
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 17 May 2023
Sec. Pharmacology of Anti-Cancer Drugs
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1192210
This article is part of the Research Topic New Mechanisms for Anti-Cancer Drugs View all 22 articles
 Kang Rong1,2†
Kang Rong1,2† Qingmin He1,2†
Qingmin He1,2† Shu Chen3†
Shu Chen3† Yong Yu1,2
Yong Yu1,2 Lu Mei1,2
Lu Mei1,2 Yang Mi1,2
Yang Mi1,2 Liufan Mu1,2
Liufan Mu1,2 Mingyang Zhu1,2
Mingyang Zhu1,2 Mengjiao Nan1,2
Mengjiao Nan1,2 Xiaoyang Zhang1,2
Xiaoyang Zhang1,2 Zhaoyang Wan1,2
Zhaoyang Wan1,2 Huang Huang1,2*
Huang Huang1,2* Pengyuan Zheng1,2*
Pengyuan Zheng1,2*Objective: Colorectal cancer (CRC) is a common cancer that cannot be detected at an early stage and is a major challenge in oncology research. Studies have shown that vitamin D3 has some anti-cancer and preventive effects on colorectal cancer, but the exact anti-cancer mechanism is not clear. We applied the relevant research methods of network pharmacology to speculate and validate the possible potential pharmacological mechanisms of vitamin D3 for the prevention of colorectal cancer, and to provide more theoretical support for the clinical anticancer effects of vitamin D3.
Methods: The relevant targets for vitamin D3 and CRC were obtained from the database of drug and disease targets, respectively. The target of vitamin D3 and the target of colorectal cancer were taken to intersect to obtain common targets. Then, the PPI network was constructed. In addition, the pathways of drug-disease interactions were predicted by GO and KEGG enrichment analysis. Finally, the obtained results were verified to ensure the reliability of the experiments.
Results: 51 targets of vitamin D3 for the prevention of colorectal cancer were obtained. The 10 core targets were obtained from the PPI network. The 10 core targets include: ALB, SRC, MMP9, PPARG, HSP90AA1, IGF1, EGFR, MAPK1, MAP2K1 and IGF1R. The core targets were further validated by molecular docking and animal experiments. The results suggest that vitamin D3 plays a key role in the prevention of CRC through core targets, PI3K-Akt pathway, HIF-1 pathway, and FoxO pathway.
Conclusion: This study will provide more theoretical support for vitamin D3 to reduce the incidence of CRC and is important to explore more pharmacological effects of vitamin D3.
Colorectal cancer (CRC) is a malignant tumor that has the third highest incidence rate among cancers and the second highest mortality rate among malignant tumors (Sung et al., 2021). In China, CRC ranks fifth in the incidence of malignant tumors in men and fourth in the incidence of malignant tumors in women. CRC ranks fifth in both male malignancy mortality and female malignancy mortality (Chen et al., 2016). Despite extensive research, the pathogenesis of CRC remains unclear (Thanikachalam and Khan, 2019). With the improvement of treatment methods and medical devices, the risk of CRC has already decreased, but the disease burden of CRC is still heavy, and more prevention strategies are needed (Schoen et al., 2012; Nishihara et al., 2013; Shaukat et al., 2013; Fakih, 2015; Doubeni et al., 2018; Levin et al., 2018). The concept of chemoprevention was first introduced in 1976. Chemoprevention of cancer usually involves the use of synthetic drugs to reduce the incidence of cancer and even reverse pre-cancerous and carcinogenic processes (Sporn, 1976). Currently, drugs that can have a chemopreventive effect on CRC include statins, drugs that target metabolic pathways, aspirin,non-aspirin non-steroidal anti-inflammatory drugs, and vitamins and minerals (Katona and Weiss, 2020). Vitamin D maintains relatively stable levels of calcium in the body’s serum, and calcium salts deposited in certain areas contribute to the formation of bone. Several epidemiological studies have shown that patients with inadequate vitamin D intake have an increased risk of developing cancer (Holick, 2004; Bikle, 2016; Wang et al., 2017). In addition, studies have shown that vitamin D has a specific inhibitory effect on cancer (Harris and Go, 2004; Fleet et al., 2012; Feldman et al., 2014; Giammanco et al., 2015). The vitamin D receptor is expressed in a variety of cells in the body. By targeting the vitamin D receptor to initiate intracellular signaling regulatory mechanisms, it regulates complex intracellular signaling and ultimately inhibits CRC (Leyssens et al., 2013; Baron JA et al., 2015). Studies have shown that patients with low levels of vitamin D in the serum are more likely to develop CRC. Thus, vitamin D supplementation has a role in improving patient prognosis and preventing the development of CRC (Terry et al., 2002; Mccullough et al., 2003; Ma et al., 2011), although these results are based primarily on studies of the distal colon and rectum (Zheng et al., 1998; Wu et al., 2002). This study reviewed the literature related to vitamin D and CRC and ultimately concluded that vitamin D has an inhibitory effect on CRC. However, the pathway is still unclear and more research is needed.
The research process is shown (Figure 1), and the databases involved are listed (Table 1).
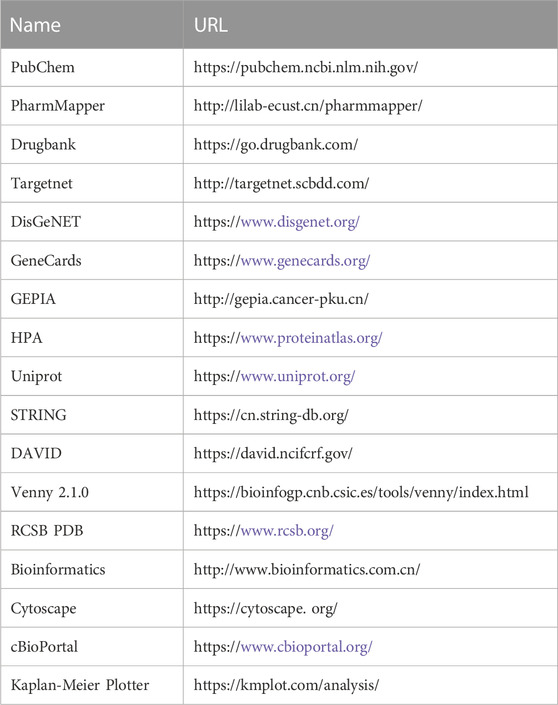
TABLE 1. Website information of the database involved in the study of vitamin D3 prevention of colorectal cancer.
Vitamin D3 is a class of steroid hormones that are usually synthesized by the body in response to UV radiation or supplemented by food. Studies have shown that vitamin D3 is necessary to maintain calcium homeostasis and bone mineralization in the body. Binding of vitamin D3 and its receptors can regulate specific gene expression and signal transduction in the body, exerting biological effects. The properties of vitamin D3 are shown (Table 2), and the data are from PubChem.
