- 1Department of Biology, Faculty of Basic Sciences, East of Tehran Branch, Islamic Azad University, Tehran, Iran
- 2Department of Microbiology, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 3Department of Veterinary medicine, Science and Research Branch, Islamic Azad University, Tehran, Iran
- 4Department Of Veterinary, Azad University Of Shahr-E Kord, Shahrekord, Iran
- 5Department of Microbiology, Saveh Branch, Islamic Azad University, Saveh, Iran
Objective: Cholera is a challenging ancient disease caused by Vibrio cholera (V. cholera). Antibiotics that prevent cell wall synthesis are among the first known antibiotic groups. Due to its high consumption, V. cholera has developed resistance to the majority of antibiotics in this class. Resistance to recommended antibiotics for the treatment of V. cholera has also increased. In light of the decrease in consumption of certain antibiotics in this group that inhibit cell wall synthesis and the implementation of new antibiotics, it is necessary to determine the antibiotic resistance pattern of V. cholera and to employ the most effective treatment antibiotic.
Method: An comprehensive systematic search for relevant articles was conducted in PubMed, Web of Science, Scopus, and EMBASE through October 2020. Stata version 17.1 utilized the Metaprop package to execute a Freeman-Tukey double arcsine transformation in order to estimate weighted pooled proportions.
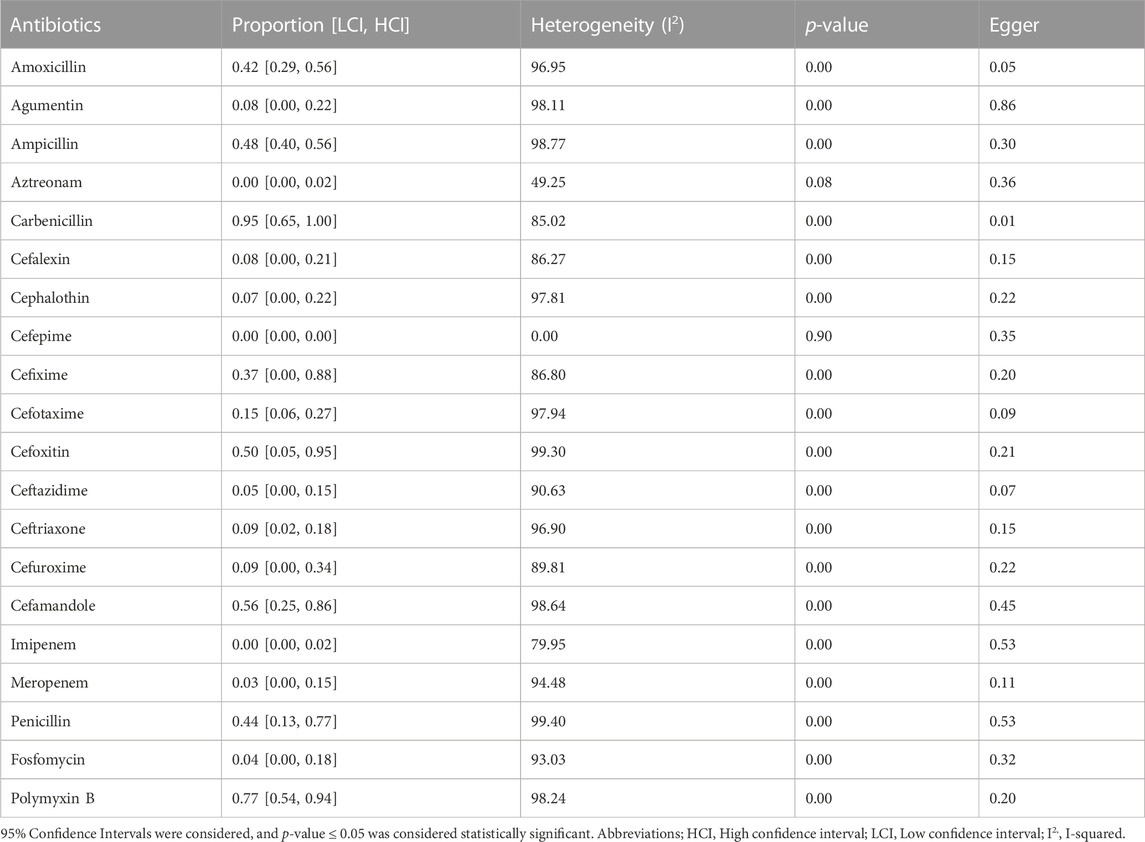
Results: A total of 131 articles were included in the meta-analysis. Ampicillin was the most investigated antibiotic. The prevalence of antibiotic resistance was in order aztreonam (0%), cefepime (0%), imipenem (0%), meropenem (3%), fosfomycin (4%), ceftazidime (5%), cephalothin (7%), augmentin (8%), cefalexin (8%), ceftriaxone (9%), cefuroxime (9%), cefotaxime (15%), cefixime (37%), amoxicillin (42%), penicillin (44%), ampicillin (48%), cefoxitin (50%), cefamandole (56%), polymyxin-B (77%), carbenicillin (95%) respectively.
Discussion: Aztreonam, cefepime, and imipenem are the most efficient V. cholera cell wall synthesis inhibitors. There has been an increase in resistance to antibiotics such as cephalothin, ceftriaxone, amoxicillin, and meropenem. Over the years, resistance to penicillin, ceftazidime, and cefotaxime, has decreased.
Introduction
Koch isolated and described the Gram-negative bacillus Vibrio cholera (V. cholera) in the late nineteenth century (Dengo-Baloi et al., 2017). As a foodborne and waterborne pathogen, V. cholera can cause an acute intestinal infection as severe watery diarrhea in humans (Mukhopadhyay et al., 1998; Yuan et al., 2022). Virulence factors include toxin-related fimbria and cholera toxins (Ngandjio et al., 2009; Awuor et al., 2020a). In areas with poor sanitation and no clean water, V. cholera can be endemic, epidemic, or pandemic (Mukhopadhyay et al., 1998). This bacterium causes 2.9 million cholera cases and 95,000 deaths annually (Onohuean et al., 2022). According to the O antigen, V. cholera species are divided into 206 serotypes. Although the O1 and O139 serotypes are linked to epidemic cholera and non-agglutinating V. cholera (NOVC), which are negative to the O1 and O139 antigens, they cause infrequent severe illnesses (Liu et al., 2022; Onohuean et al., 2022). Seventh cholera pandemics linked to O1 and O139 V. cholera serotypes. The disease is a major issue in Asia, Africa, and Latin America (Abera et al., 2010; Alam et al., 2012). V. cholera infections can be mild, moderate, or severe in endemic areas (rapidly deadly diarrhea) (Araj et al., 1994; Anand et al., 1996).
Early treatment with an oral rehydration salts (ORS) solution, including glucose, potassium chloride, sodium chloride, and trisodium citrate, are critical for cholera patients with moderate watery diarrhea (Araj et al., 1994). Severe cholera dehydration requires intravenous rehydration and appropriate antibiotics to shorten the disease’s period (Araj et al., 1994; Baddam et al., 2020).
Although antibiotic susceptibility testing (AST) of V. cholera was not suggested in the past due to the low resistance of V. cholera to common antibiotics (Bag et al., 1998; Baddam et al., 2020), the development of resistance to tetracycline, a common antibiotic used in the treatment of V. cholera infection, is becoming increasingly widespread throughout the world (Bakhshi and Pourshafie, 2009). These resistant strains have been responsible for severe epidemics in Latin America, Tanzania, Bangladesh, and Zaire (Bag et al., 1998). However, various isolates of V. cholera resistant to antibiotics have been reported worldwide. Antibiotics that target the cell wall were once the first-line treatment for infections, but their use has decreased as bacterial resistance to these antibiotics has increased over time. Globally, the prevalence of V. cholera resistance to cell wall-active antibiotics has not been thoroughly studied. Replacing less effective antibiotics is necessary for more effective treatment, which has already increased the prevalence of resistance.
The tetracycline antibiotic class has long been the most effective for treating cholera. Non-etheless, earlier research revealed a global increase in V. cholera strains resistant to tetracycline (Dengo-Baloi et al., 2017). Additionally, studies have demonstrated that fluoroquinolone resistance in V. cholera strains began to increase in July 1996 (Mukhopadhyay et al., 1998; Yuan et al., 2022). A previously published study suggested erythromycin as a tetracycline alternative in small children and pregnant women. Furazolidone and nalidixic acid have traditionally been used as cholera treatments.
Nevertheless, due to the high level of resistance found in V. cholera isolates, these antibiotics are currently less effective (Ngandjio et al., 2009; Awuor et al., 2020b). Due to the lack of meta-analyses concerning antibiotics resistant to the V. cholera cell wall, we decided to explore newer antibiotics in terms of the prevalence of resistance as well as the global resistance pattern and the resistance trend over time in all V. cholera serotypes. This study’s outcomes can potentially improve the global antimicrobial resistance situation significantly.
Methods
Search strategy
The comprehensive systematic search of relevant articles through four electronic databases, including PubMed, Web of Science, Scopus, and EMBASE, with two researchers independently until October 2020. The search was performed using “V. cholera” and “Antibiotic resistance” related keywords. Obtained articles have been merged in EndNote X20 (Thomson Reuters, NY, United States), and duplicates were removed. The search syntax is available in Supplementary Material.
Selection criteria and data extraction
The screening and selection of articles procedures were performed in Rayyan online software. Two authors (A. Sh and O. Sh) independently reviewed all records’ titles, abstracts, and full texts. They removed irrelevant articles, and the third author (M. Sh) solved disagreements. The exclusion criteria were as follows: review articles, case reports, congress abstracts, studies with ambiguous results, not the English language, sample size of fewer than three isolates, duplicate publications, and studies of antimicrobial resistance of other than V. cholera species.
The extracted information from each included study was: first author, year of publication, country, sample source (clinical or environmental isolates), serogroups, the total number of isolates (sample size), AST method, and the number of resistant isolates for each antibiotic.
Quality assessment
The quality assessment of included studies was performed by two reviewers (R. Sa, and O. Sh) independently using an adapted version of the Newcastle-Ottawa assessment scale adapted for cross-sectional studies. Each study received a score ranging from 0 to 8 (5 points or higher: high quality, three or 4: medium quality, 2 points or lower: low quality).
Statistical analysis
Due to the high number of zero prevalence in antibiotic resistance reports, the Freeman Tukey double arcsine conversion was conducted on the data using the metaprop command to estimate the weighted pooled proportion of resistance (WPR) in STATA software (version. 17.1), which range is from 0.00 to 1.00. A random-effects model was used for pooling effect size. As a measure of heterogeneity, the tau-squared and I2 were considered. The Egger regression test was used to determine the effect of small studies or publication bias. Subgroup analyses were conducted using the following variables to find the sources of variation: country, continent, country development status (World Economic Situation and Prospects, classification), publication year group (1970–2000, 2001–2010, 2011–2020), source of V. cholera isolation, AST method (Disc and Gradient methods), interpretation guideline (CLSI, Non-CLSI), and serogroups (O1/O139, Non-O1/O139).
