
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 02 September 2022
Sec. Drug Metabolism and Transport
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.985159
This article is part of the Research TopicDrug-Drug Interactions in PharmacologyView all 14 articles
 Zhize Ye1†
Zhize Ye1† Bingbing Chen1†
Bingbing Chen1† Nanyong Gao1
Nanyong Gao1 Qihui Kong1
Qihui Kong1 Xiaoqin Hu1
Xiaoqin Hu1 Zhongqiu Lu2
Zhongqiu Lu2 Jianchang Qian1
Jianchang Qian1 Guoxin Hu1*
Guoxin Hu1* Jianping Cai1*
Jianping Cai1* Bin Wu2*
Bin Wu2*This study aimed 1) to investigate the influence of CYP2D6 variants on the catalyzing of fluvoxamine, and 2) to study the interaction between fluvoxamine and apatinib. An enzymatic reaction system was setup and the kinetic profile of CYP2D6 in metabolizing fluvoxamine was determined. In vivo, drug-drug interaction was investigated using Sprague–Dawley (SD) rats. Fluvoxamine was given gavage with or without apatinib. Ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) was used to determine the concentrations of fluvoxamine and desmethyl-fluvoxamine. The results demonstrated that the relative clearance rates of CYP2D6.A5V, V104A, D337G, F164L, V342M, R440C and R497C increased significantly compared with CYP2D6.1, ranging from 153.626% ± 6.718% to 394.310% ± 33.268%. The activities of other variants reduced to different extent, or even lost function, but there was no statistical difference. The IC50 of apatinib against fluvoxamine disposition was determined, which is 0.190 μM in RLM and 6.419 μM in HLM, respectively. In vivo, apatinib can enhance the plasma exposure of fluvoxamine remarkably characterized by increased AUC, Tmax and Cmax. Meanwhile, the produce of desmethyl fluvoxamine was dramatically inhibited, both AUC and Cmax decreased significantly. Mechanistically, apatinib inhibit the generation of fluvoxamine metabolite with a mixed manner both in RLM and HLM. Furthermore, there were differences in the potency of apatinib in suppressing fluvoxamine metabolism among CYP2D6.1, 2 and 10. In conclusion, CYP2D6 gene polymorphisms and drug-drug interaction can remarkably affect the plasma exposure of fluvoxamine. The present study provides basis data for guiding individual application of fluvoxamine.
Fluvoxamine is commonly prescribed to treat depression and anxiety disorders with mechanism of selective inhibit serotonin reuptake (Yuan et al., 2020). Central nervous system symptoms, rash, gastrointestinal symptoms, and suicidal tendencies are common adverse reactions during medicine of fluvoxamine (Lenze et al., 2020). Its clinical efficacy is highly variable among individuals. Drug efficacy stratification may be the result of individual differences in blood concentrations which caused by genetic polymorphisms of metabolic enzymes and drug interactions. Cytochrome P450 family member 2D6 (CYP2D6) is a major enzyme involved in catalyzing metabolism of fluvoxamine, which produced desmethyl fluvoxamine (Hicks et al., 2015; Zastrozhin et al., 2021). However, there are genetic polymorphism of CYP2D6 resulting in large inter-individual variability in enzyme activity, further leading to subtherapeutic phenomena or severe adverse effects (Zastrozhin et al., 2021). Moreover, interactions between fluvoxamine and other drugs are frequently being reported. A study found one fatality in a woman who was taking clotiapine, 7-aminoclonazepam, propranolol, gabapentin and haloperidol alongside fluvoxamine developing antipsychotic malignant syndrome (Vignali et al., 2021). Interactions of fluvoxamine with antiepileptic or antidepressant drugs have also been issued (Mula and Trimble, 2003; Spina et al., 2016). Therefore, defining the correlation between CYP2D6 genotype and fluvoxamine metabolic phenotype, identifying drugs that could potential interact with fluvoxamine are helpful for personalized medicine.
Apatinib is a small molecule drug that targeting inhibition of angiogenesis (Wang N. et al., 2020). It’s approved to be safe and effective after failure of standard chemotherapy in advanced gastric cancer (Geng et al., 2018). Interestingly, it is worth noting that apatinib is a pan CYP inhibitor (Zhou et al., 2014). Therefore, the research on the interaction between apatinib and other CYP substrate drugs has gradually attracted people’s attention. Statistics data display that cancer patients usually complicated with various psychological symptoms, especially depression and anxiety (Wang YH. et al., 2020). This will diminish benefits of medicine and affects the quality life of patients. Therefore, the combined application of apatinib and fluvoxamine is a feasible clinical treatment option. However, the interaction between them has not been unveiled.
