
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 05 April 2022
Sec. Experimental Pharmacology and Drug Discovery
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.864336
This article is part of the Research TopicMetal-Complexed Molecules of Natural and Synthetic Origin: Pharmacological Advances and Therapeutic ApplicationsView all 5 articles
 Niaz Muhammad1*
Niaz Muhammad1* Mukhtar Ahmad1
Mukhtar Ahmad1 Muhammad Sirajuddin2*
Muhammad Sirajuddin2* Zafar Ali1
Zafar Ali1 Nikolay Tumanov3
Nikolay Tumanov3 Johan Wouters3
Johan Wouters3 Abdelbasset Chafik4,5
Abdelbasset Chafik4,5 Kübra Solak6,7
Kübra Solak6,7 Ahmet Mavi7,8
Ahmet Mavi7,8 Shabbir Muhammad9
Shabbir Muhammad9 Shaukat Shujah10
Shaukat Shujah10 Saqib Ali11
Saqib Ali11 Abdullah G. Al-Sehemi9
Abdullah G. Al-Sehemi9Four new carboxylates complexes with general formula R2SnL2 and R3SnL, where R = n-butyl (1, 3), methyl (2, 4) and L = 4-Chlorophenoxyacetate, were synthesized in significant yields. FT-IR analysis revealed a chelating (1 and 2) and a bridging bidentate (3 and 4) coordination modes for the carboxylate ligand in solid state which was further confirmed by the single crystal X-ray analysis of complex 4. The NMR data (1H, 13C and 119Sn) revealed a higher coordination number around the tin center in R2SnL2 (1 and 2) compared to R3SnL (3 and 4). A close matching was observed between the experimental and calculated structures (obtained at B3LYP/6-31G* + LANL2DZ basis set). Quantum chemical analysis indicates that the carboxylate moiety has the major contribution in the formation of filled and unfilled orbitals as well as in ligand to ligand intramolecular charge transfer during the electronic transitions. The cytotoxicity data of the screened compounds evaluated against lung cancer cell line (A549) and normal lung fibroblast cell line (MRC-5) revealed that 1, 3 and 4 have shown dose dependent cytotoxic effects while HL and 2 have shown steady and low cytotoxic activities. The antibacterial activity of complexes 1–4 is higher than that of HL. Molecular docking study showed an intercalation binding mode for complex 3 with DNA (docking score = −3.6005) involving four polar interactions. Complex 3 docking with tubulin (PDB ID 1SA0) with colchicine as a target protein resulted in three polar interactions (docking score −5.2957). Further, the docking analysis of the HL and 1–4 has shown an adequate interactions with the coronavirus SARS-CoV-2 spike protein, nucleocapsid protein and human angiotensin converting enzyme (ACE2).
Cancer is the most threating disease of the present time. In spite of extensive research in the field, only few drugs are available commercially. Cis-platin being the lead one is associated with some severe side effects like neuro-, nephro-, and ototoxicity (Mubeen and Kini, 2012; Pantelić et al., 2019) which motivates the researchers for developing of non-platinum based anticancer drugs. Organotin(IV) carboxylates are found be the most promising alternatives, as these have shown higher apoptosis inducing character compared to the platinum complexes, both in vivo and in vitro (Indumathy et al., 2007; Butt et al., 2019). In addition to the anticancer potential, organotin(IV) carboxylates also possess significant antibacterial, antifungal, insecticidal, anti-tuberculosis and antidiabetic activities (Kovala-Demertzi et al., 2009; Tzimopoulos et al., 2010; Tariq et al., 2013; Sirajuddin et al., 2014a; Sirajuddin et al., 2014b; Shah et al., 2015; Tariq et al., 2016; Debnath et al., 2019; Sirajuddin et al., 2021).
In spite of wide range available data on the anticancer activity of organotin(IV) carboxylates, exact conclusive results are not available on the cytotoxic effects of these complexes. Some possible targets identified for the anticancer activity of the organotin(IV) complexes are discussed in the form nucleotide base pairs, nitrogen atoms of DNA, phosphate groups on the backbone of DNA and the sulfhydryl groups on proteins (Gielen, 2008; Hong et al., 2015; Xiao et al., 2020).
For an effective interaction with these possible target sites and other bioactivities, the coordination expansion ability of Sn atom, number and nature of R groups bonded to Sn and nature of the carboxylate ligand are found the important controlling factors (Saxena and Huber, 1989; Hook, 1994; Gielen, 2003; Hadjikakou and Hadjiliadis, 2009; Sirajuddin et al., 2012). The nature of carboxylate ligand is quite important in deciding the bioactivity of the organotin(IV) complexes as it can alter hydro/lipophilicity of the complex and assist the transportation of complexes across the cell membrane (Cooney and Wuertz, 1989; Yenişehirli et al., 2010; Ettouhami et al., 2012). Also for the same ligand the earlier reported toxicity order is as: R3Sn > R2Sn > RSn. (Basu, 2008). In addition, the biological target (microorganism and stain, nature of the cancer cell lines) also significantly affect the activity as well.
A. Lavecchia, et al., investigated the screening results for a series of chiral phenoxyacetic acid analogues agonist activity toward the human PPARα (hPPARα) and PPARγ (hPPARγ) subtypes and found some of analogues are potent PPARα agonists as well as PPARγ agonists. Interestingly, for one of these analogues, the stereoselectivity toward PPARα was reversed which was further confirmed by docking experiments to rationalize this peculiar behavior (Fracchiolla et al., 2007). Santiago Gómez-Ruiz and Goran N. Kaluđerović et al., reported the cytotoxic activity of phenoxyacetic acid based di and triphenyltin(IV) derivatives against tumor cell lines human adenocarcinoma HeLa, human myelogenous leukemia K562, human malignant melanoma Fem-x and normal immunocompetent cells, peripheral blood mononuclear cells PBMC. It is evidenced form their study that triphenyltin(IV) complexes show higher activities than the diphenyltin(IV) derivatives and some of the invested compounds are more potent than cis-platin against all the tested cells and relative high selectivity especially on K562 cells (Gómez-Ruiz et al., 2008; Kaluđerović et al., 2010). Tariq Ali et al., studied via docking studies the interactions of 2-chlorophenyl ethanoic acid based organotin(IV) complexes against the SARS-Co-2 virus which is responsible for the most challenging disease Covid-19 and found a considerable binding interactions of compounds with the angiotensin-converting enzyme 2 (ACE2) and with the nucleocapsid protein of the SARS- CoV-2 virus.
Thus keeping in view the biological importance of the phenoxyacetic acid derivatives especially against SARS- CoV-2 virus, here we have chosen the 4-chlorophenoxyacetate for the preparation of organotin(IV) complexes. The aim of the present study is to described the synthesis, characterization, in vitro anticancer and antibacterial activity of di- and tri-n-butyl/methyltin(IV) derivatives of 4-chlorophenoxy acetic acid. The interaction of the synthesized compounds with DNA, spike protein, nucleocapsid protein of coronavirus SARS-CoV-2 and human angiotensin converting enzyme (ACE2) was also studied using molecular docking.
Trimethyltin chloride (1,498+%), tri-n-btyltin chlorides (96%), dimethyltin dichlorides (98%), di-n-butyltin dichlorides (96%), 4-chlorophenoxyacetic acid (98+%), sodium bicarbonate (99.7%) and analytical grade chloroform (99.8+%) were purchased from commercial suppliers and used as such without any further purification. However, chloroform was dried by a reported method (Armarego and Chai, 2003).
Lung cancer cell line (A549) and Normal lung fibroblast cell line (MRC-5 An1) were provided by Eastern Anatolia High Technology Application and Research Center (DAYTAM, Erzurum, Turkey) and Ministry of Agriculture and Forestry (şap institute, Turkey), respectively. 3-(4,5-dimethylthiazol-2-yl)-2.5-diphenyl tetrazolium bromide (MTT, M2128) was purchased from Sigma-Aldrich. Roswell Park Memorial Institute (RPMI) medium, Trypsin/Ethylenediaminetetra acetic acid (EDTA) solution, fetal bovine serum (FBS), L-glutamine were purchased from Gibco. Dimethylsulfoxide (DMSO) (cell culture reagent) was purchased from Santa Cruz Biotechnology.
A Gallenkamp (United Kingdom) electro thermal instrument was used to find the melting points of the synthesized compounds. Elemental analysis was done on Leco CHNS 932. FT-IR spectra (4,000–400 cm−1) were recorded on a Nicolet-6700 FT-IR spectrophotometer, Thermoscientific, United States. The NMR (1H, 13C and 119Sn) spectra were obtained at room temperature on a Bruker Advance Digital 300 MHz NMR spectrometer (Switzerland). Single-crystal X-ray diffraction data was collected using the Oxford Diffraction Gemini R Ultra diffractometer (Mo Kα, graphite monochromator, Ruby CCD area detector) at 295 (2) K.
Synthesis of the ligand salt (NaL) and complexes (1–4) is shown in Scheme 1 and is discussed as follow.
The ligand salt (NaL) was prepared by adding dropwise 20 ml aqueous sodium bicarbonate solution (20 mmol) to the 20 ml methanolic solution of 4-chlorophenoxyacetic acid, HL (20 mmol). The mixture was heated for 40 min at 60°C. The soluble salt (NaL) was recovered from the solution as a white solid by rotary evaporation and dried under vacuum.
For the synthesis of complex 1, the ligand salt (1.043 g, 5 mmol) and (n-C4H9)2SnCl2 (0.760 g, 2.5 mmol) were dissolved in 100 ml dry chloroform contained in a round bottom flask. The mixture was refluxed for 8 h. The resultant turbid mixture was cooled to a room temperature and left undisturbed for 24 h. The precipitated sodium chloride was removed from the mixture by filtration. The filtrate was evaporated under reduced pressure by rotary evaporator. The remaining solid crude product was recrystallized from chloroform and n-hexane mixture (4:1).
