- 1Department of Clinical Pharmacy, School of Basic Medicine and Clinical Pharmacy, China Pharmaceutical University, Nanjing, China
- 2Division of Clinical Pharmacology, General Clinical Research Center, Nanjing First Hospital, Nanjing Medical University, Nanjing, China
- 3Department of Pharmacy, Nanjing First Hospital, China Pharmaceutical University, Nanjing, China
- 4Department of Laboratory Medicine, Nanjing First Hospital, Nanjing Medical University, Nanjing, China
- 5H. pylori Research Key Laboratory, Nanjing Medical University, Nanjing, China
Background: The cure rates of Helicobacter pylori (H. pylori) treatment using a proton pump inhibitor (PPI) are gradually decreasing due to antibiotic resistance, poor compliance, high gastric acidity, and cytochrome P450 2C19 (CYP2C19) polymorphism, and the effects of PPI depend on metabolic enzymes, cytochrome P450 enzymes. The aim of this meta-analysis was to determine whether CYP2C19 polymorphisms affect H. pylori cure rates in patients treated with different proton pump inhibitors (PPIs) according to stratified analysis.
Materials and methods: The literature was searched with the key words “H. pylori” and “CYP2C19” in PubMed, CNKI, and Wanfang up to 31 May 2022, and the studies were limited to clinical observational or randomized controlled trials (RCTs). Finally, seven RCTs and 29 clinical observational studies met the inclusion criteria and were used for the meta-analysis via STATA version 16.
Results: The cure rates were significantly different between genotypes of homozygous extensive metabolizers (EM) and poor metabolizers (PM) (OR = 0.58, 95% CI: 0.47–0.71) and between EM and heterozygous extensive metabolizers (IM) (OR = 0.71, 95% CI: 0.59–0.86), but not between IM and PM. Moreover, there was a significantly lower H. pylori cure rate in EM subjects than that in IM subjects when treated with omeprazole (66.4% vs. 84.1%), lansoprazole (76.1% vs. 85.6%), but not rabeprazole, esomeprazole, or pantoprazole. In addition, there was a significantly lower H. pylori cure rate in EM subjects than that in IM subjects when treated with a PPIs for 7 days (77.4% vs. 82.1%), but not 14 days (85.4% vs. 90.0%).
Conclusion: Carriers of CYP2C19 loss-of-function variant alleles (IM and PM) exhibit a significantly greater cure rate of H. pylori than noncarriers (EM) regardless of other factors (84.7% vs. 79.2%). In addition, pantoprazole- and rabeprazole-based quadruple therapy for H. pylori treatment is less dependent on the CYP2C19 genotype and should be prioritized in Asian populations with H. pylori.
Introduction
Helicobacter pylori (H. pylori) infection is a major risk factor for peptic ulcer and gastritis and is also associated with mucosal-associated lymphoid tissue (MALT) lymphoma and gastric cancer (Marshall and Warren, 1984; Yamada, 1994; Marshall and Windsor, 2005; Fischbach and Malfertheiner, 2018). According to the content of the Kyoto global consensus, the H. pylori infection exceeded 50% of the general populations worldwide, and H. pylori control has become an important issue for the Centers for Disease Control and Prevention in the world (Sugano et al., 2015).
At present, triple therapy and quadruple therapies are mainly used for H. pylori treatment all over the world due to the high cure rate; however, large-scale H. pylori treatments have resulted in increasing rates of resistance to multiple antibiotics, together with factors such as drug compliance, inappropriate treatment regimens, therapy duration, intragastric acidity, and CYP2C19 genetic polymorphisms, resulting in a gradual decline in H. pylori cure rates (Zhong et al., 2022). Moreover, previous studies indicated that H. pylori cure rates may vary by the use of different proton pump inhibitors (PPIs), whose effects are affected by genetic polymorphisms of drug-metabolizing enzymes CYP2C19 (Chaudhry et al., 2008; Klotz, 2009).
The fields of medicine where clinical outcomes are particularly dependent on CYP2C19 polymorphisms are gastroenterology, cardiology, psychiatry, fungology, and oncology. CYP2C19 is involved in the metabolism of PPIs and therefore it can influence reflux therapy, ulcer prevention, and H. pylori therapy. The CYP2C19 enzyme also plays an important role in two bioactivation steps of clopidogrel, leading to a lower (CYP2C19*17 carriers) or higher (CYP2C19*2 carriers) risk of major adverse cardiovascular events. It affects antidepressant therapy and methadone replacement therapy as well as voriconazole prophylaxis. Moreover, in breast cancer patients treated with tamoxifen, the presence of the *2 allele was associated with a longer recurrence-free time or better survival, while the *17 allele was associated with a more favorable outcome (Sienkiewicz-Oleszkiewicz et al., 2018). Our study focused on the effect of the CYP2C19 genotype on the metabolism of PPIs and consequently on the cure rate of H. pylori.
CYP2C19 is a major drug-metabolizing enzyme for the clearance of the first-generation PPIs omeprazole and lansoprazole (∼80%) with a relatively lesser contribution of CYP3A4. In contrast, the second-generation PPIs esomeprazole and rabeprazole are less dependent on CYP2C19 in their metabolism, suggesting that they may be less influenced by genetic variability in CYP2C19 compared to the first-generation PPIs (Lima et al., 2021).
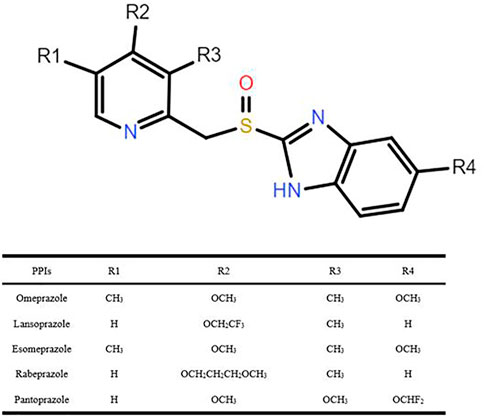
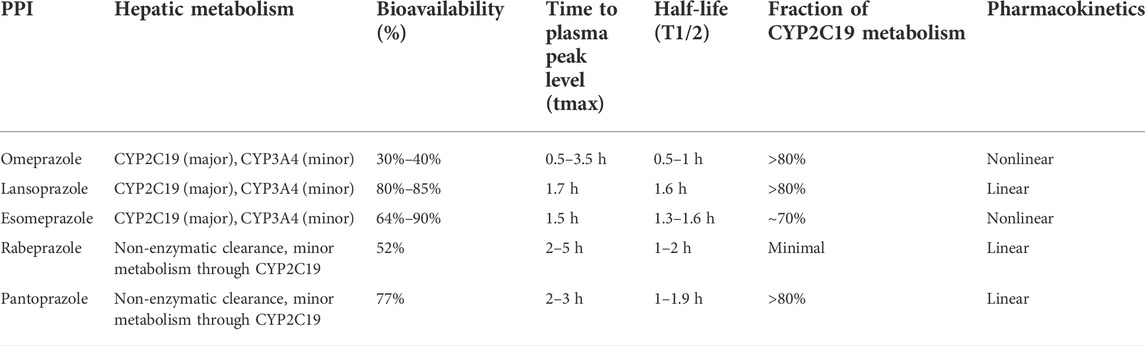
The PPIs, a class of drugs that suppress gastric acid production through irreversible inhibition of the H+/K+-ATPase (or proton pump) (Goldstein et al., 2017), have been used to treat gastric acid–associated disorders, such as gastroesophageal reflux and peptic ulcer. Six PPIs are currently approved in the United States including omeprazole, the prototype in this class, lansoprazole, dexlansoprazole, pantoprazole, rabeprazole, and esomeprazole (a stereoisomer of omeprazole) (El Rouby et al., 2018), and five of these reagents were explored in our study (i.e., omeprazole, lansoprazole, pantoprazole, rabeprazole, and esomeprazole), as shown in Figure 1. PPI metabolism has been studied in adults, and thus the PK parameters summarized in Table 1 (Li et al., 2004; Shin and Kim, 2013; Ward and Kearns, 2013; Yu et al., 2017; Savarino et al., 2018) apply to adults. There are some differences in the extent to which PPIs are metabolized by CYP2C19, leading to variability in their PK and pharmacodynamic (PD) parameters, ultimately impacting their efficacy. It is documented that CYP2C19 is responsible for > 80% of the metabolism of omeprazole, lansoprazole, and pantoprazole metabolism (Andersson et al., 1998). However, previous studies have confirmed that esomeprazole is metabolized, to less content, by CYP2C19 than omeprazole, resulting in less interindividual variation in plasma drug concentrations than omeprazole (Dent, 2003). Rabeprazole is metabolized to thioether-rabeprazole mainly via a non-enzymatic pathway, with minor involvement of CYP2C19 (Tybring et al., 1997).
Polymorphic CYP2C19 phenotypes could affect the cure rate of H. pylori because some of the PPIs are metabolized predominantly by CYP2C19, whose alleles are categorized into three groups as follows: wild-type function (e.g., CYP2C19*1), loss-of-function (e.g., CYP2C19*2 and *3), and enhanced function (e.g., CYP2C19*17) (Lima et al., 2021). Several phenotypes have been identified for CYP2C19. The extensive or normal metabolizer (EM) is carrier of the *1/*1 or *1/*17 genotypes; the intermediate metabolizer (IM) is carrier of *1/*2, *1/*3, or *2/*17 genotypes; the poor metabolizer (PM) is carrier of *2/*2, *2/*3, or *3/*3 genotypes; and the ultra-rapid metabolizer (UM) is an allele *17 homozygous carrier (*17/*17). EM and UM metabolize PPIs at a fast rate, and thus higher doses of these agents are required in EM and UM to achieve the same effect as that required in IM and PM (Arenas et al., 2019). It is important to note that most CYP2C19 studies evaluating PPIs were conducted in Asian populations, in whom the allele frequency of the CYP2C19*17 is lower than that in non-Asian populations, therefore, few studies about CYP2C19 EMs and UMs have been published to date (Lima et al., 2021). Following administration of standard doses of the first-generation PPIs, CYP2C19 IM and PM experienced higher PPI AUC (3–14 fold) and Cmax (2–6 fold) than CYP2C19 EM as a result of reduced PPI clearance via the CYP2C19 pathway (Chang et al., 1995; He et al., 2003; Qiao et al., 2006). The increased PPI exposure in CYP2C19 IM and PM has been linked to improved acid suppression (i.e., higher intragastric pH and longer time with pH > 4.0) and improved therapeutic benefits. Thus, CYP2C19 IM and PM are considered to be “therapeutically advantaged” compared to EM in terms of efficacy (Furuta et al., 1999; Shimatani et al., 2003; Kurzawski et al., 2006; Park et al., 2017).
In clinical practice, EMs produce an abundance of more active enzymes and metabolize the PPIs at a higher rate, limiting the drugs’ bioavailability and consequently lowering their antisecretory efficacies. IMs contain one wild-type allele and one mutant allele, resulting in the compromised production of the enzyme and thus slower metabolism of the PPI. In the PMs, both alleles are mutated (loss-of-function variant alleles), resulting in a much slower rate of PPI metabolism, ensuring greater bioavailability and subsequently increased antisecretory efficacy. Meanwhile, the frequency of the genotype status is highly varied among different regions (Zhao et al., 2008), which may affect the H. pylori treatment. Therefore, the effects of CYP2C19 genotypes on the H. pylori cure rates were reported extensively from different treatment regimens, among different geographic regions or ethnic populations.
There are also some factors that we overlook that can affect our results, such as the dose of PPI, antibiotic resistance, and the interleukin (IL)-1β genotype can have an impact on the cure rate of H. pylori. It has been reported in the literature that the effect of CYP2C19 genetic polymorphisms on the cure rate of H. pylori can be attenuated by increasing the dose of PPIs (omeprazole and lansoprazole) (Padol et al., 2006; Zhao et al., 2008; Tang et al., 2013). The cure rate of H. pylori also decreases significantly when metronidazole and clarithromycin resistance is observed (Houben et al., 1999; Miwa et al., 2001). IL-1β can affect the gastric acid secretion and thus mediate the cure of H. pylori, but the influence of the IL-1β genotype on treatment of H. pylori remains highly controversial (Take et al., 2003; Furuta et al., 2004).
In our study, we investigated the effects of CYP2C19 genetic polymorphisms on H. pylori treatment regardless of PPI doses, resistance to antibiotics, medication compliance, and IL-1β genotype and performed subgroup analysis to investigate whether the effects of PPI, treatment duration, treatment regimen, and geographic factors on H. pylori treatment were associated with the CYP2C19 genotype. Moreover, we indicated that the main underlying mechanism of H. pylori treatment failure is insufficiently sustained gastric acid suppression.
