
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 27 February 2023
Sec. Drug Metabolism and Transport
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1102810
This article is part of the Research Topic Drug Metabolism and Transport: The Frontier of Personalized Medicine View all 22 articles
 Zebin Jiang1†
Zebin Jiang1† Zemin Wu2†
Zemin Wu2† Ruixue Liu2
Ruixue Liu2 Qin Du2
Qin Du2 Xian Fu1
Xian Fu1 Min Li2
Min Li2 Yongjun Kuang2
Yongjun Kuang2 Shen Lin2
Shen Lin2 Jiaxuan Wu3
Jiaxuan Wu3 Weiji Xie4
Weiji Xie4 Ganggang Shi2
Ganggang Shi2 Yanqiang Peng5*
Yanqiang Peng5* Fuchun Zheng1,2*
Fuchun Zheng1,2*Dyslipidemia due to renal insufficiency is a common complication in patients with chronic kidney diseases (CKD), and a major risk factor for the development of cardiovascular events. Atorvastatin (AT) is mainly used in the treatment of dyslipidemia in patients with CKD. However, response to the atorvastatin varies inter-individually in clinical applications. We examined the association between polymorphisms in genes involved in drug metabolism and transport, and plasma concentrations of atorvastatin and its metabolites (2-hydroxy atorvastatin (2-AT), 2-hydroxy atorvastatin lactone (2-ATL), 4-hydroxy atorvastatin (4-AT), 4-hydroxy atorvastatin lactone (4-ATL), atorvastatin lactone (ATL)) in kidney diseases patients. Genotypes were determined using TaqMan real time PCR in 212 CKD patients, treated with 20 mg of atorvastatin daily for 6 weeks. The steady state plasma concentrations of atorvastatin and its metabolites were quantified using ultraperformance liquid chromatography in combination with triple quadrupole mass spectrometry (UPLC−MS/MS). Univariate and multivariate analyses showed the variant in ABCC4 (rs3742106) was associated with decreased concentrations of AT and its metabolites (2-AT+2-ATL: β = -0.162, p = 0.028 in the dominant model; AT+2-AT+4-AT: β = -0.212, p = 0.028 in the genotype model), while patients carrying the variant allele ABCC4-rs868853 (β = 0.177, p = 0.011) or NR1I2-rs6785049 (β = 0.123, p = 0.044) had higher concentrations of 2-AT+2-ATL in plasma compared with homozygous wildtype carriers. Luciferase activity was enhanced in HepG2 cells harboring a construct expressing the rs3742106-T allele or the rs868853-G allele (p < 0.05 for each) compared with a construct expressing the rs3742106G or the rs868853-A allele. These findings suggest that two functional polymorphisms in the ABCC4 gene may affect transcriptional activity, thereby directly or indirectly affecting release of AT and its metabolites from hepatocytes into the circulation.
China has one of the largest populations of patients with chronic kidney diseases (CKD) in Asia (up to 159.8 million in 2020) (Liyanage et al., 2022). Dyslipidemia due to renal insufficiency is a common complication in patients with CKD, leads to further development of kidney damage and deterioration of kidney function (Jungers et al., 1997). Both CDK and dyslipidemia are considered to be major risk factors for cardiovascular events (Sarnak et al., 2003; Kopin and Lowenstein, 2017). Therefore, for CKD patients, treatment of dyslipidemia is particularly important.
Atorvastatin (AT) is currently the first-line drug for lipid lowering and prevention of cardiovascular disease (Arca and Gaspardone, 2007). As an HMG-CoA reductase inhibitor, atorvastatin reduces cholesterol synthesis and increases the number of LDL receptors on the surface of hepatocytes, thereby reducing plasma LDL cholesterol levels, and has a stronger lipid-lowering effect in women or in patients with non-familial hyperlipidemia (Adams et al., 2015). In addition to its lipid-lowering effects, atorvastatin also protects the cardiovascular system and reduces damage to the kidney through its multiple anti-inflammatory, antioxidative, endothelial protective and anti-cell proliferative effects (Aviram et al., 1998; Mason, 2006). CKD patients who are not on dialysis or renal transplantation were are now recommended to adopt lipid-regulating therapy with statins, according to the Kidney Disease Improving Global Outcomes (KDIGO) Clinical Practice Guideline (Tonelli and Wanner, 2014).
However, there are significant individual differences in clinical responses (efficacy or toxicity) to atorvastatin among different patients. Previous studies have reported that nearly one-third of patients fail to achieve lipid-lowering goals despite dose adjustments to statins based on patient response to treatment (Mangravite et al., 2006). Observational studies have found that 10%–15% of statin users experience varying degrees of statin-related muscle side effects, including mild muscle pain, muscle cramps, muscle weakness and even the rare and serious symptom of rhabdomyolysis (Abd and Jacobson, 2011). This individual variation in pharmacodynamics is related to the level of drug in the plasma (Link et al., 2008). However, the plasma AT levels are not only influenced by clinical factors such as gender, age, BMI, co-morbidities and co-administration (Turner et al., 2020a; Hirota et al., 2020), but also are strongly associated with polymorphisms in genes related to drug absorption, distribution, metabolism and excretion (ADME) (DeGorter et al., 2013; Cruz-Correa et al., 2017; Turner et al., 2020b) which have been reported to have high inter-individual variability (45-fold) (DeGorter et al., 2013). It is also noteworthy that genetic variability may contribute to >90% of the variance in plasma AT concentrations, and mainly occurs in genes for drug metabolizing enzymes and transporters (DeGorter et al., 2013; Cruz-Correa et al., 2017; Turner et al., 2020b).
AT is administered orally as a calcium salt that is absorbed into the blood via the small intestine and taken up by the OATP transporter into hepatocytes, where it is metabolized by CYP3A4 and CYP3A5 to the partially bioactive hydroxylated derivatives (2-hydroxy atorvastatin (2-AT) and 4-AT) (Lennernäs, 2003). AT and its active metabolites undergo lactonization via an unstable acyl glucuronide intermediate to produce AT lactone (ATL), 2-ATL and 4-ATL (Prueksaritanont et al., 2002). These lactone metabolites can be hydrolyzed to the corresponding hydroxy acids by plasma paraoxonases or by pH changes (Riedmaier et al., 2011) (Figure 1). AT and its metabolites are mainly eliminated by bile, with only about 1% excreted by the kidneys (Lennernäs, 2003). Its hydroxylated metabolite was found to inhibit HMG-CoA reductase as much as AT in vitro, about 70% of the total plasma HMG-CoA reductase inhibitory activity is accounted for by active metabolites (Lea and McTavish, 1997; Prake-Davis, 2004). In addition, the active metabolites of AT also protect the cardiovascular system with pleiotropic effects such as anti-oxidation and improvement of endothelial function.[20, 21] Therefore, the plasma concentration of AT metabolites should also be of concern.
CYP3A4 and CYP3A5 enzymes are the main metabolizing enzymes for AT, playing an important role in disposition of AT. It has been reported that rs2242480, located on intron 10 of CYP3A4, is associated with the lipid-lowering efficacy of atorvastatin (Gao et al., 2008; Peng et al., 2018), and the presence of rs4646437 in CYP3A4 intron 7 affects CYP3A4 protein expression and enzymatic activity in a gender-dependent manner (Schirmer et al., 2007), but there has been no study on the effects of these polymorphisms on plasma AT concentration. CYP3A5*3 (rs776746C) in intron 3 of CYP3A5 leads to aberrant splicing of mRNA and truncation of protein, resulting in deletion of CYP3A5 protein in individuals homozygous for this allele (Kuehl et al., 2001), and has been shown to be associated with increased systemic atorvastatin in a prior bioequivalence study in healthy volunteers (Zubiaur et al., 2021).
The OATP1B1 transporter, encoded by SLC O 1B1 (König et al., 2000), mediates the absorption of drugs into hepatocytes, and is the rate-limiting step in the hepatic clearance of atorvastatin (Maeda et al., 2011). Previous studies have revealed that single coding single-nucleotide polymorphisms (SNPs, rs4149056 and rs2306283) of SLC O 1B1 are associated with the plasma AT concentration (DeGorter et al., 2013). Moreover, SLC O 1B1 rs4149056 contributes to statin-induced myopathy, and the Clinical Pharmacogenetics Implementation Consortium (CPIC) has published guidelines for the use of atorvastatin in carriers of this SNP (Cooper-DeHoff et al., 2022), but it is not specifically indicated for CKD complications.
Atorvastatin is a substrate for the efflux transporters P-glycoprotein, encoded by ABCB1, and BCRP, encoded by ABCG2, which may limit intestinal absorption and biliary clearance of AT. The common SNPs of ABCB1 (rs1045642, rs1128503 and rs2032582) have been reported to be associated with the lipid-lowering efficacy of atorvastatin (Thompson et al., 2005; Hoenig et al., 2011; Prado et al., 2018), but their association with plasma AT concentration is unknown. The ABCG2 (421C>A) variant (rs2231142) contains a replacement of glutamine with lysine at position 141 in the intracellular region of the protein, and demonstrates lower protein expression and transport capacity in cells transfected with the variant than with the wild type (Imai et al., 2002; Kondo et al., 2004). Although many studies have shown a correlation between rs2231142 and plasma concentrations of atorvastatin, the phenotypic results of the effects differ in different populations (Birmingham et al., 2015; Lee et al., 2019).
