- 1Department of Experimental and Clinical Pharmacology, College of Pharmacy, University of Minnesota, Twin Cities, MN, United States
- 2Division of Clinical Pharmacology, Toxicology and Therapeutic Innovation, Children’s Mercy Research Institute, Kansas City, MO, United States
- 3School of Medicine, University of Missouri-Kansas City, Kansas City, MO, United States
Objective: Hmong individuals represent a unique East Asian subpopulation in whom limited information concerning pharmacogenetic variation exists. The objectives of this study were to comprehensively characterize the highly polymorphic CYP2D6 gene in Hmong, estimate allele and phenotype frequencies and to compare results between two testing platforms.
Methods: DNA from 48 self-identified Hmong participants were sequenced using a targeted next-generation sequencing (NGS) panel. Star allele calls were made using Astrolabe, manual inspection of NGS variant calls and confirmatory Sanger sequencing. Structural variation was determined by long-range (XL)-PCR and digital droplet PCR (ddPCR). The consensus diplotypes were subsequently translated into phenotype utilizing the activity score system. Clinical grade pharmacogenetic testing was obtained for 12 of the 48 samples enabling an assessment of concordance between the consensus calls and those determined by clinical testing platforms.
Results: A total of 13 CYP2D6 alleles were identified. The most common alleles were CYP2D6*10 and its structural arrangements (37.5%, 36/96) and the *5 gene deletion (13.5%, 13/96). Three novel suballeles (*10.007, *36.004, and *75.002) were also identified. Phenotype frequencies were as follows: ultrarapid metabolizers (4.2%, 2/48), normal metabolizers (41.7%, 20/48) and intermediate metabolizers (52.1%, 25/48); none of the 48 participants were predicted to be poor metabolizers. Concordance of diplotype and phenotype calls between the consensus and clinical testing were 66.7 and 50%, respectively.
Conclusion: Our study to explore CYP2D6 genotypes in the Hmong population suggests that this subpopulation is unique regarding CYP2D6 allelic variants; also, a higher portion of Hmong participants (50%) are predicted to have an intermediate metabolizer phenotype for CYP2D6 compared to other East Asians which range between 27 and 44%. Results from different testing methods varied considerably. These preliminary findings underscore the importance of thoroughly interrogating unique subpopulations to accurately predict a patient’s CYP2D6 metabolizer status.
Introduction
Implementing pharmacogenetic (PGx) testing for clinical practice has been shown to positively impact patient outcomes in a variety of settings and medical conditions (Cavallari et al., 2018; Smith et al., 2019; Hulot et al., 2020). In select cases and healthcare systems, this practice has been demonstrated to be cost-effective (Tanner et al., 2020; Karamperis et al., 2021; Kim et al., 2021; Zhu et al., 2021). However, our ability to develop, validate and utilize clinical guidelines in an equitable manner for all members of society is hindered by the lack of population-specific PGx knowledge. Despite the best efforts from the Association for Molecular Pathology (AMP) Clinical Practice Committee’s Pharmacogenomics Working Group outlining which alleles within the clinically important pharmacogenes should be tested, (Pratt et al., 2018; Pratt et al., 2019; Pratt et al., 2020; Pratt et al., 2021), novel single nucleotide polymorphisms (SNPs) and novel structural variants of clinical relevance have continued to be identified in many less-well-studied populations (Montane Jaime et al., 2013; Boone et al., 2020). This critical gap in knowledge for any sub-population can lead to disparities in health outcomes. This is particularly true for individuals with limited access to health care and those more susceptible to select medical ailments. For example, only until recently, investigators (Hernandez et al., 2014; De et al., 2018) have acknowledged the critical importance to incorporate ethno-specific variants in pharmacogene unique to individuals of African descent that must be included when considering genetic-guided dosing of warfarin (Johnson et al., 2017). Hmong individuals residing in Minnesota represent another example where this under-served and under-resourced population whose life circumstances have led to limited engagement in research, especially research evaluating the safety and efficacy of medications (Park et al., 2018).
In our previous work (Wen et al., 2020) regarding the Very Important Pharmacogenes in the Hmong Community (VIP-Hmong) study, we identified significant differences in allele frequencies between Hmong and East Asians (EA) for 23% (5/22) of actionable genetic variants in eight unique VIPs (CYP2C9, CYP2C19, CYP4F2, DYPD, G6PD, SLC O 1B1, TMPT, VKORC1). These differences are even greater between Hmong and Europeans (Wen et al., 2020) and may have clinical consequences when predicting drug dose and response (Sun et al., 2020). For select pharmacogenes, key genetic variants and their frequencies can be predicted with a relatively high degree of confidence across populations. However, for highly polymorphic genes, such as CYP2D6, the reliability of pre-designed test panels is limited (Nofziger et al., 2019). This limitation is critical as it impacts our ability to accurately translate PGx knowledge into actionable recommendations for about 21% of important medications including antidepressants, antipsychotics, opioids, and chemotherapeutic agents (Saravanakumar et al., 2019). In addition, CYP2D6 is highly polymorphic within and between populations which leads to a wide range of potential activity and corresponding predicted therapeutic responses. In the absence of sub-population specific knowledge, the potential for phenotype misclassification due to incomplete genotype assessment is high. Inaccuracies in phenotype prediction can lead to potentially severe adverse drug reactions and/or an altered efficacy due to inappropriate drug selection and dosing.
To characterize the complex CYP2D6 gene locus for SNPs, small nucleotide insertions/deletions (INDELs), copy number variation (CNVs) including gene deletions and duplications, the presence of gene conversions and structural rearrangements involving the highly similar CYP2D7 pseudogene, often requires the application of several methods and/or approaches. Methodologies to determine structural variants may include NGS, Sanger sequencing, SNP/indel genotyping (Yang et al., 2017), long-read single molecule real-time (SMRT) sequencing (Qiao et al., 2016; Buermans et al., 2017), and/or allele-specific long-range PCR (ASXL-PCR) (Gaedigk et al., 2015). Additional methods to determine CNVs include TaqMan based assays or droplet digital-PCR (ddPCR) (Gaedigk et al., 2019), allele quantification-based genotyping using Pyrosequencing (Langaee et al., 2015), and multiplex ligation-dependent probe amplification (MLPA) (Qiao et al., 2019). These are either used alone or in combination (Fukunaga et al., 2020). Although there is an increasing number of clinical laboratories within a health care institution offering PGx testing (Krebs and Milani, 2019), such sophisticated methods may not always be available. Commonly, PGx testing is performed in a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory which includes CYP2D6 Tier 1 and possibly Tier 2 alleles recommended by AMP on the testing panel. Information about testing companies can be found on the Genetic Testing Registry website, National Center for Biotechnology Information, U.S. National Library of Medicine (National Center for Biotechnology Information, 2022).
In the present study, we utilized XL-PCR and ddPCR to detect CNVs and structural rearrangements (SVs) and Sanger sequencing to resolve and/or validate diplotype calls from NGS data. The first objective of this study was to comprehensively characterize CYP2D6 genetic variation in a select population of Hmong, determine allele frequencies, and estimate the consequent phenotype frequencies. The second objective was to compare these with those of a clinical-grade test report.
Materials and Methods
Study Participants
Genomic DNA from 48 Hmong adults living in either Minnesota or Wisconsin was obtained from two independently conducted studies. Hmong ancestry was based on participants self-reporting that both their parents were of Hmong descent. For the purpose of this present study, 34 samples were selected from the VIP-Hmong study (Wen et al., 2020) and 14 from the Genetics Of hyperUricemia Therapy in Hmong (GOUT-H) study (Roman et al., 2017). All participants provided informed consent and both studies were approved by the University of Minnesota Institutional Review Board (UMN IRB #1702M06041 and IRB #1408M53223, respectively). The GOUT-H study was also registered under clinicaltrial. gov: NCT02371421. To ensure that samples from the two studies represented unrelated participants, the names and date of birth of the participants were cross-checked.
DNA Isolation
Saliva was collected using ORAgene® DISCOVER kits (OGR-500, DNA Genotek Inc., Ottawa, ON, Canada) and processed per the manufacturer’s protocol. Genomic DNA was extracted using a QIAamp DNA Kit (Qiagen Inc., Germantown, MD, United States). DNA was quantified using a Qubit Fluorometer (Thermo Fisher Scientific, Waltham, MA, United States).
