- 1Department of Pediatrics, The Affiliated Hospital of Guangdong Medical University, Zhanjiang, China
- 2Graduate School of Guangdong Medical University, Zhanjiang, China
Asthma is a chronic airway inflammatory disease in children characterized by airway inflammation, airway hyperresponsiveness and airway remodeling. Childhood asthma is usually associated with allergy and atopy, unlike adult asthma, which is commonly associated with obesity, smoking, etc. The pathogenesis and diagnosis of childhood asthma also remains more challenging than adult asthma, such as many diseases showing similar symptoms may coexist and be confused with asthma. In terms of the treatment, although most childhood asthma can potentially be self-managed and controlled with drugs, approximately 5–10% of children suffer from severe uncontrolled asthma, which carries significant health and socioeconomic burdens. Therefore, it is necessary to explore the pathogenesis of childhood asthma from a new perspective. Studies have revealed that non-coding RNAs (ncRNAs) are involved in the regulation of respiratory diseases. In addition, altered expression of ncRNAs in blood, and in condensate of sputum or exhalation affects the progression of asthma via regulating immune response. In this review, we outline the regulation and pathogenesis of asthma and summarize the role of ncRNAs in childhood asthma. We also hold promise that ncRNAs may be used for the development of biomarkers and support a new therapeutic strategy for childhood asthma.
Introduction
Asthma is one of the most common chronic inflammatory disease characterized by high heterogeneity in pathogenesis (Koczulla et al., 2017; Papi et al., 2018), with symptoms including showing paroxysmal, reversible wheezing, shortness of breath, chest tightness and cough, which occur or intensify at night and/or in the early morning (Holgate et al., 2015). With the exposure of allergens and the use of antibiotics in the first year of infant life, the prevalence of asthma in children is rising (Metsälä et al., 2015). According to the data from the Centers for Disease Control and Prevention (CDC) in 2016, the prevalence of asthma in children aged 5–11 and 12–17 is respectively 9.6% and 10.5% (Haktanir Abul and Phipatanakul, 2019).
Asthma affects more than nearly 339 million people globally from childhood to old age, among which childhood asthma is more complicated than adult asthma with multiple phenotypes and variable natural course (Lambrecht and Hammad, 2015; Jat and Kabra, 2017; Qiu et al., 2019; El-Husseini et al., 2020). Evidences confirm that childhood asthma is associated with allergy and strongly driven by genetic and environmental factors which determine the susceptibility and severity of asthma (Lee et al., 2017; Tang et al., 2018a). The typical feature of childhood asthma is airway hyper-responsiveness (AHR), Th2-driven eosinophilic airway inflammation and airway remodeling (Qiu et al., 2019; Li et al., 2021). As for adult asthma, it not only shares the same features with childhood asthma, but also is strongly associated with smoking, obesity, and occupational exposures (Kirenga et al., 2020), which mainly refers to non-Th2-type asthma. Different from adult asthma, childhood asthma is usually related to environmental allergens, such as IgE-dependent Th2-type allergic reaction and viral infections and so on (Ferreira et al., 2019; Qiu et al., 2019; Hammad and Lambrecht, 2021). The onset of childhood asthma involves eosinophils, mast cells, T lymphocytes, neutrophils, airway epithelial cells and their cellular components, leading to increased airway responsiveness, and ultimately resulting in widespread and variable reversible airflow limitation (Foster et al., 2017) (Figure 1).
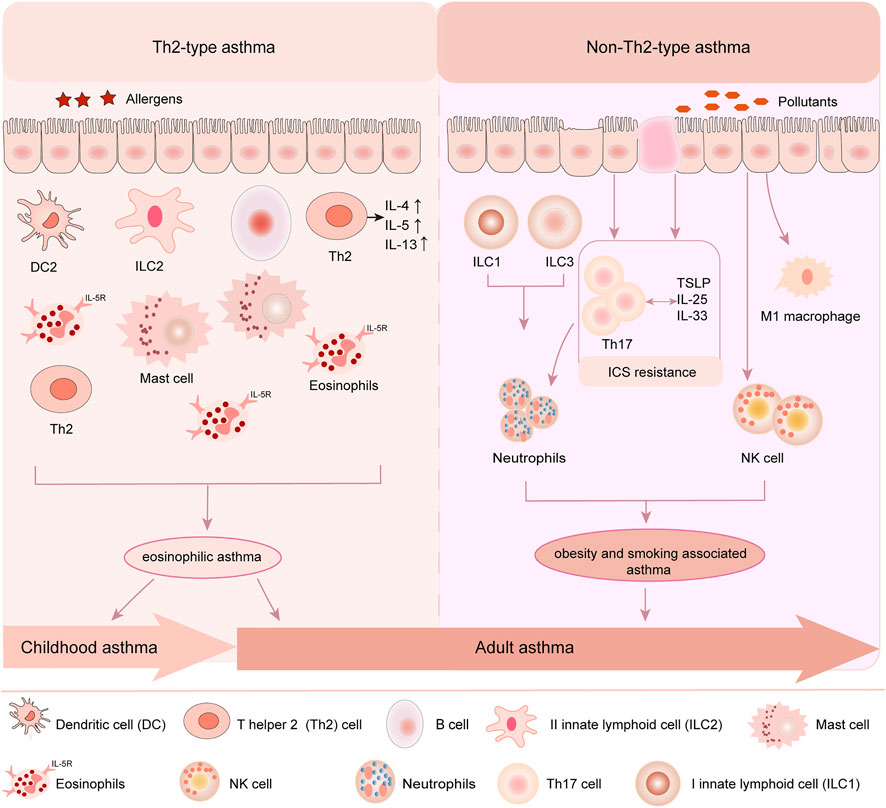
FIGURE 1. Childhood asthma and adult asthma phenotypes. Childhood asthma and adult asthma are crossed and different in phenotypes. In children, Th2-type asthma is the main common type. Allergen stimulates the recruitment of inflammatory cells such as eosinophils, the proliferation and activation of immune cells such as mast cells and DCs, and induces the injury of airway epithelial cells, which lead to the release of inflammatory factors and Th2 cytokines, such as IL4, IL5 and IL13. In adult, both Th2-type and non-Th2-type asthma are two common types. Upon pollutants stimulation, type I innate lymphoid cells (ILC1) and type III innate lymphoid cells activation (ILC3) activate neutrophils and airway epithelial cells to drive the proliferation of Th17 cells which mediates in turn neutrophil recruitment. Pollutants also contribute to M1 macrophage and NK cell recruitment to the airways, resulting in non-Th2-type asthma in adult.
Airway remodeling mainly refers to a constellation of structural changes induced by asthma, including epithelial injury, increased thickness of the basement membrane, airway angiogenesis and so on (Enomoto et al., 2009; Inoue et al., 2020). Additionally, studies confirm that vascular endothelial growth factor (VEGF), an important mediator in airway structural changes, has a proliferation-inducing effect on normal bronchial epithelial cells and bronchial smooth muscle cells and is increased in children with recurrent early wheezing (Yoshisue et al., 2007; Enomoto et al., 2009). The upregulation of VEGF and downregulation of lymphocyte lead to the development of airway remodeling in asthma (Altman et al., 2019). At present, asthma predictive index is used to judge childhood asthma with wheezing combined with clinical experience at home and abroad, but the diagnosis of asthma in children under 6 years old is still a challenging clinical problem (Lee et al., 2019a; Driscoll et al., 2020). Underdiagnosis of asthma leads to delays in the optimal timing of asthma treatment and may prompt the transition from mild asthma to severe, refractory asthma (Shi et al., 2020). Although the efficacy of pharmacotherapies including inhaled corticosteroids and leukotriene receptor antagonists may help prevent airway remodeling, they cannot reverse the established airway remodeling (Huo et al., 2021). Consequently, it is necessary to strengthen the understanding of the pathogenesis of childhood asthma in order to improve early diagnosis rate. Meanwhile, intervention in the occurrence of airway remodeling is crucial to preventing and treating asthma in children.
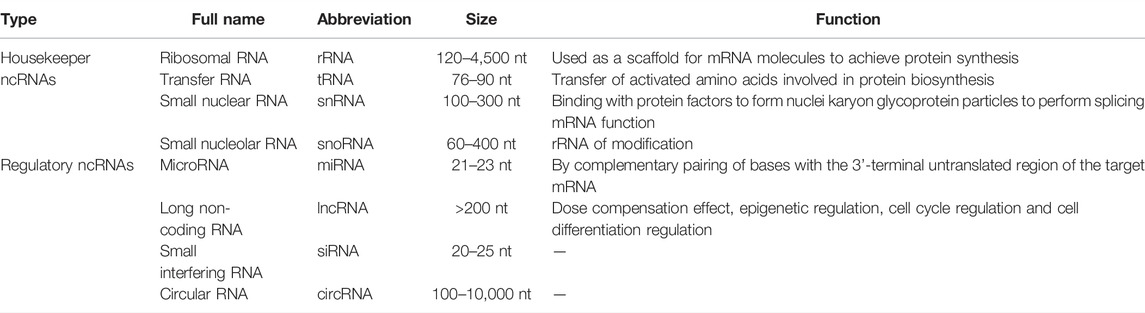
NcRNAs are non-protein-coding RNAs molecules, mainly including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), which are profoundly involved in post-transcriptional gene expression and participate in the regulation of various biological processes (Defnet et al., 2019; Zangouei et al., 2020; Wang et al., 2021). Some studies have confirmed that ncRNAs play a crucial role in the pathogenesis and regulation of asthma including childhood asthma and adult asthma (Qiu et al., 2018; Yang et al., 2022). Correctly, miRNAs are involved in the regulation of airway inflammation and airway smooth muscle proliferation. LncRNAs not only regulate airway inflammation and airway remodeling, but also affect the regulation of immune responses (Narożna et al., 2017; Elnady et al., 2020; Dai et al., 2021). Moreover, emerging evidence implicates that circRNAs are also involved in the proliferation of smooth muscles and airway remodeling in the progression of asthma (Jiang et al., 2021; Wasti et al., 2021). Hence, ncRNAs are considered as potential biomarkers and promising therapeutic targets for childhood asthma (Narożna et al., 2017; Specjalski and Jassem, 2019).
Clinically, children are special categories of patients and childhood asthma can be misdiagnosed as other diseases with similar symptoms (Abdullahi et al., 2016), which makes the diagnosis of asthma difficult. Besides, even if most childhood asthma patients are relieved after treatment in the early stages according to the international recommendations of the Global Asthma Initiative, drug failure and drug resistance often occur during asthma treatment. That’s to say, up to 5%–10% of children have severe asthma or poor asthma control (Moral et al., 2021; Ntontsi et al., 2021). Therefore, further understanding of the pathogenesis of asthma, especially the identification of differential pathogenesis of asthma between children and adults, will help to find the way of more effective diagnoses and treatments. Accordingly, mechanism study of ncRNAs on the pathogenesis of childhood asthma is worthy of attention. In this review, we mainly demonstrate the pathogenesis of childhood asthma and emerge roles of ncRNAs in asthma. Besides, we hold great promise for the discovery of new ncRNAs biomarkers and therapeutics for asthma.
Asthma-Associated Pathogenesis
Immune Factors in the Development and Regulation of Childhood Asthma
Asthma is a heterogenous disease with complex pathogenesis and various phenotypes which can be reclassified via molecular biomarkers called “endotypes” (Bond et al., 2018; Assaf and Hanania, 2019). At present, asthma endotypes are divided into T-helper-2(Th2)-high (eosinophilic) and Th2-low (non-eosinophilic). The ratio tilt of T lymphocyte subsets (Th1/Th2) is the most important pathogenesis of asthma (Tang et al., 2018b). Th2-high asthma is related to adaptive immunity and allergic asthma (Lampalo et al., 2019; Song et al., 2019). Upon allergens stimulation, dendritic cells (DCs) activate T-helper-2 (Th2) cells to secrete Th2 cytokines such as IL-4, IL-5 and IL-13 which act on airway epithelial cells, eosinophils, B lymphocytes and other inflammatory cells. This drives B cells to produce a large number of IgE that cross-linking causes degranulation of mast cells to produce a series of inflammatory mediators such as leukotrienes, endothelin, prostaglandin and thromboxane A2, etc. and eventually induce rapid onset (increased IgE) allergies and eosinophilic airway inflammation (Bégin and Nadeau, 2014; Chogtu et al., 2016; McCracken et al., 2016). Studies have shown that childhood asthma is associated with eosinophils in the airways, allergic sensitization and adaptive immunity (Kim et al., 2010). Th2 cells concretely activated by allergens via secreting Th2 cytokines IL-4, IL-5 and IL-13, amplify type II inflammation, while T helper 1 (Th1) cells by secreting Th1 cytokines such as IFN-γ, IL-2, lymphotoxin (LT)-α and tumor necrosis factor (TNF)-α and so on, limit type II inflammation and mediate type I inflammation (Foster et al., 2017; Mukherjee and Nair, 2018), which causing childhood asthma. Consequently, the imbalance of the ratio of T lymphocyte subsets (Th1/Th2) is the key mechanism of childhood asthma, whether it is innate immunity or adaptive immunity.