Therefore, the aim of our study was to provide an alternate strategy by which optimizing H. pylori treatment would use first-line treatments that show less CYP2C19 genetic polymorphism dependence on cure rates.
Materials and methods
Literature search
A computerized systematic literature search was conducted via PubMed, China National Knowledge Infrastructure (CNKI), and Wanfang, and the relevant literature was searched up to 31 May 2022 by using the key words “cytochrome P450 2C19” or “CYP2C19,” and “Helicobacter pylori” or “H. pylori,” Published English review scientific studies were included. At the same time, supplementary retrieval was carried out by browsing references included in the publications, and limited clinical trials and randomized controlled trials (RCTs) were included.
Inclusion criteria
For this meta-analysis, all searched literature followed the following inclusion criteria: 1) double, triple, and quadruple therapies for 7–14 days; 2) patients positive for H. pylori infection prior to treatment; 3) established genotypes of CYP2C19, such as EM, IM, and PM, using a standard method (i.e., polymerase chain reaction-restriction fragment length polymorphism and Taqman probe); 4) full-text articles written in English. The following two parameters were also considered: randomization and blindness (single or double blindness either to treatment or genotype groups). In order to improve the power of the meta-analysis, dropouts/withdrawals were not recorded.
Data extraction
Two evaluators (XH.Z and ZQ.Z) independently screened the literature according to the inclusion criteria (see aforementioned), extracted the data, and cross-checked them, and any discrepancies were resolved through discussion with a third reviewer (L.F) or referring back to the original articles until the two reviewers reached a consensus. The data of enrolled studies were extracted, including the first author, publication year, study group, a total number of cases, country, age, gender, basic characteristics of patients, treatment regimens, types of PPIs, therapy length, gene detection method, number of expected phenotypic cases of CYP2C19, and the cure rate of H. pylori.
Statistical analysis
After the pooled comparison, cure rates were calculated with the number needed to treat (NNT) and odds ratio (OR) with their corresponding 95% CI of each study by using STATA 16.0 software. For those studies (Sapone et al., 2003; Sugimoto et al., 2007; Miehlke et al., 2008; Burhan et al., 2010; Gawrońska-Szklarz et al., 2010; Jinda et al., 2011; Prasertpetmanee et al., 2013; Chanagune et al., 2014; Sugimoto et al., 2014; Kittichet et al., 2016; Yun-An et al., 2017; Piyakorn et al., 2019; Ke et al., 2021; Yang et al., 2021) with value 0 appeared in the four-grid table, we added 1 to replace 0 for analysis after reading the literature and discussion. Cochrane’s Q-test and I2 test were performed to evaluate heterogeneity of enrolled studies (Hoaglin, 2016). If the Cochran’s Q-test probability was 0.05, this means that the study was significantly heterogeneous, and the I2 test was used to classify heterogeneity as low (≤25%), medium (≈50%), or high (≥75%) (McNicholl et al., 2012). p < 0.05 means the difference was statistically significant. If there was a significant heterogeneity (p < 0.05), we selected a random-effects model to pool all the eligible data, otherwise, a fixed-effects model was used. Sensitivity analysis was conducted to examine the stability of the pooled results. Publication bias was assessed using the funnel plot with the Egger’s and Begg’s tests.
Results
Characteristics of included studies
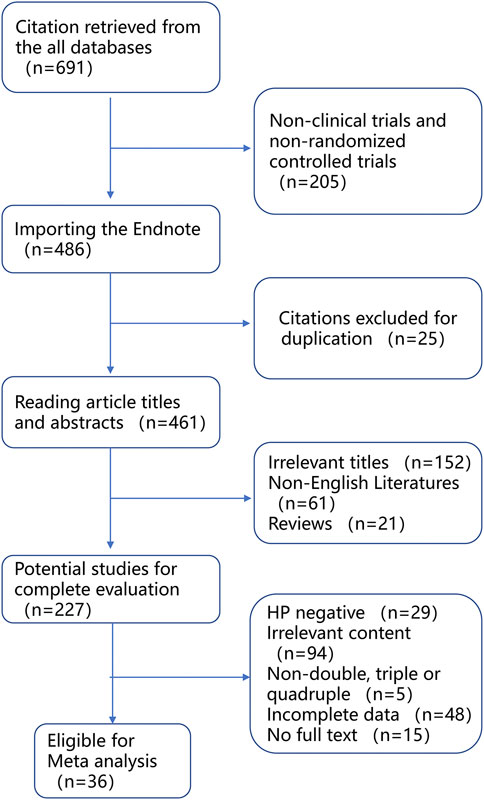
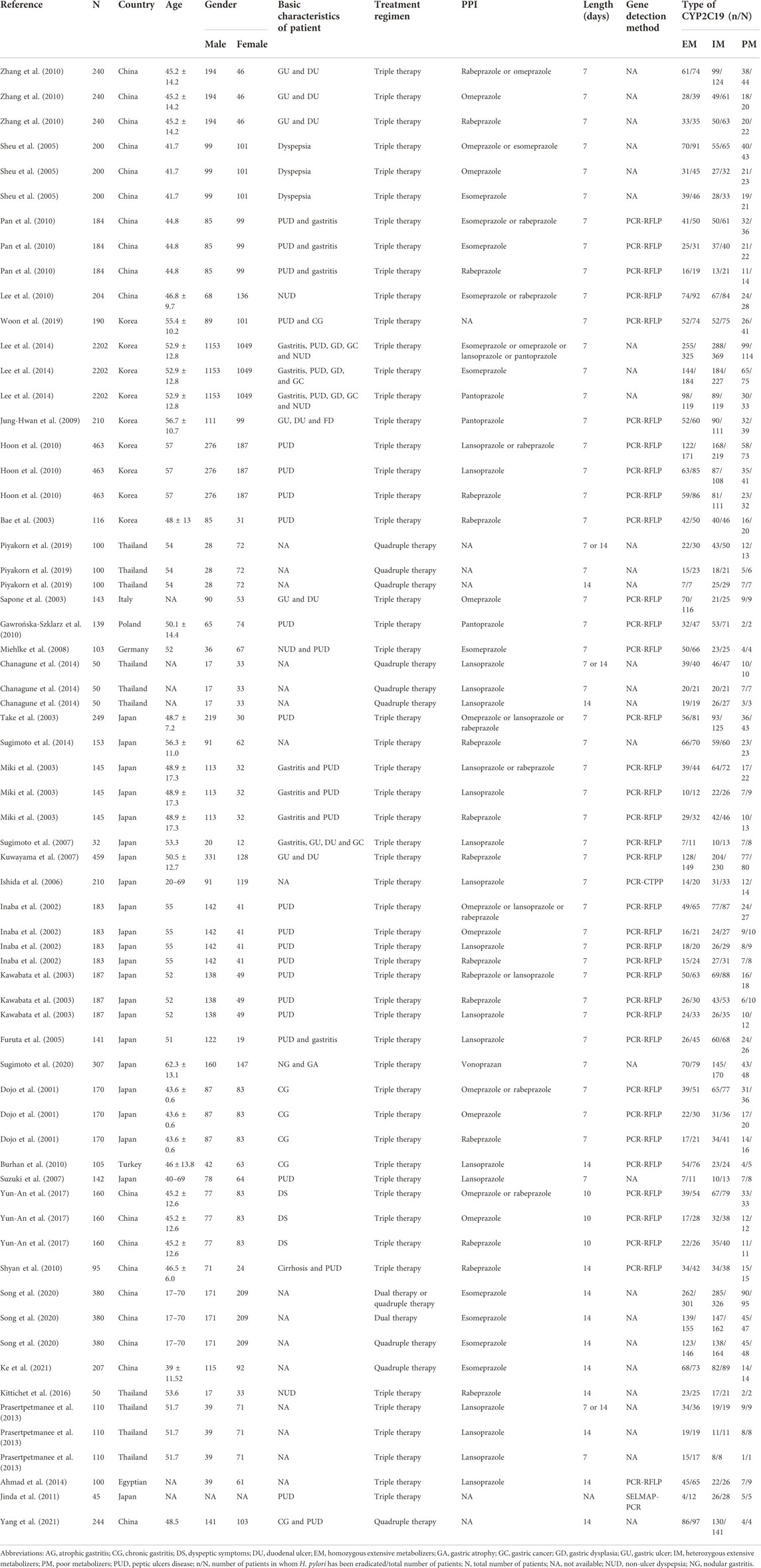
Overall, a total of 36 articles (Dojo et al., 2001; Inaba et al., 2002; Bae et al., 2003; Kawabata et al., 2003; Miki et al., 2003; Sapone et al., 2003; Take et al., 2003; Suzuki et al., 2007; Sheu et al., 2005; Ishida et al., 2006; Furuta et al., 2007; Kuwayama et al., 2007; Sugimoto et al., 2007; Miehlke et al., 2008; Jung-Hwan et al., 2009; Burhan et al., 2010; Gawrońska-Szklarz et al., 2010; Hoon et al., 2010; Lee et al., 2010; Pan et al., 2010; Shyan et al., 2010; Zhang et al., 2010; Jinda et al., 2011; Prasertpetmanee et al., 2013; Ahmad et al., 2014; Chanagune et al., 2014; Lee et al., 2014; Sugimoto et al., 2014; Kittichet et al., 2016; Yun-An et al., 2017; Piyakorn et al., 2019; Woon et al., 2019; Song et al., 2020; Sugimoto et al., 2020; Ke et al., 2021; Yang et al., 2021) were included in the meta-analysis from 691 relevant reports according to the inclusion criteria (Figure 2; Table 2). To conduct subgroup analysis, studies (Dojo et al., 2001; Inaba et al., 2002; Bae et al., 2003; Kawabata et al., 2003; Miki et al., 2003; Sapone et al., 2003; Take et al., 2003; Furuta et al., 2005; Sheu et al., 2005; Ishida et al., 2006; Furuta et al., 2007; Kuwayama et al., 2007; Sugimoto et al., 2007; Miehlke et al., 2008; Jung-Hwan et al., 2009; Burhan et al., 2010; Gawrońska-Szklarz et al., 2010; Hoon et al., 2010; Lee et al., 2010; Pan et al., 2010; Shyan et al., 2010; Zhang et al., 2010; Jinda et al., 2011; Prasertpetmanee et al., 2013; Ahmad et al., 2014; Chanagune et al., 2014; Lee et al., 2014; Sugimoto et al., 2014; Kittichet et al., 2016; Yun-An et al., 2017; Piyakorn et al., 2019; Woon et al., 2019; Song et al., 2020; Sugimoto et al., 2020; Ke et al., 2021; Yang et al., 2021) were divided into more than one group according to stratified variation, including the type of PPIs, treatment duration, regions, and treatment regimens. In the analysis of the PPI treatment subgroup, a total of 36 research articles were enrolled, which comprised the treatment of omeprazole (Dojo et al., 2001; Inaba et al., 2002; Sapone et al., 2003; Sheu et al., 2005; Zhang et al., 2010; Yun-An et al., 2017), lansoprazole (Inaba et al., 2002; Kawabata et al., 2003; Miki et al., 2003; Furuta et al., 2005; Ishida et al., 2006; Sugimoto et al., 2007; Burhan et al., 2010; Hoon et al., 2010; Prasertpetmanee et al., 2013; Chanagune et al., 2014), esomeprazole (Sheu et al., 2005; Miehlke et al., 2008; Pan et al., 2010; Lee et al., 2014; Song et al., 2020; Ke et al., 2021), rabeprazole (Dojo et al., 2001; Inaba et al., 2002; Bae et al., 2003; Kawabata et al., 2003; Miki et al., 2003; Kuwayama et al., 2007; Hoon et al., 2010; Shyan et al., 2010; Zhang et al., 2010; Sugimoto et al., 2014; Kittichet et al., 2016; Yun-An et al., 2017), and pantoprazole (Jung-Hwan et al., 2009; Gawrońska-Szklarz et al., 2010; Lee et al., 2014). One study reported the use of the PPI vonoprazan (Sugimoto et al., 2020), but meta-analysis could not be performed on a single report, which was not included in the analysis. In the subgroup analysis of the treatment duration, a total of 36 studies (Dojo et al., 2001; Inaba et al., 2002; Bae et al., 2003; Kawabata et al., 2003; Miki et al., 2003; Sapone et al., 2003; Take et al., 2003; Furuta et al., 2005; Sheu et al., 2005; Ishida et al., 2006; Furuta et al., 2007; Kuwayama et al., 2007; Sugimoto et al., 2007; Miehlke et al., 2008; Jung-Hwan et al., 2009; Burhan et al., 2010; Gawrońska-Szklarz et al., 2010; Hoon et al., 2010; Lee et al., 2010; Pan et al., 2010; Shyan et al., 2010; Zhang et al., 2010; Jinda et al., 2011; Prasertpetmanee et al., 2013; Ahmad et al., 2014; Chanagune et al., 2014; Lee et al., 2014; Sugimoto et al., 2014; Kittichet et al., 2016; Yun-An et al., 2017; Piyakorn et al., 2019; Woon et al., 2019; Song et al., 2020; Sugimoto et al., 2020; Ke et al., 2021; Yang et al., 2021) were chosen, including 27 studies (Dojo et al., 2001; Inaba et al., 2002; Bae et al., 2003; Kawabata et al., 2003; Sapone et al., 2003; Take et al., 2003; Furuta et al., 2005; Sheu et al., 2005; Ishida et al., 2006; Furuta et al., 2007; Kuwayama et al., 2007; Sugimoto et al., 2007; Miehlke et al., 2008; Jung-Hwan et al., 2009; Gawrońska-Szklarz et al., 2010; Hoon et al., 2010; Lee et al., 2010; Pan et al., 2010; Zhang et al., 2010; Prasertpetmanee et al., 2013; Chanagune et al., 2014; Lee et al., 2014; Piyakorn et al., 2019; Woon et al., 2019; Sugimoto et al., 2020) exerting 7-day treatment, and 10 studies (Burhan et al., 2010; Shyan et al., 2010; Prasertpetmanee et al., 2013; Ahmad et al., 2014; Chanagune et al., 2014; Kittichet et al., 2016; Piyakorn et al., 2019; Song et al., 2020; Ke et al., 2021; Yang et al., 2021) reporting 14-day treatment, respectively. In the subgroup analysis of the region, a total of 35 articles were included, the majority of studies were from Asia (13 in Japan, nine in China, five in South Korea, four in Thailand, and one in Turkey), followed by Europe (one in Germany, Poland, and Italy each). In subgroup analysis of regimens, including 31 studies (Dojo et al., 2001; Inaba et al., 2002; Bae et al., 2003; Kawabata et al., 2003; Miki et al., 2003; Sapone et al., 2003; Take et al., 2003; Furuta et al., 2005; Sheu et al., 2005; Ishida et al., 2006; Furuta et al., 2007; Kuwayama et al., 2007; Sugimoto et al., 2007; Miehlke et al., 2008; Jung-Hwan et al., 2009; Burhan et al., 2010; Gawrońska-Szklarz et al., 2010; Hoon et al., 2010; Lee et al., 2010; Pan et al., 2010; Shyan et al., 2010; Zhang et al., 2010; Jinda et al., 2011; Prasertpetmanee et al., 2013; Ahmad et al., 2014; Lee et al., 2014; Sugimoto et al., 2014; Kittichet et al., 2016; Yun-An et al., 2017; Woon et al., 2019; Song et al., 2020) about triple therapy, five studies (Chanagune et al., 2014; Piyakorn et al., 2019; Sugimoto et al., 2020; Ke et al., 2021; Yang et al., 2021) about quadruple therapy, and only one study (Ke et al., 2021) about double therapy met the inclusion criteria, but no analysis could be performed on a single study and it was excluded.
