is synergistic with ionizing radiation. Oncol. Lett. 15, 1728–1736. doi:10.3892/ol.2017.7523
Heer, E., Ruan, Y., Mah, B., Nguyen, T., Lyons, H., Poirier, A., et al. (2022). The efficacy of chemopreventive agents on the incidence of colorectal adenomas: a systematic review and network meta-analysis. Prev. Med. 162, 107169. doi:10.1016/j.ypmed.2022.107169
Hegazy, S. K., El-Azab, G. A., Zakaria, F., Mostafa, M. F., and El-Ghoneimy, R. A. (2022). Mebendazole; from an anti-parasitic drug to a promising candidate for drug repurposing in colorectal cancer. Life Sci. 299, 120536. doi:10.1016/j.lfs.2022.120536
Hieronymus, H., Lamb, J., Ross, K. N., Peng, X. P., Clement, C., Rodina, A., et al. (2006). Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell 10, 321–330. doi:10.1016/j.ccr.2006.09.005
Higurashi, T., and Nakajima, A. (2018). Metformin and colorectal cancer. Front. Endocrinol. 9, 622. doi:10.3389/fendo.2018.00622
Hirt, C. K., Booij, T. H., Grob, L., Simmler, P., Toussaint, N. C., Keller, D., et al. (2022). Drug screening and genome editing in human pancreatic cancer organoids identifies drug-gene interactions and candidates for off-label treatment. Cell Genom 2, 100095. doi:10.1016/j.xgen.2022.100095
Honarparvar, B., Govender, T., Maguire, G. E. M., Soliman, M. E. S., and Kruger, H. G. (2014). Integrated approach to structure-based enzymatic drug design: molecular modeling, spectroscopy, and experimental bioactivity. Chem. Rev. 114, 493–537. doi:10.1021/cr300314q
Hossain, M. T., Reza, M. S., Peng, Y., Feng, S., and Wei, Y. (2023). Identification of key genes as potential drug targets for gastric cancer. Tsinghua Sci. Technol. 28, 649–664. doi:10.26599/tst.2022.9010035
Hu, S., Liu, L., Chang, E. B., Wang, J.-Y., and Raufman, J.-P. (2015). Butyrate inhibits pro-proliferative miR-92a by diminishing c-Myc-induced miR-17-92a cluster transcription in human colon cancer cells. Mol. Cancer 14, 180. doi:10.1186/s12943-015-0450-x
Huang, C., Lan, W., Fraunhoffer, N., Meilerman, A., Iovanna, J., and Santofimia-Castaño, P. (2019). Dissecting the anticancer mechanism of trifluoperazine on pancreatic ductal adenocarcinoma. Cancers 11, 1869. doi:10.3390/cancers11121869
Huang, J., Lok, V., Ngai, C. H., Zhang, L., Yuan, J., Lao, X. Q., et al. (2021). Worldwide burden of, risk factors for, and trends in pancreatic cancer. Gastroenterology 160, 744–754. doi:10.1053/j.gastro.2020.10.007
Huggett, M. T., Jermyn, M., Gillams, A., Illing, R., Mosse, S., Novelli, M., et al. (2014). Phase I/II study of verteporfin photodynamic therapy in locally advanced pancreatic cancer. Br. J. Cancer 110, 1698–1704. doi:10.1038/bjc.2014.95
Hui, B., Shi, Y.-H., Ding, Z.-B., Zhou, J., Gu, C.-Y., Peng, Y.-F., et al. (2012). Proteasome inhibitor interacts synergistically with autophagy inhibitor to suppress proliferation and induce apoptosis in hepatocellular carcinoma. Cancer 118, 5560–5571. doi:10.1002/cncr.27586
Hulkower, K. I., and Herber, R. L. (2011). Cell migration and invasion assays as tools for drug discovery. Pharmaceutics 3, 107–124. doi:10.3390/pharmaceutics3010107
Hurle, M. R., Yang, L., Xie, Q., Rajpal, D. K., Sanseau, P., and Agarwal, P. (2013). Computational drug repositioning: from data to therapeutics. Clin. Pharmacol. Ther. 93, 335–341. doi:10.1038/clpt.2013.1
Iida, T., Mizutani, Y., Esaki, N., Ponik, S. M., Burkel, B. M., Weng, L., et al. (2022). Pharmacologic conversion of cancer-associated fibroblasts from a protumor phenotype to an antitumor phenotype improves the sensitivity of pancreatic cancer to chemotherapeutics. Oncogene 41, 2764–2777. doi:10.1038/s41388-022-02288-9
Iljin, K., Ketola, K., Vainio, P., Halonen, P., Kohonen, P., Fey, V., et al. (2009). High-throughput cell-based screening of 4910 known drugs and drug-like small molecules identifies disulfiram as an inhibitor of prostate cancer cell growth. Clin. Cancer Res. 15, 6070–6078. doi:10.1158/1078-0432.CCR-09-1035
Iorio, F., Bosotti, R., Scacheri, E., Belcastro, V., Mithbaokar, P., Ferriero, R., et al. (2010). Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl. Acad. Sci. U. S. A. 107, 14621–14626. doi:10.1073/pnas.1000138107
Iorio, F., Saez-Rodriguez, J., and di Bernardo, D. (2013). Network based elucidation of drug response: from modulators to targets. BMC Syst. Biol. 7, 139. doi:10.1186/1752-0509-7-139
Jandaghi, P., Najafabadi, H. S., Bauer, A. S., Papadakis, A. I., Fassan, M., Hall, A., et al. (2016). Expression of DRD2 is increased in human pancreatic ductal adenocarcinoma and inhibitors slow tumor growth in mice. Gastroenterology 151, 1218–1231. doi:10.1053/j.gastro.2016.08.040
Jeon, Y., Sym, S. J., Yoo, B. K., and Baek, J.-H. (2022). Long-term survival, tolerability, and safety of first-line bevacizumab and FOLFIRI in combination with ginsenoside-modified nanostructured lipid carrier containing curcumin in patients with unresectable metastatic colorectal cancer. Integr. Cancer Ther. 21, 15347354221105498. doi:10.1177/15347354221105498
Jeong, J. B., Choi, J., Baek, S. J., and Lee, S.-H. (2013). Reactive oxygen species mediate tolfenamic acid-induced apoptosis in human colorectal cancer cells. Arch. Biochem. Biophys. 537, 168–175. doi:10.1016/j.abb.2013.07.016
Ji, L., Qian, W., Gui, L., Ji, Z., Yin, P., Lin, G. N., et al. (2020). Blockade of β-catenin-induced CCL28 suppresses gastric cancer progression via inhibition of treg cell infiltration. Cancer Res. 80, 2004–2016. doi:10.1158/0008-5472.CAN-19-3074
Ji, T., Lang, J., Wang, J., Cai, R., Zhang, Y., Qi, F., et al. (2017). Designing liposomes to suppress extracellular matrix expression to enhance drug penetration and pancreatic tumor therapy. ACS Nano 11, 8668–8678. doi:10.1021/acsnano.7b01026
Jiang, F., Xing, H.-S., Chen, W.-Y., Du, J., Ruan, Y.-L., Lin, A.-Y., et al. (2019). Itraconazole inhibits proliferation of pancreatic cancer cells through activation of Bak-1. J. Cell. Biochem. 120, 4333–4341. doi:10.1002/jcb.27719
Jian-Hui, C., Er-Tao, Z., Si-Le, C., Hui, W., Kai-Ming, W., Xin-Hua, Z., et al. (2016). CD44, sonic hedgehog, and Gli1 expression are prognostic biomarkers in gastric cancer patients after radical resection. Gastroenterol. Res. Pract. 2016, 1013045. doi:10.1155/2016/1013045
Jin, H., Wang, S., Zaal, E. A., Wang, C., Wu, H., Bosma, A., et al. (2020). A powerful drug combination strategy targeting glutamine addiction for the treatment of human liver cancer. Elife 9, e56749. doi:10.7554/eLife.56749
Jin, R. U., and Mills, J. C. (2020). Tumor organoids to study gastroesophageal cancer: a primer. J. Mol. Cell Biol. 12, 593–606. doi:10.1093/jmcb/mjaa035
Jin, X., Pan, Y., Wang, L., Zhang, L., Ravichandran, R., Potts, P. R., et al. (2017). MAGE-TRIM28 complex promotes the Warburg effect and hepatocellular carcinoma progression by targeting FBP1 for degradation. Oncogenesis 6, e312. doi:10.1038/oncsis.2017.21
Joshi, S. S., and Badgwell, B. D. (2021). Current treatment and recent progress in gastric cancer. CA Cancer J. Clin. 71, 264–279. doi:10.3322/caac.21657
Kaji, K., Nishimura, N., Seki, K., Sato, S., Saikawa, S., Nakanishi, K., et al. (2018). Sodium glucose cotransporter 2 inhibitor canagliflozin attenuates liver cancer cell growth and angiogenic activity by inhibiting glucose uptake. Int. J. Cancer 142, 1712–1722. doi:10.1002/ijc.31193
Kan, J.-Y., Hsu, Y.-L., Chen, Y.-H., Chen, T.-C., Wang, J.-Y., and Kuo, P.-L. (2013). Gemifloxacin, a fluoroquinolone antimicrobial drug, inhibits migration and invasion of human colon cancer cells. Biomed. Res. Int. 2013, 159786. doi:10.1155/2013/159786
Kang, H. J., Seol, H. S., Lee, S. E., Suh, Y.-A., Kim, J., Jang, S. J., et al. (2019). Guanabenz acetate induces endoplasmic reticulum stress-related cell death in hepatocellular carcinoma cells. J. Pathol. Transl. Med. 53, 94–103. doi:10.4132/jptm.2019.01.14