NGS-Based Targeted Sequencing and Variant Calling
ADMEseq is an NGS-based panel targeting 286 genes involved in drug absorption distribution metabolism and elimination (ADME). Briefly, sequencing was performed using a MiSeq instrument (Illumina, San Diego, CA) with paired end 200 base pair reads to a total data of about 355 MB. The average read depth was about 530X over a panel target of about 660 kilo base pairs. TruSeq Libraries were prepared according to the manufacturer’s protocol: 10 cycles of PCR, followed by enrichment with the ADMEseq custom panel.
The sequence reads were aligned to human genome assembly GRCh37 and variants were detected using the DRAGEN Bio-IT platform v2.5.3 (Illumina, San Diego, CA). Variants were called with positions downsampled to 2000 reads with the following criteria: sequence quality ≥10, mapping quality ≥20, and phred-scaled confidence score ≥20.
CYP2D6 Star Allele Calling Using Bioinformatics Tools and Designations of Novel Haplotypes
Astrolabe (v0.8.7.2) (Twist et al., 2016; Twist et al., 2017) was used to call CYP2D6 star allele diplotypes, which were complemented with experimental CNV analyses described below. Parameters were set based on recommendations by the tool’s authors (see supplementary text for analysis scripts). The BAM and VCF files were used as input for Astrolabe. CYP2D6 allele designations followed those defined by the PharmVar Consortium (https://www.pharmvar.org) (Gaedigk et al., 2018). All variant calls from Astrolabe were manually inspected and analyzed. Novel haplotypes were fully characterized by XL-PCR and Sanger sequencing as described in the sections below; these were also submitted and subsequently accepted by PharmVar for allele designation.
Determination of CYP2D6 Copy Number Variation
XL-PCR was employed to detect structural variants as previously described (Bathum et al., 1998; Gaedigk et al., 2010a; Gaedigk et al., 2010b; Gaedigk et al., 2019). A brief description of each amplicon, its conventional name and size, primer sequences, and cycling conditions are summarized in Supplementary Table S1. All XL-PCR reactions were performed using 2x KAPA LongRange HotStart ReadyMix with dye (Roche Diagnostics). Amplicons were visualized on a 0.7% agarose gel with 1x final concentration of SYBR™ Safe DNA Gel Stain (Thermo Fisher Scientific, MA, United States). Samples with CNVs were confirmed with ddPCR to obtain quantitative CNV information using a QX200 Droplet Digital PCR System (Bio-Rad) as previously described (Gaedigk et al., 2019) with the following modifications: 100 ng of genomic DNA was digested with 8U of BamHI-HF (New England BioLabs, Ipswich, MA) in a 20-µL reaction at 37°C for 1.5 h, followed by 20 min at 65°C for inactivation. Ten-15 ng of digested DNA was run in a multiplex reaction using TaqMan copy number assays targeting the 5′UTR (Hs04078252_cn) and exon 9 (Hs00010001_cn) regions. To achieve optimal cluster separation, the concentration of the exon 9 assay was reduced by half (Whale et al., 2016).
Sanger Sequencing
Selected samples were Sanger sequenced to resolve ambiguous Astrolabe calls or confirm SNP calls that suggested the presence of a novel haplotype(s) (Gaedigk et al., 2015). See Supplementary Table S1 for more information on primer sequences and ASXL-PCR amplicons.
Translation of CYP2D6 Diplotypes Into Phenotype
The activity score (AS) of a CYP2D6 diplotype was retrieved from the “CYP2D6 allele definition table” and translated into phenotype according to the “CYP2D6 diplotype-phenotype table” (both available through the PharmGKB at https://www.pharmgkb.org/page/cyp2d6RefMaterials). This method, first described by Gaedigk et al (2008) is recommended by the CPIC and Dutch Pharmacogenetics Working Group (DPWG): AS > 2.25, ultrarapid metabolizer (UM); 1.25 ≤ AS ≤ 2.25, normal metabolizer (NM); 0.25 ≤ AS < 1.25, intermediate metabolizer (IM) and AS = 0, poor metabolizer (PM) (Caudle et al., 2020).
Clinical Grade CYP2D6 Pharmacogenetic Testing
Among the 34 participants from the VIP-Hmong study, 12 agreed to participate in focus group discussions pertaining to their perceptions of return of PGx results (Nelson et al., 2020). As such, a commercial RightMed® test from OneOme, LLC was obtained for these participants. The RightMed® test uses quantitative real-time PCR assays to determine copy number at four gene regions, the 3′UTR, intron 2, intron 6, and exon 9 of CYP2D6. Variants tested by the RightMed® test panel are listed in Supplementary Table S2.
Comparison of CYP2D6 Allele and Phenotype Frequencies With Other East Asian Populations
CYP2D6 allele frequencies were compared with other EA populations using the “CYP2D6 frequencies table” available through the PharmGKB at (https://www.pharmgkb.org/page/cyp2d6RefMaterials). CYP2D6 phenotype frequencies for other EA populations, including Chinese, Japanese, Korean, Thai, and Vietnamese were obtained from previous publications (Gaedigk et al., 2017).
Concordance of CYP2D6 Diplotypes and Phenotypes Among Methods
CYP2D6 diplotype and phenotype calls obtained by NGS, CNV analysis and Sanger sequencing (referred to as ‘consensus’ calls) were compared with those provided by the RightMed® clinical test reports. Because of small sample size (N=12), only descriptive statistics (count and percentage) were applied.
Results
Participant Characteristics
Forty-eight self-identified Hmong participants from VIP-Hmong and GOUT-H were included in the analysis. Demographics of the participants are presented in Table 1.
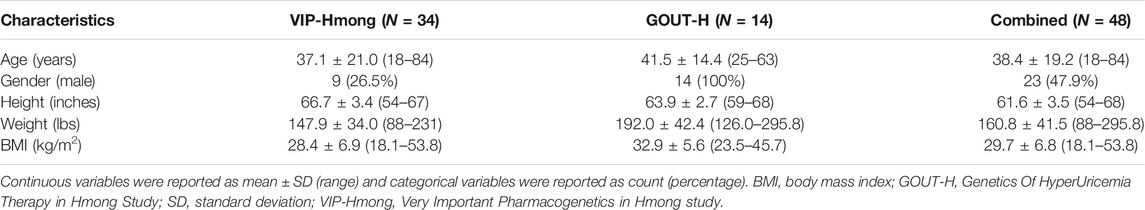
TABLE 1. Characteristics of the 48 Hmong participants from the two independent studies, VIP-Hmong and GOUT-H study.
CYP2D6 Allele and Diplotype Frequencies
The procedure used to determine CYP2D6 star allele designations and phenotype calls is summarized in Figure 1.
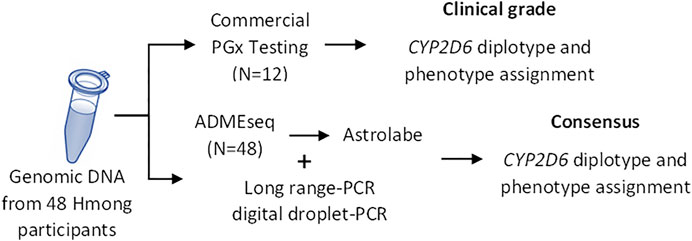
FIGURE 1. Study Overview. CYP2D6 diplotype was determined using two different approaches: commercial “clinal-grade” testing and via NGS sequencing complemented with CNV analyses (consensus). PGx, pharmacogenetics.
