- 1Department of Molecular Science and Technology, Ajou University, Suwon, South Korea
- 2S&K Therapeutics, Ajou University, Suwon, South Korea
The innate immune system is the first line of host’s defense against invading pathogens. Multiple cellular sensors that detect viral components can induce innate antiviral immune responses. As a result, interferons and pro-inflammatory cytokines are produced which help in the elimination of invading viruses. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to Coronaviridae family, and has a single-stranded, positive-sense RNA genome. It can infect multiple hosts; in humans, it is responsible for the novel coronavirus disease 2019 (COVID-19). Successful, timely, and appropriate detection of SARS-CoV-2 can be very important for the early generation of the immune response. Several drugs that target the innate immune receptors as well as other signaling molecules generated during the innate immune response are currently being investigated in clinical trials. In this review, we summarized the current knowledge of the mechanisms underlying host sensing and innate immune responses against SARS-CoV-2 infection, as well as the role of innate immune receptors in terms of their therapeutic potential against SARS-CoV-2. Moreover, we discussed the drugs undergoing clinical trials and the FDA approved drugs against SARS-CoV-2. This review will help in understanding the interactions between SARS-CoV-2 and innate immune receptors and thus will point towards new dimensions for the development of new therapeutics, which can be beneficial in the current pandemic.
1 Introduction
Coronaviridae is a family of enveloped positive-sense single-stranded RNA viruses. In the last 19 years, this virus family has been the cause of three different zoonotic outbreaks, and the latest one—caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)—resulted in the COVID-19 pandemic (Gorbalenya et al., 2020). The family of coronaviridae can be divided into four genera: alpha (α), beta (ß), gamma (γ), and delta (Δ) (Sheahan et al., 2020). Alpha- and Betacoronavirus members have been reported to infect humans. Being the seventh member of Coronaviridae involved in a zoonosis, SARS-CoV-2 belongs to the genus Betacoronavirus (Li X. et al., 2020). With a particle size of 125 nm, this ball-shaped virus has club-shaped appendages; a spike protein embedded into the envelope, giving it a crown-like appearance in an electron micrograph, hence the name coronavirus (Anderson and Schneider, 2012).
The virus was first identified in China, and the World Health Organization (WHO) used a provisional nonspecific name: “2019 novel coronavirus” (2019-nCoV). Later, based on the phylogenetic analysis of the other coronaviruses, the Coronavirus Study Group of the International Committee on Virus Taxonomy suggested renaming it as SARS-CoV-2 (Awadasseid et al., 2020).
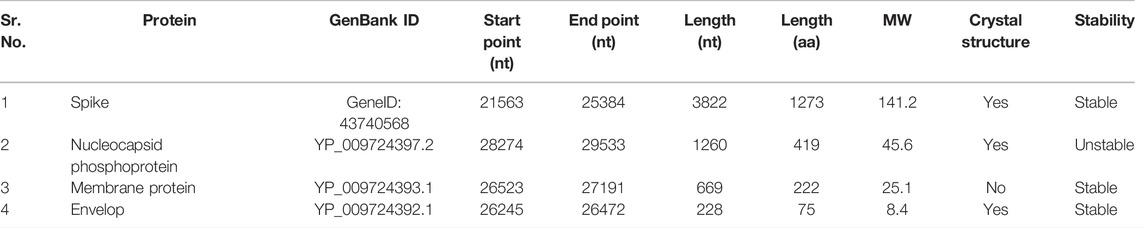
Coronaviruses have one of the largest RNA genomes, with a size ranging between 26 and 32 kb. SARS-CoV-2 has a genome length of 30 kbp, and the genome is flanked by 3′ and 5′ untranslated regions and contains 15 open reading frames (ORFs). The genome translates into a polyprotein of approximately 800 kDa. There are 4 structural proteins that are encoded by 2–14 open reading frames: Nucleocapsid (N), envelope (E) and Spike (S) and Membrane protein (M). The structural proteins play their role in virion assembly and infection; and the non-structural proteins are involved in the replication cycle of the virus (Mirza and Froeyen, 2020). The shape of the virus is maintained by the M protein. The M protein binds to Nucleocapsid, which constitutes almost 1% of the expressed protein during infection, and it binds to the viral RNA genome through its two different subunits. It helps to package the genome inside the virion (Chen Y. et al., 2020; Jackson et al., 2021a; Tugaeva et al., 2021). E and M interact to form the envelope of the virus. Receptor Binding Domain (RBD) of Spike, that is glycoprotein in nature, is responsible for interaction between virus and the host cell receptors thus helps the virus to enter the cell (Luan et al., 2020).
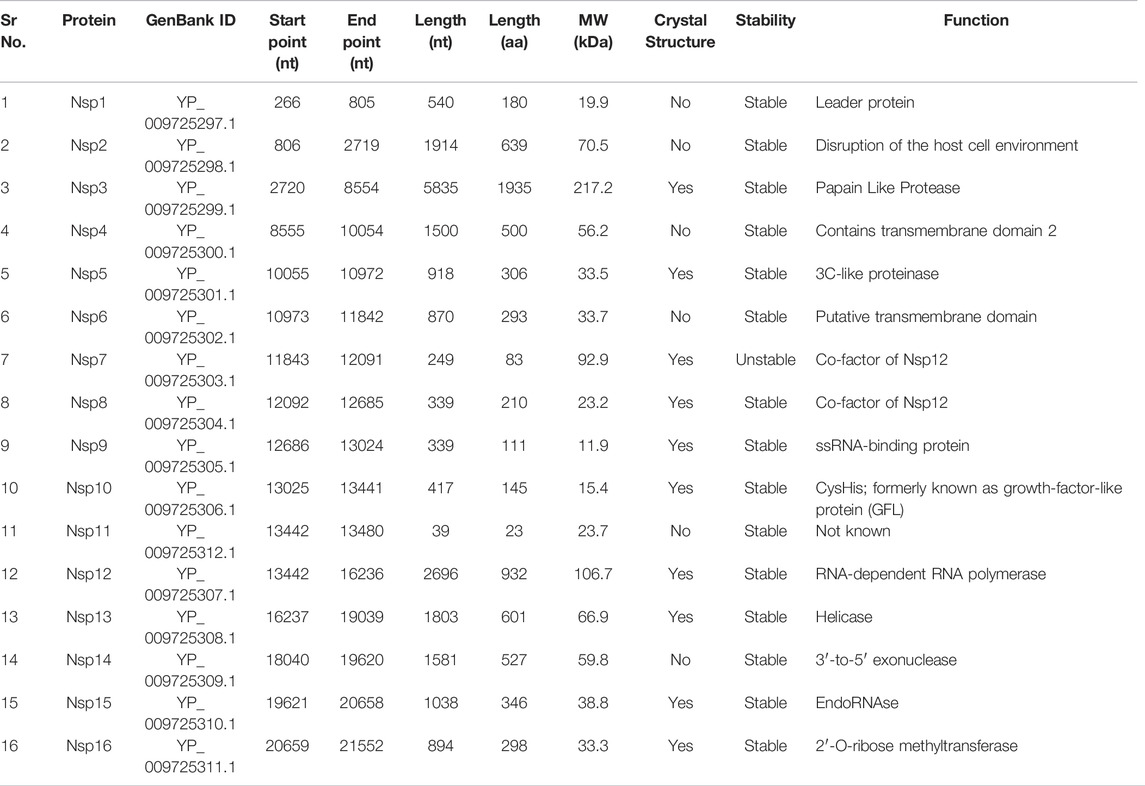
The first two ORFs encode nonstructural proteins. There are 16 of these proteins, which are Nsp1 through Nsp16. Nsp1 (protein ID: YP_009725297.1) has a length of 180 amino acid residues (aa). Nsp2 (YP_009725298.1) consists of 638 aa. Nsp3 (YP_009725299.1) consists of 1945 aa. It contains the following domains: N-terminal acidic (Ac), predicted phosphoesterase, papain-like proteinase, Y-domain, transmembrane domain 1 (TM1), and adenosine diphosphate-ribose 1″-phosphatase (ADRP). Nsp4 (YP_009725300.1) has a length of 500 aa and contains transmembrane domain 2. Nsp5 (YP_009725301.1) is 3C-like proteinase (3Cl-pro), and its length is 306 aa. Nsp6 (protein ID YP_009725302.1) is a putative transmembrane domain having a length of 290 aa. Nsp7 (YP_009725303.1) and Nsp8 (YP_009725304.1) are proposed cofactors for the polymerase. Lengths of their genes are 249 and 339 nucleotides encoding an 83 aa protein and 198 aa proteins, respectively. Nsp9 (YP_009725305.1) is a single-stranded RNA (ssRNA)-binding protein whose length is 113 aa. Nsp10 (YP_009725306.1) has a length of 139 aa and is also called CysHis; it has previously been known as growth factor-like protein (GFL). The length of Nsp11 (YP_009725295.1) is 13 aa. Nsp12 (YP_009725307.1), with a length of 932 aa, carries an RNA-dependent RNA polymerase (RdRp) domain and NIRAN domain. Nsp13 (YP_009725308.1) is helicase, which contains a zinc-binding domain (NTPase/helicase domain (HEL)) and RNA 5′-triphosphatase domain and has a length of 601 aa. Nsp14 (YP_009725309.1) is 3′-to-5′ exonuclease and has a length of 521 aa. Nsp15 (YP_009725310.1), with a length of 346 aa, is endoRNase. Nsp16 (YP_009725311) is 2′-O-ribose methyltransferase and is 298 aa long. Structural and nonstructural proteins and their details are summarized in Tables 1 and 2 (extracted from https://www.ncbi.nlm.nih.gov/gene/43740578).
The virus primarily infects the lung cells that express angiotensin-converting enzyme 2 (ACE2, which is a virus receptor) and is thus responsible for a pneumonia-like disease. ACE2, the main receptor of SARS-CoV-2, is present on various types of cells in the human body, e.g., cells in the heart, kidneys, liver, gastrointestinal tract, nervous system, endocrine system, and some skin sites (Zheng YY. et al., 2020; Pandey et al., 2020). Other receptors which also mediate viral entry include neuropilin-1, AXL, and antibody-FcγR complexes (Zhang et al., 2021b). Neuropilin-1 (NRP1), which is known to bind furin-cleaved substrates, enhances SARS-CoV-2 infectivity. NRP1 is highly expressed in the respiratory and olfactory epithelium, with endothelial and epithelial cells expressing the most. The spike protein’s furin-cleaved S1 fragment binds directly to cell surface NRP1 and disrupting its interaction with a small-molecule inhibitor or monoclonal antibodies inhibited viral infection in cell culture (Cantuti-Castelvetri et al., 2020). The other coronaviruses cause an upper-respiratory-tract infection, whereas SARS-CoV-2 causes a lower-respiratory-tract disease by infecting ciliated cells and type 2 alveolar epithelial cells in the lungs (Astuti and Ysrafil, 2020). SARS-CoV-2 has been detected in the upper respiratory tract too, and these data indicate that the nasopharynx is a site of viral replication. The Spike protein of SARS-CoV and that of SARS-CoV-2 are 76.5% similar, and the binding between the Spike of SARS-CoV-2 and ACE2 is stronger than the binding between SARS-CoV Spike and ACE2 (Zhang et al., 2020).
2 The Replication Cycle
Enveloped viruses either enter the cell through direct fusion with the cell membrane and inject their genome or use the alternative mechanism, which involves endocytic machinery. When viruses enter the cell by either of these mechanisms, an additional step of activation is required. For SARS-CoV-2, in the case of direct fusion, the activation is implemented by proteolytic cleavage of Spike, whereas in the endosomal pathway, acidic pH in the endosome is required for successful virus entry. Acidic pH helps the viral and endosomal membranes to fuse and activate proteases. Therefore, the entry of the virus through direct fusion is independent of cellular pH as opposed to the endosomal entry (Yuki et al., 2020).
SARS-CoV-2 is an RNA virus that carries the enzymes required for its replication, because host cells that do not contain enzymes for replicating and translating the RNA genome. Out of the 16 nonstructural proteins that the SARS-CoV-2 genome codes for, two proteins are viral proteases that are responsible for the cleavage of the polyprotein synthesized via translation of the viral genomic RNA. The virus then translates the nonstructural proteins that are required for the replication of the viral genome. After the synthesis of structural proteins, the virion assembly begins. RNA gets packaged inside the nucleocapsid, and the virions are released from the cell, which are now ready to infect other cells (Poduri et al., 2020).
SARS-CoV-2 spike is a homotrimeric glycoprotein of 150 kDa. The receptor binding domain (RBD) of spike binds to its receptor called human ACE2 and facilitates the entry of the virus into the host cell (Hoffmann et al., 2020). The virus is activated by the cleavage of the spike glycoprotein into two subunits—S1 and S2—by host cellular proteases, particularly TMPRSS2 (or by lysosomal cathepsins if the virus enters via the endosomal pathway) (Shang et al., 2020a; Hörnich et al., 2021). The first subunit determines cellular tropism, whereas the second one mediates the fusion of a host cell with the virus membrane. After the fusion, the virus enters the cell. Once the virus is inside the cell, it releases its genome into the cytoplasm, where the replication of the genome and its translation into the structural and nonstructural proteins take place (Shang et al., 2020b). SARS-CoV-2 assembly begins with the coating of genomic RNA by the nucleocapsid protein, forming nucleocapsid structures that bud into the endoplasmic reticulum-Golgi intermediate compartment, thereby giving rise to a lipid bilayer containing the viral spike, membrane, and envelope proteins resulting in the formation of the mature virion (Jackson et al., 2021b). The latter then leaves the cell through exocytosis as shown in Figure 1 (Mendonça et al., 2021; Yadav et al., 2021).
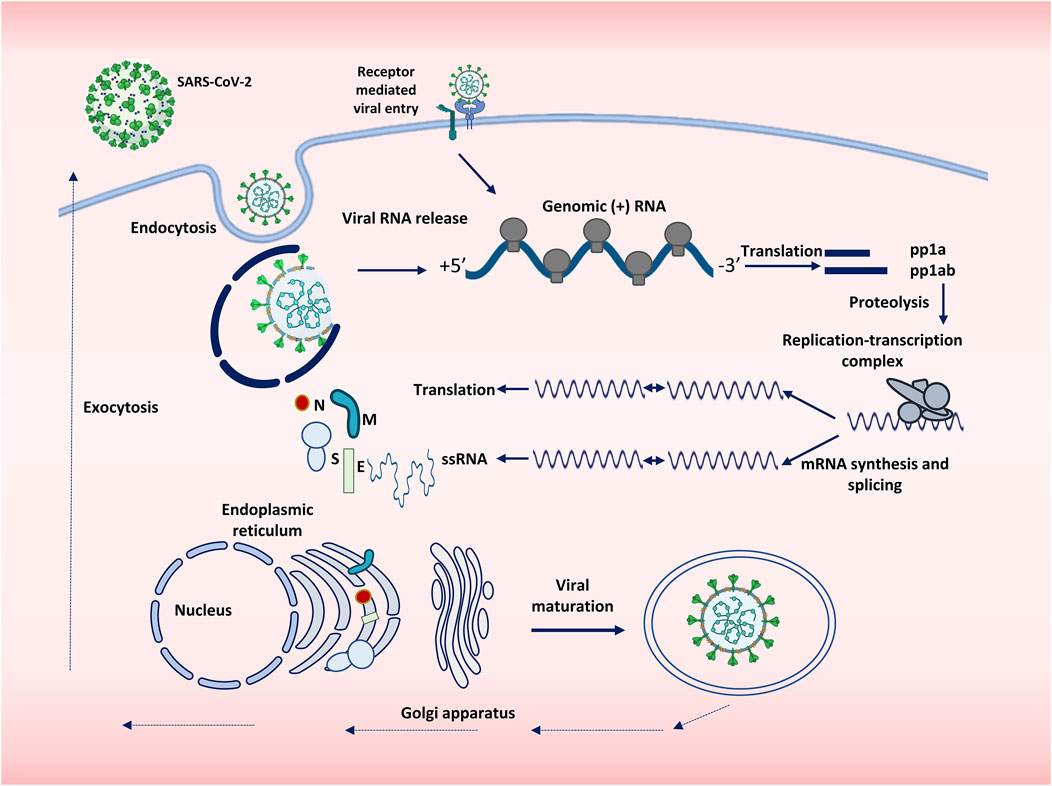
FIGURE 1. Replication cycle of SARS-CoV-2. SARS-CoV-2 enters the cell either by endocytosis or by receptor mediated membrane fusion. The genome of the virus is released and is subjected to replication and translation. Viral proteins are synthesized, and single stranded RNA is produced, both of which are assembled with the help of Endoplasmic reticulum and Golgi apparatus. The virions thus assembled are then transported outside the body by exocytosis.
The virus downregulates ACE2, as a result of which, the renin angiotensin aldosterone system is dysregulated. This problem leads to tissue injury, vasoconstriction, and increased vascular permeability. Endothelial cells are damaged because of the virus and undergo apoptosis, which results in decreased fibrinolysis and elevated thrombin production. SARS-CoV-2 excessively activates innate immunity and inhibits interferon (IFN) signaling. Consequently, the host develops cytokine release syndrome, wherein excessive production of cytokines occurs, and interleukin 1 beta (IL-1β), IL-6, and IL-10 levels increase. Lymphopenia, a typical condition in severe infections, has been detected in 60–70% of the patients (Gupta et al., 2020).
SARS-CoV-2 infection has a wide variety of clinical manifestations. They range from asymptomatic cases to mild and severe symptoms, and even death. Asymptomatic patients play an especially important role in the COVID-19 pandemic because they spread the infection further without showing any symptoms. It has been reported that 40–50% of the patients were asymptomatic (Oran and Topol, 2020; Wang et al., 2020). Five common clinical manifestations include fever, malaise, cough, sputum secretion, and dyspnea. Comparatively less common symptoms are neurological problems, myalgia, sneezing, sore throat, rhinitis, goosebumps, chest pain, diarrhea etc (da Rosa Mesquita et al., 2021). According to data published in 2020, only 5–10% of all infected individuals develop a severe disease, which includes pneumonia and acute respiratory failure (Raghu and Wilson, 2020). However, the rate of development of severe disease varies based on the age as well as gender (Poletti et al., 2021; Herrera-Esposito and de los Campos, 2022).
3 Transmission routes
SARS-CoV-2 is a respiratory virus that primarily infects lower respiratory tract along with the upper respiratory tract and gastrointestinal tract (Ludwig and Zarbock, 2020). It was noticed that when human bronchial epithelial cells were infected with SARS-CoV-2 at a biosafety level 3 lab facility, a large number of virions being produced and released by the cells could be observed by scanning electron microscopy (Ehre, 2020). The main route of transmission in air but it can also be transmitted through the respiratory droplets as well (Port et al., 2022). The main route of transmission is air, but the virus can be transmitted through respiratory droplets as well (Port et al., 2022). Aerosols are also believed to be a possible means of transmission, especially in closed environment (Tellier, 2022; Wilson et al., 2022). Some authors reported that SARS-CoV-2 can survive and maintain its infectivity in an aerosol for hours (Van Doremalen et al., 2020).
