
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol., 29 March 2023
Sec. Neuropharmacology
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1129186
Major depressive disorder (MDD) is a psychiatric disorder with increasing prevalence worldwide. It is a leading cause of disability and suicide, severely affecting physical and mental health. However, the study of depression remains at an exploratory stage in terms of diagnostics and treatment due to the complexity of its pathogenesis. MicroRNAs are endogenous short-stranded non-coding RNAs capable of binding to the 3’untranslated region of mRNAs. Because of their ability to repress translation process of genes and are found at high levels in brain tissues, investigation of their role in depression has gradually increased recently. This article summarizes recent research progress on the relationship between microRNAs and depression. The microRNAs play a regulatory role in the pathophysiology of depression, involving dysregulation of monoamines, abnormalities in neuroplasticity and neurogenesis, hyperactivity of the HPA axis, and dysregulation of inflammatory responses. These microRNAs might provide new clue for the diagnosis and treatment of MDD, and the development of antidepressant drugs.
MicroRNAs (miRNAs) are short-stranded endogenous non-coding RNA molecules with a length of 19–25 nucleotides. A single microRNA can target hundreds of mRNAs and influence the expression of many genes (Friedman et al., 2009; Lu and Rothenberg, 2018). It is now established that about 70% of the known microRNAs are expressed in the brain and play critical roles in brain development through key signaling pathways involving synapse formation, neuronal plasticity, nerve growth, etc. MicroRNAs are endogenously encoded in the mammalian genome and are transcribed in the nucleus as primary transcripts (pri-miRNAs) which are hundreds of nucleotides in length. Pri-miRNAs are then trimmed into precursor microRNAs (pre-miRNAs) within the nucleus by DiGeorge syndrome critical region 8 (DGCR8) and Drosha. After processing in the nucleus, pre-miRNA transcripts are transported to the cytoplasm via the transporter Exportin-5 (XPO5). Pre-miRNAs are further processed in the cytoplasm by the enzyme Dicer into approximately 22 nucleotide-long RNA duplexes. The RNA duplexes are incorporated into the RNA-induced gene silencing complex (RISC), and further processed to form mature microRNAs (Zurawek and Turecki, 2021). RISC binds to the 3’untranslated region (3′UTR) of target mRNAs to induce targeted mRNA degradation or translational repression, thereby controlling gene expression at the post-transcriptional level.
Major depressive disorder (MDD) is a common illness that severely limits psychosocial functioning and diminishes quality of life (Malhi and Mann, 2018). MDD causes emotional changes in patients, as well as depressed mood and anhedonia, and it can lead to several psychiatric symptoms, including cognitive impairment (Hu et al., 2017). Although, there has been considerable research looking at the pathophysiology of major depressive disorder (MDD), no single mechanism can satisfactorily and completely explain all aspects of the disorder. There are several hypotheses regarding the molecular mechanisms involved in depression, including the monoamine hypothesis, hypothalamic-pituitary-adrenal (HPA) axis, neuroplasticity and neurogenesis, epigenetics, and inflammation. The monoamine hypothesis reveals that the pathophysiological basis leading to depression is due mostly to a decrease in monoamine neurotransmitters (e.g., serotonin). Evidence from clinical trials of some tricyclic antidepressants and monoamine oxidase inhibitors (MAOIs) have provided the basis for this hypothesis (Segal et al., 1974; Delgado et al., 1990; Willner et al., 2013). Hyperactivity of the HPA axis can lead to the stimulation of glucocorticoids and cortisol secretion, which may contribute to the development of depression (Goodyer et al., 2000; Harris et al., 2000). Notably, alterations of the HPA axis have also been associated with impairment of cognitive function (Keller et al., 2017). Stress-mediated inflammation and HPA axis dysfunction can lead to an alteration in neuroplasticity at the cellular level (Egeland et al., 2015). The neurogenesis process is controlled by regulatory proteins, such as brain-derived neurotrophic factor (BDNF), and peripheral BDNF has been found to be downregulated in patients with MDD (Molendijk et al., 2014). Epigenetics, the interaction of genes and the environment, plays a role in the alteration of brain neurobiology, and the effect of epigenetics can set the stage for the development of MDD (Penner-Goeke and Binder, 2019). In addition, peripheral cytokines can directly act on neurons and support cells and subsequently contribute to the development of depression (Miller and Raison, 2016). This hypothesis is supported by a role for some non-steroidal anti-inflammatory drugs in the treatment of depression (Leonard, 2018). Patients with autoimmune diseases and severe infections both have persistent activation of the immune system, causing high levels of cytokine production in the periphery. Such changes will cause changes in the patient’s central nervous system function, which in turn will lead to the occurrence and development of depression. This mechanism may explain why individuals with autoimmune diseases and severe infections are more likely to become depressed.
Up to now, first-line antidepressant drugs and other selected drugs in the clinic have low effectiveness, variable tolerance, adverse effects, and other disadvantages. Furthermore, large variations in therapeutic effects exist among individual drugs (Malhi and Mann, 2018). Our current understanding of microRNAs is continuing to increase partly, because of their high expression levels in the brain and their role in the regulation of neuronal plasticity and other functions. Recently, researchers focused on a role for microRNAs in the etiology of MDD. In this review, we have summarized the roles and mechanisms of microRNAs-mediated gene expression in the pathophysiological process of MDD. The role of each microRNA implicated in depression will be described as it relates to the different hypotheses of depression. In addition, this review could provide an attractive clue and potential targets to help diagnose and treat depression, as well as to assist in antidepressant drug development.
Many studies have confirmed that the level of microRNAs expression is associated with the onset of depression. These studies include both human and animal experiments. Postmortem human experiments were carried out to examine the expression levels of microRNAs in the prefrontal cortex, amygdala and other regions, as well as the levels and identity of their downstream target genes and protein products (As shown in Table 1). These human studies also looked at peripheral whole blood, serum, exosomes, and other tissues. The animal experiments were performed to detect microRNAs, and their downstream target genes and protein expression in the hippocampus and other tissues in rodents with depression-like symptoms (Table 2). The depression-like symptoms were induced by chronic unpredictable mild stress (CUMS) and this successful animal model was confirmed using behavioral tests, including sucrose preference test, forced swim test, and elevated plus maze test. According to the literature, microRNAs such as miR-124-3p, miR-128-3p, miR-139-5p, and miR-144-5p have been shown to play a significant role in different pathophysiological mechanisms of depression, which will be described in the corresponding sections of the text according to their different roles.
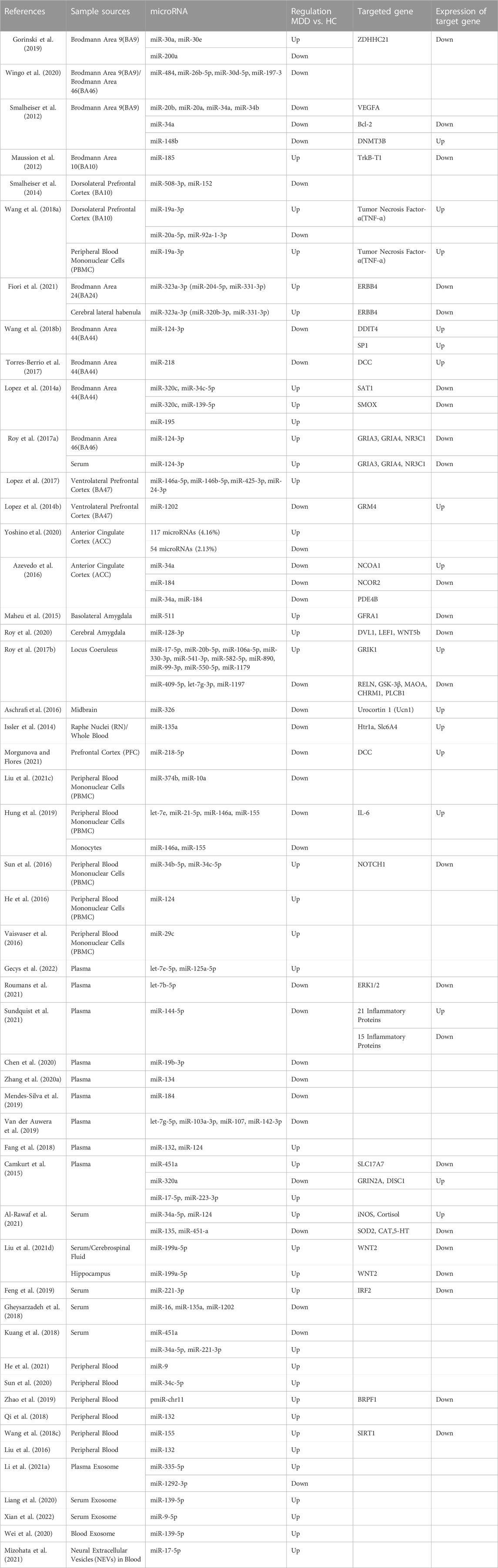
TABLE 1. Summary of researches on the changes in the levels of microRNAs and their target genes in MDD patients.
