- 1Center of Excellence in Translational Medicine, Fundeni Clinical Institute, Bucharest, Romania
- 2Digestive Diseases and Liver Transplantation Center, Fundeni Clinical Institute, Bucharest, Romania
- 3Faculty of Medicine, Carol Davila University of Medicine and Pharmacy, Bucharest, Romania
Hepatocellular carcinoma (HCC) remains a major health problem worldwide, being the leading cause of cancer-related deaths, with limited treatment options, especially in its advanced stages. Tumor resistance is closely associated with the activation of the EMT phenomenon and its reversal, being modulated by different molecules, including noncoding RNAs (ncRNAs). Noncoding RNAs have the potential to function as both tumor suppressors and oncogenic molecules, controlling the malignant potential of HCC cells. Basically, these molecules circulate in the tumor microenvironment, encapsulated in exosomes. Their impact on cell biology is more significant than originally expected, which makes related research rather complex. The temporal and spatial expression patterns, precise roles and mechanisms of specific ncRNAs encapsulated in exosomes remain primarily unknown in different stages of the disease. This review aims to highlight the recent advances in ncRNAs related to EMT and classifies the described mechanism as direct and indirect, for a better summarization. Moreover, we provide an overview of current research on the role of ncRNAs in several drug resistance-related pathways, including the emergence of resistance to sorafenib, doxorubicin, cisplatin and paclitaxel therapy. Nevertheless, we comprehensively discuss the underlying regulatory mechanisms of exosomal ncRNAs in EMT-HCC via intercellular communication pathways.
1 Introduction
Hepatocellular carcinoma (HCC) is a common lethal malignancy among patients with chronic liver disease, with approximately 800,000 deaths annually, according to the GLOBOCAN 2020 report (Sung et al., 2021). Several treatment options are available for therapeutic purposes, such as trans-arterial chemoembolization (TACE) with anthracyclines, cisplatin, and multikinase inhibitor, sorafenib (Pratama et al., 2019). However, these treatments become challenging to manage, due to the appearance of invasion, metastasis and recurrence, whose key molecular sign is EMT (Yan et al., 2018).
EMT (epithelial-mesenchymal transition) is a morphogenetic process in which epithelial cells get a mesenchymal phenotype. In early EMT, transcriptional factors (TFs) are activated to repress epithelial genes and activate the mesenchymal ones. These transcriptional changes trigger the following key events: cell-cell junction dissociations, apical-basal polarity loss, cytoskeleton architecture reorganization, the production of extracellular matrix (ECM) degradation enzymes, and cellular shape transformation. The activation of cellular pathways associates this process with proliferation, invasion, metastasis, and chemotherapy resistance (Yan et al., 2018; Dudas et al., 2020; Yang et al., 2020; Huang et al., 2022). Among these transformations, EMT is associated with numerous signaling pathways involved in inflammation, oncogenic and metabolic stress, hypoxia or apoptosis (Huang et al., 2022).
Moreover, many studies suggest that noncoding RNAs (ncRNAs), such as microRNAs (miRNAs), long-noncoding RNAs (lncRNAs) and circular RNAs (circRNAs), have been linked to both the EMT process activation and inhibition. Indeed, these types of RNAs have multiple roles in cancerous cells because one ncRNA transcript could target many molecules involved in different signaling pathways (Toden et al., 2021; Khanbabaei et al., 2022).
This review highlights ncRNAs’ significant direct and indirect signaling pathways in the EMT process and how these mechanisms are involved in HCC progression and chemoresistance. Finally, we provide an update on developing exosome-based therapies against HCC and their molecular aspects in EMT (Figure 1).
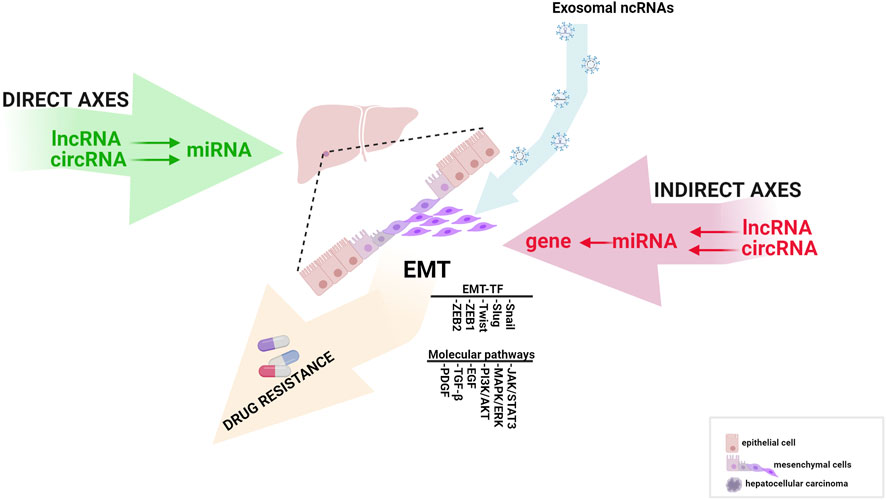
FIGURE 1. The influence of direct and indirect EMT-related ncRNA axes in HCC progression (created with biorender.com accessed on July 2023).
2 EMT-related ncRNAs mechanisms of action
As mentioned above, noncoding RNAs (ncRNAs), including microRNAs, lncRNAs and circRNAs, have oncogenic and tumor suppressor roles and regulate essential processes involved in cancer progression.
MicroRNAs (miRNAs) are noncoding single-stranded RNAs of approximately 22 nucleotides transcribed in pri-miRNA by RNA Pol II (Bartel, 2004). As described in the canonical pathway, Ribonuclease III and double-stranded-RNA-binding protein, DGCR8, recognize this structure in the nucleus, generating a pre-miRNA of ∼65 nucleotides. Pre-miRNA is exported to the cytoplasm by an Exportin 5 and Ran-GTP complex and recognized by RNase III Dicer, which forms a miRNA duplex. This mature form is incorporated into an RNA-induced silencing complex (RISC), directing RISC to complementary mRNA targets (Cai et al., 2004). In brief, miRNAs function as negative regulators of genes when binding to RNA 3′-untranslated region (3′-UTR) (Ha and Kim, 2014). Besides that, the interaction with coding sequences, gene promoters, and 5′-UTR has been proved (O'Brien et al., 2018). Because each miRNA can regulate multiple targets containing specific miRNA response elements (MREs) (Bassett et al., 2014) and play a crucial role in a variety of molecular processes, they have been studied in all cancer types (Esquela-Kerscher and Slack, 2006; Volinia et al., 2006; Nicoloso et al., 2009). In HCC, miRNAs modulate cell cycle, proliferation, apoptosis, epithelial-mesenchymal transition and metastasis (Sidhu et al., 2015). Furthermore, our previous studies have shown that miRNAs are an important tool in the prognostic and diagnostic HCC (Mjelle et al., 2019; Sorop et al., 2020).
Long noncoding RNAs (lncRNAs) are transcripts of approximately 200 nucleotides, which usually RNA Pol II transcribes, but so do RNA Pol I and RNA Pol III (Statello et al., 2021; Mattick et al., 2023). Moreover, they have a wide diversity, with an average of 100,000 human lncRNAs (Mattick et al., 2023). At first, lncRNAs were defined as transcriptional “junk” or “noise.” Still, in the past few years, more studies have shown the involvement of lncRNAs in different molecular pathways (Sun et al., 2017), indicating their interaction with DNA, RNA, or protein. The interaction mechanism could be: scaffold, decoy, guide, signal, or SINEUPs. Scaffold lncRNAs could act as archetype RNAs and are involved in the assembly of transcriptional regulators. The decoy mechanism implies acting as a competing endogenous RNA (ceRNA) or sponge of miRNAs, transcriptional factors, or RNA-binding proteins. In contrast, the guide mechanism involves the formation of a ribonucleoprotein complex, which targets a promoter or genomic loci (Rinn and Chang, 2012). Furthermore, lncRNAs could act as regulatory molecules (Nadhan et al., 2022) or SINEUPs containing SINE elements which enhance mRNAs translation (Toki et al., 2020).
Circular RNAs (circRNAs) are single-stranded RNAs with closed-loop structures and resistance to RNase R and exonucleases. They are generated from precursor RNA (pre-RNA) through back-splicing (Chen, 2016). This mechanism involves connecting a downstream donor site of a flanking downstream intron to an upstream acceptor site (Kristensen et al., 2019). Increasing research has revealed that circRNAs can sponge miRNAs, interact with proteins, interfere with transcription or splicing, or encode peptides (Zhang and Wang, 2021).
EMT plays a pivotal role in the early stage of metastasis (Bakir et al., 2020); thus, many studies have been conducted to determine the function of ncRNAs in this highly dynamic phenomenon. Therefore, this review underlines two types of mechanisms: direct and indirect.
2.1 Direct EMT-related ncRNAs’ mechanism of action
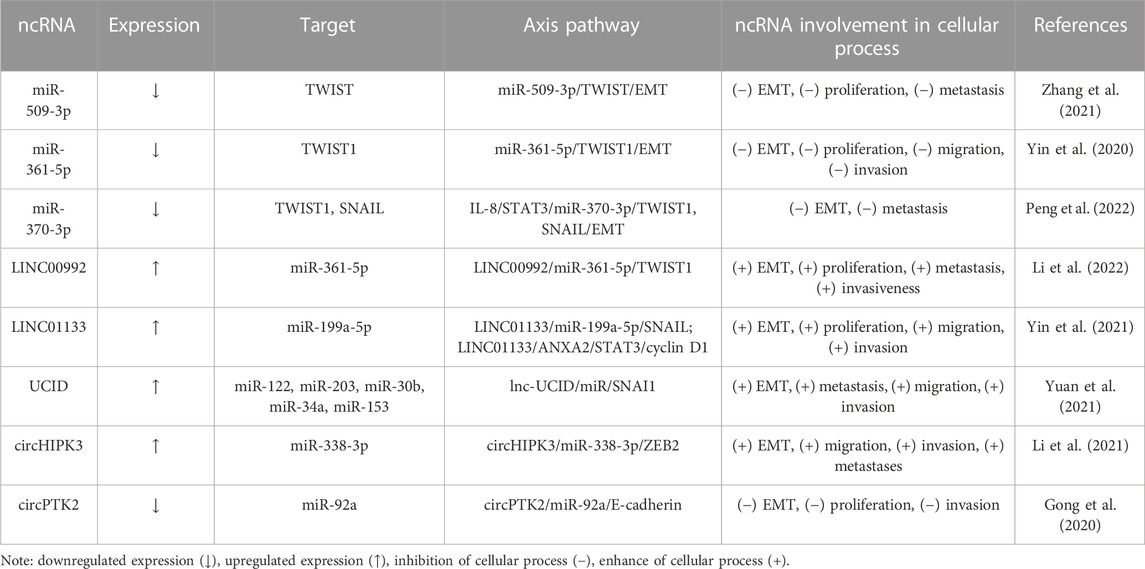
Direct mechanism involves direct interaction between miRNA and EMT-regulatory factors, such as twist family bHLH transcription factor 1 (TWIST), snail family transcriptional repressor 1 (SNAIL), or zinc finger E-box binding homeobox 1/2 (ZEB1/2) (Skovierova et al., 2018). We defined this mechanism by three crucial axes: miRNA/EMT, lncRNA/miRNA/EMT, and circRNA/miRNA/EMT.
