
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 23 November 2022
Sec. Inflammation Pharmacology
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.993498
This article is part of the Research Topic Natural Compounds Regulating Epigenetics for Treating Chronic Inflammatory Diseases View all 8 articles
 Lu Qiu1,2†
Lu Qiu1,2† Min Zhang1,3†
Min Zhang1,3† Chenghao Li1,2†
Chenghao Li1,2† Yehu Hou1,2
Yehu Hou1,2 Hao Liu1,3
Hao Liu1,3 Jia Lin1,3
Jia Lin1,3 Juan Yao1,3
Juan Yao1,3 Dong Zhu Duan4
Dong Zhu Duan4 Yi Xi Zhang1,3
Yi Xi Zhang1,3 Mi Li1,3
Mi Li1,3 Ya Ling Li1,2
Ya Ling Li1,2 Peng Wang1
Peng Wang1 Jin Tian Li2
Jin Tian Li2 Xiao Jie Jin1,2,3*
Xiao Jie Jin1,2,3* Yong Qi Liu1,2*
Yong Qi Liu1,2*Osteoarthritis (OA) is a multifactorial and chronic degenerative joint disease. Due to the adverse effects of currently used drugs, a safer and more effective therapy for treating OA is needed. Peroxisome proliferator-activated receptor-γ (PPARγ) is a key protein protecting cartilage. DNMT1-mediated hypermethylation of PPARγ promoter leads to its suppression. Therefore, DNMT1 might be an effective target for exerting cartilage protective effects by regulating the epigenetic expression of PPARγ. Dabushen decoction (DD) is a representative prescription of Dunhuang ancient medical prescription, which has a potential therapeutic effect on OA. So far, the research of the efficacy and material basis of DD in the treatment of OA remains unclear. In this study, Micro-CT, HE staining, S-O staining, and immunohistochemistry analysis were used to demonstrate that DD increased the expression of PPARγ and collagen synthesis in an OA rat model. Next, the structure of DNMT1 was used to screen the active constituents of DD by molecular docking method for treatment OA. Seven potential active constituents, including isoliquiritigenin, emodin, taxifolin, catalpol, alisol A, zingerone, and schisandrin C were hited. The protective effect of the potential active constituents to chondrocytes were evaluated by protein capillary electrophoresis, immunofluorescence assays, and ex vivo culture of rat knee cartilage. The five constituents, such as alisol A, emodin, taxifolin, isoliquiritigenin, and schisandrin C could promote the expression of PPARγ and ameliorate IL-1β-induced downregulation of collagen II and the production of MMP-13. Alisol A and Emodin could effectively mitigate cartilage damage. At last, molecular dynamics simulations with MM-GBSA method was applied to investigate the interaction pattern of the active constituents and DNMT1 complexes. The five constituents, such as alisol A, emodin, taxifolin, isoliquiritigenin, and schisandrin C achieved a stable binding pattern with DNMT1, in which alisol A has a relatively high binding free energy. In conclusion, this study elucidates that the active constituents of DD (alisol A, emodin, taxifolin, isoliquiritigenin, and schisandrin C) could ameliorate osteoarthritis via PPARγ preservation by targeting DNMT1.These findings facilitated clinical use of DD and provided a valuable strategy for developing natural epigenetic modulators from Chinese herbal formula.
Osteoarthritis (OA), a disease of joint degeneration characterized by articular cartilage (AC) loss, often causes pain, swelling, stiffness, and joint deformation (Pigeolet et al., 2021). It is a multifactorial disease that can be caused by injury, genetics, age, obesity and gender, etc (Wang et al., 2011). Numerous studies have suggested that the pathogenesis of OA mainly involves inflammatory reactions, metabolic disorders, and imbalanced oxidative stress (Barter et al., 2012; Robinson et al., 2016; Dickson et al., 2019; Ansari et al., 2020). Pharmacologic therapy of OA is mainly aimed at pain relief, including non-steroidal anti-inflammatory drugs and opioids (Abramoff and Caldera, 2020; D'Arcy et al., 2021; da Costa et al., 2021). However, the adverse effects of these drugs, including hepato and nephrotoxicity, loss of gastrointestinal motility and physical and psychological dependence emphasize the need to develop a safer and more effective therapy for treating OA.
A reported protective role for peroxisome proliferator-activated receptor-γ (PPARγ) in OA raises the possibility that upregulation of PPARγ may be beneficial in the context of preventing and treating OA (Afif et al., 2007). PPARγ is a member of ligand-activated nuclear transcription factor family and plays an essential role in cartilage health; the lack of PPARγ accelerates the onset of spontaneous OA (Vasheghani et al., 2015). PPARγ is expressed and functionally active in chondrocytes (Afif et al., 2007). Research has shown that PPARγ activators display anti-inflammatory and chondroprotective properties in vitro and improve the clinical course and histopathological features in an experimental animal model of OA (Bordji et al., 2000; Kobayashi et al., 2005; Li et al., 2016). Therefore, increasing the expression of PPARγ in cartilage might be an effective strategy to slow or prevent OA progression.
Recent studies have clearly shown that aberrant DNA methylation can lead to PPARγ suppression (Zhu et al., 2019). DNA methylation reactions are catalyzed by the DNA methyltransferase family of enzymes (DNMT1, DNMT3A and DNMT3B) (Grandi and Bhutani, 2020; Chen et al., 2021a). DNMT1 is a maintenance methyltransferase, copying methylation patterns after DNA replication (Foulks et al., 2012). It is reported that PPARγ suppression in OA cartilage can be caused by aberrantly induced DNMT1 activity and the associated promoter hypermethylation (Zhu et al., 2019). This suggests that DNMT1 might be a valuable target for protecting and preserving PPARγ activity in OA.
Traditional Chinese medicine (TCM) has been used to treat OA for many years (Lai et al., 2015; Zhang et al., 2015; Yao et al., 2021; Sun et al., 2022). “Kidney Governing Bone” is a classical theory in TCM (Jiang et al., 2022). According to TCM syndromes, modifying kidney function is regarded as the one of the main treatments for OA. Clinical studies have demonstrated that stimulating kidney function has a beneficial effect in the therapy of OA (Sun et al., 2022). Experimental studies have shown that kidney stimulant prescriptions have beneficial effects on OA animals (Gao et al., 2012; Yuan et al., 2012). It was reported that decreased collagen synthesis resulted in the degeneration of articular cartilage of the knee in several kidney deficiency rat models (Yao et al., 2013). Dabushen decoction (DD) of Dunhuang ancient medical prescription is made up of seven Chinese herbs: Rehmanniae radix praeparata, Lophatheri herba, Schisandrae chinensis fructus, Zingiberis rhizoma, Cinnamomi ramulus, Alismatis rhizoma, Glycyrrhizae radix et rhizoma. In TCM, the main function of DD is to stimulate kidney function and strengthen bones, which is believed to play an important role in the treatment of OA. However, there are few studies on the protective effect of DD in vivo. In addition, the active constituents of DD for the treatment of OA remain unclear. In this study we first established a rat model of OA to evaluate the efficacy of DD in the treatment of osteoarthritis. Based on the effectiveness, the target DNMT1 was selected to identify the active constituents of DD in ameliorating osteoarthritis, presumably by preservation of PPARγ expression. Our results provide data for the scientific formulation of DD for the treatment of OA and provide a way for the discovery of natural epigenetic modulators from Chinese herbal formula.
Rehmanniae radix praeparata, Lophatheri herba, Schisandra chinensis Baill, Zingiberis rhizoma, Cinnamomi ramulus, Alismatis rhizoma and Glycyrrhizae radix et rhizoma were obtained from affiliated Hospital of Gansu University of Chinese Medicine (Lanzhou, China).
Taxifolin, emodin, isoliquiritigenin, schisandrin C, and alisol A were purchased from PUSH (Chengdu, China). Catalpol and zingerone were purchased from MedChemExpress (NJ, United States). SGI-1027 was purchased from TOPSCIENCE (Shanghai, China). Purity of all reference standards was greater than 98%.
Papain and L-cysteine were procured from Sigma (St. Louis, United States). Fetal bovine serum (FBS) and Dulbecco’s Modified Eagle’s Medium F12 (DMEM/F12) were acquired from Gibco (NY, United States). Primary antibodies against GAPDH were purchased from Proteintech Group (Wuhan, China) and against collagen II, MMP-13 and PPARγ were purchased from Abcam (Cambridge, United Kingdom). ProteinSimple 042–206 was purchased from Bio-Techne (Minneapolis, United States). Enzyme-linked immunosorbent assay (ELISA) kits to detect prostaglandin E2 (PGE2) and tumour necrosis factor alpha (TNF-α) were purchased from Elabscience (Wuhan, China). 4% paraformaldehyde, EDTA decalcifying solution, 0.25% trypsin digestion solutions, collagenase, toluidine blue O solution, modified Safranine O-Fast Green FCF Cartilage Stain kit, RIPA buffer, penicillin-streptomycin liquid, HE Staining kit, 50× TAE Buffer, DMSO and Mayer’s Hematoxylin solution were procured from Solarbio (Beijing, China). Griess reagent was purchased from Beyotime Institute of Biotechnology (Shanghai, China). 3,3′-diaminobenzidine was purchased from BOSTER (Wuhan, China). MaxVision HRP-Polymer anti-Mouse IHC kit was purchased from MXB (Fuzhou, China). Cell counting kit-8 (CCK-8) and DAPI Fluoromount-G were purchased from Meilunbio (Dalian, China). Tissue/cell DNA extraction reagent kit was purchased from Bioteke (Wuxi, China). DNA Bisulfite Conversion kit, Methylation-specific PCR (MSP) kit and Marker (DNA) were purchased from Tiangen Biotech (Beijing, China). Agarose was purchased from YE SEN (Shanghai, China). GelstainRed was purchased from ACRO (Beijing, China). Loading buffer (DNA) was purchased from Vazyme (Nanjing, China).
All experiments involving animals were approved by the Ethics Committee of Laboratory Animal Center of Gansu University of Chinese Medicine (Lanzhou, China). A total of 27 6-week-old female Specific Pathogen Free (SPF) Sprague-Dawley rats (200 ± 20 g) were used for the in vivo experiment (9 rats for each group). All animals were housed in a clean vivarium (specific pathogen free) at room temperature and allowed free access to standard pelleted forage and tap water. All rats were fed for 1 week for acclimatization before experiments.