Kannen, V., Hintzsche, H., Zanette, D. L., Silva, W. A., Garcia, S. B., Waaga-Gasser, A. M., et al. (2012). Antiproliferative effects of fluoxetine on colon cancer cells and in a colonic carcinogen mouse model. PLoS One 7, e50043. doi:10.1371/journal.pone.0050043
Karaman, M. W., Herrgard, S., Treiber, D. K., Gallant, P., Atteridge, C. E., Campbell, B. T., et al. (2008). A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 26, 127–132. doi:10.1038/nbt1358
Karnevi, E., Said, K., Andersson, R., and Rosendahl, A. H. (2013). Metformin-mediated growth inhibition involves suppression of the IGF-I receptor signalling pathway in human pancreatic cancer cells. BMC Cancer 13, 235. doi:10.1186/1471-2407-13-235
Kaur, J., and Sanyal, S. N. (2011). Diclofenac, a selective COX-2 inhibitor, inhibits DMH-induced colon tumorigenesis through suppression of MCP-1, MIP-1α and VEGF. Mol. Carcinog. 50, 707–718. doi:10.1002/mc.20736
Keiser, M. J., Setola, V., Irwin, J. J., Laggner, C., Abbas, A. I., Hufeisen, S. J., et al. (2009). Predicting new molecular targets for known drugs. Nature 462, 175–181. doi:10.1038/nature08506
Kersten, K., de Visser, K. E., van Miltenburg, M. H., and Jonkers, J. (2017). Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol. Med. 9, 137–153. doi:10.15252/emmm.201606857
Khozin, S., Blumenthal, G. M., and Pazdur, R. (2017). Real-world data for clinical evidence generation in oncology. J. Natl. Cancer Inst. 109. doi:10.1093/jnci/djx187
Kim, H. J., and Oh, S. C. (2018). Novel systemic therapies for advanced gastric cancer. J. Gastric Cancer 18, 1–19. doi:10.5230/jgc.2018.18.e3
Kim, J., Yoo, M., Shin, J., Kim, H., Kang, J., and Tan, A. C. (2018). Systems pharmacology-based approach of connecting disease genes in genome-wide association studies with traditional Chinese medicine. Int. J. Genomics Proteomics 2018, 7697356. doi:10.1155/2018/7697356
Kim, M., Mun, H., Sung, C. O., Cho, E. J., Jeon, H.-J., Chun, S.-M., et al. (2019). Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 10, 3991. doi:10.1038/s41467-019-11867-6
Kim, S. H., Lee, H. Y., Yi, H., Ahn, Y. M., and Kim, Y. S. (2012). Haloperidol induces demethylation and expression of the dual specificity phosphatase 6 gene in MIA PaCa-2 human pancreatic cancer cells. Life Sci. 91, 1317–1322. doi:10.1016/j.lfs.2012.10.002
Knapp, S. (2018). New opportunities for kinase drug repurposing and target discovery. Br. J. Cancer 118, 936–937. doi:10.1038/s41416-018-0045-6
Kobayashi, H., Enomoto, A., Woods, S. L., Burt, A. D., Takahashi, M., and Worthley, D. L. (2019). Cancer-associated fibroblasts in gastrointestinal cancer. Nat. Rev. Gastroenterol. Hepatol. 16, 282–295. doi:10.1038/s41575-019-0115-0
Kobayashi, H., Gieniec, K. A., Wright, J. A., Wang, T., Asai, N., Mizutani, Y., et al. (2021). The balance of stromal BMP signaling mediated by GREM1 and ISLR drives colorectal carcinogenesis. Gastroenterology 160, 1224–1239. doi:10.1053/j.gastro.2020.11.011
Kobayashi, Y., Kashima, H., Rahmanto, Y. S., Banno, K., Yu, Y., Matoba, Y., et al. (2017). Drug repositioning of mevalonate pathway inhibitors as antitumor agents for ovarian cancer. Oncotarget 8, 72147–72156. doi:10.18632/oncotarget.20046
Kong, K., Guo, M., Liu, Y., and Zheng, J. (2020). Progress in animal models of pancreatic ductal adenocarcinoma. J. Cancer 11, 1555–1567. doi:10.7150/jca.37529
Kordes, S., Pollak, M. N., Zwinderman, A. H., Mathôt, R. A., Weterman, M. J., Beeker, A., et al. (2015). Metformin in patients with advanced pancreatic cancer: a double-blind, randomised, placebo-controlled phase 2 trial. Lancet Oncol. 16, 839–847. doi:10.1016/S1470-2045(15)00027-3
Kulkarni, V. S., Alagarsamy, V., Solomon, V. R., Jose, P. A., and Murugesan, S. (2023). Drug repurposing: an effective tool in modern drug discovery. Russ. J. Bioorg. Chem. 49, 157–166. doi:10.1134/S1068162023020139
Kumar, V. L., Verma, S., and Das, P. (2019). Artesunate suppresses inflammation and oxidative stress in a rat model of colorectal cancer. Drug Dev. Res. 80, 1089–1097. doi:10.1002/ddr.21590
Lai, C.-T., Chi, C.-W., Wu, S.-H., Shieh, H.-R., Yen, J.-C., and Chen, Y.-J. (2022). Midostaurin modulates tumor microenvironment and enhances efficacy of anti-PD-1 against colon cancer. Cancers 14, 4847. doi:10.3390/cancers14194847
Lally, J. S. V., Ghoshal, S., DePeralta, D. K., Moaven, O., Wei, L., Masia, R., et al. (2019). Inhibition of acetyl-CoA carboxylase by phosphorylation or the inhibitor ND-654 suppresses lipogenesis and hepatocellular carcinoma. Cell Metab. 29, 174–182. doi:10.1016/j.cmet.2018.08.020
Lan, S.-H., Wu, S.-Y., Zuchini, R., Lin, X.-Z., Su, I.-J., Tsai, T.-F., et al. (2014). Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology 59, 505–517. doi:10.1002/hep.26659
Lee, J., Lee, I., Han, B., Park, J. O., Jang, J., Park, C., et al. (2011). Effect of simvastatin on cetuximab resistance in human colorectal cancer with KRAS mutations. J. Natl. Cancer Inst. 103, 674–688. doi:10.1093/jnci/djr070
Leng, X., Yang, J., Liu, T., Zhao, C., Cao, Z., Li, C., et al. (2022). A bioinformatics framework to identify the biomarkers and potential drugs for the treatment of colorectal cancer. Front. Genet. 13, 1017539. doi:10.3389/fgene.2022.1017539
Li, J., Chen, K., Wang, F., Dai, W., Li, S., Feng, J., et al. (2017a). Methyl jasmonate leads to necrosis and apoptosis in hepatocellular carcinoma cells via inhibition of glycolysis and represses tumor growth in mice. Oncotarget 8, 45965–45980. doi:10.18632/oncotarget.17469
Li, J., Van Vranken, J. G., Pontano Vaites, L., Schweppe, D. K., Huttlin, E. L., Etienne, C., et al. (2020a). TMTpro reagents: a set of isobaric labeling mass tags enables simultaneous proteome-wide measurements across 16 samples. Nat. Methods 17, 399–404. doi:10.1038/s41592-020-0781-4
Li, J., Xu, R., Qin, S., Liu, T., Pan, H., Xu, J., et al. (2018a). Aflibercept plus FOLFIRI in Asian patients with pretreated metastatic colorectal cancer: a randomized Phase III study. Future Oncol. 14, 2031–2044. doi:10.2217/fon-2017-0669
Li, M., Fisher, W. E., Kim, H. J., Wang, X., Brunicardi, C. F., Chen, C., et al. (2005). Somatostatin, somatostatin receptors, and pancreatic cancer. World J. Surg. 29, 293–296. doi:10.1007/s00268-004-7814-5
Li, M., and Xu, H. (2019). Fear stress enhanced xenograft pancreatic tumor growth through activating epithelial-mesenchymal transition. Pancreatology 19, 377–382. doi:10.1016/j.pan.2019.01.002
Li, N., Wu, P., Shen, Y., Yang, C., Zhang, L., Chen, Y., et al. (2021a). Predictions of mortality related to four major cancers in China, 2020 to 2030. Cancer Commun. 41, 404–413. doi:10.1002/cac2.12143
Li, S., Li, J., Dai, W., Zhang, Q., Feng, J., Wu, L., et al. (2017b). Genistein suppresses aerobic glycolysis and induces hepatocellular carcinoma cell death. Br. J. Cancer 117, 1518–1528. doi:10.1038/bjc.2017.323
Li, X., Francies, H. E., Secrier, M., Perner, J., Miremadi, A., Galeano-Dalmau, N., et al. (2018). Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat. Commun. 9, 2983. doi:10.1038/s41467-018-05190-9
Li, Y., Li, Y., Li, D., Li, K., Quan, Z., Wang, Z., et al. (2020b). Repositioning of hypoglycemic drug linagliptin for cancer treatment. Front. Pharmacol. 11, 187. doi:10.3389/fphar.2020.00187
Li, Y. Y., and Jones, S. J. (2012). Drug repositioning for personalized medicine. Genome Med. 4, 27. doi:10.1186/gm326
Li, Z., Zou, L., Xiao, Z.-X., and Yang, J. (2022). Transcriptome-based drug repositioning identifies TPCA-1 as a potential selective inhibitor of esophagus squamous carcinoma cell viability. Int. J. Mol. Med. 49, 75. doi:10.3892/ijmm.2022.5131
Liao, P., Song, K., Zhu, Z., Liu, Z., Zhang, W., Li, W., et al. (2020). Propranolol suppresses the growth of colorectal cancer through simultaneously activating autologous CD8+ T cells and inhibiting tumor AKT/MAPK pathway. Clin. Pharmacol. Ther. 108, 606–615. doi:10.1002/cpt.1894
Lim, H., He, D., Qiu, Y., Krawczuk, P., Sun, X., and Xie, L. (2019). Rational discovery of dual-indication multi-target PDE/Kinase inhibitor for precision anti-cancer therapy using structural systems pharmacology. PLoS Comput. Biol. 15, e1006619. doi:10.1371/journal.pcbi.1006619