Several miRNAs, such as miR-509-3p (Zhang et al., 2021), miR-361-5p (Yin et al., 2020), and miR-370-3p (Peng et al., 2022), have been found to inhibit TWIST1 expression via targeting its 3′UTR and to abate the EMT process. Li et al. (2022) observe that LINC00992 downregulates miR-361-5p and upregulates TWIST1, thus promoting cell proliferation, migration, and invasion. In addition, miR-370-3p decreases TWIST1 and SNAIL, affecting interleukin 8 (IL-8) expression and restraining the metastasis capacity in HCC cells (Peng et al., 2022). In contrast, LINC01133 (Yin et al., 2021) and lnc-UCID (Yuan et al., 2021) increase EMT by acting as a sponge of miRNAs, increasing SNAIL expression. Furthermore, circHIPK3 promotes metastases and ZEB2 expression via inhibiting miR-338-3p (Li et al., 2021). In contrast, circPTK2 and E-cadherin compete for binding miR-92a that, aggravates proliferation and invasion, while circPTK2 suppresses miRNA’s effect in HCC cells (Gong et al., 2020), as summarized in the direct mechanism part from Table 1.
2.2 Indirect EMT-related ncRNAs’ mechanism of action
The indirect mechanism involves miRNA/mRNA, lncRNA/miR/mRNA, and circRNA/miRNA/mRNA regulatory axes that modulate an EMT molecule.
2.2.1 miRNA/mRNA axes
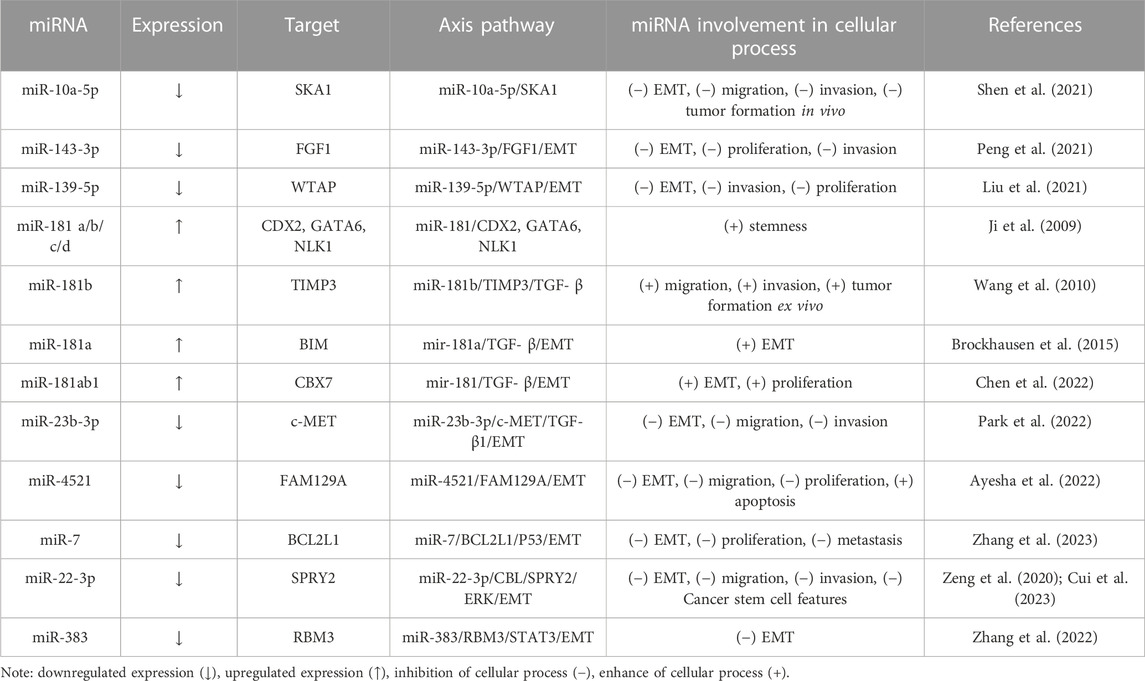
Numerous miRNA/mRNA axes have been found to be involved in the EMT process (Table 2).
For instance, Shen et al. (2021) have found that miRNA-10a-5p is downregulated in HCC tissues and decreases EMT in HCC cells by targeting spindle and kinetochore-associated complex subunit 1 (SKA1). SKA1 is upregulated in tumors, promoting cancer progression, and has a prognostic value in HCC (Chen et al., 2018; Song et al., 2022). Other oncosuppressors are miR-143-3p and miR-139-5p, which repress fibroblast growth factor 1 (FGF1) and Wilms’ tumor 1-associating protein (WTAP). Those proteins increase EMT, proliferation and invasion of HCC cells (Liu et al., 2021; Peng et al., 2021). Moreover, Zhu et al. (2021) declare that miR-139-5p is regulated by lncRNA TTN antisense RNA 1 (TTN-AS1) and inhibits Sparc/osteonectin, cwcv, and kazal-like domains proteoglycan 1 (SPOCK1), an oncogenic proteoglycan involved in EMT (Vancza et al., 2022).
Growing studies have supported the importance of transforming growth factor beta (TGF-β) in HCC via SMAD/non-SMAD-dependent signaling pathways, which induce EMT-TFs (Hao et al., 2019). Several studies have shown that the miR-181 family positively correlates with TGF-β pathways, thus increasing EMT, tumor progression and stemness (Ji et al., 2009; Wang et al., 2010; Brockhausen et al., 2015; Chen et al., 2022). In contrast, miR-23b-3p has been proven to inhibit TGF- β1-induced EMT and block invasion and migration (Park et al., 2022).
Apoptosis or programmed cell death is a complex mechanism that involves death receptors (extrinsec pathway) and mitochondria (intrinsic pathway), by which it maintains cell homeostasis (Schattenberg et al., 2011). As discussed above, EMT confers resistance to apoptosis (Valdes et al., 2002). Interestingly, miR-4521 acts as an oncosuppressor in HCC cells by modulating mechanisms involved in proliferation and apoptosis. On the one hand, miR-4521 activates two apoptosis pathways (p-FAK/p-Akt/MDM2/P53 and FAK/p-Akt/BCL-2/BAX/Cytochrome-C/Caspase-3/Caspase-9) by decreasing the expression of family with sequence similarity 129 member A (FAM129A); on the other hand, it thereby attenuates invasivity by blocking TIMP-1/MMP9/MMP2, p-FAK/p-Akt and EMT pathways (Ayesha et al., 2022).
Moreover, the miR-7/BCL2L1/P53 and miR-22-3p/CBL/SPRY2/ERK axes decrease EMT, invasion, proliferation and migration (Cui et al., 2023; Zhang et al., 2023). Another EMT inhibitor is miR-383, which negatively regulates the multi-functional RNA-binding protein (RBM3) expression. As reported, RBM3 upregulates signal transducer and activator of transcription 3 (STAT3) expression via binding to its mRNA (Zhang et al., 2022). In addition, STAT3 targets the TWIST promoter and positively regulates its transcriptional activity in HCC cells, thus inducing EMT (Zhang et al., 2015).
Moreover, many studies highlight indirect mechanisms that imply lncRNA/miRNA/mRNA and circRNA/miRNA/mRNA axes.
2.2.2 lncRNA/miRNA/mRNA axes
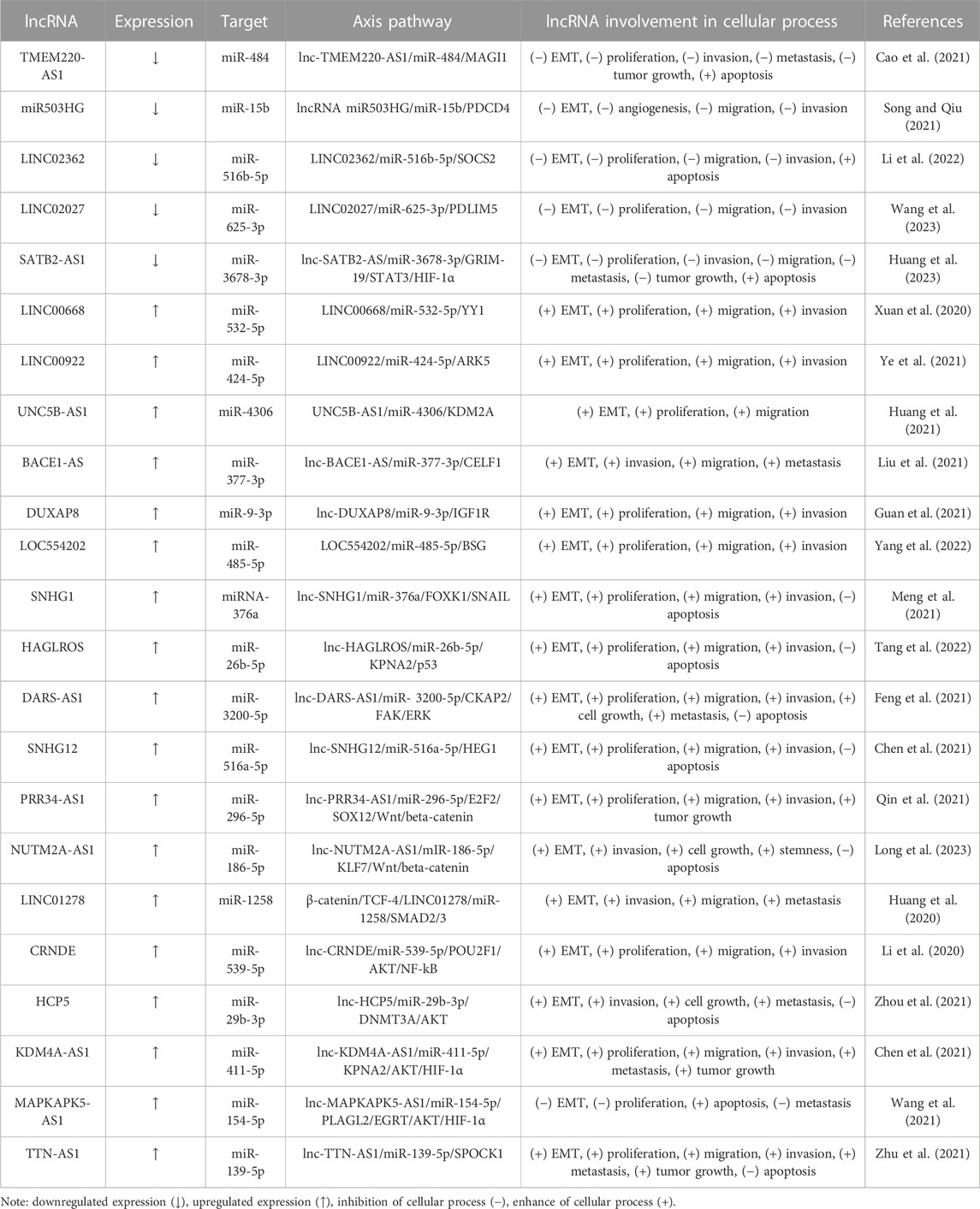
Table 3 shows the lncRNA/miRNA/mRNA axes related to EMT in HCC. According to their oncological role, lncRNAs could be classified into two groups: onco-suppressor and oncotargets. Therefore, within the last 3 years, five lncRNAs, TMEM220-AS1 (Cao et al., 2021), lncRNA miR503HG (Song and Qiu, 2021), LINC02362 (Li et al., 2022), LINC02027 (Wang et al., 2023) and SATB2-AS1 (Huang et al., 2023), have been documented to function as miRNA sponge, to decrease a gene that promotes the EMT process. For instance, Huang et al. (2023) show that SATB2-AS1 is observably reduced in HCC tissues compared to adjacent tissues and its overexpression hampers tumor growth and metastasis in vitro. Besides, SATB2-AS1 also acts as a ceRNA for miR-3678-3p. This miRNA accelerates cell proliferation and suppresses cell apoptosis by blocking GRIM-19 (gene associated with retinoic-interferon-induced mortality 19), a negative STAT3/HIF-1α pathway regulator (Huang et al., 2023).