Chemical formula: C24H30Cl2O6Sn, Molecular Mass: 604.04, Percent yield: 84%. m.p.: 85–87°C. Anal Calc. (experimental values) %: C = 47.72 (47.69); H = 5.01 (5.03). FT-IR (cm−1): 1602s υ(COOasym), 1488s υ(COOsym), 114 Δυ, 504 m υ(Sn-C), 481w υ(Sn-O). 1H-NMR (CDCl3, ppm): 4.68 (s, 4H, H-2), 7.25 (d, 4H, H-4,4′), 6.85 (d, 4H, H-5,5′), 1.78–1.63 (m, 4H, H-α), 1.78–1.63 (m, 4H, H-β), 1.44–1.28 (m, 4H, H-γ), 0.94 (t, 6H, H-δ). 13C-NMR (CDCl3, pm): 177.7, 65.34, 156.2, 115.8, 129.5, 126.7 (C-1 to C-6, respectively), 28.2, 26.6, 26.4, 13.6 (C-α, β, γ, δ, respectively). 119Sn-NMR (CDCl3, ppm): −228.
Complex 2 was synthesized and recrystallized by the same procedure as used for complex 1, just by changing (CH3)2SnCl2 (0.549 g, 2.5 mmol).
Chemical formula: C18H18Cl2O6Sn, Molecular Mass: 519.95, Percent yield: 80%. m.p.: 180–181°C. Anal Calc. (experimental values) %: C = 41.58 (41.56); H = 3.49 (3.46). FT-IR (cm−1): 1609s υ(COOasym), 1488s υ(COOsym), 121 Δυ, 583s υ(Sn-C), 445 m υ(Sn-O). 1H-NMR (CDCl3, ppm), [2J (119/117Sn, 1H) in Hz]: 4.60 (s, 4H, H-2), 7.29 (d, 4H, H-4,4′), 6.91(d, 4H, H-5,5'), 0.84 (s, 6H, H-α) (101). 13C-NMR (CDCl3, pm): 172.1, 65.7, 157.2, 116.6, 129.5, 124.9 (C-1 to C-6, respectively), 4.1 (C-α). 119Sn-NMR (CDCl3, ppm): −220.
Complex 3 was made by the same procedure as used for complex 1, except equimolar amounts of the reactants are taken, i.e., NaL (0.522 g, 2.5 mmol), (n-C4H9)3SnCl (0.814 g, 2.5 mmol).
Chemical formula: C20H33ClO3Sn, Molecular Mass: 476.11, Percent yield: 82%. m.p.: 50–51°C. Anal Calc. (experimental values)%: C = 50.50 (50.47); H = 6.99 (6.88). FT-IR (cm−1): 1580s υ(COOasym), 1409s υ(COOsym), 171 Δυ, 566 m υ(Sn-C), 499 m υ(Sn-O). 1H-NMR (CDCl3, ppm): 4.54 (s, 2H, 2), 7.18 (d, 2H, 4,4′), 6.79 (d, 2H, 5,5′), 1.59 (t, 6H, Hα), 1.37–1.24 (m, 6H, Hβ), 1.37–1.24 (m, 6H, Hγ), 0.88 (t, 9H, Hδ). 13C-NMR (CDCl3, pm), [nJ (119Sn, 13C) in Hz]: 173.3, 65.8, 156.7, 115.8, 129.2, 126.0 (C-1 to C-6, respectively), 16.9 (343), 27.7 (21), 26.9 (64), 13.6 (C-α, β, γ, δ, respectively). 119Sn-NMR (CDCl3, ppm): −50.
Complex 4 was made by the same procedure as used for complex 3, just by changing (CH3)3SnCl (0.498 g, 2.5 mmol).
Chemical formula: C11H15ClO3Sn, Molecular Mass: 349.97, Percent yield: 88%. m.p.: 160–162°C. Anal Calc. (experimental values) %: C = 37.81 (37.79); H = 4.33 (4.29). FT-IR (cm−1): 1590s υ(COOasym), 1432s υ(COOsym), 158 Δυ, 552s υ(Sn-C), 452 m υ(Sn-O). 1H-NMR (CDCl3, ppm), 2J [(119Sn, 1H), Hz]: 4.55 (s, 2H, H-2), 7.24 (d, 2H, H-4,4′), 6.84 (d, 2H, H-5,5′), 0.61 (s, 9H, H-α) (59). 13C-NMR (CDCl3, pm), 1J [(119Sn, 13C) in Hz]: 173.4, 65.8, 156.6, 115.9, 129.4, 126.2 (C-1 to C-6, respectively), −1.9 (C-α) (394). 119Sn-NMR (CDCl3, ppm): −30.
Data collection, unit cells determination and data reduction were carried out using CrysAlis PRO software package (Rigaku, 2018). Using Olex2 and shelXle, the structure was solved with the SHELXT 2015 structure solution program by Intrinsic Phasing methods and refined by full-matrix least squares on |F|2 using SHELXL-2018/3 (Sheldrick G., 1997; Sheldrick, 1997b; Dolomanov et al., 2009; Sheldrick, 2009; Hübschle, 2011; Scheldrick, 2015). Non-hydrogen atoms were refined anisotropically. Hydrogen atoms were placed on calculated positions in riding mode with temperature factors fixed at 1.2 times Ueq of the parent carbon atoms (1.5 times for methyl groups). CCDC deposition number for 4 is 2128461.
The cytotoxicity test of the HL and complexes 1–4 was performed against lung cancer cell line (A549) and normal lung fibroblast cell line (MRC-5). Cells were counted by trypan blue staining and seeded in 96 well-plate with density of 5–10 × 103 cells/well in 0.15 ml of RPMI media containing 10% fetal bovine serum and 1% L-glutamine. The cells were then incubated for 24 h at 37°C (5% CO2, 90% humidity). Cell viability was analyzed using the MTT assay after treatment for 24 and 48 h with different concentrations (0, 0.1, 0.5, 1, 2, 5, 10 and 50 μg/ml) of the screened compounds. After the removal of the culture medium the cells were subsequently treated with 10 μL of 5 mg/ml thiazolyl blue tetrazolium bromide in 90 μL of media (Pettazzoni et al., 2011; Abu-Dief et al., 2021a; Saddik et al., 2021). After 2 h, 100 μL of DMSO was used to dissolve MTT salt. The absorbance was recorded at 570 nm by a plate reader (Biotek). Values from three wells of cells from the same preparation were averaged as a single value for that experiment. Analysis of variance (ANOVA) test was used to determine the difference between the groups, p < 0.05 was considered statistically significant.
The antibacterial activity of the screened compounds was evaluated through a reported procedure (Bagamboula et al., 2003; Abu-Dief et al., 2019; Abu-Dief et al., 2020; Abu-Dief et al., 2021b). At 121°C for 30 min, micro tips, media flask, test tube, swabs, Petri plates and all other equipment were sterilized in an autoclave. 25 ml of the nutrient media was poured into each Petri plate and was then allowed to cool. Three bacterial strains (E. coli, K. pneumonia and S. aureus) were streaked on the nutrient agar plates with sterile swabs. Five wells were punched in each Petri plate with sterile borer of 6 mm distance across. Stock solutions (3 mM) of the test compounds were prepared in DMSO which were further diluted to 100, 200 and 300 μg/ml concentrations. 20 µL of each solution was added to the particular well. DMSO and Ciprofloxacin (10 μg/ml) serving as negative and positive controls were also loaded. All the plates were placed in incubator at 37°C for 24 h and zones of inhibitions were measured in mm.
All the quantum chemical calculations were executed through the use of Gaussian 16 suit of programs (Frisch et al., 2016). The B3LYP functional is used among the available DFT methods (Stephenspj and Chabalowskic, 1994). A mixed basis set (6-31G* + LANL2DZ) consisting of LANL2DZ for Sn metal atom and 6-31G* for all remaining atoms was used for all calculations (Chiodo et al., 2006). Such type of mixed basis set are found reasonable for metal complexes in several previous reports (Yang et al., 2009). The total density and partial density of states were determined using a full scale population method at the same level of theory. GaussView 6 was used for getting the 3D orbital diagrams.
The 3D molecular structures of the ligand acid HL and synthesized complexes 1–4 were drawn and optimized using Molecular Operating Environment (MOE-2016) software (Inc, 2016). From the protein data bank (www.rcsb.org/pdb) the x-ray crystallographic structures of the double stranded DNA (PDB ID: 1BNA) and tubulin ((PDB ID: 1SA0) in complex with colchicine as protein structure were retrieved as possible targets for the anticancer activity of the synthesized complexes. Also, the crystal structures of angiotensin converting enzyme of human body, nucleocapsid protein and spike protein of SARS-CoV-2 were retrieved from the same source. From the crystal structures all the water molecules were removed using MOE-2016 software (www.chemcomp.com). The 3D protonation and energy minimization were done using default parameters of Molecular Operating Environment. All the test compounds were docked with PDB files using default parameters of MOE-2016 (Method- Alpha PMI, Rigid receptor, score- Affinity dG, and poses-10).
Organotin(IV) derivatives (1–4) were made by reacting organotin(IV) chlorides with sodium-4-chlorophenoxyacetate (NaL) at a mole ratio that was appropriate. In dry chloroform, the reaction mixtures were refluxed for 8 h. As crystalline/powdered products, the complexes were produced in a high yield (>80%). At ambient temperature, the complexes are soluble in DMSO and CHCl3.