However, non-Th2-type asthma related to non-allergic asthma, which is characterized by emotion, obesity, environmental factors, such as air pollution including ozone, cigarette smoke and so on, may release cytokines such as Il-17 and IFN-γ by activation of Th1 cells, leading to neutrophilic inflammation, M1 macrophage, NKT cell recruitment to the airways and AHRs (Castan et al., 2020; Agache et al., 2021). Nevertheless, non-Th2-type asthma characterized by type I innate lymphoid cells (ILC1) and type III innate lymphoid cells activation (ILC3) is most common in adult asthma. ILC1 in asthma patients promoted eosinophil apoptosis and inhibited eosinophilic airway inflammation (Barnig et al., 2013; Barnig and Levy, 2015). ILC3 produced IL-17A and caused obesity-related AHR effects (Kim et al., 2014). Furthermore, other cytokines (such as IL-17, IL-21, IL-22) also accelerated mucus secretion of airway smooth muscle cells, increased the production of cytokines and chemokines, and promoted neutrophil recruitment by inducing CXC chemokines (Lejeune et al., 2020). In addition, IL-17 promoted airway remodeling by increasing the production of fibrotic cytokines, angiogenic factors, proteases and collagen.
Moreover, airway epithelial cells and type II innate lymphoid cells (ILC2) are also involved in adaptive immune. After allergens exposure, epithelial cells polarize macrophages, DC cells, T cells, etc. (Frati et al., 2018; Lejeune et al., 2020), releasing not only IL-4, IL-5, IL-13 and other cytokines but also pro-inflammatory cytokines such as IL-25, IL-33 and thymic stromal lymphopoietin (TSLP) activating ILC2 rather than IFN-γ and TNF-β produced by Th1 cells (Lejeune et al., 2020), which causes Th0 cells polarized into Th2 cells in asthma-specific cytokine environments, resulting in a balanced skewed Th2 cellular immune response. The immune response of Th2 cells further induced changes in the pathophysiological characteristics of asthma, including eosinophils mobilizing IgE, secreting excessive mucus, smooth muscle proliferation and airflow obstruction. Additionally, under the effect of IL-5, eosinophils entered the respiratory tract and triggered a second inflammatory response (Deschildre et al., 2017; Gao et al., 2017). Studies have shown that CD4+ T cells are essential for inducing allergic airway disease in newborns, and ILC2s are very important in the pathogenesis of allergic airway disease in adults. Mechanistically, CD4+ T cells and ILC2s regulate asthma by promoting the production of IL-13 amplifying type II inflammation (Saglani et al., 2018) (Figure 2). Therefore, understanding the role of immune factors in the development and regulation of asthma will provide opportunities for asthma treatment.
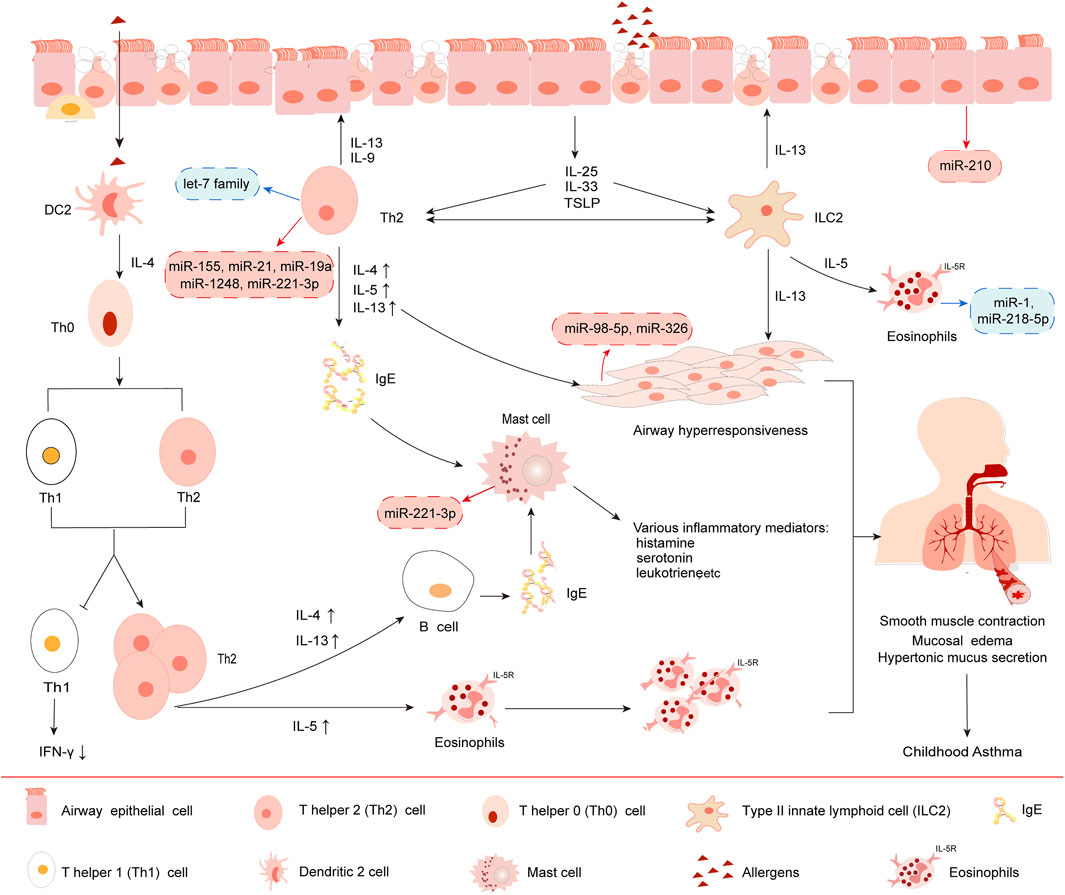
FIGURE 2. Immunopathogenesis of Th2-type asthma. 1) Under IL-4 induction, dendritic 2 (DC2) cell promotes the development of Th0 cells to Th2, resulting in Th1 (decreased secretion)/Th2 (increased secretion) cell dysfunction. 2) Upon stimulation by allergens, bronchial epithelial cells release IL-25, IL-33 and TSLP, which in turn activates group II innate lymphoid cells (ILC2) and Th2 cells. 3) Subsequently, Th2 cells release Th2 cytokines such as IL-4, IL-5 and IL-13, rather than Th1 cells produce IFN-γ and TNF-β, causing a balanced skewed Th2 cellular immune response. 4) IgE eventually induces rapid onset allergy and cytokines release induced by eosinophils, mast cells and other immune cells, leading to airway inflammation. 5) Moreover, different miRNAs have different effects on the above processes. The red boxes in the picture show typical miRNAs that exert upregulated function in asthma. The blue boxes show miRNAs that exert downregulated function.
Mitogen-Activated Protein Kinase Signaling Pathways Involved in Childhood Asthma
Neural signaling pathway also has appealing potential as an application to study pathways to childhood asthma development. Molecular studies have indicated that Mitogen-activated protein kinase (MAPK) pathways, including extracellular signal-regulated kinase 1/2 (ERK1/2), p38 mitogen-activated protein kinase and c-Jun NH2-terminal kinase (JNK), are involved in the inflammatory response and development of airway remodeling during childhood asthma (Lee et al., 2019b; Jia et al., 2019). Concretely, ERK1/2 is involved in airway remodeling (Defnet et al., 2019), while p38 MAPK and JNK are considered as anti-inflammation targets to regulate inflammatory processes via phosphorylation of downstream mediators in childhood asthma (Kim and Choi, 2010; Pulido and Lang, 2019; Theodorou et al., 2022). However, literatures about the pathogenesis of MAPK pathways in childhood asthma are limited. In this review we mainly focus on the roles of MAPK signaling pathways in the pathogenesis and related treatment of childhood asthma.
Experiment shows ERK1/2 signaling mainly acts on airway epithelial cells and smooth muscle cells of asthmatic mice (Liu et al., 2008). Concretely, ERK1/2 inducible proteins Jun b proto-oncogene (JunB) mediates ERK1/2 activation via the AP-1 complex, which increases the expression of several Th2 cytokines and drives Th2 cell differentiation, causing childhood asthma (Alam and Gorska, 2011). Besides, sprouty-2, a cytosolic adapter protein, also regulates receptor-mediated ERK1/2 activation by preventing c-Cbl-mediated degradation of EGF receptor, which also stimulates Th2 cell differentiation to amplify Th2 inflammation (Liu et al., 2008; Alam and Gorska, 2011; Sripada et al., 2021). However, further mechanism about ERK1/2 in childhood asthma is needed to explore.
In contrast to ERK1/2, p38 MAPKs and the JNK pathways favor Th1 differentiation. P38 MAPK and JNK activation contributes to the inflammatory response in asthma. Moreover, p38 MAPK is observed in the basal layer of the columnar epithelium, alveolar macrophage and bronchial epithelial, ect. driving basal metabolic processes for these cell type. That’s to say, p38 MAPK is vital for allergen induced epithelial production of IL-25 and thymic stromal lymphopoietin (TSLP), further mediating the type-2 allergic response in asthma (Yu et al., 2010; Lin et al., 2015; Southworth et al., 2018). Previous studies have confirmed that dual-specificity phosphatase1 (DUSP1) also plays vital role in anti-inflammation by deactivating MAPKs through dephosphorylation (Pulido and Lang, 2019; Xin et al., 2021; Theodorou et al., 2022; Xing and Wong, 2022). Besides, Studies have confirmed that JNK is essential for airway inflammation via modulating RAGE/β-catenin signaling (Huang et al., 2021a). Regretfully, further mechanisms between JNK and RAGE/β-catenin is deserved exploring (Figure 3).
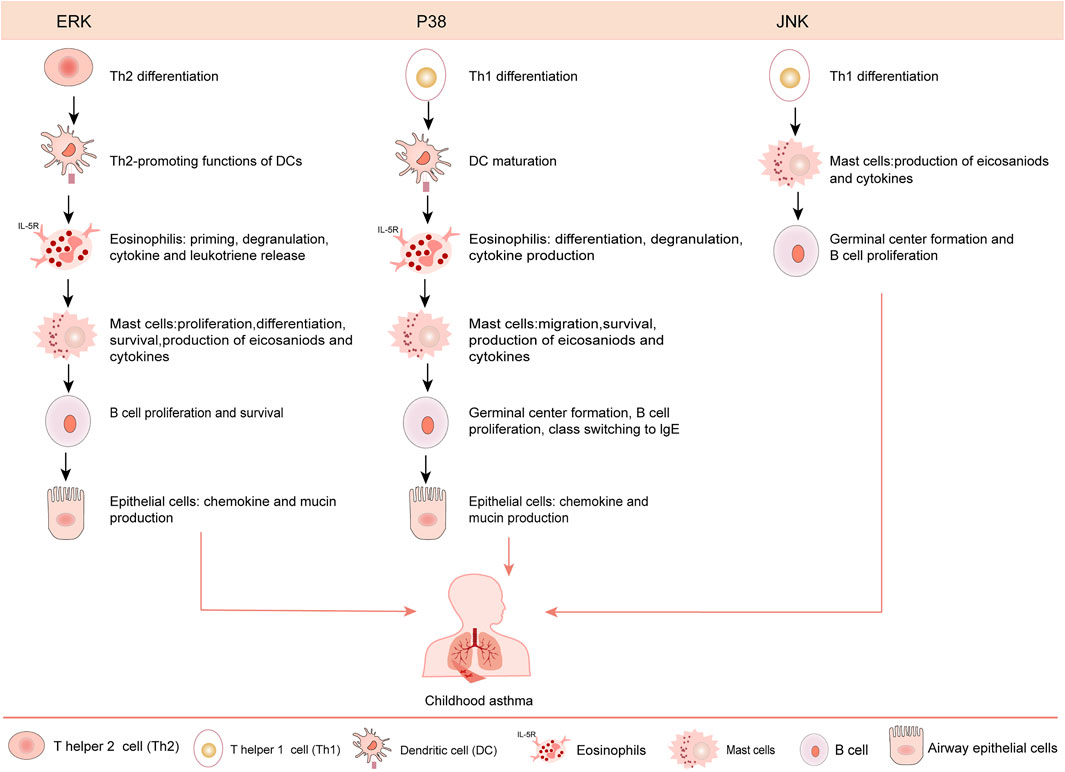
FIGURE 3. Roles of various MAPK signaling pathways in asthmatic pathogenesis. ERK favors Th2 cell differentiation, eosinophils priming, degranulation, cytokine and leukotriene production, mast cells proliferation/differentiation. The p38 MAPKs and JNK pathways regulate Th1 differentiation. The p38 MAPK contributes to eosinophil degranulation, migration and cytokine production and mast cell migration. The JNK participates in cytokine production by mast cells, regulates the proliferation of B cell, and exerts function in asthmatic pathogenesis.