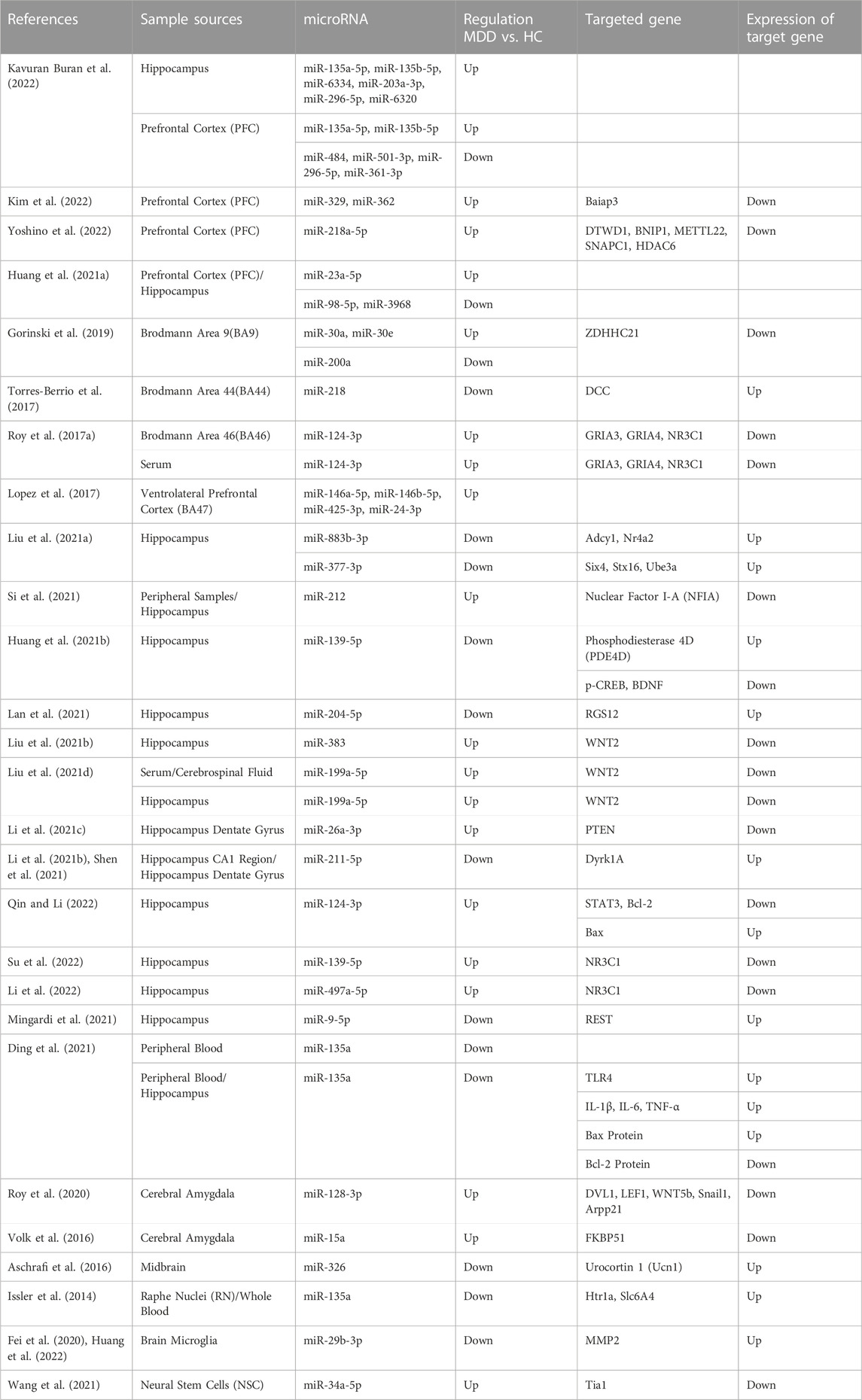
TABLE 2. Summary of researches on the changes in the levels of microRNAs and their target genes in experimental animals induced to develop depression.
Monoamine neurotransmitter (serotonin, noradrenaline, and dopamine) dysregulation is considered the most likely cause of MDD, and most of the drugs used in the clinic for the treatment of MDD are based on this principle. Monoamine-based antidepressants were the first drugs developed for the treatment of MDD (Elias et al., 2022). The monoamine hypothesis of depression has been applied for nearly six decades ago (Coppen et al., 1965) and the classical doctrine holds that monoamines are depleted and chronically below normal levels in the brains of patients with MDD (Shaw et al., 1967). This hypothesis is corroborated by the pharmacological mechanism of action of monoamine oxidase (MAO) inhibitors, tricyclic antidepressants, and selective serotonin reuptake inhibitors in MDD patients (Hillhouse and Porter, 2015). In 1996, Heninger et al. (1996) revised the monoamine doctrine to suggest that monoamine depletion may play more of a role, thereby affecting nervous system functions, or it must be present in the environment of a stressor to cause MDD. They provided a theoretical basis for investigating the role of microRNAs in MDD.
Gorinski et al. (2019) found that a decrease in miR-200a expression or an increase in miR-30a and miR-30e expression led to a decrease of ZDHHC21 expression in humans and animal models. ZDHHC21, a palmitoyl acyltransferase, was identified as the major enzyme involved in the palmitoylation of the 5HT1AR and the decrease in the palmitoylation of 5HT1AR resulted in inhibition of adenylate cyclase and subsequent decrease of cAMP levels resulting in the occurrence of MDD. The downregulated miR-135a was shown to promote the translation of the Htr1a and Slc6a4 genes in MDD patients (Issler et al., 2014) and the upregulation of the inhibitory 5HT1a receptor (5HT1AR), encoded by the Htr1a gene, and 5HT transporter (SERT), encoded by the Slc6a4 gene, contributed to aberrant monoamine neurotransmitters in patients with depression (Issler et al., 2014). DCC (Deleted in Colorectal Cancer) drives prefrontal cortex maturity by determining DA targets early in life, for example, in rats, signaling within dopamine neurons in the juvenile VTA determines the extent of innervation of the PFC (Torres-Berrio et al., 2017). Whereas miR-218 was shown to be upregulated in BA44 in MDD patients and led to a significant decrease in DCC expression levels. In rats, who had experienced chronic social defeat stress paradigms also showed the same changes (Torres-Berrio et al., 2017). MiR-1202 was found to be differentially expressed in MDD patient ventrolateral prefrontal cortices, with upregulated GRM4 expression (Lopez et al., 2014b). GRM4 is expressed throughout the brain, with predominant expression sites at presynaptic and postsynaptic membranes, where it regulates glutamatergic, dopaminergic, GABAergic, and serotonergic neurotransmission (Lopez et al., 2014b). The increased expression of miR-329 and miR-362 in the PFC of MDD patients caused downregulation of Baiap3 (brain specific angiogenesis inhibitor 1-associated protein 3), which subsequently induced defective dense core vesicles (DCVs) transport and reduced serotonin exocytosis (Kim et al., 2022). In both the central nervous system and endocrine systems, DCVs are essential for peptidergic and aminergic signaling (Persoon et al., 2018) (Figure 1).
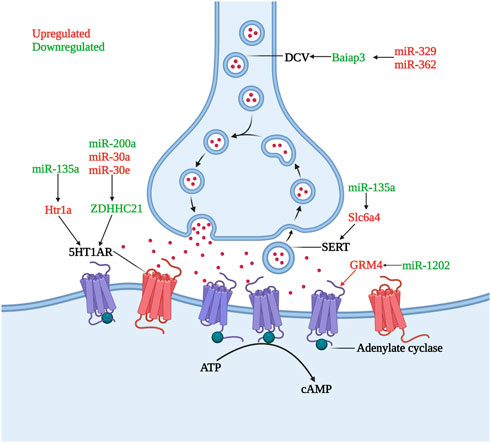
FIGURE 1. Role of microRNAs in serotonin dysregulation. The inhibitory receptor 5HT1AR is hyperactivity or increased under the influence of miR-135a, miR-200a, miR-30a, and miR-30e, lead to the occurrence and development of depression. DCV and SERT undergo quantitative abnormalities under the influence of miR-329, miR-362 and miR-135a, causing dysregulation of monoamine transmitter secretion and reuptake. GRM4, whose transcription is increased by miR-1202 downregulation, can regulate monoamine neurotransmitter transmission (Created with BioRender.com).
Neuroplasticity is a fundamental process by which the brain acquires information and produces appropriately adaptive responses in relevant environments. Thus, dysfunction in neuroplasticity and neurogenesis may contribute to the pathophysiology of MDD (Duman, 2002). Multiple signaling pathways are involved in this process. For example, Wnt signaling pathway plays a role in neurogenesis, synapse formation, synaptic transmission, and dendritic arborization in the hippocampus (Wayman et al., 2006; Gogolla et al., 2009). The mTOR signaling pathway is involved in the pathophysiology of MDD through the P70S6K/eIF4B pathway (Jernigan et al., 2011). Abnormalities in BDNF, glutamate receptors, VEGF signaling, and long-term potentiation (LTP) pathways also contribute to the pathophysiological progression of depression by affecting neuroplasticity and neurogenesis (Duric et al., 2010; Yoshii and Constantine-Paton, 2010; Gormanns et al., 2011). MicroRNAs have an influence on depression by interfering with the stability of these signaling pathways (Fan et al., 2014).
