
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 18 December 2023
Sec. Drug Metabolism and Transport
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1325932
This article is part of the Research Topic Drug Metabolism and Transport: The Frontier of Personalized Medicine Volume II View all 19 articles
The rise of fentanyl has introduced significant new challenges to public health. To improve the examination and identification of biological samples in cases of fentanyl misuse and fatalities, this study utilized a zebrafish animal model to conduct a comparative investigation of the metabolites and biotransformation pathways of fentanyl in the zebrafish’s liver and brain. A total of 17 fentanyl metabolites were identified in the positive ion mode using ultra-high-pressure liquid chromatography Q Exactive HF Hybrid Quadrupole-Orbitrap mass spectrometry (UHPLC-QE HF MS). Specifically, the zebrafish’s liver revealed 16 fentanyl metabolites, including 6 phase I metabolites and 10 phase II metabolites. Conversely, the zebrafish’s brain presented fewer metabolites, with only 8 detected, comprising 6 phase I metabolites and 2 phase II metabolites. Notably, M′4, a metabolite of dihydroxylation, was found exclusively in the brain, not in the liver. Through our research, we have identified two specific metabolites, M9-a (monohydroxylation followed by glucuronidation) and M3-c (monohydroxylation, precursor of M9-a), as potential markers of fentanyl toxicity within the liver. Furthermore, we propose that the metabolites M1 (normetabolite) and M3-b (monohydroxylation) may serve as indicators of fentanyl metabolism within the brain. These findings suggest potential strategies for extending the detection window and enhancing the efficiency of fentanyl detection, and provide valuable insights that can be referenced in metabolic studies of other new psychoactive substances.
The emergence of fentanyl analogs has significantly impacted the field of pharmacology and presented new challenges in the realm of public health (Clotz et al., 1991; Poklis, 1995; Zawilska et al., 2017; Han et al., 2019). Fentanyl is a synthetic opioid that is approximately 50–100 times more potent than morphine, its high potency, fast onset of action, and duration of the desired effect may be particularly important contributing factors to the higher risk of overdose deaths and social consequences. Additionally, it is frequently combined with other substances like heroin or cocaine, often unwittingly to the user. This scenario makes it exceedingly challenging for users to accurately assess the strength of the substances they are consuming, consequently leading to a rise in fentanyl-related overdose deaths (Henderson, 1991; Hibbs et al., 1991; Nolan et al., 2019). Survey data from New York City, West Virginia, and Australia indicate that since 2015, there has been a significant rise in the rate of fentanyl-related overdose deaths worldwide. Illicitly manufactured fentanyl and its analogues have become some of the primary substances identified in these fatalities (Colon-Berezin et al., 2019; Dai et al., 2022; Roxburgh et al., 2022).
Delving into the metabolites and biotransformation pathways of fentanyl provides a broader perspective on its toxicological impacts. Pinpointing its signature metabolites broadens the detection time frame, consequently enhancing the efficiency of tests. This is key in keeping a vigilant eye on potential fentanyl abuse. In recent years, the focus of research has increasingly centered on the qualitative and quantitative analysis of trace amounts of fentanyl and its metabolites in biological samples (Strayer et al., 2018; Busardò et al., 2019; Ott et al., 2022; Wei et al., 2022). The majority of reported fentanyl metabolites are phase I metabolites, which include N-dealkylation and hydroxylation metabolites. While phase II metabolites, which are primarily glucuronidation metabolites, are not as commonly reported. Consequently, there is a need for more research to broaden our understanding of phase II metabolism.
Zebrafish models offer several advantages over experimental rodent models, including greater time efficiency, cost-effectiveness, high-throughput capacity and sensitivity. Adhering to the internationally recognized 3Rs (replacement, reduction, refinement) principle in toxicological experiments, zebrafish models also boast a genome strikingly similar to that of humans. Furthermore, zebrafish possess a metabolic enzyme system that closely resembles that of mammals. Enzymes from the CYP450 family play a crucial role in the initial phase of drug metabolism in mammals. Goldstone et al. demonstrated that many zebrafish CYP450 enzymes have direct orthologues in humans and other mammals, underscoring the relevance of zebrafish as a model for studying drug metabolism. Notably, there are unique CYPs in fish, such as CYP1C, CYP2AE, and CYP2X, which lack human orthologs, suggesting potential differences in metabolic pathways (Goldstone et al., 2010). Furthermore, the gene expression profiles of phase II enzymes, including uridine glucuronosyltransferases (UGTs), sulfotransferases (SULTs), and methyltransferases (COMTs) in zebrafish, have been characterized and show similarities to their human counterparts (Alazizi et al., 2011; Mohammed et al., 2012; Christen et al., 2014). Consequently, zebrafish are capable of performing both phase I (oxidation, N-demethylation, O-demethylation, and N-dealkylation) and phase II (sulfation, glucuronidation, and methylation) drug metabolism reactions, paralleling human processes (Matos et al., 2020; Pesavento et al., 2022).
Zebrafish, frequently dubbed as “little mice in water” have garnered considerable interest in recent years. Their employment as biological models to investigate drug toxicity and metabolism has propelled them to the forefront of research (Xu et al., 2019; Morbiato et al., 2020; Krishna et al., 2021; Cooman et al., 2022b; Pesavento et al., 2022) conducted a study on the metabolism of fentanyl in zebrafish embryos, but no research has been reported on fentanyl metabolism in adult zebrafish. Fentanyl is a lipophilic compound, can swiftly cross the cell membrane and penetrate the blood-brain barrier to exert its medicinal effects within the brain (Yu et al., 2022). The liver, as the primary organ for drug metabolism, is abundant in drug-metabolizing enzymes.