DD was purchased from Affiliated Hospital of Gansu University of Chinese Medicine (Lanzhou, China). Rats were randomly divided into three groups: control, OA, DD-treated. On days 1, 4 and 7, the double knee joints of OA and DD-treated groups were injected with 0.2 ml 4% papain with L-cysteine, and double knee joints of control group were injected with an equal volume of sterile saline solution.
DD was purchased from Affiliated Hospital of Gansu University of Chinese Medicine (Lanzhou, China). DD-treated group rats with induced OA received 5.85 g/kg of DD (1 ml/100 g weight) orally. Rats in the control and OA groups were treated with an equal volume of Physiological saline. All groups were treated once a day for 6 consecutive weeks. Serum was collected from rats in all groups after anesthesia, and the knees were harvested after sacrifice.
At week 6 post-treatment the fixed knee samples were scanned by a high resolution Micro-CT (Micro-CT 80, SCANCO MEDICAL AG, Switzerland, Voltage = 55 kV, Current = 145 μA, Voxel size = 11.6 μm). We defined the region of interest (ROI) to cover the whole subchondral bone in tibial plateaus. Built-in software in the Micro-CT system was used for three-dimensional reconstruction. Bone volume/tissue volume (BV/TV) of tibial subchondral bone was evaluated in the Micro-CT system (Chen et al., 2019).
Knee joint samples were fixed in 4% paraformaldehyde for 24 h and decalcified in 10% EDTA solution for 4 weeks, after which the samples were dehydrated by graded alcohol, clarified, and embedded in paraffin blocks. Sections were stained with Safranin-O-Fast green (S-O) and Hematoxylin-eosin (HE). The Osteoarthritis Research Society International (OARSI) scoring system was used to evaluate the degree of pathological cartilage damage. All sections were scored by two experienced observers blinded to the study.
Immunohistochemistry sections were deparaffinized and rehydrated using a graded ethanol series and incubated overnight at 4°C with antibodies to PPARγ (1:100), collagen II (1:500), and MMP-13 (1:200). The slides were then treated with secondary antibodies, biotinylated anti-mouse IgG, and then streptavidin peroxidase complex (MXB, Fuzhou, China) for 10 min, and then 3,3′-diaminobenzidine (BOSTER, Wuhan, China) was added. The slides were counterstained with Mayer’s hematoxylin and photographed under a photomicroscope (Olympus, Tokyo, Japan). The percentage of positivity of each immunohistochemical assay was measured using ImageJ software (NIH, Bethesda, MD). Five representative images were selected for each donor (n = 3) and used for relative quantification. The negative controls staining is shown in Supplementary Figure S1.
Sandwich enzyme-linked immunosorbent assay (ELISA) kits were used to detect the levels of inflammatory factors IL-1β, PGE2, and TNF-α in the serum of each group of rats. The ELISA procedure was performed according to the manufacturer’s instructions. The NO level in serum was detected by the Griess reagent.
The structures of DD’s constituents were collected from the Traditional Chinese Medicines Systems Pharmacology (TCMSP, http://tcmspw.com/) (Ru et al., 2014) and Traditional Chinese Medicines Integrated Database (TCMID, http://www.megabionet.org/tcmid/) (Chen et al., 2006). Constituents of each herb were identified through a literature search in PubMed database (https://pubmed.ncbi.nlm.nih.gov/). A total of 906 constituents were retrieved for 7 herbs, including Rehmanniae radix praeparata (76), Lophatheri herba (4), Schisandrae chinensis fructus (129), Zingiberis rhizoma (148), Cinnamomi ramulus (221), Atractylodes lancea (49), Glycyrrhizae radix et rhizoma (279).
Molecular docking was carried out using the Glide module with the standard docking precision method (Glide SP) in Schrӧ; dinger 2020—4 (Friesner et al., 2006). Ligands were then preprocessed by LigPrep (MMFFs force fields) to generate the 3D structures and the energies of the generated conformations were minimized. The ionized states of the ligands were assigned via Epik at the pH value of 7.0 ± 2.0. The crystallographic structure of DNMT1 was obtained from Protein Data Bank (PDB ID: 3PTA (Hou et al., 2011)). Missing amino acids of the DNMT1 (PDB ID: 3PTA (Hou et al., 2011)) were added by method of the multiple templates homology modelling in the web server of SWISS-MODEL database (Waterhouse et al., 2018). The prepared structure was processed by Protein Preparation Wizard module in Maestro before calculating the docking grid, including adding hydrogen atoms and assigning protonation states and partial charges with OPLS_2005 force field. The pocket for docking was generated using the Receptor Grid Generation module by defining the S-adenosyl-l-homocysteine (SAH) as the center of the grid (Krishna et al., 2017).
Three-week-old Sprague-Dawley (SD) rats were euthanized and immersed in 75% medicinal alcohol for 15 min (Jiao et al., 2013). Cartilage collected from knee joints was minced and digested with 0.25% trypsin for 30 min (Tu et al., 2019b). Cartilage fragments were fully digested with 0.2% Collagenase for 4 h. Subsequently, cell suspensions were centrifuged (1,500 r/min, 5 min) to harvest primary chondrocytes. The isolated cells were resuspended in DMEM/F12 supplemented with 10% FBS and 1% penicillin/streptomycin solution at 37°C with 5% CO2. Toluidine blue staining and Collagenase immunofluorescence staining were utilized to identify primary rat chondrocytes. To maintain stability of the cell phenotype, the second- and third-generation cells were routinely selected for subsequent cell experiments (Tu et al., 2019a).
The cell viability of chondrocytes was measured by the CCK-8 assay according to the manufacturer’s protocol. Briefly, the chondrocytes were seeded into 96-well plates (5 × 103 cells/well) followed by treatment with different concentrations (0, 5, 10, 20, 40, 80 μM) of isoliquiritigenin, emodin, taxifolin, catalpol, alisol A, zingerone, and schisandrin C for 24 h. At the indicated time, 100 µL of DMEM/F12 containing 10 µL of CCK-8 solution was added to each well, and the cells were then incubated for another 2 h at 37°C. The absorbance of the wells was read at 450 nm by microplate reader (Bio-Rad, Hercules, CA).
For capillary electrophoresis, cells were lysed directly in the Laemmli buffer (Solarbio, Beijing, China) containing 1× protease and phosphatase inhibitor cocktail (Solarbio, Beijing, China). For protein capillary electrophoresis, the primary antibodies were used rabbit anti-PPARγ (Cambridge, United States); the secondary antibodies were used with the manufacturer’s instructions: Anti-Rabbit Secondary HRP Antibody (ProteinSimple 042–206).
Chondrocytes were seeded in 24-well plates at a density of 1 × 104 cells/well. Cells were cultured on coverslips and treated with 10 ng/ml IL-1β and with different concentrations of isoliquiritigenin, emodin, taxifolin, catalpol, alisol A, zingerone and schisandrin C for 24 h. Coverslips were washed blocked for 30 min after fixation, and incubated overnight at 4°C with the primary antibodies MMP-13, PPARγ, and collagen II. Incubation with Alexa Fluor 647-conjugated donkey anti-rabbit secondary antibodies was performed for 2 h at room temperature. The nuclei were counterstained with DAPI fluorescent probes and analyzed with a high content analysis system (PerkinElmer, Operetta CLS).
The concentration of TNF-α and PGE2 in cell culture supernatants was determined by using commercial ELISA kits (Elabscience, wuhan) according to the manufacturer’s instructions. The NO level in the culture medium was detected by the Griess reagent.
Cartilage explants were harvested from the bilateral knee joints of 3-week-old SD rats. The explants were initially cultured at 37°C with 5% CO2 in DMEM/F12 containing 10% FBS and 1% penicillin/streptomycin solution for 2 days. Cartilage explants then were transferred to 48-well plates and cultured in serum-free medium containing 10 ng/ml IL-1β with different concentrations of constituents for 3 days (n = 6 explants per condition) (Latourte et al., 2017). Subsequently, explants were collected, fixed in 4% paraformaldehyde and embedded in paraffin. Samples were cut along the coronal plane and stained with HE for cartilage surface observation and Safranin O staining for degradation of cartilage matrix (Headland et al., 2015).
The initial complex structures of the DNMT1-isoliquiritigenin, DNMT1-emodin, DNMT1-taxifolin, DNMT1-catalpol, DNMT1-alisol A, DNMT1-zingerone, and DNMT1-schisandrin C complexes were obtained from molecular docking (Yang et al.) Structure optimization and frequency calculations for the above ligand compounds were computed by Gaussian16 at the level HF/6-31G*. The restrained electrostatic potential was used to describe the partial atomic charges. The general AMBER force field (GAFF) (Wang et al., 2004), the ff14SB force field (Maier et al., 2015) and the Zinc AMBER force field (ZAFF) (Peters et al., 2010) were used for describing the active constituents, proteins and Zn2+, respectively. All systems were solvated in an atomistic TIP3P water box with 15 Å distance around solutes. After adding chloride ions to neutralize the systems, the systems were minimized, heated, and equilibrated. MD simulations (50 ns) were performed at 300 K with 1.0 atmospheric pressure in an NPT ensemble. The trajectory analysis was performed via the cpptraj module in AMBER 20 (Case et al., 2005).
The binding free energies (ΔGbind) of complexes were calculated via the endpoint molecular mechanics generalized Born surface area (MM/GBSA) approach using the following equation Eq. 1, 3: (Hou et al., 2011; Chen et al., 2016)
Furthermore, we decomposed the total binding free energy into each residue by Eq. 4 to identify the key residues responsible for the binding of constituents.
We used 200 snapshots extracted from the last 10 ns trajectories of the system to calculate binding free energy. Residue free energy decomposition was performed to identify the key residues responsible for ligand binding by splitting the total free energy into the energy contributions from individual residue-ligand pairs.
