
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 09 September 2022
Sec. Pharmacogenetics and Pharmacogenomics
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.911827
This article is part of the Research TopicRecent Developments in Pharmacogenetics and PharmacogenomicsView all 11 articles
Background: Uridine 5′-diphospho glucuronosyl transferase (UGT) is the main enzyme responsible for the glucuronide conjugation, the principal metabolic pathway of sodium valproate. The objective of the study was to explore if there was an association between the UGT2B7 genetic polymorphism and clinical efficacy and safety in paediatric epileptic patients on sodium valproate monotherapy.
Methods and materials: The cohort study included 100 pediatric epileptic patients aged 2–18 years who had been on sodium valproate monotherapy for at least 1 month. PCR-RFLP was carried out to assess the genetic polymorphism patterns of UGT2B7 (C161T, A268G, G211T). Based on the extent of seizure control throughout the 1-year follow-up, clinical outcome was assessed in terms of responders and non-responders. Hepatic, renal, and other lab parameters were assayed to determine safety. The SNPstat web software was used to calculate linkage disequilibrium.
Results: Out of 100 patients, CC (38%), CT (43%), TT (19%) pattern was observed in UGT2B7 (C161T) gene, AA (15%), AG (39%), GG (46%) in (A268G) gene and GG (80%), GT (18%), TT (02%) in (G211T) gene. There was no statistical difference in clinical outcome with distinct UGT2B7 genetic polymorphism patterns, according to the findings. With low D′ and R2 values, linkage disequilibrium between alleles was statistically insignificant. However, the associations of C161T and G211T with treatment response were significant (p = 0.014) in determining treatment response.
Conclusion: Our findings show that the genetic variation of UGT2B7 had no bearing on the clinical outcome of epilepsy. Gene interactions, on the other hand, had an impact on treatment response.
Epilepsy is a neurological illness that includes a variety of seizure types and epilepsy syndromes that are heterogeneous and complex brain disorders (Berg et al., 2010; Tejada et al., 2013). India’s overall epilepsy prevalence is estimated to be 3–11.9 per 1,000 people (Amudhan et al., 2015). After a patient has had unprovoked seizures, paediatricians and neurologists frequently prescribe long-term, anti-epileptic drugs (AEDs) (Cockerell et al., 1997). About 60–70% of newly treated patients have satisfactory seizure control (Sillanpaa and Schmidt, 2006) while the remaining 30% of patients will have uncontrolled epilepsy with recurrent seizures, adverse effects, and a significantly increased risk of mortality and morbidity if they are treated with AEDs for a long time (Sillanpaa and Schmidt, 2006; Tejada et al., 2013). The goal of treating epilepsy in children is to prevent seizures for 2 years with an anti-epileptic drug (AED) that has minimal side effects, so that the drug can be gradually taken off.
Pharmacogenetics is the study of genetic variants that impact drug metabolism, pharmacological targets, or disease pathways, resulting in variable responses to drugs in terms of efficacy or side effects. As a result, it is critical to look into the function of genetic polymorphisms in drug metabolism and antiepileptic drug effects.
Rationale: Sodium valproate is a widely used antiepileptic medication with a broad spectrum of action. Uridine-5′-diphosphate glucuronosyl transferases catalyse glucuronidation, a key route of VPA (UGTs). Changes in sodium valproate metabolism may be caused by polymorphisms in genes encoding UGT enzymes. Changes in sodium valproate’s glucuronidation rate can affect the drug’s blood level, resulting in either insufficient plasma concentrations that impair therapy effectiveness or elevated plasma concentrations that cause toxicity (Chatzistefanidis et al., 2012). The glucuronidation of sodium valproate is aided by polymorphisms in the rs7439366, rs12233719, and rs145725367 genes (Chatzistefanidis et al., 2012). The functional impacts of SNPs of UGT2B7 was investigated using computational prediction tools (Polyphen analysis and SIFT analysis). Three SNPs (rs12233719, rs7668258, rs7662029 were selected for the wet lab analysis. Although the selected variants were tolerant, studies have shown the significant role of those SNPs in different populations.
Novelty: UGT2B7 polymorphisms have previously been studied in Chinese and Japanese populations. The majority of investigations have focused on determining the pattern of genetic polymorphism and its impact on a patient’s serum concentration. However, there is very little evidence about the effect of sodium valproate on the UGT2B7 polymorphism and its relationship to clinical outcome in the Indian population.