In addition to the above transporters, MRP3, MRP4 and MRP5, which are localized in the basolateral membrane of hepatocytes, have also been found to be involved in the transport of atorvastatin in vitro (Knauer et al., 2010; Deng et al., 2021), and their polymorphisms may play an important role in the plasma concentration of AT and its metabolites. Previous studies have indicated that rs4793665 located in the ABCC3 promoter region, affects the plasma concentration of morphine and its metabolites, and it is now clear that polymorphisms in ABCC4 rs2274407, rs3742106, rs868853 and rs9561778 have a significant effect on plasma levels, drug efficacy and disease susceptibility (Anderson et al., 2006; Low et al., 2009; Venkatasubramanian et al., 2014; Tanaka et al., 2015; Sánchez-Martín et al., 2016; Chidambaran et al., 2017; Che et al., 2018). Similarly, ABCC5 variants (rs562 and rs3749438) also have been reported to be associated with severe irinotecan-induced toxicity and its plasma concentration (Chen et al., 2015; Teft et al., 2015).
The pregnane X receptor (PXR, NR1I2), a prototypical member of the nuclear receptor superfamily, can be activated by a range of steroids or exogenous drugs to regulate the transcription of target genes, and plays an important role in the regulation of the environmental homeostasis and pathophysiological processes (Rogers et al., 2021). Atorvastatin acts as a ligand for PXR and activates transcription of target genes, including CYP3A4, CYP3A5, SLC O 1B1 and ABCB1 (Marino et al., 2011; Hoffart et al., 2012), thereby affecting the metabolic process of the drug. Many studies have reported that polymorphisms in NR1I2 (rs6785049 and rs1523127) impact the pharmacokinetics of a variety of drugs, including immunosuppressive agents (Fanta et al., 2010; Mbatchi et al., 2017), antifungal agents (Zeng et al., 2020), antineoplastic agents (Liu et al., 2017) and anti-AIDS agents (Swart et al., 2012).
Accordingly, we examined the effects of reported polymorphisms in metabolic enzymes (CYP3A4 and CYP3A5) and transporters (SLC O 1B1, ABCB1 and ABCG2) associated with atorvastatin efficacy, as well as other drug transport-related gene (ABCC3, ABCC4, ABCC5 and NR1I2) variants on plasma concentrations of atorvastatin and its metabolites in patients with CKD. The study of individual variation in plasma concentrations of atorvastatin and its metabolites at the genetic level is helpful to predict the efficacy and toxicity of atorvastatin in CKD patients accurately, and has practical guiding significance for clinical individualized drug application.
A prospective study was performed with Chinese Han chronic kidney disease patients recruited from both out-patients and in-patients of the Nephrology Departments of the First and Second Affiliated Hospitals of Shantou University Medical College, from May 2014 to September 2019 (Chinese Clinical Trial Registry No. ChiCTR2000041391). According to the inclusion and exclusion criteria (Supplementary Table S1), there were 354 eligible CKD patients, 142 patients were excluded due to missing data, a total of 212 CKD patients participated in our study. All patients were prescribed atorvastatin (Pfizer, NY, America) 20 mg/day for 6 weeks. Overnight fasting blood samples were collected from patients to measure biochemical parameters and drug levels. Baseline parameters were measured the morning of the day of treatment initiation, and final parameters were measured the day the 6-week treatment was completed. Demographic and clinical information, including age, sex, body, and medical histories were recorded. Estimated glomerular filtration rate (eGFR) was calculated according to the Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equation (Levey et al., 2009). Experimental subjects were not randomized into groups and the experimenters were not blinded because these were deemed inappropriate for the design of this study. This study was approved by the ethics committees of the First and Second Affiliated Hospitals of Shantou University Medical College. All enrolled patients were informed of the purpose and other matters of the study, understood and signed the informed consent form.
DNA was isolated from EDTA-coated whole blood tubes, following patient blood collection, using a TIANamp blood DNA kit (TIANGEN, Beijing, China). The concentration and purity of extracted DNA following the manufacturer’s protocols were determined with a Nanodrop 2000.
Genotyping was performed using TaqMan® Real Time Polymerase Chain Reaction (PCR) allelic discrimination assays, with a Drug Metabolism Enzyme or predesigned probe and primer (Applied Biosystems, CA, United States), according to the manufacturer’s instructions. TaqMan drug metabolism enzyme genotyping assays (for ABCB1 rs1128503, rs2032582, rs1045642, ABCG2 rs2231142, SLC O 1B1 rs2306283, rs4149056, CYP3A5 rs776746, CYP3A4 rs2242480, ABCC4 rs3742106, rs2274407 and NR1I2 rs1523127) used different PCR conditions from the predesigned TaqMan SNP genotyping assays (for CYP3A4 rs4646437, ABCC4 rs9561778, rs868853, ABCC3 rs4793665, ABCC5 rs562, rs3749438 and NR1I2 rs6785049). Conditions for the former were as follows: 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and 60°C for 90 s. For the latter, 40 cycles of a 1 min annealing/1 min extension were used. Hardy-Weinberg equilibrium tests were performed using chi square tests, where the allele frequencies of the study population were consistent with the law of genetic equilibrium, as indicated by a p-value >0.05.
The concentrations of atorvastatin and its metabolites (2-AT, 4-AT, ATL, 2-ATL and 4-ATL) were quantified in EDTA plasma samples from the 6 week, when the concentration of atorvastatin and its metabolites were considered to remain at a steady-state level, by using an UPLC-MS/MS assay with a lower limit of quantification of 5 ng/mL. Blood samples from patients were mixed with 300ul internal standard solution (100 ug/ml methaqualone) in 2 ml polypropylene tubes, and the mixtures were vortex-mixed for 30 s then cenrtifuged at 14,000 rpm for 10 min at 4°C. A 3 μL aliquot was injected into a 2.7 µm Poroshell 120 EC-C18 column (4.6 × 100 mm, Agilent Technologies, Santa Clara, CA United States of America), and analytes were separated using gradient elution with 0.1% formic acid in water and methanol at a flow rate of 0.3 mL/min. Analyte detection was via multiple reaction monitoring (Sciex triple quadrupole 6,500 QTRAP mass spectrometer with a Turbo V electrospray source, AB Sciex, United States) using transitions of (M + H+) m/z): AT 559.3→440.2, 2-AT 575.2→440.2 4-AT 575.2→440.2, ATL 541.3→448.3, 2-ATL 557.2→448.2, 4-ATL 557.2→448.2, and methaqualone 251.2→132.2.
Human embryonic kidney 293T cells (HEK293T) and human hepatocellular carcinoma cells (HepG2) were cultured at 37°C with 5% CO2 and 95% humidity in Dulbecco’s modified Eagle’s medium (Gibco, United States) supplemented with 10% fetal bovine serum, 100 U/mL penicillin G sodium, and 100 μg/mL streptomycin sulfate (Gibco).
The 1740 bp 3′UTR of ABCC4 (NM_005845), with incorporated terminal Xba1 restriction sites, was amplified human genomic DNA with PCR primers. The fragment was then cloned into the luciferase reporter plasmid pGL3-promoter, and creation of the rs3742106 (T/G) polymorphism was achieved by site-directed mutagenesis. Similarly, the promoter of ABCC4 gene was amplified with the primers 5′-TTTCTCTATCGATAGGTACCTAGGATTATAGGCGTGAGCC-3' (forward) and 5′-CTTAGATCGCAGATCTCGAGGCTGGGGCTCCGGCCGCCACGCC-3' (reverse). The products were digested with restriction endonucleases KpnI and XhoI, and then were cloned into pGL3-Basic vector, and the rs868853 (G/A) polymorphism was generated by using PCR-based site-directed mutagenesis. The recombinant plasmids were validated by PCR, endonuclease digestion, and DNA sequencing.
Plasmid DNA was isolated with a PureLink HiPure Plasmid Midiprep kit (Invitrogen, CA, United States), and supercoiled plasmid DNA was transfected into HEK293T and HepG2 cells using Lipofectamine 2,000 transfection reagent (Invitrogen) according to the manufacturer’s protocol. The cells were plated in 12-well plates 1 day before transfection at a density of 1 × 105 cells per well, and then were transfected with 2 μg of the reporter construct, 200 ng Renilla plasmid DNA and 2 μL Lipofectamine 2,000 after reaching 70%–80% confluence. After transfection for 24 h, cells were lysed for sequential measurement with the dual luciferase assay system (Promega, United States) in the ultra-sensitive GloMax Navigator Detection System instrument (Promega). Luciferase results are expressed as the ratio of firefly luciferase activity to Renilla luciferase activity from triplicate transfections.
