
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 12 May 2022
Sec. Pharmacology of Anti-Cancer Drugs
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.902290
This article is part of the Research Topic Imaging Technology in Oncology Pharmacological Research, Volume II View all 6 articles
Cyclic adenosine 3′,5′-monophosphate (cAMP) is an important intracellular second messenger molecule downstream of many G protein-coupled receptors (GPCRs). Fluorescence imaging with bright and sensitive cAMP indicators allows not only dissecting the spatiotemporal dynamics of intracellular cAMP, but also high-content screening of compounds against GPCRs. We previously reported the high-performance circularly permuted GFP (cpGFP)-based cAMP indicator G-Flamp1. Here, we developed improved G-Flamp1 variants G-Flamp2 and G-Flamp2b. Compared to G-Flamp1, G-Flamp2 exhibited increased baseline fluorescence (1.6-fold) and larger fluorescence change (ΔF/F0) (1,300% vs. 1,100%) in HEK293T cells, while G-Flamp2b showed increased baseline fluorescence (3.1-fold) and smaller ΔF/F0 (400% vs. 1,100%). Furthermore, live cell imaging of mitochondrial matrix–targeted G-Flamp2 confirmed cytosolic cAMP was able to enter the mitochondrial matrix. G-Flamp2 imaging also showed that adipose tissue extract activated the Gi protein-coupled orphan GPCR GPR50 in HEK293T cells. Taken together, our results showed that the high-performance of G-Flamp2 would facilitate sensitive intracellular cAMP imaging and activity measurement of compounds targeting GPCR-cAMP signaling pathway during early drug development.
Cyclic adenosine 3′,5′-monophosphate (cAMP) is an important second messenger downstream of G protein-coupled receptors (GPCRs) that are coupled to Gs, Gi and Gq proteins (Massengill et al., 2021). It is produced from adenosine triphosphate (ATP) by adenylyl cyclases (ACs) and degraded to adenosine monophosphate (AMP) by phosphodiesterases (PDEs) (Cooper, 2003). cAMP signaling regulates a variety of cellular functions including cell migration, mitochondrial homeostasis, cell proliferation and cell death (Lefkimmiatis and Zaccolo, 2014). Given that cAMP can integrate signals from a diversity of GPCRs and its abnormal dynamics are associated with many diseases including cancer (Gold et al., 2013), the GPCR-cAMP signaling pathway is an attractive pharmaceutical target (Zaccolo, 2021).
The cAMP concentration is precisely controlled in space and time in living cells. cAMP and cAMP signaling could be constrained on or within subcellular structures (Zaccolo et al., 2021). Recently, cAMP and cAMP signaling in the mitochondrial matrix has gained much attention due to its key roles in regulating oxidative phosphorylation (OXPHOS) and mitochondrial biogenesis (Zhang et al., 2016). To measure the cAMP dynamics with high spatiotemporal resolution in mitochondrial matrix, optical imaging with genetically encoded fluorescent indicators (GEFIs) (Mattiazzi Usaj et al., 2016) is highly desirable.
During the past 20 years, more than 50 fluorescent protein (FP)-based indicators have been developed (Massengill et al., 2021). The majority of them are fluorescence resonance energy transfer (FRET) indicators and some are single-FP indicators. The latter usually gives much larger fluorescence change than the former (Massengill et al., 2021). However, current cAMP single-FP indicators (Flamindo2, cAMPr, Pink Flamindo and R-FlincA) exhibit small fluorescence change (|ΔF/F0| < 150%) and most are dim in mammalian cells at 37°C (Odaka et al., 2014; Harada et al., 2017; Hackley et al., 2018; Ohta et al., 2018). To address these problems, we recently developed a high-performance single-FP cAMP indicator named G-Flamp1 (Wang et al., 2022), which is created by inserting a circularly permuted GFP (cpGFP) into a bacterial cAMP-binding domain of the bacterial MlotiK1 channel (mlCNBD). G-Flamp1 exhibits a large fluorescence increase (a maximum ΔF/F0 of 1,100% in HEK293T cells), appropriate affinity (a Kd of 2.17 μM) and sub-second response kinetics. However, compared to the well-known single-FP calcium indicator GCaMP6s, the brightness of G-Flamp1 is still relatively low, which could hinder the cAMP imaging of subcellular structures such as mitochondria (Wang et al., 2022).
To further increase the signal-to-background ratio and sensitivity for cAMP imaging here we aimed to increase the brightness and dynamic range of the G-Flamp1 indicator. Using a combination of crystal structure-guided mutagenesis and library screening, we developed two variants named G-Flamp2 and G-Flamp2b with improved properties. Specifically, compared to G-Flamp1, G-Flamp2 exhibited an increased brightness (1.6-fold) and larger maximum ΔF/F0 (1300% vs. 1100% of G-Flamp1), while G-Flamp2b exhibited higher brightness (3.1-fold) but smaller ΔF/F0 (400% vs. 1100% of G-Flamp1) in HEK293T cells at 37°C. Then we demonstrated that G-Flamp2 enables sensitive monitoring of cAMP changes in mitochondrial matrix. Finally, based on G-Flamp2 imaging, we found that adipose tissue extracts can activate the Gi-coupled orphan GPCR GPR50. Taken together, these results suggest that G-Flamp2 is a powerful tool for sensitive cAMP imaging in subcellular compartments and image-based high-content screening (HCS).
The cpFP cAMP indicator G-Flamp1 was originally constructed by inserting cpGFP from GCaMP6f into mICNBD (Nimigean et al., 2004; Mukherjee et al., 2016) at the mouth of the cAMP-binding pocket (Figure 1A). To develop improved G-Flamp1 variants, we focused on both the cpGFP and the interface between cpGFP and mlCNBD. First, we incorporated beneficial mutations from the OPT (Optimized) variant of circularly permuted super folder GFP (K86N/S175R/T216S/E284V/I301T, numbering according to PDB 6M63) into G-Flamp1 and generated G-Flamp1.5 variant with brightness increased by 17% (Figure 1B, Supplementary Figures S3A, S6). Second, we performed random mutagenesis on cpGFP part of G-Flamp1.5 using error-prone PCR and were able to identify a brighter (increased by 49%) (Supplementary Figure S3B) and highly responsive (ΔF/F0 of 1810%) variant G-Flamp2 (Figure 1B, Supplementary Figures S1A, S6), which harbors K94E mutation in the cpGFP. Inspection of crystal structure of G-Flamp1 revealed that the positive residue of Arg96 with the negative residue of Glu94 formed electrostatic interaction, and thus stabilized the protein structure (Supplementary Figure S4A). Third, to further increase the brightness of G-Flamp2, crystal structure–guided mutagenesis for stabilization of the interface between cpGFP and mlCNBD and random mutagenesis of mlCNBD were employed. The improved variant G-Flamp2b with brightness increased by 45% (Supplementary Figure S3C) was finally identified and has the following mutations: F151Y/E323K/S346I in the interface (Supplementary Figure S4B), G10E/G29D/V34D in mlCNBD and R318N/V319S in linker 2 (Figure 1B, Supplementary Figure S6). G-Flamp2b had a relatively small ΔF/F0 of 440% (Supplementary Figure S1B).