The FT-IR spectra of the complexes 1–4 was compared with that of the free ligand acid (HL) for the preliminary structural identification. The lack of a wide band in the higher wave number region above 2,400 cm−1, attributable to the hydroxyl group of the COOH moiety in the free carboxylic acid, was a notable difference in the spectra of the complexes (Muhammad et al., 2009). The absence of this peak indicated the presence of deprotonated coordinated carboxylate moiety in the complexes (Muhammad et al., 2009). The presence of several additional weak/medium intensity bands in the spectra of the complexes below 500 cm−1 owing to Sn-O bond formation bolstered the claim.27 The existence of a set of two peaks in the ranges 1,609-1,580 cm−1 and 1,488-1,409 cm−1 for complexes 1–4, corresponding to asymmetric and symmetric COO vibrations of the coordinated ligand, respectively, was another notable aspect of the spectra of the complexes (Muhammad et al., 2019). In complexes 1-2 and 3-4, the difference (Δυ) of these two bands suggested a chelating and a bridging bidentate coordination mode of the carboxylate moiety, respectively (Muhammad et al., 2009). Single-crystal X-ray diffraction study backed up the structural assumptions provided by FT-IR spectra for the complex 4. The representative FT-IR spectra of complexes 1 and 4 are given in Supplementary Figures 1, 2, respectively.
1H, 13C and 119Sn NMR spectra obtained in CDCl3 and DMSO were used to analyze the solution state structural analyses of the complexes 1–4. The 1H NMR spectra showed no singlet signal above 10 ppm, indicating the existence of a deprotonated carboxylate ligand in the complexes (Muhammad et al., 2019). A singlet was observed for the -CH2- protons (at position 2) in the range 4.68–4.54 ppm. An important observation for this signal is that, in case of diorganotin (IV) derivatives (1 and 2) the signal appeared in the down field region (at 4.68 ppm for 1 and at 4.60 ppm for 2) compared to the triorganotin (IV) derivatives (at 4.55 ppm for 3 and at 4.54 ppm for 4). This observation clearly indicates the coordination mode differences between di- and triorganotin(IV) derivatives. In case of diorganotin(IV) complexes (1 and 2) a bidentate mode of coordination can be observed in solution (Muhammad et al., 2019). This makes the carbonyl carbon (Cooney and Wuertz) and subsequently the methylene group (-CH2-) more de-shielded. However, in case of triorganotin(IV) derivatives a monodentate behavior of the carboxylate ligand causes less de-shielding of the –OOC–CH2– fragment, so methylene signal appears a bit up field. The benzene ring protons of the ligand appeared as two doublets in the ranges 7.29–7.18 ppm and 6.91–6.79 ppm for 4,4′ and 5,5′, protons respectively. Complexes 2 and 4 gave clear singlets for tin bonded methyl groups at 0.84 and 0.61 ppm, respectively. The 2J (119Sn,1H) coupling constant values for these signals were found to be 101 and 59 Hz for 2 and 4 respectively. These values confirm hexa and tetra-coordinated tin cneter in solution for 2 and 4, respectively (Muhammad et al., 2019). In complexes 1 and 3, the n-butyl groups have shown a complicated pattern in form of multiplets for the –CH2–CH2–CH2 skeleton. However, clear triplets were observed for the terminal CH3 groups in both the complexes.
The 13C-NMR spectra of complexes revealed six signals in the predicted locations for the magnetically non-equivalent carbons of the carboxylate ligand. By comparing the signals of the produced complexes to those of comparable reported tin carboxylate complexes, the carbons of the tin bound alkyl groups were allocated (Muhammad et al., 2014). Four coordinated tetrahedral tin centres have been verified in complexes 3 and 4 using 1J (119Sn,13C) coupling constant values of 343 and 394 Hz, respectively (Muhammad et al., 2014). In the carbon NMR spectra of all produced compounds, no additional peak was identified. This represented the pure character of synthesised complexes.
The 119Sn NMR of the reported compounds gave a sharp single (Supplementary Figure 2) and fall in the range of hexa-coordinated (for complexes 1 and 3) and tetra-coordinated (for complexes 2 and 4) environment around the Sn atom (Sirajuddin et al., 2014a).
The representative 1H, 13C and 119Sn NMR spectra of complexes 2, 3 and 4 are given in Supplementary Figures 3–9.
The molecular structure obtained from the single-crystal X-ray diffraction analysis of complex 4 is shown in Figure 1A. The complex crystallizes in the monoclinic crystal system with a P21/c space group. The important crystallographic parameters, selected bond lengths and bond angles are presented in Tables 1, 2, respectively. Complex 4 adopted an infinite zig-zag 1-D polymeric chain structure consisting mono deprotonated bridging carboxylate ligands connecting the tin atoms as shown in Figure 1B. The Sn1-O1 and Sn1-O2 bond distances are 2.2018 (16) Å and 2.3803 (17) Å, respectively showing a slight asymmetric coordination of the carboxylate ligand to the tin atom. The shorter tin-oxygen bond is associated with a longer C-O bond and vice versa. The Sn–O bond distances are more close to the sum of the covalent radii of tin and oxygen (2.13 Å), but are considerably less than the van der Waals radii (3.69 Å), indicating a strong Sn–O bonding interactions (Barsan et al., 2008; Shujah et al., 2014; An et al., 2020). The Sn-O bonded distances are comparable to the other trimethyltin (IV) carboxylate derivatives previously reported (Iqbal et al., 2013). Sn1-C9, Sn1-C10 and Sn1-C11 bond distances are 2.110 (3)Å, 2.108 (3)Å and 2.123 (3)Å, respectively. The tin atom in complex 4 has a five coordinated trigonal bipyramidal geometry. The two axial sites of the trigonal bipyramid are occupied by the oxygen atoms of the bridging carboxylate ligands with an O2-Sn1-O1 bond angle of 171.49 (6)°, showing distorted geometry around tin center. The three tin bonded methyl groups form the equatorial plane of the trigonal bipyramid. The sum of equatorial CH3-Sn-CH3 bond angles is 359.8° which indicates that the three methyl groups are nearly coplanar and more symmetrically bonded compared to the oxygen atoms of the carboxylate ligands. The three coplanar methyl groups are slightly tilted towards the longer tin-oxygen bond (Sn1-O2) shown by the smaller C-Sn-O2 angle (mean bond angle = 88.53°) compared to the larger C-Sn-O1 angle (mean bond angle = 91.34°). The distortion in pentagonal bipyramidal geometry was also quantitatively calculated by Addison tau-parameter (τ = β–α/60, where β is the largest and α is the second largest angle around tin atom) (Addison et al., 1984). The τ value for a perfect trigonal-bipyramidal and square- pyramidal geometry is 1 and 0, respectively for five-coordinated metal centers. The calculated τ value for the complex 4 is 0.81. This indicates a distorted trigonal bipyramidal geometry around Sn center. Furthermore, the coordination polymers of the complex 4 are connected via relatively weaker C7-H7-----O1 (2.592) interactions resulting in a 2-D polymeric network as shown in Figure 1C.
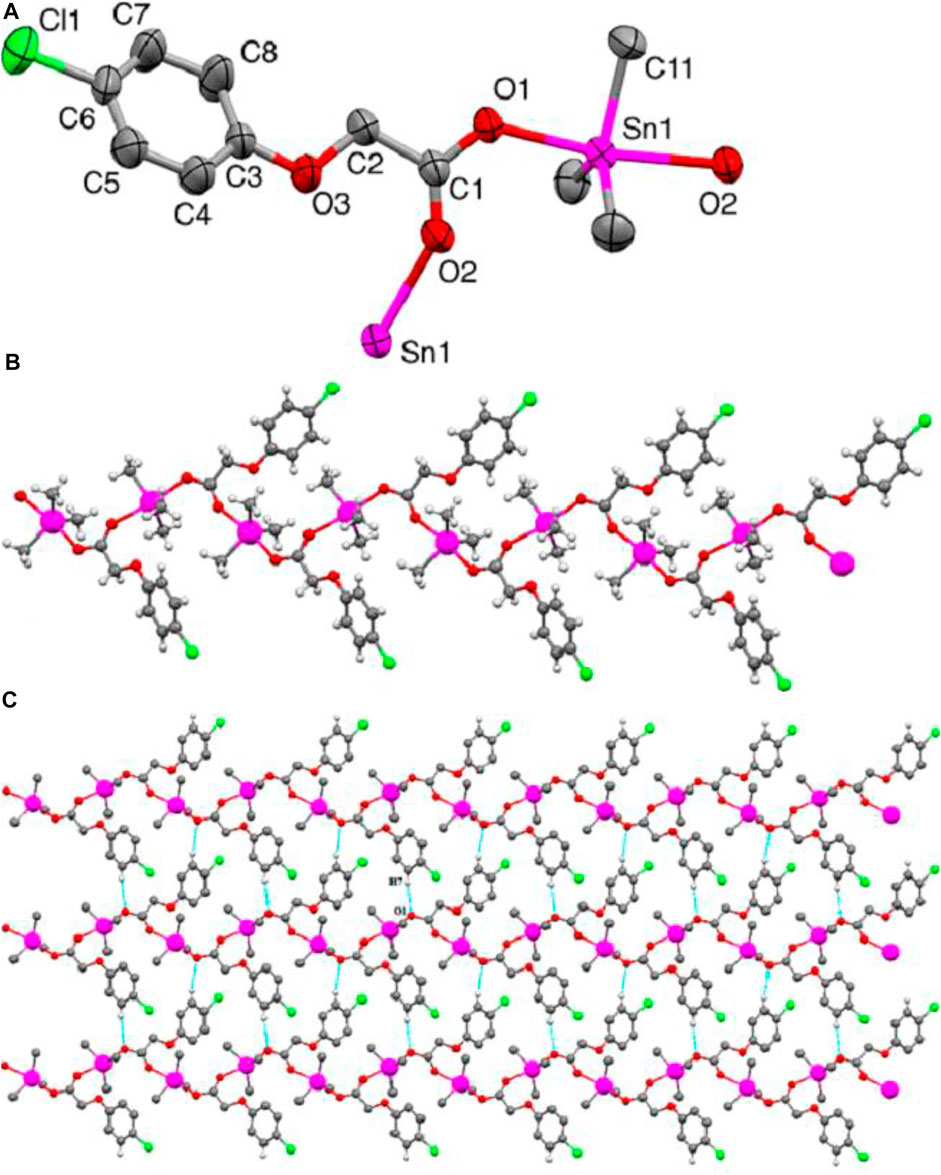
FIGURE 1. (A) ORTEP view of 4 with a 50% probability level (B) 1-D zigzag polymeric chain structure. (C) 2-D polymeric network. H-atoms non-involved in hydrogen bonding are omitted for clarity.