Results
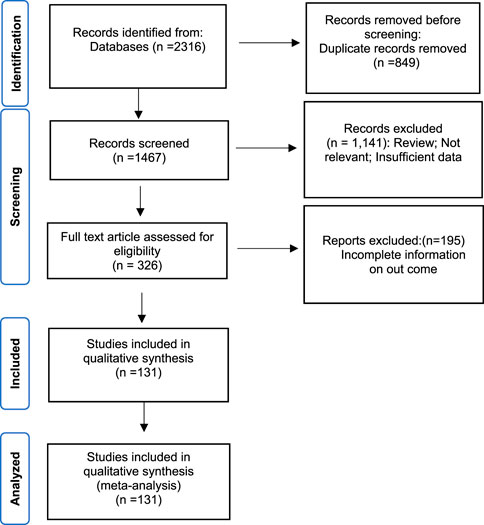
The systematic search identified two thousand three hundred sixteen records, and 849 duplicates were removed. According to titles and abstracts screening, 1,141 articles were excluded, and according to the full-text evaluation, 195 articles were considered irrelevant. Finally, 131 articles published between 1980 and 2020 were included in this meta-analysis [Abera et al. (2010), Alam et al. (2012), Amita et al. (2003), Anand et al. (1996), Araj et al. (1994), Baddam et al. (2020), Bag et al. (1998), Bakhshi et al. (2014), Bakhshi and Pourshafie (2009), Bakhshi et al. (2008), Balaji et al. (2013), Ballal and Shivananda (2002), Bani et al. (2007); Barati et al. (2015), Basu et al. (2000), Bhat et al. (2012), Bhattacharya et al. (2015), Bhattacharya et al. (2012), Bhotra et al. (2017), Bhowmick et al. (2007), Bier et al. (2015), Borkakoty et al. (2012), Campos et al. (2004), Ceccarelli et al. (2016); Ceccarelli et al. (2011), Chakraborty et al. (2001), Chandrasekhar et al. (2008), Chatterjee et al. (2007), Chhotray et al. (2002), Chomvarin et al. (2013), Chomvarin et al. (2012), Chowdhury et al. (2016), Colombo et al. (1993), Dalsgaard et al. (1998), Dalsgaard et al. (2000), Dalsgaard et al. (1996), Das et al. (2016), Das et al. (2011), Das et al. (2008), Dengo-Baloi et al., 2017, Dey et al. (2014), Dua et al. (2018), Echeverria et al. (1983), Eibach et al. (2016); Faruque et al. (2000); Fazil et al. (2011); Feglo and Sewurah (2018), Fernandez-Abreu et al. (2017), Folgosa et al. (2000), Garg et al. (2000), Goel et al. (2010), Gupta et al. (2016), Ibarra and Alvarado (2007), Iramiot et al. (2019); Islam et al. (2011); Ismail et al. (2013); Jain et al. (2016); Jaiswal et al. (2015), Kacou-N’douba et al. (2012), Kaistha et al. (2005), Kar et al. (2015), Karki et al. (2010), Kingston et al. (2009), Koley et al. (2014), Kondo et al. (2001), Krishna et al. (2006), Kumar et al. (2014), Kutar et al. (2013), Luo et al. (2013), Mandal et al. (2010), Mandal et al. (2009), Mandomando et al. (2007), Marashi et al. (2012), Mohapatra et al. (2007), Sarkar et al. (2019), Moyo et al. (2011), Mukhopadhyay et al. (1995), Okuda et al. (2007), Oyofo et al. (2002b), Oyofo et al. (2002a), Pal et al. (2006), Pal et al. (2017), Pan et al. (2008), Parveen et al. (2003), Patrick et al. (2012), Rahbar et al. (2010), Ranjbar et al. (2016), Rashed et al. (2012), Rezaie et al. (2017), Roy et al. (2012), Saidi et al. (2014), Sambe-Ba et al. (2017), Sedaghat et al. (2012), Shakya et al. (2012), Sharma et al. (2007), Talkington et al. (2011), Torane et al. (2016), Tran et al. (2012), Vijayalakshmi et al. (1997), Vinothkumar et al. (2018), Akoachere et al. (2013), Akoachere and Mbuntcha (2014), Alaoui et al. (2010), Al-Hilu et al. (2019), Amau et al. (1988), Baron et al. (2017), Baron et al. (2016), Bhanumathi et al. (2003), Bidinost et al. (2004), Bier et al. (2015), Campos et al. (2004), Ceccarelli et al. (2016), Chakraborty et al. (2001), Chomvarin et al. (2013), Dalsgaard et al. (1996), Eja et al. (2006), Falcao et al. (1998), Ibarra and Alvarado (2007), Imziln and Hassani (1994), Isaac-Márquez et al. (1998), Jagadeeshan et al. (2009), Kiiru et al. (2013), Kumar and Lalitha (2013), Li et al. (2011), Mandal et al. (2010), Mercy et al. (2014), Mishra et al. (2011), Mohapatra et al. (2007), Odjadjare and Igbinosa (2017), Onyuka et al. (2011), Osawa et al. (2020), Ottaviani et al. (2018), Prabhu et al. (2007), Radu et al. (2002), Kutar et al. (2013), Singh et al. (2018), Song et al. (2013), Wang et al. (2016), Waturangi et al. (2013), Zachariah et al. (2002), Kumaran and Citarasu (2016)].
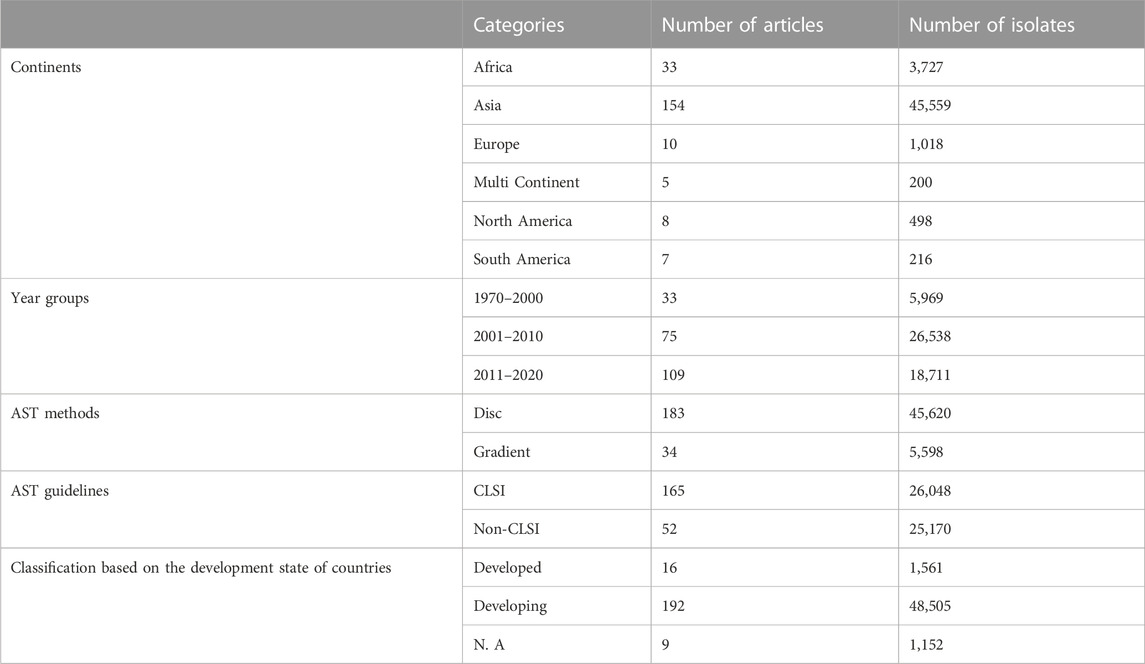
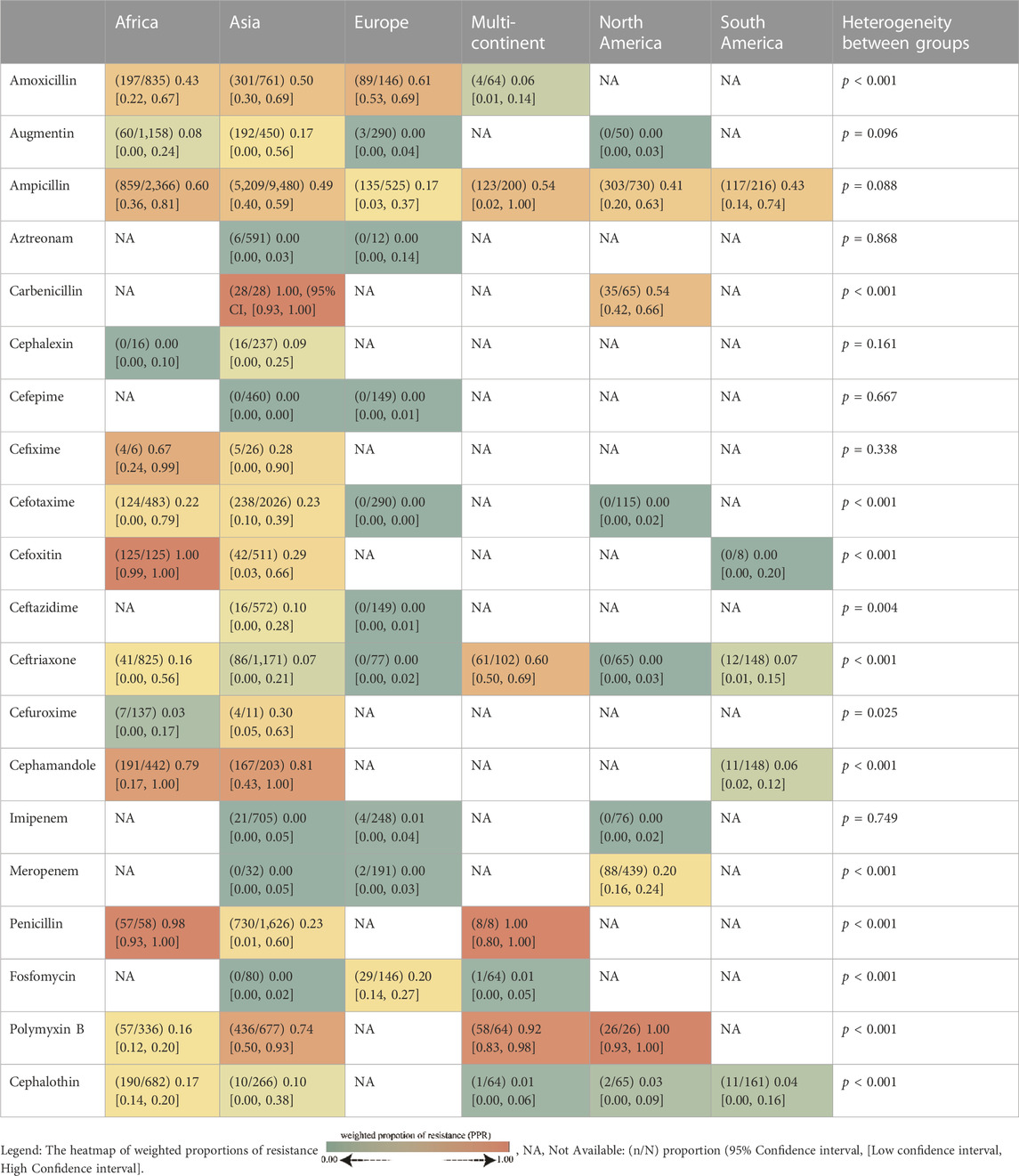
The whole search and selection prudcidure are summarized in prizma flow diagram (Figure 1). The included studies have been conducted in 40 countries. Descriptive statistics of included articles are summarized in Table 1. The WPR of V. cholera to each antibiotic and the result of the egger test are summarized in Table 2. The total number of V. cholera isolates, the number of resistant isolates, and the WPR of each antibiotic are mentioned in the forest plot (Figure 2). The V. cholera WPR to the antibiotics in each continent is summarized in Table 3. The evolution of antibiotic resistance over time is depicted in Table 4. Also, the WPR in developed and developing countries is summarized in Table 5. The search syntax and all extracted data from included articles are available in Supplementary Material.
