- 1College of Pharmacy, Al Ain University, Abu Dhabi, United Arab Emirates
- 2College of Pharmacy, Mansoura University, Mansoura, Egypt
The human microbiota produces molecules that are evolved to interact with the diverse cellular machinery of both the host and microbes, mediating health and diseases. One of the most puzzling microbiome molecules is colibactin, a genotoxin encoded in some commensal and extraintestinal microbes and is implicated in initiating colorectal cancer. The colibactin cluster was discovered more than 15 years ago, and most of the research studies have been focused on revealing the biosynthesis and precise structure of the cryptic encoded molecule(s) and the mechanism of carcinogenesis. In 2022, the Balskus group revealed that colibactin not only hits targets in the eukaryotic cell machinery but also in the prokaryotic cell. To that end, colibactin crosslinks the DNA resulting in activation of the SOS signaling pathway, leading to prophage induction from bacterial lysogens and modulation of virulence genes in pathogenic species. These unique activities of colibactin highlight its ecological role in shaping gut microbial communities and further consequences that impact human health. This review dives in-depth into the molecular mechanisms underpinning colibactin cellular targets in eukaryotic and prokaryotic cells, aiming to understand the fine details of the role of secreted microbiome chemistry in mediating host–microbe and microbe–microbe interactions. This understanding translates into a better realization of microbiome potential and how this could be advanced to future microbiome-based therapeutics or diagnostic biomarkers.
Introduction
Trillions of microbes reside in the human gut interacting with each other and the outcome of their interaction affects human health and diseases (Sharon et al., 2014; Dhanaraju and Rao 2022; de Vos et al., 2022). This interaction is mediated by evolved small molecules, of which the vast majority are still dark matter (Wilson, Zha, and Balskus 2017; Zha et al., 2022). A few examples of microbiome secreted products are reported, each with a unique activity such as the microbial genotoxin colibactin (Wernke et al., 2020). Colibactin is a hybrid polyketide–non-ribosomal peptide product encoded in the genome of some Enterobacteriaceae that belong to the phylogenetic group B. Colibactin is implicated in causing colorectal cancer (CRC) by inducing a signature mutation in DNA. Isolation and structural elucidation of colibactin has challenged the scientific field for over 15 years because of its contact-dependent synthesis, minimal expression, and chemical instability. The bulk of the research study on colibactin is centered around revealing its structure, mainly through bioinformatic analysis, comparative metabolomics, and mutational studies (Tang et al., 2022). However, to appreciate the evolved function of colibactin(s) and other microbiome products, we must understand their ecological role in enhancing the competency and fitness of producers. There are some reports on the antimicrobial-like activity of colibactin, but most of the data come from observational or associational studies without revealing the underpinning mechanism of action (T. Faïs et al., 2016). In a pioneering study, the group of Dr. Balskus revealed that colibactin induces prophage activation on a wide scale in multiple phage–bacteria systems. This activity is mediated by its ability to damage DNA activating the SOS response. Moreover, induction of prophages affects prophage-encoded genes such as some bacterial toxins, which are crucial for their pathogenicity. This finding reveals another interesting role of colibactin in mediating microbe–microbe interaction and shaping the structure of gut microbes (Silpe et al., 2022; Tronnet et al., 2020).
Our knowledge of colibactin(s) is developing and there are previous reviews covering their prospective research period (Faïs et al., 2018; Bode 2015; Balskus 2015; Williams et al., 2020; Wernke et al., 2020; Tang et al., 2022). These reviews mainly focused on summarizing advances in understanding possible biosynthetic pathways, bioinformatic prediction of colibactin and precolibactin structures, and the mechanism of DNA mutation. In this review, we analyze the unique cellular targets of colibactin(s) mediating host–microbe interactions that shape gut microbiome structures and influence host’s health. We discuss the molecular mechanisms underlying the evolved function of colibactin in hitting multiple targets in both the eukaryotic and prokaryotic cell machinery. We propose a mechanistic model for the series of events leading to colibactin-induced cancer. This knowledge is central to better advancing the use of microbiome secreted products or genes as diagnostic biomarkers or therapeutic interventions.
The latest development in understanding the biosynthesis and structure of colibactin(s)
Prevalence and significance of colibactin gene cluster in prokaryotes
The first report on the colibactin cluster is dated back to 2006 when Nougayrède et al. (2006) reported the discovery of Escherichia coli strains belonging to the phylogenetic group B2 that can block mitosis, leading to megalocytosis and eventually cell death. The authors linked this weird activity to a genomic island that encodes cryptic molecule(s), named at this time colibactin (Nougayrède et al., 2006). Interestingly, this gene cluster is prevalent in gut microbes, in particular Group B of human-associated strains of E. coli and Enterobacteriaceae, including commensal and probiotic strains such as E. coli Nissle 1917, a commercial probiotic (Mutaflor) used to improve gastrointestinal inflammatory conditions such as ulcerative colitis (Schultz 2008). The Colibactin cluster is also sequenced from other non-human associated microbes such as Pseudovibrio sp. JE062, isolated from a marine sponge (Bondarev et al., 2013) and a novel strain of Erwina oleae sp. isolated from olive tree nots caused by Pseudomonas savastanoi (Moretti et al., 2011). The Frischella perrara PEB0191 is a commensal gut microbe in honey bees. The Crawford group showed that F. perrara PEB0191 produces colibactin mimics and induces DNA breaks similar to human microbiome strains (Engel, Vizcaino, and Crawford 2015). This widespread prevalence of colibactin or its homologs indicates its potential role in a symbiotic relationship, which likely confers beneficial outcomes to the host.
Genomic organization of the colibactin cluster
The colibactin cluster encodes 54-kb hybrid nonribosomal peptide synthetase-polyketide synthase (NRPS-PKS) biosynthetic genes known as the pks or clb gene cluster (Figure 1). The cluster consisted of 19 genes, named alphabetically, clb A-S. These 19 genes encode three polyketide synthases (PKSs), three nonribosomal peptide synthases (NRPSs), two hybrids NRPS/PKS, MATE transporter, resistance gene, and other nine tailoring and accessory enzymes as per the latest update as reviewed (Tang et al., 2022). Intensive research reports confirm that all of these genes are essential for genotoxicity, except for clb M, S, and R (Nougayrède et al., 2006). Our current understanding of colibactin structure is shaped through understanding the organization of its biosynthetic genes, mutational studies, heterologous expression, and structure prediction using bioinformatics models. Over the last 16 years, there have been multiple hypotheses for the biosynthesis of colibactin, and the most recent is the prodrug activation theory (Figure 1) developed by the Balskus group (Brotherton and Balskus 2013; Balskus 2015). The genomic island of colibactin contains the clbP gene, which is a peptidase enzyme that shares structure/function homology with cleavage enzymes functioning in ribosomaly synthesized posttranslational modified peptides such as ZmaM in zwittermicin biosynthesis (Luo et al., 2011). This similarity is revealed by a plethora of confirmation analyses, including structure–activity relationship, crystallography, and mutagenesis (Cougnoux et al., 2012; Dubois et al., 2011). This fact leads Balskus and coworkers to hypothesize the prodrug activation theory for the biosynthesis of colibactin in which two NRPS modules (clbN and clbB) install the N-acyl-d-asparagine scaffold earlier in the biosynthesis, followed by clbP cleaving the amide bond and releasing the final active colibactin product (Brotherton and Balskus 2013). The Muller group added additional verification of the hypothesis by isolating the hypothesized prodrug from E. coli-Nissle 1917 (Bian et al., 2013). Further studies revealed that silencing of this peptidase enzyme (Δ clbP strain) facilitates the accumulation of precolibactins, which are more stable derivatives of the main genotoxic molecule. Δ clbP strains have a diminished genotoxic activity compared to wild type. Of note, we still lack more precise information on the genetic organization of colibactin-like clusters in other species and how the possible polymorphism might lead to functionally diverse molecules.
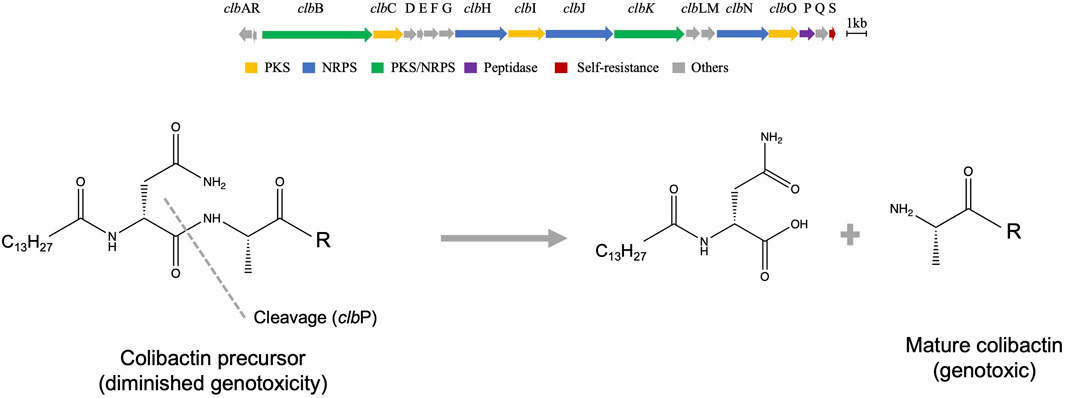
FIGURE 1. Genetic organization and possible biosynthesis of colibactin. (A) Organization of the biosynthetic genes cluster of colibactin. (B) Prodrug activation hypothesis overview shows the last required step for production of the genotoxin, which is catalyzed by peptidase enzyme.
