- Department of Nephrology, The Affiliated Hospital of Qingdao University, Qingdao, China
Renal ischemia reperfusion injury (IRI) is a leading and common cause of acute kidney injury (AKI), and inflammation is a critical factor in ischemic AKI progression. Calycosin (CAL), a major active component of Radix astragali, has been reported to have anti-inflammatory effect in multiple organs. However, whether CAL can alleviate renal IRI and its mechanism remain uncertain. In the present study, a renal IRI model is established by bilateral renal pedicles occlusion for 35 min in male C57BL/6 mice, and the effect of CAL on renal IRI is measured by serum creatinine and pathohistological assay. Hypoxia/reoxygenation (H/R) stimulated human renal tubular epithelial cells HK-2 were applied to explore the regulatory mechanisms of CAL. Luciferase reporter assay and molecular docking were applied to identify the CAL’s target protein and pathway. In the mice with renal IRI, CAL dose dependently alleviated the renal injury and decreased nuclear factor kappa B (NF-κB) mediated inflammatory response. Bioinformatics analysis and experiments showed that early growth response 1 (EGR1) increased in mice with renal IRI and promoted NF-κB mediated inflammatory processes, and CAL dose-dependably reduced EGR1. Through JASPAR database and luciferase reporter assay, peroxisome proliferator-activated receptor γ (PPARγ) was predicted to be a transcription factor of EGR1 and repressed the expression of EGR1 in renal tubular epithelial cells. CAL could increase PPARγ in a dose dependent manner in mice with renal IRI and molecular docking predicted CAL could bind stably to PPARγ. In HK-2 cells after H/R, CAL increased PPARγ, decreased EGR1, and inhibited NF-κB mediated inflammatory response. However, PPARγ knockdown by siRNA transfection abrogated the anti-inflammation therapeutic effect of CAL. CAL produced a protective effect on renal IRI by attenuating NF-κB mediated inflammatory response via PPARγ/EGR1 pathway.
Introduction
Acute kidney injury (AKI) is defined as a rapid decrease in kidney function with increased serum creatinine levels and/or decreased urine output. AKI occurs in approximately 10%–15% of hospitalized patients, which is increasingly recognized as a risk factor for chronic kidney disease (CKD) (Ronco et al., 2019; Rashid et al., 2022). However, there is no effective prevention and treatment for AKI (Zhou et al., 2020; Li et al., 2021). Renal ischemia reperfusion injury (IRI) is a leading and common cause of AKI, and delayed recovery of IRI contributes to chronic kidney disease (CKD) and even end-stage kidney disease (Pefanis et al., 2019; Husain-Syed et al., 2021). Therefore, it is important to explore effective intervention for ischemic AKI.
Previous studies indicated that renal IRI is closely associated with the oxidative stress and inflammatory response (Andrade-Oliveira et al., 2019; Sato et al., 2020), which is especially relevant to renal tubular epithelial cells (TECs) injury (Tammaro et al., 2020) that contributing to the overall renal damage. Nuclear factor kappa B (NF-κB)-mediated pro-inflammatory response plays a critical role in AKI (Song et al., 2019). The underlying mechanism study showed that NF-κB activation triggers the release of inflammatory cytokines such as interleukin-1beta (IL-1β), interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF-α) that exacerbating kidney injury (Place and Kanneganti., 2018). Therefore, targeting NF-κB pathway is a promising anti-inflammation treatment which necessary to prevent or treat ischemic AKI.
Early growth response protein 1 (EGR1), a zinc-finger transcription factor of the immediate early gene family, has been reported to promote the release of pro-inflammatory mediators by activating NF-κB and exacerbate renal damage (Ho et al., 2016). However, the effect of EGR1 on inflammation in renal IRI has not been clarified. Peroxisome proliferator-activated receptor γ (PPARγ) is an important transcriptional factor, which belongs to superfamily of nuclear receptors. Previous studies have shown a relationship between PPARγ and EGR1, among which activation PPARγ can inhibit EGR1 to suppress inflammation responses (Li and Xia, 2020). However, the regulatory relationship between EGR1 and PPARγ and their role in inflammatory response in renal IRI remain unclear.
Natural products have been considered an alternative therapy for the treatment of various renal diseases including AKI and CKD (Geng et al., 2020; Meng et al., 2020; Fang et al., 2021; Shao et al., 2021; He et al., 2022). Calycosin, an isoflavonoid phytoestrogen isolated from Radix astragali, has various pharmacological activities including anti-cancer, anti-inflammatory, anti-oxidant and neuroprotective effects (Deng et al., 2021; Wang et al., 2022). Recent research has focus on the anti-inflammatory effect of CAL, such as suppressing neuroinflammation via blocking NF-κB pathway and NLRP3 inflammasome (Chen et al., 2020). However, whether CAL has a protective effect on renal IRI and its mechanism remains elusive. In this study, we investigate CAL effect on ischemic AKI and underlying mechanisms.
Materials and methods
Drug
CAL (CAS: 20575-57-9, purity ≥98.0%, MedChem Express, NJ, United States) was suspended in 10% dimethyl sulfoxide (DMSO) and 90% corn oil for mice administration. For cell experiment, CAL was dissolved in DMSO to make a 1 mg/ml stock solution.
Animals protocol
C57BL/6 male mice (6–8weeks) were purchased from Pengyue Laboratory Animal Co., Ltd. (Jinan, China). Mice were randomly allocated into seven groups (eight per group): control, sham, IRI, IRI + 5 mg/kg CAL, IRI +10 mg/kg CAL, IRI +20 mg/kg CAL, IRI + DMSO. CAL was intragastrically given daily for 7 days before IRI modeling (Figure 1A). To establish the renal IRI model, mice were anesthetized with intraperitoneal pentobarbital and subjected to bilateral renal artery occlusion for 35 min followed by reperfusion 24 h. The kidneys were observed until the color turned bright red which confirmed reperfusion. The sham operation was identical to the treatment operation, except for renal pedicle clamping. To explore the therapeutic effect of post-surgery CAL administration on IRI, some mice was subjected to bilateral renal artery occlusion for 35 min followed by reperfusion 24 h, and then treated with CAL for 7 days. Some mice were treated with CAL intragastrically without IRI operation. Blood samples were obtained from the orbital sinus and renal cortex tissue samples were stored at −80°C until use. Animal care and experimental procedures were approved by the Laboratory Animal Welfare Ethics Committee at Qingdao University (ethics number: 202105C5730202106036).
