- 1Department of Dermatology, Affiliated Jinling Hospital, Medical School of Nanjing University, Nanjing, China
- 2Affiliated Hospital for Skin Diseases, Chinese Academy of Medical Sciences, Nanjing, China
Fungal infections have become clinically challenging owing to the emergence of drug resistance in invasive fungi and the rapid increase in the number of novel pathogens. The development of drug resistance further restricts the use of antifungal agents. Therefore, there is an urgent need to identify alternative treatments for Cryptococcus neoformans (C. neoformans). Disulfiram (DSF) has a good human safety profile and promising applications as an antiviral, antifungal, antiparasitic, and anticancer agent. However, the effect of DSF on Cryptococcus is yet to be thoroughly investigated. This study investigated the antifungal effects and the mechanism of action of DSF against C. neoformans to provide a new theoretical foundation for the treatment of Cryptococcal infections. In vitro studies demonstrated that DSF inhibited Cryptococcus growth at minimum inhibitory concentrations (MICs) ranging from 1.0 to 8.0 μg/mL. Combined antifungal effects have been observed for DSF with 5-fluorocytosine, amphotericin B, terbinafine, or ketoconazole. DSF exerts significant protective effects and synergistic effects combined with 5-FU for Galleria mellonella infected with C. neoformans. Mechanistic investigations showed that DSF dose-dependently inhibited melanin, urease, acetaldehyde dehydrogenase, capsule and biofilm viability of C. neoformans. Further studies indicated that DSF affected C. neoformans by interfering with multiple biological pathways, including replication, metabolism, membrane transport, and biological enzyme activity. Potentially essential targets of these pathways include acetaldehyde dehydrogenase, catalase, ATP-binding cassette transporter (ABC transporter), and iron-sulfur cluster transporter. These findings provide novel insights into the application of DSF and contribute to the understanding of its mechanisms of action in C. neoformans.
1 Introduction
Fungal infections are clinically challenging owing to the evolution of drug-resistant strains, emergence of new pathogens, prolonged treatment times, and limited antifungal treatment options (Wiederhold, 2017). Cryptococcus neoformans (C. neoformans) is a widespread opportunistic pathogenic fungus found in the environment. C. neoformans can enter the body through the respiratory tract, causing pulmonary infections and fatal meningitis, resulting in approximately 223,100 Cryptococcal meningitis cases and 181,100 fatalities annually (Rajasingham et al., 2017). As the coronavirus (CoV) disease 2019 pandemic has spread worldwide, cases of co-infection with C. neoformans and severe acute respiratory syndrome (SARS)-CoV-2 have been reported. These patients had high 30-day mortality rate (Bhatt et al., 2021; Chong et al., 2021; Mina et al., 2022). Cryptococcus spp. are susceptible to three classes of antifungal agents: polyenes, fluorocytosines, and azoles (Bermas and Geddes-McAlister, 2020). Nevertheless, the polyene amphotericin B (AmB) is susceptible to nephrotoxicity, anemia, and electrolyte imbalances (Klepser, 2011). Hepatotoxicity, gastrointestinal issues, and bone marrow suppression are common adverse effects of 5-fluorocytosine (5-FC), to which C. neoformans is frequently resistant (Klepser, 2011). Fluconazole (FLC) is an antifungal agent that plays a critical role in the initial treatment of Cryptococcal meningitis. However, its widespread use has resulted in frequent and persistent drug resistance (Kneale et al., 2016). Alternative treatment options for C. neoformans are urgently required considering the development of drug resistance, which severely restricts the use of antifungal medications.
Because fungi share organelles and metabolic pathways with human hosts, several antifungal medications can also harm humans. The development of antifungal drugs has presented significant challenges in the search for drugs with selective toxicity to fungi, but non-toxic or causing little harm to humans (Su et al., 2018). Toxicological and pharmacokinetic data have been established for conventional medications (Chong and Sullivan, 2007). Repurposing these medications for fungal therapy, or combining them with available antifungal drugs, is a promising strategy. For instance, the anticancer drug bortezomib inhibits the ubiquitin–proteasome pathway and capsule production in fungi (Geddes et al., 2016). Sertraline, an antidepressant, exerts antifungal effects when administered alone or in combination with FLC (Treviño-Rangel et al., 2016).
Disulfiram (DSF), often known as antabuse, is a safe dithiocarbamate well-tolerated by humans (Wright and Moore, 1990). Since 1951, when DSF was certified by the Food and Drug Administration (FDA) as an alcohol withdrawal drug, it has been used clinically for over 70 years. DSF has effective antiviral (Lee et al., 2016), antifungal (Mukherjee et al., 2006), antiparasitic (Peniche et al., 2015), and anticancer (Lu et al., 2022) properties. There are several mechanisms by which DSF inhibited bacterial growth and DSF can overcome multiple resistance barriers (Phillips et al., 1991; Thakare et al., 2019). DSF inhibits activities of many proteins, including aldehyde dehydrogenase (ALDH) (Velasco-García et al., 2003), metallo-β-lactamase (Chen et al., 2020), P-glycoprotein (P-gp), deoxyribonucleic acid (DNA)-methyltransferases, DNA polymerase (Sharma et al., 2016; Tesson et al., 2017), and phosphoglycerate dehydrogenase (Locasale et al., 2011). Disulfiram derivatives have been shown to exhibit the inhibitory effect against inflammatory scalp diseases by inhibiting carbonic anhydrase activity of Malassezia, which was an important causative pathogen for inflammatory dandruff (Vullo et al., 2017). DSF also inhibits the growth of Pythium insidiosum (Krajaejun et al., 2019). Additionally, it demonstrates significant antifungal activity against specific yeast strains, Aspergillus, and Candida albicans (Harrison et al., 2007; Khan et al., 2007). By inhibiting the function of multidrug transporter like Cdr1p, which has been shown to play a key role in azole resistance, DSF can increase the drug sensitivity of Saccharomyces cerevisiae (Shukla et al., 2004). Furthermore, due to its ability to curtail superoxide dismutase activity in C. albicans biofilm, the combination of DSF derivatives and AmB collaborates to limit biofilm activity (De Brucker et al., 2013). However, studies examining the effects of DSF on C. neoformans are still scarce.
In this study, we designed DSF sensitivity assays for 48 Cryptococcus spp. from different sources and performed combination susceptibility tests using common antifungal drugs. Galleria mellonella (G. mellonella) infected with C. neoformans was used to evaluate the in vivo antifungal activity of DSF. Mechanistically, we examined how DSF affects biofilm viability and the production of essential virulence components (melanin, urease, and capsule) in C. neoformans. Additionally, in combination with reference transcriptome sequencing, the molecular docking of DSF to protein targets was performed to investigate the mechanisms of the pathways and potential targets.
2 Materials and methods
2.1 Strains, culture, and reagents
Forty-two strains of Cryptococcus spp. were obtained from the strain banks of the Jinling Hospital (Nanjing, China). Candida parapsilosis (American Type Culture Collection 22019) was used as the quality control strain. The 5-FC stock solutions (6.4 mg/mL) were prepared from sterile ultrapure water. The FLC (6.4 mg/mL), ketoconazole (KET) (1.6 mg/mL), terbinafine (TRB) (1.6 mg/mL), AmB (1.6 mg/mL), and DSF (12.8 mg/mL) stock solutions were prepared from dimethyl sulfoxide (DMSO) and stored at −80°C. All reagents were purchased from Aladdin Biochemical Technology (Shanghai, China).
2.2 In vitro determination of minimum inhibitory concentrations of planktonic cells
Fungal solutions and drug susceptibility plates were prepared using the microdilution method in Roswell Park Memorial Institute (RPMI) 1,640 medium according to document M27-A4 published by the Clinical and Laboratory Standards Institute (CLSI). The final concentration of the fungal solution was adjusted to 5 × 103 colony forming unit (CFU)/mL. One hundred microliters of RPMI 1640 and the drug were added to all wells of the plate. Through serial dilution at a ratio of two, concentrations from 256 to 0.5 μg/mL were obtained (DSF as example) and made in each plate, from column 1 to 10. The 11th column was reserved for the microorganism growth positive control. Then, 100 μL of the fungal suspension was added to each well. The assay was performed in triplicate and incubated at 35°C for 72 h. The MIC was defined as the lowest concentration that could inhibit fungal growth (Al-Odaini et al., 2021). The prepared drug-sensitive 96-well plates (U-shaped) were incubated at 35°C for 72 h. Each drug’s minimum inhibitory concentrations (MIC) and minimal fungicidal concentration (MFC) values against Cryptococcus were determined using a microplate reader (Thermo Fisher Scientific, United States) by combining the visual/microscopic reading and the optical density at 600 nm.
2.3 Combination susceptibility test using the checkerboard method
The gradient multiplicative dilution method was used to create a combined drug sensitivity plate (de Oliveira et al., 2020), and strain B3501 was used in this study. Considering the differences in chemical properties and cellular targets between DSF and other antifungal agents, both the Bliss synergistic score (https://synergyfinder.fimm.fi) and fractional inhibitory concentration index (FICI) based on Loewe’s summation theory were used to explain the drug-drug interactions (de Oliveira et al., 2020).
2.4 In vivo survival assays
Using C. neoformans (H99)-infected G. mellonella as a model, 80 healthy larvae of uniform size (250–350 mg) were randomly selected. According to the clinically safe dose, DSF was prepared using phosphate-buffered saline (PBS) (containing 10% DMSO), and the therapeutic concentration (500 mg/70 kg) was determined. The larvae were divided into four groups (10 larvae per group): infected group, the (post-infection) DSF treatment group, DSF control group, and PBS (10% DMSO) control group. The infected and (post-infection) DSF treatment group were injected with 10 μL of H99 fungal solution. The DSF and PBS groups were injected with 10 μL of PBS (pH 7.4). Two hours later, the infected group and the PBS control group were injected with 10 μL of PBS (containing 10% DMSO) and the (post-infection) DSF treatment group and the DSF control group were injected with 10 μL of DSF treatment solution. Fatality was defined as the failure to respond to a light touch. Inoculation was achieved by injecting the inoculum into the hemocoel of the second left foot (Sudan et al., 2013). The fresh H99 fungal solution was incubated overnight in a shaker at 30°C (180 rpm) and centrifuged for 5 min at 2,800 × g (Thermo Fisher Scientific). The fungal precipitate was washed thrice with sterile PBS (pH 7.4). After resuspension, the fungal solution was counted using a cell-counting plate and diluted to 2 × 106 and 2 × 107 CFU/mL with PBS. G. mellonella were incubated in the dark at 37°C for 5–7 days, and their survival was monitored every 24 h.
The in vivo antifungal activities of 5-FU, DSF and their combination were evaluated in a higher concentration model (each injected with 106 CFU H99 in 10 μL PBS). Through repeated preliminary experimentation, the concentrations of 5-FU at 1.25 mg/kg combined with DSF at 3.5 mg/kg were used for treated. Groups treated only with 5-FU or DSF alone were dosed at 2.5 mg/kg and 7.1 mg/kg, respectively. The volume of solvent and the method of preparation were the same for all groups during the treatment.
