
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol. , 01 September 2023
Sec. Pharmacology of Anti-Cancer Drugs
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1189799
This article is part of the Research Topic Strategies in Overcoming the Chemoresistance in Colorectal Cancer View all 11 articles
Colorectal cancer (CRC) is the third most common and second most lethal type of cancer worldwide, presenting major health risks as well as economic costs to both people and society. CRC survival chances are significantly higher if the cancer is diagnosed and treated early. With the development of molecular biology, numerous initiatives have been undertaken to identify novel biomarkers for the early diagnosis of CRC. Pathological disorders can be diagnosed at a lower cost with the help of biomarkers, which can be detected in stool, blood, and tissue samples. Several lines of evidence suggest that the gut microbiota could be used as a biomarker for CRC screening and treatment. CRC treatment choices include surgical resection, chemotherapy, immunotherapy, gene therapy, and combination therapies. Targeted therapies are a relatively new and promising modality of treatment that has been shown to increase patients’ overall survival (OS) rates and can inhibit cancer cell development. Several small-molecule tyrosine kinase inhibitors (TKIs) are being investigated as potential treatments due to our increasing awareness of CRC’s molecular causes and oncogenic signaling. These compounds may inhibit critical enzymes in controlling signaling pathways, which are crucial for CRC cells’ development, differentiation, proliferation, and survival. On the other hand, only one of the approximately 42 TKIs that demonstrated anti-tumor effects in pre-clinical studies has been licensed for clinical usage in CRC. A significant knowledge gap exists when bringing these tailored medicines into the clinic. As a result, the emphasis of this review is placed on recently discovered biomarkers for early diagnosis as well as tyrosine kinase inhibitors as possible therapy options for CRC.
Colorectal cancer (CRC) is a third primary global health concern and a second leading cause of cancer-related deaths worldwide, which poses financial burdens on people and society (Morgan et al., 2023). In all clinical practice and research disciplines, CRC, including rectal and colon cancer, is treated as a single tumor type (Alzahrani et al., 2021). CRC originates from the lining of the colon or rectum and follows a specific pathological progression. In most cases, it typically begins as small growths called polyps, which can be detected during a colonoscopy. Over time, these polyps can develop into cancerous tumors, invading the surrounding tissues and potentially spreading to distant organs, frequently the liver. Over 10–15 years, this process necessitates the accumulation of genetic mutations that can be somatic or germ-line in nature (Lotfollahzadeh et al., 2022). Some common risk factors for CRC include family, genetic, geriatric, nutritional, lifestyle, and environmental variables. Inflammatory bowel conditions, including Crohn’s disease and ulcerative colitis, are additional risk factors. Moreover, issues with inactivity, obesity, smoking, and alcohol usage can be resolved (Sawicki et al., 2021).
Certain pathological features, such as adenomatous polyps or advanced stages of carcinoma, are associated with an increased disease prevalence (Akimoto et al., 2021). The incidence and mortality of CRC differ significantly by country and region worldwide. According to the data from Global Cancer Observatory (GLOBOCAN), there were around 1.9 million new cases of CRC and 930,000 fatalities in 2020 worldwide. The incidence rates were lowest in several African countries and Southern Asia, while the highest incidence rates were reported from Europe, Australia, and New Zealand regions (Morgan et al., 2023). Similar trends were observed in CRC mortality rates, with Southern Asia having the lowest rates (2.5 per 100,000 females) and Eastern Europe having the highest rates (20.2 per 100,000 males). Additionally, there was a 10-fold difference in incidence rates between males and females in all countries. Males showed higher incidence and fatality rates than females (Wong et al., 2021). By 2040, CRC is expected to cause 3.2 million new cases and 1.6 million fatalities, mostly in high-HDI (human development index) countries (Xi and Xu., 2021). The incidence and mortality of CRC have decreased, and the US now ranks among the third-highest HDI countries. In the US, stage I colon cancer has a 5-year relative survival rate of around 92%. Stage IIA and IIB exhibit rates of 87% and 65%, respectively. Surprisingly, stage IIIA and stage IIIB have slightly greater 5-year survival rates, at 90% and 72%, respectively. While stage IV, or metastatic CRC (mCRC), has a 5-year survival rate of only 12%, stage IIIC has a survival rate of 53%. With 88% for stage I, 81% for stage IIA, 50% for stage IIB, 83% for stage IIIA, 72% for stage IIIB, 58% for stage IIIC, and 13% for stage IV, the 5-year survival rates for rectal cancer seem to be slightly lower. These stages are based on the TNM system’s previous version. The unexpected increase in survival from stage II to stage III tumors can be attributed to the technique utilized to diagnose and treat different types of CRC (Rawla et al., 2019).
A higher prevalence of the disease underscores the need for effective screening programs and public awareness campaigns to promote early detection, as it significantly improves the prognosis and treatment outcomes for individuals with CRC (Stark et al., 2020). Over the past few decades, there has been remarkable progress in understanding the molecular basis of CRC, leading to the identification of novel biomarkers for early diagnosis and the development of targeted therapies, such as tyrosine kinase inhibitors (TKIs), for the treatment of CRC (Bresalier et al., 2020). Early diagnosis is critical in improving patient outcomes by enabling timely intervention and reducing mortality rates. Traditional screening methods for CRC, such as colonoscopy and fecal occult blood tests, have effectively detected early-stage tumors and precancerous lesions (Huang and Yang., 2022). However, these methods often have invasiveness, cost, and patient compliance limitations. Therefore, there is an urgent need to explore and validate novel biomarkers that can enhance the sensitivity and specificity of CRC detection while offering non-invasive and cost-effective alternatives (Shaukat and Levin., 2022).
Biomarkers have been developed to aid in identifying patient responses to cancer diagnosis, therapy, and monitoring (Oh and Joo., 2022). Biomarkers, which include genetic alterations, epigenetic modifications, and protein expression patterns, hold immense promise as tools for CRC screening, risk assessment, and prognosis prediction. These biomarkers can be detected not only in solid tissue samples but also in blood and/or stool, allowing for non-invasive and convenient testing (Shen et al., 2022). The development of high-throughput genomic technologies has revolutionized biomarker discovery and enabled the identification of candidate markers associated with CRC initiation, progression, and response to treatment. Furthermore, integrating multiple biomarkers into diagnostic algorithms can improve the accuracy and reliability of CRC screening, facilitating the implementation of personalized medicine approaches (Islam Khan et al., 2022). Moreover, liquid biopsy approaches, which involve the analysis of circulating tumor DNA (ctDNA) and microRNAs (miRNAs), have gained significant attention as non-invasive methods for CRC screening and monitoring (Zhou et al., 2022).
In addition to early diagnosis, targeted therapies have revolutionized the treatment landscape for CRC patients. Emerging evidence suggests a potential correlation between CRC and the use of TKIs. Tyrosine kinases play a crucial role in cell signaling pathways, and their dysregulation has been implicated in various types of cancers, including CRC (Thomson et al., 2022). TKIs are a class of drugs that selectively inhibit the activity of specific tyrosine kinase enzymes involved in CRC pathogenesis and progression. The aberrant activation of signaling pathways, such as the epidermal growth factor receptor (EGFR) and vascular endothelial growth factor receptor (VEGFR) pathways, has been implicated in CRC tumorigenesis and angiogenesis (Iyer et al., 2022). While TKIs have shown promising results in treating certain cancers, including gastrointestinal stromal tumors, their efficacy in CRC has been more limited. Studies have indicated that specific genetic mutations and alterations in tyrosine kinase signaling pathways may influence the response to TKIs in CRC patients (Xie et al., 2020). TKIs targeting these pathways, either as monotherapy or combined with standard chemotherapy regimens, have shown clinical efficacy in various studies (Hossain et al., 2022).
This review article will provide a comprehensive overview of the recent advancements in novel biomarkers used for early diagnosis of CRC. We will explore the potential of genetic alterations, epigenetic modifications, and other molecular markers as diagnostic tools in CRC. We will discuss the application of specific genetic markers, such as mutations in the Adenomatous polyposis coli (APC), Kirsten rat sarcoma (KRAS), and tumor protein p53 (TP53) genes, as well as epigenetic including DNA methylation patterns and histone modifications as diagnostic and prognostic indicators. We hope to contribute to the ongoing efforts to improve CRC patient outcomes and facilitate precision medicine approaches by integrating the knowledge of these emerging biomarkers and therapies. Moreover, we will also provide an in-depth analysis of the current status and future perspectives of TKIs as targeted therapies in CRC treatment.
Biomarkers are commonly used in CRC diagnostics to detect the presence of biochemical compounds that circulate in the body. These compounds may include gut microorganisms, miRNA in the blood, tumor-derived cells, tumor DNA, and proteins.
Inflammation, immunological modulation, dietary component metabolism, and exposure to genotoxic substances are the primary ways the gut microbiota contributes to cancer. Patients with CRC have a range of unique microbiomes that can be used as biomarkers for the diagnosis, prognosis, and treatment efficacy (Wong and Yu., 2019). There has been a lot of interest lately in the possible connection between gut bacteria and CRC.
Gut fungi, specifically the dysbiosis or imbalance in the fungal community, have been implicated in CRC development. Recent studies have identified specific fungal biomarkers, such as Candida tropicalis and Debaryomyces hansenii, whose overgrowth or altered abundance in the gut may serve as potential molecular indicators of CRC, offering insights into its pathogenesis and possible diagnostic strategies. The overgrowth of gut fungi in CRC patients can be weakened immune system function and change the microenvironment of the colon, creating an environment conducive to fungal proliferation and colonization (Plaza-Díaz et al., 2021). However, there is insufficient information on the fungus microbiome in CRC. The top 3 fungi highly enriched in CRC were Phanerochaete chrysosporium, Lachancea waltii, and Aspergillus rambellii. It has been established that fungi anomalies in feces are associated with CRC (Gao et al., 2022) and in healthy individuals compared to CRC patients. It was observed that the proportion of Basidiomycota or Ascomycota was higher in CRC patients than in healthy individuals.
Moreover, two fungi, Pneumocystis and Saccharomyces cerevisiae, which have beneficial effects on the gut and possess anti-inflammatory properties, were found to be reduced in CRC patients. Conversely, Malasseziomycetes (fungi) were more abundant in healthy individuals than in CRC patients (Liu et al., 2022). Furthermore, the researchers noted that patients exhibited notably elevated levels of Candida albicans, a type of yeast. According to Stary et al. (2020), individuals who are at risk for CRC or have early asymptomatic with CRC may find it helpful to use C. albicans yeast as a diagnostic marker (Starý et al., 2020).
Gut bacteria, particularly the alterations in the composition and diversity of the bacterial community, have been linked to CRC development. The molecular mechanisms underlying this association involve the production of specific metabolites, such as short-chain fatty acids and secondary bile acids, as well as the activation of pro-inflammatory pathways, which can serve as potential biomarkers for the detection and monitoring of CRC (Xie et al., 2020). In a large-scale study, researchers discovered that individuals with CRC had an increased population of certain bacteria, including Fusobacterium nucleatum, Porphyromonas asaccharolytica, Bacteroides fragilis, Parvimonas micra, Prevotella intermedia, Alistipes finegoldii, and Thermanaerovibrio acidaminovorans. These seven bacteria are potential markers for diagnosing CRC (Chang et al., 2021). Adenomas were found to have unusually elevated levels of the “m3” product, particularly those originating from Clostridium hathewayi (Ch), Fusobacterium nucleatum (Fn), and Lachnoclostridium. Only these three bacteria have been identified in feces as markers for colorectal adenomas and cancers (Chan and Liang., 2022). Actinomyces odontolyticus and Atopobium parvulum were exclusively found in polypoid adenomas and/or intramucosal carcinomas (early stage), indicating the wide availability of Fn enhanced slowly from intramucosal carcinoma to early CRC (Liu et al., 2022). This discovery raises the possibility of employing these microorganisms as stool-screening indicators.
The role of gut viruses in CRC is still being explored. Emerging evidence suggests that certain viral infections, such as high-risk human papillomavirus (HPV) and Epstein-Barr virus (EBV), may contribute to the development and progression of CRC through various molecular mechanisms, including viral integration into the host genome, dysregulation of host cell signaling pathways, and evasion of immune surveillance, providing potential avenues for viral-based biomarker identification in CRC diagnosis and treatment (Koonin et al., 2021). Most cancer-associated bacteriophages were temperate, demonstrating a connection between bacteriophage communities and CRC and the possibility that they could influence cancer progression by altering bacterial host populations (Hannigan et al., 2018). A similar study confirmed the association between viral indicators and CRC by observing a substantial rise in the variety of gut bacteriophage populations in feces from CRC patients and controls. Numerous studies have shown how closely microbes and cancer are related and how gut bacteria have opened up new possibilities for CRC detection (Handley and Devkota., 2019). However, a common microbiome biomarker has not been used to diagnose CRC because there is not yet a universally accepted standard for discovering microbiota. In order to enhance CRC diagnosis in the future, investigators must examine multiple microbiomes in patients from various ethnic groups since a microbe might not be capable of predicting CRC with adequate precision (Ding et al., 2022).
The ability to distinguish between diseases based on their “smell” has gained popularity as a research topic in recent years due to the growing interest in the “smell” of diseases. A potential early CRC screening method involves the detection of VOCs, which are non-invasive biomarkers. Multiple investigations have revealed several VOCs as CRC biomarkers (Ding et al., 2022). Furthermore, changes in gut flora have a direct impact on the profile of VOC generation.
Propan-2-ol, produced from ethyl 3-methyl-butanoate, hexane-2-one, and acetone, produced when ethanol and 3-methylbutaninoic react, positively correlates with the diagnosis of CRC, according to a 3-fecal volatile organic compound panel (Bond et al., 2019). One stool VOC contributing to CRC formation is hydrogen sulphide (HS). Microorganisms in the gut and internal enzymatic activities in the colon generate hydrogen sulfide. Higher levels of HS (over 2.4 mmol/kg) are hazardous, but lower levels are benign. The presence of higher-than-normal quantities of HS in both the lumen and the feces can throw off the equilibrium of the microbiota. For instance, this phenomenon affects patients with CRC (Lin et al., 2023). With the help of eNose Cyranose 320, patients with CRC could be characterized from controls with 85% sensitivity and 87% specificity (AUC 0.92). Similar results were attained with selected ion flow tube mass spectrometry (SIFT-MS), which separated CRC and advanced adenoma patients from healthy controls with 75% accuracy (72% sensitivity and 78% specificity). Recently, eNose Scent A1 was even more successful in many patients (Vernia et al., 2021).
