- 1Genetics and Bioinformatics Department, Dasman Diabetes Institute, Kuwait City, Kuwait
- 2Narcotic and Psychotropic Department, Ministry of Interior, Farwaniya, Kuwait
Human leukocyte antigen (HLA) proteins are present at the cellular surface of antigen-presenting cells and play a crucial role in the adaptive immune response. Class I genes, specifically certain HLA-B alleles, are associated with adverse drug reactions (ADRs) and are used as pharmacogenetic markers. Although ADRs are a common causes of hospitalization and mortality, the data on the prevalence of HLA-B pharmacogenetics markers in Arab countries are scarce. In this study, we investigated the frequencies of major HLA-B pharmacogenomics markers in the Qatari population. Next-generation sequencing data from 1,098 Qatari individuals were employed for HLA-B typing using HLA-HD version 1.4.0 and IPD-IMGT/HLA database. In addition, HLA-B pharmacogenetics markers were obtained from the HLA Adverse Drug Reaction Database. In total, 469 major HLA-B pharmacogenetic markers were identified, with HLA-B*51:01 being the most frequent pharmacogenetic marker (26.67%) in the Qatari population. Moreover, HLA-B*51:01 is associated with phenytoin- and clindamycin-induced ADRs. The second most frequent pharmacogenetic marker was the HLA-B*58:01 allele (6.56%), which is associated with allopurinol-induced ADRs. The third most frequent pharmacogenetic marker was the HLA-B*44:03 allele, which is associated with phenytoin-induced ADRs. The establishment of a pharmacogenetics screening program in Qatar for cost effective interventions aimed at preventing drug-induced hypersensitivity can be aided by the highly prevalent HLA-B pharmacogenetic markers detected here.
Introduction
Human leukocyte antigen (HLA) proteins are encoded by the HLA gene complex, which is located on the short arm of chromosome 6, and are inherited from both parents, one being paternal and the other being maternal (Choo, 2007). These molecules are present at the cellular surface of antigen-presenting cells and play a crucial role in the adaptive immune response. The HLA antigens are classified into three classes according to their coding gene locus, function, expression, and biochemical characteristics (classes I, II, and III). The classical HLA loci consist of class I molecules (HLA-A, -B, and -C); class II molecules, which are encoded by six main genes, namely, HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQB1, HLA-DRA, and HLA-DRB1; and class III antigens, which are encoded by the HLA-Bf, C2, and C4A loci (Ulvestad et al., 1994; Howell et al., 2010). Class I molecules present peptides derived from intracellular proteins to cytotoxic T cells (CD8+), whereas class II peptides present internalized exogenous proteins to T helper cells (CD4+) (Reviewed by Dendrou et al., 2018).
The HLA genes are the most polymorphic genetic region in the human genome, as they need to present a huge variety of peptides. Different populations and ethnic groups were shown to encode different HLA alleles. In addition, there is a cumulative body of evidence showing that certain HLA-B alleles variants can be used as pharmacogenomic markers to predict adverse drug reactions (ADRs) and hypersensitivities, as several drugs were shown to induce immune hypersensitivity responses through interactions with HLA-B proteins (Jung et al., 2018). Such allele variants, which are significantly associated with responses to specific types of drugs, are referred to as pharmacogenetic markers. It has been observed that pharmacogenetic markers are usually drug-specific, phenotype-specific, and population-specific markers (Supplementary Table S1).
ADRs were defined by Edwards and Aronson (2000) as “an appreciably harmful or unpleasant” reaction resulting from an intervention related to the use of a medicinal product, which predicts hazard from future administration and warrants prevention/specific treatment, alteration of the dosage regimen, or withdrawal of the product. ADRs are considered a major health issue as they are a common cause of hospitalization and mortality (Naisbitt et al., 2003; Gomes and Demoly, 2005). Traditionally, ADRs have been classified into two major types, i.e., Type A or augmented reaction, and Type B or bizarre reaction. Type A is dose dependent and is predicted based on the pharmacology of the drug, whereas Type B is idiosyncratic and is unpredicted based on pharmacology, a it is primarily determined by host genetics. Type B, although less frequent (representing approximately 10%–15% of all ADR cases) than Type A, is relatively more severe (Pirmohamed, 2010). Hypersensitivity drug reactions, which are a Type B reaction, have clinical manifestations, with cutaneous adverse drug reactions (CADRs) being the most common. Some of the CADRs can be classified as severe cutaneous adverse reactions (SCARs), including Stevens–Johnson syndrome (SJS), toxic epidermal necrolysis (TEN), drug reactions with eosinophilia and systemic symptoms (DRESS), drug-induced hypersensitivity syndrome, or hypersensitivity syndrome (HSS) (Sukasem et al., 2014). These SCARs (with no exception) are characterized by considerable rates of morbidity and mortality. However, each SCAR has its own characteristic cutaneous presentations, causative drugs, clinical courses, pathomechanisms, and possible treatment modalities (Pichler et al., 2011; Wei et al., 2012).
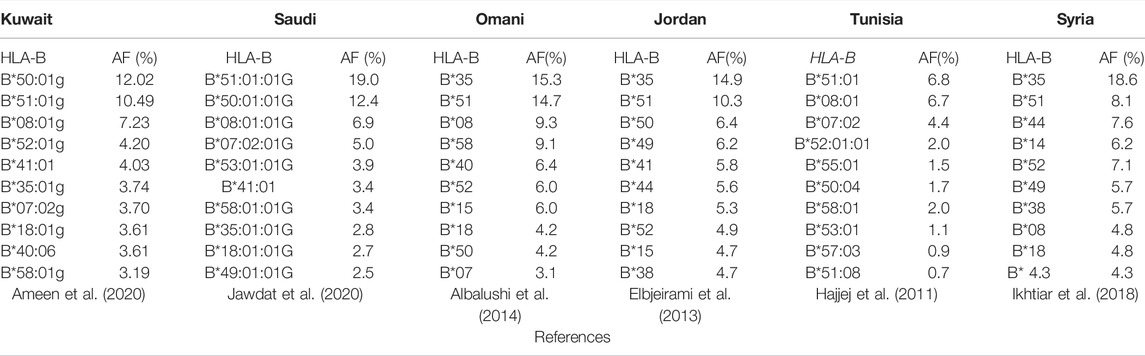
A previous study conducted on the Kuwaiti population showed that B*50:01g is one of the most common groups of HLA-B alleles, whereas in Saudi Arabia, B*51:01:01G (19.0%), and B*50:01:01G (12.3%) are the most common groups of HLA-B alleles (Ameen et al., 2020; Jawdat et al., 2020). Upper G denotes to merged group of exons 2 and 3 for HLA class I and exon 2 for HLA class II alleles with identical nucleotide sequences (Marsh et al., 2010). Lower g denotes to merged group of alleles that differ in nonsynonymous mutations outside the relevant exons, and synonymous mutations inside or outside the relevant exons (Schmidt et al., 2009; Schaefer et al., 2017). Moreover, in other Arab populations, the B*35 group of alleles was detected at high frequencies among Omanis (15.3%), Jordanians (14%), and Arab Emirati (11.1%) (Sanchez-Velasco et al., 2001; Elbjeirami et al., 2013; Albalushi et al., 2014). B*51 is another group of alleles that has been found at high frequencies among Saudis (19.3%), Omanis (14.7%), and Arab Emiratis (15.6%) (Sanchez-Velasco et al., 2001; Albalushi et al., 2014). In addition, B*50 is also a frequent allele group in most Arabs, including Saudis (18.8%) and Libyans (16.1%), together with B*08 and B*44 among the Tunisian Berbers of Zrawa (32.8%), with the latter being the highest frequency recorded worldwide (Sanchez-Velasco et al., 2001; Hajjej et al., 2011). In contrast, the B*37, 42, 46, 47, 48, 54, 59, 67, and 78 allele groups are extremely rare or virtually absent in all Arab populations (Hajjej et al., 2011). Among Syrians, the frequency of the B*35 allele group is 18.6%, whereas that of B*44 is 7.6% and that of B*51 is 8.1% (Ikhtiar et al., 2018). Table 1 shows the top 10 most frequent HLA-B alleles in the Arab population.
