- 1Department of Pharmacy, Faculty of Pharmacy, Mahidol University, Bangkok, Thailand
- 2School of Pharmacy, Walailak University, Nakhon Si Thammarat, Thailand
- 3Division of Pharmacogenomics and Personalized Medicine, Department of Pathology, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Bangkok, Thailand
- 4Laboratory for Pharmacogenomics, Clinical Pathology, Somdetch Phra Debharatana Medical Centre, Ramathibodi Hospital, Bangkok, Thailand
- 5Pharmacogenomics and Precision Medicine Clinic, Bumrungrad International Hospital, Bangkok, Thailand
- 6Bumrungrad Genomic Medicine Institute (BGMI), Bumrungrad International Hospital, Bangkok, Thailand
- 7Division of Medical Oncology, Department of Medicine, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Bangkok, Thailand
- 8Chulabhorn International College of Medicine, Thammasat University, Pathum Thani, Thailand
- 9Ramathibodi Comprehensive Cancer Center, Faculty of Medicine, Ramathibodi Hospital, Mahidol University, Bangkok, Thailand
Background: Osimertinib has shown greater efficacy than standard epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) and fewer grade 3 or higher adverse drug reactions (ADRs) in patients with advanced non-small cell lung cancer (NSCLC) harboring epidermal growth factor receptor (EGFR) mutations. However, the clinical outcomes of osimertinib treatment vary depending on the patient’s ethnicity. Therefore, further research is necessary to evaluate the impact of single nucleotide polymorphisms (SNPs) in cytochrome P450 (CYP450) and drug transporters on the therapeutic outcomes and ADRs to osimertinib in Thai patients, to provide improved pharmacological treatments for cancer patients.
Methods: This retrospective and prospective cohort study enrolled 63 Thai patients with NSCLC treated with 80 mg of osimertinib once daily as monotherapy. Seventeen SNPs in candidate genes related to drug metabolism and transport pathways were analyzed in each patient. Chi-square or Fisher’s exact tests were used to evaluate the associations between SNPs and clinical outcomes, including ADR incidence and objective response rate (ORR). In addition, the correlation between the genotype and median time to treatment failure (TTF) or progression-free survival (PFS) was assessed using Kaplan-Meier analysis and a log-rank test.
Results: We identified six SNPs (rs2231142 and rs2622604 in ABCG2, rs762551 in CYP1A2, rs1057910 in CYP2C9, rs28371759 in CYP3A4, and CYP2A6 deletion polymorphism (CYP2A6*4)) that significantly increased the incidence of ADRs. In addition, we found two SNPs (rs2069514 in CYP1A2 and rs1057910 in CYP2C9) that significantly decreased the median TTF, and two SNPs (rs28399433 in CYP2A6 and rs1057910 in CYP2C9) that significantly decreased the median progression-free survival (PFS). Specifically, we found that one of these SNPs (rs1057910 in CYP2C9) influenced ADRs, TTF, and PFS. Additionally, SNPs in the CYP2A6 heterozygous variant (non4/*4) significantly increased ADR incidence, leading to a high frequency of dose reduction (27.0%).
Conclusion: Our study demonstrated significant SNPs associated with increased ADR incidence, decreased PFS, and decreased TTF in Thai patients with NSCLC treated with osimertinib. The CYP2C9 (*3) and CYP2A6 (*4) allele frequencies differed between ethnicities and were associated with an increased incidence of ADRs. These findings highlight the importance of considering genetic factors in NSCLC treatment and may facilitate personalized medicine approaches. Moreover, our study showed a higher incidence of ADRs than the previous trials, including FLAURA and AURA2, and a higher frequency of dose reduction than reported in the AURA 3 trial, possibly due to genetic differences among the study populations.
1 Introduction
Lung cancer is the leading cause of cancer-related deaths, accounting for approximately 18% (Sung et al., 2021). Approximately 80%–85% of these cases are classified as non-small cell lung cancer (NSCLC) (Nicholson et al., 2022). In Thai patients with NSCLC, epidermal growth factor receptor (EGFR) mutations were most commonly detected, accounting for 68% of cases (Detarkom et al., 2018). Activating EGFR mutations have been identified as predictive indicators of sensitivity to first- and second-generation EGFR tyrosine kinase inhibitors (TKIs). Acquired resistance develops 9–12 months after treatment initiation (Westover et al., 2018). One common mechanism underlying acquired resistance is the substitution of threonine with methionine at amino acid position 790 (T790M) in exon 20 of the EGFR gene, which accounts for 50%–60% of cases. This mutation impairs the binding of both first- and second-generation EGFR-TKIs by enhancing the ATP-binding affinity of the kinase domain of the EGFR mutant receptor, leading to treatment resistance (Yu et al., 2013).
Osimertinib is a third-generation EGFR-TKI that irreversibly binds to cysteine-797 at the ATP-binding site of the EGFR kinase domain (Li et al., 2023). It potently inhibits EGFR phosphorylation in the cases of exon 19 deletion and exon 21 L858R substitution, with IC50 values ranging 13–54 nmol/L. Additionally, in cell lines harboring EGFR T790M mutation, osimertinib demonstrates remarkable potency with an IC50 of less than 15 nmol/L. In addition, Osimertinib exhibits high selectivity for mutated EGFR receptors over wild-type EGFR (IC50: 480–1865 nmol/L) (Remon et al., 2018), resulting in less severe gastrointestinal and skin toxicities than those elicited by the first- or second-generation EGFR-TKIs. Additionally, osimertinib has improved overall survival in previously untreated advanced NSCLC patients with EGFR mutations compared with standard EGFR-TKIs (Ramalingam et al., 2020). Osimertinib is predominantly metabolized by CYP3A4, CYP2A6, CYP2C9, CYP3A5, and CYP2E1 enzymes, which account for 44.4%, 15.5%, 12.0%, 9.6%, and 3.0% of the metabolism, respectively (Dickinson et al., 2016). It produces at least two circulating metabolites, AZ5104 and AZ7550, accounting for 10% of the parent compound (AstraZeneca, 2021). AZ7550 has a potency and selectivity profile comparable to osimertinib, whereas AZ5104 has an 8-fold greater potency against EGFR mutations (Han et al., 2021).
In a previous study, variability in the steady-state area under the plasma drug concentration-time curve (AUCs) of AZ5104 between ethnic groups was observed, with a 10%–23% decrease in Asian versus Caucasian patients; however, the underlying reason remains unclear (Brown et al., 2017). CYP450 predominantly metabolizes Osimertinib and is a substrate for P-glycoprotein (P-gp), which is encoded by ABCB1, and breast cancer resistance protein (BCRP), which is encoded by ABCG2 (AstraZeneca, 2021), which may lead to individual variations in plasma osimertinib concentrations due to genetic polymorphisms. A previous study also found a linear relationship between ADR development and osimertinib concentration, with an increased risk of rash, diarrhea, and QTc prolongation (Brown et al., 2017). Moreover, in Asian patients, the incidence of QTc prolongation was slightly higher than that reported in the FLAURA study (Soria et al., 2018; Cho et al., 2019). Furthermore, a significant association was demonstrated between the AUC0-24 of osimertinib, SNPs rs1128503 in ABCB1, and SNPs rs2231137 in ABCG2 with grade 2 or higher adverse events (Ishikawa et al., 2023). The previous studies on EGFR-TKIs found a correlation between SNPs rs762551 in CYP1A2 and the severity of erlotinib-induced skin rash, along with the development of diarrhea and SNPs rs2470890 in ABCB1 and rs776746 in CYP3A5 (Liao et al., 2020). Additionally, SNPs rs2032582 in ABCB1 were associated with afatinib-induced diarrhea (Sogawa et al., 2020), and reduced function of CYP2D6 increased the risk of gefitinib-induced rash (Suzumura et al., 2012). However, the association between SNPs in CYP450 and drug efflux transporters and efficacy outcomes remains unclear. We investigated seventeen SNPs in CYP450 and drug efflux transporters that may alter the pharmacokinetic and pharmacodynamic profile of osimertinib, intending to advance personalized medicine approaches.