Lin, C.-C., Suen, K. M., Stainthorp, A., Wieteska, L., Biggs, G. S., Leitão, A., et al. (2019). Targeting the Shc-EGFR interaction with indomethacin inhibits MAP kinase pathway signalling. Cancer Lett. 457, 86–97. doi:10.1016/j.canlet.2019.05.008
Liñares-Blanco, J., Munteanu, C. R., Pazos, A., and Fernandez-Lozano, C. (2020). Molecular docking and machine learning analysis of Abemaciclib in colon cancer. BMC Mol. Cell Biol. 21, 52. doi:10.1186/s12860-020-00295-w
Liu, H., Tao, H., Wang, H., Yang, Y., Yang, R., Dai, X., et al. (2020a). Doxycycline inhibits cancer stem cell-like properties via PAR1/FAK/PI3K/AKT pathway in pancreatic cancer. Front. Oncol. 10, 619317. doi:10.3389/fonc.2020.619317
Liu, P.-F., Tsai, K.-L., Hsu, C.-J., Tsai, W.-L., Cheng, J.-S., Chang, H.-W., et al. (2018). Drug repurposing screening identifies tioconazole as an ATG4 inhibitor that suppresses autophagy and sensitizes cancer cells to chemotherapy. Theranostics 8, 830–845. doi:10.7150/thno.22012
Liu, S.-H., Yu, J., Creeden, J. F., Sutton, J. M., Markowiak, S., Sanchez, R., et al. (2020b). Repurposing metformin, simvastatin and digoxin as a combination for targeted therapy for pancreatic ductal adenocarcinoma. Cancer Lett. 491, 97–107. doi:10.1016/j.canlet.2020.08.002
Liu, T., Li, S., Wu, L., Yu, Q., Li, J., Feng, J., et al. (2020c). Experimental study of hepatocellular carcinoma treatment by shikonin through regulating PKM2. J. Hepatocell. Carcinoma 7, 19–31. doi:10.2147/JHC.S237614
Liu, X., He, M., Li, L., Wang, X., Han, S., Zhao, J., et al. (2021). EMT and cancer cell stemness associated with chemotherapeutic resistance in esophageal cancer. Front. Oncol. 11, 672222. doi:10.3389/fonc.2021.672222
Long, Q., Ao, L., Li, K., and Li, Y. (2020). The efficacy and safety of sulindac for colorectal polyps: a protocol for systematic review and meta-analysis. Medicine 99, e22402. doi:10.1097/MD.0000000000022402
Low, Y. S., Daugherty, A. C., Schroeder, E. A., Chen, W., Seto, T., Weber, S., et al. (2017). Synergistic drug combinations from electronic health records and gene expression. J. Am. Med. Inf. Assoc. 24, 565–576. doi:10.1093/jamia/ocw161
Lübbert, M., Grishina, O., Schmoor, C., Schlenk, R. F., Jost, E., Crysandt, M., et al. (2020). Valproate and retinoic acid in combination with decitabine in elderly nonfit patients with acute myeloid leukemia: results of a multicenter, randomized, 2 × 2, phase II trial. J. Clin. Oncol. 38, 257–270. doi:10.1200/JCO.19.01053
Luhn, P., Kuk, D., Carrigan, G., Nussbaum, N., Sorg, R., Rohrer, R., et al. (2019). Validation of diagnosis codes to identify side of colon in an electronic health record registry. BMC Med. Res. Methodol. 19, 177. doi:10.1186/s12874-019-0824-7
Luo, Y., Zhao, X., Zhou, J., Yang, J., Zhang, Y., Kuang, W., et al. (2017). A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat. Commun. 8, 573. doi:10.1038/s41467-017-00680-8
Ma, M. K. F., Lau, E. Y. T., Leung, D. H. W., Lo, J., Ho, N. P. Y., Cheng, L. K. W., et al. (2017). Stearoyl-CoA desaturase regulates sorafenib resistance via modulation of ER stress-induced differentiation. J. Hepatol. 67, 979–990. doi:10.1016/j.jhep.2017.06.015
Mao, Y., Wang, W., Yang, J., Zhou, X., Lu, Y., Gao, J., et al. (2023). Drug repurposing screening and mechanism analysis based on human colorectal cancer organoids. Protein Cell, pwad038. doi:10.1093/procel/pwad038
Martinez Molina, D., Jafari, R., Ignatushchenko, M., Seki, T., Larsson, E. A., Dan, C., et al. (2013). Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87. doi:10.1126/science.1233606
Martinez-Pacheco, S., and O’Driscoll, L. (2021). Pre-clinical in vitro models used in cancer research: results of a worldwide survey. Cancers 13, 6033. doi:10.3390/cancers13236033
Massó-Vallés, D., Jauset, T., Serrano, E., Sodir, N. M., Pedersen, K., Affara, N. I., et al. (2015). Ibrutinib exerts potent antifibrotic and antitumor activities in mouse models of pancreatic adenocarcinoma. Cancer Res. 75, 1675–1681. doi:10.1158/0008-5472.CAN-14-2852
Mateus, A., Kurzawa, N., Becher, I., Sridharan, S., Helm, D., Stein, F., et al. (2020). Thermal proteome profiling for interrogating protein interactions. Mol. Syst. Biol. 16, e9232. doi:10.15252/msb.20199232
Mazumder, S., De, R., Debsharma, S., Bindu, S., Maity, P., Sarkar, S., et al. (2019). Indomethacin impairs mitochondrial dynamics by activating the PKCζ-p38-DRP1 pathway and inducing apoptosis in gastric cancer and normal mucosal cells. J. Biol. Chem. 294, 8238–8258. doi:10.1074/jbc.RA118.004415
McGuigan, A., Kelly, P., Turkington, R. C., Jones, C., Coleman, H. G., and McCain, R. S. (2018). Pancreatic cancer: a review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 24, 4846–4861. doi:10.3748/wjg.v24.i43.4846
Meyn, R. E., Krishnan, S., and Skinner, H. D. (2016). Everything old is new again: using nelfinavir to radiosensitize rectal cancer. Clin. Cancer Res. 22, 1834–1836. doi:10.1158/1078-0432.CCR-16-0024
Midgley, R. S., McConkey, C. C., Johnstone, E. C., Dunn, J. A., Smith, J. L., Grumett, S. A., et al. (2010). Phase III randomized trial assessing rofecoxib in the adjuvant setting of colorectal cancer: final results of the VICTOR trial. J. Clin. Oncol. 28, 4575–4580. doi:10.1200/JCO.2010.29.6244
Miller, K. D., Siegel, R. L., Lin, C. C., Mariotto, A. B., Kramer, J. L., Rowland, J. H., et al. (2016). Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 66, 271–289. doi:10.3322/caac.21349
Miriovsky, B. J., Shulman, L. N., and Abernethy, A. P. (2012). Importance of health information technology, electronic health records, and continuously aggregating data to comparative effectiveness research and learning health care. J. Clin. Oncol. 30, 4243–4248. doi:10.1200/JCO.2012.42.8011
Mitrović, A., and Kos, J. (2019). Nitroxoline: repurposing its antimicrobial to antitumor application. Acta Biochim. Pol. 66, 521–531. doi:10.18388/abp.2019_2904
Mizutani, Y., Iida, T., Ohno, E., Ishikawa, T., Kinoshita, F., Kuwatsuka, Y., et al. (2022). Safety and efficacy of MIKE-1 in patients with advanced pancreatic cancer: a study protocol for an open-label phase I/II investigator-initiated clinical trial based on a drug repositioning approach that reprograms the tumour stroma. BMC Cancer 22, 205. doi:10.1186/s12885-022-09272-2
Mizutani, Y., Kobayashi, H., Iida, T., Asai, N., Masamune, A., Hara, A., et al. (2019). Meflin-positive cancer-associated fibroblasts inhibit pancreatic carcinogenesis. Cancer Res. 79, 5367–5381. doi:10.1158/0008-5472.CAN-19-0454
Moffat, J. G., Rudolph, J., and Bailey, D. (2014). Phenotypic screening in cancer drug discovery - past, present and future. Nat. Rev. Drug Discov. 13, 588–602. doi:10.1038/nrd4366
Moffat, J. G., Vincent, F., Lee, J. A., Eder, J., and Prunotto, M. (2017). Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov. 16, 531–543. doi:10.1038/nrd.2017.111
Monticelli, M., Liguori, L., Allocca, M., Bosso, A., Andreotti, G., Lukas, J., et al. (2022). Drug repositioning for Fabry disease: acetylsalicylic acid potentiates the stabilization of lysosomal alpha-galactosidase by pharmacological chaperones. Int. J. Mol. Sci. 23, 5105. doi:10.3390/ijms23095105
Moore, M. J., Goldstein, D., Hamm, J., Figer, A., Hecht, J. R., Gallinger, S., et al. (2007). Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol. 25, 1960–1966. doi:10.1200/JCO.2006.07.9525
Morgan, S., Grootendorst, P., Lexchin, J., Cunningham, C., and Greyson, D. (2011). The cost of drug development: a systematic review. Health Policy 100, 4–17. doi:10.1016/j.healthpol.2010.12.002
Morton, C. L., and Houghton, P. J. (2007). Establishment of human tumor xenografts in immunodeficient mice. Nat. Protoc. 2, 247–250. doi:10.1038/nprot.2007.25
Mottini, C., Napolitano, F., Li, Z., Gao, X., and Cardone, L. (2021). Computer-aided drug repurposing for cancer therapy: approaches and opportunities to challenge anticancer targets. Semin. Cancer Biol. 68, 59–74. doi:10.1016/j.semcancer.2019.09.023