The English name of the drug was input to get its details in PubChem (https://pubchem. ncbi.nlm.nih.gov/) (Kim et al., 2021). The SDF format file of the drug was obtained from PubChem and then imported into PharmMapper (http://lilab-ecust.cn/pharmmapper/) (Liu et al., 2010). In PharmMapper, the screening criteria are as follows: Generate Conformers:Yes, Maximum Generated Conformations:300, Number of Reserved Matched Targets (Max 1,000):300. The canonical smiles of the drug was searched to obtain the target of vitamin D3 in Targetnet (http://targetnet.scbdd.com/) (Yao et al., 2016), and the English name of the drug was searched to obtain the target of vitamin D3 in Drugbank (https://go.drugbank.com/) (Wishart et al., 2018). The targets of the drug were converted into gene names in UniProt (https://www.uniprot.org/) (UniProt Consortium, 2021). Finally, the gene names were merged and de-duplicated to obtain the potential targets of vitamin D3.
The targets were obtained in DisGeNET (https://www.disgenet.org/) (Pinero et al., 2020) and GeneCards (https://www.genecards.org/) (Stelzer et al., 2016). The targets were merged and de-duplicated to be the related targets of CRC.
The targets of vitamin D3 and CRC-related targets were taken intersected in Venny 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/index.html). Finally, the intersecting targets obtained are potential targets of vitamin D3 for CRC prevention. The intersection targets are imported into String (https://cn.string-db.org/) to construct the PPI network. The tsv file of the PPI network was obtained and imported into Cytoscape 3.9.0 (https://cytoscape. org/; Version 3.9.0) (Doncheva et al., 2019) for analysis and visualization. In cytohubba, the core targets are obtained according to the importance of the nodes. The core targets were visualized.
Core targets were imported into David (https://david.ncifcrf.gov/) (Huang et al., 2007) to obtain the results of GO and KEGG enrichment analysis (Ashburner et al., 2000; Kanehisa et al., 2021). The screening criteria were p < 0.05 and the enriched signaling pathway contained multiple core targets. The first 10 entries of BP, CC and MF were selected in this study, respectively. The collated data were imported into the bioinformatics (http://www.bioinformatics. com. cn/) for visualization and analysis.
p < 0.05 was used as a screening criterion, and the top 20 signaling pathways were selected based on the ranking of p-values from smallest to largest. Data of vitamin D3, signaling pathways and core targets were made into network. xlsx and type. xlsx and the files were imported into Cytoscape for visualization. The vitamin D3-core target-pathway network was constructed. The red triangle represents vitamin D3, the blue diamond represents the core target, and the green circle represents the signaling pathway.
To explore the possible mechanisms of vitamin D3 action on CRC. The strength and pattern of interactions between vitamin D3 and core targets were assessed by molecular docking in this study. The PDB ID of the core target was obtained from the RCSB PDB and its 3D structure was downloaded in PDB format. Molecular docking was performed using Discovery studio (2019 version) and the “Prepare Ligands” module was used to obtain 3D structures in this study. After removing the water of crystallization, the “Prepare Protein” module is used to remove multiple conformations of the target protein by adding hydrogen atoms. Then, the fast molecular docking option is selected in the “LibDock” module. The software calculates the corresponding docking scores in the columns of the LibDockScore. The docking score as a judgment criterion was used to evaluate the affinity of the target protein and the drug.
To verify the reliability of molecular docking, the original ligands of 10 target protein complexes were separated and then reattached to the pockets of the complexes in this study. The root-means-square deviation (RMSD) of the ligand conformation after ligand docking and the ligand conformation in the original crystal structure were calculated. The RMSD value less than 2.0Å indicates that the docking method can well reproduce the original ligand-receptor binding mode and the docking parameters are reasonably set.
In GEPIA (http://gepia.cancer-pku.cn), the expression information of the core targets were obtained. The relevant information of core targets for different populations and different stages of cancer were obtained in the “Expression DIY” module (Tang et al., 2017).
Differences of core targets were studied at the protein expression level. The relevant data from the Human Protein Atlas database (https://www.proteinatlas.org/) (Uhlen et al., 2015) were analyzed. The core targets were analyzed and compared using the relevant data from this database to make further conclusions.
The 105 colon cancer (CPTAC - 2 prospective, cell 2019) samples in the database were analyzed using cBioPortal (https://www.cbioportal.org/) (Wu et al., 2019). We obtained the genetic information of the core targets (He et al., 2023). In addition, the mutation sites and mutation types of the core targets were obtained from this database.
In GEPIA, the “General” module was selected and the core targets were entered to obtain the distribution and expression levels of the core targets in different cancers. The obtained results were presented in the form of a dot plot.
To explore the impact of core targets on overall survival in CRC, Kaplan-Meier Plotter (https://kmplot.com/analysis/) was accessed. After the core targets were entered, the results of the overall survival of each target were obtained in this database.
Eighteen 7-week-old male C57BL/6N mice were procured from Beijing Weitong Lihua Experimental Animal Technology Co. The mice were randomly divided into control group, model group and vitamin D3 intervention group, 6 mice in each group. The experimental conditions included constant temperature, pressure and humidity, and 12 h of light per day. Mice were fed with Co60 irradiated sterilized maintenance feed. The animal experiments were approved by the Animal Ethics Committee of the Fifth Affiliated Hospital of Zhengzhou University (Ethics No: KY2023007).
Vitamin D3 (Item No.: B2065) was purchased from APExBIO(Houston,TX). Azoxymethane (AOM, item number: 0218397125) was purchased from Abe Medical Device Trading (Shanghai) Co., LTD., and Dextran sulfate sodium (DSS, item number MB5535) was purchased from Meilan Biotechnology (Dalian) Co., LTD. RNAiso Plus was purchased from Takara bio, Japan, and RIPA cell lysate and protease inhibitors were purchased from Dingguo Changsheng Biotechnology (Beijing) Co., LTD. Protein phosphatase inhibitors were purchased from bimake Biotechnology Co., LTD. HiScript®II 1st Strand cDNA Synthesis Kit and ChamQ universal SYBR qPCR Master Mix were purchased from Novizam Biotechnology (Nanjing) Co., LTD. The corresponding primers of the core target were synthesized by Sangon Biotech (Shanghai).
A completely randomized design was used to divide the mice into three groups. Physiological saline control group (Nacl): 6 mice, model group (AOM + DSS): 6 mice, vitamin D3 intervention group (AOM + DSS + vitaminD3): 6 mice. After 1 week of adaptive feeding, the mice were gavaged with vitamin D for 2 continuous weeks at a dose of 60 IU/g per week, 3 times per week. At week 4, mice of the model and vitamin D3 intervention groups were given a single intraperitoneal injection of AOM (10 mg/kg). At weeks 5, 8 and 11, the model and vitamin D3 intervention groups were given drinking water containing 2% DSS for 5 days and normal drinking water for the rest of the day. The mice were weighed twice a week. Mice were sacrificed at week 15, and colorectal tissues were collected for RT-qPCR experiments.
Gene expression of the core targets were detected in colorectal tissue by RT-qPCR. Total RNA was extracted from mouse intestinal tissue following the tissue RNA extraction protocol. The purity of RNA was detected by NanoDrop ND-2000 spectrophotometer and the ratio of A260/A280 was recorded. The reverse transcription reactions were performed on a common PCR instrument using the HiScript®II 1st Strand cDNA Synthesis Kit. Real-time fluorescence quantitative PCR was performed on a Light Cycler 480II real-time fluorescence quantitative PCR instrument. The 2−ΔΔCT method was used to calculate the expression of core targets. The corresponding primer sequences of the core targets were listed (Table 3).