The optimized geometry of complex 4 is shown in Figure 2 at B3LYP/6-31G* + LANL2DZ basis set. For comparative analysis, the single crystal structure of complex 4 is also presented along with its calculated geometry. An overview of both calculated and experimental structures indicates that there is a reasonably good agreement between them. For more detailed structural comparison, the bonds lengths of only one symmetric unit of left-hand side are compared. For instance, the most important Sn-O21 bond lengths are found to be 2.201 and 2.234 Å for experimental and calculated geometries, respectively. Similarly, other important bond lengths of Cl20-C33 are found to be 1.749 and 1.768 Å for experimental and calculated geometries, respectively.
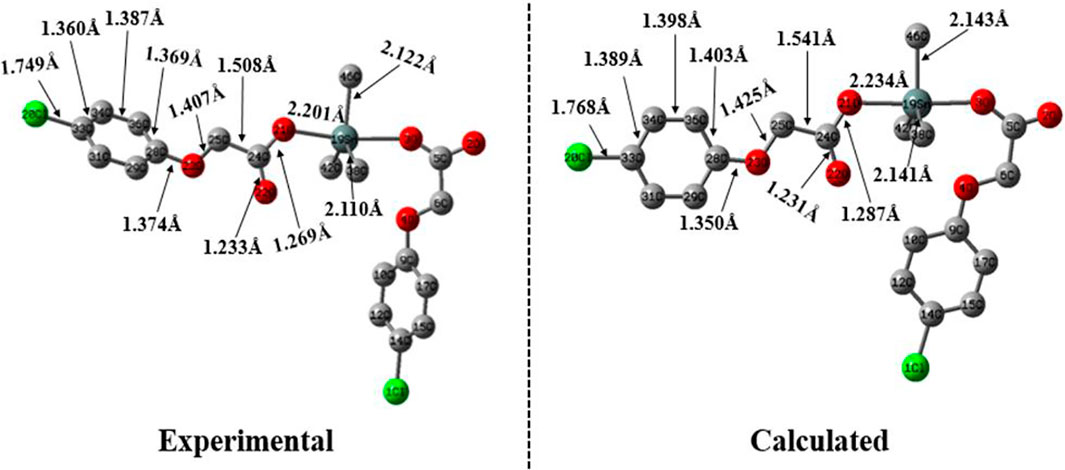
FIGURE 2. The experimental and calculated geometry of complex 4 computed B3LYP/6-31G* + LANL2DZ basis set.
In order to understand the intrinsic contributions of different moieties of complex to the total density of states, the partial density of states projected over individual moieties of complex 4 are calculated. To obtain the TDOS and PDOS diagrams, the molecules are partitioned into three fragments i.e., fragment A [methyl-2-(4-chlorophenoxy) acetate], fragment B (three methyl groups) and fragment C (Sn metal). It can be observed that Fragment A shows a major percentage contribution both in the formation of filled and unfilled orbitals. According to Figure 3, the percentage contribution of fragment B is maximum in the whole range of energy levels while Fragment B shows maximum percentage contribution in the range of about ∼6.3 to ∼8 eV. The region from −3 to −5 eV is interesting where all fragments relatively contribute including Sn metal atom too. The Sn metal contribution in formation of unfilled orbitals is more (from ∼5 to ∼6 eV) as compared to the states in filled orbitals as can be seen in Figure 3.
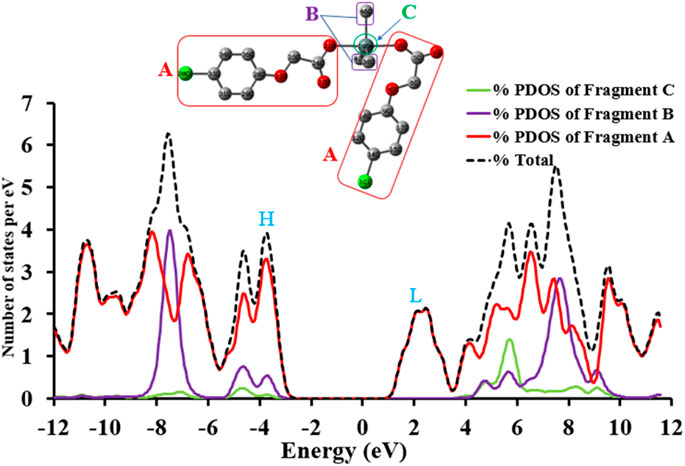
FIGURE 3. The TDOS and PDOS of complex 4 at B3LYP/6-31G* + LANL2DZ level of theory, where H and L illustrate the energy regions of HOMO and LUMO, respectively.
The Frontier molecular orbitals (FMOs) play an essential role in defining the intramolecular charge transfer properties, reactivity and optical properties of a complex. The FMOs of complex 4 are depicted in Figure 4, which include HOMO-1, HOMO, LUMO, LUMO + 1 orbitals along with their orbital energies and orbital energy gaps (Eg). In metal complexes, there are different types of intramolecular charge transfer processes including metal to ligands, ligand to metal and ligand to ligand, etc. A careful analysis of Figure 4 shows that there is ligand to ligand intramolecular charge transfer upon electronic transition. It can be visualized from Figure 4 that the HOMO is localized mainly on the methoxy acetate groups of complex 4, while LUMO is confined to chlorobenzene ring of one side ligand. There is very little and/or negligible electron density on Sn metal which indicates that the Sn metal acts as more like a bridge for intramolecular charge transfer or redistribution upon electron transitions. On the other hand, the distributions of HOMO-1 to LUMO + 1 orbitals solely involve the chlorobenzene rings with significant contributions from chlorine atoms as can be seen in Figure 4. Overall a ligand to ligand charge transfer is evident from the analysis of Frontier molecular orbital analysis which indicates that the optoelectronic properties of complex 4 might be tuned exclusively by the ancillary ligands (Vogler and Kunkely, 1997).
In vitro cytotoxic effect of the free ligand acid HL and synthesized complexes 1–4 was assessed against A549 and MRC-5 at different concentrations (0, 0.1, 0.5, 1, 2, 5, 10 and 50 μg/ml) for 24 and 48 h. The amount of DMSO used to dissolve the compounds was applied below the cytotoxicity limit (less than 0.1% range as suggested for the cell culture). The experimental findings regarding cell viability are presented in Figure 5 as well in tabulated form in Supplementary Table 1. Also, Figure 6 show the effect of studied concentrations of the tested compounds on the morphology of A549 and MRC-5 cell lines. The IC50 values are given in Table 3.
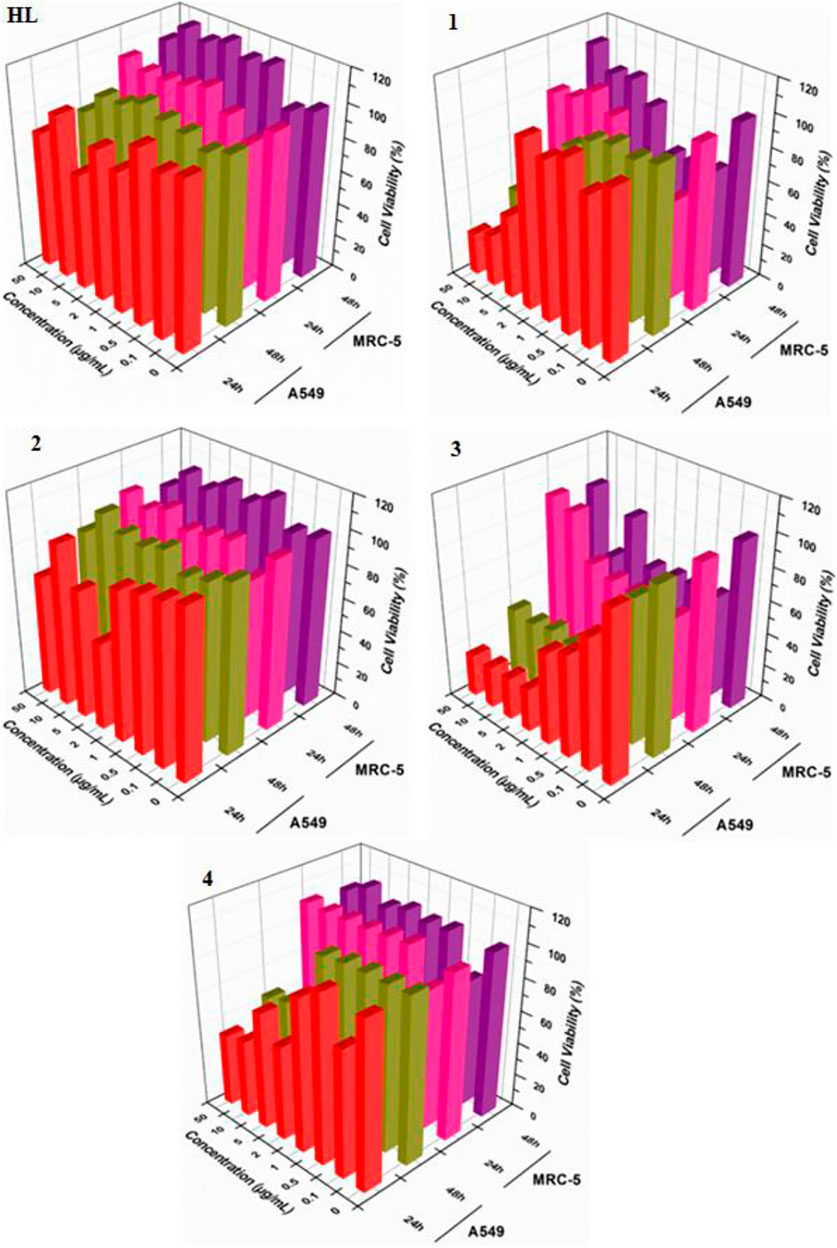
FIGURE 5. Cytotoxic effects of ligand acid (HL) and synthesized complexes (1–4) against A549 and MRC-5.