Nonetheless, the virus has other transmission pathways in addition to the respiratory route. In a study on organoids, it was proved that the virus is capable of infecting enterocytes (gut cells) that abundantly express ACE2 (the virus receptor). Viral RNA was detected in rectal swabs, suggesting that the fecal-oral route is also a possible transmission pathway (Zhou et al., 2020). Individuals infected with SARS-CoV-2 exhibit gastrointestinal symptoms including diarrhea, nausea, vomiting, and abdominal pain. This observation also points to the possibility that SARS-CoV-2 is an enteric virus (Ding and Liang, 2020; Moura et al., 2022). There is a report of persistent virus shedding by children through feces after their nasopharyngeal swabs tested negative for viral RNA (Xu et al., 2020b). Another study revealed virus shedding through feces even after the disappearance of the virus from nasopharyngeal area (Chen C. et al., 2020). It has been found that the mucosal membrane may also be involved in the transmission of SARS-CoV-2. Close contact with the eyes, nose, or mouth, whether direct or indirect, can lead to COVID-19 transmission (Lu et al., 2021).
4 Innate immune sensors and recognition of SARS-CoV-2
In order to maintain cell homeostasis and survival, all living organisms require immediate cellular responses to pathogen invasion (Buchmann, 2014). Pattern recognition receptors (PRRs) are germline-encoded cellular receptors that recognize certain patterns of ‘non-self’ and ‘danger’ molecules, referred to as ‘pathogen-associated molecular patterns’ (PAMPs) and ‘danger-associated molecular patterns’ (DAMPs) respectively (DAMPs). PAMPs or DAMPs activate PRRs in mammals, triggering innate immune responses and the production of numerous IFNs and proinflammatory cytokines (Hansen et al., 2011). Several PRRs have been discovered in recent decades, including Toll-like receptors (TLRs), nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), C-type lectin receptors (CLRs), AIM2-like receptors (ALRs), cyclic GMP-AMP synthase (cGAS) and retinoic acid-inducible gene I (RIG-I)-like receptors (Muñoz-Wolf and Lavelle, 2016). TLRs and RLRs are two of the most important receptors for recognizing RNA virus infection and activating antiviral IFN programs (Chen et al., 2017) (Figure 2).
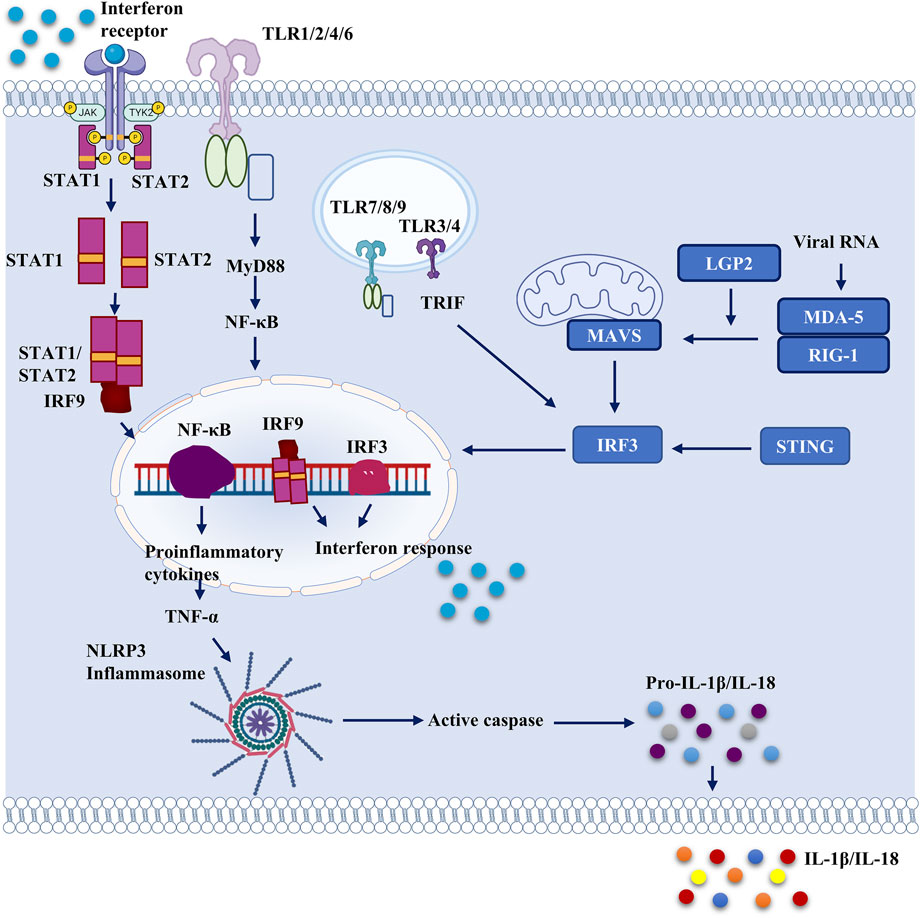
FIGURE 2. Important pattern recognition receptors and cellular proteins which can serve as potential therapeutic targets against SARS-CoV-2. Different cellular receptors can be activated by viral proteins and viral genome. Structural proteins of SARS-CoV-2 activate membrane bound toll-like receptors and they activate NF-κB by MyD88 activation. Endosomal TLRs are activated by viral structural as well as non-structural proteins, which also activate NF-κB and downstream signaling. Thus, proinflammatory cytokines are released from the cell. Proinflammatory cytokines further activate the NLRP3 inflammasome which leads to secretion of IL-1β and IL-18. NLRP3 inflammasome is activated directly too by viral proteins including ORF3a and non-structural proteins. STING expression is also dysregulated by ORF3a, and 3CLpro. Nucleocapsid and Membrane protein blocks IRF3 phosphorylation and attenuates interferon response. It also interferes with RIG-1. Various SARS-CoV-2 proteins have been reported to interfere with RIG-like receptor signaling.
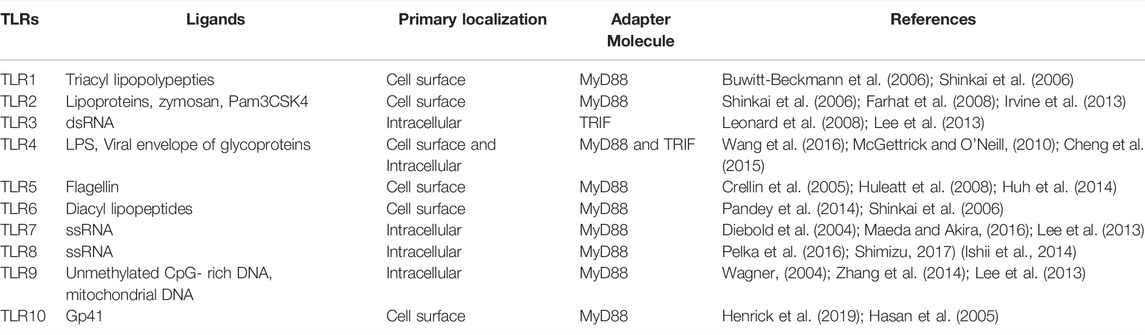
4.1 TLRs and SARS-CoV-2
Toll was first identified as an antifungal gene that is key to the Drosophila immune system. Subsequent studies further elucidated the fundamental role of TLRs in innate immune sensing in mammals (Lemaitre et al., 1996; Takeda and Akira, 2005). To date, multiple TLRs have been discovered, and their functions have been elucidated. In humans, 10 TLRs (TLR1–TLR10) have been identified, whereas in mice, 12 TLRs (TLR1–TLR9 and TLR11–TLR13) have been discovered, and their functions have been studied (Kawasaki and Kawai, 2014). Each TLR can identify PAMPs that are composed of microbial components such as nucleic acids, lipoproteins, and lipids (Kawai and Akira, 2010). Cell surface TLRs (TLR1/2, 2/6, 4, 5) detect lipids, lipoproteins, and proteins present in microbial membranes as shown in table 3. Endosomal TLRs 3,7,8 and 9 recognize bacterial and viral genomic components; TLR7/TLR8 receptors detect ssRNA, TLR3 detect double-stranded RNA (dsRNA), TLR9 recognizes double stranded DNA in endosomal compartments and recognize viruses that enter through endocytosis (Kawai and Akira, 2007). RLRs and TLRs launch antiviral defense mechanisms as soon as these receptors are engaged by RNA ligands. TLRs bind to adapter proteins such as myeloid differentiation primary response 88 (MyD88; for TLR7 and TLR8) and TIR domain-containing adapter inducing IFN-β (TRIF; for TLR3) to initiate downstream signaling cascades (Vidya et al., 2018).
4.1.1 Membrane-Bound TLRs
Studies have revealed that TLR3−/−-, TLR4−/−-, and TRIF-related adaptor molecule (TRAM)−/−- deficient mice are more vulnerable to SARS-CoV infection and show greater transient weight loss when compared to wild-type mice. Mice lacking TRIF (the adaptor protein for TLR3 and TLR4) are more susceptible to SARS-CoV infection (Devarajan and Vaseghi, 2021). Various in silico and in vitro studies point to the involvement of TLRs in the pathogenesis of COVID-19 (Choudhury and Mukherjee, 2020a; Planès et al., 2021). The injection of INNA-051 (a TLR2/6 agonist) dramatically lowers the amount of viral RNA in the nose and throat in a ferret model (Proud et al., 2021). Another paper indicated that the SARS-CoV-2 envelope protein can upregulate production of pro-inflammatory cytokine through TLR2-dependent signaling (Planès et al., 2021). It is also reported that TLR2 and MyD88 expression affects the severity of the disease. Moreover, TLR2 recognizes the SARS-CoV-2 envelope protein as its ligand and then induces the production of pro-inflammatory cytokines (Sariol and Perlman, 2021; Zheng et al., 2021). Some studies found that the spike protein of SARS-CoV-2 induces a pro-inflammatory response through a TLR-2 mediated pathway (Khan et al., 2021).
TLR4 is expressed in both immune cells and tissue resident cells. The expression of TLR4 and TLR7 was found to be significantly upregulated in mucosal membrane cells in the children with COVID-19; same study reported expression of pro-inflammatory cytokines at both the transcriptional and translational levels (Gankovskaya et al., 2021). TLR4 mediated inflammatory cytokines were found to be upregulated in another study conducted by Sohn et al. (Sohn et al., 2020). It has been proposed that the spike protein of SARS-CoV-2 upregulates TLR4, which ultimately increases the expression of ACE2 on cells and facilitates the entry of SARS-CoV-2. This hypothesis is supported by the finding that ACE2 is expressed by 1–2% of cells in the lungs. Due to ACE2 upregulation mediated by TLR-4, SARS-CoV-2 is able to infect more heart and lung cells (Heads, 2020; Aboudounya et al., 2021; Han et al., 2021; Manik et al., 2022). TAK-242, an anti-inflammatory molecule which inhibits TLR4 mediated signaling, was found to reduce the level of TNF-α and IL-6 in BV-2 mouse microglial cell line (Olajide et al., 2022).
In peripheral blood mononuclear cells, the spike protein is capable of upregulating TLR6 and MMP9 and downregulating SKAP1 and LAG3, which are involved in the control of T-cell function (Zhang et al., 2021a). One of the in silico studies suggested that a citrus flavonoid called rutin may be a promising antiviral therapeutic, binding to M-pro, TLR2, TLR6, and TLR7 (Hu et al., 2020). Another in silico study indicated that TLR4 has high affinity for the spike protein, and that TLR6 and TLR1 have comparatively lower affinity (Choudhury and Mukherjee, 2020b).
4.1.2 Endosomal TLRs
4.1.2.1 TLR3
TLR3 is an endosomal receptor that recognizes viral dsRNA, thereby inducing the production of type I IFN and inflammatory cytokines (Matsumoto and Seya, 2008). An in silico study on the binding of all mRNAs to intracellular TLRs shows that the mRNAs of Nsp10, S2, and E SARS-CoV-2 proteins are potential virus-associated molecular patterns that bind to TLR3, TLR9, and TLR7, respectively, and trigger downstream cascades (Choudhury et al., 2021). Other investigations noticed that SARS-CoV-2 activates TLR3 and TLR7; nevertheless, TLR3 mainly acts through IRF3 activation, whereas TLR7 exerts its action mainly via NF-κB activation (Bortolotti et al., 2021). It has been demonstrated that ORF9b of SARS-CoV-2 inhibits IFN production by targeting the TLR3-TRIF cascade, in addition to RIG-I/MDA-5 MAVS and cGAS-STING signaling pathways (Han et al., 2021). According to a recently published article, polyinosinic:polycytidylic acid (polyI:C; a synthetic TLR3 agonist) protects mice against COVID-19 (Tamir et al., 2022).
4.1.2.2 TLR7
TLR7 recognizes ssRNA and synthetic oligoribonucleotide analogs such as imidazoquinoline, imiquimod, and R-848 (Sun and Reddy, 2013). TLR8 is an endosomal sensor of RNA degradation products in human phagocytes and facilitates the detection of viral and bacterial infections (Moen et al., 2019). In a comparative analysis via an immunoinformatic approach, it was revealed that SARS-CoV-2 has more fragments of ssRNA than SARS-CoV does; therefore, SARS-CoV-2 is capable of hyperactivating the innate immune system by triggering of TLR7/8, resulting in acute lung injury because of the induction of a pro-inflammatory response via the TLR7/8 engagement (Moreno-Eutimio et al., 2020). A loss-of-function mutation in TLR7 increases COVID-19 severity in young males (Moreno-Eutimio et al., 2020). TLR7 agonist imiquimod has been proposed as a possible therapeutic for COVID-19 (Poulas et al., 2020). TLR4 and TLR7 over-expression mediated by SARS-CoV-2 has been detected in adipose tissue-derived mesenchymal stromal cells in response to periodic thermomechanical manipulation (Xu et al., 2021). Due to loss-of-function mutations that suppress the antiviral response to SARS-CoV-2, TLR7 variants constitute a major risk factor for severe COVID-19 in men under the age of 50 (Englmeier and Subburayalu, 2022). In a nested case study, it was found that the loss-of-function mutation of TLR7 is present in 2.1% of the patients with severe COVID-19, but in none of the asymptomatic patients (Fallerini et al., 2021).
Women have higher levels of immune-cell activation than men do, which is linked to TLR7 activation and IFN production (Webb et al., 2019). TLR7 expression is higher in females than in males, and its biallelic expression leads to enhanced immune responses and viral-infection resistance (Spiering and de Vries, 2021). Furthermore, after a viral infection, a lower amount of inflammatory IL-6 is produced in women than in men, and this sign is often associated with a longer lifespan. In addition to this, there are loci on the X chromosome that contain genes implicated in immune-cell modulation (e.g., FOXP3) and a transcription factor for regulatory T cells important for viral pathogenesis. The X chromosome affects the immune system by expressing a variety of proteins, including TLR8, CD40L, and CXCR3, all of which can be over-expressed in women and alter the immune system’s response to viral infections and immunizations. That is why women are less susceptible to viral infections than men (Conti and Younes, 2020).
Several TLR7 agonists have been proposed as drugs and vaccine adjuvants against SARS-CoV-2. TLR7 agonists have been conjugated to CoVac501, a self-adjuvanting peptide vaccine. The vaccine is manufactured entirely chemically and incorporates immunodominant peptides identified in the RBD. CoVac501 generates robust and long-lasting titers of protective neutralizing antibodies against numerous RBD mutants, original SARS-CoV-2 strain, and variants in non-human primates (Long et al., 2022).
4.1.2.3 TLR8
An in silico study on brain glial cells determined the ability of viral spike epitope peptides M1Lys60 and Ala240Glu300 to bind or interact with human TLR8, with the brain-targeted vascular cell adhesion molecule (VCAM1) protein, with zonula occludens, with glial-cell-specific protein NDRG2, and with apo-S100B. It was also found that a SARS-COV-2 spike peptide forms a heterodimeric complex with human TLR8 (Dasgupta and Bandyopadhyay, 2021). A bioinformatic analysis of the viral genome revealed hundreds of fragments that might activate TLR7/8, suggesting that products of endosomal processing of the virus can potently induce IFN-based and inflammatory responses downstream of these receptors. SARS-CoV-2 associated molecular patterns elicit MyD88-dependent lung inflammation in mice, which is characterized by over-expression of pro-inflammatory and cytotoxic mediators, immune-cell infiltration, and phenotypic maturation of splenic dendritic cells. Thus, TLR7/8 was discovered as a critical cellular sensor of SARS-CoV-2 ssRNAs that takes part in host resistance and in COVID-19 development (Salvi et al., 2021).
In another research article, it is demonstrated that in the absence of pyroptosis, GU-rich ssRNA (GU-rich RNA) derived from SARS-CoV-2, SARS-CoV-1, or HIV-1 launches a TLR8-dependent pro-inflammatory cytokine response in human macrophages, with GU-rich RNA from the SARS-CoV-2 spike gene triggering a pro-inflammatory response. Moreover, it was shown using genetic and pharmacological suppression that the production of mature IL-1β is mediated by a non-classic pathway that is dependent on caspase 1, caspase 8, the NLRP3 inflammasome, potassium ion efflux, and autophagy while being independent of TRIF (TICAM1), vitamin D3, and pyroptosis (Campbell et al., 2021).
TLR8 mRNA has turned out to be more abundant than TLR7 mRNA, which indicates the presence of monocyte-derived dendritic cells (MDDCs). In in vitro assays on MDDCs activated with ssRNA40 (a TLR8 agonist that is a positive-sense ssRNA similar to SARS-CoV-2 RNA), upregulation of sXBP1 and mRNA and protein expression of IL-1β, IL-6, and TNF was noticed. An IRE1 ribonuclease inhibitor (MKC8866) suppressed these responses. Moreover, 1) the presence of sXBP1 in nasopharyngeal swabs and BAAs, 2) sXBP1 induction by ssRNA40 in MDDCs, 3) the effect of IRE1 RNase inhibition on the cytokine induction produced by ssRNA40, and 4) the demonstration of sXBP1 binding to IL1B, IL6, and TNF promoters, all suggest that TLR8-induced XBP1 splicing potentially contributes to the viral sepsis observed in severe cases of COVID-19 (Fernández et al., 2022).
4.1.2.4 TLR9
It has been hypothesized that for COVID-19, TLR9 activation is a subtle but potent factor causing hyperinflammation and thrombotic complications in vulnerable patients. As a result, testing the efficacy of TLR9 agonists is recommended for the fight against SARS-CoV-2 (Oberemok et al., 2020).
Human mitochondrial DNA, which is derived from endosymbiont bacteria and contains unmethylated CpG motifs, is a model of a well-known DAMP that directly activates the inflammatory process via TLR9 upon tissue damage and infection (Zhang et al., 2010). In endothelial cells, SARS-CoV-2 infection affects mitochondrial function and promotes TLR9 signaling. TLR9 launches inflammatory responses that culminate in endothelial-cell dysfunction, which may contribute to the severity of COVID-19 symptoms. Targeting of mitochondrial metabolic pathways may help to devise new COVID-19 treatments (Costa et al., 2022).