Herein, we evaluated the catalytic activity of CYP2D6.1 and other 23 variants on the disposition of fluvoxamine. In addition, we used microsomes and Spragge-Dawley (SD) rats to clarify the interaction between fluvoxamine and apatinib in vitro and in vivo. The results will provide fundamental data to facilitate the precision medicine application of fluvoxamine.
Fluvoxamine maleate was bought from Shanghai Canspec Scientific & Technology Co., Ltd. Desmethyl fluvoxamine was obtained from TRC Ltd. (Toronto, Canada). Diazepam was purchased from Shanghai Xudong Haipu Pharmaceutical Co., Ltd. and used as internal standard. Apatinib was obtained from Beijing Sunflower Technology Development Co., Ltd. Sodium carboxymethyl cellulose (CMC-Na), methanol, acetonitrile (ACN) and formic acid were purchased from Merck (Darmstadt, Germany). Microsomes were purchased from Corning Life Sciences Co., Ltd. CYP2D6 and cytochrome B5 were prepared as previously issued (Cai et al., 2016).
A newly developed and validated UPLC-MS/MS method was used to detect fluvoxamine and desmethyl fluvoxamine. The analytes were separated on a BEH C18 column (2.1 × 100 mm, 1.7 μm; Waters Corp., Millipore, Bedford, MA, United States), which incubated at 40°C. The mobile phase was consisted of 0.1% formic acid and ACN, and elution at 0.40 ml/min for 3.0 min with a gradient condition. The program was set as 10%–90% ACN (0–1.0 min), 90%–10% ACN (1.0–2.0 min), and 10% ACN (2.1–3.0 min).
Incubation system was dissolved in phosphate buffered saline which contained 1 pmol CYP2D6.1 or variants, 50 μg/ml cytochrome B5, 0.5–50 μM fluvoxamine. Before the reaction, the mixture was pre-incubated at 37°C for 5 min. Subsequently, add 1 mM nicotinamide adenine dinucleotide phosphate oxidase to initiate the reaction. 20 min’ later, the reaction was terminated. Add acetonitrile twice volume as much as reaction system and 20 μl internal standard to the mixture. After vortexing and centrifugation, the supernatant was taken and subjected to UPLC-MS/MS.
Animal ethics was reviewed and approved by Wenzhou Medical University. Male rats weighed 180–220 g were supplied by Vital River Laboratories (Beijing, China), and adaptive feeding for a week. SD rats were divided into two groups. Group A served as control, dosing of vehicle (0.5% CMC-Na). Group B was administrated 40 mg/kg apatinib. 30 min’ later, fluvoxamine (10 mg/kg) was given orally to the rats. Then, the vein blood was collected at 0, 0.25, 0.5, 1, 2, 3, 4, 5, 6, 8, 10, 12, and 24 h after administration. The sample was prepared and subjected to UPLC-MS/MS examination.
The microsomes reaction system was set up as indicated in above. In briefly, the reaction was carried out in PBS. The buffer was consisted of 0.2 mg/ml RLM or HLM, 0.5–50 μM fluvoxamine. NADPH was used to initiate the reaction. To determine the half maximal inhibitory concentration (IC50), the concentration of apatinib was set at 0.01, 0.1, 1, 10, 25, 50, and 100 μM. To determine the mechanism underlied the inhibition, the concentration of fluvoxamine was set according to the Km value, while the concentration of apatinib was set at 0, 0.25, 0.5, 1 μM according to the IC50 as well. After incubation, the samples were prepared and determined by UPLC-MS/MS.
Lineweaver-Burk double reciprocal plot was performed on GraphPad Prism 5.0 software. The kinetic parameters were obtained using non-compartmental model fitting by Drug and statistics (DAS) software 3.0. The corresponding drug-time curves were drawn by Origin 8.0. All data are expressed as Mean ± SD. Statistical analysis was performed by independent samples t-test using GraphPad Prism 5.0 software. p < 0.05 indicates a significant difference.
To detect the analytes, a UPLC-MS/MS method was developed and validated. The linear range, precision, accuracy, recovery, matrix effect and stability were evaluated. The detail data was presented in the Supplementary Information. In briefly, the monitoring transitions of diazepam, fluvoxamine and desmethyl fluvoxamine were m/z 285→ 154, m/z 319.4→ 71.2 and m/z 305.13→ 228.68, respectively. As Figure 1 showed, there was no obvious endogenous interference. The retention time of diazepam, fluvoxamine and desmethyl fluvoxamine were 1.45, 1.29 and 1.24 min, accordingly.