For this study, the metabolites and biotransformation pathways of fentanyl in the liver and brain of zebrafish were examined and identified using ultra-high-pressure liquid chromatography Q Exactive HF Hybrid Quadrupole-Orbitrap mass spectrometry (UHPLC-QE HF MS), with an emphasis on the analysis of Phase II metabolites. This approach lays a solid foundation for the examination and identification of biological samples in cases of fentanyl misuse and fatalities.
Fentanyl hydrochloride standard with a purity of at least 99.5% was sourced from Shanghai Yuansi Standard Technology Co., Ltd. in Shanghai, China, and was dissolved in ultrapure water to create a 10 mgL−1 (26.8 μM) fentanyl solution for later use. Chromatographically pure acetonitrile and formic acid were purchased from Merck & Co., Inc. In New Jersey, United States.
All zebrafish experimental procedures were reviewed and approved by Zhejiang University Experimental Animal Welfare. In our experiment, we worked with adult wild-type zebrafish of the AB strain, which we sourced from the Wuhan Zebrafish Center in China. We kept them in a recirculating tank system (RTS), courtesy of Shanghai Haisheng Biotechnology. This system kept the water clean and well-oxygenated, with a pH of around 7.2, conductivity from 500 to 600 μS, and a cozy temperature between 27.5°C and 28.5°C. The zebrafish enjoyed a diet of freshly hatched brine shrimp served twice a day, and we mimicked their natural light/dark cycle with 14 h of light followed by 10 h of darkness. Before we introduced them to the bath treatment for our metabolic studies, we let them get used to the tank system for at least a week.
Eighteen adult zebrafish (4–6 months old, weighing 0.5 ± 0.1 g) were divided into two groups of nine. A couple of hours before the experiment kicked off, we transferred both groups from the RTS to Petri dishes, each containing 30 mL of RTS water. This was to help them acclimate. We put three fish in each dish. One group was chosen for drug exposure. When the experiment started, we replaced the water in their RTS with an equal volume of fentanyl aqueous solution (10 mgL−1, 26.8 μM), and the fish were exposed to this solution for 24 h. The other group served as a control, remaining in the RTS water without drug exposure for the same period.
After the bath administration period, the zebrafish were rinsed thrice with ultrapure water, then humanely euthanized in grinding tubes. The livers were then harvested, with every three livers from the same group pooled together. Next, three grinding beads and 200 µL of acetonitrile were added to each tube, followed by homogenizing the samples using a JXFSTPRP-CL fully automatic sample freezing and grinding machine (Shanghai JingXin Co., Ltd., China) at −4°C (a frequency of 60 Hz, a run time of 40 s, and a pause time of 20 s, for a total of 10 cycles). This was followed by centrifugation at 13,000 r/min and 4°C for 10 min using a Legend Micro 21R freezing high-speed centrifuge (Thermo Fisher Scientific, Waltham, MA, United States). The supernatant was collected, re-dissolved using a nitrogen blow, and then filtered through a 0.22 μm organic PTFE microporous membrane into a lined tube for subsequent instrument analysis.
The treatment was similarly administered to the zebrafish brain.
All samples were analyzed using a by ultra-high-pressure liquid chromatography Q Exactive HF Hybrid Quadrupole-Orbitrap mass spectrometry (Thermo Fisher Scientific, Waltham, MA, United States). The chromatographic column was a ACQUITY UPLC HSS T3 Column (150 mm × 2.1 mm, 1.8 μm) fitted with a VanGuard precolumn, both from Waters (Milford, MA, United States). Mobile phases were 0.1% formic acid (A) and 0.1% formic acid in acetonitrile (B) and were run in a gradient at a flow rate of 0.3 mLmin−1 starting at 1% B until 1.0 min, ramped to 99% B at 8.0 min and held until 10.0 min, then ramped down to 1% B at 10.1 min and finally re-equilibration until 12 min. The column temperature was kept at a steady temperature of 30°C, and an injection volume of 3 µL was maintained.
An electrospray ionization source (ESI) was utilized that operated at a spray voltage of 3,800 V. Nitrogen was used as the collision gas, and the atomization temperature was set at 320°C, an atomization gas pressure of 38 arb was maintained and an auxiliary gas pressure of 15 arb, with a capillary temperature of 350°C. Data acquisition was performed using the Full MS-ddMS2 scan mode under the following conditions: a Full MS resolution of 35,000, a maximum injection time of 100 m, and a scan range of m/z 70–1,000. For tandem MS data collection, the dd-MS2 resolution was 17,500, with collision energies set at 17.5, 35, and 52.5 eV.
Fentanyl, with the molecular formula C22H28N2O, exhibited an accurate mass of the protonated molecular ion [M + H]+ at m/z 337.22744 under the positive ion collection mode. The MSMS spectrum of fentanyl with characteristic fragment structures is depicted in Figure 1. Referencing Davidson’s study (Davidson et al., 2022) on fentanyl’s mass spectrometry fragmentation patterns, we observed that the fragmentation of fentanyl’s mass spectrum adheres to the patterns of m/z 337→281→188→146→134→132, 337→188→105 and 337→216. The detailed MSMS spectrometry fragmentation pathways, which further elucidate these patterns, are illustrated in Figure 2.