221 targets of vitamin D3 were obtained in PharmMapper, DrugBank and Targetnet databases. 1205 CRC-related targets were obtained in the DisGeNET and GeneCards databases. The targets of vitamin D3 and CRC-related targets were imported into Venny 2.1.0, and 51 intersecting targets were obtained (Figure 2A).
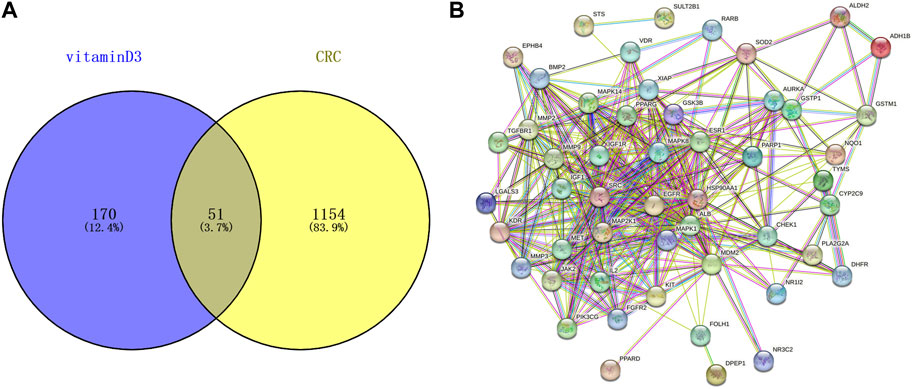
FIGURE 2. Information related to CRC chemoprevention. (A) Venn diagram of intersection targets between vitamin D3 and CRC (B) Establishment of PPI network for intersection targets.
51 intersecting targets were input to String, and a PPI network (Figure 2B) containing 51 nodes and 369 edges was established. The tsv file of the PPI network was downloaded and imported into Cytoscape 3.9.0 for visualization and analysis (Figure 3A). The intersecting targets were ranked from largest to smallest according to the degree value using the Cytohubba plugin. The top 10 intersecting targets were selected as core targets for vitamin D3 prevention of CRC. The core targets are visualized (Figure 3B). The circles represent the core targets for screening, and the redder the color of the circle, the greater the importance (Chin et al., 2014). The 10 core targets include ALB, SRC, MMP9, PPARG, HSP90AA1, IGF1, EGFR, MAPK1, MAP2K1 and IGF1R.
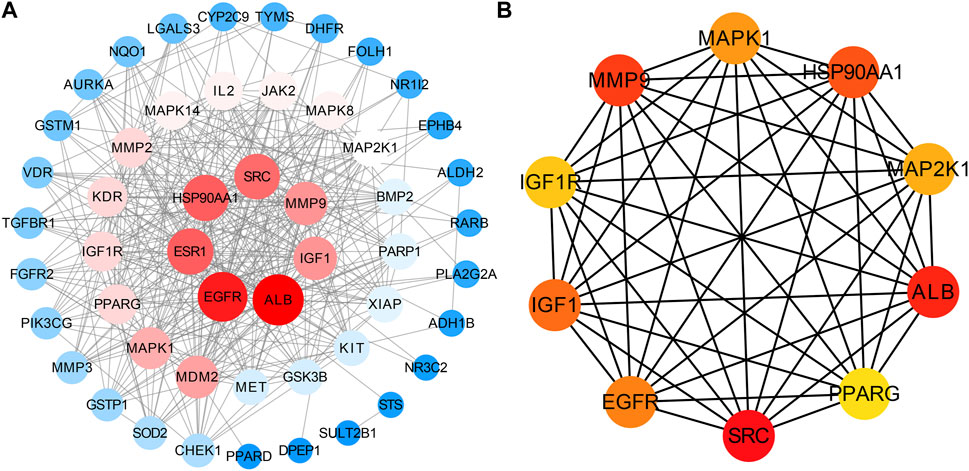
FIGURE 3. Visual results of intersection and core targets. (A) Graphical display of intersection targets (B) Visualization of core target genes.
Core targets were imported into DAVID for GO and KEGG enrichment analysis. The results of GO enrichment analysis of core targets included 134 entries, BP(Biological Process):90, CC(Cellular Component):23, MF(Molecular Function):21. The top 10 items in each category were selected for visual analysis (Figure 4A). The biological processes involved in the 10 core targets were mainly endosome transport, peptidyl-tyrosine autophosphorylation, and positive regulation of DNA binding. They mainly act on the alphav-beta3 integrin-IGF-1-IGF1R complex, dendritic growth cone. They are involved in molecular functions including insulin receptor binding, scaffold protein binding, protein tyrosine kinase activity, etc. 80 signaling pathways were identified by KEGG enrichment analysis of core targets, and 20 important signaling pathways were visualized and analyzed (Figure 4B). The main signaling pathways involved include the HIF-1 signaling pathway, PI3K-Akt signaling pathway, etc. The results of KEGG enrichment analysis suggest that vitamin D3 may prevent CRC mainly through the above pathway.
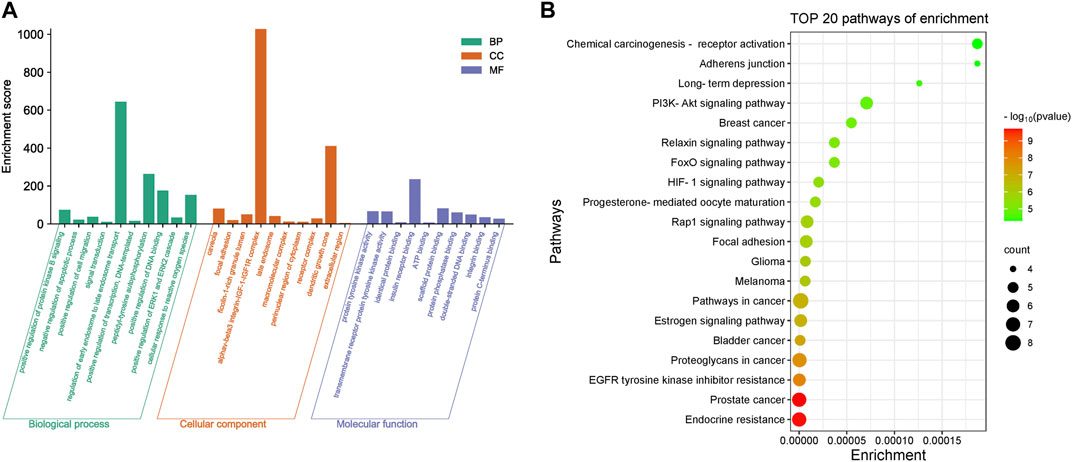
FIGURE 4. Enrichment results of core targets (A). Visualization results of GO enrichment analysis. (B) Visualization results of KEGG enrichment analysis.
The core targets, vitamin D3 and KEGG enrichment pathways were categorized and organized to construct the drug-target-pathway network in Cytoscape 3.9.0 (Figure 5). In Figure 5, the triangle represents vitamin D3, the circle represents the core target, and the diamond represents the pathway in which the core target was enriched.