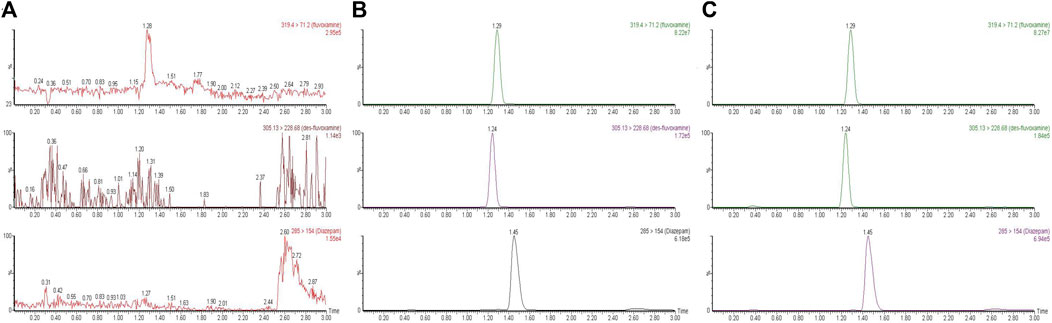
FIGURE 1. Representative chromatograms of fluvoxamine, desmethyl fluvoxamine and IS. (A) a blank plasma sample. (B) A blank plasma sample spiked with fluvoxamine, desmethyl Fluvoxamine and IS. (C) Rat’s plasma sample after dosing.
Michaelis curves and kinetic parameters of CYP2D6.1 and other variants in metabolizing fluvoxamine were shown in Figure 2 and Table 1, respectively. Based on maximum reaction velocity (Vmax), they can be divided into four groups. No significant differences were observed between CYP2D6.1 and CYP2D.2, V104, 90, C161S, D337G, E215K, R497C. Among them, CYP2D6.92 and 96 almost lost enzymatic function. Moreover, the Vmax of CYP2D6.A5V, 89 and 95 increased remarkably, ranging from 270.31% to 441.17%. In opposite, the remaining variants decreased significantly, ranging from 9.93% to 39.25%. From the michaelis constant (Km), a lot of them decreased obviously compared with CYP2D6.1, including CYP2D6.10, 97, R88P, F164L, F219S, V327M, D336N, V342M, R344Q, R440C, R497C. Besides, the other variants had no significant difference. Finally, the intrinsic clearance (Clint) and relative clearance were determined. In all, activities of seven variants, involving CYP2D6.A5V, V104A, D337G, F164L, V342M, R440C, R497C, increased compared with CYP2D6.1, ranging from 153.63% to 394.31%. The others’ The remaining variants showed no statistical difference in intrinsic clearance. In addition, CYP2D6.92 and 96 had no significant activity. The rest of the variants had different effect of reduced metabolic activity.