As shown in Figure 2. Wang et al. (2018b) found that miR-124-3p was significantly downregulated in Brodmann area 44 (BA44) of patients with MDD. Downregulation of miR-124-3p abolished its inhibition of DNA damage inducible transcript 4 protein (DDIT4) and SP1 expression, and inhibited the mTOR signaling pathway. Roy et al. (2020) demonstrated that miR-128-3p was upregulated in the amygdala of MDD patients, leading to a decreased expression of Wnt5b, LEF1 and DVL1, which are genes related to the Wnt signaling pathway. Disruption of canonical Wnt/Fz/GSK3 signaling leads to abnormal neurodevelopment that is associated with neuropsychiatric disorders (Voleti and Duman, 2012).

FIGURE 2. Role of microRNAs in neuroplasticity and neurogenesis abnormalities. MiR-124, miR-128, miR-139, miR-144 and others are involved in the regulation of neuroplasticity and neurogenesis through multiple pathways. These pathways mainly include Wnt/β-Catenin signaling pathway, mTOR signaling pathway, LTP signaling pathway, etc.
Moreover, the downregulation of Gria3 and Gria4 receptors induced by miR-124-3p had an influence on modulation of AMPA receptor, and correlated with an impaired synaptic plasticity in patients with depression (Roy et al., 2017a). In the basolateral amygdala of depressed patients, upregulated miR-511 downregulated the encoded GFRα1a specific isoform of the GFRA1 gene of the receptor (Maheu et al., 2015). The subtypes, GFRα1a and GFRα1b elicited different downstream effects and had opposing effects in some aspects of neuroplasticity. The promotion of axonal growth by GFRα1a, was downregulated, while the inhibition of axonal growth by GFRα1b, was relatively upregulated, leading to the development of depression (Maheu et al., 2015). The upregulation of miR-185 in brain BA10 of MDD patients resulted in a decrease of TrkB-T1 expression. TrkB-T1, a BDNF receptor lacking the tyrosine kinase domain, was highly expressed in astrocytes and it regulated BDNF-evoked calcium transients (Maussion et al., 2012). Importantly, downregulation of TrkB-T1 in the frontal cortex might be associated with the neurobiology of suicide (Maussion et al., 2012).
In animal models, miR-139-5p regulates the cAMP/PKA/CREB/BDNF pathway to promote hippocampal neurogenesis by targeting PDE4D. Huang et al. (2021b) demonstrated that downregulation of miR-139-5p along with upregulation of its target gene PDE4D and downregulation of p-CREB and BDNF after inducing depression-like symptoms in CUMS mice. Such alterations show a bidirectional role for microRNAs in both protection and impairment of the neurogenesis pathways. In addition, Mingardi et al. (2021) found that miR-9-5p expression decreased in the hippocampus of rats subjected to chronic mild stress and primary hippocampal cultures. This change would cause overexpression of its downstream target protein REST, which would negatively affect neuronal dendritic morphology.
Chronic stress has long been recognized to be a potential risk factor for depression, which is often associated with depression prevalence. The activity of the HPA axis is mediated by arginine vasopressin (AVP) and hypothalamic secretion of corticotropin releasing factor (CRF), which in turn activates the pituitary gland to secrete adrenocorticotropic hormone (ACTH), and finally stimulates the adrenal cortex to secrete glucocorticoids. Glucocorticoids then interact with receptors in multiple target tissues, where they directly exert negative feedback regulation on ACTH secreted by the pituitary as well as CRF secreted by the hypothalamus (Pariante and Lightman, 2008). Changes in glucocorticoid receptor (GR) expression, nuclear translocation, cofactor binding, and GR mediated gene transcription may play an important role in glucocorticoid resistance, which will lead to the development of HPA axis hyperactivity (Colla et al., 2007; Alt et al., 2010). Impaired GR function occurring in the periphery leads to the development of HPA axis hyperactivity. High glucocorticoid levels resulting from HPA axis hyperactivity may be involved in glucocorticoid-dependent hippocampal plasticity changes, causing hippocampal atrophy and reduced hippocampal neurogenesis, which in turn promotes the development of MDD (Kronenberg et al., 2009; Schmidt et al., 2009). As observed in depressed patients, HPA axis activity is the main biochemical change in addition to monoaminergic neurotransmitter disturbances (Budziszewska, 2002). MicroRNAs can influence the HPA axis activity by affecting glucocorticoid related receptors or other pathways (Uchida et al., 2008; Vreugdenhil et al., 2009).
Roy et al. (2017a) confirmed the effect of HPA axis hyperactivity on depression by examining the changes in miR-124-3p and its downstream target genes in PFC (BA46) and serum of mice with depression-like symptom after chronic CORT treatment. Furthermore, the detection of PFC (BA46) in post-mortem brains from depressed patients coincides with animal experiments (Roy et al., 2017a). In addition, upregulation of miR-124-3p in human and animal models was confirmed to be associated with downregulation of AMPA receptor family members Gria3 and Gria4, and glucocorticoid receptor NR3C1. MiR-124-3p mediated repression of NR3C1 may be central to the associated neuroendocrine response to stress (Roy et al., 2017a).
The central nervous system responses are of greater concern regarding hyperactive HPA axis responses. Al-Rawaf et al. (2021) demonstrated that the excessive cortisol activity induced by HPA axis hyperfunction was significantly correlated with decreased serotonin levels. A previous study has confirmed that the expression level of miR-124 was regulated by serotonin and demonstrated a significant negative correlation (Rajasethupathy et al., 2009). MiR-124 could control serotonin to induce synaptic function by repressing the transcription of cAMP response element binding protein (CREB), and conversely, CREB could further regulate miR-124 expression (Rajasethupathy et al., 2009). In addition, aberrant expression of miR-34a-5p and miR-451-a significantly reduced BDNF expression, and BDNF affected serotonin and cortisol expression by producing pro-neuroprotective signals (Numakawa et al., 2009; Numakawa et al., 2012; Wibrand et al., 2012).
Depression and inflammation mutually contribute to the development of each other’s pathophysiology (Kiecolt-Glaser et al., 2015). Since the study of T and B lymphocytes in psychiatric patients by Herzog et al. (1979), the exploration of the relationship between the inflammatory response and depression has gradually unfolded (Herzog et al., 1979). Over the past four decades, accumulating evidence has shown that MDD is associated with systemic immune activation, including inflammatory markers, and changes in the number of immune cells (Gibney and Drexhage, 2013). Cytokines are one of the most important components of the immune system in depression. In response to peripheral infections, innate immune cells produce pro-inflammatory cytokines that act on the brain leading to development of neuropsychiatric disorders. When the peripheral immune system is continuously activated, immune signaling to the brain leads to exacerbation of the disease, and development of depressive symptoms in patients (Dantzer et al., 2008). The traditional routes of communication between the periphery and the central involve neural and humoral pathways, which mainly include: neural pathways (Harrison et al., 2009), signaling via cerebral endothelial cells (CECs) (Rivest et al., 2000; Kobayashi, 2010), signaling via circumventricular organs (CVOs) (Ransohoff et al., 2003) and peripheral immune-cell-to-brain signaling (Geissmann et al., 2003). TNFα, IL-1β and IL-6 are the main cytokines involved in the signaling of these pathways (Dantzer et al., 2008; Capuron and Miller, 2011). Recently, communication through the gut-microbiota-to-brain rout has gained increasing attention because of its role in regulating brain function (Jenkins et al., 2016; Sherwin et al., 2016). MicroRNAs participate in the pathophysiological process of inflammation in depression by promoting the production of inflammatory factors, as shown Figure 3. Changes in cytokine levels in patients with MDD have been identified to be associated with patient mood and volition (Beurel et al., 2020).
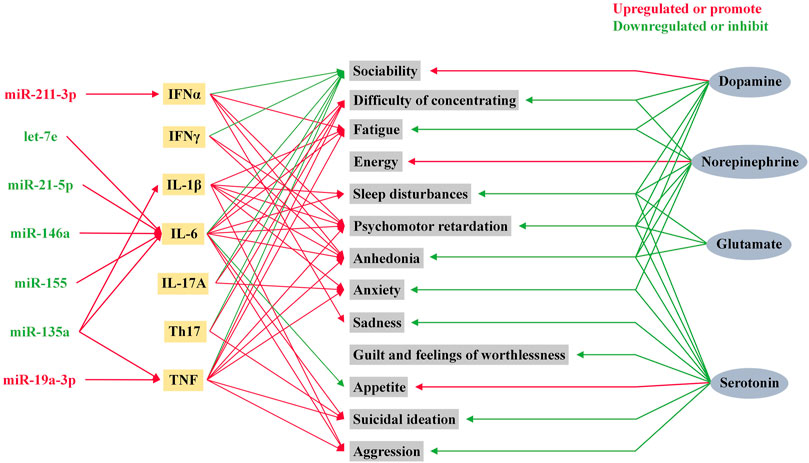
FIGURE 3. Role of microRNAs in inflammatory factors and neurotransmitters in depression. Psychiatric symptoms in humans are influenced by a variety of neurotransmitters and inflammatory factors. Among these inflammatory factors, IL-1β, IL-6, TNF, and IFNα are influenced by microRNA levels.