In the positive ion mode, a total of 17 fentanyl metabolites were detected in the zebrafish’s liver and brain, M′4 was unique that it was only found in the brain but not in the liver. All other metabolites detected in the brain were identifiable in the liver of the zebrafish. To be specific, 16 fentanyl metabolites were detected in the zebrafish’s liver, comprising 6 phase I metabolites and 10 phase II metabolites. In contrast to the liver, the brain of zebrafish contained fewer metabolites, with a total of 8 detected. This included 6 phase I metabolites and 2 phase II metabolites, detailed in Table 1. Notably, glucuronidated fentanyl (M9-a) and monohydroxy fentanyl (M3-c) were the major metabolites in the liver. In the brain, however, norfentanyl (M1) and monohydroxy fentanyl (M3-b) were more prevalent. Phase I metabolites primarily consisted of dealkylation and hydroxylation products, while the phase II metabolites were predominantly the results of methylation, glucuronidation and sulfation processes. The extracted ion chromatogram of these metabolites is depicted in Figure 3, and the detailed information can be found in Table 2, mass error for protonated molecule of all these metabolites being less than 3.46 ppm. In comparison to earlier studies on fentanyl metabolism using hepatocytes (Kanamori et al., 2018a; Kanamori et al., 2018b) or zebrafish embryos (Pesavento et al., 2022), our study revealed a more diverse fentanyl metabolites, with a particular increase in phase II metabolites. Of significant note, this study presents the first report of fentanyl’s sulfation metabolites.
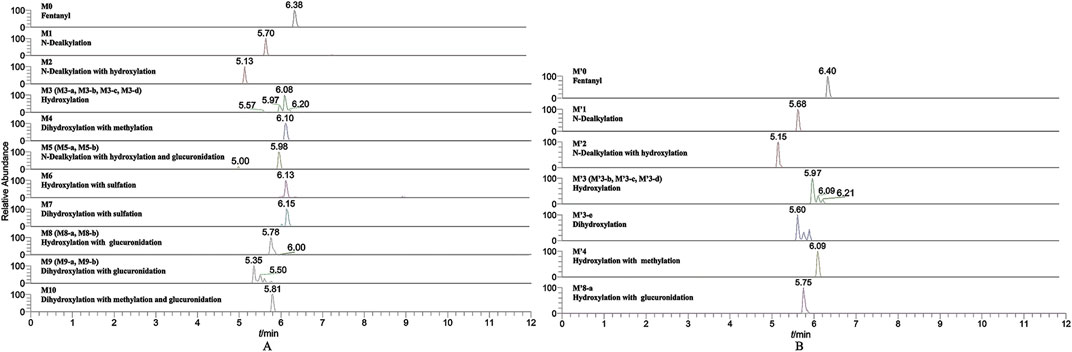
FIGURE 3. Extracted ion chromatogram for the major metabolites of fentanyl detected in the liver (A) and brain (B) of zebrafish.
Six phase I metabolites were identified in both the liver and brain of zebrafish, M3-a was exclusive to the liver, whereas M′4 was found only in the brain.
The N-dealkylation metabolite on the piperidine ring, designated as M1and also referred to as norfentanyl, has been identified as a signature metabolite of fentanyl (Wilde et al., 2019; Armenian et al., 2022), was identified in both the liver and brain of zebrafish. With a retention time of 5.70 min, M1 formed an accurate mass of the protonated molecular ion [M + H]+ at m/z 233.16484 (C14H20N2O), showing a reduction of 104 Da (-C8H8) compared to the parent drug. The main characteristic fragment ions of M1 were m/z 84.08068, 150.09119, and 177.13835, as depicted in Figure 4. The characteristic fragments ions at m/z 84 and 150 align with the corresponding parent drug. However, the fragment at m/z 177 was 104 Da less than the parent drug at m/z 281, indicating that M1 was formed as a result of N-dealkylation on the piperidine ring of the parent drug.
M2 was generated by hydroxylation of M1. M3-a, M3-b, M3-c, and M3-d were the monohydroxylation metabolites of the parent drug, while dihydroxylation results in M′3-e. M3-a was solely identified in the liver of zebrafish and not in the brain. Conversely, M′3-e was exclusively detected in the brain. M2 was observed at a retention time of 5.13 min, with an accurate mass of the protonated molecular ion [M + H]+ at m/z 249.15975, corresponding to the molecular formula C14H20N2O2. Compared to the characteristic fragments of M1, it was inferred from m/z 166.08578, 148.07497 (166→148, a loss of H2O) and 73.02837 that the hydroxylation site of M2 was located on the propionyl moiety as shown in Figure 5.
M3-a, M3-b, M3-c, and M3-d were identified at retention times of 5.57, 5.97, 6.08, and 6.20 min, respectively. These metabolites exhibited an accurate mass of the protonated molecular ion [M + H]+ at m/z 353.22235 (C22H28N2O2). The MSMS spectrum of M3 group is depicted in Figure 6. The presence of characteristic fragment ion peaks at m/z 84, 121, 162, and 204, along with the absence of m/z 335 (a loss of H2O), in both M3-a and M3-c, suggest that a monohydroxylation reaction occurred on the benzene ring of phenylethyl moiety in each metabolite.
