
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 26 October 2022
Sec. Inflammation Pharmacology
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.926317
This article is part of the Research Topic Drugging p53 for Non-Cancer Diseases View all 6 articles
CX-5461, a novel selective RNA polymerase I inhibitor, shows potential anti-inflammatory and immunosuppressive activities. However, the molecular mechanisms underlying the inhibitory effects of CX-5461 on macrophage-mediated inflammation remain to be clarified. In the present study, we attempted to identify the systemic biological processes which were modulated by CX-5461 in inflammatory macrophages. Primary peritoneal macrophages were isolated from normal Sprague Dawley rats, and primed with lipopolysaccharide or interferon-γ. Genome-wide RNA sequencing was performed. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes databases were used for gene functional annotations. Enrichment analysis was conducted using the ClusterProfiler package of R software. We found that CX-5461 principally induced a molecular signature related to cell cycle inhibition in primed macrophages, featuring downregulation of genes encoding cell cycle mediators and concomitant upregulation of cell cycle inhibitors. At the same concentration, however, CX-5461 did not induce a systemic anti-inflammatory transcriptional program, although some inflammatory genes such as IL-1β and gp91phox NADPH oxidase were downregulated by CX-5461. Our data further highlighted a central role of p53 in orchestrating the molecular networks that were responsive to CX-5461 treatment. In conclusion, our study suggested that limiting cell proliferation predominated in the inhibitory effects of CX-5461 on macrophage-mediated inflammation.
CX-5461 is a novel selective RNA polymerase I (Pol I) inhibitor which exhibits promising therapeutic efficacies in treatment of certain blood and solid tumors (Drygin et al., 2011; Haddach et al., 2012; Ferreira et al., 2020), with favorable tolerance and safety profiles in human subjects (Khot et al., 2019). Pol I mediates the synthesis of ribosomal RNA, while inhibiting Pol I activity in mammalian cells induces a distinct cellular stress response termed nucleolar stress response, which culminates in stabilization and/or activation of the p53 pathway (Boulon et al., 2010; Bywater et al., 2013; Yang et al., 2018). Specifically, CX-5461 triggers a non-canonical DNA damage response (DDR), featuring activation of the ataxia telangiectasia mutated (ATM)/ATM and Rad3-related (ATR) pathway upstream of p53 (Negi and Brown, 2015; Quin et al., 2016; Ye et al., 2017; Pang et al., 2021). Experimental evidence from our and other laboratories implies that the anti-proliferative effect of CX-5461 in p53-intact cells is mainly mediated by stimulation of the DDR-p53 axis, but not directly attributable to inhibition of ribosome biogenesis (Bywater et al., 2012; Tsai and Pederson, 2014; Quin et al., 2016; Ye et al., 2017; Sanij et al., 2020; Pang et al., 2021). However, the molecular events induced by CX-5461 in the cell leading to ATM/ATR activation is still under debate (Drygin et al., 2011; Quin et al., 2016; Xu et al., 2017; Bruno et al., 2020; Mars et al., 2020).
In addition to its anti-tumor activities, our previous studies have also revealed that CX-5461 has unique pharmacological effects on pathologic vascular remodeling and the accompanying vascular inflammation. For example, CX-5461 treatment effectively suppressed the arterial remodeling induced by orthotopic aortic transplantation and experimental pulmonary arterial hypertension (Dai et al., 2018; Xu et al., 2021); in these experiments, CX-5461 significantly reduced the number of infiltrating adventitial macrophages. In vitro, CX-5461 inhibited macrophage proliferation, maturation, and lipopolysaccharide (LPS)-induced expression of pro-inflammatory cytokines (Dai et al., 2018). Because infiltration and activation of adventitial macrophages have crucial roles in promoting the development of vascular inflammation and remodeling (Wang et al., 2018), our results suggest that the modulating effects of CX-5461 on macrophage functions may have a major contribution to its vascular protective actions.
Consistent with our experimental results, there is evidence showing that p53 activation exhibits anti-inflammatory properties under various pathologic conditions [reviewed recently in (Cui et al., 2021)]. In macrophages, p53 may produce anti-inflammatory effects by downregulating the expression of STAT-1 (signal transducer and activator of transcription 1) (Zheng et al., 2005). In addition, p53 may possibly antagonize the pro-inflammatory signaling events downstream of interleukin (IL)-1β receptor, tumor necrosis factor (TNF)-α receptor, and Toll-like receptors, by limiting functions of the master transcription factor NF-κB (nuclear factor-κB) (Wadgaonkar et al., 1999; Webster and Perkins, 1999). It has been well established that pro-inflammatory activation of macrophages has pivotal roles in orchestrating the development of inflammation, thereby making significant contributions to the pathogenesis of most acute and chronic inflammatory diseases, such as infectious sepsis, severe acute respiratory syndromes, autoimmune diseases, allergies, cardiovascular diseases, metabolic syndromes, and cancer (Austermann et al., 2022). At present, however, the molecular mechanisms underlying the inhibitory effects of CX-5461 on macrophage-mediated inflammation remain to be clarified. Based on our previous observations from cell functional studies, here we attempt to identify the systemic biological processes which are modulated by CX-5461 in LPS- and interferon (IFN)-γ-primed macrophages, using contemporary genome-wide RNA sequencing technology. These data may provide a molecular framework for understanding the mechanisms by which CX-5461 suppresses macrophage-mediated inflammation.
Normal adult Sprague Dawley rats (8–10 weeks of age) were purchased from Vital River Laboratory (Beijing, China). The use of experimental animals was approved by the institutional Animal Ethics Committee of Qilu Hospital of Shandong University. Animals were handled in accordance with the Guideline for the Care and Use of Laboratory Animals (NIH, United States). Peritoneal macrophages were elicited as described (Liu et al., 2016). For each rat, 5 ml of 3% thioglycollate broth (from Sigma, St. Louis, MO, United States) was injected into the peritoneal cavity. After 72 h, the animal was euthanized by intraperitoneal injection of an overdose of pentobarbital sodium. The peritoneal cavity was flushed with 10 ml of PBS solution containing 1 mM EDTA (pH 7.5). The solution was withdrawn using a syringe with 18G needle, and cells were harvested by centrifugation. Cells were resuspended in DMEM medium supplemented with 10% FBS and antibiotics (all from Thermo Fisher, Waltham, MA, United States), and seeded in 6-well plates. After 2 h of incubation, non-adherent cells were removed by gently washing the wells. Cells were used for experimentation when ∼70% of confluence was reached.
CX-5461 (purchased from ApexBio Technology LLC, Houston, TX, United States) was initially dissolved in DMSO, and diluted in the culture medium to the final concentration of 1 μM (treatment duration 24 h). DMSO was used as vehicle control. LPS was purchased from Solarbio (Beijing, China) and used at the final concentration of 1 μg/ml. IFN-γ was from Proteintech (Wuhan, Hubei Province, China) and used at 150 U/ml.