Mugiyanto, E., Adikusuma, W., Irham, L. M., Huang, W.-C., Chang, W.-C., and Kuo, C.-N. (2022). Integrated genomic analysis to identify druggable targets for pancreatic cancer. Front. Oncol. 12, 989077. doi:10.3389/fonc.2022.989077
Mühl, H., Paulukat, J., Höfler, S., Hellmuth, M., Franzen, R., and Pfeilschifter, J. (2004). The HIV protease inhibitor ritonavir synergizes with butyrate for induction of apoptotic cell death and mediates expression of heme oxygenase-1 in DLD-1 colon carcinoma cells. Br. J. Pharmacol. 143, 890–898. doi:10.1038/sj.bjp.0706023
Munoz, L. (2017). Non-kinase targets of protein kinase inhibitors. Nat. Rev. Drug Discov. 16, 424–440. doi:10.1038/nrd.2016.266
Murphy, J. E., Wo, J. Y., Ryan, D. P., Clark, J. W., Jiang, W., Yeap, B. Y., et al. (2019). Total neoadjuvant therapy with FOLFIRINOX in combination with losartan followed by chemoradiotherapy for locally advanced pancreatic cancer: a phase 2 clinical trial. JAMA Oncol. 5, 1020–1027. doi:10.1001/jamaoncol.2019.0892
Mussin, N., Oh, S. C., Lee, K. W., Park, M. Y., Seo, S., Yi, N. J., et al. (2017). Sirolimus and metformin synergistically inhibits colon cancer in vitro and in vivo. J. Korean Med. Sci. 32, 1385–1395. doi:10.3346/jkms.2017.32.9.1385
Nagaraj, A. B., Wang, Q. Q., Joseph, P., Zheng, C., Chen, Y., Kovalenko, O., et al. (2018). Using a novel computational drug-repositioning approach (DrugPredict) to rapidly identify potent drug candidates for cancer treatment. Oncogene 37, 403–414. doi:10.1038/onc.2017.328
Nair, V., Sreevalsan, S., Basha, R., Abdelrahim, M., Abudayyeh, A., Rodrigues Hoffman, A., et al. (2014). Mechanism of metformin-dependent inhibition of mammalian target of rapamycin (mTOR) and Ras activity in pancreatic cancer: role of specificity protein (Sp) transcription factors. J. Biol. Chem. 289, 27692–27701. doi:10.1074/jbc.M114.592576
Nakayama, G., Tanaka, C., and Kodera, Y. (2013). Current options for the diagnosis, staging and therapeutic management of colorectal cancer. Gastrointest. Tumors 1, 25–32. doi:10.1159/000354995
Nambara, S., Masuda, T., Nishio, M., Kuramitsu, S., Tobo, T., Ogawa, Y., et al. (2017). Antitumor effects of the antiparasitic agent ivermectin via inhibition of Yes-associated protein 1 expression in gastric cancer. Oncotarget 8, 107666–107677. doi:10.18632/oncotarget.22587
Neo, J. H., Malcontenti-Wilson, C., Muralidharan, V., and Christophi, C. (2007). Effect of ACE inhibitors and angiotensin II receptor antagonists in a mouse model of colorectal cancer liver metastases. J. Gastroenterol. Hepatol. 22, 577–584. doi:10.1111/j.1440-1746.2006.04797.x
Nero, T. L., Morton, C. J., Holien, J. K., Wielens, J., and Parker, M. W. (2014). Oncogenic protein interfaces: small molecules, big challenges. Nat. Rev. Cancer 14, 248–262. doi:10.1038/nrc3690
Nuevo-Tapioles, C., Santacatterina, F., Stamatakis, K., Núñez de Arenas, C., Gómez de Cedrón, M., Formentini, L., et al. (2020). Coordinate β-adrenergic inhibition of mitochondrial activity and angiogenesis arrest tumor growth. Nat. Commun. 11, 3606. doi:10.1038/s41467-020-17384-1
Nygren, P., Fryknäs, M., Agerup, B., and Larsson, R. (2013). Repositioning of the anthelmintic drug mebendazole for the treatment for colon cancer. J. Cancer Res. Clin. Oncol. 139, 2133–2140. doi:10.1007/s00432-013-1539-5
Nygren, P., and Larsson, R. (2014). Drug repositioning from bench to bedside: tumour remission by the antihelmintic drug mebendazole in refractory metastatic colon cancer. Acta Oncol. 53, 427–428. doi:10.3109/0284186X.2013.844359
Ogura, S., Yoshida, Y., Kurahashi, T., Egawa, M., Furuta, K., Kiso, S., et al. (2018). Targeting the mevalonate pathway is a novel therapeutic approach to inhibit oncogenic FoxM1 transcription factor in human hepatocellular carcinoma. Oncotarget 9, 21022–21035. doi:10.18632/oncotarget.24781
Okada, J., Yamada, E., Saito, T., Yokoo, H., Osaki, A., Shimoda, Y., et al. (2020). Dapagliflozin inhibits cell adhesion to collagen I and IV and increases ectodomain proteolytic cleavage of DDR1 by increasing ADAM10 activity. Molecules 25, 495. doi:10.3390/molecules25030495
Oliveira, L. F. S., Predes, D., Borges, H. L., and Abreu, J. G. (2022). Therapeutic potential of naturally occurring small molecules to target the wnt/β-catenin signaling pathway in colorectal cancer. Cancers 14, 403. doi:10.3390/cancers14020403
Onaciu, A., Munteanu, R., Munteanu, V. C., Gulei, D., Raduly, L., Feder, R.-I., et al. (2020). Spontaneous and induced animal models for cancer research. Diagn. (Basel) 10, 660. doi:10.3390/diagnostics10090660
Onoda, T., Ono, T., Dhar, D. K., Yamanoi, A., Fujii, T., and Nagasue, N. (2004). Doxycycline inhibits cell proliferation and invasive potential: combination therapy with cyclooxygenase-2 inhibitor in human colorectal cancer cells. J. Lab. Clin. Med. 143, 207–216. doi:10.1016/j.lab.2003.12.012
Osada, T., Chen, M., Yang, X. Y., Spasojevic, I., Vandeusen, J. B., Hsu, D., et al. (2011). Antihelminth compound niclosamide downregulates Wnt signaling and elicits antitumor responses in tumors with activating APC mutations. Cancer Res. 71, 4172–4182. doi:10.1158/0008-5472.CAN-10-3978
Overman, M., Javle, M., Davis, R. E., Vats, P., Kumar-Sinha, C., Xiao, L., et al. (2020). Randomized phase II study of the Bruton tyrosine kinase inhibitor acalabrutinib, alone or with pembrolizumab in patients with advanced pancreatic cancer. J. Immunother. Cancer 8, e000587. doi:10.1136/jitc-2020-000587
Palve, V., Liao, Y., Remsing Rix, L. L., and Rix, U. (2021). Turning liabilities into opportunities: off-target based drug repurposing in cancer. Semin. Cancer Biol. 68, 209–229. doi:10.1016/j.semcancer.2020.02.003
Pantziarka, P., Verbaanderd, C., Sukhatme, V., Rica Capistrano, I., Crispino, S., Gyawali, B., et al. (2018). ReDO_DB: the repurposing drugs in oncology database. Ecancermedicalscience 12, 886. doi:10.3332/ecancer.2018.886
Parikh, A. R., Leshchiner, I., Elagina, L., Goyal, L., Levovitz, C., Siravegna, G., et al. (2019). Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 25, 1415–1421. doi:10.1038/s41591-019-0561-9
Park, W., Amin, A. R. M. R., Chen, Z. G., and Shin, D. M. (2013). New perspectives of curcumin in cancer prevention. Cancer Prev. Res. 6, 387–400. doi:10.1158/1940-6207.CAPR-12-0410
Patel, M. M., and Patel, B. M. (2018). Repurposing of sodium valproate in colon cancer associated with diabetes mellitus: role of HDAC inhibition. Eur. J. Pharm. Sci. 121, 188–199. doi:10.1016/j.ejps.2018.05.026
Perrin, J., Werner, T., Kurzawa, N., Rutkowska, A., Childs, D. D., Kalxdorf, M., et al. (2020). Identifying drug targets in tissues and whole blood with thermal-shift profiling. Nat. Biotechnol. 38, 303–308. doi:10.1038/s41587-019-0388-4
Perrone, F., and Zilbauer, M. (2021). Biobanking of human gut organoids for translational research. Exp. Mol. Med. 53, 1451–1458. doi:10.1038/s12276-021-00606-x
Petroni, G., Bagni, G., Iorio, J., Duranti, C., Lottini, T., Stefanini, M., et al. (2020). Clarithromycin inhibits autophagy in colorectal cancer by regulating the hERG1 potassium channel interaction with PI3K. Cell Death Dis. 11, 161. doi:10.1038/s41419-020-2349-8
Pham, T.-H., Qiu, Y., Liu, J., Zimmer, S., O’Neill, E., Xie, L., et al. (2022). Chemical-induced gene expression ranking and its application to pancreatic cancer drug repurposing. Patterns (N Y) 3, 100441. doi:10.1016/j.patter.2022.100441
Pintova, S., Dharmupari, S., Moshier, E., Zubizarreta, N., Ang, C., and Holcombe, R. F. (2019). Genistein combined with FOLFOX or FOLFOX-Bevacizumab for the treatment of metastatic colorectal cancer: phase I/II pilot study. Cancer Chemother. Pharmacol. 84, 591–598. doi:10.1007/s00280-019-03886-3
