- 1National Engineering Laboratory for Breeding of Endangered Medicinal Materials, Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2China Resources Sanjiu Medical & Pharmaceutical Co., Ltd, Shenzhen, China
- 3Key Laboratory of Plant Biotechnology in Universities of Shandong Province, College of Life Sciences, Qingdao Agricultural University, Qingdao, China
- 4Engineering Research Center of Chinese Medicine Resources, Ministry of Education, Beijing, China
Bidens plants are annuals or perennials of Asteraceae and usually used as medicinal materials in China. They are difficult to identify by using traditional identification methods because they have similar morphologies and chemical components. Universal DNA barcodes also cannot identify Bidens species effectively. This situation seriously hinders the development of medicinal Bidens plants. Therefore, developing an accurate and effective method for identifying medicinal Bidens plants is urgently needed. The present study aims to use phylogenomic approaches based on organelle genomes to address the confusing relationships of medicinal Bidens plants. Illumina sequencing was used to sequence 12 chloroplast and eight mitochondrial genomes of five species and one variety of Bidens. The complete organelle genomes were assembled, annotated and analysed. Phylogenetic trees were constructed on the basis of the organelle genomes and highly variable regions. The organelle genomes of these Bidens species had a conserved gene content and codon usage. The 12 chloroplast genomes of the Bidens species were 150,489 bp to 151,635 bp in length. The lengths of the eight mitochondrial genomes varied from each other. Bioinformatics analysis revealed the presence of 50–71 simple sequence repeats and 46–181 long repeats in the organelle genomes. By combining the results of mVISTA and nucleotide diversity analyses, seven candidate highly variable regions in the chloroplast genomes were screened for species identification and relationship studies. Comparison with the complete mitochondrial genomes and common protein-coding genes shared by each organelle genome revealed that the complete chloroplast genomes had the highest discriminatory power for Bidens species and thus could be used as a super barcode to authenticate Bidens species accurately. In addition, the screened highly variable region trnS-GGA-rps4 could be also used as a potential specific barcode to identify Bidens species.
1 Introduction
Bidens plants are annuals or perennials of Asteraceae. In China, this genus includes 10 species (Shi et al., 2011), five of which are medicinal plants recorded in different local standards for Chinese medicinal materials. Bidens plants have a long history of medicinal use in China (Chen et al., 2009). There are many kinds of compounds in the Bidens plants, including flavonoids, phenylpropanoids, triterpenoids, alkaloids and organic acids, of which flavonoids are the main effective components in the medicinal Bidens plants (Wang et al., 2010). Modern pharmacological studies show that these compounds in Bidens plants have antiinflammatory, analgesic, antibacterial, antitumor, hypolipidemic, and liver protection functions (Lin et al., 2013; Shandukani et al., 2018). However, some problems exist in the records of these medicinal plants in local standards. For example, their scientific names are inconsistent with their Chinese names and records in the Flora of China. Bidens plants commonly have homonyms and synonyms. Furthermore, they are difficult to identify by using traditional identification methods because they have similar morphologies and chemical components (Bartolome et al., 2013; Chen et al., 2013; Wang et al., 2014; Yang et al., 2020). Reports on the molecular identification of Bidens species are limited. Tsai et al. (2008) (Tsai et al., 2008) used the noncoding regions of the chloroplast genome (trnL intorn and trnL-trnF) and nuclear ribosomal DNA (ITS1, 5.8S and ITS2) to identify Bidens species and found that ITS1, 5.8S, ITS2 and the trnL intron could separate only Bidens biternata and B. pilosa var. pilosa from each other. Our preliminary experiments showed that the universal DNA barcodes ITS, ITS2 and psbA-trnH were all ineffective in identifying Bidens species. The difficulties encountered in the identification of Bidens species seriously hinder the development of medicinal Bidens plants and reduce their medicinal quality. An accurate and effective method for identifying medicinal Bidens plants is urgently needed.
The main content of phylogenomic studies includes the use of large-scale molecular data to investigate the phylogenetic relationships between organisms at the genomic level and the application of evolutionary relationships to study the evolutionary mechanisms of genomes, such as the process of DNA repair and the functional annotation of unknown genes (Delsuc et al., 2005). In general, plant cells contain three kinds of genomes, namely, the chloroplast, mitochondrial and nuclear genomes. The chloroplast and mitochondrial genomes are also called organelle genomes. The relative abundances of the three genomic DNA in cells show significant differences. For example, a leaf cell of the model plant Arabidopsis thaliana contains approximately 1,000 copies of chloroplast DNA, 100 copies of mitochondrial DNA and two copies of nuclear genomic DNA (Logan 2006). Given the important role of the chloroplast and mitochondrial genomes in phylogenetic and nucleo-cytoplasmic interactions, their sequence analysis is becoming increasingly important. The chloroplast is an organelle that plays an important role in plant photosynthesis (Clegg et al., 1994). The chloroplast genome is more conserved than the nuclear genome in terms of gene content and order and contains more variations than DNA barcodes. Therefore, the chloroplast genome is widely used in species identification and plant evolution studies. Zhang et al. (Zhang et al., 2019) found that the whole chloroplast genome could be used as a super barcode to identify Dracaena species. Chen et al. (Chen et al., 2018; Chen et al., 2019) successfully identified six Ligularia and three Ephedra herbs by using the chloroplast genome as a super barcode. A growing number of works have provided support showing that identifying related species by using the molecular markers of the chloroplast genome or the complete chloroplast genome as super barcodes is practicable. The mitochondrial genome, another important organelle genome, is usually similar to the chloroplast genome, which has a circular molecular structure. The mitochondrial genome is used in species identification because it is complex and highly variable with abundant noncoding regions and introns and a relatively fixed sequence (An et al., 2017; Park and Lee, 2020; Jeong and Lee, 2021). The mitochondrial genome of angiosperms could reveal the phylogenetic relationship between species and be used to investigate intraspecific differentiation (Fujii et al., 2010). However, the number of reported mitochondrial genomes is not as large as that of chloroplast genomes.
Studying organelle genomes is an essential way to analyse the genetic information of a species. The excavation of organelle genomes is helpful for analysing the inherent properties and changes of organelle genomes and thus contributes to the genetic evolution, identification and breeding research of various species. In addition, organelle genomes are important data sources for comparative genomics, phylogenetics and population genetics (Qian 2014). Organelle genomes are smaller and easier to sequence than nuclear genomes. In this work, we sequenced and analysed the 12 chloroplast and eight mitochondrial genomes of five species and one variety of Bidens to solve the difficulties encountered in the identification of Bidens species. Finally, we constructed phylogenetic trees by using different datasets and analysed the feasibility of identifying Bidens species on the basis of organelle genomes. This research could provide a foundation for the species identification, phylogeny, medication safety and plant resource protection of Bidens.
2 Materials and Methods
2.1 DNA Sources
The DNA sources were the fresh leaves of B. biternata, B. bipinnata, B. pilosa var. pilosa, B. pilosa var. radiata, B. parviflora and B. tripartita. These species were identified by Prof. Yulin Lin from the Institute of Medicinal Plant Development (IMPLAD), Chinese Academy of Medical Sciences and Peking Union Medical College. Voucher specimens were deposited in the herbarium at IMPLAD. The ID numbers and collecting locations are shown in Supplementary Table S1. The total DNA of the species was extracted by using the DNeasy Plant Mini Kit (Qiagen Co., Germany), and DNA concentration and quality were assessed by using Nanodrop 2000C spectrophotometry and electrophoresis in 1% (w/v) agarose gel, respectively.