Unfortunately, the published data connecting the frequencies of HLA-B haplotypes and alleles with pharmacogenetics in the Arab countries are scarce. Providing such data would lead to a better understanding of the pharmacogenomics of the Arab population and would further shed light on associations between ADRs and HLA. Thus, the current study will highlight this issue.
Materials and Methods
Ethics Statement
The current study was reviewed and approved by the institutional Ethical Review Committee at the Dasman Diabetes Institute, Kuwait in accordance with the declaration of Helsinki. The genomic data of Qatari individuals used in this study are publicly available from the National Center for Biotechnology Information Sequence Read Archive. Informed consent was obtained from the recruited participants of the original studies (Fakhro et al., 2016; Rodriguez-Flores et al., 2016) under protocols approved by the Institutional Review Boards of Hamad Medical Corporation and Weill Cornell Medical College in Qatar.
Study Samples
The whole exome sequencing data of individuals living in Qatar (Fakhro et al., 2016; Rodriguez-Flores et al., 2016), as sequenced using Agilent SureSelect Human All Exon V5 and V4 kits (Agilent Technologies Inc., United States) on an Illumina HiSeq platform, are publicly available from the National Center for Biotechnology Information Sequence Read Archive, with the following accessions: SRP060765, SRP061943, and SRP061463. Only 1,000 exomes of native Qatari individuals were considered for this study as they were sequenced with the Agilent V5 kit (Agilent Technologies Inc., United States). Furthermore, 98 native Qatari individuals, whose whole genome sequence data were available, were also used in our study. In total, 1,098 individuals were enrolled in this study: 475 males (43.57%) and 623 females (57.15%) with an average age of 50 years. Three samples were excluded from this study as they failed to meet the quality control threshold resulting in untyped HLA-B alleles, whereas HLA-B loci were successfully sequenced in the samples of the remaining 1,095 individuals.
Quality Control
Furthermore, we downloaded the whole genome sequencing data from an additional eight individuals (common with whole exome samples, sharing the same sample identification number) available from the same Qatari studies (Fakhro et al., 2016; Rodriguez-Flores et al., 2016) and were used to validate the HLA typing from whole exome data.
HLA-B Typing
The FastQ files of 1,098 Qatari individuals were used as input for the HLA-HD tool version 1.4.0 (Kawaguchi et al., 2017), to determine the HLA alleles by mapping reads to a comprehensive reference panel from the IPD-IMGT/HLA database (Robinson et al., 2015) version 3.46 (2021-October) build 2d19adf. The companion website to the official repository is hla.alleles.org. The data are also accessible at https://www.ebi.ac.uk/ipd/imgt/hla/licence/.
HLA-B Pharmacogenomic Markers
Major HLA-B pharmacogenetic markers were obtained from the HLA Adverse Drug Reaction Database (HLA-ADR) website (http://www.allelefrequencies.net/).
Statistical Analysis
HLA-B allele frequency (AF) calculations were carried out by direct counting, followed by dividing the total number of occurrences of the specific allele by the total number of B alleles in a population. HLA-B AF are assessed for deviation from Hardy–Weinberg equilibrium (HWE) using the R software (https://www.R-project.org/) version 3.6.2.
Results
HLA-B Allelic Frequencies
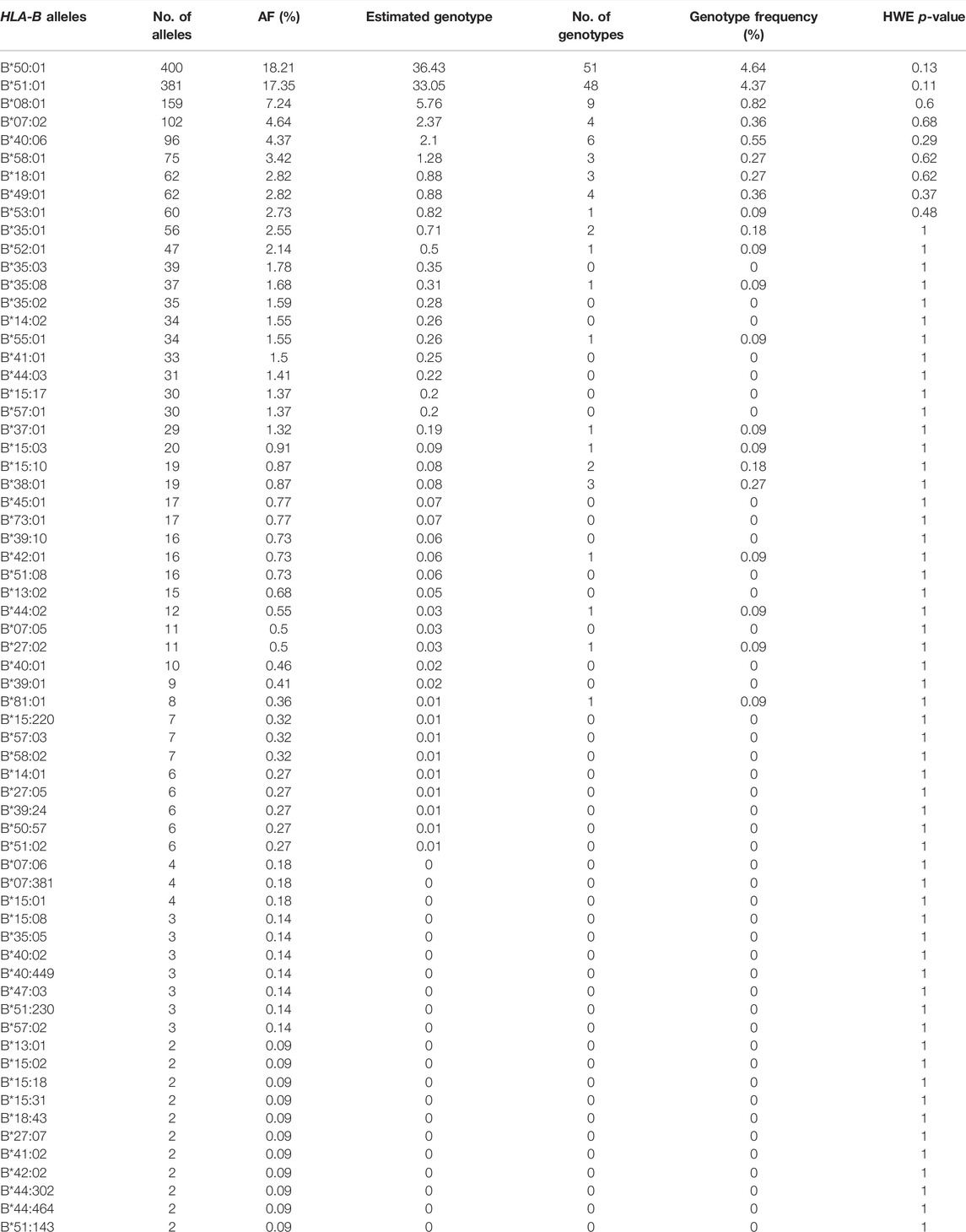
After demonstrating the effectiveness of HLA-B typing using next-generation sequencing (NGS) data (Supplementary Table S2), the frequency of the 107 different HLA-B alleles identified in the 1,098 Qatari individuals are listed in Supplementary Table S3 and Table 2 (n > 1). The most frequent HLA-B alleles were B*50:01 (18.21%), B*51:01 (17.35%), B*08:01 (7.24%), B*07:02 (4.64%), B*40:06 (4.37%), and B*58:01 (3.42%). HLA-B AF did not significantly deviate from HWE.
