
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol. , 02 September 2022
Sec. Pharmacology of Anti-Cancer Drugs
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.984453
This article is part of the Research Topic Epigenetic Regulation and Therapy Resistance in Cancer View all 5 articles
Hepatocellular carcinoma (HCC) is a highly mortal type of primary liver cancer. Abnormal epigenetic modifications are present in HCC, and RNA modification is dynamic and reversible and is a key post-transcriptional regulator. With the in-depth study of post-transcriptional modifications, RNA modifications are aberrantly expressed in human cancers. Moreover, the regulators of RNA modifications can be used as potential targets for cancer therapy. In RNA modifications, N6-methyladenosine (m6A), N7-methylguanosine (m7G), and 5-methylcytosine (m5C) and their regulators have important regulatory roles in HCC progression and represent potential novel biomarkers for the confirmation of diagnosis and treatment of HCC. This review focuses on RNA modifications in HCC and the roles and mechanisms of m6A, m7G, m5C, N1-methyladenosine (m1A), N3-methylcytosine (m3C), and pseudouridine (ψ) on its development and maintenance. The potential therapeutic strategies of RNA modifications are elaborated for HCC.
Hepatocellular carcinoma (HCC) is the fifth most prevalent cancer worldwide (Llovet et al., 2021). As an important histological subtype of liver cancer, HCC is characterized by high recurrence rates and heterogeneity and represents a major health challenge (Bosch et al., 1999). HCC has a poor prognosis, making it crucial to investigate the molecular mechanisms and therapeutic strategies underlying hepatocellular carcinogenesis (Llovet et al., 2008; Chen et al., 2015). It has been shown that hepatocellular carcinogenesis is regulated by complex genetic and epigenetic mechanisms with pathophysiological processes involving viral and non-viral factors and influenced by fatty liver, immune cell infiltration, and the tumor microenvironment (Ozen et al., 2013; Han et al., 2018; Pea et al., 2021; Braghini et al., 2022). A study using whole-genome and -exome sequencing analysis has shown that epigenetic regulation is the most unusual differential modifier in HCC, with aberrant RNA modifications leading to differential gene expression, affecting the HCC formation and progression (Schulze et al., 2015; Klingenberg et al., 2017; Chen et al., 2018). One of the characteristics of tumor cells is their dynamic adaptation to a microenvironment that is harmful to them. Therefore, modulating RNA modifications can dynamically regulate tumor cell proliferation and apoptosis, affecting the fate of both normal and cancerous cells (Haruehanroengra et al., 2020; Uddin et al., 2020).
Epigenetics is a branch in the field of genetics which explores the phenotypes that can be inherited from chromosomal changes which do not modify the nucleotide sequence of genes, of which RNA modification has been a recent hotspot of epigenetic research (Fu et al., 2014). In nature, dynamically created RNA modifications are widely present on all nucleotides (Teng et al., 2021; Willbanks et al., 2021). In the context of advances in high-throughput sequencing technologies, there has been a landmark breakthrough in the identification and characterization of human transcriptome modifications (Zhao et al., 2017). To date, over 170 different types of RNA modification have been detected, existing not only on transfer (tRNA), ribosomal (rRNA), and messenger (mRNA) RNA but also on other noncoding RNAs (ncRNAs) such as long-noncoding RNA (lncRNA) and microRNA (miRNA), and have direct or indirect functional effects on gene expression and regulate a wide range of biological activities (Boccaletto et al., 2018; Rong et al., 2021a). RNA modifications that have been subject to intensive study include N6-methyladenosine (m6A), N7-methylguanosine (m7G), 5-methylcytosine (m5C), N1-methyladenosine (m1A), N3-methylcytosine (m3C), and pseudouridine (ψ) (Delaunay and Frye, 2019; Zhang et al., 2021a). However, the biological functions and mechanisms of these RNA modifications continue to be discovered within the complex epigenomic transcriptome environment of HCC.
The epigenomic environment is highly complex. RNA modification is an important epigenetic regulatory mechanism, regulating the structure, function, and stability of RNA and controlling the expression of related genes (Roundtree et al., 2017). RNA modifications are highly relevant to biological factors including RNA decay, stability, metabolism, and binding to RNA-binding proteins (Shi et al., 2020a; Boo and Kim, 2020; Wang et al., 2021a). A large majority of works indicate that dysregulation of RNA epigenetic pathways is tightly linked to the pathogenesis of human diseases, particularly cancer (Delaunay and Frye, 2019; Yang et al., 2019; Barbieri and Kouzarides, 2020; Liang et al., 2020). Cancer is a type of disease featured by the progressive build-up of genetic and epigenetic modifications of unique cancer-causing and oncogenic genes that regulate cellular behaviors such as proliferation, differentiation, and migration. Indeed, dynamic RNA modifications are implicated in the metabolic RNA process, an important emerging regulator in cancer (Ozen et al., 2013; Delaunay and Frye, 2019; Han et al., 2021). Directing abnormal posttranscriptional modifications in carcinoma cells promises to be an effective tumor therapy (Zhao et al., 2014; Geula et al., 2015; Su et al., 2018; Janin et al., 2019; Uddin et al., 2020; Chen et al., 2021a; Han et al., 2021; Nombela et al., 2021). This review concentrates on the functions of six RNA modifications that regulate cancer progression in HCC.
First discovered in 1974, m6A is the most popular RNA modification (Figure 1A). m6A modifications are dynamically reversible and are primarily found in mRNA, rRNA, tRNA, miRNA, circular RNA (circRNA), lncRNA, and small nuclear RNA (snRNA), regulating molecular functions and participating in the pathogenesis of various diseases, including cancer (Figure 2) (Desrosiers et al., 1974; Krug et al., 1976; Horowitz et al., 1984; Björk et al., 1987; Shimba et al., 1995; Piekna-Przybylska et al., 2008; Wang and He, 2014; Alarcón et al., 2015; Ben-Haim et al., 2015; Kennedy et al., 2016; Patil et al., 2016; Bartosovic et al., 2017; Yang et al., 2017b; Coots et al., 2017; Rosa-Mercado et al., 2017; Zhou et al., 2017; Liu et al., 2018; Taketo et al., 2018; Yang et al., 2018; van Tran et al., 2019; Xiao et al., 2019; Zhang et al., 2020b; Di Timoteo et al., 2020). m6A modifications not only affect RNA stability, splicing, transcription, processing, translation, and metabolism, they regulate gene expression in a wide range of physiological processes and play an important role in cellular differentiation, apoptosis, and proliferation (Meyer et al., 2015; Yue et al., 2015; Yoon et al., 2017; Zhao et al., 2017; Chen et al., 2018).
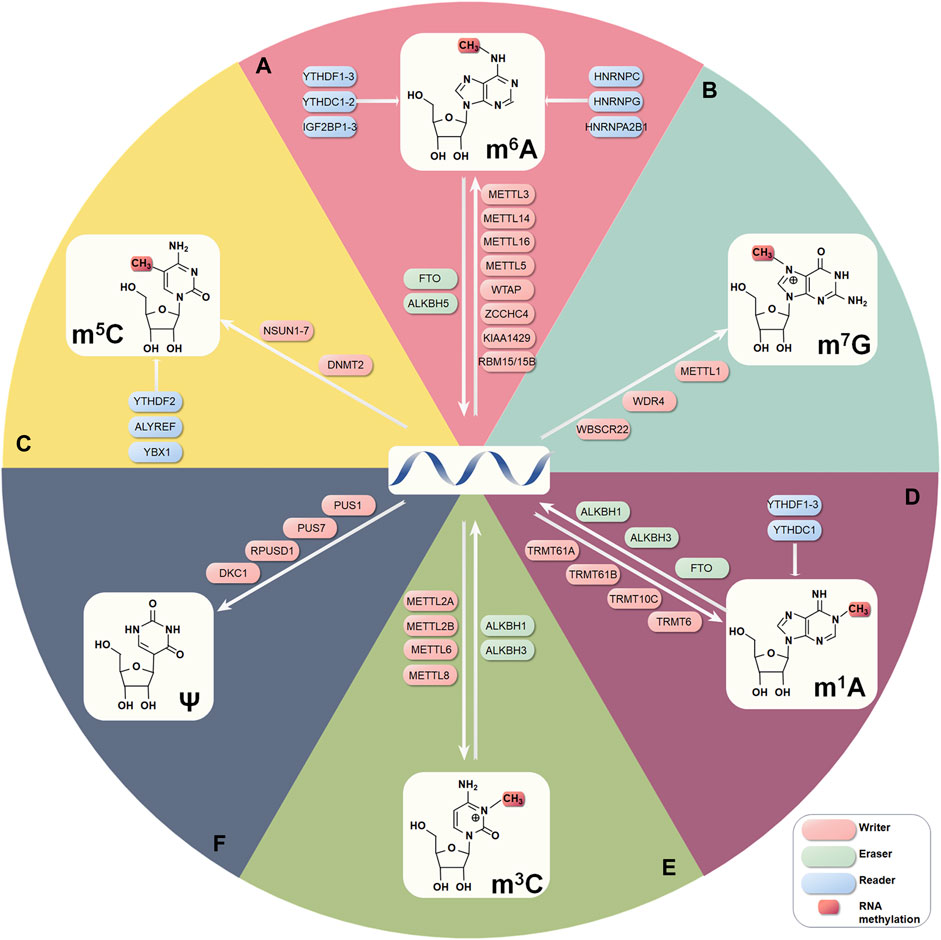
FIGURE 1. The six internal RNA modifications focused on in this review and their regulatory mechanisms. (A) m6A. (B) m7G. (C) m5C. (D) m1A. (E) m3C. (F) Ψ.
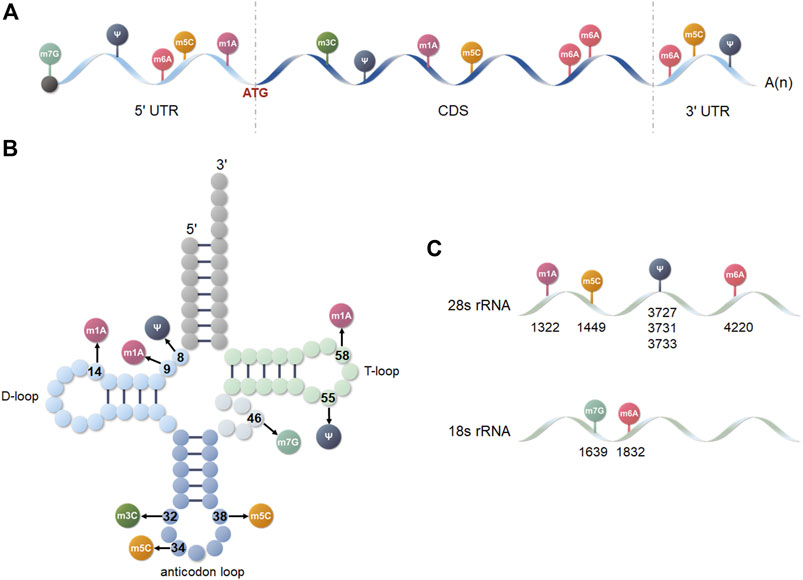
FIGURE 2. Distribution of the six posttranscriptional modifications on different RNA isoforms. (A) mRNA. (B) tRNA. (C) 18 and 28s rRNA.
Accurate detection of RNA modifications is the basis of epigenomic studies, and several types of assays for m6A modifications have been developed. Antibody-based assays include methylated RNA immunoprecipitation sequencing (m6A-MeRIP-seq) and m6A-level and isoform-characterization sequencing (m6A-LAIC-seq), enabling measurement of transcriptome-wide m6A modification levels (Dominissini et al., 2012; Meyer et al., 2012; Linder et al., 2015; Molinie et al., 2016). The digestion-based sequencing assays MazF RNase assisted sequencing (MAZTER-seq) and m6A-sensitive RNA-endoribonuclease–facilitated sequencing (m6A-REF-seq) are simple and efficient (Zhang et al., 2019b; Garcia-Campos et al., 2019; Pandey and Pillai, 2019). Ligation-based detection methods are highly sensitive and low cost, including single-base elongation- and ligation-based qPCR amplification method (SELECT) and locus-specific extension of annealed DNA probes targeting m6A and sequencing (LEAD-m6A-seq) (Xiao et al., 2018; Wang et al., 2021d). Deaminase sequencing adjacent to RNA modification targets (DART-seq) is an antibody-free m6A sequencing method based on gene editing (Meyer, 2019). In addition, the DART-seq-based single-cell assay (scDART-seq) is the first method to examine m6A sites in single cells. Nanopore RNA sequencing (RNA-seq) is a real-time sequencing technique that is simple and sensitive, greatly expanding the scope of m6A research and providing a scientific platform for the quantitative detection of m6A modifications (Garalde et al., 2018). However, due to the still high error rate of nanopore sequencing itself and the relatively small number of RNA methylation modifications, which may be below the threshold level, the detection is prone to have a high false positive rate (Leger et al., 2021). The detection of RNA modifications at single molecule level with single nucleotide resolution can be chosen as a classical sequencing tool to ensure the accuracy of detection. Overall, these m6A sequencing methods could provide potential clues for HCC diagnosis and treatment.
The m6A methylation modification is written by methyltransferase 3 (METTL3), 14 (METTL14), and 16 (METTL16), RNA binding motif proteins 15 (RBM15) and 15B (RBM15B), KIAA1429, and Wilms tumor 1-associated protein (WTAP). It is subsequently erased by the demethylases alkane hydroxylase homolog 5 (ALKBH5) and fat mass and obesity-associated protein (FTO). Proteins that read the modification include members of the YT521-B homolog (YTH) structural domain family of m6A RNA binding proteins (YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2), insulin-like growth factor 2 mRNA-binding protein family (IGF2BP1, IGF2BP2, and IGF2BP3), and heterogeneous nuclear ribonucleoprotein family (HNRNPA2B1, HNRNPC, and HNRNPG), and proline-rich and coiled-coil protein 2A (PRRC2A) (Schwartz et al., 2014b; Fu et al., 2014; Meyer and Jaffrey, 2014; Haussmann et al., 2016; Schumann et al., 2016; Schwartz, 2016; Arguello et al., 2017; Edupuganti et al., 2017; Meyer and Jaffrey, 2017; Wu et al., 2017; Müller et al., 2019). m6A methyltransferases and demethylases are associated with various diseases, including cancer (Boissel et al., 2009; Church et al., 2010; Boles and Temple, 2017; Wang et al., 2018; Chen et al., 2019b).
The addition of m6A onto mRNA, long intergenic non-coding RNA (lincRNA), and miRNA is catalyzed by the METTL3-METTL14 complex (Liu et al., 2014; Wang et al., 2016b). METTL3 is the central element of catalysis that converts adenosine to m6A through its methyltransferase structural domain, enhances mRNA translation, and affects gene expression and the pattern of alternative splicing (Dominissini et al., 2012; Schumann et al., 2016; Bartosovic et al., 2017; Ma et al., 2017; Rosa-Mercado et al., 2017). METTL14 recognizes RNA substrates as well as recruits microprocessor complexes for co-transcription on primary (pri)-miRNA transcripts (Huang et al., 2019). The combination of the two methyltransferases improves the methylation rate and has a cooperative influence, preferentially methylating RNA primers to contain the consensual sequence GGACU (Bokar et al., 1994; Liu et al., 2014; Wang et al., 2014; Wang et al., 2016a; Wang et al., 2016b; Ke et al., 2017). WTAP is the third key member as a regulatory subunit of the mammalian m6A methyltransferase enzyme complex, enhancing methyltransferase catalytic activity (Horiuchi et al., 2006; Ping et al., 2014). Indeed, the major m6A mediators have a significant role in cancer causation or inhibition in different cancer types. METTL3 is shown to be highly explored in advanced acute myeloid leukemia (AML) and has reported roles in lung cancer and HCC (Lin et al., 2016; Vu et al., 2017; Chen et al., 2018). METTL16 was shown to be required for HCC cell proliferation, while METTL3 and METTL14 have conflicting roles in HCC (Vu et al., 2017; Chen et al., 2018).
The addition of m6A is reversible with reliance on a coordinated and highly dynamic system of methyltransferases and demethylases. m6A demethylases FTO and ALKBH5 are part of the non-heme Fe(II)/α-ketoglutarate-dependent dioxygenase family (Gerken et al., 2007; Fu et al., 2010; Jia et al., 2011; Zheng et al., 2013). FTO is related to human obesity and mental development and was the first m6A mRNA demethylase identified to convert mRNA m6A to adenosine in mRNA (Dina et al., 2007; Loos and Yeo, 2014; Mauer et al., 2017; Wei et al., 2018). ALKBH5 reverses m6A to adenosine in a direct manner and influences mRNA export and RNA turnover in a demethylation-dependent way (Zheng et al., 2013). Indeed, FTO is a key factor regulating HCC, promoting migration, invasion, and proliferation of HCC cells, countering the function of ALKBH5 in HCC(Fischer et al., 2009; Ueda et al., 2017).
m6A has a major role in cell differentiation with tissue development. It can regulate the entire RNA lifecycle, in terms of RNA processing, decay, and translation, through the identification of specific binding proteins, dynamically regulating physiological and pathological processes such as tumorigenesis (Allis and Jenuwein, 2016; Shi et al., 2017; Chen et al., 2019b; Han et al., 2019b; Wu et al., 2019b). m6A writers have a common RNA-binding YTH structural domain, including YTHDC1, YTHDC2, YTHDF1, YTHDF1 YTHDF2, and YTHDF3 (Li et al., 2014; Xu et al., 2014). Eukaryotic initiation factor 3 (eIF3), HNRNPC, HNRNPA2B1, IGF2BP1, IGF2BP2, IGF2BP3, PRRC2A, and fragile X mental retardation protein (FMRP) also proved to be m6A writers (Liu et al., 2015; Liu et al., 2017). YTH structural domain proteins recognize m6A modifications specifically and manage mRNA decay, translation, and maturation (Liu et al., 2017). Heterodimers of YTH structural domain family (YTHDF) proteins are mainly found in the cytoplasm, while m6A affects mRNA decay upon the combination of three YTHDF members (Zaccara and Jaffrey, 2020).
m6A promotes the binding of regulatory proteins by altering the structure of RNA to regulate gene expression and RNA maturation (Wan et al., 2014). The IGF2BPs family of m6A readers specifically recognize the GG (m6A) C motif through its K homology (KH) structural domain, promote the structured stability and translations in most target mRNAs, and are highly expressed in various cancers and involved in many different molecular mechanisms (Huang et al., 2018). IGF2BPs act as m6A readers during post-transcriptional gene regulation and the biology of cancer (Huang et al., 2018).
There is growing evidence suggesting that abnormal changes in the levels of total m6A and its regulators in HCC tissues are strongly connected with poor clinical survival (Chen et al., 2020; Li et al., 2021c). m6A regulators of the modifications perform oncogenic or anti-tumor effects in HCC by influencing the expression of particular genes.
m6A and its regulators are closely related to oncogenic or tumor suppressor functions and play a role as oncogenes or antioncogenes in malignant tumors (Zhou et al., 2015; Lin et al., 2016; Li et al., 2017b; Xiang et al., 2017; Zhong et al., 2018; Han et al., 2019a). With the intensive study of HCC, m6A modifiers and regulators have been found to take a pivotal role in the development and progression of HCC, capable of regulating HCC cell phenotype, migration, invasion, and epithelial-mesenchymal transition (EMT) processes, and a potential therapeutic target for HCC treatment. (Figure 3). Many studies have provided data to support the expression of m6A modifications and m6A regulators in HCC via bioinformatics analysis (Liu et al., 2020a; Jiang et al., 2021). METTL3, YTHDF1, YTHDF2, YTHDF3, YTHDC1, YTHDC2, FTO, KIAA1429, HNRNPC, HNRNPA2B1, and RBM15 are all overexpressed in HCC (Liu et al., 2020a). One study found higher expression levels of FTO, ALKBH5, RBM15, and WTAP in HCC through the TCGA database (Jiang et al., 2021). Copy number variation (CNV), mutation, and clinical databases of HCC patients from The Cancer Genome Atlas (TCGA) showed differential expression of the m6A regulators in HCC, significantly associated with clinicopathological features, and correlated with sorafenib treatment effect (Jiang et al., 2021). Patients with m6A regulator mutations showed inferior overall survival (OS) and disease-free survival (DFS).
METTL3 is highly expressed in HCC, METTL3 regulates RNA expression in HCC in an m6A-dependent manner and is independent of recurrence-free survival (RFS) as a prognostic factor, promoting HCC cell proliferation, colony formation, migration, and tumorigenicity in vitro (Supplementary Table S1) (Liu et al., 2020d; Qu et al., 2020). METTL3 could serve as a potential HCC prognostic biological marker and contribute to HCC carcinogenesis, tumor progression, and drug resistance (Chen et al., 2018; Liu et al., 2021a; Pan et al., 2021). Studies have shown that overexpression of promotes HCC progression by increasing the level of m6A modification and suppressing the expression of cytokine signaling 2 (SOCS2) in HCC by a YTHDF2-dependent mechanism (Chen et al., 2018).