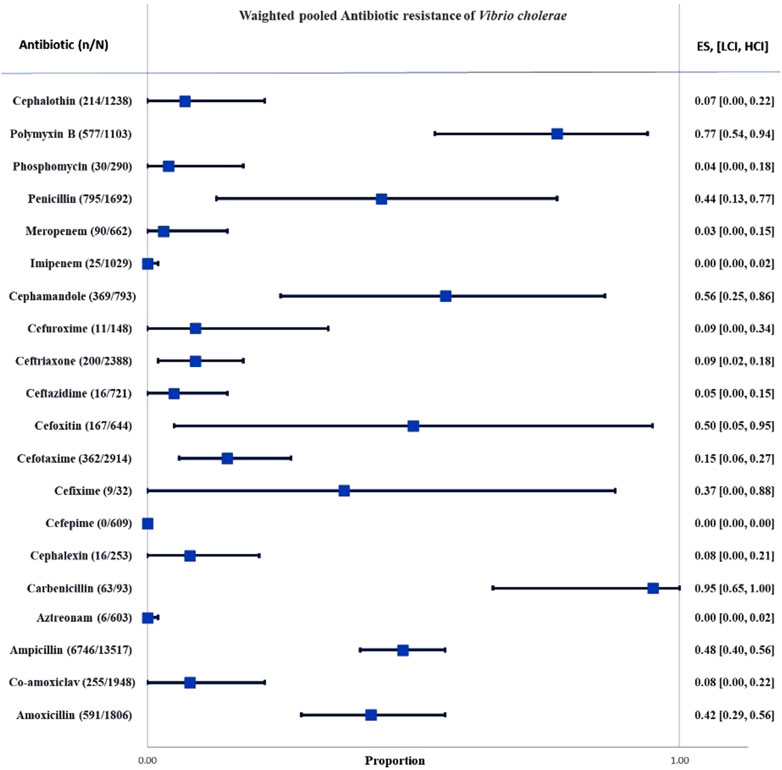
FIGURE 2. Forest plot of the weighted pooled proportion of antimicrobial-resistant Vibrio cholera. Abbreviations; Effect size or proportion of resistance (ES), number of resistant isolates (n), number of isolates (N), high confidence interval (HCI), low confidence interval (LCI).
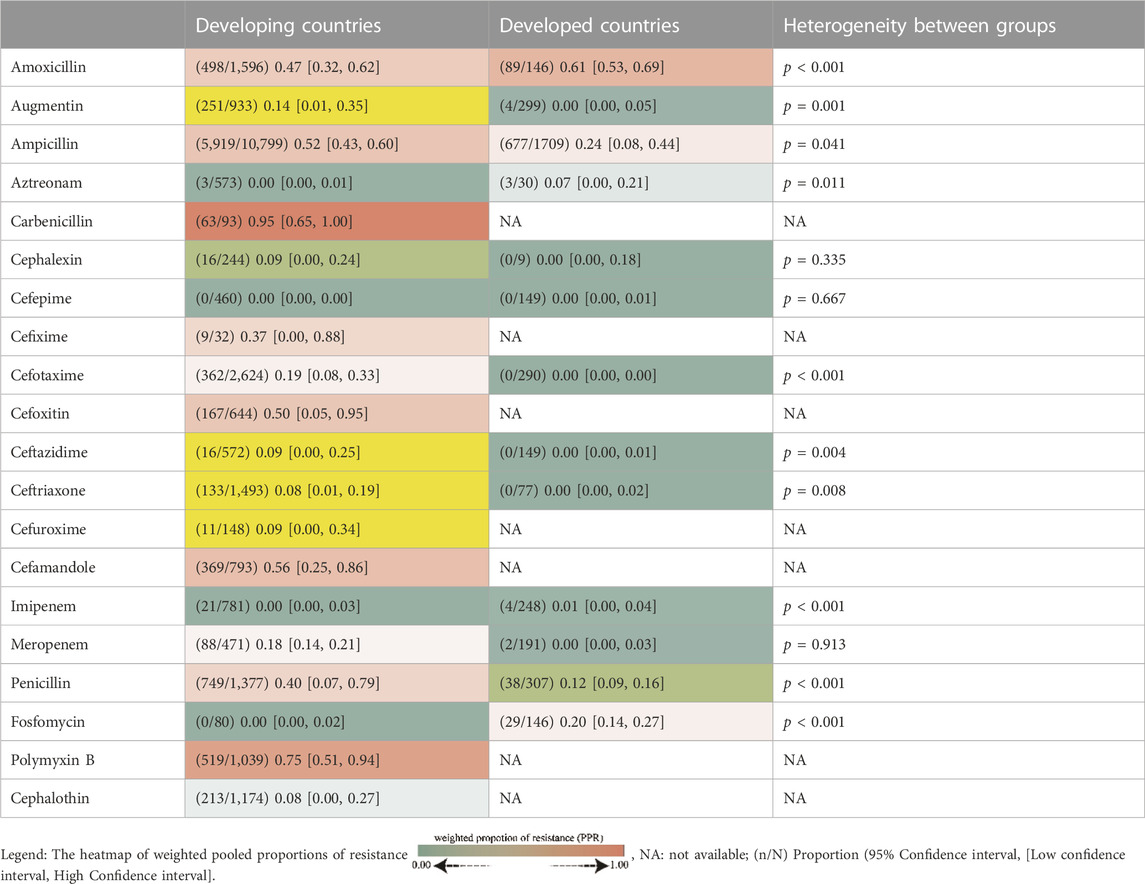
TABLE 5. Pooled proportion of Vibrio cholera antibiotic resistance in developing and developed countries.
Resistance to penicillin’s
The V. cholera WPR to penicillin is 0.44 (95% CI, (95% CI, [0.13, 0.77]), and the heterogeneity was significant (I2 = 99.40, p < 0.01). The highest WPR among countries was observed in Bangladesh (WPR; 0.14). In most studies, AST was performed by the disc diffusion method. WPR in CLSI and non-CLSI method subgroups was 0.36 and 0.84, respectively. The serogroup heterogeneity and isolate sources subgroups were insignificant (p > 0.05).
V. cholera WPR to ampicillin is 0.48 (95% CI [0.40, 0.56]), and heterogeneity was significant (I2 = 98.77, p < 0.01). The countries with the highest WPR were France, Nepal, and Iran (WPR; 1.00, 1.00, 0.97, respectively). Heterogeneity between the source of isolates, serogroups, and AST method subgroups was insignificant (p > 0.05).
The V. cholera WPR to amoxicillin is 0.42 (95% CI [0.29, 0.56]) and heterogeneity was significant (I2 = 96.95, p < 0.01). Cameroon and India have the highest WPR among countries (WPR; 0.52, 0.43, respectively). The resistance rate of clinical isolates was much higher than environmental isolates (WPR; 0.43, 0.35, respectively). The heterogeneity of subgrouping based on the AST method, AST guidelines, and serogroups were insignificant (p > 0.05).
The V. cholera WPR to carbenicillin is 0.95 (95% CI [0.65, 1.00]), and heterogeneity was significant (I2 = 85.02, p < 0.01). The highest WPR was in Malaysia and India (WPR; 1.00, 1.00, respectively). The WPR in clinical isolates was much higher than in environmental isolates (WPR; 1.00, 0.54, respectively). The WPR of the O1/O139 serogroup was higher than the non-O1/O139 serogroup (WPR; 1.00, 0.54, respectively). All the studies have used CLSI guidelines. The heterogeneity of subgrouping based on the AST method was insignificant (p > 0.05).
The V. cholera WPR to augmentin (amoxicillin/clavulanate) is 0.08 (95% CI [0.00, 0.22]), and the heterogeneity was significant (I2 = 98.11, p < 0.01). India has the highest resistance rate (WPR). The heterogeneity of subgrouping based on the isolate’s sources, serogroups, and AST guidelines was insignificant (p > 0.05).
Cephalosporins (1st gen)
Resistance to cefalexin
The V. cholera WPR to cefalexin is 0.08 (95% CI [0.0.0, 0.21]), and heterogeneity between reports was significant (I2 = 86.27, p < 0.01). Every included sample was a clinical isolate. The heterogeneity of subgrouping based on the countries, AST methods, AST guidelines, and serogroups was insignificant (p > 0.05). The V. cholera WPR to cephalothin is 0.07 (95% CI [0.00–0.22]), and heterogeneity was significant (I2 = 97.81, p < 0.01). India, Brazil, and Morocco have the highest WPR among countries (WPR; 0.62, 0.06, and 0.17, respectively). Most of the articles have used the disk diffusion method for AST. Heterogeneity between the isolated sources was insignificant (p = 0.969).
Cephalosporins (2nd gen)
The V. cholera WPR to cefoxitin is 0.50 (95% CI [0.05–0.95]), and heterogeneity was significant (I2 = 99.30, p < 0.01). The highest WPR was in Bangladesh (WPR; 0.01). The V. cholera WPR to cefuroxime was 0.09 (95% CI [0.00 to 0.34]), and the heterogeneity was significant (I2 = 89.81, p < 0.01). The countries with the highest WPR were India and Ghana (WPR; 0.30, 0.17, respectively). The WPR of clinical isolates was higher than environmental isolates (WPR; 0.16, 0.00, respectively). Heterogeneity of subgrouping based on the AST method; the AST guidelines were insignificant (p > 0.05). All studies have used CLSI, and all isolates were O1/O139 serogroup.
The WPR of V. cholera to carbenicillin was 0.95 (95% CI [0.65, 1.00]), and the heterogeneity was significant (I2 = 85.02, p < 0.01). Malaysia and India have the highest WPRs (WPR; 1.00, 1.00, respectively). All of the studies have been in developed countries. Clinical isolates had a substantially higher WPR than environmental isolates (WPR; 1.00, 0.54, respectively). The WPR in O1/O139 serogroup was higher than the non-O1/O139 serogroup (WPR; 1.00 and 0.54, respectively). All studies have used CLSI as an AST guideline. The heterogeneity of subgrouping based on the AST methods was insignificant (p = 0.52).
