- Cancer Center, The First Hospital of Jilin University, Changchun, China
Prostate cancer (PCa), bladder cancer (BC), and renal cell cancer (RCC) are the most common urologic tumours in males. N6-methyladenosine (m6A), adenosine N6 methylation, is the most prevalent RNA modification in mammals. Increasing evidence suggests that m6A plays a crucial role in cancer development. In this review, we comprehensively analyzed the influence of m6A methylation on Prostate cancer, bladder cancer, and renal cell cancer and the relationship between the expression of relevant regulatory factors and their development and occurrence, which provides new insights and approaches for the early clinical diagnosis and targeted therapy of urologic malignancies.
1 Introduction
Urological tumours, primarily including PCa, BC and RCC, are a global threat to human health. Incidence and mortality rates are increasing globally (Sung et al., 2021; Siegel et al., 2022). However, most urological tumour patients are diagnosed at advanced stages owing to the absence of distinct symptoms and signs. Although the risk factors for urological tumours are well-defined, however, the underlying molecular mechanisms remain unclear. Therefore, it is vital to understand urological tumour pathogenesis. These might provide novel targets for the early diagnosis and clinical treatment of urological tumours, which may present an objective basis for clinical decision-making (Wang et al., 2020a; Su et al., 2020; Yankova et al., 2021).
Among more than 100 chemical modifications, m6A has been identified as one of the most prevalent chemical forms in eukaryotic RNA (Fu et al., 2014; Lewis et al., 2017). It plays a significant role in various biological functions, including RNA translation, stability (degradation and stabilization), splicing, and nucleus export. This reversible m6A RNA modification is coordinated by three groups of proteins: methyltransferases (“writers”), demethylases (“erasers”), and m6A-binding proteins (“readers”) (Gilbert et al., 2016; Deng et al., 2018; Dierks et al., 2021). Recently, various studies demonstrated that aberrantly expressed m6A modifications are associated with numerous human tumours progression, including tumourigenesis, proliferation, and drug resistance (He et al., 2019; Huang et al., 2020a).
In brief, studies on the interaction between m6A and tumours havegradually entered the public view. Numerous studies have established a link between m6A variables and the development of urological cancers. However, understanding the association between m6A modifications and urological cancers remains unclear. Therefore, a systematic review of the molecular mechanisms underlying m6A regulation in PCa, BC, and RCC progression is strongly recommended.
This review outlines recent findings on m6A modification, relationships, biological activities, and mechanisms of its related proteins in the malignant progression of these three urologic tumours.
2 m6A modification
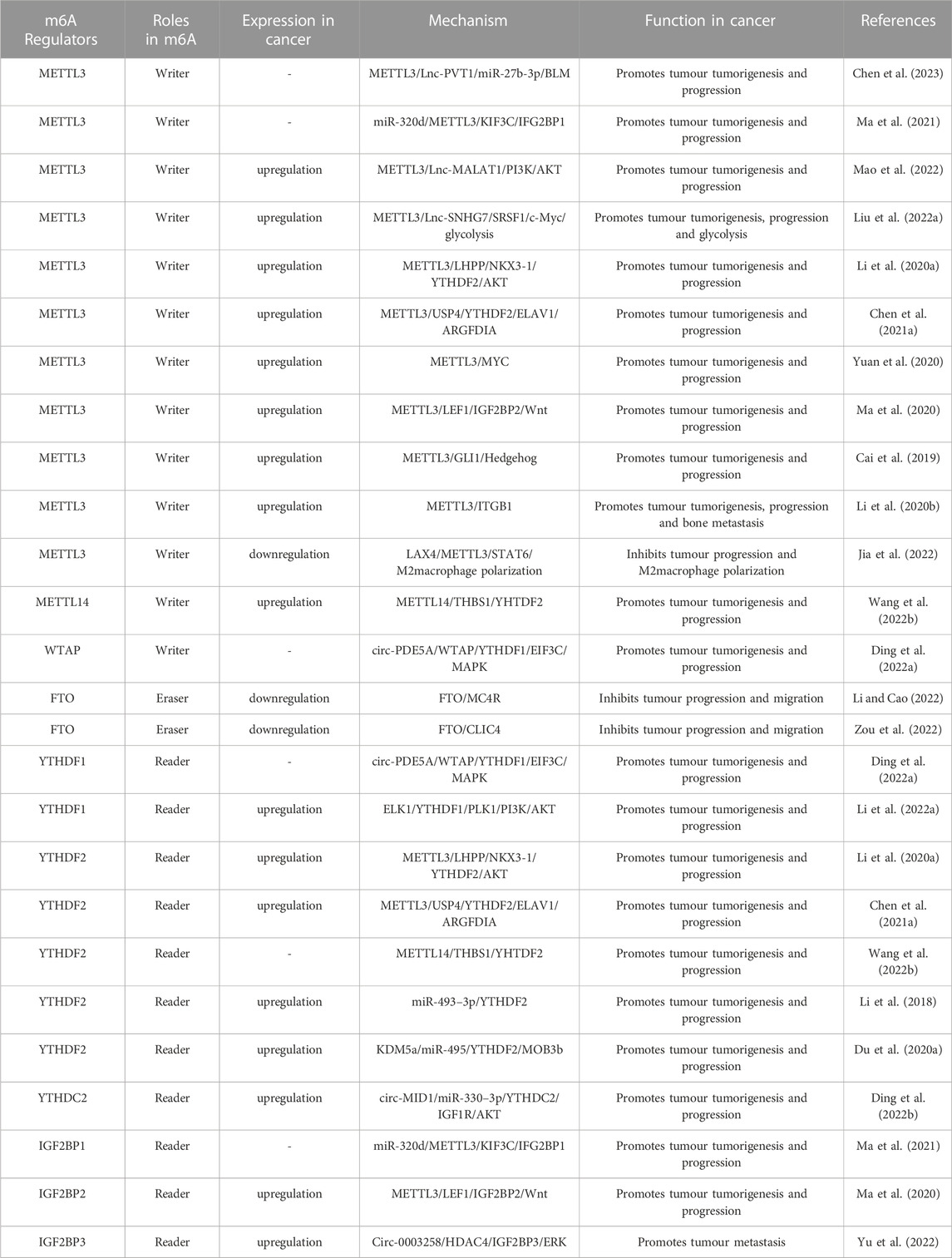
In the process of m6A modification, it, mainly occurs in adenines in the consensus motif RRACH(H = A,C or U,R = G or A) with enrichment more prone to be detected in the 3′-untranslated regions (Bodi et al., 2010; Meyer et al., 2012). The deposition, recognition and removal of m6A are carried out respectively by regulators “writers”, “readers” and “erasers”. First, the deposition of m6A in mRNA is installed and catalysed by a complex consisting of METTL3, METTL14 and WTAP (“writers”). Next, the m6A-binding proteins (“readers”), all of which can selectively recognize and bind to methylated mRNA, controlling gene expression via regulating diverse processes, including mRNA maturation, export, translation, and decay, it depends on different kinds of “readers”. Beyond this, the methylation of m6A can be removed passively from the transcriptome through degradation of modified RNA by “readers” or actively through FTO and ALKBH5 demethylases. A diagram of the methylation processes involved in m6A and their biological functions are displayed in Figure 1. This presents “writers” methylating RNA at the amino group, which is located at position N6 of the adenine nucleobase. As soon as m6A is deposited on RNA, the recruitment of m6A recognition proteins influences the fate of the RNA, including as translation effectiveness, stability (degradation and stabilization) and splicing, which ultimately affect gene expression. Additionally, “erasers” demethylate m6A to remove it from RNA.
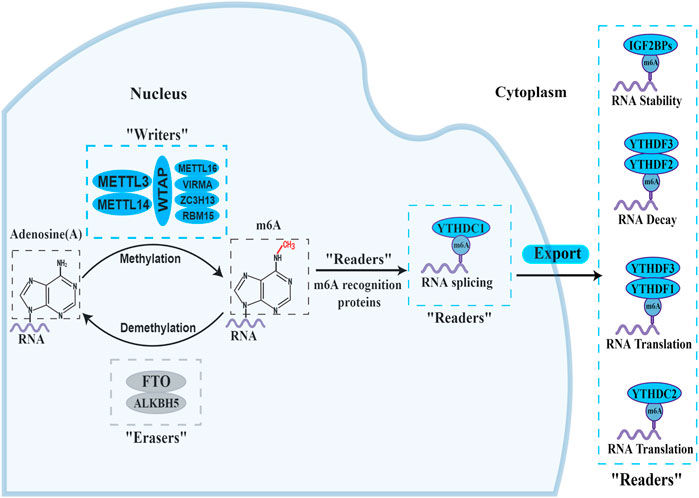
FIGURE 1. The biology functions of m6A regulators. The three kinds of m6A enzymes are as follows: “writers” (METTL3, METTL14, WTAP, METTL16, VIRMA, ZC3H13 and RBM15), “erasers” (FTO and ALKBH5), and “readers” (YTHDF1/2/3, YTHDC1/2 and IGF2BP1/2/3). They regulate the deposition of m6A modification and affect the translation, stability (degradation and stabilization) and splicing of RNA.
At present, “writers” have been extensively studied most. The m6A methylase complex comprises a range of methyltransferases. Methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), and wilm’s tumour-associated protein (WTAP) are the core components of complex (Bokar et al., 1997; Ping et al., 2014; Schwartz et al., 2014). The METTL3 and METTL14 form a heterodimer (Wang et al., 2017). With the help of WTAP, the METTL3-METTL14 heterodimer catalyzes the covalent transfer of a methyl group to adenine. In addition, several other multicomponent methyltransferase complexes have recently been identified and characterized. Such as Methyltransferase-like 16 (METTL16), vir-like m6A methyltransferase-associated protein (VIRMA), zinc finger CCCH domain-containing protein 13 (ZC3H13), and RNA binding pattern protein 15 (RBM15) (Melstrom and Chen, 2020; Oerum et al., 2021).
“Erasers", or m6A demethylases, can reverse the dynamic m6A methylation in the nucleus, including α-ketoglutarate-dependent dioxygenase homolog 5 (ALKBH5), fat mass and obesity-related protein (FTO) (Zheng et al., 2013; Zhao et al., 2014). They all belong to the α-ketoglutarate-dependent dioxygenase family. The discovery of m6A demethylases has confirmed that m6A modification is dynamically reversible.
“Readers”, or m6A recognition proteins, could recognize and bind to m6A modification sites and participate in diverse stages of RNA metabolism. Different readers have different biological functions. The most well-known m6A readers are the insulin-like growth factor 2 mRNA binding protein (IGF2BP) and the YT521-B homology (YTH) domain families. The IGF2BP family includes IGF2BP1, IGF2BP2, and IGF2BP3, which can improve the stability and translation of the target RNAs (Huang et al., 2018). YTHDF1-3 and YTHDC1-2 were the five main YTH domain proteins. YTHDC1, which is localized in the nucleus, regulates splicing and nuclear export, whereas YTHDC2 enhances the translation efficiency of target RNA. YTHDF3 coordinates with YTHDF1 to promote the translation of target RNA, whereas YTHDF3 cooperates with YTHDF2 to induce target RNA degradation (Shi et al., 2017; Einstein et al., 2021). These proteins create an effective m6A regulatory network and play a crucial role in m6A modification.
3 Role of m6A regulators in the progression of PCa
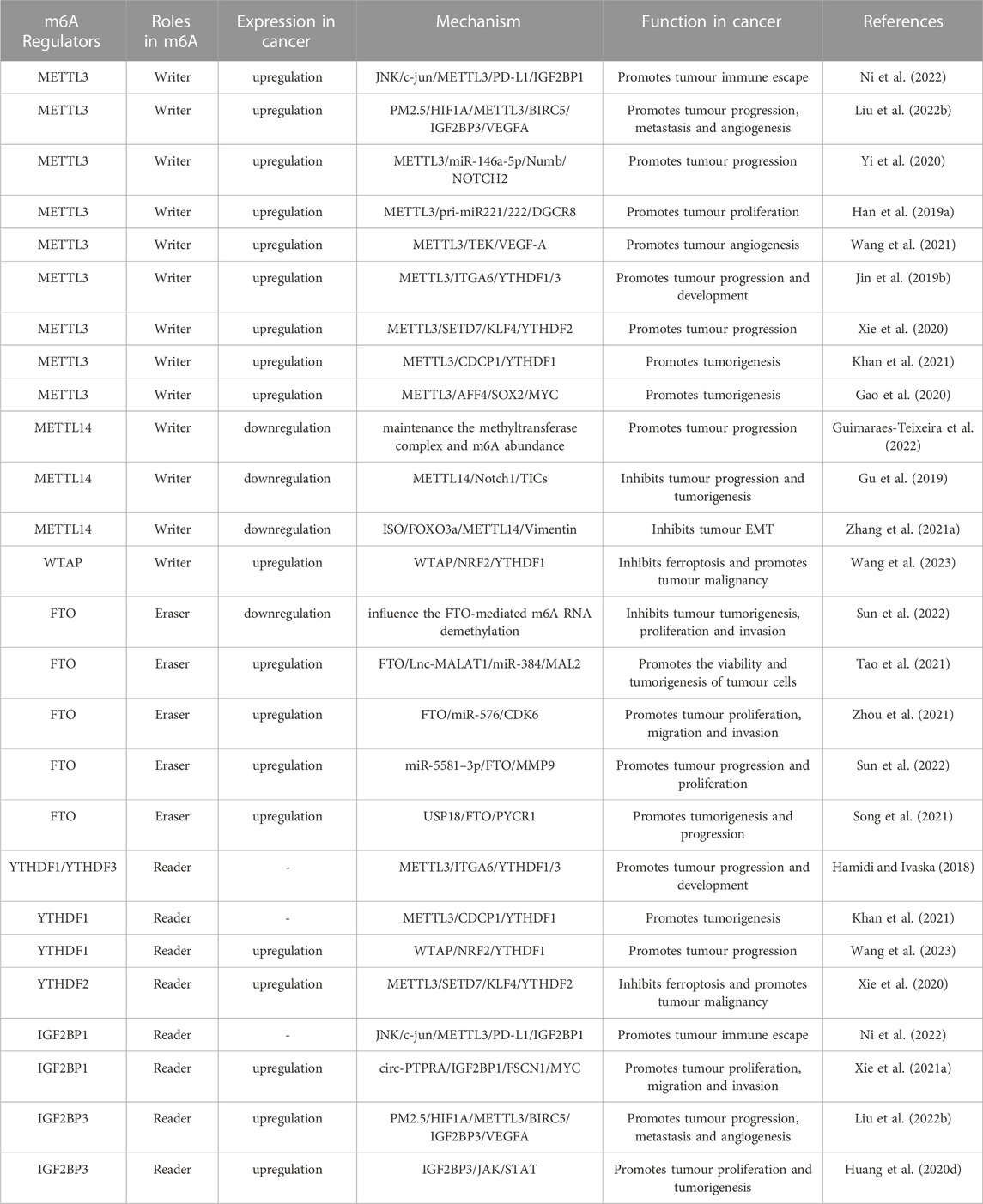
Numerous studies have demonstrated the involvement of m6A regulators in various functions of PCa, such as the metabolism of non-coding RNAs (ncRNAs) and M2 macrophage polarization. The roles and mechanisms of m6A regulators in PCa are summarized in Figure 2 and Table 1.