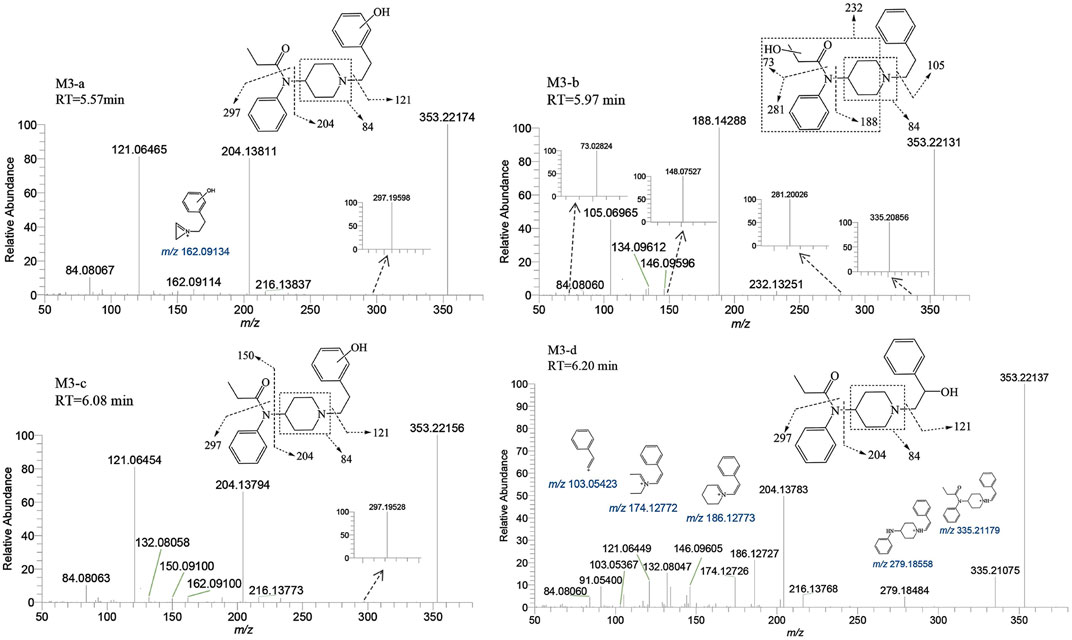
FIGURE 6. Mass spectra and assigned fragmentation patterns for monohydroxylated metabolites (M3 group).
The variation in retention times across these metabolites indicates distinct reaction sites for each hydroxylation event. The fragment ions of M3-b at m/z 73.02824, 148.07527, 232.13251, and 335.20856 indicate that the monohydroxylation reaction occurred on the propionyl moiety. For metabolite M3-d, the ion fragments adhered to a fragmentation pattern of m/z 353→204→186 and 353→335→279→186→174 (Cooman et al., 2022a). The transitions of m/z 353→335 and 204→186 were indicative of dehydration, thereby suggesting that the hydroxylation reaction site for M3-d is situated on the alkyl portion of the phenethyl moiety. Through detailed analysis of the tandem mass spectrometry data, it has become apparent that the MSMS spectrum alone is insufficient to pinpoint the exact carbon atom undergoing hydroxylation. However, considering the inherent instability of α-carbon hydroxylation in fentanyl metabolites, we infer that the hydroxylation occurs at the more stable β-carbon position. This deduction is supported by both the spectral data and the chemical logic pertaining to the likely metabolic pathways.
M’4 was exclusively observed in the brain of zebrafish at a retention time of 5.60 min. It formed an accurate mass of the protonated molecular ion [M + H]+ at m/z 369.21727 (C22H28N2O3). The absence of product ions at m/z 351 and 333, due to the loss of H2O, indicates that M′4 underwent hydroxylation on the benzene ring of fentanyl. The presence of the characteristic fragment ions at m/z 121.06461, 204.13803, and 84.08065 strongly suggest that one hydroxylation site is on the benzene ring of phenylethyl moiety. This observation exclude the possibility of a second hydroxy group being present on the phenylethyl moiety, as shown in Figure 7. Similarly, it can be inferred that another hydroxylation site could be located on the benzene ring of the aniline group from the fragment ions m/z 232.13333 and 249.16071.
Ten phase II metabolites were identified in the liver of zebrafish, contrastingly, only two (M5 and M9-a) were detected in the brain.
M5 was observed at a retention time of 6.10 min, with an accurate mass of the protonated molecular ion [M + H]+ at m/z 383.23292 (C23H30N2O3). Similar to M′4, the absence of product ions at m/z 365, 351, and 333, which would indicate the loss of H2O, was also noted for M5. The presence of characteristic fragment ions at m/z 151.07515, 234.14851, 119.04902, 192.10179, and 57.01083 suggests that the dihydroxylation reaction occurred on the benzene ring of the phenylethyl moiety. The specific fragment at m/z 192 supports the inference that a catechol metabolite was formed on the benzene ring of the phenylethyl moiety. Subsequently, a hydroxyl moiety underwent a methylation reaction formed M5. M11 was characterized by a retention time of 5.81 min with an accurate mass of the protonated molecular ion [M + H]+ at m/z 559.26501 (C29H38N2O9). From the characteristic fragment ions m/z 151.07515, 383.23221, 234.14845, and 192.10159, it can be inferred that M11 was formed by the glucuronidation of another hydroxyl moiety on M5. The MSMS spectrum of methylated metabolites are displayed in Figure 8.