On the other hand, several lncRNAs increase EMT by sponging miRNAs that target oncogenes. LncRNAs such as LINC00668 (Xuan et al., 2020), LINC00922 (Ye et al., 2021), UNC5B-AS1 (Huang et al., 2021), BACE1-AS (Liu et al., 2021), DUXAP8 (Guan et al., 2021) and LOC554202 (Yang et al., 2022) were upregulated in HCC to contribute to specific lncRNA/miR/mRNA axes induced EMT. SNHG1 is another lncRNA with high expression levels in HCC; it is negatively correlated to a poor patient prognosis. SNHG1 regulates cell proliferation and invasion via EMT through miR-376a binding to elevate forkhead box protein K1 (FOXK1) expression, a molecule that binds and upregulates SNAIL (Meng et al., 2021). HAGLROS knockdown impaired HCC tumorigenesis in vitro and in vivo. HALGROS increases the karyopherin α2 (KPNA2) level and suppresses p53 signaling to abate apoptosis by acting as a miR-26b-5p sponge (Tang et al., 2022). DARS-AS1 induces EMT via interacting with miR-3200-5p, further promoting Cytoskeleton associated protein 2 (CKAP2) expression and FAK/ERK pathway activation (Feng et al., 2021).
SNHG12 (Chen et al., 2021), PRR34-AS1 (Qin et al., 2021) and NUTM2A-AS1 (Long et al., 2023) axes induce EMT via Wnt/β-catenin signaling. Furthermore, Huang et al. (2020) point out that miR-1258 is downregulated in HCC patients. In vivo, experiments showed that the miR-1258 overexpression in nude mice impeded metastatic lung nodule formation. At the molecular level, LINC01278 acts as a sponge of miR-1258 and upregulates SMAD2/3, thus suppressing E-cadherin and enhancing vimentin expression. Moreover, transcription factor 4 (TCF-4) binds to the promoter site of LINC01278 and increases β-catenin expression, TGF-β and Wnt/β-catenin pathways, thereby activating the LINC01278/miR-1258/Samd2/Smad3 axis (Huang et al., 2020).
CRNDE and HCP5 induce Akt pathway activation by sponging miR-539-5p andmiR-29b-3p, respectively, to promote the EMT and the progression of HCC (Li et al., 2020; Zhou et al., 2021). Furthermore, two others oncogenic lncRNAs, KDM4A-AS1 and MAPKAPK5-AS1, activated by hypoxia-inducible factor 1-alpha (HIF1α), have also been found to increase protein kinase B (Akt) (Chen et al., 2021; Wang et al., 2021); their corresponding axes being listed in Table 3.
2.2.3 circRNA/miRNA/mRNA axes
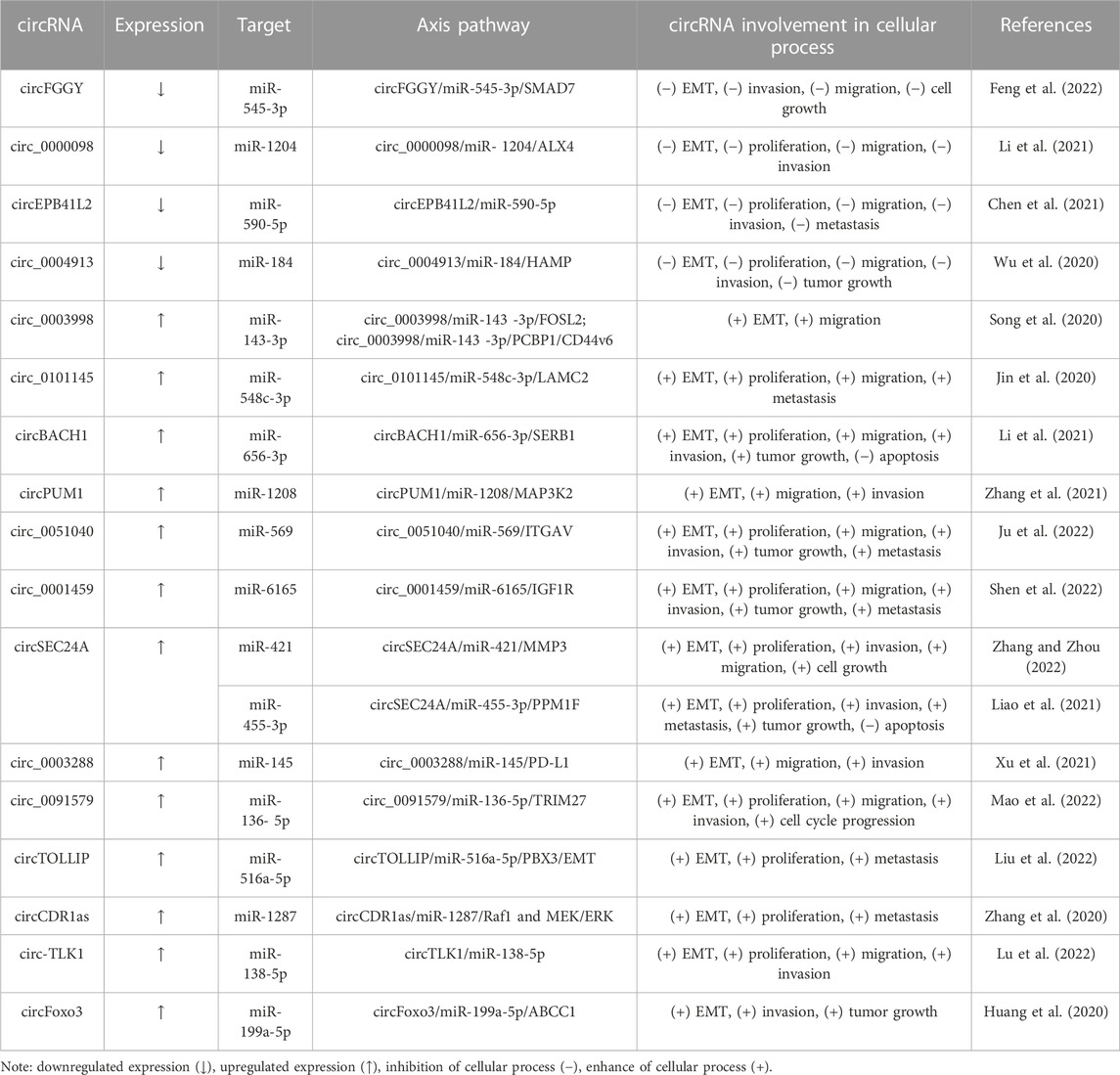
In the Table 4 there are highlighted critical pathways that involve circRNAs. In HCC, the levels of circFGGY (circ_0006633) (Feng et al., 2022), circ_0000098 (Li et al., 2021), and circEPB41L2 (Chen et al., 2021) are downregulated in tumor tissues and inhibit EMT, proliferation, migration, and invasion. In summary, authors highlight circFGGY/miR-545-3p/Smad7 (Feng et al., 2022), circ_0000098/miR-1204/ALX4 (Li et al., 2021) and circEPB41L2/miR-590-5p (Chen et al., 2021) axes as being important in HCC. Furthermore, Wu et al. (2020) revealed that circ_0004913 was downregulated in HCC tissues and that the overexpression of circ_0004913 constrained proliferation, EMT and metastasis by acting as a sponge of miR-184 and promoting hepcidin antimicrobial peptide (HAMP) expression. In brief, the circ_0004913/miR-184/HAMP axis regulates JAK2/STAT3/Akt signaling in HCC cells (Wu et al., 2020).
In contrast, six circRNAs, circ_0003998 (Song et al., 2020), circ_0101145 (Jin et al., 2020), circBACH1 (Li et al., 2021), circPUM1 (Zhang et al., 2021), circ_0051040 (Ju et al., 2022) and circ_0001459 (Shen et al., 2022), have been observed to manipulate various miR/mRNA axes to induce EMT. Besides, elevated level of circSEC24A leads to the expression of protein phosphatase, Mg2+/Mn2+ dependent 1F (PPM1F) and matrix metalloproteinase 3 (MMP3) by sponging miR-455-3p and miR-421, respectively (Liao et al., 2021; Zhang and Zhou, 2022). MMPs are a class of enzymes that degrade extracellular matrix (ECM) proteins (Klein and Bischoff, 2011). In HCC, it was reported that MMP3 promotes EMT and metastasis (Scheau et al., 2019).
Circ_0003288 is an oncogenic RNA that enhances EMT by increasing programmed death-ligand 1 (PD-L1) and Akt pathways via miR-145 sponging (Xu et al., 2021). Circ_0091579 has been demonstrated to pin HCC patients and its downregulation inhibits EMT and promotes apoptosis in vitro. Also, miR-136-5p is a direct target of circ_0091579 and its overexpression suppresses the malignant potential of HCC cells via regulating tripartite motif containing 27 (TRIM27) expression (Mao et al., 2022).
Moreover, the Toll interacting protein (TOLLIP)-derived circRNA (circTOLLIP) is also found to be involved in the EMT of HCC. CircTOLLIP is upregulated in HCC via eukaryotic translation initiation factor 4A3 (EIF4A3), an RNA-binding protein. This circRNA acts as a ceRNA for miR-516a-5p, thus upregulating PBX3 and exhibiting pro-tumor roles in vitro and in vivo (Liu et al., 2022).
CircRNA CDR1as is highly expressed in some cancers (Jiang et al., 2020). Specifically, circRNA CDR1as is overexpressed in HCC tissues and its expression positively regulates EMT, proliferation and metastasis in HCC cells via the miR-1287 sponge. This circRNA enhances Raf-1 proto-oncogene, serine/threonine kinase (RAF1) expression, a crucial molecule in the RAS/RAF/MEK/ERK pathway (Zhang et al., 2020).
3 The role of ncRNA/mRNA axes in HCC drug resistance
As discussed above, EMT is associated with chemotherapy resistance by avoiding cell death mechanisms (De Las Rivas et al., 2021). Therefore, a growing number of studies have supported the importance of EMT-related ncRNAs in molecular pathways of different therapies (He et al., 2022).
Sorafenib is the first-line FDA-approved treatment for HCC (Niu et al., 2021) and an oral multikinase inhibitor that targets vascular endothelial growth factor receptor 2 (VEGFR2), platelet-derived growth factor receptor (PDGFR), hepatocyte factor receptor (KIT), or other molecules to decrease angiogenesis. HCC cells acquire resistance to sorafenib by different molecular pathways, including EMT (Marisi et al., 2018; Tang et al., 2020). In this context, lncH19 knockdown has been reported to inhibit EMT in HCC cells by enhancing miR-675 expression, which is involved in sorafenib sensitivity. In brief, H19 promoted sorafenib resistance (Xu et al., 2020). LncRNA-POIR also has an oncogenic effect and suppresses miR-182-5p expression, inhibiting the EMT process and triggering sorafenib sensitivity (Chen et al., 2021). Additionally, small nucleolar RNA host gene 3 (SNHG3) induces EMT and CD151 expression by functioning as a ceRNA for miR-128. LncRNA-SNHG3 can induce sorafenib resistance and promote invasion in vitro (Zhang et al., 2019). In contrast, lncLIMT (LINC01089), which reppresses miR-665 expression and EMT, decreases sorafenib resistance. In addition, LIMT inhibits tumor growth in vivo in tumor nude mouse models (Sun et al., 2022). MiR-125b-5p is upregulated in sorafenib-resistant HCC cell lines and its overexpression induces EMT by repressing ataxin 1 (ATXN1) expression. Thus, it was reported that miR-125b-5p enhances sorafenib resistance in vivo (Hirao et al., 2021).
Besides Sorafenib, TACE with doxorubicin and cisplatin is used in HCC advanced patients (Lu et al., 2017; Couri and Pillai, 2019).