Assessing patterns of linkage disequilibrium (LD) of single nucleotide polymorphisms (SNPs) has become an essential aspect of both evolutionary biology and medical genetics. Within a population, LD occurs when two alleles at different loci are genetically related and show non-random association in the same chromosome. Many factors influence LD, including selection, genetic recombination rate, mutation rate, genetic drift, mating system, population structure, and genetic linkage. Because the range of potential values relies on the frequencies of the alleles it refers to, the coefficient of linkage disequilibrium (D) is not necessarily a practical measure of linkage disequilibrium.
The objective of the study was to explore the association between the UGT2B7 gene polymorphisms and clinical efficacy and safety in pediatric epileptic patients on sodium valproate monotherapy, as well as the linkage disequilibrium between the SNPs of UGT2B7 gene.
The observational cohort study was conducted from February 2018 to February 2020. Hundred pediatric epileptic patients of South Indian descent were included in the study which was conducted at the Justice K S Hegde Charitable Hospital in Mangalore, India, and the Central Research Laboratory of the K S Hegde Medical Academy in Mangalore.
Epileptic patients of 2–18 years of age were diagnosed clinically by EEG, patients on a stable dose of 20 mg/kg/day sodium valproate on a frequency of twice/thrice a day for the past 1 month.
Exclusion Criteria: Patients who were already on treatment with any other antiepileptic drugs or drugs which induce or inhibit enzymatic action of sodium valproate metabolism. History or evidence of hepatitis or impaired renal functions.
The research protocol was approved by Central Ethics committee of NITTE (Deemed to be University) (Approval number: NU/CEC/2018/0174 dated 19-01-2018, amendment in the proposal was approved by NU/CEC/2018/08 dated 26-11-2018, and approval to have house visit and telephonic data collection about the clinical pattern seizure control, recurrence and patient compliance (NU/CEC/2020/0300 dated 10-7-2020).
The research process was briefly explained orally in simple words in their native language for children in the age group of 7–12 years along with written consent from their parents. The written assent was obtained from children of 12–18 years and written consent was obtained from the participants parents in the presence of a witness (not related to patient and research team). The written consent was obtained from parents of eligible participants who are less than 6 years in the presence of a witness as mentioned above. The mentioned are the guidelines given by Indian Council of Medical Research for enrolling the participants for biomedical research (2016).
Once the patient met the requirements, 4 mL of whole blood were collected aseptically during the 6-months follow-up appointment. 2 ml of EDTA whole blood was preserved at -80°C for genetic polymorphism study, and 2 ml of whole blood in a plain vial was centrifuged to obtain serum and then stored at -80°C for biochemical parameters analysis.
At the time of enrolment and at 6 months, liver function tests (Albumin, total protein, total, direct bilirubin, SGOT, SGPT, ALP), renal function tests (Blood urea, serum creatinine), platelet count, and serum amylase were estimated.
Blood sample was centrifuged with RBC lysing solution (NH4Cl, KHCO3, Na2. EDTA). The residual RBC lysate was suspended with a cell lysing solution (50 mM Tris HCl, 50 mM EDTA, 10 mM NaCl, 1% SDS) and the cell lysates was digested overnight followed by the addition of protein precipitating solution. After centrifugation (3000 RPM, 10 min), the supernatant was collected in the 2% isopropanol and centrifuged again. The pellet which consists of DNA was washed with 70% ethanol and the DNA was allowed to precipitate. The precipitated DNA was transferred to the vials containing 50 µl of TE buffer (pH 7.5). The quantity of the DNA isolated was determined using a nanodrop spectrophotometer (Eppendorf) at 260 nm. The purity of the DNA sample was calculated by the ratio of 260/280 nm. Quantified DNA was sealed and stored at −20°C until further analysis.
The UGT2B7 polymorphisms at C161T, G211T, A268G were analyzed by the Polymerase Chain Reaction–Restriction Fragment Length Polymorphism (PCR–RFLP) method. Amplification was performed in MJ-Mini Thermal cycler (Bio-Rad, Tokyo, Japan).
Genotyping of rs-7668258 (161 C>T) was carried out using the forward primer, 5′-GATCTGTCACTGCTACTGTTC-3′ and reverse primer 5′-GTCTGAGCATGTGGATGGCCA-3′, with annealing temperature being 59°C and BstNI was the restriction enzyme (RE) used. For the genotyping of rs-12233719 (211 G>T).
Primers were F: 5′-TGCTTTAGCTCTGGGAATTGT -3′ R: 5′-TGCATGATGAAATTCTCCAAC-3´, RE MbiI, annealing temperature 59°C.