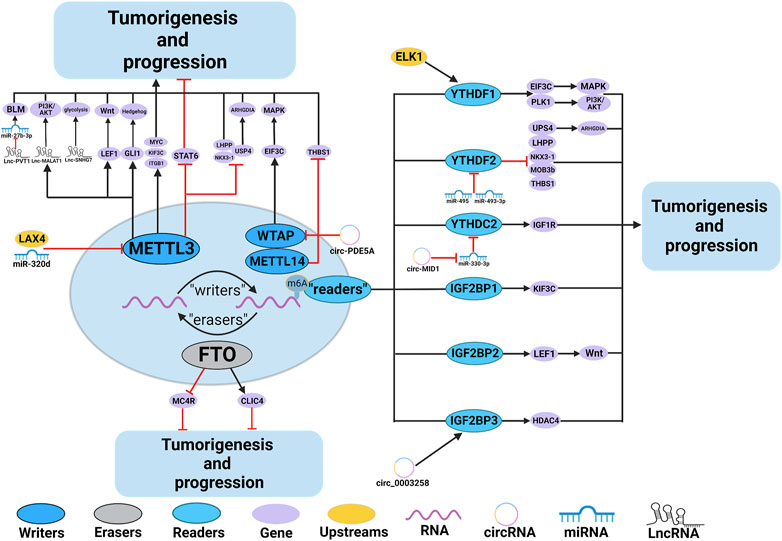
FIGURE 2. In PCa, m6A regulatory proteins contribute to tumorigenesis and progression by interacting with various RNAs. METTL3 and WTAP stimulate the progression of PCa by promoting the expression levels of Lnc-PVT1, Lnc-MALAT1, Lnc-SNHG7, MYC, LEF1, GLI1, ITGB, KIFEC and EIF3C. In addition, miR-320d and circ-PDE5A inhibit the expression of METTL3 and WTAP, leading to PCa suppression. METTL3 and METTL14 stimulate the progression of PCa by inhibiting the expression levels of USP4, NKX3-1, LHPP and THBS1. METTL3 suppresses the progression of PCa by inhibiting the expression levels of STAT6. LXA4 inhibits the expression of METTL3, leading to PCa progression. FTO suppress the progression of PCa by affecting the expression levels of MC4R and CLIC4. YTHDF1, YTHDC2, and IGF2BPs stimulate the progression of PCa by promoting the expression of EIF3C, PLK1, IGF1R, KIF3C, LEF1 and HDAC4. YTHDF2 stimulate the progression of PCa by inhibiting the expression of USP4, LHPP, NKX3-1, MOB3b and THBS1. In addition, ELK1 and circ_0003258 promote the expression of YTHDF1 and IGF2BP3, leading to PCa progression. circ-MID1 could sponge miR-330–3p to promote YTHDC2 expression.
3.1 m6A writers and PCa
The m6A regulatory factors are essential for PCa tumourigenesis and progression. Studies of writers on PCa have primarily focused on the regulatory component METTL3. Recent studies have indicated that METTL3 plays an oncogenic role in PCa (Cai et al., 2019; Li et al., 2020a; Li et al., 2020b; Ma et al., 2020; Yuan et al., 2020; Chen et al., 2021a; Ma et al., 2021; Liu et al., 2022a; Wang et al., 2022a; Mao et al., 2022; Chen et al., 2023). Only one study has suggested that it is a carcinogenic tumour suppressor (Jia et al., 2022).
Recent studies have shown that METTL3 can affect the synthesis and function of various ncRNAs, such as miRNAs and LncRNAs. Moreover, ncRNAs also have an important regulatory effect on METTL3 in PCa progression (Zeng et al., 2020). Chen et al. (Chen et al., 2023) reported that METTL3 could upregulate the Lnc-plasmacytoma variant translocation 1 (PVT1) expression in an m6A-dependent manner. Further investigation showed that PVT1 serve as a ceRNA to isolate miR-27b-3p and promote PCa progression via the METTL3/PVT1/miR-27b-3p/Bloom syndrome protein (BLM) axis. In addition, a study indicated that Kinesin Family Member 3C (KIF3C) was overexpressed in PCa and negatively correlated with patient outcomes (Ma et al., 2021). The m6A writer METTL3 can promote m6A modification of KIF3C, thus enhancing KIF3C mRNA stabilization and expression by IGF2BP1. miR-320d, as a tumour-suppressor factor and upstream target of METTL3, inhibits KIF3C expression by targeting METTL3 and curbing PCa progression. As a lncRNA, the metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is oncogenic in numerous tumours progressions (Fan et al., 2014; Jen et al., 2017). Yuan et al. (Mao et al., 2022) confirmed that METTL3 could upregulate the m6A level of LncRNA-MALAT1, which activates the PI3K/AKT signaling pathway and promotes the growth and invasion of PCa. The molecular mechanisms of m6A methylation modifications in tumour cells have recently been studied. Liu et al. (Liu et al., 2022a) reported upregulation of lncRNA small nucleolar RNA host gene 7 (SNHG7) induced by m6A. METTL3 promoted the m6A modification of SNHG7 and enhanced its stability. Upregulated SNHG7 expression could accelerate glycolysis via the serine/arginine-rich splicing factor 1 (SRSF1)/c-Myc axis and promote the progression of PCa.
Recent studies revealed that m6A modification could affect tumour formation by regulating the m6A modification in the mRNAs of oncogenes, tumour suppressors, and transcription factors (An and Duan, 2022). Li et al.demonstrated that two tumour suppressors, the phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP) and NK3 Homeobox 1 (NKX3-1), are downregulated in PCa (Li et al., 2020a). Further investigation verified that YTHDF2 could recognize the abundance of m6A-modified mRNA of LHPP and NKX3-1 induced by METTL3 and degrade them, thereby promoting AKT phosphorylation and tumour progression. Chen et al. (Chen et al., 2021a) reported that YTHDF2 could recognize and degrade the abundance of m6A-modified Ubiquitin Specific peptidase 4 (USP4) mRNA promoted by METTL3 in the PCa cells. The decreased USP4 mRNA could enhance Rho GDP dissociation inhibitor α (ARHGDIA) expression via the METTL3-USP4-ELAVL1 axis, which finally promotes the progression of PCa cells. In addition, researchers elucidated that MYC(c-myc) was a target of METTL3-mediated m6A modification and its expression was enhanced by METTL3 in PCa (Yuan et al., 2020). This leads to PCa’s oncogenic functions. Several signaling pathways and molecules involved in tumour development have been identified as m6A targets and controlled by m6A modifiers. According to Ma et al. (Ma et al., 2020), a significant upregulation of Lymphoid Enhancer-binding Factor 1 (LEF1) expression was induced by increased METTL3 in PCa patients. Increased METTL3 and LEF1 levels activate the Wnt pathway and promote tumour progression. Similarly, the increased expression of METTL3 in PCa could enhance the expression of Glioma-associated Oncogene Homolog 1 (GLI1) via an m6A catalytic activity-dependent manner (Cai et al., 2019). The GLI1 overexpression activates Hedgehog Pathway and promotes tumour growth.
Tumour microenvironments (TMEs) are a complex microenvironment of cancer, stromal, and immune cells (Binnewies et al., 2018). Recently, much research has been conducted to study the tumour microenvironment recently. However, little is known about the role of m6A in regulating the immune microenvironment of tumours. Macrophages are the most prevalent immune cells in the microenvironment of tumour (Condeelis and Pollard, 2006). Studies have revealed that the total m6A levels were considerably downregulated in macrophages induced by the downregulation of METTL3 in PCa. According to Jia et al. (Jia et al., 2022), prostate cancer cell-derived small lipid molecule lipoxin A4 (LXA4) could inhibit METTL3, and the decreased METTL3 promotes signal transducers and activators of transcription 6 (STAT6) phosphorylation, which in turn induces M2 macrophage polarization and enhances tumour progression. This indicates that METTL3 could also act as a tumour suppressor in PCa. m6A modification also accounts for TME remodeling in tumours, including remodeling in organs with tumour metastasis. Li et al. (Li et al., 2020b) reported that elevated METTL3 levels in PCa upregulated and enhanced the integrin β1 (ITGB1) mRNA’s stability and expression in an m6A-HuR-dependent manner. The overexpressed ITGB1 promotes bone metastasis by improving binding to Collagen I and tumour cell motility in PCa.
Wang et al. (Wang et al., 2022b) revealed that METTL14 is upregulated and oncogenic in PCa. Thrombospondin 1 (THBS1), a tumour suppressor, was identified as the target gene of METTL14. YTHDF2 recognizes methylated THBS1 mRNA induced by METTL14 and inhibits its expression. Circ-PDE5A is found downregulated and acts as a metastasis suppressor in PCa. Ding et al. (Ding et al., 2022a) reported that circ-PDE5A inhibited the Eukaryotic Initiation Factor 3 (EIF3C) expression by binding to WTAP and inhibiting its m6A methylation ability. Moreover, the decreased m6A levels of EIF3C mRNA also inhibited the recognition by YTHDF1, which curbed the MAPK pathway. WATP plays an oncogenic role in PCa.
3.2 m6A erasers and PCa
Studies on m6A erasers in PCa have focused on FTO. As an m6A demethylase, FTO is downregulated in PCa, and its level is negatively correlated with advanced tumour stage (Zhu et al., 2021; Li and Cao, 2022; Zou et al., 2022). Cao et al. (Li and Cao, 2022) reported that FTO downregulates the melanocortin-4 receptor (MC4R) in an m6A-dependent manner to inhibit PCa tumour progression and migration. The reduction of chloride intracellular channel 4 (CLIC4) in cancer cells is common in human tumours and signals the progression and invasion of malignant cells (Suh et al., 2007). However, the mechanism by which CLIC4 expression in PCa is regulated by m6A remains unclear. According to a recent study (Zou et al., 2022), FTO suppressed PCa proliferation and metastasis by reducing the degradation of CLIC4 mRNA in an m6A-dependent manner, which provided new insights into the mechanism of PCa.
3.3 m6A readers and PCa
In PCa, studies on the m6A readers mainly focused on YTHDF1, YTHDF2, YTHDC2, and IGF2BPs. Recent studies have reported that YTHDF1 is upregulated in PCa tissues compared to those normal tissues and is correlated with poor outcomes (Li et al., 2021; Li et al., 2022a).
Polo-like kinase 1 (PLK1) regulates mitotic processes, leading to cellular proliferation. There is a substantial relationship between PLK1 overexpression and certain human cancers (Lee et al., 2015). However, the relationship between m6A regulation and PLK1 in PCa remains unclear. Li et al. (Li et al., 2022a) reported that the transcription factor ELK1 was overexpressed in PCa tissues and enhanced the YTHDF1 transcription. Further investigation showed that PLK1 is an m6A modification downstream target of YTHDF1 and is facilitated by it. This activates the PI3K/AKT signaling and drives the progression of PCa.
Some researchers have identified the tumour-promoting effect of YTHDF2 by investigating its upstream signaling in PCa. Some of these studies have been described in the previous sections (Li et al., 2020a; Wang et al., 2022b). Several studies have shown that miRNA processing and maturation are significantly affected by m6A modifications (Chen et al., 2020a). In lieu, miRNAs can also affect m6A synthesis. miR-493–3p and miR-495, tumour suppressor miRNAs in PCa, have been reported to regulate the expression of the YTHDF2. YTHDF2 is a direct downstream target of miRNAs (Li et al., 2018; Du et al., 2020a). The down-expressed miR-493–3p in PCa could enhance the expression of YTHDF2, thereby promoting the progression of PCa by indirectly regulating m6A levels. (Li et al., 2018). Chen et al. (Du et al., 2020a) showed that miR-495 is inhibited and downregulated by lysine demethylase 5a (KDM5a), an H3K4me3 demethylase overexpressed in PCa. Further investigation proved that YTHDF2 was a downstream target of miR-495, and the upregulated YTHDF2 could inhibit the expression of MOB kinase activator 3 B (MOB3B) by recognizing m6A modification of MOB3B mRNA and inducing mRNA degradation, which consequently promoted tumour tumourigenesis and progression in PCa through an m6A-dependent way. These findings demonstrated the roles of miRNA and YTHDF2 in the m6A modification and development of PCa and described how the m6A-related regulatory factors in PCa were activated, revealing a deeper understanding of the mechanism of PCa.
Recent studies have confirmed that some circRNAs play a crucial role in the progression of PCa (Xiang et al., 2019; Kong et al., 2020), and several studies have assessed the potential molecular mechanism of circRNAs with m6A modifications in PCa. Type 1 insulin-like growth factor receptor (IGF1R), an oncogene, is considered to be involved in the tumour progression by mediating the ATK signaling pathway; however, the specific mechanism of IGF1R in PCa is unclear (Li et al., 2014; Du et al., 2020b). Ding et al. (Ding et al., 2022b) reported that circRNA midline-1 (circ-MID1) was overexpressed and functioned to sponge miR-330–3p in PCa cells. MiR-330–3p plays an anti-tumour role in many cancers (Jin et al., 2019a; Huang et al., 2020b), including PCa (Li et al., 2020c). Further investigations demonstrated that YTHDC2 and IGF1R are the downstream targets of miR-330–3p. Therefore, circ-MID1 may promote the progression of PCa by regulating the miR-330–3p/YTHDC2/IGF1R/AKT axis. Additionally, Yu et al. (Yu et al., 2022) reported that overexpressed circ-0003258 in PCa could bind to IGF2BP3, increase HDAC4 mRNA stability, and activate the ERK signaling pathway, thereby accelerating PCa metastasis.
4 Role of m6A regulators in the progression of BC
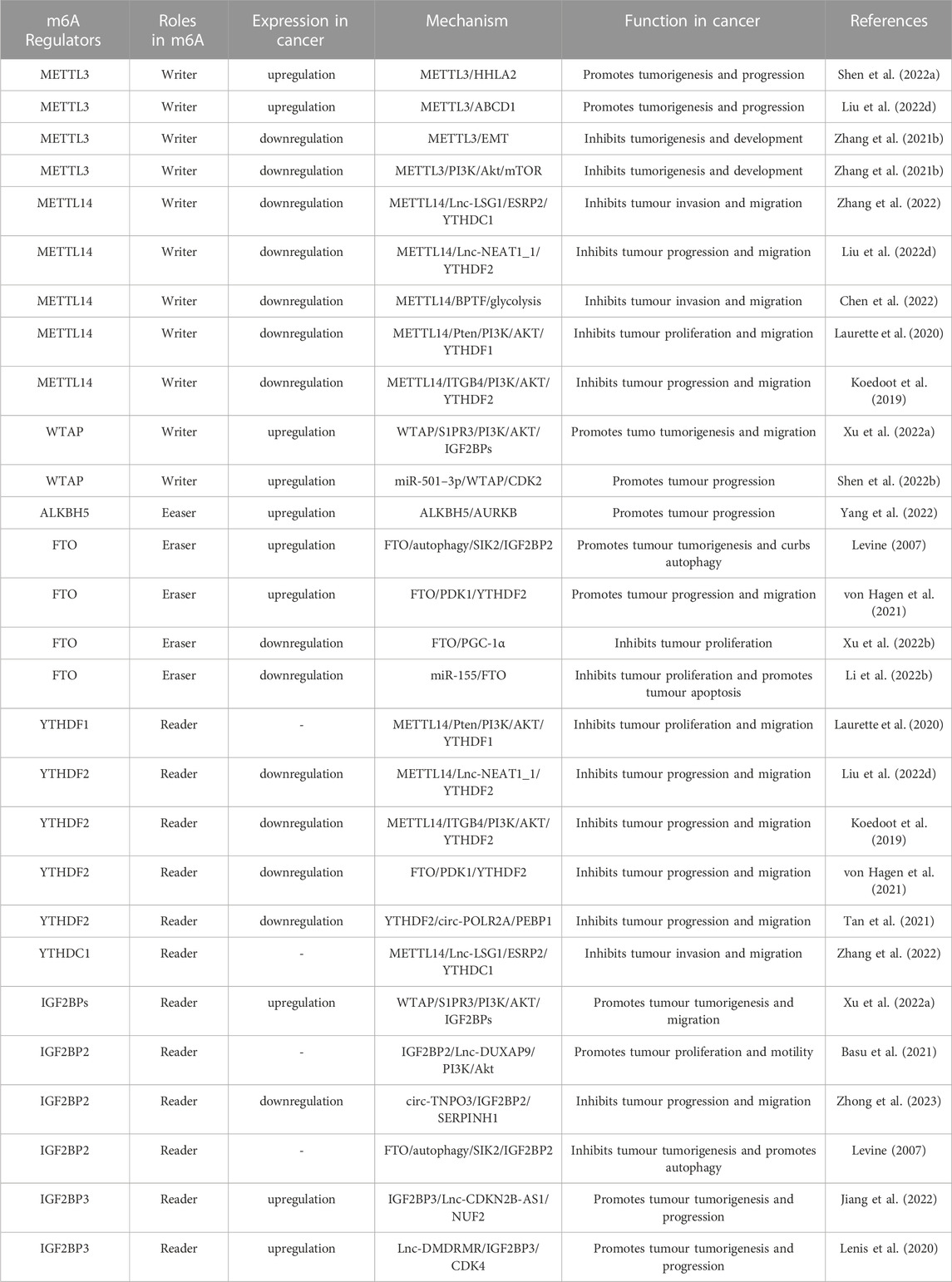
Numerous studies have demonstrated the participation of m6A regulators in BC, such as immune escape and EMT. The roles and mechanisms of m6A regulators in BC are summarized in Figure 3 and Table 2.