HLA-B Genotype Frequencies
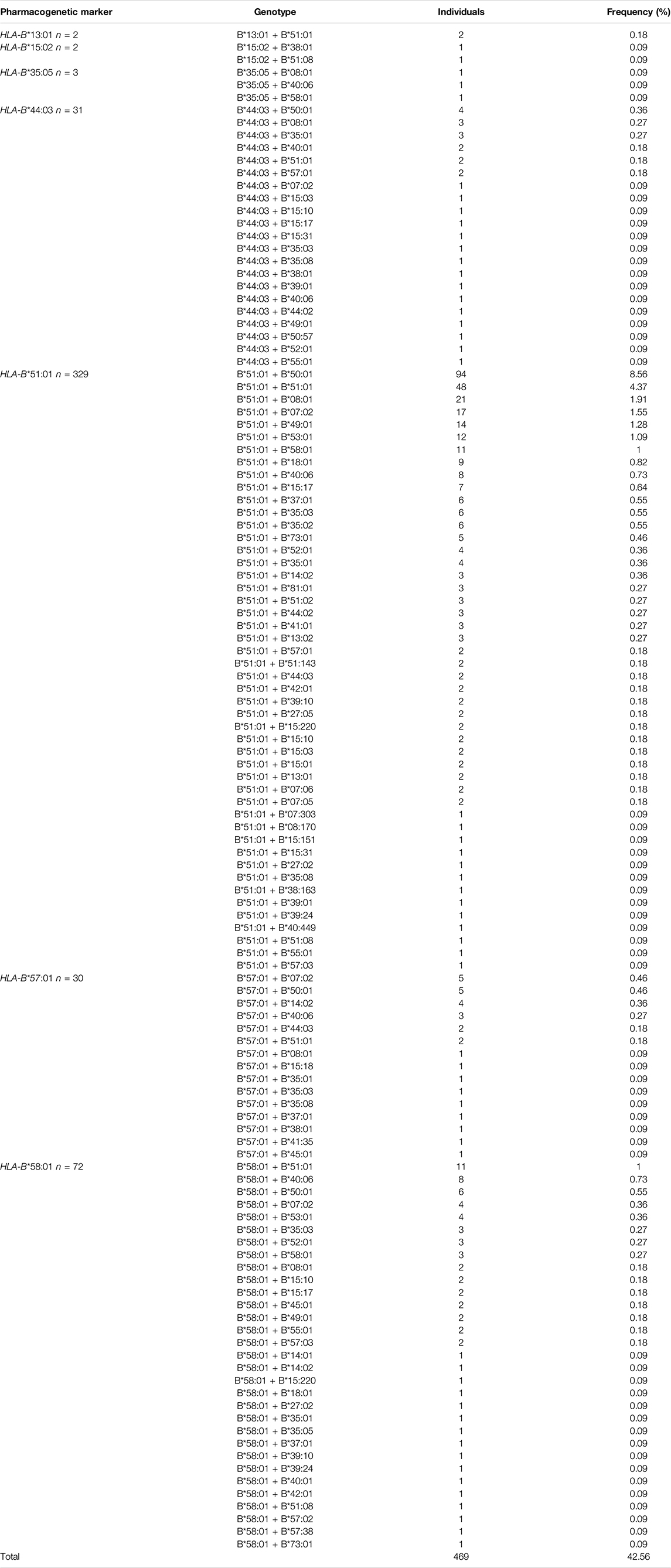
428 different HLA-B genotypes were identified in the 1,098 Qatari individuals. The top 10 most frequent HLA-B genotypes are listed in Table 3. The B*50:01 + B*51:01 genotype emerged as the most common genotype at a frequency of 8.56% in the Qatari population. The remainder of the frequent genotypes exhibited a frequency of <5% in the Qatari population.
Frequency of Major HLA-B Pharmacogenetics Markers in the Qatari Population
In total, 469 major HLA-B pharmacological markers were identified when we screened the HLA-B markers from the 1,098 Qatari individuals (Table 3). The most frequent pharmacogenetic marker in the Qatari population was HLA-B*51:01 (26.67%), with 329 individuals carrying this marker, which is associated with phenytoin- and clindamycin-induced ADRs. Among them, 55% were women and 45% were men. In contrast, only 48 Qatari individuals were shown to carry the homozygous HLA-B*51:01 genotype, with the remainder of the cohort carrying heterozygous genotypes. The second most frequent pharmacogenetic marker was the HLA-B*58:01 allele, which was carried by 72 Qatari individuals (6.56%) and is associated with allopurinol-induced CADRs; 58% of the individuals were women and 42% were men. Furthermore, only three individuals carried the homozygous HLA-B*58:01 genotype and 69 individuals carried the heterozygous B*58:01 genotype. The third most frequent pharmacogenetic marker was the HLA-B*44:03 allele, which is associated with phenytoin-induced ADRs, with 61% of the individuals who carried this allele being women and 39% being men. Moreover, none of the Qatari individuals carried the homozygous HLA-B*44:03 genotype. The fourth most frequent pharmacogenetic marker was the HLA-B*57:01 allele (2.72%), which is involved in abacavir-induced hypersensitivity syndrome (AHS); 73% of the carriers were women and 27% were men. Finally, none of the Qatari individuals carried the homozygous HLA-B*57:01 genotype.
Discussion
The Middle East constitutes to be a historic intersection of human civilizations and migrations. Qatar exhibits extensive diversity as well as genetic ancestries representing the main founding Arab genealogical lineages of Qahtanite (Peninsular Arabs) and Adnanite (General Arabs and West Eurasian Arabs) (Razali et al., 2021). A principal components analysis on genome-wide genotype data from Qatar revealed three clear clusters of genotypes, the proximity of which to other human population samples is consistent with an Arabian origin, a more eastern or Persian origin, and individuals with African admixture (Hunter-Zinck et al., 2010).