2.2 DNA Sequencing, Assembly and Annotation
The DNA was used to generate libraries with an average insert size of 350 bp and sequenced by using Illumina NovaSeq6000 in accordance with standard protocols, and the sequencing information was shown in Supplementary Tables S2, S3. Paired-end sequencing was performed to obtain 150 bp sequences at both ends of each molecule. Adapters and low-quality regions in the original data were trimmed by applying Trimmomatic software (Bolger et al., 2014). Reference organelle sequences from the family of Asteraceae were downloaded from NCBI genome resources (https://www.ncbi.nlm.nih.gov/genome). The gene sequences of each reference were extracted to build a custom database, and clean reads were mapped by using BWA 0.7.17 (Li and Durbin 2009). NOVOPlasty 4.2 (Dierckxsens et al., 2017) was used to assemble organelle genomes, which needed a sequence as the initial seed. For the chloroplasts, a read from the psbA gene was selected as the seed input, and the output chloroplast sequences were manually adjusted for the start position. For the mitochondria, seed reads from the conserved cox and nad genes were separately tested, and the output contigs were combined and manually linked to obtain the mitochondrial sequences. Then, clean reads were mapped back to the chloroplast and mitochondrial genomes and inspected in IGV (Robinson et al., 2017) to exclude any assembly error. Finally, a custom-made script that took the assembly and the bam files as input was utilised to correct ambiguous bases and generate the complete organelle genome sequences. The sequences were initially annotated by using the CPGAVAS2 software (Shi et al., 2019) and the GeSeq (Tillich et al., 2017) and corrected manually. tRNAs were annotated by using tRNAscan-SE software (Schattner et al., 2005). Genes, introns and coding region boundaries were compared with reference sequences. Then the borders of LSC, SSC and IR regions in the chloroplast genomes were validated by designing primers and polymerase chain reaction (Supplementary Table S4).
2.3 Structural Analyses
Chloroplast and mitochondrial genome maps were generated by using the Organellar Genome DRAW v1.2 (Lohse et al., 2007) and manually corrected. The CodonW software (Sharp and Li 1987) was adopted to analyse codon usage. Simple sequence repeats (SSRs) were detected by using the MISA (Beier et al., 2017) with the definition of ≥ 10 repeat units for mononucleotide SSRs, ≥ 5 repeat units for dinucleotide SSRs, ≥ 4 repeat units for trinucleotide SSRs, and ≥ 3 repeat units for tetranucleotide, pentanucleotide and hexanucleotide SSRs. Long repeated sequences were detected by using REPuter (Kurtz et al., 2001). Sequence homology analysis was carried out by using EMBOSS (Rice et al., 2000). The boundaries of the four regions of the chloroplast genomes were compared by applying IRscope (Amiryousefi et al., 2018).
2.4 Comparative and Phylogenetic Analyses
The chloroplast genomes of Bidens species were compared by using mVISTA software (Frazer et al., 2004). The nucleotide diversity (Pi) values of shared genes and intergenic spacers were calculated with DnaSP software (Librado and Rozas 2009). By combining the mVISTA results and Pi values, seven highly variable regions were screened out. The complete organelle genome sequences of Asteraceae species and common protein-coding genes shared by these genomes were used to construct maximum likelihood (ML) phylogenetic trees by utilising IQ-TREE (Nguyen et al., 2015) with a bootstrap of 1,000 repetitions. ML analysis was conducted based on the TVM + F + R4 (complete chloroplast genomes), TVM + F + I + G4 (complete mitochondrial genomes), TVM + F + R3 (common protein-coding genes shared by chloroplast genomes), and GTR + F + G4 (common protein-coding genes shared by mitochondrial genomes) models. MEGA software (Tamura et al., 2013) was used to construct Neighbor-joining (NJ) phylogenetic trees based on seven highly variable regions. NJ analysis was conducted based on the K2P model.
3 Results
3.1 Organelle Genome Structure of Bidens Species
The 12 chloroplast genomes of these Bidens species showed a typical circular tetramerous structure and included two inverted repeats (IRs), a large single copy (LSC) and a small single copy (SSC) (Figure 1A). The lengths of the chloroplast genomes of the same species were the same or similar. The total lengths of the chloroplast genomes were 150,489 bp (B. tripartita) to 151,635 bp (B. pilosa var. radiata). The sizes of the LSC regions ranged from 83,499 bp (B. tripartita) to 83,899 bp (B. biternata). The sizes of the SSC regions varied between 17,628 bp (B. tripartita) and 18,439 bp (B. pilosa var. radiata). The sizes of the IR regions varied from 24,652 bp (B. bipinnata) to 24,701 bp (B. pilosa var. radiata). The total GC contents of the 12 chloroplast genomes were all 37.5%, indicating that the base composition of the Bidens species was relatively conserved (Supplementary Table S5).
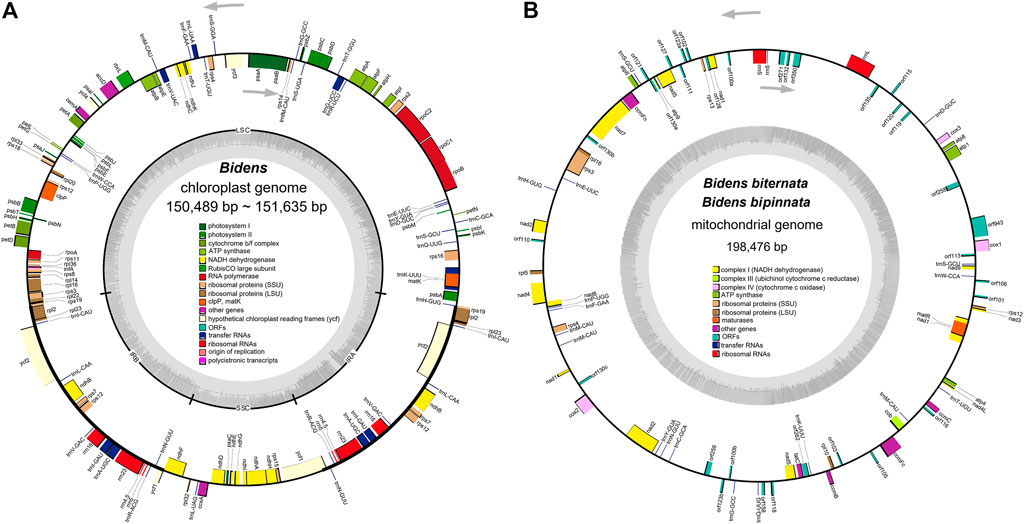
FIGURE 1. Chloroplast (A) and mitochondrial (B) genome maps of Bidens species. The transcription directions of the inner and outer genes are listed clockwise and anticlockwise, respectively, and are represented by arrows. The dark grey colour in the inner circle corresponds to the GC content, whereas the light grey colour corresponds to the AT content.
The 12 chloroplast genomes were all annotated with 130 genes, including 85 protein-coding genes, 37 tRNA genes and eight rRNA genes (Supplementary Table S6). Amongst these genes, 17 were located in IR regions. They included six protein-coding genes (ndhB, rpl2, rpl23, rps7, rps12 and ycf2), four rRNA genes (rrn4, rrn4.5, rrn5 and rrn16) and seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG and trnV-GAC). In addition, ycf1 and rps19 were annotated as pseudogenes. At the junctions, the gene positions in the boundary regions of the chloroplast genomes of the Bidens species were conserved. The difference was that the ndhF gene of B. tripartita was located at the boundary of the SSC and IRb regions, and the ndhF genes of the other five species were located in the SSC regions (Figure 2). Coding regions (protein-coding regions, tRNA genes and rRNA genes) accounted for 56.0–59.4% of the regions, and the rest were noncoding regions (pseudogenes, introns and gene spacers).