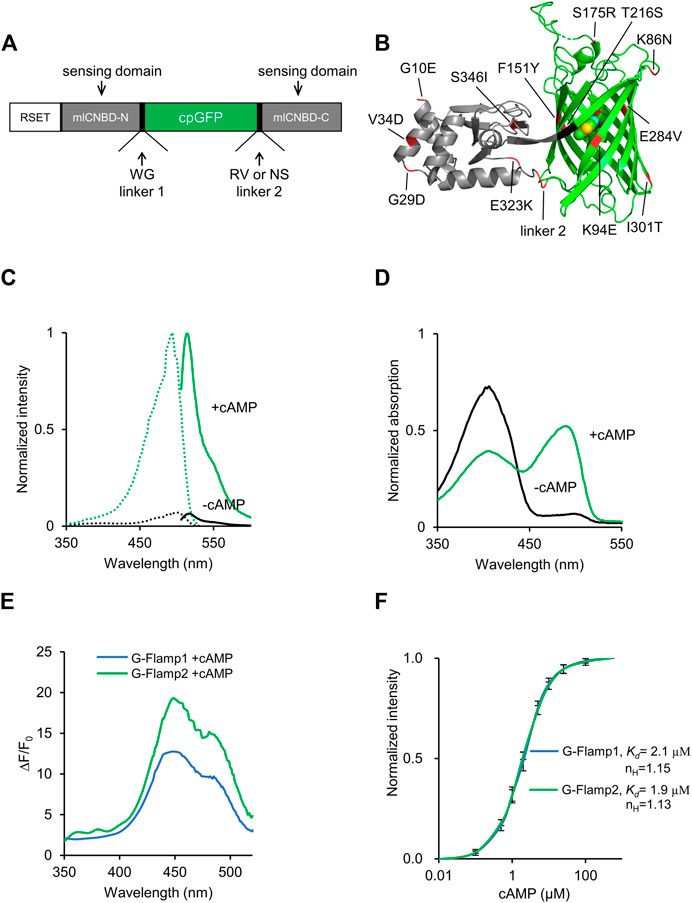
FIGURE 1. Development and in vitro characterization of G-Flamp2. (A) Schematic of G-Flamp biosensor. cpGFP was inserted into mlCNBD with linker1 and linker 2 (both have two amino acids). RSET: the N-terminal fusion peptide including a 6× His tag from the pNCS bacterial expression vector. (B) Crystal structure of G-Flamp1 (PDB 6M63). Sites mutated in G-Flamp2 and G-Flamp2b are marked in red. Linker is marked in black. (C) Excitation (dotted lines) and emission (solid lines) spectra of purified G-Flamp2 sensor in HEPES buffer (pH 7.15) with (green line, +cAMP) or without (black line, -cAMP) 500 μM cAMP. (D) Absorbance spectra of 50 μM G-Flamp2 in HEPES buffer (pH 7.15) in the presence (green line, +cAMP) or absence (black line, -cAMP) of 500 μM cAMP. (E) Excitation wavelength-dependent ΔF/F0 of G-Flamp1 and G-Flamp2 in HEPES buffer (pH 7.15) in the presence of 500 μM cAMP. (F) Binding titration curves of G-Flamp1 and G-Flamp2 biosensors to cAMP (500 μM) in HEPES buffer (pH 7.15). The data were fitted by a sigmoidal binding function to extract the dissociation constant Kd. Data are presented as mean ± standard deviation (SD) from three independent experiments.
We first investigated the fluorescence and absorption properties of purified G-Flamp2. The fluorescence excitation and emission spectra of cAMP-bound G-Flamp2 were similar to those of G-Flamp1 and peaked at 488 and 516 nm, respectively (Figure 1C). The excitation and emission peaks of cAMP-free G-Flamp2 were redder than those of cAMP-bound G-Flamp2 by 8 and 3 nm, respectively (Supplementary Table S1). G-Flamp2b had similar fluorescence spectra to those of G-Flamp2 (Supplementary Figure S2A). According to these fluorescence spectra, the calculated fluorescence change peaked at 450 nm with a maximum ΔF/F0 of 2000 and 430% for G-Flamp2 and G-Flamp2b, respectively (Figure 1E, Supplementary Figure S2B). Absorbance spectra of G-Flamp2 revealed that both cAMP-bound and cAMP-free indicators displayed two peaks with maxima at 400 and 490 nm. Moreover, the deprotonated form of cAMP-bound G-Flamp2 significantly increased, making it much brighter than deprotonated cAMP-free G-Flamp2 (Figure 1D).
The extinction coefficient (EC) of G-Flamp2 increased by 8.8-fold (5,189 mM−1cm−1 vs. 45,459 mM−1cm−1) after cAMP binding, while G-Flamp2b increased by 2.7-fold (12,528 mM−1cm−1 vs. 33,760 mM−1cm−1) (Supplementary Table S1). The quantum yield (QY) of G-Flamp2 and G-Flamp2b slightly decreased (0.38 vs. 0.33 for G-Flamp2 and 0.39 vs. 0.37 for G-Flmap2b) after cAMP binding (Supplementary Table S1). Like other single-FP probes, the fluorescence intensity of G-Flamp2 was sensitive to pH with pKa values of 8.50 and 6.97 for cAMP-free and cAMP-bound G-Flamp2, respectively (Supplementary Figure S7A, Supplementary Table S1). Moreover, the calculated ΔF/F0 peaked at pH 6.5 with a value of 1940% and remained high at pH 7.0 with a value of 1,590% (Supplementary Figure S7B), indicating that G-Flamp2 would be highly responsive in the pH environment of mammalian cells. Besides, the G-Flamp2b calculated ΔF/F0 peaked at pH 6.7 with a value of 370% and was still high at pH 7.0 with a value of 347% (Supplementary Figure S7C).
Next we examined the concentration-response relationship of G-Flamp2 and G-Flamp2b with cAMP or cGMP. The apparent dissociation constant (Kd) values of G-Flamp2 and G-Flamp2b for cAMP were 1.9 and 3.2 μM, respectively (Figure 1F, Supplementary Table S1). The apparent Kd values of G-Flamp2 and G-Flamp2b for cGMP were 43 and 32 μM, respectively (Supplementary Figure S5). Based on the Kd values for cAMP and cGMP, G-Flamp2 and G-Flamp2b have ∼23 and 10-fold higher selectivity for cAMP over cGMP (defined as Kd ratio of cGMP/cAMP), while G-Flamp1 has a ∼13-fold selectivity towards cAMP (Ohta et al., 2018). This specificity is similar or better than most reported cAMP indicators (Supplementary Table S2). Since previous studies reported that cAMP indicators with Kd between 30 nM and 10 μM successfully monitored physiological cAMP changes (Börner et al., 2011; Kitaguchi et al., 2013; Odaka et al., 2014; Mukherjee et al., 2016; Ohta et al., 2018), G-Flamp2 and G-Flamp2b would be well suitable for most applications.