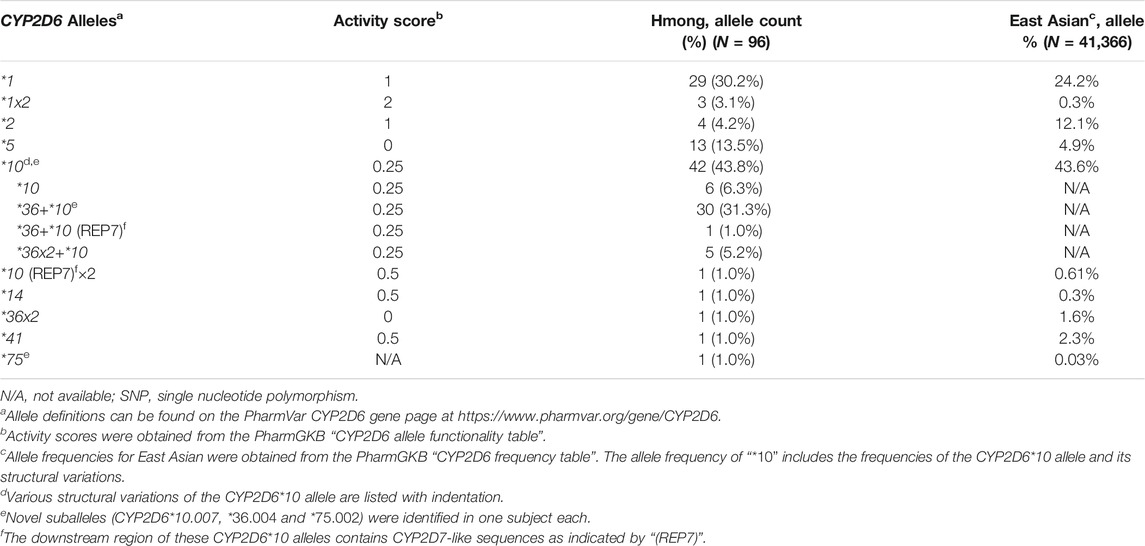
From the consensus diplotype calls, a total of 13 distinct CYP2D6 allelic variants were identified (Table 2). The most common star alleles were the decreased function CYP2D6*10 allele (6.3%, 6/96) or structural variants containing this allele, such as CYP2D6*36+*10 (31.3%, 30/96), CYP2D6*36+*10 (REP7) (1.0%, 1/96) and CYP2D6*36×2+*10 (5.2%, 5/96). REP7 indicates that the downstream CYP2D6 gene has a CYP2D7-derived repetitive element including the spacer sequence. Overall, CYP2D6*10 and its structural variants accounted for a total of 43.8% (42/96) of the observed alleles in the Hmong in comparison to allele frequency of 43.6% observed in EAs. The CYP2D6*5 gene deletion (13.5%, 13/96) was the second most common allele in the Hmong in comparison to 4.9% observed in EAs. Other alleles observed in the Hmong included the normal functioning CYP2D6*2 allele (4.2%, 4/96), the decreased function CYP2D6*14 (1%, 1/96) and *41 (1%, 1/96) alleles. The prevalence of CYP2D6*2 in Hmong was less prevalent compared to EA’s where it represented the second most prevalent allele at 12.1%.
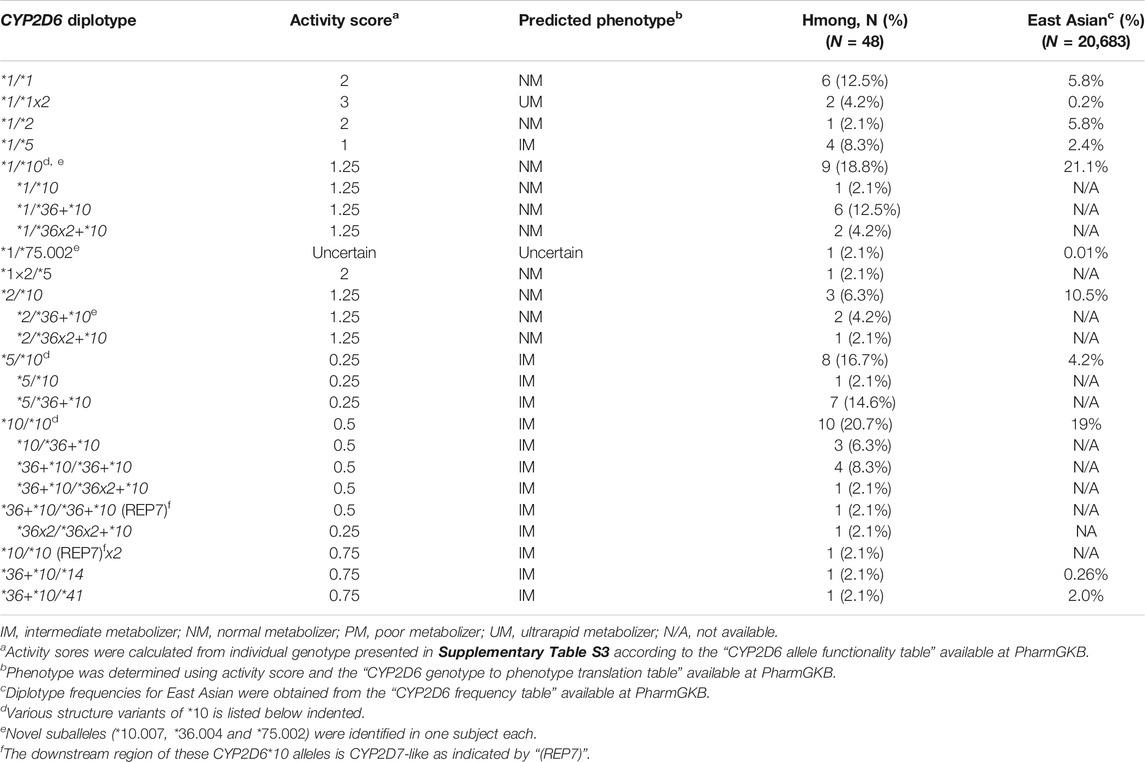
Table 3 summarizes all diplotypes identified in this Hmong population cohort and those reported in EAs. The CYP2D6*1/*10 and its structural variant, CYP2D6*1/*36+*10 and CYP2D6*1/*36x2+*10 were the most common diplotypes observed in the Hmong at 18.8% in comparison to 21.1% in an EA population. The second most common diplotype observed in Hmong included CYP2D6*5/*10 (2.1%, 1/48) and its variant CYP2D6*5/*36+*10 (14.6%, 7/48) which was less common in the EA population at 4.2%. In contrast, the second most commonly observed diplotype in EAs is CYP2D6*10/*36+*10 (19%). This compares to a less common frequency in the Hmong for CYP2D6*10/*36+*10 (6.3%) and *36+*10/*36+*10 (8.3%). Supplementary Table S3 provides individual CYP2D6 diplotype assignments for each of the 48 Hmong subjects.
It is important to note that a few diplotypes with complex structures (4 subjects, Supplementary Table S3) could not be resolved with certainty and assigned the most likely haplotypes to summarize allele and diplotype frequencies. For example, a person with a CYP2D6*36+*10/*36+*10 diplotype could also be *36×2+*10/*10. Similarly, a person with a CYP2D6*36+*10/*36+*10 (REP7) diplotype could be *10[REP7]/*36×2+*10 or *10/*36×2+*10 (REP7). These potential ambiguities in diplotype calling will nonetheless result in the same phenotype prediction regardless of how the gene copies are arranged.
Identification of Novel CYP2D6 Suballeles
Three novel suballeles (*10.007, *36.004, and *75.002) were identified in subjects 41, 21, and 10, respectively (Figure 2 and Figure 3). The novel CYP2D6*10.007 suballele was discovered in a CYP2D6*36.001+*10.007 tandem. This structure is supported by the presence of a 10.2 kb long Fragment D, the intergenic fragment C (shown to have 100C>T by genotyping), as well as quantitative ddPCR (5′UTR, 3 copies; exon 9, 2 copies). Sanger sequencing of allele-specific XL-PCR amplicons allowed us to fully characterize the novel suballele and confirm a CYP2D6*2.001 on the other chromosome (Figure 2A and Figure 3, column *10.007) of subject 41. As shown in Figure 2B, subject 21 produced a series of XL-PCR amplicons that together with ddPCR (5′UTR, 3 copies; exon 9, 2 copies) indicated the presence of a CYP2D6*36+*10 tandem. In this case, however, Sanger sequencing of allele-specific amplicons revealed a novel CYP2D6*36 suballele, which was designated *36.004 by PharmVar (Figure 3, column *36.004). The CYP2D6*10 allele was found to match the *10.002 suballele definition, and the allele on the other chromosome was determined to be a CYP2D6*1.010 suballele. The third novel suballele was CYP2D6*75.002 (Figure 3, column *75.002). As illustrated in Figure 2C, Sanger sequencing of an allele specific XL-PCR product allowed us to show that all SNPs initially identified by NGS were on the same chromosome.