Wang et al. (2018a) pointed out that the upregulated expression of miR-19a-3p was detected in dlPFC and PBMC of MDD suicide completers. Gene analysis demonstrated that the elevated miR-19a-3p upregulated the expression of TNF-α by affecting the transcription of TAR-RNA binding protein (TRBP) and HuR (Wang et al., 2018a). The upregulation of TNF-α in dlPFC and PBMC was confirmed to be associated with suicidal ideation in MDD patients (Wang et al., 2018a). Sundquist et al. (2021) demonstrated that, in 178 patients with depression, anxiety, or stress and adjustment disorders, 36 inflammatory proteins with significantly different expression in peripheral blood of patients at baseline were seen, including 21 inflammatory proteins with increased levels and 15 with decreased levels, and all were associated with changes in miR-144-5p levels. In addition, the alteration in inflammatory proteins, which occurs after receiving treatment, was demonstrated to be associated with improvement in patients’ psychiatric symptoms (Sundquist et al., 2021). CircDYM, as an endogenous miR-9 sponge, is able to inhibit the activity of miR-9. Zhang et al. (2020b), by examining peripheral blood samples from MDD patients, hippocampus and plasma samples from MDD animal models, found that circDYM levels were significantly decreased. This would lead to enhanced miR-9 activity, which in turn would cause polarization of microglia. In a recent research, Xian et al. (2022) found miR-9-5p-enriched exosomes derived from PC12 cells in the serum of MDD patients. After BV2 microglia phagocytosed miR-9-5p-enriched exosomes, they were polarized to M1 subtype microglia via the SOCS2-STAT3 axis. Since then, M1 subtype microglia has produced a large amount of IL-1β, IL-6 and TNF-α. It leads to and intensifies the damage of neurons and causes the occurrence and development of MDD. Recent studies on depression triggered by microbial dysbiosis has shed new light on the role of abnormal inflammatory responses in the pathophysiology of depression (Borre et al., 2014; Dubois et al., 2019; Rea et al., 2020). This perspective explores the link between the gut microbiota and the regulation of the brain-gut axis, immune and endocrine system activity, and neurophysiological changes. Communication between the brain and the gut occurs bidirectionally via neural, endocrine, and immune pathways. Microbiota dysbiosis and an increased intestinal permeability with subsequent immune responses seem to be at the root of chronic mild inflammation associated with neuropsychiatric disorders (Petra et al., 2015; Rea et al., 2017; Farzi et al., 2018).
MicroRNAs are recognized as key epigenetic regulators of multiple functions in the brain and play a key role in MDD pathogenesis. As research continues to deepen, the roles of microRNAs in the pathophysiology of depression are gradually being elucidated. This review summarized recent research progress focusing on the role of microRNAs in the pathophysiology of depression, including dysregulation of monoamines, abnormalities in neuroplasticity and neurogenesis, hyperactivity of the HPA axis, and dysregulation of inflammatory responses. This suggests that an indispensable role for microRNAs occurs in these pathways. Several studies looking at changes in the levels of microRNAs and their downstream target genes before and after antidepressant treatment have confirmed a role for microRNAs in depression. Clearly, there are interactions between these different pathways and this exhibits the complexity in the pathogenesis of depression.
Based on the above four pathophysiological mechanisms of depression, it can be found that MDD, whether caused by dysregulation of monoamines or hyperactivity of the HPA, have parts that interact and influence each other. It is difficult to explain by a single pathophysiological mechanism, either from the clinical presentation of MDD patients or from changes in laboratory experiments. For example, high levels of cortisol in patients with Cushing syndrome resulted in alterations of neurotransmitter function, such as reduced serotonin synthesis. This can also be detected in MDD patients with HPA axis hyperactivity induced by long-term chronic stress (Stokes, 1995). In addition, high levels of cortisol inducing loss of hippocampal dendrites, and neuronal plasticity is recognized as one of the causes of depression (Gotlib et al., 2008). In addition, miR-124 can in turn control serotonin-induced synaptic facilitation by inhibiting the transcription of CREB (Rajasethupathy et al., 2009). Taken together, neuroinflammation could contribute to the pathogenesis of depression by interacting with the dysregulation of brain monoamines, dysregulation of the HPA axis, and alterations in hippocampal dentate gyrus neurons (Troubat et al., 2021).
It is important to note that current studies based on the role of microRNAs in depression have certain limitations, especially for the relationship between microRNAs and depression. Whether protective or injurious during the development of the disease, the levels of microRNAs in the brain tissue or peripheral tissues of patients do change when compared to normal individuals. Nevertheless, it is tough to confirm which of the varied microRNAs are responsible for the pathogenesis of MDD or that the major depressive disorder causes changes in certain microRNAs. If changes in specific microRNAs can be confirmed to contribute to the development of MDD, these microRNAs could be used as biomarkers for the diagnosis of the disease. In the same way, if it can be confirmed that MDD causes changes in the expression of microRNAs, and at the same time, alterations in these microRNAs can cause changes in the expression of downstream mRNAs and then have favorable or adverse effects on patients, this finding will be very important for the potential treatment of the disease and in stopping its development.
Since the discovery of the stable presence of free microRNAs in serum in 2018 (Chen et al., 2008), studies on the determination of microRNA levels in the serum of patients with depression have also gradually increased. However, it is undeniable that such studies have limitations as microRNAs in blood samples may not accurately reflect disease pathogenesis in the brain, because blood microRNAs are a mixture of brain-derived microRNAs and other microRNAs excreted from various tissues. The identification of microRNA within exosomes secreted by brain cells into the circulation may be able to compensate for the limitations that exist.
Finally, it is clear that microRNAs play an integral role in the pathophysiology of depression and may perhaps be able to provide a reference for the diagnostics and prognostics in depression by examining microRNA levels in relevant tissues. Moreover, promoting or inhibiting the expression of microRNAs might provide new clues for the development of antidepressant drugs.
Conceptualization: SL and XJ Writing—original draft: RD, DS, and QZ Writing—review and editing: XJ, SL, RD, YW, and J-YW
This work was supported by the National Natural Science Foundation of China (No. 81971280), the Program for Innovative Talents of Science and Technology in Henan Province (No. 23HASTIT043), the Natural Science Foundation of Henan Province for Excellent Young Scholars (No. 212300410026), the Medical Science and Technology Program of Henan Province (No. SBGJ202103096), and the Program for Young Key Teacher of Henan Province (No. 2020GGJS037).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Al-Rawaf, H. A., Alghadir, A. H., and Gabr, S. A. (2021). Circulating microRNAs and molecular oxidative stress in older adults with neuroprogression disorders. Dis. Markers 2021, 4409212. doi:10.1155/2021/4409212
Alt, S. R., Turner, J. D., Klok, M. D., Meijer, O. C., Lakke, E. A., Derijk, R. H., et al. (2010). Differential expression of glucocorticoid receptor transcripts in major depressive disorder is not epigenetically programmed. Psychoneuroendocrinology 35, 544–556. doi:10.1016/j.psyneuen.2009.09.001
Aschrafi, A., Verheijen, J. M., Gordebeke, P. M., Olde Loohuis, N. F., Menting, K., Jager, A., et al. (2016). MicroRNA-326 acts as a molecular switch in the regulation of midbrain urocortin 1 expression. J. Psychiatry Neurosci. 41, 342–353. doi:10.1503/jpn.150154
Azevedo, J. A., Carter, B. S., Meng, F., Turner, D. L., Dai, M., Schatzberg, A. F., et al. (2016). The microRNA network is altered in anterior cingulate cortex of patients with unipolar and bipolar depression. J. Psychiatr. Res. 82, 58–67. doi:10.1016/j.jpsychires.2016.07.012
Beurel, E., Toups, M., and Nemeroff, C. B. (2020). The bidirectional relationship of depression and inflammation: Double trouble. Neuron 107, 234–256. doi:10.1016/j.neuron.2020.06.002
Borre, Y. E., Moloney, R. D., Clarke, G., Dinan, T. G., and Cryan, J. F. (2014). The impact of microbiota on brain and behavior: Mechanisms and therapeutic potential. Adv. Exp. Med. Biol. 817, 373–403. doi:10.1007/978-1-4939-0897-4_17
Budziszewska, B. (2002). Effect of antidepressant drugs on the hypothalamic-pituitary-adrenal axis activity and glucocorticoid receptor function. Pol. J. Pharmacol. 54, 343–349.