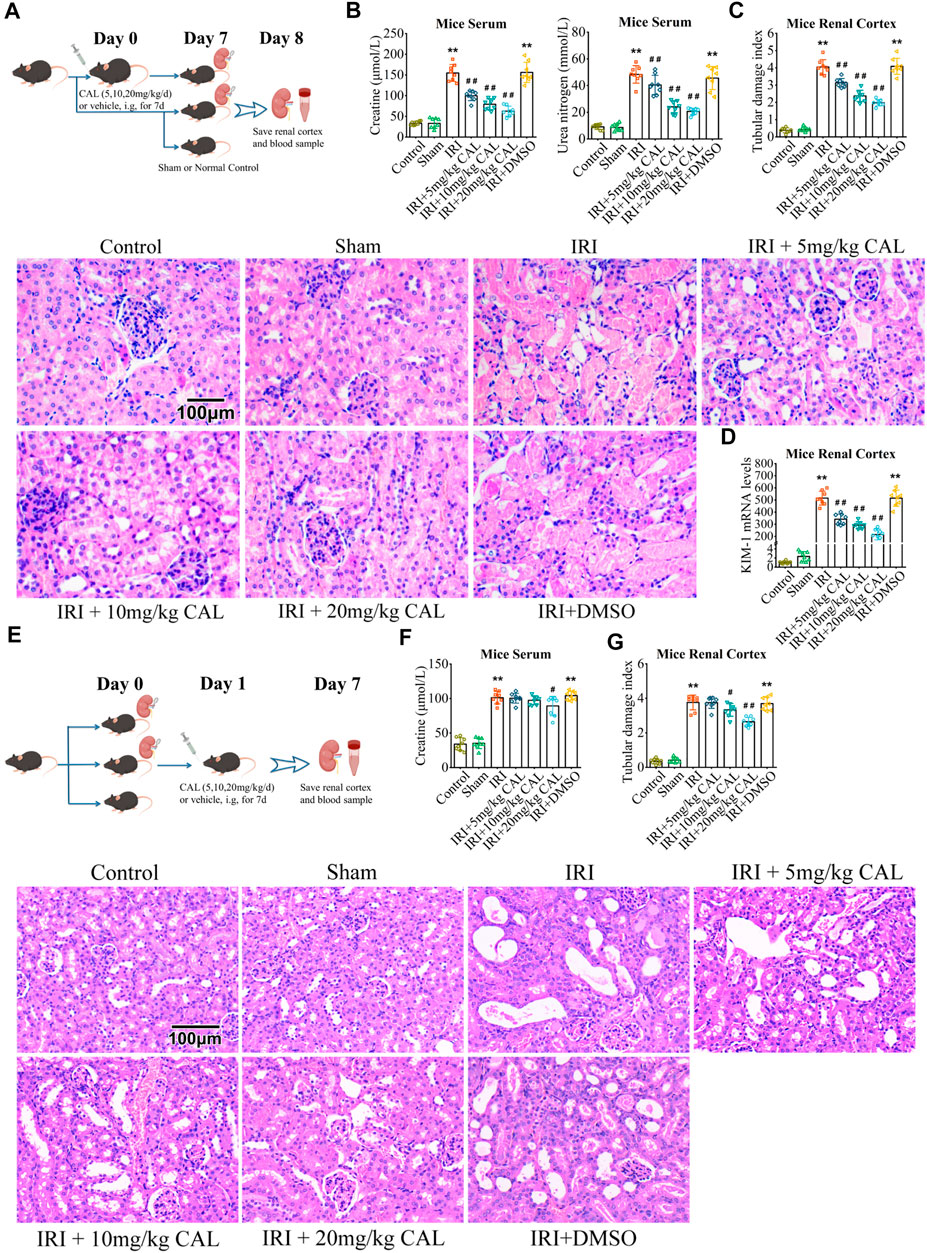
FIGURE 1. CAL protected the kidney from IRI. (A) The pattern diagram of the animal research design for CAL pre-treatment. (B) SCr and BUN levels in kidney samples of different groups. (C) Renal tubular damage index and H and E staining images of kidney tissue samples in different groups. Scale bar, 100 µm. (D) KIM-1 mRNA level in kidney samples of different groups. **p < 0.01 vs. sham group. ##p < 0.01 vs. IRI group. n = 8 per group. (E) The pattern diagram of the animal research design for CAL post-surgery administration. (F) SCr level in kidney samples of different groups. (G) Renal tubular damage index and H and E staining images of kidney tissue samples in different groups. Scale bar, 100 µm. **p < 0.01 vs. sham group. #p < 0.05, ##p < 0.01 vs. IRI group. n = 8 per group. The data are presented as the mean ± SD.
Renal function assessment
The renal function was assessed by measuring serum creatinine (SCr) and blood urea nitrogen (BUN) levels. The samples were evaluated using commercially available kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s recommended protocol.
Histopathological studies
Kidney tissue samples were collected after IRI and fixed in 10% formaldehyde overnight. The samples were embedded in paraffin, cut into 4-µm-thick sections. And then stained with hematoxylin and eosin (H & E) (Solarbio, Beijing, China) is performed. Renal tubular damage was scored according to the extent of foamy degeneration and the detachment of tubular cells on a semiquantitative scale.
Cell lines and cell treatment
Human renal tubular epithelial cells HK-2 were purchased from the Procell Life Science and Technology (Wuhan, China). HK-2 cells were cultured in DMEM/F-12 medium (Biological Industries, Kibbutz Beit Haemek, Israel) mixed with 10% fetal bovine serum (Gibco, NY, United States) and 100 × penicillin-streptomycin solution (Solarbio, Beijing, China) and incubated in a 37°C humidified incubator in an atmosphere of 5% CO2.
CAL solutions at different concentrations were prepared for incubation with HK-2 cells for 24 h, and equal volumes of DMSO were added to the vehicle groups. For Hypoxia/reoxygenation (H/R) treatment, cells were plated to 80% confluence, and exposed to 24 h of hypoxia (5% CO2, 1% O2, and 94% N2) followed by 3 h of reoxygenation. The experimental groups are as follows: control, H/R, H/R + 8 μM CAL, H/R + 16 μM CAL, H/R + 32 μMCAL, H/R + DMSO.
Cell viability detection
Cell Counting Kit-8 (CCK-8, MedChem Express, NJ, United States) was used for cell viability assessment. HK-2 cells were seeded in 96-well plates followed by incubation with graded concentrations of CAL for 24 h. Then 10 μl CCK-8 was added into each well and incubated for 2 h at 37°C. A microplate reader (Thermo Fisher Scientific Inc., MA, United States) was used for the measurement of absorbance at 450 nm.
Western blot
The mice kidney tissues and cells were lysed in RIPA lysis buffer (Elabscience Biotechnology, Wuhan, China). Protein samples were subjected to SDS-PAGE, followed by transfer to polyvinylidene fluoride membranes (PVDF, Millipore, MA, United States). After blocked, membranes were incubated at 4°C overnight with primary antibodies as follows: anti-phospho-IκBα (1:1000, Cell Signaling Technology, MA, United States), anti-IκBα (1:1000, Cell Signaling Technology, MA, United States),anti-phospho-NF-κB p65 (1:1000, Cell Signaling Technology, MA, United States),anti-NF-κB p65 (1:1000, Cell Signaling Technology, MA, United States), anti-β-actin (1:1000, Proteintech, IL, United States), anti-EGR1 (1:2000, Cell Signaling Technology, MA, United States), anti-PPARγ (1:1000, Cell Signaling Technology, MA, United States). The secondary antibody, goat anti-rabbit IgG-HRP (1:5000, Elabscience Biotechnology, Wuhan, China), was incubated at room temperature for 1 h and detected using Excellent Chemiluminescent Substrate (ECL) detection kit (Elabscience Biotechnology, Wuhan, China). The bands were subjected to gray scale analysis using ImageJ (version 1.8.0) software.
Enzyme linked immunosorbent assay
The levels of IL-1β, IL-6, and TNF-α of mice serum and cell culture supernatants were determined by an ELISA system (Elabscience Biotechnology, Wuhan, China) according to the manufacturer’s protocols.
Quantitative real-time PCR
Total RNA isolation was performed using Trizol (Invitrogen, MA, United States). cDNA was synthesized using a HiScript RT SuperMix (Vazyme Biotech, Nanjing, China). qRT-PCR was performed using ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech, Nanjing, China), and the data were normalized to the expression of β-actin. The primers used are shown in Supplementary Table S1.
Immunohistochemical
EGR1 and PPARγ immunohistochemical were performed as described previously (Singh et al., 2019). Primary antibody: anti-EGR1 antibody (1:500, Proteintech, IL, United States), anti-PPARγ antibody (1:200, Elabscience Biotechnology, Wuhan, China). EGR1 and PPARγ area ratio was analyzed using ImageJ (version 1.8.0) software.
Bioinformatics
Raw microarray data of GSE52004 (Affymetrix Mouse Gene 1.0 ST Array platform) were downloaded from GEO Datasets (https://www.ncbi.nlm.nih.gov/gds). The microarray data were contributed by Liu et al. (2014). The raw data were preprocessed using a robust multi-array averaging algorithm in R (version 4.0.4) software. The Limma package was loaded for differential expression analysis. The adjusted p value (adj. P) was calculated with the Benjamini and Hochberg method. DEGs were identified with absolute log2FC > 3 and adj. p < 0.05.
PPI information was acquired from the STRING database (http://www.string-db.org/). Medium confidence (0.4) was chosen as the minimum required interaction score to screen the interactions among DEGs. PPI network visualization was conducted by Cytoscape (version 3.9.1) software. Hub genes were selected by Maximal Clique Centrality methods with a criterion of score >100 (Chin et al., 2014).
Molecular docking was carried out by Auto Dock 4.2.6 (The Scripps Research Institute). Briefly, proteins were selected as rigid receptor molecules, while CAL was selected as the ligand for protein docking. PDB files of proteins were downloaded from the RCSB Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). The SDF file for CAL was downloaded from the NCBI PubChem Compound database (https://www.ncbi.nlm.nih.gov/pccompound). After the receptor molecule and ligand files were prepared, a grid box was set for the movement and rotation of the ligand. Finally, docking was accomplished via the AutoDock4 program. The visual simulation was conducted using the PyMOL 2.5 (DeLano Scientific LLC, CA, United States).