2 Materials and methods
2.1 Patients and study design
We recruited 63 NSCLC patients for this retrospective and prospective cohort study between June 2022 and January 2023 from the Division of Medical Oncology, Department of Medicine, Faculty of Medicine, Ramathibodi Hospital, Mahidol University, Bangkok, Thailand. The inclusion criteria were histologically confirmed EGFR mutation, monotherapy with 80 mg osimertinib once daily, age at diagnosis of >18 years, and normal baseline laboratory findings (complete blood count, renal function test, and liver function test). Patients were excluded if they received other medications that interfered with osimertinib drug levels or if toxicity was reported in the osimertinib-approved product monograph, such as itraconazole, rifampicin, simvastatin, and amiodarone (AstraZeneca, 2021). All patients provided written consent before enrolling in the study, and the study was approved by the Ethics Committee of Ramathibodi (ethics approval code: COA. MURA 2022/370).
2.2 Genotyping methods
Genomic DNA was isolated from an EDTA tube (6 mL) using an automatic DNA extraction system (MagNaPure; Roche, Mannheim, Germany). The concentration of DNA was approximately 5 ng/μL, and the A260/A280 ratio was in the range 1.70–2.10. A total of 17 SNPs were genotyped, namely, ABCB1 rs1128503, ABCG2 rs1871744, ABCG2 rs2231142, ABCG2 rs2231164, ABCG2 rs2622604, ABCG2 rs4148157, CYP1A2 rs1871744, CYP1A2 rs2069514, CYP1A2 rs762551, CYP2A6*4, CYP2A6 rs28399433, CYP2C9 rs1057910, CYP2C9 rs1799853, CYP3A4 rs28371759, CYP3A5 rs10264272, CYP3A5 rs776746, and POR rs1057868. Genotyping was conducted using real-time PCR ViiA7 (ABI, Foster City, CA, USA), following the manufacturer’s instructions. All samples were analyzed with positive and negative controls in 96-well plates to ensure the authenticity of the results. Genotyping of the candidate genes was performed using a TaqMan real-time PCR assay (ABI, Foster City, CA, USA).
2.3 Clinical endpoint assessment
We analyzed the association between the SNPs and clinical outcomes, including the incidence of adverse drug reactions (ADRs), median time to treatment failure (TTF), median progression-free survival (PFS), and objective response rate (ORR). ADRs were periodically assessed by each patient’s physician using the National Cancer Institute Common Terminology Criteria for Adverse Events version 5.0. The Naranjo algorithm was used to determine causality. TTF was the interval between initiating osimertinib treatment and a new locally directed or systemic treatment other than osimertinib monotherapy. PFS was defined as the time from osimertinib treatment initiation to disease progression or death from any cause. ORR was defined as the percentage of study patients who achieved a complete or partial response to treatment within a certain period, as assessed by each patient’s physician according to the Response Evaluation Criteria in Solid Tumors version 1.1.
2.4 Statistical analysis
Associations between SNPs and clinical outcomes, including ADRs and ORR, and those between patients’ baseline characteristics and clinical outcomes were evaluated using the appropriate chi-square test or Fisher’s exact test. Genetic polymorphisms were assessed for concordance with Hardy-Weinberg Equilibrium (HWE) using Fisher’s exact test. Linkage disequilibrium was explored using Haploview version 4.0. Univariate and multivariate logistic regression analyses were performed to identify the factors associated with clinical outcomes. The multivariate logistic regression analyses included all variables and all SNPs with a p-value of <0.1 from the univariate analysis, which is presented in Supplementary Table S2. The correlation between TTF, PFS, and genotype was assessed using Kaplan-Meier analysis and the log-rank test. Statistical analysis was performed using SPSS version 23 for Windows, and significance was set at p < 0.05. The sample size calculation was based on a prospective longitudinal observational cohort study of 53 patients with advanced NSCLC receiving osimertinib therapy, which found a significant association between SNPs rs2231137 in ABCG2 and grade ≥2 adverse events (p = 0.008). Of the ABCG2 wild-type (G/G) patients, 22 (68.75%) had grade ≥2 adverse events, while all three (100%) of the ABCG2 mutant-type (A/A) patients had grade ≥2 adverse events (Ishikawa et al., 2023). Therefore, based on the output of sample size calculation from n4Studies for a cohort study with binary outcomes (Ngamjarus, 2016), at least 58 patients were needed.
3 Results
3.1 Patient characteristics
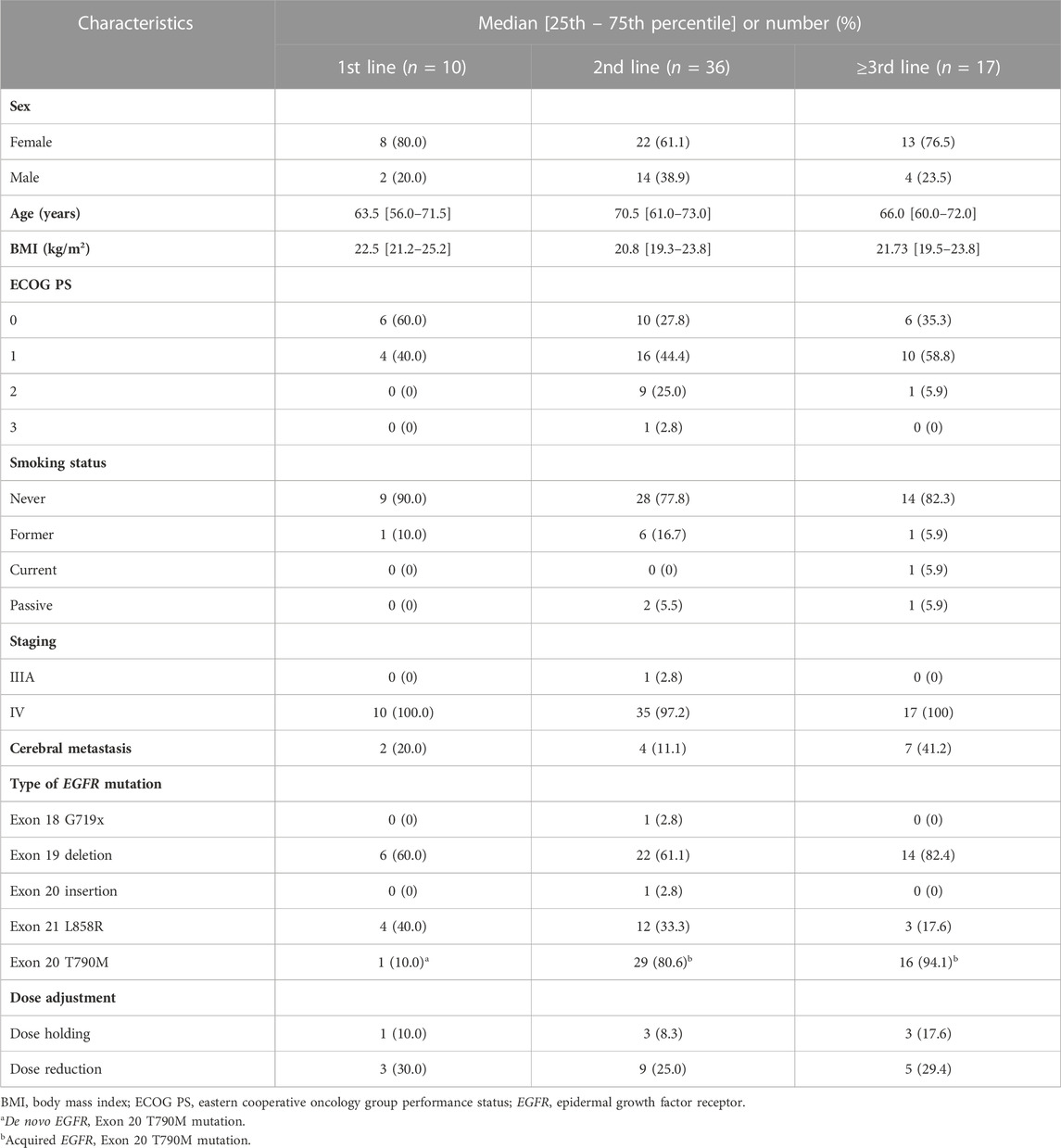
The patient characteristics are presented in Table 1. Sixty-three patients were included in the study: 20 (31.75%) men and 43 (68.25%) women, with a median age of 68 years (range 60–73). Forty-two patients (66.7%) had EGFR exon 19 deletion, nineteen patients (30.2%) had an L858R substitution, one (1.6%) had EGFR exon 20 insertion, and one (1.6%) had EGFR exon 18 G719X mutation. Forty-six patients (73%) harbored the EGFR T790M mutation. Patient characteristics were not significantly associated with the ADRs, TTF, PFS, or ORR. Ten patients (15.9%) received osimertinib as first-line therapy, whereas thirty-six patients (57.1%) and 17 patients (27.0%) received osimertinib as second- and later-line therapy, respectively. The previous systemic therapies included erlotinib (40.8%), gefitinib (35.5%), platinum-doublet chemotherapy (21.9%), and mobocertinib (1.8%). The line of treatment and previous systemic therapy were not significantly associated with the clinical outcomes (p > 0.05). All patients were administered 80 mg of osimertinib once daily. At the time of data cutoff, the mean follow-up duration was 18 (10–30) months, and 46 patients (73%) were still receiving osimertinib at the initial dosage. All patients had normal baseline laboratory test results, including those of complete blood count, liver function tests, and renal function tests.