Ponzini, F. M., Schultz, C. W., Leiby, B. E., Cannaday, S., Yeo, T., Posey, J., et al. (2023). Repurposing the FDA-approved anthelmintic pyrvinium pamoate for pancreatic cancer treatment: study protocol for a phase I clinical trial in early-stage pancreatic ductal adenocarcinoma. BMJ Open 13, e073839. doi:10.1136/bmjopen-2023-073839
Prasad, K., and Kumar, V. (2021). Artificial intelligence-driven drug repurposing and structural biology for SARS-CoV-2. Curr. Res. Pharmacol. Drug Discov. 2, 100042. doi:10.1016/j.crphar.2021.100042
Pritchard, J.-L. E., O’Mara, T. A., and Glubb, D. M. (2017). Enhancing the promise of drug repositioning through genetics. Front. Pharmacol. 8, 896. doi:10.3389/fphar.2017.00896
Pushpakom, S., Iorio, F., Eyers, P. A., Escott, K. J., Hopper, S., Wells, A., et al. (2019). Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 18, 41–58. doi:10.1038/nrd.2018.168
Qiao, X., Wang, X., Shang, Y., Li, Y., and Chen, S.-Z. (2018). Azithromycin enhances anticancer activity of TRAIL by inhibiting autophagy and up-regulating the protein levels of DR4/5 in colon cancer cells in vitro and in vivo. Cancer Commun. 38, 43. doi:10.1186/s40880-018-0309-9
Qorri, B., Mokhtari, R. B., Harless, W. W., and Szewczuk, M. R. (2022). Next generation of cancer drug repurposing: therapeutic combination of aspirin and oseltamivir phosphate potentiates gemcitabine to disable key survival pathways critical for pancreatic cancer progression. Cancers 14, 1374. doi:10.3390/cancers14061374
Rabben, H.-L., Andersen, G. T., Ianevski, A., Olsen, M. K., Kainov, D., Grønbech, J. E., et al. (2021a). Computational drug repositioning and experimental validation of ivermectin in treatment of gastric cancer. Front. Pharmacol. 12, 625991. doi:10.3389/fphar.2021.625991
Rabben, H.-L., Kodama, Y., Nakamura, M., Bones, A. M., Wang, T. C., Chen, D., et al. (2021b). Chemopreventive effects of dietary isothiocyanates in animal models of gastric cancer and synergistic anticancer effects with cisplatin in human gastric cancer cells. Front. Pharmacol. 12, 613458. doi:10.3389/fphar.2021.613458
Ramachandran, S., and Srivastava, S. K. (2020). Repurposing pimavanserin, an anti-Parkinson drug for pancreatic cancer therapy. Mol. Ther. Oncolytics 19, 19–32. doi:10.1016/j.omto.2020.08.019
Ranjan, A., German, N., Mikelis, C., Srivenugopal, K., and Srivastava, S. K. (2017). Penfluridol induces endoplasmic reticulum stress leading to autophagy in pancreatic cancer. Tumour Biol. 39, 1010428317705517. doi:10.1177/1010428317705517
Raziq, K., Cai, M., Dong, K., Wang, P., Afrifa, J., and Fu, S. (2020). Competitive endogenous network of lncRNA, miRNA, and mRNA in the chemoresistance of gastrointestinal tract adenocarcinomas. Biomed. Pharmacother. 130, 110570. doi:10.1016/j.biopha.2020.110570
Rebelo, R., Polónia, B., Santos, L. L., Vasconcelos, M. H., and Xavier, C. P. R. (2021). Drug repurposing opportunities in pancreatic ductal adenocarcinoma. Pharmaceuticals 14, 280. doi:10.3390/ph14030280
Regan-Fendt, K., Li, D., Reyes, R., Yu, L., Wani, N. A., Hu, P., et al. (2020). Transcriptomics-based drug repurposing approach identifies novel drugs against sorafenib-resistant hepatocellular carcinoma. Cancers 12, 2730. doi:10.3390/cancers12102730
Ribeiro, E., Araújo, D., Pereira, M., Lopes, B., Sousa, P., Sousa, A. C., et al. (2023). Repurposing benztropine, natamycin, and nitazoxanide using drug combination and characterization of gastric cancer cell lines. Biomedicines 11, 799. doi:10.3390/biomedicines11030799
Richmond, A., and Su, Y. (2008). Mouse xenograft models vs GEM models for human cancer therapeutics. Dis. Model. Mech. 1, 78–82. doi:10.1242/dmm.000976
Ridges, S., Heaton, W. L., Joshi, D., Choi, H., Eiring, A., Batchelor, L., et al. (2012). Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood 119, 5621–5631. doi:10.1182/blood-2011-12-398818
Rios Perez, M. V., Roife, D., Dai, B., Pratt, M., Dobrowolski, R., Kang, Y., et al. (2019). Antineoplastic effects of auranofin in human pancreatic adenocarcinoma preclinical models. Surg. Open Sci. 1, 56–63. doi:10.1016/j.sopen.2019.05.004
Ripani, P., Delp, J., Bode, K., Delgado, M. E., Dietrich, L., Betzler, V. M., et al. (2020). Thiazolides promote G1 cell cycle arrest in colorectal cancer cells by targeting the mitochondrial respiratory chain. Oncogene 39, 2345–2357. doi:10.1038/s41388-019-1142-6
Rithanya, P., and Ezhilarasan, D. (2021). Sodium valproate, a histone deacetylase inhibitor, provokes reactive oxygen species-mediated cytotoxicity in human hepatocellular carcinoma cells. J. Gastrointest. Cancer 52, 138–144. doi:10.1007/s12029-020-00370-7
Rose, J. S., and Bekaii-Saab, T. S. (2011). New developments in the treatment of metastatic gastric cancer: focus on trastuzumab. Onco. Targets. Ther. 4, 21–26. doi:10.2147/OTT.S10188
Rosenberg, D. W., Giardina, C., and Tanaka, T. (2009). Mouse models for the study of colon carcinogenesis. Carcinogenesis 30, 183–196. doi:10.1093/carcin/bgn267
Ruggeri, B. A., Camp, F., and Miknyoczki, S. (2014). Animal models of disease: pre-clinical animal models of cancer and their applications and utility in drug discovery. Biochem. Pharmacol. 87, 150–161. doi:10.1016/j.bcp.2013.06.020
Sadeghi, N., Abbruzzese, J. L., Yeung, S.-C. J., Hassan, M., and Li, D. (2012). Metformin use is associated with better survival of diabetic patients with pancreatic cancer. Clin. Cancer Res. 18, 2905–2912. doi:10.1158/1078-0432.CCR-11-2994
Sakamoto, K., and Maeda, S. (2010). Targeting NF-kappaB for colorectal cancer. Expert Opin. Ther. Targets 14, 593–601. doi:10.1517/14728221003769903
Sala, C., Dhar, N., Hartkoorn, R. C., Zhang, M., Ha, Y. H., Schneider, P., et al. (2010). Simple model for testing drugs against nonreplicating Mycobacterium tuberculosis. Mycobacterium Tuberc. Antimicrob. Agents Chemother. 54, 4150–4158. doi:10.1128/AAC.00821-10
Samaras, P., Tusup, M., Nguyen-Kim, T. D. L., Seifert, B., Bachmann, H., von Moos, R., et al. (2017). Phase I study of a chloroquine-gemcitabine combination in patients with metastatic or unresectable pancreatic cancer. Cancer Chemother. Pharmacol. 80, 1005–1012. doi:10.1007/s00280-017-3446-y
Sanomachi, T., Suzuki, S., Kuramoto, K., Takeda, H., Sakaki, H., Togashi, K., et al. (2017). Olanzapine, an atypical antipsychotic, inhibits survivin expression and sensitizes cancer cells to chemotherapeutic agents. Anticancer Res. 37, 6177–6188. doi:10.21873/anticanres.12067
Sanseau, P., Agarwal, P., Barnes, M. R., Pastinen, T., Richards, J. B., Cardon, L. R., et al. (2012). Use of genome-wide association studies for drug repositioning. Nat. Biotechnol. 30, 317–320. doi:10.1038/nbt.2151
Sarfstein, R., Friedman, Y., Attias-Geva, Z., Fishman, A., Bruchim, I., and Werner, H. (2013). Metformin downregulates the insulin/IGF-I signaling pathway and inhibits different uterine serous carcinoma (USC) cells proliferation and migration in p53-dependent or -independent manners. PLoS One 8, e61537. doi:10.1371/journal.pone.0061537
Savitski, M. M., Zinn, N., Faelth-Savitski, M., Poeckel, D., Gade, S., Becher, I., et al. (2018). Multiplexed proteome dynamics profiling reveals mechanisms controlling protein homeostasis. Cell 173, 260–274. doi:10.1016/j.cell.2018.02.030
Schultz, C. W., McCarthy, G. A., Nerwal, T., Nevler, A., DuHadaway, J. B., McCoy, M. D., et al. (2021). The FDA-approved anthelmintic pyrvinium pamoate inhibits pancreatic cancer cells in nutrient-depleted conditions by targeting the mitochondria. Mol. Cancer Ther. 20, 2166–2176. doi:10.1158/1535-7163.MCT-20-0652
Senkowski, W., Zhang, X., Olofsson, M. H., Isacson, R., Höglund, U., Gustafsson, M., et al. (2015). Three-dimensional cell culture-based screening identifies the anthelmintic drug nitazoxanide as a candidate for treatment of colorectal cancer. Mol. Cancer Ther. 14, 1504–1516. doi:10.1158/1535-7163.MCT-14-0792
Shafiee, G., Saidijam, M., Tavilani, H., Ghasemkhani, N., and Khodadadi, I. (2016). Genistein induces apoptosis and inhibits proliferation of HT29 colon cancer cells. Int. J. Mol. Cell Med. 5, 178–191.