M6-a and M6-b were glucuronidated metabolites, formed through dealkylation and hydroxylation of the piperidine ring. They were identified with retention times of 5.00 and 5.98 min, respectively. Their protonated molecular ion [M + H]+ was observed at an accurate mass of m/z 425.19184 (C20H28N2O8) as shown in Figure 9. The formation of M6-a from M2 through glucuronidation can be inferred from the characteristic fragment ions. The presence of fragment ions m/z 82.06499, 249.15921, 233.16437, 150.09111, and 100.07552 suggest that M6-b was formed via glucuronidation after the N atom of the piperidine ring underwent dealkylation and oxidation. M9 and M10 moiety were formed through glucuronidation following the hydroxylation of the parent drug (Figure 10). M9-a and M9-b were identified at the following retention times: 5.78 and 6.00 min, with an accurate mass of the protonated molecular ion [M + H]+ at m/z 529.25444 (C26H36N2O8). They were formed as a result of glucuronidation, a process that occurred after parent drug underwent monohydroxylation. The MSMS spectrum of M9-a revealed fragment ions at m/z 121.06458, 353.22153, 204.13795, 84.08066, 380.16959 and no m/z 335 (a loss of H2O), suggesting that M9-a resulted from the glucuronidation of either M3-a or M3-c. In contrast, the fragment ions of M9-b at m/z 353.22153, 204.13788, 186.12737, 335.22153, and 134.09613 infer that M9-b is a product of the glucuronidation of M3-d. Similarly, M10-a and M10-b were formed through glucuronidation following the dihydroxylation of the parent drug.
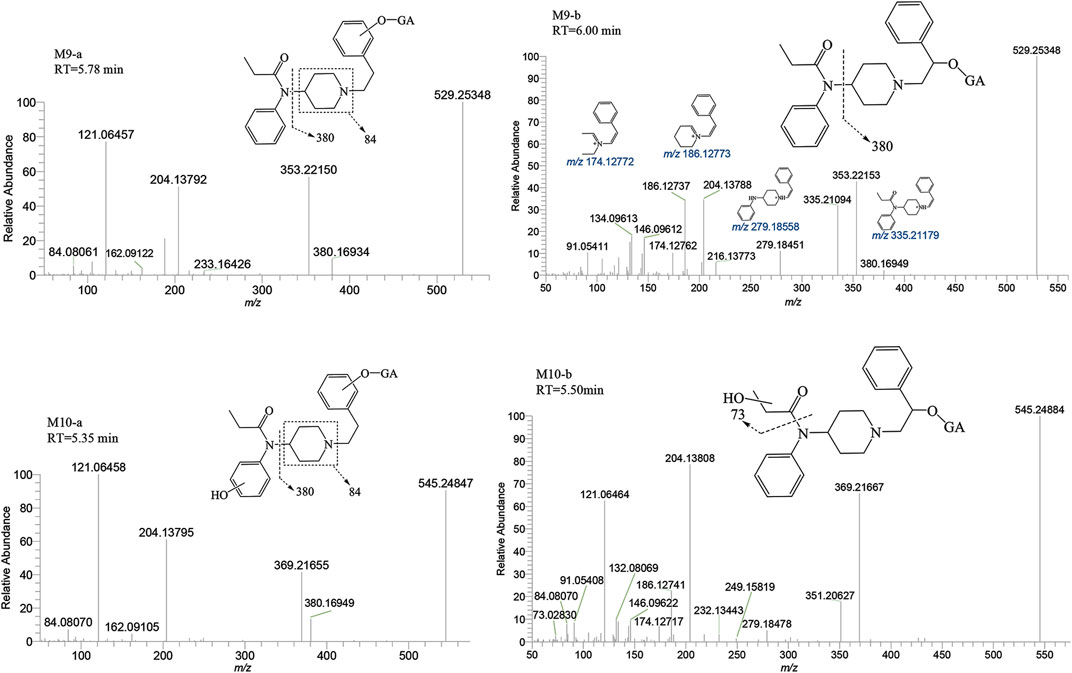
FIGURE 10. Mass spectra and assigned fragmentation patterns for glucuronidated metabolites (M9 and M10 group).
The metabolite M7 was observed with a retention time of 6.13 min, and it formed an accurate mass of the protonated molecular ion [M + H]+ at m/z 433.17917 (C22H28N2O5S). The presence of the characteristic fragment ions at m/z 353.22153, 204.13794, 121.06456, 284.09439, 162.09109 and no m/z 335 (a loss of H2O) was observed indicated that M7 was formed through sulfation, following monohydroxylation of the parent drug, a process likely carried out by M3-a or M3-c. The post-dihydroxylation sulfation product M8, presented a retention time of 6.15 min. Its protonated molecular ion [M + H]+ ion was at m/z 449.17408, corresponding to the molecular formula C22H28N2O6S. The MSMS spectrum of M8 displayed characteristic fragment ions at m/z 369.21661, 220.13290, 137.05951, 178.08575 and no m/z 431, 351, and 333 by the loss of H2O were observed indicating the presence of two hydroxy moiety at the benzene ring of phenylethyl moiety, with one of them underwent sulfation (Figure 11).
The parent drug was found in high abundance in all samples. Upon comparing the peak areas of 17 metabolites in the experiment, ranking peak areas as outlined in Table 1, the most dominant metabolite was the metabolite of monohydroxylation and glucuronidation (M9-a), followed by monohydroxylated metabolite (M3-c, precursor of M9-a) in the zebrafish’s liver, and the highest abundance of normetabolite (M1) was found in the zebrafish’s brain, followed by monohydroxylated metabolite (M3-b). The N-dealkylation metabolite on the piperidine ring, also referred to as norfentanyl, has been identified as a signature metabolite of fentanyl. M9-a, M3-b, and M3-c contains all the structural characteristics of fentanyl. In summary, we have discerned that the metabolites, specifically M9-a in conjunction with M3-c, have been identified as potential indicators of fentanyl toxicity within the hepatic system. Simultaneously, we advocate for the combined use of M1 and M3-b metabolites as the metabolic signposts for fentanyl within the cerebrum.