3.2 Genotype frequencies
The genotype status of drug-metabolizing enzymes and transporters was determined for all 63 patients (Table 2). No genotype distribution deviated from the Hardy–Weinberg equation.
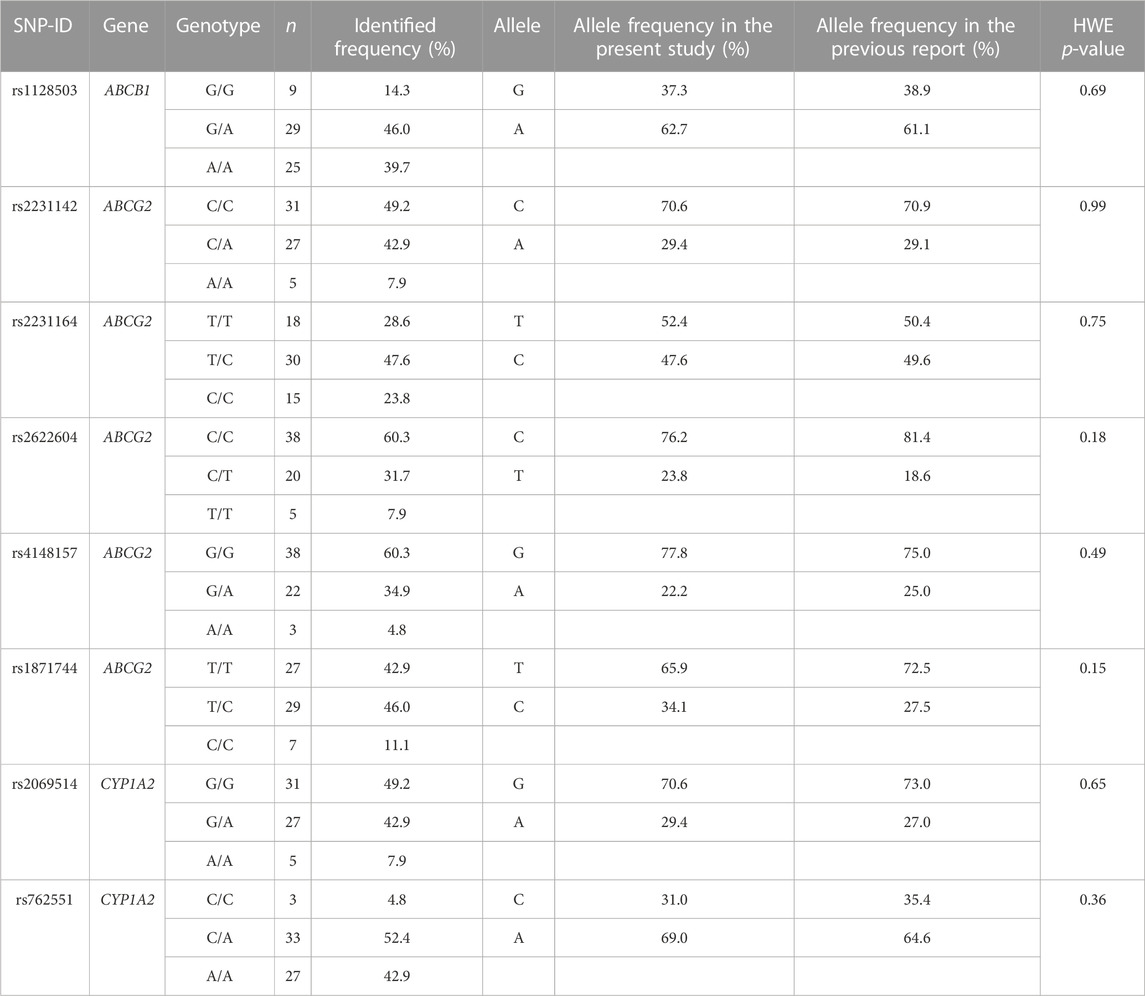
TABLE 2. Genotype and allele frequencies of SNPs were compared between the present study and a previous report (PharmGKB, 2021).