Genetic Susceptibility of Childhood Asthma
Although the immune mechanisms and signaling pathways of asthma are widely reported, complex interactions between genetic susceptibility and environmental influences also lead to childhood asthma (Thomsen, 2015). Studies have shown that the genetic susceptibility of asthma can be as high as 60–70%, which suggests that understanding the genetic basis of childhood asthma might unravel mechanisms, directing the treatment of asthma. So further genome-wide association studies (GWAS) is imminent due to the complexity and heterogeneity of asthma (Alizadeh et al., 2017; Kabesch and Tost, 2020). Consequently, it has been found that the main genetic risk factor for asthma is single nucleotide polymorphism (SNPs), which is a single-base pair, occurring on average one in 300 nucleotides (Lee et al., 2015). Asthma-associated SNPs in this locus are related to levels of mRNA expression of ORMDL3 in lymphoblastic cell lines using eQTL mapping, which associates SNPs with gene expression (El-Husseini et al., 2020). Non-synonymous SNP of The IL7R (rs6897932) and IL2RB (rs2284033) (Arenas-Ramirez et al., 2015) regulate asthma by regulating type II inflammation via activating three pathways: JAK-STAT, PI3K-Akt-mTOR (Patel and Chang, 2012), and MEK-ERK (El-Husseini et al., 2020). Approximately 88% of the disease-associated variants acquired from genome-wide association studies (GWASs) reside in non-coding regions (El-Husseini et al., 2020). Examples of pathways or networks that are implicated by GWASs in asthma are the IL33-IL1-RL1 receptor pathway, leading to eosinophilia, and the T-helper-2 (Th2) cytokine IL-5 and IL-4RA receptor, leading to eosinophilia and type 2 inflammation or viral response (CDHR3, ORMDL3) (Zhang et al., 2019a; Basnet et al., 2019). For example, genetic variation at chromosome 17q12-21 is associated with childhood asthma but not adult asthma (Ferreira et al., 2019; Pividori et al., 2019). Subsequent investigations also linked SNPs at this locus to GSDMA, GSDMB, CRKRS, ZBPB2, and IKZF2 expression in whole blood cells and lung tissue.
NcRNAs in the Regulation of Childhood Asthma
NcRNAs are a class of RNA transcripts that do not encode proteins, but they have been implicated in regulating gene expression at the epigenetic, transcriptional and post-transcriptional levels and affect the biological functions such as immune response, tissue repair and remodeling (Cech and Steitz, 2014; Cai et al., 2015; Xie and Liu, 2015; Zhang et al., 2019b). Mounting studies have shown that ncRNAs are involved in the occurrence and progression of childhood asthma. In this review, we mainly focus on the regulatory role of ncRNAs in the pathogenesis of childhood asthma and discuss the feasibility of ncRNAs as new biomarkers for the treatment of childhood asthma (Fasanaro et al., 2015) (Table 1).
Long Non-Coding RNAs in the Regulation of Childhood Asthma
LncRNAs, with a length of over 200 nt, play critical roles in regulating gene expression at multiple levels, including transcriptional, post-transcriptional levels, microRNA chelation and translation efficiency regulation and more (Hao and Zan, 2021; Wang et al., 2021). Although many studies have proved that lncRNAs and childhood asthma are inextricably linked (Wang et al., 2021), the roles of lncRNAs in childhood asthma remain unclear.
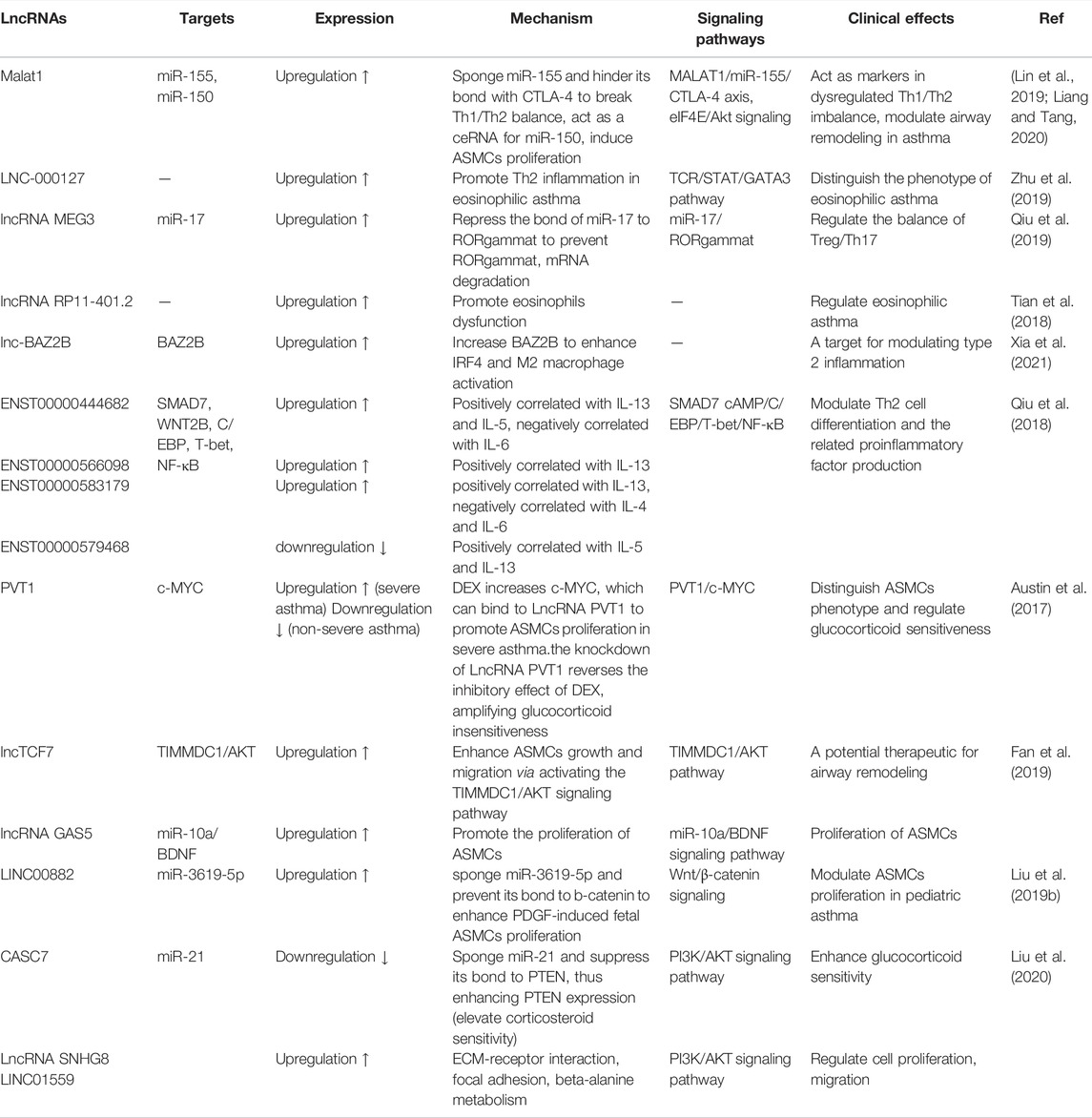
Recently, it has been reported that lncRNAs participate in regulating airway inflammation and remodeling (Ezegbunam and Foronjy, 2018; Liu et al., 2019a; Li et al., 2020a), and are helpful to further determine the biomarkers and therapeutic target of childhood asthma. Increasing numbers of studies suggest that lncRNAs affect the regulation of immune response, airway inflammation and cytokine expression (Dai et al., 2021). Moreover, lncRNAs also participate in the regulation of T helper (Th)1/Th2 imbalance, T regulatory (Treg)/Th17 imbalance, eosinophils dysfunction, macrophage polarization, airway smooth muscle cells (ASMCs) proliferation and so on (Zhu et al., 2020), to mediate childhood asthma.
Studies have found that LNC-000127 was not only closely related to Th2 inflammation but also positively regulated eosinophilic asthma. Malat1 has capability to modulate Th1/Th2 balance in asthma via MALAT1/miR-155/CTLA-4 axis. Therefore, MALAT1 and LNC-000127 could be used as a biomarker for eosinophilic asthma (Zhu et al., 2019). Additionally, Treg/Th17 imbalance was also associated with childhood asthma through neutrophil recruitment and exacerbation of airway inflammation, which was mediated by upregulating Th17-type cytokines (IL-17A, IL-17F) and downregulating Treg-type cytokines (IL-10, transforming growth factor (TGF-β) and so on (Westfall et al., 2021). LncRNA MEG3 regulated RORgammat expression by competitively sponge miR-17 and ultimately affected the balance of Treg/Th17 (Qiu et al., 2019; Hao and Zan, 2021; Wang et al., 2021). In addition, lncRNA RP11-401.2 upregulated in eosinophils dysfunction regulated eosinophilic asthma (Tian et al., 2018). Lnc-BAZ2B promoted M2 macrophage activation and was significantly upregulated in childhood asthma, whereas LncRNA PTPRE-AS1 was downregulated in macrophage polarization which mediated type II inflammation (Zeng et al., 2019). Regretfully, the detailed regulated mechanism of lncRNAs in childhood asthma is unclear. Besides, microarray analysis of CD4+ T cells of asthma patients showed that lncRNA ENST00000583179, lncRNA ENST00000579468 and lncRNA ENST00000444682 were positively correlated with the expression of IL-5 and IL-13, while lncRNA ENST00000583179 was positively correlated with the expression of IL-4 and IL-6. Interestingly, lncRNAs also regulated the expression of cytokines (IL-5 and IL-13), transcription factors (STAT5 and STAT6) and chemokines (CCL17 and CCL22), which mediated Th1 and Th2 inflammatory response in asthma. Nevertheless, its role in the pathogenesis of asthma remains to be further studied (Wang et al., 2017). Recently, lncRNAs SNHG8 and LINC01559 have been found to be directly involved in the occurrence and progression of childhood asthma through PI3K/AKT signaling pathway (Hao and Zan, 2021), and may serve as candidates for therapeutic strategies for childhood asthma (Table 2). Further elucidating the role of various lncRNAs in childhood asthma will help to understand the pathogenesis of asthma and develop potential treatment targets.
In sum, lncRNAs associated with cytokines, Th2-related transcription factors (STAT5 and STAT6) and Th2 related chemokines (CCL17 and CCL22) to affect the balance of Th1/Th2 (Wang et al., 2017). In addition, lncRNAs may act as regulator of Th1 and Th2 inflammation in the pathogenesis of asthma through lncRNA-miRNA-mRNA axis. For example, lncRNA fantom3-9230106C11 binded to miRNAs and transcription factors to mediate Th2 cell differentiation and regulated Th2 inflammation (Wang et al., 2019). However, only a small number of lncRNAs were applied to the clinical diagnosis and treatment of childhood asthma. Further research associated with lncRNAs is needed in the field of asthma therapy.
MicroRNAs Act on Target Pathways and Regulatory Mechanisms of Childhood Asthma
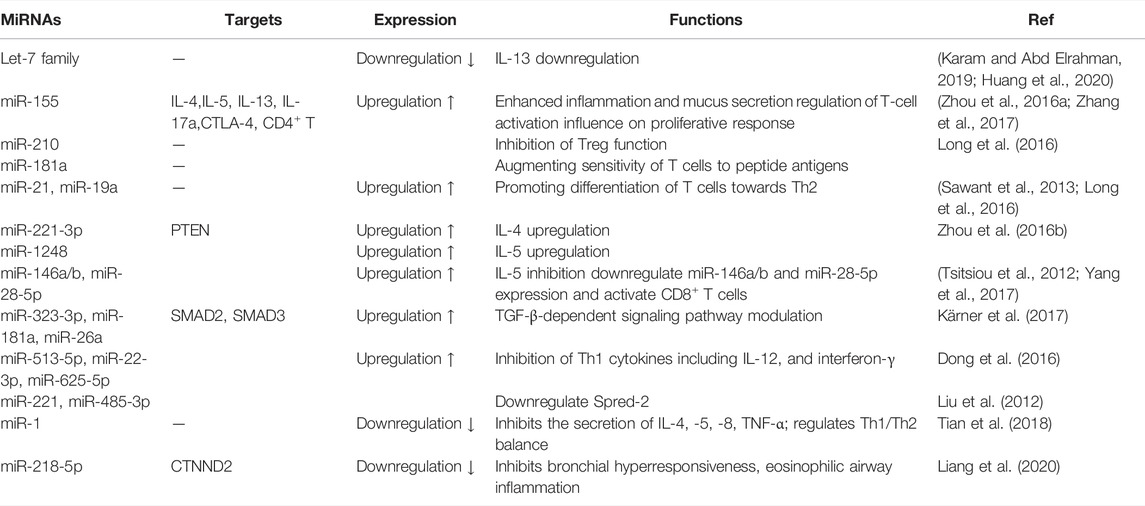
MiRNAs are a class of small ncRNAs (approximately 21–25 nucleotides) that regulate gene expression and cellular function by primarily bind to the 3′ untranslated region (3′ UTR) of mRNAs (Simpson et al., 2014; Kho et al., 2018). MiRNAs play vital roles in regulating Th2 activation, differentiation and proliferation by directly or indirectly acting on target genes (Rebane and Akdis, 2013; Qiu et al., 2018). On one hand, miRNAs regulate airway inflammation of childhood asthma by increasing Th2 cytokine secretion to decrease Th1 cytokine secretion and promote the differentiation of CD4+ T cells into Th2 (Midyat et al., 2016; Aripova et al., 2020). On the other hand, miRNAs also play roles in hyperplasia and hypertrophy of airway smooth muscle cells (Specjalski and Niedoszytko, 2020).