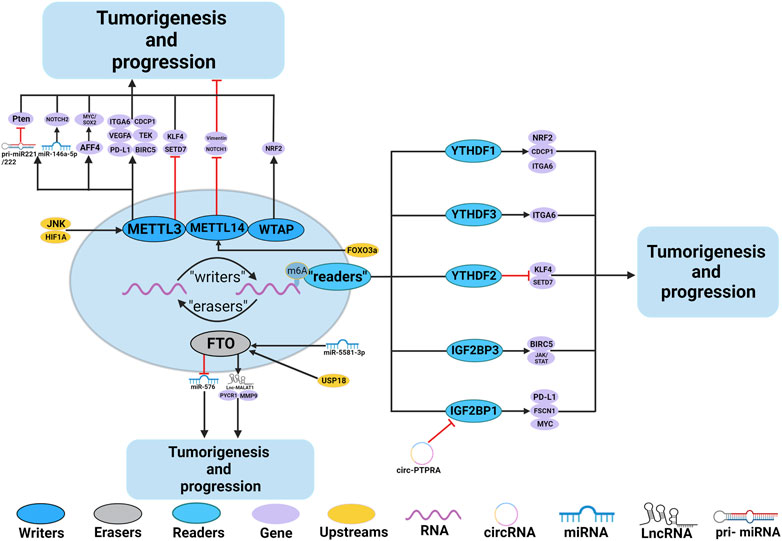
FIGURE 3. In BC, m6A regulatory proteins contribute to tumorigenesis and progression by interacting with various RNAs. METTL3 and WTAP stimulate the progression of BC by promoting the expression levels of PD-L1, BIRC5, miR-146a-5p, pri-miR221/222, TEK, VEGF-A, ITGA6, CDCP1, AFF4 and NRF2. Upstream regulators JNK and HIF1A promote the expression of METTL3, leading to BC progression. METTL3 promote the progression of BC by inhibiting the expression levels of KLF4 and SETD7. METTL14 suppresses the progression of BC by inhibiting the expression levels of Vimentin and Notch1. FTO promote the tumorigenesis and progression of BC by regulating the expression of Lnc-MALAT1, miR-576, MMP9 and PYCR1. In addition, miR-5581–3p inhibits the expression of FTO, leading to BC suppression. USP18 promotes the expression of FTO, leading to BC progression. YTHDF1, YTHDF3, IGF2BP1 and IGF2BP3 stimulate the progression of BC by promoting the expression of NRF2, CDCP1, ITGA6, PD-L1, FSCN1, MYC, BIRC5 and JAK/STAT. In addition, circ-PTPRA inhibits the expression of IGF2BP1, leading to BC suppression. YTHDF2 promote the progression of BC by inhibiting the expression levels of KLF4 and SETD7.
4.1 m6A writers and BC
m6A regulatory factors are essential for BC tumourigenesis and progression. Studies on writers of BC have mostly focused on the regulatory factor METTL3. The results showed that METTL3 was significantly upregulated in bladder cancer cell lines and was identified as an oncogene. Poor prognosis was also associated with high levels of METTL3 in BC. Knockdown of METTL3 in bladder cancer cells rendered it less proliferative, invasive, and viable (Chen et al., 2019a; Han et al., 2019a).
In recent years, the components of TME have been increasingly recognized as effective tumour immunotherapy targets (Bader et al., 2020; Sadeghi Rad et al., 2021). In addition, m6A modification has become increasingly important in anti-tumour immunotherapy (Han et al., 2019b; Wang et al., 2020b). Exploring their association and mechanisms is the premise of individualized precision treatment. Ni et al. (Ni et al., 2022) reported that activated JNK signaling is associated with increased METTL3 expression in BC, and inhibition of JNK signaling reduced the expression of METTL3 and Programmed death ligand 1 (PD-L1). Further studies showed that IGF2BP1 could recognize methylated PD-L1 mRNA induced by overexpressed METTL3 and promote its translation. The JNK/c-jun/METTL3/PD-L1/IGF2BP1 axis contributes to tumour immune escape in an m6A-dependent manner in BC.
Long-term exposure to fine particulate matter (PM2.5) is associated with multiple malignancies, including BC. However, the specific mechanisms of oncogenic processes in BC remain unclear. Liu et al. (Liu et al., 2022b) discovered a new mechanism underlying PM2.5 involvement in BC by m6A modification. They found that by hypomethylating the METTL3 promoter and increasing the affinity of HIF1A binding to its promoter, PM2.5 aberrantly upregulated METTL3 expression and increased the m6A modification of Baculoviral IAP Repeat Containing 5 (BIRC5). The upregulaion of BIRC5 expression recognized and enhanced by IGF2BP3 could then promote the tumour’s progression, metastasis, and VEGFA-regulated angiogenesis.
Methylation modifications of m6A and miRNAs are crucial in human tumours. Some new studies have identified an association between m6A modification and miRNA in various tumours (Yi et al., 2020). However, little is known about their association with BC. Yan et al. (Yan et al., 2022) reported the underlying antitumour mechanism of melittin in BC. They demonstrated that the m6A modification of miR-146a-5p was responsible for the maturation of pri-miR-146. Through the selective reduction of METTL3 in BC cells, melittin inhibits the miR-146a-5p maturation pathway, suppresses the NOTCH2 pathway, and induces BC cell apoptosis. Han et al. (Han et al., 2019a) confirmed that METTL3 could affect the binding of the microprocessor protein DGCR8 to m6A methylated miRNAs, which in turn enhanced DGCR8 recognition and binding to pri-miRNA221/222 in BC, thereby increasing pri-miRNA processing and maturation. Further functional studies showed that miR221/222 directly targeted and inhibited phosphate and tension homology (Pten) expression and promoted tumour progression, and METTL3 plays a cancerous role in BC by positively regulating pri-miR221/222 processes in an m6A-dependent way.
It is widely accepted that angiogenesis and tumour cell adhesion are essential in tumour metastasis (Carmeliet and RakeshJain, 2000; Hamidi and Ivaska, 2018). Wang et al. (Wang et al., 2021) revealed that the knockdown of METTL3 suppresses tyrosine kinase endothelial (TEK) and vascular endothelial growth factor A (VEGF-A) expression in BC. The METTL3-mediated modification of m6A is necessary to activate TEK/VEGF-A mediated tumour development and angiogenesis in BC. Jin et al. (Jin et al., 2019b) found that the adhesive molecule integrin alpha-6 (ITGA6) was a downstream target of METTL3. The methylated ITGA6 mRNA induced by METTL3 could be recognized by YTHDF1 and YTHDF3, which finally enhanced ITGA6 expression, BC adhesion ability, and malignant phenotypes in BC.
Xie et al. (Xie et al., 2020) demonstrated that SET domain containing 7 (SETD7) and Kruppel-like factor 4 (KLF4), two tumour suppressors, are downstream targets of METTL3 in BC. Further investigations revealed that YTHDF2 could recognize and degrade SETD7 mRNA and KLF4 mRNA methylation by METTL3, which consequently induces BCa progression.
As an oncogene, the CUB domain-containing protein 1 (CDCP1) has emerged as a potential biomarker and therapeutic target in various cancers (Khan et al., 2021). However, the exact mechanism underlying carcinogenesis in BC remains unclear. Yang et al. (Yang et al., 2019) reported that METTL3 and YTHDF1 increased the expression of CDCP1 in BC through a mechanism dependent on the METTL3/CDCP1/YTHDF1 axis, which plays a crucial role in BC tumourigenesis. Ying et al. (Ying et al., 2020) constructed the Cas9-M3 system and achieved targeted mRNA modification. They promote CDCP1 mRNA translation by targeting m6A installation onto the 3′untranslated region of CDCP1, facilitating BC development. This approach provides a novel method for targeted interventions for BC and diseases associated with RNA modification defects. Gao et al. (Gao et al., 2020) demonstrated that AF4/FMR2 family member 4 (AFF4) is a direct target of METTL3 and is regulated by METTL3 in an m6A-dependent manner. METTL3 enhances the AFF4 RNA’s m6A modification and expression in bladder cancer stem cells (BCSCs), which directly controls the expression of MYC and SRY-Box Transcription Factor 2 (SOX2) in BC cells to enhance tumourigenesis in BC. This suggests that AFF4 could be used as a biomarker and therapeutic target for BC patients.
As a key component of m6A RNA deposits, METTL14 is identified downregulated in BC and influences tumour aggressiveness. There are currently two opposing views regarding the role of METTL14 in BC. Gu et al. (Gu et al., 2019) found that the knockdown of METTL14 considerably promoted cell proliferation, metastasis, and tumour initiation in bladder tumour-initiating cells (TICs). A positive correlation was observed between METTL14 expression and BC outcomes. However, Catarina et al. (Guimaraes-Teixeira et al., 2022) found that the tumour size and vessel density were considerably reduced by METTL14 protein downregulation in BC cells. They suggested that METTL14 knockdown could impair the tumour aggressiveness because METTL14 could maintain the methyltransferase complex and m6A abundance.
TICs are a subpopulation of cancer cells that are indispensable for tumours. Gu et al. (Gu et al., 2019) reported a new regulatory axis in bladder TICs. As a METTL14 downstream target, Notch1 is vital in tumour oncogenesis and TICs self-renewal. The METTL14’s m6A modification of Notch1 could decrease the stability of Notch1 mRNA, resulting in the suppression of TICs in BC. The Mettl14-Notch1 pathway may be a potential site for eliminating bladder TICs. Zhang et al. (Zhang et al., 2021a) found that Isorhapontigenin (ISO) could upregulate the expression of METTL14 by activating the transcription factor FOXO3a, and then downregulating the epithelial-to-mesenchymal transition (EMT) marker vimentin in an m6A-dependant manner, which in turn inhibits EMT and invasion in BC.
Additionally, WTAP, an m6A writer, has been identified as a novel biomarker for the onset and progression of BC, thereby assisting in the diagnosis and prognosis of BC (Chen and Wang, 2018). The expression level of WTAP was significantly overexpressed in BC and associated with poor prognosis (Wei et al., 2021; Wang et al., 2023).
As a novel form of cell death, ferroptosis influences the formation of many tumours, including BC (Chen et al., 2021b; Liu et al., 2021). However, little is known about the association between ferroptosis and m6A modification in BC. The endogenous antioxidant factor nuclear factor erythroid 2-related factor 2 (NRF2) is a critical component of tumour ferroptosis. Wang et al. (Wang et al., 2023) identified NRF2 as a remarkable m6A site and a downstream target of WTAP. WTAP recognized and enhanced the stability of NRF2 mRNA in a WTAP-YTHDF1-m6A-dependent manner, which in turn suppressed the erastin-induced ferroptosis and promoted BC malignancy.
4.2 m6A erasers and BC
In BC, studies on the m6A erasers have mainly focused on the ALKBH5 and FTO. Yu et al. (Yu et al., 2021) demonstrated a reduced expression level of ALKBH5 in BC, which was strongly related to the poor outcomes in BC patients.
As a kind of m6A demethylase, the role of FTO in various tumours has been confirmed recently, such as glioblastoma, leukemia, and ovarian cancer (Li et al., 2017). FTO plays different roles in different tumour cell types. It can play an oncogenic role in glioblastoma (Cui et al., 2017); however, it acts as a tumour suppressor gene in ovarian cancer (Huang et al., 2020c). There is also controversy regarding its role in BC. Some studies have demonstrated that FTO is downregulated in BC cell lines than normal bladder cell lines. The upregulated expression of FTO can significantly decrease cell proliferation and invasion; therefore, FTO can exhibit a tumour-suppressing effect on BC (Wen et al., 2020; Yi et al., 2021). In contrast, others reported that FTO was overexpressed in BC, and the knockdown of FTO inhibited bladder cancer cell proliferation (Song et al., 2021; Tao et al., 2021; Zhou et al., 2021; Sun et al., 2022).
Recent studies have depicted that FTO can also regulate the progression of BC cells by affecting the metabolism of miRNAs and lncRNAs. A previous section elucidated that Lnc-MALAT1 could promote progression in the PCa via an m6A-dependent mechanism (Mao et al., 2022). According to Tao et al. (Tao et al., 2021), FTO can facilitate suppression of m6A modification of the Lnc-Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) 5′-end and thus prevents YTHDF2’s degradation of MALAT1 mRNA. Thereby enhancing MALAT1 mRNA half-life and expression. This study revealed that FTO promotes the viability and tumourigenesis of BC cells through the FTO/Lnc-MALAT1/miR-384/Mal,T cell differentiation protein 2 (MAL2) axis. Zhou et al. (Zhou et al., 2021) found that FTO regulates miR-576 processing via m6A-dependent mechanisms. FTO plays an oncogenic role by regulating the FTO/miR-576/CDK6 pathway in BC by inhibiting miRNA synthesis. They demonstrated that FTO is a potential diagnostic biomarker for BC. Conversely, miRNAs can also affect the FTO process during m6A modifications. Sun et al. (Sun et al., 2022) reported that miR-5581–3p is a tumour suppressor in BC, and FTO is regarded as a direct downstream target of miR-5581–3p. miR-5581–3p suppressed FTO synthesis and inhibited BC progression and proliferation via the miR-5581–3p/FTO/Matrix metallopeptidase 9 (MMP9) axis.
Ubiquitin-specific proteases (USPs) are the main members of the deubiquitinase family. Numerous studies have demonstrated that USPs affect tumour progression by regulating the proliferation and death of cancer cells (Pal et al., 2014; Young et al., 2019). Song et al. (Song et al., 2021), for the first time, reported that the post-translational deubiquitination of USP18 could upregulate FTO protein expression and stability. In contrast, FTO can facilitate BC tumourigenesis via its demethylase activity and stabilize the PYCR1 transcripts. Therefore, the USP18/FTO/PYCR1 signaling network may be a potential therapeutic target for BC.