Categorical data are presented as percentages, and continuous variables complied with normal distribution were expressed as mean ± standard deviation (M ± SD). The Shapiro-Wilk test was used for normality testing and non-normal distributed variables are presented as the median ± interquartile range (M ± Q). The correlation between plasma concentrations of atorvastatin and its metabolites were analyzed by Spearman correlation analysis as these variables were skewed even with a log transformation.
Considering that the lactonization and hydrolysis of atorvastatin and its metabolites are reversible processes, the lactone metabolisms and their corresponding hydroxyl acids were analyzed as a whole (AT + ATL, 2-AT+2-ATL, 4-AT+4-ATL). The sum of the active components of atorvastatin (AT+2-AT+4-AT) was also analyzed. Plasma concentrations of atorvastatin and its metabolites below the lower limit of detection were excluded. Outlier values (mean ± 3*SD) suggestive of errors in sampling procedure, technical measurements or data manipulation were excluded from the analysis.
Ancestral alleles from the Ensembl Genome database were defined as wild-type alleles, and the association between the genetic polymorphisms and the plasma concentration of atorvastatin and its metabolites were assessed in different genetic models (general, dominant, recessive models) by Mann–Whitney U or Kruskal–Wallis H tests. The addition concentration of atorvastatin lactone metabolites and their corresponding hydroxyl acids was log-transformed and then analyzed with clinical variables in univariate analyses. Any independent variables with a p-value of <0.1 in the univariate analysis were entered into a model of multivariable regression analysis to assess the influences of clinical variables on drug concentrations using the stepwise method. Finally, adjusting for gender, age, smoking and alcohol consumption, multivariable linear regression methods were used to evaluate the effects of genetic variation and baseline biochemical indices on plasma concentrations of atorvastatin and its metabolites. Statistical significance was defined having a two-sided p-value <0.05. Statistical analyses were performed using SPSS software (version 23, IBM, United States), and box and whisker plotting was performed using GraphPad Prism software (version 8.0.2.263, GraphPad, United States).
A total of 212 CKD patients were included in this study. The baseline clinical information of the patients is shown in Table 1. The average age of the patients was about 56 years old, and the male to female ratio was 1.2:1, 79.5% of patients had hypertensive disease, 42.6% had diabetes mellitus and 29.1% had cardiovascular disease. After 6 weeks of atorvastatin treatment, lipid levels of patients with CKD significantly improved.
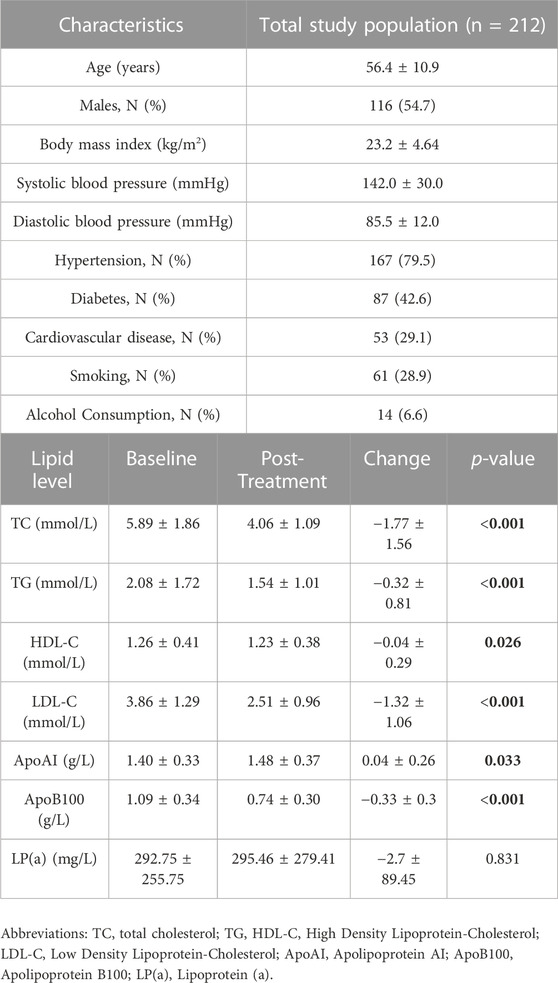
TABLE 1. Clinical and demographic characteristics of the study population for atorvastatin treatment (20 mg/day/6 weeks).
Genotyped variants and their distribution in the population studied are shown in Table 2. All genotypes tested were in Hardy-Weinberg equilibrium with the exception of CYP3A5 rs776746 (p = 0.003) and ABCB1 rs2032582 (p = 0.027), and the minor allele frequency (MAF) was close to the MAF of the Southern Chinese Han Chinese in the 1,000 Genomes database (https://www.ncbi.nlm.nih.gov/snp/) except for rs776746 and rs2032582, and are given in Supplementary Table S2.
The plasma concentrations of atorvastatin and its metabolites varied widely among individuals, with patient plasma concentrations of atorvastatin ranging from 0.097 to 16.520 ng/mL, 2-hydroxy atorvastatin from 0.062 to 19.900 ng/mL, 4-hydroxy atorvastatin from 0.179 to 9.785 ng/mL, atorvastatin lactone from 0.064 to 66.390 ng/mL, 2-hydroxy atorvastatin lactone concentrations ranged from 0.196 to 11.510 ng/mL and 4-hydroxy atorvastatin lactone concentrations ranged from 0.398 to 81.700 ng/mL. Spearman’s analysis of the correlation between atorvastatin and its metabolites showed that the concentrations of atorvastatin metabolites significantly correlated with the parent drug (r > 0.5, p < 0.0001) (Figure 2).
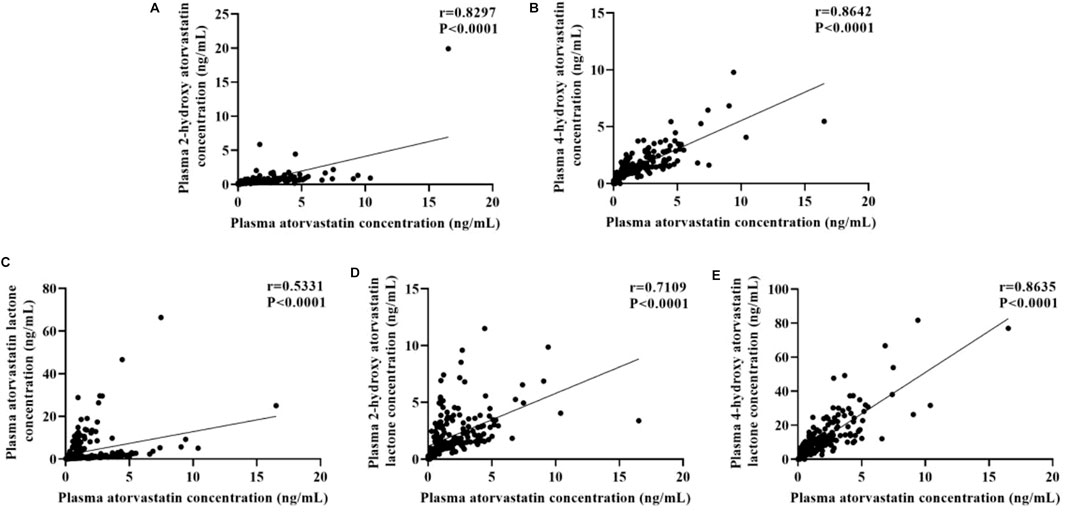
FIGURE 2. Correlations between plasma concentration of atorvastatin and its metabolites: (A) 2-hydroxy atorvastatin; (B) 4-hydroxy atorvastatin; (C) atorvastatin lactone; (D) 2-hydroxy atorvastatin lactone; (E) 4-hydroxy atorvastatin lactone.
First, the effect of baseline clinical characteristics of plasma concentrations of atorvastatin and its metabolites was evaluated by univariate and multivariate analyses. A high Apolipoprotein AI (apoAI) level was correlated with higher plasma levels of AT + ATL (β = 0.421, p = 0.003), and baseline Lipoprotein a (LPa) (β = 2.44E-04, p = 0.037) and glucose (β = −0.019, p = 0.027) had an independent influence on the concentration of 2-AT+2-ATL. In terms of the plasma level of AT+2-AT+4-AT, it depended in part on levels of apoAI (β = 0.374, p < 0.001), ALB (β = -0.013, p = 0.012), glucose (β = −0.023, p = 0.012), eGFR (β = -0.004, p = 0.031) and alcohol consumption (β = −0.303, p = 0.015), indicating exposure levels of atorvastatin and its metabolites may be affected by hepatic and renal function (Supplementary Table S3).
The effect of genetic polymorphisms on plasma concentration of atorvastatin and its metabolites was analyzed in different genetic models (Supplementary Tables S4A–C). In the general model, three SNPs of ABCC4 were significantly associated with the concentration of atorvastatin and its metabolites, showing that plasma 2-AT+2-ATL and AT+2-AT+4-AT levels were lower in individuals with the rs3742106 homozygous variant allele (CC) than the wild-type allele carriers (approximately reduced by 42% in both, p < 0.05, Table 4, Table 5), but the plasma concentration of 2-AT+2-ATL significantly increased in patients with the rs868853 heterozygous genotype (CT) when compared with the TT genotype (p = 0.015, CT vs TT) (Table 4). After correcting p-values with the Bonferroni-Dunn test, no association between rs9561778 polymorphism and plasma AT + ATL levels was observed (Table 3).