We first examine the fluorescence brightness of G-Flamp1/2/2b indicators in HEK293T cells. Under 450 nm or 480 nm excitation, G-Flamp2 and G-Flamp2b showed a 1.6-fold and 3.1-fold higher baseline fluorescence than G-Flamp1, respectively (Figure 2A, Supplementary Figure S8).
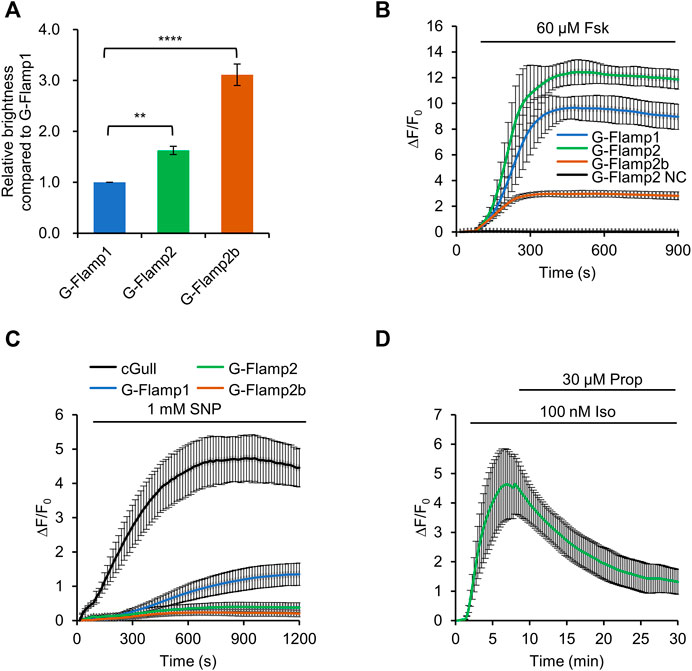
FIGURE 2. Characterization of G-Flamp2 and G-Flamp2b in HEK293T cells. (A) Relative brightness of G-Flamp1, G-Flamp2 and G-Flamp2b in resting HEK293T cells measured using a plate reader (excited at 450 nm). The IRES-mCherry-CAAX gene was attached downstream of the green sensors and the red fluorescence intensity was used to calibrate the sensor’s brightness. (B) Traces of ΔF/F0 in responses to 60 µM Fsk of G-Flamp1, G-Flamp2, G-Flamp2b and G-Flamp2 NC (G-Flamp2 R341E, cAMP-insensitive negative control) in HEK293T cells. Data are presented as mean ± standard deviation (SD) from three independent experiments. n = 7, 13, 12, 9 cells for G-Flamp1, G-Flamp2, G-Flamp2b and G-Flamp2 NC, respectively. (C) Similar to (B) except that 1 mM SNP was used. n = 15, 12, 10 cells for G-Flamp1, G-Flamp2 and G-Flamp2b, respectively. n = 5 cells for cGull. (D) Traces of ΔF/F0 in response to 100 nM Iso followed by 30 μM propranolol in HEK293T cells expressing G-Flamp2. n = 16 cells from 3 cultures. Data are presented as the mean ± standard deviation. *Significantly different from G-Flamp1, **p < 0.01, ****p < 0.0001 as measured by One-way analysis of variance (ANOVA) with post hoc Tukey’s test.
Then we investigated the fluorescence change of G-Flamp1/2/2b sensors. To this end, 60 μM Forskolin (Fsk), a potent pharmacological activator of transmembrane adenylyl cyclase (tmAC), was used to induce a large amount of cAMP in HEK293T cells. Under 450 nm excitation, G-Flamp1, G-Flamp2 and G-Flamp2b exhibited a maximum ΔF/F0 of 1,000, 1200 and 300%, respectively (Figure 2B). As a negative control, the cAMP-insensitive G-Flamp2 NC (G-Flamp2 Negative Control) sensor with the R341E mutation showed no detectable fluorescence change (Figure 2B).
Next we characterized the specificity and reversibility of G-Flamp1/2/2b indicators. We first tested the responses of indicators to cGMP, which was reported to bind the mlCNBD with a weaker affinity than cAMP. 1 mM sodium nitroprusside (SNP, a NO donor that activates soluble guanylyl cyclase) was exploited to induce a large amount of cGMP in HEK293T cells. Under 480 nm excitation, Green cGull, a single fluorescent protein (FP)-based cGMP indicator, exhibited a large fluorescence change with a peak ΔF/F0 of 500% (Figure 2C) (Ponsioen et al., 2004; Matsuda et al., 2017). Under 450 nm excitation, G-Flamp1 exhibited a significant fluorescence increase with a peak ΔF/F0 of 134% (Figure 2C), while G-Flamp2 and G-Flamp2b showed a relatively smaller peak ΔF/F0 (40 and 20%, respectively) (Figure 2C). These results indicated that G-Flamp2/2b had a high specificity toward cAMP. Regarding reversibility, HEK293T cells expressing G-Flamp2 exhibited increased fluorescence upon 100 nM Iso treatment and then returned to basal level after addition of 30 μM β-AR anti-agonist propranolol (Prop) (Figure 2D).
Taken together, the above results demonstrated that both G-Flamp2 and G-Flamp2b had higher brightness and better specificity than G-Flamp1, and G-Flamp2 also had a larger fluorescence change than G-Flamp1 in living mammalian cells. Therefore, G-Flamp2 is more suitable to monitor intracellular cAMP dynamics.
The origin of cAMP in the mitochondrial matrix is still under debate (Valsecchi et al., 2013). Since cAMP can not freely diffuse across the inner mitochondrial membrane (Acin-Perez et al., 2009), one model suggested that cAMP inside mitochondria is actively transported from cytoplasmic pool (DiPilato et al., 2004) and another one suggested that the intramitochondrial cAMP is produced by mitochondrially localized soluble AC (sAC) (Zhang et al., 2016).
To test whether cytosolic cAMP can enter mitochondria, we monitor cAMP dynamics inside the mitochondrial matrix while stimulating cAMP production in cytosol. We fused mitochondrial matrix-targeting sequence from subunit VIII of the human cytochrome oxidase (COX) at the N terminus of G-Flamp2 to generate Mito-G-Flamp2 (Figure 3A). Mito-G-Flamp2 showed good colocalization with the mitochondria marker dye Mito-Tracker Red CMXRos (Figure 3B). HeLa cells expressing Mito-G-Flamp2, G-Flamp2 and G-Flamp2 NC showed a peak ΔF/F0 of 50, 260 and 10% after 60 μM Fsk treatment, respectively (Figure 3C, Supplementary Figure S9A–C). Since Fsk only activate tmAC but not sAC (Steegborn, 2014), these results suggests that cAMP generated in the cytosol can access the mitochondrial matrix and lead to the fluorescence increase of Mito-G-Flamp2.