In vitro transfection experiment
HK-2 cells were plated into 6-well plates. When grown to 60% confluence, the cells were transfected with siRNA against target gene or control siRNA (GenePharm, Shanghai, China) using Lipofectamine 3,000 (Invitrogen, MA, United States). After 24 h, knockdown of target gene expression was validated by qRT-PCR analysis and cultures with more than 80% reduction in target gene mRNA expression comparing to the control cultures were included in further experiments.
Luciferase reporter assay
The promoter sequence of EGR1 was cloned into the pGL3 vector (Promega, WI, United States) upstream of the luciferase sequence (pGL3-EGR1). 293T cells were seeded into 24-well plates. When grown to 60% confluence, 293T cells were co-transfected with the 0.5 μg PPARγ over-expression plasmid (PPARγ-OE), 0.5 μg pGL3-EGR1 and 0.02 μg Renilla plasmid (pRL-TK) using X-tremegene HP reagent (Roche, Basel, Switzerland). And CAL (8 μM) were prepared for incubation with cells. After 48 h, the firefly and Renilla luciferase activities were measured with a Dual-Luciferase Reporter Assay System (Promega, WI, United States). The firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analyses
Values are expressed as the mean ± SD from at least three experiments. Statistical significance was analyzed by ANOVA followed by the Bonferroni post hoc test in SPSS 25.0 statistical software. A value of p < 0.05 was considered statistically significant.
Results
Calycosin improved kidney function and reduced renal inflammation responses
After IRI 24 h, mice exhibited elevated SCr and BUN levels, while CAL pretreatment significantly reduced both in a dose-dependent manner (Figure 1B). The IRI kidneys showed more renal tubular injury with high kidney pathological injury score, but 20 mg/kg CAL treatment reduced approximately half of the score (Figure 1C). Furthermore, CAL significantly decreased the level of kidney injury molecule-1 (KIM-1) in renal cortex compared with IRI group (Figure 1D). The effect of post-surgery CAL administration on IRI was explored (Figure 1E), and the results showed that CAL treatment after IRI preserved more kidney function and alleviated the renal pathological change (Figures 1F,G). To investigate the anti-inflammatory effects of CAL on IRI-induced AKI, the mRNA levels and concentrations of inflammatory cytokines, such as IL-1β, IL-6, and TNF-α, were detected. As shown in Figure 2A, the inflammatory cytokines significantly increased after IRI, whereas CAL pretreatment markedly inhibited them. Meanwhile, we found that IRI significantly increased NF-κB activation and IκBα degradation. However, treatment of CAL dose-dependently inhibited IRI-induced NF-κB activation (Figure 2B).
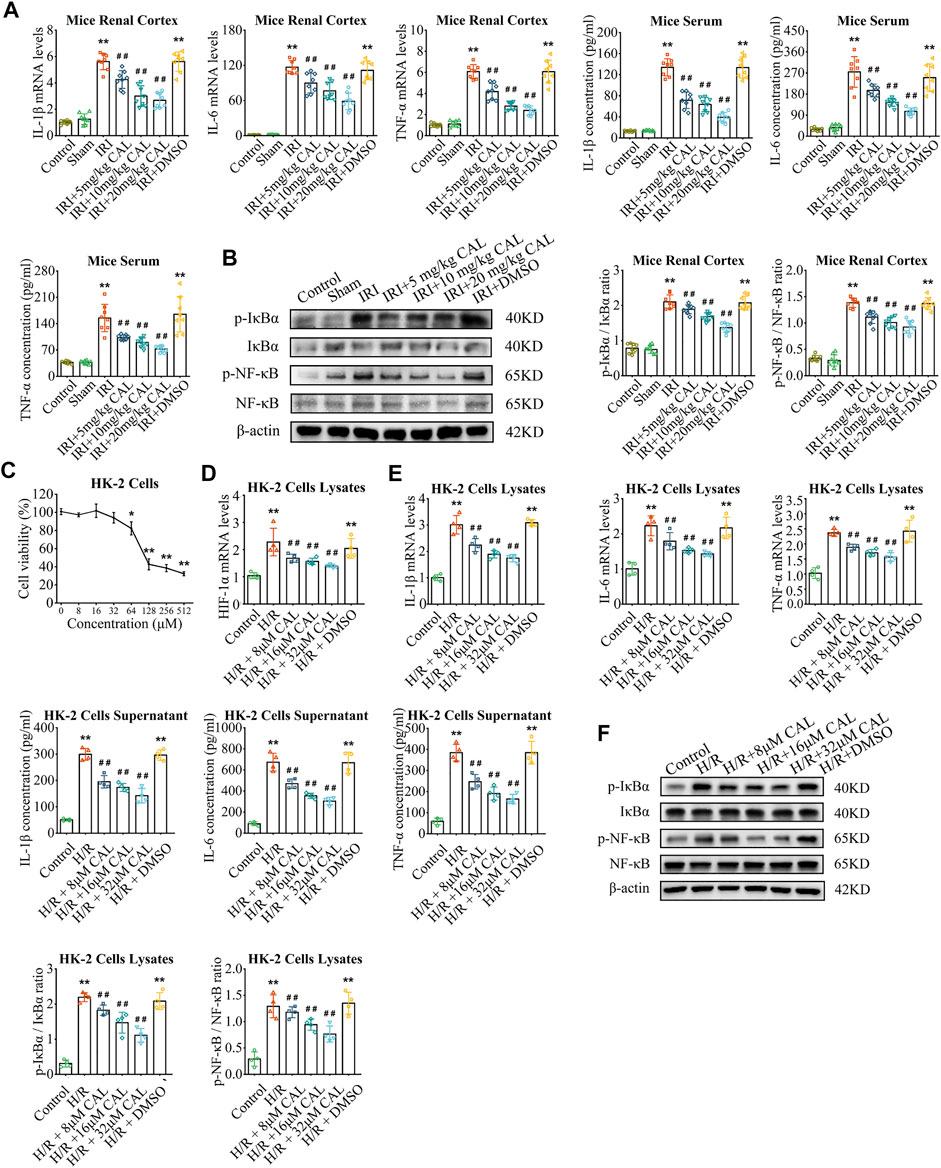
FIGURE 2. CAL ameliorated inflammation response in mice after IRI and H/R-induced HK-2 cells. (A) IL-1β, IL-6, and TNF-α mRNA levels in kidney samples and IL-1β, IL-6, and TNF-α concentrations in mice serum of different groups. (B) The protein levels of p-NF-κB/NF-κB and p-IκBα/IκBα in kidney samples of different groups. **p < 0.01 vs. sham group. ##p < 0.01 vs. IRI group. n = 8 per group. (C) Results of CCK-8 testing of different concentrations of CAL on HK-2 cells. *p < 0.05, **p < 0.01 vs. control cells. (D) The HIF-1α mRNA level in different cell groups. (E) IL-1β, IL-6, and TNF-α mRNA levels in cell lysates and IL-1β, IL-6, and TNF-α concentrations in supernatant of different groups. (F) The protein levels of p-NF-κB/NF-κB and p-IκBα/IκBα in cell lysates of different groups. **p < 0.01 vs. control group. ##p < 0.01 vs. H/R group. n = 4 per group. The data are presented as the mean ± SD.
Invitro, the toxicity of CAL was negligible at concentrations of 32 μM or less for 24 h (Figure 2C). H/R induced HK-2 cells model was applied to mimic renal IRI, and H/R treatment led to a significant increase in hypoxia-inducible factor 1α (HIF-1α) expression in HK-2 cells, whereas CAL incubation reduced the levels of HIF-1α in a dose-dependent manner (Figure 2D). Likewise, the inflammatory cytokines were induced by treatment with H/R in HK-2 cells and were decreased by incubation with CAL (Figure 2E). And CAL inhibited H/R induced NF-κB activation in HK-2 cells (Figure 2F). These results suggested that CAL alleviates renal function injury and inhibits excessive inflammation in TECs after H/R.
Early growth response 1 involved in the anti-inflammatory effect of calycosin
We identified 58 DEGs in the kidneys of IRI mice (Figures 3A,B; Supplementary Table S2), and ten hub genes were selected (Figure 3C). Among these hub genes, early growth response 1 (EGR1) had the highest score and upregulated markedly in IRI (adj. p < 0.01). In vivo, both protein and mRNA levels of EGR1 are significantly increased after IRI (Figure 3D). The EGR1 mainly expressed in TECs in the renal cortex, which increased significantly after IRI (Figure 3E), while CAL pretreatment showed a significant dose-dependent decrease of EGR1 expression in TECs. In vitro, EGR1 siRNA was transferred into HK-2 cells to knock down EGR1 (Figure 3F). Compared to the H/R group, the expression of inflammatory cytokines and NF-κB activation were suppressed after EGR1 knock down (Figures 3G,H). Taken together, EGR1 was upregulated after IRI, and involved in the inflammatory response of TECs.