The RNA processing and sequencing services were provided by LC-Bio Technology (Hangzhou, Zhejiang Province, China). Total RNA was extracted using Trizol reagent (Thermo Fisher) following the manufacturer’s instructions. The RNA quantity and purity were analyzed using Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (from Agilent, Santa Clara, CA, United States). RNA samples with RIN number > 7.0 were used for library construction. Purified mRNA was obtained from 5 μg of total RNA using Dynabeads Oligo (dT) (Thermo Fisher) with two rounds of purification; the mRNA was then fragmented using divalent cations (Magnesium RNA Fragmentation Module from New England Biolabs, Ipswich, MA, United States) under 94°C for 5 min.
The cleaved mRNAs were reverse-transcribed to cDNAs using SuperScript II Reverse Transcriptase (Thermo Fisher), which were used as templates to synthesize U-labeled second-strand DNAs with E. coli DNA polymerase I, RNase H (all from New England Biolabs) and dUTP (Thermo Fisher). After ligation with Dual-index adapters, the DNA library was treated with heat-labile UDG enzyme (New England Biolabs) to remove the U-labeled second strands, and amplified by PCR. The average insert size of the final cDNA library was 300 bp ± 50 bp. Paired-end sequencing (2 bp × 150 bp) was performed using a NovaSeq 6000 Sequencing System (Illumina, San Diego, CA, United States). High quality clean reads were obtained by filtering with Cutadapt (version 1.9) (https://cutadapt.readthedocs.io/en/stable/) and verified with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Read alignment to the Rattus norvegicus reference genome was performed using HISAT2 package (version 2.0.4) (https://daehwankimlab.github.io/hisat2). Read assembly was performed using StringTie (version 1.3.4d) (http://ccb.jhu.edu/software/stringtie). StringTie and Ballgown (http://www.bioconductor.org/packages/release/bioc/html/ballgown.html) were used to estimate the relative expression levels (expressed as fragment per kilobase per million reads/FPKM values). In each experimental group, 3 independent samples were included. Gene expression data were analyzed with DESeq2 software (version 3.14) (https://bioconductor.org/packages/release/bioc/html/DESeq2.html). Genes with p value of < 0.05 and absolute fold change ≥ 2 were included for further bioinformatics analysis. The raw data set of RNA sequencing experiment was deposited at Dryad public repository (https://doi.org/10.5061/dryad.51c59zwb7).
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases were used for gene functional annotations. Enrichment analysis was conducted using the ClusterProfiler package of R software (Yu et al., 2012), and gene clusters with false discovery rate (FDR) of < 0.05 were considered as significant.
Cells were homogenized in TRIzol Reagent (Thermo Fisher), and total RNA was isolated according to the manufacturer’s instructions. The RNA concentration was determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher). cDNA was synthesized using PrimeScript RT-PCR Kit from TaKaRa (RR037A) (Otsu, Shiga, Japan). The PCR reaction was carried out using SYBR green qPCR mix (#CW0957M, from CoWin Biosciences, Taizhou, Jiangsu Province, China) on a StepOne Real-Time PCR System (Thermo Fisher). The primer sequences used in the study were listed in Supplementary Table S1.
Cells re-suspended in culture medium were seeded in 12-well plates (3 × 105 cells per well). To inhibit p53, the cells were pretreated with pifithrin-α (20 μM) (#S5791 from Selleck Chemicals, Houston, TX, United States). After various treatments as indicated, the cells were pulse labeled with BrdU (10 μM) (from MedChemExpress, NJ, United States) for 1.5 h. BrdU incorporation was detected using immunofluorescence with a monoclonal BrdU antibody (#A20790, from ABclonal, Woburn, MA, United States) followed by Alexa Fluor 488-conjugated secondary antibody. The images were taken using a fluorescence microscope equipped with a DP74 CCD camera (Olympus, Tokyo, Japan).
The numerical data of individual genes were analyzed with two-tailed unpaired t-test for comparison between two groups, or one-way ANOVA followed by Tukey’s test for multi comparisons, using Prism software (GraphPad, San Diego, CA, United States). A value of p < 0.05 was considered as significant.
To delineate the potential anti-inflammatory activities of CX-5461, we first primed macrophage cells separately with LPS and IFN-γ, two classical pro-inflammatory stimuli with interweaving effects in macrophage (Schroder et al., 2006; Hu et al., 2008). LPS stimulation changed the expressions of 979 genes (Figure 1A; Supplementary Table S2), including several classical pro-inflammatory molecules such as IL1A (IL-1α), IL1B (IL-1β), IL6 (IL-6), NOS2 (inducible nitric oxide synthase/iNOS), CCL2 (C-C motif chemokine ligand 2; aka. monocyte chemoattractant protein 1/MCP-1), CCL3 (aka. macrophage inflammatory protein-1α/MIP-1α), CXCL2 (C-X-C motif chemokine ligand 2; aka. MIP-2α), and CCL12 (aka. MCP-5). IFN-γ stimulation changed the expressions of 285 genes (Figure 1A; Supplementary Table S3), including CCL4 and CCL6 (GO term: cellular response to interferon-gamma), which were moderately upregulated; and SLAMF1 (signaling lymphocytic activation molecule family member 1) (GO term: negative regulation of interferon-gamma production), which was highly upregulated. The upregulation of SLAMF1 gene in IFN-γ-primed macrophages was consistent with previous reports (Howie et al., 2002). Unsupervised k-means clustering analysis confirmed that LPS and IFN-γ induced distinct transcriptome signatures in the macrophage (Figure 1B).