In the dominant model, patients carrying the variant rs3742106 allele had lower levels of 2-AT+2-ATL than homozygous wild-type allele carriers (p = 0.015, AA vs AC + CC) (Table 4), and the homozygous wild-type rs9561778 allele carriers showed higher plasma concentrations of AT + ATL (p = 0.029, GG vs GT + TT). Interestingly, subjects with one or two copies of the variant ABCC4 rs868853 or NR1Ⅰ2 rs6785049 allele had increased plasma concentrations of 2-AT+2-ATL (p = 0.017, TT vs CT + TT; p = 0.048, GG vs AG + AA) (Table 4). For the CYP3A4 rs464637 polymorphism, although the plasma concentration of 4-AT+4-ATL was distributed differently in carriers of different genotypes, there were only 10 cases of the AA genotype in CKD patients, which may reduce the statistical power.
In the recessive model, the wild-type rs1128503 allele carriers had lower AT + ATL levels (p = 0.047, AA vs AG + GG) (Table 3), but plasma levels of 2-AT+2-ATL and AT+2-AT+4-AT decreased in individuals with the homozygous rs3742106 variant than wild-type allele carriers (p = 0.046; p = 0.008; CC vs AC + AA) (Table 4, Table 5). There were no significant associations between these polymorphisms and the plasma concentrations of 4-AT+4-ATL (Supplementary Table S4).
Furthermore, multivariate linear regression analysis showed that the rs3742106 and rs868853 variants of ABCC4 and the NR1Ⅰ2 rs6785049 polymorphism were predictors of plasma 2-AT+2-ATL levels after adjustment for gender, age, smoking and alcohol consumption. In addition, baseline levels of lipoprotein (a) and glucose were correlated to the plasma 2-AT+2-ATL concentration, which together explained 14% of the variance in the plasma 2-AT+2-ATL concentration (Table 6; Figure 3). The ABCC4 rs3742106CC genotype was a predictor of lower AT+2-AT+4-AT levels (β = −0.212, p = 0.028), and explained 19% of the variance in the plasma levels of this component with other significantly non-genetic clinical factors (Table 6).
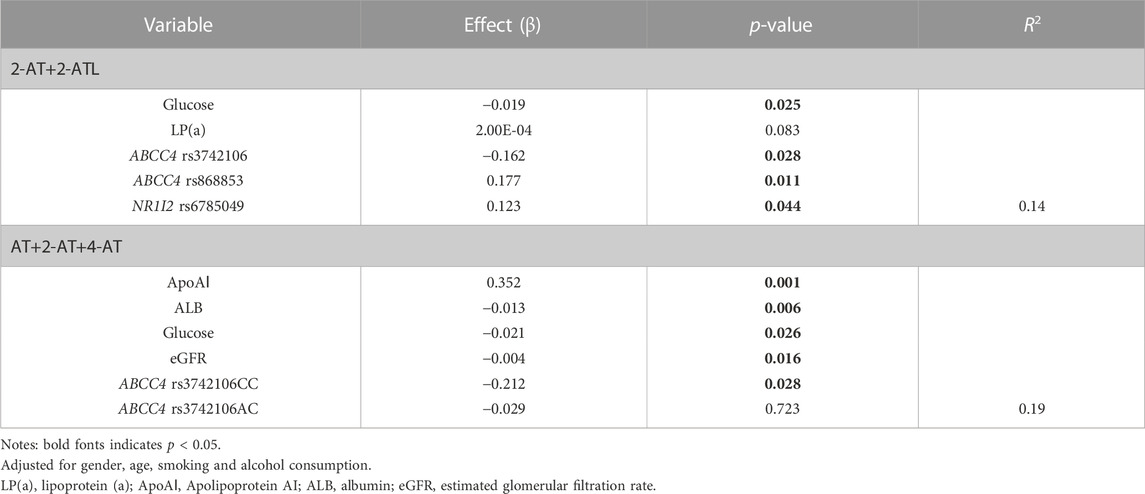
TABLE 6. Plasma atorvastatin and its metabolite concentrations-linear regression model coefficients.
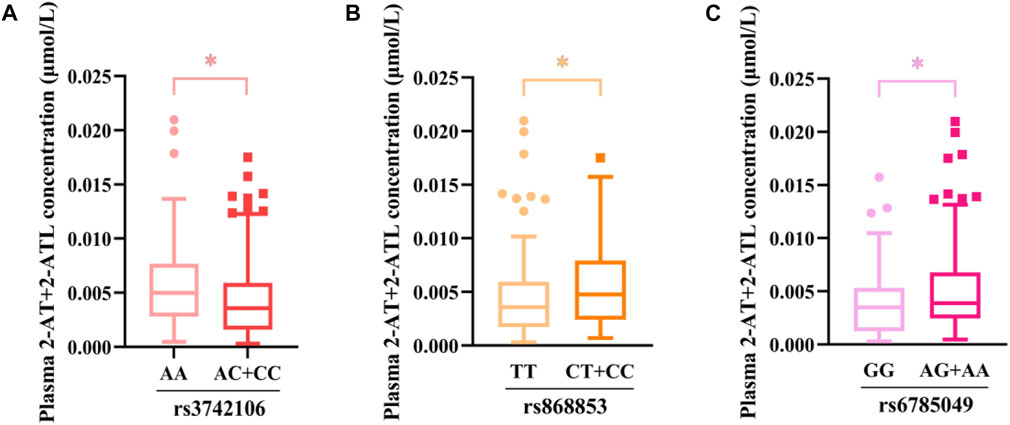
FIGURE 3. Association between genetic variants and the plasma concentration of 2-AT+2-ATL as determined using the Mann–Whitney U test. *p < 0.05. (A) rs3742106; (B)rs868853; (C) rs6785049.
The clinical trial showed significant associations between ABCC4 and NR1Ⅰ2 polymorphisms (rs868853, rs3742106 and rs6785049) with plasma levels of atorvastatin and its metabolites, so we further investigated the functional effects of these genetic variants in vitro. Since rs6785049 is located in intron of NR1Ⅰ2, we only studied ABCC4 polymorphisms, in the promoter and 3′ untranslated (3′UTR) regions, using a luciferase assay in vitro. The luciferase constructs and information of the polymorphism sites are shown in Figures 4A,D. The constructs, which were comprised of the ABCC4 sequence (promoter or 3′UTR region) and a luciferase reporter gene, were transfected into HEK293T and HepG2 cells. Functional studies showed variant alleles of rs868853-A had significantly lower luciferase activities than rs868853-G in both cell lines (HEK293T p = 0.0026, HeG2 p = 0.022), rs3742106-G had significantly lower luciferase activities than rs3742106-T in HepG2 cells lines (p = 0.0031), but not significantly in HEK293T (p = 0.31). (Figure 4).
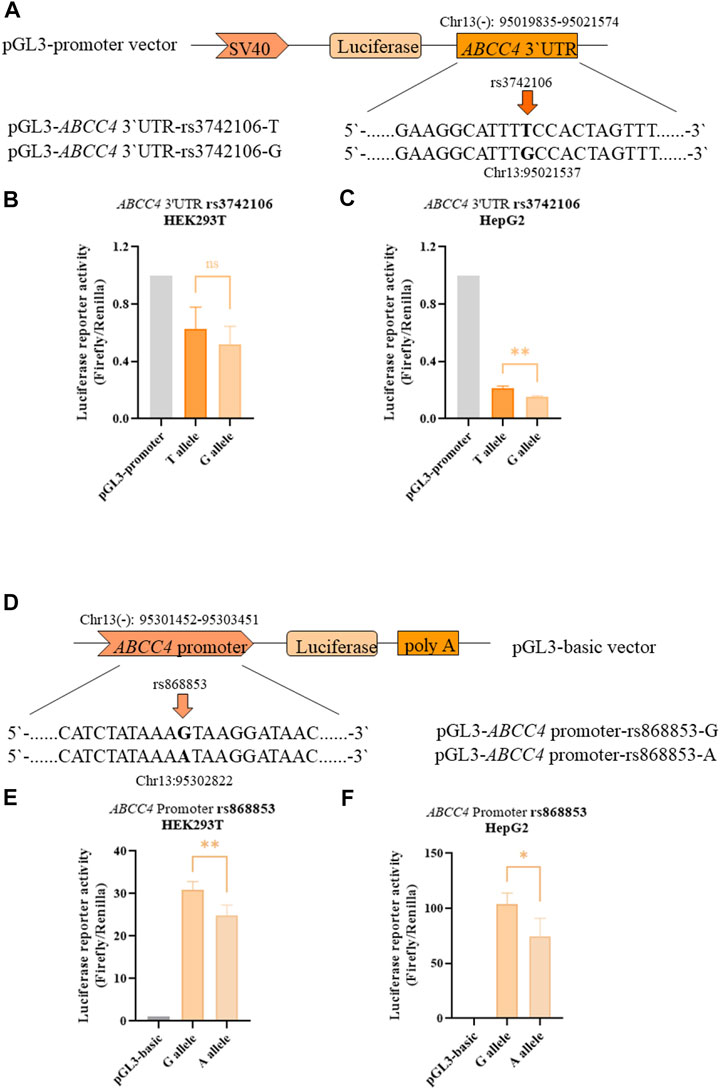
FIGURE 4. ABCC4 rs3742106 and rs868853 affect luciferase reporter activity. (A) Schematic representation of the ABCC4 3′UTR expression plasmid and the position of corresponding mutant sites. Firefly luciferase constructs contained the SV40 promoter, luciferase coding region and a fragment of the ABCC4 3′UTR. The rs3742106 SNP polymorphism site is indicated by the arrow. Plasmid constructs were transfected into HEK293T (B) and HepG2 cells (C). (D) Schematic representation of the ABCC4 promoter expression plasmid and the position of the corresponding mutant sites. Firefly luciferase constructs contained 2 kb fragment of the ABCC4 promoter, luciferase coding region and SV40 poly (A). The rs868853SNP polymorphism site is indicated by the arrow. Plasmid constructs were transfected into HEK293T (E) and HepG2 cells (F). Data represent the mean values of four independent experiments ±SD. Each experiment was conducted in triplicate. Ns indicates statistically non-significant; *p < 0.05, **p < 0.01 were considered as significant.