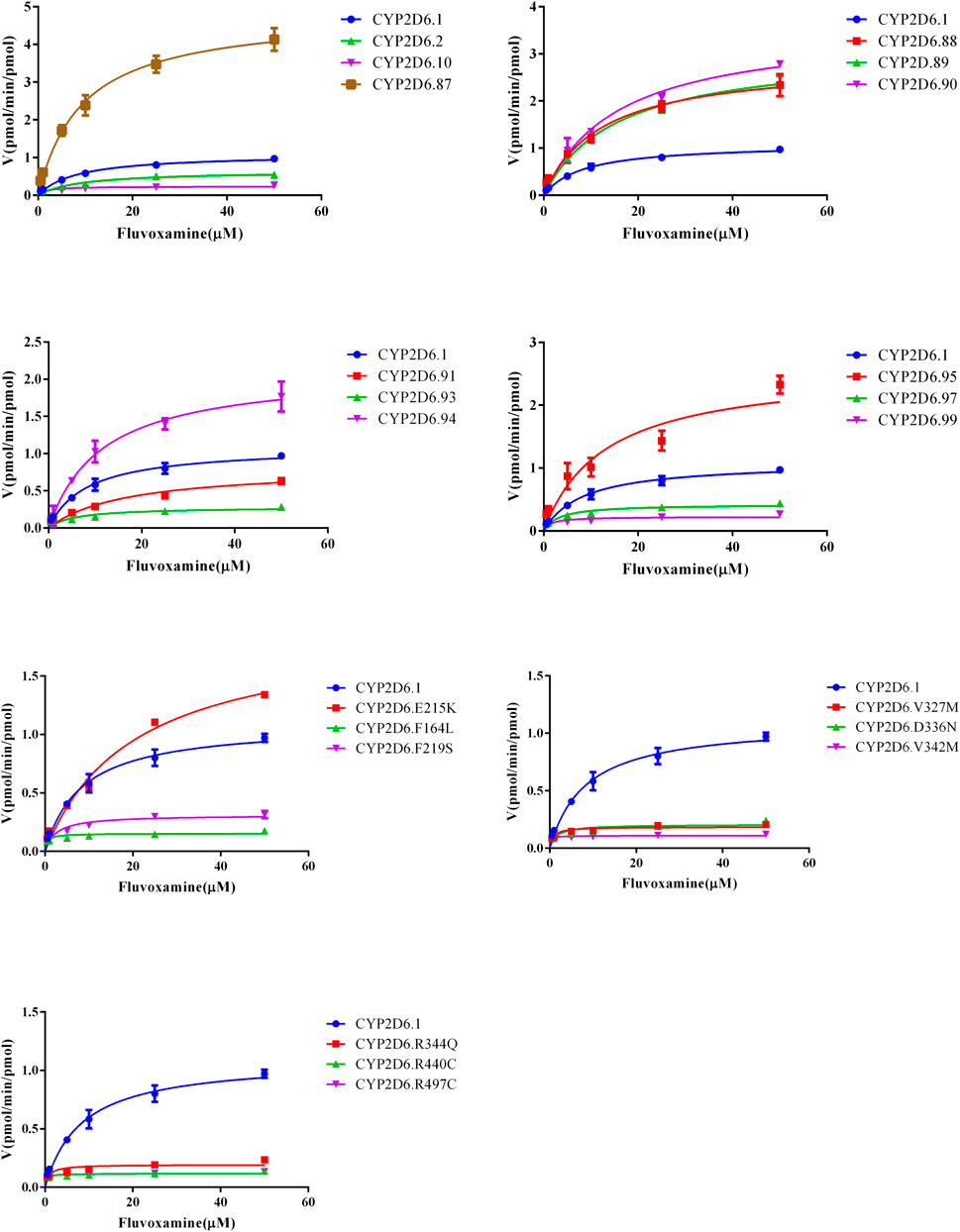
FIGURE 2. Michaelis–Menten curves of CYP2D6 in disposition of fluvoxamine. The reaction was performed as indicated in the section of method, n = 3.
As Figure 3A showed, when co-administration of fluvoxamine with apatinib, the production of desmethyl fluvoxamine was inhibited dramatically. The peak of the Y-axis of the time-concentration curve decreased, and the curve shifted to the right with significantly enhanced in Tmax. The AUC, t1/2, and Cmax reduced, but no significant difference was found, Table 2. Accordingly, the concentration of fluvoxamine increased remarkably after combination, Figure 3B. The AUC(0-t), AUC(0-∞) and Cmax values of fluvoxamine increased by 8.52-, 2.78-, 2.90-time, respectively. In addition, Tmax is prolonged by 3.80 times, CLz/F is reduced by about one time, and t1/2z is reduced by nearly one time, Table 3.
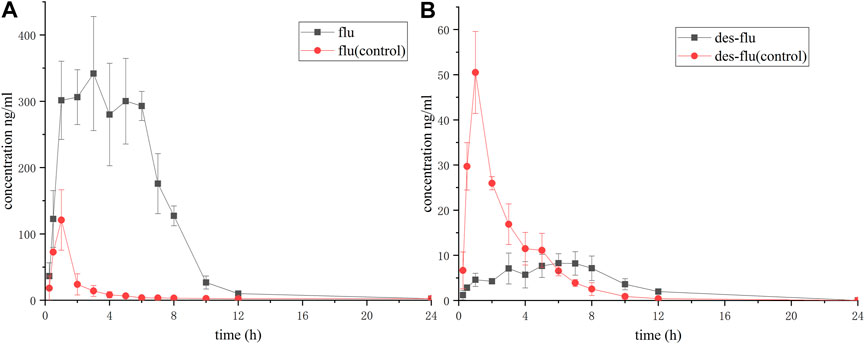
FIGURE 3. Concentration-time curve of fluvoxamine and its metabolite. The rats were administrated with fluvoxamine via gavage. As follows, the tail vein blood was collected, and was subjected to UPLC-MS/MS assay. (A) Desmethyl fluvoxamine, (B) Fluvoxamine. The curve was plotted using Prism 5, n = 6.
