
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 16 January 2023
Sec. Inflammation Pharmacology
Volume 14 - 2023 | https://doi.org/10.3389/fphar.2023.1096816
This article is part of the Research TopicAntibody-Drugs and Natural Medicines for Autoimmune DiseasesView all 5 articles
Aims: Genetic variants increase the susceptibility to anti-drug antibodies (ADA) in response to anti-TNF therapy in chronic inflammatory diseases. However, little is known about genetic variants in Chinese populations. This study aimed to identify genetic variants contributing to the risk of the development of antibodies to infliximab (ATI) in Chinese patients with Crohn’s disease (CD).
Methods: CD patients (n = 104) treated with infliximab (IFX) during the maintenance therapy were enrolled in this cross-sectional study. ATI was assessed by an in-house developed drug-tolerant ELISA method. ATI titers of 1:20 and ≥1:60 were considered a low titer and a high titer, respectively. Thirteen types of single nucleotide polymorphisms (SNPs) within 13 genes involved in the immune process, the susceptibility to chronic inflammatory diseases, cytokines and apoptosis pathways were investigated.
Results: The median trough levels of infliximab (TLI) in patients with clinical remission (CR) were higher than those in patients without CR (3.80 vs. 1.50 μg/mL, p < .001). The median TLI in patients with high-titer ATI was significantly lower than that in ATI-negative patients (1.15 vs. 4.48 μg/mL, p < .001) or those with low-titer ATI (1.15 vs. 2.95 μg/mL, p = .03). The HLA-DQA1*05 rs2097432 GG and GA genotypes were more frequent in patients with ATI (GG and AG vs. AA, 27/38 = 71.05% vs. 29/66 = 43.94%, OR 2.94, 95% CI 1.19–7.30, p = .02). Patients carrying the CC and AC genotypes of rs396991 in FCGR3A were associated with a higher frequency of ATI formation (CC and AC vs. AA, 37/57 = 64.91% vs. 19/47 = 40.43%, OR 2.94, 95% CI 1.24–6.96, p = .01). According to the number of variants in rs2097432 and rs393991, patients with two variants had a higher proportion of producing ATI (two variants vs. no variant, 17/21 = 80.95% vs. 9/30 = 30.00%, OR 9.92, 95% CI 2.59–37.87, p = .001; single variant vs. no variant, 30/53 = 56.60% vs. 9/30 = 30.00%, OR 3.04, 95% CI 1.18–7.88, p = .02). No association was found between other SNPs and ATI production.
Conclusion: Rs2097432 in HLA-DQA1*05 and rs396991 in FCGR3A are associated with ATI production in Chinese patients with CD. A pharmacogenomic strategy could help with the clinical management of CD.
Crohn’s disease (CD) exhibits periods of remission and aggravated inflammations in the gastrointestinal tract. Infliximab (IFX), a chimeric monoclonal antibody against tumor necrosis factor alpha (TNF-α), has profoundly improved therapeutic outcomes (Torres et al., 2020). Many studies have demonstrated the correlations between trough levels of infliximab (TLI) and therapeutic targets (Ward et al., 2017; Yarur et al., 2017; Papamichael et al., 2018). Up to 30% of individuals with CD experience loss of response in the maintenance phase due to an inadequate TLI (Zhang et al., 2019; Greuter et al., 2020). A major contributor to subtherapeutic levels of IFX is immunogenicity, which refers to the development of anti-drug antibodies (ADA) (Hemperly and Vande Casteele, 2018).
A number of factors were identified for ADA development, including genetic predisposition, antibiotic use differentially and formation of drug-target complexes (Moss et al., 2013; Bar-Yoseph et al., 2019; Gorelik et al., 2022). Increasing evidence suggests that the variation in the HLA-DQA1 gene involved in aberrant adaptive immune responses is relevant to the development of antibodies to infliximab (ATI) (Sazonovs et al., 2020; Wilson et al., 2020). Due to the differences in genetic backgrounds of the populations, the association between genetic variants and ATI production in Chinese populations needs to be confirmed.