Revealing the structure of colibactin derivatives (precolibactins)
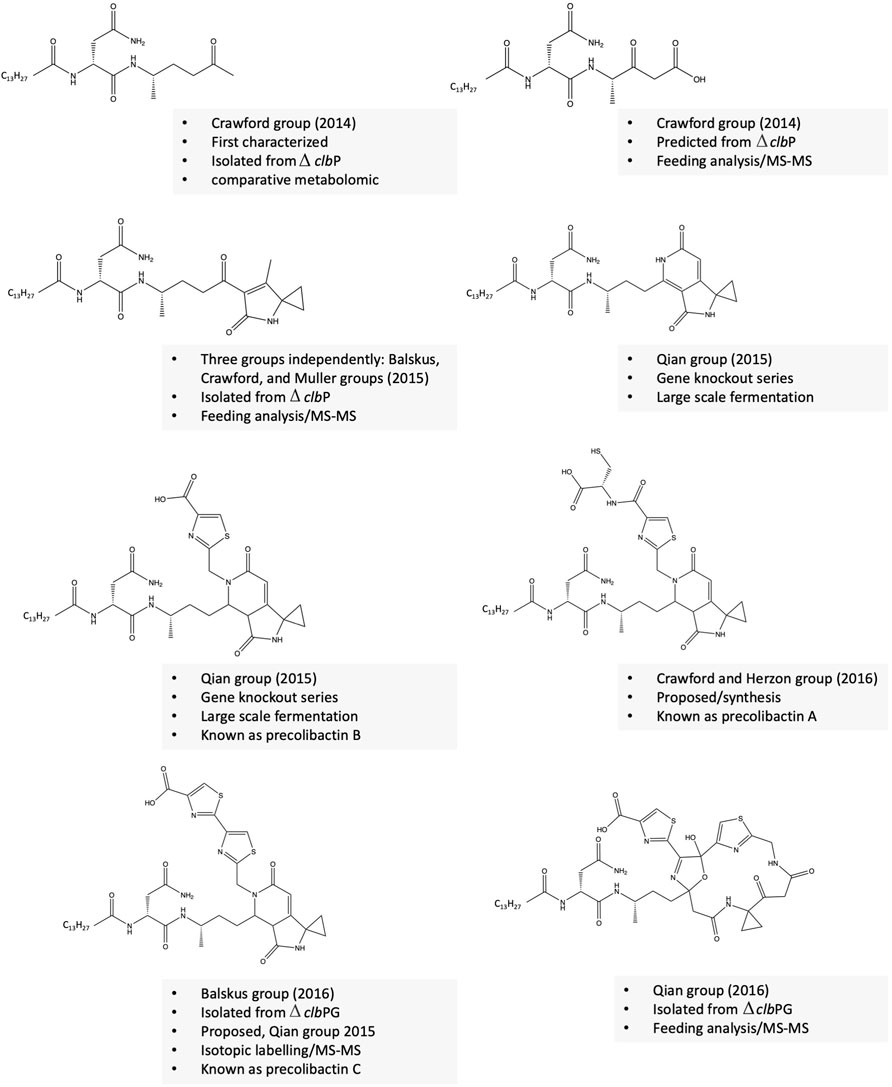
The precise structure of the colibactin molecule has remained unknown for over a decade. Purification of colibactin from native or heterologous hosts is challenging due to its minimal production upon fermentation, instability, and contact or inflammation-dependent gene expression. To overcome this challenge, several research groups developed alternative strategies to characterize the more stable derivatives or precursors of colibactin, named precolibactins (Vizcaino et al., 2014; Li et al., 2019; Vizcaino and Crawford 2015; Brotherton et al., 2015; Bian et al., 2015; Li et al., 2015; Li et al., 2016; Healy, Vizcaino, et al., 2016b). These strategies involve: 1) mutating the clbP gene, an essential enzyme required to produce active genotoxic colibactin molecules, and the generation of a series of other mutants; 2) heterologous expression of the mutated gene clusters; 3) comparative metabolomic analysis of mutants and wild-type strains; 4) tandem MS–MS fragmentation analysis coupled with isotopic feeding experiments; and 5) synthesis of potential precolibactin analogues. Following these approaches, around 40 precolibactins have been predicted, isolated, or synthesized. Some structurally unique examples of precolibactins are shown in Figure 2. Isolated or predicted precolibactins might be biosynthetically related, and for example, theoretically, precolibactin C could be biosynthesized from A via cyclodehydration as proposed (Healy 2017). Healy et al. proposed that precolibactins A-C may not be the precursors for colibactin as previously thought but rather alternative products produced only when the cibP is nonfunctional by a double cyclodehydration pathway (Healy et al., 2016a; Healy et al., 2016b). This hypothesis is based on some evidence that colibactin alkylates DNA by the formation of unsaturated imines, which could not be generated from the pyridone ring in precolibactins A-C (Healy et al., 2016a). Interestingly, some precolibactins share a structural similarity with yatakemycin and duramycin, mainly the aminocyclopropane moiety, which attacks DNA via nucleophilic ring-opening (Tichenor and Boger 2008). Precolibactin A weakly cross-links DNA in vitro while a precolibactin derivative lacking the spiro bicyclic structure was inactive (Vizcaino and Crawford 2015). Further evidence suggests that the two precolibactins isolated are products of the intact colibactin cluster.
Revealing the structure of colibactin(s)
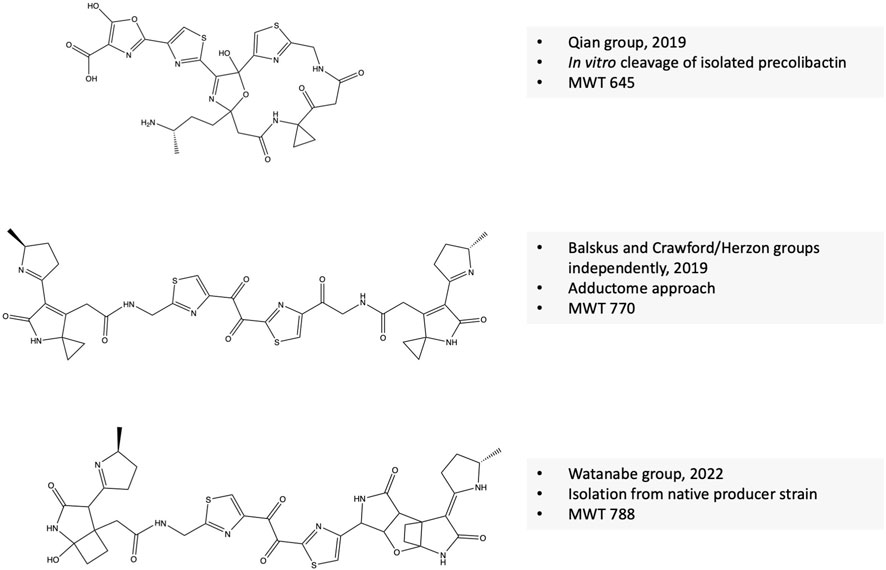
In 2019, the Qian group elucidated the structure of a colibactin molecule based on in vitro cleavage of an isolated precolibactin with an MWT of 969 to yield colibactin with an MWT of 645 (Li et al., 2019). Further evidence confirms that this structure is produced from the native PKS + strain. This evidence includes the presence of the product in the extract of native colibactin-harboring E. coli CFT073 as detected by MS–MS analysis and the ability of the compound to induce the characteristic DNA double-strand breaks (Li et al., 2019). In the same year, Crawford/Herzon and Balskus groups independently elucidated the structure of another colibactin molecule with an MWT of 770 and validated its genotoxic activity (Xue et al., 2019; Jiang et al., 2019). Crawford/Herzon predicted the structure by a DNA adductome approach followed by total synthesis. The Balskus group predicted the structure by studying clbP/clbL double mutants, labeling experiments, and an adductome approach. The clbL is predicted to encode amidase functions by hydrolysis of an amide bond; however, the authors validated that this enzyme is rather the final step in the production of intact precolibactin (Jiang et al., 2019). In 2022, the Watanabe group isolated a colibactin molecule with an MWT of 788 from the pks + E. coli strain-50, a strain isolated from colorectal cancer tissues that produces 26-fold more colibactin than Nissle 1917 (Hirayama et al., 2019; Zhou et al., 2021). Of note, the same group has developed an activity-based fluorescent probe to identify high colibactin producers from clinical samples 3 years before elucidating the first-isolation-based structure of colibactin (Hirayama et al., 2019). At first, the authors obtained two fragments of the compound whose structures are suggestive of spontaneous hydrolysis of the candidate molecule at the 1,2-diketone moiety as previously described by the Herzon group (Healy et al., 2019). Next, they managed to protect the compound from hydrolysis by adding o-phenylenediamine to convert the 1,2-diketone moiety into quinoxaline, which is not subjected to hydrolysis and isolated a few micrograms of colibactin sufficient for further NMR spectroscopic analysis (Zhou et al., 2021). The three revealed structures of colibactin are shown in Figure 3.