3.3 SNPs associated with osimertinib-induced ADRs
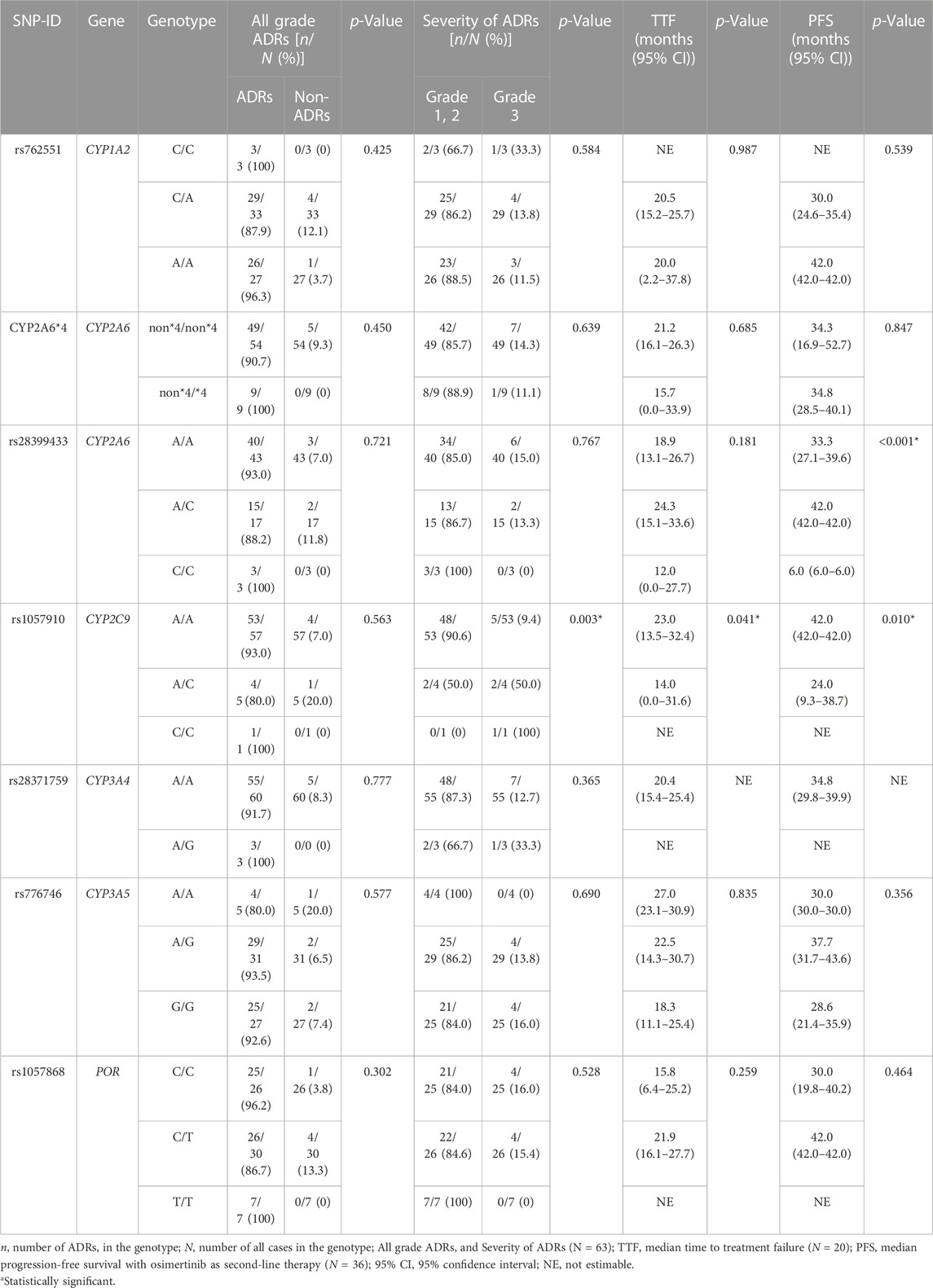
Table 3 shows the association between genetic polymorphisms and the overall ADR incidence. We found that SNP rs1057910 in CYP2C9 was significantly associated with an increased incidence of grade 3 ADRs (p = 0.003). Additionally, we identified several SNPs that were significantly associated with an increased incidence of specific ADRs, including rs2622604 in ABCG2 mutant-type (T/T) and CYP2A6 heterozygous variant (non*4/*4), which were significantly associated with diarrhea (p = 0.011 and p = 0.046, respectively). Furthermore, SNPs rs2231142 in ABCG2 mutant-type (A/A), rs1057910 in CYP2C9 mutant-type (C/C), rs28371759 in CYP3A4 heterozygous variant (A/G), and rs762551 in CYP1A2 wild-type (C/C) were identified. These SNPs were associated with myalgia, grade 3 acneiform rash, QTc prolongation, and bullous dermatitis, respectively (p = 0.007, p = 0.012, p = 0.001, and p = 0.006, respectively; Table 4).
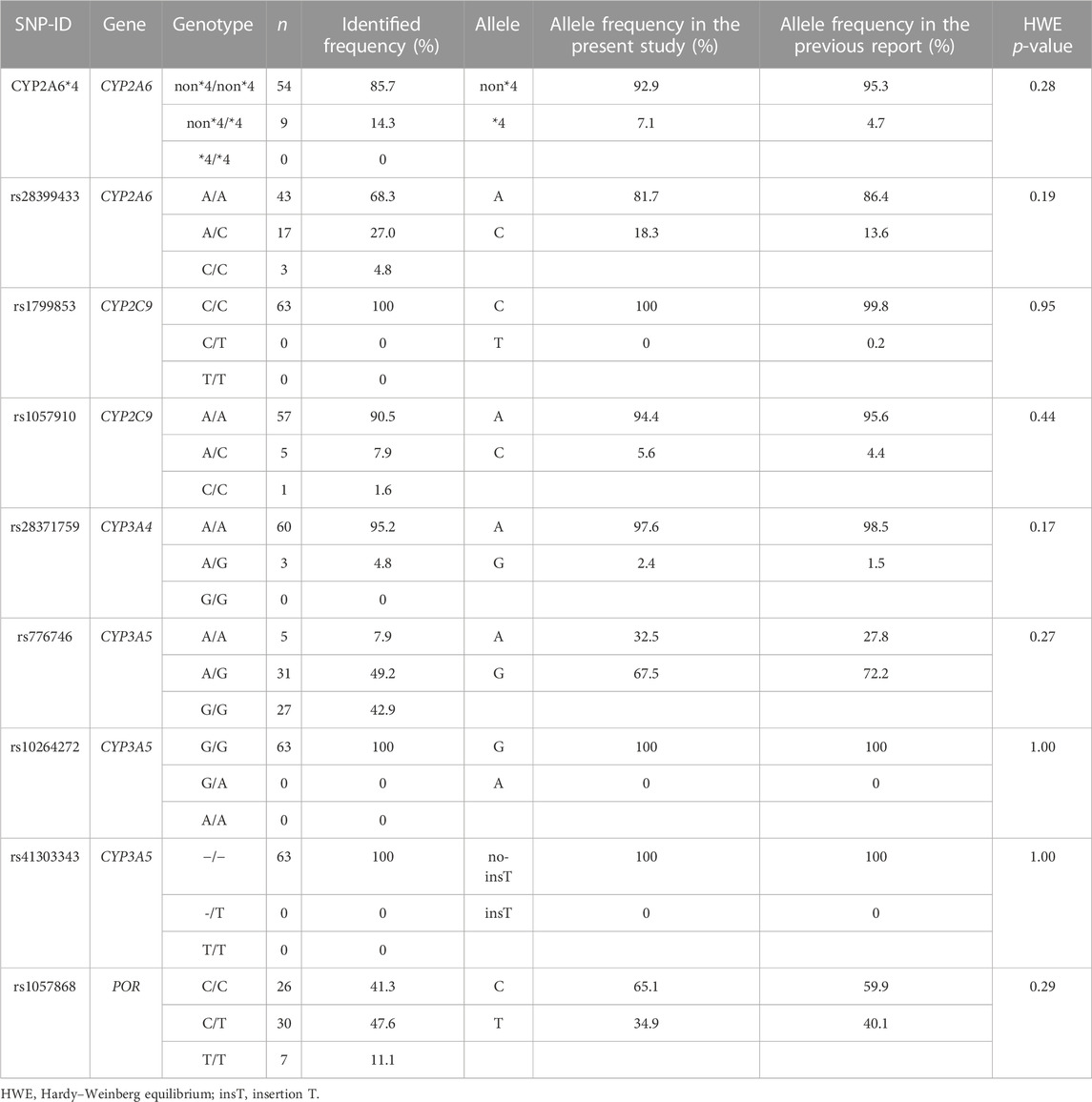
TABLE 3. Genotype and allele frequencies of SNPs were compared between the present study and a previous report (PharmGKB, 2021) (cont.).
3.4 SNPs associated with osimertinib efficacy outcomes
After initiating osimertinib, 31 patients (49.2%) had an objective response, including one patient (1.6%) with a complete response and 30 patients (47.6%) with a partial response. In the non-response group, 32 patients (50.8%) did not respond, including 30 patients (47.6%) with stable disease and two patients (3.2%) with progressive disease.
The median TTF was 19 (10.3–29.0) months. In addition, SNPs rs2069514 in CYP1A2 mutant-type (A/A) and rs1057910 in CYP2C9 heterozygous variant (A/C) were significantly associated with decreased TTF with p < 0.001 and 0.041, respectively. These findings are presented in Table 5 and Figure 1.