In the current study, we employed the publicly available NGS data for the Qatari population, with the majority being from the whole exome of 1,098 individuals, to report the HLA-B locus diversity in the Qatari population for the first time.
Several previous studies were conducted on the HLA-B AF, covering different GCC countries, including Oman, Saudi Arabia, Kuwait, and the United Arab Emirates (Williams et al., 2001; Hajeer et al., 2013; Ameen et al., 2020; Jawdat et al., 2020). In addition, similar studies were conducted in other Arabic countries, including Libya, Tunisia, Syria, and Jordan (Elbjeirami et al., 2013; Hajjej et al., 2015; Jazairi et al., 2016; Ikhtiar et al., 2018). However, there is a lack of data regarding AF in Qatar compared with the remainder of the Arabic countries (Table 1). Thus, our study was the first to shed light on HLA diversity in this population.
The analysis of the obtained NGS data has demonstrated a good coverage across the major histocompatibility complex (MHC) region, which resulted in high-resolution HLA-B typing (three fields). Therefore, this can be used to discriminate between samples (Supplementary Table S2) specifically from whole exome data.
In our study, we identified 107 allele types from the delineated NGS data that were generated from 1,098 Qatari individuals; furthermore, by analyzing the frequencies of those alleles we showed that they passed the tests for HWE (Table 4). The most frequently observed HLA-B alleles were B*50:01 (18.21%), B*51:01 (17.35%), B*08:01 (7.24%), and B*07:02 (4.64%). These most frequent HLA-B alleles and genotype were very similar to the findings reported for the Kuwaiti (Ameen et al., 2020), Saudi Arabian (Hajeer et al., 2013; Jawdat et al., 2020), and Omani (Williams et al., 2001) populations, and exhibited comparable frequencies to those of the surrounding Arab populations, such as the Jordanian (Elbjeirami et al., 2013), Tunisian (Hajjej et al., 2015), and Syrian (Jazairi et al., 2016; Ikhtiar et al., 2018) populations. This was not the case regarding HLA-B allele studies in other populations from Thailand (Puangpetch et al., 2015), China (Middleton et al., 2004), Singapore (Williams et al., 2001), Malaysia (Jinam et al., 2010), and European American (Creary et al., 2019), which are not close geographically. The sharing of the most frequent alleles among GCC countries compared with other Arab countries could be mainly attributed to a similar gene pool; the close geographical location of the GCC countries; and the sharing of similar culture, language, religion, history, and ancestors.
In addition, to the best of our knowledge, none of the previous studies of HLA-B AF in Arab countries (Williams et al., 2001; Elbjeirami et al., 2013; Hajjej et al., 2015; Ikhtiar et al., 2018; Ameen et al., 2020; Jawdat et al., 2020) investigated or reported pharmacogenomic data pertaining to genetic markers for genetic disorders, such as drug hypersensitivity. Thus, our study was the first of its kind to investigate and report genetic markers that were associated with ADRs. Moreover, obtaining such data by screening patients for HLA-B alleles is very useful, as it is a cost effective intervention for the prevention of ADRs.
Our data (Table 3) showed that HLA-B*51:01 was the most prevalent pharmacogenetic marker allele in the studied Qatari population. In the Han Chinese population, this genetic marker has been shown to be strongly associated with CADRs related to clindamycin, which is an antibiotic that is used for the treatment of several bacterial infections, including bone or joint infections, pelvic inflammatory disease, strep throat, pneumonia, middle ear infections, and endocarditis (Guay, 2007; Yang et al., 2017). Moreover, in the Thai population, it is also significantly associated with SCARs (such as SJS/TEN and DRESS) related to phenytoin (PHT), which is sold under the brand name Dilantin among others and is an anti-seizure medication that is useful for the prevention of tonic–clonic seizures (also known as grand mal seizures) and focal seizures (Tassaneeyakul et al., 2016). In addition, a very recent study demonstrated the association between HLA-B*51:01, HLA-B*55:01, and CYP2C9*3 and phenytoin-induced CADRs in the South Indian Tamil population (John et al., 2021). Another phenytoin-induced ADR allele is HLA-B*15:02, which also has been associated with other antiepileptic drugs, such as carbamazepine, in populations with Asian ancestry (Sukasem et al., 2018) and in the Spanish population (Ramirez et al., 2017); however, it had a rare prevalence in our study.
The HLA-B*57:01 and HLA-B*35:05 alleles, which have been found to be associated with antiviral-drug-induced hypersensitivity, were detected in 30 and 3 Qatari individuals, respectively. The HLA-B*57:01 allele was found to be associated with abacavir in Caucasians (Mallal et al., 2008), Western Australians (Mallal et al., 2002), and other populations (Hetherington et al., 2002; Hughes et al., 2004; Martin et al., 2004; Phillips et al., 2005). Abacavir (ABC) is an antiretroviral drug of the nucleoside reverse transcriptase inhibitor class, which is commonly combined with antiretroviral therapy for treating human immunodeficiency virus (HIV) infection. Abacavir acts as a potent guanosine nucleoside inhibitor of reverse transcriptase. However, approximately 5%–8% of patients experience a hypersensitivity reaction within the first 6 weeks of treatment, which can be severe and potentially life threatening (Ma et al., 2010). The other allele that has been shown to be associated with antiviral-drug-induced hypersensitivity is HLA-B*35:05, which was shown to be associated with nevirapine in the Thai population (Chantarangsu et al., 2009). Nevirapine (NVP) is sold under the brand name of Viramune and is used to treat and prevent HIV infection, specifically HIV-1. It is a non-nucleoside reverse transcriptase inhibitor that works by blocking the function of reverse transcriptase. The most common adverse effect of nevirapine is the development of a mild or moderate rash (13%). SCARs, including SJS/TEN and hypersensitivity, have been observed in 1.5% of patients (Pawar et al., 2015).
Moreover, our study showed that the HLA-B*58:01 allele had a high prevalence (72 individuals) in the Qatari population. This allele was shown to be associated with allopurinol-induced SJS/TEN in Asian populations, such as the Taiwanese (Ko et al., 2015), Korean (Kang et al., 2011), Japanese (Kaniwa et al., 2008), Thai (Tassaneeyakul et al., 2009), and some European (such as the Portuguese) (Goncalo et al., 2013) populations. This medication is used to decrease the elevated blood uric acid levels triggered by chemotherapy and to prevent gout and specific types of kidney stones (Jung et al., 2015).
The HLA-B*44:03 allele, which was carried by 2.8% of Qataris in our study, has been associated with cold-medicine-induced SJS/TEN in the Brazilian (Wakamatsu et al., 2015) and Japanese (Ueta et al., 2014) populations. Cold medicines include non-steroidal anti-inflammatory drugs and multi-ingredient cold medications. In another study, Park et al. (2016) stated that HLA-B*44:03 may be associated with lamotrigine-induced SJS/TEN among Koreans. Lamotrigine, which is sold under the brand name of lamictal, is used to treat epilepsy, including focal seizures, tonic–clonic seizures, and seizures in Lennox–Gastaut syndrome, and to prevent the recurrence of depressive–manic episodes in patients with bipolar disorder (Lorberg et al., 2009).