Cephalosporins (3rd gen)
The WPR of V. cholera to cefixime is 0.37 (95% CI [0.00–0.88]), and the heterogeneity was significant (I2 = 86.80, p < 0.01). Nigeria had the highest WPRs among countries (WPR; 0.67).
The heterogeneity between countries and sources of isolates was significant (p < 0.05). The heterogeneity between the AST method and serogroups was insignificant (p = 0.072). All studies used CLSI guidelines.
The WPR of V. cholera to cefotaxime is 0.15 (95% CI [0.06, 0.27]), and the heterogeneity was significant (I2 = 97.94, p < 0.01). The countries with the highest WPR were India, Bangladesh, and Germany (WPR; 0.21, 0.12, and 0.00, respectively). Heterogeneity of subgrouping based on the. the isolate sources, AST method, AST guidelines, and serogroups were insignificant (p > 0.05). The V. cholera WPR to ceftazidime is 0.05 (95% CI [0.00–0.15]), and the heterogeneity was significant (I2 = 90.63, p < 0.01). Malaysia and India have the highest WPR (WPR; 0.80, 0.12, respectively). The WPR in clinical isolates was higher than in environmental isolates (WPR; 0.02, 0.00, respectively). Most studies have investigated AST by disk diffusion method (WPR; 0.10). All studies have used CLSI as an AST guideline. The WPR of the O1/O139 serogroup was higher than the non-O1/O139 serogroup (WPR; 0.10 and 0.00, respectively). The V. cholera WPR to ceftriaxone is 0.09 (95% CI [0.02–0.18]), and the heterogeneity was significant (I2 = 96.90, p < 0.01). The countries with the highest WPR were Iran, Brazil, and India (WPR; 1.00, 0.07, 0.04, respectively). The WPR of clinical isolates was much more than environmental isolates (WPR; 0.14, 0.03, respectively). The heterogeneity of subgrouping based on the AST guidelines and serotypes was insignificant (p > 0.05).
Cephalosporins (4th gen)
The V. cholera WPR to cefepime is 0.00 (95% CI [0.00, 0.00]), and the heterogeneity was insignificant (I2 = 0.00, p = 0.90).
Resistance to carbapenems
The V. cholera WPR to imipenem is 0.00 (95% CI [0.00–0.02]), and heterogeneity was significant (I2 = 79.95, p < 0.01). India and Germany had the highest WPR (0.05 and 0.02, respectively). CLSI was utilized as a guideline in all studies. The heterogeneity of subgrouping based on the continents, AST method, AST guideline, isolated sources, and serogroups were insignificant (p > 0.05).
The V. cholera WPR to meropenem is 0.03 (95% CI [0.00, 0.15], and the heterogeneity was significant (I2 = 94.48, p < 0.01). Germany had the lowest WPR among countries (WPR; 0.00). The WPR in O1/O139 serogroup was higher than non-O1/O139 serogroup (WPR; 0.18 0.00 respectively).
Resistance to aztreonam
The WPR of V. cholera to aztreonam is 0.00 (95% CI [0.00, 0.02]). The heterogeneity was insignificant (I2 = 49.25, p = 0.08).
Resistance to fosfomycin
The V. cholera WPR to fosfomycin is 0.04 (95% CI [0.00–0.18]), and the heterogeneity was significant (I2 = 93.03, p < 0.01). Spain had the highest WPR among countries (WPR; 0.20), and Iran reported the lowest resistance rate. (WPR; 0.00). The WPR of clinical isolates was higher than environmental isolates (WPR; 0.00, 0.17, respectively). The WPR of the non-CLSI group was higher than the CLSI group (WPR; 0.00 and 0.20, respectively).
Resistance to polymyxin B
The V. cholera WPR to polymyxin B is 0.77 (95% CI [0.54, 0.94]), and the heterogeneity was significant (I2 = 98.24, p < 0.01). The countries with the highest WPR were India and Iran (WPR; 0.86, 0.65, respectively). The WPR of the CLSI subgroup was higher than the non-CLSI group (WPR; 0.78, 0.16, respectively). The heterogeneity of subgrouping based on the serogroups and isolate sources was insignificant (p > 0.05).
Discussion
Cholera is a historical, unresolved problem and a severe health-threatening infection, particularly in developing countries. This disease has been endemic throughout South Asia, notably in Bangladesh and India’s Ganges delta region (18). This condition expanded outside the Indian subcontinent via trade channels, resulting in worldwide pandemics with significant fatality rates (millions of deaths) (19). Antibiotics alone are ineffective against severe cholera. However, fluid replacement and antibiotic treatment are combined because antibiotics can reduce cholera bacteria in the feces and shorten the duration of the disease (19).
Due to V. cholera’s low resistance, the AST of these microorganisms was avoided in the past. (14, 15). However, the emergence of β-lactam-resistant species has had severe repercussions on managing infectious diseases globally (16). Tetracyclines have long been the antibiotic for treating severe cholera (15). Conversely, tetracycline-resistant V. cholera strains are spreading internationally (17). Severe epidemics in Latin America, Tanzania, Bangladesh, and Zaire have been linked to tetracycline-resistant strains (15). A meta-analysis with 52 studies has estimated the tetracycline and doxycycline resistance rate of V. cholera serotype O1 over 50% and 28%, respectively (20). This escalating resistance rate underscores the importance of regulating antibiotic prescriptions and discovering effective alternatives.
The current study investigated V. cholera cell wall-targeting antibiotic resistance patterns. This study revealed that carbenicillin (95%), and polymyxin B (77%) have the highest resistance rates. The ampicillin, penicillin, and amoxicillin resistance rates were 48%, 44%, and 42%, respectively. Adding clavulanate acid to amoxicillin reduced the proportion of resistant bacteria to 8%, which was more effective than amoxicillin alone.
The rates of resistance to the first generation of cephalosporins, such as cefalexin and cephalothin, were 8% and 7%, respectively. The percentages of resistance to the second generation of cephalosporins, including cefamandole, cefoxitin, and cefuroxime, were 50%, 56%, and 9%, respectively. Compared to the first generation of cephalosporins, V. cholera was more resistant to the second generation. Cefixime (37%), cefotaxime (15%), ceftazidime (5%), and ceftriaxone (9%) As members of the third generation of cephalosporins are more effective than second-generation cephalosporins. Cefepime is the only member of the fourth generation of cephalosporins studied in four articles evaluating 609 V. cholera isolates, and none of them were resistant to it.
The rate of aztreonam resistance was 0%, while the rate of fosfomycin resistance was 4%. Cefepime, aztreonam, and imipenem were the most efficient antibiotics against V. cholera. Non-etheless, the heterogeneity between reported imipenem resistance rates was substantial, and it appears necessary to perform AST before prescribing antibiotics to ensure the isolate’s susceptibility.
Considering sulfonamides are commonly used to treat HIV, TB, malaria, pneumonia, and febrile illness, Onohuean et al. (2022) estimated the prevalence of quinolone, tetracycline, and sulfonamide resistance genes to be 32.97 percent (95% CI [0.18–0.55]). This percentage is lower than that of polymyxin and higher than that of cephalosporins. An additional study was analyzed from a total of 139 articles involving 24,062 isolates of V. cholera O1/O139. Asia was the location of origin for 102 out of the total research. The WPR was calculated as follows: azithromycin had a 1%, erythromycin 36%, ciprofloxacin 3%, cotrimoxazole 79%, doxycycline 7%, and tetracycline had a success rate of 20% WPR. Between the years 1980 and 2020, there was a growth in drug resistance to cotrimoxazole, ciprofloxacin, and tetracycline (Liu et al., 2022).
According to the findings of Xin-hui Yuan’s research, there has been an increase in drug resistance in recent years, particularly to nalidixic acid, cotrimoxazole, furazolidone, and tetracycline. Between 2000 and 2020, however, resistance to antibiotics such as amoxicillin, ciprofloxacin, erythromycin, chloramphenicol, ampicillin, streptomycin, and ceftriaxone decreased. The frequency of doxycycline and ciprofloxacin resistance in V. cholera O1/O139 isolates significantly reduced from 2011 to 2020 compared to the frequency of these resistances from 2001 to 2010 (p < 0.05) (Yuan et al., 2022).
Based on subgroup analysis of continents, Africa and Asia had the highest proportion of resistant individuals. Patterns of resistance vary between developing and developed nations, and the resistance rate in developing nations was significantly higher than in developed nations. Even though developed countries had more resistant bacteria to amoxicillin, aztreonam, imipenem, and fosfomycin, developing countries had more resistant bacteria to augmentin, penicillin, ampicillin, cefotaxime, ceftazidime, and ceftriaxone.
Antibiotics to which V. cholera is highly resistant in developed nations have not been studied in developing countries. Cefixime, cefoxitin, cefuroxime, cefamandole, polymyxin B, cephalothin, and carbenicillin appear to have been taken off the list of V. cholera treatments. There has been an increase in resistance to antibiotics such as cephalothin, ceftriaxone, amoxicillin, and meropenem. Over the years, resistance to penicillin, ceftazidime, and cefotaxime has decreased. Because cholera was historically treated with numerous antibiotics, resistance to these antibiotics grew over time. For instance, penicillin resistance reached 98% during a specific time frame. After this increase in V. cholera infection treatment, it appears that this antibiotic was no longer considered. In recent years, the resistance to this antibiotic has decreased significantly, reaching 19% due to the lack of prescribing and use.
Conclusion
Our findings indicate the importance of prescribing antibiotics accurately to control and prevent V. cholera antibiotic resistance. Following the disappointing emergence of antibiotic resistance to certain antibiotics, such as penicillin, these antibiotics were utilized less frequently, which resulted in a decrease in antibiotic resistance to these antibiotics in general. This event increases optimism that using old antibiotics will be effective if antibiotic use is controlled. In that order, the antibiotics with the least resistance to V. cholera were cefepime, aztreonam, imipenem, and meropenem. Due to the vastly different patterns of antibiotic resistance of V. cholera to these antibiotics in various geographic locations, it appears necessary to investigate the antibiotic resistance of the isolates.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2023.1027277/full#supplementary-material
References
Awuor, S. O., Omwenga, E. O., and Daud, (2020a). Geographical distribution and antibiotics susceptibility patterns of toxigenic Vibrio cholerae isolates from Kisumu County, Kenya. Afr. J. Prim. Health Care Fam. Med. 12, e1–e6. doi:10.4102/phcfm.v12i1.2264
Abera, B., Bezabih, B., and Dessie, A. (2010). Antimicrobial suceptibility of V. cholerae in north west, Ethiopia. Ethiop. Med. J. 48, 23–28.