Doxorubicin (Adriamycin, DOX) is an anthracycline drug used as an antineoplastic agent. The most known mechanism of action involves the interaction with topoisomerase IIα (TOP2A) (Tewey et al., 1984) and the activation of apoptosis (Roos and Kaina, 2013). Anthracycline drug resistance is caused by the incapability of DOX to accumulate in the nucleus (Cox and Weinman, 2016). For instance, Zhang et al. (2021) reported that overexpression of linc-ROR (long intergenic non-protein coding RNA (linc)-regulator of reprogramming) increases DOX resistance in HCC cell lines by TWIST upregulation. Also, circFoxo3 has higher expression in adriamycin-resistant patients. It has been shown that circFoxo3 via miR-199a enhances ABCC1 expression, a known protein involved in drug resistance. Moreover, the downregulation of miR-199a promoted EMT signaling in HCC cells and reversed circFoxo3 inhibition effects (Huang et al., 2020).
Li et al. (2020) identified that circ_0003998 downregulation facilitated DOX-sensitivity by E2F Transcription Factor 3 (E2F3) regulation. They further identified circ_0003998 as a sponge of miR-218-5p and Eukaryotic initiation factor 5A2 (EIF5A2) as a direct target of miR (Li et al., 2020). Moreover, EIF5A2 is involved in genistein resistance, an essential anti-tumoral phytoestrogen that promotes apoptosis (Sarkar and Li, 2002) and inhibits EMT and stemness. MiR-1275 is a tumor suppressor that can bind 3′-UTR EIF5A2 as a protein that upregulated PI3K/Akt and EMT pathways. MiR-1275 was expressed at a higher level by genistein treatment (Yang et al., 2022). Furthermore, it has been shown that miR-140-5p is involved in drug resistance in HCC cells. In brief, miR-140-5p improves DOX sensitivity through PIN1 depletion (Gao et al., 2021) and catalpol sensitivity through EMT suppression (Wu et al., 2021).
Cisplatin is a chemotherapeutic that inhibits transcription and replication, inducing apoptosis and necrosis in HCC cells (Ishikawa, 2009). It has been shown that miR-9 increases cisplatin sensitivity in vitro and in vivo by targeting EIF5A2 and EMT process. Besides that, EIF5A2 depletion decreases vimentin expression and increases E-cadherin in HCC cell lines (Bao et al., 2020). Another ncRNA involved in cisplatin sensitivity is miR-138 by its direct target, enhancer of zeste homolog 2 (EZH2). This miRNA upregulates EMT markers; therefore, the miR-138/EZH2/EMT axis could regulate cisplatin resistance (Zeng et al., 2021), also involved in radiosensitivity. Bai et al. (2022) show that miR-138 is downregulated in HCC tissue and its expression is indirectly correlated with EZH2 expression, which is a direct target of miR-138-5p. By RNA-seq, they observed that miR-138-5p upregulation inhibits HIF-1α and EMT (Bai et al., 2022). Moreover, Lu et al. (2022) reported that miR-138-5p is negatively regulated by circ-TLK1.
Paclitaxel—a microtubule-stabilizing molecule, induces cell death (Weaver, 2014). As mentioned above, paclitaxel (PTX) is another drug whose resistance could be caused by different signaling pathways, including ncRNAs and EMT (Ashrafizadeh et al., 2021). Liu et al. (2020) pointed out circ-BIRC6 (circRNA baculoviral IAP repeat-containing 6) as an inhibitor of PTX sensitivity by sponging miR-8 77-5p to enhance tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta (YWHAZ) expression. Its role in drug resistance has been reported in ovarian cancer (Hong et al., 2018), bladder cancer (Yu et al., 2019), and gastric cancer (Zhao et al., 2021). Furthermore, miR-212-3p is decreased in PTX-resistant cells. This miRNA can bind to 3′UTR ZEB2, thus mediating chemoresistance in HCC cells. Transfection of miR-212-3p in resistant cells inhibited ZEB2 expression, reversing EMT (Yang et al., 2020). Figure 2 summarizes the ncRNAs axes involved in HCC drug resistance.
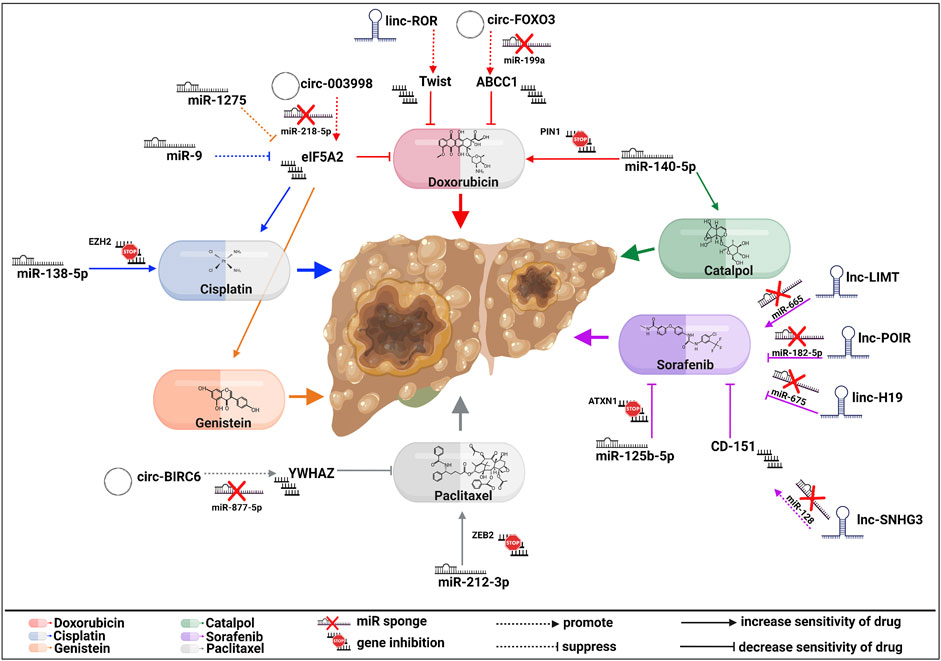
FIGURE 2. Molecular actions of EMT-related ncRNA axes in HCC drug resistance. Multiple regulatory components either increase or decrease sensitivity to sorafenib, paclitaxel, genistein, cisplatin, doxorubicin, or catalpol, affecting HCC progression. The signaling pathways of every drug are represented by different colors, as seen above (created with biorender.com accessed on July 2023).
These investigations show the complex and dual role of ncRNAs in EMT. The exact mechanism by which every ncRNA is involved in the HCC will be difficult to decode because of its functions in many hepatocellular processes. One way to start is by classifying the miRNAs based on their direct or indirect impact on the EMT process. Undoubtedly, future studies are necessary to report new miRNAs associated with HCC-EMT and to map their function in this process, which can lead to the development of novel therapies.
Therefore, to translate ncRNAs in a therapeutic situation, tools must be developed to analyze these ncRNA axes functionally and to devise therapy strategies, so as to overcome off-target and toxicity consequences.
4 EMT-associated exosomal ncRNAs in HCC
Exosomes can be found in all human body fluids (blood, urine, saliva, ascites, cerebrospinal and synovial fluids) (Jiang et al., 2022). They are extracellular 30–100 nm vesicles (EVs) having a lipid bilayer; they are generated from the luminal membranes of multivesicular bodies (MVBs) and released into the extracellular matrix after MVBs fusion with the cell membrane (Kim et al., 2020). The primary physiological role of exosomes is to mediate cell-cell communication by transferring bioactive molecules, such as proteins or nucleic acids (Chen et al., 2021), thus being one of the most studied tools for the interchange of substances between tumor cells and the tumor microenvironment (Jiang et al., 2022).
In the last decade, more studies have highlighted the regulatory effects of different bioactive molecules delivered by exosomes, such as ncRNAs, in the EMT process in various types of cancers, including HCC. Interestingly, they can promote or suppress the EMT phenomena in HCC cells.
According to RNAseq investigation, exosomal miR-92a-3p expression level increases in two established high-metastatic HCC cell lines (97 hm and Huhm). Besides, treatment with high-metastatic HCC-derived exosomal miR-92a-3p facilitates the aggressiveness of HCC cells via PTEN inhibition and Akt/Snail signaling activation, promoting EMT (Yang et al., 2020). Similarly, high levels of miR-4800-3p were found in Huh7 cell-derived exosomes. Thus, Lin et al. (2022) demonstrated that exosomal miR-4800-3p heightened the progression of HCC by regulating the Hippo signaling pathway and targeting STK25 in both in vitro and in vivo experiments. Moreover, the treatment of low metastatic HCC cells with exosomal miR-4800-3p downregulates the expression of E-cadherin and ZO-1 and increases the expression of N-cadherin, activating the EMT process (Lin et al., 2022).
Interestingly, M2 macrophages can influence tumor development by secreting various cytokines and exosomes that can be loaded with specific miRNAs. For instance, miR-660-5p-loaded M2 exosomes augmented EMT and enhanced the tumorigenic ability in HCC cells through downregulating Kruppel-like factor 3 (KLF3) expression (Tian et al., 2021).
Human umbilical cord mesenchymal stem cells (hucMSCs) have low immunogenicity and high proliferation and differentiation potential. Additionally, the treatment of HCC cells with hucMSC-Exo upregulates miR-451a. This miRNA inhibits a disintegrin and metalloprotease 10 (ADAM10), thus reducing EMT and aggressive phenotypes of HCC (Xu et al., 2021).
Several studies showed that TGF-β treatment induces EMT (Miyazono, 2009; Lin et al., 2020; Kim et al., 2021) and treatment with exosomes derived from these cells increases proliferation and metastasis in HCC cells (Lin et al., 2020) through intercellular communication. Lin et al. (2020) reported that 119 miRNAs are upregulated, such as miR-125b-5p, 374a-5p, miR-24-3p, miR-200b-3p, and miR-21-5p, and 186 are downregulated in EMT-Hep3B-derived exosomes (EMT-Hep3B exo), as compared to Hep3B exo. Moreover, treatment with EMT-Hep3B exo with miR-374a-5p interference inhibits hepatocellular metastasis by upregulation of growth arrest and DNA damage 45-alpha (GADD45A), a cell growth suppressor (Lin et al., 2020). In contrast, Huh7 cell-derived exosomes loaded with miR-125b (Exo-125b) blocks EMT and suppresses metastatic potential via inhibiting TGF-β1/SMAD pathways (Kim et al., 2021). Similarly, miR-374c-5p was found to be downregulated in the EMT model and transferred by exosomes derived from bone marrow mesenchymal stem cells (BMSC) suppresses EMT via targeting LIM domain kinase 1 (LIMK1) and inhibiting Wnt/β-catenin and TGF-β1 axes in HCC cells (Ding et al., 2023).
Yao et al. (2022) identified that lncRNA THEMIS2-211 is upregulated in plasma-derived exosomes from HCC patients. Knockdown of THEMIS2-211 increases E-cadherin and decreases N-cadherin and vimentin in HCC cells. Mechanistically, they showed that THEMIS2-211 is an oncogene that promotes proliferation, migration, invasion, and EMT by sponging miR-940 and increasing SPOCK1 expression (Yao et al., 2022).
Circ-0004277 and lncRNA PRR34-AS1 transfer via exosomes to human hepatic cells increases the malignant phenotype (Zhu et al., 2020; Zhang et al., 2022). Thus, PRR34-AS1 enhanced Rab27a expression to increase the exosome secretion of VEGF and TGF-β in HCC cells and transmitted them into the human liver epithelial (THLE-3) cells (Zhang et al., 2022).
In summary, these studies prove that exosomes act as ncRNAs cargo for tumor cells and have distinct regulatory effects on the EMT process in HCC and various underlying processes. Although exosomes are promising therapy in cancer, improvement of their purification, and additional studies on the interaction and mechanisms with other types of cells remain the main problems to be solved in their uses.