MiRNAs are involved in the pathogenesis of asthma by regulating inflammatory reaction. Specifically speaking, a core set of miRNAs were involved in childhood asthma including the downregulated let-7 family and upregulated miR-155, miR-21, miR-142-5p, miR-142-3p, miR-223, and miR-146a/b, etc. (Specjalski and Niedoszytko, 2020; van den Berge and Tasena, 2019). Let-7 microRNAs belongs to a family of highly conserved microRNA and comprises the most abundant miRNAs in lungs (Karam and Abd Elrahman, 2019; Cañas et al., 2020). Kumar et al. (2014) showed that let-7 family members were decreased in ovalbumin-sensitized animal models (Huang et al., 2020; Weidner et al., 2021), playing a proinflammatory role in asthma. Furthermore, let-7 family inhibited the secretion of IL-13 by directly targeting IL-13 transcript (Huang et al., 2020; Xu et al., 2020). Among numerous miRNAs reviewed in this field, miR-146, miR-155, and miR-223 have been identified as inflammatory response miRNAs that are up-regulated by NF-κB (Boldin and Baltimore, 2012; Kumar et al., 2014). MiRNA-21 is involved in the pathogenesis of asthma by limiting the activation of IL-12/IFN-γ and the differentiation of Th1 and Th2. It has been described that lack of miR-21 in mice may cause the increasion of levels of IFN-γ secreted by Th1 cells and the decreasion of pulmonary eosinophils, hence inhibiting the inflammation (Das et al., 2014).
MiRNAs are also involved in airway remodeling in asthmatic mice model. Ras homolog family member A (RhoA) of the Rho family GTPases regulated airway remodeling through regulating mesenchymal stem cell (MSC) differentiation. MiR-133a negatively regulated RhoA expression and bronchial smooth muscle cells (BSMCs) contraction (Chiba, 2020) to influence airway remodeling. Moreover, miR-26a induced human airway smooth muscle cells (HASMCs) hypertrophy (Mohamed et al., 2010). MiR-10a reduced mitogen-induced HASMCs proliferation (Hu et al., 2014) to regulate airway remodeling.
MiRNAs are not only the regulators in asthma pathogenesis, but also the targets of asthma therapeutics. Further studies illustrated miRNAs inhibited asthma through downregulation or antagonism of disease-related miRNA (Wang, 2010). Growing evidence showed that up-regulated miRNAs could be inhibited by miRNAs inhibitors or synthetic miRNAs oligonucleotides against miRNAs activity, and the administration of miRNAs inducers that increased tissue-specific miRNAs expression might be another treatment for asthma (Lukiw, 2013; He et al., 2014). For example, miR-155 knockout and miR-106a knockdown alleviated asthma though diminishing airway inflammation, mucus hypersecretion and airway Th2 cytokine levels (Mattes et al., 2009; Huang et al., 2014; Malmhäll et al., 2014). MiRNA-221 blockade suppressed airway inflammation. Moreover, Let-7 miRNA inhibition reduced airway hyperresponsiveness and subepithelial fibrosis by decreasing IL-13 levels to suppress airway inflammation and attenuate mucus metaplasia (Chiba et al., 2009; Kumar et al., 2011) (Table 3).
CircRNAs in the Regulation of Childhood Asthma
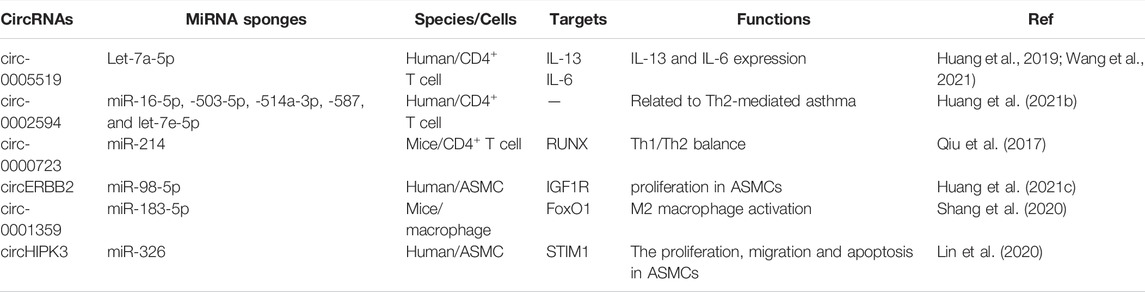
CircRNAs are a special class of ncRNAs that function as miRNA sponges to indirectly regulate downstream mRNA expression and epigenetically influence various biological processes, especially in cancers (Zhao et al., 2019; Huang et al., 2021b). However, the contribution of circRNAs to childhood asthma progression remains unknown. In recent years, circRNAs have attracted extensive attention in the pathogenesis of childhood asthma.
Huang et al. (2019) showed that circ-0005519 could regulate the secretion of IL-13/IL-6 by competitively sponging let-7a-5p in CD4+ T cells from asthma. Previous studies also found that circ-0002594 was a proinflammatory factor in Th2-mediated asthma (Huang et al., 2021b). Circ-0000723 could sponge miR-214 to impact the balance of Th1/Th2 by runt-related transcription factor (RUNX) signal transduction. Moreover, circRNAs also play vital roles in airway smooth muscle cells (ASMCs). Mounting evidence showed that circERBB2 and circHIPK3 could sponge miR-98-5p and miR-326, respectively, and promoted the proliferation of ASMCs by targeting IGF1R and STIM1 (Lin et al., 2020; Huang et al., 2021c). Circ-0001359 could attenuate airway remodeling by targeting FoxO1-dependent M2-like macrophage activation, with sponging miR-183-5p (Shang et al., 2020; Mathis et al., 2021) (Table 4). At present, although there are many studies on the competitively sponging of circRNAs to miRNAs, the specific effect of these circRNAs on asthma is not completely clear. So, the pathogenesis of circRNAs in childhood asthma should be further studied in order to seek for potential diagnostic and therapeutic targets of childhood asthma, which can be used as a new direction of targeted drug therapy.
NcRNAs Function as Targets for Asthma Diagnosis and Treatment
Although high-dose inhaled corticosteroids and long-acting β2 agonists have been improved for the treatment of asthma, none of these treatments have been shown to alter the natural history of the asthma, and some patients are still failing in these treatments (Sweeney et al., 2012). NcRNAs play an essential role in the treatment and prognosis of asthma. Hence, asthma has been turned to lncRNAs, miRNAs and circRNAs in search of new breakthroughs (Zampetaki et al., 2012; Milagro et al., 2013). Evidences have implicited that lncRNAs, miRNAs and circRNAs can be used as biomarkers of sensitivity and early diagnosis of asthma (Zampetaki et al., 2012). For instance, the expression of lncRNA CASC2 in serum was at a lower level in asthma children than healthy individuals, which suggested that lncRNA CASC2 might be involved in childhood asthma (Yang et al., 2022). Moreover, lnc-BAZ2B also played a crucial role in exacerbating the progression of childhood asthma (Xia et al., 2021). It can be inferred that lncRNA CASC2 and lnc-BAZ2B may serve as potential diagnostic biomarkers, and are expected to become new targets for childhood asthma treatment in the near future. Besides, many surveys have identified that miRNAs such as let-7a, miR-146b-5p, miR-21, miR-532-5p, miR-155 and so on, are promising to be used as diagnostic biomarkers and therapeutic targets in childhood asthma (Kho et al., 2018; Li et al., 2020b; Xu et al., 2020). CircRNAs also play crucial roles to regulate childhood asthma through circRNA-miRNA-mRNA regulatory network and can be served as potential biomarkers and therapeutic targets in childhood asthma (Chen et al., 2021; Wang et al., 2022). Although siRNAs are one of the regulatory ncRNAs, seldom studies have demonstrated the roles of siRNAs in the pathogenesis of childhood asthma. Recently, only several studies have implicated the therapeutic effects of synthetic siRNA in allergen-induced asthma models (Miyamoto et al., 2011; Chen et al., 2017). Consequently, research of drugs related to ncRNAs, especially lncRNAs, miRNAs and circRNAs, may become a new direction in the field of targeted asthma therapy.
Conclusion and Perspective
In this review, we mainly discuss childhood asthma. Children are special categories of patients and the symptoms of many other diseases are similar with asthma, which makes the diagnosis of asthma difficult. This review provides a novel strategy to diagnose and treat childhood asthma by targeting ncRNAs. Despite the treatment of asthma by inhaling corticosteroids, long acting β agonists, and leukotriene modifiers could reduce symptoms, the burden of asthma remains high. As far as childhood asthma, there is no evidence that early treatment decreases the risk of subsequent asthma or alters its natural history (Depner et al., 2014; Ducharme et al., 2014). It is widely quoted that 5–10% of the asthmatic population have severe asthma, suffering a significant health and socioeconomic burden (Papi et al., 2018; Agache et al., 2021). Facing these challenges, biomarker-directed therapy is more and more attractive and biomarkers for asthma have potential utility for distinguishing the inflammatory endotype, predicting responsiveness to specific treatments, monitoring success of a selected treatment option, and assessing the risk of disease progression (Agache and Rogozea, 2017; Cosmi et al., 2017). Accordingly, the discovery of lncRNAs, miRNAs and circRNAs offers a new opportunity for understanding the pathogenesis of childhood asthma (Rundell et al., 2015) and it is necessary to develop ncRNAs as new therapeutic targets for asthma.
Clinically, we still lack effective treatment measures for refractory asthma. NcRNAs have been considered to be one of the most promising and novel therapeutic targets for childhood asthma. Correctly, lncRNAs contributing to the regulation of airway remodeling and glucocorticoid sensitivity during transcription make it a potential biomarker for the preclinical identification, diagnosis, prognosis, phenotypes of asthma as well as therapeutic targets. However, there is no research to discuss the relationship between lncRNAs and Th2 cell and T follicular helper cells, which might be the focus of future research. In the same way, miRNAs also have multiple potential targets that may coordinate or antagonize each other’s functions. Restoring normal physiological levels of miRNAs in asthma, such as miRNA mimics, inhibitors, might have the potential to improve clinical outcomes (Ameis et al., 2017). Actually, ncRNAs still exist limitations, for example, how to accurately locate at a specific target or a certain organ, and whether through specific chemical modification of nucleic acid drugs or not, possible off-target effects of nucleic acid drug, etc. which is still worth exploring. Besides, there are still some unexpected side effects on ncRNAs, such as the disruption of the immune response and incompleteness during the treatment because of individual differences and so on. Luckily, if given appropriate immunotherapy and individualized treatment, these side effects may be reduced.
Nevertheless, due to complicated crosstalks between ncRNAs and inflammatory pathways in asthma, the expression of single lncRNAs, miRNAs and circRNAs may not be a truly reliable biomarker. Thus, several attempts have been made in finding asthma associated with pathways to provide a new avenue to treat asthma. For example, the metastatic-associated lung adenocarcinoma transcript 1 (MALAT1)/miR-155/CTLA-4 axis has the ability to regulate Th1/Th2 balance in asthma. MALAT1 is upregulated in the blood of asthmatic patients, while the miR-155 is downregulated, and the Th1/Th2 ratio is decreased, which suggests that Th1 inflammation is weakened and Th2 inflammation is amplified, so it can be used as a marker of inflammatory disorders in asthma (Liang and Tang, 2020). Thus, future studies should be more focused on human settings. Application of lncRNAs, miRNAs and circRNAs as non-invasive biomarkers should be investigated with an emphasis of possible determination of disease endotype and predicting treatment effects. Recently, the roles of all ncRNAs in the pathogenesis of childhood asthma have not been clarified, which should be further studied in order to seek for potential diagnostic and therapeutic targets of childhood asthma.
Author Contributions
JL wrote the manuscript. JL, X-HL, X-MC, X-LS drew manuscript figures. WL initiated this study and helped with writing-review and editing. WL and YH monitored the whole progress of this project.