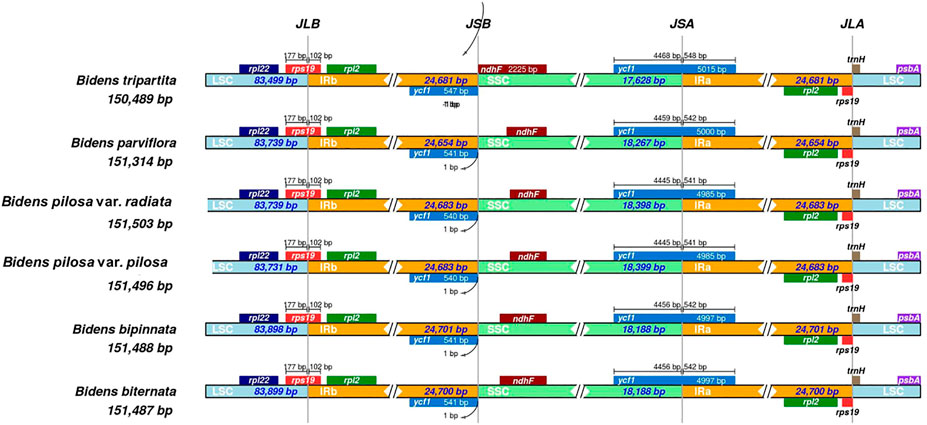
FIGURE 2. Comparison amongst the borders of LSC, SSC and IR regions in the chloroplast genomes of Bidens species. The number above the gene features indicates the distance between the ends of genes and the border sites. These features are not to scale. JLB: junction of LSC/IRb; JSB: junction of IRb/SSC; JSA: junction of SSC/IRa; JLA: junction of IRa/LSC.
The eight mitochondrial genomes of these Bidens species all had a circular molecular structure (Figure 1B; Supplementary Figure S1) that ranged in length from 183 kb (B. pilosa var. pilosa) to 216 kb (B. tripartita). The GC contents were between 45.4 and 45.8%. The mitochondrial genomes of B. biternata and B. bipinnata were annotated with 83 genes, including 30 protein-coding genes, 18 tRNA genes, three rRNA genes and 32 open reading frames (ORFs). A total of 81, 87, 90 and 82 genes were annotated in the mitochondrial genomes of B. pilosa var. pilosa, B. pilosa var. radiata, B. parviflora and B. tripartita, respectively. The total proportion of all genes in the mitochondrial genomes of the Bidens species was between 24.4 and 28.6% (Supplementary Table S7).
The genes annotated in the mitochondrial genomes of the Bidens species could be divided into 11 categories (Supplementary Table S8). The similar gene types and numbers of complex I, complex III, complex IV, complex V, rRNA genes and maturation enzyme genes indicated that these genes were highly conserved in the mitochondrial genomes. The number of tRNA genes ranged from 18 to 20, and the number of ORFs was quite different.
3.2 Relative Synonymous Codon Usage
The relative synonymous codon usage (RSCU) of the chloroplast and mitochondrial genomes of the Bidens species was calculated on the basis of all protein-coding genes (Figure 3). The results showed that the chloroplast and mitochondrial genomes of the Bidens species contained 64 types of codons encoding 20 amino acids. Leucine, serine and arginine all had six types of codons. Amongst all amino acids, leucine had the highest number of codons, whereas cysteine had the lowest.
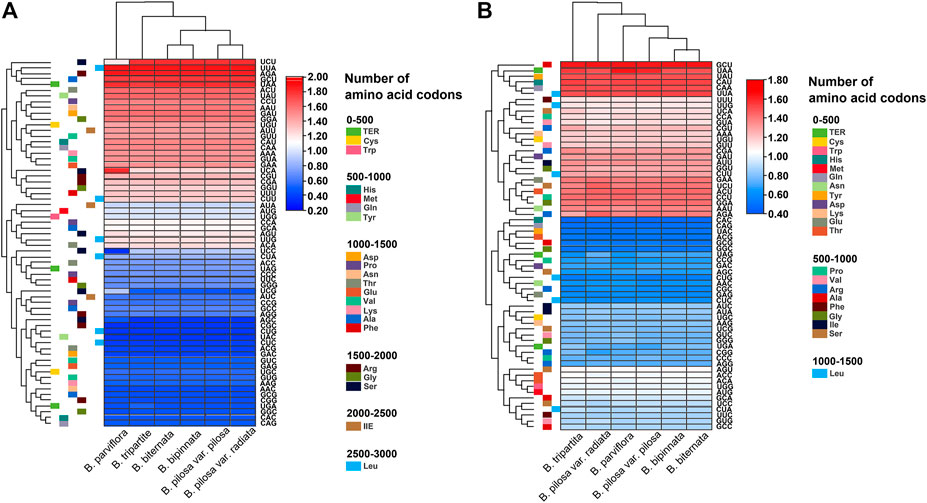
FIGURE 3. Heat map of the RSCU values of the chloroplast (A) and mitochondrial (B) genomes of Bidens species.
Given that methionine and tryptophan possess only one codon each, no codon usage bias was found, and the RSCU value was 1. Codon usage bias was found for the rest of the amino acid codons. In the chloroplast genomes, 30 codons were found with RSCU > 1, of which 29 were A/U-ending codons, and 34 codons were found with RSCU ≤ 1, of which 31 were G/C-ending codons. The highest and lowest RSCU values were recorded for UUA and CGC, which encoded leucine and arginine, respectively. In the mitochondrial genomes, 30 codons were found with RSCU > 1, of which 28 were A/U-ending codons, and 34 codons were found with RSCU ≤ 1, of which 30 were G/C-ending codons. The highest and lowest RSCU values were recorded for GCU and CAC, which encoded alanine and histidine, respectively. These results indicated that the chloroplast and mitochondrial genomes exhibited a higher bias towards A/U-ending codons than towards G/C-ending codons.
3.3 Simple Sequence Repeats and Long Repetitive Sequences in the Organelle Genomes of Bidens Species
In this study, a total of 56 (B. parviflora) to 71 (B. tripartita) SSRs were detected in the chloroplast genomes of these Bidens species. The distribution of SSRs in the three samples of B. biternata was the same. It was also the same in the four samples of B. bipinnata. Bidens bipinnata had one more mononucleotide repeat than B. biternata, and the remaining SSR types and numbers were the same. SSR distribution differed between the two B. pilosa var. radiata samples. The MW551955 sample had two more mononucleotide repeats, one more dinucleotide repeat and one more trinucleotide repeat than the MW551952 sample, indicating obvious SSR polymorphism in the chloroplast genome of B. pilosa var. radiata. Mononucleotide to hexanucleotide repeats were present in the Bidens species, most of which were mononucleotide repeats, followed by dinucleotide and tetranucleotide repeats (Supplementary Table S9). The SSRs of the mitochondrial genomes of the Bidens species were also analysed. A total of 50 (B. pilosa var. pilosa) to 62 (B. pilosa var. radiata) SSRs were detected in the mitochondrial genomes of the Bidens species. Six types of SSR were detected in the mitochondrial genomes of B. biternata, B. bipinnata, B. pilosa var. radiata and B. parviflora, whereas only five types were detected in B. pilosa var. pilosa and B. tripartita (Supplementary Table S10).
Some repetitive sequences with length ≥ 30 bp and sequence similarity ≥ 90% were found. These sequences were called long repetitive sequences, namely, forward (F), palindrome (P), reverse (R) and complement (C). In the chloroplast genomes of Bidens species, our analysis revealed 46 (B. biternata and B. bipinnata) to 114 (B. pilosa var. radiata) long repeats, most of which were F and P repeats. The long repeats mainly had lengths of 30–39 bp and included four types. Repeats that were larger than 40 bp were mainly F and P repeats. No repeats with lengths of 50–59 and 60–69 bp were found in B. biternata, B. bipinnata and B. parviflora, and no repeat with lengths of 60–69 bp was found in B. tripartita (Supplementary Table S11). The mitochondrial genomes of Bidens species all contained F and P repeats. The mitochondrial genomes of B. biternata, B. bipinnata, B. pilosa var. pilosa, B. pilosa var. radiata and B. parviflora contained R repeats. Only the mitochondrial genomes of B. biternata, B. bipinnata and B. pilosa var. pilosa contained C repeats. The mitochondrial genome of B. tripartita did not contain R and C repeats. The F and P repeats accounted for more than 90% of the long repeats in the mitochondrial genomes of the Bidens species. The number of long repeats was the highest in B. pilosa var. radiata and B. tripartita, with 106 and 108 F repeats, respectively. In these species, the number of repeats with lengths of 30–39 bp was the largest, followed by that of repeats with lengths of 40–99 bp; repeats with length ≥ 1 kb were rare (Supplementary Table S12).