Variations in genes coding for TNF, TNF receptor superfamily 1A (TNFRSF1A), TNF receptor superfamily 1B (TNFRSF1B) and a receptor for the Fc portion of IgG (FcγRIIIa) were recently associated with the response to anti-TNF-α drugs in CD (Louis et al., 2006; Linares-Pineda et al., 2018; Matsuoka et al., 2018; Romero-Cara et al., 2018; Curci et al., 2021). The association between genetic polymorphisms in the ATG16L1 gene contributing to CD risk and response to anti-TNF-α treatment suggests that genetic variants involved in the susceptibility to chronic inflammatory diseases plays an important role in the drug response (Koder et al., 2015). Genetic variations in these aforementioned genes may affect response to IFX through ADA formation. None of all these single nucleotide polymorphisms (SNPs) within the relevant genes involved in the susceptibility to chronic inflammatory diseases, cytokines and apoptosis pathways have yet been investigated at the same time with the aim of identifying patients who develop ATI. Based on the frequency of SNPs in Chinese populations, 13 SNPs in 13 genes were selected to investigate the association between genetic variants and development of ATI.
The present study assessed the association between genetic variants and ATI production during the IFX maintenance therapy in Chinese patients with CD, offering evidence of the risk of ATI production as well as new opportunities for stratifying patients based on their genetic makeup prior to initiation of IFX therapy.
Patients with CD were enrolled between October 2020 and April 2022 at the Department of Gastroenterology, the First Affiliated Hospital of Soochow University (Suzhou, China). Participants (>18 years old) had a histopathological diagnosis of CD and received scheduled therapy. The treatment regimen included the administration of 5 mg/kg IFX at weeks 0, 2, and 6 and then every 8 weeks during maintenance therapy. A stable dose of 5-aminosalicylic acid (5-ASA) was allowed for at least 4 weeks. The study excluded patients who had been administered IFX for more than 2 years and had comedications including immunomodulators (azathioprine, 6-mercaptopurine) and steroids. Electronic medical records were used to collect information about weight, age, sex, smoking history, disease phenotype and faecal calprotectin (fCal) values. Within the 24 h before IFX infusion, a total of 2 mL of peripheral whole blood was collected for genetic analysis as well as TLI and ATI measurements. Meanwhile, the Crohn’s disease activity index (CDAI) was recorded. Clinical remission (CR) was defined as a CDAI less than 150 points. The normalization of fCal was defined as less than 250 μg/g. Patients provided written and informed consent. The research was approved by the Research Ethics Committee of the First Affiliated Hospital of Soochow University.
Separation of plasma from whole blood was performed to detect TLI. TLI were analyzed with an in-house developed and validated ELISA method. High binding 96-well plates were coated overnight with TNF-α (300-01A, PeproTech, United Kingdom) at 4°C. Plasma samples were diluted in PBS and incubated for 2 h at 37°C in a shaker. Human anti-infliximab, clone AbD19376_hIgG1 (HCA216P, Bio-Rad, United Kingdom), was used as the detecting agent. The lower and upper limits of quantification were 0.50 μg/mL and 40.00 μg/mL, respectively.
ATI was detected by a bridging ELISA method. The analytical method was validated according to current industry practice (Myler et al., 2021). Plasma samples and controls were diluted in glycine (100 mM, pH 2.5) at a minimum dilution of 1:20 for 30 min at RT in a shaker. Then, 50 μL of the diluted samples or controls and 95 μL master mixture containing 0.1 μg/mL each of biotin-IFX, HRP-IFX and 5 μL of Tris (1.5 M, pH 9.0) were added to a 96-well polypropylene plate and then incubated for 2 h at RT in a shaker. Immediately after, 100 μL of a mixture was moved to a streptavidin matrix coated 96-well plate and incubated at RT for 1 h in a shaker. The plate was washed three times, followed by the addition of 3,3’,5,5’-tetramethylbenzidine substrate and 2 M H2SO4. A negative control (NC) of pooled plasma from IFX-naïve CD patients as well as low and high positive controls containing 100 and 20, 000 ng/mL rabbit anti-IFX polyclonal antibodies prepared in NC, was included in each plate. The screening cut point based on a panel of plasma samples from 51 IFX-naïve CD patients, was determined to be 1.16 (S/N). Based on the analysis of 51 IFX-naïve plasma samples from CD patients without or with IFX (20 μg/mL), the confirmatory cut point was determined to be a decrease in signal of more than 17.21%. The titer cut point was determined to be 1.34. The minimum significant ratio was determined to be 3. ATI titers of 1:20 and ≥1:60 were considered a low titer and a high titer, respectively. Drug-tolerance at low positive control was ≤12.50 μg/mL IFX. The relative sensitivity of this assay was determined to be 50 ng/mL (for validation parameters, see Supplementary Table S1).