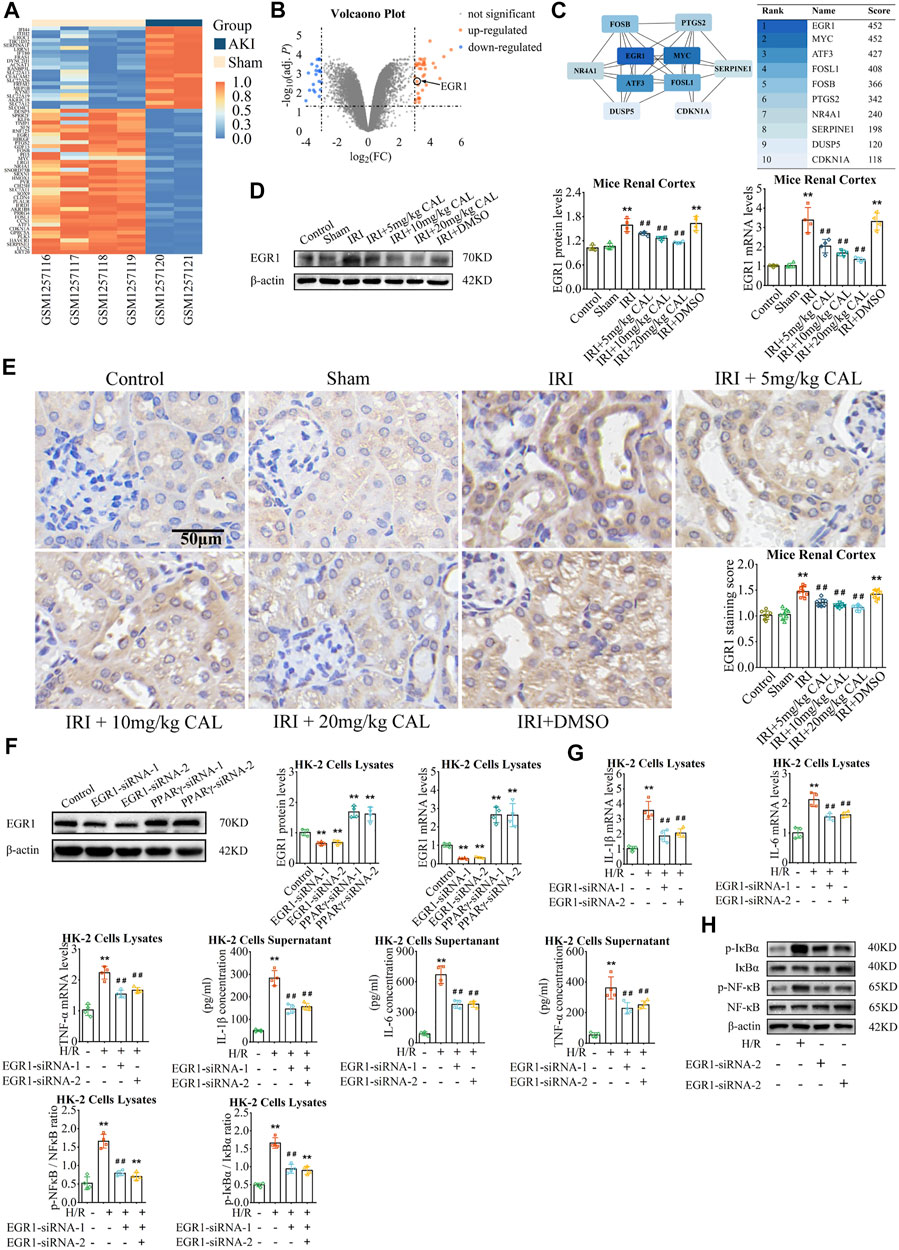
FIGURE 3. EGR1 involved in the anti-inflammatory effect of CAL. (A) Heatmap of GSE52004. Absolute log2FC > 3 with adj. p < 0.05 was considered the threshold of DEGs (orange: high expression; blue: low expression). (B) Volcanic plot of the DEGs (orange: up-regulated; blue: down-regulated). (C) PPI network of top 10 hub genes selected by the Maximal Clique Centrality method via Cytoscape. (D) The EGR1 protein and mRNA levels in kidney samples of different groups. (E) Immunohistochemistry showed the expression of EGR1 of kidney tissue samples in different groups. Scale bar, 50 µm. **p < 0.01 vs. sham group. ##p < 0.01 vs. IRI group. n = 8 per group. (F) The protein and mRNA expression of EGR1 in HK-2 cells transfected with EGR1 siRNA or PPARγ siRNA. **p < 0.01 vs. control group (G) IL-1β, IL-6, and TNF-α mRNA levels in cell lysates and IL-1β, IL-6, and TNF-α concentrations in cell supernatant of different groups. (H) The protein levels of p-NF-κB/NF-κB and p-IκBα/IκBα in cell lysates of different groups. **p < 0.01 compared with control group. ##p < 0.01 compared with the H/R group. n = 4 per group. The data are presented as the mean ± SD.
Peroxisome proliferator-activated receptor γ targeted and inhibited early growth response 1
In order to study the upstream of EGR1, we explored the transcriptional factor that might bind to the promoter region of EGR1. As the result, PPARγ is one of the factors that targeting the EGR1 with a relative score = 0.766 (Figure 4A). In vivo, PPARγ expressed in renal cortex and was significantly reduced after IRI compared with the sham group, but CAL promote PPARγ expression (Figures 4B,C). In vitro, PPARγ siRNA significantly increased the expression of EGR1 (Figures 3F, 4D), indicating that PPARγinhibitEGR1 in tubular cells. Moreover, luciferase reporter assay confirmed that PPARγ interact with EGR1 promoter, as EGR1 increased after EGR1 promoter plasmid transfection but decreased after PPARγ overexpression (Figure 4E). Meanwhile, the repressive effect of PPARγ on the EGR1 promoter was more obvious upon CAL incubation (Figure 4E). Taken together, these results indicated that PPARγ inhibited EGR1 expression and represses the activity of the EGR1 promoter.
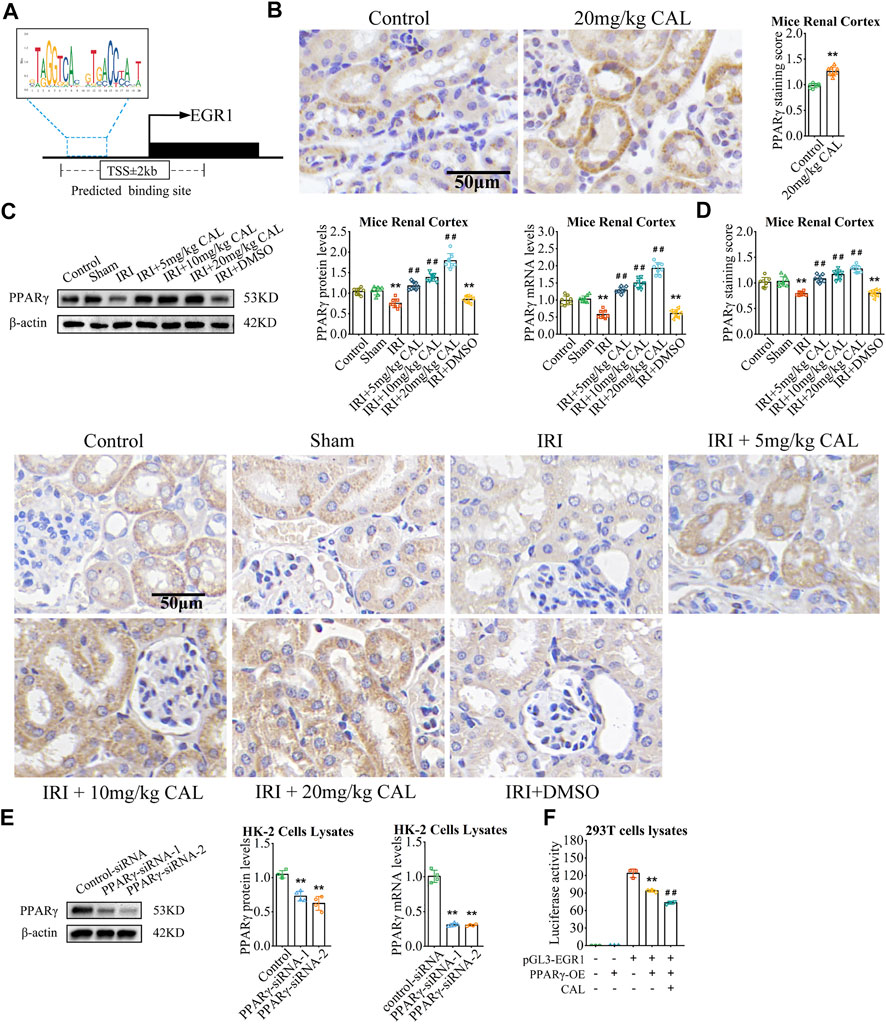
FIGURE 4. PPARγ targeted and inhibited EGR1. (A) The binding sites of PPARγ between EGR1 promoter region predicted by JASPAR website. (B) Immunohistochemistry showed the expression level PPARγ of kidney tissue samples in normal control group and 20 mg/kg CAL group. Scale bar, 50 µm. **p < 0.01 vs. control group. n = 3 per group. (C) The PPARγ mRNA and protein levels in kidney samples of different groups. (D) Immunohistochemistry showed the expression level PPARγ of kidney tissue samples in different groups. Scale bar, 50 µm. **p < 0.01 vs. sham group. ##p < 0.01 vs. IRI group. n = 8 per group. (E) The mRNA and protein levels of PPARγ in HK-2 cells transfected with PPARγ siRNA or control siRNA. **p < 0.01 vs. control siRNA group. n = 4 per group. (F) Luciferase reporter assay showed the effect of CAL and PPARγ on the activities of EGR1 promoters. **p < 0.01 vs. pGL3-EGR1 without PPARγ-OE group. ##p < 0.01 vs. pGL3-EGR1 with PPARγ-OE group. n = 3 per group. The data are presented as the mean ± SD.