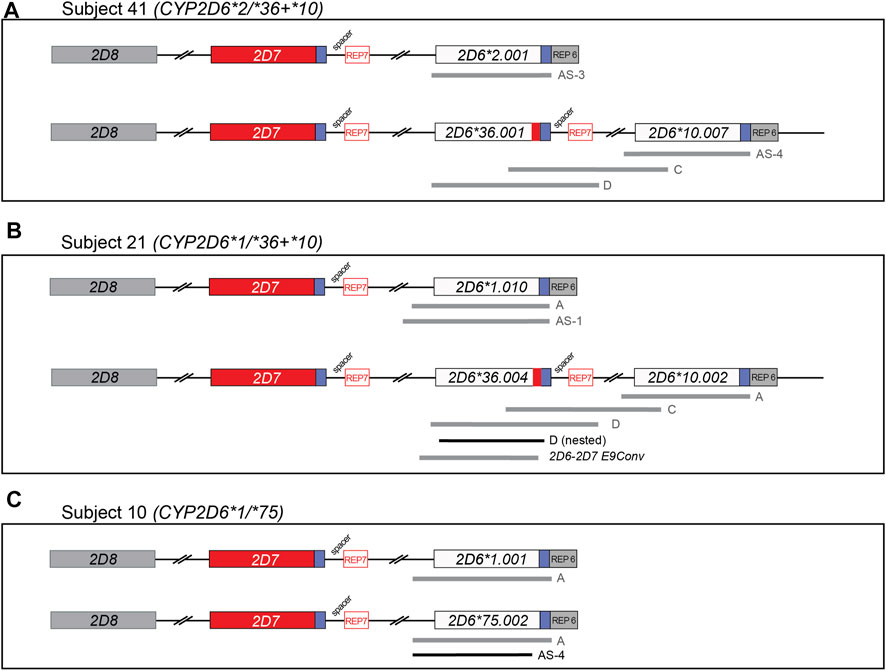
FIGURE 2. Graphical overview of long-range (XL) PCR fragments generated to characterize gene copy number arrangements and novel allelic variants. Regions amplified from genomic DNA using XL-PCR are represented by gray bars. Selected amplicons were subsequently used as templates to generate sufficient materials for Sanger sequencing (shown in black). Blue boxes downstream of CYP2D6 and CYP2D7 denote highly similar regions; those labeled ‘REP’ contain repetitive sequences. CYP2D7-derived downstream regions are characterized by the presence of a 1.6 kb “spacer” sequence. See Supplementary Table S1 for a comprehensive list of PCR products generated in this study to characterize the alleles. Panel (A–C) describe the summary of XL-PCR products generated to characterize Subject 41, 21, and 10, respectively. All sequence variants found on each of the novel haplotypes are detailed in Figure 3.
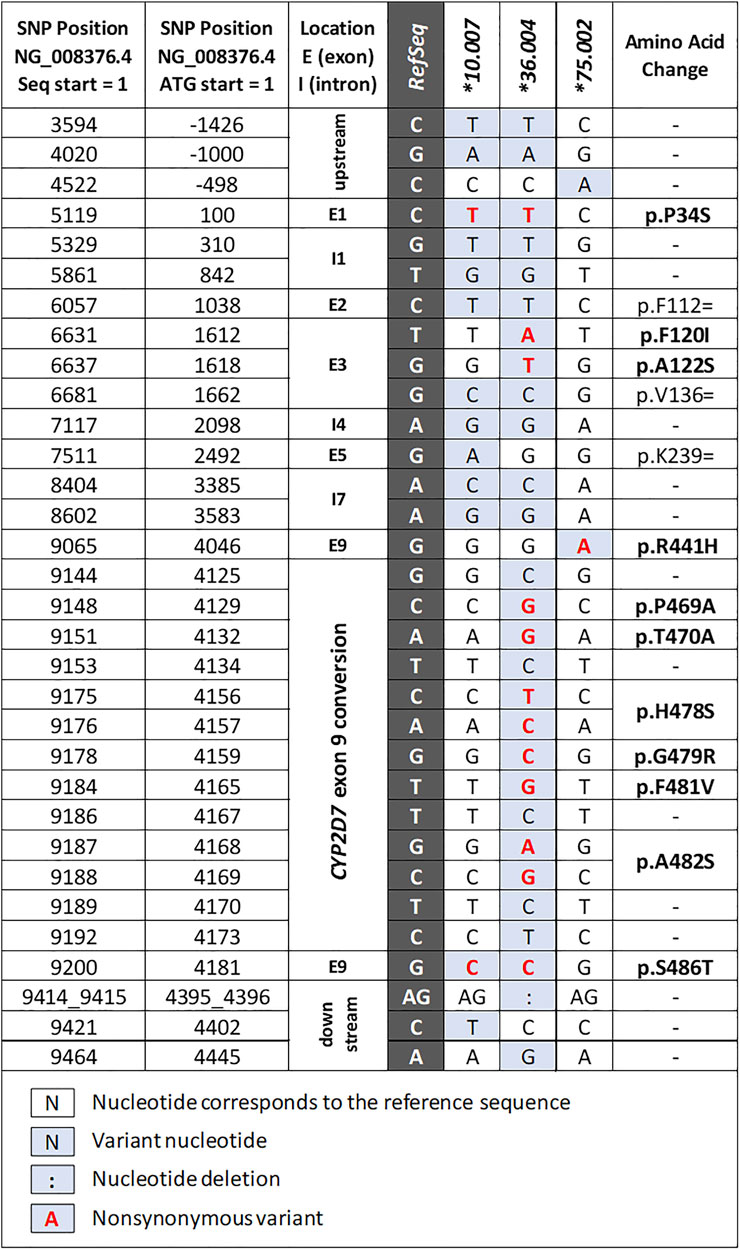
FIGURE 3. Summary of the novel allele and suballeles identified in the study. The columns to the left provide nucleotide positions counting from the start of the NG_008376.4 reference sequence and the translation start (ATG = +1), respectively. The dark gray column indicates the reference nucleotide of NG_008376.4 which corresponds to the CYP2D6*1.001 allele definition. “Exon 9 conversion” denotes a CYP2D7-derived region containing several SNPs (details can be found in the structural variant document available on the PharmVar CYP2D6 gene page at https://www.pharmvar.org/gene/CYP2D6).
CYP2D6 Phenotype Frequencies
Phenotype was assigned based on the AS for each diplotype (Table 3). In this Hmong population, 4.2% (2/48) were classified as UMs, 41.7% (20/48) as NMs, and 52.1% (25/48) as IMs. No PMs were observed. Phenotype could not be determined for one subject (2.4%). Since function is uncertain for the CYP2D6*75 allele, the CYP2D6*1/*75 diplotype remains ‘undetermined’; however, due to the presence of a CYP2D6*1 allele, this subject is most likely an IM or NM. (Table 4). AS and phenotype assignments for all 48 Hmong participants using different methods are presented in Supplementary Table S3. Comparisons of phenotype distributions between Hmong, EA, and other EA populations are summarized in Table 4. PM, IM and NM phenotype frequencies in the Hmong were similar to those reported for Thai and Vietnamese populations but the Hmong exhibited a higher frequency of UMs compared to the Thai and Vietnamese.
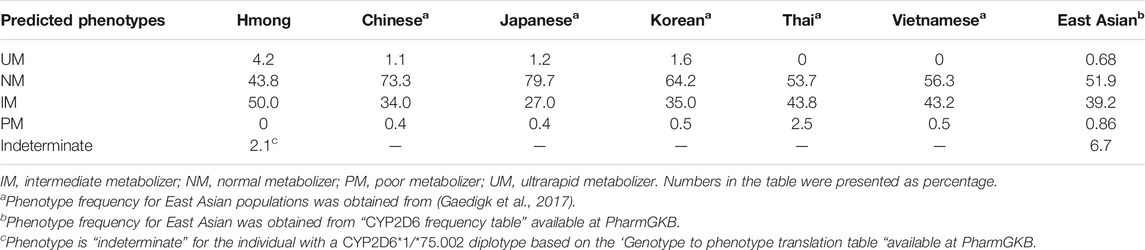
TABLE 4. Comparison of CYP2D6 predicted phenotypes frequencies with other East Asian sub-populations.
Concordance of CYP2D6 Star Alleles and Phenotypes
Concordance for diplotype and phenotype calls between the consensus and clinical (RightMed® test) results was 66.7% (8/12) and 50% (6/12), respectively (Supplementary Table S3).