To investigate the interaction between the 10 core targets and vitamin D3, molecular docking was used. The relevant information of 10 core target proteins was downloaded from the PDB database (Table 4). First, the 3D structure file of vitaminD3 was uploaded to Discovery Studio 2019; then, its docking potential with ALB, SRC, MMP9, PPARG, HSP90AA1, IGF1, EGFR, MAPK1, MAP2K1 and IGF1R were analyzed. The results of molecular docking scores were obtained. The root-mean-square deviation (RMSD) of the ligand conformation after docking and the ligand conformation of the original crystal structure were calculated. It is generally considered that when the RMSD value is less than 2.0, it proves that the docking method can better reproduce the original ligand-receptor binding mode and the docking parameters are reasonably set (Table 5). All results of RMSD are displayed. The results of core target docking with vitamin D3 are shown, the relationship between vitamin D3 and docking sites was represented in two and three dimensions, respectively (Figure 6A). RMSD validates the molecular docking reliability. In the 2D and 3D structure diagrams, we can observe the type of interaction forces between vitamin D3 and amino acid residues, and the length of the bonds. Validation of the reliability of molecular docking results (Figure 6B). The higher the degree of overlap of different conformations of the same protein, the more reliable the molecular docking results. The RMSD value is generally considered to be less than 2, and molecular docking is more reliable.
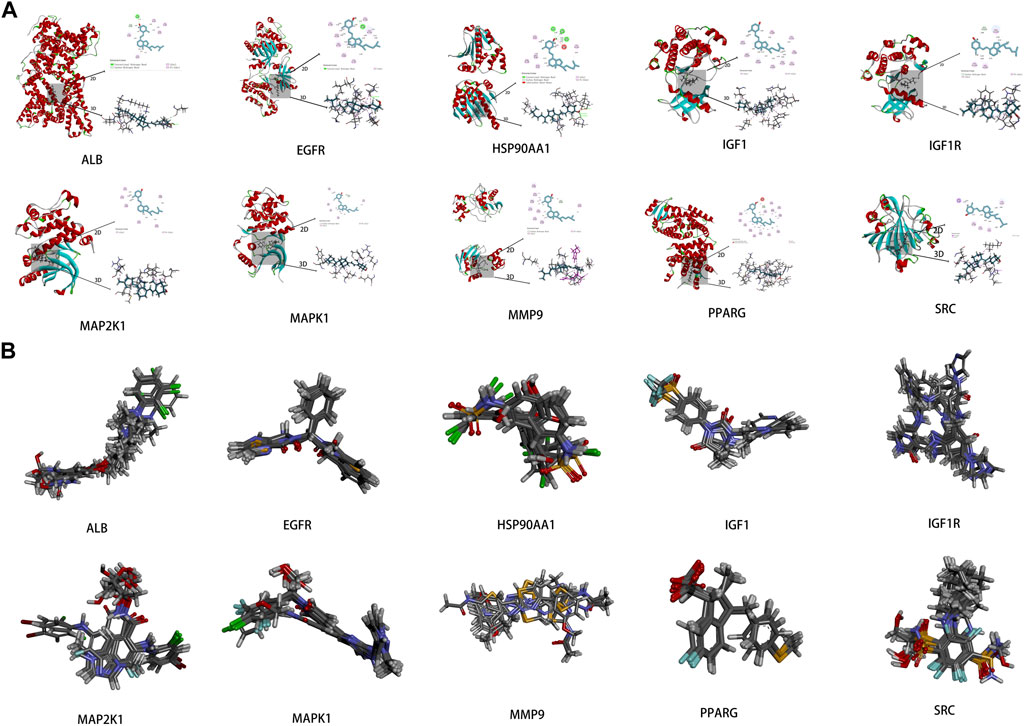
FIGURE 6. Results of molecular docking (A). Schematic diagram of molecular docking of the core targets with vitamin D3 (B). Visualization results of molecular docking RMSD validation.
The results showed that the mRNA expression levels of PPARG, HSP90AA1and MMP9 were higher than normal in CRC(p < 0.05). The mRNA expression level of IGF1 was lower than normal in CRC (p < 0.05) (Figure 7A). The mRNA expression levels of EGFR and SRC correlated with the stages of CRC(p < 0.05) (Figure 7B).
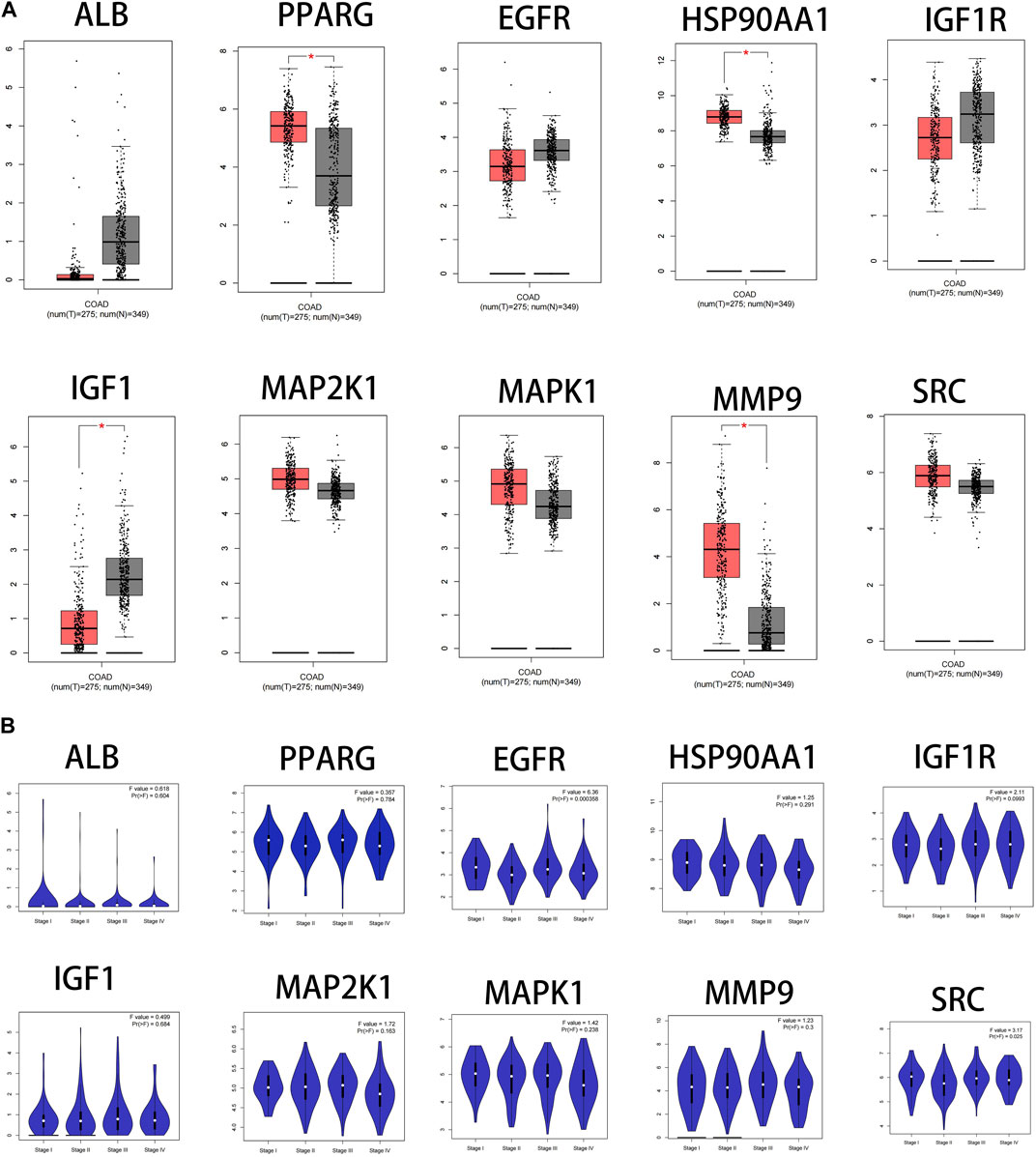
FIGURE 7. Fact sheet of the core targets on the GEPIA website. (A) Boxplot results of mRNA expression levels of core targets. Tumor tissue is shown in red and normal tissue is shown in gray. (B) Diagram of the relationship between core target gene expression and CRC cancer stage.