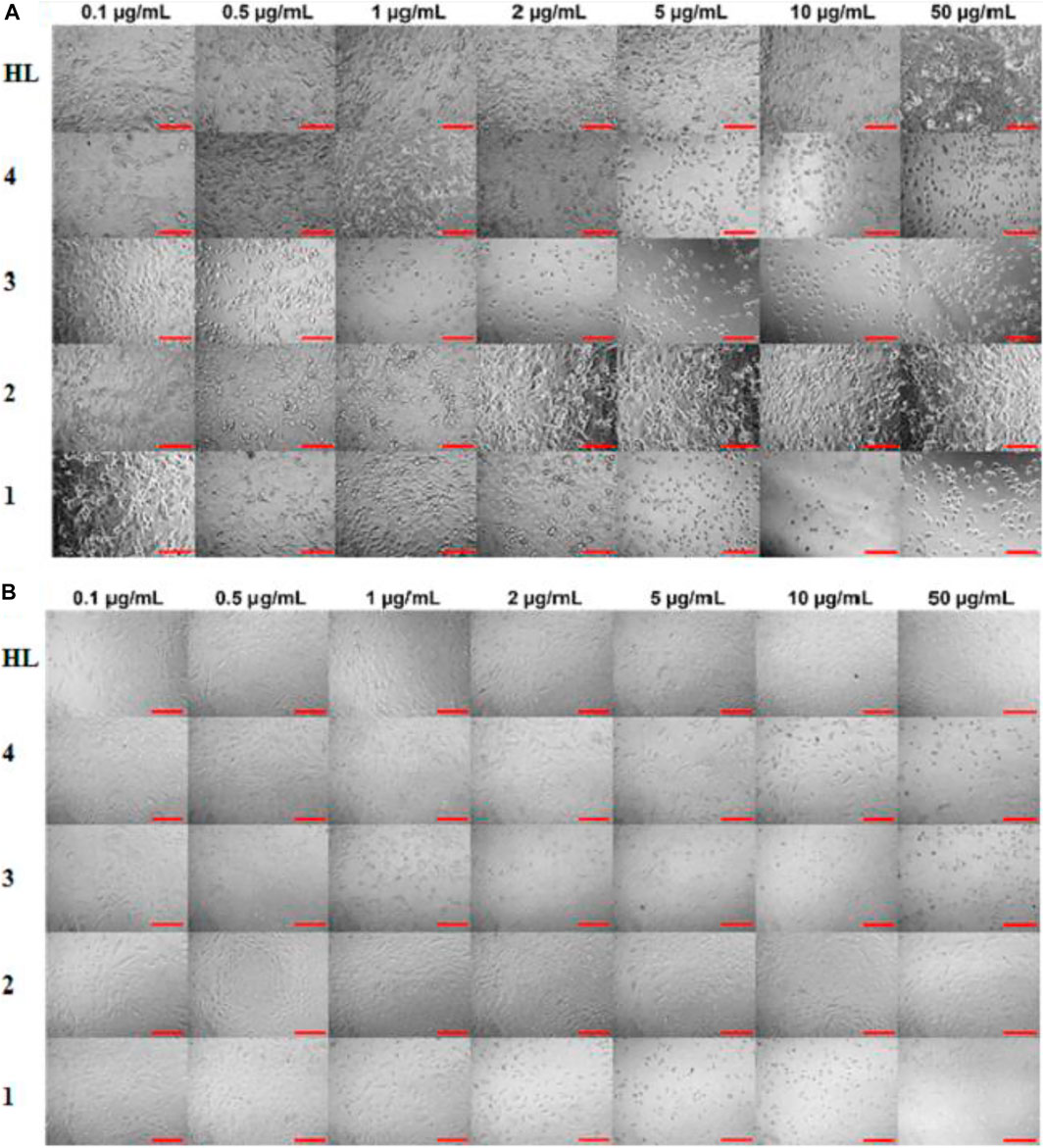
FIGURE 6. Effect of ligand acid (HL) and complexes 1–4 on morphology of (A) A549 and (B) MRC-5 for 48 h. The scale bar is 100 µm.
All the tested compounds showed an anticancer activity against A549 cell line. In a dose dependent manner, the anticancer activity of complexes 1, 3 and 4, increased by increasing their concentrations at 24 h. These complexes (1, 3 and 4) were found particularly more active anticancer agents against A549 at higher concentrations. Interestingly, these complexes are not toxic at higher concentrations against MRC-5 cell line. However, complexes 1 and 3 exhibited cytotoxicity against MRC-5 at lower concentrations. The anticancer activity of the complexes 1, 3 and 4 slightly decreased after 48 h. The free ligand acid HL and complex 2 showed a lower and stable anticancer activity against A549 at all studied concentrations, at both 24 and 48 h. Also the toxic effect of the free ligand acid HL and complex 2 against MRC-5 was relatively very low at all studied concentrations.
The experimental results suggested a metal-based activity, as complexation increased the anticancer activity of the free ligand acid HL. The activity was also affected by the length of the alkyl group, as complexes 1 and 3 with bulky n-butyl groups were found more active than complexes 2 and 4 with smaller methyl groups. In fact, among the tested complexes, the complex 3 showed higher anticancer activity. This reflects the importance of both lipophilicity as well as tin coordination number in anticancer activity of the synthesized complexes. In addition, the presence of three nonpolar n-butyl groups in complex 3 makes it more lipophilic. So, the movement of the complex through the cellular membrane of the target cell to reach the action site is easy. Also, the low coordinated tin atom (coordination number = 4 as confirmed by NMR spectra) in complex 3 is more exposed for interaction with the donor atoms of the biomolecule of the target cell. The argument is further supported by the lower cytotoxicity of complex 2, where, in this complex two smaller methyl groups and a higher coordination number of six as confirmed by NMR spectra offer lower lipophilicity and lesser exposed tin center, respectively. This makes the complex 2 a relatively weaker anticancer agent.
Table 4 describes the antibacterial activity of the ligand acid (HL) and synthesized complexes 1–4, assessed against five bacterial strains namely E. coli, S. aureus, P. aeregionosa, K. pneumonia and B. cereus. The nature of the bacterial strain seems to be the dominant factor affecting the antibacterial efficiency of the tested compounds. Generally the activity of the complexes 1–4 is higher than that of the free ligand acid (HL). Comparing the activity of the complexes 1–4 with the standards, Streptomycin and Ampicillin, it can be seen from the data their activity is very slightly smaller than the standards against the tested strains. Among the tested complexes, n-butyltin (IV) derivatives (1 and 3) are more active than methyltin (IV) derivatives (2 and 4). The bulky n-butyl groups offer more lipophilic (organic) character to the complexes and thus making them more active in the present study.
To have a theoretical insight about the anticancer activity of the synthesized complexes, the DNA interactions were performed for the most active complex 3. The complex 3 has shown interaction with DNA in form of intercalation. The complex 3 show four polar interactions (docking score = −3.6005), two H-donor with DG 16, DA 17 and two π-H with DC 9, DG 10 active sites of DNA as shown in Figure 7A.
The interacting ability of the complex 3 was also assessed with tubulin (PDB ID 1SA0) in complex with colchicine as a target protein structure. Complex 3 was stabilized in the colchicine binding cavity by three polar interactions (docking score −5.2957), one H-donor with LEU 195, one H- acceptor with TRP 407 and one H-π with PHE 93 active sites of the tubulin as shown in Figure 7B. The docking analysis has shown that complex 3 offers best dock pose with tubulin in colchicine binding cavity compared to DNA.
The ligand acid and its organotin(IV) derivatives were docked with spike protein, nucleocapsid protein of corona virus SARS-CoV-2 and angiotensin converting enzyme (ACE2) of human. The docking analysis have shown reasonable interactions of the screened compounds with the active sites of the targets. The ligand acid HL (docking score = −3.6103) has shown four polar interactions, two H-acceptor, one π-H and one H-donor interaction with VAL 389, LYS 390, LYS 390 and GLU 37 active sites residues of spike protein as shown in Figure 8A. Complex 1 (docking score = −7.6506) has best docked pose with three polar H-donor interactions with SER 985, GLY 981 active residues of the spike protein as shown in Figure 8B. Complex 2 (docking score = −5.1539) has shown two polar interactions, one H-acceptor and one π-H with ILE 995 and ALA 998 active residues of spike protein as shown in Figure 8C. Complex 3 (docking score = −5.5353) has shown three polar interactions, two are H-π and one are H-acceptor with TRP 868, TYR 1029 and LYS 1020 active residues of spike protein as shown in Figure 8D. Complex 4 (docking score = −3.7082) has shown three polar interactions, one H-donor, one H-acceptor and one π-π interaction with LYS 1020, TRP 868 and TYR 1029 active residues of the spike protein. These polar interactions reflect that the synthesized complexes can interact spike protein and affect its ability to develop any interaction with the host receptor.