In order to study the upstream of EGR1, we explored the transcriptional factor that might bind to the promoter region of EGR1. As the result, PPARγ is one of the factors that targeting the EGR1 with a relative score = 0.766 (Figure 4A). In vivo, CAL administration improved PPARγ expression in renal cortex (Figure 4B). Meanwhile, PPARγ was significantly reduced after IRI compared with the sham group, but CAL promoted PPARγ expression (Figures 4C,D). In vitro, PPARγ siRNA significantly increased the expression of EGR1 (Figures 3F, 4E), indicating that PPARγ inhibit EGR1 in tubular cells. Moreover, luciferase reporter assay confirmed that PPARγ interact with EGR1 promoter, as EGR1 increased after EGR1 promoter plasmid transfection but decreased after PPARγ overexpression (Figure 4F). Meanwhile, the repressive effect of PPARγ on the EGR1 promoter was more obvious upon CAL incubation (Figure 4F). Taken together, these results indicated that PPARγ inhibited EGR1 expression and represses the activity of the EGR1 promoter.
Calycosin inhibited inflammation via peroxisome proliferator-activated receptor γ/early growth response 1
As a ligand, CAL could be embedded into the orthosteric pocket of PPARγ with a binding energy of -6.46 kcal/mol, which is exactly the binding pocket of thiazolidinedione agonists represented by troglitazone (Figures 5A,B; Supplementary Table S3). Interestingly, CAL formed hydrogen bonds with the residues Ser342, Ile281, and Glu291, in the ligand binding cavity of PPARγ, whereas troglitazone with the residues Ser342 and Ser289 (Figure 5C). This suggests that CAL probably activate PPARγ through a canonical thiazolidinedione ligand-binding cavity in a similar manner with troglitazone.
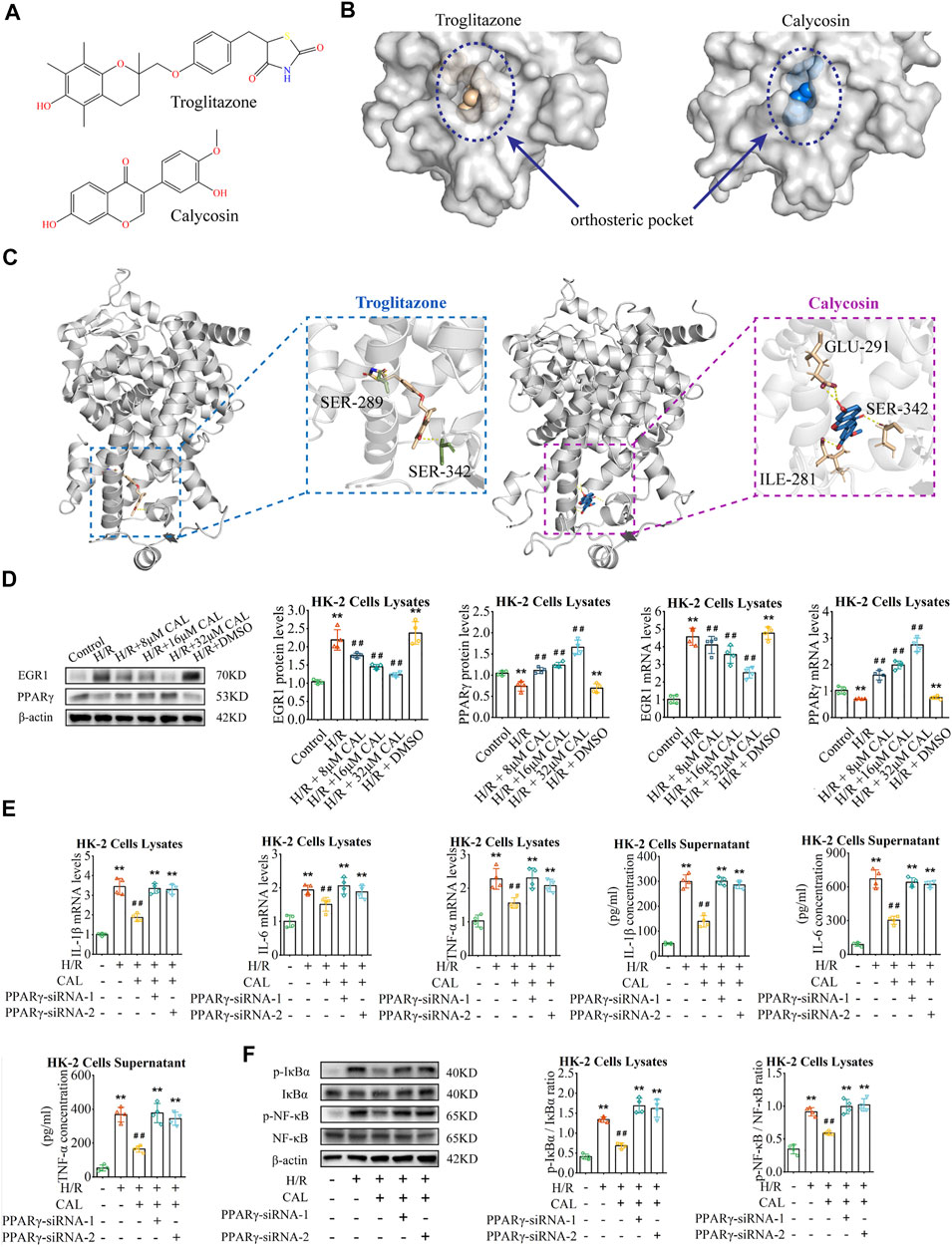
FIGURE 5. CAL inhibited inflammation via PPARγ/EGR1. (A) Chemical structure of troglitazone and CAL. (B) Structure of PPARγ revealed troglitazone and CAL is located in the orthosteric pocket of PPARγ. Troglitazone and CAL are shown as the spheres structure, and PPARγ is shown as the surfaces structure. (C) Troglitazone and CAL (ligand) bound to PPARγ (receptor) via hydrogen bonds. Ligand are shown as the stick structure, and PPARγ is shown as the cartoon structure. Hydrogen bonds are shown as dotted lines (yellow). ILE, isoleucine; SER, serine; GLU, glutamate. (D) The PPARγ and EGR1 mRNA and protein levels in different cell groups. (E) IL-1β, IL-6, and TNF-α mRNA levels in cell lysates and IL-1β, IL-6, and TNF-α concentrations in supernatant of different groups. (F) The protein levels of p-NF-κB/NF-κB and p-IκBα/IκBα in cell lysates of different groups. **p < 0.01 vs. control group. ##p < 0.01 vs. H/R group. n = 4 per group. The data are presented as the mean ± SD.