To understand the expression of core targets in CRC, the relevant immunohistochemical images were analyzed by HPA database. The protein levels of PPARG and MMP9 were found to be high in CRC by comparative analysis. The protein expression of ALB, IGF1R, and MAP2K1 was higher in normal tissues than in CRC. The protein expression of EGFR, HSP90AA1, and MAPK1 was equal in normal and CRC. Immunohistochemical data of IGF1 and SRC were not found in the HPA database (Figure 8).
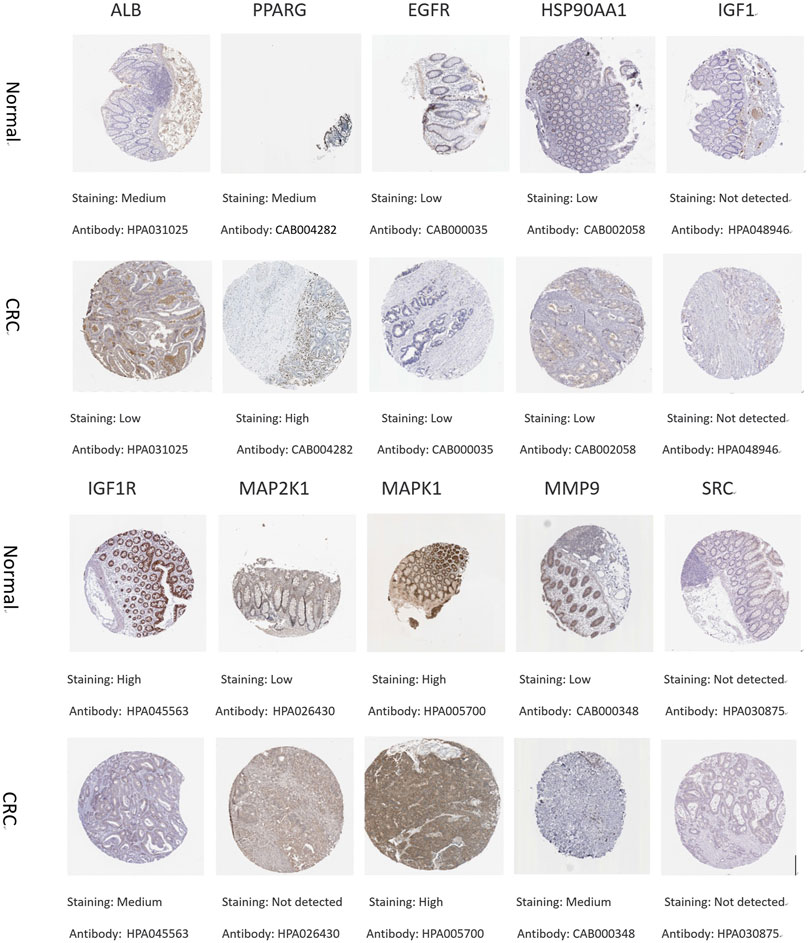
FIGURE 8. Immunohistochemical pictures of core target proteins in normal and CRC patients from the HPA database.
Among the samples with mutation and CNA data (105 samples/patient), a total of 36 samples (34%) had mutations in the relevant genes, including 10 core targets (Figure 9A). It also shows the site and type of mutations in the core target genes in CRC patients (Figure 9B). Some details of the mutations in the core target genes are shown (Table 6).
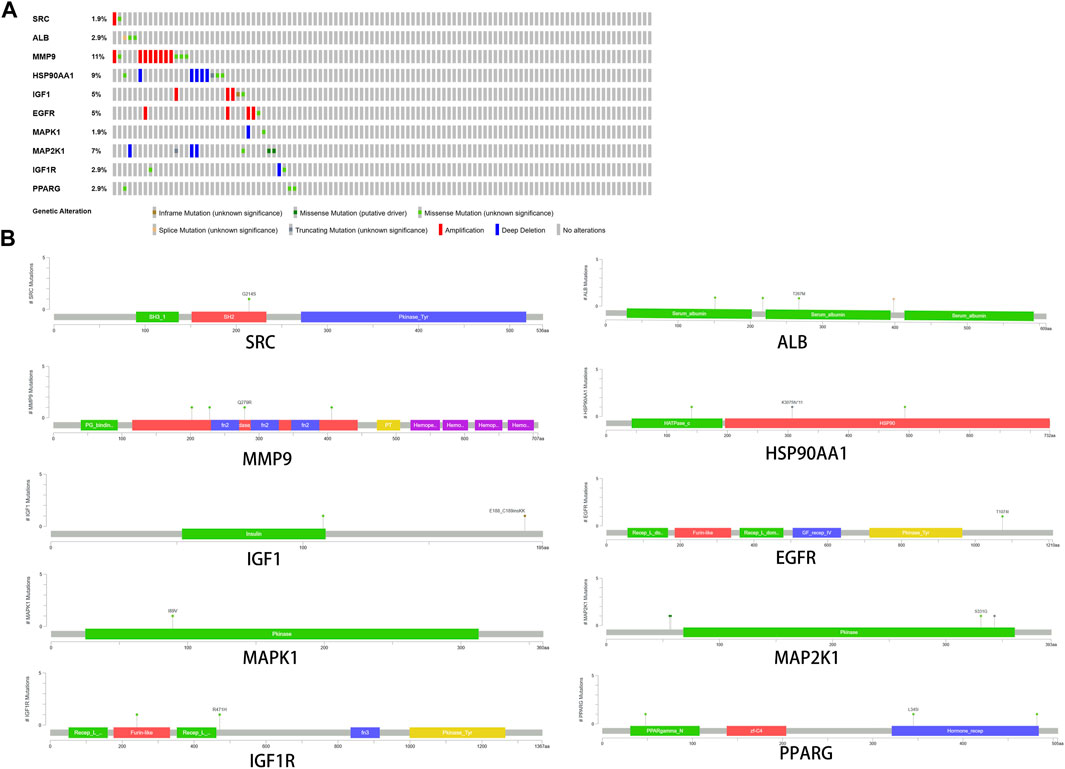
FIGURE 9. Genetic data on core targets. (A) The data suggested that 34 of 105 patients (36%) had a heritable variation on core target genes. (B) Information on the site and type of gene mutations on core targets in CRC patients.