CpG 1018, which is a T helper 1 (Th1)-biasing synthetic TLR9 agonist, has been chosen as an adjuvant candidate to improve immunogenicity and attenuate possible vaccine-induced immunopathology. Cytokines generated by splenocytes of mice vaccinated with CpG 1018 and alum show a strong Th1-dominant response. In a dose-ranging trial, rats subjected to single- or double-dose regimens of S-2P coupled with CpG 1018 alone or CpG 1018 plus alum manifested no vaccine-related significant adverse effects. These findings support the development of a COVID-19 vaccine candidate based on CHO-derived S-2P formulated with CpG 1018 and alum (Kuo et al., 2020).
In another study, several combinations of PRR agonists and aluminum hydroxide (AH) were compared, and it was discovered that combining CpG oligodeoxynucleotides (TLR9 agonists) with AH and RBD significantly improves immune responses to RBD in young mice on a prime-boost immunization schedule. In aged mice, the AH:CpG-adjuvanted RBD vaccination evoked a strong anti-RBD immune response, with an extra boost dose generating an anti-RBD antibody response comparable to that in young adult mice and conferring complete protection against a live-SARS-CoV-2 challenge. In addition, it was observed that the AH:CpG adjuvant formulation has unique immunological capabilities yielding human-leukocyte activation in vitro. Overall, the thorough adjuvant comparison analysis showed that co-adjuvanting AH:CpG can overcome both RBD immunogenicity and immunosenescence (Nanishi et al., 2021).
Haabeth et al. (2021) looked into the development of a new mRNA vaccine that involves charge-altering releasable transporters as a delivery mechanism. It was proposed that vaccination immunogenicity could be customized by including co-formulated adjuvants such as oligodeoxynucleotides with CpG motifs (CpG-ODN) in these intrinsically nonimmunogenic vehicles. Mice given the mRNA-charge-altering-releasable transporter vaccine acquired therapeutically relevant levels of RBD-specific neutralizing antibodies in the blood and bronchial secretions. Furthermore, the vaccination induced RBD-specific Th1 T-cell responses, including CD4+ and CD8+ T-cell memory, that were strong and long-lasting (Haabeth et al., 2021).
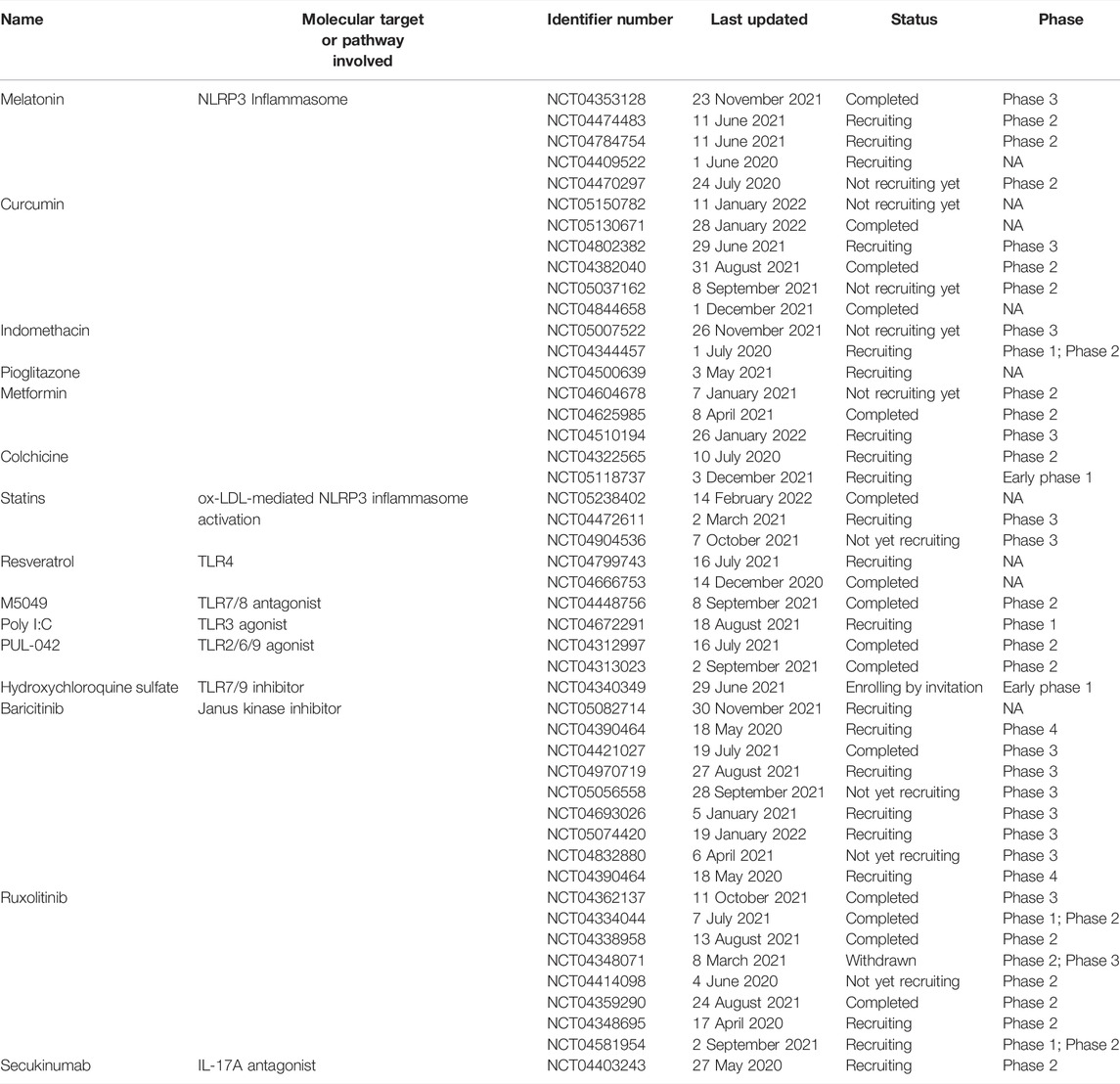
Various drugs which target toll-like receptors are being evaluated in clinical trials. Some of them have been listed in table 4.
4.2 RIG-Like Receptors (RLRs) and SARS-CoV-2
RLRs, unlike TLRs, are crucial cytoplasmic virus sensors that detect intracellular non-self RNAs with different secondary-structure patterns or metabolic changes (Lässig and Hopfner, 2017). RLRs are RNA helicases that include the Asp-Glu-Ala-Asp (DEAD) box, and this family includes three members: RIG-I, melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2). All RLRs have an RNA helicase domain with a C-terminal domain, which is responsible for RNA binding. Although RIG-I and MDA5 both contain tandem caspase activation and recruitment domains (CARDs) for downstream signal transduction, LGP2 does not have a CARD and therefore acts as a modulator of RIG-I and MDA5 (Yoo et al., 2014). RIG-I and MDA5 have similar as well as separate mechanisms for recognizing non-self RNAs. As mammalian cells lack an RdRp, dsRNA is a characteristic non-self RNA that is normally absent in uninfected cells. An artificial dsRNA called polyI:C can activate both RIG-I and MDA5 (Schlee and Hartmann, 2016). Of note, RIG-I and MDA5 activation by dsRNA is regulated differently depending on the length of the dsRNA. The shortest dsRNA found to activate RIG-I is 13 bp, which is the same as the shortest length required for the assembly of a 2-RIG-I/dsRNA dimer (Brisse and Ly, 2019). Recent research uncovered several biochemical properties of RLR-activating RNA species, including 1) 5′-triphosphate in RNAs having a secondary structure, 2) 5′-diphosphate and the absence of a cap, and 3) the presence of an unmethylated 5′-end nucleotide at the 2′-O position; such RNAs are typically generated during the replication of RNA viruses, including coronaviruses (Rehwinkel and Gack, 2020).
RLRs undergo conformational changes driven by ATPase/helicase activity, from an inactive “closed” state to an active “open” state. The CARD-CARD interaction allows activated RLRs to release CARDs, which then come into contact with a signaling adaptor molecule called mitochondria antiviral signaling protein (MAVS) (Leung and Amarasinghe, 2012). Signaling adapters MyD88, TRIF, and MAVS then recruit several ubiquitin ligases, such as TNF receptor-associated factor (TRAF) 3 and TRAF6, which associate with antiviral kinases like TANK-binding kinase 1 (TBK1), IκB kinase (IKK), and the IKK complex, to coordinate downstream signaling pathways. As a result of the activation of transcription factors IRF3 and IRF7 as well as NF-κB, type I IFN and pro-inflammatory cytokines are produced, allowing host antiviral IFN systems to function freely (Konno et al., 2009; Kawasaki and Kawai, 2014). It was found in vitro experiments that IFNs inhibit the replication cycle of SARS-CoV-2 in a dose-dependent manner (Busnadiego et al., 2020; Vanderheiden et al., 2020).
Yin et al. (2021) reported the involvement of MDA5 and LGP2 (members of the RLR family) in the induction of an IFN response as a consequence of SARS-CoV-2 infection. They also identified IRF3, IRF7, and NF-κB as the key transcription factors responsible for the regulation of IFN production in lung epithelial cells during SARS-CoV-2 infection (Yin et al., 2021). Another group demonstrated that the MDA5 protein mediates IFN production in lung cells as a result of COVID-19; however, the enhanced IFN response could not control the viral replication in the lung cells (Rebendenne et al., 2021). Another research group proposed that the RNA sensing through MDA5 is the key driver of inflammation in lung cells. They also suggested that the inhibition of innate immune sensing of SARS-CoV-2 may alleviate the inflammation caused by COVID-19 (Thorne et al., 2021). Different research groups have reported that different proteins of SARS-CoV-2 interfere with RIG1 and MDA5 or with either RIG1 or MDA5 signaling. One group reported that PL-pro antagonizes ISG15-dependent activation of MDA5 by de-ISGylation. MDA5 oligomerization is triggered by ISGylation of CARDs, and this process triggers innate immunity against a variety of viruses such as coronaviruses, flaviviruses, and picornaviruses. Accordingly, this mechanism helps the virus to evade the immune response (Liu et al., 2021b). By contrast, other studies reported that the M protein of SARS-CoV-2 suppresses type I and type III IFN responses by preventing the phosphorylation and activation of IRF3 through interaction with RIG-I, MAVS, and TBK1 (Zheng Y. et al., 2020). The nucleocapsid protein inhibits IFN secretion by blocking IRF3 phosphorylation and by downregulating Tyk2 and STAT2 (Kumar et al., 2021).
Recently, it was observed that MDA5 is over-expressed in asymptomatic and mildly symptomatic patients. Type I IFN was also found to be significantly upregulated in nasopharyngeal cells of the asymptomatic patients. This result points to a protective role of MDA5 involving upregulation of type I IFN to protect the patient from possible severe tissue damage caused by SARS-CoV-2 (Mohanty et al., 2021). The nucleocapsid protein of SARS-CoV-2 has been reported to inhibit the interaction between tripartite motif protein 25 (TRIM25) and RIG-I, as well as to suppress nuclear translocation of IFN-regulatory factor 3, thereby attenuating IFN production (Oh and Shin, 2021). Other components of SARS-CoV-2 have also been found to inhibit the induction of an IFN response. ORF3b, ORF6, ORF7a, ORF8, and Nsp14 in addition to the Nucleocapsid protein of SARS-CoV-2 are potent inhibitors of an IFN response (Li JY. et al., 2020; Cao et al., 2021; Hsu et al., 2021; Redondo et al., 2021).
4.3 NLRs and SARS-CoV-2
Inflammasomes are multiprotein complexes composed of members of the NLR family and of the pyrin and HIN domain (PYHIN) family. Inflammasomes’ biochemical purpose is to trigger caspase 1, which induces the maturation of IL-1β and IL-18 as well as the activation of pyroptosis, a type of cell death. The roles of the NLRP3 inflammasome in human immunity and pathologies have been proven extensively. The NLRP3 inflammasome is a crucial mediator of host immune responses because it activates caspase 1 and IL-1β and/or IL-18 (He et al., 2016).
The NLRP3 inflammasome, unlike other inflammasomes, can be activated by a variety of stimuli. The NLRP3 inflammasome is thought to sense cellular homeostasis rather than directly recognize a common motif present in its inducers; many physiological signals, e.g., K+ efflux, Ca2+ signaling, mitochondrial malfunction, and lysosomal rupture, have been hypothesized to drive its activation (He et al., 2016). Infectious microorganisms such as the Sendai virus, influenza virus, adenovirus, fungi Saccharomyces cerevisiae and Candida albicans, and bacteria (such as Staphylococcus aureus, Listeria monocytogenes, and Shigella flexneri) are also able to activate the NLRP3 inflammasome. In some cases, specific microbial components or products that activate the NLRP3 inflammasome have been identified, such as bacterial and viral RNA, malaria-causing parasite hemozoin crystals, and various bacterial pore-forming toxins (Chengcheng et al., 2010; Zhao N. et al., 2021). The ORF3a-encoded SARS-CoV-2 viroporin—a type of tiny hydrophobic protein encoded by viruses that oligomerizes in host cell membranes resulting in the formation of hydrophilic pores—activates the NLRP3 inflammasome (Nieva et al., 2012). ORF3a activates the inflammasome via either an ASC-dependent or ASC-independent mechanism by triggering IL-1β expression. ORF3a-mediated inflammasome activation necessitates K+ efflux and NEK7-NLRP3 oligomerization (Xu et al., 2022). MCC950, a small-molecule inhibitor of NLRP3’s ATPase activity, lowers the production of IL-18 in response to these viral proteins (Rousseau et al., 2021). It was observed in another study that MCC950 can block ORF3a-mediated inflammasome activation. ORF3a key residues needed for the virus release and inflammasome activation are conserved among SARS-CoV-2 isolates across continents, suggesting that ORF3a and NLRP3 are prime targets for a therapeutic intervention (Xu et al., 2020a).
Additionally, NLRP3 leads to the release of several biologically active DAMPs. This action sets off a chain of events that result in amplification of the innate immune system’s reaction and trigger the complement cascade (ComC). In addition to DAMPs, mannan-binding lectin (MBL), which binds to SARS-CoV-2 proteins, has been shown to directly launch the ComC. Notably, activation of the ComC via the MBL-MASP-2 protease complex also triggers the coagulation cascade (CoaC), and the launch of the latter is associated with a worse prognosis in patients infected with SARS-CoV-2. This observation explains why ComC and CoaC inhibition are individually being investigated as a therapeutic option (Ratajczak and Kucia, 2020). It has been demonstrated that NLRP3 is activated by SARS-CoV-2 at an early stage of the infection and induces cytokine release syndrome. These data suggest that NLRP3 can serve as an effective target for reducing organ damage (Marchetti et al., 2021).
Yalcinkaya et al. (2021) reported that the E protein prevents inflammasome priming and NLRP3 inflammasome activation in cultured macrophages. Similarly, in animals transfected with the E protein and treated with polyI:C mimicking the effects of viral RNA, the E protein reduces pro-IL-1β expression, IL-1β and IL-18 levels in bronchoalveolar lavage fluid, and macrophage infiltration of the lungs in an NLRP3-dependent manner. Macrophages were treated with both LPS and polyI:C to simulate the consequences of a more advanced infection. Under these conditions, the E protein boosted the activation of the NLRP3 inflammasome in both murine and human macrophages. As a consequence, the SARS-CoV-2 E protein may reduce the host NLRP3 inflammasome response to viral RNA at early stages of the infection while possibly enhancing NLRP3 inflammasome responses later on. The mentioned study suggests that targeting the SARS-CoV-2 E protein, particularly in the early stages of the infection, may be a unique way to treat COVID-19 (Yalcinkaya et al., 2021).
Two non-structural proteins, Nsp1 and Nsp13, suppress caspase 1-mediated IL-1β activation, according to a screening of a complementary DNA (cDNA) library containing 28 SARS-CoV-2 ORFs. Caspase 1 inhibition was attributed to Nsp1 amino acid residues involved in the silencing of host translation and to Nsp13 domains responsible for helicase activity. Both Nsp1 and Nsp13 attenuated NLRP3 inflammasome-induced caspase 1 activity and IL-1β production in THP-1 cells. These data suggest that SARS-CoV-2 Nsp1 and Nsp13 are effective NLRP3 inflammasome antagonists (Kim et al., 2021b).
A recent mutational analysis of SARS-CoV-2 implies that Nsp6 is an important indicator of viral pathogenicity. Inflammasome-related NOD-like receptor signaling is activated in SARS-CoV-2-infected lung epithelial cells and COVID-19 patients’ lung tissues, according to transcriptome analysis. The induction of inflammasomes/pyroptosis in patients with severe COVID-19 was validated by means of serological markers. Nsp6 over-expression resulted in NLRP3/ASC-dependent caspase 1 activation, maturation of IL-1β and IL-18 or either IL-1β or IL-18, and pyroptosis of lung epithelial cells. Nsp6 inhibited autophagic flux by impairing lysosome acidification, which was reversed by 1,25-dihydroxyvitamin D3, metformin, or polydatin, which prevented Nsp6-induced pyroptosis. Nsp6 interacts directly with ATP6AP1 (a proton pump component of vacuolar ATPase) and reduces its cleavage-mediated activation. Reduced binding to ATP6AP1 and a diminished ability to impede lysosome acidification for promoting pyroptosis were revealed in the L37F Nsp6 variant, which was linked with asymptomatic COVID-19. Infection of cultured lung epithelial cells with live SARS-CoV-2 caused autophagic-flux stagnation, inflammasome activation, and pyroptosis in a consistent manner. Overall, the mentioned research suggests that SARS-CoV-2 Nsp6 can cause inflammatory cell death of lung epithelial cells, and this effect could be exploited therapeutically through pharmacological correction of autophagic flux (Sun et al., 2022).
In a confocal immunofluorescence analysis, the SARS-CoV-2 nucleocapsid was found to co-localize with neurons, astrocytes, oligodendrocytes, and microglia. In cerebral cortical tissues, the viral nucleocapsid was discovered alongside ACE2 (cell entry receptor) and the NLRP3 inflammasome (Cama et al., 2021). In another study, it was reported that the N protein interacts directly with the NLRP3 protein and facilitates NLRP3 inflammasome assembly by promoting NLRP3 binding to ASC (Pan et al., 2021).
The spike protein of SARS-CoV-2 has also been found to activate the NLRP3 inflammasome (Kucia et al., 2020). In another research article, it is reported that in cultured murine alveolar macrophages, ultramicronized palmitoylethanolamide inhibits NLRP3 inflammasome expression and the pro-inflammatory response activated by the SARS-CoV-2 Spike protein (Del Re et al., 2021). It was also observed that the infected cells undergo pyroptosis mediated by inflammasome activation involving NLRP3 and AIM2. Moreover, tissue-resident macrophages show inflammasome activation, but this phenomenon is not observed in infected epithelial cells (Junqueira et al., 2021). The SARS-CoV-2 spike protein triggers inflammasome activation and an IL-1β release in macrophages obtained from COVID-19 patients but not in macrophages from healthy SARS-CoV-2-naive controls. Ex vivo, chemical NLRP3 suppression inhibits spike protein-induced IL-1β production (Theobald et al., 2021). Another study indicated that ORF3a is required for the assembly of the NLRP3 inflammasome and for the secretion of mature IL-18. Albeit to a lesser extent, Nsp2 and ORF3b (among 26 tested viral proteins) enhanced NLPR3 inflammasome activity and the production of mature IL-18. In human-ACE2 transgenic mice, specific suppression of the NLRP3 inflammasome by MCC950 reduces excessive lung inflammation and hence COVID-19-like pathology (Zeng et al., 2022).