Colibactin hits unique molecular targets in the eukaryotic cells
Our gut microbes secrete a plethora of molecules with diverse structures and functions. These molecules can either promote host health such as butyrate or trigger diseases. In addition to colibactin, several microbiome-derived molecules are noted as mediators of diseases, such as the autism-promoting molecule 4-ethylphenylsulfate and the gut genotoxin cyclomodulin (Hsiao et al., 2013; Buc et al., 2013). Colibactin might hit other targets in the eukaryotic machinery than what we currently appreciate. A study shows that some synthetic precolibactins exhibited antagonist activity on the mammalian brain receptors such as serotonin 5-hydroxytryptamine type 7 (5-HT7) in vitro (Vizcaino et al., 2014). Of note, 5-HT7 is expressed in dendritic cells. Studies show that 5-HT7 receptor antagonists such as SB-269970 decrease both acute and chronic inflammation in a mouse model of colitis (Kim et al., 2013). Synthetic precolibactin also showed antagonist activity against dopamine 5 receptors (Vizcaino et al., 2014). Dopamine receptors are reported to be located in the CNS and gut (Z. S. Li et al., 2006).
Mechanistic insights into colibactin signature mutation
Colibactin alkylates DNA in the eukaryotic cell leading to colorectal cancer. Overall, colibactin causes an inter-strand DNA cross-link, leading to double-strand breaks in the DNA triggering cell-cycle arrest and further to CRC. The mechanism of colibactin mutation is detailed below and illustrated in Figure 4. DNA double-strand breaks are prevalent in more than half of the patients suffering from inflammatory bowel diseases and CRC (Buc et al., 2013). The colibactin-DNA signature adduct is detected in both human and animal cells and in vivo in experimental animals (Wilson et al., 2019). A study conducted on mouse models of invasive carcinoma revealed that inoculation with pks+ E. coli NC101 increases tumor and enables metastasis (Arthur et al., 2012). This tumor-promoting activity is diminished with the deletion of the colibactin gene cluster. However, this deletion does not affect the inflammatory status (Arthur et al., 2012). The DNA alkylation or formation of the covalent bond between the electrophilic warhead of colibactin (azospiro 2,4 bicyclic ring) and the nucleophilic DNA results in the formation of a colibactin–DNA adduct and creates a second electrophilic center that further reacts with DNA to form a cross-link. DNA alkylation is mediated by the electrophilic cyclopropane moiety of colibactin. Initially, colibactin forms an unstable adduct, which is considered a biomarker signature for colibactin (Wilson et al., 2019).
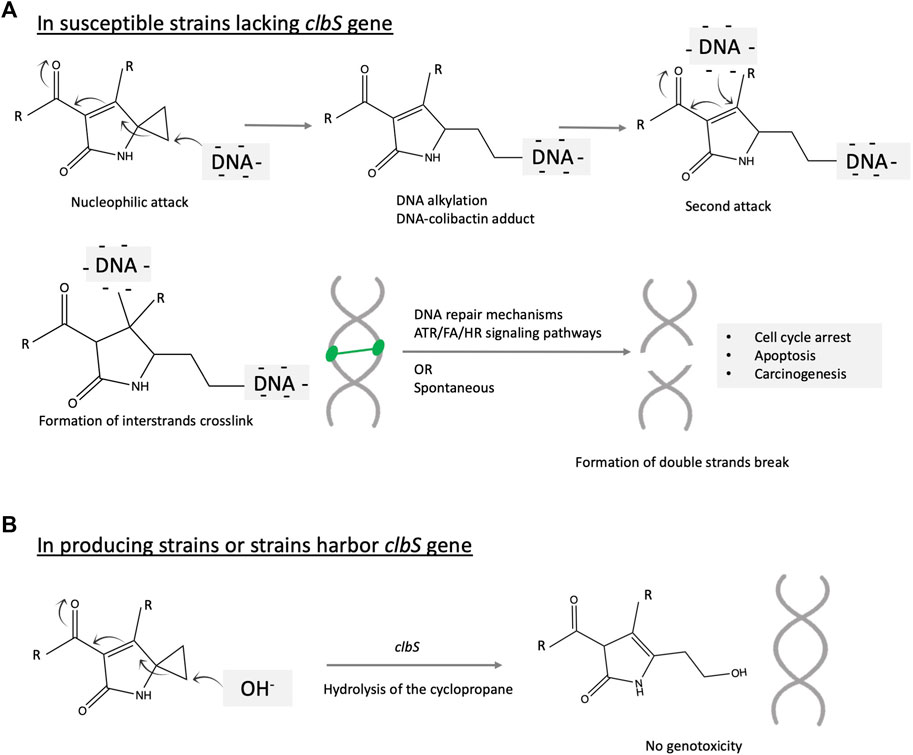
FIGURE 4. Illustration of the proposed mechanism of colibactin mutagenesis on the eukaryotic cell. The key structural scaffolds mediating this mechanism is the azospirobicyclic ring (warhead). The process is initiated by a nucleophilic attack on the electrophilic ring, which results in DNA alkylation and creates another electrophilic center subjected to a second alkylation reaction. Two alkylation events cross-link the DNA, which further activate multiple and complex DNA repair mechanisms including Ataxia telangiectasia mutated-and Rad3-related (ATR), Fanconi anemia (FA), homologous recombination (HR) signaling pathways. The DNA double strands break may be the product of the repair system or spontaneously formed. Accumulation of this mutation might initiate colorectal cancer.
Cross-linked DNA adducts trigger multiple DNA repair signaling pathways, leading to the formation of double-strand breaks and further carcinogenesis (Figure 4). Another research study proposed an alternative mechanism of formation of this characteristic DNA mutation involving cupper-mediated oxidative damage (Li et al., 2019). DNA alkylating agents constitute a major class of chemotherapeutics that cause cytotoxic DNA damage and collateral mutagenic damage, leading to cell death. This damaging process is defended by several cellular mechanisms, including base excision repair, mismatch repair, and direct DNA damage reversal. The response to alkylating agents required delicate coordination between repair pathways and show much variability between cells and individuals as reviewed (Fu et al., 2012). Alkylating agents attack the nitrogen or oxygen atoms in the DNA nitrogenous base. Alkylating agents could be monofunctional if they only have one active moiety and can attack a single site on DNA or bifunctional if they have two reactive moieties and can bind to two separate bases and consequently crosslink DNA. In 2016, Healy et al. validated the hypothesis that colibactin alkylates the DNA by cyclopropane ring opening via the formation of unsaturated imines. The authors synthesized 13 structural mimics of colibactin and tested their DNA alkylation potential (Healy et al., 2016a). In 2019, Wilson et al. used chemical synthesis coupled with an untargeted DNA adductomic approach to reveal DNA alkylation in HeLa cell lines incubated with a colibactin-producing strain of E. coli (Wilson et al., 2019). The same reaction is observed in vivo in mice (Wilson et al., 2019). The authors reveal that this covalent DNA alkylation is mediated by the electrophilic cyclopropane moiety of colibactin (Wilson et al., 2019). Other data show that colibactin cross-links DNA. The study employed NMR spectroscopy and bioinformatics-guided isotopic labeling to characterize the colibactin warhead (Vizcaino and Crawford 2015). The synthetic colibactin mimic contains a spiro bicyclic structure and can cross-links duplex DNA in vitro. The authors proposed that colibactin alkylates DNA via a homo-Michael addition reaction, turning colibactin into a second Michael receptor. Thereafter, intramolecular Michael addition will generate a DNA cross-link (Vizcaino and Crawford 2015). This hypothesis is supported by pieces of evidence. First, is the observation that precolibactins do not form much of the higher molecular weight adduct product when reacting with linearized plasmid DNA. Second, synthetic colibactin inspired by the biosynthetic information showed that the cyclopropane ring could be a target for thiol nucleophile attack and is essential to shearing the DNA (Healy et al., 2016a). Further results based on gel electrophoresis reveal that, upon artificial dimerization, these colibactin mimics cross-link the DNA.