Akoachere, J. F. T. K., Masalla, T. N., and Njom, H. A. (2013). Multi-drug resistant toxigenic Vibrio cholerae O1 is persistent in water sources in New Bell-Douala. Cameroon 13. doi:10.1186/1471-2334-13-366
Akoachere, J. F. T. K., and Mbuntcha, C. K. P. (2014). Water sources as reservoirs of Vibrio cholerae O1 and non-O1 strains in Bepanda, Douala (Cameroon): Relationship between isolation and physico-chemical factors. BMC Infect. Dis. 14, 421. doi:10.1186/1471-2334-14-421
AL-Hilu, S. A., AL-Mohana, A, Jaber, Z., and Hinthong, W. (2019). Molecular epidemiology of antimicrobial resistance and virulence profiles of Escherichia coli, Salmonella spp., and Vibrio spp. isolated from coastal seawater for aquaculture. Antibiotics 11, 1688. doi:10.3390/antibiotics11121688
Alam, M., Islam, M. T., Rashed, S. M., Johura, F. T., Bhuiyan, N. A., Delgado, G., et al. (2012). Vibrio cholerae classical biotype strains reveal distinct signatures in Mexico. J. Clin. Microbiol. 50, 2212–2216. doi:10.1128/JCM.00189-12
Alaoui, H. L., Oufdou, K., and Mezrioui, N.-E. (2010). Determination of several potential virulence factors in non-o1 Vibrio cholerae, Pseudomonas aeruginosa, faecal coliforms and streptococci isolated from Marrakesh groundwater. Water Sci. Technol. 61, 1895–1905. doi:10.2166/wst.2010.263
Amau, A., Pujalte, M. J., Amaro, C., and Garay, E. (1988). The annual cycle of zooplankton-associated Vibrio cholerae and related vibrios in Albufera lake and its coastal surrounding waters (Valencia, Spain). PRONADISA React. de calidad internacional made Spain 185.
Amita, M., Chowdhury, S. R., Thungapathra, M., Ramamurthy, T., Nair, G. B., and Ghosh, A. (2003). Class I integrons and SXT elements in El Tor strains isolated before and after 1992 Vibrio cholerae O139 outbreak, Calcutta, India. Emerg. Infect. Dis. 9, 500–502. doi:10.3201/eid0904.020317
Anand, V. K., Arora, S., Patwari, A. K., Agarwal, G. D., and Dewan, N. (1996). Multidrug resistance in Vibrio cholerae. Indian Pediatr. 33, 774–777.
Araj, G. F., Uwaydah, M. M., and Alami, S. Y. (1994). Antimicrobial susceptibility patterns of bacterial isolates at the American University Medical Center in Lebanon. Diagnostic Microbiol. Infect. Dis. 20, 151–158. doi:10.1016/0732-8893(94)90109-0
Awuor, S. O., Omwenga, E. O., and Daud, I. I. (2020b). Geographical distribution and antibiotics susceptibility patterns of toxigenic Vibrio cholerae isolates from Kisumu County, Kenya. Afr. J. Prim. Health Care Fam. Med. 12, 1–6. doi:10.4102/phcfm.v12i1.2264
Baddam, R., Sarker, N., Ahmed, D., Mazumder, R., Abdullah, A., Morshed, R., et al. (2020). Genome dynamics of Vibrio cholerae isolates linked to seasonal outbreaks of cholera in dhaka, Bangladesh. mBio 11, 033399–e3419. doi:10.1128/mBio.03339-19
Bag, P. K., Maiti, S., Sharma, C., Ghosh, A., Basu, A., Mitra, R., et al. (1998). Rapid spread of the new clone of Vibrio cholerae O1 biotype El Tor in cholera endemic areas in India. Epidemiol. Infect. 121, 245–251. doi:10.1017/s0950268898001423
Bakhshi, B., Eftekhari, N., and Pourshafie, M. R. (2014). Genetic elements associated with antimicrobial resistance among intestinal bacteria. Jundishapur J. Microbiol. 7, e9924. doi:10.5812/jjm.9924
Bakhshi, B., and Pourshafie, M. R. (2009). Assessing clonality of Vibrio cholerae strains isolated during four consecutive years (2004 - 2007) in Iran. Scand. J. Infect. Dis. 41, 256–262. doi:10.1080/00365540902767049
Bakhshi, B., Pourshafie, M. R., Navabakbar, F., Tavakoli, A., Shahcheraghi, F., Salehi, M., et al. (2008). Comparison of distribution of virulence determinants in clinical and environmental isolates of Vibrio cholera. Iran. Biomed. J. 12, 159–165.
Balaji, K., Okonjo, P. A., Thenmozhi, R., and Karutha Pandian, S. (2013). Virulence and multidrug resistance patterns of Vibrio cholerae O1 isolates from diarrheal outbreaks of South India during 2006-2009. Microb. Drug Resist 19, 198–203. doi:10.1089/mdr.2012.0127
Ballal, M., and Shivananda, P. G. (2002). Rotavirus and enteric pathogens in infantile diarrhoea in Manipal, South India. Indian J. Pediatr. 69, 393–396. doi:10.1007/BF02722628
Bani, S., Mastromarino, P. N., Ceccarelli, D., LE VAN, A., Salvia, A. M., Ngo Viet, Q. T., et al. (2007). Molecular characterization of ICE Vch Vie0 and its disappearance in Vibrio cholerae O1 strains isolated in 2003 in Vietnam. FEMS Microbiol. Lett. 266, 42–48. doi:10.1111/j.1574-6968.2006.00518.x
Barati, H., Moradi, G., Rasouli, M. A., and Mohammadi, P. (2015). Epidemiologic and drug resistance pattern of Vibrio cholerae O1 biotype el tor, serotype ogawa, in the 2011 cholera outbreak, in alborz province, Iran. Jundishapur J. Microbiol. 8, e23477. doi:10.5812/jjm.23477
Baron, S., Larvor, E., Chevalier, S., Jouy, E., Kempf, I., Granier, S. A., et al. (2017). Antimicrobial susceptibility among urban wastewater and wild shellfish isolates of non-O1/Non-O139 Vibrio cholerae from La Rance Estuary (Brittany, France). Front. Microbiol. 8, 1637. doi:10.3389/fmicb.2017.01637
Baron, S., Lesne, J., Jouy, E., Larvor, E., Kempf, I., Boncy, J., et al. (2016). Antimicrobial susceptibility of autochthonous aquatic Vibrio cholerae in Haiti. Front. Microbiol. 7, 1671. doi:10.3389/fmicb.2016.01671
Basu, A., Garg, P., Datta, S., Chakraborty, S., Bhattacharya, T., Khan, A., et al. (2000). Vibrio cholerae O139 in Calcutta, 1992-1998: Incidence, antibiograms, and genotypes. Emerg. Infect. Dis. 6, 139–147. doi:10.3201/eid0602.000206
Bhanumathi, R., Sabeena, F., Isac, S. R., Shukla, B. N., and Singh, D. V. (2003). Molecular characterization of Vibrio cholerae O139 bengal isolated from water and the aquatic plant Eichhornia crassipes in the River Ganga, Varanasi, India. Appl. Environ. Microbiol. 69, 2389–2394. doi:10.1128/AEM.69.4.2389-2394.2003
Bhat, T., Yousuf, I., Wani, M. A., Naqash, M., Roshan, R., and Malik, S. (2012). Anti microbial sensitivity pattern of cholera epidemic in kashmir valley. JK-Practitioner 17, 26–28.
Bhattacharya, D., Dey, S., Roy, S., Parande, M. V., Telsang, M., Parande, A. V., et al. (2015). Multidrug-resistant Vibrio cholerae O1 was responsible for a cholera outbreak in 2013 in Bagalkot, North Karnataka. Jpn. J. Infect. Dis. 68, 347–350. doi:10.7883/yoken.JJID.2014.257
Bhattacharya, D., Sayi, D. S., Thamizhmani, R., Bhattacharjee, H., Bharadwaj, A. P., Roy, A., et al. (2012). Emergence of multidrug-resistant Vibrio cholerae O1 biotype el tor in port blair, India. Am. J. Trop. Med. Hyg. 86, 1015–1017. doi:10.4269/ajtmh.2012.11-0327
Bhotra, T., Das, M., Pal, B., and Singh, D. (2017). Genomic profile of antibiotic resistant, classical ctxB positive Vibrio cholerae O1 biotype el tor isolated in 2003 and 2005 from puri, India: A retrospective study. Indian J. Med. Microbiol. 34, 462–470. doi:10.4103/0255-0857.195356
Bhowmick, T. S., Das, M., Roy, N., and Sarkar, B. L. (2007). Phenotypic and molecular typing of Vibrio cholerae O1 and O139 isolates from India. J. Infect. 54, 475–482. doi:10.1016/j.jinf.2006.09.018
Bidinost, C., Saka, H. A., Aliendro, O., Sola, C., Panzetta-Duttari, G., Carranza, P., et al. (2004). Virulence factors of non-O1 non-O139 Vibrio cholerae isolated in Cordoba, Argentina. Rev. Argent. Microbiol. 36, 158–163.
Bier, N., Schwartz, K., Guerra, B., and Strauch, E. (2015). Survey on antimicrobial resistance patterns in Vibrio vulnificus and Vibrio cholerae non-O1/non-O139 in Germany reveals carbapenemase-producing Vibrio cholerae in coastal waters. Front. Microbiol. 6, 1179. doi:10.3389/fmicb.2015.01179
Borkakoty, B., Biswas, D., Devi, U., Yadav, K., and Mahanta, J. (2012). Emergence of classical ctxB genotype 1 and tetracycline resistant strains of Vibrio cholerae O1 El Tor in Assam, India. Trans. R. Soc. Trop. Med. Hyg. 106, 382–386. doi:10.1016/j.trstmh.2012.03.005
Campos, L., Zahner, V., Avelar, K., Alves, R., Pereira, D., Brazil, J. V., et al. (2004). Genetic diversity and antibiotic resistance of clinical and environmental Vibrio cholerae suggests that many serogroups are reservoirs of resistance. Epidemiol. Infect. 132, 985–992. doi:10.1017/s0950268804002705
Ceccarelli, D., Alam, M., Huq, A., and Colwell, R. R. (2016). Reduced susceptibility to extended-spectrum beta-lactams in Vibrio cholerae isolated in Bangladesh. Front. Public Health 4, 231. doi:10.3389/fpubh.2016.00231
Ceccarelli, D., Spagnoletti, M., Bacciu, D., Cappuccinelli, P., and Colombo, M. M. (2011). New V. cholerae atypical El Tor variant emerged during the 2006 epidemic outbreak in Angola. BMC Microbiol. 11, 130. doi:10.1186/1471-2180-11-130
Chakraborty, S., Deokule, J., Garg, P., Bhattacharya, S., Nandy, R., Nair, G. B., et al. (2001). Concomitant infection of EnterotoxigenicEscherichia coli in an outbreak of cholera caused byVibrio cholerae O1 and O139 in ahmedabad, India. J. Clin. Microbiol. 39, 3241–3246. doi:10.1128/JCM.39.9.3241-3246.2001
Chandrasekhar, M. R., Krishna, B. V., and Patil, A. B. (2008). Changing characteristics of Vibrio cholerae: Emergence of multidrug resistance and non-O1, non-O139 serogroups. Southeast Asian J. Trop. Med. Public Health 39, 1092–1097.