2.5 Determination of fungal burden
An additional 160 healthy larvae of uniform size (250–350 mg) were randomly selected. Four groups of randomly selected larvae (20 for each group) were established as described above and incubated at 35°C for 4 days (Kong et al., 2020). During incubation, five larvae from each group were randomly selected daily on days 1–4. A high-speed homogenizer (Powteq GT300, Grinder Instrument, Beijing, China) was used to thoroughly grind the larvae for 30 min (1,600 rpm) after adding 0.7 mL PBS and sterile magnetic beads (3 mm, Beyotime, Haimen, China). We used PBS to dilute the homogenate in a gradient and inoculated 5 μL of samples with different concentrations on Sabouraud dextrose agar (SDA) at 37°C for 48 h.
2.6 Histological examination
Histological analysis was performed to observe the effects of DSF on G. mellonella larvae infected with C. neoformans (H99). Another 40 healthy larvae of uniform size (250–350 mg) were randomly selected. Four groups of randomly selected larvae were injected with fungal inocula and DSF, as described above. Three G. mellonella in each group were taken for pathological section (20 μm) and periodic acid-Schiff staining (Solarbio, Beijing, China). The staining results were observed under a light microscope (Zeiss Axioscope 5, ZEISS, Germany) at different magnifications (Sangalli-Leite et al., 2016).
2.7 Melanin production assay
Fresh H99 (C. neoformans var. grubii), ZYB24 (C. gattii), B3501 (C. neoformans var. neoforman), and D2A (C. neoformans var. neoforman) solutions were washed thrice with sterile PBS to reduce the melanin background. A simplified dopa-asparagine agar (with MgSO4 at 10 mM, KH2PO4 at 29.4 mM, glycine at 13 mM, vitamin B1 at 3.0 µM, L-asparagine at 7.6 mM, and L-dopa at 1 mM) was prepared (Teymuri et al., 2021). Different concentrations of DSF (range 0.5–8 μg/mL) were applied to dopa-asparagine agar before being inoculated with 10 μL of the fungus solution protected from light for 2–7 days at 35°C. The inoculum concentration was 1 × 107 CFU/mL. Images of the plates were captured daily (García-Rodas et al., 2015).
2.8 Urease activity assay
Urea slant agar mediums containing different concentrations of DSF (0.5–8 μg/mL) were prepared. Agar was prepared with a urease agar base (Hi-tech, Qingdao, China) and sterile urea solution according to Christensen’s formula (Christensen, 1946). The mediums were inoculated with fresh fungal solutions (10 μL), and color changes were observed for 2–10 days. Urease activity judgment index: The medium turned completely purple-red (+++) or pink (++); only the colony and surroundings turned pink (+); and no color change (−). Rapid urea broth (RUH broth), developed by Roberts (Roberts et al., 1978) and adapted by Kwon-Chung (Kwon-Chung et al., 1987) was used to detect urease activity for quantitative statistics. Fungal suspension (100 μL) was mixed with an equivalent amount of RUH broth (two times concentration) and shaken for 10 h at 37°C. The absorbance (560 nm) of the supernatant of equal fungal amounts was measured using a microplate reader (Feder et al., 2015).
2.9 Determination of capsule size
The H99 and B3501 strains were inoculated into SDA and capsule induction medium [10% SDA, 3-(N-morpholino) propanesulfonic acid (MOPS)] at a 5 × 105 CFU/mL concentration (Zaragoza and Casadevall, 2004). Positive control and DSF groups (MICs concentration) were shaken for 18–24 h at 37°C. The mixture of fungal solution (4 μL) and India ink (6 μL, Yuanye Biotechnology, Shanghai, China) was added dropwise on a slide and photographed by the MShot Image Analysis System.
2.10 Biofilm assay
Fresh H99 and B3501 fungal solutions were resuspended in RPMI 1640 (MOPS, pH 7.4) at a concentration of 1 × 107 cells/mL. To investigate the degradation effect of DSF on biofilms, two hundred microliters of fungal broths of H99 or B3501 were added to each well of the sterile 96-well plates (Clear polystyrene microplates, Corning, NY, United States) and incubated at 30°C for 2 h. The solutions were aspirated and softly washed with PBS before being replaced with RPMI 1640 (Sigma, Cleveland, United States) of different DSF gradient concentrations (0.25–128 μg/mL) and then incubated for 48 h. Furthermore, the medium and non-adherent cells were discarded, and the biofilms were stained with 0.1% crystal violet (Yuanye Biotechnology, Shanghai, China). Following the washing and drying of the biofilm, 150 μL of 50% acetic acid was added to each well. The absorbance was measured at 530 nm using a microplate reader (Thermo Fisher Scientific) to determine the level of biofilm degradation (Segev-Zarko and Shai, 2017; Song et al., 2019; Huang et al., 2023). The data for the drug-treated group was normalized relative to the control group.
Cellular activity within the biofilm was quantified based on the capacity of living cells to reduce the methyl thiazolyl tetrazolium (MTT, Beyotime, Haimen, China) yellow dye to purple formazan. The new batch of biofilm was incubated (1 × 106 cells/mL) for 48 h before being replaced with RPMI 1640 containing DSF for another 24 h. Afterwards, the MTT formazan product was dissolved in 150 μL DMSO and the absorbance was measured using the multiplate reader (da Silva et al., 2016; Huang et al., 2023).
2.11 Transcriptome sequencing and real-time quantitative PCR (RT-qPCR)
The PBS-washed H99 solution was inoculated in fresh yeast extract peptone dextrose medium at a final concentration of 1 × 106 CFU/mL for 4 h. Control and DSF (4 μg/mL) groups were prepared. Total ribonucleic acid (RNA) was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States) and evaluated using a NanoDrop 2000 spectrophotometer (Thermo Scientific). RNA integrity was assessed using the Agilent 2,100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). Libraries were then constructed using the VAHTS Universal V6 RNA sequencing (RNA-seq) Library Prep Kit and sequenced on an Illumina NovaSeq 6000 platform. Raw reads in fastq format were first processed using fastp (Chen et al., 2018), and clean reads were mapped to the reference genome using HISAT2 (Kim et al., 2015). The differential expression (DE) analysis was performed using DESeq2 (Love et al., 2014). DE genes (DEGs) were identified between DSF-treated and control groups. Based on the hypergeometric distribution, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of the DEGs were performed to screen for significantly enriched terms using R (v 3.2.0).
The expression of mRNAs was verified by RT-qPCR (Yi et al., 2022). RNA was used to synthesize cDNA using the PrimeScript RT Reagent Kit (Vazyme, Nanjing, China). RT-qPCR was performed with the help of SYBR GREEN PCR MasterMix (Vazyme) and an Applied Biosystems 7500 real-time machine. Actin was used as a housing-keeping gene. The primer sequences are detailed in Supplementary Table SA5.
2.12 Homology modeling of C. neoformans proteins and molecular docking of DSF
The amino acid sequences of DEGs were imported into the Protein Data Bank (PDB) database. Based on Sequence Identity and E-Value, as well as functional and species annotations, homologous proteins with great matches were chosen, and their crystal structures were downloaded. Structural information of the DSF molecules was obtained from the PubChem database. The corresponding protein structures of H99 predicted using AlphaFold were obtained from the UniProt database. The structures of the Cryptococcus proteins and homologous proteins were imported into the PyMol software, and the alignment command was executed. After executing the Autogrid program in the AutoDockTools software for acceptors and ligands, the AutoDock Vina program was run to perform calculations to obtain multiple docking models. Finally, the structure of the ligand-acceptor complex was imported into Discovery Studio 2021 to visualize the chemical bonds.
2.13 ALDH activity assay
To analyze acetate production by C. neoformans, an ALDH activity assay was performed (Ammar, 2019). Fresh H99 strains cultured overnight in yeast extract peptone dextrose medium were expanded to 5 mL of yeast nitrogen base (Thermo Fisher Scientific) medium with 2% glucose at a concentration of 1 × 104 CFU/mL. The control and DSF (2 μg/mL) groups were incubated at 30°C for 48 h. The final concentration of each fungal solution was adjusted to the same level (OD600 = 0.65) at the time of measurement and was centrifuged at 10,000 × g (Thermo Fisher Scientific). Once all supernatants were collected, the concentration of acetic acid, a metabolite catalyzed by ALDH (Pronk et al., 1996), was measured using an ACETIC ACID Kit (K-ACET, Megazyme, Ireland).
2.14 Statistical analysis
All experiments were performed in triplicate unless specified otherwise. Statistical analysis was performed using SPSS Statistics 23.0. The statistical procedure utilized was the Student’s t-test. Survival curves were plotted using the Kaplan–Meier method and statistical analysis was performed using the log-rank test. Each experiment was performed in triplicate and repeated at least thrice. The value p < 0.05 was considered significantly different (p < 0.05 for *, p < 0.01 for **, and p < 0.001 for ***).
3 Results
3.1 DSF demonstrated broad-spectrum and strong susceptibility to C. neoformans and C. gattii in vitro
The 42 Cryptococcus isolates in this study, covering the four serotypes A, B, C, and D, showed MICs ranging from 1 to 8 μg/mL (Figure 1A). As shown in Supplementary Table SA1, the FLC MICs for these strains ranged from 0.5 to 16.0 μg/mL. These findings suggest that DSF efficiently suppressed non-FLC-resistant Cryptococcus strains of various serotypes. The results for the quality control strain ATCC 22019 were in accordance with those of CLSI-M27-A4. The MIC of DSF was 2 μg/mL against B3501 and 4 μg/mL against H99 (Figure 1B).
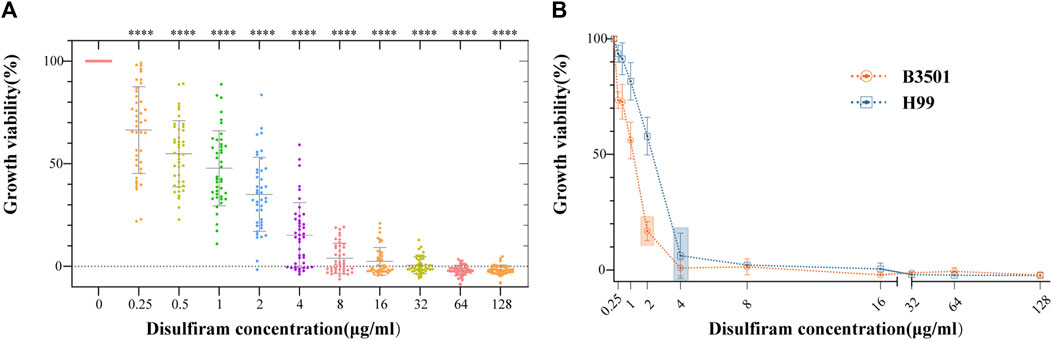
FIGURE 1. In vitro sensitivity assays of DSF against 42 strains of Cryptococcus. (A) Growth viability (%) of different Cryptococcus serotypes exposed to different DSF concentrations (ranging from 0 to 128 μg/mL). ****p < 0.0001, vs. positive controls. (B) Growth of B3501 and H99 sensitive to DSF at 2 and 4 μg/mL (boxed area), respectively.