Using a pattern of 15 VOCs identified with gas-chromatography mass spectrometry (GC-MS), Altomare et al. differentiated between CRC patients and healthy controls with an accuracy of more than 80% (Altomare et al., 2015). A study isolated 4 volatile organic compounds: methyl octane, ethanol, ethyl acetate, and acetone, 4-. Acetone and ethyl acetate levels were more significant in patients with CRC (94% specificity and 85% sensitivity) and an accuracy of 91% (Zhang et al., 2021). The VOC samples from CRC patients also had considerably higher levels of 3-hydroxy-2,4,4-trimethylpentyl, trans-2-dodecen-1-ol, ethylaniline, cyclooctylmethanol, 4-ethyl-1-octyn-3-ol, 2,2-dimethyl decane, Cyclohexanone, and dodecane. But much lower levels of 2-methylpropanoate and 6-t-butyl-2,2,9,9 tetramethyl-3,5-decadien-7-yne (Amal et al., 2016).
In a larger sample of 562 people, the diagnostic efficacy of urine VOCs detected by Field Asymmetric Ion Mobility Spectrometry (FAIMS) was less accurate than that of the fecal immunochemical test (FIT) (80% sensitivity and 93% specificity vs. 63% sensitivity and 63% specificity, respectively). One study found that CRC patients had significantly higher concentrations of 2-methyl-3-phenyl-2-propenal, 2,7-dimethyl-quinoline, and 1,4,5-trimethyl-naphthalene (Vernia et al., 2021).
CDH17 is a glycoprotein that spans the cell membrane and requires calcium to function properly. Its primary purpose is to aid tissues in preserving their typical structure under normal conditions. The immunohistochemistry marker CDH17 helps identify primary and metastatic colorectal adenocarcinomas (Tournier et al., 2023). According to reports, 100% of metastatic and 96%–100% of primary CRC express CDH17. CDH17 and SATB2 were excellent potential biomarkers for diagnosing metastatic colorectal adenocarcinoma and pulmonary enteric malignancy (Neri et al., 2020). The expression of CDH17 in CRC tissues and plasma gradually increased as the disease progressed to more advanced stages. There is a link between liver metastasis, high CDH17 expression, and a bad prognosis for CRC patients (Tsoi et al., 2019).
GPA33, a transmembrane protein overexpressed in CRC, has potential molecular mechanisms involving cell adhesion, immune evasion, and tumor growth, highlighting its significance as a biomarker and therapeutic target; however, its current clinical application as a diagnostic or prognostic tool in CRC is still under investigation and requires further validation. The tumor-associated antigen human GPA33 is expressed in approximately 95% of primary and mCRC (Markandeywar et al., 2023). It is a surface-localized, extremely persistent, and inert protein. PGA 33 has a 95.9% sensitivity and an 85.4% specificity for CRC. A33 has been the target of clinical-stage antibodies used to treat CRC (Wei et al., 2020). The new anti-A33 antibody may prevent the development of mouse CRC lung metastases, and A33-expressing murine adenocarcinoma cells may be destroyed by antibody-dependent cell-mediated cytotoxicity (ADCC) (Murer et al., 2020). The A33 had a sensitivity comparable to Caudal-type homeobox transcription factor 2 (CDX2). Still, it had a specificity that was significantly greater than that of CDX2 as an immunomarker of CRC, according to the findings of an immunohistological investigation that compared A33 with CDX2 (Davidsen, 2020).
CKs are proteins found in the cytoskeleton and located in intermediate filaments. It is a member of a group of approximately 20 cytoskeletal proteins frequently used as immune-histochemical markers in diagnosing CRC in tumors that have been generated from epithelia. In most cases, neoplastic cells maintain CK expression; specialized anti-CK antibodies are frequently used in histopathology diagnoses to trace tumor origins, especially in metastases (Hrudka et al., 2021). Cytokeratins, a group of intermediate filament proteins, have potential molecular mechanisms involving epithelial cell differentiation, tumor invasion, and metastasis, making them valuable biomarkers for cancer diagnosis and prognosis; currently, cytokeratin-based assays, such as CK19 for detecting disseminated tumor cells, are utilized in clinical practice for assessing the presence of minimal residual disease and predicting treatment response in various cancer types, including CRC. Two enzymes, CK7 and CK20, are frequently involved in CRC. Different types of glandular and ductal epithelia contain CK7. Simultaneously, CK20 is abundantly expressed in mucosal cells of the gastrointestinal and urinary tracts (Hrudka et al., 2022). Tissue expression of CK15 was significantly linked with CRC subtype and stage. A higher level of CK18 expression is found in CRC tumors compared to the normal colorectal tissue surrounding them (Sun et al., 2018). It is a single predictor of long-term survival in CRC patients when CK18 expression is upregulated in tumor tissue. The viability, migration, and invasion of CRC cells were decreased by downregulating CK18 expression (Jelski and Mroczko., 2020). All three of the detected cytokeratins 8, 18, and 19 can potentially be helpful biomarkers for the early diagnosis of CRC (Luo et al., 2020).
A telomerase ribonucleoprotein increases the number of TTAGGG repeats at the ends of chromosomes to preserve telomere length. Intrinsic RNA serves as a scaffold for reverse transcription in telomerase. The telomere controls chromosomal stability and cell life span. The telomerase enzyme is present in human cancer cells (80%–90%) and differentiated cells, such as germ-line cells (Kibriya et al., 2022). Telomere length in cancer tissue was substantially shorter than in normal mucosa. Advanced CRC (stage II–IV) cancers have longer telomeres than stage I tumors (Ye et al., 2021). According to Taheri et al. (2022), CRC tissue had reduced hTERT expression levels. Since telomerase is present in healthy and malignant intestinal epithelial cells, measuring hTERT alone may underestimate its prevalence. Numerous studies have demonstrated that telomerase has a high-level telomere-specific reverse transcriptase (hTERT), which improves Nuclear factor erythroid 2-related factor 2 (NRF2) synthesis by recruiting Y-box binding protein 1 (YBX1) to trigger the NRF2 promoter, promoting CRC proliferation and migration. These findings provide a new conceptual underpinning for CRC treatment (Gong et al., 2021).
SATB2 belongs to the group of transcription factors that bind to matrix attachment regions and control the development of the skeleton. The appendix and colon epithelium both had significant levels of SATB2 expression (Huang and Yang, 2022). SATB2, a transcription factor involved in gene regulation, has potential molecular mechanisms related to cell differentiation, cell migration, and tumor metastasis, suggesting its significance as a biomarker. However, SATB2 expression has been identified as a useful marker for CRC diagnosis and distinguishing primary colorectal tumors from metastatic tumors, and further research is needed to establish its full clinical utility and potential therapeutic implications. SABT2 has recently been identified as a biomarker for CRC, and various hereditary disorders connected to SABT2 have been reported (Mezheyeuski et al., 2020). SATB2 was positive in 83.7% of stage III/IV, 91.4% of stage II, and 92.4% of phase I colorectal adenocarcinomas, according to Dabir et al. (2018). According to multiple studies, when paired with conventional panels of CDX2, CK20, CK7, and cytokeratin 20, SATB2 is a highly specialized marker for CRC (Oh and Joo, 2020).
Intestinal epithelial cells express the homeobox protein CDX2 in their nuclei. As a trustworthy and accurate immunomarker for CRC, CDX2 is frequently used. It is believed that CDX2 is a tumor suppressor gene because it does not manifest itself in instances of CRC. By overexpressing CDX2 using an hTERT (hypoxia-inducible human telomerase reverse transcriptase) promoter-driven plasmid, colon cancer cells were prevented from progressing malignantly (Al-Duhaidahawi., 2023). The CDX2 gene promoter region’s methylation has been associated with a higher risk of CRC. The CDX2 gene promoter area was methylated in 78.5% of the CRC tissue. CDX2 downregulation was associated with high-grade, advanced cancers with liver metastases (Aasebø et al., 2020). Furthermore, disease-free and overall survival (OS) were considerably poorer in people with stage T4 CRC and low CDX2 expression (Choi et al., 2022).
At every stage of carcinogenesis, from polyps to colorectal adenocarcinomas, hypermethylation drives the transcriptional silence or downregulation of suppressor genes, which renders tumor suppressor genes inactive. In CRC, numerous genes, particularly those in the promoter region, including HLTF, CDH1, SEPT, VIM, TIMP3, CDK2A, SFRP2, SFRP1, MGMT, MLH1, and APC, are methylated (Mo et al., 2023). Suppression of histone deacetylation and demethylation are used in CRC cells to increase Syndecan-2 (SDC2) expression because the SDC2 promotor region is typically hypermethylated in CRC. Methylated SDC2 for the non-invasive diagnosis of CRC has reasonable specificity (88.2%–98%) and sensitivity (77.4%–90.2%) (Siri et al., 2022). The zinc finger protein 625, LON peptidase N terminal domain and ring finger 2, WD repeat domain 17, and syndecan 2 CpG island promoters were methylation in both cancer and laterally spreading tumor non-granular (LST NG). This indicates that the LST NG phase may be the first stage of colorectal carcinogenesis (Iwaizumi et al., 2023). N-Myc downstream-regulated gene 4 (NDRG4) influences cell development and differentiation. In CRC, the NDRG4 expression is downregulated (Cao et al., 2020).
Biomarkers can be detected using immunohistochemistry or blood-based protein quantification techniques. Blood-based markers can serve as a convenient screening tool for CRC, as blood donation or collection is a relatively simple process.
CTCs are epithelial cancer cells in peripheral blood after they have spread from the primary tumor or metastases and entered the circulatory system. They can be utilized as biomarkers to detect CRC or as knowledge-dissemination pathways, enabling therapy decisions (Chan et al., 2023). In early-stage cancers, circulating CTCs vary from 1 to 10 cells/10 mL of blood and may be less. CTCs can be differentiated from normal blood cells by differences in size and shape. More research is needed to validate the findings. Necrosis releases circulating-free DNA (cfDNA) as significantly bigger fragments in tumor cells (Li et al., 2023). This resulted in promising findings when circulating cfDNAs were quantitatively examined using the ratio of longer to shorter DNA fragments or when the cfDNA integrity number was measured after CRC diagnosis. Although CTCs have been shown to have predictive value in CRC, their use in screening is controversial (Hendricks et al., 2021).
ctDNA (circulating tumor DNA) and cfDNA have emerged as promising biomarkers in CRC. The presence of ctDNA and cfDNA in the bloodstream allows for non-invasive detection of specific genetic alterations, such as mutations and methylation patterns, providing valuable information about tumor burden, treatment response, and disease progression, thereby enabling personalized medicine approaches in the management of CRC patients (Lyu et al., 2022). ctDNA has received extensive evaluation as a promising indicator for liquid biopsy in detecting and assessing therapeutic responses for CRC. ctDNA released from necrotic or apoptotic tumor cells. Although normal nontumor cells also shed cfDNA into the bloodstream, the cfDNA from tumor cells (i.e., ctDNA) only accounts for less than 1% of total cfDNA in the blood (Peng et al., 2021). Tests that rely on the detection of circulating tumor DNA (ctDNA), also known as liquid biopsies, are susceptible to vulnerabilities. A study conducted by Min et al. (2023) in 2023 demonstrated that quantitative analysis of ctDNA and qualitative investigation of SEPT9 methylation effectively diagnose CRC. According to a study conducted by Perdyan et al. (2020) in 2020, ctDNA demonstrated an accuracy of 87.2% and a precision of 99.2% in identifying clinically significant KRAS gene mutations in a group of 206 patients with mCRC. Several studies have reported elevated levels of cfDNA in cancer patients. According to Raunkilde et al. (2022), most cfDNA fragments of 180–200 base pairs in length originate from tumor cells that have undergone necrosis or cell death (Raunkilde et al., 2022). Tumor-specific genomic changes, including microsatellite instability, loss of heterozygosity, methylation, and mutations, are present in cfDNA.
Small non-coding RNAs called microRNAs (miRNAs) control the expression of genes by attaching them to mRNA. C-miRNA has contributed to diagnosis. The dysregulation of miRNA activity causes a variety of diseases, including cancer. Diagnostic panels that combined single miRNAs as a CRC marker with combinations of those detected in serum or plasma miRNA indicators have been investigated recently (Abo-Elela et al., 2023). In comparison to serum, plasma has higher levels of miRNA. Hemolysis must be regulated in samples during the preanalytical stage of the experiment because it can change the amounts of circulating miRNA in samples by rupturing erythrocytes that transport miRNA. The only miRNAs that seem suitable as clinical markers are those with severe up- or downregulation (Liu et al., 2021). Nearly 2/3 of miRNAs were downregulated in CRC compared to normal mucosa. According to one study, the four-stage change from colorectal adenocarcinoma via high- and low-grade dysplasia in adenoma involved differential expression of 230 miRNAs (Sur et al., 2022).
Long non-coding RNAs (lncRNAs), made up of more than 200 nucleotides and cannot be translated into proteins, are involved in several biological processes, including differentiation, immunological responses, and chromosomal dynamics. Because lncRNAs can pass across cell membranes, they can be discovered in various bodily fluids, including blood, plasma/serum, and urine (He and Wu., 2023). Numerous lncRNAs are linked to the development of CRC and carcinogenesis at all stages. The WNT/-β catenin, PI3K/Akt, EGFR, NOTCH, mTOR, and TP53 signaling pathways are only a few carcinogenic signaling cascades that their changed expression can affect (Cai et al., 2019). The extracellular phospholipid-enclosed vesicles, known as apoptotic bodies, microvesicles, and exosomes, travel with lncRNAs in the blood. Exosomes, one of the three forms of extracellular vesicles, have the largest concentrations of long micro RNAs (lmiRNAs), which aid in tumor spreading, immunomodulation, and chemoresistance (Lulli et al., 2022). The first indicators discovered to have significantly higher expression in the plasma of CRC patients compared to healthy people were HOTAIR and CCAT1. Numerous more circulating lncRNAs, including NEAT1 variants 1 and 2, MEG3, PVT-1, 91H, Nbla12061, RP11-462C24.1, and LOC285194 have been identified as possible biomarkers for the identification of CRC (Zygulska and Pierzchalski., 2022).
PKM2 is found in healthy and cancerous cells and is involved in energy metabolism. When PKM2 regulates the rate-limiting stage of glycolysis, tumor cells produce lactate rather than the normal respiratory chain for glucose metabolism (Cruz et al., 2021). There have been reports of PKM2 overexpression in colon adenomas, gastric cancer, and CRC. With its excellent sensitivity, PKM2 is adequate blood and fecal biomarker for CRC screening (Zahra et al., 2020). The study found that the fecal Tumor pyruvate kinase M2 isoform (tM2-PK) test had a 100% sensitivity and a 68% specificity in the tumor group. Specificity and sensitivity for the polyp group were 68% and 87%, respectively. The tM2-PK test is recommended as a non-invasive method to identify CRC and adenomatous polyps (Rigi et al., 2020).