Chatterjee, S., Ghosh, K., Raychoudhuri, A., Pan, A., Bhattacharya, M. K., Mukhopadhyay, A. K., et al. (2007). Phenotypic and genotypic traits and epidemiological implication of Vibrio cholerae O1 and O139 strains in India during 2003. J. Med. Microbiol. 56, 824–832. doi:10.1099/jmm.0.46982-0
Chhotray, G. P., Pal, B. B., Khuntia, H. K., Chowdhury, N. R., Chakraborty, S., Yamasaki, S., et al. (2002). Incidence and molecular analysis of Vibrio cholerae associated with cholera outbreak subsequent to the super cyclone in Orissa, India. Epidemiol. Infect. 128, 131–138. doi:10.1017/s0950268801006720
Chomvarin, C., Johura, F. T., Mannan, S. B., Jumroenjit, W., Kanoktippornchai, B., Tangkanakul, W., et al. (2013). Drug response and genetic properties of Vibrio cholerae associated with endemic cholera in north-eastern Thailand, 2003-2011. J. Med. Microbiol. 62, 599–609. doi:10.1099/jmm.0.053801-0
Chomvarin, C., Jumroenjit, W., Wongboot, W., Kanoktippornchai, B., Chaimanee, P., Jamjane, O., et al. (2012). Molecular analysis and antimicrobial resistance of Vibrio cholerae O1 in northeastern Thailand. Southeast Asian J. Trop. Med. Public Health 43, 1437–1446.
Chowdhury, G., Bhadra, R. K., Bag, S., Pazhani, G. P., Das, B., Basu, P., et al. (2016). Rugose atypical Vibrio cholerae O1 El Tor responsible for 2009 cholera outbreak in India. J. Med. Microbiol. 65, 1130–1136. doi:10.1099/jmm.0.000344
Colombo, M. M., Francisco, M., Ferreira, B. D., Rubino, S., and Cappuccinelli, P. (1993). The early stage of the recurrent cholera epidemic in Luanda, Angola. Eur. J. Epidemiol. 9, 563–565. doi:10.1007/BF00209537
Dalsgaard, A., Forslund, A., Petersen, A., Brown, D. J., Dias, F., Monteiro, S., et al. (2000). Class 1 integron-borne, multiple-antibiotic resistance encoded by a 150-kilobase conjugative plasmid in epidemic vibrio cholerae O1 strains isolated in Guinea-Bissau. J. Clin. Microbiol. 38, 3774–3779. doi:10.1128/JCM.38.10.3774-3779.2000
Dalsgaard, A., Mortensen, H. F., Molbak, K., Dias, F., Serichantalergs, O., and Echeverria, P. (1996). Molecular characterization of Vibrio cholerae O1 strains isolated during cholera outbreaks in Guinea-Bissau. J. Clin. Microbiol. 34, 1189–1192. doi:10.1128/jcm.34.5.1189-1192.1996
Dalsgaard, A., Serichantalergs, O., Forslund, A., Pitarangsi, C., and Echeverria, P. (1998). Phenotypic and molecular characterization of Vibrio cholerae O1 isolated in Samutsakorn, Thailand before, during and after the emergence of V. cholerae O139. Epidemiol. Infect. 121, 259–268. doi:10.1017/s0950268898001125
Das, M. M., Bhotra, T., Zala, D., and Singh, D. V. (2016). Phenotypic and genetic characteristics of Vibrio cholerae O1 carrying Haitian ctxB and attributes of classical and El Tor biotypes isolated from Silvassa, India. J. Med. Microbiol. 65, 720–728. doi:10.1099/jmm.0.000282
Das, S., Choudhry, S., Saha, R., Ramachandran, V. G., Kaur, K., and Sarkar, B. L. (2011). Emergence of multiple drug resistance Vibrio cholerae O1 in East Delhi. J. Infect. Dev. Ctries. 5, 294–298. doi:10.3855/jidc.1251
Das, S., Saha, R., and Kaur, I. R. (2008). Trend of antibiotic resistance of Vibrio cholerae strains from East Delhi. Indian J. Med. Res. 127, 478–482.
Dengo-Baloi, L. C., Sema-Baltazar, C. A., Manhique, L. V., Chitio, J. E., Inguane, D. L., and Langa, J. P. (2017). Antibiotics resistance in El Tor Vibrio cholerae 01 isolated during cholera outbreaks in Mozambique from 2012 to 2015. PLoS One 12, e0181496. doi:10.1371/journal.pone.0181496
Dey, S., Parande, M. V., Parande, A. M., Lakkannavar, S. L., Rathore, P. K., Mantur, B. G., et al. (2014). Twin outbreak of cholera in rural North Karnataka, India. Indian J. Med. Res. 140, 420–426.
Dua, P., Karmakar, A., and Ghosh, C. (2018). Virulence gene profiles, biofilm formation, and antimicrobial resistance of Vibrio cholerae non-O1/non-O139 bacteria isolated from West Bengal, India. Heliyon 4, e01040. doi:10.1016/j.heliyon.2018.e01040
Echeverria, P., Pitarangsi, C., Eampokalap, B., Vibulbandhitkit, S., Boonthai, P., and Rowe, B. (1983). A longitudinal study of the prevalence of bacterial enteric pathogens among adults with diarrhea in Bangkok, Thailand. Diagnostic Microbiol. Infect. Dis. 1, 193–204. doi:10.1016/0732-8893(83)90018-4
Eibach, D., Herrera-Leon, S., Gil, H., Hogan, B., Ehlkes, L., Adjabeng, M., et al. (2016). Molecular epidemiology and antibiotic susceptibility of Vibrio cholerae associated with a large cholera outbreak in Ghana in 2014. PLoS Negl. Trop. Dis. 10, e0004751. doi:10.1371/journal.pntd.0004751
Eja, M. E., Etok, C. A., Asikong, B. E., Mboto, C. I., and Arikpo, G. E. (2006). Incidence of enteric bacterial pathogens in water found at the bottom of commercial freezers in Calabar, Southeastern Nigeria. Southeast Asian J. Trop. Med. Public Health 37, 394–399.
Falcao, D. P., Lustri, W. R., and Bauab, T. M. (1998). Incidence of non-01 Vibrio cholerae and Aeromonas spp. in fresh water in Araraquara, Brazil. Curr. Microbiol. 37, 28–31. doi:10.1007/s002849900332
Faruque, S. M., Saha, M. N., Bag, P. K., Bhadra, R. K., Bhattacharya, S., Sack, R. B., et al. (2000). Genomic diversity among Vibrio cholerae O139 strains isolated in Bangladesh and India between 1992 and 1998. FEMS Microbiol. Lett. 184, 279–284. doi:10.1111/j.1574-6968.2000.tb09027.x
Fazil, M. H., Bhanumathi, R., Pandey, H. P., and Singh, D. V. (2011). Characterization of Vibrio cholerae O139 belonging to multiple ribotypes and isolated from diarrhoeal patients in Kerala, southern India. Infect. Genet. Evol. 11, 454–459. doi:10.1016/j.meegid.2010.12.008
Feglo, P. K., and Sewurah, M. (2018). Characterization of highly virulent multidrug resistant Vibrio cholerae isolated from a large cholera outbreak in Ghana. BMC Res. Notes 11, 45. doi:10.1186/s13104-017-2923-z
Fernandez-Abreu, A., Bravo-Farinas, L., Rivero-Navea, G., Cabrera-Cantelar, N., Nunez-Fernandez, F. A., Cruz-Infante, Y., et al. (2017). Determinants of virulence and antimicrobial susceptibility in non-O1, non-O139 Vibrio cholerae isolates. MEDICC Rev. 19, 21. doi:10.37757/MR2017.V19.N4.6
Folgosa, E., Mastrandrea, S., Cappuccinelli, P., Uzzau, S., Rappelli, P., Brian, M., et al. (2000). Molecular identification of pathogenicity genes and ERIC types in Vibrio cholerae O1 epidemic strains from Mozambique. Epidemiol. Infect. 127, 17–25. doi:10.1017/s0950268801005623
Garg, P., Chakraborty, S., Basu, I., Datta, S., Rajendran, K., Bhattacharya, T., et al. (2000). Expanding multiple antibiotic resistance among clinical strains of Vibrio cholerae isolated from 1992–7 in Calcutta, India. Epidemiol. Infect. 124, 393–399. doi:10.1017/s0950268899003957
Goel, A. K., Jain, M., Kumar, P., and Jiang, S. C. (2010). Molecular characterization of Vibrio cholerae outbreak strains with altered El Tor biotype from southern India. World J. Microbiol. Biotechnol. 26, 281–287. doi:10.1007/s11274-009-0171-7
Gupta, P. K., Pant, N. D., Bhandari, R., and Shrestha, P. (2016). Cholera outbreak caused by drug resistant Vibrio cholerae serogroup O1 biotype ElTor serotype Ogawa in Nepal; a cross-sectional study. Antimicrob. Resist Infect. Control 5, 23. doi:10.1186/s13756-016-0122-7
Ibarra, J., and Alvarado, D. (2007). Antimicrobial resistance of clinical and environmental strains of Vibrio cholerae isolated in Lima-Peru during epidemics of 1991 and 1998. Braz. J. Infect. Dis. 11, 100–105. doi:10.1590/s1413-86702007000100022
Imziln, B., and Hassani, L. (1994). Antimicrobial susceptibility of non-O1 Vibrio cholerae isolated from wastewater stabilization ponds in Marrakesh, Morocco. World J. Microbiol. Biotechnol. 10, 230–231. doi:10.1007/BF00360895
Iramiot, J. S., Rwego, I. B., Kansiime, C., and Asiimwe, B. B. (2019). Epidemiology and antibiotic susceptibility of Vibrio cholerae associated with the 2017 outbreak in Kasese district, Uganda. BMC Public Health 19, 1405. doi:10.1186/s12889-019-7798-6
Isaac-Márquez, A. P., Lezama-Dávila, C. M., Eslava-Campos, C., Navarro-Ocaña, A., and Cravioto-Quintana, A. (1998). Serotypes of Vibrio cholerae non-O1 isolated from water supplies for human consumption in Campeche, Mexico and their antibiotic susceptibility pattern. Memórias do Inst. Oswaldo Cruz 93, 17–22. doi:10.1590/s0074-02761998000100004
Islam, M. S., Mahmud, Z. H., Ansaruzzaman, M., Faruque, S. M., Talukder, K. A., Qadri, F., et al. (2011). Phenotypic, genotypic, and antibiotic sensitivity patterns of strains isolated from the cholera epidemic in Zimbabwe. J. Clin. Microbiol. 49, 2325–2327. doi:10.1128/JCM.00432-11
Ismail, H., Smith, A. M., Tau, N. P., Sooka, A., Keddy, K. H., Group For Enteric, R., et al. (2013). Cholera outbreak in south Africa, 2008-2009: Laboratory analysis of Vibrio cholerae O1 strains. J. Infect. Dis. 208 (1), S39–S45. doi:10.1093/infdis/jit200
Jagadeeshan, S., Kumar, P., Abraham, W. P., and Thomas, S. (2009). Multiresistant Vibrio cholerae non-O1/non-O139 from waters in south India: Resistance patterns and virulence-associated gene profiles. J. Basic Microbiol. 49, 538–544. doi:10.1002/jobm.200900085
Jain, M., Kumar, P., and Goel, A. K. (2016). Emergence of tetracycline resistant Vibrio cholerae O1 biotype el tor serotype ogawa with classical ctxB gene from a cholera outbreak in odisha, eastern India. J. Pathog. 2016, 1695410. doi:10.1155/2016/1695410
Jaiswal, A., Sarkar, S., Das, P., Nandy, S., Koley, H., and Sarkar, B. (2015). Trends in the genomic epidemiology of Vibrio cholerae O1 isolated worldwide since 1961. Int. J. Antimicrob. agents 46, 460–464. doi:10.1016/j.ijantimicag.2015.06.012
Kacou-N'Douba, A., Anne, J. C., Okpo, L. S., Elogne-Kouame, C., Koffi, S., Koffi, V., et al. (2012). Antimicrobial resistance of Vibrio cholerae O1 isolated during a cholera epidemic in 2011 in dry season in Cote d'Ivoire. J. Infect. Dev. Ctries. 6, 595–597. doi:10.3855/jidc.2001
Kaistha, N., Mehta, M., Gautam, V., and Gupta, V. (2005). Outbreak of cholera in and around Chandigarh during two successive years (2002, 2003). Indian J. Med. Res. 122, 404–407.