In infected human monocytes, SARS-CoV-2 infection activates the NLRP3 inflammasome and inhibits caspase 1, GSDMD cleavage, and pyroptosis. in vitro and in vivo experiments, the SARS-CoV-2 nucleocapsid protein interacts with GSDMD in cells and suppresses GSDMD cleavage. The Nucleocapsid protein attaches to the GSDMD linker region, thus preventing caspase 1 from processing this protein (Ma et al., 2021). In human microglial cells, NLRP3 could be primed and activated by the Spike protein of SARS-CoV-2 in an NF-κB-dependent as well as ACE2-dependent manner (Albornoz et al., 2022). Thus, it becomes very clear that NLRP3 is an attractive target to be probed against SARS-CoV-2. Drugs which are under clinical trials have been listed in table 4.
4.4 CLRs and SARS-CoV-2
CLRs, which belong to the superfamily of membrane-bound phagocytic PRRs, recognize carbohydrates on the surface of pathogens in a Ca2+-dependent manner and hence are termed “C-type lectin receptors.” CLRs play a substantial part in the regulation of adaptive immune responses by managing the induction of signaling pathways. CLRs can regulate adaptive immunity on multiple levels by initiating signaling on their own, through interaction with other PRRs, or by triggering carbohydrate-specific signaling cascades (Geijtenbeek and Gringhuis, 2009; Li and Wu, 2021). They can either modulate TLR signaling or directly induce gene expression. CLRs can be soluble or transmembrane receptors (Kerrigan and Brown, 2010), and are expressed by antigen-presenting cells such as dendritic cells, macrophages, monocytes, and Langerhans cells (Bermejo-Jambrina et al., 2018). The participation of CLRs in virus recognition is complicated. They can detect and respond efficiently to a wide spectrum of viruses and thus can shape antiviral immune responses (Monteiro and Lepenies, 2017).
Several C-type lectins (DC-SIGN, L-SIGN, LSECtin, ASGR1, and CLEC10A) serve as glycan-dependent binding partners of the spike protein of SARS-CoV-2. These substances interact with spike through the region around the RBD (Lu et al., 2021). Recently, it was demonstrated that attachment to such receptors as CLRs, DC-SIGN, L-SIGN, and sialic-acid-binding immunoglobulin-like lectin 1 (SIGLEC1) enhances ACE2-mediated infection and modulates the neutralizing activity of distinct types of spike-specific antibodies (Rahimi, 2020; Amraei et al., 2021; Lempp et al., 2021). Another study revealed that DC-SIGN, L-SIGN, langerin, and macrophage galactose-type C-type lectin can interact with the spike protein of SARS-CoV-2, and that DC/L-SIGN facilitates the virus entry into the cell through ACE2. Nonetheless, by means of a glycol-mimetic compound designed against DC-SIGN, trans-infection between different cell types can be inhibited (Thépaut et al., 2021). Human liver sinusoidal endothelial cells and lymph node lymphatic endothelial cells express L-SIGN abundantly, whereas blood endothelial cells do not. SARS-CoV-2 proteins have been found within liver sinusoidal endothelial cells in liver autopsy samples from COVID-19 patients by high-resolution confocal microscopy. In comparison with control cells, both a pseudo-typed virus wrapped in the SARS-CoV-2 spike protein and the genuine SARS-CoV-2 virus infect L-SIGN-expressing cells. Furthermore, inhibition of L-SIGN function reduces SARS-CoV-2 infection. These findings suggest that L-SIGN is a receptor mediating SARS-CoV-2 infection (Kondo et al., 2021).
4.5 cGAS-STING Pathway
Cyclic GMP-AMP (cGAMP) synthase (cGAS) cytosolic DNA sensor triggers innate immune responses by producing cGAMP, the second messenger, which activates the interferon gene adaptor stimulator (STING). The cGAS-STING system mediates protective immune protection against a wide range of DNA-containing pathogens (Chen et al., 2016). Increasing data suggest that RNA viruses can elicit cGAS-STING responses and be blocked by cGAS-STING activation in general (Ishikawa and Barber, 2008). Bats are believed to be a natural host for many coronaviruses, and all bats studied so far have STING molecules that are faulty in inducing type-I IFN and have limited antiviral efficacy (Xie et al., 2018). The duration of coronavirus infection in bats has been associated with faulty STING activation. During cross-species transmission, SARS-CoV-2 appears to have retained or regained its ability to block the STING function. SARS-CoV-2 inhibits cGAS-STING activity with at least two viral proteins. SARS-CoV-2 3CLpro and ORF3a can antagonize various vertebrate STING functions, including those of humans, mice, and chickens, which is consistent with the fact that SARS-CoV-2 can infect a wide spectrum of species (Rui et al., 2021).
Through transcriptome analysis, SARS-CoV-2 was found to be responsible for the activation of the cGAS-STING pathway and NF-κB activation. This suggests that inhibition of STING can ameliorate the upregulated cytokine production (Neufeldt et al., 2022). Furthermore, within cells expressing ACE2 and the SARS-CoV-2 spike protein, viral infection can cause the development of syncytia, resulting in the creation of micronuclei. Micronucleus translocation cGAS, as well as overexpression of their downstream target genes, revealed that these micronuclei had a high level of activation of both the DNA damage response and cGAS-STING signaling. (Ren et al., 2021). The STING small-molecule agonist has been tested against SARS-Cov-2 infection in vitro and preclinical studies and has been found to stimulate interferon signaling and antiviral state against the virus. Intranasal delivery of diABZI-4, small molecule agonist of STING, was found to activate STING and proinflammatory cytokines (Humphries et al., 2021). In another study, diABZI significantly inhibited SARS-CoV-2 infection by inducing interferon signaling both in vitro and in vivo experiments (Li et al., 2021). These studies suggest the importance of cGAS-STING pathway in interferon signaling in response to SARS-CoV-2 thus pointing out to the need of conducting clinical trials for further validation of using STING agonist as a clinical drug.
5 Inflammatory Cytokines and the Cytokine storm in COVID-19
Immunopathological manifestations of COVID-19 include lymphopenia, dysregulated macrophages and monocytes, neutrophilia, antibody-dependent enhancement, a reduced or delayed IFN response, and a cytokine storm (Yang et al., 2021). The latter is defined as an immunological state marked by rapid proliferation and over-activation of T cells, macrophages, and natural killer cells as well as over-production of more than 150 inflammatory cytokines and chemical mediators released by the immune cells and nonimmune cells (Rabaan et al., 2021) The term “cytokine storm” was first used in 1933, in reference to acute graft-versus-host disease in the context of hematopoietic stem cell transplantation (Kim et al., 2021a). After the successful recognition of a virus by the immune system in the human body, the virus is presented to natural killer cells and CD8+ cells in the form of viral antigens by antigen-presenting cells, which subsequently activate immune responses (Liskova et al., 2021). Cytokine and chemokine responses are a part of innate as well as adaptive immune responses because they play a crucial role in communication between different cells (Lacy, 2015). It has been reported that in a large proportion of cases, the severity of COVID-19 is explained mainly by hyper-inflammation and the cytokine storm (Kim et al., 2021a). The latter also contributes to deterioration of the health condition in a large number of COVID-19 patients by promoting acute respiratory distress syndrome, multiple organ dysfunction, and coagulation deficits (Chen et al., 2021).
Secretion of multiple cytokines, also known as cytokine release syndrome, is linked with the emergence of clinical symptoms. For example, IFN-γ can cause fever, chills, headache, dizziness, and fatigue. TNF can induce not only flu-like symptoms such as fever, general malaise, and fatigue, but also can cause vascular leakage, cardiomyopathy, lung injury, and acute-phase protein synthesis. IL-6, a key target in adoptive-cell therapy-induced cytokine release syndrome, can cause vascular leakage, complement activation, and triggering of the coagulation cascade, leading to severe symptoms of cytokine release syndrome such as diffuse intravascular coagulation and multiple organ failure (Shimabukuro-Vornhagen et al., 2018). It is worth noting that IL-6 may cause cardiomyopathy by promoting cardiac dysfunction, which is common in patients with cytokine release syndrome (Sun et al., 2020). It is reported that patients with COVID-19 symptoms have higher levels of cytokines and inflammatory markers such as IL-6, IL-8, IL-1β, IL-10, IL-12, and IFN-γ (Costela-Ruiz et al., 2020). At present, there is a strong link between higher levels of pro-inflammatory cytokines—as potential markers of a cytokine storm—and the severity of COVID-19 as well as poor outcomes in hospitalized patients (Del Valle et al., 2020). Elevated IL-6 concentration is recognized as one of the most important markers of unfavorable outcomes and serves as an indicator of disease progression (Santa Cruz et al., 2021).
A stressed or infected cell activates a vast number of white blood cells, including B cells, T cells, natural killer cells, macrophages, dendritic cells, and monocytes, through receptor-ligand interactions. Inflammatory cytokines are released, which stimulate additional white blood cells in a positive feedback loop. After a primary infection, a cytokine storm begins locally and spreads throughout the body via systemic circulation. Calour (heat), dolour (pain), rubor (redness), tumor (swelling or edema), and loss of function (functio laesa) are all classic indicators of inflammation (Fara et al., 2020a). Initially, the localized response involves defensive mechanisms such as increased blood flow, facilitation of leucocyte extravasation and supply of plasma proteins to the site of injury, an increase in body temperature (advantageous in the case of bacterial infections), and pain triggering (Askenase and Sansing, 2016). Soon after the pathogenic trigger, repair mechanisms are initiated. There are two likely outcomes of these events: 1) organ function is progressively restored and 2) healing occurs via fibrosis, which can lead to chronic organ dysfunction (Fara et al., 2020b).
Several drugs that act via the blockade of the upregulated cytokines are currently in clinical trials against COVID-19. Some of them are listed in Table 5.
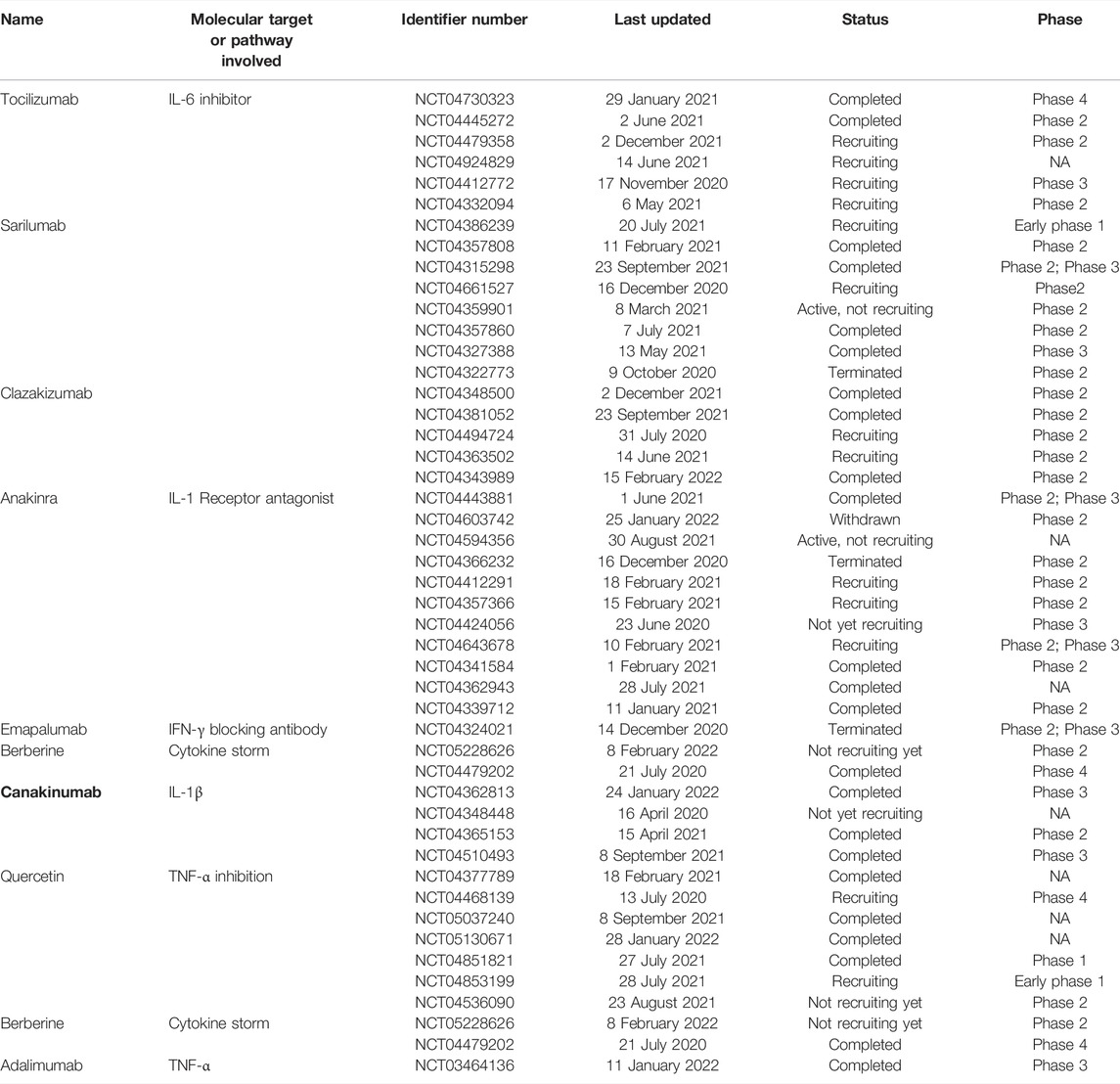
TABLE 5. Clinical trials of the drugs targeting cytokines different signaling molecules involved in the immune response against SARS-CoV-2.
6 Therapeutic targets involving viral proteins
As far as blocking the virus by inhibiting viral proteins is concerned, therapeutic targets can be generally divided into two main categories.
✓ Inhibition of viral entry
✓ Inhibition of replication machinery
6.1 Inhibition of viral entry
As stated in section 3, SARS-CoV-2 enters the cell through its spike protein by interacting with the host cell receptor (ACE2) and host cell proteases that are involved in the activation of spike. Inhibiting the viral entry may be an efficient way to stop further infection. Multiple strategies are being utilized to block the Spike protein (Dai and Gao, 2020). The most efficient among them is the use of convalescent plasma, which contains anti-SARS-CoV-2 antibodies produced by a recovered COVID-19 patient. Convalescent plasma is being used and tested in a clinical trial (NCT04356534). An anti-spike monoclonal antibody against SARS-CoV-2 is being assessed in a phase 3 clinical trial (NCT04452318).
Blockage of TMRPSS2 is one of the best ways to inhibit the virus entry because this protein is indispensable for the viral pathogenesis. Camostat mesilate and enzalutamide have been reported to inhibit TMPRSS2 in vitro, and at present, they are in phase 2 clinical trials (NCT04455815 and NCT04475601, respectively). Nafamostat mesilate also inhibits TMPRSS2 and is being tested in a phase 3 clinical trial (NCT04390594).
Blocking ACE2 is another choice for blocking the virus entry. It has been demonstrated that ACE2 blockage through derivative peptides can inhibit the infectivity by up to 74% (Yang et al., 2020). Today, several drugs that block ACE2 are in clinical trials. Drugs which inhibit angiotensin receptor have also been evaluated to inhibit viral entry. One of them (losartan) is being evaluated in a phase 4 trial (NCT04394117). Another drug—telmisartan—which also blocks ACE2, has been tested in phase 4 clinical trial (NCT04355936). Other drugs that block ACE2 have been tested in clinical trials include bromhexine with or without hydroxychloroquine and valsartan (NCT04355026 and NCT04335786, respectively). Furin inhibitors have been proved to inhibit viral entry and replication, and are further studied for their potential to be used as a therapeutic agent against SARS-CoV-2 (Cheng et al., 2020; Devi et al., 2022).
6.2 Inhibition of Replication Machinery
6.2.1 3Cl-Pro
3Cl-pro, also known as Main protease (M-pro), is a chymotrypsin-like protease that is 306 aa long and has a molecular weight of 33.8 kDa. Phylogenetic analysis of 3Cl-pro of SARS-CoV-2 revealed that it shares 96% structural similarity with that of SARS-CoV. 3Cl-pro is first cleaved from the polyprotein between Nsp4 and Nsp6 by autocleavage, and then it cleaves the polyprotein at 11 sites. It also participates in the maturation of the nonstructural proteins and thus gives rise to multiple proteins crucial for viral replication. These proteins include RdRp, helicase, exonuclease, and endoRNase. The fact that this protease is essential for the formation of proteins crucial for viral replication makes it an attractive drug target (Wu et al., 2020).
Lopinavir in combination with ritonavir has been approved by the FDA against HIV. Lopinavir is a protease inhibitor, whereas ritonavir inhibits metabolic inactivation of lopinavir by suppressing cytochrome P450 3A. Thus, ritonavir serves as a metabolic enhancer for lopinavir while lopinavir acts as a protease inhibitor and hence as the main antiviral in this combination (Tobaiqy et al., 2020). As for COVID-19, the main target of lopinavir is 3Cl-pro (Nukoolkarn et al., 2008). Currently, the combination is in a phase 4 trial (NCT04255017). Some toxic and adverse effects of lopinavir/ritonavir have been reported; therefore, these drugs need further research. PF-07321332 is a 3CLpro inhibitor with strong antiviral activity in vitro against SARS-CoV-2 and other coronaviruses and it has been tested by in silico studies and clinical trial as well (NCT04756531) (Ahmad et al., 2021). Another protease inhibitor that is being evaluated in clinical trials is danoprevir in combination with ritonavir. Danoprevir was approved by China’s FDA in 2018 for the treatment of HCV infection. It targets the NS3 protease in HCV and has a half-maximal inhibitory concentration of 0.29 µM (Chen et al.). It has been tested against COVID-19 in a phase 4 trial (NCT04345276). Lately, it is being reported that sitagliptin and daclatasvir target PL-pro, whereas lycorine HCl and nelfinavir mesylate inhibit SARS-CoV-2 M-pro (Narayanan et al., 2022).
PL-pro is the papain-like protease encoded by the nsp3 gene. It acts on the polyprotein at the N terminus and cleaves it into three nonstructural proteins that are indispensable for the replication cycle. Aside from functioning as the protease, it not only inhibits the innate immune response of the cell by suppressing IFN production but also recognizes the C terminus of ubiquitin and serves as deubiquitinase (Ullrich and Nitsche, 2020).
Therefore, being pivotal for viral replication, PL-pro is an appealing target for therapeutic regimens. Nevertheless, today, there is no FDA-approved drug that targets PL-pro. Possible adverse effects may be a reason for the absence of FDA approval because this drug mimics host deubiquitinases and hence may have a huge number of adverse effects and can be cytotoxic.
6.2.2 RdRp
This is an enzyme that is most crucial for the viral life cycle and is the most conserved enzyme among various viruses including hepatitis C virus, Zika virus, and coronaviruses. Because RdRp structure is conserved among many species, its function and mechanism of action are conserved too. For successful replication of the virus, it first synthesizes RdRp so that positive-sense ssRNA can be produced. Owing to the similarity of structure and function among RdRps of different RNA viruses, it is believed that the broad-spectrum antivirals that act on RdRp may inhibit replication of many different RNA viruses.