DNA extraction from human chondrocytic SW1353 cells was performed with a tissue/cell DNA extraction reagent kit (Bioteke, China) according to the manufacturer’s instructions. Bisulfite modification of DNA was carried out using the DNA Bisulfite Conversion Kit (Tiangen Biotech, China) following the manufacturer’s instructions. Primers used to amplify a bisulfite converted DNA fragment as follow: Methylated primer TTAATTTATTTTGGATAGGTTACGA (forward) and TCTTAACAATATTTCTAACACCGAA (reverse); unmethylated primer TTAATTTATTTTGGATAGGTTATGA (forward) and TTAAATTTCTTAACAATATTTCTAACACCA (reverse), which amplify the –232/–76 locus. For internal control (Input), the genomic DNA was PCR amplified simultaneously with forward primer TAGTTTTAGGAAGGTAAAGGGAGTG and reverse primer AAATCCCAAAAAAAACACAACAAA. The PCR reaction was performed with a Methylation-specific PCR (MSP) kit (Tiangen Biotech, China) and confirmed by electrophoresis in a 2% agarose gel, visualized under ultraviolet light, and densitometry analysis of each PCR band was analyzed using the Image J program. The relative levels of PCR products were first normalized with input PCR and then presented as ratio of methylated or unmethylated PCR over total PCR products.
All data are expressed as the mean ± SD for each group. One-way analysis of variance (ANOVA) followed by LSD post hoc test was used to compare the significance of the group differences. Comparisons were considered significant for p < 0.05. Each experiment consisted of at least three replicates per condition.
Subchondral bone and articular cartilage make up a functional joint unit, which plays a complementary role in the weight-bearing capacity of the joint (Pan et al., 2009). To explore the effects of DD on the tibia subchondral bone of OA rats, the 2D images and reconstructed 3D images obtained from the Micro-CT were used to assess the microarchitecture of the rat subchondral bone. As revealed in Figure 1A, the tibial subchondral bone showed significant changes between control and OA groups, which displayed osteophyte development and the hardened subchondral bone, resulting in cartilage damage in the OA rats. Meanwhile, we observed that DD treatment markedly alleviated the destruction of the tibial subchondral bone. As revealed in Figure 1B, significant differences were observed in the BV/TV of the tibial subchondral bone between the control (0.691 ± 0.027) (p < 0.05) and OA group (0.727 ± 0.005). The average value of BV/TV was 0.693 ± 0.009 in DD-treated group, which was lower than that of OA group (0.727 ± 0.005) (p < 0.05). These results show that DD treatment prominently alleviated tibial subchondral bone damage relative to the OA group.
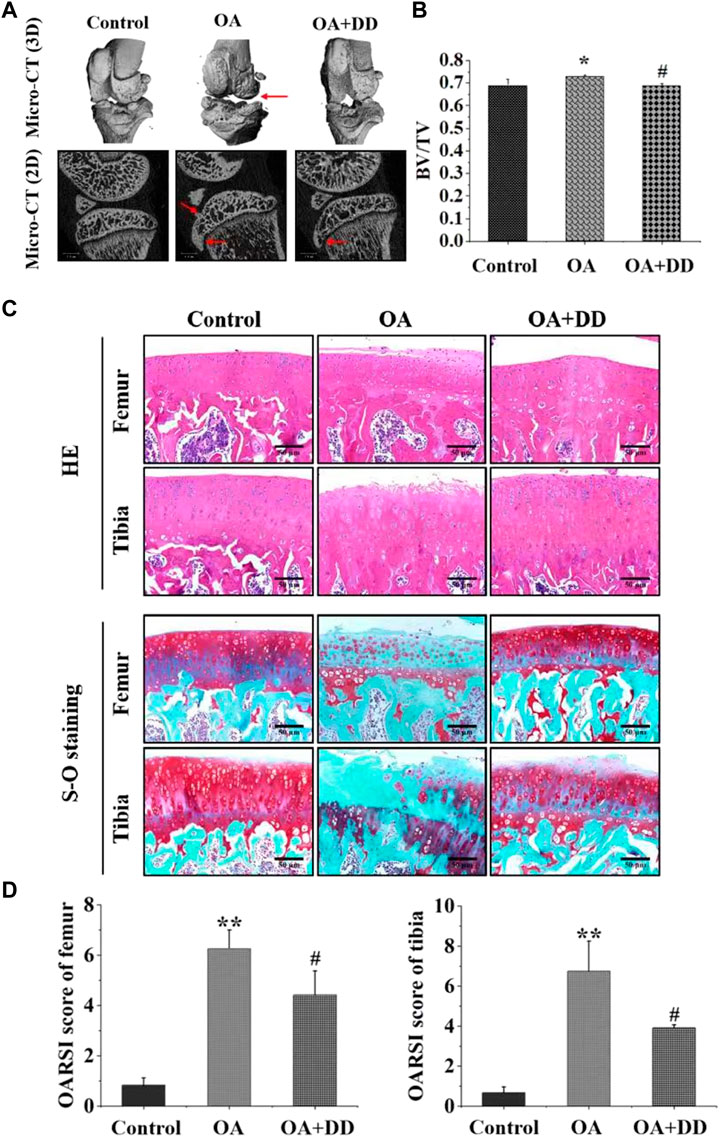
FIGURE 1. DD alleviated tibial subchondral bone and Cartilage damage. (A) Micro-CT was used to analyze 3D images and 2D representative images of DD treated the OA rats, with arrows showing the medial side of the tibial plateau. (B) Quantitative micro-CT analysis of tibial subchondral bone with trabecular bone volume per total volume. (C, D) Representative images of the knee joint stained with HE staining and S-O staining and OARSI scores after 6 weeks of treatment. The data are presented as means ± SD (n = 3). *p < 0.05, **p < 0.01 compared with the control group; #p < 0.05 compared with the OA group.
To explore the effects of DD on the articular cartilage of OA group, histological analysis of OA was conducted by S-O and HE staining. As shown in Figure 1C, HE staining showed that the OA group exhibited a reduction in chondrocytes and articular cartilage thickness with an irregular morphological structure, while articular cartilage in the control and DD-treated group had a regular morphological structure. Meanwhile, S-O staining showed decreased proteoglycans and stromal disturbances in OA group (Figure 1C). By contrast, the matrix degradation in the DD-treated group was less than in the OA group. This was further confirmed by OARSI scores, which was significantly decreased in the DD-treated group compared with the OA group (Figure 1D). Taken together, these results reinforced the protective effect of DD on OA by alleviating cartilage damage in the rat OA models.
PPARγ affects inflammation and extracellular matrix remodeling in OA by regulating transcriptional activity (Afif et al., 2007). We found that the rat OA models demonstrated decreased expression of PPARγ in the articular cartilage. DD inhibited the reduction in PPARγ expression in chondrocytes of OA model rats. MMP-13 and collagen II are key proteins for assessing OA chondrocyte matrix degradation (Wang et al., 2013; Tiku and Madhan, 2016). These results suggest that DD promotes OA matrix formation and inhibits its degradation (Figures 2A–D). Furthermore, we detected the expression of inflammatory cytokines (IL-1β, PGE2, NO, and TNF-α) in the serum by ELISA. As shown in Figure 2E, the production of IL-1β, PGE2, NO, and TNF-α increased significantly in the OA group. In DD-treated group, the release of IL-1β, PGE2, NO, and TNF-α were decreased sharply in the serum. These results demonstrate that DD inhibited the reduction of PPARγ in OA chondrocytes and alleviate cartilage degeneration and inflammation.
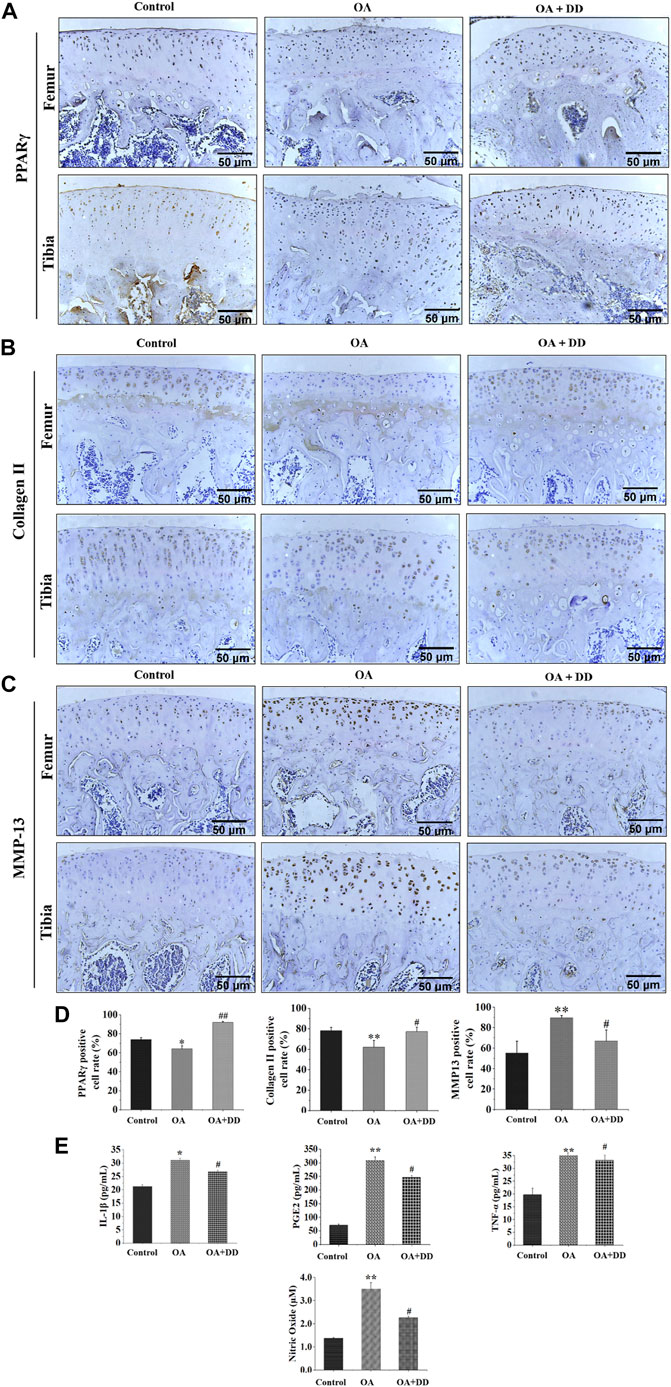
FIGURE 2. DD reduced cartilage ECM degradation and inhibited inflammatory factor production in OA rats. (A) The expression of PPARγ in sections of cartilage; (B) The expression of Collage II in sections of cartilage; (C) The expression of MMP-13 in sections of cartilage; (D) Quantification of histology positive cells of PPARγ, Collage II, and MMP-13; (E) The effect of DD on IL-1β, PGE2, TNF-α, and NO inflammatory cytokines. The data are presented as mean ± SD (n = 3). *p < 0.05, **p < 0.01 compared with the control group; #p < 0.05, ##p < 0.01 compared with the OA group.