4.3 m6A readers and BC
In BC, studies on the m6A readers have mainly focused on YTHDF1, YTHDF2, YTHDF3, IGF2BP1, and IGF2BP3. Several databases have indicated that YTHDF1 and YTHDF2 mRNA levels were significantly higher in BC tissues than in normal tissues and correlated with poor outcomes (Hu et al., 2021; Zheng et al., 2021). In addition, there was no significant difference in the expression levels of YTHDC1 between tumour and normal tissues; nevertheless, high expression levels of YTHDC1 were associated with a favorable prognosis in BC patients (Chen et al., 2021c). Some studies on YTHDF1-3 in BC have been described in the previous sections (Jin et al., 2019b; Yang et al., 2019; Xie et al., 2020).
According to research, IGF2BPs have a close relationship with BC. IGF2BP1 mRNA levels were significantly higher in BC tissues than in nearby noncancerous tissues and were positively correlated with tumour size and advanced clinical stages of BC (Xie et al., 2021a). By analyzing the TCGA database, Ding et al. (Ding et al., 2022c) demonstrated that IGF2BP2-AS1 expression was elevated in BC tissues and was significantly associated with immune-related factors, indicating that IGF2BP2-AS1 is a potential therapeutic target for immune-related conditions. While other studies have proposed that the expression of IGF2BP2 and IGF2BP3 is significantly higher in BC tissues than in normal bladder tissues and is related to poor prognoses (Huang et al., 2020d; Wu et al., 2021). Xie et al. (Xie et al., 2021a) found that IGF2BP1 was blocked from recognizing downstream m6A-modified RNA by circ-PTPRA. By targeting the IGF2BP1/MYC and IGF2BP1/FSCN1 axis, circ-PTPRA inhibits BC progression. As a potential endogenous blocker, circ-PTPRA is expected to broaden the choice for the curative management of BC. Huang et al. (Huang et al., 2020d) reported that IGF2BP3 has an oncogenic effect on human BC progression. By regulating JAK/STAT signaling, IGF2BP3 increases cell proliferation and the cell cycle and inhibits apoptosis in BC cells. There is potential for gene therapy to improve bladder cancer prognosis by targeting IGF2BP3.
5 Role of m6A regulators in the progression of RCC
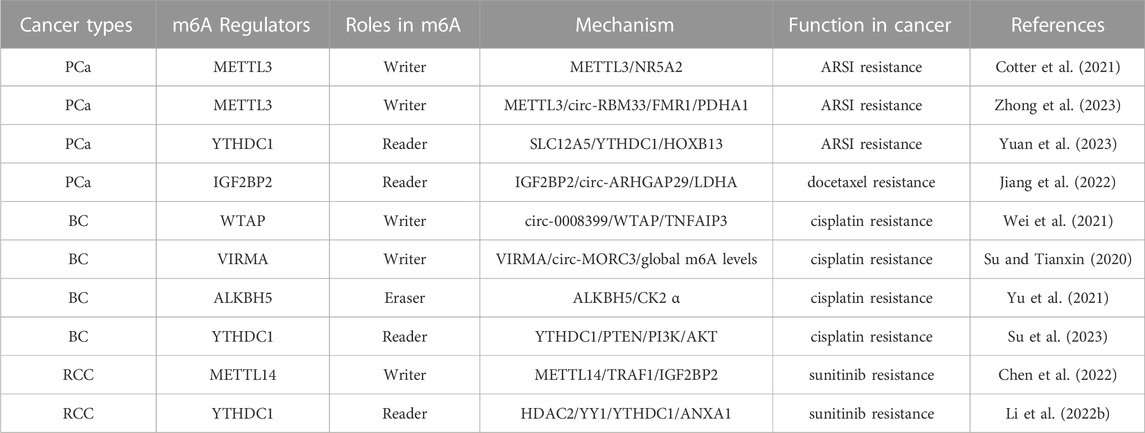
Numerous studies have demonstrated the participation of m6A regulators in various functions in the RCC, such as dysregulation of signaling pathways and regulation of autophagy. The roles and mechanisms of m6A regulators in RCC are summarized in Figure 4 and Table 3.
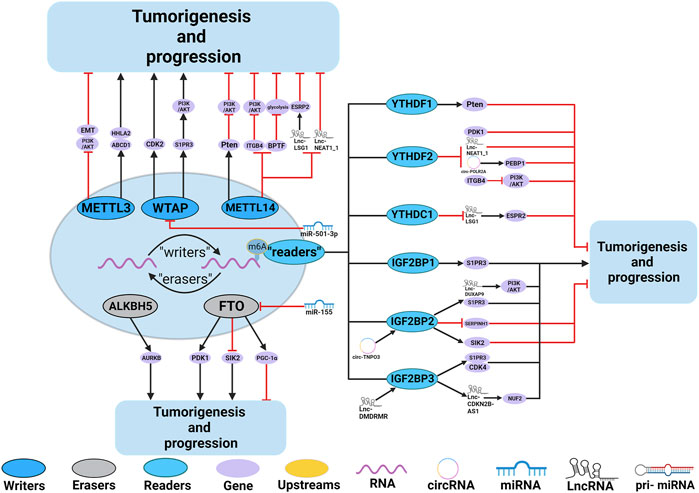
FIGURE 4. In RCC, m6A regulatory proteins contribute to tumorigenesis and progression by interacting with various RNAs. METTL3, METTL14 and WTAP stimulate the progression of RCC by promoting the expression levels of HHLA2, ABCD1, S1PR3 and CDK2. miR-501–3p inhibit the expression of WATP, leading to RCC suppression. METTL14 suppresses the progression of RCC by promoting the expression levels of Pten. METTL3 and METTL14 suppress the progression of RCC by inhibiting the expression levels of EMT, PI3K, BPTF, ITGB4, Lnc-LSG1 and Lnc-NEAT1_1. FTO and ALKBH5 stimulate/inhibit the progression of RCC by affecting the expression levels of AURKB, PDK1, SIK2, and PGC-1α. In addition,miR-155 inhibits the expression of FTO, leading to RCC suppression. IGF2BP1, IGF2BP2 and IGF2BP3 promote the tumorigenesis and progression of RCC by regulating the expression levels of S1PR3, CDK4, Lnc-DUXAP9 and Lnc-CDKN2B-AS1. Lnc-DMDRMR bounds the IGF2BP3 expression and helps stabilize the cell-cycle kinase CDK4, thereby supporting the malignancy state of RCC. YTHDF1, YTHDF2, YTHDC1 and IGF2BP2 inhibit the tumorigenesis and progression of RCC by regulating the expression levels of Pten, ITGB4, PDK1, Lnc-NEAT1_1, Lnc-LSG1, circ-POLR2A, SERPINH1 and SIK2. In addition, circ-TNPO3 interacts with IGF2BP2 to destabilize the mRNA of SERPINH1, thereby inhibiting the tumorigenesis and progression of RCC.
5.1 m6A writers and RCC
Studies on RCC have mostly focused on the regulatory factors METTL3 and METTL14. Numerous studies have indicated that METTL3 has two roles in RCC. Most studies found that METTL3, an oncogene, was upregulated in RCC compared to normal tissues and correlated with poor outcomes (Zhang et al., 2020a; Chen et al., 2020b; Wang et al., 2020c; Zhao et al., 2020). An oncogene, the human endogenous retrovirus-H long terminal repeat-associating protein 2 (HHLA2), is an important prognostic predictor in RCC (Chen et al., 2019b). According to Zhu et al. (Zhu et al., 2022), through m6A modification of HHLA2 mRNA, METTL3 regulates and promotes HHLA2 expression, thereby enabling RCC growth and invasion. Shi et al. (Shi et al., 2021) reported that ABCD1 (an ATP-binding cassette transporter of fatty acids) is a key downstream target of METTL3 in RCC, and the knockdown of ABCD1 in RCC cells decreased cancer cell migration and spheroid formation. METTL3 enhances ABCD1 translation in an m6A-dependent manner to promote RCC.
Although most studies provide evidence that METTL3 contributes to the proliferation and progression of RCC, reports have shown a tumour suppressor role of METTL3. Li et al. (Xiao et al., 2017) indicated that METLL3 acts as a tumour suppressor, and the knockdown of METTL3 considerably promotes RCC proliferation. According to their findings, EMT pathways may contribute to the underlying mechanism of RCC and upregulated METTL3 may reverse EMT progression. In addition, they demonstrated that through its negative impact on PI3K/Akt/mTOR signaling, METTL3 might influence RCC progression.
Like BC, the precise role of METTL14 in RCC remains controversial. Most studies have demonstrated that lower METTL14 expression in RCC tissues is negatively associated with a poor prognosis. METTL14 can act as a tumour suppressor (Zhang et al., 2021b; Shen et al., 2022a; Liu et al., 2022c; Liu et al., 2022d; Zhang et al., 2022). However, some researchers have opposite conclusions (Chen et al., 2022).
Methylation modifications of lncRNAs and m6A play an important role in tumours. Studies have demonstrated that METTL14 can also regulate the growth of RCC cells by influencing the metabolism of lncRNAs. According to a new study in 2022 (Shen et al., 2022a), the Lnc-LSG1 was identified as a downstream target of METTL14, and it could promote RCC progression and metastasis by binding to ESRP2 and promoting ESRP2 ubiquitination and degradation. METTL14 inhibited the interaction between epithelial splicing regulatory protein 2 (ESRP2) and Lnc-LSG1 by increasing the binding of YTHDC1 to the m6A site in Lnc-LSG1. The overexpression of METTL14 considerably decreases the migration and invasion abilities of RCC cells. Similarly, Liu et al. (Liu et al., 2022c) found that RCC tissues showed higher levels of Lnc-NEAT1_1, which was negatively correlated with METTL14 expression. Further studies have demonstrated that METTL14 suppresses the expression of NEAT1_1 in an m6A-dependent manner via YTHDF2-dependent RNA degradation, thereby inhibiting the progression and metastasis of RCC. These studies presented the first insight into how m6A-modified lncRNAs function and may set the stage for novel biomarkers and approaches in RCC.
The bromodomain PHD finger transcription factor (BPTF) plays a significant role in the invasion and progression of cancers (Koedoot et al., 2019; Laurette et al., 2020). Nevertheless, very few studies have investigated the transcriptional or post-transcriptional regulation of BPTF, particularly m6A modification. In a groundbreaking study, Zhang et al. (Zhang et al., 2021b) found a negative correlation between the stability of BPTF mRNA and the modification of m6A in the BPTF 3′UTR regions by METTL14 in RCC. The deficiency of METTL14 increased the expression of BPTF, which accelerated RCC metastasis. A subsequent study has demonstrated that the METTL14/BPTF axis contributes to RCC metastasis by increasing glycolysis. They indicated that BPTF inhibitor AU1 may be a promising therapeutic candidate for overexpressing BPTF in RCC.
Many signaling pathways associated with tumour development have been recognized as m6A targets and are controlled by m6A modifiers. The PI3K/AKT pathway plays an essential role in various cellular processes, including the growth and survival of RCC cells (Sato et al., 2013; Zhu et al., 2020). Recently, the mechanism of METTL14 and WTAP methylation effects on tumour proliferation through the PI3K-AKT pathways in RCC has been demonstrated. According to Zhang et al. (Zhang et al., 2022), Pten is a downstream target of METTL14 in RCC. By regulating Pten mRNA for m6A modification via a YTHDF1-dependent mechanism, the downregulation of METTL14 in RCC inhibited Pten expression, leading to the progression of tumours through PI3K/AKT signaling. The Integrinβ4 (ITGB4) is a transmembrane receptor involved in the tumourigenesis and the invasiveness of several malignancies (Lipscomb et al., 2005; Ruan et al., 2020). Nonetheless, the underlying mechanisms in RCC remain unclear. Liu et al. (Liu et al., 2022d) reported that significant overexpression of ITGB4 in RCC tissues and metastases, together with poor prognosis, is associated with high levels of ITGB4. Further investigations revealed that a METTL14-mediated modification of m6A negatively regulates ITGB4 expression. During this process, YTHDF2 preferentially interacts with the ITGB4 mRNA and promotes its degradation. Downregulation of METTL14 and overexpression of ITGB4 promote tumour invasion, metastasis, and the PI3K/AKT signaling in RCC.
WTAP is a crucial gene involved in many tumour diseases. A relationship between WTAP and RCC was also identified. According to recent studies, WTAP is significantly overexpressed in RCC cell lines and tissues, and higher WTAP levels lead to a poor prognosis in patients with RCC (Tang et al., 2018; He et al., 2021; Ying et al., 2021). Ying et al. (Ying et al., 2021) reported that WTAP and IGF2BP proteins enhanced S1PR3 stability in an m6A-dependent manner and regulated its PI3K/AKT pathway, which promoted tumourigenesis and migration. He et al. Reported that the interaction of miRNA with the WTAP affects RCC proliferation and development (He et al., 2021). Studies have indicated that miR-501–3p is a latent suppressor that is downregulated in RCC. Further investigation revealed that WTAP is a direct downstream target gene of miR-501–3p and is inhibited by it. The overexpressed WTAP may play an oncogenic role through the miR-501–3p/WTAP/Cyclin Dependent Kinase 2 (CDK2) axis in RCC.
5.2 m6A eraser and RCC
Studies of m6A erasers in the RCC have focused on ALKBH5 and FTO. Recent studies have provided insight into the regulation of ALKBH5 methylation in RCC. They concluded that ALKBH5 had higher expression levels in both RCC tumour tissues and cell lines. (Chen et al., 2020b; Guimaraes-Teixeira et al., 2021). Zhang et al. (Zhang et al., 2020b). reported that ALKBH5 could play an oncogenic role in the tumourigenesis of RCC, and that Aurora Kinase B (AURKB) expression was positively correlated with the expression of ALKBH5 in RCC tissues. By demethylating the mRNA of AURKB mRNA, ALKBH5 enhances the AURKB stability and promotes RCC cell proliferation in an m6A-dependent manner.
Depending on the context, FTO is believed to play both tumour-suppressing and cancer-promoting roles in an m6A-dependent manner in solid tumours. Among the four studies discussed in detail in the subsequent sections, two identified FTO as carcinogenic (Xu et al., 2022a; Shen et al., 2022b), whereas the other two suggested that it is a carcinogenic tumour suppressor (Zhuang et al., 2019; Yang et al., 2022).
Autophagy has been implicated in various pathophysiological processes, including tumourigenesis, proliferation, and cell death (Levine, 2007). Excitingly, Xu et al. reported that the m6A modification participates in autophagy (Xu et al., 2022a). Salt-inducible kinase 2 (SIK2) is identified as a functional target of m6A-mediated autophagy in RCC. FTO demethylates SIK2, inhibits the stability of SIK2 through an m6A-IGF2BP2-dependent mechanism, and plays a crucial role in inhibiting autophagy and promoting carcinogenicity in RCC. High levels of FTO were also found in the tumour tissues and databases of RCC patients. (Shen et al., 2022b). By protecting Pyruvate Dehydrogenase Kinase 1 (PDK1) mRNA from degradation caused by YTHDF2, FTO promotes the progression and migration of RCC in an m6A-dependent manner. RCC may benefit from the therapeutic targeting of the FTO/YTHDF2/PDK1/m6A axis.