Dot plots of core target protein distribution were obtained in different cancers in the GEPIA database. The results showed that the expression levels of core target proteins differed in different cancers (Figure 10).
The core targets were entered into Kaplan-Meier Plotter for analysis. The effect of expression levels of 10 targets on the overall survival of CRC patients were obtained (Figure 11). The results of survival analysis showed that patients with high expression of ALB,HSP90AA1, PPARG and MAPK1 had a longer survival period than those with low expression, and the difference was statistically significant. Patients with low expression of EGFR,IGF1, IGF1R and SRC in CRC patients had a longer survival time than those with high expression, and the difference was statistically significant. Patients with higher expression of MAP2K1 and MMP9 in CRC had longer survival, but the difference was not statistically significant.
Through the experiment, the following findings were obtained:SRC, IGF1, IGF1R, HSP90AA1, MAPK1 and MMP9 are high expression in CRC tissues and after the intervention of vitamin D3, the expression of these genes was reduced in cancer tissues, and the differences were statistically significant (p < 0.05). The expression of EGFR is high in CRC tissues and decreased in CRC tissues after vitamin D3 intervention, but the difference was not statistically significant (p > 0.05). ALB, MAP2K1, and PPARG are low expression in CRC tissues, and their expression levels increased after the intervention of vitamin D3, which indicates that the treatment of vitamin D3 is effective and the difference is statistically significant (p < 0.05). The results of statistical analysis of RT-qPCR experiments are shown (Figure 12).
The disease burden of CRC has been high and has increased in Asia in recent years (Onyoh et al., 2019). Vitamin D3 is a small molecule compound. More and more epidemiological studies have shown that vitamin D3 has a more prominent role in preventing cancer, especially CRC. The chemopreventive effect of vitamin D3 on CRC has been reported (Lamprecht and Lipkin, 2003). However, the pathogenesis of cancer is complex and the chemical nature of the drug is not fully clear, so the molecular mechanism of the interaction between vitamin D3 and CRC needs to be further investigated.
In this study, we further investigated the molecular mechanism of vitamin D3 in the prevention of CRC through network pharmacology and animal experimental studies. The results of the enrichment analysis suggest that vitamin D3 may play a role in the prevention of CRC through the PI3K-Akt signaling pathway and the HIF signaling pathway. A study showed that vitamin D3 can inhibit cell proliferation, invasion, and metastasis and promote apoptosis through the PI3K-Akt pathway (Songyang et al., 2021). Vitamin D3 can also interfere with the metabolism of cancer cells, inhibit the growth and proliferation of cancer cells, and play an anti-cancer role through the HIF signaling pathway (Gkotinakou et al., 2022).
The mechanisms of action of vitamin D include inhibition of VDR-mediated IGF1, HIF1, NF-κB and PI3K/AKT activities, or activation of MAPK family-related genes, downregulation of cell cycle progression, anti-inflammation, inhibition of angiogenesis, induction of apoptosis, and other anti-tumor effects (Skrajnowska and Bobrowska-Korczak, 2019). Solid tumor cells usually exhibit reduced oxygen levels depending on the rate of cell proliferation, vascular abnormalities, and the distance to the area surrounding the oxygen-containing vessels (Semenza, 2021). The growth and reproduction of cancer cells depend on a moderate supply of oxygen, so a hypoxic microenvironment would significantly affect the biological behavior of cancer cells during their life course. Under hypoxic conditions, the HIF signaling pathway is activated, causing transcription of the corresponding target genes and promoting the development of tumors. Thus, overexpression of HIF-1α can induce increased mortality in many common cancers (Keith et al., 2011). Activation of HIFs induces a series of cascade responses, including alteration of the metabolism of cancer cells and inhibition of the apoptosis of cancer cells. Under the combined effect of these factors, the proliferation, angiogenesis, immune escape, epithelial-mesenchymal transition and the ability of extracellular matrix remodeling of cancer cells are enhanced (Mylonis et al., 2021; Semenza, 2021). The existing evidence suggests that ossifying triol can act on the signaling process of HIF pathway, blocking signal transduction, promoting apoptosis of cancer cells and exerting tumor suppressive effects (Gkotinakou et al., 2022). Mitogen-activated protein kinase (MAPK) is an important member of the serine-threonine kinase family and is the main cell proliferation signaling pathway from the cell surface to the nucleus (Dong et al., 2002).
A growing number of evidences suggest that activation of ERK MAPK pathway is closely associated with the occurrence, development and oncogenic behavior of human CRC (Wang et al., 2004). The antitumor effects of vitamin D in CRC are achieved through various pathways such as promoting cell cycle arrest in the G1 phase, reducing the synthesis of vascular endothelial growth factor (VEGF), and decreasing the ability of cell migration and angiogenesis. Meanwhile, IGF1 and IGF1R can induce cell proliferation, promote the transcription and expression of vascular endothelial growth factor genes, promote angiogenesis, provide more nutrients for tumor growth, and promote the further development of CRC to advanced stages. The effect of vitamin D on the G1/S cell cycle transition is opposite to that of IGF-1 and also has antagonistic effects on angiogenesis and the tumor microenvironment. Therefore, this evidence may provide new ideas on the mechanism of the prevention of vitamin D in CRC (Ciulei et al., 2020). The results of RT-qPCR from animal experiments in this study also indicated that vitamin D has a therapeutic and preventive effect on CRC.
These results suggest that vitamin D may be an effective chemical for the prevention of CRC. In summary, ALB, SRC, MMP9, PPARG, HSP90AA1, IGF1, EGFR, MAPK1, MAP2K1 and IGF1R play important roles in the pathogenesis of CRC. Therefore, based on the network pharmacological analysis, it is expected to discover more novel compounds that can prevent cancer or play an anti-cancer role. This study provides additional theoretical basis for the prevention of CRC with vitamin D3.
In this study, vitamin D was found to be a multi-target medicine for the prevention of CRC. We predict that the main mechanism of vitamin D for the prevention of CRC is the regulation of tumor cell proliferation, apoptosis, migration and angiogenesis through signaling pathways such as HIF-1, FoxO and PI3K-Akt, thus playing a role in the prevention of CRC.
Based on network pharmacological analysis, this study predicted 10 potential core targets of vitamin D3 for the prevention of CRC, suggesting that vitamin D3 is a single-component, multi-target, multi-pathway small molecule compound. Network pharmacological analysis suggests that vitamin D3 may play a role in CRC chemoprevention by regulating different targets, such as MAPK, IGF1 and IGF1R. Graphene oxide analysis of the core targets ALB, SRC, MMP9, PPARG, HSP90AA1, IGF1, EGFR, MAPK1, MAP2K1 and IGF1R showed that vitamin D3 exerts pharmacological effects by affecting different biological processes, such as endosomal transport and positive regulation of protein kinase B signaling. The analysis of KEGG pathway in this study suggests that vitamin D3 may exert pharmacological effects by simultaneously regulating different CRC-related signaling pathways, such as FoxO signaling pathway, HIF-1 signaling pathway, PI3K-Akt signaling pathway, etc. These results support that vitamin D3 may mediate the inhibition of HIF-1 through inhibition of the upstream PI3K-AKT pathway.