The HLA-B*13:01 allele was observed at an extremely low frequency in our study (two Qatari individuals) and was shown by others to be associated with Dapsone-induced SJS, TEN, and DRESS in Asian populations (Zhang et al., 2013; Tempark et al., 2017). Dapsone is used for the treatment of infection and inflammation, including leprosy, Pneumocystis jiroveci pneumonia, or Toxoplasma gondii encephalitis, in HIV prophylaxis, neutrophilic dermatoses, dermatitis herpetiformis, and autoimmune bullous disease (Tangamornsuksan and Lohitnavy, 2018). Dapsone hypersensitivity syndrome (DHS), or dapsone-induced hypersensitivity reactions, is a life threatening drug reaction that usually manifests itself between the 4th and 6th weeks after the initiation of treatment. Approximately 0.5%–3.6% of patients treated with dapsone have been reported to develop DHS, with a mortality rate of 9.9% (Fan et al., 2017; Tangamornsuksan and Lohitnavy, 2018).
The current study had several limitations, one of which was the absence of association studies between HLA-B alleles and medical drugs in the Middle East. As a result, some of the pharmacogenetic markers discussed in our study might be ethnicity specific based on the genetic background. For instance, in the Chinese (Chung et al., 2004), Thai (Tassaneeyakul et al., 2010), and European (Lonjou et al., 2008) populations, the association between the HLA-B*15:02 allele and carbamazepine-induced ADRs was significant, which was not the case in the Korean population (Kim et al., 2011). Although the NGS-based HLA typing has good resolution and we used duplicate samples (whole genome and whole exome) that showed consistent results, and despite the high sensitivity and specificity of the bioinformatics tools used here (Kawaguchi et al., 2017; Liu et al., 2021), the high variability of the MHC region can render NGS-based HLA typing prone to accuracy issues, as demonstrated in our study (Supplementary Table S2) and other studies (Major et al., 2013; Wittig et al., 2015; Larjo et al., 2017).
Conclusion
In the current study, we observed similarities in the HLA-B alleles and genotypes in the Qatari population compared with other GCC countries. The prevalent alleles were also found to be associated with different drug-induced hypersensitivities in many other populations. Thus, we recommend performing a selected drug testing for some of the individuals that have been screened for HLA-B pharmacogenetic markers in a controlled clinical setting for the populations of the GCC countries. This is because such studies would be considered as a cost effective intervention for the prevention of drug-induced hypersensitivity.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by The current study was reviewed and approved by the institutional Ethical Review Committee at Dasman Diabetes Institute, Kuwait in accordance with the declaration of Helsinki. The genomic data of Qatari individuals used in this study are publicly available from the National Center for Biotechnology Information Sequence Read Archive. Informed consent has been obtained from the recruited participants of the original studies (Rodriguez-Flores et al., 2016; Fakhro et al., 2016) under protocols approved by the Institutional Review Boards of Hamad Medical Corporation and Weill Cornell Medical College in Qatar. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
MD, FA-M, and TAT designed and performed the study and wrote the manuscript. MD and AA-M performed the data analyses. AC and PH participated in data analysis. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors acknowledge the institutional funding by the Kuwait Foundation for Advancement of the Sciences (KFAS).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.891838/full#supplementary-material
References
Albalushi, K. R., Sellami, M. H., AlRiyami, H., Varghese, M., Boukef, M. K., and Hmida, S. (2014). The Investigation of the Evolutionary History of the Omani Population by Analysis of HLA Class I Polymorphism. Anthropol. 18, 205–210. doi:10.1080/09720073.2014.11891537
Ameen, R., Al Shemmari, S. H., and Marsh, S. G. E. (2020). HLA Haplotype Frequencies and Genetic Profiles of the Kuwaiti Population. Med. Princ. Pract. 29, 39–45. doi:10.1159/000499593
Bellón, E. T., Tong, H. Y., Borobia, A. M., de Abajo, F. J., Lerma, V., Moreno Hidalgo, M. A., et al. (2017). Significant HLA Class I Type Associations with Aromatic Antiepileptic Drug (AED)-induced SJS/TEN Are Different from Those Found for the Same AED-Induced DRESS in the Spanish Population. Pharmacol. Res. 115, 168–178. doi:10.1016/j.phrs.2016.11.027
Chantarangsu, S., Mushiroda, T., Mahasirimongkol, S., Kiertiburanakul, S., Sungkanuparph, S., Manosuthi, W., et al. (2009). HLA-B*3505 Allele Is a Strong Predictor for Nevirapine-Induced Skin Adverse Drug Reactions in HIV-Infected Thai Patients. Pharmacogenet Genomics 19, 139–146. doi:10.1097/FPC.0b013e32831d0faf
Choo, S. Y. (2007). The HLA System: Genetics, Immunology, Clinical Testing, and Clinical Implications. Yonsei Med. J. 48, 11–23. doi:10.3349/ymj.2007.48.1.11
Chung, W. H., Hung, S. I., Hong, H. S., Hsih, M. S., Yang, L. C., Ho, H. C., et al. (2004). Medical Genetics: a Marker for Stevens-Johnson Syndrome. Nature 428, 486. doi:10.1038/428486a
Coutinho, M. I., Teixeira, V., Gameiro, A. R., Brites, M. M., Nunes, R., and Martinho, A. (2013). HLA-B*58:01 Is a Risk Factor for Allopurinol-Induced DRESS and Stevens-Johnson Syndrome/toxic Epidermal Necrolysis in a Portuguese Population. Br. J. Dermatol 169, 660–665. doi:10.1111/bjd.12389
Creary, L. E., Gangavarapu, S., Mallempati, K. C., Montero-Martín, G., Caillier, S. J., Santaniello, A., et al. (2019). Next-generation Sequencing Reveals New Information about HLA Allele and Haplotype Diversity in a Large European American Population. Hum. Immunol. 80, 807–822. doi:10.1016/j.humimm.2019.07.275
Dendrou, C. A., Petersen, J., Rossjohn, J., and Fugger, L. (2018). HLA Variation and Disease. Nat. Rev. Immunol. 18, 325–339. doi:10.1038/nri.2017.143
Edwards, I. R., and Aronson, J. K. (2000). Adverse Drug Reactions: Definitions, Diagnosis, and Management. Lancet 356, 1255–1259. doi:10.1016/S0140-6736(00)02799-9