The efficacy of CYP2C19 polymorphisms on the overall cure rates of H. pylori
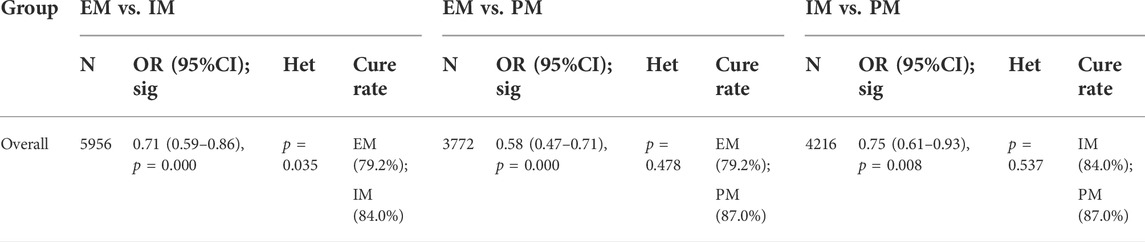
The results revealed that, regardless of the type of PPIs, treatment duration regions, and treatment regimens, there was a significant difference in the cure rate of H. pylori between EM and IM genotypes (79.2% vs. 84.0%, OR = 0.71, 95% CI: 0.59–0.86, p = 0.000, pHeterogeneity = 0.035), between EM and PM (79.2% vs. 87.0%, OR = 0.58, 95% CI: 0.47–0.71, p = 0.000, pHeterogeneity = 0.478), and between IM and PM (84.0% vs. 87%, OR = 0.75, 0.95% CI: 0.61–0.93, p = 0.008, pHeterogeneity = 0.537) as shown in Figure 3; Table 3.
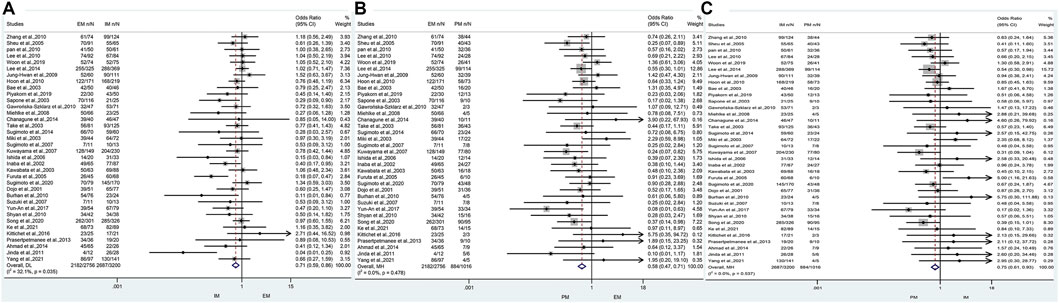
FIGURE 3. (A) Forest plot of EM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype (p = 0.035). (B) Forest plot of EM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype (p = 0.478). (C) Forest plot of IM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype (p = 0.537). p = 0.05, significant heterogeneity, or statistically significant, when appropriate. p < 0.05, random-effects model; p > 0.05, random-effects model. Symbol:  , single studies included in the meta-analysis;
, single studies included in the meta-analysis;  , sample size of single studies;
, sample size of single studies;  , confidence interval (CI);
, confidence interval (CI);  , overall pool estimated;
, overall pool estimated;  , tendency;
, tendency;  , overall pool OR.
, overall pool OR.
The efficacy of CYP2C19 polymorphisms on the cure rates of H. pylori in the treatment regimens containing different proton pump inhibitors
In the subgroup of omeprazole-based therapy, there was a significant difference in the H. pylori cure rate between EM and IM (66.4% vs. 84.1%, OR = 0.42, 95% CI: 0.26–0.88, p = 0.000, pHeterogeneity = 0.947) and between EM and PM (66.4% vs. 90.5%, OR = 0.25, 95% CI: 0.12–0.51, p = 0.000, pHeterogeneity = 0.906), but not between IM and PM (84.1% vs. 90.5%, OR = 0.59, 95% CI: 0.28–1.25, p = 0.171, pHeterogeneity = 0.945) as shown in Figure 4; Table 4.
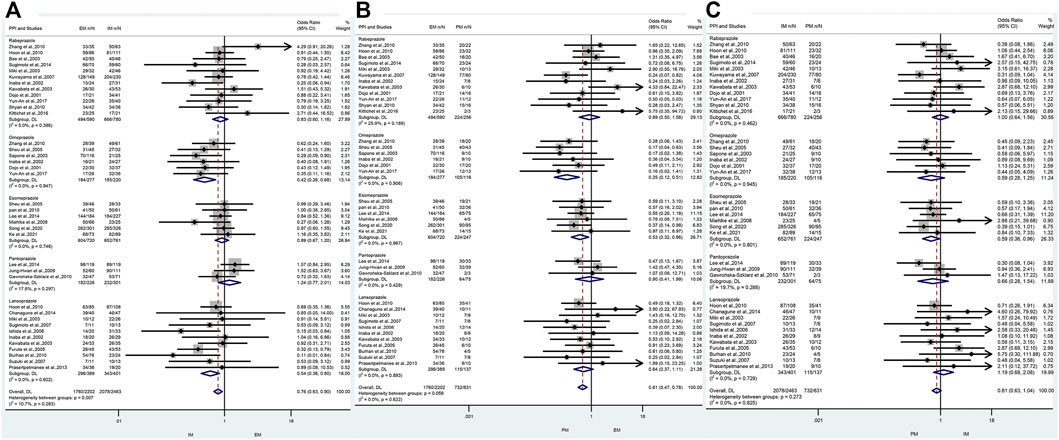
FIGURE 4. (A) Forest plot of EM vs. IM in relation to the H. pylori cure rate in the CYP2C19 genotype in different PPIs (p = 0.283). (B) Forest plot of EM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype in different PPIs (p = 0.622). (C) Forest plot of IM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype in different PPIs (p = 0.825). p = 0.05, significant heterogeneity or statistically significant, when appropriate. p < 0.05, random-effects model; p > 0.05, random-effects model. Symbol:  , single studies included in the meta-analysis;
, single studies included in the meta-analysis;  , sample size of single studies;
, sample size of single studies;  , confidence interval (CI);
, confidence interval (CI);  , overall pool estimated;
, overall pool estimated;  , tendency;
, tendency;  , overall pool OR.
, overall pool OR.
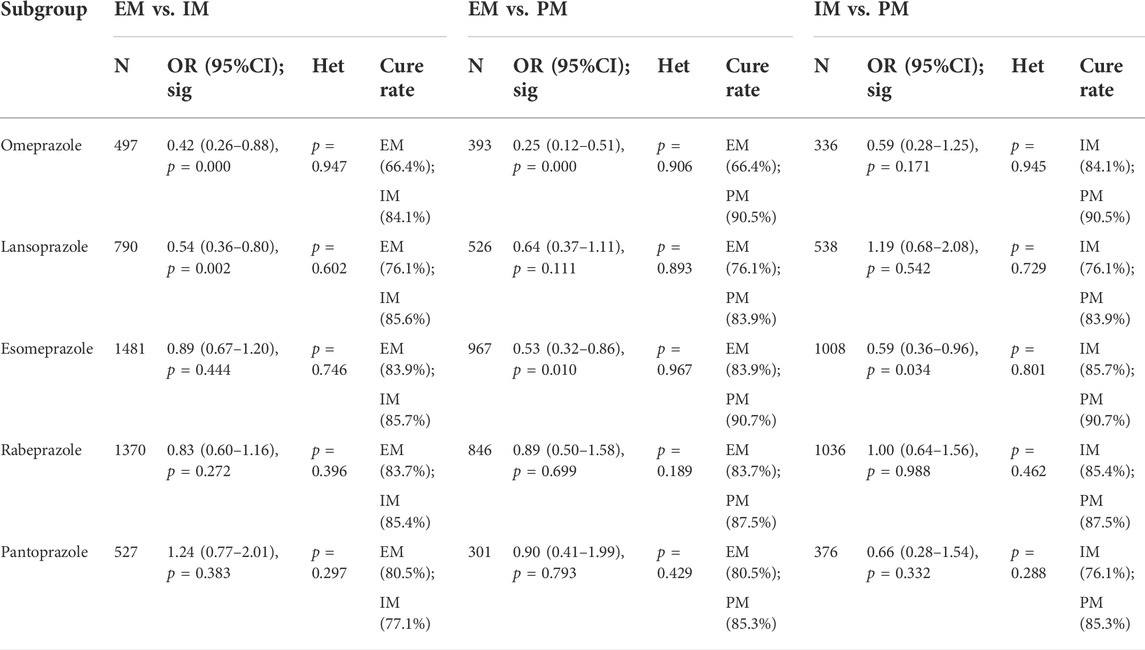
TABLE 4. Efficacy of CYP2C19 polymorphisms on the cure rates of H. pylori in the treatment regimens containing different PPIs.
Interestingly, in subgroup analysis of lansoprazole-based therapy, the pooled result showed that there was a significant difference in the H. pylori cure rate between EM and IM (76.1% vs. 84.1%, OR = 0.42, 95% CI: 0.36–0.80, p = 0.002, pHeterogeneity = 0.602), but neither between EM and PM (76.1% vs. 83.9%, OR = 0.64, 95% CI: 0.37–1.11, p = 0.111, pHeterogeneity = 0.893) nor between IM and PM (76.1% vs. 83.9%, OR = 1.19, 95% CI: 0.68–2.08, p = 0.542, pHeterogeneity = 0.729) as shown in Figure 3; Table 3. In the subgroup of esomeprazole-based therapy, there was a significant difference in the H. pylori cure rate between EM and PM (83.9% vs. 90.7%, OR = 0.53, 95% CI: 0.32–0.86, p = 0.010, pHeterogeneity = 0.967) and between IM and PM (85.7% vs. 90.7%, OR = 0.59, 95% CI: 0.36–0.96, p = 0.034, pHeterogeneity = 0.801), but not between EM and IM (83.9% vs. 85.7%, OR = 0.89, 95% CI: 0.67–1.20, p = 0.444, pHeterogeneity = 0.746) as shown in Figure 4; Table 4.