Camkurt, M. A., Acar, S., Coskun, S., Gunes, M., Gunes, S., Yilmaz, M. F., et al. (2015). Comparison of plasma MicroRNA levels in drug naive, first episode depressed patients and healthy controls. J. Psychiatr. Res. 69, 67–71. doi:10.1016/j.jpsychires.2015.07.023
Capuron, L., and Miller, A. H. (2011). Immune system to brain signaling: Neuropsychopharmacological implications. Pharmacol. Ther. 130, 226–238. doi:10.1016/j.pharmthera.2011.01.014
Chen, X., Ba, Y., Ma, L., Cai, X., Yin, Y., Wang, K., et al. (2008). Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell. Res. 18, 997–1006. doi:10.1038/cr.2008.282
Chen, Y., Shi, J., Liu, H., Wang, Q., Chen, X., Tang, H., et al. (2020). Plasma microRNA array analysis identifies overexpressed miR-19b-3p as a biomarker of bipolar depression distinguishing from unipolar depression. Front. Psychiatry 11, 757. doi:10.3389/fpsyt.2020.00757
Colla, M., Kronenberg, G., Deuschle, M., Meichel, K., Hagen, T., Bohrer, M., et al. (2007). Hippocampal volume reduction and HPA-system activity in major depression. J. Psychiatr. Res. 41, 553–560. doi:10.1016/j.jpsychires.2006.06.011
Coppen, A., Shaw, D. M., and Malleson, A. (1965). Changes in 5-hydroxytryptophan metabolism in depression. Br. J. Psychiatry 111, 105–107. doi:10.1192/bjp.111.470.105
Dantzer, R., O'connor, J. C., Freund, G. G., Johnson, R. W., and Kelley, K. W. (2008). From inflammation to sickness and depression: When the immune system subjugates the brain. Nat. Rev. Neurosci. 9, 46–56. doi:10.1038/nrn2297
Delgado, P. L., Charney, D. S., Price, L. H., Aghajanian, G. K., Landis, H., and Heninger, G. R. (1990). Serotonin function and the mechanism of antidepressant action. Reversal of antidepressant-induced remission by rapid depletion of plasma tryptophan. Arch. Gen. Psychiatry 47, 411–418. doi:10.1001/archpsyc.1990.01810170011002
Ding, Y., Zhong, M., Qiu, B., Liu, C., Wang, J., and Liang, J. (2021). Abnormal expression of miR-135a in patients with depression and its possible involvement in the pathogenesis of the condition. Exp. Ther. Med. 22, 726. doi:10.3892/etm.2021.10158
Dubois, T., Reynaert, C., Jacques, D., Lepiece, B., and Zdanowicz, N. (2019). Role of gut microbiota in the interaction between immunity and psychiatry: A literature review. Psychiatr. Danub 31, 381–385.
Duman, R. S. (2002). Pathophysiology of depression: The concept of synaptic plasticity. Eur. Psychiatry 17 (3), 306–310. doi:10.1016/s0924-9338(02)00654-5
Duric, V., Banasr, M., Licznerski, P., Schmidt, H. D., Stockmeier, C. A., Simen, A. A., et al. (2010). A negative regulator of MAP kinase causes depressive behavior. Nat. Med. 16, 1328–1332. doi:10.1038/nm.2219
Egeland, M., Zunszain, P. A., and Pariante, C. M. (2015). Molecular mechanisms in the regulation of adult neurogenesis during stress. Nat. Rev. Neurosci. 16, 189–200. doi:10.1038/nrn3855
Elias, E., Zhang, A. Y., and Manners, M. T. (2022). Novel pharmacological approaches to the treatment of depression, 12. Basel: Life. doi:10.3390/life12020196
Fan, H. M., Sun, X. Y., Guo, W., Zhong, A. F., Niu, W., Zhao, L., et al. (2014). Differential expression of microRNA in peripheral blood mononuclear cells as specific biomarker for major depressive disorder patients. J. Psychiatr. Res. 59, 45–52. doi:10.1016/j.jpsychires.2014.08.007
Fang, Y., Qiu, Q., Zhang, S., Sun, L., Li, G., Xiao, S., et al. (2018). Changes in miRNA-132 and miR-124 levels in non-treated and citalopram-treated patients with depression. J. Affect Disord. 227, 745–751. doi:10.1016/j.jad.2017.11.090
Farzi, A., Frohlich, E. E., and Holzer, P. (2018). Gut microbiota and the neuroendocrine system. Neurotherapeutics 15, 5–22. doi:10.1007/s13311-017-0600-5
Fei, M., Wang, H., Zhou, M., Deng, C., Zhang, L., and Han, Y. (2020). Podoplanin influences the inflammatory phenotypes and mobility of microglia in traumatic brain injury. Biochem. Biophys. Res. Commun. 523, 361–367. doi:10.1016/j.bbrc.2019.12.003
Feng, J., Wang, M., Li, M., Yang, J., Jia, J., Liu, L., et al. (2019). Serum miR-221-3p as a new potential biomarker for depressed mood in perioperative patients. Brain Res. 1720, 146296. doi:10.1016/j.brainres.2019.06.015
Fiori, L. M., Kos, A., Lin, R., Theroux, J. F., Lopez, J. P., Kuhne, C., et al. (2021). miR-323a regulates ERBB4 and is involved in depression. Mol. Psychiatry 26, 4191–4204. doi:10.1038/s41380-020-00953-7
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi:10.1101/gr.082701.108
Gecys, D., Dambrauskiene, K., Simonyte, S., Patamsyte, V., Vilkeviciute, A., Musneckis, A., et al. (2022). Circulating hsa-let-7e-5p and hsa-miR-125a-5p as possible biomarkers in the diagnosis of major depression and bipolar disorders. Dis. Markers 2022, 3004338. doi:10.1155/2022/3004338
Geissmann, F., Jung, S., and Littman, D. R. (2003). Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 19, 71–82. doi:10.1016/s1074-7613(03)00174-2
Gheysarzadeh, A., Sadeghifard, N., Afraidooni, L., Pooyan, F., Mofid, M. R., Valadbeigi, H., et al. (2018). Serum-based microRNA biomarkers for major depression: MiR-16, miR-135a, and miR-1202. J. Res. Med. Sci. 23, 69. doi:10.4103/jrms.JRMS_879_17
Gibney, S. M., and Drexhage, H. A. (2013). Evidence for a dysregulated immune system in the etiology of psychiatric disorders. J. Neuroimmune Pharmacol. 8, 900–920. doi:10.1007/s11481-013-9462-8
Gogolla, N., Galimberti, I., Deguchi, Y., and Caroni, P. (2009). Wnt signaling mediates experience-related regulation of synapse numbers and mossy fiber connectivities in the adult hippocampus. Neuron 62, 510–525. doi:10.1016/j.neuron.2009.04.022
Goodyer, I. M., Herbert, J., Tamplin, A., and Altham, P. M. (2000). Recent life events, cortisol, dehydroepiandrosterone and the onset of major depression in high-risk adolescents. Br. J. Psychiatry 177, 499–504. doi:10.1192/bjp.177.6.499
Gorinski, N., Bijata, M., Prasad, S., Wirth, A., Abdel Galil, D., Zeug, A., et al. (2019). Attenuated palmitoylation of serotonin receptor 5-HT1A affects receptor function and contributes to depression-like behaviors. Nat. Commun. 10, 3924. doi:10.1038/s41467-019-11876-5
Gormanns, P., Mueller, N. S., Ditzen, C., Wolf, S., Holsboer, F., and Turck, C. W. (2011). Phenome-transcriptome correlation unravels anxiety and depression related pathways. J. Psychiatr. Res. 45, 973–979. doi:10.1016/j.jpsychires.2010.12.010
Gotlib, I. H., Joormann, J., Minor, K. L., and Hallmayer, J. (2008). HPA axis reactivity: A mechanism underlying the associations among 5-HTTLPR, stress, and depression. Biol. Psychiatry 63, 847–851. doi:10.1016/j.biopsych.2007.10.008
Harris, T. O., Borsanyi, S., Messari, S., Stanford, K., Cleary, S. E., Shiers, H. M., et al. (2000). Morning cortisol as a risk factor for subsequent major depressive disorder in adult women. Br. J. Psychiatry 177, 505–510. doi:10.1192/bjp.177.6.505
Harrison, N. A., Brydon, L., Walker, C., Gray, M. A., Steptoe, A., Dolan, R. J., et al. (2009). Neural origins of human sickness in interoceptive responses to inflammation. Biol. Psychiatry 66, 415–422. doi:10.1016/j.biopsych.2009.03.007
He, C., Bai, Y., Wang, Z., Fan, D., Wang, Q., Liu, X., et al. (2021). Identification of microRNA-9 linking the effects of childhood maltreatment on depression using amygdala connectivity. Neuroimage 224, 117428. doi:10.1016/j.neuroimage.2020.117428
He, S., Liu, X., Jiang, K., Peng, D., Hong, W., Fang, Y., et al. (2016). Alterations of microRNA-124 expression in peripheral blood mononuclear cells in pre- and post-treatment patients with major depressive disorder. J. Psychiatr. Res. 78, 65–71. doi:10.1016/j.jpsychires.2016.03.015
Heninger, G. R., Delgado, P. L., and Charney, D. S. (1996). The revised monoamine theory of depression: A modulatory role for monoamines, based on new findings from monoamine depletion experiments in humans. Pharmacopsychiatry 29, 2–11. doi:10.1055/s-2007-979535
Herzog, P., Dvorakova, M., and Zvolsky, P. (1979). T and B lymphocytes in psychotic patients (author's transl). Cesk Psychiatr. 75, 153–159.