Discussion
To our knowledge, this is the first comprehensive characterization of the CYP2D6 gene locus in Hmong. Although California has the highest population of Hmong, at over 98,000, Minnesota and Wisconsin represent two states which have the second (73,110) and third (49,240) highest populations of Hmong residing in the US (United States Census Bureau, 2016), respectively. There is a growing prevalence of medical conditions within the Hmong community for which the prescribed medications are predominately metabolized by CYP2D6 (Lee et al., 2010; Bart et al., 2012; Lee and Chang, 2012; Lee, 2013; Arcan et al., 2014; Thao et al., 2015). For instance, the Hmong have higher rates of mental health conditions and cervical cancer with lower rates of treatment success (Lee et al., 2010; Lee and Chang, 2012; Lee, 2013; Oyenuga et al., 2018) when compared to general US populations; clinicians have expressed challenges with finding effective medications for the Hmong. The findings from this study indicate that Hmong exhibit distinct differences in allele frequencies and predicted phenotypes relative to other EA populations (Table 4). This implies that the standard medication dose used for patients of European as well as other EA ancestries may not be appropriate for the Hmong patients. This latter comparison is not instinctively intuitive. Knowledge of the unique prevalence or distribution of genetic variations in the Hmong could help clinicians optimize drug therapy (choice of drug and/or dosage), and thus reduce disparities in the quality of treatment while improving health by avoiding predictable, harmful side effects.
In our cohort of Hmong described herein, CYP2D6*10 and its structural variants represented the most common alleles observed at 43.8%. This is consistent with the previously reported frequency of 43.6% in EA populations. However, in the present study, the combined use of quantitative and qualitative CNV analyses enabled us to discriminate between CYP2D6*36+*10 (31.3%) and CYP2D6*10 (6.3%) in the Hmong. It is important to note that the majority of published studies and many commercial tests fail to distinguish between the two. The activity of both alleles is deemed equivalent and therefore, a value of 0.25 is used to calculate the AS of diplotypes containing these alleles. Our observation of the high frequency of the CYP2D6*36+*10 tandem is also consistent with existing data, which suggests that this tandem is exclusively seen in subjects of Asian ancestry including Japanese (Kiyotani et al., 2010), Korean (Kim et al., 2012), and Kinh Vietnamese (Nguyen et al., 2019) populations while the CYP2D6*10 allele on its own is found across populations, but most frequently in EAs (Nofziger et al., 2019). Our findings are also in line with previous studies, which reported almost half of all CYP2D6*10 alleles were found in CYP2D6*36+*10 tandems (Kiyotani et al., 2010; Kim et al., 2012; Qiao et al., 2016; Nguyen et al., 2019). Although it is not necessary to distinguish CYP2D6*10 from CYP2D6*36+*10 for phenotype prediction, the presence of CYP2D6*36+*10 tandems in diplotypes having two or more copies of CYP2D6*10 and/or *36 complicates CNV testing and data interpretation. Thus, it is important to understand which structures are present in a population and at what frequency they occur. This allows for strategic implementation of customized testing approaches to accurately predict CYP2D6 phenotype from genotype data. Additionally, the CYP2D7-like REP7 was observed downstream of a CYP2D6*10 allele in two subjects. The presence of the REP7 is likely not impacting the allele’s function, and thus a value of 0.25 was used for AS calculations. Despite not affecting CYP2D6 activity, the observation of the CYP2D7-like downstream region with CYP2D6*10 suggests that both hybrid and tandem arrangements could be observed in other alleles within the Hmong population.
Although the sample size of our study was modest, thereby limiting the generalizability of the CYP2D6 allele and diplotype frequencies to the Hmong community at large, we nonetheless discovered the presence of the CYP2D6*75 allele in this population sample. This allele was first described in Han populations from mainland China (Qin et al., 2008). Notably, the CYP2D6*75 allele found in this study had an additional variant (−498C > A) and was thus designated as a novel suballele (Figure 2) by PharmVar. Moreover, there is not yet enough knowledge regarding the function of this allele; CPIC is currently listing its function as ‘uncertain’. We believe that finding this relatively rare allele in our limited Hmong cohort suggests that there are other “rare alleles” that previously have only been discovered in Chinese or other EA populations which may not only be present in the Hmong but be more prevalent.
We also discovered a novel CYP2D6*10 suballele which has a synonymous SNP in exon 5 (2492G>A; p. K239=). This SNP has not been found in any other haplotype defined by PharmVar and is currently unique to CYP2D6*10.007. The function of this allele is predicted to be equivalent to those of other CYP2D6*10 alleles.
The third novel haplotype identified was designated CYP2D6*36.004. We would like to highlight that this haplotype has several SNPs in addition to 100C>T and the exon 9 conversion which causes the CYP2D6*36 hybrid to be nonfunctional. Due to the presence of these variants, this novel haplotype was designated as a CYP2D6*36 suballele by PharmVar. Additional SNPs observed were: 1612T>A (p.F120I) and 1618G>T (p.A122S), which are the defining core SNPs of CYP2D6*53. Note that 1612T>A is also a core SNP of CYP2D6*49. Due to the presence of 1612T>A and 1618G>T Astrolabe returned a CYP2D6*53 call and CNV analysis suggested the presence of a CYP2D6*36+*10 tandem (the initial tentative diplotype call was CYP2D6*36+*10/*53). However, follow-up studies using XL-PCR and Sanger sequencing revealed that both CYP2D6*53 core SNPs were on the upstream CYP2D6*36 gene copy in the tandem. The diplotype of this subject was ultimately resolved as CYP2D6*1.010/*36.004+*10.002, which can be collapsed to CYP2D6*1/*36+*10 for reporting. First discovered in a Japanese population (Ebisawa et al., 2005), the CYP2D6*53 allele and its activity were found to be comparable to that of CYP2D6*1 (CPIC assigned normal function to CYP2D6*53). Recent studies suggest, however, that CYP2D6*53 may have increased activity based on in vitro data (Muroi et al., 2014; Glass et al., 2018). This underscores the necessity to discriminate between CYP2D6*36.004 and *53 when 1612T>A and 1618G>T are detected; these alleles are functionally different (no function vs. normal or possibly increased function). Accurate diplotype determination is even further complicated as commercial tests may only test for 1612T>A (present in CYP2D6*49 and *53) but not 1618G>T (present in CYP2D6*36.004 and *53). This may lead to a false-positive CYP2D6*49 call and decreased function assignment.
Taken together, this study demonstrated the complexity and challenges of accurately calling CYP2D6 star alleles and diplotypes and corresponding phenotype assignments despite the availability of NGS data and quantitative CNV testing. Additionally, although the CYP2D6 enzyme activity of the three novel suballeles (*10.007, *36.004, and *75.002) are expected to be consistent with their major alleles, a dedicated phenotyping study should be conducted to assert their impact on the phenotype.
The concordance of diplotype calls and respective AS between the consensus calls and the clinical RightMed® test results was 66.7% (8/12). Furthermore, the concordance for predicted phenotypes between the two approaches was only 50% (6/12). The observed discordance was mainly attributed to the differences in technology used between the platforms. The utilization of NGS, XL-PCR and Sanger sequencing allowed us to identify copy number and structural variants and all SNPs present in the individuals. Lower concordance between phenotypes in contrast to diplotype calls can be attributed to using different methods for translating diplotype to phenotype. For example, a diplotype with an AS of 1.25 is categorized as IM to NM by RightMed® test while the CPIC recommended genotype to phenotype translation method categorized this AS as NM. Likewise, an AS of 0.25 is categorized by the RightMed® test as PM to IM while CPIC categorizes this AS as IM. Using different phenotype assignments based on the same genotype can cause confusion for prescribers who are selecting medications based on PGx results.
The increasing number of CPIC guidelines related to CYP2D6 (Crews et al., 2015; Hicks et al., 2015; Bell et al., 2017; Hicks et al., 2017; Goetz et al., 2018) highlights the critical need to understand CYP2D6 genetic variation and individual activity of this enzyme in special populations, such as the Hmong. This perspective was specifically identified by participants’ engaged in focus group meetings conducted as part of the VIP-Hmong study (Culhane-Pera et al., 2017; Holzer et al., 2020). Specifically themes from focus groups questioning how clinicians decide on drug and dosage selection for Hmong individuals based on guidance and data generated from studies of Western medications (Culhane-Pera et al., 2017) conducted in non-Hmong, raise prescient questions germane to this study. Our study findings could serve to motivate clinicians and payers to adopt and support individualized PGx testing for all individuals.