Colibactin-induced events leading to inflammation and cancer
The interplay between genotoxic molecules such as colibactin and inflammation is not clear. It is speculated that DNA damage initiates tumor formation while inflammatory cytokines and cells promote it by creating a microenvironment that enables more DNA damage (Balkwill and Mantovani 2001; Ullman and Itzkowitz 2011) by, for example, inducing gene expression of genotoxic molecules. Previous studies show that microbial colonization enhances the development of cancer in mice that are genetically susceptible to inflammation (Ullman and Itzkowitz 2011). A shift in gut microbes was observed during inflammation, with unknown directionality, characterized by enrichment in genera of E. coli, Akkermansia, Shigella, and Bacteroides and a sharp reduction in Lachnospiraceae, Muribaculaceae, and Lactobacilli (Lang et al., 2020). Another study suggested that the microbial shift is associated with inflammation in the first place and not with cancer in the colitis-susceptible Il10−/− mouse strain. Among the sifted species, E. coli showed a 100-fold increase in colitis (Ullman and Itzkowitz 2011). Additionally, E. coli NC101 causes profound colitis in germ-free mice with 80% of the subjects developing adenocarcinoma (Ullman and Itzkowitz 2011), again questioning the directionality of the association. Another study used in vitro 3-D cell culture models to examine the host factors that might contribute to susceptibility to colibactin-induced cancer. The authors reported that the presence of the expression of mucin genes in forming the mucous layer that adheres to the intestinal epithelia decreased the genotoxic effect of colibactin. Moreover, the removal of the mucous layer in other models restored the genotoxicity (Reuter et al., 2018). Since inflammation is associated with degradation of the mucous layer and a decrease in the tight junction protein, also known as “leaky gut” that might be another factor to add to the colibactin–inflammation–cancer equation. Inflammation also allows leakage of microbes and other microbial products, not only colibactin, which flares up an even more intense immune response (Ahmad et al., 2017; Kidane et al., 2014). Of note is that the microbial composition in the presence of colibactin is altered, which might promote the abundance of pro-inflammatory bacteria. The interplay between colibactin-induced inflammation and cancer is illustrated in Figure 5.
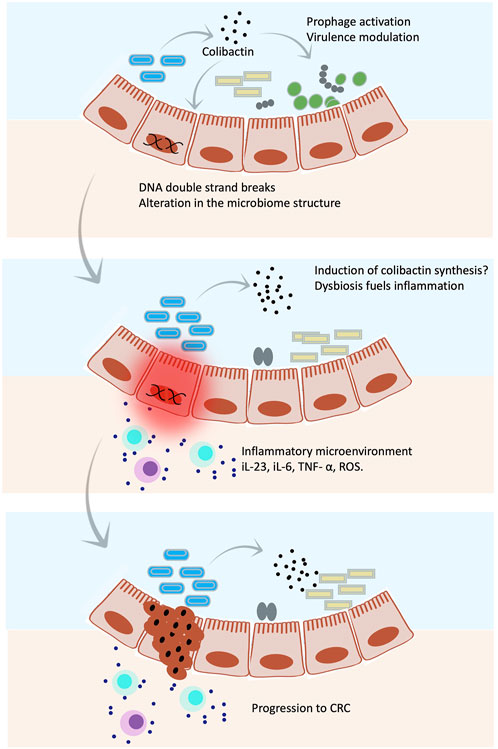
FIGURE 5. Colibactin hits targets in both eukaryotic and prokaryotic cells. Illustrations show an overall mechanism by which colibactin mediates host–microbe interaction. Colibactin shapes the gut microbial community by selective killing of some species through induction of prophages and/or stimulation of the virulence factor expression. Meanwhile, colibactin induces double strand breaks in host DNA. The altered microbial community together with the genetic abnormalities caused by colibactin triggers an inflammatory microenvironment, which might induce synthesis of more colibactin, leading to even greater damage that might result in CRC.
Colibactin hits targets of the prokaryotic cells controlling microbiome structure
The genomic organization of the colibactin cluster suggests a potential antimicrobial activity
Microbes are known to produce microbial toxins such as antibiotics to kill competing microbes and gain significant ecological competency. To protect themselves, the toxin-producing strains mostly harbor self-resistance genes. The presence of a self-resistance gene in the colibactin cluster suggests that colibactin is not only affecting cellular target in the human cell but also the bacterial cell. The biosynthetic gene cluster of colibactin ends with a resistance gene encoding a 170-amino acid protein named clbS. clbS encodes a cyclopropane hydrolase, resulting in the ring-opening of the DNA alkylating scaffolds, conferring self-resistance to producing strains (Tripathi et al., 2017) (Figure 4B). This gene is not only present in colibactin-producing strains but also in some other strains that lack the entire colibactin cluster, suggesting that this gene is transferable within the bacterial community by horizontal gene transfer to gain an ecological advantage. clbS-like genes with at least 50% sequence similarity have been found in members of the human gut microbiome, namely, Escherichia albertii, Kluyvera intestini, and Metakosakonia sp. and the honeybee microbiome, namely, Snodgrasella alvi (Silpe et al., 2022). Heterologous expression of these four clbS-like genes resulted in attenuated DNA damage and phage lysis when co-cultured with a colibactin-producing strain, suggesting that acquisition of clbS provides immunity against colibactin-induceddamage. Interestingly, clbS protects against colibactin-specific induction of prophages but provides no protection against other inducing agents such as MMC. This resistance is intra-cellular and is not shared between cells (Silpe et al., 2022). Other studies show that deletion of the clbS gene does not kill the strain but its growth will be dependent on the DNA repair mechanism (Bossuet-Greif et al., 2016).
Molecular targets behind colibactin activity on prokaryotes
Colibactin belongs to the PKS/NRPS natural products, which include many antibiotics such as daptomycin and β-lactams (Walsh 2004). Earlier studies suggested that colibactin might have antibiotic-like activity but the precise mechanism was not clear. A study showed that E. coli harboring a colibactin cluster exhibits growth inhibition against multi-resistant Staphylococcus aureus in the agar diffusion method and growth competition assay. This activity is observed in 95% of the tested strains, while the ΔclbP E. coli lost this activity (T. Faïs et al., 2016). The authors reported that this activity requires live culture, suggesting it is an inducible trait. Another study reported that inoculation of pregnant mice with a colibactin harboring strain resulted in a decreased abundance of firmicutes and a significant alteration in microbial diversity in pups, especially after 35 days of birth (Tronnet et al., 2020). Interestingly, the authors reported increased activity in DNA repair pathways, suggesting that colibactin not only modulates the structure of gut microbes but also their function (Tronnet et al., 2020). Another research study shows that the colibactin biosynthetic gene (clbA) might affect siderophore biosynthesis enhancing its producer fitness (Martin et al., 2013).
In 2022, the Balskus group revealed an interesting mechanism beyond the observed activities of colibactin in shaping the microbial population (Silpe et al., 2022). They discovered that colibactin-induced damage activates SOS signaling pathways, leading to the induction of prophages exerting lethal action on their host bacteria. The authors show that the effect of colibactin on prophages extends to a wide range of phages residing in phylogenetically distinct bacteria, including Salmonella typhimurium, Staphylococcus aureus, Citrobacter rodentium, and Enterococcus faecium (Silpe et al., 2022). Interestingly, there is a significant increase in Shiga toxin production upon co-culture of C. rodentium (harboring Stx genes) with pks + E. coli (Silpe et al., 2022). Induction of prophages enables selective lethal action against other members of the microbial community, providing an elegant ecological advantage and enhancing the competitiveness of the producing strain.
Molecular mechanism of prophage induction following DNA damage in the host
Lytic activation of prophages in bacterial lysogens is normally induced by DNA damage, such as after exposure to UV radiation or DNA damaging agents such as mitomycin C (MMC) (Lee et al., 2006; Humphrey, Stanton, and Jensen 1995). In addition to DNA damage, other internal and external stimuli might lead to prophage induction, such as pH, heat, reactive oxygen species (ROS), or even spontaneous induction (Nanda, Thormann, and Frunzke 2015). Maintenance of lysogeny is tightly controlled by the activity of the λ CI repressor proteins. Two dimers of CI bind to the operator regions of phage genes and suppress their expression (Hochschild 2002; Little, Shepley, and Wert 1999). The CI protein is a molecular switch that controls lysogeny in lambda phages. CI binds to operator regions of phage genes and represses their expression while inducing its own expression. DNA damage activates the master regulator RecA, resulting in de-repression of SOS genes mediated by cleavage of their transcriptional repressor LexA (Gimble and Sauer 1989; Little and Michalowski 2010; Thomason et al., 2021). Activation of SOS genes and RecA leads to the cleavage of the CI protein resulting in the de-repression of phage genes to enter the lytic cycle (Hochschild 2002; Little, Shepley, and Wert 1999). This molecular mechanism is simply illustrated in Figure 6. Genetic mutations affecting dimerization of lambda repressor increase the rate of its cleavage, while repressor dimer stabilized by covalent disulfide bond resists cleavage (Gimble and Sauer 1989). An exception is the prophages in Salmonella genomes, which uses another inductive strategy based on the use of anti-repressor molecules that bind the dimer without cleavage, leading to its dissociation from the DNA. Production of these anti-repressors is under the direct control of LexA (Lemire, Figueroa-Bossi, and Bossi 2011).