Hillhouse, T. M., and Porter, J. H. (2015). A brief history of the development of antidepressant drugs: From monoamines to glutamate. Exp. Clin. Psychopharmacol. 23, 1–21. doi:10.1037/a0038550
Hu, Z., Jiang, Y., Huo, X., Yang, Y., Davies, H., Botchway, B. O. A., et al. (2017). Prospective role of MicroRNAs in depression. Curr. Med. Chem. 24, 3508–3521. doi:10.2174/0929867324666170714112620
Huang, C., Wang, Y., Wu, Z., Xu, J., Zhou, L., Wang, D., et al. (2021a). miR-98-5p plays a critical role in depression and antidepressant effect of ketamine. Transl. Psychiatry 11, 454. doi:10.1038/s41398-021-01588-0
Huang, P., Wei, S., Luo, M., Tang, Z., Lin, Q., Wang, X., et al. (2021b). MiR-139-5p has an antidepressant-like effect by targeting phosphodiesterase 4D to activate the cAMP/PKA/CREB signaling pathway. Ann. Transl. Med. 9, 1594. doi:10.21037/atm-21-5149
Huang, X., Yang, Q., Xie, L., and Lei, S. (2022). Histone methyltransferase enhancer of zeste 2 polycomb repressive complex 2 subunit exacerbates inflammation in depression rats by modulating microglia polarization. Bioengineered 13, 5509–5524. doi:10.1080/21655979.2022.2036892
Hung, Y. Y., Wu, M. K., Tsai, M. C., Huang, Y. L., and Kang, H. Y. (2019). Aberrant Expression of Intracellular let-7e, miR-146a, and miR-155 Correlates with Severity of Depression in Patients with Major Depressive Disorder and Is Ameliorated after Antidepressant Treatment. Cells 8, 647. doi:10.3390/cells8070647
Issler, O., Haramati, S., Paul, E. D., Maeno, H., Navon, I., Zwang, R., et al. (2014). MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron 83, 344–360. doi:10.1016/j.neuron.2014.05.042
Jenkins, T. A., Nguyen, J. C., Polglaze, K. E., and Bertrand, P. P. (2016). Influence of tryptophan and serotonin on mood and cognition with a possible role of the gut-brain Axis. Nutrients 8, 56. doi:10.3390/nu8010056
Jernigan, C. S., Goswami, D. B., Austin, M. C., Iyo, A. H., Chandran, A., Stockmeier, C. A., et al. (2011). The mTOR signaling pathway in the prefrontal cortex is compromised in major depressive disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry 35, 1774–1779. doi:10.1016/j.pnpbp.2011.05.010
Kalueff, A. V., and Nutt, D. J. (2007). Role of GABA in anxiety and depression. Depress Anxiety 24, 495–517. doi:10.1002/da.20262
Kavuran Buran, I., Onalan Etem, E., and Tektemur, A. (2022). Inhibition of TRPC1, TRPM4 and CHRNA6 ion channels ameliorates depression-like behavior in rats. Behav. Brain Res. 423, 113765. doi:10.1016/j.bbr.2022.113765
Keller, J., Gomez, R., Williams, G., Lembke, A., Lazzeroni, L., Murphy, G. M., et al. (2017). HPA axis in major depression: Cortisol, clinical symptomatology and genetic variation predict cognition. Mol. Psychiatry 22, 527–536. doi:10.1038/mp.2016.120
Kiecolt-Glaser, J. K., Derry, H. M., and Fagundes, C. P. (2015). Inflammation: Depression fans the flames and feasts on the heat. Am. J. Psychiatry 172, 1075–1091. doi:10.1176/appi.ajp.2015.15020152
Kim, H., Kim, J., Lee, H., Shin, E., Kang, H., Jeon, J., et al. (2022). Baiap3 regulates depressive behaviors in mice via attenuating dense core vesicle trafficking in subsets of prefrontal cortex neurons. Neurobiol. Stress 16, 100423. doi:10.1016/j.ynstr.2021.100423
Kobayashi, Y. (2010). The regulatory role of nitric oxide in proinflammatory cytokine expression during the induction and resolution of inflammation. J. Leukoc. Biol. 88, 1157–1162. doi:10.1189/jlb.0310149
Kronenberg, G., Kirste, I., Inta, D., Chourbaji, S., Heuser, I., Endres, M., et al. (2009). Reduced hippocampal neurogenesis in the GR(+/-) genetic mouse model of depression. Eur. Arch. Psychiatry Clin. Neurosci. 259, 499–504. doi:10.1007/s00406-009-0036-y
Kuang, W. H., Dong, Z. Q., Tian, L. T., and Li, J. (2018). MicroRNA-451a, microRNA-34a-5p, and microRNA-221-3p as predictors of response to antidepressant treatment. Braz J. Med. Biol. Res. 51, e7212. doi:10.1590/1414-431x20187212
Lan, T., Li, Y., Fan, C., Wang, L., Wang, W., Chen, S., et al. (2021). MicroRNA-204-5p reduction in rat hippocampus contributes to stress-induced pathology via targeting RGS12 signaling pathway. J. Neuroinflammation 18, 243. doi:10.1186/s12974-021-02299-5
Leonard, B. E. (2018). Inflammation and depression: A causal or coincidental link to the pathophysiology? Acta Neuropsychiatr. 30, 1–16. doi:10.1017/neu.2016.69
Li, L. D., Naveed, M., Du, Z. W., Ding, H., Gu, K., Wei, L. L., et al. (2021a). Abnormal expression profile of plasma-derived exosomal microRNAs in patients with treatment-resistant depression. Hum. Genomics 15, 55. doi:10.1186/s40246-021-00354-z
Li, X., Sun, X., Xie, J., and Wan, H. (2022). CircDYM ameliorates CUMS mice depressive-like behavior and inhibits hippocampal neurons injury via miR-497a-5p/NR3C1 axis. Brain Res. 1787, 147911. doi:10.1016/j.brainres.2022.147911
Li, Y., Fan, C., Gao, R., Lan, T., Wang, W., and Yu, S. Y. (2021b). Hippocampal miR-211-5p regulates neurogenesis and depression-like behaviors in the rat. Neuropharmacology 194, 108618. doi:10.1016/j.neuropharm.2021.108618
Li, Y., Fan, C., Wang, L., Lan, T., Gao, R., Wang, W., et al. (2021c). MicroRNA-26a-3p rescues depression-like behaviors in male rats via preventing hippocampal neuronal anomalies. J. Clin. Investig. 131, e148853. doi:10.1172/JCI148853
Liang, J. Q., Liao, H. R., Xu, C. X., Li, X. L., Wei, Z. X., Xie, G. J., et al. (2020). Serum exosome-derived miR-139-5p as a potential biomarker for major depressive disorder. Neuropsychiatr. Dis. Treat. 16, 2689–2693. doi:10.2147/NDT.S277392
Liu, L., Wang, H., Chen, X., Zhang, Y., Li, W., Rao, X., et al. (2021a). Integrative analysis of long non-coding RNAs, messenger RNAs, and MicroRNAs indicates the neurodevelopmental dysfunction in the Hippocampus of gut microbiota-dysbiosis mice. Front. Mol. Neurosci. 14, 745437. doi:10.3389/fnmol.2021.745437
Liu, S., Liu, Q., Ju, Y., and Liu, L. (2021b). Downregulation of miR-383 reduces depression-like behavior through targeting Wnt family member 2 (Wnt2) in rats. Sci. Rep. 11, 9223. doi:10.1038/s41598-021-88560-6
Liu, W., Zhang, F., Zheng, Y., He, S., Zhang, T., Guo, Q., et al. (2021c). The role of circulating blood microRNA-374 and microRNA-10 levels in the pathogenesis and therapeutic mechanisms of major depressive disorder. Neurosci. Lett. 763, 136184. doi:10.1016/j.neulet.2021.136184
Liu, Y., Yang, X., Zhao, L., Zhang, J., Li, T., and Ma, X. (2016). Increased miR-132 level is associated with visual memory dysfunction in patients with depression. Neuropsychiatr. Dis. Treat. 12, 2905–2911. doi:10.2147/NDT.S116287
Liu, Z., Yang, J., Fang, Q., Shao, H., Yang, D., Sun, J., et al. (2021d). MiRNA-199a-5p targets WNT2 to regulate depression through the CREB/BDNF signaling in hippocampal neuron. Brain Behav. 11, e02107. doi:10.1002/brb3.2107
Lopez, J. P., Fiori, L. M., Cruceanu, C., Lin, R., Labonte, B., Cates, H. M., et al. (2017). MicroRNAs 146a/b-5 and 425-3p and 24-3p are markers of antidepressant response and regulate MAPK/Wnt-system genes. Nat. Commun. 8, 15497. doi:10.1038/ncomms15497