3.2 DSF potentiated the activity of clinical antifungal drugs
Figures 2A–E show the Bliss scores for the efficacy of DSF in combination with the common antifungal agents 5-FC, TRB, KET, AmB, and FLC against C. neoformans. For instance, when 5-FU and DSF were combined at 2 and 0.5 μg/mL, respectively, a high synergistic score of 45.75 was obtained (Figure 2F). Synergistic activity with DSF was observed in a broader concentration range of 5-FC, TRB, KET, and AmB. For instance, in the combined concentration range of DSF from 0.25 to 1 μg/mL and 5-FC from 1 to 4 μg/mL, a Bliss score of 37.64 was calculated. The TRB, KET, AmB, and FLC scores were 46.38 (synergistic), 30.06 (synergistic), 22.20 (synergistic), and 8.06 (no interaction), respectively. This was in accordance with the findings obtained from the FICI scores (Supplementary Table SA2).
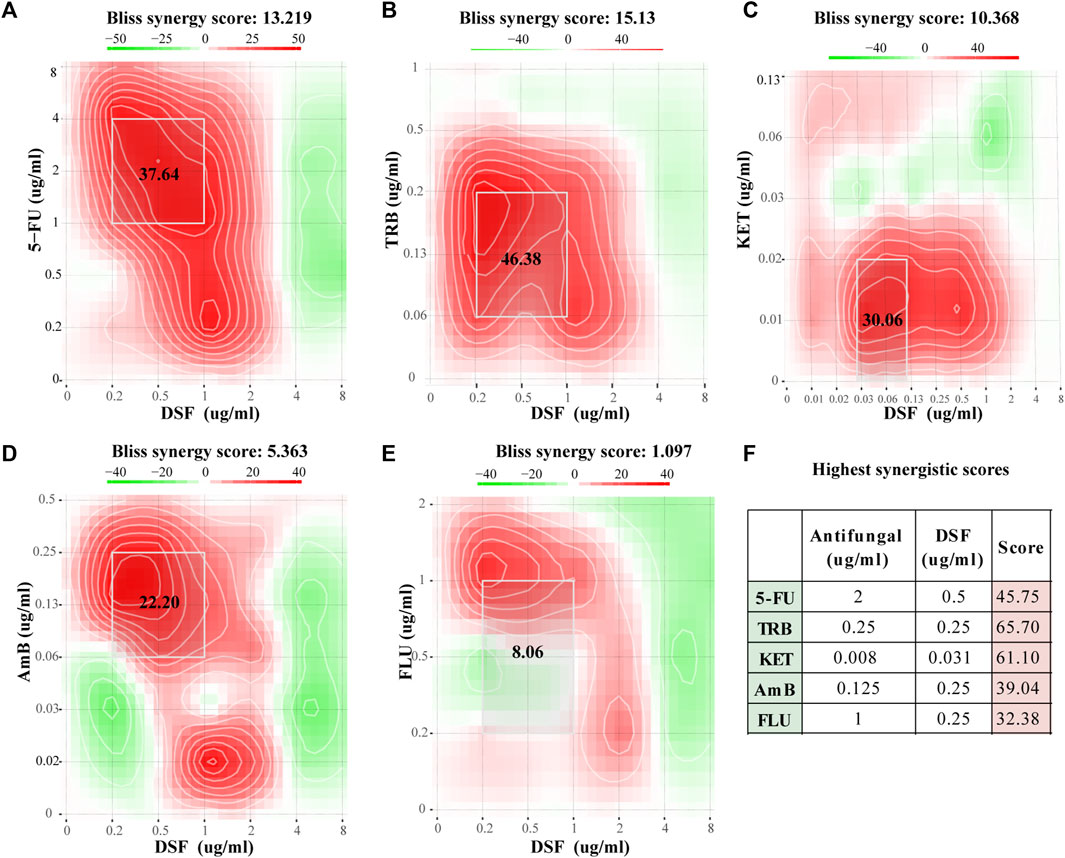
FIGURE 2. Checkerboard assay results for DSF against C. neoformans. (A–E) Bliss scores combined with 5-FC, TRB, KET, AmB, and FLC. (F) Highest score for each group. Scores less than −10 indicate that the interaction between the two drugs was antagonistic. Scores from −10 to 10 suggest an additive effect, and scores above 10 indicate synergism.
3.3 DSF protected G. mellonella from H99 infection in vivo
In the present study, G. mellonella larvae were infected with strain H99 as an experimental model and larval mortality was observed daily for 4–7 days. The larval survival rate on day 7 was 0% in the infected groups at a concentration of 1 × 104 CFU/mL (104-infected groups) and 60% in the DSF-treated group (Figure 3A), with a significant difference between the groups (p < 0.01). The survival rate of the 105-infected group was 0% on day 5 and increased to 50% after DSF treatment (Figure 3B, p < 0.01). This indicates that DSF has antifungal efficacy in vivo and significantly improves the survival rate of H99-infected larvae.
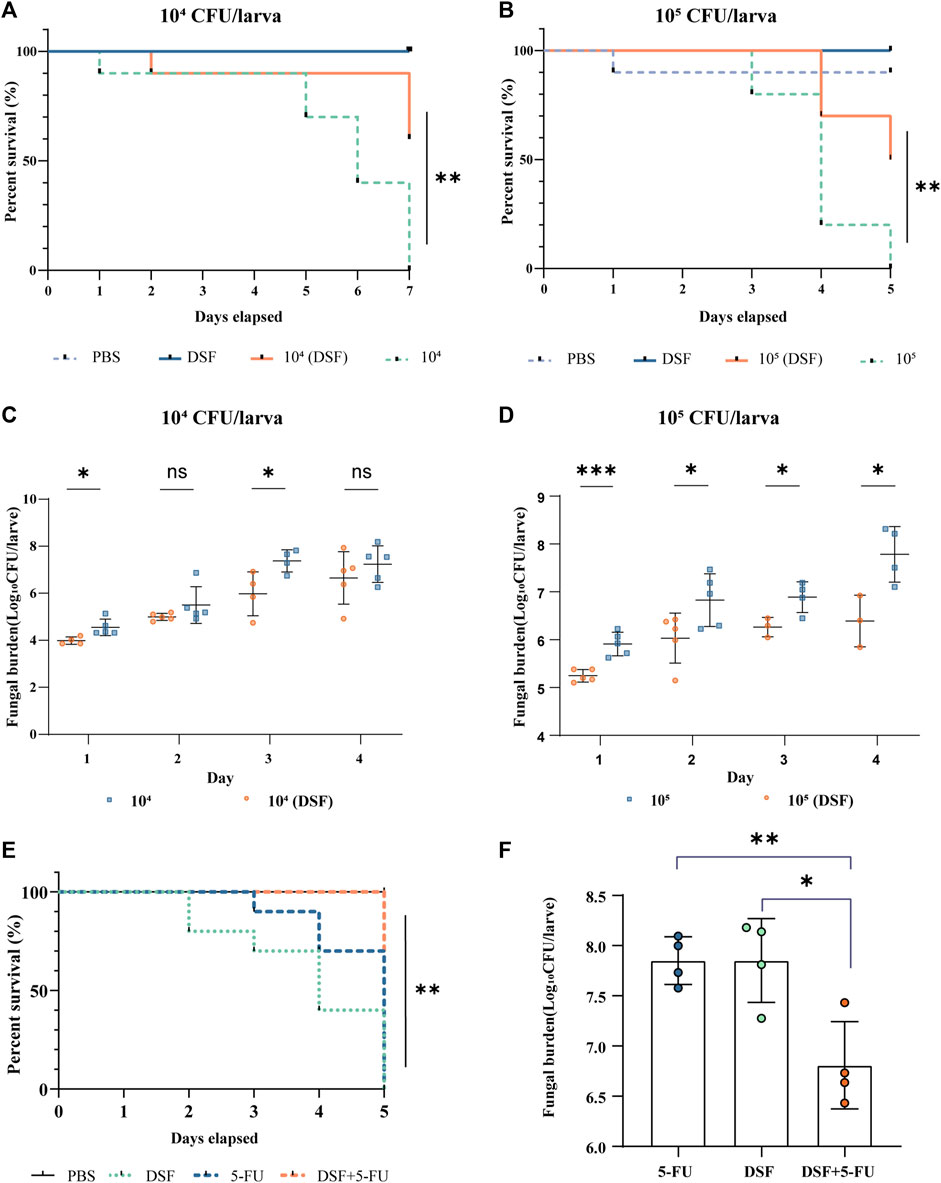
FIGURE 3. Protective effects of DSF against H99-infected G. mellonella. (A,B) Survival curves at different infection concentrations. (C,D) Fungal burden of infected larvae. The treatment groups were administered DSF (7.1 mg/kg) at 3 h after infection. The PBS and DSF groups were used as negative controls. Compared with the infected group, *p < 0.05, **p < 0.01, and ***p < 0.001. (E) Survival curves of different treatments on infected larvae. (F) Fungal burden of different treatments on infected larvae. Each group was treated with either PBS (10% DMSO), 5-FU (2.5 mg/kg), DSF (7.1 mg/kg), or 5-FU (1.25 mg/kg) plus DSF (3.5 mg/kg). *p < 0.05, **p < 0.01, compared with the 5-FU-treated group. 5-FU, 5-fluorocytosine; DSF, disulfiram.
3.4 Determination of fungal burden
Five larvae per group were selected, homogenized, inoculated, and counted daily using fungal load analysis. The fungal burden gradually increased with the duration of infection. The fungal burden was lower in the DSF-treated group than that in the respective infection groups at different H99 concentrations (Figures 3C, D). In particular, DSF treatment significantly reduced the fungal burden on the first day in the 105-infected group (p < 0.001).
3.5 Histological examination
On day 3, the protective effect of DSF on G. mellonella was observed by histopathology. The periodic acid-Schiff (PAS)-stained sections were photographed at different magnifications (×40, ×100, and ×400). Compared with the PBS and DSF control groups (Figures 4A, D), the infected larvae (Figures 4B, C) displayed extensive tissue destruction, with many dense capsule antigens stained in a typical purple-red color and surrounded by inflammatory cells. A significant reduction in the number of C. neoformans cells was observed in the DSF-treated groups (Figures 4E, F), and larval tissue damage was considerably reduced.