Primers used for rs-7662029 (268 A>G) were F: 5′-TCCAACTGATTGTTATGGTAGAT-3′
R: 5′-GCTGTTCCTTTCTGTCATTTCTC-3′ at an annealing temperature of 54°C, RE BglII.
The primers were obtained from Bionova Suppliers, chemicals required for DNA isolation and other procedures were procured from Merck and Sigma laboratory (Analytical grade). PCR was carried out with an initial denaturation enzyme activation step at 95°C for 5 min, amplification step for 35 cycles at 95°C for 30 s, annealing temperature for 30 s, treatment at 72°C for 30 s, final extension step at 72°C for 5 min. The amplified product of DNA was confirmed on a 2% agarose gel with ethidium bromide.
Amplified PCR product was digested with respective restriction enzymes (BstNI, MbiI, BglII) respectively for UGT2B7 (C161T, G211T, A268G) genes. The restricted fragment was separated on a 3% agarose gel with ethidium bromide. The genotype was assigned based on the results of the analysis of the digestion patterns. Trough level serum concentration of Sodium valproate was measured by HPLC analysis (Agillent Technologies, 1,260 Infinity). Standard Sodium valproate was procured from sigma laboratory for HPLC estimation.
Parents were asked to maintain diary in which seizure attack, duration of the attack, and any changes in the pattern from the prior episode, untoward response (loss of appetite, nausea, vomiting, discoloration of eye, pain abdomen, rashes) to the treatment at any point of time and missed medication dates to be mentioned. Were mentioned and shared with the treating pediatrician. Any of the above untoward effects and missing more than two doses of medication in a month was considered non-compliance.
The clinical outcome was evaluated in terms of sodium valproate efficacy and safety. The drug’s efficacy was assessed in terms of treatment responders and non-responders. Patients who had no recurrence or only two bouts of seizure attack in 6 months of follow-up were considered responders and sodium valproate monotherapy was well tolerated with no side effects. Non-responders were defined as individuals who had two or more seizure attacks in 6 months or experienced any adverse drug effect after starting therapy (Carpay and Arts, 1996). Tolerance was determined by comparing biochemical markers at baseline and 6 months later. If any patients developed jaundice, pancreatitis, or showed symptoms of thrombocytopenia, their tolerability was examined.
Taking SD of drug concentration as 23 μg/ml from the referred article [75] and fixing the margin of error = 5, the sample size would be 82. Z1-α/2 2 n = E x S2 n = 1.96 two x (23)2 5 n = 82 Considering 15% as attrition, the calculated total sample size is 95. The sample size calculated was 82. Expecting an attrition of 15%, the final sample size was 95. However because of COVID-19 pandemic we could obtain all the parameters of 75 patients only.
The obtained data was reported as frequency, percentage for qualitative data and mean and SD for quantitative data Using SPSS software version 16.0 statistical analysis was carried out. The Chi square test was used to find the association of UGT 2B7 and clinical responsiveness. A paired 't' test was used to compare the biochemical markers between baseline and 6 months follow-up in different age groups. Statistical significance was defined as a p value of less than 0.05.
D′ and r2 were calculated to determine the extent of LD in paired combinations of SNPs. The SHESIS plus and SNPstat online programme platforms were used to assess the LD between UGT2B7 haplotypes (C161T, A268G, and G211T). The analysis excluded haplotypes with frequencies of less than 0.03.
The mean age of our patients was 8.5 ± 4.3 years (2.2–17.3 years), and the average BMI was 16.5 ± 4.3 (7.81–32.84). The study subjects included 57 boys and 43 females. Between baseline and 1 year, there was a significant difference in sodium valproate concentration, adjusted sodium valproate concentration, creatinine, and platelet values. At different time intervals, there was no significant variation in hepatic and renal function test parameters. In UGT2B7 (C161T) gene, CC (38 percent), CT (43 percent), TT (19 percent) pattern was detected, AA (15 percent), AG (39 percent), GG (46 percent) in (A268G) gene, and GG (80 percent), GT (18 percent), TT (02 percent) in (G211T) gene (Figure 1).