DNA was extracted from 250 μL of whole blood using a DNA Extraction Kit (Takara, Japan). The concentration and purity of DNA were calculated with the OD-1000 (OneDropTM, China). Thirteen primer pools of SNPs, namely, HLA-DQA1 (rs2097432), FCGR3A (rs396991), FCGR2A (rs1801274), PTPRC (rs10919563), KLRC1 (rs7301582), HLA-E (rs1264457), IL-17RA (rs4819554), ATG16L1 (rs10210302), TRAF1 (rs3761847), TNF (rs1800629), TNFRSF1A (rs767455), TNFRSF1B (rs1061622) and CCNY (rs12777960) (Supplementary Table S2), were designed. Library preparation was performed using a two-step polymerase chain reaction (PCR). These reaction mixtures contained 1 μL of DNA, 0.4 mM dNTPs, 3 μM of each primer, Taq Buffer (with MgCL2) and 0.04 U Taq DNA polymerase. Twenty-five μL of reaction mixture was used for PCR. The first PCR conditions were 95°C for 5 min, followed by 10 cycles of 94°C for 30 s, 63°C for 30 s, and 72°C for 30 s. The second PCR conditions were 30 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 10 min. All PCR reagents were provided by ThermoFisher (Waltham, MA) and Sangon Biotech (Shanghai, China). Paired-end sequencing of the library was performed on HiSeq XTen sequencers (Illumina, San Diego, CA).
Statistical analysis was performed using GraphPad Prism® version 9.0 and IBM SPSS Statistics for Mac, version 26.0. The TLI below the lower limits of quantification were imputed by 0.25 μg/mL. The mean and standard deviation or the median and interquartile range (IQR) were used to describe continuous clinical and demographic variables. Categorical variables were expressed as percentages. The Mann‒Whitney U test (two groups) and the Kruskal–Wallis H test (more than two groups) were used to compare continuous non-normality variables. The distribution of 13 SNPs within 13 genes was tested for Hardy-Weinberg equilibrium using the chi-squared goodness-of-fit test. For all SNPs, two genetic models were used: the additive model, which supposes that the effect of the risk allele increases with the addition of each allele copy; and the dominant model, which analyzes the heterosis. A chi-squared test and logistic regression were performed to evaluate the association and multiplicative interactions between SNPs and ATI formation, respectively. All reported p values were 2-sided, and a significant p < .05 was considered a statistically significant difference.
A total of 104 patients with CD were included. The patients’ demographic and clinical characteristics are shown in Table 1. A total of 73.08% of patients were male. The median duration of IFX treatment was 10 months (IQR: 5–16.5). The median CDAI point was 87.6 (IQR: 68.2–123.5). The median fCal value was 64.5 (IQR: 30–344.5). The median TLI in CD patients was 2.89 μg/mL (IQR: 1.58–5.07). The TLI in 10 patients was less than 0.50 μg/mL (10/104, 9.62%).
Eighty-three of the 104 patients (79.81%) achieved clinical remission (CR). The median TLI in patients with CR was higher than that in patients without CR (3.80 vs. 1.50 μg/mL, p < .001, Figure 1A). The median TLI in patients who achieved CR and fCal normalization was higher than that in patients who did not (4.40 vs. 1.90 μg/mL, p = .02, Figure 1B).
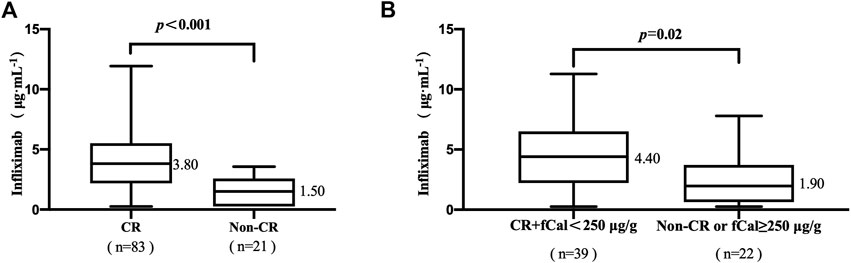
FIGURE 1. (A) Association between trough levels of infliximab (TLI) and clinical remission (CR). (B) The association of TLI with CR and faecal calprotectin (fCal) normalization. Clinical remission (CR) was defined by a Crohn’s disease activity index (CDAI) < 150 points. Normalization of faecal calprotectin (fCal) was defined if below 250 μg/g stools. Data for fCal were not available for all patients included in this study.