HCC correlates with ubiquitination of METTL3, METTL3 SUMOylation was increased in HCC, and the UBC9/SUMOylated METTL3/Snail axis is a novel mediator of sumoylation (SUMO) pathway involvement in HCC progression (Xu et al., 2020). In addition, inhibition of METTL3 to inhibit glycolysis is also a potential strategy for HCC treatment (Lin et al., 2020a). Another study showed that overexpression of METTL3 promoted invasion, migration, and proliferation of HCC cells by upregulating USP7 expression through m6A methylation (Li et al., 2021d). Furthermore, hepatitis B virus X-interacting protein (HBXIP)-mediated METTL3 upregulation driven by hypoxia-inducible factor 1-α (HIF-1α) promotes metabolic reprogramming in HCC cells (Yang et al., 2021).
Sorafenib is a first-line drug approved for treating advanced HCC. However, sorafenib resistance reduces its efficacy in HCC(Vogel et al., 2021). METTL3 is found to be significantly decreased in sorafenib-resistant HCC, and depletion of METTL3 under conditions of hypoxia promotes sorafenib tolerance and angiogenic gene expression in HCC cells and activates autophagy-related pathways (Lin et al., 2020b). Overexpression of forkhead box O3 (FOXO3) rescues the METTL3 depletion-induced sorafenib resistance phenotype. FOXO3 is an essential marker of m6A modification in HCC in its resistance to sorafenib (Lin et al., 2020b). lincRNA 1,273 (LINC01273) is significantly overexpressed in sorafenib-resistant HCC tissues, binds complementarily to miR-600, promotes miR-600-targeted downregulation of METTL3 expression levels, reduces m6A modification in HCC, and promotes HCC progression. The LINC01273/miR-600/METTL3 feedback regulatory axis is a promising intervention pathway for patients with sorafenib-resistant HCC (Kong et al., 2022).
The METTL3/IGF2BP1 complex enhanced the stability of NIFK-AS1 mRNA by increasing the expression level of m6A, promoted the growth and invasion of HCC cells, and promoted the progression of HCC through the AKT1-MMP7/9 axis (Chen et al., 2021d). In addition, METTL3-mediated m6A modification down-regulates the expression level of maternally expressed 3 (MEG3), which targets miRNA 544b (miR-544b), miR-544b promotes HCC proliferation, invasion, and metastasis by suppressing BTG2 expression (Wu et al., 2021b). Interleukin enhancer-binding factor 3 (IlF3) antisense RNA 1 (ILF3-AS1) promotes HCC cell proliferation, migration, and invasion increases ILF3 m6A levels in an m6A-dependent manner via METTL3, and is an important oncogene in HCC progression, providing a new therapeutic target for HCC (Bo et al., 2021). METTL3 mediates m6A modification to upregulate the expression of lincRNA 958 (LINC00958), which promotes HCC production and development by binding to miRNA 3619-5p (miR-3619-5p) to upregulate the expression of HCC growth factor (HDGF) (Zuo et al., 2020). Moreover, METTL3 accelerates HCC progression by upregulating the m6A level of circRNA hsa_circ_0058493 and promoting its binding to YTHDC1 (Wu et al., 2021a). METTL3 mediates the expression of m6A to promote the formation of circRNA HPS5 (circHPS5), which can act as a miRNA 370 (miR-370) sponge to regulate the expression of high-mobility group AT-hook 2 (HMGA2) and further accelerate tumorigenesis in HCC cells, providing a potential prognostic marker and therapeutic target (Rong et al., 2021b). Hepatitis B virus (HBV) infection is one of the main risk elements for HCC. METTL3 mediates lncRNA mitogen-activated protein kinase activation of protein kinase five antisense RNA 1 (MAAS) targeting c-Myc to promote the proliferation of HBV + HCC cells (Tao et al., 2022). It has also been shown that HBV x protein (HBx) upregulates METTL3 expression, increases m6A modification of circRNA ARL3 (circ-ARL3), and circ-ARL3 antagonizes the repressive effect of miRNA 1,305 (miR-1305) on oncogenes, thereby promoting HBV + HCC progression, and targeting this pathway is a promising approach for treating HBV + HCC patients (Rao et al., 2021).
METTL14 is a major gene for aberrant m6A modification that is downregulated and can act as a poor element for recurrence-free survival associated with tumor metastasis in HCC (Ma et al., 2017; Shi et al., 2020c). Overexpression of METTL14 inhibits HCC malignant progression by upregulating m6A expression levels of genes critical for prognosis in HCC patients, such as cysteine sulfite decarboxylase, glutamate-oxaloacetate transaminase 2, and cytokine signaling inhibitor 2 (Li et al., 2020). METTL14 is lowly expressed in HCC, and overexpression of METTL14 is able to promote m6A by increasing the binding of the microprocessor protein DiGeorge syndrome critical region 8 (DGCR8) and pri-miR126 to inhibit HCC progression (Ma et al., 2017). METTL14 regulates the epidermal growth factor receptor (EGFR)/phosphoinositide 3-kinase (PI3K)/AKT signaling pathway to inhibit migration, invasion, and EMT of HCC cells (Shi et al., 2020c). METTL14 mediates downregulation of hepatocyte nuclear factor 3γ (HNF3γ) expression levels, promotes cell differentiation to inhibit HCC progression, and is sensitive to sorafenib-induced growth inhibition and apoptosis in HCC cells (Zhou et al., 2020). Hypoxia triggers METTL14 inhibition in a HIF-1α-dependent manner, effectively eliminating iron death in HCC cells. METTL14 induces degradation of solute carrier family seven member 11 (SLC7A11) in a YTHDF2-dependent pathway, and the HIF-1α/METTL14/YTHDF2/SLC7A11 axis is a potential therapeutic target for HCC interventional embolization therapy. therapeutic target (Fan et al., 2021b). Additionally, METTL14-mediated m6A modification maintains sirtuin 6 (SIRT6) stability by regulating the expression of ubiquitin-specific peptidase 48 (USP48) and promoting the tumor suppressive function of glycolysis to regulate the metabolic activity of HCC (Du et al., 2021).
WTAP is expressed at a high level in HCC, promotes HCC development, is associated with poor prognosis, and is shown to be an independent predictor for survival in HCC (Chen et al., 2019c). Knockdown of WTAP can upregulate and promote autophagy in HCC, promotes liver kinase B1 (LKB1) expression, increases adenosine 5′monophosphate-activated protein kinase (AMPK) phosphorylation, and inhibits HCC proliferation (Li et al., 2021a). WTAP also promotes the proliferative capacity and growth of HCC cells (Chen et al., 2019c). ETS proto-oncogene 1 (ETS1) is a downstream effector of WTAP and WTAP-regulated m6A modification leads to post-transcriptional repression of ETS1, which promotes HCC progression through the human antigen R (HuR)-ETS1-p21/p27 axis and is a potential target for HCC therapy (Chen et al., 2019c). Zinc finger CCCH domain-containing protein 13 (ZC3H13) is a potential HCC target for biomarker and therapy, is significantly downregulated in HCC, and downregulation of ZC3H13 mediated by miRNA 362-3p (miR-362-3p) and miRNA 5425-5p (miR-425-5p) is associated with poor prognosis and cancer immune invasion in HCC (Wu et al., 2022). As the largest known element of the m6A methyltransferase, KIAA1429 is shown to be highly upregulated in HCC tissues and related to poor prognosis in HCC patients (Cheng et al., 2019; Lan et al., 2019). KIAA1429 promotes the malignant phenotype of HCC cells by inducing m6A methylation of GATA binding protein 3 (GATA3) to downregulate its expression, thus providing a new strategy for HCC treatment (Lan et al., 2019). In addition, KIAA1429 inhibits the inhibitor of DNA binding 2 (ID2) mRNA by upregulating its m6A modification, promoting HCC migration and invasion (Cheng et al., 2019). Overexpression of circ_KIAA1429 promotes HCC migration, invasion, and EMT processes and accelerates HCC progression through the m6A/YTHDF3/Zinc finger E box binding homeobox1 (ZEB1) pathway (Wang et al., 2020a). It has been shown that circRNA DLC1 (circDLC1) is downregulated in HCC tissues, and knockdown of KIAA1429 is able to increase the expression level of circDLC1 and reduce the interaction between HuR and matrix metallopeptidase 1 (MMP1), thereby inhibiting MMP1 expression and ultimately contributing to the inhibition of HCC progression (Liu et al., 2021b). Recent research has shown that is aberrantly overexpressed in HCC, is associated with poor prognosis, promotes tumorigenesis by regulating eIF3a/b expression, and could be a potential new target for HCC therapy (Su et al., 2022).
Reduced expression of FTO in HCC cells and tissues leads to the accumulation of m6A in HCC and is associated with poor patient prognosis, and it may serve as a novel biomarker for HCC (Zhao et al., 2019a). One study demonstrated that overexpression of FTO was able to block the proliferation of HCC cells and inhibit HCC progression by targeting the downregulation of CUL4A expression (Mittenbühler et al., 2020). Deacetylase silencing information regulator 1 (SIRT1) promotes HCC progression by downregulating the expression of FTO, increasing the expression level of m6A, and decreasing the expression level of G protein subunit αO1 (GNA O 1) (Liu et al., 2020b). Transarterial chemoembolization (TACE) has dramatically improved the OS of some patients with unresectable HCC (Liu et al., 2021c). A study found that YTHDC2 expression levels were upregulated in HCC tissues while FTO expression levels were decreased, and functional single nucleotide polymorphisms (SNPs) in YTHDC2 and FTO were found to have prognostic value in TACE-treated HCC patients (Liu et al., 2021c).
ALKBH5 expression is downregulated in HCC and inhibits the proliferation as well as the invasive ability in HCC cells, which is an indicator of an independent prognosis in HCC patients. ALKBH5-mediated m6A demethylation in HCC cells attenuates LY6/PLAUR domain containing 1 (LYPD1) expression and inhibits the malignant progression of HCC (Chen et al., 2020). In HBV-HCC tissues, depletion of ALKBH5 significantly inhibited HBV-driven tumor cell growth and migration. HBV could upregulate ALKBH5 through the HBx-WDR5-H3K4me3 axis and promote the progression of HCC (Fang and Chen, 2020; Qu et al., 2021). ALKBH5 can upregulate nuclear paraspeckle assembly transcript 1 (NEAT1) expression by inhibiting m6A enrichment, and NEAT1 promotes cell proliferation and migration of HCC by sponging miRNA 214 (miR-214) (Yeermaike et al., 2022). ALKBH5 mediates circular cleavage and polyadenylation-specific factor 6 (circCPSF6) demethylation, promoting the malignant development of HCC through aberrant activation of the circCPSF6 and Yes1 associated transcriptional regulator (YAP1) axis, providing new insights into the regulation of circRNAs by m6A modification together with epigenetic reprogramming in HCC (Chen et al., 2022). Both FTO and ALKBH5 may serve as potential targets for the treatment of HCC. Although the above studies demonstrated that ALKBH5 can be a therapeutic target for HCC and overexpression of ALKBH5 can inhibit the progression of HCC. However, other studies have shown that ALKBH5 is involved in DNA repair and that overexpression of ALKBH5 can promote DNA alkylation repair (Akula et al., 2021). Since DNA repair may prolong the telomere length of cancer cells and allow them to continue to proliferate, there is a need to balance the therapeutic effects of ALKBH5 on cancers such as HCC with the promotion of cancer cells through DNA repair (Roake and Artandi, 2016). Anticancer agents that target the DNA repair pathway, such as PARP inhibitors, have been developed to promote apoptosis in cancer cells by inhibiting DNA damage repair in cancer cells (Gavande et al., 2016; Huang and Zhou, 2021). This type of inhibitor can be utilized synergistically with ALKBH5 to enhance the efficacy of targeting ALKBH5 for the treatment of HCC.
The expression of YTHDF1 was elevated in HCC and showed a positive correlation with the pathological HCC stage (Zhao et al., 2018a). Knockdown YTHDF1 remarkably inhibited the proliferation, migration, and invasion of HCC (Zhao et al., 2018a).
YTHDF1 mediates frizzled class receptor 5 (FZD5) expression to regulate the WNT/β-catenin axis to exert oncogenic function as a potential HCC therapeutic strategy (Liu et al., 2020e). YTHDF1 promotes the malignant phenotype of HCC by activating AKT/glycogen synthase kinase (GSK)/3β/β-catenin signaling and promoting EMT (Bian et al., 2020). YTHDF1 can also promote HCC progression by activating the PI3K/AKT/mammalian target of rapamycin (mTOR) signaling route to induce EMT (Luo et al., 2021). It has been shown that overexpression of YTHDF1 depends on high expression of m6A modifications in HCC, inhibits cellular infiltration of CD3+ and CD8+ T, and promotes malignant progression of HCC (Li et al., 2021b). YTHDF1 deficiency inhibits HCC metastasis, growth, and autophagy (Li et al., 2021c). HIF-1α-regulated YTHDF1 expression enhanced the translation of autophagy-associated 2A (ATG2A) and 14 (ATG14) in an m6A-dependent manner to drive hypoxia-induced HCC autophagy It was shown that radiofrequency ablation (IRFA)-deficient sublethal heat stress promotes HCC progression by upregulating EGFR expression levels through an N6-methyladenosine mRNA methylation-dependent mechanism (Su et al., 2021). The m6A/YTHDF1/EGFR axis promotes HCC progression after IRFA, providing a rationale for YTHDF1 binding to EGFR to regulate HCC progression (Su et al., 2021). Furthermore, circRNA MAP2K4 (circMAP2K4) was validated to promote HCC cell proliferation by binding with miRNA 139-5p (miR-139-5p) to promote YTHDF1 expression, providing clues for HCC therapeutic targets (Chi et al., 2021). Moreover, circRNA RHBDD1 (circRHBDD1) uses YTHDF1 to accelerate the translation of phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1) in an m6A-dependent manner and plays an important role in the metabolism of HCC. Inhibition of circRHBDD1 enhances the treatment of HCC with anti-programmed cell death protein 1 (PD-1) (Cai et al., 2022). Thus, YTHDF1 is a potential biomarker of prognosis and a target for treatment (Li et al., 2021c).
YTHDF2 is negatively correlated with survival in HCC patients and YTHDF2 upregulates octamer-binding transcription factor 4 (OCT4) expression in an m6A-dependent manner, promoting HCC progression (Zhang et al., 2020a). miRNA 145 (miR-145) regulates m6A expression levels by regulating the 3′-UTR of YTHDF2 mRNA in HCC cells to regulate m6A expression levels (Yang et al., 2017c). It was shown that hypoxia-specific downregulates YTHDF2 expression in HCC cells and YTHDF2 binding to the m6A site modified by EGFR 3′-UTR promotes EGFR mRNA degradation in HCC cells and inhibits proliferation of HCC cells (Zhong et al., 2019). YTHDF2 can regulate serpin family E member 2 (SERPINE2), interleukin 11 (IL11)decay, and vascular abnormalities in HCC, and inhibits the malignant progression of HCC (Hou et al., 2019). It was shown that YTHDF2 inhibited the expression of lncRNA AY (lncAY) and suppressed HCC progression through the BMI1 proto-oncogene polysaccharide ring finger (BMI1)/WNT/β-catenin axis form (Chen et al., 2021b). It was also shown that lysine demethylase 5B (KDM5B) regulates the axis formed by YTHDF3 and integrin subunit alpha 6 (ITGA6) axis by suppressing miRNA 448 (miR-448) expression and promoting HCC progression (Guo et al., 2021).
RBM15 is highly expressed in HCC, and RBM15 promotes HCC progression through the IGF2BP1/YES1/MAPK axis (Cai et al., 2021). Microwave ablation is a therapeutic approach for HCC, and studies have shown that sublethal heat treatment promotes HCC migration and EMT transformation through induction of METTL3 and CD47, that METTL3 mediates HCC migration through CD47 in an m 6 A-dependent manner, and that the METTL3/IGF2BP1/CD47 axis is a target for potential treatment by microwave ablation, providing clues to the link between RNA methylation and incomplete thermal ablation (Fan et al., 2021a). It was shown that IGF2BP1 can enhance c-Myc and Ki-67 protein translation and promote HCC progression by binding and stabilizing proliferation markers C-Myc and Ki-67 (MKI67) (Gutschner et al., 2014). LincRNA 1,093 (LINC01093) inhibits HCC progression by promoting attenuation of GLI family zinc finger 1 (GLI1) mRNA through interaction with IGF2BP1 (He et al., 2019). The oncogene IGF2BP2 is highly expressed in HCC and can be used for prognosis prediction (Pu et al., 2020). IGF2BP1 promotes the translation of circRNA MAP3K4 (circMAP3K4) into circMAP3K4-455aa, inhibiting allograft inflammatory factor 1 (AIF1) cleavage and nuclear distribution, preventing cisplatin-induced apoptosis, and promoting HCC progression (Duan et al., 2022). It has also been shown that pre-genomic RNA (pgRNA) promotes cancer progression in HCC and upregulates IGF2BP3 expression at the post-transcriptional level (Ding et al., 2021). The above studies suggest that m6A-modification writers, erasers, and readers are involved in HCC progression and that regulator-mediated m6A modifications can influence HCC progression.
m7G modifications are commonly found in eubacteria, eukaryotes, and some archaea (Zhang et al., 2019a; Wiener and Schwartz, 2021) and are one of the few methylation modifications that introduce positively charged or amphiphilic ions into nucleobases (Figure 1B). m7G has a broad impact on mRNA, tRNA, and rRNA, playing a crucial function in various biological processes including transcriptional elongation, pre-mRNA splicing, and mRNA translation, as well as serving as an important indicator for disease diagnosis (Figure 2) (Alexandrov et al., 2002; Leulliot et al., 2008; Haag et al., 2015a; Lin et al., 2018; Liu et al., 2020c; Song et al., 2020; Blersch et al., 2021; Zhao et al., 2021).
Precise identification of RNA modification sites is essential to investigate their functional effects in the control of gene expression and to clarify their relevance in different physiological processes (Cao et al., 2021). In response to the easy conversion of m7G to abasic sites, single-base sequencing methods have been developed to detect m7G in RNA (Boulias and Greer, 2019). Mainly includes tRNA reduction and cleavage sequencing (TRAC-seq)、AlkAline-seq、m7G-MAP-seq、m7G-seq and m7G individual-nucleotide-resolution cross-linking and immunoprecipitation with sequencing (m7G-miCLIP-seq) (Lin et al., 2018; Zhang et al., 2019a; Lin et al., 2019; Malbec et al., 2019; Marchand et al., 2021). These sequencing methods provide a scientific platform for detecting the expression levels of m7G modifications.
The main enzymes that perform m7G modification on RNA are methyltransferase-like 1 (METTL1), WD repeats structural domain 4 (WDR4), and Williams-Beuren syndrome chromosome region 22 protein (WBSCR22, also known as BUD23) (Deng et al., 2020). m7G modification of mRNA, tRNA, and miRNA is primarily mediated by METTL1 and WDR4, while WBSCR22 mediates m7G modification of rRNA (Létoquart et al., 2014). METTL1 adds m7G modifications in GA-rich sequences in mammalian internal mRNAs (Chen et al., 2021e; Zhao et al., 2021). In tRNAs, m7G is added by METTL1 and WDR4 in the AG (m7G)H motif (Malbec et al., 2019). There is growing evidence that impaired tRNA m7G modification is associated with various diseases (Braun et al., 2018). m7G tRNA modification mediated by METTL1 and WDR4 has critical functions in determining cell fate and growth (Lin et al., 2018). (Michaud et al., 2000; Shaheen et al., 2015; Lin et al., 2018; Ma et al., 2021; Orellana et al., 2021). A recent study showed that dysregulation of METTL1 is associated with viability and sensitivity to chemotherapy in colon and cervical cancer cells, suggesting a potential link between m7G tRNA modifications and cancer development (Cartlidge et al., 2005; Okamoto et al., 2014; Liu et al., 2019). METTL1 regulates cell proliferation, with potentially oncogenic effects on tRNA but tumor-suppressive activity in miRNA maturation, and is amplified explicitly in HCC and is correlated with poor prognosis (Pandolfini et al., 2019).