RdRp is an essential enzyme catalyzing viral replication by means of the 3′ polyA end of the positive-sense ssRNA as a template to synthesize the complementary strand. Two mechanisms have been proposed to explain the initiation of genome replication. The first one is primer-dependent initiation of synthesis, and the second mechanism is primer-independent synthesis also known as de novo synthesis. In case of primer-dependent synthesis, new nucleotides are added to the already present 3′ hydroxyl end provided by the bound oligonucleotide or the protein primer. By contrast, in the case of the de novo synthesis, genomic-RNA synthesis is catalyzed by the polymerase itself or by co-factors of the polymerase without any pre-existing 3′ hydroxyl end (Flint et al., 2009). As for SARS-CoV-2, Nsp7 and Nsp8 serve as these co-factors. Nsp8 can synthesize a primer of up to 6 nucleotides (Hillen et al., 2020). The elongation complex consists of 14 bases in the template strand and 11 bases in the primer strand, but for the initiation of the polymerization reaction, the first one or two nucleotides of the primer are very important (Romano et al., 2020).
Targeting RdRp by an antiviral can be effective because stopping this enzyme will inhibit viral replication, and thus the infection cycle will be blocked (Zhao J. et al., 2021; Jukič et al., 2021). Moreover, RdRp has no host cell homolog. Nevertheless, the challenges associated with protein expression, purification, and catalytic activity in vitro systems tend to be limitations of RdRp as a target of an antiviral (Zhu et al., 2020).
To date, several drugs that target RdRp have been proposed as COVID-19 therapeutics, and many of them are being tested in clinical trials. At present, remdesivir, favipiravir, and maraviroc are such drugs. Remdesivir, an adenosine analog, has been found to be consistently effective against four families of viruses: Paramyxoviridae, Filoviridae, Pneumoviridae, and Coronaviridae (Pardo et al., 2020). It is reported to be effective against SARS-CoV-2 in vitro studies, and at present, it has been tested in phase 3 clinical trial (NCT04409262).
Another promising drug is favipiravir. It is a guanine analog that inhibits RdRp. It was initially approved by the FDA in 2014 and is used against influenza A and B. It has been proposed as a drug against influenza strains that are resistant to neuraminidase inhibitors (Li H. et al., 2020). Favipiravir was recently tested against COVID-19 and was found effective, with an excellent half-maximal effective concentration of 61.88 µM. It has been tested in phase 3 clinical trial (NCT04464408).
Ribavirin has been employed in combination with IFN-α against HCV (Hadziyannis et al., 2004). It is a guanosine analog and has been found to exert broad-spectrum antiviral action against various DNA and RNA viruses (Te et al., 2007). Today, ribavirin in combination with IFN-β-1b is being assessed in a phase 2 clinical trial (NCT04494399). Besides, ribavirin is being tested in combination with sofosbuvir in another clinical study currently in phase 3 trial (NCT04460443).
6.3 Accessory proteins
6.3.1 Exonuclease
Some of the most widely used antivirals are the nucleotide analogs that block viral replication by getting incorporated into the elongated genome, consequently leading to termination of the genome synthesis, thereby inhibiting the viral replication cycle. This approach is particularly useful against RNA viruses because RNA polymerase does not possess a proofreading activity. This drawback makes RNA viruses more susceptible to mutations and to nucleotide-based inhibitors (Modrow et al., 2013). Nonetheless, coronaviruses are an exception in this context because they possess exonuclease and endonuclease. The N terminus of Nsp14 serves as the exonuclease. A nucleotide analog incorporated in the growing strand is readily removed from the strand. Hence, the virus continues its replication without any hinderance (Gurung, 2020). Moreover, guanine-N7 methyltransferase, which is involved in the capping of SARS-CoV-2 is also being investigated for targeting in order to interfere with viral pathogenesis (Selvaraj et al., 2021). In an in silico study, Saquinavir, Hypericin, Baicalein and Bromocriptine have been found to interact with Nsp14 (Liu et al., 2021a). Therefore, Nsp14 can serve as a promising target, particularly when RdRp is being suppressed with nucleotide analogs.
6.3.2 Helicase
Helicases are multifunctional proteins of Coronaviridae and have been conserved over the course of evolution. The helicase of SARS-CoV-2 belongs to helicase superfamily 1 (Shu et al., 2020). It contains three main domains: an N-terminal domain, metal-binding domain, and helicase domain. Helicase hydrolyzes nucleotides of DNA as well as RNA to unwind the helix in the 5′-to-3′ direction and is a crucial component of viral replication machinery (Wu et al., 2020). Therefore, this enzyme has been suggested as an effective druggable target. Some authors reported that vapreotide and atazanavir are candidate drugs against the helicase of SARS-CoV-2 (Francis Borgio et al., 2020). Atazanavir is currently being evaluated in a clinical trial against COVID-19 (NCT04468087).
6.3.3 Virulence Factors
Apart from participating in replication, viral proteins interfere with the host immune system and either antagonize or enhance the immune response depending upon the context. Such proteins can also be a target for drug design.
Nsp1, also known as the leader peptide, is encoded by the first nonstructural-protein gene in the SARS-CoV-2 genome. Nsp1 of SARS-CoV and that of SARS-CoV-2 are 100% similar in amino acid sequence. Some authors reported that Nsp1 of SARS-CoV inhibits IFN-β expression; furthermore, it induces the degradation of host endogenous mRNAs (Kamitani et al., 2006). It has also been observed that SARS-CoV-2 ORF9b interacts with TOM70 and inhibits IFN-β production, and hence no antiviral response from the host machinery is induced (Jiang et al., 2020). Moreover, viral protein ORF9b participates in immune evasion too, by influencing the interaction of MAVS with TOM70 (Konno et al., 2020; Ribero et al., 2020).
7 Concluding Remarks
SARS-CoV-2 has been causing great harm since the beginning of the pandemic. As reviewed above, multiple innate-immunity receptors play a part in the pathogenicity of SARS-CoV-2. Despite the availability of vaccines and drugs, this virus still poses a great danger to humanity. Although it is not very common for RNA viruses to possess exonuclease activity, SARS-CoV-2 has the exonuclease activity as it belongs to Nidovirales order. Despite the exonuclease activity, the emergence of multiple variants has made it clear that the viral proteins can keep on mutating (thereby making the available drugs inefficacious against SARS-CoV-2). This phenomenon points to the importance of exploring the therapeutic potential of human endogenous immune receptors against COVID-19. Innate immune system is body’s first line of defense against any invading pathogen. A wide variety of innate-immunity receptors including TLRs, RLRs, CLRs, and their accessory signaling molecules play an important role in virus sensing and in the development of an optimal immune response. Once the viral components are detected by the cellular innate immune receptors, the body mounts an optimum immune response against SARS-CoV-2. The key features of the antiviral immune response include interferon generation, cytokine production and cell death. However, viral proteins interfere the immune response and create trouble for the body by hyperactivating the immune system which leads to cytokine storm. As a result of cytokine storm and other co-morbidities, the disease complications and severity increases, thus causing death of the host.
The present review has summarized current knowledge about innate-immunity receptors and about signaling molecules in relation to COVID-19 pathophysiology as well as the possibility of their use as druggable targets. Our review also summarizes the knowledge about currently available drugs as well as the drugs under clinical trials which are be being tested these days. Various drugs which target immune receptors and other signaling molecules have also been summarized in the review. Antibodies targeting cytokines and their receptors, including Canakinumab, Anakinra, Emapalumab, Sarilumab, and Tocilizumab, are under clinical trials. Although the success rate is relatively low when it comes to the application of innate-immunity receptors for the therapeutic purposes, the high mutation rate of RNA viruses makes these receptors a promising option.
Author Contributions
MF, AK, and BA conceived the scope of the manuscript and wrote the manuscript. MF, AK, BA, and SC revised the manuscript. Project administration and funding acquisition is done by SC and MK. All authors read and approved the final manuscript.
Funding
This research was supported by South Korea Drug Development Fund funded by Ministry of Science and ICT, Ministry of Trade, Industry, and Energy, and Ministry of Health and Welfare (HN21C1058). This work was also supported by the National Research Foundation of Korea (NRF-2022M3A9G1014520, 2019M3D1A1078940 and 2019R1A6A1A11051471).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aboudounya, M. M., Holt, M. R., and Heads, R. J. (20212021). SARS-CoV-2 Spike S1 Glycoprotein Is a TLR4 Agonist, Upregulates ACE2 Expression and Induces Pro-inflammatory M1 Macrophage Polarisation. bioRxiv. doi:10.1101/2021.08.11.455921
Ahmad, B., Batool, M., Ain, Q. u., Kim, M. S., and Choi, S. (2021). Exploring the Binding Mechanism of PF-07321332 SARS-CoV-2 Protease Inhibitor through Molecular Dynamics and Binding Free Energy Simulations. Ijms 22, 9124. doi:10.3390/ijms22179124
Albornoz, E., Amarilla, A. A., Modhiran, N., Parker, S., Li, X. X., Wijesundara, D. K., et al. (20222022). SARS-CoV-2 Drives NLRP3 Inflammasome Activation in Human Microglia through Spike-ACE2 Receptor Interaction. bioRxiv. doi:10.1101/2022.01.11.475947
Amraei, R., Yin, W., Napoleon, M. A., Suder, E. L., Berrigan, J., Zhao, Q., et al. (2021). CD209L/L-SIGN and CD209/DC-SIGN Act as Receptors for SARS-CoV-2. ACS Cent. Sci. 7, 1156–1165. doi:10.1021/ACSCENTSCI.0C01537/SUPPL_FILE/OC0C01537_SI_001.PDF
Anderson, L. J., and Schneider, E. (2012). Coronaviruses. Philadelphia, PA: Goldman’s Cecil Med., 2102–2104. doi:10.1016/b978-1-4377-1604-7.00374-2
Askenase, M. H., and Sansing, L. H. (2016). Stages of the Inflammatory Response in Pathology and Tissue Repair after Intracerebral Hemorrhage. Semin. Neurol. 36, 288–297. doi:10.1055/S-0036-1582132
Astuti, I., and Ysrafil, (2020). Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An Overview of Viral Structure and Host Response. Diabetes Metab. Syndr. 14, 407–412. doi:10.1016/j.dsx.2020.04.020
Awadasseid, A., Wu, Y., Tanaka, Y., and Zhang, W. (2020). Initial Success in the Identification and Management of the Coronavirus Disease 2019 (COVID-19) Indicates Human-To-Human Transmission in Wuhan, China. Int. J. Biol. Sci. 16, 1846–1860. doi:10.7150/ijbs.45018
Bermejo-Jambrina, M., Eder, J., Helgers, L. C., Hertoghs, N., Nijmeijer, B. M., Stunnenberg, M., et al. (2018). C-Type Lectin Receptors in Antiviral Immunity and Viral Escape. Front. Immunol. 9, 590. doi:10.3389/FIMMU.2018.00590/BIBTEX
Borgio, J. F., Alsuwat, H. S., Al Otaibi, W. M., Ibrahim, A. M., Almandil, N. B., Al Asoom, L. I., et al. (2020). State-of-the-art Tools Unveil Potent Drug Targets Amongst Clinically Approved Drugs to Inhibit Helicase in SARS-CoV-2. Arch. Med. Sci. 16, 508–518. doi:10.5114/aoms.2020.94567
Bortolotti, D., Gentili, V., Rizzo, S., Schiuma, G., Beltrami, S., Strazzabosco, G., et al. (2021). Tlr3 and Tlr7 Rna Sensor Activation during Sars-Cov-2 Infection. Microorganisms 9, 1820. doi:10.3390/MICROORGANISMS9091820/S1
Brisse, M., and Ly, H. (2019). Comparative Structure and Function Analysis of the RIG-I-like Receptors: RIG-I and MDA5. Front. Immunol. 10, 1586. doi:10.3389/FIMMU.2019.01586/BIBTEX
Buchmann, K. (2014). Evolution of Innate Immunity: Clues from Invertebrates via Fish to Mammals. Front. Immunol. 5, 459. doi:10.3389/FIMMU.2014.00459/BIBTEX
Busnadiego, I., Fernbach, S., Pohl, M. O., Karakus, U., Huber, M., Trkola, A., et al. (2020). Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2. MBio 11, 1–10. doi:10.1128/MBIO.01928-20
Buwitt-Beckmann, U., Heine, H., Wiesmüller, K. H., Jung, G., Brock, R., Akira, S., et al. (2006). TLR1- and TLR6-independent Recognition of Bacterial Lipopeptides. J. Biol. Chem. 281, 9049–9057. doi:10.1074/JBC.M512525200
Cama, V. F., Marín-Prida, J., Acosta-Rivero, N., Acosta, E. F., Díaz, L. O., Casadesús, A. V., et al. (2021). The Microglial NLRP3 Inflammasome Is Involved in Human SARS-CoV-2 Cerebral Pathogenicity: A Report of Three Post-mortem Cases. J. Neuroimmunol. 361, 577728. doi:10.1016/J.JNEUROIM.2021.577728
Campbell, G. R., To, R. K., Hanna, J., and Spector, S. A. (2021). SARS-CoV-2, SARS-CoV-1, and HIV-1 Derived ssRNA Sequences Activate the NLRP3 Inflammasome in Human Macrophages through a Non-classical Pathway. iScience 24, 102295. doi:10.1016/J.ISCI.2021.102295
Cantuti-Castelvetri, L., Ojha, R., Pedro, L. D., Djannatian, M., Franz, J., Kuivanen, S., et al. (2020). Neuropilin-1 Facilitates SARS-CoV-2 Cell Entry and Infectivity. Science 370, 856–860. doi:10.1126/SCIENCE.ABD2985/SUPPL_FILE/ABD2985_MDAR_REPRODUCIBILITY_CHECKLIST.PDF
Cao, Z., Xia, H., Rajsbaum, R., Xia, X., Wang, H., and Shi, P.-Y. (20212021). Ubiquitination of SARS-CoV-2 ORF7a Promotes Antagonism of Interferon Response. Cell. Mol. Immunol. 18 (18), 746–748. doi:10.1038/s41423-020-00603-6
Chen, C., Gao, G., Xu, Y., Pu, L., Wang, Q., Wang, L., et al. (2020a). SARS-CoV-2-Positive Sputum and Feces after Conversion of Pharyngeal Samples in Patients with COVID-19. Ann. Intern. Med. 172, 832–834. doi:10.7326/M20-0991
Chen, H., Zhang, Z., Wang, L., Huang, Z., Gong, F., Li, X., et al. (2016). First Clinical Study Using HCV Protease Inhibitor Danoprevir to Treat Naïve and Experienced COVID-19 Patients. doi:10.1101/2020.03.22.20034041
Chen, N., Xia, P., Li, S., Zhang, T., Wang, T. T., and Zhu, J. (2017). RNA Sensors of the Innate Immune System and Their Detection of Pathogens. IUBMB Life 69, 297–304. doi:10.1002/IUB.1625
Chen, R., Lan, Z., Ye, J., Pang, L., Liu, Y., Wu, W., et al. (2021). Cytokine Storm: The Primary Determinant for the Pathophysiological Evolution of COVID-19 Deterioration. Front. Immunol. 12, 1409. doi:10.3389/FIMMU.2021.589095/BIBTEX
Chen, Y., Liu, Q., and Guo, D. (2020b). Emerging Coronaviruses: Genome Structure, Replication, and Pathogenesis. J. Med. Virol. 92, 2249–2423. doi:10.1002/jmv.26234
Cheng, Y. W., Chao, T. L., Li, C. L., Chiu, M. F., Kao, H. C., Wang, S. H., et al. (2020). Furin Inhibitors Block SARS-CoV-2 Spike Protein Cleavage to Suppress Virus Production and Cytopathic Effects. Cell. Rep. 33, 108254. doi:10.1016/J.CELREP.2020.108254
Cheng, Z., Taylor, B., Ourthiague, D. R., and Hoffmann, A. (2015). Distinct Single-Cell Signaling Characteristics Are Conferred by the MyD88 and TRIF Pathways during TLR4 Activation. Sci. Signal. 8. doi:10.1126/SCISIGNAL.AAA5208/SUPPL_FILE/