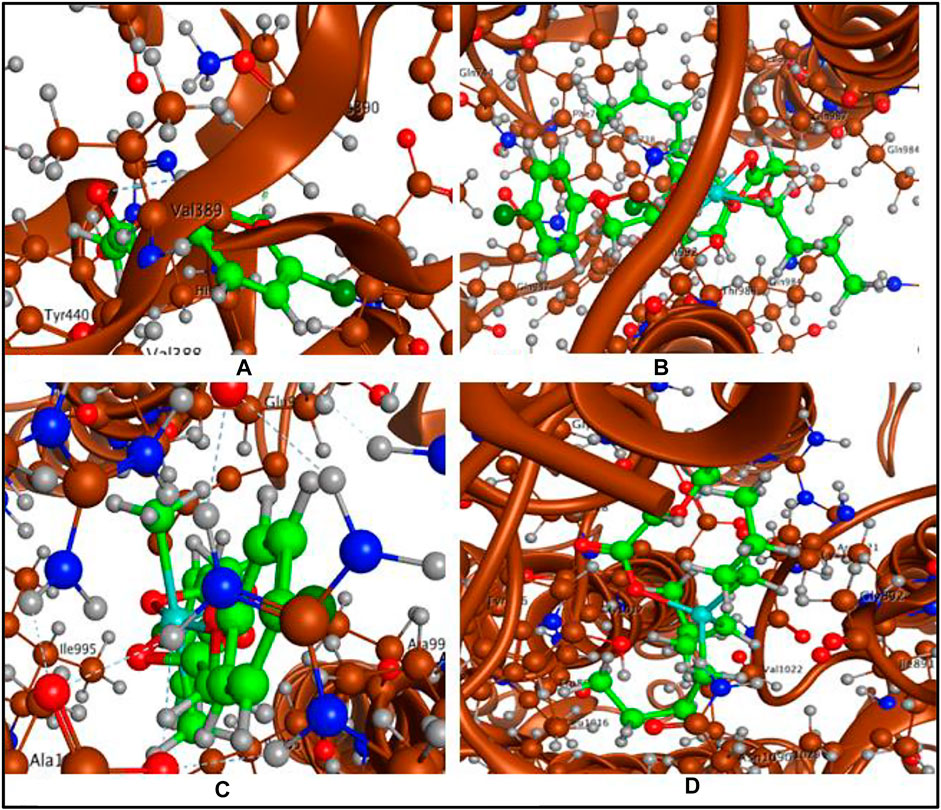
FIGURE 8. (A) Ligand acid (HL)-spike protein interaction, (B) Complex 1-spike protein interaction, (C) Complex 2-spike protein interaction and (D) Complex 3-spike protein interaction.
The ligand acid and synthesized complexes have also shown interactions with the nucleocapsid protein of the corona virus SARS-CoV-2. Complex 1 has shown best dock pose (−5.7402) with two polar interactions, one H-donor and one π-H interaction with GLU 63 and TRP 53 active site residues, respectively of the nucleocapsid protein as shown in Figure 9. The ligand acid HL (−3.2769), complex 2 (−3.1275), complex 3 (−3.6005) and complex 4 (−2.2132) have shown three, two, four and two polar interactions, respectively in form of H-donor/acceptor, π-H and H-π polar interactions as mentioned in Table 5 (Supplementary Figures 10–13). The higher docking scores of the test compounds with spike protein compared to the nucleocapsid protein show that the test compounds offer best dock poses for interactions with spike protein as target for the corona virus SARS-CoV-2 inhibition.
Molecular docking studies with angiotensin converting enzyme also reflected best dock pose for the complex 1 (docking score = −6.1678) with four polar interactions, one H-donor and three H-acceptor interactions with GLN 442, PHE 438, ARG 518 and THR 371 active sites as shown in Figure 10 (a). Complex 2 (docking score = −5.6760) has shown six polar interactions, two H-donor and one H-π and three π-H interactions with GLU 406, ASP 367, HIS 374, THR 371, and LYS 441 active sites as shown in Figure 10B. Complex 3 (docking score = −5.1188), complex 4 (docking score = −3.5105) and ligand acid HL (docking score = −3.4990) have shown one, three and two polar interactions respectively, with the active site residues of the angiotensin converting enzyme as mentioned in Table 6 (Supplementary Figures 14–17). The docking analysis shows that the free ligand acid and its complexes can interact with the three possible targets sites involved in corona disease. The complex 1 with relatively higher docking scores offered the best dock poses for the interaction with the active site residues of all the three targets.
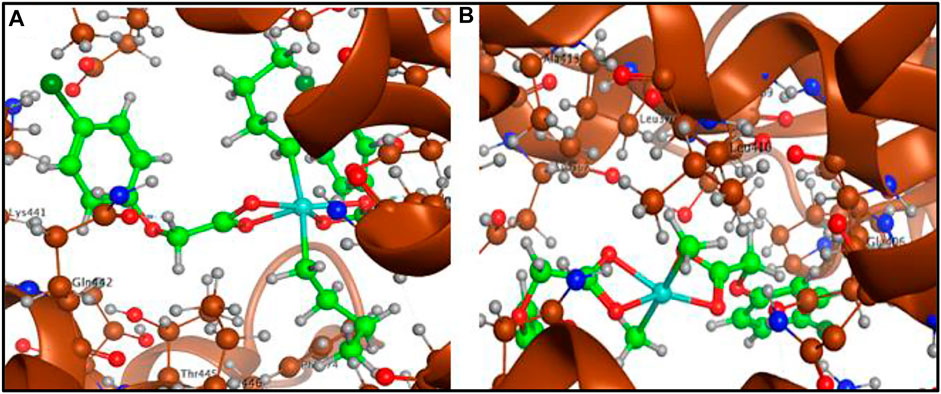
FIGURE 10. (A) Complex 1-angiotensin converting enzyme interaction and (B) Complex 2-angiotensin converting enzyme interaction.
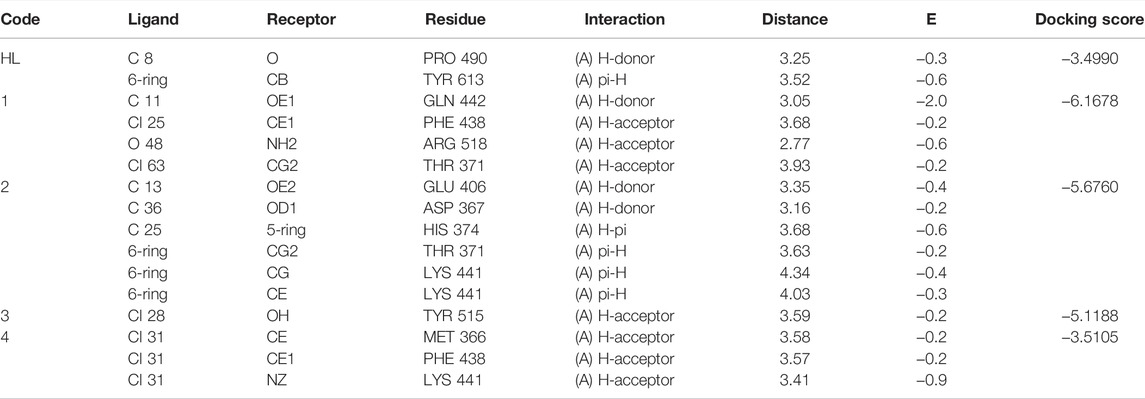
TABLE 6. Ligand acid (HL) and complexes 1–4 interaction report with angiotensin converting enzyme (ACE2).