Studies of m7G have shown that m7G modifications and its regulators METTL1, WDR4, and WBSCR22 are significantly upregulated in HCC and are associated with poor prognosis and chemotherapy resistance (Figure 3). Studies have shown that WBSCR22 is highly expressed in HCC and promotes the proliferation of HCC cells, making it a possible therapeutic target for HCC (Stefanska et al., 2014). Another study showed that METTL1 is shown to be upregulated in HCC and promotes HCC migration and proliferation through the phosphatase and tensin homolog (PTEN) as well as the AKT signaling pathway (Tian et al., 2019). METTL1 exerts oncogenic activity by inhibiting PTEN signaling, and the METTL1/PTEN axis has potential for the treatment of HCC. METTL1-mediated tRNA m7G modification has also been proved to accelerate the translation of target mRNAs with higher m7G-associated codon frequencies, accelerating HCC progression and tumorigenesis (Chen et al., 2021e). Another study showed that upregulated WDR4 expression significantly increased m7G methylation levels in HCC and conferred a poor prognosis. WDR4 can enhance HCC progression by promoting cyclin B1 (CCNB1) mRNA stability and translation and is a candidate HCC therapeutic target (Xia et al., 2021). These findings suggest that RNA m7G methylation modifications mediated by METTL1, WDR4, and WBSCR22 have important roles in HCC.
The RNA 5-methylcytosine (m5C) is mainly distributed in archaea, prokaryotes, and eukaryotes, but is only conserved in Saccharomyces cerevisiae (Figure 1C) (Brzezicha et al., 2006; Edelheit et al., 2013; Chen et al., 2021c). It occurs in mRNA, tRNA, rRNA, ncRNA, and enhancer RNA (eRNA), regulating RNA in metabolic procedures, in terms of RNA translation, structure, stability, and mRNA export (Figure 2) (Haag et al., 2015b; Bourgeois et al., 2015; Tuorto et al., 2015; Haag et al., 2016; Amort et al., 2017; Yang et al., 2017a; Natchiar et al., 2017; García-Vílchez et al., 2019; Van Haute et al., 2019).
In recent years, advances in m5C detection methods have led to more extensive analysis of m5C. m5C-RIP-seq is an RNA immunoprecipitation method based on m5C antibodies (Edelheit et al., 2013). miCLIP-seq is a single nucleotide resolution protein-based method of immunoprecipitation (Hussain et al., 2013). AZA-immunoprecipitation-based sequencing (AZA-IP-seq) is a single-nucleotide method based on RCMT antibodies and 5-azacytidine (5-azaC) (Khoddami and Cairns, 2013, 2014). TET-assisted peroxotungstate oxidation sequencing (TAWO-seq) is a method that uses 5-azaC or peroxy chemically-dependent sequencing (Yuan et al., 2019). Nanopore sequencing is also capable of providing high-throughput sequencing of m5C (Garalde et al., 2018; Rang et al., 2018). Nanopore sequencing is also capable of providing high-throughput sequencing of m5C.
m5C modifications are distributed in mRNA, enriched near the 5′UTR and 3′UTR, and also present in tRNA and rRNA (Trixl and Lusser, 2019). Several enzymes are capable of dynamically regulating m5C levels, including members of the NOP2/Sun RNA methyltransferase family (NSUNs), DNA methyltransferase family (DNMTs), Aly/REF export factor (ALYREF), tRNA aspartic acid methyltransferase family (TRDMTs), and AlkB homolog one histone H2A dioxygenase (ALKBH1) and Y-box binding protein 1 (YBX1), and TET methylcytosine dioxygenase family (TETs) (Reid et al., 1999; Goll et al., 2006; Yang et al., 2017a; Chen et al., 2019a; Bohnsack et al., 2019; Chellamuthu and Gray, 2020). NSUN1、NNSUN4, and NSUN5 methylated rRNAs.NSUN2, DNMT2, NSUN3, and NSUN6 methylated cytoplasmic tRNA, and NSUN2 also modifies ncRNA and mRNA. ALYREF, YBX1, and YTHDF2 can read m5C (Yang et al., 2017a). ALYREF is the most critical RNA m5C-binding protein known to participate in the mRNA exocytosis process in concert with NSUN2 by binding to m5C sites on mRNA. In eukaryotes, the most active m5C methyltransferases are DNMT2 and NSUN2 (Okamoto et al., 2012; Squires et al., 2012). NSUN2 m5C methylation of tRNA is associated with the survival, differentiation, and development of eukaryotes (Khan et al., 2012; Martinez et al., 2012; Blanco et al., 2014; Blanco et al., 2016). DNMT2 regulates tRNA differentiation, survival, and translation accuracy (Schaefer et al., 2010; Tuorto et al., 2012; Durdevic et al., 2013).
m5C modifications have important implications for tumor progression and can act as an effective target for cancer treatment (He et al., 2020b; Nombela et al., 2021). The expression of m5C in HCC tissues was significantly higher than that in adjacent tissues (He et al., 2020a). m5C modifications and their regulators can regulate HCC cell proliferation, migration, and invasion, regulating HCC development and progression (Figure 3). It has been shown that m5C expression levels and regulators NSUN4, NSUN5, DNMT1, TET2, and ALYREF are significantly upregulated in HCC and are related to poor prognosis in HCC patients (Blanco et al., 2016; He et al., 2020a; He et al., 2020b; He et al., 2020c; Du et al., 2020;Zhang et al., 2020e; Cui et al., 2021b; He et al., 2021; Liu et al., 2022). DNMT1 is an independent risk factor, highly expressed in HCC tissues and positively associated with many tumor microenvironments (TME) infiltrating immune cells, promoting the malignant progression of HCC and strongly associated with poor prognosis in HCC patients (Gu et al., 2021). Another study found that overexpression of ALYREF and upregulation of eukaryotic translation initiation factor 4A3 (eIF4A3) were related to poor prognosis in HCC patients (Xue et al., 2021). The expression of NSUN2 is upregulated in HCC, which promotes tumor progression by increasing the expression level of H19 imprinted maternally expressed transcript (H19) and promoting the binding of H19 to G3BP stress granule assembly factor 1 (G3BP1) (Sun et al., 2020). Another study found that NSUN2 overexpression increased the stability of Fizzy-related-1 (FZR1) mRNA and promoted the proliferation of HCC cells (Zhai et al., 2021). These findings indicate that m5C-related regulatory genes, such as NSUN2, NSUN4, and ALYREF, can be used as potential biomarkers for HCC diagnosis.
First discovered in 1961, m1A is widely distributed in bacteria, archaea, and eukaryotes and is evolutionarily conserved (Figure 1D) (Dunn, 1961; Dominissini et al., 2016; Shi et al., 2020a). m1A is a methylation modification present in tRNA, mRNA, rRNA, and lncRNA, and mitochondrial RNA that is positively charged under physiological conditions. It can alter RNA structure, disrupt base-pairing specificity, affect the tertiary structure of ribosomes, and also regulates gene expression, controls cell fate, and influences disease onset and progression (Figure 2) (Agris, 1996; Delatte et al., 2016; Liu et al., 2016; Li et al., 2017a; Bhan et al., 2017; Oerum et al., 2017; Zhao et al., 2019b).
m1A sequencing has enabled a comprehensive exploration of m1A methylation modifications in the transcriptome, providing an important tool for exploring m1A methylation function. m1A blocks Watson-Crick pairing, effectively blocking RT and inducing truncation or mutation of RT products (Yang et al., 2020). Based on the properties of m1A, the commonly used m1A high-throughput sequencing methods include m1A-seq-TGIRT、m1A-MAP、m1A-IP-seq, and m1A-quant-seq (Safra et al., 2017b; Dai et al., 2018; Zhou et al., 2019a). These methods provide the basis for in-depth studies of m1A and can contribute to a detailed exploration of the biological role of m1A.
m1A occurs in mRNA, tRNA, rRNA, and mitochondrial RNA transcripts. m1A modification has been extensively studied and is regulated by enzymes, including tRNA methyltransferases (TRMT6, TRMT10C, TRMT61A, and TRMT61B), readers (YTHDF1, YTHDF2, YTHDF3, and YTHDC1) and demethylases (ALKBH1, ALKBH3) (Dominissini et al., 2016; Dai et al., 2018; Scheitl et al., 2020; Esteve-Puig et al., 2021; Shafik et al., 2021). TRMT6 and TRMT61A are responsible for cytoplasmic tRNA and some nuclear mRNA m1A methylation with GUUCRA tRNA-like motifs. Underexpression of TRMT6 and TRMT61 reduces proliferation and death of C6 glioma cells, while their overexpression biological processes encode mRNAs involved in tumorigenic processes (Macari et al., 2016). The m1A modification at tRNA position 58 (m1A58) is conserved in many organisms (Hamdane et al., 2014). m1A58 modification mediated by TRMT6/61A has important roles in the replication of retroviral HIV-1 in vivo and tRNA modification (Fukuda et al., 2021). ALKB protein family members ALKBH1 and ALKBH3 are demethylases of m1A (Chujo and Suzuki, 2012). The YTH structural domain-containing proteins YTHDF1, YTHDF2, YTHDF3, and YTHDC1 are readers of m1A in RNA and can interact directly with m1A-containing RNA (Seo and Kleiner, 2020). YTDHF proteins recognize and bind multiple m1A-modified sequence motifs, demonstrating m1A destabilization of RNA transcripts and demonstrating its function in post-transcriptional gene regulation.
The contribution of m1A modifications to the epigenetic regulation of gene expression during HCC development and progression has been progressively demonstrated. m1A methylation modifications and regulators can effectively regulate HCC progression (Figure 3). Using bioinformatics analysis found that m1A-related regulatory genes are altered in HCC and affect clinicopathological features and prognosis (Wang et al., 2019; Shi et al., 2020b). TRMT6, TRMT10C, and YTHDF1 are expressed at higher levels in HCC than in normal tissues and are associated with poor prognosis in HCC and could be used as prognostic biomarkers in HCC (Wang et al., 2019; Shi et al., 2020b). The MYC pathway and PI3K/AKT signaling pathway could be engaged in controlling m1A in HCC cells (Shi et al., 2020b). TRMT6/TRMT61A increases m 1 A methylation in tRNA subsets to increase peroxisome proliferator-activated receptor-delta (PPARδ) translation, which in turn triggers cholesterol synthesis to activate Hedgehog signaling. Therefore, the TRMT6/TRMT61A complex can be a potential target for HCC to provide a more effective treatment strategy for HCC patients (Wang et al., 2021b). Another study showed that YTHDF1, YTHDF2, YTHDF3, and ALKBH3 expression levels were upregulated in HCC using a raw letter analysis and were closely associated with HCC metabolism, providing new insights into m1A modification and metabolic heterogeneity in HCC (Tong et al., 2022). These studies demonstrate that m1A RNA epigenetic modifications take a crucial part in regulating HCC development.
m3C is less abundant and is primarily found in eukaryotic tRNA and mRNA, and some rRNA (Figures 1E, 2) (Hall, 1963; Clark et al., 2016). m3C modifications are highly conserved and specific modifications that affect tRNA structure, ribosome binding affinity, decoding activity, and maintain anticodon folding and pairing functions, playing a key role in many biological processes (Auffinger and Westhof, 2001; Olejniczak and Uhlenbeck, 2006; D'Silva et al., 2011; Han et al., 2017; Li et al., 2022).
The m3C modification significantly destabilizes Watson-Crick C: G pairing. High-throughput detection methods for m3C based on this feature include ALKB-facilitated RNA methylation sequencing (ARM-seq)、AlkAniline-seq and Hydrazine-aniline cleavage sequencing (HAC-seq) (Cozen et al., 2015; Marchand et al., 2018; Cui et al., 2021a; Marchand et al., 2021). These methods provide a research basis for exploring the biochemical and biomedical potential of m3C.
The four known m3C modifying enzymes, methyltransferase methylcytidine 2A (METTL2A), 2B (METTL2B), 6 (METTL6), and 8 (METTL8), all contain conserved S-adenosylmethionine (SAM) binding domains. ALKBH1 and ALKBH3 demethylate m3C in tRNAs, affecting RNA stability and preventing degradation (Ougland et al., 2004; Ueda et al., 2017). METTL2 and METTL6 regulate m3C in specific tRNAs to regulate m3C. METTL8 regulates m3C in mRNAs (Xu et al., 2017) and has recently been shown to be a mitochondrial tRNA m3C32 methyltransferase (Arimbasseri et al., 2016; Schöller et al., 2021). The formation of m3C in human mt-tRNAs depends on METTL8 (Towns and Begley, 2012; Suzuki et al., 2020; Schöller et al., 2021; Lentini et al., 2022), which can promote tumorigenesis by affecting genetic organization within or close to the nucleolus and regulatory R-loop formation through its m3C methyltransferase activity (Zhang et al., 2020d). ALKBH1 can demethylate m3C in mammalian cell mRNA and tRNA, providing m3C a single-stranded substrate preference in RNA and DNA and promoting proliferation, migration, and invasion of cancer cells (Aas et al., 2003; Sundheim et al., 2006; Chen et al., 2019d). Expression of ALKBH3 is associated with tumor development and modification of protein synthesis, indicating that m3C may play a prominent part in cancer biology (Chen et al., 2019d).
Studies have shown that m3C and its regulators play an important role in HCC (Figure 3). Bioinformatic analysis of METTL6 in HCC revealed that METTL6 mRNA expression levels were significantly upregulated in HCC and were closely associated with poorer patient survival outcomes. In contrast, METTL6 downregulation inhibited HCC progression by suppressing cell adhesion molecules and could be a potential therapeutic target for HCC (Bolatkan et al., 2022). METTL6-mediated m3C tRNA methylation regulates gene expression, cellular homeostasis, translation, and tumor cell growth (Ignatova et al., 2020; Bolatkan et al., 2022). Downregulation of METTL6 attenuates cell proliferation, invasion, and adhesion in HCC by inhibiting cell adhesion molecules and suppresses the malignant phenotype of HCC (Bolatkan et al., 2022). METTL6 deficiency decreases the metabolic level of hepatic cells, inhibits HCC proliferation, and is significantly associated with a low survival rate in patients with HCC (Ignatova et al., 2020). Another research indicated that m3C levels in mRNA in HCC tissues were remarkably decreased compared to normal tumor-adjacent tissues, potentially due to increased ALKBH1 expression and decreased METTL8 expression in HCC tissues (Ma et al., 2019).
Ψ is a five-carbon isomer of uridine, that is the first post-transcriptional modification to be identified, known as the “fifth nucleoside” of RNA, and found in tRNA, rRNA, snRNA, small nucleolar RNA (snoRNA), telomerase RNA, lncRNA, and polyadenylated mRNA (Figures 1F, 2) (Cohn, 1951, 1960; Fedorov and Bogomazov, 1969; Becker et al., 1998; Kim et al., 2010; Machnicka et al., 2013; Schwartz et al., 2014a; Carlile et al., 2014; Li et al., 2015). When bound to RNA, Ψ could change the structure of RNA, and improving base pairing, act in RNA folding, secondary structure, stability, and translation, and is regulated dynamically by the environment, which adds a potential mechanism for regulating RNA fate (Arnez and Steitz, 1994; Davis, 1995; Yu et al., 1998; Yarian et al., 1999; Zebarjadian et al., 1999; Newby and Greenbaum, 2001; Lecointe et al., 2002; Newby and Greenbaum, 2002; Nobles et al., 2002; King et al., 2003; Cabello-Villegas and Nikonowicz, 2005; Karikó et al., 2008; Bilbille et al., 2009; Karijolich and Yu, 2011; Fernández et al., 2013; Gu et al., 2013; Zhao and He, 2015; Hoernes et al., 2016; Safra et al., 2017a; Sloan et al., 2017; Bohnsack and Sloan, 2018; Zhao et al., 2018b; Guzzi et al., 2018; Penzo and Montanaro, 2018; Eyler et al., 2019).
As research has progressed, several methods for Ψ have been developed. Mainly include Ψ-seq、PSI-seq、Pseudo-seq、CeU-Seq、HydraPsi-Seq and Nanopore RNA sequencing (Schwartz et al., 2014a; Carlile et al., 2014; Lovejoy et al., 2014; Li et al., 2015; Marchand et al., 2020; Begik et al., 2021; Thomas et al., 2021). These methods provide a research basis for exploring the biological role of Ψ.
Ψ is the most abundant type of post-transcriptional modification in ncRNA. An essential difference between pseudouridylation and methylation is that pseudouridylation occurs as an irreversible modification in mammals (Rasmuson and Björk, 1995; Sibert and Patton, 2012). The formation of pseudouridine is promoted by Ψ synthase (PUS or Ψ synthase), which is responsible for catalyzing Ψ in various RNA substrates, including mRNA, tRNA, rRNA, 5S RNA, snRNA, and snoRNA. Pseudouridylation can be achieved by two different and independent mechanisms, RNA-independent and RNA-dependent pseudouridylation. Pseudouridine obeys an RNA-independent mechanism in bacteria and humans, and an RNA-dependent mechanism in eukaryotes and archaea (Rintala-Dempsey and Kothe, 2017). RNA-independent pseudouridylation is catalyzed by PUS, which simultaneously performs substrate recognition and catalyzes uridine isomerization to Ψ in the absence of the RNA template strand, primarily modifying tRNA, rRNA, and snRNA (Becker et al., 1997a; Becker et al., 1997b; Hamma and Ferré-D'Amaré, 2006; Hur et al., 2006; Roovers et al., 2006; Rintala-Dempsey and Kothe, 2017). The RNA-dependent mechanism is mediated by the RNA-protein complex family of cassette H/ACA small ribonucleoproteins (snoRNPs), which consists of dyskerin Ψ synthase 1 (DKC1) and ribonucleoproteins NHP2, NOP10, and GAR1.
Ψ was the first RNA modification shown to be associated with cancer, and Ψ modifications and regulators have been proved to take an important part in HCC (Figure 3) (Zhao et al., 2004; Jana et al., 2017; Sperling et al., 2017; Ji et al., 2020). The study demonstrates serum Ψ concentration was increased in HCC patients (Amuro et al., 1988; Tamura et al., 1988). Studies have shown that HCC cells lacking snoRNA H/ACA box 24 (SNORA24)-guided Ψ modification has increased translation error coding and stop codon read-through frequencies and are associated with poor patient survival (McMahon et al., 2019). Another study used bioinformatics to analyze the expression and role of the H/ACA snoRNP gene family in HCC. The expression of the H/ACA snoRNP gene family was higher in HCC tissues than in normal or adjacent tissues, and their differential expression was strongly associated with poor prognosis and multiple immune cell infiltration in HCC patients and could be considered as biomarkers for the treatment of HCC(Zhang et al., 2021b). It has also been shown that overexpression of DKC1 can serve as a marker of the proliferative potential of HCC cells and is an unfavorable prognostic factor for HCC (Liu et al., 2012). One study found that oxidatively modified cytoplasmic protein disulfide bond isomerase 3 (PDIA3) in HCC promoted DKC1-mediated survival of HCC and that DKC1 is a worthwhile target for HCC prediction and treatment (Ko et al., 2018). Except for DKC1, Ψ 5′-phosphatase (PUDP) has been identified as a potential oncogene for HCC (Yu et al., 2022). PUDP positively correlates with tumor immune cell infiltration, and immune checkpoint expression and causes poor prognosis and a poor response to immunotherapy in HCC patients (Yu et al., 2022). These studies suggest that Ψ and its modulators can be a potential biomarker and treatment target in HCC treatment.
Currently, aberrant post-transcriptional modifications in cancer cells are gradually being studied, and there are many assays to detect aberrant RNA epigenetic modifications in tumors (Table 1). Studies on RNA modifications in HCC have mainly used Quantitative Real-time PCR (qPCR) and western blot to detect the expression levels of major regulators of RNA modifications (Chen et al., 2018; Qiao et al., 2021). Northern blotting (Northern blot) was used to verify the trend of RNA expression levels (Chen et al., 2021e). High-throughput sequencing techniques such as RNA-seq or methylated RNA Immunoprecipitation (meRIP) were used to detect the expression levels of RNA modifications (Chen et al., 2018; Chen et al., 2019c; Lan et al., 2019; Lin et al., 2020b; Liu et al., 2020b; Chen et al., 2020; Wu et al., 2021a; Xia et al., 2021). Liquid chromatography-tandem mass spectrometry (LC-MS) was used to detect the expression levels of RNA modifications (Chen et al., 2021e; Qiao et al., 2021; Xia et al., 2021). For m6A modifications, some studies have used kits such as EpiQuik m6A RNA Methylation Quantification Kit to quantify the expression of m6A modifications based on absorbance or fluorescence intensity, and m6A-IP-qPCR to quantify the enriched RNA (Chen et al., 2019c; Liu et al., 2020b; Chen et al., 2020; Li et al., 2021d; Yang et al., 2021). For m7G modification and m5C modification, the study examined the expression levels of both RNA modifications using tRNA m7G reduction and cleavage sequencing (TRAC-seq) and bisulfite pyrophosphate sequencing, respectively (Sun et al., 2020). After determining the differential expression of RNA modifications in tumors, most of them were detected using RNA Binding Protein Immunoprecipitation Assay (RIP) technique, Co-Immunoprecipitation (Co-IP), and Chromatin Isolation by RNA Purification (CHIRP-Seq) and other methods to study the regulatory network of post-transcriptional modifications in tumors (Chen et al., 2019c; Lan et al., 2019; Wang et al., 2020a; Lin et al., 2020b; Chen et al., 2020; Sun et al., 2020; Wu et al., 2021a; Bo et al., 2021; Xia et al., 2021). These technical tools can confirm the interactions with RNA modifications and related proteins, and search for upstream or downstream genes that regulate tumor progression (Chen et al., 2018). The use of these assays can provide an in-depth study of the relationship between the abnormal expression of tumor-associated RNA modifications and tumor progression, investigate the mechanism of tumorigenesis, and lay the scientific foundation for tumor treatment.