Funding
This study was supported by the Natural Science Foundation of China (32100602), Scientific Research Start-up from Affiliated Hospital of Guangdong Medical University (1057Z20210004), the Guangdong Natural Science Foundation of China(2018A030307043) and Clinical research project of the Affiliated Hospital of Guangdong Medical University(LCYJ2020DL01).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdullahi, M., Ranjbaran, R., Alyasin, S., Keshavarz, Z., Ramezani, A., Behzad-Behbahani, A., et al. (2016). Expression of Basophil Activation Markers in Pediatric Asthma. Iran J. Immunol. 13 (1), 27–36.
Agache, I., Eguiluz-Gracia, I., Cojanu, C., Laculiceanu, A., Del Giacco, S., Zemelka-Wiacek, M., et al. (2021). Advances and Highlights in Asthma in 2021. Allergy 76 (11), 3390–3407. doi:10.1111/all.15054
Agache, I., and Rogozea, L. (2017). Asthma Biomarkers: Do They Bring Precision Medicine Closer to the Clinic? Allergy Asthma Immunol. Res. 9 (6), 466–476. doi:10.4168/aair.2017.9.6.466
Alam, R., and Gorska, M. M. (2011). Mitogen-activated Protein Kinase Signalling and ERK1/2 Bistability in Asthma. Clin. Exp. Allergy 41 (2), 149–159. doi:10.1111/j.1365-2222.2010.03658.x
Alizadeh, Z., Mortaz, E., Adcock, I., and Moin, M. (2017). Role of Epigenetics in the Pathogenesis of Asthma. Iran J. Allergy Asthma Immunol. 16 (2), 82–91.
Altman, M. C., Gill, M. A., Whalen, E., Babineau, D. C., Shao, B., Liu, A. H., et al. (2019). Transcriptome Networks Identify Mechanisms of Viral and Nonviral Asthma Exacerbations in Children. Nat. Immunol. 20 (5), 637–651. doi:10.1038/s41590-019-0347-8
Ameis, D., Khoshgoo, N., Iwasiow, B. M., Snarr, P., and Keijzer, R. (2017). MicroRNAs in Lung Development and Disease. Paediatr. Respir. Rev. 22, 38–43. doi:10.1016/j.prrv.2016.12.002
Arenas-Ramirez, N., Woytschak, J., and Boyman, O. (2015). Interleukin-2: Biology, Design and Application. Trends Immunol. 36 (12), 763–777. doi:10.1016/j.it.2015.10.003
Aripova, A., Akparova, A., and Bersimbaev, R. (2020). Moderate Bronchial Asthma. Microrna. doi:10.2174/2211536609666201221122715
Assaf, S. M., and Hanania, N. A. (2019). Biological Treatments for Severe Asthma. Curr. Opin. Allergy Clin. Immunol. 19 (4), 379–386. doi:10.1097/ACI.0000000000000549
Austin, P. J., Tsitsiou, E., Boardman, C., Jones, S. W., Lindsay, M. A., Adcock, I. M., et al. (2017). Transcriptional Profiling Identifies the Long Noncoding RNA Plasmacytoma Variant Translocation (PVT1) as a Novel Regulator of the Asthmatic Phenotype in Human Airway Smooth Muscle. J. Allergy Clin. Immunol. 139 (3), 780–789. doi:10.1016/j.jaci.2016.06.014
Barnig, C., Cernadas, M., Dutile, S., Liu, X., Perrella, M. A., Kazani, S., et al. (2013). Lipoxin A4 Regulates Natural Killer Cell and Type 2 Innate Lymphoid Cell Activation in Asthma. Sci. Transl Med. 5 (174), 174ra26. doi:10.1126/scitranslmed.3004812
Barnig, C., and Levy, B. D. (2015). Innate Immunity Is a Key Factor for the Resolution of Inflammation in Asthma. Eur. Respir. Rev. 24 (135), 141–153. doi:10.1183/09059180.00012514
Basnet, S., Bochkov, Y. A., Brockman-Schneider, R. A., Kuipers, I., Aesif, S. W., Jackson, D. J., et al. (2019). CDHR3 Asthma-Risk Genotype Affects Susceptibility of Airway Epithelium to Rhinovirus C Infections. Am. J. Respir. Cel Mol Biol 61 (4), 450–458. doi:10.1165/rcmb.2018-0220OC
Bégin, P., and Nadeau, K. C. (2014). Epigenetic Regulation of Asthma and Allergic Disease. Allergy Asthma Clin. Immunol. 10 (1), 27. doi:10.1186/1710-1492-10-27
Boldin, M. P., and Baltimore, D. (2012). MicroRNAs, New Effectors and Regulators of NF-Κb. Immunol. Rev. 246 (1), 205–220. doi:10.1111/j.1600-065X.2011.01089.x
Bond, S., Léguillette, R., Richard, E. A., Couetil, L., Lavoie, J. P., Martin, J. G., et al. (2018). Equine Asthma: Integrative Biologic Relevance of a Recently Proposed Nomenclature. J. Vet. Intern. Med. 32 (6), 2088–2098. doi:10.1111/jvim.15302
Cai, Z., Zeng, W., Tao, K., E, Z., Wang, B., and Yang, Q. (2015). Chaperone-mediated Autophagy: Roles in Neuroprotection. Neurosci. Bull. 31 (4), 452–458. doi:10.1007/s12264-015-1540-x
Cañas, J. A., Rodrigo-Muñoz, J. M., Sastre, B., Gil-Martinez, M., Redondo, N., and Del Pozo, V. (2020). MicroRNAs as Potential Regulators of Immune Response Networks in Asthma and Chronic Obstructive Pulmonary Disease. Front. Immunol. 11, 608666. doi:10.3389/fimmu.2020.608666
Castan, L., Bøgh, K. L., Maryniak, N. Z., Epstein, M. M., Kazemi, S., O'Mahony, L., et al. (2020). Overview of In Vivo and Ex Vivo Endpoints in Murine Food Allergy Models: Suitable for Evaluation of the Sensitizing Capacity of Novel Proteins? Allergy 75 (2), 289–301. doi:10.1111/all.13943
Cech, T. R., and Steitz, J. A. (2014). The Noncoding RNA Revolution-Trashing Old Rules to Forge New Ones. Cell 157 (1), 77–94. doi:10.1016/j.cell.2014.03.008
Chen, D., Wu, W., Yi, L., Feng, Y., Chang, C., Chen, S., et al. (2021). A Potential circRNA-miRNA-mRNA Regulatory Network in Asthmatic Airway Epithelial Cells Identified by Integrated Analysis of Microarray Datasets. Front. Mol. Biosci. 8, 703307. doi:10.3389/fmolb.2021.703307
Chen, H., Xu, X., Cheng, S., Xu, Y., Xuefei, Q., Cao, Y., et al. (2017). Small Interfering RNA Directed against microRNA-155 Delivered by a Lentiviral Vector Attenuates Asthmatic Features in a Mouse Model of Allergic Asthma. Exp. Ther. Med. 14 (5), 4391–4396. doi:10.3892/etm.2017.5093
Chiba, Y. (2020). Non-coding RNAs and Bronchial Smooth Muscle Hyperresponsiveness in Allergic Bronchial Asthma. Nihon Yakurigaku Zasshi 155 (6), 364–368. doi:10.1254/fpj.20053
Chiba, Y., Tanabe, M., Goto, K., Sakai, H., and Misawa, M. (2009). Down-regulation of miR-133a Contributes to Up-Regulation of Rhoa in Bronchial Smooth Muscle Cells. Am. J. Respir. Crit. Care Med. 180 (8), 713–719. doi:10.1164/rccm.200903-0325OC
Chogtu, B., Bhattacharjee, D., and Magazine, R. (2016). Epigenetics: The New Frontier in the Landscape of Asthma. Scientifica (Cairo) 2016, 4638949. doi:10.1155/2016/4638949
Cosmi, L., Liotta, F., Maggi, L., and Annunziato, F. (2017). Role of Type 2 Innate Lymphoid Cells in Allergic Diseases. Curr. Allergy Asthma Rep. 17 (10), 66. doi:10.1007/s11882-017-0735-9
Dai, B., Sun, F., Cai, X., Li, C., Liu, F., and Shang, Y. (2021). Long Noncoding RNA PTTG3P/miR-192-3p/CCNB1 axis Is a Potential Biomarker of Childhood Asthma. Int. Immunopharmacol 101 (Pt B), 108229. doi:10.1016/j.intimp.2021.108229
Das, A., Ganesh, K., Khanna, S., Sen, C. K., and Roy, S. (2014). Engulfment of Apoptotic Cells by Macrophages: a Role of microRNA-21 in the Resolution of Wound Inflammation. J. Immunol. 192 (3), 1120–1129. doi:10.4049/jimmunol.1300613
Defnet, A. E., Huang, W., Polischak, S., Yadav, S. K., Kane, M. A., Shapiro, P., et al. (2019). Effects of ATP-Competitive and Function-Selective ERK Inhibitors on Airway Smooth Muscle Cell Proliferation. FASEB J. 33 (10), 10833–10843. doi:10.1096/fj.201900680R
Depner, M., Fuchs, O., Genuneit, J., Karvonen, A. M., Hyvärinen, A., Kaulek, V., et al. (2014). Clinical and Epidemiologic Phenotypes of Childhood Asthma. Am. J. Respir. Crit. Care Med. 189 (2), 129–138. doi:10.1164/rccm.201307-1198OC
Deschildre, A., Pichavant, M., Engelmann, I., Langlois, C., Drumez, E., Pouessel, G., et al. (2017). Virus-triggered Exacerbation in Allergic Asthmatic Children: Neutrophilic Airway Inflammation and Alteration of Virus Sensors Characterize a Subgroup of Patients. Respir. Res. 18 (1), 191. doi:10.1186/s12931-017-0672-0
Dong, X., Xu, M., Ren, Z., Gu, J., Lu, M., Lu, Q., et al. (2016). Regulation of CBL and ESR1 Expression by microRNA-22-3p, 513a-5p and 625-5p M-ay I-mpact the P-athogenesis of D-ust M-ite-I-nduced P-ediatric A-sthma. Int. J. Mol. Med. 38 (2), 446–456. doi:10.3892/ijmm.2016.2634
Driscoll, A. J., Arshad, S. H., Bont, L., Brunwasser, S. M., Cherian, T., Englund, J. A., et al. (2020). Does Respiratory Syncytial Virus Lower Respiratory Illness in Early Life Cause Recurrent Wheeze of Early Childhood and Asthma? Critical Review of the Evidence and Guidance for Future Studies from a World Health Organization-Sponsored Meeting. Vaccine 38 (11), 2435–2448. doi:10.1016/j.vaccine.2020.01.020
Ducharme, F. M., Tse, S. M., and Chauhan, B. (2014). Diagnosis, Management, and Prognosis of Preschool Wheeze. Lancet 383 (9928), 1593–1604. doi:10.1016/S0140-6736(14)60615-2
El-Husseini, Z. W., Gosens, R., Dekker, F., and Koppelman, G. H. (2020). The Genetics of Asthma and the Promise of Genomics-Guided Drug Target Discovery. Lancet Respir. Med. 8 (10), 1045–1056. doi:10.1016/S2213-2600(20)30363-5
Elnady, H. G., Sherif, L. S., Kholoussi, N. M., Ali Azzam, M., Foda, A. R., Helwa, I., et al. (2020). Aberrant Expression of Immune-Related MicroRNAs in Pediatric Patients with Asthma. Int. J. Mol. Cel Med 9 (4), 246–255. doi:10.22088/IJMCM.BUMS.9.4.246