DNMT1-mediated hypermethylation of PPARγ promoter in chondrocytes is a key mechanism for promoting OA (Ball et al., 2022). To identify the active constituents of DD for the treatment of OA by targeting DNMT1, 906 constituents from DD were docked into the pocket for DNMT1. In this study, the constituents with a docking score ≤ −5 kcal/mol were defined as potentially active constituents (Kun-Yi et al., 2013). The total number of constituents with potential DNMT1 inhibitory activities was 608 in DD (Supplementary Table S1). Each herb had many potential inhibitory constituents (Supplementary Table S1). Therefore, according to the Pharmacopoeia of the People’s Republic of China (2020) and the literature of on herb main constituents, seven constituents, including isoliquiritigenin, emodin, taxifolin, catalpol, alisol A, zingerone, and schisandrin C which showed high docking scores were considered for further analysis (Table 1).
Chondrocytes were extracted from 3-week-old SD rats. Newly isolated chondrocytes appeared round, whereas most cells cultured for 24 h showed adherent growth. At 7 days of cell culture, chondrocytes showed uniform polygonal or short spindle shapes microscopically (Figure 3A1, A2). Toluidine blue staining and collagen II immunofluorescence staining were utilized to identify primary rat chondrocytes (Figure 3A3, A4).
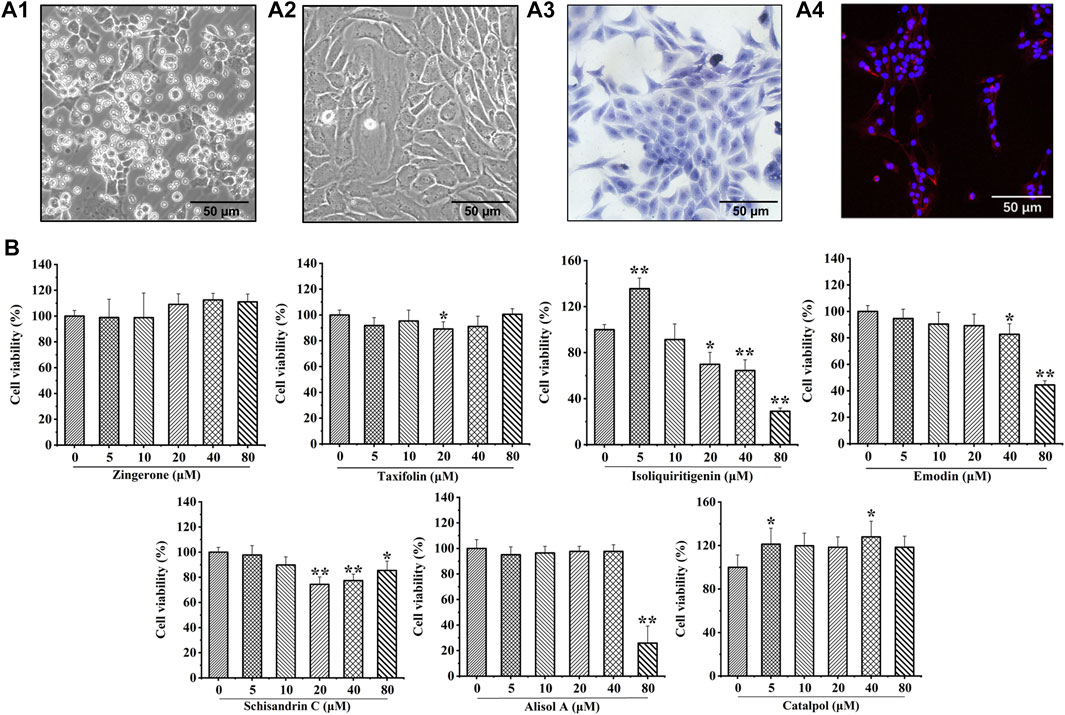
FIGURE 3. Culture and identification of rat chondrocytes, and the effects of seven potential active constituents (Isoliquiritigenin, Emodin, Taxifolin, Catalpol, Alisol A, Zingerone, and Schisandrin C) on rat chondrocytes viability. (A1) Primary rat chondrocytes cultured for 24 h; (A2) Primary rat chondrocytes cultured for 7 days; (A3) Toluidine blue staining of primary rat chondrocytes; (A4) Immunofluorescence staining of collagen II of primary rat chondrocytes; (B) Effect of seven potential active constituents (Isoliquiritigenin, Emodin, Taxifolin, Catalpol, Alisol A, Zingerone, and Schisandrin C) on chondrocytes viability at different concentrations (0, 5, 10, 20, 40, and 80 μM) for 24 h. *p < 0.05, **p < 0.01 compared with the control group.
The cytotoxic effect of isoliquiritigenin, emodin, taxifolin, catalpol, alisol A, zingerone, and schisandrin C were evaluated after treatment with different concentrations (0, 5, 10, 20, 40, and 80 μM) for 24 h using a CCK-8 assay. As shown in Figure 3B, isoliquiritigenin (10 µM), emodin (20 µM), taxifolin (10 µM), catalpol (10 µM), alisol A (20 µM), zingerone (20 µM), and schisandrin C (10 µM) did not affect the viability of chondrocytes. Hence, these concentrations of potential active constituents were chosen for subsequent experiments.
The results of in vivo experiments confirmed that DD can promote the expression of PPARγ in OA chondrocytes. To explore the effect of above seven potential active constituents on PPARγ expression, we used protein capillary electrophoresis to test the expression of PPARγ in chondrocytes after treatment with IL-1β. As shown in Figure 4A, most of the seven constituents reversed the decline of PPARγ in chondrocytes IL-1β-treated.
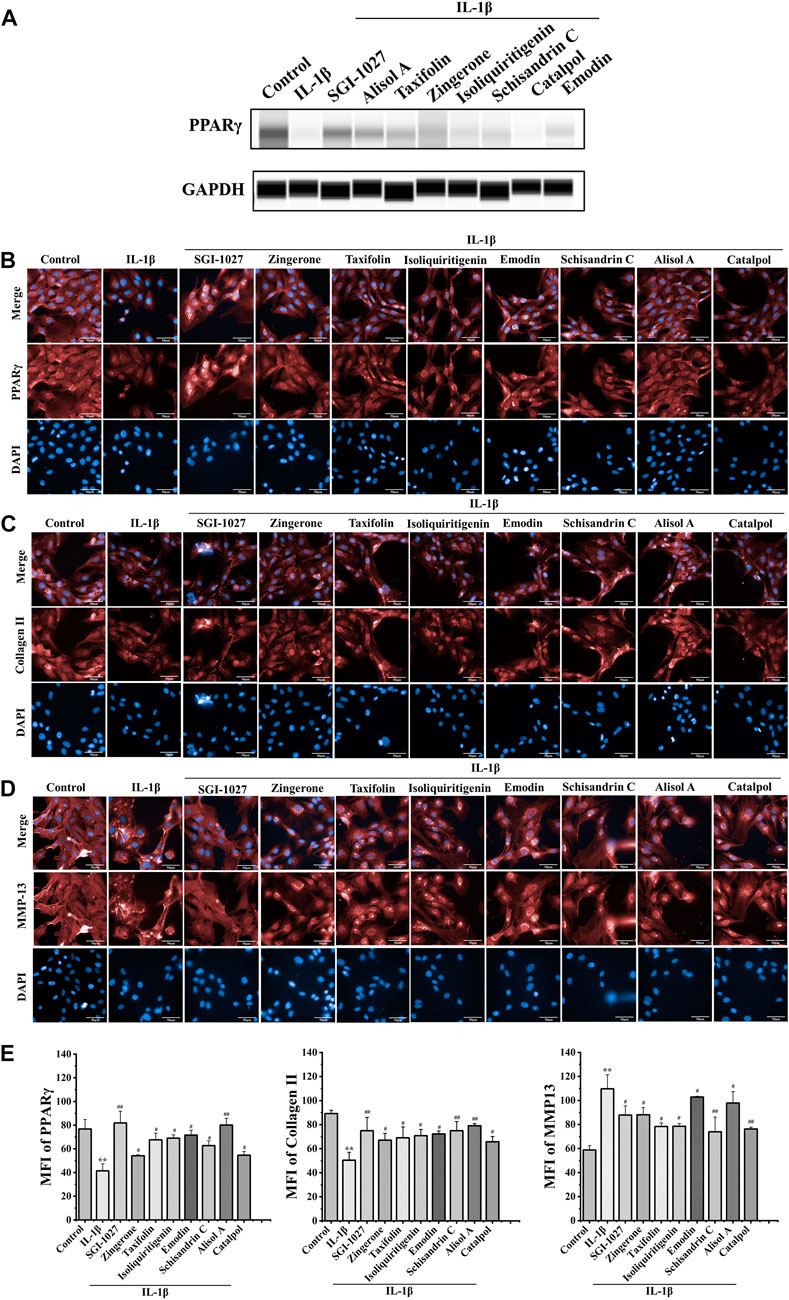
FIGURE 4. Effect of seven potential active constituents (Isoliquiritigenin, Emodin, Taxifolin, Catalpol, Alisol A, Zingerone, and Schisandrin C) on PPARγ, collagen II, and MMP-13 in IL-1β-treated chondrocytes. (A) The expression of PPARγ in IL-1β-treated chondrocytes by protein capillary electrophoresis; (B) PPARγ (red) was detected by immunofluorescence; (C) Collagen II (red) was detected by immunofluorescence; (D) MMP-13 (red) was detected by immunofluorescence; (E) Quantification analysis of the immunofluorescence of PPARγ, collagen II, and MMP-13. **p < 0.01 compared with the control group; #p < 0.05, ##p < 0.01 compared with the IL-1β group.
PPARγ plays a role in the regulation of matrix decomposition (Afif et al., 2007). Previous reports showed that collagen II is a major component of the cartilage matrix (Handa et al., 2021). MMP-13 can hydrolyze collagen II, which is considered as a significant biomarker to assess OA therapeutic effects and OA progression (Corciulo et al., 2017). To further verify the effect of DD active constituents on PPARγ expression in chondrocytes, we measured the extent to which the active constituents promoted PPARγ expression in IL-1β-induced chondrocytes by a high content-laser confocal system (Figures 4B,E). These potential active constituents of DD could also inhibit IL-1β-induced downregulation of collagen II and the production of MMP-13 (Figures 4C,D,E). Based on the experimental results of the above methods, five active constituents, including alisol A, emodin, taxifolin, isoliquiritigenin, and schisandrin C inhibited the changes of these proteins in IL-1β-treated chondrocytes. These five active constituents should be the representative active compounds in DD which can ameliorate OA via PPARγ preservation.