The TLI ranged from undetectable to 11.93 μg/mL. The occurrence of ATI was 53.85% (56/104). The titers of ATI showed a wide range from low to high titers. The relationship between TLI and titers of ATI was evaluated. The median TLI in patients with high-titer ATI (1.15 μg/mL, IQR: 0.25–2.39, Figure 2) was significantly lower than that in patients with low-titer ATI (2.95 μg/mL, IQR: 2.25–4.28, p = .03, Figure 2). A significant difference in TLI was also observed between the high-titer group and the negative-ATI group (4.48 μg/mL, IQR: 2.65–6.64, p < .001, Figure 2). No significant differences in TLI were observed between the negative-ATI group and the low-titer group (p = .17, Figure 2).
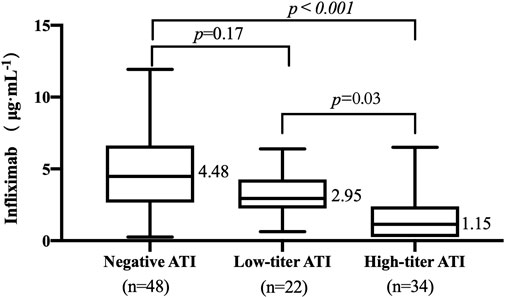
FIGURE 2. Relationship between trough levels of infliximab (TLI) and antibodies to infliximab (ATI) titer. ATI titers of 1:20 and ≥1:60 were considered a low titer and a high titer, respectively.
The frequencies of genotypes for 13 SNPs within 13 genes in 104 patients were in line with Hardy-Weinberg equilibrium (HWE, p > .05, Supplementary Table S3 in the Supplement). The association between 13 SNPs and ATI formation under the dominant model and additive model are shown in Supplementary Table S4. Variant carriers of the SNP (rs2097432 in HLA-DQA1*05) under the dominant model had an higher rate of ATI formation (GG and AG: 27/38 = 71.05%, AA: 29/66 = 43.94%, p = .01, Table 2). There was also a significant association between rs396991 in FCGR3A and ATI production under the dominant model. Carriers of the variant SNP (rs396991 in FCGR3A) had a higher probability of producing ATI (CC and AC:37/57 = 64.91%, AA: 19/47 = 40.43%, p = .01, Table 2). No association was found between other SNPs and ATI production (Table 2; Supplementary Table S5). In logistic regression, the HLA-DQA1*05 rs2097432 GG and GA genotypes increased the risk of ATI formation compared to the AA genotype (GG and AG vs. AA, OR 2.94, 95% CI 1.19–7.30, p = .02, Table 3). Patients carrying the CC and AC genotypes of rs396991 in FCGR3A were associated with a higher frequency of ATI formation (CC and AC vs. AA, OR 2.94, 95% CI 1.24–6.96, p = .01, Table 