Elbjeirami, W. M., Abdel-Rahman, F., and Hussein, A. A. (2013). Probability of Finding an HLA-Matched Donor in Immediate and Extended Families: The Jordanian Experience. Biol. Blood Marrow Transpl. 19, 221–226. doi:10.1016/j.bbmt.2012.09.009
Fakhro, K. A., Staudt, M. R., Ramstetter, M. D., Robay, A., Malek, J. A., Badii, R., et al. (2016). The Qatar Genome: a Population-specific Tool for Precision Medicine in the Middle East. Hum. Genome Var. 3, 16016. doi:10.1038/hgv.2016.16
Fan, W. L., Shiao, M. S., Hui, R. C., Su, S. C., Wang, C. W., Chang, Y. C., et al. (2017). HLA Association with Drug-Induced Adverse Reactions. J. Immunol. Res. 2017, 3186328. doi:10.1155/2017/3186328
Gomes, E. R., and Demoly, P. (2005). Epidemiology of Hypersensitivity Drug Reactions. Curr. Opin. Allergy Clin. Immunol. 5, 309–316. doi:10.1097/01.all.0000173785.81024.33
Guay, D. (2007). Update on Clindamycin in the Management of Bacterial, Fungal and Protozoal Infections. Expert Opin. Pharmacother. 8, 2401–2444. doi:10.1517/14656566.8.14.2401
Hajeer, A. H., Al Balwi, M. A., Aytül Uyar, F., Alhaidan, Y., Alabdulrahman, A., Al Abdulkareem, I., et al. (2013). HLA-A, -B, -C, -DRB1 and -DQB1 Allele and Haplotype Frequencies in Saudis Using Next Generation Sequencing Technique. Tissue Antigens 82, 252–258. doi:10.1111/tan.12200
Hajjej, A., Almawi, W. Y., Hattab, L., El-Gaaied, A., and Hmida, S. (2015). HLA Class I and Class II Alleles and Haplotypes Confirm the Berber Origin of the Present Day Tunisian Population. Plos One 10, e0136909. doi:10.1371/journal.pone.0136909
Hajjej, A., Sellami, M. H., Kaabi, H., Hajjej, G., El-Gaaied, A., Boukef, K., et al. (2011). HLA Class I and Class II Polymorphisms in Tunisian Berbers. Ann. Hum. Biol. 38, 156–164. doi:10.3109/03014460.2010.504195
Hetherington, S., Hughes, A. R., Mosteller, M., Shortino, D., Baker, K. L., Spreen, W., et al. (2002). Genetic Variations in HLA-B Region and Hypersensitivity Reactions to Abacavir. Lancet 359, 1121–1122. doi:10.1016/S0140-6736(02)08158-8
Howell, W. M., Carter, V., and Clark, B. (2010). The HLA System: Immunobiology, HLA Typing, Antibody Screening and Crossmatching Techniques. J. Clin. Pathol. 63, 387–390. doi:10.1136/jcp.2009.072371
Hughes, A. R., Mosteller, M., Bansal, A. T., Davies, K., Haneline, S. A., Lai, E. H., et al. (2004). Association of Genetic Variations in HLA-B Region with Hypersensitivity to Abacavir in Some, but Not All, Populations. Pharmacogenomics 5, 203–211. doi:10.1517/phgs.5.2.203.27481
Hunter-Zinck, H., Musharoff, S., Salit, J., Al-Ali, K. A., Chouchane, L., Gohar, A., et al. (2010). Population Genetic Structure of the People of Qatar. Am. J. Hum. Genet. 87, 17–25. doi:10.1016/j.ajhg.2010.05.018
Ikhtiar, A. M., Jazairi, B., Khansa, I., and Othman, A. (2018). HLA Class I Alleles Frequencies in the Syrian Population. BMC Res. Notes 11, 324. doi:10.1186/s13104-018-3427-1
Jawdat, D., Uyar, F. A., Alaskar, A., Müller, C. R., and Hajeer, A. (2020). HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 Allele and Haplotype Frequencies of 28,927 Saudi Stem Cell Donors Typed by Next-Generation Sequencing. Front. Immunol. 11, 544768. doi:10.3389/fimmu.2020.544768
Jazairi, B., Khansaa, I., Ikhtiar, A., and Murad, H. (2016). Frequency of HLA-DRB1 and HLA-DQB1 Alleles and Haplotype Association in Syrian Population. Immunol. Invest 45, 172–179. doi:10.3109/08820139.2015.1131293
Ji, E.-Y. K.-H., Kim, H.-J., Jung, H.-E., Cha, E.-Y., and Shin, J.-G. (2016). HLA-A*24:02/B*51:01haplotype and Lamotrigine-Induced Cutaneous Adverse Drug Reactions in Koreans. Transl. Clin. Pharmacol. 24, 143–146. doi:10.12793/tcp.2016.24.3.143
Jinam, T. A., Saitou, N., Edo, J., Mahmood, A., and Phipps, M. E. (2010). Molecular Analysis of HLA Class I and Class II Genes in Four Indigenous Malaysian Populations. Tissue Antigens 75, 151–158. doi:10.1111/j.1399-0039.2009.01417.x
John, S., Balakrishnan, K., Sukasem, C., Anand, T. C. V., Canyuk, B., and Pattharachayakul, S. (2021). Association of HLA-B*51:01, HLA-B*55:01, CYP2C9*3, and Phenytoin-Induced Cutaneous Adverse Drug Reactions in the South Indian Tamil Population. J. Personalized Med. 11, 737. doi:10.3390/jpm11080737
Jung, J. W., Kim, D. K., Park, H. W., Oh, K. H., Joo, K. W., Kim, Y. S., et al. (2015). An Effective Strategy to Prevent Allopurinol-Induced Hypersensitivity by HLA Typing. Genet. Med. 17, 807–814. doi:10.1038/gim.2014.195
Jung, J. W., Kim, J. Y., Park, I. W., Choi, B. W., and Kang, H. R. (2018). Genetic Markers of Severe Cutaneous Adverse Reactions. Korean J. Intern Med. 33, 867–875. doi:10.3904/kjim.2018.126
Kang, H. R., Jee, Y. K., Kim, Y. S., Lee, C. H., Jung, J. W., Kim, S. H., et al. (2011). Positive and Negative Associations of HLA Class I Alleles with Allopurinol-Induced SCARs in Koreans. Pharmacogenet Genomics 21, 303–307. doi:10.1097/FPC.0b013e32834282b8
Kaniwa, N., Saito, Y., Aihara, M., Matsunaga, K., Tohkin, M., Kurose, K., et al. (2008). HLA-B Locus in Japanese Patients with Anti-epileptics and Allopurinol-Related Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis. Pharmacogenomics 9, 1617–1622. doi:10.2217/14622416.9.11.1617
Kawaguchi, S., Higasa, K., Shimizu, M., Yamada, R., and Matsuda, F. (2017). HLA-HD: An Accurate HLA Typing Algorithm for Next-Generation Sequencing Data. Hum. Mutat. 38, 788–797. doi:10.1002/humu.23230
Kim, S. H., Lee, K. W., Song, W. J., Kim, S. H., Jee, Y. K., Lee, S. M., et al. (2011). Carbamazepine-induced Severe Cutaneous Adverse Reactions and HLA Genotypes in Koreans. Epilepsy Res. 97, 190–197. doi:10.1016/j.eplepsyres.2011.08.010
Ko, T. M., Tsai, C. Y., Chen, S. Y., Chen, K. S., Yu, K. H., Chu, C. S., et al. (2015). Use of HLA-B*58:01 Genotyping to Prevent Allopurinol Induced Severe Cutaneous Adverse Reactions in Taiwan: National Prospective Cohort Study. BMJ 351, h4848. doi:10.1136/bmj.h4848
Larjo, A., Eveleigh, R., Kilpeläinen, E., Kwan, T., Pastinen, T., Koskela, S., et al. (2017). Accuracy of Programs for the Determination of Human Leukocyte Antigen Alleles from Next-Generation Sequencing Data. Front. Immunol. 8, 1815. doi:10.3389/fimmu.2017.01815
Liu, P., Yao, M. Y., Gong, Y., Song, Y. J., Chen, Y. A., Ye, Y. Z., et al. (2021). Benchmarking the Human Leukocyte Antigen Typing Performance of Three Assays and Seven Next-Generation Sequencing-Based Algorithms. Front. Immunol. 12, 652258. doi:10.3389/fimmu.2021.652258