Shameer, K., Readhead, B., and Dudley, J. T. (2015). Computational and experimental advances in drug repositioning for accelerated therapeutic stratification. Curr. Top. Med. Chem. 15, 5–20. doi:10.2174/1568026615666150112103510
Shantikumar, S., Satheeshkumar, N., and Srinivas, R. (2015). Pharmacokinetic and protein binding profile of peptidomimetic DPP-4 inhibitor - teneligliptin in rats using liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 1002, 194–200. doi:10.1016/j.jchromb.2015.08.023
Shen, P.-W., Chou, Y.-M., Li, C.-L., Liao, E.-C., Huang, H.-S., Yin, C.-H., et al. (2021). Itraconazole improves survival outcomes in patients with colon cancer by inducing autophagic cell death and inhibiting transketolase expression. Oncol. Lett. 22, 768. doi:10.3892/ol.2021.13029
Sherif, D. A., Makled, M. N., and Suddek, G. M. (2021). The HIV reverse transcriptase Inhibitor Tenofovir suppressed DMH/HFD-induced colorectal cancer in Wistar rats. Fundam. Clin. Pharmacol. 35, 940–954. doi:10.1111/fcp.12679
Shi, J., Zhou, L., Huang, H.-S., Peng, L., Xie, N., Nice, E., et al. (2022a). Repurposing oxiconazole against colorectal cancer via PRDX2-mediated autophagy arrest. Int. J. Biol. Sci. 18, 3747–3761. doi:10.7150/ijbs.70679
Shi, W., Li, C., Wartmann, T., Kahlert, C., Du, R., Perrakis, A., et al. (2022b). Sensory ion channel candidates inform on the clinical course of pancreatic cancer and present potential targets for repurposing of FDA-approved agents. J. Pers. Med. 12, 478. doi:10.3390/jpm12030478
Shi, X.-N., Li, H., Yao, H., Liu, X., Li, L., Leung, K.-S., et al. (2015). Adapalene inhibits the activity of cyclin-dependent kinase 2 in colorectal carcinoma. Mol. Med. Rep. 12, 6501–6508. doi:10.3892/mmr.2015.4310
Shitara, K., Doi, T., Nagano, O., Imamura, C. K., Ozeki, T., Ishii, Y., et al. (2017). Dose-escalation study for the targeting of CD44v+ cancer stem cells by sulfasalazine in patients with advanced gastric cancer (EPOC1205). Gastric Cancer 20, 341–349. doi:10.1007/s10120-016-0610-8
Simian, M., and Bissell, M. J. (2017). Organoids: a historical perspective of thinking in three dimensions. J. Cell Biol. 216, 31–40. doi:10.1083/jcb.201610056
Sinn, D. H., Choi, G.-S., Park, H. C., Kim, J. M., Kim, H., Song, K. D., et al. (2019). Multidisciplinary approach is associated with improved survival of hepatocellular carcinoma patients. PLoS One 14, e0210730. doi:10.1371/journal.pone.0210730
Sivaraman, A., Leach, J. K., Townsend, S., Iida, T., Hogan, B. J., Stolz, D. B., et al. (2005). A microscale in vitro physiological model of the liver: predictive screens for drug metabolism and enzyme induction. Curr. Drug Metab. 6, 569–591. doi:10.2174/138920005774832632
Sloane, D. A., Trikic, M. Z., Chu, M. L. H., Lamers, M. B. A. C., Mason, C. S., Mueller, I., et al. (2010). Drug-resistant aurora A mutants for cellular target validation of the small molecule kinase inhibitors MLN8054 and MLN8237. ACS Chem. Biol. 5, 563–576. doi:10.1021/cb100053q
Smith, S. B., Dampier, W., Tozeren, A., Brown, J. R., and Magid-Slav, M. (2012). Identification of common biological pathways and drug targets across multiple respiratory viruses based on human host gene expression analysis. PLoS One 7, e33174. doi:10.1371/journal.pone.0033174
Sobko, T., Reinders, C. I., Jansson, E., Norin, E., Midtvedt, T., and Lundberg, J. O. (2005). Gastrointestinal bacteria generate nitric oxide from nitrate and nitrite. Nitric Oxide 13, 272–278. doi:10.1016/j.niox.2005.08.002
Son, D.-S., Lee, E.-S., and Adunyah, S. E. (2020). The antitumor potentials of benzimidazole anthelmintics as repurposing drugs. Immune Netw. 20, e29. doi:10.4110/in.2020.20.e29
Son, K., Fujioka, S., Iida, T., Furukawa, K., Fujita, T., Yamada, H., et al. (2009). Doxycycline induces apoptosis in PANC-1 pancreatic cancer cells. Anticancer Res. 29, 3995–4003.
Stastna, M., Janeckova, L., Hrckulak, D., Kriz, V., and Korinek, V. (2019). Human colorectal cancer from the perspective of mouse models. Genes 10, 788. doi:10.3390/genes10100788
Stewart, A. K., Jacobus, S., Fonseca, R., Weiss, M., Callander, N. S., Chanan-Khan, A. A., et al. (2015). Melphalan, prednisone, and thalidomide vs melphalan, prednisone, and lenalidomide (ECOG E1A06) in untreated multiple myeloma. Blood 126, 1294–1301. doi:10.1182/blood-2014-12-613927
Streicher, S. A., Yu, H., Lu, L., Kidd, M. S., and Risch, H. A. (2014). Case-control study of aspirin use and risk of pancreatic cancer. Cancer Epidemiol. Biomarkers Prev. 23, 1254–1263. doi:10.1158/1055-9965.EPI-13-1284
Sud, A., Kinnersley, B., and Houlston, R. S. (2017). Genome-wide association studies of cancer: current insights and future perspectives. Nat. Rev. Cancer 17, 692–704. doi:10.1038/nrc.2017.82
Sugarbaker, P. H. (2005). Strategies for the prevention and treatment of peritoneal carcinomatosis from gastrointestinal cancer. Cancer Invest. 23, 155–172. doi:10.1081/cnv-50478
Sun, W., Weingarten, R. A., Xu, M., Southall, N., Dai, S., Shinn, P., et al. (2016). Rapid antimicrobial susceptibility test for identification of new therapeutics and drug combinations against multidrug-resistant bacteria. Emerg. Microbes Infect. 5, e116. doi:10.1038/emi.2016.123
Sun, X., Sun, G., Huang, Y., Hao, Y., Tang, X., Zhang, N., et al. (2020). 3-Bromopyruvate regulates the status of glycolysis and BCNU sensitivity in human hepatocellular carcinoma cells. Biochem. Pharmacol. 177, 113988. doi:10.1016/j.bcp.2020.113988
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Tabatabai, E., Khazaei, M., Asgharzadeh, F., Nazari, S. E., Shakour, N., Fiuji, H., et al. (2021). Inhibition of angiotensin II type 1 receptor by candesartan reduces tumor growth and ameliorates fibrosis in colorectal cancer. EXCLI J. 20, 863–878. doi:10.17179/excli2021-3421
Takahashi, M., Kobayashi, H., Mizutani, Y., Hara, A., Iida, T., Miyai, Y., et al. (2021). Roles of the mesenchymal stromal/stem cell marker meflin/islr in cancer fibrosis. Front. Cell Dev. Biol. 9, 749924. doi:10.3389/fcell.2021.749924
Tan, X.-P., He, Y., Yang, J., Wei, X., Fan, Y.-L., Zhang, G.-G., et al. (2023). Blockade of NMT1 enzymatic activity inhibits N-myristoylation of VILIP3 protein and suppresses liver cancer progression. Signal Transduct. Target Ther. 8, 14. doi:10.1038/s41392-022-01248-9
Teixeira Farinha, H., Digklia, A., Schizas, D., Demartines, N., Schäfer, M., and Mantziari, S. (2022). Immunotherapy for esophageal cancer: state-of-the art in 2021. Cancers 14, 554. doi:10.3390/cancers14030554
Tentler, J. J., Tan, A. C., Weekes, C. D., Jimeno, A., Leong, S., Pitts, T. M., et al. (2012). Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 9, 338–350. doi:10.1038/nrclinonc.2012.61
Tomi-Andrino, C., Pandele, A., Winzer, K., King, J., Rahman, R., and Kim, D.-H. (2022). Metabolic modeling-based drug repurposing in Glioblastoma. Sci. Rep. 12, 11189. doi:10.1038/s41598-022-14721-w
Tomita, H., Takaishi, S., Menheniott, T. R., Yang, X., Shibata, W., Jin, G., et al. (2011). Inhibition of gastric carcinogenesis by the hormone gastrin is mediated by suppression of TFF1 epigenetic silencing. Gastroenterology 140, 879–891. doi:10.1053/j.gastro.2010.11.037
Toschi, E., Sgadari, C., Malavasi, L., Bacigalupo, I., Chiozzini, C., Carlei, D., et al. (2011). Human immunodeficiency virus protease inhibitors reduce the growth of human tumors via a proteasome-independent block of angiogenesis and matrix metalloproteinases. Int. J. Cancer 128, 82–93. doi:10.1002/ijc.25550