Individual differences in response to atorvastatin exist between patients. In this study, we investigated the association between genetic variants in drug metabolism- and transportation-related genes, and plasma concentrations of atorvastatin and its metabolites in patients with chronic kidney disease. Eighteen polymorphisms in 9 genes (CYP3A4, CYP3A5, SLC O 1B1, ABCB1, ABCG2, ABCC3, ABCC4, ABCC5 and NR1I2) were identified and displayed non-significant deviation from Hardy-Weinberg equilibrium, except for rs776746 and rs2032582, indicating that the study population is representative. However, the reason why two SNPs were not in equilibrium in our population is unclear.
DeGorter et al. demonstrated a high, 45-fold inter-individual variability in circulating atorvastatin levels following treatment with the same dose (DeGorter et al., 2013). Similarly, we found large individual differences in plasma concentrations of atorvastatin and its metabolites, especially for its lactone metabolites (over 80-fold). Considering that lactonization and hydrolysis of atorvastatin is reversible with a change in pH (Jemal and Xia, 2000), the lactone metabolites and their corresponding hydroxyl acids were analyzed as a whole, avoiding conversion that might be affected by environmental factors in the disposal of blood samples, and correlated with genetic polymorphisms. The results from univariate and multivariate analyses showed that the ABCC4 rs3742106 polymorphism was associated with plasma concentrations of AT and its metabolite, indicating patients carrying the wild-type allele (C) have higher 2-AT+2-ATL and AT+2-AT+4-AT levels than variant allele (A) carriers. We further show rs3742106, located in the region of ABCC4 encoding the 3′UTR, is an independent factor affecting plasma concentrations of AT and its metabolites.
ABCC4 is located on chromosome 13q32.1, contains 31 exons and encodes the multidrug resistance-associated protein MRP4, which is localized in the basolateral membrane of cells in the liver, but is more highly expressed in the parietal membrane of renal tubular cells in the kidney (Russel et al., 2008). ABCC4 polymorphisms have mostly been reported to correlate with tumor prognosis and the efficacy of anti-HIV drugs. Anderson et al. reported that plasma lamivudine triphosphate concentrations were 20% higher in patients carrying the rs3742106 variant allele C than in carriers with the homozygous AA genotype (Anderson et al., 2006). Similarly, Rungtivasuwan et al. also found the plasma concentrations of tenofovir were 30% higher in patients carrying variant allele than in the AA genotype carriers infected with HIV (Rungtivasuwan et al., 2015), which is opposite to our findings. However, these seemingly paradoxical results can be explained on the basis of the metabolic characteristics of the drug and the localization of MRP4. Anti-HIV drugs are mainly excreted by the kidneys, thus the level of MRP4 efflux transport in the parietal membrane of renal tubular cells can directly influence the plasma level of anti-HIV drugs. Thus, rs3742106C may reduce MRP4 protein expression, resulting in reduced efflux of drugs in renal tubular cells (Anderson et al., 2006), allowing an increase in drug plasma concentrations. In contrast, atorvastatin is metabolized in the liver and mostly eliminated by the bile, with only about 1% excreted by the kidneys (Lennernäs, 2003). Combining the correlation between rs3742106 and plasma concentrations of AT and its metabolites with the reporter gene results, it is hypothesized that rs3742106C also decreases hepatic MPR4 protein expression, leading to a decrease in drug excretion from hepatocytes into the hepatic sinusoids and into the circulation, resulting in a decrease in plasma levels of AT and its metabolites. These results further validate Anderson’s first hypothetical description of rs3742106 affecting the expression of ABCC4.
Variant allele carriers of rs868853(C), which is located in the ABCC4 promoter, exhibited higher 2-AT+2-ATL levels than homozygous wild-type carriers (TT), and rs868853 was found to be an independent influence on plasma concentrations of 2-AT+2-ATL after adjusting for non-genetic factors by multiple linear regression. Palikhe et al. showed asthma patients with the rs868853G allele had higher concentrations of asthma severity markers in blood and urine, and that the rs868853G SNP increased ABCC4 promoter transcriptional activity, suggesting that rs868853C may be associated with the transport of metabolites from immune cells (Palikhe et al., 2017). Additionally, carriers of the rs868853T allele were found to have a lower susceptibility to Kawasaki disease in a southern Chinese population (Che et al., 2018). However, there has been no reported on the association between this SNP and the pharmacokinetics or pharmacodynamics of drugs. To the best of our knowledge, this is the first report on the influence of rs868853 on plasma concentrations of AT and its metabolites. Interestingly, luciferase assays demonstrated the ABCC4 rs868853G allele enhanced transcription compared with the rs868853A allele, which is consistent with Palikhe’s results. Taken together, the rs868853(C/G) variant allele may enhance the transcriptional activity of ABCC4, resulting in enhanced expression of MRP4, leading to increased efflux of AT and its metabolites from the basal membrane of the hepatocyte into the circulation.
It was reported that rs9561778, located in an intron of ABCC4, was associated with cyclophosphamide-induced adverse effects (gastrointestinal toxicity and leukopenia or neutropenia) in patients with breast cancer, and FASTSNP suggested that rs9561778 may be located within transcription factor binding sites or intron enhancer sequences, and is a pathogenic variant affecting gene expression (Low et al., 2009). In our study, we found patients carrying the rs9561778 (T) variant allele had decreased concentrations of AT + ATL compared to homozygous wild-type (GG) carriers, but there was no statistical difference in our multiple linear regression analysis, suggesting that rs9561778 is not an independent factor affecting plasma concentrations of AT and its metabolites. Therefore, the association between the rs9561778 and plasma concentration of AT and its metabolites still needs to be further explored. Our results explain the clinical and cellular evidence that the polymorphisms of ABCC4 are associated with the circulating levels of AT and its metabolites, which changed the expression of the transporter encoded by ABCC4.
The pregnane X receptor (PXR), is encoded by NR1I2 on chromosome 3q12/13.3, and contains nine exons, but only exons two to nine are involved in encoding protein (Zhang et al., 2001). PXR is widely expressed in a variety of tissues, mainly in the liver and intestine (Lamba et al., 2005). Genetic polymorphisms in PXR may explain the variation in the expression of the target genes (Lamba et al., 2005), which may affect the pharmacokinetics or pharmacodynamics of atorvastatin. Several studies have demonstrated a relationship between rs6785049 and the plasma concentrations of various drugs (Miura et al., 2008; Mbatchi et al., 2017; Zeng et al., 2020). When we considered polymorphisms in NR1I2, we found rs6785049A was associated with increased concentration of 2-AT+2-ATL. Additionally, active metabolites (2-AT and 4-AT) of atorvastatin also are ligands of PXR, and 4-AT reduced induction of genes regulated by PXR compared to 2-AT and AT in vitro, which may be due to the weaker ability of the 4-AT to induce expression of target genes as the result of the retention of co-repressors during ligand activation (Hoffart et al., 2012). We show a genetic variant in PXR is associated with the plasma concentration of 2-AT+2-ATL, which may partly be explained in part by the metabolism of atorvastatin, i.e. since active metabolites are produced by CYP3A, acting as ligand to activate PXR regulates CYP3A4/CYP3A5, thereby producing more hydroxylated metabolites especially of 2-AT. Moreover, it has been reported that individuals with the GG genotype have twice the expression levels of CYP3A in enterocytes, than rs6785049A carriers, after the activation of PXR (Zhang et al., 2001). Taken together, these results imply that multiple pathways act together to cause differences in plasma 2-AT+2-ATL concentrations in patients carrying different genotypes.