Lopez, J. P., Fiori, L. M., Gross, J. A., Labonte, B., Yerko, V., Mechawar, N., et al. (2014a). Regulatory role of miRNAs in polyamine gene expression in the prefrontal cortex of depressed suicide completers. Int. J. Neuropsychopharmacol. 17, 23–32. doi:10.1017/S1461145713000941
Lopez, J. P., Lim, R., Cruceanu, C., Crapper, L., Fasano, C., Labonte, B., et al. (2014b). miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat. Med. 20, 764–768. doi:10.1038/nm.3582
Lu, T. X., and Rothenberg, M. E. (2018). MicroRNA. J. Allergy Clin. Immunol. 141, 1202–1207. doi:10.1016/j.jaci.2017.08.034
Maheu, M., Lopez, J. P., Crapper, L., Davoli, M. A., Turecki, G., and Mechawar, N. (2015). MicroRNA regulation of central glial cell line-derived neurotrophic factor (GDNF) signalling in depression. Transl. Psychiatry 5, e511. doi:10.1038/tp.2015.11
Malhi, G. S., and Mann, J. J. (2018). Depression. Lancet 392, 2299–2312. doi:10.1016/S0140-6736(18)31948-2
Maussion, G., Yang, J., Yerko, V., Barker, P., Mechawar, N., Ernst, C., et al. (2012). Regulation of a truncated form of tropomyosin-related kinase B (TrkB) by Hsa-miR-185* in frontal cortex of suicide completers. PLoS One 7, e39301. doi:10.1371/journal.pone.0039301
Mendes-Silva, A. P., Fujimura, P. T., Silva, J., Teixeira, A. L., Vieira, E. M., Guedes, P. H. G., et al. (2019). Brain-enriched MicroRNA-184 is downregulated in older adults with major depressive disorder: A translational study. J. Psychiatr. Res. 111, 110–120. doi:10.1016/j.jpsychires.2019.01.019
Miller, A. H., and Raison, C. L. (2016). The role of inflammation in depression: From evolutionary imperative to modern treatment target. Nat. Rev. Immunol. 16, 22–34. doi:10.1038/nri.2015.5
Mingardi, J., La Via, L., Tornese, P., Carini, G., Trontti, K., Seguini, M., et al. (2021). miR-9-5p is involved in the rescue of stress-dependent dendritic shortening of hippocampal pyramidal neurons induced by acute antidepressant treatment with ketamine. Neurobiol. Stress 15, 100381. doi:10.1016/j.ynstr.2021.100381
Mizohata, Y., Toda, H., Koga, M., Saito, T., Fujita, M., Kobayashi, T., et al. (2021). Neural extracellular vesicle-derived miR-17 in blood as a potential biomarker of subthreshold depression. Hum. Cell. 34, 1087–1092. doi:10.1007/s13577-021-00553-9
Molendijk, M. L., Spinhoven, P., Polak, M., Bus, B. A., Penninx, B. W., and Elzinga, B. M. (2014). Serum BDNF concentrations as peripheral manifestations of depression: Evidence from a systematic review and meta-analyses on 179 associations (N=9484). Mol. Psychiatry 19, 791–800. doi:10.1038/mp.2013.105
Morgunova, A., and Flores, C. (2021). MicroRNA regulation of prefrontal cortex development and psychiatric risk in adolescence. Semin. Cell. Dev. Biol. 118, 83–91. doi:10.1016/j.semcdb.2021.04.011
Numakawa, T., Adachi, N., Richards, M., Chiba, S., and Kunugi, H. (2012). The influence of glucocorticoids on neuronal survival and synaptic function. Biomol. Concepts 3, 495–504. doi:10.1515/bmc-2012-0012
Numakawa, T., Kumamaru, E., Adachi, N., Yagasaki, Y., Izumi, A., and Kunugi, H. (2009). Glucocorticoid receptor interaction with TrkB promotes BDNF-triggered PLC-gamma signaling for glutamate release via a glutamate transporter. Proc. Natl. Acad. Sci. U. S. A. 106, 647–652. doi:10.1073/pnas.0800888106
Pariante, C. M., and Lightman, S. L. (2008). The HPA axis in major depression: Classical theories and new developments. Trends Neurosci. 31, 464–468. doi:10.1016/j.tins.2008.06.006
Penner-Goeke, S., and Binder, E. B. (2019). Epigenetics and depression. Dialogues Clin. Neurosci. 21, 397–405. doi:10.31887/DCNS.2019.21.4/ebinder
Persoon, C. M., Moro, A., Nassal, J. P., Farina, M., Broeke, J. H., Arora, S., et al. (2018). Pool size estimations for dense-core vesicles in mammalian CNS neurons. EMBO J. 37, e99672. doi:10.15252/embj.201899672
Petra, A. I., Panagiotidou, S., Hatziagelaki, E., Stewart, J. M., Conti, P., and Theoharides, T. C. (2015). Gut-microbiota-brain Axis and its effect on neuropsychiatric disorders with suspected immune dysregulation. Clin. Ther. 37, 984–995. doi:10.1016/j.clinthera.2015.04.002
Qi, S., Yang, X., Zhao, L., Calhoun, V. D., Perrone-Bizzozero, N., Liu, S., et al. (2018). MicroRNA132 associated multimodal neuroimaging patterns in unmedicated major depressive disorder. Brain 141, 916–926. doi:10.1093/brain/awx366
Qin, G., and Li, Z. (2022). Effects of miR-124-3p silencing on neuronal damage in the Hippocampus of depression rats by regulating STAT3 gene. Comput. Math. Methods Med. 2022, 3733656. doi:10.1155/2022/3733656
Rajasethupathy, P., Fiumara, F., Sheridan, R., Betel, D., Puthanveettil, S. V., Russo, J. J., et al. (2009). Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 63, 803–817. doi:10.1016/j.neuron.2009.05.029
Ransohoff, R. M., Kivisakk, P., and Kidd, G. (2003). Three or more routes for leukocyte migration into the central nervous system. Nat. Rev. Immunol. 3, 569–581. doi:10.1038/nri1130
Rea, K., Dinan, T. G., and Cryan, J. F. (2020). Gut microbiota: A perspective for psychiatrists. Neuropsychobiology 79, 50–62. doi:10.1159/000504495
Rea, K., Dinan, T. G., and Cryan, J. F. (2017). The brain-gut Axis contributes to neuroprogression in stress-related disorders. Mod. Trends Pharmacopsychiatry 31, 152–161. doi:10.1159/000470813
Rivest, S., Lacroix, S., Vallieres, L., Nadeau, S., Zhang, J., and Laflamme, N. (2000). How the blood talks to the brain parenchyma and the paraventricular nucleus of the hypothalamus during systemic inflammatory and infectious stimuli. Proc. Soc. Exp. Biol. Med. 223, 22–38. doi:10.1046/j.1525-1373.2000.22304.x
Roumans, S., Sundquist, K., Memon, A. A., Hedelius, A., Sundquist, J., and Wang, X. (2021). Association of circulating let-7b-5p with major depressive disorder: A nested case-control study. BMC Psychiatry 21, 616. doi:10.1186/s12888-021-03621-4
Roy, B., Dunbar, M., Agrawal, J., Allen, L., and Dwivedi, Y. (2020). Amygdala-based altered miRNome and epigenetic contribution of miR-128-3p in conferring susceptibility to depression-like behavior via Wnt signaling. Int. J. Neuropsychopharmacol. 23, 165–177. doi:10.1093/ijnp/pyz071
Roy, B., Dunbar, M., Shelton, R. C., and Dwivedi, Y. (2017a). Identification of MicroRNA-124-3p as a putative epigenetic signature of major depressive disorder. Neuropsychopharmacology 42, 864–875. doi:10.1038/npp.2016.175
Roy, B., Wang, Q., Palkovits, M., Faludi, G., and Dwivedi, Y. (2017b). Altered miRNA expression network in locus coeruleus of depressed suicide subjects. Sci. Rep. 7, 4387. doi:10.1038/s41598-017-04300-9
Schmidt, M. V., Sterlemann, V., Wagner, K., Niederleitner, B., Ganea, K., Liebl, C., et al. (2009). Postnatal glucocorticoid excess due to pituitary glucocorticoid receptor deficiency: Differential short- and long-term consequences. Endocrinology 150, 2709–2716. doi:10.1210/en.2008-1211
Segal, D. S., Kuczenski, R., and Mandell, A. J. (1974). Theoretical implications of drug-induced adaptive regulation for a biogenic amine hypothesis of affective disorder. Biol. Psychiatry 9, 147–159.