In another study, FTO was found to be downregulated in Von Hippel-Lindau (VHL)-deficient RCC tissue and correlated with tumour severity and poor prognosis (Zhuang et al., 2019). Further investigation confirmed that PPARg coactivators-1α (PGC-1α) was a functionally important target of FTO. Overexpression of FTO in VHL-deficient RCC restored mitochondrial activity, induced oxidative stress and ROS production, and restrained tumour proliferation by increasing the expression of PGC-1α. Therefore, identifying a novel FTO/PGC-1 axis represents a potential therapeutic target for VHL-deficient RCC.
Interactions between m6A modification of FTO and miRNAs in RCC have been demonstrated. Zhuang et al. (Yang et al., 2022) observed that miR-155 expression was considerably higher in RCC tissues than in adjacent normal tissues. Further studies confirmed that miR-155 inhibited FTO expression and increased the global m6A levels while promoting tumour cell progression and decreasing apoptosis in an FTO-dependent manner.
5.3 m6A reader and RCC
Studies of m6A readers in RCC have mainly focused on the YTHDF1, YTHDF2, YTHDC1, and IGF2BPs. Several aspects of the role and mechanism of YTHDF1, YTHDF2, YTHDC1, and IGF2BPs in RCC have been explained in the previous sections (Ying et al., 2021; Shen et al., 2022a; Xu et al., 2022a; Shen et al., 2022b; Chen et al., 2022; Liu et al., 2022c; Liu et al., 2022d; Zhang et al., 2022). Recent studies have reported that YTHDF1-3 could act as tumour markers in RCC. Felix et al. (von Hagen et al., 2021) found that YTHDF1-3 were significantly downregulated in RCC compared to normal tissue and were correlated with patients’ clinical M stage. Xu et al. (Xu et al., 2022b) reported that circ-POLR2A serves as an oncogene by enhancing the ubiquitination and degradation of Phosphatidylethanolamine Binding Protein 1 (PEBP1). YTHDF2 played a suppressive part in the expression of circ-POLR2A in an m6A-dependent manner.
YTHDC1 was downregulated in RCC tissues compared to that in normal tissues and had a predictive value for the risk of RCC. Low levels of YTHDC1 been linked to poor outcomes. This highlights the diagnostic and prognostic value of YTHDC1 in RCC (von Hagen et al., 2021; Li et al., 2022b).
IGF2BPs are principal members of the reader protein of the N6-Methyladenosine RNA Methylation family; however, their function and content in RCC remain controversial. Studies suggest that IGF2BP1-3 were overexpressed in RCC compared to adjacent normal controls and were associated with poor patient prognoses (Zhang et al., 2021c; Ying et al., 2021; Xing et al., 2022). Recent studies have also identified the interactions of lncRNAs and miRNAs with the IGF2BPs in RCC. Tan et al. (Tan et al., 2021) reported that Lnc-Double Homeobox A Pseudogene 9 (DUXAP9) is significantly upregulated in localized RCC and is closely associated with poor prognosis. IGF2BP2 binds to DUXAP9 through its m6A modification sites, which act as m6A readers and promote the stability of DUXAP9. They concluded that Lnc-DUXAP9 might promote tumour proliferation via IGF2BP2/Lnc-DUXAP9/PI3K/Akt axis. Pan et al. (Pan et al., 2022) reported that circ-ransportin-3 (TNPO3) was significantly downregulated in RCC using TCGA database and in vivo experiments. Further tests proved that cicr-TNPO3 could downregulate Serpin family H member 1 (SERPINH1) expression by interacting with IGF2BP2 through m6A modification, thereby inhibiting RCC metastasis. Xie et al. (Xie et al., 2021b) showed that IGF2BP3 promotes Ndc80 kinetochore complex component (NUF2) expression and RCC progression by enhancing the stability of cyclin-dependent kinase inhibitor 2 B antisense 1 lncRNA (CDKN2B-AS1). Gu et al. (Gu et al., 2021) reported that a high co-expression level of DMDRMR (a novel DNA methylation-deregulated and RNA m6A reader-cooperating lncRNA) and IGF2BP3 is associated with poor prognosis in RCC patients. IGF2BP3 which bind Lnc-DMDRMR to stabilize the cell-cycle kinase CDK4 in an m6A-dependent manner, ultimately enhancing the G1-S transition and promoting tumour progression in RCC.
6 Role of m6A regulators in urologic tumours drug resistance
Increasing studies have shown a significant correlation between m6A modification and drug resistance in urologic tumours, suggesting the potential of targeting m6A in urologic tumours with therapeutic resistance. The regulation for m6A levels in urologic tumours maybe a cut-point for chemotherapy, targeted therapy and other drug therapy. We summarizes current studies showing that m6A regulators are closely related to drug resistance and aims to provide deeper insights into tumourigenesis or the drug resistance mechanism of urologic tumours. The role of m6A modification in urologic tumours drug resistance are summarized in Table 4.
6.1 m6A modification and drug resistance in PCa
Androgen deprivation therapy (ADT) is the mainstream therapy strategy for PCa patients. Although initially effective, most of patients progress to castration-resistant prostate cancer (CRPC) after a few years of treatment. CRPC finally develops resistance to second generation of anti-androgen agents. Therefore, identification of the mechanism underlying androgen receptor signaling inhibitors (ARSI) may provide new effective treatments for PCa. Yuan et al. (Yuan et al., 2023) reported that SLC12A5 (a neuron-specific potassium-chloride co-transporter) is significantly upregulated in CRPC compared to prostate adenocarcinoma samples and normal counterparts and promotes castration resistance. Further mechanism studies elucidated that SLC12A5 bind with m6A reader YTHDC1 and in turn promotes the expression of transcription factor HOXB13 to promote tumour progression and castration resistance in an m6A-dependent manner. Cotter et al. (Cotter et al., 2021) found that METTL3 is downregulated in advanced metastatic CRPC compared with benign prostate tissue. They then demonstrated a negative correlation between METTL3 and NR5A2 (a hepatocyte nuclear factor) in CRPC patients. It has been speculated that the upregulation of NR5A2 promote CRPC cells resistant to androgen receptor antagonists via an androgen receptor–independent mechanism. This result provides a new mechanism for m6A in regulating drug resistance of ARSI.
Unlike normal cells, cancer cells are more dependent on glycolysis even under aerobic conditions. However, a latest study indicated that oxidative phosphorylation is substantially increased in CPRC cells compared to androgen sensitive PCa cells (Basu et al., 2021). According to Zhong et al. (Zhong et al., 2023), circ-RBM33 was upregulated and played a oncogenic role in PCa. The knockdown of circ-RBM33 increased the response sensitivity of PCa cells to ARSI treatment. Mechanistically, METTL3 methylated circ-RBM33 and promoted the interaction between circ-RBM33 and FMR1 in an m6A manner, which enhanced mitochondrial respiration by regulating the expression of Pyruvate dehydrogenase alpha 1 (PDHA1). Therefore, METTL3 and circ-RBM33 may function as potential targets for future CPRC therapy.
Docetaxel (DTX) is the first-line chemotherapy for metastatic prostate cancer. However, its therapeutic effect could be limited due to acquired drug resistance in PCa and recurrence may appear after several years of treatment. Jiang et al. (Jiang et al., 2022) showed that circ-ARHGAP29 expression was markedly elevated in PCa docetaxel-resistant cells compared to docetaxel-sensitive cells. The circARHGAP29 could promote the expression of Lactate dehydrogenase A (LDHA) and glycolysis by interacting with m6A reader IGF2BP2, thus increasing docetaxel resistance.
6.2 m6A modification and drug resistance in BC
Chemotherapy is the first-line treatment for muscle-invasive and metastatic bladder cancer. However, resistance to chemotherapy often results in treatment failure in BC (Lenis et al., 2020). Therefore, the mechanism of resistance to chemotherapy in BC requires further investigation.
A recent study reported that YTHDC1 plays a pivotal role in cisplatin resistance of bladder cancer. Su et al. (Su et al., 2023) revealed that the expression of YTHDC1 in cisplatin-resistant BC tissues is lower compared to the cisplatin-sensitive cancer tissues. In BC, YTHDC1 could promote the PTEN mRNA stability and expression by interacting with it and, in turn, inhibting the PI3K/AKT signalling. Wei et al. (Wei et al., 2021) reported that circ-0008399 could bind to WTAP to promote the constitution of the m6A methyltransferase complex and then increase the mRNA stability of TNF alpha-induced protein 3 (TNFAIP3) in an m6A-dependent manner. They confirmed that the circ-0008399/WTAP/TNFAIP3 axis might be crucial in developing cisplatin resistance in BC. Su et al. (Su and Tianxin, 2020) found a positive correlation between upregulated circ-MORC3 and cisplatin-resistance in BC tissues. circ-MORC3 could bind to the m6A reader VIRMA and increase global m6A levels, which then promote cisplatin resistance by enhancing DNA repair. According to a recent study (Yu et al., 2021), ALKBH5 could hinder the glycolytic pathway by reducing the stability of casein kinase 2 α(CK2 α) mRNA in an m6A-dependent manner, thus inhibiting the progression of BC and promoting BC cell sensitivity to cisplatin.
6.3 m6A modification and drug resistance in RCC
The mechanism of tumour resistance involves numerous factors in RCC, including angiogenic switching, gene mutations, and modifications of the tumour microenvironment (Makhov et al., 2018). In RCC, the METTL14 is involved in drug resistance by upregulating the tumour necrosis factor receptor-associated factor 1 (TRAF1) expression (Chen et al., 2022). Chen et al. identified that METTL14 was upregulated and acted as an oncogene in RCC, with a distinct role in other known studies. Moreover, a positive correlation was observed between METTL14 and TRAF1 expression in sunitinib-resistant tissues. Further investigation showed that METTL14-mediated m6A modification enhanced the stability of TRAF1 mRNA in an IGF2BP2-dependent manner and positively modulated sunitinib resistance by regulating apoptosis and angiogenesis.
The association between Annexin-A1 (ANXA1) and the chemosensitivity of tyrosine kinase inhibitors has been illustrated by many recent studies (Chuang et al., 2021; Xiong et al., 2021). Interestingly, Li et al. reported that the reader YTHDC1 regulates ANXA1 in RCC (Li et al., 2022b). ANXA1 is recognized as a downstream target of YTHDC1 in RCC. By inhibiting ANXA1, YTHDC1 suppressed RCC MAPK signaling and inhibited RC cell proliferation. Further studies confirmed that YTHDC1-ANXA1 axis could control sunitinib sensitivity in RCC. The YY1/HDAC2 complex could inhibit the expression of YTHDC1 and promot sunitinib sensitivity via HDAC2/YY1/YTHDC1/ANXA1 axis in RCC. ANXA1 is an important target of m6A modification in developing drug resistance to sunitinib therapy in RCC.
7 Discussion
The role of m6A modification in tumour mechanisms and treatment has become a hot topic in epigenetics. It has been found that m6A modification affects every aspect of urologic tumours. This review comprehensively summarized the m6A methylation modifications in PCa, BC, and RCC based on tumour mechanisms and developmental aspects. It should be noted that the roles of m6A regulation in urologic tumours are complex and even inconsistent. For example, Zhu et al. (Zhu et al., 2022)reported that the expression of METTL3 is upregulated in RCC tissues compared with normal tissues. Poor prognosis was associated with high levels of METTL3 in patients with RCC. By contrast, Li et al. (Xiao et al., 2017)showed that METTL3 inhibits RCC progression through its negative impact on PI3K/Akt/mTOR signaling in an m6A-dependent manner. In addition, The same kind of m6A regulators can exert different functions in different tumours. These contradicting results can be attributed to tumour heterogeneity, differences in experimental setups and different ethnic. For instance, Lewis et al. (Lewis et al., 2010) reported the rs9939609 in FTO was negatively associated with risk of PCa in white European cohort. Beyond this, the target gene expression modified by m6A should depend not only on the “writers” and “erasers” but also on the functions of “readers”. These may explain the contradictory results between “writers” and “erasers” previously reported. In the future, more studies on the regulatory mechanisms of m6A are quite needed.
Apparently, notable progress has been achieved in exploring the role of m6A modification of urologic tumours. m6A regulators have been reported to serve as potential biomarkers for the early diagnosis and prognosis of urologic tumours. For instance, as has been previously noted, METTL3 was significantly upregulated in BC and associated with poor prognosis, it may be a novel promising prognostic biomarker for BC. Among the studies summarized, we supposed that the global m6A levels serve as biomarker for diagnosis of urologic tumours are limited and unreliable. Because we surprisingly found that both “writers” (METTL3, METTL14 and WTAP) and “erasers” (ALKBH5 and FTO) could abnormally overexpressed and exert oncogenic functions or downregulated and play tumour suppressor roles in urologic tumours. In comparison, the target genes modified by m6A are more appropriate to serve as biomarkers. However, although with the advancement of high-throughput sequencing mass spectrometry technology, increasing target genes have been identified in m6A modification of tumours (Dominissini et al., 2012; Meyer et al., 2012; Linder et al., 2015). The m6A modification detection technology is still characterized by large amounts of RNA requirement, high cost and low precision. Therefore, continuous refinement of m6A detection methods and analytical techniques are warranted. It would be helpful to further explore and clarify the use of m6A target transcripts as potential diagnosis biomarkers of urologic tumours. As m6A detection techniques and research methods continue to progress, we believe in the future the improved methods of m6A modification would make it possible to screening early tumour, thus reducing urologic tumours mortality.
The critical roles of m6A modification in urologic tumours also showed that m6A regulators have an enormous potential as novel antitumour therapeutic targets. Small molecule inhibitors and activators promise to treat urologic tumours by inhibiting or compensating the expression of abnormal m6A regulators. Discussion to date, there have been a few studies that reported the development of inhibitors that target m6A regulators in urologic tumours. UZH2 is recently developed and is a potent and selective inhibitor for METTL3. It exhibited interesting antitumours activity in PCa cell lines (Dolbois et al., 2021). Meclofenamic acid is a highly selective FTO inhibitor, it could be combined with nanoplatform based on gold nanorods and promote photothermal immunotherapy in PCa (Liu et al., 2022e). Future studies should work toward to the development of more specific and potent m6A regulatiors inhibitors and activators by high throuput screening, structure-based virtual screening and ligand-based virtual screening. In addition, just as the abovementioned, dysregulation of m6A regulators also serves a key role in urologic tumours drug resistance, which could provide foundations for treating cancer resistant research in the future.
There are still some issues that need to be resolved since m6A modification in urologic tumours has only recently gained attention as a critical epigenetic regulator. For instance, most of the available urologic tumour studies have focused on popular methyltransferases and m6A binding proteins, such as METTL3, METTL14, YTHDFs and IGF2BPs. However, only a small amount of research has conveyed the directions for future exploration of “erasers”. Moreover, the role and aberrant expression of some m6A regulators in tumours remain controversial. Further investigations are required to fully resolve these issues. Thirdly, most of the research have been molecular mechanisms and basic research of m6A regulators, with a lack of clinical studies and applications. Moreover, the probable side effects of m6A modification in urologic tumours are unclear and should be clarified with further detailed studies.
8 Conclusion
m6A modification as a research hotspot in epigenetics have been rapidly developed during recent years. The recent research highlight the important role of m6A modification not only in tumorigenesis and progression, but also in regulating drug resistance. However, the specific and detailed molecular mechanisms of urologic tumours remain unclear, related studies to m6A modification are still in preliminary stage. Undeniably, m6A modification show great potential in the treatment of tumours, which requires more advanced technology and studies. Previous studies on m6A modification have paved the way for future research. It is essential to understand the abnormal characteristics of urologic tumours by elucidating the mechanisms of post-transcriptional m6A modifications. By clarifying these issues, we will gain a more in-depth understanding of the pathogenic mechanisms of PCa, BC, and RCC and provide more feasible treatment strategies targeting m6A modification.