CYP3A5 is another atorvastatin metabolizing enzyme, and its genetic polymorphisms have an important impact on the distribution of drugs (Zubiaur et al., 2021), although it has been found that CYP3A5 contributes less than 15% to atorvastatin metabolism in vitro (Park et al., 2008). CYP3A5*3 (rs776746C) is the major CYP3A5 allele that found in all ethnic groups (Zhou et al., 2017) and is functionally deficient (Kuehl et al., 2001). In our study, we found patients with the homozygous CC variant genotype have higher levels of 2-AT+2-ATL compared to the wild-type allele carriers. A previous study by Kim et al. found that after a single dose of simvastatin, the AUC0-12 was 3.3-fold higher in healthy individuals with the CC genotype than in those with the TT genotype (Kim et al., 2007). On the contrary, Zubiaur et al. indicated individuals carrying the C allele had lower plasma concentration of atorvastatin than the TT genotype carriers (Zubiaur et al., 2021). Significantly, the CYP3A5 rs776746 polymorphism was not in equilibrium in our population. Thus, further association analysis between rs776746 and plasma concentrations of AT and its metabolites, in large representative groups, is needed to clarify the reason for the different conclusions.
The P-glycoprotein protein encoded by ABCB1 plays an important role in the absorption of oral drugs in the intestine, and polymorphisms of this gene have been reported to be related to the lipid-lowering effect of atorvastatin (Thompson et al., 2005; Rebecchi et al., 2009; Hoenig et al., 2011; Kadam et al., 2016). Although rs1045642, located in exon 26 of ABCB1, does not change the expression of mRNA and protein, it may change of P-glycoprotein folding (Kimchi-Sarfaty et al., 2007). Non-etheless, there is a lack of studies into the effect of ABCB1-rs1045642 on the metabolism of patients to atorvastatin. ABCB1 rs1045642AA was associated with higher plasma concentrations of AT + ATL in this study. Therefore, this association still needs to be further verified.
Although a number of polymorphisms have been suggested as candidates for the pharmacokinetic variability of atorvastatin, the present study is the first to indicate the involvement of ABCC4-rs3742106 and rs868853, as well as NR1I2-rs6785049. While the current study demonstrates the contribution of genetic polymorphisms to the plasma concentrations of atorvastatin and its metabolites, there are a number of limitations. First, the sample size of this study is limited, and of a single ethnic population. Secondly, although drugs that may interact with atorvastatin have been excluded from the inclusion criteria, patients may have taken other medications during the 6 weeks of statin use, which may have had an impact on atorvastatin, and some drugs may have undetected effects on the statin, so factors of drug combination should be carefully analyzed in future research. In addition, it has been reported that chronic kidney disease affects the pharmacokinetics of drugs cleared by non-renal mechanisms (Yeung et al., 2014), thus the results of this study are only applicable to the CKD population.
In conclusion, the genetic variants in drug metabolism- and transportation-related genes and their effects on plasma concentrations of atorvastatin and its metabolites were identified and characterized in CKD patients. The present findings suggest the CC genotype of ABCC4-rs3742106 is associated with decreased concentrations of AT and its metabolites, whereas patients carrying the ABCC4-rs868853 or NR1I2-rs6785049 variant had higher concentrations of 2-AT+2-ATL in plasma compared with homozygous wild-type carriers with CKD. In addition, we also found that two ABCC4 SNPs may affect transcriptional activity, thereby affecting release of AT and its metabolites from hepatocytes into the circulation. These novel findings increase the panel of potential genetic biomarkers related to atorvastatin metabolism, and the results also help to improve the efficacy and toxicity prediction of atorvastatin in patients with CKD.
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the ethics committees of the First and Second Affiliated Hospitals of Shantou University Medical College. All enrolled patients were informed of the purpose and other matters of the study, understood and signed the informed consent form. The patients/participants provided their written informed consent to participate in this study.
FZ, GS, and YP designed the study, ZJ and ZW performed the experiments, wrote the manuscript and analyzed the data. RL, QD, XF, ML, YK, SL, JW, and WX helped collect the sample and recorded the information.
This study was supported by First Affiliated Hospital of Shantou University Medical College (NO: 2016067223-21).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2023.1102810/full#supplementary-material
Abd, T. T., and Jacobson, T. A. (2011). Statin-induced myopathy: A review and update. Expert Opin. Drug Saf. 10 (3), 373–387. doi:10.1517/14740338.2011.540568
Adams, S. P., Tsang, M., and Wright, J. M. (2015). Lipid-lowering efficacy of atorvastatin. Cochrane Database Syst. Rev. 2015 (3), Cd008226. doi:10.1002/14651858.CD008226.pub3
Anderson, P. L., Lamba, J., Aquilante, C. L., Schuetz, E., and Fletcher, C. V. (2006). Pharmacogenetic characteristics of indinavir, zidovudine, and lamivudine therapy in HIV-infected adults: A pilot study. J. Acquir Immune Defic. Syndr. 42 (4), 441–449. doi:10.1097/01.qai.0000225013.53568.69
Arca, M., and Gaspardone, A. (2007). Atorvastatin efficacy in the primary and secondary prevention of cardiovascular events. Drugs 67, 29–42. doi:10.2165/00003495-200767001-00004
Aviram, M., Rosenblat, M., Bisgaier, C. L., and Newton, R. S. (1998). Atorvastatin and gemfibrozil metabolites, but not the parent drugs, are potent antioxidants against lipoprotein oxidation. Atherosclerosis 138 (2), 271–280. doi:10.1016/s0021-9150(98)00032-x
Birmingham, B. K., Bujac, S. R., Elsby, R., Azumaya, C. T., Wei, C., Chen, Y., et al. (2015). Impact of ABCG2 and SLCO1B1 polymorphisms on pharmacokinetics of rosuvastatin, atorvastatin and simvastatin acid in caucasian and asian subjects: A class effect? Eur. J. Clin. Pharmacol. 71 (3), 341–355. doi:10.1007/s00228-014-1801-z
Che, D., Pi, L., Fang, Z., Xu, Y., Cai, M., Fu, L., et al. (2018). ABCC4 variants modify susceptibility to Kawasaki disease in a southern Chinese population. Dis. Markers 2018, 8638096. doi:10.1155/2018/8638096
Chen, S., Villeneuve, L., Jonker, D., Couture, F., Laverdière, I., Cecchin, E., et al. (2015). ABCC5 and ABCG1 polymorphisms predict irinotecan-induced severe toxicity in metastatic colorectal cancer patients. Pharmacogenet Genomics 25 (12), 573–583. doi:10.1097/fpc.0000000000000168
Chidambaran, V., Venkatasubramanian, R., Zhang, X., Martin, L. J., Niu, J., Mizuno, T., et al. (2017). ABCC3 genetic variants are associated with postoperative morphine-induced respiratory depression and morphine pharmacokinetics in children. Pharmacogenomics J. 17 (2), 162–169. doi:10.1038/tpj.2015.98
Cooper-DeHoff, R. M., Niemi, M., Ramsey, L. B., Luzum, J. A., Tarkiainen, E. K., Straka, R. J., et al. (2022). The clinical pharmacogenetics implementation Consortium guideline for SLCO1B1, ABCG2, and CYP2C9 genotypes and statin-associated musculoskeletal symptoms. Clin. Pharmacol. Ther. 111, 1007–1021. doi:10.1002/cpt.2557
Cruz-Correa, O. F., León-Cachón, R. B., Barrera-Saldaña, H. A., and Soberón, X. (2017). Prediction of atorvastatin plasmatic concentrations in healthy volunteers using integrated pharmacogenetics sequencing. Pharmacogenomics 18 (2), 121–131. doi:10.2217/pgs-2016-0072
DeGorter, M. K., Tirona, R. G., Schwarz, U. I., Choi, Y. H., Dresser, G. K., Suskin, N., et al. (2013). Clinical and pharmacogenetic predictors of circulating atorvastatin and rosuvastatin concentrations in routine clinical care. Circ. Cardiovasc Genet. 6 (4), 400–408. doi:10.1161/circgenetics.113.000099
Deng, F., Tuomi, S. K., Neuvonen, M., Hirvensalo, P., Kulju, S., Wenzel, C., et al. (2021). Comparative hepatic and intestinal efflux transport of statins. Drug Metab. Dispos. 49 (9), 750–759. doi:10.1124/dmd.121.000430
Fanta, S., Jönsson, S., Karlsson, M. O., Niemi, M., Holmberg, C., Hoppu, K., et al. (2010). Long-term changes in cyclosporine pharmacokinetics after renal transplantation in children: Evidence for saturable presystemic metabolism and effect of NR1I2 polymorphism. J. Clin. Pharmacol. 50 (5), 581–597. doi:10.1177/0091270009348223
Gao, Y., Zhang, L., and Fu, Q. (2008). CYP3A4*1G polymorphism is associated with lipid-lowering efficacy of atorvastatin but not of simvastatin. Eur. J. Clin. Pharmacol. 64 (9), 877–882. doi:10.1007/s00228-008-0502-x
Hirota, T., Fujita, Y., and Ieiri, I. (2020). An updated review of pharmacokinetic drug interactions and pharmacogenetics of statins. Expert Opin. Drug Metab. Toxicol. 16 (9), 809–822. doi:10.1080/17425255.2020.1801634
Hoenig, M. R., Walker, P. J., Gurnsey, C., Beadle, K., and Johnson, L. (2011). The C3435T polymorphism in ABCB1 influences atorvastatin efficacy and muscle symptoms in a high-risk vascular cohort. J. Clin. Lipidol. 5 (2), 91–96. doi:10.1016/j.jacl.2011.01.001
Hoffart, E., Ghebreghiorghis, L., Nussler, A. K., Thasler, W. E., Weiss, T. S., Schwab, M., et al. (2012). Effects of atorvastatin metabolites on induction of drug-metabolizing enzymes and membrane transporters through human pregnane X receptor. Br. J. Pharmacol. 165, (5), 1595–1608. doi:10.1111/j.1476-5381.2011.01665.x
Imai, Y., Nakane, M., Kage, K., Tsukahara, S., Ishikawa, E., Tsuruo, T., et al. (2002). C421A polymorphism in the human breast cancer resistance protein gene is associated with low expression of Q141K protein and low-level drug resistance. Mol. Cancer Ther. 1 (8), 611–616.