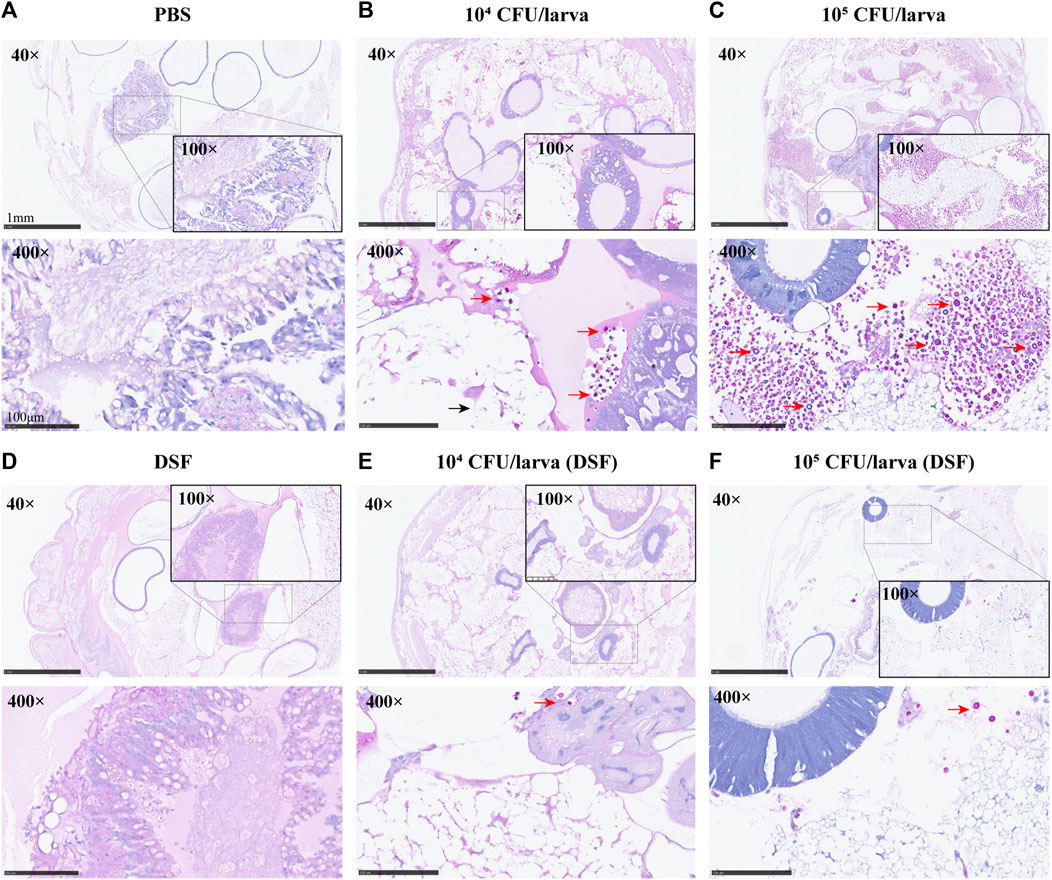
FIGURE 4. Histological examination (×40, ×100, and ×400) of G. mellonella larvae. (A) PBS (10% DMSO) control group. (B,C) Group infected with concentrations of H99 1 × 104 or 105 CFU/larva. (D) DSF control. (E,F) DSF-treated infected larvae group. Sections were stained with the PAS reagent; red arrows indicate C. neoformans and black arrows indicate tissue destruction.
3.6 Synergistic effect of DSF combined with 5-FU against C. neoformans in vivo
The in vivo antifungal activities of 5-FU, DSF and their combination were evaluated in an infected G. mellonella model. The concentrations of 5-FU at 1.25 mg/kg combined with DSF at 3.5 mg/kg were used, showing a strong antifungal activity against H99 in vivo, compared with only use of 5-FU (2.5 mg/kg) or DSF (7.1 mg/kg). These concentrations of the drugs used in this experiment meet the requirements for safe human dosages. The survival assay (Figure 3E) showed that when DSF was added in combination with 5-FU, the survival rate of infected larvae of a higher concentration model was improved and the fungal burden in day 2 (Figure 3F) was significantly induced (p < 0.05). These results indicated that DSF in combination with 5-FU exhibited a significant synergistic antifungal effect against C. neoformans in vivo.
3.7 DSF inhibited melanin production by Cryptococcus spp. in a dose-dependent manner
Cryptococcus produces virulence factors, such as polysaccharide capsules, melanin, and extracellular enzymes (including phospholipases, proteases, and urease), and tolerates a 37°C environment (Alspaugh, 2015). These factors protect cells from desiccation and oxidative stress damage and disrupt the host immune response (Nosanchuk et al., 2015; Lee et al., 2019). We examined the melanin-producing ability of different serotypes of Cryptococcus on dopa-asparagine agar at 30°C (Figure 5A). Colonies grew at all time points, and the positive controls produced varying degrees of melanin. As the concentration of DSF gradually increased (0.5–8 μg/mL), the melanin production of each strain progressively decreased (from dark brown to coffee or from light coffee to white). This suggests that DSF exerts a dose-dependent inhibitory effect on the production of Cryptococcal melanin.
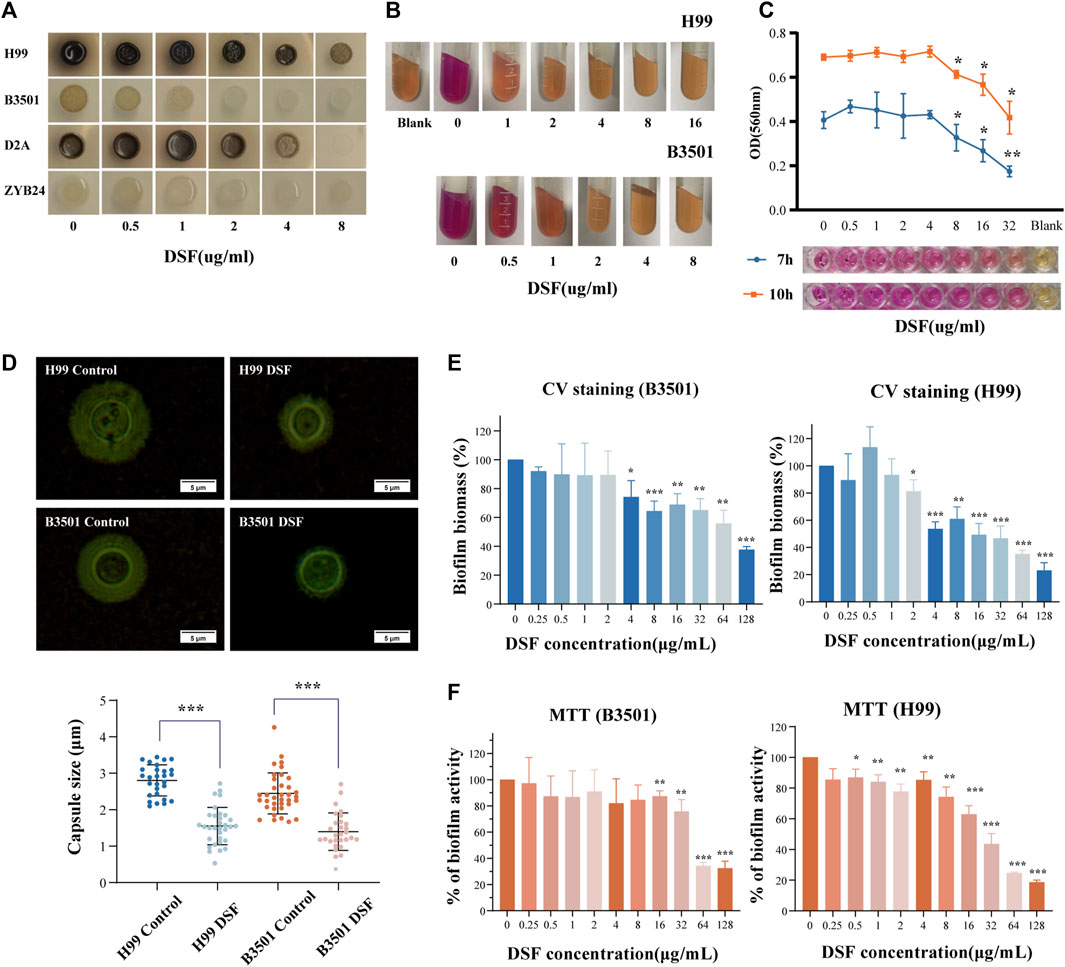
FIGURE 5. Effect of DSF on virulence factors of C. neoformans. (A) Melanin production on agar plates with different doses of DSF; (B) Urease activity on agar; (C) Variation in urease activity in RUH liquid medium. The rightmost well contained the RUH medium control group. (D) India ink staining after induction. Capsule thickness is defined as the distance between the cell wall and outer boundary of the capsule. (E) Biofilm degradation; (F) Viability of cells within the biofilm. *p < 0.05, **p < 0.01, ***p < 0.001, vs. controls.
3.8 DSF inhibited the urease activity of C. neoformans
Once the urea substrate is broken down by urease, the alkaline product discolors the medium. The positive control exhibited a strong urease activity (+++). With increasing concentration of DSF in the medium (Figure 5B), no color change (−) occurred in all groups except for the 0.5 μg/mL and 1 μg/mL groups, which showed (++/+) changes. The modified RUH experiment monitored changes in urease activity in a shorter time (Figure 5C) and controlled the interference of the fungal load. When DSF was increased to 8 μg/mL, the activity was obviously reduced compared to that in the non-dosed group (p < 0.05). This showed that C. neoformans urease activity was inhibited by DSF as the drug concentration increased.
3.9 DSF restricted capsule formation of C. neoformans
The capsules stained with ink were photographed and counted (Figure 5D). The capsule thickness was 2.803 ± 0.127 μm [mean ± standard deviation (SD)] in the H99-control group and 1.551 ± 0.513 μm in the H99-DSF group (p < 0.001 compared to the control). The capsule thickness was 2.448 ± 0.563 μm (mean ± SD) in the B3501-control group and 1.399 ± 0.512 μm in the B3501-DSF group (p < 0.001). The inhibition of capsule formation of C. neoformans by DSF contributed to the reduction in fungal resistance and invasiveness.
3.10 Biofilm inhibition effects of DSF
Biofilm increases the virulence of Cryptococcus and facilitates drug resistance. Biofilms were prepared using a fungal solution. After incubation period to form biofilms, we measured the optical density of each well and found consistent values. Compared to the control, H99 showed a statistical difference at the DSF concentration up to 2 µg/mL and B3501 at 4 µg/mL compared to the control (Figure 5E). This suggests that DSF had a degradation effect on the formed biofilm. Another group of prepared 48 h-biofilms was incubated for 24 h with DSF. The number of living cells within the biofilm decreased as drug concentration increased. Compared to the control, a statistical difference was observed at 0.5 µg/mL DSF concentration for H99 and at 16 µg/mL for B3501 (Figure 5F). The data for the drug-treated group was normalized relative to the control group.
3.11 DSF controlled pathways related to cell division and biological enzyme activity of H99
To further investigate the mechanism of action of DSF against C. neoformans, we analyzed the regulation of the gene expression at the transcriptional level. A1-A3 (parallel replicates) were H99 control groups, and B1–B3 were H99 treated with 4 μg/mL of DSF. A total of 319 DEGs were identified. Cluster analysis showed that 98 DEGs were upregulated and 221 were downregulated in the DSF group compared to those in the control group (Figure 6A). The overall distribution of DEGs was observed in a volcano plot (Figure 6B).