5 Conclusion and future perspectives
The development of transcriptomics approaches in the last decade has highlighted the essential roles of ncRNAs in cancer (Slack and Chinnaiyan, 2019; Winkle et al., 2021). The formation of ncRNA axes starts to become an essential tool in various cellular mechanisms, and its role in the progression of HCC is decisive (Wong et al., 2018). Furthermore, it will be crucial to comprehend how ncRNA axes regulate migration, proliferation, and EMT in HCC cells, so as to generate cutting-edge therapeutic medications based on ncRNAs, to prevent and manage HCC.
Taking together these observations, we find that defining ncRNA pathways in direct and indirect mechanisms could map a precise road to a therapeutic target as close to a clinical necessity as possible. The EMT-related miRNAs’ direct mechanism of action could be a promissive path in developing new therapies against metastasis. However, more research is needed to understand how these miRNA axes work and to determine which transcripts are valuable targets. Undoubtedly, since a single miRNA could have several targets and can affect more therapeutic drugs, its use as a new therapy in cancer requires an in-depth study of the mechanisms involved.
Author contributions
A-VG: Writing–original draft, Writing–review and editing, Conceptualization. AS: Writing–original draft, Writing–review and editing, Conceptualization. SOD: Project administration, Supervision, Writing–review and editing, Funding acquisition.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was founded by “Carol Davila” University of Medicine and Pharmacy Bucharest, Romania through Contract no. 33PFE/30.12.2021 funded by the Ministry of Research and Innovation within PNCDI III, Program 1–Development of the National RD system, Subprogram 1.2–Institutional Performance–RDI excellence funding projects; EEA Financial Mechanism 2009–2014 under the project contract number 4SEE/30.06.2014; and by a grant from the Ministry of Research, Innovation and Digitization, CCCDI-UEFISCDI, project number PN-III-P2-2.1-PED-2021-3180, contract number 629PED/2022, within PNCDI III.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ashrafizadeh, M., Mirzaei, S., Hashemi, F., Zarrabi, A., Zabolian, A., Saleki, H., et al. (2021). New insight towards development of paclitaxel and docetaxel resistance in cancer cells: EMT as a novel molecular mechanism and therapeutic possibilities. Biomed. Pharmacother. 141, 111824. doi:10.1016/j.biopha.2021.111824
Ayesha, M., Majid, A., Zhao, D., Greenaway, F. T., Yan, N., Liu, Q., et al. (2022). MiR-4521 plays a tumor repressive role in growth and metastasis of hepatocarcinoma cells by suppressing phosphorylation of FAK/AKT pathway via targeting FAM129A. J. Adv. Res. 36, 147–161. doi:10.1016/j.jare.2021.05.003
Bai, B., Liu, Y., Fu, X. M., Qin, H. Y., Li, G. K., Wang, H. C., et al. (2022). Dysregulation of EZH2/miR-138-5p Axis contributes to radiosensitivity in hepatocellular carcinoma cell by downregulating hypoxia-inducible factor 1 alpha (HIF-1α). Oxid. Med. Cell Longev. 2022, 7608712. doi:10.1155/2022/7608712
Bakir, B., Chiarella, A. M., Pitarresi, J. R., and Rustgi, A. K. (2020). EMT, MET, plasticity, and tumor metastasis. Trends Cell Biol. 30 (10), 764–776. doi:10.1016/j.tcb.2020.07.003
Bao, Y., Zhang, Y., Lu, Y., Guo, H., Dong, Z., Chen, Q., et al. (2020). Overexpression of microRNA-9 enhances cisplatin sensitivity in hepatocellular carcinoma by regulating EIF5A2-mediated epithelial-mesenchymal transition. Int. J. Biol. Sci. 16 (5), 827–837. doi:10.7150/ijbs.32460
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 (2), 281–297. doi:10.1016/s0092-8674(04)00045-5
Bassett, A. R., Azzam, G., Wheatley, L., Tibbit, C., Rajakumar, T., McGowan, S., et al. (2014). Understanding functional miRNA-target interactions in vivo by site-specific genome engineering. Nat. Commun. 5, 4640. doi:10.1038/ncomms5640
Brockhausen, J., Tay, S. S., Grzelak, C. A., Bertolino, P., Bowen, D. G., d'Avigdor, W. M., et al. (2015). miR-181a mediates TGF-beta-induced hepatocyte EMT and is dysregulated in cirrhosis and hepatocellular cancer. Liver Int. 35 (1), 240–253. doi:10.1111/liv.12517
Cai, X., Hagedorn, C. H., and Cullen, B. R. (2004). Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10 (12), 1957–1966. doi:10.1261/rna.7135204
Cao, C., Li, J., Li, G., Hu, G., Deng, Z., Huang, B., et al. (2021). Long non-coding RNA TMEM220-AS1 suppressed hepatocellular carcinoma by regulating the miR-484/MAGI1 Axis as a competing endogenous RNA. Front. Cell Dev. Biol. 9, 681529. doi:10.3389/fcell.2021.681529
Chen, B. W., Zhou, Y., Wei, T., Wen, L., Zhang, Y. B., Shen, S. C., et al. (2021). lncRNA-POIR promotes epithelial-mesenchymal transition and suppresses sorafenib sensitivity simultaneously in hepatocellular carcinoma by sponging miR-182-5p. J. Cell Biochem. 122 (1), 130–142. doi:10.1002/jcb.29844
Chen, F., He, L., Qiu, L., Zhou, Y., Li, Z., Chen, G., et al. (2021). Circular RNA CircEPB41L2 functions as tumor suppressor in hepatocellular carcinoma through sponging miR-590-5p. Cancer Manag. Res. 13, 2969–2981. doi:10.2147/CMAR.S291682
Chen, H., Wang, L., Zeng, X., Schwarz, H., Nanda, H. S., Peng, X., et al. (2021). Exosomes, a new star for targeted delivery. Front. Cell Dev. Biol. 9, 751079. doi:10.3389/fcell.2021.751079
Chen, J., Zhao, Y., Zhang, F., Li, J., Boland, J. A., Cheng, N. C., et al. (2022). Liver-specific deletion of miR-181ab1 reduces liver tumour progression via upregulation of CBX7. Cell Mol. Life Sci. 79 (8), 443. doi:10.1007/s00018-022-04452-6
Chen, L. L. (2016). The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 17 (4), 205–211. doi:10.1038/nrm.2015.32
Chen, P. P., Zhang, Z. S., Wu, J. C., Zheng, J. F., and Lin, F. (2021). LncRNA SNHG12 promotes proliferation and epithelial mesenchymal transition in hepatocellular carcinoma through targeting HEG1 via miR-516a-5p. Cell Signal 84, 109992. doi:10.1016/j.cellsig.2021.109992
Chen, T., Liu, R., Niu, Y., Mo, H., Wang, H., Lu, Y., et al. (2021). HIF-1α-activated long non-coding RNA KDM4A-AS1 promotes hepatocellular carcinoma progression via the miR-411-5p/KPNA2/AKT pathway. Cell Death Dis. 12 (12), 1152. doi:10.1038/s41419-021-04449-2
Chen, Y., Zhao, J., Jiao, Z., Wang, W., Wang, D., Yu, X., et al. (2018). SKA1 overexpression is associated with poor prognosis in hepatocellular carcinoma. BMC Cancer 18 (1), 1240. doi:10.1186/s12885-018-5119-6
Couri, T., and Pillai, A. (2019). Goals and targets for personalized therapy for HCC. Hepatol. Int. 13 (2), 125–137. doi:10.1007/s12072-018-9919-1
Cox, J., and Weinman, S. (2016). Mechanisms of doxorubicin resistance in hepatocellular carcinoma. Hepat. Oncol. 3 (1), 57–59. doi:10.2217/hep.15.41
Cui, S., Chen, Y., Guo, Y., Wang, X., and Chen, D. (2023). Hsa-miR-22-3p inhibits liver cancer cell EMT and cell migration/invasion by indirectly regulating SPRY2. PLoS One 18 (2), e0281536. doi:10.1371/journal.pone.0281536
De Las Rivas, J., Brozovic, A., Izraely, S., Casas-Pais, A., Witz, I. P., and Figueroa, A. (2021). Cancer drug resistance induced by EMT: Novel therapeutic strategies. Arch. Toxicol. 95 (7), 2279–2297. doi:10.1007/s00204-021-03063-7
Ding, B., Lou, W., Fan, W., and Pan, J. (2023). Exosomal miR-374c-5p derived from mesenchymal stem cells suppresses epithelial-mesenchymal transition of hepatocellular carcinoma via the LIMK1-Wnt/β-catenin axis. Environ. Toxicol. 38 (5), 1038–1052. doi:10.1002/tox.23746
Dudas, J., Ladanyi, A., Ingruber, J., Steinbichler, T. B., and Riechelmann, H. (2020). Epithelial to mesenchymal transition: A mechanism that fuels cancer radio/chemoresistance. Cells 9 (2), 428. doi:10.3390/cells9020428
Esquela-Kerscher, A., and Slack, F. J. (2006). Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer 6 (4), 259–269. doi:10.1038/nrc1840
Feng, K. L., Diao, N., Zhou, Z. W., Fang, C. K., Wang, J. N., Zhang, Y., et al. (2022). CircFGGY inhibits cell growth, invasion and epithelial-mesenchymal transition of hepatocellular carcinoma via regulating the miR-545-3p/smad7 Axis. Front. Cell Dev. Biol. 10, 850708. doi:10.3389/fcell.2022.850708
Feng, Y., Wei, G., Zhang, L., Zhou, H., Wang, W., Guo, P., et al. (2021). LncRNA DARS-AS1 aggravates the growth and metastasis of hepatocellular carcinoma via regulating the miR-3200-5p-Cytoskeleton associated protein 2 (CKAP2) axis. Bioengineered 12 (1), 8217–8232. doi:10.1080/21655979.2021.1982272
Gao, X., Jiang, Y., and Li, Y. (2021). Inhibitory effect of miR-140-5p on doxorubicin resistance of hepatocellular carcinoma. Exp. Ther. Med. 21 (5), 507. doi:10.3892/etm.2021.9938
Gong, T. T., Sun, F. Z., Chen, Y. J., Liu, J. F., Yan, Y., Li, D., et al. (2020). The circular RNA circPTK2 inhibits EMT in hepatocellular carcinoma by acting as a ceRNA and sponging miR-92a to upregulate E-cadherin. Eur. Rev. Med. Pharmacol. Sci. 24 (18), 9333–9342. doi:10.26355/eurrev_202009_23015