Lonjou, C., Borot, N., Sekula, P., Ledger, N., Thomas, L., Halevy, S., et al. (2008). A European Study of HLA-B in Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis Related to Five High-Risk Drugs. Pharmacogenet Genomics 18, 99–107. doi:10.1097/FPC.0b013e3282f3ef9c
Lorberg, B., Youssef, N. A., and Bhagwagar, Z. (2009). Lamotrigine-associated Rash: to Rechallenge or Not to Rechallenge? Int. J. Neuropsychopharmacol. 12, 257–265. doi:10.1017/S1461145708009504
Ma, J. D., Lee, K. C., and Kuo, G. M. (2010). HLA-B*5701 Testing to Predict Abacavir Hypersensitivity. PLoS Curr. 2, RRN1203. doi:10.1371/currents.RRN1203
Major, E., Rigó, K., Hague, T., Bérces, A., and Juhos, S. (2013). HLA Typing from 1000 Genomes Whole Genome and Whole Exome Illumina Data. Plos One 8, e78410. doi:10.1371/journal.pone.0078410
Mallal, S., Nolan, D., Witt, C., Masel, G., Martin, A. M., Moore, C., et al. (2002). Association between Presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and Hypersensitivity to HIV-1 Reverse-Transcriptase Inhibitor Abacavir. Lancet 359, 727–732. doi:10.1016/s0140-6736(02)07873-x
Mallal, S., Phillips, E., Carosi, G., Molina, J. M., Workman, C., Tomazic, J., et al. (2008). HLA-B*5701 Screening for Hypersensitivity to Abacavir. N. Engl. J. Med. 358, 568–579. doi:10.1056/NEJMoa0706135
Marsh, S. G. E., Albert, E. D., Bodmer, W. F., Bontrop, R. E., Dupont, B., Erlich, H. A., et al. (2010). Nomenclature for Factors of the HLA System, Update December 2009. Tissue Antigens 76, 81–85. doi:10.1111/j.1399-0039.2010.01462.x
Martin, A. M., Nolan, D., Gaudieri, S., Almeida, C. A., Nolan, R., James, I., et al. (2004). Predisposition to Abacavir Hypersensitivity Conferred by HLA-B*5701 and a Haplotypic Hsp70-Hom Variant. Proc. Natl. Acad. Sci. U. S. A. 101, 4180–4185. doi:10.1073/pnas.0307067101
Middleton, D., Hawkins, B. R., Williams, F., Meenagh, A., Moscoso, J., Zamora, J., et al. (2004). HLA Class I Allele Distribution of a Hong Kong Chinese Population Based on High-Resolution PCR-SSOP Typing. Tissue Antigens 63, 555–561. doi:10.1111/j.0001-2815.2004.00234.x
Naisbitt, D. J., Pirmohamed, M., and Park, B. K. (2003). Immunopharmacology of Hypersensitivity Reactions to Drugs. Curr. Allergy Asthma Rep. 3, 22–29. doi:10.1007/s11882-003-0006-9
Nguyen, D. V., Chu, H. C., Nguyen, D. V., Phan, M. H., Craig, T., Baumgart, K., et al. (2015). HLA-B*1502 and Carbamazepine-Induced Severe Cutaneous Adverse Drug Reactions in Vietnamese. Asia Pac Allergy 5, 68–77. doi:10.5415/apallergy.2015.5.2.68
Park, H. J., Kim, Y. J., Kim, D. H., Kim, J., Park, K. H., Park, J. W., et al. (2016). HLA Allele Frequencies in 5802 Koreans: Varied Allele Types Associated with SJS/TEN According to Culprit Drugs. Yonsei Med. J. 57, 118–126. doi:10.3349/ymj.2016.57.1.118
Pawar, M. P., Pore, S. M., Pradhan, S. N., Burute, S. R., Bhoi, U. Y., and Ramanand, S. J. (2015). Nevirapine: Most Common Cause of Cutaneous Adverse Drug Reactions in an Outpatient Department of a Tertiary Care Hospital. J. Clin. Diagn Res. 9, FC17–20. doi:10.7860/JCDR/2015/13672.6768
Phillips, E. J., Wong, G. A., Kaul, R., Shahabi, K., Nolan, D. A., Knowles, S. R., et al. (2005). Clinical and Immunogenetic Correlates of Abacavir Hypersensitivity. Aids 19, 979–981. doi:10.1097/01.aids.0000171414.99409.fb
Pichler, W. J., Naisbitt, D. J., and Park, B. K. (2011). Immune Pathomechanism of Drug Hypersensitivity Reactions. J. Allergy Clin. Immunol. 127, S74–S81. doi:10.1016/j.jaci.2010.11.048
Pirmohamed, M. (2010). Pharmacogenetics of Idiosyncratic Adverse Drug Reactions. Handb. Exp. Pharmacol. 2010, 477–491. doi:10.1007/978-3-642-00663-0_17
Puangpetch, A., Koomdee, N., Chamnanphol, M., Jantararoungtong, T., Santon, S., Prommas, S., et al. (2015). HLA-B Allele and Haplotype Diversity Among Thai Patients Identified by PCR-SSOP: Evidence for High Risk of Drug-Induced Hypersensitivity. Front. Genet. 5, 478. doi:10.3389/fgene.2014.00478
Razali, R. M., Rodriguez-Flores, J., Ghorbani, M., Naeem, H., Aamer, W., Aliyev, E., et al. (2021). Thousands of Qatari Genomes Inform Human Migration History and Improve Imputation of Arab Haplotypes. Nat. Commun. 12, 5929. doi:10.1038/s41467-021-25287-y
Robinson, J., Halliwell, J. A., Hayhurst, J. D., Flicek, P., Parham, P., and Marsh, S. G. (2015). The IPD and IMGT/HLA Database: Allele Variant Databases. Nucleic Acids Res. 43, D423–D431. doi:10.1093/nar/gku1161
Rodriguez-Flores, J. L., Fakhro, K., Agosto-Perez, F., Ramstetter, M. D., Arbiza, L., Vincent, T. L., et al. (2016). Indigenous Arabs Are Descendants of the Earliest Split from Ancient Eurasian Populations. Genome Res. 26, 151–162. doi:10.1101/gr.191478.115
Sánchez-Velasco, P., Karadsheh, N. S., García-Martín, A., Ruíz de Alegría, C., and Leyva-Cobián, F. (2001). Molecular Analysis of HLA Allelic Frequencies and Haplotypes in Jordanians and Comparison with Other Related Populations. Hum. Immunol. 62, 901–909. doi:10.1016/s0198-8859(01)00289-0
Satapornpong, P., Pratoomwun, J., Rerknimitr, P., Klaewsongkram, J., Nakkam, N., Rungrotmongkol, T., et al. (2021). HLA-B*13 :01 Is a Predictive Marker of Dapsone-Induced Severe Cutaneous Adverse Reactions in Thai Patients. Front. Immunol. 12, 661135. doi:10.3389/fimmu.2021.661135
Schaefer, C., Schmidt, A. H., and Sauter, J. (2017). Hapl-o-Mat: Open-Source Software for HLA Haplotype Frequency Estimation from Ambiguous and Heterogeneous Data. Bmc Bioinforma. 18, 284. doi:10.1186/s12859-017-1613-0
Schmidt, A. H., Baier, D., Solloch, U. V., Stahr, A., Cereb, N., Wassmuth, R., et al. (2009). Estimation of High-Resolution HLA-A, -B, -C, -DRB1 Allele and Haplotype Frequencies Based on 8862 German Stem Cell Donors and Implications for Strategic Donor Registry Planning. Hum. Immunol. 70, 895–902. doi:10.1016/j.humimm.2009.08.006
Sukasem, C., Chaichan, C., Nakkrut, T., Satapornpong, P., Jaruthamsophon, K., Jantararoungtong, T., et al. (2018). Association between HLA-B Alleles and Carbamazepine-Induced Maculopapular Exanthema and Severe Cutaneous Reactions in Thai Patients. J. Immunol. Res. 2018, 2780272. doi:10.1155/2018/2780272
Sukasem, C., Puangpetch, A., Medhasi, S., and Tassaneeyakul, W. (2014). Pharmacogenomics of Drug-Induced Hypersensitivity Reactions: Challenges, Opportunities and Clinical Implementation. Asian Pac J. Allergy Immunol. 32, 111–123.