In vitro, compared with H/R induced cells, PPARγ increased and EGR1 decreased considerably in the CAL incubation groups (Figure 5D). However, the protective effect of CAL on H/R induced inflammation was absent when PPARγ knockdown, with increase of inflammatory cytokines and NF-κB activation (Figures 5E, F). These results indicated the PPARγ plays a role in CAL protection against H/R induced cells. Thus, we suggested that CAL might ameliorate renal IRI and inhibit inflammation via PPARγ/EGR1.
Discussion
The present study provided a novel approach and mechanism for the treatment of ischemic AKI. The main findings are as follows: 1) CAL alleviated renal injury and suppressed excessive inflammation response in renal IRI. 2) The protective effect of CAL was accessed by targeting the PPARγ/EGR1 pathway.
Oxidative stress and inflammation play a vital role in the pathophysiology of various diseases, such as senescence, cardiovascular disease, cancer and AKI (Li et al., 2020; Ikeda et al., 2021; Radovanovic et al., 2021; Zhang. et al., 2021; Wen et al., 2022). The activation of NF-κB signaling pathway has been identified as a major contributor of inflammation in the injured kidneys (Zhu et al., 2021). Consistent with this, in the present study, activation of NF-κB and inflammatory cytokines such as IL-1β, IL-6, and TNF-α are significantly increased in IRI mice, while CAL has reported to effectively inhibit inflammation by inhibiting NF-κB in various organ models (Zhang et al., 2019; Zhu et al., 2021). Of note, CAL treatment showed a good anti-inflammatory effect with the decrease of proinflammatory cytokines and inhibition of NF-κB activation in kidney after IRI.
To investigate the potential mechanisms of CAL, we applied differential gene analysis and PPI network analysis, and the results showed that EGR1 as a hub gene is significantly elevated in ischemic AKI. EGR1, a zinc-finger transcription factor of the immediate early gene family, is known to modulate inflammation responses in various tissues (Pritchard et al., 2010; Yan et al., 2021). Particularly, Ho et al. (2016) demonstrated that EGR1 aggravates tubulointerstitial nephritis by promotes NF-κB/NLRP3 in tubular cells. Consistent with this, we found that EGR1 expression in TECs was elevated after IRI stimulation and H/R stimulation, and proinflammatory cytokines were reduced and NF-κB activation was reduced when EGR1 knockdown, suggesting that EGR1 is involved in promoting the inflammatory response in TECs. In the IRI mice model, CAL dose-dependently decreased EGR1 in TECs after IRI, suggesting that the anti-inflammatory effect of CAL is related to EGR1.
PPARγ is a transcriptional factor belonging to nuclear receptors (NRs) superfamily (Wang et al., 2016; Gao et al., 2021). Previous studies have found that the activation of PPARγ inhibits EGR1 to suppress inflammation responses in brain and lung (Li et al., 2020; Yan et al., 2021), but the specific mechanism remains unclear. In the present study, EGR1 was upregulated significantly in HK-2 cells after PPARγ knockdown. Furthermore, luciferase reporter assay suggested that PPARγ repressed the activity of the EGR1 promoter, which provided a possibility to explain the regulatory relationship between PPAR and EGR1. In IRI stimulated mice and H/R induced HK-2 cells, the expression of PPARγ decreased, which are consistent with the previous findings (Singh et al., 2019; Wei et al., 2019). One of the possible reasons is that pro-inflammatory stimulation causes a decrease in the expression of PPARγ (Korbecki et al., 2019). A regulatory feedback loop between the PPARγ and TLR4 pathways may partly explain this phenomenon. When stimulated by LPS or other stimuli, TLR4 acts as an initial signal transduction, triggering the activation of pro-inflammatory signaling pathways, and preventing anti-inflammatory activity by negatively regulating PPARγ (Cao et al., 2018). Activation of PPARγ can negatively regulate the expression of TLR4 gene and attenuate the activation of downstream inflammatory signaling pathways (Mateu et al., 2015). Besides, prostaglandin (PG)H2 is specifically catalyzed to PGE2 by mPGES-1 under inflammatory conditions. The pro-inflammatory PEG2 can also inhibit the expression of PPARγ through pathways involving PI3K and Akt signaling (Kapoor et al., 2007; García-Alonso et al., 2013).
Extensive studies have demonstrated that many natural compounds are PPARγ agonist (Bernardo et al., 2021; Ji et al., 2021; Zhang et al. (2021), Zhang et al., 2020; Zhu et al., 2020). CAL as an isoflavonoid is reported to be an effective PPARγ agonist (Shen et al., 2006). This phenomenon was supported by molecular docking evidence, showing that CAL bind PPARγ stably by forming hydrogen bonds with residues Ser342, Ile281, and Glu291 in the orthosteric pocket of PPARγ. The binding patterns of agonists of PPARγ affect the recruitment of co-activators and co-repressors, playing an important role in modulating target gene expression (Jang et al., 2017). identified that interaction with Ser342 could be essential for transrepression of hypoxia inducible factor-1alpha (HIF-1α) by ligand-activated PPARγ, since Ser342 mutation abolished the transrepression effect of PPARγ on HIF-α by recruiting the co-repressors SMRT (Zhang et al., 2018). The mechanism by which CAL binds to PPARγ affects the expression of downstream target genes is still unclear, but the results of molecular docking provided a potential direction for further research.
Furthermore, although CAL may facilitate the activation PPARγ resulting in inhibiting inflammation, the potential role of CAL regulation of PPARγ expression in this context could not be excluded. We found that CAL has increased the expression of PPARγ in kidney after IRI and tubular cells after H/R. Increased protein abundance of PPARγ may facilitate its nuclear transcriptional activity. We thus speculated that CAL suppression of inflammation could also be partially attributed to upregulation of PPARγ expression. The siRNA-mediated knockdown of PPARγ expression could validate this speculation.
However, there are still some limitations to this study. The effect of CAL on the transcriptional activity of PPARγ and the binding site of PPARγ regulating the EGR1 promoter still require further experiments to explore. In addition, the regulation of inflammation by PPARγ involves multiple signaling pathways, such as transrepressing of NF-κB expression, inhibiting of NF-κB p65 activation, suppressing of AP-1 activation and inhibiting the AP-1 DNA binding activity, increasing SOCS3 expression to disrupt JAK2/STAT3 activation and many more (Pascual et al., 2005; Yu et al., 2008; Wang et al., 2021). In this work, we only explored that CAL inhibits inflammatory responses through the PPARγ/EGR1 pathway, but CAL may also exert anti-inflammatory effects through other signaling pathways, and further studies are needed.
In summary, our study demonstrated that CAL could significantly modulate inflammation by targeting the PPARγ/EGR1 pathway, contributing to the alleviation of renal IRI (Supplementary Figure S1). CAL, as a novel PPARγ activator, may serve as a potential candidate for pharmaceutic prevention and therapy of ischemic AKI. While the current in vivo and in vitro models supported the effect of CAL to attenuate ischemic AKI, additional studies will be required to determine whether this strategy could also be used in clinical settings.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by Laboratory Animal Welfare Ethics Committee at Qingdao University (ethics number: 202105C5730202106036).
Author contributions
NZ performed the design, experiments, and thesis writing. CG and ZL performed the animal experiments and experimental guidance. CL and CY performed the bioinformatics analysis and data analysis. LX and MN performed the cell experiments and data analysis. LZ, BZ, LC, and YW performed the design and experimental guidance. YX performed the design, experimental guidance, and data analysis.
Funding
This work was funded by the National Natural Science Foundation of China (81770679 and 81970582), Qingdao Municipal Science and Technology Bureau (20-3-4-36-nsh), and Qingdao Key Health Discipline Development Fund. This work was also supported by Clinical Medicine + X project of the Affiliated Hospital of Qingdao University and Medical Science and Technology Development Project of Shandong Province (202103050716).