Choudhury, A., Das, N. C., Patra, R., and Mukherjee, S. (2021). In Silico analyses on the Comparative Sensing of SARS-CoV-2 mRNA by the Intracellular TLRs of Humans. J. Med. Virol. 93, 2476–2486. doi:10.1002/JMV.26776
Choudhury, A., and Mukherjee, S. (2020a). In Silico studies on the Comparative Characterization of the Interactions of SARS-CoV-2 Spike Glycoprotein with ACE-2 Receptor Homologs and Human TLRs. J. Med. Virol. 92, 2105–2113. doi:10.1002/JMV.25987
Choudhury, A., and Mukherjee, S. (2020b). In Silico studies on the Comparative Characterization of the Interactions of SARS-CoV-2 Spike Glycoprotein with ACE-2 Receptor Homologs and Human TLRs. J. Med. Virol. 92, 2105–2113. doi:10.1002/JMV.25987
Conti, P., and Younes, A. (2020). Coronavirus COV-19/sars-CoV-2 Affects Women Less Than Men: Clinical Response to Viral Infection. J. Biol. Regul. Homeost. Agents 34, 339–343. doi:10.23812/EDITORIAL-CONTI-3
Costa, T. J., Potje, S. R., Fraga-Silva, T. F. C., da Silva-Neto, J. A., Barros, P. R., Rodrigues, D., et al. (2022). Mitochondrial DNA and TLR9 Activation Contribute to SARS-CoV-2-Induced Endothelial Cell Damage. Vasc. Pharmacol. 142, 106946. doi:10.1016/J.VPH.2021.106946
Costela-Ruiz, V. J., Illescas-Montes, R., Puerta-Puerta, J. M., Ruiz, C., and Melguizo-Rodríguez, L. (2020). SARS-CoV-2 Infection: The Role of Cytokines in COVID-19 Disease. Cytokine Growth Factor Rev. 54, 62–75. doi:10.1016/J.CYTOGFR.2020.06.001
Crellin, N. K., Garcia, R. V., Hadisfar, O., Allan, S. E., Steiner, T. S., and Levings, M. K. (2005). Human CD4+ T Cells Express TLR5 and its Ligand Flagellin Enhances the Suppressive Capacity and Expression of FOXP3 in CD4+CD25+ T Regulatory Cells. J. Immunol. 175, 8051–8059. doi:10.4049/JIMMUNOL.175.12.8051
da Rosa Mesquita, R., Francelino Silva Junior, L. C., Santos Santana, F. M., Farias de Oliveira, T., Campos Alcântara, R., Monteiro Arnozo, G., et al. (2021). Clinical Manifestations of COVID-19 in the General Population: Systematic Review. Wien. Klin. Wochenschr. 133, 377–382. doi:10.1007/S00508-020-01760-4/TABLES/2
Dai, L., and Gao, G. F. (20202020). Viral Targets for Vaccines against COVID-19. Nat. Rev. Immunol. 21, 73–82. doi:10.1038/s41577-020-00480-0
Dasgupta, S., and Bandyopadhyay, M. (2021). Molecular Docking of SARS-COV-2 Spike Epitope Sequences Identifies Heterodimeric Peptide-Protein Complex Formation with Human Zo-1, TLR8 and Brain Specific Glial Proteins. Med. Hypotheses 157, 110706. doi:10.1016/J.MEHY.2021.110706
Del Re, A., Corpetti, C., Pesce, M., Seguella, L., Steardo, L., Palenca, I., et al. (2021). Ultramicronized Palmitoylethanolamide Inhibits NLRP3 Inflammasome Expression and Pro-inflammatory Response Activated by SARS-CoV-2 Spike Protein in Cultured Murine Alveolar Macrophages. Metabolites 202111, 592. Page 592 11. doi:10.3390/METABO11090592
Del Valle, D. M., Kim-Schulze, S., Huang, H. H., Beckmann, N. D., Nirenberg, S., Wang, B., et al. (20202020). An Inflammatory Cytokine Signature Predicts COVID-19 Severity and Survival. Nat. Med. 26, 1636–1643. doi:10.1038/s41591-020-1051-9
Devarajan, A., and Vaseghi, M. (2021). Hydroxychloroquine Can Potentially Interfere with Immune Function in COVID-19 Patients: Mechanisms and Insights. Redox Biol. 38, 101810. doi:10.1016/J.REDOX.2020.101810
Devi, K. P., Pourkarim, M. R., Thijssen, M., Sureda, A., Khayatkashani, M., Cismaru, C. A., et al. (2022). A Perspective on the Applications of Furin Inhibitors for the Treatment of SARS-CoV-2. Pharmacol. Rep. 74, 425–430. doi:10.1007/S43440-021-00344-X/FIGURES/1
Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S., and Reis E Sousa, C. (2004). Innate Antiviral Responses by Means of TLR7-Mediated Recognition of Single-Stranded RNA. Science 303, 1529–1531. doi:10.1126/SCIENCE.1093616/SUPPL_FILE/DIEBOLD.SOM.PDF
Ding, S., and Liang, T. J. (2020). Is SARS-CoV-2 Also an Enteric Pathogen with Potential Fecal-Oral Transmission? A COVID-19 Virological and Clinical Review. Gastroenterology 159, 53–61. doi:10.1053/j.gastro.2020.04.052
Ehre, C. (2020). SARS-CoV-2 Infection of Airway Cells. N. Engl. J. Med. 383, 969. doi:10.1056/NEJMicm2023328
Englmeier, L., and Subburayalu, J. (2022). What’s Happening where when SARS-CoV-2 Infects: Are TLR7 and MAFB Sufficient to Explain Patient Vulnerability? Immun. Ageing 19, 1–8. doi:10.1186/S12979-022-00262-3/FIGURES/4
Fallerini, C., Daga, S., Mantovani, S., Benetti, E., Picchiotti, N., Francisci, D., et al. (2021). Association of Toll-like Receptor 7 Variants with Life-Threatening COVID-19 Disease in Males: Findings from a Nested Case-Control Study. Elife 10. doi:10.7554/ELIFE.67569
Fara, A., Mitrev, Z., Rosalia, R. A., and Assas, B. M. (2020a). Cytokine Storm and COVID-19: a Chronicle of Pro-inflammatory Cytokines. Open Biol. 10, 200160. doi:10.1098/RSOB.200160
Fara, A., Mitrev, Z., Rosalia, R. A., and Assas, B. M. (2020b). Cytokine Storm and COVID-19: a Chronicle of Pro-inflammatory Cytokines. Open Biol. 10, 200160. doi:10.1098/RSOB.200160
Farhat, K., Riekenberg, S., Heine, H., Debarry, J., Lang, R., Mages, J., et al. (2008). Heterodimerization of TLR2 with TLR1 or TLR6 Expands the Ligand Spectrum but Does Not Lead to Differential Signaling. J. Leukoc. Biol. 83, 692–701. doi:10.1189/JLB.0807586
Fernández, J. J., Mancebo, C., Garcinuño, S., March, G., Alvarez, Y., Alonso, S., et al. (20222022). IRE1α-XBP1 Activation Elicited by Viral Singled Stranded RNA via TLR8 May Modulate Lung Cytokine Induction in SARS-CoV-2 Pneumonia. medRxiv. doi:10.1101/2022.01.26.22269752
Flint, S. J., Enquist, L. W., Racaniello, V. R., and Skalka, A. M. (2009). Principles of Virology: Volume II, 419.Pathogenesis Control
Gankovskaya, L. V., Zinina, E. V., Zinina, E. V., Gankovskii, V. A., and Semushkin, A. R. (2021). Changes in the Expression of Toll-like Receptors, Cytokines and Chemokines in the Cells of the Mucous Membrane of the Oropharynx in Children with COVID-19. Immunologiya 42, 222–231. doi:10.33029/0206-4952-2021-42-3-222-231
Geijtenbeek, T. B., and Gringhuis, S. I. (20092009). Signalling through C-type Lectin Receptors: Shaping Immune Responses. Nat. Rev. Immunol. 9, 465–479. doi:10.1038/nri2569
Gorbalenya, A. E., Baker, S. C., Baric, R. S., de Groot, R. J., Drosten, C., Gulyaeva, A. A., et al. (2020). The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-nCoV and Naming it SARS-CoV-2. Nat. Microbiol. 5, 536–544. doi:10.1038/s41564-020-0695-z
Gupta, A., Madhavan, M. V., Sehgal, K., Nair, N., Mahajan, S., Sehrawat, T. S., et al. (2020). Extrapulmonary Manifestations of COVID-19. Nat. Med. 26, 1017–1032. doi:10.1038/s41591-020-0968-3
Gurung, A. B. (2020). In Silico structure Modelling of SARS-CoV-2 Nsp13 Helicase and Nsp14 and Repurposing of FDA Approved Antiviral Drugs as Dual Inhibitors. Gene Rep. 21, 100860. doi:10.1016/j.genrep.2020.100860
Haabeth, O. A. W., Lohmeyer, J. J. K., Sallets, A., Blake, T. R., Sagiv-Barfi, I., Czerwinski, D. K., et al. (2021). An mRNA SARS-CoV-2 Vaccine Employing Charge-Altering Releasable Transporters with a TLR-9 Agonist Induces Neutralizing Antibodies and T Cell Memory. ACS Cent. Sci. 7, 1191–1204. doi:10.1021/ACSCENTSCI.1C00361/SUPPL_FILE/OC1C00361_SI_001
Hadziyannis, S. J., Sette, H., Morgan, T. R., Balan, V., Diago, M., Marcellin, P., et al. (2004). Peginterferon-alpha2a and Ribavirin Combination Therapy in Chronic Hepatitis C: a Randomized Study of Treatment Duration and Ribavirin Dose. Ann. Intern. Med. 140, 346–355. doi:10.7326/0003-4819-140-5-200403020-00010
Han, L., Zhuang, M. W., Deng, J., Zheng, Y., Zhang, J., Nan, M. L., et al. (2021). SARS-CoV-2 ORF9b Antagonizes Type I and III Interferons by Targeting Multiple Components of the RIG-I/MDA-5-MAVS, TLR3-TRIF, and cGAS-STING Signaling Pathways. J. Med. Virol. 93, 5376–5389. doi:10.1002/JMV.27050
Hansen, J. D., Vojtech, L. N., and Laing, K. J. (2011). Sensing Disease and Danger: A Survey of Vertebrate PRRs and Their Origins. Dev. Comp. Immunol. 35, 886–897. doi:10.1016/J.DCI.2011.01.008
Hasan, U., Chaffois, C., Gaillard, C., Saulnier, V., Merck, E., Tancredi, S., et al. (2005). Human TLR10 Is a Functional Receptor, Expressed by B Cells and Plasmacytoid Dendritic Cells, Which Activates Gene Transcription through MyD88. J. Immunol. 174, 2942–2950. doi:10.4049/JIMMUNOL.174.5.2942
He, Y., Hara, H., and Núñez, G. (2016). Mechanism and Regulation of NLRP3 Inflammasome Activation. Trends biochem. Sci. 41, 1012–1021. doi:10.1016/J.TIBS.2016.09.002
Heads, D. R. (2020). COVID-19 and Toll-like Receptor 4 (TLR4): SARS-CoV-2 May Activate TLR4 to Increase ACE2 Expression for Entry and Cause Abnormal Inflammatory Signalling: A Cardio-Respiratory Focus. Authorea Prepr. doi:10.22541/AU.160010865.58124057
Henrick, B. M., Yao, X. D., Zahoor, M. A., Abimiku, A., Osawe, S., and Rosenthal, K. L. (2019). TLR10 Senses HIV-1 Proteins and Significantly Enhances HIV-1 Infection. Front. Immunol. 10, 482. doi:10.3389/FIMMU.2019.00482/BIBTEX
Herrera-Esposito, D., and de los Campos, G. (2022). Age-specific Rate of Severe and Critical SARS-CoV-2 Infections Estimated with Multi-Country Seroprevalence Studies. BMC Infect. Dis. 22, 311–314. doi:10.1186/S12879-022-07262-0/TABLES/4
Hillen, H. S., Kokic, G., Farnung, L., Dienemann, C., Tegunov, D., and Cramer, P. (2020). Structure of Replicating SARS-CoV-2 Polymerase. Nature 584, 154–156. doi:10.1038/s41586-020-2368-8
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 181, 271–e8. e8. doi:10.1016/J.CELL.2020.02.052
Hörnich, B. F., Großkopf, A. K., Schlagowski, S., Tenbusch, M., Kleine-Weber, H., Neipel, F., et al. (2021). SARS-CoV-2 and SARS-CoV Spike-Mediated Cell-Cell Fusion Differ in Their Requirements for Receptor Expression and Proteolytic Activation. J. Virol. 95, 2–21. doi:10.1128/JVI.00002-21
Hsu, J. C. C., Laurent-Rolle, M., Pawlak, J. B., Wilen, C. B., and Cresswell, P. (2021). Translational Shutdown and Evasion of the Innate Immune Response by SARS-CoV-2 NSP14 Protein. Proc. Natl. Acad. Sci. U. S. A. 118. doi:10.1073/PNAS.2101161118/SUPPL_FILE/PNAS.2101161118.SAPP.PDF
Hu, X., Cai, X., Song, X., Li, C., Zhao, J., Luo, W., et al. (2020). Possible SARS-Coronavirus 2 Inhibitor Revealed by Simulated Molecular Docking to Viral Main Protease and Host Toll-like Receptor. Future Virol. 15, 359–368. doi:10.2217/FVL-2020-0099/ASSET/IMAGES/LARGE/FIGURE5.JPEG
Huh, J. W., Shibata, T., Hwang, M., Kwon, E. H., Jang, M. S., Fukui, R., et al. (2014). UNC93B1 Is Essential for the Plasma Membrane Localization and Signaling of Toll-like Receptor 5. Proc. Natl. Acad. Sci. U. S. A. 111, 7072–7077. doi:10.1073/PNAS.1322838111/-/DCSUPPLEMENTAL
Huleatt, J. W., Nakaar, V., Desai, P., Huang, Y., Hewitt, D., Jacobs, A., et al. (2008). Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine 26, 201–214. doi:10.1016/J.VACCINE.2007.10.062
Humphries, F., Shmuel-Galia, L., Jiang, Z., Wilson, R., Landis, P., Ng, S.-L., et al. (2021). A Diamidobenzimidazole STING Agonist Protects against SARS-CoV-2 Infection. Sci. Immunol. 6. doi:10.1126/SCIIMMUNOL.ABI9002
Irvine, K. L., Hopkins, L. J., Gangloff, M., and Bryant, C. E. (2013). The Molecular Basis for Recognition of Bacterial Ligands at Equine TLR2, TLR1 and TLR6. Vet. Res. 44, 50–11. doi:10.1186/1297-9716-44-50/FIGURES/5
Ishii, N., Funami, K., Tatematsu, M., Seya, T., and Matsumoto, M. (2014). Endosomal Localization of TLR8 Confers Distinctive Proteolytic Processing on Human Myeloid Cells. J. Immunol. 193, 5118–5128. doi:10.4049/JIMMUNOL.1401375/-/DCSUPPLEMENTAL
Ishikawa, H., and Barber, G. N. (20082008). STING Is an Endoplasmic Reticulum Adaptor that Facilitates Innate Immune Signalling. Nature 455, 674–678. doi:10.1038/nature07317
Jackson, C. B., Farzan, M., Chen, B., and Choe, H. (2021a2021). Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell. Biol. 23, 3–20. doi:10.1038/s41580-021-00418-x
Jackson, C. B., Farzan, M., Chen, B., and Choe, H. (2021b2021). Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell. Biol. 23, 3–20. doi:10.1038/s41580-021-00418-x
Jiang, H. W., Zhang, H. N., Meng, Q. F., Xie, J., Li, Y., Chen, H., et al. (2020). SARS-CoV-2 Orf9b Suppresses Type I Interferon Responses by Targeting TOM70. Cell. Mol. Immunol. 17, 998–1000. doi:10.1038/s41423-020-0514-8
Jin, C., Flavell, R. A., Jin, C., and Flavell, R. A. (20102010). Molecular Mechanism of NLRP3 Inflammasome Activation. J. Clin. Immunol. 30, 628–631. doi:10.1007/S10875-010-9440-3
Jukič, M., Janežič, D., and Bren, U. (2021). Potential Novel Thioether-Amide or Guanidine-Linker Class of SARS-CoV-2 Virus RNA-dependent RNA Polymerase Inhibitors Identified by High-Throughput Virtual Screening Coupled to Free-Energy Calculations. Int. J. Mol. Sci. 22. doi:10.3390/IJMS222011143
Junqueira, C., Crespo, Â., Ranjbar, S., Lewandrowski, M., Ingber, J., de Lacerda, L. B., et al. (2021). SARS-CoV-2 Infects Blood Monocytes to Activate NLRP3 and AIM2 Inflammasomes, Pyroptosis and Cytokine Release. Res. Sq. doi:10.21203/RS.3.RS-153628/V1
Kamitani, W., Narayanan, K., Huang, C., Lokugamage, K., Ikegami, T., Ito, N., et al. (2006). Severe Acute Respiratory Syndrome Coronavirus Nsp1 Protein Suppresses Host Gene Expression by Promoting Host mRNA Degradation. Proc. Natl. Acad. Sci. U. S. A. 103, 12885–12890. doi:10.1073/pnas.0603144103
Kawai, T., and Akira, S. (2007). Antiviral Signaling through Pattern Recognition Receptors. J. Biochem. 141, 137–145. doi:10.1093/JB/MVM032
Kawai, T., and Akira, S. (20102010). The Role of Pattern-Recognition Receptors in Innate Immunity: Update on Toll-like Receptors. Nat. Immunol. 11 (11), 373–384. doi:10.1038/ni.1863
Kawasaki, T., and Kawai, T. (2014). Toll-like Receptor Signaling Pathways. Front. Immunol. 5, 461. doi:10.3389/FIMMU.2014.00461/BIBTEX
Kerrigan, A. M., and Brown, G. D. (2010). Syk-coupled C-type Lectin Receptors that Mediate Cellular Activation via Single Tyrosine Based Activation Motifs. Immunol. Rev. 234, 335–352. doi:10.1111/J.0105-2896.2009.00882.X
Khan, S., Shafiei, M. S., Longoria, C., Schoggins, J. W., Savani, R. C., and Zaki, H. (2021). SARS-CoV-2 Spike Protein Induces Inflammation via TLR2-dependent Activation of the NF-Κb Pathway. Elife 10. doi:10.7554/eLife.68563
Kim, J. S., Lee, J. Y., Yang, J. W., Lee, K. H., Effenberger, M., Szpirt, W., et al. (2021a). Immunopathogenesis and Treatment of Cytokine Storm in COVID-19. Theranostics 11, 316–329. doi:10.7150/THNO.49713
Kim, N.-E., Kim, D.-K., and Song, Y.-J. (2021b2021). SARS-CoV-2 Nonstructural Proteins 1 and 13 Suppress Caspase-1 and the NLRP3 Inflammasome Activation. Microorganisms 9, 494. doi:10.3390/MICROORGANISMS9030494
Kondo, Y., Larabee, J. L., Gao, L., Shi, H., Shao, B., Hoover, C. M., et al. (2021). L-SIGN Is a Receptor on Liver Sinusoidal Endothelial Cells for SARS-CoV-2 Virus. JCI Insight 6. doi:10.1172/JCI.INSIGHT.148999
Konno, H., Yamamoto, T., Yamazaki, K., Gohda, J., Akiyama, T., Semba, K., et al. (2009). TRAF6 Establishes Innate Immune Responses by Activating NF-kappaB and IRF7 upon Sensing Cytosolic Viral RNA and DNA. PLoS One 4, e5674. doi:10.1371/JOURNAL.PONE.0005674
Konno, Y., Kimura, I., Uriu, K., Fukushi, M., Irie, T., Koyanagi, Y., et al. (2020). SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant. Cell. Rep. 32, 108185. doi:10.1016/j.celrep.2020.108185