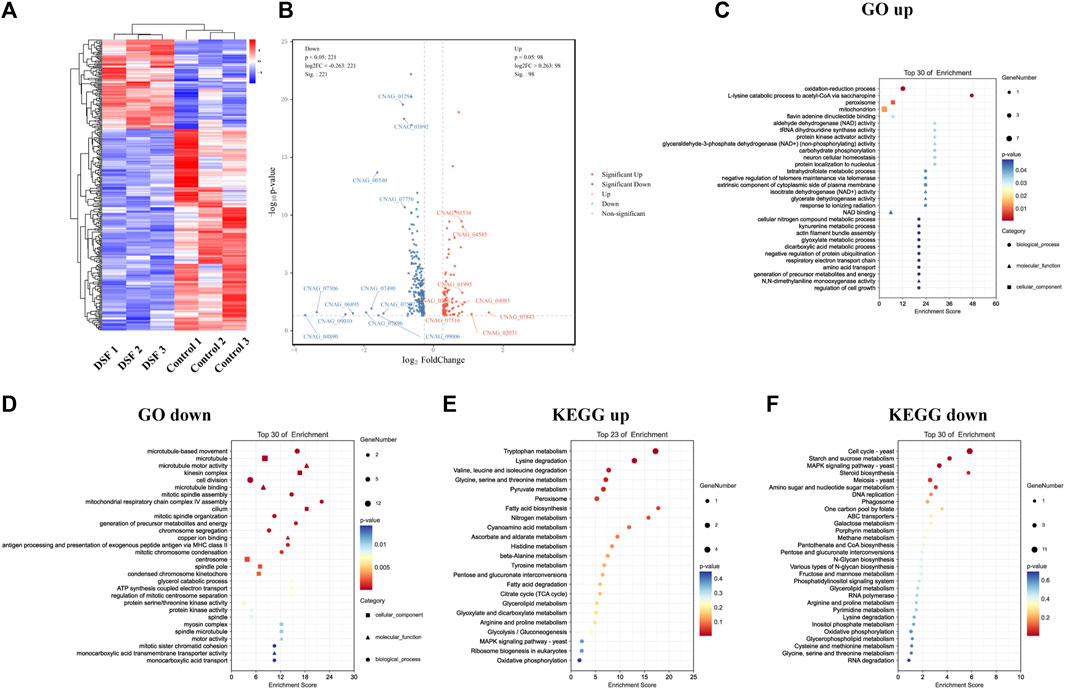
FIGURE 6. Transcriptome sequencing profile of H99 cells after DSF treatment. (A) Heat map of DEGs clustering. Red, highly expressed genes; blue, low-expression genes. (B) Volcanic map. Gray: non-significant DEGs; red and blue: significant DEGs. (C,D) Bubble map of GO enrichment. (E,F) Bubble map of KEGG enrichment.
Enrichment analysis was performed using the GO database, and 10 items with a respective number of upregulated (Figure 6C) and downregulated DEGs (Figure 6D) greater than or equal to two under subcategories were screened for bubble plots. DEGs were mainly enriched in the oxidation-reduction process (GO:0055114), peroxisomes (GO:0005777), mitochondria (GO:0005739), and nicotinamide adenine dinucleotide (NAD+) binding (GO:0051287). Downregulation was highly significant, with the DEGs mainly enriched in microtubule-based movements (GO:0099098), cell division (GO:0051301), microtubule motor activity (GO:0003774), protein serine/threonine kinase activity (GO:0004674), protein kinase activity (GO:0004672), and adenosine triphosphate hydrolase activity (GO:0042623). During this process, DSF regulates cell division, important biological enzyme activities, and reduction-oxidation (redox)-related pathways in C. neoformans.
3.12 DSF affected membrane transport function and multiple metabolic processes of H99
The KEGG database was also used for the enrichment analysis of DEGs, and 2,387 genes were annotated to 36 KEGG pathways (Figures 6E, F). The downregulated pathways were highly significant (Figure 6F) and involved cell growth and death, transport and catabolism, membrane transport, signal transduction, folding/sorting and degradation, amino acid metabolism, carbohydrate metabolism, energy metabolism, metabolism of cofactors and vitamins, and nucleotide metabolism. In conclusion, we found that DSF affects membrane transport function and multiple metabolic processes in C. neoformans. We verified the up and downregulation of eight DEGs (upregulated: CNAG_04085, CNAG_01995, CNAG_04585, CNAG_02863; downregulated: CNAG_05752, CNAG_07756, CNAG_00540, CNAG_00735) by q-PCR. The q-PCR data were in agreement with RNA-seq data (Supplementary Figure S2).
3.13 Homology modeling of ALDH (NAD+) and molecular docking of DSF
Based on the confirmed mechanism of DSF, we screened H99 functional proteins, including ALDH (NAD+), catalase (CAT), ABC multidrug transporter AFR2, iron-sulfur clusters transporter ATM1, based on the essential differential expression pathways suggested by RNA-seq. They serve as critical biological enzymes or transport-related proteins and are closely related to the growth, metabolism, virulence, drug resistance, and oxidation-reduction processes of C. neoformans. They were then docked to DSF ligands for calculations.
DSF was docked to the Alpha-Fold predicted structure of H99’s ALDH (NAD+) (UniProt ID: J9VME7, open reading form (ORF) name: CNAG_00735), with a complex binding affinity of −4.4 kcal/mol (Figure 7A). The optimal conformational affinity and root-mean-square deviation of the receptor pocket are shown in Supplementary Table SA3. In the ligand-receptor complex, ALDH interacted with DSF through five amino acid residues in the active pocket, including PHE-467, VAL-304, PHE-298, ALA-219, and PHE-220 (Figure 7C). The best-matched human protein for ALDH (NAD+) (PDB ID 4X0T) and matching parameters are shown in Supplementary Table SA4. By importing the optimal DSF docking conformation containing the coordinate information, we found that the position of the DSF molecule coincided with the active substrate NAD+ of ALDH (4X0T). Moreover, DSF was located at the center of four cysteine (Cys) residues, with the closest residue at position 302 (Figure 7B). Moreover, ALDH of H99 showed significant downregulation results (p < 0.05) in the KEGG Pathway significance analysis. The q-PCR results (Figure 7E) also verified this result.
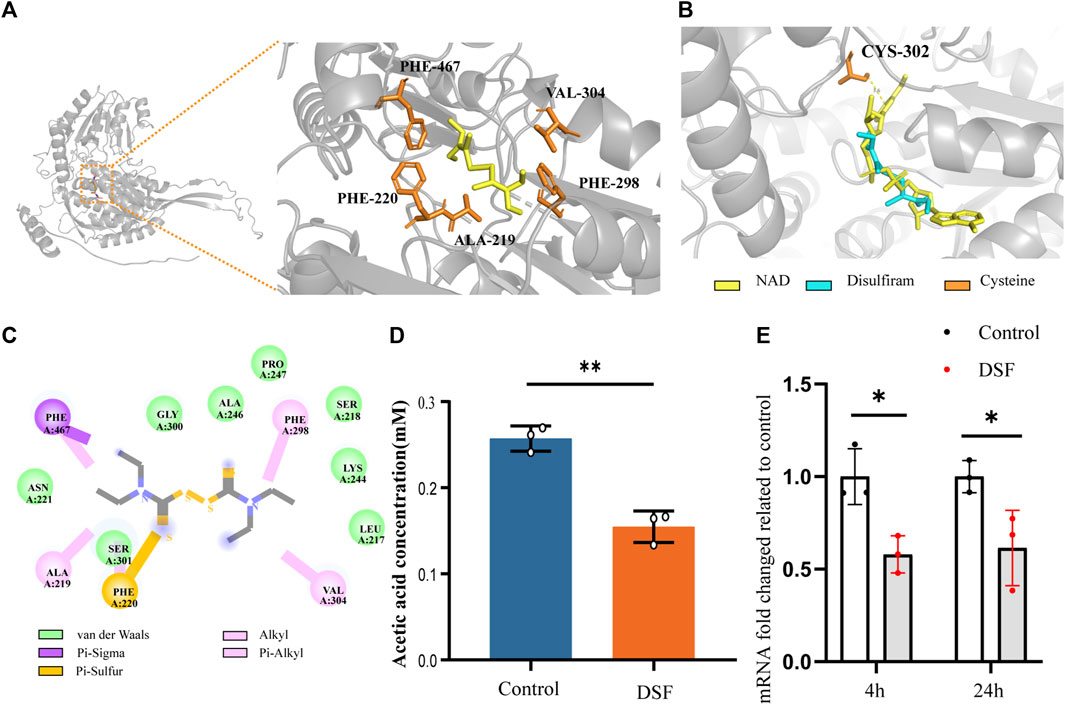
FIGURE 7. DSF disrupts acetic acid metabolism by affecting mRNA expression and protein structure of ALDH (NAD+) from C. neoformans. (A) The complexes ribbon structure and enlarged molecular docking images of the DSF ligand with predicted Alpha-Fold 3D structures of ALDH (NAD+) from H99. (B) The optimal DSF conformation containing coordinate information of the homologous protein structure. (C) 2D interactions between DSF and the predicted binding sites of H99 ALDH (i.e., conventional H-bond, carbon-hydrogen bond, alkyl or Pi-alkyl bond, Pi-sulfur bond, Pi-sigma bond, and Van der Waals bond). (D) Biochemical effects of DSF on ALDH in H99 cells. (E) RT-qPCR results of ALDH (NAD+) of different time, *p < 0.05, **p < 0.01, vs. controls.
3.14 DSF inhibited the ability of C. neoformans to metabolize aldehydes
ALDH, an essential metabolic enzyme in the yeast fungus, transforms harmful aldehyde metabolites into acetic acid, which is then excreted. C. neoformans isolated from the lungs of infected mice expresses high levels of ALDH transcripts, and ALDH expression is important for protecting Cryptococcus cells from phagocytosis or destruction by macrophages after engulfment (Hu et al., 2008). ALDH knockout mutants have partially compromised cell walls and exhibit reduced recovery after phagocytosis and significantly reduced virulence in G. mellonella infection model (Ammar, 2019). To determine whether DSF interferes with the catalytic pathway involving ALDH, we assayed the biochemical activity of acetic acid in DSF-induced culture media using K-ACET (Figure 7D). Compared to the positive control, the DSF group (2 μg/mL) induced a significant decrease in acetic acid content in the medium at 48 h (p < 0.01). This assay indirectly indicates that DSF diminishes the amount or activity of ALDH, thus inhibiting the ability of C. neoformans to metabolize aldehydes.