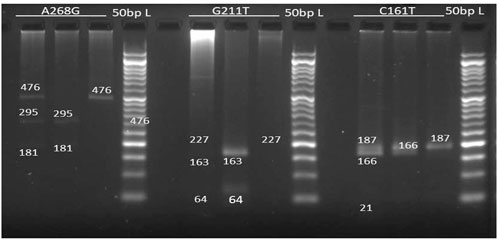
FIGURE 1. Electrophoresis gel image patterns of UGT2B7 (A268G, G211T, C161T) gene after digestion with BglII, MbiI, BstNI enzymes respectively (L): 50bp Ladder, Lane 1): homozygosity for variant allele, Lane(2): homozygosity for common allele, Lane 3): heterozygosity.
Frequencies of alleles and chi-square values of Hardy-Weinberg equilibrium are represented in (Table 1). After anti-epileptic drug therapy, the clinical outcome (seizure frequency) was monitored at 6-months and 1-year intervals (Figure 2). Responders (total seizure control from the day of enrolment to the day of assessment) and non-responders (two episodes of seizures from the day of enrollment) were compared.
The G211T gene polymorphism in the UGT2B7 gene exhibited a lower frequency of mutant carrier allele type than the wild type. The mutant carrier allele types C161T and A268G were found to be more common. Eighty patients (80%) reacted effectively to the treatment with no seizure relapses, while 20 patients (20%) experienced a seizure relapse with two or more occurrences. Other anti-epileptic medicines were prescribed for these patients. There was no statistically significant association between UGT2B7 (C161T, A268G, G211T) gene polymorphism and clinical efficacy (seizure control) (Table 2). Eighty-three patients (83%) had generalised tonic-clonic seizures (GTCS), while 17 patients (17%) had complex partial seizures (CPS).
There was no statistically significant association found between seizure type UGT2B7 (C161T, A268G, G211T) gene polymorphism and seizure type (Table 3).
During the baseline, 6-month, and 1-year follow-ups, all of the children had normal liver function values and renal function parameters (Table 4). Between baseline and 1 year, there was a significant difference in sodium valproate concentration, adjusted sodium valproate concentration, creatinine, and platelet values. At different time intervals, there was no significant variation in hepatic and renal function test parameters (Table 4).
During baseline and 1-year follow-up, all children exhibited normal liver function, hematological, and renal function parameters, with the exception of two cases. Acute pancreatitis developed in one patient, who had increased serum lipase and USG findings indicative of pancreatitis. This patient was given de-challenge, and he recovered totally within a week. Levetiracetam was prescribed for the patient. The Naranjo algorithm was used to assess causality, and a score of +6 was obtained, indicating that ADR was most likely caused by sodium valproate. The use of sodium valproate has been associated to the development of acute pancreatitis.
Analysis of combined genotyping data for patients and controls was used to look for LD between pairs of SNPs. Controls were paediatric epileptics who reacted to VPA monotherapy, whereas non-responders were considered cases. The global statistic R2 was created, as well as the statistic D′, which accounts for the constraints on R imposed by differing allele frequencies of the marker pair.
The three SNPs of UGT2B7 showed a weak LD between A268G and G211T of UGT2B7, as suggested by low D′ values, 0.3, 0.73, and 0.35 between three alleles. Low R2 values (0.06,0.03 and 0.01 respectively) do not support the co-inheritance of the above alleles. Low R2 values observed could be due to the low allele frequencies. Closer the D’ and R2 values to 1, a stronger disequilibrium between the alleles is suggested which imply strong chance of co-inheritance of the alleles. If the values are closer or equal to zero, it is suggestive of independence of alleles. None of the gene interactions and their association with the response to therapy were statistically significant as shown by binary analysis (Tables 5, 6). However the association of the interactions of C161T and G211T with the response to therapy were significant (p = 0.014) (Table 7).
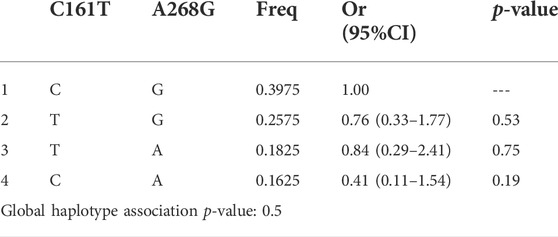
TABLE 5. Haplotype association of UGT2B7 C161T and A268G with response (n = 100, adjusted by Age + Sex).
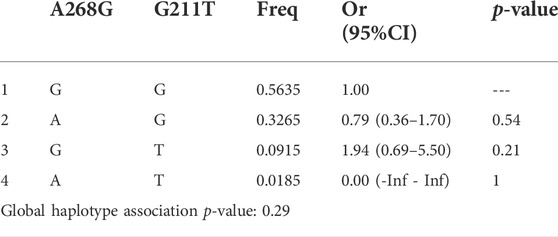
TABLE 6. Haplotype association of UGT2B7 A268G and G211T with response (n = 100, adjusted by Age + Sex).