3.4 Variation in the Organelle Genomes of Bidens Species
Consistent with the chloroplast genome analysis above, the global comparison of the chloroplast genomes of the Bidens species showed that the seven chloroplast genomes of B. biternata and B. bipinnata were highly similar. A high degree of similarity was found between the samples of B. pilosa var. pilosa and B. pilosa var. radiata. The difference between B. tripartita and the other five species was the largest, and a mutation locus was found in the intergene region of ndhF-rpl32. In addition, the results revealed that the variation in the noncoding region was considerably greater than that in the coding region. Most of the variation was located in the LSC and SSC regions, and slight variation occurred in the IR regions. The rRNA genes of these species were highly conserved with little variation (Figure 4).
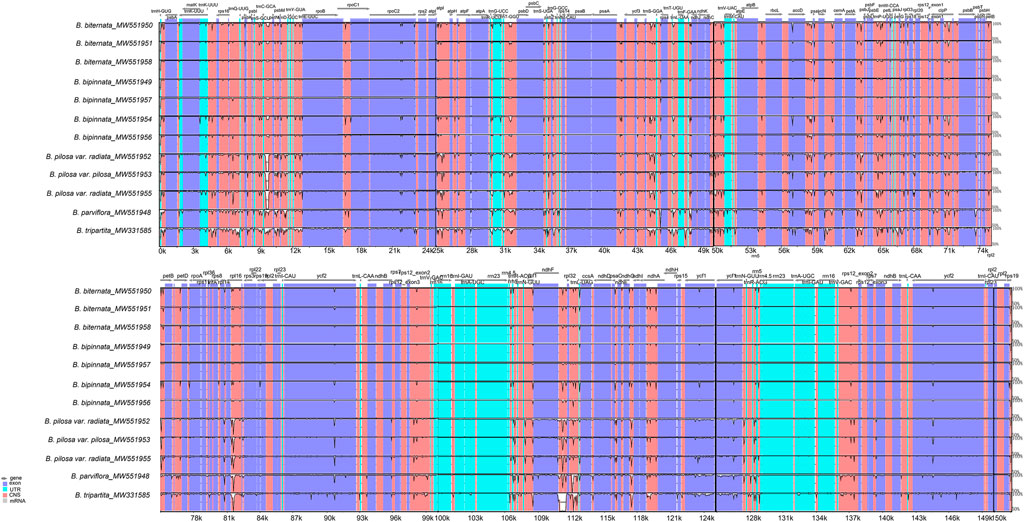
FIGURE 4. Global alignment of the chloroplast genomes of Bidens species. The x-axis represents the coordinates in the chloroplast genome. The y-axis indicates the average percent identity of sequence similarity, which ranged between 50 and 100%, in the aligned regions.
The Pi values of the shared genes and intergenic spacers of the chloroplast genomes of the Bidens species were calculated. Figure 5 shows the intergenic spacers and genes with Pi > 0. Intergenic spacers had more polymorphisms than gene regions, and these results were consistent with the mVISTA analysis results. By combining the mVISTA and Pi results, seven candidate highly variable regions (Pi > 0.018; length > 200 bp) were screened out for species identification and relationship studies.
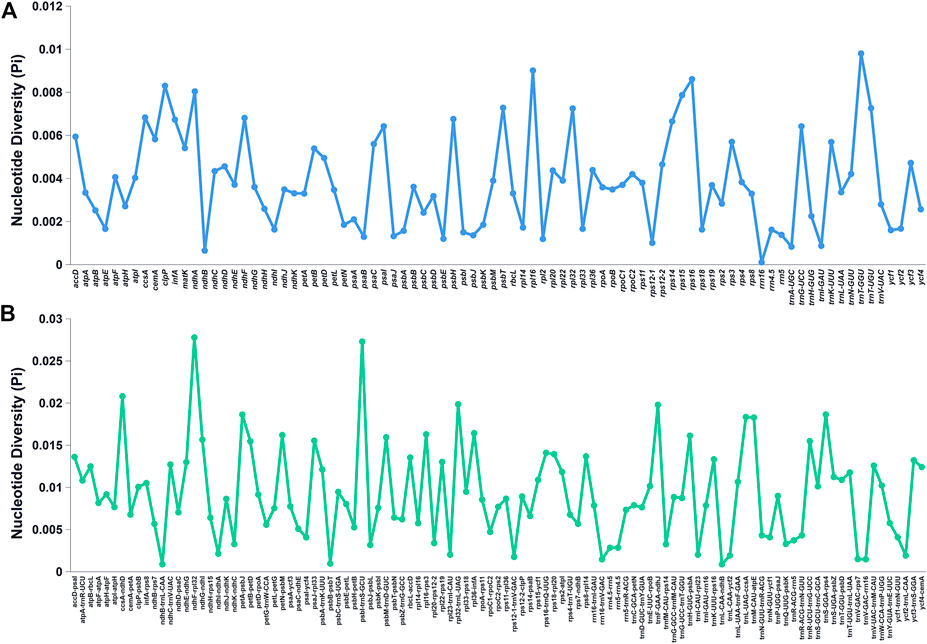
FIGURE 5. Nucleotide diversity of various shared regions in the chloroplast genomes of Bidens species. (A) Pi values in the gene regions. (B) Pi values in the intergenic spacer regions.
Consistent with the result based on chloroplast genomes, the results of collinearity and homology analysis showed that the mitochondrial genomes of B. biternata and B. bipinnata had high homology. The mitochondrial genomes of B. pilosa var. pilosa, B. pilosa var. radiata, B. parviflora and B. tripartita were quite different from those of B. biternata and B. bipinnata (Figure 6). The dot plot of the mitochondrial genome sequences between B. biternata and B. bipinnata showed an evident line on the diagonal, indicating that the chloroplast genome sequences of B. biternata and B. bipinnata had high homology (Supplementary Figure S2). In the dot plot of mitochondrial genome sequences between B. pilosa var. pilosa and B. pilosa var. radiata, only several diagonal lines made up of marker points were parallel to the diagonal lines, which represent the same substring of two sequences. This result indicated that the mitochondrial genome sequences of B. pilosa var. pilosa and B. pilosa var. radiata have low homology (Supplementary Figure S3).
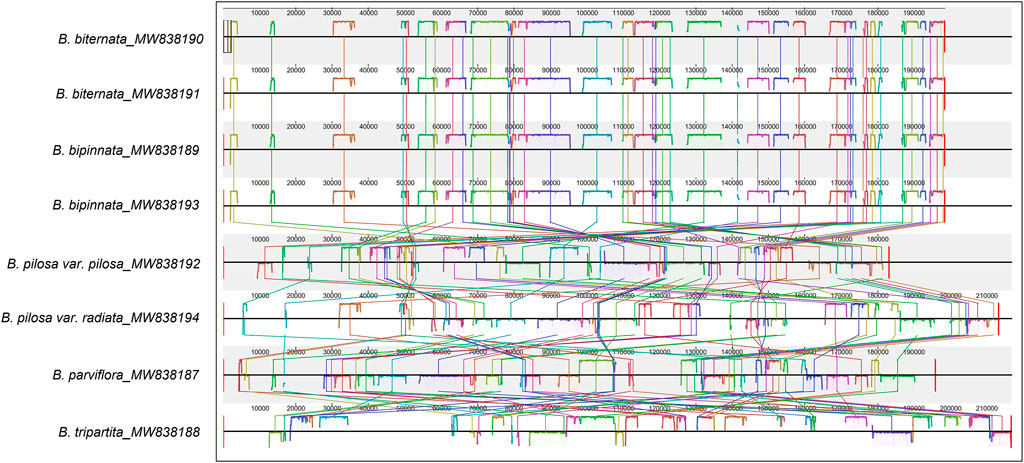
FIGURE 6. Collinearity analysis of the chloroplast genomes of Bidens species. Local collinear blocks are represented by blocks of the same colour connected by lines.