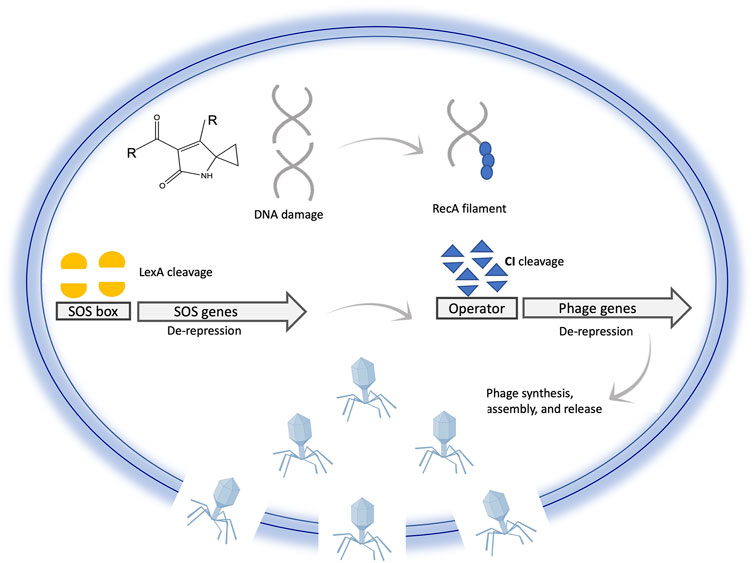
FIGURE 6. Illustration of prophage activation following DNA damage inside a bacterial cell. DNA damage induces cleavage of phage repressor genes mediated by SOS signaling. Cleavage of the phage repressor leads to exciton of prophage from the bacterial chromosome, synthesis of phage-related genes and proteins, assembly, cell lysis, and phage release. This illustration is simplified to communicate the main idea; for example CI binds to two distant regions and not only to one site as shown for simplicity of the illustration.
Prophages contribute to the ecological fitness of harboring strains
Viral genes make up to 20% of microbial genomes, either as prophage-like elements, phage remnants, or fully prophages that could be induced into the lytic life cycle (Wang et al., 2010). These prophages contribute to the ecological fitness and virulence of the lysogens. Reports show that lysogens can use prophages as a survival strategy (Bossi et al., 2003; Gama et al., 2013). For example, in a mixed culture, Salmonella enterica induces its prophages, leading to the killing of some self-cells while the others are converted back to lysogens. The released prophages are then used as a weapon to kill competitors although it results in some self-destruction. This strategy will also benefit the phage by spreading its genes (Bossi et al., 2003; Gama et al., 2013). Another example is the induction of prophages in Streptococcus pneumoniae to wipe off S. aureus via the production of hydrogen peroxide-inducing SOS genes. Induction of SOS leads to activation of prophages in S. aureus, which are mostly lysogens while S. pneumoniae are immune, giving S. pneumoniae an ecological advantage to dominate (Selva et al., 2009; Pericone et al., 2003). A study shows that deletion of nine cryptic prophages from E-coli suppresses fitness by rendering the bacteria more susceptible to sublethal concentrations of β-lactam and quinolone antibiotics. Moreover, this deletion rendered the bacteria less able to form biofilms and more prone to damage caused by acid or osmotic pressure (Wang et al., 2010). A study shows that prophages enable biofilm formation in Pseudomonas aeruginosa, contributing to its virulence and. when cells die in the biofilm, the prophage is converted to a super infective form (Rice et al., 2009). Biofilm is a group behavior in which bacterial cells adhere to one another and solid surfaces using a sticky matrix made of extracellular proteins, carbohydrates, and DNA molecules (Madsen et al., 2012). The formation of biofilm is a virulence strategy that helps enable bacteria to resist antibiotics and host immune defense. Prophage induction in a biofilm will lead to the accumulation of extracellular DNA promoting horizontal gene transfer and enhancing diversification of the microbial community within the biofilm (Molin and Tolker-Nielsen 2003). The role of prophages on biofilm formation and further consequences on host fitness is reviewed (Nanda, Thormann, and Frunzke 2015). The production of toxins is a crucial virulence trait for pathogens (Brown et al., 2006). Some toxins are encoded by bacteriophages, including cholera toxin (Ctx) and Shiga toxins (Stx), required for the virulence of Vibrio cholerae and E. coli, respectively (Waldor and Mekalanos 1996; Neely and Friedman 1998). For example, Shiga toxin (Stx) in enterohemorragic E. coli enables bacterial cells to attach and colonize the gut epithelia (Robinson et al., 2006). Similarly, some cells of E. faecalis V583 and Streptococcus mitis induce prophages to express phage genes to help the remaining cells adhere to human platelets, leading to systemic infection (Matos et al., 2013; Seo et al., 2010). This regulatory activity of microbiome-secreted molecules has been noted before. For example, quinolone antibiotics increase the production of Stx toxin via activation of prophages via SOS response (Pleguezuelos-Manzano et al., 2020). Co-culture of colibactin with Citrobacter rodentium increases the production of Stx (Silpe et al., 2022). Another interesting activity of colibactin was reported by Marcq et al. (2014). Driven by the association between pks+ E. coli strains and septicemia, the authors investigated the effect of colibactin-harboring strains on lymphopenia and sepsis. The results show that colibactin-producing E. coli causes the characteristic double-strand break in the DNA of lymphocytes, leading to an exaggerated lymphopenia and subsequent low survival rate from bacteremia in mice (Marcq et al., 2014).
Conclusion
Colibactin demonstrates the potential of microbiome secreted chemistry to hit multiple targets spanning prokaryotic and eukaryotic cell machinery. It is interesting to speculate that colibactin gives the producing species an ecological advantage in occupying specific niches such as the human gut by controlling other competitive microbes. Understanding how precisely colibactin-producing strains alter gut microbiome composition and their further indirect impact on human health and diseases seems exciting. Of note, we still do not appreciate all the microbiome species that we host in our bodies or their cryptic genes and encoded chemistry. Revealing the microbiome products that exert microbial control and affect population dynamics will help to advance better strategies to tackle the antibiotic resistance crisis. Microbiome chemistry is certainly a new uncharted Frontier for understanding mediators of human conditions and developing innovative therapeutics or diagnostic biomarkers. For example, delivering colibactin mimics or genes to induce the lethal killing of a cancer cell or as an antimicrobial for life-threatening biofilm infection.
Is colibactin a harmful molecule?
Host-associated microbes produce molecules that help to increase their ecological fitness and competitiveness and to establish a long-term, mostly symbiotic, relationship with the host. From an ecological perspective, we can imagine colibactin as a beneficial microbial product that is produced to shape the microbial community and confer beneficial traits to the host, such as decreasing inflammation as long as the mutualistic relationship is established. Evidence for this hypothesis came from the research on Nissle 1917. The efficacy of probiotic E-coli Nissle 1917 in treating colitis and preventing further remission is even comparable to that of mesalazine, the gold standard drug in the treatment of colitis (Kruis et al., 2004). The mechanism of action involves modulation of cytokine expression. Interestingly, further investigation on Nissle 1917 activity suggests that its beneficial anti-inflammatory activity is dependent on the expression of the colibactin biosynthetic pathway (Olier et al., 2012). Knockout strains that lost the ability to express the colibactin cluster not only lost the anti-inflammatory activity but also resulted in more inflammation (Olier et al., 2012). Moreover, inflammation is thought to be a triggering signal for further expression of the colibactin gene cluster. Chronic inflammation and accumulation of DNA damage will eventually lead to cancer development. The question now is, do these microbiome strains, or more specifically colibactin, initiate cancer or evolved to combat cancer and act as an immunomodulin based on the activity profile of E. coli Nissle 1917? One can postulate that it produces colibactin to kill transformative cells at the beginning of inflammation in a similar way to chemotherapeutic agents and more inflammation triggers more synthesis of the mysterious molecule(s). This bidirectional dependency is puzzling and poses an outstanding question on the directionality of microbiome diseases association and warns of the urgent need for a holistic deep understanding of the microbiome secreted chemistry evolved to hit multiple cellular targets under very tight and complex regulation resulting in a variety of negative and positive outcomes for the host. Another interesting hypothesis is that the colibactin biosynthetic cluster encodes or supplies precursor molecules with immunomodulatory activity as originally proposed (Olier et al., 2012). Support for this hypothesis comes from the strong association between colitis and microbial dysbiosis, so it is interesting to speculate on the role of colibactin in shaping microbiome structure and whether it may restore the balanced composition to the pre-colitis status. However, the members of Enterobacteriacea are often considered proinflammatory, and some reports show significant enrichment in Enterobacteriacea with the onset of gastrointestinal inflammatory conditions (Garrett et al., 2010) and the dominant presence of adherent-invasive strains of E. coli such as LF82 (Carvalho et al., 2008), belonging to the same phylogenetic group as Nissle 1917, although it lacks the colibactin genomic cluster. Taken together, our knowledge of microbiome mediators and their evolved function is still in its infancy. Despite the explosion of research on the microbiome, the diversity and richness in species of the microbiome, their cryptic genes, and secreted molecules are still a dark matter.
A proposed model for colibactin–microbe–host interaction and health outcome
Host-associated microbes produce toxic molecules with the primary function of killing competitors and gaining an ecological advantage to dominate a particular niche. However, these toxins might also hit molecular targets in the host. Considering that the priority function of colibactin is microbial related, we can propose that the reasonable order of events leading to CRC might be as follows: 1) colibactin shapes microbial composition in the gut to modulate inflammation, especially during microbial dysbiosis; 2) chronic dysbiosis triggers inflammation, which leads to leaky gut; 3) colibactin diffuses inside the eukaryotic cell and causes DNA damage eliciting the repair mechanism; 4) chronic inflammatory microenvironment further advances the microbial dysbiosis, and gut permeability resulted in more diffusion of colibactin to eukaryotic cells (or maybe an induction of its synthesis); and 5) under high load of colibactin, accumulation of DNA damage might occur leading to colorectal cancer (Figure 7). This situation gets worse with a genetic predisposition to inflammation or defect in the DNA repair pathways. Considering this model, chronic microbial dysbiosis and inflammation are crucial for colibactin-induced colorectal cancer and might serve as a risk factor to predicting CRC, especially if this is combined with genetic disorders in genes related to the DNA repair pathways or immune function. This understanding is not only helpful in predicting risks but also in taking preventative measures such as implementing an anti-inflammatory diet for high-risk groups to decrease the incidence of CRC.