Kar, S. K., Pal, B. B., Khuntia, H. K., Achary, K. G., and Khuntia, C. P. (2015). Emergence and spread of tetracycline resistant Vibrio cholerae O1 El Tor variant during 2010 cholera epidemic in the tribal areas of Odisha, India. Int. J. Infect. Dis. 33, 45–49. doi:10.1016/j.ijid.2014.12.025
Karki, R., Bhatta, D. R., Malla, S., and Dumre, S. P. (2010). Cholera incidence among patients with diarrhea visiting National Public Health Laboratory, Nepal. Jpn. J. Infect. Dis. 63, 185–187. doi:10.7883/yoken.63.185
Kiiru, J., Mutreja, A., Mohamed, A. A., Kimani, R. W., Mwituria, J., Sanaya, R. O., et al. (2013). A study on the geophylogeny of clinical and environmental Vibrio cholerae in Kenya. PLoS One 8, e74829. doi:10.1371/journal.pone.0074829
Kingston, J. J., Thavachelvam, K., Tuteja, U., James, T., Janardhanan, B., and Batra, H. V. (2009). Antimicrobial susceptibility and molecular characterization of Vibrio cholerae from cholera outbreaks in Chennai. Indian J. Microbiol. 49, 84–88. doi:10.1007/s12088-009-0007-z
Koley, H., Ray, N., Chowdhury, G., Barman, S., Mitra, S., Ramamurthy, T., et al. (2014). Outbreak of cholera caused by Vibrio cholerae O1 El Tor variant strain in Bihar, India. Jpn. J. Infect. Dis. 67, 221–226. doi:10.7883/yoken.67.221
Kondo, S., Kongmuang, U., Kalnauwakul, S., Matsumoto, C., Chen, C. H., and Nishibuchi, M. (2001). Molecular epidemiologic analysis of Vibrio cholerae O1 isolated during the 1997-8 cholera epidemic in southern Thailand. Epidemiol. Infect. 127, 7–16. doi:10.1017/s0950268801005660
Krishna, B. V., Patil, A. B., and Chandrasekhar, M. R. (2006). Fluoroquinolone-resistant Vibrio cholerae isolated during a cholera outbreak in India. Trans. R. Soc. Trop. Med. Hyg. 100, 224–226. doi:10.1016/j.trstmh.2005.07.007
Kumar, P., Mishra, D., Deshmukh, D., Jain, M., Zade, A., Ingole, K., et al. (2014). Haitian variant ctxB producing V ibrio cholerae O1 with reduced susceptibility to ciprofloxacin is persistent in Y avatmal, M aharashtra, I ndia, after causing a cholera outbreak. Clin. Microbiol. Infect. 20, O292–O293. doi:10.1111/1469-0691.12393
Kumar, R., and Lalitha, K. V. (2013). Prevalence and molecular characterization of Vibrio cholerae O1, non-O1 and non-O139 in tropical seafood in Cochin, India. Foodborne Pathog. Dis. 10, 278–283. doi:10.1089/fpd.2012.1310
Kumaran, T., and Citarasu, T. (2016). Isolation and characterization of Vibrio species from shrimp and Artemia culture and evaluation of the potential virulence factor. Intel. Prop. Rights 4. doi:10.4172/2375-4516.1000153
Kutar, B. M., Rajpara, N., Upadhyay, H., Ramamurthy, T., and Bhardwaj, A. K. (2013). Clinical isolates of Vibrio cholerae O1 el tor ogawa of 2009 from Kolkata, India: Preponderance of SXT element and presence of Haitian ctxB variant. PLoS One 8, e56477. doi:10.1371/journal.pone.0056477
Li, B. S., Tan, H. L., Wang, D. C., Deng, X. L., Chen, J. D., Zhong, H. J., et al. (2011). Phenotypic and genotypic characterization Vibrio cholerae O139 of clinical and aquatic isolates in China. Curr. Microbiol. 62, 950–955. doi:10.1007/s00284-010-9802-3
Liu, C., Wang, Y., Azizian, K., Omidi, N., Hassan Kaviar, V., Kouhsari, E., et al. (2022). Antimicrobial resistance in Vibrio cholerae O1/O139 clinical isolates: A systematic review and meta-analysis. Expert Rev. Anti Infect. Ther. 20, 1217–1231. doi:10.1080/14787210.2022.2098114
Luo, Y., Ye, J., Jin, D., Ding, G., Zhang, Z., Mei, L., et al. (2013). Molecular analysis of non-O1/non-O139 Vibrio cholerae isolated from hospitalised patients in China. BMC Microbiol. 13, 52. doi:10.1186/1471-2180-13-52
Mandal, S., Debmandal, M., and Pal, N. K. (2010). Plasmid mediated antibiotic resistance of Vibrio cholerae O1 biotype El Tor serotype Ogawa associated with an outbreak in Kolkata, India. Asian Pac. J. Trop. Med. 3, 637–641. doi:10.1016/s1995-7645(10)60154-x
Mandal, S., Pal, N. K., Chowdhury, I. H., and Debmandal, M. (2009). Antibacterial activity of ciprofloxacin and trimethoprim, alone and in combinittion, against Vibrio cholerae O1 biotype El Tor serotype Ogawa isolates. Pol. J. Microbiol. 58, 57–60.
Mandomando, I., Espasa, M., Valles, X., Sacarlal, J., Sigauque, B., Ruiz, J., et al. (2007). Antimicrobial resistance of Vibrio cholerae O1 serotype ogawa isolated in manhica district hospital, southern Mozambique. J. Antimicrob. Chemother. 60, 662–664. doi:10.1093/jac/dkm257
Marashi, S. M. A., Esfahani, B. N., Tavakoli, A., Moghim, S., Pourshafie, M. R., and Salehi, M. (2012). Simultaneous detection of integrase and antibiotic resistance genes within SXT Constin in Vibrio cholerae O1 El Tor strains isolated from Iran using multiplex-PCR. Iran. J. basic Med. Sci. 15, 885–889.
Mercy, N., Mohamed, A. A., Zipporah, N., Chowdhury, G., Pazhani, G. P., Ramamurthy, T., et al. (2014). Phenotypic and genetic characterization of Vibrio cholerae O1 isolated from various regions of Kenya between 2007 and 2010. Pan Afr. Med. J. 8, 8. doi:10.11604/pamj.2014.19.8.2496
Mishra, A., Taneja, N., and Sharma, M. (2011). Demonstration of viable but nonculturable Vibrio cholerae O1 in fresh water environment of India using ciprofloxacin DFA–DVC method. Lett. Appl. Microbiol. 53, 124–126. doi:10.1111/j.1472-765X.2011.03077.x
Mohapatra, S. S., Ramachandran, D., Mantri, C. K., and Singh, D. V. (2007). Characterization of the genetic background of Vibrio cholerae O1 biotype El Tor serotype Inaba strains isolated in Trivandrum, southern India. J. Med. Microbiol. 56, 260–265. doi:10.1099/jmm.0.46868-0
Moyo, S. J., Gro, N., Matee, M. I., Kitundu, J., Myrmel, H., Mylvaganam, H., et al. (2011). Age specific aetiological agents of diarrhoea in hospitalized children aged less than five years in Dar es Salaam, Tanzania. BMC Pediatr. 11, 19–26. doi:10.1186/1471-2431-11-19
Mukhopadhyay, A., Garg, S., Nair, G. B., Kar, S., Ghosh, R., Pajni, S., et al. (1995). Biotype traits and antibiotic susceptibility of Vibrio cholerae serogroup O1 before, during and after the emergence of the O139 serogroup. Epidemiol. Infect. 115, 427–434. doi:10.1017/s0950268800058581
Mukhopadhyay, A. K., Basu, I., Bhattacharya, S., Bhattacharya, M., and Balakrish Nair, G. (1998). Emergence of fluoroquinolone resistance in strains of Vibrio cholerae isolated from hospitalized patients with acute diarrhea in Calcutta, India. Antimicrob. agents Chemother. 42, 206–207. doi:10.1128/AAC.42.1.206
Ngandjio, A., Tejiokem, M., Wouafo, M., Ndome, I., Yonga, M., Guenole, A., et al. (2009). Antimicrobial resistance and molecular characterization of Vibrio cholerae O1 during the 2004 and 2005 outbreak of cholera in Cameroon. Foodborne pathogens Dis. 6, 49–56. doi:10.1089/fpd.2008.0127
Odjadjare, E. E., and Igbinosa, E. O. (2017). Multi-drug resistant Vibrio species isolated from abattoir effluents in Nigeria. J. Infect. Dev. Ctries. 11, 373–378. doi:10.3855/jidc.8097
Okuda, J., Ramamurthy, T., and Yamasaki, S. (2007). The potent antibacterial activity of Sitafloxacin against fluoroquinolone-resistant clinical isolates of Vibrio cholerae O1. Microbiol. Immunol. 51, 467–469. doi:10.1111/j.1348-0421.2007.tb03921.x
Onohuean, H., Agwu, E., and Nwodo, U. (2022). Systematic review and meta-analysis of environmental Vibrio species–antibiotic resistance. Heliyon 8, e08845. doi:10.1016/j.heliyon.2022.e08845
Onyuka, J. H. O., Kakai, R., Onyango, D. M., Arama, P. F., Gichuki, J., and Ofulla, A. V. O. (2011). Prevalence and antimicrobial susceptibility patterns of enteric bacteria isolated from water and fish in lake victoria basin of western Kenya.