3.5 Identification and Phylogenetic Analysis of Bidens Species
In the current study, the complete chloroplast genome sequences of five species and one variety of Bidens and 21 other Asteraceae species were used to construct a phylogenetic tree with Magnolia officinalis and Nicotiana tabacum as the outgroups (Figure 7). The results showed that the Bidens species clustered in a big branch and that different samples of the same species clustered together. The three samples of B. biternata clustered in one branch, which was sister to the cluster comprising the four samples of B. bipinnata. Bidens pilosa var. radiata and B. pilosa var. pilosa clustered together. Bidens tripartita and B. parviflora clustered in a single branch, respectively. In addition, the species in Ligularia were close to those in Bidens.
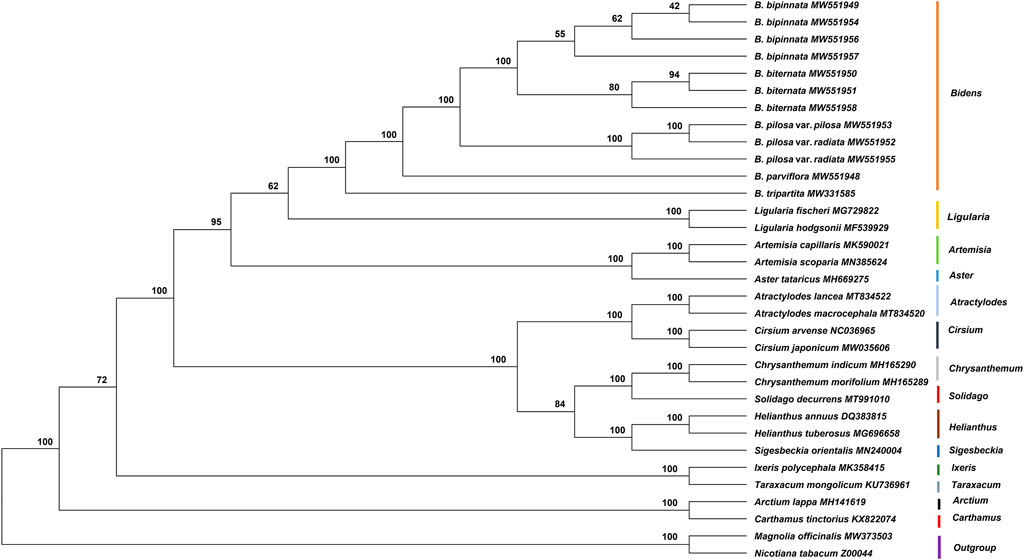
FIGURE 7. Phylogenetic tree constructed by using the ML method based on the complete chloroplast genome sequences of Asteraceae species. The numbers at the nodes are the values for bootstrap support.
Then, the common protein-coding genes shared in these chloroplast genomes were applied to construct the ML phylogenetic tree (Supplementary Figure S4). The result was slightly different from the finding based on the complete chloroplast genome sequences. The four samples of B. bipinnata did not cluster together. For the seven candidate highly variable regions, only trnS-GGA-rps4 showed the capability to identify Bidens species (Supplementary Figure S5). The species of B. bipinnata, B. biternata and B. pilosa clustered in different branches, respectively. In contrast to the tree based on the complete chloroplast genome, B. parviflora was sister to the B. pilosa species but with a low bootstrap value.
The ML phylogenetic tree was constructed by using the complete mitochondrial genomes and common protein-coding genes shared by the mitochondrial genomes of Asteraceae species, including the Bidens species (Supplementary Figures S6, S7). The Bidens species did not cluster together in the tree based on the complete mitochondrial genomes, whereas the Bidens species clustered together in the tree based on the common protein-coding genes shared by these mitochondrial genomes. However, species within the genus Bidens were indistinguishable from each other. The results showed that in contrast to the chloroplast genome, the mitochondrial genome was unsuitable for the identification and phylogenetic analysis of Bidens species.
4 Discussion
The lengths of the chloroplast genomes of these Bidens species were similar to those of other reported Asteraceae species, such as Ligularia species (151,118–151,253 bp) (Chen et al., 2018), Artemisia frigida (151,076 bp) (Liu et al., 2013) and Stilpnolepis centiflora (151,017 bp) (Shi and Xie 2020). The GC contents of the chloroplast genomes of the Bidens species were all 37.5%, which was similar to those of Carthamus tinctorius (Lu et al., 2016), Saussurea involucrata (Xie et al., 2017) and Arctium lappa (Xing et al., 2019). The majority of the chloroplast genomes of Asteraceae species, such as Soroseris umbrella (Lv et al., 2020), Saussurea inversa and Saussurea medusa (Wang et al., 2021), as well as Pertya phylicoides (Wang et al., 2020), contained approximately 130 genes, which included approximately 113 unique genes. The global comparison of the chloroplast genomes showed that the variation of the noncoding region was considerably larger than that of the coding region, most of the variation was located in the LSC and SSC regions, and very little variation occurred in the IR regions. The same findings were also found for Arctium lappa (Nie et al., 2020), Ligularia species (Chen et al., 2018) and Artemisia species (Liu et al., 2013). Moreover, we compared the CDS of the chloroplast genomes of 11 Asteraceae medicinal plants, including the Bidens species in this study, Bidens frondose (Knope et al., 2020), Arctium lappa (Nie et al., 2020), Atractylodes lancea (Shi et al., 2021), Aster tataricus (Shen et al., 2018) and Artemisia annua (Shen et al., 2017). The results showed that although the gene composition of the chloroplast genome of Asteraceae medicinal plants was highly conserved, slight variations were still present. The chloroplast genome of A. annua, but not the chloroplast genomes of other 10 medicinal plants, contained the photosystem II gene psbG. Only the chloroplast genome of B. frondose did not contain the ycf1 gene. The chloroplast genomes of 11 medicinal plants contained the ycf2 gene, whereas in the chloroplast genome of A. tataricus, ycf2 gene did not repeat in the IR regions. The ycf15 gene existed only in the chloroplast genome of B. frondose and A. annua and not in other medicinal plants (Supplementary Figure S8). Bioinformatics analysis revealed the presence of SSRs and long repeats in the chloroplast genome of Bidens species. Given that long repeats are abundant in the chloroplast genomes of some highly recombinant algae and angiosperms, especially at the end of the recombinant site, they are considered to be one of the main reasons for promoting the recombination of chloroplast genomes. However, in the chloroplast genome without recombination, the role of these repeats remains unclear (Qian 2014). The SSRs and long repeats contained in the chloroplast genome can be used as important sources of molecular markers for the development of research on Bidens species.