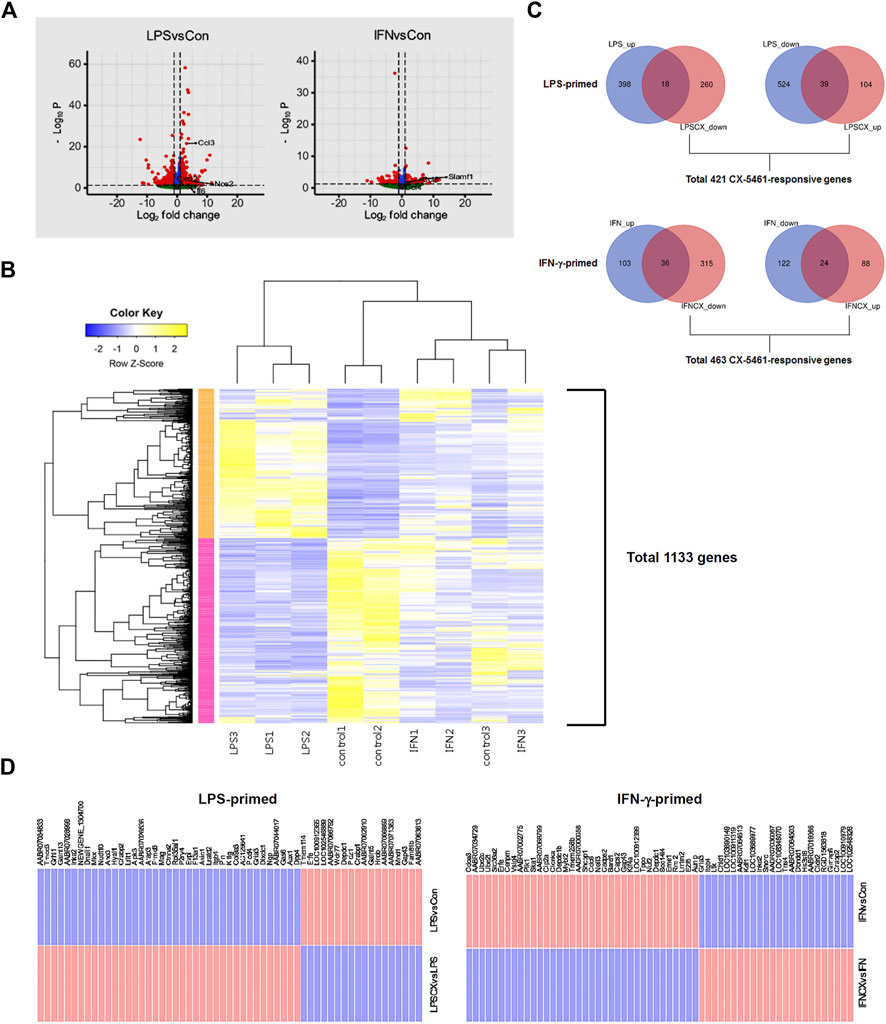
FIGURE 1. CX-5461 did not directly reverse the transcriptome signatures induced by LPS or IFN-γ in macrophages. (A) Volcano plots (log normalized p values versus log normalized fold change values) showing the genome-wide transcriptional effects of LPS and IFN-γ stimulation in primary macrophages. (B) Result of unsupervised k-means clustering analysis confirming that LPS and IFN-γ induced distinct transcriptome signatures in the macrophage (each treatment condition had 3 biological replications). (C) Venn diagrams showing that only a minor part of the transcriptome signature induced by LPS or IFN-γ was directly reversed by CX-5461 treatment. LPS_up: number of upregulated genes LPS treatment versus resting cells; LPSCX_down: downregulated genes LPS + CX-5461 versus LPS alone; LPS_down: downregulated genes LPS treatment versus resting cells; LPSCX_up: upregulated genes LPS + CX-5461 versus LPS alone. Grouping annotations were the same for IFN-γ-primed cells. The total numbers of CX-5461 responsive genes in LPS-primed and IFN-γ-primed cells were shown below the graphs respectively. (D) Lists of LPS- and IFN-γ-regulated genes which were directly reverted by CX-5461 treatment.
According to the observations from our previous studies (Ye et al., 2017; Dai et al., 2018; Pang et al., 2021; Xu et al., 2021; Pan et al., 2022), activation of the p53 pathway following Pol I inhibition (rather than reduced productions of rRNAs and ribosomes) appears to have a predominant role in mediating CX-5461 effects. This phenomenon has also been recognized in cancer cells (Ferreira et al., 2020). Hence we first confirmed that CX-5461 at 1 μM (as established by our previous studies) effectively reduced the pre-rRNA level and stimulated p53 phosphorylation (on Ser15) under the present experimental settings (see Supplementary Figure S1). Using this concentration of CX-5461, we showed that of the 416 genes which were upregulated in LPS-primed macrophages (increased in LPS group as compared to control group), 18 were reverted by CX-5461 treatment (i.e., decreased in CX + LPS group as compared to LPS alone group); of the 563 genes which were downregulated in LPS-primed cells, 39 were reverted by CX-5461 (Figures 1C,D). Of the 139 genes which were upregulated in IFN-γ-primed cells, 36 were reverted by CX-5461; of the 146 genes which were downregulated in IFN-γ-primed cells, 24 were reverted by CX-5461 (Figures 1C,D; Supplementary Table S4). Overall, CX-5461 treatment reverted 6% of the genes which were changed by LPS, and 21% of those changed by IFN-γ. A special family of cytokines known as ELR+ CXCL cytokines (so named for their N-terminal glutamate, leucine, and arginine tripeptide motif preceding the C-X-C chemokine motif) have been shown to have important pro-inflammatory and pro-angiogenic effects (Giuliano et al., 2014), including CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL7 and CXCL8. Our study showed that, however, none of these ELR+ CXCL cytokines or their cognate receptors were significantly changed by CX-5461 treatment in macrophages.
In LPS-primed cells, CX-5461 caused changes in expression of 421 genes; in IFN-γ-primed cells, CX-5461 changed the expression of 463 genes (see Figure 1C, Figure 2A; Supplementary Table S5, S6). It is noted that of these differentially expressed genes, 235 genes were identical under the two experimental settings (account for 56% in LPS-primed cells and 51% in IFN-γ-primed cells) (Figure 2A). K-means clustering analysis confirmed that CX-5461 treatment resulted in overlapping transcriptome signatures in LPS- and IFN-γ-primed macrophages (Figure 2B). GO and KEGG analyses on the CX-5461-responsive genes similarly identified enrichment in biological processes related to cell cycle regulation (GO: cell cycle phase transition; KEGG: cell cycle), which were highly statistically significant (Table 1). Consistent with previous data suggesting that activation of the p53 pathway had a pivotal role in mediating the pharmacological effects of CX-5461 (Bywater et al., 2012; Tsai and Pederson, 2014; Ye et al., 2017; Cui et al., 2021; Pang et al., 2021; Xu et al., 2021), we also showed that a cluster of 6 genes associated with p53 signaling (KEGG: p53 signaling pathway) were significantly altered by CX-5461 in both LPS- and IFN-γ-primed cells. These p53-related genes included CDKN1A (cyclin-dependent kinase inhibitor 1/p21Cip1), CDK1 (cyclin-dependent kinase 1/Cdk1), CCNE2 (cyclin E2), CCNE1 (cyclin E1), RRM2 (ribonucleotide reductase regulatory subunit M2) and CCNB1 (cyclin B1). Except for RRM2, these genes were also present in the functional clusters of “cell cycle phase transition” and “cell cycle” as mentioned above (Figure 3A). Because CDKN1A, CDK1, CCNE2, CCNE1 and CCNB1 were all key regulators of cell cycle, we plotted the numerical expression data for these genes, and demonstrated that CX-5461 upregulated CDKN1A (cell cycle inhibitor) and downregulated CDK1, CCNE2, CCNE1 and CCNB1 (mediators of cell cycle) in both LPS- and IFN-γ-primed cells (Figures 3B,C), supporting that CX-5461 predominantly inhibited cell cycle progression in macrophages. Because the group size (n = 3 each) in the sequencing experiment was relatively small (restricted by the total cost), we further confirmed the key findings shown in Figures 3B,C using real-time PCR assays. We demonstrated that CX-5461 treatment significantly upregulated CDKN1A, and downregulated CDK1, CCNE2, CCNE1 and CCNB1, in both LPS- and IFN-γ-primed cells (Supplementary Figure S2).