Inflammation is causally involved in OA pathogenesis (Kraus et al., 2015). To further analyze the anti-inflammatory effects of active constituents, we measured the levels of major inflammatory mediators TNF-α, NO, and PGE2 in cell culture media. The active constituents decrease TNF-α, NO, and PGE2 secretion in IL-1β-treated chondrocytes. These results demonstrated that active constituents of DD mentioned above have the effect of inhibiting IL-1β-induced inflammation (Figure 5).
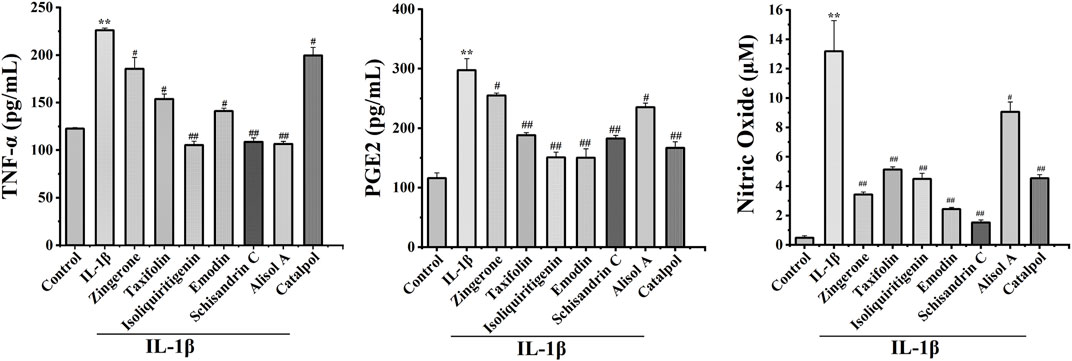
FIGURE 5. Effects of seven potential active constituents (Isoliquiritigenin, Emodin, Taxifolin, Catalpol, Alisol A, Zingerone, and Schisandrin C) on IL-1β-induced PGE2, TNF-α, and NO synthesis in rat chondrocytes. The culture supernatants were then harvested for further detection. **p < 0.01 compared with the control group; #p < 0.05, ##p < 0.01 compared with the IL-1β group.
To manifest the effects of active constituents on IL-1β-treated rat cartilage explants, an ex vivo model culture of rat cartilage explants was established (Tu et al., 2019b). Active constituents at 20 μM were selected for cartilage explants intervention. The cartilage matrix degradation was assessed by safranin O staining analysis of samples cultured for 3 days with or without IL-1β plus compound treatment. Compared with the control group, IL-1β treatment of explants enhanced matrix degradation of the cartilage explants (Figure 6). The active constituents in DD alleviated this change compared with the IL-1β-treated alone rat cartilage explants. In the alisol A, isoliquiritigenin, and emodin intervention groups, cartilage explants matrix degradation was alleviated. The extent of cartilage damage was assessed by HE analysis has a similar trend (Supplementary Figure S4, Supplementary Figure S5). These results suggested that the five constituents, such as alisol A, emodin, taxifolin, isoliquiritigenin, and schisandrin C mitigated matrix degradation and cartilage damage in rat cartilage explants.
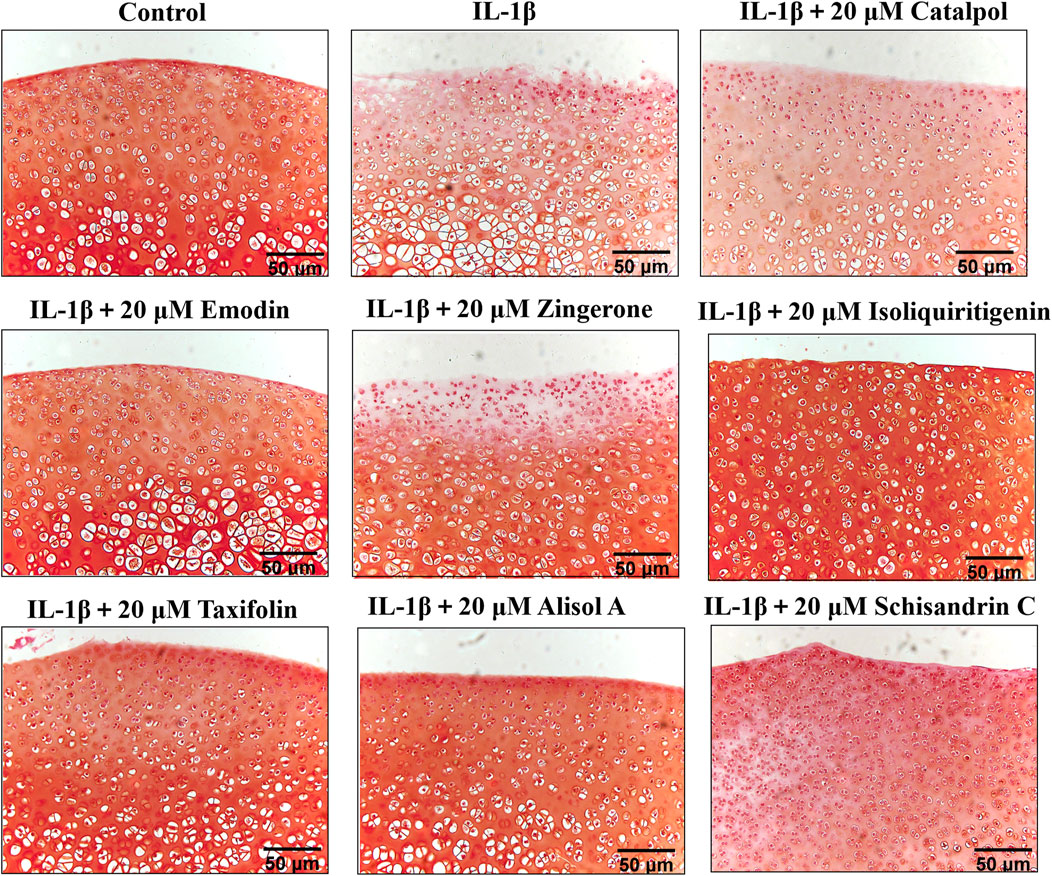
FIGURE 6. Rat cartilage explants were exposed to 20 μM active constituents with IL-1β (10 ng/ml) for 3 days and stained with Safranin O staining.
The above experimental results show that the active constituents of DD appear to be able to ameliorate osteoarthritis by targeting DNMT1 to preserve PPARγ expression. To further study the interaction of DNMT1-isoliquiritigenin, DNMT1-emodin, DNMT1-taxifolin, DNMT1-catalpol, DNMT1-alisol A, DNMT1-zingerone, and DNMT1-schisandrin C complexes at the molecular level, molecular dynamics simulations with the MM-GBSA method was applied. After acquiring the trajectory of molecular dynamic simulation, the structural stability of all complexes in 50 ns simulation were evaluated by root means square deviation (RMSD) values of Cα atoms of protein and heavy atoms of ligand. As Figures 7A,B showed that all the simulations were well equilibrated during 50 ns and the ligand did not undergo a remarkable conformational change.
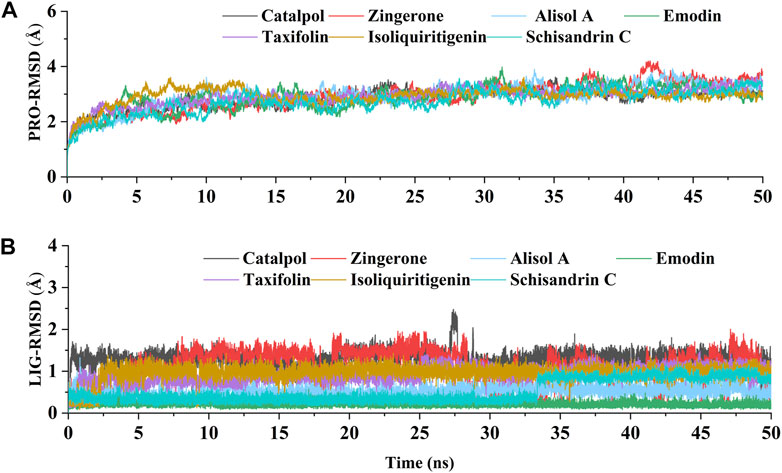
FIGURE 7. The RMSD values relative to the initial structure. (A) RMSD of the backbone atoms of DNMT1 in each constituents-DNMT1 complex; (B) RMSD of the heavy atoms of seven potential active constituents in each constituents-DNMT1 complex.
For most enzyme systems, the bioactivities of inhibitors are largely related to binding affinities (Wang et al., 2019). In this study, the binding free energies between the seven potential active constituents (isoliquiritigenin, emodin, taxifolin, catalpol, alisol A, zingerone, and schisandrin C) and DNMT1 complexes were calculated by the MM-GBSA method, and the results were showed that all seven constituents have good binding affinity (Table 2). Thereinto, the constituents with relatively good binding free energies are alisol A (-40.05 kcal/mol), emodin (−27.56 kcal/mol), taxifolin (−27.35 kcal/mol), isoliquiritigenin (−20.61 kcal/mol), and schisandrin C (−25.18 kcal/mol). Preliminary results showed that, alisol A, emodin, taxifolin, isoliquiritigenin, and schisandrin C achieved a stable binding with DNMT1, which was highly consistent with the results of cell and explant experiments.