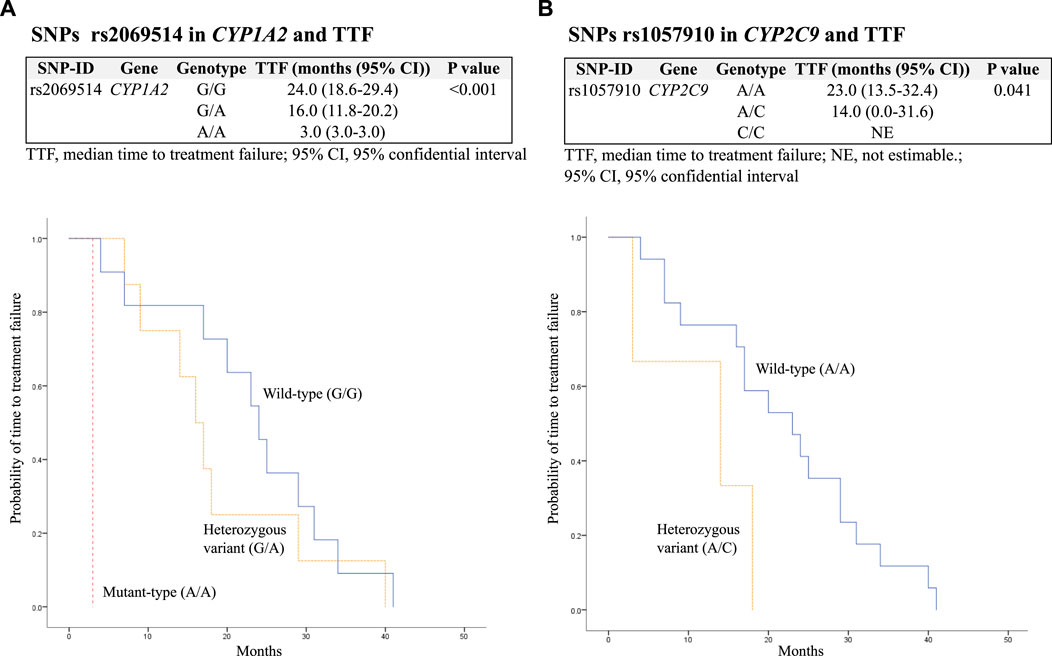
FIGURE 1. Kaplan–Meier estimates and log-rank tests for median time to treatment failure (TTF) associated with SNPs. (A) Association between SNPs rs2069514 in CYP1A2 and TTF; (B) association between SNPs rs1057910 in CYP2C9 and TTF.
The median PFS was significantly decreased in patients with SNPs rs28399433 in CYP2A6 mutant-type (C/C) and rs1057910 in CYP2C9 heterozygous variant (A/C), with p = 0.023 and <0.001, respectively. In addition, among patients who received osimertinib as second-line therapy (N = 36), SNPs rs28399433 in CYP2A6 mutant-type (C/C) and rs1057910 in CYP2C9 heterozygous variant (A/C) were also significantly associated with decreased PFS, with p values of 0.001 and 0.010, respectively (Table 6; Figure 2).
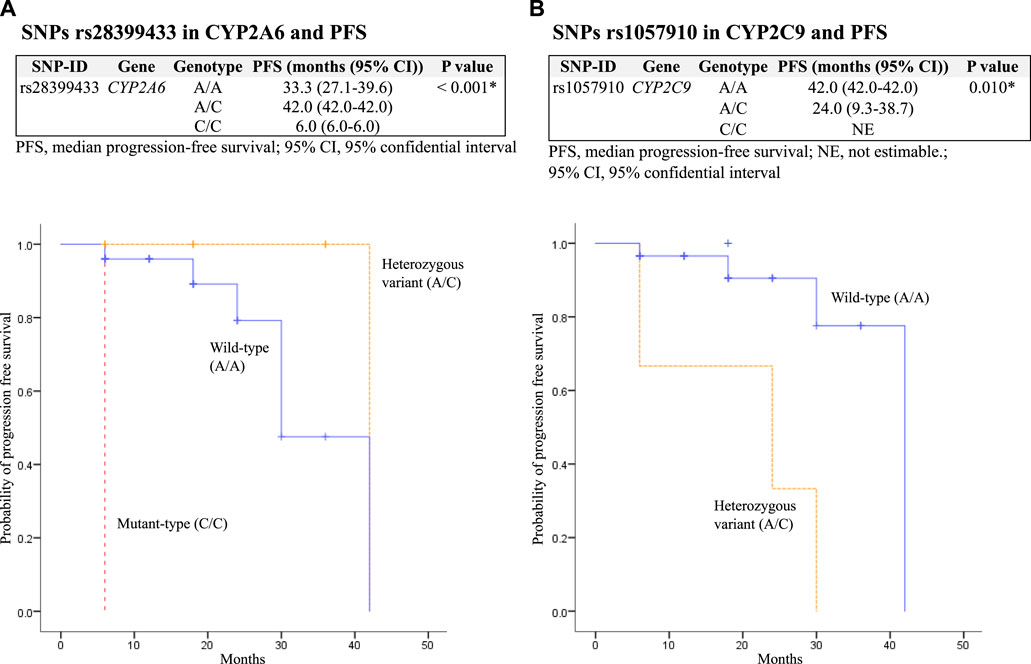
FIGURE 2. Kaplan–Meier estimates and log-rank tests for median progression-free survival (PFS) associated with SNPs among patients who received osimertinib as second-line therapy. (A) association between SNPs rs28399433 in CYP2A6 and PFS; and (B) association between SNPs rs1057910 in CYP2C9 and PFS.
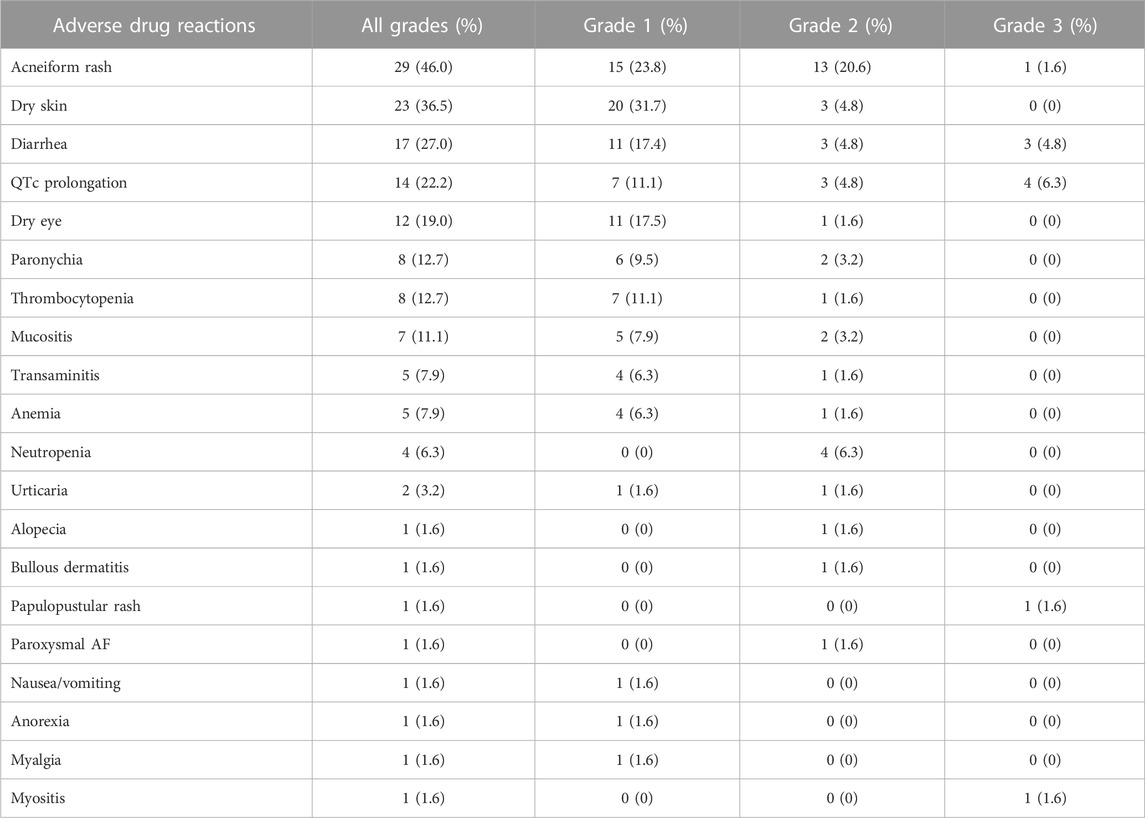
3.5 Incidence of ADRs
The incidence of ADRs is shown in Table 7. Six patients (9.5%) required a dose hold, and seventeen patients (27.0%) required a dose reduction from the standard prescription owing to ADRs, which included diarrhea in four patients (6.3%), acneiform rash in three patients (4.8%), neutropenia in three patients (4.8%), thrombocytopenia in one patient (1.6%), bullous dermatitis in one patient (1.6%), myositis in one patient (1.6%), transaminitis in one patient (1.6%), QTc prolongation in one patient (1.6%), mucositis in one patient (1.6%), and alopecia in one patient (1.6%). Dose reduction included reducing the dose to 80 mg every other day in 10 patients (15.9%), 80 mg three times a week in 5 patients (7.9%), 80 mg five times a week in 1 patient (1.6%), and 40 mg once daily in 1 patient (1.6%).