Guan, Q., Yuan, B., Zhang, X., Yan, T., Li, J., and Xu, W. (2021). Long non-coding RNA DUXAP8 promotes tumorigenesis by regulating IGF1R via miR-9-3p in hepatocellular carcinoma. Exp. Ther. Med. 22 (1), 755. doi:10.3892/etm.2021.10187
Ha, M., and Kim, V. N. (2014). Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15 (8), 509–524. doi:10.1038/nrm3838
Hao, Y., Baker, D., and Ten Dijke, P. (2019). TGF-beta-Mediated epithelial-mesenchymal transition and cancer metastasis. Int. J. Mol. Sci. 20 (11), 2767. doi:10.3390/ijms20112767
He, Q., Guo, P., Bo, Z., Yu, H., Yang, J., Wang, Y., et al. (2022). Noncoding RNA-mediated molecular bases of chemotherapy resistance in hepatocellular carcinoma. Cancer Cell Int. 22 (1), 249. doi:10.1186/s12935-022-02643-6
Hirao, A., Sato, Y., Tanaka, H., Nishida, K., Tomonari, T., Hirata, M., et al. (2021). MiR-125b-5p is involved in sorafenib resistance through ataxin-1-mediated epithelial-mesenchymal transition in hepatocellular carcinoma. Cancers (Basel) 13 (19), 4917. doi:10.3390/cancers13194917
Hong, L., Chen, W., Xing, A., Wu, D., and Wang, S. (2018). Inhibition of tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ) overcomes drug resistance and tumorigenicity in ovarian cancer. Cell Physiol. Biochem. 49 (1), 53–64. doi:10.1159/000492839
Huang, J., Yang, Y., Zhao, F., Zhang, Z., Deng, J., Lu, W., et al. (2023). LncRNA SATB2-AS1 overexpression represses the development of hepatocellular carcinoma through regulating the miR-3678-3p/GRIM-19 axis. Cancer Cell Int. 23 (1), 82. doi:10.1186/s12935-023-02901-1
Huang, W., Huang, F., and Feng, C. (2020). CircFoxo3 promotes adriamycin resistance through regulation of miR-199a-5p/ATP binding cassette subfamily C member 1 Axis in hepatocellular carcinoma. Onco Targets Ther. 13, 5113–5122. doi:10.2147/OTT.S243571
Huang, W. J., Tian, X. P., Bi, S. X., Zhang, S. R., He, T. S., Song, L. Y., et al. (2020). The beta-catenin/TCF-4-LINC01278-miR-1258-Smad2/3 axis promotes hepatocellular carcinoma metastasis. Oncogene 39 (23), 4538–4550. doi:10.1038/s41388-020-1307-3
Huang, X., Pan, J., Wang, G., Huang, T., Li, C., Wang, Y., et al. (2021). UNC5B-AS1 promotes the proliferation, migration and EMT of hepatocellular carcinoma cells via regulating miR-4306/KDM2A axis. Cell Cycle 20 (20), 2114–2124. doi:10.1080/15384101.2021.1962632
Huang, Y., Hong, W., and Wei, X. (2022). The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J. Hematol. Oncol. 15 (1), 129. doi:10.1186/s13045-022-01347-8
Ishikawa, T. (2009). Future perspectives on the treatment of hepatocellular carcinoma with cisplatin. World J. Hepatol. 1 (1), 8–16. doi:10.4254/wjh.v1.i1.8
Ji, J., Yamashita, T., Budhu, A., Forgues, M., Jia, H. L., Li, C., et al. (2009). Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology 50 (2), 472–480. doi:10.1002/hep.22989
Jiang, C., Zeng, X., Shan, R., Wen, W., Li, J., Tan, J., et al. (2020). The emerging picture of the roles of CircRNA-CDR1as in cancer. Front. Cell Dev. Biol. 8, 590478. doi:10.3389/fcell.2020.590478
Jiang, J., Li, J., Zhou, X., Zhao, X., Huang, B., and Qin, Y. (2022). Exosomes regulate the epithelial-mesenchymal transition in cancer. Front. Oncol. 12, 864980. doi:10.3389/fonc.2022.864980
Jin, J., Liu, H., Jin, M., Li, W., Xu, H., and Wei, F. (2020). Silencing of hsa_circ_0101145 reverses the epithelial-mesenchymal transition in hepatocellular carcinoma via regulation of the miR-548c-3p/LAMC2 axis. Aging (Albany NY) 12 (12), 11623–11635. doi:10.18632/aging.103324
Ju, L., Yao, M., Lu, R., Cao, Y., Wang, H., Yuan, L., et al. (2022). Circular RNA hsa_circ_0051040 promotes hepatocellular carcinoma progression by sponging miR-569 and regulating ITGAV expression. Cells 11 (22), 3571. doi:10.3390/cells11223571
Khanbabaei, H., Ebrahimi, S., Garcia-Rodriguez, J. L., Ghasemi, Z., Pourghadamyari, H., Mohammadi, M., et al. (2022). Non-coding RNAs and epithelial mesenchymal transition in cancer: Molecular mechanisms and clinical implications. J. Exp. Clin. Cancer Res. 41 (1), 278. doi:10.1186/s13046-022-02488-x
Kim, H., Lee, S., Shin, E., Seong, K. M., Jin, Y. W., Youn, H., et al. (2020). The emerging roles of exosomes as EMT regulators in cancer. Cells 9 (4), 861. doi:10.3390/cells9040861
Kim, H. S., Kim, J. S., Park, N. R., Nam, H., Sung, P. S., Bae, S. H., et al. (2021). Exosomal miR-125b exerts anti-metastatic properties and predicts early metastasis of hepatocellular carcinoma. Front. Oncol. 11, 637247. doi:10.3389/fonc.2021.637247
Klein, T., and Bischoff, R. (2011). Physiology and pathophysiology of matrix metalloproteases. Amino Acids 41 (2), 271–290. doi:10.1007/s00726-010-0689-x
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20 (11), 675–691. doi:10.1038/s41576-019-0158-7
Li, D., Zhou, T., Li, Y., Xu, Y., Cheng, X., Chen, J., et al. (2022). LINC02362 attenuates hepatocellular carcinoma progression through the miR-516b-5p/SOSC2 axis. Aging (Albany NY) 14 (1), 368–388. doi:10.18632/aging.203813
Li, G., Du, P., He, J., and Li, Y. (2021). CircRNA circBACH1 (hsa_circ_0061395) serves as a miR-656-3p sponge to facilitate hepatocellular carcinoma progression through increasing SERBP1 expression. Biochem. Biophys. Res. Commun. 556, 1–8. doi:10.1016/j.bbrc.2021.03.136
Li, M., Yue, W., Li, Q., Yu, W., Li, Y., and Cao, X. (2021). Circular RNA Circ_0000098 elevates ALX4 expression via adsorbing miR-1204 to inhibit the progression of hepatocellular carcinoma. Front. Oncol. 11, 696078. doi:10.3389/fonc.2021.696078
Li, N. L., Xiao, G., Jin, Y. Y., Deng, Y. Y., Liu, Y. J., and Yin, L. C. (2022). Long non-coding RNA LINC00992 promotes hepatocellular carcinoma cell proliferation, metastasis, and invasiveness by downregulating MicroRNA miR-361-5p expression to increase levels of the transcription factor twist1. Pathol. Res. Pract. 238, 154115. doi:10.1016/j.prp.2022.154115
Li, W., Xue, H., Li, Y., Li, P., Ma, F., Liu, M., et al. (2021). HIPK3 circular RNA promotes metastases of HCC through sponging miR-338-3p to induce ZEB2 expression. Dig. Dis. Sci. 66 (10), 3439–3447. doi:10.1007/s10620-020-06688-3
Li, X., He, J., Ren, X., Zhao, H., and Zhao, H. (2020). Circ_0003998 enhances doxorubicin resistance in hepatocellular carcinoma by regulating miR-218-5p/EIF5A2 pathway. Diagn Pathol. 15 (1), 141. doi:10.1186/s13000-020-01056-1
Li, Z., Wu, G., Li, J., Wang, Y., Ju, X., and Jiang, W. (2020). lncRNA CRNDE promotes the proliferation and metastasis by acting as sponge miR-539-5p to regulate POU2F1 expression in HCC. BMC Cancer 20 (1), 282. doi:10.1186/s12885-020-06771-y
Liao, Y., Yang, D., Luo, H., Yang, G., Wen, X., Zhang, K., et al. (2021). CircSEC24A promotes tumor progression through sequestering miR-455-3p in hepatocellular carcinoma. Neoplasma 2021, 210305N285. doi:10.4149/neo_2021_210305N285
Lin, H., Peng, J., Zhu, T., Xiong, M., Zhang, R., and Lei, L. (2022). Exosomal miR-4800-3p aggravates the progression of hepatocellular carcinoma via regulating the Hippo signaling pathway by targeting STK25. Front. Oncol. 12, 759864. doi:10.3389/fonc.2022.759864
Lin, Q., Zhou, C. R., Bai, M. J., Zhu, D., Chen, J. W., Wang, H. F., et al. (2020). Exosome-mediated miRNA delivery promotes liver cancer EMT and metastasis. Am. J. Transl. Res. 12 (3), 1080–1095.
Liu, C., Wang, H., Tang, L., Huang, H., Xu, M., Lin, Y., et al. (2021). LncRNA BACE1-AS enhances the invasive and metastatic capacity of hepatocellular carcinoma cells through mediating miR-377-3p/CELF1 axis. Life Sci. 275, 119288. doi:10.1016/j.lfs.2021.119288
Liu, W., Gao, X., Chen, X., Zhao, N., Sun, Y., Zou, Y., et al. (2021). miR-139-5p loss-mediated WTAP activation contributes to hepatocellular carcinoma progression by promoting the epithelial to mesenchymal transition. Front. Oncol. 11, 611544. doi:10.3389/fonc.2021.611544
Liu, Y., Guo, J., Shen, K., Wang, R., Chen, C., Liao, Z., et al. (2020). Paclitaxel suppresses hepatocellular carcinoma tumorigenesis through regulating circ-BIRC6/miR-877-5p/YWHAZ Axis. Onco Targets Ther. 13, 9377–9388. doi:10.2147/OTT.S261700
Liu, Y., Song, J., Zhang, H., Liao, Z., Liu, F., Su, C., et al. (2022). EIF4A3-induced circTOLLIP promotes the progression of hepatocellular carcinoma via the miR-516a-5p/PBX3/EMT pathway. J. Exp. Clin. Cancer Res. 41 (1), 164. doi:10.1186/s13046-022-02378-2
Long, J., Liu, L., Yang, X., Zhou, X., Lu, X., and Qin, L. (2023). LncRNA NUTM2A-AS1 aggravates the progression of hepatocellular carcinoma by activating the miR-186-5p/KLF7-mediated Wnt/beta-catenin pathway. Hum. Cell 36 (1), 312–328. doi:10.1007/s13577-022-00802-5
Lu, J., Wang, X. Z., Zhang, T. Q., Huang, X. Y., Yao, J. G., Wang, C., et al. (2017). Prognostic significance of XRCC4 expression in hepatocellular carcinoma. Oncotarget 8 (50), 87955–87970. doi:10.18632/oncotarget.21360