DDK3, a tumor suppressor gene, and IGFBP2 a growth factor regulator, have potential molecular mechanisms involving cell cycle control, growth inhibition, and modulation of the IGF signaling pathway, highlighting their potential as biomarkers. However, further studies are needed to determine their clinical utility and application in cancer diagnosis, prognosis, and targeted therapies (Zygulska and Pierzchalski, 2022). The biomarker model can identify early-stage CRC with 95% specificity, 57% sensitivity for stage I, and 76% sensitivity for stage II. As a result, this panel of biomarkers recommends being used as a non-invasive blood screening and/or diagnostic test. It is comparable to a fecal occult blood test (FOBT) and FIT in CRC detection (Zygulska and Pierzchalski., 2022).
DDK3 (DNA-damage-inducible 3) and DDK1 (Dickkopf-1) are potential biomarkers in CRC. DDK3 expression has been associated with tumor suppressive effects, and its downregulation is often observed in CRC, suggesting its potential as a diagnostic or prognostic biomarker. On the other hand, DDK, an antagonist of the Wnt signaling pathway, is frequently overexpressed in CRC, and its elevated levels may serve as a biomarker for disease progression and therapeutic response in CRC patients (Akhlaghipour et al., 2021). The human Dickkopf family includes the proteins DDK-1, DDK-2, DDK-3, and DDK-4, and the specific protein Soggy (Sgy) related to DDK-3 is all TEMs (tumor endothelium markers). The tumor endothelium of CRC tissues displays more pronounced expression levels of the TEMs, a group of 46 genes (Safari et al., 2018). Wnt blocker genes are epigenetically inhibited in CRC, among several other factors. During cancer development, the Wnt signaling pathway is triggered. These antagonistic genes include DDK genes, which are hypermethylated in the promoter of CRC cells and epigenetically silenced (Kaur et al., 2012). According to one study, CRC that was DDK-3 positive had a considerably greater mean microvessel count (9.70 vessels) than cancer that was DDK-3 negative. Therefore, it is believed that DDK-3 is a pro-angiogenic mediator in the growth of neovascularization during the progression of CRC (Soheilifar et al., 2019).
IGFBP-2 is an extracellular protein that binds insulin growth factor 2 (IGF-2), which is involved in the development and spread of cancer through the action of heat shock protein 27. In patients with colon cancer, higher serum concentrations of IGFBP-2 are associated with neoplastic changes in the higher levels of carcinoembryonic antigen (CEA) and colon. Consequently, it has been suggested that monitoring patients with CRC includes measuring IGFBP-2 levels as a diagnostic indicator (Chen et al., 2021). Thus, by preventing cell division, IGFBP-2 overproduction during colorectal carcinogenesis slows the formation of tumors. The sensitivity may be improved by combining IGFBP-2 with additional biomarkers, such as CEA (Gligorijević et al., 2022). Zhu et al. (2019) found that IGFBP2 overexpression promoted CRC cell proliferation and migration by suppressing E-cadherin expression and enhancing cell growth. Additionally, more significant tumor sizes and lower OS rates were linked to higher plasma IGFBP-2 levels, demonstrating that IGFBP-2 may function as a prognostic and diagnostic biomarker for CRC.
The SEPT gene family in humans consists of 13 genes (SEPT1-SEPT13). SEPT9 methylation DNA is the most well-known blood biomarker. The molecular mechanism of SEPT9 gene methylation in CRC involves the aberrant addition of methyl groups to CpG islands within the gene’s promoter region. This hypermethylation leads to the silencing of the SEPT9 gene and subsequent loss of septin protein expression. The disrupted septin function contributes to defective cytokinesis, abnormal cell morphology, and increased genomic instability, promoting the development and progression of CRC (Baharudin et al., 2022). The detection rate for those with CRC stages 0-I using this method ranges from 57% to 64% (Zhao et al., 2020). According to the meta-analysis, individuals with advanced CRC cases were more likely to test positive for methylated SEPT 9 (mSEPT9) than those with early-stage CRC, and the opposite was true for people with early-stage CRC (Min et al., 2023). According to the latest meta-analysis released in 2020, the SEPT9 assay has a specificity of 92% and a sensitivity of 69% for CRC diagnoses (Hariharan and Jenkins, 2020).
Stool samples are more suitable for the early detection of CRC than blood tests because exfoliating tumor cells appear in the large intestine or rectal lumen during colorectal carcinogenesis far earlier than the beginning of tumor cell vascular penetration.
The molecular mechanism of stool DNA methylation in CRC involves detecting aberrant DNA methylation patterns in the stool samples of patients. Abnormal methylation of specific genes associated with CRC can serve as a non-invasive biomarker for the early detection and screening of the disease (Mueller and Győrffy., 2022). The human genome makes up less than 0.01% of the total DNA in stools; the other 99.99% comes from gut bacteria or food. The DNA of tumor cells expelled with feces contains abnormal genetic and epigenetic changes, which may serve as biomarkers for the detection of cancer (Gao et al., 2023). Several genes such as WIF1, VIM, TFPI2, SFRP2, RASSF2A, NDRG4, MGMT, MLH1, MINT31, MINT1, KRAS, ITGA4, IRF8, ID4, HLTF, GSTP1, GATA4, ESR1, CXCL21, CRBP1, CDH13, CDKN2A, CDH1, BMP3, ATM, and APC have all been studied for CRC diagnosis (Park et al., 2017). There is a Multitarget stool DNA (mt-sDNA) test (Cologuard, which combines hemoglobin, NDRG4, KRAS mutations, and BMP3 DNA methylation) and a plasma SEPT9 DNA methylation test (Epi proColon) that has been utilized more extensively in clinical settings. In asymptomatic people, mt-sDNA testing has a sensitivity of 90% for detecting CRC. DNA testing has a specificity range of 86.6%–98% (Ladabaum et al., 2020). A colonoscopy is a next stage in diagnosing a colorectal tumor in the event of a positive mtsDNA test. Asymptomatic participants in an intriguing study endured CT colonography and an mt-sDNA test (FDA-approved). Overall, CT colonography screening had a considerably higher detection rate for advanced neoplastic lesions (5%) than the mt-sDNA test (2.7%). There were 0.31% and 0.23% overall detection rates for CRC (Pickhardt et al., 2020).
The FOBT is modified into the FIT, which checks for blood that digestive proteolytic enzymes have broken down. Even though early CRC detection is crucial for reducing CRC mortality, there is limited data to support the stage-specific sensitivity of the FIT in CRC detection. The FIT detected stage I cancers with a sensitivity of 68% (95% CI, 57%–78%), stage II cancers with a sensitivity of 92% (95% CI, 87%–96%), stage III cancers with a sensitivity of 82% (95% CI, 73%–89%), and stage IV cancers with a sensitivity of 89% (95% CI, 80%–95%) (Niedermaier et al., 2020). FIT offers an extensive spectrum of susceptibility for all stages of CRC, ranging from 25% to 79% (Van Doorn et al., 2015). T3 sensitivity was 83%, and T1 sensitivity was 40% in those with greater severity of CRC (Hijos-Mallada et al., 2021).
The diagnostic tools, such as RNA- or DNA-based testing, studied in a community-based population were found to increase the efficacy of the FIT procedure. Another study found that mt-sDNA testing was more effective than FIT at identifying advanced adenomas and sessile serrated polyps (Tanriver and Kocagoncu., 2023). However, mt-sDNA had a lower overall specificity for detecting all lesions than FIT. According to reports, a DNA-FIT test boosted detection sensitivity for CRC to 97.5% and advanced adenomas to 53.1% (Xu et al., 2022).
In CRC, ncRNAs are abnormally produced, and based on the genes or pathways they control downstream, they may act as oncogenes (oncomiRs) or tumor suppressors (tsmiRs). A novel therapeutic approach and testing biomarkers have been developed due to the possible discrepancy among miRNA profiles of CRC and the normal intestinal mucosa (Fonseca et al., 2021). The study of stool miRNA has some drawbacks. First, it can be challenging to standardize procedures due to daily changes in fecal characteristics (density, volume). Second, it is essential to distinguish between the three types of feces miRNAs: fecal colonocyte miRNAs, exosomal miRNAs from fecal exosomes, and cell-free miRNAs from fecal homogenates (Ahmed et al., 2019). In the feces of CRC patients, the miR-145 and miR-143 were downregulated, while miR135, the miR17-92 cluster, miR-106a, miR-92a, miR144, and miR-21 were upregulated (Sattar et al., 2022).
Urine biomarkers can be obtained non-invasively without forcing the patients to attend the clinic. Urine containing various components is considered the most effective and ideal sample for medical examination. Additionally, because the urinary tract is highly clean physiologically, substances found in urine may be less contaminated by germs than those in feces. Metabolomics has been routinely employed to identify metabolic abnormalities in CRC patients’ tissue, serum, and urine materials. A recent metabolomic study discovered that CRC patients have a unique metabolic phenotype characterized by dysregulated expression of metabolites in glycolysis, the tricarboxylic acid (TCA) cycle, the urea cycle, tryptophan, arginine, proline, pyrimidine, polyamine, lactate, fatty acid, and amino acid metabolism, as well as gut microbial metabolism (Zhu et al., 2022; Qiu et al., 2023). Therefore, identifying urine biomarkers is desirable for diagnosing various cancers, including CRC (Iwasaki et al., 2019). ProstaglandinE2 and MicroRNA have demonstrated value in CRC detection. According to the available data, VOCs may be a possible biomarker for identifying CRC (Chandrapalan and Arasaradnam, 2020). Recently, researchers identified urinary metabolite biomarkers N1, N12-diacetyl spermine, hippurate, p-hydroxy hippurate, and glutamate as the best metabolites to discriminate CRC patients via low-cost point-of-care (POC) screening test (Zhou et al., 2022).
Early detection of colorectal tumors allows for successful management with first-line therapies such as surgery, radiation, or traditional chemotherapy. The 5-year OS rate for patients is 88%–92%, while it drops to 58%–72% for patients with stage IIIC. Even though traditional chemotherapy has a remarkable influence on cancer treatment, it is nevertheless significantly hindered by its nonspecific toxicity toward rapidly dividing cells (Mou et al., 2021). By interacting with particular genes or proteins involved in cell growth or apoptosis resistance, highly targeted medications that aim to eradicate cancer cells have been made possible thanks to the amazing advancements in molecular oncology in this field. Highly effective cancer medicines are helpful for cancer treatment since they have improved tumor selectivity and have fewer adverse effects than traditional cancer treatments (Xie et al., 2020). As a result of a better understanding of the mechanisms involved in the evolution and proliferation of cancer cells, targeted therapies and medications with action focused on these pathways/features have been developed.
An earlier meta-analysis examined chemotherapy’s effectiveness and safety outcomes with bevacizumab, panitumumab, or cetuximab in mCRC. It demonstrated that bevacizumab was more effective in treating right-sided mCRC. In contrast, cetuximab was more successful in treating left-sided RAS wild-type (WT) mCRC (Cai et al., 2022). Both cetuximab and panitumumab, two different monoclonal antibodies (mAbs) that target the epidermal growth factor receptor (EGFR), are frequently used either alone or in conjunction with chemotherapy to treat people with mCRC that has the RAS wild-type. Despite being often regarded as interchangeable, the two antibodies possess distinctive molecular compositions and can function therapeutically significant in various ways (Shim, 2011). While there is less research on cetuximab or panitumumab as first-line therapies for older patients with mCRC, these drugs may still be an option for those with the wild-type KRAS mutation. Cetuximab, either alone or in combination with irinotecan, had a benign toxicity profile in elderly patients with severely pretreated mCRC, and the efficacy was comparable to that reported in younger patients, according to two retrospective studies (Bouchahda et al., 2008; Fornaro et al., 2011). Another study suggests combining bevacizumab and capecitabine is a safe and effective treatment for elderly individuals with mCRC (Sastre et al., 2012). The preliminary results that are currently available show that patients with mCRC who receive cetuximab or panitumumab treatment had higher response rates and longer PFS when KRAS mutations are absent (Kim, 2015).
Small-molecule, oral drugs that target particular tumor-causing proteins have been available to treat colorectal malignancies since the turn of the millennium. These proteins that cause tumors are known as tyrosine kinases, and over 90 are in the human genome. Based on their structure, activity, and localization, these 58 can be further separated into the two significant classes of receptor tyrosine kinases (RTKs) and non-receptor tyrosine kinases (NRTKs) (Natoli et al., 2010). RTKs and NRTKs have both been linked to the emergence of CRC. Drugs that target these proteins have several distinct benefits over conventional chemotherapy. Blocking these enzymes can help prevent cancer cell development because they may be overactive or abundant in cancer cells (Piawah and Venook., 2019).
The structural characteristics of the RTK superfamily of cell membrane proteins include an extracellular ligand-binding domain, a transmembrane region, and a cytoplasmic region, including ATP-binding and catalytic kinase domains. Based on similar receptor characteristics and/or shared ligands, at least 20 subfamilies of the approximately 60 RTKs have been discovered (Schlessinger, 2014). When peptide-based ligands transmit extracellular signals, these proteins are crucial for recognizing and transforming those signals. Their signals regulate cellular functions such as cell division, proliferation, and life span (Figure 1). RTKs function as monomeric transmembrane proteins when they are dormant. These proteins dimerize and create oligomeric pairs after becoming active. The receptors’ enzymatic domains must be activated to produce receptor oligomers, and their intracellular region’s tyrosine residues must be autophosphorylated (Figures 1A, B) (Diwanji et al., 2019). When ATP attaches to a specific receptor region, tyrosine residues on the receptor and effector proteins are phosphorylated. Many effector proteins involved in multiple signal transduction cascades associated with these receptors have been suggested to dock to the receptor’s phosphorylated tyrosines (Trenker and Jura., 2020). After docking, the receptor can activate these effector proteins via various phosphorylation processes. RTK activation requires the binding of ATP. If the ability of these receptors to bind and use ATP is impaired, their function will be significantly reduced. This is crucial for targeted therapeutic interventions (Figure 1C).
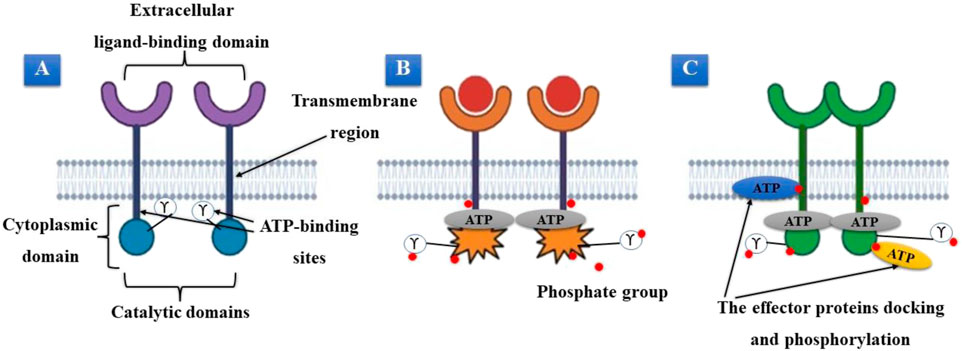
FIGURE 1. (A) Inactive tyrosine kinase receptor. (B) Activation of receptors, dimerization, and ATP binding. (C) The receptor’s phosphorylated tyrosines serve as a docking site for effector proteins.