Author contributions
WZ, RZ, XG, and XW conceived the review. WZ wrote the manuscript. XW revised the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Acknowledgments
The authors would like to thank Home for Researchers (www.home-for-researchers.com) for providing English editing services during the preparation of this manuscript and acknowledge the BioRender (www.biorender.com), as figures in this review were created with Home for Researchers and BioRender platform.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
An, Y., and Duan, H. (2022). The role of m6A RNA methylation in cancer metabolism. Mol. Cancer 21 (1), 14. doi:10.1186/s12943-022-01500-4
Bader, J. E., Voss, K., and Rathmell, J. C. (2020). Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol. Cell 78 (6), 1019–1033. doi:10.1016/j.molcel.2020.05.034
Basu, H. S., Wilganowski, N., Robertson, S., Reuben, J. M., Cohen, E. N., Zurita, A., et al. (2021). Prostate cancer cells survive anti-androgen and mitochondrial metabolic inhibitors by modulating glycolysis and mitochondrial metabolic activities. Prostate 81 (12), 799–811. doi:10.1002/pros.24146
Binnewies, M., Roberts, E. W., Kersten, K., Chan, V., Fearon, D. F., Merad, M., et al. (2018). Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 24 (5), 541–550. doi:10.1038/s41591-018-0014-x
Bodi, Z., Button, J. D., Grierson, D., and Fray, R. G. (2010). Yeast targets for mRNA methylation. Nucleic Acids Res. 38 (16), 5327–5335. doi:10.1093/nar/gkq266
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G., and Rottman, F. M. (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3 (11), 1233–1247.
Cai, J., Yang, F., Zhan, H., Situ, J., Li, W., Mao, Y., et al. (2019). RNA m(6)A methyltransferase METTL3 promotes the growth of prostate cancer by regulating Hedgehog pathway. Onco Targets Ther. 12, 9143–9152. doi:10.2147/OTT.S226796
Carmeliet, P., and RakeshJain, K. (2000). Angiogenesis in cancer and other diseases. NATURE 407, 249–257. doi:10.1038/35025220
Chen, B., Liu, C., Long, H., Bai, G., Zhu, Y., and Xu, H. (2023). N(6)-methyladenosine-induced long non-coding RNA PVT1 regulates the miR-27b-3p/BLM axis to promote prostate cancer progression. Int. J. Oncol. 62 (1), 16. doi:10.3892/ijo.2022.5464
Chen, F., Wang, Q., and Zhou, Y. (2021). The construction and validation of an RNA binding protein-related prognostic model for bladder cancer. BMC Cancer 21 (1), 244. doi:10.1186/s12885-021-07930-5
Chen, J., Yu, K., Zhong, G., and Shen, W. (2020). Identification of a m(6)A RNA methylation regulators-based signature for predicting the prognosis of clear cell renal carcinoma. Cancer Cell Int. 20, 157. doi:10.1186/s12935-020-01238-3
Chen, L., and Wang, X. (2018). Relationship between the genetic expression of WTAP and bladder cancer and patient prognosis. Oncol. Lett. 16 (6), 6966–6970. doi:10.3892/ol.2018.9554
Chen, L., Zhu, D., Feng, J., Zhou, Y., Wang, Q., Feng, H., et al. (2019). Overexpression of HHLA2 in human clear cell renal cell carcinoma is significantly associated with poor survival of the patients. Cancer Cell Int. 19, 101. doi:10.1186/s12935-019-0813-2
Chen, M., Nie, Z. Y., Wen, X. H., Gao, Y. H., Cao, H., and Zhang, S. F. (2019). m6A RNA methylation regulators can contribute to malignant progression and impact the prognosis of bladder cancer. Biosci. Rep. 39 (12). doi:10.1042/BSR20192892
Chen, X., Kang, R., Kroemer, G., and Tang, D. (2021). Broadening horizons: The role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. 18 (5), 280–296. doi:10.1038/s41571-020-00462-0
Chen, Y., Lin, Y., Shu, Y., He, J., and Gao, W. (2020). Interaction between N(6)-methyladenosine (m(6)A) modification and noncoding RNAs in cancer. Mol. Cancer 19 (1), 94. doi:10.1186/s12943-020-01207-4
Chen, Y., Lu, Z., Qi, C., Yu, C., Li, Y., Huan, W., et al. (2022). N(6)-methyladenosine-modified TRAF1 promotes sunitinib resistance by regulating apoptosis and angiogenesis in a METTL14-dependent manner in renal cell carcinoma. Mol. Cancer 21 (1), 111. doi:10.1186/s12943-022-01549-1
Chen, Y., Pan, C., Wang, X., Xu, D., Ma, Y., Hu, J., et al. (2021). Silencing of METTL3 effectively hinders invasion and metastasis of prostate cancer cells. Theranostics 11 (16), 7640–7657. doi:10.7150/thno.61178
Chuang, M. C., Lung, J. H., Chen, Y. C., Lin, Y. C., Li, Y. C., and Hung, M. S. (2021). The association of Annexin A1 and chemosensitivity to osimertinib in lung cancer cells. Cancers (Basel) 13 (16), 4106. doi:10.3390/cancers13164106
Condeelis, J., and Pollard, J. W. (2006). Macrophages: Obligate partners for tumor cell migration, invasion, and metastasis. Cell 124 (2), 263–266. doi:10.1016/j.cell.2006.01.007
Cotter, K. A., Gallon, J., Uebersax, N., Rubin, P., Meyer, K. D., Piscuoglio, S., et al. (2021). Mapping of m(6)A and its regulatory targets in prostate cancer reveals a METTL3-low induction of therapy resistance. Mol. Cancer Res. 19 (8), 1398–1411. doi:10.1158/1541-7786.MCR-21-0014
Cui, Q., Shi, H., Ye, P., Li, L., Qu, Q., Sun, G., et al. (2017). m(6 A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18 (11), 2622–2634. doi:10.1016/j.celrep.2017.02.059
Deng, X., Su, R., Weng, H., Huang, H., Li, Z., and Chen, J. (2018). RNA N(6)-methyladenosine modification in cancers: Current status and perspectives. Cell Res. 28 (5), 507–517. doi:10.1038/s41422-018-0034-6
Dierks, D., Garcia-Campos, M. A., Uzonyi, A., Safra, M., Edelheit, S., Rossi, A., et al. (2021). Multiplexed profiling facilitates robust m6A quantification at site, gene and sample resolution. Nat. Methods 18 (9), 1060–1067. doi:10.1038/s41592-021-01242-z
Ding, K., Zheng, Z., Han, Y., and Huang, X. (2022). Prognostic values of the immune microenvironment-related non-coding RNA IGF2BP2-AS1 in bladder cancer. Cell Cycle 21, 2533–2549. doi:10.1080/15384101.2022.2103898
Ding, L., Wang, R., Zheng, Q., Shen, D., Wang, H., Lu, Z., et al. (2022). circPDE5A regulates prostate cancer metastasis via controlling WTAP-dependent N6-methyladenisine methylation of EIF3C mRNA. J. Exp. Clin. Cancer Res. 41 (1), 187. doi:10.1186/s13046-022-02391-5
Ding, Y., Wang, M., and Yang, J. (2022). Circular RNA midline-1 (circMID1) promotes proliferation, migration, invasion and glycolysis in prostate cancer. Bioengineered 13 (3), 6293–6308. doi:10.1080/21655979.2022.2037367
Dolbois, A., Bedi, R. K., Bochenkova, E., Muller, A., Moroz-Omori, E. V., Huang, D., et al. (2021). 1,4,9-Triazaspiro[5.5]undecan-2-one derivatives as potent and selective METTL3 inhibitors. J. Med. Chem. 64 (17), 12738–12760. doi:10.1021/acs.jmedchem.1c00773
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 (7397), 201–206. doi:10.1038/nature11112
Du, C., Lv, C., Feng, Y., and Yu, S. (2020). Activation of the KDM5A/miRNA-495/YTHDF2/m6A-MOB3B axis facilitates prostate cancer progression. J. Exp. Clin. Cancer Res. 39 (1), 223. doi:10.1186/s13046-020-01735-3
Du, P., Liu, F., Liu, Y., Shao, M., Li, X., and Qin, G. (2020). Linc00210 enhances the malignancy of thyroid cancer cells by modulating miR-195-5p/IGF1R/Akt axis. J. Cell Physiol. 235 (2), 1001–1012. doi:10.1002/jcp.29016
Li, E., Wei, B., Wang, X., and Kang, R. METTL3 enhances cell adhesion through stabilizing integrin β1 mRNA via an m6A-HuR-dependent mechanism in prostatic carcinoma. Am. J. Cancer Res. 2020;10(3):1012–1025.
Einstein, J. M., Perelis, M., Chaim, I. A., Meena, J. K., Nussbacher, J. K., Tankka, A. T., et al. (2021). Inhibition of YTHDF2 triggers proteotoxic cell death in MYC-driven breast cancer. Mol. Cell 81 (15), 3048–3064.e9. doi:10.1016/j.molcel.2021.06.014
Fan, Y., Shen, B., Tan, M., Mu, X., Qin, Y., Zhang, F., et al. (2014). TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 20 (6), 1531–1541. doi:10.1158/1078-0432.CCR-13-1455
Fu, Y., Dominissini, D., Rechavi, G., and He, C. (2014). Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat. Rev. Genet. 15 (5), 293–306. doi:10.1038/nrg3724
Gao, Q., Zheng, J., Ni, Z., Sun, P., Yang, C., Cheng, M., et al. (2020). The m(6)A methylation-regulated AFF4 promotes self-renewal of bladder cancer stem cells. Stem Cells Int. 2020, 8849218. doi:10.1155/2020/8849218
Gilbert, W. V., Bell, T. A., and Schaening, C. (2016). Messenger RNA modifications: Form, distribution, and function. Science 352 (6292), 1408–1412. doi:10.1126/science.aad8711
Gu, C., Wang, Z., Zhou, N., Li, G., Kou, Y., Luo, Y., et al. (2019). Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N(6)-methyladenosine of Notch1. Mol. Cancer 18 (1), 168. doi:10.1186/s12943-019-1084-1
Gu, Y., Niu, S., Wang, Y., Duan, L., Pan, Y., Tong, Z., et al. (2021). DMDRMR-mediated regulation of m(6)a-modified CDK4 by m(6)A reader IGF2BP3 drives ccRCC progression. Cancer Res. 81 (4), 923–934. doi:10.1158/0008-5472.CAN-20-1619
Guimaraes-Teixeira, C., Barros-Silva, D., Lobo, J., Soares-Fernandes, D., Constancio, V., Leite-Silva, P., et al. (2021). Deregulation of N6-methyladenosine RNA modification and its erasers FTO/ALKBH5 among the main renal cell tumor subtypes. J. Pers. Med. 11 (10), 996. doi:10.3390/jpm11100996
Guimaraes-Teixeira, C., Lobo, J., Miranda-Goncalves, V., Barros-Silva, D., Martins-Lima, C., Monteiro-Reis, S., et al. (2022). Downregulation of m(6) A writer complex member METTL14 in bladder urothelial carcinoma suppresses tumor aggressiveness. Mol. Oncol. 16 (9), 1841–1856. doi:10.1002/1878-0261.13181
Hamidi, H., and Ivaska, J. (2018). Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 18 (9), 533–548. doi:10.1038/s41568-018-0038-z
Han, D., Liu, J., Chen, C., Dong, L., Liu, Y., Chang, R., et al. (2019). Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature 566 (7743), 270–274. doi:10.1038/s41586-019-0916-x
Han, J., Wang, J. Z., Yang, X., Yu, H., Zhou, R., Lu, H. C., et al. (2019). METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer 18 (1), 110. doi:10.1186/s12943-019-1036-9
He, L., Chen, S., Ying, Y., Xie, H., Li, J., Ma, X., et al. (2021). MicroRNA-501-3p inhibits the proliferation of kidney cancer cells by targeting WTAP. Cancer Med. 10 (20), 7222–7232. doi:10.1002/cam4.4157
He, L., Li, H., Wu, A., Peng, Y., Shu, G., and Yin, G. (2019). Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 18 (1), 176. doi:10.1186/s12943-019-1109-9
Hu, J., Qiu, D., Yu, A., Hu, J., Deng, H., Li, H., et al. (2021). YTHDF1 is a potential pan-cancer biomarker for prognosis and immunotherapy. Front. Oncol. 11, 607224. doi:10.3389/fonc.2021.607224
Huang, H., Wang, Y., Kandpal, M., Zhao, G., Cardenas, H., Ji, Y., et al. (2020). FTO-dependent N (6)-methyladenosine modifications inhibit ovarian cancer stem cell self-renewal by blocking cAMP signaling. Cancer Res. 80 (16), 3200–3214. doi:10.1158/0008-5472.CAN-19-4044
Huang, H., Weng, H., and Chen, J. (2020). m6A modification in coding and non-coding RNAs: Roles and therapeutic implications in cancer. Cancer Cell 37 (3), 270–288. doi:10.1016/j.ccell.2020.02.004
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20 (3), 285–295. doi:10.1038/s41556-018-0045-z
Huang, W., Li, Y., Zhang, C., Zha, H., Zhou, X., Fu, B., et al. (2020). IGF2BP3 facilitates cell proliferation and tumorigenesis via modulation of JAK/STAT signalling pathway in human bladder cancer. J. Cell Mol. Med. 24 (23), 13949–13960. doi:10.1111/jcmm.16003
Huang, Y., Sun, H., Ma, X., Zeng, Y., Pan, Y., Yu, D., et al. (2020). HLA-F-AS1/miR-330-3p/PFN1 axis promotes colorectal cancer progression. Life Sci. 254, 117180. doi:10.1016/j.lfs.2019.117180
Li, J., Meng, S., Xu, M., Wang, S., He, L., Xu, X., et al. (2018). Downregulation of N6-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N6-methyladenosine levels. Oncotarget 9, (3), 3752–3764. doi:10.18632/oncotarget.23365
Jen, J., Tang, Y. A., Lu, Y. H., Lin, C. C., Lai, W. W., and Wang, Y. C. (2017). Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol. Cancer 16 (1), 104. doi:10.1186/s12943-017-0674-z
Jia, G., Wang, X., Wu, W., Zhang, Y., Chen, S., Zhao, J., et al. (2022). LXA4 enhances prostate cancer progression by facilitating M2 macrophage polarization via inhibition of METTL3. Int. Immunopharmacol. 107, 108586. doi:10.1016/j.intimp.2022.108586
Jiang, X., Guo, S., Wang, S., Zhang, Y., Chen, H., Wang, Y., et al. (2022). EIF4A3-Induced circARHGAP29 promotes aerobic glycolysis in docetaxel-resistant prostate cancer through IGF2BP2/c-Myc/LDHA signaling. Cancer Res. 82 (5), 831–845. doi:10.1158/0008-5472.CAN-21-2988
Jin, H., Ying, X., Que, B., Wang, X., Chao, Y., Zhang, H., et al. (2019). N(6)-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine 47, 195–207. doi:10.1016/j.ebiom.2019.07.068