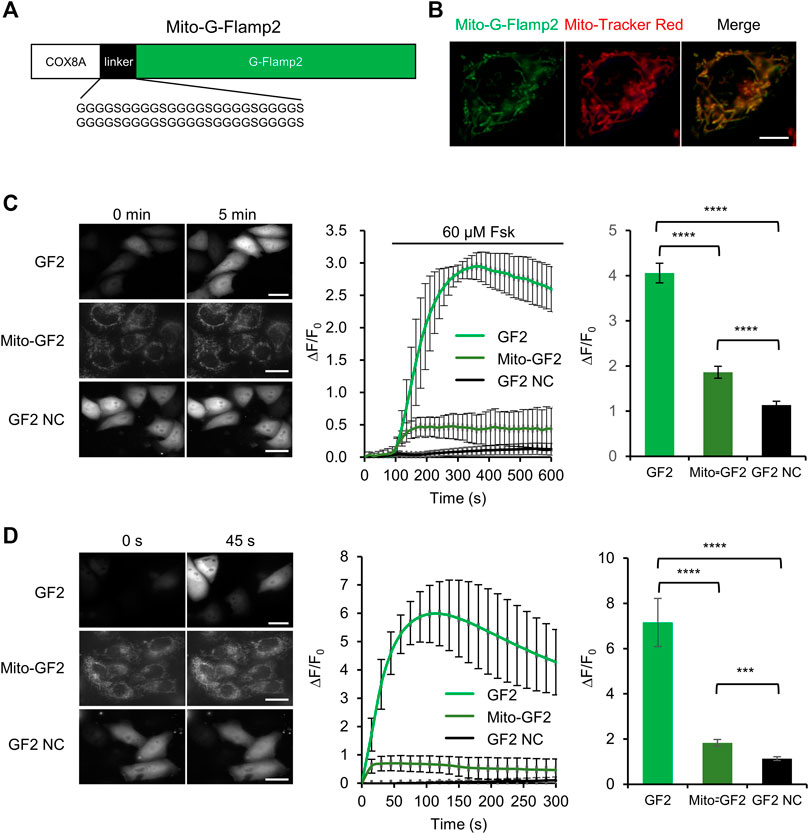
FIGURE 3. cAMP diffusion from cytosol to mitochondrial matrix in HeLa cells. (A) Schematic of mitochondrial matrix-localized G-Flamp2 (Mito-G-Flamp2) biosensor. The mitochondrial-targeting sequence from subunit VIII of the human cytochrome oxidase (COX) was fused to the N-terminus of G-Flamp2 with a flexible (G4S)10 linker. (B) Epifluorescence images of Mito-G-Flamp2 (Green) and Mito-Tracker (Red) in HeLa cells. (Scale bars, 5 μm). (C) Epifluorescence images of G-Flamp2 (GF2), Mito-G-Flamp2 (Mito-GF2) and G-Flamp2 NC (GF2 NC) to 60 µM Fsk stimulation in HeLa cells. Response of GF2, Mito-GF2 and GF2 NC to 60 µM Fsk stimulation. Quantification of peak ∆F/F0 are on the right. n = 11, 12 and 11 cells for GF2, Mito-GF2 and GF2 NC from 3 cultures for each sensor. (Scale bars, 10 μm). (D) Epifluorescence images of GF2, Mito-GF2 and GF2 NC to cAMP that is generated by bPACm in HeLa cells. The absorption peak of purified bPAC with and without light is at 453 and 441 nm, respectively, which is coincidentally consistent with the spectral-dependent G-Flamp sensors that the excitation peak at ∼ 450 nm. Response of GF2, Mito-GF2 and GF2 NC to cAMP that is generated by bPAC mutation. Quantification of peak ∆F/F0 are on the right. n = 12, 14 and 12 cells for GF2, Mito-GF2 and GF2 NC from 3 cultures for each sensor. (Scale bars, 10 μm). Data are presented as the mean ± standard deviation. *Significantly different from GF2 NC, ***p < 0.001, ****p < 0.0001 as measured by one-way analysis of variance (ANOVA) with post hoc Tukey’s test. See Supplementary Figure S9.
A potent blue light-activatable optogenetic adenylyl cyclase bPACm (a bPAC variant with much lower dark activity) (Yang et al., 2021), was also used to specifically increase the cytosolic cAMP concentration. Mito-G-Flamp2, G-Flamp2 or G-Flamp2 NC were co-expressed with bPACm-mCherry in HeLa cells and the 450 nm light was exploited to activate bPACm and excite the indicators simultaneously. Mito-G-Flamp2, G-Flamp2 or G-Flamp2 NC exhibited a peak ΔF/F0 of 60, 590 and 20%, respectively (Figure 3D, Supplementary Figure S9D–F). These results confirmed that cytosolic cAMP can enter mitochondria.
GPR50 is an orphan receptor of the GPCR family and its endogenous ligand has not been identified. It has been shown that GPR50 interacts with Gi and inhibits cAMP production in GPR50-overexpressing HEK293T cells (Levoye et al., 2006; Hand, 2012). Interestingly, GPR50 is down-regulated in breast, cervical, ovarian and lung cancers while up-regulated in liver cancer (Wojciech et al., 2018; Saha et al., 2020). Hence, GPR50-cAMP signaling pathway is a promising cancer drug target and screening ligands for GPR50 is of great importance.
Here we take the advantage of high spatiotemporal resolution of G-Flamp2 imaging to explore the dynamics of cAMP production inhibition by overexpression of GPR50 and analyze the effect of adipose tissue extracts on GPR50 activation. We first established stable 293T cells overexpressing GPR50 using the lentivirus and confirmed the expression of GPR50 at the mRNA and protein levels (Supplementary Figure S10). Then we used G-Flamp2 sensor to detect the dynamics of cAMP in living HEK293T cells. Upon 60 μM Fsk stimulation, G-Flamp2 in GPR50-expressing HEK293T cells exhibited a smaller peak fluorescence increase (ΔF/F0 of 350% vs. 680%) and larger half-rise time (400 vs. 300 s) than in control cells (Figures 4A,B). Measurements of total cAMP in cell lysate using a commercial cAMP parameter assay kit confirmed that GPR50-expressing cells produced only half amount of cAMP in control cells after a 15 min treatment of 60 μM Fsk (Figure 4C). The above results using G-Flamp2 validate that GPR50 is Gi-coupled and provide the basis for subsequent screening of GPR50 ligands by G-Flamp2.