Acknowledgments
We thank all the research subjects and their families for their generous participation in this study. We thank all those colleagues who provided support and help in various ways to this study. Supplementary Figure S1is created by Figdraw (www.figdraw.com).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.970616/full#supplementary-material
References
Andrade-Oliveira, V., Foresto-Neto, O., Watanabe, I. K. M., Zatz, R., and Câmara, N. O. S. (2019). Inflammation in renal diseases: New and old players. Front. Pharmacol. 10, 1192. doi:10.3389/fphar.2019.01192
Bernardo, A., Plumitallo, C., De Nuccio, C., Visentin, S., and Minghetti, L. (2021). Curcumin promotes oligodendrocyte differentiation and their protection against TNF-α through the activation of the nuclear receptor PPAR-γ. Sci. Rep. 11 (1), 4952. doi:10.1038/s41598-021-83938-y
Cao, X. J., Zhang, M. J., Zhang, L. L., Yu, K., Xiang, Y., Ding, X., et al. (2018). TLR4 mediates high-fat diet induced physiological changes in mice via attenuating PPARγ/ABCG1 signaling pathway. Biochem. Biophys. Res. Commun. 503 (3), 1356–1363. doi:10.1016/j.bbrc.2018.07.048
Chen, C., Cui, J., Ji, X., and Yao, L. (2020). Neuroprotective functions of calycosin against intracerebral hemorrhage-induced oxidative stress and neuroinflammation. Future Med. Chem. 12 (7), 583–592. doi:10.4155/fmc-2019-0311
Chin, C. H., Chen, S. H., Wu, H. H., Ho, C. W., Ko, M. T., and Lin, C. Y. (2014). cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8, S11. doi:10.1186/1752-0509-8-s4-s11
Deng, M., Chen, H., Long, J., Song, J., Xie, L., and Li, X. (2021). Calycosin: A review of its pharmacological effects and application prospects. Expert Rev. anti. Infect. Ther. 19 (7), 911–925. doi:10.1080/14787210.2021.1863145
Fang, C. Y., Lou, D. Y., Zhou, L. Q., Wang, J. C., Yang, B., He, Q. J., et al. (2021). Natural products: Potential treatments for cisplatin-induced nephrotoxicity. Acta Pharmacol. Sin. 42 (12), 1951–1969. doi:10.1038/s41401-021-00620-9
Gao, Z., Xu, X., Li, Y., Sun, K., Yang, M., Zhang, Q., et al. (2021). Mechanistic insight into PPARγ and Tregs in atherosclerotic immune inflammation. Front. Pharmacol. 12, 750078. doi:10.3389/fphar.2021.750078
García-Alonso, V., López-Vicario, C., Titos, E., Morán-Salvador, E., González-Périz, A., Rius, B., et al. (2013). Coordinate functional regulation between microsomal prostaglandin E synthase-1 (mPGES-1) and peroxisome proliferator-activated receptor γ (PPARγ) in the conversion of white-to-brown adipocytes. J. Biol. Chem. 288 (39), 28230–28242. doi:10.1074/jbc.M113.468603
Geng, X. Q., Ma, A., He, J. Z., Wang, L., Jia, Y. L., Shao, G. Y., et al. (2020). Ganoderic acid hinders renal fibrosis via suppressing the TGF-β/Smad and MAPK signaling pathways. Acta Pharmacol. Sin. 41 (5), 670–677. doi:10.1038/s41401-019-0324-7
He, J. Y., Hong, Q., Chen, B. X., Cui, S. Y., Liu, R., Cai, G. Y., et al. (2022). Ginsenoside Rb1 alleviates diabetic kidney podocyte injury by inhibiting aldose reductase activity. Acta Pharmacol. Sin. 43 (2), 342–353. doi:10.1038/s41401-021-00788-0
Ho, L. C., Sung, J. M., Shen, Y. T., Jheng, H. F., Chen, S. H., Tsai, P. J., et al. (2016). Egr-1 deficiency protects from renal inflammation and fibrosis. J. Mol. Med. 94 (8), 933–942. doi:10.1007/s00109-016-1403-6
Husain-Syed, F., Villa, G., Wilhelm, J., Samoni, S., Matt, U., Vadász, I., et al. (2021). Renal markers for monitoring acute kidney injury transition to chronic kidney disease after COVID-19. Nephrol. Dial. Transpl. 36 (11), 2143–2147. doi:10.1093/ndt/gfab235
Ikeda, Y., Taniguchi, K., Nagase, N., Tsuji, A., Kitagishi, Y., and Matsuda, S. (2021). Reactive oxygen species may influence on the crossroads of stemness, senescence, and carcinogenesis in a cell via the roles of APRO family proteins. Explor. Med. 2, 443–454. doi:10.37349/emed.2021.00062
Jang, J. Y., Koh, M., Bae, H., An, D. R., Im, H. N., Kim, H. S., et al. (2017). Structural basis for differential activities of enantiomeric PPARγ agonists: Binding of S35 to the alternate site. Biochim. Biophys. Acta. Proteins Proteom. 1865 (6), 674–681. doi:10.1016/j.bbapap.2017.03.008
Ji, Z., Li, J., and Wang, J. (2021). Jujuboside B inhibits neointimal hyperplasia and prevents vascular smooth muscle cell dedifferentiation, proliferation, and migration via activation of AMPK/PPAR-γ signaling. Front. Pharmacol. 12, 672150. doi:10.3389/fphar.2021.672150
Kapoor, M., Kojima, F., Qian, M., Yang, L., and Crofford, L. J. (2007). Microsomal prostaglandin E synthase-1 deficiency is associated with elevated peroxisome proliferator-activated receptor gamma: Regulation by prostaglandin E2 via the phosphatidylinositol 3-kinase and Akt pathway. J. Biol. Chem. 282 (8), 5356–5366. doi:10.1074/jbc.M610153200
Korbecki, J., Bobiński, R., and Dutka, M. (2019). Self-regulation of the inflammatory response by peroxisome proliferator-activated receptors. Inflamm. Res. 68 (6), 443–458. doi:10.1007/s00011-019-01231-1
Li, H., and Xia, N. (2020). The role of oxidative stress in cardiovascular disease caused by social isolation and loneliness. Redox Biol. 37, 101585. doi:10.1016/j.redox.2020.101585
Li, Y. Y., Guo, J. H., Liu, Y. Q., Dong, J. H., and Zhu, C. H. (2020). PPARγ activation-mediated egr-1 inhibition benefits against brain injury in an experimental ischaemic stroke model. J. Stroke Cerebrovasc. Dis. 29 (12), 105255. doi:10.1016/j.jstrokecerebrovasdis.2020.105255
Li, Z. L., Wang, B., Lv, L. L., Tang, T. T., Wen, Y., Cao, J. Y., et al. (2021). FIH-1-modulated HIF-1α C-TAD promotes acute kidney injury to chronic kidney disease progression via regulating KLF5 signaling. Acta Pharmacol. Sin. 42 (12), 2106–2119. doi:10.1038/s41401-021-00617-4
Liu, J., Krautzberger, A. M., Sui, S. H., Hofmann, O. M., Chen, Y., Baetscher, M., et al. (2014). Cell-specific translational profiling in acute kidney injury. J. Clin. Invest. 124 (3), 1242–1254. doi:10.1172/jci72126
Mateu, A., Ramudo, L., Manso, M. A., and De Dios, I. (2015). Cross-talk between TLR4 and PPARγ pathways in the arachidonic acid-induced inflammatory response in pancreatic acini. Int. J. Biochem. Cell Biol. 69, 132–141. doi:10.1016/j.biocel.2015.10.022
Meng, J., Sai-Zhen, W., He, J. Z., Zhu, S., Huang, B. Y., Wang, S. Y., et al. (2020). Ganoderic acid A is the effective ingredient of Ganoderma triterpenes in retarding renal cyst development in polycystic kidney disease. Acta Pharmacol. Sin. 41 (6), 782–790. doi:10.1038/s41401-019-0329-2
Pascual, G., Fong, A. L., Ogawa, S., Gamliel, A., Li, A. C., Perissi, V., et al. (2005). A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature 437 (7059), 759–763. doi:10.1038/nature03988
Pefanis, A., Ierino, F. L., Murphy, J. M., and Cowan, P. J. (2019). Regulated necrosis in kidney ischemia-reperfusion injury. Kidney Int. 96 (2), 291–301. doi:10.1016/j.kint.2019.02.009
Place, D. E., and Kanneganti, T. D. (2018). Recent advances in inflammasome biology. Curr. Opin. Immunol. 50, 32–38. doi:10.1016/j.coi.2017.10.011