Conclusion
Our ‘first in Hmong’ CYP2D6 study suggests that the Hmong represent a population that exhibit unique CYP2D6 allelic variation in that a sizable portion of subjects have decreased CYP2D6 activity. Results utilizing different platforms also illustrated the extent and nature of important sources of variation. These preliminary findings underscore the importance of thoroughly investigating and acknowledging unique populations to have relevant genetic variations which have the potential effect on phenotype prediction accuracy, therapeutic drug response and possibly important clinical outcomes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.pharmvar.org/haplotype/1720, https://www.pharmvar.org/haplotype/1725, https://www.pharmvar.org/haplotype/1728.
Ethics Statement
The studies involving human participants were reviewed and approved by the University of Minnesota Institutional Review Board (UMN IRB #1702M06041 and IRB #1408M53223). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Conceptualization: YFW, AG, and RJS. Methodology: YFW, AG, ECB, WYW, and RJS. Data Analysis: YFW, AG, ECB, and WYW. Funding Acquisition: RJS. Writing–Original Draft Preparation: YFW, Writing–Review and Editing: YFW, AG, ECB, WYW, and RJS; Supervision: AG and RJS.
Funding
Grand Challenge Exploratory Research Award, Office of the Executive Vice President and Provost, University of Minnesota and National Institutes of Health’s National Center for Advancing Translational Sciences, grant UL1TR002494.
Conflict of Interest
AG was the Director of PharmVar.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors wish to thank the support from all the participants and the Hmong Genomic Community. In addition, we appreciate Kathleen A Culhane-Pera, Muaj Lo, Txia Xiong and Kerui Peng, and Koobmeej Lee for their efforts recruiting the study participants, the Genomic Medicine Center at the Children’s Mercy Research Institute, Kansas City for performing next-generation sequencing and Karim Pirani for technical assistance with Sanger sequencing. We also thank scientists at OneOme, LLC for their support in providing RightMed® test; and Adam Herman and Marissa Macchietto at Minnesota Supercomputing Institute, University of Minnesota for providing critical assistance in sequencing data analysis. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health’s National Center for Advancing Translational Sciences.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.867331/full#supplementary-material
References
Arcan, C., Larson, N., Bauer, K., Berge, J., Story, M., and Neumark-Sztainer, D. (2014). Dietary and Weight-Related Behaviors and Body Mass index Among Hispanic, Hmong, Somali, and White Adolescents. J. Acad. Nutr. Diet. 114, 375–383. doi:10.1016/j.jand.2013.11.019
Bart, G., Wang, Q., Hodges, J. S., Nolan, C., and Carlson, G. (2012). Superior Methadone Treatment Outcome in Hmong Compared with Non-Hmong Patients. J. Subst. Abuse Treat. 43, 269–275. doi:10.1016/j.jsat.2011.12.006
Bathum, L., Johansson, I., Ingelman-Sundberg, M., Hørder, M., and Brøsen, K. (1998). Ultrarapid Metabolism of Sparteine: Frequency of Alleles with Duplicated CYP2D6 Genes in a Danish Population as Determined by Restriction Fragment Length Polymorphism and Long Polymerase Chain Reaction. Pharmacogenetics 8, 119–123. doi:10.1097/00008571-199804000-00004
Bell, G. C., Caudle, K. E., Whirl-Carrillo, M., Gordon, R. J., Hikino, K., Prows, C. A., et al. (2017). Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 Genotype and Use of Ondansetron and Tropisetron. Clin. Pharmacol. Ther. 102, 213–218. doi:10.1002/cpt.598
Boone, E. C., Wang, W. Y., Gaedigk, R., Cherner, M., Bérard, A., Leeder, J. S., et al. (2020). Long-Distance Phasing of a Tentative "Enhancer" Single-Nucleotide Polymorphism with CYP2D6 Star Allele Definitions. Front. Pharmacol. 11, 486. doi:10.3389/fphar.2020.00486
Buermans, H. P., Vossen, R. H., Anvar, S. Y., Allard, W. G., Guchelaar, H. J., White, S. J., et al. (2017). Flexible and Scalable Full-Length CYP2D6 Long Amplicon PacBio Sequencing. Hum. Mutat. 38, 310–316. doi:10.1002/humu.23166
Caudle, K. E., Sangkuhl, K., Whirl-Carrillo, M., Swen, J. J., Haidar, C. E., Klein, T. E., et al. (2020). Standardizing CYP2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin. Transl Sci. 13, 116–124. doi:10.1111/cts.12692
Cavallari, L. H., Lee, C. R., Beitelshees, A. L., Cooper-Dehoff, R. M., Duarte, J. D., Voora, D., et al. (2018). Multisite Investigation of Outcomes with Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy after Percutaneous Coronary Intervention. JACC Cardiovasc. Interv. 11, 181–191. doi:10.1016/j.jcin.2017.07.022
Crews, K. R., Caudle, K. E., Dunnenberger, H. M., Sadhasivam, S., and Skaar, T. C. (2015). Considerations for the Utility of the CPIC Guideline for CYP2D6 Genotype and Codeine Therapy. Clin. Chem. 61, 775–776. doi:10.1373/clinchem.2014.237412
Culhane-Pera, K. A., Moua, M., Vue, P., Xiaaj, K., Lo, M. X., and Straka, R. J. (2017). Leaves Imitate Trees: Minnesota Hmong Concepts of Heredity and Applications to Genomics Research. J. Community Genet. 8, 23–34. doi:10.1007/s12687-016-0284-2
De, T., Alarcon, C., Hernandez, W., Liko, I., Cavallari, L. H., Duarte, J. D., et al. (2018). Association of Genetic Variants With Warfarin-Associated Bleeding Among Patients of African Descent JAMA 320 (16), 1670–1677. doi:10.1001/jama.2018.14955
Ebisawa, A., Hiratsuka, M., Sakuyama, K., Konno, Y., Sasaki, T., and Mizugaki, M. (2005). Two Novel Single Nucleotide Polymorphisms (SNPs) of the CYP2D6 Gene in Japanese Individuals. Drug Metab. Pharmacokinet. 20, 294–299. doi:10.2133/dmpk.20.294
Fukunaga, K., Hishinuma, E., Hiratsuka, M., Kato, K., Okusaka, T., Saito, T., et al. (2020). Determination of Novel CYP2D6 Haplotype Using the Targeted Sequencing Followed by the Long-Read Sequencing and the Functional Characterization in the Japanese Population. J. Hum. Genet. 66 (2), 1–11. doi:10.1038/s10038-020-0815-x
Gaedigk, A., Fuhr, U., Johnson, C., Bérard, L. A., Bradford, D., and Leeder, J. S. (2010a). CYP2D7-2D6 Hybrid Tandems: Identification of Novel CYP2D6 Duplication Arrangements and Implications for Phenotype Prediction. Pharmacogenomics 11, 43–53. doi:10.2217/pgs.09.133