Tseng, C.-N., Huang, C.-F., Cho, C.-L., Chang, H.-W., Huang, C.-W., Chiu, C.-C., et al. (2013). Brefeldin a effectively inhibits cancer stem cell-like properties and MMP-9 activity in human colorectal cancer Colo 205 cells. Molecules 18, 10242–10253. doi:10.3390/molecules180910242
Vacante, F., Senesi, P., Montesano, A., Paini, S., Luzi, L., and Terruzzi, I. (2019). Metformin counteracts HCC progression and metastasis enhancing KLF6/p21 expression and downregulating the IGF Axis. Int. J. Endocrinol. 2019, 7570146. doi:10.1155/2019/7570146
Valastyan, S., and Weinberg, R. A. (2011). Tumor metastasis: molecular insights and evolving paradigms. Cell 147, 275–292. doi:10.1016/j.cell.2011.09.024
van de Wetering, M., Francies, H. E., Francis, J. M., Bounova, G., Iorio, F., Pronk, A., et al. (2015). Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945. doi:10.1016/j.cell.2015.03.053
Verduin, M., Hoeben, A., De Ruysscher, D., and Vooijs, M. (2021). Patient-derived cancer organoids as predictors of treatment response. Front. Oncol. 11, 641980. doi:10.3389/fonc.2021.641980
Veschi, S., De Lellis, L., Florio, R., Lanuti, P., Massucci, A., Tinari, N., et al. (2018). Effects of repurposed drug candidates nitroxoline and nelfinavir as single agents or in combination with erlotinib in pancreatic cancer cells. J. Exp. Clin. Cancer Res. 37, 236. doi:10.1186/s13046-018-0904-2
Veschi, S., Ronci, M., Lanuti, P., De Lellis, L., Florio, R., Bologna, G., et al. (2020). Integrative proteomic and functional analyses provide novel insights into the action of the repurposed drug candidate nitroxoline in AsPC-1 cells. Sci. Rep. 10, 2574. doi:10.1038/s41598-020-59492-4
Vickers, N. J. (2017). Animal communication: when I’m calling you, will you answer too? Curr. Biol. 27, R713–R715. doi:10.1016/j.cub.2017.05.064
Villarruel-Melquiades, F., Hernandez-Gallegos, E., Solano-Agama, C., Mendoza-Garrido, M. E., and Camacho, J. (2023). The combination sorafenib-raloxifene-loratadine as a novel potential therapeutic approach against human liver cancer. Vivo 37, 1156–1163. doi:10.21873/invivo.13190
Walrath, J. C., Hawes, J. J., Van Dyke, T., and Reilly, K. M. (2010). Genetically engineered mouse models in cancer research. Adv. Cancer Res. 106, 113–164. doi:10.1016/S0065-230X(10)06004-5
Wang, J., Ren, X.-R., Piao, H., Zhao, S., Osada, T., Premont, R. T., et al. (2019). Niclosamide-induced Wnt signaling inhibition in colorectal cancer is mediated by autophagy. Biochem. J. 476, 535–546. doi:10.1042/BCJ20180385
Wang, S.-T., Ho, H. J., Lin, J.-T., Shieh, J.-J., and Wu, C.-Y. (2017). Simvastatin-induced cell cycle arrest through inhibition of STAT3/SKP2 axis and activation of AMPK to promote p27 and p21 accumulation in hepatocellular carcinoma cells. Cell Death Dis. 8, e2626. doi:10.1038/cddis.2016.472
Wang, W., McLeod, H. L., and Cassidy, J. (2003). Disulfiram-mediated inhibition of NF-kappaB activity enhances cytotoxicity of 5-fluorouracil in human colorectal cancer cell lines. Int. J. Cancer 104, 504–511. doi:10.1002/ijc.10972
Wang, X., Wu, X., Zhang, Z., Ma, C., Wu, T., Tang, S., et al. (2018a). Monensin inhibits cell proliferation and tumor growth of chemo-resistant pancreatic cancer cells by targeting the EGFR signaling pathway. Sci. Rep. 8, 17914. doi:10.1038/s41598-018-36214-5
Wang, Y., Xu, W., Yan, Z., Zhao, W., Mi, J., Li, J., et al. (2018b). Metformin induces autophagy and G0/G1 phase cell cycle arrest in myeloma by targeting the AMPK/mTORC1 and mTORC2 pathways. J. Exp. Clin. Cancer Res. 37, 63. doi:10.1186/s13046-018-0731-5
Wang, Y.-S., Huang, N.-K., Lin, Y.-C., Chang, W.-C., and Huang, W.-C. (2022). Aspirin and Sulindac act via different mechanisms to inhibit store-operated calcium channel: implications for colorectal cancer metastasis. Biomed. Pharmacother. 145, 112476. doi:10.1016/j.biopha.2021.112476
Wang, Z.-Y., and Zhang, H.-Y. (2013). Rational drug repositioning by medical genetics. Nat. Biotechnol. 31, 1080–1082. doi:10.1038/nbt.2758
Wei, L., Sun, J., Zhang, N., Shen, Y., Wang, T., Li, Z., et al. (2021). Novel implications of MicroRNAs, long non-coding RNAs and circular RNAs in drug resistance of esophageal cancer. Front. Cell Dev. Biol. 9, 764313. doi:10.3389/fcell.2021.764313
Wei, W.-Q., Mosley, J. D., Bastarache, L., and Denny, J. C. (2013). Validation and enhancement of a computable medication indication resource (MEDI) using a large practice-based dataset. AMIA Annu. Symp. Proc. 2013, 1448–1456.
Westphalen, C. B., and Olive, K. P. (2012). Genetically engineered mouse models of pancreatic cancer. Cancer J. 18, 502–510. doi:10.1097/PPO.0b013e31827ab4c4
Wilkinson, G. F., and Pritchard, K. (2015). In vitro screening for drug repositioning. J. Biomol. Screen. 20, 167–179. doi:10.1177/1087057114563024
Willyard, C. (2018). New human gene tally reignites debate. Nature 558, 354–355. doi:10.1038/d41586-018-05462-w
Wong, L. H., Sinha, S., Bergeron, J. R., Mellor, J. C., Giaever, G., Flaherty, P., et al. (2016). Reverse chemical genetics: comprehensive fitness profiling reveals the spectrum of drug target interactions. PLoS Genet. 12, e1006275. doi:10.1371/journal.pgen.1006275
Wu, M., and Swartz, M. A. (2014). Modeling tumor microenvironments in vitro. J. Biomech. Eng. 136, 021011. doi:10.1115/1.4026447
Wu, X., Cao, Y., Xiao, H., Li, C., and Lin, J. (2016). Bazedoxifene as a novel GP130 inhibitor for pancreatic cancer therapy. Mol. Cancer Ther. 15, 2609–2619. doi:10.1158/1535-7163.MCT-15-0921
Würth, R., Thellung, S., Bajetto, A., Mazzanti, M., Florio, T., and Barbieri, F. (2016). Drug-repositioning opportunities for cancer therapy: novel molecular targets for known compounds. Drug Discov. Today 21, 190–199. doi:10.1016/j.drudis.2015.09.017
Xavier, C. P. R., Castro, I., Caires, H. R., Ferreira, D., Cavadas, B., Pereira, L., et al. (2021). Chitinase 3-like-1 and fibronectin in the cargo of extracellular vesicles shed by human macrophages influence pancreatic cancer cellular response to gemcitabine. Cancer Lett. 501, 210–223. doi:10.1016/j.canlet.2020.11.013
Xiao, Y., Liu, Q., Peng, N., Li, Y., Qiu, D., Yang, T., et al. (2022). Lovastatin inhibits RhoA to suppress canonical wnt/β-catenin signaling and alternative wnt-YAP/TAZ signaling in colon cancer. Cell Transpl. 31, 9636897221075749. doi:10.1177/09636897221075749
Xie, H., Qiang, P., Wang, Y., Xia, F., Liu, P., and Li, M. (2022). Discovery and mechanism studies of a novel ATG4B inhibitor Ebselen by drug repurposing and its anti-colorectal cancer effects in mice. Cell Biosci. 12, 206. doi:10.1186/s13578-022-00944-x
Xie, J., Xia, L., Xiang, W., He, W., Yin, H., Wang, F., et al. (2020). Metformin selectively inhibits metastatic colorectal cancer with the KRAS mutation by intracellular accumulation through silencing MATE1. Proc. Natl. Acad. Sci. U. S. A. 117, 13012–13022. doi:10.1073/pnas.1918845117
Xing, Y.-X., Li, M.-H., Tao, L., Ruan, L.-Y., Hong, W., Chen, C., et al. (2018). Anti-cancer effects of emodin on HepG2 cells as revealed by 1H NMR based metabolic profiling. J. Proteome Res. 17, 1943–1952. doi:10.1021/acs.jproteome.8b00029
Xiong, L., Lou, Y., and Wang, L. (2021). Effect of bevacizumab combined with first-line chemotherapy on metastatic colorectal cancer. Am. J. Transl. Res. 13, 3609–3617.