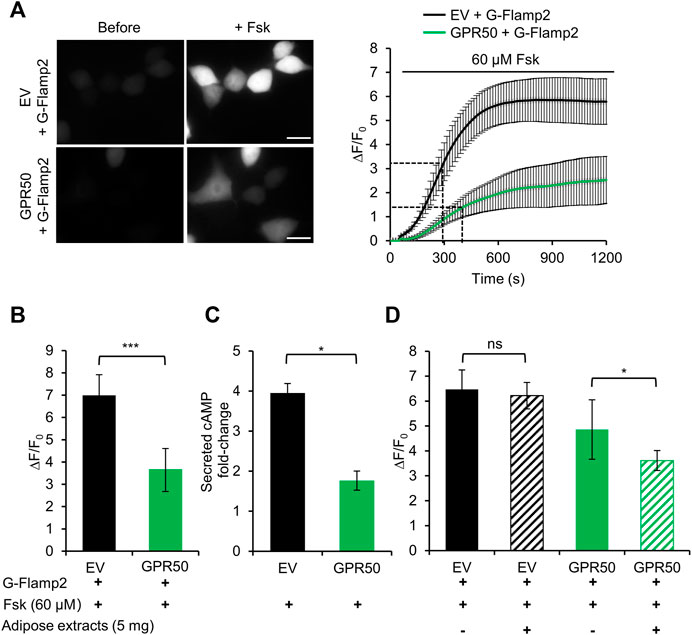
FIGURE 4. Adipose tissue extract activates the orphan GPCR GPR50 in HEK293T cells. (A) Representative fluorescence images (left) and traces of ΔF/F0 in response to 60 μM Fsk of G-Flamp2 in HEK239T cells stably expressing Empty Vector (EV) or GPR50 receptor (GPR50) (right). n = 14 and 10 cells for EV and GPR50, respectively. (B) Peak ΔF/F0 in response to 60 μM Fsk in HEK293T cells stably expressing EV or GPR50. (C) Quantification of intracellular cAMP levels in EV and GPR50 cells after 20 min in stimulation of 60 μM n = 3 cultures for both. (D) Peak ΔF/F0 in response to 5 mg/ml adipose extracts pretreatment for 5 min with 60 μM Fsk in HEK293T cells stably expressing EV or GPR50. Scale bars: 50 μm. Data are presented as the mean ± standard deviation (SD). *p < 0.05, ***p < 0.001 as measured by a Two-tailed Student’s t-test.
Next, we used this system to identify agonists or tissue fluid against GPR50. Our previous experiments demonstrated that GPR50 is highly expressed in adipose tissues (Yao et al., 2019) and adipose tissue extracts were tested here. G-Flamp2 in GPR50-expressing HEK293T cells with a 5 min pretreatment with adipose tissue extracts showed a smaller peak ΔF/F0 (360 vs. 485%) than in cells without pretreatment (Figure 4D), which suggested that GPR50 may be activated by certain compound(s) in the adipose tissue extract. To further identify the exact compound(s) activating GPR50, fractions separated by chromatography of adipose tissue extracts could be tested by G-Flamp2 imaging method. Taken together, the above results demonstrated that G-Flamp2 has high sensitivity in monitoring the inhibition effects of GPR50 on the total amount and rate of cAMP production and can be used to screen compounds against GPCR-cAMP signaling.
In this study, we engineered two G-Flamp1 variants G-Flamp2 and G-Flamp2b with improved brightness. G-Flamp2 also showed larger maximum ΔF/F0 than G-Flamp1 in HEK293T cells (ΔF/F0 ∼ 1,300% vs. 1,100%), which is also the largest among the cAMP fluorescent sensors reported. G-Flamp2/G-Flamp2b can be paired with spectrally distinct indicators (e.g. red calcium sensor) for multiplex imaging, since the single-FP indicator only occupy one fluorescence channel, which provides the possibility for simultaneous labeling and visualization of multiple molecule activities in living cells. Thus, G-Flamp2 and G-Flamp2b could be robust tools for basic cell biology research and early drug screening.
Using mitochondria-localized G-Flamp2, we observed increase of cAMP level in the mitochondrial matrix when cytosolic cAMP was increased by high concentration of Fsk or activated bPACm (a potent blue light-activatable AC) in HeLa cells, which is similar to the result in DiPilato et al. (DiPilato et al. 2004) and indicated that cytosolic cAMP can enter mitochondrial matrix. However, other studies such as Di Benedetto et al and Lefkimmiatis et al demonstrated that cAMP generated in the cytosol did not enter the mitochondrial matrix, except during mitochondrial permeability transition (Di Benedetto et al., 2013; Lefkimmiatis et al., 2013). These different observations might be due to different sensitivities of the indicators used (Wang et al., 2022). For those biological systems in which cytosolic cAMP can enter mitochondrial matrix, further studies are required to dissect the underlying mechanism.
Using the tumor-related Gi-coupled GPCR (GPR50) signaling pathway as a model, our G-Flamp2 living cell imaging experiments demonstrated that adipose tissue extract activated the orphan GPCR GPR50. These data indicated that G-Flamp2 could be a useful fluorescent probe for image-based high-content screening (HCS), which emerges as a powerful technique in both quantitative cell biology studies and drug discovery. Recently, the single-FP cAMP indicator cADDis was used to detect cAMP level changes upon the stimulation of the ligand of Gs or Gi protein-coupled GPCRs using automated microscope methods (Tewson et al., 2016; Clayton, 2018), suggesting its potential capabilities in HCS. Since G-Flamp2 has a higher affinity for cAMP (Kd of 1.9 μM vs. 10–100 μM) and larger fluorescence change (ΔF/F0 of 13 vs. −0.55) than cADDis (Klausen et al., 2019), and a relatively high brightness, G-Flamp2 would be more suitable for HCS to identify and evaluate the effect of candidate compounds targeting GPCR-cAMP signaling pathways in drug discovery.
cAMP-Na (Cat. No. A6885), cGMP-Na (Cat. No. G6129) and 3-isobutyl-1-methylxanthin (IBMX) (Cat. No. I5879) were purchased from Sigma-Aldrich (Darmstadt, Germany). cAMP (Cat. No. C107047), isoproterenol HCl (Cat. No. I129810) and propranolol (Cat. No. S133437) were purchased from Aladdin (Shanghai, China). Forskolin (Cat. No. S1612), puromycin (Cat. No. ST551), SNP (Cat. No. S0015), Mito-Tracker Red CMXRos (Cat. No. C1049B) and RIPA lysis buffer (Cat. No. P0013B) were purchased from Beyotime Biotechnology (Shanghai, China).