Four organotin(IV) derivatives (n-C4H9)2SnL2 (1), (CH3)2SnL2 (2), (n-C4H9)3SnL (3) and (CH3)3SnL (4) of 4-chlorophenoxyacetic acid (HL) were synthesized and characterized. The ligand has shown a chelating (1 and 2) and bridging (3 and 4) coordination modes to the tin centre in solid state. The NMR (1H, 13C and 119Sn) results have shown higher coordination number for tin in diorganotin(IV) derivatives (1 and 2) compared to the triorganotin(IV) derivatives (3 and 4) in solution. The calculated structure of complex 4 at B3LYP/6-31G* + LANL2DZ level of theory indicated a good resemblance with the experimentally determined X-ray single crystal structure. Quantum chemical analysis indicated the significant involvement of the 4-chlorophenoxyacetate in the formation of Frontier molecular orbitals of the complex 4. The in vitro cytotoxic effect of the HL against A549 and MRC-5 has increased after complexation with organotin(IV) moieties in the complexes 1–4. Complex 3 with more lipophilic character and low coordinated tin atom offered higher cytotoxicity. The in vitro antibacterial activity order HL > n-butyltin(IV) derivatives (1 and 3) > methyltin(IV) deivaitves (2 and 4) against the tested bacterial strains reflected an organic mode of action for the tested compounds. The docking study (for complex 3) has suggested tubulin in complex with colchicine (docking score −5.2957) as better anticancer target compared to DNA (docking score = −3.6005). The docking interactions of the synthesized complexes (especially 1) with all the three possible targets (spike and nucleocapsid protein of corona virus SARS-CoV-2 and angiotensin converting enzyme (ACE2) of human) make the organotin(IV) carboxylates as potential candidates for further studies in corona virus treatment in future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
NM: Methodology, Conceptualization and Writing–original draft MA: Writing–original draft MS: Methodology, Conceptualization and Writing–original draft ZA: Writing–review and editing NT: Formal analysis JW: Formal analysis AC: Formal analysis KS: Anticancer activity AM: Anticancer activity SM: Writing–review and editing SS: Writing–review and editing SA: Writing–review and editing AA-S: Formal analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Financial support from Higher Education Commission, Pakistan in the form of NRPU Project # 20-3092/R&D/HEC/13 638 is acknowledged by NM. The authors SM and AA-S acknowledge the financial support of Institute of Research and Consulting Studies at King Khalid University for supporting this research through grant number 2-N-20/22 and the support of Research Center for Advanced Materials Science is highly acknowledged. KS also thankful for The Council of Higher Education (CoHE, 100/2000) Ph.D. Scholarship Program, Turkey.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.864336/full#supplementary-material
Abu Dief, A. M., El Sagher, H. M., and Shehata, M. R. (2019). Fabrication, Spectroscopic Characterization, Calf Thymus DNA Binding Investigation, Antioxidant and Anticancer Activities of Some Antibiotic Azomethine Cu (II), Pd (II), Zn (II) and Cr (III) Complexes. Appl. Organomet. Chem. 33, e4943. doi:10.1002/aoc.4943
Abu-Dief, A. M., Abdel-Rahman, L. H., Abdelhamid, A. A., Marzouk, A. A., Shehata, M. R., Bakheet, M. A., et al. (2020). Synthesis and Characterization of New Cr(III), Fe(III) and Cu(II) Complexes Incorporating Multi-Substituted Aryl Imidazole Ligand: Structural, DFT, DNA Binding, and Biological Implications. Spectrochim Acta A. Mol. Biomol. Spectrosc. 228, 117700. doi:10.1016/j.saa.2019.117700
Abu-Dief, A. M., El-Khatib, R. M., Sayed, S. M. E., Alzahrani, S., Alkhatib, F., El-Sarrag, G., et al. (2021a). Tailoring, Structural Elucidation, DFT Calculation, DNA Interaction and Pharmaceutical Applications of Some Aryl Hydrazone Mn(II), Cu(II) and Fe(III) Complexes. J. Mol. Struct. 1244, 131017. doi:10.1016/j.molstruc.2021.131017
Abu-Dief, A. M., El-Metwaly, N. M., Alzahrani, S. O., Alkhatib, F., Abualnaja, M. M., El-Dabea, T., et al. (2021b). Synthesis and Characterization of Fe(III), Pd(II) and Cu(II)-thiazole Complexes; DFT, Pharmacophore Modeling, Iin-Vvitro Assay and DNA Binding Studies. J. Mol. Liquids 326, 115277. doi:10.1016/j.molliq.2021.115277
Addison, A., Rao, T. N., Reedijk, J., Rijin, J., and Verschoor, G. C. (1984). Dalton Trans. J. Chem. Soc. 1349.
An, B. H., Zhang, R. F., Du, X. M., Li, Q. L., Cheng, S., Huang, H. L., et al. (2020). Novel Triorganotin Complexes Based on Phosphonic Acid Ligands: Syntheses, Structures and In Vitro Cytostatic Activity. J. Inorg. Biochem. 206, 111022. doi:10.1016/j.jinorgbio.2020.111022
Bagamboula, C. F., Uyttendaele, M., and Debevere, J. (2003). Antimicrobial Effect of Spices and Herbs on Shigella Sonnei and Shigella Flexneri. J. Food Prot. 66, 668–673. doi:10.4315/0362-028x-66.4.668
Barsan, M. M., Wharf, I., Muhammad, N., Ali, S., Meetsma, A., and Butler, I. S. (2008). Synthesis, Spectroscopic Characterization, and crystal Structures of Two Chlorodiorganotin (IV) 4-(2-methoxyphenyl) Piperazine-1-Carbodithioates. Inorg. Chim. Acta 361, 3322–3326. doi:10.1016/j.ica.2008.05.006
Basu Baul, T. S. (2008). Antimicrobial Activity of Organotin(IV) Compounds: a Review. Appl. Organometal. Chem. 22, 195–204. doi:10.1002/aoc.1378
Butt, A. F., Ahmed, M. N., Bhatti, M. H., Choudhary, M. A., Ayub, K., Tahir, M. N., et al. (2019). Synthesis, Structural Properties, DFT Studies, Antimicrobial Activities and DNA Binding Interactions of Two Newly Synthesized Organotin(IV) Carboxylates. J. Mol. Struct. 1191, 291–300. doi:10.1016/j.molstruc.2019.04.066
Chiodo, S., Russo, N., and Sicilia, E. (2006). LANL2DZ Basis Sets Recontracted in the Framework of Density Functional Theory. J. Chem. Phys. 125, 104107. doi:10.1063/1.2345197
Cooney, J. J., and Wuertz, S. (1989). Toxic Effects of Tin Compounds on Microorganisms. J. Ind. Microbiol. 4, 375–402. doi:10.1007/bf01569539
Debnath, P., Das, A., Singh, K. S., Yama, T., Singh, S. S., Butcher, R. J., et al. (2019). Synthesis, Structural Characterization and Antimicrobial Activities of triorganotin(IV)azo-carboxylates Derived from Ortho/para-Amino Benzoic Acids and β-naphthol. Inorg. Chim. Acta 498, 119172. doi:10.1016/j.ica.2019.119172
Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K., and Puschmann, H. (2009). OLEX2: a Complete Structure Solution, Refinement and Analysis Program. J. Appl. Cryst. 42, 339–341. doi:10.1107/s0021889808042726
Ettouhami, A., Yahyi, A., El Mejdoubi, A., El Bali, B., Siddiq, S., Noureen, S., et al. (2012). Diorganotin (IV) Derivative of 2-thiophene Acetic Acid: Characterizations and Biological Activities of {[n-Bu2SnO2C-CH2-C4h3s]2o}2. Med. Chem. Res. 21, 3607–3614. doi:10.1007/s00044-011-9916-6
Fracchiolla, G., Laghezza, A., Piemontese, L., Carbonara, G., Lavecchia, A., Tortorella, P., et al. (2007). Synthesis, Biological Evaluation, and Molecular Modeling Investigation of Chiral Phenoxyacetic Acid Analogues with PPARalpha and PPARgamma Agonist Activity. ChemMedChem 2, 641–654. doi:10.1002/cmdc.200600307
Frisch, M., Trucks, G., Schlegel, H., Scuseria, G., Robb, M., Cheeseman, J., et al. (2016). Gaussian 16. Revision A. Wallingford, 310.
Gielen, M. (2003). An Overview of Forty Years Organotin Chemistry Developed at the Free Universities of Brussels ULB and VUB. J. Braz. Chem. Soc. 14, 870–877. doi:10.1590/s0103-50532003000600003
Gielen, M. (2008). Tin Chemistry: Fundamentals, Frontiers, and Applications. West Sussex, UK: John Wiley & Sons.
Gómez-Ruiz, S., Kaluđerović, G. N., Prashar, S., Hey-Hawkins, E., Erić, A., Žižak, Ž., et al. (2008). Study of the cytotoxic activity of di and triphenyltin (IV) carboxylate complexes. J. Inorg. Biochem. 102, 2087–2096. doi:10.1016/j.jinorgbio.2008.07.009
Hadjikakou, S., and Hadjiliadis, N. (2009). Antiproliferative and Anti-tumor Activity of Organotin Compounds. Coord. Chem. Rev. 253, 235–249. doi:10.1016/j.ccr.2007.12.026
Hong, M., Yang, Y., Li, C., Xu, L., Li, D., and Li, C.-Z. (2015). Study of the Effect of Molecular Structure and Alkyl Groups Bound with Tin(iv) on Their Cytotoxicity of Organotin(iv) 2-Phenyl-4-Selenazole Carboxylates. RSC Adv. 5, 102885–102894. doi:10.1039/c5ra18445b
Hook, J. M., Linahan, B. M., Taylor, R. L., Tiekink, E. R. T., Gorkom, L. V., and Webster, L. K. (1994). Phenyltin Diethyldithiocarbamates: Solid State and Solution Structures and in vitro Anti-tumour Activity. Main Group Met. Chem. 17, 293–312. doi:10.1515/MGMC.1994.17.5.293
Hübschle, S., Sheldrick, G. M., and Dittrich, B. (2011). ShelXle: a Qt Graphical User Interface for SHELXL. J. Appl. Cryst. 44, 1281–1284. doi:10.1107/S0021889811043202
Indumathy, R., Radhika, S., Kanthimathi, M., Weyhermuller, T., and Unni Nair, B. (2007). Cobalt Complexes of Terpyridine Ligand: crystal Structure and Photocleavage of DNA. J. Inorg. Biochem. 101, 434–443. doi:10.1016/j.jinorgbio.2006.11.002
Iqbal, M., Ali, S., Muhammad, N., Parvez, M., Langer, P., and Villinger, A. (2013). Synthesis, Characterization, crystal Structures and Electrochemical Studies of Organotin(IV) Carboxylates. J. Organomet. Chem. 723, 214–223. doi:10.1016/j.jorganchem.2012.10.006
Kaluđerović, G. N., Kommera, H., Hey-Hawkins, E., Paschke, R., and Gómez-Ruiz, S. (2010). Synthesis and Biological Applications of Ionic Triphenyltin(iv) Chloride Carboxylate Complexes with Exceptionally High Cytotoxicity. Metallomics 2, 419–428. doi:10.1039/c0mt00007h
Kovala-Demertzi, D., Dokorou, V., Primikiri, A., Vargas, R., Silvestru, C., Russo, U., et al. (2009). Organotin Meclofenamic Complexes: Synthesis, crystal Structures and Antiproliferative Activity of the First Complexes of Meclofenamic Acid - Novel Anti-tuberculosis Agents. J. Inorg. Biochem. 103, 738–744. doi:10.1016/j.jinorgbio.2009.01.014
Mubeen, M., and Kini, S. G. (2012). A Review on the Design and Development of EGFR Tyrosine Kinase Inhibitors in Cancer Therapy. Int. J. Ther. Appl. 5, 29–37.