4 Discussion
A recent population pharmacokinetic study showed a linear correlation between exposure to osimertinib, measured using the AUCs of the parent compound and two active metabolites (AZ5104 and AZ7550), and the incidence of ADRs (Brown et al., 2017). AZ5104, in particular, demonstrated an 8-fold greater potency against EGFR mutations (Han et al., 2021). While AZ7550 and AZ5104 are present in approximately 10% of the parent compound (Brown et al., 2017), a decrease in AZ5104 AUCs of 10%–23% in Asian patients compared with that of Caucasian patients may influence clinical outcomes. In addition, a study in Asian populations reported a significant association between SNPs rs1128503 in ABCB1 and rs2231137 in ABCG2 and osimertinib-induced grade ≥2 adverse events (Ishikawa et al., 2023). However, the association between SNPs and the efficacy of osimertinib remains unclear, and the specific SNPs that influence the pharmacokinetics and clinical effects of osimertinib in NSCLC remain unknown.
This is the first study to analyze a large number of genetic polymorphisms in candidate genes involved in the osimertinib pharmacokinetic pathway to enable the assessment of both efficacy and safety endpoints. All allele frequencies were consistent with the Hardy–Weinberg equilibrium (PharmGKB, 2021). In the present study, the frequency of dose reduction (27.0%) was higher than that reported in the AURA 3 trial (16.5%), and the incidence of ADRs was higher than that in the FLAURA, AURA2, and AURA3 trials (Goss et al., 2016; Mok et al., 2017; Soria et al., 2018). This may be because of genetic differences between patient populations; Asian patients who were administered osimertinib in these studies accounted for only 62%, 63%, and 65% of the patients, respectively. The frequency of the ABCG2 rs2231164 (C) allele, known as a loss-of-function variant, was 23.72% in the South Asian population and only 12.12% in the European population (Whirl-Carrillo et al., 2021). Furthermore, the frequency of the CYP2A6*4 allele, a slower metabolizer, also differed significantly between ethnic groups (Pang et al., 2015), potentially leading to higher plasma concentrations of osimertinib in the Thai population. These genetic differences may have contributed to the observed differences in dose reductions and ADRs between our study and previous clinical trials. Additionally, we found that the ORR in our study was 49.2%, which was lower than that reported in the AURA 3 trial (71%) (Mok et al., 2017). This difference in response rates may be because of the differences in patient populations between the two studies. In the AURA 3 trial, patients received osimertinib as second-line therapy and 96% of cases and later-line therapy in 4%. In contrast, in our study, 57.1% of patients received osimertinib as second-line therapy, and 27.0% received osimertinib as later-line therapy.
In our study, we identified six SNPs that were significantly associated with the incidence of ADRs, namely, rs2231142 in ABCG2 mutant-type (A/A), rs2622604 in ABCG2 mutant-type (T/T), CYP2A6 heterozygous variant (non*4/*4), rs1057910 in CYP2C9 mutant-type (C/C), rs28371759 in CYP3A4 heterozygous variant (A/G) and rs762551 in CYP1A2 wild-type (C/C). These findings are consistent with previous studies reporting significant associations between genetic polymorphisms and ADR risk. For example, rs2231142 in the ABCG2 mutant-type (A/A) is significantly associated with sunitinib-induced severe thrombocytopenia (Low et al., 2016), whereas rs2622604 in the ABCG2 mutant-type (T/T) is significantly associated with irinotecan-induced severe myelosuppression (Cha et al., 2009). Similarly, the CYP2A6 (*4) allele is significantly associated with letrozole-induced ADRs (Desta et al., 2011), whereas the CYP3A4 *18 (G) allele is significantly associated with tacrolimus-induced ADRs (Bruckmueller et al., 2015). Notably, in contrast to other genes, rs762551 in the CYP1A2 wild-type (C/C) was significantly related to osimertinib-induced ADRs. This result may be attributable to the higher enzyme activity observed in the presence of an inducer, such as smoking or heavy coffee consumption, which leads to higher enzyme activity in the CYP1A2 mutant-type (A/A) (Djordjevic et al., 2010; Wang et al., 2012), and a lower incidence of ADRs was observed in this variant. With regard to efficacy outcomes, we identified two SNPs (rs2069514 in CYP1A2 and rs1057910 in CYP2C9) that were significantly associated with the median TTF and two SNPs (rs28399433 in CYP2A6 and rs1057910 in CYP2C9) that were significantly associated with the median PFS. Notably, one of these SNPs (rs1057910 in CYP2C9) was significantly associated with ADRs, TTF, and PFS.
The SNPs in CYP450 and the drug efflux transporters discussed above were significantly associated with ADRs, TTF, and PFS. Because osimertinib is a substrate of CYP450, ABCB1, and ABCG2 (AstraZeneca, 2021), polymorphisms in these genes may affect the distribution and pharmacokinetics of this drug. Therefore, we hypothesized that mutations causing the decreased function of CYP450, ABCB1, and ABCG2 may affect the tissue distribution and accumulation of osimertinib. An in vivo study suggested that ABCB1 and ABCG2 are involved in the tissue accumulation of other TKIs (Al-Shammari et al., 2019). As the active metabolite of osimertinib (AZ5104) is more potent than the parent compound against EGFR mutations (Remon et al., 2018) and accounts for approximately 10% of the parent compound (Brown et al., 2017), the accumulation of osimertinib may lead to a significantly increased incidence of ADRs but significantly decreased TTF and PFS. For example, SNP rs1057910 (C), located in the CYP2C9 gene, typically encodes the amino acid leucine at position 359, and the resulting allele is also known as CYP2C9*3, which is a decreased function variant (VCV000008408.12 - ClinVar - NCBI, 2013). This variant may lead to poor metabolism of osimertinib in patients carrying the CYP2C9*3 allele, which increases the risk of osimertinib-induced ADRs due to higher osimertinib exposure but also decreases survival outcomes due to lower exposure to its active metabolite (AZ5104).