In contrast, there were no significant differences in the H. pylori cure rates between the all genotypes for those patients treated with rabeprazole- or pantoprazole-based regimen as shown in Figure 4; Table 4.
The efficacy of CYP2C19 polymorphisms on the cure rates of H. pylori in 7-day therapy and 14-day therapy
Only one study (Yun-An et al., 2017) was just treated for 10 days, but no analysis could be performed on a single study and it was excluded. For those studies with the therapy of 7 days, there was a significant difference in the H. pylori cure rate between EM and IM (77.4% vs. 82.1%, OR = 0.75, 95% CI: 0.62–0.91, p = 0.004, pHeterogeneity = 0.170) and between EM and PM (77.4% vs. 85.5%, OR = 0.63, 95% CI: 0.50–0.79, p = 0.000, pHeterogeneity = 0.640), but not between IM and PM (82.1% vs. 85.5%, OR = 0.82, 95% CI: 0.65–1.04, p = 0.099, pHeterogeneity = 0.533). In contrast, when the use of a PPI was maintained for 14 days, there were no significant differences in H. pylori cure rates between the three genotypes as shown in Figure 5; Table 5. These results indicated that the CYP2C19 genetic polymorphisms exhibit less important effects on H. pylori cure rates in patients treated with a PPI for 2 weeks than those for 1 week in three genotypes.
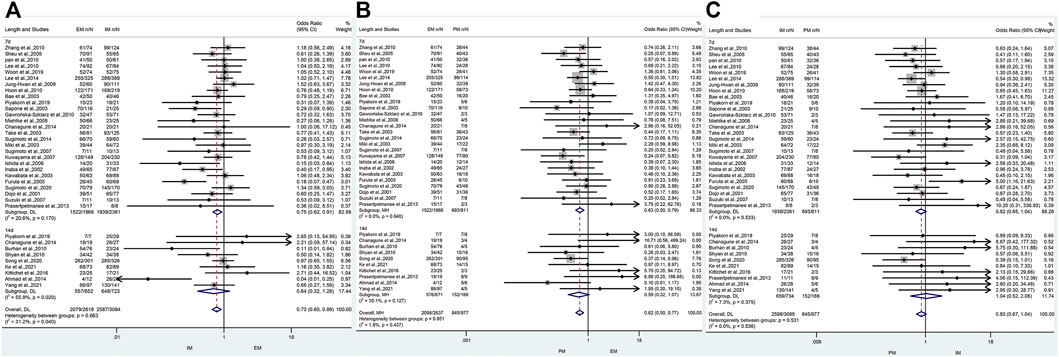
FIGURE 5. (A) Forest plot of EM vs. IM in relation to the H. pylori cure rate in the CYP2C19 genotype in different treatment duration (p = 0.040). (B) Forest plot of EM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype in different treatment duration (p = 0.437). (C) Forest plot of IM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype in different treatment duration (p = 0.536). p = 0.05, significant heterogeneity or statistically significant, when appropriate. p < 0.05, random-effects model; p > 0.05, random-effects model. Symbol:  , single studies included in the meta-analysis;
, single studies included in the meta-analysis;  , sample size of single studies;
, sample size of single studies;  , confidence interval (CI);
, confidence interval (CI);  , overall pool estimated;
, overall pool estimated;  , tendency;
, tendency;  , overall pool OR.
, overall pool OR.
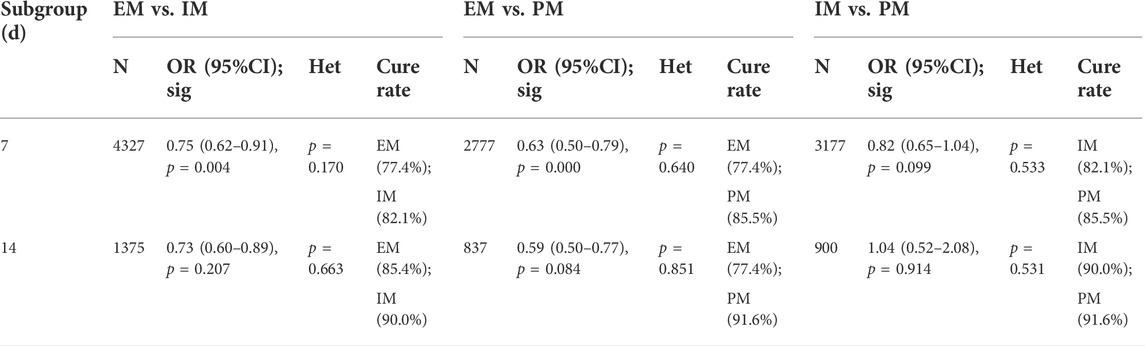
TABLE 5. Efficacy of CYP2C19 polymorphisms on the cure rates of H. pylori in 7-day therapy and 14-day therapy.
The efficacy of CYP2C19 polymorphisms on the cure rates of H. pylori in different geographical location of patients
In Asia, there was a significant difference in the H. pylori cure rate between the all genotypes.
(EM and IM: 80.6% vs. 84.1%, OR = 0.75, 95% CI: 0.61–0.91, p = 0.003, PHeterogeneity = 0.043; EM and PM: 80.6% vs. 87.2%, OR = 0.63, 95% CI: 0.49–0.79, p = 0.000, pHeterogeneity = 0.388; IM and PM: 84.1% vs. 87.2%, OR = 0.79, 95% CI: 0.63–0.99, p = 0.039, pHeterogeneity = 0.443). However, in Africa, there was a significant difference between EM and IM (66.4% vs. 80.2%, OR = 0.46, 95% CI: 0.24–0.90, p = 0.023, pHeterogeneity = 0.323), but neither between EM and PM (66.4% vs. 83.3%, OR = 0.47, 95% CI: 0.13–1.74, p = 0.258, pHeterogeneity = 0.466) nor between IM and PM (80.2% vs. 83.3%, OR = 1.27, 95% CI: 0.31–5.25, p = 0.743, pHeterogeneity = 0.665) as shown in Figure 6; Table 6. Therefore, it showed that effects of CYP2C19 genetic polymorphisms influence on the H. pylori cure rates could be of greater clinical implication in the Asian populations.
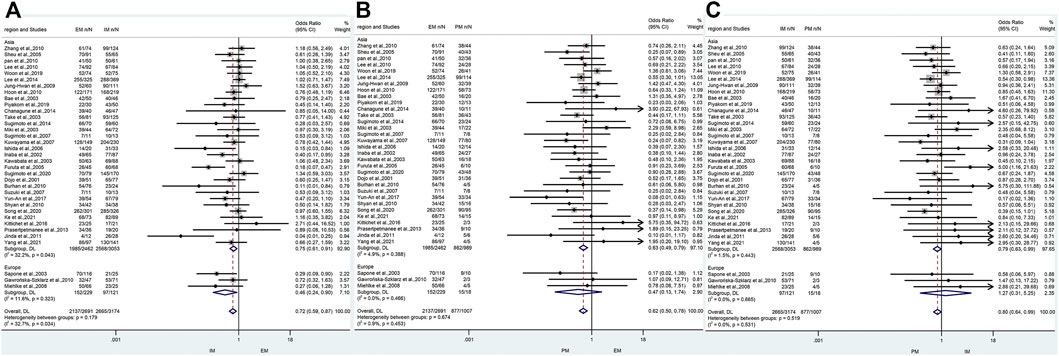
FIGURE 6. (A) Forest plot of EM vs. IM in relation to the H. pylori cure rate in the CYP2C19 genotype in different regions (p = 0.034). (B) Forest plot of EM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype in different regions (p = 0.453). (C) Forest plot of IM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype in different regions (p = 0.531). p = 0.05, significant heterogeneity, or statistically significant, when appropriate. p < 0.05, random-effects model; p > 0.05, random-effects model. Symbol:  , single studies included in the meta-analysis;
, single studies included in the meta-analysis;  , sample size of single studies;
, sample size of single studies;  , confidence interval (CI);
, confidence interval (CI);  , overall pool estimated;
, overall pool estimated;  , tendency;
, tendency;  , overall pool OR.
, overall pool OR.
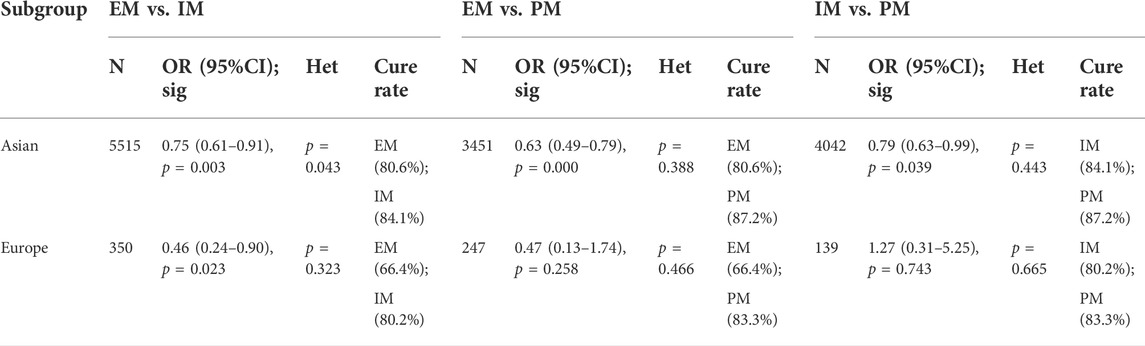
TABLE 6. Efficacy of CYP2C19 polymorphisms on the cure rates of H. pylori in different geographical location of patients.
The efficacy of CYP2C19 polymorphism on the cure rates of H. pylori in triple therapy and quadruple therapy
Only one study (Ke et al., 2021) about double therapy met the inclusion criteria, but no analysis could be performed for a single study and thus it was excluded. There was a significant difference in H. pylori cure rates among all genotypes for triple therapy (EM vs. IM: 77.8% vs. 82.9%, OR = 0.69, 95% CI: 0.56–0.85, p = 0.000, pHeterogeneity = 0.019; EM vs. PM: 77.8% vs. 86.7%, OR = 0.60, 95% CI: 0.48–0.76, p = 0.000, pHeterogeneity = 0.449; IM vs. PM: 82.9% vs. 86.7%, OR = 0.80, 95% CI: 0.63–1.00, p = 0.050, pHeterogeneity = 0.455). On the contrary, there was no significant difference in H. pylori cure rates among all genotypes for quadruple therapy (EM vs. IM: 87.6% vs. 89.4%, OR = 0.83, 95% CI: 0.54–1.26, p = 0.382, pHeterogeneity = 0.440; EM vs. PM: 87.6% vs. 92.4%, OR = 0.63, 95% CI: 0.26–1.55, p = 0.312, pHeterogeneity = 0.928; IM vs. PM: 89.4% vs. 92.4%, OR = 0.77, 95% CI: 0.31–1.94, p = 0.578, pHeterogeneity = 0.346) as shown in Figure 7; Table 7. Moreover, overall H. pylori cure rates were better with quadruple therapy than those with triple therapy (EM: 87.6% vs. 77.8%, IM: 89.4% vs. 82.9%; PM: 92.4% vs. 86.7%). This might indicate that the cure rate of H. pylori with bismuth-containing quadruple therapy is less influenced by the CYP2C19 genotype and a desirable cure rate can be achieved.
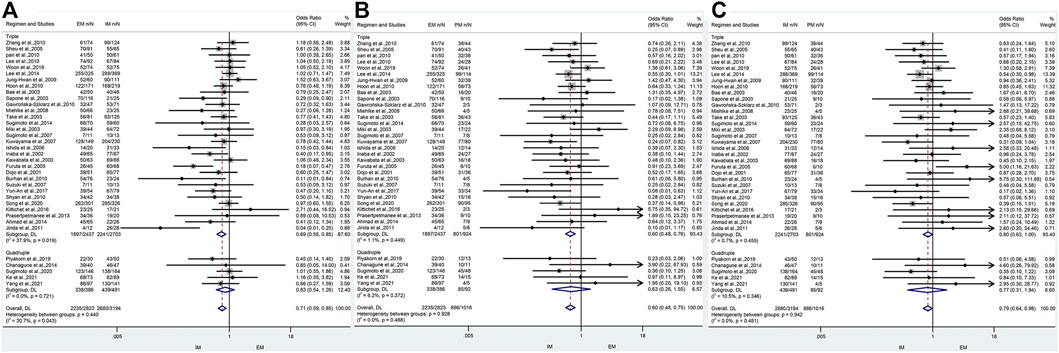
FIGURE 7. (A) Forest plot of EM vs. IM in relation to the H. pylori cure rate in the CYP2C19 genotype in different treatment regimens (p = 0.043). (B) Forest plot of EM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype in different treatment regimens (p = 0.488). (C) Forest plot of IM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype in different treatment regimens (p = 0.481). p = 0.05, significant heterogeneity or statistically significant, when appropriate. p < 0.05, random-effects model; p > 0.05, random-effects model. Symbol:  , single studies included in the meta-analysis;
, single studies included in the meta-analysis;  , sample size of single studies;
, sample size of single studies;  , confidence interval (CI);
, confidence interval (CI);  , overall pool estimated;
, overall pool estimated;  , tendency;
, tendency;  , overall pool OR.