With the advancement of gene editing technology, gene knockdown of tumor-related genes using gene editing systems such as CRISPR/Cas9 has been widely studied (Doyle et al., 2012; Wu et al., 2019a; Brommage et al., 2019). Currently, studies have been conducted to target RNA-modified enzymes for gene knockout and prepare knockout mouse models for studying the treatment of diseases such as tumors. One study used a lentivirus-based clustered regularly interspaced short palindromic repeat (CRISPR) gene editing system (lentiCRISPR v2) to knock out METTL3 in the HCC cell line Huh-7, and subsequently injected the knocked-out METTL3 cell line in situ into the left liver lobe of nude mice using in situ transplantation experiments, and found that knocking out METTL3 significantly inhibited the growth of HCC tumors in situ in the liver of nude mice (Chen et al., 2018). Another study using CRISPR/Cas9 technology to construct liver-specific METTL1 knockout mice showed a significant reduction in HCC tumor lesions and a significant decrease in tumor load (Chen et al., 2021e). In the study of HCC, some studies have used gene editing technology to construct knockout mouse models to knock out RNA-modified regulators, which can more scientifically validate the role of regulators in HCC (Chen et al., 2018; Li et al., 2021c; Chen et al., 2021e). These animal models have played an important role in studying the mechanism of action of m6A in HCC.
RNA modifications play a key role in regulating cell fate, an important regulator of HCC, in tumorigenesis and disease progression (Han et al., 2018; Pea et al., 2021; Braghini et al., 2022). The m6A, m7G, m5C, m1A, m3C, and ψ RNA modifications dynamically regulate the development of HCC, providing new strategies and possibilities for selecting possible therapeutic targets and investigating precisely directed intervention strategies for future HCC therapy (Tian et al., 2019; Bolatkan et al., 2022; Liu et al., 2022; Ren et al., 2022; Tong et al., 2022; Yu et al., 2022). As the most intensively studied RNA modification, m6A, and its regulators regulate the proliferation, migration, invasion, and EMT processes of HCC cells and are promising targets for HCC therapy (Zhou et al., 2019b; Liu et al., 2020a; Wang et al., 2020b; Zhang et al., 2020c; Qu et al., 2020; Wu et al., 2020; Zhu et al., 2020; Li et al., 2021e; Wei, 2021; Gu et al., 2022; Ren et al., 2022). Although m6A in mammalian RNA has been extensively studied, there is currently no evidence for DNA N6 -methyladenine in mammals (Douvlataniotis et al., 2020). It suggests that m6A may regulate mammalian life activities only through modification of RNA, and that regulation of m6A modification in RNA can effectively mitigate the progression of diseases such as HCC. m7G modifications and their regulators play an integral part in the growth, and invasion of HCC cells and are associated with poor patient prognosis and chemotherapy resistance, representing valuable markers for clinical diagnosis and poor prognosis (Dai et al., 2021). m5C modifications and their regulators can regulate HCC cells (Xue et al., 2021). m1A, m3C, and Ψ methylation modifications and their regulators can also effectively regulate HCC progression (McMahon et al., 2019; Shi et al., 2020b; Bolatkan et al., 2022). The differential expression of RNA modifications in HCC can be quantified by RNA technologies to investigate their precise biological role in HCC occurrence and progression (Zheng et al., 2020; Cao et al., 2021). With advances in sequencing technology, increasing numbers of RNA modifications can be localized and quantified at a single nucleotide resolution (Edelheit et al., 2013; Schwartz et al., 2014a; Cozen et al., 2015; Linder et al., 2015; Li et al., 2017a; Zheng et al., 2020; Cao et al., 2021). Existing sequencing technologies cannot achieve 100% accuracy and sensitivity, and there is no suitable sequencing technology to detect several RNA modifications in the same transcript simultaneously (Wang et al., 2021c). The most advanced sequencing technology available is nanopore sequencing, extending the scope of RNA modification studies, but it still has limitations (Workman et al., 2019; Pratanwanich et al., 2021; Wang et al., 2021e). Therefore, the development of technologies for identifying and quantifying RNA modifications is critical for advancing understanding of their role in cancer development processes in HCC. Although unusual expression of RNA modifying enzymes has now been described in the development of HCC, further investigation is required for the exact contribution of these enzymes and the corresponding modifications to HCC metastasis, and resistance requires further investigation.
In conclusion, although RNA modifications and differential expression of associated regulatory genes have been explored in most physiological processes of HCC and development, their precise role and effects on tumorigenesis, proliferation, metastasis, and resistance still require further investigation. In this review, we have summarized existing knowledge on the important roles and regulatory mechanisms of m6A, m7G, m5C, m1A, m3C, and ψ in HCC, suggesting that targeting aberrant post-transcriptional modifications in cancer cells has the potential to be an effective tool for HCC treatment.
QF, DW, and DL wrote the manuscript. QF, DW, TX, CL, YG, LS, YJ, and DL collected the references and prepared figures. All authors reviewed the manuscript.
This work was supported by the Natural Science Foundation of Jilin Province (20210101310JC), Central University Basic Scientific Research Fund (2019JCKT70), Scientific Research Project of Jilin University Key Laboratory ([2019]004), Jilin Science and Technology Department Program 20220505033ZP, 202002006JC and 202101010JC.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.984453/full#supplementary-material
Aas, P. A., Otterlei, M., Falnes, P. O., Vågbø, C. B., Skorpen, F., Akbari, M., et al. (2003). Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421 (6925), 859–863. doi:10.1038/nature01363
Agris, P. F. (1996). The importance of being modified: Roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid. Res. Mol. Biol. 53, 79–129. doi:10.1016/s0079-6603(08)60143-9
Akula, D., O'Connor, T. R., and Anindya, R. (2021). Oxidative demethylase ALKBH5 repairs DNA alkylation damage and protects against alkylation-induced toxicity. Biochem. Biophys. Res. Commun. 534, 114–120. doi:10.1016/j.bbrc.2020.12.017
Alarcón, C. R., Lee, H., Goodarzi, H., Halberg, N., and Tavazoie, S. F. (2015). N6-methyladenosine marks primary microRNAs for processing. Nature 519 (7544), 482–485. doi:10.1038/nature14281
Alexandrov, A., Martzen, M. R., and Phizicky, E. M. (2002). Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8 (10), 1253–1266. doi:10.1017/S1355838202024019
Allis, C. D., and Jenuwein, T. (2016). The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17 (8), 487–500. doi:10.1038/nrg.2016.59
Amort, T., Rieder, D., Wille, A., Khokhlova-Cubberley, D., Riml, C., Trixl, L., et al. (2017). Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 18 (1), 1. doi:10.1186/s13059-016-1139-1
Amuro, Y., Nakaoka, H., Shimomura, S., Fujikura, M., Yamamoto, T., Tamura, S., et al. (1988). Serum pseudouridine as a biochemical marker in patients with hepatocellular carcinoma. Clin. Chim. Acta. 178 (2), 151–158. doi:10.1016/0009-8981(88)90221-5
Arguello, A. E., DeLiberto, A. N., and Kleiner, R. E. (2017). RNA chemical proteomics reveals the N(6)-methyladenosine (m(6)A)-Regulated protein-RNA interactome. J. Am. Chem. Soc. 139 (48), 17249–17252. doi:10.1021/jacs.7b09213
Arimbasseri, A. G., Iben, J., Wei, F. Y., Rijal, K., Tomizawa, K., Hafner, M., et al. (2016). Evolving specificity of tRNA 3-methyl-cytidine-32 (m3C32) modification: A subset of tRNAsSer requires N6-isopentenylation of A37. Rna 22 (9), 1400–1410. doi:10.1261/rna.056259.116
Arnez, J. G., and Steitz, T. A. (1994). Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 33 (24), 7560–7567. doi:10.1021/bi00190a008
Auffinger, P., and Westhof, E. (2001). An extended structural signature for the tRNA anticodon loop. Rna 7 (3), 334–341. doi:10.1017/s1355838201002382
Barbieri, I., and Kouzarides, T. (2020). Role of RNA modifications in cancer. Nat. Rev. Cancer 20 (6), 303–322. doi:10.1038/s41568-020-0253-2
Bartosovic, M., Molares, H. C., Gregorova, P., Hrossova, D., Kudla, G., and Vanacova, S. (2017). N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic Acids Res. 45 (19), 11356–11370. doi:10.1093/nar/gkx778
Becker, H. F., Motorin, Y., Florentz, C., Giegé, R., and Grosjean, H. (1998). Pseudouridine and ribothymidine formation in the tRNA-like domain of turnip yellow mosaic virus RNA. Nucleic Acids Res. 26 (17), 3991–3997. doi:10.1093/nar/26.17.3991
Becker, H. F., Motorin, Y., Planta, R. J., and Grosjean, H. (1997a). The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of psi55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res. 25 (22), 4493–4499. doi:10.1093/nar/25.22.4493
Becker, H. F., Motorin, Y., Sissler, M., Florentz, C., and Grosjean, H. (1997b). Major identity determinants for enzymatic formation of ribothymidine and pseudouridine in the T psi-loop of yeast tRNAs. J. Mol. Biol. 274 (4), 505–518. doi:10.1006/jmbi.1997.1417
Begik, O., Lucas, M. C., Pryszcz, L. P., Ramirez, J. M., Medina, R., Milenkovic, I., et al. (2021). Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat. Biotechnol. 39 (10), 1278–1291. doi:10.1038/s41587-021-00915-6
Ben-Haim, M. S., Moshitch-Moshkovitz, S., and Rechavi, G. (2015). Fto: Linking m6A demethylation to adipogenesis. Cell Res. 25 (1), 3–4. doi:10.1038/cr.2014.162
Bhan, A., Soleimani, M., and Mandal, S. S. (2017). Long noncoding RNA and cancer: A new paradigm. Cancer Res. 77 (15), 3965–3981. doi:10.1158/0008-5472.Can-16-2634
Bian, S., Ni, W., Zhu, M., Song, Q., Zhang, J., Ni, R., et al. (2020). Identification and validation of the N6-methyladenosine RNA methylation regulator YTHDF1 as a novel prognostic marker and potential target for hepatocellular carcinoma. Front. Mol. Biosci. 7, 604766. doi:10.3389/fmolb.2020.604766
Bilbille, Y., Vendeix, F. A., Guenther, R., Malkiewicz, A., Ariza, X., Vilarrasa, J., et al. (2009). The structure of the human tRNALys3 anticodon bound to the HIV genome is stabilized by modified nucleosides and adjacent mismatch base pairs. Nucleic Acids Res. 37 (10), 3342–3353. doi:10.1093/nar/gkp187
Björk, G. R., Ericson, J. U., Gustafsson, C. E., Hagervall, T. G., Jönsson, Y. H., and Wikström, P. M. (1987). Transfer RNA modification. Annu. Rev. Biochem. 56, 263–287. doi:10.1146/annurev.bi.56.070187.001403
Blanco, S., Bandiera, R., Popis, M., Hussain, S., Lombard, P., Aleksic, J., et al. (2016). Stem cell function and stress response are controlled by protein synthesis. Nature 534 (7607), 335–340. doi:10.1038/nature18282
Blanco, S., Dietmann, S., Flores, J. V., Hussain, S., Kutter, C., Humphreys, P., et al. (2014). Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. Embo J. 33 (18), 2020–2039. doi:10.15252/embj.201489282
Blersch, K. F., Burchert, J. P., August, S. C., Welp, L., Neumann, P., Köster, S., et al. (2021). Structural model of the M7G46 methyltransferase TrmB in complex with tRNA. RNA Biol. 18 (12), 2466–2479. doi:10.1080/15476286.2021.1925477
Bo, C., Li, N., He, L., Zhang, S., and An, Y. (2021). Long non-coding RNA ILF3-AS1 facilitates hepatocellular carcinoma progression by stabilizing ILF3 mRNA in an m(6)A-dependent manner. Hum. Cell 34 (6), 1843–1854. doi:10.1007/s13577-021-00608-x
Boccaletto, P., Machnicka, M. A., Purta, E., Piatkowski, P., Baginski, B., Wirecki, T. K., et al. (2018). Modomics: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46 (D1), D303–d307. doi:10.1093/nar/gkx1030
Bohnsack, K. E., Höbartner, C., and Bohnsack, M. T. (2019). Eukaryotic 5-methylcytosine (m⁵C) RNA methyltransferases: Mechanisms, cellular functions, and links to disease. Genes (Basel) 10 (2), E102. doi:10.3390/genes10020102
Bohnsack, M. T., and Sloan, K. E. (2018). Modifications in small nuclear RNAs and their roles in spliceosome assembly and function. Biol. Chem. 399 (11), 1265–1276. doi:10.1515/hsz-2018-0205
Boissel, S., Reish, O., Proulx, K., Kawagoe-Takaki, H., Sedgwick, B., Yeo, G. S., et al. (2009). Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am. J. Hum. Genet. 85 (1), 106–111. doi:10.1016/j.ajhg.2009.06.002
Bokar, J. A., Rath-Shambaugh, M. E., Ludwiczak, R., Narayan, P., and Rottman, F. (1994). Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 269 (26), 17697–17704. doi:10.1016/s0021-9258(17)32497-3
Bolatkan, A., Asada, K., Kaneko, S., Suvarna, K., Ikawa, N., Machino, H., et al. (2022). Downregulation of METTL6 mitigates cell progression, migration, invasion and adhesion in hepatocellular carcinoma by inhibiting cell adhesion molecules. Int. J. Oncol. 60 (1), 4. doi:10.3892/ijo.2021.5294
Boles, N. C., and Temple, S. (2017). Epimetronomics: m6A marks the tempo of corticogenesis. Neuron 96 (4), 718–720. doi:10.1016/j.neuron.2017.11.002
Boo, S. H., and Kim, Y. K. (2020). The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 52 (3), 400–408. doi:10.1038/s12276-020-0407-z
Bosch, F. X., Ribes, J., and Borràs, J. (1999). Epidemiology of primary liver cancer. Semin. Liver Dis. 19 (3), 271–285. doi:10.1055/s-2007-1007117
Boulias, K., and Greer, E. L. (2019). Put the pedal to the METTL1: Adding internal m(7)G increases mRNA translation efficiency and augments miRNA processing. Mol. Cell 74 (6), 1105–1107. doi:10.1016/j.molcel.2019.06.004
Bourgeois, G., Ney, M., Gaspar, I., Aigueperse, C., Schaefer, M., Kellner, S., et al. (2015). Eukaryotic rRNA modification by yeast 5-methylcytosine-methyltransferases and human proliferation-associated antigen p120. PLoS One 10 (7), e0133321. doi:10.1371/journal.pone.0133321
Braghini, M. R., Lo Re, O., Romito, I., Fernandez-Barrena, M. G., Barbaro, B., Pomella, S., et al. (2022). Epigenetic remodelling in human hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 41 (1), 107. doi:10.1186/s13046-022-02297-2
Braun, D. A., Shril, S., Sinha, A., Schneider, R., Tan, W., Ashraf, S., et al. (2018). Mutations in WDR4 as a new cause of Galloway-Mowat syndrome. Am. J. Med. Genet. A 176 (11), 2460–2465. doi:10.1002/ajmg.a.40489
Brommage, R., Powell, D. R., and Vogel, P. (2019). Predicting human disease mutations and identifying drug targets from mouse gene knockout phenotyping campaigns. Dis. Model. Mech. 12 (5), dmm038224. doi:10.1242/dmm.038224
Brzezicha, B., Schmidt, M., Makalowska, I., Jarmolowski, A., Pienkowska, J., and Szweykowska-Kulinska, Z. (2006). Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res. 34 (20), 6034–6043. doi:10.1093/nar/gkl765
Cabello-Villegas, J., and Nikonowicz, E. P. (2005). Solution structure of psi32-modified anticodon stem-loop of Escherichia coli tRNAPhe. Nucleic Acids Res. 33 (22), 6961–6971. doi:10.1093/nar/gki1004
Cai, J., Chen, Z., Zhang, Y., Wang, J., Zhang, Z., Wu, J., et al. (2022). CircRHBDD1 augments metabolic rewiring and restricts immunotherapy efficacy via m(6)A modification in hepatocellular carcinoma. Mol. Ther. Oncolytics 24, 755–771. doi:10.1016/j.omto.2022.02.021
Cai, X., Chen, Y., Man, D., Yang, B., Feng, X., Zhang, D., et al. (2021). RBM15 promotes hepatocellular carcinoma progression by regulating N6-methyladenosine modification of YES1 mRNA in an IGF2BP1-dependent manner. Cell Death Discov. 7 (1), 315. doi:10.1038/s41420-021-00703-w
Cao, J., Shu, X., Feng, X. H., and Liu, J. (2021). Mapping messenger RNA methylations at single base resolution. Curr. Opin. Chem. Biol. 63, 28–37. doi:10.1016/j.cbpa.2021.02.001
Carlile, T. M., Rojas-Duran, M. F., Zinshteyn, B., Shin, H., Bartoli, K. M., and Gilbert, W. V. (2014). Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515 (7525), 143–146. doi:10.1038/nature13802
Cartlidge, R. A., Knebel, A., Peggie, M., Alexandrov, A., Phizicky, E. M., and Cohen, P. (2005). The tRNA methylase METTL1 is phosphorylated and inactivated by PKB and RSK in vitro and in cells. Embo J. 24 (9), 1696–1705. doi:10.1038/sj.emboj.7600648
Chellamuthu, A., and Gray, S. G. (2020). The RNA methyltransferase NSUN2 and its potential roles in cancer. Cells 9 (8), E1758. doi:10.3390/cells9081758
Chen, H., Yao, J., Bao, R., Dong, Y., Zhang, T., Du, Y., et al. (2021a). Cross-talk of four types of RNA modification writers defines tumor microenvironment and pharmacogenomic landscape in colorectal cancer. Mol. Cancer 20 (1), 29. doi:10.1186/s12943-021-01322-w
Chen, J., Jin, R., Zhao, J., Liu, J., Ying, H., Yan, H., et al. (2015). Potential molecular, cellular and microenvironmental mechanism of sorafenib resistance in hepatocellular carcinoma. Cancer Lett. 367 (1), 1–11. doi:10.1016/j.canlet.2015.06.019
Chen, M. H., Fu, L. S., Zhang, F., Yang, Y., and Wu, X. Z. (2021b). LncAY controls BMI1 expression and activates BMI1/Wnt/β-catenin signaling axis in hepatocellular carcinoma. Life Sci. 280, 119748. doi:10.1016/j.lfs.2021.119748
Chen, M., Wei, L., Law, C. T., Tsang, F. H., Shen, J., Cheng, C. L., et al. (2018). RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67 (6), 2254–2270. doi:10.1002/hep.29683
Chen, X., Li, A., Sun, B. F., Yang, Y., Han, Y. N., Yuan, X., et al. (2019a). 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 21 (8), 978–990. doi:10.1038/s41556-019-0361-y