RTKs are crucial for controlling numerous cellular activities in a normal state. Still, when they express themselves abnormally, they can lead to uncontrollable cell division, contributing to cancer’s pathobiology (Wheeler and Yarden., 2014). Activating a particular subclass of RTKs subclass 1, or ERBB, is aberrant in epithelial cancers. Additional RTKs, including the tumor metastasis-promoting Platelet-derived growth factor (PDGF), VEGFR, and VEGFR2 receptors, appear essential for tumor growth (García-Aranda and Redondo, 2019).
The nucleus, the inner surface of the plasma membrane, and other cell components all include NRTKs, a sizable class of cytosolic proteins. By participating in cellular signaling cascades, these proteins play an essential role in controlling survival, migration, differentiation, cellular proliferation, and metabolism. Given the role of NRTKs in cells, it is unsurprising that the cell tightly regulates their activity (Siveen et al., 2018). When these proteins, like their receptor counterparts, fail to function correctly due to genetic mutation, resulting in overexpression, loss of autoregulatory processes, or abnormal signaling, they can lead to the pathophysiology of various cancer types. Therefore, it is unsurprising that this protein family has become a crucial therapeutic focus in the fight against cancer (Fox et al., 2019).
After the Food and Drug Administration (FDA) gave its approval for the TKI imatinib in 2001, there was a rise in people’s interest in protein kinase inhibitors (Huang et al., 2020). Imatinib’s anticancer activities have been confirmed by numerous in vitro studies against CRC, and in vivo experiments may validate these findings. Imatinib’s anticancer effects in CRC were synergistic and pleiotropic (Dhiman et al., 2020). The development of small kinase inhibitors is based on the structure and sequence of the kinase catalytic core, which is defined by the presence of a smaller N-terminal subdomain (N-lobe) made up of a long α-helix and a β-sheet. A big C-terminal subdomain (C-lobe) with a primarily α-helical structure and an ATP binding site that serves as a hinge during structural adjustments (Adnan et al., 2022). A highly conserved Asp-Phe-Gly (DFG) motif, a component of the ATP-binding site that regulates magnesium binding, follows the activation loop (A-loop), which controls kinase activity (Bhullar et al., 2018).
Kinase inhibitors are divided into two major categories based on how they work: In addition to competing for the primary ATP-binding domain of the kinase catalytic core in the active form, type-I and type-II small kinase inhibitors are designed to bind to an additional allosteric pocket close to the ATP-binding site in the inactive state. Even so, type-II inhibitors are more focused than type-I inhibitors (Zhao et al., 2021). Type-I and type-II inhibitors prevent the specific protein kinase from phosphorylating a substrate molecule and deactivating downstream signaling. Kinase inhibitors may inhibit unregulated cell growth or apoptosis inhibition because dysregulated kinases can lead to defective signaling that can cause uncontrolled cell growth and proliferation (Roskoski, 2015).
Tyrosine kinases influence cell growth, migration, differentiation, apoptosis, and death by phosphorylating certain amino acids on substrate enzymes. This alters the downstream signal transduction triggered by TKs Du and Lovly. (2018). Dysregulated signal pathways can result from mutations or other constitutive activation or inhibition processes, which can cause cancer. Therefore, blocking these early signals with TKIs can avoid abnormal behavior of mutant or dysfunctional TKs. In tandem with the development of targeted monoclonal antibodies, a greater understanding of the molecular underpinnings and oncogenic signaling of CRC growth has led to testing TKIs (Yang et al., 2022). These substances can inhibit key enzymes that regulate signaling pathways crucial for cell survival, proliferation, differentiation, and development. Several RTKs, or the pathways through which these kinases function, have prospective therapeutic targets that have been enhanced or altered in CRC. Numerous small molecule TKIs have been discovered and examined for their potential to treat CRC cancer (Jiao et al., 2018).
The US FDA has authorized more than 50 TKIs, though most of these TKIs exhibit encouraging results in CRC pre-clinical testing. In the clinic, most patients fail (Tauriello and Batlle., 2016). Various causes include the absence of complicated predictive pre-clinical models, a lack of understanding of the pharmacodynamics and pharmacokinetics of TKIs, and a lack of information on the tumor mutational background and heterogeneity, which can cause clinical failure. Despite being a primary contributing factor in CRC metastasis and a therapeutic target, the TME remains unclear, contributing to the discrepancy between pre-clinical and clinical results (Tolba, 2020).
The 14 TKIs that had passed pre-clinical monotherapy testing were also investigated in a clinical trial for mCRC. Four TKIs (Lenvatinib, cediranib, cabozantinib, and apatinib) were identified as promising in non-randomized phase I/II studies, and two (fruquintinib and regorafenib) indicated therapeutic value in a randomized phase III study. 13 of them shown noticeable anti-cancer benefits in a pre-clinical setting (Han et al., 2020). Regorafenib is an oral multikinase inhibitor, that is, now approved for use in third-line mCRC therapy. It prevents tumor angiogenesis, oncogenesis, metastasis, and immunology by inhibiting tyrosine kinase receptors (Calvo-Garcia et al., 2022). Apatinib, an s-SRC, c-Kit, and VEGFR2, a relatively specific inhibitor, had a robust pro-apoptotic effect in vitro in animal CRC cell lines and humans. Patients with refractory CRC who did not have liver metastases responded well to apatinib alone, with a PFS of 3.9 months (Srivastava et al., 2022). A selective VEGFR1,-2,-3 inhibitors called fruquintinib has received approval in China to treat people with mCRC who have already failed at least two courses of systemic anti-neoplastic therapy. It is expected to be the 2nd TKI authorized for mCRC after receiving FDA fast-track approval for mCRC patients (Zhang et al., 2019).
Cediranib and cabozantinib, two TKIs that inhibit several kinases, produced apoptosis and antiproliferative activity in culture and slowed the growth of tumors in vivo. For CRC patients, it was not given any empirical investigation (Qin et al., 2019). Bosutinib is successful in vitro and in vivo in one analysis; however, its efficacy was only moderate in a following phase I clinical experiment (Isakoff et al., 2014). Numerous other TKIs also demonstrated encouraging anti-tumor pre-clinical effects, such as vandetanib (patient-derived cells), gefitinib (CRC cell lines), dasatinib (CRC cell lines and xenograft models), erlotinib (patient-derived xenografts), and linifanib (CRC cell lines, 3D micro tumor). However, none of these treatments worked effectively in the clinic as monotherapy (Table 1) (Lyer et al., 2022).
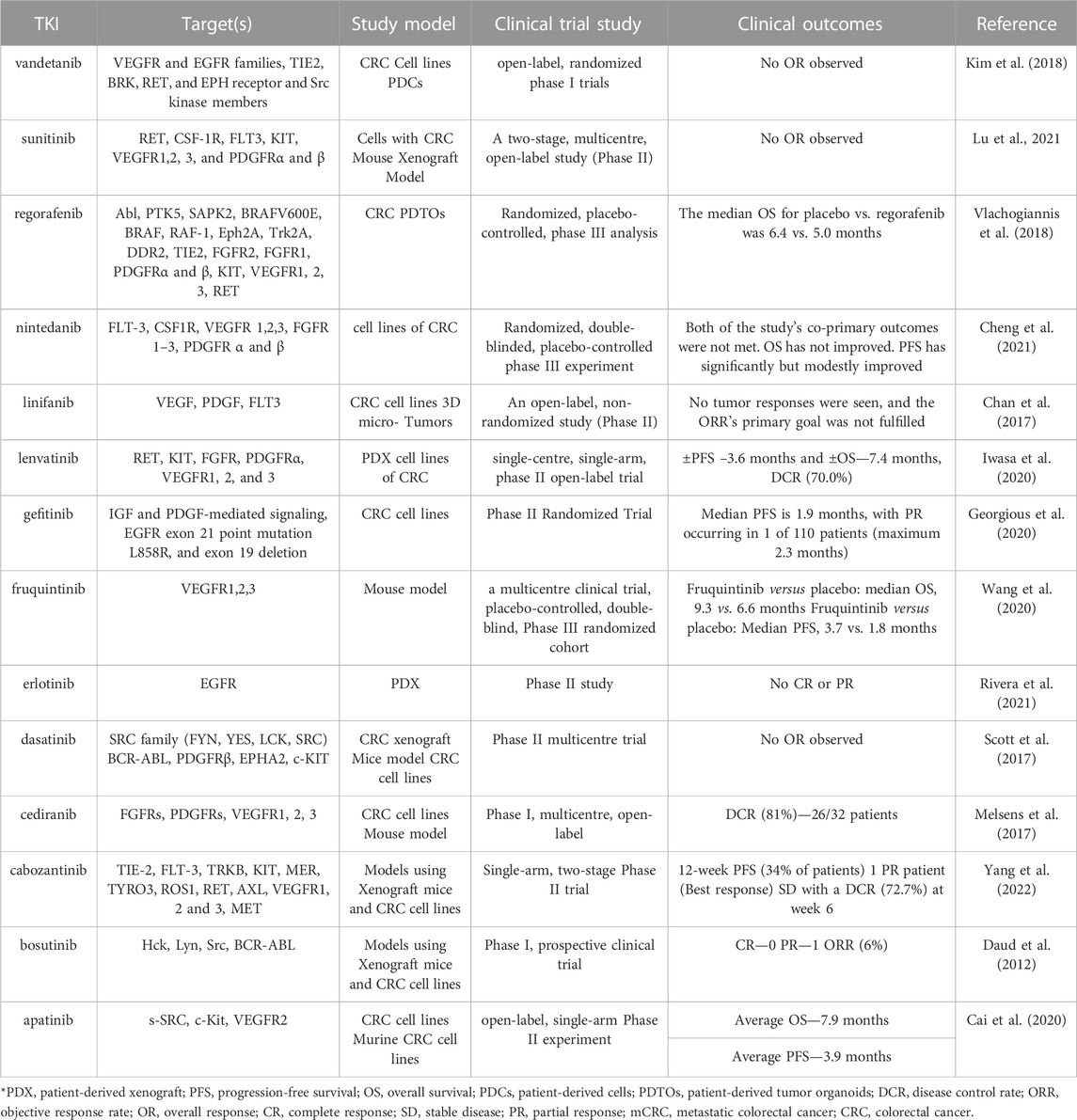
TABLE 1. The clinical results and pre-clinical efficacy of specific TKIs in monotherapy. The TKIs shown in bold have either demonstrated promising outcomes or have been effectively adapted for use in the clinic.
TKIs have been explored in CRC treatment combinations, and pre-clinical studies with 17 TKIs have been successful. Compared to the related pre-clinical investigations, several clinical trials employed various chemotherapy agents or antibodies with similar sites (Chen H. et al., 2022a; Chen R. et al., 2022b).
Targeting the EGFR/mitogen-activated protein kinase (MAPK) pathway, TKIs are combined with antibodies. In a previous study, cabozantinib was combined with the anti-EGFR antibody cetuximab to treat CRC cell lines, and it was found that this combination helped overcome cetuximab tolerance (Strickler et al., 2021). Furthermore, when TKIs are combined with an anti-VEGF antibody, such as bevacizumab or imatinib, a greater degree of vascular normalization has been observed without activation of extracellular matrix (ECM) deposition (Schiffmann et al., 2017).
When CT-26 isografts were treated with lenvatinib, pembrolizumab, and an anti-PD-1 antibody, tumor growth in vivo was significantly inhibited. Regorafenib and nivolumab had a synergistic immune-modulatory effect on CRC cells in another instance (Kato et al., 2019). Anti-PD-1 antibody nivolumab was combined with regorafenib to promote anti-tumor activity (Doleschel et al., 2021). Combinations of regorafenib and ICI were the focus of subsequent research. However, the findings revealed no therapeutic value. It evaluated the potential effectiveness of the combination of pembrolizumab and lenvatinib in patients with advanced non-MSI-H mCRC (Wang et al., 2020).
Encouraging results have been reported for the pre-clinical trials of cediranib in combination with radiation in CRC. In addition, vandetanib, irinotecan, and radiation significantly diminished tumor size in human colorectal xenograft models (Meyerhardt et al., 2012). However, the combination of vandetanib with cetuximab and irinotecan did not demonstrate any improvement in effectiveness compared to earlier results in patients with mCRC who had undergone therapy (Table 2) (Wachsberger et al., 2009).
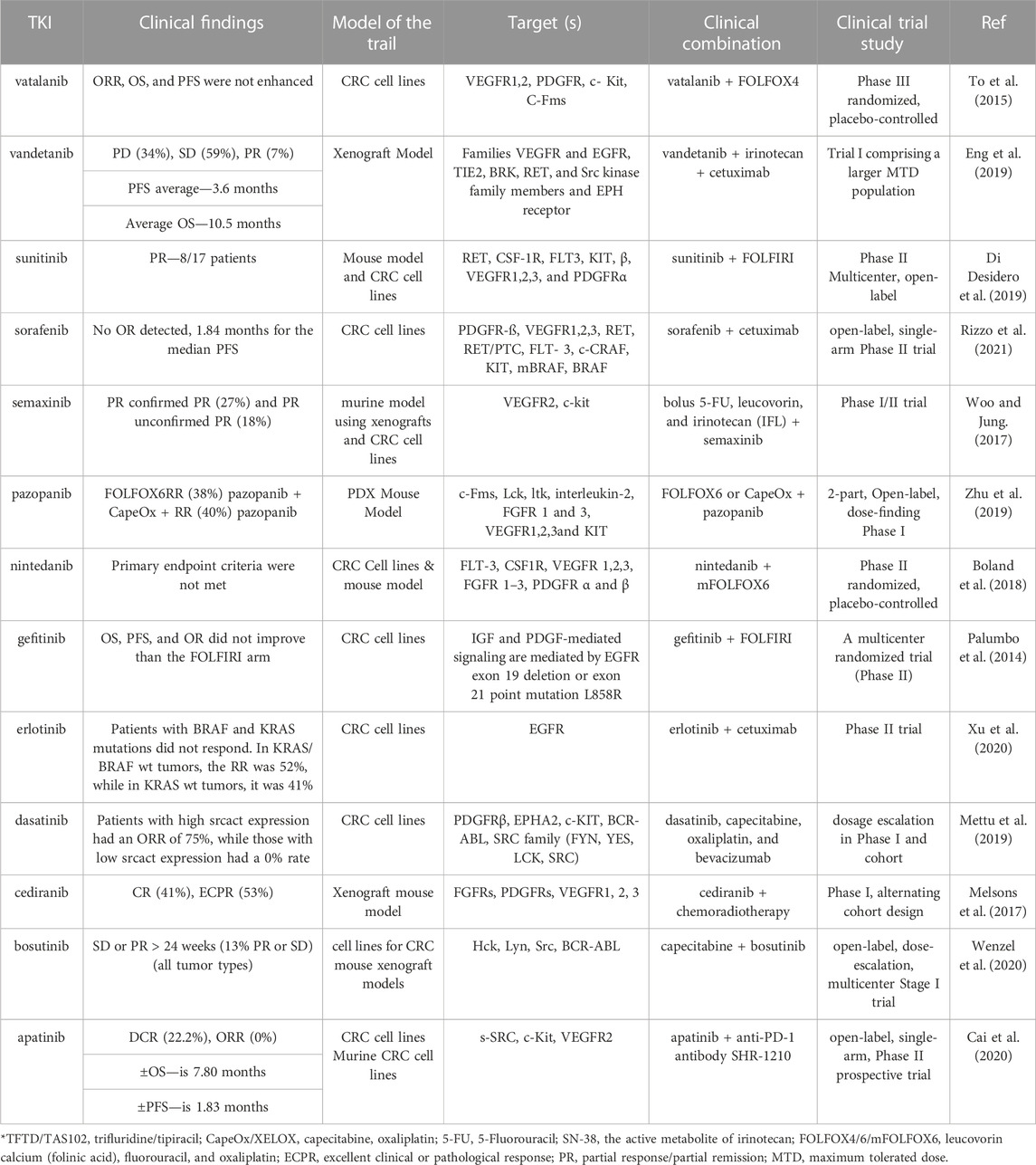
TABLE 2. Clinical results of additional TKI selected from combination therapies in pre-clinical studies.