, overall pool OR.
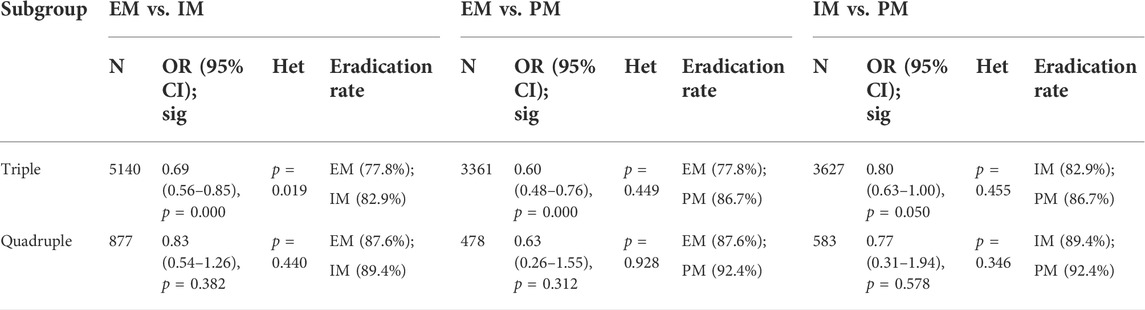
TABLE 7. Efficacy of CYP2C19 polymorphism on the cure rates of H. pylori in triple therapy and quadruple therapy.
Heterogeneity and sensitivity analysis of overall
For the meta-analysis comparing EM vs. IM in relation to the H. pylori cure rate in the CYP2C19 genotype, there was a significant heterogeneity across the enrolled studies (pHeterogeneity = 0.035, I2 = 32.1%), but neither in EM vs. PM (pHeterogeneity = 0.478, I2 = 0%) nor in IM vs. PM (pHeterogeneity = 0.537, I2 = 0%). In addition, to assess the stability of the pooled results, sensitivity analysis was carried out to assess the influence of individual studies and sources of heterogeneity on the overall effects for EM vs. IM through repeating the meta-analysis after sequentially omitting each study. As shown in Figure 8, for EM vs. IM, the heterogeneity analysis suggested that a study by Lee et al. (2014), Furuta et al. (2005), and Song et al. (2020) were the main source of heterogeneity. After deleting these three studies (Furuta et al., 2005; Lee et al., 2014; Song et al., 2020), the results were still not changed (EM vs. IM: OR = 0.71, 95% CI: 0.61–0.83, PHeterogeneity = 0.180, I2 = 18.2%).
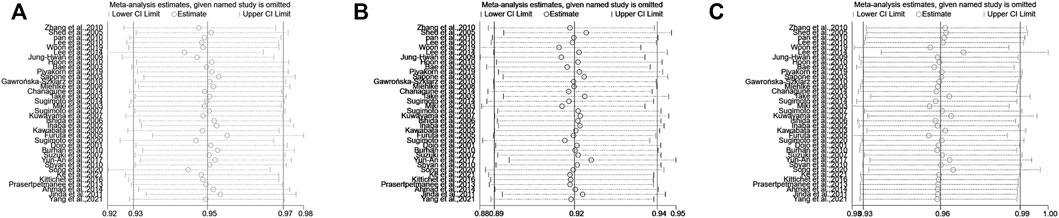
FIGURE 8. (A) Sensitivity analysis of EM vs. IM in relation to the H. pylori cure rate in the CYP2C19 genotype. (B) Sensitivity analysis of EM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype. (C) Sensitivity analysis of IM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype.
Publication bias
To test the publication bias of the included studies, the funnel plot asymmetry test was used. The funnel plots comparing EM vs. PM with H. pylori cure rates in the CY2C19 genotype were symmetrical, indicating no publication bias of these studies was presented (EM vs. PM: t = −0.34, p = 0.737); however, for EM vs. IM and IM vs. PM, the shape of the funnel plot was obviously asymmetrical and Egger’s test also provided statistical evidence of funnel plot asymmetry (t = −3.15, p = 0.003; Figure 8; IM vs. PM: t = 2.19, p = 0.035; Figure 9). To adjust these publication biases, a trim-and-fill analysis method (Duval and Tweedie, 2000) was performed. Data demonstrated that the results were stable for EM vs. PM before and after the use of this analysis, but those nine studies and seven studies needed to be filled for EM vs. IM and IM vs. PM, respectively (Figure 10).
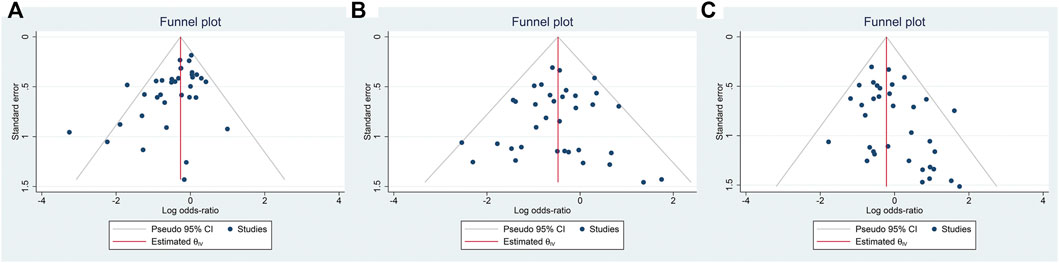
FIGURE 9. (A) Publication bias analysis of EM vs. IM in relation to the H. pylori cure rate in the CYP2C19 genotype (t = −3.15, p = 0.003). (B) Sensitivity analysis of EM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype (t = −0.34, p = 0.737). (C) Sensitivity analysis of IM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype (t = 2.19, p = 0.035). Publication bias, p = 0.05.

FIGURE 10. (A) Trim-and-fill analysis of EM vs. IM in relation to the H. pylori cure rate in the CYP2C19 genotype. Nine studies needed to be filled. (B) Trim-and-fill analysis of IM vs. PM in relation to the H. pylori cure rate in the CYP2C19 genotype. Seven studies needed to be filled.
Discussion
In our study, we found that the H. pylori cure rate of individuals with the EM genotype was significantly lower than that of those with IM or PM genotypes. Our results also revealed that the CYP2C19 genotype status affected the efficacy of omeprazole-based therapy, but not of pantoprazole- and rabeprazole-based therapy. In addition, a significant difference was observed between CYP2C19 genotypes and H. pylori cure rates in therapy of 7-day, but not in the 14-day therapy regimen. In Asia, H. pylori cure rates were influenced by the CYP2C19 genotype. But in Europe, whether H. pylori cure rates could be influenced by the CYP2C19 genotype would need to be explored in detail with an expanded sample size. Finally, we also found significant differences in H. pylori cure rates among the three genotypes in triple therapy, but no significant differences in H. pylori cure rates were observed among the three genotypes in quadruple therapy.
To date, the published data showed inconsistent results on the effects of the CYP2C19 genotype on the cure rates of H. pylori infections. Our pooled result showed that the CYP2C19 genotype could affect the cure rate of H. pylori, and the cure rate of EM genotype was significantly lower than that of the IM or PM genotype, consistent with the results reported by Ormeci et al. (2016) and Fu et al. (2021). Actually, the relative enzyme activity of the EM was roughly 2-fold higher than that of the IM (0.15 vs. 0.08, respectively), which in turn was double as that of the PM (0.08 vs. 0.04) (Kuo et al., 2014), indicating a gene-dosage effect. Individuals with an EM phenotype clear PPIs at a higher rate (Franciosi et al., 2018). However, there was no significant difference in the cure rate of H. pylori between the IM genotype and PM genotype in our study.
Clinical research studies have demonstrated that the use of PPIs is the determinant in enhancing the cure rate of H. pylori (Kim et al., 2019). PPIs are primarily inactivated in the liver by the microsomal enzyme CYP2C19, and genetic variation in the CYP2C19 gene determines enzyme activity (Zamani et al., 2018). However, some studies provided conflicting evidence for the impact of PPI-related CYP2C19 genetic polymorphisms on PPI-treated cure rates of H. pylori. For example, Shah et al. (2021) categorized omeprazole, pantoprazole, and lansoprazole as PPIs that were predominantly metabolized by CYP2C19, but esomeprazole and rabeprazole are minimally or not metabolized by CYP2C19. McNicholl et al. (2012) observed that the clinical efficacy of some new-generation PPIs (esomeprazole and rabeprazole) was not affected by CYP2C19 genetic polymorphisms and that there were higher cure rates for the use of the first-generation PPIs (omeprazole, lansoprazole, and pantoprazole) in EM patients. Padol et al. (2006) indicated that the H. pylori cure rate was not affected by CYP2C19 genetic polymorphisms when using lansoprazole or rabeprazole. Moreover, Arenas et al. (2019) described phenotypes of CYP2C19 genetic polymorphisms that did not affect the H. pylori cure rate. These controversies could be explained by none of these studies taking into account the effects of antibiotic sensitivity, doses of PPIs, geographic differences, patient compliance, and more. In fact, most of the studies that supported the association between CYP2C19 genetic polymorphisms and H. pylori cure rates were based on Asian populations, with a greater proportion of the PM genotype (Arenas et al., 2019). Our meta-analysis showed that the cure rate of H. pylori of PPIs, such as omeprazole was effected by CYP2C19 genotypes, consistent with previous reports among the main PPIs, the various influences of the CYP2C19 polymorphisms on metabolic parameters were different (omeprazole > lansoprazole > pantoprazole > rabeprazole) (Zhang et al., 2020). However, in our study, we observed that there was no significant difference in the cure rates between all genotypes with either pantoprazole- or rabeprazole-based double, triple, or quadruple therapies. Rabeprazole has been reported to be mainly metabolized (approximately 85%) via a non-enzymatic pathway to thioether-rabeprazole, with only minor involvement of CYP2C19 and CYP3A4 (Hu et al., 2005; Wang et al., 2011), consistent with the results derived from our meta-analysis, suggesting that rabeprazole-based triple or quadruple therapies can be used to eradicate H. pylori infection for all patients, with no need in considering the status of CYP2C19 genetic polymorphisms. However, pantoprazole is mainly metabolized by different metabolic enzymes (CYP2C19 and CYP3A4) (Zhang et al., 2020), inconsistent with our results, and the explanations for the discrepancy may be the small numbers of studies (Jung-Hwan et al., 2009; Gawrońska-Szklarz et al., 2010; Lee et al., 2014) and subjects (602 of 5496) included in the meta-analysis. Therefore, rabeprazole and pantoprazole treatment programs were superior to omeprazole in the case of ignoring other influencing factors, such as side effects, cost of treatment, other metabolizer genes affecting other drugs, and more. Moreover, the positioning of potassium-competitive acid blockers (P-CABs) is also worth considering, particularly because vonoprazan, which is not currently approved outside certain Asia-Pacific and South/Central American countries/regions and is actively being investigated for the use in the United States and Europe for conditions necessitating gastric acid suppression (Shah et al., 2021). Vonoprazan is metabolized mainly by CYP3A4/5 and partially by CYP2B6, CYP2C19, and CYP2D6, and its pharmacokinetics and pharmacodynamics may be influenced by genetic variations in the respective genes (Sugimoto et al., 2020). However, our study showed a significant difference only in EM vs. IM in lansoprazole-based therapy, but not in EM vs. PM or IM vs. PM, a significant difference was observed only in EM vs. PM in esomeprazole-based therapy, but not in EM vs. IM or IM vs. PM. Previous reports suggested that the efficacy of lansoprazole is dependent on the CYP2C19 gene status (Fu et al., 2021), but Padol et al. (2006) showed that H. pylori therapies with omeprazole are dependent on the CYP2C19 genotype while therapies with lansoprazole and rabeprazole are not. Thus, the conclusion drawn is ambiguous and requires further research. Meanwhile, the articles used in this study were based on the difference between PPI dosages and antibiotic susceptibility, which may affect the conclusion of this study. Therefore, the choice of different PPIs and/or doses should be individualized based on the pharmacogenetics background of each patient.