Lu, Y., Liu, Y., Zhang, K., and Jiang, L. (2022). Circular RNA TLK1 exerts oncogenic functions in hepatocellular carcinoma by acting as a ceRNA of miR-138-5p. J. Oncol. 2022, 2415836. doi:10.1155/2022/2415836
Mao, Y., Ding, Z., Jiang, M., Yuan, B., Zhang, Y., and Zhang, X. (2022). Circ_0091579 exerts an oncogenic role in hepatocellular carcinoma via mediating miR-136-5p/TRIM27. Biomed. J. 45 (6), 883–895. doi:10.1016/j.bj.2021.12.009
Marisi, G., Cucchetti, A., Ulivi, P., Canale, M., Cabibbo, G., Solaini, L., et al. (2018). Ten years of sorafenib in hepatocellular carcinoma: Are there any predictive and/or prognostic markers? World J. Gastroenterol. 24 (36), 4152–4163. doi:10.3748/wjg.v24.i36.4152
Mattick, J. S., Amaral, P. P., Carninci, P., Carpenter, S., Chang, H. Y., Chen, L. L., et al. (2023). Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 24 (6), 430–447. doi:10.1038/s41580-022-00566-8
Meng, F., Liu, J., Lu, T., Zang, L., Wang, J., He, Q., et al. (2021). SNHG1 knockdown upregulates miR-376a and downregulates FOXK1/Snail axis to prevent tumor growth and metastasis in HCC. Mol. Ther. Oncolytics 21, 264–277. doi:10.1016/j.omto.2021.02.002
Miyazono, K. (2009). Transforming growth factor-beta signaling in epithelial-mesenchymal transition and progression of cancer. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85 (8), 314–323. doi:10.2183/pjab.85.314
Mjelle, R., Dima, S. O., Bacalbasa, N., Chawla, K., Sorop, A., Cucu, D., et al. (2019). Comprehensive transcriptomic analyses of tissue, serum, and serum exosomes from hepatocellular carcinoma patients. BMC Cancer 19 (1), 1007. doi:10.1186/s12885-019-6249-1
Nadhan, R., Isidoro, C., Song, Y. S., and Dhanasekaran, D. N. (2022). Signaling by LncRNAs: Structure, cellular homeostasis, and disease pathology. Cells 11 (16), 2517. doi:10.3390/cells11162517
Nicoloso, M. S., Spizzo, R., Shimizu, M., Rossi, S., and Calin, G. A. (2009). MicroRNAs--the micro steering wheel of tumour metastases. Nat. Rev. Cancer 9 (4), 293–302. doi:10.1038/nrc2619
Niu, M., Yi, M., Li, N., Wu, K., and Wu, K. (2021). Advances of targeted therapy for hepatocellular carcinoma. Front. Oncol. 11, 719896. doi:10.3389/fonc.2021.719896
O'Brien, J., Hayder, H., Zayed, Y., and Peng, C. (2018). Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne) 9, 402. doi:10.3389/fendo.2018.00402
Park, N. R., Cha, J. H., Sung, P. S., Jang, J. W., Choi, J. Y., Yoon, S. K., et al. (2022). MiR-23b-3p suppresses epithelial-mesenchymal transition, migration, and invasion of hepatocellular carcinoma cells by targeting c-MET. Heliyon 8 (10), e11135. doi:10.1016/j.heliyon.2022.e11135
Peng, J., Wu, H. J., Zhang, H. F., Fang, S. Q., and Zeng, R. (2021). miR-143-3p inhibits proliferation and invasion of hepatocellular carcinoma cells by regulating its target gene FGF1. Clin. Transl. Oncol. 23 (3), 468–480. doi:10.1007/s12094-020-02440-5
Peng, S., Chen, Y., Li, T., Mao, J., Yang, P., Zou, B., et al. (2022). Hsa-microRNA-370-3p targeting Snail and Twist1 suppresses IL-8/STAT3-driven hepatocellular carcinoma metastasis. Cancer Sci. 113 (12), 4120–4134. doi:10.1111/cas.15571
Pratama, M. Y., Pascut, D., Massi, M. N., and Tiribelli, C. (2019). The role of microRNA in the resistance to treatment of hepatocellular carcinoma. Ann. Transl. Med. 7 (20), 577. doi:10.21037/atm.2019.09.142
Qin, M., Meng, Y., Luo, C., He, S., Qin, F., Yin, Y., et al. (2021). lncRNA PRR34-AS1 promotes HCC development via modulating Wnt/β-catenin pathway by absorbing miR-296-5p and upregulating E2F2 and SOX12. Mol. Ther. Nucleic Acids 25, 37–52. doi:10.1016/j.omtn.2021.04.016
Rinn, J. L., and Chang, H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. doi:10.1146/annurev-biochem-051410-092902
Roos, W. P., and Kaina, B. (2013). DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 332 (2), 237–248. doi:10.1016/j.canlet.2012.01.007
Sarkar, F. H., and Li, Y. (2002). Mechanisms of cancer chemoprevention by soy isoflavone genistein. Cancer Metastasis Rev. 21 (3-4), 265–280. doi:10.1023/a:1021210910821
Schattenberg, J. M., Schuchmann, M., and Galle, P. R. (2011). Cell death and hepatocarcinogenesis: Dysregulation of apoptosis signaling pathways. J. Gastroenterol. Hepatol. 26 (1), 213–219. doi:10.1111/j.1440-1746.2010.06582.x
Scheau, C., Badarau, I. A., Costache, R., Caruntu, C., Mihai, G. L., Didilescu, A. C., et al. (2019). The role of matrix metalloproteinases in the epithelial-mesenchymal transition of hepatocellular carcinoma. Anal. Cell Pathol. (Amst) 2019, 9423907. doi:10.1155/2019/9423907
Shen, D., Zhao, H. Y., Gu, A. D., Wu, Y. W., Weng, Y. H., Li, S. J., et al. (2021). miRNA-10a-5p inhibits cell metastasis in hepatocellular carcinoma via targeting SKA1. Kaohsiung J. Med. Sci. 37 (9), 784–794. doi:10.1002/kjm2.12392
Shen, D., Zhao, H., Zeng, P., Ge, M., Shrestha, S., and Zhao, W. (2022). Circular RNA circ_0001459 accelerates hepatocellular carcinoma progression via the miR-6165/IGF1R axis. Ann. N. Y. Acad. Sci. 1512 (1), 46–60. doi:10.1111/nyas.14753
Sidhu, K., Kapoor, N. R., Pandey, V., and Kumar, V. (2015). The "macro" world of microRNAs in hepatocellular carcinoma. Front. Oncol. 5, 68. doi:10.3389/fonc.2015.00068
Skovierova, H., Okajcekova, T., Strnadel, J., Vidomanova, E., and Halasova, E. (2018). Molecular regulation of epithelial-to-mesenchymal transition in tumorigenesis (Review). Int. J. Mol. Med. 41 (3), 1187–1200. doi:10.3892/ijmm.2017.3320
Slack, F. J., and Chinnaiyan, A. M. (2019). The role of non-coding RNAs in oncology. Cell 179 (5), 1033–1055. doi:10.1016/j.cell.2019.10.017
Song, G. Q., He, T. L., Ji, K. J., Duan, Y. M., Zhang, J. W., and Hu, G. Q. (2022). SKA1/2/3 is a biomarker of poor prognosis in human hepatocellular carcinoma. Front. Oncol. 12, 1038925. doi:10.3389/fonc.2022.1038925
Song, L. N., Qiao, G. L., Yu, J., Yang, C. M., Chen, Y., Deng, Z. F., et al. (2020). Hsa_circ_0003998 promotes epithelial to mesenchymal transition of hepatocellular carcinoma by sponging miR-143-3p and PCBP1. J. Exp. Clin. Cancer Res. 39 (1), 114. doi:10.1186/s13046-020-01576-0
Song, S., and Qiu, X. (2021). LncRNA miR503HG inhibits epithelial-mesenchymal transition and angiogenesis in hepatocellular carcinoma by enhancing PDCD4 via regulation of miR-15b. Dig. Liver Dis. 53 (1), 107–116. doi:10.1016/j.dld.2020.09.008
Sorop, A., Iacob, R., Iacob, S., Constantinescu, D., Chitoiu, L., Fertig, T. E., et al. (2020). Plasma small extracellular vesicles derived miR-21-5p and miR-92a-3p as potential biomarkers for hepatocellular carcinoma screening. Front. Genet. 11, 712. doi:10.3389/fgene.2020.00712
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22 (2), 96–118. doi:10.1038/s41580-020-00315-9
Sun, J., Zheng, X., Wang, B., Cai, Y., Zheng, L., Hu, L., et al. (2022). LncRNA LIMT (LINC01089) contributes to sorafenib chemoresistance via regulation of miR-665 and epithelial to mesenchymal transition in hepatocellular carcinoma cells. Acta Biochim. Biophys. Sin. (Shanghai) 54 (2), 261–270. doi:10.3724/abbs.2021019
Sun, W., Yang, Y., Xu, C., and Guo, J. (2017). Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet. 216-217, 105–110. doi:10.1016/j.cancergen.2017.06.003
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Tang, G., Zhao, H., Xie, Z., Wei, S., and Chen, G. (2022). Long non-coding RNA HAGLROS facilitates tumorigenesis and progression in hepatocellular carcinoma by sponging miR-26b-5p to up-regulate karyopherin α2 (KPNA2) and inactivate p53 signaling. Bioengineered 13 (3), 7829–7846. doi:10.1080/21655979.2022.2049472
Tang, W., Chen, Z., Zhang, W., Cheng, Y., Zhang, B., Wu, F., et al. (2020). The mechanisms of sorafenib resistance in hepatocellular carcinoma: theoretical basis and therapeutic aspects. Signal Transduct. Target Ther. 5 (1), 87. doi:10.1038/s41392-020-0187-x
Tewey, K. M., Rowe, T. C., Yang, L., Halligan, B. D., and Liu, L. F. (1984). Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science 226 (4673), 466–468. doi:10.1126/science.6093249
Tian, B., Zhou, L., Wang, J., and Yang, P. (2021). miR-660-5p-loaded M2 macrophages-derived exosomes augment hepatocellular carcinoma development through regulating KLF3. Int. Immunopharmacol. 101, 108157. doi:10.1016/j.intimp.2021.108157
Toden, S., Zumwalt, T. J., and Goel, A. (2021). Non-coding RNAs and potential therapeutic targeting in cancer. Biochim. Biophys. Acta Rev. Cancer 1875 (1), 188491. doi:10.1016/j.bbcan.2020.188491
Toki, N., Takahashi, H., Zucchelli, S., Gustincich, S., and Carninci, P. (2020). Synthetic in vitro transcribed lncRNAs (SINEUPs) with chemical modifications enhance target mRNA translation. FEBS Lett. 594 (24), 4357–4369. doi:10.1002/1873-3468.13928
Valdes, F., Alvarez, A. M., Locascio, A., Vega, S., Herrera, B., Fernandez, M., et al. (2002). The epithelial mesenchymal transition confers resistance to the apoptotic effects of transforming growth factor Beta in fetal rat hepatocytes. Mol. Cancer Res. 1 (1), 68–78.