Jin, Z., Jia, B., Tan, L., and Liu, Y. (2019). miR-330-3p suppresses liver cancer cell migration by targeting MAP2K1. Oncol. Lett. 18 (1), 314–320. doi:10.3892/ol.2019.10280
Khan, T., Kryza, T., Lyons, N. J., He, Y., and Hooper, J. D. (2021). The CDCP1 signaling hub: A target for cancer detection and therapeutic intervention. Cancer Res. 81 (9), 2259–2269. doi:10.1158/0008-5472.CAN-20-2978
Koedoot, E., Fokkelman, M., Rogkoti, V. M., Smid, M., van de Sandt, I., de Bont, H., et al. (2019). Uncovering the signaling landscape controlling breast cancer cell migration identifies novel metastasis driver genes. Nat. Commun. 10 (1), 2983. doi:10.1038/s41467-019-11020-3
Kong, Z., Wan, X., Lu, Y., Zhang, Y., Huang, Y., Xu, Y., et al. (2020). Circular RNA circFOXO3 promotes prostate cancer progression through sponging miR-29a-3p. J. Cell Mol. Med. 24 (1), 799–813. doi:10.1111/jcmm.14791
Laurette, P., Coassolo, S., Davidson, G., Michel, I., Gambi, G., Yao, W., et al. (2020). Chromatin remodellers Brg1 and Bptf are required for normal gene expression and progression of oncogenic Braf-driven mouse melanoma. Cell Death Differ. 27 (1), 29–43. doi:10.1038/s41418-019-0333-6
Lee, K. S., Burke, T. R., Park, J. E., Bang, J. K., and Lee, E. (2015). Recent advances and new strategies in targeting Plk1 for anticancer therapy. Trends Pharmacol. Sci. 36 (12), 858–877. doi:10.1016/j.tips.2015.08.013
Lenis, A. T., Lec, P. M., Chamie, K., and Mshs, M. D. (2020). Bladder cancer: A review. JAMA 324 (19), 1980–1991. doi:10.1001/jama.2020.17598
Lewis, C. J., Pan, T., and Kalsotra, A. (2017). RNA modifications and structures cooperate to guide RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 18 (3), 202–210. doi:10.1038/nrm.2016.163
Lewis, S. J., Murad, A., Chen, L., Davey Smith, G., Donovan, J., Palmer, T., et al. (2010). Associations between an obesity related genetic variant (FTO rs9939609) and prostate cancer risk. PLoS One 5 (10), e13485. doi:10.1371/journal.pone.0013485
Li, J., Xie, H., Ying, Y., Chen, H., Yan, H., He, L., et al. (2020). YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol. Cancer 19 (1), 152. doi:10.1186/s12943-020-01267-6
Li, P., Shi, Y., Gao, D., Xu, H., Zou, Y., Wang, Z., et al. (2022). ELK1-mediated YTHDF1 drives prostate cancer progression by facilitating the translation of Polo-like kinase 1 in an m6A dependent manner. Int. J. Biol. Sci. 18 (16), 6145–6162. doi:10.7150/ijbs.75063
Li, Q., Wang, W., Zhang, M., Sun, W., Shi, W., and Li, F. (2020). Circular RNA circ-0016068 promotes the growth, migration, and invasion of prostate cancer cells by regulating the miR-330-3p/BMI-1 Axis as a competing endogenous RNA. Front. Cell Dev. Biol. 8, 827. doi:10.3389/fcell.2020.00827
Li, S., and Cao, L. (2022). Demethyltransferase FTO alpha-ketoglutarate dependent dioxygenase (FTO) regulates the proliferation, migration, invasion and tumor growth of prostate cancer by modulating the expression of melanocortin 4 receptor (MC4R). Bioengineered 13 (3), 5598–5612. doi:10.1080/21655979.2021.2001936
Li, W., Chen, G., Feng, Z., Zhu, B., Zhou, L., Zhang, Y., et al. (2021). YTHDF1 promotes the proliferation, migration, and invasion of prostate cancer cells by regulating TRIM44. Genes Genomics 43 (12), 1413–1421. doi:10.1007/s13258-021-01175-z
Li, W., Ye, K., Li, X., Liu, X., Peng, M., Chen, F., et al. (2022). YTHDC1 is downregulated by the YY1/HDAC2 complex and controls the sensitivity of ccRCC to sunitinib by targeting the ANXA1-MAPK pathway. J. Exp. Clin. Cancer Res. 41 (1), 250. doi:10.1186/s13046-022-02460-9
Li, X. X., Huang, L. Y., Peng, J. J., Liang, L., Shi, D. B., Zheng, H. T., et al. (2014). Klotho suppresses growth and invasion of colon cancer cells through inhibition of IGF1R-mediated PI3K/AKT pathway. Int. J. Oncol. 45 (2), 611–618. doi:10.3892/ijo.2014.2430
Li, Z., Weng, H., Su, R., Weng, X., Zuo, Z., Li, C., et al. (2017). FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell 31 (1), 127–141. doi:10.1016/j.ccell.2016.11.017
Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., and Jaffrey, S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12 (8), 767–772. doi:10.1038/nmeth.3453
Lipscomb, E. A., Simpson, K. J., Lyle, S. R., Ring, J. E., Dugan, A. S., and Mercurio, A. M. (2005). The alpha6beta4 integrin maintains the survival of human breast carcinoma cells in vivo. Cancer Res. 65 (23), 10970–10976. doi:10.1158/0008-5472.CAN-05-2327
Liu, H., Gu, J., Huang, Z., Han, Z., Xin, J., Yuan, L., et al. (2022). Fine particulate matter induces METTL3-mediated m(6)A modification of BIRC5 mRNA in bladder cancer. J. Hazard Mater 437, 129310. doi:10.1016/j.jhazmat.2022.129310
Liu, J., Ma, H., Meng, L., Liu, X., Lv, Z., Zhang, Y., et al. (2021). Construction and external validation of a ferroptosis-related gene signature of predictive value for the overall survival in bladder cancer. Front. Mol. Biosci. 8, 675651. doi:10.3389/fmolb.2021.675651
Liu, J., Song, Y., Wang, Y., Han, M., Wang, C., and Yan, F. (2022). Cyclodextrin-functionalized gold nanorods loaded with meclofenamic acid for improving N6-methyladenosine-mediated second near-infrared photothermal immunotherapy. ACS Appl. Mater. Interfaces 14 (36), 40612–40623. doi:10.1021/acsami.2c09978
Liu, J., Yuan, J. F., and Wang, Y. Z. (2022). METTL3-stabilized lncRNA SNHG7 accelerates glycolysis in prostate cancer via SRSF1/c-Myc axis. Exp. Cell Res. 416 (1), 113149. doi:10.1016/j.yexcr.2022.113149
Liu, T., Wang, H., Fu, Z., Wang, Z., Wang, J., Gan, X., et al. (2022). Methyltransferase-like 14 suppresses growth and metastasis of renal cell carcinoma by decreasing long noncoding RNA NEAT1. Cancer Sci. 113 (2), 446–458. doi:10.1111/cas.15212
Liu, Z., Sun, T., Piao, C., Zhang, Z., and Kong, C. (2022). METTL14-mediated N(6)-methyladenosine modification of ITGB4 mRNA inhibits metastasis of clear cell renal cell carcinoma. Cell Commun. Signal 20 (1), 36. doi:10.1186/s12964-022-00831-5
Ma, Z., Zhong, H., Facai, Q., and Hou, J. (2021). METTL3-mediated m6A modification of KIF3C-mRNA promotes prostate cancer progression and is negatively regulated by miR-320d. Aging 13 (18), 22332–22344. doi:10.18632/aging.203541
Makhov, P., Joshi, S., Ghatalia, P., Kutikov, A., Uzzo, R. G., and Kolenko, V. M. (2018). Resistance to systemic therapies in clear cell renal cell carcinoma: Mechanisms and management strategies. Mol. Cancer Ther. 17 (7), 1355–1364. doi:10.1158/1535-7163.MCT-17-1299
Mao, Y., Li, W., Weng, Y., Hua, B., Gu, X., Lu, C., et al. (2022). METTL3-Mediated m(6)A modification of lncRNA MALAT1 facilitates prostate cancer growth by activation of PI3K/AKT signaling. Cell Transpl. 31, 9636897221122997. doi:10.1177/09636897221122997
Melstrom, L., and Chen, J. (2020). RNA N(6)-methyladenosine modification in solid tumors: New therapeutic frontiers. Cancer Gene Ther. 27 (9), 625–633. doi:10.1038/s41417-020-0160-4
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149 (7), 1635–1646. doi:10.1016/j.cell.2012.05.003
Ni, Z., Sun, P., Zheng, J., Wu, M., Yang, C., Cheng, M., et al. (2022). JNK signaling promotes bladder cancer immune escape by regulating METTL3-mediated m6A modification of PD-L1 mRNA. Cancer Res. 82 (9), 1789–1802. doi:10.1158/0008-5472.CAN-21-1323
Oerum, S., Meynier, V., Catala, M., and Tisne, C. (2021). A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 49 (13), 7239–7255. doi:10.1093/nar/gkab378
Pal, A., Young, M. A., and Donato, N. J. (2014). Emerging potential of therapeutic targeting of ubiquitin-specific proteases in the treatment of cancer. Cancer Res. 74 (18), 4955–4966. doi:10.1158/0008-5472.CAN-14-1211
Pan, X., Huang, B., Ma, Q., Ren, J., Liu, Y., Wang, C., et al. (2022). Circular RNA circ-TNPO3 inhibits clear cell renal cell carcinoma metastasis by binding to IGF2BP2 and destabilizing SERPINH1 mRNA. Clin. Transl. Med. 12 (7), e994. doi:10.1002/ctm2.994
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24 (2), 177–189. doi:10.1038/cr.2014.3
Ruan, S., Lin, M., Zhu, Y., Lum, L., Thakur, A., Jin, R., et al. (2020). Integrin β4-targeted cancer immunotherapies inhibit tumor growth and decrease metastasis. Cancer Res. 80 (4), 771–783. doi:10.1158/0008-5472.CAN-19-1145
Sadeghi Rad, H., Monkman, J., Warkiani, M. E., Ladwa, R., O'Byrne, K., Rezaei, N., et al. (2021). Understanding the tumor microenvironment for effective immunotherapy. Med. Res. Rev. 41 (3), 1474–1498. doi:10.1002/med.21765
Sato, Y., Yoshizato, T., Shiraishi, Y., Maekawa, S., Okuno, Y., Kamura, T., et al. (2013). Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 45 (8), 860–867. doi:10.1038/ng.2699
Schwartz, S., Mumbach, M. R., Jovanovic, M., Wang, T., Maciag, K., Bushkin, G. G., et al. (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 8 (1), 284–296. doi:10.1016/j.celrep.2014.05.048
Shen, D., Ding, L., Lu, Z., Wang, R., Yu, C., Wang, H., et al. (2022). METTL14-mediated Lnc-LSG1 m6A modification inhibits clear cell renal cell carcinoma metastasis via regulating ESRP2 ubiquitination. Mol. Ther. Nucleic Acids 27, 547–561. doi:10.1016/j.omtn.2021.12.024
Shen, H., Ying, Y., Ma, X., Xie, H., Chen, S., Sun, J., et al. (2022). FTO promotes clear cell renal cell carcinoma progression via upregulation of PDK1 through an m(6)A dependent pathway. Cell Death Discov. 8 (1), 356. doi:10.1038/s41420-022-01151-w
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27 (3), 315–328. doi:10.1038/cr.2017.15
Shi, Y., Dou, Y., Zhang, J., Qi, J., Xin, Z., Zhang, M., et al. (2021). The RNA N6-methyladenosine methyltransferase METTL3 promotes the progression of kidney cancer via N6-methyladenosine-dependent translational enhancement of ABCD1. Front. Cell Dev. Biol. 9, 737498. doi:10.3389/fcell.2021.737498
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics, 2022. CA Cancer J. Clin. 72 (1), 7–33. doi:10.3322/caac.21708
Song, W., Yang, K., Luo, J., Gao, Z., and Gao, Y. (2021). Dysregulation of USP18/FTO/PYCR1 signaling network promotes bladder cancer development and progression. Aging 13 (3), 3909–3925. doi:10.18632/aging.202359
Su, R., Dong, L., Li, Y., Gao, M., Han, L., Wunderlich, M., et al. (2020). Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell 38 (1), 79–96. doi:10.1016/j.ccell.2020.04.017
Su, Y., and Tianxin, L. T. (2020). circMORC3 contributes to cisplatin-resistance by affecting m6A modification on DNA damage response related genes in bladder cancer. Eur. Urology Open Sci. 19, e421–e422. doi:10.1016/s2666-1683(20)32843-3
Su, Y., Wang, B., Huang, J., Huang, M., and Lin, T. (2023). YTHDC1 positively regulates PTEN expression and plays a critical role in cisplatin resistance of bladder cancer. Cell Prolif., e13404. doi:10.1111/cpr.13404
Suh, K. S., Crutchley, J. M., Koochek, A., Ryscavage, A., Bhat, K., Tanaka, T., et al. (2007). Reciprocal modifications of CLIC4 in tumor epithelium and stroma mark malignant progression of multiple human cancers. Clin. Cancer Res. 13 (1), 121–131. doi:10.1158/1078-0432.CCR-06-1562
Sun, J., Ma, X., Ying, Y., Wang, W., Shen, H., Wang, S., et al. (2022). SMAD3 and FTO are involved in miR-5581-3p-mediated inhibition of cell migration and proliferation in bladder cancer. Cell Death Discov. 8 (1), 199. doi:10.1038/s41420-022-01010-8
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Tan, L., Tang, Y., Li, H., Li, P., Ye, Y., Cen, J., et al. (2021). N6-Methyladenosine modification of LncRNA DUXAP9 promotes renal cancer cells proliferation and motility by activating the PI3K/AKT signaling pathway. Front. Oncol. 11, 641833. doi:10.3389/fonc.2021.641833
Tang, J., Wang, F., Cheng, G., Si, S., Sun, X., Han, J., et al. (2018). Wilms' tumor 1-associating protein promotes renal cell carcinoma proliferation by regulating CDK2 mRNA stability. J. Exp. Clin. Cancer Res. 37 (1), 40. doi:10.1186/s13046-018-0706-6
Tao, L., Mu, X., Chen, H., Jin, D., Zhang, R., Zhao, Y., et al. (2021). FTO modifies the m6A level of MALAT and promotes bladder cancer progression. Clin. Transl. Med. 11 (2), e310. doi:10.1002/ctm2.310
von Hagen, F., Gundert, L., Strick, A., Klumper, N., Schmidt, D., Kristiansen, G., et al. (2021). N(6) -Methyladenosine (m(6) A) readers are dysregulated in renal cell carcinoma. Mol. Carcinog. 60 (5), 354–362. doi:10.1002/mc.23297
Wang, D., Wang, X., Huang, B., Zhao, Y., Tu, W., Jin, X., et al. (2022). METTL3 promotes prostate cancer progression by regulating miR-182 maturation in m6A-dependent manner. Andrologia 54 (7), 1581–1591. doi:10.1111/and.14422
Wang, G., Dai, Y., Li, K., Cheng, M., Xiong, G., Wang, X., et al. (2021). Deficiency of Mettl3 in bladder cancer stem cells inhibits bladder cancer progression and angiogenesis. Front. Cell Dev. Biol. 9, 627706. doi:10.3389/fcell.2021.627706