Osawa, K., Shigemura, K., Kitagawa, K., Kuntaman, K., Mertaniasih, N. M., Setyarini, W., et al. (2020). Difference of phenotype and genotype between human and environmental: Isolated Vibrio cholerae in surabaya, Indonesia. Indian J. Microbiol. 60, 230–238. doi:10.1007/s12088-020-00861-y
Ottaviani, D., Medici, L., Talevi, G., Napoleoni, M., Serratore, P., Zavatta, E., et al. (2018). Molecular characterization and drug susceptibility of non-O1/O139 V. cholerae strains of seafood, environmental and clinical origin, Italy. Food Microbiol. 72, 82–88. doi:10.1016/j.fm.2017.11.011
Oyofo, B. A., Lesmana, M., Subekti, D., Tjaniadi, P., Larasati, W., Putri, M., et al. (2002a). Surveillance of bacterial pathogens of diarrhea disease in Indonesia. Diagn Microbiol. Infect. Dis. 44, 227–234. doi:10.1016/s0732-8893(02)00454-6
Oyofo, B. A., Subekti, D., Tjaniadi, P., Machpud, N., Komalarini, S., Setiawan, B., et al. (2002b). Enteropathogens associated with acute diarrhea in community and hospital patients in Jakarta, Indonesia. FEMS Immunol. Med. Microbiol. 34, 139–146. doi:10.1111/j.1574-695X.2002.tb00615.x
Pal, B. B., Khuntia, H. K., Samal, S. K., Das, S. S., and Chhotray, G. P. (2006). Emergence of Vibrio cholerae O1 biotype El Tor serotype Inaba causing outbreaks of cholera in Orissa, India. Jpn. J. Infect. Dis. 59, 266–269.
Pal, B. B., Nayak, S. R., and Khuntia, H. K. (2017). Epidemiology and antibiogram profile of Vibrio cholerae isolates between 2004-2013 from Odisha, India. Japanese journal of infectious diseases. JJID 71, 193. doi:10.7883/yoken.JJID.2017.193
Pan, J. C., Ye, R., Wang, H. Q., Xiang, H. Q., Zhang, W., Yu, X. F., et al. (2008). Vibrio cholerae O139 multiple-drug resistance mediated by Yersinia pestis pIP1202-like conjugative plasmids. Antimicrob. Agents Chemother. 52, 3829–3836. doi:10.1128/AAC.00375-08
Parveen, S., Farrah, S. R., Gonzalez-Bonilla, C., Zamudio, A. V., and Tamplin, M. L. (2003). Characterization of a clinical Vibrio cholerae O139 isolate from Mexico. Can. J. Microbiol. 49, 65–70. doi:10.1139/w03-004
Patrick, G., Nishibuchi, M., Tunung, R., and Son, R. (2012). Molecular characterization of clinical isolate of Vibrio cholerae isolated from outbreaks cases in Malaysia. Int. Food Res. J. 19, 1267.
Prabhu, D. I., Pandian, R. S., and Vasan, P. T. (2007). Pathogenicity, antibiotic susceptibility and genetic similarity of environmental and clinical isolates of Vibrio cholerae. Indian J. Exp. Biol. 45, 817–823.
Radu, S., Vincent, M., Apun, K., Abdul-Rahim, R., Benjamin, P. G., Yuherman, , et al. (2002). Molecular characterization of Vibrio cholerae O1 outbreak strains in Miri, Sarawak (Malaysia). Acta Trop. 83, 169–176. doi:10.1016/s0001-706x(02)00110-9
Rahbar, M., Zahraei, M., Omidvarnia, A., Afshani, M. T., Glami, M., Sabourian, R., et al. (2010). Survey of epidemiology and bacteriology features of cholera in Iran. Asian Pac. J. Trop. Med. 3, 45–47. doi:10.1016/s1995-7645(10)60030-2
Ranjbar, R., Sadeghy, J., Shokri Moghadam, M., and Bakhshi, B. (2016). Multi-locus variable number tandem repeat analysis of Vibrio cholerae isolates from 2012 to 2013 cholera outbreaks in Iran. Microb. Pathog. 97, 84–88. doi:10.1016/j.micpath.2016.05.023
Rashed, S. M., Mannan, S. B., Johura, F. T., Islam, M. T., Sadique, A., Watanabe, H., et al. (2012). Genetic characteristics of drug-resistant Vibrio cholerae O1 causing endemic cholera in Dhaka, 2006-2011. J. Med. Microbiol. 61, 1736–1745. doi:10.1099/jmm.0.049635-0
Rezaie, N., Bakhshi, B., and Najar-Peerayeh, S. (2017). Distribution of resistance genetic determinants among Vibrio cholerae isolates of 2012 and 2013 outbreaks in IR Iran. Microb. Pathog. 104, 12–16. doi:10.1016/j.micpath.2017.01.005
Roy, S., Parande, M. V., Mantur, B. G., Bhat, S., Shinde, R., Parande, A. M., et al. (2012). Multidrug-resistant Vibrio cholerae O1 in belgaum, south India. J. Med. Microbiol. 61, 1574–1579. doi:10.1099/jmm.0.049692-0
Saidi, S. M., Chowdhury, N., Awasthi, S. P., Asakura, M., Hinenoya, A., Iijima, Y., et al. (2014). Prevalence of Vibrio cholerae O1 El Tor variant in a cholera-endemic zone of Kenya. J. Med. Microbiol. 63, 415–420. doi:10.1099/jmm.0.068999-0
Sambe-Ba, B., Diallo, M. H., Seck, A., Wane, A. A., Constantin DE Magny, G., Boye, C. S., et al. (2017). Identification of atypical el TorV. cholerae O1 ogawa hosting SXT element in Senegal, Africa. Front. Microbiol. 8, 748. doi:10.3389/fmicb.2017.00748
Sarkar, A., Morita, D., Ghosh, A., Chowdhury, G., Mukhopadhyay, A. K., Okamoto, K., et al. (2019). Altered integrative and conjugative elements (ICEs) in recent Vibrio cholerae O1 isolated from cholera cases, Kolkata, India. Front. Microbiol. 10, 2072. doi:10.3389/fmicb.2019.02072
Sedaghat, M., Rahimi, F., Talebi, M., and Pourshafie, M. (2012). Serotyping, antibiotic susceptibility pattern and detection of hlyA gene among Cholera patients in Iran. Jundishapur J. Microbiol. 6, 20–23. doi:10.5812/jjm.4709
Shakya, G., Kim, D. W., Clemens, J. D., Malla, S., Upadhyaya, B. P., Dumre, S. P., et al. (2012). Phenotypic and genetic characterization of Vibrio cholerae O1 clinical isolates collected through national antimicrobial resistance surveillance network in Nepal. World J. Microbiol. Biotechnol. 28, 2671–2678. doi:10.1007/s11274-012-1077-3
Sharma, N., Mandal, P., Dhillon, R., and Jain, M. (2007). Changing profile of Vibrio cholerae O1, O139 in Delhi and its periphery (2003-2005). Indian J. Med. Res. 125, 633–640.
Singh, B., Tyagi, A., Billekallu Thammegowda, N. K., and Ansal, M. D. (2018). Prevalence and antimicrobial resistance of vibrios of human health significance in inland saline aquaculture areas. Aquac. Res. 49, 2166–2174. doi:10.1111/are.13672
Song, Y., Yu, P., Li, B., Pan, Y., Zhang, X., Cong, J., et al. (2013). The mosaic accessory gene structures of the SXT/R391-like integrative and conjugative elements derived from Vibrio spp. isolated from aquatic products and environment in the Yangtze River Estuary, China. BMC Microbiol. 13, 214. doi:10.1186/1471-2180-13-214
Talkington, D., Bopp, C., Tarr, C., Parsons, M. B., Dahourou, G., Freeman, M., et al. (2011). Characterization of toxigenic Vibrio cholerae from Haiti, 2010-2011. Emerg. Infect. Dis. 17, 2122–2129. doi:10.3201/eid1711.110805
Torane, V., Kuyare, S., Nataraj, G., Mehta, P., Dutta, S., and Sarkar, B. (2016). Phenotypic and antibiogram pattern of V. cholerae isolates from a tertiary care hospital in Mumbai during 2004-2013: A retrospective cross-sectional study. BMJ Open 6, e012638. doi:10.1136/bmjopen-2016-012638
Tran, H. D., Alam, M., Trung, N. V., VAN Kinh, N., Nguyen, H. H., Pham, V. C., et al. (2012). Multi-drug resistant Vibrio cholerae O1 variant El Tor isolated in northern Vietnam between 2007 and 2010. J. Med. Microbiol. 61, 431–437. doi:10.1099/jmm.0.034744-0
Vijayalakshmi, N., Rao, R., and Badrinath, S. (1997). Minimum inhibitory concentration (MIC) of some antibiotics against Vibrio cholerae O139 isolates from Pondicherry. Epidemiol. Infect. 119, 25–28. doi:10.1017/s0950268897007553
Vinothkumar, K., Bhalara, S. R., Shah, A., Ramamurthy, T., Niyogi, S. K., Kumar, G. N., et al. (2018). Involvement of topoisomerase mutations and qnr and aac(6')Ib-cr genes in conferring quinolone resistance to clinical isolates of Vibrio and Shigella spp. from Kolkata, India (1998-2009). J. Glob. Antimicrob. Resist 13, 85–90. doi:10.1016/j.jgar.2017.10.013
Wang, R., Yu, D., Yue, J., and Kan, B. (2016). Variations in SXT elements in epidemic Vibrio cholerae O1 El Tor strains in China. Sci. Rep. 6, 22733. doi:10.1038/srep22733
Waturangi, D. E., Wennars, M., Suhartono, M. X., and Wijaya, Y. F. (2013). Edible ice in Jakarta, Indonesia, is contaminated with multidrug-resistant Vibrio cholerae with virulence potential. J. Med. Microbiol. 62, 352–359. doi:10.1099/jmm.0.048769-0
Yuan, X.-H., Li, Y.-M., Vaziri, A. Z., Kaviar, V. H., Jin, Y., Jin, Y., et al. (2022). Global status of antimicrobial resistance among environmental isolates of Vibrio cholerae O1/O139: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 11, 62–11. doi:10.1186/s13756-022-01100-3
Zachariah, R., Harries, A. D., Arendt, V., Nchingula, D., Chimtulo, F., Courteille, O., et al. (2002). Characteristics of a cholera outbreak, patterns of Vibrio cholerae and antibiotic susceptibility testing in rural Malawi. Trans. R. Soc. Trop. Med. Hyg. 96, 39–40. doi:10.1016/s0035-9203(02)90233-6
Keywords: antibiotic resistance, resistance, Vibrio cholera, V. cholera, meta-analysis
Citation: Nateghizad H, Sajadi R, Shivaee A, Shirazi O, Sharifian M, Tadi DA and Amini K (2023) Resistance of Vibrio cholera to antibiotics that inhibit cell wall synthesis: A systematic review and meta-analysis. Front. Pharmacol. 14:1027277. doi: 10.3389/fphar.2023.1027277
Received: 24 August 2022; Accepted: 10 January 2023;
Published: 20 March 2023.
Edited by:
Tao Li, Shanghai Veterinary Research Institute (CAAS), ChinaReviewed by:
Jitendraa Vashistt, Jaypee University of Information Technology, IndiaDurg Vijai Singh, Central University of South Bihar, India
Jianmin Zhang, South China Agricultural University, China
Copyright © 2023 Nateghizad, Sajadi, Shivaee, Shirazi, Sharifian, Tadi and Amini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kumarss Amini, RHJfa3VtYXJzc19hbWluaUB5YWhvby5jb20=
 Hossein Nateghizad1
Hossein Nateghizad1 Kumarss Amini
Kumarss Amini