3). According to the number of the variants in rs2097432 and rs393991, a greater proportion of patients with two variants showed ATI production (two variants vs. no variant, 17/21 = 80.95% vs. 9/30 = 30.00%, OR 9.92, 95% CI 2.59–37.87, p = .001; single variant vs. no variant, 30/53 = 56.60% vs. 9/30 = 30.00%, OR 3.04, 95% CI 1.18–7.88, p = .02, Figure 3 and Table 3).
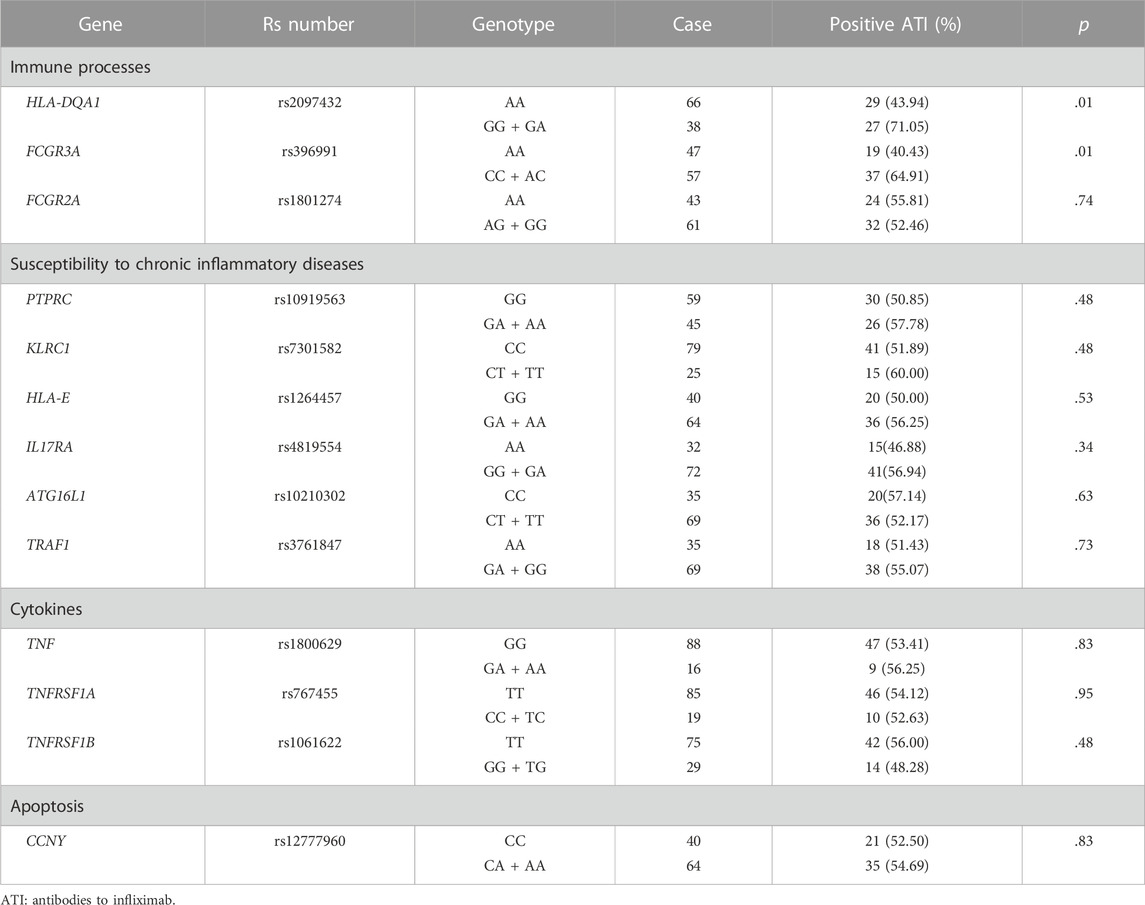
TABLE 2. Patients who developed antibodies to infliximab according to single nucleotide polymorphisms (SNPs) under IKthe dominant model.
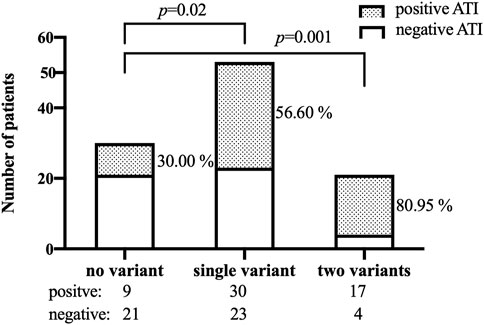
FIGURE 3. Patients who developed ATI according to the number of the variants in both SNPs (rs2097432 and rs393991). ATI: antibodies to infliximab.
The production of ATI constitutes an important cause of subtherapeutic levels of IFX. Therefore, patients at risk of ATI formation should be identified. In this cross-sectional study, the association between ATI production and the SNPs in the genes involved in several pathways was assessed in Chinese patients with CD, demonstrating that patients with two of the variants in rs2097432 and rs393991 had an almost 10-fold higher risk of producing ATI than those with no variant.
Using a drug-tolerant ATI assay, the present study showed that HLA-DQA1*05 and FCGR3A genetic variation are associated with ATI formation in Chinese patients with CD, in line with the previous studies in Europe and Canada (Romero-Cara et al., 2018; Sazonovs et al., 2020; Wilson et al., 2020; Curci et al., 2021). The HLA-DQA1*05 genotype is also associated with the development of antibodies to adalimumab, IFX’s sister TNF antagonist (Powell Doherty et al., 2020). HLA may affect antigen binding and immune cell activation downstream (Choo, 2007). The mechanisms of HLA-DQA1*05 variants in ATI formation should be further investigated. Another gene involved in the immune process, the Fc-gamma receptors (FcγR) type IIIA (FCGR3A), is able to remove antigen-antibody complexes from the circulation. FCGR3A variant carriers have a higher binding affinity of FcγRs to IgG as well as greater antibody-dependent cellular cytotoxicity activity (Moroi et al., 2013).