Wang, J., Zhang, C., He, W., and Gou, X. (2020). Effect of m(6)A RNA methylation regulators on malignant progression and prognosis in renal clear cell carcinoma. Front. Oncol. 10, 3. doi:10.3389/fonc.2020.00003
Wang, K., Wang, G., Li, G., Zhang, W., Wang, Y., Lin, X., et al. (2023). m6A writer WTAP targets NRF2 to accelerate bladder cancer malignancy via m6A-dependent ferroptosis regulation. Apoptosis 28, 627–638. doi:10.1007/s10495-023-01817-5
Wang, L., Hui, H., Agrawal, K., Kang, Y., Li, N., Tang, R., et al. (2020). m(6 A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J. 39 (20), e104514. doi:10.15252/embj.2020104514
Wang, T., Kong, S., Tao, M., and Ju, S. (2020). The potential role of RNA N6-methyladenosine in Cancer progression. Mol. Cancer 19 (1), 88. doi:10.1186/s12943-020-01204-7
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2017). Corrigendum: Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 542 (7640), 260. doi:10.1038/nature21073
Wang, Y., Chen, J., Gao, W. Q., and Yang, R. (2022). METTL14 promotes prostate tumorigenesis by inhibiting THBS1 via an m6A-YTHDF2-dependent mechanism. Cell Death Discov. 8 (1), 143. doi:10.1038/s41420-022-00939-0
Wei, W., Sun, J., Zhang, H., Xiao, X., Huang, C., Wang, L., et al. (2021). Circ0008399 interaction with WTAP promotes assembly and activity of the m(6)A methyltransferase complex and promotes cisplatin resistance in bladder cancer. Cancer Res. 81 (24), 6142–6156. doi:10.1158/0008-5472.CAN-21-1518
Wen, L., Pan, X., Yu, Y., and Yang, B. (2020). Down-regulation of FTO promotes proliferation and migration, and protects bladder cancer cells from cisplatin-induced cytotoxicity. BMC Urol. 20 (1), 39. doi:10.1186/s12894-020-00612-7
Wu, Y., Liu, Z., Wei, X., Feng, H., Hu, B., Liu, B., et al. (2021). Identification of the functions and prognostic values of RNA binding proteins in bladder cancer. Front. Genet. 12, 574196. doi:10.3389/fgene.2021.574196
Ma, X.-X., Cao, Z.-G., and Zhao, S.-L. m6A methyltransferase METTL3 promotes the progression of prostate cancer via m6A-modified LEF1. Eur. Rev. Med. Pharmacol. Sci. 2020 24:3565–3571. doi:10.26355/eurrev_202004_20817
Xiang, Z., Xu, C., Wu, G., Liu, B., and Wu, D. (2019). CircRNA-UCK2 increased TET1 inhibits proliferation and invasion of prostate cancer cells via sponge MiRNA-767-5p. Open Med. (Wars) 14, 833–842. doi:10.1515/med-2019-0097
Xiao, L., Tang, J., Huang, W., Wang, F., Pu, L., Qin, C., et al. (2017). The M6A methyltransferase METTL3: Acting as a tumor suppressor in renal cell carcinoma. oncotarget 8 (56), 96103–96116. doi:10.18632/oncotarget.21726
Xie, F., Huang, C., Liu, F., Zhang, H., Xiao, X., Sun, J., et al. (2021). CircPTPRA blocks the recognition of RNA N(6)-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression. Mol. Cancer 20 (1), 68. doi:10.1186/s12943-021-01359-x
Xie, H., Li, J., Ying, Y., Yan, H., Jin, K., Ma, X., et al. (2020). METTL3/YTHDF2 m(6) A axis promotes tumorigenesis by degrading SETD7 and KLF4 mRNAs in bladder cancer. J. Cell Mol. Med. 24 (7), 4092–4104. doi:10.1111/jcmm.15063
Xie, X., Lin, J., Fan, X., Zhong, Y., Chen, Y., Liu, K., et al. (2021). LncRNA CDKN2B-AS1 stabilized by IGF2BP3 drives the malignancy of renal clear cell carcinoma through epigenetically activating NUF2 transcription. Cell Death Dis. 12 (2), 201. doi:10.1038/s41419-021-03489-y
Xing, Q., Jiaochen, L., Shouyong, L., Limin, M., and Wang, Y. (2022). Six RNA binding proteins (RBPs) related prognostic model predicts overall survival for clear cell renal cell carcinoma and is associated with immune infiltration. Bosn. J. Basic Med. Sci. 22 (3), 435–452. doi:10.17305/bjbms.2021.6097
Xiong, W., Zhang, B., Yu, H., Zhu, L., Yi, L., and Jin, X. (2021). RRM2 regulates sensitivity to sunitinib and PD-1 blockade in renal cancer by stabilizing ANXA1 and activating the AKT pathway. Adv. Sci. (Weinh) 8 (18), e2100881. doi:10.1002/advs.202100881
Xu, Y., Zhou, J., Li, L., Yang, W., Zhang, Z., Zhang, K., et al. (2022). FTO-mediated autophagy promotes progression of clear cell renal cell carcinoma via regulating SIK2 mRNA stability. Int. J. Biol. Sci. 18 (15), 5943–5962. doi:10.7150/ijbs.77774
Xu, Z., Chen, S., Liu, R., Chen, H., Xu, B., Xu, W., et al. (2022). Circular RNA circPOLR2A promotes clear cell renal cell carcinoma progression by facilitating the UBE3C-induced ubiquitination of PEBP1 and, thereby, activating the ERK signaling pathway. Mol. Cancer 21 (1), 146. doi:10.1186/s12943-022-01607-8
Yan, R., Dai, W., Wu, R., Huang, H., and Shu, M. (2022). Therapeutic targeting m6A-guided miR-146a-5p signaling contributes to the melittin-induced selective suppression of bladder cancer. Cancer Lett. 534, 215615. doi:10.1016/j.canlet.2022.215615
Yang, F., Jin, H., Que, B., Chao, Y., Zhang, H., Ying, X., et al. (2019). Dynamic m(6)A mRNA methylation reveals the role of METTL3-m(6)A-CDCP1 signaling axis in chemical carcinogenesis. Oncogene 38 (24), 4755–4772. doi:10.1038/s41388-019-0755-0
Yang, W., Xie, L., Wang, P., and Zhuang, C. (2022). MiR-155 regulates m(6)A level and cell progression by targeting FTO in clear cell renal cell carcinoma. Cell Signal 91, 110217. doi:10.1016/j.cellsig.2021.110217
Yankova, E., Blackaby, W., Albertella, M., Rak, J., De Braekeleer, E., Tsagkogeorga, G., et al. (2021). Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593 (7860), 597–601. doi:10.1038/s41586-021-03536-w
Yi, W., Yu, Y., Li, Y., Yang, J., Gao, S., and Xu, L. (2021). The tumor-suppressive effects of alpha-ketoglutarate-dependent dioxygenase FTO via N6-methyladenosine RNA methylation on bladder cancer patients. Bioengineered 12 (1), 5323–5333. doi:10.1080/21655979.2021.1964893
Yi, Y. C., Chen, X. Y., Zhang, J., and Zhu, J. S. (2020). Novel insights into the interplay between m(6)A modification and noncoding RNAs in cancer. Mol. Cancer 19 (1), 121. doi:10.1186/s12943-020-01233-2
Ying, X., Jiang, X., Zhang, H., Liu, B., Huang, Y., Zhu, X., et al. (2020). Programmable N6-methyladenosine modification of CDCP1 mRNA by RCas9-methyltransferase like 3 conjugates promotes bladder cancer development. Mol. Cancer 19 (1), 169. doi:10.1186/s12943-020-01289-0
Ying, Y., Ma, X., Fang, J., Chen, S., Wang, W., Li, J., et al. (2021). EGR2-mediated regulation of m(6)A reader IGF2BP proteins drive RCC tumorigenesis and metastasis via enhancing S1PR3 mRNA stabilization. Cell Death Dis. 12 (8), 750. doi:10.1038/s41419-021-04038-3
Young, M. J., Hsu, K. C., Lin, T. E., Chang, W. C., and Hung, J. J. (2019). The role of ubiquitin-specific peptidases in cancer progression. J. Biomed. Sci. 26 (1), 42. doi:10.1186/s12929-019-0522-0
Yu, H., Yang, X., Tang, J., Si, S., Zhou, Z., Lu, J., et al. (2021). ALKBH5 inhibited cell proliferation and sensitized bladder cancer cells to cisplatin by m6A-ck2α-mediated glycolysis. Mol. Ther. Nucleic Acids 23, 27–41. doi:10.1016/j.omtn.2020.10.031
Yu, Y. Z., Lv, D. J., Wang, C., Song, X. L., Xie, T., Wang, T., et al. (2022). Hsa_circ_0003258 promotes prostate cancer metastasis by complexing with IGF2BP3 and sponging miR-653-5p. Mol. Cancer 21 (1), 12. doi:10.1186/s12943-021-01480-x
Yuan, S., He, S. H., Li, L. Y., Xi, S., Weng, H., Zhang, J. H., et al. (2023). A potassium-chloride co-transporter promotes tumor progression and castration resistance of prostate cancer through m(6)A reader YTHDC1. Cell Death Dis. 14 (1), 7. doi:10.1038/s41419-022-05544-8
Yuan, Y., Du, Y., Wang, L., and Liu, X. (2020). The M6A methyltransferase METTL3 promotes the development and progression of prostate carcinoma via mediating MYC methylation. J. Cancer 11 (12), 3588–3595. doi:10.7150/jca.42338
Zeng, C., Huang, W., Li, Y., and Weng, H. (2020). Roles of METTL3 in cancer: Mechanisms and therapeutic targeting. J. Hematol. Oncol. 13 (1), 117. doi:10.1186/s13045-020-00951-w
Zhang, C., Chen, L., Liu, Y., Huang, J., Liu, A., Xu, Y., et al. (2021). Downregulated METTL14 accumulates BPTF that reinforces super-enhancers and distal lung metastasis via glycolytic reprogramming in renal cell carcinoma. Theranostics 11 (8), 3676–3693. doi:10.7150/thno.55424
Zhang, L., Luo, X., and Qiao, S. (2022). METTL14-mediated N6-methyladenosine modification of Pten mRNA inhibits tumour progression in clear-cell renal cell carcinoma. Br. J. Cancer 127 (1), 30–42. doi:10.1038/s41416-022-01757-y
Zhang, N., Hua, X., Tu, H., Li, J., Zhang, Z., and Max, C. (2021). Isorhapontigenin (ISO) inhibits EMT through FOXO3A/METTL14/VIMENTIN pathway in bladder cancer cells. Cancer Lett. 520, 400–408. doi:10.1016/j.canlet.2021.07.041
Zhang, Q. J., Luan, J. C., Song, L. B., Cong, R., Ji, C. J., Zhou, X., et al. (2020). m6A RNA methylation regulators correlate with malignant progression and have potential predictive values in clear cell renal cell carcinoma. Exp. Cell Res. 392 (1), 112015. doi:10.1016/j.yexcr.2020.112015
Zhang, X., Wang, F., Wang, Z., Yang, X., Yu, H., Si, S., et al. (2020). ALKBH5 promotes the proliferation of renal cell carcinoma by regulating AURKB expression in an m(6)A-dependent manner. Ann. Transl. Med. 8 (10), 646. doi:10.21037/atm-20-3079
Zhang, Y., Yao, Y., Qi, X., Li, J., Liu, M., Che, X., et al. (2021). Identification of a new prognostic risk signature of clear cell renal cell carcinoma based on N(6)-methyladenosine RNA methylation regulators. J. Immunol. Res. 2021, 6617841. doi:10.1155/2021/6617841
Zhao, X., Yang, Y., Sun, B. F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24 (12), 1403–1419. doi:10.1038/cr.2014.151
Zhao, Y., Tao, Z., and Chen, X. (2020). Identification of a three-m6A related gene risk score model as a potential prognostic biomarker in clear cell renal cell carcinoma. PeerJ 8, e8827. doi:10.7717/peerj.8827
Zheng, B., Wang, J., Zhao, G., Chen, X., Yao, Z., Niu, Z., et al. (2021). A new m6A methylation-related gene signature for prognostic value in patient with urothelial carcinoma of the bladder. Biosci. Rep. 41 (4). doi:10.1042/BSR20204456
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49 (1), 18–29. doi:10.1016/j.molcel.2012.10.015
Zhong, C., Long, Z., Yang, T., Wang, S., Zhong, W., Hu, F., et al. (2023). M6A-modified circRBM33 promotes prostate cancer progression via PDHA1-mediated mitochondrial respiration regulation and presents a potential target for ARSI therapy. Int. J. Biol. Sci. 19 (5), 1543–1563. doi:10.7150/ijbs.77133
Zhou, G., Yan, K., Liu, J., Gao, L., Jiang, X., and Fan, Y. (2021). FTO promotes tumour proliferation in bladder cancer via the FTO/miR-576/CDK6 axis in an m6A-dependent manner. Cell Death Discov. 7 (1), 329. doi:10.1038/s41420-021-00724-5
Zhu, D., Liu, Y., Chen, J., Wang, Q., Li, Y., Zhu, Y., et al. (2022). The methyltransferase METTL3 promotes tumorigenesis via mediating HHLA2 mRNA m6A modification in human renal cell carcinoma. J. Transl. Med. 20 (1), 298. doi:10.1186/s12967-022-03496-3
Zhu, K., Li, Y., and Xu, Y. (2021). The FTO m(6)A demethylase inhibits the invasion and migration of prostate cancer cells by regulating total m(6)A levels. Life Sci. 271, 119180. doi:10.1016/j.lfs.2021.119180
Zhu, Q., Zhong, A. L., Hu, H., Zhao, J. J., Weng, D. S., Tang, Y., et al. (2020). Acylglycerol kinase promotes tumour growth and metastasis via activating the PI3K/AKT/GSK3β signalling pathway in renal cell carcinoma. J. Hematol. Oncol. 13 (1), 2. doi:10.1186/s13045-019-0840-4
Zhuang, C., Zhuang, C., Luo, X., Huang, X., Yao, L., Li, J., et al. (2019). N6-methyladenosine demethylase FTO suppresses clear cell renal cell carcinoma through a novel FTO-PGC-1α signalling axis. J. Cell Mol. Med. 23 (3), 2163–2173. doi:10.1111/jcmm.14128
Zou, L., Chen, W., Zhou, X., Yang, T., Luo, J., Long, Z., et al. (2022). N6-methyladenosine demethylase FTO suppressed prostate cancer progression by maintaining CLIC4 mRNA stability. Cell Death Discov. 8 (1), 184 doi:10.1038/s41420-022-01003-7
Glossary
Keywords: urologic tumours, N6-methyladenosine, epitranscriptome, posttranscriptional modification, coding RNAs, non-coding RNAs
Citation: Zhu W, Zhao R, Guan X and Wang X (2023) The emerging roles and mechanism of N6-methyladenosine (m6A) modifications in urologic tumours progression. Front. Pharmacol. 14:1192495. doi: 10.3389/fphar.2023.1192495
Received: 23 March 2023; Accepted: 09 May 2023;
Published: 22 May 2023.
Edited by:
Mingyang Li, Fourth Military Medical University, ChinaReviewed by:
Tianqi Xu, Air Force Medical University, ChinaYanru Yang, Air Force Medical University, China
Qiong Wang, Southern Medical University, China
Copyright © 2023 Zhu, Zhao, Guan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu Wang, d194dUBqbHUuZWR1LmNu
 Wenhao Zhu
Wenhao Zhu Renshan Zhao
Renshan Zhao Xiaomin Guan
Xiaomin Guan