To study the mechanism underlied drug-drug interaction, the enzymatic reaction was performed using RLM, HLM. As shown in Figure 4, the Km of fluvoxamine metabolizing was 4.738 μM in RLM and 13.54 μM in HLM. To evaluate the inhibitory potency, the IC50 was determined. Fluvoxamine was dose-dependently inhibited by apatinib in RLM with IC50 of 0.19 μM, Figure 5A. In HLM, it’s 6.419 μM, Figure 5A. Mechanistically, apatinib inhibited fluvoxamine metabolism with a mixed manner in the RLM. The Ki is 0.05 μM, Figure 5B. Meanwhile, it is the same in HLM with Ki of 2.23 μM, Figure 5C. To further investigate differences in inhibitory activity among different CYP2D6 variants, IC50 was determined using CYP2D6.1, CYP2D6.2 and CYP2D6.10. It’s 17.58, 14.46 and 3.673 μM accordingly, Figure 6A. The relative metabolic rates of fluvoxamine were 44.32%, 54.03%, and 57.69%, respectively.
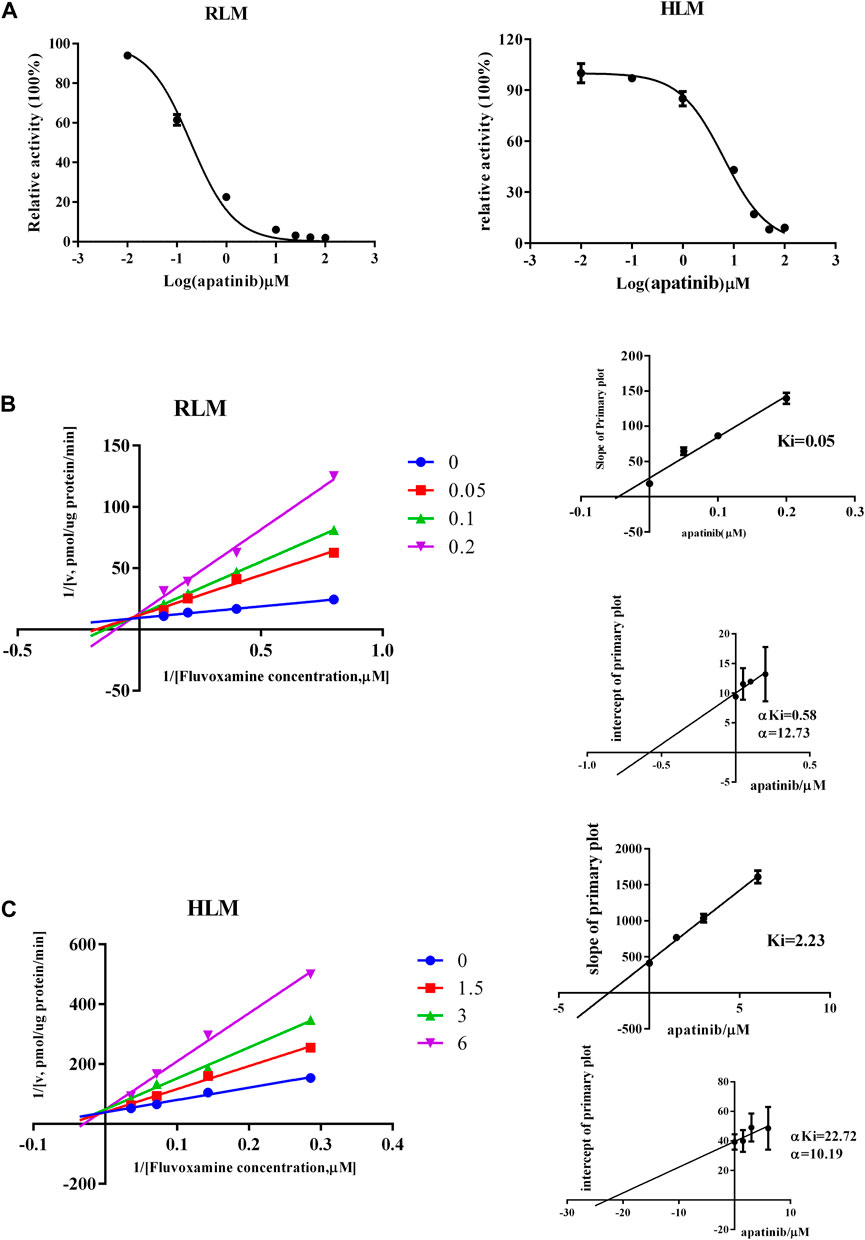
FIGURE 5. Apatinib inhibited the metabolism of fluvoxamine with a mixed mechanism both in RLM and HLM. (A) The effect of apatinib on inhibiting fluvoxamine metabolism. (B,C) Lineweaver-Burk plot and the secondary plot for Ki in the inhibition of fluvoxamine catalyzing, n = 3.