5-fluorouracil (5-FU) treatment has enhanced survival in several cancers. The drug’s most significant effect has been documented in CRC. Active metabolites of 5-FU interfere with DNA and RNA synthesis via the folate metabolic pathway (Pardini et al., 2011). Patients with mCRC treated with oxaliplatin as a single drug showed limited efficacy, with response rates (RR) ranging from 10% to 24%. In contrast, the combination of oxaliplatin with 5FU has demonstrated RRs that vary from 20% to more than 50% due to a synergistic effect with 5FU (Comella et al., 2009). A camptothecin derivative known as irinotecan hydrochloride has anticancer efficacy against several tumor types. Irinotecan’s active metabolite is SN-38, which is synthesized by the enzyme carboxylesterase in the body. Survival has significantly increased since introducing irinotecan for treating CRC around the beginning of the 20th century. The overall survival time has been extended to more than 30 months due to the combination of irinotecan with 5-fluorouracil, oxaliplatin, and numerous molecularly targeted anticancer medications (Fujita et al., 2015).
The synthetic drug semaxanib, which inhibits VEGFR-1 and -2 tyrosine kinases, is a small and lipophilic molecule. For 28 patients with mCRC, semaxanib at two different dose levels in combination with fluorouracil and leucovorin showed a promising response of 31.6% as the first-line therapy (Rosen et al., 1999). A multicenter MABEL (Minimum anticipated biological effect level) trial studied the combination of cetuximab and CPT-11 at a dose and schedule as a pre-study in 1123 patients with mCRC exhibiting detectable EGFR. 9.2 months was the anticipated median survival, although at the expense of a tolerable toxicity profile. The efficacy and safety of C225 with CPT-11 observed in earlier studies were validated by MABEL in a larger context (Wilke et al., 2006). Gefitinib (ZD 1839) inhibits the EGFR tyrosine kinase selectively and has a 100-fold more effective potency against EGFR than other tyrosine or serine/threonine kinases. Gefitinib, unlike cetuximab, does not cause EGFR internalization or destruction in CRC cells, nor does it decrease EGF binding sites or EGFR protein levels (MacKenzie et al., 2005). Gefitinib monotherapy has shown anticancer activity in various CRC cell lines in both in vitro and in vivo investigations. Gefitinib, on the other hand, showed little activity in phase I/II clinical trials in individuals with mCRC (Rothenberg et al., 2004).
The most developed monoclonal antibody against EGFR currently under clinical development is cetuximab. A phase II trial of cetuximab with irinotecan was conducted in patients with EGFR-positive colorectal cancer who were refractory to both 5-fluorouracil (5-FU) and Irinotecan because preclinical and early clinical research indicate that cetuximab might reverse irinotecan resistance in CRC both in vitro and in vivo. The overall response rate for the 120 patients who received this regimen was 22.5% (Saltz et al., 2001). Kuo et al. reported results from a phase II research that included one cycle of FOLFOX-4, followed by further cycles of FOLFOX-4 with 500 mg/d gefitinib in 27 patients with proven progressing CRC following at least one chemotherapy regimen (generally irinotecan-based). 33% of patients experienced objective responses, whereas 48% maintained stable conditions over a prolonged period. The number of prior regimens exhibited no effect on response rates. The median event-free survival period was 5.4 months, and the total survival period was 12 months (Kuo et al., 2005).
CRC is a common cancer that substantially increases cancer mortality rates. Due to the complexity of colorectal carcinogenesis, the CRC survival rates of individual patients vary. It is thus beneficial to determine accurate and useful molecular biomarkers that contribute to CRC detection and management. Current studies have focused on finding precise, specialized biomarkers for CRC diagnosis and treatment success. This review article offered an overview of the latest CRC diagnostic biomarkers. Multiple signalling pathways are activated in CRC, making it impossible to address the disease with a single therapy. Combining conventional treatments with novel inhibitors that target multiple pathways is essential. Small-molecule TKIs are among the most recent additions to the vast range of cancer-treating drugs. TKI is a useful pharmacological strategy for treating a variety of malignancies, but it is not a solution. Protein tyrosine kinases appear to accelerate the growth and onset of CRC. The development of more effective biomarkers and the success of tailored medicines offer hope for the future management of CRC, but first, we need to learn more about the disease. Standardizing protocols, including extraction and quantification procedures and normalizing approaches, will be complicated in the future. Biomarker translation into the therapeutic context is also critical. Lastly, complicated laboratory equipment should be avoided for simple, inexpensive, quick solutions. We believed that developing additional novel targeted medicines could reduce the burden of CRC cancer.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported by the Shaoxing Health Science and Technology Plan (No. 2022SY018 to SZ).
We are thankful to the Shaoxing Health Science and Technology Plan for supporting this project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aasebø, K., Dragomir, A., Sundström, M., Mezheyeuski, A., Edqvist, P. H., Eide, G. E., et al. (2020). CDX2: A prognostic marker in metastatic colorectal cancer defining a better BRAF mutated and a worse KRAS mutated subgroup. Front. Oncol. 10, 8. doi:10.3389/fonc.2020.00008
Abo-Elela, D. A., Salem, A. M., Swellam, M., and Hegazy, M. G. (2023). Potential diagnostic role of circulating miRNAs in colorectal cancer. Int. J. Immunopathol. Pharmacol. 37, 03946320221144565. doi:10.1177/03946320221144565
Adnan, M., Koli, S., Mohammad, T., Siddiqui, A. J., Patel, M., Alshammari, N., et al. (2022). Searching for novel anaplastic lymphoma kinase inhibitors: structure-guided screening of natural compounds for a tyrosine kinase therapeutic target in cancers. OMICS A J. Integr. Biol. 26 (8), 461–470. doi:10.1089/omi.2022.0067
Ahmed, F. E., Gouda, M. M., Ahmed, N. C., and Hussein, L. (2019). Quantification of microRNAs by absolute Dpcr for the diagnostic screening of colon cancer. J. Colon Rectal Cancer 1 (3), 10–37. doi:10.14302/issn.2471-7061.jcrc-18-2526
Akhlaghipour, I., Bina, A. R., Abbaszadegan, M. R., and Moghbeli, M. (2021). Methylation as a critical epigenetic process during tumor progressions among Iranian population: an overview. Genes Environ. 43, 14. doi:10.1186/s41021-021-00187-1
Akimoto, N., Ugai, T., Zhong, R., Hamada, T., Fujiyoshi, K., Giannakis, M., et al. (2021). Rising incidence of early-onset colorectal cancer—a call to action. Nat. Rev. Clin. Oncol. 18 (4), 230–243. doi:10.1038/s41571-020-00445-1
Al-Duhaidahawi, M. (2023). Role of CDX2 marker in patients with colorectal cancer. Biomed. Chem. Sci. 2 (1), 11–15. doi:10.48112/bcs.v2i1.321
Altomare, D. F., Di Lena, M., Porcelli, F., Travaglio, E., Longobardi, F., Tutino, M., et al. (2015). Effects of curative colorectal cancer surgery on exhaled volatile organic compounds and potential implications in clinical follow-up. Ann. Surg. 262 (5), 862–866; discussion 866-867. doi:10.1097/SLA.0000000000001471
Alzahrani, S. M., Al Doghaither, H. A., and Al-Ghafari, A. B. (2021). General insight into cancer: an overview of colorectal cancer. Mol. Clin. Oncol. 15 (6), 1–8. doi:10.3892/mco.2021.2433
Amal, H., Leja, M., Funka, K., Lasina, I., Skapars, R., Sivins, A., et al. (2016). Breath testing as potential colorectal cancer screening tool. Int. J. Cancer 138 (1), 229–236. doi:10.1002/ijc.29701
Baharudin, R., Ishak, M., Muhamad Yusof, A., Saidin, S., Syafruddin, S. E., Wan Mohamad Nazarie, W. F., et al. (2022). Epigenome-wide DNA methylation profiling in colorectal cancer and normal adjacent colon using Infinium human methylation 450K. Diagnostics 12 (1), 198. doi:10.3390/diagnostics12010198
Bhullar, K. S., Lagarón, N. O., McGowan, E. M., Parmar, I., Jha, A., Hubbard, B. P., et al. (2018). Kinase-targeted cancer therapies: progress, challenges, and future directions. Mol. cancer 17, 48–20. doi:10.1186/s12943-018-0804-2
Boland, P. M., Fakih, M., Lim, D., Attwood, K., Tan, W., Mastri, M., et al. (2018). A phase I/II study of nintedanib and capecitabine in refractory metastatic colorectal cancer. J. Clin. Oncol. 36 (15). doi:10.1200/JCO.2018.36.15_suppl.3552
Bond, A., Greenwood, R., Lewis, S., Corfe, B., Sarkar, S., O'Toole, P., et al. (2019). Volatile organic compounds emitted from faeces as a biomarker for colorectal cancer. Alimentary Pharmacol. Ther. 49 (8), 1005–1012. doi:10.1111/apt.15140
Bouchahda, M., Macarulla, T., Spano, J. P., Bachet, J. B., Lledo, G., Andre, T., et al. (2008). Cetuximab efficacy and safety in a retrospective cohort of elderly patients with heavily pretreated metastatic colorectal cancer. Crit. Rev. oncology/hematology 67 (3), 255–262. doi:10.1016/j.critrevonc.2008.02.003
Bresalier, R. S., Grady, W. M., Markowitz, S. D., Nielsen, H. J., Batra, S. K., and Lampe, P. D. (2020). Biomarkers for early detection of colorectal cancer: the early detection research network, a framework for clinical translation. Cancer Epidemiol. Biomarkers Prev. 29 (12), 2431–2440. doi:10.1158/1055-9965.EPI-20-0234
Cai, C., Luo, Q., Liu, Y., Peng, Y., Zhang, X., Jiang, Z., et al. (2022). The optimal first-line treatment for patients with left-sided RAS wild-type metastatic colorectal cancer: double-drug regimen or triple-drug regimen therapy. Front. Pharmacol. 13, 1015510. doi:10.3389/fphar.2022.1015510
Cai, J., Zuo, X., Chen, Z., Zhang, Y., Wang, J., Wang, J., et al. (2019). Long non-coding RNAs serve as potential diagnostic biomarkers for colorectal cancer. J. Cancer 10 (3), 611–619. doi:10.7150/jca.28780
Cai, X., Wei, B., Li, L., Chen, X., Yang, J., Li, X., et al. (2020). Therapeutic potential of apatinib against colorectal cancer by inhibiting VEGFR2-mediated angiogenesis and β-catenin signaling. OncoTargets Ther. 13, 11031–11044. doi:10.2147/OTT.S266549
Calvo-García, A., Pérez Abánades, M., Ruíz-García, S., Fernández Román, A. B., Letellez Fernández, J., Candel García, B., et al. (2022). Effectiveness, toxicity, and survival predictors of regorafenib in metastatic colorectal cancer: A multicenter study of routinely collected data. Oncol. (08909091) 36 (12), 732–738. doi:10.46883/2022.25920981
Cao, L., Hu, T., Lu, H., and Peng, D. (2020). N-MYC downstream-regulated gene 4 (NDRG4), a frequent downregulated gene through DNA hypermethylation, plays a tumor-suppressive role in esophageal adenocarcinoma. Cancers 12 (9), 2573. doi:10.3390/cancers12092573
Chan, E., Goff, L. W., Cardin, D. B., Ancell, K., Smith, S. J., Whisenant, J. G., et al. (2017). Phase II study of the multi-kinase inhibitor of angiogenesis, linifanib, in patients with metastatic and refractory colorectal cancer expressing mutated KRAS. Investig. New Drugs 35, 491–498. doi:10.1007/s10637-017-0458-8
Chan, H. T., Nagayama, S., Otaki, M., Chin, Y. M., Fukunaga, Y., Ueno, M., et al. (2023). Tumor-informed or tumor-agnostic circulating tumor DNA as a biomarker for risk recurrence in resected colorectal cancer patients. Front. Oncol., 7079. doi:10.3389/fonc.2022.1055968
Chan, S. C. H., and Liang, J. Q. (2022). Advances in tests for colorectal cancer screening and diagnosis. Expert Rev. Mol. Diagnostics 22 (4), 449–460. doi:10.1080/14737159.2022.2065197
Chandrapalan, S., and Arasaradnam, R. P. (2020). Urine as a biological modality for colorectal cancer detection. Expert Rev. Mol. diagnostics 20 (5), 489–496. doi:10.1080/14737159.2020.1738928
Chang, H., Mishra, R., Cen, C., Tang, Y., Ma, C., Wasti, S., et al. (2021). Metagenomic analyses expand bacterial and functional profiling biomarkers for colorectal cancer in a hainan cohort, China. Curr. Microbiol. 78, 705–712. doi:10.1007/s00284-020-02299-3
Chen, H., Lu, B., and Dai, M. (2022a). Colorectal cancer screening in China: status, challenges, and prospects—China, 2022. China CDC Wkly. 4 (15), 322–328. doi:10.46234/ccdcw2022.077
Chen, H. M., Lin, C. C., Chen, W. S., Jiang, J. K., Yang, S. H., Chang, S. C., et al. (2021). Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) is a prognostic biomarker and associated with chemotherapy responsiveness in colorectal cancer. Int. J. Mol. Sci. 22 (13), 6940. doi:10.3390/ijms22136940
Chen, R., Li, Q., Xu, S., Ye, C., Tian, T., Jiang, Q., et al. (2022b). Modulation of the tumour microenvironment in hepatocellular carcinoma by tyrosine kinase inhibitors: from modulation to combination therapy targeting the microenvironment. Cancer Cell Int. 22 (1), 73. doi:10.1186/s12935-021-02435-4
Cheng, G., Li, Y., Liu, Z., and Song, X. (2021). The microRNA-429/DUSP4 axis regulates the sensitivity of colorectal cancer cells to nintedanib. Mol. Med. Rep. 23 (4), 1–1. doi:10.3892/mmr.2021.11867
Choi, H. B., Pyo, J. S., Son, S., Kim, K., and Kang, G. (2022). Diagnostic and prognostic roles of CDX2 immunohistochemical expression in colorectal cancers. Diagnostics 12 (3), 757. doi:10.3390/diagnostics12030757
Comella, P., Casaretti, R., Sandomenico, C., Avallone, A., and Franco, L. (2009). Role of oxaliplatin in the treatment of colorectal cancer. Ther. Clin. risk Manag. 5, 229–238. doi:10.2147/tcrm.s3583
Cruz, A., Carvalho, C. M., Cunha, A., Crespo, A., Iglesias, Á., García-Nimo, L., et al. (2021). Faecal diagnostic biomarkers for colorectal cancer. Cancers 13 (21), 5568. doi:10.3390/cancers13215568
Dabir, P. D., Svanholm, H., and Christiansen, J. J. (2018). SATB2 is a supplementary immunohistochemical marker to CDX2 in the diagnosis of colorectal carcinoma metastasis in an unknown primary. Apmis 126 (6), 494–500. doi:10.1111/apm.12854
Daud, A. I., Krishnamurthi, S. S., Saleh, M. N., Gitlitz, B. J., Borad, M. J., Gold, P. J., et al. (2012). Phase I study of bosutinib, a src/abl tyrosine kinase inhibitor, administered to patients with advanced solid tumors. Clin. Cancer Res. 18 (4), 1092–1100. doi:10.1158/1078-0432.CCR-11-2378
Davidsen, J. (2020). The role of CDX2 in colon cancer development and progression. Denmark: Doctoral dissertation, Department of Science and Environment, Roskilde University.