In addition, we reported for the first time a subgroup analysis regarding treatment duration (7 versus 14 days) and revealed a significantly different cure rate of H. pylori for CYP2C19 genotypes (EM vs. PM) in those studies of 7 days, but not of 14 days, indicating that effects of the CYP2C19 genotype on the cure rate of H. pylori could be reduced with prolonging of treatment time. The cure rate of H. pylori was higher in 14-day therapy than that in 7-day therapy (88.1% vs. 80.8%), Hwang et al. (2015) reported that 14-day therapy was a more effective second-line treatment as compared to the 7-day therapy for H. pylori infection in South Korea (Hwang et al., 2015), indicating that the 14-day therapy may prioritize patients with all genotypes, regardless of the effect of other factors. Therefore, the cost effectiveness of a general recommendation of 14-day therapy has to be confirmed by pharmacoeconomic analysis.
Different geographical locations and genetic backgrounds of CYP2C19 could affect the cure rate of H. pylori. We collected data about H. pylori treatment from several countries and found that the cure rates of the EM genotype and IM genotype were significantly different in Asia, but not between the EM genotype and PM genotype. Therefore, a more detailed subgroup analysis stratified by country or geographical location, including Mainland China, China Taiwan, Japan, South Korea, India, Thailand, and Turkey, should be performed. However, CYP2C19 genetic polymorphism was not associated with the cure rate of H. pylori by PPIs-based therapy in Europe. Why are there regional differences? The distribution of PM was reported to show a considerable interethnic variation. For example, East Asian people, including Chinese, South Koreans, and Japanese, have 13%–23% PMs, whereas Caucasians and African-Americans have only less than 6% (Küpfer et al., 1984; Wedlund et al., 1984; Jacqz et al., 1988; Horai et al., 1989; Sanz et al., 1989; Bertilsson et al., 1992; Sohn et al., 1992; Edeki et al., 1996; Kubota et al., 1996), indicating that the frequency of the PM genotype is significantly higher in the Asian population than in those of other ethnic descents (Adachi et al., 2000). In addition, there were differences in dietary habits and economic levels between Asia and Europe, as well as differences in therapeutic schedules and treatment levels. Therefore, it is more clinically relevant to investigate the effect of the CYP2C19 genotype on H. pylori treatment in Asia.
Finally, we analyzed the influence of CYP2C19 genotypes on the cure rate of H. pylori in patients who received PPI-based triple therapy and quadruple therapies. We found that H. pylori treatment by triple therapy was influenced by the CYP2C19 genotype, but quadruple therapy was not. In addition, Fu et al. (2021) also showed that the quadruple therapy was not affected by CYP2C19 genetic polymorphism, consistent with our results. This could be interpreted as bismuth is mainly metabolized by the kidney, and its inhibitory effect on H. pylori is mainly through the inhibition of proteases, urokinase, and phospholipase produced by H. pylori, all of which are not affected by CYP2C19. Therefore, bismuth-containing quadruple regimens and PPIs (e.g., rabeprazole, esomeprazole, and pantoprazole) are less affected by CYP2C19 genetic polymorphisms and may be more appropriate solutions when only the effects of single genetic polymorphisms are considered (Fu et al., 2021).
Although the effect of the CYP2C19 genotype on the cure rate of H. pylori had been reported, our analysis was more comprehensive and the data more detailed than other meta-analyses. Tang et al. (2013) showed that the treatment of H. pylori by omeprazole and lansoprazole were affected by CYP2C19 genetic polymorphisms, while esomeprazole and rabeprazole were not affected, consistent with our results, but Tang et al. did not explore whether H. pylori treatment by pantoprazole was affected by CYP2C19 genetic polymorphisms due to the inclusion of less data on pantoprazole, and only triple therapy was included, so subgroup analysis of treatment regimens could not be performed, furthermore, subgroup analysis of geography and treatment duration were also not conducted. Recently, Fu et al. (2021) considered the effect of geographical factors on the CYP2C19 genotype, demonstrated the H. pylori cure rates of Mainland Chinese were influenced by CYP2C19 genetic polymorphism, whereas China Taiwanese was not affected, they attributed it to possible difference in dietary habits and treatment protocols.
In addition, to investigate whether there were crossover patients in the clinical trial studies, we pooled the country, institution, time, number of patients, and type of the study for each study and found that each study was conducted at a different institution and at a different time, so there were no crossover patients (Table 8).
This study may have the following limitations. First, the sample size of the included studies was not uniform, and the between-study difference may be large. Some studies had a small sample size (at least 32 cases) and the largest sample size (at most 2202) with a 68-fold difference in between. Second, there were limitations of the included literature. In this study, reviewed English scientific journals were included and other language literature works were excluded, which may cause a certain bias to the research results. Third, there was only published literature included. There may still be some documents with negative results that have not been published and included in the analysis, which would also affect the results of this study. Fourth, documents with incomplete data are excluded, which may lead to selection bias. Finally, the effects of dose of PPIs, antibiotic sensitivity, patient compliance, and other genotypes (IL-1β and CYP3A4) on H. pylori cure rates were not considered.
Taken together, our study concluded that there is a significant difference in the cure rate of H. pylori between EM and PM/IM genotypes, especially for the treatment with omeprazole, 7-day therapy, and triple therapy. Therefore, to overcome or minimize the effect of the CYP2C19 genotype on H. pylori cure rates, the appropriate PPIs and treatment plan should be selected according to the genotypes of CYP2C19.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author contributions
XZ and ZZ contributed the central idea, analyzed most of the data, and wrote the initial draft of the manuscript. The remaining authors contributed to refining the ideas, carrying out additional analyses, and finalizing this manuscript.
Funding
This work was supported by the Jiangsu Provincial Key Research and Development Plan (BE2019614).
Acknowledgments
We are grateful to my respected supervisor Mr. Hongguang Xie, whose guidance and advice have ensured the accomplishment of this thesis. During the epidemic, he still meticulously review the amendments, and putting forward valuable comments until this article was formed. Finally, we would also like to thank all the authors for their contributions to this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adachi, K., KaTsube, T., KawAmurA, A., Takashima, T., YukiM., , , Amano, K., et al. (2000). CYP2C19 genotype status and intragastric pH during dosing with lansoprazole or rabeprazole. Aliment. Pharmacol. Ther. 14, 1259–1266. doi:10.1046/j.1365-2036.2000.00840.x
Ahmad, S., Fathy, A. A., Rizk, E.-B., and Amr, G. (2014). Cure rate of Helicobacter pylori infection in Egyptian children related to CYP2C19 gene polymorphism. Indian J. Gastroenterol. 33, 330–335. doi:10.1007/s12664-014-0450-6
Andersson, T., Holmberg, J., Röhss, K., and Walan, A. (1998). Pharmacokinetics and effect on caffeine metabolism of the proton pump inhibitors, omeprazole, lansoprazole, and pantoprazole. Br. J. Clin. Pharmacol. 45, 369–375. doi:10.1046/j.1365-2125.1998.t01-1-00702.x
Arenas, A., Serrano, C., Quinones, L., Harris, P., SandovalM., , , LavanderosM., , et al. (2019). High prevalence of clarithromycin resistance and effect on Helicobacter pylori eradication in a population from santiago, Chile: Cohort study and meta-analysis. Sci. Rep. 9, 20070. doi:10.1038/s41598-019-56399-7
Bae, L. S., Park, S. J., Ryu, J. K., Lee, J. K., Kim, H. J., Bae, J. S., et al. (2003). Efficacy of triple therapy with rabeprazole for Helicobacter pylori infection in relation to CYP2C19 genotype. Korean J. gastroenterology = Taehan Sohwagi Hakhoe chi 42, 468–475.
Bertilsson, L., Lou, Y. Q., Du, Y. L., Liu, Y., Kuang, T. Y., Liao, X. M., et al. (1992). Pronounced differences between native Chinese and Swedish populations in the polymorphic hydroxylations of debrisoquin and S-mephenytoin. Clin. Pharmacol. Ther. 51, 388–397. doi:10.1038/clpt.1992.38
Burhan, O., Akkiz, H., Bayram, S., Bekar, A., Akgollu, E., and Sandikci, M. (2010). Influence of CYP2C19 functional polymorphism on Helicobacter pylori eradication. Turk. J. Gastroenterol. 21, 23–28.
Chanagune, S., Sith, S., Arti, W.-u., Varocha, M., and Ratha-korn, V. (2014). Improved eradication rate of standard triple therapy by adding bismuth and probiotic supplement for Helicobacter pylori treatment in Thailand. Asian pac. J. Cancer Prev. 15, 9909–9913. doi:10.7314/apjcp.2014.15.22.9909
Chang, M., TybrinG, G., Dahl, M. L., Gotharson, E., SagarM., , , Seensalu, R., et al. (1995). Interphenotype differences in disposition and effect on gastrin levels of omeprazole-suitability of omeprazole as a probe for CYP2C19. Br. J. Clin. Pharmacol. 39, 511–518. doi:10.1111/j.1365-2125.1995.tb04488.x
Chaudhry, A. S., Kochhar, R., and Kohli, K. K. (2008). Genetic polymorphism of CYP2C19 & therapeutic response to proton pump inhibitors. Indian J. Med. Res. 127, 521–530.
Dent, J. (2003). Review article: Pharmacology of esomeprazole and comparisons with omeprazole. Aliment. Pharmacol. Ther. 17 (1), 5–9. doi:10.1046/j.1365-2036.17.s1.2.x
Dojo, M., Azuma, T., SaiTo, T., OhtaniM., , , MurAmAtsu, A., and KuriyaMaM., (2001). Effects of CYP2C19 gene polymorphism on cure rates for Helicobacter pylori infection by triple therapy with proton pump inhibitor (omeprazole or rabeprazole), amoxycillin and clarithromycin in Japan. Dig. Liver Dis. 33, 671–675. doi:10.1016/s1590-8658(01)80043-8
Duval, S., and Tweedie, R. (2000). Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 56, 455–463. doi:10.1111/j.0006-341x.2000.00455.x
Edeki, T. I., Goldstein, J. A., de Morais, S. M., HajiLoo, L., ButlerM., , , ChaPdelaine, P., et al. (1996). Genetic polymorphism of S-mephenytoin 4'-hydroxylation in african-Americans. Pharmacogenetics 6, 357–360. doi:10.1097/00008571-199608000-00009
El Rouby, N., Lima, J. J., and Johnson, J. A. (2018). Proton pump inhibitors: From CYP2C19 pharmacogenetics to precision medicine. Expert Opin. Drug Metab. Toxicol. 14, 447–460. doi:10.1080/17425255.2018.1461835
Fischbach, W., and Malfertheiner, P. (2018). Helicobacter pylori infection. Dtsch. Arztebl. Int. 115, 429–436. doi:10.3238/arztebl.2018.0429