bPAC mutation is a gift from Shiqiang Gao, who works at Department of Biology, Institute for Molecular Plant Physiology and Biophysics, Biocenter, ulius-Maximilians-University of Würzburg, Julius-von-Sachs-Platz 2, 97,082 Würzburg, Germany. Plasmids were made using the Infusion method (Takara Bio United States, Inc.). PCR fragments were amplified using PrimerStar (normal PCR or site-directed mutagenesis) or Taq (random mutagenesis) DNA polymerases. When needed, overlap PCR was exploited to generate the intact DNA fragment for Infusion. All PCR primers were purchased from Sangon Biotechnology Co., Ltd. (Shanghai, China). DNA sequence for Green cGull and mitochondrial targeting peptide was synthesized by Genscript. To express fluorescent proteins or sensors in bacterial or mammalian cells, cDNAs of FPs or sensors were subcloned into pNCS or pCAG vector (Chu et al., 2016). The pNCS and pCAG-mEGFP were kept in our lab. For mitochondrial localization of G-Flamp2, DNA coding sequences for mitochondrial targeting peptide of COX8A (G4S)10 and G-Flamp2 was overlapped and cloned into pCAG vector. All constructs were confirmed by DNA sequencing (Sangon Biotechnology Co., Ltd., Shanghai, China).
The DNA ligation products for site-directed or random mutagenesis were transformed into DH5α cells lacking adenylate cyclase gene CyaA(DH5α-ΔCyaA). After overnight growth at 34°C, the colonies with different fluorescence intensities on the LB agar plates were screened by eye in a BlueView Transilluminator (Vernier) with the 400–500 nm excitation light and a yellow acrylic long-pass filter, or by fluorescence imaging in a home-made imaging system with 480/20 nm excitation and 520/20 nm emission filters. To quantitatively compare the brightness of selected variants, bacterial patches on the agar plates cultured overnight at 34°C were imaged in the home-made system mentioned above and analyzed by ImageJ software (National Institutes of Health). The fluorescence change (ΔF/F0) of the cAMP sensor mutants were tested as previously described (Wang et al., 2022). Briefly, selected bacterial colonies were patched on LB agar plate and grew at 25°C for 3 days. Then the bacteria from each patch were collected, suspended in HEPES buffer (pH = 7.15) and lysed by sonication. The clear lysates were then used for measuring the ΔF/F0 in response to 500 μM cAMP with an Infinite M1000 PRO fluorometer (Tecan).
Expression, purification and characterization of G-Flamp biosensor with a N-terminal 6×His tag were performed as previously reported (Wang et al., 2022). Briefly, the sensors were transformed into DH5αΔCyaA cells and cultured on SOB agar plates for 3 days at room temperature. The bacterial cells were harvested and suspended in HEPES buffer and lysed by sonication. The proteins in the clear lysates were purified using HisPur Cobalt Resin (Pierce, Rockford, IL) and desalted with HEPES buffer using the gel filtration column (Bio-Rad).
The concentrations of purified sensors were measured by the BCA assay (Bio-Rad, Hercules, CA). Excitation and emission spectra were measured with an Infinite M1000 PRO fluorometer (Tecan). Extinction coefficients were calculated using the previously described ‘base denatured chromophore’ method (Chu et al., 2016). Quantum yields were determined using mEGFP as a standard (QY = 0.60). pH titrations were performed by mixing 10 μL concentrated protein solution with 110 μL buffers of different pH ranging from 2 to 11 and the pKa was determined as described previously (Wang et al., 2022). cAMP titrations were performed by mixing 1 μM of purified protein in HEPES buffer with varying concentrations of cAMP (0.001, 0.01, 0.1, 0.5, 1, 2, 5, 10, 25, 100 and 500 μM) or cGMP (0.01, 0.1, 0.5, 1, 2, 5, 10, 25, 100, 500, 1,000 and 2000 μM) and Kd and Hill coefficient were determined as described previously (Wang et al., 2022).
HEK293T and HeLa cells were cultured in high glucose Dulbecco’s modified Eagle’s medium (Gibco, Grand Island, NY) supplemented with 10% (v/v) fetal bovine serum (Gibco, Grand Island, NY) and 100 units/mL penicillin/streptomycin (Gibco, Grand Island, NY) in a humidified incubator at 37°C with 5% CO2. Plasmid transfections of cultured cells on 6 or 12-well plates and glass-bottom coverslips were performed according to the manufacturer’s manuals of Lipofectamine 2000.
HEK293T cells grown in 12-well plates were transfected with pCAG-G-Flamp1-IRES-mCherry, pCAG-G-Flamp2-IRES-mCherry, pCAG-G-Flamp2b-IRES-mCherry construct separately using Lipofectamine 2000 48 h later, they were washed once with PBS, suspended in live cell imaging solution and transferred to a clear flat-bottom 96-well plate. The green and red fluorescence was recorded under 480 nm excitation and 560 nm excitation, respectively.
Wide-filed fluorescence imaging was carried out on an Olympus IX83 microscope equipped with a 60 ×1.4 numerical aperture (NA) objective (for Figures 2, 3) or 40 ×1.25 NA objective (for Figure 4). Briefly, cells grown on glass-bottom dishes (Cat. No. #FD35-100, World Precision Instruments) were transfected with indicated plasmids and cultured for 24 h or 48 h. Then the cells were serum starved for 4 h and culture medium was replaced with live cell imaging solution (Cat. No. A14291DJ, Invitrogen) right before imaging. The excitation and emission filters used for different sensors or fluorescent proteins were as follows: ex 480/30 nm and em 530/30 nm for Green cGull, ex 568/20 nm and em 630/50 nm for mCherry, and ex 441/20 nm and em 530/30 nm for G-Flamp1, G-Flamp2 and G-Flamp2b. Images were acquired every 15 s for Forskolin stimulation (for Figures 2, 3C, 4) and 5 s for mitochondria-targeted indicators (for Figure 3D).
Lentiviruses were used for establishing of stable cell lines expressing GPR50. Briefly, HEK293T cells were seeded into 6-well plates at a density of 1 × 106 cells/well and cultured for 24 h, infected with PLV7-GPR50 lentivirus and or PLV7 (control) lentivirus separately for 24 h and then replaced with normal medium. 72 h later, medium containing 2 μg/ml puromycin was used for stable cell line selection.
0.5 g epididymal adipose tissue from 8-week old male mice was washed with cold PBS containing gentamicin (50 mg/L) three times. Then the tissue was cut into small pieces and homogenized to grind the tissue. Lipid components were extracted with Soxhlet extractor (Palmisciano Bedê et al., 2015) and dissolved in DMSO.
Total RNA from cultured cells was extracted with TRIzol Reagent (Invitrogen, Carlsbad, CA). The qPCR reactions were performed according to the SYBR Premix Ex Taq II (Takara, Japan) protocol. RNA levels were calculated using the ΔΔCT method, where CT is the cycle threshold. The amount of β-actin mRNA was used as a reference. Forward and reverse primers for GPR50 were 5′, GCTCCGAAATTCTGGCAACAT and 5′TCAAAGGGTAGGGGTAGATGG, respectively. Forward and reverse primers for β-actin were: 5′ GTATCCATGAAATAAGTGGTTACAGG and 5′GCAGTACATAATTTACACAGCAAT, respectively.