Chen, X. Y., Zhang, J., and Zhu, J. S. (2019b). The role of m(6)A RNA methylation in human cancer. Mol. Cancer 18 (1), 103. doi:10.1186/s12943-019-1033-z
Chen, Y., Ling, Z., Cai, X., Xu, Y., Lv, Z., Man, D., et al. (2022). Activation of YAP1 by N6-methyladenosine-modified circCPSF6 drives malignancy in hepatocellular carcinoma. Cancer Res. 82 (4), 599–614. doi:10.1158/0008-5472.Can-21-1628
Chen, Y., Peng, C., Chen, J., Chen, D., Yang, B., He, B., et al. (2019c). WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer 18 (1), 127. doi:10.1186/s12943-019-1053-8
Chen, Y. S., Yang, W. L., Zhao, Y. L., and Yang, Y. G. (2021c). Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip. Rev. RNA 12 (4), e1639. doi:10.1002/wrna.1639
Chen, Y. T., Xiang, D., Zhao, X. Y., and Chu, X. Y. (2021d). Upregulation of lncRNA NIFK-AS1 in hepatocellular carcinoma by m(6)A methylation promotes disease progression and sorafenib resistance. Hum. Cell 34 (6), 1800–1811. doi:10.1007/s13577-021-00587-z
Chen, Y., Zhao, Y., Chen, J., Peng, C., Zhang, Y., Tong, R., et al. (2020). ALKBH5 suppresses malignancy of hepatocellular carcinoma via m(6)A-guided epigenetic inhibition of LYPD1. Mol. Cancer 19 (1), 123. doi:10.1186/s12943-020-01239-w
Chen, Z., Qi, M., Shen, B., Luo, G., Wu, Y., Li, J., et al. (2019d). Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 47 (5), 2533–2545. doi:10.1093/nar/gky1250
Chen, Z., Zhu, W., Zhu, S., Sun, K., Liao, J., Liu, H., et al. (2021e). METTL1 promotes hepatocarcinogenesis via m(7) G tRNA modification-dependent translation control. Clin. Transl. Med. 11 (12), e661. doi:10.1002/ctm2.661
Cheng, X., Li, M., Rao, X., Zhang, W., Li, X., Wang, L., et al. (2019). KIAA1429 regulates the migration and invasion of hepatocellular carcinoma by altering m6A modification of ID2 mRNA. Onco. Targets. Ther. 12, 3421–3428. doi:10.2147/ott.S180954
Chi, F., Cao, Y., and Chen, Y. (2021). Analysis and validation of circRNA-miRNA network in regulating m(6)A RNA methylation modulators reveals CircMAP2K4/miR-139-5p/YTHDF1 Axis involving the proliferation of hepatocellular carcinoma. Front. Oncol. 11, 560506. doi:10.3389/fonc.2021.560506
Chujo, T., and Suzuki, T. (2012). Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. Rna 18 (12), 2269–2276. doi:10.1261/rna.035600.112
Church, C., Moir, L., McMurray, F., Girard, C., Banks, G. T., Teboul, L., et al. (2010). Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 42 (12), 1086–1092. doi:10.1038/ng.713
Clark, W. C., Evans, M. E., Dominissini, D., Zheng, G., and Pan, T. (2016). tRNA base methylation identification and quantification via high-throughput sequencing. Rna 22 (11), 1771–1784. doi:10.1261/rna.056531.116
Cohn, W. E. (1960). Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: Isolation, structure, and chemical characteristics. J. Biol. Chem. 235, 1488–1498. doi:10.1016/s0021-9258(18)69432-3
Cohn, W. E. (1951). Some results of the applications of ion-exchange chromatography to nucleic acid chemistry. J. Cell. Physiol. Suppl. 38 (1), 21–40. doi:10.1002/jcp.1030380405
Coots, R. A., Liu, X. M., Mao, Y., Dong, L., Zhou, J., Wan, J., et al. (2017). m(6)A Facilitates eIF4F-Independent mRNA Translation. Mol. Cell 68 (3), 504–514.e7. e507. doi:10.1016/j.molcel.2017.10.002
Cozen, A. E., Quartley, E., Holmes, A. D., Hrabeta-Robinson, E., Phizicky, E. M., and Lowe, T. M. (2015). ARM-Seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 12 (9), 879–884. doi:10.1038/nmeth.3508
Cui, J., Liu, Q., Sendinc, E., Shi, Y., and Gregory, R. I. (2021a). Nucleotide resolution profiling of m3C RNA modification by HAC-seq. Nucleic Acids Res. 49 (5), e27. doi:10.1093/nar/gkaa1186
Cui, M., Qu, F., Wang, L., Liu, X., Yu, J., Tang, Z., et al. (2021b). m5C RNA methyltransferase-related gene NSUN4 stimulates malignant progression of hepatocellular carcinoma and can be a prognostic marker. Cancer Biomark. 33, 389–400. doi:10.3233/cbm-210154
D'Silva, S., Haider, S. J., and Phizicky, E. M. (2011). A domain of the actin binding protein Abp140 is the yeast methyltransferase responsible for 3-methylcytidine modification in the tRNA anti-codon loop. Rna 17 (6), 1100–1110. doi:10.1261/rna.2652611
Dai, X., Wang, T., Gonzalez, G., and Wang, Y. (2018). Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA. Anal. Chem. 90 (11), 6380–6384. doi:10.1021/acs.analchem.8b01703
Dai, Z., Liu, H., Liao, J., Huang, C., Ren, X., Zhu, W., et al. (2021). N(7)-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol. Cell 81 (16), 33393339–33393355.e8. doi:10.1016/j.molcel.2021.07.003
Davis, D. R. (1995). Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 23 (24), 5020–5026. doi:10.1093/nar/23.24.5020
Delatte, B., Wang, F., Ngoc, L. V., Collignon, E., Bonvin, E., Deplus, R., et al. (2016). RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351 (6270), 282–285. doi:10.1126/science.aac5253
Delaunay, S., and Frye, M. (2019). RNA modifications regulating cell fate in cancer. Nat. Cell Biol. 21 (5), 552–559. doi:10.1038/s41556-019-0319-0
Deng, Y., Zhou, Z., Ji, W., Lin, S., and Wang, M. (2020). METTL1-mediated m(7)G methylation maintains pluripotency in human stem cells and limits mesoderm differentiation and vascular development. Stem Cell Res. Ther. 11 (1), 306. doi:10.1186/s13287-020-01814-4
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 71 (10), 3971–3975. doi:10.1073/pnas.71.10.3971
Di Timoteo, G., Dattilo, D., Centrón-Broco, A., Colantoni, A., Guarnacci, M., Rossi, F., et al. (2020). Modulation of circRNA metabolism by m(6)A modification. Cell Rep. 31 (6), 107641. doi:10.1016/j.celrep.2020.107641
Dina, C., Meyre, D., Gallina, S., Durand, E., Körner, A., Jacobson, P., et al. (2007). Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 39 (6), 724–726. doi:10.1038/ng2048
Ding, W. B., Wang, M. C., Yu, J., Huang, G., Sun, D. P., Liu, L., et al. (2021). HBV/Pregenomic RNA increases the stemness and promotes the development of HBV-related HCC through reciprocal regulation with insulin-like growth factor 2 mRNA-binding protein 3. Hepatology 74 (3), 1480–1495. doi:10.1002/hep.31850
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 (7397), 201–206. doi:10.1038/nature11112
Dominissini, D., Nachtergaele, S., Moshitch-Moshkovitz, S., Peer, E., Kol, N., Ben-Haim, M. S., et al. (2016). The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530 (7591), 441–446. doi:10.1038/nature16998
Douvlataniotis, K., Bensberg, M., Lentini, A., Gylemo, B., and Nestor, C. E. (2020). No evidence for DNA N (6)-methyladenine in mammals. Sci. Adv. 6 (12), eaay3335. doi:10.1126/sciadv.aay3335
Doyle, A., McGarry, M. P., Lee, N. A., and Lee, J. J. (2012). The construction of transgenic and gene knockout/knockin mouse models of human disease. Transgenic Res. 21 (2), 327–349. doi:10.1007/s11248-011-9537-3
Du, E., Li, J., Sheng, F., Li, S., Zhu, J., Xu, Y., et al. (2020). A pan-cancer analysis reveals genetic alterations, molecular mechanisms, and clinical relevance of m(5) C regulators. Clin. Transl. Med. 10 (5), e180. doi:10.1002/ctm2.180
Du, L., Li, Y., Kang, M., Feng, M., Ren, Y., Dai, H., et al. (2021). USP48 is upregulated by Mettl14 to attenuate hepatocellular carcinoma via regulating SIRT6 stabilization. Cancer Res. 81 (14), 3822–3834. doi:10.1158/0008-5472.Can-20-4163
Duan, J. L., Chen, W., Xie, J. J., Zhang, M. L., Nie, R. C., Liang, H., et al. (2022). A novel peptide encoded by N6-methyladenosine modified circMAP3K4 prevents apoptosis in hepatocellular carcinoma. Mol. Cancer 21 (1), 93. doi:10.1186/s12943-022-01537-5
Dunn, D. B. (1961). The occurrence of 1-methyladenine in ribonucleic acid. Biochim. Biophys. Acta 46, 198–200. doi:10.1016/0006-3002(61)90668-0
Durdevic, Z., Hanna, K., Gold, B., Pollex, T., Cherry, S., Lyko, F., et al. (2013). Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep. 14 (3), 269–275. doi:10.1038/embor.2013.3
Edelheit, S., Schwartz, S., Mumbach, M. R., Wurtzel, O., and Sorek, R. (2013). Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 9 (6), e1003602. doi:10.1371/journal.pgen.1003602
Edupuganti, R. R., Geiger, S., Lindeboom, R. G. H., Shi, H., Hsu, P. J., Lu, Z., et al. (2017). N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24 (10), 870–878. doi:10.1038/nsmb.3462
Esteve-Puig, R., Climent, F., Piñeyro, D., Domingo-Domènech, E., Davalos, V., Encuentra, M., et al. (2021). Epigenetic loss of m1A RNA demethylase ALKBH3 in Hodgkin lymphoma targets collagen, conferring poor clinical outcome. Blood 137 (7), 994–999. doi:10.1182/blood.2020005823
Eyler, D. E., Franco, M. K., Batool, Z., Wu, M. Z., Dubuke, M. L., Dobosz-Bartoszek, M., et al. (2019). Pseudouridinylation of mRNA coding sequences alters translation. Proc. Natl. Acad. Sci. U. S. A. 116 (46), 23068–23074. doi:10.1073/pnas.1821754116
Fan, Z., Gao, Y., Zhang, W., Yang, G., Liu, P., Xu, L., et al. (2021a). METTL3/IGF2BP1/CD47 contributes to the sublethal heat treatment induced mesenchymal transition in HCC. Biochem. Biophys. Res. Commun. 546, 169–177. doi:10.1016/j.bbrc.2021.01.085
Fan, Z., Yang, G., Zhang, W., Liu, Q., Liu, G., Liu, P., et al. (2021b). Hypoxia blocks ferroptosis of hepatocellular carcinoma via suppression of METTL14 triggered YTHDF2-dependent silencing of SLC7A11. J. Cell. Mol. Med. 25 (21), 10197–10212. doi:10.1111/jcmm.16957
Fang, Q., and Chen, H. (2020). The significance of m6A RNA methylation regulators in predicting the prognosis and clinical course of HBV-related hepatocellular carcinoma. Mol. Med. 26 (1), 60. doi:10.1186/s10020-020-00185-z
Fedorov, N. A., and Bogomazov, M. J. (1969). Urinary excretion of purine bases and pseudouridine normal human and in cancer patients before and after radiotherapy. 1. Radiobiol. Radiother. 10 (5), 605–608.
Fernández, I. S., Ng, C. L., Kelley, A. C., Wu, G., Yu, Y. T., and Ramakrishnan, V. (2013). Unusual base pairing during the decoding of a stop codon by the ribosome. Nature 500 (7460), 107–110. doi:10.1038/nature12302
Fischer, J., Koch, L., Emmerling, C., Vierkotten, J., Peters, T., Brüning, J. C., et al. (2009). Inactivation of the Fto gene protects from obesity. Nature 458 (7240), 894–898. doi:10.1038/nature07848
Fu, Y., Dai, Q., Zhang, W., Ren, J., Pan, T., and He, C. (2010). The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew. Chem. Int. Ed. Engl. 49 (47), 8885–8888. doi:10.1002/anie.201001242
Fu, Y., Dominissini, D., Rechavi, G., and He, C. (2014). Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat. Rev. Genet. 15 (5), 293–306. doi:10.1038/nrg3724
Fukuda, H., Chujo, T., Wei, F. Y., Shi, S. L., Hirayama, M., Kaitsuka, T., et al. (2021). Cooperative methylation of human tRNA3Lys at positions A58 and U54 drives the early and late steps of HIV-1 replication. Nucleic Acids Res. 49 (20), 11855–11867. doi:10.1093/nar/gkab879
Garalde, D. R., Snell, E. A., Jachimowicz, D., Sipos, B., Lloyd, J. H., Bruce, M., et al. (2018). Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15 (3), 201–206. doi:10.1038/nmeth.4577
Garcia-Campos, M. A., Edelheit, S., Toth, U., Safra, M., Shachar, R., Viukov, S., et al. (2019). Deciphering the "m(6)A code" via antibody-independent quantitative profiling. Cell 178 (3), 731–747.e16. e716. doi:10.1016/j.cell.2019.06.013
García-Vílchez, R., Sevilla, A., and Blanco, S. (2019). Post-transcriptional regulation by cytosine-5 methylation of RNA. Biochim. Biophys. Acta. Gene Regul. Mech. 1862 (3), 240–252. doi:10.1016/j.bbagrm.2018.12.003
Gavande, N. S., VanderVere-Carozza, P. S., Hinshaw, H. D., Jalal, S. I., Sears, C. R., Pawelczak, K. S., et al. (2016). DNA repair targeted therapy: The past or future of cancer treatment? Pharmacol. Ther. 160, 65–83. doi:10.1016/j.pharmthera.2016.02.003
Gerken, T., Girard, C. A., Tung, Y. C., Webby, C. J., Saudek, V., Hewitson, K. S., et al. (2007). The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318 (5855), 1469–1472. doi:10.1126/science.1151710
Geula, S., Moshitch-Moshkovitz, S., Dominissini, D., Mansour, A. A., Kol, N., Salmon-Divon, M., et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347 (6225), 1002–1006. doi:10.1126/science.1261417
Goll, M. G., Kirpekar, F., Maggert, K. A., Yoder, J. A., Hsieh, C. L., Zhang, X., et al. (2006). Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311 (5759), 395–398. doi:10.1126/science.1120976
Gu, B. W., Ge, J., Fan, J. M., Bessler, M., and Mason, P. J. (2013). Slow growth and unstable ribosomal RNA lacking pseudouridine in mouse embryonic fibroblast cells expressing catalytically inactive dyskerin. FEBS Lett. 587 (14), 2112–2117. doi:10.1016/j.febslet.2013.05.028
Gu, X., Zhou, H., Chu, Q., Zheng, Q., Wang, J., and Zhu, H. (2021). Uncovering the association between m(5)C regulator-mediated methylation modification patterns and tumour microenvironment infiltration characteristics in hepatocellular carcinoma. Front. Cell Dev. Biol. 9, 727935. doi:10.3389/fcell.2021.727935
Gu, Z., Du, Y., Zhao, X., and Wang, C. (2022). Diagnostic, therapeutic, and prognostic value of the m(6)A writer complex in hepatocellular carcinoma. Front. Cell Dev. Biol. 10, 822011. doi:10.3389/fcell.2022.822011
Guo, J. C., Liu, Z., Yang, Y. J., Guo, M., Zhang, J. Q., and Zheng, J. F. (2021). KDM5B promotes self-renewal of hepatocellular carcinoma cells through the microRNA-448-mediated YTHDF3/ITGA6 axis. J. Cell. Mol. Med. 25 (13), jcmm.16342–5962. doi:10.1111/jcmm.16342
Gutschner, T., Hämmerle, M., Pazaitis, N., Bley, N., Fiskin, E., Uckelmann, H., et al. (2014). Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) is an important protumorigenic factor in hepatocellular carcinoma. Hepatology 59 (5), 1900–1911. doi:10.1002/hep.26997
Guzzi, N., Cieśla, M., Ngoc, P. C. T., Lang, S., Arora, S., Dimitriou, M., et al. (2018). Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173 (5), 1204–1216.e26. e1226. doi:10.1016/j.cell.2018.03.008
Haag, S., Kretschmer, J., and Bohnsack, M. T. (2015a). WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. Rna 21 (2), 180–187. doi:10.1261/rna.047910.114
Haag, S., Sloan, K. E., Ranjan, N., Warda, A. S., Kretschmer, J., Blessing, C., et al. (2016). NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. Embo J. 35 (19), 2104–2119. doi:10.15252/embj.201694885
Haag, S., Warda, A. S., Kretschmer, J., Günnigmann, M. A., Höbartner, C., and Bohnsack, M. T. (2015b). NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. Rna 21 (9), 1532–1543. doi:10.1261/rna.051524.115
Hall, R. H. (1963). Isolation of 3-METHYLURIDINE and 3-METHYLCYTIDINE from solubleribonucleic acid. Biochem. Biophys. Res. Commun. 12, 361–364. doi:10.1016/0006-291x(63)90105-0
Hamdane, D., Guelorget, A., Guérineau, V., and Golinelli-Pimpaneau, B. (2014). Dynamics of RNA modification by a multi-site-specific tRNA methyltransferase. Nucleic Acids Res. 42 (18), 11697–11706. doi:10.1093/nar/gku820
Hamma, T., and Ferré-D'Amaré, A. R. (2006). Pseudouridine synthases. Chem. Biol. 13 (11), 1125–1135. doi:10.1016/j.chembiol.2006.09.009
Han, D., Liu, J., Chen, C., Dong, L., Liu, Y., Chang, R., et al. (2019a). Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature 566 (7743), 270–274. doi:10.1038/s41586-019-0916-x
Han, J., Wang, J. Z., Yang, X., Yu, H., Zhou, R., Lu, H. C., et al. (2019b). METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer 18 (1), 110. doi:10.1186/s12943-019-1036-9
Han, L., Marcus, E., D'Silva, S., and Phizicky, E. M. (2017). S. cerevisiae Trm140 has two recognition modes for 3-methylcytidine modification of the anticodon loop of tRNA substrates. Rna 23 (3), 406–419. doi:10.1261/rna.059667.116
Han, T. S., Ban, H. S., Hur, K., and Cho, H. S. (2018). The epigenetic regulation of HCC metastasis. Int. J. Mol. Sci. 19 (12), E3978. doi:10.3390/ijms19123978
Han, X., Wang, M., Zhao, Y. L., Yang, Y., and Yang, Y. G. (2021). RNA methylations in human cancers. Semin. Cancer Biol. 75, 97–115. doi:10.1016/j.semcancer.2020.11.007
Haruehanroengra, P., Zheng, Y. Y., Zhou, Y., Huang, Y., and Sheng, J. (2020). RNA modifications and cancer. RNA Biol. 17 (11), 1560–1575. doi:10.1080/15476286.2020.1722449
Haussmann, I. U., Bodi, Z., Sanchez-Moran, E., Mongan, N. P., Archer, N., Fray, R. G., et al. (2016). m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540 (7632), 301–304. doi:10.1038/nature20577
He, J., Zuo, Q., Hu, B., Jin, H., Wang, C., Cheng, Z., et al. (2019). A novel, liver-specific long noncoding RNA LINC01093 suppresses HCC progression by interaction with IGF2BP1 to facilitate decay of GLI1 mRNA. Cancer Lett. 450, 98–109. doi:10.1016/j.canlet.2019.02.033
He, Y., Shi, Q., Zhang, Y., Yuan, X., and Yu, Z. (2020a). Transcriptome-wide 5-methylcytosine functional profiling of long non-coding RNA in hepatocellular carcinoma. Cancer Manag. Res. 12, 6877–6885. doi:10.2147/cmar.S262450
He, Y., Yu, X., Li, J., Zhang, Q., Zheng, Q., and Guo, W. (2020b). Role of m(5)C-related regulatory genes in the diagnosis and prognosis of hepatocellular carcinoma. Am. J. Transl. Res. 12 (3), 912–922.
He, Y., Yu, X., Zhang, M., and Guo, W. (2021). Pan-cancer analysis of m(5)C regulator genes reveals consistent epigenetic landscape changes in multiple cancers. World J. Surg. Oncol. 19 (1), 224. doi:10.1186/s12957-021-02342-y
He, Y., Zhang, Q., Zheng, Q., Yu, X., and Guo, W. (2020c). Distinct 5-methylcytosine profiles of circular RNA in human hepatocellular carcinoma. Am. J. Transl. Res. 12 (9), 5719–5729.