3.15 Homology modeling of CAT, AFR2, and ATM1 and molecular docking of DSF
By combining CAT (UniProt ID: J9VNU0, ORF name: CNAG_05015), AFR2 (UniProt ID: J9VPA2, ORF name: CNAG_00869), and ATM1 (UniProt ID: J9VWU3, ORF name: CNAG_04358) of the Alpha-Fold conformation with DSF (Figures 8A, D, G), the optimal conformation binding affinities were −3.7, −3.8, and −3.8 kcal/mol, respectively. The best homologous protein to which CAT matched was the CAT of Komagataella pastoris (PDB ID 6RJN). The best docking conformation of DSF overlapped with the position of NDP (the active substrate of 6RJN), indicating that DSF successfully occupied the active NDP pocket (Figure 8B). The protein most homologous to AFR2 was derived from S. cerevisiae S288C (PDB ID 7P05). The best docking conformation of DSF, closing to Cys in the 1041th position of H99’s AFR2, was only 3.7 Å away from active substrate ADP of 7P05. Moreover, the ATP substrate was close to multiple active Cys residues, including the 129th, 199th, 325th, 331th, 369th, and 381th positions of 7P05 (Figure 8E), ranging from 6.9 to 11.5 Å. The most homologous protein (PDB ID: 4MYH) matching ATM1 of H99 was derived from S288C. The optimal docking conformation of DSF was only 3.7 Å away from the active substrate GSH position (Figure 8H).
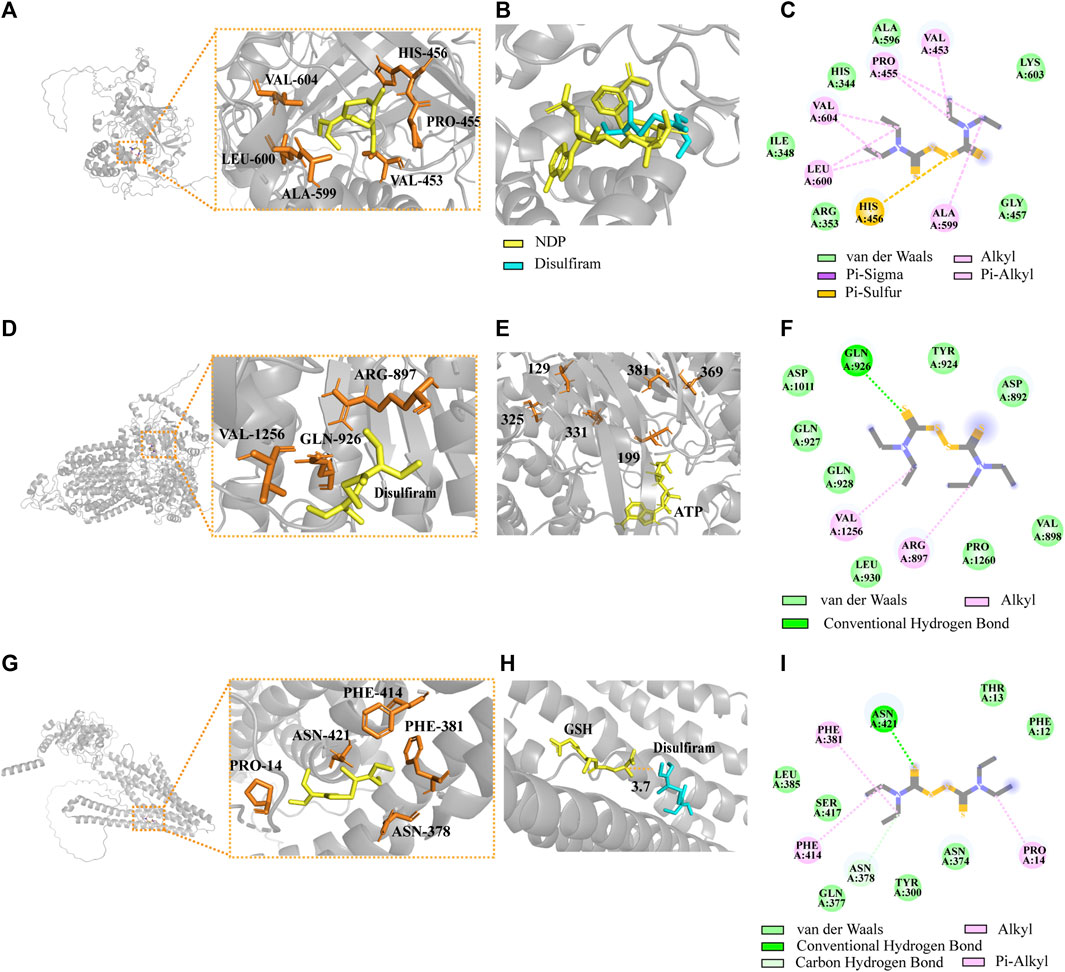
FIGURE 8. Complexes of the DSF ligand with predicted Alpha-Fold 3D structures of CAT, AFR2, and ATM1. (A) Structural models of CAT docking with DSF. (B) Introduction of the optimal DSF conformation for the homologous protein structure of CAT. (C) The 2D interactions between DSF and predicted binding sites of CAT. (D) Structural models of AFR2 docking with DSF. (E) Introduction of the optimal DSF conformation for the homologous protein structure of AFR2. (F) The 2D interactions between DSF and AFR2. (G) Structural models of ATM1 docking with DSF. (H) Introduction of the optimal DSF conformation for ATM1. (I) The 2D interactions between DSF and ATM1.
4 Discussion and conclusion
Fungi pose a serious threat to humanity, with at least 300 million people suffering from severe fungal infections (Bongomin et al., 2017). In recent years, it has been found that patients coinfected with C. neoformans and SARS-CoV-2 are at a significant risk of death during the first 30 days. While new drugs are costly to develop and have long approval cycles, “Repurposed old drugs” bring new ideas and savings (Chong and Sullivan, 2007). To the best of our knowledge, this is the first systematic study assessing the effects of DSF on Cryptococcus spp.
In this study, 42 Cryptococcus strains of various serotypes were selected, all with excellent DSF MIC. This indicates that DSF has a broad-spectrum rather than individual inhibitory effect on Cryptococcus. Unfortunately, these strains did not include FLC-resistant strains. The combined sensitivity test revealed that DSF and AmB/5-FC/TRB/KET exerted synergistic inhibitory effects on C. neoformans. This suggests that DSF may possess pathways other than conventional antifungal drugs, or that there are unique targets that interfere with drug resistance. These combinations may reduce the toxic side effects of conventional antifungal agents.
The antimicrobial mechanism of DSF can be understood from three perspectives: synergism with copper, affinity for the cysteine mercapto group of target proteins, and immunomodulatory functions. Combined with the existing studies on DSF antifungals, this implies a focus on significant proteins containing cysteine near active sites associated with membrane transport function (especially copper) and redox. Transcriptome sequencing corroborated this hypothesis. Therefore, in this study, molecular docking calculations were performed between these important proteins and DSF based on computer simulations. Molecular docking calculations were used to assess the degree of interaction between the ligand and receptor (Morris and Lim-Wilby, 2008).
The inhibition of ALDH by DSF is the primary basis for its alcohol-withdrawal effect. DSF inhibits activated cysteine by forming a thiocarbamate-thiol structure, which irreversibly inhibits ALDH activity in the human liver. In vitro, the inhibition of hepatic mitochondrial ALDH is caused by the formation of intramolecular disulfide bonds between two of the three adjacent cysteines within the active site. In vivo, DSF can form a carbamoyl derivative with the Cys302 of ALDH (Shen et al., 2000). ALDH is an important metabolic enzyme in yeast. Throughout their growth, fungi are continuously subjected to various stresses; in particular, the metabolic products ethanol and acetaldehyde can accumulate to toxic levels (Ingram and Buttke, 1984; Jones, 1990). Yeasts use a specialized ALDH pathway to convert toxic aldehydes into less hazardous chemicals. By converting toxic byproducts into acetate, this pathway lowers the toxicity of byproducts and makes it easy for pathogens to adapt to their surroundings (Marchitti et al., 2008; Black et al., 2009).
ALDH in H99 showed significant downregulation (p < 0.05) in the KEGG Pathway significance analysis. To investigate how DSF affects cryptococcal ALDH activity at the molecular level, we aligned the target and homologous proteins. The distance between the homologous protein Cys302 and DSF was only 3.6 Å. At the same time, the Cys302 corresponded to the 356th Cys of H99 ALDH. The pocket where the DSF was located was also at the center of the adjacent Cys 96, 356, and 278 of H99 ALDH. In contrast, the spatial location of the optimally predicted DSF coincided with the human-derived ALDH active substrate, NAD+. This high similarity indicated that the disulfide bond of DSF reacted with cysteine in both ways, thereby affecting the functional activation of H99 ALDH.
Catalase (UniProt ID: J9VNU0, ORF name: CNAG_05015) is an important scavenger of reactive oxygen species in peroxisomes, protecting cells and lipids from the toxic effects of hydrogen peroxide (Gómez et al., 2019). CAT was significantly upregulated in the KEGG Pathway analysis. CAT interacts with DSF through six amino acid residues near the active pocket: VAL-604, HIS-456, PRO-455, VAL-453, ALA-599, and LEU-600 (Figure 8C). DSF overlaps with the active substrate NDP in the H99 CAT pocket, suggesting that DSF may structurally interfere with protein function. An earlier study also showed a significant reduction in catalase activity in the livers of rats fed high doses of DSF (1.0–4.5 g per kg/d) (Koivusalo, 1959). We hypothesized that during the early stages, DSF disturbs the redox homeostasis of H99 cells, leading to enhanced levels of intracellular oxidative stress and increased levels of free radicals, thus causing a compensatory negative feedback upregulation at the CAT transcriptome level. However, the exact regulatory mechanism remains to be explored.
DSF can reverse cerebellar degeneration related 1 (Cdr1)-mediated Candida resistance by interacting with the ATP/substrate-binding site of the transporter protein and forming a disulfide bridge (Prasad et al., 1995; Tsao et al., 2009). The functional region of AFR2 (845–1.087 bp), a multipotent drug-resistant efflux pump of H99, was highly similar to that of Cdr1 (859–1,103 bp) (Supplementary Figure S1). AFR2 (UniProt ID: J9VPA2; ORF name: CNAG_00869) was significantly downregulated in the transcriptome following DSF treatment. AFR2 interacts with DSF via ARG-897, GLN-926, and VAL1256 (Figure 8F). In the docking results, the six closest Cys near the ATP active site were only 6.9–11.5 Å from DSF (Sauna et al., 2004). DSF, within 8 Å from Cys, can interfere with ATP binding site activity by assisting cysteines to form intramolecular disulfide bonds. Whether this is related to the synergistic effect of DSF needs to be verified through further experiments.
The iron-sulfur cluster transporter ATM1 (UniProt ID: J9VWU3, ORF name: CNAG_04358) plays an important role in the production of cytoplasmic iron-sulfur proteins (Do et al., 2018). ATM1 depends on copper-dependent transcription factor 1 (CUF1), which functions in an environment of copper stress and increases susceptibility to copper toxicity (Garcia-Santamarina et al., 2017). Transcriptome analysis revealed that CUF1 was significantly downregulated. Studies have shown that DSF treatment decreases GSH levels, leading to increased oxidized glutathione (GSSG) levels, which interferes with the GSH/GSSG oxidoreduction state (Nagendra et al., 1994). ATM1 interacts with DSF via PHE-414, PHE381, ASN-2378, PRO-14, and ASN-421 in the active pocket (Figure 8I). DSF is only 3.7 Å away from the homologous protein substrate GSH. DSF consumes GSH and disrupts the oxidation-reduction environment, which may prevent GSH from forming an iron-sulfur conjugation complex with ATM1.