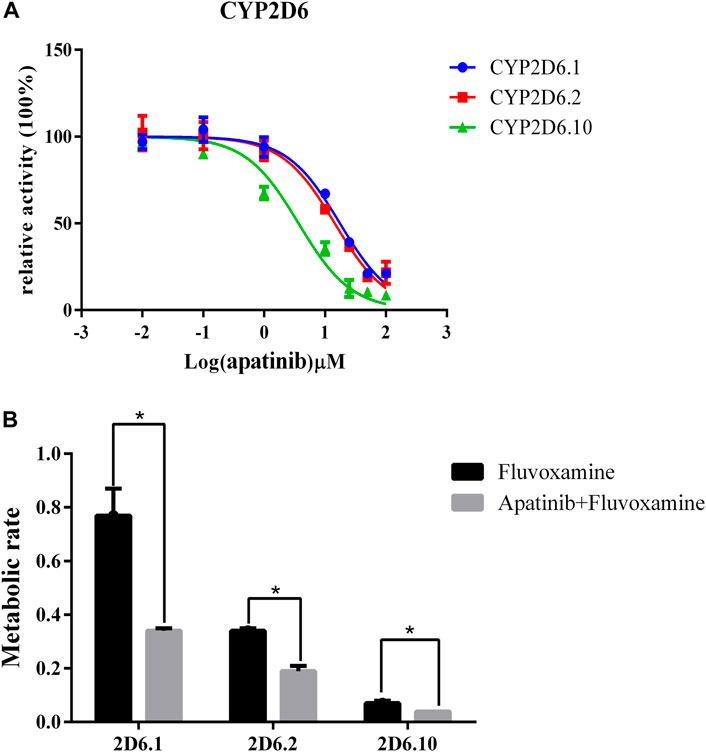
FIGURE 6. The different effect of apatinib on fluvoxamine metabolism in three CYP2D6 variants. (A) The catalytic profile of fluvoxamine in CYP2D6.1, 2 and 10. (B) The effect of apatinib on the metabolic rate of fluvoxamine in CYP2D6.1, 2, and 10, n = 3, *p < 0.05.
CYP2D6 accounts for about 2% of total CYP abundance in liver, and catalyzing metabolism of 20%–30% therapeutic drugs (Zhou et al., 2016; Taylor et al., 2020). To date, 149 CYP2D6 alleles have been reported (LLerena et al., 2014). However, the activities of a vast majority of variants are still unclear. Therefore, elucidating the effect of mutation on enzyme activity will help to understand the metabolic characteristics of CYP2D6 substrate drugs. Moreover, it will provide the basic data for precise medicine.
Fluvoxamine is almost completely absorbed through the gastrointestinal tract, and is mainly metabolized into desmethyl fluvoxamine in the liver through CYP2D6 and CYP1A2 pathway (van Harten, 1995). Interestingly, CYP2D6 is rarely being induced, but has abundance gene polymorphism. Therefore, genetic polymorphisms of CYP2D6 are likely to cause differences in plasma exposure of fluvoxamine. This study demonstrated that CYP2D6.A5V, V104A, D337G, F164L, V342M, R440C, R497C showed higher catalytic activity compared with CYP2D6.1. The patients carry these mutations would probably be sub-therapied. In contrast, CYP2D6*92 and *96 almost lost function in catalyzing fluvoxamine. CYP2D6*10 is predominantly distributed in East-Asia (Lin et al., 2016). It has been reported that its activity is significantly reduced. In the present study, we found that the Vmax of CYP2D6 decreased, the Km was also decreased accordingly. Therefore, the relative clearance is nearly the equal to CYP2D6.1. We think CYP2D6 has a certain selectivity for substrate drugs (Ingelman-Sundberg, 2005). The structure of the compound and the affinity of compound to the enzyme determine the characteristics of reaction (Zhou et al., 2009). However, these speculations require further study. Taken together, our data suggest that CYP2D6 gene polymorphisms indeed have varying degrees of impact on fluvoxamine metabolism.