Kucia, M., Bujko, K., Ciechanowicz, A., Cymer, M., Sielatycka, K., Ratajczak, J., et al. (2020). The ACE2 Receptor for COVID-19 Entry Is Expressed on the Surface of Hematopoietic Stem/Progenitor Cells and Endothelial Progenitors as Well as Their Precursor Cells and Becomes Activated in Nlrp3 Inflammasome-dependent Manner by Virus Spike Protein - a Potential Pathway Leading to a "Cytokine Storm". Blood 136, 8. doi:10.1182/BLOOD-2020-137083
Kumar, A., Ishida, R., Strilets, T., Cole, J., Lopez-Orozco, J., Fayad, N., et al. (2021). SARS-CoV-2 Nonstructural Protein 1 Inhibits the Interferon Response by Causing Depletion of Key Host Signaling Factors. J. Virol. 95. doi:10.1128/JVI.00266-21/ASSET/0F4B9D93-FCCC-415C-9A34-B4D699001681/ASSETS/IMAGES/LARGE/JVI.00266-21-F0008.JPG
Kuo, T. Y., Lin, M. Y., Coffman, R. L., Campbell, J. D., Traquina, P., Lin, Y. J., et al. (2020). Development of CpG-Adjuvanted Stable Prefusion SARS-CoV-2 Spike Antigen as a Subunit Vaccine against COVID-19. Sci. Rep. 10, 20085–20110. doi:10.1038/s41598-020-77077-z
Lacy, P. (2015). Editorial: Secretion of Cytokines and Chemokines by Innate Immune Cells. Front. Immunol. 6, 1. doi:10.3389/FIMMU.2015.00190
Lässig, C., and Hopfner, K.-P. (2017). Discrimination of Cytosolic Self and Non-self RNA by RIG-I-like Receptors. J. Biol. Chem. 292, 9000–9009. doi:10.1074/JBC.R117.788398
Lee, B. L., Moon, J. E., Shu, J. H., Yuan, L., Newman, Z. R., Schekman, R., et al. (20132013). UNC93B1 Mediates Differential Trafficking of Endosomal TLRs. Elife 2. doi:10.7554/ELIFE.00291
Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J. M., and Hoffmann, J. A. (1996). The Dorsoventral Regulatory Gene Cassette spätzle/Toll/cactus Controls the Potent Antifungal Response in Drosophila Adults. Cell. 86, 973–983. doi:10.1016/S0092-8674(00)80172-5
Lempp, F. A., Soriaga, L. B., Montiel-Ruiz, M., Benigni, F., Noack, J., Park, Y.-J., et al. (2021). Lectins Enhance SARS-CoV-2 Infection and Influence Neutralizing Antibodies. Nature 598, 342–347. doi:10.1038/s41586-021-03925-1
Leonard, J. N., Ghirlando, R., Askins, J., Bell, J. K., Margulies, D. H., Davies, D. R., et al. (2008). The TLR3 Signaling Complex Forms by Cooperative Receptor Dimerization. Proc. Natl. Acad. Sci. U. S. A. 105, 258–263. doi:10.1073/PNAS.0710779105
Leung, D. W., and Amarasinghe, G. K. (2012). Structural Insights into RNA Recognition and Activation of RIG-I-like Receptors. Curr. Opin. Struct. Biol. 22, 297–303. doi:10.1016/J.SBI.2012.03.011
Li, D., and Wu, M. (20212021). Pattern Recognition Receptors in Health and Diseases. Sig Transduct. Target Ther. 6, 1–24. doi:10.1038/s41392-021-00687-0
Li, H., Yang, L., Liu, F. F., Ma, X. N., He, P. L., Tang, W., et al. (2020a). Overview of Therapeutic Drug Research for COVID-19 in China. Acta Pharmacol. Sin. 41, 1133–1140. doi:10.1038/s41401-020-0438-y
Li, J. Y., Liao, C. H., Wang, Q., Tan, Y. J., Luo, R., Qiu, Y., et al. (2020b). The ORF6, ORF8 and Nucleocapsid Proteins of SARS-CoV-2 Inhibit Type I Interferon Signaling Pathway. Virus Res. 286, 198074. doi:10.1016/J.VIRUSRES.2020.198074
Li, M., Ferretti, M., Ying, B., Descamps, H., Lee, E., Dittmar, M., et al. (2021). Pharmacological Activation of STING Blocks SARS-CoV-2 Infection. Sci. Immunol. 6. doi:10.1126/SCIIMMUNOL.ABI9007
Li, X., Song, Y., Wong, G., and Cui, J. (2020c). Bat Origin of a New Human Coronavirus: There and Back Again. Sci. China Life Sci. 63, 461–462. doi:10.1007/s11427-020-1645-7
Liskova, A., Samec, M., Koklesova, L., Samuel, S. M., Zhai, K., Al-Ishaq, R. K., et al. (2021). Flavonoids against the SARS-CoV-2 Induced Inflammatory Storm. Biomed. Pharmacother. 138, 111430. doi:10.1016/j.biopha.2021.111430
Liu, C., Zhu, X., Lu, Y., Zhang, X., Jia, X., and Yang, T. (2021a). Potential Treatment with Chinese and Western Medicine Targeting NSP14 of SARS-CoV-2. J. Pharm. Anal. 11, 272–277. doi:10.1016/J.JPHA.2020.08.002
Liu, G., Lee, J.-H., Parker, Z. M., Acharya, D., Chiang, J. J., van Gent, M., et al. (2021b2021). ISG15-dependent Activation of the Sensor MDA5 Is Antagonized by the SARS-CoV-2 Papain-like Protease to Evade Host Innate Immunity. Nat. Microbiol. 6 (6), 467–478. doi:10.1038/s41564-021-00884-1
Long, Y., Sun, J., Song, T.-Z., Liu, T., Tang, F., Zhang, X., et al. (2022). CoVac501, a Self-Adjuvanting Peptide Vaccine Conjugated with TLR7 Agonists, against SARS-CoV-2 Induces Protective Immunity. Cell. Discov. 8 (8), 1–16. doi:10.1038/s41421-021-00370-2
Lu, Q., Liu, J., Zhao, S., Gomez Castro, M. F., Laurent-Rolle, M., Dong, J., et al. (2021). SARS-CoV-2 Exacerbates Proinflammatory Responses in Myeloid Cells through C-type Lectin Receptors and Tweety Family Member 2. Immunity 54, 1304–e9. e9. doi:10.1016/J.IMMUNI.2021.05.006
Luan, J., Lu, Y., Jin, X., and Zhang, L. (2020). Spike Protein Recognition of Mammalian ACE2 Predicts the Host Range and an Optimized ACE2 for SARS-CoV-2 Infection. Biochem. Biophys. Res. Commun. doi:10.1016/j.bbrc.2020.03.047
Ludwig, S., and Zarbock, A. (2020). Coronaviruses and SARS-CoV-2: A Brief Overview. Anesth. Analg. 131, 93–96. doi:10.1213/ANE.0000000000004845
Luwei, C. W. L., Liu, X. F., and Jia, Z. F. (2020). 2019-nCoV Transmission through the Ocular Surface Must Not Be Ignored. Lancet 395, e39. doi:10.1016/S0140-6736(20)30313-5
Ma, J., Zhu, F., Zhao, M., Shao, F., Yu, D., Ma, J., et al. (2021). SARS-CoV-2 Nucleocapsid Suppresses Host Pyroptosis by Blocking Gasdermin D Cleavage. EMBO J. 40, e108249. doi:10.15252/EMBJ.2021108249
Maeda, K., and Akira, S. (2016). TLR7 Structure: Cut in Z-Loop. Immunity 45, 705–707. doi:10.1016/J.IMMUNI.2016.10.003
Manik, M., Singh, R. K., Singh, K., and Singh, R. K. (2022). Role of Toll-like Receptors in Modulation of Cytokine Storm Signaling in SARS-CoV-2-Induced COVID-19. J. Med. Virol. 94, 869–877. doi:10.1002/JMV.27405
Marchetti, C., Mould, K., Tengesdal, I. W., Janssen, W. J., and Dinarello, C. A. (2021). Targeting of the NLRP3 Inflammasome for Early COVID-19. Cold Spring Harbor, New York: bioRxiv, 432734. doi:10.1101/2021.02.24.432734
Matsumoto, M., and Seya, T. (2008). TLR3: Interferon Induction by Double-Stranded RNA Including poly(I:C). Adv. Drug Deliv. Rev. 60, 805–812. doi:10.1016/J.ADDR.2007.11.005
McGettrick, A. F., and O'Neill, L. A. (2010). Localisation and Trafficking of Toll-like Receptors: an Important Mode of Regulation. Curr. Opin. Immunol. 22, 20–27. doi:10.1016/J.COI.2009.12.002
Mendonça, L., Howe, A., Gilchrist, J. B., Sheng, Y., Sun, D., Knight, M. L., et al. (20212021). Correlative Multi-Scale Cryo-Imaging Unveils SARS-CoV-2 Assembly and Egress. Nat. Commun. 12, 1–10. doi:10.1038/s41467-021-24887-y
Mirza, M. U., and Froeyen, M. (2020). Structural Elucidation of SARS-CoV-2 Vital Proteins: Computational Methods Reveal Potential Drug Candidates against Main Protease, Nsp12 Polymerase and Nsp13 Helicase. J. Pharm. Anal. 10, 320–328. doi:10.1016/j.jpha.2020.04.008
Modrow, S., Falke, D., Truyen, U., and Schätzl, H. (2013). Molecular Virology. Springer-Verlag Berlin Heidelberg. doi:10.1007/978-3-642-20718-1
Moen, S. H., Ehrnström, B., Kojen, J. F., Yurchenko, M., Beckwith, K. S., Afset, J. E., et al. (2019). Human Toll-like Receptor 8 (TLR8) Is an Important Sensor of Pyogenic Bacteria, and Is Attenuated by Cell Surface TLR Signaling. Front. Immunol. 10, 1209. doi:10.3389/FIMMU.2019.01209/BIBTEX
Mohanty, M. C., Varose, S. Y., Sawant, U. P., and Fernandes, M. M. (2021). Expression of Innate Immune Response Genes in Upper Airway Samples of SARS-CoV-2 Infected Patients: A Preliminary Study. Indian J. Med. Res. 153, 677–683. doi:10.4103/IJMR.IJMR_131_21
Monteiro, J. T., and Lepenies, B. (2017). Myeloid C-type Lectin Receptors in Viral Recognition and Antiviral Immunity. Viruses 9. doi:10.3390/V9030059
Moreno-Eutimio, M. A., López-Macías, C., and Pastelin-Palacios, R. (2020). Bioinformatic Analysis and Identification of Single-Stranded RNA Sequences Recognized by TLR7/8 in the SARS-CoV-2, SARS-CoV, and MERS-CoV Genomes. Microbes Infect. 22, 226–229. doi:10.1016/J.MICINF.2020.04.009
Moura, I. B., Buckley, A. M., and Wilcox, M. H. (2022). Can SARS-CoV-2 Be Transmitted via Faeces? Curr. Opin. Gastroenterol. 38, 26–29. doi:10.1097/MOG.0000000000000794
Muñoz-Wolf, N., and Lavelle, E. C. (2016). Innate Immune Receptors. Methods Mol. Biol. 1417, 1–43. doi:10.1007/978-1-4939-3566-6_1
Nanishi, E., Borriello, F., O’Meara, T. R., McGrath, M. E., Saito, Y., Haupt, R. E., et al. (2021). An Aluminum hydroxide:CpG Adjuvant Enhances Protection Elicited by a SARS-CoV-2 Receptor-Binding Domain Vaccine in Aged Mice. Sci. Transl. Med. 14, eabj5305. doi:10.1126/SCITRANSLMED.ABJ5305/SUPPL_FILE/SCITRANSLMED.ABJ5305_MDAR_REPRODUCIBILITY_CHECKLIST.ZIP
Narayanan, A., Narwal, M., Majowicz, S. A., Varricchio, C., Toner, S. A., Ballatore, C., et al. (20222022). Identification of SARS-CoV-2 Inhibitors Targeting Mpro and PLpro Using In-Cell-Protease Assay. Commun. Biol. 5 (5), 1–17. doi:10.1038/s42003-022-03090-9
Neufeldt, C. J., Cerikan, B., Cortese, M., Frankish, J., Lee, J.-Y., Plociennikowska, A., et al. (20222022). SARS-CoV-2 Infection Induces a Pro-inflammatory Cytokine Response through cGAS-STING and NF-Κb. Commun. Biol. 5 (5), 1–15. doi:10.1038/s42003-021-02983-5
Nieva, J. L., Madan, V., and Carrasco, L. (20122012). Viroporins: Structure and Biological Functions. Nat. Rev. Microbiol. 10 (10), 563–574. doi:10.1038/nrmicro2820
Nukoolkarn, V., Lee, V. S., Malaisree, M., Aruksakulwong, O., and Hannongbua, S. (2008). Molecular Dynamic Simulations Analysis of Ritonavir and Lopinavir as SARS-CoV 3CL(pro) Inhibitors. J. Theor. Biol. 254, 861–867. doi:10.1016/j.jtbi.2008.07.030
Oberemok, V. V., Laikova, K. V., Yurchenko, K. A., Marochkin, N. A., Fomochkina, I. I., and Kubyshkin, A. V. (2020). SARS-CoV-2 Will Constantly Sweep its Tracks: a Vaccine Containing CpG Motifs in 'lasso' for the Multi-Faced Virus. Inflamm. Res. 69, 801–812. doi:10.1007/S00011-020-01377-3/TABLES/2
Oh, S. J., and Shin, O. S. (20212021). SARS-CoV-2 Nucleocapsid Protein Targets RIG-I-like Receptor Pathways to Inhibit the Induction of Interferon Response. Cells 10, 530. doi:10.3390/CELLS10030530
Olajide, O. A., Iwuanyanwu, V. U., Adegbola, O. D., and Al-Hindawi, A. A. (2022). SARS-CoV-2 Spike Glycoprotein S1 Induces Neuroinflammation in BV-2 Microglia. Mol. Neurobiol. 59, 445–458. doi:10.1007/S12035-021-02593-6/FIGURES/9
Oran, D. P., and Topol, E. J. (2020). Prevalence of Asymptomatic SARS-CoV-2 Infection : A Narrative Review. Ann. Intern. Med. 173, 362–367. doi:10.7326/M20-3012
Pan, P., Shen, M., Yu, Z., Ge, W., Chen, K., Tian, M., et al. (20212021). SARS-CoV-2 N Protein Promotes NLRP3 Inflammasome Activation to Induce Hyperinflammation. Nat. Commun. 12, 1–17. doi:10.1038/s41467-021-25015-6
Pandey, P., Rane, J. S., Chatterjee, A., Kumar, A., Khan, R., Prakash, A., et al. (2020). Targeting SARS-CoV-2 Spike Protein of COVID-19 with Naturally Occurring Phytochemicals: an In Silico Study for Drug Development. J. Biomol. Struct. Dyn. 1–11, 1–11. doi:10.1080/07391102.2020.1796811
Pandey, S. P., Chandel, H. S., Srivastava, S., Selvaraj, S., Jha, M. K., Shukla, D., et al. (2014). Pegylated Bisacycloxypropylcysteine, a Diacylated Lipopeptide Ligand of TLR6, Plays a Host-Protective Role against Experimental Leishmania Major Infection. J. Immunol. 193, 3632–3643. doi:10.4049/JIMMUNOL.1400672
Pardo, J., Shukla, A. M., Chamarthi, G., and Gupte, A. (2020). The Journey of Remdesivir: from Ebola to COVID-19. Drugs Context 9. doi:10.7573/dic.2020-4-14
Pelka, K., Shibata, T., Miyake, K., and Latz, E. (2016). Nucleic Acid-Sensing TLRs and Autoimmunity: Novel Insights from Structural and Cell Biology. Immunol. Rev. 269, 60–75. doi:10.1111/IMR.12375
Planès, R., Bert, J.-B., Tairi, S., Benmohamed, L., and Bahraoui, E. (2021). SARS-CoV-2 Envelope Protein (E) Binds and Activates TLR2: A Novel Target for COVID-19 Interventions. Cold Spring Harbor, New York: bioRxiv. doi:10.1101/2021.11.10.468173
Poduri, R., Joshi, G., and Jagadeesh, G. (2020). Drugs Targeting Various Stages of the SARS-CoV-2 Life Cycle: Exploring Promising Drugs for the Treatment of Covid-19. Cell. Signal. 74, 109721. doi:10.1016/j.cellsig.2020.109721
Poletti, P., Tirani, M., Cereda, D., Trentini, F., Guzzetta, G., Sabatino, G., et al. (2021). Association of Age with Likelihood of Developing Symptoms and Critical Disease Among Close Contacts Exposed to Patients with Confirmed SARS-CoV-2 Infection in Italy. JAMA Netw. Open 4, e211085. doi:10.1001/JAMANETWORKOPEN.2021.1085
Port, J. R., Yinda, C. K., Avanzato, V. A., Schulz, J. E., Holbrook, M. G., van Doremalen, N., et al. (20222022). Increased Small Particle Aerosol Transmission of B.1.1.7 Compared with SARS-CoV-2 Lineage A In Vivo. Nat. Microbiol. 7 (7), 213–223. doi:10.1038/s41564-021-01047-y
Poulas, K., Farsalinos, K., and Zanidis, C. (2020). Activation of TLR7 and Innate Immunity as an Efficient Method against COVID-19 Pandemic: Imiquimod as a Potential Therapy. Front. Immunol. 11, 1373. doi:10.3389/FIMMU.2020.01373/BIBTEX
Proud, P. C., Tsitoura, D., Watson, R. J., Chua, B. Y., Aram, M. J., Bewley, K. R., et al. (2021). Prophylactic Intranasal Administration of a TLR2/6 Agonist Reduces Upper Respiratory Tract Viral Shedding in a SARS-CoV-2 Challenge Ferret Model. eBioMedicine 63, 103153. doi:10.1016/J.EBIOM.2020.103153
Rabaan, A. A., Al-Ahmed, S. H., Muhammad, J., Khan, A., Sule, A. A., Tirupathi, R., et al. (2021). Role of Inflammatory Cytokines in COVID-19 Patients: A Review on Molecular Mechanisms, Immune Functions, Immunopathology and Immunomodulatory Drugs to Counter Cytokine Storm. Vaccines 9, 436. doi:10.3390/VACCINES9050436
Raghu, G., and Wilson, K. C. (2020). COVID-19 Interstitial Pneumonia: Monitoring the Clinical Course in Survivors. doi:10.1016/s2213-2600(20)30349-0
Rahimi, N. (20202021). C-Type Lectin CD209L/L-SIGN and CD209/DC-SIGN: Cell Adhesion Molecules Turned to Pathogen Recognition Receptors. Biology 10, 1. doi:10.3390/BIOLOGY10010001
Ratajczak, M. Z., and Kucia, M. (2020). SARS-CoV-2 Infection and Overactivation of Nlrp3 Inflammasome as a Trigger of Cytokine "storm" and Risk Factor for Damage of Hematopoietic Stem Cells. Leukemia 34, 1726–1729. doi:10.1038/s41375-020-0887-9
Rebendenne, A., Chaves Valadão, A. L., Tauziet, M., Maarifi, G., Bonaventure, B., McKellar, J., et al. (2021). SARS-CoV-2 Triggers an MDA-5-dependent Interferon Response Which Is Unable to Control Replication in Lung Epithelial Cells. J. Virol. 95. doi:10.1128/JVI.02415-20/ASSET/6D37141D-124F-4749-A48E-1968D7E81198/ASSETS/20-F0009.JPG
Redondo, N., Zaldívar-López, S., Garrido, J. J., and Montoya, M. (2021). SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns. Front. Immunol. 12, 2698. doi:10.3389/FIMMU.2021.708264/BIBTEX
Rehwinkel, J., and Gack, M. U. (2020). RIG-I-like Receptors: Their Regulation and Roles in RNA Sensing. Nat. Rev. Immunol. 20, 537–551. doi:10.1038/S41577-020-0288-3
Ren, H., Ma, C., Peng, H., Zhang, B., Zhou, L., Su, Y., et al. (2021). Micronucleus Production, Activation of DNA Damage Response and cGAS-STING Signaling in Syncytia Induced by SARS-CoV-2 Infection. Biol. Direct 16, 20–10. doi:10.1186/S13062-021-00305-7/FIGURES/4
Romano, M., Ruggiero, A., Squeglia, F., Maga, G., and Berisio, R. (2020). A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells 9, 1267. doi:10.3390/cells9051267
Rousseau, K. E., Figueroa, A., Massaccesi, G., and Chattergoon, M. . (2021). SARS-CoV-2 ORF3A Activates the Nlrp3 Inflammasome | Topics in Antiviral Medicine; 29(1):71, 2021. | EMBASE. Top. Antivir. Med., 71.