To identify the key amino acids for the protein-ligand interaction, residue energy decomposition of the binding free energies was calculated and the binding modes of all above complex were investigated by the representative conformations extracted from the MD trajectories. In this study, the total binding free energy was decomposed and the top 15 amino acids with the largest contributions to ligand binding were recorded (Figure 8). The results showed that Phe 648, Glu 698, and Glu 1,168 are the important amino acids in the DNMT1-alisol A complex system (Figure 8A), and a hydrogen bond was found to be formed between alisol A and amino acids Glu 1,168 (Figures 9A,B). Phe 648, Glu 698, and Glu 1,168 were the important amino acids in the DNMT1-emodin complex system, and the hydrogen bond was found to be formed between emodin and amino acids Glu 1,168 and Met 1,169 (Figure 8B, Figure 9C). In the DNMT1-taxifolin complex system, Pro 1,225 and Glu 1,266 were the important amino acids, and a hydrophobic were formed between Glu 1,266, Arg 1,312, and taxifolin (Figure 8C, Figure 9D). Interestingly, the active residues here, such as Met 1,169, Glu 1,266, Glu 1,168, and Pro 1,225, are the same as those reported by Liang et al. (Liang et al., 2020). It is indicated that the binding pockets of above-mentioned active constituents have partial overlap with the allosteric pockets (Zhu et al., 2021). Phe 1,145, Met 1,169, Gly 1,223, and Pro 1,225 were the important amino acids in the DNMT1-isoliquiritigenin complex system (Figure 8D, Figure 9E). In the DNMT1-schisandrin C complex systems, Phe 648, Glu 698, and Pro 1,225 were the important amino acids (Figure 8E, Figure 9F). Targeted hot spot amino acids common to the above constituents are Phe 648, Glu 698, and Glu 1,168 might were key interacting amino acids for binding of DNMT1 inhibitors. Besides, the five constituents, such as alisol A, emodin, taxifolin, isoliquiritigenin, and schisandrin C had similar combined amino acids and binding patterns (Figure 9G).
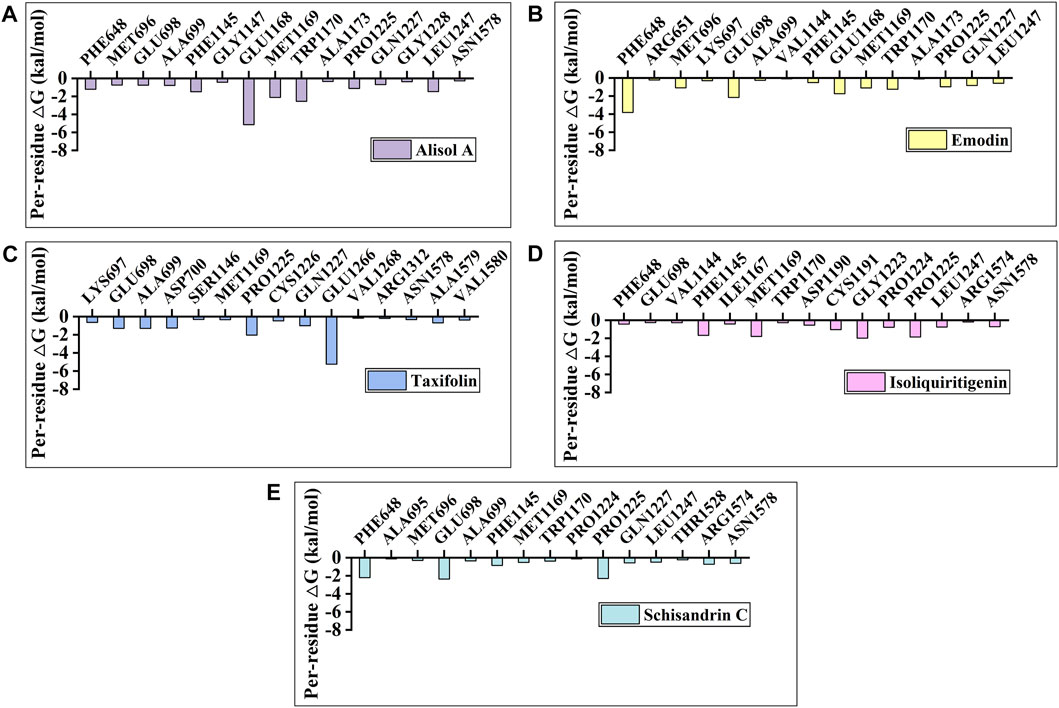
FIGURE 8. Per-residue binding free energy decomposition of (A) Alisol A; (B) Emodin; (C) Taxifolin; (D) Isoliquiritigenin; and (E) Schisandrin C binding to DNMT1.
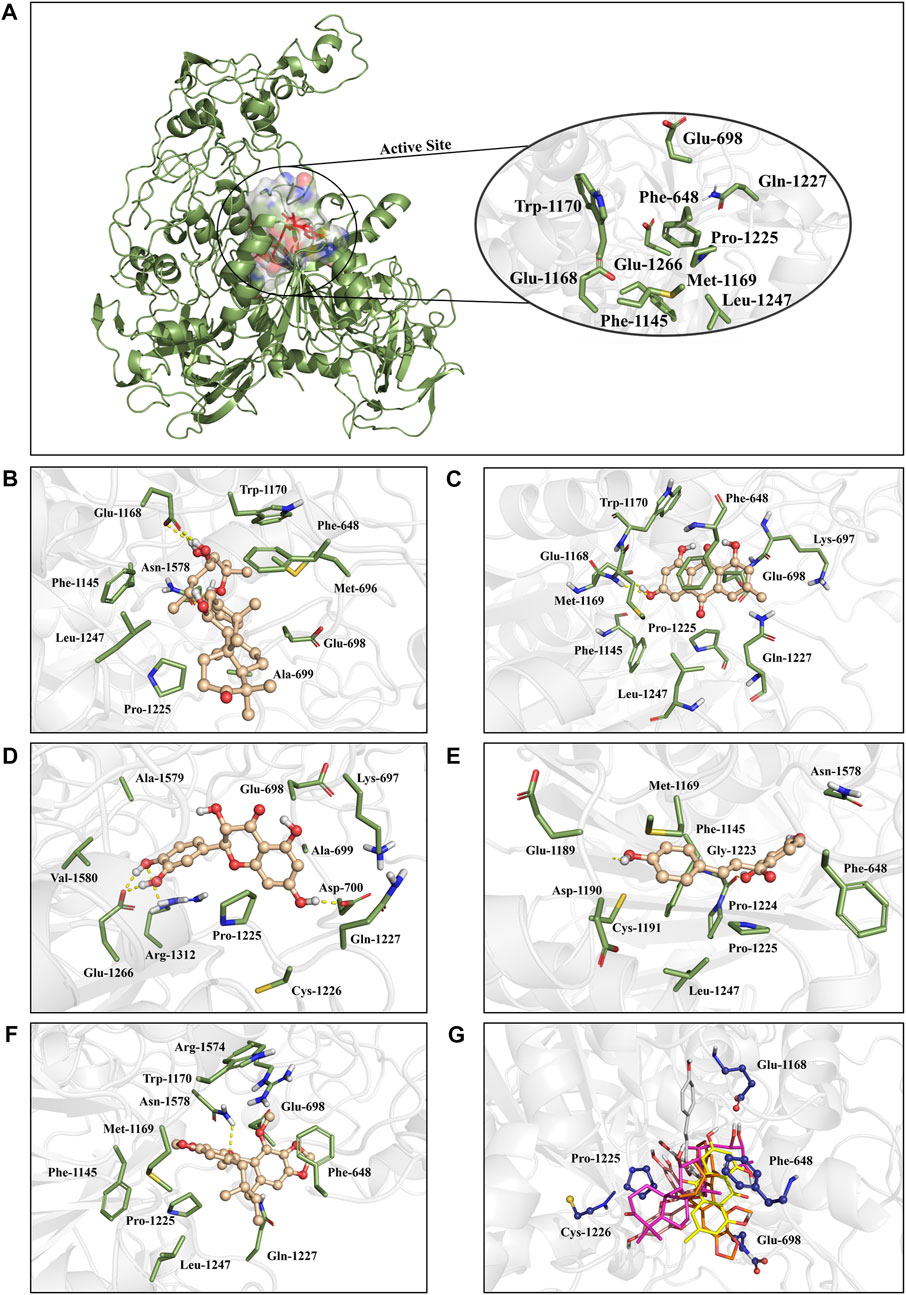
FIGURE 9. The positions of the active constituents are anchored by the interaction with DNMT1. (A) The complex structure and active site of DNMT1. The active site of DNMT1 was shown in rainbow surface. (B) Interaction mode of alisol A with the DNMT1; (C) Interaction mode of emodin with the DNMT1; (D) Interaction mode of taxifolin with the DNMT1; (E) Interaction mode of isoliquiritigenin with the DNMT1; (F) Interaction mode of schisandrin C; and (G) above five constituents with the DNMT1.
Based on the calculation results, alisol A may have a strong targeted inhibition effect with a relatively high binding free energy (-40.05 kcal/mol), which showed a similar binding energy as a DNMT1 inhibitor DC-05 (Xie et al., 2019) (Tu et al., 2019b). Alisol A also had a significant promoted effect on PPARγ expression in IL-1β-treated chondrocytes by protein capillary electrophoresis and immunofluorescence assays. To assess whether alisol A can reverse PPARγ suppression by targeting DNMT1, the effect of alisol A on methylation of PPARγ promoter was conducted by MSP method. The results show that alisol A can reverse the methylation of PPARγ promoter, which was similar to that of DNMT1 inhibitor SGI-1027 (Figure S3A-B) (Zhu et al., 2019). These results suggested that alisol A could be developed as a natural epigenetic regulator derived from Chinese herbal formula.
Inflammation-induced destruction of articular cartilage is the most prominent feature of OA (Kraus et al., 2015; Quicke et al., 2022). At present, the treatment of OA is mainly to relieve pain, and there is a lack of targeted drugs to slow down the inflammatory destruction of cartilage (Goldring and Otero, 2011). The destruction of articular cartilage is closely related to the degradation of the cartilage matrix induced by inflammation (Goldring and Otero, 2011; Chen et al., 2021b), and TCM has an original theoretical system for maintaining the physiological stability of bone (Sun et al., 2022). In this study, we first confirmed by in vivo experiments that DD reduced papain-induced osteochondral injury and significantly decreased chondrocyte matrix degradation. Therefore, our study indicates that DD exerts its protective effect on cartilage mainly by decreasing the degradation of the osteochondral matrix.
Recent studies found that PPARγ played a key role in the degradation and protection of osteochondral matrix in OA (Afif et al., 2007; Fahmi et al., 2011). We also found that PPARγ expression decreased in the osteochondral matrix in OA model rats, while DD increased the expression of PPARγ. The hypermethylation of PPARγ promoter is one of the reasons for the decrease of PPARγ in OA (Zhu et al., 2019). Therefore, DD is likely to participate in the methylation regulation of PPARγ promoter and preserve the expression of PPARγ. The results of molecular docking suggest that isoliquiritigenin, emodin, taxifolin, catalpol, alisol A, zingerone, and schisandrin C in DD have high docking affinity to DNMT1, and should be the main potentially active constituents to preserve the expression of PPARγ in DD (Chen et al., 2021a).