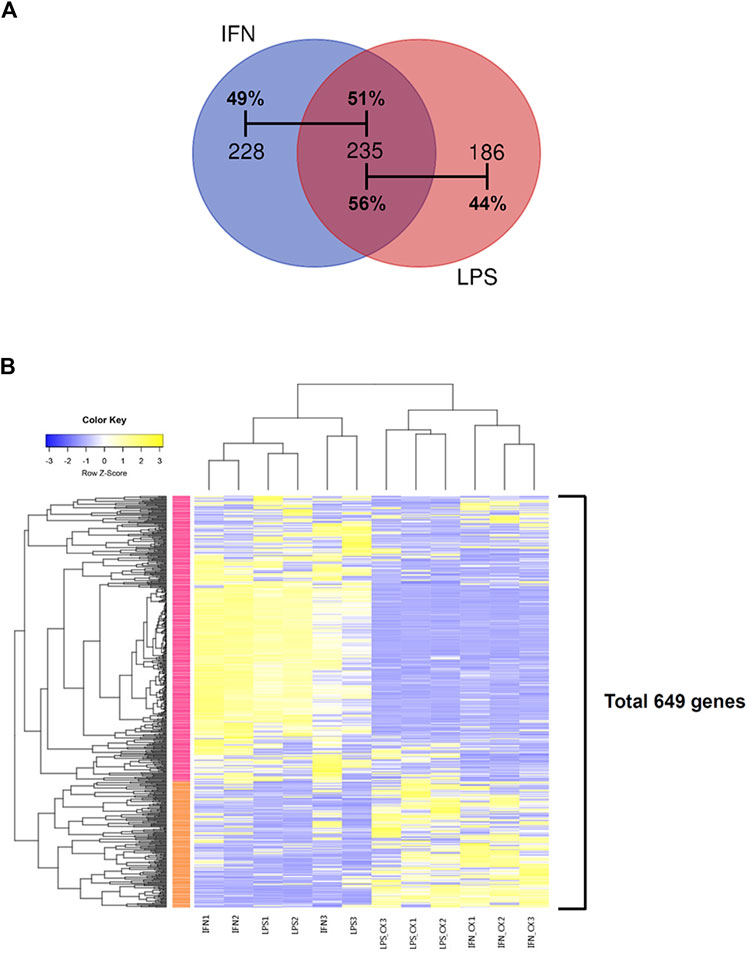
FIGURE 2. CX-5461 treatment resulted in overlapping transcriptome signatures in LPS- and IFN-γ-primed macrophages. (A) Venn diagrams showing that over 50% of the differentially regulated genes by CX-5461 were identical between LPS-primed and IFN-γ-primed cells. (B) Result of k-means clustering analysis confirming that CX-5461 treatment resulted in overlapping transcriptome signatures in LPS- and IFN-γ-primed macrophages.
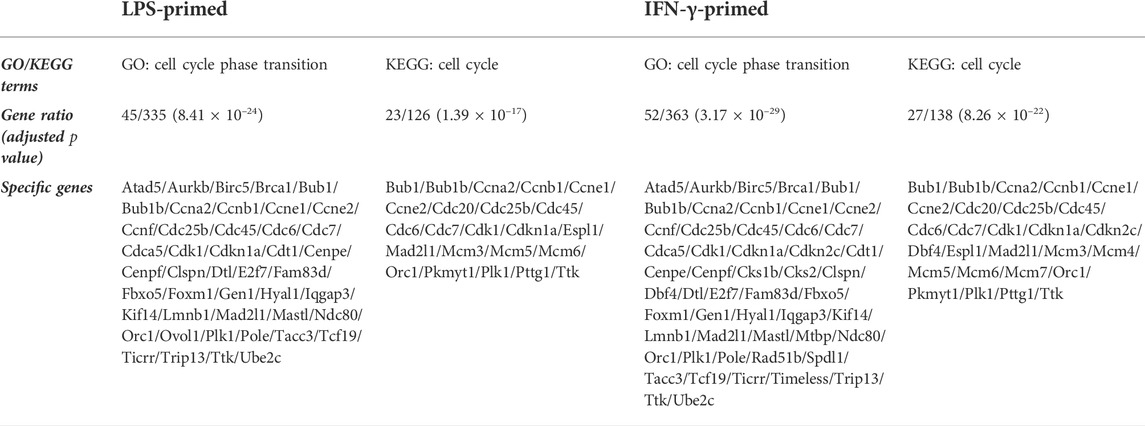
TABLE 1. Functional enrichment analysis showing that CX-5461 changed the expression of a cluster of genes associated with cell cycle regulation in macrophages.