Enomoto, Y., Orihara, K., Takamasu, T., Matsuda, A., Gon, Y., Saito, H., et al. (2009). Tissue Remodeling Induced by Hypersecreted Epidermal Growth Factor and Amphiregulin in the Airway after an Acute Asthma Attack. J. Allergy Clin. Immunol. 124 (5), 913–917. doi:10.1016/j.jaci.2009.08.044
Ezegbunam, W., and Foronjy, R. (2018). Posttranscriptional Control of Airway Inflammation. Wiley Interdiscip. Rev. RNA 9 (1). doi:10.1002/wrna.1455
Fan, M., Xu, J., Xiao, Q., Chen, F., and Han, X. (2019). Long Non-coding RNA TCF7 Contributes to the Growth and Migration of Airway Smooth Muscle Cells in Asthma through Targeting TIMMDC1/Akt axis. Biochem. Biophys. Res. Commun. 508 (3), 749–755. doi:10.1016/j.bbrc.2018.11.187
Fasanaro, P., D'Alessandra, Y., Magenta, A., Pompilio, G., and Capogrossi, M. C. (2015). microRNAs: Promising Biomarkers and Therapeutic Targets of Acute Myocardial Ischemia. Curr. Vasc. Pharmacol. 13 (3), 305–315. doi:10.2174/15701611113119990011
Ferreira, M. A. R., Mathur, R., Vonk, J. M., Szwajda, A., Brumpton, B., Granell, R., et al. (2019). Genetic Architectures of Childhood- and Adult-Onset Asthma Are Partly Distinct. Am. J. Hum. Genet. 104 (4), 665–684. doi:10.1016/j.ajhg.2019.02.022
Foster, P. S., Maltby, S., Rosenberg, H. F., Tay, H. L., Hogan, S. P., Collison, A. M., et al. (2017). Modeling TH 2 Responses and Airway Inflammation to Understand Fundamental Mechanisms Regulating the Pathogenesis of Asthma. Immunol. Rev. 278 (1), 20–40. doi:10.1111/imr.12549
Frati, F., Salvatori, C., Incorvaia, C., Bellucci, A., Di Cara, G., Marcucci, F., et al. (2018). The Role of the Microbiome in Asthma: The Gut⁻Lung Axis. Int. J. Mol. Sci. 20 (1), 123. doi:10.3390/ijms20010123
Gao, H., Ying, S., and Dai, Y. (2017). Pathological Roles of Neutrophil-Mediated Inflammation in Asthma and its Potential for Therapy as a Target. J. Immunol. Res. 2017, 3743048. doi:10.1155/2017/3743048
Haktanir Abul, M., and Phipatanakul, W. (2019). Severe Asthma in Children: Evaluation and Management. Allergol. Int. 68 (2), 150–157. doi:10.1016/j.alit.2018.11.007
Hammad, H., and Lambrecht, B. N. (2021). The Basic Immunology of Asthma. Cell 184 (6), 1469–1485. doi:10.1016/j.cell.2021.02.016
Hao, M., and Zan, J. (2021). The Identification of Childhood Asthma Progression-Related lncRNAs and mRNAs Suitable as Biomarkers Using Weighted Gene Coexpression Network Analysis. Genet. Res. (Camb) 2021, 5511507. doi:10.1155/2021/5511507
He, X., Jing, Z., and Cheng, G. (2014). MicroRNAs: New Regulators of Toll-like Receptor Signalling Pathways. Biomed. Res. Int. 2014, 945169. doi:10.1155/2014/945169
Holgate, S. T., Wenzel, S., Postma, D. S., Weiss, S. T., Renz, H., and Sly, P. D. (2015). Asthma. Nat. Rev. Dis. Primers 1 (1), 15025. doi:10.1038/nrdp.2015.25
Hu, R., Pan, W., Fedulov, A. V., Jester, W., Jones, M. R., Weiss, S. T., et al. (2014). MicroRNA-10a Controls Airway Smooth Muscle Cell Proliferation via Direct Targeting of the PI3 Kinase Pathway. FASEB J. 28 (5), 2347–2357. doi:10.1096/fj.13-247247
Huang, F., Jia, H., Zou, Y., Yao, Y., and Deng, Z. (2020). Exosomes: an Important Messenger in the Asthma Inflammatory Microenvironment. J. Int. Med. Res. 48 (2), 300060520903220. doi:10.1177/0300060520903220
Huang, G., Su, J., Zhao, W., Deng, Z., Wang, P., Dong, H., et al. (2021). JNK Modulates RAGE/β-catenin Signaling and Is Essential for Allergic Airway Inflammation in Asthma. Toxicol. Lett. 336, 57–67. doi:10.1016/j.toxlet.2020.10.002
Huang, J. Q., Wang, F., Wang, L. T., Li, Y. M., Lu, J. L., and Chen, J. Y. (2021). Circular RNA ERBB2 Contributes to Proliferation and Migration of Airway Smooth Muscle Cells via miR-98-5p/IGF1R Signaling in Asthma. J. Asthma Allergy 14, 1197–1207. doi:10.2147/JAA.S326058
Huang, Y., Liu, Y., Li, L., Su, B., Yang, L., Fan, W., et al. (2014). Involvement of Inflammation-Related miR-155 and miR-146a in Diabetic Nephropathy: Implications for Glomerular Endothelial Injury. BMC Nephrol. 15, 142. doi:10.1186/1471-2369-15-142
Huang, Z., Cao, Y., Zhou, M., Qi, X., Fu, B., Mou, Y., et al. (2019). Hsa_circ_0005519 Increases IL-13/IL-6 by Regulating Hsa-Let-7a-5p in CD4+ T Cells to Affect Asthma. Clin. Exp. Allergy 49 (8), 1116–1127. doi:10.1111/cea.13445
Huang, Z., Fu, B., Qi, X., Xu, Y., Mou, Y., Zhou, M., et al. (2021). Diagnostic and Therapeutic Value of Hsa_circ_0002594 for T Helper 2-Mediated Allergic Asthma. Int. Arch. Allergy Immunol. 182 (5), 388–398. doi:10.1159/000511612
Huo, R., Tian, X., Chang, Q., Liu, D., Wang, C., Bai, J., et al. (2021). Targeted Inhibition of β-catenin Alleviates Airway Inflammation and Remodeling in Asthma via Modulating the Profibrotic and Anti-inflammatory Actions of Transforming Growth Factor-Β1. Ther. Adv. Respir. Dis. 15, 1753466620981858. doi:10.1177/1753466620981858
Inoue, H., Akimoto, K., Homma, T., Tanaka, A., and Sagara, H. (2020). Airway Epithelial Dysfunction in Asthma: Relevant to Epidermal Growth Factor Receptors and Airway Epithelial Cells. J. Clin. Med. 9 (11), 3698. doi:10.3390/jcm9113698
Jat, K. R., and Kabra, S. K. (2017). Awareness about Childhood Asthma. Indian J. Med. Res. 145 (5), 581–583. doi:10.4103/ijmr.IJMR_420_17
Jia, X. X., Zhu, T. T., Huang, Y., Zeng, X. X., Zhang, H., and Zhang, W. X. (2019). Wnt/β-catenin Signaling Pathway Regulates Asthma Airway Remodeling by Influencing the Expression of C-Myc and Cyclin D1 via the P38 MAPK-dependent Pathway. Exp. Ther. Med. 18 (5), 3431–3438. doi:10.3892/etm.2019.7991
Jiang, Y., Guo, X., and Qin, J. (2021). Silencing of circHIPK3 Hampers Platelet-Derived Growth Factor-Induced Proliferation and Migration in Airway Smooth Muscle Cells through the miR-375/MMP-16 axis. Cytotechnology 73 (4), 629–642. doi:10.1007/s10616-021-00483-2
Kabesch, M., and Tost, J. (2020). Recent Findings in the Genetics and Epigenetics of Asthma and Allergy. Semin. Immunopathol 42 (1), 43–60. doi:10.1007/s00281-019-00777-w
Karam, R. A., and Abd Elrahman, D. M. (2019). Differential Expression of miR-155 and Let-7a in the Plasma of Childhood Asthma: Potential Biomarkers for Diagnosis and Severity. Clin. Biochem. 68, 30–36. doi:10.1016/j.clinbiochem.2019.04.007
Kärner, J., Wawrzyniak, M., Tankov, S., Runnel, T., Aints, A., Kisand, K., et al. (2017). Increased microRNA-323-3p in IL-22/IL-17-producing T Cells and Asthma: a Role in the Regulation of the TGF-β Pathway and IL-22 Production. Allergy 72 (1), 55–65. doi:10.1111/all.12907
Kho, A. T., McGeachie, M. J., Moore, K. G., Sylvia, J. M., Weiss, S. T., and Tantisira, K. G. (2018). Circulating microRNAs and Prediction of Asthma Exacerbation in Childhood Asthma. Respir. Res. 19 (1), 128. doi:10.1186/s12931-018-0828-6
Kim, E. K., and Choi, E. J. (2010). Pathological Roles of MAPK Signaling Pathways in Human Diseases. Biochim. Biophys. Acta 1802 (4), 396–405. doi:10.1016/j.bbadis.2009.12.009
Kim, H. Y., DeKruyff, R. H., and Umetsu, D. T. (2010). The many Paths to Asthma: Phenotype Shaped by Innate and Adaptive Immunity. Nat. Immunol. 11 (7), 577–584. doi:10.1038/ni.1892
Kim, H. Y., Lee, H. J., Chang, Y. J., Pichavant, M., Shore, S. A., Fitzgerald, K. A., et al. (2014). Interleukin-17-producing Innate Lymphoid Cells and the NLRP3 Inflammasome Facilitate Obesity-Associated Airway Hyperreactivity. Nat. Med. 20 (1), 54–61. doi:10.1038/nm.3423
Kirenga, B., Chakaya, J., Yimer, G., Nyale, G., Haile, T., Muttamba, W., et al. (2020). Phenotypic Characteristics and Asthma Severity in an East African Cohort of Adults and Adolescents with Asthma: Findings from the African Severe Asthma Project. BMJ Open Respir. Res. 7 (1), e000484. doi:10.1136/bmjresp-2019-000484
Koczulla, A. R., Vogelmeier, C. F., Garn, H., and Renz, H. (2017). New Concepts in Asthma: Clinical Phenotypes and Pathophysiological Mechanisms. Drug Discov. Today 22 (2), 388–396. doi:10.1016/j.drudis.2016.11.008
Kumar, M., Ahmad, T., Sharma, A., Mabalirajan, U., Kulshreshtha, A., Agrawal, A., et al. (2011). Let-7 microRNA-Mediated Regulation of IL-13 and Allergic Airway Inflammation. J. Allergy Clin. Immunol. 128 (5), 1077–1110. doi:10.1016/j.jaci.2011.04.034
Kumar, V., Palermo, R., Talora, C., Campese, A. F., Checquolo, S., Bellavia, D., et al. (2014). Notch and NF-kB Signaling Pathways Regulate miR-223/FBXW7 axis in T-Cell Acute Lymphoblastic Leukemia. Leukemia 28 (12), 2324–2335. doi:10.1038/leu.2014.133
Lambrecht, B. N., and Hammad, H. (2015). The Immunology of Asthma. Nat. Immunol. 16 (1), 45–56. doi:10.1038/ni.3049
Lampalo, M., Majer, M., Ferara, N., Milošević, M., Barišić Kutija, M., and Jukić, I. (2019). Gender Differences in Relationship between Body Mass Index and Asthma. Psychiatr. Danub 31 (Suppl. 5), 786–791.