The lengths of the mitochondrial genomes of these Bidens species were similar to those of most land plants reported in the organelle database of NCBI (Supplementary Figure S9) and was similar to that of Tanacetum vulgare (Won et al., 2018). The total GC content of the mitochondrial genomes of the Bidens species was between 45.4 and 45.8% and were similar to that of Helianthus annuus (Grassa et al., 2016). The number of CDS in the mitochondrial genomes of the Bidens species was approximately 30 genes and was similar to that of most of reported plants in the NCBI organelle genome databases (30–45). In contrast to those of the conserved chloroplast genome, the sizes and structures of the mitochondrial genomes of angiosperms vary greatly (Levings and Brown 1989; Adams et al., 2002). Studies have shown that the plant mitochondrial genome is a mixture of DNA molecules with different shapes (Kubo and Newton 2008). The mitochondrial genomes of chrysanthemum and sunflower are circular (Makarenko et al., 2019; Wynn and Christensen 2019), whereas those of wheat, rape and cucumber comprise multiple rings (Handa 2003; Ogihara et al., 2005; Alverson et al., 2011). The mitochondrial genomes of plants are considerably larger than those of animals and range from 200 to 2,500 kb with a variation of more than 10 times (Levings and Brown 1989). In most angiosperms, the size of the mitochondrial genome is concentrated within the range of 300–600 kb (Levings and Brown 1989). The secondary structure of the mitochondrial genome is complex and changeable, and gene recombination frequently occurs in the mitochondrial genomes of angiosperms (Kubo and Newton 2008; Gualberto and Newton 2017). Mitochondrial genomes are generally used for high-level classification, such as intergenus and interfamily classification (Qiu et al., 2010).
In this study, phylogenetic trees were constructed on the basis of the complete chloroplast genomes, complete mitochondrial genomes, common protein-coding genes shared by each organelle genome, and seven selected highly variable regions. The complete chloroplast genomes showed the best capability for the identification and phylogenetic analysis of Bidens species. The chloroplast genome is an ideal material for species authentication and phylogenetic studies because it can be maternally inherited, usually does not undergo genetic recombination and has highly conserved gene content and order (Verma and Daniell 2007; Jansen and Ruhlman 2012). In fact, chloroplast genomes have been successfully used as a super barcode to identify numerous species and individuals (Doorduin et al., 2011; Kane et al., 2012; Chen et al., 2019; Wu et al., 2020). The phylogenetic trees constructed in this study demonstrated that complete chloroplast genome sequences can also be used as a super barcode for the identification of Bidens species. Bidens pilosa var. pilosa and B. pilosa var. radiata had similar morphological characteristics. In the Flora of China database, B. pilosa var. radiata and B. pilosa var. pilosa have been merged into one species named B. pilosa, and the Latin name B. pilosa var. radiata has been listed as a synonym of B. pilosa. In this study, the chloroplast genomes of B. pilosa var. pilosa and B. pilosa var. radiata were similar to each other. The ML phylogenetic tree showed that B. pilosa var. radiata and B. pilosa var. pilosa clustered together with a bootstrap value of 100%. Therefore, the analysis results of this study provided support for the merging of B. pilosa var. pilosa and B. pilosa var. radiata into the same species.
Conclusion
In this study, 12 chloroplast and eight mitochondrial genomes of five species and one variety of Bidens were analysed. Then, identification and phylogenetic analysis were performed by constructing phylogenetic trees on the basis of the complete chloroplast genomes, complete mitochondrial genomes, common protein-coding genes shared by the chloroplast genomes, common protein-coding genes shared by the mitochondrial genomes, and seven selected highly variable regions. The results of phylogenetic trees based on the complete chloroplast genomes and trnS-GGA-rps4 showed that different samples of the same species clustered together. This work indicated that complete chloroplast genomes could be used as a super barcode to authenticate Bidens species accurately, and the screened highly variable region trnS-GGA-rps4 could be used as a potential specific barcode to identify Bidens species.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: The 12 chloroplast genomes and eight mitochondrial genomes of the six Bidens species assembled in this study were deposited in GenBank with the accession numbers MW551948-MW551958, MW331585 (chloroplast genomes), and MW838187-MW838194 (mitochondrial genomes).
Author Contributions
LW, LN, and QW performed the experiments. SG and TG assembled and annotated the organelle genomes. LW, LN, and SG analysed the data. LW and LN wrote the manuscript. ZW, YL, YW, and BL collected plant material. HY, BL, and TG acquired the funding. HY conceived the research and revised the manuscript. All authors read and approved the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Science & Technology Fundamental Resources Investigation Program of China (No. 2018FY100700), National Natural Science Foundation of China (No. 81903748), and Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (CIFMS) (No. 2021-I2M-1-071).
Conflict of Interest
Author SG and ZW are employed by China Resources Sanjiu Medical & Pharmaceutical Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.842131/full#supplementary-material
References
Adams, K. L., Qiu, Y. L., Stoutemyer, M., and Palmer, J. D. (2002). Punctuated Evolution of Mitochondrial Gene Content: High and Variable Rates of Mitochondrial Gene Loss and Transfer to the Nucleus during Angiosperm Evolution. Proc. Natl. Acad. Sci. U S A. 99 (15), 9905–9912. doi:10.1073/pnas.042694899
Alverson, A. J., Rice, D. W., Dickinson, S., Barry, K., and Palmer, J. D. (2011). Origins and Recombination of the Bacterial-Sized Multichromosomal Mitochondrial Genome of Cucumber. Plant Cell 23 (7), 2499–2513. doi:10.1105/tpc.111.087189
Amiryousefi, A., Hyvönen, J., and Poczai, P. (2018). IRscope: an Online Program to Visualize the junction Sites of Chloroplast Genomes. Bioinformatics 34 (17), 3030–3031. doi:10.1093/bioinformatics/bty220
An, S. M., Kim, S. Y., Noh, J. H., and Yang, E. C. (2017). Complete Mitochondrial Genome of Skeletonema marinoi (Mediophyceae, Bacillariophyta), a Clonal Chain Forming Diatom in the West Coast of Korea. Mitochondrial DNA A. DNA Mapp. Seq. Anal. 28 (1), 19–20. doi:10.3109/19401736.2015.1106523
Bartolome, A. P., Villaseñor, I. M., and Yang, W. C. (2013). Bidens pilosa L. (Asteraceae): Botanical Properties, Traditional Uses, Phytochemistry, and Pharmacology. Evid. Based Complement. Alternat Med. 2013, 340215. doi:10.1155/2013/340215
Beier, S., Thiel, T., Münch, T., Scholz, U., and Mascher, M. (2017). MISA-web: a Web Server for Microsatellite Prediction. Bioinformatics 33 (16), 2583–2585. doi:10.1093/bioinformatics/btx198
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30 (15), 2114–2120. doi:10.1093/bioinformatics/btu170
Chen, F., Wu, F., Wang, D., and He, X. (2009). Pharmacognosy Identification of Bidens bipinnata L. Lishizhen Med. Materia Med. Res. 20 (2), 329–330. 10.1155/2013/340215.