Jemal, M., and Xia, Y. Q. (2000). Bioanalytical method validation design for the simultaneous quantitation of analytes that may undergo interconversion during analysis. J. Pharm. Biomed. Anal. 22 (5), 813–827. doi:10.1016/s0731-7085(00)00245-4
Jungers, P., Massy, Z. A., Nguyen Khoa, T., Fumeron, C., Labrunie, M., Lacour, B., et al. (1997). Incidence and risk factors of atherosclerotic cardiovascular accidents in predialysis chronic renal failure patients: A prospective study. Nephrol. Dial. Transpl. 12 (12), 2597–2602. doi:10.1093/ndt/12.12.2597
Kadam, P., Ashavaid, T. F., Ponde, C. K., and Rajani, R. M. (2016). Genetic determinants of lipid-lowering response to atorvastatin therapy in an Indian population. J. Clin. Pharm. Ther. 41 (3), 329–333. doi:10.1111/jcpt.12369
Kim, K. A., Park, P. W., Lee, O. J., Kang, D. K., and Park, J. Y. (2007). Effect of polymorphic CYP3A5 genotype on the single-dose simvastatin pharmacokinetics in healthy subjects. J. Clin. Pharmacol. 47 (1), 87–93. doi:10.1177/0091270006295063
Kimchi-Sarfaty, C., Oh, J. M., Kim, I. W., Sauna, Z. E., Calcagno, A. M., Ambudkar, S. V., et al. (2007). A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science 315 (5811), 525–528. doi:10.1126/science.1135308
Knauer, M. J., Urquhart, B. L., Meyer zu Schwabedissen, H. E., Schwarz, U. I., Lemke, C. J., Leake, B. F., et al. (2010). Human skeletal muscle drug transporters determine local exposure and toxicity of statins. Circ. Res. 106 (2), 297–306. doi:10.1161/circresaha.109.203596
Kondo, C., Suzuki, H., Itoda, M., Ozawa, S., Sawada, J., Kobayashi, D., et al. (2004). Functional analysis of SNPs variants of BCRP/ABCG2. Pharm. Res. 21 (10), 1895–1903. doi:10.1023/b:pham.0000045245.21637.d4
König, J., Cui, Y., Nies, A. T., and Keppler, D. (2000). A novel human organic anion transporting polypeptide localized to the basolateral hepatocyte membrane. Am. J. Physiology-Gastrointestinal Liver Physiology 278 (1), G156–G164. doi:10.1152/ajpgi.2000.278.1.G156
Kopin, L., and Lowenstein, C. (2017). Dyslipidemia. Ann. Intern Med. 167 (11), Itc81–itc96. doi:10.7326/aitc201712050
Kuehl, P., Zhang, J., Lin, Y., Lamba, J., Assem, M., Schuetz, J., et al. (2001). Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat. Genet. 27 (4), 383–391. doi:10.1038/86882
Lamba, J., Lamba, V., and Schuetz, E. (2005). Genetic variants of PXR (NR1I2) and CAR (NR1I3) and their implications in drug metabolism and pharmacogenetics. Curr. Drug Metab. 6 (4), 369–383. doi:10.2174/1389200054633880
Lea, A. P., and McTavish, D. (1997). Atorvastatin. A review of its pharmacology and therapeutic potential in the management of hyperlipidaemias. Drugs 53 (5), 828–847. doi:10.2165/00003495-199753050-00011
Lee, N., Maeda, K., Fukizawa, S., Ieiri, I., Tomaru, A., Akao, H., et al. (2019). Microdosing clinical study to clarify pharmacokinetic and pharmacogenetic characteristics of atorvastatin in Japanese hypercholesterolemic patients. Drug Metab. Pharmacokinet. 34 (6), 387–395. doi:10.1016/j.dmpk.2019.08.004
Lennernäs, H. (2003). Clinical pharmacokinetics of atorvastatin. Clin. Pharmacokinet. 42 (13), 1141–1160. doi:10.2165/00003088-200342130-00005
Levey, A. S., Stevens, L. A., Schmid, C. H., Zhang, Y. L., Castro, A. F., Feldman, H. I., et al. (2009). A new equation to estimate glomerular filtration rate. Ann. Intern Med. 150 (9), 604–612. doi:10.7326/0003-4819-150-9-200905050-00006
Link, E., Parish, S., Armitage, J., Bowman, L., Heath, S., Matsuda, F., et al. (2008). SLCO1B1 variants and statin-induced myopathy-a genomewide study. N. Engl. J. Med. 359 (8), 789–799. doi:10.1056/NEJMoa0801936
Liu, J., Chen, Z., Chen, H., Hou, Y., Lu, W., He, J., et al. (2017). Genetic polymorphisms contribute to the individual variations of imatinib mesylate plasma levels and adverse reactions in Chinese GIST patients. Int. J. Mol. Sci. 18 (3), 603. doi:10.3390/ijms18030603
Liyanage, T., Toyama, T., Hockham, C., Ninomiya, T., Perkovic, V., Woodward, M., et al. (2022). Prevalence of chronic kidney disease in Asia: A systematic review and analysis. BMJ Glob. Health 7 (1), e007525. doi:10.1136/bmjgh-2021-007525
Low, S. K., Kiyotani, K., Mushiroda, T., Daigo, Y., Nakamura, Y., and Zembutsu, H. (2009). Association study of genetic polymorphism in ABCC4 with cyclophosphamide-induced adverse drug reactions in breast cancer patients. J. Hum. Genet. 54 (10), 564–571. doi:10.1038/jhg.2009.79
Maeda, K., Ikeda, Y., Fujita, T., Yoshida, K., Azuma, Y., Haruyama, Y., et al. (2011). Identification of the rate-determining process in the hepatic clearance of atorvastatin in a clinical cassette microdosing study. Clin. Pharmacol. Ther. 90 (4), 575–581. doi:10.1038/clpt.2011.142
Mangravite, L. M., Thorn, C. F., and Krauss, R. M. (2006). Clinical implications of pharmacogenomics of statin treatment. Pharmacogenomics J. 6 (6), 360–374. doi:10.1038/sj.tpj.6500384
Marino, M., di Masi, A., Trezza, V., Pallottini, V., Polticelli, F., and Ascenzi, P. (2011). Xenosensors CAR and PXR at work: Impact on statin metabolism. Curr. Drug Metab. 12 (3), 300–311. doi:10.2174/138920011795101859
Mason, R. P. (2006). Molecular basis of differences among statins and a comparison with antioxidant vitamins. Am. J. Cardiol. 98, 34p–41p. doi:10.1016/j.amjcard.2006.09.018
Mbatchi, L. C., Gassiot, M., Pourquier, P., Goberna, A., Mahammedi, H., Mourey, L., et al. (2017). Association of NR1I2, CYP3A5 and ABCB1 genetic polymorphisms with variability of temsirolimus pharmacokinetics and toxicity in patients with metastatic bladder cancer. Cancer Chemother. Pharmacol. 80 (3), 653–659. doi:10.1007/s00280-017-3379-5
Miura, M., Satoh, S., Inoue, K., Kagaya, H., Saito, M., Inoue, T., et al. (2008). Influence of CYP3A5, ABCB1 and NR1I2 polymorphisms on prednisolone pharmacokinetics in renal transplant recipients. Steroids 73 (11), 1052–1059. doi:10.1016/j.steroids.2008.04.002
Palikhe, S., Uuganbayar, U., Trinh, H. K. T., Ban, G. Y., Yang, E. M., Park, H. S., et al. (2017). A role of the ABCC4 gene polymorphism in airway inflammation of asthmatics. Mediat. Inflamm. 2017, 3549375. doi:10.1155/2017/3549375
Park, J. E., Kim, K. B., Bae, S. K., Moon, B. S., Liu, K. H., and Shin, J. G. (2008). Contribution of cytochrome P450 3A4 and 3A5 to the metabolism of atorvastatin. Xenobiotica 38 (9), 1240–1251. doi:10.1080/00498250802334391
Peng, C., Ding, Y., Yi, X., Shen, Y., Dong, Z., Cao, L., et al. (2018). Polymorphisms in CYP450 genes and the therapeutic effect of atorvastatin on ischemic stroke: A retrospective cohort study in Chinese population. Clin. Ther. 40 (3), 469–477.e2. doi:10.1016/j.clinthera.2018.02.002
Prado, Y., Zambrano, T., and Salazar, L. A. (2018). Transporter genes ABCG2 rs2231142 and ABCB1 rs1128503 polymorphisms and atorvastatin response in Chilean subjects. J. Clin. Pharm. Ther. 43 (1), 87–91. doi:10.1111/jcpt.12607
Prake-Davis (2004). Prake-davis, Product information: Lipitor (atorvastatin calcium). Ann Arbor, MI: Prake-Davis(Divisioin of Pfizer Inc.