Hoernes, T. P., Clementi, N., Faserl, K., Glasner, H., Breuker, K., Lindner, H., et al. (2016). Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 44 (2), 852–862. doi:10.1093/nar/gkv1182
Horiuchi, K., Umetani, M., Minami, T., Okayama, H., Takada, S., Yamamoto, M., et al. (2006). Wilms' tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc. Natl. Acad. Sci. U. S. A. 103 (46), 17278–17283. doi:10.1073/pnas.0608357103
Horowitz, S., Horowitz, A., Nilsen, T. W., Munns, T. W., and Rottman, F. M. (1984). Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc. Natl. Acad. Sci. U. S. A. 81 (18), 5667–5671. doi:10.1073/pnas.81.18.5667
Hou, J., Zhang, H., Liu, J., Zhao, Z., Wang, J., Lu, Z., et al. (2019). YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer 18 (1), 163. doi:10.1186/s12943-019-1082-3
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20 (3), 285–295. doi:10.1038/s41556-018-0045-z
Huang, H., Weng, H., Zhou, K., Wu, T., Zhao, B. S., Sun, M., et al. (2019). Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 567 (7748), 414–419. doi:10.1038/s41586-019-1016-7
Huang, R., and Zhou, P. K. (2021). DNA damage repair: Historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduct. Target. Ther. 6 (1), 254. doi:10.1038/s41392-021-00648-7
Hur, S., Stroud, R. M., and Finer-Moore, J. (2006). Substrate recognition by RNA 5-methyluridine methyltransferases and pseudouridine synthases: A structural perspective. J. Biol. Chem. 281 (51), 38969–38973. doi:10.1074/jbc.R600034200
Hussain, S., Sajini, A. A., Blanco, S., Dietmann, S., Lombard, P., Sugimoto, Y., et al. (2013). NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 4 (2), 255–261. doi:10.1016/j.celrep.2013.06.029
Ignatova, V. V., Kaiser, S., Ho, J. S. Y., Bing, X., Stolz, P., Tan, Y. X., et al. (2020). METTL6 is a tRNA m(3)C methyltransferase that regulates pluripotency and tumor cell growth. Sci. Adv. 6 (35), eaaz4551. eaaz4551. doi:10.1126/sciadv.aaz4551
Jana, S., Hsieh, A. C., and Gupta, R. (2017). Reciprocal amplification of caspase-3 activity by nuclear export of a putative human RNA-modifying protein, PUS10 during TRAIL-induced apoptosis. Cell Death Dis. 8 (10), e3093. doi:10.1038/cddis.2017.476
Janin, M., Ortiz-Barahona, V., de Moura, M. C., Martínez-Cardús, A., Llinàs-Arias, P., Soler, M., et al. (2019). Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. 138 (6), 1053–1074. doi:10.1007/s00401-019-02062-4
Ji, P., Ding, D., Qin, N., Wang, C., Zhu, M., Li, Y., et al. (2020). Systematic analyses of genetic variants in chromatin interaction regions identified four novel lung cancer susceptibility loci. J. Cancer 11 (5), 1075–1081. doi:10.7150/jca.35127
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7 (12), 885–887. doi:10.1038/nchembio.687
Jiang, H., Ning, G., Wang, Y., and Lv, W. (2021). Identification of an m6A-related signature as biomarker for hepatocellular carcinoma prognosis and correlates with sorafenib and anti-PD-1 immunotherapy treatment response. Dis. Markers 2021, 5576683. doi:10.1155/2021/5576683
Karijolich, J., and Yu, Y. T. (2011). Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474 (7351), 395–398. doi:10.1038/nature10165
Karikó, K., Muramatsu, H., Welsh, F. A., Ludwig, J., Kato, H., Akira, S., et al. (2008). Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16 (11), 1833–1840. doi:10.1038/mt.2008.200
Ke, S., Pandya-Jones, A., Saito, Y., Fak, J. J., Vågbø, C. B., Geula, S., et al. (2017). m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31 (10), 990–1006. doi:10.1101/gad.301036.117
Kennedy, E. M., Bogerd, H. P., Kornepati, A. V., Kang, D., Ghoshal, D., Marshall, J. B., et al. (2016). Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 19 (5), 675–685. doi:10.1016/j.chom.2016.04.002
Khan, M. A., Rafiq, M. A., Noor, A., Hussain, S., Flores, J. V., Rupp, V., et al. (2012). Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet. 90 (5), 856–863. doi:10.1016/j.ajhg.2012.03.023
Khoddami, V., and Cairns, B. R. (2013). Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 31 (5), 458–464. doi:10.1038/nbt.2566
Khoddami, V., and Cairns, B. R. (2014). Transcriptome-wide target profiling of RNA cytosine methyltransferases using the mechanism-based enrichment procedure Aza-IP. Nat. Protoc. 9 (2), 337–361. doi:10.1038/nprot.2014.014
Kim, N. K., Theimer, C. A., Mitchell, J. R., Collins, K., and Feigon, J. (2010). Effect of pseudouridylation on the structure and activity of the catalytically essential P6.1 hairpin in human telomerase RNA. Nucleic Acids Res. 38 (19), 6746–6756. doi:10.1093/nar/gkq525
King, T. H., Liu, B., McCully, R. R., and Fournier, M. J. (2003). Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell 11 (2), 425–435. doi:10.1016/s1097-2765(03)00040-6
Klingenberg, M., Matsuda, A., Diederichs, S., and Patel, T. (2017). Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J. Hepatol. 67 (3), 603–618. doi:10.1016/j.jhep.2017.04.009
Ko, E., Kim, J. S., Ju, S., Seo, H. W., Chang, Y., Kang, J. A., et al. (2018). Oxidatively modified protein-disulfide isomerase-associated 3 promotes dyskerin pseudouridine synthase 1-mediated malignancy and survival of hepatocellular carcinoma cells. Hepatology 68 (5), 1851–1864. doi:10.1002/hep.30039
Kong, H., Sun, J., Zhang, W., Zhang, H., and Li, H. (2022). Long intergenic non-protein coding RNA 1273 confers sorafenib resistance in hepatocellular carcinoma via regulation of methyltransferase 3. Bioengineered 13 (2), 3108–3121. doi:10.1080/21655979.2022.2025701
Krug, R. M., Morgan, M. A., and Shatkin, A. J. (1976). Influenza viral mRNA contains internal N6-methyladenosine and 5'-terminal 7-methylguanosine in cap structures. J. Virol. 20 (1), 45–53. doi:10.1128/jvi.20.1.45-53.1976
Lan, T., Li, H., Zhang, D., Xu, L., Liu, H., Hao, X., et al. (2019). KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer 18 (1), 186. doi:10.1186/s12943-019-1106-z
Lecointe, F., Namy, O., Hatin, I., Simos, G., Rousset, J. P., and Grosjean, H. (2002). Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J. Biol. Chem. 277 (34), 30445–30453. doi:10.1074/jbc.M203456200
Leger, A., Amaral, P. P., Pandolfini, L., Capitanchik, C., Capraro, F., Miano, V., et al. (2021). RNA modifications detection by comparative Nanopore direct RNA sequencing. Nat. Commun. 12 (1), 7198. doi:10.1038/s41467-021-27393-3
Lentini, J. M., Bargabos, R., Chen, C., and Fu, D. (2022). Methyltransferase METTL8 is required for 3-methylcytosine modification in human mitochondrial tRNAs. J. Biol. Chem. 298. doi:10.1016/j.jbc.2022.101788
Létoquart, J., Huvelle, E., Wacheul, L., Bourgeois, G., Zorbas, C., Graille, M., et al. (2014). Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc. Natl. Acad. Sci. U. S. A. 111 (51), E5518–E5526. doi:10.1073/pnas.1413089111
Leulliot, N., Chaillet, M., Durand, D., Ulryck, N., Blondeau, K., and van Tilbeurgh, H. (2008). Structure of the yeast tRNA m7G methylation complex. Structure 16 (1), 52–61. doi:10.1016/j.str.2007.10.025
Li, F., Zhao, D., Wu, J., and Shi, Y. (2014). Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 24 (12), 1490–1492. doi:10.1038/cr.2014.153
Li, G., Deng, L., Huang, N., Cui, Z., Wu, Q., Ma, J., et al. (2021a). m(6)A mRNA methylation regulates LKB1 to promote autophagy of hepatoblastoma cells through upregulated phosphorylation of AMPK. Genes (Basel) 12 (11), 1747. doi:10.3390/genes12111747
Li, J., Wang, W., Zhou, Y., Liu, L., Zhang, G., Guan, K., et al. (2021b). m6A regulator-associated modification patterns and immune infiltration of the tumor microenvironment in hepatocarcinoma. Front. Cell Dev. Biol. 9, 687756. doi:10.3389/fcell.2021.687756
Li, Q., Ni, Y., Zhang, L., Jiang, R., Xu, J., Yang, H., et al. (2021c). HIF-1α-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct. Target. Ther. 6 (1), 76. doi:10.1038/s41392-020-00453-8
Li, S., Zhou, H., Liao, S., Wang, X., Zhu, Z., Zhang, J., et al. (2022). Structural basis for METTL6-mediated m3C RNA methylation. Biochem. Biophys. Res. Commun. 589, 159–164. doi:10.1016/j.bbrc.2021.12.013
Li, X., Xiong, X., Zhang, M., Wang, K., Chen, Y., Zhou, J., et al. (2017a). Base-resolution mapping reveals distinct m(1)A methylome in nuclear- and mitochondrial-encoded transcripts. Mol. Cell 68 (5), 993–1005. doi:10.1016/j.molcel.2017.10.019
Li, X., Zhu, P., Ma, S., Song, J., Bai, J., Sun, F., et al. (2015). Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 11 (8), 592–597. doi:10.1038/nchembio.1836
Li, Y., Cheng, X., Chen, Y., Zhou, T., Li, D., and Zheng, W. V. (2021d). METTL3 facilitates the progression of hepatocellular carcinoma by modulating the m6A level of USP7. Am. J. Transl. Res. 13 (12), 13423–13437.
Li, Y., Qi, D., Zhu, B., and Ye, X. (2021e). Analysis of m6A RNA methylation-related genes in liver hepatocellular carcinoma and their correlation with survival. Int. J. Mol. Sci. 22 (3), 1474. doi:10.3390/ijms22031474
Li, Z., Li, F., Peng, Y., Fang, J., and Zhou, J. (2020). Identification of three m6A-related mRNAs signature and risk score for the prognostication of hepatocellular carcinoma. Cancer Med. 9 (5), 1877–1889. doi:10.1002/cam4.2833
Li, Z., Weng, H., Su, R., Weng, X., Zuo, Z., Li, C., et al. (2017b). FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell 31 (1), 127–141. doi:10.1016/j.ccell.2016.11.017
Liang, W., Lin, Z., Du, C., Qiu, D., and Zhang, Q. (2020). mRNA modification orchestrates cancer stem cell fate decisions. Mol. Cancer 19 (1), 38. doi:10.1186/s12943-020-01166-w
Lin, S., Choe, J., Du, P., Triboulet, R., and Gregory, R. I. (2016). The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62 (3), 335–345. doi:10.1016/j.molcel.2016.03.021
Lin, S., Liu, Q., Jiang, Y. Z., and Gregory, R. I. (2019). Nucleotide resolution profiling of m(7)G tRNA modification by TRAC-Seq. Nat. Protoc. 14 (11), 3220–3242. doi:10.1038/s41596-019-0226-7
Lin, S., Liu, Q., Lelyveld, V. S., Choe, J., Szostak, J. W., and Gregory, R. I. (2018). Mettl1/Wdr4-Mediated m 7 G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol. Cell 71 (2), 244–255.e5. e245. doi:10.1016/j.molcel.2018.06.001
Lin, Y., Wei, X., Jian, Z., and Zhang, X. (2020a). METTL3 expression is associated with glycolysis metabolism and sensitivity to glycolytic stress in hepatocellular carcinoma. Cancer Med. 9 (8), 2859–2867. doi:10.1002/cam4.2918
Lin, Z., Niu, Y., Wan, A., Chen, D., Liang, H., Chen, X., et al. (2020b). RNA m(6) A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. Embo J. 39 (12), e103181. doi:10.15252/embj.2019103181
Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., and Jaffrey, S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12 (8), 767–772. doi:10.1038/nmeth.3453
Liu, B., Zhang, J., Huang, C., and Liu, H. (2012). Dyskerin overexpression in human hepatocellular carcinoma is associated with advanced clinical stage and poor patient prognosis. PLoS One 7 (8), e43147. doi:10.1371/journal.pone.0043147
Liu, F., Clark, W., Luo, G., Wang, X., Fu, Y., Wei, J., et al. (2016). ALKBH1-Mediated tRNA demethylation regulates translation. Cell 167 (3), 816–828.e16. e816. doi:10.1016/j.cell.2016.09.038
Liu, G. M., Zeng, H. D., Zhang, C. Y., and Xu, J. W. (2021a). Identification of METTL3 as an adverse prognostic biomarker in hepatocellular carcinoma. Dig. Dis. Sci. 66 (4), 1110–1126. doi:10.1007/s10620-020-06260-z
Liu, H., Lan, T., Li, H., Xu, L., Chen, X., Liao, H., et al. (2021b). Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics 11 (3), 1396–1411. doi:10.7150/thno.53227
Liu, J., Eckert, M. A., Harada, B. T., Liu, S. M., Lu, Z., Yu, K., et al. (2018). m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 20 (9), 1074–1083. doi:10.1038/s41556-018-0174-4
Liu, J., Sun, G., Pan, S., Qin, M., Ouyang, R., Li, Z., et al. (2020a). The Cancer Genome Atlas (TCGA) based m(6)A methylation-related genes predict prognosis in hepatocellular carcinoma. Bioengineered 11 (1), 759–768. doi:10.1080/21655979.2020.1787764
Liu, J., Wang, D., Zhou, J., Wang, L., Zhang, N., Zhou, L., et al. (2021c). N6-methyladenosine reader YTHDC2 and eraser FTO may determine hepatocellular carcinoma prognoses after transarterial chemoembolization. Arch. Toxicol. 95 (5), 1621–1629. doi:10.1007/s00204-021-03021-3
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10 (2), 93–95. doi:10.1038/nchembio.1432
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518 (7540), 560–564. doi:10.1038/nature14234
Liu, N., Zhou, K. I., Parisien, M., Dai, Q., Diatchenko, L., and Pan, T. (2017). N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45 (10), 6051–6063. doi:10.1093/nar/gkx141
Liu, X., Liu, J., Xiao, W., Zeng, Q., Bo, H., Zhu, Y., et al. (2020b). SIRT1 regulates N(6) -methyladenosine RNA modification in hepatocarcinogenesis by inducing RANBP2-dependent FTO SUMOylation. Hepatology 72 (6), 2029–2050. doi:10.1002/hep.31222
Liu, X., Liu, Z., Mao, X., and Li, Q. (2020c). m7GPredictor: An improved machine learning-based model for predicting internal m7G modifications using sequence properties. Anal. Biochem. 609, 113905. doi:10.1016/j.ab.2020.113905
Liu, X., Qin, J., Gao, T., Li, C., Chen, X., Zeng, K., et al. (2020d). Analysis of METTL3 and METTL14 in hepatocellular carcinoma. Aging (Albany NY) 12 (21), 21638–21659. doi:10.18632/aging.103959
Liu, X., Qin, J., Gao, T., Li, C., He, B., Pan, B., et al. (2020e). YTHDF1 facilitates the progression of hepatocellular carcinoma by promoting FZD5 mRNA translation in an m6A-dependent manner. Mol. Ther. Nucleic Acids 22, 750–765. doi:10.1016/j.omtn.2020.09.036
Liu, Y., Yang, C., Zhao, Y., Chi, Q., Wang, Z., and Sun, B. (2019). Overexpressed methyltransferase-like 1 (METTL1) increased chemosensitivity of colon cancer cells to cisplatin by regulating miR-149-3p/S100A4/p53 axis. Aging (Albany NY) 11 (24), 12328–12344. doi:10.18632/aging.102575
Liu, Y., Zheng, S., Wang, T., Fang, Z., Kong, J., and Liu, J. (2022). Identification of the expression patterns and potential prognostic role of 5-methylcytosine regulators in hepatocellular carcinoma. Front. Cell Dev. Biol. 10, 842220. doi:10.3389/fcell.2022.842220
Llovet, J. M., Kelley, R. K., Villanueva, A., Singal, A. G., Pikarsky, E., Roayaie, S., et al. (2021). Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 7 (1), 6. doi:10.1038/s41572-020-00240-3
Llovet, J. M., Ricci, S., Mazzaferro, V., Hilgard, P., Gane, E., Blanc, J. F., et al. (2008). Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359 (4), 378–390. doi:10.1056/NEJMoa0708857
Loos, R. J., and Yeo, G. S. (2014). The bigger picture of FTO: The first GWAS-identified obesity gene. Nat. Rev. Endocrinol. 10 (1), 51–61. doi:10.1038/nrendo.2013.227
Lovejoy, A. F., Riordan, D. P., and Brown, P. O. (2014). Transcriptome-wide mapping of pseudouridines: Pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 9 (10), e110799. doi:10.1371/journal.pone.0110799
Luo, X., Cao, M., Gao, F., and He, X. (2021). YTHDF1 promotes hepatocellular carcinoma progression via activating PI3K/AKT/mTOR signaling pathway and inducing epithelial-mesenchymal transition. Exp. Hematol. Oncol. 10 (1), 35. doi:10.1186/s40164-021-00227-0
Ma, C. J., Ding, J. H., Ye, T. T., Yuan, B. F., and Feng, Y. Q. (2019). AlkB homologue 1 demethylates N(3)-methylcytidine in mRNA of mammals. ACS Chem. Biol. 14 (7), 1418–1425. doi:10.1021/acschembio.8b01001
Ma, J., Han, H., Huang, Y., Yang, C., Zheng, S., Cai, T., et al. (2021). METTL1/WDR4-mediated m(7)G tRNA modifications and m(7)G codon usage promote mRNA translation and lung cancer progression. Mol. Ther. 29 (12), 3422–3435. doi:10.1016/j.ymthe.2021.08.005
Ma, J. Z., Yang, F., Zhou, C. C., Liu, F., Yuan, J. H., Wang, F., et al. (2017). METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology 65 (2), 529–543. doi:10.1002/hep.28885
Macari, F., El-Houfi, Y., Boldina, G., Xu, H., Khoury-Hanna, S., Ollier, J., et al. (2016). TRM6/61 connects PKCα with translational control through tRNAi(met) stabilization: Impact on tumorigenesis. Oncogene 35 (14), 1785–1796. doi:10.1038/onc.2015.244
Machnicka, M. A., Milanowska, K., Osman Oglou, O., Purta, E., Kurkowska, M., Olchowik, A., et al. (2013). Modomics: A database of RNA modification pathways--2013 update. Nucleic Acids Res. 41, D262–D267. Database issue). doi:10.1093/nar/gks1007
Malbec, L., Zhang, T., Chen, Y. S., Zhang, Y., Sun, B. F., Shi, B. Y., et al. (2019). Dynamic methylome of internal mRNA N(7)-methylguanosine and its regulatory role in translation. Cell Res. 29 (11), 927–941. doi:10.1038/s41422-019-0230-z
Marchand, V., Ayadi, L., Ernst, F. G. M., Hertler, J., Bourguignon-Igel, V., Galvanin, A., et al. (2018). AlkAniline-Seq: Profiling of m(7) G and m(3) C RNA modifications at single nucleotide resolution. Angew. Chem. Int. Ed. Engl. 57 (51), 16785–16790. doi:10.1002/anie.201810946