Lee, E., Lee, S. H., Kwon, J. W., Kim, Y. H., Yoon, J., Cho, H. J., et al. (2017). Persistent Asthma Phenotype Related with Late-Onset, High Atopy, and Low Socioeconomic Status in School-Aged Korean Children. BMC Pulm. Med. 17 (1), 45. doi:10.1186/s12890-017-0387-5
Lee, J. U., Kim, J. D., and Park, C. S. (2015). Gene-Environment Interactions in Asthma: Genetic and Epigenetic Effects. Yonsei Med. J. 56 (4), 877–886. doi:10.3349/ymj.2015.56.4.877
Lee, J. W., Min, J. H., Kim, M. G., Kim, S. M., Kwon, O. K., Oh, T. K., et al. (2019). Pistacia Weinmannifolia Root Exerts a Protective Role in Ovalbumin-induced L-ung I-nflammation in a M-ouse A-llergic A-sthma M-odel. Int. J. Mol. Med. 44 (6), 2171–2180. doi:10.3892/ijmm.2019.4367
Lee, Y. J., Fujisawa, T., and Kim, C. K. (2019). Biomarkers for Recurrent Wheezing and Asthma in Preschool Children. Allergy Asthma Immunol. Res. 11 (1), 16–28. doi:10.4168/aair.2019.11.1.16
Lejeune, S., Deschildre, A., Le Rouzic, O., Engelmann, I., Dessein, R., Pichavant, M., et al. (2020). Childhood Asthma Heterogeneity at the Era of Precision Medicine: Modulating the Immune Response or the Microbiota for the Management of Asthma Attack. Biochem. Pharmacol. 179, 114046. doi:10.1016/j.bcp.2020.114046
Li, B., Sun, W. X., Zhang, W. Y., Zheng, Y., Qiao, L., Hu, Y. M., et al. (2021). The Transcriptome Characteristics of Severe Asthma from the Prospect of Co-expressed Gene Modules. Front. Genet. 12, 765400. doi:10.3389/fgene.2021.765400
Li, J., Panganiban, R., Kho, A. T., McGeachie, M. J., Farnam, L., Chase, R. P., et al. (2020). Circulating MicroRNAs and Treatment Response in Childhood Asthma. Am. J. Respir. Crit. Care Med. 202 (1), 65–72. doi:10.1164/rccm.201907-1454OC
Li, X., Ye, S., and Lu, Y. (2020). Long Non-coding RNA NEAT1 Overexpression Associates with Increased Exacerbation Risk, Severity, and Inflammation, as Well as Decreased Lung Function through the Interaction with microRNA-124 in Asthma. J. Clin. Lab. Anal. 34 (1), e23023. doi:10.1002/jcla.23023
Liang, Y., Feng, Y., Wu, W., Chang, C., Chen, D., Chen, S., et al. (2020). microRNA-218-5p Plays a Protective Role in Eosinophilic Airway Inflammation via Targeting δ-catenin, a Novel Catenin in Asthma. Clin. Exp. Allergy 50 (1), 29–40. doi:10.1111/cea.13498
Liang, Z., and Tang, F. (2020). The Potency of lncRNA MALAT1/miR-155/CTLA4 axis in Altering Th1/Th2 Balance of Asthma. Biosci. Rep. 40 (2), BSR20190397. doi:10.1042/BSR20190397
Lin, C. H., Hong, Y. C., and Kao, S. H. (2015). Aeroallergen Der P 2 Induces Apoptosis of Bronchial Epithelial BEAS-2B Cells via Activation of Both Intrinsic and Extrinsic Pathway. Cell Biosci 5, 71. doi:10.1186/s13578-015-0063-5
Lin, J., Feng, X., and Zhang, J. (2020). Circular RNA circHIPK3 Modulates the Proliferation of Airway Smooth Muscle Cells by miR-326/STIM1 axis. Life Sci. 255, 117835. doi:10.1016/j.lfs.2020.117835
Lin, L., Li, Q., Hao, W., Zhang, Y., Zhao, L., and Han, W. (2019). Upregulation of LncRNA Malat1 Induced Proliferation and Migration of Airway Smooth Muscle Cells via miR-150-eIF4E/Akt Signaling. Front. Physiol. 10, 1337. doi:10.3389/fphys.2019.01337
Liu, F., Qin, H. B., Xu, B., Zhou, H., and Zhao, D. Y. (2012). Profiling of miRNAs in Pediatric Asthma: Upregulation of miRNA-221 and miRNA-485-3p. Mol. Med. Rep. 6 (5), 1178–1182. doi:10.3892/mmr.2012.1030
Liu, J. H., Li, C., Zhang, C. H., and Zhang, Z. H. (2020). LncRNA-CASC7 Enhances Corticosteroid Sensitivity via Inhibiting the PI3K/AKT Signaling Pathway by Targeting miR-21 in Severe Asthma. Pulmonology 26 (1), 18–26. doi:10.1016/j.pulmoe.2019.07.001
Liu, W., Liang, Q., Balzar, S., Wenzel, S., Gorska, M., and Alam, R. (2008). Cell-specific Activation Profile of Extracellular Signal-Regulated Kinase 1/2, Jun N-Terminal Kinase, and P38 Mitogen-Activated Protein Kinases in Asthmatic Airways. J. Allergy Clin. Immunol. 121 (4), 893–e2. doi:10.1016/j.jaci.2008.02.004
Liu, X., Zhang, Y., Jiang, H., Jiang, N., and Gao, J. (2019). Integrative Analysis of the Contribution of mRNAs and Long Non-coding RNAs to the P-athogenesis of A-sthma. Mol. Med. Rep. 20 (3), 2617–2624. doi:10.3892/mmr.2019.10511
Liu, Z., Mei, L., and He, Z. (2019). Long Non-coding RNA00882 Contributes to Platelet-Derived Growth Factor-Induced Proliferation of Human Fetal Airway Smooth Muscle Cells by Enhancing Wnt/β-Catenin Signaling via Sponging miR-3619-5p. Biochem. Biophys. Res. Commun. 514 (1), 9–15. doi:10.1016/j.bbrc.2019.04.106
Long, C. M., Lukomska, E., Marshall, N. B., Nayak, A., and Anderson, S. E. (2016). Potential Inhibitory Influence of miRNA 210 on Regulatory T Cells during Epicutaneous Chemical Sensitization. Genes (Basel) 8 (1), 9. doi:10.3390/genes8010009
Lukiw, W. J. (2013). Antagonism of NF-Κb-Up-Regulated Micro RNAs (miRNAs) in Sporadic Alzheimer's Disease (AD)-anti-NF-κB vs. Anti-miRNA Strategies. Front. Genet. 4, 77. doi:10.3389/fgene.2013.00077
Malmhäll, C., Alawieh, S., Lu, Y., Sjöstrand, M., Bossios, A., Eldh, M., et al. (2014). MicroRNA-155 Is Essential for T(H)2-mediated Allergen-Induced Eosinophilic Inflammation in the Lung. J. Allergy Clin. Immunol. 133 (5), 1429–1438. doi:10.1016/j.jaci.2013.11.008
Mathis, B. J., Kusumoto, M., Zaboronok, A., and Hiramatsu, Y. (2021). Packaging and Delivery of Asthma Therapeutics. Pharmaceutics 14 (1), 92. doi:10.3390/pharmaceutics14010092
Mattes, J., Collison, A., Plank, M., Phipps, S., and Foster, P. S. (2009). Antagonism of microRNA-126 Suppresses the Effector Function of TH2 Cells and the Development of Allergic Airways Disease. Proc. Natl. Acad. Sci. U S A. 106 (44), 18704–18709. doi:10.1073/pnas.0905063106
McCracken, J. L., Tripple, J. W., and Calhoun, W. J. (2016). Biologic Therapy in the Management of Asthma. Curr. Opin. Allergy Clin. Immunol. 16 (4), 375–382. doi:10.1097/ACI.0000000000000284
Metsälä, J., Lundqvist, A., Virta, L. J., Kaila, M., Gissler, M., and Virtanen, S. M. (2015). Prenatal and post-natal Exposure to Antibiotics and Risk of Asthma in Childhood. Clin. Exp. Allergy 45 (1), 137–145. doi:10.1111/cea.12356
Midyat, L., Gulen, F., Karaca, E., Ozkinay, F., Tanac, R., Demir, E., et al. (2016). MicroRNA Expression Profiling in Children with Different Asthma Phenotypes. Pediatr. Pulmonol 51 (6), 582–587. doi:10.1002/ppul.23331
Milagro, F. I., Miranda, J., Portillo, M. P., Fernandez-Quintela, A., Campión, J., and Martínez, J. A. (2013). High-throughput Sequencing of microRNAs in Peripheral Blood Mononuclear Cells: Identification of Potential Weight Loss Biomarkers. PloS one 8 (1), e54319. doi:10.1371/journal.pone.0054319
Miyamoto, S., Hattori, N., Senoo, T., Onari, Y., Iwamoto, H., Kanehara, M., et al. (2011). Intra-airway Administration of Small Interfering RNA Targeting Plasminogen Activator Inhibitor-1 Attenuates Allergic Asthma in Mice. Am. J. Physiol. Lung Cel Mol Physiol 301 (6), L908–L916. doi:10.1152/ajplung.00115.2011
Mohamed, J. S., Lopez, M. A., and Boriek, A. M. (2010). Mechanical Stretch Up-Regulates microRNA-26a and Induces Human Airway Smooth Muscle Hypertrophy by Suppressing Glycogen Synthase Kinase-3β. J. Biol. Chem. 285 (38), 29336–29347. doi:10.1074/jbc.M110.101147
Moral, L., Asensi Monzó, M., Juliá Benito, J. C., Ortega Casanueva, C., Paniagua Calzón, N. M., Pérez García, M. I., et al. (2021). Pediatric Asthma: The REGAP Consensus. Anales de pediatria 95 (2), 125. doi:10.1016/j.anpede.2021.02.007
Mukherjee, M., and Nair, P. (2018). Autoimmune Responses in Severe Asthma. Allergy Asthma Immunol. Res. 10 (5), 428–447. doi:10.4168/aair.2018.10.5.428
Narożna, B., Langwiński, W., and Szczepankiewicz, A. (2017). Non-Coding RNAs in Pediatric Airway Diseases. Genes 8 (12), 348. doi:10.3390/genes8120348
Ntontsi, P., Photiades, A., Zervas, E., Xanthou, G., and Samitas, K. (2021). Genetics and Epigenetics in Asthma. Int. J. Mol. Sci. 22 (5), 2412. doi:10.3390/ijms22052412
Papi, A., Brightling, C., Pedersen, S. E., and Reddel, H. K. (2018). Asthma. The Lancet 391 (10122), 783–800. doi:10.1016/s0140-6736(17)33311-1
Patel, E. S., and Chang, L. J. (2012). Synergistic Effects of Interleukin-7 and Pre-T Cell Receptor Signaling in Human T Cell Development. J. Biol. Chem. 287 (40), 33826–33835. doi:10.1074/jbc.M112.380113
Pividori, M., Schoettler, N., Nicolae, D. L., Ober, C., and Im, H. K. (2019). Shared and Distinct Genetic Risk Factors for Childhood-Onset and Adult-Onset Asthma: Genome-wide and Transcriptome-wide Studies. Lancet Respir. Med. 7 (6), 509–522. doi:10.1016/S2213-2600(19)30055-4
Pulido, R., and Lang, R. (2019). Dual Specificity Phosphatases: From Molecular Mechanisms to Biological Function. Int. J. Mol. Sci. 20 (18), 4372. doi:10.3390/ijms20184372
Qiu, L., Zhang, Y., Do, D. C., Ke, X., Zhang, S., Lambert, K., et al. (2018). miR-155 Modulates Cockroach Allergen- and Oxidative Stress-Induced Cyclooxygenase-2 in Asthma. J. Immunol. 201 (3), 916–929. doi:10.4049/jimmunol.1701167
Qiu, Y. Y., Wu, Y., Lin, M. J., Bian, T., Xiao, Y. L., and Qin, C. (2019). LncRNA-MEG3 Functions as a Competing Endogenous RNA to Regulate Treg/Th17 Balance in Patients with Asthma by Targeting microRNA-17/RORγt. Biomed. Pharmacother. 111, 386–394. doi:10.1016/j.biopha.2018.12.080
Qiu, Y. Y., Zhang, Y. W., Qian, X. F., and Bian, T. (2017). miR-371, miR-138, miR-544, miR-145, and miR-214 Could Modulate Th1/Th2 Balance in Asthma through the Combinatorial Regulation of Runx3. Am. J. Transl Res. 9 (7), 3184–3199.