The structural analysis of various metabolites in the liver and brain of zebrafish has revealed the biotransformation pathways of fentanyl. Metabolites were formed through several processes: N-dealkylation (M1) followed by hydroxylation (M2) and glucuronidation (M6-a); N-dealkylation followed by oxidation at the N atom of the piperidine ring and glucuronidation (M6-b); monohydroxylation (M3-a, M3-b, M3-c, M3-d) followed by glucuronidation (M9-a, M9-b) or sulfation (M7); dihydroxylation (M′4) followed by glucuronidation (M10-a, M10-b) or sulfation (M8); and dihydroxylation at the benzene ring of phenylethyl moiety followed by methylation of one hydroxyl moiety (M5) and glucuronidation of another hydroxyl moiety (M11). Figure 12 shows the proposed biotransformation pathway of fentanyl in the zebrafish’s liver and brain.
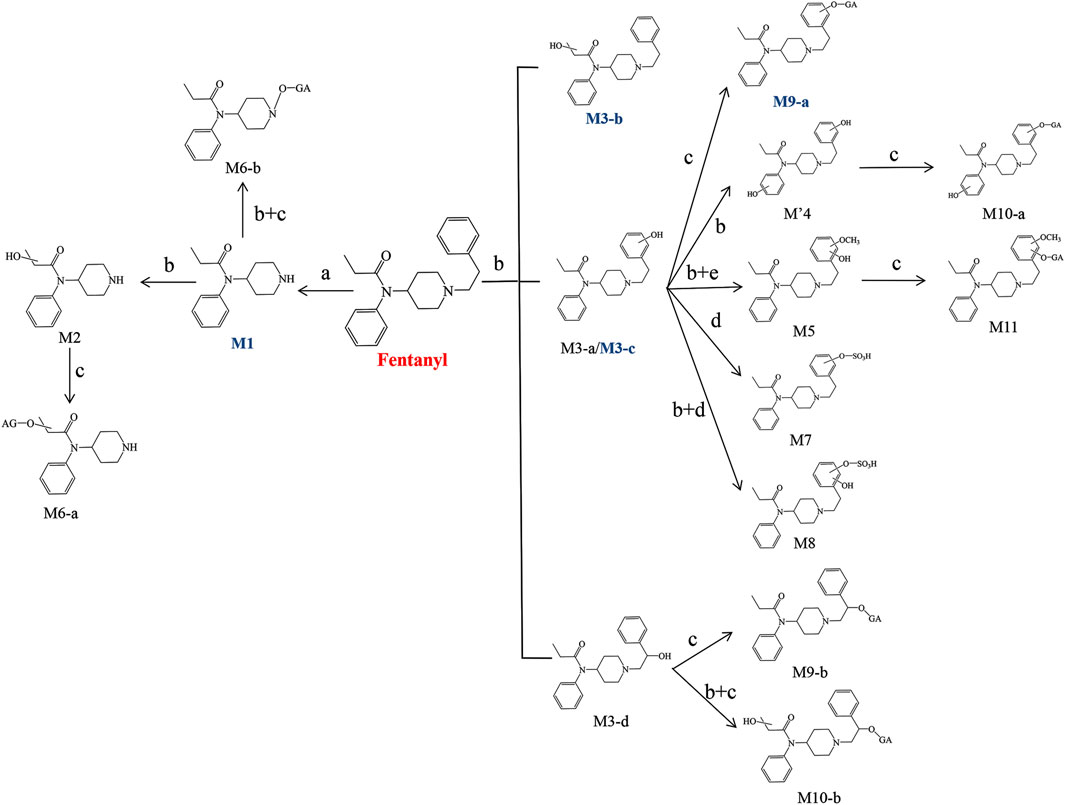
FIGURE 12. Proposed biotransformation pathways of fentanyl in the liver and brain of zebrafish (a. N-Dealkylation, b. Hydroxylation, c. Glucuronidation, d. Sulfation, e. Methylation). Major metabolites are marked by blue bold.
Given the increasing influence of fentanyl on public health, enhancing research into fentanyl metabolism is crucial. In this study, we used a zebrafish animal model, a method that is efficient in terms of time, cost, and sensitivity, in conjunction with UHPLC-QE HF MS, to perform a comparative analysis of the metabolites and biotransformation pathways of fentanyl in the zebrafish’s liver and brain. Our research identified 17 unique metabolites of fentanyl in the liver and brain of the zebrafish, including 7 phase I metabolites and 10 phase II metabolites. Fewer metabolites were detected in the brain than in the liver, the primary metabolic organ. M′4 was found exclusively in the brain, not in the liver. M9-a (monohydroxylation followed by glucuronidation) and M3-c (monohydroxylation) have been discerned as potential harbingers of fentanyl-induced hepatotoxicity. Concurrently, M1 (normetabolite) and M3-b (monohydroxylation) have been identified as cerebral metabolic markers for fentanyl. Notably, the diversity of metabolites identified in this study exceeded prior reports, particularly regarding phase II metabolites, and the sulfation metabolite was identified for the first time. This study provides an in-depth analysis of fentanyl metabolism. These insights could provide valuable evidence for the examination and identification of biological samples in instances of fentanyl misuse and related fatalities.