Shaw, D. M., Camps, F. E., and Eccleston, E. G. (1967). 5-Hydroxytryptamine in the hind-brain of depressive suicides. Br. J. Psychiatry 113, 1407–1411. doi:10.1192/bjp.113.505.1407
Shen, J., Zhang, P., Li, Y., Fan, C., Lan, T., Wang, W., et al. (2021). Neuroprotective effects of microRNA-211-5p on chronic stress-induced neuronal apoptosis and depression-like behaviours. J. Cell. Mol. Med. 25, 7028–7038. doi:10.1111/jcmm.16716
Sherwin, E., Rea, K., Dinan, T. G., and Cryan, J. F. (2016). A gut (microbiome) feeling about the brain. Curr. Opin. Gastroenterol. 32, 96–102. doi:10.1097/MOG.0000000000000244
Si, L., Wang, Y., Liu, M., Yang, L., and Zhang, L. (2021). Expression and role of microRNA-212/nuclear factor I-A in depressive mice. Bioengineered 12, 11520–11532. doi:10.1080/21655979.2021.2009964
Smalheiser, N. R., Lugli, G., Rizavi, H. S., Torvik, V. I., Turecki, G., and Dwivedi, Y. (2012). MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS One 7, e33201. doi:10.1371/journal.pone.0033201
Smalheiser, N. R., Lugli, G., Zhang, H., Rizavi, H., Cook, E. H., and Dwivedi, Y. (2014). Expression of microRNAs and other small RNAs in prefrontal cortex in schizophrenia, bipolar disorder and depressed subjects. PLoS One 9, e86469. doi:10.1371/journal.pone.0086469
Stokes, P. E. (1995). The potential role of excessive cortisol induced by HPA hyperfunction in the pathogenesis of depression. Eur. Neuropsychopharmacol. 5 (1), 77–82. doi:10.1016/0924-977x(95)00039-r
Su, B., Cheng, S., Wang, L., and Wang, B. (2022). MicroRNA-139-5p acts as a suppressor gene for depression by targeting nuclear receptor subfamily 3, group C, member 1. Bioengineered 13, 11856–11866. doi:10.1080/21655979.2022.2059937
Sun, N., Lei, L., Wang, Y., Yang, C., Liu, Z., Li, X., et al. (2016). Preliminary comparison of plasma notch-associated microRNA-34b and -34c levels in drug naive, first episode depressed patients and healthy controls. J. Affect Disord. 194, 109–114. doi:10.1016/j.jad.2016.01.017
Sun, N., Yang, C., He, X., Liu, Z., Liu, S., Li, X., et al. (2020). Impact of expression and genetic variation of microRNA-34b/c on cognitive dysfunction in patients with major depressive disorder. Neuropsychiatr. Dis. Treat. 16, 1543–1554. doi:10.2147/NDT.S247787
Sundquist, K., Memon, A. A., Palmer, K., Sundquist, J., and Wang, X. (2021). Inflammatory proteins and miRNA-144-5p in patients with depression, anxiety, or stress- and adjustment disorders after psychological treatment. Cytokine 146, 155646. doi:10.1016/j.cyto.2021.155646
Torres-Berrio, A., Lopez, J. P., Bagot, R. C., Nouel, D., Dal Bo, G., Cuesta, S., et al. (2017). DCC confers susceptibility to depression-like behaviors in humans and mice and is regulated by miR-218. Biol. Psychiatry 81, 306–315. doi:10.1016/j.biopsych.2016.08.017
Troubat, R., Barone, P., Leman, S., Desmidt, T., Cressant, A., Atanasova, B., et al. (2021). Neuroinflammation and depression: A review. Eur. J. Neurosci. 53, 151–171. doi:10.1111/ejn.14720
Uchida, S., Nishida, A., Hara, K., Kamemoto, T., Suetsugi, M., Fujimoto, M., et al. (2008). Characterization of the vulnerability to repeated stress in fischer 344 rats: Possible involvement of microRNA-mediated down-regulation of the glucocorticoid receptor. Eur. J. Neurosci. 27, 2250–2261. doi:10.1111/j.1460-9568.2008.06218.x
Vaisvaser, S., Modai, S., Farberov, L., Lin, T., Sharon, H., Gilam, A., et al. (2016). Neuro-epigenetic indications of acute stress response in humans: The case of MicroRNA-29c. PLoS One 11, e0146236. doi:10.1371/journal.pone.0146236
Van Der Auwera, S., Ameling, S., Wittfeld, K., D'harcourt Rowold, E., Nauck, M., Volzke, H., et al. (2019). Association of childhood traumatization and neuropsychiatric outcomes with altered plasma micro RNA-levels. Neuropsychopharmacology 44, 2030–2037. doi:10.1038/s41386-019-0460-2
Voleti, B., and Duman, R. S. (2012). The roles of neurotrophic factor and Wnt signaling in depression. Clin. Pharmacol. Ther. 91, 333–338. doi:10.1038/clpt.2011.296
Volk, N., Pape, J. C., Engel, M., Zannas, A. S., Cattane, N., Cattaneo, A., et al. (2016). Amygdalar MicroRNA-15a is essential for coping with chronic stress. Cell. Rep. 17, 1882–1891. doi:10.1016/j.celrep.2016.10.038
Vreugdenhil, E., Verissimo, C. S., Mariman, R., Kamphorst, J. T., Barbosa, J. S., Zweers, T., et al. (2009). MicroRNA 18 and 124a down-regulate the glucocorticoid receptor: Implications for glucocorticoid responsiveness in the brain. Endocrinology 150, 2220–2228. doi:10.1210/en.2008-1335
Wang, H., Ma, Z., Shen, H., Wu, Z., Liu, L., Ren, B., et al. (2021). Early life irradiation-induced hypoplasia and impairment of neurogenesis in the dentate gyrus and adult depression are mediated by MicroRNA- 34a-5p/T-cell intracytoplasmic antigen-1 pathway. Cells 10, 2476. Cells. doi:10.3390/cells10092476
Wang, Q., Roy, B., Turecki, G., Shelton, R. C., and Dwivedi, Y. (2018a). Role of complex epigenetic switching in tumor necrosis factor-alpha upregulation in the prefrontal cortex of suicide subjects. Am. J. Psychiatry 175, 262–274. doi:10.1176/appi.ajp.2017.16070759
Wang, Q., Zhao, G., Yang, Z., Liu, X., and Xie, P. (2018b). Downregulation of microRNA1243p suppresses the mTOR signaling pathway by targeting DDIT4 in males with major depressive disorder. Int. J. Mol. Med. 41, 493–500. doi:10.3892/ijmm.2017.3235
Wang, X., Wang, B., Zhao, J., Liu, C., Qu, X., and Li, Y. (2018c). MiR-155 is involved in major depression disorder and antidepressant treatment via targeting SIRT1. Biosci. Rep. 38. doi:10.1042/BSR20181139
Wayman, G. A., Impey, S., Marks, D., Saneyoshi, T., Grant, W. F., Derkach, V., et al. (2006). Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron 50, 897–909. doi:10.1016/j.neuron.2006.05.008
Wei, Z. X., Xie, G. J., Mao, X., Zou, X. P., Liao, Y. J., Liu, Q. S., et al. (2020). Exosomes from patients with major depression cause depressive-like behaviors in mice with involvement of miR-139-5p-regulated neurogenesis. Neuropsychopharmacology 45, 1050–1058. doi:10.1038/s41386-020-0622-2
Wibrand, K., Pai, B., Siripornmongcolchai, T., Bittins, M., Berentsen, B., Ofte, M. L., et al. (2012). MicroRNA regulation of the synaptic plasticity-related gene Arc. PLoS One 7, e41688. doi:10.1371/journal.pone.0041688
Willner, P., Scheel-Kruger, J., and Belzung, C. (2013). The neurobiology of depression and antidepressant action. Neurosci. Biobehav Rev. 37, 2331–2371. doi:10.1016/j.neubiorev.2012.12.007
Wingo, T. S., Yang, J., Fan, W., Min Canon, S., Gerasimov, E. S., Lori, A., et al. (2020). Brain microRNAs associated with late-life depressive symptoms are also associated with cognitive trajectory and dementia. NPJ Genom Med. 5, 6. doi:10.1038/s41525-019-0113-8
Xian, X., Cai, L. L., Li, Y., Wang, R. C., Xu, Y. H., Chen, Y. J., et al. (2022). Neuron secrete exosomes containing miR-9-5p to promote polarization of M1 microglia in depression. J. Nanobiotechnology 20, 122. doi:10.1186/s12951-022-01332-w
Yoshii, A., and Constantine-Paton, M. (2010). Postsynaptic BDNF-TrkB signaling in synapse maturation, plasticity, and disease. Dev. Neurobiol. 70, 304–322. doi:10.1002/dneu.20765
Yoshino, Y., Roy, B., and Dwivedi, Y. (2020). Altered miRNA landscape of the anterior cingulate cortex is associated with potential loss of key neuronal functions in depressed brain. Eur. Neuropsychopharmacol. 40, 70–84. doi:10.1016/j.euroneuro.2020.06.004
Yoshino, Y., Roy, B., and Dwivedi, Y. (2022). Corticosterone-mediated regulation and functions of miR-218-5p in rat brain. Sci. Rep. 12, 194. doi:10.1038/s41598-021-03863-y
Zhang, H. P., Liu, X. L., Chen, J. J., Cheng, K., Bai, S. J., Zheng, P., et al. (2020a). Circulating microRNA 134 sheds light on the diagnosis of major depressive disorder. Transl Psychiatry, 10, 95. doi:10.1038/s41398-020-0773-2
Zhang, Y., Du, L., Bai, Y., Han, B., He, C., Gong, L., et al. (2020b). CircDYM ameliorates depressive-like behavior by targeting miR-9 to regulate microglial activation via HSP90 ubiquitination. Mol. Psychiatry 25, 1175–1190. doi:10.1038/s41380-018-0285-0
Zhao, L., Yang, X., Cui, L., Wei, J., Ni, P., Li, M., et al. (2019). Increased expression of a novel miRNA in peripheral blood is negatively correlated with hippocampal volume in patients with major depressive disorder. J. Affect Disord. 245, 205–212. doi:10.1016/j.jad.2018.10.363
Keywords: microRNA, depression, brain, biomarker, MDD
Citation: Ding R, Su D, Zhao Q, Wang Y, Wang J-Y, Lv S and Ji X (2023) The role of microRNAs in depression. Front. Pharmacol. 14:1129186. doi: 10.3389/fphar.2023.1129186
Received: 21 December 2022; Accepted: 21 March 2023;
Published: 29 March 2023.
Edited by:
Guillaume Lucas, INSERM U1215 Neurocentre Magendie, FranceReviewed by:
Cortney Ann Turner, University of Michigan, United StatesCopyright © 2023 Ding, Su, Zhao, Wang, Wang, Lv and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuangyu Lv, c2h1YW5neXVsdkAxNjMuY29t; Xinying Ji, MTAxOTAwOTZAdmlwLmhlbnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.