Rebane, A., and Akdis, C. A. (2013). MicroRNAs: Essential Players in the Regulation of Inflammation. J. Allergy Clin. Immunol. 132 (1), 15–26. doi:10.1016/j.jaci.2013.04.011
Rundell, K. W., Anderson, S. D., Sue-Chu, M., Bougault, V., and Boulet, L. P. (2015). Air Quality and Temperature Effects on Exercise-Induced Bronchoconstriction. Compr. Physiol. 5 (2), 579–610. doi:10.1002/cphy.c130013
Saglani, S., Gregory, L. G., Manghera, A. K., Branchett, W. J., Uwadiae, F., Entwistle, L. J., et al. (2018). Inception of Early-Life Allergen-Induced Airway Hyperresponsiveness Is Reliant on IL-13+CD4+ T Cells. Sci. Immunol. 3 (27), eaan4128. doi:10.1126/sciimmunol.aan4128
Sawant, D. V., Wu, H., Kaplan, M. H., and Dent, A. L. (2013). The Bcl6 Target Gene microRNA-21 Promotes Th2 Differentiation by a T Cell Intrinsic Pathway. Mol. Immunol. 54 (3-4), 435–442. doi:10.1016/j.molimm.2013.01.006
Shang, Y., Sun, Y., Xu, J., Ge, X., Hu, Z., Xiao, J., et al. (2020). Exosomes from Mmu_circ_0001359-Modified ADSCs Attenuate Airway Remodeling by Enhancing FoxO1 Signaling-Mediated M2-like Macrophage Activation. Mol. Ther. Nucleic Acids 19, 951–960. doi:10.1016/j.omtn.2019.10.049
Shi, H. L., Lan, Y. H., Hu, Z. C., Yan, Z. N., Liu, Z. Z., Kadier, X., et al. (2020). Microecology Research: a New Target for the Prevention of Asthma. Chin. Med. J. (Engl) 133 (22), 2712–2720. doi:10.1097/CM9.0000000000001127
Simpson, L. J., Patel, S., Bhakta, N. R., Choy, D. F., Brightbill, H. D., Ren, X., et al. (2014). A microRNA Upregulated in Asthma Airway T Cells Promotes TH2 Cytokine Production. Nat. Immunol. 15 (12), 1162–1170. doi:10.1038/ni.3026
Song, J., Lim, H. X., Lee, A., Kim, S., Lee, J. H., and Kim, T. S. (2019). Staphylococcus Succinus 14BME20 Prevents Allergic Airway Inflammation by Induction of Regulatory T Cells via Interleukin-10. Front. Immunol. 10, 1269. doi:10.3389/fimmu.2019.01269
Southworth, T., Mason, S., Bell, A., Ramis, I., Calbet, M., Domenech, A., et al. (2018). PI3K, P38 and JAK/STAT Signalling in Bronchial Tissue from Patients with Asthma Following Allergen challenge. Biomark Res. 6, 14. doi:10.1186/s40364-018-0128-9
Specjalski, K., and Jassem, E. (2019). MicroRNAs: Potential Biomarkers and Targets of Therapy in Allergic Diseases? Arch. Immunol. Ther. Exp. (Warsz) 67 (4), 213–223. doi:10.1007/s00005-019-00547-4
Specjalski, K., and Niedoszytko, M. (2020). MicroRNAs: Future Biomarkers and Targets of Therapy in Asthma? Curr. Opin. Pulm. Med. 26 (3), 285–292. doi:10.1097/MCP.0000000000000673
Sripada, A., Sirohi, K., Michalec, L., Guo, L., McKay, J. T., Yadav, S., et al. (2021). Sprouty2 Positively Regulates T Cell Function and Airway Inflammation through Regulation of CSK and LCK Kinases. Plos Biol. 19 (3), e3001063. doi:10.1371/journal.pbio.3001063
Sweeney, J., Brightling, C. E., Menzies-Gow, A., Niven, R., Patterson, C. C., and Heaney, L. G. (2012). Clinical Management and Outcome of Refractory Asthma in the UK from the British Thoracic Society Difficult Asthma Registry. Thorax 67 (8), 754–756. doi:10.1136/thoraxjnl-2012-201869
Tang, H. H., Teo, S. M., Belgrave, D. C., Evans, M. D., Jackson, D. J., Brozynska, M., et al. (2018). Trajectories of Childhood Immune Development and Respiratory Health Relevant to Asthma and Allergy. eLife 7, e35856. doi:10.7554/eLife.35856
Tang, Y., Huang, W., Song, Q., Zheng, X., He, R., and Liu, J. (2018). Paeonol Ameliorates Ovalbumin-Induced Asthma through the Inhibition of TLR4/NF-Κb and MAPK Signaling. Evid. Based Complement. Alternat Med. 2018, 3063145. doi:10.1155/2018/3063145
Theodorou, J., Nowak, E., Böck, A., Salvermoser, M., Beerweiler, C., Zeber, K., et al. (2022). Mitogen-activated Protein Kinase Signaling in Childhood Asthma Development and Environment-Mediated protection. Pediatr. Allergy Immunol. 33 (1), e13657. doi:10.1111/pai.13657
Thomsen, S. F. (2015). Genetics of Asthma: an Introduction for the Clinician. Eur. Clin. Respir. J. 2. doi:10.3402/ecrj.v2.24643
Tian, M., Zhou, Y., Jia, H., Zhu, X., and Cui, Y. (2018). The Clinical Significance of Changes in the Expression Levels of MicroRNA-1 and Inflammatory Factors in the Peripheral Blood of Children with Acute-Stage Asthma. Biomed. Res. Int. 2018, 7632487. doi:10.1155/2018/7632487
Tsitsiou, E., Williams, A. E., Moschos, S. A., Patel, K., Rossios, C., Jiang, X., et al. (2012). Transcriptome Analysis Shows Activation of Circulating CD8+ T Cells in Patients with Severe Asthma. J. Allergy Clin. Immunol. 129 (1), 95–103. doi:10.1016/j.jaci.2011.08.011
van den Berge, M., and Tasena, H. (2019). Role of microRNAs and Exosomes in Asthma. Curr. Opin. Pulm. Med. 25 (1), 87–93. doi:10.1097/MCP.0000000000000532
Wang, S. Y., Fan, X. L., Yu, Q. N., Deng, M. X., Sun, Y. Q., Gao, W. X., et al. (2017). The lncRNAs Involved in Mouse Airway Allergic Inflammation Following Induced Pluripotent Stem Cell-Mesenchymal Stem Cell Treatment. Stem Cel Res Ther 8 (1), 2. doi:10.1186/s13287-016-0456-3
Wang, X., Chen, H., Liu, J., Gai, L., Yan, X., Guo, Z., et al. (2021). Emerging Advances of Non-coding RNAs and Competitive Endogenous RNA Regulatory Networks in Asthma. Bioengineered 12 (1), 7820–7836. doi:10.1080/21655979.2021.1981796
Wang, X., Xu, C., Cai, Y., Zou, X., Chao, Y., Yan, Z., et al. (2022). CircZNF652 Promotes the Goblet Cell Metaplasia by Targeting the miR-452-5p/JAK2 Signaling Pathway in Allergic Airway Epithelia. J. Allergy Clin. Immunol. S0091-6749 (22), 00129–134. doi:10.1016/j.jaci.2021.10.041
Wang, Z., Ji, N., Chen, Z., Wu, C., Sun, Z., Yu, W., et al. (2019). Next Generation Sequencing for Long Non-coding RNAs Profile for CD4+ T Cells in the Mouse Model of Acute Asthma. Front. Genet. 10, 545. doi:10.3389/fgene.2019.00545
Wang, Z. (2010). MicroRNA: A Matter of Life or Death. World J. Biol. Chem. 1 (4), 41–54. doi:10.4331/wjbc.v1.i4.41
Wasti, B., Liu, S. K., and Xiang, X. D. (2021). Role of Epigenetics in the Pathogenesis, Treatment, Prediction, and Cellular Transformation of Asthma. Mediators Inflamm. 2021, 9412929. doi:10.1155/2021/9412929
Weidner, J., Bartel, S., Kılıç, A., Zissler, U. M., Renz, H., Schwarze, J., et al. (2021). Spotlight on microRNAs in Allergy and Asthma. Allergy 76 (6), 1661–1678. doi:10.1111/all.14646
Westfall, S., Caracci, F., Zhao, D., Wu, Q.-l., Frolinger, T., Simon, J., et al. (2021). Microbiota Metabolites Modulate the T Helper 17 to Regulatory T Cell (Th17/Treg) Imbalance Promoting Resilience to Stress-Induced Anxiety- and Depressive-like Behaviors. Brain Behav. Immun. 91, 350–368. doi:10.1016/j.bbi.2020.10.013
Xia, L., Wang, X., Liu, L., Fu, J., Xiao, W., Liang, Q., et al. (2021). lnc-BAZ2B Promotes M2 Macrophage Activation and Inflammation in Children with Asthma through Stabilizing BAZ2B Pre-mRNA. J. Allergy Clin. Immunol. 147 (3), 921–e9. doi:10.1016/j.jaci.2020.06.034
Xie, N., and Liu, G. (2015). ncRNA-Regulated Immune Response and its Role in Inflammatory Lung Diseases. Am. J. Physiol. Lung Cel Mol Physiol 309 (10), L1076–L1087. doi:10.1152/ajplung.00286.2015
Xin, Y., Tang, L., Chen, J., Chen, D., Wen, W., and Han, F. (2021). Inhibition of miR-101-3p P-rotects against S-epsis-induced M-yocardial I-njury by I-nhibiting MAPK and NF-κB P-athway A-ctivation via the U-pregulation of DUSP1. Int. J. Mol. Med. 47 (3), 20. doi:10.3892/ijmm.2021.4853
Xing, Y., and Wong, G. W. (2022). Environmental Influences and Allergic Diseases in the Asia-Pacific Region: What Will Happen in Next 30 years? Allergy Asthma Immunol. Res. 14 (1), 21–39. doi:10.4168/aair.2022.14.1.21
Xu, L., Yi, M., Tan, Y., Yi, Z., and Zhang, Y. (2020). A Comprehensive Analysis of microRNAs as Diagnostic Biomarkers for Asthma. Ther. Adv. Respir. Dis. 14, 1753466620981863. doi:10.1177/1753466620981863
Yang, Y., Sun, Z., Ren, T., and Lei, W. (2022). Differential Expression of lncRNA CASC2 in the Serum of Childhood Asthma and its Role in Airway Smooth Muscle Cells Proliferation and Migration. J. Asthma Allergy 15, 197–207. doi:10.2147/JAA.S337236
Yang, Y., Yin, X., Yi, J., and Peng, X. (2017). MiR-146a Overexpression Effectively Improves Experimental Allergic Conjunctivitis through Regulating CD4+CD25-T Cells. Biomed. Pharmacother. 94, 937–943. doi:10.1016/j.biopha.2017.07.157
Yoshisue, H., Kirkham-Brown, J., Healy, E., Holgate, S. T., Sampson, A. P., and Davies, D. E. (2007). Cysteinyl Leukotrienes Synergize with Growth Factors to Induce Proliferation of Human Bronchial Fibroblasts. J. Allergy Clin. Immunol. 119 (1), 132–140. doi:10.1016/j.jaci.2006.08.028
Yu, H. S., Angkasekwinai, P., Chang, S. H., Chung, Y., and Dong, C. (2010). Protease Allergens Induce the Expression of IL-25 via Erk and P38 MAPK Pathway. J. Korean Med. Sci. 25 (6), 829–834. doi:10.3346/jkms.2010.25.6.829
Zampetaki, A., Willeit, P., Drozdov, I., Kiechl, S., and Mayr, M. (2012). Profiling of Circulating microRNAs: from Single Biomarkers to Re-wired Networks. Cardiovasc. Res. 93 (4), 555–562. doi:10.1093/cvr/cvr266
Zangouei, A. S., Rahimi, H. R., Mojarrad, M., and Moghbeli, M. (2020). Non Coding RNAs as the Critical Factors in Chemo Resistance of Bladder Tumor Cells. Diagn. Pathol. 15 (1), 136. doi:10.1186/s13000-020-01054-3
Zeng, H., Wang, Y., Gu, Y., Wang, J., Zhang, H., Gao, H., et al. (2019). Polydatin Attenuates Reactive Oxygen Species-Induced Airway Remodeling by Promoting Nrf2-Mediated Antioxidant Signaling in Asthma Mouse Model. Life Sci. 218, 25–30. doi:10.1016/j.lfs.2018.08.013
Zhang, J. H., Yang, X., Chen, Y. P., Zhang, J. F., and Li, C. Q. (2019). Nrf2 Activator RTA-408 Protects against Ozone-Induced Acute Asthma Exacerbation by Suppressing ROS and γδT17 Cells. Inflammation 42 (5), 1843–1856. doi:10.1007/s10753-019-01046-6
Zhang, P., Wu, W., Chen, Q., and Chen, M. (2019). Non-Coding RNAs and Their Integrated Networks. J. Integr. Bioinform 16 (3), 20190027. doi:10.1515/jib-2019-0027
Zhang, Y., Sun, E., Li, X., Zhang, M., Tang, Z., He, L., et al. (2017). miR-155 Contributes to Df1-Induced Asthma by Increasing the Proliferative Response of Th Cells via CTLA-4 Downregulation. Cell Immunol 314, 1–9. doi:10.1016/j.cellimm.2017.01.005
Zhao, X., Cai, Y., and Xu, J. (2019). Circular RNAs: Biogenesis, Mechanism, and Function in Human Cancers. Int. J. Mol. Sci. 20 (16), 3926. doi:10.3390/ijms20163926
Zhou, H., Li, J., Gao, P., Wang, Q., and Zhang, J. (2016). miR-155: A Novel Target in Allergic Asthma. Int. J. Mol. Sci. 17 (10), 1773. doi:10.3390/ijms17101773
Zhou, Y., Yang, Q., Xu, H., Zhang, J., Deng, H., Gao, H., et al. (2016). miRNA-221-3p Enhances the Secretion of Interleukin-4 in Mast Cells through the Phosphatase and Tensin Homolog/p38/Nuclear Factor-kappaB Pathway. PloS one 11 (2), e0148821. doi:10.1371/journal.pone.0148821
Zhu, X., Wei, Y., and Dong, J. (2020). Long Noncoding RNAs in the Regulation of Asthma: Current Research and Clinical Implications. Front. Pharmacol. 11, 532849. doi:10.3389/fphar.2020.532849
Keywords: childhood asthma, ncRNAs, lncRNAs, miRNAs, circRNAs
Citation: Liang J, Liu X-H, Chen X-M, Song X-L, Li W and Huang Y (2022) Emerging Roles of Non-Coding RNAs in Childhood Asthma. Front. Pharmacol. 13:856104. doi: 10.3389/fphar.2022.856104
Received: 16 January 2022; Accepted: 11 April 2022;
Published: 17 May 2022.
Edited by:
Yasuhiko Koga, Gunma University, JapanReviewed by:
Wan Ezumi Mohd Fuad, Universiti Sains Malaysia Health Campus, MalaysiaMohammad Reza Raoufy, Tarbiat Modares University, Iran
Copyright © 2022 Liang, Liu, Chen, Song, Li and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen Li, bGl3ZW40MTBAMTYzLmNvbQ==; Yuge Huang, R2RtY2Vya2VAaG90bWFpbC5jb20=
 Juan Liang
Juan Liang Xiao-Hua Liu
Xiao-Hua Liu Xue-Mei Chen1,2
Xue-Mei Chen1,2 Wen Li
Wen Li Yuge Huang
Yuge Huang