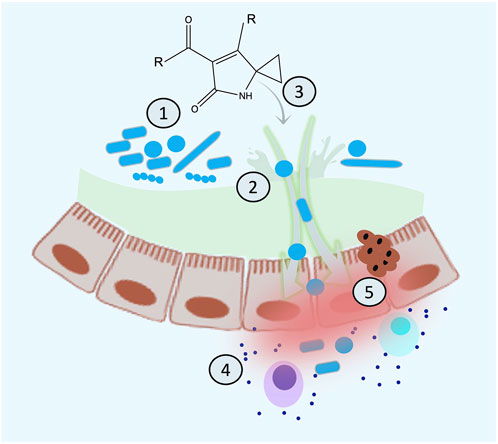
FIGURE 7. Proposed mechanistic model for the sequence of the events of colibactin-induced cancer. (A) Colibactin induces dysbiosis, (B) dysbiosis results in inflammation and leaky gut, (C) colibactin diffuses inside the cell resulting in DNA damage, (D) chronic inflammation increases the load of colibactin inside the cell and more DNA damage, and (E) accumulating DNA damage leads to CRC.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmad, R., Sorrell, M. F., Batra, S. K., Dhawan, P., Singh, A. B., and Dhawan, P. (2017). Gut permeability and mucosal inflammation: Bad, good or context dependent. Mucosal Immunol. 10 (2), 307–317. doi:10.1038/mi.2016.128
Arthur, Janelle C., Perez-Chanona, Ernesto, Mühlbauer, Marcus, Tomkovich, Sarah, Uronis, Joshua M., Fan, Ting-Jia, et al. (2012). Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338 (6103), 120–123. doi:10.1126/science.1224820
Balkwill, F., and Mantovani, A. (2001). Inflammation and cancer: Back to virchow? Lancet (London, Engl. 357 (9255), 539–545. doi:10.1016/S0140-6736(00)04046-0
Balskus, Emily P. (2015). Colibactin: Understanding an elusive gut bacterial genotoxin. Nat. Prod. Rep. 32 (11), 1534–1540. doi:10.1039/c5np00091b
Bian, Xiaoying, Fu, Jun, Plaza, Alberto, Herrmann, Jennifer, Pistorius, Dominik, Zhang, Youming, et al. (2013). In vivo evidence for a prodrug activation mechanism during colibactin maturation. Chembiochem 14 (10), 1194–1197. doi:10.1002/cbic.201300208
Bian, Xiaoying, Plaza, Alberto, Zhang, Youming, and Müller, Rolf (2015). Two more pieces of the colibactin genotoxin puzzle from Escherichia coli show incorporation of an unusual 1-aminocyclopropanecarboxylic acid moiety. Chem. Sci. 6 (5), 3154–3160. doi:10.1039/C5SC00101C
Bode, Helge B. (2015). The microbes inside us and the race for colibactin. Angew. Chem. Int. Ed. Engl. 54 (36), 10408–10411. doi:10.1002/anie.201505341
Bondarev, Vladimir, Richter, Michael, Romano, Stefano, Piel, Jörn, Schwedt, Anne, and Schulz-Vogt, Heide N. (2013). The genus Pseudovibrio contains metabolically versatile bacteria adapted for symbiosis. Environ. Microbiol. 15 (7), 2095–2113. doi:10.1111/1462-2920.12123
Bossi, Lionello, Fuentes, Juan A., Guido, Mora, and Figueroa-Bossi, Nara (2003). Prophage contribution to bacterial population dynamics. J. Bacteriol. 185 (21), 6467–6471. doi:10.1128/jb.185.21.6467-6471.2003
Bossuet-Greif, Nadège, Dubois, Damien, Petit, Claude, Tronnet, Sophie, Martin, Patricia, Bonnet, Richard, et al. (2016). Escherichia coli ClbS is a colibactin resistance protein. Mol. Microbiol. 99 (5), 897–908. doi:10.1111/mmi.13272
Brotherton, Carolyn A., and Balskus, Emily P. (2013). A prodrug resistance mechanism is involved in colibactin biosynthesis and cytotoxicity. J. Am. Chem. Soc. 135 (9), 3359–3362. doi:10.1021/ja312154m
Brotherton, Carolyn A., Wilson, Matthew, Byrd, Gary, and Balskus, Emily P. (2015). Isolation of a metabolite from the pks island provides insights into colibactin biosynthesis and activity. Org. Lett. 17 (6), 1545–1548. doi:10.1021/acs.orglett.5b00432
Brown, Nat F., Wickham, Mark E., Coombes, Brian K., and Finlay, B. Brett (2006). Crossing the line: Selection and evolution of virulence traits. PLoS Pathog. 2 (5), e42. doi:10.1371/journal.ppat.0020042
Buc, Emmanuel, Dubois, Damien, Sauvanet, Pierre, Raisch, Jennifer, Delmas, Julien, Darfeuille-Michaud, Arlette, et al. (2013). High prevalence of mucosa-associated E. Coli producing cyclomodulin and genotoxin in colon cancer. PloS One 8 (2), e56964. doi:10.1371/journal.pone.0056964
Carvalho, Frédéric A., Barnich, Nicolas, Sauvanet, Pierre, Darcha, Claude, Gelot, Agathe, and Darfeuille-Michaud, Arlette (2008). Crohn’s disease-associated Escherichia coli LF82 aggravates colitis in injured mouse colon via signaling by flagellin. Inflamm. Bowel Dis. 14 (8), 1051–1060. doi:10.1002/ibd.20423
Cougnoux, Antony, Gibold, Lucie, Robin, Frederic, Dubois, Damien, Pradel, Nathalie, Darfeuille-Michaud, Arlette, et al. (2012). Analysis of structure-function relationships in the colibactin-maturating enzyme ClbP. J. Mol. Biol. 424 (3–4), 203–214. doi:10.1016/j.jmb.2012.09.017
de Vos, Willem M., Herbert, Tilg, Van Hul, Matthias, and Cani, Patrice D. (2022). Gut microbiome and health: Mechanistic insights. Gut 71 (5), 1020–1032. doi:10.1136/gutjnl-2021-326789
Dhanaraju, Rajkumar, and Rao, Desirazu N. (2022). The human microbiome: An acquired organ? Reson. 27 (2), 247–272. doi:10.1007/s12045-022-1312-7
Dubois, Damien, Baron, Olivier, Antony, Cougnoux, Delmas, Julien, Pradel, Nathalie, Boury, Michèle, et al. (2011). ClbP is a prototype of a peptidase subgroup involved in biosynthesis of nonribosomal peptides. J. Biol. Chem. 286 (41), 35562–35570. doi:10.1074/jbc.M111.221960
Engel, Philipp, Vizcaino, Maria I., and Crawford, Jason M. (2015). Gut symbionts from distinct hosts exhibit genotoxic activity via divergent colibactin biosynthesis pathways. Appl. Environ. Microbiol. 81 (4), 1502–1512. doi:10.1128/AEM.03283-14
Faïs, T., Cougnoux, A., Dalmasso, G., Laurent, F., Delmas, J., and Bonnet, R. (2016). Antibiotic activity of Escherichia coli against multiresistant Staphylococcus aureus. Antimicrob. Agents Chemother. 60 (11), 6986–6988. doi:10.1128/AAC.00130-16
Faïs, Tiphanie, Delmas, Julien, Barnich, Nicolas, Bonnet, Richard, and Dalmasso, Guillaume (2018). Colibactin: More than a new bacterial toxin. Toxins 10 (4), E151. doi:10.3390/toxins10040151
Fu, Dragony, Calvo, Jennifer A., and Samson, Leona D. (2012). Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer 12 (2), 104–120. doi:10.1038/nrc3185
Gama, João Alves, Reis, Ana Maria, Domingues, Iolanda, Mendes-Soares, Helena, Matos, Ana Margarida, and Dionisio, Francisco (2013). Temperate bacterial viruses as double-edged swords in bacterial warfare. PloS One 8 (3), e59043. doi:10.1371/journal.pone.0059043
Garrett, Wendy S., Gallini, Carey A., Yatsunenko, Tanya, Michaud, Monia, DuBois, Andrea, Delaney, Mary L., et al. (2010). Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8 (3), 292–300. doi:10.1016/j.chom.2010.08.004
Gimble, F. S., and Sauer, R. T. (1989). Lambda repressor mutants that are better substrates for RecA-mediated cleavage. J. Mol. Biol. 206 (1), 29–39. doi:10.1016/0022-2836(89)90521-4
Healy, Alan R., Herman, Nikolayevskiy, Patel, Jaymin R., Crawford, Jason M., and Herzon, Seth B. (2016a). A mechanistic model for colibactin-induced genotoxicity. J. Am. Chem. Soc. 138 (48), 15563–15570. doi:10.1021/jacs.6b10354
Healy, Alan R., Vizcaino, Maria I., Crawford, Jason M., and Herzon, Seth B. (2016b). Convergent and modular synthesis of candidate precolibactins. Structural revision of precolibactin A. J. Am. Chem. Soc. 138 (16), 5426–5432. doi:10.1021/jacs.6b02276
Healy, Alan R., Wernke, Kevin M., Kim, Chung Sub, Lees, Nicholas R., Crawford, Jason M., and Herzon, Seth B. (2019). Synthesis and reactivity of precolibactin 886. Nat. Chem. 11 (10), 890–898. doi:10.1038/s41557-019-0338-2