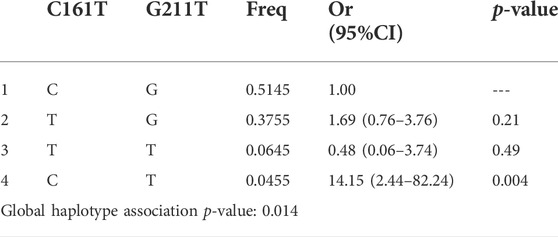
TABLE 7. Haplotype association of UGT2B7 C161T and G211T with response (n = 100, adjusted by Age + Sex).
A haplotype analysis of all the genes was performed using haplotype frequencies predicted by the Shesisplus for association with the response to therapy. Of the pairwise haplotypes association, none were statistically significant except for a haplotype TAACGT, with a chi square value of 7.77, p = 0.005, with OR 0.121 [95% CI = 0.021–0.689]. This suggests a good response to therapy among these haplotypes.
The association between UGT2B7 gene patterns and clinical outcomes were investigated in this study. Interindividual variability in pharmacokinetics and pharmacodynamics in paediatric epileptic patients on sodium valproate monotherapy may be influenced by gene polymorphism. Many researchers have looked into the effect of gene polymorphisms on sodium valproate pharmacokinetics and pharmacodynamics. When compared to the Chinese and Japanese populations, the pattern of UGT2B7 gene polymorphism was similar for UGT2B7 (C161T, A268G, G211T type) (Table 8) (Hung et al., 2011; Ma et al., 2013; Inoue et al., 2014; Yin-xiang et al., 2015; Du et al., 2016). The prevalent genetic polymorphism of UGT2B7 (C161T) pattern reported in Chinese paediatric epileptic population (Hung et al., 2011) was wild type, whereas the predominant pattern of UGT2B7 (C161T) gene observed in our and Japanese study populations was mutant carrier allele type. However, no genetic differences between our patients and Japanese pediatric epileptic patients were found (Inoue et al., 2014).
Several investigations in Chinese juvenile epileptics found that UGT2B7 (A268G) had a greater population of mutant carrier allele type than wild type, while UGT2B7 (G211T) had a lower population of mutant carrier allele than wild type.
To date, research on the impact of UGT2B7 polymorphism on substrate drug metabolism have been conflicting. Guo et al. (Guo et al., 2012) found that polymorphisms had no effect on serum valproate concentrations. UGT2B7 (A268G) plays a significant function in sodium valproate metabolism, according to Hongying et al. and Zhongliang et al. (Ma et al., 2013; Du et al., 2016).
Patients with the UGT2B7 (C161T) genotype had lower adjusted plasma valproic acid concentrations than those with the mutant carrier allele type, according to Inoue et al. (Inoue et al., 2014). To yet, none of the existing literatures have described the impact of UGT genetic variations on sodium valproate monotherapy clinical outcomes. Hung et al. tentatively tackled it, explaining that patients with strong medication response (seizure free/good seizure management) were evaluated for numerous genetic influences on VPA pharmacokinetics, including the UGT1A6 gene (Hung et al., 2011). However, we found no association between distinct polymorphism patterns of UGT2B7 and clinical efficacy or tolerability in our research.
Our study has a limitation due to the small sample size. Only a metacentric investigation including multiple regions could explain our limitation and add quality to the data in terms of genetic pattern in the current context of modified protocol in the treatment of pediatric epilepsy (preference of levetiracetam over sodium valproate).
Our findings showed that varied patterns of UGT2B7 genetic polymorphisms have no effect on the efficacy and tolerability of sodium valproate in the treatment of epilepsy. We also came to the conclusion that sodium valproate was well tolerated by pediatric epilepsy patients and might be employed as an effective anti-epileptic drug.
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by the Central ethics committee, Nitte DU. Written informed consent to participate in this study was provided by the participants’ parent/legal guardian.
1. NP: Collected samples, processed samples, evaluated the results and helped in data analysis 2. SA: Helped in receiving funds under Nitte (Deemed to be University), Manuscript writing and supervised the work. 3. UA: Concept and designed the study 4. VS: Provided pediatric epileptic cases. VS: Enrolled patients and supervised clinical follow-up of patients.