The seven identified active constituents have been identified as the main constituents of DD that target DNMT1. Specifically, alisol A, emodin, taxifolin, isoliquiritigenin, and schisandrin C promoted PPARγ expression and cartilage matrix formation. The prominent roles of these constituents in promoting cartilage matrix formation are consistent with the in vivo experiment of DD. It was further confirmed that these active constituents showed protective effect against IL-1β-induced inflammatory injury by culturing rat cartilage explants. Among them, alisol A and emodin could effectively mitigate cartilage damage. Finally, through molecular dynamics simulations with the MM-GBSA method, alisol A, emodin, taxifolin, isoliquiritigenin, and schisandrin C were found to achieve a stable binding pattern. The common targeted hot spot residues of the above constituents were Phe 648, Glu 698, and Glu 1,168. Therefore, alisol A with a relatively high binding free energy was considered a key constituent in DD to reverse PPARγ demethylation by targeting DNMT1 inhibition.
In conclusion, in this work, combined with in vivo experiments and virtual screening, cell and explant experiments, the protective effect of DD on OA and the active constituents maintaining matrix stability of cartilage through epigenetic regulation of PPARγ have been systematically elucidated. The five active constituents consist, such as alisol A, emodin, taxifolin, isoliquiritigenin, and schisandrin C in DD could promote the expression of PPARγ and the formation of cartilage matrix. Moreover, molecular dynamics simulations with MM-GBSA method indicated the five active constituents also achieved a stable binding with DNMT1, an effective target for regulating the epigenetic expression of PPARγ. This work demonstrates that DD has material basis for its therapeutic role in OA through epigenetic regulation of PPARγ and the active constituents, such as alisol A, deserves further investigation as an epigenetic modulator in the treatment of OA.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by Ethics Committee of Laboratory Animal Center of Gansu University of Chinese Medicine. Written informed consent was obtained from the owners for the participation of their animals in this study.
YQL, XJJ, LQ, MZ and CHL generated the conception/design of the study. YHH, LQ, HL, YXZ, CHL performed some of the lab work and data analysis. DZD, YLL, JY, PW, JTL supported the overall data analysis and provided constructive discussion. JL and ML collected the compounds of DD and performed molecular docking. MZ and ML performed molecular dynamic simulation. All authors were responsible for reviewing data. LQ, MZ, CHL, XJJ and YQL wrote the manuscript. All authors have read and approved the final manuscript.
This study was supported by the National Natural Science Foundation of China (No. 81960823) and Provincial University Industry Support Project in Gansu (2020C-15).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.993498/full#supplementary-material
Abramoff, B., and Caldera, F. E. (2020). Osteoarthritis: Pathology, diagnosis, and treatment options. Med. Clin. North Am. 104, 293–311. doi:10.1016/j.mcna.2019.10.007
Afif, H., Benderdour, M., Mfuna-Endam, L., Martel-Pelletier, J., Pelletier, J. P., Duval, N., et al. (2007). Peroxisome proliferator-activated receptor gamma1 expression is diminished in human osteoarthritic cartilage and is downregulated by interleukin-1beta in articular chondrocytes. Arthritis Res. Ther. 9, R31. doi:10.1186/ar2151
Ansari, M. Y., Ahmad, N., and Haqqi, T. M. (2020). Oxidative stress and inflammation in osteoarthritis pathogenesis: Role of polyphenols. Biomed. Pharmacother. 129, 110452. doi:10.1016/j.biopha.2020.110452
Ball, H. C., Alejo, A. L., Samson, T. K., and Safadi, F. F. (2022). Epigenetic regulation of chondrocytes and subchondral bone in osteoarthritis. Life (Basel) 12, 582. doi:10.3390/life12040582
Barter, M. J., Bui, C., and Young, D. A. (2012). Epigenetic mechanisms in cartilage and osteoarthritis: DNA methylation, histone modifications and microRNAs. Osteoarthr. Cartil. 20, 339–349. doi:10.1016/j.joca.2011.12.012
Bordji, K., Grillasca, J. P., Gouze, J. N., Magdalou, J., ScHoHn, H., Keller, J. M., et al. (2000). Evidence for the presence of peroxisome proliferator-activated receptor (PPAR) alpha and gamma and retinoid Z receptor in cartilage PPARgamma activation modulates the effects of interleukin-1beta on rat chondrocytes. J. Biol. Chem. 275, 12243–12250. doi:10.1074/jbc.275.16.12243
Case, D. A., Cheatham, T. E3R. D., Darden, T., Gohlke, H., Luo, R., Merz, K. M., et al. (2005). The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688. doi:10.1002/jcc.20290
Chen, F., Liu, H., Sun, H., Pan, P., Li, Y., Li, D., et al. (2016). Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein-protein binding free energies and re-rank binding poses generated by protein-protein docking. Phys. Chem. Chem. Phys. 18, 22129–22139. doi:10.1039/c6cp03670h
Chen, X., Zhou, H., Liu, Y. B., Wang, J. F., Li, H., Ung, C. Y., et al. (2006). Database of traditional Chinese medicine and its application to studies of mechanism and to prescription validation. Br. J. Pharmacol. 149, 1092–1103. doi:10.1038/sj.bjp.0706945
Chen, Y., Shao, X., Zhao, X., Ji, Y., Liu, X., Li, P., et al. (2021a). Targeting protein arginine methyltransferase 5 in cancers: Roles, inhibitors and mechanisms. Biomed. Pharmacother. 144, 112252. doi:10.1016/j.biopha.2021.112252
Chen, Y., Yuan, S., Cao, Y., Kong, G., Jiang, F., Li, Y., et al. (2021b). Gasotransmitters: Potential therapeutic molecules of fibrotic diseases. Oxid. Med. Cell. Longev. 2021, 3206982. doi:10.1155/2021/3206982
Chen, Z., Zhong, H., Wei, J., Lin, S., Zong, Z., Gong, F., et al. (2019). Inhibition of Nrf2/HO-1 signaling leads to increased activation of the NLRP3 inflammasome in osteoarthritis. Arthritis Res. Ther. 21, 300. doi:10.1186/s13075-019-2085-6
Corciulo, C., Lendhey, M., Wilder, T., Schoen, H., Cornelissen, A. S., Chang, G., et al. (2017). Endogenous adenosine maintains cartilage homeostasis and exogenous adenosine inhibits osteoarthritis progression. Nat. Commun. 8, 15019. doi:10.1038/ncomms15019
D'Arcy, Y., Mantyh, P., Yaksh, T., Donevan, S., Hall, J., Sadrarhami, M., et al. (2021). Treating osteoarthritis pain: Mechanisms of action of acetaminophen, nonsteroidal anti-inflammatory drugs, opioids, and nerve growth factor antibodies. Postgrad. Med. 133, 879–894. doi:10.1080/00325481.2021.1949199
Da Costa, B. R., Pereira, T. V., Saadat, P., Rudnicki, M., Iskander, S. M., Bodmer, N. S., et al. (2021). Effectiveness and safety of non-steroidal anti-inflammatory drugs and opioid treatment for knee and hip osteoarthritis: Network meta-analysis. Bmj 375, n2321. doi:10.1136/bmj.n2321
Dickson, B. M., Roelofs, A. J., Rochford, J. J., Wilson, H. M., and De Bari, C. (2019). The burden of metabolic syndrome on osteoarthritic joints. Arthritis Res. Ther. 21, 289. doi:10.1186/s13075-019-2081-x
Fahmi, H., Martel-Pelletier, J., Pelletier, J. P., and Kapoor, M. (2011). Peroxisome proliferator-activated receptor gamma in osteoarthritis. Mod. Rheumatol. 21, 1–9. doi:10.1007/s10165-010-0347-x
Foulks, J. M., Parnell, K. M., Nix, R. N., Chau, S., Swierczek, K., Saunders, M., et al. (2012). Epigenetic drug discovery: Targeting DNA methyltransferases. J. Biomol. Screen. 17, 2–17. doi:10.1177/1087057111421212
Friesner, R. A., Murphy, R. B., Repasky, M. P., Frye, L. L., Greenwood, J. R., Halgren, T. A., et al. (2006). Extra precision Glide: Docking and scoring incorporating a model of hydrophobic enclosure for ProteinLigand complexes. J. Med. Chem. 49, 6177–6196. doi:10.1021/jm051256o
Gao, G., Wu, H., Tian, J., Du, J., Xie, X., and Gao, J. (2012). Clinical efficacy of bushen huoxue qubi decoction on treatment of knee-osteoarthritis and its effect on hemarheology, anti-inflammation and antioxidation. Zhongguo Zhong Yao Za Zhi 37, 390–396.