Marchand, V., Bourguignon-Igel, V., Helm, M., and Motorin, Y. (2021). Mapping of 7-methylguanosine (m(7)G), 3-methylcytidine (m(3)C), dihydrouridine (D) and 5-hydroxycytidine (ho(5)C) RNA modifications by AlkAniline-Seq. Methods Enzymol. 658, 25–47. doi:10.1016/bs.mie.2021.06.001
Marchand, V., Pichot, F., Neybecker, P., Ayadi, L., Bourguignon-Igel, V., Wacheul, L., et al. (2020). HydraPsiSeq: A method for systematic and quantitative mapping of pseudouridines in RNA. Nucleic Acids Res. 48 (19), e110. doi:10.1093/nar/gkaa769
Martinez, F. J., Lee, J. H., Lee, J. E., Blanco, S., Nickerson, E., Gabriel, S., et al. (2012). Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J. Med. Genet. 49 (6), 380–385. doi:10.1136/jmedgenet-2011-100686
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m(6)A(m) in the 5' cap controls mRNA stability. Nature 541 (7637), 371–375. doi:10.1038/nature21022
McMahon, M., Contreras, A., Holm, M., Uechi, T., Forester, C. M., Pang, X., et al. (2019). A single H/ACA small nucleolar RNA mediates tumor suppression downstream of oncogenic RAS. Elife 8, e48847. doi:10.7554/eLife.48847
Meyer, K. D. (2019). DART-Seq: An antibody-free method for global m(6)A detection. Nat. Methods 16 (12), 1275–1280. doi:10.1038/s41592-019-0570-0
Meyer, K. D., and Jaffrey, S. R. (2017). Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342. doi:10.1146/annurev-cellbio-100616-060758
Meyer, K. D., and Jaffrey, S. R. (2014). The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15 (5), 313–326. doi:10.1038/nrm3785
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5' UTR m(6)A promotes cap-independent translation. Cell 163 (4), 999–1010. doi:10.1016/j.cell.2015.10.012
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149 (7), 1635–1646. doi:10.1016/j.cell.2012.05.003
Michaud, J., Kudoh, J., Berry, A., Bonne-Tamir, B., Lalioti, M. D., Rossier, C., et al. (2000). Isolation and characterization of a human chromosome 21q22.3 gene (WDR4) and its mouse homologue that code for a WD-repeat protein. Genomics 68 (1), 71–79. doi:10.1006/geno.2000.6258
Mittenbühler, M. J., Saedler, K., Nolte, H., Kern, L., Zhou, J., Qian, S. B., et al. (2020). Hepatic FTO is dispensable for the regulation of metabolism but counteracts HCC development in vivo. Mol. Metab. 42, 101085. doi:10.1016/j.molmet.2020.101085
Molinie, B., Wang, J., Lim, K. S., Hillebrand, R., Lu, Z. X., Van Wittenberghe, N., et al. (2016). m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat. Methods 13 (8), 692–698. doi:10.1038/nmeth.3898
Müller, S., Glaß, M., Singh, A. K., Haase, J., Bley, N., Fuchs, T., et al. (2019). IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 47 (1), 375–390. doi:10.1093/nar/gky1012
Natchiar, S. K., Myasnikov, A. G., Kratzat, H., Hazemann, I., and Klaholz, B. P. (2017). Visualization of chemical modifications in the human 80S ribosome structure. Nature 551 (7681), 472–477. doi:10.1038/nature24482
Newby, M. I., and Greenbaum, N. L. (2001). A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. Rna 7 (6), 833–845. doi:10.1017/s1355838201002308
Newby, M. I., and Greenbaum, N. L. (2002). Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc. Natl. Acad. Sci. U. S. A. 99 (20), 12697–12702. doi:10.1073/pnas.202477199
Nobles, K. N., Yarian, C. S., Liu, G., Guenther, R. H., and Agris, P. F. (2002). Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding. Nucleic Acids Res. 30 (21), 4751–4760. doi:10.1093/nar/gkf595
Nombela, P., Miguel-López, B., and Blanco, S. (2021). The role of m(6)A, m(5)C and Ψ RNA modifications in cancer: Novel therapeutic opportunities. Mol. Cancer 20 (1), 18. doi:10.1186/s12943-020-01263-w
Oerum, S., Dégut, C., Barraud, P., and Tisné, C. (2017). m1A Post-Transcriptional Modification in tRNAs. Biomolecules 7 (1), E20. doi:10.3390/biom7010020
Okamoto, M., Fujiwara, M., Hori, M., Okada, K., Yazama, F., Konishi, H., et al. (2014). tRNA modifying enzymes, NSUN2 and METTL1, determine sensitivity to 5-fluorouracil in HeLa cells. PLoS Genet. 10 (9), e1004639. doi:10.1371/journal.pgen.1004639
Okamoto, M., Hirata, S., Sato, S., Koga, S., Fujii, M., Qi, G., et al. (2012). Frequent increased gene copy number and high protein expression of tRNA (cytosine-5-)-methyltransferase (NSUN2) in human cancers. DNA Cell Biol. 31 (5), 660–671. doi:10.1089/dna.2011.1446
Olejniczak, M., and Uhlenbeck, O. C. (2006). tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie 88 (8), 943–950. doi:10.1016/j.biochi.2006.06.005
Orellana, E. A., Liu, Q., Yankova, E., Pirouz, M., De Braekeleer, E., Zhang, W., et al. (2021). METTL1-mediated m(7)G modification of Arg-TCT tRNA drives oncogenic transformation. Mol. Cell 81 (16), 33233323–33233338.e14. doi:10.1016/j.molcel.2021.06.031
Ougland, R., Zhang, C. M., Liiv, A., Johansen, R. F., Seeberg, E., Hou, Y. M., et al. (2004). AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol. Cell 16 (1), 107–116. doi:10.1016/j.molcel.2004.09.002
Ozen, C., Yildiz, G., Dagcan, A. T., Cevik, D., Ors, A., Keles, U., et al. (2013). Genetics and epigenetics of liver cancer. N. Biotechnol. 30 (4), 381–384. doi:10.1016/j.nbt.2013.01.007
Pan, F., Lin, X. R., Hao, L. P., Chu, X. Y., Wan, H. J., and Wang, R. (2021). The role of RNA methyltransferase METTL3 in hepatocellular carcinoma: Results and perspectives. Front. Cell Dev. Biol. 9, 674919. doi:10.3389/fcell.2021.674919
Pandey, R. R., and Pillai, R. S. (2019). Counting the cuts: MAZTER-seq quantifies m(6)A levels using a methylation-sensitive ribonuclease. Cell 178 (3), 515–517. doi:10.1016/j.cell.2019.07.006
Pandolfini, L., Barbieri, I., Bannister, A. J., Hendrick, A., Andrews, B., Webster, N., et al. (2019). METTL1 promotes let-7 MicroRNA processing via m7G methylation. Mol. Cell 74 (6), 1278–1290.e9. e1279. doi:10.1016/j.molcel.2019.03.040
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537 (7620), 369–373. doi:10.1038/nature19342
Pea, A., Jamieson, N. B., and Braconi, C. (2021). Biology and clinical application of regulatory RNAs in hepatocellular carcinoma. Hepatology 73 (1), 38–48. doi:10.1002/hep.31225
Penzo, M., and Montanaro, L. (2018). Turning uridines around: Role of rRNA pseudouridylation in ribosome biogenesis and ribosomal function. Biomolecules 8 (2), E38. doi:10.3390/biom8020038
Piekna-Przybylska, D., Decatur, W. A., and Fournier, M. J. (2008). The 3D rRNA modification maps database: With interactive tools for ribosome analysis. Nucleic Acids Res. 36, D178–D183. doi:10.1093/nar/gkm855
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24 (2), 177–189. doi:10.1038/cr.2014.3
Pratanwanich, P. N., Yao, F., Chen, Y., Koh, C. W. Q., Wan, Y. K., Hendra, C., et al. (2021). Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore. Nat. Biotechnol. 39 (11), 1394–1402. doi:10.1038/s41587-021-00949-w
Pu, J., Wang, J., Qin, Z., Wang, A., Zhang, Y., Wu, X., et al. (2020). IGF2BP2 promotes liver cancer growth through an m6A-FEN1-dependent mechanism. Front. Oncol. 10, 578816. doi:10.3389/fonc.2020.578816
Qiao, K., Liu, Y., Xu, Z., Zhang, H., Zhang, H., Zhang, C., et al. (2021). RNA m6A methylation promotes the formation of vasculogenic mimicry in hepatocellular carcinoma via Hippo pathway. Angiogenesis 24 (1), 83–96. doi:10.1007/s10456-020-09744-8
Qu, N., Qin, S., Zhang, X., Bo, X., Liu, Z., Tan, C., et al. (2020). Multiple m(6)A RNA methylation modulators promote the malignant progression of hepatocellular carcinoma and affect its clinical prognosis. BMC Cancer 20 (1), 165. doi:10.1186/s12885-020-6638-5
Qu, S., Jin, L., Huang, H., Lin, J., Gao, W., and Zeng, Z. (2021). A positive-feedback loop between HBx and ALKBH5 promotes hepatocellular carcinogenesis. BMC Cancer 21 (1), 686. doi:10.1186/s12885-021-08449-5
Rang, F. J., Kloosterman, W. P., and de Ridder, J. (2018). From squiggle to basepair: Computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 19 (1), 90. doi:10.1186/s13059-018-1462-9
Rao, X., Lai, L., Li, X., Wang, L., Li, A., and Yang, Q. (2021). N(6) -methyladenosine modification of circular RNA circ-ARL3 facilitates Hepatitis B virus-associated hepatocellular carcinoma via sponging miR-1305. IUBMB Life 73 (2), 408–417. doi:10.1002/iub.2438
Rasmuson, T., and Björk, G. R. (1995). Urinary excretion of pseudouridine and prognosis of patients with malignant lymphoma. Acta Oncol. 34 (1), 61–67. doi:10.3109/02841869509093640
Reid, R., Greene, P. J., and Santi, D. V. (1999). Exposition of a family of RNA m(5)C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. 27 (15), 3138–3145. doi:10.1093/nar/27.15.3138
Ren, S. H., Qin, Y. F., Qin, H., Wang, H. D., Li, G. M., Zhu, Y. L., et al. (2022). N6-Methyladenine-Related signature for immune microenvironment and response to immunotherapy in hepatocellular carcinoma. Int. J. Gen. Med. 15, 3525–3540. doi:10.2147/ijgm.S351815
Rintala-Dempsey, A. C., and Kothe, U. (2017). Eukaryotic stand-alone pseudouridine synthases - RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 14 (9), 1185–1196. doi:10.1080/15476286.2016.1276150
Roake, C. M., and Artandi, S. E. (2016). DNA repair: Telomere-lengthening mechanism revealed. Nature 539 (7627), 35–36. doi:10.1038/nature19483
Rong, D., Sun, G., Wu, F., Cheng, Y., Sun, G., Jiang, W., et al. (2021a). Epigenetics: Roles and therapeutic implications of non-coding RNA modifications in human cancers. Mol. Ther. Nucleic Acids 25, 67–82. doi:10.1016/j.omtn.2021.04.021
Rong, D., Wu, F., Lu, C., Sun, G., Shi, X., Chen, X., et al. (2021b). m6A modification of circHPS5 and hepatocellular carcinoma progression through HMGA2 expression. Mol. Ther. Nucleic Acids 26, 637–648. doi:10.1016/j.omtn.2021.09.001
Roovers, M., Hale, C., Tricot, C., Terns, M. P., Terns, R. M., Grosjean, H., et al. (2006). Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res. 34 (15), 4293–4301. doi:10.1093/nar/gkl530
Rosa-Mercado, N. A., Withers, J. B., and Steitz, J. A. (2017). Settling the m(6)A debate: Methylation of mature mRNA is not dynamic but accelerates turnover. Genes Dev. 31 (10), 957–958. doi:10.1101/gad.302695.117
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell 169 (7), 1187–1200. doi:10.1016/j.cell.2017.05.045
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27 (3), 315–328. doi:10.1038/cr.2017.15
Safra, M., Nir, R., Farouq, D., Vainberg Slutskin, I., and Schwartz, S. (2017a). TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res. 27 (3), 393–406. doi:10.1101/gr.207613.116
Safra, M., Sas-Chen, A., Nir, R., Winkler, R., Nachshon, A., Bar-Yaacov, D., et al. (2017b). The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551 (7679), 251–255. doi:10.1038/nature24456
Schaefer, M., Pollex, T., Hanna, K., Tuorto, F., Meusburger, M., Helm, M., et al. (2010). RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 24 (15), 1590–1595. doi:10.1101/gad.586710
Scheitl, C. P. M., Ghaem Maghami, M., Lenz, A. K., and Höbartner, C. (2020). Site-specific RNA methylation by a methyltransferase ribozyme. Nature 587 (7835), 663–667. doi:10.1038/s41586-020-2854-z
Schöller, E., Marks, J., Marchand, V., Bruckmann, A., Powell, C. A., Reichold, M., et al. (2021). Balancing of mitochondrial translation through METTL8-mediated m(3)C modification of mitochondrial tRNAs. Mol. Cell 81 (23), 48104810–48104825.e12. doi:10.1016/j.molcel.2021.10.018
Schulze, K., Imbeaud, S., Letouzé, E., Alexandrov, L. B., Calderaro, J., Rebouissou, S., et al. (2015). Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 47 (5), 505–511. doi:10.1038/ng.3252
Schumann, U., Shafik, A., and Preiss, T. (2016). METTL3 gains R/W access to the epitranscriptome. Mol. Cell 62 (3), 323–324. doi:10.1016/j.molcel.2016.04.024
Schwartz, S., Bernstein, D. A., Mumbach, M. R., Jovanovic, M., Herbst, R. H., León-Ricardo, B. X., et al. (2014a). Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159 (1), 148–162. doi:10.1016/j.cell.2014.08.028
Schwartz, S., Mumbach, M. R., Jovanovic, M., Wang, T., Maciag, K., Bushkin, G. G., et al. (2014b). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 8 (1), 284–296. doi:10.1016/j.celrep.2014.05.048
Seo, K. W., and Kleiner, R. E. (2020). YTHDF2 recognition of N(1)-methyladenosine (m(1)A)-Modified RNA is associated with transcript destabilization. ACS Chem. Biol. 15 (1), 132–139. doi:10.1021/acschembio.9b00655
Shafik, A. M., Zhou, H., Lim, J., Dickinson, B., and Jin, P. (2021). Dysregulated mitochondrial and cytosolic tRNA m1A methylation in Alzheimer's disease. Hum. Mol. Genet. 31, 1673–1680. doi:10.1093/hmg/ddab357
Shaheen, R., Abdel-Salam, G. M., Guy, M. P., Alomar, R., Abdel-Hamid, M. S., Afifi, H. H., et al. (2015). Mutation in WDR4 impairs tRNA m(7)G46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol. 16, 210. doi:10.1186/s13059-015-0779-x
Shi, H., Chai, P., Jia, R., and Fan, X. (2020a). Novel insight into the regulatory roles of diverse RNA modifications: Re-Defining the bridge between transcription and translation. Mol. Cancer 19 (1), 78. doi:10.1186/s12943-020-01194-6
Shi, Q., Xue, C., Yuan, X., He, Y., and Yu, Z. (2020b). Gene signatures and prognostic values of m1A-related regulatory genes in hepatocellular carcinoma. Sci. Rep. 10 (1), 15083. doi:10.1038/s41598-020-72178-1
Shi, Y., Zhuang, Y., Zhang, J., Chen, M., and Wu, S. (2020c). METTL14 inhibits hepatocellular carcinoma metastasis through regulating EGFR/PI3K/AKT signaling pathway in an m6A-dependent manner. Cancer Manag. Res. 12, 13173–13184. doi:10.2147/cmar.S286275
Shimba, S., Bokar, J. A., Rottman, F., and Reddy, R. (1995). Accurate and efficient N-6-adenosine methylation in spliceosomal U6 small nuclear RNA by HeLa cell extract in vitro. Nucleic Acids Res. 23 (13), 2421–2426. doi:10.1093/nar/23.13.2421
Sibert, B. S., and Patton, J. R. (2012). Pseudouridine synthase 1: A site-specific synthase without strict sequence recognition requirements. Nucleic Acids Res. 40 (5), 2107–2118. doi:10.1093/nar/gkr1017
Sloan, K. E., Warda, A. S., Sharma, S., Entian, K. D., Lafontaine, D. L. J., and Bohnsack, M. T. (2017). Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14 (9), 1138–1152. doi:10.1080/15476286.2016.1259781
Song, B., Tang, Y., Chen, K., Wei, Z., Rong, R., Lu, Z., et al. (2020). m7GHub: deciphering the location, regulation and pathogenesis of internal mRNA N7-methylguanosine (m7G) sites in human. Bioinformatics 36 (11), 3528–3536. doi:10.1093/bioinformatics/btaa178
Sperling, A. S., Gibson, C. J., and Ebert, B. L. (2017). The genetics of myelodysplastic syndrome: From clonal haematopoiesis to secondary leukaemia. Nat. Rev. Cancer 17 (1), 5–19. doi:10.1038/nrc.2016.112
Squires, J. E., Patel, H. R., Nousch, M., Sibbritt, T., Humphreys, D. T., Parker, B. J., et al. (2012). Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40 (11), 5023–5033. doi:10.1093/nar/gks144
Stefanska, B., Cheishvili, D., Suderman, M., Arakelian, A., Huang, J., Hallett, M., et al. (2014). Genome-wide study of hypomethylated and induced genes in patients with liver cancer unravels novel anticancer targets. Clin. Cancer Res. 20 (12), 3118–3132. doi:10.1158/1078-0432.Ccr-13-0283
Su, R., Dong, L., Li, C., Nachtergaele, S., Wunderlich, M., Qing, Y., et al. (2018). R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell 172 (1-2), 90–105.e23. e123. doi:10.1016/j.cell.2017.11.031
Su, R., Dong, L., Li, Y., Gao, M., He, P. C., Liu, W., et al. (2022). METTL16 exerts an m(6)A-independent function to facilitate translation and tumorigenesis. Nat. Cell Biol. 24 (2), 205–216. doi:10.1038/s41556-021-00835-2
Su, T., Huang, M., Liao, J., Lin, S., Yu, P., Yang, J., et al. (2021). Insufficient radiofrequency ablation promotes hepatocellular carcinoma metastasis through N6-methyladenosine mRNA methylation-dependent mechanism. Hepatology 74 (3), 1339–1356. doi:10.1002/hep.31766
Sun, Z., Xue, S., Zhang, M., Xu, H., Hu, X., Chen, S., et al. (2020). Aberrant NSUN2-mediated m(5)C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene 39 (45), 6906–6919. doi:10.1038/s41388-020-01475-w
Sundheim, O., Vågbø, C. B., Bjørås, M., Sousa, M. M., Talstad, V., Aas, P. A., et al. (2006). Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. Embo J. 25 (14), 3389–3397. doi:10.1038/sj.emboj.7601219
Suzuki, T., Yashiro, Y., Kikuchi, I., Ishigami, Y., Saito, H., Matsuzawa, I., et al. (2020). Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 11 (1), 4269. doi:10.1038/s41467-020-18068-6
Taketo, K., Konno, M., Asai, A., Koseki, J., Toratani, M., Satoh, T., et al. (2018). The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 52 (2), 621–629. doi:10.3892/ijo.2017.4219
Tamura, S., Fujioka, H., Nakano, T., Amuro, Y., Hada, T., Nakao, N., et al. (1988). Urinary pseudouridine as a biochemical marker in the diagnosis and monitoring of primary hepatocellular carcinoma. Am. J. Gastroenterol. 83 (8), 841–845.