In summary, vitamin D3 may be a new safe, effective, multi-targeted medicine for the prevention of CRC. This study concludes from a network pharmacological analysis that vitamin D3 may play a preventive role in CRC by regulating cell metabolism, proliferation and apoptosis through multiple targets, pathways and biological processes. However, the clinical efficacy of vitamin D3 in the prevention of CRC and its mechanism of action need to be further validated.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by the Ethics Committee of the Fifth Affiliated Hospital of Zhengzhou University (KY2023007). Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Conception and design KR and SC; Administrative support HH; Provision of study materials or patients: KR and SC; Collection and assembly of data KR, SC, LM, and MN; Data analysis and interpretation KR, SC, and LM; Manuscript writing. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Medical Science and Technology Research Program of Henan Province (No: 2018020228).
We like to thank the editors and the reviewers for their valuable comments and suggestions to improve the quality of the paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2023.1192210/full#supplementary-material
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25 (1), 25–29. doi:10.1038/75556
Baron, J. .A., Barry, E. L., Mott, L. A., Rees, J. R., Sandler, R. S., Snover, D. C., et al. (2015). A trial of calcium and vitamin D for the prevention of colorectal adenomas. N. Engl. J. Med. 373 (16), 1519–1530. doi:10.1056/NEJMoa1500409
Bikle, D. D. (2016). Extraskeletal actions of vitamin D. Ann. N. Y. Acad. Sci. 1376 (1), 29–52. doi:10.1111/nyas.13219
Chen, W., Zheng, R., Baade, P. D., Zhang, S., Zeng, H., Bray, F., et al. (2016). Cancer statistics in China, 2015. CA Cancer J. Clin. 66 (2), 115–132. doi:10.3322/caac.21338
Chin, C. H., Chen, S. H., Wu, H. H., et al. (2014). cytoHubba: identifying hub objects and sub-networks from complex interactome [J]. BMC Syst. Biol. 8 (Suppl. 4), s11.
Ciulei, G., Orasan, O. H., Coste, S. C., Cozma, A., Negrean, V., and Procopciuc, L. M. (2020). Vitamin D and the insulin-like growth factor system: Implications for colorectal neoplasia. Eur. J. Clin. Invest. 50 (9), e13265. doi:10.1111/eci.13265
Doncheva, N. T., Morris, J. H., Gorodkin, J., and Jensen, L. J. (2019). Cytoscape StringApp: Network analysis and visualization of proteomics data. J. Proteome Res. 18 (2), 623–632. doi:10.1021/acs.jproteome.8b00702
Dong, C., Davis, R. J., and Flavell, R. A. (2002). MAP kinases in the immune response. Annu. Rev. Immunol. 20, 55–72. doi:10.1146/annurev.immunol.20.091301.131133
Doubeni, C. A., Corley, D. A., Quinn, V. P., Jensen, C. D., Zauber, A. G., Goodman, M., et al. (2018). Effectiveness of screening colonoscopy in reducing the risk of death from right and left colon cancer: A large community-based study. Gut 67 (2), 291–298. doi:10.1136/gutjnl-2016-312712
Fakih, M. G. (2015). Metastatic colorectal cancer: Current state and future directions. J. Clin. Oncol. 33 (16), 1809–1824. doi:10.1200/JCO.2014.59.7633
Feldman, D., Krishnan, A. V., Swami, S., Giovannucci, E., and Feldman, B. J. (2014). The role of vitamin D in reducing cancer risk and progression. Nat. Rev. Cancer. 14 (5), 342–357. doi:10.1038/nrc3691
Fleet, J. C., Desmet, M., Johnson, R., and Li, Y. (2012). Vitamin D and cancer: A review of molecular mechanisms. Biochem. J. 441 (1), 61–76. doi:10.1042/BJ20110744
Giammanco, M., Di Majo, D., La Guardia, M., Aiello, S., Crescimannno, M., Flandina, C., et al. (2015). Vitamin D in cancer chemoprevention. Pharm. Biol. 53 (10), 1399–1434. doi:10.3109/13880209.2014.988274
Gkotinakou, I. M., Mylonis, I., and Tsakalof, A. (2022). Vitamin D and hypoxia: Points of interplay in cancer. Cancers (Basel) 14 (7), 1791. doi:10.3390/cancers14071791
Harris, D. M., and Go, V. L. (2004). Vitamin D and colon carcinogenesis. J. Nutr. 134, 3463S–3471S. doi:10.1093/jn/134.12.3463S
He, Q., Liu, C., Wang, X., Rong, K., Zhu, M., Duan, L., et al. (2023). Exploring the mechanism of curcumin in the treatment of colon cancer based on network pharmacology and molecular docking. Front. Pharmacol. 14, 1102581. doi:10.3389/fphar.2023.1102581
Holick, M. F. (2004). Sunlight and vitamin D for bone health and prevention of autoimmune diseases, cancers, and cardiovascular disease. Am. J. Clin. Nutr. 80, 1678S–88S. doi:10.1093/ajcn/80.6.1678S
Huang, D. W., Sherman, B. T., Tan, Q., Kir, J., Liu, D., Bryant, D., et al. (2007). DAVID bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35, W169–W175. doi:10.1093/nar/gkm415
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M., and Tanabe, M. (2021). KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 49 (D1), D545–D551. doi:10.1093/nar/gkaa970
Katona, B. W., and Weiss, J. M. (2020). Chemoprevention of colorectal cancer. Gastroenterology 158 (2), 368–388. doi:10.1053/j.gastro.2019.06.047
Keith, B., Johnson, R. S., and Simon, M. C. (2011). HIF1α and HIF2α: Sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer. 12 (1), 9–22. doi:10.1038/nrc3183
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., et al. (2021). PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 49 (D1), D1388–D1395. doi:10.1093/nar/gkaa971
Lamprecht, S. A., and Lipkin, M. (2003). Chemoprevention of colon cancer by calcium, vitamin D and folate: Molecular mechanisms. Nat. Rev. Cancer. 3 (8), 601–614. doi:10.1038/nrc1144
Levin, T. R., Corley, D. A., Jensen, C. D., Schottinger, J. E., Quinn, V. P., Zauber, A. G., et al. (2018). Effects of organized colorectal cancer screening on cancer incidence and mortality in a large community-based population. Gastroenterology 155 (5), 1383–1391. doi:10.1053/j.gastro.2018.07.017
Leyssens, C., Verlinden, L., and Verstuyf, A. (2013). Antineoplastic effects of 1,25(OH)2D3 and its analogs in breast, prostate and colorectal cancer. Endocr. Relat. Cancer 20 (2), R31–R47. doi:10.1530/ERC-12-0381