Our study did not find the association between ATI production and polymorphisms in the genes involved in the susceptibility to chronic inflammatory diseases, cytokines and apoptosis pathways. First, the limited sample size may not provide sufficient statistical power to determine the effect of 11 SNPs on the ATI formation. Perhaps in a larger sample study, we can find obvious associations between those SNPs and ATI production. Second, some other mechanisms of these 11 SNPs in the IFX response should be investigated. Third, the distribution of the TNFRSF1A and IL17RA genotypes in Chinese populations differed from that in European populations (Supplementary Table S3).
The rates of ATI production across different assays have varied widely from 0% to 79% (Gorovits et al., 2018). Most ATI detection assays are drug sensitive (Barrau et al., 2022). Using a drug-tolerant ATI assay, the positive rate of ATI increased from 21% (drug-sensitive ATI assay) to 63% (Van Stappen et al., 2018). Therefore, among these studies, drug-sensitive ATI assays may lead to an underestimated the rate of ATI formation (Romero-Cara et al., 2018; Wilson et al., 2020; Curci et al., 2021). The dissociation of drug/anti-drug antibodies complexes with acid and subsequent detection of ADA have been successfully applied to improve drug tolerance (Patton et al., 2005; Sickert et al., 2008; Mikulskis et al., 2011). Using acid dissociation in most assays, though others using anti-lambda chain antibody, or more rarely heat-dissociation allows the assay to be drug-tolerant. In the present study the dissociation of IFX/ATI complexes using glycine and subsequent detection of ATI by bridging assays improved drug tolerance. The drug tolerance in our assay was determined to be 12.5 μg/mL when the low positive control was at 100 ng/mL, which is higher than the highest TLI (11.93 μg/mL). This method has the ability to detect multivalent monospecific antibodies such as IgG or IgM antibodies but not bispecific antibodies such as IgG4.
There are limitations that need to be considered. The results were based on a cross-sectional study, and a prospective cohort study with a larger sample size is therefore needed. Additionally, as well as the presence or absence of ATI, the duration of the response (transient, persistent) should also be considered. Transient vs. sustained ATI were not differentiated in this work. Transient ATI may become negative with no clinically meaningful impact. Although these limitations exist, the discovery of rs2097432 in HLA-DQA1*05 and rs396991 in FCGR3A provides further evidence that genotyping may optimize IFX treatment in Chinese patients with CD.
In summary, rs2097432 in HLA-DQA1*05 and rs396991 in FCGR3A are significantly associated with the development of ATI. Patients with two variants (rs2097432 and rs393991) had an almost 10-fold risk of producing ATI than those with no variants. To achieve personalized therapy in CD patients treated with IFX, genetic testing in genes involved in ATI formation prior to initiation of IFX therapy, such as rs2097432 in HLA-DQA1*05 and rs396991 in FCGR3A, could be suggested.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://www.ensembl.org/index.html, rs396991 and rs2097432, https://www.ncbi.nlm.nih.gov/snp/, rs396991 and rs2097432.
The studies involving human participants were reviewed and approved by the Research Ethics Committee of the First Affiliated Hospital of Soochow University. The patients/participants provided their written informed consent to participate in this study.
KZ, XD, and LM designed and performed the study; ZC helped with genetic typing; WC participated in the study design; KZ drafted the manuscript; XD and LM reviewed the manuscript; and QX and XP enrolled patients and interpreted the medical results.
This study was financed by the Suzhou Health Leading Talent (No. GSWS2019001), Key R&D Program of Jiangsu Province (BE2021644), the National Clinical Research Center for Hematologic Diseases, the First Affiliated Hospital of Soochow University (2020WSC07) and the Priority Academic Program Development of the Jiangsu Higher Education Institutes (PAPD).