Xiong, T., Li, Z., Huang, X., Lu, K., Xie, W., Zhou, Z., et al. (2019). TO901317 inhibits the development of hepatocellular carcinoma by LXRα/Glut1 decreasing glycometabolism. Am. J. Physiol. Gastrointest. Liver Physiol. 316, G598–G607. doi:10.1152/ajpgi.00061.2018
Xiong, W., Li, W.-H., Jiang, Y.-X., Liu, S., Ai, Y.-Q., Liu, R., et al. (2015). Parecoxib: an enhancer of radiation therapy for colorectal cancer. Asian pac. J. Cancer Prev. 16, 627–633. doi:10.7314/apjcp.2015.16.2.627
Xu, D., Jin, J., Yu, H., Zhao, Z., Ma, D., Zhang, C., et al. (2017). Chrysin inhibited tumor glycolysis and induced apoptosis in hepatocellular carcinoma by targeting hexokinase-2. J. Exp. Clin. Cancer Res. 36, 44. doi:10.1186/s13046-017-0514-4
Xu, H., Aldrich, M. C., Chen, Q., Liu, H., Peterson, N. B., Dai, Q., et al. (2015). Validating drug repurposing signals using electronic health records: a case study of metformin associated with reduced cancer mortality. J. Am. Med. Inf. Assoc. 22, 179–191. doi:10.1136/amiajnl-2014-002649
Xu, M., Lee, E. M., Wen, Z., Cheng, Y., Huang, W.-K., Qian, X., et al. (2016). Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 22, 1101–1107. doi:10.1038/nm.4184
Xu, X., Wang, J., Han, K., Li, S., Xu, F., and Yang, Y. (2018). Antimalarial drug mefloquine inhibits nuclear factor kappa B signaling and induces apoptosis in colorectal cancer cells. Cancer Sci. 109, 1220–1229. doi:10.1111/cas.13540
Yadav, A. K., Srikrishna, S., and Gupta, S. C. (2016). Cancer drug development using Drosophila as an in vivo tool: from bedside to bench and back. Trends Pharmacol. Sci. 37, 789–806. doi:10.1016/j.tips.2016.05.010
Yamasaki, D., Kawabe, N., Nakamura, H., Tachibana, K., Ishimoto, K., Tanaka, T., et al. (2011). Fenofibrate suppresses growth of the human hepatocellular carcinoma cell via PPARα-independent mechanisms. Eur. J. Cell Biol. 90, 657–664. doi:10.1016/j.ejcb.2011.02.005
Yang, C., Zhang, H., Chen, M., Wang, S., Qian, R., Zhang, L., et al. (2022). A survey of optimal strategy for signature-based drug repositioning and an application to liver cancer. Elife 11, e71880. doi:10.7554/eLife.71880
Yang, J., Jin, X., Yan, Y., Shao, Y., Pan, Y., Roberts, L. R., et al. (2017). Inhibiting histone deacetylases suppresses glucose metabolism and hepatocellular carcinoma growth by restoring FBP1 expression. Sci. Rep. 7, 43864. doi:10.1038/srep43864
Yu, M., Tong, X., Qi, B., Qu, H., Dong, S., Yu, B., et al. (2014). Berberine enhances chemosensitivity to irinotecan in colon cancer via inhibition of NF-κB. Mol. Med. Rep. 9, 249–254. doi:10.3892/mmr.2013.1762
Yu, Q.-S., Xin, H.-R., Qiu, R.-L., Deng, Z.-L., Deng, F., and Yan, Z.-J. (2020). Niclosamide: drug repurposing for human chondrosarcoma treatment via the caspase-dependent mitochondrial apoptotic pathway. Am. J. Transl. Res. 12, 3688–3701.
Yuan, M., Shong, K., Li, X., Ashraf, S., Shi, M., Kim, W., et al. (2022). A gene Co-expression network-based drug repositioning approach identifies candidates for treatment of hepatocellular carcinoma. Cancers 14, 1573. doi:10.3390/cancers14061573
Yumimoto, K., Sugiyama, S., Mimori, K., and Nakayama, K. I. (2019). Potentials of C-C motif chemokine 2-C-C chemokine receptor type 2 blockers including propagermanium as anticancer agents. Cancer Sci. 110, 2090–2099. doi:10.1111/cas.14075
Zamami, Y., Imanishi, M., Takechi, K., and Ishizawa, K. (2017). Pharmacological approach for drug repositioning against cardiorenal diseases. J. Med. Invest. 64, 197–201. doi:10.2152/jmi.64.197
Zhang, D., Ma, Q., Shen, S., and Hu, H. (2009). Inhibition of pancreatic cancer cell proliferation by propranolol occurs through apoptosis induction: the study of beta-adrenoceptor antagonist’s anticancer effect in pancreatic cancer cell. Pancreas 38, 94–100. doi:10.1097/MPA.0b013e318184f50c
Zhang, D., Ma, Q.-Y., Hu, H.-T., and Zhang, M. (2010). β2-adrenergic antagonists suppress pancreatic cancer cell invasion by inhibiting CREB, NFκB and AP-1. Cancer Biol. Ther. 10, 19–29. doi:10.4161/cbt.10.1.11944
Zhang, J., Gao, M., Niu, Y., and Sun, J. (2022a). From DNMT1 degrader to ferroptosis promoter: drug repositioning of 6-Thioguanine as a ferroptosis inducer in gastric cancer. Biochem. Biophys. Res. Commun. 603, 75–81. doi:10.1016/j.bbrc.2022.03.026
Zhang, J., Gao, M., Niu, Y., and Sun, J. (2022b). Identification of a novel ferroptosis inducer for gastric cancer treatment using drug repurposing strategy. Front. Mol. Biosci. 9, 860525. doi:10.3389/fmolb.2022.860525
Zhang, J., Jiang, K., Lv, L., Wang, H., Shen, Z., Gao, Z., et al. (2015). Use of genome-wide association studies for cancer research and drug repositioning. PLoS One 10, e0116477. doi:10.1371/journal.pone.0116477
Zhang, X., Hu, P., Ding, S.-Y., Sun, T., Liu, L., Han, S., et al. (2019). Induction of autophagy-dependent apoptosis in cancer cells through activation of ER stress: an uncovered anti-cancer mechanism by anti-alcoholism drug disulfiram. Am. J. Cancer Res. 9, 1266–1281.
Zhang, X., and Luo, H. (2018). Effects of thalidomide on growth and VEGF-A expression in SW480 colon cancer cells. Oncol. Lett. 15, 3313–3320. doi:10.3892/ol.2017.7645
Zhang, X., Wu, T., Cai, X., Dong, J., Xia, C., Zhou, Y., et al. (2022c). Neoadjuvant immunotherapy for MSI-H/dMMR locally advanced colorectal cancer: new strategies and unveiled opportunities. Front. Immunol. 13, 795972. doi:10.3389/fimmu.2022.795972
Zhao, P., Shen, Y., Li, M., Dan, H., Zhao, Z., and Zhang, J. (2022). Integration of transcriptomics and metabolomics reveals the antitumor mechanism underlying tadalafil in colorectal cancer. Front. Pharmacol. 13, 793499. doi:10.3389/fphar.2022.793499
Zhao, Y., He, M., Liang, R., Li, Q., and Shi, M. (2021). Evaluation of antiemetic therapy for hepatic arterial infusion chemotherapy with oxaliplatin, fluorouracil, and leucovorin. Ther. Clin. Risk Manag. 17, 73–77. doi:10.2147/TCRM.S283192
Zheng, H.-C. (2017). The molecular mechanisms of chemoresistance in cancers. Oncotarget 8, 59950–59964. doi:10.18632/oncotarget.19048
Zheng, S., Wu, Y. X., Wang, J. Y., Li, Y., Liu, Z. J., Liu, X. G., et al. (2020). Identifying the characteristics of patients with cervical degenerative disease for surgical treatment from 17-year real-world data: retrospective study. JMIR Med. Inf. 8, e16076. doi:10.2196/16076
Zhou, L., Gao, W., Wang, K., Huang, Z., Zhang, L., Zhang, Z., et al. (2019a). Brefeldin A inhibits colorectal cancer growth by triggering Bip/Akt-regulated autophagy. FASEB J. 33, 5520–5534. doi:10.1096/fj.201801983R
Zhou, S., Wu, H., Ning, W., Wu, X., Xu, X., Ma, Y., et al. (2021). Ivermectin has new application in inhibiting colorectal cancer cell growth. Front. Pharmacol. 12, 717529. doi:10.3389/fphar.2021.717529
Zhou, Y., Zhou, Y., Yang, M., Wang, K., Liu, Y., Zhang, M., et al. (2019b). Digoxin sensitizes gemcitabine-resistant pancreatic cancer cells to gemcitabine via inhibiting Nrf2 signaling pathway. Redox Biol. 22, 101131. doi:10.1016/j.redox.2019.101131
Zhuang, S., Jian, Y.-M., and Sun, Y.-N. (2017). Inhibition of N-methyl-N-nitrosourea-induced gastric tumorigenesis by Liuwei Dihuang Pill in db/db mice. World J. Gastroenterol. 23, 4233–4242. doi:10.3748/wjg.v23.i23.4233
Keywords: gastrointestinal cancers, therapeutic strategies, drug repurposing, colorectal cacner, pancreatic cancer, liver cancer
Citation: Fatemi N, Karimpour M, Bahrami H, Zali MR, Chaleshi V, Riccio A, Nazemalhosseini-Mojarad E and Totonchi M (2024) Current trends and future prospects of drug repositioning in gastrointestinal oncology. Front. Pharmacol. 14:1329244. doi: 10.3389/fphar.2023.1329244
Received: 28 October 2023; Accepted: 11 December 2023;
Published: 04 January 2024.
Edited by:
Onur Bender, Ankara University, TürkiyeReviewed by:
Ferda Kaleagasioglu, University of Istinye, TürkiyeCopyright © 2024 Fatemi, Karimpour, Bahrami, Zali, Chaleshi, Riccio, Nazemalhosseini-Mojarad and Totonchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Riccio, YW5kcmVhLnJpY2Npb0B1bmljYW1wYW5pYS5pdA==; Mehdi Totonchi, bS50b3RvbmNoaUByb3lhbmluc3RpdHV0ZS5vcmc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.