Prueksaritanont, T., Subramanian, R., Fang, X., Ma, B., Qiu, Y., Lin, J. H., et al. (2002). Glucuronidation of statins in animals and humans: A novel mechanism of statin lactonization. Drug Metab. Dispos. 30 (5), 505–512. doi:10.1124/dmd.30.5.505
Rebecchi, I. M., Rodrigues, A. C., Arazi, S. S., Genvigir, F. D., Willrich, M. A., Hirata, M. H., et al. (2009). ABCB1 and ABCC1 expression in peripheral mononuclear cells is influenced by gene polymorphisms and atorvastatin treatment. Biochem. Pharmacol. 77 (1), 66–75. doi:10.1016/j.bcp.2008.09.019
Riedmaier, S., Klein, K., Winter, S., Hofmann, U., Schwab, M., and Zanger, U. M. (2011). Paraoxonase (PON1 and PON3) polymorphisms: Impact on liver expression and atorvastatin-lactone hydrolysis. Front. Pharmacol. 2, 41. doi:10.3389/fphar.2011.00041
Rogers, R. S., Parker, A., Vainer, P. D., Elliott, E., Sudbeck, D., Parimi, K., et al. (2021). The interface between cell signaling pathways and pregnane X receptor. Cells 10 (11), 3262. doi:10.3390/cells10113262
Rungtivasuwan, K., Avihingsanon, A., Thammajaruk, N., Mitruk, S., Burger, D. M., Ruxrungtham, K., et al. (2015). Influence of ABCC2 and ABCC4 polymorphisms on tenofovir plasma concentrations in Thai HIV-infected patients. Antimicrob. Agents Chemother. 59 (6), 3240–3245. doi:10.1128/aac.04930-14
Russel, F. G., Koenderink, J. B., and Masereeuw, R. (2008). Multidrug resistance protein 4 (MRP4/ABCC4): A versatile efflux transporter for drugs and signalling molecules. Trends Pharmacol. Sci. 29 (4), 200–207. doi:10.1016/j.tips.2008.01.006
Sánchez-Martín, A., Cabrera Figueroa, S., Cruz, R., Porras-Hurtado, L., Calvo-Boyero, F., Rasool, M., et al. (2016). Gene-gene interactions between DRD3, MRP4 and CYP2B6 polymorphisms and its influence on the pharmacokinetic parameters of efavirenz in HIV infected patients. Drug Metab. Pharmacokinet. 31 (5), 349–355. doi:10.1016/j.dmpk.2016.06.001
Sarnak, M. J., Levey, A. S., Schoolwerth, A. C., Coresh, J., Culleton, B., Hamm, L. L., et al. (2003). Kidney disease as a risk factor for development of cardiovascular disease: A statement from the American heart association councils on kidney in cardiovascular disease, high blood pressure research, clinical cardiology, and Epidemiology and prevention. Circulation 108 (17), 2154–2169. doi:10.1161/01.Cir.0000095676.90936.80
Schirmer, M., Rosenberger, A., Klein, K., Kulle, B., Toliat, M. R., Nürnberg, P., et al. (2007). Sex-dependent genetic markers of CYP3A4 expression and activity in human liver microsomes. Pharmacogenomics 8 (5), 443–453. doi:10.2217/14622416.8.5.443
Swart, M., Whitehorn, H., Ren, Y., Smith, P., Ramesar, R. S., and Dandara, C. (2012). PXR and CAR single nucleotide polymorphisms influence plasma efavirenz levels in South African HIV/AIDS patients. BMC Med. Genet. 13, 112. doi:10.1186/1471-2350-13-112
Tanaka, Y., Manabe, A., Fukushima, H., Suzuki, R., Nakadate, H., Kondoh, K., et al. (2015). Multidrug resistance protein 4 (MRP4) polymorphisms impact the 6-mercaptopurine dose tolerance during maintenance therapy in Japanese childhood acute lymphoblastic leukemia. Pharmacogenomics J. 15 (4), 380–384. doi:10.1038/tpj.2014.74
Teft, W. A., Welch, S., Lenehan, J., Parfitt, J., Choi, Y. H., Winquist, E., et al. (2015). OATP1B1 and tumour OATP1B3 modulate exposure, toxicity, and survival after irinotecan-based chemotherapy. Br. J. Cancer 112 (5), 857–865. doi:10.1038/bjc.2015.5
Thompson, J. F., Man, M., Johnson, K. J., Wood, L. S., Lira, M. E., Lloyd, D. B., et al. (2005). An association study of 43 SNPs in 16 candidate genes with atorvastatin response. Pharmacogenomics J. 5 (6), 352–358. doi:10.1038/sj.tpj.6500328
Tonelli, M., and Wanner, C.Kidney Disease: Improving Global Outcomes Lipid Guideline Development Work Group Members (2014). Lipid management in chronic kidney disease: Synopsis of the kidney disease: Improving global Outcomes 2013 clinical practice guideline. Ann. Intern Med. 160 (3), 182. doi:10.7326/m13-2453
Turner, R. M., Fontana, V., FitzGerald, R., Morris, A. P., and Pirmohamed, M. (2020a). Investigating the clinical factors and comedications associated with circulating levels of atorvastatin and its major metabolites in secondary prevention. Br. J. Clin. Pharmacol. 86 (1), 62–74. doi:10.1111/bcp.14133
Turner, R. M., Fontana, V., Zhang, J. E., Carr, D., Yin, P., FitzGerald, R., et al. (2020b). A genome-wide association study of circulating levels of atorvastatin and its major metabolites. Clin. Pharmacol. Ther. 108 (2), 287–297. doi:10.1002/cpt.1820
Venkatasubramanian, R., Fukuda, T., Niu, J., Mizuno, T., Chidambaran, V., Vinks, A. A., et al. (2014). ABCC3 and OCT1 genotypes influence pharmacokinetics of morphine in children. Pharmacogenomics 15 (10), 1297–1309. doi:10.2217/pgs.14.99
Yeung, C. K., Shen, D. D., Thummel, K. E., and Himmelfarb, J. (2014). Effects of chronic kidney disease and uremia on hepatic drug metabolism and transport. Kidney Int. 85 (3), 522–528. doi:10.1038/ki.2013.399
Zeng, G., Wang, L., Shi, L., Li, H., Zhu, M., Luo, J., et al. (2020). Variability of voriconazole concentrations in patients with hematopoietic stem cell transplantation and hematological malignancies: Influence of loading dose, procalcitonin, and pregnane X receptor polymorphisms. Eur. J. Clin. Pharmacol. 76 (4), 515–523. doi:10.1007/s00228-020-02831-1
Zhang, J., Kuehl, P., Green, E. D., Touchman, J. W., Watkins, P. B., Daly, A., et al. (2001). The human pregnane X receptor: Genomic structure and identification and functional characterization of natural allelic variants. Pharmacogenetics 11 (7), 555–572. doi:10.1097/00008571-200110000-00003
Zhou, Y., Ingelman-Sundberg, M., and Lauschke, V. M. (2017). Worldwide distribution of cytochrome P450 alleles: A meta-analysis of population-scale sequencing projects. Clin. Pharmacol. Ther. 102 (4), 688–700. doi:10.1002/cpt.690
Keywords: atorvastatin, genetic polymorphism, drug-metabolizing enzymes, transporters, plasma concentration, ABCC4
Citation: Jiang Z, Wu Z, Liu R, Du Q, Fu X, Li M, Kuang Y, Lin S, Wu J, Xie W, Shi G, Peng Y and Zheng F (2023) Effect of polymorphisms in drug metabolism and transportation on plasma concentration of atorvastatin and its metabolites in patients with chronic kidney disease. Front. Pharmacol. 14:1102810. doi: 10.3389/fphar.2023.1102810
Received: 19 November 2022; Accepted: 15 February 2023;
Published: 27 February 2023.
Edited by:
Junmin Zhang, Lanzhou University, ChinaReviewed by:
Guoping Yang, Central South University, ChinaCopyright © 2023 Jiang, Wu, Liu, Du, Fu, Li, Kuang, Lin, Wu, Xie, Shi, Peng and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuchun Zheng, emhlbmdmdWNodW5zaEAxNjMuY29t; Yanqiang Peng, cGVuZ3lxZ3pzdW1zQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.