These findings are similar to those of a previous study on the osimertinib exposure-response relationship, which found that the mortality rate was significantly higher in the high osimertinib drug level group (Rodier et al., 2022). Additionally, the probability of developing rashes, diarrhea, or QTc prolongation increased with exposure, and a linear relationship between adverse event development and osimertinib levels was identified (Brown et al., 2017). The mechanism that supports EGFR-TKI-induced ADRs is the inhibition of EGFR1 and EGFR2 (HER2) signaling, leading to a reduction in growth and impaired healing of the epithelium where EGFR is expressed. This subsequently causes alterations in keratinocyte proliferation and differentiation, reduced growth, impaired healing of the intestinal epithelium, and alterations in myocyte growth (Giovannini et al., 2009; Hirsh et al., 2014; Ikebe et al., 2020).
Our study has several limitations. First, the sample size was small, and some clinical data were retrospectively collected from medical records. Nevertheless, no genotype distribution deviated from the Hardy–Weinberg equation (PharmGKB, 2021), and the clinical data were confirmed by the patient’s physician. Second, our inability to control for other confounding factors, such as compliance with osimertinib treatment caused by home oral medication but we had oncology pharmacist pill count measures of compliance at every visit, and all patients maintained a 100% compliance rate. Additionally, there were no observed drug interactions that influenced osimertinib drug levels. However, we did not restrict patient coffee consumption, which may have an impact on higher enzyme activity in the SNP rs762551 in CYP1A2 mutant-type (A/A). This enzyme was inducible by heavy coffee consumption and has been associated with a lower incidence of ADRs (Djordjevic et al., 2010; Wang et al., 2012). Finally, a pharmacokinetic analysis was not included in this study. However, the association between SNPs and clinical outcomes was consistent with that in previous reports.
In conclusion, our study identified significant SNPs associated with increased ADRs incidence, decreased TTF, and decreased PFS in Thai patients with NSCLC treated with osimertinib. The findings can potentially guide treatment decisions and help optimize individualized therapy for patients with NSCLC harboring EGFR mutations. However, more extensive studies with analysis of osimertinib and its active metabolite drug levels must be conducted to confirm these findings.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by The Ethics Committee of Ramathibodi (ethics approval code: COA. MURA 2022/370). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
Conception and design: TM, JM, TR, CS and CA; Sample process: TR, CS, JM, NT, TM, TT and PC; Analysis: TR, CS, TM, NT, JM, TT RS and CA; Interpretation of data: TR, CS, JM, TM, TT, NT and PC; Conclusion and suggestion: TR, CS and JM; Drafting and Editing manuscript: TM, JM, TR and CS. Supervising project: TM, JM, TR and CS. TM was the first author, TR was the essentially intellectual contributor, and JM was the corresponding author. All authors contributed to the article and approved the submitted version.
Funding
The Doctor Kasem Pangsrivongse Foundation (Thailand) provided a research grant (02-002-65) to fund this study, which was established to support professional development in public healthcare. The authors were solely responsible for the preparation of the study protocol, data analysis, and writing and submission of the manuscript, ensuring the independence and objectivity of the study’s results.
Acknowledgments
The success of this study required the assistance and contributions of many individuals. We would like to express our sincere gratitude to all the patients who agreed to participate in this study and the staff who provided invaluable support throughout the research process.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2023.1222435/full#supplementary-material
Abbreviations
ADR, Adverse drug reactions; BCRP, Breast cancer resistance protein; EGFR, Epidermal growth factor receptor; NSCLC, Non-small cell lung cancer; ORR, Objective response rate; PFS, Progression-free survival; SNP, Single nucleotide polymorphisms; TKI, Tyrosine kinase inhibitors; TTF, Time to treatment failure.
References
Al-Shammari, A. H., Masuo, Y., Fujita, K. I., Yoshikawa, Y., Nakamichi, N., Kubota, Y., et al. (2019). Influx and efflux transporters contribute to the increased dermal exposure to active metabolite of regorafenib after repeated oral administration in mice. J. Pharm. Sci. 108 (6), 2173–2179. doi:10.1016/j.xphs.2019.01.018
AstraZeneca (2021). TAGRISSO (osimertinib) [Product monograph]. Mississauga, Ontario: AstraZeneca Canada Inc. Available from: https://www.astrazeneca.ca/content/dam/az-ca/downloads/productinformation/tagrisso-product-monograph-en.pdf.
Brown, K., Comisar, C., Witjes, H., Maringwa, J., de Greef, R., Vishwanathan, K., et al. (2017). Population pharmacokinetics and exposure-response of osimertinib in patients with non-small cell lung cancer. Br. J. Clin. Pharmacol. 83 (6), 1216–1226. doi:10.1111/bcp.13223
Bruckmueller, H., Werk, A. N., Renders, L., Feldkamp, T., Tepel, M., Borst, C., et al. (2015). Which genetic determinants should be considered for tacrolimus dose optimization in kidney transplantation? A combined analysis of genes affecting the CYP3A locus. Ther. Drug Monit. 37 (3), 288–295. doi:10.1097/ftd.0000000000000142
Cha, P. C., Mushiroda, T., Zembutsu, H., Harada, H., Shinoda, N., Kawamoto, S., et al. (2009). Single nucleotide polymorphism in ABCG2 is associated with irinotecan-induced severe myelosuppression. J. Hum. Genet. 54 (10), 572–580. doi:10.1038/jhg.2009.80
Cho, B. C., Chewaskulyong, B., Lee, K. H., Dechaphunkul, A., Sriuranpong, V., Imamura, F., et al. (2019). Osimertinib versus standard of care EGFR TKI as first-line treatment in patients with EGFRm advanced NSCLC: FLAURA asian subset. J. Thorac. Oncol. 14 (1), 99–106. doi:10.1016/j.jtho.2018.09.004
CV000008408.12 - ClinVar - NCBI (2013). VCV000008408.12 - ClinVar - NCBI. Available from: https://www.ncbi.nlm.nih.gov/clinvar/variation/8408/.