Research Grant No: NUFR2/2018/10/15, NITTE (Deemed to be University), Mangalore, India.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Amudhan, S., Gururaj, G., and Satishchandra, P. (2015). Epilepsy in India I: Epidemiology and public health. Ann. Indian Acad. Neurol. 18, 263–277. doi:10.4103/0972-2327.160093
Berg, A. T., Berkovic, S. F., Brodie, M. J., Buchhalter, J., Cross, J. H., van Emde Boas, W., et al. (2010). Revised terminology and concepts for organization of seizures and epilepsies: Report of the ILAE commission on classification and terminology, 2005-2009. Epilepsia 51, 676–685. doi:10.1111/j.1528-1167.2010.02522.x
Carpay, H., and Arts, W. (1996). Outcome assessment in epilepsy: Available rating scales for adults and methodological issues pertaining to the development of scales for childhood epilepsy. Epilepsy Res. 24, 127–136. doi:10.1016/0920-1211(96)00013-7
Chatzistefanidis, D., Georgiou, I., Kyritsis, A. P., and Markoula, S. (2012). Functional impact and prevalence of polymorphisms involved in the hepatic glucuronidation of valproic acid. Pharmacogenomics 13, 1055–1071. doi:10.2217/pgs.12.78
Cockerell, O. C., Johnson, A. L., Sander, J. W., and Shorvon, S. D. (1997). Prognosis of epilepsy: A review and further analysis of the first nine years of the British national general practice study of epilepsy, a prospective population-based study. Epilepsia 38, 31–46. doi:10.1111/j.1528-1157.1997.tb01075.x
Du, Z., Jiao, Y., and Shi, L. (2016). Association of UGT2B7 and UGT1A4 polymorphisms with serum concentration of antiepileptic drugs in children. Med. Sci. Monit. 22, 4107–4113. doi:10.12659/msm.897626
Guo, Y., Hu, C., He, X., Qiu, F., and Zhao, L. (2012). Effects of UGT1A6, UGT2B7, and CYP2C9 genotypes on plasma concentrations of valproic acid in Chinese children with epilepsy. Drug Metab. Pharmacokinet. 27, 536–542. doi:10.2133/dmpk.dmpk-11-nt-144
Hung, C. C., Ho, J. L., Chang, W. L., Tai, J. J., Hsieh, T. J., Hsieh, Y. W., et al. (2011). Association of genetic variants in six candidate genes with valproic acid therapy optimization. Pharmacogenomics 12, 1107–1117. doi:10.2217/pgs.11.64
Inoue, K., Suzuki, B. S., Yazawa, R., Yamamoto, Y., Takahashi, T., Takahashi, Y., et al. (2014). Influence of uridine diphosphate glucuronosyl transferase 2B7-161C>T polymorphism on the concentration of valproic acid in pediatric epilepsy patients. Ther. Drug Monit. 36, 406–409. doi:10.1097/FTD.0000000000000012
Ma, H., Zhang, T., Gong, Z., Zhou, B., Zou, M., Xiao, S., et al. (2013). Effect of UGT2B7 genetic variants on serum valproic acid concentration. J. Central South Univ. Med. Sci. 38 (8), 766–772. doi:10.3969/j.issn.1672-7347.2013.08.002
Sillanpaa, M., and Schmidt, D. (2006). Natural history of treated childhood-onset epilepsy: Prospective, long-term population-based study. Brain. 129, 617–624. doi:10.1093/brain/awh726
Tejada, J., Costa, K. M., Bertti, P., and Garcia-Cairasco, N. (2013). The epilepsies: Complex challenges needing complex solutions. Epilepsy Behav. 26, 212–228. doi:10.1016/j.yebeh.2012.09.029
Keywords: pediatric epilepsy, genetic polymorphism, sodium valproate, clinical outcome, linkage disequilibrium
Citation: Adiga S, PB N, Adiga U and Shenoy V (2022) UGT2B7 gene polymorphism and linkage disequilibrium in pediatric epileptic patients and their influence on sodium valproate monotherapy: A cohort study. Front. Pharmacol. 13:911827. doi: 10.3389/fphar.2022.911827
Received: 03 April 2022; Accepted: 26 July 2022;
Published: 09 September 2022.
Edited by:
Francisco Lopez-Munoz, Camilo José Cela University, SpainReviewed by:
Shakir Ullah, Khyber Medical University, PakistanCopyright © 2022 Adiga, PB, Adiga and Shenoy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Usha Adiga, YWRpZ2FwaGFybWFAbml0dGUuZWR1Lmlu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.