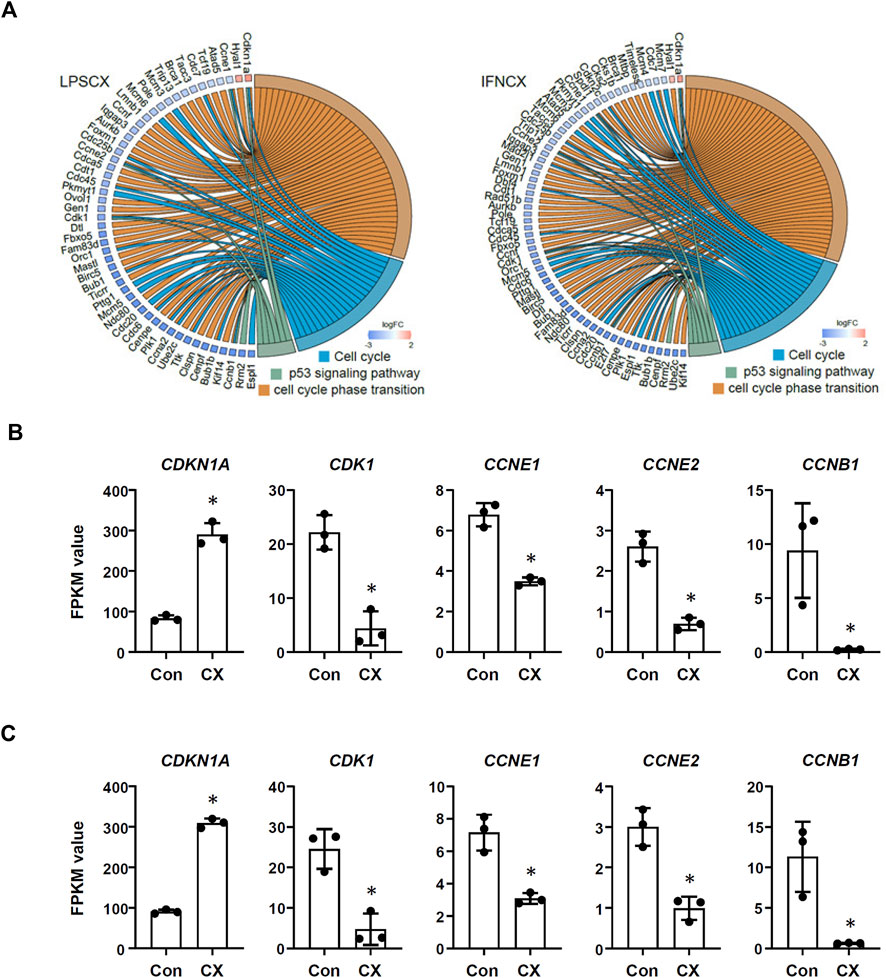
FIGURE 3. CX-5461 induced a molecular signature associated with cell cycle inhibition in macrophages. (A) Circos plots showing that CX-5461 regulated a cluster of genes which were related to cell cycle regulation in LPS- and IFN-γ-primed macrophages as identified by functional enrichment analyses. Some of these genes were linked to p53 signaling. (B,C) Numerical analysis of the expression levels of various genes involved in cell cycle regulation in LPS- and IFN-γ-primed macrophages respectively. The relative expression levels were expressed as fragment per kilobase per million reads (FPKM) values. CX, CX-5461. Bars represented mean ± standard deviation. *p < 0.05 versus vehicle control (Con), unpaired t-test.
Previous results from our and other groups suggested that induction of DDR featuring ATM/ATR pathway activation was a prominent characteristic of the CX-5461 actions in tumor cell lines and in cardiovascular cells (Negi and Brown, 2015; Quin et al., 2016; Ye et al., 2017; Pang et al., 2021). Hence we examined whether CX-5461 treatment in macrophages was transcriptionally linked to DDR pathways. We found that in both LPS- and IFN-γ-primed macrophages, CX-5461 altered the expression pattern of a gene cluster associated with DDR (GO: DNA repair) (see Table 2). This functional cluster included 45 genes in LPS-primed cells and 56 in IFN-γ-primed cells, of which the majority (41 genes) were identical between the two groups. Among these CX-5461-responsive genes, there were multiple well-established factors involved in double-strand break repair/homologous recombination, such as RAD51, BRCA1 (breast cancer type 1 susceptibility protein), RAD51AP1 (Rad51 associated protein 1), and GEN1 (GEN1 Holliday Junction 5′ flap endonuclease). CX-5461 also changed several genes which were essential for DNA replication, such as CDC45 (cell division cycle 45) and MCM (minichromosome maintenance complex component) members 3, 5 and 6. However, further quantitative analysis demonstrated that CX-5461 consistently downregulated the expression of these genes in macrophage (see Supplementary Table S5, S6).
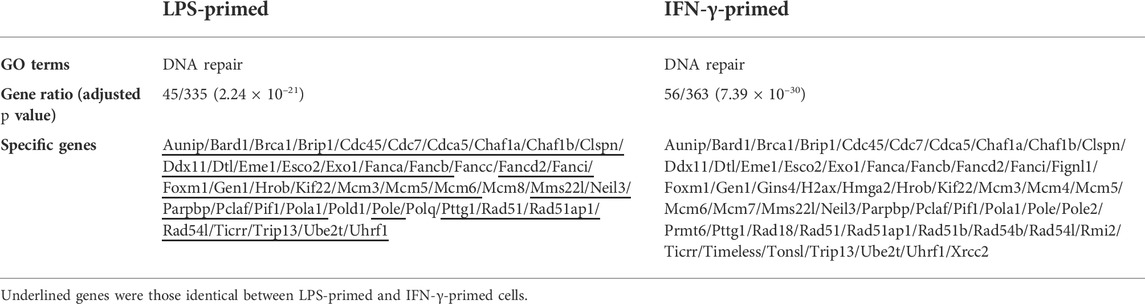
TABLE 2. Functional enrichment analysis showing that CX-5461 changed the expression of a cluster of genes associated with DNA damage response in macrophages.
As mentioned above, treatment with CX-5461 did not induce a distinct anti-inflammatory transcriptional signature in LPS- or IFN-γ-primed macrophages. Therefore, we examined the changes of several individual genes with critical pro-inflammatory functions in the macrophage, including NOS2, IL6, IL1B, TNF (TNF-α), CCL2, and PTGS1 (prostaglandin-endoperoxide synthase 1; aka. cyclooxygenase-1/COX-1) and PTGS2 (aka COX-2), in LPS-primed cells. It was found that the effects of CX-5461 on these genes were variable (Figure 4); only the expression of IL1B was decreased by CX-5461 (p = 0.0462). In addition, we examined the effects of CX-5461 on CYBA (cytochrome b-245 α chain; aka. p22phox) and CYBB (cytochrome b-245 β chain; aka. gp91phox), two membrane-bound subunits of the phagocytic NADPH oxidase. CX-5461 decreased the expression of CYBB (p = 0.0262), but not CYBA (Figure 4). We further examined the effects of CX-5461 on the expression of these pro-inflammatory genes using real-time PCR. Confirming our genomics data, these experiments demonstrated variable effects of CX-5461 (see Supplementary Figure S3), supporting the notion that the anti-inflammatory action of CX-5461 in macrophages is unlikely to be attributable to a systemic repression of major pro-inflammatory genes.
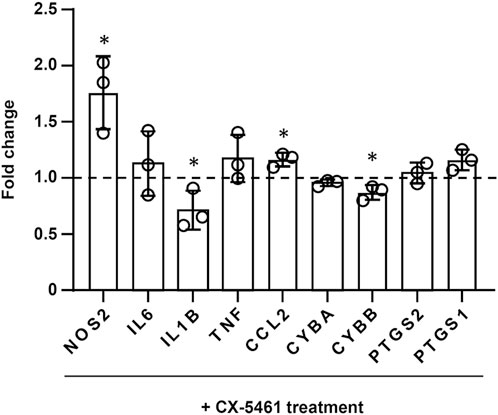
FIGURE 4. Effects of CX-5461 treatment on the expression of individual inflammatory genes in LPS-primed macrophages. FPKM values of CX-5461-treated cells were normalized to corresponding control cells and expressed as fold of control. Bars represented mean ± standard deviation. *p < 0.05 versus control, unpaired t-test.