Desta, Z., Kreutz, Y., Nguyen, A. T., Li, L., Skaar, T., Kamdem, L. K., et al. (2011). Plasma letrozole concentrations in postmenopausal women with breast cancer are associated with CYP2A6 genetic variants, body mass index, and age. Clin. Pharmacol. Ther. 90 (5), 693–700. doi:10.1038/clpt.2011.174
Detarkom, S., Incharoen, P., Jinawat, A., Trachu, N., Kamprerasart, K., Prasongsook, N., et al. (2018). P3.09-08 tumor heterogeneity and molecular profile of NSCLC in Thai population. J. Thorac. Oncol. 13 (10), S949–S950. doi:10.1016/j.jtho.2018.08.1777
Dickinson, P. A., Cantarini, M. V., Collier, J., Frewer, P., Martin, S., Pickup, K., et al. (2016). Metabolic disposition of osimertinib in rats, dogs, and humans: insights into a drug designed to bind covalently to a cysteine residue of epidermal growth factor receptor. Drug Metabolism Dispos. 44 (8), 1201–1212. doi:10.1124/dmd.115.069203
Djordjevic, N., Ghotbi, R., Jankovic, S., and Aklillu, E. (2010). Induction of CYP1A2 by heavy coffee consumption is associated with the CYP1A2 −163C>A polymorphism. Eur. J. Clin. Pharmacol. 66 (7), 697–703. doi:10.1007/s00228-010-0823-4
Giovannini, M., Gregorc, V., Belli, C., Roca, E., Lazzari, C., Viganò, M. G., et al. (2009). Clinical significance of skin toxicity due to EGFR-targeted therapies. J. Oncol. 2009, 849051–849058. doi:10.1155/2009/849051
Goss, G., Tsai, C. M., Shepherd, F. A., Bazhenova, L., Lee, J. S., Chang, G. C., et al. (2016). Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol. 17 (12), 1643–1652. doi:10.1016/s1470-2045(16)30508-3
Han, L., Zhang, X., Wang, Z., Zhang, X., Zhao, L., Fu, W., et al. (2021). SH-1028, an irreversible third-generation EGFR TKI, overcomes t790m-mediated resistance in non-small cell lung cancer. Front. Pharmacol. 12, 665253. doi:10.3389/fphar.2021.665253
Hirsh, V., Blais, N., Burkes, R., Verma, S., and Croitoru, K. (2014). Management of diarrhea induced by epidermal growth factor receptor tyrosine kinase inhibitors. Curr. Oncol. 21 (6), 329–336. doi:10.3747/co.21.2241
Ikebe, S., Amiya, R., Minami, S., Ihara, S., Higuchi, Y., and Komuta, K. (2020). Osimertinib-induced cardiac failure with QT prolongation and torsade de pointes in a patient with advanced pulmonary adenocarcinoma. Int. Cancer Conf. J. 10 (1), 68–71. doi:10.1007/s13691-020-00450-2
Ishikawa, E., Yokoyama, Y., Chishima, H., Kasai, H., Kuniyoshi, O., Kimura, M., et al. (2023). Population pharmacokinetics, Pharmacogenomics, and adverse events of osimertinib and its two active metabolites, AZ5104 and AZ7550, in Japanese patients with advanced non-small cell lung cancer: a prospective observational study. Investig. New Drugs 41 (1), 122–133. doi:10.1007/s10637-023-01328-9
Li, Y., Mao, T., Wang, J., Zheng, H., Hu, Z., Cao, P., et al. (2023). Toward the next generation EGFR inhibitors: an overview of osimertinib resistance mediated by EGFR mutations in non-small cell lung cancer. Cell Commun. Signal. 21 (1), 71. doi:10.1186/s12964-023-01082-8
Liao, D., Liu, Z., Zhang, Y., Liu, N., Yao, D., Cao, L., et al. (2020). Polymorphisms of drug-metabolizing enzymes and transporters contribute to the individual variations of erlotinib steady state trough concentration, treatment outcomes, and adverse reactions in epidermal growth factor receptor–mutated non-small cell lung cancer patients. Front. Pharmacol. 11, 664. doi:10.3389/fphar.2020.00664
Low, S. K., Fukunaga, K., Takahashi, A., Matsuda, K., Hongo, F., Nakanishi, H., et al. (2016). Association study of a functional variant on ABCG2 gene with sunitinib-induced severe adverse drug reaction. PLOS ONE 11 (2), e0148177. doi:10.1371/journal.pone.0148177
Mok, T. S., Wu, Y. L., Ahn, M. J., Garassino, M. C., Kim, H. R., Ramalingam, S. S., et al. (2017). Osimertinib or platinum–pemetrexed in EGFR t790m–positive lung cancer. N. Engl. J. Med. 376 (7), 629–640. doi:10.1056/nejmoa1612674
Ngamjarus, C. (2016). n4Studies: sample size calculation for an epidemiological study on a smart device | siriraj medical journal. n4Studies: sample size calculation for an epidemiological study on a smart device | siriraj medical journal. Available from: https://he02.tci-thaijo.org/index.php/sirirajmedj/article/view/58342.
Nicholson, A. G., Tsao, M. S., Beasley, M. B., Borczuk, A. C., Brambilla, E., Cooper, W. A., et al. (2022). The 2021 WHO classification of lung tumors: impact of advances since 2015. J. Thorac. Oncol. 17 (3), 362–387. doi:10.1016/j.jtho.2021.11.003
Pang, C., Liu, J. H., Xu, Y. S., Chen, C., and Dai, P. G. (2015). The allele frequency of CYP2A6*4 in four ethnic groups of China. Exp. Mol. Pathology 98 (3), 546–548. doi:10.1016/j.yexmp.2015.03.040
PharmGKB (2021). PharmGKB. Available from: https://www.pharmgkb.org/.
Ramalingam, S. S., Vansteenkiste, J., Planchard, D., Cho, B. C., Gray, J. E., Ohe, Y., et al. (2020). Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N. Engl. J. Med. 382 (1), 41–50. doi:10.1056/nejmoa1913662
Remon, J., Steuer, C., Ramalingam, S., and Felip, E. (2018). Osimertinib and other third-generation EGFR TKI in EGFR-mutant NSCLC patients. Ann. Oncol. 29, i20–i27. doi:10.1093/annonc/mdx704
Rodier, T., Puszkiel, A., Cardoso, E., Balakirouchenane, D., Narjoz, C., Arrondeau, J., et al. (2022). Exposure–response analysis of osimertinib in patients with advanced non-small-cell lung cancer. Pharmaceutics 14 (9), 1844. doi:10.3390/pharmaceutics14091844
Sogawa, R., Nakashima, C., Nakamura, T., Takeuchi, K., Kimura, S., Komiya, K., et al. (2020). Association of genetic polymorphisms with afatinib-induced diarrhoea. Vivo 34 (3), 1415–1419. doi:10.21873/invivo.11922
Soria, J. C., Ohe, Y., Vansteenkiste, J., Reungwetwattana, T., Chewaskulyong, B., Lee, K. H., et al. (2018). Osimertinib in UntreatedEGFR-mutated advanced non–small-cell lung cancer. N. Engl. J. Med. 378 (2), 113–125. doi:10.1056/nejmoa1713137
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Suzumura, T., Kimura, T., Kudoh, S., Umekawa, K., Nagata, M., Matsuura, K., et al. (2012). Reduced CYP2D6 function is associated with gefitinib-induced rash in patients with non-small cell lung cancer. BMC Cancer 12 (1), 568. doi:10.1186/1471-2407-12-568
Wang, L., Hu, Z., Deng, X., Wang, Y., Zhang, Z., and Cheng, Z. N. (2012). Association between CommonCYP1A2Polymorphisms and theophylline metabolism in non-smoking healthy volunteers. Basic & Clin. Pharmacol. Toxicol. 112 (4), 257–263. doi:10.1111/bcpt.12038
Westover, D., Zugazagoitia, J., Cho, B., Lovly, C., and Paz-Ares, L. (2018). Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann. Oncol. 29, i10–i19. doi:10.1093/annonc/mdx703
Whirl-Carrillo, M., Huddart, R., Gong, L., Sangkuhl, K., Thorn, C. F., Whaley, R., et al. (2021). An evidence-based framework for evaluating Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 110 (3), 563–572. doi:10.1002/cpt.2350
Keywords: non-small cell lung cancer, pharmacogenetics, single nucleotide polymorphisms (SNPs), osimertinib, drug-metabolizing enzymes, transporters
Citation: Majam T, Sukasem C, Reungwetwattana T, Chansriwong P, Atasilp C, Trachu N, Thamrongjirapat T, Sukprasong R and Meanwatthana J (2023) CYP450 and drug efflux transporters polymorphism influence clinical outcomes of Thai osimertinib-treated non-small cell lung cancer patients. Front. Pharmacol. 14:1222435. doi: 10.3389/fphar.2023.1222435
Received: 14 May 2023; Accepted: 23 October 2023;
Published: 06 November 2023.
Edited by:
Yen-Chen Anne Feng, National Taiwan University, TaiwanReviewed by:
Lucia Guadalupe Taja Chayeb, National Institute of Cancerology (INCAN), MexicoPrem Prakash Kushwaha, Case Western Reserve University, United States
Cyprian Onyeji, University of Nigeria, Nigeria
Copyright © 2023 Majam, Sukasem, Reungwetwattana, Chansriwong, Atasilp, Trachu, Thamrongjirapat, Sukprasong and Meanwatthana. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennis Meanwatthana, amVubmlzLm1lYUBtYWhpZG9sLmFjLnRo
 Teerapat Majam
Teerapat Majam Chonlaphat Sukasem
Chonlaphat Sukasem Thanyanan Reungwetwattana
Thanyanan Reungwetwattana Phichai Chansriwong7
Phichai Chansriwong7 Rattanaporn Sukprasong
Rattanaporn Sukprasong Jennis Meanwatthana
Jennis Meanwatthana