Hirayama, Yuichiro, Tsunematsu, Yuta, Yoshikawa, Yuko, Tamafune, Ryota, Matsuzaki, Nobuo, Iwashita, Yuji, et al. (2019). Activity-based probe for screening of high-colibactin producers from clinical samples. Org. Lett. 21 (12), 4490–4494. doi:10.1021/acs.orglett.9b01345
Hochschild, Ann. (2002). The λ switch: CI closes the gap in autoregulation. Curr. Biol. 12 (3), R87–R89. doi:10.1016/S0960-9822(02)00667-X
Hsiao, Elaine Y., McBride, Sara W., Hsien, Sophia, Gil, Sharon, Hyde, Embriette R., Tyler, McCue, et al. (2013). Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155 (7), 1451–1463. doi:10.1016/j.cell.2013.11.024
Humphrey, S. B., Stanton, T. B., and Jensen, N. S. (1995). Mitomycin C induction of bacteriophages from serpulina hyodysenteriae and serpulina innocens. FEMS Microbiol. Lett. 134 (1), 97–101. doi:10.1111/j.1574-6968.1995.tb07921.x
Jiang, Yindi, Stornetta, Alessia, Villalta, Peter W., Wilson, Matthew R., Boudreau, Paul D., Zha, L., et al. (2019). Reactivity of an unusual amidase may explain colibactin’s DNA cross-linking activity. J. Am. Chem. Soc. 141 (29), 11489–11496. doi:10.1021/jacs.9b02453
Kidane, Dawit, Chae, Wook Jin, Czochor, Jennifer, Eckert, Kristin A., Glazer, Peter M., Bothwell, Alfred L. M., et al. (2014). Interplay between DNA repair and inflammation, and the link to cancer. Crit. Rev. Biochem. Mol. Biol. 49 (2), 116–139. doi:10.3109/10409238.2013.875514
Kim, Janice J., Bridle, Byram W., Ghia, Jean-Eric, Wang, Huaqing, Syed, Shahzad N., Manocha, Marcus M., et al. (2013). Targeted inhibition of serotonin type 7 (5-HT7) receptor function modulates immune responses and reduces the severity of intestinal inflammation. J. Immunol. 190 (9), 4795–4804. doi:10.4049/jimmunol.1201887
Kruis, W., Fric, P., Pokrotnieks, J., Lukás, M., Fixa, B., Kascák, M., et al. (2004). Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53 (11), 1617–1623. doi:10.1136/gut.2003.037747
Lang, Michaela, Baumgartner, Maximilian, Rożalska, Aleksandra, Frick, Adrian, Riva, Alessandra, Jarek, Michael, et al. (2020). Crypt residing bacteria and proximal colonic carcinogenesis in a mouse model of lynch syndrome. Int. J. Cancer 147 (8), 2316–2326. doi:10.1002/ijc.33028
Lee, Young-Ju, Park, Su-Jung, Ciccone, Samantha L. M., Kim, Chong-Rak, and Lee, Suk-Hee (2006). An in vivo analysis of MMC-induced DNA damage and its repair. Carcinogenesis 27 (3), 446–453. doi:10.1093/carcin/bgi254
Lemire, Sébastien, Figueroa-Bossi, Nara, and Bossi, Lionello (2011). Bacteriophage crosstalk: Coordination of prophage induction by trans-acting antirepressors. PLoS Genet. 7 (6), e1002149. doi:10.1371/journal.pgen.1002149
Li, Zhi Shan, Schmauss, Claudia, Cuenca, Abigail, Ratcliffe, Elyanne, and Gershon, Michael D. (2006). Physiological modulation of intestinal motility by enteric dopaminergic neurons and the D2 receptor: Analysis of dopamine receptor expression, location, development, and function in wild-type and knock-out mice. J. Neurosci. 26 (10), 2798–2807. doi:10.1523/JNEUROSCI.4720-05.2006
Li, Zhong-Rui, Li, Jie, Cai, Wenlong, Lai, Jennifer Y. H., McKinnie, Shaun M. K., Zhang, Wei-Peng, et al. (2019). Macrocyclic colibactin induces DNA double-strand breaks via copper-mediated oxidative cleavage. Nat. Chem. 11 (10), 880–889. doi:10.1038/s41557-019-0317-7
Li, Zhong-Rui, Li, Jie, Gu, Jin-Ping, Lai, Jennifer Y. H., Duggan, Brendan M., Zhang, Wei-Peng, et al. (2016). Divergent biosynthesis yields a cytotoxic aminomalonate-containing precolibactin. Nat. Chem. Biol. 12 (10), 773–775. doi:10.1038/nchembio.2157
Li, Zhong-Rui, Li, Yongxin, Lai, Jennifer Y. H., Tang, Jianqiang, Wang, Bin, Lu, Liang, et al. (2015). Critical intermediates reveal new biosynthetic events in the enigmatic colibactin pathway. Chembiochem 16 (12), 1715–1719. doi:10.1002/cbic.201500239
Little, John W., Shepley, Donald P., and Wert, David W. (1999). Robustness of a gene regulatory circuit. EMBO J. 18 (15), 4299–4307. doi:10.1093/emboj/18.15.4299
Little, John W., and Michalowski, Christine B. (2010). Stability and instability in the lysogenic state of phage lambda. J. Bacteriol. 192 (22), 6064–6076. doi:10.1128/JB.00726-10
Luo, Yi, Ruan, Li-Fang, Zhao, Chang-Ming, Wang, Cheng-Xian, Peng, Dong-Hai, and Sun, Ming (2011). Validation of the intact zwittermicin A biosynthetic gene cluster and discovery of a complementary resistance mechanism in Bacillus thuringiensis. Antimicrob. Agents Chemother. 55 (9), 4161–4169. doi:10.1128/AAC.00111-11
Madsen, Jonas Stenløkke, Burmølle, Mette, Hansen, Lars Hestbjerg, and Sørensen, Søren Johannes (2012). The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 65 (2), 183–195. doi:10.1111/j.1574-695X.2012.00960.x
Marcq, Ingrid, Martin, Patricia, Payros, Delphine, Cuevas-Ramos, Gabriel, Boury, Michèle, Watrin, Claude, et al. (2014). The genotoxin colibactin exacerbates lymphopenia and decreases survival rate in mice infected with septicemic Escherichia coli. J. Infect. Dis. 210 (2), 285–294. doi:10.1093/infdis/jiu071
Martin, Patricia, Marcq, Ingrid, Magistro, Giuseppe, Penary, Marie, Garcie, Christophe, Payros, Delphine, et al. (2013). Interplay between siderophores and colibactin genotoxin biosynthetic pathways in Escherichia coli. PLoS Pathog. 9 (7), e1003437. doi:10.1371/journal.ppat.1003437
Matos, R. C., Lapaque, N., Rigottier-Gois, L., Debarbieux, L., Meylheuc, T., Gonzalez-Zorn, B., et al. (2013). Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet. 9 (6), e1003539. doi:10.1371/journal.pgen.1003539
Molin, Søren, and Tolker-Nielsen, Tim (2003). Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14 (3), 255–261. doi:10.1016/s0958-1669(03)00036-3
Moretti, C., Hosni, T., Vandemeulebroecke, K., Brady, C., De Vos, P., Buonaurio, R., et al. (2011). Erwinia Oleae sp. Nov., isolated from olive knots caused by Pseudomonas savastanoi pv. Savastanoi. Int. J. Syst. Evol. Microbiol. 61 (11), 2745–2752. doi:10.1099/ijs.0.026336-0
Nanda, Arun M., Thormann, Kai, and Julia, Frunzke (2015). Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J. Bacteriol. 197 (3), 410–419. doi:10.1128/jb.02230-14
Neely, Melody N., and Friedman, David I. (1998). Arrangement and functional identification of genes in the regulatory region of lambdoid phage H-19B, a carrier of a shiga-like toxin. Gene 223 (1–2), 105–113. doi:10.1016/s0378-1119(98)00236-4
Nougayrède, Jean-Philippe, Homburg, Stefan, Taieb, Frédéric, Boury, Michèle, Brzuszkiewicz, Elzbieta, Gottschalk, Gerhard, et al. (2006). Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Sci. (New York, N.Y.) 313 (5788), 848–851. doi:10.1126/science.1127059
Olier, M., Marcq, I., Salvador-Cartier, C., Secher, T., Ulrich, D., Boury, M., et al. (2012). Genotoxicity of Escherichia coli Nissle 1917 strain cannot Be dissociated from its probiotic activity. Gut Microbes 3 (6), 501–509. doi:10.4161/gmic.21737