Rui, Y., Su, J., Shen, S., Hu, Y., Huang, D., Zheng, W., et al. (20212021). Unique and Complementary Suppression of cGAS-STING and RNA Sensing- Triggered Innate Immune Responses by SARS-CoV-2 Proteins. Sig Transduct. Target Ther. 6, 1–11. doi:10.1038/s41392-021-00515-5
Sa Ribero, M., Jouvenet, N., Dreux, M., and Nisole, S. (2020). Interplay between SARS-CoV-2 and the Type I Interferon Response. PLoS Pathog. 16, e1008737. doi:10.1371/journal.ppat.1008737
Salvi, V., Nguyen, H. O., Sozio, F., Schioppa, T., Gaudenzi, C., Laffranchi, M., et al. (2021). SARS-CoV-2-associated ssRNAs Activate Inflammation and Immunity via TLR7/8. JCI Insight 6. doi:10.1172/JCI.INSIGHT.150542
Santa Cruz, A., Mendes-Frias, A., Oliveira, A. I., Dias, L., Matos, A. R., Carvalho, A., et al. (2021). Interleukin-6 Is a Biomarker for the Development of Fatal Severe Acute Respiratory Syndrome Coronavirus 2 Pneumonia. Front. Immunol. 12, 263. doi:10.3389/FIMMU.2021.613422/BIBTEX
Sariol, A., and Perlman, S. (2021). SARS-CoV-2 Takes its Toll. Nat. Immunol. 22, 801–802. doi:10.1038/s41590-021-00962-w
Schlee, M., and Hartmann, G. (20162016). Discriminating Self from Non-self in Nucleic Acid Sensing. Nat. Rev. Immunol. 16, 566–580. doi:10.1038/nri.2016.78
Selvaraj, C., Dinesh, D. C., Panwar, U., Abhirami, R., Boura, E., and Singh, S. K. (2021). Structure-based Virtual Screening and Molecular Dynamics Simulation of SARS-CoV-2 Guanine-N7 Methyltransferase (Nsp14) for Identifying Antiviral Inhibitors against COVID-19. J. Biomol. Struct. Dyn. 39, 4582–4593. doi:10.1080/07391102.2020.1778535/SUPPL_FILE/TBSD_A_1778535_SM4594.DOCX
Severe Acute Respiratory Syndrome (2021). Health-Topics. Available at: https://www.who.int/health-topics/severe-acute-respiratory-syndrome#tab=tab_1 (Accessed March 4, 2022).
Shang, J., Ye, G., Shi, K., Wan, Y., Luo, C., Aihara, H., et al. (2020a). Structural Basis of Receptor Recognition by SARS-CoV-2. Nature 581, 221–224. doi:10.1038/s41586-020-2179-y
Shang, J., Ye, G., Shi, K., Wan, Y., Luo, C., Aihara, H., et al. (2020b). Structural Basis of Receptor Recognition by SARS-CoV-2. Nature 581, 221–224. doi:10.1038/s41586-020-2179-y
Sheahan, T. P., Sims, A. C., Zhou, S., Graham, R. L., Pruijssers, A. J., Agostini, M. L., et al. (2020). An Orally Bioavailable Broad-Spectrum Antiviral Inhibits SARS-CoV-2 in Human Airway Epithelial Cell Cultures and Multiple Coronaviruses in Mice. Sci. Transl. Med. 12. doi:10.1126/scitranslmed.abb5883
Shimabukuro-Vornhagen, A., Gödel, P., Subklewe, M., Stemmler, H. J., Schlößer, H. A., Schlaak, M., et al. (2018). Cytokine Release Syndrome. J. Immunother. Cancer 6, 56. doi:10.1186/S40425-018-0343-9
Shimizu, T. (2017). Structural Insights into Ligand Recognition and Regulation of Nucleic Acid-Sensing Toll-like Receptors. Curr. Opin. Struct. Biol. 47, 52–59. doi:10.1016/J.SBI.2017.05.010
Shinkai, H., Tanaka, M., Morozumi, T., Eguchi-Ogawa, T., Okumura, N., Muneta, Y., et al. (2006). Biased Distribution of Single Nucleotide Polymorphisms (SNPs) in Porcine Toll-like Receptor 1 (TLR1), TLR2, TLR4, TLR5, and TLR6 Genes. Immunogenetics 58, 324–330. doi:10.1007/S00251-005-0068-Z/FIGURES/4
Shu, T., Huang, M., Wu, D., Ren, Y., Zhang, X., Han, Y., et al. (2020). SARS-Coronavirus-2 Nsp13 Possesses NTPase and RNA Helicase Activities that Can Be Inhibited by Bismuth Salts. Virol. Sin. 35, 321–329. doi:10.1007/s12250-020-00242-1
Sohn, K. M., Lee, S. G., Kim, H. J., Cheon, S., Jeong, H., Lee, J., et al. (2020). COVID-19 Patients Upregulate Toll-like Receptor 4-mediated Inflammatory Signaling that Mimics Bacterial Sepsis. J. Korean Med. Sci. 35, e343. doi:10.3346/JKMS.2020.35.E343
Spiering, A. E., and de Vries, T. J. (2021). Why Females Do Better: The X Chromosomal TLR7 Gene-Dose Effect in COVID-19. Front. Immunol. 12, 756262. doi:10.3389/FIMMU.2021.756262
Sun, X., Liu, Y., Huang, Z., Xu, W., Hu, W., Yi, L., et al. (20222021). SARS-CoV-2 Non-structural Protein 6 Triggers NLRP3-dependent Pyroptosis by Targeting ATP6AP1. Cell. Death Differ. 29, 1240–1254. doi:10.1038/s41418-021-00916-7
Sun, X., Wang, T., Cai, D., Hu, Z., Chen, J., Liao, H., et al. (2020). Cytokine Storm Intervention in the Early Stages of COVID-19 Pneumonia. Cytokine Growth Factor Rev. 53, 38–42. doi:10.1016/J.CYTOGFR.2020.04.002
Sun, Y., and Reddy, P. (2013). Intracellular Sensors of Immunity and Allogeneic Hematopoietic Stem Cell Transplantation. Immune Biol. Allogeneic hematop. Stem Cell. Transpl., 425–447. doi:10.1016/B978-0-12-416004-0.00018-5
Takeda, K., and Akira, S. (2005). Toll-like Receptors in Innate Immunity. Int. Immunol. 17, 1–14. doi:10.1093/INTIMM/DXH186
Tamir, H., Melamed, S., Erez, N., Politi, B., Yahalom-Ronen, Y., Achdout, H., et al. (20222022). Induction of Innate Immune Response by TLR3 Agonist Protects Mice against SARS-CoV-2 Infection. Viruses 14, 189. Page 189 14. doi:10.3390/V14020189
Te, H. S., Randall, G., and Jensen, D. M. (2007). Mechanism of Action of Ribavirin in the Treatment of Chronic Hepatitis C. Gastroenterol. Hepatol. (N Y) 3, 218–225. Available at:/pmc/articles/PMC3099343/?report=abstract. (Accessed September 18, 2020).
Tellier, R. (2022). COVID-19: the Case for Aerosol Transmission. Interface Focus 12, 20210072. doi:10.1098/RSFS.2021.0072
Theobald, S. J., Simonis, A., Georgomanolis, T., Kreer, C., Zehner, M., Eisfeld, H. S., et al. (2021). Long-lived Macrophage Reprogramming Drives Spike Protein-Mediated Inflammasome Activation in COVID-19. EMBO Mol. Med. 13, e14150. doi:10.15252/EMMM.202114150
Thépaut, M., Luczkowiak, J., Vivès, C., Labiod, N., Bally, I., Lasala, F., et al. (2021). DC/L-SIGN Recognition of Spike Glycoprotein Promotes SARS-CoV-2 Trans-infection and Can Be Inhibited by a Glycomimetic Antagonist. PLOS Pathog. 17, e1009576. doi:10.1371/JOURNAL.PPAT.1009576
Thorne, L. G., Reuschl, A. K., Zuliani-Alvarez, L., Whelan, M. V. X., Turner, J., Noursadeghi, M., et al. (2021). SARS-CoV-2 Sensing by RIG-I and MDA5 Links Epithelial Infection to Macrophage Inflammation. EMBO J. 40, e107826. doi:10.15252/EMBJ.2021107826
Tobaiqy, M., Qashqary, M., Al-Dahery, S., Mujallad, A., Hershan, A. A., Kamal, M. A., et al. (2020). Therapeutic Management of Patients with COVID-19: a Systematic Review. Infect. Prev. Pract. 2, 100061. doi:10.1016/j.infpip.2020.100061
Tugaeva, K. V., Hawkins, D. E. D. P., Smith, J. L. R., Bayfield, O. W., Ker, D. S., Sysoev, A. A., et al. (2021). The Mechanism of SARS-CoV-2 Nucleocapsid Protein Recognition by the Human 14-3-3 Proteins. J. Mol. Biol. 433, 166875. doi:10.1016/J.JMB.2021.166875
Ullrich, S., and Nitsche, C. (2020). The SARS-CoV-2 Main Protease as Drug Target. Bioorg Med. Chem. Lett. 30, 127377. doi:10.1016/j.bmcl.2020.127377
Van Doremalen, N., Bushmaker, T., Morris, D. H., Holbrook, M. G., Gamble, A., Williamson, B. N., et al. (2020). Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 382, 1564–1567. doi:10.1056/NEJMc2004973
Vanderheiden, A., Ralfs, P., Chirkova, T., Upadhyay, A. A., Zimmerman, M. G., Bedoya, S., et al. (2020). Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J. Virol. 94. doi:10.1128/JVI.00985-20
Vidya, M. K., Kumar, V. G., Sejian, V., Bagath, M., Krishnan, G., and Bhatta, R. (2018). Toll-like Receptors: Significance, Ligands, Signaling Pathways, and Functions in Mammals. Int. Rev. Immunol. 37, 20–36. doi:10.1080/08830185.2017.1380200
Wagner, H. (2004). The Immunobiology of the TLR9 Subfamily. Trends Immunol. 25, 381–386. doi:10.1016/J.IT.2004.04.011
Wang, Y., Kang, H., Liu, X., and Tong, Z. (2020). Asymptomatic Cases with SARS-CoV-2 Infection. J. Med. Virol. 92, 1401–1403. doi:10.1002/jmv.25990
Wang, Y., Su, L., Morin, M. D., Jones, B. T., Whitby, L. R., Surakattula, M. M., et al. (2016). TLR4/MD-2 Activation by a Synthetic Agonist with No Similarity to LPS. Proc. Natl. Acad. Sci. U. S. A. 113, E884–E893. doi:10.1073/PNAS.1525639113/-/DCSUPPLEMENTAL
Webb, K., Peckham, H., Radziszewska, A., Menon, M., Oliveri, P., Simpson, F., et al. (2019). Sex and Pubertal Differences in the Type 1 Interferon Pathway Associate with Both X Chromosome Number and Serum Sex Hormone Concentration. Front. Immunol. 9, 3167. doi:10.3389/FIMMU.2018.03167/FULL
Wilson, A. M., Sleeth, D. K., Schaefer, C., and Jones, R. M. (2022). Transmission of Respiratory Viral Diseases to Health Care Workers: COVID-19 as an Example. doi:10.1146/ANNUREV-PUBLHEALTH-052120-110009
Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., et al. (2020). Analysis of Therapeutic Targets for SARS-CoV-2 and Discovery of Potential Drugs by Computational Methods. Acta Pharm. Sin. B 10, 766–788. doi:10.1016/j.apsb.2020.02.008
Xie, J., Li, Y., Shen, X., Goh, G., Zhu, Y., Cui, J., et al. (2018). Dampened STING-dependent Interferon Activation in Bats. Cell. Host Microbe 23, 297–e4. e4. doi:10.1016/J.CHOM.2018.01.006
Xu, H., Akinyemi, I. A., Chitre, S. A., Loeb, J. C., Lednicky, J. A., McIntosh, M. T., et al. (2022). SARS-CoV-2 Viroporin Encoded by ORF3a Triggers the NLRP3 Inflammatory Pathway. Virology 568, 13–22. doi:10.1016/J.VIROL.2022.01.003
Xu, H., Chitre, S. A., Akinyemi, I. A., Loeb, J. C., Lednicky, J. A. 4, Mcintosh, M. T., et al. (2020a2020). SARS-CoV-2 Viroporin Triggers the NLRP3 Inflammatory Pathway. bioRxiv. doi:10.27.35773110.1101/2020.10.27.357731
Xu, X., Nie, Y., Wang, W., Ma, N., and Lendlein, A. (2021). Periodic Thermomechanical Modulation of Toll-like Receptor Expression and Distribution in Mesenchymal Stromal Cells. MRS Commun. 11, 425–431. doi:10.1557/S43579-021-00049-5/FIGURES/3
Xu, Y., Li, X., Zhu, B., Liang, H., Fang, C., Gong, Y., et al. (2020b). Characteristics of Pediatric SARS-CoV-2 Infection and Potential Evidence for Persistent Fecal Viral Shedding. Nat. Med. 26, 502–505. doi:10.1038/s41591-020-0817-4
Yadav, R., Chaudhary, J. K., Jain, N., Chaudhary, P. K., Khanra, S., Dhamija, P., et al. (2021). Role of Structural and Non-structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 10, 821. doi:10.3390/CELLS10040821
Yalcinkaya, M., Liu, W., Islam, M. N., Kotini, A. G., Gusarova, G. A., Fidler, T. P., et al. (2021). Modulation of the NLRP3 Inflammasome by Sars-CoV-2 Envelope Protein. Sci. Rep. 11, 1–12. doi:10.1038/s41598-021-04133-7
Yang, J., Petitjean, S. J. L., Koehler, M., Zhang, Q., Dumitru, A. C., Chen, W., et al. (2020). Molecular Interaction and Inhibition of SARS-CoV-2 Binding to the ACE2 Receptor. Nat. Commun. 11, 4541. doi:10.1038/s41467-020-18319-6
Yang, L., Xie, X., Tu, Z., Fu, J., Xu, D., and Zhou, Y. (20212021). The Signal Pathways and Treatment of Cytokine Storm in COVID-19. Sig Transduct. Target Ther. 6, 1–20. doi:10.1038/s41392-021-00679-0
Yin, X., Riva, L., Pu, Y., Martin-Sancho, L., Kanamune, J., Yamamoto, Y., et al. (2021). MDA5 Governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell. Rep. 34, 108628. doi:10.1016/J.CELREP.2020.108628
Yoo, J. S., Kato, H., and Fujita, T. (2014). Sensing Viral Invasion by RIG-I like Receptors. Curr. Opin. Microbiol. 20, 131–138. doi:10.1016/J.MIB.2014.05.011
Yuki, K., Fujiogi, M., and Koutsogiannaki, S. (2020). COVID-19 Pathophysiology: A Review. Clin. Immunol. 215, 108427. doi:10.1016/j.clim.2020.108427
Zeng, J., Xie, X., Feng, X. L., Xu, L., Han, J. B., Yu, D., et al. (2022). Specific Inhibition of the NLRP3 Inflammasome Suppresses Immune Overactivation and Alleviates COVID-19 like Pathology in Mice. eBioMedicine 75, 103803. doi:10.1016/J.EBIOM.2021.103803
Zhang, H., Penninger, J. M., Li, Y., Zhong, N., and Slutsky, A. S. (2020). Angiotensin-converting Enzyme 2 (ACE2) as a SARS-CoV-2 Receptor: Molecular Mechanisms and Potential Therapeutic Target. Intensive Care Med. 46, 586–590. doi:10.1007/s00134-020-05985-9
Zhang, J. Z., Liu, Z., Liu, J., Ren, J. X., and Sun, T. S. (2014). Mitochondrial DNA Induces Inflammation and Increases TLR9/NF-Κb Expression in Lung Tissue. Int. J. Mol. Med. 33, 817–824. doi:10.3892/ijmm.2014.1650
Zhang, Q., Meng, Y., Wang, K., Zhang, X., Chen, W., Sheng, J., et al. (2021a). Inflammation and Antiviral Immune Response Associated with Severe Progression of COVID-19. Front. Immunol. 12, 631226. doi:10.3389/FIMMU.2021.631226
Zhang, Q., Raoof, M., Chen, Y., Sumi, Y., Sursal, T., Junger, W., et al. (2010). Circulating Mitochondrial DAMPs Cause Inflammatory Responses to Injury. Nature 464, 104–107. doi:10.1038/nature08780
Zhang, Q., Xiang, R., Huo, S., Zhou, Y., Jiang, S., Wang, Q., et al. (2021b2021). Molecular Mechanism of Interaction between SARS-CoV-2 and Host Cells and Interventional Therapy. Sig Transduct. Target Ther. 6, 1–19. doi:10.1038/s41392-021-00653-w
Zhao, J., Guo, S., Yi, D., Li, Q., Ma, L., Zhang, Y., et al. (2021a). A Cell-Based Assay to Discover Inhibitors of SARS-CoV-2 RNA Dependent RNA Polymerase. Antivir. Res. 190, 105078. doi:10.1016/J.ANTIVIRAL.2021.105078
Zhao, N., Di, B., and Xu, L. L. (2021b). The NLRP3 Inflammasome and COVID-19: Activation, Pathogenesis and Therapeutic Strategies. Cytokine Growth Factor Rev. 61, 2–15. doi:10.1016/J.CYTOGFR.2021.06.002
Zheng, M., Karki, R., Williams, E. P., Yang, D., Fitzpatrick, E., Vogel, P., et al. (2021). TLR2 Senses the SARS-CoV-2 Envelope Protein to Produce Inflammatory Cytokines. Nat. Immunol. 22, 829–838. doi:10.1038/s41590-021-00937-x
Zheng, Y. Y., Ma, Y. T., Zhang, J. Y., and Xie, X. (2020a). COVID-19 and the Cardiovascular System. Nat. Rev. Cardiol. 17, 259–260. doi:10.1038/s41569-020-0360-5
Zheng, Y., Zhuang, M.-W., Han, L., Zhang, J., Nan, M.-L., Zhan, P., et al. (2020b). Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Membrane (M) Protein Inhibits Type I and III Interferon Production by Targeting RIG-I/MDA-5 Signaling. Sig Transduct. Target Ther. 5 (5), 1–13. doi:10.1038/s41392-020-00438-7
Zhou, J., Li, C., Liu, X., Chiu, M. C., Zhao, X., Wang, D., et al. (2020). Infection of Bat and Human Intestinal Organoids by SARS-CoV-2. Nat. Med. 26, 1077–1083. doi:10.1038/s41591-020-0912-6
Zhu, W., Chen, C. Z., Gorshkov, K., Xu, M., Lo, D. C., and Zheng, W. (2020). RNA-dependent RNA Polymerase as a Target for COVID-19 Drug Discovery. SLAS Discov. 25, 1141–1151. doi:10.1177/2472555220942123
Keywords: SARS-CoV-2, innate immune receptor, toll-like receptor, RIG-like receptor, antiviral response
Citation: Farooq M, Khan AW, Ahmad B, Kim MS and Choi S (2022) Therapeutic Targeting of Innate Immune Receptors Against SARS-CoV-2 Infection. Front. Pharmacol. 13:915565. doi: 10.3389/fphar.2022.915565
Received: 08 April 2022; Accepted: 15 June 2022;
Published: 30 June 2022.
Edited by:
Gerhard Wolber, Freie Universität Berlin, GermanyReviewed by:
Budheswar Dehury, Regional Medical Research Center (ICMR), IndiaMarko Jukic, University of Maribor, Slovenia
Copyright © 2022 Farooq, Khan, Ahmad, Kim and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sangdun Choi, c2FuZ2R1bmNob2lAYWpvdS5hYy5rcg==
 Mariya Farooq
Mariya Farooq Abdul Waheed Khan
Abdul Waheed Khan Bilal Ahmad
Bilal Ahmad Moon Suk Kim
Moon Suk Kim Sangdun Choi
Sangdun Choi