To further confirm that p53 activation was functionally involved in the inhibitory effect of CX-5461 on macrophage proliferation, we pretreated the cells with the selective p53 inhibitor pifithrin-α. We demonstrated that pifithrin-α blunted the inhibitory effect of CX-5461 on macrophage proliferation (Supplementary Figure S4).
In the present study, we attempt to delineate the molecular mechanisms underlying the observed anti-inflammatory effects of CX-5461 in macrophages, by characterizing the transcriptome signatures in response to CX-5461 treatment. The most consistent finding from our study is that in both LPS- and IFN-γ-primed macrophages, CX-5461 predominantly triggers a transcriptional program favoring cell cycle inhibition, featuring downregulations of genes encoding cell cycle mediators and concomitant upregulations of cell cycle inhibitors. On the other hand, our data do not support that, under the present experimental settings, CX-5461 can directly induce a distinct anti-inflammatory transcriptional program in macrophages. Therefore, it is suggested that the most important mechanism by which CX-5461 regulates macrophage-mediated inflammation is to modulate the expansion of the macrophage cell population. This conclusion is consistent with our previous observation that CX-5461 can significantly inhibit macrophage proliferation both in vitro and in vivo (Dai et al., 2018). Moreover, our bioinformatics results further corroborate the central role of p53 in mediating the cell cycle regulatory effect of CX-5461 in macrophages.
It is well recognized that chemotactic migration of monocytes from circulation to the inflamed tissue, and in situ differentiation of the recruited monocytes to mature macrophages, constitute the major pathway leading to accumulation of inflammatory macrophages and release of pro-inflammatory cytokines at the site of inflammation (Haskill et al., 1992; Ohashi et al., 2015). However, emerging evidence suggests that in addition to the mobilization of monocytes/macrophages, localized macrophage proliferation may also have a crucial contribution to the maintenance of inflammatory reaction. This mechanism may be of particular relevance in the chronic inflammatory conditions associated with blood vessels. For example, multiple studies have shown that macrophage proliferation facilitated by the local micro-environment in the vessel wall is a predominant contributor to the development of atherosclerosis (Robbins et al., 2013; Tang et al., 2015; Tanaka et al., 2016). This notion is also supported by our previous results indicating that inhibiting macrophage proliferation by CX-5461 may at least partly contribute to the amelioration of aortic transplantation-induced vascular inflammation and remodeling (Dai et al., 2018). Apart from vascular inflammation, there is evidence showing that local macrophage proliferation is also important for parasite-induced Th2 type inflammation (Jenkins et al., 2011).
Independent lines of evidence suggest that induction of ATM/ATR-mediated DDR is a prominent upstream signaling event of CX-5461-induced cellular effects, which may result in p53-dependent or p53-independent cytostatic actions in proliferating cells (Negi and Brown, 2015; Quin et al., 2016; Ye et al., 2017; Sanij et al., 2020; Pang et al., 2021). In the present study, we have shown that CX-5461 treatment in macrophages is indeed associated with a transcriptome signature related to DNA repair. Specifically, CX-5461 downregulates the expression of multiple genes involved in homologous recombination, an important pathway responsible for repairing double-strand breaks and maintaining genome stability (Spies et al., 2021). These data seem to be paradoxical to the observed activation of DDR in CX-5461-treated cells, and the physiological significance of CX-5461-induced downregulations of the DDR genes remain elusive. Nevertheless, our data are exactly consistent with previous studies in which p53-dependent downregulations of RAD51, BRCA1 and MCM5 expression have been reported (Arizti et al., 2000; Arias-Lopez et al., 2006; Agarwal et al., 2007), further highlighting p53 in orchestrating the molecular networks that are responsive to CX-5461 treatment.
Although the available data regarding the role of p53 in modulating inflammatory reactions appear to be controversial, a mass of evidence suggests that p53 has anti-inflammatory activities in certain pathological contexts (see Cui et al., 2021). In a recent study we have shown that CX-5461 can inhibit T cell-mediated immunity functions via p53-DUSP5 (dual-specificity phosphatase 5) axis and subsequent antagonism of the Erk1/2 mitogen-activated protein kinase activity, thereby preventing the occurrence of acute transplant rejection (Pan et al., 2022). These results suggest that CX-5461 has independent modulatory activities on inflammatory/immune signalings in immune cells apart from regulation of the cell cycle. Supporting this notion, different studies have demonstrated negative correlations between p53 and the expression of prototypic inflammatory genes including IL6 (Santhanam et al., 1991), IL1B (Yamanishi et al., 2002), NOS2 (Huang et al., 2014) and TNF (Tang et al., 2011). Our present genomic data also confirm that CX-5461 downregulates expressions of IL-1β and gp91phox (the catalytic subunit of superoxide-producing NADPH oxidase). However, the effects of CX-5461 on the expression of other inflammatory genes appear to be variable; the relatively weak effects of CX-5461 on macrophage inflammatory gene expression in comparison to our previous data might be due to the lower concentration of CX-5461 used in the present study.
In conclusion, CX-5461 principally induces a molecular signature related to cell cycle inhibition in macrophages, featuring downregulation of genes encoding cell cycle mediators and concomitant upregulation of cell cycle inhibitors. On the other hand, at a concentration which is effective for regulating macrophage proliferation, CX-5461 does not necessarily induce a distinct anti-inflammatory transcriptional program, suggesting that limiting cell proliferation predominates in the inhibitory effects of CX-5461 on macrophage-mediated inflammation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://datadryad.org/stash/dataset/doi:10.5061/dryad.51c59zwb7.
The animal study was reviewed and approved by The Institutional Animal Ethics Committee of Qilu Hospital of Shandong University.
JW was involved in cell experiments and data collection; ZZ was involved in genomic data analysis and bioinformatics mining; XC was involved in cell experiments and data interpretation; CD was involved in cell experiments and data collection; JL was involved in cell experiments; QZ was involved in genomic data analysis; MC was involved in study design, data interpretation, and project management; FJ was involved in study conception, data interpretation, manuscript writing, revision and approval.
This study was partially supported by research grants from National Natural Science Foundation of China (82070265 for FJ) and Shandong Science and Technology Development Project (2017G006041 for MC).