Pericone, Christopher D., Park, Sunny, James, A. Imlay, and Weiser, Jeffrey N. (2003). Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the fenton reaction. J. Bacteriol. 185 (23), 6815–6825. doi:10.1128/jb.185.23.6815-6825.2003
Pleguezuelos-Manzano, Cayetano, Puschhof, Jens, Arne van Hoeck, , Wood, Henry M., Nomburg, Jason, Gurjao, Carino., et al. (2020). Mutational signature in colorectal cancer caused by genotoxic Pks+ E. Coli. Nature 580 (7802), 269–273. doi:10.1038/s41586-020-2080-8
Reuter, Christian, Alzheimer, Mona, Walles, Heike, and Oelschlaeger, Tobias A. (2018). An adherent mucus layer attenuates the genotoxic effect of colibactin. Cell. Microbiol. 20 (2), e12812. doi:10.1111/cmi.12812
Rice, Scott A., Tan, Chuan Hao, Kung, Vanderlene, Woo, Jerry, Tay, Martin, Hauser, Alan, et al. (2009). The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 3 (3), 271–282. doi:10.1038/ismej.2008.109
Robinson, Cory M., Sinclair, James F., Smith, Michael J., and O’Brien, Alison D. (2006). Shiga toxin of enterohemorrhagic Escherichia coli type O157: H7 promotes intestinal colonization. Proc. Natl. Acad. Sci. U. S. A. 103 (25), 9667–9672. doi:10.1073/pnas.0602359103
Schultz, Michael (2008). Clinical use of E. Coli Nissle 1917 in inflammatory bowel disease. Inflamm. Bowel Dis. 14 (7), 1012–1018. doi:10.1002/ibd.20377
Selva, Laura, Viana, David, Regev-Yochay, Gili, Trzcinski, Krzysztof, Lasa, Inigo, Novick, Richard P., et al. (2009). Killing niche competitors by remote-control bacteriophage induction. Proc. Natl. Acad. Sci. U. S. A. 106 (4), 1234–1238. doi:10.1073/pnas.0809600106
Seo, Ho Seong, Xiong, Yan Q., Mitchell, Jennifer, Seepersaud, Ravin, Bayer, Arnold S., and Sullam, Paul M. (2010). Bacteriophage lysin mediates the binding of Streptococcus mitis to human platelets through interaction with fibrinogen. PLoS Pathog. 6 (8), e1001047. doi:10.1371/journal.ppat.1001047
Sharon, Gil, Garg, Neha, Debelius, Justine, Knight, Rob, Dorrestein, Pieter C., and Mazmanian, Sarkis K. (2014). Specialized metabolites from the microbiome in health and disease. Cell Metab. 20 (5), 719–730. doi:10.1016/j.cmet.2014.10.016
Silpe, Justin E., Joel, W. H., Wong, Siân V. Owen, Balskus, Emily P., and Balskus, E. P. (2022). The bacterial toxin colibactin triggers prophage induction. Nature 603 (7900), 315–320. doi:10.1038/s41586-022-04444-3
Tang, Jian-Wei, Liu, Xin, Ye, Wei, Li, Zhong-rui, and Qian, Pei-Yuan (2022). Biosynthesis and bioactivities of microbial genotoxin colibactins. Nat. Product. Rep. 39 (5), 991–1014. doi:10.1039/D1NP00050K
Thomason, Lynn C., Schiltz, Carl J., Court, Carolyn, Hosford, Christopher J., Chappie, Myfanwy C. Adams, Joshua S., Court, Donald L., et al. (2021). Bacteriophage λ RexA and RexB functions assist the transition from lysogeny to lytic growth. Mol. Microbiol. 116 (4), 1044–1063. doi:10.1111/mmi.14792
Tichenor, Mark S., and Boger, Dale L. (2008). Yatakemycin: Total synthesis, DNA alkylation, and biological properties. Nat. Prod. Rep. 25 (2), 220–226. doi:10.1039/b705665f
Tripathi, Prabhanshu, Shine, Emilee E., Healy, Alan R., Kim, Chung Sub, Herzon, Seth B., Bruner, Steven D., et al. (2017). ClbS is a cyclopropane hydrolase that confers colibactin resistance. J. Am. Chem. Soc. 139 (49), 17719–17722. doi:10.1021/jacs.7b09971
Tronnet, Sophie, Floch, Pauline, Lucarelli, Laetitia, Gaillard, Deborah, Martin, Patricia, Serino, Matteo, et al. (2020). The genotoxin colibactin shapes gut microbiota in mice. MSphere 5 (4), e00589–20. doi:10.1128/mSphere.00589-20
Ullman, Thomas A., and Itzkowitz, Steven H. (2011). Intestinal inflammation and cancer. Gastroenterology 140 (6), 1807–1816. doi:10.1053/j.gastro.2011.01.057
Vizcaino, Maria I., and Crawford, Jason M. (2015). The colibactin warhead crosslinks DNA. Nat. Chem. 7 (5), 411–417. doi:10.1038/nchem.2221
Vizcaino, Maria I., Engel, Philipp, Trautman, Eric, Crawford, Jason M., and Crawford, J. M. (2014). Comparative metabolomics and structural characterizations illuminate colibactin pathway-dependent small molecules. J. Am. Chem. Soc. 136 (26), 9244–9247. doi:10.1021/ja503450q
Waldor, Matthew K., and Mekalanos, John J. (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272 (5270), 1910–1914. doi:10.1126/science.272.5270.1910
Walsh, Christopher T. (2004). Polyketide and nonribosomal peptide antibiotics: Modularity and versatility. Sci. (New York, N.Y.) 303 (5665), 1805–1810. doi:10.1126/science.1094318
Wang, Xiaoxue, Kim, Younghoon, Ma, Qun, Hong, Seok Hoon, Pokusaeva, Karina, Sturino, Joseph M., et al. (2010). Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 1 (1), 147–149. doi:10.1038/ncomms1146
Wernke, Kevin M., Xue, Mengzhao, Tirla, Alina, Kim, Chung Sub, Crawford, Jason M., and Herzon, Seth B. (2020). Structure and bioactivity of colibactin. Bioorg. Med. Chem. Lett. 30 (15), 127280. doi:10.1016/j.bmcl.2020.127280
Williams, Peyton C., Wernke, Kevin M., Tirla, Alina, and Herzon, Seth B. (2020). Employing chemical synthesis to study the structure and function of colibactin, a ‘dark matter’ metabolite. Nat. Prod. Rep. 37 (11), 1532–1548. doi:10.1039/d0np00072h
Wilson, Matthew R., Jiang, Yindi, Villalta, Peter W., Stornetta, Alessia, Boudreau, Paul D., Carrá, Andrea, et al. (2019). The human gut bacterial genotoxin colibactin alkylates DNA. Science 363 (6428), eaar7785. doi:10.1126/science.aar7785
Wilson, Matthew R., Zha, Li, and Balskus, Emily P. (2017). Natural product discovery from the human microbiome. J. Biol. Chem. 292 (21), 8546–8552. doi:10.1074/jbc.R116.762906
Xue, Mengzhao, Kim, Chung Sub, Healy, Alan R., Wernke, Kevin M., Wang, Zhixun, Frischling, Madeline C., et al. (2019). Structure elucidation of colibactin and its DNA cross-links. Science 365 (6457), eaax2685. doi:10.1126/science.aax2685
Zha, Yuguo, Chong, Hui, Yang, Pengshuo, and Kang, Ning (2022). Microbial dark matter: From discovery to applications. Genomics Proteomics Bioinforma. S1672-0229 (22), 00037–7. doi:10.1016/j.gpb.2022.02.007
Zhou, Tao, Hirayama, Yuichiro, Tsunematsu, Yuta, Suzuki, Nanami, Tanaka, Seiji, Uchiyama, Nahoko, et al. (2021). Isolation of new colibactin metabolites from wild-type Escherichia coli and in situ trapping of a mature colibactin derivative. J. Am. Chem. Soc. 143 (14), 5526–5533. doi:10.1021/jacs.1c01495
Keywords: colibactin, phage induction, genotoxicity, human microbiome, colorectal cancer, inflammation, colitis, DNA damage
Citation: Mousa WK (2022) The microbiome-product colibactin hits unique cellular targets mediating host–microbe interaction. Front. Pharmacol. 13:958012. doi: 10.3389/fphar.2022.958012
Received: 31 May 2022; Accepted: 18 August 2022;
Published: 12 September 2022.
Edited by:
Fareeha Anwar, Riphah International University (Lahore), PakistanReviewed by:
Irum Shahzadi, COMSATS University Islamabad, PakistanJun-Seob Kim, Incheon National University, South Korea
Copyright © 2022 Mousa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Walaa K. Mousa, d2FsYWEubW91c2FAYWF1LmFjLmFl
 Walaa K. Mousa
Walaa K. Mousa