In conclusion, DSF possesses excellent in vitro and in vivo antifungal activity against C. neoformans and this effect is mediated through multiple mechanisms. DSF is an FDA-approved drug that can be repurposed as a potential treatment option for C. neoformans.
Data availability statement
The data presented in the study are deposited in the Gene Expression Omnibus repository, accession number GSE248987 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE248987).
Ethics statement
The animal study was reviewed and approved by the Laboratory Animal Ethics Committee of Jinling Hospital (2022DZSJJDWLS-006).
Author contributions
MP: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing–original draft. CZ: Data curation, Methodology, Writing–review and editing. Y-YD: Writing–review and editing, Formal Analysis. H-BL: Writing–review and editing. X-YP: Writing–review and editing. QW: Writing–review and editing, Data curation, Formal Analysis. Q-YC: Writing–review and editing. HS: Writing–review and editing, Project administration. Q-TK: Project administration, Writing–review and editing, Funding acquisition, Resources.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Jiangsu Dermatology Innovation Team Foundation (Grant number CXTDA2017038).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2023.1268649/full#supplementary-material
References
Al-Odaini, N., Li, X. Y., Li, B. K., Chen, X. C., Huang, C. Y., Lv, C. Y., et al. (2021). In vitro antifungal susceptibility profiles of Cryptococcus neoformans var. grubii and Cryptococcus gattii clinical isolates in guangxi, southern China. Front. Microbiol. 12, 708280. doi:10.3389/fmicb.2021.708280
Alspaugh, J. A. (2015). Virulence mechanisms and Cryptococcus neoformans pathogenesis. Fungal Genet. Biol. 78, 55–58. doi:10.1016/j.fgb.2014.09.004
Ammar, M. A. N. (2019). “Role of Cryptococcus neoformans pyruvate decarboxylase and aldehyde dehydrogenase enzymes in acetate production and virulence,” (South Carolina: Clemson University). [Doctoral dissertation].
Bermas, A., and Geddes-McAlister, J. (2020). Combatting the evolution of antifungal resistance in Cryptococcus neoformans. Mol. Microbiol. 114 (5), 721–734. doi:10.1111/mmi.14565
Bhatt, K., Agolli, A., Patel, M. H., Garimella, R., Devi, M., Garcia, E., et al. (2021). High mortality co-infections of COVID-19 patients: mucormycosis and other fungal infections. Discoveries 9 (1), e126. doi:10.15190/d.2021.5
Black, W. J., Stagos, D., Marchitti, S. A., Nebert, D. W., Tipton, K. F., Bairoch, A., et al. (2009). Human aldehyde dehydrogenase genes: alternatively spliced transcriptional variants and their suggested nomenclature. Pharmacogenet Genomics 19 (11), 893–902. doi:10.1097/FPC.0b013e3283329023
Bongomin, F., Gago, S., Oladele, R. O., and Denning, D. W. (2017). Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi (Basel) 3 (4), 57. doi:10.3390/jof3040057
Chen, C., Yang, K. W., Wu, L. Y., Li, J. Q., and Sun, L. Y. (2020). Disulfiram as a potent metallo-β-lactamase inhibitor with dual functional mechanisms. Chem. Commun. (Camb) 56 (18), 2755–2758. doi:10.1039/c9cc09074f
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (17), i884–i890. doi:10.1093/bioinformatics/bty560
Chong, C. R., and Sullivan, D. J. (2007). New uses for old drugs. Nature 448 (7154), 645–646. doi:10.1038/448645a
Chong, W. H., Saha, B. K., Ananthakrishnan, R., and Chopra, A. (2021). State-of-the-art review of secondary pulmonary infections in patients with COVID-19 pneumonia. Infection 49 (4), 591–605. doi:10.1007/s15010-021-01602-z
Christensen, W. B. (1946). Urea decomposition as a means of differentiating Proteus and paracolon cultures from each other and from Salmonella and Shigella types. J. Bacteriol. 52 (4), 461–466. doi:10.1128/jb.52.4.461-466.1946
da Silva, A. R., de Andrade Neto, J. B., da Silva, C. R., Campos Rde, S., Costa Silva, R. A., Freitas, D. D., et al. (2016). Berberine antifungal activity in fluconazole-resistant pathogenic yeasts: action mechanism evaluated by flow cytometry and biofilm growth inhibition in Candida spp. Antimicrob. Agents Chemother. 60 (6), 3551–3557. doi:10.1128/aac.01846-15
De Brucker, K., Bink, A., Meert, E., Cammue, B. P., and Thevissen, K. (2013). Potentiation of antibiofilm activity of amphotericin B by superoxide dismutase inhibition. Oxid. Med. Cell Longev. 2013, 704654. doi:10.1155/2013/704654
de Oliveira, H. C., Joffe, L. S., Simon, K. S., Castelli, R. F., Reis, F. C. G., Bryan, A. M., et al. (2020). Fenbendazole controls in vitro growth, virulence potential, and animal infection in the Cryptococcus model. Antimicrob. Agents Chemother. 64 (6), e00286. doi:10.1128/aac.00286-20
Do, E., Park, S., Li, M. H., Wang, J. M., Ding, C., Kronstad, J. W., et al. (2018). The mitochondrial ABC transporter Atm1 plays a role in iron metabolism and virulence in the human fungal pathogen Cryptococcus neoformans. Med. Mycol. 56 (4), 458–468. doi:10.1093/mmy/myx073
Feder, V., Kmetzsch, L., Staats, C. C., Vidal-Figueiredo, N., Ligabue-Braun, R., Carlini, C. R., et al. (2015). Cryptococcus gattii urease as a virulence factor and the relevance of enzymatic activity in cryptococcosis pathogenesis. Febs J. 282 (8), 1406–1418. doi:10.1111/febs.13229
García-Rodas, R., Trevijano-Contador, N., Román, E., Janbon, G., Moyrand, F., Pla, J., et al. (2015). Role of Cln1 during melanization of Cryptococcus neoformans. Front. Microbiol. 6, 798. doi:10.3389/fmicb.2015.00798
Garcia-Santamarina, S., Uzarska, M. A., Festa, R. A., Lill, R., and Thiele, D. J. (2017). Cryptococcus neoformans iron-sulfur protein biogenesis machinery is a novel layer of protection against Cu stress. mBio 8 (5), e01742. doi:10.1128/mBio.01742-17
Geddes, J. M., Caza, M., Croll, D., Stoynov, N., Foster, L. J., and Kronstad, J. W. (2016). Analysis of the protein kinase A-regulated proteome of Cryptococcus neoformans identifies a role for the ubiquitin-proteasome pathway in capsule formation. mBio 7 (1), e01862–e01815. doi:10.1128/mBio.01862-15
Gómez, S., Navas-Yuste, S., Payne, A. M., Rivera, W., López-Estepa, M., Brangbour, C., et al. (2019). Peroxisomal catalases from the yeasts Pichia pastoris and Kluyveromyces lactis as models for oxidative damage in higher eukaryotes. Free Radic. Biol. Med. 141, 279–290. doi:10.1016/j.freeradbiomed.2019.06.025
Harrison, J. J., Turner, R. J., and Ceri, H. (2007). A subpopulation of Candida albicans and Candida tropicalis biofilm cells are highly tolerant to chelating agents. FEMS Microbiol. Lett. 272 (2), 172–181. doi:10.1111/j.1574-6968.2007.00745.x
Hu, G., Cheng, P. Y., Sham, A., Perfect, J. R., and Kronstad, J. W. (2008). Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol. Microbiol. 69 (6), 1456–1475. doi:10.1111/j.1365-2958.2008.06374.x
Huang, J., Lei, J., Ge, A., Xiao, W., Xin, C., Song, Z., et al. (2023). Antifungal effect of vitamin D(3) against Cryptococcus neoformans coincides with reduced biofilm formation, compromised cell wall integrity, and increased generation of reactive oxygen species. J. Fungi (Basel) 9 (7), 772. doi:10.3390/jof9070772
Ingram, L. O., and Buttke, T. M. (1984). Effects of alcohols on micro-organisms. Adv. Microb. Physiol. 25, 253–300. doi:10.1016/s0065-2911(08)60294-5
Jones, R. P. (1990). Roles for replicative deactivation in yeast-ethanol fermentations. Crit. Rev. Biotechnol. 10 (3), 205–222. doi:10.3109/07388559009038208
Khan, S., Singhal, S., Mathur, T., Upadhyay, D. J., and Rattan, A. (2007). Antifungal potential of disulfiram. Nihon Ishinkin Gakkai Zasshi 48 (3), 109–113. doi:10.3314/jjmm.48.109
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 (4), 357–360. doi:10.1038/nmeth.3317
Klepser, M. (2011). The value of amphotericin B in the treatment of invasive fungal infections. J. Crit. Care 26 (2), 225.e1–10. doi:10.1016/j.jcrc.2010.08.005
Kneale, M., Bartholomew, J. S., Davies, E., and Denning, D. W. (2016). Global access to antifungal therapy and its variable cost. J. Antimicrob. Chemother. 71 (12), 3599–3606. doi:10.1093/jac/dkw325
Koivusalo, M. (1959). Effect of disulfiram (tetraethylthiuram disulphide) on the metabolism of methanol in rat liver homogenates. Acta Physiol. Scand. 45, 271–277. doi:10.1111/j.1748-1716.1959.tb01697.x
Kong, Q., Cao, Z., Lv, N., Zhang, H., Liu, Y., Hu, L., et al. (2020). Minocycline and fluconazole have a synergistic effect against Cryptococcus neoformans both in vitro and in vivo. Front. Microbiol. 11, 836. doi:10.3389/fmicb.2020.00836
Krajaejun, T., Lohnoo, T., Yingyong, W., Rujirawat, T., Kumsang, Y., Jongkhajornpong, P., et al. (2019). The repurposed drug disulfiram inhibits urease and aldehyde dehydrogenase and prevents in vitro growth of the oomycete Pythium insidiosum. Antimicrob. Agents Chemother. 63 (8), e00609. doi:10.1128/aac.00609-19
Kwon-Chung, K. J., Wickes, B. L., Booth, J. L., Vishniac, H. S., and Bennett, J. E. (1987). Urease inhibition by EDTA in the two varieties of Cryptococcus neoformans. Infect. Immun. 55 (8), 1751–1754. doi:10.1128/iai.55.8.1751-1754.1987