Gaedigk, A., Ingelman-Sundberg, M., Miller, N. A., Leeder, J. S., Whirl-Carrillo, M., Klein, T. E., et al. (2018). The Pharmacogene Variation (PharmVar) Consortium: Incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin. Pharmacol. Ther. 103, 399–401. doi:10.1002/cpt.910
Gaedigk, A., Jaime, L. K., Bertino, J. S., Bérard, A., Pratt, V. M., Bradfordand, L. D., et al. (2010b). Identification of Novel CYP2D7-2D6 Hybrids: Non-Functional and Functional Variants. Front. Pharmacol. 1, 121. doi:10.3389/fphar.2010.00121
Gaedigk, A., Riffel, A. K., and Leeder, J. S. (2015). CYP2D6 Haplotype Determination Using Long Range Allele-Specific Amplification: Resolution of a Complex Genotype and a Discordant Genotype Involving the CYP2D6*59 Allele. J. Mol. Diagn. 17, 740–748. doi:10.1016/j.jmoldx.2015.06.007
Gaedigk, A., Sangkuhl, K., Whirl-Carrillo, M., Klein, T., and Leeder, J. S. (2017). Prediction of CYP2D6 Phenotype from Genotype across World Populations. Genet. Med. 19, 69–76. doi:10.1038/gim.2016.80
Gaedigk, A., Simon, S. D., Pearce, R. E., Bradford, L. D., Kennedy, M. J., and Leeder, J. S. (2008). The CYP2D6 Activity Score: Translating Genotype Information Into A Qualitative Measure Of Phenotype Clin. Pharmacol. Ther. 83 (2), 234–242. doi:10.1038/sj.clpt.6100406
Gaedigk, A., Turner, A., Everts, R. E., Scott, S. A., Aggarwal, P., Broeckel, U., et al. (2019). Characterization of Reference Materials for Genetic Testing of CYP2D6 Alleles: A GeT-RM Collaborative Project. J. Mol. Diagn. 21, 1034–1052. doi:10.1016/j.jmoldx.2019.06.007
Glass, S. M., Martell, C. M., Oswalt, A. K., Osorio-Vasquez, V., Cho, C., Hicks, M. J., et al. (2018). CYP2D6 Allelic Variants *34, *17-2, *17-3, and *53 and a Thr309Ala Mutant Display Altered Kinetics and NADPH Coupling in Metabolism of Bufuralol and Dextromethorphan and Altered Susceptibility to Inactivation by SCH 66712. Drug Metab. Dispos 46, 1106–1117. doi:10.1124/dmd.117.079871
Goetz, M. P., Sangkuhl, K., Guchelaar, H. J., Schwab, M., Province, M., Whirl-Carrillo, M., et al. (2018). Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin. Pharmacol. Ther. 103, 770–777. doi:10.1002/cpt.1007
Hernandez, W., Gamazon, E. R., Aquino-Michaels, K., Patel, S., O'Brien, T. J., Harralson, A. F., et al. (2014). Ethnicity-specific Pharmacogenetics: The Case of Warfarin in African Americans Pharmacogenomics J. 14 (3), 223–228. doi:10.1038/tpj.2013.34
Hicks, J. K., Bishop, J. R., Sangkuhl, K., Müller, D. J., Ji, Y., Leckband, S. G., et al. (2015). Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin. Pharmacol. Ther. 98, 127–134. doi:10.1002/cpt.147
Hicks, J. K., Sangkuhl, K., Swen, J. J., Ellingrod, V. L., Müller, D. J., Shimoda, K., et al. (2017). Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2D6 and CYP2C19 Genotypes and Dosing of Tricyclic Antidepressants: 2016 Update. Clin. Pharmacol. Ther. 102, 37–44. doi:10.1002/cpt.597
Holzer, K., Culhane-Pera, K. A., Straka, R. J., Wen, Y. F., Lo, M., Lee, K., et al. (2020). Hmong Participants' Reactions to Return of Individual and Community Pharmacogenetic Research Results: "A Positive Light for Our Community". J. Community Genet. 12 (1), 53–65. doi:10.1007/s12687-020-00475-3
Hulot, J. S., Chevalier, B., Belle, L., Cayla, G., Khalife, K., Funck, F., et al. (2020). Routine CYP2C19 Genotyping to Adjust Thienopyridine Treatment after Primary PCI for STEMI: Results of the GIANT Study. JACC Cardiovasc. Interv. 13, 621–630. doi:10.1016/j.jcin.2020.01.219
Johnson, J. A., Caudle, K. E., Gong, L., Whirl-Carrillo, M., Stein, C. M., Scott, S. A., et al. (2017). Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clin. Pharmacol. Ther. 102, 397–404. doi:10.1002/cpt.668
Karamperis, K., Koromina, M., Papantoniou, P., Skokou, M., Kanellakis, F., Mitropoulos, K., et al. (2021). Economic Evaluation in Psychiatric Pharmacogenomics: a Systematic Review. Pharmacogenomics J. 21, 533–541. doi:10.1038/s41397-021-00249-1
Kim, J., Lee, S. Y., and Lee, K. A. (2012). Copy Number Variation and Gene Rearrangements in CYP2D6 Genotyping Using Multiplex Ligation-dependent Probe Amplification in Koreans. Pharmacogenomics 13, 963–973. doi:10.2217/pgs.12.58
Kim, J. H., Tan, D. S., and Chan, M. Y. Y. (2021). Cost-Effectiveness of CYP2C19-Guided Antiplatelet Therapy for Acute Coronary Syndromes in Singapore. Pharmacogenomics J. 21, 243–250. doi:10.1038/s41397-020-00204-6
Kiyotani, K., Shimizu, M., Kumai, T., Kamataki, T., Kobayashi, S., and Yamazaki, H. (2010). Limited Effects of Frequent CYP2D6*36-*10 Tandem Duplication Allele on In Vivo Dextromethorphan Metabolism in a Japanese Population. Eur. J. Clin. Pharmacol. 66, 1065–1068. doi:10.1007/s00228-010-0876-4
Krebs, K., and Milani, L. (2019). Translating Pharmacogenomics into Clinical Decisions: Do Not Let the Perfect Be the Enemy of the Good. Hum. Genomics 13, 39. doi:10.1186/s40246-019-0229-z
Langaee, T., Hamadeh, I., Chapman, A. B., Gums, J. G., and Johnson, J. A. (2015). A Novel Simple Method for Determining CYP2D6 Gene Copy Number and Identifying Allele(s) with Duplication/multiplication. PLoS One 10, e0113808. doi:10.1371/journal.pone.0113808
Lee, H. Y., Lytle, K., Yang, P. N., and Lum, T. (2010). Mental Health Literacy in Hmong and Cambodian Elderly Refugees: A Barrier to Understanding, Recognizing, and Responding to Depression. Int. J. Aging Hum. Dev. 71, 323–344. doi:10.2190/AG.71.4.d
Lee, S., and Chang, J. (2012). Mental Health Status of the Hmong Americans in 2011: Three Decades Revisited. J. Soc. Work Disabil. Rehabil. 11, 55–70. doi:10.1080/1536710X.2012.648117
Lee, S. E. (2013). Mental Health of Hmong Americans: A Metasynthesis of Academic Journal Article Findings. Hmong Stud. J. 14, 1–31.
Montané Jaime, L. K., Lalla, A., Steimer, W., and Gaedigk, A. (2013). Characterization of the CYP2D6 Gene Locus and Metabolic Activity in Indo- and Afro-Trinidadians: Discovery of Novel Allelic Variants. Pharmacogenomics 14, 261–276. doi:10.2217/pgs.12.207
Muroi, Y., Saito, T., Takahashi, M., Sakuyama, K., Niinuma, Y., Ito, M., et al. (2014). Functional Characterization of Wild-Type and 49 CYP2D6 Allelic Variants for N-Desmethyltamoxifen 4-Hydroxylation Activity. Drug Metab. Pharmacokinet. 29, 360–366. doi:10.2133/dmpk.dmpk-14-rg-014
National Center for Biotechnology Information (2022). Genetic Test Registry CYP2D6 Cytochrome P450 Family 2 Subfamily D Member 6 [Online]. U.S. National Library of Medicine. Available at: https://www.ncbi.nlm.nih.gov/gtr/genes/1565/ (Accessed January 24, 2021).