Tangamornsuksan, W., and Lohitnavy, M. (2018). Association Between HLA-B*1301 and Dapsone-Induced Cutaneous Adverse Drug Reactions: A Systematic Review and Meta-Analysis. JAMA Dermatol 154, 441–446. doi:10.1001/jamadermatol.2017.6484
Tassaneeyakul, W., Jantararoungtong, T., Chen, P., Lin, P. Y., Tiamkao, S., Khunarkornsiri, U., et al. (2009). Strong Association between HLA-B*5801 and Allopurinol-Induced Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis in a Thai Population. Pharmacogenet Genomics 19, 704–709. doi:10.1097/FPC.0b013e328330a3b8
Tassaneeyakul, W., Prabmeechai, N., Sukasem, C., Kongpan, T., Konyoung, P., Chumworathayi, P., et al. (2016). Associations between HLA Class I and Cytochrome P450 2C9 Genetic Polymorphisms and Phenytoin-Related Severe Cutaneous Adverse Reactions in a Thai Population. Pharmacogenet Genomics 26, 225–234. doi:10.1097/FPC.0000000000000211
Tassaneeyakul, W., Tiamkao, S., Jantararoungtong, T., Chen, P., Lin, S. Y., Chen, W. H., et al. (2010). Association between HLA-B*1502 and Carbamazepine-Induced Severe Cutaneous Adverse Drug Reactions in a Thai Population. Epilepsia 51, 926–930. doi:10.1111/j.1528-1167.2010.02533.x
Tempark, T., Satapornpong, P., Rerknimitr, P., Nakkam, N., Saksit, N., Wattanakrai, P., et al. (2017). Dapsone-induced Severe Cutaneous Adverse Drug Reactions Are Strongly Linked with HLA-B*13: 01 Allele in the Thai Population. Pharmacogenet Genomics 27, 429–437. doi:10.1097/FPC.0000000000000306
Ueta, M., Kaniwa, N., Sotozono, C., Tokunaga, K., Saito, Y., Sawai, H., et al. (2014). Independent Strong Association of HLA-A*02:06 and HLA-B*44:03 with Cold Medicine-Related Stevens-Johnson Syndrome with Severe Mucosal Involvement. Sci. Rep. 4, 4862. doi:10.1038/srep04862
Ulvestad, E., Williams, K., Bø, L., Trapp, B., Antel, J., and Mørk, S. (1994). HLA Class II Molecules (HLA-DR, -DP, -DQ) on Cells in the Human CNS Studied In Situ and In Vitro. Immunology 82, 535–541.
Wakamatsu, T. H., Ueta, M., Loureiro, R. R., Costa, K. A., Sallum, J. M. F., Sotozono, C., et al. (2015). Association of HLA-B(star)44:03 with Stevens-Johnson Syndrome in Brazilian Patients. Investigative Ophthalmol. Vis. Sci. 56, 5993.
Wei, C. Y., Ko, T. M., Shen, C. Y., and Chen, Y. T. (2012). A Recent Update of Pharmacogenomics in Drug-Induced Severe Skin Reactions. Drug Metab. Pharmacokinet. 27, 132–141. doi:10.2133/dmpk.dmpk-11-rv-116
Williams, F., Meenagh, A., Darke, C., Acosta, A., Daar, A. S., Gorodezky, C., et al. (2001). Analysis of the Distribution of HLA-B Alleles in Populations from Five Continents. Hum. Immunol. 62, 645–650. doi:10.1016/s0198-8859(01)00247-6
Wittig, M., Anmarkrud, J. A., Kässens, J. C., Koch, S., Forster, M., Ellinghaus, E., et al. (2015). Development of a High-Resolution NGS-Based HLA-Typing and Analysis Pipeline. Nucleic Acids Res. 43, E70–U11. doi:10.1093/nar/gkv184
Yang, Y., Chen, S., Yang, F., Zhang, L., Alterovitz, G., Zhu, H., et al. (2017). HLA-B*51:01 Is Strongly Associated with Clindamycin-Related Cutaneous Adverse Drug Reactions. Pharmacogenomics J. 17, 501–505. doi:10.1038/tpj.2016.61
Keywords: HLA-B alleles, pharmacogenetics, drug hypersensitivity, HLA typing class I, Qatari population
Citation: Dashti M, Al-Matrouk A, Channanath A, Hebbar P, Al-Mulla F and Thanaraj TA (2022) Distribution of HLA-B Alleles and Haplotypes in Qatari: Recommendation for Establishing Pharmacogenomic Markers Screening for Drug Hypersensitivity. Front. Pharmacol. 13:891838. doi: 10.3389/fphar.2022.891838
Received: 08 March 2022; Accepted: 14 June 2022;
Published: 08 August 2022.
Edited by:
Chonlaphat Sukasem, Mahidol University, ThailandReviewed by:
Yoshiro Saito, National University of Health Sciences, United StatesJames Robinson, Anthony Nolan Research Institute, United Kingdom
Copyright © 2022 Dashti, Al-Matrouk, Channanath, Hebbar, Al-Mulla and Thanaraj. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fahd Al-Mulla, ZmFoZC5hbG11bGxhQGRhc21hbmluc3RpdHV0ZS5vcmc=; Thangavel Alphonse Thanaraj, YWxwaG9uc2UudGhhbmdhdmVsQGRhc21hbmluc3RpdHV0ZS5vcmc=
 Mohammed Dashti
Mohammed Dashti Abdullah Al-Matrouk
Abdullah Al-Matrouk Arshad Channanath
Arshad Channanath Prashantha Hebbar
Prashantha Hebbar Fahd Al-Mulla
Fahd Al-Mulla Thangavel Alphonse Thanaraj
Thangavel Alphonse Thanaraj