Liu, X., Ouyang, S., Yu, B., Liu, Y., Huang, K., Gong, J., et al. (2010). PharmMapper server: A web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 38, W609–W614. doi:10.1093/nar/gkq300
Ma, Y., Zhang, P., Wang, F., Yang, J., Liu, Z., and Qin, H. (2011). Association between vitamin D and risk of colorectal cancer: A systematic review of prospective studies. J. Clin. Oncol. 29 (28), 3775–3782. doi:10.1200/JCO.2011.35.7566
Mccullough, M. L., Robertson, A. S., Rodriguez, C., Jacobs, E. J., Chao, A., Carolyn, J., et al. (2003). Calcium, vitamin D, dairy products, and risk of colorectal cancer in the Cancer Prevention Study II Nutrition Cohort (United States). Cancer Causes Control 14 (1), 1–12. doi:10.1023/a:1022591007673
Mylonis, I., Chachami, G., and Simos, G. (2021). Specific inhibition of HIF activity: Can peptides lead the way? Cancers (Basel) 13 (3), 410. doi:10.3390/cancers13030410
Nishihara, R., Wu, K., Lochhead, P., Morikawa, T., Liao, X., Qian, Z. R., et al. (2013). Long-term colorectal-cancer incidence and mortality after lower endoscopy. N. Engl. J. Med. 369 (12), 1095–1105. doi:10.1056/NEJMoa1301969
Onyoh, E. F., Hsu, W. F., Chang, L. C., Lee, Y. C., Wu, M. S., and Chiu, H. M. (2019). The rise of colorectal cancer in Asia: Epidemiology, screening, and management. Curr. Gastroenterol. Rep. 21 (8), 36. doi:10.1007/s11894-019-0703-8
Pinero, J., Ramirez-Anguita, J. M., Sauch-Pitarch, J., Ronzano, F., Centeno, E., Sanz, F., et al. (2020). The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 48 (D1), D845–D855. doi:10.1093/nar/gkz1021
Schoen, R. E., Pinsky, P. F., Weissfeld, J. L., Yokochi, L. A., Church, T., Laiyemo, A. O., et al. (2012). Colorectal-cancer incidence and mortality with screening flexible sigmoidoscopy. N. Engl. J. Med. 366 (25), 2345–2357. doi:10.1056/NEJMoa1114635
Semenza, G. L. (2021). Heritable disorders of oxygen sensing. Am. J. Med. Genet. A 185 (8), 2576–2581. doi:10.1002/ajmg.a.62250
Shaukat, A., Mongin, S. J., Geisser, M. S., Lederle, F. A., Bond, J. H., Mandel, J. S., et al. (2013). Long-term mortality after screening for colorectal cancer. N. Engl. J. Med. 369 (12), 1106–1114. doi:10.1056/NEJMoa1300720
Skrajnowska, D., and Bobrowska-Korczak, B. (2019). Potential molecular mechanisms of the anti-cancer activity of vitamin D. Anticancer Res. 39 (7), 3353–3363. doi:10.21873/anticanres.13478
Songyang, Y., Song, T., Shi, Z., Li, W., Yang, S., and Li, D. (2021). Effect of vitamin D on malignant behavior of non-small cell lung cancer cells. Gene 768, 145309. doi:10.1016/j.gene.2020.145309
Sporn, M. B. (1976). Approaches to prevention of epithelial cancer during the preneoplastic period. Cancer Res. 36 (7 PT 2), 2699–2702.
Stelzer, G., Rosen, N., Plaschkes, I., Zimmerman, S., Twik, M., Fishilevich, S., et al. (2016). The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinforma. 54, 1.30.1–1.30.33. doi:10.1002/cpbi.5
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Tang, Z., Li, C., Kang, B., Gao, G., Li, C., and Zhang, Z. (2017). GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45 (W1), W98–W102. doi:10.1093/nar/gkx247
Terry, P., Baron, J. .A., Bergkvist, L., Holmberg, L., and Wolk, A. (2002). Dietary calcium and vitamin D intake and risk of colorectal cancer: A prospective cohort study in women. Nutr. Cancer. 43 (1), 39–46. doi:10.1207/S15327914NC431_4
Thanikachalam, K., and Khan, G. (2019). Colorectal cancer and nutrition. Nutrients 11 (1), 164. doi:10.3390/nu11010164
Uhlen, M., Fagerberg, L., Hallstrom, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347 (6220), 1260419. doi:10.1126/science.1260419
UniProt Consortium (2021). UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 49 (D1), D480–D489. doi:10.1093/nar/gkaa1100
Wang, X., Wang, Q., Hu, W., and Evers, B. M. (2004). Regulation of phorbol ester-mediated TRAF1 induction in human colon cancer cells through a PKC/RAF/ERK/NF-kappaB-dependent pathway. Oncogene 23 (10), 1885–1895. doi:10.1038/sj.onc.1207312
Wang, H., Chen, W., Li, D., Yin, X., Zhang, X., Olsen, N., et al. (2017). Vitamin D and chronic diseases. Aging Dis. 8 (3), 346–353. doi:10.14336/AD.2016.1021
Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., et al. (2018). DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 46 (D1), D1074–D1082. doi:10.1093/nar/gkx1037
Wu, K., Willett, W. C., Fuchs, C. S., Colditz, G. A., and Giovannucci, E. L. (2002). Calcium intake and risk of colon cancer in women and men. J. Natl. Cancer Inst. 94 (6), 437–446. doi:10.1093/jnci/94.6.437
Wu, P., Heins, Z. J., Muller, J. T., Katsnelson, L., de Bruijn, I., Abeshouse, A. A., et al. (2019). Integration and analysis of CPTAC proteomics data in the context of cancer genomics in the cBioPortal. Mol. Cell. Proteomics. 18 (9), 1893–1898. doi:10.1074/mcp.TIR119.001673
Yao, Z. J., Dong, J., Che, Y. J., Zhu, M. F., Wen, M., Wang, N. N., et al. (2016). TargetNet: A web service for predicting potential drug-target interaction profiling via multi-target SAR models. J. Comput. Aided Mol. Des. 30 (5), 413–424. doi:10.1007/s10822-016-9915-2
Zheng, W., Anderson, K. E., Kushi, L. H., Sellers, T. A., Greenstein, J., Hong, C. P., et al. (1998). A prospective cohort study of intake of calcium, vitamin D, and other micronutrients in relation to incidence of rectal cancer among postmenopausal women. Cancer Epidemiol. Biomarkers Prev. 7 (3), 221–225.
Keywords: vitaminD3, CRC, chemoprevention, network pharmacology, molecular docking
Citation: Rong K, He Q, Chen S, Yu Y, Mei L, Mi Y, Mu L, Zhu M, Nan M, Zhang X, Wan Z, Huang H and Zheng P (2023) The mechanism of vitamin D3 in preventing colorectal cancer through network pharmacology. Front. Pharmacol. 14:1192210. doi: 10.3389/fphar.2023.1192210
Received: 23 March 2023; Accepted: 05 May 2023;
Published: 17 May 2023.
Edited by:
Ayaz Shahid, Western University of Health Sciences, United StatesCopyright © 2023 Rong, He, Chen, Yu, Mei, Mi, Mu, Zhu, Nan, Zhang, Wan, Huang and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huang Huang, aGFvaHVhbmdodWFuZ0AxMjYuY29t; Pengyuan Zheng, cHl6aGVuZ0B6enUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.