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
The animal study was approved by The Zhejiang University Experimental Animal Welfare and Ethical Review Committee approved the study (Ethical approval number: ZJU20220147). The study was conducted in accordance with the local legislation and institutional requirements.
ML: Writing–original draft, Writing–review and editing, Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Supervision. JH: Investigation, Resources, Writing–original draft. SZ: Conceptualization, Investigation, Methodology, Writing–original draft. BW: Conceptualization, Methodology, Writing–original draft. HZ: Project administration, Resources, Writing–review and editing. YL: Conceptualization, Project administration, Resources, Supervision, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Research Funding Support Program by Academician YL (2022-YZ-02), Zhejiang Provincial Natural Science Foundation of China (LGF21B050002), and the Open Project of Zhejiang Provincial Key Laboratory of Drug Prevention and Control Technology Research (2020013).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alazizi, A., Liu, M. Y., Williams, F. E., Kurogi, K., Sakakibara, Y., Suiko, M., et al. (2011) Identification, characterization, and ontogenic study of a catechol O-methyltransferase from zebrafish. Aquat. Toxicol. 102, 18–23. doi:10.1016/j.aquatox.2010.12.016
Armenian, P., Vo, K. T., Barr-Walker, J., and Lynch, K. L. (2018). Fentanyl, fentanyl analogs and novel synthetic opioids: a comprehensive review. Neur 134, 121–132. doi:10.1016/j.neuropharm.2017.10.016
Busardò, F. P., Carlier, J., Giorgetti, R., Tagliabracci, A., Pacifici, R., Gottardi, M., et al. (2019). Ultra-high-performance liquid chromatography-tandem mass spectrometry assay for quantifying fentanyl and 22 analogs and metabolites in whole blood, urine, and hair. Front. Chem. 7:184. doi:10.3389/fchem.2019.00184
Christen, V., and Fent, K. (2014) Tissue-sex- and development-specific transcription profiles of eight UDP-glucuronosyltransferase genes in zebrafish (Danio rerio) and their regulation by activator of aryl hydrocarbon receptor. Aqual Toxicol. 150, 93–102. doi:10.1016/j.aquatox.2014.02.019
Clotz, M. A., and Nahata, M. C. (1991). Clinical uses of fentanyl, sufentanil, and alfentanil. Am. J. Health-Syst. Ph. 48 (9), 2021–2024. doi:10.1093/ajhp/48.9.2021
Colon-Berezin, C., Nolan, M. L., Blachman-Forshay, J., and Paone, D. (2019). Overdose deaths involving fentanyl and fentanyl analogs—New York City, 2000-2017. Morb. Mortal. Wkly. Rep. 68, 37–40. doi:10.15585/mmwr.mm6802a3
Cooman, T., Bergeron, S. A., Coltogirone, R., Horstick, E., and Arroyo, L. (2022). Evaluation of fentanyl toxicity and metabolism using a zebrafish model. J. Appl. Toxicol. 42(4), 706–714. doi:10.1002/jat.4253
Cooman, T., Hoover, B., Sauvé, B., Bergeron, S. A., Quinete, N., Gardinali, P., et al. (2022). The metabolism of valerylfentanyl using human liver microsomes and zebrafish larvae. Drug Test. Anal. 14(6), 1116–1129. doi:10.1002/dta.3233
Dai, Z., Abate, M. A., Smith, G. S., Kraner, J. C., and Mock, A. R. (2019). Fentanyl and fentanyl-analog involvement in drug-related deaths. Drug Alcohol Depend. 196, 1–8. doi:10.1016/j.drugalcdep.2018.12.004
Davidson, J. T., Sasiene, Z. J., and Jackson, G. P. (2020). The characterization of isobaric product ions of fentanyl using multi-stage mass spectrometry, high-resolution mass spectrometry and isotopic labeling. Drug Test. Anal. 12(4), 496–503. doi:10.1002/dta.2758
Goldstone, J. V., McArthur, G. A., Kubota, A., Zanette, J., Parente, T., Jönsson, M. E., et al. (2010) Identification and developmental expression of the full complement of Cytochrome P450 genes in Zebrafish. BMC Genom 11, 643. doi:10.1186/1471-2164-11-643
Han, Y., Yan, W., Zheng, Y., Khan, M. Z., Yuan, K., and Lu, L. (2019). The rising crisis of illicit fentanyl use, overdose, and potential therapeutic strategies. Transl. Psychiatry 9, 282. doi:10.1038/s41398-019-0625-0
Henderson, G. L. (1991). Fentanyl-related deaths: demographics, circumstances, and toxicology of 112 cases. J. Forensic Sci. 36, 13045J–13433J. doi:10.1520/jfs13045j
Hibbs, J., Perper, J., and Winek, C. L. (1991). An outbreak of designer drug-related deaths in Pennsylvania. JAMA 265, 1011–1013. doi:10.1001/jama.265.8.1011
Kanamori, T., Togawa-Iwata, Y., Segawa, H., Yamamuro, T., Kuwayama, K., Tsujikawa, K., et al. (2018). Metabolism of fentanyl and acetylfentanyl in human-induced pluripotent stem cell-derived hepatocytes. Biol Phar Bull 41(1), 106–114. doi:10.1248/bpb.b17-00709