Tao, L., Li, D., Mu, S., Tian, G., and Yan, G. (2022). LncRNA MAPKAPK5_AS1 facilitates cell proliferation in hepatitis B virus -related hepatocellular carcinoma. Lab. Invest. 102, 494–504. doi:10.1038/s41374-022-00731-9
Teng, P. C., Liang, Y., Yarmishyn, A. A., Hsiao, Y. J., Lin, T. Y., Lin, T. W., et al. (2021). RNA modifications and epigenetics in modulation of lung cancer and pulmonary diseases. Int. J. Mol. Sci. 22 (19), 10592. doi:10.3390/ijms221910592
Thomas, N. K., Poodari, V. C., Jain, M., Olsen, H. E., Akeson, M., and Abu-Shumays, R. L. (2021). Direct nanopore sequencing of individual full length tRNA strands. ACS Nano 15 (10), 16642–16653. doi:10.1021/acsnano.1c06488
Tian, Q. H., Zhang, M. F., Zeng, J. S., Luo, R. G., Wen, Y., Chen, J., et al. (2019). METTL1 overexpression is correlated with poor prognosis and promotes hepatocellular carcinoma via PTEN. J. Mol. Med. 97 (11), 1535–1545. doi:10.1007/s00109-019-01830-9
Tong, C., Wang, W., and He, C. (2022). m1A methylation modification patterns and metabolic characteristics in hepatocellular carcinoma. BMC Gastroenterol. 22 (1), 93. doi:10.1186/s12876-022-02160-w
Towns, W. L., and Begley, T. J. (2012). Transfer RNA methytransferases and their corresponding modifications in budding yeast and humans: Activities, predications, and potential roles in human health. DNA Cell Biol. 31 (4), 434–454. doi:10.1089/dna.2011.1437
Trixl, L., and Lusser, A. (2019). The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip. Rev. RNA 10 (1), e1510. doi:10.1002/wrna.1510
Tuorto, F., Herbst, F., Alerasool, N., Bender, S., Popp, O., Federico, G., et al. (2015). The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. Embo J. 34 (18), 2350–2362. doi:10.15252/embj.201591382
Tuorto, F., Liebers, R., Musch, T., Schaefer, M., Hofmann, S., Kellner, S., et al. (2012). RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 19 (9), 900–905. doi:10.1038/nsmb.2357
Uddin, M. B., Wang, Z., and Yang, C. (2020). Dysregulations of functional RNA modifications in cancer, cancer stemness and cancer therapeutics. Theranostics 10 (7), 3164–3189. doi:10.7150/thno.41687
Ueda, Y., Ooshio, I., Fusamae, Y., Kitae, K., Kawaguchi, M., Jingushi, K., et al. (2017). AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci. Rep. 7, 42271. doi:10.1038/srep42271
Van Haute, L., Lee, S. Y., McCann, B. J., Powell, C. A., Bansal, D., Vasiliauskaitė, L., et al. (2019). NSUN2 introduces 5-methylcytosines in mammalian mitochondrial tRNAs. Nucleic Acids Res. 47 (16), 8720–8733. doi:10.1093/nar/gkz559
van Tran, N., Ernst, F. G. M., Hawley, B. R., Zorbas, C., Ulryck, N., Hackert, P., et al. (2019). The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47 (15), 7719–7733. doi:10.1093/nar/gkz619
Vogel, A., Qin, S., Kudo, M., Su, Y., Hudgens, S., Yamashita, T., et al. (2021). Lenvatinib versus sorafenib for first-line treatment of unresectable hepatocellular carcinoma: Patient-reported outcomes from a randomised, open-label, non-inferiority, phase 3 trial. Lancet. Gastroenterol. Hepatol. 6 (8), 649–658. doi:10.1016/s2468-1253(21)00110-2
Vu, L. P., Pickering, B. F., Cheng, Y., Zaccara, S., Nguyen, D., Minuesa, G., et al. (2017). The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23 (11), 1369–1376. doi:10.1038/nm.4416
Wang, X., and He, C. (2014). Dynamic RNA modifications in posttranscriptional regulation. Mol. Cell 56 (1), 5–12. doi:10.1016/j.molcel.2014.09.001
Wan, Y., Qu, K., Zhang, Q. C., Flynn, R. A., Manor, O., Ouyang, Z., et al. (2014). Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505 (7485), 706–709. doi:10.1038/nature12946
Wang, M., Yang, Y., Yang, J., Yang, J., and Han, S. (2020a). circ_KIAA1429 accelerates hepatocellular carcinoma advancement through the mechanism of m(6)A-YTHDF3-Zeb1. Life Sci. 257, 118082. doi:10.1016/j.lfs.2020.118082
Wang, P., Doxtader, K. A., and Nam, Y. (2016a). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63 (2), 306–317. doi:10.1016/j.molcel.2016.05.041
Wang, P., Wang, X., Zheng, L., and Zhuang, C. (2020b). Gene signatures and prognostic values of m6A regulators in hepatocellular carcinoma. Front. Genet. 11, 540186. doi:10.3389/fgene.2020.540186
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016b). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534 (7608), 575–578. doi:10.1038/nature18298
Wang, X., Ma, R., Zhang, X., Cui, L., Ding, Y., Shi, W., et al. (2021a). Crosstalk between N6-methyladenosine modification and circular RNAs: Current understanding and future directions. Mol. Cancer 20 (1), 121. doi:10.1186/s12943-021-01415-6
Wang, Y., Huang, Q., Deng, T., Li, B. H., and Ren, X. Q. (2019). Clinical significance of TRMT6 in hepatocellular carcinoma: A bioinformatics-based study. Med. Sci. Monit. 25, 3894–3901. doi:10.12659/msm.913556
Wang, Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., and Zhao, J. C. (2014). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16 (2), 191–198. doi:10.1038/ncb2902
Wang, Y., Li, Y., Yue, M., Wang, J., Kumar, S., Wechsler-Reya, R. J., et al. (2018). N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 21 (2), 195–206. doi:10.1038/s41593-017-0057-1
Wang, Y., Wang, J., Li, X., Xiong, X., Wang, J., Zhou, Z., et al. (2021b). N(1)-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat. Commun. 12 (1), 6314. doi:10.1038/s41467-021-26718-6
Wang, Y., Zhang, X., Liu, H., and Zhou, X. (2021c). Chemical methods and advanced sequencing technologies for deciphering mRNA modifications. Chem. Soc. Rev. 50 (24), 13481–13497. doi:10.1039/d1cs00920f
Wang, Y., Zhang, Z., Sepich-Poore, C., Zhang, L., Xiao, Y., and He, C. (2021d). LEAD-m(6) A-seq for locus-specific detection of N(6) -methyladenosine and quantification of differential methylation. Angew. Chem. Int. Ed. Engl. 60 (2), 873–880. doi:10.1002/anie.202007266
Wang, Y., Zhao, Y., Bollas, A., Wang, Y., and Au, K. F. (2021e). Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 39 (11), 1348–1365. doi:10.1038/s41587-021-01108-x
Wei, J., Liu, F., Lu, Z., Fei, Q., Ai, Y., He, P. C., et al. (2018). Differential m(6)A, m(6)A(m), and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71 (6), 973–985.e5. e975. doi:10.1016/j.molcel.2018.08.011
Wei, Q. (2021). Bioinformatical identification of key genes regulated by IGF2BP2-mediated RNA N6-methyladenosine and prediction of prognosis in hepatocellular carcinoma. J. Gastrointest. Oncol. 12 (4), 1773–1785. doi:10.21037/jgo-21-306
Wiener, D., and Schwartz, S. (2021). The epitranscriptome beyond m(6)A. Nat. Rev. Genet. 22 (2), 119–131. doi:10.1038/s41576-020-00295-8
Willbanks, A., Wood, S., and Cheng, J. X. (2021). RNA epigenetics: Fine-tuning chromatin plasticity and transcriptional regulation, and the implications in human diseases. Genes (Basel) 12 (5), 627. doi:10.3390/genes12050627
Workman, R. E., Tang, A. D., Tang, P. S., Jain, M., Tyson, J. R., Razaghi, R., et al. (2019). Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods 16 (12), 1297–1305. doi:10.1038/s41592-019-0617-2
Wu, A., Hu, Y., Xu, Y., Xu, J., Wang, X., Cai, A., et al. (2021a). Methyltransferase-like 3-mediated m6A methylation of Hsa_circ_0058493 accelerates hepatocellular carcinoma progression by binding to YTH domain-containing protein 1. Front. Cell Dev. Biol. 9, 762588. doi:10.3389/fcell.2021.762588
Wu, B., Li, L., Huang, Y., Ma, J., and Min, J. (2017). Readers, writers and erasers of N(6)-methylated adenosine modification. Curr. Opin. Struct. Biol. 47, 67–76. doi:10.1016/j.sbi.2017.05.011
Wu, J., Pang, R., Li, M., Chen, B., Huang, J., and Zhu, Y. (2021b). m6A-Induced LncRNA MEG3 suppresses the proliferation, migration and invasion of hepatocellular carcinoma cell through miR-544b/BTG2 signaling. Onco. Targets. Ther. 14, 3745–3755. doi:10.2147/ott.S289198
Wu, S., Liu, S., Cao, Y., Chao, G., Wang, P., and Pan, H. (2022). Downregulation of ZC3H13 by miR-362-3p/miR-425-5p is associated with a poor prognosis and adverse outcomes in hepatocellular carcinoma. Aging (Albany NY) 14 (5), 2304–2319. doi:10.18632/aging.203939
Wu, W., Yang, Y., and Lei, H. (2019a). Progress in the application of CRISPR: From gene to base editing. Med. Res. Rev. 39 (2), 665–683. doi:10.1002/med.21537
Wu, X., Zhang, X., Tao, L., Dai, X., and Chen, P. (2020). Prognostic value of an m6A RNA methylation regulator-based signature in patients with hepatocellular carcinoma. Biomed. Res. Int. 2020, 2053902. doi:10.1155/2020/2053902
Wu, Y., Yang, X., Chen, Z., Tian, L., Jiang, G., Chen, F., et al. (2019b). m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol. Cancer 18 (1), 87. doi:10.1186/s12943-019-1014-2
Xia, P., Zhang, H., Xu, K., Jiang, X., Gao, M., Wang, G., et al. (2021). MYC-targeted WDR4 promotes proliferation, metastasis, and sorafenib resistance by inducing CCNB1 translation in hepatocellular carcinoma. Cell Death Dis. 12 (7), 691. doi:10.1038/s41419-021-03973-5
Xiang, Y., Laurent, B., Hsu, C. H., Nachtergaele, S., Lu, Z., Sheng, W., et al. (2017). RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 543 (7646), 573–576. doi:10.1038/nature21671
Xiao, S., Cao, S., Huang, Q., Xia, L., Deng, M., Yang, M., et al. (2019). The RNA N(6)-methyladenosine modification landscape of human fetal tissues. Nat. Cell Biol. 21 (5), 651–661. doi:10.1038/s41556-019-0315-4
Xiao, Y., Wang, Y., Tang, Q., Wei, L., Zhang, X., and Jia, G. (2018). An elongation- and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-specific N(6) -methyladenosine modification. Angew. Chem. Int. Ed. Engl. 57 (49), 15995–16000. doi:10.1002/anie.201807942
Xu, C., Wang, X., Liu, K., Roundtree, I. A., Tempel, W., Li, Y., et al. (2014). Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10 (11), 927–929. doi:10.1038/nchembio.1654
Xu, H., Wang, H., Zhao, W., Fu, S., Li, Y., Ni, W., et al. (2020). SUMO1 modification of methyltransferase-like 3 promotes tumor progression via regulating Snail mRNA homeostasis in hepatocellular carcinoma. Theranostics 10 (13), 5671–5686. doi:10.7150/thno.42539
Xu, L., Liu, X., Sheng, N., Oo, K. S., Liang, J., Chionh, Y. H., et al. (2017). Three distinct 3-methylcytidine (m(3)C) methyltransferases modify tRNA and mRNA in mice and humans. J. Biol. Chem. 292 (35), 14695–14703. doi:10.1074/jbc.M117.798298
Xue, C., Zhao, Y., Li, G., and Li, L. (2021). Multi-omic analyses of the m(5)C regulator ALYREF reveal its essential roles in hepatocellular carcinoma. Front. Oncol. 11, 633415. doi:10.3389/fonc.2021.633415
Yang, D., Qiao, J., Wang, G., Lan, Y., Li, G., Guo, X., et al. (2018). N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 46 (8), 3906–3920. doi:10.1093/nar/gky130
Yang, N., Wang, T., Li, Q., Han, F., Wang, Z., Zhu, R., et al. (2021). HBXIP drives metabolic reprogramming in hepatocellular carcinoma cells via METTL3-mediated m6A modification of HIF-1α. J. Cell. Physiol. 236 (5), 3863–3880. doi:10.1002/jcp.30128
Yang, S., Wei, J., Cui, Y. H., Park, G., Shah, P., Deng, Y., et al. (2019). m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 10 (1), 2782. doi:10.1038/s41467-019-10669-0
Yang, W., Meng, J., Liu, J., Ding, B., Tan, T., Wei, Q., et al. (2020). The N(1)-methyladenosine methylome of petunia mRNA. Plant Physiol. 183 (4), 1710–1724. doi:10.1104/pp.20.00382
Yang, X., Yang, Y., Sun, B. F., Chen, Y. S., Xu, J. W., Lai, W. Y., et al. (2017a). 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 27 (5), 606–625. doi:10.1038/cr.2017.55
Yang, Y., Fan, X., Mao, M., Song, X., Wu, P., Zhang, Y., et al. (2017b). Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 27 (5), 626–641. doi:10.1038/cr.2017.31
Yang, Z., Li, J., Feng, G., Gao, S., Wang, Y., Zhang, S., et al. (2017c). MicroRNA-145 modulates N(6)-methyladenosine levels by targeting the 3'-untranslated mRNA region of the N(6)-methyladenosine binding YTH domain family 2 protein. J. Biol. Chem. 292 (9), 3614–3623. doi:10.1074/jbc.M116.749689
Yarian, C. S., Basti, M. M., Cain, R. J., Ansari, G., Guenther, R. H., Sochacka, E., et al. (1999). Structural and functional roles of the N1- and N3-protons of psi at tRNA's position 39. Nucleic Acids Res. 27 (17), 3543–3549. doi:10.1093/nar/27.17.3543
Yeermaike, A., Gu, P., Liu, D., and Nadire, T. (2022). LncRNA NEAT1 sponges miR-214 to promoted tumor growth in hepatocellular carcinoma. Mamm. Genome 33, 525–533. doi:10.1007/s00335-022-09952-1
Yoon, K. J., Ringeling, F. R., Vissers, C., Jacob, F., Pokrass, M., Jimenez-Cyrus, D., et al. (2017). Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell 171 (4), 877–889.e17. e817. doi:10.1016/j.cell.2017.09.003
Yu, J., Zhang, W., Ding, D., Hu, Y., Guo, G., Wang, J., et al. (2022). Bioinformatics analysis combined with experiments predicts PUDP as a potential prognostic biomarker for hepatocellular carcinoma through its interaction with tumor microenvironment. Front. Oncol. 12, 830174. doi:10.3389/fonc.2022.830174
Yu, Y. T., Shu, M. D., and Steitz, J. A. (1998). Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. Embo J. 17 (19), 5783–5795. doi:10.1093/emboj/17.19.5783
Yuan, F., Bi, Y., Siejka-Zielinska, P., Zhou, Y. L., Zhang, X. X., and Song, C. X. (2019). Bisulfite-free and base-resolution analysis of 5-methylcytidine and 5-hydroxymethylcytidine in RNA with peroxotungstate. Chem. Commun. 55 (16), 2328–2331. doi:10.1039/c9cc00274j
Yue, Y., Liu, J., and He, C. (2015). RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29 (13), 1343–1355. doi:10.1101/gad.262766.115
Zaccara, S., and Jaffrey, S. R. (2020). A unified model for the function of YTHDF proteins in regulating m(6)a-modified mRNA. Cell 181 (7), 1582–1595.e18. e1518. doi:10.1016/j.cell.2020.05.012
Zebarjadian, Y., King, T., Fournier, M. J., Clarke, L., and Carbon, J. (1999). Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol. 19 (11), 7461–7472. doi:10.1128/mcb.19.11.7461
Zhai, C. T., Tian, Y. C., Tang, Z. X., and Shao, L. J. (2021). RNA methyltransferase NSUN2 promotes growth of hepatocellular carcinoma cells by regulating fizzy-related-1 in vitro and in vivo. Kaohsiung J. Med. Sci. 37 (11), 991–999. doi:10.1002/kjm2.12430
Zhang, C., Huang, S., Zhuang, H., Ruan, S., Zhou, Z., Huang, K., et al. (2020a). YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene 39 (23), 4507–4518. doi:10.1038/s41388-020-1303-7
Zhang, L., Hou, C., Chen, C., Guo, Y., Yuan, W., Yin, D., et al. (2020b). The role of N(6)-methyladenosine (m(6)A) modification in the regulation of circRNAs. Mol. Cancer 19 (1), 105. doi:10.1186/s12943-020-01224-3
Zhang, L. H., Zhang, X. Y., Hu, T., Chen, X. Y., Li, J. J., Raida, M., et al. (2020d). The SUMOylated METTL8 induces R-loop and tumorigenesis via m3C. iScience 23 (3), 100968. doi:10.1016/j.isci.2020.100968
Zhang, L., Qiao, Y., Huang, J., Wan, D., Zhou, L., Lin, S., et al. (2020c). Expression pattern and prognostic value of key regulators for m6A RNA modification in hepatocellular carcinoma. Front. Med. 7, 556. doi:10.3389/fmed.2020.00556
Zhang, L. S., Liu, C., Ma, H., Dai, Q., Sun, H. L., Luo, G., et al. (2019a). Transcriptome-wide mapping of internal N(7)-methylguanosine methylome in mammalian mRNA. Mol. Cell 74 (6), 1304–1316.e8. e1308. doi:10.1016/j.molcel.2019.03.036
Zhang, M., Song, J., Yuan, W., Zhang, W., and Sun, Z. (2021a). Roles of RNA methylation on tumor immunity and clinical implications. Front. Immunol. 12, 641507. doi:10.3389/fimmu.2021.641507
Zhang, M., Zhao, W., Liu, S., Liu, H., Liu, L., Peng, Q., et al. (2021b). H/ACA snoRNP gene family as diagnostic and prognostic biomarkers for hepatocellular carcinoma. Pharmgenomics. Pers. Med. 14, 1331–1345. doi:10.2147/pgpm.S333838
Zhang, Q., Zheng, Q., Yu, X., He, Y., and Guo, W. (2020e). Overview of distinct 5-methylcytosine profiles of messenger RNA in human hepatocellular carcinoma and paired adjacent non-tumor tissues. J. Transl. Med. 18 (1), 245. doi:10.1186/s12967-020-02417-6
Zhang, Z., Chen, L. Q., Zhao, Y. L., Yang, C. G., Roundtree, I. A., Zhang, Z., et al. (2019b). Single-base mapping of m(6)A by an antibody-independent method. Sci. Adv. 5 (7), eaax0250. doi:10.1126/sciadv.aax0250
Zhao, B. S., and He, C. (2015). Pseudouridine in a new era of RNA modifications. Cell Res. 25 (2), 153–154. doi:10.1038/cr.2014.143
Zhao, B. S., Roundtree, I. A., and He, C. (2017). Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18 (1), 31–42. doi:10.1038/nrm.2016.132
Zhao, X., Chen, Y., Mao, Q., Jiang, X., Jiang, W., Chen, J., et al. (2018a). Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 21 (4), 859–868. doi:10.3233/cbm-170791
Zhao, X., Patton, J. R., Davis, S. L., Florence, B., Ames, S. J., and Spanjaard, R. A. (2004). Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol. Cell 15 (4), 549–558. doi:10.1016/j.molcel.2004.06.044
Zhao, X., Yang, Y., Sun, B. F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24 (12), 1403–1419. doi:10.1038/cr.2014.151
Zhao, Y., Dunker, W., Yu, Y. T., and Karijolich, J. (2018b). The role of noncoding RNA pseudouridylation in nuclear gene expression events. Front. Bioeng. Biotechnol. 6, 8. doi:10.3389/fbioe.2018.00008
Zhao, Y., Kong, L., Pei, Z., Li, F., Li, C., Sun, X., et al. (2021). m7G methyltransferase METTL1 promotes post-ischemic angiogenesis via promoting VEGFA mRNA translation. Front. Cell Dev. Biol. 9, 642080. doi:10.3389/fcell.2021.642080
Zhao, Y., You, S., Yu, Y. Q., Zhang, S., Li, P. T., Ye, Y. H., et al. (2019a). Decreased nuclear expression of FTO in human primary hepatocellular carcinoma is associated with poor prognosis. Int. J. Clin. Exp. Pathol. 12 (9), 3376–3383.
Zhao, Y., Zhao, Q., Kaboli, P. J., Shen, J., Li, M., Wu, X., et al. (2019b). m1A regulated genes modulate PI3K/AKT/mTOR and ErbB pathways in gastrointestinal cancer. Transl. Oncol. 12 (10), 1323–1333. doi:10.1016/j.tranon.2019.06.007
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49 (1), 18–29. doi:10.1016/j.molcel.2012.10.015
Zheng, H. X., Zhang, X. S., and Sui, N. (2020). Advances in the profiling of N(6)-methyladenosine (m(6)A) modifications. Biotechnol. Adv. 45, 107656. doi:10.1016/j.biotechadv.2020.107656
Zhong, L., Liao, D., Zhang, M., Zeng, C., Li, X., Zhang, R., et al. (2019). YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 442, 252–261. doi:10.1016/j.canlet.2018.11.006
Zhong, X., Yu, J., Frazier, K., Weng, X., Li, Y., Cham, C. M., et al. (2018). Circadian clock regulation of hepatic lipid metabolism by modulation of m(6)A mRNA methylation. Cell Rep. 25 (7), 18161816–18161828.e4. doi:10.1016/j.celrep.2018.10.068
Zhou, C., Molinie, B., Daneshvar, K., Pondick, J. V., Wang, J., Van Wittenberghe, N., et al. (2017). Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 20 (9), 2262–2276. doi:10.1016/j.celrep.2017.08.027
Zhou, H., Rauch, S., Dai, Q., Cui, X., Zhang, Z., Nachtergaele, S., et al. (2019a). Evolution of a reverse transcriptase to map N(1)-methyladenosine in human messenger RNA. Nat. Methods 16 (12), 1281–1288. doi:10.1038/s41592-019-0550-4
Zhou, J., Wan, J., Gao, X., Zhang, X., Jaffrey, S. R., and Qian, S. B. (2015). Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526 (7574), 591–594. doi:10.1038/nature15377
Zhou, T., Li, S., Xiang, D., Liu, J., Sun, W., Cui, X., et al. (2020). m6A RNA methylation-mediated HNF3γ reduction renders hepatocellular carcinoma dedifferentiation and sorafenib resistance. Signal Transduct. Target. Ther. 5 (1), 296. doi:10.1038/s41392-020-00299-0
Zhou, Y., Yin, Z., Hou, B., Yu, M., Chen, R., Jin, H., et al. (2019b). Expression profiles and prognostic significance of RNA N6-methyladenosine-related genes in patients with hepatocellular carcinoma: Evidence from independent datasets. Cancer Manag. Res. 11, 3921–3931. doi:10.2147/cmar.S191565
Zhu, G. Q., Yu, L., Zhou, Y. J., Du, J. X., Dong, S. S., Wu, Y. M., et al. (2020). Genetic alterations and transcriptional expression of m(6)A RNA methylation regulators drive a malignant phenotype and have clinical prognostic impact in hepatocellular carcinoma. Front. Oncol. 10, 900. doi:10.3389/fonc.2020.00900
Keywords: hepatocellular carcinoma, RNA modification, m6A, m7G, m5C, m1A, m3C, ψ
Citation: Feng Q, Wang D, Xue T, Lin C, Gao Y, Sun L, Jin Y and Liu D (2022) The role of RNA modification in hepatocellular carcinoma. Front. Pharmacol. 13:984453. doi: 10.3389/fphar.2022.984453
Received: 02 July 2022; Accepted: 11 August 2022;
Published: 02 September 2022.
Edited by:
Yongxia Zhu, Sichuan Cancer Hospital, ChinaReviewed by:
Andrew Shafik, Emory University, United StatesCopyright © 2022 Feng, Wang, Xue, Lin, Gao, Sun, Jin and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dianfeng Liu, Y2NsZGZAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.