Due to the existence of various complications, cancer patients usually take multiple drugs in combination, which can easily lead to drug-drug interaction (Flepisi et al., 2014; Moghaddas et al., 2021). Identifying potential drug interactions will help guide rational drug use and improve patients’ quality of life. Anxiety is a common complication in cancer patients (Kapfhammer, 2015). Therefore, combining apatinib with fluvoxamine is an effective dosing strategy. Many studies had shown that apatinib interacts with antipsychotics like buspirone and venlafaxine (Bao et al., 2018; Zhang et al., 2020). In this study, the data demonstrated that it can also inhibit the metabolizing of fluvoxamine via suppression the activities of microsomes system, especially CYP2D6. This inhibitory effect was similar even with different CYP2D6 alleles. Although this study preliminarily demonstrated differences in the rates of mutants metabolizing fluvoxamine through in vitro experiments, the affinity of the substrate to the enzyme was not determined, as well as in vivo experiments. Therefore, further data cannot be used to explain the in vivo situation, which has certain limitations in guiding clinical drug treatment. Since fluvoxamine and apatinib may be used clinically in combination, in this study we combined in vivo and in vitro experiments to demonstrate the interaction between fluvoxamine and apatinib. At the same time, the relevant experiments of HLM in vitro confirmed that apatinib may have a certain inhibitory effect on fluvoxamine in humans. In all, the present study provides basic data for the clinical application of fluvoxamine, especially in cancer patients. This prescription needs to be vigilant and prevent the occurrence of adverse reactions. Although prolonging lifespan is the primary goal for cancer patients, maximizing the quality of life is an urgent clinical problem, and this study provides limited data support for this goal.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by Ethics Committee of Wenzhou Medical University.
GH, JC, ZL, and BW contributed to the literature search and study design. BC, JQ, and ZY participated in drafting of the article. ZY, QK, XH, and NG carried out the experiments and analysis. GH and JQ revised the manuscript. All authors contributed to data analysis, drafting, or revising the article; agreed on the journal to which the article will be submitted; and have agreed to be accountable for all aspects of the work.
This work was supported by the Natural Science Foundation of Zhejiang Province (LGF20H310001), the National Key Research and Development Program of China (2020YFC2008301), and the National Natural Science Foundation of China (81973397).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.985159/full#supplementary-material
Bao, S. S., Wen, J., Lin, Q. M., Li, Y. H., Huang, Y. G., Zhou, H. Y., et al. (2018). Evaluation of the effects of apatinib on the pharmacokinetics of venlafaxine and O-desmethylvenlafaxine in SD male rats by UPLC-MS/MS. Basic Clin. Pharmacol. Toxicol. 123, 721–726. doi:10.1111/bcpt.13081
Cai, J., Dai, D. P., Geng, P. W., Wang, S. H., Wang, H., Zhan, Y. Y., et al. (2016). Effects of 22 novel CYP2D6 variants found in the Chinese population on the bufuralol and dextromethorphan metabolisms in vitro. Basic Clin. Pharmacol. Toxicol. 118, 190–199. doi:10.1111/bcpt.12478
Flepisi, B. T., Bouic, P., Sissolak, G., and Rosenkranz, B. (2014). Drug-drug interactions in HIV positive cancer patients. Biomed. Pharmacother. = Biomedecine Pharmacother. 68, 665–677. doi:10.1016/j.biopha.2014.04.010
Geng, R., Song, L., Li, J., and Zhao, L. (2018). The safety of apatinib for the treatment of gastric cancer. Expert Opin. Drug Saf. 17, 1145–1150. doi:10.1080/14740338.2018.1535592
Hicks, J. K., Bishop, J. R., Sangkuhl, K., Müller, D. J., Ji, Y., Leckband, S. G., et al. (2015). Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 98, 127–134. doi:10.1002/cpt.147
Ingelman-Sundberg, M. (2005). Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): Clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 5, 6–13. doi:10.1038/sj.tpj.6500285
Kapfhammer, H. P. (2015). Comorbid depressive and anxiety disorders in patients with cancer. Nervenarzt 86 (294-298), 291300–292291. doi:10.1007/s00115-014-4156-x