Nelson, J. M., Wen, Y.-F., Culhane-Pera, K. A., Lo, M., Xiong, T., Lee, K., et al. (2020). Abstract 234: Pharmacogenomics as a Guide to Improve Antidepressant Drug Selection and Dosing in Minnesota Hmong. 2020 ACCP Annual Meeting (Virtual) October 19 - 30, 2020. Jaccp: J. Am. Coll. Clin. Pharm. 3, 1525–1689. doi:10.1002/jac5.1351
Nguyen, H. H., Ma, T. T. H., Vu, N. P., Bach, Q. T. N., Vu, T. H., Nguyen, T. D., et al. (2019). Single Nucleotide and Structural Variants of CYP2D6 Gene in Kinh Vietnamese Population. Medicine (Baltimore) 98, e15891. doi:10.1097/MD.0000000000015891
Nofziger, C., Turner, A. J., Sangkuhl, K., Whirl-Carrillo, M., Agundez, J. A. G., Black, J. L., et al. (2019). PharmVar GeneReview: CYP2D6. Clin. Pharmacol. Ther. 107 (1), 154–170. doi:10.1002/cpt.1643
Oyenuga, M., Yang, J. K., Prizment, A. E., Bushhouse, S., Demerath, E. W., and Spector, L. G. (2018). Cancer Patterns in Hmong in Minnesota, 2000 to 2012. Cancer 124, 3560–3566. doi:10.1002/cncr.31592
Park, L., Schwei, R. J., Xiong, P., and Jacobs, E. A. (2018). Addressing Cultural Determinants of Health for Latino and Hmong Patients with Limited English Proficiency: Practical Strategies to Reduce Health Disparities. J. Racial Ethn. Health Disparities 5, 536–544. doi:10.1007/s40615-017-0396-3
Pratt, V. M., Cavallari, L. H., Del Tredici, A. L., Hachad, H., Ji, Y., Kalman, L. V., et al. (2020). Recommendations for Clinical Warfarin Genotyping Allele Selection: A Report of the Association for Molecular Pathology and the College of American Pathologists. J. Mol. Diagn. 22, 847–859. doi:10.1016/j.jmoldx.2020.04.204
Pratt, V. M., Cavallari, L. H., Del Tredici, A. L., Hachad, H., Ji, Y., Moyer, A. M., et al. (2019). Recommendations for Clinical CYP2C9 Genotyping Allele Selection: A Joint Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 21, 746–755. doi:10.1016/j.jmoldx.2019.04.003
Pratt, V. M., Del Tredici, A. L., Hachad, H., Ji, Y., Kalman, L. V., Scott, S. A., et al. (2018). Recommendations for Clinical CYP2C19 Genotyping Allele Selection: A Report of the Association for Molecular Pathology. J. Mol. Diagn. 20, 269–276. doi:10.1016/j.jmoldx.2018.01.011
Pratt, V. M., Cavallari, L. H., Del Tredici, A. L., Gaedigk, A., Hachad, H., Ji, Y., et al. (2021). Recommendations for Clinical CYP2D6 Genotyping Allele Selection: A Joint Consensus Recommendation of the Association for Molecular Pathology, College of American Pathologists, Dutch Pharmacogenetics Working Group of the Royal Dutch Pharmacists Association, and European Society for Pharmacogenomics and Personalized Therapy. J. Mol. Diagn. 23 (9), 1047–1064. doi:10.1016/j.jmoldx.2021.05.013
Qiao, W., Martis, S., Mendiratta, G., Shi, L., Botton, M. R., Yang, Y., et al. (2019). Integrated CYP2D6 Interrogation for Multiethnic Copy Number and Tandem Allele Detection. Pharmacogenomics 20, 9–20. doi:10.2217/pgs-2018-0135
Qiao, W., Yang, Y., Sebra, R., Mendiratta, G., Gaedigk, A., Desnick, R. J., et al. (2016). Long-Read Single Molecule Real-Time Full Gene Sequencing of Cytochrome P450-2D6. Hum. Mutat. 37, 315–323. doi:10.1002/humu.22936
Qin, S., Shen, L., Zhang, A., Xie, J., Shen, W., Chen, L., et al. (2008). Systematic Polymorphism Analysis of the CYP2D6 Gene in Four Different Geographical Han Populations in Mainland China. Genomics 92, 152–158. doi:10.1016/j.ygeno.2008.05.004
Roman, Y. M., Culhane-Pera, K., Lo, M., Yang, S., Yang, J., Lo, M., et al. (2017). The Impact of Rs505802 for SLC22A12 on Oxipurinol and Uric Acid Disposition in Hmong Patients on Allopurinol from the Genetics of Hyperuricemia Therapy in Hmong (GOUT-H) Study. Clin. Pharmacol. Ther. 101, S5–S99. doi:10.1002/cpt.570
Saravanakumar, A., Sadighi, A., Ryu, R., and Akhlaghi, F. (2019). Physicochemical Properties, Biotransformation, and Transport Pathways of Established and Newly Approved Medications: A Systematic Review of the Top 200 Most Prescribed Drugs vs. The FDA-Approved Drugs between 2005 and 2016. Clin. Pharmacokinet. 58, 1281–1294. doi:10.1007/s40262-019-00750-8
Smith, D. M., Weitzel, K. W., Elsey, A. R., Langaee, T., Gong, Y., Wake, D. T., et al. (2019). CYP2D6-Guided Opioid Therapy Improves Pain Control in CYP2D6 Intermediate and Poor Metabolizers: A Pragmatic Clinical Trial. Genet. Med. 21, 1842–1850. doi:10.1038/s41436-018-0431-8
Sun, B., Wen, Y. F., Culhane-Pera, K. A., Lo, M., Xiong, T., Lee, K., et al. (2020). Differences in Predicted Warfarin Dosing Requirements between Hmong and East Asians Using Genotype-Based Dosing Algorithms. Pharmacotherapy 41 (3), 265–276. doi:10.1002/phar.2487
Tanner, J. A., Davies, P. E., Overall, C. C., Grima, D., Nam, J., and Dechairo, B. M. (2020). Cost-Effectiveness of Combinatorial Pharmacogenomic Testing for Depression from the Canadian Public Payer Perspective. Pharmacogenomics 21, 521–531. doi:10.2217/pgs-2020-0012
Thao, K. K., Arndt, B., Tandias, A., and Hanrahan, L. (2015). The Prevalence of Type 2 Diabetes Mellitus in a Wisconsin Hmong Patient Population. WMJ 114, 190–195.
Twist, G. P., Gaedigk, A., Miller, N. A., Farrow, E. G., Willig, L. K., Dinwiddie, D. L., et al. (2016). Constellation: A Tool for Rapid, Automated Phenotype Assignment of a Highly Polymorphic Pharmacogene, CYP2D6, from Whole-Genome Sequences. NPJ Genom Med. 1, 15007. doi:10.1038/npjgenmed.2015.7
Twist, G. P., Gaedigk, A., Miller, N. A., Farrow, E. G., Willig, L. K., Dinwiddie, D. L., et al. (2017). Erratum: Constellation: A Tool for Rapid, Automated Phenotype Assignment of a Highly Polymorphic Pharmacogene, CYP2D6, from Whole-Genome Sequences. NPJ Genom Med. 2, 16039. doi:10.1038/npjgenmed.2016.39
United States Census Bureau (2016). American Community Survey 1-Year Estimates [Online]. Available at: https://factfinder.census.gov/faces/tableservices/jsf/pages/productview.xhtml?pid=ACS_16_1YR_S0201&prodType=table (Accessed March, 2018).
Wen, Y. F., Culhane-Pera, K. A., Thyagarajan, B., Bishop, J. R., Zierhut, H., Lo, M., et al. (2020). Potential Clinical Relevance of Differences in Allele Frequencies Found within Very Important Pharmacogenes between Hmong and East Asian Populations. Pharmacotherapy 40, 142–152. doi:10.1002/phar.2360
Whale, A. S., Huggett, J. F., and Tzonev, S. (2016). Fundamentals of Multiplexing with Digital PCR. Biomol. Detect. Quantif 10, 15–23. doi:10.1016/j.bdq.2016.05.002
Yang, Y., Botton, M. R., Scott, E. R., and Scott, S. A. (2017). Sequencing the CYP2D6 Gene: From Variant Allele Discovery to Clinical Pharmacogenetic Testing. Pharmacogenomics 18, 673–685. doi:10.2217/pgs-2017-0033
Zhu, Y., Moriarty, J. P., Swanson, K. M., Takahashi, P. Y., Bielinski, S. J., Weinshilboum, R., et al. (2021). A Model-Based Cost-Effectiveness Analysis of Pharmacogenomic Panel Testing in Cardiovascular Disease Management: Preemptive, Reactive, or None? Genet. Med. 23, 461–470. doi:10.1038/s41436-020-00995-w
Keywords: CYP2D6, pharmacogenetics, minority health, population genetics, ethnic variability, targeted exome sequencing, Hmong
Citation: Wen YF, Gaedigk A, Boone EC, Wang WY and Straka RJ (2022) The Identification of Novel CYP2D6 Variants in US Hmong: Results From Genome Sequencing and Clinical Genotyping. Front. Pharmacol. 13:867331. doi: 10.3389/fphar.2022.867331
Received: 01 February 2022; Accepted: 21 February 2022;
Published: 21 March 2022.
Edited by:
Travis J. OBrien, George Washington University, United StatesReviewed by:
Pajaree Chariyavilaskul, Chulalongkorn University, ThailandPedro Dorado, University of Extremadura, Spain
Ingrid Fricke-Galindo, Instituto Nacional de Enfermedades Respiratorias-México (INER), Mexico
Copyright © 2022 Wen, Gaedigk, Boone, Wang and Straka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert J. Straka, c3RyYWswMDFAdW1uLmVkdQ==
 Ya Feng Wen
Ya Feng Wen Andrea Gaedigk
Andrea Gaedigk Erin C. Boone
Erin C. Boone Wendy Y. Wang
Wendy Y. Wang Robert J. Straka
Robert J. Straka