Chen, J., Wei, J. H., Cai, S. F., Zhang, H. J., Zhang, X. H., Liang, W. J., et al. (2013). Chemical Constituents in Whole Herb of Bidens pilosa var. radiata. Zhong Yao Cai 36 (3), 410–413. doi:10.13863/j.issn1001-4454.2013.03.022
Chen, X., Cui, Y., Nie, L., Hu, H., Xu, Z., Sun, W., et al. (2019). Identification and Phylogenetic Analysis of the Complete Chloroplast Genomes of Three Ephedra Herbs Containing Ephedrine. Biomed. Res. Int. 2019, 5921725. doi:10.1155/2019/5921725
Chen, X., Zhou, J., Cui, Y., Wang, Y., Duan, B., and Yao, H. (2018). Identification of Ligularia Herbs Using the Complete Chloroplast Genome as a Super-barcode. Front. Pharmacol. 9, 695. doi:10.3389/fphar.2018.00695
Clegg, M. T., Gaut, B. S., Learn, G. H., and Morton, B. R. (1994). Rates and Patterns of Chloroplast DNA Evolution. Proc. Natl. Acad. Sci. U S A. 91 (15), 6795–6801. doi:10.1073/pnas.91.15.6795
Delsuc, F., Brinkmann, H., and Philippe, H. (2005). Phylogenomics and the Reconstruction of the Tree of Life. Nat. Rev. Genet. 6 (5), 361–375. doi:10.1038/nrg1603
Dierckxsens, N., Mardulyn, P., and Smits, G. (2017). NOVOPlasty: De Novo Assembly of Organelle Genomes from Whole Genome Data. Nucleic Acids Res. 45 (4), e18. doi:10.1093/nar/gkw955
Doorduin, L., Gravendeel, B., Lammers, Y., Ariyurek, Y., Chin-A-Woeng, T., and Vrieling, K. (2011). The Complete Chloroplast Genome of 17 Individuals of Pest Species Jacobaea Vulgaris: SNPs, Microsatellites and Barcoding Markers for Population and Phylogenetic Studies. DNA Res. 18 (2), 93–105. doi:10.1093/dnares/dsr002
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). VISTA: Computational Tools for Comparative Genomics. Nucleic Acids Res. 32, W273–W279. doi:10.1093/nar/gkh458
Fujii, S., Kazama, T., Yamada, M., and Toriyama, K. (2010). Discovery of Global Genomic Re-organization Based on Comparison of Two Newly Sequenced rice Mitochondrial Genomes with Cytoplasmic Male Sterility-Related Genes. BMC Genomics 11, 209. doi:10.1186/1471-2164-11-209
Grassa, C. J., Ebert, D. P., Kane, N. C., and Rieseberg, L. H. (2016). Complete Mitochondrial Genome Sequence of Sunflower (Helianthus annuus L.). Genome Announc 4 (5), e00981–00916. doi:10.1128/genomeA.00981-16
Gualberto, J. M., and Newton, K. J. (2017). Plant Mitochondrial Genomes: Dynamics and Mechanisms of Mutation. Annu. Rev. Plant Biol. 68, 225–252. doi:10.1146/annurev-arplant-043015-112232
Handa, H. (2003). The Complete Nucleotide Sequence and RNA Editing Content of the Mitochondrial Genome of Rapeseed (Brassica napus L.): Comparative Analysis of the Mitochondrial Genomes of Rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 31 (20), 5907–5916. doi:10.1093/nar/gkg795
Jeong, Y., and Lee, J. (2021). The Complete Mitochondrial Genome of the Benthic Diatom Pleurosigma inscriptura. Mitochondrial DNA B Resour. 6 (9), 2584–2586. doi:10.1080/23802359.2021.1945970
Kane, N., Sveinsson, S., Dempewolf, H., Yang, J. Y., Zhang, D., Engels, J. M., et al. (2012). Ultra-barcoding in Cacao (Theobroma spp.; Malvaceae) Using Whole Chloroplast Genomes and Nuclear Ribosomal DNA. Am. J. Bot. 99 (2), 320–329. doi:10.3732/ajb.1100570
Knope, M. L., Bellinger, M. R., Datlof, E. M., Gallaher, T. J., and Johnson, M. A. (2020). Insights into the Evolutionary History of the Hawaiian Bidens (Asteraceae) Adaptive Radiation Revealed through Phylogenomics. J. Hered. 111 (1), 119–137. doi:10.1093/jhered/esz066
Kubo, T., and Newton, K. J. (2008). Angiosperm Mitochondrial Genomes and Mutations. Mitochondrion 8 (1), 5–14. doi:10.1016/j.mito.2007.10.006
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., and Giegerich, R. (2001). REPuter: the Manifold Applications of Repeat Analysis on a Genomic Scale. Nucleic Acids Res. 29 (22), 4633–4642. doi:10.1093/nar/29.22.4633
Levings, C. S., and Brown, G. G. (1989). Molecular Biology of Plant Mitochondria. Cell 56 (2), 171–179. doi:10.1016/0092-8674(89)90890-8
Li, H., and Durbin, R. (2009). Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 25 (14), 1754–1760. doi:10.1093/bioinformatics/btp324
Librado, P., and Rozas, J. (2009). DnaSP V5: a Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 25 (11), 1451–1452. doi:10.1093/bioinformatics/btp187
Lin, M., Chen, F., Ge, J., Tang, J., and Ni, W. (2013). Protective Effects of Total Flavonoids of Bidens bipinnata Lon Acute Inflammation and Possible Mechanism. Chin. J. Clin. Pharmacol. Ther. 18 (6), 614–620. doi:10.1211/jpp/60.10.0016
Liu, Y., Huo, N., Dong, L., Wang, Y., Zhang, S., Young, H. A., et al. (2013). Complete Chloroplast Genome Sequences of Mongolia Medicine Artemisia frigida and Phylogenetic Relationships with Other Plants. PLoS One 8 (2), e57533. doi:10.1371/journal.pone.0057533
Logan, D. C. (2006). The Mitochondrial Compartment. J. Exp. Bot. 57 (6), 1225–1243. doi:10.1093/jxb/erj151
Lohse, M., Drechsel, O., and Bock, R. (2007). OrganellarGenomeDRAW (OGDRAW): a Tool for the Easy Generation of High-Quality Custom Graphical Maps of Plastid and Mitochondrial Genomes. Curr. Genet. 52 (5-6), 267–274. doi:10.1007/s00294-007-0161-y
Lu, C., Shen, Q., Yang, J., Wang, B., and Song, C. (2016). The Complete Chloroplast Genome Sequence of Safflower (Carthamus tinctorius L.). Mitochondrial DNA A. DNA Mapp. Seq. Anal. 27 (5), 3351–3353. doi:10.3109/19401736.2015.1018217
Lv, Z. Y., Zhang, J. W., Chen, J. T., Li, Z. M., and Sun, H. (2020). The Complete Chloroplast Genome of Soroseris umbrella (Asteraceae). Mitochondrial DNA B Resour. 5 (1), 637–638. doi:10.1080/23802359.2019.1711223
Makarenko, M. S., Usatov, A. V., Tatarinova, T. V., Azarin, K. V., Logacheva, M. D., Gavrilova, V. A., et al. (2019). Characterization of the Mitochondrial Genome of the MAX1 Type of Cytoplasmic Male-Sterile sunflower. BMC Plant Biol. 19, 51. doi:10.1186/s12870-019-1637-x
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi:10.1093/molbev/msu300
Nie, L., Cui, Y., Chen, X., Xu, Z., Sun, W., Wang, Y., et al. (2020). Complete Chloroplast Genome Sequence of the Medicinal Plant Arctium lappa. Genome 63 (1), 53–60. doi:10.1139/gen-2019-0070
Ogihara, Y., Yamazaki, Y., Murai, K., Kanno, A., Terachi, T., Shiina, T., et al. (2005). Structural Dynamics of Cereal Mitochondrial Genomes as Revealed by Complete Nucleotide Sequencing of the Wheat Mitochondrial Genome. Nucleic Acids Res. 33 (19), 6235–6250. doi:10.1093/nar/gki925
Park, S. I., and Lee, J. (2020). The Complete Mitochondrial Genome of Pyropia pulchra (Bangiophyceae, Rhodophyta). Mitochondrial DNA B Resour. 5 (3), 3157–3158. doi:10.1080/23802359.2020.1806132
Qiu, Y. L., Li, L., Wang, B., Xue, J. Y., Hendry, T. A., Li, R. Q., et al. (2010). Angiosperm Phylogeny Inferred from Sequences of Four Mitochondrial Genes. J. Syst. Evol. 48 (6), 391–425. doi:10.1111/j.1759-6831.2010.00097.×
Qian, J. (2014). Study on Chloroplast and Mitochondrial Genomes of Salvia Miltiorrhiza. Beijing: Chinese Academy of Medical Sciences & Peking Union Medical College.