Lenze, E. J., Mattar, C., Zorumski, C. F., Stevens, A., Schweiger, J., Nicol, G. E., et al. (2020). Fluvoxamine vs placebo and clinical deterioration in outpatients with symptomatic COVID-19: A randomized clinical trial. Jama 324, 2292–2300. doi:10.1001/jama.2020.22760
Lin, G., Zhang, K., Yi, L., Han, Y., Xie, J., and Li, J. (2016). National prociency testing result of CYP2D6*10 genotyping for adjuvant tamoxifen therapy in China. PloS one 11, e0162361. doi:10.1371/journal.pone.0162361
LLerena, A., Naranjo, M. E., Rodrigues-Soares, F., Penas, L. E. M., Fariñas, H., and Tarazona-Santos, E. (2014). Interethnic variability of CYP2D6 alleles and of predicted and measured metabolic phenotypes across world populations. Expert Opin. Drug Metab. Toxicol. 10, 1569–1583. doi:10.1517/17425255.2014.964204
Moghaddas, A., Adib-Majlesi, M., Sabzghabaee, A. M., Hajigholami, A., and Riechelmann, R. (2021). Potential drug-drug interactions in hospitalized cancer patients: A report from the middle-east. J. Oncol. Pharm. Pract. 27, 46–53. doi:10.1177/1078155220910209
Mula, M., and Trimble, M. R. (2003). Pharmacokinetic interactions between antiepileptic and antidepressant drugs. World J. Biol. Psychiatry 4, 21–24. doi:10.3109/15622970309167906
Spina, E., Pisani, F., and de Leon, J. (2016). Clinically significant pharmacokinetic drug interactions of antiepileptic drugs with new antidepressants and new antipsychotics. Pharmacol. Res. 106, 72–86. doi:10.1016/j.phrs.2016.02.014
Taylor, C., Crosby, I., Yip, V., Maguire, P., Pirmohamed, M., and Turner, R. M. (2020). A review of the important role of CYP2D6 in pharmacogenomics. Genes (Basel) 11, E1295. doi:10.3390/genes11111295
van Harten, J. (1995). Overview of the pharmacokinetics of fluvoxamine. Clin. Pharmacokinet. 29 (1), 1–9. doi:10.2165/00003088-199500291-00003
Vignali, C., Moretti, M., Quaiotti, J., Freni, F., Tajana, L., Osculati, A. M. M., et al. (2021). Distribution of fluvoxamine and identification of the main metabolite in a fatal intoxication. J. Anal. Toxicol. 45, e1–e5. doi:10.1093/jat/bkaa084
Wang, N., Chen, S., Liu, D., Guo, J., Sun, Y., Zhang, J., et al. (2020a). Therapeutic effect of small molecule targeting drug apatinib on gastric cancer and its role in prognosis and anti-infection mechanism. Saudi J. Biol. Sci. 27, 606–610. doi:10.1016/j.sjbs.2019.11.036
Wang, Y. H., Li, J. Q., Shi, J. F., Que, J. Y., Liu, J. J., Lappin, J. M., et al. (2020b). Depression and anxiety in relation to cancer incidence and mortality: A systematic review and meta-analysis of cohort studies. Mol. Psychiatry 25, 1487–1499. doi:10.1038/s41380-019-0595-x
Yuan, Z., Chen, Z., Xue, M., Zhang, J., and Leng, L. (2020). Application of antidepressants in depression: A systematic review and meta-analysis. J. Clin. Neurosci. 80, 169–181. doi:10.1016/j.jocn.2020.08.013
Zastrozhin, M., Skryabin, V., Smirnov, V., Zastrozhina, A., Grishina, E., Ryzhikova, K., et al. (2021). Effect of genetic polymorphism of the CYP2D6 gene on the efficacy and safety of fluvoxamine in major depressive disorder. Am. J. Ther. 29, e26–e33. doi:10.1097/MJT.0000000000001388
Zhang, X. D., Li, Y. H., Chen, D. X., You, W. W., Hu, X. X., Chen, B. B., et al. (2020). The effect of apatinib on pharmacokinetic profile of buspirone both in vivo and in vitro. J. Pharm. Pharmacol. 72, 1405–1411. doi:10.1111/jphp.13320
Zhou, H. Y., Gu, E. M., Chen, Q. L., Zhan, Y. Y., Wang, S. H., Liang, B. Q., et al. (2016). Effects of 22 CYP2D6 genetic variations newly identified in Chinese population on olanzapine metabolism in vitro. Pharmacology 98, 124–133. doi:10.1159/000446807
Zhou, S. F., Liu, J. P., and Chowbay, B. (2009). Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab. Rev. 41, 89–295. doi:10.1080/03602530902843483
Keywords: CYP2D6, fluvoxamine, apatinib, interaction, metabolism
Citation: Ye Z, Chen B, Gao N, Kong Q, Hu X, Lu Z, Qian J, Hu G, Cai J and Wu B (2022) CYP2D6 gene polymorphism and apatinib affect the metabolic profile of fluvoxamine. Front. Pharmacol. 13:985159. doi: 10.3389/fphar.2022.985159
Received: 03 July 2022; Accepted: 11 August 2022;
Published: 02 September 2022.
Edited by:
Simona Pichini, National Institute of Health (ISS), ItalyReviewed by:
Niyaz Ahmad, Imam Abdulrahman Bin Faisal University, Saudi ArabiaCopyright © 2022 Ye, Chen, Gao, Kong, Hu, Lu, Qian, Hu, Cai and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoxin Hu, aGd4QHdtdS5lZHUuY24=; Jianping Cai, Y2FpanA2MUB2aXAuc2luYS5jb20=; Bin Wu, ZHJ3enl5d2JAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.