Franciosi, J. P., Mougey, E. B., Williams, A., Gomez Suarez, R. A., Thomas, C., Creech, C. L., et al. (2018). Association between CYP2C19 extensive metabolizer phenotype and childhood anti-reflux surgery following failed proton pump inhibitor medication treatment. Eur. J. Pediatr. 177, 69–77. doi:10.1007/s00431-017-3051-4
Fu, J., Sun, C. F., He, H. Y., Ojha, S. C., Shi, H., Deng, C. L., et al. (2021). The effect of CYP2C19 gene polymorphism on the eradication rate of Helicobacter pylori by proton pump inhibitors-containing regimens in asian populations: A meta-analysis. Pharmacogenomics 22, 859–879. doi:10.2217/pgs-2020-0127
Furuta, T., Ohashi, K., Kosuge, K., Zhao, X. J., TakashiMaM., , , KiMuraM., , et al. (1999). CYP2C19 genotype status and effect of omeprazole on intragastric pH in humans. Clin. Pharmacol. Ther. 65, 552–561. doi:10.1016/S0009-9236(99)70075-5
Furuta, T., Sagehashi, Y., Shirai, N., Sugimoto, M., Nakamura, A., Kodaira, M., et al. (2005). Influence of CYP2C19 polymorphism and Helicobacter pylori genotype determined from gastric tissue samples on response to triple therapy for H pylori infection. Clin. Gastroenterol. Hepatol. 3, 564–573. doi:10.1016/s1542-3565(04)00779-7
Furuta, T., Shirai, N., Xiao, F., El-Omar, E. M., Rabkin, C. S., Sugimura, H., et al. (2004). Polymorphism of interleukin-1beta affects the eradication rates of Helicobacter pylori by triple therapy. Clin. Gastroenterol. Hepatol. 2, 22–30. doi:10.1016/s1542-3565(03)00288-x
Furuta, T., Sugimoto, M., Shirai, N., and Ishizaki, T. (2007). CYP2C19 pharmacogenomics associated with therapy of Helicobacter pylori infection and gastro-esophageal reflux diseases with a proton pump inhibitor. Pharmacogenomics 8, 1199–1210. doi:10.2217/14622416.8.9.1199
Gawrońska-Szklarz, B., Siuda, A., Kurzawski, M., Bielicki, D., Marlicz, W., and Drozdzik, M. (2010). Effects of CYP2C19 , MDR1 , and interleukin 1-B gene variants on the eradication rate of Helicobacter pylori infection by triple therapy with pantoprazole, amoxicillin, and metronidazole. Eur. J. Clin. Pharmacol. 66, 681–687. doi:10.1007/s00228-010-0818-1
Goldstein, F. C., Steenland, K., Zhao, L., Wharton, W., Levey, A. I., and Hajjar, I. (2017). Proton pump inhibitors and risk of mild cognitive impairment and dementia. J. Am. Geriatr. Soc. 65, 1969–1974. doi:10.1111/jgs.14956
He, N., Huang, S. L., Zhu, R. H., Tan, Z. R., Liu, J., Zhu, B., et al. (2003). Inhibitory effect of troleandomycin on the metabolism of omeprazole is CYP2C19 genotype-dependent. Xenobiotica. 33, 211–221. doi:10.1080/0049825021000023996
Hoaglin, D. C. (2016). Misunderstandings about Q and 'Cochran's Q test' in meta-analysis. Stat. Med. 35, 485–495. doi:10.1002/sim.6632
Hoon, L. J., Jung, H. Y., Choi, K. D., Song, H. J., Lee, G. H., and Kim, J. H. (2010). The influence of CYP2C19 polymorphism on eradication of Helicobacter pylori: A prospective randomized study of lansoprazole and rabeprazole. Gut Liver 4, 201–206. doi:10.5009/gnl.2010.4.2.201
Horai, Y., NakanoM., , , Ishizaki, T., IshiKawa, K., Zhou, H. H., Zhou, B. I., et al. (1989). Metoprolol and mephenytoin oxidation polymorphisms in far eastern oriental subjects: Japanese versus mainland Chinese. Clin. Pharmacol. Ther. 46, 198–207. doi:10.1038/clpt.1989.126
Houben, M. H., van De Beek, D., Hensen, E. F., de Craen, A. J., Rauws, E. A., and Tytgat, G. N. (1999). A systematic review of Helicobacter pylori eradication therapy-the impact of antimicrobial resistance on eradication rates. Aliment. Pharmacol. Ther. 13, 1047–1055. doi:10.1046/j.1365-2036.1999.00555.x
Hu, Y.-m., Xu, J.-m., Mei, Q., Xu, X.-h., and Xu, S.-y. (2005). Pharmacodynamic effects and kinetic disposition of rabeprazole in relation to CYP2C19 genotype in healthy Chinese subjects. Acta Pharmacol. Sin. 26, 384–388. doi:10.1111/j.1745-7254.2005.00047.x
Hwang, J. J., Lee, D. H., Lee, A. R., Yoon, H., Shin, C. M., Park, Y. S., et al. (2015). Efficacy of 14-d vs 7-d moxifloxacin-based triple regimens for second-line Helicobacter pylori eradication. World J. Gastroenterol. 21, 5568–5574. doi:10.3748/wjg.v21.i18.5568
Inaba, T., Mizuno, M., Kawai, K., Yokota, K., Oguma, K., Miyoshi, M., et al. (2002). Randomized open trial for comparison of proton pump inhibitors in triple therapy for Helicobacter pylori infection in relation to CYP2C19 genotype. J. Gastroenterol. Hepatol. 17, 748–753. doi:10.1046/j.1440-1746.2002.02790.x
Ishida, Y., Goto, Y., Kondo, T., Kurata, M., Nishio, K., Kawai, S., et al. (2006). Eradication rate of Helicobacter pylori according to genotypes of CYP2C19, IL-1B, and TNF-A. Int. J. Med. Sci. 3, 135–140. doi:10.7150/ijms.3.135
Jacqz, E., Dulac, H., and Mathieu, H. (1988). Phenotyping polymorphic drug metabolism in the French Caucasian population. Eur. J. Clin. Pharmacol. 35, 167–171. doi:10.1007/BF00609247
Jinda, S., Nakatani, K., Nishioka, J., Yasuda, K., Soya, Y., Hayashi, A., et al. (2011). Personalized treatment in the eradication therapy for Helicobacter pylori. Int. J. Mol. Med. 27, 255–261. doi:10.3892/ijmm.2010.569
Jung-Hwan, O., Dong, M. S., Choi, M. G., Yoo, H. W., Lee, S. B., Park, Y. I., et al. (2009). Effects of CYP2C19 and MDR1 genotype on the eradication rate of Helicobacter pylori infection by triple therapy with pantoprazole, amoxycillin and clarithromycin. J. Gastroenterol. Hepatol. 24, 294–298. doi:10.1111/j.1440-1746.2008.05605.x
Kawabata, H., Habu, Y., Tomioka, H., Kutsumi, H., KobayashiM., , , Oyasu, K., et al. (2003). Effect of different proton pump inhibitors, differences in CYP2C19 genotype and antibiotic resistance on the eradication rate of Helicobacter pylori infection by a 1-week regimen of proton pump inhibitor, amoxicillin and clarithromycin. Aliment. Pharmacol. Ther. 17, 259–264. doi:10.1046/j.1365-2036.2003.01406.x
Ke, H., Li, J., Lu, B., Yang, C., Wang, J., Wang, Z., et al. (2021). The appropriate cutoff gastric pH value for Helicobacter pylori eradication with bismuth-based quadruple therapy. Helicobacter 26, e12768. doi:10.1111/hel.12768
Kim, D., Jung, S. W., Lee, D. W., Lee, C. M., Kim, S. Y., Hyun, J. J., et al. (2019). Efficacy of low dose proton pump inhibitor-based therapy to eradicate Helicobacter pylori in patients with subtotal gastrectomy. J. Clin. Med. 8, E1933. doi:10.3390/jcm8111933
Kittichet, P., Vilaichone, R. K., Siramolpiwat, S., Tangaroonsanti, A., Chonprasertsuk, S., Bhanthumkomol, P., et al. (2016). Effect of IL-1 polymorphisms, CYP2C19 genotype and antibiotic resistance on Helicobacter pylori eradication comparing between 10-day sequential therapy and 14-day standard triple therapy with four-times-daily-dosing of amoxicillin in Thailand: A prospective randomized study. Asian pac. J. Cancer Prev. 17, 1903–1907. doi:10.7314/apjcp.2016.17.4.1903
Klotz, U. (2009). Impact of CYP2C19 polymorphisms on the clinical action of proton pump inhibitors (PPIs). Eur. J. Clin. Pharmacol. 65, 1–2. doi:10.1007/s00228-008-0571-x
Kubota, T., Chiba, K., and Ishizaki, T. (1996). Genotyping of S-mephenytoin 4'-hydroxylation in an extended Japanese population. Clin. Pharmacol. Ther. 60, 661–666. doi:10.1016/S0009-9236(96)90214-3
Kuo, C.-H., Lu, C. Y., Shih, H. Y., Liu, C. J., Wu, M. C., Hu, H. M., et al. (2014). CYP2C19 polymorphism influences Helicobacter pylori eradication. World J. Gastroenterol. 20, 16029–16036. doi:10.3748/wjg.v20.i43.16029
Küpfer, A., Preisig, R., and Kupfer, A. (1984). Pharmacogenetics of mephenytoin: A new drug hydroxylation polymorphism in man. Eur. J. Clin. Pharmacol. 26, 753–759. doi:10.1007/BF00541938
Kurzawski, M., Gawronska-Szklarz, B., Wrzesniewska, J., Siuda, A., Starzynska, T., and Drozdzik, M. (2006). Effect of CYP2C19*17 gene variant on Helicobacter pylori eradication in peptic ulcer patients. Eur. J. Clin. Pharmacol. 62, 877–880. doi:10.1007/s00228-006-0183-2
Kuwayama, H., AsakaM., , , Sugiyama, T., Fukuda, Y., AoyamaN., , Hirai, Y., et al. (2007). Rabeprazole-based eradication therapy for Helicobacter pylori: A large-scale study in Japan. Aliment. Pharmacol. Ther. 25, 1105–1113. doi:10.1111/j.1365-2036.2007.03298.x
Lee, J. Y., Kim, N., Kim, M. S., Choi, Y. J., Lee, J. W., Yoon, H., et al. (2014). Factors affecting first-line triple therapy of Helicobacter pylori including CYP2C19 genotype and antibiotic resistance. Dig. Dis. Sci. 59, 1235–1243. doi:10.1007/s10620-014-3093-7
Lee, V. W., Chau, T. S., Chan, A. K. W., Lee, K. K. C., Waye, M. M. Y., Ling, T. K. W., et al. (2010). Pharmacogenetics of esomeprazole or rabeprazole-based triple therapy in Helicobacter pylori eradication in Hong Kong non-ulcer dyspepsia Chinese subjects. J. Clin. Pharm. Ther. 35, 343–350. doi:10.1111/j.1365-2710.2009.01088.x
Li, X.-Q., Andersson, T. B., Ahlström, M., and Weidolf, L. (2004). Comparison of inhibitory effects of the proton pump-inhibiting drugs omeprazole, esomeprazole, lansoprazole, pantoprazole, and rabeprazole on human cytochrome P450 activities. Drug Metab. Dispos. 32, 821–827. doi:10.1124/dmd.32.8.821
Lima, J. J., Thomas, C. D., Barbarino, J., Desta, Z., Van Driest, S. L., El Rouby, N., et al. (2021). Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2C19 and proton pump inhibitor dosing. Clin. Pharmacol. Ther. 109, 1417–1423. doi:10.1002/cpt.2015
Marshall, B. J., and Warren, J. R. (1984). Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1, 1311–1315. doi:10.1016/s0140-6736(84)91816-6
Marshall, B. J., and Windsor, H. M. (2005).The relation of Helicobacter pylori to gastric adenocarcinoma and lymphoma: Pathophysiology, epidemiology, screening, clinical presentation, treatment, and preventionMed. Clin. North Am., 89. viii, 313–344. doi:10.1016/j.mcna.2004.09.001
McNicholl, A. G., Linares, P. M., Nyssen, O. P., Calvet, X., and Gisbert, J. P. (2012). Meta-analysis: Esomeprazole or rabeprazole vs. first-generation pump inhibitors in the treatment of Helicobacter pylori infection. Aliment. Pharmacol. Ther. 36, 414–425. doi:10.1111/j.1365-2036.2012.05211.x
Miehlke, S., Schneider-Brachert, W., Kirsch, C., Morgner, A., Madisch, A., Kuhlisch, E., et al. (2008). One-week once-daily triple therapy with esomeprazole, moxifloxacin, and rifabutin for eradication of persistent Helicobacter pylori resistant to both metronidazole and clarithromycin. Helicobacter 13, 69–74. doi:10.1111/j.1523-5378.2007.00588.x
Miki, I., Aoyama, N., Sakai, T., Shirasaka, D., Wambura, C. M., Maekawa, S., et al. (2003). Impact of clarithromycin resistance and CYP2C19 genetic polymorphism on treatment efficacy of Helicobacter pylori infection with lansoprazole- or rabeprazole-based triple therapy in Japan. Eur. J. Gastroenterol. Hepatol. 15, 27–33. doi:10.1097/00042737-200301000-00006
Miwa, H., Misawa, H., Yamada, T., Naga, A., Ohtaka, K., and SatoN., (2001). Clarithromycin resistance, but not CYP2C-19 polymorphism, has a major impact on treatment success in 7-day treatment regimen for cure of H. pylori infection: A multiple logistic regression analysis. Dig. Dis. Sci. 46, 2445–2450. doi:10.1023/a:1012371702918
Ormeci, A., Emrence, Z., Baran, B., Gokturk, S., Soyer, O. M., Evirgen, S., et al. (2016). Effect of cytochrome P450 2C19 polymorphisms on the Helicobacter pylori eradication rate following two-week triple therapy with pantoprazole or rabeprazole. Eur. Rev. Med. Pharmacol. Sci. 20, 879–885.