Muhammad, N., Shah, A., Zia-ur-Rehman, S., Shuja, S., Ali, S., Qureshi, R., et al. (2009). Organotin(IV) 4-nitrophenylethanoates: Synthesis, Structural Characteristics and Intercalative Mode of Interaction with DNA. J. Organomet. Chem. 694, 3431–3437. doi:10.1016/j.jorganchem.2009.06.036
Muhammad, N., Shah, N. A., Ali, S., Elahi, S. N., Rehman, W., Shujah, S., et al. (2019). Theoretical and Experimental In Vitro Antifungal and Antitumor Activities of Organotin(IV) Derivatives of 3-(4-Nitrophenyl)-2-Methylacrylic Acid. Pharm. Chem. J. 53, 689–696. doi:10.1007/s11094-019-02064-2
Muhammad, N., Zia-Ur, R, Shujah, S., Ali, S., Shah, A., and Meetsma, A. (2014). Synthesis and Structural Characterization of Monomeric and Polymeric Supramolecular Organotin(IV) 4-chlorophenylethanoates. J. Coord. Chem. 67, 1110–1120. doi:10.1080/00958972.2014.898755
Pantelić, N. Đ., Zmejkovski, B. B., Žižak, Ž., Banjac, N. R., Božić, B. Đ., Stanojković, T. P., et al. (2019). Design and In Vitro Biological Evaluation of a Novel Organotin (IV) Complex with 1-(4-Carboxyphenyl)-3-Ethyl-3-Methylpyrrolidine-2, 5-dione. J. Chem., 1–8. doi:10.1155/2019/2905840
Pettazzoni, P., Pizzimenti, S., Toaldo, C., Sotomayor, P., Tagliavacca, L., Liu, S., et al. (2011). Induction of Cell Cycle Arrest and DNA Damage by the HDAC Inhibitor Panobinostat (LBH589) and the Lipid Peroxidation End Product 4-hydroxynonenal in Prostate Cancer Cells. Free Radic. Biol. Med. 50, 313–322. doi:10.1016/j.freeradbiomed.2010.11.011
Saddik, M. S., Elsayed, M. M. A., Abdelkader, M. S. A., El-Mokhtar, M. A., Abdel-Aleem, J. A., Abu-Dief, A. M., et al. (2021). Novel green Biosynthesis of 5-fluorouracil Chromium Nanoparticles Using Harpullia Pendula Extract for Treatment of Colorectal Cancer. Pharmaceutics 13, 226. doi:10.3390/pharmaceutics13020226
Saxena, A. K., and Huber, F. (1989). Organotin Compounds and Cancer Chemotherapy. Coord. Chem. Rev. 95, 109–123. doi:10.1016/0010-8545(89)80003-7
Scheldrick, G. (2015). SHELXT-integrated Space-Group and Crystals-Structure Determination. Acta Crystallogr. Sect. A: Found. Adv. 71, 3–8. doi:10.1107/S2053273314026370
Shah, F. A., Sabir, S., Fatima, K., Ali, S., Qadri, I., and Rizzoli, C. (2015). Organotin(IV) Based Anti-HCV Drugs: Synthesis, Characterization and Biochemical Activity. Dalton Trans. 44, 10467–10478. doi:10.1039/c5dt00862j
Sheldrick, G. (1997a). SHELXS-97, Program for Crystal Structure Determination. Germany: University of Göttingen.
Sheldrick, G. M. (1997b). SHELXL-97-2, Program for Crystal Structure Refinement. Germany: University of Göttingen.
Shujah, S., Zia-ur-Rehman, N., Muhammad, N., Shah, A., Ali, S., Meetsma, A., et al. (2014). Homobimetallic Organotin(IV) Complexes with Hexadentate Schiff Base: Synthesis, crystal Structure and Antimicrobial Studies. J. Organomet. Chem. 759, 19–26. doi:10.1016/j.jorganchem.2014.02.010
Sirajuddin, M., Ali, S., Mckee, V., Sohail, M., and Pasha, H. (2014a). Potentially Bioactive Organotin(IV) Compounds: Synthesis, Characterization, In Vitro Bioactivities and Interaction with SS-DNA. Eur. J. Med. Chem. 84, 343–363. doi:10.1016/j.ejmech.2014.07.028
Sirajuddin, M., Ali, S., Haider, A., Shah, N. A., Shah, A., and Khan, M. R. (2012). Synthesis, Characterization, Biological Screenings and Interaction with Calf Thymus DNA as Well as Electrochemical Studies of Adducts Formed by Azomethine [2-((3,5-dimethylphenylimino) methyl) phenol] and Organotin (IV) Chlorides. Polyhedron 40, 19–31. doi:10.1016/j.poly.2012.03.048
Sirajuddin, M., Ali, S., Mckee, V., Zaib, S., and Iqbal, J. (2014b). Organotin(iv) Carboxylate Derivatives as a New Addition to Anticancer and Antileishmanial Agents: Design, Physicochemical Characterization and Interaction with Salmon Sperm DNA. RSC Adv. 4, 57505–57521. doi:10.1039/c4ra10487k
Sirajuddin, M., Ali, S., and Tahir, M. N. (2021). Organotin(IV) Derivatives Based on 2-((2-methoxyphenyl)carbamoyl)benzoic Acid: Synthesis, Spectroscopic Characterization, Assessment of Antibacterial, DNA Interaction, Anticancer and Antileishmanial Potentials. J. Mol. Struct. 1229, 129600. doi:10.1016/j.molstruc.2020.129600
Stephenspj, D., and Chabalowskic, F.(1994). Abinitio calculation of vibrational absorption and circular Dichroismspectrausingdensityfunctionalforcefields. TheJournalofPhysicalChemistry 98 (45), 1162311627.
Tariq, M., Sirajuddin, M., Ali, S., Khalid, N., Tahir, M. N., Khan, H., et al. (2016). Pharmacological Investigations and Petra/Osiris/Molinspiration (POM) Analyses of Newly Synthesized Potentially Bioactive Organotin(IV) Carboxylates. J. Photochem. Photobiol. B 158, 174–183. doi:10.1016/j.jphotobiol.2016.02.028
Tariq, M., Muhammad, N., Sirajuddin, M., Ali, S., Shah, N. A., Khalid, N., et al. (2013). Synthesis, Spectroscopic Characterization, X-ray Structures, Biological Screenings, DNA Interaction Study and Catalytic Activity of Organotin(IV) 3-(4-Flourophenyl)-2-Methylacrylic Acid Derivatives. J. Organomet. Chem. 723, 79–89. doi:10.1016/j.jorganchem.2012.09.011
Tzimopoulos, D., Sanidas, I., Varvogli, A. C., Czapik, A., Gdaniec, M., Nikolakaki, E., et al. (2010). On the Bioreactivity of Triorganotin Aminobenzoates. Investigation of Trialkyl and Triarylyltin(IV) Esters of 3-amino and 4-aminobenzoic Acids. J. Inorg. Biochem. 104, 423–430. doi:10.1016/j.jinorgbio.2009.12.006
Vogler, A., and Kunkely, H. (1997). Inorganic Chromophores: Optical Charge Transfer in Coordination Compounds. Comments Inorg. Chem. 19, 283–306. doi:10.1080/02603599708032742
Xiao, X., Wang, Z., Zhao, M., Li, J., Hu, M., He, Y., et al. (2020). A 24-membered Macrocyclic Triorganotin(IV) Carboxylate: Synthesis, Characterization, crystal Structure, and Antitumor Activity. J. Mol. Struct. 1216, 128255. doi:10.1016/j.molstruc.2020.128255
Yang, Y., Weaver, M. N., and Merz, K. M. (2009). Assessment of the "6-31+G** + LANL2DZ" Mixed Basis Set Coupled with Density Functiona Theory Methods and the Effective Core Potential: Prediction of Heats of Formation and Ionization Potentials for First-Row-Transition-Metal Complexes. J. Phys. Chem. A. 113, 9843–9851. doi:10.1021/jp807643p
Keywords: organotins, single crystal XRD, antibacterial activity, anticancer activity, molecular docking studies
Citation: Muhammad N, Ahmad M, Sirajuddin M, Ali Z, Tumanov N, Wouters J, Chafik A, Solak K, Mavi A, Muhammad S, Shujah S, Ali S and Al-Sehemi AG (2022) Synthesis, Characterization, Biological Activity and Molecular Docking Studies of Novel Organotin(IV) Carboxylates. Front. Pharmacol. 13:864336. doi: 10.3389/fphar.2022.864336
Received: 28 January 2022; Accepted: 17 March 2022;
Published: 05 April 2022.
Edited by:
Ranjan K. Mohapatra, Government College of Engineering, Keonjhar, IndiaReviewed by:
Ahmed M. Abu-Dief, Sohag University, EgyptCopyright © 2022 Muhammad, Ahmad, Sirajuddin, Ali, Tumanov, Wouters, Chafik, Solak, Mavi, Muhammad, Shujah, Ali and Al-Sehemi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Niaz Muhammad, ZHJuaWF6QGF3a3VtLmVkdS5waw==; Muhammad Sirajuddin, bS5zaXJhajA5QGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.