Lee, D., Jang, E. H., Lee, M., Kim, S. W., Lee, Y., Lee, K. T., et al. (2019). Unraveling melanin biosynthesis and signaling networks in Cryptococcus neoformans. mBio 10 (5), e02267. doi:10.1128/mBio.02267-19
Lee, Y. M., Duh, Y., Wang, S. T., Lai, M. M., Yuan, H. S., and Lim, C. (2016). Using an old drug to target a new drug site: application of disulfiram to target the Zn-site in HCV NS5A protein. J. Am. Chem. Soc. 138 (11), 3856–3862. doi:10.1021/jacs.6b00299
Locasale, J. W., Grassian, A. R., Melman, T., Lyssiotis, C. A., Mattaini, K. R., Bass, A. J., et al. (2011). Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 43 (9), 869–874. doi:10.1038/ng.890
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi:10.1186/s13059-014-0550-8
Lu, Y., Pan, Q., Gao, W., Pu, Y., Luo, K., He, B., et al. (2022). Leveraging disulfiram to treat cancer: mechanisms of action, delivery strategies, and treatment regimens. Biomaterials 281, 121335. doi:10.1016/j.biomaterials.2021.121335
Marchitti, S. A., Brocker, C., Stagos, D., and Vasiliou, V. (2008). Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin. Drug Metab. Toxicol. 4 (6), 697–720. doi:10.1517/17425255.4.6.697
Mina, S., Yaakoub, H., Annweiler, C., Dubée, V., and Papon, N. (2022). COVID-19 and Fungal infections: a double debacle. Microbes Infect. 24 (8), 105039. doi:10.1016/j.micinf.2022.105039
Morris, G. M., and Lim-Wilby, M. (2008). Molecular docking. Methods Mol. Biol. 443, 365–382. doi:10.1007/978-1-59745-177-2_19
Mukherjee, P. K., Mohamed, S., Chandra, J., Kuhn, D., Liu, S., Antar, O. S., et al. (2006). Alcohol dehydrogenase restricts the ability of the pathogen Candida albicans to form a biofilm on catheter surfaces through an ethanol-based mechanism. Infect. Immun. 74 (7), 3804–3816. doi:10.1128/iai.00161-06
Nagendra, S. N., Shetty, K. T., Rao, K. M., and Rao, B. S. (1994). Effect of disulfiram administration on rat brain glutathione metabolism. Alcohol 11 (1), 7–10. doi:10.1016/0741-8329(94)90004-3
Nosanchuk, J. D., Stark, R. E., and Casadevall, A. (2015). Fungal melanin: what do we know about structure? Front. Microbiol. 6, 1463. doi:10.3389/fmicb.2015.01463
Peniche, A. G., Renslo, A. R., Melby, P. C., and Travi, B. L. (2015). Antileishmanial activity of disulfiram and thiuram disulfide analogs in an ex vivo model system is selectively enhanced by the addition of divalent metal ions. Antimicrob. Agents Chemother. 59 (10), 6463–6470. doi:10.1128/aac.05131-14
Phillips, M., Malloy, G., Nedunchezian, D., Lukrec, A., and Howard, R. G. (1991). Disulfiram inhibits the in vitro growth of methicillin-resistant staphylococcus aureus. Antimicrob. Agents Chemother. 35 (4), 785–787. doi:10.1128/aac.35.4.785
Prasad, R., De Wergifosse, P., Goffeau, A., and Balzi, E. (1995). Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet. 27 (4), 320–329. doi:10.1007/bf00352101
Pronk, J. T., Yde Steensma, H., and Van Dijken, J. P. (1996). Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12 (16), 1607–1633. doi:10.1002/(sici)1097-0061(199612)12:16<1607::aid-yea70>3.0.co;2-4
Rajasingham, R., Smith, R. M., Park, B. J., Jarvis, J. N., Govender, N. P., Chiller, T. M., et al. (2017). Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect. Dis. 17 (8), 873–881. doi:10.1016/s1473-3099(17)30243-8
Roberts, G. D., Horstmeier, C. D., Land, G. A., and Foxworth, J. H. (1978). Rapid urea broth test for yeasts. J. Clin. Microbiol. 7 (6), 584–588. doi:10.1128/jcm.7.6.584-588.1978
Sangalli-Leite, F., Scorzoni, L., Alves de Paula, E. S. A. C., da Silva, J. F., de Oliveira, H. C., de Lacorte Singulani, J., et al. (2016). Synergistic effect of pedalitin and amphotericin B against Cryptococcus neoformans by in vitro and in vivo evaluation. Int. J. Antimicrob. Agents 48 (5), 504–511. doi:10.1016/j.ijantimicag.2016.07.025
Sauna, Z. E., Peng, X. H., Nandigama, K., Tekle, S., and Ambudkar, S. V. (2004). The molecular basis of the action of disulfiram as a modulator of the multidrug resistance-linked ATP binding cassette transporters MDR1 (ABCB1) and MRP1 (ABCC1). Mol. Pharmacol. 65 (3), 675–684. doi:10.1124/mol.65.3.675
Segev-Zarko, L. A., and Shai, Y. (2017). Methods for investigating biofilm inhibition and degradation by antimicrobial peptides. Methods Mol. Biol. 1548, 309–322. doi:10.1007/978-1-4939-6737-7_22
Sharma, V., Verma, V., Lal, N., Yadav, S. K., Sarkar, S., Mandalapu, D., et al. (2016). Disulfiram and its novel derivative sensitize prostate cancer cells to the growth regulatory mechanisms of the cell by re-expressing the epigenetically repressed tumor suppressor-estrogen receptor β. Mol. Carcinog. 55 (11), 1843–1857. doi:10.1002/mc.22433
Shen, M. L., Lipsky, J. J., and Naylor, S. (2000). Role of disulfiram in the in vitro inhibition of rat liver mitochondrial aldehyde dehydrogenase. Biochem. Pharmacol. 60 (7), 947–953. doi:10.1016/s0006-2952(00)00435-4
Shukla, S., Sauna, Z. E., Prasad, R., and Ambudkar, S. V. (2004). Disulfiram is a potent modulator of multidrug transporter Cdr1p of Candida albicans. Biochem. Biophys. Res. Commun. 322 (2), 520–525. doi:10.1016/j.bbrc.2004.07.151
Song, Y. J., Yu, H. H., Kim, Y. J., Lee, N. K., and Paik, H. D. (2019). Anti-biofilm activity of grapefruit seed extract against Staphylococcus aureus and Escherichia coli. J. Microbiol. Biotechnol. 29 (8), 1177–1183. doi:10.1041/jmb.1905.05022
Su, H., Han, L., and Huang, X. (2018). Potential targets for the development of new antifungal drugs. J. Antibiot. (Tokyo) 71 (12), 978–991. doi:10.1038/s41429-018-0100-9
Sudan, A., Livermore, J., Howard, S. J., Al-Nakeeb, Z., Sharp, A., Goodwin, J., et al. (2013). Pharmacokinetics and pharmacodynamics of fluconazole for cryptococcal meningoencephalitis: implications for antifungal therapy and in vitro susceptibility breakpoints. Antimicrob. Agents Chemother. 57 (6), 2793–2800. doi:10.1128/aac.00216-13
Tesson, M., Anselmi, G., Bell, C., and Mairs, R. (2017). Cell cycle specific radiosensitisation by the disulfiram and copper complex. Oncotarget 8 (39), 65900–65916. doi:10.18632/oncotarget.19539
Teymuri, M., Shams-Ghahfarokhi, M., and Razzaghi-Abyaneh, M. (2021). Inhibitory effects and mechanism of antifungal action of the natural cyclic depsipeptide, aureobasidin A against Cryptococcus neoformans. Bioorg Med. Chem. Lett. 41, 128013. doi:10.1016/j.bmcl.2021.128013
Thakare, R., Shukla, M., Kaul, G., Dasgupta, A., and Chopra, S. (2019). Repurposing disulfiram for treatment of Staphylococcus aureus infections. Int. J. Antimicrob. Agents 53 (6), 709–715. doi:10.1016/j.ijantimicag.2019.03.024
Treviño-Rangel, R. J., Villanueva-Lozano, H., Hernández-Rodríguez, P., Martínez-Reséndez, M. F., García-Juárez, J., Rodríguez-Rocha, H., et al. (2016). Activity of sertraline against Cryptococcus neoformans: in vitro and in vivo assays. Med. Mycol. 54 (3), 280–286. doi:10.1093/mmy/myv109
Tsao, S., Rahkhoodaee, F., and Raymond, M. (2009). Relative contributions of the Candida albicans ABC transporters Cdr1p and Cdr2p to clinical azole resistance. Antimicrob. Agents Chemother. 53 (4), 1344–1352. doi:10.1128/aac.00926-08
Velasco-García, R., Chacón-Aguilar, V. M., Hervert-Hernández, D., and Muñoz-Clares, R. A. (2003). Inactivation of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa and Amaranthus hypochondriacus L. leaves by disulfiram. Chem. Biol. Interact. 143, 149–158. doi:10.1016/s0009-2797(02)00199-0
Vullo, D., Del Prete, S., Nocentini, A., Osman, S. M., AlOthman, Z., Capasso, C., et al. (2017). Dithiocarbamates effectively inhibit the β-carbonic anhydrase from the dandruff-producing fungus Malassezia globosa. Bioorg Med. Chem. 25 (3), 1260–1265. doi:10.1016/j.bmc.2016.12.040
Wiederhold, N. P. (2017). Antifungal resistance: current trends and future strategies to combat. Infect. Drug Resist 10, 249–259. doi:10.2147/idr.S124918
Wright, C., and Moore, R. D. (1990). Disulfiram treatment of alcoholism. Am. J. Med. 88 (6), 647–655. doi:10.1016/0002-9343(90)90534-k
Yi, C., Liu, J., Deng, W., Luo, C., Qi, J., Chen, M., et al. (2022). Macrophage elastase (MMP12) critically contributes to the development of subretinal fibrosis. J. Neuroinflammation 19 (1), 78. doi:10.1186/s12974-022-02433-x
Keywords: Cryptococcus neoformans, disulfiram, drug repurposing, antifungal, drug resistance
Citation: Peng M, Zhang C, Duan Y-Y, Liu H-B, Peng X-Y, Wei Q, Chen Q-Y, Sang H and Kong Q-T (2024) Antifungal activity of the repurposed drug disulfiram against Cryptococcus neoformans. Front. Pharmacol. 14:1268649. doi: 10.3389/fphar.2023.1268649
Received: 28 July 2023; Accepted: 27 December 2023;
Published: 11 January 2024.
Edited by:
Jian-Cheng Jin, Zhongyuan University of Technology, ChinaReviewed by:
Javiera Alejandra Norambuena, Universidad de Playa Ancha, ChileShizhu Zhang, Nanjing Normal University, China
Copyright © 2024 Peng, Zhang, Duan, Liu, Peng, Wei, Chen, Sang and Kong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Sang, c2FuZ2hvbmdAbmp1LmVkdS5jbg==; Qing-Tao Kong, bmp1a3F0QDE2My5jb20=
†These authors have contributed equally to this work
 Min Peng
Min Peng Chen Zhang
Chen Zhang Yuan-Yuan Duan2
Yuan-Yuan Duan2 Xin-Yuan Peng
Xin-Yuan Peng Hong Sang
Hong Sang Qing-Tao Kong
Qing-Tao Kong