Dhiman, D. K., Sanyal, S. N., and Vaish, V. (2020). Imatinib exhibit synergistic pleiotropy in the prevention of colorectal cancer by suppressing proinflammatory, cell survival and angiogenic signaling. Cell. Signal. 76, 109803. doi:10.1016/j.cellsig.2020.109803
Di Desidero, T., Orlandi, P., Fioravanti, A., Alì, G., Cremolini, C., Loupakis, F., et al. (2019). Chemotherapeutic and antiangiogenic drugs beyond tumor progression in colon cancer: evaluation of the effects of switched schedules and related pharmacodynamics. Biochem. Pharmacol. 164, 94–105. doi:10.1016/j.bcp.2019.04.001
Ding, Q., Kong, X., Zhong, W., Liu, W., and Ye, Q. (2022). Fecal biomarkers: non-invasive diagnosis of colorectal cancer. Front. Oncol. 12, 812663. doi:10.3389/fonc.2022.812663
Diwanji, D., Thaker, T., and Jura, N. (2019). More than the sum of the parts: toward full-length receptor tyrosine kinase structures. IUBMB life 71 (6), 706–720. doi:10.1002/iub.2060
Doleschel, D., Hoff, S., Koletnik, S., Rix, A., Zopf, D., Kiessling, F., et al. (2021). Regorafenib enhances anti-PD1 immunotherapy efficacy in murine colorectal cancers, and its combination prevents tumor regrowth. J. Exp. Clin. Cancer Res. 40 (1), 1–14. doi:10.1186/s13046-021-02043-0
Du, Z., and Lovly, C. M. (2018). Mechanisms of receptor tyrosine kinase activation in cancer. Mol. cancer 17, 58–13. doi:10.1186/s12943-018-0782-4
Eng, C., Kim, T. W., Bendell, J., Argilés, G., Tebbutt, N. C., Di Bartolomeo, M., et al. (2019). Atezolizumab with or without cobimetinib versus regorafenib in previously treated metastatic colorectal cancer (IMblaze370): A multicentre, open-label, phase 3, randomised, controlled trial. lancet Oncol. 20 (6), 849–861. doi:10.1016/S1470-2045(19)30027-0
Fonseca, A., Ramalhete, S. V., Mestre, A., das Neves, R. P., Marreiros, A., Castelo-Branco, P., et al. (2021). Identification of colorectal cancer associated biomarkers: an integrated analysis of miRNA expression. Aging (Albany NY) 13 (18), 21991–22029. doi:10.18632/aging.203556
Fornaro, L., Baldi, G. G., Masi, G., Allegrini, G., Loupakis, F., Vasile, E., et al. (2011). Cetuximab plus irinotecan after irinotecan failure in elderly metastatic colorectal cancer patients: clinical outcome according to KRAS and BRAF mutational status. Crit. Rev. oncology/hematology 78 (3), 243–251. doi:10.1016/j.critrevonc.2010.06.003
Fox, M., Crafter, C., and Owen, D. (2019). The non-receptor tyrosine kinase ACK: regulatory mechanisms, signalling pathways and opportunities for attACKing cancer. Biochem. Soc. Trans. 47 (6), 1715–1731. doi:10.1042/BST20190176
Fujita, K. I., Kubota, Y., Ishida, H., and Sasaki, Y. (2015). Irinotecan a key chemotherapeutic drug for metastatic colorectal cancer. World J. gastroenterology 21 (43), 12234–12248. doi:10.3748/wjg.v21.i43.12234
Gao, H. L., Lv, L. B., Zhao, W. F., Lu, Q. W., and Fan, J. Q. (2023). Diagnostic accuracy of the multi-target stool DNA test in detecting colorectal cancer: A hospital-based study. World J. Gastrointest. Oncol. 15 (1), 102–111. doi:10.4251/wjgo.v15.i1.102
Gao, R., Xia, K., Wu, M., Zhong, H., Sun, J., Zhu, Y., et al. (2022). Alterations of gut mycobiota profiles in adenoma and colorectal cancer. Front. Cell. Infect. Microbiol. 119, 839435. doi:10.3389/fcimb.2022.839435
García-Aranda, M., and Redondo, M. (2019). Targeting receptor kinases in colorectal cancer. Cancers 11 (4), 433. doi:10.3390/cancers11040433
Georgiou, A., Stewart, A., Cunningham, D., Banerji, U., and Whittaker, S. R. (2020). Inactivation of NF1 promotes resistance to EGFR inhibition in KRAS/NRAS/brafv600-wild-type colorectal cancer. Mol. Cancer Res. 18 (6), 835–846. doi:10.1158/1541-7786.MCR-19-1201
Gligorijević, N., Dobrijević, Z., Šunderić, M., Robajac, D., Četić, D., Penezić, A., et al. (2022). The insulin-like growth factor system and colorectal cancer. Life 12 (8), 1274. doi:10.3390/life12081274
Gong, C., Yang, H., Wang, S., Liu, J., Li, Z., Hu, Y., et al. (2021). hTERT promotes CRC proliferation and migration by recruiting YBX1 to increase NRF2 expression. Front. Cell Dev. Biol. 9, 658101. doi:10.3389/fcell.2021.658101
Han, Y., Zhu, L., Wu, W., Zhang, H., Hu, W., Dai, L., et al. (2020). Small molecular immune modulators as anti-cancer agents. Adv. Exp. Med. Biol. 1248, 547–618. doi:10.1007/978-981-15-3266-5_22
Handley, S. A., and Devkota, S. (2019). Going viral: A novel role for bacteriophage in colorectal cancer. MBio 10 (1), 02618–02626. doi:10.1128/mBio.02626-18
Hannigan, G. D., Duhaime, M. B., Ruffin IV, M. T., Koumpouras, C. C., and Schloss, P. D. (2018). Diagnostic potential and interactive dynamics of the colorectal cancer virome. MBio 9 (6), 02218–02248. doi:10.1128/mBio.02248-18
Hariharan, R., and Jenkins, M. (2020). Utility of the methylated SEPT9 test for the early detection of colorectal cancer: A systematic review and meta-analysis of diagnostic test accuracy. BMJ Open Gastroenterol. 7 (1), e000355. doi:10.1136/bmjgast-2019-000355
He, J., Wu, W., Tang, H. C., Liu, B., and Bu, L. L. (2023). Comprehensive landscape and future perspectives of long non-coding RNAs (lncRNAs) in colorectal cancer (CRC): based on bibliometric analysis. Non-coding RNA Res. 8 (1), 33–92. doi:10.1016/bs.ircmb.2022.11.002
Hendricks, A., Dall, K., Brandt, B., Geisen, R., Röder, C., Schafmayer, C., et al. (2021). Longitudinal analysis of circulating tumor cells in colorectal cancer patients by a cytological and molecular approach: feasibility and clinical application. Front. Oncol. 11, 646885. doi:10.3389/fonc.2021.646885
Hijos-Mallada, G., Lué, A., Velamazan, R., Saura, N., Abril, C., Lorenzo, M., et al. (2021). The addition of other fecal biomarkers does not improve the diagnostic accuracy of immunochemical fecal occult blood test alone in a colorrectal cancer screening cohort. Front. Med. 8, 665786. doi:10.3389/fmed.2021.665786
Hossain, M. S., Karuniawati, H., Jairoun, A. A., Urbi, Z., Ooi, D. J., John, A., et al. (2022). Colorectal cancer: A review of carcinogenesis, global epidemiology, current challenges, risk factors, preventive and treatment strategies. Cancers 14 (7), 1732. doi:10.3390/cancers14071732
Hrudka, J., Fišerová, H., Jelínková, K., Matěj, R., and Waldauf, P. (2021). Cytokeratin 7 expressions as a predictor of an unfavorable prognosis in colorectal carcinoma. Sci. Rep. 11 (1), 1–10.
Hrudka, J., Matěj, R., Nikov, A., Tomyak, I., Fišerová, H., Jelínková, K., et al. (2022). Loss of SATB2 expression correlates with cytokeratin 7 and PD-L1 tumor cell positivity and aggressiveness in colorectal cancer. Sci. Rep. 12 (1), 19152. doi:10.1038/s41598-022-22685-0
Huang, L., Jiang, S., and Shi, Y. (2020). Tyrosine kinase inhibitors for solid tumors in the past 20 years (2001–2020). J. Hematol. Oncol. 13, 1–23. doi:10.1186/s13045-020-00977-0
Huang, Z., and Yang, M. (2022). Molecular network of colorectal cancer and current therapeutic options. Front. Oncol. 12, 852927. doi:10.3389/fonc.2022.852927
Isakoff, S. J., Wang, D., Campone, M., Calles, A., Leip, E., Turnbull, K., et al. (2014). Bosutinib plus capecitabine for selected advanced solid tumours: results of a phase 1 dose-escalation study. Br. J. Cancer 111 (11), 2058–2066. doi:10.1038/bjc.2014.508
Islam Khan, M. Z., Tam, S. Y., and Law, H. K. W. (2022). Advances in high throughput proteomics profiling in establishing potential biomarkers for gastrointestinal cancer. Cells 11 (6), 973. doi:10.3390/cells11060973
Iwaizumi, M., Taniguchi, T., Kurachi, K., Osawa, S., Sugimoto, K., Baba, S., et al. (2023). Methylation of CpG island promoters at ZNF625, LONRF2, SDC2, and WDR17 in a patient with numerous non-granular type laterally spreading tumors and colorectal cancer: A case report. Oncol. Lett. 25 (1), 14–18. doi:10.3892/ol.2022.13600
Iwasa, S., Okita, N., Kuchiba, A., Ogawa, G., Kawasaki, M., Nakamura, K., et al. (2020). Phase II study of lenvatinib for metastatic colorectal cancer refractory to standard chemotherapy: the LEMON study (NCCH1503). ESMO open 5 (4), e000776. doi:10.1136/esmoopen-2020-000776
Iwasaki, H., Shimura, T., and Kataoka, H. (2019). Current status of urinary diagnostic biomarkers for colorectal cancer. Clin. Chim. Acta 498, 76–83. doi:10.1016/j.cca.2019.08.011
Iyer, K. K., van Erp, N. P., Tauriello, D. V., Verheul, H. M., and Poel, D. (2022). Lost in translation: revisiting the use of tyrosine kinase inhibitors in colorectal cancer. Cancer Treat. Rev. 110, 102466. doi:10.1016/j.ctrv.2022.102466
Jelski, W., and Mroczko, B. (2020). Biochemical markers of colorectal cancer–present and future. Cancer Manag. Res. 12, 4789–4797. doi:10.2147/CMAR.S253369
Jiao, Q., Bi, L., Ren, Y., Song, S., Wang, Q., and Wang, Y. S. (2018). Advances in studies of tyrosine kinase inhibitors and their acquired resistance. Mol. cancer 17 (1), 36–12. doi:10.1186/s12943-018-0801-5
Kato, Y. U., Tabata, K., Kimura, T., Yachie-Kinoshita, A., Ozawa, Y., Yamada, K., et al. (2019). Lenvatinib plus anti-PD-1 antibody combination treatment activates CD8+ T cells through reduction of tumor-associated macrophage and activation of the interferon pathway. PloS one 14 (2), e0212513. doi:10.1371/journal.pone.0212513
Kaur, P., Mani, S., Cros, M. P., Scoazec, J. Y., Chemin, I., Hainaut, P., et al. (2012). Epigenetic silencing of sFRP1 activates the canonical Wnt pathway and contributes to increased cell growth and proliferation in hepatocellular carcinoma. Tumor Biol. 33, 325–336. doi:10.1007/s13277-012-0331-5
Kibriya, M. G., Raza, M., Kamal, M., Haq, Z., Paul, R., Mareczko, A., et al. (2022). Relative telomere length change in colorectal carcinoma and its association with tumor characteristics, gene expression and microsatellite instability. Cancers 14 (9), 2250. doi:10.3390/cancers14092250
Kim, J. H. (2015). Chemotherapy for colorectal cancer in the elderly. World J. Gastroenterology WJG 21 (17), 5158–5166. doi:10.3748/wjg.v21.i17.5158
Kim, S. Y., Oh, S. O., Kim, K., Lee, J., Kang, S., Kim, K. M., et al. (2018). NCOA4-RET fusion in colorectal cancer: therapeutic challenge using patient-derived tumor cell lines. J. Cancer 9 (17), 3032–3037. doi:10.7150/jca.26256
Koonin, E. V., Dolja, V. V., and Krupovic, M. (2021). The healthy human virome: from virus–host symbiosis to disease. Curr. Opin. virology 47, 86–94. doi:10.1016/j.coviro.2021.02.002
Kuo, T., Cho, C. D., Halsey, J., Wakelee, H. A., Advani, R. H., Ford, J. M., et al. (2005). Phase II study of gefitinib, fluorouracil, leucovorin, and oxaliplatin therapy in previously treated patients with metastatic colorectal cancer. J. Clin. Oncol. 23 (24), 5613–5619. doi:10.1200/JCO.2005.08.359
Ladabaum, U., Dominitz, J. A., Kahi, C., and Schoen, R. E. (2020). Strategies for colorectal cancer screening. Gastroenterology 158 (2), 418–432. doi:10.1053/j.gastro.2019.06.043
Li, M., Wu, S., Zhuang, C., Shi, C., Gu, L., Wang, P., et al. (2023). Metabolomic analysis of circulating tumor cells derived liver metastasis of colorectal cancer. Heliyon 9 (1), e12515. doi:10.1016/j.heliyon.2022.e12515
Lin, H., Yu, Y., Zhu, L., Lai, N., Zhang, L., Guo, Y., et al. (2023). Implications of hydrogen sulfide in colorectal cancer: mechanistic insights and diagnostic and therapeutic strategies. Redox Biol. 59, 102601. doi:10.1016/j.redox.2023.102601
Liu, N. N., Jiao, N., Tan, J. C., Wang, Z., Wu, D., Wang, A. J., et al. (2022). Multi-kingdom microbiota analyses identify bacterial–fungal interactions and biomarkers of colorectal cancer across cohorts. Nat. Microbiol. 7 (2), 238–250. doi:10.1038/s41564-021-01030-7
Liu, T., Liu, D., Guan, S., and Dong, M. (2021). Diagnostic role of circulating MiR-21 in colorectal cancer: A update meta-analysis. Ann. Med. 53 (1), 87–102. doi:10.1080/07853890.2020.1828617
Lotfollahzadeh, S., Recio-Boiles, A., and Cagir, B. (2022). Colon cancer. United States: StatPearls Publishing. StatPearls [Internet].