GPR50-293T or Empty Vector-293T cells cultured on coverslips for 24 h were washed with PBS twice and fixed with 4% paraformaldehyde solution. Plasma membranes were permeabilized with 1% Triton X-100 solution for 5 min and 5% Bovine Serum Albumin (BSA, Sangon Biotech, China) was used for blocking for 30 min. Cells were then incubated with diluted primary antibody against GPR50 (#19762-AP, Proteintech, China) at 4°C overnight. Cells were then washed with PBS for three times. Then cells were incubated with Alexa-488-Donkey anti-rabbit (A21206, Invitrogen, CA) for 2 h and stained by DAPI staining solution (Beyotime, China) for 5 min in the dark at room temperature. The fluorescence images were acquired with a Olympus IX71 microscope equipped with 20 × 1.25 NA objective.
GPR50-293T or Empty Vector-293T cells were seeded into 6-well plates at a density of 1 × 106 cells/well and cultured for 24 h. Then the cells were pre-treated with 200 μM IBMX for 30 min and then stimulated with 60 μM forskolin for another 30 min to induce cAMP production. Next the cells were washed with cold PBS three times, and the amount of cAMP in cells were measured using the Parameter cAMP assay kit (R&D Systems, United Kingdom).
Analysis of the fluorescent images were performed using ImageJ software (NIH software). Background-subtracted fluorescence was used to calculate fluorescence change ΔF/F0 that is defined as (F-F0)/F0, where F0 is the baseline signal before stimulation. The statistical significances between groups were determined using the two-tailed unpaired Student’s t-test (Figure 4) or one-way ANOVA (Figures 2, 3) with GraphPad Prism 7.0 software (La Jolla, CA). *p < 0.05, **p < 0.01, ***p < 0.001 and NS (not significant) for p > 0.05.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by Ethics Committee for Animal Research, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences.
JC, PR and LW designed research; WL and CL performed experiments and analyzed the data. All authors wrote the manuscript.
This work was supported by National Key Research and Development Program of China (2017YFA0700403, 2021YFA0719303), National Natural Science Foundation of China (Grant 31670872, 21874145, 2018M633180, 32000732), Shenzhen Science and Technology Program (JCYJ20200109115441918).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.902290/full#supplementary-material
Acin-Perez, R., Salazar, E., Kamenetsky, M., Buck, J., Levin, L. R., and Manfredi, G. (2009). Cyclic AMP Produced inside Mitochondria Regulates Oxidative Phosphorylation. Cell Metab 9 (3), 265–276. doi:10.1016/j.cmet.2009.01.012
Börner, S., Schwede, F., Schlipp, A., Berisha, F., Calebiro, D., Lohse, M. J., et al. (2011). FRET Measurements of Intracellular cAMP Concentrations and cAMP Analog Permeability in Intact Cells. Nat. Protoc. 6 (4), 427–438. doi:10.1038/nprot.2010.198
Chu, J., Oh, Y., Sens, A., Ataie, N., Dana, H., Macklin, J. J., et al. (2016). A Bright Cyan-Excitable orange Fluorescent Protein Facilitates Dual-Emission Microscopy and Enhances Bioluminescence Imaging In Vivo. Nat. Biotechnol. 34 (7), 760–767. doi:10.1038/nbt.3550
Clayton, J. (2018). Characterizing Gs-And Gi-dependent Regulation of cAMP Levels in Real Time with cADDis Biosensor and Automated Live Cell Imaging.
Cooper, D. M. (2003). Regulation and Organization of Adenylyl Cyclases and cAMP. Biochem. J. 375 (Pt 3), 517–529. doi:10.1042/BJ20031061
Di Benedetto, G., Scalzotto, E., Mongillo, M., and Pozzan, T. (2013). Mitochondrial Ca²⁺ Uptake Induces Cyclic AMP Generation in the Matrix and Modulates Organelle ATP Levels. Cel Metab 17 (6), 965–975. doi:10.1016/j.cmet.2013.05.003
DiPilato, L. M., Cheng, X., and Zhang, J. (2004). Fluorescent Indicators of cAMP and Epac Activation Reveal Differential Dynamics of cAMP Signaling within Discrete Subcellular Compartments. Proc. Natl. Acad. Sci. U S A. 101 (47), 16513–16518. doi:10.1073/pnas.0405973101
Gold, M. G., Gonen, T., and Scott, J. D. (2013). Local cAMP Signaling in Disease at a Glance. J. Cel Sci 126 (Pt 20), 4537–4543. doi:10.1242/jcs.133751
Hackley, C. R., Mazzoni, E. O., Blau, J., and cAMPr, (2018). cAMPr: A Single-Wavelength Fluorescent Sensor for Cyclic AMP. Sci. Signal. 11 (520). doi:10.1126/scisignal.aah3738
Hand, L. (2012). The Biology of GPR50, the Melatonin-Related Receptor. Manchester: University of Manchester.