The authors thank Ye Chen and Guopin Pan for providing various technical assistances.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.926317/full#supplementary-material
Agarwal, M. K., Ruhul Amin, A. R., and Agarwal, M. L. (2007). DNA replication licensing factor minichromosome maintenance deficient 5 rescues p53-mediated growth arrest. Cancer Res. 67, 116–121. doi:10.1158/0008-5472.CAN-06-2835
Arias-Lopez, C., Lazaro-Trueba, I., Kerr, P., Lord, C. J., Dexter, T., Iravani, M., et al. (2006). p53 modulates homologous recombination by transcriptional regulation of the RAD51 gene. EMBO Rep. 7, 219–224. doi:10.1038/sj.embor.7400587
Arizti, P., Fang, L., Park, I., Yin, Y., Solomon, E., Ouchi, T., et al. (2000). Tumor suppressor p53 is required to modulate BRCA1 expression. Mol. Cell. Biol. 20, 7450–7459. doi:10.1128/mcb.20.20.7450-7459.2000
Austermann, J., Roth, J., and Barczyk-Kahlert, K. (2022). The good and the bad: Monocytes' and macrophages' diverse functions in inflammation. Cells 11, 1979. doi:10.3390/cells11121979
Boulon, S., Westman, B. J., Hutten, S., Boisvert, F. M., and Lamond, A. I. (2010). The nucleolus under stress. Mol. Cell 40, 216–227. doi:10.1016/j.molcel.2010.09.024
Bruno, P. M., Lu, M., Dennis, K. A., Inam, H., Moore, C. J., Sheehe, J., et al. (2020). The primary mechanism of cytotoxicity of the chemotherapeutic agent CX-5461 is topoisomerase II poisoning. Proc. Natl. Acad. Sci. U. S. A. 117, 4053–4060. doi:10.1073/pnas.1921649117
Bywater, M. J., Pearson, R. B., Mcarthur, G. A., and Hannan, R. D. (2013). Dysregulation of the basal RNA polymerase transcription apparatus in cancer. Nat. Rev. Cancer 13, 299–314. doi:10.1038/nrc3496
Bywater, M. J., Poortinga, G., Sanij, E., Hein, N., Peck, A., Cullinane, C., et al. (2012). Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 22, 51–65. doi:10.1016/j.ccr.2012.05.019
Cui, X., Pan, G., Chen, Y., Guo, X., Liu, T., Zhang, J., et al. (2021). The p53 pathway in vasculature revisited: A therapeutic target for pathological vascular remodeling? Pharmacol. Res. 169, 105683. doi:10.1016/j.phrs.2021.105683
Dai, C., Sun, M., Wang, F., Zhu, J., Wei, Y., Guo, X., et al. (2018). The selective RNA polymerase I inhibitor CX-5461 mitigates neointimal remodeling in a modified model of rat aortic transplantation. Transplantation 102, 1674–1683. doi:10.1097/TP.0000000000002372
Drygin, D., Lin, A., Bliesath, J., Ho, C. B., O'brien, S. E., Proffitt, C., et al. (2011). Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 71, 1418–1430. doi:10.1158/0008-5472.CAN-10-1728
Ferreira, R., Schneekloth, J. S., Panov, K. I., Hannan, K. M., and Hannan, R. D. (2020). Targeting the RNA polymerase I transcription for cancer therapy comes of age. Cells 9, E266. doi:10.3390/cells9020266
Giuliano, S., Guyot, M., Grépin, R., and Pagès, G. (2014). The ELR(+)CXCL chemokines and their receptors CXCR1/CXCR2: A signaling axis and new target for the treatment of renal cell carcinoma. Oncoimmunology 3, e28399. doi:10.4161/onci.28399
Haddach, M., Schwaebe, M. K., Michaux, J., Nagasawa, J., O'brien, S. E., Whitten, J. P., et al. (2012). Discovery of CX-5461, the first direct and selective inhibitor of RNA polymerase I, for cancer therapeutics. ACS Med. Chem. Lett. 3, 602–606. doi:10.1021/ml300110s
Haskill, S., Yurochko, A. D., and Isaacs, K. L. (1992). Regulation of macrophage infiltration and activation in sites of chronic inflammation. Ann. N. Y. Acad. Sci. 664, 93–102. doi:10.1111/j.1749-6632.1992.tb39752.x
Howie, D., Okamoto, S., Rietdijk, S., Clarke, K., Wang, N., Gullo, C., et al. (2002). The role of SAP in murine CD150 (SLAM)-mediated T-cell proliferation and interferon gamma production. Blood 100, 2899–2907. doi:10.1182/blood-2002-02-0445
Hu, X., Chakravarty, S. D., and Ivashkiv, L. B. (2008). Regulation of interferon and Toll-like receptor signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms. Immunol. Rev. 226, 41–56. doi:10.1111/j.1600-065X.2008.00707.x
Huang, Y., Yu, P., Li, W., Ren, G., Roberts, A. I., Cao, W., et al. (2014). p53 regulates mesenchymal stem cell-mediated tumor suppression in a tumor microenvironment through immune modulation. Oncogene 33, 3830–3838. doi:10.1038/onc.2013.355
Jenkins, S. J., Ruckerl, D., Cook, P. C., Jones, L. H., Finkelman, F. D., Van Rooijen, N., et al. (2011). Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science 332, 1284–1288. doi:10.1126/science.1204351
Khot, A., Brajanovski, N., Cameron, D. P., Hein, N., Maclachlan, K. H., Sanij, E., et al. (2019). First-in-Human RNA polymerase I transcription inhibitor CX-5461 in patients with advanced hematologic cancers: Results of a phase I dose-escalation study. Cancer Discov. 9, 1036–1049. doi:10.1158/2159-8290.CD-18-1455
Liu, F., Cheng, W., Bi, X., Zhang, Y., Zhao, Y., and Jiang, F. (2016). Stage-dependent effects of exogenous TRAIL on atherogenesis: Role of ER stress-mediated sensitization of macrophage apoptosis. Clin. Exp. Pharmacol. Physiol. 43, 543–551. doi:10.1111/1440-1681.12561
Mars, J. C., Tremblay, M. G., Valere, M., Sibai, D. S., Sabourin-Felix, M., Lessard, F., et al. (2020). The chemotherapeutic agent CX-5461 irreversibly blocks RNA polymerase I initiation and promoter release to cause nucleolar disruption, DNA damage and cell inviability. Nar. Cancer 2, zcaa032. doi:10.1093/narcan/zcaa032
Negi, S. S., and Brown, P. (2015). rRNA synthesis inhibitor, CX-5461, activates ATM/ATR pathway in acute lymphoblastic leukemia, arrests cells in G2 phase and induces apoptosis. Oncotarget 6, 18094–18104. doi:10.18632/oncotarget.4093
Ohashi, W., Hattori, K., and Hattori, Y. (2015). Control of macrophage dynamics as a potential therapeutic approach for clinical disorders involving chronic inflammation. J. Pharmacol. Exp. Ther. 354, 240–250. doi:10.1124/jpet.115.225540