It is with great gratitude that the authors acknowledge the patients for consenting to participate in this study. We would like to thank the nurses and medical staff of the Department of Gastroenterology at the First Affiliated Hospital of Soochow University for their assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2023.1096816/full#supplementary-material
Bar-Yoseph, H., Pressman, S., Blatt, A., Vainberg, S. G., Maimon, N., Starosvetsky, E., et al. (2019). Infliximab–tumor necrosis factor complexes elicit formation of anti-drug antibodies. Gastroenterology 157, 1338–1351. e8. doi:10.1053/j.gastro.2019.08.009
Barrau, M., Duprat, M., Veyrard, P., Tournier, Q., Williet, N., Phelip, J. M., et al. (2022). A systematic review on the interest of drug tolerant assay in the monitoring of inflammatory bowel disease. J. Crohns Colitis, jjac164. doi:10.1093/ecco-jcc/jjac164
Choo, S. Y. (2007). The HLA system: Genetics, immunology, clinical testing, and clinical implications. Yonsei Med. J. 48, 11–23. doi:10.3349/ymj.2007.48.1.11
Curci, D., Lucafò, M., Cifù, A., Fabris, M., Bramuzzo, M., Martelossi, S., et al. (2021). Pharmacogenetic variants of infliximab response in young patients with inflammatory bowel disease. Clin. Transl. Sci. 14, 2184–2192. doi:10.1111/cts.13075
Gorelik, Y., Freilich, S., Gerassy-Vainberg, S., Pressman, S., Friss, C., Blatt, A., et al. (2022). Antibiotic use differentially affects the risk of anti-drug antibody formation during anti-tnfα therapy in inflammatory bowel disease patients: A report from the epi-IIRN. Gut 71, 287–295. doi:10.1136/gutjnl-2021-325185
Gorovits, B., Baltrukonis, D. J., Bhattacharya, I., Birchler, M. A., Finco, D., Sikkema, D., et al. (2018). Immunoassay methods used in clinical studies for the detection of anti-drug antibodies to adalimumab and infliximab. Clin. Exp. Immunol. 192, 348–365. doi:10.1111/cei.13112
Greuter, T., Maillard, M. H., Juillerat, P., Michetti, P., Seibold, F., Mottet, C., et al. (2020). Therapeutic drug monitoring to guide clinical decision making in inflammatory bowel disease patients with loss of response to anti-TNF: A delphi technique-based consensus. Digestion 101, 683–691. doi:10.1159/000501930
Hemperly, A., and Vande Casteele, N. (2018). Clinical pharmacokinetics and pharmacodynamics of infliximab in the treatment of inflammatory bowel disease. Clin. Pharmacokinet. 57, 929–942. doi:10.1007/s40262-017-0627-0
Koder, S., Repnik, K., Ferkolj, I., Pernat, C., Skok, P., Weersma, R. K., et al. (2015). Genetic polymorphism in ATG16L1 gene influences the response to adalimumab in Crohn’s disease patients. Pharmacogenomics 16, 191–204. doi:10.2217/pgs.14.172
Linares-Pineda, T. M., Cañadas-Garre, M., Sánchez-Pozo, A., and Calleja-Hernández, M. Á. (2018). Pharmacogenetic biomarkers of response in Crohn's disease. Pharmacogenomics 18 (1), 1–13. doi:10.1038/tpj.2017.27
Louis, E. J., Watier, H. E., Schreiber, S., Hampe, J., Taillard, F., Olson, A., et al. (2006). Polymorphism in IgG Fc receptor gene FCGR3A and response to infliximab in Crohn’s disease: A subanalysis of the ACCENT I study. Pharmacogenet. Genomics 16, 911–914. doi:10.1097/01.fpc.0000230421.12844.fd
Matsuoka, K., Hamada, S., Shimizu, M., Nanki, K., Mizuno, S., Kiyohara, H., et al. (2018). Factors predicting the therapeutic response to infliximab during maintenance therapy in Japanese patients with Crohn's disease. PLoS One 13 (10), e0204632. doi:10.1371/journal.pone.0204632
Mikulskis, A., Yeung, D., Subramanyam, M., and Amaravadi, L. (2011). Solution ELISA as a platform of choice for development of robust, drug tolerant immunogenicity assays in support of drug development. J. Immunol. Methods 365, 38–49. doi:10.1016/j.jim.2010.11.011
Moroi, R., Endo, K., Kinouchi, Y., Shiga, H., Kakuta, Y., Kuroha, M., et al. (2013). FCGR3A-158 polymorphism influences the biological response to infliximab in Crohn’s disease through affecting the ADCC activity. Immunogenetics 65, 265–271. doi:10.1007/s00251-013-0679-8
Moss, A. C., Brinks, V., and Carpenter, J. F. (2013). Review article: Immunogenicity of anti-TNF biologics in IBD - the role of patient, product and prescriber factors. Aliment. Pharmacol. Ther. 38, 1188–1197. doi:10.1111/apt.12507