Goldring, M. B., and Otero, M. (2011). Inflammation in osteoarthritis. Curr. Opin. Rheumatol. 23, 471–478. doi:10.1097/BOR.0b013e328349c2b1
Grandi, F. C., and Bhutani, N. (2020). Epigenetic therapies for osteoarthritis. Trends Pharmacol. Sci. 41, 557–569. doi:10.1016/j.tips.2020.05.008
Handa, A., Grigelioniene, G., and Nishimura, G. (2021). Radiologic features of type II and type XI collagenopathies. Radiographics. 41, 192–209. doi:10.1148/rg.2021200075
Headland, S. E., Jones, H. R., Norling, L. V., Kim, A., Souza, P. R., Corsiero, E., et al. (2015). Neutrophil-derived microvesicles enter cartilage and protect the joint in inflammatory arthritis. Sci. Transl. Med. 7, 315ra190. doi:10.1126/scitranslmed.aac5608
Hou, T., Wang, J., Li, Y., and Wang, W. (2011). Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 51, 69–82. doi:10.1021/ci100275a
Jiang, L., Xu, P., Zhang, D., Lu, J., Chang, T., Zhang, Y., et al. (2022). Treatment of myasthenia gravis with the method of tonifying spleen and replenishing qi in traditional Chinese medicine: A protocol for systematic review and meta-analysis. Med. Baltim. 101, e28530. doi:10.1097/MD.0000000000028530
Jiao, K., Zhang, J., Zhang, M., Wei, Y., Wu, Y., Qiu, Z. Y., et al. (2013). The identification of CD163 expressing phagocytic chondrocytes in joint cartilage and its novel scavenger role in cartilage degradation. PLoS One 8, e53312. doi:10.1371/journal.pone.0053312
Kobayashi, T., Notoya, K., Naito, T., Unno, S., Nakamura, A., Martel-Pelletier, J., et al. (2005). Pioglitazone, a peroxisome proliferator-activated receptor gamma agonist, reduces the progression of experimental osteoarthritis in Guinea pigs. Arthritis Rheum. 52, 479–487. doi:10.1002/art.20792
Kraus, V. B., Blanco, F. J., Englund, M., Karsdal, M. A., and Lohmander, L. S. (2015). Call for standardized definitions of osteoarthritis and risk stratification for clinical trials and clinical use. Osteoarthr. Cartil. 23, 1233–1241. doi:10.1016/j.joca.2015.03.036
Krishna, S., Shukla, S., Lakra, A. D., Meeran, S. M., and Siddiqi, M. I. (2017). Identification of potent inhibitors of DNA methyltransferase 1 (DNMT1) through a pharmacophore-based virtual screening approach. J. Mol. Graph. Model. 75, 174–188. doi:10.1016/j.jmgm.2017.05.014
Kun-Yi, H., Samik, G., and Hiroaki, K. (2013). Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology. PLoS One 8, e83922. doi:10.1371/journal.pone.0083922
Lai, N., Zhang, Z., Wang, B., Miao, X., Guo, Y., Yao, C., et al. (2015). Regulatory effect of traditional Chinese medicinal formula Zuo-Gui-Wan on the Th17/Treg paradigm in mice with bone loss induced by estrogen deficiency. J. Ethnopharmacol. 166, 228–239. doi:10.1016/j.jep.2015.03.011
Latourte, A., Cherifi, C., Maillet, J., Ea, H. K., Bouaziz, W., Funck-Brentano, T., et al. (2017). Systemic inhibition of IL-6/Stat3 signalling protects against experimental osteoarthritis. Ann. Rheum. Dis. 76, 748–755. doi:10.1136/annrheumdis-2016-209757
Li, Y., Zhang, Y., Chen, C., Zhang, H., Ma, C., and Xia, Y. (2016). Establishment of a rabbit model to study the influence of advanced glycation end products accumulation on osteoarthritis and the protective effect of pioglitazone. Osteoarthr. Cartil. 24, 307–314. doi:10.1016/j.joca.2015.08.001
Liang, Z., Zhu, Y., Long, J., and Ye, F. (2020). Both intra and inter-domain interactions define the intrinsic dynamics and allosteric mechanism in DNMT1s. Comput. Struct. Biotechnol. J. 18, 749–764. doi:10.1016/j.csbj.2020.03.016
Maier, J. A., Martinez, C., Kasavajhala, K., Wickstrom, L., Hauser, K. E., and Simmerling, C. (2015). ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713. doi:10.1021/acs.jctc.5b00255
Pan, J., Zhou, X., Li, W., Novotny, J. E., Doty, S. B., and Wang, L. (2009). In situ measurement of transport between subchondral bone and articular cartilage. J. Orthop. Res. 27, 1347–1352. doi:10.1002/jor.20883
Peters, M. B., Yang, Y., Wang, B., Fusti-Molnar, L., Weaver, M. N., and Merz, K. M. (2010). Structural survey of Zinc containing proteins and the development of the Zinc AMBER force field (ZAFF). J. Chem. Theory Comput. 6, 2935–2947. doi:10.1021/ct1002626
Pigeolet, M., Jayaram, A., Park, K. B., and Meara, J. G. (2021). Osteoarthritis in 2020 and beyond. Lancet 397, 1059–1060. doi:10.1016/S0140-6736(21)00208-7
Quicke, J. G., Conaghan, P. G., Corp, N., and Peat, G. (2022). Osteoarthritis year in review 2021: Epidemiology & therapy. Osteoarthr. Cartil. 30, 196–206. doi:10.1016/j.joca.2021.10.003
Robinson, W. H., Lepus, C. M., Wang, Q., Raghu, H., Mao, R., Lindstrom, T. M., et al. (2016). Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 12, 580–592. doi:10.1038/nrrheum.2016.136
Ru, J., Li, P., Wang, J., Zhou, W., Li, B., Huang, C., et al. (2014). Tcmsp: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 6, 13. doi:10.1186/1758-2946-6-13
Sun, Z., Su, W., Wang, L., Cheng, Z., and Yang, F. (2022). Clinical effect of bushen huoxue method combined with platelet-rich plasma in the treatment of knee osteoarthritis and its effect on IL-1, IL-6, VEGF, and PGE-2. J. Healthc. Eng. 2022, 9491439. doi:10.1155/2022/9491439
Tiku, M. L., and Madhan, B. (2016). Preserving the longevity of long-lived type II collagen and its implication for cartilage therapeutics. Ageing Res. Rev. 28, 62–71. doi:10.1016/j.arr.2016.04.011
Tu, C., Huang, X., Xiao, Y., Song, M., Ma, Y., Yan, J., et al. (2019a). Schisandrin A inhibits the IL-1β-induced inflammation and cartilage degradation via suppression of mapk and NF-κB signal pathways in rat chondrocytes. Front. Pharmacol. 10, 41. doi:10.3389/fphar.2019.00041
Tu, C., Ma, Y., Song, M., Yan, J., Xiao, Y., and Wu, H. (2019b). Liquiritigenin inhibits IL-1β-induced inflammation and cartilage matrix degradation in rat chondrocytes. Eur. J. Pharmacol. 858, 172445. doi:10.1016/j.ejphar.2019.172445
Vasheghani, F., Zhang, Y., Li, Y. H., Blati, M., Fahmi, H., Lussier, B., et al. (2015). PPARγ deficiency results in severe, accelerated osteoarthritis associated with aberrant mTOR signalling in the articular cartilage. Ann. Rheum. Dis. 74, 569–578. doi:10.1136/annrheumdis-2014-205743
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., and Case, D. A. (2004). Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174. doi:10.1002/jcc.20035
Wang, M., Sampson, E. R., Jin, H., Li, J., Ke, Q. H., Im, H. J., et al. (2013). MMP13 is a critical target gene during the progression of osteoarthritis. Arthritis Res. Ther. 15, R5. doi:10.1186/ar4133
Wang, M., Shen, J., Jin, H., Im, H. J., Sandy, J., and Chen, D. (2011). Recent progress in understanding molecular mechanisms of cartilage degeneration during osteoarthritis. Ann. N. Y. Acad. Sci. 1240, 61–69. doi:10.1111/j.1749-6632.2011.06258.x
Wang, Q., Li, Y., Xu, J., Wang, Y., Shi, D., Liu, L., et al. (2019). Computational study on the selective inhibition mechanism of MS402 to the first and second bromodomains of BRD4. Proteins 87, 3–11. doi:10.1002/prot.25611
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–w303. doi:10.1093/nar/gky427
Xie, T., Yu, J., Fu, W., Wang, Z., Xu, L., Chang, S., et al. (2019). Insight into the selective binding mechanism of DNMT1 and DNMT3A inhibitors: A molecular simulation study. Phys. Chem. Chem. Phys. 21, 12931–12947. doi:10.1039/c9cp02024a
Yang, J., Lin, X., Xing, N., Zhang, Z., Zhang, H., Wu, H., et al. (2021). Structure-based discovery of novel nonpeptide inhibitors targeting SARS-CoV-2 M pro. J. Chem. Inf. Model. 61, 3917–3926. doi:10.1021/acs.jcim.1c00355
Yao, C. F., Zhao, Y. J., Niu, K., Sun, Y. l., Li, C. g., Tang, D. z., et al. (2013). Experimental study on lumbar intervetebral disc degeneration model with kidney deficiency by ovariectomizing. Zhongguo Gu Shang 26, 1015–1022.
Yao, N., Chen, G. C., Lu, Y. Y., Xu, X. M., Zhao, C. X., Huang, X. J., et al. (2021). Bushen Qiangjin capsule inhibits the Wnt/α-catenin pathway to ameliorate papain-induced knee osteoarthritis in rat. J. Tradit. Chin. Med. 41, 935–942. doi:10.19852/j.cnki.jtcm.2021.06.010
Yuan, Q., Kan, W. B., Song, P. F., Zhao, J., Yu, W. G., and Wang, Y. J. (2012). Influence of Bushen Huoxue decoction on beta-catenin, MMP-7 of synoviocytes in rats with knee osteoarthritis. Zhongguo Gu Shang 25, 761–765.
Zhang, C., Huang, Y., Zhang, Q. Z., Ji, Y. C., and Zhang, Y. Z. (2015). Effect of Bushen Gujin Recipe on serum and synovia interleukin-1 and tumor necrosis factor-alpha of knee osteoarthritis model rabbits. Zhongguo Zhong Xi Yi Jie He Za Zhi 35, 355–358.
Zhu, X., Chen, F., Lu, K., Wei, A., Jiang, Q., and Cao, W. (2019). PPARγ preservation via promoter demethylation alleviates osteoarthritis in mice. Ann. Rheum. Dis. 78, 1420–1429. doi:10.1136/annrheumdis-2018-214940
Keywords: traditional Chinese medicine, Dabushen decoction, osteoarthritis, epigenetics, Dnmt1, PPARγ
Citation: Qiu L, Zhang M, Li C, Hou Y, Liu H, Lin J, Yao J, Duan DZ, Zhang YX, Li M, Li YL, Wang P, Li JT, Jin XJ and Liu YQ (2022) Deciphering the active constituents of Dabushen decoction of ameliorating osteoarthritis via PPARγ preservation by targeting DNMT1. Front. Pharmacol. 13:993498. doi: 10.3389/fphar.2022.993498
Received: 13 July 2022; Accepted: 01 November 2022;
Published: 23 November 2022.
Edited by:
Qianqian Wang, Dalian University, ChinaReviewed by:
Guang Hu, Soochow University, ChinaCopyright © 2022 Qiu, Zhang, Li, Hou, Liu, Lin, Yao, Duan, Zhang, Li, Li, Wang, Li, Jin and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao Jie Jin, amlubG92ZWRyZWFtQDE2My5jb20=; Yong Qi Liu, bGl1eW9uZ3FpNzNAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.