Rice, P., Longden, I., and Bleasby, A. (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16 (6), 276–277. doi:10.1016/s0168-9525(00)02024-2
Robinson, J. T., Thorvaldsdóttir, H., Wenger, A. M., Zehir, A., and Mesirov, J. P. (2017). Variant Review with the Integrative Genomics Viewer. Cancer Res. 77 (21), e31–e34. doi:10.1158/0008-5472.Can-17-0337
Schattner, P., Brooks, A. N., and Lowe, T. M. (2005). The tRNAscan-SE, Snoscan and snoGPS Web Servers for the Detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, W686–W689. doi:10.1093/nar/gki366
Shandukani, P. D., Tshidino, S. C., Masoko, P., and Moganedi, K. M. (2018). Antibacterial activity and In Situ efficacy of Bidens pilosa Linn and Dichrostachys cinerea Wight et Arn extracts against common diarrhoea-causing waterborne bacteria. BMC Complement. Altern. Med. 18 (1), 171. doi:10.1186/s12906-018-2230-9
Sharp, P. M., and Li, W. H. (1987). The Codon Adaptation Index--a Measure of Directional Synonymous Codon Usage Bias, and its Potential Applications. Nucleic Acids Res. 15 (3), 1281–1295. doi:10.1093/nar/15.3.1281
Shen, X., Guo, S., Yin, Y., Zhang, J., Yin, X., Liang, C., et al. (2018). Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Aster tataricus. Molecules 23 (10), 2426. doi:10.3390/molecules23102426
Shen, X., Wu, M., Liao, B., Liu, Z., Bai, R., Xiao, S., et al. (2017). Complete Chloroplast Genome Sequence and Phylogenetic Analysis of the Medicinal Plant Artemisia annua. Molecules 22 (8), 1330. doi:10.3390/molecules22081330
Shi, L., Chen, H., Jiang, M., Wang, L., Wu, X., Huang, L., et al. (2019). CPGAVAS2, an Integrated Plastome Sequence Annotator and Analyzer. Nucleic Acids Res. 47 (W1), W65–W73. doi:10.1093/nar/gkz345
Shi, M., Xie, H., Zhao, C., Shi, L., Liu, J., and Li, Z. (2021). The Complete Chloroplast Genome of Atractylodes japonica Koidz. Ex Kitam. And its Phylogenetic Inference. Mitochondrial DNA B Resour. 6 (7), 2038–2040. doi:10.1080/23802359.2021.1927217
Shi, X., and Xie, K. (2020). The Complete Chloroplast Genome Sequence of Stilpnolepis centiflora (Asteraceae), an Endemic Desert Species in Northern China. Mitochondrial DNA B Resour. 5 (3), 3545–3546. doi:10.1080/23802359.2020.1829127
Shi, Z., Chen, Y. L., Chen, Y. S., Lin, Y. R., Liu, S. W., Ge, X. J., et al. (2011). Flora of China. Beijing: Science Press. Vols. 20–21.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30 (12), 2725–2729. doi:10.1093/molbev/mst197
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq - Versatile and Accurate Annotation of Organelle Genomes. Nucleic Acids Res. 45 (W1), W6–W11. doi:10.1093/nar/gkx391
Tsai, L. C., Wang, J. C., Hsieh, H. M., Liu, K. L., Linacre, A., and Lee, J. C. (2008). Bidens Identification Using the Noncoding Regions of Chloroplast Genome and Nuclear Ribosomal DNA. Forensic Sci. Int. Genet. 2 (1), 35–40. doi:10.1016/j.fsigen.2007.07.005
Verma, D., and Daniell, H. (2007). Chloroplast Vector Systems for Biotechnology Applications. Plant Physiol. 145 (4), 1129–1143. doi:10.1104/pp.107.106690
Wang, B., Zhao, Q., Wang, X. H., and Fu, Z. X. (2020). The Complete Chloroplast Genome of Pertya phylicoides (Asteraceae, Pertyeae): a Shurby Endemic Species from China. Mitochondrial DNA B Resour. 5 (1), 963–964. doi:10.1080/23802359.2020.1722763
Wang, J., He, R., Zhang, H., Hu, Y., Wang, J., Wang, L., et al. (2021). Complete Chloroplast Genome of Saussurea inversa (Asteraceae) and Phylogenetic Analysis. Mitochondrial DNA B 6 (1), 8–9. doi:10.1080/23802359.2020.1845108
Wang, R., Wu, Q. X., and Shi, Y. P. (2010). Polyacetylenes and Flavonoids from the Aerial Parts of Bidens pilosa. Planta Med. 76 (9), 893–896. doi:10.1055/s-0029-1240814
Wang, X. Y., Chen, G. R., Deng, Z. Y., Zhao, J., Ge, J. F., Li, N., et al. (2014). Chemical Constituents from Bidens bipinnata. Zhongguo Zhong Yao Za Zhi 39 (10), 1838–1844. doi:10.4268/cjcmm20141017
Won, S. Y., Jung, J. A., and Kim, J. S. (2018). The Complete Mitochondrial Genome Sequence of Chrysanthemum boreale (Asteraceae). Mitochondrial DNA B Resour. 3 (2), 529–530. doi:10.1080/23802359.2018.1468226
Wu, L., Nie, L., Xu, Z., Li, P., Wang, Y., He, C., et al. (2020). Comparative and Phylogenetic Analysis of the Complete Chloroplast Genomes of Three Paeonia Section Moutan Species (Paeoniaceae). Front. Genet. 11, 980. doi:10.3389/fgene.2020.00980
Wynn, E. L., and Christensen, A. C. (2019). Repeats of Unusual Size in Plant Mitochondrial Genomes: Identification, Incidence and Evolution. G3 (Bethesda) 9 (2), 549–559. doi:10.1534/g3.118.200948
Xie, Q., Shen, K. N., Hao, X., Nam, P. N., Ngoc Hieu, B. T., Chen, C. H., et al. (2017). The Complete Chloroplast Genome of Tianshan Snow Lotus (Saussurea involucrata), a Famous Traditional Chinese Medicinal Plant of the Family Asteraceae. Mitochondrial DNA A. DNA Mapp. Seq. Anal. 28 (2), 294–295. doi:10.3109/19401736.2015.1118086
Xing, Y.-P., Xu, L., Chen, S.-Y., Liang, Y.-M., Wang, J.-H., Liu, C.-S., et al. (2019). Comparative Analysis of Complete Chloroplast Genomes Sequences of Arctium lappa and A. tomentosum. Biol. Plant. 63, 565–574. doi:10.32615/bp.2019.101
Yang, X., Bai, Z. F., Zhang, D. W., Zhang, Y., Cui, H., and Zhou, H. L. (2020). Enrichment of Flavonoid-Rich Extract from Bidens bipinnata L. By Macroporous Resin Using Response Surface Methodology, UHPLC-Q-TOF MS/MS-assisted Characterization and Comprehensive Evaluation of its Bioactivities by Analytical Hierarchy Process. Biomed. Chromatogr. 34 (11), e4933. doi:10.1002/bmc.4933
Keywords: bidens, species identification, super barcode, organelle genomes, quality control
Citation: Wu L, Nie L, Guo S, Wang Q, Wu Z, Lin Y, Wang Y, Li B, Gao T and Yao H (2022) Identification of Medicinal Bidens Plants for Quality Control Based on Organelle Genomes. Front. Pharmacol. 13:842131. doi: 10.3389/fphar.2022.842131
Received: 23 December 2021; Accepted: 18 January 2022;
Published: 14 February 2022.
Edited by:
Jian-Bo Yang, National Institutes for Food and Drug Control (China), ChinaReviewed by:
Komwit Surachat, Prince of Songkla University, ThailandBo Wang, Hubei Provincial Institute of Drug Control, China
Xu-Mei Wang, Xi’an Jiaotong University, China
Copyright © 2022 Wu, Nie, Guo, Wang, Wu, Lin, Wang, Li, Gao and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Yao, c2NhdXlhb2hAc2luYS5jb20=
†These authors have contributed equally to this work and share first authorship
 Liwei Wu1†
Liwei Wu1† Baoli Li
Baoli Li Ting Gao
Ting Gao Hui Yao
Hui Yao