Pritchard, M. T., Cohen, J. I., Roychowdhury, S., Pratt, B. T., and Nagy, L. E. (2010). Early growth response-1 attenuates liver injury and promotes hepatoprotection after carbon tetrachloride exposure in mice. J. Hepatol. 53 (4), 655–662. doi:10.1016/j.jhep.2010.04.017
Radovanovic, J., Banjac, K., Obradovic, M., and Isenovic, E. R. (2021). Antioxidant enzymes and vascular diseases. Explor. Med. 2 (6), 544–555. doi:10.37349/emed.2021.00070
Rashid, I., Katravath, P., Tiwari, P., D'Cruz, S., Jaswal, S., and Sahu, G. (2022). Hyperuricemia-a serious complication among patients with chronic kidney disease: A systematic review and meta-analysis. Explor. Med. 3 (3), 249–259. doi:10.37349/emed.2022.00089
Ronco, C., Bellomo, R., and Kellum, J. A. (2019). Acute kidney injury. Lancet 394 (10212), 1949–1964. doi:10.1016/s0140-6736(19)32563-2
Sato, Y., Takahashi, M., and Yanagita, M. (2020). Pathophysiology of AKI to CKD progression. Semin. Nephrol. 40 (2), 206–215. doi:10.1016/j.semnephrol.2020.01.011
Shao, M., Ye, C., Bayliss, G., and Zhuang, S. (2021). New insights into the effects of individual Chinese herbal medicines on chronic kidney disease. Front. Pharmacol. 12, 774414. doi:10.3389/fphar.2021.774414
Shen, P., Liu, M. H., Ng, T. Y., Chan, Y. H., and Yong, E. L. (2006). Differential effects of isoflavones, from Astragalus membranaceus and Pueraria thomsonii, on the activation of PPARalpha, PPARgamma, and adipocyte differentiation in vitro. J. Nutr. 136 (4), 899–905. doi:10.1093/jn/136.4.899
Singh, A. P., Singh, N., Pathak, D., and Bedi, P. M. S. (2019). Estradiol attenuates ischemia reperfusion-induced acute kidney injury through PPAR-γ stimulated eNOS activation in rats. Mol. Cell. Biochem. 453 (1-2), 1–9. doi:10.1007/s11010-018-3427-4
Song, N., Thaiss, F., and Guo, L. (2019). NFκB and kidney injury. Front. Immunol. 10, 815. doi:10.3389/fimmu.2019.00815
Tammaro, A., Kers, J., Scantlebery, A. M. L., and Florquin, S. (2020). Metabolic flexibility and innate immunity in renal ischemia reperfusion injury: The fine balance between adaptive repair and tissue degeneration. Front. Immunol. 11, 1346. doi:10.3389/fimmu.2020.01346
Wang, J., Xu, X., Li, P., Zhang, B., and Zhang, J. (2021). HDAC3 protects against atherosclerosis through inhibition of inflammation via the microRNA-19b/PPARγ/NF-κB axis. Atherosclerosis 323, 1–12. doi:10.1016/j.atherosclerosis.2021.02.013
Wang, S., Dougherty, E. J., and Danner, R. L. (2016). PPARγ signaling and emerging opportunities for improved therapeutics. Pharmacol. Res. 111, 76–85. doi:10.1016/j.phrs.2016.02.028
Wang, X., Li, W., Zhang, Y., Sun, Q., Cao, J., Tan, N., et al. (2022). Calycosin as a novel PI3K activator reduces inflammation and fibrosis in heart failure through AKT-IKK/STAT3 Axis. Front. Pharmacol. 13, 828061. doi:10.3389/fphar.2022.828061
Wei, L., Qin, Y., Jiang, L., Yu, X., and Xi, Z. (2019). PPARγ and mitophagy are involved in hypoxia/reoxygenation-induced renal tubular epithelial cells injury. J. Recept. Signal Transduct. Res. 39 (3), 235–242. doi:10.1080/10799893.2019.1660894
Wen, L., Tao, S. H., Guo, F., Li, L. Z., Yang, H. L., Liang, Y., et al. (2022). Selective EZH2 inhibitor zld1039 alleviates inflammation in cisplatin-induced acute kidney injury partially by enhancing RKIP and suppressing NF-κB p65 pathway. Acta Pharmacol. Sin. 43 (8), 2067–2080. doi:10.1038/s41401-021-00837-8
Yan, C., Chen, J., Ding, Y., Zhou, Z., Li, B., Deng, C., et al. (2021). The crucial role of pparγ-egr-1-pro-inflammatory mediators Axis in IgG immune complex-induced acute lung injury. Front. Immunol. 12, 634889. doi:10.3389/fimmu.2021.634889
Yu, J. H., Kim, K. H., and Kim, H. (2008). SOCS 3 and PPAR-gamma ligands inhibit the expression of IL-6 and TGF-beta1 by regulating JAK2/STAT3 signaling in pancreas. Int. J. Biochem. Cell Biol. 40 (4), 677–688. doi:10.1016/j.biocel.2007.10.007
Zhang, F., Lu, S., He, J., Jin, H., Wang, F., Wu, L., et al. (2018). Ligand activation of PPARγ by ligustrazine suppresses pericyte functions of hepatic stellate cells via SMRT-mediated transrepression of HIF-1α. Theranostics 8 (3), 610–626. doi:10.7150/thno.22237
Zhang, W., Qi, R., Li, T., Zhang, X., Shi, Y., Xu, M., et al. (2021). Kidney organoids as a novel platform to evaluate lipopolysaccharide-induced oxidative stress and apoptosis in acute kidney injury. Front. Med. 8, 766073. doi:10.3389/fmed.2021.766073
Zhang, Y. Y., Tan, R. Z., Zhang, X. Q., Yu, Y., and Yu, C. (2019). Calycosin ameliorates diabetes-induced renal inflammation via the NF-κB pathway in vitro and in vivo. Med. Sci. Monit. 25, 1671–1678. doi:10.12659/msm.915242
Zhang, Y., Zhang, Y., Li, Y., Zhang, L., and Yu, S. (2021). Preclinical investigation of alpinetin in the treatment of cancer-induced cachexia via activating PPARγ. Front. Pharmacol. 12, 687491. doi:10.3389/fphar.2021.687491
Zhang, Z. H., He, J. Q., Zhao, Y. Y., Chen, H. C., and Tan, N. H. (2020). Asiatic acid prevents renal fibrosis in UUO rats via promoting the production of 15d-PGJ2, an endogenous ligand of PPAR-γ. Acta Pharmacol. Sin. 41 (3), 373–382. doi:10.1038/s41401-019-0319-4
Zhou, Y., Ren, Q., Chen, G., Jin, Q., Cui, Q., Luo, H., et al. (2020). Chronic kidney diseases and acute kidney injury in patients with COVID-19: Evidence from a meta-analysis. Front. Med. 7, 588301. doi:10.3389/fmed.2020.588301
Zhu, D., Yu, H., Liu, P., Yang, Q., Chen, Y., Luo, P., et al. (2021). Calycosin modulates inflammation via suppressing TLR4/NF-κB pathway and promotes bone formation to ameliorate glucocorticoid-induced osteonecrosis of the femoral head in rat. Phytotherapy Res. 35, 2824–2835. doi:10.1002/ptr.7028
Zhu, Y. D., Guan, X. Q., Chen, J., Peng, S., Finel, M., Zhao, Y. Y., et al. (2020). Neobavaisoflavone induces bilirubin metabolizing enzyme UGT1A1 via PPARα and PPARγ. Front. Pharmacol. 11, 628314. doi:10.3389/fphar.2020.628314
Keywords: calycosin, acute kidney injury, ischemia/reperfusion injury, peroxisome proliferator-activated receptor γ, early growth response 1
Citation: Zhang N, Guan C, Liu Z, Li C, Yang C, Xu L, Niu M, Zhao L, Zhou B, Che L, Wang Y and Xu Y (2022) Calycosin attenuates renal ischemia/reperfusion injury by suppressing NF-κB mediated inflammation via PPARγ/EGR1 pathway. Front. Pharmacol. 13:970616. doi: 10.3389/fphar.2022.970616
Received: 16 June 2022; Accepted: 26 September 2022;
Published: 07 October 2022.
Edited by:
Jianping Chen, Shenzhen Traditional Chinese Medicine Hospital, ChinaReviewed by:
Lei Yu, University of Michigan, United StatesMasashi Mukohda, Okayama University of Science, Japan
Copyright © 2022 Zhang, Guan, Liu, Li, Yang, Xu, Niu, Zhao, Zhou, Che, Wang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Xu, eHV5YW5AcWR1LmVkdS5jbg==
 Ningxin Zhang
Ningxin Zhang Chen Guan
Chen Guan Zengying Liu
Zengying Liu Chenyu Li
Chenyu Li Chengyu Yang
Chengyu Yang Lingyu Xu
Lingyu Xu Meng Niu
Meng Niu Long Zhao
Long Zhao Bin Zhou
Bin Zhou Lin Che
Lin Che Yanfei Wang
Yanfei Wang Yan Xu
Yan Xu