Lulli, M., Napoli, C., Landini, I., Mini, E., and Lapucci, A. (2022). Role of non-coding RNAs in colorectal cancer: focus on long non-coding RNAs. Int. J. Mol. Sci. 23 (21), 13431. doi:10.3390/ijms232113431
Luo, H., Shen, K., Li, B., Li, R., Wang, Z., and Xie, Z. (2020). Clinical significance and diagnostic value of serum NSE, CEA, CA19-9, CA125, and CA242 levels in colorectal cancer. Oncol. Lett. 20 (1), 742–750. doi:10.3892/ol.2020.11633
MacKenzie, M. J., Hirte, H. W., Glenwood, G., Jean, M., Goel, R., Major, P. P., et al. (2005). A phase II trial of ZD1839 (Iressa™) 750 mg per day, an oral epidermal growth factor receptor-tyrosine kinase inhibitor, in patients with metastatic colorectal cancer. Investig. new drugs 23, 165–170. doi:10.1007/s10637-005-5862-9
Markandeywar, T. S., Narang, R. K., Singh, D., and Rai, V. K. (2023). Targeted delivery of doxorubicin as a potential chemotherapeutic agent. Curr. Drug Deliv. 20, 904–918. doi:10.2174/1567201819666220714101952
Melsens, E., Verberckmoes, B., Rosseel, N., Vanhove, C., Descamps, B., Pattyn, P., et al. (2017). The VEGFR inhibitor cediranib improves the efficacy of fractionated radiotherapy in a colorectal cancer xenograft model. Eur. Surg. Res. 58 (3-4), 95–108. doi:10.1159/000452741
Mettu, N. B., Niedzwiecki, D., Rushing, C., Nixon, A. B., Jia, J., Haley, S., et al. (2019). A phase I study of gemcitabine+ dasatinib (gd) or gemcitabine+ dasatinib+ cetuximab (GDC) in refractory solid tumors. Cancer Chemother. Pharmacol. 83, 1025–1035. doi:10.1007/s00280-019-03805-6
Meyerhardt, J. A., Ancukiewicz, M., Abrams, T. A., Schrag, D., Enzinger, P. C., Chan, J. A., et al. (2012). Phase I study of cetuximab, irinotecan, and vandetanib (ZD6474) as therapy for patients with previously treated metastastic colorectal cancer. PLoS One 7 (6), e38231. doi:10.1371/journal.pone.0038231
Mezheyeuski, A., Ponten, F., Edqvist, P. H., Sundström, M., Thunberg, U., Qvortrup, C., et al. (2020). Metastatic colorectal carcinomas with high SATB2 expression are associated with better prognosis and response to chemotherapy: A population-based scandinavian study. Acta Oncol. 59 (3), 284–290. doi:10.1080/0284186X.2019.1691258
Min, L., Chen, J., Yu, M., and Liu, D. (2023). Using circulating tumor DNA as a novel biomarker to screen and diagnose colorectal cancer: A meta-analysis. J. Clin. Med. 12 (2), 408. doi:10.3390/jcm12020408
Mo, S., Dai, W., Wang, H., Lan, X., Ma, C., Su, Z., et al. (2023). Early detection and prognosis prediction for colorectal cancer by circulating tumour DNA methylation haplotypes: A multicentre cohort study. EClinicalMedicine 55, 101717. doi:10.1016/j.eclinm.2022.101717
Morgan, E., Arnold, M., Gini, A., Lorenzoni, V., Cabasag, C. J., Laversanne, M., et al. (2023). Global burden of colorectal cancer in 2020 and 2040: incidence and mortality estimates from GLOBOCAN. Gut 72 (2), 338–344. doi:10.1136/gutjnl-2022-327736
Mou, L., Tian, X., Zhou, B., Zhan, Y., Chen, J., Lu, Y., et al. (2021). Improving outcomes of tyrosine kinase inhibitors in hepatocellular carcinoma: new data and ongoing trials. Front. Oncol. 11, 752725. doi:10.3389/fonc.2021.752725
Mueller, D., and Győrffy, B. (2022). DNA methylation-based diagnostic, prognostic, and predictive biomarkers in colorectal cancer. Biochimica Biophysica Acta (BBA)-Reviews Cancer 1877, 188722. doi:10.1016/j.bbcan.2022.188722
Murer, P., Plüss, L., and Neri, D. (2020). A novel human monoclonal antibody specific to the A33 glycoprotein recognizes colorectal cancer and inhibits metastasis. MAbs 12, 1714371. doi:10.1080/19420862.2020.1714371
Natoli, C., Perrucci, B., Perrotti, F., Falchi, L., and Iacobelli, S.Consorzio Interuniversitario Nazionale per Bio-Oncologia CINBO (2010). Tyrosine kinase inhibitors. Curr. cancer drug targets 10 (5), 462–483. doi:10.2174/156800910791517208
Neri, G., Arpa, G., Guerini, C., Grillo, F., Lenti, M. V., Giuffrida, P., et al. (2020). Small bowel adenocarcinomas featuring special AT-rich sequence-binding protein 2 (SATB2) expression and a colorectal cancer-like immunophenotype: A potential diagnostic pitfall. Cancers 12 (11), 3441. doi:10.3390/cancers12113441
Niedermaier, T., Tikk, K., Gies, A., Bieck, S., and Brenner, H. (2020). Sensitivity of fecal immunochemical test for colorectal cancer detection differs according to stage and location. Clin. Gastroenterology Hepatology 18 (13), 2920–2928. doi:10.1016/j.cgh.2020.01.025
Oh, H. H., and Joo, Y. E. (2020). Novel biomarkers for the diagnosis and prognosis of colorectal cancer. Intestinal Res. 18 (2), 168–183. doi:10.5217/ir.2019.00080
Palumbo, I., Piattoni, S., Valentini, V., Marini, V., Contavalli, P., Calzuola, M., et al. (2014). Gefitinib enhances the effects of combined radiotherapy and 5-fluorouracil in a colorectal cancer cell line. Int. J. colorectal Dis. 29, 31–41. doi:10.1007/s00384-013-1754-1
Pardini, B., Kumar, R., Naccarati, A., Novotny, J., Prasad, R. B., Forsti, A., et al. (2011). 5-Fluorouracil-based chemotherapy for colorectal cancer and MTHFR/MTRR genotypes. Br. J. Clin. Pharmacol. 72 (1), 162–163. doi:10.1111/j.1365-2125.2010.03892.x
Park, S. K., Baek, H. L., Yu, J., Kim, J. Y., Yang, H. J., Jung, Y. S., et al. (2017). Is methylation analysis of SFRP2, TFPI2, NDRG4, and BMP3 promoters suitable for colorectal cancer screening in the Korean population? Intestinal Res. 15 (4), 495–501. doi:10.5217/ir.2017.15.4.495
Peng, L., Jiang, J., Chen, H. N., Zhou, L., Huang, Z., Qin, S., et al. (2021). Redox-sensitive cyclophilin A elicits chemoresistance through realigning cellular oxidative status in colorectal cancer. Cell Rep. 37 (9), 110069. doi:10.1016/j.celrep.2021.110069
Perdyan, A., Spychalski, P., Kacperczyk, J., Rostkowska, O., and Kobiela, J. (2020). Circulating Tumor DNA in KRAS positive colorectal cancer patients as a prognostic factor–a systematic review and meta-analysis. Crit. Rev. Oncology/Hematology 154, 103065. doi:10.1016/j.critrevonc.2020.103065
Piawah, S., and Venook, A. P. (2019). Targeted therapy for colorectal cancer metastases: A review of current methods of molecularly targeted therapy and the use of tumor biomarkers in the treatment of metastatic colorectal cancer. Cancer 125 (23), 4139–4147. doi:10.1002/cncr.32163
Pickhardt, P. J., Graffy, P. M., Weigman, B., Deiss-Yehiely, N., Hassan, C., and Weiss, J. M. (2020). Diagnostic performance of multi-target stool DNA and CT colonography for non-invasive colorectal cancer screening. Radiology 297 (1), 120–129. doi:10.1148/radiol.2020201018
Plaza-Díaz, J., Solis-Urra, P., Aragón-Vela, J., Rodríguez-Rodríguez, F., Olivares-Arancibia, J., and Álvarez-Mercado, A. I. (2021). Insights into the impact of microbiota in the treatment of NAFLD/NASH and its potential as a biomarker for prognosis and diagnosis. Biomedicines 9 (2), 145. doi:10.3390/biomedicines9020145
Qin, S., Li, A., Yi, M., Yu, S., Zhang, M., and Wu, K. (2019). Recent advances in anti-angiogenesis receptor tyrosine kinase inhibitors in cancer therapy. J. Hematol. Oncol. 12, 1–11. doi:10.1186/s13045-019-0718-5
Qiu, S., Cai, Y., Yao, H., Lin, C., Xie, Y., Tang, S., et al. (2023). Small molecule metabolites: discovery of biomarkers and therapeutic targets. Signal Transduct. Target. Ther. 8 (1), 132. doi:10.1038/s41392-023-01399-3
Raunkilde, L., Hansen, T. F., Andersen, R. F., Havelund, B. M., Thomsen, C. B., and Jensen, L. H. (2022). NPY gene methylation in circulating tumor DNA as an early biomarker for treatment effect in metastatic colorectal cancer. Cancers 14 (18), 4459. doi:10.3390/cancers14184459
Rawla, P., Sunkara, T., and Barsouk, A. (2019). Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Gastroenterol. Review/Przegląd Gastroenterol. 14 (2), 89–103. doi:10.5114/pg.2018.81072
Rigi, F., Jannatabad, A., Izanloo, A., Roshanravan, R., Hashemian, H. R., and Kerachian, M. A. (2020). Expression of tumor pyruvate kinase M2 isoform in plasma and stool of patients with colorectal cancer or adenomatous polyps. BMC Gastroenterol. 20 (1), 241–247. doi:10.1186/s12876-020-01377-x
Rivera, M., Fichtner, I., Wulf-Goldenberg, A., Sers, C., Merk, J., Patone, G., et al. (2021). Patient-derived xenograft (PDX) models of colorectal carcinoma (CRC) as a platform for chemosensitivity and biomarker analysis in personalized medicine. Neoplasia 23 (1), 21–35. doi:10.1016/j.neo.2020.11.005
Rizzo, G., Bertotti, A., Leto, S. M., and Vetrano, S. (2021). Patient-derived tumor models: A more suitable tool for pre-clinical studies in colorectal cancer. J. Exp. Clin. Cancer Res. 40 (1), 178. doi:10.1186/s13046-021-01970-2
Rosen, L., Rosen, P., Amado, R., Chang, D., Mulay, M., Parson, M., et al. (1999). “November). A Phase I/II study of SU5416 in combination with 5-FU/leucovorin in patients with metastatic colorectal cancer,” in Clinical cancer research (BIRMINGHAM, AL 35202 USA: AMER ASSOC CANCER RESEARCH), 3731S–3732S. PO BOX 11806.
Roskoski, R. (2015). A historical overview of protein kinases and their targeted small molecule inhibitors. Pharmacol. Res. 100, 1–23. doi:10.1016/j.phrs.2015.07.010
Rothenberg, M. L., Lafleur, B., Washington, M. K., Levy, D. E., Morgan-Meadows, S. L., Ramanathan, R. K., et al. (2004). Changes in epidermal growth factor receptor signaling in serum and tumor biopsies obtained from patients with progressive metastatic colorectal cancer (MCRC) treated with gefitinib (ZD1839): an Eastern Cooperative Oncology Group study. J. Clin. Oncol. 22, 3000. doi:10.1200/jco.2004.22.90140.3000
Safari, E., Mosayebi, G., and Khorram, S. (2018). Dkk-3 as a potential biomarker for diagnosis and prognosis of colorectal cancer. Med. J. Islamic Repub. Iran 32, 86. doi:10.14196/mjiri.32.86
Saltz, L., Rubin, M., Hochster, H., Tchekmeydian, N. S., Waksal, H., and Needle, M. (2001). Cetuximab plus irinotecan is active in CPT-11-refractory colorectal cancer that expresses epidermal growth factor receptors. Proc. Am. Soc. Clin. Oncol. 20.