Pan, G., Zhang, J., Han, Y., Chen, Y., Guo, X., Cui, X., et al. (2022). CX-5461 is a potent immunosuppressant which inhibits T cell-mediated alloimmunity via p53-DUSP5. Pharmacol. Res. 177, 106120. doi:10.1016/j.phrs.2022.106120
Pang, S., Chen, Y., Dai, C., Liu, T., Zhang, W., Wang, J., et al. (2021). Anti-fibrotic effects of p53 activation induced by RNA polymerase I inhibitor in primary cardiac fibroblasts. Eur. J. Pharmacol. 907, 174303. doi:10.1016/j.ejphar.2021.174303
Quin, J., Chan, K. T., Devlin, J. R., Cameron, D. P., Diesch, J., Cullinane, C., et al. (2016). Inhibition of RNA polymerase I transcription initiation by CX-5461 activates non-canonical ATM/ATR signaling. Oncotarget 7, 49800–49818. doi:10.18632/oncotarget.10452
Robbins, C. S., Hilgendorf, I., Weber, G. F., Theurl, I., Iwamoto, Y., Figueiredo, J. L., et al. (2013). Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat. Med. 19, 1166–1172. doi:10.1038/nm.3258
Sanij, E., Hannan, K. M., Xuan, J., Yan, S., Ahern, J. E., Trigos, A. S., et al. (2020). CX-5461 activates the DNA damage response and demonstrates therapeutic efficacy in high-grade serous ovarian cancer. Nat. Commun. 11, 2641. doi:10.1038/s41467-020-16393-4
Santhanam, U., Ray, A., and Sehgal, P. B. (1991). Repression of the interleukin 6 gene promoter by p53 and the retinoblastoma susceptibility gene product. Proc. Natl. Acad. Sci. U. S. A. 88, 7605–7609. doi:10.1073/pnas.88.17.7605
Schroder, K., Sweet, M. J., and Hume, D. A. (2006). Signal integration between IFNgamma and TLR signalling pathways in macrophages. Immunobiology 211, 511–524. doi:10.1016/j.imbio.2006.05.007
Spies, J., Polasek-Sedlackova, H., Lukas, J., and Somyajit, K. (2021). Homologous recombination as a fundamental genome surveillance mechanism during DNA replication. Genes (Basel) 12, 1960. doi:10.3390/genes12121960
Tanaka, S., Matsumoto, T., Matsubara, Y., Harada, Y., Kyuragi, R., Koga, J. I., et al. (2016). BubR1 insufficiency results in decreased macrophage proliferation and attenuated atherogenesis in apolipoprotein E-deficient mice. J. Am. Heart Assoc. 5, e004081. doi:10.1161/JAHA.116.004081
Tang, J., Lobatto, M. E., Hassing, L., Van Der Staay, S., Van Rijs, S. M., Calcagno, C., et al. (2015). Inhibiting macrophage proliferation suppresses atherosclerotic plaque inflammation. Sci. Adv. 1, e1400223. doi:10.1126/sciadv.1400223
Tang, X., O'reilly, A., Asano, M., Merrill, J. C., Yokoyama, K. K., and Amar, S. (2011). p53 peptide prevents LITAF-induced TNF-alpha-mediated mouse lung lesions and endotoxic shock. Curr. Mol. Med. 11, 439–452. doi:10.2174/156652411796268731
Tsai, R. Y., and Pederson, T. (2014). Connecting the nucleolus to the cell cycle and human disease. Faseb J. 28, 3290–3296. doi:10.1096/fj.14-254680
Wadgaonkar, R., Phelps, K. M., Haque, Z., Williams, A. J., Silverman, E. S., and Collins, T. (1999). CREB-binding protein is a nuclear integrator of nuclear factor-kappaB and p53 signaling. J. Biol. Chem. 274, 1879–1882. doi:10.1074/jbc.274.4.1879
Wang, J., Wang, Y., Wang, J., Guo, X., Chan, E. C., and Jiang, F. (2018). Adventitial activation in the pathogenesis of injury-induced arterial remodeling: Potential implications in transplant vasculopathy. Am. J. Pathol. 188, 838–845. doi:10.1016/j.ajpath.2017.12.002
Webster, G. A., and Perkins, N. D. (1999). Transcriptional cross talk between NF-kappaB and p53. Mol. Cell. Biol. 19, 3485–3495. doi:10.1128/mcb.19.5.3485
Xu, H., Di Antonio, M., Mckinney, S., Mathew, V., Ho, B., O'neil, N. J., et al. (2017). CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 8, 14432. doi:10.1038/ncomms14432
Xu, X., Feng, H., Dai, C., Lu, W., Zhang, J., Guo, X., et al. (2021). Therapeutic efficacy of the novel selective RNA polymerase I inhibitor CX-5461 on pulmonary arterial hypertension and associated vascular remodelling. Br. J. Pharmacol. 178, 1605–1619. doi:10.1111/bph.15385
Yamanishi, Y., Boyle, D. L., Pinkoski, M. J., Mahboubi, A., Lin, T., Han, Z., et al. (2002). Regulation of joint destruction and inflammation by p53 in collagen-induced arthritis. Am. J. Pathol. 160, 123–130. doi:10.1016/S0002-9440(10)64356-8
Yang, K., Yang, J., and Yi, J. (2018). Nucleolar stress: Hallmarks, sensing mechanism and diseases. Cell Stress 2, 125–140. doi:10.15698/cst2018.06.139
Ye, Q., Pang, S., Zhang, W., Guo, X., Wang, J., Zhang, Y., et al. (2017). Therapeutic targeting of RNA polymerase I with the small-molecule CX-5461 for prevention of arterial injury-induced neointimal hyperplasia. Arterioscler. Thromb. Vasc. Biol. 37, 476–484. doi:10.1161/ATVBAHA.116.308401
Yu, G., Wang, L. G., Han, Y., and He, Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287. doi:10.1089/omi.2011.0118
Keywords: RNA polymerase I inhibitor, CX-5461, inflammation, macrophage, transcriptome sequencing, cell cycle, systems biology
Citation: Wang J, Zheng Z, Cui X, Dai C, Li J, Zhang Q, Cheng M and Jiang F (2022) A transcriptional program associated with cell cycle regulation predominates in the anti-inflammatory effects of CX-5461 in macrophage. Front. Pharmacol. 13:926317. doi: 10.3389/fphar.2022.926317
Received: 16 May 2022; Accepted: 12 October 2022;
Published: 26 October 2022.
Edited by:
Gerard Bannenberg, Global Organization for EPA and DHA Omega-3s, United StatesReviewed by:
Denis Drygin, Regulus Therapeutics Inc., United StatesCopyright © 2022 Wang, Zheng, Cui, Dai, Li, Zhang, Cheng and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fan Jiang, ZmppYW5nQHNkdS5lZHUuY24=; Mei Cheng, am5jbTY1QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.