Harada, K., Ito, M., Wang, X., Tanaka, M., Wongso, D., Konno, A., et al. (2017). Red Fluorescent Protein-Based cAMP Indicator Applicable to Optogenetics and In Vivo Imaging. Sci. Rep. 7 (1), 7351. doi:10.1038/s41598-017-07820-6
Kitaguchi, T., Oya, M., Wada, Y., Tsuboi, T., and Miyawaki, A. (2013). Extracellular Calcium Influx Activates Adenylate Cyclase 1 and Potentiates Insulin Secretion in MIN6 Cells. Biochem. J. 450 (2), 365–373. doi:10.1042/BJ20121022
Klausen, C., Kaiser, F., Stüven, B., Hansen, J. N., and Wachten, D. (2019). Elucidating Cyclic AMP Signaling in Subcellular Domains with Optogenetic Tools and Fluorescent Biosensors. Biochem. Soc. Trans. 47 (6), 1733–1747. doi:10.1042/BST20190246
Lefkimmiatis, K., Leronni, D., and Hofer, A. M. (2013). The Inner and Outer Compartments of Mitochondria Are Sites of Distinct cAMP/PKA Signaling Dynamics. J. Cel Biol 202 (3), 453–462. doi:10.1083/jcb.201303159
Lefkimmiatis, K., and Zaccolo, M. (2014). cAMP Signaling in Subcellular Compartments. Pharmacol. Ther. 143 (3), 295–304. doi:10.1016/j.pharmthera.2014.03.008
Levoye, A., Dam, J., Ayoub, M. A., Guillaume, J. L., Couturier, C., Delagrange, P., et al. (2006). The Orphan GPR50 Receptor Specifically Inhibits MT1 Melatonin Receptor Function through Heterodimerization. EMBO J. 25 (13), 3012–3023. doi:10.1038/sj.emboj.7601193
Massengill, C. I., Ceballos, C. C., Cebul, E. R., Qin, M., Ye, B., Mao, T., et al. (2021). Highly Sensitive Genetically-Encoded Sensors for Population and Subcellular Imaging of cAMP in Vivo. bioRxiv. doi:10.1101/2021.08.27.457999 [pre-print]
Matsuda, S., Harada, K., Ito, M., Takizawa, M., Wongso, D., Tsuboi, T., et al. (2017). Generation of a cGMP Indicator with an Expanded Dynamic Range by Optimization of Amino Acid Linkers between a Fluorescent Protein and PDE5α. ACS Sens 2 (1), 46–51. doi:10.1021/acssensors.6b00582
Mattiazzi Usaj, M., Styles, E. B., Verster, A. J., Friesen, H., Boone, C., and Andrews, B. J. (2016). High-Content Screening for Quantitative Cell Biology. Trends Cel Biol 26 (8), 598–611. doi:10.1016/j.tcb.2016.03.008
Mukherjee, S., Jansen, V., Jikeli, J. F., Hamzeh, H., Alvarez, L., Dombrowski, M., et al. (2016). A Novel Biosensor to Study cAMP Dynamics in Cilia and Flagella. Elife 5. doi:10.7554/elife.14052
Nimigean, C. M., Shane, T., and Miller, C. (2004). A Cyclic Nucleotide Modulated Prokaryotic K+ Channel. J. Gen. Physiol. 124 (3), 203–210. doi:10.1085/jgp.200409133
Odaka, H., Arai, S., Inoue, T., and Kitaguchi, T. (2014). Genetically-encoded Yellow Fluorescent cAMP Indicator with an Expanded Dynamic Range for Dual-Color Imaging. PLoS One 9 (6), e100252. doi:10.1371/journal.pone.0100252
Ohta, Y., Furuta, T., Nagai, T., and Horikawa, K. (2018). Red Fluorescent cAMP Indicator with Increased Affinity and Expanded Dynamic Range. Sci. Rep. 8 (1), 1866. doi:10.1038/s41598-018-20251-1
Palmisciano Bedê, T., Pascoal, A. C., Hauaji Facó, L., de Salvo Castro, E., Mattoso, V., Furtado Dias, J., et al. (2015). Effect of the Intake of Liquids Rich in Polyphenols on Blood Pressure and Fat Liver Deposition in Rats Submitted to High-Fat Diet. Nutr. Hosp. 31 (6), 2539–2545. doi:10.3305/nh.2015.31.6.8655
Ponsioen, B., Zhao, J., Riedl, J., Zwartkruis, F., van der Krogt, G., Zaccolo, M., et al. (2004). Detecting cAMP-Induced Epac Activation by Fluorescence Resonance Energy Transfer: Epac as a Novel cAMP Indicator. EMBO Rep. 5 (12), 1176–1180. doi:10.1038/sj.embor.7400290
Saha, S. K., Choi, H. Y., Yang, G. M., Biswas, P. K., Kim, K., Kang, G. H., et al. (2020). GPR50 Promotes Hepatocellular Carcinoma Progression via the Notch Signaling Pathway through Direct Interaction with ADAM17. Mol. Ther. Oncolytics 17, 332–349. doi:10.1016/j.omto.2020.04.002
Steegborn, C. (2014). Structure, Mechanism, and Regulation of Soluble Adenylyl Cyclases - Similarities and Differences to Transmembrane Adenylyl Cyclases. Biochim. Biophys. Acta 1842 (12 Pt B), 2535–2547. doi:10.1016/j.bbadis.2014.08.012
Tewson, P. H., Martinka, S., Shaner, N. C., Hughes, T. E., and Quinn, A. M. (2016). New DAG and cAMP Sensors Optimized for Live-Cell Assays in Automated Laboratories. J. Biomol. Screen. 21 (3), 298–305. doi:10.1177/1087057115618608
Valsecchi, F., Ramos-Espiritu, L. S., Buck, J., Levin, L. R., and Manfredi, G. (2013). cAMP and Mitochondria. Physiology (Bethesda) 28 (3), 199–209. doi:10.1152/physiol.00004.2013
Wang, L., Wc, C., Peng, W., Zhou, Z., Zeng, J., Li, X., et al. (2022). A High-Performance Genetically Encoded Fluorescent Indicator for In Vivo cAMP Imaging. bioRxiv. doi:10.1101/2022.02.27.482140 [pre-print]
Wojciech, S., Ahmad, R., Belaid-Choucair, Z., Journé, A. S., Gallet, S., Dam, J., et al. (2018). The Orphan GPR50 Receptor Promotes Constitutive TGFβ Receptor Signaling and Protects against Cancer Development. Nat. Commun. 9 (1), 1216. doi:10.1038/s41467-018-03609-x
Yang, S., Constantin, O. M., Sachidanandan, D., Hofmann, H., Kunz, T. C., Kozjak-Pavlovic, V., et al. (2021). PACmn for Improved Optogenetic Control of Intracellular cAMP. BMC Biol. 19 (1), 227. doi:10.1186/s12915-021-01151-9
Yao, Z., Liu, C., Yu, X., Meng, J., Teng, B., Sun, Y., et al. (2019). Microarray Profiling and Coexpression Network Analysis of Long Noncoding RNAs in Adipose Tissue of Obesity-T2dm Mouse. Obesity (Silver Spring) 27 (10), 1644–1651. doi:10.1002/oby.22590
Zaccolo, M., Zerio, A., and Lobo, M. J. (2021). Subcellular Organization of the cAMP Signaling Pathway. Pharmacol. Rev. 73 (1), 278–309. doi:10.1124/pharmrev.120.000086
Zaccolo, M. (2021). cAMP Buffering via Liquid-Liquid Phase Separation. Function (Oxf) 2 (1), zqaa048.
Keywords: single-FP indicator, fluorescence imaging, circular permutation, cAMP, GPCR, drug discovery, high-content screening
Citation: Liu W, Liu C, Ren P-G, Chu J and Wang L (2022) An Improved Genetically Encoded Fluorescent cAMP Indicator for Sensitive cAMP Imaging and Fast Drug Screening. Front. Pharmacol. 13:902290. doi: 10.3389/fphar.2022.902290
Received: 23 March 2022; Accepted: 18 April 2022;
Published: 12 May 2022.
Edited by:
Qi Zeng, Xidian University, ChinaReviewed by:
Zhixing Chen, Peking University, ChinaCopyright © 2022 Liu, Liu, Ren, Chu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pei-Gen Ren, cGcucmVuQHNpYXQuYWMuY24=; Jun Chu, anVuLmNodUBzaWF0LmFjLmNu; Liang Wang, bGlhbmcud2FuZzFAc2lhdC5hYy5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.