Sastre, J., Grávalos, C., Rivera, F., Massuti, B., Valladares-Ayerbes, M., Marcuello, E., et al. (2012). First-line cetuximab plus capecitabine in elderly patients with advanced colorectal cancer: clinical outcome and subgroup analysis according to KRAS status from a Spanish TTD group study. Oncol. 17 (3), 339–345. doi:10.1634/theoncologist.2011-0406
Sattar, R. S. A., Verma, R., Kumar, A., Dar, G. M., Sharma, A. K., Kumari, I., et al. (2022). Diagnostic and prognostic biomarkers in colorectal cancer and the potential role of exosomes in drug delivery. Cell. Signal. 99, 110413. doi:10.1016/j.cellsig.2022.110413
Sawicki, T., Ruszkowska, M., Danielewicz, A., Niedźwiedzka, E., Arłukowicz, T., and Przybyłowicz, K. E. (2021). A review of colorectal cancer in terms of epidemiology, risk factors, development, symptoms, and diagnosis. Cancers 13 (9), 2025. doi:10.3390/cancers13092025
Schiffmann, L. M., Brunold, M., Liwschitz, M., Goede, V., Loges, S., Wroblewski, M., et al. (2017). A combination of low-dose bevacizumab and imatinib enhances vascular normalisation without inducing extracellular matrix deposition. Br. J. Cancer 116 (5), 600–608. doi:10.1038/bjc.2017.13
Schlessinger, J. (2014). Receptor tyrosine kinases: legacy of the first two decades. Cold Spring Harb. Perspect. Biol. 6 (3), a008912. doi:10.1101/cshperspect.a008912
Scott, A. J., Song, E. K., Bagby, S., Purkey, A., McCarter, M., Gajdos, C., et al. (2017). Evaluation of the efficacy of dasatinib, a Src/Abl inhibitor, in colorectal cancer cell lines and explant mouse model. PLoS One 12 (11), e0187173. doi:10.1371/journal.pone.0187173
Shaukat, A., and Levin, T. R. (2022). Current and future colorectal cancer screening strategies. Nat. Rev. Gastroenterology Hepatology 19 (8), 521–531. doi:10.1038/s41575-022-00612-y
Shen, L., Lu, W., Huang, Y., He, J., Wang, Q., Zheng, X., et al. (2022). SNORD15B and SNORA5C: novel diagnostic and prognostic biomarkers for colorectal cancer. BioMed Res. Int. 2022, 8260800. doi:10.1155/2022/8260800
Shim, H. (2011). One target, different effects: A comparison of distinct therapeutic antibodies against the same targets. Exp. Mol. Med. 43 (10), 539–549. doi:10.3858/emm.2011.43.10.063
Siri, G., Alesaeidi, S., Dizghandi, S. E., Alani, B., Mosallaei, M., and Soosanabadi, M. (2022). Analysis of SDC2 gene promoter methylation in whole blood for non-invasive early detection of colorectal cancer. J. Cancer Res. Ther. 18 (Suppl. 2), S354–S358. doi:10.4103/jcrt.jcrt_1072_22
Siveen, K. S., Prabhu, K. S., Achkar, I. W., Kuttikrishnan, S., Shyam, S., Khan, A. Q., et al. (2018). Role of non-receptor tyrosine kinases in hematological malignancies and its targeting by natural products. Mol. cancer 17 (1), 1–21.
Soheilifar, M. H., Grusch, M., Neghab, H. K., Amini, R., Maadi, H., Saidijam, M., et al. (2019). Angioregulatory microRNAs in colorectal cancer. Cancers 12 (1), 71. doi:10.3390/cancers12010071
Srivastava, A., Rai, S., Bisht, D., Sachan, M., Jit, B. P., and Srivastava, S. (2022). “Targeting the altered tyrosine kinases in colorectal cancer: from inhibitors to drugs,” in Protein kinase inhibitors (Cambridge: Academic Press), 361–391.
Stark, U. A., Frese, T., Unverzagt, S., and Bauer, A. (2020). What is the effectiveness of various invitation methods to a colonoscopy in the early detection and prevention of colorectal cancer? Protocol of a systematic review. Syst. Rev. 9 (1), 1–7. doi:10.1186/s13643-020-01312-x
Starý, L., Mezerová, K., Vysloužil, K., Zbořil, P., Skalický, P., Stašek, M., et al. (2020). Candida albicans culture from a rectal swab can be associated with newly diagnosed colorectal cancer. Folia Microbiol. 65, 989–994. doi:10.1007/s12223-020-00807-3
Strickler, J. H., Rushing, C. N., Uronis, H. E., Morse, M. A., Niedzwiecki, D., Blobe, G. C., et al. (2021). Cabozantinib and Panitumumab for RAS wild-type metastatic colorectal cancer. Oncol. 26 (6), 465–e917. doi:10.1002/onco.13678
Sun, S., Xiao, J., Huo, J., Geng, Z., Ma, K., Sun, X., et al. (2018). Targeting ectodysplasin promotor by CRISPR/dCas9-effector effectively induces reprogramming of human bone marrow-derived mesenchymal stem cells into sweat gland-like cells. Stem Cell Res. Ther. 9 (1), 1–10. doi:10.1186/s13287-017-0758-0
Sur, D., Advani, S., and Braithwaite, D. (2022). MicroRNA panels as diagnostic biomarkers for colorectal cancer: A systematic review and meta-analysis. Front. Med. 9, 915226. doi:10.3389/fmed.2022.915226
Taheri, M., Ghafouri-Fard, S., Najafi, S., Kallenbach, J., Keramatfar, E., Atri Roozbahani, G., et al. (2022). Hormonal regulation of telomerase activity and hTERT expression in steroid-regulated tissues and cancer. Cancer Cell Int. 22 (1), 1–17. doi:10.1186/s12935-022-02678-9
Tanriver, G., and Kocagoncu, E. (2023). Additive pre-diagnostic and diagnostic value of routine blood-based biomarkers in the detection of colorectal cancer in the UK Biobank cohort. Sci. Rep. 13 (1), 1367. doi:10.1038/s41598-023-28631-y
Tauriello, D. V., and Batlle, E. (2016). Targeting the microenvironment in advanced colorectal cancer. Trends cancer 2 (9), 495–504. doi:10.1016/j.trecan.2016.08.001
Thomson, R. J., Moshirfar, M., and Ronquillo, Y. (2022). “Tyrosine kinase inhibitors,” in StatPearls (United States: StatPearls Publishing).
To, K. K., Poon, D. C., Wei, Y., Wang, F., Lin, G., and Fu, L. W. (2015). Vatalanib sensitizes ABCB1 and ABCG2-overexpressing multidrug-resistant colon cancer cells to chemotherapy under hypoxia. Biochem. Pharmacol. 97 (1), 27–37. doi:10.1016/j.bcp.2015.06.034
Tolba, M. F. (2020). Revolutionizing the landscape of colorectal cancer treatment: the potential role of immune checkpoint inhibitors. Int. J. Cancer 147 (11), 2996–3006. doi:10.1002/ijc.33056
Tournier, B., Aucagne, R., Truntzer, C., Fournier, C., Ghiringhelli, F., Chapusot, C., et al. (2023). Integrative clinical and DNA methylation analyses in a population-based cohort identifies CDH17 and LRP2 as risk recurrence factors in stage II colon cancer. Cancers 15 (1), 158. doi:10.3390/cancers15010158
Trenker, R., and Jura, N. (2020). Receptor tyrosine kinase activation: from the ligand perspective. Curr. Opin. Cell Biol. 63, 174–185. doi:10.1016/j.ceb.2020.01.016
Tsoi, H., Wong, K. F., Luk, J. M., and Staunton, D. (2019). Clinical utility of CDH17 biomarker in tumor tissues and liquid biopsies for detection and prognostic staging of colorectal cancer (CRC). J. Glob. Oncol. 5, 53. doi:10.1200/JGO.2019.5.suppl.53
Van Doorn, S. C., Stegeman, I., Stroobants, A. K., Mundt, M. W., De Wijkerslooth, T. R., Fockens, P., et al. (2015). Fecal immunochemical testing results and characteristics of colonic lesions. Endoscopy 47 (11), 1011–1017. doi:10.1055/s-0034-1392412
Vernia, F., Valvano, M., Fabiani, S., Stefanelli, G., Longo, S., Viscido, A., et al. (2021). Are volatile organic compounds accurate markers in assessing colorectal cancer and inflammatory bowel diseases? A review. Cancers 13 (10), 2361. doi:10.3390/cancers13102361
Vlachogiannis, G., Hedayat, S., Vatsiou, A., Jamin, Y., Fernández-Mateos, J., Khan, K., et al. (2018). Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359 (6378), 920–926. doi:10.1126/science.aao2774
Wachsberger, P., Burd, R., Ryan, A., Daskalakis, C., and Dicker, A. P. (2009). Combination of vandetanib, radiotherapy, and irinotecan in the LoVo human colorectal cancer xenograft model. Int. J. Radiat. Oncology* Biology* Phys. 75 (3), 854–861. doi:10.1016/j.ijrobp.2009.06.016
Wang, C., Chevalier, D., Saluja, J., Sandhu, J., Lau, C., and Fakih, M. (2020). Regorafenib and nivolumab or pembrolizumab combination and circulating tumor DNA response assessment in refractory microsatellite stable colorectal cancer. Oncol. 25 (8), e1188–e1194. doi:10.1634/theoncologist.2020-0161
Wei, D., Tao, Z., Shi, Q., Wang, L., Liu, L., She, T., et al. (2020). Selective photokilling of colorectal tumors by near-infrared photoimmunotherapy with a GPA33-targeted single-chain antibody variable fragment conjugate. Mol. Pharm. 17 (7), 2508–2517. doi:10.1021/acs.molpharmaceut.0c00210
Wenzel, T., Büch, T., Urban, N., Weirauch, U., Schierle, K., Aigner, A., et al. (2020). Restoration of MARCKS enhances chemosensitivity in cancer. J. Cancer Res. Clin. Oncol. 146, 843–858. doi:10.1007/s00432-020-03149-2
Wheeler, D. L., and Yarden, Y. (2014). Receptor tyrosine kinases: Structure, functions, and role in human disease. Heidelberg: Springer.
Wilke, H., Glynne-Jones, R., Thaler, J. M. A. B. E. L., Adenis, A., Preusser, P., Aranda Aguilar, E., et al. (2006). MABEL—a large multinational study of cetuximab plus irinotecan in irinotecan-resistant metastatic colorectal cancer. J. Clin. Oncol. 24, 3549. doi:10.1200/jco.2006.24.18_suppl.3549
Wong, M. C., Huang, J., Lok, V., Wang, J., Fung, F., Ding, H., et al. (2021). Differences in incidence and mortality trends of colorectal cancer worldwide based on sex, age, and anatomic location. Clin. Gastroenterology Hepatology 19 (5), 955–966.e61. doi:10.1016/j.cgh.2020.02.026
Wong, S. H., and Yu, J. (2019). Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat. Rev. Gastroenterology Hepatology 16 (11), 690–704. doi:10.1038/s41575-019-0209-8
Woo, I. S., and Jung, Y. H. (2017). Metronomic chemotherapy in metastatic colorectal cancer. Cancer Lett. 400, 319–324. doi:10.1016/j.canlet.2017.02.034
Xi, Y., and Xu, P. (2021). Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 14 (10), 101174. doi:10.1016/j.tranon.2021.101174
Xie, Y. H., Chen, Y. X., and Fang, J. Y. (2020). Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct. Target. Ther. 5 (1), 22. doi:10.1038/s41392-020-0116-z
Xu, H., Chen, H., Hu, J., Xiong, Z., Li, D., Wang, S., et al. (2022). Feasibility of quantification based on novel evaluation with stool DNA and fecal immunochemical test for colorectal cancer detection. BMC Gastroenterol. 22 (1), 384–388. doi:10.1186/s12876-022-02470-z
Xu, H., Zhou, L., Lu, Y., Su, X., Cheng, P., Li, D., et al. (2020). Dual targeting of the epidermal growth factor receptor using combination of nimotuzumab and erlotinib in advanced non-small-cell lung cancer with leptomeningeal metastases: A report of three cases. OncoTargets Ther. 13, 647–656. doi:10.2147/OTT.S230399
Yang, Y., Li, S., Wang, Y., Zhao, Y., and Li, Q. (2022). Protein tyrosine kinase inhibitor resistance in malignant tumors: molecular mechanisms and future perspective. Signal Transduct. Target. Ther. 7 (1), 329. doi:10.1038/s41392-022-01168-8
Ye, X., Li, J., Song, C., and Chen, W. (2021). Telomere in colorectal cancer associated with distant metastases and predicted a poor prognosis. Transl. Cancer Res. 10 (6), 2906–2917. doi:10.21037/tcr-20-3341
Zahra, K., Dey, T., Mishra, S. P., and Pandey, U. (2020). Pyruvate kinase M2 and cancer: the role of PKM2 in promoting tumorigenesis. Front. Oncol. 10, 159. doi:10.3389/fonc.2020.00159
Zhang, J., Tian, Y., Luo, Z., Qian, C., Li, W., and Duan, Y. (2021). Breath volatile organic compound analysis: an emerging method for gastric cancer detection. J. Breath Res. 15 (4), 044002. doi:10.1088/1752-7163/ac2cde
Zhang, Y., Zou, J. Y., Wang, Z., and Wang, Y. (2019). Fruquintinib: A novel antivascular endothelial growth factor receptor tyrosine kinase inhibitor for the treatment of metastatic colorectal cancer. Cancer Manag. Res. 11, 7787–7803. doi:10.2147/CMAR.S215533
Zhao, G., Liu, X., Liu, Y., Li, H., Ma, Y., Li, S., et al. (2020). Aberrant DNA methylation of SEPT9 and SDC2 in stool specimens as an integrated biomarker for colorectal cancer early detection. Front. Genet. 11, 643. doi:10.3389/fgene.2020.00643
Zhao, Y., Bilal, M., Raza, A., Khan, M. I., Mehmood, S., Hayat, U., et al. (2021). Tyrosine kinase inhibitors and their unique therapeutic potentialities to combat cancer. Int. J. Biol. Macromol. 168, 22–37. doi:10.1016/j.ijbiomac.2020.12.009
Zhou, H., Zhu, L., Song, J., Wang, G., Li, P., Li, W., et al. (2022). Liquid biopsy at the frontier of detection, prognosis, and progression monitoring in colorectal cancer. Mol. Cancer 21 (1), 86. doi:10.1186/s12943-022-01556-2
Zhu, C., Huang, F., Li, Y., Zhu, C., Zhou, K., Xie, H., et al. (2022). Distinct urinary metabolic biomarkers of human colorectal cancer. Dis. Markers 2022, 1758113. doi:10.1155/2022/1758113
Zhu, H., Zhang, Y., Geng, Y., Lu, W., Yin, J., Li, Z., et al. (2019). IGFBP2 promotes the EMT of colorectal cancer cells by regulating E-cadherin expression. Int. J. Clin. Exp. pathology 12 (7), 2559–2565.
Keywords: colorectal cancer, diagnosis, tyrosine kinase inhibitors, biomarker, therapies
Citation: Jiang H, Zhou S and Li G (2023) Novel biomarkers used for early diagnosis and tyrosine kinase inhibitors as targeted therapies in colorectal cancer. Front. Pharmacol. 14:1189799. doi: 10.3389/fphar.2023.1189799
Received: 20 March 2023; Accepted: 14 August 2023;
Published: 01 September 2023.
Edited by:
Sona Vodenkova, Institute of Experimental Medicine (ASCR), CzechiaReviewed by:
Radka Václavíková, National Institute of Public Health (NIPH), CzechiaCopyright © 2023 Jiang, Zhou and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Li, TGVlZ2FubmdAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.