Kanamori, T., Togawa-Iwata, Y., Segawa, H., Yamamuro, T., Kuwayama, K., Tsujikawa, K., et al. (2018). Use of hepatocytes isolated from a liver-humanized mouse for studies on the metabolism of drugs: application to the metabolism of fentanyl and acetylfentanyl. Forensic Toxicol. 36(2), 467–475. doi:10.1007/s11419-018-0425-x
Krishna, T., Claudia, E., Ksenia, J., Julita, S.-M., Rik, I. L., Sc, K., et al. (2021). Zebrafish early life stages as an alternative model to study 'designer drugs': concordance with mammals in response to opioids. Toxicol. Appl. Phar 419, 115483. doi:10.1016/j.taap.2021.115483
Mohammed, Y. I., Kurog, K., Shaban, A. A., Xu, Z., Liu, M. Y., Williams, F. E., et al. (2012) Identification and characterization of zebrafish SULT1 ST9, SULT3 ST4, and SULT3 ST5. Aquat. Toxicol. 112-113, 11–18. doi:10.1016/j.aquatox.2012.01.015
Morbiato, E., Bilel, S., Tirri, M., Arfè, R., Fantinati, A., Savchuk, S., et al. (2020). Potential of the zebrafish model for the forensic toxicology screening of NPS: a comparative study of the effects of APINAC and methiopropamine on the behavior of zebrafish larvae and mice. Neur Toxicol. 78, 36–46. doi:10.1016/j.neuro.2020.02.003
Nolan, M. L., Shamasunder, S., Colon-Berezin, C., Kunins, H. V., and Paone, D. (2019). Increased presence of fentanyl in cocaine-involved fatal overdoses: implications for prevention. J. Urban Health 96, 49–54. doi:10.1007/s11524-018-00343-z
Ott, C. E., Perez-Estebanez, M., Hernandez, S., Kelly, K., Dalzell, K. A., Arcos-Martinez, M. J., et al. (2022). Forensic identification of fentanyl and its analogs by electrochemical-surface enhanced Raman spectroscopy (EC SERS) for the screening of seized drugs of abuse. Front. Anal. Sci. 2:834820. doi:10.3389/frans.2022.834820
Pesavento, S., Bilel, S., Murari, M., Gottardo, R., Arfè, R., Tirri, M., et al. (2022). Zebrafish larvae: a new model to study behavioural effects and metabolism of fentanyl, in comparison to a traditional mice model. Med, Sci Law 62(3), 188–198. doi:10.1177/00258024221074568
Poklis, A. (1995). Fentanyl: a review for clinical and analytical toxicologists. J. Toxicol. Clin. Toxicol. 33, 439–447. doi:10.3109/15563659509013752
Roxburgh, A., and Nielsen, S. (2022). Twenty-year trends in pharmaceutical fentanyl and illicit fentanyl deaths, Australia 2001-2021. Int. J. Drug Policy 109, 103854. doi:10.1016/j.drugpo.2022.103854
Strayer, K. E., Antonides, H. M., Juhascik, M. P., Daniulaityte, R., and Sizemore, I. E. (2018). LC-MS/MS-Based method for the multiplex detection of 24 fentanyl analogues and metabolites in whole blood at sub ng mL-1 concentrations. ACS Omega 3(1), 514–523. doi:10.1021/acsomega.7b01536
Wei, Q., and Su, F. H. (2022). Determination of nine fentanyl drugs in hair samples by GC-MS/MS and LC-MS/MS. ACS Omega 7(23), 19176–19182. doi:10.1021/acsomega.2c00087
Wilde, M., Pichini, S., Pacifici, R., Tagliabracci, A., Busardò, F. P., Auwärter, V., et al. (2019). Metabolic pathways and potencies of new fentanyl analogs. Front. Pharmacol. 10, 238. doi:10.3389/fphar.2019.00238
Xu, D., Zhang, W., Li, J., Wang, J., Qin, S., and Lu, J. (2019). Analysis of AMB-FUBINACA biotransformation pathways in human liver microsome and zebrafish systems by liquid chromatography-high resolution mass spectrometry. Front. Chem. 7, 240. doi:10.3389/fchem.2019.00240
Yu, C. C., Yuan, M., Yang, H. Y., Zhuang, X. M., and Li, H. (2022). P-glycoprotein on blood-brain barrier plays a vital role in fentanyl brain exposure and respiratory toxicity in rats. Toxicol. Sci. 164(1), 353–362. doi:10.1093/toxsci/kfy093
Keywords: fentanyl, metabolites, biotransformation pathways, the liver and brain of zebrafish, HR-MS
Citation: Liu M, Huang J, Zhao S, Wang B-j, Zhou H and Liu Y (2023) Comparative analysis of the metabolites and biotransformation pathways of fentanyl in the liver and brain of zebrafish. Front. Pharmacol. 14:1325932. doi: 10.3389/fphar.2023.1325932
Received: 22 October 2023; Accepted: 04 December 2023;
Published: 18 December 2023.
Edited by:
Junmin Zhang, Lanzhou University, ChinaReviewed by:
Xilin Li, National Center for Toxicological Research (FDA), United StatesCopyright © 2023 Liu, Huang, Zhao, Wang, Zhou and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meng Liu, bGl1bWVuZ0B6anBjLmVkdS5jbg==; Yao Liu, bGl1eWFvMTEyM0BhbGl5dW4uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.