Padol, S., Yuan, Y., Thabane, M., Padol, I. T., and Hunt, R. H. (2006). The effect of CYP2C19 polymorphisms on H. pylori eradication rate in dual and triple first-line PPI therapies: A meta-analysis. Am. J. Gastroenterol. 101, 1467–1475. doi:10.1111/j.1572-0241.2006.00717.x
Pan, X., Li, Y., Qiu, Y., Tang, Q., Qian, B., Yao, L., et al. (2010). Efficacy and tolerability of first-line triple therapy with levofloxacin and amoxicillin plus esomeprazole or rabeprazole for the eradication of Helicobacter pylori infection and the effect of CYP2C19 genotype: A 1-week, randomized, open-label study in Chinese adults. Clin. Ther. 32, 2003–2011. doi:10.1016/j.clinthera.2010.11.005
Park, S., Hyun, Y. J., Kim, Y. R., Lee, J. H., Ryu, S., Kim, J. M., et al. (2017). Effects of CYP2C19 genetic polymorphisms on PK/PD responses of omeprazole in Korean healthy volunteers. J. Korean Med. Sci. 32, 729–736. doi:10.3346/jkms.2017.32.5.729
Piyakorn, P., Peranart, C., and Ratha-Korn, V. (2019). High effective of 14-day high-dose PPI- bismuth-containing quadruple therapy with probiotics supplement for Helicobacter pylori eradication: A double blinded-randomized placebo-controlled study. Asian pac. J. Cancer Prev. 20, 2859–2864. doi:10.31557/APJCP.2019.20.9.2859
Prasertpetmanee, S., Mahachai, V., and Vilaichone, R. K. (2013). Improved efficacy of proton pump inhibitor - amoxicillin - clarithromycin triple therapy for Helicobacter pylori eradication in low clarithromycin resistance areas or for tailored therapy. Helicobacter 18, 270–273. doi:10.1111/hel.12041
Qiao, H.-L., Hu, Y. R., Tian, X., Jia, L. J., Gao, N., Zhang, L. R., et al. (2006). Pharmacokinetics of three proton pump inhibitors in Chinese subjects in relation to the CYP2C19 genotype. Eur. J. Clin. Pharmacol. 62, 107–112. doi:10.1007/s00228-005-0063-1
Sanz, E. J., Villén, T., Alm, C., and Bertilsson, L. (1989). S-mephenytoin hydroxylation phenotypes in a Swedish population determined after coadministration with debrisoquin. Clin. Pharmacol. Ther. 45, 495–499. doi:10.1038/clpt.1989.63
Sapone, A., Vaira, D., Trespidi, S., PernaF., , , Gatta, L., Tampieri, A., et al. (2003). The clinical role of cytochrome p450 genotypes in Helicobacter pylori management. Am. J. Gastroenterol. 98, 1010–1015. doi:10.1111/j.1572-0241.2003.07427.x
Savarino, V., Marabotto, E., Zentilin, P., Furnari, M., Bodini, G., De Maria, C., et al. (2018). The appropriate use of proton-pump inhibitors. Minerva Med. 109, 386–399. doi:10.23736/S0026-4806.18.05705-1
Shah, S. C., Tepler, A., Chung, C. P., Suarez, G., Peek, R. M., Hung, A., et al. (2021). Host genetic determinants associated with Helicobacter pylori eradication treatment failure: A systematic review and meta-analysis. Gastroenterology 161, 1443–1459. doi:10.1053/j.gastro.2021.07.043
Sheu, B. S., Kao, A. W., Cheng, H. C., Hunag, S. F., Chen, T. W., Lu, C. C., et al. (2005). Esomeprazole 40 mg twice daily in triple therapy and the efficacy of Helicobacter pylori eradication related to CYP2C19 metabolism. Aliment. Pharmacol. Ther. 21, 283–288. doi:10.1111/j.1365-2036.2005.02281.x
Shimatani, T., InoueM., , , Kuroiwa, T., Horikawa, Y., Mieno, H., and NakaMuraM., (2003). Effect of omeprazole 10 mg on intragastric pH in three different CYP2C19 genotypes, compared with omeprazole 20 mg and lafutidine 20 mg, a new H2-receptor antagonist. Aliment. Pharmacol. Ther. 18, 1149–1157. doi:10.1046/j.1365-2036.2003.01804.x
Shin, J. M., and Kim, N. (2013). Pharmacokinetics and pharmacodynamics of the proton pump inhibitors. J. Neurogastroenterol. Motil. 19, 25–35. doi:10.5056/jnm.2013.19.1.25
Shyan, L. C., Rong, L. J., and Lin, J. R. (2010). Correlation of CYP2C19 genetic polymorphisms with Helicobacter pylori eradication in patients with cirrhosis and peptic ulcer. J. Chin. Med. Assoc. 73, 188–193. doi:10.1016/S1726-4901(10)70039-3
Sienkiewicz-Oleszkiewicz, B., Wiela-Hojeńska, A., and WielA-HojenskA, A. (2018). CYP2C19 polymorphism in relation to the pharmacotherapy optimization of commonly used drugs. Pharmazie 73, 619–624. doi:10.1691/ph.2018.8689
Sohn, D. R., KusakaM., , , Ishizaki, T., Shin, S. G., Jang, I. J., Shin, J. G., et al. (1992). Incidence of S-mephenytoin hydroxylation deficiency in a Korean population and the interphenotypic differences in diazepam pharmacokinetics. Clin. Pharmacol. Ther. 52, 160–169. doi:10.1038/clpt.1992.125
Song, Z., Zhou, L., Xue, Y., Suo, B., Tian, X., and Niu, Z. (2020). A comparative study of 14-day dual therapy (esomeprazole and amoxicillin four times daily) and triple plus bismuth therapy for first-line Helicobacter pylori infection eradication: A randomized trial. Helicobacter 25, e12762. doi:10.1111/hel.12762
Sugano, K., Tack, J., Kuipers, E. J., Graham, D. Y., El-Omar, E. M., Miura, S., et al. (2015). Kyoto global consensus report on Helicobacter pylori gastritis. Gut 64, 1353–1367. doi:10.1136/gutjnl-2015-309252
Sugimoto, M., Furuta, T., Shirai, N., Kodaira, C., Nishino, M., Ikuma, M., et al. (2007). Evidence that the degree and duration of acid suppression are related to Helicobacter pylori eradication by triple therapy. Helicobacter 12, 317–323. doi:10.1111/j.1523-5378.2007.00508.x
Sugimoto, M., Hira, D., Murata, M., Kawai, T., and Terada, T. (2020). Effect of antibiotic susceptibility and CYP3A4/5 and CYP2C19 genotype on the outcome of vonoprazan-containing Helicobacter pylori eradication therapy. Antibiot. (Basel) 910, 645. doi:10.3390/antibiotics9100645
Sugimoto, M., Uotani, T., Sahara, S., Ichikawa, H., Yamade, M., Sugimoto, K., et al. (2014). Efficacy of tailored Helicobacter pylori eradication treatment based on clarithromycin susceptibility and maintenance of acid secretion. Helicobacter 19, 312–318. doi:10.1111/hel.12128
Suzuki, T, Matsuo, K, Sawaki, A, Wakai, K, Hirose, K, Ito, H, et al. (2007). Influence of smoking and CYP2C19 genotypes on H. pylori eradication success. Epidemiol. Infect. 135 (1), 171–6. doi:10.1017/S0950268806006613
Take, S., Mizuno, M., Ishiki, K., Nagahara, Y., Yoshida, T., Inaba, T., et al. (2003). Interleukin-1beta genetic polymorphism influences the effect of cytochrome P 2C19 genotype on the cure rate of 1-week triple therapy for Helicobacter pylori infection. Am. J. Gastroenterol. 98, 2403–2408. doi:10.1111/j.1572-0241.2003.07707.x
Tang, H. L., Li, Y., Hu, Y. F., Xie, H. G., and Zhai, S. D. (2013). Effects of CYP2C19 loss-of-function variants on the eradication of H. pylori infection in patients treated with proton pump inhibitor-based triple therapy regimens: A meta-analysis of randomized clinical trials. PLoS One 8, e62162. doi:10.1371/journal.pone.0062162
Tybring, G., Böttiger, Y., Widén, J., and Bertilsson, L. (1997). Enantioselective hydroxylation of omeprazole catalyzed by CYP2C19 in Swedish white subjects. Clin. Pharmacol. Ther. 62, 129–137. doi:10.1016/S0009-9236(97)90060-6
Wang, Y., Yuan, Y., Meng, L., Fan, H., Xu, J., Zhang, H., et al. (2011). Study of the pharmacokinetics and intragastric pH of rabeprazole given as successive intravenous infusion to healthy Chinese volunteers. Eur. J. Clin. Pharmacol. 67, 25–31. doi:10.1007/s00228-010-0949-4
Ward, R. M., and Kearns, G. L. (2013). Proton pump inhibitors in pediatrics : Mechanism of action, pharmacokinetics, pharmacogenetics, and pharmacodynamics. Paediatr. Drugs 15, 119–131. doi:10.1007/s40272-013-0012-x
Wedlund, P. J., Aslanian, W. S., McAllister, C. B., Wilkinson, G. R., and Branch, R. A. (1984). Mephenytoin hydroxylation deficiency in Caucasians: Frequency of a new oxidative drug metabolism polymorphism. Clin. Pharmacol. Ther. 36, 773–780. doi:10.1038/clpt.1984.256
Woon, C. Y., Ko, W. J., Oh, C. H., Park, Y. M., Oh, S. J., Moon, J. R., et al. (2019). Clarithromycin resistance and female gender affect Helicobacter pylori eradication failure in chronic gastritis. Korean J. Intern. Med. 34, 1022–1029. doi:10.3904/kjim.2018.054
Yamada, T., Searle, J. G., Ahnen, D., Aipers, D. H., Greenberg, H. B., Gray, M., et al. (1994). NIH consensus conference. Helicobacter pylori in peptic ulcer disease. NIH consensus development panel on Helicobacter pylori in peptic ulcer disease. JAMA 272, 65–69. doi:10.1001/jama.1994.03520010077036
Yang, L., Zou, A., Wu, H., Guo, H., Zhang, F., Zou, B., et al. (2021). Application of visual gene clip-based tailored therapy for the eradication of Helicobacter pylori. BioMed Res. Int. 2021, 1–6. doi:10.1155/2021/6150628
Yu, L.-Y., Sun, L. N., Zhang, X. H., Li, Y. Q., Yu, L., Yuan, Z. Q. Y., et al. (2017). A review of the novel application and potential adverse effects of proton pump inhibitors. Adv. Ther. 34, 1070–1086. doi:10.1007/s12325-017-0532-9
Yun-An, L., Wang, H., Gu, Z. J., Wang, W. J., Zeng, X. Y., Du, Y. L., et al. (2017). Effect of CYP2C19 gene polymorphisms on proton pump inhibitor, amoxicillin, and levofloxacin triple therapy for eradication of Helicobacter pylori. Med. Sci. Monit. 23, 2701–2707. doi:10.12659/msm.901514
Zamani, M., EbrahimtabarF., , , Zamani, V., Miller, W. H., Alizadeh-Navaei, R., Shokri-Shirvani, J., et al. (2018). Systematic review with meta-analysis: The worldwide prevalence of Helicobacter pylori infection. Aliment. Pharmacol. Ther. 47, 868–876. doi:10.1111/apt.14561
Zhang, H.-J., Zhang, X. H., Liu, J., Sun, L. N., Shen, Y. W., Zhou, C., et al. (2020). Effects of genetic polymorphisms on the pharmacokinetics and pharmacodynamics of proton pump inhibitors. Pharmacol. Res. 152, 104606. doi:10.1016/j.phrs.2019.104606
Zhang, L., Mei, Q., Li, Q. S., Hu, Y. M., and Xu, J. M. (2010). The effect of cytochrome P2C19 and interleukin-1 polymorphisms on H. pylori eradication rate of 1-week triple therapy with omeprazole or rabeprazole, amoxycillin and clarithromycin in Chinese People. J. Clin. Pharm. Ther. 35, 713–722. doi:10.1111/j.1365-2710.2009.01140.x
Zhao, F., Wang, J., Yang, Y., Wang, X., Shi, R., Xu, Z., et al. (2008). Effect of CYP2C19 genetic polymorphisms on the efficacy of proton pump inhibitor-based triple therapy for Helicobacter pylori eradication: A meta-analysis. Helicobacter 13, 532–541. doi:10.1111/j.1523-5378.2008.00643.x
Keywords: Helicobacter pylori, eradication rate, genetic polymorphisms, CYP2C19, meta- analysis
Citation: Zhao X, Zhang Z, Lu F, Xiong M, Jiang L, Tang K, Fu M, Wu Y and He B (2022) Effects of CYP2C19 genetic polymorphisms on the cure rates of H. pylori in patients treated with the proton pump inhibitors: An updated meta-analysis. Front. Pharmacol. 13:938419. doi: 10.3389/fphar.2022.938419
Received: 07 May 2022; Accepted: 12 September 2022;
Published: 06 October 2022.
Edited by:
Carsten Wrenger, University of São Paulo, BrazilReviewed by:
Junji Umeno, Kyushu University, JapanYogan Khatri, Cayman Chemical, Ann Arbor, United States
Ali Saffaei, Shahid Beheshti University of Medical Sciences, Iran
Copyright © 2022 Zhao, Zhang, Lu, Xiong, Jiang, Tang, Fu, Wu and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bangshun He, YmhlQG5qbXUuZWR1LmNu
†These authors have contributed equally to this work
 Xianghong Zhao
Xianghong Zhao Zhongqiu Zhang1,3†
Zhongqiu Zhang1,3† Bangshun He
Bangshun He