Myler, H., Pedras-Vasconcelos, J., Phillips, K., Hottenstein, C. S., Chamberlain, P., Devanaryan, V., et al. (2021). Anti-drug antibody validation testing and reporting harmonization. Aaps J. 24, 4. doi:10.1208/s12248-021-00649-y
Papamichael, K., Rakowsky, S., Rivera, C., Cheifetz, A. S., and Osterman, M. T. (2018). Association between serum infliximab trough concentrations during maintenance therapy and biochemical, endoscopic, and histologic remission in Crohn’s disease. Inflamm. Bowel Dis. 24, 2266–2271. doi:10.1093/ibd/izy132
Patton, A., Mullenix, M. C., Swanson, S. J., and Koren, E. (2005). An acid dissociation bridging ELISA for detection of antibodies directed against therapeutic proteins in the presence of antigen. J. Immunol. Methods 304, 189–195. doi:10.1016/j.jim.2005.06.014
Powell Doherty, R. D., Liao, H., Satsangi, J. J., and Ternette, N. (2020). Extended analysis identifies drug-specific association of 2 distinct HLA class II haplotypes for development of immunogenicity to adalimumab and infliximab. Gastroenterology 159, 784–787. doi:10.1053/j.gastro.2020.03.073
Romero-Cara, P., Torres-Moreno, D., Pedregosa, J., Vílchez, J. A., García-Simón, M. S., Ruiz-Merino, G., et al. (2018). A FCGR3A polymorphism predicts anti-drug antibodies in chronic inflammatory bowel disease patients treated with anti-TNF. Int. J. Med. Sci. 15, 10–15. doi:10.7150/ijms.22812
Sazonovs, A., Kennedy, N. A., Moutsianas, L., Heap, G. A., Rice, D. L., Reppell, M., et al. (2020). HLA-DQA1*05 carriage associated with development of anti-drug antibodies to infliximab and adalimumab in patients with Crohn’s disease. Gastroenterology 158, 189–199. doi:10.1053/j.gastro.2019.09.041
Sickert, D., Kroeger, K., Zickler, C., Chokote, E., Winkler, B., Grenet, J.-M., et al. (2008). Improvement of drug tolerance in immunogenicity testing by acid treatment on Biacore. J. Immunol. Methods 334, 29–36. doi:10.1016/j.jim.2008.01.010
Torres, J., Bonovas, S., Doherty, G., Kucharzik, T., Gisbert, J. P., Raine, T., et al. (2020). ECCO guidelines on therapeutics in Crohn’s disease: Medical treatment. J. Crohns Colitis 14, 4–22. doi:10.1093/ecco-jcc/jjz180
Van Stappen, T., Vande Casteele, N., Van Assche, G., Ferrante, M., Vermeire, S., and Gils, A. (2018). Clinical relevance of detecting anti-infliximab antibodies with a drug-tolerant assay: Post hoc analysis of the TAXIT trial. Gut 67, 818–826. doi:10.1136/gutjnl-2016-313071
Ward, M. G., Warner, B., Unsworth, N., Chuah, S.-W., Brownclarke, C., Shieh, S., et al. (2017). Infliximab and adalimumab drug levels in Crohn’s disease: Contrasting associations with disease activity and influencing factors. Aliment. Pharmacol. Ther. 46, 150–161. doi:10.1111/apt.14124
Wilson, A., Peel, C., Wang, Q., Pananos, A. D., and Kim, R. B. (2020). HLADQA1*05 genotype predicts anti-drug antibody formation and loss of response during infliximab therapy for inflammatory bowel disease. Aliment. Pharmacol. Ther. 51, 356–363. doi:10.1111/apt.15563
Yarur, A. J., Kanagala, V., Stein, D. J., Czul, F., Quintero, M. A., Agrawal, D., et al. (2017). Higher infliximab trough levels are associated with perianal fistula healing in patients with Crohn’s disease. Aliment. Pharmacol. Ther. 45, 933–940. doi:10.1111/apt.13970
Keywords: infliximab, anti-drug antibodies, genetic variants, drug-tolerant enzyme immunoassay, Crohn’s disease
Citation: Zhu K, Ding X, Chen Z, Xi Q, Pang X, Chen W and Miao L (2023) Association between genetic variants and development of antibodies to infliximab: A cross-sectional study in Chinese patients with Crohn’s disease. Front. Pharmacol. 14:1096816. doi: 10.3389/fphar.2023.1096816
Received: 12 November 2022; Accepted: 03 January 2023;
Published: 16 January 2023.
Edited by:
Tao Zhang, Xian Jiaotong University, ChinaReviewed by:
Luis Andrés López-Fernández, Instituto de Investigación Sanitaria Gregorio Marañón, SpainCopyright © 2023 Zhu, Ding, Chen, Xi, Pang, Chen and Miao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weichang Chen, d2VpY2hhbmdjaGVuQDEyNi5jb20=; Liyan Miao, bWlhb2x5c3V6aG91QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.