Vancza, L., Karaszi, K., Peterfia, B., Turiak, L., Dezso, K., Sebestyen, A., et al. (2022). SPOCK1 promotes the development of hepatocellular carcinoma. Front. Oncol. 12, 819883. doi:10.3389/fonc.2022.819883
Volinia, S., Calin, G. A., Liu, C. G., Ambs, S., Cimmino, A., Petrocca, F., et al. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U. S. A. 103 (7), 2257–2261. doi:10.1073/pnas.0510565103
Wang, B., Hsu, S. H., Majumder, S., Kutay, H., Huang, W., Jacob, S. T., et al. (2010). TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene 29 (12), 1787–1797. doi:10.1038/onc.2009.468
Wang, J., Zhu, Y., Ai, X., Wan, H., Jia, W., Chu, J., et al. (2023). Long noncoding RNA 02027 inhibits proliferation, migration and invasion of hepatocellular carcinoma via miR-625-3p/PDLIM5 pathway. J. Gene Med. 25 (6), e3485. doi:10.1002/jgm.3485
Wang, L., Sun, L., Liu, R., Mo, H., Niu, Y., Chen, T., et al. (2021). Long non-coding RNA MAPKAPK5-AS1/PLAGL2/HIF-1α signaling loop promotes hepatocellular carcinoma progression. J. Exp. Clin. Cancer Res. 40 (1), 72. doi:10.1186/s13046-021-01868-z
Weaver, B. A. (2014). How Taxol/paclitaxel kills cancer cells. Mol. Biol. Cell 25 (18), 2677–2681. doi:10.1091/mbc.E14-04-0916
Winkle, M., El-Daly, S. M., Fabbri, M., and Calin, G. A. (2021). Noncoding RNA therapeutics - challenges and potential solutions. Nat. Rev. Drug Discov. 20 (8), 629–651. doi:10.1038/s41573-021-00219-z
Wong, C. M., Tsang, F. H., and Ng, I. O. (2018). Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 15 (3), 137–151. doi:10.1038/nrgastro.2017.169
Wu, L., Li, H., Chen, S., Wu, X., Chen, X., and Wang, F. (2021). Catalpol inhibits the proliferation, migration and metastasis of HCC cells by regulating miR-140-5p expression. Mol. Med. Rep. 23 (1), 29. doi:10.3892/mmr.2020.11667
Wu, M., Sun, T., and Xing, L. (2020). Circ_0004913 inhibits cell growth, metastasis, and glycolysis by absorbing miR-184 to regulate HAMP in hepatocellular carcinoma. Cancer Biother Radiopharm. 2020. doi:10.1089/cbr.2020.3779
Xu, G., Zhang, P., Liang, H., Xu, Y., Shen, J., Wang, W., et al. (2021). Circular RNA hsa_circ_0003288 induces EMT and invasion by regulating hsa_circ_0003288/miR-145/PD-L1 axis in hepatocellular carcinoma. Cancer Cell Int. 21 (1), 212. doi:10.1186/s12935-021-01902-2
Xu, Y., Lai, Y., Cao, L., Li, Y., Chen, G., Chen, L., et al. (2021). Human umbilical cord mesenchymal stem cells-derived exosomal microRNA-451a represses epithelial-mesenchymal transition of hepatocellular carcinoma cells by inhibiting ADAM10. RNA Biol. 18 (10), 1408–1423. doi:10.1080/15476286.2020.1851540
Xu, Y., Liu, Y., Li, Z., Li, H., Li, X., Yan, L., et al. (2020). Long non-coding RNA H19 is involved in sorafenib resistance in hepatocellular carcinoma by upregulating miR-675. Oncol. Rep. 44 (1), 165–173. doi:10.3892/or.2020.7608
Xuan, W., Zhou, C., and You, G. (2020). LncRNA LINC00668 promotes cell proliferation, migration, invasion ability and EMT process in hepatocellular carcinoma by targeting miR-532-5p/YY1 axis. Biosci. Rep. 40 (5). doi:10.1042/BSR20192697
Yan, L., Xu, F., and Dai, C. L. (2018). Relationship between epithelial-to-mesenchymal transition and the inflammatory microenvironment of hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 37 (1), 203. doi:10.1186/s13046-018-0887-z
Yang, B., Feng, X., Liu, H., Tong, R., Wu, J., Li, C., et al. (2020). High-metastatic cancer cells derived exosomal miR92a-3p promotes epithelial-mesenchymal transition and metastasis of low-metastatic cancer cells by regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene 39 (42), 6529–6543. doi:10.1038/s41388-020-01450-5
Yang, J., Antin, P., Berx, G., Blanpain, C., Brabletz, T., Bronner, M., et al. (2020). Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21 (6), 341–352. doi:10.1038/s41580-020-0237-9
Yang, J., Cui, R., and Liu, Y. (2020). MicroRNA-212-3p inhibits paclitaxel resistance through regulating epithelial-mesenchymal transition, migration and invasion by targeting ZEB2 in human hepatocellular carcinoma. Oncol. Lett. 20 (4), 23. doi:10.3892/ol.2020.11884
Yang, L., Deng, W. L., Zhao, B. G., Xu, Y., Wang, X. W., Fang, Y., et al. (2022). FOXO3-induced lncRNA LOC554202 contributes to hepatocellular carcinoma progression via the miR-485-5p/BSG axis. Cancer Gene Ther. 29 (3-4), 326–340. doi:10.1038/s41417-021-00312-w
Yang, X., Jiang, W., Kong, X., Zhou, X., Zhu, D., and Kong, L. (2022). Genistein restricts the epithelial mesenchymal transformation (EMT) and stemness of hepatocellular carcinoma via upregulating miR-1275 to inhibit the EIF5A2/PI3K/Akt pathway. Biol. (Basel) 11 (10), 1383. doi:10.3390/biology11101383
Yao, J., Hua, X., Shi, J., Hu, X., Lui, K., He, K., et al. (2022). LncRNA THEMIS2-211, a tumor-originated circulating exosomal biomarker, promotes the growth and metastasis of hepatocellular carcinoma by functioning as a competing endogenous RNA. FASEB J. 36 (4), e22238. doi:10.1096/fj.202101564R
Ye, Z., He, Q., Wang, Q., Lin, Y., Cen, K., and Chen, X. (2021). LINC00922 promotes the proliferation, migration, invasion and EMT process of liver cancer cells by regulating miR-424-5p/ARK5. Mol. Cell Biochem. 476 (10), 3757–3769. doi:10.1007/s11010-021-04196-0
Yin, D., Hu, Z. Q., Luo, C. B., Wang, X. Y., Xin, H. Y., Sun, R. Q., et al. (2021). LINC01133 promotes hepatocellular carcinoma progression by sponging miR-199a-5p and activating annexin A2. Clin. Transl. Med. 11 (5), e409. doi:10.1002/ctm2.409
Yin, L. C., Xiao, G., Zhou, R., Huang, X. P., Li, N. L., Tan, C. L., et al. (2020). MicroRNA-361-5p inhibits tumorigenesis and the EMT of HCC by targeting Twist1. Biomed. Res. Int. 2020, 8891876. doi:10.1155/2020/8891876
Yu, C. C., Li, C. F., Chen, I. H., Lai, M. T., Lin, Z. J., Korla, P. K., et al. (2019). YWHAZ amplification/overexpression defines aggressive bladder cancer and contributes to chemo-/radio-resistance by suppressing caspase-mediated apoptosis. J. Pathol. 248 (4), 476–487. doi:10.1002/path.5274
Yuan, S., Si, W., Zhuang, K., Li, Y., Zhang, Y., Liu, J., et al. (2021). LncRNA UCID promotes hepatocellular carcinoma metastasis via stabilization of snail. Onco Targets Ther. 14, 725–736. doi:10.2147/OTT.S277951
Zeng, T., Luo, L., Huang, Y., Ye, X., and Lin, J. (2021). Upregulation of miR-138 increases sensitivity to cisplatin in hepatocellular carcinoma by regulating EZH2. Biomed. Res. Int. 2021, 6665918. doi:10.1155/2021/6665918
Zeng, Z., Dong, J., Li, Y., Dong, Z., Liu, Z., Huang, J., et al. (2020). The expression level and diagnostic value of microRNA-22 in HCC patients. Artif. Cells Nanomed Biotechnol. 48 (1), 683–686. doi:10.1080/21691401.2019.1703723
Zhang, B., Li, F., Zhu, Z., Ding, A., and Luo, J. (2020). CircRNA CDR1as/miR-1287/raf1 Axis modulates hepatocellular carcinoma progression through MEK/ERK pathway. Cancer Manag. Res. 12, 8951–8964. doi:10.2147/CMAR.S252679
Zhang, B., and Zhou, J. (2022). CircSEC24A (hsa_circ_0003528) interference suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells via miR-421/MMP3 axis. Bioengineered 13 (4), 9049–9062. doi:10.1080/21655979.2022.2057761
Zhang, C., Guo, F., Xu, G., Ma, J., and Shao, F. (2015). STAT3 cooperates with Twist to mediate epithelial-mesenchymal transition in human hepatocellular carcinoma cells. Oncol. Rep. 33 (4), 1872–1882. doi:10.3892/or.2015.3783
Zhang, H., Liu, S., Chen, L., Sheng, Y., Luo, W., and Zhao, G. (2021). MicroRNA miR-509-3p inhibit metastasis and epithelial-mesenchymal transition in hepatocellular carcinoma. Bioengineered 12 (1), 2263–2273. doi:10.1080/21655979.2021.1932210
Zhang, L., Zhang, Y., Shen, D., Chen, Y., Feng, J., Wang, X., et al. (2022). RNA binding motif protein 3 promotes cell metastasis and epithelial-mesenchymal transition through STAT3 signaling pathway in hepatocellular carcinoma. J. Hepatocell. Carcinoma 9, 405–422. doi:10.2147/JHC.S351886
Zhang, N., Wang, F., Zhu, L., Chang, R., Mok, S. R. S., Peixoto, R. D., et al. (2023). Molecular mechanism of the miR-7/BCL2L1/P53 signaling axis regulating the progression of hepatocellular carcinoma. Ann. Transl. Med. 11 (1), 12. doi:10.21037/atm-22-5929
Zhang, P. F., Wang, F., Wu, J., Wu, Y., Huang, W., Liu, D., et al. (2019). LncRNA SNHG3 induces EMT and sorafenib resistance by modulating the miR-128/CD151 pathway in hepatocellular carcinoma. J. Cell Physiol. 234 (3), 2788–2794. doi:10.1002/jcp.27095
Zhang, Y., Wang, D., Zhu, T., Yu, J., Wu, X., Lin, W., et al. (2021). CircPUM1 promotes hepatocellular carcinoma progression through the miR-1208/MAP3K2 axis. J. Cell Mol. Med. 25 (1), 600–612. doi:10.1111/jcmm.15998
Zhang, Y., and Wang, Y. (2021). Circular RNAs in hepatocellular carcinoma: Emerging functions to clinical significances. Front. Oncol. 11, 667428. doi:10.3389/fonc.2021.667428
Zhang, Y., Wu, W., Sun, Q., Ye, L., Zhou, D., and Wang, W. (2021). linc-ROR facilitates hepatocellular carcinoma resistance to doxorubicin by regulating TWIST1-mediated epithelial-mesenchymal transition. Mol. Med. Rep. 23 (5), 340. doi:10.3892/mmr.2021.11979
Zhang, Z., Zhou, Y., Jia, Y., Wang, C., Zhang, M., and Xu, Z. (2022). PRR34-AS1 promotes exosome secretion of VEGF and TGF-beta via recruiting DDX3X to stabilize Rab27a mRNA in hepatocellular carcinoma. J. Transl. Med. 20 (1), 491. doi:10.1186/s12967-022-03628-9
Zhao, J., Fu, X., Chen, H., Min, L., Sun, J., Yin, J., et al. (2021). G3BP1 interacts with YWHAZ to regulate chemoresistance and predict adjuvant chemotherapy benefit in gastric cancer. Br. J. Cancer 124 (2), 425–436. doi:10.1038/s41416-020-01067-1
Zhou, Y., Li, K., Dai, T., Wang, H., Hua, Z., Bian, W., et al. (2021). Long non-coding RNA HCP5 functions as a sponge of miR-29b-3p and promotes cell growth and metastasis in hepatocellular carcinoma through upregulating DNMT3A. Aging (Albany NY) 13 (12), 16267–16286. doi:10.18632/aging.203155
Zhu, C., Su, Y., Liu, L., Wang, S., Liu, Y., and Wu, J. (2020). Circular RNA hsa_circ_0004277 stimulates malignant phenotype of hepatocellular carcinoma and epithelial-mesenchymal transition of peripheral cells. Front. Cell Dev. Biol. 8, 585565. doi:10.3389/fcell.2020.585565
Keywords: hepatocellular carcinoma, noncoding RNA, epithelial-mesenchymal transition, chemoresistance, exosomes
Citation: Ghionescu A-V, Sorop A and Dima SO (2023) The pivotal role of EMT-related noncoding RNAs regulatory axes in hepatocellular carcinoma. Front. Pharmacol. 14:1270425. doi: 10.3389/fphar.2023.1270425
Received: 31 July 2023; Accepted: 30 August 2023;
Published: 11 September 2023.
Edited by:
Sujit Nair, Viridis BioPharma Pvt. Ltd., IndiaReviewed by:
Thushara Thamban, HaystackAnalytics Private Limited, IndiaNikita Jadhav, University of Mumbai, India
Copyright © 2023 Ghionescu, Sorop and Dima. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simona Olimpia Dima, ZGltYS5zaW1vbmFAZ21haWwuY29t
†These authors have contributed equally to this work and share first authorship
 Alina-Veronica Ghionescu
Alina-Veronica Ghionescu Andrei Sorop
Andrei Sorop Simona Olimpia Dima
Simona Olimpia Dima